Abstract
Pectin is a major component of the plant cell wall, forming a network that contributes to cell wall integrity and flexibility. Pectin methylesterase (PME) catalyzes the removal of methylester groups from the homogalacturonan backbone, the most abundant pectic polymer, and contributes to intercellular adhesion during plant development and different environmental stimuli stress. In this study, we identified and characterized an Arabidopsis type-II PME, PME53, which encodes a cell wall deposited protein and may be involved in the stomatal lineage pathway and stomatal functions. We demonstrated that PME53 is expressed explicitly in guard cells as an abscisic acid (ABA)-regulated gene required for stomatal movement and thermotolerance. The expression of PME53 is significantly affected by the stomatal differentiation factors SCRM and MUTE. The null mutation in PME53 results in a significant increase in stomatal number and susceptibility to ABA-induced stomatal closure. During heat stress, the pme53 mutant highly altered the activity of PME and significantly lowered the expression level of the calmodulin AtCaM3, indicating that PME53 may be involved in Ca2+-pectate reconstitution to render plant thermotolerance. Here, we present evidence that the PME53-mediated de-methylesterification status of pectin is directed toward stomatal development, movement, and regulation of the flexibility of the guard cell wall required for the heat response.
Introduction
Plants are sessile organisms that have to cope with the challenge of extreme environmental conditions, including various abiotic and biotic stresses, all of which exert adverse effects on plant growth and development. The most likely negative impact of these changes is an increase in temperature, which directly impacts DNA, proteins, cell membranes, and metabolism processes, severely limiting plant growth, development, and productivity. As a result, plants have devised a variety of strategies to deal with temperature extremes. Plant thermotolerance entails converting a moderate temperature increase into molecular defenses against subsequent extreme temperatures and preventing and repairing heat-labile protein and membrane damage (Larkindale and Vierling, 2008). The induction of molecular chaperone heat shock proteins (HSPs) is an essential part of the universally conserved heat response, allowing organisms to survive stress conditions such as elevated temperatures that cause protein unfolding (Lindquist and Craig, 1988).
Apart from the well-known heat stress response, little is known about how the dynamics of plant cell wall changes in wall composition and architecture are associated with various stresses. The plant cell wall is a sophisticated structure formed by a complex mixture of polysaccharide-rich proteins and other polymers assembled into a rigid, flexible, and dynamically organized network (Wolf et al., 2009; Wu et al., 2018). When cells are stressed, specific transcriptional responses, some profitable cell wall proteins, and critical changes in cell wall architecture are affected (Klis et al., 2006). In plant cells, the integrity of the cell wall is tightly controlled and closely coordinated with the stress response. We previously discussed key components of cell wall reconstruction during heat stress, such as pectin, pectin methylesterase (PME), and apoplastic Ca2+ (Wu and Jinn, 2010; Wu et al., 2010, 2018). Therefore, the maintenance of cell wall integrity is essential for cell viability and function during stress (Hamann et al., 2009; Wu et al., 2018).
Pectin is the most complex polysaccharide in the cell wall, which is synthesized and methylesterified in the Golgi and then secreted into the cell wall in a highly methylesterified state. Pectin is important for both cellular adhesion and cell wall plasticity, thus controlling cell and organ growth during plant development (Willats et al., 2001; Palin and Geitmann, 2012). Homogalacturonan (HGA), a major component of pectin, consists of linear α-1,4-linked D-galacturonic acid (D-GalA) residues and has conformational flexibility, which can be influenced by growth, development, and environmental cues (Willats et al., 2001). A critical feature of HGA that influences its properties is the methylesterification of C6 and acetylesterification on C2 or C3, which occurs on GalA during the synthesis of the backbone.
Subsequently, highly esterified HGAs exported into cell walls were de-methylesterified by the mature pectin methylesterases (EC 3.1.1.11; PMEs), which belong to class 8 (CE8) of the carbohydrate esterases (CAZy website)1 (Cantarel et al., 2009), which converts methoxyl groups into carboxyl groups on the HGA backbone, resulting in the release of methanol and protons. Plant PMEs belong to a multigene family whose members may play role in a broad range of physiological processes. For instance, 66 PME-related genes are predicted in Arabidopsis thaliana, 89 in Populus trichocarpa, 81 in Gossypium raimondii, 79 in Lycopersicon esculentum, 105 in Linum usitatissimum, 43 in Oryza sativa and Zea mays (Louvet et al., 2006; Liu et al., 2013; Pinzón-Latorre and Deyholos, 2013; Horowitz and Ospina-Giraldo, 2015; Jeong et al., 2015; Zhang et al., 2019). The precise biological functions of PMEs remain unclear although various roles have been proposed.
In higher plants, the PME gene encodes the individual PRE-PRO-region, which is considered a signature of PMEs. PMEs are led to the wall by the PRE domain, and the conserved PME domain, which is the active part of the protein, is preceded by the PRO region, which shares similarities with PME inhibitors (PMEIs) (Pelloux et al., 2007). Therefore, PMEs can be based on the presence or absence of the PRO region and are classified into type I and type II. Type I PMEs contain PRO regions, whereas type II PMEs do not. The type II PMEs sequences are similar to those of phytopathogenic organisms (fungi and bacteria) and are associated with cell wall softening and loosening during pathogen infection (Micheli, 2001).
The fine control of the degree of methylesterification is a key factor in HGAs and is potentially involved in the regulation of cell wall architecture, which determines that the methylesterification status of pectin is regulated by the action of PMEs. In recent years, PMEs have attracted public attention because of their roles in many aspects. Over the past few years, only several specific PME genes have been described and have revealed multiple roles in Arabidopsis. For example, overexpression of Arabidopsis PME5 and PMEI3 resulted in softer and harder shoot apical meristem cell walls, respectively, when compared with the wild-type plants (Peaucelle et al., 2011). Arabidopsis PME35 is explicitly expressed in the basal part of the inflorescence stem and provides mechanical support (Hongo et al., 2012). Highly methyl esterified seeds (HMS; PME6) is abundant during mucilage secretion, acting on embryo morphology, and mucilage extrusion, both of which are involved in embryo development (Levesque-Tremblay et al., 2015). In stress responses, Arabidopsis PME41 and PME48 are associated with chilling/freezing tolerance through the regulation of brassinosteroid signaling and pollen grain germination, respectively (Qu et al., 2011; Leroux et al., 2015). The cyst nematode (Heterodera schachtii) parasitism can be reduced by inhibiting Arabidopsis PME3 to interact with the cellulose-binding protein of the cyst nematode (Hewezi et al., 2008). PME17 in Arabidopsis performs a linear (blockwise) de-methylesterification that promotes the formation of egg-box structures, which aids in Botrytis cinerea resistance (Del Corpo et al., 2020).
Stomata are pores on the surface of leaves, formed by a pair of curved guard cells with unique cell wall compositions that regulate gas and water exchange by opening and closing the stomata. The opening and closure of stomata pores are mediated by changes in the turgor pressure of the two guard cells, which can be affected by various signals, including CO2, humidity, red/blue light, abscisic acid (ABA), auxins, calcium (Ca2+), and extracellular calmodulin (Jones et al., 2003; Hõrak et al., 2017). In addition, guard cells have remarkable elasticity and can act reversibly during stomatal opening and closing due to differential thickening and the orientation of cellulose microfibrils. The shape of stomata is limited by the mechanical properties of cells, which largely depend on the structure and composition of cell walls that allow them to undergo repeated swelling and deflation. Although it has been known that the anisotropic nature of guard cell walls, showing differential wall thickness and the orientation of cellulose microfibrils determined the stomatal opening and closure, the role of cell wall mechanical properties remains an open question.
In Arabidopsis, the walls of guard cells are rich in un-methylesterified pectin, but the high-methylesterified HGA and Ca2+ crosslinked de-methylesterified HGA are absent from guard cell walls (Amsbury et al., 2016). This suggests that the stomatal function of guard cells may be influenced by the cell wall properties of guard cells. PME also appears to play a role in the regulation of plant development by influencing the mechanical properties of plant cell walls, according to some studies. The guard cell wall of Arabidopsis pme6 mutant, for example, was rich in methylesterified pectin and had a narrower dynamic range in response to stomatal opening/closure triggers, implying that stomatal function loss is due to a mechanical change in the guard cell wall (Amsbury et al., 2016). Therefore, the function of stomata requires pectin de-methylesterification of guard cell walls through the regulation of PMEs. In our previous study, we identified PME34 which plays a role in regulating guard cell wall flexibility to control stomatal aperture involved in mediating heat response in Arabidopsis (Huang et al., 2017; Wu et al., 2017). Therefore, changes in cell wall metabolism and cell wall-modifying enzyme activity in controlling guard cell wall plasticity are important physiological mechanisms of plants to heat stress (Wu et al., 2017, 2018).
Guard cells can inflate and deflate repeatedly, and the formation of guard cell pores necessitates the modification of pectin. It causes pore initiation and enlargement by de-methylesterification of HGA (Rui et al., 2019). Although PME6 and PME34 have been shown to play a role in stomatal movement (Amsbury et al., 2016; Huang et al., 2017), little is known about how pectin modification affects stomatal development and downstream signaling pathways. It is known that stomatal development is regulated by a group of basic helix-loop-helix (bHLH) transcription factors, including SPEECHLESS (SPCH), MUTE, and FAMA, which act together and are critical in mediating guard cell differentiation at different steps of the stomatal lineage in Arabidopsis (Davies and Bergmann, 2014). Despite extensive investigations of the bHLH factors involved in stomatal development, the correlation between the mechanical properties of stomatal guard cells and the regulation of bHLH factors remains elusive.
In a previous study, we verified that maintaining apoplastic Ca2+ homeostasis through PME activity has a pronounced effect on plant growth and heat response (Wu et al., 2010). Using a genetic approach, we characterized a type I PME, PME34, which plays an important role in controlling stomatal aperture to regulate the transpiration rate during the heat response (Huang et al., 2017). In this study, we revealed a guard cell-specific type II PME, PME53, required for stomatal density, movement, and heat response control. Transcript accumulation of PME53 is induced by exogenous ABA treatment, and the PME53 promoter mediates strong induction of GUS reporter expression in guard cells. Using a trans-activation assay, we showed that the expression level of PME53 is significantly affected by SCRM and MUTE, which function as master regulators of stomatal development. This work highlighted that PME53 is involved in fine-tuning pectin methylesterification, modulating stomatal cell fate and patterning, and aiding in environmental adaption.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild type (Col-0) and homozygous T-DNA insertion mutants PME53 (SALK_150305) and SCRM (SALK_003115) were obtained from the Biological Resource Center (ABRC). Sterile seeds were placed on solid half-strength Murashige and Skoog basal medium (1/2 MS; Sigma M5519) (Murashige and Skoog, 1962) containing 1% sucrose and 0.8% phytagel (Sigma). Plants were grown in growth chambers at 22 to 24°C under an 8-h dark/16-h light cycle at an intensity of 80 to 100 μmol m2 s–1. Transgenic plants of PME53-promoter:GUS with pCAMBIA1391Z vector in Col background and CaMV 35S:PME53-3xFLAG overexpression plants with pCAMBIA3300 vector (Karimi et al., 2002) in a pme53 mutant background were generated by Agrobacterium tumefaciens GV3101-mediated transformation and by the floral dip method (Clough and Bent, 1998).
RNA Preparation, cDNA Synthesis, and Quantitative Real-Time PCR
Seven-day-old seedlings were ground with liquid nitrogen and suspended with TRIZOL reagent (Invitrogen) for RNA preparation and removed the contaminating DNA involved with TURBO DNA-free kit (Applied Biosystems). cDNA synthesis was performed with the high-capacity cDNA reverse transcription kit (Applied Biosystems). PCR primers were designed using the Primer32. Quantitative real-time PCR (q-PCR) reactions were analyzed by the 7500 Fast Real-Time PCR System (Applied Biosystems) with the q-PCR mix of SYBR Green Supermix (BIO-RAD). The internal control for normalization was PP2AA3 (PP2A; At1g13320) (Czechowski et al., 2005).
Constructs of Transactivation Assay and amiRNA-Mediated Gene Suppression
A 1.974-kb promoter region of PME53 was amplified and cloned into the pGreenII 0800-LUC vector through BamHI and HindIII sites for PME53-promoter:LUC (Firefly luciferase) fusion then used for transactivation assay. The automated design of artificial miRNAs (amiRNA) was according to the WMD3 (Web MicroRNA Designer)3. The 21mer amiRNA of the tester was cloned into the pRS300 vector which contains the miR319a precursor (Schwab et al., 2006). Sequence-verified amiRNA was cloned into the Gateway vector pCR8™/GW/TOPO and was recombinant into a destination vector pCAMBIA3300-CHF-DEST which was regulated by the 35S promoter. The amiRNA-containing vector was used for suppression of tester gene in transactivation assay (Karimi et al., 2002).
Pectin Methylesterase Activity Assay
Fifty of 7-day-old seedlings were ground and suspended with 100 μL extraction buffer (1 M NaCl, 0.1 M citric acid, and 0.2 M Na2HPO4, pH 5.0) for 5 min and centrifuged at 13,800 × g for 15 min. The supernatant was collected and cell wall protein concentrations were determined according to the method of Bradford (1976). The enzymatic PME activity was quantified by a gel diffusion assay as described (Downie et al., 1998; Bethke et al., 2014; Lionetti, 2015) with some modification. 10 μL of 3.75 μg protein extract was loaded into the 0.3 mm well on the 20 mL gel prepared in McIlvaine buffer adjusted to pH levels of 4 to 8, which contains 2% agarose and 0.1% of high methylesterified pectin (≥85% esterified pectin from citrus fruit; P9561, Sigma-Aldrich). After 16 h incubation at 28°C, the gels were stained with 10 mL of 0.05% (w/v) ruthenium red (Sigma-Aldrich) for 1 h and de-stained with distilled water. The calibration curve of PME activity was established with a detection range from 2 to 8 μg/mL of cell-wall proteins. The PME activity was calculated by measuring the stained area (cm2) by ImageJ software4.
Stomatal Aperture Measurement
The stomata tape-peel method was used for guard cell sample preparation as described (Lawrence et al., 2018). Fully expanded leaves of 4-week-old soil-grown plants were used per line for each treatment. Briefly, mature leaves (7th and 8th) were selected and attached to two pieces of tapes to both sides, then gently peeled apart the two pieces of tapes. The peels were incubated in the stomata opening buffer (50 mM KCl, 10 mM MES-KOH, pH 6.2) at 22°C for 2.5 h with light for equilibration and stomata opening. Afterward, the peels were further treated with 10 μM ABA for the appropriate treatment time. After treatment, the stomata aperture was observed by a light microscope and measured by ImageJ software4. The data were presented in comparison to the corresponding controls.
Determination of Water Loss Assay
Water loss and standardized water contents in the full opened leaf by following the procedure described previously (Fujita et al., 2005). Briefly, 40 detached 7th and 8th rosette leaves from 4-week-old soil-grown seedlings were kept in a petri dish. The transpiration (water loss) measurement was standardized (%) and then calculated as [(FWi-DW)/(FWo-DW)] × 100, where FWi and FWo are fresh weight for any given interval and original fresh weight, respectively, and DW is dry weight. These tests were conducted on the laboratory bench at 24 to 26°C and 60 to 70% relative humidity.
Histochemical Glucuronidase Staining
A 1.974-kb promoter region of PME53 was fused with reporter β-glucuronidase (GUS) gene in pCAMBIA1391Z (CAMBIA) and used for the tissue-specific expression analysis, and the GUS staining method was performed as described previously (Weigel and Glazebrook, 2002). The provided images of GUS-stained tissues represent the typical results of at least six independent transgenic lines.
Protoplast Isolation, Transient Expression and Transactivation Assay
Protoplast isolation and transactivation assay were carried out as previously described (Yoo et al., 2007). For the transactivation assay, 4 × 104 protoplasts were transfected with 5 μg plasmid DNA or 10 μg amiRNA-containing plasmid in a total 20 μL reaction were conducted. Transfected protoplasts were incubated at 22 to 24°C under a light condition at 80 to 100 μmol m–2 s–1 for 14 to 16 h. The YFP signal was detected by Zeiss LSM780 confocal microscope. The dual-luciferase activity was measured by quantifying LUC and REN (Renilla luciferase) activities with the microplate reader Infinite 200 PRO (TECAN), according to the manufacturer’s instructions (Promega). The 35S promoter-driven REN gene in the pGreen0800-LUC vector is used as an internal control.
Subcellular Localization in Onion Epidermal Cells and Agroinfiltration-Based Transient Gene Expression in Tobacco
Transgene 35S:PME53-YFP with pEarleyGate101 vector was transformed into onion epidermal cells by particle bombardment as described previously (Guan et al., 2004; Huang et al., 2017). Approximately 2.5 μg of DNA was coated onto 0.6 μm gold particles (Bio-Rad) and transiently introduced into onion epidermal cells with a helium biolistic particle-delivery system (PDS-1000; Bio-Rad). The bombarded cells were kept at 26°C in the dark for 16 h. The plasmolysis of onion epidermal cells was induced by addition of 0.8 M mannitol solution for 30 min. 4-week-old tobacco (Nicotiana benthamiana) plants were infiltrated 100 μL recombinant Agrobacteria per spot by needless syringe then keep plants at 22 to 24°C with a 16 h/8 h light/dark photoperiod for 2 to 3 days (Sparkes et al., 2006). The signals were then observed and photographed by Zeiss LSM780 confocal microscope.
Heat Treatments
Thermotolerance assays of seven-day-old seedlings were performed as previously described (Charng et al., 2006; Hsu et al., 2010; Huang et al., 2016). For the acquired thermotolerance test, plates (20-mL 1/2 MS) were preheated at a 37°C sub-lethal heat shock for 1 h and allowed to recover at 22°C for 2 h before a 44°C lethal heat shock for 170 to 180 min. Healthy growing seedlings were counted 10 to 14 days after the heat treatment.
Statistical Analysis
All experiments were repeated independently at least three times. Statistical analysis involved Student’s t-test and ANOVA with Tukey’s HSD post hoc test. P < 0.05 was considered statistically significant.
Primers and Accession Numbers
Primers and accession numbers are given in Supplementary Table 1.
Results
Expression of PME53 in Response to Abiotic Stress and in Developing Tissues
In the Arabidopsis microarray database, the expression profile of the predicted PME genes varied by ∼75% in response to abiotic and biotic stresses (Pelloux et al., 2007). Here, the guard-cell-specific microarray analyses side by side with a mesophyll-cell-specific microarray (Yang et al., 2008), 59 PME genes were assayed under ABA treatment. We showed that the expression of PME53 increased significantly after treatment with 100 μM ABA for 1 h in guard cells, but not in mesophyll cells (Supplementary Figure 1). The Arabidopsis PME34 gene, an ABA-regulated stomatal gene, was used as reference (Huang et al., 2017).
Seven-day-old Arabidopsis seedlings subjected to ABA, salt, and osmotic treatments were also conducted, and PME53 expression levels were analyzed by q-PCR (Figure 1A and Supplementary Figure 2). After treatment with 100 μM ABA for 1 h, the expression levels of PME53 increased 15-fold compared with the untreated plants (Figure 1A). In addition, treatment with 150 mM NaCl and 300 mM mannitol for 6 h resulted in significantly downregulated PME53 expression (Supplementary Figure 2). Arabidopsis RAD29B, an ABA, salt, and osmotic-responsive gene, was used as reference.
FIGURE 1
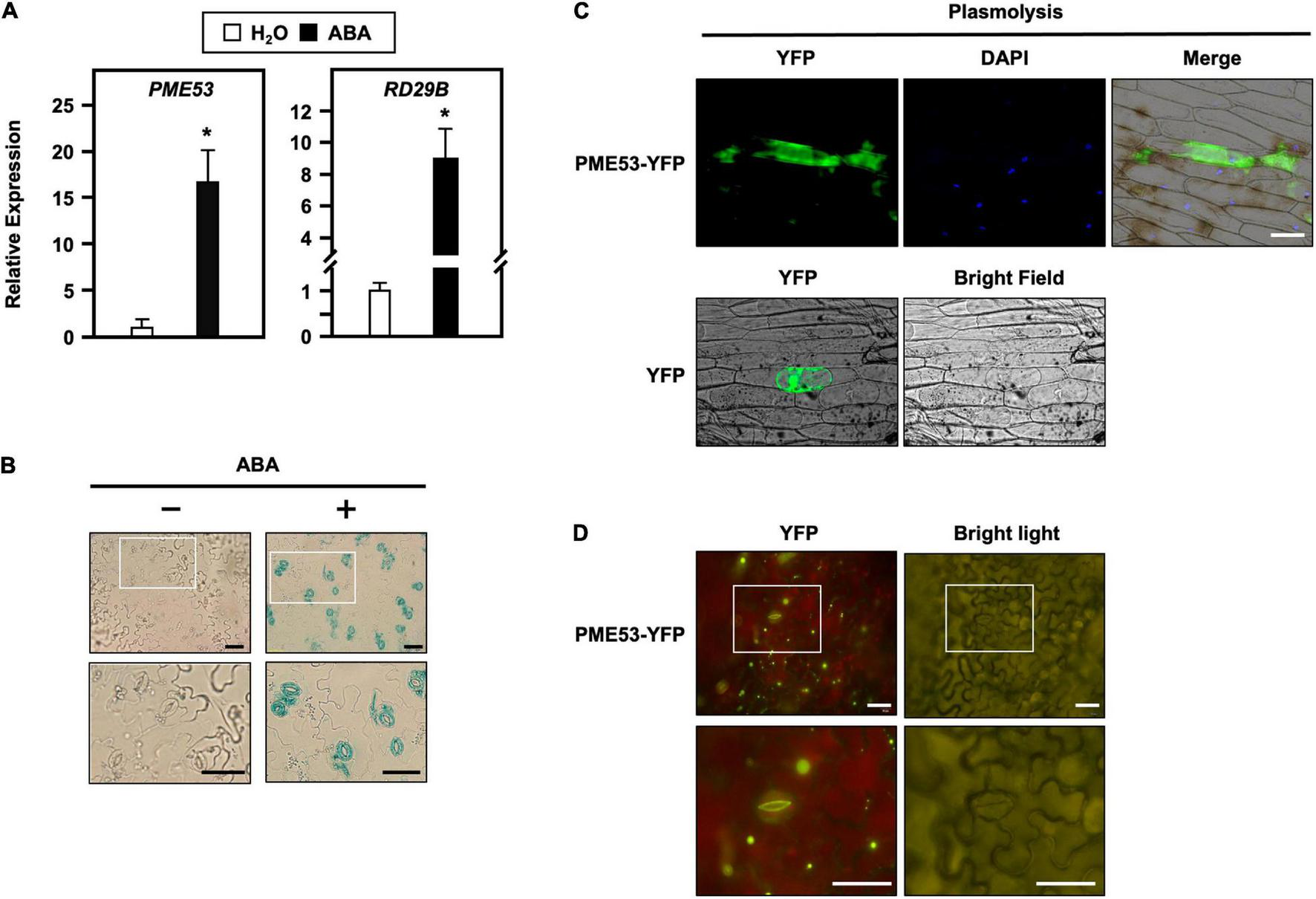
Transcriptional level, tissue-specific expression and subcellular localization of PME53. (A) 7-day-old seedlings were treated without (H2O control) or with 100 μM ABA for 1 h. The expression levels of PME53 were analyzed by q-PCR. RD29B, an ABA-responsive gene, was used as reference. The fold change expression was normalized relative to level of the H2O control. Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the H2O treatment. (B) Cotyledons of the same PME53-promoter:GUS transgenic plants treated without (-) or with (+) 30 μM ABA for 3 h, and then specimens were immersed together in a staining solution. The magnification of the insert (white frame) is shown. Bars = 40 μm. (C) The onion epidermal cells were treated with mannitol for the plasmolysis assay. Subcellular localization analyzes of PME53-YFP by confocal microscopy. YFP was used as a control. Blue shows nuclei stained with 4′,6-diamino-phenylindole (DAPI). Bar = 200 μm. (D) Tobacco cells were infiltrated with Agrobacterium harboring 35S:PME53-YFP transgene. The magnification of the insert (white frame) shows that PME53 is mainly localized in the inner wall layer of guard cells. Bars = 40 μm.
The potential cis-elements in the 2-kb promoter region of PME53 show multiple potential ABA response elements (ABREs), with the Plant Promoter Analysis Navigator (PlantPAN3)5 (Chow et al., 2019; Supplementary Figure 3). Therefore, the tissue-specific expression of PME53 was analyzed in transgenic plants harboring a 1.974-kb promoter region of PME53 fused with a reporter GUS gene. The PME53-promoter:GUS transgenic lines indicated that PEM53 was ubiquitously expressed in the analyzed tissues (Supplementary Figure 4).
Notably, it was strongly upregulated after 3 h of treatment with 30 μM ABA in guard cells (Figure 1B). Together these results imply that PME53 may play a role in the regulation of ABA-mediated stomatal responses.
PME53 Resides in Apoplast and Inner Wall Layer of Guard Cell
Arabidopsis PME53 protein (UniProt: Q8VYZ3) belongs to a type-II PME that is predicted to have a signal peptide (SP; amino acids 1 to 28) along with the conserved PME domain (amino acids 29 to 383) according to the UniPort database6 (Supplementary Figure 5). Using the CaMV 35S:PME53-YFP transgene, the onion epidermal cells were submitted to 0.8 M mannitol for 30 min to induce plasmolysis, indicating that the PME53 reporter was located in the apoplast (Figure 1C). Tobacco cells were infiltrated with Agrobacterium cells expressing the 35S:PME53-YFP transgene, demonstrating that PME53 is primarily found in the inner-wall layer of guard cells (Figure 1D). These findings suggest that PME53 is a guard cell wall-localized protein that may aid in the opening and closing of stomatal pores in guard cells.
Characterization of PME53 T-DNA Insertion and PME53-Overexpression Lines
Thus, the role of PME53 physiological has mainly been studied through its T-DNA insertion and overexpression lines (Figure 2). One PME53 homozygous T-DNA insertion mutant was screened by PCR-based genotyping and the null-mutant lines were confirmed by RT-PCR (Figure 2A). PME53 fused with a 3xFLAG tag to the C-terminus and driven by a 35S promoter to generate the PME53-overexpression (OE) lines in a pme53 mutant background (PME53-OE). Three independent OE lines, PME53-OE1 to OE3, were obtained and characterized by q-PCR (Figure 2B) and western blotting with an α-FLAG antibody (Figure 2C, top). These three PME53-OE lines showed an increased PME activates compared with the wild-type (Col) plants (Figure 2C, bottom), then the PME53-OE2 plants used for the following experiments.
FIGURE 2
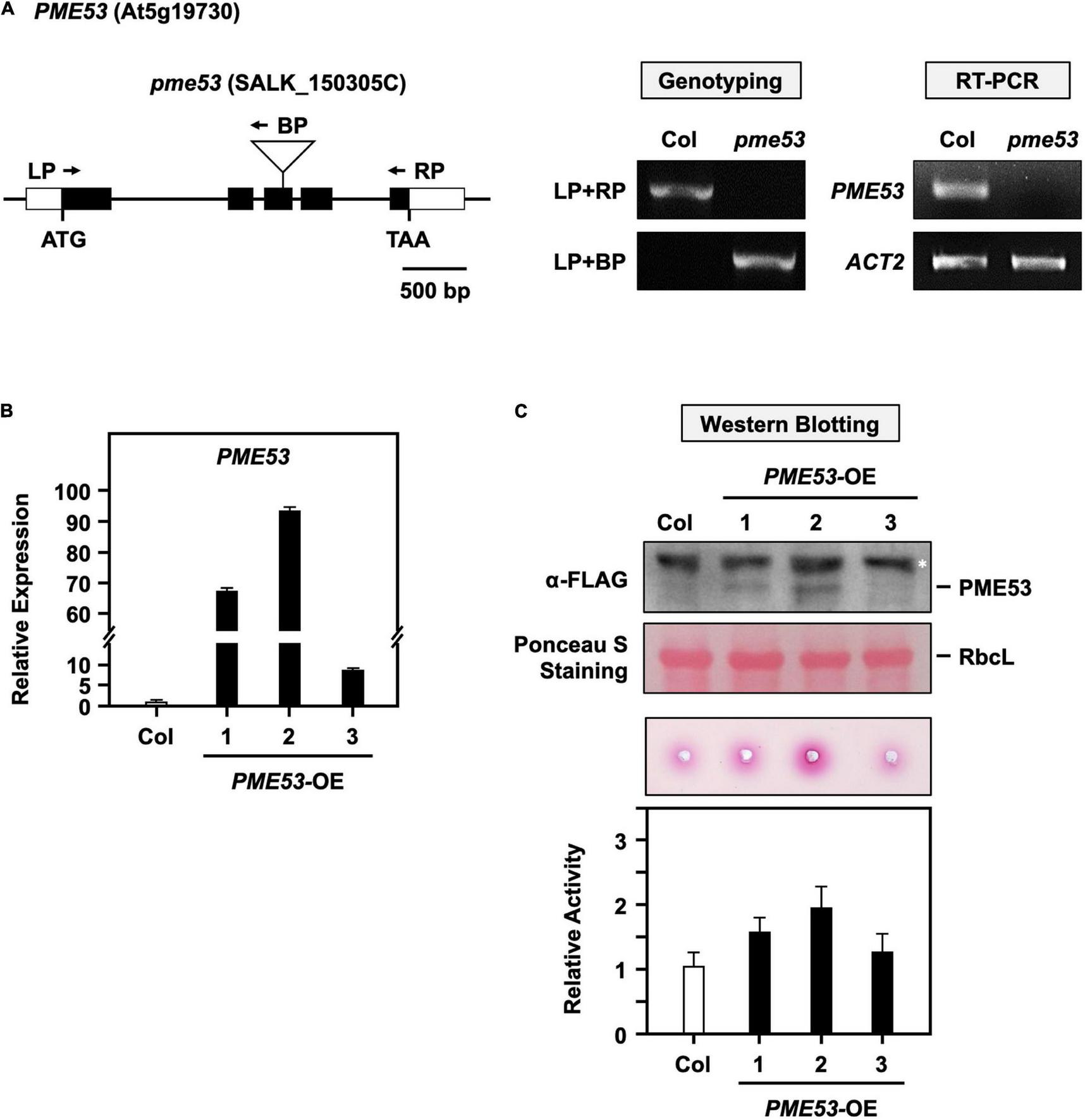
Characterization of PME53 T-DNA insertion and its overexpression lines. (A) Schematic map of PME53 gene and T-DNA insertion site. The white box, black box, and solid line indicate the UTR region, exon, and intron, respectively. ATG are shown as initiation codons. T-DNA insertion site is indicated as triangle, and primers for genotyping are indicated as arrows. RT-PCR involved the PME53 gene-specific primers. Actin2 (ACT2) was used as an input control. (B) The expression levels of PME53 in three independent lines, PME53-OE1 to OE3, were analyzed by q-PCR. The fold change expression was normalized relative to level of the Col. (C) 7-day-old seedlings were used for PME53 protein level (top) and activity (bottom) analysis. Immunoblot stained with Ponceau S and the ribulose bisphosphate carboxylase large subunit (RbcL) was as a loading control. Western blotting analysis using anti-FLAG antibody. *Indicates non-specific immune signals. The fold change PME activity was normalized relative to level of the Col.
PME53 Is a Functional Pectin Methylesterase With an Optimal Activity at Acidic pH Value
The evidence indicates that apoplastic pH is an important determinant of PME activity and contributes to the de-methylesterification of cell wall pectins (Bosch and Hepler, 2005). The PME activity was measured by a gel diffusion assay (Supplementary Figure 6, top), and the calibration curve of PME activity was linear (R2 = 0.9831) over the range from 2 to 8 μg/mL of cell wall proteins (Supplementary Figure 6, bottom). A 3.75 μg protein extract used for the following PME activity assay.
Here, we demonstrated that pme53 showed a significant reduction in PME activity as compared with the Col (Figure 3A) but significantly increased PME activity in PME53-OE2 plants (Figure 3B). The optimal PME activity at different pH levels in the Col and PME53-OE2 plants was analyzed (Figure 3C). PME53 activity was highest at acidic pH levels of 4 to 5 and decreased with alkaline pH of 8 (Figure 3D). This result indicated that PME53 protein is a functional PME with optimal PME activity at a more acidic pH value.
FIGURE 3
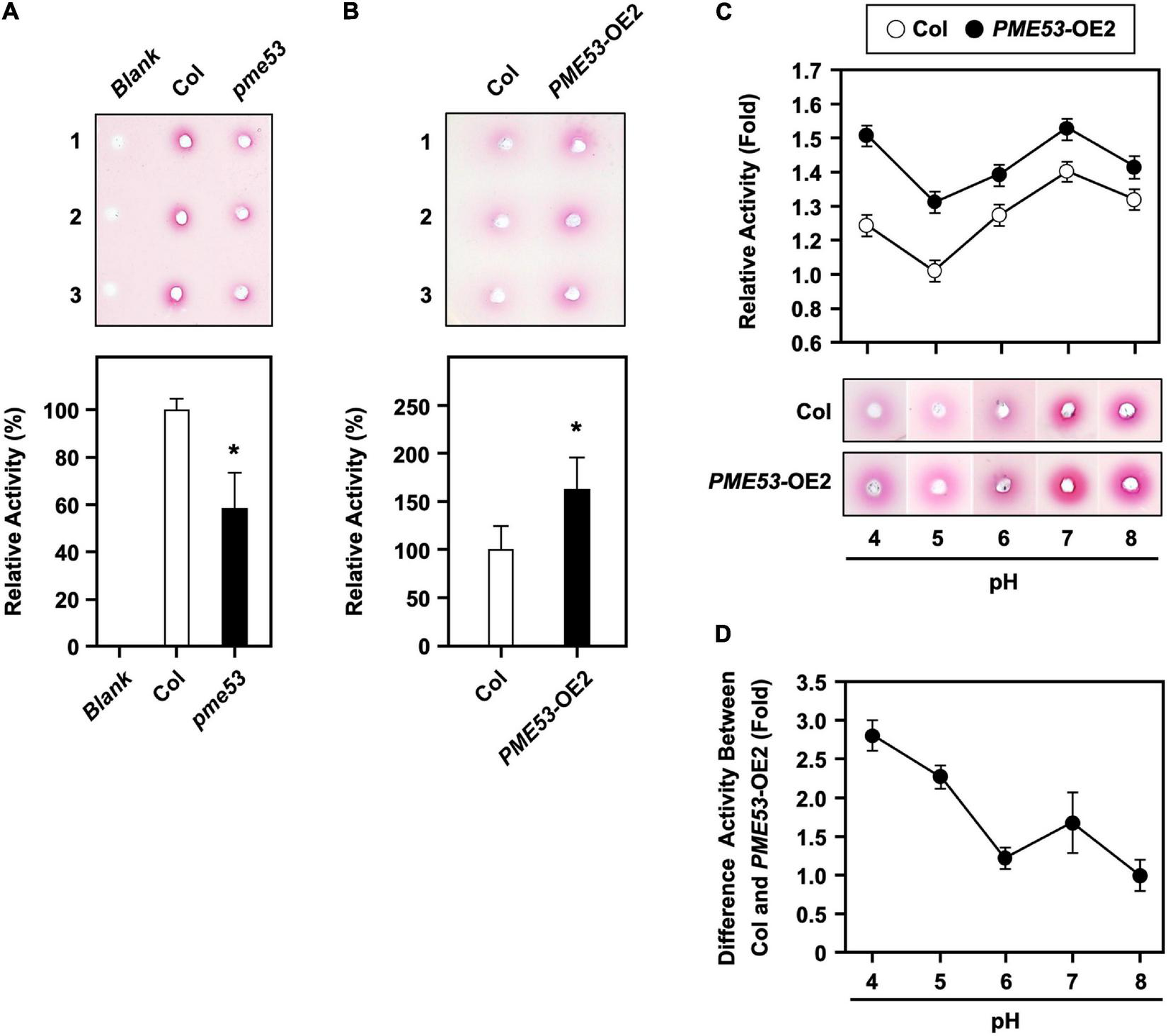
PME activity assay in pme53 mutant and PME53-overexpression plants. PME activity was analyzed (A) in Col and pme53 and (B) in Col and PME53-OE2 plants. The ruthenium red-stained zone diameters resulting from the hydrolysis of esterified pectin in an agarose gel by diffusion were photographed and measured. The fold change PME activity was normalized relative to level of the Col. Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the Col. Blank, the buffer used only. (C) PME activity analyzed at pH levels of 4 to 8. PME activity was normalized to the weakest activity at pH 5 of the Col. (D) The optimal PME53 activity at different pH levels was summarized. Data are mean ± SE of three biological replicates.
Phenotyping of pme53 Mutant Plants
A comparison of the rosette leaf index showed that the leaf width to length ratio in the leaves of pme53 was significantly smaller than that of the Col (Figure 4A). In particular, the size of the cotyledon (cm2) of 7-day-old pme53 appeared smaller and shorter than that of the Col.
FIGURE 4
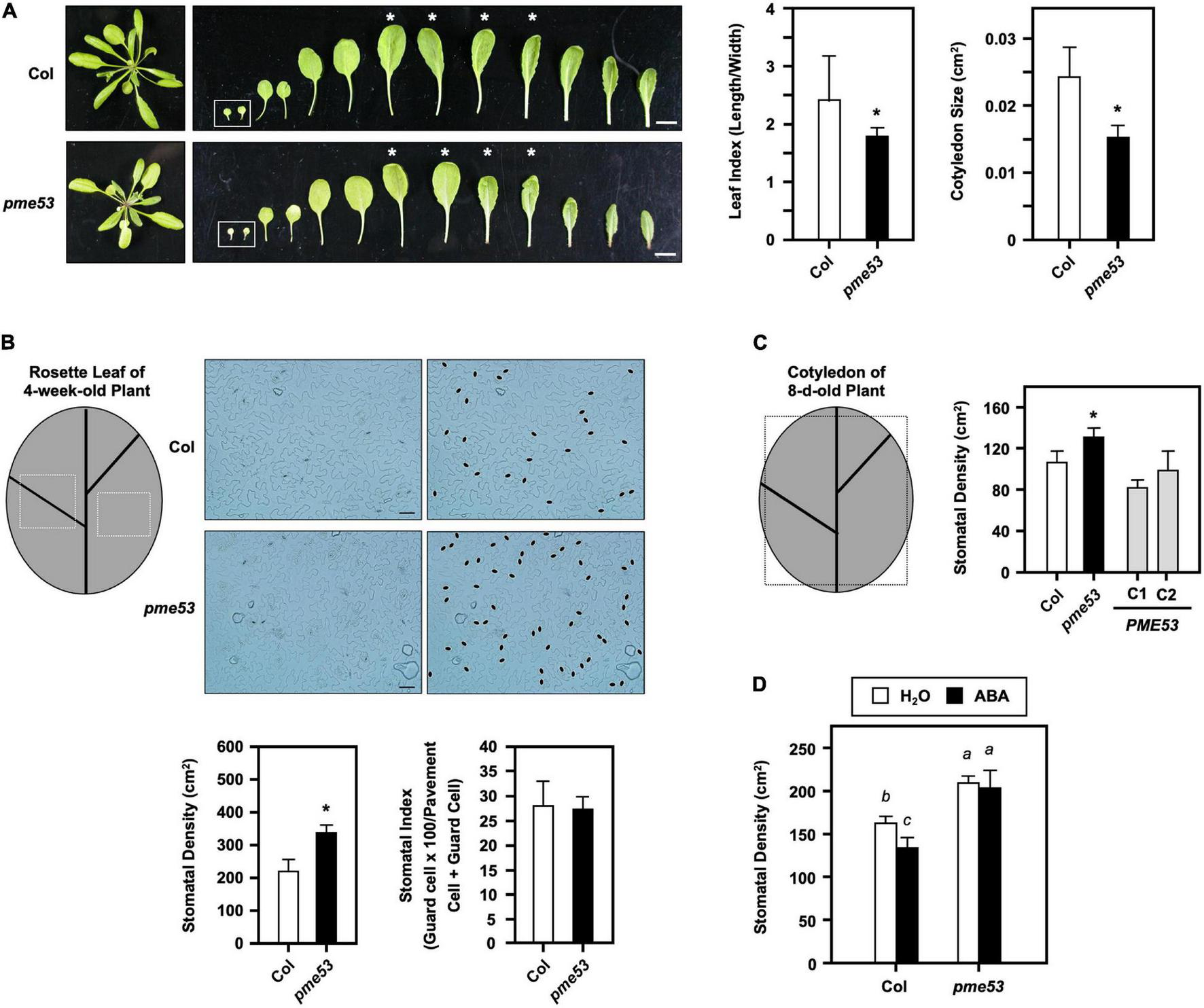
Phenotyping of pme53 mutant plants. (A) 4-week-old plants and leaves were photographed. Cotyledons are shown in the inset. Leaf index (length/width) was determined by the ratio of the length to width of the 3rd and 4th pair of rosette leaves (asterisk indicated) and the size of the 7-day-old cotyledon (cm2) was measured. n = 50 cotyledons. Bars = 1 cm. (B) Epidermal cells on the abaxial surface of 3rd and 4th pair leaves of 4-week-old plants were photographed. Stomata were highlighted in black ovals. Bars = 50 μm. Stomatal density (stomata/cm2) and stomatal index (guard cell × 100/pavement cell + guard cell) were measured. (C) Stomatal density in the abaxial surface of the cotyledon of 8-day-old plants were measured. PME53-C1 and -C2 are PME53-complemented lines generated in pme53 background. The frames are indicated that the regions used for the stomatal density measurement. *Significant at P < 0.05 compared with the Col. n > 10 leaves and cotyledons. (D) Seedlings were grown together then applied with H2O (control) or 2.5 mL 10 μM ABA, at day 4 after planting. Stomatal development (stomatal/cm2) in cotyledons was analyzed on day 7. Data are mean ± SE of three biological replicates. n > 10 cotyledons. Statistical significances among groups are indicated using different letters (Tukey’s HSD test).
The GUS staining results showed that the PME53 was highly expressed in the vascular tissue of cotyledons (Supplementary Figure 4), and therefore, the vascular network complexity of cotyledons was analyzed (Supplementary Figure 7). According to previous studies, the number of closed areoles (2, 3, or 4) formed by secondary veins and the number of vein branches/incomplete areoles in the proximal (closest to the petiole) part of the cotyledon (Supplementary Figure 7, top) were used to classify vein complexity patterns in cotyledons (Roschzttardtz et al., 2014). In comparison to the Col, pme53 plants had more areoles of 4.1, 4.0, and 3.1, but fewer areoles of 2.2, 3.0, and 2.1 (Supplementary Figure 7, bottom). As a result, pme53 seedlings with 7-day-old cotyledons had abnormal vein networks. These data imply that PME53 may determine the continuity and pattern formation of vascular tissues in Arabidopsis.
Stomatal Density in pme53 Mutant Plants
The stomatal density (number of stomata/cm2), stomatal index (guard cell × 100/pavement cell + guard cell), and pavement cell size were analyzed on the leaf epidermal layers of pme53 (Figure 4B). The stomatal number and pavement cell size were largely affected and were analyzed on the 7th and 8th leaves of 4-week-old pme53 compared with the Col (Figure 4B). Higher stomatal density was found in pme53; however, the stomatal index was not significantly affected (Figure 4B). Notably, two complementation of pme53 mutation lines, PME53-C1 and C2, restored the high stomatal density phenotype in pme53 to that of the Col (Figure 4C).
It is known that ABA treatment reduces the number of stomata cells in Col plants (Tanaka et al., 2013). Four-day-old pme53 plants were treated with H2O (control) or 10 μM ABA for 3 days, and then the stomatal density of cotyledon was measured. The effect of ABA on reduced stomatal density was not found in pme53 compared with that in Col plants (Figure 4D).
Stomatal Size and Movements Control in pme53 Mutant Plants
Phenotyping of the stomata in pme53 was conducted to measure the structural features, such as stomatal opening length and width (Figure 5). After incubation in the opening buffer, we showed that stomata lengths were longer and wider in pme53 than in Col plants (Figure 5A). Notably, pme53 showed statistically significant decreases in stomatal aperture compared with the Col (Figure 5A). However, the stomatal aperture in three PME53-OE lines was similar to that in pme53 plants (Supplementary Figure 8).
FIGURE 5
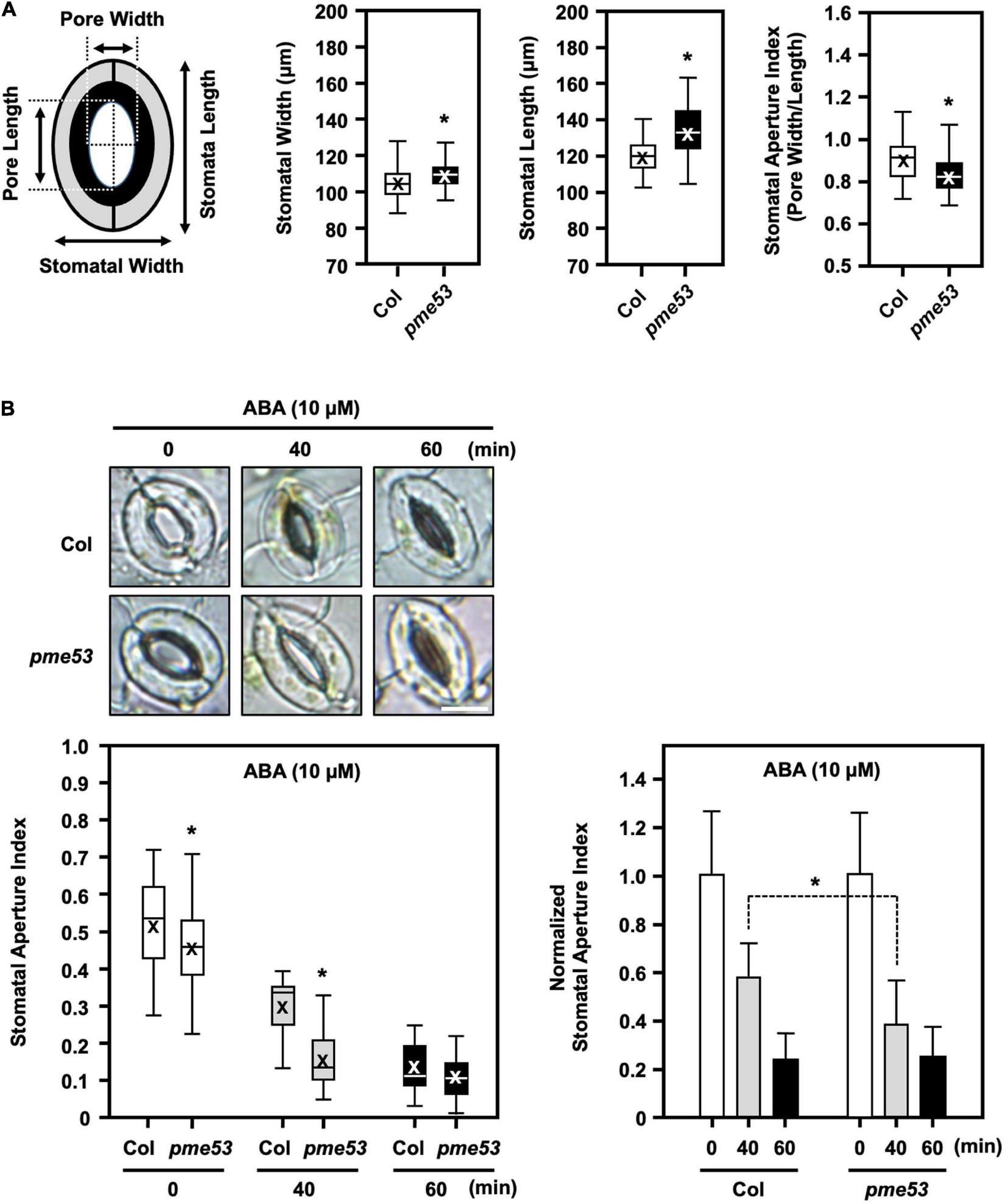
Stomata maximal-opening ability and movement in pme53 mutant plants in response to ABA. Schematic representation of stomata pore measurement. (A) Epidermis peels of the fully expended 4-week-old plants incubated in an opening buffer under light for 3 h then the stomata maximal-opening ability was analyzed stomatal length (μm) and width (μm), as well as the stomatal aperture index (pore width/length), were measured. n > 80 stomata. (B) Stomatal movement in response to 10 μM ABA for 40 or 60 min was measured. The stomata aperture index and the normalized stomata aperture index were measured. n > 80 stomata. Representative images of stomata were shown. Bars = 10 μm. Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the Col (ANOVA Tukey-Kramer post hoc test).
In response to 10 μM ABA treatment, pme53 showed more sensitivity to stomatal closure than Col plants (Figure 5B). Our results suggest that PME53 plays a predominant role in the regulation of stomatal patterning and movement control.
PME53 Controls in Stomata Development
Abscisic acid-mediated regulation of stomatal density acts upstream to regulate the expression of SCREAM (SCRM, also known as ICE1) and MUTE genes, which are master regulators for stomatal formation in Arabidopsis. One SCRM-knockout mutant (scrm; ice1-2) was confirmed by q-PCR analysis (Figure 6A, left) and used to elucidate the requirement of the PME53 gene expression. In the scrm mutant, PME53 expression was reduced (Figure 6A, middle), whereas MUTE expression was increased (Figure 6A, right).
FIGURE 6
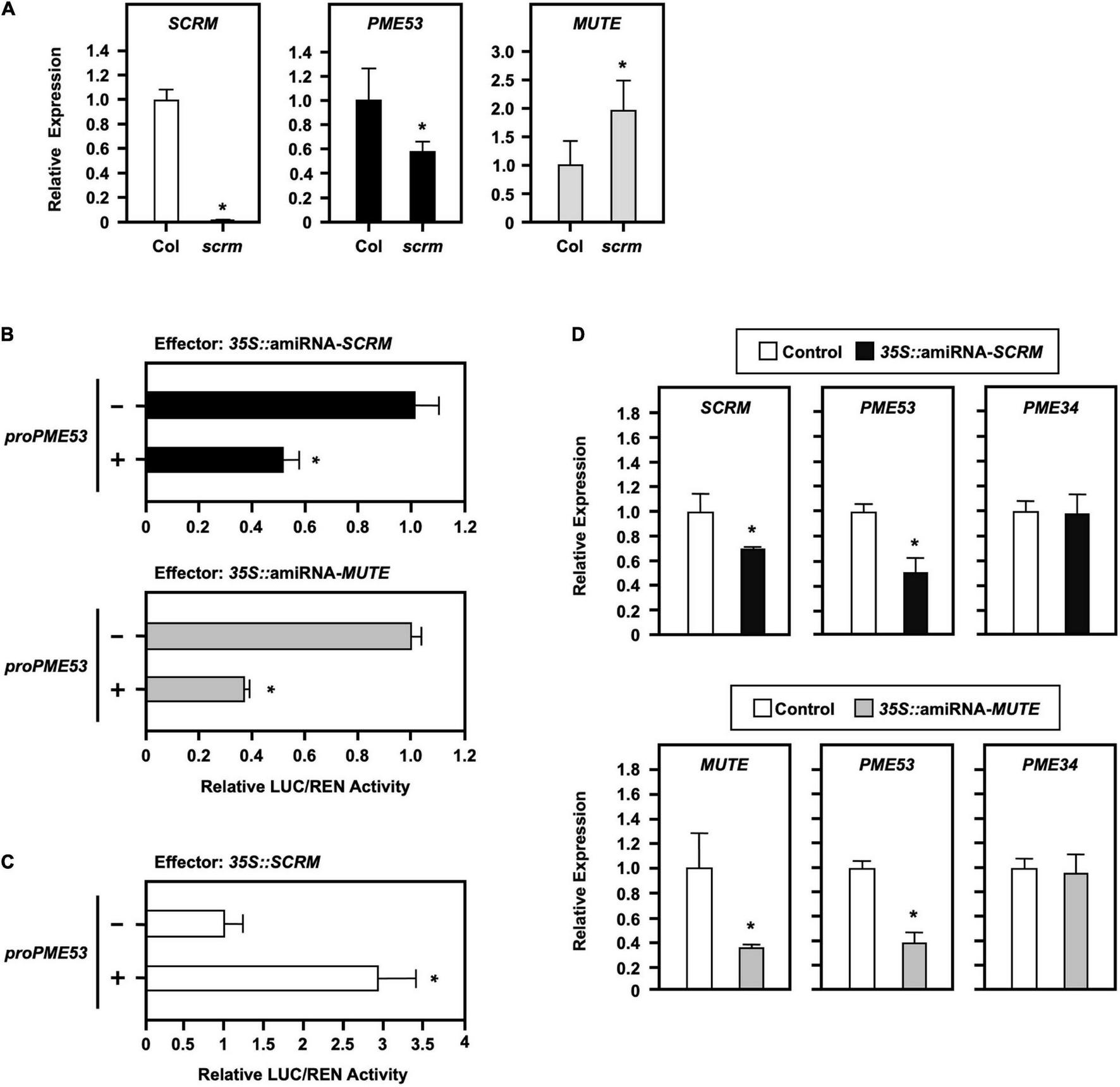
Effect of the stomatal-positive master regulator SCRM on PME53 expression. (A) The expression levels of SCRM, PME53, and MUTE in 7-day-old seedlings of Col and scrm (the null-mutant line) were analyzed by q-PCR. The fold change expression was normalized relative to the level of Col. (B,C) The 35S:Effector of artificial miRNA mediated the suppression of SCRM and MUTE expression (amiRNA-SCRM and amiRNA-MUTE) and 35S:SCRM, were used for the trans-activation analysis of PME53-promoter:LUC reporter in Arabidopsis protoplasts, respectively. The fold change expression was normalized relative to the activity of LUC/REN using the dual-luciferase system. *Significant at P < 0.05 compared with the mock control (-). (D) The expression levels of SCRM, MUTE, PME53, and PME34 were analyzed by q-PCR. PME34 was used as a negative reference control. Data are mean ± SE of three biological replicates.
Furthermore, a 1.974-kb promoter region of PME53 fused with a reporter Firefly luciferase (LUC) and co-transfected with the effector of a 35S-driven artificial miRNA, amiRNA-SCRM and amiRNA-MUTE, to mediate the downregulation of SCRM and MUTE analyzed in Arabidopsis protoplasts (Figure 6B). Renilla luciferase (REN) activity was used as the input control, and the activity of LUC/REN normalized the fold of expression. The results of transactivation assays highlighted that the overexpression of amiRNA-SCRM and amiRNA-MUTE led to the downregulated expression of PME53 (Figure 6B). In addition, overexpression of SCRM exerted positive effects on the expression of PME53 (Figure 6C). The overexpression of amiRNA-SCRM and amiRNA-MUTE suppressed the expression of SCRM and MUTE, which led to the downregulation of PME53 gene expression (Figure 6D). Arabidopsis PME34, a stomatal-density-independent gene, was used as a negative reference control (Huang et al., 2017).
pme53 Mutant Plants in Response to Heat Stress
Stomata regulate gas exchange by allowing water vapor to leave the plant and CO2 to enter to keep the leaf temperature stable. Although the pme53 had a higher stomatal density and decreased in stomatal aperture, preliminary data indicated that pme53 did not significantly influence water transpiration rates during 2 h recovery from a 1-h 37°C mild heat shock treatment (Supplementary Figure 9). It has been reported that PME34 is required for proper heat response due to its role in promoting stomatal movements, as we previously reported (Huang et al., 2017). To gain insight into whether PME53 plays a role in the heat response, we showed that the expression of PME53 gene was slightly downregulated on 1-h 37°C heat shock treatment (Figure 7A). However, PME53 expression was significantly increased after a 44°C-lethal heat shock (LHS) and 3-h recovery from the LHS (LHSR) (Figure 7A). HSP18.1, an HS-responsive marker gene, was used as reference. Notably, the pme53 exhibited a less heat-sensitive phenotype with a higher survival rate compared with the Col (Figure 7B), while complementation lines, PME53-C1 and C2, restored the defective heat-responsive phenotype similar to that of Col plants. hsp101, a heat-sensitive mutant, was used as reference.
FIGURE 7
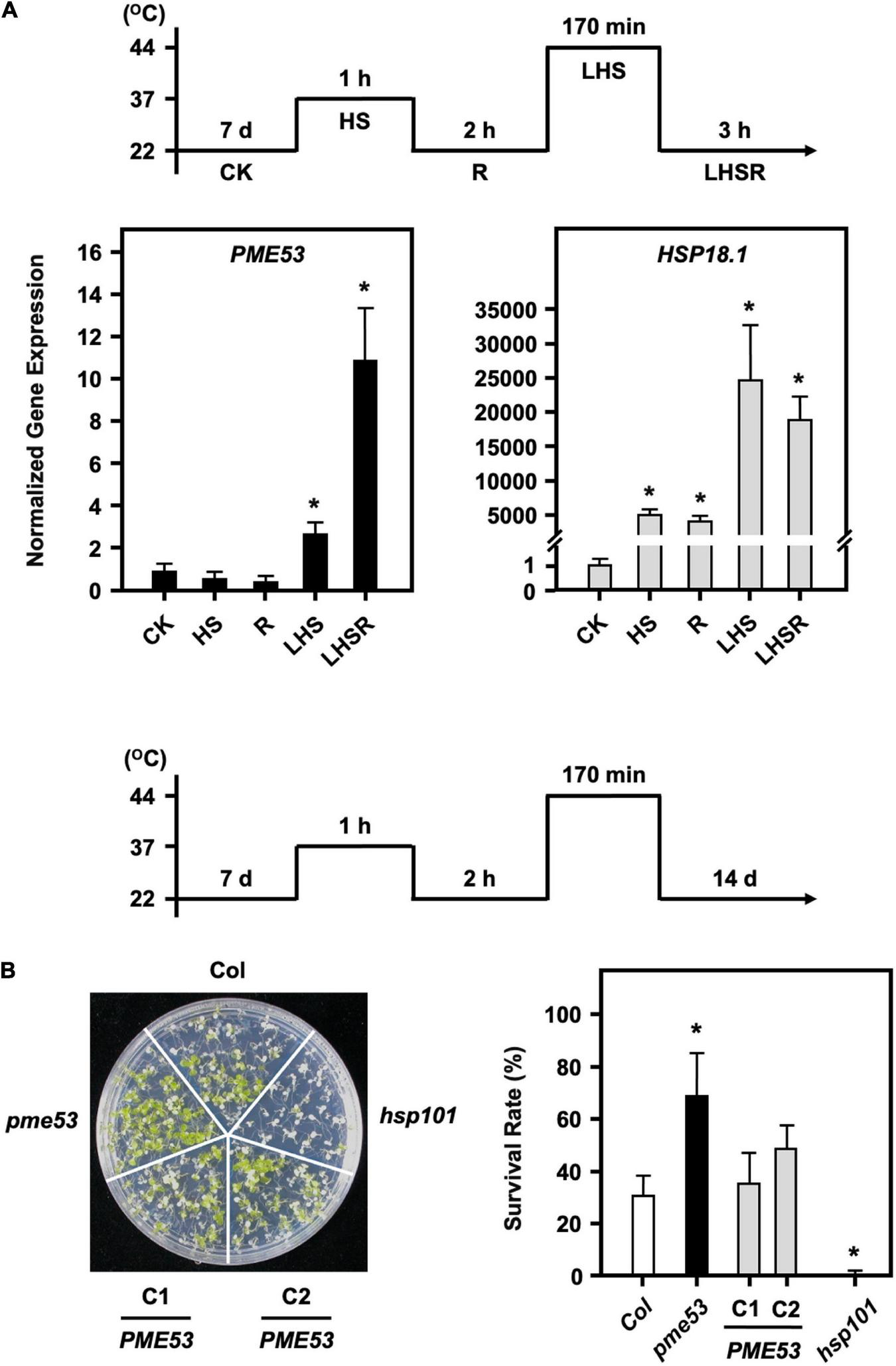
pme53 mutant and its complementation lines in response to heat stress. The pictogram shows the heat stress regime. (A) The expression levels of PME53 in 7-day-old seedlings were analyzed by q-PCR. HSP18.1, a heat-responsive gene, was used as reference. The fold change expression was normalized relative to the level of control (CK). Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the control. (B) Seedlings were photographed and survival was measured at 14 days after the LHS treatment. PME53-C1 and -C2 are PME53-complemented lines generated in pme53 background. hsp101, a heat-sensitive mutant, was used as reference. Data are mean ± SE of three biological replicates (n = 50 seedlings). *Significant at P < 0.05 compared with the Col.
We showed that pme53 was properly perceived and responsive to the heat shock at 37°C for 1 h. In agreement with the expression levels of major heat-responsive genes, including HSP101, HSP90, HSP70, and HSP18.1, were unaffected compared with the Col (Supplementary Figure 10).
Status of De-Methylesterified Pectins in pme53 Mutant Plants in Response to Heat Stress
To assess the correlation between patterns of pectic enzyme production and heat response, ruthenium red (RR), a cationic reagent, has been used to visualize polysaccharides on the plant cell wall (Wu et al., 2010; Suzuki et al., 2011). Firstly, we characterized pectic substances in the cotyledon of 7-day-old seedlings by RR staining in situ, pme53 showed a less RR stain-abled demethylesterified pectin than the Col after a 44°C LHS and 3-h recovery from the LHS (LHSR) (Supplementary Figure 11A).
Our previous studies confirmed that the inward movement of Ca2+ and the recovery of heat shock-triggered released apoplastic Ca2+ accompanied by PME activity are required for thermotolerance (Wu et al., 2010). Thus, the total PME activity in pme53 was analyzed under heat shock (Figure 8). The PME activities in Col were induced in response to heat shock treatments at 37°C and 44°C, while that of pme53 was significantly reduced under normal growth conditions, response to 37°C heat shock, and recovery form 37°C heat shock compared with the Col (Figure 8, HS and R). Notably, the total PME activity was activated in pme53 during 44°C-LHS treatment similar to the Col (Figure 8, LHS), we suggest that other unclarified PMEs were activated to compensate for the PME53 activity in pme53. Also, the PME53-OE2 has been shown a similar PME activity pattern as that of Col plants in response to heat stress (Supplementary Figure 11B).
FIGURE 8
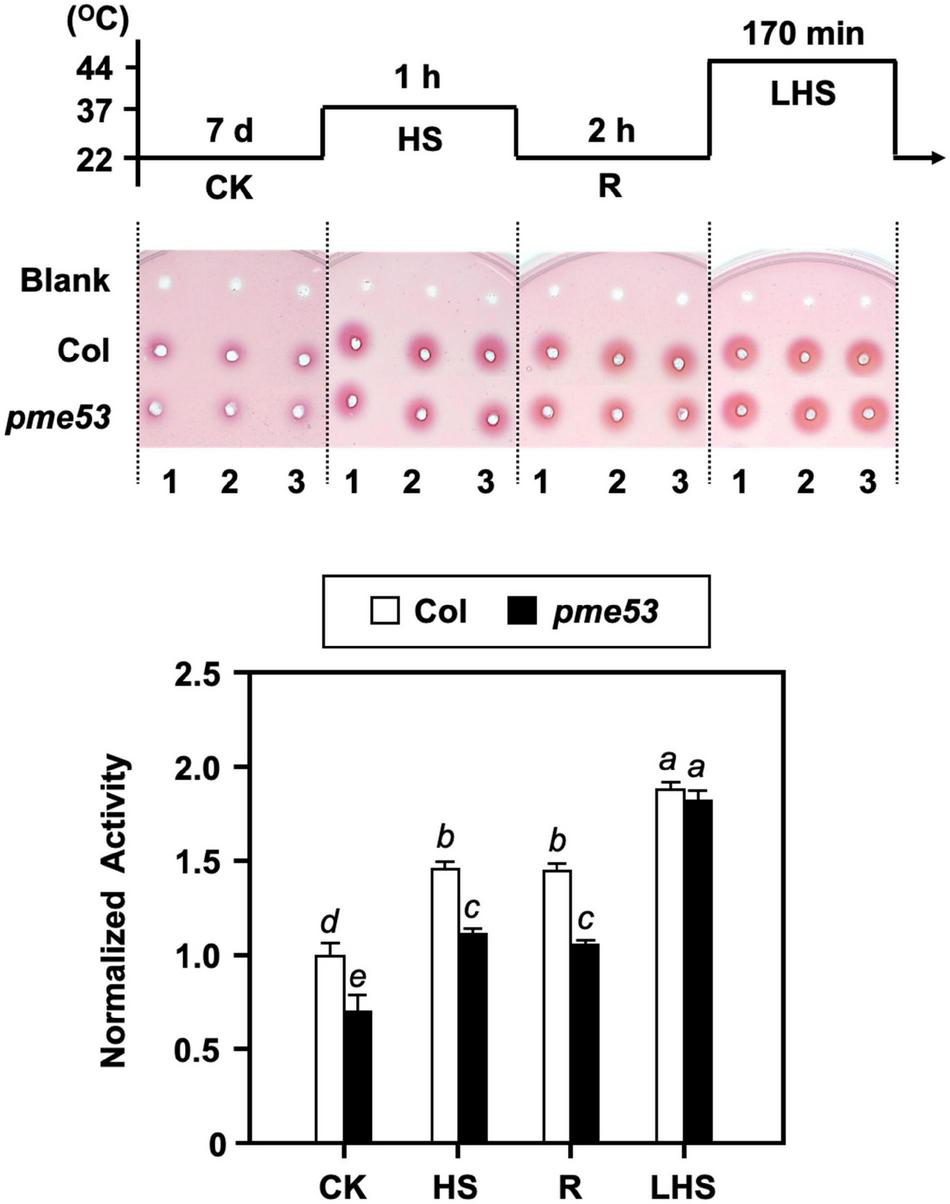
PME activity in pme53 mutant plants in response to heat stress. The pictogram shows the heat stress regime. PME activity in 7-day-old seedlings was analyzed. The ruthenium red-stained zone diameters resulting from the hydrolysis of esterified pectin in an agarose gel by diffusion were photographed and measured. The fold change PME activity was normalized relative to level of the Col control (CK). Blank, the buffer used only. Data are mean ± SE of three biological replicates. Statistical significances among groups are indicated using different letters (Tukey’s HSD test).
PME53 May Be Involved in Pectin Methylesterase-Mediated Inward Movement of Ca2+
To confirm that the thermotolerant phenotype of pme53 is related to the PME-mediated inward movement of Ca2+. 7-day-old seedlings were treated with 20 μM EDTA or 20 μM EDTA + 15 mM CaCl2 before heat stress (Figure 9A). Our results demonstrated that the EDTA-treated seedlings in pme53 showed a thermoresponsive phenotype similar to that of Col plants. Meanwhile, treatment with Ca2+ to antagonize the effect of EDTA restored the less heat-sensitivity phenotype of pme53 that was similar to H2O control treatment.
FIGURE 9
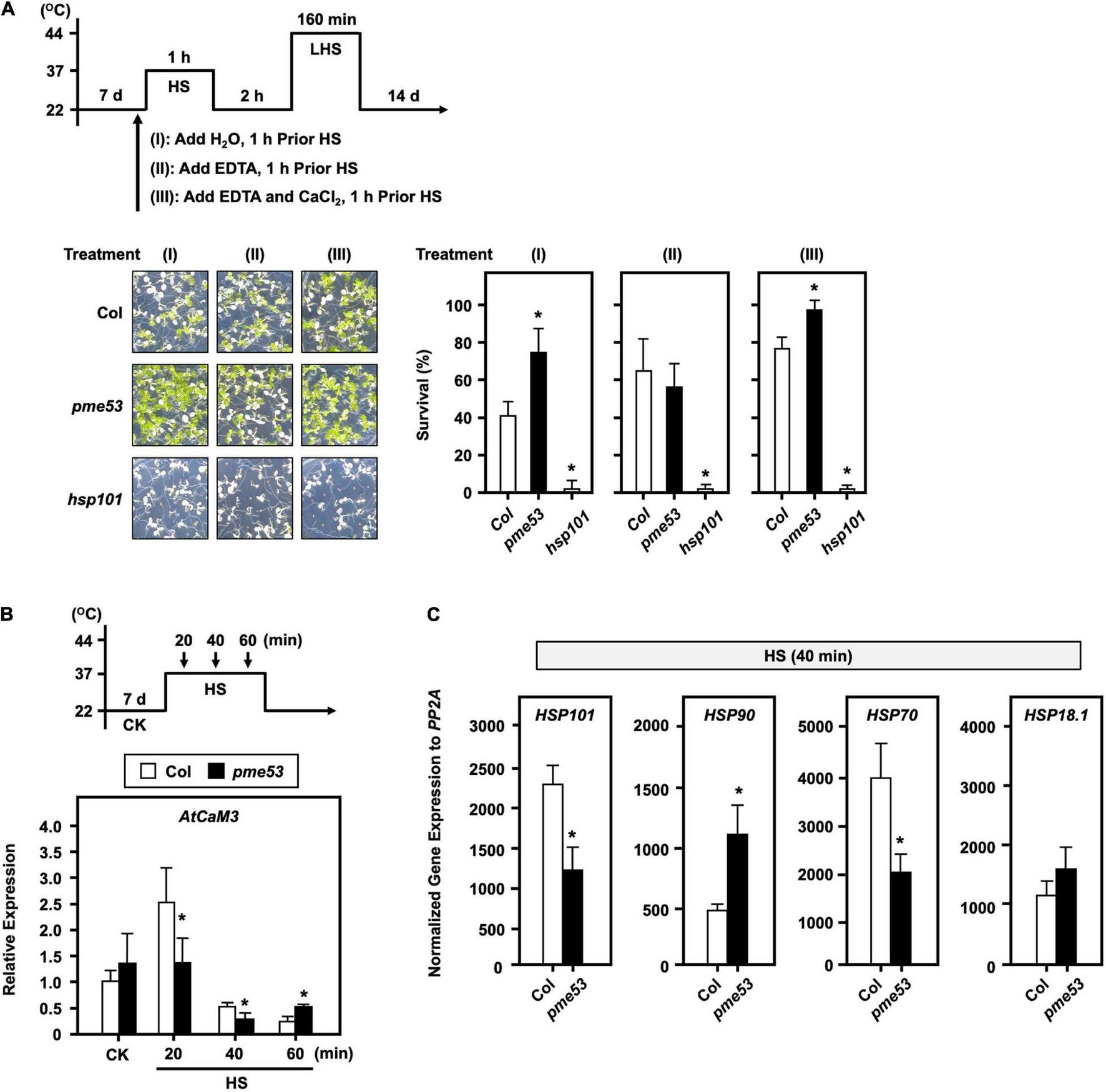
The effect of EDTA and Ca2+ on the phenotype of pme53 plants and expression levels of AtCaM3 in response to heat stress. The pictogram shows the heat stress regime. (A) Seedlings were applied with 2.5 mL H2O, 20 μM EDTA, or 20 μM EDTA with 15 mM CaCl2 solution, 1 h before HS treatment. Seedlings were photographed and survival was measured at 14 days after 44°C-LHS treatment. hsp101, a heat-sensitive mutant, was used as reference. Data are mean ± SE of three biological replicates (n = 50 seedlings). *Significant at P < 0.05 compared with the Col. (B) 7-day-old seedlings in response to 37°C HS for 20 to 60 min. The expression levels of AtCaM3 were analyzed by q-PCR. The fold change expression was normalized relative to the level of Col control (CK). (C) The expression levels of heat-responsive genes after 40-min 37°C HS were analyzed by q-PCR. Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the Col.
In addition, Arabidopsis calmodulin AtCaM3 is a key member of the Ca2+-mediated heat stress signaling pathway (Zhang et al., 2009). The Ca2+-induced AtCaM3 gene expression reached its maximum level after 20 min 37°C heat stress in Col; however, it significantly reduced expression in pme53, which was much lower than that of Col (Figure 9B). The expression level of HSP90 was increased, while HSP70 and HSP101 were decreased in pme53 significantly than that of Col at the early 40 min 37°C heat stress (Figure 9C), indicating that PME53 may coordinate with heat stress signaling, which is involved in Ca2+-pectate reconstitution to render thermotolerance in plants.
Abscisic Acid Pre-treated pme53 Mutant Plants in Response to Heat Stress
To verify that ABA signaling is required for the expression of PME53, we examined ABA-deficient (aba2-1) mutant and ABA-insensitive (abi1-1) mutant plants by ABA treatment. The expression level of PME53 was significantly affected in abi1-1 (Figure 10A). We also showed that the pme53 were more sensitive to 0.25 and 1.0 μM ABA treatments, with lower post-germination growth rates than the Col (Figure 10B). These results indicate that ABA perception and response are required for the appropriate expression of PME53. Arabidopsis ABA-insensitive 4 mutant (abi4-1) was used as a reference control (Söderman et al., 2000).
FIGURE 10
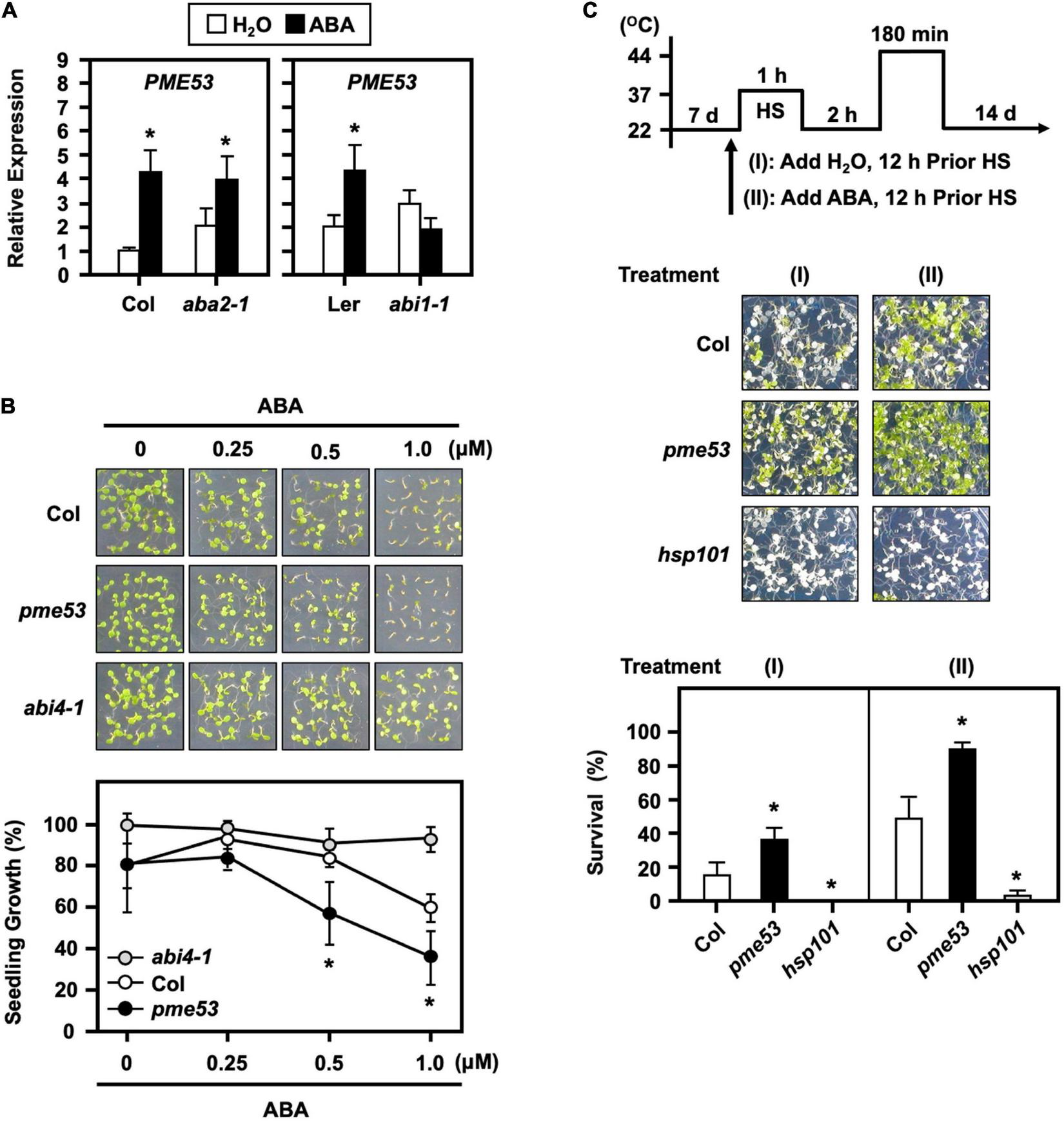
Transcriptional levels of PME53 in ABA-deficient (aba2-1) and ABA-insensitive (abi1-1) mutant plants and the effect of ABA on pme53 mutant post-germination growth and in response to heat stress. (A) 7-day-old seedlings were treated without (H2O control) or with 30 μM ABA for 3 h. The expression levels of PME53 were analyzed by q-PCR. The fold change expression was normalized relative to their corresponding wild-type (Col or Ler) plants. Data are mean ± SE of three biological replicates. *Significant at P < 0.05 compared with the Col or Ler. (B) Seedling growth without or with 0.25 to 1 μM ABA on plates was photographed and measured at day 7 after planting. abi4-1, an ABA-insensitive mutant, was used as reference. (C) The pictogram shows the heat stress regime. Plates were incubated with 2.5 mL H2O or 10 μM ABA solution, 12 h before HS treatment. Seedlings were photographed and survival was measured at 14 days after the LHS treatment. hsp101, a heat-sensitive mutant, was used as reference. Data are mean ± SE of three biological replicates (n = 50 seedlings). *Significant at P < 0.05 compared with the Col.
The effects of pre-treatment with ABA on plants responses to heat have been investigated, and it has been shown that plants have a better survival rate than untreated (Huang et al., 2016). Here, a 10 μM ABA applied 12 h before the heat stress conferred an increased thermotolerance in the Col plants (Figure 10C). Notably, the ABA-pretreated pme53 showed a higher survival rate than that of the untreated plants. The results implied that the PME53 gene was necessary and not functionally redundant in the ABA-mediated heat response.
Discussion
Cell wall enzymatic modification of the pectin network in the functional properties of guard cell walls has an important effect on normal stomatal function. Guard cell walls in pme6 plants are enriched in methylesterified pectin and decrease the response to trigger stomatal movement (Amsbury et al., 2016). This indicates that the mechanical change in the guard cell wall requires de-methylesterification of pectin, indicating the significant role of PMEs in stomatal function. In the current study, we identified a PME gene, PME53, which is an ABA-responsive gene highly expressed in guard cells, and plays a significant role in stomatal development through the regulation of the core stomatal SCRM and MUTE transcription factors, and modulating the flexibility of the guard cell wall for elevated temperature adaptation. Hence, we determined that the function of PME53 showed the significance of ABA-mediated changes in guard cell walls involving the inflation characteristics of guard cell walls. Thus, the integration of ABA and PME activity is important for controlling stomatal functions by controlling guard cell wall plasticity in Arabidopsis.
PME53, a Guard-Cell-Specific Gene, Is Involved ABA-Induced Stomatal Aperture
The molecular mechanisms underlying ABA-mediated stomatal aperture, thereby inducing adaptation to stress conditions, have been extensively investigated (Wasilewska et al., 2008; Hubbard et al., 2010; Lee and Luan, 2012). In addition, signaling by stress phytochrome ABA is involved in the activation of heat-responsive genes and acquired thermotolerance (Baron et al., 2012; Huang et al., 2016; Zandalinas et al., 2016). It has been shown that ABA changes heat-induced stomatal opening toward a higher threshold temperature, and this opening at elevated temperatures was fully reversible. It can be concluded that elevated temperatures can stimulate ABA-mediated stomatal opening, which has an opposite role to ABA in promoting stomatal closure under drought stress (Feller, 2006; Reynolds-Henne et al., 2010). Multiple ABRE domains were found in the promoter of PME53 (Supplementary Figure 3), which was highly induced by exogenous ABA treatment (Figure 1A). PME53 exhibited a tissue-specific pattern that was highly expressed in guard cells (Supplementary Figure 1 and Figure 1B), indicating that PME53 acts as a positive regulator of ABA signaling in guard cells.
The role of ABA in the regulation of guard cell wall structure, which determines stomatal aperture changes in response to stress, is still unknown. We found that the stomatal aperture in pme53 was smaller than in Col plants (Figure 5A). Furthermore, in response to ABA treatment, the stomatal movement was more sensitive in pme53 than in Col plants (Figure 5B), implying a strong and positive correlation between ABA-mediated stomatal aperture and PME53 function. ABA production and stomatal sensitivity to ABA were drastically reduced in aba2-1 and abi1-1 (Koornneef et al., 1982, 1984). Comparison of sensitivity to ABA inhibition of germination on pme53 greatly increased ABA levels and significantly lowered the germination rate compared to Col and ABA-insensitive 4 (abi4-1) mutant plants (Figure 10B). As a result, PME53 may contribute to the initiation of stomatal aperture control under unfavorable stress conditions as an ABA-inducible gene.
PME53 Involves in Stomatal Density Control and Stomatal Lineage Pathway
It is known that cell shape, cell size, and cell division of plant cell walls can be affected by intra- and extracellular cues; however, the relationship between pectin HGA methylesterification and epidermal cell growth remains elusive. The shape of pavement cells (non-stomatal cells) due to tension in the periclinal walls and their local reinforcement is linked to pectin de-methylesterification (Altartouri et al., 2019). The undulatory-shaped lobe of pavement cells can be affected by de-methylesterification of HGA with PME activity and interaction with Ca2+ in Arabidopsis epidermal cells (Haas et al., 2020). Epidermal pavement cells predominantly exhibit de-methylesterified pectin patterns, which can increase the stiffness of cell walls by crosslinking with Ca2+ (Bidhendi et al., 2019). Therefore, the formation of lobes in the epidermal pavement cells of leaves is associated with increased cell wall thickness, radial microfibril distribution, and HGA de-methylesterification (Majda et al., 2017). In Arabidopsis, un-methylesterified HGA is the predominant form of pectin in the guard cell wall, but highly methylesterified and Ca2+ cross-linked de-methylesterified HGA are present in the epidermal pavement and mesophyll cells, indicating that the mechanical properties of guard cells are largely dependent on the composition and structure of guard cell walls (Amsbury et al., 2016). Here, we showed that the leaf and pavement cells on the epidermis appeared to be different in pme53 compared to Col (Figure 4A and Supplementary Figure 7). Our data indicated that PME53 may be involved in the morphogenesis of pavement cells by regulating the methylesterification status of pectin. In addition, the results of this study may provide crucial evidence underlying the initiation mechanism of cell shaping interrelated with the roles of de-methylesterified pectin of PMEs during the developmental process of the undulations of pavement cells.
Variation in stomatal density may arise due to genetic factors and/or growth to adapt to various environmental stresses (Bertolino et al., 2019). In addition, stomatal density directly affects stomatal efficiency; therefore, the development of stomata is tightly controlled by developmental and environmental cues to ensure precise stomatal lineage and patterning (Zhu et al., 2020). The stomatal apertures of pme53 and PME-OEs plants were smaller than those of Col (Figure 5 and Supplementary Figure 8). The stomatal densities of pme53 were significantly higher than those of Col plants (Figure 5B). An ABA-deficient aba2-2 mutant had an increased stomata density within a smaller cotyledon, and reduced expansion of pavement cells, indicating that ABA negatively regulation in the initiation of stomatal development and ABA action on the enlargement of pavement cells in Arabidopsis cotyledons (Tanaka et al., 2013). The pme53 exhibited an increased stomatal density and significantly reduced expansion of pavement cells. We suggest that PME53 may be involved in the ABA-mediated regulation of stomatal density and pavement cell expansion. Interestingly, the application of exogenous ABA did not reduce the stomatal density in pme53 plants as that of the Col (Figure 4D), which is a different feature compared with the aba2 mutant (Tanaka et al., 2013). Additionally, the expression levels of PME53 were induced under ABA treatment in aba2-1 but not abi1-1 (Figure 10A), indicating that PME53 plays a role in ABA perception and signaling. Besides, the pme53 conferred a better survival rate than Col under heat stress (Figure 7B); therefore, we suggest that pme53 exhibited a significantly higher stomatal density, correlating with remarkable resistance to temperature elevation.
Stomatal cell fate and patterning, which are controlled by key bHLH transcription factors, including SPCH, MUTE, FAMA, and their partners ICE1 and SCRM2, play a critical role in regulating sequential developmental decisions in Arabidopsis guard cell differentiation at different steps (Hofmann, 2008; Davies and Bergmann, 2014). Since SCRM form dimers with MUTE, FAMA, and SPCH, to regulate the formation of stomata (Hofmann, 2008). MUTE marks a subset of meristemoids that are differentiating into guard mother cells (Pillitteri et al., 2007). Additionally, PME6 mRNA was highly accumulated in the scrm-D mutant, which has an excess of mature guard cells, and epidermal cell differentiation was blocked at the pavement cell stage (Amsbury et al., 2016). Transcriptome analysis revealed that PME53 expression was also upregulated in scrm-D mute-double mutant plants (Pillitteri et al., 2011). Based on literatures, the present study focused on the analysis of SCRM and MUTE genes, which are strongly linked to pectin de-methylesterification of the guard cell wall. The results of the transactivation assay and RNA level highlighted the propensity of PME53 to be positively regulated by SCRM and MUTE (Figure 6), indicating that PME53 may function as a critical element for stomatal development at a relatively late stage of differentiation.
Abscisic acid might be involved in stomatal initiation and differentiation by repressing SPCH and MUTE (Le et al., 2014), even though our data indicated that PME53 was positively regulated by ABA and SCRM, as well as MUTE. SCRM is also an inducer of CBF expression 1 (ICE1), a master regulator of freezing tolerance, mediating the cold stress signal via an ABA-independent pathway (Kanaoka et al., 2008; Liang and Yang, 2015). To the current our knowledge, little is known about the heterodimerization between SCRM and MUTE is involved in ABA-regulated which still need to be investigated. With limited studies on the divergent regulation of ABA, how ABA-mediated stomatal development through the regulation of PME53 has yet to be determined.
The Defective of PME53 With Altered the Status of Pectin Methylation Has an Impaired Stomatal Function in Response to Heat Stress
Pectins have a conserved role in stomata, and the modification of the composition and structure of guard cell walls can directly affect stomatal functions. In the guard cell wall of pme6, methylesterified pectins are enriched, and the response to stomatal opening/closure triggers may be due to a mechanical change in the guard cell wall (Amsbury et al., 2016). The STOMATAL CARPENTER1 (SCAP1) mutant, the pectin de-methylesterification in the ventral wall of guard cell walls was suppressed and showed an abnormal shape of stomata and reduced efficiency of stomatal movement (Negi et al., 2013). The degrees of de-methylesterified pectin on pme53 and cotyledons differed from that of Col, indicating that their PME activity was affected (Figure 3A). Thus, this finding suggests that stomatal function is involved in the de-methylesterification of pectin in the guard cell wall. Our results may establish a link between the genes of guard cells and their cell wall properties with the corresponding effect of PME de-methylesterification on stomatal function.
During PMEs acting on the HGA chain, the effects can be classified as linear or non-block-wise de-methylesterification. The non-blockwise removal of methyl groups of HGA by PME following PG activity catalyzes the degradation of pectin and thus loosens the cell wall. In addition, it is hypothesized that the optimal PMEs at acidic pH tend to be randomly active in the HGA chain. AtPME2 participates in pectin de-methylesterification by randomly acting on the HGA chains, leading to the creation of substrates for PGs and consequent de-structuration of HGA; therefore, pme2 showed high methylesterification of pectins, which prevented the hydrolysis by PGs and reduced elongation by stiffening the walls (Hocq et al., 2021). In this study, the activity of PME53 appeared to change with pH, indicating a pronounced pH dependence of its activity. PME53 exhibited optimum activity in the acidic context of the cell wall, suggesting that PME53 might mediate the random de-methylesterification of pectin (Figures 3C,D). That is, PME53 may contribute to loosening the cell wall by mediating the de-methylesterification of HGA randomly at acidic pH, leading to the formation of demethylated stretches. We suggest that the enzymatic activity of PME53 has dramatic consequences on cell wall texture and mechanical properties.
Apoplastic Ca2+ is essential for controlling cell integrity, cell wall cohesion, and plasma membrane permeability in plant cells. The recovery of heat shock-released Ca2+ is essential for acquiring thermoprotection to mitigate lethal heat injury in both soybean and rice seedlings (Wu and Jinn, 2010; Wu et al., 2010). EDTA treatment chelates the apoplastic Ca2+ and hence lowers the thermotolerance of pme53 (Figure 9A). This suggests that the thermoprotection of pme53 might be due to the different levels of extracellular Ca2+, which can be transported into the cell as stress-induced pathway signals. As a result, de-methylesterification caused by PMEs in the cell wall can directly impact the properties of pectin’s cell wall in terms of extracellular free Ca2+. During heat shock, the [Ca2+]cyt signature changes by initiating biphasic elevation after 10 min in rice (Oryza sativa) (Wu et al., 2012). In pme53, the expression of AtCaM3 which is a key member participating in the heat signal pathway (Zhang et al., 2009), was significantly reduced after 20 and 40 min of heat treatment (Figure 9B). At 37°C heat shock for 40 min, the expression of HSP70 and HSP101 is altered in pme53 compared to Col (Figure 9C), indicating that PME53 may coordinate with heat stress signaling.
Based on our previous studies (Wu and Jinn, 2010; Wu et al., 2010, 2012, 2017, 2018), the cleavage of apoplastic Ca2+ bridges between pectic carboxyl groups by the activity of PMEs is important for cell wall remodeling to retain cell integrity under heat stress. Accordingly, we propose that the Ca2+ distribution in cells of pme53 may be altered during heat shock, and thereby the AtCaM3 perception of Ca2+ signal was affected in pme53. Ca2+ bridging is less effective on a pectic domain when PME53 acts randomly as non-blockwise de-methylesterification; other PMEs may be activated/compensated to work on linearly promoting the formation of Ca2+-mediated cross-linking which may confer the less heat-sensitive phenotype in pme53. The genetic evidence showed that PME53 is required for heat response; however, the underlying molecular mechanism of PME53 remains to be elucidated.
Conclusion
To sum up our findings, we conclude with Figure 11. As demonstrated in this study, the expression levels of PME53 were significantly affected through the expression of SCRM and MUTE, ABA signaling, and recovery from heat stress, therefore we propose that they likely act upstream of PME53, indicating that cell wall modifying enzymes are required to guard cell wall formation appropriately during normal stomatal development and heat response. But the action of ABA on the regulation of SCRM and MUTE interaction remains equivocal. The pme53 exhibited a significantly high stomatal density, we suggest that is correlating with remarkable resistance to temperature elevation. Additionally, the role of PME53 combined with other PMEs in cell wall remodeling involved in Ca2+-pectate reconstitution may play a decisive role in thermotolerance to plants. Present evidence shows that the PME53-mediated de-methylesterification status of pectin is directed toward the effect on stomatal development, movements, and regulating the flexibility of the guard cell wall required for proper heat response.
FIGURE 11
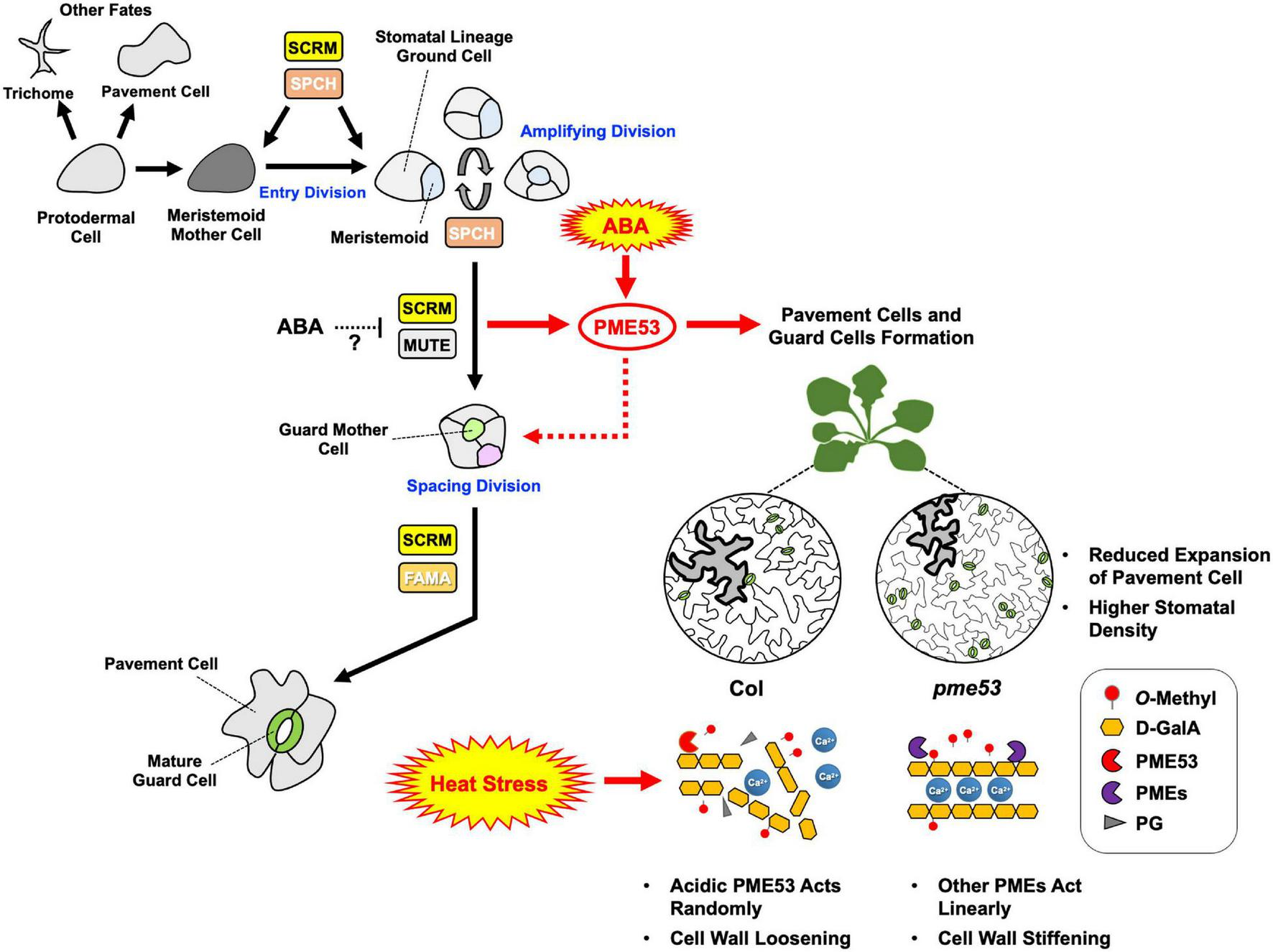
Arabidopsis PME53, a guard-cell specific gene, involves in stomatal development and stress response. The stomatal lineage consists of five major cell types: meristemoid mother cells (MMCs), meristemoids, stomatal lineage ground cells (SLGCs), guard mother cells (GMCs), and guard cells (GCs). GCs initiate from MMCs which are derived from a subset of protodermal cells. MMCs carry out asymmetric entry divisions to produce smaller meristemoids and larger SLGCs. Meristemoids undergo one to three rounds of asymmetric amplifying divisions before differentiating into GMCs. GMCs can undergo symmetric division to form a pair of GCs. Stomatal development is regulated by a group of bHLH transcription factors, including SPCH, MUTE, and FAMA, and the interacting factor SCRM, which are critical to mediate guard cell differentiation at different steps of stomatal lineage in Arabidopsis (Davies and Bergmann, 2014). PME53 may involve in stomatal cell lineage through the regulation of SCRM and MUTE transcription factors. ABA might be involved in stomatal development by repressing SPCH and MUTE. Whether the heterodimerization between SCRM and MUTE is involved in ABA-regulated, still need to be investigated. ABA may act upstream of PME53 to regulate the function of PME53: a null mutation in PME53 results in a significantly increased number of stomata as well as affected stomatal movement in response to heat as compared with the Col. During non-lethal heat stress, acidic PME53 acts randomly on pectin. Absence of PME53 may be led to active other PMEs which work on linearly promoting the formation of Ca2+ mediated cross-linking to confer the less heat-sensitive phenotype in pme53. Black arrows indicate that stomatal lineage signaling pathways have been thoroughly studied. Red arrows represent the new findings in this study, indicating that PME activity is governed by a network of signaling pathways related to ABA regulation and heat stress response.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the Ministry of Science and Technology (MOST), Taiwan (Grant Nos. 107-2923-B-002-003-MY3 and 109-2311-B-002-022-MY3) and National Taiwan University (Grants No. NTU-CC-110L893602) to T-LJ. This work was also supported by MOST and Ministry of Education, Taiwan (Grant Nos. 107-2923-B-002-003-MY3 and PAG107012) to H-CW.
Acknowledgments
We would like to thank the NTU Confocal Microscope Laboratory for performing fluorescence imaging.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.836151/full#supplementary-material
Footnotes
3.^http://wmd3.weigelworld.org/cgi-bin/webapp.cgi
4.^https://imagej.nih.gov/ij/download.html
References
1
AltartouriB.BidhendiA. J.TaniT.SuzukiJ.ConradC.ChebliY.et al (2019). Pectin chemistry and cellulose crystallinity govern pavement cell morphogenesis in a multi-step mechanism.Plant Physiol.181127–141. 10.1104/pp.19.00303
2
AmsburyS.HuntL.ElhaddadN.BaillieA.LundgrenM.VerhertbruggenY.et al (2016). Stomatal function requires pectin de-methyl-esterification of the guard cell wall.Curr. Biol.262899–2906. 10.1016/j.cub.2016.08.021
3
BaronK. N.SchroederD. F.StasollaC. (2012). Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana.Plant Sci.188–18948–59. 10.1016/j.plantsci.2012.03.001
4
BertolinoL. T.CaineR. S.GrayJ. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world.Front. Plant Sci.10:225. 10.3389/fpls.2019.00225
5
BethkeG.GrundmanR. E.SreekantaS.TrumanW.KatagiriF.GlazebrookJ. (2014). Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae.Plant Physiol.1641093–1107. 10.1104/pp.113.227637
6
BidhendiA. J.AltartouriB.GosselinF. P.GeitmannA. (2019). Mechanical stress initiates and sustains the morphogenesis of wavy leaf epidermal cells.Cell Rep.281237–1250.e6. 10.1016/j.celrep.2019.07.006
7
BoschM.HeplerP. K. (2005). Pectin methylesterases and pectin dynamics in pollen tubes.Plant Cell173219–3226. 10.1105/tpc.105.037473
8
BradfordM. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding.Anal. Biochem.72248–254. 10.1006/abio.1976.9999
9
CantarelB. L.CoutinhoP. M.RancurelC.BernardT.LombardV.HenrissatB. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics.Nucleic Acids Res.37D233–D238. 10.1093/nar/gkn663
10
CharngY. Y.LiuH. C.LiuN. Y.HsuF. C.KoS. S. (2006). Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation.Plant Physiol.1401297–1305. 10.1104/pp.105.074898
11
ChowC. N.LeeT. Y.HungY. C.LiG. Z.TsengK. C.LiuY. H.et al (2019). Plantpan3.0: a new and updated resource for reconstructing transcriptional regulatory networks from chip-seq experiments in plants.Nucleic Acids Res.47D1155–D1163. 10.1093/nar/gky1081
12
CloughS. J.BentA. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana.Plant J.16735–743.
13
CzechowskiT.StittM.AltmannT.UdvardiM. K.ScheibleW. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis.Plant Physiol.1395–17. 10.1104/pp.105.063743
14
DaviesK. A.BergmannD. C. (2014). Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential.Proc. Natl. Acad. Sci. U.S.A.11115585–15590. 10.1073/pnas.1411766111
15
Del CorpoD.FulloneM. R.MieleR.LafondM.PontiggiaD.GriselS.et al (2020). AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea.Mol. Plant Pathol.211620–1633. 10.1111/mpp.13002
16
DownieB. D.DirkL. M.HadfieldK. A.WilkinsT. A.BennettA. B.BradfordK. J. (1998). A gel diffusion assay for quantification of pectin methylesterase activity.Anal. Biochem.264149–157. 10.1006/abio.1998.2847
17
FellerU. (2006). Stomatal opening at elevated temperature: an underestimated regulatory mechanism.Gen. Appl. Plant Physiol.3219–31. 10.1007/s12374-010-9097-8
18
FujitaY.FujitaM.SatohR.MaruyamaK.ParvezM. M.SekiM.et al (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis.Plant Cell173470–3488. 10.1105/tpc.105.035659
19
GuanJ. C.JinnT. L.YehC. H.FengS. P.ChenY. M.LinC. Y. (2004). Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.).Plant Mol. Biol.56795–809. 10.1007/s11103-004-5182-z
20
HaasK. T.WightmanR.MeyerowitzE. M.PeaucelleA. (2020). Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells.Science3671003–1007. 10.1126/science.aaz5103
21
HamannT.BennettM.MansfieldJ.SomervilleC. (2009). Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses.Plant J.571015–1026. 10.1111/j.1365-313X.2008.03744.x
22
HeweziT.HoweP.MaierT. R.HusseyR. S.MitchumM. G.DavisE. L.et al (2008). Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism.Plant Cell203080–3093. 10.1105/tpc.108.063065
23
HocqL.HabryloO.VoxeurA.Pau-RoblotC.SafranJ.SénéchalF.et al (2021). Arabidopsis AtPME2 has a pH-dependent processivity and control cell wall mechanical properties.bioRxiv [Preprint]. 10.1101/2021.03.03.433777
24
HofmannN. R. (2008). They all scream for ICE1/SCRM2: core regulatory units in stomatal development.Plant Cell201732–1732. 10.1105/tpc.108.200711
25
HongoS.SatoK.YokoyamaR.NishitaniK. (2012). De-methylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem.Plant Cell242624–2634. 10.1105/tpc.112.099325
26
HõrakH.KollistH.MeriloE. (2017). Fern stomatal responses to ABA and CO2 depend on species and growth conditions.Plant Physiol.174672–679. 10.1104/pp.17.00120
27
HorowitzB. B.Ospina-GiraldoM. D. (2015). The pectin methylesterase gene complement of Phytophthora sojae: structural and functional analyses, and the evolutionary relationships with its oomycete homologs.PLoS One10:e0142096. 10.1371/journal.pone.0142096
28
HsuS. F.LaiH. C.JinnT. L. (2010). Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis.Plant Physiol.153773–784. 10.1104/pp.109.151225
29
HuangY. C.NiuC. Y.YangC. R.JinnT. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses.Plant Physiol.1721182–1199. 10.1104/pp.16.00860
30
HuangY. C.WuH. C.WangY. D.LiuC. H.LinC. C.LuoD. L.et al (2017). PECTIN METHYLESTERASE34 contributes to heat tolerance through its role in promoting stomatal movement.Plant Physiol.174748–763. 10.1104/pp.17.00335
31
HubbardK. E.NishimuraN.HitomiK.GetzoffE. D.SchroederJ. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions.Genes Dev.241695–1708. 10.1101/gad.1953910
32
JeongH. Y.NguyenH. P.LeeC. (2015). Genome-wide identification and expression analysis of rice pectin methylesterases: implication of functional roles of pectin modification in rice physiology.J. Plant Physiol.18323–29. 10.1016/j.jplph.2015.05.001
33
JonesL.MilneJ. L.AshfordD.McQueen-MasonS. J. (2003). Cell wall arabinan is essential for guard cell function.Proc. Natl. Acad. Sci. U.S.A.10011783–11788. 10.1073/pnas.1832434100
34
KanaokaM. M.PillitteriL. J.FujiiH.YoshidaY.BogenschutzN. L.TakabayashiJ.et al (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation.Plant Cell201775–1785. 10.1105/tpc.108.060848
35
KarimiM.InzéD.DepickerA. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation.Trends Plant Sci.7193–195.
36
KlisF. M.BoorsmaA.De GrootP. W. (2006). Cell wall construction in Saccharomyces cerevisiae.Yeast23185–202.
37
KoornneefM.JornaM. L.Brinkhorst-Van Der SwanD. L.KarssenC. M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh.Theor. Appl. Genet.61385–393. 10.1007/BF00272861
38
KoornneefM.ReulingG.KarssenC. M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana.Physiol. Plant.61377–383.
39
LarkindaleJ.VierlingE. (2008). Core genome responses involved in acclimation to high temperature.Plant Physiol.146748–761. 10.1104/pp.107.112060
40
LawrenceS.IIPangQ.KongW.ChenS. (2018). Stomata tape-peel: an improved method for guard cell sample preparation.J. Vis. Exp.137:e57422. 10.3791/57422
41
LeJ.ZouJ.YangK.WangM. (2014). Signaling to stomatal initiation and cell division.Front. Plant Sci.5:297. 10.3389/fpls.2014.00297
42
LeeS. C.LuanS. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses.Plant Cell Environ.3553–60. 10.1111/j.1365-3040.2011.02426.x
43
LerouxC.BoutonS.Kiefer-MeyerM. C.FabriceT. N.MareckA.GuéninS.et al (2015). PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination.Plant Physiol.167367–380. 10.1104/pp.114.250928
44
Levesque-TremblayG.MüllerK.MansfieldS. D.HaughnG. W. (2015). HIGHLY METHYL ESTERIFIED SEEDS is a pectin methyl esterase involved in embryo development.Plant Physiol.167725–737. 10.1104/pp.114.255604
45
LiangC. H.YangC. C. (2015). Identification of ICE1 as a negative regulator of ABA-dependent pathways in seeds and seedlings of Arabidopsis.Plant Mol. Biol.88459–470. 10.1007/s11103-015-0335-9
46
LindquistS.CraigE. A. (1988). The heat-shock proteins.Annu. Rev. Genet.22631–677.
47
LionettiV. (2015). PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses.Front. Plant Sci.6:331. 10.3389/fpls.2015.00331
48
LiuQ.TalbotM.LlewellynD. J. (2013). Pectin methylesterase and pectin remodelling differ in the fibre walls of two Gossypium species with very different fibre properties.PLoS One8:e65131. 10.1371/journal.pone.0065131
49
LouvetR.CavelE.GutierrezL.GuéninS.RogerD.GilletF.et al (2006). Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana.Planta224782–791. 10.1007/s00425-006-0261-9
50
MajdaM.GronesP.SintornI. M.VainT.MilaniP.KrupinskiP.et al (2017). Mechanochemical polarization of contiguous cell walls shapes plant pavement cells.Dev. Cell43290–304.e4. 10.1016/j.devcel.2017.10.017
51
MicheliF. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology.Trends Plant Sci.6414–419. 10.1016/s1360-1385(01)02045-3
52
MurashigeT.SkoogF. A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant15, 473–497. 10.1111/j.1399-3054.1962.tb08052.x
53
NegiJ.MoriwakiK.KonishiM.YokoyamaR.NakanoT.KusumiK.et al (2013). A dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis.Curr. Biol.23479–484. 10.1016/j.cub.2013.02.001
54
PalinR.GeitmannA. (2012). The role of pectin in plant morphogenesis.Biosystems109397–402. 10.1016/j.biosystems.2012.04.006
55
PeaucelleA.BraybrookS. A.Le GuillouL.BronE.KuhlemeierC.HöfteH. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis.Curr. Biol.211720–1726. 10.1016/j.cub.2011.08.057
56
PellouxJ.RustérucciC.MellerowiczE. J. (2007). New insights into pectin methylesterase structure and function.Trends Plant Sci.12267–277. 10.1016/j.tplants.2007.04.001
57
PillitteriL. J.PetersonK. M.HorstR. J.ToriiK. U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis.Plant Cell233260–3275. 10.1105/tpc.111.088583
58
PillitteriL. J.SloanD. B.BogenschutzN. L.ToriiK. U. (2007). Termination of asymmetric cell division and differentiation of stomata.Nature445501–505. 10.1038/nature05467
59
Pinzón-LatorreD.DeyholosM. K. (2013). Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum).BMC Genomics14:742. 10.1186/1471-2164-14-742
60
QuT.LiuR.WangW.AnL.ChenT.LiuG.et al (2011). Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress.Cryobiology63111–117. 10.1016/j.cryobiol.2011.07.003
61
Reynolds-HenneC. E.LangeneggerA.ManiJ.SchenkN.ZumstegA.FellerU. (2010). Interactions between temperature, drought and stomatal opening in legumes.Environ. Exp. Bot.6837–43. 10.1016/j.envexpbot.2009.11.002
62
RoschzttardtzH.Paez-ValenciaJ.DittakaviT.JaliS.ReyesF. C.BaisaG.et al (2014). The VASCULATURE COMPLEXITY AND CONNECTIVITY gene encodes a plant-specific protein required for embryo provasculature development.Plant Physiol.166889–902. 10.1104/pp.114.246314
63
RuiY.ChenY.YiH.PurzyckiT.PuriV. M.AndersonC. T. (2019). Synergistic pectin degradation and guard cell pressurization underlie stomatal pore formation.Plant Physiol.18066–77. 10.1104/pp.19.00135
64
SchwabR.OssowskiS.RiesterM.WarthmannN.WeigelD. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis.Plant Cell181121–1133. 10.1105/tpc.105.039834
65
SödermanE. M.BrocardI. M.LynchT. J.FinkelsteinR. R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks.Plant Physiol.1241752–1765. 10.1104/pp.124.4.1752
66
SparkesI. A.RunionsJ.KearnsA.HawesC. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants.Nat. Protoc.12019–2025. 10.1038/nprot.2006.286
67
SuzukiN.MillerG.MoralesJ.ShulaevV.TorresM. A.MittlerR. (2011). Respiratory burst oxidases: the engines of ROS signaling.Curr. Opin. Plant Biol.14691–699. 10.1016/j.pbi.2011.07.014
68
TanakaY.NoseT.JikumaruY.KamiyaY. (2013). ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves.Plant J.74448–457. 10.1111/tpj.12136
69
WasilewskaA.VladF.SirichandraC.RedkoY.JammesF.ValonC.et al (2008). An update on abscisic acid signaling in plants and more.Mol. Plant1198–217. 10.1093/mp/ssm022
70
WeigelD.GlazebrookJ. (2002). Arabidopsis: a Laboratory Manual.Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
71
WillatsW. G. T.MccartneyL.MackieW.KnoxJ. P. (2001). “Pectin: cell biology and prospects for functional analysis,” in Plant Cell Walls, edsCarpitaN. C.CampbellM.TierneyM. (Dordrecht: Springer), 9–27. 10.1007/978-94-010-0668-2_2
72
WolfS.MouilleG.PellouxJ. (2009). Homogalacturonan methyl-esterification and plant development.Mol. Plant2851–860. 10.1093/mp/ssp066
73
WuH. C.BulgakovV. P.JinnT. L. (2018). Pectin methylesterases: cell wall remodeling proteins are required for plant response to heat stress.Front. Plant Sci.9:1612. 10.3389/fpls.2018.01612
74
WuH. C.HsuS. F.LuoD. L.ChenS. J.HuangW. D.LurH. S.et al (2010). Recovery of heat shock-triggered released apoplastic Ca2+ accompanied by pectin methylesterase activity is required for thermotolerance in soybean seedlings.J. Exp. Bot.612843–2852. 10.1093/jxb/erq121
75
WuH. C.HuangY. C.StracovskyL.JinnT. L. (2017). Pectin methylesterase is required for guard cell function in response to heat.Plant Signal. Behav.12:e1338227. 10.1080/15592324.2017.1338227
76
WuH. C.JinnT. L. (2010). Heat shock-triggered Ca2+ mobilization accompanied by pectin methylesterase activity and cytosolic Ca2+ oscillation are crucial for plant thermotolerance.Plant Signal. Behav.51252–1256. 10.4161/psb.5.10.12607
77
WuH. C.LuoD. L.VignolsF.JinnT. L. (2012). Heat shock-induced biphasic Ca2+ signature and OsCaM1-1 nuclear localization mediate downstream signalling in acquisition of thermotolerance in rice (Oryza sativa L.).Plant Cell Environ.351543–1557. 10.1111/j.1365-3040.2012.02508.x
78
YangY.CostaA.LeonhardtN.SiegelR. S.SchroederJ. I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool.Plant Methods4:6. 10.1186/1746-4811-4-6
79
YooS. D.ChoY. H.SheenJ. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis.Nat. Protoc.21565–1572. 10.1038/nprot.2007.199
80
ZandalinasS. I.BalfagónD.ArbonaV.Gómez-CadenasA.InupakutikaM. A.MittlerR. (2016). ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress.J. Exp. Bot.675381–5390. 10.1093/jxb/erw299
81
ZhangP.WangH.QinX.ChenK.ZhaoJ.ZhaoY.et al (2019). Genome-wide identification, phylogeny and expression analysis of the PME and PMEI gene families in maize.Sci. Rep.9:19918. 10.1038/s41598-019-56254-9
82
ZhangW.ZhouR. G.GaoY. J.ZhengS. Z.XuP.ZhangS. Q.et al (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis.Plant Physiol.1491773–1784. 10.1104/pp.108.133744
83
ZhuJ.ParkJ. H.LeeS.LeeJ. H.HwangD.KwakJ. M.et al (2020). Regulation of stomatal development by stomatal lineage miRNAs.Proc. Natl. Acad. Sci. U.S.A.1176237–6245. 10.1073/pnas.1919722117
Summary
Keywords
ABA response, guard cell, pectin, pectin methylesterase, stomatal-lineage pathway, thermotolerance
Citation
Wu H-C, Yu S-Y, Wang Y-D and Jinn T-L (2022) Guard Cell-Specific Pectin METHYLESTERASE53 Is Required for Abscisic Acid-Mediated Stomatal Function and Heat Response in Arabidopsis. Front. Plant Sci. 13:836151. doi: 10.3389/fpls.2022.836151
Received
15 December 2021
Accepted
28 January 2022
Published
21 February 2022
Volume
13 - 2022
Edited by
Yifeng Wang, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), China
Reviewed by
Ruibo Hu, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CAS), China; Gaojie Hong, Zhejiang Academy of Agricultural Sciences, China; Zhoufei Wang, South China Agricultural University, China
Updates
Copyright
© 2022 Wu, Yu, Wang and Jinn.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Chen Wu, huichenwu@mail.nutn.edu.twTsung-Luo Jinn, jinnt@ntu.edu.tw
†These authors have contributed equally to this work
This article was submitted to Plant Development and EvoDevo, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.