- 1School of Life Science and Technology, Henan Institute of Science and Technology, Xinxiang, China
- 2Faculty of Agrotechnologies and Natural Resource Management, Sumy National Agrarian University, Sumy, Ukraine
- 3Life Science College, Yuncheng University, Yuncheng, China
The toxic heavy metal cadmium (Cd) is easily absorbed and accumulated in crops and affects human health through the food chains. Sunflower (Helianthus annuus L.) is a globally important oil crop. In this study, two sunflower cultivars 62\3 (high Cd) and JB231AC (low Cd), were chosen to compare physiological and transcriptomic responses at different Cd concentrations (0, 25, 50, and 100 μM). The results showed that JB231AC had better Cd tolerance than 62\3. The contents of H2O2 and MDA (malondialdehyde) in 62\3 were lower than that in JB231AC under Cd stress, but the activities of SOD (superoxide dismutase) and POD (peroxidase) in JB231AC were higher than in 62\3, which indicated that JB231AC had a strong ability to remove reactive oxygen species (ROS)-induced toxic substances. Many deferentially expressed ABC (ATP-binding cassette) and ZIP (Zn-regulated transporter, Iron-regulated transporter-like protein) genes indicated that the two gene families might play important roles in different levels of Cd accumulation in the two cultivars. One up-regulated NRAMP (Natural resistance-associated macrophage protein) gene was identified and had a higher expression level in 62\3. These results provide valuable information to further understand the mechanism of Cd accumulation and provide insights into breeding new low Cd sunflower cultivars.
Introduction
Cadmium (Cd) is a non-essential heavy metal element in plants and animals, which inhibits plant growth and development, and seriously threatens human health though the food chains (Maria Celeste et al., 2013). With the rapid development of human activities, such as mining, industrial, and agricultural production, heavy metal soil pollution has become increasingly serious because of the release of industrial wastewater, waste gas discharge, sewage irrigation, and misuse of chemical fertilizers and pesticides (Yan et al., 2021). Cd is easily absorbed and accumulated in plants (Shahid et al., 2017; Chen et al., 2018), and soil Cd pollution seriously affects crop quality and safety processes in production. In order to reduce soil Cd pollution in crops, breeders have carried out genetic improvement screening programs to identify soil metal hyperaccumulators and low Cd accumulation cultivars.
Currently, Sedum alfredii (Hu et al., 2019), Solanum nigrum (Dou et al., 2020), and Bidens pilosa (Dou et al., 2019) have been studied extensively as hyperaccumulators based on their ability to grow in Cd-rich soils and to accumulate large amounts of Cd. In addition, cultivating low Cd crop varieties is one of the most effective ways of reducing the human health risk, and a great deal of research has been conducted on crops, such as rice (Liu C. et al., 2020) and wheat (Liu N. et al., 2020). However, there is only limited understanding of the mechanism of Cd detoxification and accumulation in plants, which hinders further development and application of hyperaccumulators and low Cd accumulation cultivars.
At specific concentrations, Cd toxicity can severely affect plant metabolism, respiration, photosynthesis, transport, and growth, which results in reduced root activity, slow seedling development, small and yellow leaves, and eventually leads to plant death (Zhang et al., 2014; Ahmad et al., 2015). Plants respond to Cd stress by adjusting their own physiological and biochemical processes, among which the accumulation and subsequent detoxification of reactive oxygen species (ROS) caused by heavy metals is an important defense response (Zhang F. et al., 2020). In order to reduce the oxidative damage caused by excessive ROS induced by Cd stress, plants have evolved antioxidant enzyme and non-enzyme systems over a long-term phylogenetic process. Key antioxidant enzymes include superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and glutathione reductase (GR). Non-enzymatic systems mainly consist of reduced glutathione (GSH) and ascorbic acid (AsA) (Wang et al., 2018). Under the regulation of antioxidant enzyme defense systems, plants can maintain normal growth and development within a certain range of heavy metal concentration.
With the advent of next-generation sequencing technologies, many transcriptomic studies on heavy metal stress have focused mainly on rice (Oono et al., 2016), wheat (Zhou et al., 2019), rapeseed (Zhang et al., 2018a), Sedum alfredii (Yang et al., 2017), and mustard (Zhang D. et al., 2021), which have revealed the molecular mechanism of transport, accumulation, and detoxification of Cd in different plants. Two rapeseed genotypes were chosen to investigate the Cd translocation mechanism by transcriptomic comparison, and the results showed that BnNramp2;1 and BnNramp4;2 were two main Cd transporters (Wang et al., 2019). Hu et al. (2019) compared the transcriptomes of different tissues of Sedum alfredii under Cd stress, and the results showed that ATP-binding cassette (ABC) transporters exhibited significant enrichment and accounted for approximately one-third of total selected differentially expressed genes.
Sunflower (Helianthus annuus L.) belongs to the Asteraceae family and is an important food and bioenergy product (Bashir et al., 2021). It is used in phytoremediation research because of its large biomass, fast growth, and high tolerance to heavy metals (Tang et al., 2003; Bayat et al., 2021; Benavides et al., 2021). However, few Cd accumulation-related studies in physiology and transcriptomics have been conducted with sunflower. In our study, two sunflower cultivars 62\3 and JB231AC were chosen as high/low Cd accumulators and physiological and transcriptomic analyses were conducted under different levels of Cd stress. The study provides important information for further research on Cd accumulation and detoxification mechanisms in sunflower.
Materials and Methods
Plant Material and Screening Conditions
Two genotype accessions of sunflower, 62\3 and JB231AC, with high and low Cd accumulation capability were identified in our previous work (data unpublished). The two cultivars were obtained from Sumy National Agrarian University in Ukraine.
In 2020, the healthy seeds of each accession were sterilized by 15% H2O2 for 30 min, rinsed in distilled water three or more times, and then soaked in deionized water at room temperature for 4 h. Then, they were sown in a germination box (32 cm × 25.5 cm × 11 cm) containing vermiculite and moistened with deionized water. Seeds were incubated in a culture room with 16 h light (28°C, 5,000 Lux) and 8 h dark (25°C) photoperiod for 6 days. Following germination, seedlings were transferred to plastic pots (19 cm × 13 cm × 12 cm) filled with 10 L of 1/4 strength modified Hoagland nutrient solution and grown for 7 days. Then, the nutrient solution was increased to 1/2 strength for 7 days. After 20 days of Cd-free growth, seedlings of uniform size were randomly assigned to four different Cd treatments: 0, 25, 50, and 100 μM CdCl2⋅2.5H2O for 7 days. The 1/2 strength nutrient solution contained 2.5 mmol/L Ca(NO3)2, 1 mmol/L MgSO4, 0.5 mmol/L NH4H2PO4, 2.5 mmol/L KCL, 2 mmol/L NaCl, 0.2 μmol/L CuSO4, 1 μmol/L ZnSO4, 0.1 mmol/L EDTA-FeNa, 0.02 mmol/L H3BO3, 5 nmol/L (NH4)6Mo7O24, and, 1 μmol/L MnSO4. Finally, the seedlings were harvested; half were used to measure morphological indexes and Cd content, and the remaining half were quickly isolated, frozen in liquid nitrogen, stored at –80°C, and then used to measure physiological indicators and transcriptome analysis. All the treatments were replicated three times.
Identification of Growth Index and Cadmium Content
For the presentation and evaluation of Cd treatment results, the plants were photographed, and height and fresh weight were measured manually. The roots and shoots of seedlings were oven-dried at 80°C until constant weight, and then weighed for dry weight.
To measure Cd content, the roots of seedlings were rinsed with deionized water at least three times to remove surface ions. Then, roots and shoots were harvested separately. The samples were dried at 105°C for 30 min, and then at 80°C in an oven until completely dry. The dry samples were then ground to a powder. The dry powder for each sample was digested in 5 ml HNO3 overnight (at least for 3 h) at room temperature, then 2 ml H2O2 was added, then further digested for approximately 3 h at 180°C. The digested solution was diluted to 25 mL, and then investigated by an atomic absorption spectrophotometer (ICP-OES, Optima 2100DV, Perkin Elmer, United States).
Investigation of Antioxidant Systems and the Ascorbate-Glutathione Cycle
To further investigate and compare the physiological characteristics of Cd tolerance of 62\3 and JB231AC, the concentration of hydrogen peroxide (H2O2) and malondialdehyde (MDA) in the leaves of each fresh sample was measured. Antioxidant enzyme systems were evaluated by detecting the activities of SOD and POD. The Ascorbate-Glutathione (AsA-GSH) cycle was investigated by detecting the activity of glutathione S-transferase (GST) and the concentration of AsA. All physiological indexes were carried out with the corresponding assay kits (Jiancheng Biotechnology, Nanjing, China).
RNA-Seq Analysis and Library Assembly
Based on the phenotypic results of 62\3 and JB231AC under Cd gradients combined with previous studies, two treatments (0 and 50 μM CdCl2⋅2.5H2O) of the two cultivars were selected for further transcriptomic analysis. The total RNAs were extracted from leaves for analysis of sequencing libraries by NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States), and the library quality was assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina NovaSeq 6000 platform after clustering on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) in Biomarker Biotechnology Corporation (Beijing, China). To obtain high quality clean data, reads containing adapter, reads containing ploy-N, and low quality reads were removed from raw data. Transcriptome was assembled based on the left.fq and right.fq using Trinity (Grabherr et al., 2011). Fragments per kilobase of transcript per million mapped reads (FPKM) (Trapnell et al., 2010) were used to estimate gene expression level in RNA sequencing.
Screening of Differential Expression Genes
The DESeq R package (1.10.1) (Love et al., 2014) was used for differential expression analyses of two conditions/groups. Differential expression genes (DEGs) were identified according to the threshold values: FDR < 0.01 and | log2 (fold change)| > 2. Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the topGO R packages based on the Kolmogorov–Smirnov test. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment was analyzed using KOBAS software to test the statistical enrichment of the DEGs (Mao et al., 2005).
Validation of Gene Expression by Quantitative Real Time PCR
Twelve RNA samples obtained from RNA-seq experiments were reverse transcribed into cDNA. The qRT-PCR reactions were performed using cDNA as a template in three biological and three technical replicates. The PCR reaction conditions were denaturation at 95°C for 5 min followed by 40 cycles at 95°C for 5s and 60°C for 30s. The specific primers of candidate genes for qRT-PCR were designed by Primer Premier 6.0 software, and the reference gene was 18s rRNA (gene accession No: HM638219) of sunflower (Supplementary Table 1). Relative expression was analyzed using the cycle threshold 2–ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
Transfer factor (TF) is the ratio of the concentration of Cd in the shoot to that in the root. All data were statistically analyzed using GraphPad Prism 8. Two-way ANOVA was performed on data sets, and the mean and standard deviation (SD) of each treatment was calculated. Multiple comparisons with Sidak’s test were mainly used to compare the mean values between treatments (p < 0.05).
Results
Comparison of Phenotype Characteristics to Cadmium Stress Between High/Low Cadmium Sunflower Cultivars
Plant growth parameters were investigated in order to examine the plant growth differences between 62\3 and JB231AC under Cd stress. With an increase in Cd concentration, the seedlings of both cultivars showed gradually weaker growth potential and became yellow (Figure 1A), and 62\3 almost wilted and died under 100 μM Cd stress. The result showed that 62\3 was sensitive to Cd toxicity; i.e., JB231AC had a higher Cd tolerance than 62\ 3.
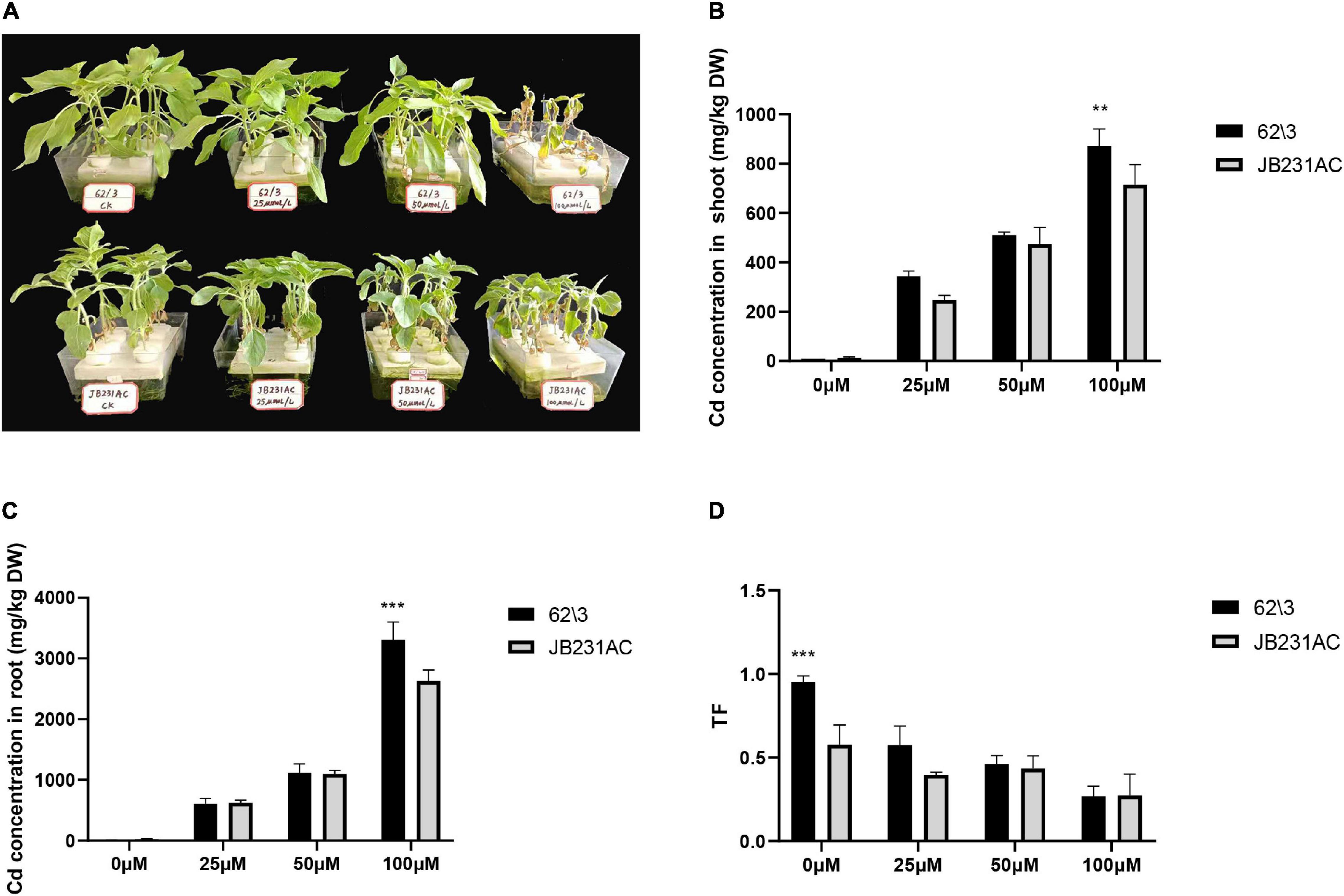
Figure 1. Performance of growth and Cd accumulation of 62\3 and JB231AC genotypes. (A) Growth performance of 62\3 and JB231AC grown hydroponically under Cd stress for 7 days. (B) Cd concentration in the shoots of the seedlings. (C) Cd concentration in the roots of the seedlings. (D) TF (Translocation factor), the ratio of Cd concentration in shoots to that in roots. For (B–D) the statistical analyses were conducted using all the performance data of 62\3 and JB231AC under Cd stress. Data presented are the means (n = 3), and error bars denote the standard deviations. The asterisk represent the significant difference between 62\3 and JB231AC under one same Cd stress. **P < 0.01, ***P < 0.001.
The Cd concentration in the roots was much higher than in the shoots of both cultivars, and the Cd concentration in the roots and shoots of 62\3 was higher than in JB231AC, particularly under 100 μM Cd stress (Figures 1B,C). The Cd TF value of 62\3 was higher than JB231AC in the 0–50 μM range of Cd concentration, and there was no significant difference between the two cultivars under the same concentration of Cd stress (Figure 1D). The result showed that 62\3 was a high Cd accumulation cultivar and had a higher root-to-shoot Cd translocation ratio than JB231AC.
The seedling height and fresh weight of 62\3 and JB231AC decreased with an increase in Cd concentration, but there was no difference between the two cultivars at the same concentration of Cd stress (Figures 2A,B). The dry weight of shoots and roots of 62\3 and JB231AC decreased with an increase in Cd stress, but the genotype JB231AC had a larger biomass than 62\3 at the same concentration of Cd stress (Figures 2C,D).
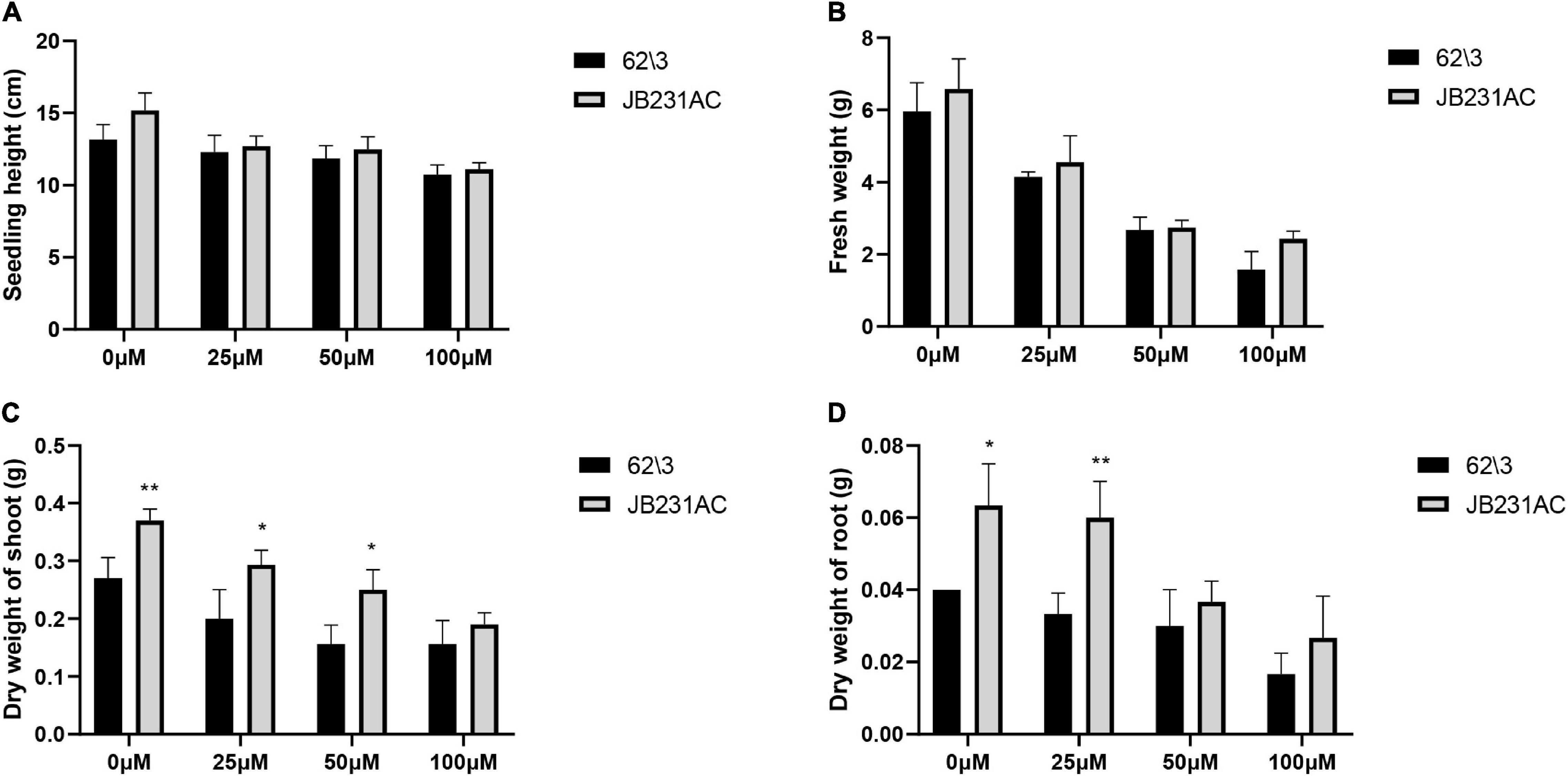
Figure 2. Vegetative performance of 62\3 and JB231AC. (A–D) Growth-related indices of the seedlings, seedling height (A), fresh weigh (B), dry weight of shoot (C) and dry weight of root (D). For (A–D) the statistical analyses were conducted using all the performance data of 62\3 and JB231AC under Cd stress. Data presented are the means (n = 3), and error bars denote the standard deviations. The asterisk represent the significant difference between 62\3 and JB231AC under one same Cd stress. *P < 0.05, **P < 0.01.
Physiological Responses to Cadmium Stress in 62\3 and JB231AC
The contents of H2O2 and MDA were measured to investigate the level of antioxidant reaction to Cd stress. The H2O2 content was higher in JB231AC than in 62\3 in the 0–50 μM range of Cd stress; it increased with an increase in Cd concentration, and then decreased under 100 μM Cd stress (Figure 3A). The content of MDA increased with an increase in Cd concentration and was higher in JB231AC than in 62\3 in the 0–50 μM range of Cd stress (Figure 3B). The contents of H2O2 and MDA were not significantly different between 62\3 and JB231AC under the same concentration of Cd stress.
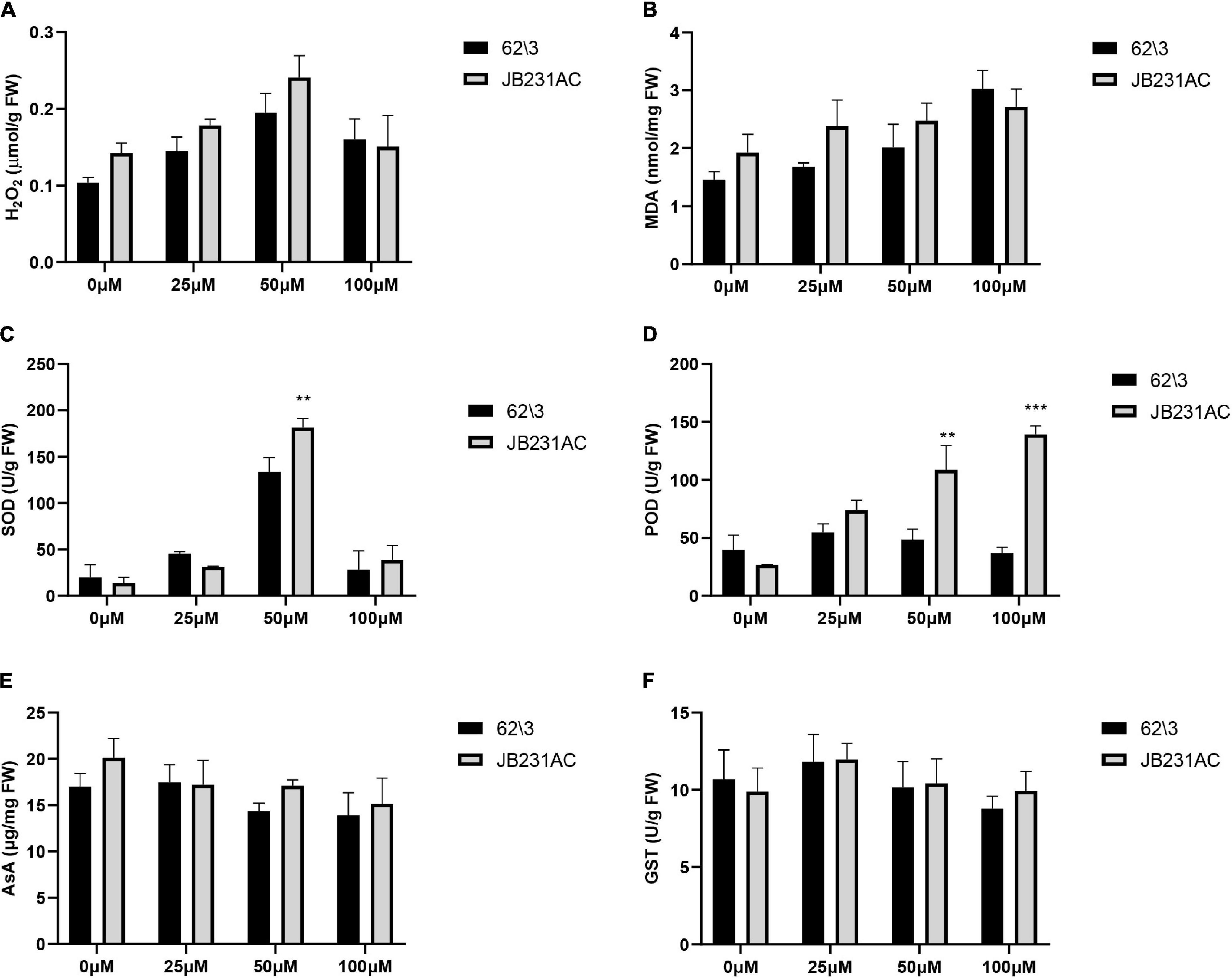
Figure 3. Antioxidant systems of 62\3 and JB231AC under Cd stress. (A) H2O2 content. (B) MDA, Malondialdehyde. (C) SOD, Superoxide dismutase. (D) POD, Peroxidase. (E) AsA, Ascorbic acid. (F) GST, Glutathione S-transferase. For (A–F) the statistical analyses were conducted using all the performance data of 62\3 and JB231AC under Cd stress. Data presented are the means (n = 3), and error bars denote the standard deviations. The asterisk represent the significant difference between 62\3 and JB231AC under one same Cd stress. **: P < 0.01, ***: P < 0.001.
The activities of SOD and POD were important indices for showing Cd tolerance levels in antioxidant enzyme systems. With an increase in Cd concentration, the activity of SOD first increased quickly and then decreased quickly, and reached a maximum value under 50 μM Cd stress with significant differences seen between 62\3 and JB231AC (Figure 3C). This result indicated that the activity of SOD was strongly stimulated under 50 μM Cd stress. With an increase in Cd concentration, the activity of POD in 62\3 first increased and then decreased, and reached a maximum value under 25 μM Cd stress (Figure 3D). However, the activity of POD in JB231AC increased with an increased Cd concentration. The activity of POD was higher in JB231AC than in 62\3, and showed highly significant differences under 50 μM and 100 μM Cd stress. This result indicated that the regulation response of POD to Cd stress was different in the two cultivars.
The changes in concentration of AsA and the activities of GST were almost unaffected at different Cd concentrations compared to the control, and there was no significant difference between cultivars (Figures 3E,F). This result suggested that there was negligible AsA and GST response to Cd stress in this study.
RNA-Seq Analyses of 62\3 and JB231AC Under Cadmium Stress
The leaves of the no-Cd control and Cd-treated (50 μM CdCl2⋅2.5H2O) plants were collected to investigate molecular responses at the transcriptional level by RNA-seq. Twelve samples (containing three replicates per treatment) were processed for mRNA sequencing, and 93.62 Gb clean data with Q30 ≥ 93.90% was obtained (Supplementary Table 2). The data was deposited in the NCBI database (The accession number of Bioproject is PRJNA797513 and the accession number of Biosample is SAMN25008009). A total of 78,975 unigenes with a total length of 59,469,664 nt and an average length of 1,340 nt were obtained after sequence assembly (Supplementary Table 3). Length distribution of unigenes and saturation test of RNA-seq data were all perfect (Supplementary Figures 1, 2). The result of sequencing and assembly showed that the mRNA-seq Library was of high quality and could be used for transcriptome data analyses.
Identification of Cadmium-Regulated Differential Expression Genes
To further elucidate the molecular mechanisms involved in Cd tolerance, the high-throughput DEGs sequencing between 62\3 and JB231AC under Cd stress was performed by DESeq R package (1.10.1). Fold change (FC) ≥ 2 and the criteria of false discovery rate (FDR) < 0.01 were the threshold values. A total of 8,818 DEGs were identified, and among them, 3,196, 4,769, and 4,764 DEGs were identified in the three comparison groups 62\3 (CK vs. 50 μM), JB231AC (CK vs. 50 μM), and 62\3 vs. JB231AC (50 μM Cd), respectively (Figure 4). There were 650 co-expressed DEGs in the three groups. More DEGs were identified in JB231AC (CK vs. 50 μM) than in 62\3 (CK vs. 50 μM), and the two cultivars had more up-regulated DEGs than down-regulated. Comparing 62\3 with JB231AC under 50 μM Cd stress, there were approximately equal numbers of up- and down-regulated DEGs.
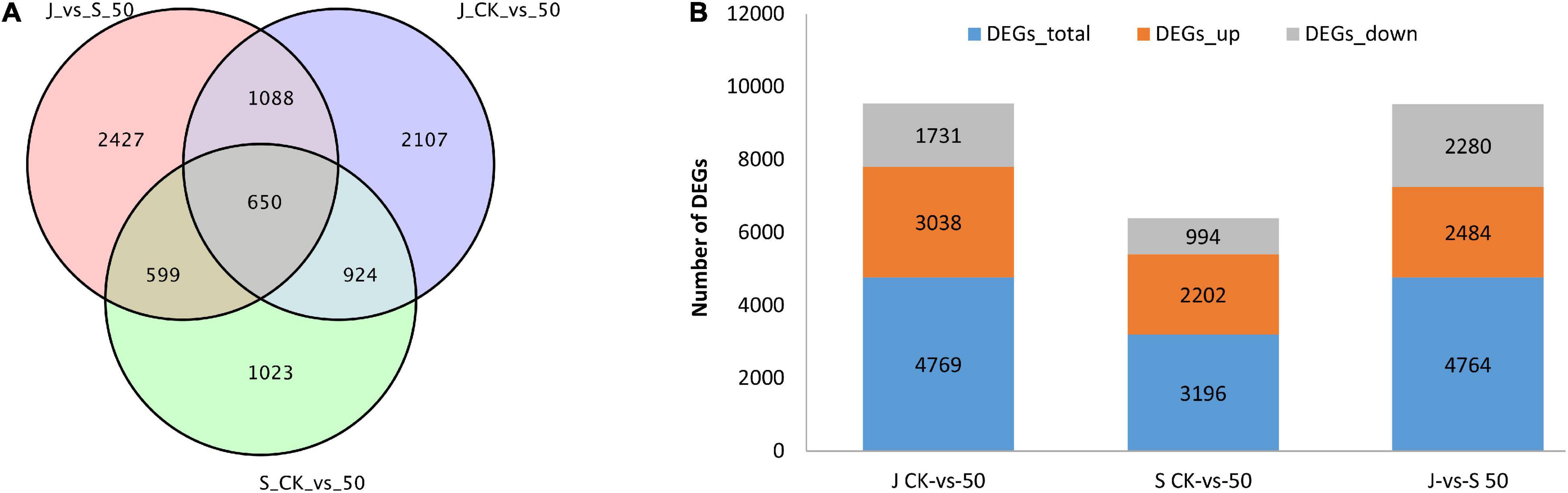
Figure 4. DEGs comparison of 62\3 and JB231AC. (A) Venn diagrams showing all DEGs shared among the three groups. (B) Number of DEGs that were up or down-regulated in the three groups. S: 62\3, J: JB231AC, CK: 0 μM CdCl2⋅2.5H2O, 50: 50 μM CdCl2⋅2.5H2O, vs., versus.
Functional Annotations of Cadmium-Regulated Differential Expression Genes
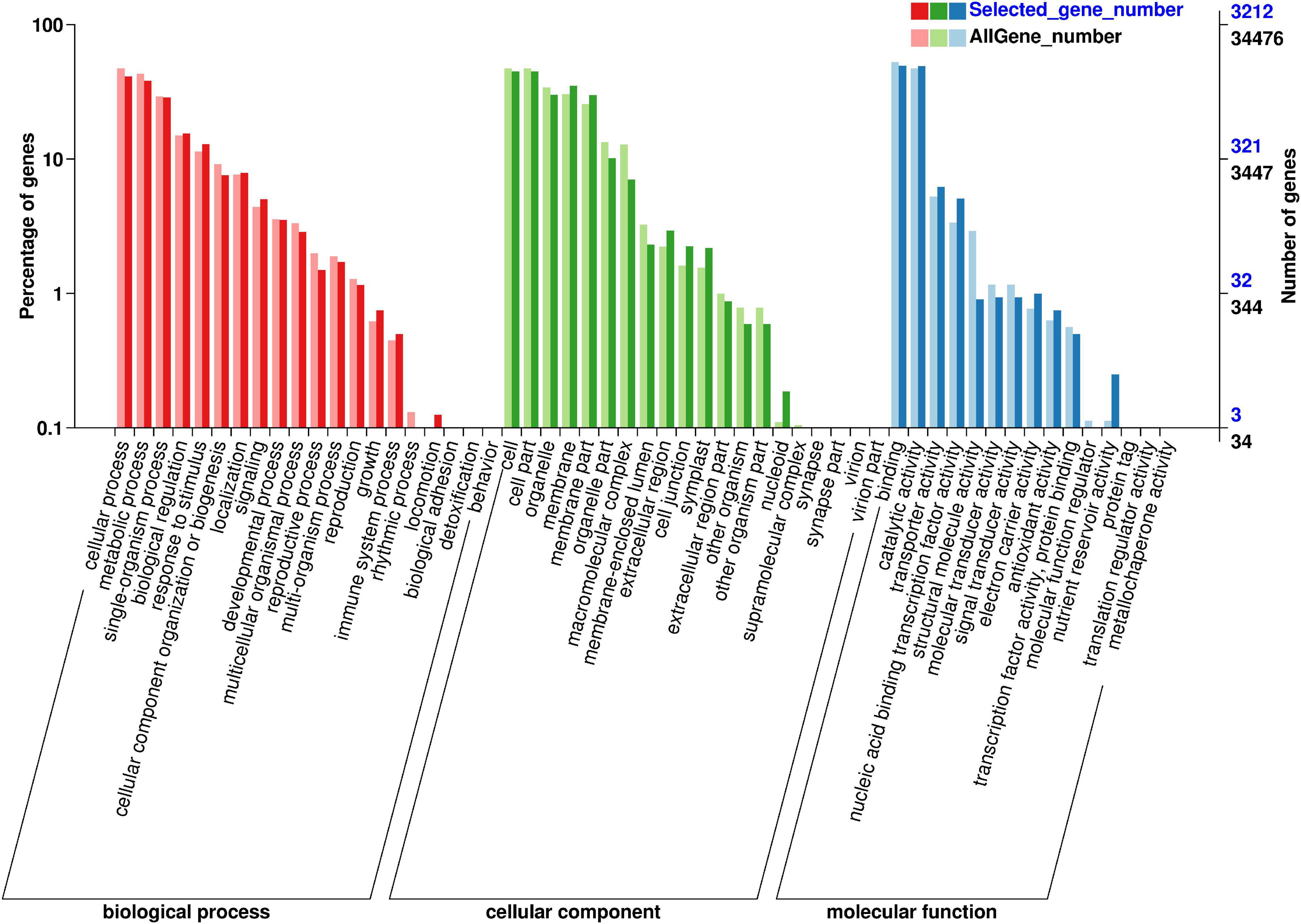
GO class analysis was used to identify the function annotation of 4,764 DEGs between 62\3 and JB231AC under 50 μM Cd stress (Figure 5 and Supplementary Table 4). The result showed that 55 GO terms were grouped into three categories (biological process, cellular component, and molecular function). Binding and catalytic activity had the highest DEG number, followed by cell, cell part, cellular process, and metabolic process.
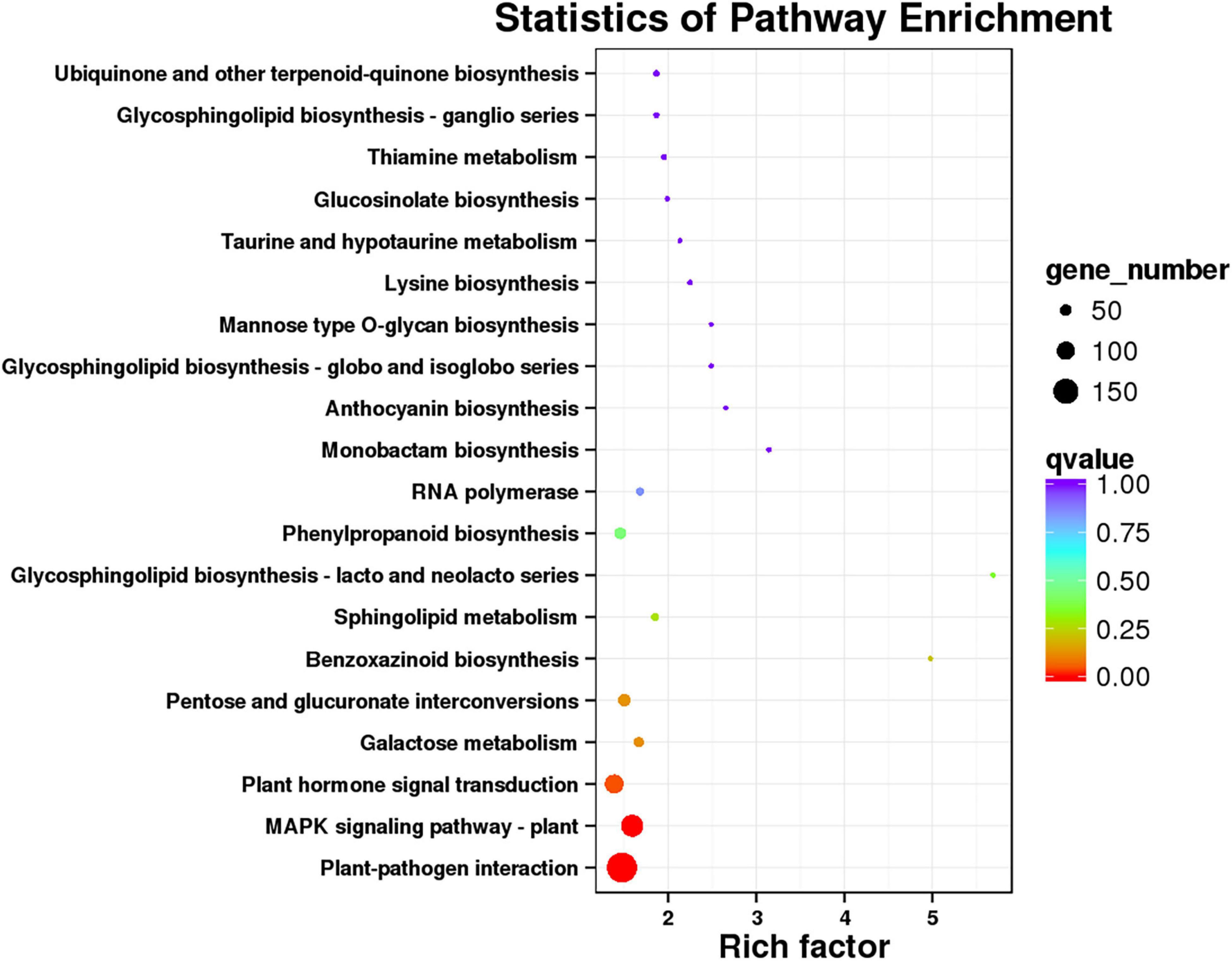
In order to identify the metabolic pathway enrichment between 62\3 and JB231AC under 50 μM Cd stress, 4,764 DEGs were analyzed by KEGG. A total of 130 KEGG pathways were enriched and 20 KEGG pathways were significantly enriched (Figure 6 and Supplementary Table 5). Among them, plant-pathogen interaction, MAPK signaling pathway-plant, plant hormone signal transduction, galactose metabolism, and pentose and glucuronate interconversions were listed in the top five KEGG pathways.
Differential Expression Genes Related to Cadmium Detoxification, Transport, and Accumulation
A total of 79 DEGs related to Cd detoxification, transport, or accumulation were identified. Among them, 36 DEGs ABC, 24 up-regulated, and 12 down-regulated, were identified between 62\3 and JB231AC under 50 μM Cd stress, accounting for 43.4% of all selected Cd related genes (Figure 7A and Supplementary Table 6). Both DEGs, c50541. graph_c4 (log2FC = 4.987981385) and c53176. graph_c10 (log2FC = 4.848310244), were up-regulated and had a higher differential expression in 62\3 than in JB231AC.
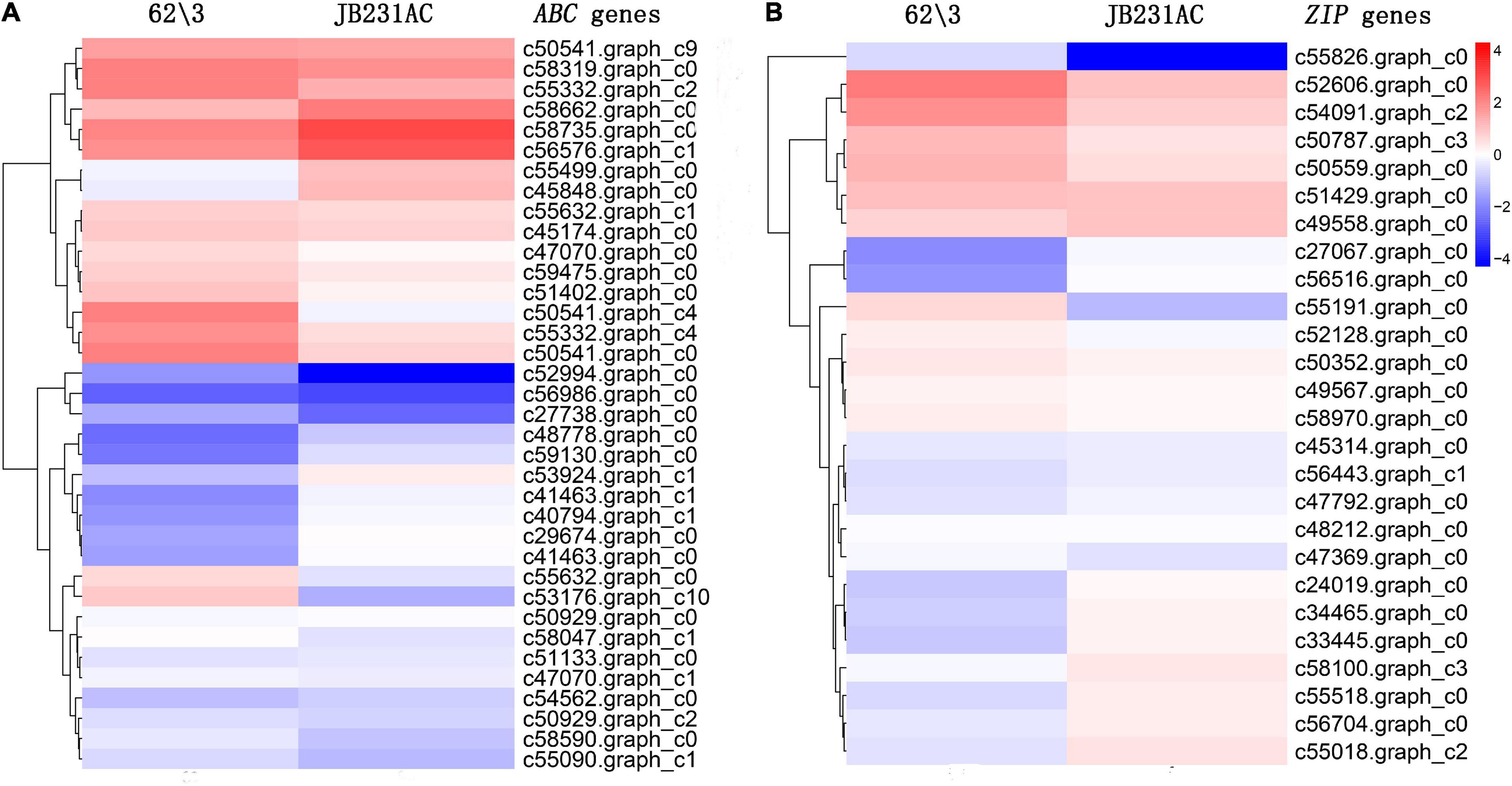
Figure 7. Cluster heat maps of DEG expression of 62\3 and JB231AC under 50 μM Cd stress. (A) Predicted ATP-binding cassette (ABC) gene expression. (B) Predicted ZIP (Zn-regulated transporter, Iron-regulated transporter-like protein) gene expression. Expression values of two cultivars were presented as log2-transformed normalized FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values, and FPKM value was average of three replicates. Additional details are presented in Supplementary Tables 6, 7.
Twenty-six DEGs Zn-regulated transporter, Iron-regulated transporter-like protein (ZIP), 15 up-regulated, and 11 down-regulated, were identified in 62\3 and JB231AC under 50 μM Cd stress, accounting for 31.3% of all selected Cd related genes (Figure 7B and Supplementary Table 7). Among them, the genes c55191.graph_c0 (log2FC = 6.693782781) and c55826.graph_c0 (log2FC = 4.498795) were up-regulated and had higher differential expressions in 62\3 than in JB231AC.
In addition, Cd-related DEGs, including HIPP (Heavy metal associated isoprenylated plant protein), MTP (Metal tolerance protein), YSL (Metal-nicotianamine transporter), HMA (Heavy metal ATPase), and NRAMP (Natural resistance-associated macrophage protein), were screened out, and the number of DEGs for each was 7, 4, 3, 2, and 1, respectively (Supplementary Table 8). Among them, there were 12 up-regulated and 5 down-regulated, with a total of 17. The up-regulated gene c37848.graph_c0, belonging to the HIPP family, had a large differential expression level (Log2FC = 4.28146756), and its expression level in 62\3 was higher than in JB231AC. One up-regulated NRAMP gene, c53780.graph_c0 (Log2FC = 4.207403041), was identified and had a higher expression level in 62\3. Two up-regulated HMA genes, c55845.graph_c0 and c49575.graph_c0, were screened out and both had a higher expression levels in 62\3 than in JB231AC.
Validation of Differential Expression Genes by Quantitative Real Time PCR
To verify the expression levels of DEGs identified by RNA-seq, five Cd-induced key genes used for Quantitative Real Time PCR (qRT-PCR) verification were randomly selected from the comparison group of the two cultivars with Cd treatment (50 μM). As a whole, the results showed that the expression profile of qRT-PCR of 5 candidate genes were consistent with the trend of their transcriptome data, most up-regulated expression (Figure 8). Therefore, The RNA-seq data and transcriptome results are mostly reliable and contribute to the identification of DEGs related to Cd.
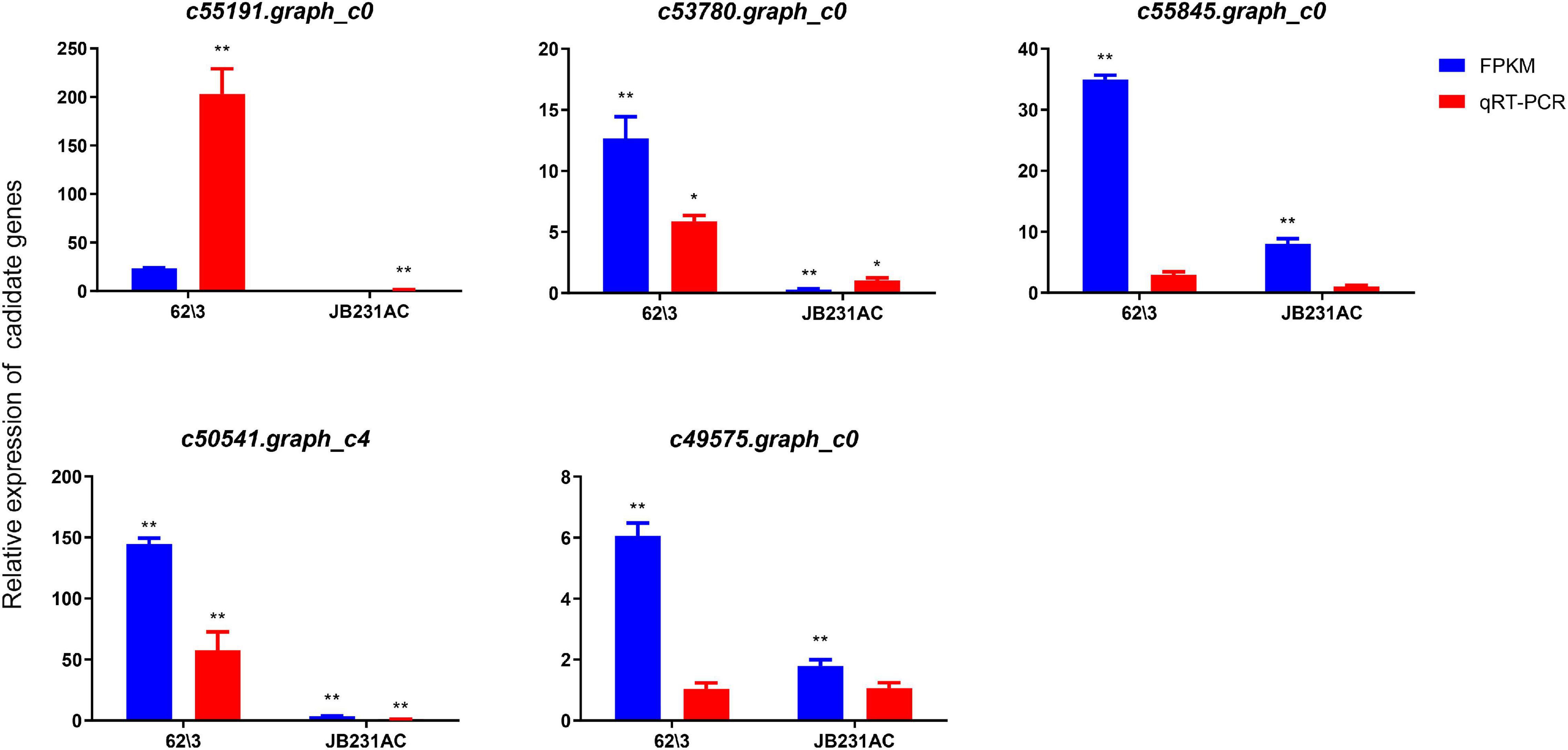
Figure 8. RNA-Seq and qRT-PCR were used to compare DEGs in 62\3 and JB231AC. The relative gene expression level was calculated by 2–ΔΔCT in qRT-PCR. *P < 0.05, **P < 0.01.
Discussion
Growth and Physiological Response of Two Sunflower Cultivars to Cadmium Stress
In this study, the two sunflower cultivars 62\3 and JB231AC were selected as high and low Cd accumulation cultivars for physiological study under Cd stress. The Cd concentration in both roots and shoots of 62\3 was higher than that in JB231AC, and root-to-shoot Cd translocation was stronger in 62\3 than in JB231AC (Figures 1B–D). With increasing Cd stress, JB231AC cultivars became weak and stunted and the leaves turned slightly yellow (Figure 1A). Cultivar 62\3 was sensitive to Cd toxicity and almost withered and died under 100 μM Cd stress. The results show that JB231AC had some tolerance to Cd toxicity. Chen et al. (2021) showed that after 10 days of Cd treatment, plant height and biomass of kenaf seedlings were significantly inhibited under 10 mg L–1 Cd stress compared to the control. Saidi et al. (2021) also showed that plant growth was significantly impacted by an increasing Cd dose, and was severely suppressed at the highest Cd concentrations (50 and 100 μM) after Cd treatment for 4 days. Therefore, a 50 μM Cd dose can be considered as an important candidate or reference concentration for studying short-term Cd treatment at the seedling stage.
Cd is an environmental pollutant which increases ROS production, and this leads to excessive accumulation of O2, H2O2, and hydroxyl radicals. This, in turn, leads to oxidation bursts (Heyno et al., 2008) and eventually causes high toxicity in plants (Li et al., 2012; Unsal et al., 2020). Plants avoid or reduce heavy metal-induced oxidative damage by a series of complex enzymatic and non-enzymatic antioxidant defense mechanisms (Yu et al., 2013; Liu J. et al., 2019); i.e., Cd induces membrane lipid peroxidation and regulates the expression of antioxidant defense systems (Chen et al., 2010; Imran et al., 2020). Li et al. (2012) showed that H2O2 and MDA contents are important indexes to evaluate the degree of peroxidation damage under Cd stress due to ROS-induced membrane lipid peroxidation. In our study, the contents of H2O2 and MDA in 62\3 was lower than in JB231AC, which showed that JB231AC produces more toxic peroxidation substances (Figures 3A,B). The contents of H2O2 and MDA in both cultivars increased with an increase in Cd concentration, indicating that oxidative damage was increasingly severe; our findings are similar to the results of other studies (Shi et al., 2013; Kanu et al., 2019; Khan et al., 2020).
SOD and POD are two important antioxidant enzymes that remove ROS (Wang et al., 2019). In our study, the activities of SOD and POD in JB231AC were higher than in 62\3, indicating that JB231AC had a pronounced ability to remove ROS-induced toxic substances. JB231AC had a better tolerance to Cd toxicity and had a significantly higher dry weight of roots and stems than 62\3 (Figures 2C,D). With an increase in Cd concentration, the SOD activity first increased rapidly and then decreased (Figure 3C), which might indicate that SOD in the two sunflower cultivars had a strong response to Cd stress and played an important role in removing harmful products. The variation trend of POD activity was different (Figure 3D), and the regulation mechanism of POD should be different in the two cultivars. The result showed that antioxidant enzymes played important roles in Cd accumulation and tolerance. However, Zhang et al. (2015) found that the antioxidant enzymes governing Cd detoxification were not found to be active in castor leaves under two different Cd concentrations (2 mg L–1 and 5 mg L–1) over 10 days. The roles of antioxidant enzymes and the AsA-GSH cycle in different materials are very different under Cd stress (Wu et al., 2015). In our study, no differences were found in the AsA content or the activity of GST between the two cultivars under the same Cd treatments (Figures 3E,F), suggesting that the AsA-GSH cycle was not the key factor affecting the differences in Cd accumulation and tolerance between the two cultivars.
Differential Expression Gene Annotation Functions of Two Sunflower Genotypes Under Cadmium Stress
Many genes are regulated and expressed under Cd stress, and the regulations of Cd-related genes in different Cd accumulation materials are different, which may be the reason for the different Cd accumulations observed. A total of 3,321 and 2,221 DEGs were regulated in the two tolerant and sensitive Cd-treated kenaf cultivars compared to the controls (Chen et al., 2021). The transcriptomic analysis revealed 883 DEGs between control and Cd-stressed plants in Nicotiana rustica (high Cd), while 2,119 DEGs were found in Nicotiana tabacum (low Cd) (Zhang Y. et al., 2021). In our study, 4,764 genes were differentially expressed by DEGseq between 62\3 and JB231AC under 50 μM Cd stress (Figure 4). GO class showed that most DEGs were classed in binding, catalytic activity, and cell at second level (Figure 5 and Supplementary Table 4), and KEGG enrichment pathways were mostly plant-pathogen interaction, MAPK (mitogen-activated protein kinase) signaling pathway-plant, and plant hormone signal transduction (Figure 6 and Supplementary Table 5). The results indicate that the differences in Cd accumulation between the two cultivars may be caused mainly by DEGs regulation in the above annotations or pathways.
Key Candidate Genes May Be Involved in Cadmium Detoxification, Transport, and Accumulation
The ABC transporter is located on the vacuole membrane, and can sequester Cd and its complexes in the vacuole (Mendoza-Cózatl et al., 2011). ABC transporters involved in Cd or Cd conjugated transport have been found in some plants, such as Arabidopsis (Kim et al., 2007; Park et al., 2012), rice (Oda et al., 2011; Cai et al., 2021), and rapeseed (Zhang et al., 2018b). Spatial expression analysis showed that OsABCG36 was expressed in the root tip and the mature root region, and knockout of OsABCG36 increased Cd accumulation in root cell sap and enhanced Cd sensitivity (Fu et al., 2019). Overexpressing OsABCG48 in rice lowers root Cd accumulation which prevents further transport of Cd to the shoots and seeds (Cai et al., 2021). In our study, of 36 ABC DEGs, two out of three genes up-regulated, and were identified in 62\3 and JB231AC under 50 μM Cd stress, accounting for 43.4% of all selected Cd related genes (Figure 7A and Supplementary Table 6). The result showed that ABC transporters probably play a significant role in the regulation of different Cd accumulation in both cultivars.
The plant ZIP family proteins, which belong to integral membrane transporters, exist widely in plants and have the function of transporting various metal elements, such as Zn2+, Ca2+, Fe2+, and Cd2+ (Guerinot, 2000; Mills et al., 2005). OsZIP1 is a metal-detoxified transporter preventing excess Cd accumulation in rice. It is located in the endoplasmic reticulum and plasma membrane, it is abundantly expressed in roots, and it is a metal-detoxified transporter by preventing excess Zn, Cu, and Cd accumulation in rice (Liu X. S. et al., 2019). Zheng et al. (2018) showed that there might be a feedback regulation of OsZIP1 in roots to prevent increasing Cd uptake from soil. Overexpressing OsZIP1 improves rice growth under excess metal stress but accumulates less of the metals (Liu X. S. et al., 2019). The expression of OsZIP1 and OsZIP3 in yeast leads to increased Cd sensitivity and Cd accumulation (Zheng et al., 2018). OsZIP7, expressed in parenchyma cells of vascular bundles in roots and nodes, plays an important role in root xylem load and inter-node vascular transport, and its expression promotes Cd transport to shoots (Tan et al., 2019). In our study, of 26 ZIP DEGs, 57.7% genes up-regulated, and were identified in 62\3 and JB231AC under 50 μM Cd stress, accounting for 31.3% of all selected Cd related genes (Figure 7B and Supplementary Table 7). Therefore, the ZIP family could play an important role in our findings.
In previous studies, HMA (Takahashi et al., 2012; Liu C. L. et al., 2020) and NRAMP (Takahashi et al., 2011; Yang et al., 2019) have been shown to be two important Cd transport and detoxification related genes in crops. OsHMA2 is localized in the pericycle of roots and the phloem of diffuse vascular bundles of nodules, and it is the main transporter of Zn and Cd from the roots to the shoots (Yamaji et al., 2013). OsHMA3, expressed in root cell vacuoles, confers high root-to-shoot Cd translocation rates (Miyadate et al., 2011) and it restricts Cd translocation from roots to above-ground tissues by selectively isolating Cd into root vacuoles (Ueno et al., 2010; Miyadate et al., 2011). NRAMP family genes are responsible for the transport of divalent cations (Fe2+, Zn2+, Mn2+, and Cd2+) (Nevo and Nelson, 2006). SaNRAMP1 is localized at the plasma membrane, and overexpressing of SaNRAMP1 in tobacco (Nicotiana sp.) significantly increases Cd, Zn, and Mn concentration in the shoots (Zhang J. et al., 2020). In our study, two up-regulated HMA genes and one up-regulated NRAMP gene were screened out, and they all had a higher expression level in 62\3 than in JB231AC. It is speculated that HMA and NRAMP might make certain contributes in the transport and accumulation of Cd in the two sunflower cultivars (Supplementary Table 8).
HIPP plays crucial roles in metal homeostasis and detoxification as metal-binding metallochaperones (Khan et al., 2019). HIPP1-V was up-regulated in H. villosa under Cd stress, and overexpressing HIPP1-V showed enhanced Cd tolerance in wheat (Zhang H. et al., 2020). MTPs are divalent cation transporters which are essential for metal homeostasis and tolerance in plants, sequestrating Zn and Cd in vacuoles in cucumber (Migocka et al., 2015). SnYSL3 is a transporter delivering metal-nicotianamine complexes, and SnYSL3 transgenic plants increase the translocation ratios of Fe and Cd from roots to shoots (Feng et al., 2017). In our study, DEGs related to Cd transport or detoxification, including HIPP, MTP, and YSL were screened out, with 7, 4, and 3, respectively (Supplementary Table 8). This result suggests that HIPP and MTP might perform Cd detoxification and tolerance in the two sunflower cultivars, and YSL might be involved in Cd translocation.
In conclusion, JB231AC has the characteristics of good Cd tolerance and low Cd accumulation, and it can be used as a good germplasm material for hybridization in order to improve the Cd tolerance of sunflower. In follow-up work, the functions of candidate genes related to Cd physiology would be further validated by the genetic transformation, and the functional molecular markers could be developed for the marker-assisted selection of sunflower in future breeding programs.
Conclusion
Two sunflower cultivars, 62\3 and JB231AC, were verified as high and low Cd accumulators, respectively, and 62\3 had the stronger root-to-shoot Cd translocation ability. Comparative physiological and transcriptomic analyses between the two cultivars revealed that JB231AC had better Cd tolerance. The contents of H2O2 and MDA in 62\3 were lower than that in JB231AC under Cd stress, which showed that JB231AC produces more toxic peroxidation substances. However, the activities of SOD and POD in JB231AC were higher than in 62\3, indicating that JB231AC had a very strong ability to remove ROS-induced toxic substances. The activity of SOD was simultaneously induced quickly with an increase in Cd stress in the two cultivars, but the variation trend of POD activity was different. The transcriptomic analysis showed that many ABC and ZIP genes were differentially expressed, indicating that the two kinds of genes might play an important role in different Cd accumulation in the two cultivars. Two up-regulated HMA (Heavy metal ATPase) genes and one up-regulated NRAMP (Natural resistance-associated macrophage protein) genes were screened out.
Data Availability Statement
The data was deposited in the NCBI database (The accession number of Bioproject is PRJNA797513 and the accession number of Biosample is SAMN25008009).
Author Contributions
YF wrote the manuscript. HZ, YL, and QL performed the experiments. CL and VT designed the experiment. All authors commented on the manuscript.
Funding
This research was supported by the Science Technology Project of Henan Province (202102110017) and the Key Discipline Project of Food Science and Engineering of Yuncheng University (XK-2021013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.854386/full#supplementary-material
Supplementary Figure 1 | Length distribution of unigenes.
Supplementary Figure 2 | Express gene saturation test of RNA-Seq data.
Supplementary Table 1 | Primers used for qRT-PCR.
Supplementary Table 2 | Quality assessment of the raw data.
Supplementary Table 3 | Quality of assembled unigenes.
Supplementary Table 4 | All GO classifications of DEGs between 62\3 and JB231AC under 50μM Cd stress.
Supplementary Table 5 | All KGEE enrichment pathways of DEGs between 62\3 and JB231AC under 50μM Cd stress.
Supplementary Table 6 | Details of DEGs predicted to belong to the ATP-binding cassette (ABC) transporter family.
Supplementary Table 7 | Details of DEGs predicted to belong to the Zn-regulated transporter, iron-regulated transporter-like protein (ZIP) family.
Supplementary Table 8 | Details of differentially-expressed genes (DEGs) predicted to the other Cd-related genes.
References
Ahmad, P., Sarwat, M., Bhat, N. A., Wani, M. R., Kazi, A. G., and Tran, L. S. (2015). Alleviation of cadmium toxicity in Brassica juncea L. (Czern. & Coss.) by calcium application involves various physiological and biochemical strategies. PLoS One 10:e0114571. doi: 10.1371/journal.pone.0114571
Bashir, S., Qayyum, M. A., Husain, A., Bakhsh, A., Ahmed, N., Hussain, M. B., et al. (2021). Efficiency of different types of biochars to mitigate Cd stress and growth of sunflower (Helianthus; L.) in wastewater irrigated agricultural soil. Saudi. J. Biol. Sci. 28, 2453–2459. doi: 10.1016/j.sjbs.2021.01.045
Bayat, M., Faramarzi, A., Ajalli, J., Abdi, M., and Nourafcan, H. (2021). Bioremediation of potentially toxic elements of sewage sludge using sunflower (Heliantus annus L.) in greenhouse and field conditions. Environ. Geochem. Health [Epub ahead of print]. doi: 10.1007/s10653-021-01018-6
Benavides, B. J., Drohan, P. J., Spargo, J. T., Maximova, S. N., Guiltinan, M. J., and Miller, D. A. (2021). Cadmium phytoextraction by Helianthus annuus (sunflower), Brassica napus cv Wichita (rapeseed), and Chyrsopogon zizanioides (vetiver). Chemosphere 265:129086. doi: 10.1016/j.chemosphere.2020.129086
Cai, X., Wang, M., Jiang, Y., Wang, C., and Ow, D. W. (2021). Overexpression of OsABCG48 lowers cadmium in rice (Oryza sativa L.). Agronomy 11:918. doi: 10.3390/agronomy11050918
Chen, F., Wang, F., Wu, F. B., Mao, W. H., Zhang, G. P., and Zhou, M. X. (2010). Modulation of exogenous glutathione in antioxidant defense system against Cd stress in the two barley genotypes differing in Cd tolerance. Plant Physiol. Biochem. 48, 663–672. doi: 10.1016/j.plaphy.2010.05.001
Chen, H. P., Zhang, W. W., Yang, X. P., Wang, P., McGrath Steve, P., and Zhao, F. J. (2018). Effective methods to reduce cadmium accumulation in rice grain. Chemosphere 207, 699–707. doi: 10.1016/j.chemosphere.2018.05.143
Chen, P., Li, Z., Luo, D., Jia, R., Lu, H., Tang, M., et al. (2021). Comparative transcriptomic analysis reveals key genes and pathways in two different cadmium tolerance kenaf (Hibiscus cannabinus L.) cultivars. Chemosphere 263:128211. doi: 10.1016/j.chemosphere2020.128211
Dou, X., Dai, H., Skuza, L., and Wei, S. (2019). Bidens pilosa L. hyperaccumulating Cd with different species in soil and the role of EDTA on the hyperaccumulation. Environ. Sci. Pollut. Res. Int. 26, 25668–25675. doi: 10.1007/s11356-019-05831-6
Dou, X., Dai, H., Skuza, L., and Wei, S. (2020). Strong accumulation capacity of hyperaccumulator Solanum nigrum L. for low or insoluble Cd compounds in soil and its implication for phytoremediation. Chemosphere 260:127564. doi: 10.1016/j.chemosphere.2020.127564
Feng, S., Tan, J., Zhang, Y., Liang, S., Xiang, S., Wang, H., et al. (2017). Isolation and characterization of a novel cadmium-regulated Yellow Stripe-Like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 36, 281–296. doi: 10.1007/s00299-016-2079-7
Fu, S., Lu, Y., Zhang, X., Yang, G., Chao, D., Wang, Z., et al. (2019). The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 70, 5909–5918. doi: 10.1093/jxb/erz335
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guerinot, M. L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465, 190–198. doi: 10.1016/s0005-2736(00)00138-3
Heyno, E., Klose, C., and Krieger-Liszkay, A. (2008). Origin of cadmium-induced reactive oxygen species production: mitochondrial electron transfer versus plasma membrane NADPH oxidase. New Phytol. 179, 687–699. doi: 10.1111/j.1469-8137.2008.02512.x
Hu, Y., Xu, L., Tian, S., Lu, L., and Lin, X. (2019). Site-specific regulation of transcriptional responses to cadmium stress in the hyperaccumulator, Sedum alfredii: based on stem parenchymal and vascular cells. Plant Mol. Biol. 99, 347–362. doi: 10.1007/s11103-019-00821-1
Imran, M., Hussain, S., ElEsawi Mohamed, A., Rana Muhammad, S., Saleem Muhammad, H., Riaz, M., et al. (2020). Molybdenum supply alleviates the cadmium toxicity in fragrant rice by modulating oxidative stress and antioxidant gene expression. Biomolecules 10:1582.
Kanu, A. S., Ashraf, U., Mo, Z. W., Sabir, S.-U.-R., Baggie, I., Charley, C. S., et al. (2019). Calcium amendment improved the performance of fragrant rice and reduced metal uptake under cadmium toxicity. Environ. Sci. Pollut. Res. 26, 24748–24757. doi: 10.1007/s11356-019-05779-7
Khan, I. U., Rono, J. K., Zhang, B. Q., Liu, X. S., Wang, M. Q., Wang, L. L., et al. (2019). Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotoxicol. Environ. Saf. 175, 8–18. doi: 10.1016/j.ecoenv.2019.03.040
Khan, Z. S., Rizwan, M., Hafeez, M., Ali, S., Adrees, M., Qayyum, M. F., et al. (2020). Effects of silicon nanoparticles on growth and physiology of wheat in cadmium contaminated soil under different soil moisture levels. Environ. Sci. Pollut. Res. Int. 27, 4958–4968. doi: 10.1007/s11356-019-06673-y
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Li, X. M., Zhang, L. H., Li, Y. Y., Ma, L. J., Bu, N., and Ma, C. Y. (2012). Changes in photosynthesis, antioxidant enzymes and lipid peroxidation in soybean seedlings exposed to UV-B radiation and/or Cd. Plant Soil 352, 377–387. doi: 10.1007/s11104-011-1003-8
Liu, C., Ding, S., Zhang, A., Hong, K., Jiang, H., Yang, S., et al. (2020). Development of nutritious rice with high zinc/selenium and low cadmium in grains through QTL pyramiding. J. Integr. Plant Biol. 62, 349–359. doi: 10.1111/jipb.12909
Liu, C. L., Gao, Z. Y., Shang, L. G., Yang, C. H., Ruan, B. P., Zeng, D. L., et al. (2020). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 62, 314–329. doi: 10.1111/jipb.12794
Liu, J., Hou, H., Zhao, L., Sun, Z., Lu, Y., and Li, H. (2019). Mitigation of Cd accumulation in rice from Cd-contaminated paddy soil by foliar dressing of S and P. Sci. Total Environ. 690, 321–328. doi: 10.1016/j.scitotenv.2019.06.332
Liu, N., Huang, X., Sun, L., Li, S., Chen, Y., Cao, X., et al. (2020). Screening stably low cadmium and moderately high micronutrients wheat cultivars under three different agricultural environments of China. Chemosphere 241:125065. doi: 10.1016/j.chemosphere.2019.125065
Liu, X. S., Feng, S. J., Zhang, B. Q., Wang, M. Q., Cao, H. W., Rono, J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 19:283. doi: 10.1186/s12870-019-1899-3
Livak, K. J., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Maria Celeste, D., Cristina, M., José, M. P., Carlos, C., Berta, G., and Conceição, S. (2013). Cadmium toxicity affects photosynthesis and plant growth at different levels. Acta Physiol. Plant. 35, 1281–1289. doi: 10.1007/s11738-012-1167-8
Mendoza-Cózatl, D. G., Jobe, T. O., Hauser, F., and Schroeder, J. I. (2011). Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr. Opin. Plant Biol. 14, 554–562. doi: 10.1016/j.pbi.2011.07.004
Migocka, M., Kosieradzka, A., Papierniak, A., Maciaszczyk-Dziubinska, E., Posyniak, E., Garbiec, A., et al. (2015). Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 66, 1001–1015. doi: 10.1093/jxb/eru459
Mills, R. F., Francini, A., Ferreira da Rocha, P. S., Baccarini, P. J., Aylett, M., Krijger, G. C., et al. (2005). The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett. 579, 783–791. doi: 10.1016/j.febslet.2004.12.040
Miyadate, H., Adachi, S., Hiraizumi, A., Tezuka, K., Nakazawa, N., Kawamoto, T., et al. (2011). OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199. doi: 10.1111/j.1469-8137.2010.03459.x
Nevo, Y., and Nelson, N. (2006). The NRAMP family of metal-ion transporters. Biochim. Biophys. Acta. 1763, 609–620. doi: 10.1016/j.bbamcr.2006.05.007
Oda, K., Otani, M., Uraguchi, S., Akihiro, T., and Fujiwara, T. (2011). Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci. Biotechnol. Biochem. 75, 1211–1213. doi: 10.1271/bbb.110193
Oono, Y., Yazawa, T., Kanamori, H., Sasaki, H., Mori, S., Handa, H., et al. (2016). Genome-wide transcriptome analysis of cadmium stress in rice. Biomed. Res. Int. 2016:9739505. doi: 10.1155/2016/9739505
Park, J., Song, W. Y., Ko, D., Eom, Y., Hansen, T. H., Schiller, M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Saidi, I., Guesmi, F., Kharbech, O., Hfaiedh, N., and Djebali, W. (2021). Gallic acid improves the antioxidant ability against cadmium toxicity: impact on leaf lipid composition of sunflower (Helianthus annuus) seedlings. Ecotoxicol. Environ. Saf. 210:111906. doi: 10.1016/j.ecoenv.2021.111906
Shahid, M., Dumat, C., Khalid, S., Niazi, N. K., and Antunes, P. M. C. (2017). Cadmium bioavailability, uptake, toxicity and detoxification in soil-plant system. Rev. Environ. Contam. Toxicol. 241, 73–137. doi: 10.1007/398_2016_8
Shi, J., Pan, G. X., Xia, Y. S., Zhang, S. Y., and Zhang, N. M. (2013). Effects of Cd on different rice growth and antioxidant enzyme system. Ecol. Environ. Sci. 22, 832–837.
Takahashi, R., Bashir, K., Ishimaru, Y., Nishizawa, N. K., and Nakanishi, H. (2012). The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 7, 1605–1607. doi: 10.4161/psb.22454
Takahashi, R., Ishimaru, Y., Senoura, T., Shimo, H., Ishikawa, S., Arao, T., et al. (2011). The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 62, 4843–4850. doi: 10.1093/jxb/err136
Tan, L., Zhu, Y. X., Fan, T., Peng, C., Wang, J. R., Sun, L., et al. (2019). OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 512, 112–118. doi: 10.1016/j.bbrc.2019.03.024
Tang, S., Xi, L., Zheng, J., and Li, H. (2003). Response to elevated CO2 of Indian mustard and sunflower growing on copper contaminated soil. Bull. Environ. Contam. Toxicol. 71, 988–997. doi: 10.1007/s00128-003-0224-9
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. PNAS 107, 16500–16505. doi: 10.1073/pnas.1005396107
Unsal, V., Dalkıran, T., Çiçek, M., and Kölükçü, E. (2020). The role of natural antioxidants against reactive oxygen species produced by cadmium toxicity: a review. Adv. Pharm. Bull. 10, 184–202. doi: 10.34172/apb.2020.023
Wang, S., Sun, J., Li, S., Lu, K., Meng, H., Xiao, Z., et al. (2019). Physiological, genomic and transcriptomic comparison of two Brassica napus cultivars with contrasting cadmium tolerance. Plant Soil 441, 71–87. doi: 10.1007/s11104-019-04083-0
Wang, X., Zhang, X., Chen, J., Wang, X., Cai, J., Zhou, Q., et al. (2018). Parental drought-priming enhances tolerance to post-anthesis drought in offspring of wheat. Front. Plant Sci. 9:261. doi: 10.3389/fpls.2018.00261
Wu, Z. C., Zhao, X. H., Sun, X. C., Tan, Q. L., Tang, Y. F., Nie, Z. J., et al. (2015). Antioxidant enzyme systems and the ascorbate–glutathione cycle as contributing factors to cadmium accumulation and tolerance in two oilseed rape cultivars (Brassica napus L.) under moderate cadmium stress. Chemosphere 138, 526–536. doi: 10.1016/j.chemosphere.2015.06.080
Yamaji, N., Xia, J., Mitani-Ueno, N., Yokosho, K., and Feng, M. J. (2013). Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 162, 927–939. doi: 10.1104/pp.113.216564
Yan, Y., Sun, Q. Q., Yang, J. J., and Zhang, X. W. (2021). Source attributions of cadmium contamination in rice grains by cadmium isotope composition analysis: a field study. Ecotoxicol. Environ. Saf. 210:111865. doi: 10.1016/j.ecoenv.2020.111865
Yang, C. H., Zhang, Y., and Huang, C. F. (2019). Reduction in cadmium accumulation in japonica rice grains by CRISPR/Cas9-mediated editing of OsNRAMP5. J. Integr. Agric. 18, 688–697.
Yang, Q., Shohag, M. J. I., Feng, Y., He, Z., and Yang, X. (2017). Transcriptome comparison reveals the adaptive evolution of two contrasting ecotypes of Zn/Cd hyperaccumulator Sedum alfredii hance. Front. Plant. Sci. 8:425. doi: 10.3389/fpls.2017.00425
Yu, F., Liu, K., Li, M., Zhou, Z., Deng, H., and Chen, B. (2013). Effects of cadmium on enzymatic and non-enzymatic antioxidative defences of rice (Oryza sativa L.). Int. J. Phytoremediation 15, 513–521. doi: 10.1080/15226514.2012.702807
Zhang, D., Du, Y., He, D., Zhou, D., Wu, J., Peng, J., et al. (2021). Use of comparative transcriptomics combined with physiological analyses to identify key factors underlying cadmium accumulation in Brassica juncea L. Front. Genet. 12:655885. doi: 10.3389/fgene.2021.655885
Zhang, F., Xiao, X., and Wu, X. (2020). Physiological and molecular mechanism of cadmium (Cd) tolerance at initial growth stage in rapeseed (Brassica napus L.). Ecotoxicol. Environ. Saf. 197:110613. doi: 10.1016/j.ecoenv.2020.110613
Zhang, H., Guo, Q., Yang, J., Shen, J., Chen, T., Zhu, G., et al. (2015). Subcellular cadmium distribution and antioxidant enzymatic activities in the leaves of two castor (Ricinus communis L.) cultivars exhibit differences in Cd accumulation. Ecotoxicol. Environ. Saf. 120, 184–192.
Zhang, H., Zhang, X., Liu, J., Niu, Y., and Wang, X. (2020). Characterization of the Heavy-metal-associated isoprenylated plant protein (HIPP) gene family from Triticeae species. Int. J. Mol. Sci. 21:6191.
Zhang, J., Zhang, M., Song, H., Zhao, J., and Yang, X. (2020). A novel plasma membrane-based NRAMP transporter contributes to Cd and Zn hyperaccumulation in Sedum alfredii Hance. Environ. Exp. Bot. 176:104121.
Zhang, X. D., Sun, J. Y., You, Y. Y., Song, J. B., and Yang, Z. M. (2018a). Identification of Cd-responsive RNA helicase genes and expression of a putative BnRH 24 mediated by miR158 in canola (Brassica napus). Ecotoxicol. Environ. Saf. 157, 159–168. doi: 10.1016/j.ecoenv.2018.03.081
Zhang, X. D., Zhao, K. X., and Yang, Z. M. (2018b). Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene 664, 139–151. doi: 10.1016/j.gene.2018.04.060
Zhang, X., Gao, B., and Xia, H. (2014). Effect of cadmium on growth, photosynthesis, mineral nutrition and metal accumulation of bana grass and vetiver grass. Ecotoxicol. Environ. Saf. 106, 102–108. doi: 10.1016/j.ecoenv.2014.04.025
Zhang, Y., Chao, J., Li, X., Zhang, C., Khan, R., Du, S., et al. (2021). Comparative transcriptome combined with biochemical and physiological analyses provide new insights toward cadmium accumulation with two contrasting Nicotiana species. Physiol. Plant 173, 369–383. doi: 10.1111/ppl.13431
Zheng, X., Chen, L., and Li, X. (2018). Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot. Stud. 59:22. doi: 10.1186/s40529-018-0238-6
Keywords: antioxidant, cadmium accumulation, cadmium stress, cadmium tolerance, differential expression genes, transcriptome
Citation: Fu Y, Zhatova H, Li Y, Liu Q, Trotsenko V and Li C (2022) Physiological and Transcriptomic Comparison of Two Sunflower (Helianthus annuus L.) Cultivars With High/Low Cadmium Accumulation. Front. Plant Sci. 13:854386. doi: 10.3389/fpls.2022.854386
Received: 13 January 2022; Accepted: 15 March 2022;
Published: 09 May 2022.
Edited by:
Baohua Wang, Nantong University, ChinaReviewed by:
Hongxian Mei, Henan Academy of Agricultural Sciences (HNAAS), ChinaLuming Yang, Henan Agricultural University, China
Copyright © 2022 Fu, Zhatova, Li, Liu, Trotsenko and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volodymyr Trotsenko, dnRyb3RzZW5rb0B1a3IubmV0; Chengqi Li, bGljaHEyMDEwQDEyNi5jb20=
 Yuanzhi Fu1,2
Yuanzhi Fu1,2 Yuqing Li
Yuqing Li Chengqi Li
Chengqi Li