Abstract
Drought stress has severely hampered maize production, affecting the livelihood and economics of millions of people worldwide. In the future, as a result of climate change, unpredictable weather events will become more frequent hence the implementation of adaptive strategies will be inevitable. Through utilizing different genetic and breeding approaches, efforts are in progress to develop the drought tolerance in maize. The recent approaches of genomics-assisted breeding, transcriptomics, proteomics, transgenics, and genome editing have fast-tracked enhancement for drought stress tolerance under laboratory and field conditions. Drought stress tolerance in maize could be considerably improved by combining omics technologies with novel breeding methods and high-throughput phenotyping (HTP). This review focuses on maize responses against drought, as well as novel breeding and system biology approaches applied to better understand drought tolerance mechanisms and the development of drought-tolerant maize cultivars. Researchers must disentangle the molecular and physiological bases of drought tolerance features in order to increase maize yield. Therefore, the integrated investments in field-based HTP, system biology, and sophisticated breeding methodologies are expected to help increase and stabilize maize production in the face of climate change.
Introduction
The average global temperature is rising, and the environment is becoming more unpredictable, with severe drought occurring (IPCC, 2013; EPA, 2021). As per estimates, water scarcity is expected to affect 1.5–1.7 billion people in South Asia by 2050 (World Bank Group, 2021). Agriculture is the most vulnerable sector to this rapid climate change. Among many unprecedented challenges, drought is the major menace to crop production, worldwide. Drought is one of the most serious abiotic stresses, affecting human health and crop productivity in about a third of the world’s population. According to FAO estimates, drought caused a direct loss of USD 29 billion to agriculture in developing countries from 2005 to 2015 (FAO, 2021). As a result, it has become a major element in undermining the positive impacts of technological interventions, fertilization, and other contemporary developments made in recent past (Lobell et al., 2011). Hence, it is critical to develop crop varieties that, in addition to delivering higher yields, can withstand abiotic stresses like drought (Fita et al., 2015; Raza et al., 2019). Being the major biofuel and biotech (as 92% genetically modified corn in United States) crop of world, maize (Zea mays L.) has been anticipated to become the most important crop as its importance goes beyond food and feed (Schwietzke et al., 2009; Yadava et al., 2017). Drought is one of the major constraints in maize production as nearly 80% of wet-season maize is grown in rainfed conditions and dry season maize suffers severe water shortage (Trachsel et al., 2016). Depending on the intensity or duration of drought stress and crop stage, the maize yield losses vary from 30 to 90%, severely affecting the flowering and grain filling stages (Sah et al., 2020). In most of the countries, maize is cultivated under rainfed areas with 300–500 mm of precipitation, which is lower than the critical level to obtain decent yield (Sah et al., 2020). Since, genetic solutions are unlikely to cover more than 30% of the gap between potential and realized yield under water stress hence, the understanding of genetics and further applying it to improve drought tolerance is a key component to stabilize global maize production. The advanced molecular breeding approaches like marker-assisted selection (MAS), transgenics, and genome-editing also holds great promises to improve maize production against drought stress.
Recent studies have projected that by 2050 the global average temperature will rise and probably exceed by 2°C under the current high emission scenario (Tesfaye et al., 2018). It will cause additional maize yield loss of 10 million tons per year with increasing temperature and changing rainfall patterns (Tesfaye et al., 2018). Hence, the novel molecular approaches in genomics era have mainly targeted the dissection and manipulation of drought stress related traits, genes/quantitative trait loci (QTLs), and molecular pathways (Deikman et al., 2012; Kumar A. et al., 2022). In addition, different omics platforms also facilitate the extensive mining of the transcriptomes, proteomes and metabolomes involved in drought tolerant mechanisms (Yu D. et al., 2021). Therefore, the objective of this review is to assess the recent progress and potential benefits of novel breeding technologies made in maize to develop drought tolerance. Untangling the molecular and physiological mechanisms for drought tolerant associated traits is essential to improve crops through integrating advanced breeding tools with biotechnology.
Morphological, Physiological, and Molecular Responses of Maize to Drought
Drought stress has a multifaceted impact on plant organization, resulting in complex and often spontaneous physiological and cellular responses. The vegetative, silking (flowering), and ear stages (grain filling) of maize are the most susceptible to drought stress, with yield losses of up to 25, 50, and 21%, respectively (Sah et al., 2020). It interferes with various molecular and physiological processes involved in plant growth and development, including photosynthetic activity, abscisic acid (ABA) accumulation, osmotic adjustment, and antioxidant capacity (Gong et al., 2014). Physiologically, when plants are stressed, they consume a large amount of adenosine triphosphate (ATP), draining their energy, and causing irreversible damage or even plant death. Plants, on the other hand, have certain adaptative mechanisms of “metabolic flexibility” that protect plants by activating apyrophosphate, which is required for transitioning poly (ADP-ribose) into ATP molecules (Hakme et al., 2008), or alternative glycolytic reactions override ATP-requiring steps, and finally the salvage pathways are induced to counteract the effects of ATP depletion (Dobrota, 2006). In response to a water deficit, adaptive traits such as osmotic adjustment, dehydration tolerance, and a reduction in photosynthetic activity emerge. Reduced photosynthetic activity results in stomatal closure and decreased photosynthetic enzyme production. Drought stress-related physiological changes at the cellular level include turgor loss and changes in membrane fluidity and composition. During drought stress, the ABA hormone concentration got increased which triggers an internal signal transduction cascade that led to reduce the osmotic potential of guard cells via loss of K+ and Cl–, resulting in stomatal closure (Anami et al., 2009).
Morphologically, plant growth and development are hampered due to disruptions in the cell cycle machinery (Carneiro et al., 2021). The perception of abiotic stress activates signaling cascades that stimulate cell cycle checkpoints, resulting in an impaired G1 to S transition, slowed DNA replication, and/or delayed entry into mitosis (Kadota et al., 2005; Qi and Zhang, 2020). Water stress causes meristem shortening in wheat and maize leaves and increases cell cycle duration due to decreased CDK activity (Granier et al., 2000). Changes in root and shoot growth, leading to increased root/shoot ratio, tissue water storage capacity, cuticle thickness, and water permeability are also potentially important in the long run, with root growth changes being the most important for crop plants (Verslues et al., 2006). In early days, corn plants that are severely water-stressed (WS) respond by rolling their leaves. Spikelet development is delayed, resulting in higher anthesis-silking interval (ASI), silk senescence, and pollen abortion (Lu et al., 2011; Edmeades, 2013). Stomatal closure, which reduces transpiration and CO2 absorption and subsequently leads to a decrease in photosynthetic activity, is one of the first reactions to drought stress (Wang et al., 2018). Severe stress during the flowering stage induces full ear abortion, and the plant becomes barren. Drought disrupts developmental stages by limiting leaf growth, plant height, and tassel architectural characteristics, which in turn reduce crop yield since plants require adequate photosynthate to acquire a necessary stature (Aslam et al., 2015). Many plants, including maize, respond to water scarcity by shifting growth and dry matter accumulation away from the shoot and toward the root (Maazou et al., 2016).
At metabolic level, many amino acids in the sap of WS plants increases transiently, and got accumulated only under severe water stress. Alterations in solute concentration, as well as protein–protein and protein–lipid interactions, are all metabolic changes linked to drought stress (Valliyodan and Nguyen, 2006). The synthesis of osmo-protectants and osmolytes is associated with the plant’s defense response to drought stress. As a response to water scarcity, adaptation mechanisms such as phytoalexin production, activation of the general phenylpropanoid pathway, and induction of lignin biosynthesis have evolved. Salicylic acid, methyl salicylate, jasmonic acid, methyl jasmonate, and other small molecules produced as a result of stress can also function as signaling molecules stimulating systemic defense and adaptability retorts (Nadeem et al., 2019), whereas others protect plants from oxidative damage caused by a variety of stresses, such as ascorbic acid, glutathione, tocopherols, anthocyanins, and carotenoids by scrounging the active oxygen intermediates produced. Several genes and transcription factors (TFs) have been involved and altered in order to fine-tune plant cellular metabolism in response to stress, either through osmotic homeostasis or damage control or repair, or by regulating gene expression levels. The cellular ionic, osmotic, and hormonal (ABA) signaling activates a number of stress-responsive genes which regulate gene expression patterns and provide osmotolerance and protection to plants as well as play a role in signal transduction for stress response (Lata et al., 2015; Figure 1).
FIGURE 1
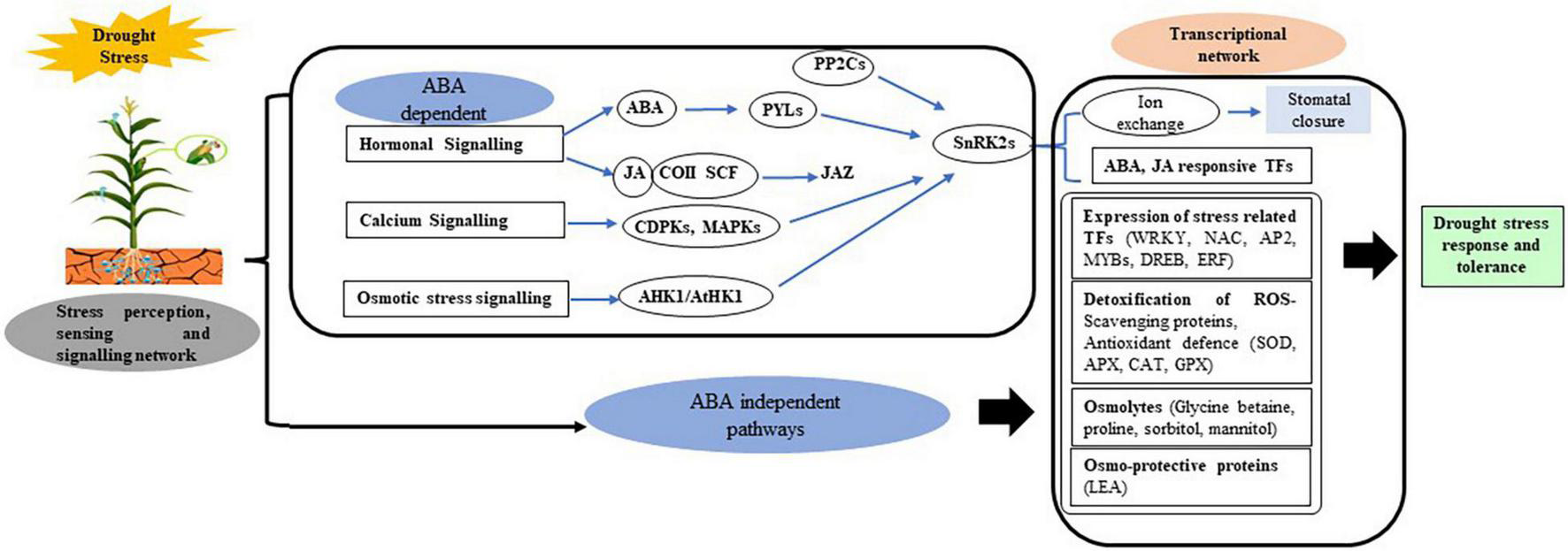
Drought stress sensing, perception, and signaling modulate transcription factors (TFs) that enhances reactive oxygen species (ROS) scavenging, protein turnover, osmotic regulation, and photosynthesis processes. ABA, abscisic acid; AP2, APETALA; Apx, ascorbate peroxidase; CAT, catalase; CDPK1, calcium-dependent protein kinase 1; DREB, dehydration responsive element binding gene; ERF, ethylene responsive factor; GPx, glutathione peroxidase; JA, jasmonic acid; LEA, late embryogenesis abundant; MAPK, mitogen-activated protein kinase; MYB, myeloblastosis oncogene; NAC, (NAM, ATAF 1/2, and CUC2) domain proteins; PP2Cs, 2C-type protein phosphatases; ROS, reactive oxygen species; SnRK2, SNF-related protein kinase 2; SOD, superoxide dismutase; TF, transcription factor; WRKY, family denoted by protein domain composed of a conserved WRKYGQK motif and a zinc-finger domain.
Selection Criteria for Drought Tolerance in Maize
Because drought is a multi-dimensional trait, phenotyping for drought tolerance traits is time-consuming and resource-intensive, requiring years of testing at multiple places to accurately portray the linked features (Khadka et al., 2020). Under drought stress, the direct selection for yield is often inefficient (Ziyomo and Bernardo, 2013). The practice of drought-controlled environments and deployment of secondary traits has been effective for accurate phenotyping to improve drought tolerance related traits.
The indirect selection via secondary traits with higher heritability and genetic correlation becomes more effective than direct selection (Ziyomo and Bernardo, 2013). The traits related to early vigor, root architecture, flowering time, carbon isotope discrimination, stomatal conductance, canopy temperature, ABA concentration, osmotic adjustment and stay green, etc., are the potential suitable secondary traits to be targeted to analyze drought stress tolerance (Kosova et al., 2014). ASI is an excellent key trait which is widely used as selection criteria in maize for drought tolerance. Generally, male and female flowering is usually synchronized within 2–3 days while under stress conditions it may be prolonged due to delay in silking and ultimately reproductive failure. Therefore, there is need to select the genotypes which have shorter ASI in case of drought stress (Araus et al., 2012). To achieve large genetic gain, the testing environment must be specifically selected. In terms of soil qualities, rainfall distribution, water distribution profiles, and possible evapotranspiration rates, the testing environment should ideally be identical to the target environment (Ribaut et al., 2009). The use of a controlled environment, as well as rigorous experimental designs and statistical analysis, will aid to increase the precision of genotypic means. To assess the G × E interactions, which is crucial for drought tolerance breeding, a multi-location evaluation is required. Recently, molecular markers related with grain yield (GY) and other traits in response to stress tolerance have been used to identify critical genes/QTLs engaged in the metabolic pathway in order to make successful selection for drought stress (Younis et al., 2020).
Genetic Dissection of Drought Related Traits in Maize
The advancement in high-throughput genotyping and the release of reference genome dataset has made a substantial increment in conducting studies to dissect economically important traits. There are mainly two methods, i.e., linkage mapping and association mapping to widely identify complex traits associated with drought tolerance. However, detecting candidate genes remain a challenge, which has led to the development of new methods such as Mendelian randomization (MR) analysis and transcriptome-wide association studies (TWAS) to evaluate expression-trait associations and identify candidate genes linked to a trait based on gene expression variation (Zhu et al., 2016).
Rapid Trait/Quantitative Trait Locus Mapping Through Linkage Mapping
Identification of the QTLs is an important strategy for effective selection through MAS. Since last decades, numerous studies have been undertaken to map genes or QTL associated to drought tolerance in maize (Ribaut et al., 1997; Tuberosa et al., 2002; Lu et al., 2006; Liu et al., 2011; Rahman et al., 2011; Zhu et al., 2011; Zhao et al., 2018). The advanced sequencing tools have enabled the revelation of ultra-high density genetic maps to discover massive molecular markers and precisely locate the position of QTLs. There have been multiple reports of QTLs linked to distinct phenotypes in maize under drought stress across various mapping populations (Table 1). In the series, long back Frova et al. (1999) detected QTLs for blooming time and plant height under both well-watered (WW) and WS circumstances. Nikolić et al. (2011) identified 43 QTLs for GY and morphological characteristics on all maize chromosomes except chromosome 9. They identified five QTLs for GY, which explained 0.1–15.86% of the variation. A total of 12 QTLs affecting leaf width were identified while 15 QTLs for plant height, ear height and only 1 QTL for leaf number was detected on chromosome 6. Almeida et al. (2013), identified 83 and 62 genomic regions responsible for GY and ASI, respectively, across multiple environments and diverse genetic backgrounds. Zhao et al. (2018) identified 69 QTLs for plant height, ear height, ASI, ear weight, cob weight, 100-kernel weight, and ear length under both drought-stressed and control conditions, explaining 4.0–17.2% of the phenotypic variation. Linkage mapping is a well-known method to locate QTLs but only a few QTLs are usually detected via this approach. Additionally, for QTL fine mapping and cloning of the genes present within it, generally large populations are required to get high map resolution (Dinka et al., 2007), making it more resource- and time-consuming process as well as the 85% of maize repetitive sequences (Schnable et al., 2009) further slows down the QTL fine mapping and cloning. The genetic dissection of drought tolerance in maize have been broadly reported but accounts of successful application of these identified QTL in maize improvement programs have been scarce. It is due to lack of validation of identified QTLs across genetic and environmental backgrounds, including genetic complexity, epistasis, profound QTL × environment interactions (QEI), population-specific QTLs, and use of agronomically poor donor lines (Vargas et al., 2006; Liu et al., 2011). Hence, meta-QTL (MQTL) analysis is adopted as another innovative tool to detect real and large effect QTL across different populations and environment. It enables the identification of genomic areas responsible for GY under both WS and WW circumstances in a variety of genotypes (Swamy et al., 2011). Zhao et al. (2018) reported 36 MQTLs in 26 populations under WW and WS conditions, as well as 39 candidate genes within those regions. Almeida et al. (2013) used MQTL analysis to find seven genomic regions for GY and one for ASI on chromosomes 1, 4, 5, 7, and 10, which were constitutively expressed in both WS and WW environments.
TABLE 1
| S. No. | Parents | Population type | Marker | QTL identified | QTL identified per traits | PVE (%) | References |
| 1. | SD34 × SD35 | F3 | RFLP | 11 | Grain yield (5), anthesis silking interval (3), number of ears per plants (3) | 9.4–49.6 | Agrama and Moussa, 1996 |
| 2. | Ac7643 × Ac7729/TZSRW | F2 | RFLP | 20 | Male flowering (10), female flowering (10) | 4.19–12.9 | Ribaut et al., 1997 |
| 3. | Zong 3 × 87-1 | RIL | SSR | 17 | Ear length (6), kernel number per row (5), 100-kernal weight (2), kernel weight per plant (4) | 1.94–10.32 | Lu et al., 2006 |
| 4. | Ac7643 × Ac7729/TZSRW | F2:3 | RFLP | 6 | Anthesis silking interval (6) | 1.91–7.02 | Vargas et al., 2006 |
| 5. | Zong 3 × 87-1 | RIL | SSR | 9 | Relative shoot fresh weight (5), leaf temperature difference (4) | 6.2–13.1 | Liu et al., 2011 |
| 6. | D5 × 7924 | F2:3 | SSR | 25 | Anthesis silking interval (4), grain yield (8), plant height (5), ear height (4), ear setting (4) | 5.39–15.64 | Zhu et al., 2011 |
| 7. | DTP79 × B73 | F2:3 | SSR | 21 | Sugar concentration (1), grain yield (3), leaf abscisic acid content (1), osmotic potential (4), relative water content (1), root density (1), root dry weight (1), total biomass (1), leaf surface area (8) | 0.2–52.2 | Rahman et al., 2011 |
| 8. | DTP79 × B73 | F2 | SSR | 45 | Grain yield (5), number of rows per ear (5), number of kernels per row (7), kernel weight (5), ear length (5), ear diameter (5), kernel length (2), Kernel width (6), kernel thickness (5) | 0.1–32.08 | Nikolić et al., 2013 |
| 9. | Langhuang × TS141 | F2:3 | SSR | 16 | Plant height (1), ear height (4), anthesis silking interval (2), ear weight (4), cob weight (3), 100-kernal weight (1), ear length (1) | 4–15.77 | Zhao et al., 2018 |
| 10. | Chang 7-2 × TS141 | F2:3 | SSR | 17 | Plant height (1), ear height (4), anthesis silking interval (1), ear weight (3), cob weight (3), 100-kernal weight (2), ear length (3) |
Genetic dissection for drought tolerance related traits in maize.
Genome-Wide Association Studies for Drought Stress Tolerance in Maize
Conventional methods captured limited allelic variation for drought tolerance traits hence, genome-wide association studies (GWAS) approach has been practiced to identify favorable alleles for drought tolerance in maize. Unlike linkage mapping, it utilizes diverse populations for QTL fine mapping by harnessing the historical recombination (Schaid et al., 2018). GWAS was first applied as a candidate gene association study in maize at the beginning of the 21st century (Thornsberry et al., 2001; Beló et al., 2008). Presently, with the release of the reference genome (Schnable et al., 2009), it has been widely utilized to dissect numerous economically important traits under drought stress as mentioned in Table 2. The recent advances suggest GWAS as a prevailing tool to effectively and efficiently categorize genotype and phenotype associations. It also facilitates mapping of expression QTL (eQTL) using the transcriptomic variation, and has demonstrated the genetic basis for gene expression traits by capturing the large proportion of phenotypic variation. Fu et al. (2013), analyzed the expression of total 14,375 genes with averagely more than 15% of phenotypic variance explained by each trait per eQTL. Drought being a highly complex and influential trait makes it difficult to dissect using GWAS also. Hence, researchers use the correlated metabolic traits or seedling survival rate under drought stressed conditions and a number of association studies to detect genes involved in imparting drought tolerance in maize (Setter et al., 2011; Liu et al., 2013; Xue et al., 2013; Farfan et al., 2015; Mao et al., 2015; Zhang X. et al., 2016). In this direction, several studies have been carried to identify the loci significantly associated with tolerance related traits/genes such as accumulation of carbohydrates and ABA metabolites (Setter et al., 2011) functional dehydration responsive element binding gene (DREB) genes and other stress related genes (Liu et al., 2013; Thirunavukkarasu et al., 2014; Wallace et al., 2016) under drought. Wang et al. (2016) identified 83 genetic variants for drought resistance and located the ZmVPP1 gene through GWAS which is demonstrated to confer drought resistance.
TABLE 2
| Population | Sample size | Traits | Markers | Marker-trait associations (MTAs) identified | References |
| IAP | 95 | Agronomic traits-7 | 1K | 29 SNPs | Hao et al., 2011 |
| IAP | 368 | Agronomic traits-14 | 525K | 83 genetic variants, 42 candidate genes, and 1 peak SNP located on ZmVPP1 gene | Wang et al., 2016 |
| IAP | 513 | Agronomic traits-17 | 560K | 1 candidate gene | Mao et al., 2015 |
| IAP | 318 | Agronomic, metabolic, and physiological traits | 157K | 123 significant SNPs | Zhang X. et al., 2016 |
| IAP | 368 | Seedling stage traits | 525K | 1 candidate gene | Liu et al., 2013 |
| IAP | 346 | Agronomic traits | 60K | 10 QTVs | Farfan et al., 2015 |
| IAP | 240 | Agronomic traits-7 | 30K | 61 SNPs | Thirunavukkarasu et al., 2014 |
| MAGIC | 420 | Seedling and germination traits | 0.95K | 28 significant SNPs | Rida et al., 2021 |
| IAP | 300 | Grain yield and secondary traits | 381K | 1549 significant SNPs, 46 candidate genes | Yuan et al., 2019 |
| IAP | 224 | Agronomic traits | 1288K | 73,573 eQTL and 97 candidate genes | Liu et al., 2020 |
| IAP | 350 | Agronomic traits-9 | 56K | 42 SNPs | Xue et al., 2013 |
| IAP | 166 | Seedling traits | – | 1 candidate gene | Zhang et al., 2020 |
| IAP | 209 | Seedling trait–seminal root length | 56K | 7 candidate genes | Guo et al., 2020 |
| IAP | 309 | Grain yield and related traits | 58K | 22 significant trait–marker associations for grain yield per plant and yield-related traits | Ma and Cao, 2021 |
Genome wide associated studies (GWAS) carried out in maize to dissect drought tolerant traits.
eQTLs, expression quantitative trait loci; IAP, inbred association panel; MAGIC, multi-parent advanced generation inter-cross; QTVs, quantitative trait variants.
Omics Approaches to Dissect Drought Tolerance in Maize
Recent advancements in plant omics, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics have facilitated decryption of the complex mechanisms of drought stress tolerance (Budak et al., 2015; Figure 2). As drought tolerant related traits are complex involving several regulations at gene, post-translational modifications, and protein interactions. The omics studies help to get more valuable and quality information to develop drought-tolerant crop cultivars (Wu et al., 2017). It has been used to identify the genes/QTLs, gene networks, and the regulatory pathways associated with drought stress tolerance in maize (Table 3). Genomics tools have been utilized to detect gene functions and their interactions in regulatory networks. The information related to RNA (transcriptomics), protein (proteomics), and metabolite (metabolomics) can be measured to map traits of interest onto a segregating population. Utilizing omics tools, the drought responses of maize in seeds, leaves, and roots has been extensively studied. Huang et al. (2012) investigated into maize seed desiccation tolerance and revealed differentially expressed proteins (HSP3, EMB564, and stress response protein) that could play a role in drought resistance during embryogenesis and germination. To better understand how ABA modulates the maize proteome in response to drought, Hu et al. (2011, 2012) compared the proteomic differences between the ABA-deficient maize mutant vp5 and the wild-type Vp5 under drought stress. Furthermore, several QTLs or genes associated with kernel desiccation were discovered to be involved in ABA synthesis using QTL mapping (Capelle et al., 2010) and oligo-microarray (Luo et al., 2010) analyses. Many drought tolerance related proteins or genes, such as ZmTPA (Huang et al., 2012), ZmRFP1 (Xia et al., 2012), and ZmCPK4 (Jiang et al., 2013), were also found to be induced by ABA or in an ABA-dependent manner. In the series, Bonhomme et al. (2012) also identified 3664 unique phosphorylation sites on 2496 proteins that influence epigenetic control, transcriptional regulation, cell cycle-dependent processes, phytohormone-mediated responses, cell cycle control, histone modification, DNA methylation, and ABA-, ethylene-, auxin-, or jasmonate-related responses under drought. Benešová et al. (2012) studied the drought-induced changes in the maize leaf proteome. Zhang et al. (2021) detected the role of Bx12 and ZmGLK44 genes in regulating metabolite biosynthesis and drought tolerance in maize while Gaffney et al. (2021) revealed several drought related biomarkers such as neophaseic acid, hydroxyabscisic acid, methyl itaconate, several phospholipids and lysolecithin, etc. The omics studies revealed that the proteins involved in maize drought response mainly include protective proteins (such as heat shock proteins, HSPs) (Hu et al., 2010; Benešová et al., 2012; Wang X. et al., 2019), stress response-related proteins (such as NBS LRR resistance-like protein) (Hu et al., 2011), 14-3-3-like proteins (Li et al., 2009; Huang et al., 2012), late embryogenesis abundant proteins (LEAs) (Huang et al., 2012; Wang X. et al., 2019), phytohormone-related proteins, and signaling proteins (such as auxin repressed protein and serine/threonine protein kinase) (Luo et al., 2010; Hu et al., 2011; Bonhomme et al., 2012; Dong et al., 2020; Li H. et al., 2021). Most of the omics studies are carried out in laboratory conditions only but there are huge differences in protein markers involved in similar metabolic pathways under field and lab conditions. Furthermore, the majority of stress treatments did not take into account the drought responses mechanism of adaptation, injury, and recovery in crops which is a major concern in such experimental designs (Lyon et al., 2016). As a result, many studies contribute only little to knowledge beyond adding some species to the list of previously discovered stress responsive proteins. Therefore, due to a lack of system biology data from field trials and huge G × E interactions in complex and unknown ways, the systematic mechanism of drought responses in crops and its application in drought tolerance development remain mainly unknown. Omics have been a valuable approach to discover the gene expressions, TFs, proteins, and pathways involved to improve drought tolerance. But the huge omics data developed for any crop drought tolerance has yet to be practically utilized at field level from laboratory. Hence, there is need to investigate the omics related data for drought responses under field conditions.
FIGURE 2
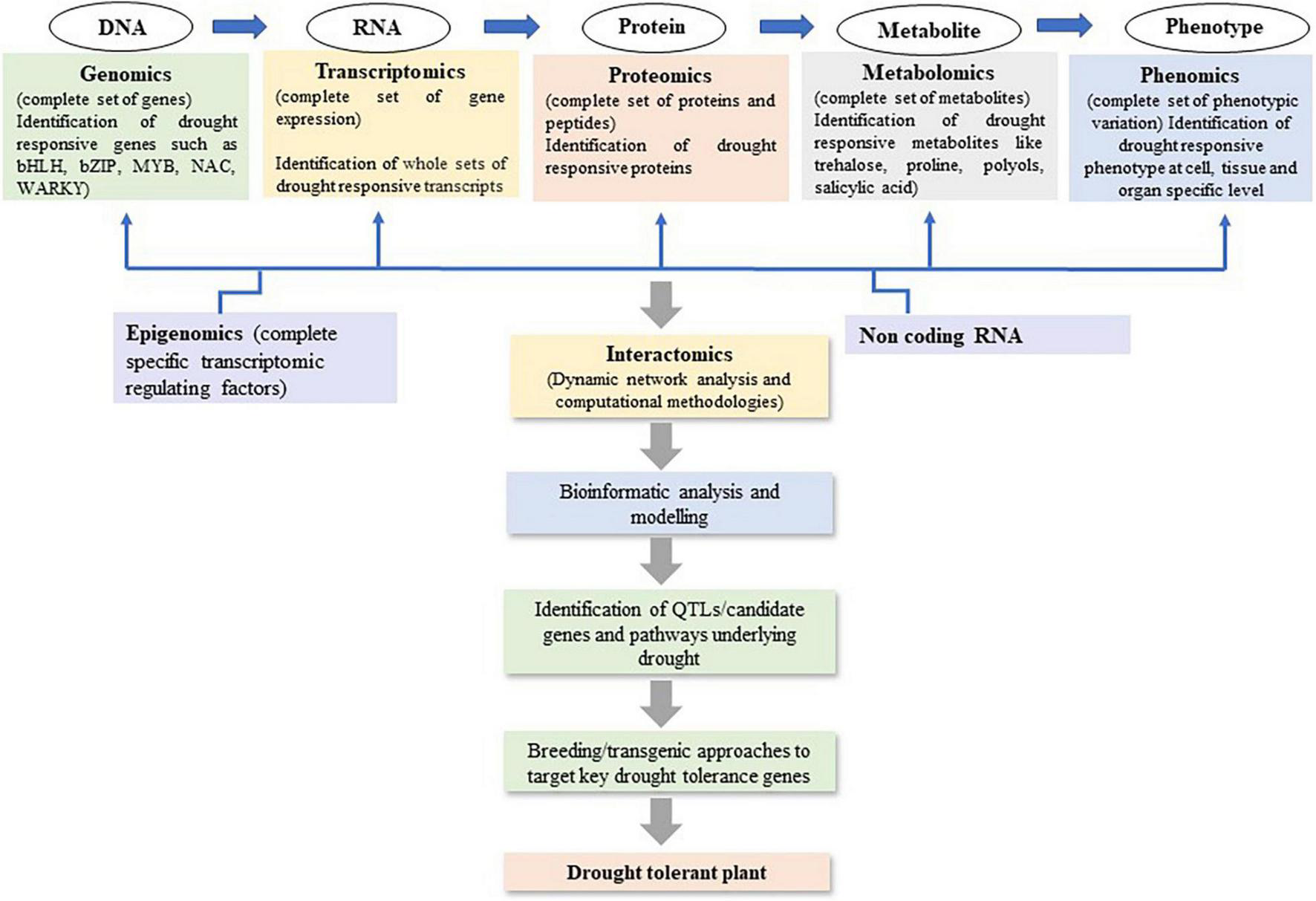
System biology-based omics approaches to strategies future basic research to decipher and develop drought tolerance maize cultivars.
TABLE 3
| Genotype | Tissue and developmental stage | Technique | Comments | References |
| 209 maize inbred lines | Seminal roots | RNA-seq, qRT-PCR | 343 differentially expressed genes (DEGs) were identified | Guo et al., 2020 |
| Two maize RILs | Leaves | qRT-PCR | Total 613 DEGs identified at different stages in drought tolerant RIL in addition the stable expression observed of cell cycle genes, ABA-, and programmed cell death-related genes | Min et al., 2016 |
| YE8112 (tolerant) and MO17 (sensitive) | Three-leaf-stage seedlings | RNA-seq, qRT-PCR | Upregulated LEA proteins, heat stress transcriptional factor B-2b, MYB-related TF96, pyrophosphate fructose-6-phosphate 1-phosphotransferase subunit alpha 1 (PFP ALPHA 1), chaperons, etc. | Zenda et al., 2019 |
| 287M (tolerant), 753F (sensitive) | Four-leaf stage | RNA-seq | Upregulated carotenoid cleavage dioxygenase 8, MYB-IF35, WRKY70, WRKY35, and ZFP2 | Zheng et al., 2020 |
| Zhongdi 175 | Seedling stage | RNA-seq, qRT-PCR | 7837 DEGs identified for drought tolerance including for sucrose metabolism and cell growth, MYB-flavonoid or MYB-bHLH-flavonoid biosynthetic pathways, ARF-Aux/IAA, bZIP/NAC-ABA, and GRAS-GA/auxin pathways | Li H. et al., 2021 |
| Zhengdan 958 | Eight-leaf stage | Multiplex iTRAQ and LC-MS/MS | Upregulated superoxide dismutase, ascorbate peroxidase, and glutathione reductase enzymes activity | Zhao et al., 2016 |
| ND476 (tolerant), ZX978 (sensitive) | Kernel | iTRAQ, tandem mass spectrometry (MS/MS), qRT-PCR | Upregulated oxidoreductase, peroxidase and hydrolytic enzyme activities, and elevated expression of stress defense proteins | Dong et al., 2020 |
| YE8112 (tolerant), MO17 (sensitive) | Kernel filling stage | iTRAQ Analysis, Strong Cation Exchange (SCX) and LC-MS/MS, qRT-PCR | 11 up-regulated proteins under drought stress including HSPs, chaperons, late embryogenesis abundant (LEA) protein, defensin-like protein, and catalase | Wang X. et al., 2019 |
| YE8112 (tolerant), MO17 (sensitive) | Seedling stage | iTRAQ Analysis, qRT-PCR | Total 721 differentially abundant proteins (DAPs) identified, up-regulated chlorophyll a–b binding proteins, lipid metabolism related proteins, abscisic acid stress ripening 1 (ASR1) protein | Zenda et al., 2018 |
| B73 | Leaves at seedling stage | iTRAQ, qRT-PCR, antioxidants assays | 200 DAPS involved in drought signal transduction, ROS scavenging, osmotic regulation, protein synthesis, cell structure modulation, as well as other metabolisms were identified | Jiang et al., 2019 |
| Chang 7-2 (tolerant), TS141 (sensitive) | Three-leaf stage and roots at seedling | iTRAQ with LC-MS/MS, qRT-PCR | 1243 significantly differentially expressed proteins associated with associated with ribosome pathway, glycolysis/gluconeogenesis pathway, and amino sugar, and nucleotide sugar metabolism were identified | Zeng et al., 2019 |
| 10 maize hybrids | Leaves | Gas chromatography-mass spectroscopy | AA-glycine and myoinositol key metabolite for drought tolerance | Obata et al., 2015 |
| DY 606 | Leaves | 1H NMR spectroscopy | AA-alanine, lipids-triacylglyceride and OM: malate, glutamate, formate involved as key metabolites | Sun et al., 2015 |
| 385 inbred | Leaves | LC-MS | Detected Bx12 and ZmGLK44 gene’s role in regulating metabolite biosynthesis and drought tolerance in maize | Zhang et al., 2021 |
| B73 | Whole plant tissues | LC-MS, high-resolution mass spectrometry | Neophaseic acid, hydroxyabscisic acid, methyl itaconate, several phospholipids, and lysolecithin detected as drought related biomarkers | Gaffney et al., 2021 |
List of studies using transcriptomics/proteomics/metabolomics approaches to unravel drought stress tolerance mechanism in maize.
AA, amino acid; 1H-NMR, nuclear magnetic resonance; iTRAQ, isobaric tags for relative and absolute quantitation; LC-MS, liquid chromatography–mass spectrometry; OM, other metabolites; qRT-PCR, quantitative real time polymerase chain reaction.
Phenomics: High-Throughput and Precision Phenotyping for Drought Stress Traits
Phenomics is a new transdisciplinary area for obtaining extensive measurements of large number of plants that result in phenotypic variations over the course of their life span (Egea-Cortines and Doonan, 2018). High-throughput phenotyping (HTP) enables accelerated accurate, labor-, and cost-effective phenotyping of complex morpho-physiological traits to detect drought stress at preliminary stage (Khadka et al., 2020). According to Yang et al. (2020), advanced plant phenomics will allow for more efficient use of genetic data, leading to novel gene discovery and improved crop output and quality in the field. To make genetic progress for drought tolerance, managed stress environments (MSEs) are required, in which the severity and timing of drought stress are controlled in a way that is relevant to target environment conditions. In the absence of rain, precise water management allows stress intensity to be adjusted, maximizing the expression of genetic variability for essential secondary traits and repeating the stress pattern, which is targeted at appropriate growth phases (White et al., 2012).
The phenotypes under drought stress are dynamic and clearly defined by a series of response curves to environmental stimuli (Tardieu, 2012). The HTP systems enable the smooth running and standardization of reliable phenotypic data in glasshouse conditions by automating procedures (Li D. et al., 2021). The phenomics tools such as digital imaging, robotics and computing equipment facilitate uninterrupted measurements of thousands of plants automatically and non-destructively (Tuberosa, 2012; Fahlgren et al., 2015). For various traits such as canopy features (Fiorani et al., 2012), root architectural traits (Blouin et al., 2007), digital imaging can be used as it provides inexpensive and rapid analysis of plant features (Richardson et al., 2018). In addition, it allows to accurately analyze the root growth process at higher resolutions (Armengaud et al., 2009; Guimarães et al., 2017). Through remote-sensing via near-infrared spectroscopy and spectral reflectance, integrative traits should be collected with high temporal resolution (Bai et al., 2019). It helps to quantify the agronomic parameters like leaf area, yield, crop cover, biomass, etc., with evolving understanding about leaf reflectance, leaf emittance, leaf thickness, canopy shape, leaf age, nutrient status and, importantly, water status (Kumar S. et al., 2022). Dar et al. (2021) utilized HTP technique to identify drought tolerant maize lines possessing favorable traits for drought stress tolerance. Under stress, the data revealed a robust link between canopy temperature and above-ground biomass. Recent improvements in low-cost field HTPPs, high-capacity data recording, scoring, and processing, and non-invasive remote sensing approaches, together with automated environmental data gathering, aid in evaluating maize yield and other quality characteristics in drought-stricken conditions. The study of essential plant components and traits may supplement direct phenotyping in the field and advancements in user-friendly data management strategies with a more powerful interpretation of results, which have expanded the usage of field HTPPs for bumper maize production. As a result, enhancing the efficiency of crop genetic modification to fulfill the needs of future generations’ foods is based on the principles and comprehensive set of references suitable for phenotyping practices management. However, the installation of phenomics facility is highly expensive and still in infancy stage in developing countries.
Epigenetic Modifications to Improve Drought Tolerance in Maize
Epigenetics has emerged as a novel research area to detect the biochemical changes in DNA that modulate gene expression without any change in the DNA sequence. Epigenetic changes are particularly common in response to environmental changes such as drought, and these changes can be long-lasting being heritable and through plant stress memory.
Epigenetic modifications via small RNA (miRNA or siRNA) for post-transcriptional gene silencing, DNA methylation, and histone changes affect gene activity and play an important role in gene expression to cope with environmental stresses (Thiebaut et al., 2019; Wojtyla et al., 2020). These modifications improve stress resistance in plants by upregulating and downregulating sRNAs (miRNAs, siRNAS) to downregulate negative regulators, upregulate positive regulators, and regulate plant hormones, reactive oxygen species (ROS), and transcriptional factors (Banerjee et al., 2017). Many advances have been made in the quantification of epigenetic variations and their impact on plant growth and development, resulting in increased yield and quality (Cortijo et al., 2014). Post-translational and post-transcriptional changes such as phosphorylation, ubiquitination, sumoylation, DNA methylation, RNA interference, histone posttranslational modifications, functional proteins, and chromatin modifications are examples of epigenetic regulation (Thiebaut et al., 2019). Using traditional breeding principles, the selection of epigenetic phenotypes can be beneficial for breeding stress-tolerant plants at various levels (metabolites, simple traits, and polygenic). Regulatory and epigenetic factors play an important role in gene expression and multi-trait development in selection process for genotype yield and quality (Verkest et al., 2015). Epigenetic changes can alter gene transcription and are an important mechanism for controlling gene expression during development in response to environmental stimulation. Basic mechanistic studies of these modifications will provide proper insight into the various genes and specific regions within them that are responsible for adapting to different abiotic stresses, resulting in a better understanding of the pathway to be targeted for crop improvement (Sun et al., 2021).
DNA methylation is a critical epigenetic modification that has been linked to plant development and stress responses (Huang et al., 2019). Various enzymes (DNA methyltransferase) that are targeted by different plant regulatory pathway systems participate in the process of catalyzing DNA methylation for a better and faster response to biotic and abiotic stresses (Zhang et al., 2018). Wang et al. (2021) generated MeDIP-Seq data to profile the DNA methylation map of four inbred lines and results showed that there were distinct drought stress responses of DNA methylation among drought-tolerant and drought-sensitive inbred lines. It was also observed that DNA methylation is involved in not only gene expression inhibition but also alternative splicing in response to drought stress. Under water stress, there was an increase in DNA methylation levels and the methylome of drought-tolerant inbred lines was much more stable than those of drought-sensitive inbred lines. Taken together, our findings may aid in deciphering the roles of DNA methylation in plant drought tolerance variations and emphasizing its role in alternative splicing.
Breeding for Drought Stress Tolerance in Maize
Conventional Breeding for Drought Stress Tolerance
It has been really challenging to improve drought tolerance through conventional breeding. Various drought tolerance cultivars such as Oh605, 16 tropically adapted inbred lines (TZEI 1 to TZEI 16), ND2005 and ND2006, TZE-W Pop DT STR C4, and TZE-Y Pop DT STR C4 have been successfully developed using conventional breeding methods such as recurrent selection, inbreeding, pedigree breeding, backcrossing, and hybridization (Ashraf, 2010). The success of conventional breeding depends on the germplasm diversity availability and identification of a suitable donor for tolerant genes. Hence, various germplasm sources tolerant to drought stress have been identified in maize (Cairns et al., 2013). However, the implementation of conventional breeding is highly time-consuming and cost- and labor-intensive approach. It requires repetitive number of selection and breeding cycles. In addition, it also transfers some undesirable genes causing linkage drag. The multiple genes controlling drought tolerance and their strong interactions with yield related traits make the conventional breeding methods limitedly accessible (Ahmar et al., 2020). Hence, now with progressions in next-generation sequencing, a number of advanced approaches have been implemented successfully to tailor maize for enhanced drought tolerance.
Molecular Breeding Approaches for Drought Stress Tolerance
The application of molecular markers has improved the efficiency of breeding drought tolerant crops significantly (Andjelković and Ignjatović-Micić, 2011; Mwamahonje et al., 2021). Unlike conventional methods, genomics provides broader chances for dissecting quantitative traits into specific genetic determinants, paving the door for MAS and, eventually, cloning QTLs and manipulating them directly through genetic engineering (Tuberosa and Salvi, 2006; Oladosu et al., 2019).
Advanced genomics-assisted breeding techniques including MAS, marker-assisted back crossing (MABC), marker-assisted recurrent selection (MARS), and genomic selection (GS) have opened up new possibilities for drought tolerance enhancement. Under WS circumstances, Ribaut and Ragot (2007) employed MABC to introduce five QTLs associated to GY and flowering traits into a drought-prone maize line, observing increased grain and lower ASI. Only large effect QTLs that have been identified and validated across a wide range of genetic backgrounds are introduced into an elite variety through MABC (Bernardo, 2008). Unlike MABC, MARS uses a subset of markers that are significantly related with target traits to accumulate a high number of medium effect QTLs in a given population (Bernardo, 2008; Ribaut et al., 2010). Beyene et al. (2016) studied the genetic gains in maize GY using 10 biparental MARS populations and demonstrated its potential in increasing genetic gain under WS conditions. Another marker-based approach, i.e., GS utilizes all available significant or insignificant marker information simultaneously to envisage the genetic value of progenies for selection (Lorenz, 2013). Using GS, Beyene et al. (2015) assessed genetic gains in GY from eight bi-parental maize populations under managed drought stress environments. The study showed that GS is a more effective method for enhancing genetic gains under drought stress in tropical maize than pedigree-based traditional phenotypic selection. Through “breeding by design” strategy, all identified QTLs and trans genes relevant to yield and drought tolerance traits can be accumulated into elite genotypes. Breeders would be able to strategize new ideotype crops by analyzing allelic variations and the genetic basis of secondary traits involved in regulating drought tolerance.
The cloning of genes associated to drought tolerant QTLs is another important application of molecular breeding. By mapping of known stress responsive genes, different techniques are being employed to clone candidate genes (Salvi and Tuberosa, 2005; Tondelli et al., 2006). Masle et al. (2005), for example, cloned the ERECTA gene in Arabidopsis thaliana, which is a DNA sequence that is beyond a QTL for transpiration efficiency. However, there is no report in the literature on the cloning of genes underlying QTLs in any crop species, or determining a suitable QTL for gene cloning is still difficult (Hu et al., 2021). By compiling all of the aforementioned data, it is clear that molecular breeding research has not progressed beyond the detection of a specific trait under drought stressed conditions. However, to evaluate whether QTLs found in a mapping population can improve drought tolerance in high yielding elite genotypes after introduction remains a difficult task for researchers. Although the application of MAS appears to be more promising and meaningful but its contribution to the development of drought-tolerant cultivars has been modest so far with only a few successful studies available (Zhao et al., 2008; Beyene et al., 2016).
Transgenic Approaches to Develop Drought Tolerance in Maize
The rapid advancement in biotechnology with genome sequencing has resulted in a diverse set of techniques for manipulating and identifying genes of interest involved in certain processes, such as drought responses. The current research on engineering drought-tolerant plants is primarily based on the transferring of one or more genes involved in signaling and regulatory pathways, or that encode enzymes involved in pathways leading to the synthesis of functional and structural protectants like osmolytes and antioxidants or stress tolerance conferring proteins (Vinocur and Altman, 2005; Khan et al., 2019). Genetic engineering facilitates wider range of solutions to improve drought tolerance more rapidly with reduced time (Martignago et al., 2020). Through genetic engineering techniques, a number of natural and synthetic genes and TFs have been incorporated into maize to enhance drought tolerance (Parmar et al., 2017; Table 4). A large number of drought responsive genes and TFs have been identified such as TsVP and BetA from Thellungiella halophila and Escherichia coli, ZmPLC1, ZmVPP1, ZmTIP1, and ZmNAC111 from maize (Wang et al., 2008, 2016; Mao et al., 2015; Zhang et al., 2020), SbER1–1 and SbER2–1 from sorghum (Li et al., 2019), LOS5 from Arabidopsis (Lu et al., 2013), TsCBF1 from T. halophila (Zhang et al., 2010), and many more, whose overexpression result in drought tolerance development in maize via transgenic approach (Jia et al., 2020; Muppala et al., 2021; Yu H. et al., 2021). Generally, the DREB proteins, i.e., a subfamily of APETALA 2/ethylene-responsive element binding factor (AP2/ERF) family and mitogen activated proteins are targeted for maize drought tolerance (Bidhan et al., 2011). Shou et al. (2004) incorporated Nicotiana Protease Kinase (NPK1) via genetic transformation to develop transgenic maize which induces HSPs and glutathione-S-transferases (GSTs) to protect photosynthetic machinery during drought stress conditions. Quan et al. (2004) transformed an elite maize inbred line DH4866 with a key E. coli enzyme encoding choline dehydrogenase, which has been linked to maize abiotic stress. At the germination and young seedling stage, transgenic maize plants accumulated more glycine betaine and exhibited improved GY and drought resistance than non-transgenic plants. Amara et al. (2013) improved drought tolerance in transgenic maize plants by over-expressing the group LEA Rab28 candidate gene, which leads to increased Rab28 protein accumulation and stability and improved water stress tolerance. He et al. (2018) reported that the overexpression of ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13 genes played important role to impart drought resistance in transgenic plants by enhancing ABA signaling, proline, and other drought related marker genes. A transgenic gene-silencing approach was used to modulate the ethylene biosynthesis in maize and determine its effect on GY under drought stress. The results showed that there was significantly higher GY, decreased and an increase in kernel number/ear in transgene-positive events compared to nulls (Habben et al., 2014). Similarly, Liu et al. (2015) cloned maize ZmSDD1 gene and dissected its functions and performance to drought stress. The over-expression of ZmSDD1 in transformed maize lines converts these lines in drought-resistant lines. The expression level of ZmSDD1 in transgenic plants was 6.68 times higher than that in wild types with reduction of stomatal density and 80% higher survival rate than that of wild type. In this direction, Monsanto company in alliance with BASF, developed the first biotechnology-derived drought tolerant maize variety, i.e., MON 87460 expressing bacterial cold shock protein B (CspB) and obtained 6% increased GY under drought conditions (Nemali et al., 2015). In maize ZmNF-YB2, the homolog of AtNF-YB, was found to be constitutively expressed in transgenic plants and outperformed Arabidopsis NF-YBs in terms of survival. Transgenic plants were also drought resistant in field trials, due to a number of higher stomatal conductance, cooler leaf temperatures, higher chlorophyll content, and delayed onset of senescence (Nelson et al., 2007). Recently, Muppala et al. (2021) has introduced rpk and nced genes into elite maize inbred lines from LeaP and SalT promoters to develop stable transgenic lines. The results showed more accumulation of ABA in transgenic plants and significant survival capacity over the control plants under stress. The improvement of drought stress tolerance is a multi-tool approach by integrating breeding, biotechnological and agronomic practices. Hence, in an attempt to develop drought tolerant maize cultivars, the research conducted utilizing transgenics declares it as one of the potential tools to manage drought stress associated risks.
TABLE 4
| Targeted genes | Source organism | Gene expression | Traits | Status | References |
| Transgenics | |||||
| CspA, CspB | Escherichia coli | Overexpression | High chlorophyll content, improved photosynthetic rate, and reduced leaf area during vegetative growth | Best performing lines commercialized as Genuity DroughtGard | Nemali et al., 2015 |
| NF-YB1, NF-YB2 | Arabidopsis, maize | Overexpression | Higher stomatal conductance and chlorophyll content, and delayed senescence | 50% yield increment under severe drought conditions. These lines not assessed in the field and never introduced to the market | Nelson et al., 2007 |
| TPS/TPP (T6P phosphatase, TPP) | Rice | Overexpression | Altered carbon allocation and improved yield in both well-watered and water-limited field trials | Drought tolerance effects assessed in extensive field trials with yield improvement under severe drought | Nuccio et al., 2015 |
| TsVP | Thellungiella halophila | Constitutive expression | Increased total soluble sugars and proline under osmotic stress. Improvements in dehydration tolerance | In a small-scale field experiment, these transgenic plants showed a higher yield under drought conditions than the control plants | Li et al., 2008 |
| Zm-ARGOS1 a, Zm-ARGOS8 | Maize | Overexpression | Improve dehydration avoidance in both plant species by reducing ethylene sensitivity | Reduced ethylene sensitivity and enhanced maize yields under both drought stress and well-watered conditions | Shi et al., 2015 |
| nced and rpk | Maize | Overexpression | Play key roles in the abscisic acid pathway and upstream component in ABA signaling | Introducing two genes involved in the ABA pathway and developed stable transgenic plants with desired characters | Muppala et al., 2021 |
| sod, apx, gr, dhar, and mdhar | – | Overexpression | Play key roles in the ascorbate–glutathione pathway | Fertile putative transgenic maize plants produced | Sreenu et al., 2016 |
| Genome editing | |||||
| ARGOS8 | Arabidopsis | Constitutive expression CRISPR/Cas9 | Improved drought tolerance in a field trial under stress conditions without affecting yield in well-watered control experiments | Commercialization of these lines is under evaluation | Shi et al., 2017 |
| ahb2 | Maize | Gene knockout | Quicker stomatal closure in response to dehydration stress | Three independent homozygous lines (i1, d2, and d35) tolerant to drought stress obtained | Liu et al., 2020 |
Application of transgenic and genome editing tools to improve drought tolerance in maize.
Genome Editing: Recent Progress to Improve Drought Tolerance in Maize
Recently, the genome editing technologies have emerged that enable rapid and precise manipulation of DNA sequences by editing major genes for development of drought-tolerant germplasm. The genome editing tools such as mega nucleases, zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease protein 9 (Cas9) system, have provided targeted gene modification in plants (Li et al., 2013; Čermák et al., 2015; Khalil, 2020). The CRISPR-Cas9 system has revolutionized both basic and applied research of plants and animals as a powerful genome editing tool (Shan et al., 2013; Wang T. et al., 2019). The approach of genome editing has been widely adopted in many plant species, such as Arabidopsis (Fauser et al., 2014), rice (Hu et al., 2016), wheat (Zhang Y. et al., 2016), potato (Andersson et al., 2018), tomato, and many fruit crops (Wang T. et al., 2019) to improve various traits like yield improvement, biotic and abiotic stress management. CRISPR/Cas9 based genome editing has been applied to increase crop disease resistance and also to improve tolerance to major abiotic stresses like drought and salinity (Wang T. et al., 2019). It helps to identify the important role/function of genes under different biotic and abiotic stresses through creating the particular gene knock-out or knock-down. In maize, recently a few studies have been carried out to improve drought tolerance by utilizing CRISPR-Cas technology (Table 4). Shi et al. (2017) employed a CRISPR-Cas aided advanced breeding technology to generate novel variants of maize ARGOS8 which is a negative regulator of ethylene responses. The results showed that in comparison to wild types, the ARGOS8 variants increased GY under drought stressed conditions. It demonstrated that modifying the expression patterns of a single native gene responsible for drought tolerance under stressed conditions can maintain yield potential. Plants derived from this method could be classified as non-GM crops. To engineer drought tolerance, multiple genes associated with complex metabolic pathways needs to be manipulated. As a result, the practical realization of drought tolerance is dependent on the use of molecular techniques capable of manipulating multiple genes at the same time. Recently, using Golden Gate cloning or the Gibson assembly method, multiple sgRNAs driven by independent promoters have been multiplexed into a single CRISPR/Cas9 expression vector (Silva and Patron, 2017). In combination with haploid induction, genome editing will be another important development in improving the breeding speed of stress tolerant maize variants in the future. In this method, the HI-Edit (haploid induction editing technology) or IMGE (haploid-inducer mediated genome editing), i.e., the haploid inducer line is also the editor line. Following the HI inducer cross, the desired genome edit occurs in the zygote, which is followed by a uniparental genome elimination (haploidization) (Malenica et al., 2021). This strategy generates edits to elite breeding lines faster, avoiding selfing or backcrossing if necessary to eliminate the transgene encoding the editing machinery. CRISPR-edited crops, on the other hand, confront socio-political obstacles, such as public acceptability and government regulation (Scheben and Edwards, 2017).
Progress to Develop Drought Tolerant Maize Cultivars: Mega Projects
There are a number of international and national level initiatives undertaken to develop drought tolerant maize cultivars such as drought tolerant maize for Africa (DTMA), water efficient maize for Africa (WEMA), and affordable, accessible Asian (AAA) drought tolerance maize project. Under DTMA project nearly 160 drought tolerant maize varieties superior to the commercial maize varieties have been released between 2007 and 2013. These cultivars have been tested in experimental trails at farmers’ fields, and disseminated among farmers in 13 African countries through different national agricultural research systems and private seed companies (Fisher et al., 2015). As per reports, more than 2 million farmers in sub-Saharan Africa are growing these new varieties and reporting 20–30% higher yield than traditional varieties, even under moderate drought conditions. It benefited 30–40 million people and gained $160–200 million worth by increased grain production each year in drought-affected areas of sub-Saharan Africa (CIMMYT, 2021).
Another project, WEMA was in pipeline from 2008 to 2018 to develop drought-tolerant maize for smallholder farmers in Asia. Similarly, a project is ongoing at Indian Council of Agricultural Research (ICAR)-Indian Institute of Maize Research (IIMR), India for genetic enhancement of maize for the development of high yielding and climate resilient hybrids with the objective to develop drought tolerance new productive inbreds and hybrids (IIMR, 2019). Various seed companies have also released drought tolerant hybrids utilizing both traditional and transgenic breeding technologies such as Pioneer Optimum AQUAmax™, Syngenta Artesian™, and DroughtGard™ (Adee et al., 2016). Since, last few decades considering the severity of drought stress, a number of drought tolerant maize varieties have been developed under such projects throughout the globe (Table 5).
TABLE 5
| S. No. | Germplasm/variety for drought tolerance | Strategy/method | Traits targeted | References |
| 1. | CML 562, CML 563, CML 564, CML 565, CML 566, CML 567 | Inbred evaluation and doubled haploidy | Drought tolerance traits | CIMMYT, 2015 |
| 3. | GDRM-187 | Participatory plant breeding | Extra-early maturity | Witcombe et al., 2003 |
| 4. | ZM 309, 401, 423, 521, 623, 625, 721, KDVI 1, 4, 6, WS 103, Melkassa 4, WH 403, 502, 504, and ZMS 402, 737 | Conventional breeding | – | Dar et al., 2016 |
| 6. | KSC720, KSC 710GT, and KSC 700 | Field screening | Grain yield and drought tolerant indices | Naghavi et al., 2013 |
| 7. | TZEE Y POP STR QPM C0 and EVDT W 99 STR QPM CO | Mother and baby trials at field | Yield related traits | Buah et al., 2013 |
| 9. | M1227-17, M0826-3, and M1124-18 | Field screening | Grain yield, ASI | Meseka et al., 2018 |
| 11. | La Posta Sequia, Pool 26 Sequia, Pool 18 Sequia, Pool 16 Sequia, DTPW, DTPY, TuxpenoSequia | FS/S1/S2 breeding schemes breeding schemes | – | Edmeades et al., 1997 |
| 26. | Malawi hybrids 30, 31, and 32 | Conventional breeding | High grain yield and flint grains | CIMMYT, 2013 |
| 14. | AMDROUT1(DT-Tester) c1F2, AMDROUT2(Ac)c1F2, AMDROUT3, AMDROUT4, AMDROUT (5 × 6), MARS7 to MARS12, G16BNSEQ-C3, DTPY-C9 | Genomic selection | Genetic gains per year | Vivek et al., 2017 |
| 15. | 70 May 80, Aaccel, and Indaco | Drought indices | Grain yield | Barutcular et al., 2016 |
| 16. | ADL47 × EXL15, ADL41 × EXL15, and EXL02 × ADL47 | Evaluation under drought stress | Grain yield | Adebayo and Menkir, 2014 |
| 17. | 9011-30, STR-EV-IWD, and IYFD-C0 | Evaluation under drought stress | Ears/plant and kernels/ear | Kamara et al., 2003 |
| 18. | SAMMAZ-26 (DTSTR WC1) | Improvement breeding | High grain yield | NSPI, 2022 |
| TL 98, 99, and 01 | MABC | ASI, flowering traits, yield | Ribaut and Ragot, 2007 | |
| 19. | Hybrid TA5084 | Gene introgression | Yield | Syngenta Foundation, 2021 |
| 20. | MON87460 | Transgenic | Yield | Nemali et al., 2015 |
| 21. | 2004 TZE-WDT STR C4, 2013 DTE STR-Y Syn, DT-Y STR Synthetic, 2009 TZE-WDT STR, 2008 DTMA-Y STR, 2012 TZE-WDT C4 STR C5, 2014 TZE-WDT STR, 2011 TZE-Y DT STR | Genetic improvement breeding | Ear height, plant height, kernel row number, ear weight, grain yield | Oluwaranti and Ajani, 2016 |
| 25. | Longe 1, Longe 4, Longe 5, MM3, Longe 7H, Longe 9H, Longe 10H, Longe 11H, UH5051, UH5052, UH5053, PAN 67, WE 2101, WE 2103, WE 2104, WE 2106, WE 2114, WE 2115 | Modern conventional methods | Early maturity, yield | Simtowe et al., 2019 |
Drought tolerant maize cultivars/genotypes developed utilizing conventional and molecular breeding tools.
Conclusion and Future Perspectives
Currently, the agriculture sector is facing the biggest challenge of climatic change with frequent spells of drought. In addition, drought being a complex trait further complicates the speedily development of drought tolerant maize cultivars. A number of advanced QTL mapping, genomic-assisted breeding methods, genetic engineering, and genome editing tools have been implemented successfully to harness the commercial stress-tolerant maize varieties. With the deployment of recent advanced technologies, researchers have provided some insight on the genetic architecture and regulatory pathways of drought tolerance mechanism in maize. The rapid trait mining has detected several genomic regions regulating drought-associated traits that can be utilized for direct selection. But still the candidate genes involved to create genetic variants for these traits are unexplored. Hence, it is highly important to execute the multi-omics studies, genetic designs, and pertinent analytical methods in integration to clearly understand the molecular regulatory mechanisms of drought response. The other approaches viz., transgenics and genome editing have widened the range to improve desired traits in shortest period of time. Among crop plants, Arabidopsis, Brachypodium distachyon, and rice are the excellent model plants to study the drought stress responses. It provides a useful heterologous system to test new genes discovered in cereal crops like maize and their relative molecular responses. As a wild grass, Brachypodium has never been subjected to human selection, which is why it has a very promising framework for tracking drought-tolerance mechanisms in grasses that may have been lost during temperate cereal crop’s domestication. Hence, the research work done in these model plants should be transferred to maize by using an accurate experimental design, including high throughput phenotyping platforms to thoroughly investigate tissue- and cell type-specific drought responses. It is crucial to strengthen the coordination among different research groups and institutions working to develop drought tolerance in maize. In addition, partnership among public and private sector could not only prove instrumental to provide promising results, but also help in designing potential drought-tolerant strategies that might have a huge impact on global maize production.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
SeS and YK wrote the manuscript. SK and ShS contributed in structuring the manuscript. RK conceptualized the idea. RK and SR edited the manuscript thoroughly. All authors read and finalized the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Adebayo M. A. Menkir A. (2014). Assessment of hybrids of drought tolerant maize (Zea mays L.) inbred lines for grain yield and other traits under stress managed conditions.Nigerian J. Genet.2819–23. 10.1016/j.nigjg.2015.06.004
2
Adee E. Roozeboom K. Balboa G. R. Schlegel A. Ciampitti I. A. (2016). Drought-tolerant corn hybrids yield more in drought-stressed environments with no penalty in non-stressed environments.Front. Plant Sci.7:1534. 10.3389/fpls.2016.01534
3
Agrama H. A. Moussa M. E. (1996). Mapping QTLs in breeding for drought tolerance in maize (Zea mays L.).Euphytica9189–97. 10.1007/bf00035278
4
Ahmar S. Gill R. A. Jung K. H. Faheem A. Qasim M. U. Mubeen M. et al (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook.Int. J Mol. Sci.21:2590. 10.3390/ijms21072590
5
Almeida G. D. Makumbi D. Magorokosho C. Nair S. Borém A. Ribaut J. M. et al (2013). QTL mapping in three tropical maize populations reveals a set of constitutive and adaptive genomic regions for drought tolerance.Theor. Appl. Genet.126583–600. 10.1007/s00122-012-2003-7
6
Amara I. Capellades M. Ludevid M. D. Pagès M. Goday A. (2013). Enhanced water stress tolerance of transgenic maize plants over-expressing LEA Rab28 gene.J. Plant Physiol.170864–873. 10.1016/j.jplph.2013.01.004
7
Anami S. De Block M. Machuka J. Van Lijsebettens M. (2009). Molecular improvement of tropical maize for drought stress tolerance in sub-Saharan Africa.Crit. Rev. Plant Sci.2816–35. 10.1080/07352680802665305
8
Andersson M. Turesson H. Olsson N. Fält A. S. Ohlsson P. Gonzalez M. N. et al (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery.Physiol. Plant.164378–384. 10.1111/ppl.12731
9
Andjelković V. Ignjatović-Micić D. (2011). ESTs analysis in maize developing kernels exposed to single and combined water and heat stresses.Sci. Agric.68353–360. 10.1590/s0103-90162011000300013
10
Araus J. L. Serret M. D. Edmeades G. (2012). Phenotyping maize for adaptation to drought.Front. Physiol.3:305. 10.3389/fphys.2012.00305
11
Armengaud P. Zambaux K. Hills A. Sulpice R. Pattison R. J. Blatt M. R. et al (2009). EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture.Plant J.57945–956. 10.1111/j.1365-313X.2008.03739.x
12
Ashraf M. (2010). Inducing drought tolerance in plants: recent advances.Biotechnol. Adv.28169–183. 10.1016/j.biotechadv.2009.11.005
13
Aslam M. Maqbool M. A. Cengiz R. (2015). Drought Stress in Maize (Zea Maysl.) Effects, Resistance Mechanisms, Global Achievements.Cham: Springer.
14
Bai G. Ge Y. Scoby D. Leavitt B. Stoerger V. Kirchgessner N. et al (2019). NU-Spidercam: a large-scale, cable-driven, integrated sensing and roboticsystem for advanced phenotyping, remote sensing, and agronomic research.Comp. Electron. Agric.16071–81. 10.1016/j.compag.2019.03.009
15
Banerjee S. Sirohi A. Ansari A. A. Gill S. S. (2017). Role of small RNAs in abiotic stress responses in plants.Plant Gene11180–189. 10.1016/j.plgene.2017.04.005
16
Barutcular C. Sabagh A. E. Konuskan O. Saneoka H. Yoldash K. M. (2016). Evaluation of maize hybrids to terminal drought stress tolerance by defining drought indices.J. Exp. Biol. Agric. Sci.4610–616. 10.18006/2016.4(issue6).610.616
17
Beló A. Zheng P. Luck S. Shen B. Meyer D. J. Li B. et al (2008). Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize.Mol. Genet. Genom.2791–10. 10.1007/s00438-007-0289-y
18
Benešová M. Hola D. Fischer L. Jedelský P. L. Hnilièka F. Wilhelmová N. et al (2012). The physiology and proteomics of drought tolerance in maize: early stomatal closure as a cause of lower tolerance to short-term dehydration?PLoS One7:e38017. 10.1371/journal.pone.0038017
19
Bernardo R. (2008). Molecular markers and selection for complex traits in plants: learning from the last 20 years.Crop Sci.481649–1664. 10.2135/cropsci2008.03.0131
20
Beyene Y. Semagn F. K. Mugo S. N. Tarekegne A. T. Babu R. Meisel B. et al (2015). Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress.Crop Sci.55154–163. 10.2135/cropsci2014.07.0460
21
Beyene Y. Semagn K. Crossa J. Mugo S. Atlin G. Tarekegne A. T. et al (2016). Improving maize grain yield under drought stress and non-stress environments in sub-Saharan Africa using marker-assisted recurrent selection.Crop Sci.56344–353. 10.2135/cropsci2015.02.0135
22
Bidhan R. Noren S. K. Mandal A. B. Basu A. K. (2011). Genetic engineering for abiotic stress tolerance in agricultural crops.Biotechnology101–22. 10.1016/b978-0-12-813066-7.00001-2
23
Blouin M. Barot S. Roumet C. (2007). A quick method to determine root biomass distribution in diameter classes.Plant Soil290371–381. 10.1007/s11104-006-9169-1
24
Bonhomme L. Valot B. Tardieu F. Zivy M. (2012). Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves.Mol. Cell Proteom.11957–972. 10.1074/mcp.M111.015867
25
Buah S. S. J. Kombiok J. M. Kanton R. A. L. Denwar N. N. Haruna A. Wiredu A. N. et al (2013). Participatory evaluation of drought tolerant maize varieties in the guinea savanna of ghana using mother and baby trial design.J. Sci. Tech.3312–23. 10.4314/just.v33i2.2
26
Budak H. Hussain B. Khan Z. Ozturk N. Z. Ullah N. (2015). From genetics to functional genomics: improvement in drought signaling and tolerance in wheat.Front. Plant Sci.6:1012. 10.3389/fpls.2015.01012
27
Cairns J. E. Crossa J. Zaidi P. H. Grudloyma P. Sanchez C. Araus J. L. et al (2013). Identification of drought, heat, and combined drought and heat tolerant donors in maize.Crop Sci.531335–1346. 10.2135/cropsci2012.09.0545
28
Capelle V. Remoue C. Moreau L. Reyss A. Mahé A. Massonneau A. et al (2010). QTLs and candidate genes for desiccation and abscisic acid content in maize kernels.BMC Plant Biol.10:2. 10.1186/1471-2229-10-2
29
Carneiro A. K. Montessoro P. D. F. Fusaro A. F. Araújo B. G. Hemerly A. S. (2021). Plant CDKs—driving the cell cycle through climate change.Plants10:1804. 10.3390/plants10091804
30
Čermák T. Baltes N. J. Čegan R. Zhang Y. Voytas D. F. (2015). High-frequency, precise modification of the tomato genome.Genome Biol.16:232. 10.1186/s13059-015-0796-9
31
CIMMYT (2013). Available online at:https://www.cimmyt.org/news/three-new-drought-tolerant-maize-hybrids-released-in-malawi/(accessed October 30, 2021).
32
CIMMYT (2015). Available online at:https://www.cimmyt.org/news/16-new-cimmyt-maize-lines-released/#:~:text=CIMMYT%20is%20pleased%20to%20announce,Africa%2C%20Latin%20America%20and%20Asia(accessed January 29, 2022).
33
CIMMYT (2021). Available online at:https://www.cimmyt.org/projects/drought-tolerant-maize-for-africa-dtma/(accessed January 28, 2022).
34
Cortijo S. Wardenaar R. Colomé-Tatché M. Gilly A. Etcheverry M. Labadie K. et al (2014). Mapping the epigenetic basis of complex traits.Science343:1145. 10.1126/science.1248127
35
Dar Z. A. Dar S. A. Khan J. A. Lone A. A. Langyan S. Lone B. A. et al (2021). Identification for surrogate drought tolerance in maize inbred lines utilizing high-throughput phenomics approach.PLoS One16:e0254318. 10.1371/journal.pone.0254318
36
Dar Z. A. Lone A. A. Alie B. A. Makdoomi M. I. Wani G. A. Gazal S. H. A. et al (2016). Development of stress resilient maize cultivars for sustainability.Adv. Life Sci.5349–355.
37
Deikman J. Petracek M. Heard J. E. (2012). Drought tolerance through biotechnology: improving translation from the laboratory to farmers’ fields.Curr. Opin. Biotechnol.23243–250. 10.1016/j.copbio.2011.11.003
38
Dinka S. J. Campbell M. A. Demers T. Raizada M. N. (2007). Predicting the size of the progeny mapping population required to positionally clone a gene.Genetics1762035–2054. 10.1534/genetics.107.074377
39
Dobrota C. (2006). Energy dependent plant stress acclimation.Rev. Environ. Sci. Biotechnol.5243–251. 10.1007/s11157-006-0012-1
40
Dong A. Yang Y. Liu S. Zenda T. Liu X. Wang Y. et al (2020). Comparative proteomics analysis of two maize hybrids revealed drought-stress tolerance mechanisms.Biotechnol. Biotechnol. Equip.34763–780. 10.1080/13102818.2020.1805015
41
Edmeades G. O. (2013). Progress in Achieving and Delivering Drought Tolerance in Maize: An Update.Ithaca: ISAAA.
42
Edmeades G. O. Banziger M. Beck D. (1997). “Development and per se performance of CIMMYT maize populations as drought tolerant sources,” in Developing Drought and Low N-Tolerant Maize, ed.EdmeadesG. O. (El Batan: CIMMYT), 254–262.
43
Egea-Cortines M. Doonan J. H. (2018). Phenomics.Front. Plant Sci.9:678. 10.3389/fpls.2018.00678
44
EPA (2021). Available:https://www.epa.gov/climate-indicators/weather-climate(accessed January 25, 2022).
45
Fahlgren N. Gehan M. A. Baxter I. (2015). Lights, camera, action: high-throughput plant phenotyping is ready for a close-up.Curr. Opin. Plant Biol.2493–99. 10.1016/j.pbi.2015.02.006
46
FAO (2021). The Impact of Disasters and Crises on Agriculture and Food Security.Rome: Food and agriculture organization of the United Nations.
47
Farfan I. D.B. De La Fuente G. N. Murray S. C. Isakeit T. Huang P. C. Warburton M. et al (2015). Genome wide association study for drought, aflatoxin resistance, and important agronomic traits of maize hybrids in the sub-tropics.PLoS One10:e0117737. 10.1371/journal.pone.0117737
48
Fauser F. Schiml S. Puchta H. (2014). Both CRISPR/C as-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana.Plant J.79348–359. 10.1111/tpj.12554
49
Fiorani F. Rascher U. Jahnke S. Schurr U. (2012). Imaging plants dynamics in heterogenic environments.Curr. Opin. Biotech.23227–235. 10.1016/j.copbio.2011.12.010
50
Fisher M. Abate T. Lunduka R. W. Asnake W. Alemayehu Y. Madulu R. B. (2015). Drought tolerant maize for farmer adaptation to drought in sub-Saharan Africa: determinants of adoption in eastern and southern Africa.Clim. Change133283–299. 10.1007/s10584-015-1459-2
51
Fita A. Rodríguez-Burruezo A. Boscaiu M. Prohens J. Vicente O. (2015). Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production.Front. Plant Sci.6:978. 10.3389/fpls.2015.00978
52
Frova C. Krajewski P. Di Fonzo N. Villa M. Sari-Gorla M. (1999). Genetic analysis of drought tolerance in maize by molecular markers II. Plant height and flowering.Theor. Appl. Genet.99289–295. 10.1007/s001220051234
53
Fu J. Cheng Y. Linghu J. Yang X. Kang L. Zhang Z. et al (2013). RNA sequencing reveals the complex regulatory network in the maize kernel.Nat. Commun.4:2832. 10.1038/ncomms3832
54
Gaffney I. Sallach J. B. Wilson J. Bergström E. Thomas-Oates J. (2021). Metabolomic approaches to studying the response to drought stress in corn (Zea mays) cobs.Metabolites11:438. 10.3390/metabo11070438
55
Gong F. Yang L. Tai F. Hu X. Wang W. (2014). “Omics” of maize stress response for sustainable food production: opportunities and challenges.Omics J. Integr. Biol.18714–732. 10.1089/omi.2014.0125
56
Granier C. Inze D. Tardieu F. (2000). Spatial distribution of cell division rate can be deduced from that of p34cdc2 kinase activity in maize leaves grown at contrasting temperatures and soil water conditions.Plant Physiol.1241393–1402. 10.1104/pp.124.3.1393
57
Guimarães C. M. Stone L. F. Brito G. G. D. Heuert J. (2017). Evaluation of water-stress tolerance of Acala SJ 2 and Auburn 2 cotton cultivars in a phenotyping platform.Rev. Ambient. Água12629–642. 10.4136/ambi-agua.2105
58
Guo J. Li C. Zhang X. Li Y. Zhang D. Shi Y. et al (2020). Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress.Plant Sci.292:110380. 10.1016/j.plantsci.2019.110380
59
Habben J. E. Bao X. Bate N. J. DeBruin J. L. Dolan D. Hasegawa D. et al (2014). Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions.Plant Biotech. J.12685–693. 10.1111/pbi.12172
60
Hakme A. Wong H.-K. Dantzer F. Schreiber V. (2008). The expanding field of polylation (ADP-ribosy).EMBO Rep.91094–1100. 10.1038/embor.2008.191
61
Hao Z. Li X. Xie C. Weng J. Li M. Zhang D. et al (2011). Identification of functional genetic variations underlying drought tolerance in maize using SNP markers. J. Integr. Plant Biol.53, 641–652. 10.1111/j.1744-7909.2011.01051.x
62
He Z. Zhong J. Sun X. Wang B. Terzaghi W. Dai M. (2018). The maize ABA receptors ZmPYL8, 9, and 12 facilitate plant drought resistance.Front. Plant Sci.9:422. 10.3389/fpls.2018.00422
63
Hu X. Li Y. Li C. Yang H. Wang W. Lu M. (2010). Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress.J. Plant Growth Regul.29455–464. 10.1007/s00344-010-9157-9
64
Hu X. Lu M. Li C. Liu T. Wang W. Wu J. et al (2011). Differential expression of proteins in maize roots in response to abscisic acid and drought.Acta Physiol. Plant332437–2446. 10.1007/s11738-011-0784-y
65
Hu X. Wang C. Fu Y. Liu Q. Jiao X. Wang K. (2016). Expanding the range of CRISPR/Cas9 genome editing in rice.Mol. Plant9943–945. 10.1016/j.molp.2016.03.003
66
Hu X. Wang G. Du X. Zhang H. Xu Z. Wang J. et al (2021). QTL analysis across multiple environments reveals promising chromosome regions associated with yield-related traits in maize under drought conditions.Crop J.9759–766. 10.1016/j.cj.2020.10.004
67
Hu X. Wu X. Li C. Lu M. Liu T. Wang Y. et al (2012). Abscisic acid refines the synthesis of chloroplast proteins in maize (Zea mays) in response to drought and light.PLoS One7:e49500. 10.1371/journal.pone.0049500
68
Huang H. Liu R. Niu Q. Tang K. Zhang B. Zhang H. et al (2019). Global increase in DNA methylation during orange fruit development and ripening.Proc. Natl. Acad. Sci. U.S.A.1161430–1436. 10.1073/pnas.1815441116
69
Huang H. Mølle I. M. Song S. Q. (2012). Proteomics of desiccation tolerance during development and germination of maize embryos.J. Proteomics751247–1262. 10.1016/j.jprot.2011.10.036
70
IIMR (2019). Available online at:https://iimr.icar.gov.in/genetic-enhancement-of-maize-for-the-development-of-high-yielding-and-climate-resilient-hybrids/(accessed October 30, 2021).
71
IPCC (2013). “Policymakers,” in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, edsStockerT. F.QinD.PlattnerG.-K.TignorM.AllenS. K.BoschungJ.et al (Cambridge, MA: Cambridge University Press).
72
Jia T. J. Jing-Jing L. I. Wang L. F. Cao Y. Y. Juan M. A. Hao W. A. N. G. et al (2020). Evaluation of drought tolerance in ZmVPP1-overexpressing transgenic inbred maize lines and their hybrids.J. Integr. Agric.192177–2187. 10.1016/s2095-3119(19)62828-5
73
Jiang S. Zhang D. Wang L. Pan J. Liu Y. Kong X. et al (2013). A maize calcium dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis.Plant Physiol. Biochem.71112–120. 10.1016/j.plaphy.2013.07.004
74
Jiang Z. Jin F. Shan X. Li Y. (2019). iTRAQ-based proteomic analysis reveals several strategies to cope with drought stress in maize seedlings.Int. J. Mol. Sci.20:5956. 10.3390/ijms20235956
75
Kadota Y. Watanabe T. Fujii S. Maeda Y. Ohno R. Higashi K. et al (2005). Cell cycle dependence of elicitor-induced signal transduction in tobacco BY-2 cells.Plant Cell Physiol.46156–165. 10.1093/pcp/pci008
76
Kamara A. Y. Menkir A. Badu-Apraku B. Ibikunle O. (2003). The influence of drought stress on growth, yield and yield components of selected maize genotypes.J. Agric. Sci.14143–50. 10.1017/s0021859603003423
77
Khadka K. Earl H. J. Raizada M. N. Navabi A. (2020). A physio-morphological trait-based approach for breeding drought tolerant wheat.Front. Plant Sci.11:715. 10.3389/fpls.2020.00715
78
Khalil A. M. (2020). The genome editing revolution.J. Genet. Eng. Biotechnol.181–16. 10.1093/oso/9780198766834.003.0001
79
Khan S. Anwar S. Yu S. Sun M. Yang Z. Gao Z. Q. (2019). Development of drought-tolerant transgenic wheat: achievements and limitations.Int. J. Mol. Sci.20:3350. 10.3390/ijms20133350
80
Kosova K. Vitamvas P. Urban M. O. Kholova J. Prášil I. T. (2014). Breeding for enhanced drought resistance in barley and wheat-drought-associated traits, genetic resources and their potential utilization in breeding programmes.Czech J. Genet. Plant Breed.50247–261. 10.17221/118/2014-cjgpb
81
Kumar A. Carter A. Mir R. R. Sehgal D. Agarwal P. Paterson A. H. (eds) (2022). Genetics and genomics to enhance crop production, towards food security.Front. Genet.12:798308. 10.3389/fgene.2021.798308
82
Kumar S. Meena R. S. Sheoran S. Jangir C. K. Jhariya M. K. Banerjee A. et al (2022). “Remote sensing for agriculture and resource management,” in Natural Resources Conservation and Advances for Sustainability, edsJhariyaM. K.MeenaR. S.BanerjeeA.MeenaS. N. (Amsterdam: Elsevier), 91–135.
83
Lata C. Muthamilarasan M. Prasad M. (2015). “Drought stress responses and signal transduction in plants,” in Elucidation of Abiotic Stress Signaling in Plants, ed.PandeyG. K. (New York, NY: Springer), 195–225. 10.1007/978-1-4939-2540-7_7
84
Li B. Wei A. Song C. Li N. Zhang J. (2008). Heterologous expression of the TsVP gene improves the drought resistance of maize.Plant Biotechnol. J.6146–159. 10.1111/j.1467-7652.2007.00301.x
85
Li D. Quan C. Song Z. Li X. Yu G. Li C. et al (2021). High-throughput plant phenotyping platform (HT3P) as a novel tool for estimating agronomic traits from the lab to the field.Front. Bioeng. Biotechnol.8:623705. 10.3389/fbioe.2020.623705
86
Li H. Han X. Liu X. Zhou M. Ren W. Zhao B. et al (2019). A leucine-rich repeat-receptor-like kinase gene SbER2–1 from sorghum (Sorghum bicolor L.) confers drought tolerance in maize.BMC Genom.20:737. 10.1186/s12864-019-6143-x
87
Li H. Yue H. Xie J. Bu J. Li L. Xin X. et al (2021). Transcriptomic profiling of the high-vigour maize (Zea mays L.) hybrid variety response to cold and drought stresses during seed germination.Sci. Rep.111–16. 10.1038/s41598-021-98907-8
88
Li H. Y. Huang S. H. Shi Y. S. Song Y. C. Zhong Z. B. Wang G. Y. et al (2009). Isolation and analysis of drought-induced genes in maize roots.Agric. Sci. China8129–136. 10.1016/s1671-2927(09)60019-5
89
Li J. F. Norville J. E. Aach J. McCormack M. Zhang D. Bush J. et al (2013). Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotianabenthamiana using guide RNA and Cas9.Nat. Biotechnol.31688–691. 10.1038/nbt.2654
90
Liu S. Li C. Wang H. Wang S. Yang S. Liu X. et al (2020). Mapping regulatory variants controlling gene expression in drought response and tolerance in maize.Genom. Biol.211–22. 10.1186/s13059-020-02069-1
91
Liu S. Wang X. Wang H. Xin H. Yang X. Yan J. et al (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L.PLoS Genet.9:e1003790. 10.1371/journal.pgen.1003790
92
Liu Y. Qin L. Han L. Xiang Y. Zhao D. (2015). Overexpression of maize SDD1 (ZmSDD1) improves drought resistance in Zea mays L. by reducing stomatal density.Plant Cell Tissue Organ Cult.122147–159. 10.1007/s11240-015-0757-8
93
Liu Y. Subhash C. Yan J. Song C. Zhao J. Li J. (2011). Maize leaf temperature responses to drought: thermal imaging and quantitative trait loci (QTL) mapping.Enviorn. Exp. Bot.71158–165. 10.1016/j.envexpbot.2010.11.010
94
Lobell D. B. Schlenker W. Costa-Roberts J. (2011). Climate trends and global crop production since 1980.Science333616–620. 10.1126/science.1204531
95
Lorenz A. J. (2013). Resource allocation for maximizing prediction accuracy and genetic gain of genomic selection in plant breeding: a simulation experiment.G3 Genes Genomes Genet.3481–491. 10.1534/g3.112.004911
96
Lu G. H. Tang J. H. Yan J. B. Ma X. Q. Li J. S. Chen S. J. et al (2006). Quantitative trait loci mapping of maize yield and its components under different water treatments at flowering time.J. Integr. Plant Biol.481233–1243. 10.1111/j.1744-7909.2006.00289.x
97
Lu Y. Hao Z. Xie C. Crossa J. Araus J. L. Gao S. et al (2011). Large-scale screening for maize well-watered environments.Field Crop Res.12437–45. 10.1016/j.fcr.2011.06.003
98
Lu Y. Li Y. Zhang J. Xiao Y. Yue Y. Duan L. et al (2013). Overexpression of Arabidopsis molybdenum cofactor sulfurase gene confers drought tolerance in maize (Zea mays L.).PLoS One8:e52126. 10.1371/journal.pone.0052126
99
Luo M. Liu J. Lee R. D. Scully B. T. Guo B. (2010). Monitoring the expression of maize genes in developing kernels under drought stress using oligo-microarray.J. Integr. Plant Biol.521059–1074. 10.1111/j.1744-7909.2010.01000.x
100
Lyon D. Castillejo M. A. Mehmeti-Tershani V. Staudinger C. Kleemaier C. Wienkoop S. (2016). Drought and recovery: independently regulated processes highlighting the importance of protein turnover dynamics and translational regulation in Medicago truncatula.Mol. Cell. Proteom.15, 1921–1937.
101
Ma J. Cao Y. (2021). Genetic dissection of grain yield of maize and yield-related traits through association mapping and genomic prediction.Front. Plant Sci.12:690059. 10.3389/fpls.2021.690059
102
Maazou A. R. S. Tu J. Qiu J. Liu Z. (2016). Breeding for drought tolerance in maize (Zea mays L.).Am. J. Plant Sci.7:1858.
103
Malenica N. Dunić J. A. Vukadinović L. Cesar V. Šimić D. (2021). Genetic approaches to enhance multiple stress tolerance in maize.Genes12:1760. 10.3390/genes12111760
104
Mao H. Wang H. Liu S. Li Z. Yang X. Yan J. et al (2015). Qin A transposable element in a NAC gene is associated with drought tolerance in maize seedlings.Nat. Commun.6:8326. 10.1038/ncomms9326
105
Martignago D. Rico-Medina A. Blasco-Escámez D. Fontanet-Manzaneque J. B. Caño-Delgado A. I. (2020). Drought resistance by engineering plant tissue-specific responses.Front. Plant Sci.10:1676. 10.3389/fpls.2019.01676
106
Masle J. Gilmore S. R. Farquhar G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis.Nature436866–870. 10.1038/nature03835
107
Meseka S. Menkir A. Bossey B. Mengesha W. (2018). Performance assessment of drought tolerant maize hybrids under combined drought and heat stress.Agronomy8:274. 10.3390/agronomy8120274
108
Min H. Chen C. Wei S. Shang X. Sun M. Xia R. et al (2016). Identification of drought tolerant mechanisms in maize seedlings based on transcriptome analysis of recombination inbred lines.Front. Plant Sci.7:1080. 10.3389/fpls.2016.01080
109
Muppala S. Gudlavalleti P. K. Malireddy K. R. Puligundla S. K. Dasari P. (2021). Development of stable transgenic maize plants tolerant for drought by manipulating ABA signaling through agrobacterium-mediated transformation.J. Genet. Eng. Biotechnol.191–14. 10.1186/s43141-021-00195-2
110
Mwamahonje A. Eleblu J. S. Y. Ofori K. Deshpande S. Feyissa T. Tongoona P. (2021). Drought tolerance and application of marker-assisted selection in sorghum.Biology10:1249. 10.3390/biology10121249
111
Nadeem M. Li J. Yahya M. Sher A. Ma C. Wang X. et al (2019). Research progress and perspective on drought stress in legumes: a review.Int. J. Mol. Sci.20:2541. 10.3390/ijms20102541
112
Naghavi M. R. Aboughadareh A. P. Khalili M. (2013). Evaluation of drought tolerance indices for screening some of corn (Zea mays L.) cultivars under environmental conditions.Not. Sci. Biol.5388–393. 10.15835/nsb539049
113
Nelson D. E. Repetti P. P. Adams T. R. Creelman R. A. Wu J. Warner D. C. et al (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres.Proc. Natl. Acad. Sci. U.S.A.10416450–16455. 10.1073/pnas.0707193104
114
Nemali K. S. Bonin C. Dohleman F. G. Stephens M. Reeves W. R. Nelson D. E. et al (2015). Physiological responses related to increased grain yield under drought in the first biotechnology-derived drought-tolerant maize.Plant Cell Environ.381866–1880. 10.1111/pce.12446
115
Nikolić A. Anðelković V. Dodig D. Ignjatović-Micić D. (2011). Quantitative trait loci for yield and morphological traits in maize under drought stress.Genetika43263–276. 10.2298/gensr1102263n
116
Nikolić A. Anđelković V. Dodig D. Mladenović-Drinić S. Kravić N. Ignjatović-Micić D. (2013). Identification of QTL-s for drought tolerance in maize, II: yield and yield components.Genetika45341–350. 10.2298/gensr1302341n
117
NSPI (2022). Nigerian Seed Portal Initiative. Available online at:https://www.seedportal.org.ng/(accessed January 29, 2022).
118
Nuccio M. L. Wu J. Mowers R. Zhou H.-P. Meghji M. Primavesi L. F. et al (2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions.Nat. Biotechnol.33:862. 10.1038/nbt3277
119
Obata T. Witt S. Lisec J. Palacios-Rojas N. Florez-Sarasa I. Yousfi S. et al (2015). Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield.Plant Physiol.1692665–2683. 10.1104/pp.15.01164
120
Oladosu Y. Rafii M. Y. Samuel C. Fatai A. Magaji U. Kareem I. et al (2019). Drought resistance in rice from conventional to molecular breeding: a review.Int. J. Mol. Sci.20:3519. 10.3390/ijms20143519
121
Oluwaranti A. Ajani O. T. (2016). Evaluation of drought tolerant maize varieties under drought and rain-fed conditions: a rainforest location.J. Agric. Sci.8153–162. 10.5539/jas.v8n7p153
122
Parmar N. Singh K. H. Sharma D. Singh L. Kumar P. Nanjundan J. et al (2017). Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review.Biotechnology71–35. 10.1007/s13205-017-0870-y
123
Qi F. Zhang F. (2020). Cell cycle regulation in the plant response to stress.Front. Plant Sci.10:1765. 10.3389/fpls.2019.01765
124
Quan R. Shang M. Zhang H. Zhao Y. Zhang J. (2004). Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize.Plant Biotechnol. J.2477–486. 10.1111/j.1467-7652.2004.00093.x
125
Rahman H. Pekic S. Lazic-Jancic V. Quarrie S. A. Shah S. M. Pervez A. et al (2011). Molecular mapping of quantitative trait loci for drought tolerance in maize plants.Genet. Mol. Res.10889–901. 10.4238/vol10-2gmr1139
126
Raza A. Razzaq A. Mehmood S. S. Zou X. Zhang X. Lv Y. et al (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: a review.Plants8:34. 10.3390/plants8020034
127
Ribaut J. M. Betran J. Monneveux P. Setter T. (2009). “Drought tolerance in maize,” in Handbook of Maize: Its Biology, edsBennetzenJ.HakeS. (New York, NY: Springer), 311–344. 10.1007/978-0-387-79418-1_16
128
Ribaut J. M. De Vicente M. C. Delannay X. (2010). Molecular breeding in developing countries: challenges and perspectives.Curr. Opin. Plant Biol.13213–218. 10.1016/j.pbi.2009.12.011
129
Ribaut J. M. Jiang C. Gonzalez-de-Leon D. Edmeades G. O. Hoisington D. A. (1997). Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis-silking interval.Theor. Appl. Genet.92905–914. 10.1007/BF00221905
130
Ribaut J. M. Ragot M. (2007). Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives.J. Exp. Bot.58351–360. 10.1093/jxb/erl214
131
Richardson A. D. Hufkens K. Milliman T. Aubrecht D. M. Chen M. Gray J. M. et al (2018). Tracking vegetation phenology across diverse North American biomes using PhenoCam imagery.Sci. Data51–24. 10.1038/sdata.2018.28
132
Rida S. Maafi O. López-Malvar A. Revilla P. Riache M. Djemel A. (2021). Genetics of germination and seedling traits under drought stress in a MAGIC population of maize.Plants10:1786. 10.3390/plants10091786
133
Sah R. P. Chakraborty M. Prasad K. Pandit M. Tudu V. K. Chakravarty M. K. et al (2020). Impact of water deficit stress in maize: phenology and yield components.Sci. Rep.101–5. 10.1038/s41598-020-59689-7
134
Salvi S. Tuberosa R. (2005). To clone or not to clone plant QTLs: present and future challenges.Trends Plant Sci.10297–304. 10.1016/j.tplants.2005.04.008
135
Schaid D. J. Chen W. Larson N. B. (2018). From genome-wide associations to candidate causal variants by statistical fine-mapping.Nat. Rev. Genet.19491–504. 10.1038/s41576-018-0016-z
136
Scheben A. Edwards D. (2017). Genome editors take on crops.Science3551122–1123. 10.1126/science.aal4680
137
Schnable P. S. Ware D. Fulton R. S. Stein J. C. Wei F. Pasternak S. et al (2009). The B73 maize genome: complexity, diversity, and dynamics.Science3261112–1115. 10.1126/science.1178534
138
Schwietzke S. Kim Y. Ximenez E. Mosier N. Ladisch M. (2009). Molecular Genetic Approaches to Maize Improvement.Berlin: Springer- Verlag, 347–364.
139
Setter T. L. Yan J. Warburton M. Ribaut J. M. Xu Y. Sawkins M. et al (2011). Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought.J. Exp. Bot.62701–716. 10.1093/jxb/erq308
140
Shan Q. Wang Y. Li J. Zhang Y. Chen K. Liang Z. et al (2013). Targeted genome modification of crop plants using a CRISPR-Cas system.Nat. Biotechnol.31686–688. 10.1038/nbt.2650
141
Shi J. Gao H. Wang H. Lafitte H. R. Archibald R. L. Yang M. et al (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions.Plant Biotechnol. J.15207–216. 10.1111/pbi.12603
142
Shi J. Habben J. E. Archibald R. L. Drummond B. J. Chamberlin M. A. Williams R. W. et al (2015). Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize.Plant Physiol.169266–282. 10.1104/pp.15.00780
143
Shou H. Bordallo P. Wang K. (2004). Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize.J. Exp. Bot.551013–1019. 10.1093/jxb/erh129
144
Silva N. V. Patron N. J. (2017). CRISPR-based tools for plant genome engineering.Emerg. Top. Life Sci.1135–149. 10.1042/ETLS20170011
145
Simtowe F. Marenya P. Amondo E. Worku M. Rahut D. B. Erenstein O. (2019). Heterogeneous seed access and information exposure: implications for the adoption of drought-tolerant maize varieties in Uganda.Agric. Food Econ.71–23.
146
Sreenu P. Sateesh Kumar P. Reddy M. K. Sailaja D. Pavan Kumar G. (2016). Resourceful and high efficiency Agrobacterium mediated transformation of maize (Zea mays L.) using coleoptilar nodal explants.Int. J. Curr. Res. Biosci. Plantbiol.31–9. 10.20546/ijcrbp.2016.312.001
147
Sun C. Ali K. Yan K. Fiaz S. Dormatey R. Bi Z. et al (2021). Exploration of epigenetics for improvement of drought and other stress resistance in crops: a review.Plants1012–26. 10.3390/plants10061226
148
Sun C. Gao X. Fu J. Zhou J. Wu X. (2015). Metabolic response of maize (Zea mays L.) plants to combined drought and salt stress.Plant Soil38899–117. 10.1007/s11104-014-2309-0
149
Swamy B. P. Vikram P. Dixit S. Ahmed H. U. Kumar A. (2011). Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus.BMC Genom.12:319. 10.1186/1471-2164-12-319
150
Syngenta Foundation (2021). Affordable, Accessible, Asian (“AAA”) Drought-Tolerant Maize Overview of a Collaborative Program Between CIMMYT and Syngenta, Supported and Coordinated by Syngenta Foundation for Sustainable Agriculture.Basel: Syngenta Foundation.
151
Tardieu F. (2012). Any trait or trait–related allele can confer drought tolerance: just design the right drought scenario.J. Exp. Bot.6325–31. 10.1093/jxb/err269
152
Tesfaye K. Kruseman G. Cairns J. E. Zaman-Allah M. Wegary D. Zaidi P. H. et al (2018). Potential benefits of drought and heat tolerance for adapting maize to climate change in tropical environments.Clim. Risk Manag.19106–119. 10.1016/j.crm.2017.10.001
153
Thiebaut F. Hemerly A. S. Ferreira P. C. G. (2019). A role for epigenetic regulation in the adaptation and stress responses of non-model plants.Front. Plant Sci.10:246. 10.3389/fpls.2019.00246
154
Thirunavukkarasu N. Hossain F. Arora K. Sharma R. Shiriga K. Mittal S. et al (2014). Functional mechanisms of drought tolerance in subtropical maize (Zea mays L.) identified using genome-wide association mapping.BMC Genom.15:1182. 10.1186/1471-2164-15-1182
155
Thornsberry J. M. Goodman M. M. Doebley J. Kresovich S. Nielsen D. Buckler E. S. (2001). Dwarf8 polymorphisms associate with variation in flowering time.Nat. Genet.28286–289. 10.1038/90135
156
Tondelli A. Francia E. Barabaschi D. Aprile A. Skinner J. S. Stockinger E. J. et al (2006). Mapping regulatory genes as candidates for cold and drought stress tolerance in barley.Theor. Appl. Genet.112445–454. 10.1007/s00122-005-0144-7
157
Trachsel S. Sun D. SanVicente F. M. Zheng H. Atlin G. N. Suarez E. A. et al (2016). Identification of QTL for early vigor and stay-green conferring tolerance to drought in two connected advanced backcross populations in tropical maize (Zea mays L.).PLoS One11:e0149636. 10.1371/journal.pone.0149636
158
Tuberosa R. (2012). Phenotyping for drought tolerance of crops in the genomics era.Front. Physiol.3:347. 10.3389/fphys.2012.00347
159
Tuberosa R. Salvi S. (2006). Genomics-based approaches to improve drought tolerance of crops.Trends Plant Sci.11405–412. 10.1016/j.tplants.2006.06.003
160
Tuberosa R. Salvi S. Sanguineti M. C. Landi P. Maccaferri M. Conti S. (2002). Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize.Ann. Bot.89941–963. 10.1093/aob/mcf134
161
Valliyodan B. Nguyen H. T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants.Curr. Opin. Plant Biol.9189–195. 10.1016/j.pbi.2006.01.019
162
Vargas M. van Eeuwijk F. A. Crossa J. Ribaut J. M. (2006). Mapping QTLs and QTL x environment interaction for CIMMYT maize drought stress program using factorial regression and partial least squares methods.Theor. Appl. Genet.1121009–1023. 10.1007/s00122-005-0204-z
163
Verkest A. Byzova M. Martens C. Willems P. Verwulgen T. Slabbinck B. et al (2015). Selection for improved energy use efficiency and drought tolerance in canola results in distinct transcriptome and epigenome changes.Plant Physiol.1681338–1350. 10.1104/pp.15.00155
164
Verslues P. E. Agarwal M. Katiyar-Agarwal S. Zhu J. Zhu J.-K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status.Plant J.45523–539. 10.1111/j.1365-313X.2005.02593.x
165
Vinocur B. Altman A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations.Curr. Opin. Biotechnol.16123–132. 10.1016/j.copbio.2005.02.001
166
Vivek B. Krishna G. Vengadessan V. Babu R. Zaidi P. H. Mandal S. S. et al (2017). Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize.Plant Genom.101–8. 10.3835/plantgenome2016.07.0070
167
Wallace J. G. Zhang X. Beyene Y. Semagn K. Olsen M. Prasanna B. M. et al (2016). Genome-wide association for plant height and flowering time across 15 tropical maize populations under managed drought stress and well-watered conditions in sub-saharan africa.Crop Sci.562365–2378. 10.2135/cropsci2015.10.0632
168
Wang C. R. Yang A. F. Yue G. D. Gao Q. Yin H. Y. Zhang J. R. (2008). Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize.Planta2271127–1140. 10.1007/s00425-007-0686-9
169
Wang Q. Xu J. Pu X. Lv H. Liu Y. Ma H. et al (2021). Maize DNA methylation in response to drought stress is involved in target gene expression and alternative splicing.Int. J. Mol. Sci.2282–85. 10.3390/ijms22158285
170
Wang T. Zhang H. Zhu H. (2019). CRISPR technology is revolutionizing the improvement of tomato and other fruit crops.Hortic. Res.61–13. 10.1038/s41438-019-0159-x
171
Wang X. Wang H. Liu S. Ferjani A. Li J. Yan J. et al (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings.Nat. Genet.481233–1241. 10.1038/ng.3636
172
Wang X. Zenda T. Liu S. Liu G. Jin H. Dai L. et al (2019). Comparative proteomics and physiological analyses reveal important maize filling-kernel drought-responsive genes and metabolic pathways.Int. J. Mol. Sci.20:3743. 10.3390/ijms20153743
173
Wang Z. Li G. Sun H. Ma L. Guo Y. Zhao Z. et al (2018). Effects of drought stress on photosynthesis and photosynthetic electron transport chain in young apple tree leaves.Biol. Open7:bio035279. 10.1242/bio.035279
174
White J. W. Andrade-Sanchez P. Gore M. A. (2012). Field-based phenomics for plant genetics research.Field Crop Res.133101–112.
175
Witcombe J. R. Joshi A. Goyal S. N. (2003). Participatory plant breeding in maize: a case study from Gujarat, India.Euphytica130413–422.
176
Wojtyla Ł Paluch-Lubawa E. Sobieszczuk-Nowicka E. Garnczarska M. (2020). “Drought stress memory and subsequent drought stress tolerance in plants,” in Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants, edsHossainM. A.LiuF.BurrittD. J.FujitaM.HuangB. (Amsterdam: Elsevier), 115–131. 10.1016/b978-0-12-817892-8.00007-6
177
World Bank Group (2021). Available online at:https://moderndiplomacy.eu/2021/06/04/to-slow-himalayan-glacier-melt-curbing-air-pollution-is-key/(accessed January 25, 2022).
178
Wu S. Ning F. Zhang Q. Wu X. Wang W. (2017). Enhancing omics research of crop responses to drought under field conditions.Front. Plant Sci.8:174. 10.3389/fpls.2017.00174
179
Xia Z. Liu Q. Wu J. Ding J. (2012). ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize.Gene495146–153. 10.1016/j.gene.2011.12.028
180
Xue Y. Warburton M. L. Sawkins M. Zhang X. Setter T. Xu Y. et al (2013). Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions.Theor. Appl. Genet.1262587–2596. 10.1007/s00122-013-2158-x
181
Yadava P. Abhishek A. Singh R. Singh I. Kaul T. Pattanayak A. et al (2017). Advances in maize transformation technologies and development of transgenic maize.Front. Plant Sci.7:1949. 10.3389/fpls.2016.01949
182
Yang W. Feng H. Zhang X. Zhang J. Doonan J. H. Batchelor W. D. et al (2020). Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives.Mol. Plant13187–214. 10.1016/j.molp.2020.01.008
183
Younis A. Ramzan F. Ramzan Y. Zulfiqar F. Ahsan M. Lim K. B. (2020). Molecular markers improve abiotic stress tolerance in crops: a review.Plants9:1374. 10.3390/plants9101374
184
Yu D. Ke L. Zhang D. Wu Y. Sun Y. Mei J. et al (2021). Multi-omics assisted identification of the key and species-specific regulatory components of drought-tolerant mechanisms in Gossypium stocksii.Plant Biotechnol. J.19:1690. 10.1111/pbi.13655
185
Yu H. Qu J. Guo X. Li L. Zhang X. Yang Q. et al (2021). Overexpression of vacuolar H+-pyrophosphatase (H+-PPase) gene from Ammopiptanthusnanus enhances drought tolerance in maize.J. Agron. Crop Sci.[Epub ahead of print].
186
Yuan Y. Cairns J. E. Babu R. Gowda M. Makumbi D. Magorokosho C. et al (2019). Genome-wide association mapping and genomic prediction analyses reveal the genetic architecture of grain yield and flowering time under drought and heat stress conditions in maize.Front. Plant Sci.9:1919. 10.3389/fpls.2018.01919
187
Zenda T. Liu S. Wang X. Jin H. Liu G. Duan H. (2018). Comparative proteomic and physiological analyses of two divergent maize inbred lines provide more insights into drought-stress tolerance mechanisms.Int. J. Mol. Sci.19:3225. 10.3390/ijms19103225
188
Zenda T. Liu S. Wang X. Liu G. Jin H. Dong A. et al (2019). Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting inbred lines.Int. J. Mol. Sci.20:1268. 10.3390/ijms20061268
189
Zeng W. Peng Y. Zhao X. Wu B. Chen F. Ren B. et al (2019). Comparative proteomics analysis of the seedling root response of drought-sensitive and drought-tolerant maize varieties to drought stress.Int. J. Mol. Sci.20:2793. 10.3390/ijms20112793
190
Zhang F. Wu J. Sade N. Wu S. Egbaria A. Fernie A. R. et al (2021). Genomic basis underlying the metabolome-mediated drought adaptation of maize.Genom. Biol.221–26. 10.1186/s13059-021-02481-1
191
Zhang H. Lang Z. Zhu J.-K. (2018). Dynamics and function of DNA methylation in plants.Nat. Rev. Mol. Cell Biol.19489–506. 10.1038/s41580-018-0016-z
192
Zhang S. Li N. Gao F. Yang A. Zhang J. (2010). Over-expression of TsCBF1 gene confers improved drought tolerance in transgenic maize.Mol. Breed.26455–465. 10.1007/s11032-009-9385-5
193
Zhang X. Mi Y. Mao H. Liu S. Chen L. Qin F. (2020). Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize.Plant Biotechnol. J.181271–1283. 10.1111/pbi.13290
194
Zhang X. Warburton M. L. Setter T. Liu H. Xue Y. Yang N. et al (2016). Genome-wide association studies of drought-related metabolic changes in maize using an enlarged SNP panel.Theor. Appl. Genet.1291449–1463. 10.1007/s00122-016-2716-0
195
Zhang Y. Liang Z. Zong Y. Wang Y. Liu J. Chen K. et al (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA.Nat. Commun.71–8. 10.1038/ncomms12617
196
Zhao C. X. Guo L. Y. Jaleel C. A. Shao H. B. Yang H. B. (2008). Prospective for applying molecular and genetic methodology to improve wheat cultivars in drought environments.C. R. Biol.331579–586. 10.1016/j.crvi.2008.05.006
197
Zhao F. Zhang D. Zhao Y. Wang W. Yang H. Tai F. et al (2016). The difference of physiological and proteomic changes in maize leaves adaptation to drought, heat, and combined both stresses.Front. Plant Sci.7:1471. 10.3389/fpls.2016.01471
198
Zhao X. Peng Y. Zhang J. Fang P. Wu B. (2018). Identification of QTLs and Meta-QTLs for seven agronomic traits in multiple maize populations under well-watered and water-stressed conditions.Crop Sci.58507–520. 10.2135/cropsci2016.12.0991
199
Zheng H. Yang Z. Wang W. Guo S. Li Z. Liu K. et al (2020). Transcriptome analysis of maize inbred lines differing in drought tolerance provides novel insights into the molecular mechanisms of drought responses in roots.Plant Physiol. Biochem.14911–26. 10.1016/j.plaphy.2020.01.027
200
Zhu J. J. Wang X. P. Sun C. X. Zhu X. M. Meng L. I. Zhang G. D. et al (2011). Mapping of QTL associated with drought tolerance in a semi-automobile rain shelter in maize (Zea mays L.).Agric. Sci. China10987–996.
201
Zhu Z. Zhang F. Hu H. Bakshi A. Robinson M. R. Powell J. E. et al (2016). Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets.Nat. Genet.48481–487. 10.1038/ng.3538
202
Ziyomo C. Bernardo R. (2013). Drought tolerance in maize: indirect selection through secondary traits versus genome wide selection.Crop Sci.531269–1275. 10.2135/cropsci2012.11.0651
Summary
Keywords
drought, genome editing, high-throughput phenotyping, maize, omics, QTL mapping
Citation
Sheoran S, Kaur Y, Kumar S, Shukla S, Rakshit S and Kumar R (2022) Recent Advances for Drought Stress Tolerance in Maize (Zea mays L.): Present Status and Future Prospects. Front. Plant Sci. 13:872566. doi: 10.3389/fpls.2022.872566
Received
09 February 2022
Accepted
26 April 2022
Published
30 May 2022
Volume
13 - 2022
Edited by
Amol N. Nankar, Center of Plant Systems Biology and Biotechnology, Bulgaria
Reviewed by
Olubukola Oluranti Babalola, North-West University, South Africa; Blanca E. Barrera-Figueroa, Universidad del Papaloapan, Mexico
Updates
Copyright
© 2022 Sheoran, Kaur, Kumar, Shukla, Rakshit and Kumar.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramesh Kumar, rk.phagna@gmail.com
†Present address: Seema Sheoran, Indian Council of Agricultural Research-Indian Agricultural Research Institute, Regional Station, Karnal, India
This article was submitted to Technical Advances in Plant Science, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.