- 1Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
- 2Division of Microbial Technology, Biotechnology Center of Ho Chi Minh City, Ho Chi Minh City, Vietnam
- 3Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
- 4College of Education, Ibaraki University, Mito, Japan
- 5Sugadaira Research Station, Mountain Science Center, University of Tsukuba, Ueda, Japan
This research focused on the incidence and population genetics of coffee leaf rust (CLR) fungus, Hemileia vastatrix, to estimate the possible original source(s) and subsequent migration pathways of wind-borne and human-aided spores in three main coffee production regions (Northwest, Central Highlands, and Southeast) in Vietnam. In southern Vietnam (Central Highlands and Southeast), Coffea canephora covers the majority area, while Catimor lines of C. arabica accounts for 95% of the coffee plantations in northwestern Vietnam. Field surveys conducted at eighty-five plantations, show coffee leaf samples infected by the rust fungus across forty-one plantations. Catimor varieties exhibited high levels of susceptibility with severe rust symptoms, while robusta varieties had varying degrees of susceptibility. We analyzed 863−869 base pairs of internal transcribed spacer (ITS) region from 83 samples (41 sequences from Vietnam, 2 from Thailand, and the remaining 40 from American countries); and fifty-two haplotypes consisting of 123 polymorphic sites were detected. Although the analysis of molecular variance (AMOVA) indicates significant genetic differentiation in the H. vastatrix populations in Vietnam, there was no clear genetic structure with respect to the three geographic areas surveyed. Based on the haplotype network, NeighborNet analysis, and geographical distribution patterns of the haplotypes, five haplotypes were identified as early established, from which most other haplotypes in Vietnam were derived. The early established haplotypes were found in the highest frequency in Northwest Vietnam. This finding corresponds to the earliest record of CLR in Vietnam. The phylogenetic network analysis also illustrated that H. vastatrix had expanded from the northwest to southern Vietnam. Pairwise genetic distance analysis and the geophylogenetic tree also suggests that CLR was first established in the Northwest. In addition, some scattered individuals on the principal coordinate analysis (PCoA) diagram and several separated haplotypes in the phylogenetic networks indicated that other branches of CLR in Vietnam were initiated in the Central Highlands. Hemileia vastatrix from these branches have been spreading in southern Vietnam.
Introduction
The rust fungi belong to the order Pucciniales (Class: Pucciniomycetes, Division: Basidiomycota). They are obligate parasites of vascular plants and cause rust diseases in several important crops worldwide (Kolmer et al., 2009). Management of rust disease is difficult because the life cycle of most rust fungi is complicated, with up to five different spore stages, including basidiospore, spermatium, aeciospore, urediniospore, and teliospore, which may occur on the same host (autoecious life cycle) or on two taxonomically unrelated hosts (heteroecious life cycle) (Cummins and Hiratsuka, 2003; Kolmer et al., 2009). Among them, Hemileia (Zaghouaniaceae P. Syd. & Syd.) (Aime and McTaggart, 2020) is characterized by its unique uredinial anamorph (Ritschel, 2005) and comprises 42 species, found mainly in plants of Rubiaceae and Apocynaceae families. Among these plants, species of Coffea (Rubiaceae) produce coffee beans—the most valuable and traded tropical agricultural product, which is severely affected by the rust disease (Talhinhas et al., 2017). Even if coffee trees do not die from early defoliation by heavy rust infections, the fungal infection will cause a severe decrease in berry yield and quality. This disease in coffee trees causes a rapid decline in the trees’ vigor and makes them more vulnerable to other pathogenic fungi and pests in successive years. CLR has been recorded to cause approximately 35% yield losses, resulting in annual losses of one to two billion US dollars (Talhinhas et al., 2017). Therefore, it is essential to develop effective and practical measures to manage the CLR disease.
The best strategy to control this disease is by using resistant varieties (Silva et al., 2006). Hibrido de Timor (HdT) is a natural hybrid between Coffea arabica and C. canephora, which was resistant to all races of the coffee rust fungus, Hemileia vastatrix, in the 1950s. The hybrids Catimor and Sachimor have also been considered resistant to H. vastatrix (Talhinhas et al., 2017). Planting disease-resistant coffee varieties was considered an effective strategy for preventing or decreasing the infection of H. vastatrix for a long time. However, in recent years, there have been reports of the outbreak of CLR in most coffee-growing areas worldwide, including regions planting rust-resistant varieties (Maia et al., 2013; Cabral et al., 2016; Talhinhas et al., 2017). Thus, to develop effective and practical management programs, we must know the current pathogenic diversity of the rust populations, understand the underlying genetic variations, and predict the tendency for variations in future.
Currently, H. vastatrix and H. coffeicola Maubl. and Roger are the known causative agents of the CLR disease (Thirumalachar and Narasimhan, 1947; Ritschel, 2005; Talhinhas et al., 2017). Hemileia vastatrix is the major agent causing substantial yield loss in worldwide coffee production. On the contrary, H. coffeicola occurs only in West and Central Africa on C. canephora (Ritschel, 2005). Previously, the global outbreaks of CLR have always been related to H. vastatrix. The native range of the fungus is suggested to be a geographic range spanning the Great Lake region, Ethiopia, and the eastern half of the Congo River basin in East Africa (McCook, 2019). After the domestication of C. arabica in Yemen in the fifteenth century, the fungus and its cultivated host had spread through Ceylon and southern India in 1869. The reasons for the first explosion of CLR were monsoon, shipment of infected coffee seedlings or goods, and the attached rust spores on the clothes of British soldiers returning from Ethiopia. Thereafter, the CLR pandemic exploded in the Indian Ocean basin and the Pacific from 1875 to 1920. Besides other routes related to wind, trade, and communication, the migration of coffee planters from infected areas to the Indian Ocean basin and the Pacific could also be considered the causal factor. Wind, infected materials, and anthropogenic activities also explain the CLR outburst in West Africa during 1950−1970 and in America during 1970−1989 (McCook, 2006, 2019).
Coffea arabica was first cultivated in northern Vietnam in 1857 by French missionaries (D’haeze et al., 2004; Anonymous, 2019). Some decades after that, in October 1890, H. vastatrix was first collected on C. arabica in some plantations (Hennings, 1895). Several years later, C. arabica spread to Southern Vietnam. However, when diseases, including coffee rust, caused yield losses in the coffee plantations, C. canephora was introduced to replace C. arabica, and it rapidly spread in the southern parts of Vietnam in the early 1900s (D’haeze et al., 2003). In the meantime, the Northwest of Vietnam also switched to Catimor derivatives due to its ability to resist H. vastatrix (Anonymous, 2019). After World War II, the introduction of C. canephora cultivars enabled Vietnam to become the world’s second-largest coffee producer, after Brazil. There are three main coffee-growing regions in Vietnam—the Southeast, the Central highlands, and the Northwest. Topographic and climatic differences affect seasonal disease occurrences and their severity in the three major coffee-growing regions. Annually, the disease begins from March to April and from July to September in the northern part of Vietnam. By contrast, the rust disease symptoms become apparent in the wet season between April and September in the southern part. CLR has been reported periodically in Vietnam. However, the actual occurrence and distribution of the disease in Vietnam and the origin of CLR have not been investigated. Therefore, the objectives of this research were to (i) investigate the occurrence of CLR in the main coffee-producing regions in Vietnam, (ii) evaluate the current genetic diversity and population structure of the coffee rust fungus based on sequencing of internal transcribed spacer (ITS) region, and (iii) estimate the geographic region where H. vastatrix first established as well as the migration direction of CLR between main coffee-growing areas in Vietnam.
Materials and Methods
Field Investigation and Sample Collection
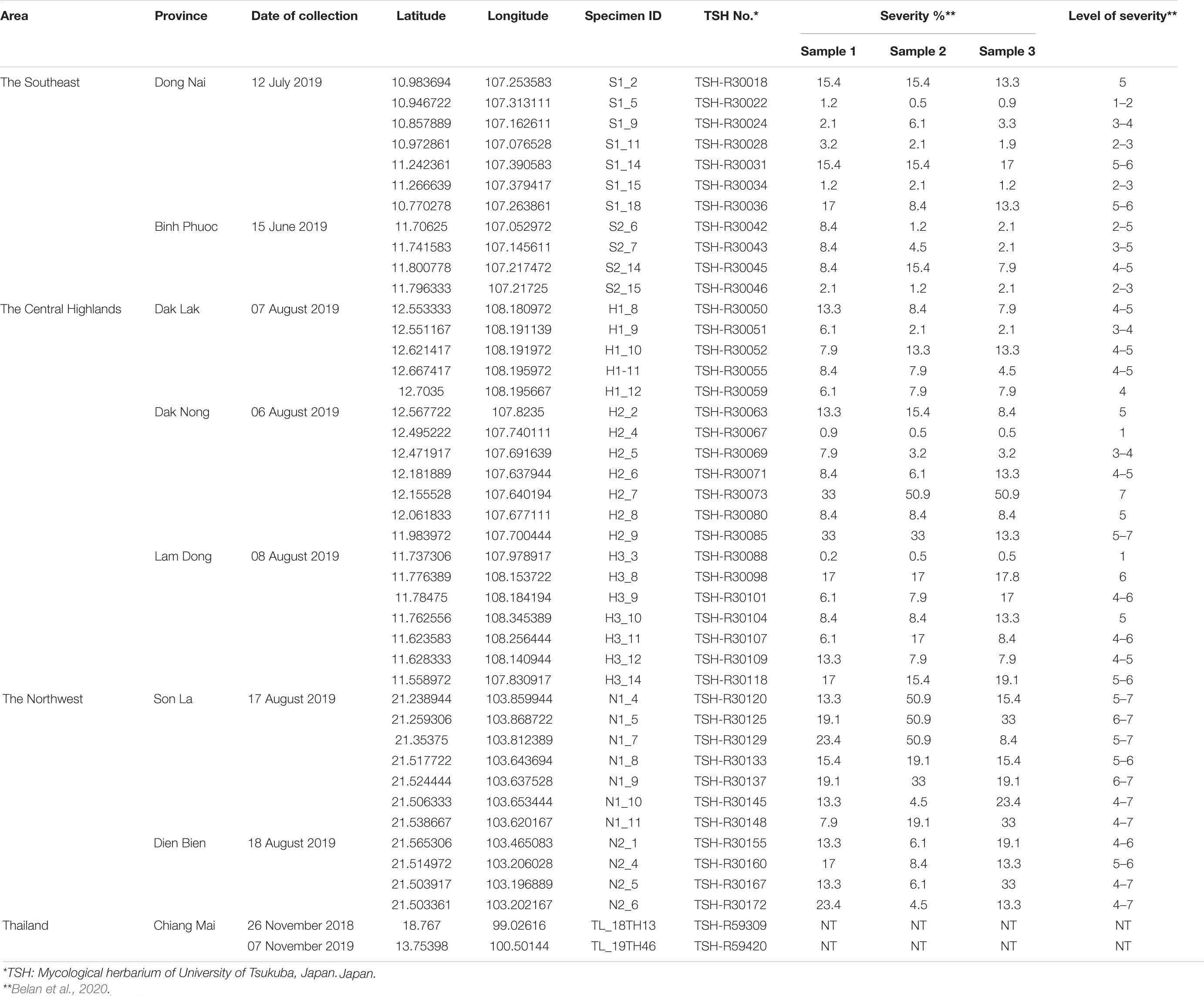
Coffee species were identified based on morphology (flower, bract, apex, leaf, fruit, etc.), according to the descriptions of Backer and Bakhuizen Van Den Brink (1965). Three coffee regions, including the Southeast (Dong Nai and Binh Phuoc provinces), the Central Highlands (Dak Lak, Dak Nong, and Lam Dong provinces), and the Northwest (Son La and Dien Bien provinces), were investigated for the occurrence of CLR. The accessed plantations were along the street and were more than 1 km apart (Supplementary Figure S1). In each plantation, two rows of trees around the edges were examined for H. vastatrix. Ten coffee leaves with orange-colored rust sori were collected from three trees at each collection site, packed in a plastic bag, and referred to as one sample. Photographs of the rust-infected leaves were also taken, and the severity of the coffee leaf rust estimated using the method of Belan et al. (2020). The leaves were then dried by pressing them between dry blotting papers and maintained at room temperature (25 ± 2°C) at the microbiology laboratory of the Biotechnology Center of Ho Chi Minh City. Dried specimens were then deposited in the Mycological Herbarium of the University of Tsukuba (TSH), Japan. In addition to the Vietnamese samples, two coffee leaf specimens from Thailand, deposited in TSH (TSH-R59309 and TSH-R59420), were also used for this research. All samples were collected between July and November in 2019, except for one sample collected in November 2018 (Table 1).
DNA Extraction, Internal Transcribed Spacer Region Amplification, and Sequencing
DNA extraction from herbarium specimens requires special methods, in which only a small amount of spores must be picked to avoid possible cross-contamination. The thermal-shock method given by Fraczek et al. (2019) was used for DNA extraction, and BPS buffer was substituted with buffer-1 (10 mM Tris-Cl pH 8.3, 1.5 mM MgCl2, 50 mM KCl). The protocol can be summarized as follows: A coffee leaf with fully matured, seemingly clean sori on the lower surface was chosen for DNA extraction from each sample (one sample consisted of ten leaves from three different trees). Urediniospores were collected from several pustules on a single leaf and immersed in a 200 μl tube containing 30 μl buffer-1. The tubes were then vortexed and incubated at 95°C for 15 min and immediately transferred to a deep freezer (–80°C) for 10 min. Subsequently, the spore suspension was defrosted at room temperature (25 ± 2°C) and centrifugated at 15,000 rpm for 1 min. Finally, the supernatant was transferred to a new 200 μl tube. The nucleic acid concentration and 260/280 and 260/230 ratios were estimated using a DS-11 spectrophotometer (DeNovix, Wilmington, DE, United States).
The ITS region was amplified using ITS5 and ITS4 primers (White et al., 1990). A 25 μl reaction mixture containing 1–2 μl DNA template (30−50 ng/μl), 0.2 mM of each primer (2.5 μl), 12.5 μl Gene RED PCR Mix Plus (Nippon Gene, Tokyo, Japan), and 5.5–6.5 μl autoclaved distilled water, were prepared. Polymerase chain reaction (PCR) was conducted using a TaKaRa PCR Thermal Cycler Dice® Touch (TaKaRa, Tokyo, Japan). The PCR process was as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 54°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min.
The amplification was confirmed using electrophoresis. The amplified products were purified using FastGene™ Gel/PCR extraction kit (Nippon Genetics, Tokyo, Japan) and consigned to Eurofins Genomics (Tokyo, Japan) for sequencing. The obtained sequences were edited and assembled using ATGC software (Genetyx Co., Tokyo, Japan). Finally, the sequences were aligned and compared with other sequences in the National Center for Biotechnology Information database using the BLASTn program1.
Population Genetic Analysis
Besides sequences from Vietnam and two sequences from Thailand, 40 ITS sequences of H. vastatrix from a few Central and South American countries (Mexico, Brazil, and Colombia), and those from Portugal’s Coffee Rust Research Centre (CIFC: Centro de Investigação de Ferrugens do Cafeeiro) were downloaded from the NCBI Genbank (Supplementary Table S1) and added to the data, for identifying the original source(s) of CLR in Vietnam. Rust fungus samples from the same coffee-growing area (province or city) in different coffee-growing regions (Northwest, Central Highlands, and Southeast in Vietnam, and Thailand) were designated as a single population. Rust samples represented by ITS sequences downloaded from GenBank were treated similarly. Totally, 20 populations of H. vastatrix were examined in this study, including Southeast-Dong Nai, Southeast-Binh Phuoc, Central Highlands-Dak Lak, Central Highlands-Dak Nong, Central Highlands-Lam Dong, Northwest-Son La, and Northwest-Dien Bien populations from Vietnam; Chiang Mai population from Thailand; Veracruz, Chiapas, Oaxaca, and Puebla populations from Mexico; Minas Gerais-Coimbra, Minas Gerais-Capinopolis, Minas Gerais-São Sebastião do Paraíso, Minas Gerais-Senhora de Oliveira, and Espírito Santo-Venda Nova do Imigrante populations from Brazil; Quindío-Buenavista and Caldas-Chinchiná populations from Colombia; and the CIFC population from Portugal.
The obtained sequences were aligned using Clustal X version 2.0 (Larkin et al., 2007) (Supplementary Material S4). The number of haplotypes, haplotype diversity (Hd) (Nei and Li, 1979), and nucleotide diversity (Pi) of each population were calculated using the Dnasp program version 6 (Librado and Rozas, 2009). To visualize the correlation between nucleotide diversity and geographical pattern, the regression statistics between nucleotide diversity and latitude was analyzed using PAST v4.03 (Hammer et al., 2001), and Microsoft Excel was used to generate a scatter diagram. The demographic pattern in each population and in the whole H. vastatrix population was evaluated by conducting Tajima’s neutrality test (Tajima, 1989) using Arlequin version 3.5 (Excoffier and Lischer, 2010).
To evaluate the genetic structure of H. vastatrix populations, analyses of molecular variance (AMOVA) within each population and among different populations were calculated using GenAlEx 6.51b2 (Peakall and Smouse, 2012). Next, the matrices of pairwise genetic distance (ΦPT) (Weir and Cockerham, 1984) between populations and individuals based on the AMOVA were calculated using GenAlEx 6.51b2. These matrices were used as input data for principal coordinate analysis (PCoA) to visualize the relationships between populations and the genetic structure of the whole H. vastatrix population.
To evaluate the genetic relationship among the haplotypes and to predict the ancestors of the populations in Vietnam, we employed parsimony and NeighborNet network approaches (Bryant and Moulton, 2013). Firstly, Dnasp program version 6 was used to generate a raw nexus file that was manually prepared to form an input trait file for PopART software version 1.7 (Leigh and Bryant, 2015). Next, a Median-joining haplotype network was constructed using PopART version 1.7. Meanwhile, the NeighborNet network (Bryant and Moulton, 2013) was generated using Splitstree 5 software (Huson and Bryant, 2006). Moreover, a map-based phylogenetic tree was generated to estimate the migration routes and the influence of geographic patterns on the similarities or differences among the H. vastatrix populations. First, a map figure was generated using GeoMapApp 3.6.122 (Ryan et al., 2009). Next, a phylogenetic tree of the H. vastatrix populations was built using the program MEGA X (Kumar et al., 2018), based on the maximum likelihood statistical method with 1000 bootstrap replications, Generalized time-reversible model (Tavaré, 1986) with the rate among sites Gamma distributed with invariant sites (G + I) was used. Lastly, a 3D graphical interface of the phylogeographic tree was generated by combining data from the map, phylogenic tree, and locations of collected samples using GenGIS2 software (Parks et al., 2013).
Results
Field Investigation
Field investigations showed that C. canephora is commonly planted in southern Vietnam, while C. arabica cv. Catimor is planted in the Northwest (Table 1 and Supplementary Table S2). In the Southeast of Vietnam, thirty-three plantations in Dong Nai and Binh Phuoc provinces were investigated, of which 26.7% (4/15) plantations in Binh Phuoc and 38.9% (7/18) plantations in Dong Nai were infected by the rust fungus. Simultaneously, thirty-five plantations in the Central Highlands were investigated. The incidence of coffee rust disease in the three provinces was variable, i.e., 41.7% (5/12) in Dak Lak, 77.8% (7/9) in Dak Nong, and 50.0% (7/14) in Lam Dong province. Our investigation at 17 plantations in the mountainous area of North Vietnam (Northwest) revealed that coffee rust disease incidence in this region was high—63.6% (7/11) and 66.7% (4/6) in Son La and Dien Bien provinces, respectively. After thorough field investigations at 85 coffee plantations in Vietnam, a total of 41 CLR samples were collected from 41 plantations in the three regions (Table 1, Figure 1, Supplementary Figure S1, and Supplementary Table S2).
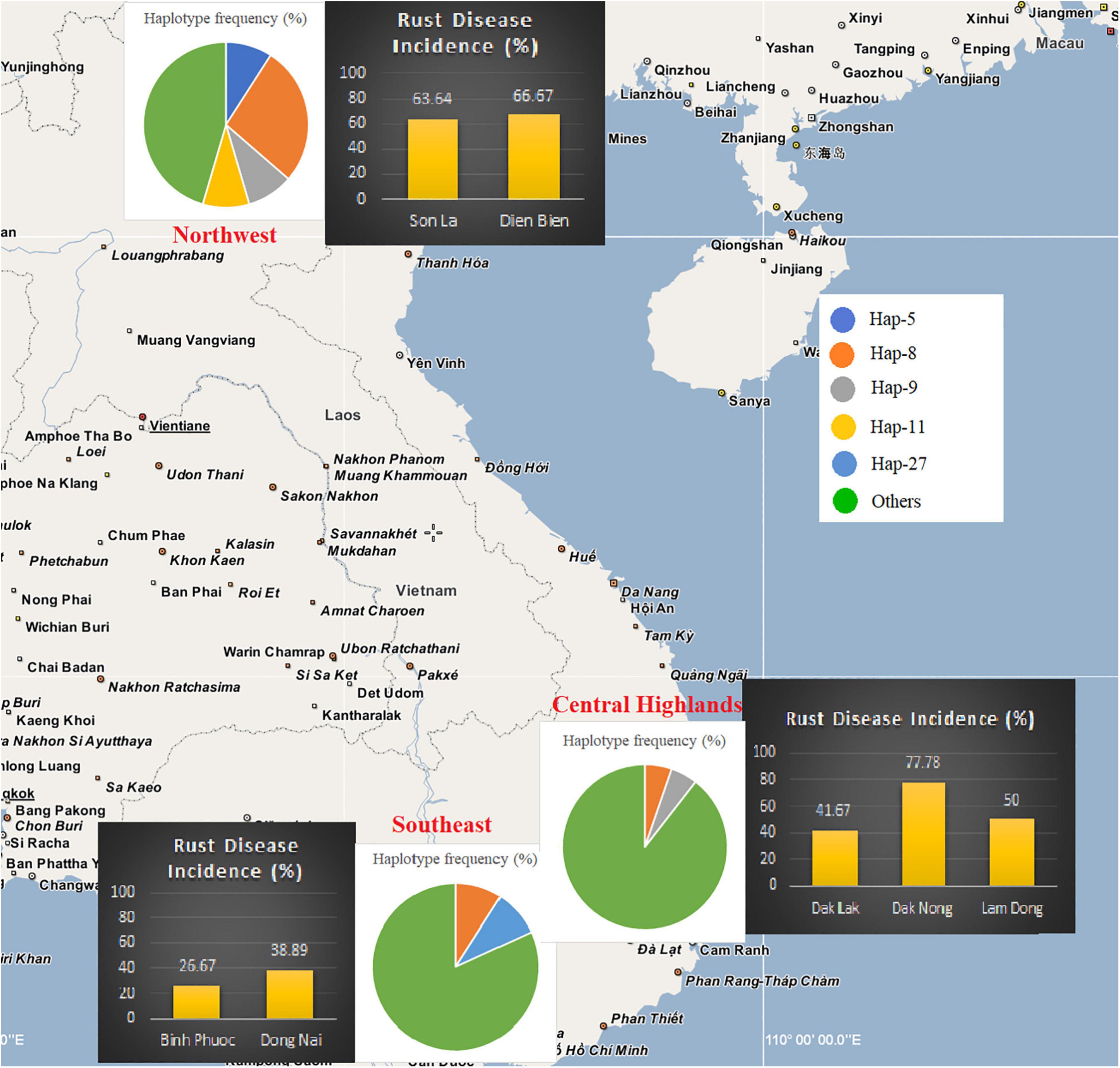
Figure 1. Rust disease incidence and the frequency of ancestral Hemileia vastatrix haplotypes in different geographical sample areas of Vietnam. Disease incidence in the areas of each province is indicated using bar charts and Hemileia vastatrix haplotype frequency in the sampling areas is shown using pie charts.
The severity of coffee leaf rust on C. arabica in the Northwest and C. canephora in the Central Highlands and Southeast of Vietnam was assessed using individual survey sites and the method described by Belan et al. (2020) (Supplementary Figure S2 and Table 1). Overall, the severity of the disease in Vietnam ranged from 1–7. Coffee plantations in Son La and Dien Bien provinces (Northwest) had the highest rust severity (up to level 7). While in southern Vietnam the disease appeared less severe than that in northern Vietnam, except at two plantations in Dak Nong province (Central Highlands); however, these two plantations had been abandoned many years prior to our sampling. All plantations in Lam Dong province had moderate rust disease severity (levels 4-6) except plantation 3 (level 1) which is in a high mountainous area. The severity of rust disease in Southeast Vietnam was found to be lower than in the other areas, except for plantations 14 and 18 in Dong Nai province (levels 5-6) where coffee is intercropped with black pepper. Arabica varieties at all sites in the Northwest exhibited high susceptibility as the number and size of the rust lesions was greater when compared with other regions, while robusta varieties in the Central Highlands and the Southeast exhibited varying degrees of susceptibility. It was noticeable that those in Dak Nong and Dong Nai were highly variable among survey sites, ranging from highly resistant to highly susceptible (Table 1 and Supplementary Figure S2).
Population Genetic Analysis of Hemileia vastatrix
The DNA sequences of the ITS region from all samples were 863−869 bp and had 98–100% similarity with H. vastatrix (Supplementary Table S3) (Genbank accession numbers: LC682363−LC682405). From the 86 sequences (41 sequences from Vietnam), 123 polymorphic (segregating) sites (77 singleton variable sites and 46 parsimony informative sites) were detected. In total, 52 haplotypes (36 haplotypes from Vietnam) were identified (Table 2), with 0.952 ± 0.00022 Hd and 0.00779 nucleotide diversity (Pi). The nucleotide diversity was highest (0.02651) in the Southeast (Binh Phuoc population), and the genetic variation was absent in the Veracruz (Mexico) population. Populations from Dak Nong (Central Highlands), Lam Dong (Central Highlands), and Dong Nai (Southeast) in Vietnam, and Chiang Mai in Thailand had higher diversity (0.02527, 0.01452, 0.01175, and 0.01153, respectively) (Supplementary Table S5) than the remaining populations. We detected a significant negative correlation between latitude and nucleotide diversity when we focused on the populations in Vietnam and Thailand (p < 0.05). The value of Tajima’s neutrality test (D) for the whole population of H. vastatrix was −0.19518. Although almost all populations in Vietnam had a negative value of Tajima’s D (except for the Central Highlands-Lam Dong population); however, the values were not significant (Supplementary Table S6).

Table 2. List of haplotypes obtained from sequences of coffee leaf rust fungus in Vietnam, Thailand, Portugal, and some American countries (Brazil, Colombia, Mexico).
Haplotype diversity of most of the populations was high (> 0.900) and did not show any geographic patterns, except the Caldas-Chinchiná population from Colombia, which showed a much lower value (0.500) than others (Supplementary Table S5). When we focused on the haplotype distribution, the most common and major haplotype was Hap 8 (18.1%; detected in all populations except for the Thailand population), followed by Haps 17 [10.8%; Mexico (Veracruz, Chiapas, and Puebla), CIFC-Portugal, Brazil (Minas Gerais-São Sebastião do Paraíso)], Hap 32 [8.4%; Colombia and Brazil (Minas Gerais-Coimbra)], and three haplotypes, namely, Hap 9 (the Northwest and the Central Highlands of Vietnam), Hap 24 [Mexico (Chiapas and Oaxaca)], and Hap 27 (Colombia and the Southeast of Vietnam). The remaining haplotypes were singleton (Table 2 and Figure 2).
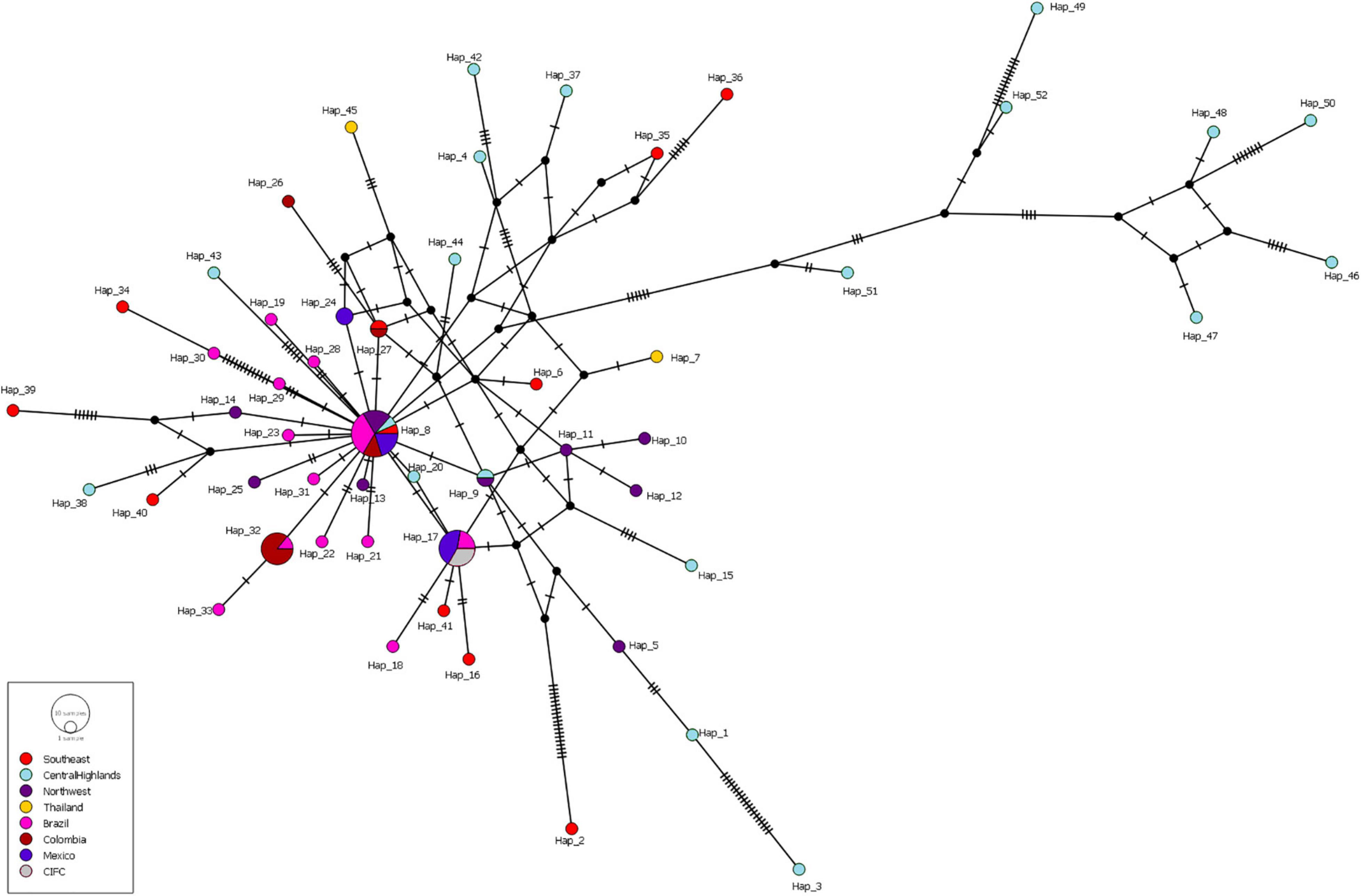
Figure 2. Median-joining haplotype network inferred from the ITS regions of Hemileia vastatrix sampled from Vietnam, Thailand, Portugal, and other countries in America (Brazil, Colombia, Mexico). Circles represent haplotypes, with bright red corresponding to samples collected from Southeast Vietnam, light turquoise the Central Highlands of Vietnam, purple the Northwest of Vietnam, while yellow indicates Thailand, pink Brazil, brick red Colombia, dark blue Mexico, and grey CIFC (Portugal). The size of the circles is proportional to the number of sequences in each haplotype, one transverse line represents simple mutations, and black nodes are non-sampled haplotypes.
The AMOVA showed that genetic differentiation among the twenty H. vastatrix populations from the six countries (Vietnam, Thailand, Brazil, Colombia, Mexico, and Portugal) was low (FST = 0.175) but significant (p < 0.01). Only 17% of the total variation was found among populations, while 83% of the differences were among individuals within a population (Table 3). The genetic differentiation (ΦPT) between the Vietnamese–Thai and Central and South American populations, and the Brazilian and Caldas-Chinchiná population (Colombia) was high (Supplementary Table S7). Meanwhile, the genetic differentiation between the Mexican and Vietnamese–Thai populations was lower than that observed between other populations (Supplementary Table S7). The PCoA chart indicated that the Vietnamese–Thai and the Mexican populations (Puebla, Chiapas, Veracruz, and Oaxaca) had closer genetic distance than combinations with the Brazilian–Colombian populations (Supplementary Figure S3).
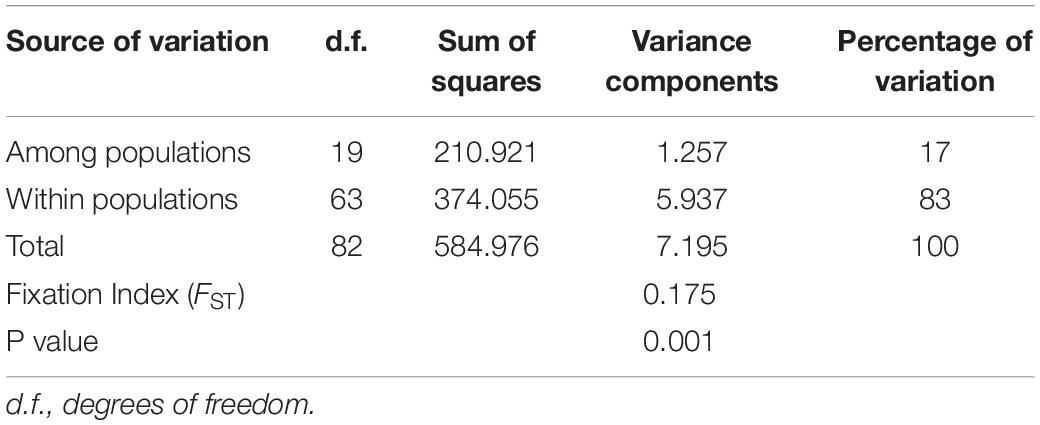
Table 3. Analysis of molecular variance (AMOVA) based on ITS sequences of the Hemileia vastatrix populations.
Analysis of the genetic structure of the whole H. vastatrix population (by studying the genetic distance between individuals) revealed that most individuals were closely gathered, while some from the Central Highlands and Southeast were distantly scattered (TSH-R30034, 30045, 30046, 30051, 30067, 30071, 30080, 30085, 30088, 30101, and 30118) (Figure 3). Haplotype (Figure 2) and NeighborNet (Figure 4) networks showed three groups in the H. vastatrix populations. The first group included Haps 13, 14, 19, 20, 21, 22, 23, 25, 28, 29, 30, 31, 32, 33, and 34, which were derived from Hap 8, and were mostly from Central and South American countries and the Northwest of Vietnam. There were 19 haplotypes in group 2, namely, Haps 1, 2, 3, 4, 5, 6, 7, 9, 10, 11, 12, 15, 16, 17, 18, 38, 40, 41, and 45 from Vietnam, Thailand, Brazil, and CIFC-Portugal. The remaining haplotypes (Haps 26, 27, 35, 36, 37, 39, 42, 43, 44, 46, 47, 48, 49, 50, 51, and 52) were included in the third group and were mainly from the Central Highlands and the Southeast of Vietnam.
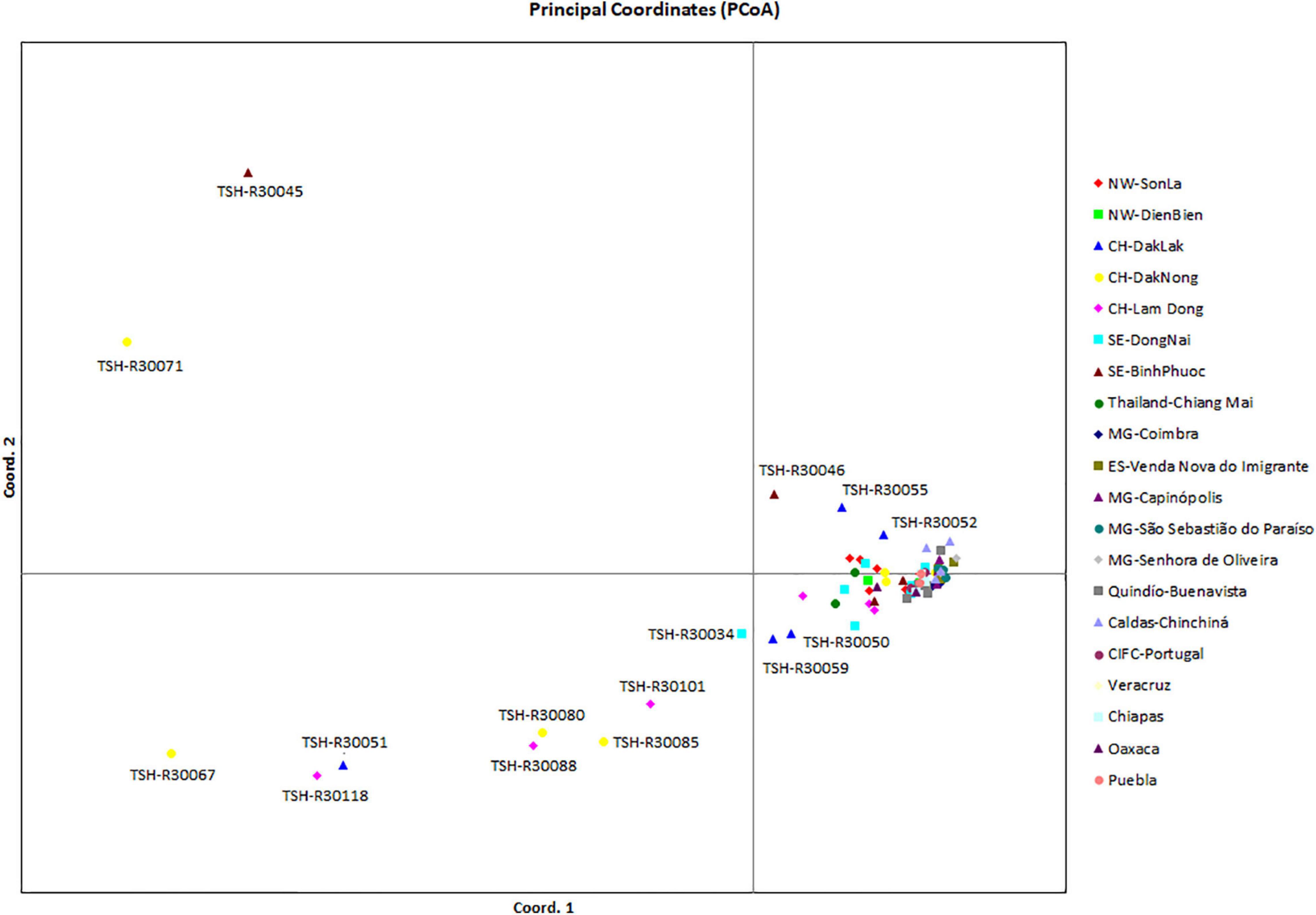
Figure 3. Principal coordinate analysis (PCoA) of the genetic (nucleotide) diversity of Hemileia vastatrix in Vietnam, Thailand, Portugal, and some countries in the Americas (Brazil, Colombia, Mexico). Individuals that are closer together have smaller genetic distances. NW: Northwest (Vietnam), CH: Central Highands (Vietnam), ST: Southeast (Vietnam), CIFC: Centro de Investigação de Ferrugens do Cafeeiro (Portugal), MG: Minas Gerais (Brazil), ES: Espírito Santo (Brazil).
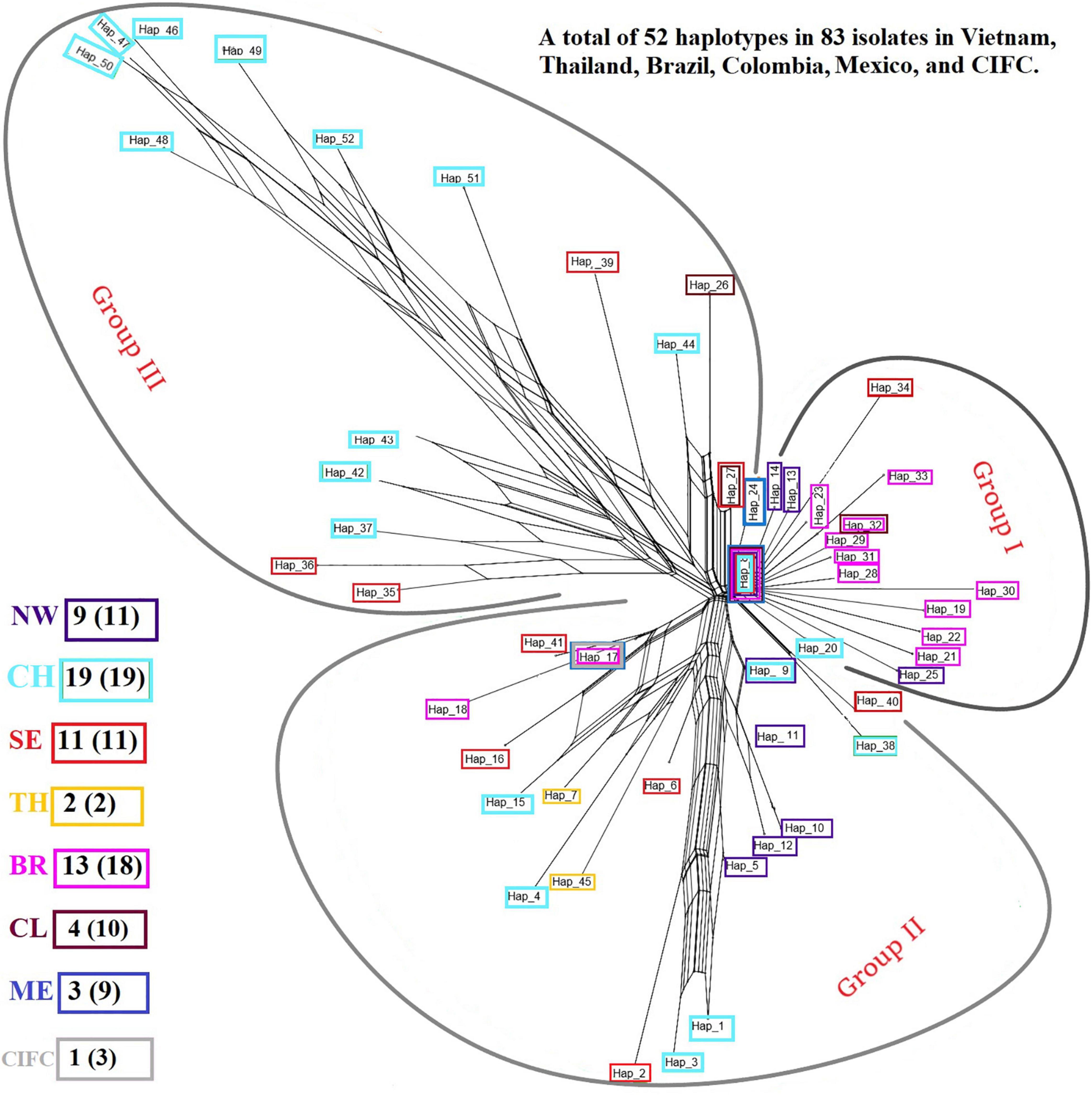
Figure 4. NeighborNet graph constructed from the pairwise FST values of Hemileia vastatrix samples from Vietnam, Thailand, Portugal, and other American countries (Brazil, Colombia, Mexico) based on their ITS regions. The haplotypes detected in seven areas are framed in boxes with different colors: purple indicates the Northwest (NW) of Vietnam, light turquoise the Central Highlands (CH) of Vietnam, bright red the Southeast (SE) of Vietnam, while yellow indicates Thailand (TH), pink Brazil (BR), brick red Colombia (CL), blue Mexico (ME), and grey the Centro de Investigação de Ferrugens do Cafeeiro (CIFC)-Portugal. After the abbreviation of each region the number of haplotypes detected in each area is stated, and the number in the parentheses indicates the number of sequences obtained from the region.
The map-based phylogenetic tree highlighted the close genetic relationship of some populations in Mexico and Brazil with the Northwest (Vietnam) populations. Populations from Oaxaca, Chiapas, and Puebla (Mexico) were directly related to populations from Son La and Dien Bien (Northwest of Vietnam) (Figure 5A). Besides that, some individuals from Minas Gerais and Espírito Santo (Brazil) had close relationships with some individuals from the Northwest of Vietnam (Figure 5A). On the contrary, a population from Oaxaca had a closer relationship with some individuals from Dong Nai (Southeast of Vietnam) (Figure 5A). Figure 5B shows the close relationship of the Chiang Mai (Thailand) population with populations in the Central Highlands and the Southeast (Dong Nai) of Vietnam. Moreover, all populations in the Central Highlands and the Southeast were connected with the Northwest populations (Figure 5B).
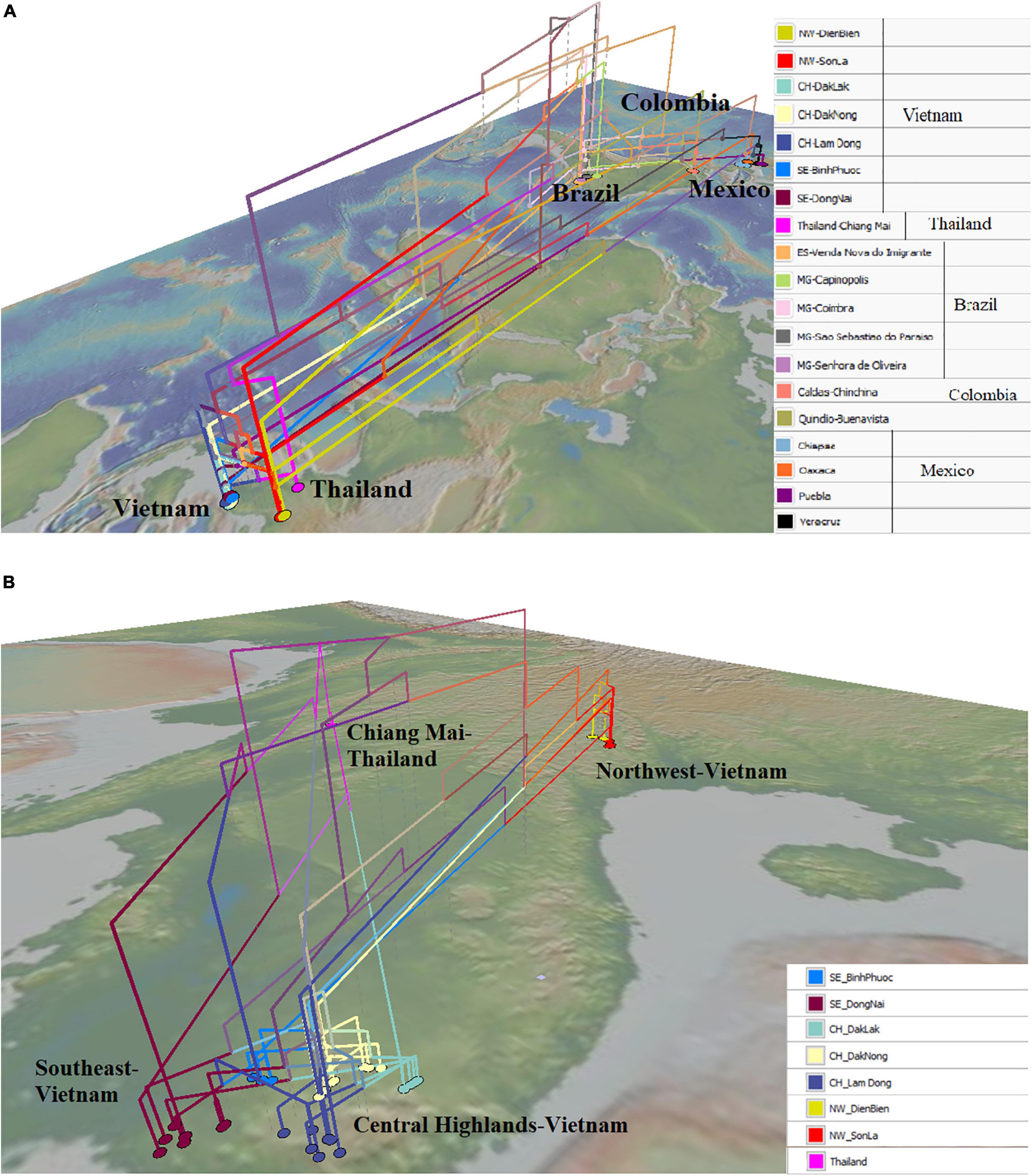
Figure 5. Geophylogeny of Hemileia vastatrix from Vietnam, Thailand, and other American countries (Brazil, Colombia, Mexico) using ITS sequences. Colored nodes on the map correspond to sampling locations. (A) Geophylogeny of Hemileia vastatrix from Vietnam, Thailand, and Central and South American countries, (B) Geophylogeny of Hemileia vastatrix from areas in Vietnam and Chiang Mai-Thailand. NW: Northwest (Vietnam), CH: Central Highands (Vietnam), ST: Southeast (Vietnam), CIFC: Centro de Investigação de Ferrugens do Cafeeiro (Portugal), MG: Minas Gerais (Brazil), ES: Espírito Santo (Brazil).
Discussion
Catimor Derivatives of Coffee in Vietnam Are No Longer Resistant to Rust Disease?
The results of field investigations show that the leaf rust disease is present in all coffee growing regions in Vietnam, from South to North, and from low altitudes to high mountainous areas. These observations suggest that all coffee species and cultivars planted in Vietnam now are susceptible to rust disease. Previously, Coffea arabica cv. Catimor, HdT, and Sachimor were known to be rust-resistant (Talhinhas et al., 2017). However, our survey shows that this cultivar can be infected by rust fungus. The number and size of the rust lesions was large on the coffee tree leaves in the Northwest plantations, where Catimor varieties are predominately planted (Table 1 and Supplementary Figure S2). The detection of rust-infected leaves in regions planting rust-resistant coffee varieties is not unique to this research; numerous previous researches have revealed that the rust disease existed in HdT growing areas of Brazil and Colombia (Maia et al., 2013; Cabral et al., 2016; Talhinhas et al., 2017). This raises concerns about whether all rust-resistant coffee varieties have become susceptible to H. vastatrix or whether the situation is only present in a few countries or areas. The most probable reasons for the loss of rust resistance are mutations in the genome of H. vastatrix or the appearance of new pathogen races (Talhinhas et al., 2017). However, other factors that could affect the rust resistance of these coffee varieties must also be studied because climatic conditions and cultural practices can also promote disease outbreaks (Avelino et al., 2015). For example, there were high levels of rust severity found in the abandoned coffee plantations of the Central Highlands (Table 1 and Supplementary Figure S2). Therefore, further research on such factors is required to control CLR disease in Vietnam.
Hemileia vastatrix in Vietnam Has Low Genetic Divergence and Unstructured Population but High Haplotype Diversity
Genetic diversity and population structure analyses disclosed some actualities of the H. vastatrix populations in Vietnam. Firstly, the genetic differentiation of H. vastatrix in Vietnam is low, as shown in the calculated AMOVA (Table 3 and Supplementary Table S5). The percentage of variation within populations surpasses that among populations (Table 3), indicating that the genetic divergence among populations is small. Moreover, the nucleotide diversity values of populations in Vietnam are low (less than 0.1; Supplementary Table S5). Secondly, H. vastatrix populations in Vietnam are unstructured. The PCoA diagram (Figure 3) illustrates that some individuals are scattered from others. Dispersed haplotypes are also well illustrated as they are divided into groups distinguished from two others (Figures 2, 4); however, these groups are divided without relation to the regions where CLR samples were collected. Lastly, the Hd of H. vastatrix in Vietnam is high. This is not only because of their high values showed in Supplementary Table S5 but also because many groups of haplotype are divided into networks (Figures 2, 4). Besides that, the linear regression (Figure 6) shows that the nucleotide diversity has a negative relationship with latitude; the higher the latitude, the lower the nucleotide diversity value. Most haplotypes in Vietnam are unique. Mutations in the H. vastatrix populations of Vietnam are likely to be spontaneous without being affected by geographical factors or the host’s resistance genes. Nonetheless, random alterations due to environmental changes, such as warmer climate or increased annual rainfall, could result in the formation of new virulent races (Ali et al., 2014). Furthermore, the unstructured population of H. vastatrix could result from asexual reproduction (Ramírez-Camejo et al., 2021). The results of this study are similar to those from studies in the coffee-growing regions of Peru and Brazil (Maia et al., 2013; Cabral et al., 2016; Quispe-Apaza et al., 2017, 2021).
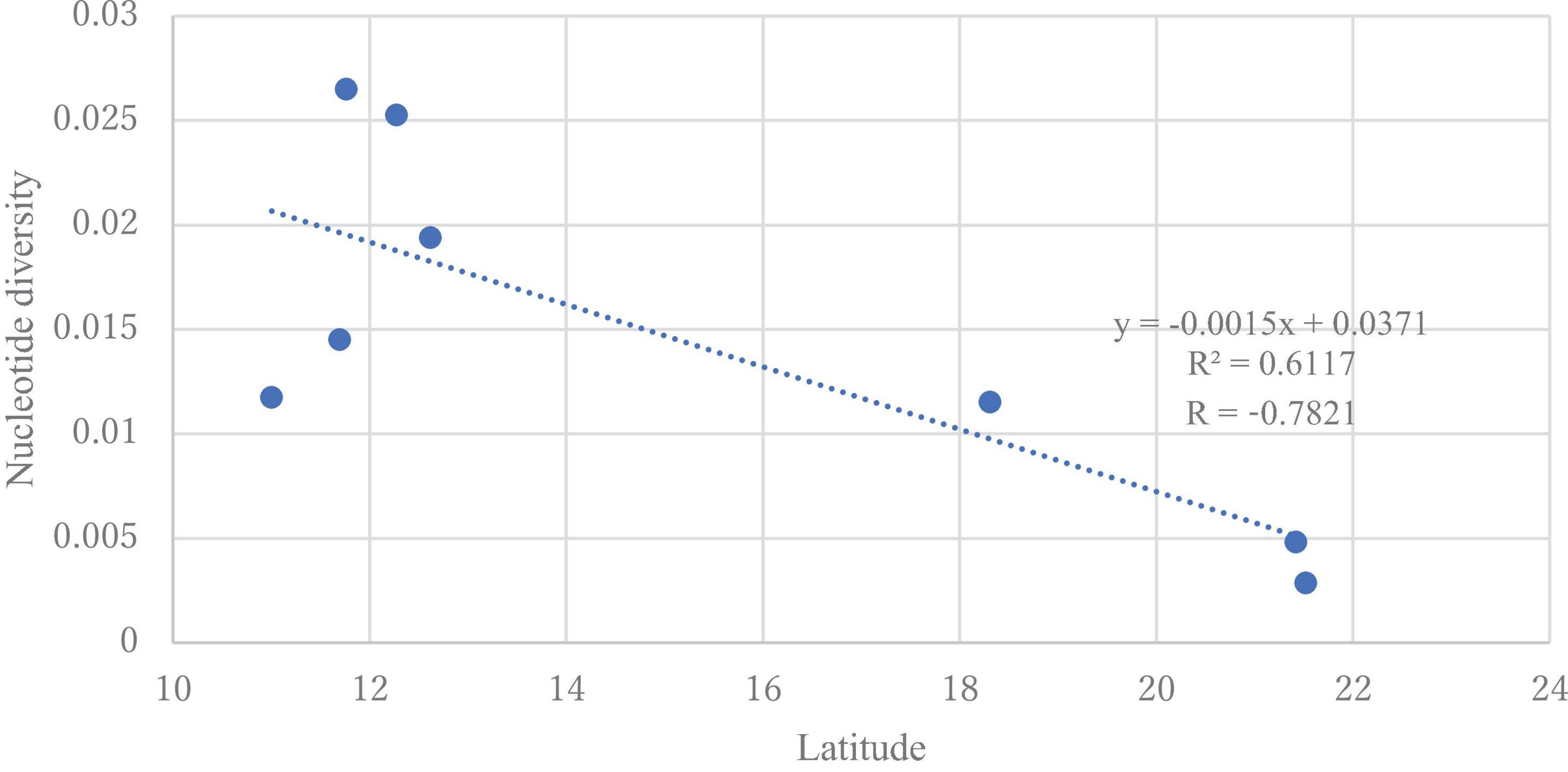
Figure 6. Linear regression model for the latitude and nucleotide diversity of Hemileia vastatrix populations from Vietnam and Thailand. The dotted line represents the regression line with equation y = −0.0015x + 0.0371.
The Hypothesized Original Sources and the Migration Direction of Hemileia vastatrix in Vietnam
Based on the population genetic analyses, a hypothetical interpretation of the first establishment of CLR disease and the successive spore transportation pathways through coffee-growing areas in Vietnam is possible. The frequency of Haps 5, 8, 9, 11, 17, 27, and 32 in the whole H. vastatrix population and their positions in the phylogenetic analyses (Figures 1, 2, 4) indicate that these haplotypes are shared in most populations and steady in the reproductive process. Thus, these haplotypes are referred to as the ‘early established haplotypes’. Among them, Hap 32 originated from Hap 8; and Haps 11 and 5 are descendants of Hap 9. Haps 8, 17, 27, and 32 are shared between Brazil, Colombia, CIFC-Portugal, and Mexico. In addition, the pairwise genetic distance matrices (Supplementary Table S7), PCoA diagram (Supplementary Figure S3), and phylogeographic tree (Figure 5A) highlight the close genetic distances and genetic similarities between some populations in Mexico and Brazil and the Northwest population in Vietnam. These results prove that H. vastatrix populations from Vietnam (mostly from the Northwest) and those from Central and South American countries (Mexico and Brazil) have very close relationships. However, we cannot ascertain that the H. vastatrix population in Vietnam spread from these countries just because the first rust detection in Vietnam was in 1890, while the first confirmation of rust establishment in Brazil and Mexico was in the 1970s and 1980s, respectively (McCook, 2006, 2019). On second thought, H. vastatrix might have migrated to Vietnam from African countries because Vietnam and these countries were colonized by the French government during World War I. Hemileia vastatrix was first identified in Vietnam in 1890, the exact timeline of the global CLR pandemic (1875−1920). At that time, rust-infected coffee seedlings or materials from infected areas in Africa (where H. vastatrix is native) might have been carried to Vietnam by the French. However, confirming the origin of CLR in Vietnam necessitates further studies with larger rust samples from other countries, especially African countries. For these reasons, this discussion only focused on the first establishment of CLR in the three main coffee growing regions in Vietnam. Three main inferences regarding the origin and migration route of H. vastatrix can be gathered based on the present study. Firstly, because ΦPT was previously measured only based on the genetic drift (Hedrick, 2005), the low genetic distance recorded between populations in Vietnam (Supplementary Table S7 and Supplementary Figure S3) proves that the probability of migration between these regions was high. Secondly, most ancestral haplotypes (Haps 5, 8, 9, 11) are present in the Northwest, and the percentage of those ancestral haplotypes in the Northwest is larger than that in other areas (Figure 1). Thirdly, the phylogeographic tree shows that H. vastatrix from Brazil and Mexico have genetic similarity with populations in the Northwest of Vietnam (Figure 5A); all populations from southern Vietnam had a close genetic relationship with the Northwest population (Figure 5B). Therefore, we can infer that H. vastatrix existed in the north of Vietnam in the early years of introducing coffee, then spread to the Southeast and the Central Highlands as coffee cultivation was extended to these areas. Although C. arabica was cut down for Catimor coffee in northern Vietnam (Anonymous, 2019), urediniospores of the rust fungus could still exist by parasitizing coffee plants in other plantations (the replacement of coffee varieties was not synchronized well and did not occur at the same time in all plantations of a region) or wild coffee plants and infected Catimor coffee, making this coffee variety susceptible to coffee rust. Hap 8 is one of the first ancestors that spread in northern Vietnam. After that, this haplotype became widespread in coffee plantations in the Northwest and formed Haps 13, 14, and 25. Later, it was transported to the South via human activities or infected materials and formed Haps 20, 27, and 34. Hap 27 is one of the ancestral haplotypes in the Southeast. Meanwhile, another ancestor, Hap 9, is widespread and formed Haps 5, 10, 11, and 12. This ancestor and some haplotypes (Haps 5 and 11) were then carried to the Central Highlands of Vietnam and formed Haps 1, 3, and 15 (Figures 2, 4). Thus, Haps 5, 8, 9, 11, and 27 are the ancestral haplotypes of the Vietnamese H. vastatrix.
On the other hand, data from the population genetic analyses (Figures 2, 3, 4) suggest that coffee rust in Vietnam has another origin besides spreading from the Northwest. This is indicated in the PCoA diagram (Figure 3): some individuals from the Central Highlands and the Southeast are scattered distantly from others. Figures 2, 4 also pointed out that some haplotypes (Haps 46–52) are genetically distant from others and form a different group. Information from the history of coffee in Vietnam consolidates this hypothesis. Coffea canephora was introduced in the Central Highlands of Vietnam to replace C. arabica in the early 1900s (D’haeze et al., 2003; Anonymous, 2019). When C. canephora spread to southern Vietnam, they might have also brought H. vastatrix to this region, broadened, and formed other branches of CLR in Vietnam. The appearance of absent haplotype (black node) in the haplotype network and the NeighborNet (Figures 2, 4) suggest that other haplotypes are yet to be discovered, and some of them are likely to be ancestral for haplotypes found in this study.
This study sheds new light on the genetic diversity of the CLR fungus, H. vastatrix, in Vietnam and in Central and South America. However, all analyses in this study are based on only one locus (ITS region). Because of this limitation, we could only discover two possible migration pathways of H. vastatrix in Vietnam. In future, we intend to conduct population genomic studies on H. vastatrix in Vietnam to reveal more information regarding the genetic structure, population demography, origins, and migration pathways of H. vastatrix in Vietnam.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CL collected the samples, designed and performed the experiments, and wrote the manuscript. YO, IO, YT, YY, and CL conceived and directed the study. CL, YT, IO, and YO analyzed and biologically interpreted the data. YO, YT, and YY revised and approved the final version of the manuscript. All authors contributed greatly to the present manuscript.
Funding
This research was financially supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 20H03006 (IO) and the fund for student research from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
Author Disclaimer
All the specimens in this study were collected under the permission of the Vietnamese Government. In this study, all experiments were performed with the current laws of Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Board of Directors of Biotechnology Center of Ho Chi Minh City, Dinh A. H., Nguyen T. T. D., Tran C. H., and Nguyen T. N. B. for their assistance during collecting samples in Vietnam.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.872877/full#supplementary-material
Supplementary Figure S1 | Locations in Vietnam and Thailand where coffee leaf rust samples were collected. The 3D map was generated using Microsoft Excel. Purple dots represent plantations without rust disease and blue dots represent plantations with rust disease.
Supplementary Figure S2 | Coffee leaf rust samples collected from Coffea canephora and Coffea arabica cv. Catimor in Vietnam.
Supplementary Figure S3 | Principal coordinate analysis (PCoA) of Hemileia vastatrix distance matrix between populations. Populations that are closer together have smaller divergence. NW, Northwest; CH, Central Highlands; ST, Southeast; CIFC, Centro de Investigação de Ferrugens do Cafeeiro; MG, Minas Gerais; ES, Espírito Santo.
Supplementary Table S1 | Internal transcribed spacer (ITS) sequences for coffee leaf rust fungus dowloaded from Genbank NCBI.
Supplementary Table S2 | Coffee leaf rust samples collected in Vietnam and Thailand.
Supplementary Table S3 | Information including query cover, % identity, name, and accession of the query sequences after using the NCBI BLAST tool for Vietnamese and Thailand coffee leaf rust fungus sequences.
Supplementary Table S5 | Number of haplotypes, haplotype diversity, and nucleotide diversity of the Hemileia vastatrix populations.
Supplementary Table S6 | Tajima’s D value for the Hemileia vastatrix populations.
Supplementary Table S7 | Genetic differentiation (measured by ϕPT) of the Hemileia vastatrix populations between Vietnam and other countries.
Supplementary Material S4 | Alignment dataset used for the population genetic analyses.
Footnotes
References
Aime, M. C., and McTaggart, A. R. (2020). A higher-rank classification for rust fungi, with notes on genera. Fungal Syst. Evol. 7, 21–47. doi: 10.3114/fuse.2021.07.02
Ali, S., Gladieux, P., Leconte, M., Gautier, A., Justesen, A. F., Hovmøller, M. S., et al. (2014). Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PLoS Pathog. 10:e1003903. doi: 10.1371/journal.ppat.1003903
Avelino, J., Cristancho, M., Georgiou, S., Imbach, P., Aguilar, L., Bornemann, G., et al. (2015). The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Sec. 7, 303–321. doi: 10.1007/s12571-015-0446-9
Backer, C. A., and Bakhuizen Van Den Brink, R. C. (1965). Flora of Java (Spermatophytes only) (Vol. 2). Angiospermae, Families 111–160, (Groningen: N. V. P. Noordhoff), 322–323.
Belan, L. L., Belan, L. L., Rafael, A. M., Gomes, C. A. G., Alves, F. R., Junior, W. C. J., et al. (2020). Standard area diagram with color photographs to estimate the severity of coffee leaf rust in Coffea canephora. Crop Prot. 130:105077. doi: 10.1016/j.cropro.2020.105077
Bryant, D., and Moulton, V. (2013). Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. 21, 255–265. doi: 10.1093/molbev/msh018
Cabral, P. G. C., Maciel-Zambolim, E., Oliveira, S. A. S., Caixeta, E. T., and Zambolim, L. (2016). Genetic diversity and structure of Hemileia vastatrix populations on Coffea spp. Plant Pathol. 65. 196–204. doi: 10.1111/ppa.12411
Cummins, G. B., and Hiratsuka, Y. (2003). Illustrated Genera Of Rust Fungi. 3 rd Edn. St. Paul, MN: American Phytophathological Society, APS Press.
D’haeze, D., Deckers, J., Raes, D., Tran, A. P., and Nguyen, D. M. C. (2003). Over-irrigation of Coffea canephora in the central highlands of vietnam revisited simulation of soil moisture dynamics in Rhodic Ferralsols. Agric. Water Manag. 63, 185–202. doi: 10.1016/S0378-3774(03)00181-1
D’haeze, D., Deckers, J., Raes, D., Tran, A. P., and Loi, H. V. (2004). Environmental and socio-economic impacts of institutional reforms on the agricultural sector of Vietnam Land suitability assessment for Robusta coffee in the Dak Gan region. Agric. Ecosyst. Environ. 105, 59–76. doi: 10.1016/j.agee.2004.05.009
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fraczek, M. G., Zhao, C., Dineen, L., Lebedinec, R., Bowyer, P., Bromley, M., et al. (2019). Fast and reliable PCR amplification from Aspergillus fumigatus spore suspension without traditional DNA extraction. Curr. Protoc. Microbiol. 54:e89. doi: 10.1002/cpmc.89
Hammer, Ø, Harper, D. A. T., and Paul, D. R. (2001). Past: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, Available online at: http://palaeo-electronica.org/2001_1/past/issue1_01.html*.
Hedrick, P. W. (2005). A standardized genetic differentiation measure. Evolution 59:1638. doi: 10.1111/j.0014-3820.2005.tb01814.x
Hennings, P. (1895). Neue und interessante Pilze aus dem Königl. Botanischen Museum in Berlin. III. Hedwigia 34, 10–13.
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Kolmer, J. A., Ordonez, M. E., and Groth, J. V. (2009). The Rust Fungi. In: Encyclopedia of Life Sciences (ELS). Chichester: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0021264
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Maia, T. A., Maciel-Zambolim, E., Caixeta, E. T., Mizubuti, E. S. G., and Zambolim, L. (2013). The population structure of Hemileia vastatrix in Brazil inferred from AFLP. Aust. Plant Pathol. 42, 533–542. doi: 10.1007/s13313-013-0213-3
McCook, S. (2006). Global rust belt: Hemileia vastatrix and the ecological integration of world coffee production since 1850. J. Glob. History 1, 177–195. doi: 10.1017/S174002280600012X
McCook, S. (2019). Coffee Is Not Forever: A Global History Of Coffee Leaf Rust. Athens, OH: Ohio University Press, 67–68.
Nei, M., and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. doi: 10.1073/pnas.76.10.5269
Parks, D. H., Mankowski, T., Zangooei, S., Porter, M. S., Armanini, D. G., Baird, D. J., et al. (2013). GenGIS 2: Geospatial analysis of traditional and genetic biodiversity, with new gradient algorithms and an extensible plugin framework. PLoS One 8:e69885. doi: 10.1371/journal.pone.0069885
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Quispe-Apaza, C. S., Mansilla-Samaniego, R. C., López-Bonilla, C. F., Espejo-Joya, R., Villanueva-Caceda, J., and Monzón, C. (2017). Genetic diversity of Hemileia vastatrix of two coffee producing areas in Peru. Rev. Mex. Fitopatol. 35, 418–436. doi: 10.18781/R.MEX.FIT.1612-7
Quispe-Apaza, C., Mansilla-Samaniego, R., Espejo-Joya, R., Bernacchia, G., Yabar-Larios, M., and López-Bonilla, C. (2021). Spatial and temporal genetic diversity and population structure of Hemileia vastatrix from Peruvian Coffee Plantations. Plant Pathol. J. 37, 280–290. doi: 10.5423/PPJ.OA.10.2020.0192
Ramírez-Camejo, L. A., Eamvijarn, A., Díaz-Valderrama, J. R., Karlsen-Ayala, E., Johnson, E., Pruvot-Woehl, S., et al. (2021). Global analysis of Hemileia vastatrix populations shows clonal reproduction for the coffee leaf rust pathogen throughout most of its range. Phytopathology doi: 10.1094/PHYTO-06-21-0255-R
Ritschel, A. (2005). “Monograph of the genus Hemileia (Uredinales),” in Bibliotheca Mycologica, Vol. 200, eds A. Bresinsky, H. Butin, and P. Tudzinski (Stuttgart: J. Cramer).
Ryan, W. B. F., Carbotte, S. M. J., Coplan, S., O’Hara, A., Melkonian, R., Arko, R. A., et al. (2009). Global Multi-Resolution Topography (GMRT) synthesis data set. Geochem. Geophys. Geosyst. 10:Q03014. doi: 10.1029/2008GC002332
Silva, M. C., Várzea, V., Guerra-Guimarães, L., Azinheira, H. G., Fernandez, D., Petitot, A., et al. (2006). Coffee resistance to the main diseases: leaf rust and coffee berry disease. Braz. J. Plant Physiol. 18, 119–147. doi: 10.1590/S1677-04202006000100010
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Talhinhas, P., Batista, D., Diniz, I., Vieira, A., Silva, D. N., Loureiro, A., et al. (2017). The coffee leaf rust pathogen Hemileia vastatrix: one and a half centuries around the tropics. Mol. Plant Pathol. 18, 1039–1051. doi: 10.1111/mpp.12512
Tavaré, S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences (PDF). Lect. Math. Life Sci. 17, 57–86.
Thirumalachar, M. J., and Narasimhan, M. J. (1947). Studies on the morphology and parasitism of hemileia species on rubiaceae in Mysore. Ann. Bot. 11, 77–89. doi: 10.1093/oxfordjournals.aob.a083149
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide To Methods And Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Keywords: coffee leaf rust disease, genetic diversity, Hemileia vastatrix, ITS, Southeast Asia
Citation: Le CTM, Okane I, Ono Y, Tsuda Y and Yamaoka Y (2022) Incidence of Coffee Leaf Rust in Vietnam, Possible Original Sources and Subsequent Pathways of Migration. Front. Plant Sci. 13:872877. doi: 10.3389/fpls.2022.872877
Received: 10 February 2022; Accepted: 08 March 2022;
Published: 05 April 2022.
Edited by:
Prem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), IndiaReviewed by:
Jaime A. Collazo, North Carolina State University, United StatesJaspal Kaur, Punjab Agricultural University, India
Copyright © 2022 Le, Okane, Ono, Tsuda and Yamaoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cham Thi Mai Le, bHRtY2hhbS5zbm5AdHBoY20uZ292LnZu; bHRtYWljaGFtLmJpb0BnbWFpbC5jb20=
 Cham Thi Mai Le
Cham Thi Mai Le Izumi Okane
Izumi Okane Yoshitaka Ono
Yoshitaka Ono Yoshiaki Tsuda
Yoshiaki Tsuda Yuichi Yamaoka
Yuichi Yamaoka