- 1College of Life Sciences, Zhejiang Sci-Tech University, Hangzhou, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 3College of Pharmacy, Zhejiang Chinese Medical University, Hangzhou, China
Nowadays, because of the great benefit to human health, more and more efforts have been made to increase the production of alkaloids in Pinellia ternata (Thunb.) Breit. Phosphate (Pi) plays a critical role in plant growth and development, as well as secondary metabolism. However, its effect and regulation mechanism of Pi signaling on alkaloid biosynthesis call for further exploration. Here, we reported that Pi starvation could induce alkaloid accumulation in P. ternata. We cloned a cDNA sequence encoding PtPHR1 from P. ternata, which was further identified by nuclear localization, transcription activity, and binding ability to the PHR1-binding sequence. We found that the transformation of PtPHR1 into the Arabidopsis phr1 mutant (designated as PtPHR1OE/phr1) led to the rescue of the phenotype of the phr1 mutant to that of the wild-type, including the expression level of Pi starvation-induced genes and anthocyanin accumulation. The combination of these biochemical and genetic experiments indicated that PtPHR1 was intended to have a role similar to that of AtPHR1 in Pi signaling and metabolic responses. Interestingly, we found that Pi starvation also induced the production of benzoic acid, an intermediate in the biosynthetic pathway of phenylpropylamino alkaloids. Furthermore, this induction effect was impaired in the phr1 mutant but partly recovered in PtPHR1OE/phr1 plants. Together, our data suggest that Pi starvation promoted benzoic acid-derived alkaloid biosynthesis in P. ternata under the control of PtPHR1. Our finding that PtPHR1 is involved in the regulation of Pi signaling on alkaloid biosynthesis shows a direct link between the Pi nutrient supply and secondary metabolism.
Introduction
Pinellia ternata (Thunb.) Breit, a member of the Araceae family, is an essential Chinese traditional herb with a long history of use for medical treatment (Mao and He, 2020). The dried tuber of this herb, called “banxia” in Chinese, is the major part of the plant used for medicine. P. ternata has broad pharmacological and clinical properties such as, wound healing, cough soothing, anti-spasmodic, anti-tumoral, and lipid-lowering effects (Wang et al., 2009; Wu et al., 2013; Ji et al., 2014; Mao and He, 2020). P. ternata is abundant in secondary metabolites (i.e., alkaloids, iridoids, iridoid glycosides, anthraquinones, anthraquinone glycosides, sterols, amino acids, and fatty acids), as well as their derivatives (Oshio et al., 1978; Ge and Hao, 2009; Sun et al., 2018). Among them, alkaloids are the main biologically active compounds with anti-tumoral and anti-viral activities (Xu et al., 2007; Ji et al., 2014). According to the Chinese Pharmacopoeia Commission (2005), ephedrine and purine, two main active ingredients of alkaloids, are quality markers of P. ternata. It is apparent that the supply of alkaloids in P. ternata cannot keep up with the growing demand for them any longer (Liu et al., 2015; Xue et al., 2019; Mao and He, 2020). Therefore, a deeper and more comprehensive understanding of the alkaloid synthesis pathway and its regulatory mechanism would aid in improving the supply.
To date, two potential pathways of phenylpropylamino alkaloids biosynthesis have been characterized and identified, namely, the β-oxidative and non-β-oxidative routes (Boatright et al., 2004; Zhang et al., 2016; Duan et al., 2019). Respectively, the main enzymes involved in the β-oxidative pathway are phenylalanine ammonia lyase (PAL), cinnamate: CoA ligase (CNL), cinnamoyl CoA hydratase-dehydrogenase (CHD), and 3-ketoacyl-CoA thiolase (KAT) (Klempien et al., 2012). In the non-β-oxidative pathway, sequential catalyzation by 3-hydroxyisobutyryl-CoA hydrolase (CHY), benzaldehyde dehydrogenase (BALDH) and aldehyde oxidase 4 (AO4) (Van et al., 2009) was responsible for the formation of benzoic acid (BA). Followed by ThDP-dependent pyruvate decarboxylase (ThPDC) and acetolactate synthase (AHAS), the BA was further converted into 1-phenylpropane-1,2-dione (Müller et al., 2009). Using RNA-seq data, a comprehensive genome of P. ternata has been provided (Zhang et al., 2016; Tian et al., 2022). However, the genes and signaling pathways involved in the regulation of alkaloid biosynthesis need to be further elucidated.
The essential element phosphorus is indispensable in and critical for plant growth and development (Cong et al., 2020). In soil, the concentration of soluble inorganic phosphate (Pi) is relatively low, and as a result, Pi starvation seriously impacts and limits plant growth and yield (Neumann and Römheld, 2012). To adapt to Pi starvation, plants change the architecture of their root systems so they have a shorter primary root and higher root density, enhance the root's exudation, and increase the expression of Pi transporter genes and they also use other strategies to facilitate Pi acquisition (Yuan and Dong, 2008). In recent decades, the Pi signaling pathway has been well elucidated and most components involved have been identified (Yuan and Dong, 2008; Crombez et al., 2019). Among them, phosphate starvation response proteins (PHRs) serve as the key regulators of the Pi starvation signal. As reported, PHRs can recognize a specific DNA motif (P1BS, sequence GNATATNC) and regulate most Pi starvation-induced (PSI) genes (Rubio et al., 2001). The SPX protein family, made up of a single SYG1/Pho81/XPR1 (SPX) domain, suppresses Pi starvation signaling by interacting with PHRs and inhibiting their transcriptional activities (Lv et al., 2014; Ruan et al., 2019).
Until now, several studies have implicated Pi starvation signaling in the biosynthesis of secondary metabolites. In Arabidopsis, the phr1 mutant ablates Pi starvation-induced anthocyanin production whereas the spx1spx2 double mutant accumulates more anthocyanins in response to Pi starvation (Rubio et al., 2001; Puga et al., 2014). Most recently, we reported that SPX4 could interact with both PHR1 and PAP1 to regulate Pi starvation-mediated flavonoid biosynthesis (He et al., 2021). Pi deficiency also affects the accumulation of indole glucosinolates in Arabidopsis co-cultivated with Colletotrichum tofieldiae (Frerigmann et al., 2021). In addition, the Pi supply could affect chicoric acid accumulation in the hairy roots of Echinacea purpurea (Salmanzadeh et al., 2020). However, the involvement of Pi signaling in the regulation of alkaloid biosynthesis and its molecular mechanism has yet to be reported.
Herein, we demonstrated that Pi starvation increased the transcript level of alkaloid biosynthetic genes and thereby promoted alkaloid accumulation. Based on the cloning and characterizing of the PtPHR1 sequence of P. ternata, we identified its nuclear localization, transcriptional activity, and recognition of the P1BS element. Through heterologous expression of PtPHR1 in the Arabidopsis phr1 mutant, we further demonstrated that PtPHR1 regulated Pi-starvation induced BA biosynthesis. Together, our results revealed that Pi signaling mediated alkaloid biosynthesis, which was under the control of PtPHR1.
Materials and methods
Plant materials
Tubers of P. ternata were washed, sliced, and cultured at 25°C with a 16 h/8 h-light/dark photoperiod as described (Liu et al., 2010). For Pi starvation, 30-day-old P. ternata (Figure 1A) was cultured in half-strength liquid MS (Murashige and Skoog) medium. The medium was prepared with MS without phosphate (MSP11-50LT; Caisson Laboratories, USA), 0.05% (w/v) MES, and 0.6% (w/v) sucrose. In addition, the high Pi medium was supplemented by 10 mM KH2PO4, 10 μM KH2PO4, and equimolar amounts of KCl (Khan et al., 2016; Kong et al., 2021) were added to the low Pi medium. During treatment, the nutrient solution was replaced every 3 days.
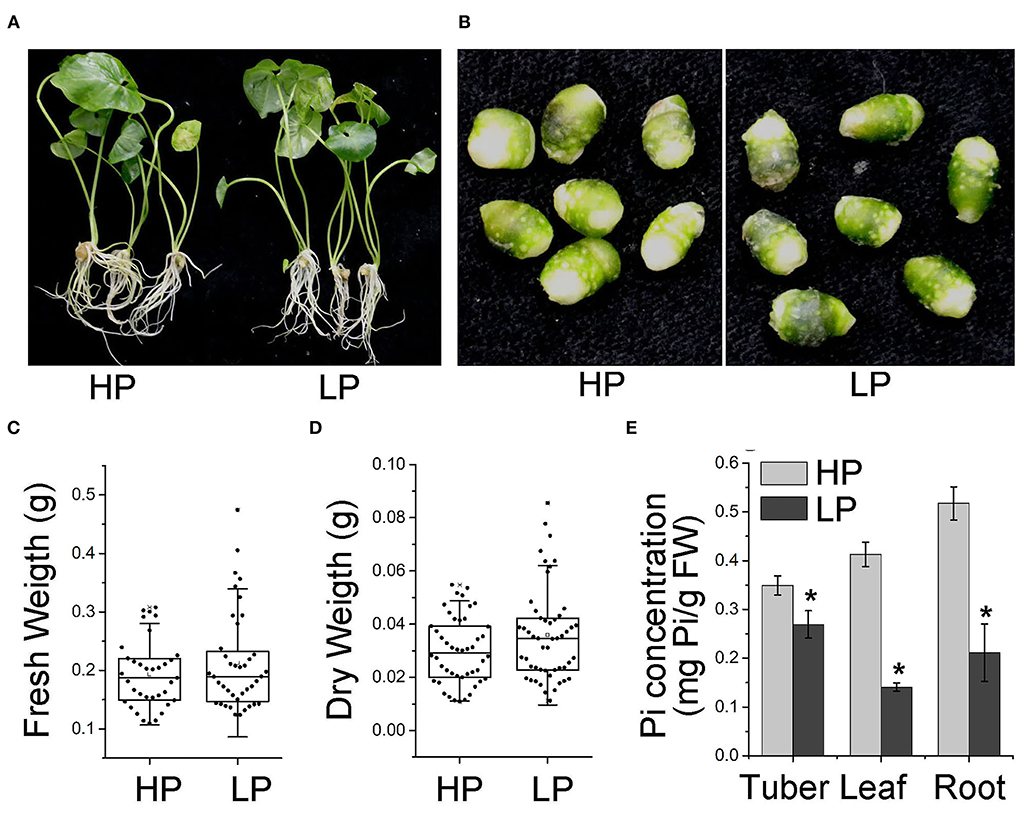
Figure 1. Phenotype of P. ternata in conditions of high and Low Pi. Phenotype of P. ternata whole plants (A) and tubers (B) cultured in HP and LP medium for 7 days. (C,D) Effect of Pi starvation on the tubers of P. ternata in terms of wet (C) and dry (D) weight. (E) Pi concentration in the tubers, leaves, and roots of P. ternata upon Pi starvation treatment for 7 days. Values are means ± SD of three biological replicates. *P < 0.05, Student's t-test. HP: high Pi, 10 mM. LP: low Pi, 10 μM.
Surface-sterilized Arabidopsis thaliana ecotype Columbia-0 (Col-0) and phr1 mutants were germinated on half-strength MS and grown in a growth chamber at 22°C with a 16 h/8 h-light/dark cycle. To generate the PtPHR1-myc constructor, the full-length ORF of PtPHR1 was cloned and inserted into a modified pCAMBIA1300 vector with an N-terminal fusion 6xmyc tag under the control of the 35S promoter (He et al., 2021). The PtPHR1-myc was then introduced into phr1 mutants by the Agrobacterium-mediated flower-dip method to generate PtPHR1OE/phr1 plants. Hygro-F/R (hygromycin B phosphotransferase detection) and PtPHR1-detect-F/R were used to identify homozygous lines, and anti-myc antibody was used to measure the PtPHR1-myc protein level. The primers used in this study are listed in Supplementary Table S1.
Determination of Pi content
The tubers, leaves, and roots of P. ternata grown in a high and low Pi medium for 7 days were collected respectively for Pi measurement, according to the procedure previously described (Zhou et al., 2008).
Total RNA extraction and RT-qPCR
A total of 1 μg of RNA was used to synthesize the first-strand cDNA, using the HiScript II Q RT for qPCR (+gDNA viper) kit (Vazyme, China). Then RT-qPCR (Hieff qPCR SYBR Green Master Mix, Yeasen, China) was performed on a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Singapore), following the manufacturer's instructions. Pt18S was used as a reference for normalization, and expression levels were analyzed by the comparative Ct method (2−ΔΔCt method). At least three biological replicate samples were included. The primers used in this study are listed in Supplementary Table S1.
Determination of alkaloid and flavonoid content
Standard chemicals, including ephedrine hydrochloride (171241-201809), guanosine (111977-201501), adenosine (110879-201703), and uridine (110887-202104) were purchased from the National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China, and BA (PHR1050), validated by 1H Nuclear Magnetic Resonance (NMR, D2O) spectroscopy on Bruker AVANCE III 500 MHz with TMS, were purchased from Sigma–Aldrich, Shanghai, China.
Tubers of P. ternata grown under high and low Pi conditions were completely dried at 60°C until they reached a constant weight. The dried tubers were ground into powder and then used for alkaloid extraction. The total alkaloid content was determined by using a UV–visible spectrophotometer at a wavelength of 416 nm according to a previous protocol (Liu et al., 2010; Duan et al., 2019). Guanosine, adenosine, and uridine were determined at 240 nm (Ji et al., 2013), and BA at 230 nm (Iwakoshi et al., 2019). High-performance liquid chromatography (HPLC) analysis was performed using a Shimadzu Prominence-I LC-2030C 3D Plus with a 5 μm C18 column (five-particles, 4.6 × 150 mm).
The total flavonoids were extracted and measured by the Flavonoid Extraction Kit (LHT-1-G, Cominbio, China). The anthocyanin content was extracted and determined at the valve of A657-A530.
Isolation of PtPHR1 and sequence analysis
The PtPHR1 ORF sequence was amplified using the rapid amplification of the cDNA ends method. The conserved domains were analyzed using the NCBI database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Multiple alignments were analyzed using ClustalW (http://clustalw.ddbj.nig.ac.jp/). Phylogenetic and molecular evolutionary analysis was conducted using MEGA 6.0 software.
Subcellular localization analysis
The PtPHR1 ORF was cloned into the pCV-GFP-N1 binary vector to generate the pCV:PtPHR1-GFP construct. The pCV:PtPHR1-GFP was transiently expressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens infiltration (He et al., 2020). Using a TCS SP5 confocal laser scanning microscope system, the fluorescence signal was detected (Leica Microsystems, Bannockburn, IL, USA) at 40–44 h after infiltration.
Transcriptional activation
The transcriptional activation assay was conducted as described (Qiao et al., 2017). The ORFs of PtPHR1 and AtPHR1 were cloned and inserted into the vector pGBKT7 (BD) to obtain BD-PtPHR1 and BD-AtPHR1, respectively. The BD-AtPHR1 was used as a positive control and the empty BD vector as a negative control. All these vectors were transformed into AH109, grown on –Trp medium, and then selected on the –Trp/–His medium with 3-amino-1,2,4-triazole (3-AT). The primers used are listed in Supplementary Table S1.
Y1H assay
For the Y1H assay, the 4×P1BS sequence was inserted into the pHis2.1 vector. The ORFs of PtPHR1 and AtPHR1 were cloned and co-transformed with the SmaI-linearized pGADT7-Rec2 vector into the yeast strain Y1HGold, respectively. Th colonies were incubated at 30°C on SD medium lacking Leu and Trp, and then spotted on –His/–Leu/–Trp minimal medium with 0, 5, 50 mM 3-AT, respectively. The primers used are provided in Supplementary Table S1.
Statistical analysis
For analyzing differences between two variables, the Student's t-test was used. When variables were over two, ANOVA with Fisher's least significant difference test was adopted. A p-value < 0.05 was considered statistically significant. All analysis was performed using ORIGIN 8 software.
Results
Extracellular phosphate limiting resulted in intracellular Pi deficiency
To investigate the effect of Pi starvation on P. ternata, we cultured P. ternata in liquid media containing low Pi (10 μM, HP) and high Pi (10 mM, LP) (Khan et al., 2016), respectively. After 7 days, there was no sign of toxicity phenotypes at the high Pi level or Pi deficiency phenotypes at the low Pi level (Figures 1A,B). No significant difference was observed in the fresh and dry tuber weight between plants grown in Pi-sufficient and Pi-deficient conditions (Figures 1C,D). However, the Pi concentrations in the tubers, leaves, and roots of P. ternata grown in low Pi media were obviously lower than those grown in a high-Pi medium (Figure 1E). These results suggested that the growth of P. ternata was not altered during a short period of Pi deficiency.
The induction of the alkaloid biosynthetic pathway by Pi starvation
It has been well known that Pi starvation enhances the expression of flavonoid biosynthesis genes and thereby promotes flavonoid accumulation (Rubio et al., 2001; Puga et al., 2014; He et al., 2021). Consistently, we found that the gene expression involved in flavonoid biosynthesis, such as that of PtCHS, PtCHI, and PtF3H (Zhang et al., 2016; Xue et al., 2019), significantly increased in Pi-starved P. ternata compared with the high-Pi control (Supplementary Figures S1A–C), which resulted in a higher total flavonoid accumulation (Supplementary Figure S1D). To explore the effects of Pi on alkaloid biosynthesis, we analyzed the expression of alkaloid biosynthetic genes using RT-qPCR. In P. ternata tubers, the expression levels of PtCNL, PtCHD, PtKAT, PtPDC, PtAO4, and PtAHAS (Zhang et al., 2016) were significantly induced by 3- and 7-day Pi starvation (Figures 2A,B, Supplementary Figure S2). The up-regulation of PtCHY and PtBALDH was only observed in plants during the 7-day treatment, not during the 3-day (Figures 2A,B). The consistent induction pattern of Pi starvation on alkaloid biosynthetic genes in the leaves and roots of P. ternata was also observed (Figures 2C–F).
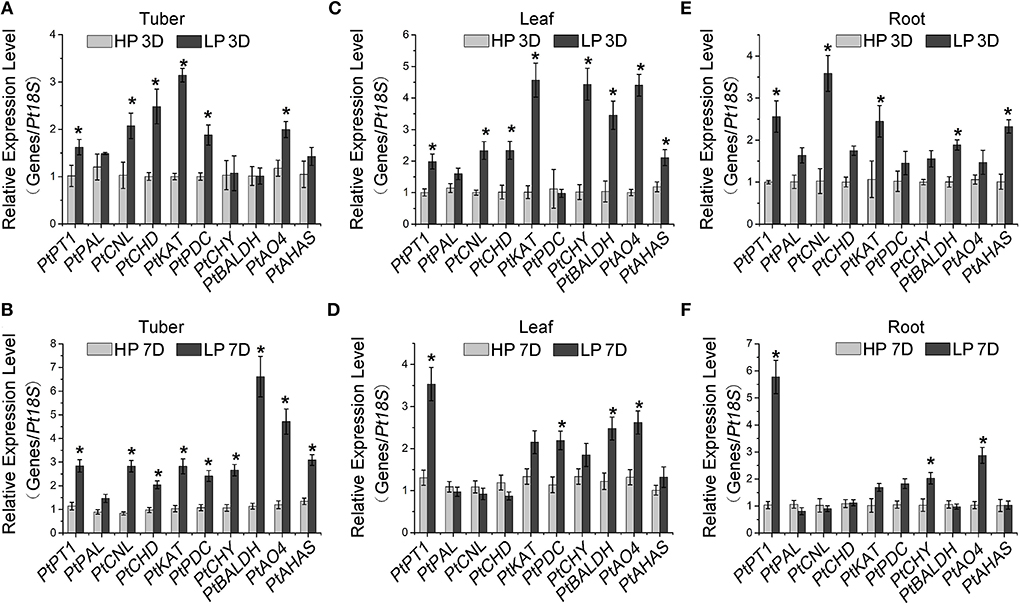
Figure 2. Induction of alkaloid-responsive gene expression by Pi starvation. RT-qPCR analysis of alkaloid-responsive genes in P. ternata tubers (A,B), leaves (C,D), and roots (E,F) upon low Pi treatment for 3 days (A,C,E) and 7 days (B,D,F). PtPT1 is a marker gene for Pi starvation. Values are means ± SD of three biological replicates. *Indicates a significant difference between high/low Pi treatment at P < 0.05 by Student's t-test. HP: high Pi, 10 mM. LP: low Pi, 10 μM.
To confirm whether the alteration of alkaloid biosynthetic gene expression led to the change in alkaloid production, the alkaloid content in the tubers of Pi-starved plants was measured. The results showed that the total alkaloid content was higher in tubers from plants grown in low Pi conditions than in those from high Pi conditions (Figure 3A). In addition, the content of ephedrine hydrochloride and BA, as well as three purine alkaloids (including adenosine, guanosine, and uridine) was significantly increased to different degrees in Pi-starved conditions (Figures 3B–F, Supplementary Figures S3, S4). These results together suggest that Pi starvation activates the alkaloid pathway.
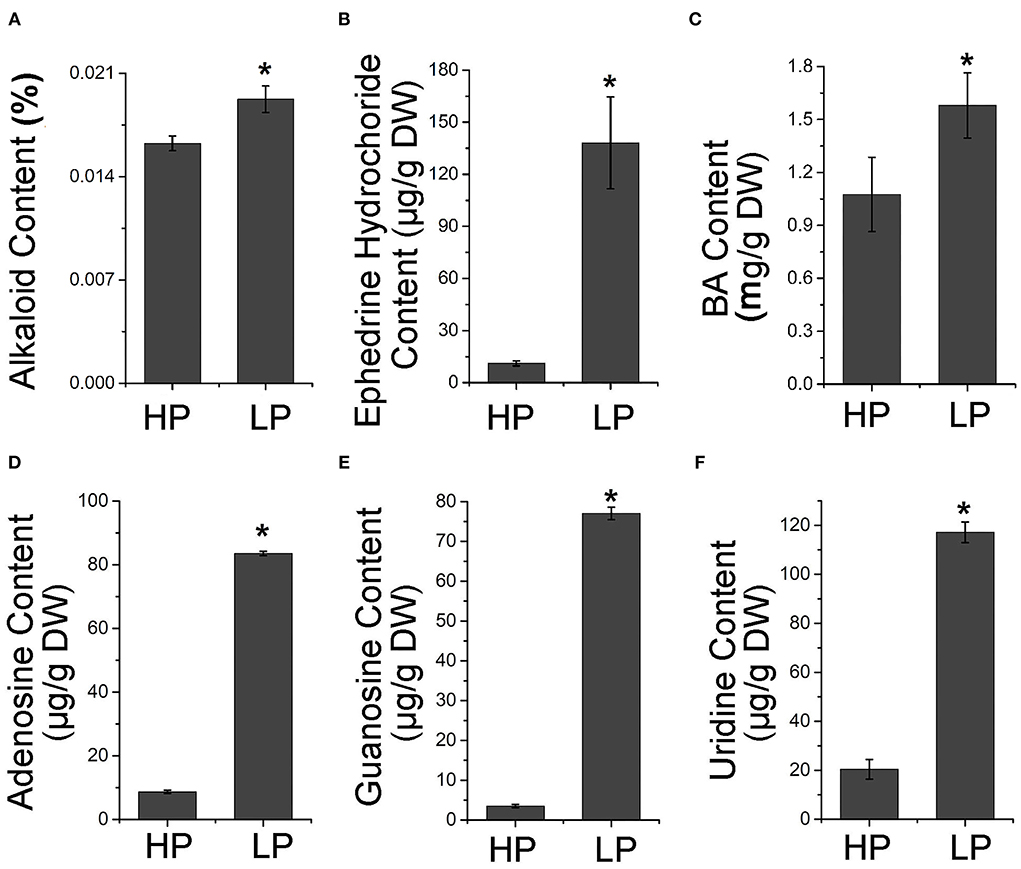
Figure 3. Accumulation of alkaloid compounds by Pi starvation treatment. Contents of total alkaloid (A), ephedrine hydrochloride (B), BA (C), guanosine (D), adenosine (E), and uridine (F) in tubers of P. ternata grown in high and low Pi for 7 days. Values are means ± SD of three biological replicates. *Indicates a significant difference between high/low Pi treatment at P < 0.05 by Student's t-test. DW, dry weight. HP: high Pi, 10 mM. LP: low Pi, 10 μM.
Cloning and characterization of PtPHR1 gene
We cloned and obtained the full-length cDNA of PtPHR1 (GenBank no. ON075805, https://www.ncbi.nlm.nih.gov/), encoding a predicted protein of 477 amino acids (Supplementary Figure S5). Multiple alignments with AtPHR1 (Rubio et al., 2001) and OsPHR2 (Zhou et al., 2008) revealed that a conserved MYB DNA-binding domain existed in the N-terminal of PtPHR1, and that a coiled–coil (CC) domain was present in its C-terminal (Figure 4A). Phylogenetic tree analysis revealed that PHR1 proteins from P. ternata and Colocasia esculenta were grouped together, both of which belonged to an Asteraceae cluster (Figure 4B). To characterize its features as a transcription factor, we carried out a subcellular localization analysis, showing that the PtPHR1-GFP fusion protein was localized in the nucleus of N. benthamiana epidermal cells (Figure 4C). Then, the PtPHR1 was inserted into pGBKT7 (BD) and the BD-PtPHR1 constructor was transformed into the yeast strain AH109 (Qiao et al., 2017). The results showed that PtPHR1 possessed transcription activity, as did the positive control AtPHR1 (Figure 4D). To examine the binding ability of PtPHR1 to the P1BS cis-element, we co-transformed pH is 2.1–4× P1BS (bait) and pGADT7-Rec2-PtPHR1 (prey) into AH109 yeast cells. When spotted on selective media lacking Leu, the colonies grew well with an addition of 3-AT, irrespective of 5 or 50 mM. These results demonstrated that PtPHR1, behaving like AtPHR1, could bind to the P1BS cis-element in yeast (Figure 4E). Furthermore, we measured the expression of PtPHR1 after Pi deficiency and found that the transcript level of PtPHR1 in the tuber, roots, and leaves was not changed between high and low Pi conditions for 3- or 7-day culture (Supplementary Figure S6).
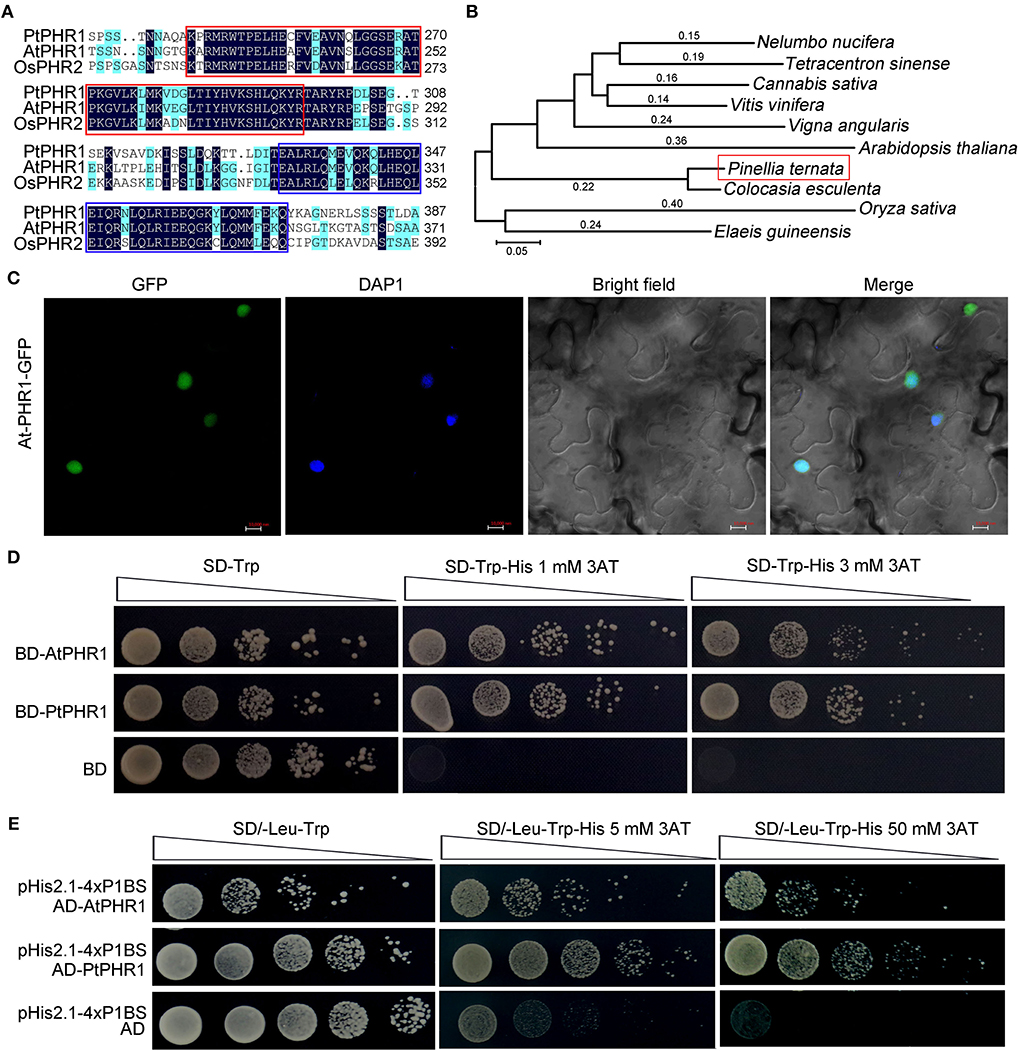
Figure 4. Characterization of PtPHR1 structure, evolution, localization, and activity. (A) Multiple alignments of PtPHR1, AtPHR1, and OsPHR2. Similar residues are shaded blue and identical residues are shaded black. The conserved MYB-domain (red) and predicted CC domain (purple) are shown. (B) Phylogenetic analysis of PtPHR1 amino acid sequences used the neighbor-joining method. The accession numbers of sequences used are as follows: Arabidopsis thaliana, NP_194590.2; Oryza sativa, XP_015647735.1; Nelumbo nucifera, XP_010264912.1; Tetracentron sinense, KAF8394238.1; Cannabis sativa, XP_030498437; Vitis vinifera, XP_002270511.1; Vigna angularis, XP_017411143.1; Colocasia esculenta, MQM19898.1 and Elaeis guineensis, XP_010920302.1. (C) Nuclear localization of PtPHR1-GFP. Bars = 10 μm. (D) PtPHR1 has transcriptional activation activity in yeast. The ORF of PtPHR1 was cloned into the vector pGBKT7 (BD). AtPHR1 was used as a positive control. Transformed yeasts were serially diluted and selected on media lacking Trp and His containing 3-aminotriazole (3AT). (E) Yeast one-hybrid assays showing interaction between PtPHR2 and P1BS cis-element. AtPHR1 was used as a positive control and empty pGADT7-Rec2 (pAD) vectors were used as a negative control.
PtPHR1 regulates Pi starvation response in Arabidopsis
To further investigate the role of PtPHR1 in Pi signaling, we transformed PtPHR1 fused with myc-tag into the phr1 mutant to generate PtPHR1OE/phr1 transgenic plants (Figure 5A, Supplementary Figure S7). We observed that the phenotype of the phr1 mutants was partly rescued with the transformation of the PtPHR1-myc plasmid when grown in Pi starvation conditions (Figure 5A). We then analyzed the transcript level of the PSI genes and anthocyanin accumulation in phr1 and PtPHR1/phr1 plants grown on low P media. The results showed that the expression of AtIPS1 and AtPT2 were elevated by Pi starvation (Figures 5B,C). Besides, AtDFR expression in the PtPHR1OE/phr1 plants was more responsive to Pi starvation compared to that in the phr1 mutant (Figure 5D). Consistently, the Pi starvation-induced anthocyanin accumulation was recovered in PtPHR1OE/phr1 plants (Figure 5E). These results indicated that heterologous expression of PtPHR1 could modulate Pi signaling and anthocyanin biosynthesis in Arabidopsis.
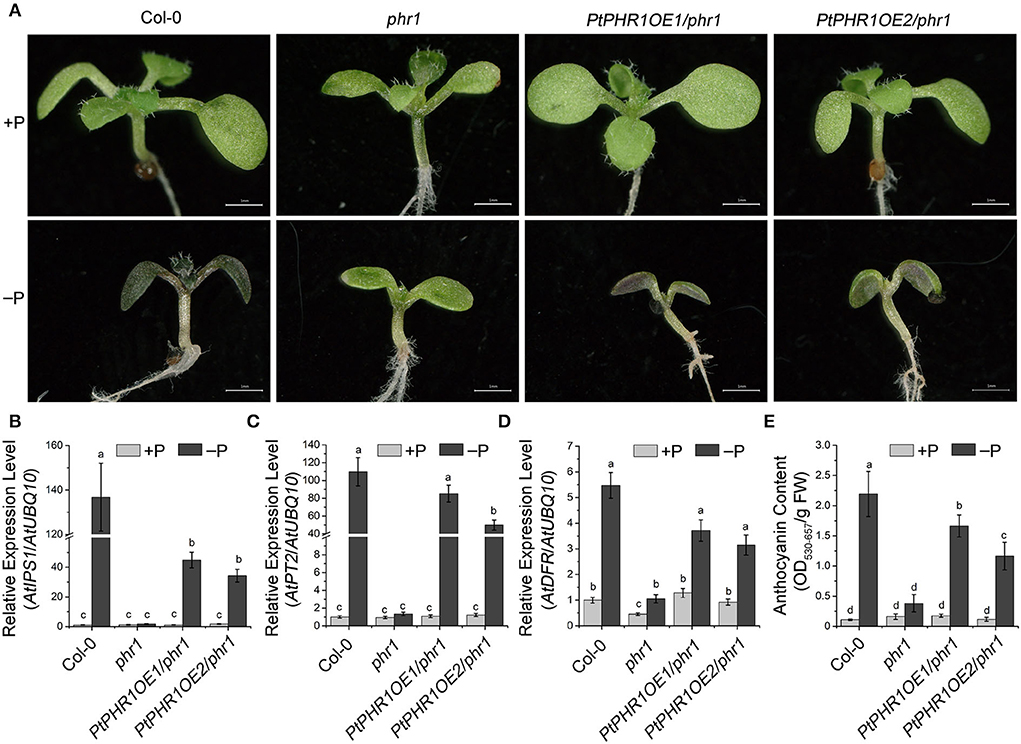
Figure 5. PtPHR1 regulates PSI gene induction and anthocyanin accumulation. (A) Images of the 9-day-old Col-0, phr1, and PtPHR1OE/phr1 seedlings grown in +P and –P media. A high-level accumulation of anthocyanins is indicated by the purple color. Bars, 500 mm. (B–D) Pi signaling gene and AtDFR expression induction levels in the Col-0, phr1, and phr1PtPHR1 seedlings grown in the –P conditions relative to those in +P conditions for 9 days; the transcript levels were analyzed by qRT-PCR. Values are means ± SD of three biological replicates. Different letters indicate significant differences at P < 0.05 by Fisher's least significant difference tests. (E) Anthocyanin content in the Col-0, phr1, and phr1PtPHR1 plants are grown in the +P/–P condition for 9 days. Values are means ± SD of three biological replicates. P < 0.05, Fisher's least significant difference test.
Involvement of PtPHR1 in Pi starvation-induced BA accumulation in Arabidopsis
In Arabidopsis, the homologs of AtPAL, AtCHY1, and ARABIDOPSIS ALDEHYDE OXIDASE4 (AtAAO4) are involved in BA biosynthesis (Facchini et al., 2004; Ibdah and Pichersky, 2009; Ibdah et al., 2009; Krizevski et al., 2010; Fraser and Chapple, 2011). Given that Pi starvation enhanced BA accumulation in P. ternata (Figure 3B), we investigated the impact of Pi signaling and PtPHR1 on alkaloid biosynthesis. Among Col-0, phr1, and PtPHR1OE/phr1 plants, the transcript level of AtPAL and AtAAO4 was strongly increased in Col-0 and PtPHR1OE/phr1 seedlings grown in Pi-starved medium for 9 days compared with those grown in Pi-sufficient medium, and no obvious induction was observed in phr1 mutant seedlings (Figure 6A). However, the expression pattern of AtCHY1 exhibited relatively constant among different genotypes, irrespective of Pi supply (Supplementary Figure S8). Consistently, BA accumulation was enhanced in Col-0 and PtPHR1OE/phr1 plants but not in the phr1 mutant (Figure 6B, Supplementary Figure S9). These results demonstrated that PtPHR1, at least partly, regulated Pi signaling-mediated BA biosynthesis.
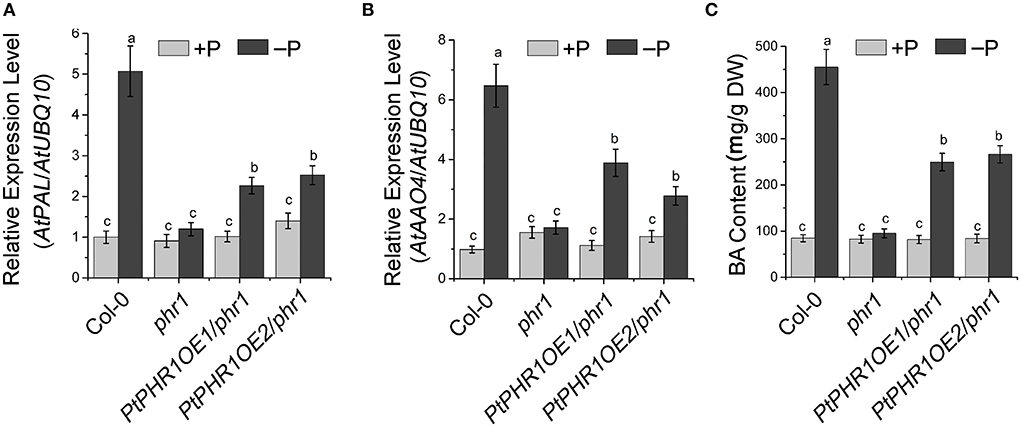
Figure 6. PHR1 is involved in Pi starvation-induced BA biosynthesis. Expression level of AtPAL (A) and AtAAO4 (B) and BA content (C) in 9-day-old Col-0, phr1, and PtPHR1OE/phr1 seedlings grown in +P and –P conditions. The transcript levels were analyzed by qRT-PCR. Values are means ± SD of three biological replicates. Different letters indicate significant difference at P < 0.05 by Fisher's least significant difference test.
Discussion
It has been widely reported that in plants, the regulation of metabolism by Pi signaling is involved in the adaptation to environmental stress (Baek et al., 2017). Pi starvation promotes sugar accumulation, and in turn sucrose elevates the transcript levels of Pi transporters and PSI genes (Hammond and White, 2008). Pi starvation also affects plant hormone biosynthesis, through the effects of auxin (Pérez-Torres et al., 2008), cytokinin (López-Bucio et al., 2002), and gibberellins (Jiang et al., 2007) in modifying the root system architecture formation; and through the effects of jasmonate acid in preventing attacks from insect herbivory and pathogen (Khan et al., 2016; Kong et al., 2021). In addition, Pi starvation results in increased flavonoid accumulation, especially excessive anthocyanin (Bustos et al., 2010; Khan et al., 2016; He et al., 2021). Here, we also observed that Pi-starvation enhanced the expression of flavonoid biosynthesis genes and the accumulation of flavonoids in P. ternata (Supplementary Figure S1). Importantly, we demonstrated that Pi starvation could promote alkaloid metabolism in P. ternata (Figures 2, 3).
In this work, we characterized the full-length sequence of the PtPHR1 gene. There were several common features shared by PtPHR1 and other PHR homologs (Rubio et al., 2001; Zhou et al., 2008). Along with the existence of both the MYB domain and CC domain, PtPHR1 was further suggested as a member of the MYB-CC family by sequence comparison analysis (Figure 4A). Based on the chloroplast genome sequence analysis of P. ternata and C. esculenta, PtPHR1 was identified as a distinct member of the Asteraceae via phylogenetic tree analysis (Figure 4B) (Han et al., 2016). In addition, we demonstrated that PtPHR1 was localized to the nucleus and possessed transcription activity (Figures 4C,D), and we pointed out that PtPHR1 could directly bind to the P1BS cis-element in yeast (Figure 4E). Furthermore, the steady expression of PtPHR1 was not very responsive to Pi deprivation (Supplementary Figure S6), consistent with previously reported findings (Rubio et al., 2001; Zhou et al., 2008). Together, these data indicated that PtPHR1 was conserved as plant PHRs and might play an important role in P. ternata during the Pi starvation response.
In Arabidopsis, AtPHR1 has been identified as the core mediator in Pi signaling and as a regulator of PSI genes (Bustos et al., 2010). It has been reported that the mutation of AtPHR1 could lead to the impaired responsiveness of PSI genes to Pi starvation (Rubio et al., 2001; He et al., 2021). Here, we expressed PtPHR1 in the phr1 mutant to generate PtPHR1OE/phr1 plants, which showed that heterologous expression of PtPHR1 could partially rescue the phenotype of phr1 in P-starved conditions (Figure 5). We also observed that the Pi starvation elevated PSI gene expression and anthocyanin content in PtPHR1OE/phr1 was partially recovered to the level of wild-type plants (Figure 5). These data together indicated that PtPHR1 is a functional homolog of AtPHR1, and this is consistent with the conception that the transcriptional component of the Pi homeostasis regulatory network may be conserve in plants (Zhou et al., 2008). In this study, we demonstrated that PtPHR1 was involved in Pi starvation-induced alkaloid biosynthesis. We found that Pi starvation increased the expression level of AtAAO4 and the production of BA, whereas these activities were impaired by the loss-function of PHR1 (Figure 6). The findings indicated that Pi starvation-mediated BA biosynthesis was controlled by AtPHR1. Furthermore, suppression of BA biosynthesis in phr1 mutant could be rescued by the heterologous expression of PtPHR1 (Figure 6), which suggested that PtPHR1 behaved in a manner similar to that of AtPHR1 in the regulation of BA biosynthesis. It has been demonstrated that BA is an intermediate in the formation of alkaloids (Krizevski et al., 2010), and PtAO4 is also reported to be involved in BA and ephedrine biosynthesis in P. ternata (Zhang et al., 2016). Thus, we speculated that PtPHR1 mediated Pi starvation-induced BA-derived alkaloid biosynthesis in P. ternata.
Because of its extensive pharmacological activity, increasing concerns have been focused on improving alkaloid production. As the main bioactive compound in P. ternata, the research progress in the regulatory of alkaloid metabolism has been described (Miao et al., 2013). Tissue culture materials, such as calluses and protocorm-like bodies, accumulate greater amounts of alkaloid than that of field-grown tubers, and different combinations of 6-benzyladenine, kinetin, α-naphthaleneacetic acid, and 2,4-dichlorophenoxyacetic acid also impaired alkaloid metabolism (Liu et al., 2010, 2015). Application of salicylic acid or methyl jasmonate enhances alkaloids accumulation in P. ternata grown in field and suspension tuber, as well as in in vitro cultured microtubers (Duan et al., 2017, 2019). In this work, we demonstrated that Pi starvation played a positive role in alkaloid accumulation in P. ternata. We also found that Pi starvation-induced BA accumulation in both P. ternata and Arabidopsis (Figure 6). BA not only severs as intermediate compounds for alkaloid but also is the backbone of numerous compounds in plants, including taxol, cocaine, methylbenzoate, and benzylbenzoate (Ibdah et al., 2009; Del Olmo et al., 2017). BA and its derivatives with high antibacterial and antifungal activity, are commonly used as preservatives and medicines (Del Olmo et al., 2017). Thus, the activation of the BA, as well as alkaloid biosynthesis, by Pi starvation would have a broad ecological and evolutionary consequence on P. ternata or closely related species. Also, the regulation of the alkaloid accumulation by the PHRs would provide new insights into the linkage between secondary metabolism and nutrient supply.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number ON075805.
Author contributions
GH and YH conceived the project, designed the experiments, and wrote the manuscript. HW, JH, and YH carried out the experiments with assistance from LL, XZ, HZ, ZL, and QS. HW, JH, LL, XZ, HZ, ZL, QS, YH, and GH analyzed and discussed the results. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Zhejiang Provincial Natural Science Foundation of China (LR22C020003), the Major Science and Technology Special Project of Variety Breeding of Zhejiang Province (2021C02067-7), National Natural Science Foundation of China (32000234, 31670291, and 31800249), and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2021DG700024-KF202102).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.914648/full#supplementary-material
References
Baek, D., Chun, H. J., Yun, D. J., and Min, C. K. (2017). Cross-talk between phosphate starvation and other environmental stress signaling pathways in plants. Mol. Cells 40, 697–705. doi: 10.14348/molcells.2017.0192
Boatright, J., Negre, F., Chen, X., Kish, C. M., Wood, B., Peel, G., et al. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135, 1993–2011. doi: 10.1104/pp.104.045468
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Pérez-Pérez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate ptarvation in Arabidopsis. PLoS Genet. 6, e1001102. doi: 10.1371/journal.pgen.1001102
Chinese Pharmacopoeia Commission (2005). Pharmacopoeia of the People's Republic of China. Beijing: People's Medical Publishing House.
Cong, W., Suriyagoda, L., and Lambers, H. (2020). Tightening the phosphorus cycle through phosphorus-efficient crop genotypes. Trends Plant Sci. 25, 967–975. doi: 10.1016/j.tplants.2020.04.013
Crombez, H., Motte, H., and Beeckman, T. (2019). Tackling plant phosphate starvation by the roots. Dev. Cell 48, 599–615. doi: 10.1016/j.devcel.2019.01.002
Del Olmo, A., Calzada, J., and Nuñez, M. (2017). Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 57, 3084–3103. doi: 10.1080/10408398.2015.1087964
Duan, Y., Zhang, H., Meng, X., Huang, M., Zhang, Z., Huang, C., et al. (2019). Accumulation of salicylic acid-elicited alkaloid compounds in in vitro cultured Pinellia ternata microtubers and expression profiling of genes associated with benzoic acid-derived alkaloid biosynthesis. Plant Cell Tissue Organ Culture (PCTOC) 139, 317–325. doi: 10.1007/s11240-019-01685-5
Duan, Y. B., Fang, L. U., Cui, T. T., Zhao, F. L., Teng, J. T., Sheng, W., et al. (2017). Effects of abiotic elicitors MeJA and SA on alkaloids accumulation and related enzymes metabolism in Pinellia ternata suspension cell cultures. Chin. J. Inform. Tradit. Chin. Med. 24, 87–90. doi: 10.3969/j.issn.1005-5304.2017.01.021
Facchini, P. J., Bird, D. A., and St-Pierre, B. (2004). Can Arabidopsis make complex alkaloids? Trends Plant Sci. 9, 116–122. doi: 10.1016/j.tplants.2004.01.004
Fraser, C. M., and Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9, e0152. doi: 10.1199/tab.0152
Frerigmann, H., Piotrowski, M., Lemke, R., Bednarek, P., and Schulze-Lefert, P. (2021). A network of phosphate starvation and immune-related signaling and metabolic pathways controls the interaction between Arabidopsis thaliana and the Beneficial Fungus Colletotrichum tofieldiae. Mol. Plant-Microbe Interact. 34, 560–570. doi: 10.1094/MPMI-08-20-0233-R
Ge, X., and Hao, W. (2009). Phytochemical properties and quality evaluation methods of Pinelliae ternate. China Pharm. 18, 3. doi: 10.3969/j.issn.1006-4931.2009.09.002
Hammond, J., and White, P. (2008). Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J. Exp. Bot. 59, 93. doi: 10.1093/jxb/erm221
Han, L., Chen, C., Wang, B., and Wang, Z. (2016). The complete chloroplast genome sequence of medicinal plant Pinellia ternata. Mitochondrial DNA 27, 2921–2922. doi: 10.3109/19401736.2015.1060441
He, Y., Hong, G., Zhang, H., Tan, X., Li, L., Kong, Y., et al. (2020). The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4. Plant Cell 32, 2806–2822. doi: 10.1105/tpc.19.00499
He, Y., Zhang, X., Li, L., Sun, Z., Li, J., Chen, X., et al. (2021). SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 230, 205–217. doi: 10.1111/nph.17139
Ibdah, M., Chen, Y., Wilkerson, C., and Pichersky, E. (2009). An aldehyde oxidase in developing seeds of Arabidopsis converts benzaldehyde to benzoic acid. Plant Physiol. 150, 416–423. doi: 10.1104/pp.109.135848
Ibdah, M., and Pichersky, E. (2009). Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds. Plant Biol. 11, 574–581. doi: 10.1111/j.1438-8677.2008.00160.x
Iwakoshi, K., Shiozawa, Y., Yamajima, Y., Baba, I., Monma, K., Kobayashi, C., et al. (2019). Determination of nine preservatives in processed foods using a modified QuEChERS extraction and quantified by HPLC-PDA. Food Addit. Contam.: Part A 36, 1020–1031. doi: 10.1080/19440049.2019.1615644
Ji, J., Hu, J., Chen, S., Liu, R., Wang, L., Cheng, J., et al. (2013). Development and application of a method for determination of nucleosides and nucleobases in Mactra veneriformis. Pharmacogn. Mag. 9, 96–102. doi: 10.4103/0973-1296.111241
Ji, X., Huang, B., Wang, G., and Zhang, C. (2014). The ethnobotanical, phytochemical and pharmacological profile of the genus Pinellia. Fitoterapia 93, 1–17. doi: 10.1016/j.fitote.2013.12.010
Jiang, C., Gao, X., Liao, L., Harberd, N. P., and Fu, X. (2007). Phosphate starvation root architecture and anthocyanin cccumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470. doi: 10.1104/pp.107.103788
Khan, G. A., Vogiatzaki, E., Glauser, G., and Poirier, Y. (2016). Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 171, 632–644. doi: 10.1104/pp.16.00278
Klempien, A., Kaminaga, Y., Qualley, A., Nagegowda, D. A., Widhalm, J. R., Orlova, I., et al. (2012). Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell 24, 2015–2130. doi: 10.1105/tpc.112.097519
Kong, Y., Wang, G., Chen, X., Li, L., Zhang, X., Chen, S., et al. (2021). OsPHR2 modulates phosphate starvation-induced OsMYC2 signaling and resistance to Xanthomonas oryzae pv. oryzae. Plant Cell Environ. 44, 3432–3444. doi: 10.1111/pce.14078
Krizevski, R., Bar, E., Shalit, O., Sitrit, Y., Ben-Shabat, S., and Lewinsohn, E. (2010). Composition and stereochemistry of ephedrine alkaloids accumulation in Ephedra sinica Stapf. Phytochemistry 71, 895–903. doi: 10.1016/j.phytochem.2010.03.019
Liu, Y., Liang, Z., and Zhang, Y. (2010). Induction and in vitro alkaloid yield of calluses and protocorm-like bodies (PLBs) from Pinellia ternata. In Vitro Cell. Dev. Biol. – Plant 46, 239–245. doi: 10.1007/s11627-009-9268-9
Liu, Y., Liu, W., and Liang, Z. (2015). Endophytic bacteria from Pinellia ternata, a new source of purine alkaloids and bacterial manure. Pharm. Biol. 53, 1545–1548. doi: 10.3109/13880209.2015.1016580
López-Bucio, J., Hernandezabreu, E., Sanchezcalderon, L., Nietojacobo, M. F., Simpson, J., and Herreraestrella, L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. doi: 10.1104/pp.010934
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through Its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. doi: 10.1105/tpc.114.123208
Mao, R., and He, Z. (2020). Pinellia ternata (Thunb.) Breit: a review of its germplasm resources, genetic diversity and active components. J. Ethnopharmacol. 263, 113252. doi: 10.1016/j.jep.2020.113252
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Müller, M., Gocke, D., and Pohl, M. (2009). Thiamin diphosphate in biological chemistry: exploitation of diverse thiamin diphosphate-dependent enzymes for asymmetric chemoenzymatic synthesis. FEBS J. 276, 2894–2904. doi: 10.1111/j.1742-4658.2009.07017.x
Neumann, G., and Römheld, V. (2012). “Rhizosphere chemistry in relation to plant nutrition,” in Marschner's Mineral Nutrition of Higher Plants, 3rd Edn, ed H. Marschner (London: Academic Press). 347–368. doi: 10.1016/B978-0-12-384905-2.00014-5
Oshio, H., Tsukui, M., and Matsuoka, T. (1978). Isolation of l-ephedrine from Pinelliae tuber. Chem. Pharm. Bull. 26, 2096–2097. doi: 10.1248/cpb.26.2096
Pérez-Torres, C. A., Lopez-Bucio, J., Cruz-Ramirez, A., Ibarra-Laclette, E., Dharmasiri, S., Estelle, M., et al. (2008). Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 Auxin receptor. Plant Cell 20, 3258–3272. doi: 10.1105/tpc.108.058719
Puga, M. I., Isabel, M., Rajulu, C., Zhiye, W., Franco-Zorrilla, J. M., Laura, D. L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29, 292–309. doi: 10.1105/tpc.16.00611
Ruan, W., Guo, M., Wang, X., Guo, Z., Xu, Z., Xu, L., et al. (2019). Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant 12, 1060–1074. doi: 10.1016/j.molp.2019.04.003
Rubio, V., Linhares, F., Solano, R., Martín, A., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Salmanzadeh, M., Sabet, M. S., Moieni, A., and Homaee, M. (2020). Heterologous expression of an acid phosphatase gene and phosphate limitation leads to substantial production of chicoric acid in Echinacea purpurea transgenic hairy roots. Planta 251, 31. doi: 10.1007/s00425-019-03317-w
Sun, L., Zhang, B., Wang, Y., He, H., and Chen, X. (2018). Metabolomic analysis of raw Pinelliae Rhizoma and its alum-processed products via UPLC-MS and their cytotoxicity. Biomed. Chromatogr. 33, e4411. doi: 10.1002/bmc.4411
Tian, C., Zhang, Z., Huang, Y., Xu, J., Liu, Z., Xiang, Z., et al. (2022). Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco. Plant Physiol. Biochem. 177, 1–9. doi: 10.1016/j.plaphy.2022.02.017
Van, M., Schauvinhold, I., Pichersky, E., Haring, M., and Schuurink, R. (2009). A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 60, 292–302. doi: 10.1111/j.1365-313X.2009.03953.x
Wang, J., Wang, Q., Wang, J., Lu, Y., Xiao, X., Gong, W., et al. (2009). Effect of different plant growth regulators on micro-tuber induction and plant regeneration of Pinellia ternate (Thunb) Briet. Physiol. Mol. Biol. Plants 15, 359–365. doi: 10.1007/s12298-009-0040-8
Wu, F., Qin, Z., Li, Y., Chen, T., Pei, B., and Wei, P. (2013). Progress of the anti-tumor effect on chemical constituents of Pinellia ternate. Chin. Arch. Tradit. Chin. Med. 30, 270–272.
Xu, W., Zhang, B., Mian, L. I., and Liu, G. H. (2007). Determination of ephedrine in rhizoma pinelliae, rhizoma typhonii flagelliformis and their processed products by HPLC. Lishizhen Medicine and Meteria Medica Research 18, 884–885
Xue, T., Zhang, H., Zhang, Y., Wei, S., Chao, Q., Zhu, Y., et al. (2019). Full-length transcriptome analysis of shade-induced promotion of tuber production in Pinellia ternata. BMC Plant Biol. 19, 565. doi: 10.1186/s12870-019-2197-9
Yuan, H., and Dong, D. (2008). Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 50, 849–859. doi: 10.1111/j.1744-7909.2008.00709.x
Zhang, G., Jiang, N., Song, W., Ma, C., Yang, S., and Chen, J. (2016). De novo sequencing and tsranscriptome analysis of Pinellia ternata identify the candidate genes involved in the biosynthesis of benzoic acid and ephedrine. Front. Plant Sci. 7, 1209. doi: 10.3389/fpls.2016.01209
Keywords: Pinellia ternata (Thunb.) Berit, alkaloid metabolism, benzoic acid (BA), phosphate signaling, PtPHR1
Citation: Wang H, Hu J, Li L, Zhang X, Zhang H, Liang Z, Sheng Q, He Y and Hong G (2022) Involvement of PtPHR1 in phosphates starvation-induced alkaloid biosynthesis in Pinellia ternata (Thunb.) Breit. Front. Plant Sci. 13:914648. doi: 10.3389/fpls.2022.914648
Received: 07 April 2022; Accepted: 11 July 2022;
Published: 10 August 2022.
Edited by:
Naveen C. Bisht, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Yasuyuki Yamada, Kobe Pharmaceutical University, JapanVineeta Tripathi, Central Drug Research Institute (CSIR), India
Copyright © 2022 Wang, Hu, Li, Zhang, Zhang, Liang, Sheng, He and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing He, eXFoZV8xMjNAMTI2LmNvbQ==; Gaojie Hong, Z2pob25nQDEyNi5jb20=
†These authors have contributed equally to this work
 Huihui Wang
Huihui Wang Jitao Hu1,2†
Jitao Hu1,2† Linying Li
Linying Li Yuqing He
Yuqing He Gaojie Hong
Gaojie Hong