- 1School of Biomedical Sciences, Huaqiao University, Quanzhou, China
- 2Crops Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 3Root Biology Center, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Fujian Polytechnic Normal University, Fuqing, China
- 5Department of Cardiovascular Surgery, First Hospital of Quanzhou Affiliated to Fujian Medical University, Quanzhou, China
Baphicacanthus cusia (Nees) Bremek (B. cusia) is an important medicinal plant. Its effective substances including indigo and indirubin are metabolites in indoleacetate metabolic pathway. Based on a previous transcriptome sequencing analysis, a WRKY transcription factor, BcWRKY1, in B. cusia was identified, showing significant correlation with effective substances from B. cusia. In this study, BcWRKY1 was cloned by reverse transcription-polymerase chain reaction (RT-PCR). Further analysis showed that the BcWRKY1 gene was 916 bp in length, containing three exons and two introns. The open reading frame (ORF) of BcWRKY1 was 534 bp in length and encoded a WRKY domain-containing protein with 177 amino acids residues. Subcellular localization showed that BcWRKY1 protein was mainly localized in the nucleus. It could bind to the W-box motif and its role in transcriptional activation was confirmed in yeast. The function of BcWRKY1 was investigated by overexpressing BcWRKY1 in Arabidopsis thaliana. Metabolic profiles in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana were analyzed with LC-MS. Results showed that the metabolic profile was significantly changed in BcWRKY1-OX1 transgenic Arabidopsis thaliana compared with wild type. Furthermore, indole-related metabolites were significantly increased in BcWRKY1-OX1 transgenic Arabidopsis thaliana, and the metabolic pathway analysis showed that flavonoid biosynthesis was significantly enriched. Overexpression of BcWRKY1 significantly changed flavonoid and indole metabolism and indole-related metabolites were significantly upregulated. We postulated that the BcWRKY1 transcription factor might be involved in the regulation of effective substances metabolism in B. cusia.
Introduction
Baphicacanthus cusia(Nees) Bremek (B. cusia) (Supplementary Figure 1), also known as south isatis root, belongs to the Acanthaceae family (Huang et al., 2009). It has many medicinal properties (Lin et al., 2019). The leaves and stems of the plant are used to extract Indigo naturalis (Qing-Dai), while the roots are used for the production of traditional Chinese medicine (TCM), which has been recorded in the Chinese Pharmacopeia (China Pharmacopoeia Commission, 2020). Qing-Dai (indigo and indirubin) has the functions of purging fire, clearing heat, and detoxification, as well as analgesic and anti-inflammatory effects (Li et al., 2011). The roots of B. cusia and Isatis indigotica Fortune are termed as south and north isatis root, respectively, exhibiting antiviral, antibiosis and anti-inflammatory properties (Shen, 2009). Previous study has shown the effective substances in B. cusia. For example, indirubin can treat chronic myelogenous leukemia (Wang et al., 2014a), while its derivative 6-bromo indirubin-3′-oxime can effectively inhibit the growth of ovarian cancer cells (Yu and Zhao, 2015; Zeng and Diao, 2016). Other effective substances, such as indigo and tryptanthrin in B. cusia, were demonstrated to exhibit anti-inflammatory activities in vitro (Ishihara et al., 2000; Danz et al., 2001; Liu et al., 2014; Huang et al., 2017). Indigo and indirubin are belonging to indole alkaloids. It has been reported that the synthesis of indole alkaloids in plants is controlled by DXR, SLS, G10H, TAA1, YUC1 and so on (Han et al., 2007; Wang et al., 2015). Flavonoids are isolated from the ethanol extract of B. cusia (Liu et al., 2009). Flavonoids are also important secondary metabolites with effective substance in medicinal plant, which is usually regulated by PAL, CHS, CHI, ANR, FLS and FTH (Qiao et al., 2009; Zou et al., 2016; Zhang et al., 2017). Taken together, B. cusia is an important TCM herb of Chinese traditional medicine and has been widely used to treat various diseases.
Transcription factors (TFs) are proteins that regulate gene expressions at the transcriptional level. It is reported that TFs participate in the regulation of essential physiological metabolism (Liu et al., 2001; Mitsuda and Ohme-Takagi, 2009; Ma et al., 2015). TFs regulate the expression of target genes through binding to cis-acting elements to activate or repress the expression of downstream genes (Priest et al., 2009). WRKY TF is one of the largest TF families in higher plants, which contain a conserved WRKYGQK sequence in the DNA-binding domain (Eulgem et al., 2000; Rushton et al., 2010; Wen et al., 2017b). It has been reported that WRKY TFs specifically bind to the cis-acting element W-box [(T)TGAC(C/T)] in the promoter region of the target gene through the WRKY domain, thus activating or inhibiting transcription and regulating the expression of downstream genes (Bakshi and Oelmüller, 2014; Huang et al., 2019). It is believed that WRKY TFs are widely involved in the responses to various biotic and abiotic stresses, and hormonal signaling, regulating plant growth and development (Xiong et al., 2014; Gu et al., 2015; Wani et al., 2016; Jiang et al., 2017; Finatto et al., 2018). Besides, more and more research has shown that WRKY TFs also participate in secondary metabolic regulation such as indole alkaloid production in plants (Xu et al., 2004; Suttipanta et al., 2011). Moreover, the preliminary results from our group indicated a significant correlation of BcWRKY1 with indole metabolic pathway in B. cusia (Huang, 2017). Therefore, we postulated that BcWRKY1 might be involved in the metabolism of effective substance in B. cusia. In this study, BcWRKY1, which showed a high association with indole metabolic pathway in B. cusia, was cloned, and its function was validated in plant Arabidopsis model, especially its influence on metabolite profiles and indole metabolic pathway in Arabidopsis was investigated. These results conferred the understanding of the role of BcWRKY1 in regulating the effective substance in B. cusia.
Materials and Methods
Plant Material
The plant material, B. cusia, was collected from the Field experimental station of Huqiao University in Shufeng town of Fujian Province, China (25°25′N 118°39′E).
Primer Design
Based on the sequencing data of B. cusia transcriptome (accession number SRR4428209) and the full length of BcWRKY1 gene (c16427_g1_i1), the degenerate primers WRKY-F-XhoI and WRKY-R-BamHI were designed using Premier Premier 5.0 (Supplementary Table S2). The primer sequence was synthesized by Sunya Biotechnology Company (Fuzhou, China).
Total RNA Extraction and cDNA Synthesis
Total RNA of B. cusia was extracted with TRIzol method according to the following protocol: A total of 0.1 g leaves of B. cusia were homogenized with liquid nitrogen. The homogenate was mixed with an additional 1 mL TRIzol and incubated at room temperature for 1 min. Subsequently, 200 μL chloroform was added to the mixture and incubated at room temperature for 3–5 min. The mixture was centrifuged at 12,000 ×g for 15 min at 4 °C; 500 μL of the supernatant was mixed with 500 μL isopropyl alcohol. Then 1 mL of 75% ethanol was added after discarding the supernatant and the mixture was centrifuged at 12, 000 ×g for 5 min at 4 °C, twice. The precipitate was air-dried, solubilized in 25–30 μL ddH2O, and maintained at 4 °C for 1–2 h (Ma et al., 2008). Finally, the RNA was analyzed by 1 % agarose gel electrophoresis and the quality and concentration were determined by Nanodrop 2000 UV-vis Spectrophotometer (Thermo Scientific, Wilmington, USA). The remaining was stored at −80 °C until utilized for cDNA synthesis by reverse transcription using Trans Script One-Step gDNA Removal and cDNA Synthesis Super Mix (Beijing TransGen Biotech Co., Ltd.) according to the manufacturer's instructions.
PCR Amplification
The PCR reaction system included 25 μL 2 × Buffer, 5 μL dNTPs, 2 μL F1 primer, 2 μL R1 primer, 4 μL cDNA, and 1 μL enzyme, with 11 μL ddH2O in a reaction volume of 50 μL. The PCR reaction conditions were as follows: initial denaturation for 5 min at 95 °C, followed by 40 cycles of denaturation at 94 °C for 20 s, annealing at 58 °C for 20 s, and extension at 68 °C for 40 s. The final extension was carried out for 5 min at 68 °C. The PCR products were detected by 1 % agarose gel electrophoresis.
Cloning and Sequencing of BcWRKY1
The target PCR fragment was recovered using Easy Pure Quick Gel Extraction Kit (Beijing TransGen Biotech Co., Ltd.). The DNA was eluted, and the recovered fragment was ligated to the pEASY-Blunt vector (Beijing TransGen Biotech Co., Ltd.) and transformed into Trans1-T1 (Beijing TransGen Biotech Co., Ltd.). The positive clones were sent to BoShang Biotechnology (Fuzhou, China) for sequencing.
Bioinformatics Analysis
The ORF finder (NCBI) was used for identifying the ORF of BcWRKY1 (Gao et al., 2009). Protparam (www.expasy.org/tools/protparam.html) was used for predicting the physical and chemical properties of the BcWRKY1 amino acid sequence (Wang et al., 2013). Eslpred (Bhasin and Raghava, 2004) (http://www.imtech.res.in/raghava/eslpred/) was used for predicting the subcellular localization of BcWRKY1 protein. DAS.TMfilter (https://mendel.imp.ac.at/DAS/) was used for analyzing the transmembrane protein structure (Cserzö et al., 2002). Signal P4.1 (http://www.cbs.dtu.dk/services/SignalP/) was applied to predict the signal peptide (Shi et al., 2012). Protein functional sites were analyzed by MotifScan (https://myhits.sib.swiss/cgi-bin/motif_scan). NPS@Network Protein Sequence Analysis (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) was used to predict the secondary structure of the protein (Lv et al., 2016). SWISS-MODEL was used to predict the three-dimensional structure of BcWRKY1 (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006). The search for protein sequence similarities of BcWRKY1 was conducted using BLAST algorithm at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). The neighbor-joining method (Mega 5.10) was used for constructing a phylogenetic tree (Shang et al., 2013; Kumar et al., 2016).
Subcellular Localization
BcWRKY1 was amplified from the cDNA of B. cusia with primers WRKY-for-5941GFP-F and WRKY-for-5941GFP-R (Table S2), cloned into the intermediate 5941-35S-GFP vector. The 5941-35S-BcWRKY1-GFP plasmid was constructed by inserting the BcWRKY1 to the AscI restrict sites of the 5941-35S-GFP vector, and 5941-35S-OsSPX1-RFP was used a marker (Wang et al., 2014b). The positive clone was introduced into Agrobacterium EHA105, cultured, and injected into 4-week-old tobacco leaves, followed by culturing at 22–28 °C for 48 h. Then, the infected leaves were observed under a confocal laser scanning microscope.
DNA Binding Assay
For the verification of DNA binding activity of BcWRKY1 as a WRKY family transcription factor, the Electrophoretic Mobility Shift Assay (EMAS) assay was employed. First, the BcWRKY1 was sub cloned to the pGEX-4T-1 prokaryotic expression vector using EcoRI restriction sites with primer pair GST-WRKY-F and GST-WRKY-R (Supplementary Table S2). and then the resulting vector GST-WRKY was transformed into the BL21 (DE3) cells. The expression and purification of recombinant protein was conducted according to the study by Lv et al. (2014). The classic WRKY binding motif W-box was synthesized and labeled with biotin by SUNYA company, and the sequence of ploynucleatides including W-box was listed in Supplementary Table S2. EMSA was performed using LightShift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer's instructions. The biotin-labeled probes were detected using chemiluminescence substrate (Thermo Scientific) and the ChemDoc XRS system (Bio-Rad).
Transcriptional Activation Activity
The BcWRKY1 gene was amplified by PCR using primers WRKY-R-for-BD-F and WRKY-F-FOR-BD-R, with 40 amplification cycles as previously described (Supplementary Table S2). The pGBKT7-BcWRKY1 vector was constructed, and the pGBKT7-BcWRKY1 and pGBKT7 plasmids were respectively transformed into the AH109 competent cells, and then invertedly cultured at 30 °C for 48–72 h. Yeast plaques on SD/-trp medium were observed and photos were taken.
Real-Time Fluorescence Quantitative PCR
Total RNA were extracted from wild type and transgenic Arabidopsis overexpressing BcWRKY1 gene using RNAiso Plus reagent (TaKaRa Bio). The cDNA was further synthesized through reverse transcription reaction and quantitative PCR using TransStart® Tip Green qPCR SuperMix (TransGen Biotech, Beijing), according to the instruction of the manufacturer. Real-time fluorescence quantitative PCR was carried out on the LightCycler96 (Roche Diagnostics) PCR system. Additionally, qPCR reactions were carried out in 25 μL reaction system containing 10 μL SYBR Premix, 0.4 μL forward and reverse primers, 4 μL cDNA template, and 0.4 μL Rox, using ddH2O to adjust to 25 μL. The PCR program were set as 94 °C for 5 min, followed by 44 cycles of 94 °C 20 s, 58 °C 20 s, and 72 °C 20 s. Totally, there were three replicates for each biological analysis. The Actin gene was selected as a reference gene (Sun et al., 2007). The expression of BcWRKY1 was calculated using the methods described by Livak and Schmittgen (2001). Primers used for qPCR are listed in Supplementary Table S2.
Gene Function Verification
BcWRKY1 overexpression transgenic Arabidopsis thaliana line was obtained by transforming recombinant plasmid 5941-35S-BcWRKY1-GFP into Rdr6 wild type Arabidopsis thaliana using Agrobacterium-mediated floral dipping method (Bent, 2006). The collected T1 generation seeds on MS medium containing Glufosinate were cultured in an artificial growth chamber and the positive transgenic seedlings were further identified with fluorescence microscopy (22 °C 18 h in the day, 20 °C 6 h at night). The metabolites in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana (T2 generation) were analyzed by UPLC-MS at the Novogene Institute (Beijing, China). The offline data (raw-data) file was imported into the CD (Compound Discoverer TM2.0) search software to simply screen the retention time and mass-charge ratio, followed by peak alignment of different samples using a retention time deviation of 0.2 min and a mass deviation of 5 ppm (Thevenot et al., 2015). Peak extraction was carried out according to a set quality deviation of 5 ppm, a signal strength deviation of 30 %, the signal noise ratio, the minimum signal strength (100, 000), adduct ion and other information (Fraga et al., 2010; Chen et al., 2013; Zhu et al., 2013). At the same time, peak area was quantified, target ions were integrated, molecular formula was predicted and compared with mzCloud database (Ruttkies et al., 2016), background ions were removed by blank samples, quantitative results were normalized by QC samples, and finally the data identification and quantitative results were obtained. The experimental samples and QC samples were extracted respectively, and the corresponding peaks were obtained. After Pareto-scaling treatment, the data were analyzed by PCA. Partial Least Squares Discrimination Analysis (PLS-DA) was used to establish the PLS-DA model for each group (Chen et al., 2009; Tang et al., 2014). The Variable Importance in the Projection (VIP) value of the first principal component of PLS-DA model was used, and the P value obtained by T-test analysis was used to screen the differential metabolites between the two experimental groups (Thevenot et al., 2015), and the differential metabolites analysis table was made. A volcano map was plotted according to log2 (fold change) and -log10 (p-value). Considering KEGG Pathway as a unit, hypergeometric test was used to find out significantly enriched pathways associated with differential metabolites. The most important biochemical metabolic pathways and signal transduction pathways involved in differential metabolites could be determined by Pathway significant enrichment, and the KEGG pathway enrichment analysis map was drawn (Kanehisa and Goto, 2000; Wen et al., 2017a).
Statistical Analysis
Means and SE (Standard Error) values were calculated using GraphPad Prism version 7.0 (GraphPad Software Inc., San Diego, CA, USA; https://www.graphpad.com). The significantly differential metabolites between two groups were analyzed with DEseq2 package in R with absolute log2Foldchange > 1. Adjusted P value was calculated with FDR (False discovery rate). The two-tailed student's t test was used to compare the two samples.
Results
Cloning and Characterization of BcWRKY1
For the cloning of the BcWRKY1, the primers specifically for BcWRKY1 were designed according to the sequence of BcWRKY1 obtained through RNA-seq (Huang, 2017). PCR was performed with cDNA of B. cusia leaves using specific primer pair designed. To investigate the structure of BcWRKY1, PCR was also performed with genomic DNA of B. cusia. The results showed that the length of BcWRKY1 gene was 916 bp, containing a 534 bp ORF (Supplementary Figure 2A). BcWRKY1 contained three exons, size 257 bp, 151 bp, and 126 bp, respectively and two introns, 154 bp and 228 bp, respectively (Supplementary Figure 2B). BcWRKY1 gene encoded a protein with 177 amino acids residues (Supplementary Figure 2C). The composition and physicochemical characteristics of the deduced BcWRKY1 protein are compared in Supplementary Table S1.
Eslpred software prediction showed that BcWRKY1 protein was localized in the nucleus. DAS.TMfilter was further used to predict the transmembrane structure of the BcWRKY1 protein. The results showed that this protein did not have any transmembrane domains. Signal P4.1 prediction showed that BcWRKY1 did not contain a signal peptide. Motif Scan analysis showed that the protein contained a conserved WRKY domain (115-174 bits, E-value 2.6e-38). Network Protein Sequence Analysis (NPS) was used to predict the secondary structure of the protein, and the results demonstrated the presence of alpha helix (20.79 %), extended strand (15.73 %), and random coil (62.92 %) components (Supplementary Figure 3). These findings indicated that the secondary structure of BcWRKY1 primarily consisted of random coil and alpha-helix. The three-dimensional structure of BcWRKY1 obtained by SWISS-MODEL is shown in Supplementary Figure 2D.
Phylogenetic Analysis
Genes with close homology to BcWRKY1 were searched in NCBI and their protein sequences were downloaded to construct a phylogenetic tree with MEGA 5.10 software. ClustalX was used for multiple alignments and the Neighbor-Joining method with 1000 replicates of bootstrap testing (Saitou and Nei, 1987; Shang et al., 2013). The phylogenetic tree of BcWRKY1 showed that the BcWRKY1 of B. cusia was closely related to WRKY in Helianthus annuus (Supplementary Figure 2E). The results of multiple sequence alignment showed that the BcWRKY1 protein was highly conserved compared with its homologous proteins in Helianthus annuus, Ziziphus jujuba, Olea europaea var. Sylvestris, Prunus persica and Phoenix dactylifera, with a homology of 72.62, 73.17, 71.91, 73.68, and 75.34%, respectively (Supplementary Figure 4).
Subcellular Localization of the BcWRKY1 Protein
The vector containing 5941-35S-BcWRKY1-GFP and the empty vector containing 5941-35S-RFP were transformed into tobacco leaves for transient expressions of 5941-35S-BcWRKY1-GFP and 5941-35S-RFP. Then the tobacco leaves were cultured for 48 h, and observed under a confocal laser scanning microscope with GFP and RFP double channels. The results showed that 5941-35S-RFP was localized in both the nucleus and cytoplasm of tobacco epidermal cells (bright points), and 5941-35S-BcWRKY1-GFP was localized in the RFP bright spot where the nucleus was labeled (Figure 1A). This finding was in line with the protein subcellular localization prediction results with Eslpred, which suggested that BcWRKY1 protein was most likely to be in the nucleus. Meanwhile, BcWRKY1-GFP was further co-labeled with 5941-35S-OsSPX1-RFP that was reported to be specially localized in the nucleus (Wang et al., 2014b). In addition, OsSPX1-RFP was detected in the nucleus, perfectly overlapping with the signal of BcWRKY1-GFP (Figure 1A). Taken together, these results demonstrated that BcWRKY1 protein was localized in the nucleus.
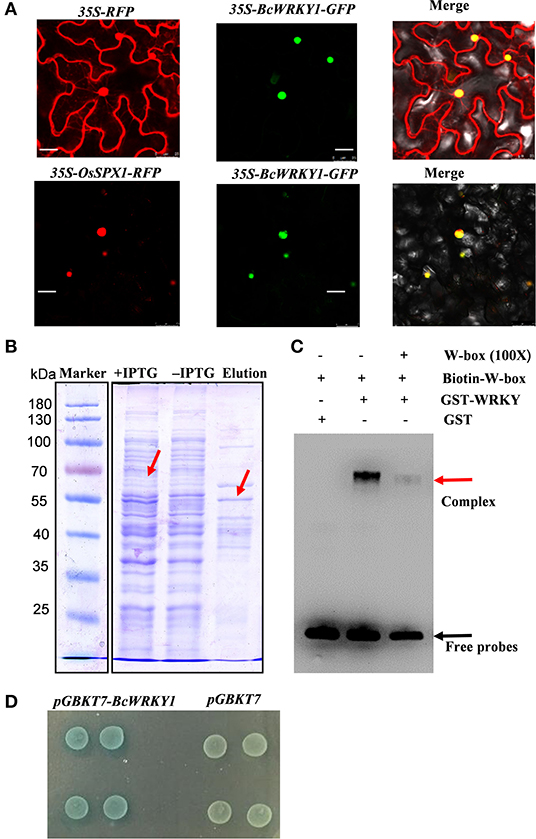
Figure 1. Expression and functional characterization of BcWRKY1 transcription factor. (A) Subcellular localization of 35S-BcWRKY1-GFP and 35S-RFP and subcellular co-localization of 35S-BcWRKY1-GFP and 35S-OsSPX1-RFP. (B) The results of recombinant protein GST-WRKY1 expression and purification. The molecular weight of the recombinant protein is indicated using protein markers. (C) The result of DNA binding activity of GST-WRKY1 to the DNA probe including W-box using EMSA assay. (D) Transcriptional activation activity of BcWRKY1 in yeast.
DNA Binding and Transcriptional Activation Activity of BcWRKY1
Since BcWRKY1 was a putative WRKY TF, it was further investigated whether BcWRKY1 could bind to the DNA sequence including classic W-box motif. Recombinant protein GST-BcWRKY1 was expressed and purified in E. coil BL21 (DE3). The results showed that GST-BcWRKY1 expression was significantly induced by the addition of IPTG (Isopropyl-β-D-thiogalactoside), and the GST-BcWRKY1 recombinant protein was successfully purified (Figure 1B). Further analysis with EMSA showed that GST-BcWRKY1 could specially bind to the DNA probe including the W-box motif, and the binding of GST-BcWRKY1 to the biotin-labeled probe could be specially attenuated by the addition of unlabeled competitor DNA probe (Figure 1C). Furthermore, to investigate whether BcWRKY1 is a transcriptional repressor or an activator. Plasmids containing pGBKT7-BcWRKY1 and pGBKT7 were respectively transformed into yeast AH109 competent cells, and then cultured at 30 °C for 48–72 h. The plaque of AH109 (pGBKT7-BcWRKY1) and AH109 (pGBKT7) were similar in size on SD/-trp medium. The results showed that pGBKT7-BcWRKY1 fusion protein had no toxicity to AH109 cells. Besides, yeast AH109 (pGBKT7-BcWRKY1) showed blue plaque (Figure 1D), suggesting that BcWRKY1 protein could promote gene transcriptional activities.
Overexpression of BcWRKY1 in Arabidopsis thaliana
To investigate the function of BcWRKY1 protein, BcWRKY1 was over-expressed in Arabidopsis thaliana. Positive transgenic Arabidopsis plants that grew normally on MS medium containing Glufosinate were then transplanted into organic soil spiked with vermiculite, and cultured in a growth chamber for a period of time. The plants with resistance to Glufosinate were sampled and observed under a fluorescence microscope. The positive transgenic plants were further identified by GFP signal detection in the leaves (Figure 2A). Two independent transgenic lines showing strong GFP signal were further selected to determine the expression levels of BcWRKY1. The results showed that the expression of BcWRKY1 was significantly higher than that in the control line (Figure 2B). Hence, the transgenic Arabidopsis plant overexpressing BcWRKY1-OX1 with strong GFP signal was selected for the following research.
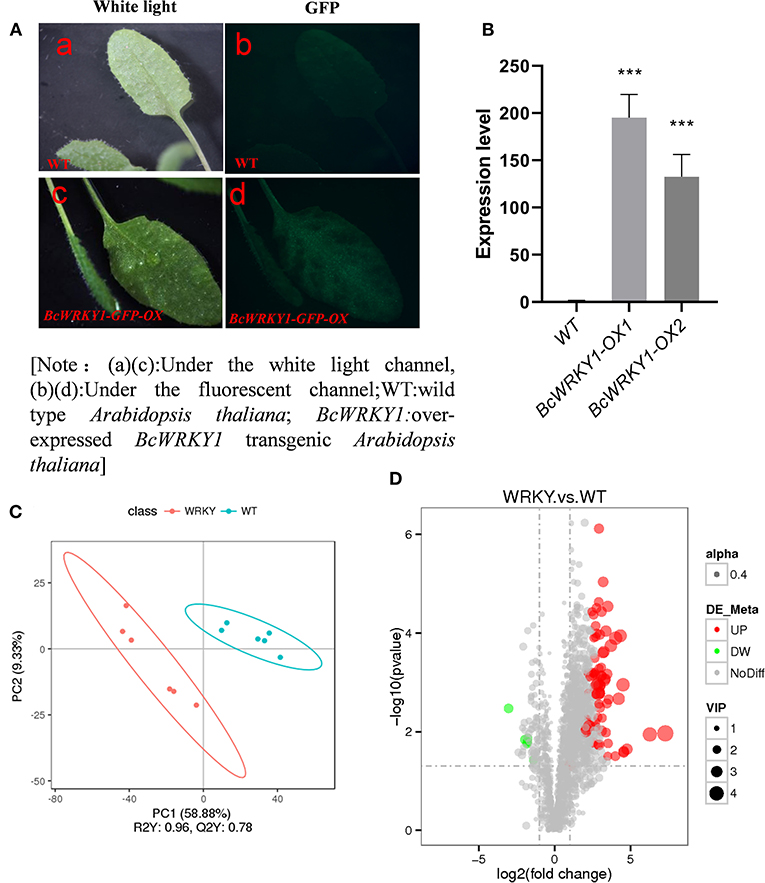
Figure 2. BcWRKY1 overexpression influences the metabolic profile in Arabidopsis thaliana. (A) Phenotypic overexpression of BcWRKY1-GFP in Arabidopsis thaliana; [Note: (a, c) Under the white light channel, (b, d) Under the fluorescent channel; WT: wild type Arabidopsis thaliana; BcWRKY1: BcWRKY1 overexpression transgenic Arabidopsis thaliana]. (B) The quantitation of expression levels of BcWRKY1 in transgenic Arabidopsis thaliana using qRT-PCR. (C) PLS-DA of differential metabolites between wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana in positive ion mode; (D) Volcano Plots of differential metabolites between wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana in positive ion mode, drawn by DEseq2 R package using a absolute log2Foldchange > 1 and an adjusted P value < 0.05 (FDR).
Differential Metabolic Profile Between Wild Type and BcWRKY1-OX1 Transgenic Arabidopsis thaliana
To investigate the function of BcWRKY1 in Arabidopsis thaliana metabolism, the metabolic profile in Arabidopsis thaliana was determined and compared between the wild type (WT) and BcWRKY1-OX1 transgenic lines. The results from the Principal Component Analysis (PCA) showed that the metabolites in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana could be clearly separated by PC1 with an explanatory degree of 58.88%, followed by PC2, with an explanatory degree of 9.33% (Figure 2C). These results suggested that overexpression of BcWRKY1 significantly changed the metabolic profile in Arabidopsis thaliana.
Metabolites Regulated by the Expression of BcWRKY1
The Variable Importance in Projection (VIP) P-value of the first principal component in the PLS-DA model was used to identify differential metabolites by t-test. The threshold was set as VIP > 2.0, and the difference multiple FC was set to be > 2.0 or < 0.5. A P-value < 0.05 was used to identify the differential metabolites in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana.
The total number of identified metabolic compounds in both lines was 2,131. In Volcano Plots, gray points represented the metabolites without significant differences (NoDiff), and the total number of significantly different metabolites between wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana was 80 (Figure 2D and Supplementary Table S3). Red points represented up-regulated metabolites in BcWRKY1-OX1 transgenic line (UP), and the number of significantly up-regulated metabolites was 74. Green points represented down-regulated metabolites in BcWRKY1-OX1 transgenic line (DW), and the number of significantly down-regulated metabolites was 6. The size of the dot represented the VIP value, and alpha (0.4) represented the transparency of points. Compared with wild type Arabidopsis thaliana, the number of up-regulated metabolites in BcWRKY1-OX1 transgenic Arabidopsis thaliana was significantly higher than that of down-regulated metabolites (Figure 2D). In addition, significantly altered metabolites in WT and BcWRKY1-OX1 transgenic lines were further analyzed and clustered, and the results showed that the differential metabolites between the two plant lines varied (Supplementary Figure 5). Furthermore, some differential metabolites exhibited significant correlation with each other (Supplementary Figure 6).
KEGG Pathway Enrichment Analysis in Wild Type and BcWRKY1-OX1 Transgenic Arabidopsis thaliana
In order to investigate which metabolic pathway these differential metabolites were enriched in, KEGG analysis was performed. It was clear that the differential metabolites in wild type and BcWRKY1-OX1 transgenic lines participated in the main biochemical metabolic pathways and signal transduction pathways. KEGG Pathway enrichment analysis in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana are shown in Supplementary Table S4.
The results showed that the abundance of indole-related metabolites were significantly increased in BcWRKY1-OX1 line (Figures 3A–E) (Supplementary Table S5). Besides, there were more differential metabolites enriched in flavonoid biosynthesis and phenylpropanoid biosynthesis pathways in BcWRKY1-OX1 transgenic line (Figure 3F). Compared with wild type Arabidopsis thaliana, the most obvious difference in BcWRKY1-OX1 transgenic line was the enrichment of flavonoid biosynthesis (Figure 4F). Compared with wild type Arabidopsis thaliana, baimaside of flavone and flavonol biosynthesis in BcWRKY1-OX1 transgenic Arabidopsis thaliana were significantly up-regulated (Supplementary Figure 7). Although tryptophan metabolism did not exhibit a significant change, the abundance of indoleacetate in the tryptophan metabolic pathway in BcWRKY1-OX1 transgenic Arabidopsis thaliana was significantly increased (Supplementary Figure 8).
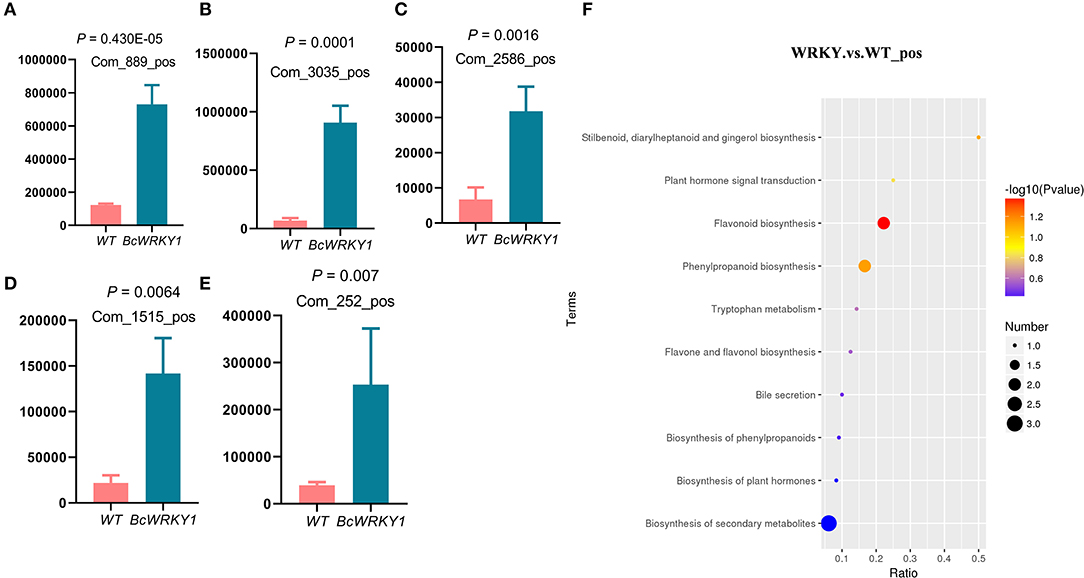
Figure 3. Indole-related metabolites are significantly increased in BcWRKY1-OX1 transgenic Arabidopsis thaliana line. (A–E) Abundance of indole-related metabolites in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana line. The P value < 0.05 indicates a significant difference between the two groups determined using student's t test. (F) KEGG pathway enrichment analysis in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana in positive ion mode.
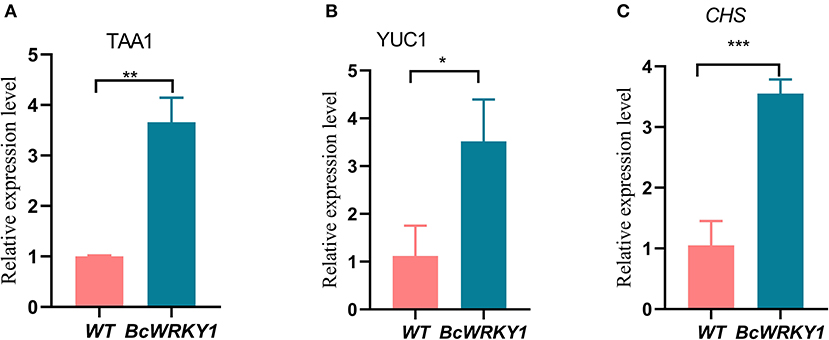
Figure 4. Expression analysis of key genes in the flavonoid and indole acetic acid biosynthesis pathway. (A) Expression analysis of TAA1 in the biosynthesis of indole acetic acid pathway by qRT-PCR. (B) Expression analysis of YUC1 in the biosynthesis of indole acetic acid pathway by qRT-PCR. (C) Expression analysis of CHS in the biosynthesis of flavonoid pathway by qRT-PCR.
To further understand why BcWRKY1 overexpression changed flavonoid and indole relative metabolites in Arabidopsis thaliana, we further detected the expression levels of key genes in the flavonoid and indole acetic acid biosynthesis pathways. Expression levels of key genes in the flavonoid and indole acetic acid biosynthesis pathway (such as TAA1, YUC1 and CHS) were detected with qRT-PCR. Results showed that the expression level of TAA1 and YUC1 in indole acetic acid biosynthesis pathways in BcWRKY1 overexpression line were significantly increased by comparison with wild type Arabidopsis. Similarly, expression of CHS in flavonoid biosynthesis in BcWRKY1 overexpression line was also significantly increased (Figure 4). Taken together, our results suggested that BcWRKY1 might regulated the flavonoid and indole metabolism, through regulation the expression of key genes in these two biosynthesis pathway.
Discussion
The development of molecular biological techniques has opened the avenue of the in-depth study of genes, which not only focuses on the gene function but also on mapping the genes in various plant systemic networks (Li and Zhou, 2014). Along with genome sequencing of different species, the WRKY gene family has also been identified in several species. WRKY is a kind of transcription factor with specific roles in plants (Cheng et al., 2012; Chi et al., 2013). Besides, our preliminary study suggested that the WRKY transcription factor exhibited a significant correlation with indole metabolic pathway in B. cusia (Huang, 2017). And the main effective substances including indirubin and indigo in B. cusia were the secondary metabolites derived from indole metabolism in B. cusia (Huang, 2017). More and more research has shown that WRKY TFs also participate in the secondary metabolic regulation such as indole alkaloid production in plants (Xu et al., 2004; Suttipanta et al., 2011). Therefore, based on the previous studies and the transcriptome sequencing database of B. cusia, the full length sequence of WRKY in B. cusia (BcWRKY1) was successfully cloned using RT-PCR, and bioinformatics analysis was performed (Supplementary Figure 2). Compared to the conventional laboratory-based experimental research, bioinformatics can obtain more reliable results in less time and was cost-effective. In subcellular localization studies, we found that BcWRKY1 protein was mainly localized in the nucleus, which was consistent with its function as a transcription factor. This result was also in line with the previous Eslpred prediction. The transcriptional regulation domain determined whether it could activate or inhibit gene expression (Dong et al., 2018). In the study, we found that BcWRKY1 protein could bind to the WRKY-binding motif W-box, acting as a transcriptional activator (Figures 1B–D), which was consistent with previous studies (Rushton et al., 1996). However, some WRKY proteins also play a role as transcriptional suppressor. For example, Bo (2016) found that tomato SIDRW1 gene encoding the WRKY transcription factor might function as a transcriptional suppressor. In addition, the WRKY protein in some species, such as AtWRKY6, could play a dual role as transcriptional activator or repressor depending on the regulation process (Xie et al., 2005). Metabolomics is an effective method to comprehensively explore the distribution of compounds in plants, including differential metabolites (Tikunov et al., 2005). Target analysis method has high accuracy, but useful information may be missed in metabolic phenotype analysis (Wang et al., 2018). Non-targeted metabolomics has been applied to investigate the differential metabolites and analyze the enriched pathways, especially in studies on the discovery of metabolic markers. Due to high throughput characteristics and a wide coverage of metabolites (Duan et al., 2016), all cellular metabolites can be detected without bias, more comprehensively reflecting the overall metabolic state of cells (Patti et al., 2012). Non-targeted metabolomics LC-MS was used to analyze the metabolic profiles in wild type and BcWRKY1-OX1 transgenic Arabidopsis thaliana. The results showed that flavonoid biosynthesis pathway was significantly influenced by BcWRKY1 overexpression (Figure 3F). Besides, although tryptophan metabolism did not show a significant change (Figure 3F), some indole-related metabolites were significantly increased in BcWRKY1-OX1 transgenic line (Figures 3A–E). These results suggested that BcWRKY1 was involved in the regulation of indole metabolic pathways in Arabidopsis thaliana plant model. In addition, qRT-PCR suggested that indeed some genes in the flavonoid and indole biosynthesis pathway were induced in the BcWRKY1-OX1 over-expression line. And indeed, the most homologous WRKY in Arabidopsis thaliana is AtWRKY50, which has been proved in regulation of sinapic metabolism in Arabidopsis thaliana (Hussain et al., 2018). However, whether BcWRKY1 directly or indirectly regulated these processes needs further investigation. Besides, our findings indicated that BcWRKY1 might regulate the corresponding metabolism pathways in B. cusia. Since the main effective substances in B. cusia were indigo and indirubin, both are products of indoles metabolism in the Tryptophan metabolismin B. cusia (Huang, 2017). Taken together, BcWRKY1 might participate in effective substances metabolism in B. cusia. These findings may lay a good foundation for further research about the effect of BcWRKY1 transcription factor on B. cusia metabolism.
Conclusion
Effective substances metabolic pathway in B. cusia is still unknown. We identified a BcWRKY1 involved in this pathway. We combined the use of bioinformatics analysis, transgenic plant model and metabolomics technology to study the function of BcWRKY1 protein in plant metabolism regulation. Overexpression of BcWRKY1 significantly changed flavonoid and indole-related metabolism and indole-related metabolites were also significantly upregulated. We postulated that BcWRKY1 transcription factor might be involved in the regulation of effective substances metabolism in B. cusia.
Data Availability Statement
The raw data presented in the study are deposited in the MetaboLights repository, accession number (MTBLS4819).
Author Contributions
MZ and YD conceived and designed the study. MZ performed the experiments and wrote the manuscript. MZ and YZ performed data analysis. MZ, ZG, HY, HZ, and LZ revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China (Association of Science and Technology Cooperation Across the Taiwan Straits, Grant No. U1405215), Natural Science Foundation of China (Grant No. 82003903), Science and Technology Planning Project of Quanzhou (Grant No. 2019N031), and Science and Technology Planning Project of Quanzhou (Grant No. 2020C061). The role of the funding body in the design of the study, the collection, analysis and interpretation of data, and in writing the manuscript is managed and supervised.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the teachers and graduate students of the Root Biology Center, Fujian Agriculture and Forestry University for their help in the experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.919071/full#supplementary-material
References
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22, 195–201. doi: 10.1093/bioinformatics/bti770
Bakshi, M., and Oelmüller, R. (2014). WRKY transcription factors:Jack of many trades in plants. Plant Signal. Behav. 9, e27700. doi: 10.4161/psb.27700
Bent, A.. (2006). Arabidopsis thaliana floral dip transformation method. Agrobacterium Protocols. Methods in Molecular Biology. Humana Press. vol 343. p. 87–104. doi: 10.1385/1-59745-130-4:87
Bhasin, M., and Raghava, G. (2004). ESLpred:SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucl. Acids Res. 32:W414–W419. doi: 10.1093/nar/gkh350
Bo, L.. (2016). Functional Analysis of Tomato WRKY Transcription Factor SIDRW1 and NAC Transcription Factor SISRN1 in the Regulation of Defense Response. Hangzhou: Zhejiang university.
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molec. Plant. 6, 1769–1780. doi: 10.1093/mp/sst080
Chen, Y., Zhang, R., Song, Y., He, J., Sun, J., Bai, J., et al. (2009). RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst. 134, 2003–2011. doi: 10.1039/b907243h
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y., Zhou, J., Chen, J., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant. 6, 287–300. doi: 10.1093/mp/sst026
China Pharmacopoeia Commission (2020). The Chinese Pharmacopoeia 2020 (Volume I). Beijing: Chinese Medical Science and Technology Press. p. 256–257.
Cserzö, M., Eisenhaber, F., Eisenhaber, B., and Simon, I. (2002). On filtering false positive transmembrane protein predictions. Protein Eng. 15,745–752. doi: 10.1093/protein/15.9.745
Danz, H., Stoyanova, S., Wippich, P., Brattström, A., and Hamburger, M. (2001). Identification and isolation of the cyclooxygenase-2 inhibitory principle in Isatis tinctoria. Planta Med. 67, 411–416. doi: 10.1055/s-2001-15805
Dong, Y., Zhang, P., Zhang, X., Gao, Y., and Wen, P. (2018). Construction and expression of yeast two-hybrid bait expression vector pGBKT7-ufgt. Molec. Plant Breed. 16, 1147–1153. doi: 10.13271/j.mpb.016.001147
Duan, L., Dai, Y., Sun, C., and Chen, S. (2016). Metabolomics research of medicinal plants. China J. Chin. Mater. Medica. 41, 4090–4095. doi: 10.4268/cjcmm20162202
Eulgem, T., Rushton, P., Robatzek, S., and Somssich, I. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Finatto, T., Viana, V., Woyann, L., Busanello, C., da Maia, L., and de Oliveria, A. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Molec. Biol. 41, 533–544. doi: 10.1590/1678-4685-gmb-2017-0232
Fraga, C., Clowers, B., Moore, R., and Zink, E. (2010). Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Analyt Chem. 82, 4165–4173. doi: 10.1021/ac1003568
Gao, Y., Zhao, Y., Li, T., Liu, Y., Ren, C., and Wang, M. (2009). Molecular cloning and expression analysis of an F-box protein gene responsive to plant hormones in Brassica napus. Mol. Biol. Rep. 37, 1037–1044. doi: 10.1007/s11033-009-9822-x
Gu, Y., Ji, Z., Chi, F., Qiao, Z., Xu, C., and Zhang, J. (2015). Bioinformatics and expression analysis of the WRKY gene family in apple. Sci. Agric. Sinica. 48, 3221–3238. doi: 10.3864/j.issn.0578-1752.2015.16.012
Guex, N., and Peitsch, M. (1997). SWISS-MODEL and the swiss-pdb viewer: an environment for comparative protein modeling. Electrophoresis. 18, 2714–2723. doi: 10.1002/elps.1150181505
Han, M., Zhao, B., An, Z., Thomas, R., and Zu, Y. (2007). Cloning and expression of cDNA encoding key enzymes (DXR, SLS, G10H and STR) in terpene indole alkaloids biosynthesis pathway from catharanthus roseus. Bull. Botan. Res. 27, 564–567. doi: 10.3969/j.issn.1673-5102.2007.05.013
Huang, X., Ding, F., Peng, H., Pan, J., He, X., Xu, J., et al. (2019). Research progress on family of plant WRKY transcription factors. Biotechnol. Bull. 35, 129–143. doi: 10.13560/j.cnki.biotech.bull.1985.2019-0626
Huang, Y.. (2017). Exploration and Functional Research of the Key Genes Related to Pharmacodynamics Substances in Baphicacanthus cusia. Quanzhou: Huaqiao University.
Huang, Y., Pan, D., Wang, Z., Ning, W., and Zhou, Y. (2009). Effects of different growing period on Baphicacanthus cusia (Nees) Bremek medicine constituents. Chin. Agric. Sci. Bull. 25, 75–78.
Huang, Y., Tan, H., Yu, J., Chen, Y., Guo, Z., Wang, G., et al. (2017). Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under hormonal stimuli and UV irradiation, and in different plant organs. Front. Plant Sci. 8, 668. doi: 10.3389/fpls.2017.00668
Hussain, R., Kim, H., Khurshid, M., Akhtar, M., and Linthorst, H. (2018). Overexpression of AtWRKY50 is correlated with enhanced production of sinapic derivatives in Arabidopsis. Metabolomics. 14, 25. doi: 10.1007/s11306-018-1317-0
Ishihara, T., Kohno, K., Ushio, S., Iwaki, K., Ikeda, M., and Kurimoto, M. (2000). Tryptanthrin inhibits nitric oxide and prostaglandin E2 synthesis by murine macrophages. Eur. J. Pharmacol. 407, 197–204. doi: 10.1016/S0014-2999(00)00674-9
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., and Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. Journal of Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molec. Biol. Evolut. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, D., Wu, Y., Wang, C., and Sun, Q. (2011). Pharmacokinetics research on anti-inflammatory effect and analgesic effect of indigo naturalis. China J. Exper. Tradit. Med. Formulae. 17, 137–140. doi: 10.13422/j.cnki.syfjx.2011.13.059
Li, K., and Zhou, C. (2014). Research progress in WRKY transcription factors in plants. Plant Physiol. J. 50, 1329–1335. doi: 10.13592/j.cnki.ppj.2014.1019
Lin, W., Huang, W., Ning, S., Gong, X., Ye, Q., and Wei, D. (2019). Comparative transcriptome analyses revealed differential strategies of roots and leaves from methyl jasmonate treatment Baphicacanthus cusia(Nees) Bremek and differentially expressed genes involved in tryptophan biosynthesis. PLoS ONE. 14, e0212863. doi: 10.1371/journal.pone.0212863
Liu, L., Wang, Y., and Li, J. (2014). Study of indigo on ulcerative colitis inflammatory cell model in vitro. China J. Tradit. Chin. Med. Pharmacy. 29, 1666–1669.
Liu, Q., Zhang, G., and Chen, S. (2001). Structure and regulatory function of plant transcription factors. Chinese Sci. Bull. 46, 271–278. doi: 10.1007/BF03187184
Liu, Y, Ou, Y., Yu, H., Li, L., Wang, N., and Yao, X. (2009). Chemical constituents in the leaves of Baphicacanthus cusia(Nees) Bremek. Chinese Journal of Medicinal Chemistry 19, 273–275.
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, A., Li, X., Liu, H., Wu, C., Zeng, Q., and Liu, X. (2016). Cloning and bioinformatics analysis of full-Length cDNA Sequence of ScHTD2 gene from sugarcane. Chin. J. Trop. Crops. 37, 1133–1140. doi: 10.3969/j.issn.1000-2561.2016.06.014
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). SPX4 negatively regulates Phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 26, 1586–1597. doi: 10.1105/tpc.114.123208
Ma, H., Jenny, Y., and Cherng, S. (2008). RNA, DNA and protein isolation using Trizol reagent. Nat. Sci. 6, 66–75.
Ma, J., Zhang, D., Gao, X., and Shao, Y. (2015). Identification and analysis of WRKY transcription factors in triticum urartu. Acta Agronomica Sinica. 41, 900–909. doi: 10.3724/SP.J.1006.2015.00900
Mitsuda, N., and Ohme-Takagi, M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi: 10.1093/pcp/pcp075
Patti, G., Yanes, O., and Siuzdak, G. (2012). Innovation: metabolomics: the apogee of the omics trilogy. Nature Reviews. Molec. Cell Biol. 13, 263–269. doi: 10.1038/nrm3314
Priest, H., Filichkin, S., and Mockler, T. (2009). Cis-regulatory elements in plant cell signaling. Curr. Opin Plant Biol. 12, 643–649. doi: 10.1016/j.pbi.2009.07.016
Qiao, X., Ma, C., and Chen, L. (2009). Plant flavonoid biosynthesis pathway and regulation of its important genes. Nat. Product Res. Develop. 21:354–360. doi: 10.3969/j.issn.1001-6880.2009.02.040
Rushton, P., Somssich, I., Ringler, P., and Shen, Q. (2010). WRKY transcription factors. Trends Plant Sci.15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, P., Torres, J., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 gene. EMBO J. 15, 5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x
Ruttkies, C., Schymanski, E., Wolf, S., Hollender, J., and Neumann, S. (2016). MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminf. 8, 3. doi: 10.1186/s13321-016-0115-9
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. (2003). SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res. 31, 3381–3385. doi: 10.1093/nar/gkg520
Shang, J., Chen, A., and Pan, W. (2013). Cloning and bioinformation analysis of chalcone synthase gene from Lyciumbarb arum in Ningxia. Chin. Tradit. Herbal Drugs. 44, 2898–2903. doi: 10.7501/j.issn.0253-2670.2013.20.020
Shen, Q.. (2009). Differences between south and north isatis root. Capital Med. 41–42. doi: 10.3969/j.issn.1005-8257.2009.22.033
Shi, H., Yin, X., Wu, M., and Tang, C. (2012). Cloning and bioinformatics analysis of an endoglucanase gene (Aucel12A) from Aspergillus usamii and its functional expression in Pichia pastoris. J. Ind. Microbiol. Biotechnol. 39, 347–357. doi: 10.1007/s10295-011-1039-z
Sun, J., Jiang, H., Xu, Y., Li, H., Wu, X., Xie, Q., et al. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48, 1148–1158. doi: 10.1093/pcp/pcm088
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tang, J., Liao, X., Tong, H., and Gao, J. (2014). GC-MS combined with PLS-DA to discriminate the varieties of XinJiang lavender essential oil. Comput. Appl. Chem. 31, 701–704. doi: 10.11719/com.app.chem20140613
Thevenot, E., Roux, A., Xu, Y., Ezan, E., and Junot, C. (2015). Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335. doi: 10.1021/acs.jproteome.5b00354
Tikunov, Y., Lommen, A., Vos, C., Verhoeven, H., Bino, R., Hall, R., et al. (2005). A novel approach for nontargeted data analysis for metabolomics.Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139, 1125–1137. 105.068130 doi: 10.1104/pp.105.068130
Wang, B., Chu, J., Yu, T., Xu, Q., Sun, X., Yuan, J., et al. (2015). Tryptophan- independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 112, 4821–4826. doi: 10.1073/pnas.1503998112
Wang, X., Gong, M., Tang, L., Zheng, S., Lou, J., Ou, L., et al. (2013). Cloning, bioinformatics and the enzyme activity analyses of a phenylalanine ammonia-lyase gene involved in dragon's blood biosynthesis in Dracaena cambodiana, Mol. Biol. Rep. 40, 97–107. doi: 10.1007/s11033-012-2032-y
Wang, X., Ming, M., Tian, Y., Li, Y, Lian, L., and Lou, D. (2018). Investigation on metabolite profiling of rice seed by non-targeted metabolomics method. Journal of Instrumental Analysis. 37, 341–345. doi: 10.3969/j.issn.1004-4957.2018.03.014
Wang, Y., Sa, F., Chan, Y., and Lee, M. (2014a). Research progress on effects of indirubin and its structurally compounds on anticancer and neuroprotection. Chin. Tradit. Herbal Drugs. 45, 2404–2411. doi: 10.7501/j.issn.0253-2670.2014.16.024
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014b). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wani, S., Kumar, V., Shriram, V., and Sah, S. (2016). Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. The Crop Journal. 4, 162–176. doi: 10.1016/j.cj.2016.01.010
Wen, B., Mei, Z., Zeng, C., and Liu, S. (2017a). MetaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics. 18, 183. doi: 10.1186/s12859-017-1579-y
Wen, F., Wu, X., Liao, L., and Liu, S. (2017b). Research advances on physiological function of WRKY transcription factor in plant stress resistance. Guihaia. 37, 69–79. doi: 10.11931/guihaia.gxzw201512022
Xie, Z., Zhang, Z., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xiong, H., Zhang, X., Wu, X., and Xie, X. (2014). Expression analysis of WRKY transcription factors related to stress response in cultivated tobacco. Acta Tabacaria Sinica. 20, 144–149. doi: 10.3969/j.issn.1004-5708.2014.06.023
Xu, Y., Wang, J., Wang, S., Wang, J., and Chen, X. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene(+)-δ-cadinene synthase-A1. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yu, A., and Zhao, L. (2015). Effects of the GSK-3β inhibitor (2Z, 3E)-6- bromoindirubin-3-oxime upon ovarian cancer cells. Tumour Biol. 37, 4857–4864. doi: 10.1007/s13277-015-4344-8
Zeng, M., and Diao, Y. (2016). Secondary metabolites of Baphicacanthus cusia (NeeS) Bremek. Chin. Agric. Sci. Bull. 32, 30–34.
Zhang, X., Abrahan, C., Colquhoun, T. A., and Liu, C. J. (2017). A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in arabidopsis. Plant Cell. 29, 1157–1174. doi: 10.1105/tpc.16.00855
Zhu, Z., Schultz, A., Wang, J., Johnson, C., Yannone, S., Patti, G., et al. (2013). Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protocols. 8, 451–460. doi: 10.1038/nprot.2013.004
Keywords: Baphicacanthus cusia (Nees) Bremek, BcWRKY1, bioinformatics analysis, metabolites, indole-related metabolism, flavonoid-related metabolism
Citation: Zeng M, Zhong Y, Guo Z, Yang H, Zhu H, Zheng L and Diao Y (2022) Expression and Functional Study of BcWRKY1 in Baphicacanthus cusia (Nees) Bremek. Front. Plant Sci. 13:919071. doi: 10.3389/fpls.2022.919071
Received: 13 April 2022; Accepted: 01 June 2022;
Published: 01 July 2022.
Edited by:
Yong Wang, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Jiaotong Yang, Guizhou University of Traditional Chinese Medicine, ChinaKoji Miyamoto, Teikyo University, Japan
Copyright © 2022 Zeng, Zhong, Guo, Yang, Zhu, Zheng and Diao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liling Zheng, emxsQGZqbXUuZWR1LmNu; Yong Diao, ZGlhb3lvbmdAaHF1LmVkdS5jbg==
 Meijuan Zeng
Meijuan Zeng Yongjia Zhong3
Yongjia Zhong3 Haisheng Zhu
Haisheng Zhu Yong Diao
Yong Diao