- 1College of Life Sciences and Agronomy, Zhoukou Normal University, Zhoukou, China
- 2School of Mechanical and Electrical Engineering, Zhoukou Normal University, Zhoukou, China
- 3College of Biological Engineering, Henan University of Technology, Zhengzhou, China
Soil salinity is an important abiotic stress factor that seriously affects the crop growth and yield. Use of plant-derived microorganisms is a promising strategy to alleviate salt stress. In a previous study, the endophytic strain Bacillus altitudinis WR10 isolated from wheat roots showed high salt resistance. In this study, we investigated the efficacy of WR10 in improving the salt tolerance of wheat and its potential mechanisms using a hydroponic test. Under salt stress, WR10 inoculation significantly increased the lengths and dry weights of the roots and shoots, indicating that WR10 improves wheat salt tolerance at the seedling stage. WR10 inoculation significantly reduced Na+ accumulation and enhanced K+, P, and Ca2+ uptake in salt-stressed plants, which can be attributed to the upregulated gene expression of H+-ATPase as well as the P-solubilizing and biofilm-producing characteristics of WR10. At the transcriptional level, L-ascorbate peroxidase (APX), glutathione (GSH) synthetase related to GSH biosynthesis, and phenylpropanoid biosynthesis genes (CYP73A, 4CL, and CAD) were significantly upregulated, whereas those of GSH metabolism genes (glutathione S-transferase and gamma-glutamyltranspeptidase) were significantly downregulated in WR10-applied wheat roots under salt stress. These changes increased the APX activity and GSH levels and resulted in a decrease in hydrogen peroxide levels. Additionally, a decrease in proline content was observed in WR10-inoculated plants under salt stress because of WR10-induced upregulation of proline dehydrogenase gene expression. These results provide supporting evidence that WR10 improves wheat salt tolerance via more than one mechanism and open a window of opportunity for WR10 application in salinized soil.
Introduction
Soil salinization has become a serious global problem owing to climate change, seawater intrusion, improper irrigation, and other factors. More than 833 million hectares of soil and 10% of farmland worldwide are under the threat of salinization (Food and Agriculture Organization [FAO], 2021). By 2050, the salinity of farmlands is suggested to increase to 50% (Shrivastava and Kumar, 2015). High salinity induces ionic toxicity, osmotic stress, oxidative stress, and nutrient depletion, which seriously endanger the crop growth and decrease the crop yield (Muranaka et al., 2002; Lodeyro and Carrillo, 2015; Zhu, 2016). The annual economic loss caused by salt stress-induced crop yield exceeds 27.3 billion dollars (Qadir et al., 2014). Therefore, salt stress poses a major challenge to global food security. With the rapid increase in world population (estimated to reach 9.1 billion in 2050) and food demand (estimated to increase by 70%), this challenge will become more severe (Food and Agriculture Organization [FAO], 2009). Therefore, improving crop salt tolerance and alleviating the adverse effects of salt stress are paramount for sustainable agricultural development and global food security.
Wheat (Triticum aestivum L.) is an important cereal crop that showed 760.92 million tonnes global production in 2020. It is the source of approximately 20% of total dietary calories and proteins worldwide (Shiferaw et al., 2013). Most wheat cultivars are moderately tolerant or sensitive to salt stress (Quan et al., 2021). When wheat plants are grown in soil with 8–10 dS/m electrical conductivity (moderate salinity), 20–28% yield loss is observed (Satir and Berberoglu, 2016). Substantial efforts to develop salt-tolerant wheat varieties via conventional breeding and genetic engineering methods have been made during the last few decades, but only limited success has been achieved because of the time constraints, intricate procedures, and ethical issues (Ashraf and Akram, 2009).
A large number of plant growth promoting bacteria (PGPB) are found in the rhizosphere and roots of plants. Many PGPB belonging to the genera Bacillus, Halomonas, Enterobacter, Moraxella, Pseudomonas, Zhihengliuella, Staphylococcus, Oceanobacillus, Thalassobacillus, Halobacillus, Dietzia, Marinibacillus, Planococcus, and Promicromonospora sp. are tolerant to high salt conditions and play important roles in improving plant salt tolerance (Raheem and Ali, 2015; Bharti et al., 2016; Orhan, 2016; Mukherjee et al., 2019; Orhan and Demirci, 2020). For example, Bacillus subtilis BERA 71 enhances plant biomass and synthesis of photosynthetic pigments in chickpea exposed to saline conditions (Abd_Allah et al., 2018). Inoculation with Hallobacillus sp. SL3 and Bacillus halodentinicans PU62 significantly increases the root length and dry weight of wheat seedlings under salt stress (Ramadoss et al., 2013). Pseudomonas sp. UW3 + UW and Pseudomonas corrugate CMH3 significantly increase the stem biomass of oats grown in saline field soils (Chang et al., 2014). Hence, use of PGPB may be a desirable strategy to mitigate the adverse effects of soil salinity.
In a previous study, we isolated a wheat endophytic PGPB strain Bacillus altitudinis WR10. It shows many plant growth-promoting traits, such as ACC deaminase production, indole-3-acetic acid secretion, and phosphate solubilization, and improves the ability of the host to withstand numerous abiotic stresses (Sun et al., 2017; Yue et al., 2019, 2021). WR10 is a halotolerant strain that tolerates up to 12% NaCl and increases seed germination of wheat under salt stress (Yue et al., 2019). Due to its inherent tolerance to NaCl, multiple plant growth-promoting traits, and intimate interaction with wheat, we speculated that WR10 may affect plant salt stress tolerance.
Therefore, this study aimed to evaluate the efficacy of WR10 in alleviating salt stress by determining the lengths and dry weights of the roots and shoots of wheat. PGPB can use multiple mechanisms, including production of phytohormones, regulation of Na+ homeostasis, improved nutrient uptake, increased number of active oxygen scavengers, and osmolyte accumulation, to improve plant tolerance (Qin et al., 2016; Mishra et al., 2021). We hypothesized that the salinity-alleviating property of WR10 could be attributed to one or more of these mechanisms. To further clarify how WR10 interacts with its wheat host, root transcriptome and biochemical experiments (including proline, nutrients, and antioxidant compounds) were performed. This study provides a theoretical basis for the application of WR10 in wheat under salt stress.
Materials and methods
Seed germination, bacterial culture, and preparation of bacterial suspension
Seeds of Zhoumai 36, a new variety cultivated by the Zhoukou Academy of Agricultural Sciences (Zhoukou, China), were surface-sterilized for 15 min using 5% sodium hypochlorite. After washing thrice with sterile deionized water, the seeds were placed in a plastic basin (21.3 cm × 16.3 cm × 6.8 cm) with sterile water and germinated for two days in the dark at 25°C.
Bacillus altitudinis WR10 preserved in 30% glycerol was inoculated into 5 mL of sterile Luria-Bertani (LB) broth and cultured for 24 h at 37°C and 150 rpm. Then, 1 mL of the bacterial suspension was added to 100 mL of LB broth and cultured for another 24 h under the same conditions. Finally, the bacterial suspension was centrifuged for 10 min at 9,000 × g and the bacterial pellet was collected. After washing twice with sterile deionized water, the pellet was resuspended in sterile deionized water to prepare a bacterial suspension with a final concentration of 3 × 1010 cfu/mL.
Effect of WR10 on wheat growth under NaCl stress condition
A total of 240 seeds with consistent germination were selected and randomly assigned to four groups: the control group without NaCl and WR10, WR10 group with WR10 and without NaCl, NaCl group with NaCl and without WR10, and WR10 + NaCl group with NaCl and WR10. Each treatment group contained 60 seeds. The seeds were placed in glass beakers containing 600 mL of 1/2 Hoagland’s medium with or without 150 mmol/L NaCl and 3 × 107 cfu/mL of WR10. NaCl concentration was selected based on a salt tolerance experiment by Zhoumai 36 (Supplementary Figure 1). To avoid osmotic shock in wheat seedlings, salinity was increased gradually in increments of 50 mmol/L every two days to reach a final salt concentration of 150 mmol/L. In groups inoculated with WR10, 0.6 mL bacterial suspension was added to each beaker, whereas groups without WR10 were supplemented with an equal volume of sterile solution. After two weeks of growth at 22 ± 1°C, 60% relative humidity, and a photoperiod of 14 h light/10 h dark, all wheat seedlings were harvested. During plant growth, the culture solutions were renewed every two days. Some seedlings were randomly selected to measure the root and shoot lengths. Then, these seedlings were placed in an oven and dried for 72 h at 75°C. The dried roots and shoots were cut off to determine the dry weight and element content. The roots and shoots of other seedlings were separated; the shoots were used for proline detection and the roots were used for transcriptome sequencing after quick freezing in liquid nitrogen.
Determination of proline content
The proline content was determined using a proline colorimetric assay kit (Elabscience Biotechnology Co., Ltd., Wuhan, China). Briefly, 0.1 g of fresh shoot was homogenized with 1 mL of extracting solution from the kit and centrifuged at 10,000 × g for 15 min. The supernatant was collected and the proline content was determined according to the manufacturer’s instructions.
Measurement of elements in wheat tissues
Dry roots or shoots were digested as described in our previous study (Sun et al., 2021). For potassium (K) and phosphorus (P) determination, 0.1 g of ground tissues were digested using 5 mL H2SO4 and 2 mL H2O2, and the digestion solutions were then diluted with 20 mL deionized water and neutralized with 10 M NaOH. For sodium (Na), calcium (Ca), and magnesium (Mg) determination, 0.1 g of ground tissues were extracted for 2 h by adding 20 mL of 0.5 mol/L HNO3 at 37°C and 200 rpm. The extracted solutions were centrifuged at 10,000 × g for 5 min and the supernatants were collected. Na content was measured using flame atomic absorption spectrophotometry (A3AFG, Persee, Beijing, China). K, P, Ca and Mg contents were determined using commercial biochemical assay kits (Elabscience, China). All absorbance measurements were performed on a SpectraMax i3x microplate reader (Molecular Devices, Sunnyvale, CA, United States).
Root RNA isolation and sequencing
Total RNA was extracted from root samples using the PureLink Plant RNA reagent (Invitrogen, United States), according to the manufacturer’s protocol. RNA concentration was measured using a Nanodrop 2000c ultra-micro spectrophotometer (Thermo Fisher, Waltham, MA, United States). RNA integrity was assessed via 1% agarose gel electrophoresis using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, United States). High-quality RNA was used to construct cDNA libraries using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, United States). Sequencing was performed using the Illumina Novaseq 6000 system (Illumina, United States) by Majorbio Biotech Co., Ltd. (Shanghai, China).
Raw reads were filtered to remove the adapter, low-quality, higher N-ratio (>10%), and <50 bp reads, and clean reads were obtained. Using the TopHat2 software, high-quality clean reads were aligned with the wheat reference genome (version IWGSC). The resulting mapped reads were further assembled using the StringTie software. Functional annotation of genes or transcripts was performed using the NR, Swiss-Prot, Pfam, EggNOG, GO, and KEGG databases. All raw data were submitted to the NCBI Sequence Read Archive under the accession numbers SRR17951558-SRR17951569.
Differentially expressed gene screening and enrichment analysis
The expression levels of genes were analyzed via the transcripts per million reads (TPM) method using the RSEM software. Differential gene expression between different treatment groups was analyzed using the DESeq2 software. The screening criterion for differentially expressed genes (DEGs) was p value adjusted with Benjamini/Hochberg’s approach <0.05, and | log2 (fold change, FC)| ≥ 1. GO and KEGG enrichment analyses for DEGs were performed using the Goatools and KOBAS software, respectively. Using Venny 2.1 software, a Venn diagram was produced. Using the XLSTAT software, principal component analysis (PCA) was conducted to assess the relationship between biological repetitions in all groups.
Measurement of hydrogen peroxide levels, L-ascorbate peroxidase activity, peroxidase activity, and glutathione levels
Hydrogen peroxide (H2O2) levels, L-ascorbate peroxidase (APX) activity, and peroxidase (POD) activity in roots, and glutathione (GSH) levels in roots and shoots were measured using colorimetric assay kits (Elabscience, China), according to the manufacturer’s instructions. H2O2 levels, APX activity, POD activity, and GSH levels were expressed as mmoL/g FW, U/g FW, U/mg protein, and μmoL/mg protein, respectively.
Statistical analysis
Data were expressed as the mean ± standard deviation (SD). Before testing for differences between the two groups (Control vs. NaCl; NaCl vs. WR10 + NaCl), tests of normality (Kolmogorov–Smirnov) and equal variance (Levene median) were performed. Student’s t-test (parametric) was used when the conditions of normality and equal variance were met. When normality test failed or unequal variances were detected, the Mann-Whitney rank sum test (non-parametric) was used. All statistical analyses were performed using IBM SPSS Statistics 19.0 (IBM, Armonk, NY, United States). p < 0.05 was considered to be statistically significant.
Results
Bacillus altitudinis WR10 improves wheat salt tolerance
Compared to the control group, salt stress significantly inhibited the growth of wheat seedlings, whereas inoculation with WR10 alleviated the damage caused by salt stress in wheat (Figure 1A). Specifically, WR10 inoculation significantly increased the root length, shoot length, root dry weight, and shoot dry weight by 1. 56-, 1. 12-, 1. 56-, and 1.44-fold, respectively, compared to the WR10 uninoculated group under salt stress (Figures 1B–C).
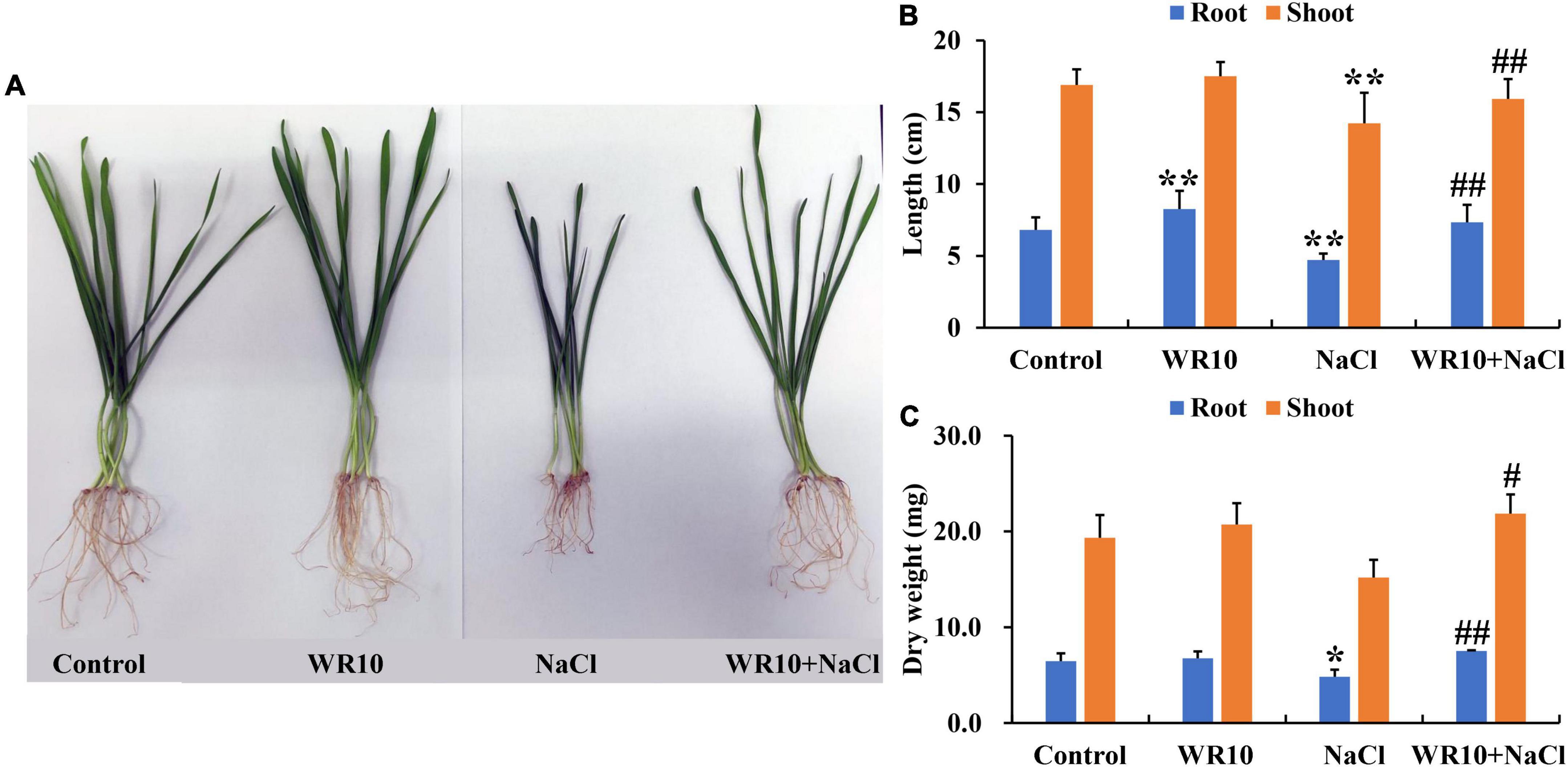
Figure 1. Bacillus altitudinis WR10 alleviates the adverse effects of salt stress on wheat seedlings. (A) Photograph of wheat seedlings; (B) lengths of roots and shoots; (C) dry weights of roots and shoots. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Data are expressed as the means ± SD (n = 12). *p < 0.05, **p < 0.01 (vs. control); #p < 0.05, ##p < 0.01 (vs. NaCl).
Bacillus altitudinis WR10 reduces Na+ accumulation and promotes nutrient absorption in wheat seedlings under salt stress
Compared to the control group, Na+ content of roots and shoots in salt-stressed wheat seedlings significantly increased by 1.46- and 1.36-fold, respectively (Figure 2A). In addition, salt stress significantly decreased the K+ content in shoots by 0.30-fold, P content in roots and shoots by 0.51- and 0.27-fold, respectively, and Ca2+ content in roots and shoots by 0.19- and 0.21-fold, respectively (Figures 2B–D). In contrast, WR10 inoculation significantly reduced Na+ content in roots and shoots by 0.79- and 0.89-fold, respectively, but increased K+ content in roots and shoots by 2.24- and 1.89-fold, P content in roots and shoots by 2.42- and 1.68-fold, and Ca2+ content in roots by 1.20-fold, respectively, compared to the WR10 uninoculated group under salt stress (Figures 2B–D). In addition, the content of Mg2+ did not change significantly in any treatment group (Figure 2E).
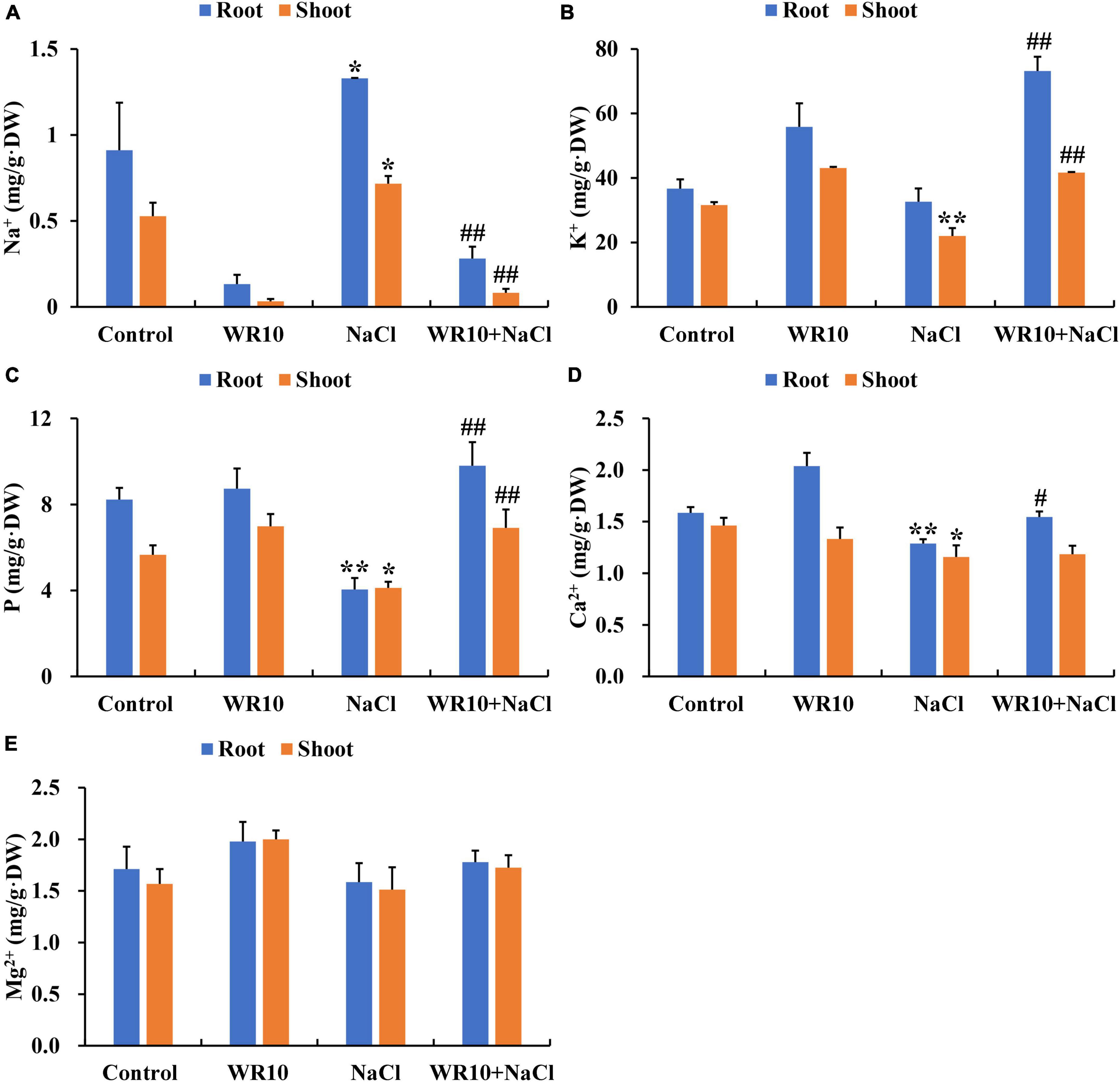
Figure 2. Effects of B. altitudinis WR10 on the levels of Na+ (A), K+ (B), P (C), Ca2+ (D), and Mg2+ (E) in the roots and shoots of wheat seedlings under salt stress. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Data are expressed as the means ± SD (n = 6). *p < 0.05, **p < 0.01 (vs. control); #p < 0.05, ##p < 0.01 (vs. NaCl).
Bacillus altitudinis WR10 alters the gene expression levels in wheat roots under salt stress
RNA-sequencing (RNA-seq) of 12 root samples produced 807.6 million raw reads. After quality control, a total of 798.7 million clean reads were obtained and mapped to the wheat reference genome with 78.75–83.22% mapped rate (Supplementary Tables 1, 2). These mapped reads were assembled into 86781 genes, 99.72% of which were successfully annotated (Supplementary Table 3).
PCA analysis showed that the samples of the control and NaCl groups and NaCl and NaCl + WR10 groups significantly separated from each other, indicating different gene expression patterns (Figures 3A–B). Further differential expression analysis revealed a total of 10,563 genes differentially expressed in NaCl-treated wheat compared with the control group, of which 5,571 genes were significantly upregulated and 4,993 genes were significantly downregulated. Moreover, 2744 DEGs were observed between NaCl and NaCl + WR10 groups. Among these DEGs, 1,515 were significantly upregulated and 1,229 were significantly downregulated in the NaCl + WR10 group (Figure 3C). Using GO enrichment analysis, DEGs in NaCl vs NaCl + WR10 were significantly enriched into 239 GO terms, including “peroxidase (POD) activity” (GO:0004601), “antioxidant activity” (GO:0016209), “response to oxidative stress” (GO:0006979), “reactive oxygen species (ROS) metabolic process” (GO:0072593), “H2O2 catabolic process” (GO:0042744), “H2O2 metabolic process” (GO:0042743), “GSH metabolic process” (GO:0006749), “GSH binding” (GO:0043295), “phenylpropanoid metabolic process” (GO:0009698), “phenylpropanoid biosynthetic process” (GO:0009699), and “phenylpropanoid catabolic process” (GO:0046271) (Figure 3D). Moreover, KEGG enrichment analysis showed that DEGs in NaCl vs NaCl + WR10 were significantly enriched in four pathways, in which “phenylpropanoid biosynthesis” (map00940) and “GSH metabolism” had the most DEGs (Figure 3E).
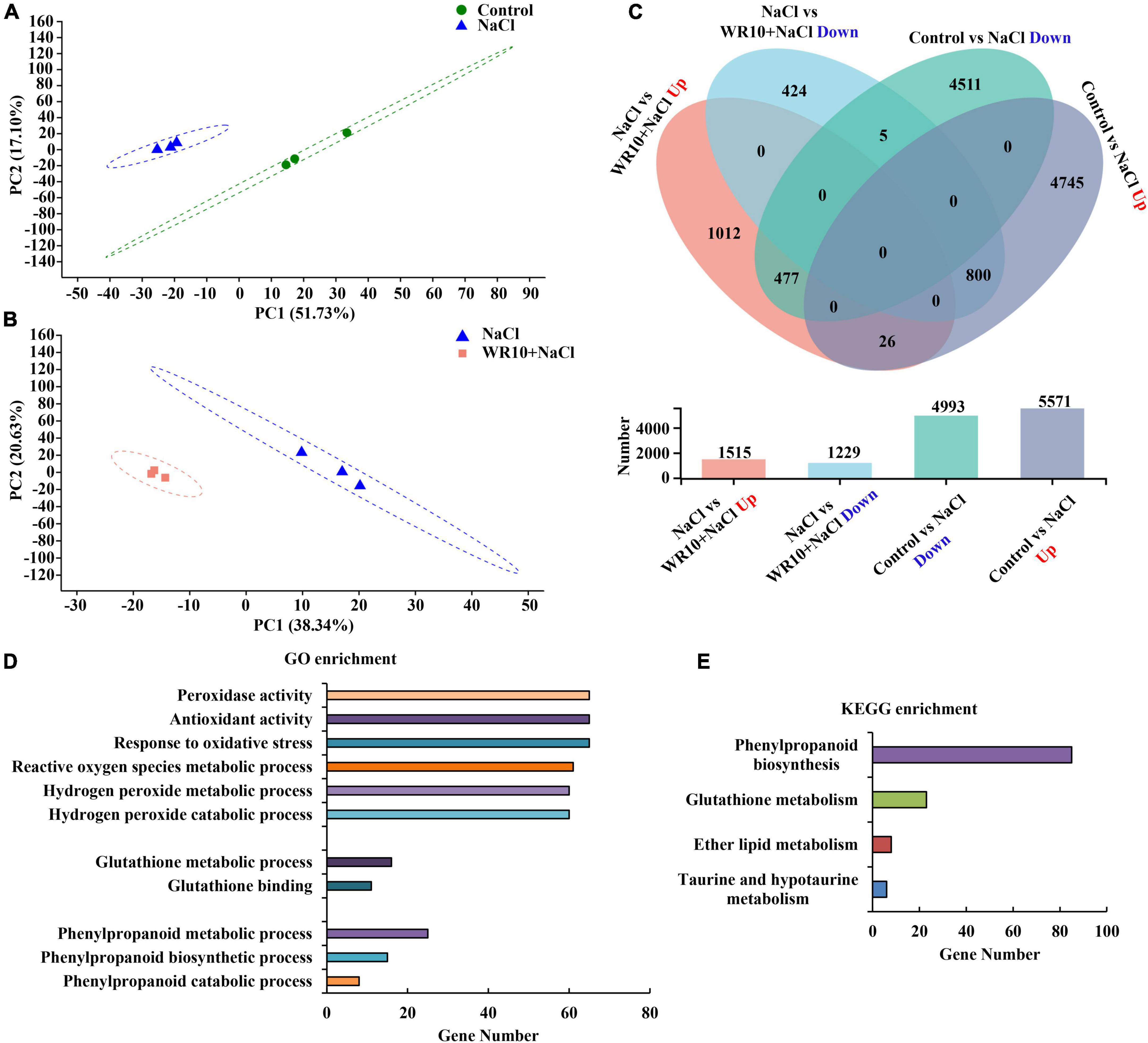
Figure 3. RNA-sequencing analyses of wheat roots. (A) Principal component analysis (PCA) analyses of Control and NaCl groups. (B) PCA analyses of NaCl and WR10 + NaCl groups. (C) Venn diagram displaying the differentially expressed genes (DEGs) in Control vs. NaCl and NaCl vs. WR10 + NaCl. (D,E) GO and KEGG enrichment analyses of DEGs in NaCl vs WR10 + NaCl. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10.
Bacillus altitudinis WR10 increases APX activity and reduces H2O2 levels under salt stress
Interestingly, 60 identical DEGs were enriched in the GO terms, GO:0004601, GO:0016209, GO:0006979, GO:0072593, GO:0042744, and GO:0042743. Among these DEGs, 59 DEGs, including 23 upregulated and 36 downregulated DEGs, were annotated as POD, while 1 upregulated DEG was annotated as APX. Subsequently, we measured the H2O2 levels, POD activity, and APX activity in the roots of wheat seedlings. Under salt stress, H2O2 levels in root were significantly increased by 1.86-fold, whereas inoculation with WR10 significantly decreased the accumulation of H2O2 by 0.18-fold (Figure 4A). In contrast, APX activity significantly increased by 1.25-fold in the roots of WR10-inoculated seedlings compared to WR10-free plants under salt stress (Figure 4B). In addition, there was no difference in POD activity between the NaCl and NaCl + WR10 groups (Supplementary Figure 2).
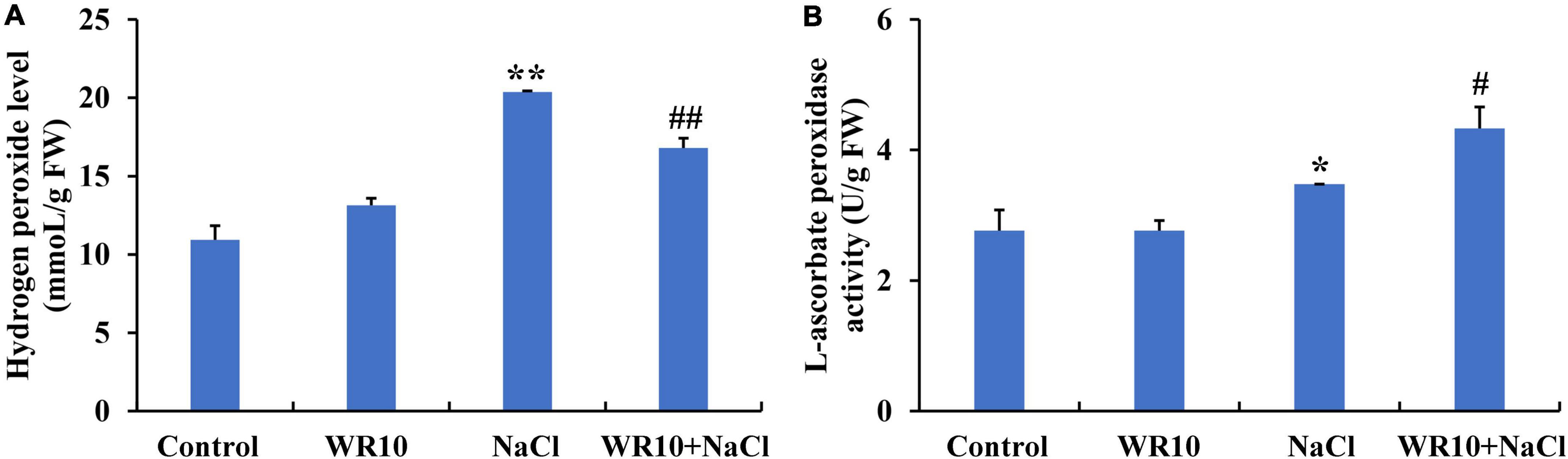
Figure 4. Bacillus altitudinis WR10 decreases H2O2 levels and increases APX activity in the roots of wheat seedlings under salt stress. (A) H2O2 levels in the roots of wheat seedlings; (B) APX activity in the roots of wheat seedlings. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Data are expressed as the means ± SD (n = 6). *p < 0.05, **p < 0.01 (vs. control); #p < 0.05, ##p < 0.01 (vs. NaCl).
Bacillus altitudinis WR10 affects GSH metabolism and increases GSH levels under salt stress
In the GSH metabolism pathway, WR10 inoculation significantly upregulated the expression levels of glutathione synthase (GSS), but significantly downregulated the expressions levels of glutathione S-transferase (GST) and gamma-glutamyltranspeptidase (GGT) under salt stress (Table 1). GSH levels in roots were not affected, but those in the shoots were significantly increased by 1.25-fold after inoculation with WR10 under salt stress (Figure 5).
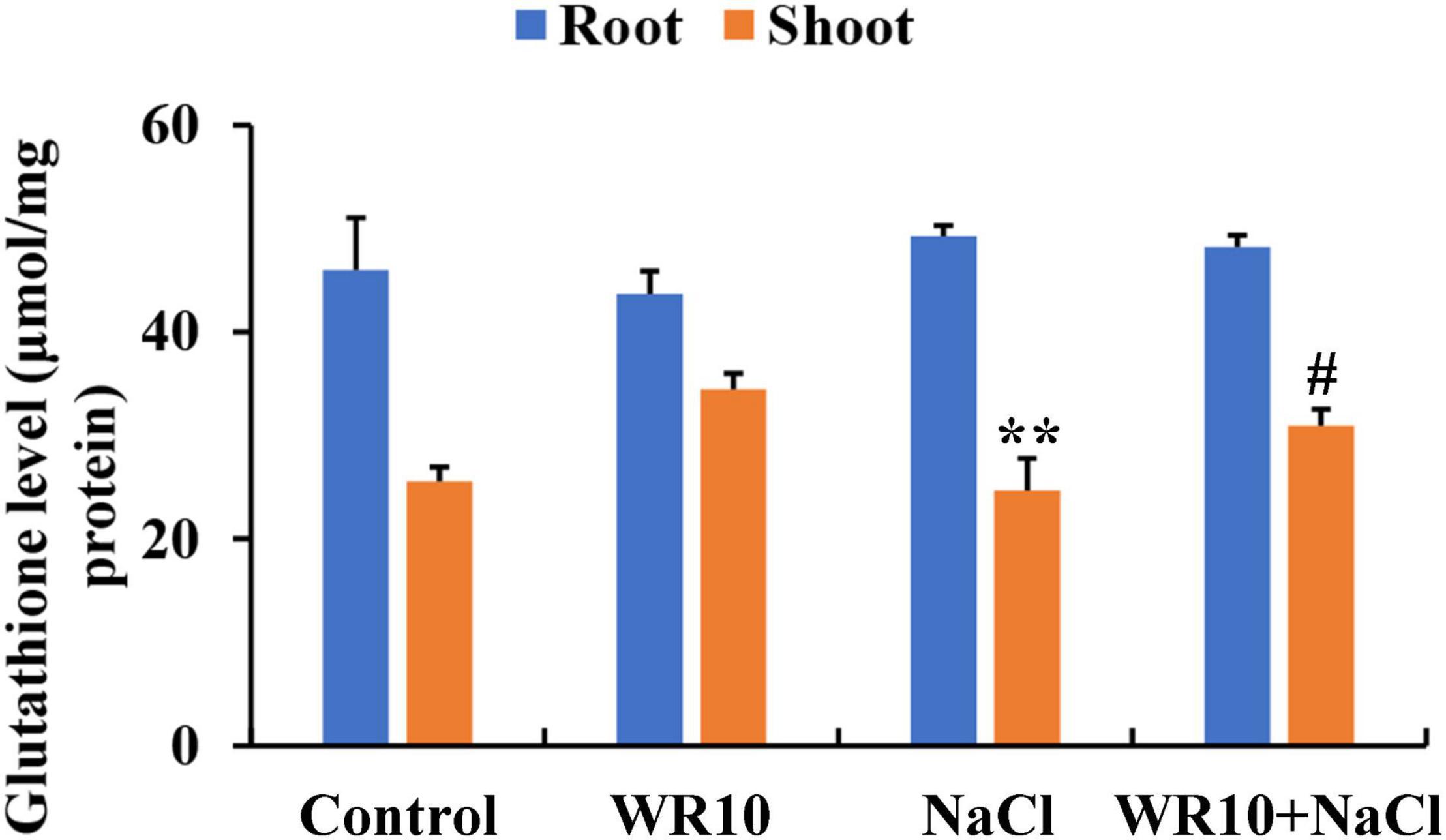
Figure 5. Bacillus altitudinis WR10 increases GSH levels in wheat seedlings under salt stress. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Data are expressed as the means ± SD (n = 6). **p < 0.01 (vs. control); #p < 0.05 (vs. NaCl).
Bacillus altitudinis WR10 activates phenylpropanoid biosynthesis under salt stress
In the phenylpropanoid biosynthesis pathway, the expression levels of trans-cinnamate 4-monooxygenase (CYP73A, EC 1.14.14.91), 4-coumarate-CoA ligase (4CL, EC 6.2.1.12), and cinnamyl alcohol dehydrogenase (CAD, EC 1.1.1.195) were significantly upregulated, while those of ferulate-5-hydroxylase (F5H, EC 1.14.-.-) were significantly downregulated. The levels of other genes, including shikimate O-hydroxycinnamoyltransferase (HCT, EC 2.3.1.133), cinnamoyl-CoA reductase (CCR, EC 1.2.1.44), and peroxidase (POD, EC 1.11.1.7), were either up- or downregulated (Figure 6).
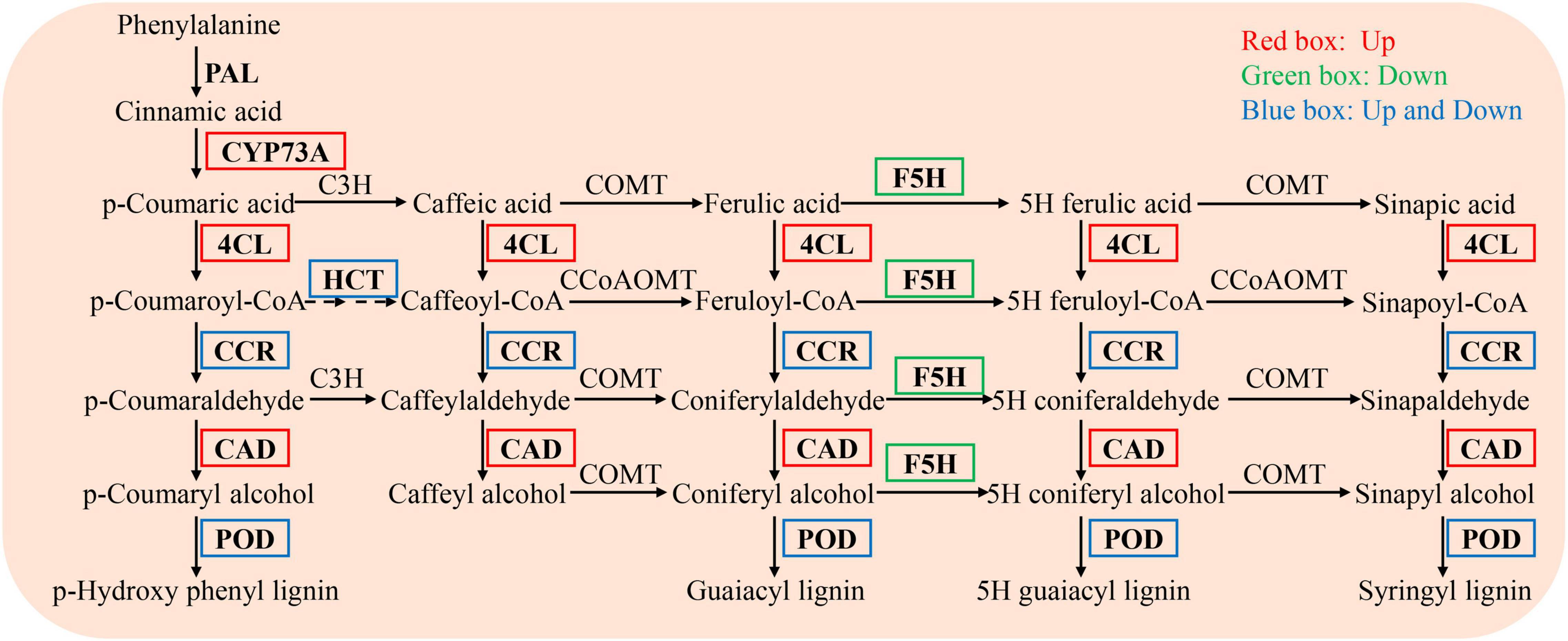
Figure 6. DEGs enriched in the phenylpropanoid biosynthesis pathway in the NaCl vs WR10 + NaCl. NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Red boxes, significantly up-regulated genes; Green boxes, significantly down-regulated genes; Blue boxes, significantly up- and downregulated genes.
Bacillus altitudinis WR10 decreases proline content under salt stress
Under salt stress, proline content increased significantly by 11.7-fold in the shoots of wheat seedlings. In contrast, WR10 inoculation significantly reduced the proline content in salt-stressed wheat plants by 0.61-fold compared to the NaCl group without WR10 inoculation (Figure 7A). RNA-seq analysis showed that the gene expression levels of proline dehydrogenase (PDH) were significantly downregulated in salt-stressed wheat plants compared with those in the control group, but were significantly upregulated in WR10-inoculated plants under salt stress compared with those in the NaCl group without WR10 inoculation (Figure 7B). In addition, delta-1-pyrroline-5-carboxylate synthase (P5CS) and pyrroline-5-carboxylate reductase (P5CR), which are involved in proline biosynthesis, were not affected by WR10 inoculation (Supplementary Table 4).
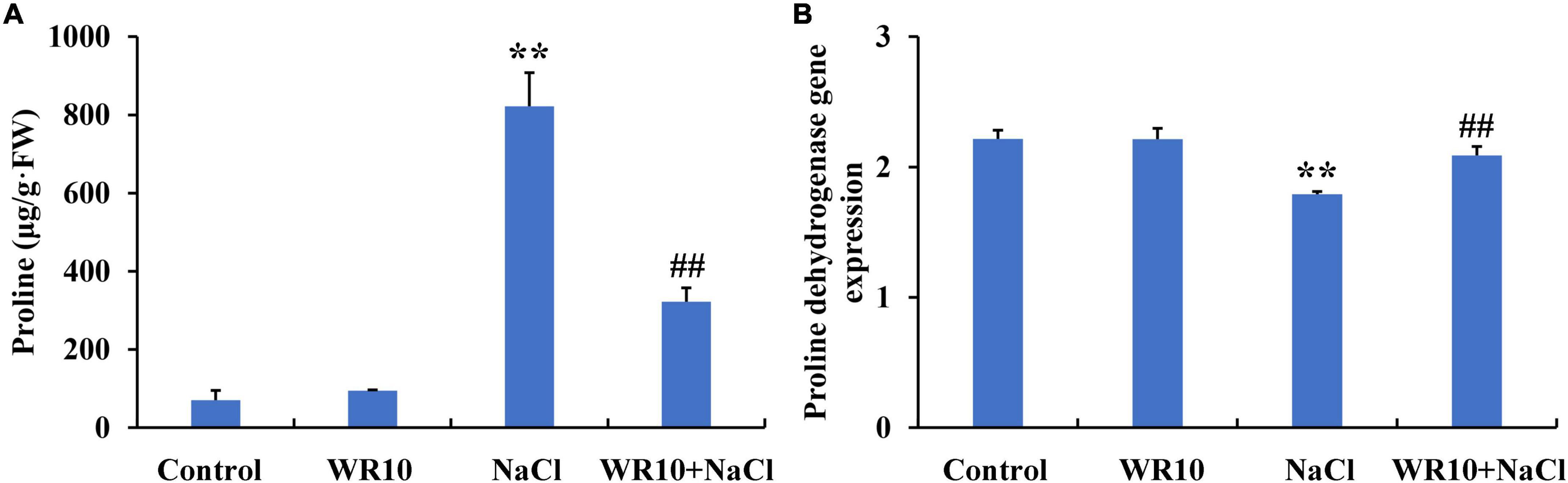
Figure 7. Bacillus altitudinis WR10 decreases the proline content (A) in the shoots and increases the proline dehydrogenase gene expression (B) in the roots of wheat seedlings under salt stress. Control, without NaCl and WR10; WR10, with WR10 and without NaCl; NaCl, with NaCl and without WR10; WR10 + NaCl, with NaCl and WR10. Data are expressed as the means ± SD (n = 6). **p < 0.01 (vs. control); ##p < 0.01 (vs. NaCl).
Discussion
Increased soil salinization seriously decreases the crop yield and endangers food security; it is one of the most threatening abiotic stresses. Wheat is moderately sensitive to salinity, and high salinity often results in growth delays and yield reduction (Royo and Abio, 2003). In this study, the growth parameters of wheat (cv. Zhoumai 36), including root length, shoot length, and root dry weight at the seedling stage, were inhibited via treatment with 150 mmol/L NaCl. This is consistent with other wheat varieties that are grown under salt stress (Quan et al., 2021). Many strategies have been developed to alleviate the adverse effects of salt stress in wheat. Use of plant-derived microbes to improve salt stress tolerance in wheat may be an excellent strategy because of the sustainability, eco-friendly nature, and economic significance of these microbes (Miransari and Smith, 2019). B. altitudinis WR10 is a salt-enduring endophyte that enhances the seed germination rate of wheat under salt stress (Yue et al., 2019). In this study, we investigated the effect of WR10 on the growth of salt-treated wheat seedlings and found that WR10 inoculation significantly improved the seedling growth in terms of root and shoot lengths and dry weights. This indicated that WR10 enhanced wheat salt tolerance at the seedling stage. Similar results were observed for plants inoculated with other Bacillus strains. For instance, Bacillus methylotrophicus M4-1-inoculated wheat shows increased plant height, root length, and dry weight under salt stress (Ji et al., 2020). Inoculation with Bacillus licheniformis HSW-16 significantly increases the root length, shoot length, and dry weight of salt-treated wheat (Singh and Jha, 2016b).
Generally, salt stress results in the accumulation of excess Na+ accompanied by a decrease in K+ levels and an imbalance in the Na+/K+ ratio, which is regarded as one of the main reasons for plant growth inhibition (Schachtman and Liu, 1999). In this study, salt-stressed wheat plants inoculated with WR10 showed lower Na+ and higher K + contents than uninoculated plants. In agreement with our results, a decrease in Na+ content and increase in K+ content was observed in wheat plants inoculated with Bacillus velezensis JC-K3 and Klebsiella sp. SBP-8 (Singh et al., 2015; Ji et al., 2021). We observed that two genes encoding plasma membrane H + -ATPase (AHA) were significantly upregulated in WR10-inoculated wheat roots under salt stress. Similar results were found in maize plants inoculated with Serratia liquefaciens KM4 (El-Esawi et al., 2018). Fan et al. (2018) revealed that Arabidopsis plants overexpressing AHA showed lower Na+ and higher K+ contents under salt stress. Therefore, WR10-induced upregulation of AHA expression may contribute to a lower Na+/K+ ratio in salt-stressed wheat. In addition to Na+ and K+, many studies have reported that PGPB also affects the uptake of other nutrients in plants under salt stress, such as P and Ca2+. In the present study, significant increases in P and Ca2+ levels were also found in salt-stressed wheat plants after WR10 inoculation. In a previous study, P-solubilizing bacteria Bacillus sp. L55, Pseudomonas fluorescens PMT1, Pantoea sp., and Acinetobacter sp. L176, which has phosphatase activity, has been reported to increase P uptake in maize and groundnut under salt stress conditions (Anzuay et al., 2017). WR10 was previously shown to have the ability to solubilize Ca3(PO4)2 and Ca-phytate because of its phosphatase and phytase activity (Yue et al., 2019). Therefore, the enhanced uptake of P could be closely associated with the P solubilizing characteristics of WR10. Another study reported an increased Ca2+ content in tomatoes under salt stress after Glutamicibacter halophytocola KLBMP 5180 inoculation. As a secondary messenger signaling molecule, Ca2+ plays an important role in plant growth, and its accumulation can increase plant resistance to salt stress (Yang et al., 2016). Collectively, these results show that WR10 may alleviate salt stress in wheat partly by improving Na+ exclusion and K+, P, and Ca2+ uptake.
To further explore the underlying mechanisms by which WR10 improves salt tolerance in wheat, transcriptome sequencing was performed. RNA-seq data showed that WR10 inoculation affected several DEGs involved in oxidative stress, ROS metabolism, and antioxidants under salt stress. It is well known that salt stress causes ROS accumulation and oxidative stress in plants (Tang et al., 2015). Some PGPB have been reported to alleviate salt stress by enhancing antioxidant enzymes. For instance, B. licheniformis promotes sunflower growth under saline conditions by increasing the expression of superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPX) (Yasmeen et al., 2020). Serratia marcescens CDP-13 protects wheat plants from salt stress by enhancing the enzymes SOD, CAT, and POD (Singh and Jha, 2016a). Rice plants inoculated with Bacillus pumilus JPVS11 show an increase in SOD and CAT under salt stress (Kumar et al., 2021). Unlike these studies, no obvious changes in SOD, CAT, GPX, or POD were observed in our study. Notably, the gene expression and activity of APX were significantly increased in salt-treated wheat inoculated with WR10. APX is an important antioxidant enzyme in plant cells that plays a pivotal role in scavenging H2O2 (Sharma et al., 2012). Overexpression of APX in tomatoes and rice enhances APX activity and leads to a reduction in H2O2 under salt stress (Wang et al., 2005; Zhang et al., 2013). Therefore, WR10 may reduce H2O2 levels under salt conditions via the activation of APX. Indeed, we observed a decrease in H2O2 levels in the roots of salt-treated wheat plants after inoculation with WR10.
In addition to enzymatic processes, plants produce many non-enzymatic components to withstand oxidative stress. GSH is a crucial antioxidant for ROS scavenging including H2O2 (Gill and Tuteja, 2010). In this study, increased GSH levels, upregulated GSS, and downregulated GST and GGT were found in WR10-inoculated plants under salt stress. GSS is a key enzyme responsible for GSH synthesis and catalyzes the formation of GSH from γGC and glycine (Ogawa, 2005). GST and GGT are important enzymes involved in GSH metabolism, in which GST catalyzes the conjugation of GSH to electrophilic xenobiotic substrates and GGT is responsible for GSH degradation (Labrou et al., 2015; Bachhawat and Kaur, 2017). Therefore, the increase in GSH synthesis and decrease in GSH metabolism induced by WR10 led to an increase in GSH levels in salt-treated wheat. Similarly, increased GSH levels were observed in salt-stressed pea plants after inoculation with B. subtilis and P. fluorescens (Sofy et al., 2021). We suggest that WR10 may accelerate the removal of H2O2 by elevating GSH levels.
KEGG enrichment analysis showed that WR10 inoculation affected many genes involved in phenylpropanoid biosynthesis under salt stress, indicating that this pathway may play an important role in alleviating the effects of WR10 on salt stress. The phenylpropanoid pathway is one of the most important metabolic pathways in plants and produces more than 8000 metabolites (Zhang and Liu, 2015). In this pathway, phenylalanine ammonia lyase (PAL) is the initial enzyme that catalyzes the formation of trans-cinnamic acid from phenylalanine. Cinnamic acid is then hydroxylated by cinnamic acid 4-hydroxylase (CYP73A) and the resultant p-coumaric acid is further catalyzed by 4-coumarate-CoA ligase (4CL) to generate p-coumaroyl-CoA (Dong and Lin, 2021). In our study, significantly upregulated CYP73A and 4CL were found in salt-stressed plants after inoculation with WR10, indicating that WR10 may increase the p-coumaroyl-CoA content. p-coumaroyl-CoA is important precursor for all downstream metabolites in the phenylpropanoid pathway, including phenolics and flavonoids (Vogt, 2010; Sharma et al., 2019). This increase may provide more substrate for these compounds protecting plants against abiotic stress. Li et al. (2021) indicated that N deficiency induces phenolic and flavonoid accumulation by upregulating the gene expression of PAL, CYP73A, and 4CL in Coreopsis tinctoria. Kim et al. (2014) reported that overexpression of CYP73A and 4CL promotes flavone accumulation in Scutellaria baicalensis roots. Therefore, activation of the phenylpropanoid pathway may be another strategy for WR10 to improve the salt tolerance of wheat.
Proline is an important osmolyte in plants, and its accumulation often occurs when plants are exposed to abiotic stress (Hayat et al., 2012). Many plants, including wheat, produce high levels of proline under salt stress. Surprisingly, WR10-inoculation resulted in significantly lower proline levels in salt-stressed wheat plants. Similar results were observed in Pseudomonas azotoformans CHB 1107-inoculated tomato plants and Azotobacter chroococcum-inoculated maize plants under salt stress (Rojas-Tapias et al., 2012; Liu et al., 2021). The proline content is controlled by proline biosynthesis and degradation. In this study, the gene expression of P5CS and P5CR related to proline biosynthesis was not affected by WR10 inoculation under salt stress, whereas PDH mRNA levels were significantly upregulated. PDH is a key enzyme involved in proline degradation, and degrades proline to P5C (Wang et al., 2015). Therefore, we speculated that upregulated PDH expression contributed to the reduction of proline.
In addition to affecting plants, our previous study found that WR10 relieved the adverse effects of salt stress on wheat seed germination through its inherent tolerance to NaCl, biofilm formation, and production of ACC deaminase (Yue et al., 2019). Biofilms enhance bacterial surface attachment in roots and contain many exopolysaccharides (EPS), which chelate dissociated Na+ and restrict Na+ importation into the roots (Ashraf et al., 2004; Dodd and Pérez-Alfocea, 2012). ACC deaminase is an endogenous enzyme in endophytes that catalyzes the conversion of ACC, a precursor of ethylene biosynthesis, into ammonia and α-ketobutyric acid. Many ACC deaminase-producing bacteria can reduce plant-produced ACC, thereby inhibiting ethylene production and mitigating salt stress in plants (Misra and Chauhan, 2020; Orozco-Mosqueda et al., 2020). Therefore, we believe that the salinity-alleviating properties of WR10 in wheat can also be attributed to the biofilm and ACC deaminase produced by WR10.
Conclusion
In conclusion, this study confirmed that the halotolerant strain, B. altitudinis WR10, improved the salt tolerance of wheat at the seedling stage. Therefore, this endophyte has the potential to improve crop growth under high-salt stress as a novel bioinoculant. Furthermore, we revealed that the mitigating effects of WR10 can be attributed to the following aspects: (1) its inherent plant growth-promoting traits and salt resistance; (2) improved Na+/K+ homeostasis and increased uptake of nutrients such as P and Ca2+; (3) enhanced H2O2 scavenging via increased APX activity and GSH levels; and (4) activation of the phenylpropanoid pathway. These findings provide a theoretical basis for the application of WR10 in saline soil. However, further studies are necessary to verify the salinity-alleviating properties of WR10 as a bioinoculant in actual fields, considering the complexity of the soil environment in these fields.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZY designed the experiments and wrote the draft manuscript. YW, LZ, QZ, YL, and CH performed the experiments. YC and KM analyzed the results. CC uploaded RNA-seq data. ZS provided the raw materials and financial support and completed the final manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32071478).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.941388/full#supplementary-material
References
Abd_Allah, E. F., Alqarawi, A. A., Hashem, A., Radhakrishnan, R., Al-Huqail, A. A., Al-Otibi, F. O. N., et al. (2018). Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 13, 37–44. doi: 10.1080/17429145.2017.1414321
Anzuay, M. S., Ciancio, M. G. R., Ludueña, L. M., Angelini, J. G., Barros, G., Pastor, N., et al. (2017). Growth promotion of peanut (Arachis hypogaea L.) and maize (Zea mays L.) plants by single and mixed cultures of efficient phosphate solubilizing bacteria that are tolerant to abiotic stress and pesticides. Microbiol. Res. 199, 98–109. doi: 10.1016/j.micres.2017.03.006
Ashraf, M., and Akram, N. A. (2009). Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnol. Adv. 27, 744–752. doi: 10.1016/j.biotechadv.2009.05.026
Ashraf, M., Hasnain, S., Berge, O., and Mahmood, T. (2004). Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 40, 157–162. doi: 10.1007/s00374-004-0766-y
Bachhawat, A. K., and Kaur, A. (2017). Glutathione degradation. Antioxid. Redox Signal. 27, 1200–1216. doi: 10.1089/ars.2017.7136
Bharti, N., Pandey, S. S., Barnawal, D., Patel, V. K., and Kalra, A. (2016). Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6:34768. doi: 10.1038/srep34768
Chang, P., Gerhardt, K. E., Huang, X. D., Yu, X. M., Glick, B. R., Gerwing, P. D., et al. (2014). Plant growth-promoting bacteria facilitate the growth of barley and oats in salt-impacted soil: implications for phytoremediation of saline soils. Int. J. Phytoremed. 16, 1133–1147. doi: 10.1080/15226514.2013.821447
Dodd, I. C., and Pérez-Alfocea, F. (2012). Microbial amelioration of crop salinity stress. J. Exp. Bot. 636, 3415–3428. doi: 10.1093/jxb/ers033
Dong, N. Q., and Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
El-Esawi, M. A., Alaraidh, I. A., Alsahli, A. A., Alzahrani, S. M., Ali, H. M., Alayafi, A. A., et al. (2018). Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 19:3310. doi: 10.3390/ijms19113310
Fan, Y., Wan, S., Jiang, Y., Xia, Y., Chen, X., Gao, M., et al. (2018). Over-expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis. Protoplasma 255, 1827–1837. doi: 10.1007/s00709-018-1275-4
Food and Agriculture Organization [FAO] (2009). High Level Expert Forum-How to Feed the World in 2050. Rome: Economic and Social Development, Food and Agricultural Organization of the United Nations.
Food and Agriculture Organization [FAO] (2021). Global Map of Salt-Affected Soils. Available online at: https://www.fao.org/soils-portal/data-hub/soil-maps-and-databases/global-map-of-salt-affected-soils/en/ (accessed April 12, 2022).
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Ji, C., Wang, X., Song, X., Zhou, Q., Li, C., and Chen, Z. (2021). Effect of Bacillus velezensis JC-K3 on endophytic bacterial and fungal diversity in wheat under salt stress. Front. Microbiol. 12:802054. doi: 10.3389/fmicb.2021.802054
Ji, C., Wang, X., Tian, H., Hao, L., Wang, C., Zhou, Y., et al. (2020). Effects of Bacillus methylotrophicus M4-1 on physiological and biochemical traits of wheat under salinity stress. J. Appl. Microbiol. 129, 695–711. doi: 10.1111/jam.14644
Kim, Y. S., Kim, Y. B., Kim, Y., Lee, M. Y., and Park, S. U. (2014). Overexpression of cinnamate 4-hydroxylase and 4-coumaroyl CoA ligase prompted flavone accumulation in Scutellaria baicalensis hairy roots. Nat. Prod. Commun. 9, 803–807.
Kumar, A., Singh, S., Mukherjee, A., Rastogi, R. P., and Verma, J. P. (2021). Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 to enhance plant growth attributes of rice and improve soil health under salinity stress. Microbiol. Res. 242:126616. doi: 10.1016/j.micres.2020.126616
Labrou, N. E., Papageorgiou, A. C., Pavli, O., and Flemetakis, E. (2015). Plant GSTome: structure and functional role in xenome network and plant stress response. Curr. Opin. Biotechnol. 32, 186–194. doi: 10.1016/j.copbio.2014.12.024
Li, Z., Jiang, H., Qin, Y., Yan, H., Jiang, X., and Qin, Y. (2021). Nitrogen deficiency maintains the yield and improves the antioxidant activity of Coreopsis tinctoria Nutt. Biosci. Biotechnol. Biochem. 85, 1492–1505. doi: 10.1093/bbb/zbab048
Liu, C. H., Siew, W., Hung, Y. T., Jiang, Y. T., and Huang, C. H. (2021). 1-Aminocyclopropane-1-carboxylate (ACC) deaminase gene in Pseudomonas azotoformans is associated with the amelioration of salinity stress in tomato. J. Agric. Food Chem. 69, 913–921. doi: 10.1021/acs.jafc.0c05628
Lodeyro, A. F., and Carrillo, N. (2015). “Salt stress in higher plants: mechanisms of toxicity and defensive responses,” in Stress Responses in Plants, eds B. Nath and T. M. Müller (Cham: Springer), 1–33. doi: 10.1007/978-3-319-13368-3_1
Miransari, M., and Smith, D. (2019). Sustainable wheat (Triticum aestivum L.) production in saline fields: a review. Crit. Rev. Biotechnol. 39, 999–1014. doi: 10.1080/07388551.2019.1654973
Mishra, P., Mishra, J., and Arora, N. K. (2021). Plant growth promoting bacteria for combating salinity stress in plants - Recent developments and prospects: a review. Microbiol. Res. 252:126861. doi: 10.1016/j.micres.2021.126861
Misra, S., and Chauhan, P. S. (2020). ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech 10:119. doi: 10.1007/s13205-020-2104-y
Mukherjee, P., Mitra, A., and Roy, M. (2019). Halomonas rhizobacteria of Avicennia marina of indian sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 10:1207. doi: 10.3389/fmicb.2019.01207
Muranaka, S., Shimizu, K., and Kato, M. (2002). Ionic and osmotic effects of salinity on single-leaf photosynthesis in two wheat cultivars with different drought tolerance. Photosynthetica 40, 201–207. doi: 10.1023/A:1021337522431
Ogawa, K. (2005). Glutathione-associated regulation of plant growth and stress responses. Antioxid. Redox Signal. 7, 973–981. doi: 10.1089/ars.2005.7.973
Orhan, F. (2016). Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Braz. J. Microbiol. 47, 621–627. doi: 10.1016/j.bjm.2016.04.001
Orhan, F., and Demirci, A. (2020). Salt stress mitigating potential of halotolerant/halophilic plant growth promoting. Geomicrobiol. J. 37, 663–669. doi: 10.1080/01490451.2020.1761911
Orozco-Mosqueda, M. D. C., Glick, B. R., and Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol. Res. 235:126439. doi: 10.1016/j.micres.2020.126439
Qadir, M., Quillérou, E., Nangia, V., Murtaza, G., Singh, M., Thomas, R. J., et al. (2014). Economics of salt-induced land degradation and restoration. Nat. Resour. Forum. 38, 282–295. doi: 10.1111/1477-8947.12054
Qin, Y., Druzhinina, I. S., Pan, X., and Yuan, Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34, 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Quan, X., Liang, X., Li, H., Xie, C., He, W., and Qin, Y. (2021). Identification and characterization of wheat germplasm for salt tolerance. Plants 10:268. doi: 10.3390/plants10020268
Raheem, A., and Ali, B. (2015). Halotolerant rhizobacteria: beneficial plant metabolites and growth enhancement of Triticum aestivum L. in salt-amended soils. Arch. Agron. Soil Sci. 61, 1691–1705. doi: 10.1080/03650340.2015.1036044
Ramadoss, D., Lakkineni, V. K., Bose, P., Ali, S., and Annapurna, K. (2013). Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. Springerplus 2:6. doi: 10.1186/2193-1801-2-6
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., and Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Royo, A., and Abio, D. (2003). Salt tolerance in durum wheat cultivars. Span. J. Agric. Res. 1, 27–35. doi: 10.5424/sjar/2003013-32
Satir, O., and Berberoglu, S. (2016). Crop yield prediction under soil salinity using satellite derived vegetation indices. Field Crop. Res. 192, 134–143. doi: 10.1016/j.fcr.2016.04.028
Schachtman, D. P., and Liu, W. (1999). Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 4, 281–287. doi: 10.1016/S1360-1385(99)01428-4
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., and Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24:2452. doi: 10.3390/molecules24132452
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:217037. doi: 10.1155/2012/217037
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., and Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Sec. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Singh, R. P., Jha, P., and Jha, P. N. (2015). The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 184, 57–67. doi: 10.1016/j.jplph.2015.07.002
Singh, R. P., and Jha, P. N. (2016a). The multifarious PGPR Serratia marcescens CDP-13 augments induced systemic resistance and enhanced salinity tolerance of wheat (Triticum aestivum L.). PLoS One 11:e0155026. doi: 10.1371/journal.pone.0155026
Singh, R. P., and Jha, P. N. (2016b). A halotolerant bacterium Bacillus licheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Front. Plant Sci. 7:1890. doi: 10.3389/fpls.2016.01890
Sofy, M. R., Aboseidah, A. A., Heneidak, S. A., and Ahmed, H. R. (2021). ACC deaminase containing endophytic bacteria ameliorate salt stress in Pisum sativum through reduced oxidative damage and induction of antioxidative defense systems. Environ. Sci. Pollut. Res. 28, 40971–40991. doi: 10.1007/s11356-021-13585-3
Sun, Z., Liu, K., Zhang, J., Zhang, Y., Xu, K., Yu, D., et al. (2017). IAA producing Bacillus altitudinis alleviates iron stress in Triticum aestivum L. seedling by both bioleaching of iron and up-regulation of genes encoding ferritins. Plant Soil 419, 1–11. doi: 10.1007/s11104-017-3218-9
Sun, Z., Yue, Z., Liu, H., Ma, K., and Li, C. (2021). Microbial-assisted wheat iron biofortification using endophytic Bacillus altitudinis WR10. Front. Nutr. 8:704030. doi: 10.3389/fnut.2021.704030
Tang, X., Mu, X., Shao, H., Wang, H., and Brestic, M. (2015). Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 35, 425–437. doi: 10.3109/07388551.2014.889080
Wang, H., Tang, X., Wang, H., and Shao, H. B. (2015). Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 6:792. doi: 10.3389/fpls.2015.00792
Wang, Y., Wisniewski, M., Meilan, R., Cui, M., Webb, R., and Fuchigami, L. (2005). Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J. Am. Soc. Hortic. Sci. 130, 167–173. doi: 10.21273/JASHS.130.2.167
Yang, H., Hu, J., Long, X., Liu, Z., and Rengel, Z. (2016). Salinity altered root distribution and increased diversity of bacterial communities in the rhizosphere soil of Jerusalem artichoke. Sci. Rep. 6:20687. doi: 10.1038/srep20687
Yasmeen, T., Ahmad, A., Arif, M. S., Mubin, M., Rehman, K., Shahzad, S. M., et al. (2020). Biofilm forming rhizobacteria enhance growth and salt tolerance in sunflower plants by stimulating antioxidant enzymes activity. Plant Physiol. Bioch. 156, 242–256. doi: 10.1016/j.plaphy.2020.09.016
Yue, Z. H., Chen, Y. J., Chen, C., Ma, K. S., Tian, E. L., Wang, Y., et al. (2021). Endophytic Bacillus altitudinis WR10 alleviates Cu toxicity in wheat by augmenting reactive oxygen species scavenging and phenylpropanoid biosynthesis. J. Hazard. Mater. 405:124272. doi: 10.1016/j.jhazmat.2020.124272
Yue, Z. H., Shen, Y. H., Chen, Y. J., Liang, A. W., Chu, C. W., Chen, C., et al. (2019). Microbiological insights into the stress-alleviating property of an endophytic Bacillus altitudinis WR10 in wheat under low-phosphorus and high-salinity stresses. Microorganisms 7:508. doi: 10.3390/microorganisms7110508
Zhang, X., and Liu, C. J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 8, 17–27. doi: 10.1016/j.molp.2014.11.001
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS One 8:e57472. doi: 10.1371/journal.pone.0057472
Keywords: Bacillus, salt stress, wheat, antioxidant enzyme, hydrogen peroxide, glutathione, phenylpropanoid biosynthesis
Citation: Yue Z, Chen Y, Wang Y, Zheng L, Zhang Q, Liu Y, Hu C, Chen C, Ma K and Sun Z (2022) Halotolerant Bacillus altitudinis WR10 improves salt tolerance in wheat via a multi-level mechanism. Front. Plant Sci. 13:941388. doi: 10.3389/fpls.2022.941388
Received: 11 May 2022; Accepted: 27 June 2022;
Published: 14 July 2022.
Edited by:
Habib-ur-Rehman Athar, Bahauddin Zakariya University, PakistanReviewed by:
Asma Imran, National Institute for Biotechnology and Genetic Engineering, PakistanNoshin Ilyas, Pir Mehr Ali Shah Arid Agriculture University, Pakistan
Hassan Etesami, University of Tehran, Iran
Furkan Orhan, Ağrı İbrahim Çeçen University, Turkey
Copyright © 2022 Yue, Chen, Wang, Zheng, Zhang, Liu, Hu, Chen, Ma and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongke Sun, c3VuemhAZGFhZC1hbHVtbmkuZGU=; Keshi Ma, emtudW1rc0AxNjMuY29t
 Zonghao Yue1
Zonghao Yue1 Chunhong Hu
Chunhong Hu Can Chen
Can Chen Zhongke Sun
Zhongke Sun