- 1Liaoning Key Laboratory for Biological Invasions and Global Changes, College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, China
- 2Chinese Academy of Science (CAS) Key Laboratory of Tropical Plant Resources and Sustainable Use, CAS Center for Excellence in Molecular Plant Sciences, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Science, Kunming, China
Horticultural plants such as vegetables, fruits, and ornamental plants are crucial to human life and socioeconomic development. Gibberellins (GAs), a class of diterpenoid compounds, control numerous developmental processes of plants. The roles of GAs in regulating growth and development of horticultural plants, and in regulating significant progress have been clarified. These findings have significant implications for promoting the quality and quantity of the products of horticultural plants. Here we review recent progress in determining the roles of GAs (including biosynthesis and signaling) in regulating plant stature, axillary meristem outgrowth, compound leaf development, flowering time, and parthenocarpy. These findings will provide a solid foundation for further improving the quality and quantity of horticultural plants products.
Introduction
Gibberellins (GAs) are numerous plant hormones that are all consist of a large number of diterpenoid compounds (Hedden, 2020; Zhang et al., 2020). GA was first characterized from the pathogenic fungus Gibberella fujikuroi by Japanese scientists (Yabuta, 1938; Binenbaum et al., 2018), and it causes severe disease with the symptom of excessive internode elongation known as ‘foolish seedling disease’ in Oryza sativa (rice). More than 130 GAs have been discovered in plants, fungi, and bacteria (Yamaguchi, 2008; Hedden, 2020), and they are named GAn (for example GA4) in the order of discovery (MacMillan and Takahashi, 1968). Among GAs, only GA1, GA3, GA4, and GA7 are bioactive, controlling multiple developmental processes in plants (Yamaguchi, 2008; Hedden, 2020; Zhang et al., 2020).
It is important to study the biosynthesis and signaling pathways of GAs to determine their biological functions. GA biosynthesis in plants consists of a series of complicated oxidation-reduction reactions, including reactions from geranylgeranyl diphosphate (GGDP) to bioactive GAs (Yamaguchi, 2008; Hedden, 2020). This process is usually classified into three stages in Arabidopsis (Figure 1A; Yamaguchi, 2008; Zhang et al., 2020; Cheng et al., 2021; Shohat et al., 2021). The first stage is the synthesis of GGDP to ent-kaurene using GGDP as starting material in plastids, which is consecutively catalyzed and oxidated by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) (Figure 1A; Sun and Kamiya, 1997; Helliwel et al., 2001; Hedden, 2020). The second stage is the conversion from ent-kaurene to GA12 in six steps. The first three steps are oxidized by ent-kaurene oxidase (KO) and the remaining steps by ent-kaurenic acid oxidase (KAO) (Figure 1A; Regnault et al., 2014). KO is located in the plastid membrane while KAO is in the endoplasmic reticulum (Yamaguchi, 2008). GA biosynthesis from GA12 to bioactive GAs is the last stage, which is divided into the 13-hydroxylated pathway (from GA53 to GA1) and the non-13-hydroxylated cascade (from GA12 to GA4). In the beginning, partial GA12 is converted to GA53 catalyzed by GA 13-oxidases (GA13ox) (Magome et al., 2013; He et al., 2019a). Then, GA12 and GA53 are converted to bioactive GAs in the cytosol after a series of oxygenation reactions catalyzed mainly by GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox; Figure 1A; Tenreira et al., 2017; Zhang et al., 2020).
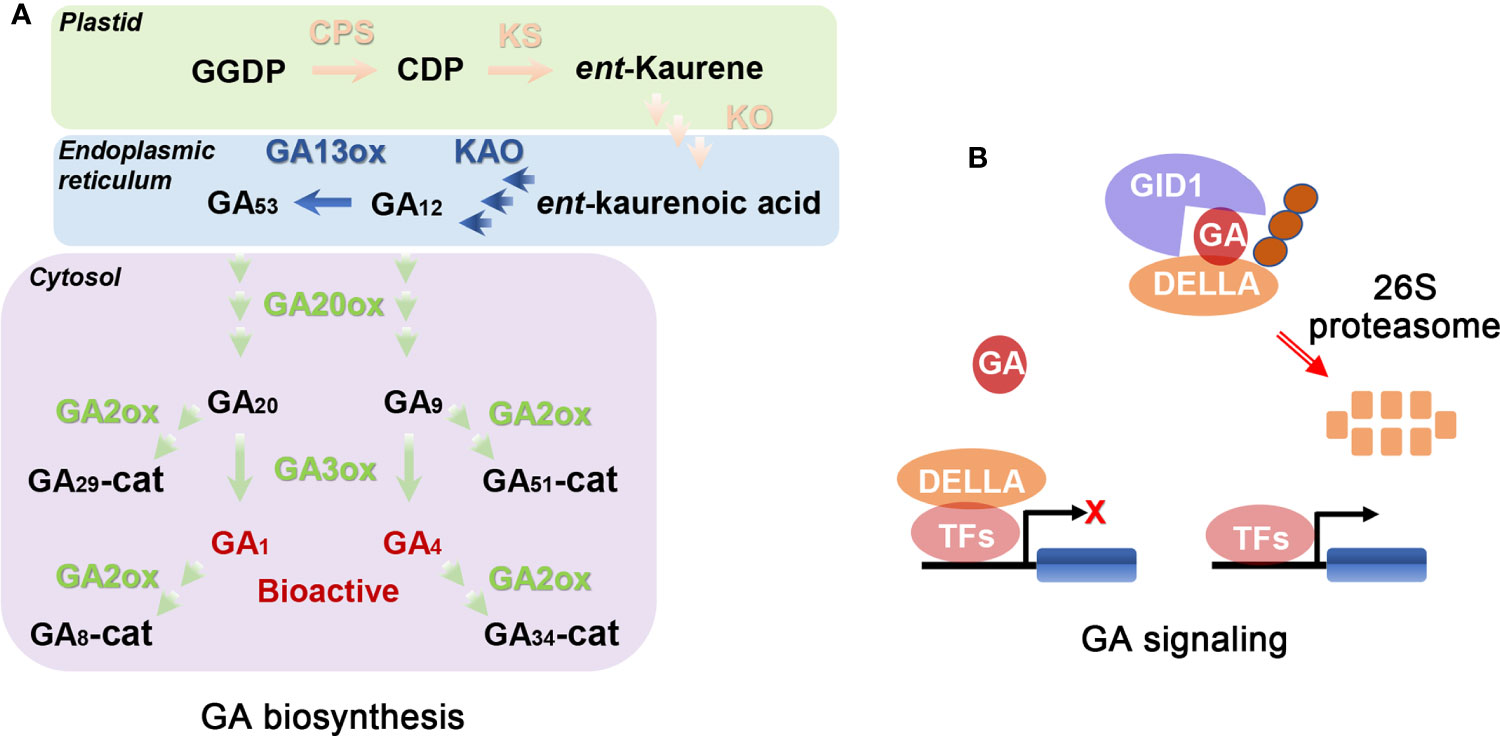
Figure 1 Schematic representations of GA biosynthesis (A) and signal (B) pathways (Yamaguchi, 2008; Binenbaum et al., 2018; Wu et al., 2020). (A) GGDP, CDP, and cat represent geranylgeranyl diphosphate, ent-copalyl diphosphate, and catabolite, respectively. CPS, KS, KO, and KAO represent ent-copalyl diphosphate synthase, ent-kaurene synthase, ent-kaurene oxidase, and ent-kaurenoic acid oxidase, respectively. GA13ox, GA20ox, GA3ox, and GA2ox represent GA13-oxidases, GA20-oxidases, GA3-oxidases, and GA2-oxidases, respectively. CPS, KS, KO, KAO, GA13ox, GA20ox, and GA3ox are the key enzymes in GA biosynthetic pathway. GA2ox is GA deactivated enzyme. Enzymes locate in plastid, endoplasmic reticulum, and cytosol using pink font, green font, and blue font, respectively. Bioactive metabolic products (GA1 and GA4) use red font. The remaining metabolic products use black font. (B) When bioactive GAs are exhausted or limited, DELLAs (consisting of the following proteins: GA-INSENSITIVE, GAI; REPRESSOR OF GA1-3, RGA; RGA-LIKE1, RGL1; RGL2 and RGL3; GRAS-domain proteins) interact with transcription factors (TFs) inhibiting expression of corresponding downstream genes and therefore growth. When endogenous GA concentration is increased, a complex of GA-INSENSITIVE DWARF1 (GID1; the GA receptor)-GA-DELLA is formed. Then, the complex is recognized by the F-box proteins, leading to degradation of DELLA by ubiquitin-26S proteasome system and thus releasing the growth inhibition. GID1 and DELLAs are the core components of GA signal pathway. The orange circles represent F-box proteins, such as SLEEPY1 (SLY1).
Among the genes encoding the above-mentioned GA biosynthetic enzymes, CPS, KS, and KO are single copy genes, while the others are multi-copy genes. Among the above-mentioned enzymes of GA biosynthesis, GA20ox is a rate-limiting enzyme (Yamaguchi, 2008). Mutations of GA biosynthesis related genes decrease plant concentrations of bioactive GAs, leading to multiple defective phenotypes (Zhang et al., 2020). These defective phenotypes can be recovered by addition of exogenous bioactive GAs, and hence these mutants are called GA-deficient mutants. Besides GA biosynthesis, GA deactivation is also important for modifying the concentrations of bioactive GAs in plant tissues (Schomburg et al., 2003; Zhu et al., 2006). For example, GA 2-oxidases (GA2ox) can catalyze GA20, GA9, GA1, and GA4 to inactive GAs (Figure 1A). Loss of function of GA deactivated genes often leads to tissues with high concentrations of bioactive GAs.
The GA signaling pathway has also been characterized in Arabidopsis. The current model of the GA signaling pathway consists of several key components, including bioactive GAs, GIBBERELLIN INSENSITIVE DWARF1 (GID1, the GA receptor), DELLAs (consisting of the following proteins: GA-INSENSITIVE, GAI; REPRESSOR OF GA1-3, RGA; RGA-LIKE1, RGL1; RGL2 and RGL3; GRAS-domain proteins), and two F-box proteins (SLEEPY1; SLY1 and SNEEZY; SNZ) (Davière and Achard, 2014; Shohat et al., 2021). When bioactive GAs are exhausted or limited, DELLAs interact with multiple transcription factors (TFs), inhibiting expression of corresponding downstream genes, and thus repressing almost all GA-related growth responses (Davière and Achard, 2014; Shohat et al., 2021; Chai et al., 2022; Jin et al., 2022). When bioactive GA concentration is increased, a GA-binding pocket in GID1 captures bioactive GAs and H2O, causing conformational change of the flexible N-terminal extension from the pocket and thus closing the GA-binding pocket (Murase et al., 2008; Shimada et al., 2008). Subsequently, DELLAs bind with the N-terminal extension of GID1 via its TVHYNP regions forming a GID1-GA-DELLA complex, and the recruited F-box proteins immediately bind with DELLA (Shohat et al., 2021). SLY1 (a F-box protein) is a component of SKP1, CULLIN, F-BOX (SCF) E3 ubiquitin-ligase complexes and the SCF complex contributes to subsequent degradation of DELLAs. Once the GID1-GA-DELLA-SCFSLY1 complex is formed, DELLAs are subsequently degraded by the 26S proteasome, releasing the growth inhibition (Figure 1B; Davière and Achard, 2014; Shohat et al., 2021; Chai et al., 2022). DELLAs exert a core role in the GA signaling pathway. Mutations of GA perception and signal transduction can lead to mutant phenotypes in plants even if they have high concentrations of GAs. This kind of defective phenotype of mutant plants cannot be recovered using exogenous bioactive GAs, and they are hence called GA-insensitive mutants.
Both GA-deficient and GA-insensitive mutants generally exhibit typical dwarf phenotypes (Davière and Achard, 2014; Hedden, 2020; Zhang et al., 2020). Among these mutants, mutations of CPS, KS, or KO always lead to severe dwarfism because they are all single-copy genes (Sun and Kamiya, 1997; Helliwel et al., 2001; Regnault et al., 2014). Mutations of other GA-biosynthesis and -signaling pathway related genes often display a semi-dwarf phenotype as they are all multi-copy genes (Griffiths et al., 2006; Magome et al., 2013). In comparison, mutants related to constitutively active GA exhibit a spindly internode, such as elongated uppermost internode (eui; a deactivated gene; Zhu et al., 2006). The semi-dwarf1 (sd1) and Reduced height-1(Rht-1) mutants, caused by mutations of GA20ox2 and DELLA, respectively, prevent excessive internode growth and lodging, and therefore grain yield loss caused by wind and overuse of chemical fertilizer (Peng et al., 1999; Sasaki et al., 2002). The dramatically increased yield from semi-dwarf crops has saved countless lives, and is referred to as the ‘Green Revolution’. Recently, the GROWTH-REGULATING FACTOR4 (GRF4)-DELLA-NITROGEN-MEDIATED TILLER GROWTH RESPONSE 5 (NGR5) module in rice has shown potential to improve nitrogen use-efficiency (NUE) and increase tiller number in semi-dwarf ‘Green Revolution’ varieties (Li et al., 2018a; Wu et al., 2020). This discovery is regarded as a new Green Revolution in the 21st century.
Apart from biosynthetic and signaling pathways of GAs, the local accumulation maxima of bioactive GAs in the organ formation zone is a prerequisite for normal organ development (Binenbaum et al., 2018). Synthesis sites of bioactive GAs are generally the sites where GAs function. However, this is not always the case (Ragni et al., 2011; Lacombe and Achard, 2016; Camut et al., 2019). Hence, transportation of GAs from synthesis sites to the sites where they function is essential (Katsumi et al., 1983). Some precursors of bioactive GAs or their deactivated metabolites can also be transported (Hu et al., 2008; Regnault et al., 2015; Lange and Lange, 2016; Camut et al, 2019). Like other hormones, GAs can possibly move in both directions in vascular tissues (Binenbaum et al., 2018; Zhang et al., 2020). The transportation of GAs can be classified into short- and long-distance movements. The short-distance movement of GAs has been demonstrated in Cucumis sativus (cucumber; Hu et al., 2008; Lange and Lange, 2016). GA9 (a precursor of GA4; Figure 1A) is synthesized in cucumber ovaries where GA20ox has relatively high expression, while GA9 is converted to GA4 in sepals and petals where a relatively high expression level of GA3ox is detected (Lange and Lange, 2016). Thus, the short-distance movement of GA9 is essential for flower development in cucumber.
Many studies in Arabidopsis have demonstrated that the long-range transportation of endogenous GA12 enhances the ability to adapt to adverse environments (Regnault et al., 2015; Camut et al, 2019). The long-distance movement of GAs can even be root-to-shoot and shoot-to-root transportation (Regnault et al., 2015; Camut et al, 2019). It is generally believed that precursors of bioactive GAs are dominating forms of movement, although the mobility of bioactive GAs was identified approximately 40 years ago (Katsumi et al., 1983). A previous study showed that bioactive GA3 could be transported in grafts between normal and mutant seedlings in Zea mays (maize; Katsumi et al., 1983). Obviously, the movements of GAs are involved in controlling plant growth and development. But it is still not clear how GAs are transported from cell to cell and what their receptors are in cell membranes in horticultural plants and other species.
Horticultural plants include numerous species, such as Solanum lycopersicum (tomato), Capsicum annuum (peppers), Pisum sativum (pea), Brassica rapa L. ssp. Pekinensis (Chinese cabbage), Lactuca sativa (lettuce), Malus pumila (apple), Vitis vinifera (grape), Rosa chinensis (rose), and Panax ginseng (ginseng). The products of horticultural plants are not only served as foods, but also provide dietary intake of vitamins and minerals. Moreover, some secondary metabolites from horticultural plants are often used to treat human diseases. Horticultural plants are becoming more and more important for human beings under population increases, global environmental changes, and land degradation. Fortunately, genomic data of numerous horticultural species have been published in recent years (Sun et al., 2021), which is advantageous for horticultural researchers. However, studies on the roles of GAs in manipulating growth and development of horticultural plants are lacking compared with rice and Arabidopsis. In this review, we focus on GA biosynthesis and GA signaling in horticultural plants and discuss how GA regulates vegetative and reproductive growth of horticultural plants, with the purpose of helping horticultural researchers understand the genetic and molecular mechanisms of GA functions.
Vegetative growth
Plant stature
An ideal plant architecture is important in cereal crops but is also critical to horticultural plants. Shoot architecture is mainly influenced by stem elongation (Guo et al., 2020; Zhang et al., 2020). In the 1960s, the use of semi-dwarf cereal crop varieties contributed to great increases in crop yield (Peng et al., 1999; Sasaki et al., 2002). Subsequently, related mutation genes (GA20ox2 and DELLA) were identified in rice and Triticum aestivum (wheat), and GAs were shown to be involved in the semi-dwarf trait (Peng et al., 1999; Sasaki et al., 2002). Similar results were also found in horticultural plants (Schrager-Lavelle et al., 2019; Sun et al., 2020).
By bulked segregant analysis (BSA) and mapping, the gene responsible for ‘w106 (dwarf)’ in Citrullus lanatus (watermelon) was identified as Cla015407 encoding ClGA3ox (Sun et al., 2020). Similar results were found at the same time by Gebremeskel et al. (2019). The tomato internode elongated-1 (tie1) mutants exhibit the internode elongation phenotype and this is a loss-of-function mutation of SlGA2ox7 (a GA catabolic gene; Schrager-Lavelle et al., 2019). Paclobutrazol (PAC), a GA biosynthesis inhibitor, can restore the tie1 defective phenotype (Schrager-Lavelle et al., 2019). The mutation of Non-Heading Mutant (NHM1; encoding KS enzyme) leads to the non-heading phenotype in Chinese cabbage (Gao et al., 2020). Reverse genetic screening results also verify that GAs positively control internode elongation. Editing PROCERA/SlDELLA in tomato and MaGA20ox2 in Musa acuminate (banana) using CRISPR-Cas9 results in shortened internodes (a dominant mutation and a loss-of-function mutation; Tomlinson et al., 2019; Shao et al., 2020). Overexpression of PsGA3ox1 and PpGA2ox1 lead to longer and more compact internodes in pea and Nicotiana tabacum (tobacco), respectively (Reinecke et al., 2013; Cheng et al., 2021). GAs and PAC application also obviously change the internode length of two commercial grapevine cultivars (Acheampong et al., 2015).
All of the genes discussed above (Cla015407, TIE1, NHM1, MaGA20ox2, PsGA3ox1, and PROCERA) are classified into GA synthesis or signal genes directly influencing endogenous GA in tissues, and abnormal concentrations of endogenous GAs affect development of tissues, such as stem. We assume that this is also a conservative mechanism of horticultural species as in model plants where GAs positively regulate stem elongation (Tomlinson et al., 2019; Shao et al., 2020; Cheng et al., 2021; Kou et al., 2021; Liu et al., 2022). Internode/stem is made up of millions of cells, and cell fate determines internode elongation. Endogenous GAs influence cell fate (elongation or/and cell proliferation). Studies of shade avoidance syndrome provided solid evidence of how GAs precisely regulate cell fate (Roig-Villanova and Martínez-García, 2016; Lee et al., 2018). When endogenous GAs were elevated, Xyloglucan endotransglucosylase/hydrolases (XTHs), Expansins (EXPs), and Cyclin-dependent kinases (CDKs) were subsequently up-regulated. These genes acidize and soften cell walls, benefiting stem cell elongation and proliferation. In other words, internode elongation or shortening in horticultural plants are determined by cell development (elongation, division, and expansion), and cell fate is accurately regulated by fluctuations in endogenous GAs.
Although a few GA synthesis and metabolism genes have been verified to be involved in internode length, regulatory factors of GA synthesis and signal transduction are still poorly understood at present for horticultural plants. For example, MCMl AGAMOUS DEFICIENS SRF4 (MADS)-box and basic helix-loop-helix (bHLH) TFs, which have been frequently found in Arabidopsis and rice, are still lacking in horticultural plants. Identifying regulatory factors in GA biosynthesis and signaling pathways is a necessary future research topic in horticultural plants.
Axillary bud outgrowth
Axillary bud outgrowth also significantly affects the quality and quantity of the products of horticultural plants, as it is another core factor determining plant architecture (Wang et al., 2018a; Zhang et al., 2020; Feng et al., 2021). The lateral axillary meristem in the axil of leaves of horticultural species first develops into axillary buds (namely, axillary bud initiation), and then the axillary bud develops into dormant buds, shoot branching/branch crown, or specialized tissues (runners etc.; Wang et al., 2018a; Zhang et al., 2020; Guo et al., 2021).
Clonal propagation (runners) produces offspring with the same genetic background (Tenreira et al., 2017) as their parents in Fragaria vesca (a diploid model plant of strawberry) or Fragaria × ananassa (a cultivated strawberry). It is important to analyze how the runners form. This process includes axillary bud initiation and outgrowth, which are both strictly controlled by internal factors such as GAs and TFs and external factors such as photoperiod (Mouhu et al., 2013; Tenreira et al., 2017; Caruana et al., 2018; Li et al., 2018b; Andrés et al., 2021; Feng et al., 2021; Guo et al., 2021; Liang et al., 2022). LOSS OF AXILLARY MERISTEMS (LAM, a GRAS gene) is highly expressed in axillary meristem. Mutation of LAM leads to decreased runner number due to a failure of axillary bud formation, indicating that LAM can provoke bud initiation in F. vesca (Feng et al., 2021). GA application only stimulates the remaining axillary bud outgrowth in lam plants but fails to recover the defect of axillary bud decrease, indicating that GAs exclusively induced runner development instead of bud initiation in F. vesca (Feng et al., 2021). A previous study demonstrated that FveGA20ox4 is exclusively expressed in axillary meristem and the developmental runners (Tenreira et al., 2017). Mutation of GA20ox4 leads to reduced runner number and the defective phenotype of the r mutant is rescued by exogenous GA3 in F. vesca (Tenreira et al., 2017). Apart from GA synthesis genes, GA signal transduction genes are also involved in axillary bud outgrowth. Loss of function or downregulation of RGA1 (one of the five DELLAs in strawberry) increases runner number in F. vesca, which has been demonstrated by different research groups (Caruana et al., 2018; Li et al., 2018b).
In addition, TFs are also essential in runner formation. FaHANABA TARANU (FaHAN; a GATA TF) promotes runner development via enhancing GA concentration in F. ananassa (Liang et al., 2022). Surprisingly, the GA20ox1 transcript is accumulated in FaHAN-OE transgenic lines, but the GA20ox4 gene is not in F. ananassa (Liang et al., 2022). Long-day and 18°C are also beneficial to runner development in F. vesca (Mouhu et al., 2013; Andrés et al., 2021). Long-day positively regulates the FvFlowering Locus T (FT)–FvSUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) cascade, and FvSOC1 directly up-regulates FvGA20ox4 expression in F. vesca. These processes lead to increased bioactive GA concentration, and therefore promote runner development. Andrés et al. (2021) further found that there may be a FvSOC1-indpendent way in F. vesca, which controls runner development at temperature of 22°C.
In contrast, silencing the SlGA2ox gene inhibits shoot branching, and the role of GAs is verified by application of GAs and PAC in tomato, which proves that GAs play a negative role in shoot branching in tomato (Martínez-Bello et al., 2015). In garlic bulb, however, GAs promote axillary meristem initiation and determine the number of cloves (Liu et al., 2019; Liu et al., 2020). Application of GA3 results in downregulation of AsGA20ox and upregulation of AsHistidine kinase (AsAHK), which positively regulate the initiation of axillary meristem (Liu et al., 2020).
In conclusion, GAs positively regulate runner development in strawberry, whereas they negatively control shoot branching in tomato. In these species, GAs determine axillary bud outgrowth without influencing axillary bud initiation. Unlike the functions in strawberry and tomato, GAs promote axillary meristem initiation in garlic bulb. In addition, GAs act as pleiotropic regulators in two developmental stages of axillary meristem in many other horticultural species. TEOSINTE BRANCHED1/BRANCHED 1 (TB1/BRC1) is the key hub in repressing axillary bud development in different species (Zhang et al., 2020). GAs and FaHAN can reduce this gene expression, leading to runner outgrowth in strawberry (Liang et al., 2022), while GAs possibly stimulate this gene expression via the cytokinin (CK) signal pathway, increasing shoot branching number in tomato (Xia et al., 2021). These results indicate that BRC1 may be regulated differently in strawberry and tomato, and that the functions of GAs are also different in strawberry and tomato. Lateral Suppressor/LATERAL SUPPRESSOR/MONOCULM 1 (Ls/LAS/MOC1) determine axillary bud initiation (Feng et al., 2021). We predict that GAs maybe directly or indirectly influence the ortholog Ls/LAS/MOC1 expression level in garlic bulb, and thus axillary bud initiation.
Compound leaf
Leaf is the main plant photosynthetic organ (Du et al., 2018). There are three steps in leaf development: leaf primordium initiation, differentiation, and morphogenesis (Bar and Ori, 2015). Tomato has been used as a model species to study compound leaf development for decades and some progress has been achieved. Hormones are key factors regulating compound leaf development, and numerous studies have shown that GAs promote leaf differentiation, decreasing leaf complexity in tomato (Jasinski et al., 2008; Israeli et al., 2021; Su et al., 2022). For example, application of exogenous GAs or mutation of PROCERA/DELLA reduces the number of leaflets and makes leaf margins smoother and petioles longer (Jasinski et al., 2008; Fleishon et al., 2011; Yanai et al., 2011). However, CKs promote leaf morphogenesis and maintenance of organogenic activity, increasing leaflet number in tomato (Fleishon et al., 2011; Shwartz et al., 2016; Israeli et al., 2021). Overexpression of ISOPENTENYL TRANSFERASE 7 (IPT7; a CK synthetic gene) increases leaf complexity (Fleishon et al., 2011). Compound leaf development is a complex process, so it is interesting to see how these two hormones maintain balance during compound leaf development.
Class I KNOX (KNOXI) helps to maintain meristem activity, and delays leaf differentiation in many species (Hake et al., 2004; Cruz et al., 2020; He et al., 2020). Mutation of TKN2 (a KNOXI gene) decreases leaf complexity in tomato, and the balance between GAs and CKs (organogenesis and differentiation) in leaflet formation is elaborately regulated by TKN2 (Figure 2). In this model, KNOXI changes endogenous GA concentration by down-regulating GA20ox1 and up-regulating GA2ox (Bolduc and Hake, 2009; Fleishon et al., 2011; Israeli et al., 2021). KNOXI also activates IPT7, leading to accumulation of CKs (Jasinski et al., 2005; Yanai et al., 2005; Fleishon et al., 2011; Israeli et al., 2021).
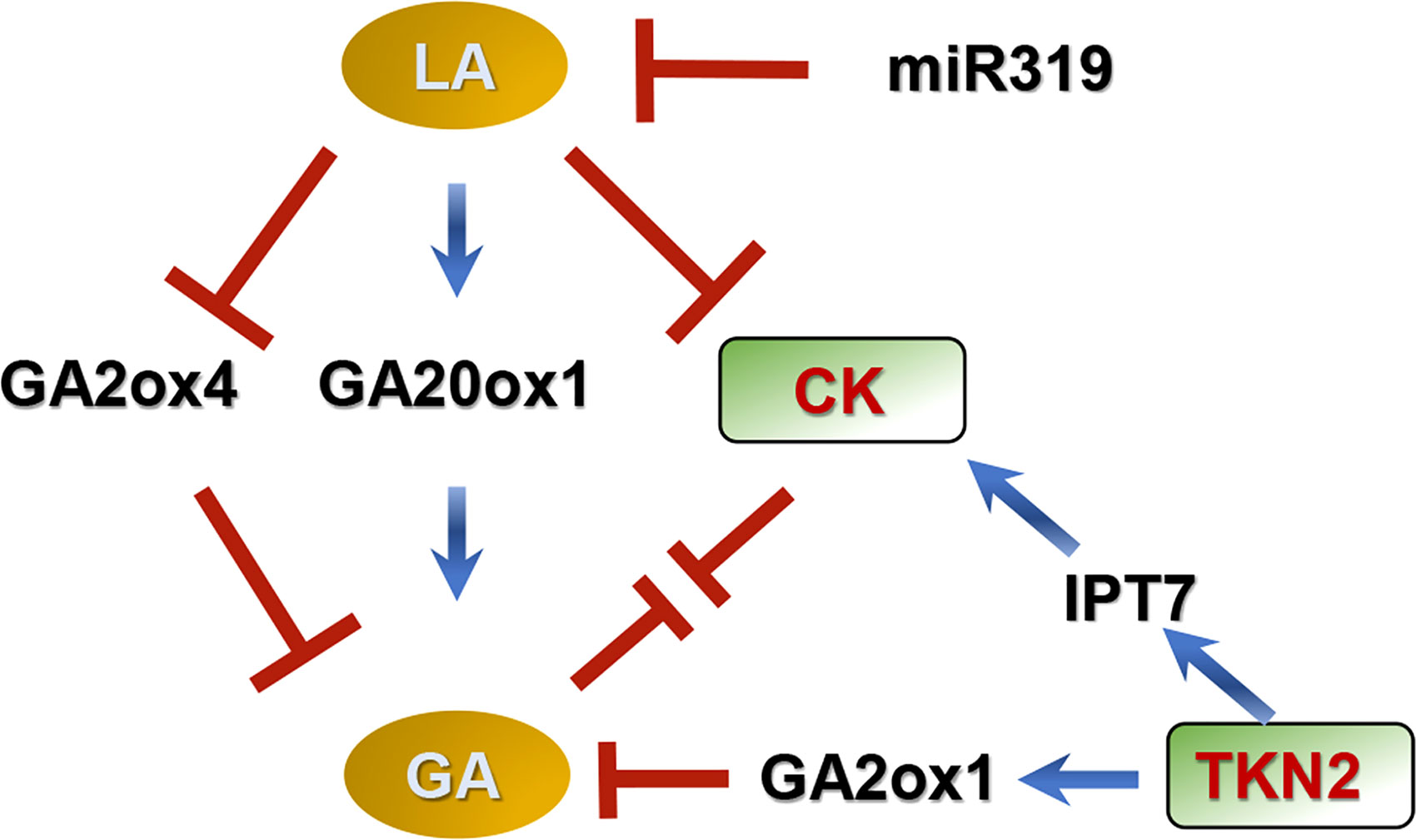
Figure 2 Model for compound leaf development regulated by GAs in tomato (Jasinski et al., 2005; Bolduc and Hake, 2009; Israeli et al., 2021). In this model, GAs promote differentiation and delay morphogenesis, decreasing leaf complexity, while CKs promote morphogenesis and delay differentiation, increasing leaflet number in tomato. GAs interact antagonistically with CKs during early leaflet origination. LANCEOLATE (LA) and TKN2 (a Class I KNOTTED1-LIKE HOMEOBOX protein) coordinate the balance between GAs and CKs during leaf development. LA is negatively regulated by MicroRNA319 (miR319). LA increases GA concentration by up-regulating GA20ox1 and down-regulating GA2ox4, meanwhile this protein decreases CK concentration. TKN2 increases CK concentration by stimulating expression of ISOPENTENYL TRANSFERASE 7 (a CK biosynthetic gene), and reduces GA concentration by motivating GA2ox1 (a GA deactivated gene) expression. ↓ represents positive regulation. ⊥ represents negative regulation.
The MicroRNA319 (miR319)-LANCEOLATE (LA; a CIN-TCP TF) cascade also controls compound leaf development through coordinating GAs and CKs (Figure 2), in which miR319 promotes morphogenesis and LA promotes differentiation (Ori et al., 2007; Jasinski et al., 2008; Yanai et al., 2011; Challa et al., 2019; Israeli et al., 2021). LA is negatively controlled by miR319 during the early stages of leaf development, and overexpression of miR319 causes an indeterminate growth phenotype of compound leaf (Ori et al., 2007; Jasinski et al., 2008; Yanai et al., 2011; Challa et al., 2019). The expression levels of GA20ox1 and GA2ox4 are respectively increased and decreased in the semi-dominant mutant La, indicating that LA increases GA levels (Yanai et al., 2011; Israeli et al., 2021). In addition, LA limits CK activity based on genetic experiments (Israeli et al., 2021).
In conclusion, early leaflet origination is a complex and dynamically balanced process. GAs interact antagonistically with CKs during this process, and promote the differentiation stage and shorten the morphogenetic stage, decreasing leaflet number. In contrast, CKs prolong morphogenesis and delay differentiation, increasing leaflet number. Multiple TFs such as TKN2 and LA delicately and fully alter concentrations of these hormones, changing the ratio of differentiation/morphogenetic stages and eventually influencing leaflet formation (Figure 2). The current model is a simple framework derived from limited existing data. Other TFs and hormones may also function in leaf development. A good example is CLAUSA (CLAU; a MYB TF), which works as LA in a mostly parallel pathway (Israeli et al., 2021). Overexpression of CLAU can rescue LA deficiency and vice versa. CLAU also medicate CK-GA balance in compound leaf development (Israeli et al., 2021). Auxins and brassinosteroids (BRs) can also coordinate with GAs, regulating compound leaf development in tomato. SlBES1.8, a key regulator of BR signaling, directly represses SlGA2ox2, SlGA2ox6 and SlGID1b-1, influencing leaf morphogenesis (Su et al., 2022). Moreover, the specific molecular mechanism underlying compound leaf development may be different among different species (Bar and Ori, 2015). In pea, GAs prolong the morphogenetic stage, promoting leaf formation, which is different from the situation in tomato (Goliber et al., 1999). Current related studies have mostly focused on tomato; thus, it is important to study compound leaf development using other horticultural plants.
Reproductive growth
Flowering time
Transformation from vegetative to reproductive growth, which is one of the most important events in the plant life cycle, is a complex process regulated by many internal and external factors. The molecular mechanism underlying flowering has been studied extensively for decades, and there are five main pathways controlling floral induction in Arabidopsis (Fornara et al., 2010; Jin and Ahn, 2021). Among these five pathways, GAs accelerate floral induction via up-regulating SOC1 in Arabidopsis, which is the so-called GA pathway (Mutasa-Gottgens and Hedden, 2009). This hormone is also involved in floral transition in horticultural plants. The difference in flowering time directly influences yield and quality of horticultural species (Randoux et al., 2012; Ghosh and Halder, 2018). Hence, it is important to explore the mechanism underlying how GAs regulate flowering transition of horticultural plants.
In tomato, DELLA coordinates microRNA156 (miR156)-SQUAMOSA PROMOTER BINDING–LIKE (SPL/SBP) cascade and miR319-LA cascade in determining flowering time through changing SINGLE FLOWER TRUSS (SFT) expression level in leaves and APETALA1 (AP1) expression level in shoot meristem (Silva et al., 2019). Silva et al. (2019) showed that DELLA promotes flowering in tomato. In other words, GAs delay floral induction via reducing DELLA accumulation. Application of bioactive GAs decreases the expression levels of AP1 and FT (known as a florigen gene) in Citrus reticulata Blanco × Citrus temple Hort. Ex Y. Tanaka (citrus) (Goldberg-Moeller et al., 2013), delaying flowering time. Once treated with GAs, RoTERMINAL FLOWER1 (RoTFL1) transcripts are accumulated in once-flowering rose, down-regulating expression of RoFT, RoSOC1, and RoAP1, and inhibiting floral induction (Randoux et al., 2012). GAs also stimulate expression of MdTFL1 in apple (Zhang et al., 2019). In contrast, the expression levels of RsFT and RsSOC1-1 are up-regulated after GA treatment in early-flowering (NH-JS2) Raphanus sativus (radish; Jung et al., 2020). Recently, a GA-DELLA (SLR1)-VvmiR159c-VvGIBBERELLIN MYB GENE (GAMYB) cascade was reported to modulate grape floral development (Wang et al., 2018b). This result indicates that GAs also have a negative role in floral development in grape. In Brassica campestris L. ssp. chinensis var. utilis Tsen (flowering Chinese cabbage), GA treatment accelerates flowering and uniconazole (GA biosynthetic inhibitor) inhibits floral induction (Song et al., 2019).
Overall, GAs work as a negative factor in flowering transformation for tomato, rose, apple, and citrus, whereas they are a positive regulator in flowering Chinese cabbage, radish, and grape. GAs possibly influence, directly or indirectly, the expression of some core downstream floral integrator genes such as AP1, SOC1, and FT, determining the transformation of flowering (negative or positive). GAs thus have different roles in flowering transition among different horticultural species.
Some horticultural species, for example radish, are evolutionarily similar to Arabidopsis in effects of GAs on flowering time, while others such as tomato have evolved different or specific characteristics in regulation of flowering time. Because floral transition is controlled by a precise regulatory network, the difference in the effects of GAs on flowering time among different horticultural species may be associated with their different natural habitats and experimental conditions (Wang et al., 2021a). For example, GA treatment inhibits flowering in once-flowering rose in mid-March (short-day), but not in summer (long-day; Randoux et al., 2012). The seasonal differences in light period and temperature may be the reasons for the different role of GAs. These results provide clues for further study of the role of GAs on flowering time using different horticultural plant species. Tomato, citrus, and flowering Chinese cabbage are all short-day plants, while Arabidopsis is a long-day plant, which may be one of the reasons for the interspecific difference in the role of GAs. The mechanism underlying the effects of GAs on flowering time is largely unknown in various horticultural species, and thus more studies on flowering regulatory networks are needed, especially in economically important species.
Parthenocarpy
Early fruit development of horticultural plants is a complex process, which is regulated by multiple phytohormones (Hu et al., 2018; Tyagi et al., 2021; Zhou et al., 2021; He and Yamamuro, 2022; Sharif et al., 2022). This process can be divided into three consecutive stages: fruit initiation stage, cell division stage controlled by auxins, and cell expansion stage regulated by GAs (Shinozaki et al., 2015). After fertilization, the developing seeds induce accumulation of GAs and auxins. Then, ovary or other tissues will receive GA and auxin signals from the immature seeds and activate the fruit initiation process (Hu et al., 2018).
Fruit formation without fertilization is also a common phenomenon, namely parthenocarpy. Parthenocarpy is an ideal agronomic trait for many horticultural species, and is popular with consumers. Recent studies show that numerous hormones are involved in parthenocarpic fruit formation (Martínez-Bello et al., 2015; Shinozaki et al., 2015; Hu et al., 2018; Liu et al., 2018; Zhou et al., 2021). A mass of cell cycle and cell expansion genes are continuously expressed in the ovary walls after treatment with phytohormones (Bermejo et al., 2016; Mesejo et al., 2016; Liu et al., 2018). Application of auxins or GAs can induce parthenocarpic fruit in tomato, cucumber, strawberry, and grape (Kang et al., 2013; Lu et al., 2016; Hu et al., 2018; Qian et al., 2018). These results demonstrate that auxins and GAs positively control parthenocarpy.
As parthenocarpic fruit growth is complicated, it is important to know how auxins coordinate with GAs during this process. AUX/IAA (IAA) proteins interact with AUXIN RESPONSE FACTOR (ARF) proteins to repress auxin signaling. In tomato, concentrations of bioactive GAs in parthenocarpic fruits increase in iaa9 mutants (Serrani et al., 2008; Mignolli et al., 2015). Moreover, the numbers of transcripts of SlGA20ox1 and SlGA3ox1, two GA synthesis genes, are increased in SlARF7 RNAi transgenic plants (Hu et al., 2018). SlARF7 can directly bind to the promotors of SlGA20ox1 and SlGA3ox1, and negatively regulate expression of these genes. PROCERA (DELLA) can also positively control expression of these genes (Hu et al., 2018). These results indicate that the interaction among PROCERA, SlARF7, and SlIAA9 coordinates the functions of auxins and GAs during parthenocarpic fruit growth. In strawberry, application of auxins also stimulates expression of FveGA20ox and FveGA3ox during growth of the parthenocarpic fruit formed from receptacle (Liao et al., 2018). Based on these results, we can conclude that auxins lead to GA accumulation by changing expression of GA biosynthesis genes in early stages of fruit development (Serrani et al., 2008; Mignolli et al., 2015; Hu et al., 2018; Zhou et al., 2021).
Although both auxins and GAs can stimulate parthenocarpic fruit formation, there are still some differences in the development process of the parthenocarpic fruits. For example, compared with normal fruits of strawberry the parthenocarpic fruits induced by auxins and GAs are rounder and longer, respectively (Liao et al., 2018). Auxins and GAs may regulate different genes related to cell division and expansion during parthenocarpic fruit development, leading to different fruit shapes (Liao et al., 2018; Chen et al., 2020).
Apart from auxins and GAs, CKs and ethylenes (ETHs) have also been reported to be involved in parthenocarpic fruit formation (Fos et al., 2000; Pascual et al., 2009; Shinozaki et al., 2015). CKs increase auxin concentrations, leading to parthenocarpic fruit in Pyrus spp. (pear) (Cong et al., 2020), while ETH decreases bioactive GA concentrations, inhibiting parthenocarpic fruit formation in tomato (Fos et al., 2000; Pascual et al., 2009; Shinozaki et al., 2015). However, a clear and systematic framework underlying the effects of various hormones on parthenocarpic fruit growth is still lacking. A recent review concluded that auxins may have a core role during this process (Sharif et al., 2022), but more evidence is needed to support this viewpoint in horticultural plants.
A view to the future
Although some significant progress has been achieved regarding the roles of GAs in horticultural species, there are still many problems to be studied. Efficient genetic transformation and plant regeneration systems are still lacking for most horticultural species, limiting studies on the molecular mechanisms underlying the effects of GAs on growth and development in horticultural plants. It is worth constructing systems in multiple horticultural species in the future, which will certainly promote progress in relevant studies.
There are many GA synthetic or signaling mutants in horticultural plants, which is helpful not only to explore the roles of GAs, but also alter plant stature or other economic traits. However, unfavorable traits such as male sterility, poor germination, or low nitrogen-use efficiency are also found in some mutants (Zhang et al., 2014; Li et al., 2018a; He et al., 2019b), reducing their commercial value. There are two strategies for breeders to solve this challenge for horticultural species. First, cis-regulatory or coding sequences of a target GA synthetic or signaling gene can be edited using the CRISPR-Cas9 technique (Park et al., 2014; Wang et al., 2021b), and a series of alleles from weak to strong effects could be obtained (Zhang et al., 2020). The mild allele in plant height and other traits may be useful in altering agronomic traits of horticultural plants. Second, promoters with development stage- or tissue-specificity are urgently needed. A target GA synthetic or signaling gene can be driven by a stem-specific promoter, which may improve stem-related traits without effects on stamen or lateral roots development (Gupta et al., 2020). That is probably regarded as a prerequisite for the precision technique of manipulating traits of horticultural plants. In addition, the results obtained from laboratories should be tested in greenhouses or even in crop fields, accelerating use in practice.
Author contributions
XZ and YS wrote the original manuscript. BZ provided help and advice. YF corrected the manuscript. All authors read and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31971557 and 32171666) and the Scientific Research Project of Shenyang Agricultural University (880421034).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acheampong, A. K., Hu, J., Rotman, A., Zheng, C., Halaly, T., Takebayashi, Y., et al. (2015). Functional characterization and developmental expression profiling of gibberellin signaling components in vitis vinifera. J. Exp. Bot. 66 (5), 1463–1476. doi: 10.1093/jxb/eru504
Andrés, J., Caruana, J., Liang, J., Samad, S., Monfort, A., Liu, Z., et al. (2021). Woodland strawberry axillary bud fate is dictated by a crosstalk of environmental and endogenous factors. Plant Physiol. 187 (3), 1221–1234. doi: 10.1093/plphys/kiab421
Bar, M., Ori, N. (2015). Compound leaf development in model plant species. Curr. Opin. Plant Biol. 23, 61–69. doi: 10.1016/j.pbi.2014.10.007
Bermejo, A., Martínez-Alcántara, B., Martínez-Cuenca, M., Yuste, R., Mesejo, C., Reig, C., et al. (2016). Biosynthesis and contents of gibberellins in seeded and seedless sweet orange (Citrus sinensis l. osbeck) cultivars. J. Plant Growth Regul. 35, 1036–1048. doi: 10.1007/s00344-016-9602-5
Binenbaum, J., Weinstain, R., Shani, E. (2018). Gibberellin localization and transport in plants. Trends Plant Sci. 23 (5), 410–421. doi: 10.1016/j.tplants.2018.02.005
Bolduc, N., Hake, S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene GA2ox1. Plant Cell. 21 (6), 1647–1658. doi: 10.1105/tpc.109.068221
Camut, L., Regnault, T., Sirlin-Josserand, M., Sakvarelide-Achaed, L., Carrera, E., Zumsteg, J., et al. (2019). Root-derived GA12 contributes to temperature-induced shoot growth in arabidopsis. Nat. PLANTS 5, 1216–1221. doi: 10.1038/s41477-019-0568-8
Caruana, J. C., Sittmann, J. W., Wang, W., Liu, Z. (2018). Suppressor of runnerless encodes a DELLA protein that controls runner formation for asexual reproduction in strawberry. Mol. Plant 11 (1), 230–233. doi: 10.1016/j.molp.2017.11.001
Chai, Z., Fang, J., Yao, W., Zhao, Y., Cheng, G., Akbar, S., et al. (2022). ScGAIL, a sugarcane n-terminal truncated DELLA-like protein, participates in gibberellin signaling in arabidopsis. J. Exp. Bot. 73 (11), 3462–3476. doi: 10.1093/jxb/erac056
Challa, K. R., Rath, M., Nath, U. (2019). The CIN-TCP transcription factors promote commitment to differentiation in arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PloS Genet. 15 (2), e1007988. doi: 10.1371/journal.pgen.1007988
Cheng, J., Ma, J., Zheng, X., Lv, H., Zhang, M., Tan, B., et al. (2021). Functional analysis of the gibberellin 2-oxidase gene family in peach. Front. Plant Sci. 12, 619158. doi: 10.3389/fpls.2021.619158
Chen, S., Wang, X., Tan, G., Zhou, W., Wang, G. (2020). Gibberellin and the plant growth retardant paclobutrazol altered fruit shape and ripening in tomato. PROTOPLASMA 257 (3), 853–861. doi: 10.1007/s00709-019-01471-2
Cong, L., Wu, T., Liu, H., Wang, H., Zhang, H., Zhao, G., et al. (2020). CPPU may induce gibberellin-independent parthenocarpy associated with PbRR9 in 'Dangshansu' pear. Hortic. Res. 7, 68. doi: 10.1038/s41438-020-0285-5
Cruz, R., Melo-de-Pinna, G. F. A., Vasco, A., Prado, J., Ambrose, B. A. (2020). Class I KNOX is related to determinacy during the leaf DEVELOPMENT of the fern mickelia scandens (Dryopteridaceae). Int. J. Mol. Sci. 21 (12), 4295. doi: 10.3390/ijms21124295
Davière, J., Achard, P. (2014). Gibberellin signaling in plants. DEVELOPMENT 140 (6), 1147–1151. doi: 10.1242/dev.087650
Du, F., Guan, C., Jiao, Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11 (9), 1117–1134. doi: 10.1016/j.molp.2018.06.006
Feng, J., Cheng, L., Zhu, Z., Yu, F., Dai, C., Liu, Z., et al. (2021). GRAS transcription factor LOSS OF AXILLARY MERISTEMS is essential for stamen and runner formation in wild strawberry. Plant Physiol. 186 (4), 1970–1984. doi: 10.1093/plphys/kiab184
Fleishon, S., Shani, E., Ori, N., Weiss, D. (2011). Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 190 (3), 609–617. doi: 10.1111/j.1469-8137.2010.03616.x
Fornara, F., Montaigu, A. D., Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. CELL 141 (3), 550–550.e1-2. doi: 10.1016/j.cell.2010.04.024
Fos, M., Nuez, F., García-Martínez, J. L. (2000). The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol. 122 (2), 471–480. doi: 10.1104/pp.122.2.471
Gao, Y., Huang, S., Qu, G., Fu, W., Zhang, M., Liu, Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa l. ssp. pekinensis). Hortic. Res. 7 (1), 178. doi: 10.1038/s41438-020-00399-6
Gebremeskel, H., Dou, J., Li, B., Zhao, S., Muhammad, U., Lu, X., et al. (2019). Molecular mapping and candidate gene analysis for GA3 responsive short internode in watermelon (Citrullus lanatus). Int. J. Mol. Sci. 21 (1), 290. doi: 10.3390/ijms21010290
Ghosh, S., Halder, S. (2018). Effect of different kinds of gibberellin on temperate fruit crops: A review. J. Pharm. Innov. 7 (3), 315–319.
Goldberg-Moeller, R., Shalom, L., Shlizerman, L., Samuels, S., Zur, N., Ophir, R., et al. (2013). Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in citrus buds. Plant Sci. 198, 46–57. doi: 10.1016/j.plantsci.2012.09.012
Goliber, T., Kessler, S., Chen, J.J., Bharathan, G., Sinha, N. (1999). Genetic, molecular, and morphological analysis of compound leaf development. Curr. Top. Dev. Biol. 43, 259–290. doi: 10.1016/s0070-2153(08)60384-1
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z., Powers, S. J., et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in arabidopsis. Plant Cell. 18 (12), 3399–3414. doi: 10.1105/tpc.106.047415
Guo, L., Plunkert, M., Luo, X., Liu, Z. (2021). Developmental regulation of stolon and rhizome. Curr. Opin. Plant Biol. 59, 101970. doi: 10.1016/j.pbi.2020.10.003
Guo, S., Zhang, X., Bai, Q., Zhao, W., Fang, Y., Zhou, S., et al. (2020). Cloning and functional analysis of dwarf gene mini plant 1 (MNP1) in medicago truncatula. Int. J. Mol. Sci. 21 (14), 4968. doi: 10.3390/ijms21144968
Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. SCIENCE 368 (6488), 266–269. doi: 10.1126/science.aaz7614
Hake, S., Smith, H. M. S., Holtan, H., Magnani, E., Mele, G., Ramirez, J. (2004). The role of Knox genes in plant DEVELOPMENT. Annu. Rev. Cell Dev. Biol. 20, 125–151. doi: 10.1146/annurev.cellbio.20.031803.093824
He, J., Chen, Q., Xin, P., Yuan, J., Ma, Y., Wang, X., et al. (2019a). CYP72A enzymes catalyse 13-hydrolyzation of gibberellins. Nat. Plants 5 (10), 1057–1065. doi: 10.1038/s41477-019-0511-z
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61 (11), 1832–1849. doi: 10.1093/pcp/pcaa092
He, L., Liu, Y., He, H., Liu, Y., Qi, J., Zhang, X., et al. (2020). A molecular framework underlying the compound leaf pattern of medicago truncatula. Nat. Plants 6 (5), 511–521. doi: 10.1038/s41477-020-0642-2
Helliwell, C. A., Sullivan, J. A., Mould, R. M., Gray, J. C., Peacock, W. J., Dennis, E. S. (2001). A plastid envelope location of arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 28 (2), 201–208. doi: 10.1046/j.1365-313x.2001.01150.x
He, H., Yamamuro, C. (2022). Interplays between auxin and GA signaling coordinate early fruit development. Hortic. Res. 9, uhab078. doi: 10.1093/hr/uhab078
He, Y., Ye, Z., Ying, Q., Ma, Y., Zang, Y., Wang, H., et al. (2019b). Glyoxylate cycle and reactive oxygen species metabolism are involved in the improvement of seed vigor in watermelon by exogenous GA3. Sci. Hortic. 247, 184–194. doi: 10.1016/j.scienta.2018.12.016
Hu, J., Israeli, A., Ori, N., Sun, T. P. (2018). The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell. 30 (8), 1710–1728. doi: 10.1105/tpc.18.00363
Hu, J., Mitchum, M. G., Barnaby, N., Ayele, B. T., Ogawa, M., Nam, E., et al. (2008). Potential sites of bioactive gibberellin production during reproductive growth in arabidopsis. Plant Cell. 20 (2), 320–336. doi: 10.1105/tpc.107.057752
Israeli, A., Burko, Y., Shleizer-Burko, S., Zelnik, I. D., Sela, N., Hajirezaei, M. R., et al. (2021). Coordinating the morphogenesis-differentiation balance by tweaking the cytokinin-gibberellin equilibrium. PloS Genet. 17 (4), e1009537. doi: 10.1371/journal.pgen.1009537
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. CURRT Biol. 15 (17), 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jasinski, S., Tattersall, A., Piazza, P., Hay, A., Martinez-Garcia, J. F., Schmitz, G., et al. (2008). PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 56 (4), 603–612. doi: 10.1111/j.1365-313X.2008.03628.x
Jin, S., Ahn, J. H. (2021). Regulation of flowering time by ambient temperature: repressing the repressors and activating the activators. New Phytol. 230 (3), 938–942. doi: 10.1111/nph.17217
Jin, Y., Song, X., Chang, H., Zhao, Y., Cao, C., Qiu, X., et al. (2022). The GA-DELLA-OsMS188 module controls male reproductive development in rice. New Phytol. 233 (6), 2629–2642. doi: 10.1111/nph.17939
Jung, H., Jo, S. H., Jung., W. Y., Park, H. J., Lee, A., Moon, J. S., et al. (2020). Gibberellin promotes bolting and flowering via the floral integrators RsFT and RsSOC1-1 under marginal vernalization in radish. Plants (BASEL) 9 (5), 594. doi: 10.3390/plants9050594
Kang, C., Darwish, O., Geretz, A., Shahan, R., Alkharouf, N., Liu, Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry fragaria vesca. Plant Cell. 25 (6), 1960–1978. doi: 10.1105/tpc.113.111732
Katsumi, M., Foard, D. E., Pjinney, B. O. (1983). Evidence for the translocation of gibberellin A3 and gibberellin-like substances in grafts between normal, Dwarf1 and Dwarf5 seedlings of zea mays l. Plant AND Cell Physiol. 24 (3), 379–388. doi: 10.1093/oxfordjournals.pcp.a076527
Kou, E., Huang, X., Zhu, Y., Su, W., Liu, H., Sun, G., et al. (2021). Crosstalk between auxin and gibberellin during stalk elongation in flowering Chinese cabbage. Sci. Rep. 11 (1), 3976. doi: 10.1038/s41598-021-83519-z
Lacombe, B., Achard, P. (2016). Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 34, 1–8. doi: 10.1016/j.pbi.2016.06.007
Lange, M. J. P., Lange, T. (2016). Ovary-derived precursor gibberellin A9 is essential for female flower development in cucumber. DEVELOPMENT 143 (23), 4425–4429. doi: 10.1242/dev.135947
Lee, Y. K., Rhee, J. Y., Lee, S. H., Chung, G. C., Park, S. J., Segami, S., et al. (2018). Functionally redundant LNG3 and LNG4 genes regulate turgor-driven polar cell elongation through activation of XTH17 and XTH24. Plant Mol. Biol. 97 (1-2), 23–36. doi: 10.1007/s11103-018-0722-0
Liang, J., Wu, Z., Zheng, J., Koskela, E. A., Fan, L., Fan, G., et al. (2022). The GATA factor HANABA TARANU promotes runner formation by regulating axillary bud initiation and outgrowth in cultivated strawberry. Plant J. 110 (5), 1237–1254. doi: 10.1111/tpj.15759
Liao, X., Li, M., Liu, B., Yan, M., Yu, X., Zi, H., et al. (2018). Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. U. S. A. 115 (49), E11542–E11550. doi: 10.1073/pnas.1812575115
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018a). Modulating plant growth-metabolism coordination for sustainable agriculture. NATURE 560 (7720), 595–600. doi: 10.1038/s41586-018-0415-5
Liu, H., Deng, R., Huang, C., Cheng, Z., Meng, H. (2019). Exogenous gibberellins alter morphology and nutritional traits of garlic (Allium sativum l.) bulb. Sci. HORTIC-AMSTERDAM 246, 298–306. doi: 10.1016/j.scienta.2018.11.003
Liu, J., Gao, P., Liu, H., Wang, X., Ma, S., Wang, J., et al. (2022). Genetic analysis and mapping of a short internode gene (cladw) in watermelon (Citrullus lanatus l.). RESEARCH SQUARE. Preprints. doi: 10.21203/rs.3.rs-1515332/v1
Liu, L., Wang, Z., Liu, J., Liu, F., Zhai, R., Zhu, C., et al. (2018). Histological, hormonal and transcriptomic reveal the changes upon gibberellin-induced parthenocarpy in pear fruit. Hortic. Res. 5, 1. doi: 10.1038/s41438-017-0012-z
Liu, H., Wen, Y., Cui, M., Qi, X., Deng, R., Gao, J., et al. (2020). Histological, physiological and transcriptomic analysis reveal gibberellin-induced axillary meristem formation in garlic (Allium sativum). Plants (BASEL) 9 (8), 970. doi: 10.3390/plants9080970
Li, W., Zhang, J., Sun, H., Wang, S., Chen, K., Liu, Y., et al. (2018b). FveRGA1, encoding a DELLA protein, negatively regulates runner production in fragaria vesca. PLANTA 247 (4), 941–951. doi: 10.1007/s00425-017-2839-9
Lu, L., Liang, J., Zhu, X., Xiao, K., Li, T., Hu, J. (2016). Auxin-and cytokinin-induced berries set in grapevine partly rely on enhanced gibberellin biosynthesis. Tree Genet. Genomes 12 (3), 1–12. doi: 10.1007/s11295-016-0980-4
MacMillan, J., Takahashi, N. (1968). Proposed procedure for the allocation of trivial names to the gibberellins. NATURE 217 (5124), 170–171. doi: 10.1038/217170a0
Magome, H., Nomura, T., Hanada, A., Takeda-Kamiya, N., Ohnishi, T., Shinma, Y., et al. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. U. S. A. 110 (5), 1947–1952. doi: 10.1073/pnas.1215788110
Martínez-Bello, L., Moritz, T., López-Díaz, I. (2015). Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 66 (19), 5897–5910. doi: 10.1093/jxb/erv300
Mesejo, C., Yuste, R., Reig, R., Martínez-Fuentes, A., Iglesias, D. J., Muñoz-Fambuena, N., et al. (2016). Gibberellin reactivates and maintains ovary-wall cell division causing fruit set in parthenocarpic citrus species. Plant Sci. 247, 13–24. doi: 10.1016/j.plantsci
Mignolli, F., Vidoz, M. L., Mariotti, L., Lombardi, L., Picciarelli, P. (2015). Induction of gibberellin 20-oxidases and repression of gibberellin 2β-oxidases in unfertilized ovaries of entire tomato mutant, leads to accumulation of active gibberellins and parthenocarpic fruit formation. Plant Growth Regul. 75 (2), 415–425. doi: 10.1007/s10725-014-0002-1
Mouhu, K., Kurokura, T., Koskela, E. A., Albert, V. A., Elomaa, P., Hytönen, T. (2013). The fragaria vesca homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 represses flowering and promotes vegetative growth. Plant Cell. 25 (9), 3296–3310. doi: 10.1105/tpc.113.115055
Murase, K., Hirano, Y., Sun, T. P., Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. NATURE 456 (7221), 459–463. doi: 10.1038/nature07519
Mutasa-Göttgens, E., Hedden, P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60 (7), 1979–1989. doi: 10.1093/jxb/erp040
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 9 (6), 787–791. doi: 10.1038/ng2036
Park, S. J., Jiang, K., Tal, L., Yichie, Y., Gar, O., Zamir, A., et al. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46 (12), 1337–1342. doi: 10.1038/ng.3131
Pascual, L., Blanca, J. M., Canizares, J., Nuez, F. (2009). Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biol. 9, 67. doi: 10.1186/1471-2229-9-67
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. NATURE 400 (6741), 256–261. doi: 10.1038/22307
Qian, C., Ren, N., Wang, J., Xu, Q., Chen, X., Qi, X. (2018). Effects of exogenous application of CPPU, NAA and GA4+7 on parthenocarpy and fruit quality in cucumber (Cucumis sativus l.). Food Chem. 243, 410–413. doi: 10.1016/j.foodchem.2017.09.150
Ragni, L., Nieminen, K., Pacheco-Villalobos, D., Sibout, R., Schwechheimer, C., Hardtke, C. S., et al. (2011). Mobile gibberellin directly stimulates arabidopsis hypocotyl xylem expansion. Plant Cell. 23 (4), 1322–1336. doi: 10.1105/tpc.111.084020
Randoux, M., Jeauffre, J., Thouroude, T., Vasseur, F., Hamama, L., Juchaux, M., et al. (2012). Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot. 63 (18), 6543–6554. doi: 10.1093/jxb/ers310
Regnault, T., Davière, J. M., Wild, M., Sakvarelidze-Achard, L., Heintz, D., Bergua, E. C., et al. (2015). The gibberellin precursor GA12 acts as a long-distance growth signal in arabidopsis. Nat. Plants 1, 15073. doi: 10.1038/nplants.2015.73
Regnault, T., Davière, J. M., Heintz, D., Lange, T., Achard, P. (2014). The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 80 (3), 462–474. doi: 10.1111/tpj.12648
Reinecke, D. M., Wickramarathna, A. D., Ozga, J. A., Kurepin, L. V., Jin, A. L., Pharis, R. P., et al. (2013). Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and DEVELOPMENT in pea. Plant Physiol. 163 (2), 929–945. doi: 10.1104/pp.113.225987
Roig-Villanova, I., Martínez-García, J. F. (2016). Plant responses to vegetation proximity: A whole life avoiding shade. Front. Plant Sci. 7, 236. doi: 10.3389/fpls.2016.00236
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). A mutant gibberellin-synthesis gene in rice. NATURE 416 (6882), 701–702. doi: 10.1038/416701a
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A., Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 15 (1), 151–163. doi: 10.1105/tpc.005975
Schrager-Lavelle, A., Gath, N. N., Devisetty, U. K., Carrera, E., Blázquez, M. A., Maloof, J. N., et al. (2019). The role of a class III gibberellin 2-oxidase in tomato internode elongation. Plant J. 97 (3), 603–615. doi: 10.1111/tpj.14145
Serrani, J. C., Ruiz-Rivero, O., Fos, M., García-Martínez, J. L. (2008). Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 56 (6), 922–934. doi: 10.1111/j.1365-313X.2008.03654.x
Shao, X., Wu, S., Dou, T., Zhu, H., Hu, C., Huo, H., et al. (2020). Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 18 (1), 17–19. doi: 10.1111/pbi.13216
Sharif, R., Su, L., Chen, X., Qi, X. (2022). Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 9, uhab024. doi: 10.1093/hr/uhab024
Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T., Nakajima, M., Naoe, Y., Ohmiya, H., et al. (2008). Structural basis for gibberellin recognition by its receptor GID1. NATURE 456 (7221), 520–523. doi: 10.1038/nature07546
Shinozaki, Y., Hao, S., Kojima, M., Sakakibara, H., Ozeki-Iida, Y., Zheng, Y., et al. (2015). Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83 (2), 237–251. doi: 10.1111/tpj.12882
Shohat, H., Eliaz, N. I., Weiss, D. (2021). Gibberellin in tomato: metabolism, signaling and role in drought responses. Mol. Hortic. 1 (1), 1–12. doi: 10.1186/s43897-021-00019-4
Shwartz, I., Levy, M., Ori, N., Bar, M. (2016). Hormones in tomato leaf development. Dev. BIO. 419 (1), 132–142. doi: 10.1016/j.ydbio.2016.06.023
Silva, G. F., Silva, E. M., Correa, J. P., Vicente, M. H., Jiang, N., Notini, M. M., et al. (2019). Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 221 (3), 1328–1344. doi: 10.1111/nph.15492
Song, S., Lei, Y., Huang, X., Su, W., Chen, Y., Hao, Y. (2019). Crosstalk of cold and gibberellin effects on bolting and flowering in flowering Chinese cabbage. J. Integr. AGR. 18 (5), 992–1000. doi: 10.1016/S2095-3119(18)62063-5
Sun, T., Kamiya, Y. (1997). Regulation and cellular localization of ent-kaurene synthesis. Physiol. PLANTARUM. 101 (4), 701–708. doi: 10.1111/j.1399-3054.1997.tb01054.x
Sun, Y., Shang, L., Zhu, Q. H., Fan, L., Guo, L. (2021). Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. S1360-1385 (21), 00281–00288. doi: 10.1016/j.tplants.2021.10.006
Sun, Y., Zhang, H., Fan, M., He, Y., Guo, P. (2020). A mutation in the intron splice acceptor site of a GA3ox gene confers dwarf architecture in watermelon (Citrullus lanatus l.). Sci. Rep. 10 (1), 14915. doi: 10.1038/s41598-020-71861-7
Su, D., Xiang, W., Liang, Q., Wen, L., Shi, Y., Song, B., et al. (2022). Tomato SlBES1.8 influences leaf morphogenesis by mediating gibberellin metabolism and signaling. Plant Cell Physiol. 63 (4), 535–549. doi: 10.1093/pcp/pcac019
Tenreira, T., Lange, M. J. P., Lange, T., Bres, C., Labadie, M., Monfort, A., et al. (2017). A specific gibberellin 20-oxidase dictates the flowering-runnering decision in diploid strawberry. Plant Cell. 29 (9), 2168–2182. doi: 10.1105/tpc.16.00949
Tomlinson, L., Yang, Y., Emenecker, Y., Smoker, M., Taylor, J., Perkins, S., et al. (2019). Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 17 (1), 132–140. doi: 10.1111/pbi.12952
Tyagi, K., Maoz, I., Kochanek, B., Sela, N., Lerno, L., Ebeler, S. E., et al. (2021). Cytokinin but not gibberellin application had major impact on the phenylpropanoid pathway in grape. Hortic. Res. 8 (1), 51. doi: 10.1038/s41438-021-00488-0
Wang, X., Aguirre, L., Rodríguez-Leal, D., Benoit, B., Lippman, Z. B. (2021b). Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat. Plants 7 (4), 419–427. doi: 10.1038/s41477-021-00898-x
Wang, S., An, H., Tong, C., Jiang, S. (2021a). Flowering and flowering genes: from model plants to orchids. Hortic. Environ. BIOTE. 62 (2), 135–148. doi: 10.1007/s13580-020-00309-8
Wang, C., Jogaiah, S., Zhang, W., Abdelrahman, M., Fang, J. (2018b). Spatio-temporal expression of miRNA159 family members and their GAMYB target gene during the modulation of gibberellin-induced grapevine parthenocarpy. J. Exp. Bot. 69 (15), 3639–3650. doi: 10.1093/jxb/ery172
Wang, B., Smith, S. M., Li, J. (2018a). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. SCIENCE 367 (6478), eaaz2046. doi: 10.1126/science.aaz2046
Xia, X., Dong, H., Yin, Y., Song, X., Gu, X., Sang, K., et al. (2021). Brassinosteroid signaling integrates multiple pathways to release apical dominance in tomato. Proc. Natl. Acad. Sci. U. S. A. 118 (11), e2004384118. doi: 10.1073/pnas.2004384118
Yabuta, T. (1938). On the crystal of gibberellin, a substance to promote plant growth. J. Agric. Chem. Soc 14, 1526.
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 (17), 1566–1571. doi: 10.1016/j.cub.2005.07.060
Yanai, O., Shani, E., Russ, D., Ori, N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68 (4), 571–582. doi: 10.1111/j.1365-313X.2011.04716.x
Zhang, S., Gottschalk, C., van Nocker, S. (2019). Genetic mechanisms in the repression of flowering by gibberellins in apple (Malus × domestica borkh.). BMC GENOMIC. 20 (1), 747. doi: 10.1186/s12864-019-6090-6
Zhang, X., He, L., Zhao, B., Zhou, S., Li, Y., He, H., et al. (2020). Dwarf and increased branching 1 controls plant height and axillary bud outgrowth in. Medicago truncatula. J. Exp. Bot. 71 (20), 6355–6365. doi: 10.1093/jxb/eraa364
Zhang, Y., Liu, B., Yang, S., An, J., Chen, C., Zhang, X., et al. (2014). A cucumber DELLA homolog CsGAIP may inhibit staminate DEVELOPMENT through transcriptional repression of b class floral homeotic genes. PloS One 9 (3), e91804. doi: 10.1371/journal.pone.0091804
Zhou, J., Sittmann, J., Guo, L., Xiao, Y., Huang, X., Pulapaka, A., et al. (2021). Gibberellin and auxin signaling genes RGA1 and ARF8 repress accessory fruit initiation in diploid strawberry. Plant Physiol. 185 (3), 1059–1075. doi: 10.1093/plphys/kiaa087
Keywords: horticultural plants, gibberellins, plant stature, flowering time, parthenocarpy
Citation: Zhang X, Zhao B, Sun Y and Feng Y (2022) Effects of gibberellins on important agronomic traits of horticultural plants. Front. Plant Sci. 13:978223. doi: 10.3389/fpls.2022.978223
Received: 25 June 2022; Accepted: 20 September 2022;
Published: 04 October 2022.
Edited by:
Junbo Du, Sichuan Agricultural University, ChinaCopyright © 2022 Zhang, Zhao, Sun and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yibo Sun, c3VueWlibzAzNjFAMTYzLmNvbQ==; Yulong Feng, ZnlsQHN5YXUuZWR1LmNu
 Xiaojia Zhang
Xiaojia Zhang Baolin Zhao
Baolin Zhao Yibo Sun
Yibo Sun Yulong Feng
Yulong Feng