Abstract
Polygonum hydropiper, is a plant of the Persicaria genus, which is commonly used to treat various diseases, including gastrointestinal disorders, neurological disorders, inflammation, and diarrhea. However, because of different local standards of P. hydropiper, people often confuse it with Polygonum lapathifolium L. and other closely related plants. This poses a serious threat to the safety and efficacy of the clinical use of P. hydropiper. This study aims to determine the six active ingredients of P. hydropiper and P. lapathifolium. Then the endophytic fungi and rhizosphere soil of the two species were sequenced by Illumina Miseq PE300. The results show significant differences between the community composition of the leaves, stems, and roots of the P. hydropiper and the P. lapathifolium in the same soil environment. Of the six secondary metabolites detected, five had significant differences between P. hydropiper and P. lapathifolium. Then, we evaluated the composition of the significantly different communities between P. hydropiper and P. lapathifolium. In the P. hydropiper, the relative abundance of differential communities in the leaves was highest, of which Cercospora dominated the differential communities in the leaves and stem; in the P. lapathifolium, the relative abundance of differential community in the stem was highest, and Cladosporium dominated the differential communities in the three compartments. By constructing the interaction network of P. hydropiper and P. lapathifolium and analyzing the network nodes, we found that the core community in P. hydropiper accounted for 87.59% of the total community, dominated by Cercospora; the core community of P. lapathifolium accounted for 19.81% of the total community, dominated by Sarocladium. Of these core communities, 23 were significantly associated with active ingredient content. Therefore, we believe that the community from Cercospora significantly interferes with recruiting fungal communities in P. hydropiper and affects the accumulation of secondary metabolites in the host plant. These results provide an essential foundation for the large-scale production of P. hydropiper. They indicate that by colonizing specific fungal communities, secondary metabolic characteristics of host plants can be helped to be shaped, which is an essential means for developing new medicinal plants.
Introduction
Polygonum hydropiper is a plant of the Persicaria genus in the family Polygonaceae. The whole plant of P. hydropiper is commonly used to treat various diseases, including gastrointestinal disorders, neurological disorders, inflammation, and diarrhea (Xiang and Ming, 2020). Due to the low requirements for growth conditions, it has abundant wild plant resources what is more, it is widely distributed in China in Sichuan, Guangdong, Guangxi, and other places (Hong and Hanshen, 2013). Many studies have shown that P. hydropiper is rich in various chemical components, including flavonoids, terpenes, and organic acids, with antimicrobial, antioxidant, antiviral, insecticidal, and other biological activities (Rahman et al., 2002; Ayaz et al., 2014; Sharif et al., 2014; Shahed-Al-Mahmud and Lina, 2017). Due to different local standards, the large-scale commercial cultivation of P. hydropiper has not yet been achieved. People are often confused by plants, such as Polygonum lapathifolium and P. orientale (Hong et al., 2019). This poses a serious threat to the safety and efficacy of the clinical use of P. hydropiper. Therefore, clarifying the differences between P. hydropiper and other obfuscated species is crucial to developing and improving the production of P. hydropiper.
Our preliminary research found that as one of the drugs for the treatment of enteritis, the role of flavonoids in it cannot be ignored (Zhang et al., 2021). Quercetin, kaempferol, isorhamnetin, hyperoside, catechins, and chlorogenic acid are the main active ingredients of P. hydropiper in treating enteritis (Yue, 2005; Wei et al., 2021). It is worth noting that Tian et al. (2014) found the potential of flavonoids to act as signaling molecules between endophytic fungi and plants. Meanwhile, Tang et al. (2020) found that the endophyte isolated from Conyza blinii H. Lév could produce flavonoids with high yield and excellent biological activity. Endophytic fungi are present in all plants and, together with host plants, determine the production of secondary metabolites (Waqas et al., 2012; Adeleke and Babalola, 2021). With the development of next-generation sequencing, there is increasing evidence that the compositional pattern of endophytic fungi is related to the production of specific secondary metabolites (Lunardelli Negreiros de Carvalho et al., 2016; Dang et al., 2021; Ribeiro et al., 2021). In some crops and medicinal plants, it is also recognized that the specific community composition pattern can reflect the quality and yield of the host plant (Song et al., 2010; Vergara et al., 2018; Cao et al., 2021; Martins et al., 2021).
There are abundant researches of chemical composition and pharmacological effect research of P. hydropiper (Hong and Hanshen, 2013). However, its endophytic fungi research is still blank. Thus, this experiment uses Illumina Miseq PE300 sequencing technology to sequence the endophytic fungi of P. hydropiper and P. lapathifolium. At the same time, we measured the content of 6 flavonoids of two species to reveal the relationship between the endophytic fungal community composition and the accumulation of active ingredients. Furthermore, determining the composition of the core microbiota related to active ingredients to improve the production of P. hydropiper provides key fundamental data.
Methods
Sampling
In July 2021, the whole plant and rhizosphere soil of P. hydropiper (ZP) and P. lapathifolium (WP) were sampled from Huoba Village in Jianyang City, China. At each sample point, four repeats of the whole plant and three repeats of the rhizosphere soil were collected. The rhizosphere soil was collected by the jitter root method. The plant residue and gravel were selected and discarded, and the samples were collected by the quartering method and loaded into sterile centrifuge tubes. To ensure the representativeness of the samples and avoid edge effects, we set the sample point away from the field ridge at least 25 m and each sample point interval at least 2 m. Samples are frozen with liquid nitrogen after on-site sampling and quickly transported back to the laboratory on dry ice for preservation at −80°C (Table 1).
Table 1
| Samples | Type | Date | Site |
|---|---|---|---|
| ZY-1 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZY-2 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZY-3 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZY-4 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZJ-1 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZJ-2 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZJ-3 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZJ-4 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZG-1 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZG-2 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZG-3 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZG-4 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| WY-1 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| WY-2 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| WY-3 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| WY-4 | Leaf | 2020/7/6 | Huoba Village in Jianyang City, China |
| WJ-1 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| WJ-2 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| WJ-3 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| WJ-4 | Stem | 2020/7/6 | Huoba Village in Jianyang City, China |
| WG-1 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| WG-2 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| WG-3 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| WG-4 | Root | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZT-1 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZT-2 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
| ZT-3 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
| WT-1 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
| WT-2 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
| WT-3 | Rhizosphere soil | 2020/7/6 | Huoba Village in Jianyang City, China |
Samples information.
Determination of active ingredients content
Chemicals and reagents
Six commercial standards of HPLC grade, including catechins, chlorogenic acid, hyperoside, quercetin, kaempferol, and isorhamnetin, were purchased from Chengdu Pufei De Biotech Co., Ltd. (China). Other HPLC grade reagents used were acetonitrile and methanol from Thermo Fisher Scientific Co., Ltd. (USA). The Watsons distilled water was applied to prepare for samples and the mobile phase. All other reagents were just met for an analytical grade.
Sample preparation
We precisely weighed 0.1 g of PL sample, added it to 15 mL of 60% ethanol in an Erlenmeyer flask, and dissolved it by sonication at 50°C for 30 min. Then filtering the sample, and evaporated the filtrate. The methanol was added to the residue until the volume to 5 mL, and the solution was filtered with a microporous filter membrane of 0.22 μm.
UPLC-QQQ-MS/MS analysis
Ultra performance liquid chromatograph LC-20A and triple quadrupole mass spectrometer LCMS-8045 were purchased from Shimadzu Co., Ltd. (Japan). A Shim-pack Velox C18 column (2.1 × 1,000 mm, 2.7 μm) was employed at a column temperature of 30°C. The mobile phase consisted of 0.3% carboxylic acid in water (A) and methanol (B), and the flow rate was 0.2 mL/min. The gradient elution parameters were set as follows: 0–6 min, 15% B; 6–15 min, 15–45% B; 15–20 min, 45%– 47% B; 20–22 min, 47%−50% B; 22–25 min, 50%−15% B. The injection volume was 2 μL.
Ionization method: Electrospray ion source (ESI); Multi-reaction monitoring mode (MRM); Curtain Gas (CUR) flow rate: 40 L·min−1; Atomized Gas (GS1) flow rate: 55 L·min−1; Auxiliary gas (GS2) flow rate: 55 L·min−1; ionization temperature (TEM): 550°C; Spray voltage (IS): 4,500 V in positive ion mode, −4,500 V in negative ion mode. The optimized MS/MS parameters of ZP are shown in Table 1.
Method validation
The six reference standards were weighed accurately and dissolved with methanol comparable to sample extracts. Calibration curves were constructed by measuring the signal intensity (peak area) of MRM transitions for at least six appropriate concentrations of each compound. Intra-day and inter-day precisions were evaluated by calculating the relative standard deviations (RSDs) of retention time and signal intensity during a single day and on three successive days, respectively. Repeatability was evaluated by calculating the RSDs of retention time and signal intensity of six tested solutions made from the same sample on a single day. Stability was evaluated by calculating the RSDs of retention time and signal intensity of the same tested solution during a single day at 0, 2, 4, 8, 12, and 24 h. Recovery experiments were done by spiking authentic standards into samples directly.
DNA extraction and library construction
Plants and soil samples were finely ground to powder in liquid nitrogen using a tissue grinder separately, and 0.5 g was taken for DNA extraction using the FastDNA®SPIN Kit (MP Biomedicals, US). Three repeats per sample were required. The DNA bands, concentration, and purity of the extract were detected using a 1% agarose gel electrophoresis and an accounting analyzer, and samples with a concentration of ≥20 ng/μL were selected and sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd for Polymerase Chain Reaction (PCR) amplification, DNA sequencing, and library construction. Fungal ITSregion was amplified using the forward primer ITS1F (CTTGGTCATTTAGAGGAAGTAA) and reverse primer ITS2R (GCTGCGTCTTCATCGATGCGC). The PCR reaction mixture, including 2 μL 10 × buffer, 2 μL 2.5 mM dNTPs, 0.8 μL each primer (5 μM), 0.2 μL rTaq polymerase, 0.2 μL BSA, 10 ng of template DNA, and ddH2O to a final volume of 20 μL. PCR amplification cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 4°C. All samples were amplified in triplicate. The PCR product was extracted from a 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer's instructions and quantified using Quantus™ Fluorometer (Promega, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) database repository, accession number SRR20897189-SRR20897218.
Data processing
Clean reads were obtained by filtering the raw sequences using a microbial ecological quantitative analysis pipeline (QIIME, version 1.9.1, USAU). Low-quality sequences (such as uncertain nucleotide sequences, three nucleotides with a Q-value of < 20, and unmatched barcode sequences) were removed. The QIIME v1.9.0 was used for quality control to obtain valid data, and the Uchime algorithm and gold database were used to remove delusion. These sequences were grouped into operational taxonomic units (OTUs) based on 97% sequence identity using UPARSE (V7.0.1090). Each row was annotated by comparing the Ribosomal Database Project (RDP) classifier (V2.11) against the unite8.0 database using a comparison threshold of 70%. Resampling was carried out with the smallest amount of data in the sample as the standard to make the uniform treatment for each sample. Mothur (version 1.30.2) 1 was used for diversity analysis. R 3.6.0 was used to perform various data conversions. Fungi Functional Guild (FUNGuild) was used for function prediction.
Statistical analysis
All statistical analyses were performed in R (v4.0.3) (Team, 2020). Hellinger transformation was first used to convert microbiota data. The Alpha diversity and Principal Component Analysis (PCA) analysis were generated by the vegan package in R (Oksanen et al., 2013) and plotted by ggplot2 (Wickham, 2011). The ternary diagram was drawn using the ggtern in R (Hamilton and Ferry, 2018). The significant difference between the functional composition of ZP and WP were detected by the Wilcoxon test. The significant different taxa between ZP and WP were detected by the MetagenomeSeq package (Wickham, 2011). Then, we performed the Pearson correlation analysis between the core community and the ingredients. Draw the heatmap of the correlation analysis using the Pheatmap package (Kolde and Kolde, 2015), and remove the community not significantly.
Interaction network analysis
We first use the iGraph package (Csardi and Nepusz, 2006) to analyze the network structure of the endophytic fungi community of P. hydropiper and P. lapathifolium. Then we use the weighted gene co-expression network analysis (WGCNA) package (Langfelder and Horvath, 2008) to calculate the correlation and P-values. And use the false discovery rates (FDR) to correct the P-values. The interaction network was generated by retaining the edges of R ≥ ±0.4 and P ≤ 0.05. Then we import the interaction network into Cytoscape (Shannon et al., 2003) to analyze the module. We choose the fast greedy algorithm to calculate the module, and use the GuImerà Amaral NeTwork (GIANT) package (Cumbo et al., 2014) to analyze the module's connectivity and the connectivity between modules. Finally, we use the ggplot2 package to plot the results.
Results
Determination of active ingredient content
Method validation
A preliminary optimization of the UPLC method (flow rate, gradient, injection volume, etc.) successfully achieved a well-separated peak for every standard compound, as shown in Figure 1. Moreover, the performance validation of the method was evaluated, including linearity, precision, reproducibility, stability, and recovery (Table 2). The linearity of the method was measured by analyzing standards over a linear range suitable for quantifying the corresponding analytes. Good linearity between concentration and signal intensity was obtained, and correlation coefficients of all compounds were calculated to be more than 0.999. Additionally, good reproducibility, stability, and precision were revealed in the results.
Figure 1
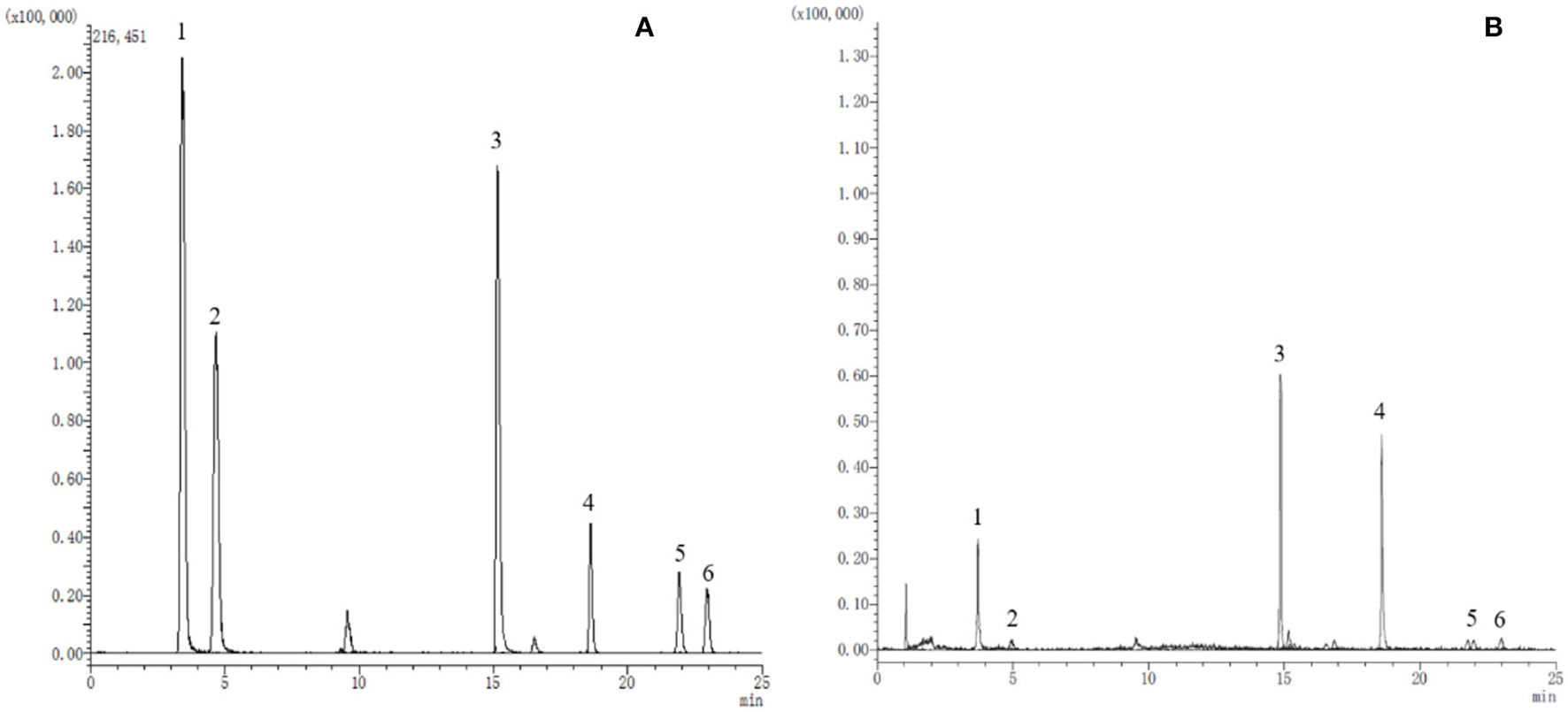
The extracted ion chromatograms (EICs) of reference substances (A) and sample (B). 1. Catechin; 2. Chlorogenic acid; 3. Hyperoside; 4. Quercetin; 5. Kaempferol; 6. Isorhamnetin.
Table 2
| Compound | Liner equations | R | Precision (RSD/%) | Repeatability RSD/% | Stability RSD/% | Recovery | ||
|---|---|---|---|---|---|---|---|---|
| Intra-day (n = 6) | Inter-day (n = 9) | Mean/% | RSD/% | |||||
| Catechins | Y = 55091.2X + 1252.94 | 0.9991 | 2.5 | 2.59 | 2.96 | 1.86 | 92.02 | 2.14 |
| Chlorogenic acid | Y = 17261.2X + 22440.6 | 0.9996 | 2.73 | 2.42 | 2.58 | 2.58 | 102.45 | 2.75 |
| Hyperoside | Y = 2982.38X + 29865.3 | 0.9992 | 2.87 | 2.57 | 0.60 | 0.11 | 100.62 | 2.41 |
| Quercetin | Y = 4467.8X + 11024.8 | 0.9995 | 2.72 | 2.66 | 2.17 | 1.53 | 102.47 | 2.65 |
| Kaempferol | Y = 12145.9X + 18648.7 | 0.9993 | 2.15 | 2.20 | 2.04 | 1.35 | 97.37 | 2.96 |
| Isorhamnetin | Y = 28372.1X + 13477.73 | 0.9993 | 1.43 | 2.76 | 1.88 | 2.02 | 97.52 | 2.06 |
Liner equations, precision, repeatability, stability, and average recovery rates of quantification of six components.
The above results suggest that the established method is sensitive, rapid, and reliable in identifying and quantifying phenolic compounds. The MRM mode of the UPLC-QQQ-MS/MS system is an effective and efficient tool for natural product analysis in complex matrices.
Sample content determination
The content determination results of catechins, chlorogenic acid, hyperoside, quercetin, kaempferol, and isorhamnetin are shown in four batches of ZP and four batches of WP in Table 3. In addition, we used student's t-test to detect the significant difference between the content of the active compounds in the two groups of ZP and WP, and the results are shown in Table 4.
Table 3
| Catechins (μg·g−1) | Chlorogenic acid (μg·g−1) | Hyperoside (μg·g−1) | Quercetin (μg·g−1) | Kaempferol (μg·g−1) | Isorhamnetin (μg·g−1) | |
|---|---|---|---|---|---|---|
| Z1 | 17.3281 | 3.5837 | 501.1931 | 366.8620 | 30.9806 | 24.3484 |
| Z2 | 17.5247 | 3.7231 | 507.4762 | 389.4847 | 30.7550 | 25.9680 |
| Z3 | 17.4783 | 2.7570 | 508.8720 | 361.1795 | 22.5175 | 25.7283 |
| Z4 | 16.7874 | 2.5801 | 519.0559 | 334.8053 | 18.7093 | 24.1923 |
| W1 | 29.2366 | 1.1047 | 225.8444 | 184.4371 | 15.9720 | 13.5882 |
| W2 | 33.4035 | 1.1563 | 215.0943 | 190.2311 | 14.0855 | 10.4824 |
| W3 | 36.5700 | 1.3370 | 234.7462 | 197.7751 | 23.6914 | 13.6166 |
| W4 | 37.6574 | 1.2769 | 204.4158 | 184.9176 | 19.7679 | 7.8025 |
The content of the six compounds in ZP and WP.
Table 4
| Compounds | ZP | WP | P-value | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Catechins | 17.2796 | 0.3387 | 34.2169 | 3.7789 | 0.0028 |
| Chlorogenic acid | 3.1610 | 0.5760 | 1.2187 | 0.1069 | 0.0057 |
| Hyperoside | 509.1493 | 7.4009 | 220.0252 | 13.1471 | 0.0000 |
| Quercetin | 363.0829 | 22.4691 | 189.3402 | 6.2059 | 0.0003 |
| Kaempferol | 25.7406 | 6.1218 | 18.3792 | 4.2575 | 0.1016 |
| Isorhamnetin | 25.0593 | 0.9184 | 11.3724 | 2.7978 | 0.0012 |
Student's t-test of component content between ZP and WP.
For ZP, the catechins content was between 16.7874 and 17.5247 μg g−1, the chlorogenic acid content was between 2.5801 and 3.7231 μg g−1, the content of hyperoside was between 501.1931 and 519.0559 μg g−1, the quercetin content was between 334.8053 and 389.4847 μg g−1, and the content of kaempferol was between 18.7093 and 30.9806 μg g−1. The content of isorhamnetin was between 24.1923 and 25.9680 μg g−1; for WP, the content of catechins was between 29.2366 and 37.6574 μg g−1; the chlorogenic acid content was between 1.1047 and 1.3370 μg g−1; the content of hyperoside was between 204.4158 and 234.7462 μg g−1; the quercetin content as between 184.4371 and 197.7751 μg g−1; the content of kaempferol was between 14.0855 and 23.6914 μg g−1, and the content of isorhamnetin was between 7.8025 and 13.6166 μg·g−1.
Student's t-test results (Table 4) showed that the contents of chlorogenic acid, hyperoside, quercetin, and isorhamnetin in ZP were significantly higher than those in WP, and the catechins in WP were considerably higher than those in ZP, while there was no significant difference in kaempferol between the two species.
Statistics of ITS amplicon sequencing
We sequenced 30 plant and soil samples, and a total of 2,321,991 reads were obtained, with a total base number of 696,597,300 bp. After filtering, the clean reads were 2,321,991, the total number of bases was 540,788,432 bp, and the average length was 232 bp. The number of sequences assigned to each sample was 47,618–169,166, with a median of 75,882 (Table 5). Based on the minimum sample sequence size, 1,642 OTUs were obtained at a similarity of 97%. The rarefaction curve showed sufficient sequencing depth (Figure 2), and the Good's coverage also shows good taxa coverage for each sample (Table 5).
Table 5
| Compartment | No. | Sequence number | Sequence base number | Average length | Min length | Max Length | Coverage | |
|---|---|---|---|---|---|---|---|---|
| ZP | Leaf | 1 | 81671 | 18659755 | 228.47 | 150 | 514 | 99.98% |
| Leaf | 2 | 75151 | 17275810 | 229.88 | 149 | 514 | 99.96% | |
| Leaf | 3 | 67298 | 15158745 | 225.25 | 144 | 473 | 99.98% | |
| Leaf | 4 | 80201 | 18570459 | 231.55 | 152 | 524 | 99.97% | |
| Rhizosphere soil | 1 | 52439 | 12505425 | 238.48 | 165 | 517 | 99.84% | |
| Rhizosphere soil | 2 | 60256 | 13933186 | 231.23 | 143 | 518 | 99.83% | |
| Rhizosphere soil | 3 | 54748 | 13127476 | 239.78 | 140 | 522 | 99.84% | |
| Stem | 1 | 47618 | 11099330 | 233.09 | 164 | 515 | 99.99% | |
| Stem | 2 | 69632 | 16378460 | 235.21 | 173 | 505 | 99.99% | |
| Stem | 3 | 63497 | 14935152 | 235.21 | 171 | 523 | 99.99% | |
| Stem | 4 | 57766 | 13408123 | 232.11 | 172 | 423 | 99.99% | |
| Root | 1 | 64740 | 16161710 | 249.64 | 173 | 494 | 99.93% | |
| Root | 2 | 57239 | 16812244 | 293.72 | 146 | 395 | 99.94% | |
| Root | 3 | 76613 | 17766891 | 231.90 | 145 | 349 | 99.93% | |
| Root | 4 | 169166 | 42516049 | 251.32 | 150 | 438 | 99.99% | |
| WP | Leaf | 1 | 69738 | 16457499 | 235.99 | 141 | 321 | 99.99% |
| Leaf | 2 | 91724 | 20805391 | 226.83 | 169 | 504 | 99.99% | |
| Leaf | 3 | 80418 | 18451726 | 229.45 | 157 | 321 | 99.98% | |
| Leaf | 4 | 88068 | 20608782 | 234.01 | 181 | 393 | 99.98% | |
| Rhizosphere soil | 1 | 72261 | 16224948 | 224.53 | 142 | 525 | 99.77% | |
| Rhizosphere soil | 2 | 66798 | 15601070 | 233.56 | 141 | 433 | 99.79% | |
| Rhizosphere soil | 3 | 63371 | 14296112 | 225.59 | 141 | 528 | 99.76% | |
| Stem | 1 | 94776 | 20978893 | 221.35 | 154 | 531 | 99.99% | |
| Stem | 2 | 96825 | 21836843 | 225.53 | 169 | 526 | 99.99% | |
| Stem | 3 | 82421 | 18990185 | 230.40 | 187 | 528 | 99.95% | |
| Stem | 4 | 76992 | 17647509 | 229.21 | 191 | 522 | 99.96% | |
| Root | 1 | 85574 | 19244983 | 224.89 | 155 | 498 | 99.98% | |
| Root | 2 | 76856 | 17077185 | 222.20 | 177 | 366 | 99.98% | |
| Root | 3 | 100743 | 22772037 | 226.04 | 168 | 529 | 99.99% | |
| Root | 4 | 97391 | 21486454 | 220.62 | 169 | 507 | 99.99% |
Sequencing statistics.
Figure 2
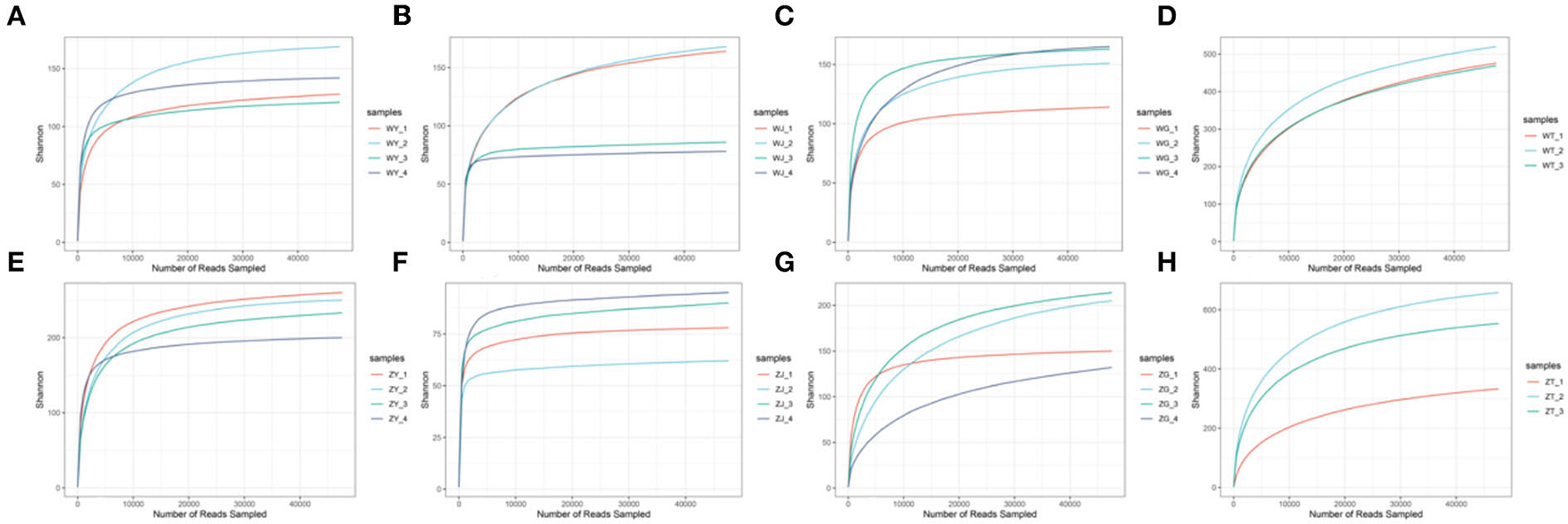
The rarefaction curve [(A) the leaf of WP; (B) the stem of WP; (C) the root of WP; (D) the rhizosphere soil of WP; (E) the leaf of ZP; (F) The stem of ZP; (G) the root of ZP; (H) the rhizosphere soil of ZP].
Analysis of the diversity and composition of the fungal communities
To compare the differences in fungal communities of two plants from a macroscopic perspective. First, we compared the difference in the alpha diversity index of all samples of the two species. As can be seen from Figure 3A, there are no significant differences in the ACE, Shannon, and Simpson indices between the two species. Then, the difference in the alpha diversity of the same compartment of the two species was compared, and it can be seen from Figure 3B that there is a significant difference in the ACE index between ZP and WP in the leaves.
Figure 3
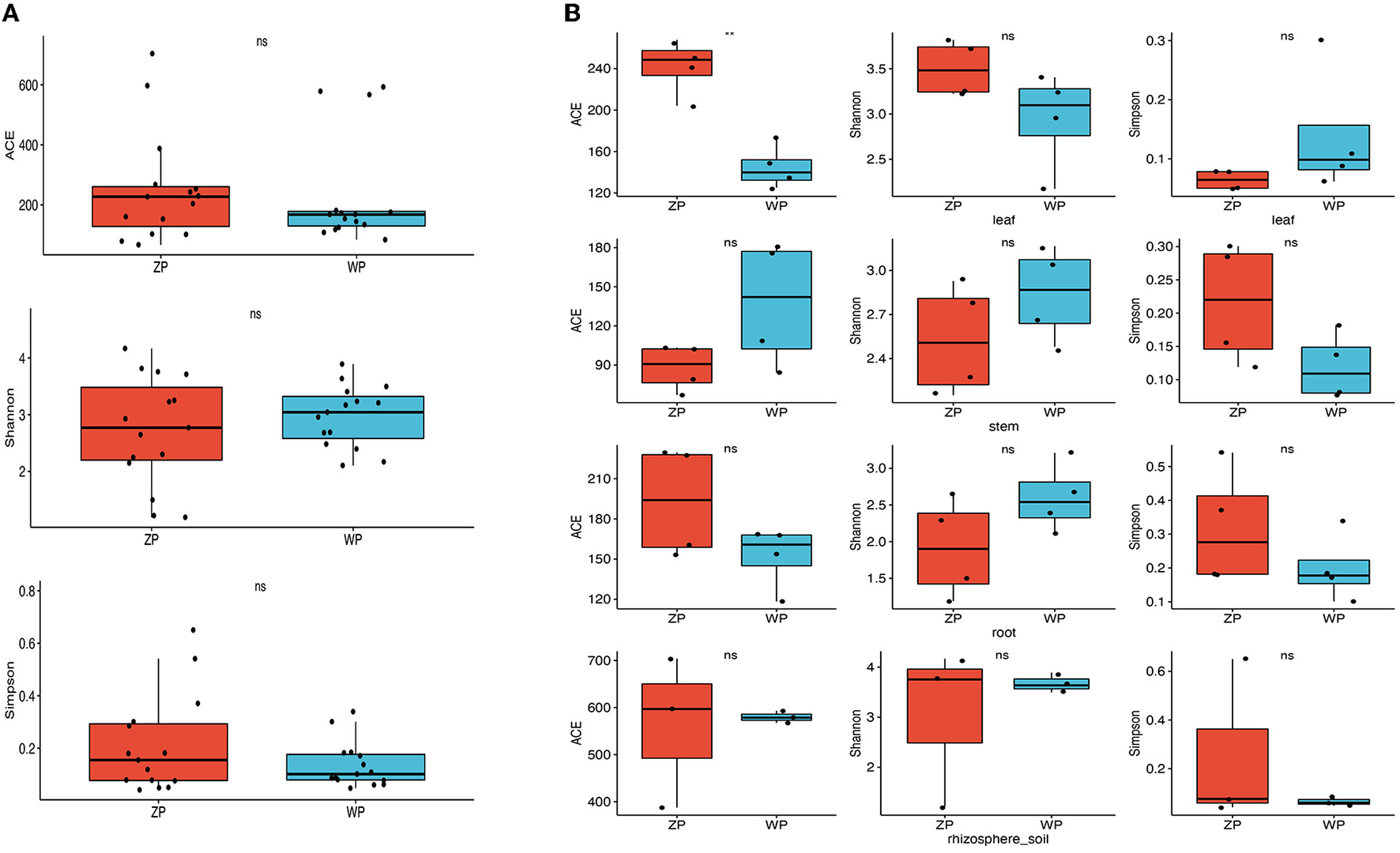
Alpha diversity of fungal communities; (A) Difference analysis between whole plant of two species; (B) Difference analysis between three compartments of two species. *Means P-value < 0.05, **means P-value < = 0.01, ***means p-value < = 0.001, ****means P-value < = 0.0001, ns mean P-value > 0.05.
If we look at the composition of all the communities of the two species, as can be seen from Figure 4A, both ZP and WP are dominated by Ascomycota, accounting for an average of 87.83% of the entire community. As the sampling compartment ranges from rhizosphere soil and rhizome to leaf, the relative abundance of Ascomycota in ZP and WP increases in quantity. The Adonis analysis of community composition in different parts of ZP and WP shows significant differences in leaves, stems, and roots (Figure 5).
Figure 4
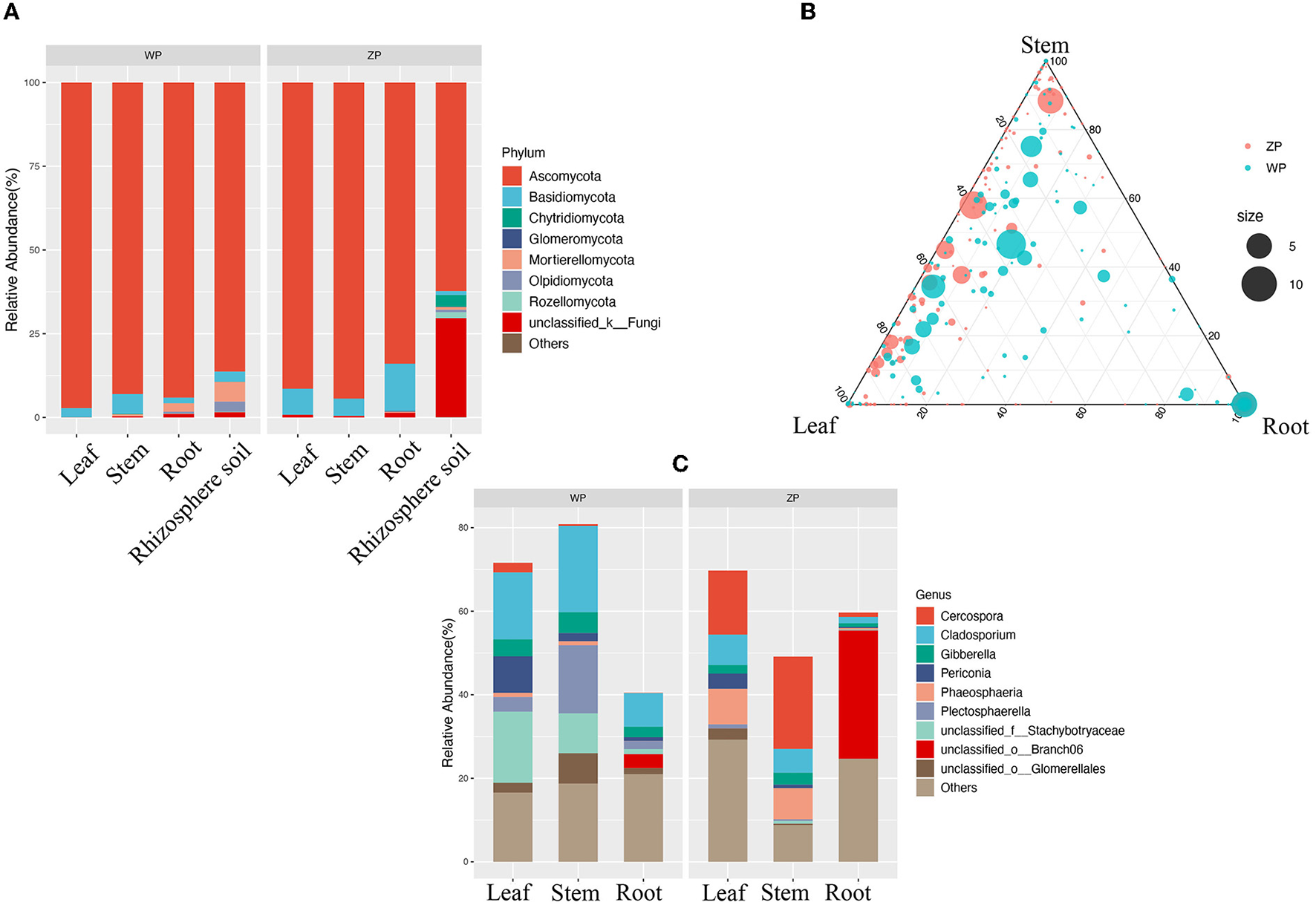
(A) The community composition of ZP and WP at the phylum level; (B) The ternary phase diagram of community in ZP and WP; (C) The significant differential communities between ZP and WP.
Figure 5
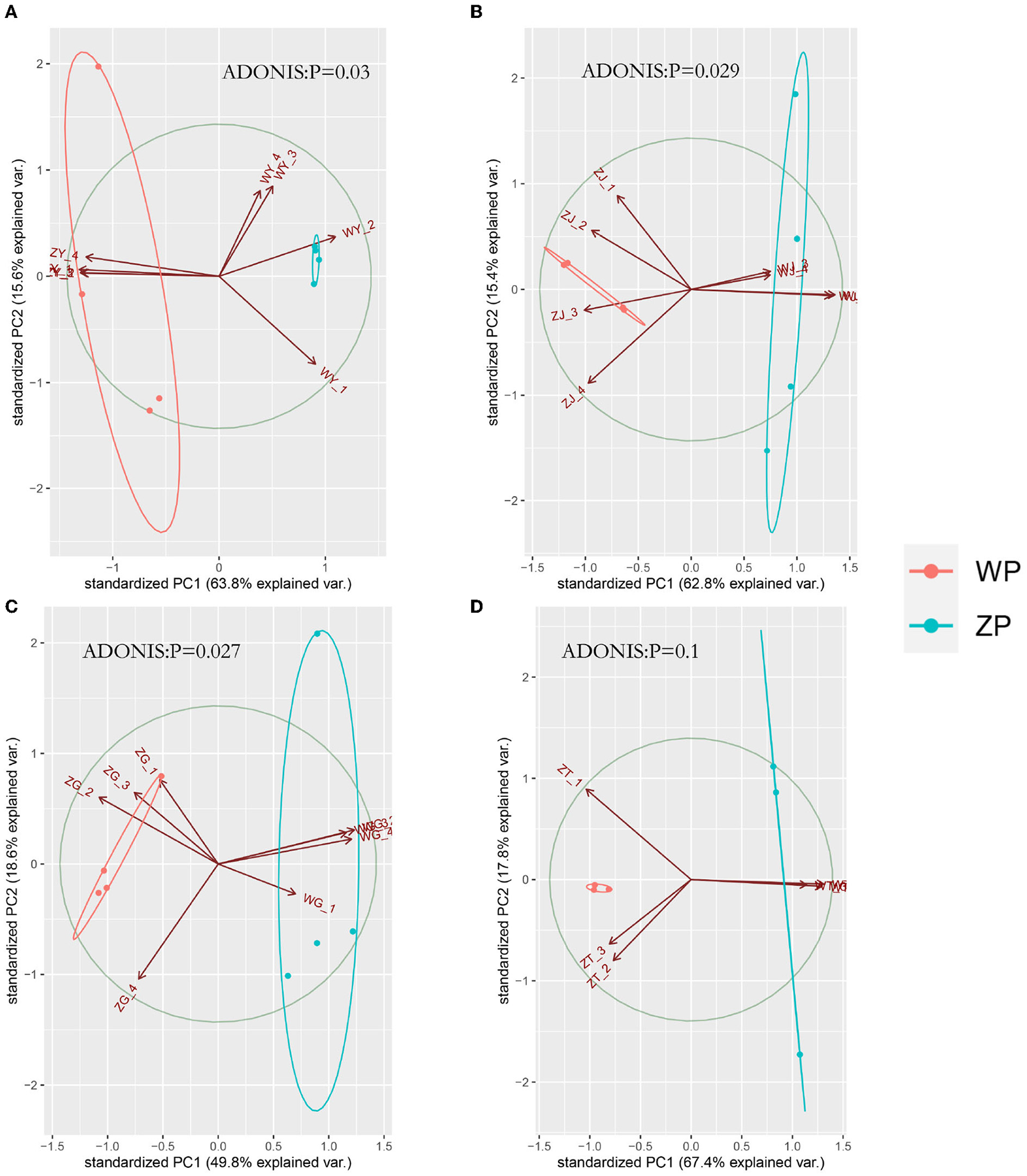
PCA analysis of three compartments between ZP and WP [(A) leaf, (B) stem, (C) root, (D) rhizosphere soil].
Furthermore, we performed the ternary phase diagram analysis of the leaves, stems, and roots of two plants (Figure 4B) to determine the distribution of the community in various compartments. We found that a large amount of OUT was shared between stems and leaves. Compared to ZP, the three compartments of WP shared more OTUs, while shared OTUs in ZP tended to occupy a higher proportion of stems and leaves. Based on MetagenomeSeq, the leaves, stems, and roots of ZP and WP were analyzed (Figure 4C). A significant difference in OTUs was found (88) in leaves, 38 in stems, and 71 in roots. We observed the composition of these OTUs at the genus level. In ZP, Cercospora (15.34 and 22.11%), Phaeosphaeria (8.48, 7.45%), and Cladosporium (7.29 and 5.68%) dominate the leaf and stem community, while Branch06 (30.69%) dominates the root community. In WP, Cladosporium (16.02, 20.68, and 8.04%) and Stachybotryaceae (17.02, 9.55, and 1.23%) dominate the community of leaves, stems, and roots. In addition, Plectosphaerella has a significantly higher abundance in WP than in ZP, especially in stems; 16.29% in WP, compared with only 0.44% in ZP.
Interaction network analysis of endophytic fungal communities
The interaction networks of ZP and WP fungal communities were constructed, respectively, and the results are shown in Figure 6. The overall module connectivity of the ZP network is 0.38, and the overall module connectivity of the WP network is 0.34. Still, the nodes of ZP show special aggregation characteristics, while the nodes of WP are more widely distributed, and there is no obvious aggregation. We performed characteristic annotations to the nodes in the network to determine the core community in the endophytic fungal community (Figure 7). This study defines the node annotated as the module node and the connectors as the core community. In ZP, three module hub nodes were identified, including OUT1551 (Teratosphaeriaceae), OUT1542 (Phaeosphaeriaceae), and OUT1374 (Basidiomycota), in addition to 32 connection nodes. In the WP, 74 connection nodes are identified. Observing the community composition at the genus level (Figure 8), the relative abundance of key communities in the three compartments in WP was close, accounting for 19.81% of the total community, dominated by Sarocladium in stems and roots and dominated by Cercospora in leaves. In ZP, the relative abundance of key communities in the three compartments was significantly different, accounting for 87.59% of the community. The relative abundance of community in stems is highest, dominated mainly by Sarocladium and Cercospora, in the leaves by Cercospora, and in the roots by Zopfiella.
Figure 6
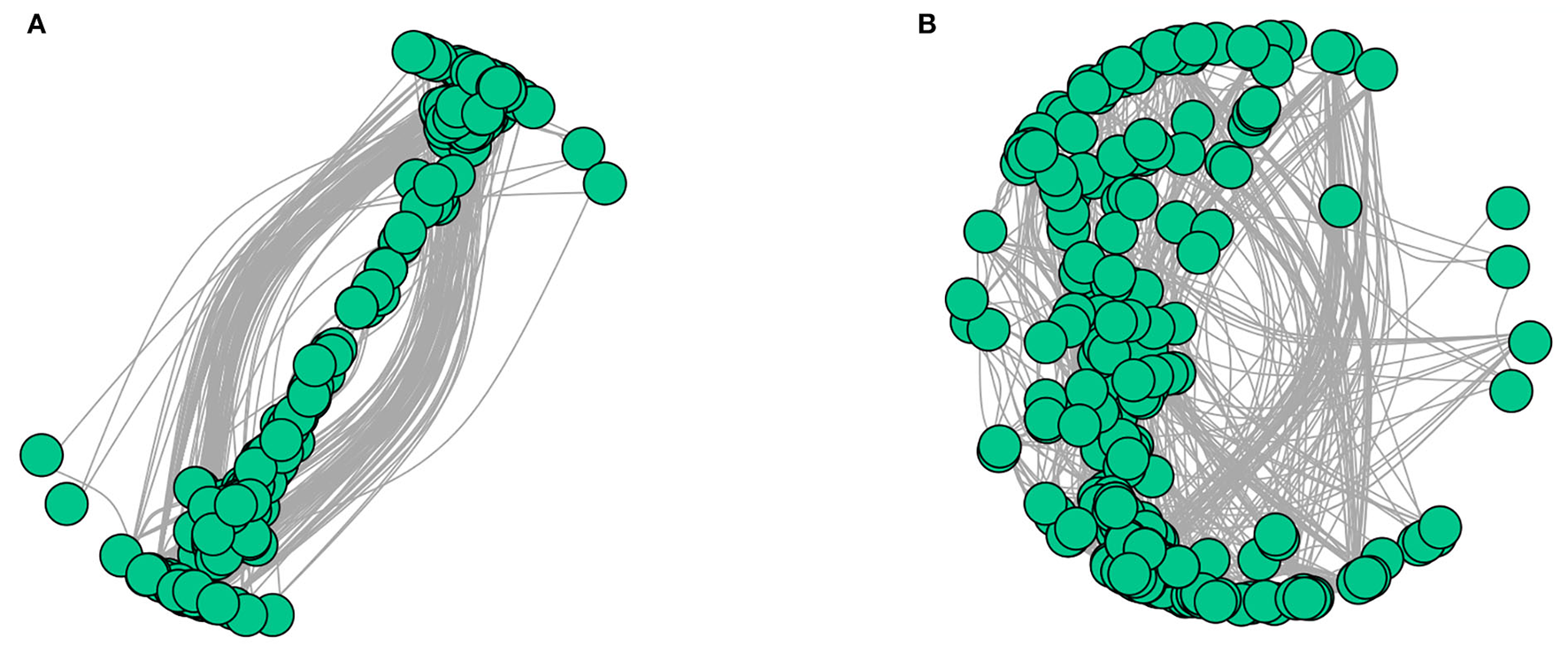
Interaction network diagram of ZP and WP [(A) ZP, (B) WP].
Figure 7

The characteristic annotation analysis of nodes in the interaction network. (A) ZP, (B) WP.
Figure 8
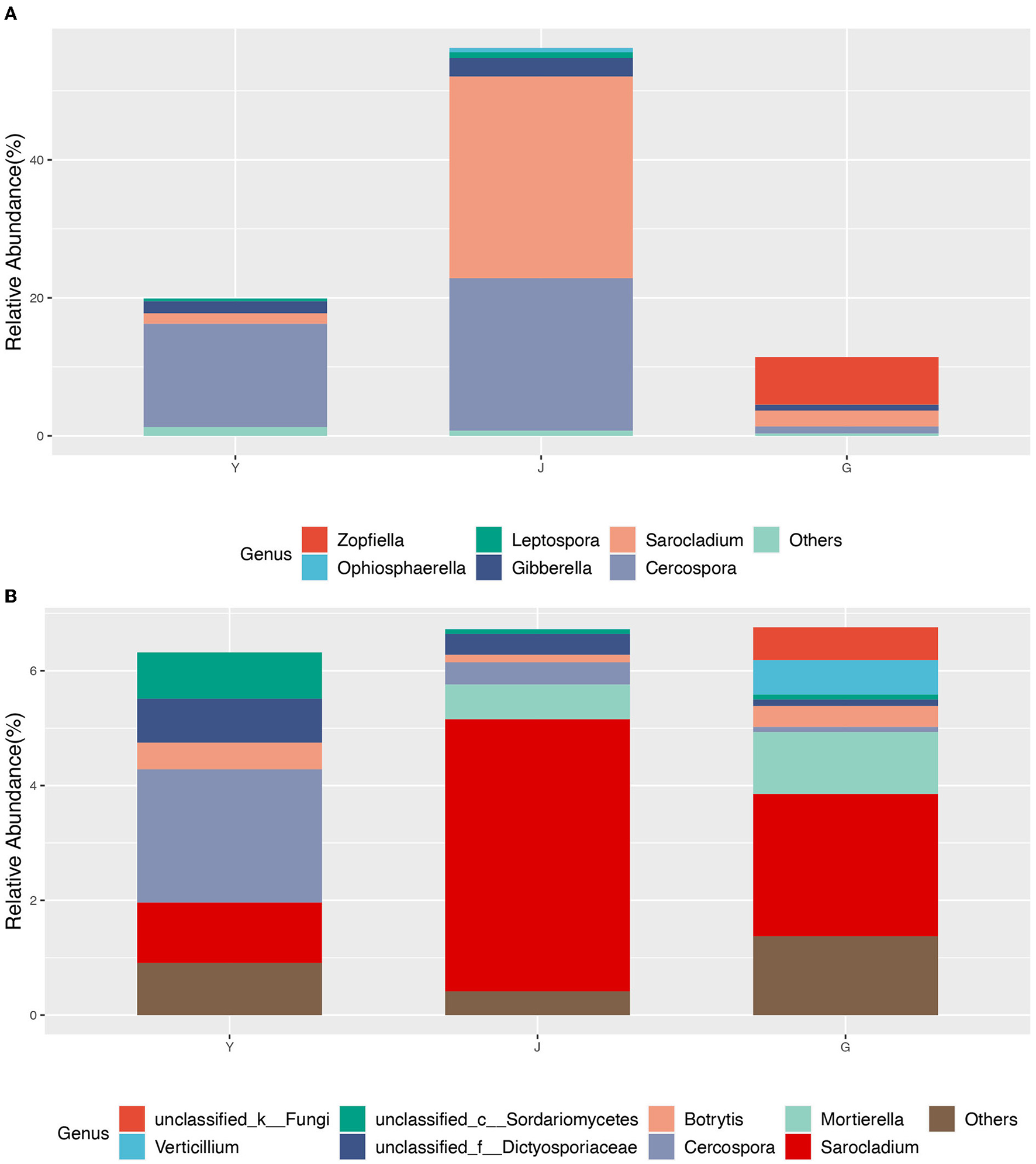
The core community composition at the genus level. (A) ZP, (B) WP.
Functional composition analysis of endophytic fungi
To detect the differences in function community composition in two plants, we used FUNGuild to perform the functional annotations. A total of 10 trophic types are annotated (Figure 9).
Figure 9
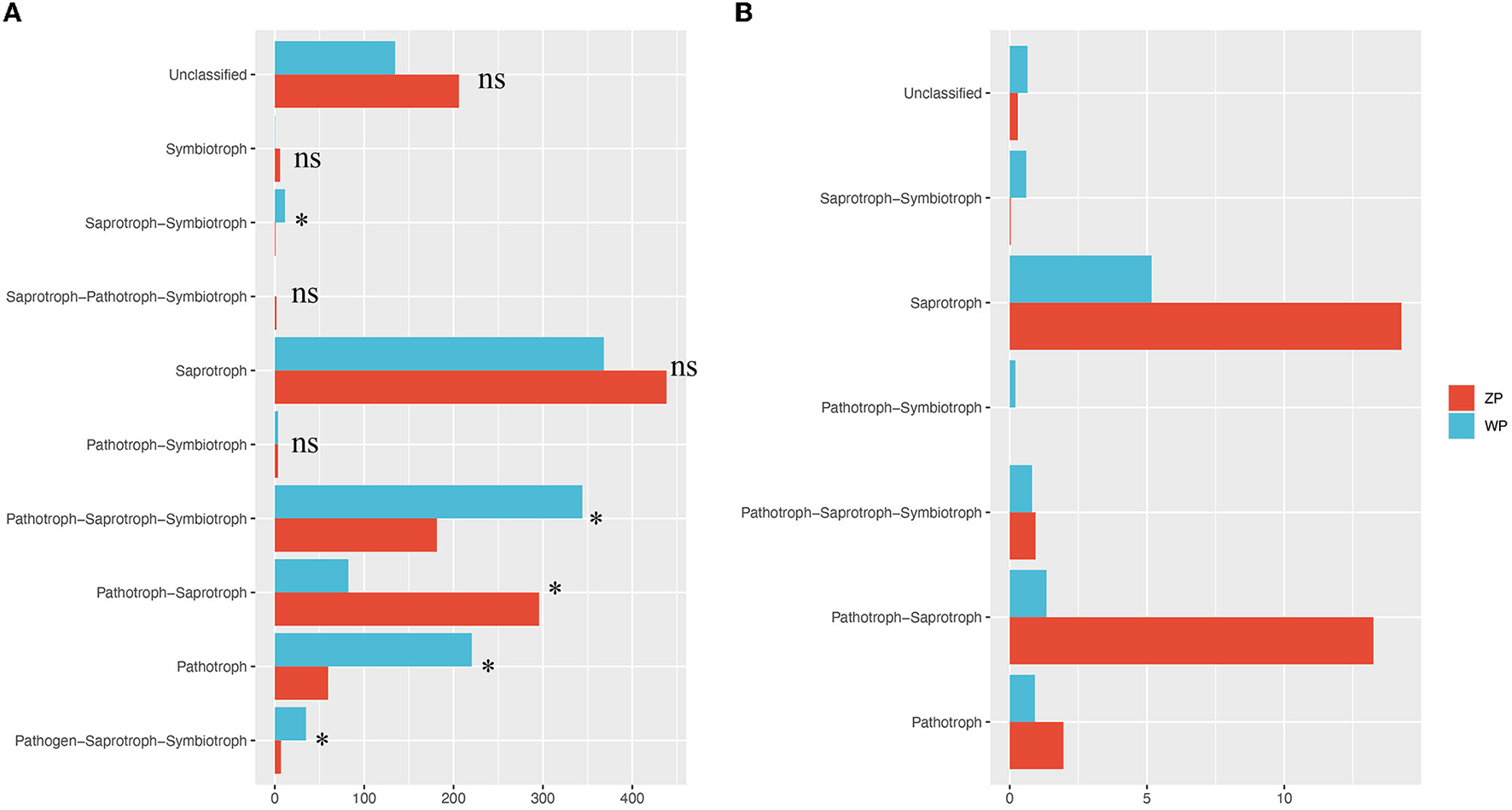
Functional community composition. (A) The functional composition of all compartments, (B) The functional composition of core community. *P-value < 0.05.
We found significant differences in the relative abundance of trophic types between ZP and WP, including pathogen-saprotroph-symbiotroph, pathotroph, pathotroph-saprotroph-saprotroph, pathotroph-saprotroph-symbiotroph, and saprotroph-symbiotroph. Further, a functional analysis of the core community (Figure 10) was performed. In the dominant functional community, saprotrophs and pathotroph-saprotrophs in ZP were significantly higher than in the WP group.
Figure 10
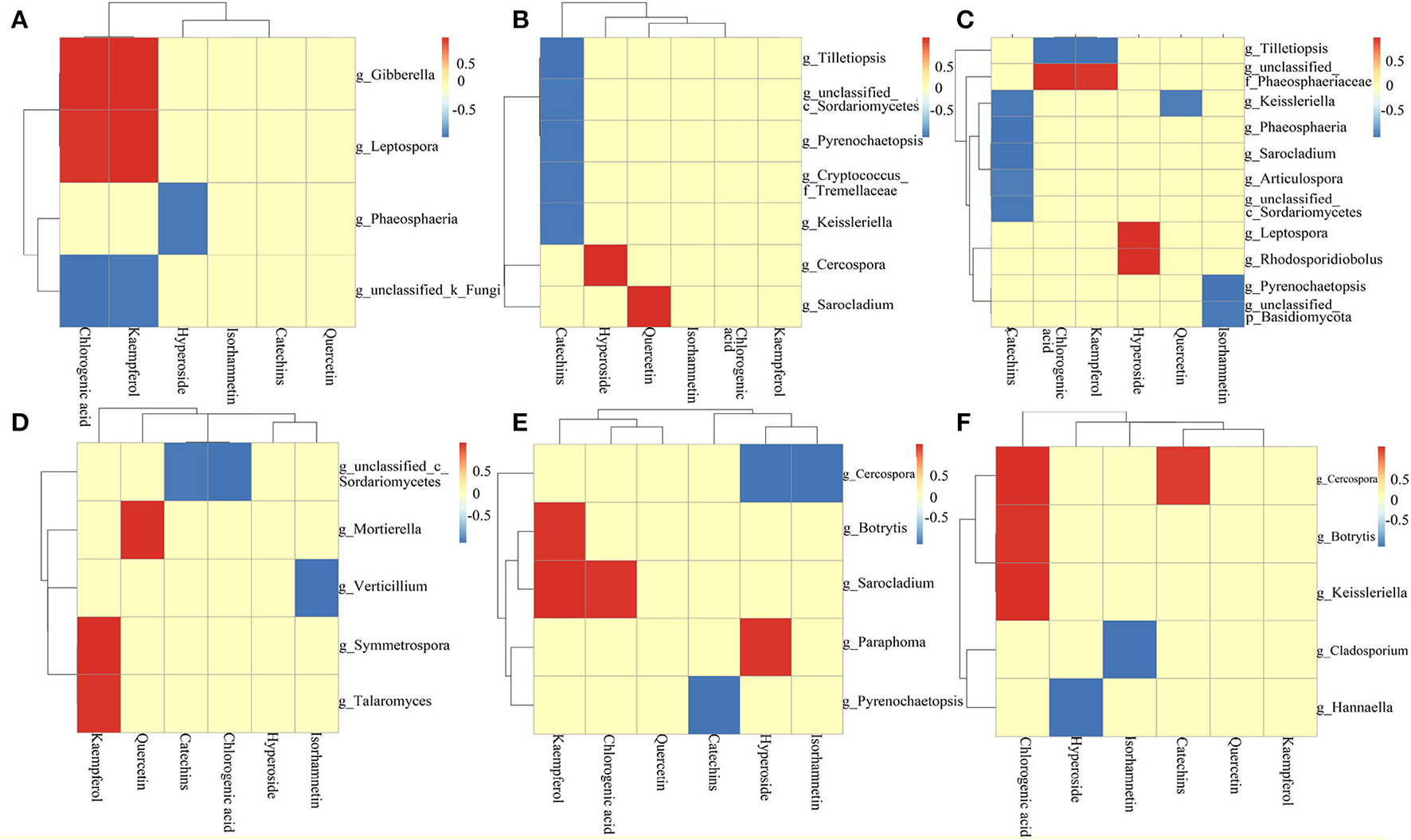
Analysis of the correlation between core community and active ingredient [(A) ZP root; (B) ZP stem; (C) ZP leaf; (D) WP root; (E) WP stem; (F) WP leaf].
Correlation analysis of fungal communities with active ingredients
The Pearson-related analysis of core community and active ingredient content in ZP and WP was performed. In the roots of ZP (Figure 10A), Gibberella and Leptospora were significantly positively correlated with chlorogenic acid and kaempferol. Phaeosphaeria was significantly inversely associated with hyperoside. In the stem of ZP (Figure 10B), Tilletiopsis, c_Sordariomycetes, Pyrenochaetopsis, Cryptococcus_f_Tremellacea, and Keissleriella were significantly negatively correlated with catechins; Cercospora was significantly positively correlated with hyperoside; Sarocladium was significantly positively correlated with quercetin. In the leaves of ZP (Figure 10C), Keissleriella, Phaeosphaeria, Sarocladium, Articulospora, and c_Sordariomycetes were significantly negatively correlated with catechins; Tilletiopsis was significantly negatively correlated with chlorogenic acid and kaempferol; f_Phaeosphaeriaceae positively correlated with chlorogenic acid and kaempferol; Leptospora and Rhodosporidiobolus were positively correlated with hyperoside; Keissleriella was negatively correlated with quercetin; Pyrenochaetopsis and p_Basidiomycota were negatively correlated with Isorhamnetin significance.
In the roots of the WP (Figure 10D), Mortierella, Symmetrospora, and Taromyces were significantly positively correlated with kaempferol. Mortierella was positively correlated with quercetin; c_Sordariomycetes was significantly inextricably linked to catechins and chlorogenic acids; Verticillium was significantly inversely associated with Isorhamnetin. In the stem of WP (Figure 10E), Botrytis and Sarocladium were significantly positively correlated with kaempferol; Sarocladium was significantly positively correlated with chlorogenic acid; Paraphoma was significantly positively correlated with Hyperoside; Cercospora was significantly negatively correlated with Hyperoside and Isorhamnetin; Pyrenochaetopsis was significantly and negatively correlated with catechins. In the leaves of WP (Figure 10F), Cercospora, Botrytis, and Keissleriella were significantly positively correlated with chlorogenic acid; Cercospora was significantly positively correlated with catechins; Cladosporium was significantly negatively correlated with Isorhamnetin; Hannaella was significantly negatively correlated with hyperoside.
Discussion
The colonization of cercospora interferes with the assembly process of the endophytic fungi of P. Hydropiper
In this study, we found a high degree of overlap in the taxa and distribution areas of ZP and WP, and there was no significant difference in the overall alpha diversity between the two. The similarity of fungal communities in rhizosphere soil confirms that both acquire fungi from the same soil species pool. Moreover, the interaction network analysis shows that the connectivity of the endophytic fungi between ZP and WP is very close. These results indicate that the two plants are similar in their ability to shape the composition and structure of endophytic fungal communities. However, we also observed significant leaves, stems, and roots differences between the two plants. This suggests that the assembly process of fungal communities is the key factor influencing the composition of the community. Therefore, we compared the community composition of the three compartments in ZP and WP and assessed the community composition that differed significantly between the two. In ZP, the relative abundance of differential communities in leaves is highest, with Cercospora dominating the differential communities in stems and leaves. At the same time, the ACE index of the community in the leaves of ZP was significantly higher than that of WP, indicating a significant enrichment of the community in the leaves of ZP. Studies have shown that Cercospora usually colonizes leaves (Khan et al., 2008). Interestingly, the core community in ZP was also dominated by Cercospora, accounting for 38.12% of the total community. This suggests that after Cercospora colonizes the leaves of ZP, it significantly affects the assembly process of the endophytic fungi. In WP, the relative abundance of differential communities in stems was highest, and the differential community at all three compartments was dominated by Cladosporium, which is generally considered to colonize the roots of plants (Hamayun et al., 2009; Chen et al., 2022); this suggests that the assembly process of endophytic fungal communities in WP is still dominated by fungi recruited in the soil.
Endophytic fungi communitites significantly affect host secondary metabolic processes
We found significantly different communities between ZP and WP, including Cercospora, Phaeosphaeria, Branch06, Stachybotryaceae, and Plectosphaerella. Most of the community from Cercospora is thought to be able to cause diseases in plants. Interestingly, Martínez et al. (2017) found that chlorogenic acid can effectively inhibit the growth of fungi such as Cercospora. In this study, the chlorogenic acid content of ZP was significantly higher than that of WP, and the relative abundance of this type of fungi in ZP was also considerably higher than WP, suggesting that it may be due to the extensive colonization of Cercospora, resulting in host stress producing more chlorogenic acid. For Pheosphaeria, there have been many studies that demonstrate it is rich in secondary metabolites, including polyketone derivatives, pyrazine alkaloids, isocoumarins, perylenequinonoid, anthraquinone, diterpenes, and cyclic peptides (El-Demerdash, 2018; Xiao et al., 2022). This may illustrate the reason that the content of secondary metabolites in ZP is significantly higher than in WP. Fungi, mainly plant pathogens or saprophytic, such as the widely studied Cladosporium fulvum, are considered to be the leading cause of moldy tomato leaves. However, many species have also been found in this genus that promotes plant growth, such as through the production of gibberellin (Hamayun et al., 2009, 2010), protein hydrolysates (Răut et al., 2021), and volatile substances (Paul and Park, 2013). In addition, some species from Cladosporium can also produce substances such as brefeldin A (Wang et al., 2007) and Cladosins LO (Pan et al., 2020) to play an anti-pathogenic fungus and bacterial activity. It is worth noting that after Ullah et al. (2019) found that the stem of Poplars was infected with Plectosphaerella populi, the content of catechins and proanthocyanidins increased significantly. In this experiment, the community of Plectosphaerella was observed in the stem of WP to be considerably higher than that of ZP. The catechin content of WP was also significantly higher than that of ZP, suggesting that Plectosphaerella is likely to be an essential potential community involved in or promoting catechin synthesis and indicating that the synthetic compartment of catechin in the plant may be the stem.
Moreover, we analyzed the functional composition of these communities. In ZP, it is dominated by saprotroph and pathotroph-saprotrophs. In WP, the community of pathotrophs, pathogen-saprotroph-symbiotrophs, and saprotroph-symbiotrophs. Moreover, the relative abundance of the ZP-led functional community was significantly higher than that of WP, and the relative abundance of the dominant community in WP was significantly higher than that of ZP. This indicates a significant difference in the functional community composition between the two. Further, in the functional composition analysis of the core community, it was also found that saprotroph and sathotroph-saprotrophs had significant differences between ZP and WP. These results suggest a difference in the status of endophytic fungi involved in the secondary metabolism of ZP and WP. Further research is needed on the function of these communities in plants.
The core communities are the essential members involved in the secondary metabolic processes of host plants
We constructed the interaction network of ZP and WP endophytic fungal communities, 3 module hub nodes and 35 connector nodes were found in ZP, and 74 connector nodes were found in WP. The two core communities present different compositions at the genus level, with ZP dominated by Cercospora and Zopfiella. In the WP, it is mainly dominated by Mortierella and Cercospora. Kemp et al. (2020) found that Sarocladium zeae from this genus acted as systemic endophytic fungi in wheat and as a biological control agent in the host. Since the Cercospora and Sarocladium play a dominant role in both ZP and WP, therefore, it may indicate that the Cercospora has a potential role in regulating the structure of the community. In addition, Mortierella is often thought to promote plant growth and increase biomass (Li, S. J. et al., 2018; Ozimek and Hanaka, 2020; Zhang et al., 2020). For Zopfiella, Sun et al. (2020) found that species from this genus are capable of producing sesquiterpenes and α-pyranone derivatives.
In the analysis of the correlation between the core community and the active ingredient, 23 distinctly associated communities were found, many of which were thought to promote plant growth and participate in secondary metabolic processes, including Mortierella (Li, F. et al., 2018; Ozimek and Hanaka, 2020; Zhang et al., 2020), Talaromyces (Naraghi et al., 2012; Lan and Wu, 2020), Sordariomycetes (Camarena-Pozos et al., 2021), Verticillium (You et al., 2009; Li, N., et al., 2018), Sarocladium (Kemp et al., 2020; Błaszczyk et al., 2021; Salvatore et al., 2021), Gibberella (Brian et al., 1954; Geng et al., 2014), Tilletiopsis (Klecan et al., 1990), and Phaeosphaeriaceae (Xiao et al., 2022; Xu et al., 2022), In addition, some plant pathogens are still present in these communities, including Botrytis, Paraphoma, Cercospora, and Pyrenochaetopsis. These communities potentially affect the secondary metabolic processes of the host, and it is necessary to further identify these communities and their functions in subsequent studies. Therefore, these results demonstrate that the core community of the interaction network plays a vital role in the secondary metabolism of ZP and WP.
Conclusion
The results showed that the endophytic fungi community assembly processes of P. hydropiper and P. lapathifolium are significantly different. Since the two acquired fungi from the same soil species pool and have the same ability to shape the composition and structure of endophytic fungal communities, we believe that this difference was mainly due to the infestation of Cercospora. In addition, the results also found that the core community was an important member involved in the secondary metabolism of P. hydropiper and P. lapathifolium. The infection of Cercospora provided an opportunity to shape a specific core community. The infection process exerted selective pressure on recruiting other fungi, resulting in a higher proportion of the core community of P. hydropiper in the community as a whole than that of P. lapathifolium. This increases the odds of the host interacting with the core community. Therefore, it can enhance the communication between host plants and endophytic fungi, affecting the content of secondary metabolites of P. hydropiper. Although our study is limited to the two current species, more in-depth studies are needed to elucidate the mechanism by which Cercospora infestation affects the process of plant endophytic fungal community assembly. This study has important practical implications for inoculating specific communities in production to improve the yield of crops or medicinal plants. It also provides ideas for the study of the assembly process of other plant endophytic fungi.
Funding
This work was financially supported by the Sichuan Science and Technology Department (2021ZYD0109 and 2022YFS0430). We are indebted to our alma mater, Chengdu University of Traditional Chinese Medicine, for the convenience of collecting documents and experiments. Thanks for all the help from everyone in our lab.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) database repository, https://www.ncbi.nlm.nih.gov/, accession number SRR20897189-SRR20897218.
Author contributions
XZ, HL, QH, and JW conceived and designed the study. XZ, HL, MT, ZD, and QF performed the experiments. XZ and HL processed the data and wrote the article. All authors read and approved the manuscript.
Conflict of interest
Author JS was employed by Sichuan Fuzheng Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.984483/full#supplementary-material
References
1
Adeleke B. S. Babalola O. O. (2021). The endosphere microbial communities, a great promise in agriculture. Int. Microbiol.24, 1–17. 10.1007/s10123-020-00140-2
2
Ayaz M. Junaid M. Subhan F. Ullah F. Sadiq A. Ahmad S. et al . (2014). Heavy metals analysis, phytochemical, phytotoxic and anthelmintic investigations of crude methanolic extract, subsequent fractions and crude saponins from Polygonum hydropiper L. BMC Complem. Altern. Med.14, 1–9. 10.1186/1472-6882-14-465
3
Błaszczyk L. Waśkiewicz A. Gromadzka K. Mikołajczak K. Chełkowski J. (2021). Sarocladium and Lecanicillium associated with maize seeds and their potential to form selected secondary metabolites. Biomolcules11, 98. 10.3390/biom11010098
4
Brian P. Elson G. Hemming H. Radley M. (1954). The plant-growth-promoting properties of gibberellic acid, a metabolic product of the fungus Gibberella fujikuroi. J. Sci. Food Agric.5, 602–612. 10.1002/jsfa.2740051210
5
Camarena-Pozos D. A. Flores-Núñez V. M. López M. G. Partida-Martínez L. P. (2021). Fungal volatiles emitted by members of the microbiome of desert plants are diverse and capable of promoting plant growth. Environ. Microbiol.23, 2215–2229. 10.1111/1462-2920.15395
6
Cao J. Liu B. Xu X. Zhang X. Zhu C. Li Y. et al . (2021). Plant endophytic fungus extract ZNC improved potato immunity, yield, and quality. Front. Plant Sci. 12, 707256. 10.3389/fpls.2021.707256
7
Chen H. Chen J. Qi Y. Chu S. Ma Y. Xu L. et al . (2022). Endophytic fungus Cladosporium tenuissimum DF11, an efficient inducer of tanshinone biosynthesis in Salvia miltiorrhiza roots. Phytochemistry194, 113021. 10.1016/j.phytochem.2021.113021
8
Csardi G. Nepusz T. (2006). The igraph software package for complex network research. Int. J. Complex Syst.1695, 1–9.
9
Cumbo F. Paci P. Santoni D. Di Paola L. Giuliani A. (2014). GIANT: a cytoscape plugin for modular networks. PLoS ONE.9, e105001–e105001. 10.1371/journal.pone.0105001
10
Dang H. Zhang T. Wang Z. Li G. Zhao W. Lv X. et al . (2021). Succession of endophytic fungi and arbuscular mycorrhizal fungi associated with the growth of plant and their correlation with secondary metabolites in the roots of plants. BMC Plant Biol.21, 1–16. 10.1186/s12870-021-02942-6
11
El-Demerdash A. (2018). Chemical diversity and biological activities of Phaeosphaeria fungi genus: a systematic review. J. Fungi.4, 130. 10.3390/jof4040130
12
Geng Z. Zhu W. Su H. Zhao Y. Zhang K. Q. Yang J. (2014). Recent advances in genes involved in secondary metabolite synthesis, hyphal development, energy metabolism and pathogenicity in Fusarium graminearum (teleomorph Gibberella zeae). Biotechnol. Adv. 32, 390–402. 10.1016/j.biotechadv.2013.12.007
13
Hamayun M. Afzal Khan S. Ahmad N. Tang D. S. Kang S. M. Na C. I. et al . (2009). Cladosporium sphaerospermum as a new plant growth-promoting endophyte from the roots of Glycine max (L.) Merr. World J. Microbiol. Biotechnol.25, 627–632. 10.1007/s11274-009-9982-9
14
Hamayun M. Khan S. A. Khan A. L. Rehman G. Kim Y. H. Iqbal I. et al . (2010). Gibberellin production and plant growth promotion from pure cultures of Cladosporium sp. MH-6 isolated from cucumber (Cucumis sativus L.). Mycologia.102, 989–995. 10.3852/09-261
15
Hamilton N. E. Ferry M. (2018). ggtern: Ternary diagrams using ggplot2. J. Stat. Softw. 87, 1–17. 10.18637/jss.v087.c03
16
Hong H. H. Hanshen Z. (2013). Research progress of Chinese herbal medicine spicy indigo in recent years. Chin. J. Ethnomed. Ethnoph.41, 38–40.
17
Hong L. L. Wei X. Long W. Kun J. W. (2019). Analysis of Polygonum hydropiper and its promiscuous speceis in same genus. Collections. 8.
18
Kemp N. D. Vaughan M. M. McCormick S. P. Brown J. A. Bakker M. G. (2020). Sarocladium zeae is a systemic endophyte of wheat and an effective biocontrol agent against Fusarium head blight. Biol. Control.149, 104329. 10.1016/j.biocontrol.2020.104329
19
Khan J. Rio L. D. Nelson R. Rivera-Varas V. Secor G. A. Khan M. F. R. (2008). Survival, dispersal, and primary infection site for Cercospora beticola in sugar beet. Plant Dis.92, 741–745. 10.1094/PDIS-92-5-0741
20
Klecan A. Hippe S. Somerville S. (1990). Reduced growth of Erysiphe graminis f. sp. hordei induced by Tilletiopsis pallescens. Phytopathology80, 325–331. 10.1094/Phyto-80-325
21
Kolde R. Kolde M. R. (2015). Package “pheatmap”. R Package 1, 790. 10.1016/j.funeco.2018.04.004
22
Lan D. Wu B. (2020). Chemistry and bioactivities of secondary metabolites from the Genus Talaromyces. Chem. Biodivers.17, e2000229. 10.1002/cbdv.202000229
23
Langfelder P. Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform.9, 559–559. 10.1186/1471-2105-9-559
24
Li F. Chen L. Redmile-Gordon M. Zhang J. Zhang C. Ning Q. et al . (2018). Mortierella elongata 's roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev.29, 1642–1651. 10.1002/ldr.2965
25
Li N. Wang W. Bitas V. Subbarao K. Liu X. Kang S. (2018). Volatile compounds emitted by diverse Verticillium species enhance plant growth by manipulating auxin signaling. Mol. Plant Microbe Interact.31, 1021–1031. 10.1094/MPMI-11-17-0263-R
26
Li S. J. Zhang X. Wang X. H. Zhao C. Q. (2018). Novel natural compounds from endophytic fungi with anticancer activity. Eur. J. Med. Chem.156, 316–343. 10.1016/j.ejmech.2018.07.015
27
Lunardelli Negreiros de Carvalho P. de Oliveira Silva E. Aparecida Chagas-Paula D. Honorata Hortolan Luiz J. Ikegaki M. (2016). Importance and implications of the production of phenolic secondary metabolites by endophytic fungi: a mini-review. Mini Rev. Med. Chem. 16, 259–271. 10.2174/1389557515666151016123923
28
Martínez G. Regente M. Jacobi S. Rio M. D. Pinedo M. Laura D. (2017). Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Physiol.140, 30–35. 10.1016/j.pestbp.2017.05.012
29
Martins F. Mina D. Pereira J. A. Baptista P. (2021). Endophytic fungal community structure in olive orchards with high and low incidence of olive anthracnose. Sci. Rep. 11, 1–11. 10.1038/s41598-020-79962-z
30
Naraghi L. Heydari A. Rezaee S. Razavi M. (2012). Biocontrol agent talaromyces flavus stimulates the growth of cotton and potato. J. Plant Growth Regul.31, 471–477. 10.1007/s00344-011-9256-2
31
Oksanen J. Blanchet F. G. Kindt R. Legendre P. Minchin P. R. O'hara R. et al . (2013). Package ‘vegan'. Commun. Ecol. Pack.2, 1–295.
32
Ozimek E. Hanaka A. (2020). Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture11, 7. 10.3390/agriculture11010007
33
Pan F. El-Kashef D. H. Kalscheuer R. Müller W. E. G. Lee J. Feldbrügge M. et al . (2020). Cladosins L–O, new hybrid polyketides from the endophytic fungus Cladosporium sphaerospermum WBS017. Eur. J. Med. Chem.191, 112159. 10.1016/j.ejmech.2020.112159
34
Paul D. Park K. S. (2013). Identification of volatiles produced by Cladosporium cladosporioides CL-1, a fungal biocontrol agent that promotes plant growth. Sens. Basel13, 13969–13977. 10.3390/s131013969
35
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at: https://www.R-project.org/.
36
Rahman E. Goni S. Rahman M. Ahmed M. (2002). Antinociceptive activity of Polygonum hydropiper. Fitoterapia. 73, 704–706. 10.1016/S0367-326X(02)00239-3
37
Răut I. Călin M. Capră L. Gurban A. M. Doni M. Radu N. et al . (2021). Cladosporium sp. isolate as fungal plant growth promoting agent. Agronomy11, 392. 10.3390/agronomy11020392
38
Ribeiro B. A. da Mata T. B. Canuto G. A. Silva E. O. (2021). Chemical diversity of secondary metabolites produced by Brazilian endophytic fungi. Curr. Microbiol. 78, 33–54. 10.1007/s00284-020-02264-0
39
Salvatore M. M. Andolfi A. Nicoletti R. (2021). The genus Cladosporium: a rich source of diverse and bioactive natural compounds. Molecules26, 3959. 10.3390/molecules26133959
40
Shahed-Al-Mahmud M. Lina S. M. M. (2017). Evaluation of sedative and anxiolytic activities of methanol extract of leaves of Persicaria hydropiper in mice. Clin. Phytosci.3, 1–12. 10.1186/s40816-017-0056-5
41
Shannon P. Markiel A. Ozier O. Baliga N. S. Wang J. T. Ramage D. et al . (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504. 10.1101/gr.1239303
42
Sharif S. Ahmed T. Haque M. A. Bhuiyan M. Shahriar M. (2014). Phytochemical screenings, thrombolytic activity, membrane stabilizing activity and cytotoxic properties of Polygonum hydropiper. Res. J. Med. Plant.8, 92–98. 10.3923/rjmp.2014.92.98
43
Song W. L. Dai C. C. Liu X. Z. Cai X. Z. (2010). The effect of different endophytic fungi on Chrysanthemum morifolium output and quality. J. Chin. Med. Mater. 33, 4–7.
44
Sun L. T. Chen Y. Yang H. X. Li Z. H. Liu J. K. Wang G. K. et al . (2020). Bisabolane sesquiterpenes and α-pyrone derivative from endophytic fungus Zopfiella sp. Phytochem. Lett.37, 29–32. 10.1016/j.phytol.2020.03.008
45
Tang Z. Wang Y. Yang J. Xiao Y. Cai Y. Wan Y. et al . (2020). Isolation and identification of flavonoid-producing endophytic fungi from medicinal plant Conyza blinii H. Lév that exhibit higher antioxidant and antibacterial activities. PeerJ. 8, e8978. 10.7717/peerj.8978
46
Tian Y. Amand S. Buisson D. Kunz C. Hachette F. Dupont J. et al . (2014). The fungal leaf endophyte Paraconiothyrium variabile specifically metabolizes the host-plant metabolome for its own benefit. Phytochemistry108, 95–101. 10.1016/j.phytochem.2014.09.021
47
Ullah C. Unsicker S. B. Reichelt M. Gershenzon J. Hammerbacher A. (2019). Accumulation of catechin and proanthocyanidins in black poplar stems after infection by Plectosphaerella populi: hormonal regulation, biosynthesis and antifungal activity. Front. Plant Sci.10, 1441. 10.3389/fpls.2019.01441
48
Vergara C. Araujo K. E. Urquiaga S. Santa-Catarina C. Schultz N. da Silva Araújo E. et al . (2018). Dark septate endophytic fungi increase green manure-15N recovery efficiency, N contents, and micronutrients in rice grains. Front. Plant Sci. 9, 613. 10.3389/fpls.2018.00613
49
Wang F. Jiao R. Cheng A. Tan S. Song Y. (2007). Antimicrobial potentials of endophytic fungi residing in Quercus variabilis and brefeldin A obtained from Cladosporium sp. World J. Microbiol. Biotechnol.23, 79–83. 10.1007/s11274-006-9195-4
50
Waqas M. Khan A. L. Kamran M. Hamayun M. Kang S. M. Kim Y. H. et al . (2012). Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules17, 10754–10773. 10.3390/molecules170910754
51
Wei Y. H. Yu L. Z. Junru L. (2021). Effects of 4 different TCM monomers on TNF-α, IL-1β, IL-6 levels and PKR/p-PKR in norovirus infectious enteritis mice. J. Liaoning Univ. TCM23, 24–27.
52
Wickham H. (2011). ggplot2. Wiley Interdiscip. Rev. Comput. Stat.3, 180–185. 10.1002/wics.147
53
Xiang L. Ming L. (2020). Progression of pharmacological effects of Polygonum hydropipe L. Int. J. Tradit. Chin. Med. 40, 196–198.
54
Xiao Y. Liang W. Zhang Z. Wang Y. Zhang S. Liu J. et al . (2022). Polyketide derivatives from the endophytic fungus Phaeosphaeria sp. LF5 isolated from huperzia serrata and their acetylcholinesterase inhibitory activities. J. Fungi8, 232. 10.3390/jof8030232
55
Xu Z. L. Yan D. J. Tan X. M. Niu S. B. Yu M. Sun B. D. et al . (2022). Phaeosphspirone (1/1′), a pair of unique polyketide enantiomers with an unusual 6/5/5/6 tetracyclic ring from the desert plant endophytic fungus Phaeosphaeriaceae sp. Phytochemistry194, 112969. 10.1016/j.phytochem.2021.112969
56
You F. Han T. Wu J. z Huang B. k. et al . (2009). Antifungal secondary metabolites from endophytic Verticillium sp. Biochem. Syst. Ecol.37, 162–165. 10.1016/j.bse.2009.03.008
57
Yue Y. (2005). Ameliorating effect of green tea catechin on inflammatory bowel disease caused by trinitrobenzenesulfonic acid. For. Med. Sci.27, 314–315.
58
Zhang K. Bonito G. Hsu C. M. Hameed K. Vilgalys R. Liao H. L. (2020). Mortierella elongata increases plant biomass among non-leguminous crop species. Agronomy10, 754. 10.3390/agronomy10050754
59
Zhang X. R. Ren Y. Y. Zeng Y. J. Li T. L. Hong Y. L. Ren C. Y. et al . (2021). Characterization of the bioactive compounds with efficacy against enteritis in Polygonum hydropiper L. by UHPLC-Q-Orbitrap HRMS combined with network pharmacological analysis. ChemistrySelect.6, 10336–47. 10.1002/slct.202102886
Summary
Keywords
Polygonum hydropiper L., endophytic fungi, community assembly process, flavonoids, core community
Citation
Zhang X, Lv H, Tian M, Dong Z, Fu Q, Sun J, Huang Q and Wang J (2022) Colonization characteristics of fungi in Polygonum hydropipe L. and Polygonum lapathifolium L. and its effect on the content of active ingredients. Front. Plant Sci. 13:984483. doi: 10.3389/fpls.2022.984483
Received
02 July 2022
Accepted
18 August 2022
Published
28 September 2022
Volume
13 - 2022
Edited by
Waheed Akram, University of the Punjab, Pakistan
Reviewed by
Shihong Luo, Shenyang Agricultural University, China; Bartholomew Saanu Adeleke, Ondo State University of Science and Technology, Nigeria
Updates
Copyright
© 2022 Zhang, Lv, Tian, Dong, Fu, Sun, Huang and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinwan Huang huangqinwan@cdutcm.edu.cnJin Wang wangjin0816@126.com
†These authors share first authorship
This article was submitted to Plant Symbiotic Interactions, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.