- College of Environmental Science and Engineering, Guilin University of Technology, Guilin, China
Thiocyanate (SCN−) can find its way into cultivated fields, which might hamper the harmony in carbon and nitrogen metabolism (CNM) of plants, ebbing their quality and productivity. In the current study, we investigated the role of the exogenous application of 2-oxoglutarate (2-OG) in maintaining homeostasis of CNM in rice seedlings under SCN− stress. Results showed that SCN− exposure significantly repressed the gene expression and activities of CNM-related enzymes (e.g., phosphoenolpyruvate carboxylase, NADP-dependent isocitrate dehydrogenases, and isocitrate dehydrogenases) in rice seedlings, thereby reducing their relative growth rate (RGR). Exogenous application of 2-OG effectively mitigated the toxic effects of SCN− on rice seedlings, judged by the aforementioned parameters. The co-expression network analysis showed that genes activated in CNM pathways were categorized into four modules (Modules 1–4). In order to identify the key module activated in CNM in rice seedlings exposed to SCN−, the results from real-time quantitative PCR (RT-qPCR) tests were used to calculate the possibility of the occurrence of genes grouped in four different modules. Notably, Module 3 showed the highest occurrence probability, which is mainly related to N metabolism and 2-OG synthesis. We can conclude that exogenous application of 2-OG can modify the imbalance of CNM caused by SCN− exposure through regulating N metabolism and 2-OG synthesis in rice seedlings.
1. Introduction
Carbon (C) and nitrogen (N) are the two primary nutrient species, and their adequate supply and dynamic balance of both elements should be essential for regulating cellular functions during plant growth and development (Zheng, 2009; Naseeruddin et al., 2018). It is well known that C-rich biomolecules (e.g., sucrose, glucose, and fructose) provide the majority of energy and C-skeletons for ammonium (NH4+) assimilation, while N-containing compounds are parts of organic (e.g., amino acids and proteins) and simple inorganic compounds (e.g., nitrate [NO3−] and NH4+), which can be synthesized through the incorporation of NH4+ into the C-skeletons (Zheng, 2009). At the enzymatic level, nitrate reductase (NR), glutamine synthetase (GS), sucrose-phosphate synthase (SPS), trehalose-6-phosphate synthase (TPS), and glutamyl tRNA synthetase (ERS) play a dominant role in regulating the carbon and nitrogen metabolism (CNM) in plants (Coruzzi and Zhou, 2001). However, various environmental stimuli, such as pollutants, drought, salinity, fertilization, and extreme temperature, can influence and destabilize CNM-associated enzymes, thereby weakening the yield and quality of crops (Xin et al., 2019; Alves et al., 2021; Guo et al., 2021).
Thiocyanate (SCN−), being part and parcel of many industrial activities (e.g., manufacturing of chemical insecticide and herbicide, production of thiourea, metal separation, and gold mining), is continuously marking its imprint in a clean environment (Yu and Zhang, 2013). Notably, gold ore processing generates a large amount of SCN− because of the lixiviant cyanide complexed with the reduced sulfur species in the gold-bearing ore (Gao et al., 2022). Even mine waste is treated before being discharged, with the aim to convert cyanide into SCN− (Gould et al., 2012). Different governing bodies have issued standards regarding the discharge of cyanide-rich effluent in the environment considering its environmental risk and health hazard (Mudder and Botz, 2004); however, discharge of SCN− in effluent has not been restricted by standards, ultimately raising SCN− level in the effluent (Gould et al., 2012). Indeed, the levels of SCN− at 1,000 mg SCN/L were detected in gold tailings wastewaters (Gao et al., 2022). Persistence of higher levels of SCN− in soils, sediments, rivers, and aquatic biota in nearby areas of gold mines has been observed, which eventually makes its entry into the food chain and poses a threat to all living organisms (Bhunia et al., 2000; Yu and Zhang, 2013; Sun et al., 2018; Lin et al., 2020). Indeed, accumulation of SCN− in plants can cause serious damage to plant growth and development by decreasing nutrient balance and transpiration, degrading photosynthetic pigments, changing the free amino acid composition, and inhibiting the activities of antioxidant enzymes (Hansson et al., 2008; Yu and Zhang, 2013). Our previous studies at the physio-biochemical and molecular levels also indicated that SCN− exposure is able to result in the dysfunction of chloroplast (Yang et al., 2021). These studies suggested that the negative effects of SCN− exposure on the CNM in rice seedlings are detectable.
In recent years, the application of plant growth regulators has been suggested to curtail the negative impact imposed by various abiotic factors (Yang et al., 2021). It is evident that 2-oxoglutarate (2-OG) is a key organic acid involved in the processes of CNM in plants (Yue et al., 2018; Ji et al., 2020). Specifically, the GS initially converts NH4+ into glutamine in an ATP-dependent reaction; afterward, the glutamate synthase (GOGAT) catalyzes the conversion of glutamine and 2-OG into two molecules of glutamate. Clearly, there is a mandatory interaction between N metabolism and C metabolism (Gálvez et al., 1999). In addition, exogenous application of 2-OG can enhance the activities of phosphoenolpyruvate carboxylase (PEPC), GS, and NADP-dependent isocitrate dehydrogenases (NADP-ICDH) in roots of rice (Yuan et al., 2007). Feeding of exogenous 2-OG can also improve the transcripts of N metabolism-related genes in plants (Araújo et al., 2014). These studies suggested the positive feedback of exogenous 2-OG on the CNM in plants. To date, little is known about the role of exogenous 2-OG in regulating the imbalance of CNM induced by SCN− in plants.
Rice (Oryza sativa L.) is one of the most important staple food crops worldwide, especially in eastern Asia countries (Mostofa et al., 2014). Nowadays, agricultural crops suffer from various environmental issues. The SCN− is a typical N-containing pollutant that can be assimilated by rice plants. Therefore, in the present study, we hypothesized that SCN− can disturb the CNM in rice plants during the detoxification of exogenous SCN−, while the application of exogenous 2-OG can maintain homeostasis of CNM in rice seedlings in response to SCN− exposure. To prove this hypothesis, the following works were performed: 1) we estimated the relative growth rate and percentage of carbon/nitrogen of rice seedlings under SCN− exposure with or without exogenous 2-OG, 2) we analyzed the effects of exogenous 2-OG on CNM-related genes and enzymes under SCN− stress, and 3) we clarified the strategies of exogenous 2-OG to regulate the imbalance of CNM in rice plants under SCN− exposure by merging the occurrence possibility into a co-expression module analysis. Overall, this study provides new evidence to expand our understanding of exogenous 2-OG to regulate the imbalance of CNM in rice plants during SCN− exposure.
2. Methods and materials
2.1. Plant growth and experiment design
The seeds of a regular medium-maturing indica rice (O. sativa L. XZX 45) were sowed in river sand after being soaked in distilled water for 24 h and then moved inside an artificial climate chamber with a controlled temperature of 25°C ± 0.5°C at a relative humidity of 60% ± 2% Zhang et al., 2022. The rice seedlings were irrigated daily with a modified 8692 nutrient solution, which was described in our previous work (Yang et al., 2021). The modified 8692 nutrient solution with KNO3 (39.5 mg N/L) was used. After 16-day growth, rice seedlings of similar size were collected and incubated in a MES-Tris solution (pH = 6.0) for 4 h to remove additional ions from the root surface and the apparent free space. These pretreated seedlings were transferred into a nutrient solution spiked with SCN− and utilized in subsequent experiments. Two treatment series were conducted:
(1) SCN− treatments: SCN− spiked solutions at concentrations of 0 (control 1), 24.0, 96.0, and 300.0 mg SCN/L. Control 1 refers to the nutrient solution without SCN− and exogenous 2-OG.
(2) “SCN− + 2-OG” treatments: seedlings were pretreated with a 2-OG solution at a concentration of 4 mmol/L for 4 h (Fritz et al., 2006), and then seedlings were exposed to SCN− solution at 0 (control 2), 24.0, 96.0, and 300.0 mg SCN/L. Control 2 refers to the nutrient solution without SCN−, but with exogenous 2-OG.
Exposure concentrations of SCN− used were based on three different effective concentrations (ECs), i.e., EC20, EC50, and EC75, referring to the 20%, 50%, and 75% inhibition of relative growth rates of rice seedlings, respectively (Lin et al., 2020). All seedlings were placed in the plant growth chamber for a 3-day exposure. Potassium thiocyanate (KSCN) of analytical grade purity with 98.5% purity was purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). α-Ketoglutaric acid (2-OG) of analytical grade purity with 98.0% purity was obtained from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). To minimize water loss and prevent algae growth, each flask was covered with aluminum foil. Each treatment concentration was performed with four independent replicates.
2.2. Analysis of growth parameter
The relative growth rate (RGR) is one of the most crucial parameters to reflect the performance of plants under various stresses (Lin et al., 2020). The RGR (%) was calculated using the biomass change of young seedlings during SCN− exposure, as follows:
where W(I) and W(F) are the initial and final fresh weights of rice seedlings, respectively.
2.3. Measurement of total C and N contents in rice tissues
After exposure to the SCN− solution for 3 days, rice seedlings were harvested and separated into roots and shoots. After being washed with double-distilled water, plant materials were oven dried at 90°C for 48 h and weighed. Then, 0.010 g of oven-dried plant materials was grained into fine powder. Total C and N (%) were measured by a vario elemental analyzer (vario EL) (Brown et al., 2007).
2.4. Measurements of activities of CNM-related enzymes in rice tissues
Activities of enzymes related to C metabolism, including PEPC (Osuna et al., 1996),ERS (Ratinaud et al., 1983), TPS (Goddijn et al., 1997), and SPS (Feng et al., 2019) in rice tissues were assayed (detailed information is shown in Supplementary material M1).
Activities of enzymes activated in N metabolism, namely, NR (Ahanger et al., 2021), nitrite reductase (NiR) (Lin et al., 2022a), and GS (Hou et al., 2019) in rice tissues were determined (detailed information is shown in Supplementary material M1).
Activities of enzymes involved in 2-OG biosynthesis, i.e., NADP-ICDH (Gálvez et al., 1994), isocitrate dehydrogenases (NAD-IDH) (Gálvez et al., 1994), and glutamate dehydrogenases (GDH) (Turano et al., 1996), were also measured (detailed information is shown in Supplementary material M1).
2.5. RNA extraction and RT-qPCR analysis
Real-time quantitative PCR (RT-qPCR) was used to quantify the expression levels of CNM-related enzymes in rice seedlings after SCN− exposure. Total RNA was extracted from both the root and shoot of all rice samples by using an Ultrapure RNA Kit (CWBio, Taizhou, China). DNase I (CWBio, Taizhou, China) was used to remove genomic DNA contamination, if any, from RNA extract. Then, the total RNA was purified by an RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). Each sample was prepared in four independent biological replicates.
A total of 40 genes encoding enzymes or proteins activated in the CNM pathways were searched from the databases, including RGAP (http://rice.plantbiology.msu.edu/analyses_search_blast.shtml), NCBI (https://www.ncbi.nlm.nih.gov/), and RAPDB (http://rapdb.dna.affrc.go.jp/). Expression of genes was assayed after SCN− exposure by RT-qPCR analysis, including PEPC (Osppc1, Osppc2a, Osppc3, and Osppc4), ERS (OsERS1, OsERS2, and OsERS3), TPS (OsTPS1, OsTPS4, OsTPS5, OsTPS8, and OsTPS9), SPS (OsSPS1, OsSPS2, OsSPS4, OsSPS5, and OsSPS6), NR (OsNIA1, OsNIA2, and OsNR1), NiR (OsNiR1, OsNiR2, and OsNiR3), GS (OsGS1;1, OsGS1;2, OsGS1;3, and OsGS2), NADP-ICDH (OsICDH1, OsICDH2, OsICDH3, and OsICDH4), NAD-IDH (OsIDHc;2, OsIDHc;1, OsIDHa, and OsIDH1), and GDH (OsGDH1, OsGDH2, OsGDH3, and OsGDH4). All genes primer sequences are listed in Table S1. RT-qPCR cycling conditions were as follows: 1) denaturation at 95°C for 10 s, 2) annealing at 58°C for 30 s, and 3) extension at 72°C for 32 s. This cycle was imitated 40 times. The RT-qPCR analysis was executed using the 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) and SYBR green chemistry. Rice GAPDH (glyceraldehyde-3-phosphate dehydrogenase, LOC_Os08g03290.1) was selected as the housekeeping gene (Yang et al., 2021). The standard 2−ΔΔCT method was used to calculate the relative expression of each of the targeted genes (Schmittgen and Livak, 2008). All values were represented as cumulative means ± standard deviation of four independent replicates.
2.6. Identification of key regulatory genes in the CNM regulatory module
2.6.1. Co-expression network analysis
In order to establish the CNM regulatory module with statistical significance, all CNM-related genes were uploaded to the STRING (https://version-10-5.string-db.org/) software, and the protein–protein interaction (PPI) networks (combined score >0.4) were constructed. Then, the modules (resolution = 0.8) with higher visualization were performed by the program Gephi 0.9.2 (Lin et al., 2022b).
2.6.2. Estimation of the normcdf of CNM-related genes
In order to identify the key module activated in CNM in rice seedlings exposed to SCN−, the results from PCR tests were used to calculate the possibility of the occurrence of genes grouped in four different modules. We first converted the data through the functions of “COMPUTE x_new=SQRT(X)” or “COMPUTE x_new = LN(x)” in the SPSS software since they were non-normally distributed. Then, the normcdf was calculated statistically.
2.7. Data analysis
Tukey’s multiple range tests were used to assess the statistical significance at the level of 0.01 or 0.05. Different letters refer to the significant difference between the treatments and control (p< 0.05). The asterisk symbol refers to the significant difference between SCN−-treated and “SCN− + 2-OG”-treated seedlings (p< 0.05).
3. Results
3.1. Relative growth rate of rice seedlings
A remarkable (p< 0.05) reduction in RGR of rice seedlings was observed at all SCN− treatments after 3-day exposure in comparison to the control (Figure 1A). Similarly, in the case of “SCN− + 2-OG” treatments in rice seedlings, a decrease in RGR that was visible in all treated plants reversed to that of control (p< 0.05). However, the RGR of rice seedlings under “SCN− + 2-OG” treatments was significantly (p< 0.05) higher than that of SCN− treatments, suggesting that the inoculation of 2-OG could mitigate the negative effect of SCN− on plant biomass growth.
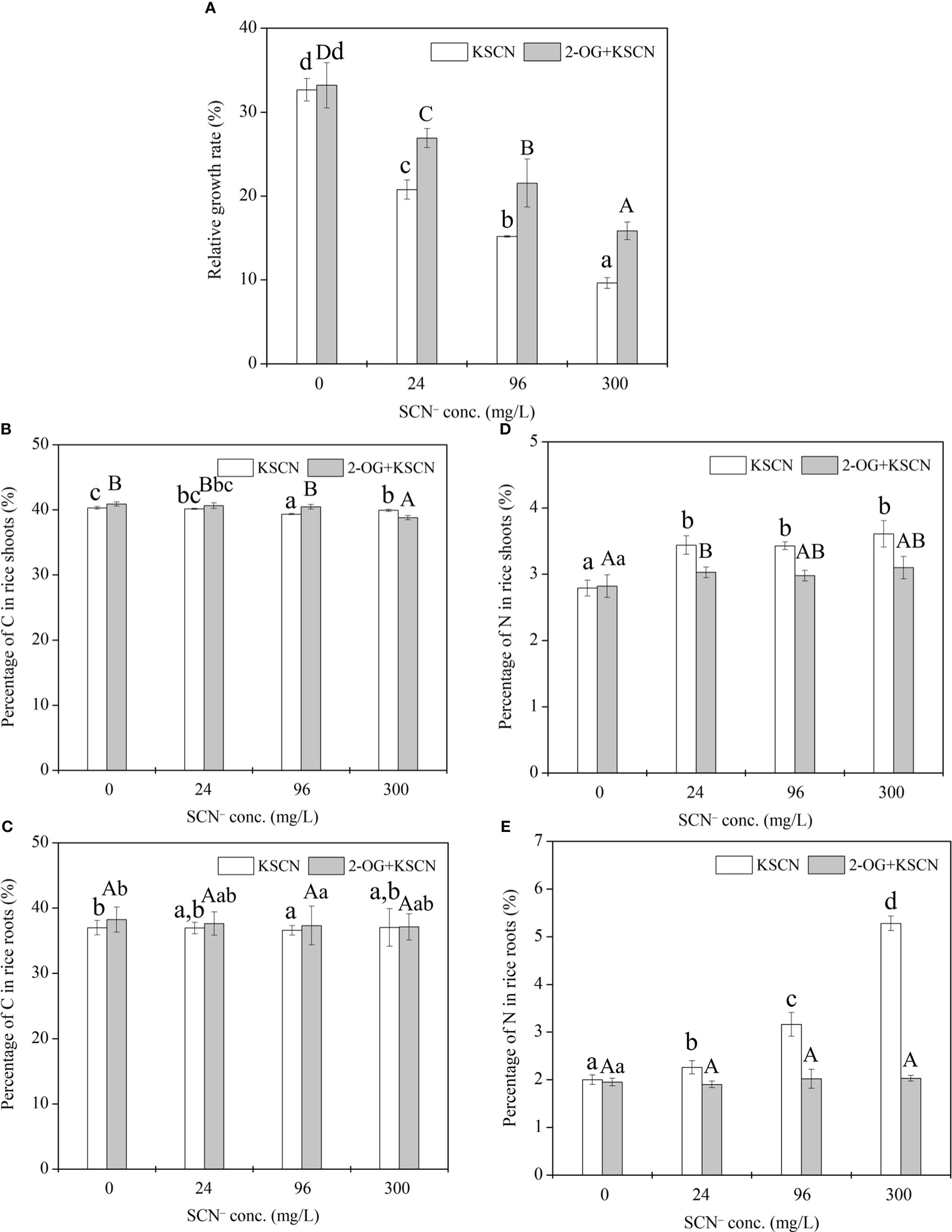
Figure 1 (A) Relative growth rate of rice seedlings under SCN− exposure in the presence or absence of 2-OG. (B) The percentage of C in rice shoots. (C) The percentage of C in rice roots. (D) The percentage of N in rice shoots. (E) The percentage of N in rice roots. Values are the mean of four independent biological replicates ± standard deviation. Different letters refer to the significant difference between treatment and control (p< 0.05). 2-OG, 2-oxoglutarate.
3.2. The total amount of C and N in rice seedlings
The C% in rice roots (shoots) was 36.99%, 36.94%, 36.58%, and 37.04% (40.31%, 40.15%, 39.35%, and 39.93%, respectively) under 0, 24, 96, and 300 mg SCN/L treatments, respectively. The application of exogenous 2-OG caused a negligible effect on the C% in rice tissues compared with their respective SCN− treatments (Figures 1B, C). The N% in rice roots (shoots) was 2.0%, 2.26%, 3.16%, and 5.28% (2.79%, 3.44%, 3.43%, and 3.61%, respectively) under 0, 24, 96, and 300 mg SCN/L treatments, respectively, while application of exogenous 2-OG significantly decreased the N% in rice tissues compared with their respective SCN− treatments (Figures 1D, E).
3.3. Response of CNM-related genes in rice plants
Mostly, more than one isogene was encoded with the specific enzyme in plants, in which the activity of the enzyme was regulated and/or governed by these isogenes together. However, each specific isogene does not carry the same weight during the regulation process, wherein there is always a master regulator gene (Yang et al., 2021), which chiefly controls the enzyme activity. Here, the upregulated master regulator genes in rice seedlings were described, while downregulated genes are not described in the following sections.
3.3.1. Response of C metabolism-related genes
As shown in Figure 2A, PEPC, upregulated genes in roots were Osppc4, Osppc2a, and Osppc2b, at all SCN− treatments, while Osppc1, Osppc2a, and Osppc2b were upregulated in shoots. In roots of rice seedlings from the “SCN− + 2-OG” treatments, Osppc4 and Osppc3 were upregulated, while Osppc4, Osppc1, and Osppc2a showed remarkable expression in shoots.
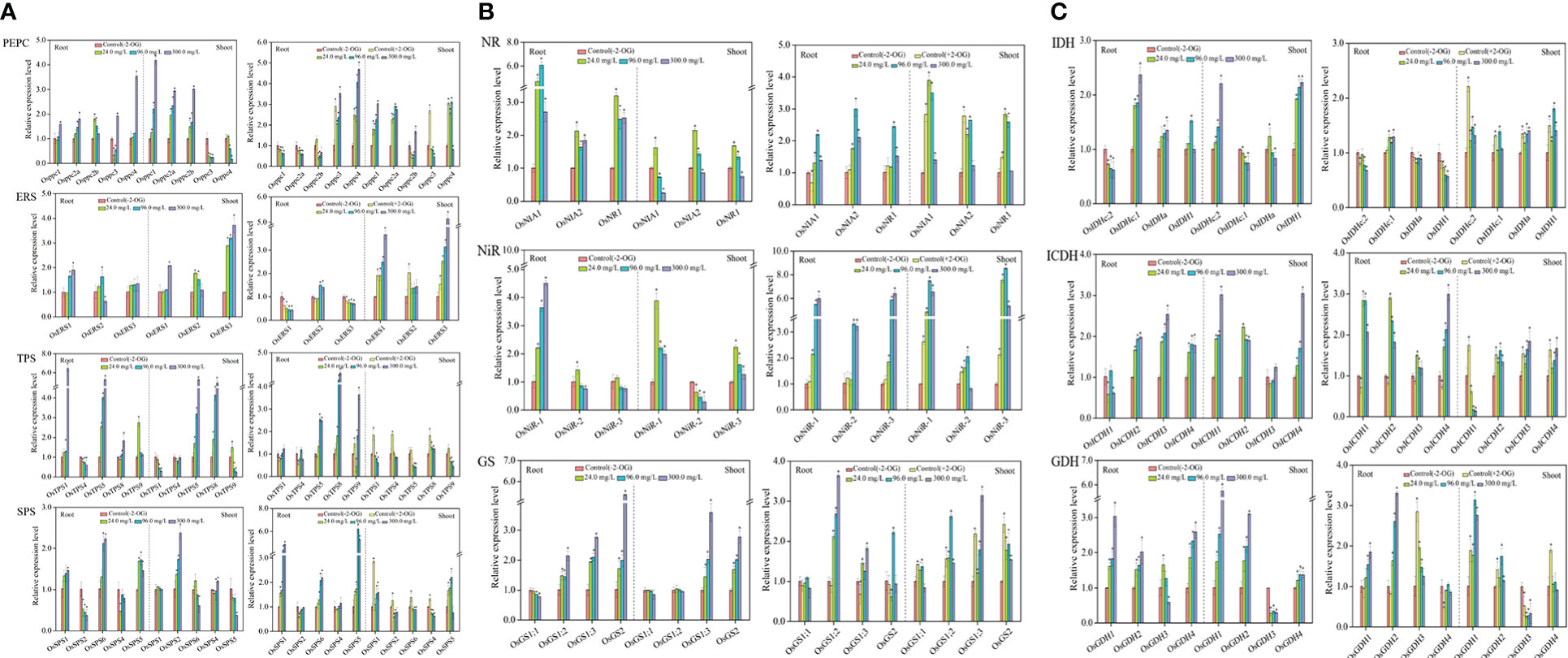
Figure 2 Response of CNM-related genes in rice roots and shoots under SCN− stress in the presence or absence of 2-OG. (A) Response of C metabolism-related genes. (B) Response of N metabolism-related genes. (C) 2-OG synthesis-related genes. The asterisk symbol refers to the significant difference between SCN−-treated (24.0, 96.0, and 300.0 mg SCN/L) and SCN−+2-OG-treated (0, 24.0, 96.0, and 300.0 mg SCN/L) seedlings and control 1 (p< 0.05). CNM, carbon and nitrogen metabolism; 2-OG, 2-oxoglutarate.
Expression of ERS-related genes is shown in Figure 2A, ERS, wherein OsERS1, OsERS2, and OsERS3 were generally upregulated in SCN−-exposed rice parts, i.e., in roots and shoots. However, the expression levels of OsERS1, OsERS2, and OsERS3 showed raised pattern in shoots of rice plants from the “SCN− + 2-OG” treatments.
Figure 2A, TPS, depicts that higher expression levels of TPS-related genes of OSTPS5 and OSTPS1 were prominent in roots in SCN− treatments, while OSTPS5 and OSTPS8 had greater expression in shoots. Interestingly, when rice seedlings were pretreated with 2-OG, OSTPS5, OSTPS8, and OSTPS9 were upregulated in roots. However, the expression levels of five TPS isogenes in shoots conferred a variance tendency, which reinforced at 0 mg SCN/L and then declined at 24 mg SCN/L.
Upregulation of three SPS isogenes (OsSPS1, OsSPS3, and OsSPS5) was observed in roots in all SCN− treatments (Figure 2A, SPS), while only OsSPS2 was upregulated in shoots. Differential expression patterns were found in SPS-related genes of roots in “SCN− + 2-OG” treatments. The expression levels of OsSPS1, OsSPS3, and OsSPS5 in roots from the “SCN− + 2-OG” treatments were higher than those of SCN− treatments. OsSPS1 and OsSPS5 were upregulated in shoots of “SCN− + 2-OG” treatments, which differed from SCN− treatments.
3.3.2. Response of N metabolism relative genes
Positive expressions of NR genes, i.e., OsNIA1, OsNIA2, and OsNR1, were observed in roots after SCN− exposure (Figure 2B, NR). However, the expression levels of OsNIA1, OsNIA2, and OsNR1 in shoots showed a disparate trend with an initial escalation from 24 mg SCN/L and then dropped at 96 mg SCN/L. In the case of rice seedlings from the “SCN− + 2-OG” treatments, higher expression levels of OsNIA1, OsNIA2, and OsNR1 were observed in rice shoots.
Within NiR genes, only OsNiR-1 was upregulated in roots at all SCN− treatments (Figure 2B, NiR), while upregulation of OsNiR-1 and OsNiR-3 was detected in shoots. Interestingly, the expression levels of OsNiR-1, OsNiR-2, and OsNiR-3 conferred an accelerating pattern in both roots and shoots of the “SCN− + 2-OG” treatments.
Differential expression of GS-related genes was observed between roots and shoots, with significant upregulation of OsGS2, OsGS1;2, and OsGS1;3 in roots and shoots (OsGS2 and OsGS1;3) (Figure 2B, GS). However, the expression levels of OsGS2, OsGS1;2, OsGS1;1, and OsGS1;3 showed linear inclination with increasing SCN− concentrations in both rice tissues by inoculation of 2-OG.
3.3.3. Genes involved in the biosynthesis of 2-OG
Transcriptional changes of NAD-IDH genes are shown in Figure 2C, IDH. As apparent from the figures, OsIDHa and OsIDHc;1 were upregulated in roots, while OsIDHc;2 and OsIDH1 were overexpressed in shoots. When rice seedlings were pretreated with 2-OG, upregulation of OsIDHc;1 was observed in roots, and OsIDHa, OsIDHc;2, and OsIDH1 were upregulated in shoots.
As presented in Figure 2C, ICDH, OsICDH2, OsICDH3, and OSICDH4 were significantly (p< 0.05) upregulated in roots after SCN− exposure, while positive expressions (p< 0.05) of OsICDH1, OsICDH2, and OsICDH4 were observed in shoots. Interestingly, OsICDH1, OsICDH2, and OsICDH4 in roots presented an upregulated pattern in the “SCN− + 2-OG” treatments and the expression levels of OsICDH2, OsICDH3, and OsICDH4 were remarkable in shoots.
Upregulation of OsGDH1, OsGDH2, and OsGDH4 was observed in both rice tissues after SCN− exposure (Figure 2C, GDH), while significantly (p< 0.05) higher expressions of OsGDH1, OsGDH2, and OsGDH3 were observed in roots, and significant upregulation of OsGDH1, OsGDH2, and OsGDH4 was detected in shoots of the “SCN− + 2-OG” treatments.
3.4. Response of CNM-related enzyme activities
3.4.1. Response of C metabolism-related enzyme activities
The activities of CNM-related enzymes were assayed in SCN− and “SCN− + 2-OG” treatment plants (Figure 3). The activity of PEPC in roots was affirmatively increased (p< 0.05) after SCN− exposure compared with the control, while the activity of PEPC in shoots presented a downward tendency. The activity of ERS in roots was significantly inhibited (p< 0.05) after SCN− exposure in comparison with the control, while the activity of ERS was increased in shoots. Activities of TSP and SPS presented significant increasing patterns in both roots and shoots in the presence of SCN− stress (p< 0.05). Under “SCN− + 2-OG” treatments, activities of PEPC, ERS, TSP, and SPS intensified in roots compared to control (p > 0.05), while activities of ERS, TSP, and SPS showed a decrement in shoots unlike control (p > 0.05).
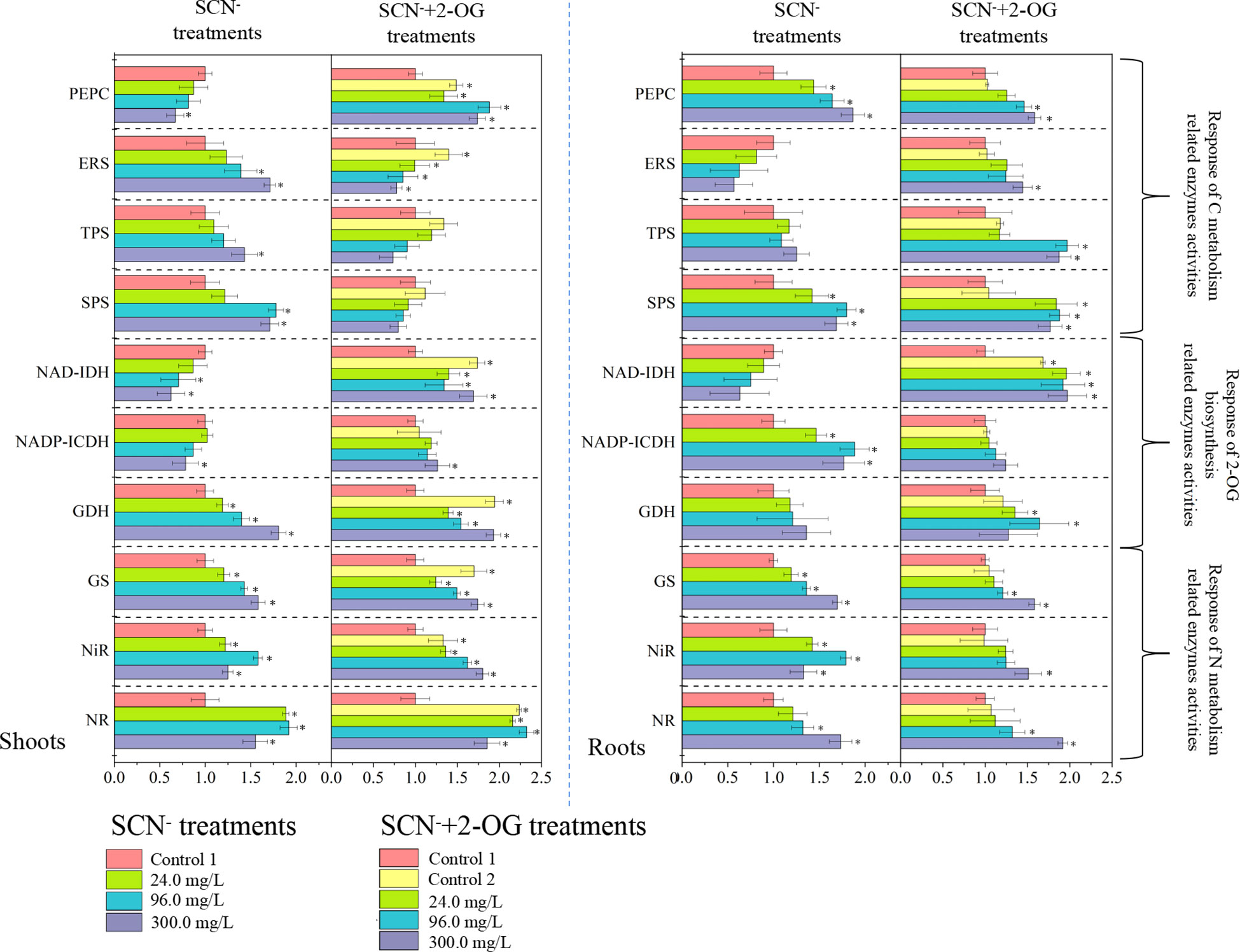
Figure 3 Response of CNM-related enzyme activities in rice roots and shoots under SCN− stress in the presence or absence of 2-OG. The asterisk symbol refers to the significant difference between SCN−-treated (24.0, 96.0, and 300.0 mg SCN/L) and SCN−+2-OG-treated (0, 24.0, 96.0, and 300.0 mg SCN/L) seedlings and control 1 (p< 0.05). CNM, carbon and nitrogen metabolism; 2-OG, 2-oxoglutarate.
3.4.2. Response of N metabolism-related enzyme activities
Activities of NR and NiR in roots presented an exponential emulate compared with the control (p< 0.05), while activities of NR and NiR in shoots illustrated an inverted “U” shape curve under SCN− stress (Figure 3). The analogous activity of GS was noticed in both roots and shoots of SCN− stress (p< 0.05), following linearity with stress concentration. Under “SCN− + 2-OG” treatments, activities of NR, NiR, and GS showed a similar pattern of increment in both roots and shoots (p< 0.05).
Overall, the activities of all selected enzymes in “SCN− + 2-OG” treatments were generally higher than those of SCN− treatments. Exogenous 2-OG had a pronounced impact on enzyme activities in shoots, unlike its counterpart. To reveal the regulation mechanism of exogenous 2-OG on the CNM in rice plants under SCN− exposure, we distinguished the effects of exogenous 2-OG on C and N metabolism in rice tissues.
3.4.3. Response of 2-OG biosynthesis-related enzyme activities
The activity of NAD-IDH was inhibited significantly (p< 0.05) in both roots and shoots under SCN− exposure compared with the control (Figure 3). The activity of NADP-ICDH in SCN−-exposed roots was prominently enhanced; nevertheless, the scenario was reversed in the case of shoots (p< 0.05). Elevated activity of GDH was observed in both roots and shoots in the presence of SCN− (p< 0.05). Under “SCN− + 2-OG” treatments, the activities of NAD-IDH, NADP-ICDH, and GDH were generally increased in rice tissues, except for GDH in shoots of rice seedlings. These results indicated that the modification mechanism of exogenous 2-OG on CNM-related enzymes in rice seedlings under SCN− exposure varied greatly.
3.5. Identification of key regulatory genes in the CNM regulatory module
3.5.1. Co-expression analysis of CNM-related genes
Plants have evolved the coordinated actions responsible for their diverse physiological processes via either direct or indirect gene connections. In order to elucidate the functional module of genes activated in the CNM process, a co-expression network was performed by the STRING program, and four modules were obtained. Detailed information on gene interaction strengths in these four modules is given in Table S2. Interestingly, all modules had similar interaction contributions, namely Module 1 (25.0%), Module 2 (25.0%), Module 3 (27.5%), and Module 4 (22.5%) (Figure 4A). We also noticed that the genes grouped in Modules 1 and 2 were involved in C metabolism (11 genes) and biosynthesis of 2-OG (10 genes), respectively; genes categorized in Module 3 were responsible for N metabolism (five genes) and biosynthesis of 2-OG (five genes). In addition, genes grouped in Module 4 were activated in C metabolism (one gene) and N metabolism (five genes) and biosynthesis of 2-OG (three genes) (Figure 4B).
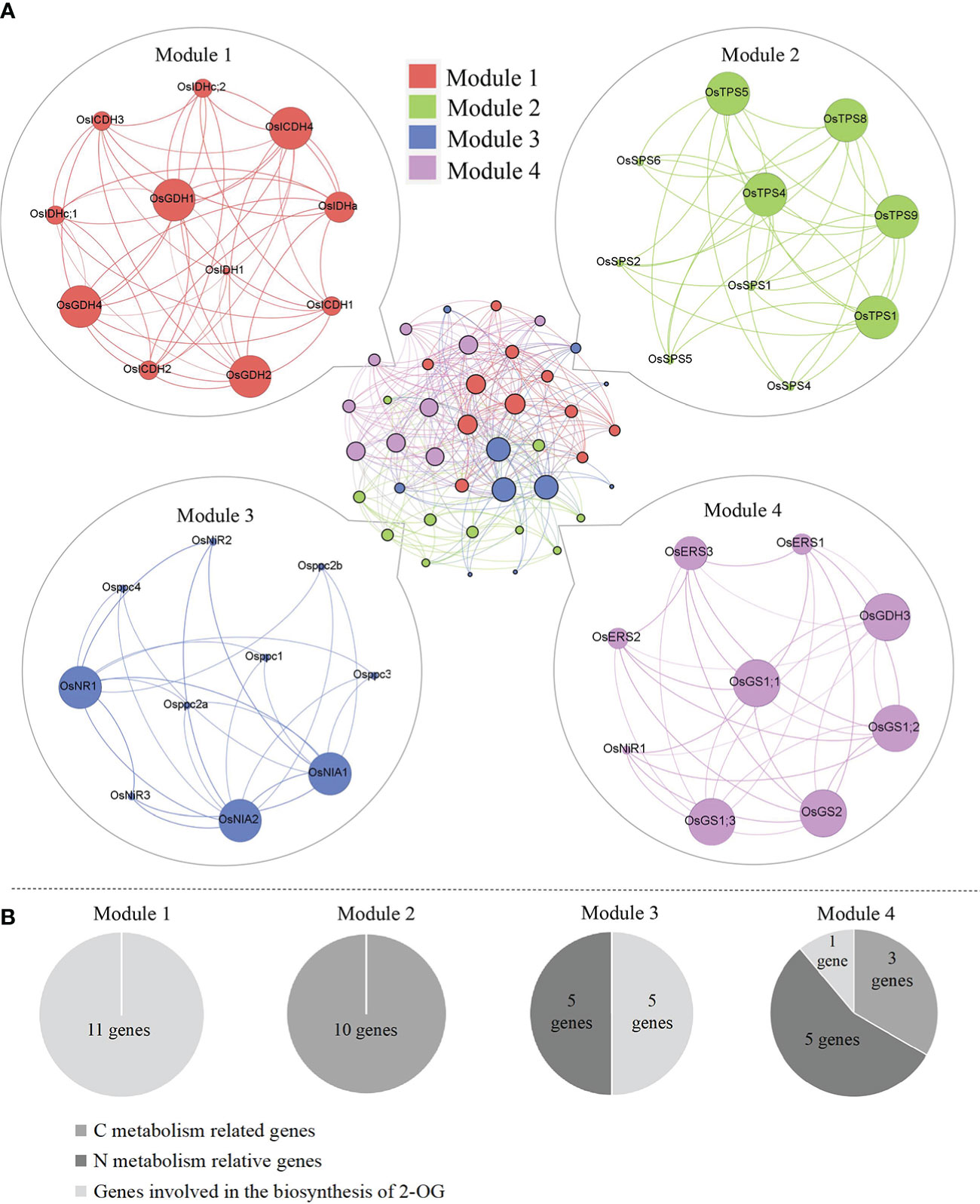
Figure 4 (A) Genetic matrix integrative analysis of CNM-related genes. (B) The number of CNM genes in each matrix. CNM, carbon, and nitrogen metabolism.
3.5.2. The occurrence probability of CNM-related genes
The normcdf of rice shoots was quite different between under SCN− and SCN−+2-OG treatments, based on the non-linear regression (Figure 5). Herein, the threshold for the highest occurrence probability was set, p > 0.75. Therefore, Module 3 showed the highest occurrence probability, suggesting that the exogenous application of 2-OG mainly regulated the expression of genes activated in the N metabolism and 2-OG synthesis to modify the imbalance of CNM in rice plants imposed by SCN− exposure. In fact, a similar conclusion was also reached in the analysis of C and N fractions in rice tissues. We noticed that the change of C fraction in rice shoots was almost constant (Figures 1B, C), while the change of N in rice shoots was evident between SCN− treatments and “SCN− + 2-OG” treatments (Figures 1D, E).
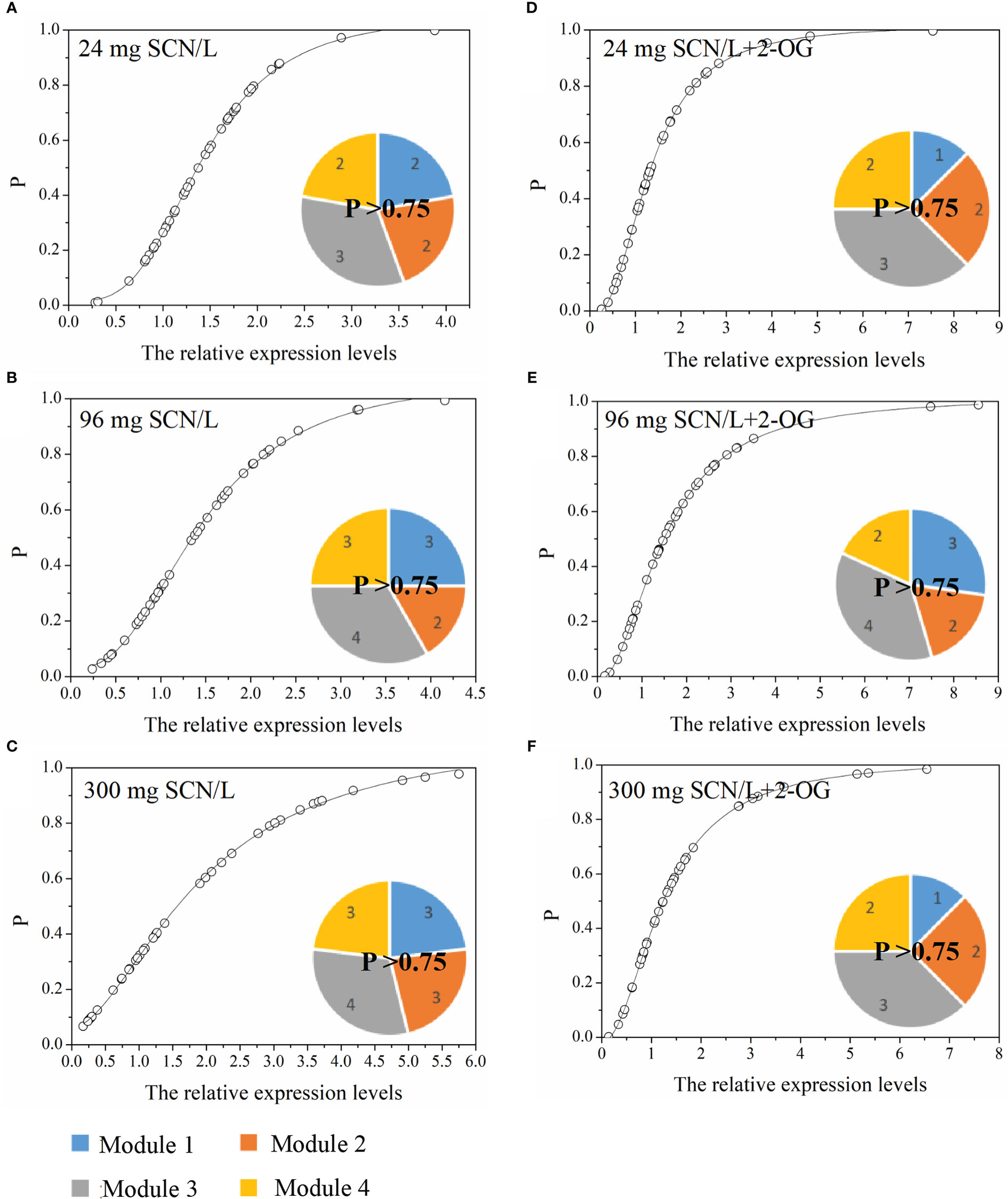
Figure 5 Occurrence P (p > 0.75) of CNM-related genetic expression in rice shoots under (A) 24 mg SCN/L, (B) 96 mg SCN/L, and (C) 300 mg SCN/L treatments and under (D) 24 mg SCN/L + 2-OG, (E) 96 mg SCN/L + 2-OG, and (F) 300 mg SCN/L + 2-OG treatments. CNM, carbon and nitrogen metabolism; 2-OG, 2-oxoglutarate.
4. Discussion
4.1. Exogenous 2-OG promotes plant growth via regulating CNM during SCN− exposure
The growth and development of plants are tightly coordinated with the balance of cellular CNM (Zheng, 2009). Once plants suffer from environmental stresses, the CNM in plants could be disrupted, thereby causing an imbalance of CNM and eventually resulting in a reduction in plant growth (Reddy et al., 2004). In this current study, the imbalance of CNM in rice seedlings due to SCN− exposure was judged by the relative growth rate (Figure 1A), in which SCN− exposure led to a significant reduction in RGR of rice seedlings (p< 0.05), indicating a severe impact on the balance of CNM in rice seedlings under SCN− stress. Also, we found that SCN− exposure significantly affected the percentage of N in rice seedlings (Figures 1D, E). This is because SCN− exposure can result in the dysfunction of chloroplast (Yang et al., 2021) and repress the activities of NR, GS, and glutamate synthase (GOGAT) in rice seedlings (Lin et al., 2022a). In addition, SCN−-treated rice seedlings with 2-OG supplied had significantly (p< 0.05) higher RGR than those without exogenous 2-OG, suggesting a positive effect of exogenous 2-OG on the RGR of rice seedlings corresponding to SCN− exposure. It is known that 2-OG is a decisive chemical involved in the homeostasis of CNM in higher plants (Yuan et al., 2007). Also, exogenous 2-OG enhances photosynthesis and increases the levels of C-skeletons in rice plants, thus affecting the N metabolism (Yuan et al., 2007; Zheng, 2009). Therefore, the imbalance of CNM in rice seedlings due to SCN− exposure could be positively modified by exogenous 2-OG.
4.2. Modification of 2-OG in balancing CNM in SCN−-treated rice plants
Herein, the main N source present in the nutrient solution to support plant growth is NO3−. In plants, only a small fraction of NO3− is assimilated in roots after uptake, and a greater part is translocated into shoots and assimilated into NH4+ and amino acids. Photosynthesis in the chloroplast is a major process for C metabolism in plants during their entire period of life (Lemaitre et al., 2007). Therefore, modification of 2-OG on CNM-related genes and enzyme activities in rice shoots will be discussed accordingly.
4.2.1. Effects of SCN− on innate 2-OG synthesis
It is known that there are three innate routines for the biosynthesis of 2-OG in plants, in which the PEPC pathway and the NADP-ICDH/NAD-IDH pathway belong to C metabolism, and the GDH pathway is mainly involved in N metabolism. In this study, we focused on correlating and perceiving the most competent pathway in controlling the generation of 2-OG and regulating the imbalance of CNM in rice seedlings caused by SCN− exposure.
The PEPC pathway in plants is an anaplerotic reaction to replenish the tricarboxylic acid (TCA) cycle with intermediates that are withdrawn for different biosynthesis pathways and N metabolism (Lemaitre et al., 2007). For example, PEPC is able to catalyze phosphoenolpyruvic acid (PEP) into 2-OG, and 2-OG synthesis from malate can be suppressed by the knockdown of Osppc4, therefore causing a decrease in plant growth and leaf area (Masumoto et al., 2010), suggesting that Osppc4 is crucial for the growth of rice plants. In the present study, significant downregulation of Osppc4 was observed in shoots of rice seedlings after SCN− exposure (Figure 2C). The activity of PEPC was decreased in shoots after SCN− treatments (Figure 3), suggesting that the synthesis of 2-OG in shoots from the PEPC pathway was repressed by SCN− exposure.
The second pathway of 2-OG generation is the NADP-ICDH/NAD-IDH pathway (Ferrario-Mery et al., 2001), in which citrate can be either exported from mitochondria to cytosol for 2-OG synthesis by cytosolic enzymes aconitase NADP-ICDH or transformed into 2-OG in mitochondria by TCA cycle enzyme aconitase NAD-IDH (Yuan et al., 2007). Indeed, NAD-IDH is often regarded as a major governing point in plants (Lemaitre et al., 2007), which is encoded with one single gene OsIDHa in rice plants (Lancien et al., 1998). Herein, significant downregulation of OsIDHa was detected in shoots after SCN− exposure (Figure 2C), suggesting that SCN− exposure could inhibit the expression of OsIDHa in shoots. A significant correlation was obtained in the enzymatic assay of NAD-IDH, wherein a decrease in the activity of NAD-IDH was observed in shoots of rice seedlings exposed to SCN− (Figure 3). Results from both C-related pathways indicated that SCN− exposure significantly repressed both pathways to produce 2-OG, thereby causing a severe impact on the C metabolism and breaking the balance of CNM.
The third source to produce 2-OG is from the GDH pathway, in which the oxidative deamination of glutamate (Glu) into 2-OG is catalyzed by GDH. It has been reported that exogenous 2-OG increased the activities of GDH in wheat seedlings and promoted yield productivity under drought stress (Lancien et al., 1998). Additionally, GDH plays a unique role in the formation of NH4+ and 2-OG during the assimilation of Glu (Lodwig et al., 2003). We found that the expression levels of OsGDH1 and OsGDH2 were significantly upregulated in shoots after SCN− treatments (Figure 2C). The change of GDH activity in shoots was constructive against SCN− exposure (Figure 3). These results indicated that SCN− exposure does not disturb the conversion of Glu into 2-OG through the activation of GDH. Combined with the results from C-related pathways of 2-OG, we have sufficient reasons to conclude that the imbalance of CNM in rice seedlings was evident due to SCN− exposure through repressing the two C-related pathways.
4.2.2. Effects of SCN− on innate 2-OG synthesis in the presence of exogenous 2-OG
Compared with SCN− treatments (Figure 2C), the expression levels of Osppc4 in shoots of rice seedlings under “SCN− + 2-OG” treatments were significantly upregulated, and the activity of PEPC in shoots was also positively responsive, suggesting that the application of 2-OG enhances the enzyme activity of PEPC and might stimulate the generation of 2-OG. A similar conclusion was also predicted in the second pathway of 2-OG generation due to the application of exogenous 2-OG, wherein a correlation between upregulated expression of OsIDHa and increases in NAD-IDH activity was obtained. Additionally, the expression levels of OsGDH1 and OsGDH2 in shoots of rice seedlings fed with 2-OG were significantly upregulated, and an increase of GDH activity in shoots was also detected (Figure 3), suggesting that the conversion of Glu into 2-OG was independent of the application of 2-OG. Co-expression network analysis showed that the GDH-related genes in Module 1 had a higher connection degree with others (Figure 4A). Apparently, these results indicated that the two C-related pathways were significantly activated due to the application of 2-OG, in which sufficient 2-OG in plant cells was able to modify the imbalance of CNM in rice seedlings caused by SCN− exposure, subsequently decreasing the negative impact on rice seedlings, which was judged by a measurable increase in biomass growth of rice seedlings from the SCN−-treated rice seedlings with 2-OG, compared with the SCN−-treated rice seedlings without 2-OG.
4.3. Responses of other CNM-related enzymes and genes in rice plants after SCN− exposure
4.3.1. Effects of SCN− on C metabolism in rice plants
The TPS and SPS are primary targeted cytosolic enzymes involved in the C metabolism (Coruzzi and Zhou, 2001). Previous studies indicated that the expressions of OsTPS2, OsTPS5, and OsTPS6 in rice were negatively correlated with sucrose starvation (Wang et al., 2007). The expression of SPS genes was positively correlated with non-structure carbohydrate content in the leaf, wherein OsSPS1 expression and SPS activity were affirmatively corresponding to spike number and grain yield (Li and Cui, 2018). However, another study showed that mRNA levels of OsSPS1 and OsSPS6 were negatively correlated with sucrose concentrations (Yonekura et al., 2013). In the present study, upregulation of OsTPS5 was detected in both rice tissues under SCN− stress (Figure 2). In addition, upregulated OsSPS1 was also evident in both roots and shoots. Meanwhile, increases in TPS and SPS activities in rice tissues were detectable (Figure 3). These results indicated that SCN− exposure stimulated the expression of C metabolism-related master regulation genes, thus regulating the enzyme activities. Enzyme ERS has an important role in maintaining the physiological homeostasis of amino acids and C metabolism as well as redox status (Yang et al., 2018). Like TPS and SPS, upregulation of OsERS1 was distinguished in both rice tissues after SCN− stress (Figure 2). Also, we observed that responses of OsERS1 to SCN− exposure were identical to the enzyme activity of ERS in rice tissues (Figure 2).
4.3.2. Effects of SCN− on N metabolism in rice plants
NR, NiR, and GS are three key enzymes involved in N assimilation. The conversion of NO3− into NH4+ is catalyzed by the enzymes NR and NiR, which is a rate-limiting step in NO3− assimilation. In the present study, three isogenes of NR (OsNIA1, OsNIA12, and OsNR1) and NiR (OsNiR-1, OsNiR-2, and OsNiR-3) showed a declining expression pattern in shoots after SCN− exposure, of which SCN− treatments at 24.0 and 96.0 mg/L positively regulated transcriptional changes in OsNR1, and 300.0 mg SCN/L treatment demonstrated a negative response (Figure 2). Enzymatic assay showed that activities of NR and NiR had a positive correlation with gene expression (Figure 3), suggesting that low-to-moderate concentrations of SCN− exposure might stimulate the conversion of NO3−, and higher SCN− concentrations had a negative effect on this process, whereas a similar conclusion was also reached by Lin et al. (2022a). Another crucial enzyme in N metabolism is GS, which is responsible for converting NH4+ into glutamine. In this study, almost all GS genes were upregulated in the SCN− treatments (Figure 2). It is established that GS in most plants occurs as GS2 in plastids and GS1 in the cytosol. The role of GS1 is to assimilate NH4+ in roots and reassimilate NH4+ in leaves, whereas GS2 is mainly responsible for assimilating NH4+ derived from NO3− reduction in plastids (Lin et al., 2022a). The enzymatic assay also showed that activities of GS had a positive correlation to gene expression (Figure 3), suggesting that SCN− treatments do not inhibit the activity of GS and subsequently increase the conversion of NH4+.
4.3.3. Effects of exogenous 2-OG on CNM in rice plants under SCN− stress
Compared with SCN− treatments (Figure 2), exogenous 2-OG decreased the expression of OsTPS5 in shoots of rice seedlings after SCN− exposure. Similarly, the downregulation of OsSPS6 and OsERS1 was also detectable. Enzymatic assays indicated that the application of 2-OG decreased the activities of TPS, SPS, and ERS in shoots of rice seedlings (Figure 3). A co-expression network analysis showed that the C metabolism-related genes in Module 2 and Module 4 had a lower connection degree with others (Figure 4A). However, we noticed that NR and NiR genes were upregulated in SCN−-treated rice seedlings inoculated with 2-OG, wherein enzymes of NR and NiR were positively responsive to 2-OG application, indicating that the application of 2-OG had a positive impact on the conversion of NO3−. A previous study also reported that feeding of 2-OG increased transcripts of the NR gene in tobacco leaf (Ferrario-Mery et al., 2001). Additionally, under “SCN− + 2-OG” treatments, the expression levels of OsGS1;2 were significantly higher than those under SCN− treatments, suggesting that the application of 2-OG also increases the conversion of NH4+ derived from the main N source of NO3− supplied. A co-expression network analysis showed that the N metabolism-related genes in Module 3 and Module 4 had a higher connection degree with others (Figure 4A). These results indicated that sufficient 2-OG in plant cells can modify the imbalance of N metabolism-related genes in rice seedlings caused by SCN− exposure.
5. Conclusion
The balance of CNM in rice seedlings can be broken by SCN− exposure, resulting in a significant reduction in the biomass growth of rice seedlings. The application of exogenous 2-OG showed a positive regulatory effect on the imbalance of CNM in rice seedlings under SCN− stress. Higher connection degrees of genes in each module in rice plants under SCN− and “SCN− + 2-OG” treatments are marked in Figure 6. Although our findings provide new insight into the role of exogenous 2-OG in minimizing the negative effect of SCN− exposure on rice plants through regulating the pathways involved in CNM, the global molecular map of regulatory genes involved in CNM remains unclear. Further comprehensive studies are needed to experimentally prove the influence of exogenous 2-OG as a chemical regulator on the quality and quantity of agricultural crops through the “omics” technology, such as transcriptome, proteome, and metabolome.
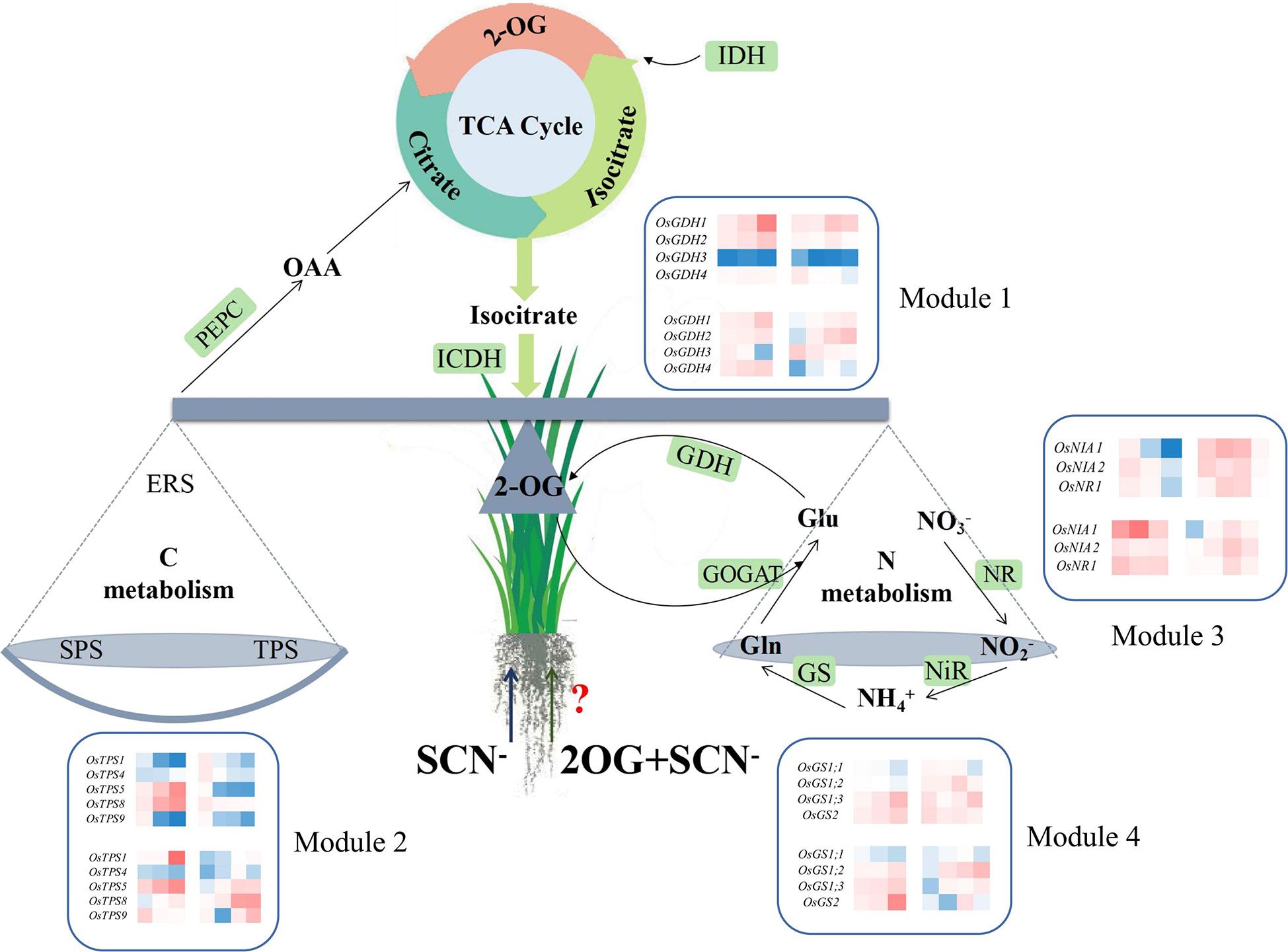
Figure 6 Overview of exogenous 2-OG-mediated CNM in rice tissues under SCN− stress. 2-OG, 2-oxoglutarate; CNM, carbon and nitrogen metabolism.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author/s.
Author contributions
X-ZY: conceptualization, methodology, supervision, writing-reviewing and editing, and funding acquisition. Y-XF: writing-original draft preparation and visualization. LY: investigation. Y-JL: investigation, data analysis, visualization, and software. YS: data analysis, visualization, and software. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 41761094) and the Postdoctoral Research Foundation of China (2021MD703821).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1086098/full#supplementary-material
References
Ahanger, M. A., Qi, M., Huang, Z., Xu, X., Begum, N., Qin, C., et al. (2021). Improving growth and photosynthetic performance of drought stressed tomato by application of nano-organic fertilizer involves up-regulation of nitrogen, antioxidant and osmolyte metabolism. Ecotoxicol. Environ. Saf. 216, 112195. doi: 10.1016/j.ecoenv.2021.112195
Alves, H., Matiolli, C. C., Soares, R. C., Almadanim, M. C., Oliveira, M. M., Abreu, I. A. (2021). Carbon/nitrogen metabolism and stress response networks-calcium-dependent protein kinases as the missing link? J. Exp. Bot. 72, 4190–4201. doi: 10.1093/jxb/erab136
Araújo, W. L., Martins, A. O., Fernie, A. R., Tohge., T. (2014). 2-oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 5, 552. doi: 10.3389/fpls.2014.00552
Bhunia, F., Saha, N. C., Kaviraj, A. (2000). Toxicity of thiocyanate to fish, plankton, worm and aquatic system. Bull. Environ. Contam. Toxicol. 64, 197–204. doi: 10.1007/s001289910030
Brown, S., Devolder, P. S., Compton, H., Henry, C. (2007). Effect of amendment C:N ratio on plant richness, cover and metal content for acidic Pb and zn mine tailings in leadville, Colorado. Environ. Pollut. 149, 165–172. doi: 10.1016/j.envpol.2007.01.008
Coruzzi, G. M., Zhou, L. (2001). Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Curr. Opin. Plant Biol. 4, 247–253. doi: 10.1016/S1369-5266(00)00168-0
Feng, Y. X., Yu, X. Z., Mo, C. H., Lu, C. J. (2019). Regulation network of sucrose metabolism in response to trivalent and hexavalent chromium in Oryza sativa. J. Agric. Food Chem. 67, 9738–9748. doi: 10.1021/acs.jafc.9b01720
Ferrario-Mery, S., Masclaux, C., Suzuki, A., Valadier, M. H., Hirel, B., Foyer, C. H. (2001). Glutamine and a-ketoglutarate are metabolite signals involved in nitrate reductase gene transcription in untransformed and transformed tobacco plants deficient in ferredoxin-glutamine-a-ketoglutarate aminotransferase. Planta 213, 265–271. doi: 10.1007/s004250000504
Fritz, C., Mueller, C., Matt, P., Feil, R., Stitt, M. (2006). Impact of the c-n status on the amino acid profile in tobacco source leaves. Plant Cell Environ. 29, 2055–2076. doi: 10.1111/j.1365-3040.2006.01580.x
Gálvez, S., Bismuth, E., Sarda, C., Gadal, P. (1994). Purification and characterization of chloroplastic NADP-isocitrate dehydrogenase from mixotrophic tobacco cells. comparison with the cytosolic isoenzyme. Plant Physiol. 105, 593–600. doi: 10.1104/pp.105.2.593
Gálvez, S., Lancien, M., Hodges, M. (1999). Are isocitrate dehydrogenases and 2-oxoglutarate involved in the regulation of glutamate synthesis? Trends Plant Sci. 4, 484–490. doi: 10.1016/s1360-1385(99)01500-9
Gao, J. J., Wang, B., Li, Z. J., Xu, J., Fu, X. Y., Han, H. J., et al. (2022). Metabolic engineering of Oryza sativa for complete biodegradation of thiocyanate. Sci. Total Environ. 820, 153283. doi: 10.1016/j.scitotenv.2022.153283
Goddijn, O. J., Verwoerd, T. C., Voogd, E., Krutwagen, R. W., De Graff, P. T. H. M., Poels, J., et al. (1997). Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol. 113, 181–190. doi: 10.1104/pp.113.1.181
Gould, W. D., King, M., Mohapatra, B. R., Cameron, R., Kapoor, A., Koren, D. W. (2012). A critical review on destruction of thiocyanate in mining effluents. Miner. Eng. 34, 38–47. doi: 10.1016/j.mineng.2012.04.009
Guo, L. Y., Lu, Y. Y., Bao, S. Y., Zhang, Q., Geng, Y. Q., Shao, X. W. (2021). Carbon and nitrogen metabolism in rice cultivars affected by salt-alkaline stress. Crop Pasture Sci. 72, 372–382. doi: 10.1071/CP20445
Hansson, D., Morra, M. J., Borek, V., Snyder, A. J., Johnsonmaynard, J. L., Thill, D. C. (2008). Ionic thiocyanate (SCN-) production, fate, and phytotoxicity in soil amended with Brassicaceae seed meals. J. Agric. Food Chem. 56, 3912–3917. doi: 10.1021/jf800104x
Hou, W. F., Xue, X. X., Li, X. K., Khan, M. R., Yan, J. Y., Ren, T., et al. (2019). Interactive effects of nitrogen and potassium on: Grain yield, nitrogen uptake and nitrogen use efficiency of rice in low potassium fertility soil in China. Field Crop Res. 236, 14–23. doi: 10.1016/j.fcr.2019.03.006
Ji, J., Shi, Z., Xie, T., Zhang, X., Chen, W., Du, C., et al. (2020). Responses of gaba shunt coupled with carbon and nitrogen metabolism in poplar under NaCl and CdCl2 stresses. Ecotoxicol. Environ. Saf. 193, 110322. doi: 10.1016/j.ecoenv.2020.110322
Lancien, M., Gadal, P., Hodges, M. (1998). Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: Evidence for a heteromeric structure by the complementation of yeast mutants. Plant J. 16, 325–333. doi: 10.1046/j.1365-313x.1998.00305.x
Lemaitre, T., Urbanczyk-Wochniak, E., Flesch, V., Bismuth, E., Fernie, A. R., Hodges, M. (2007). NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol. 144, 1546–1558. doi: 10.1104/pp.107.100677
Li, G. H., Cui, K. H. (2018). The effect of nitrogen on leaf sucrose phosphate synthase and its relationships with assimilate accumulation and yield in rice. Plant Physiol. J. 54, 1195–1204. doi: 10.13592/j.cnki.ppj.2018.0257
Lin, Y. J., Feng, Y. X., Yu, X. Z. (2022a). The importance of utilizing nitrate (NO3–) over ammonium (NH4+) as nitrogen source during detoxification of exogenous thiocyanate (SCN-) in Oryza sativa. Environ. Sci. Pollut. Res. 29, 5622–5633. doi: 10.1007/s11356-021-15959-z
Lin, Y. J., Feng, Y. X., Zhang, Q., Yu, X. Z. (2022b). Proline-mediated modulation on DNA repair pathway in rice seedlings under chromium stress by integrating gene chip and co-expression network analysis. Ecotoxicology 31, 1266–1275. doi: 10.1007/s10646-022-02586-8
Lin, Y. J., Yu, X. Z., Li, Y. H., Yang, L. (2020). Inhibition of the mitochondrial respiratory components (Complex I and complex III) as stimuli to induce oxidative damage in Oryza sativa L. under thiocyanate exposure. Chemosphere 243, 125472. doi: 10.1016/j.chemosphere.2019.125472
Lodwig, E. M., Hosie, A. H., Bourdès, A., Findlay, K., Allaway, D., Karunakaran, R., et al. (2003). Amino-acid cycling drives nitrogen fixation in the legume–rhizobium symbiosis. Nature 422, 722. doi: 10.1038/nature01527
Masumoto, C., Miyazawa, S. I., Ohkawa, H., Fukuda, T., Taniguchi, Y., Murayama, S., et al. (2010). Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc. Natl. Acad. Sci. U.S.A. 107, 5226–5231. doi: 10.1073/pnas.0913127107
Mostofa, M. G., Seraj, Z. I., Fujita, M. (2014). Exogenous sodium nitroprusside and glutathione alleviate copper toxicity by reducing copper uptake and oxidative damage in rice (Oryza sativa L.) seedlings. Protoplasma 25, 1373–1386. doi: 10.1007/s00709-014-0639-7
Mudder, T. I., Botz, M. (2004). Cyanide and society: a critical review. Eur. J. Miner. Process. Environ. Prot. 4, 62–74.
Naseeruddin, R., Sumathi, V., Prasad, T.N.V.K.V., Sudhakar, P., Chandrika, V., Reddy, B. R. (2018). Unprecedented synergistic effects of nanoscale nutrients on growth, productivity of sweet sorghum [Sorghum bicolor (L.) moench] and nutrient biofortification. J. Agric. Food Chem. 66, 1075–1084. doi: 10.1021/acs.jafc.7b04467
Osuna, L., Gonzalez, M. C., Cejudo, F. J., Vidal, J., Echevarria, C. (1996). In vivo and in vitro phosphorylation of the phosphoenolpyruvate carboxylase from wheat seeds during germination. Plant Physiol. 111, 551–558. doi: 10.1104/pp.111.2.551
Ratinaud, M. H., Thomes, J. C., Julien, R. (1983). Glutamyl-tRNA synthetases from wheat. isolation and characterization of three dimeric enzymes. Eur. J. Biochem. 135, 471–477. doi: 10.1111/j.1432-1033.1983.tb07675.x
Reddy, A. R., Chaitanya, K. V., Vivekanandan, M. (2004). Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 161, 1189–1202. doi: 10.1016/j.jplph.2004.01.013
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Sun, Z., Xie, X., Wang, P., Hu, Y., Cheng, H. (2018). Heavy metal pollution caused by small-scale metal ore mining activities: A case study from a polymetallic mine in south China. Sci. Total. Environ. 639:217–227 doi: 10.1016/j.scitotenv.2018.05.176.
Turano, F. J., Dashner, R., Upadhyaya, A., Caldwell, C. R. (1996). Purification of mitochondrial glutamate dehydrogenase from darkgrown seedlings. Plant Physiol. 112, 1357–1364. doi: 10.1104/pp.112.3.1357
Wang, H. J., Wan, A. R., Hsu, C. M., Lee, K. W., Yu, S. M., Jauh, G. Y. (2007). Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol. Biol. 63, 441–463. doi: 10.1007/s11103-006-9100-4
Xin, W., Zhang, L., Zhang, W., Gao, J., Yi, J., Zhen, X., et al. (2019). An integrated analysis of the rice transcriptome and metabolome reveals root growth regulation mechanisms in response to nitrogen availability. Int. J. Mol. Sci. 20, 5893. doi: 10.3390/ijms20235893
Yang, L., Feng, Y. X., Lin, Y. J., Yu, X. Z. (2021). Comparative effects of sodium hydrosulfide and proline on functional repair in rice chloroplast through the D1 protein and thioredoxin system under simulated thiocyanate pollution. Chemosphere 284, 131389. doi: 10.1016/j.chemosphere.2021.131389
Yang, X., Li, G., Tian, Y., Song, Y., Liang, W., Zhang, D. A. (2018). Rice glutamyl-tRNA synthetase modulates early anther cell division and patterning. Plant Physiol. 177, 728–744. doi: 10.1104/pp.18.00110
Yonekura, M., Aoki, N., Hirose, T., Onai, K., Ishiura, M., Okamura, M., et al. (2013). The promoter activities of sucrose phosphate synthase genes in rice, OsSPS1 and OsSPS11, are controlled by light and circadian clock, but not by sucrose. Front. Plant Sci. 4, 31. doi: 10.3389/fpls.2013.00031
Yu, X. Z., Zhang, F. Z. (2013). Effects of exogenous thiocyanate on mineral nutrients, antioxidative responses and free amino acids in rice seedlings. Ecotoxicology 22, 752–760. doi: 10.1007/s10646-013-1069-6
Yuan, Y., Ou, J., Wang, Z., Zhang, C., Zhou, Z., Lin, Q. (2007). Regulation of carbon and nitrogen metabolisms in rice roots by 2-oxoglutarate at the level of hexokinase. Physiol. Plantarum 129, 296–306. doi: 10.1111/j.1399-3054.2006.00806.x
Yue, J., Du, C., Ji, J., Xie, T., Chen, W., Chang, E., Chen, L., Jiang, Z., Shi, S. (2018). Inhibition of α-ketoglutarate dehydrogenase activity affects adventitious root growth in poplar via changes in GABA shunt. Planta 248, 963–979. doi: 10.1007/s00425-018-2929-3
Zhang, Q., Feng, Y. X., Lin, Y. J., Tian, P., Yu, X. Z. (2022). Proline-mediated regulation on jasmonate signals repressed anthocyanin accumulation through the MYB-bHLH-WDR complex in rice under chromium exposure. Front. Plant Sci 13, 13953398. doi: 10.3389/fpls.2022.953398
Keywords: rice, thiocyanate, 2-OG, carbon metabolism, nitrogen metabolism
Citation: Feng Y-X, Yang L, Lin Y-J, Song Y and Yu X-Z (2023) Merging the occurrence possibility into gene co-expression network deciphers the importance of exogenous 2-oxoglutarate in improving the growth of rice seedlings under thiocyanate stress. Front. Plant Sci. 14:1086098. doi: 10.3389/fpls.2023.1086098
Received: 01 November 2022; Accepted: 06 February 2023;
Published: 23 February 2023.
Edited by:
Walid Soufan, King Saud University, Saudi ArabiaCopyright © 2023 Feng, Yang, Lin, Song and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Zhang Yu, eHp5dUBnbHV0LmVkdS5jbg==
†These authors have contributed equally to this work
‡ORCID: Xiao-Zhang Yu, orcid.org/0000-0001-7846-5017
 Yu-Xi Feng
Yu-Xi Feng Li Yang†
Li Yang† Xiao-Zhang Yu
Xiao-Zhang Yu