- 1College of Mechanical and Electronic Engineering, Nanjing Forestry University, Nanjing, Jiangsu, China
- 2Peking University Institute of Advanced Agricultural Sciences / Shandong Laboratory of Advanced Agricultural Sciences at Weifang, Weifang, Shandong, China
- 3Department of Crop and Soil Sciences, University of Georgia, Griffin, GA, United States
Deep learning methods for weed detection typically focus on distinguishing weed species, but a variety of weed species with comparable plant morphological characteristics may be found in turfgrass. Thus, it is difficult for deep learning models to detect and distinguish every weed species with high accuracy. Training convolutional neural networks for detecting weeds susceptible to herbicides can offer a new strategy for implementing site-specific weed detection in turf. DenseNet, EfficientNet-v2, and ResNet showed high F1 scores (≥0.986) and MCC values (≥0.984) to detect and distinguish the sub-images containing dollarweed, goosegrass, old world diamond-flower, purple nutsedge, or Virginia buttonweed growing in bermudagrass turf. However, they failed to reliably detect crabgrass and tropical signalgrass due to the similarity in plant morphology. When training the convolutional neural networks for detecting and distinguishing the sub-images containing weeds susceptible to ACCase-inhibitors, weeds susceptible to ALS-inhibitors, or weeds susceptible to synthetic auxin herbicides, all neural networks evaluated in this study achieved excellent F1 scores (≥0.995) and MCC values (≥0.994) in the validation and testing datasets. ResNet demonstrated the fastest inference rate and outperformed the other convolutional neural networks on detection efficiency, while the slow inference of EfficientNet-v2 may limit its potential applications. Grouping different weed species growing in turf according to their susceptibility to herbicides and detecting and distinguishing weeds by herbicide categories enables the implementation of herbicide susceptibility-based precision herbicide application. We conclude that the proposed method is an effective strategy for site-specific weed detection in turf, which can be employed in a smart sprayer to achieve precision herbicide spraying.
Introduction
Turfgrass is widely grown in urban landscapes, including institutional and residential lawns, parks, or athletic fields (Potter and Braman, 1991). The total turfgrass area in the United States is 163,812 km2, which accounts for approximately 1.9% of the whole terrestrial land of the country (Milesi et al., 2005). Weed control is a challenging task for turfgrass management. Weeds compete with the turfgrass for sunlight, moisture, and soil nutrients, reducing turf aesthetics, surface quality, and functionality (Hamuda et al., 2016; Liu and Bruch, 2020). Weed management in turfgrass landscapes has relied heavily on broadcast herbicide application (McElroy and Martins, 2013), although weeds almost always present in non-uniform and patchy distributions (Dai et al., 2019; Yu et al., 2019a). Excessive application of synthetic herbicides could potentially pose a risk to human health and cause environmental pollution (Slaughter et al., 2008; Dai et al., 2019; Yu et al., 2019b; Mennan et al., 2020). Moreover, the application of synthetic herbicides represents a significant variable cost in turf weed management (Davis and Frisvold, 2017). These concerns have led to legal regulations regarding herbicide usage in several countries. For example, the European Union encourages spot-spraying to reduce the herbicide input (Busey, 2003; Marchand and Robin, 2019). Additionally, spot-spraying could effectively minimize the amount reaching off-target areas (Melland et al., 2016). In the United States, Environmental Protection Agency has proposed a series of measures, including prohibiting aerial applications for all atrazine labels to reduce their chance of runoff from the managed fields (Pimentel and Burgess, 2012; McCullough et al., 2015).
Site-specific weed management is a promising solution for sustainable weed control (Chen et al., 2022). Precision spraying a particular type or volume of herbicide onto susceptible weed species can significantly reduce herbicide input and weed control costs (Liakos et al., 2018). Site-specific weed management relies on the accurate identification and localization of weeds (Fennimore et al., 2016; Wang et al., 2019). Previous researchers explored various visual characteristics, such as color (Tang et al., 2016), morphological (Perez et al., 2000), hyper- or multi-spectral (Pantazi et al., 2016; Jiang et al., 2020), and textural features (Bakhshipour et al., 2017), for weed detection. However, crops and weeds may exhibit similar visual characteristics, thus detection and classification of weeds in crops are inherently challenging (Hasan et al., 2021). In turf, weed detection is challenging due to the presence of a variety of weed species growing with turfgrass.
In recent years, deep learning, a subfield of artificial intelligence, has demonstrated remarkable capability in speech recognition (Hinton et al., 2012; LeCun et al., 2015), natural language processing (Collobert and Weston, 2008; Collobert et al., 2011), and computer vision (Gu et al., 2018; Shi et al., 2020; Zhou et al., 2020). Deep learning technologies exhibit a tremendous ability to learn representations from raw data and extract complex features from images with a high accuracy level (Jordan and Mitchell, 2015; He et al., 2020; Yang et al., 2022a). Moreover, the improvements in graphics processing units (GPUs) have facilitated the use of deep convolutional neural networks (Bao et al., 2017; Bao et al., 2021; Ngo et al., 2021). Recent studies have investigated the feasibility of using deep learning in various agricultural domains, including plant disease detection (Martinelli et al., 2015; Saleem et al., 2019), crop yield prediction (Khaki and Wang, 2019; Van Klompenburg et al., 2020), plant phenotyping (Atefi et al., 2021; Zhang et al., 2022), and weed detection (Jin et al., 2021; Peng et al., 2022; Razfar et al., 2022). For example, Abbas et al. proposed a deep learning-based method for tomato disease detection. The trained neural network achieved a best 5-class classification accuracy of 99.51 (Abbas et al., 2021). Subeesh et al. compared four convolutional neural networks, including AlexNet, GoogLeNet, InceptionV3, and Xception for detecting various weeds growing in bell peppers (Capsicum annum L.) and found InceptionV3 achieved the highest accuracy (97.7%) (Subeesh et al., 2022). For image-based weed detection and discrimination, previous findings suggest that deep learning methods generally outperform other methods (Fennimore et al., 2016; Kamilaris and Prenafeta-Boldú, 2018).
Several studies have investigated the use of image classification or object detection neural networks for detecting and distinguishing various weed species in turfgrass (Yu et al., 2019a; Yu et al., 2019b; Yu et al., 2019c; Yu et al., 2020). Jin et al. demonstrated that VGGNet effectively detected and distinguished dallisgrass (Paspalum dilataum Poir.), purple nutsedge (Cyperus rotundus L.), and white clover (Trifolium repens L.) growing in bermudagrass [Cynodon dactylon (L.) Pers.] turf, while RegNet is well-performed in discriminating common dandelion (Taraxacum officinale Web.) (Jin et al., 2022). In another study, Yu et al. developed effective deep convolutional neural networks to detect weeds in turf. The authors reported that the image classification neural network VGGNet reliably classified broadleaf and grassy weeds growing in bermudagrass turf. In addition, the object detection neural network DetectNet achieved high overall accuracy at detecting cutleaf evening-primrose (Oenothera laciniata Hill) growing in bahiagrass (Paspalum notatum Flugge) (Yu et al., 2019b; Yu et al., 2019c).
Different weed species exhibit varying susceptibility to a particular herbicide category (McElroy and Martins, 2013; Yu et al., 2018). For example, acetolactate synthase (ALS)-inhibiting herbicides generally provide a narrow weed control spectrum (Yu and Boyd, 2018); ACCase-inhibiting herbicides are only effective for controlling grassy weeds (McElroy and Martins, 2013); nonselective herbicides, such as glyphosate and glufosinate, could nonselectively control all weeds (Johnson, 1977); and synthetic auxin herbicides, such as 2,4-D, dicamba, and MCPA, are only effective for controlling broadleaf weeds (McElroy and Martins, 2013). Therefore, precision spraying herbicides based on the susceptibility of different weed species to the herbicides can significantly reduce herbicide input and improve herbicide use efficiency. Although deep learning has been well-performed in weed detection and discrimination, previous studies have generally focused on distinguishing different weed species and did not establish a direct connection between weeds and herbicides. Moreover, a variety of weed species with comparable plant morphological characteristics may be found in turfgrass, thus it is difficult for the deep learning models to detect and distinguish every weed species with high accuracy. In the present research work, in addition to the detection and discrimination of individual weed species, different weed species growing in bermudagrass turf were grouped according to their susceptibility to herbicides, and weeds were detected and distinguished by herbicide categories. The proposed method would allow precision herbicide application based on susceptibility and thereby effectively reduce herbicide input while achieving the same level of weed control as the broadcast herbicide application. The objectives of this paper were to (1) investigate the feasibility of utilizing deep learning for herbicide susceptibility-based weed detection in bermudagrass turf, and 2) evaluate and compare the performance of different convolutional neural networks for distinguishing individual weed species.
Materials and methods
Overview
The image classification convolutional neural networks, including DenseNet (Huang et al., 2017), EfficientNet (Tan and Le, 2019), and ResNet(He et al., 2016), were selected for evaluating the feasibility of using the convolutional neural networks for detecting and distinguishing individual weed species growing in bermudagrass turf or detecting and distinguishing weeds susceptible to herbicides. DenseNet is a convolutional neural network that computes dense and multi-scale features from the convolutional layers. For each layer, it obtains additional inputs from all preceding layers and passes on its feature maps to all subsequent layers. EfficientNet uses a set of fixed scaling coefficients to uniformly scales all dimensions of depth, width, and resolution in a principled way. The EfficientNet achieves state-of-the-art accuracy with 10× better efficiency by utilizing this novel scaling method. ResNet introduced the concept of residual learning. It employs an identity-based skip connection in each residual unit. ResNet eases the flow of information across units and thus can gain accuracy from very deep networks. In this study, these three convolutional neural networks were trained and evaluated with the ultimate goal of site-specific herbicide application.
Image acquisition
The training images of crabgrass (D.igitaria ischaemum L.), dollarweed (Hydrocotyle spp.), old world diamond-flower (Hedyotis cormybosa L.), and tropical signalgrass [Urochloa distachya (L.) T.Q. Nguyen] were acquired at several golf courses in Bradenton (27.49°N, 82.47°W), Riverview (27.86°N, 82.32°W), Sun City (27.71°N, 82.35°W), and Tampa (27.95°N, 82.45°W), Florida, while the testing images were acquired at several golf courses and institutional lawns in Lakeland, Florida (28.03°N, 81.94°W). The training images of goosegrass (Eleusine indica L.) and Virginia buttonweed (Diodia virginiana L.) growing in bermudagrass turf were acquired at the University of Georgia Griffin Campus in Griffin, Georgia, United States (33.26°N, 84.28°W), while the testing images were acquired at several golf courses in Peachtree City, Georgia, United States (33.39°N, 84.59°W). The training images of purple nutsedge were acquired at sod farms in Jiangning District, Nanjing, Jiangsu, China (31.95°N, 118.85°E), while the testing images were acquired at sod farms in Shuyang, Jiangsu, China (34.12°N, 118.79°E). The training and testing images of crabgrass, dollarweed, goosegrass, old world diamond-flower, tropical signalgrass, and Virginia buttonweed were captured multiple times from April to November 2018 using a digital camera (DSC-HX1, SONY®, Cyber-Shot Digital Still Camera, SONY Corporation, Minato, Tokyo, Japan). The training and testing images of purple nutsedge were captured in spring 2021 using a digital camera (Panasonic® DMC-ZS110, Xiamen, Fujian, China). The original resolution of the training and testing images was 1,920 × 1,080 pixels. To enrich the diversity of the training dataset, images were captured under various illumination conditions, including partly cloudy, cloudy, or sunny days.
Training and testing
Images containing crabgrass, dollarweed, goosegrass, old world diamond-flower, purple nutsedge, tropical signalgrass, and Virginia buttonweed growing in bermudagrass turf were selected to constitute the training or testing datasets. Images containing a single weed species were selected for training and testing neural networks. All images were cropped into 40 equal-sized sub-images by a 5 rows × 8 columns division. Each sub-image was 240 × 216 pixels. Sub-images of crabgrass, goosegrass, and tropical signalgrass (Figure 1), purple nutsedge (Figure 2), dollarweed, old world diamond-flower, and Virginia buttonweed (Figure 3) at different growth stages and densities, and sub-images of bermudagrass at varying mowing heights and surface conditions (Figure 4) were utilized for training and testing the neural networks. Figure 5 outlines the sequence diagram of image processing and training and testing the convolutional neural networks for detecting and discriminating individual weed species or weeds susceptible to ACCase-inhibitor, ALS-inhibitor, synthetic auxin herbicides, or bermudagrass without weed infestation (no herbicide).

Figure 1 The training and testing sub-images of crabgrass (A), goosegrass (B), and tropical signalgrass (C) at different growth stages and densities.

Figure 2 The training and testing sub-images of dollarweed (A), old world diamond-flower (B), and Virginia buttonweed (C) at different growth stages and densities.

Figure 3 The training and testing sub-images of purple nutsedge at different growth stages and densities.

Figure 4 The training and testing sub-images of bermudagrass at different turfgrass management regimes, mowing heights, and surface conditions.
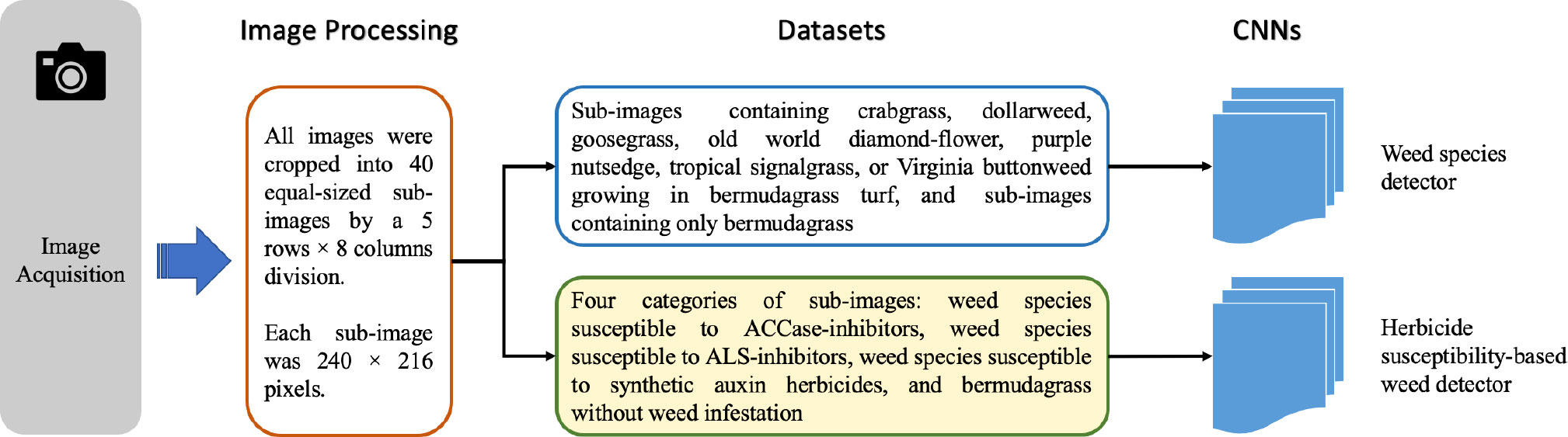
Figure 5 Flow diagram illustrates the sequence of image processing and training and testing the convolutional neural networks.
The convolutional neural networks for detecting and distinguishing weed species were trained utilizing a total of 21,000 true positive sub-images (3,000 sub-images for each weed species) containing crabgrass, dollarweed, goosegrass, old world diamond-flower, purple nutsedge, tropical signalgrass, or Virginia buttonweed growing in bermudagrass turf, while a total of 9,000 sub-images containing only bermudagrass were utilized as the true negative images. To establish the validation or testing dataset, a total of 3,500 sub-images (500 images for each weed species) containing crabgrass, dollarweed, goosegrass, old world diamond-flower, purple nutsedge, tropical signalgrass, or Virginia buttonweed growing in bermudagrass were utilized as the true positive images, while a total of 1,500 sub-images containing only bermudagrass were utilized as the true negative images.
The convolutional neural networks for detecting and distinguishing weeds susceptible to various herbicides were trained using a dataset containing four categories of sub-images: weed species susceptible to ACCase-inhibitors, weed species susceptible to ALS-inhibitors, weed species susceptible to synthetic auxin herbicides, and bermudagrass without weed infestation. To establish the training, validation, or testing dataset, the sub-images containing crabgrass, goosegrass, or tropical signalgrass, the sub-images containing purple nutsedge, the sub-images containing dollarweed, old world diamond-flower, or Virginia buttonweed, and the sub-images containing bermudagrass only were grouped and labeled as ACCase-inhibiting herbicides, ALS-inhibiting herbicides, synthetic auxin herbicides and no herbicide, respectively (Table 1).
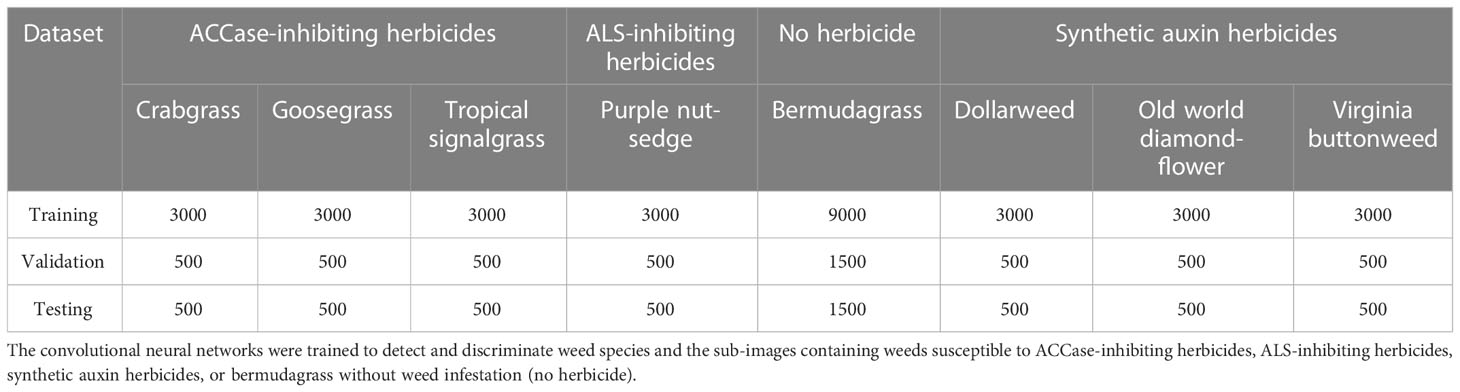
Table 1 The number of sub-images used to establish the training, validation, and testing datasets of the convolutional neural networks.
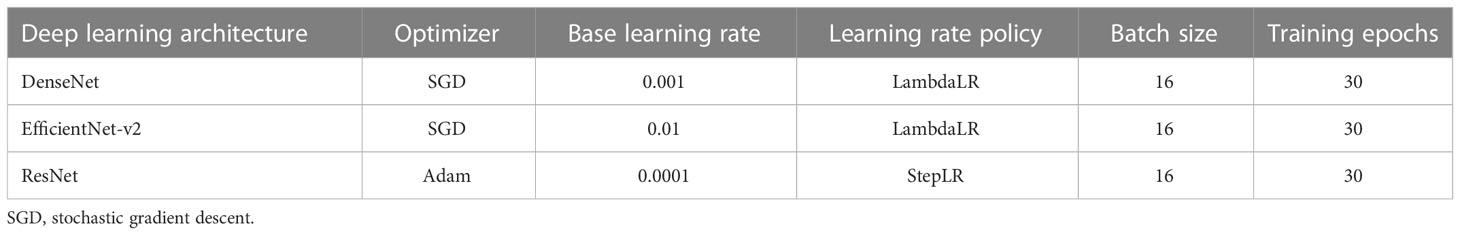
The training and testing of the convolutional neural networks were performed in PyTorch (version 1.8.1) deep learning environment (Facebook, San Jose, California, United States) with an NVIDIA GeForce RTX 2080 Ti graphic processing unit (GPU). Transfer learning seeks to use previously acquired knowledge while addressing one problem and applying it to a different but similar problem (Lu et al., 2015). The convolutional neural networks were pre-trained with the ImageNet dataset to initialize the weights and bias through the transfer learning technology. To ensure fair comparisons among the evaluated deep learning models, default values of hyper-parameters for each neural network were adopted and used (Table 2).
A binary classification confusion matrix with four conditions, including the true positive (tp), false positive (fp), true negative (tn), and false negative (fn), was used to present the training and testing results of the convolutional neural networks. The performances of the convolutional neural networks were evaluated and compared against each other in terms of precision, recall, F1 score, and Matthews Correlation Coefficient (MCC).
Precision is the ability of the neural networks to detect the susceptible weed species and was calculated using the tp and fp (Sokolova and Lapalme, 2009):
Recall is the effectiveness of the neural networks to detect the susceptible weed species and was computed using the tp and fn (Sokolova and Lapalme, 2009):
The F1 score is a commonly used metric for measuring the overall performance of the neural networks, which was defined using the following equation (Sokolova and Lapalme, 2009):
The MCC is the correlation between ground truth labels and predictions, which was determined using the following equation (Chicco and Jurman, 2020):
Frames per second (FPS) measures the number of images, also known as frames processed by the neural networks each second. A higher FPS value indicates faster image processing. The FPS value was adopted as a quantitative metric to evaluate the computational efficiency of the neural networks.
Results
Detection and discrimination of weed species
When the convolutional neural networks were trained for detecting and distinguishing weed species growing in bermudagrass turf, DenseNet, EfficientNet-v2, and ResNet exhibited excellent performances and achieved high F1 scores (≥0.995) and MCC values (≥0.994) in the validation datasets for detecting and distinguishing the sub-images containing dollarweed, goosegrass, purple nutsedge, and the sub-images containing bermudagrass only (Table 3). In general, a slight reduction in weed detection performance of all neural networks was observed in the testing datasets compared to the validation datasets.
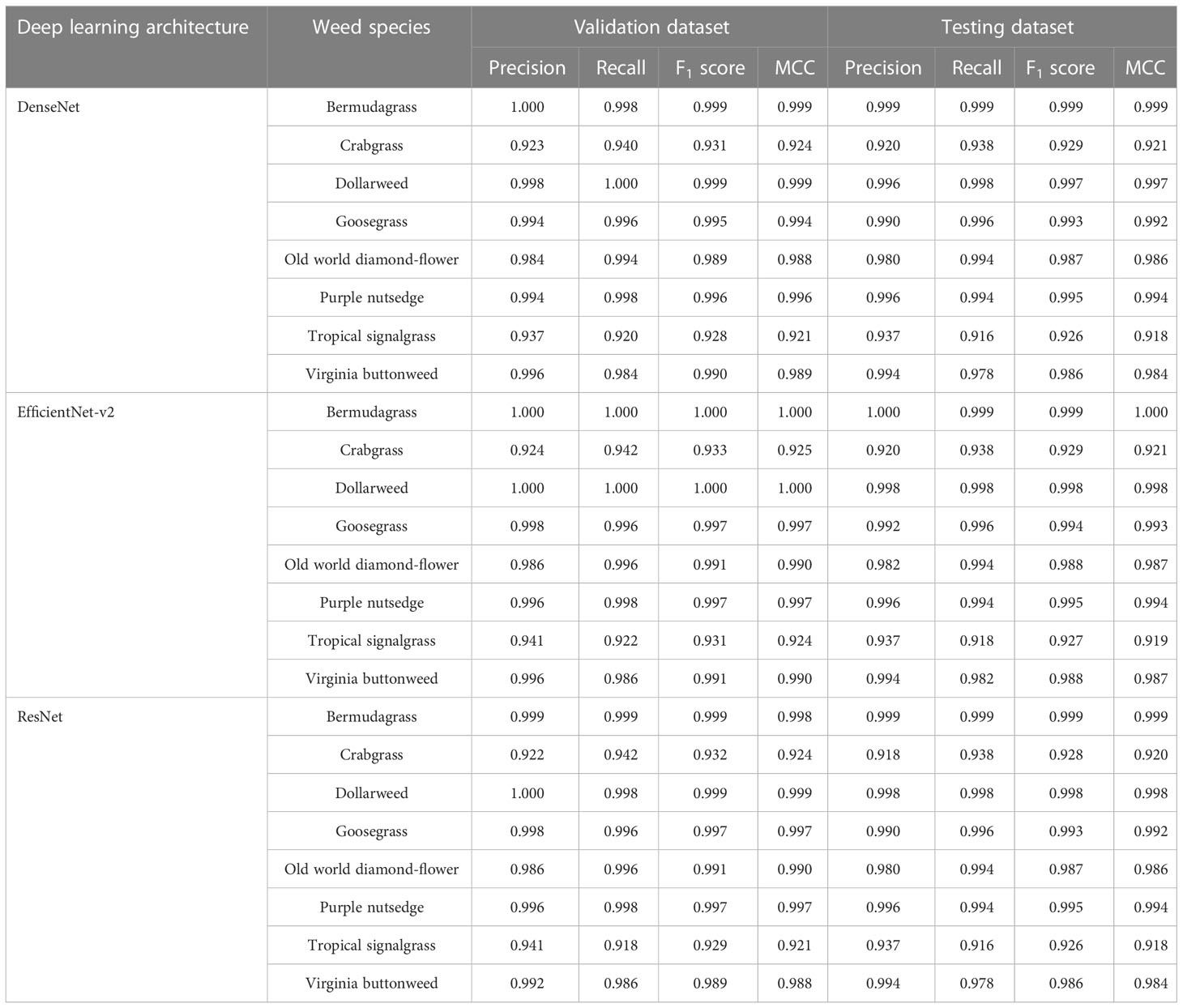
Table 3 Weed species detection and discrimination training results using various convolutional neural networks.
For the detection of old world diamond-flower, the recall values of DenseNet in the validation and testing datasets were 0.994, while the precision values were 0.984 and 0.980, respectively, in predicting the correct weed species labels. For the detection of Virginia buttonweed, the precision values of DenseNet were 0.996 and 0.994, respectively, while the recall values were 0.984 and 0.978, respectively. Similar trends were observed in the validation and testing datasets for EfficientNet-v2 and ResNet.
All three neural networks performed poorly at detecting and distinguishing crabgrass and tropical signalgrass growing in bermudagrass turf. Because of low precision and recall values, the F1 scores and MCC values of DenseNet, EfficientNet-v2, and ResNet never exceeded 0.918, 0.919, and 0.918, respectively, in the validation and testing datasets. The low F1 scores and MCC values indicate that the neural networks are more likely to mistakenly classify crabgrass as tropical signalgrass (or vice versa). This finding could likely attribute to the similarity in plant morphology between crabgrass and tropical signalgrass.
Detection and discrimination of weeds susceptible to herbicides
No obvious differences were observed among DenseNet, EfficientNet-v2, and ResNet for detecting and distinguishing weeds susceptible to ACCase-inhibitors, ALS-inhibitors, synthetic auxin herbicides, or bermudagrass without weed infestation (no herbicide) (Table 4).

Table 4 Training and testing results of various convolutional neural networks for detecting and discriminating the sub-images containing weeds susceptible to herbicides, or bermudagrass without weed infestation (no herbicide).
DenseNet, EfficientNet-v2, and ResNet achieved high F1 scores and MCC values (≥0.997) with high precision (≥0.996) and recall (≥ 0.997) in the validation datasets. All neural networks had slightly reduced precision and recall values in the testing datasets, but the F1 scores and MCC values never fell below 0.994.
These results suggest that convolutional neural networks can reliably detect and distinguish weeds susceptible to particular herbicides. Furthermore, it can be inferred that training the neural networks based on the susceptibility of weed species to herbicides could probably minimize the morphological similarity issue and hence improve weed detection accuracy.
Inference time of the convolutional neural networks
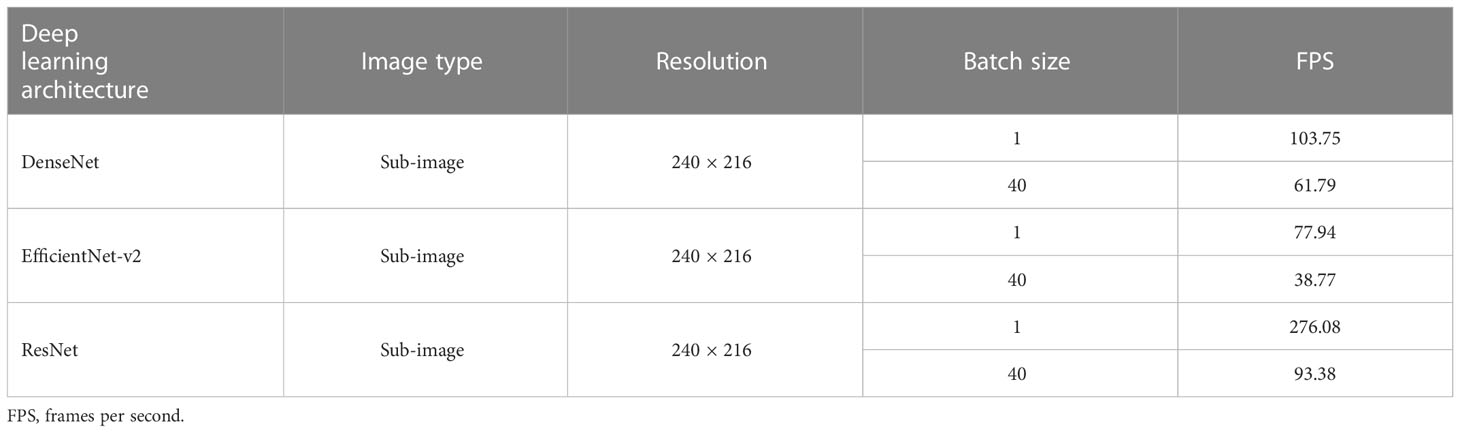
In addition to the weed detection accuracy, the inference time of the convolutional neural networks is also critical for real-time precision herbicide application. The FPS values of DenseNet, EfficientNet-v2, and ResNet were calculated by averaging the inference time of images from the testing dataset. Since the original images were captured at a resolution of 1,920 × 1,080 pixels, the detection speed with the full images was measured by processing the sub-images (240 × 216 pixels) with a batch size value of 40 (for simultaneously processing 40 sub-images).
All convolutional neural networks, including DenseNet, EfficientNet-v2, and ResNet, had an excellent detection speed (≥77.94fps) when detecting and distinguishing the sub-images with a batch size value of 1 (Table 5). DenseNet, with 61.79 full images detected per second, was 31.59 slower than ResNet but noticeably faster than EfficientNet-v2 when setting the batch size value as 40. ResNet demonstrated the fastest inference rate and outperformed the other convolutional neural networks on detection efficiency. However, the slow detection of EfficientNet-v2 may limit its potential applications.
Discussion
Deep learning methods for weed detection typically focus on distinguishing weed species, but various weed species with comparable plant morphological features may be found in the turfgrass. Thus, it is difficult for neural networks to achieve high accuracy of detection and discrimination for every weed species. Distinguishing different categories of weed species growing in turf based on their susceptibility to herbicides reduces the complexity of weed detection. By training the neural networks according to the susceptibility of weed species to herbicides, we achieved an excellent performance in weed detection. Moreover, this strategy allows the use of specific herbicides for precision spraying susceptible weeds, thus saving more herbicides.
When training convolutional neural networks for detecting weeds susceptible to herbicides, weed vegetation was grouped and labeled into three categories: weeds susceptible to ACCase-inhibitors, weeds susceptible to ALS-inhibitors, and weeds susceptible to synthetic auxin herbicides. ACCase-inhibitors, such as diclofop-methyl, can be applied in bermudagrass turf for POST control of various grassy weeds, while sethoxydim (cyclohexanedione), another ACCase-inhibitor, is used for POST control of grassy weeds growing in centipedegrass [Eremochloa ophiuroides (Munro) Hack.] (Neal et al., 1990; Tate et al., 2021). Synthetic auxin herbicides, such as 2,4-D and mecoprop, are POST herbicides that selectively control broadleaf weeds in bermudagrass turf (Grichar et al., 2008; Reed et al., 2013). ALS-inhibitors (e.g. halosulfuron, imazaquin, and trifloxysulfuron-sodium) can effectively control nutsedge weeds. However, it should be noted that certain ALS-inhibitors, such as halosulfuron and trifloxysulfuron-sodium, can also suppress or effectively control broadleaf weeds (McElroy and Martins, 2013). In this context, broadleaf and nutsedge weeds could be grouped into the same category when training the neural network for precision spraying the ALS-inhibitors that are effective for controlling both broadleaves and nutsedges growing in bermudagrass turf.
Deep learning neural networks, including image classification and object detection neural networks, can be developed and potentially integrated into the machine vision sub-system of a smart sprayer. Nevertheless, it should be noted that image classification neural networks alone do not localize weeds on the input images. Consequently, when utilizing image classification neural networks for weed detection, a smart sprayer likely generates a considerably larger spraying output area than the area covered by weeds. In the present work, localizing weeds with image classification neural networks could be realized by cropping the input image into multiple grid cells (sub-images) and identifying the grid cells containing weeds.
In the present study, original images (1,920 × 1,080 pixels) were divided into 40 grid cells (sub-images with a resolution of 240 × 216 pixels) for training and testing the image classification neural networks. Spraying areas can be localized by detecting if the grid cells contain weeds. When developing a precision spraying system, custom software can be programmed to generate grid cell maps on the input images and realize precision herbicide application by detecting if the grid cells contain weeds susceptible to the herbicides. To realize precision herbicide spraying, a binary (on/off) input command can be implemented via a nozzle control system to turn off the spray nozzles over the weed-free cells while the nozzles corresponding to the grid cells containing weeds need to be turned on.
While the convolutional neural networks achieved high classification rates for detecting and distinguishing weeds susceptible to herbicides, it should be noted that when weeds susceptible to different herbicides are grown too close or occluded, the neural networks would not effectively distinguish weed categories based on their susceptibility to the herbicides because the grid cell contains multiple targets. Although such a case may result in missed detection, this is hardly an issue in field applications because the weed infestation zone has been detected, and one of the herbicides will be sprayed onto the susceptible weeds.
It was reported that the training image size could significantly affect the reliability of image classification neural networks for weed detection (Zhuang et al., 2021; Yang et al., 2022b). For example, Zhuang et al. observed increased classification accuracy (high recall values) with AlexNet and VGGNet when they were trained with images of 200 × 200 pixels than 300 × 300 or 400 × 400 pixels; however, increasing training image quantities diminished the differences in detection accuracy (Zhuang et al., 2021). In the present study, each sub-image (240 × 216 pixels) represented a physical size of 10 cm × 9 cm. When the convolutional neural networks are integrated into the machine vision sub-system of smart sprayers for precision herbicide application, the nozzles should generate the same or slightly larger spraying outputs to cover the grid cells. An additional investigation is needed to investigate the implications of training image sizes and quantities on the performances of neural networks for weed detection in turf.
Conclusions
The present research demonstrated the reliability and effectiveness of using convolutional neural networks to detect and distinguish weeds growing in bermudagrass turf based on their susceptibility to herbicides. All convolutional neural networks, including DenseNet, EfficientNet-v2, and ResNet achieved excellent F1 scores (≥ 0.995) and MCC values (≥ 0.994) in the validation and testing datasets to detect and distinguish weeds susceptible to ACCase-inhibitors, ALS-inhibitors, and synthetic auxin herbicides, or bermudagrass turf without weed infestation (no herbicide). In addition, DenseNet, EfficientNet-v2, and ResNet had an excellent detection speed (≥77.94fps) when detecting and distinguishing the sub-images with a resolution of 240 × 216 pixels. For detecting the original/full images (1,920 × 1,080 pixels), ResNet demonstrated the fastest inference rate and outperformed the other convolutional neural networks on detection efficiency (93.38fps). Effective detection and discrimination of weeds susceptible to herbicides enable the smart sprayer to spray particular herbicides to control susceptible weeds, thereby significantly reducing herbicide input. Based on the high-level performance, we conclude that the proposed method is highly suitable for integrating into the machine vision sub-system of smart sprayers for the precision control of weeds while growing in turf.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
XJ conceived the research ideas and designed the experiments under the guidance of YC and JY. TL, PM, and JY collected the data and conducted the data analysis. XJ drafted the manuscript. PM, YC, and JY edited and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32072498) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (Grant No. KYCX22_1051).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, A., Jain, S., Gour, M., Vankudothu, S. (2021). Tomato plant disease detection using transfer learning with c-GAN synthetic images. Comput. Electron. Agric. 187, 106279. doi: 10.1016/j.compag.2021.106279
Atefi, A., Ge, Y., Pitla, S., Schnable, J. (2021). Robotic technologies for high-throughput plant phenotyping: contemporary reviews and future perspectives. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.611940
Bakhshipour, A., Jafari, A., Nassiri, S. M., Zare, D. (2017). Weed segmentation using texture features extracted from wavelet sub-images. Biosyst. Eng. 157, 1–12. doi: 10.1016/j.biosystemseng.2017.02.002
Bao, W., Yang, B., Chen, B. (2021). 2-hydr_ensemble: lysine 2-hydroxyisobutyrylation identification with ensemble method. Chemom. Intell. Lab. Syst. 215, 104351. doi: 10.1016/j.chemolab.2021.104351
Bao, W., Yuan, C.-A., Zhang, Y., Han, K., Nandi, A. K., Honig, B., et al. (2017). Mutli-features prediction of protein translational modification sites. IEEE/ACM Trans. Comput. Biol. Bioinform. 15 (5), 1453–1460. doi: 10.1109/TCBB.2017.2752703
Busey, P. (2003). Cultural management of weeds in turfgrass: a review. Crop Sci. 43 (6), 1899–1911. doi: 10.2135/cropsci2003.1899
Chen, D., Lu, Y., Li, Z., Young, S. (2022). Performance evaluation of deep transfer learning on multi-class identification of common weed species in cotton production systems. Comput. Electron. Agric. 198, 107091. doi: 10.1016/j.compag.2022.107091
Chicco, D., Jurman, G. (2020). The advantages of the matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21 (1), 1–13. doi: 10.1186/s12864-019-6413-7
Collobert, R., Weston, J. (2008). “A unified architecture for natural language processing: Deep neural networks with multitask learning,” in Proceedings of the 25th international conference on Machine learning (Helsinki, Finland:ACM). 160–167.
Collobert, R., Weston, J., Bottou, L., Karlen, M., Kavukcuoglu, K., Kuksa, P. (2011). Natural language processing (almost) from scratch. J. Mach. Learn. Res. 12 (ARTICLE), 2493–2537.
Dai, X., Xu, Y., Zheng, J., Song, H. (2019). Analysis of the variability of pesticide concentration downstream of inline mixers for direct nozzle injection systems. Biosyst. Eng. 180, 59–69. doi: 10.1016/j.biosystemseng.2019.01.012
Davis, A. S., Frisvold, G. B. (2017). Are herbicides a once in a century method of weed control? Pest Manage. science. 73 (11), 2209–2220. doi: 10.1002/ps.4643
Fennimore, S. A., Slaughter, D. C., Siemens, M. C., Leon, R. G., Saber, M. N. (2016). Technology for automation of weed control in specialty crops. Weed Technol. 30 (4), 823–837. doi: 10.1614/WT-D-16-00070.1
Grichar, W. J., Baumann, P. A., Baughman, T. A., Nerada, J. D. (2008). Weed control and bermudagrass tolerance to imazapic plus 2, 4-d. Weed Technol. 22 (1), 97–100. doi: 10.1614/WT-07-097.1
Gu, J., Wang, Z., Kuen, J., Ma, L., Shahroudy, A., Shuai, B., et al. (2018). Recent advances in convolutional neural networks. Pattern Recogn. 77, 354–377. doi: 10.1016/j.patcog.2017.10.013
Hamuda, E., Glavin, M., Jones, E. (2016). A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric. 125, 184–199. doi: 10.1016/j.compag.2016.04.024
Hasan, A. M., Sohel, F., Diepeveen, D., Laga, H., Jones, M. G. (2021). A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 184, 106067. doi: 10.1016/j.compag.2021.106067
He, T., Liu, Y., Yu, Y., Zhao, Q., Hu, Z. (2020). Application of deep convolutional neural network on feature extraction and detection of wood defects. Measurement 152, 107357. doi: 10.1016/j.measurement.2019.107357
He, K., Zhang, X., Ren, S., Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition (Las Vegas, USA:IEEE). 770–778.
Hinton, G., Deng, L., Yu, D., Dahl, G. E., Mohamed, A., Jaitly, N., et al. (2012). Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process. Magazine. 29 (6), 82–97. doi: 10.1109/MSP.2012.2205597
Huang, G., Liu, Z., van der Maaten, L., Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition (Las Vegas, USA:IEEE). 4700–4708.
Jiang, H., Jiang, X., Ru, Y., Wang, J., Xu, L., Zhou, H. (2020). Application of hyperspectral imaging for detecting and visualizing leaf lard adulteration in minced pork. Infrared Phys. Technology. 110, 103467. doi: 10.1016/j.infrared.2020.103467
Jin, X., Bagavathiannan, M., McCullough, P. E., Chen, Y., Yu, J. (2022). A deep learning-based method for classification, detection, and localization of weeds in turfgrass. Pest Manage. Sci 78(11), 4809–4821. doi: 10.1002/ps.7102
Jin, X., Che, J., Chen, Y. (2021). Weed identification using deep learning and image processing in vegetable plantation. IEEE Access. 9, 10940–10950. doi: 10.1109/ACCESS.2021.3050296
Johnson, B. (1977). Winter annual weed control in dormant bermudagrass turf. Weed Sci. 25 (2), 145–150. doi: 10.1017/S0043174500033142
Jordan, M. I., Mitchell, T. M. (2015). Machine learning: Trends, perspectives, and prospects. Science 349 (6245), 255–260. doi: 10.1126/science.aaa8415
Kamilaris, A., Prenafeta-Boldú, F. X. (2018). Deep learning in agriculture: A survey. Comput. Electron. Agric. 147, 70–90. doi: 10.1016/j.compag.2018.02.016
Khaki, S., Wang, L. (2019). Crop yield prediction using deep neural networks. Front. Plant science. 10, 621. doi: 10.3389/fpls.2019.00621
LeCun, Y., Bengio, Y., Hinton, G. (2015). Deep learning. Nature 521 (7553), 436–444. doi: 10.1038/nature14539
Liakos, K. G., Busato, P., Moshou, D., Pearson, S., Bochtis, D. (2018). Machine learning in agriculture: A review. Sensors 18 (8), 2674. doi: 10.3390/s18082674
Liu, B., Bruch, R. (2020). Weed detection for selective spraying: a review. Curr. Robotics Rep. 1 (1), 19–26. doi: 10.1007/s43154-020-00001-w
Lu, J., Behbood, V., Hao, P., Zuo, H., Xue, S., Zhang, G. (2015). Transfer learning using computational intelligence: A survey. Knowledge-Based Systems. 80, 14–23. doi: 10.1016/j.knosys.2015.01.010
Marchand, P. A., Robin, D. (2019). Evolution of directive (EC) no 128/2009 of the European parliament and of the council establishing a framework for community action to achieve the sustainable use of pesticides. J. Regul. Sci., 7, 1–7. doi: 10.21423/jrs-v07marchand
Martinelli, F., Scalenghe, R., Davino, S., Panno, S., Scuderi, G., Ruisi, P., et al. (2015). Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 35 (1), 1–25. doi: 10.1007/s13593-014-0246-1
McCullough, P. E., Yu, J., Shilling, D. G., Czarnota, M. A., Johnston, C. R. (2015). Biochemical effects of imazapic on bermudagrass growth regulation, broomsedge (Andropogon virginicus) control, and MSMA antagonism. Weed Sci. 63 (3), 596–603. doi: 10.1614/WS-D-14-00183.1
McElroy, J., Martins, D. (2013). Use of herbicides on turfgrass. Planta daninha. 31, 455–467. doi: 10.1590/S0100-83582013000200024
Melland, A. R., Silburn, D. M., McHugh, A. D., Fillols, E., Rojas-Ponce, S., Baillie, C., et al. (2016). Spot spraying reduces herbicide concentrations in runoff. J. Agric. Food Chem. 64 (20), 4009–4020. doi: 10.1021/acs.jafc.5b03688
Mennan, H., Jabran, K., Zandstra, B. H., Pala, F. (2020). Non-chemical weed management in vegetables by using cover crops: A review. Agronomy 10 (2), 257. doi: 10.3390/agronomy10020257
Milesi, C., Elvidge, C., Dietz, J., Tuttle, B., Nemani, R., Running, S. (2005). A strategy for mapping and modeling the ecological effects of US lawns. J. Turfgrass Manage. 1 (1), 83–97.
Neal, J. C., Bhowmik, P. C., Senesac, A. F. (1990). Factors influencing fenoxaprop efficacy in cool-season turfgrass. Weed Technol. 4 (2), 272–278. doi: 10.1017/S0890037X00025380
Ngo, T. D., Bui, T. T., Pham, T. M., Thai, H. T., Nguyen, G. L., Nguyen, T. N. (2021). Image deconvolution for optical small satellite with deep learning and real-time GPU acceleration. J. Real-Time Image Processing. 18 (5), 1697–1710. doi: 10.1007/s11554-021-01113-y
Pantazi, X.-E., Moshou, D., Bravo, C. (2016). Active learning system for weed species recognition based on hyperspectral sensing. Biosyst. Eng. 146, 193–202. doi: 10.1016/j.biosystemseng.2016.01.014
Peng, H., Li, Z., Zhou, Z., Shao, Y. (2022). Weed detection in paddy field using an improved RetinaNet network. Comput. Electron. Agric. 199, 107179. doi: 10.1016/j.compag.2022.107179
Perez, A., Lopez, F., Benlloch, J., Christensen, S. (2000). Colour and shape analysis techniques for weed detection in cereal fields. Comput. Electron. Agric. 25 (3), 197–212. doi: 10.1016/S0168-1699(99)00068-X
Pimentel, D., Burgess, M. (2012). Small amounts of pesticides reaching target insects. Springer, 14, 1–2. doi: 10.1007/s10668-011-9325-5
Potter, D. A., Braman, S. K. (1991). Ecology and management of turfgrass insects. Annu. Rev. Entomol. 36 (1), 383–406. doi: 10.1146/annurev.en.36.010191.002123
Razfar, N., True, J., Bassiouny, R., Venkatesh, V., Kashef, R. (2022). Weed detection in soybean crops using custom lightweight deep learning models. J. Agric. Food Res. 8, 100308. doi: 10.1016/j.jafr.2022.100308
Reed, T. V., Yu, J., McCullough, P. E. (2013). Aminocyclopyrachlor efficacy for controlling Virginia buttonweed (Diodia virginiana) and smooth crabgrass (Digitaria ischaemum) in tall fescue. Weed Technol. 27 (3), 488–491. doi: 10.1614/WT-D-12-00159.1
Saleem, M. H., Potgieter, J., Arif, K. M. (2019). Plant disease detection and classification by deep learning. Plants 8 (11), 468. doi: 10.3390/plants8110468
Shi, J., Li, Z., Zhu, T., Wang, D., Ni, C. (2020). Defect detection of industry wood veneer based on NAS and multi-channel mask r-CNN. Sensors 20 (16), 4398. doi: 10.3390/s20164398
Slaughter, D. C., Giles, D. K., Downey, D. (2008). Autonomous robotic weed control systems: A review. Comput. Electron. Agric. 61 (1), 63–78. doi: 10.1016/j.compag.2007.05.008
Sokolova, M., Lapalme, G. (2009). A systematic analysis of performance measures for classification tasks. Inf. Process. management. 45 (4), 427–437. doi: 10.1016/j.ipm.2009.03.002
Subeesh, A., Bhole, S., Singh, K., Chandel, N., Rajwade, Y., Rao, K., et al. (2022). Deep convolutional neural network models for weed detection in polyhouse grown bell peppers. Artif. Intell. Agriculture. 6, 47–54. doi: 10.1016/j.aiia.2022.01.002
Tang, J.-L., Chen, X.-Q., Miao, R.-H., Wang, D. (2016). Weed detection using image processing under different illumination for site-specific areas spraying. Comput. Electron. Agric. 122, 103–111. doi: 10.1016/j.compag.2015.12.016
Tan, M., Le, Q. (2019). “Efficientnet: Rethinking model scaling for convolutional neural networks,” in International Conference on Machine Learning. PMLR (Long Beach, USA:PMLR). 6105–6114.
Tate, T. M., McCullough, P. E., Harrison, M. L., Chen, Z., Raymer, P. L. (2021). Characterization of mutations conferring inherent resistance to acetyl coenzyme a carboxylase-inhibiting herbicides in turfgrass and grassy weeds. Crop Sci. 61 (5), 3164–3178. doi: 10.1002/csc2.20511
Van Klompenburg, T., Kassahun, A., Catal, C. (2020). Crop yield prediction using machine learning: A systematic literature review. Comput. Electron. Agric. 177, 105709. doi: 10.1016/j.compag.2020.105709
Wang, A., Zhang, W., Wei, X. (2019). A review on weed detection using ground-based machine vision and image processing techniques. Comput. Electron. Agric. 158, 226–240. doi: 10.1016/j.compag.2019.02.005
Yang, J., Bagavathiannan, M., Wang, Y., Chen, Y., Yu, J. (2022b). A comparative evaluation of convolutional neural networks, training image sizes, and deep learning optimizers for weed detection in alfalfa. Weed Technol. 36 (4), 512–522. doi: 10.1017/wet.2022.46
Yang, B., Bao, W., Wang, J. (2022a). Active disease-related compound identification based on capsule network. Brief. Bioinform. 23 (1), bbab462. doi: 10.1093/bib/bbab462
Yu, J., Boyd, N. S. (2018). Weed control and tolerance of sulfonylurea herbicides in caladium. Weed Technol. 32 (4), 424–430. doi: 10.1017/wet.2018.30
Yu, J., McCullough, P. E., Czarnota, M. A. (2018). Annual bluegrass (Poa annua) biotypes exhibit differential levels of susceptibility and biochemical responses to protoporphyrinogen oxidase inhibitors. Weed Sci. 66 (5), 574–580. doi: 10.1017/wsc.2018.30
Yu, J., Schumann, A. W., Cao, Z., Sharpe, S. M., Boyd, N. S. (2019a). Weed detection in perennial ryegrass with deep learning convolutional neural network. Front. Plant science. 10. doi: 10.3389/fpls.2019.01422
Yu, J., Schumann, A. W., Sharpe, S. M., Li, X., Boyd, N. S. (2020). Detection of grassy weeds in bermudagrass with deep convolutional neural networks. Weed Sci. 68 (5), 545–552. doi: 10.1017/wsc.2020.46
Yu, J., Sharpe, S. M., Schumann, A. W., Boyd, N. S. (2019b). Deep learning for image-based weed detection in turfgrass. Eur. J. Agron. 104, 78–84. doi: 10.1016/j.eja.2019.01.004
Yu, J., Sharpe, S. M., Schumann, A. W., Boyd, N. S. (2019c). Detection of broadleaf weeds growing in turfgrass with convolutional neural networks. Pest Manage. Science. 75 (8), 2211–2218. doi: 10.1002/ps.5349
Zhang, H., Ge, Y., Xie, X., Atefi, A., Wijewardane, N. K., Thapa, S. (2022). High throughput analysis of leaf chlorophyll content in sorghum using RGB, hyperspectral, and fluorescence imaging and sensor fusion. Plant Methods 18 (1), 1–17. doi: 10.1186/s13007-022-00892-0
Zhou, H., Zhuang, Z., Liu, Y., Liu, Y., Zhang, X. (2020). Defect classification of green plums based on deep learning. Sensors 20 (23), 6993. doi: 10.3390/s20236993
Keywords: deep learning, convolutional neural networks, weed detection, herbicide susceptibility, precision herbicide application
Citation: Jin X, Liu T, McCullough PE, Chen Y and Yu J (2023) Evaluation of convolutional neural networks for herbicide susceptibility-based weed detection in turf. Front. Plant Sci. 14:1096802. doi: 10.3389/fpls.2023.1096802
Received: 12 November 2022; Accepted: 23 January 2023;
Published: 01 February 2023.
Edited by:
Mehedi Masud, Taif University, Saudi ArabiaReviewed by:
Wenzheng Bao, Xuzhou University of Technology, ChinaYanbo Huang, United States Department of Agriculture (USDA), United States
Copyright © 2023 Jin, Liu, McCullough, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Chen, Y2hlbnlvbmdqc25qQDE2My5jb20=; Jialin Yu, amlhbGluLnl1QHBrdS1pYWFzLmVkdS5jbg==
 Xiaojun Jin
Xiaojun Jin Teng Liu
Teng Liu Patrick E. McCullough3
Patrick E. McCullough3