- 1Key Laboratory for Biology of Horticultural Plants, Ministry of Education, College of Horticulture & Forestry Sciences, Huazhong Agricultural University, Wuhan, China
- 2School of Life Sciences, Guizhou Normal University, Guiyang, China
Pyrethrins, synthesized in the perennial plant Tanacetum cinerariifolium, are a class of terpene mixtures with high insecticidal activity and low human toxicity, which are widely used in plant-derived pesticides. Numerous studies have identified multiple pyrethrins biosynthesis enzymes, which can be enhanced by exogenous hormones such as methyl jasmonate (MeJA). However, the mechanism by which hormone signaling regulates pyrethrins biosynthesis and the potential involvement of certain transcription factors (TFs) remain unclear. In this study, we found that the expression level of a TF in T. cinerariifolium was significantly increased after treatment with plant hormones (MeJA, abscisic acid). Subsequent analysis identified this TF as a member of the basic region/leucine zipper (bZIP) family and was thus named TcbZIP60. TcbZIP60 was localized in the nucleus, suggesting that it is involved in the transcription process. The expression profiles of TcbZIP60 were similar to those of pyrethrins synthesis genes in different flower organs and at different flowering stages. Furthermore, TcbZIP60 could directly bind to the E-box/G-box motifs in the promoters of the pyrethrins synthesis genes TcCHS and TcAOC to activate their expression. Transient overexpression of TcbZIP60 increased the expression levels of pyrethrins biosynthesis genes, leading to the significant accumulation of pyrethrins. Silencing of TcbZIP60 significantly downregulated pyrethrins accumulation and the expression of related genes. Overall, our results reveal a novel TF, TcbZIP60, that regulates both the terpenoid and jasmonic acid pathways of pyrethrins biosynthesis in T. cinerariifolium.
1 Introduction
Tanacetum cinerariifolium is an economically important horticultural ornamental plant in the Asteraceae family because it produces a class of insecticidal compounds called pyrethrins (Casida, 1973). Pyrethrins are widely used as plant-derived pesticides owing to their broad-spectrum and highly effective insecticidal activity, easy decomposition into harmless substances under light exposure, and low toxicity to mammals (Casida and Quistad, 1995). T. cinerariifolium has been grown as an environmentally friendly commercial crop in more than a dozen countries in Africa, Asia, Europe, and South America (Lybrand et al., 2020).
Natural pyrethrins are composed of six types of monoterpene esters with similar structures, including pyrethrin I and II, jasmolin I and II, and cinerin I and II (Ramirez et al., 2013). Pyrethrins are obtained by esterification between an acid moiety (pyrethric acid or chrysanthemic acid) and an alcohol moiety (pyrethrolone, cinerolone, or jasomolone) (Freemont et al., 2016; Khan et al., 2017). Monoterpene acids are derived from the methylerythritol-4-phosphate pathway. The first step of the reaction is the formation of chrysanthemyl diphosphate catalyzed by chrysanthemyl diphosphate synthase (CDS) from dimethylallyl diphosphate, which in turn is catalyzed by CDS to trans-chrysanthemol. CDS is a bifocal enzyme that is also known as chrysanthemol synthase (CHS) (Rivera et al., 2001; Yang et al., 2014; Hu et al., 2018; Lybrand et al., 2020). Subsequently, pyrethric acid is formed under the catalysis of alcohol dehydrogenase and aldehyde dehydrogenase (ALDH). Ketols are derived from the oxylipin alcohol pathway. In this way, linolenic acid is used as a substrate to form ketol through lipoxygenase, allene oxide synthase, allene oxide cyclase (AOC), oxo-phytodienoic acid reductase, jasmone hydroxylase, and pyrethrolone synthase (Song et al., 1993; Creelman and Mullet, 1997; Tijet and Brash, 2002; Ramirez et al., 2013; Li et al., 2018). Finally, pyrethrins are synthesized by the two precursor compounds under the catalysis of GDSL lipase-like protein (GLIP) (Kikuta et al., 2012). Although pyrethrins synthesis pathways have been largely clarified, there are relatively few studies on the mechanisms of pyrethrins synthesis at the transcriptional level.
Although pyrethrins are insecticidal compounds, on average, the pyrethrins content of the leaves of plants is only approximately 0.1% (dry weight), which is much lower than that of the flowers (1–2% dry weight) (Varga et al., 2021). A variety of methods have been evaluated to increase the yield of pyrethrins, including optimizing the cultivation mode or modifying the breeding strategy (Li et al., 2014). However, these strategies have not led to a major increase in the pyrethrins content of the leaves. Pyrethrins biosynthesis of the alcohol moiety derives from the jasmonic acid (JA) pathway (Hu et al., 2018). In particular, methyl jasmonate (MeJA) stimulates the overaccumulation of many secondary metabolites in plants. In a previous study, MeJA treatment was shown to induce the accumulation of pyrethrins for a short period, possibly via regulating multiple pyrethrins biosynthesis-related genes (Li et al., 2018). However, persistently treating MeJA to plants is not a robust strategy to maintain pyrethrins production.
In the process of plant evolution, many new transcription factor (TF) families have emerged during adaptation to changing environments or in response to exogenous hormones, further affecting the synthesis of secondary metabolites in plants by regulating the expression of target genes (Li et al., 2020; Chen et al., 2022). Therefore, the application of various hormones to plants is a feasible strategy to discover the critical TFs involved in pyrethrins biosynthesis. Basic leucine zipper (bZIP) is one of the most diverse TF families of plants, playing an important role in plant stress signal transduction, pathogen defense, and flower development (Wigge et al., 2005; Tang and Page, 2013; Sagor et al., 2015; Chen et al., 2021). All bZIP TFs possess a conserved bZIP domain, which is usually composed of 60–80 amino acids, including a basic DNA-binding region and a leucine (Leu) zipper domain that can recognize and combine cis-acting elements such as E-box (5′-CANNTG-3′), G-box (5′-CACGTG-3′), and ACGT-box (5′-ACGT-3′) in the gene promoters (Jakoby et al., 2002; Wolfgang et al., 2018).
Many studies have confirmed that bZIP TFs are involved in the response to signaling pathways and abiotic/biotic stress, including light signaling, abscisic acid (ABA) signaling, drought, and pathogen infections (Uno et al., 2000; Sornaraj et al., 2016; Wolfgang et al., 2018). Moreover, bZIP TFs also regulate the biosynthesis of numerous secondary metabolites in plants. For example, in Artemisia annua, the bZIP TF AaHY5 directly affects artemisinin biosynthesis through interaction with AaCOP1 (Hao et al., 2019). In addition, AabZIP1 was reported to activate ADS and CYP71AV1 gene expression under ABA treatment to promote artemisinin biosynthesis (Zhang et al., 2015). Along with a role in the biosynthesis of terpenoids, bZIP TFs also positively or negatively regulate flavonoids and alkaloids biosynthesis by activating the target genes in each biosynthesis pathway (Sibéril et al., 2001; Zhang et al., 2011; Pal et al., 2015; Mao et al., 2021). However, the downstream target genes of TFs are mainly involved in consecutive steps of metabolites synthesis or the same precursor-derived pathway. bZIP TFs regulating genes independently in different biosynthesis pathways are rarely reported.
In this study, we found that MeJA treatment activates reporter gene expression driven by pyrethrins biosynthesis-related gene promoters to increase the pyrethrins content of T. cinerariifolium. Based on these findings and the MeJA-treated transcriptome, we successfully identified a novel bZIP TF, named TcbZIP60. Further experiments demonstrated that TcbZIP60 responds to not only MeJA but also to a variety of other plant hormone signals, and can directly bind to the promoter region of the pyrethrins biosynthesis genes TcCHS and TcAOC. The transient overexpression and virus-induced gene silencing (VIGS) of TcbZIP60 in T. cinerariifolium leaves further confirmed the positive role of TcbZIP60 in regulating the biosynthesis of both terpenoid- and JA-derived pyrethrins.
2 Materials and methods
2.1 Plant materials and growth conditions
Tanacetum cinerariifolium ‘W99’ plants grown in the flower base of Huazhong Agricultural University, Wuhan, China were used in this study. This cultivar has the advantages of easily rooting cuttings, rapid growth, and high pyrethrins content and, therefore, was suitable for the present experiments. W99 seedlings were subcultured in half-strength Murashige and Skoog medium for 1 month (25°C, 16 h light/8 h dark) prior to transient treatment with MeJA or ABA. The rooted cuttings were sprayed with 5 ml of 2 mM MeJA/ABA solution as a single foliar application. Leaves were sampled in triplicate at 0 (control), 2, 4, 6, 8, 12, and 24 h after treatment, immediately placed in liquid nitrogen, and stored at −80°C until further analysis.
Flower heads and leaves were harvested at seven flowering stages: S1, well-developed closed buds; S2, ray floret limb in vertical position; S3, ray floret limb in horizontal position and disc florets of the outermost whorl open; S4, three whorls of disc florets open; S5, all disc florets open; S6, termed the early overblown condition, the disc floret color is faded but the ray florets remain intact; S7, termed the late overblown condition, the disc florets retain little color and the ray florets are desiccated. Three biological replicates were collected for each sample, which were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
2.2 Exogenous MeJA effect on TcCHS and TcGLIP promoter activity
Our previously established transgenic chrysanthemum (Chrysanthemum × morifolium) harboring TcCHS-promoter-driven GFP and tobacco (Nicotiana tabacum) harboring TcGLIP-promoter-driven GUS were treated with MeJA (Sultana et al., 2015). The plants were sprayed with 5 ml of 300 μM MeJA dissolved in 0.8% ethanol as a single foliar application. Leaves were collected in triplicate at 0 (control) and 12 h after treatment, and were immediately frozen in liquid nitrogen. The GFP expression level was determined by quantitative real-time PCR (qRT-PCR) analysis. The GUS activity was measured as previously described (Luo et al., 2013).
2.3 Gene cloning and bioinformatic analysis
Total RNA was extracted from the collected samples using the standard phenol–chloroform extraction method. The first-strand cDNA was synthesized using the EasyScript® One-step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China), in accordance with the manufacturer’s instructions, using the total RNA extracts as the template. The TcbZIP60 gene sequence of T. cinerariifolium was identified from the inflorescence transcriptome database determined by our laboratory. The Primer Premier software was used to design specific primers for amplifying the open reading frame (ORF) fragment. The primers are listed in Supplementary Table S1. The cis-acting elements of associated genes (TcCHS, TcAOC, TcALDH, and TcGLIP) were determined using the PlantCARE database. The cis-regulatory E-box and G-box elements were determined using PlantPAN 3.0 (Chow et al., 2019).
2.4 Real-time quantitative PCR
Total RNA was extracted using the Ultrapure RNA Kit (CWBIO, Beijing, China), and the EasyScript One-step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech) was used to reverse-transcribe the RNA into cDNA. A qRT-PCR analysis was performed using a LightCycler® 96 Real-Time PCR System (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions, with SYBR Premix Ex Taq II (Takara, Kusatsu, Japan) and sequence-specific primers (listed in Supplementary Table S1). Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was used as the internal reference gene (Li et al., 2019). The relative expression levels were calculated using the 2−ΔΔCt method with three biological and three technical replicates (Livak and Schmittgen, 2001). Student’s two-tailed t-test was used to determine statistical significance. Differences at P < 0.05 were considered to be significant and those at P < 0.01 were considered to be highly significant.
2.5 Phylogenetic analysis of TcbZIP60
The ORF of TcbZIP60 was cloned from T. cinerariifolium cDNA using the sequence-specific primers TcbZIP60-F and TcbZIP60-R (Supplementary Table S1). Arabidopsis sequences homologous to the TcbZIP60 sequence were identified by a Basic Local Alignment Search Tool (BLAST) search against The Arabidopsis Information Resource (TAIR) database, and homologous sequences from other plant species were identified by a BLAST search against the National Center for Biotechnology Information (NCBI) database. A phylogenetic tree was constructed with the maximum likelihood method using MEGA X software with 1000 bootstrap replications (Kumar et al., 2018). The GenBank accession numbers of the genes used in the analysis are listed in Supplementary Tables S2 and S3.
2.6 Subcellular localization of TcbZIP60
The TcbZIP60 coding sequence was amplified by PCR using sequence-specific primers (Supplementary Table S1). The gene was cloned using the ClonExpress® II One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The full-length sequence of TcbZIP60 (without the stop codon) was spliced into the pSuper-1300 GFP vector. The recombinant product was transferred into Escherichia coli strain DH5α, then sequenced, and the correct plasmid was extracted and constructed. The resultant plasmid was introduced into Agrobacterium tumefaciens strain GV3101, and then co-infiltrated into Nicotiana benthamiana leaves together with the RFP-NLS plasmid (a nuclear marker). The empty vector was used as a control. After 72 h of weak light exposure, fluorescence signals were observed with a confocal laser scanning microscope (TCS-SP8, Leica, Wetzlar, Germany).
2.7 Yeast one-hybrid assay
The full-length cDNA of TcbZIP60 was cloned into the pGADT7 vector used for homologous recombination, and the promoter sequences of TcCHS and TcAOC were cloned separately into the pHis2.1 vector. The resultant pGADT7:TcbZIP60 plasmid was co-transformed into yeast strain Y187, together with pHis2.1:TcCHS or pHis2.1:TcAOC, using the Super Yeast Transformation Kit (Coolaber, Beijing, China). The transformed yeast cells were selected on DDO (SD/−Leu/−Trp) medium and interreacted with TDO (SD/−Leu/−Trp/−His) medium at 30°C for 3 days.
2.8 Dual-luciferase reporter assay
To generate reporter constructs, and in accordance with the ORF sequence of the cloned TcbZIP60 gene and the map of the plant expression vector, the downstream sequences of the gene promoter region (for TcCHS and TcAOC) were cloned into the linearized pGreenII0800-LUC vector. To generate effector constructs, the coding sequence of TcbZIP60 was cloned into the linearized pGreenII62-SK vector under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The resultant vectors were transiently co-expressed in N. benthamiana leaves. Luminescence was detected using the LB 985 Nightshade system (Berthold, Bad Wildbad, Germany). Introduction of the pGreen-62-SK empty vector and the pGreen-62-SK/pGreenII-0800-LUC : TcCHS/TcAOC constructs served as negative controls. Three biological replicates per treatment were measured.
The CDS of TcbZIP60 was ligated into pGreenII62-SK vector to generate an effector plasmid, while TcCHS/TcAOC were fused into the vector pGreenII0800-LUC to produce the reporter plasmids. The effector, each of the two reporter constructs were co-transformed into A. tumefaciens GV3101, respectively. Transient expression assay in N. benthamiana leaves was as described previously (Geng and Liu, 2018). The activities of firefly luciferase (LUC) and Renilla luciferase (REN) were measured using the Dual-Luciferase® Reporter Assay System (Promega, WI, USA). The promoter activity was expressed as the ratio of LUC to REN (Supplementary Figure S1).
2.9 Electrophoretic mobility shift assay
The ORF without the TcbZIP60 terminator codon was used to generate the pET6×HN-C vector protein, which was fused into the N-terminal frame of 6×His, and the vector pET6×HN-C-TcbZIP60 was then transformed into E. coli strain Rosetta (DE3). For the induced recombinant protein, 0.5 mM IPTG was used, and the cultures were incubated at 18°C for 16 h. Then, Ni2+–nitrilotriacetic acid was used to purify the recombinant proteins. For the electrophoretic mobility shift assay (EMSA), the promoter fragments of TcCHS and TcAOC containing E-box or G-box cis-regulatory elements labeled with fluorescein amidite (FAM) as probes, the same but unlabeled DNA fragments, and cis-element mutant DNA fragments were used as competitors in the assay. After performing the EMSA assays, FAM-labeled DNA was detected from the chemiluminescent signal.
2.10 Transient overexpression of TcbZIP60 in T. cinerariifolium leaves
The full-length TcbZIP60 coding sequence was cloned into the HindIII-linearized pGreenII62-SK vector downstream of the CaMV 35S promoter using gene-specific primers (Supplementary Table S1). The experimental method followed a previously described procedure (Jung et al., 2015). The pGreenII62-SK : TcbZIP60 vector was co-transformed together with the helper plasmid pSoup19 into A. tumefaciens strain GV3101. The pSoup19-transformed Agrobacterium cells were inoculated into YEB liquid medium containing 100 mg/l kanamycin and cultured on a rotating shaker at 28°C for 12 h. The supernatant was removed and resuspended in MES (containing 100 μM acetosyringone) to attain the final optical density (OD600 = 0.6). Leaves of T. cinerariifolium were placed in a 500 ml beaker containing a suspension of A. tumefaciens, and the beaker was then placed in a vacuum chamber at 0.23 ATM for 5 min. The soaked leaves were then dried with filter paper and stored in a petri dish lined with moist filter paper in the base. After 4 days of culture, leaves were sampled for qRT-PCR and high-performance liquid chromatography (HPLC) analyses. To determine the effect of transient expression, transient overexpression of GUS in T. cinerariifolium leaves was verified by staining with X-Gluc reagent (Supplementary Figure S2).
2.11 VIGS assay
The Tobacco rattle virus (TRV)-based vectors pTRV1 and pTRV2 were used for the VIGS assay (Supplementary Table S1). The pTRV2:TcbZIP60 and pTRV1 plasmids were transformed into chemically active A. tumefaciens strain GV3101 cells using a liquid nitrogen freeze–thaw method. The positive transformant cells were inoculated into YEB liquid medium containing 100 mg/l kanamycin, incubated overnight on a shaker at 28°C, then reactivated in an infiltrating buffer (pH = 5.6) containing 10 mM MgCl2, 10 mM MES, and 100 µM acetoeugenone, and adjusted to OD600 = 0.6. After standing for 3 h, Agrobacterium cells containing the pTRV2 plasmid and pTRV1 plasmid were mixed (1:1, v/v), and the bacterial solution was injected through the adaxial epidermis of T. cinerariifolium leaves using a needle-free syringe. The leaves were then incubated in a culture room in the dark for 3 days and under light for 11 days. The pTRV1 and pTRV2 vectors were used as the control group. The method followed a previously described procedure (Senthil-Kumar and Mysore, 2014). The VIGS assay was conducted with three biological replicates. The empty vectors were used as controls. After 14 days, the samples were subjected to qRT-PCR and HPLC analyses.
2.12 Pyrethrins quantitation by HPLC
Transiently transformed leaf samples were dried for 48 h in an oven at 50°C to constant dry weight. The dried leaves were ground to a fine powder, then 100 mg powder was placed in a screw-capped glass tube, and dissolved in 600 μl n-hexane. After vortex-oscillation for 30 s, the sample was extracted in an ultrasonic water bath for a further 10 min, followed by vortex-oscillation for 30 s. The extracted samples were filtered with a 0.22 μm filter and analyzed by HPLC. The pyrethrins content was determined using a Waters HPLC system equipped with a photodiode array detector as described previously (Hu et al., 2018). Three biological replicates were analyzed for each sample, and commercial pyrethrum extract (Sigma-Aldrich, St. Louis, MO, USA) was used as the standard. Pyrethrins standard solution mother liquor was obtained by absorbing 3.0 μl pyrethrins standard and dissolving it in a small amount of n-hexane, and finally the volume was standardized to 1.00 ml. The mother liquor of 200, 100, 50, 25, 12.5, and 6.25 μl was standardized to 250 μl with methanol to obtain the pyrethrins standard sample solutions with concentrations of 1.15, 0.575, 0.288, 0.144, 0.072, and 0.036 mg/ml, respectively. After absorbing 20 μl of each standard sample solution, the samples were analyzed by HPLC and a standard curve for the pyrethrins standard samples was generated with Microsoft Excel.
2.13 Statistical analysis
All experiments were repeated using at least three biological replicates. The data were analyzed with one-way ANOVA and Student’s t-test using SPSS 18 software. Significant differences were determined with Student’s t-test, with p < 0.05 considered to be statistically significant.
3 Results
3.1 Exogenous MeJA positively regulates pyrethrins biosynthesis
Pyrethrins are derived from two independent pathways: the terpenoid biosynthesis pathway and the JA biosynthesis pathway (Figure 1A). Most pyrethrins biosynthesis enzymes have been identified to date, including those encoded by the key regulatory genes CHS, AOC, GLIP, and ALDH (Kikuta et al., 2012; Xu et al., 2018a). Therefore, we first analyzed the promoter elements of these key genes involved in pyrethrins synthesis using the PlantCARE and PlantPAN3.0 databases (Figure 1B), revealing several hormone-responsive elements, including those responding to MeJA, ABA, and salicylic acid (SA). This analysis indicated that MeJA treatment might play a role in pyrethrins synthesis.
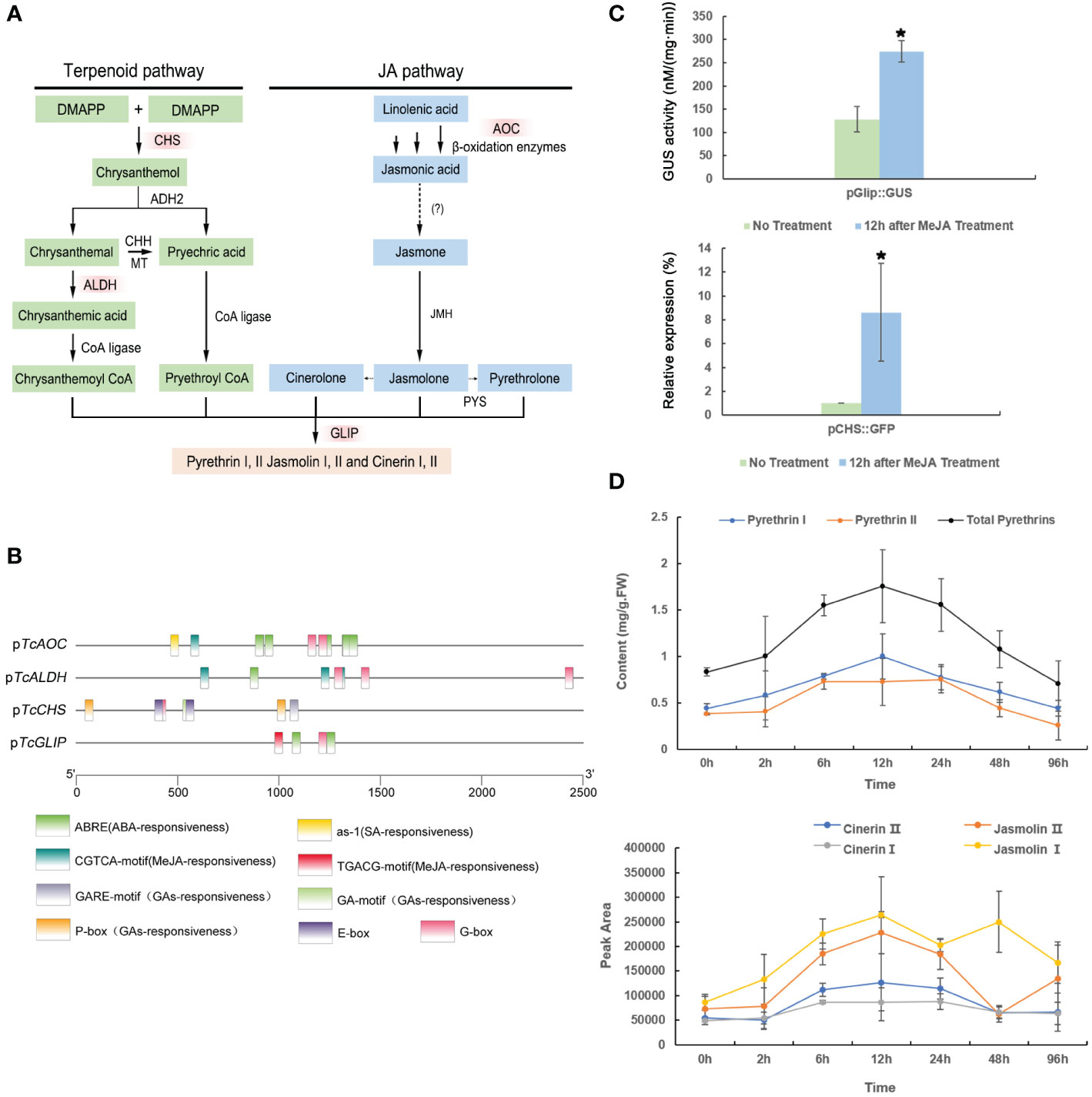
Figure 1 The pathway of pyrethrin biosynthesis and bioanalysis of promoters. (A) The biosynthesis of pyrethrins. The red boxes show the key genes involved in the pathway. (B) Analysis of the cis-regulatory elements in promoters of the key pyrethrin biosynthesis genes. (C) Effect of methyl jasmonate (MeJA) treatment on CHS and GLIP promoter expression. MeJA induced the pCHS::GFP gene in transgenic chrysanthemum. GFP gene expression was detected by real-time PCR at 12 h after spraying with 300 μM MeJA. MeJA induced the pGlip::GUS gene in transgenic tobacco. GUS activity was determined at 12 h after spraying 300 μM MeJA. Data are presented as mean ± SE. *P < 0.05 (ANOVA followed by Duncan’s multiple range test). (D) Effect of MeJA treatment on pyrethrins production in Tanacetum cinerariifolium plants. Leaves were collected at 2, 6, 12, 24, 48, and 96 h after spraying 300 μM MeJA. Samples at 0 h were considered the control. The pyrethrins content was quantified by high-performance liquid chromatography using pyrethrin external standards. Data indicate mean ± SE (n = 3).
Using our previously established transgenic platform for CHSpro-green fluorescent protein (GFP) and GLIPpro-b-glucuronidase (GUS) reporter genes (Sultana et al., 2015), we evaluated the MeJA-induced upregulation on the CHS and GLIP promoters individually. After treatment with 300 μM MeJA, the specific GFP expression level was measured by real-time polymerase chain reaction (PCR) and GUS activity was measured. Consequently, the GFP gene driven by the CHS promoter and the GUS gene driven by the GLIP promoter were both significantly activated at 12 h after MeJA treatment (Figure 1C), confirming that the CHS and GLIP promoters are MeJA-inducible, which will ultimately lead to enhanced pyrethrins production.
Indeed, in our previous study, we found that the pyrethrins content increased for a short time under high-concentration (2 mM) treatment of MeJA (Zeng et al., 2022). However, treatment of such a high concentration of MeJA leads to a dramatic decline in the pyrethrins content after 6 h due to disturbed homeostasis or metabolites feedback. Thus, in the present study, we treated T. cinerariifolium plants with a reduced concentration of MeJA of 300 μM. As expected, T. cinerariifolium plants showed a higher yield of pyrethrins after MeJA treatment, including all six constituents. The pyrethrins content increased by 2-fold to 0.99 mg/g per fresh weight at 12 h after MeJA treatment, and this accumulation level was maintained for 4 days (Figure 1D). These results strongly suggested that pyrethrins biosynthesis genes are MeJA-inducible and that the optimal working concentration of MeJA treatment could maintain a high pyrethrins yield for several days.
3.2 Cloning and characterization of bZIP60 in T. cinerariifolium
Considering that many E-box and G-box elements that bind to members of the bZIP TF family were identified in the promoter regions of pyrethrins biosynthesis genes, we searched for genes encoding putative bZIP TFs in the transcriptome data of T. cinerariifolium. Since we found that MeJA treatment can activate the CHS and GLIP promoter-driven reporter genes and improve pyrethrins production, we selected Unigene27160, which showed the highest expression level induced by MeJA, as the candidate bZIP TF (Figure 2A).
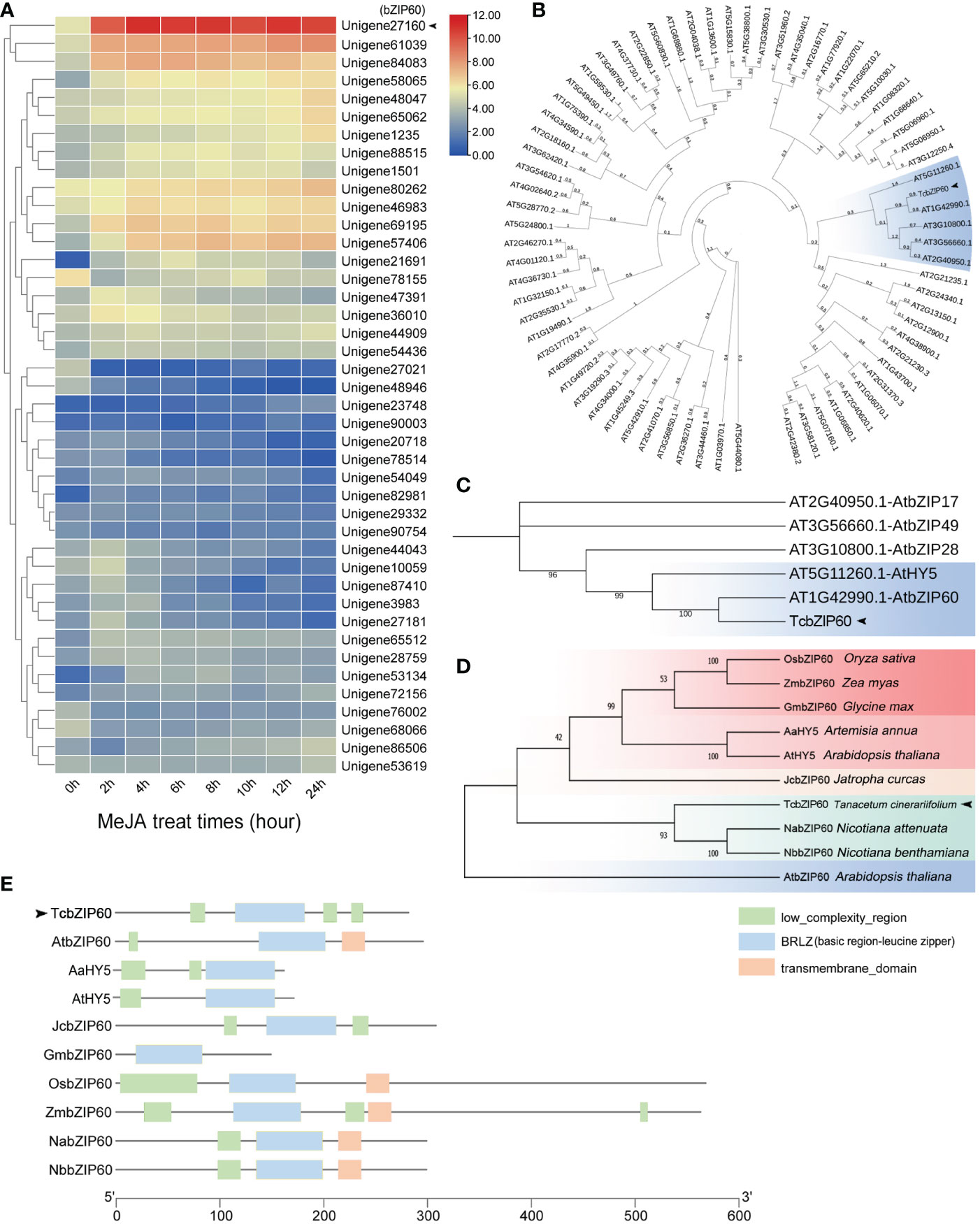
Figure 2 Expression patterns, phylogenetic analysis, and domain of the bZIP transcription factor family. (A) Heat maps of differentially expressed genes in the transcriptome with methyl jasmonate (MeJA) treatment to the T. cinerariifolium leaves at different times. The Pearson distance measure and Ward clustering algorithm were used. (B) Phylogenetic analysis of TcbZIP60 and Arabidopsis thaliana bZIP family protein sequences obtained from the PfamA domain database. (C) Partial enlargement of the phylogenetic tree in (B). (D) Phylogenetic analysis of TcbZIP60 and bZIP family protein sequences of 12 other plants obtained from the NCBI database. (E) BRLZ domain of TcbZIP60 and its homologs in other species.
To identify the Unigene27160 gene in the T. cinerariifolium genome, the sequence was input as a query object in The Arabidopsis Information Resource (TAIR). In the phylogenetic comparison between Unigene27160 and members of the Arabidopsis bZIP TF family, Unigene27160 clustered with Arabidopsis AtbZIP60 and AtHY5 in the same evolutionary branch with high homology, suggesting that these genes exhibit similar functions (Figure 2B, blue box). Unigene27160 showed the closest relationship with AtbZIP60 (Figure 2C). Moreover, 10 bZIP60 proteins from different plant species were selected for further comparative sequence analysis. These proteins clustered into five branches, represented by boxes with different colors in Figure 2D. Unigene27160 clustered with NabZIP60, NbbZIP60, and AtbZIP60, and contained a conserved bZIP domain similar to that of other bZIP proteins (Figure 2E). Thus, Unigene27160 was named TcbZIP60, and its open reading frame region was cloned for further analysis.
3.3 Expression profiles and subcellular localization of TcbZIP60
Since TcbZIP60 expression was most strongly induced by MeJA, we confirmed this result by real-time PCR and further investigated whether other hormones could also induce the expression of TcbZIP60. As expected, treatment with both MeJA and ABA strongly induced TcbZIP60 expression, and high-level expression was maintained for 24 h (Figures 3A, B). To confirm that TcbZIP60 is involved in pyrethrins biosynthesis, the relative expression levels of TcbZIP60 and other pyrethrins biosynthesis genes were analyzed at different flowering stages. The expression profiles were similar, with high expression detected in the first two stages (Figure 3C). Based on these results, we speculated that the TF TcbZIP60 regulates pyrethrins synthesis through interaction with the pyrethrins synthase gene.
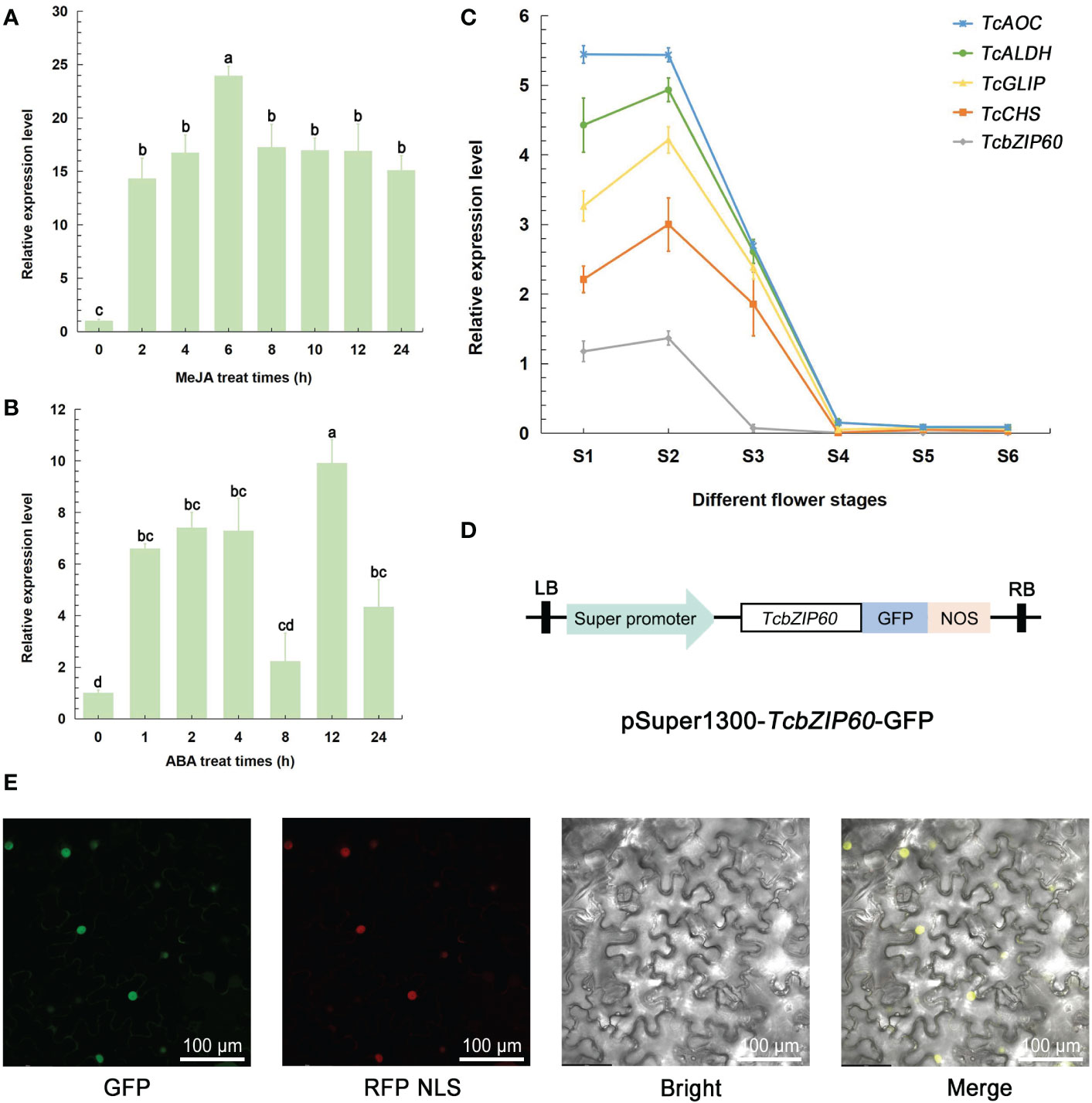
Figure 3 Expression profile and subcellular localization of TcbZIP60. (A) RT-qPCR analysis of the transcript abundance of TcbZIP60 induced in the leaves of 1-month-old tissue culture seedlings treated with methyl jasmonate (MeJA); three biological replicates were established. (B) RT-qPCR analysis of the transcript abundance of TcbZIP60 induced in the leaves of 1-month-old tissue culture seedlings treated with abscisic acid (ABA); three biological replicates were established. (C) RT-qPCR analysis of TcbZIP60 transcript abundance from stages S1 to S6. (D) Diagram of the pSuper1300-TcbZIP60-GFP construct. (E) Subcellular localization of TcbZIP60 in N. benthamiana leaves. Scale bars = 100 μm.
To determine the subcellular localization of TcbZIP60, the coding sequence of TcbZIP60 was fused into the GFP framework under control of the CaMV35S promoter. When TcbZIP60-GFP was instantaneously expressed in Nicotiana benthamiana leaves, strong and specific fluorescence was observed in the nucleus (Figure 3D). In addition, the co-localization of the GFP signal and red fluorescent protein (as a nuclear localization signal) resulted in a fused yellow fluorescence signal (Figure 3E). These results suggested that TcbZIP60 is a nuclear-localized protein, further supporting its potential role as a TF.
3.4 TcbZIP60 directly binds to the TcCHS and TcAOC promoters
The binding sites of bZIP TFs were predicted in the promoter sequence of the pyrethrins synthesis genes (Figure 1B). In T. cinerariifolium, similar expression profiles were observed between TcbZIP60 and pyrethrins synthesis genes during different flowering stages (Figure 3C). Therefore, we further speculated that TcbZIP60 can regulate the expression of these genes by directly binding to the corresponding promoters. To test this hypothesis, we performed a yeast monohybrid (Y1H) experiment. The promoters of pyrethrins synthesis genes were cloned and inserted into the pHis2.1 vector to generate reporter genes. TcbZIP60 was fused to the GAL4 activation domain to generate the effector construct pGADT7-TcbZIP60 (Figure 4A).
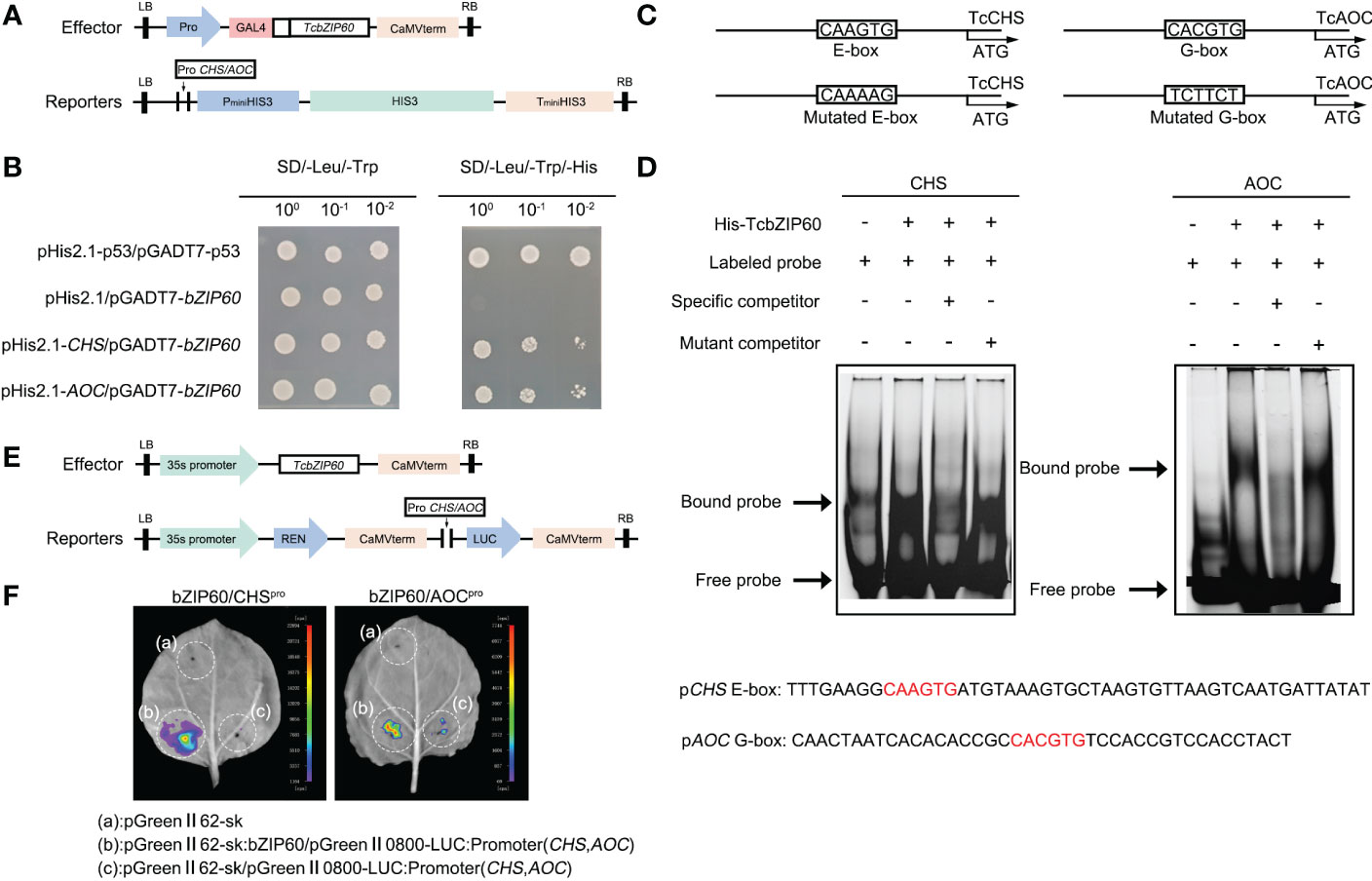
Figure 4 TcbZIP60 directly binds to and activates the promoters of the TcCHS and TcAOC genes. (A) Diagram of vectors used in the yeast one hybrid (Y1H) assay. Reporters: the promoter of TcCHS/TcAOC was introduced into the pHis2.1 vector. Effector: TcbZIP60 was introduced into the pGADT7 vector. (B) Y1H assay showing TcbZIP60 binding to TcCHS/TcAOC promoter fragments. (C) Schematic diagram of the TcCHS/TcAOC promoter showing the potential TcbZIP60 binding sites. The predicted E-box/G-box and mutated E-box/G-box sites and sequences are indicated with black boxes. (D) Electrophoretic mobility shift assay (EMSA) of interactions between TcbZIP60 and the promoters of the TcCHS and TcAOC genes. The FAM-labeled probe (10 μM) was competed with a 50-fold excess of unlabeled wild-type cold probes and unlabeled mutant-type cold probes. (E) Diagram of dual-LUC vectors. Effector: TcbZIP60 was introduced into the pGreen-62-SK vector, which was driven by the 35S promoter. Reporters: the promoter of TcCHS/TcAOC was introduced into the pGreen II-0800-LUC vector, driving the translation of LUC. (F) Dual-LUC reporter assay showing TcbZIP60 binding to the TcCHS/TcAOC promoters and promotion of LUC expression.
In the interaction experiments with several pyrethrins synthesis gene promoters, yeast clones were observed on solid synthesized Leu, Trp, and His (SD/-Leu-Trp-His) media only when pGADT7-TcbZIP60 was transferred into yeast cells expressing pHis2.1-TcCHS and pHis2.1-TcAOC, but not in those expressing pHis2.1. This suggested that TcbZIP60 binds directly to the promoters of TcCHS and TcAOC (Figure 4B). In addition, we hypothesized that TcbZIP60 directly binds to the motifs on the promoters of the two genes, which was further verified by electrophoretic mobility shift assays (EMSAs).
We predicted that TcbZIP60 binds to the potential E-box and G-box sites on the two promoters, and designed the corresponding mutation sites (Figure 4C). The migration bands were detected in the presence of the purified and concentrated TcbZIP60 protein and the labeled probe of the E-box containing the TcCHS promoter or the G-box containing the TcAOC promoter (Figure 4D). When 50 times the concentration of the cold probe (unlabeled probe) was added, the intensity of the migration band decreased. When the mutant probe (unlabeled probe) was added, the intensity of the migration band recovered.
To further verify the interaction between TcbZIP60 and the TcCHS or TcAOC promoter in plants, we performed a transient dual-luciferase (dual-LUC) assay with a reporter structure (TcCHS/TcAOC Pro : LUC) and effector structure (Figure 4E). Strong LUC activity was observed in tobacco leaves co-transformed by TcCHS/TcAOC Pro : LUC and 35S:TcbZIP60. However, almost no fluorescence signal was detected in tobacco leaves co-transformed with TcCHS/TcAOC Pro : LUC and an empty carrier (Figure 4F). These results suggested that the TF TcbZIP60 directly activates promoters of the TcCHS and TcAOC genes and regulates pyrethrins biosynthesis in T. cinerariifolium.
3.5 Activation of pyrethrins biosynthesis by TcbZIP60
Previous studies demonstrated that bZIP TFs in A. annua bind to gene promoters and regulate artemisinin biosynthesis (Hao et al., 2019; Lv et al., 2019). Similarly, we confirmed that TcbZIP60 can directly bind to the promoters of the pyrethrins biosynthesis genes TcCHS and TcAOC. Therefore, to further verify the role of TcbZIP60 in regulating pyrethrins biosynthesis, we constructed TcbZIP60-overexpressing plants (TcbZIP60-OE) and TcbZI60-suppressed plants (pTRV2-TcbZIP60) in T. cinerariifolium. Transient overexpression and transient RNA interference-mediated silencing of TcbZIP60 were performed in the leaves of T. cinerariifolium, and the expression of the pyrethrins biosynthesis genes was detected by reverse transcription-quantitative PCR after transformation. Compared with wild-type (Mock) plants of pGreenII 62-SK, the transcription level of TcbZIP60 in TcbZIP60-OE plants significantly increased by 49.65 times, and the expression levels of the pyrethrins synthesis genes TcCHS and TcAOC also increased by 1.76 times and 1.88 times, respectively. The other two pyrethrins biosynthesis genes, TcALDH and TcGLIP, were also upregulated with TcbZIP60 overexpression (Figure 5A). By contrast, in the TcbZIP60-silenced plants, the transcription level of TcbZIP60 decreased by 0.8-fold (Figure 5B) and the expression levels of four pyrethrins synthesis genes also decreased (Figure 5C).
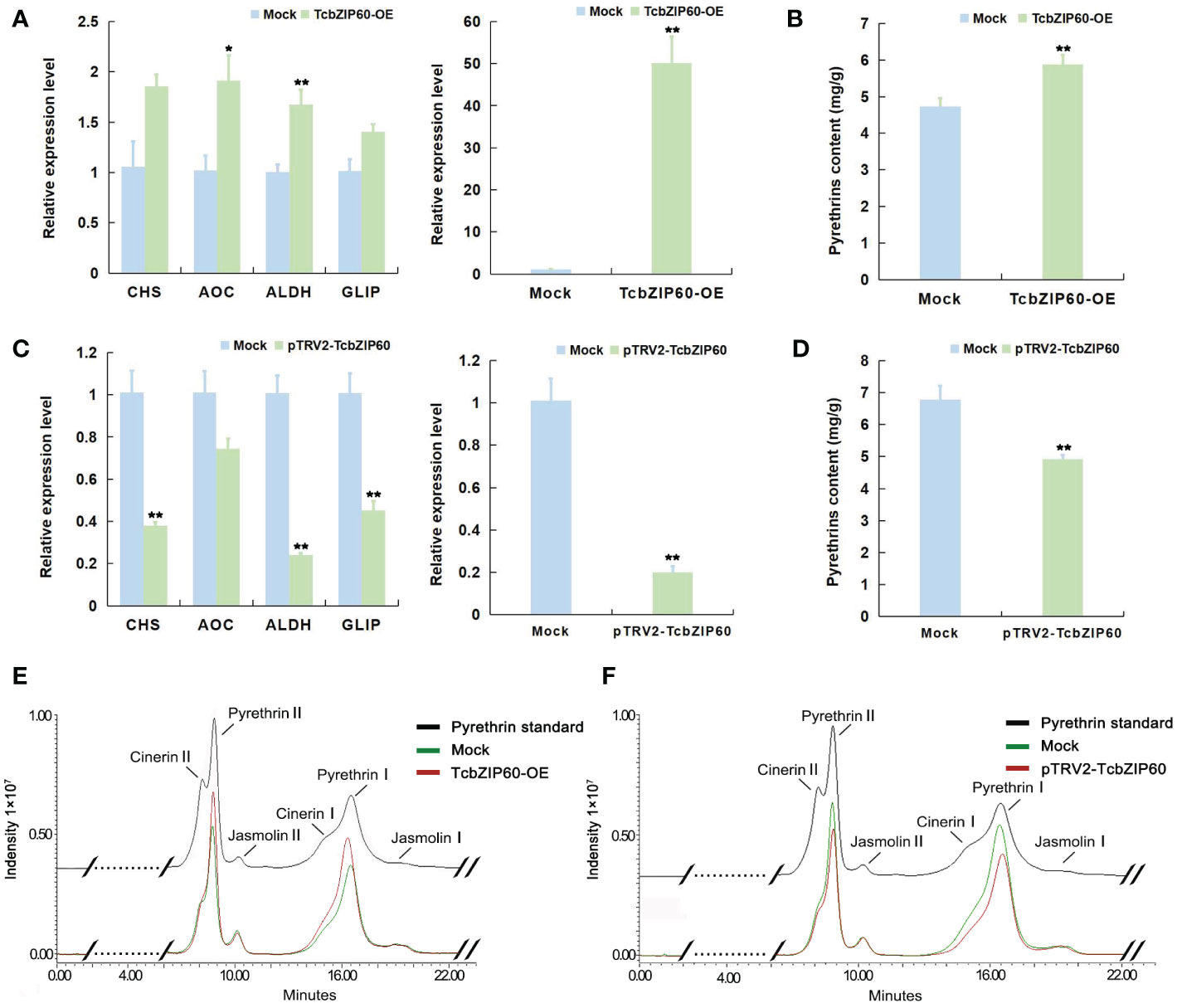
Figure 5 Transient overexpression and TRV-VIGS treatment of TcbZIP60 in T. cinerariifolium leaves. (A) Relative expression levels of pyrethrin biosynthesis genes in T. cinerariifolium leaves at 4 days after transient overexpression of TcbZIP60. Mock: transient overexpression of the empty vector; TcbZIP60-OE: transient overexpression of pGreen-62-SK-TcbZIP60. The value for the Mock was set to 1. (B) Quantity of pyrethrins in T. cinerariifolium leaves after transient TcbZIP60 overexpression at 4 days. The ordinate represents the pyrethrins content measured by high-performance liquid chromatography (HPLC) and the abscissa represents the six major constituent pyrethrins. (C) Relative expression levels of pyrethrins biosynthesis genes in T. cinerariifolium leaves at 4 days after TRV-VIGS treatment of TcbZIP60 at 14 days. Mock: VIGS empty pTRV2 vector; pTRV2-TcbZIP60: TcbZIP60 silencing of leaves. The value for Mock was set to 1. (D) Quantity of pyrethrins in T. cinerariifolium leaves after TRV-VIGS treatment of TcbZIP60. The ordinate represents pyrethrins content measured by HPLC and the abscissa represents the six major constituent pyrethrins. (E) Representative chromatogram of total pyrethrins showing the peaks of six different constituents (cinerarin II, pyrethrin II, jasmoline II, cinerarin I, pyrethrin I, and jasmoline I) in a standard sample (black line), TcbZIP60-OE (red line), and Mock (green line). (F) Representative chromatogram of total pyrethrins showing the peaks of six different constituents in a standard sample (black line), Mock (green line), and pTRV2-TcbZIP60 (red line). Asterisks indicate that the value is significantly different from that of the control (**P<0.01, *P < 0.05).
Subsequently, we measured the contents of pyrethrins in the transient overexpression and RNA interference plants. We used the pyrethrins standard to compare and identify the six main components of pyrethrins in the sample as shown by the arrow and the black line in Figures 5E, F, whereas the red and green lines represent the HPLC results of the experimental group and the control group, respectively. Compared with the pGreenII 62-SK Mock plants in the blank control, the total amount of pyrethrins in the overexpression plants significantly increased by 1.239 times (Figures 5B, E) and decreased by 0.273 times in the TcbZIP60 interference plants (Figures 5D, F). These results showed that TcZIP60 can positively regulate pyrethrins accumulation by activating pyrethrins biosynthesis genes.
4 Discussion
Pyrethrins represent unique metabolites produced in pyrethrums that can provide plants with an effective endogenous chemical defense against insect and fungal diseases (Zeng et al., 2021). Since the 19th century, pyrethrins products have been gradually used as household and agricultural insecticides, and as insect-borne disease prevention agents (Orenstein, 1913; Lange and Akesson, 1973; Katsuda, 1999). However, in the extraction process for agricultural production, pyrethrins are obtained from the dry flowers of T. cinerariifolium and the yield of pyrethrins per plant is low, only accounting for 0.10–1.35% of the dry weight of flowers (Varga et al., 2021). Therefore, increasing the amount of pyrethrins produced in the leaves is the key to breaking through the bottleneck of production.
Previously, we reported that MeJA can upregulate the expression of pyrethrins biosynthesis genes as well as induce pyrethrins accumulation (Zeng et al., 2022). However, the high working concentration of MeJA used in the previous study cannot be sustained to constituently increase the pyrethrins content. In general, under stress/MeJA treatment, plant growth and metabolites synthesis will be halted to invest in defense. In the present study, treatment of a lower concentration of MeJA maintained the increase in the pyrethrins content in the leaves for more than 3 days. This was considered to be due to activation of the promoters of pyrethrins biosynthesis genes by MeJA. To further investigate the mechanism by which MeJA regulates pyrethrins biosynthesis, we identified the TF TcbZIP60 by the global expression profile, which was confirmed to be induced by exogenous MeJA, suggesting that TcbZIP60 might be involved in MeJA-regulated pyrethrins biosynthesis.
In addition to MeJA, ABA also plays an important role in regulating the biosynthesis of terpenoids (Jing et al., 2009). TFs involved in mediating ABA stimulation of terpenoids biosynthesis, including AaHY5, AaTGA6, and AabZIP1, respond to JA, SA, and ABA treatment and regulate artemisinin biosynthesis in A. annua (Hao et al., 2019; Lv et al., 2019; Shu et al., 2022). Similarly, we found that the transcription level of TcbZIP60 was regulated by ABA treatment. Cross-talk between ABA and MeJA is a universal phenomenon, and some TFs involved in ABA signaling also participate in MeJA signaling in Arabidopsis and tobacco (Lackman et al., 2011). For example, AaMYC2, which is involved in JA signaling, was suggested to function as the bridge of JA signaling to the ABA signaling pathway (Abe et al., 2003). AaMYC2 expression could be upregulated by ABA-induced AabZIP1 through direct interaction to subsequently activate AaALDH1 transcription, thereby governing artemisinin biosynthesis (Shu et al., 2022). In T. cinerariifolium, TcMYC2 was also identified as a MeJA-induced TF that positively regulates pyrethrins biosynthesis by upregulating TcCHS, TcAOC, and TcGLIP gene expression (Zeng et al., 2022). Thus, it is strongly speculated that TcbZIP60 might also interact with TcMYC2 to co-regulate pyrethrins biosynthesis. This could explain why we detected upregulation of TcGLIP expression even when its promoter was not directly activated by TcbZIP60. Therefore, whether more TFs in T. cinerariifolium function as a network to cooperatively regulate pyrethrins biosynthesis is worthy of further study.
Through the development of metabolic engineering, there has been significant progress in producing pyrethrins in other model plants by introducing pyrethrins biosynthesis genes into heterologous systems. However, to date, only the precursors of pyrethrins have been synthesized in tobacco and tomato (Xu et al., 2018b; Xu et al., 2019), because not all of the genes in the full pathway have been cloned. T. cinerariifolium remains the only available resource for pyrethrins production. Because pyrethrins biosynthesis is derived from two independent pathways involving multiple enzymes, overexpression of one or two enzymes would not be efficient in increasing pyrethrins biosynthesis, and it is even harder to co-express enzymes from different pathways simultaneously. Members of the bZIP TF family typically target the key steps in the biosynthesis of secondary metabolites. bZIP TFs contain a conserved alkaline region consisting of 16 amino acid residues with a constant N-X7-R/K motif, which can recognize and bind to the specific elements on the DNA sequence on the promoters, thereby affecting the multiple genes involved in the synthesis of secondary metabolites. Thus, overexpression of bZIP TFs leads to the improvement of secondary metabolites production. For example, in A. annua, AabZIP1 binds to both the AaADS and AaCYP71AV1 promoters, and increases the expression of AaADS and AaCYP71AV1 as well as the artemisinin content (Zhang et al., 2015). Overexpression of AaHY5 and other upstream transcriptional regulatory genes significantly increased the transcriptional levels of the downstream genes AaADS, AaCYP71AV1, AaDBR2, and AaALDH1, and increased the synthesis and accumulation of secondary metabolites (Hao et al., 2019). In Oryza sativa, bZIP72 binds to the AOC promoter G-box, which strengthens the AOC transcription level and endogenous JA level (Wang et al., 2020). In T. cinerariifolium, CHS and AOC are upstream genes that are more likely to become rate-limiting factors for pyrethrins synthesis (Figure 1A). AOC is considered to be the first key enzyme in JA synthesis, affecting the overall biosynthesis rate (Yoeun et al., 2018), and CHS is the first key enzyme in the biosynthesis of pyrethrins (Hu et al., 2018). Our results demonstrated that TcbZIP60 could bind to the E-box and G-box cis-elements on the promoters of both TcCHS and TcAOC, and activated the transcription of these genes in vitro. Moreover, to further confirm the function of TcbZIP60 in pyrethrins production, we transiently overexpressed the TcbZIP60 gene in T. cinerariifolium leaves, which upregulated the expression of the main pyrethrins biosynthesis genes TcCHS, TcAOC, TcALDH, and TcGLIP, consequently leading to the significant accumulation of pyrethrins. As expected, interference of TcbZIP60 expression in T. cinerariifolium decreased the production of pyrethrins.
Based on these results, we propose a model of the role of TcbZIP60 in the regulation of pyrethrins biosynthesis in T. cinerariifolium (Figure 6). This study thus identified TcbZIP60 as a novel and important positive regulator that could be used in engineering pyrethrins synthesis to improve its production in T. cinerariifolium.
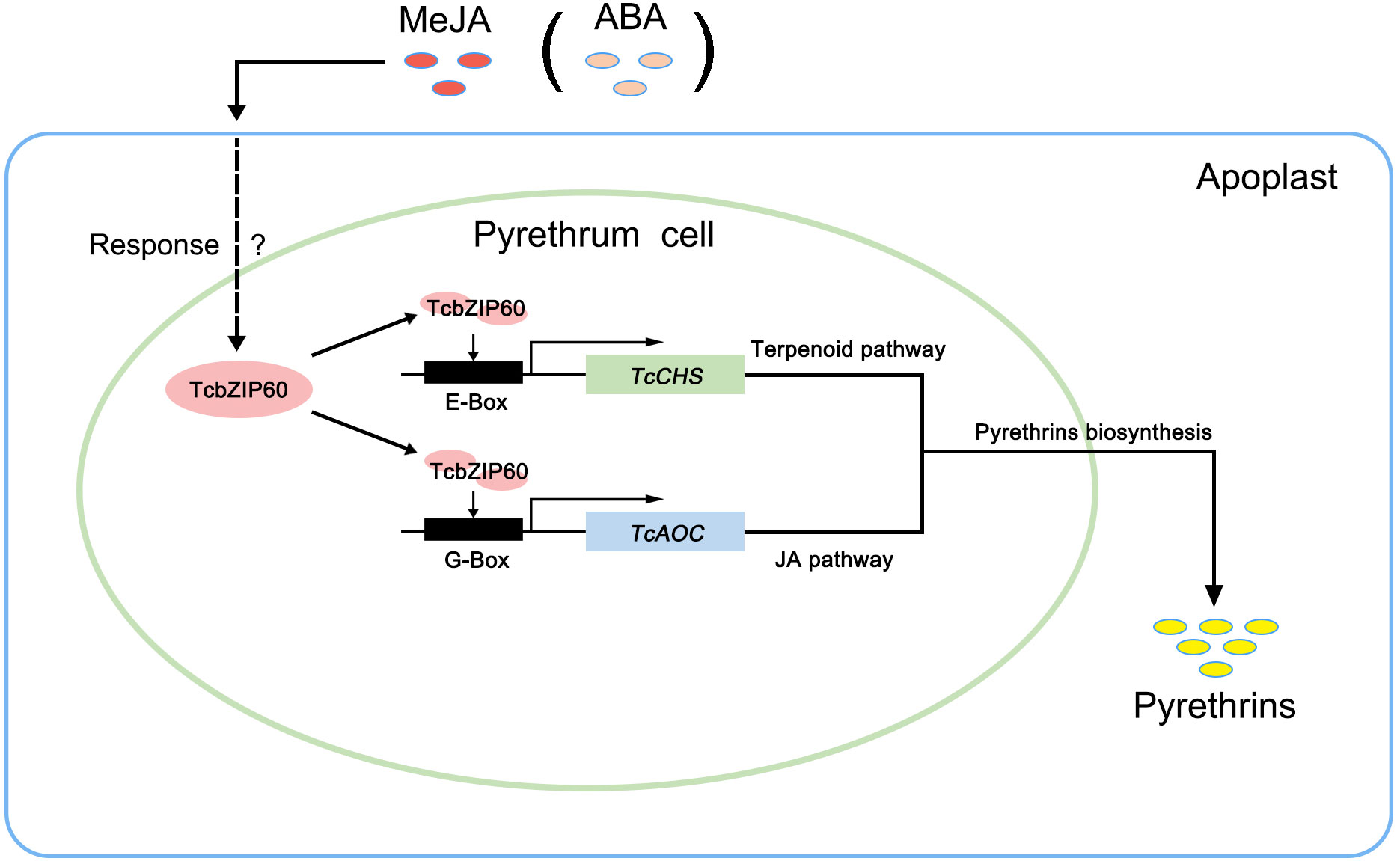
Figure 6 Model illustrating the involvement of TcbZIP60 in regulating pyrethrins biosynthesis in T. cinerariifolium. The arrows indicate TcbZIP60, regulated by methyl jasmonate (MeJA) or abscisic acid (ABA) signals, activating the TcCHS and TcAOC genes by binding to their promoters and subsequently regulating pyrethrins biosynthesis. JA, jasmonic acid.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZX, TZ, JiaL, CW, and HH conceived the project. ZX, TZ, JinjL, and HH conducted the experiments. ZX, TZ, JiaL, LZ, JinjL, RZ, JLU, YW, CW, and HH discussed the experiment. ZX, TZ, JiaL, and HH analyzed and interpreted the data. ZX and HH prepared the figures. ZX and HH prepared the manuscript draft. ZX and TZ wrote the article with input from all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key National R & D Projects During the 14th Five-Year Plan Period (2019YFD1001500), China Postdoctoral Science Foundation (2022T150243, 2018 M64072), National Natural Science Foundation of China (31902051, 32160718), and Natural Science Research Project of Guizhou (KY [2022]170, ZK [2022]301).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1133912/full#supplementary-material
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 15, 63–78. doi: 10.1105/tpc.006130
Casida, J. E., Quistad, G. B. (1995). Pyrethrum flowers: Production, chemistry,toxicology, and uses (New York: Oxford University Press).
Chen, S. Y., Ma, T., Song, S. R., Li, X. L., Fu, P. N., Wu, W., et al. (2021). Arabidopsis downy mildew effector HaRxLL470 suppresses plant immunity by attenuating the DNA-binding activity of bZIP transcription factor HY5. New Phytol. 230, 1562–1577. doi: 10.1111/nph.17280
Chen, K., Tang, W., Zhou, Y., Chen, J., Xu, Z., Ma, R., et al. (2022). AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 170, 287–295. doi: 10.1016/j.plaphy.2021.12.014
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G.-Z., Tseng, K.-C., Liu, Y.-H., et al. (2019). PlantPAN3. 0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163. doi: 10.1093/nar/gky1081
Creelman, R. A., Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Freemont, J. A., Littler, S. W., Hutt, O. E., Mauger, S., Duggan, P. J. (2016). Molecular markers for pyrethrin autoxidation in stored pyrethrum crop: analysis and structure determination. J. Agric. Food Chem. 64, 7134–7141. doi: 10.1021/acs.jafc.6b02959
Geng, J., Liu, J. H. (2018). The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 69, 2677–2692. doi: 10.1093/jxb/ery065
Hao, X., Zhong, Y., Nützmann, H.-W., Fu, X., Yan, T., Shen, Q., et al. (2019). Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 60, 1747–1760. doi: 10.1093/pcp/pcz084
Hu, H., Li, J. J., Delatte, T., Vervoort, J., Gao, L., Verstappen, F., et al. (2018). Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids. Plant Biotechnol. J. 16, 1434–1445. doi: 10.1111/pbi.12885
Jakoby, M., Weisshaar, B., Drge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jing, F., Zhang, L., Li, M., Tang, Y., Wang, Y., Wang, Y., et al. (2009). Abscisic acid (ABA) treatment increases artemisinin content in artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia. 64, 319–323. doi: 10.2478/s11756-009-0040-8
Jung, S.-K., McDonald, K. A., Dandekar, A. M. (2015). Effect of leaf incubation temperature profiles on Agrobacterium tumefaciens-mediated transient expression. Biotechnol. Progr. 31, 783–790. doi: 10.1002/btpr.2077
Katsuda, Y. (1999). Development of and future prospects for pyrethroid chemistry. Pesticide Sci. 55, 775–782. doi: 10.1002/(sici)1096-9063(199908)55:8<775::Aid-ps27>3.0.Co;2-n
Khan, S. A., Verma, P., Banerjee, S., Chaterjee, A., Tandon, S., Kalra, A., et al. (2017). Pyrethrin accumulation in elicited hairy root cultures of Chrysanthemum cinerariaefolium. Plant Growth Regul. 81, 365–376. doi: 10.1007/s10725-016-0213-8
Kikuta, Y., Ueda, H., Takahashi, M., Mitsumori, T., Yamada, G., Sakamori, K., et al. (2012). Identification and characterization of a GDSL lipase-like protein that catalyzes the ester-forming reaction for pyrethrin biosynthesis in Tanacetum cinerariifolium- a new target for plant protection. Plant J. 71, 183–193. doi: 10.1111/j.1365-313X.2012.04980.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lackman, P., González-Guzmán, M., Tilleman, S., Carqueijeiro, I., Pérez, A. C., Moses, T., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in arabidopsis and tobacco. Proc. Natl. Acad. Sci. 108, 5891–5896. doi: 10.1073/pnas.1103010108
Lange, W. H., Akesson, N. B. (1973). “Chapter 14,” in Pyrethrum for control of agricultural insects (Amsterdam: Elsevier Inc).
Li, J., Li, Y., Wang, Z., Wang, C. (2014). Effect of three growth regulators on pyrethrin content and the traits related to pyrethrin yield. Chin. J. Trop. Crops. 35, 1067–1070.
Li, J. J., Liu, H., Yang, C., Wang, J., Yang, G. J., Si, P., et al. (2020). Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus l. Indust. Crops Prod. 143, 111924. doi: 10.1016/j.indcrop.2019.111924
Li, W., Lybrand, D. B., Zhou, F., Last, R. L., Pichersky, E. (2019). Pyrethrin biosynthesis: the cytochrome P450 oxidoreductase CYP82Q3 converts jasmolone to pyrethrolone. Plant Physiol. 181, 934–944. doi: 10.1104/pp.19.00499
Li, W., Zhou, F., Pichersky, E. (2018). Jasmone hydroxylase, a key enzyme in the synthesis of the alcohol moiety of pyrethrin insecticides. Plant Physiol. 177, 1498–1509. doi: 10.1104/pp.18.00748
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. 408, 402–408. doi: 10.1006/meth.2001.1262
Luo, J., Ma, N., Pei, H., Chen, J., Li, J., Gao, J. (2013). A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. J. Exp. Botany. 64, 5075–5084. doi: 10.1093/jxb/ert296
Lv, Z., Guo, Z., Zhang, L., Zhang, F., Jiang, W., Shen, Q., et al. (2019). Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in artemisia annua. J. Exp. Botany. 70, 3969–3979. doi: 10.1093/jxb/erz166
Lybrand, D. B., Xu, H., Last, R. L., Pichersky, E. (2020). How plants synthesize pyrethrins: Safe and biodegradable insecticides. Trends Plant Sci. 25, 1240–1251. doi: 10.1016/j.tplants.2020.06.012
Mao, Z., Jiang, H., Wang, S., Wang, Y., Yu, L., Zou, Q., et al. (2021). The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 306, 110848. doi: 10.1016/j.plantsci.2021.110848
Orenstein, A. J. (1913). Mosquito catching in dwellings in the prophylaxis of malaria. Am. J. Public Health 3, 106. doi: 10.2105/AJPH.3.2.106
Pal, T., Malhotra, N., Chanumolu, S. K., Chauhan, R. S. (2015). Next-generation sequencing (NGS) transcriptomes reveal association of multiple genes and pathways contributing to secondary metabolites accumulation in tuberous roots of Aconitum heterophyllum wall. Planta. 242, 239–258. doi: 10.1007/s00425-015-2304-6
Ramirez, A. M., Saillard, N., Yang, T., Franssen, M. C., Bouwmeester, H. J., Jongsma, M. A. (2013). Biosynthesis of sesquiterpene lactones in pyrethrum (Tanacetum cinerariifolium). PloS One 8, e65030. doi: 10.1371/journal.pone.0065030
Rivera, S. B., Swedlund, B. D., King, G. J., Bell, R. N., Hussey, C. E., Shattuck-Eidens, D. M., et al. (2001). Chrysanthemyl diphosphate synthase: Isolation of the gene and characterization of the recombinant non-head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. Proc. Natl. Acad. Sci. U. S. A. 98, 4373–4378. doi: 10.1073/pnas.071543598
Sagor, G., Chawla, P., Kim, D. W., Berberich, T., Kojima, S., Niitsu, M., et al. (2015). The polyamine spermine induces the unfolded protein response via the MAPK cascade in arabidopsis. Front. Plant Sci. 6, 687. doi: 10.3389/fpls.2015.00687
Senthil-Kumar, M., Mysore, K. S. (2014). Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Shu, G., Tang, Y., Yuan, M., Wei, N., Zhang, F., Yang, C., et al. (2022). Molecular insights into AabZIP1-mediated regulation on artemisinin biosynthesis and drought tolerance in Artemisia annua. Acta Pharmaceut. Sin. B. 12, 1500–1513. doi: 10.1016/j.apsb.2021.09.026
Sibéril, Y., Benhamron, S., Memelink, J., Giglioli-Guivarc'h, N., Thiersault, M., Boisson, B., et al. (2001). Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol. Biol. 45, 477–488. doi: 10.1023/A:1010650906695
Song, W. C., Funk, C. D., Brash, A. R. (1993). Molecular cloning of an allene oxide synthase: a cytochrome-p450 specialized for the metabolism of fatty-acid hydroperoxides. Proc. Natl. Acad. Sci. U. S. A. 90, 8519–8523. doi: 10.1073/pnas.90.18.8519
Sornaraj, P., Luang, S., Lopato, S., Hrmova., M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta Gen. Subj. 1860, 46–56. doi: 10.1016/j.bbagen.2015.10.014
Sultana, S., Hu, H., Gao, L., Mao, J., Luo, J., Jongsma, M. A., et al. (2015). Molecular cloning and characterization of the trichome specific chrysanthemyl diphosphate/chrysanthemol synthase promoter from Tanacetum cinerariifolium. Scient. Horticult. 185, 193–199. doi: 10.1016/j.scienta.2015.01.032
Tang, W., Page, M. (2013). Transcription factor AtbZIP60 regulates expression of Ca2+-dependent protein kinase genes in transgenic cells. Mol. Biol. Rep. 40, 2723–2732. doi: 10.1007/s11033-012-2362-9
Tijet, N., Brash, A. R. (2002). Allene oxide synthases and allene oxides. Prostagland. Other Lipid Mediat. 68-9, 423–431. doi: 10.1016/s0090-6980(02)00046-1
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U. S. A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Varga, F., Jeran, N., Satovic, Z., Biosic, M., Grdisa, M. (2021). High diversity of natural Dalmatian pyrethrum based on pyrethrin composition at intra- and interpopulation level. Phytochemistry. 192, 11. doi: 10.1016/j.phytochem.2021.112934
Wang, Y., Hou, Y., Qiu, J., Wang, H., Wang, S., Tang, L., et al. (2020). Abscisic acid promotes jasmonic acid biosynthesis via a 'SAPK10-bZIP72-AOC' pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 228, 1336–1353. doi: 10.1111/nph.16774
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in arabidopsis. Science. 309, 1056–1059. doi: 10.1126/science.1114358
Wolfgang, D. L., Snoek, B. L., Berend, S., Christoph, W. (2018). The arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Xu, H., Li, W., Schilmiller, A. L., van Eekelen, H., de Vos, R. C. H., Jongsma, M. A., et al. (2019). Pyrethric acid of natural pyrethrin insecticide: complete pathway elucidation and reconstitution in Nicotiana benthamiana. New Phytol. 223, 751–765. doi: 10.1111/nph.15821
Xu, H., Lybrand, D., Bennewitz, S., Tissier, A., Last, R. L., Pichersky, E. (2018b). Production of trans-chrysanthemic acid, the monoterpene acid moiety of natural pyrethrin insecticides, in tomato fruit. Metab. Eng. 47, 271–278. doi: 10.1016/j.ymben.2018.04.004
Xu, H., Moghe, G. D., Wiegert-Rininger, K., Schilmiller, A. L., Barry, C. S., Last, R. L., et al. (2018a). Coexpression analysis identifies two oxidoreductases involved in the biosynthesis of the monoterpene acid moiety of natural pyrethrin insecticides in Tanacetum cinerariifolium. Plant Physiol. 176, 524–537. doi: 10.1104/pp.17.01330
Yang, T., Gao, L., Hu, H., Stoopen, G., Wang, C., Jongsma, M. A. (2014). Chrysanthemyl diphosphate synthase operates in planta as a bifunctional enzyme with chrysanthemol synthase activity. J. Biol. Chem. 289, 36325–36335. doi: 10.1074/jbc.M114.623348
Yoeun, S., Cho, K., Han, O. (2018). Structural evidence for the substrate channeling of rice allene oxide cyclase in biologically analogous nazarov reaction. Front. Chem. 6, 500. doi: 10.3389/fchem.2018.00500
Zeng, T., Li, J.-W., Xu, Z.-Z., Zhou, L., Li, J.-J., Yu, Q., et al. (2022). TcMYC2 regulates pyrethrin biosynthesis in Tanacetum cinerariifolium. Hortic. Res. 9, uhac178. doi: 10.1093/hr/uhac178
Zeng, T., Li, J.-W., Zhou, L., Xu, Z.-Z., Li, J.-J., Hu, H., et al. (2021). Transcriptional responses and GCMS analysis for the biosynthesis of pyrethrins and volatile terpenes in Tanacetum coccineum. Int. J. Mol. Sci. 22, 13005. doi: 10.3390/ijms222313005
Zhang, F., Fu, X., Lv, Z., Lu, X., Shen, Q., Zhang, L., et al. (2015). A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 8, 163–175. doi: 10.1016/j.molp.2014.12.004
Keywords: Tanacetum cinerariifolium, pyrethrins biosynthesis, exogenous hormone, transcription factor, TcbZIP60
Citation: Xu Z, Zeng T, Li J, Zhou L, Li J, Luo J, Zheng R, Wang Y, Hu H and Wang C (2023) TcbZIP60 positively regulates pyrethrins biosynthesis in Tanacetum cinerariifolium. Front. Plant Sci. 14:1133912. doi: 10.3389/fpls.2023.1133912
Received: 29 December 2022; Accepted: 06 February 2023;
Published: 20 February 2023.
Edited by:
Moonhyuk Kwon, Gyeongsang National University, Republic of KoreaCopyright © 2023 Xu, Zeng, Li, Zhou, Li, Luo, Zheng, Wang, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Hu, aGFvaHVAbWFpbC5oemF1LmVkdS5jbg==; Caiyun Wang, d2FuZ2N5QG1haWwuaHphdS5lZHUuY24=
†These authors have contributed equally to this work
 Zhizhuo Xu
Zhizhuo Xu Tuo Zeng1,2†
Tuo Zeng1,2† Jing Luo
Jing Luo Riru Zheng
Riru Zheng Yuanyuan Wang
Yuanyuan Wang Caiyun Wang
Caiyun Wang