- 1School of Life Science, Jiangsu University, Zhenjiang, Jiangsu, China
- 2Institute of Food Crops, Jiangsu Academy of Agricultural Sciences/Key Laboratory of Germplasm Innovation in Downstream of Huaihe River Ministry of Agriculture and Rural Affairs, Nanjing, Jiangsu, China
- 3Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, Yangzhou University, Yangzhou, China
FLOURY ENDOSPERM 2 (FLO2), encoding a tetratricopeptide repeat domain (TPR)-containing protein located in the nucleus, is considered to be a regulatory protein that controls the biosynthesis of seed storage substances. The diversity of flo2 allele is attributable for the variations in grain appearance, amylose content (AC), and physicochemical properties, influencing the eating and cooking quality (ECQ) of rice. In this study, we used CRISPR/Cas9 to introduce loss-of-function mutations into the FLOURY ENDOSPERM 2 gene in Suken118 (SK118), a widely cultivated elite japonica rice variety in Jiangsu, China. Physiochemical analyses of the flo2 mutants were congruent with previous studies, exhibiting lowered AC and viscosity, risen gel consistency (GC) and gelatinization temperature (GT) values, which were all instrumental to the improvement of ECQ. However, the wrinkled opaque appearance and the decrease in grain width, grain thickness and grain weight imply trade-offs in grain yield. Despite the ex-ante estimation for low yielding, the superior ECQ in these novel genotypes generated by using genome editing approach may have the potential for formulating high value specialty food.
Introduction
Rice (Oryza sativa L.), one of the most important crops worldwide, provides staple food for feeding half of the world’s population. The steady increase in grain yield, coupled with the rapid improvements in human living conditions, has fostered ever increasing public demands for foods of high quality and high versatility (Concepcion et al., 2015; Huo et al., 2017). Consequently, the rice quality, as measured by a range of standardized parameters, including milling, appearance, nutrition and eating and cooking quality (ECQ), has been commonly used to select for and evaluate the breeding lines prior to commercial release (Zhang et al., 2018; Huang X. et al., 2021; Hu et al., 2021; Zhang H. et al., 2021). Amylose and amylopectin constitute starch that is the dominant component in rice grains and accounts for about 90% of the weight of the rice endosperm (Zhou et al., 2002; Wang et al., 2018). As the most important quality trait, ECQ is affected by multiple factors such as amylose content (AC) and an eclectic list of physicochemical properties of endosperm (Lu et al., 2013; Chun et al., 2015; Van Hung et al., 2016). Despite the recent increase in the awareness of the health benefits of high AC rice, it appears that a seemingly polarized market has emerged, especially for the formulation of specialty food, where the low AC trait is desired as it could well be attributed for the favorable food texture and taste (Sagare et al., 2020; Hu et al., 2021). Further, some rice cultivars, such as Nanjing 46 and Nanjing 9108, with relatively high ECQ and low AC (10-15%) are popular with consumers in China and the broad East Asian region, which are soft yet not sticky after cooking, and are defined as soft japonica rice (Li et al., 2018; Hu et al., 2021; Zhang C. et al., 2021). Therefore, there is impetus to develop rice genotypes with low AC to meet market requirements.
FLOURY ENDOSPERM 2 (FLO2), a member of a conserved gene family in plants, encodes a tetratricopeptide repeat domain (TPR)-containing protein located in the nucleus (She et al., 2010; Wu et al., 2015; Kolli et al., 2019; Suzuki et al., 2020). It has been reported that FLOURY ENDOSPERM 2 could not only modulate the expression of starch synthesis-related genes including OsAGPL2, OsAGPS2b, OsGBSSI, OsBEI, OsBEIIb, OsISA1 and OsPUL, but also directly impact on the amylose biosynthesis by manipulating the splicing efficiency of Wx pre-mRNA (Wu et al., 2015; Cai et al., 2022; Feng et al., 2022; Sreenivasulu et al., 2022). Previous studies have shown that flo2, the loss-of-function mutant of FLOURY ENDOSPERM 2, produced two distinct phenotypes, one with white and floury endosperm, and the other with dull grain (She et al., 2010; Hunt et al., 2013; Boehlein et al., 2018). In the former, starch granules were small, spherical and loosely packed with large air space (Malinova et al., 2017; Jiang et al., 2018; Suzuki et al., 2020). The FLOURY ENDOSPERM 2 also affected grain size as demonstrated by both overexpression and null mutation approaches (She et al., 2010; Zhang et al., 2013). In addition, flo2 mutant was also endowed with alterations in grain physicochemical properties including grain breakdown, setback and consistency (Wu et al., 2015; Zhang et al., 2017). Therefore, FLOURY ENDOSPERM 2 plays an important role in the determination of grain quality by regulating the accumulation of storage substances in the endosperm. Previous methods to generate flo2 mutants by chemical mutagens such as EMS have the disadvantages of safety risk, low efficiency and uncontrollable direction (Wu et al., 2015; Kihira et al., 2017; Wu et al., 2019).
In the last few years, as one of the most advanced systems for genome editing, CRISPR/Cas has been commonly used to make precise and predictable genome modifications in crop plants to obtain desired traits (Zhu et al., 2020; Gao, 2021). This is especially true for rice, where impressive progress has been made and a plethora of traits have been modified by applying the CRISPR/Cas9 editing system, by virtue of rice’s economic significance and the ease of transgenic regeneration (Zhou et al., 2014; Ma et al., 2021). For grain quality improvement, Wx is a key gene that encodes granule binding starch synthase I (GBSSI) involved in amylose biosynthesis. It has been genetically edited to manipulate AC and/or improve ECQ in a number of recent studies in rice (V.M. Butardo et al., 2017; Bello et al., 2019; Fei et al., 2019; Huang et al., 2020; Zeng et al., 2020; Huang L. et al., 2021; Xu et al., 2021). Likewise, a number of other key genes involved in amylopectin fine structure, such as SSII-2, SBEIIb, BEIIb and ISA1, have been edited for ECQ improvements (Crofts et al., 2015; Sun et al., 2017; Chao et al., 2019; Tappiban et al., 2022).
In this study, to induce AC and improve ECQ of a widely cultivated elite japonica variety, Suken118 (SK118), two loss-of-function mutants of FLOURY ENDOSPERM 2 gene were generated by CRISPR/Cas9 technology. Our results show that knockout of FLOURY ENDOSPERM 2 gene can significantly reduce the content of straight chain powder. The physicochemical properties of flo2 mutants showed that lower AC and viscosity, higher gelation degree and gelatinization temperature were helpful to improve the edible and cooking quality of rice. Therefore, FLOURY ENDOSPERM 2 has potential application value in green and healthy rice breeding.
Results
Generation and identification of flo2 mutants in rice
To generate flo2 mutants with the expectation to produce null mutations, sgRNA was designed in the coding region of FLOURY ENDOSPERM 2. The constructed expression vector flo2 gRNA was expressed and driven by the OsU3 promoter, and the Cas9 cassette was driven by the ubiquitin promoter (Figure 1A). We took advantage of these CRISPR/Cas expression vectors to transform rice variety Sk118 by Agrobacterium tumefaciens infecting rice calli (Figure 1B). Positive T0 transgenic plants were identified by PCR amplification of a fragment of the Hyg gene that was used as the selection marker. The target genomic region of FLOURY ENDOSPERM 2 was amplified by a pair of primers (FLO2-TF/TR; Table S1) flanking the target site and sequenced. In addition, we failed to find any mutations in any of the potential off-target sites (Figure 1C). The sequencing results of T0 plants showed that the mutation rate was as high as 74.19%, among which the percentages of mutation by inserting the base “T” and “A” were the highest, being 30.43% and 19.57%, respectively. Two homozygous T1 mutant lines free of T-DNAs, named d29-2 and d29-3, were selected for further research, featuring the insertion of a single base of “T” and “A”, respectively, resulting in frame shift and premature termination of translation (Figure 2C).
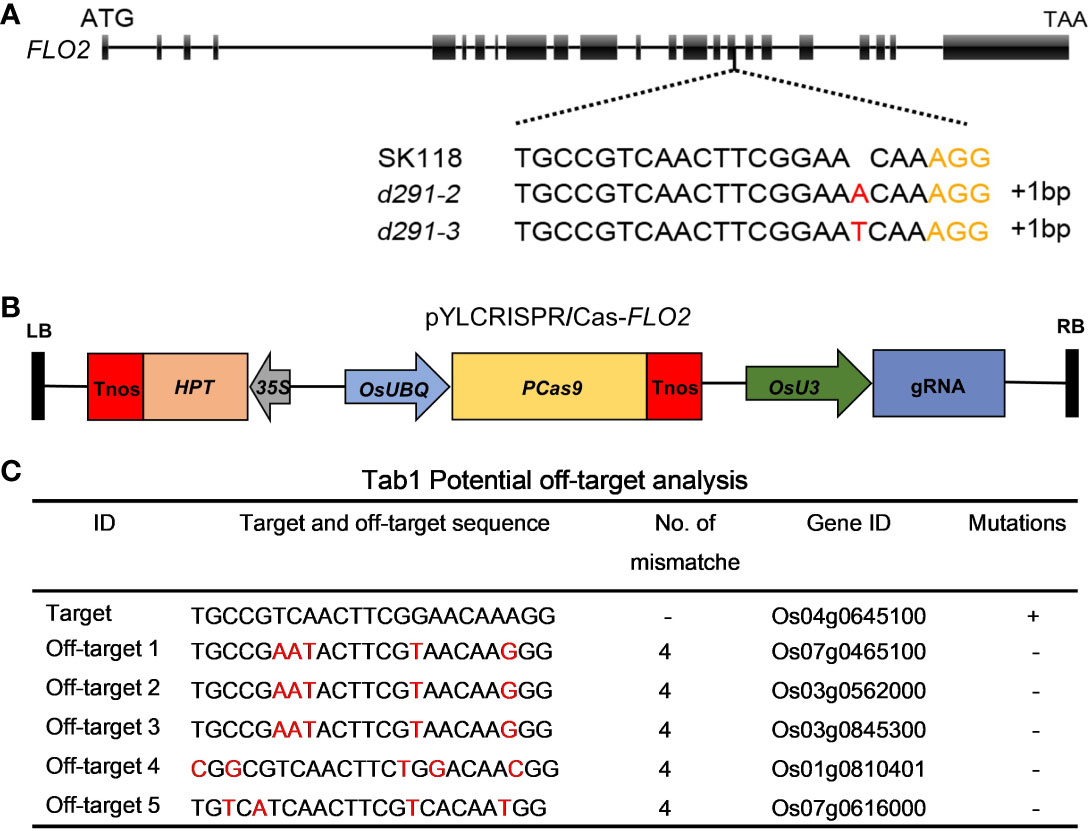
Figure 1 CRISPR/Cas9-induced mutations in the FLOURY ENDOSPERM 2 genes. (A) Schematic of the FLOURY ENDOSPERM 2 gene structures and target sites. Exons and introns are indicated with black rectangles and black lines, respectively. The spacer and PAM sequences were marked in black and orange. (B) The expression vector of pYLCRISPR/Cas9-Flo2. The expression of Cas9 is driven by the maize ubiquitin promoter (OsUBQ); the expression of the sgRNA scaffold is driven by the rice OsU3 small nuclear RNA promoter; the expression of hygromycin (HPT) is driven by CaMV35S promoter. Tnos, the terminator; LB and RB, left border and right border, respectively. (C) Potential miss analysis. Red fonts indicate different bases.
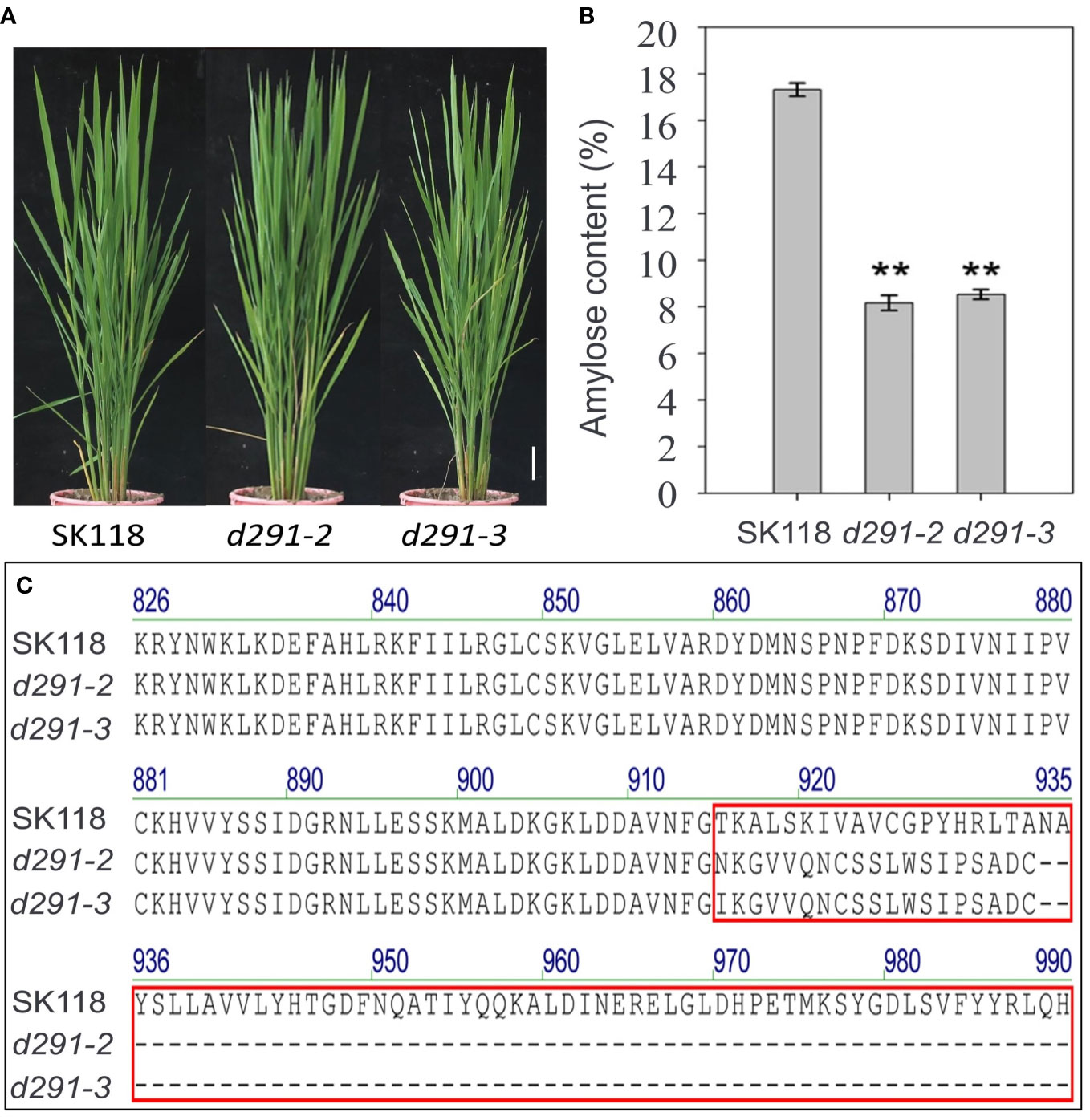
Figure 2 Phenotype and amino acid variations assays. (A) Phenotypes of flo2 mutant lines (d291-2 and d291-3) in SK118 background. Bar = 5 cm. (B) Amylose content (%). Data are given as the mean ± S.D. (n ≥ 3). Student’s t test: *P < 0.05; **P < 0.01; ns, no significance. (C) Amino acid variations of the FLO2 protein in the flo2 mutant. Wild-type, SK118. Homozygous mutations identified at the target sites of d291-2 and d291-3 mutant lines in the T1 generation. Amino acid variations were indicated in red box, with the number representing the order in the proteins. The ellipsis dots represent premature stop codons.
Knockout of FLOURY ENDOSPERM 2 gene has multiple effects on agronomic traits
All the plants were grown in the experimental field in Jiangsu Academy of Agricultural Sciences, Nanjing, China, during normal rice-growing seasons. The edited plants derived from d29-2 and d29-3 did not exhibit discernible variations from wild type (WT) control in grain length, plant height, the panicle length, the number of grains per panicle and tiller numbers per plant (Figure 2A, Figures 3A, B, E–H). In contrast, the grain width and grain thickness of the flo2 mutants were significantly lesser than WT control, which is congruent with the previous studies on chemically induced flo2 mutants (She et al., 2010); (Figures 3C, D). Conceivably, the 1000-grain weights of d29-2 and d29-3 were reduced by 26.50% and 20.90%, respectively (Figure 3I).
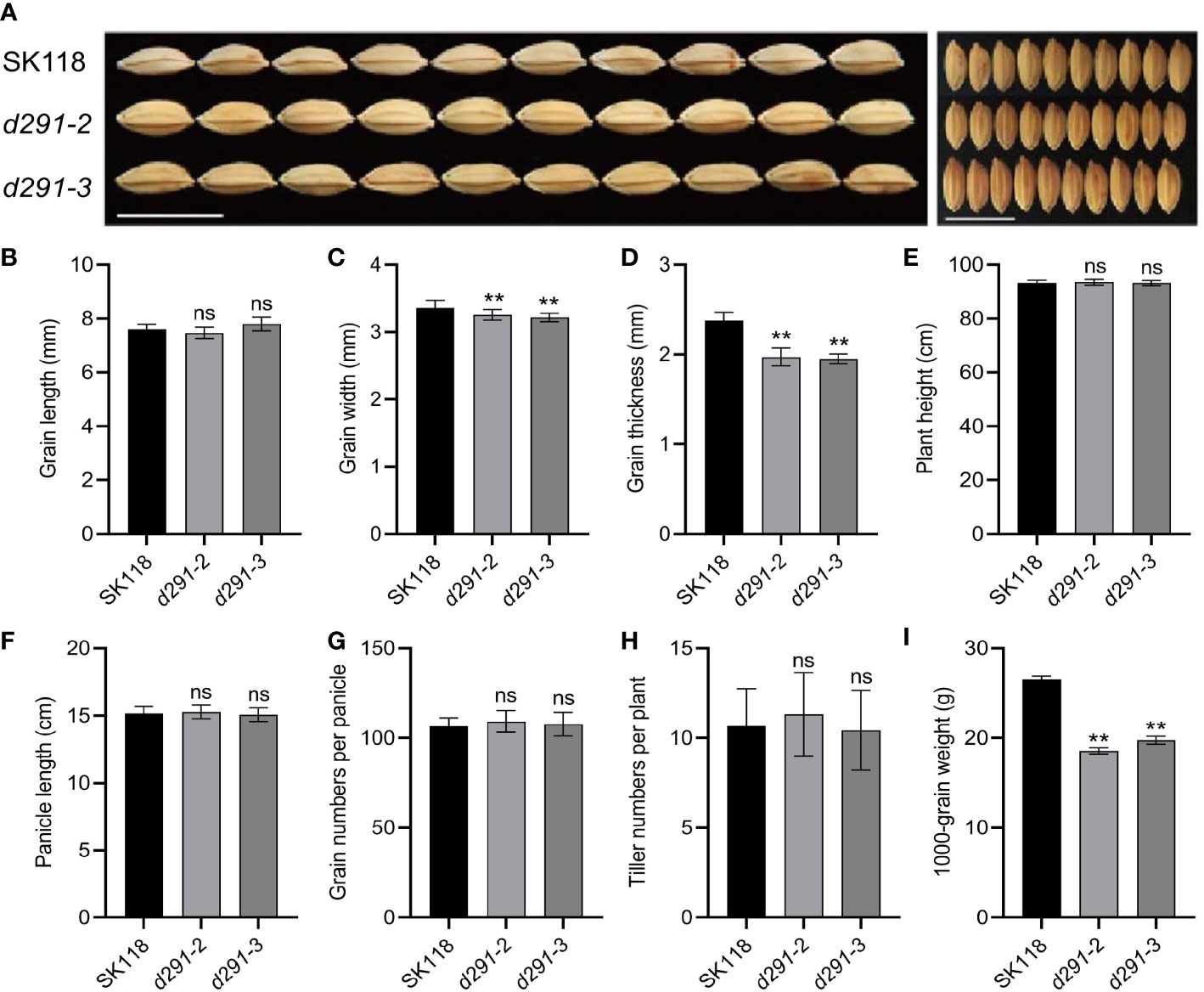
Figure 3 Knockout of FLOURY ENDOSPERM 2 gene has multiple effects on agronomic traits. (A) Grain phenotypes of flo2 mutant lines (d291-2 and d291-3) in SK118 background. Bar = 1 cm. (B) grain length (mm). (C) grain width (mm). (D) grain thickness (mm). (E) plant height(cm). (F) panicle length (cm). (G) grain numbers per panicle. (H) tiller numbers per plant. (I) 1000-grain weight (G). Data are given as the mean ± S.D. (n ≥ 20). Student’s t test: **P < 0.01; ns, not significance.
Rice yield and grain appearance quality are complex quantitative characters, which are influenced by genetic background and environmental factors. Although FLOURY ENDOSPERM 2 gene knockout mutants have multiple negative effects on grain appearance and yield, rice with low amylose content and high cooking quality has a huge market demand. In future studies, FLOURY ENDOSPERM 2 gene and yield trait genes can be combined, and the mutual balance between yield and trait can be realized by using the interaction effect between different genes.
Improvement of ECQ and taste value of flo2 transgenic rice
To further assess the effects of flo2 mutants on rice ECQ, we performed a number of assays on the grain. The gel consistency (GC) in d29-2 and d29-3 increased significantly relative to WT by 11.6 mm and 18.0 mm, respectively (Figure 4A). The gelatinization temperature (GT) of the mutant lines was about 22°C higher than WT (Figure 4D). The increases in GC and GT in flo2 mutant relative to WT are favorable for improving ECQ in rice (Zhang et al., 2020; Sreenivasulu et al., 2022). The RVA pasting profiles of the flo2 mutants were also distinct from WT, specifically, the peak paste viscosity (PKV), hot paste viscosity (HPV) and cold paste viscosity (CPV) of the mutants were significantly lower than those of WT (Figure 4B). The protein content (PC) was only increased of 1.68% and 1.64% in d29-2 and d29-3 mutants, respectively (Figure 4C), which had no significant effect on ECQ (Sreenivasulu et al., 2022). Generally, the increase of gel consistency is conducive to improving the taste quality of rice. Taken together, these results manifest the complex changes in the physicochemical properties of cooked rice grains in the flo2 mutants, which are generally conducive to ECQ improvements.
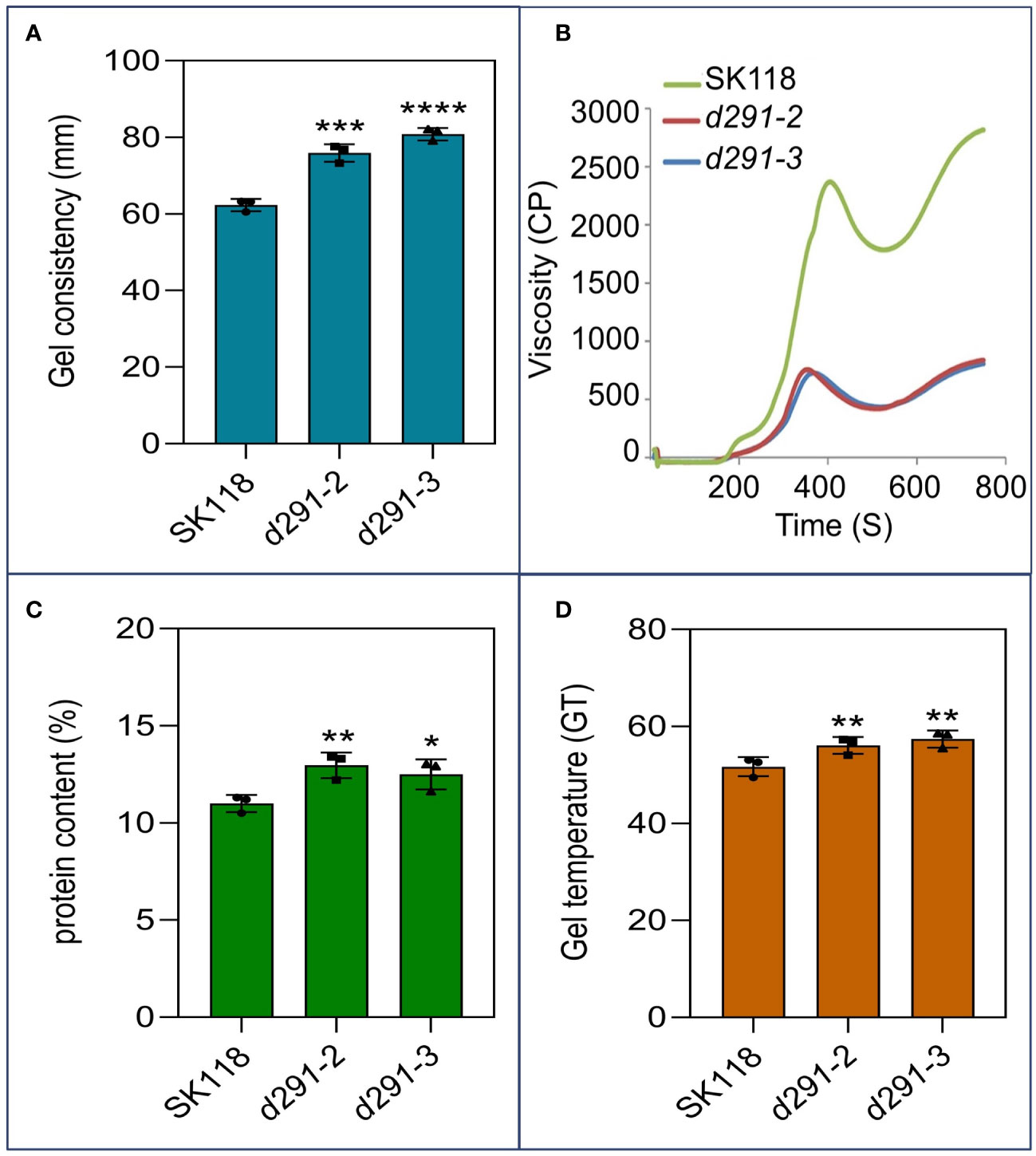
Figure 4 Improvement of ECQ and taste value of flo2 transgenic rice. (A) Gel consistency (mm). (B) RVA spectra of rice flours. (C) Total protein content of the rice flours (%). (D) Gelatinization temperature (GT) of rice flours from SK118, d291-2 and d291-3. Data are given as the mean ± S.D. (n ≥ 3). Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Rice starch with a higher amylose content always shows faster retrogradation, leading to higher setback viscosity (SBV) of the starch, while starch with a lower amylose content has a higher relative crystallinity and short-range molecular order, resulting in higher swelling power, PKV and breakdown viscosity (BDV) (Wang et al., 2017; Cai et al., 2022). Previous studies showed that AC was positively correlated with PKV, HPV, CPV in a significant manner, whereas it was negatively correlated with adhesiveness, cohesiveness, and GT (Kong et al., 2015). Our results in this study on flo2 lines that were endowed with low AC was concurrent with the reductions in these RVA pasting values, corroborating the previous reports. GC is an important indicator for distinguishing rice grains that do not undergo retrogradation (i.e., rice remains soft after cooking and cooling) (Zhang et al., 2020; Sreenivasulu et al., 2022). GT is also a crucial factor that affects rice ECQ. It is generally recognized that the rice grains with high GT would need more time to cook, and the texture of the cooked rice tends to be less sticky, especially when the cooked rice is cooled (Li et al., 2021; Sreenivasulu et al., 2022). The enhancements in GC and GT values in the flo2 mutants, as unraveled in this study, are clearly favorable for ECQ amelioration.
Opaque appearance phenotype of flo2 mutants rice grain
The polished rice grains derived from d29-2 and d29-3 were featured with distinctive opaque and shriveled appearance (Figure 5A). There might be some phenotypic variations in grain appearance between the FLO-edited lines and the flo2 mutants that were reported in previous studies, which could well be attributed to the different genetic backgrounds and cultivation practices used between different studies (She et al., 2010; Wu et al., 2015). The images of transverse sections showed that the endosperm of the flo2 mutants were more opaque and powdery compared to WT at the center of the endosperm, but such differences were diminished away from the center region (Figure 5B).

Figure 5 Appearance phenotype analysis of flo2 mutants rice grain. (A) Appearance of polished rice from flo2 mutant lines (d291-2 and d291-3) in SK118 background. Bar = 1 cm. (B) Transverse sections of rice grains. Bar = 300 μm. (C) Rice grains stained with KI-I2. Bar = 300 μm.
We speculate that this is mainly due to the fact that there are many gaps between the poorly filled starch bodies in the endosperm. When the light shines on the endosperm in the internal area, the light diffuses between the gaps, causing the endosperm to appear powdery. When the inner amyloplasts are closely arranged and there is no gap, the light can directly pass through and present transparent endosperm. Such spatial variation in endosperm between flo2 and WT is intriguing and warrants further investigation.
Genome editing of FLOURY ENDOSPERM 2 decreases amylose content significantly
Amylose has high binding capacity with iodine. The higher the AC is, the deeper the staining in the starch-iodine reaction becomes (Agasimani et al., 2013). As expected, the iodine staining of the endosperms in the transverse sections of flo2 seeds were overtly lighter than WT, indicative of AC reduction in the mutants (Figure 5C). The measurement of AC by using AA3 continuous flow analyzer showed that the AC of d29-2 and d29-3 were 8.20% and 8.56%, respectively, which are in sharp contrast to that of WT (17.35%) (Figure 2B). Rather, AC in the flo2 mutants resemble that of a typical soft rice (Wu et al., 2015). Starch is the first kind of storage material in rice. The reduction of 1000 grain weight of flo2 mutants is mainly caused by the reduction of total starch content in endosperm of seeds. Furthermore, we detected the transcript levels of FLOURY ENDOSPERM 2 in the WT (SK118), d291-2 and d291-3 plants. The transcript levels of FLOURY ENDOSPERM 2 were decreased in the d291-2 and d291-3 plants, consistent with the previous study (Figure S1) (She et al., 2010).
Morphology of starch granules in endosperm cells of flo2 mutants
To explore the causes of endosperm changes, we observed the endosperm of wild type seeds and two mutant seeds with scanning electron microscopy. Further, scanning electron microscopy images showed that the starch grains of the flo2 mutants were irregularly spherical and loosely arranged, contrary to the regular spherical shape and uniformity in size, and tight arrangement (Figure 6A). The loose arrangement of flo2 mutants starch granules is the main reason for its opaque appearance.

Figure 6 Morphology of starch granules in endosperm cells of flo2 mutants. (A) Scanning electron microscopy images of the transverse section of flo2 mutants and wild-type. 25×, Bars = 200μm. 500×, Bars = 20 μm. 1000×, Bars = 10 μm.
We speculated that the starch accumulation in flo2 mutants was poor, which led to the loose endosperm structure. During the ripening process of rice, the powder region of endosperm is easy to be broken under external pressure such as water loss, forming sub grains. A large number of cavities were left between loose starch grains, which led to diffuse reflection of light to form a silty, opaque endosperm phenotype.
Discussion
In summary, in this study we have generated low AC/high ECQ rice genotypes through targeted mutagenesis of FLOURY ENDOSPERM 2 by using CRISPR/Cas9 approach and demonstrated that editing of FLOURY ENDOSPERM 2 could be an effective route to reduce AC and accelerate rice breeding to achieve desired high ECQ trait. We have also analyzed the physicochemical properties of the flo2 mutants, which have not only verified the improvements in rice ECQ, but also provided useful data for industrial evaluation of these potentially useful germplasm. Undesired, the mutations of FLOURY ENDOSPERM 2 have led to a significant reduction in seed weight, which may imply a possible reduction in grain yield. This necessitates further evaluation for their agronomic performance, especially in the field conditions. Nevertheless, there might be a net gain in its output value given that the high value niche market for high-ECQ grains is rapidly expanding. Future studies could also be directed to the incorporation of high yielding traits, for example, by simultaneous targeting a number of negative regulatory genes, such as GS3 (Zeng et al., 2019), TGW6 (Hang et al., 2018; Zeng et al., 2019) and GS6 (Sun et al., 2013), leading to an optimal balance between quality and yield traits.
Materials and methods
Plant materials and growth conditions
Suken118 (SK118) is an elite japonica rice variety with good resistance and quality, which is cultivated widely in the central of Jiangsu Province, China (http://ricedata.cn). The 20bp long target sites were chosen in the FLOURY ENDOSPERM 2 codon region. The CRISPR/Cas9 vector was constructed as previously described then the cassette was transferred into SK118 callus by Agrobacterium tumefaciens-mediated transformation using the strain EHA105 (Hiei et al., 1997; Ma et al., 2015). Transgenic rice lines were grown in greenhouse and test fields in Jiangsu Academy of Agricultural Sciences in Nanjing, China, during normal rice-growing seasons.
Genotype assays and phenotype
Positive T0 transgenic plants were identified by PCR amplification of a fragment of the Hyg gene that was used as a selection marker. The target genomic region of FLOURY ENDOSPERM 2 was amplified by a pair of primers (FLO2-TF/TR; Table S1) flanking the target site and sequenced. Seeds were collected for each plant from WT and homozygous mutant lines. Grain length, plant height, the panicle length, the number of grains per panicle and tiller numbers per plant were measured in the fields. The 1,000 grains weight was measured after the oven at 45°C until constant weight. The grain length, width, and thickness were measured using a vernier caliper.
Grain phenotype and iodine staining of endosperm
The hulls of rice seeds were removed to observe the external appearance of the grain in both WT and mutant lines. Grains were cut through the center to expose the endosperm; 1μL 0.1% iodine-potassium iodide solution was dropped on the endosperm surface and photographs were taken after 3-5 min by VHX-500FE.
RNA isolation and qPCR
Total RNAs were extracted from the rice endosperm of rice using a TRIzol kit according to the user’s manual (Invitrogen) for expression analysis. And then reverse-transcribed into cDNA from 1 μg of total RNA following the manufacturer’s instructions (Fastking RT Kit; TIANGEN). qRT-PCR analysis was performed using a Light Cycler 480 system (Roche) and SYBR Green Real-time PCR Master Mix. The relative expression levels of FLOURY ENDOSPERM 2 were normalized to the rice UBIQUITIN (Os03g0234200) gene, which was used as an internal control. All date were measured by three individual replicates. The transcript levels were calculated by the 2-ΔΔCT method. The primer sequences are listed in Table S1.
Determination of AC and total protein content
Amylose content (AC) was measured using an AA3 continuous flow analyzer set. Flour from WT, mutant lines and four rice samples with known apparent amylose content was taken 0.05 g to determine. AA3 continuous flow analyzer set was also used to determine total protein content by measuring 0.2 g of flour from WT and mutant lines. The conversion coefficient is 5.95. All samples were repeated three times.
Evaluation of grain GC, GT and viscosity property
Gel consistency (GC) was measured following the procedure described in GB/T 22294-2008/ISO 11747:2012. Gelatinization temperature (GT) was indirectly estimated via the alkali digestion test. Rice viscosity properties were determined using a Rapid Visco Analyser (RVA-TecMaster, Newport Scientific, Warriewood, Australia). Rice flour (3g, 12% m.b.) was mixed with 25 g of double-deionized water in the RVA sample can. Thrice measurements were performed for each sample.
Scanning electron microscopy of seed cross-section
After drying, the mature seeds with shelling were frozen in liquid nitrogen for 5S then the cross-section of the samples was manually snapped and sputter-coated with gold palladium on copper studs. Magnifications of about 25×, 500×, and 1000× were used to observe endosperm cross-section and starch granule morphology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
JY, GL and YF designed and supervised the research. XS, ZC, XD, FW and JW performed most experiments. YX and WL analyzed date. XS, BL, YT and YL wrote the paper. X-LT and YZ provided resources. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Project of Seeds innovation in Jiangsu (GBGS [2021]001), Natural Science Foundation of Jiangsu Province (BK20190255) and the Exploratory Project of the Jiangsu Academy of Agricultural Sciences (ZX (21)1201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1138523/full#supplementary-material
Abbreviations
FLO2, FLOURY ENDOSPERM 2; TPR, tetratricopeptide repeat domain; AC, amylose content; ECQ, eating and cooking quality; SK118 Suken118; GC, gel consistency; GT, gelatinization temperature; Wx, waxy gene; SSII-2, soluble starch synthase ii-2; SBEIIb, starch branching enzyme IIb; BEIIb, branching enzyme IIb; ISA1, isoamylose 1; WT, wild type; Hyg, hygromycin; MNU, nitrogen methyl nitroso groupurea; EMS, methyl sulfonic acid ethyl ester; RVA, rapid visco analyser; PKV, peak paste viscosity; HPV, hot paste viscosity; CPV, cool paste viscosity; GS3, grain size 3; TGW6, thousand-grain weight 6; GS6, grain size 6.
References
Agasimani, S., Selvakumar, G., Joel, A. J., Ganesh Ram, S. (2013). A simple and rapid single kernel screening method to estimate amylose content in rice grains. Phytochem. Anal. 24 (6), 569–573. doi: 10.1002/pca.2433
Bello, B. K., Hou, Y., Zhao, J., Jiao, G., Wu, Y., Li, Z., et al. (2019). NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa l.). Plant Biotechnol. J. 17 (7), 1222–1235. doi: 10.1111/pbi.13048
Boehlein, S. K., Shaw, J. R., Boehlein, T. J., Boehlein, E. C., Hannah, L. C. (2018). Fundamental differences in starch synthesis in the maize leaf, embryo, ovary and endosperm. Plant J. 96 (3), 595–606. doi: 10.1111/tpj.14053
Cai, Y., Zhang, W., Fu, Y., Shan, Z., Xu, J., Wang, P., et al. (2022). Du13 encodes a C2 H2 zinc-finger protein that regulates Wx(b) pre-mRNA splicing and microRNA biogenesis in rice endosperm. Plant Biotechnol. J. 20 (7), 1387–1401. doi: 10.1111/pbi.13821
Chao, S., Yicong, C., Baobing, F., Guiai, J., Zhonghua, S., Ju, L., et al. (2019). Editing of rice isoamylase gene ISA1 provides insights into its function in starch formation. Rice Sci. 26 (2), 77–87. doi: 10.1016/j.rsci.2018.07.001
Chun, A., Lee, H. J., Hamaker, B. R., Janaswamy., S. (2015). Effects of ripening temperature on starch structure and gelatinization, pasting, and cooking properties in rice (Oryza sativa). J. Agric. Food Chem. 63 (12), 3085–3093. doi: 10.1021/jf504870p
Concepcion, J., Ouk, M., Zhao, D., Fitzgerald, M. A. (2015). The need for new tools and investment to improve the accuracy of selecting for grain quality in rice. Field Crops Res. 182, 60–67. doi: 10.1016/j.fcr.2015.05.003
Crofts, N., Abe, N., Oitome, N. F., Matsushima, R., Hayashi, M., Tetlow, I. J., et al. (2015). N. fujita. amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 66 (15), 4469–4482. doi: 10.1093/jxb/erv212
Fei, Y., Jie, Y., Fangquan, W., Fangjun, F., Wenqi, L., Jun, W., et al. (2019). Production of two elite glutinous rice varieties by editing Wx gene. Rice Sci. 26 (2), 118–124. doi: 10.1016/j.rsci.2018.04.007
Feng, T., Wang, L., Li, L., Liu, Y., Chong, K., Theissen, G., et al. (2022). OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm. New Phytol. 234 (1), 77–92. doi: 10.1111/nph.17990
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184 (6), 1621–1635. doi: 10.1016/j.cell.2021.01.005
Han, Y., Luo, D., Usman, B., Nawaz, G., Zhao, N., Liu, F., et al. (2018). Development of high yielding glutinous cytoplasmic Male sterile rice (Oryza sativa l.) lines through CRISPR/Cas9 based mutagenesis of Wx and TGW6 and proteomic analysis of anther. Agronomy 8, 290–311. doi: 10.3390/agronomy8120290
Hiei, Y., Komari, T., Kubo, T. (1997). Transformation of rice mediated by agrobacterium tumefaciens. Plant Mol. Biol. 35 (1), 205–218. doi: 10.1023/A:1005847615493
Hu, Y., Cong, S., Zhang, H. (2021). Comparison of the grain quality and starch physicochemical properties between japonica rice cultivars with different contents of amylose, as affected by nitrogen fertilization. Agriculture 11 (7), 616–630. doi: 10.3390/agriculture11070616
Huang, L., Gu, Z., Chen, Z., Yu, J., Chu, R., Tan, H., et al. (2021). Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 106 (4-5), 419–432. doi: 10.1007/s11103-021-01162-8
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 18 (11), 2164–2166. doi: 10.1111/pbi.13391
Huang, X., Su, F., Huang, S., Mei, F., Niu, X., Ma, C., et al. (2021). Novel Wx alleles generated by base editing for improvement of rice grain quality. J. Integr. Plant Biol. 63 (9), 1632–1638. doi: 10.1111/jipb.13098
Hunt, H. V., Moots, H. M., Graybosch, R. A., Jones, H., Parker, M., Romanova, O., et al. (2013). Waxy phenotype evolution in the allotetraploid cereal broomcorn millet: Mutations at the GBSSI locus in their functional and phylogenetic context. Mol. Biol. Evol. 30 (1), 109–122. doi: 10.1093/molbev/mss209
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8 (1), 1497. doi: 10.1038/s41467-017-01501-8
Jiang, J. Z., Kuo, C. H., Chen, B. H., Chen, M. K., Lin, C. S., Ho, S. L. (2018). Effects of OsCDPK1 on the structure and physicochemical properties of starch in developing rice seeds. Int. J. Mol. Sci. 19 (10), 3247–3262. doi: 10.3390/ijms19103247
Kihira, M., Taniguchi, K., Kaneko, C., Ishii, Y., Aoki, H., Koyanagi, A., et al. (2017). Arabidopsis thaliana FLO2 is involved in efficiency of photoassimilate translocation, which is associated with leaf growth and aging, yield of seeds and seed quality. Plant Cell Physiol. 58 (3), 440–450. doi: 10.1093/pcp/pcw217
Kolli, R., Soll, J., Carrie, C. (2019). OXA2b is crucial for proper membrane insertion of COX2 during biogenesis of complex IV in plant mitochondria. Plant Physiol. 179 (2), 601–615. doi: 10.1104/pp.18.01286
Kong, X., Zhu, P., Sui, Z., Bao., J. (2015). Physicochemical properties of starches from diverse rice cultivars varying in apparent amylose content and gelatinisation temperature combinations. Food Chem. 172, 433–440. doi: 10.1016/j.foodchem.2014.09.085
Li, Q. F., Huang, L. C., Chu, R., Li, J., Jiang, M. Y., Zhang, C. Q., et al. (2018). Down-regulation of SSSII-2 gene expression results in novel low-amylose rice with soft, transparent grains. J. Agric. Food Chem. 66 (37), 9750–9760. doi: 10.1021/acs.jafc.8b02913
Li, H., Yan, S., Yang, L., Xu, M., Ji, J., Mao, H., et al. (2021). Starch gelatinization in the surface layer of rice grains is crucial in reducing the stickiness of parboiled rice. Food Chem. 341, 128202–128221. doi: 10.1016/j.foodchem.2020.128202
Lu, S., Cik, T.-T., Lii, C.-y., Lai, P., Chen., H.-H. (2013). Effect of amylose content on structure, texture and α-amylase reactivity of cooked rice. LWT - Food Sci. Technol. 54 (1), 224–228. doi: 10.1016/j.lwt.2013.05.028
Ma, Z., Wei, M., Zhang, Y., Qin, G., Liu, C., Li, Z., et al. (2021). Development of CRISPR_Cas9 genome editing system and its application in rice molecular breeding. In Vitro Cell. Dev. Biol. - Plant 57 (4), 700–708. doi: 10.1007/s11627-021-10203-2
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8 (8), 1274–1284. doi: 10.1016/j.molp.2015.04.007
Malinova, I., Alseekh, S., Feil, R., Fernie, A. R., Baumann, O., Schottler, M. A., et al. (2017). Starch synthase 4 and plastidal phosphorylase differentially affect starch granule number and morphology. Plant Physiol. 174 (1), 73–85. doi: 10.1104/pp.16.01859
Sagare, D. B., Abbai, R., Jain, A., Jayadevappa, P. K., Dixit, S., Singh, A. K., et al. (2020). More and more of less and less: Is genomics-based breeding of dry direct-seeded rice (DDSR) varieties the need of hour? Plant Biotechnol. J. 18 (11), 2173–2186. doi: 10.1111/pbi.13454
She, K. C., Kusano, H., Koizumi, K., Yamakawa, H., Hakata, M., Imamura, T., et al. (2010). A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22 (10), 3280–3294. doi: 10.1105/tpc.109.070821
Sreenivasulu, N., Zhang, C., Tiozon, R. N., Jr., Liu, Q. (2022). Post-genomics revolution in the design of premium quality rice in a high-yielding background to meet consumer demands in the 21st century. Plant Commun. 3 (3), 100271. doi: 10.1016/j.xplc.2021.100271
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8, 298. doi: 10.3389/fpls.2017.00298
Sun, L., Li, X., Fu, Y., Zhu, Z., Tan, L., Liu, F., et al. (2013). GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J. Integr. Plant Biol. 55 (10), 938–949. doi: 10.1111/jipb.12062
Suzuki, R., Imamura, T., Nonaga, Y., Kusano, H., Teramura, H., Sekine, K. T., et al. (2020). H. shimada. A novel FLOURY ENDOSPERM2 (FLO2)-interacting protein, is involved in maintaining fertility and seed quality in rice. Plant Biotechnol. (Tokyo) 37 (1), 47–55. doi: 10.5511/plantbiotechnology.19.1212b
Tappiban, P., Hu, Y., Deng, J., Zhao, J., Ying, Y., Zhang, Z., et al. (2022). Relative importance of branching enzyme isoforms in determining starch fine structure and physicochemical properties of indica rice. Plant Mol. Biol. 108 (4-5), 399–412. doi: 10.1007/s11103-021-01207-y
Van Hung, P., Chau, H. T., Phi, N. T. (2016). In vitro digestibility and in vivo glucose response of native and physically modified rice starches varying amylose contents. Food Chem. 191, 74–80. doi: 10.1016/j.foodchem.2015.02.118
V.M. Butardo, Anacleto, R., Parween, S., Samson, I., de Guzman, K., Alhambra, C. M., et al. (2017). N. sreenivasulu. systems genetics identifies a novel regulatory domain of amylose synthesis. Plant Physiol. 173 (1), 887–906. doi: 10.1104/pp.16.01248
Wang, J., Hu, P., Lin, L., Chen, Z., Liu, Q., Wei, C. (2018). Gradually decreasing starch branching enzyme expression is responsible for the formation of heterogeneous starch granules. Plant Physiol. 176 (1), 582–595. doi: 10.1104/pp.17.01013
Wang, S., Li, P., Yu, J., Guo, P., Wang, S. (2017). Multi-scale structures and functional properties of starches from indica hybrid, japonica and waxy rice. Int. J. Biol. Macromol 102, 136–143. doi: 10.1016/j.ijbiomac.2017.04.020
Wu, Y. P., Pu, C. H., Lin, H. Y., Huang, H. Y., Huang, Y. C., Hong, C. Y., et al. (2015). Three novel alleles of FLOURY ENDOSPERM2 (FLO2) confer dull grains with low amylose content in rice. Plant Sci. 233, 44–52. doi: 10.1016/j.plantsci.2014.12.011
Wu, M., Ren, Y., Cai, M., Wang, Y., Zhu, S., Zhu, J., et al. (2019). Rice FLOURY ENDOSPERM10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol. 223 (2), 736–750. doi: 10.1111/nph.15814
Xu, Y., Lin, Q., Li, X., Wang, F., Chen, Z., Wang, J., et al. (2021). Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 19 (1), 11–13. doi: 10.1111/pbi.13433
Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., et al. (2020). Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18 (12), 2385–2387. doi: 10.1111/pbi.13427
Zeng, Y., Wen, J., Zhao, W., Wang, Q., Huang., W. (2019). Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR-Cas9 system. Front. Plant Sci. 10, 1663. doi: 10.3389/fpls.2019.01663
Zhang, C., Chen, S., Ren, X., Lu, Y., Liu, D., Cai, X., et al. (2017). Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OsGBSSI activity. J. Agric. Food Chem. 65 (10), 2222–2232. doi: 10.1021/acs.jafc.6b05448
Zhang, A., Gao, Y., Li, Y., Ruan, B., Yang, S., Liu, C., et al. (2020). Genetic analysis for cooking and eating quality of super rice and fine mapping of a novel locus qGC10 for gel consistency. Front. Plant Sci 11, 342. doi: 10.3389/fpls.2020.00342
Zhang, H., Xu, H., Jiang, Y., Zhang, H., Wang, S., Wang, F., et al. (2021). Genetic control and high temperature effects on starch biosynthesis and grain quality in rice. Front. Plant Sci. 12, 757997. doi: 10.3389/fpls.2021.757997
Zhang, C., Yang, Y., Chen, Z., Chen, F., Pan, L., Lu, Y., et al. (2020). Characteristics of grain physicochemical properties and the starch structure in rice carrying a mutated ALK/SSIIa gene. J. Agric. Food Chem. 68 (47), 13950–13959. doi: 10.1021/acs.jafc.0c01471
Zhang, C., Yang, Y., Chen, S., Liu, X., Zhu, J., Zhou, L., et al. (2021). A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 63 (5), 889–901. doi: 10.1111/jipb.13010
Zhang, Y. C., Yu, Y., Wang, C. Y., Li, Z. Y., Liu, Q., Xu, J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31 (9), 848–852. doi: 10.1038/nbt.2646
Zhang, J., Zhang, H., Botella, J. R., Zhu, J.-K. (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 60 (5), 369–375. doi: 10.1111/jipb.12620
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H., Yang, B. (2014). Large Chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42 (17), 10903–10914. doi: 10.1093/nar/gku806
Zhou, Z., Robards, K., Helliwell, S., Blanchard, C. (2002). Composition and functional properties of rice. Int. J. Food Sci. Technol. 37 (8), 849–868. doi: 10.1046/j.1365-2621.2002.00625.x
Keywords: FLOURY ENDOSPERM 2, amylose content (AC), eating and cooking quality (ECQ), japonica rice, CRISPR/Cas9, genome editing
Citation: Song X, Chen Z, Du X, Li B, Fei Y, Tao Y, Wang F, Xu Y, Li W, Wang J, Liang G, Zhou Y, Tan X, Li Y and Yang J (2023) Generation of new rice germplasms with low amylose content by CRISPR/CAS9-targeted mutagenesis of the FLOURY ENDOSPERM 2 gene. Front. Plant Sci. 14:1138523. doi: 10.3389/fpls.2023.1138523
Received: 05 January 2023; Accepted: 20 February 2023;
Published: 13 March 2023.
Edited by:
Peng Wang, Institute of Botany (CAS), ChinaReviewed by:
Ming Zheng, Oil Crops Research Institute (CAAS), ChinaKumar Paritosh, University of Delhi, India
Copyright © 2023 Song, Chen, Du, Li, Fei, Tao, Wang, Xu, Li, Wang, Liang, Zhou, Tan, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Yang, eWFuZ2ppZTE2OEBhbGl5dW4uY29t
†These authors have contributed equally to this work
 Xiaohong Song
Xiaohong Song Zhihui Chen2,3†
Zhihui Chen2,3† Yang Xu
Yang Xu Wenqi Li
Wenqi Li Guohua Liang
Guohua Liang Yong Zhou
Yong Zhou Xiaoli Tan
Xiaoli Tan Yulong Li
Yulong Li Jie Yang
Jie Yang