- 1State Key Laboratory of Crop Biology/Agronomy College, Shandong Agricultural University, Taian, China
- 2College of Life Sciences, Qingdao Agricultural University, Key Lab of Plant Biotechnology in Universities of Shandong Province, Qingdao, China
- 3College of Tropical Crops, Hainan University, Haikou, China
- 4Department of Agricultural Sciences, College of Agriculture and Environmental Sciences, Faculty of Life Sciences, Government College University Faisalabad, Faisalabad, Pakistan
- 5Institute of Industrial Crops, Shandong Academy of Agricultural Sciences, Jinan, China
The HVA22 family of genes, induced by abscisic acid and stress, encodes a class of stress response proteins with a conserved TB2/DP1/HVA22 domain that are unique among eukaryotes. Previous studies have shown that HVA22s play an important role in plant responses to abiotic stresses. In the present study, 34, 32, 16, and 17 HVA22s were identified in G. barbadense, G. hirsutum, G. arboreum, and G. raimondii, respectively. These HVA22 genes were classified into nine subgroups, randomly distributed on the chromosomes. Synteny analysis showed that the amplification of the HVA22s were mainly due to segmental duplication or whole genome replication (WGD). Most HVA22s promoter sequences contain a large number of drought response elements (MYB), defense and stress response elements (TC-rich repeats), and hormone response elements (ABRE, ERE, SARE, etc.), suggesting that HVA22s may respond to adversity stresses. Expression profiling demonstrated that most GhHVA22s showed a constitutive expression pattern in G. hirsutum and could respond to abiotic stresses such as salt, drought, and low temperature. Overexpression of GhHVA22E1D (GH_D07G0564) in Arabidopsis thaliana enhances salt and drought tolerance in Arabidopsis. Virus-induced gene silencing of GhHVA22E1D reduced salt and drought tolerance in cotton. This indicates that GhHVA22E1D plays an active role in the plant response to salt stress and drought stress. GhHVA22E1D may act in plant response to adversity by altering the antioxidant capacity of plants. This study provides valuable information for the functional genomic study of the HVA22 gene family in cotton. It also provides a reference for further elucidation of the functional studies of HVA22 in plant resistance to abiotic stress response.
Introduction
Plants are easily affected by environmental stresses during growth due to their immobility, and salt stress and drought stress are the two major abiotic stress factors that seriously affect plant growth (Magwanga et al., 2019a; Magwanga et al., 2019b). Plants have evolved a range of regulatory mechanisms to cope in response to environmental stresses, for example, osmotic regulation, antioxidant defense regulation, signal transduction mechanisms, etc. The abscisic acid (ABA) signaling pathway plays an important part in the response of plants to abiotic stresses (Campos-Rivero et al., 2017; Liu et al., 2020). ABA regulates plant physiological and metabolic responses by controlling the expression levels of many stress-responsive genes, enabling plants to tolerate stresses (Ma et al., 2018; Zhang et al., 2021). HVA22 is a unique ABA-induced gene, which was first discovered in Hordeum vulgare L. aleurone cells (Shen et al., 1993). There was a high sequence similarity between plant HVA22s and human TB2/DP1 proteins. HVA22 family proteins with conserved TB2/DP1/HVA22 domains were ubiquitous in eukaryotes, but were not found in prokaryotes(Sharon, 2017). HvHVA22 helps to regulate vesicular transport in stressed cells and reduces the non-essential secretion of stressed cells, which results in improved plant resistance (Guo and David Ho, 2008).
In previous studies, multiple functions of HVA22s have been identified. HvHVA22 negatively regulates GA (Gibberellic acid)-mediated programmed cell death and vacuolation in barley dextrin cells (Guo and David Ho, 2008). Deletion of AtHVA22d leads to enhanced autophagy and impaired flower development in Arabidopsis thaliana (Chen et al., 2009). Overexpression of OsHLP1 (HVA22-like protein 1) significantly enhances blast disease resistance by impairing endoplasmic reticulum homeostasis in Oryza sativa (Meng et al., 2022). Furthermore, numerous works have shown that HVA22s has abiotic stress response function. HvHVA22s and AtHVA22s were up-regulated in in barley and A. thaliana, respectively, to cope with various environmental stresses, such as salinity, drought, cold, and exogenous ABA (Shen et al., 1993; Shen et al., 2001; Chen et al., 2002; Collin et al., 2020). The ZmHVA22 in Zea mays was down-regulated significantly under high salt, simulated drought and cold stress, while the ZmHVA22 was up-regulated to varying degrees under high temperature stress, ethylene induction, and ABA(Chen et al., 2014). The expression of SaHVA22 in Spartina alterniflora was significantly higher when treated with a salt concentration of 68 mM than other salt concentration treatments (0 mM, 137 mM, 205 mM) (Courtney et al., 2016). Moreover, the expression of HVA22s was up-regulated in Vicia faba under cold stress (Lyu et al., 2021), G. hirsutum under salinity stress (Yuan et al., 2019) and Solanum tuberosum under drought stress (Benny et al., 2019).
In previous studies, the potential of HVA22s in abiotic stress had been emphasized, and systematic family analysis of HVA22s could help to comprehend their functional properties better. Up to now, the HVA22 family has been systematically analyzed merely in Arabidopsis thaliana, Solanum lycopersicum, Citrus clementina and Citrus sinensis, and 5, 15, 6 and 6 HVA22s had been identified, respectively (Chen et al., 2002; Gomes Ferreira et al., 2019; Wai et al., 2022). The five AtHVA22s in A. thaliana could be divided into two subfamilies; thereinto, the sequences of AtHVA22d and AtHVA22e were closer to HVA22 in barley (Chen et al., 2002). The HVA22s of tomato and citrus were divided into four subfamilies according to the sequence similarity with AtHVA22s (Gomes Ferreira et al., 2019; Wai et al., 2022).
Cotton is a vital source of natural fiber crops in the world and one of the main sources of plant protein and plant oil, and it is one of the important strategic materials in China (Liu et al., 2022). Around 1.6 million years ago, the A0-genome of an extinct diploid cotton and the D5-genome of the diploid cotton Gossypium raimondiid were crossed and doubled to form a heterotetraploid species (Grover et al., 2015; Huang et al., 2020; Peng et al., 2022). This tetraploid specie then diverged subsequently into seven tetraploid cottons namely G. hirsutum, G. barbadense, G. tomentosum, G. mustelinum, G. darwinii, G. ekmanianum, and G. stephensii, and the genomes of these seven tetraploid cotton species were labeled as (AD)1 to (AD)7 (Chen et al., 2020b; Peng et al., 2022). Among them, G. hirsutum and G. barbadense were domesticated as cultivated species, the two cultivated allotetraploid cotton species with quite different traits in morphology, yield, fiber quality, environmental adaptability, and genomic sequences (Hu et al., 2019). G. herbaceum (A1) and G. arboreum (A2) were formed about 0.7 million years ago by A0-genome divergence (Huang et al., 2020). At present, the genomes of G. barbadense, G. hirsutum, G. arboreum, and G. raimondii had been well sequenced (Paterson et al., 2012; Du et al., 2018; Hu et al., 2019; Huang et al., 2020); this greatly facilitates the determination of HVA22 gene family members in cotton and clarification of their evolutionary relationships.
In this study, the HVA22s was identified from G. raimondii, G. arboreum, G. hirsutum, and G. barbadense. The characteristics of gene structure, subcellular localization, conserved motifs and domains, evolution, synteny relationship, chromosome localization, and expression patterns were systematically characterized and analyzed. Furthermore, the distribution of cis-acting elements in the promoters of the HVA22 family was also analyzed, which has an important role in our further understanding of the function of HVA22s. Previous studies had identified the GhHVA22E1D gene as a differentially expressed gene under salt stress through a whole-genome association and differential expression analysis of salt tolerance in G. hirsutum during germination (Yuan et al., 2019). This study has shown that overexpression of GhHVA22E1D, the HVA22 family member of G. hirsutum, improved the drought resistance and salt tolerance in A. thaliana. Meanwhile, silencing of GhHVA22E1D reduced salt tolerance and drought resistance in cotton. In general, the results of this research provided a reference for further studies to understand the action of HVA22s in cotton.
Materials and methods
Database and sequence retrieval
The genome and annotation files of G. barbadense (H7124, ZJU), G. hirsutum (TM-1, ZJU), G. raimondii (JGI), and G. arboreum (Shiyaxi1 CRI) were downloaded from CottonFGD (https://cottonfgd.net/). A. thaliana HVA22 protein sequences (Chen et al., 2002) were downloaded from TAIR online website (http://www.arabidopsis.org/index.jsp).
Identification and genetic characterization of HVA22 family members
All the five A. thaliana HVA22 protein sequences were taken as reference sequences for comparison. BLASTP search was used to scan the whole-genome protein sequences of the four cotton species. The Hidden Markov Model (HMM) profile of the HVA22 domain (PF03134) was obtained from the Pfam website (http://pfam.xfam.org/). The HMM search was used to identify HVA22. The NCBI Conserved Domain Database (CDD: https://www.ncbi.nlm.nih.gov/cdd), using an automated model and default parameters (maximum number of hits = 500, threshold = 0.01), was used to subject the putative HVA22 protein sequences to HVA22 conserved structural domain validation, removing the putative HVA22 protein sequence that did not contain the TB2/DP1/HVA22 domain.
The basic information and physical and chemical properties of all HVA22s, including physical location, length, strand, molecular weight (Mw), the number of amino acids, charge, grand average of hydropathy (GRAVY), and isoelectric point (pI), were obtained in CottonFGD through the feature analysis function (Zhu et al., 2017). Finally, the chromosome localization of HVA22s was mapped by TBtools and genome annotation gff3 files (Chen et al., 2020a).
Phylogenetic analysis of HVA22s
The phylogenetic tree of G. hirsutum, G. barbadense, G. raimondii, G. arboreum, and A. thaliana HVA22s was built with Mega X (Kumar et al., 2018). The ClustalW with default parameters was used to align the protein sequences, and then Poisson model was employed to construct a maximum likelihood tree, and 1000 replicates bootstrap were performed. Finally, ITOL (https://itol.embl.de/) was applied to colorize this tree. All HVA22s were named uniformly based on evolutionary clustering results and physical location on chromosomes.
Duplication and synteny analysis of HVA22s
A one-to-one comparison of the HVA22s protein sequences of G. hirsutum, G. barbadense, G. raimondii, and G. arboreum was performed by BLAST. Next, the MCScanX (http://chibba.pgml.uga.edu/duplication/) was used to calculate the four cotton species of the synteny examination of paralogous genes.
Gene structure analysis and identification of conserved motifs
The coding sequence and full gene length of the HVA22s were submitted to the Gene Structure Display Server (GSDS 2.0, http://gsds.gao-lab.org/) for gene structure analysis. Multiple Em for Motif Elicitation (MEME v5.1.0, http://meme-suite.org/tools/meme) was carried out to identify the conserved protein motifs of HVA22s. The parameter was set as follow: znr (the occurrences of each functional domain in each sequence was variable) as the distribution type of the structure domain in the sequence, the width of motifs was 6–50, and 15 motifs were calculated (Bailey et al., 2009). Finally, the final data visualization was plotted by TBtools.
Cis-acting regulatory elements analysis
A 2000 bp sequence of each HVA22 gene upstream of the start codon was extracted from the CottonFGD database and predict the cis-acting elements contained in these sequences using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002); in addition, the visualization of cis-acting elements associated with stress response and hormonal regulation by TBtools.
Expression profiling of GhHVA22s
All expression data of HVA22s under different stresses (blank, drought, salt, high temperature, and low temperature) were acquired from the gene expression database of TM-1(Hu et al., 2019). The expression profiles of HVA22s from different organs (leaf, stem, root, torus, petal, sepal, calycle, upper stamens, lower stamens, 0–5dpa unseparated ovules, and fibers, 10–25 dpa fiber, 10–20 dpa ovule) were also obtained. The expression of HVA22s was normalized to log2(FPKM), and heat maps were created with TBtools for visualization.
A 4-week-old salt-tolerant upland cotton variety Han682 was watered with 250 mM NaCl and water and was used as control group. The whole growing process of cotton keeps 16 h light/8 h dark, 25°C. At 0, 3, 6, 12, and 24 h after treatment, root tissue samples were taken for RNA extraction using the OminiPlant RNA Kit (DNase I) (CWBIO), then reverse transcribed into cDNA for qRT-PCR verification of GhHVA22s. Each sample was taken from two cotton seedlings, and three replicates of samples were taken at each point. The NCBI primer design website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design primers for GhHVA22s. GhUBQ14 was used as internal control to standardize the expression of target genes. Gene expression was computed by 2-ΔΔCt, and Ct was the cycling threshold. The primers are listed in Table S7.
Expression characterization of GhHVA22E1D
GhHVA22E1D expression characterization was performed using 4-week-old Han 682 cotton. Blank control plants were watered with water until the soil was saturated. During salt stress treatment, NaCl (250mM) solution was watered until soil saturation. For drought treatment, the soil was saturated with PEG6000 (10%) solution. The leaves were sprayed with 500 μM ABA solution for exogenous ABA treatment. Root, stem, and leaf samples were collected at 0, 3, 6, 12, and 24 h after treatment, respectively. RNA extraction, reverse transcription, and qPCR analysis were then performed, as above. The primers are listed in Table S7.
Construction of plant expression vector
The GhHVA22E1D coding region was amplified from Han 682 using KOD polymerase (Toyobo, Shanghai, China). After double digestion with KpnI and SacI, the vector was connected to the plant overexpression vector pCAMbia2300, and 35S:: GhHVA22E1D vector was obtained (Figure S3).
The conserved sequence of GhHVA22E1D was amplified from Hand682 using KOD polymerase. The CLCrV-GhHVA22E1D vector was obtained by double digestion with SpeI and AscI and ligation to the VIGS expression vector CLCrV-00 (Gu et al., 2014). The amplification primers used are listed in Table S7.
Evaluation of stress tolerance in Arabidopsis thaliana overexpressing GhHVA22E1D
A. thaliana plants overexpressing GhHVA22E1D were obtained by infecting Col-0 with flower dip method (Clough and Bent, 1998). Positive materials were obtained after kanamycin screening and PCR identification. The transformed event OE22 with the highest GhHVA22E1D expression was selected for purification screening, and pure T3 generation seeds were obtained for subsequent experiments. A light cycle of 16 h light/8 h dark was maintained throughout the reproductive period, and the temperature was always kept at 23°C.
To evaluate salt tolerance in overexpressing GhHVA22E1D A. thaliana, NaCl solution (200 mM) was used to water 30-day-old A. thaliana daily, and the relative electrical conductivity and the total chlorophyll content of rosette leaves were measured after 7 days. For drought treatment, 70 seeds each of WT and OE A. thaliana were sown on both sides in the same pot. After 3 weeks of normal culture, the soil was saturated with water and then naturally dried for 7 days. Then re-watering was performed, and the survival rate was counted 2 days after re-watering.
VIGS of GhHVA22E1D in G. hirsutum and evaluation of salt tolerance and drought resistance
Cotyledons of 7-day-old cotton (Han 682) were infected with A. tumefaciens LBA4404 carrying the pCLCrV-GhHVA22E1D plasmid. The plants injected with pCLCrV-CLA1 were taken as positive control, and plants injected with pCLCrV-00 were taken as negative control. When positive control plants exhibited an albino phenotype, qPCR was performed on leaf samples injected with the pCLCrV-GhHVA22E1D vector to detect the silencing effect. Next, the silenced plants and negative control plants were treated with salt stress and drought stress, respectively. For salt resistance evaluation, plants were soaked with 400 mM NaCl for 10 days, then the activities of peroxidase (POD) and superoxide dismutase (SOD) and the content of malondialdehyde (MDA) in roots and leaves were determined, respectively. During the drought treatment, the watering was stopped for 2 weeks after the soil absorbed water to saturation. Next, re-watering treatment was performed and after 2 days of re-watering the survival rate was calculated.
Determination of SOD and POD activity and MDA content
SOD activity in plant tissues was measured by photoreduction with the nitroblue tetrazolium method, the activity of POD by the guaiacol colorimetric method, and the MDA content determined by colorimetric method (Ullah et al., 2018). During the measurements, technical replicates were performed three times for each plant sample.
Data statistics and analysis
For the sake of experimental reliability, all biological replicates and sample tests in this study were repeated three times. Finally, three replicate means and standard deviations were given.
Results
Genome-wide identification and characterization of the HVA22 family in four cotton species
A total of 16 GaHVA22s, 17 GrHVA22s, 32 GhHVA22s, and 34 GbHVA22s from four cotton species were identified. The gene ID and physicochemical characteristics of all HVA22 genes are shown in Table S1. The length of HVA22 proteins ranged from 140 to 418 amino acids (aa) in diploid cotton, 140 to 519 aa in G. barabadense, and 140 to 562 aa in G. hirsutum. The molecular weight ranged from 16.381 to 47.83 kDa, 16.401 to 47.779 kDa, 16.381 to 64.649 kDa, and 16.381 to 59.603 kDa, and the theoretical isoelectric point (pI) between 5.692 and 9.915, 6.262 and 9.915, 5.029 and 9.915, and 6.512 and 9.75 in GaHVA22s, GrHVA22s, GhHVA22s, and GbHVA22s, respectively.
Among all four species of cotton, the amount of HVA22s in the A- and D-genomes was equal or differed by one. The number of HVA22s in the A-genome of G. arboreum, G. barbadense, and G. hirsutum was 16, 17, and16, respectively; and the number of HVA22s in the D-genome of G. raimondii, G. barbadense and G. hirsutum was 17, 17, and 16, respectively. In the A-genomes of G. barbadense and G. hirsutum, there were 1, 3, 2, 1, 2, 1, 2, 1, 2, and 1 HVA22 genes located on chromosomes A01–A13, respectively, except A02, A08, and A11. Among them, there was no HVA22 gene on chromosomes A02, A08, and GhA11, and there was one gene on chromosome GbA11. In the D-genomes of G. barbadense and G. hirsutum, except D04 and D08, there were 1, 2, 1, 1, 2, 1, 2, 1, 1, 2 and 1 HVA22 genes located on chromosomes D01–D13, respectively. Among them, GhD04 has one gene, GbD04 has two genes, and there was no HVA22 gene on D08. In G. arboreum, there were 1, 1, 2, 2, 1, 2, 1, 1, 1, 1, 1, 1, 2, and 1 GaHVA22s distributed on 12 chromosomes except Chr8, respectively. There were two HVA22 genes on each of the four chromosomes and one HVA22 gene on each of the seven chromosomes in G. raimondii. The HVA22 gene in G. raimondii was absent on Chr4, but had two genes on Chr8, which was opposite to the distribution of the previous three cotton species. The distribution of HVA22s on chromosomes is shown in Supplementary Figure S1.
Phylogenetic analysis and subcellular localization prediction of HVA22 genes in four cotton species
In the phylogenetic tree constructed based on the combination of all HVA22 protein sequences of Arabidopsis thaliana and four cotton species, all cotton HVA22s were classified into nine lineages (A, C, E, F, G, H, I, J, and K) without B and D lineages (Figure 1).The number of HVA22 genes varied greatly among different lineages, with the J lineage containing the most genes (23 genes) and the F lineage containing the fewest genes (five genes). In the terminal branch of the phylogenetic tree, three genes from the A-genome or D-genome were clustered together, with the exception of one different branch in each of the F, G, and K lineages. The three genes in each group were from two tetraploid species and one diploid species. This is consistent with the previous finding that the A-genome and D-genome of tetraploid cotton were formed by hybridization and doubling of two diploid cotton species (Wendel and Cronn, 2003; Huang et al., 2020). All HVA22 genes were named uniformly according to their physical location and lineage classification (Table S1).
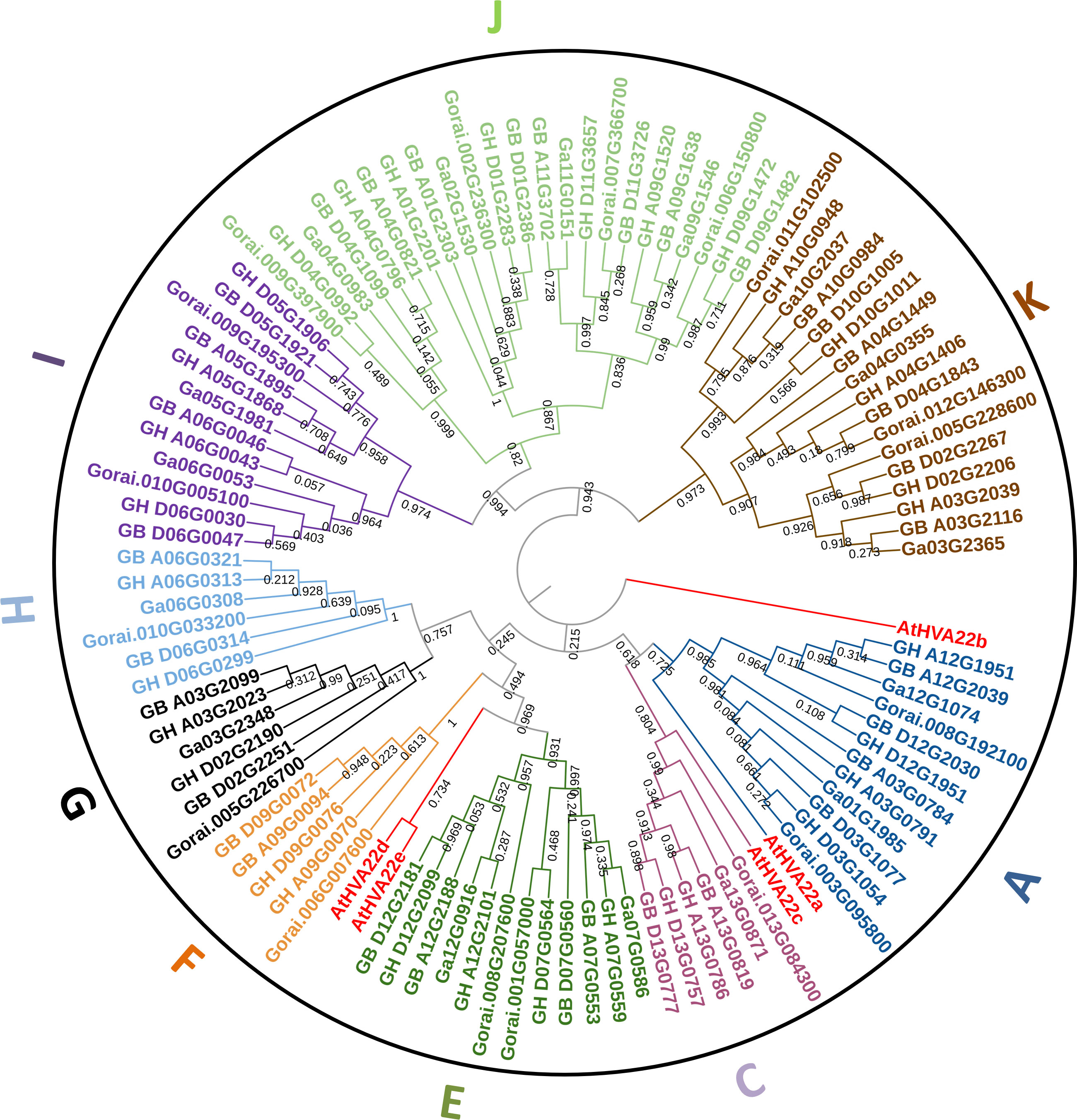
Figure 1 The phylogenetic tree of HVA22s from A. thaliana and four cotton species. The bootstrap value on the branch lines were from 0.03 to 1.
The predicted results of subcellular localization of HVA22 protein are listed in Table S1. Most cotton HVA22s were predicted to be localized to the plasma membrane, while a minority was localized to the cytoplasmic, extracellular space, chloroplast, nuclear, and mitochondria. The proteins in the A, C, E, F, and H lineages were all localized on the plasma membrane, while the proteins of the G lineage proteins were all localized on extracellular space, while the I, J, and K lineages had 8, 5, and 3 proteins with multiple localization sites, respectively. The differences of subcellular localization implied that there might be a diversity of functions of HVA22s.
Synteny and duplication analysis of HVA22s
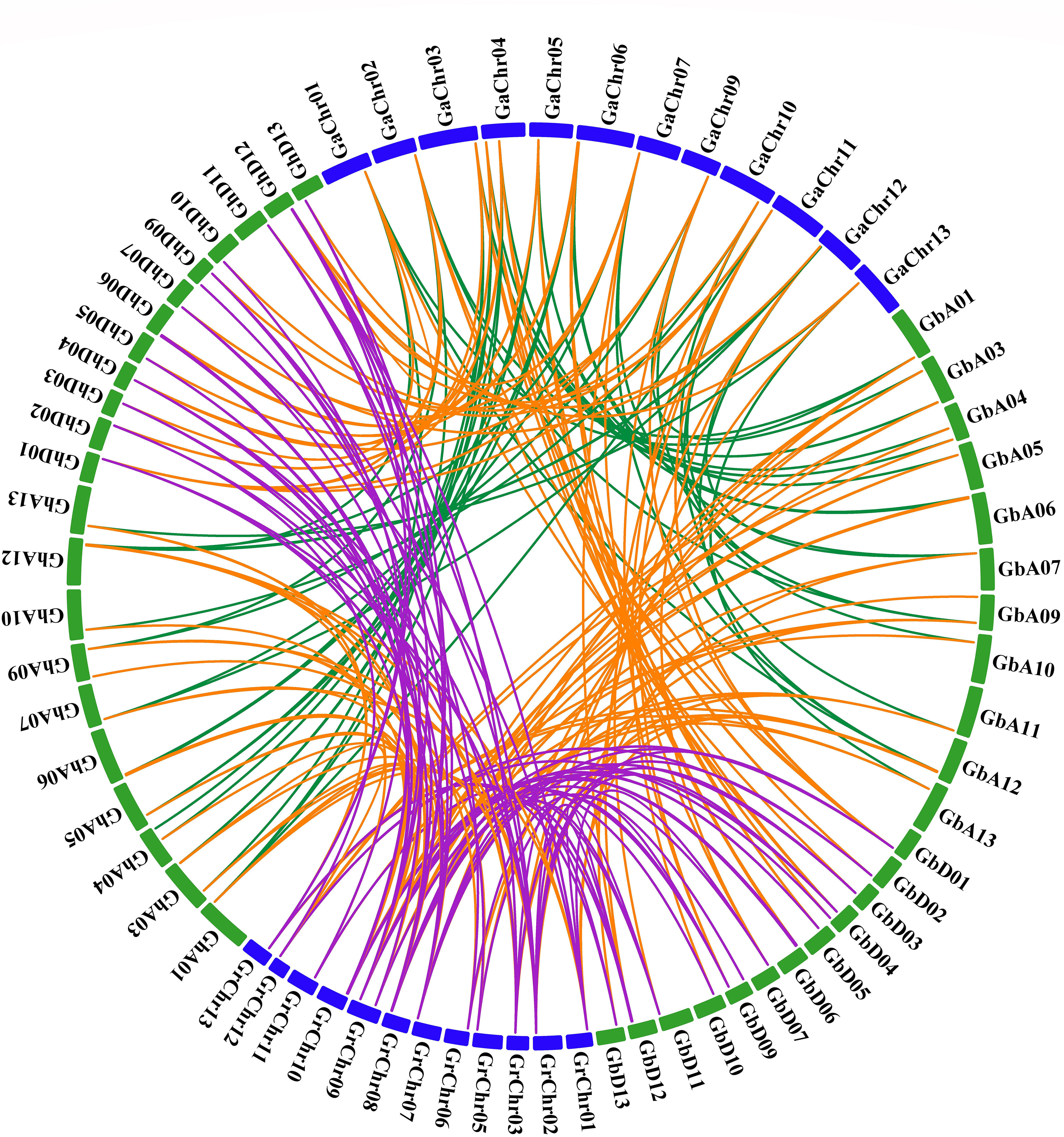
In order to show the synteny relationship, all HVA22s of the four cotton species were compared and aligned for homology, and a synteny relationship plot was constructed (Figure 2). There were 30, 32, and 32 genes homologous to 32 genes of G. hirsutum in G. barbadense, G. raimondii, and G. arboreum, respectively, and there were 30 GhHVA22s that were common homologous in all three cotton varieties (Table S2).
During the evolution of cotton, the included proximal, duplicate mechanisms, dispersed and tandem duplications, and segmental duplications or whole-genome duplications (WGD) played a crucial role in the expanded membership of gene families (Chen et al., 2019; Wang et al., 2019). The duplications of HVA22s were classified as segmental repeats or WGD in both tetraploid cotton (G. hirsutum and G. barbadense) (Table S3), but in diploids (G. raimondii and G. arboreum), three GaHVA22s (Ga01G1985, Ga04G0983, and Ga10G2037) and two GrHVA22s (Gorai.006G007600 and Gorai.011G102500) were identified as dispersed duplications, and the rest were segmental duplications or WGD. This suggests that the expansion and evolution of the HVA22 gene family in cotton may be primarily driven by segmental duplications or WGD.
In order to understand the co-linearity of all HVA22 genes among the four cotton species, all linked gene pairs were identified (Figure 2). Corresponding to G. arboreum, 26 and 25 co-linear gene pairs were characterized in the A-genome of two tetraploids (G. barbadense and G. hirsutum), respectively, and 27 co-linear gene pairs were characterized in the d-genome of both tetraploids. Furthermore, in contrast to G. raimondii, there were 36 and 31 co-linear gene pairs in the two tetraploid A-genomes and 39 and 37 co-linear gene pairs in the D-genome, respectively. Moreover, 33 collinear gene pairs were co-linear between the two diploids and 109 pairs were identified between the two tetraploids.
Structure and conserved motif of cotton HVA22s
The structural map of the HVA22s indicated the distribution of the coding regions (Figure 3). Introns were present in the HVA22s of all four cotton species. There were 10.1% of HVA22s with four exons, which were scattered in C, E, and G lineages. Five exons were found in 36.4% HVA22s, mostly concentrated in A, C, F, E, and H lineages. There were 32.3% of HVA22s that had six exons. These were concentrated in I and J lineages. The remaining 21.2% of the genes had six or more exons. All genes of the K lineages had this characteristic. In addition, untranslated regions (UTRs) were present in all 17 HVA22 genes of G. raimondii. The gff3 files provided by the genomic databases used in this report for G. hirsutum, G. barbadense, and G. arboreum had no UTR annotations, so the distribution of their UTRs was not shown here.

Figure 3 Gene structure and conserved motif of HVA22s in G. hirsutum, G. barbadense, G. arboreum, G. raimondii, and A. thaliana.
Conserved motif analysis of the full-length sequence of the HVA22 protein revealed that 15 amino acid conserved motifs were identified in all HVA22s (Table S4 and Figure 3).The conserved motifs of cotton HVA22 totaled three to nine, and motif 1 was a conserved motif shared by all HVA22 proteins. Furthermore, motif 2 and motif 15 were conserved in the A, C, E, F, G, and H lineages, and motif 12 was specific to the J lineages. The number and type of conserved motifs of genes were similar in the same lineage. This supported the reliability of the evolutionary tree that was constructed in this paper.
Analysis of the cis-acting element of the cotton HVA22s
The promoter prediction results showed that numerous drought-responsive elements (MYB) and light-responsive elements (BOX 4, Sp1, I-box, and G-box) were found in all HVA22s promoter regions (Table S5 and Figure 4). In addition, heat maps were used to visualize cis-acting elements associated with hormone regulation and stress response. There were a variety of phytohormone responsive elements in the HVA22 genes promoter, including the ABA response element (ABRE), ethylene response element (ERE), Methyl Jasmonate response element (CGGTA-motif and TGACG-motif), gibberellin response element (GARE-motif, P-box and TATC-box), and salicylic acid-responsive elements (SARE and TCA-element), which showed that the HVA22s expression was subject to the regulation of different phytohormones. Some stress-responsive elements were also found, such as low temperature response element (LTR), defense and stress response elements (TC-rich repeats), anaerobic inducible element (ARE), and wound response elements (WUN and WRE3 motifs). In addition, several growth and development elements were predicted, including meristem expression elements (CCGTCC-motif and CAT-box) and endosperm expression elements (GCN4-motif). This result showed that HVA22s were involved in the growth of cotton and its response to environmental stresses. Different lineages had disparate gene promoter characteristics. For example, the promoters of the C and E lineages generally contained a large number of osmotic stress response elements (STRE), while the promoters of J and K lineages had a great deal of ethylene response elements (ERs). This suggested that the expression of HVA22s in different subgroups was subject to disparate regulatory factors.
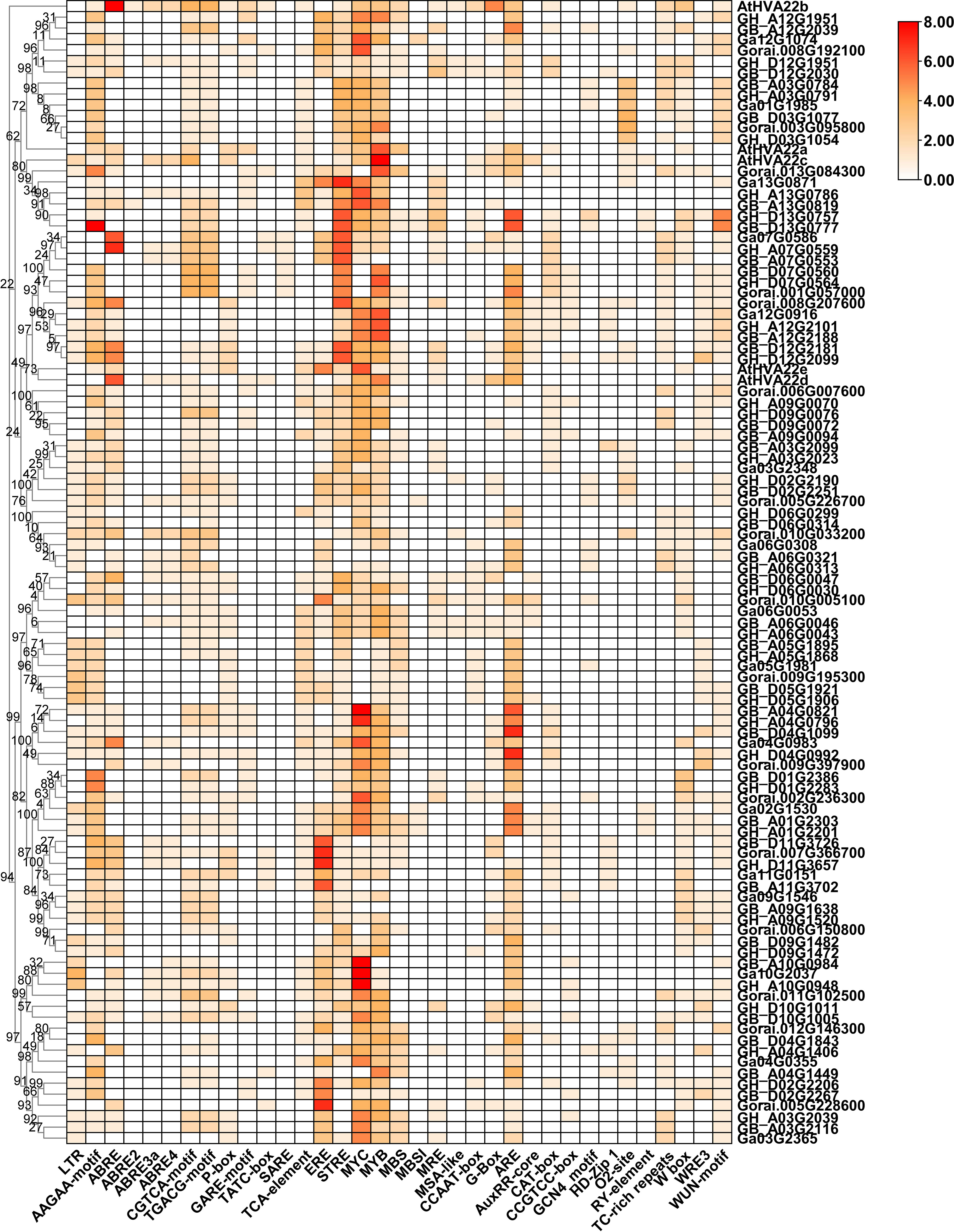
Figure 4 Predicted hormone response, stress response, and growth-related cis-elements in the promoter region of HVA22 genes.
Expression profiling of GhHVA22s under various stress and in different tissues
Heat map of gene expression shows that the expression patterns of homologous genes were relatively similar under stress conditions, except for the two homologous genes of the C lineage (Figure 5; Table S6). Genes in the A lineage were all highly expressed, while most genes in the F and J lineages were less expressed. The response patterns of different HVA22s genes to environmental stress were not necessarily the same. For example, the GhHVA22F1A (GH_A09G0070) gene in the F lineage showed a down-regulated expression pattern in response to stress, while the GhHVA22E1D (GH_D07G0564) gene in the E lineage showed an up-regulated expression pattern under stress conditions. The expression of the same gene was not the same in different plant tissues. For instance, the GhHVA22E1D gene was more highly expressed in tissues such as rhizomes, leaves, and flowers, and less highly expressed in ovules and fibers.
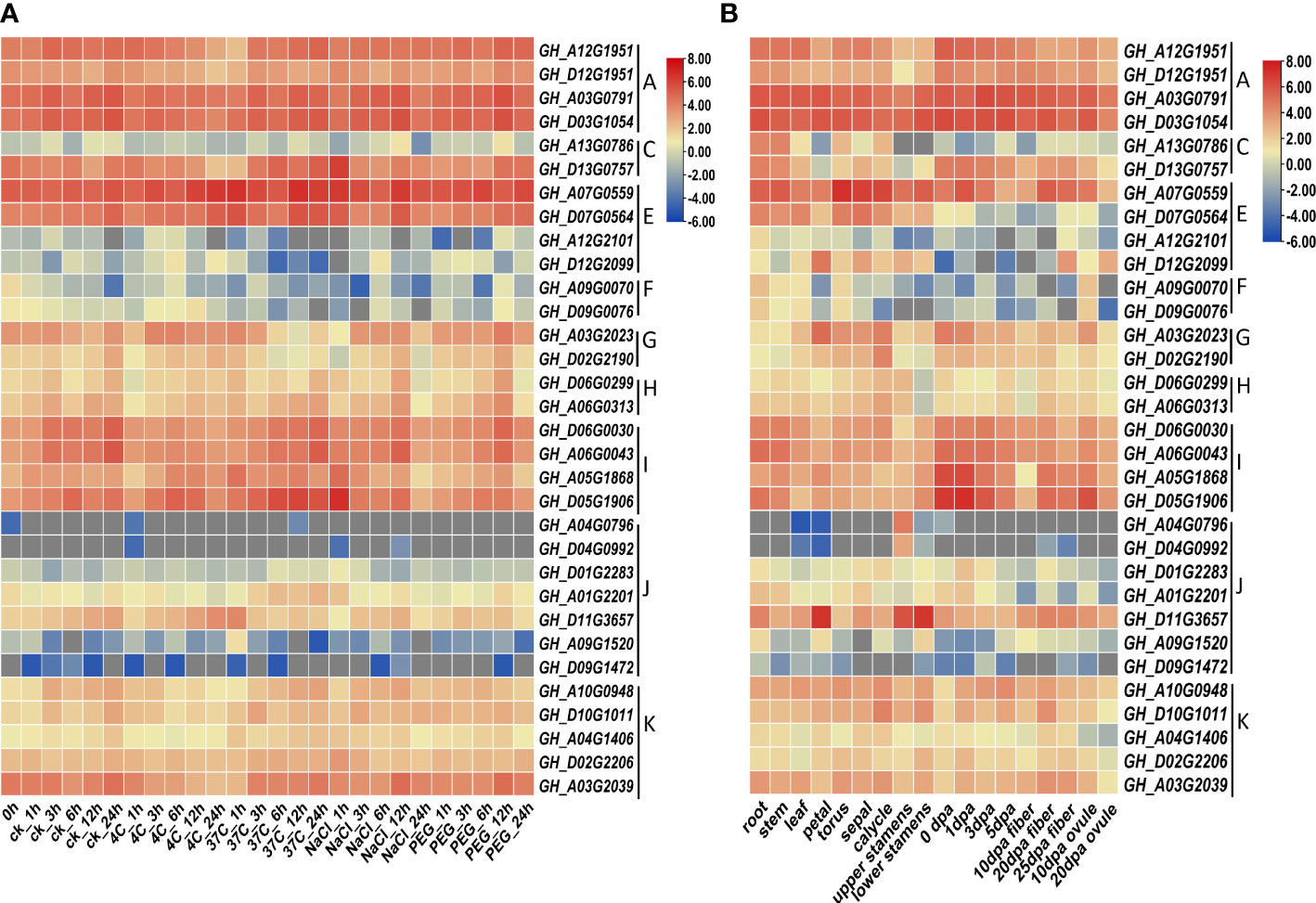
Figure 5 Expression pattern analysis of GhHVA22. (A) Expression pattern analysis of GhHVA22 under control, low temperature, high temperature, salt and drought stress conditions, expressed as CK, 4°C, 37°C, 400mM NaCl and 20% PEG, respectively. (B) Analysis of the expression pattern of GhHVA22 in different organs, where 0-5 dpa represents ovules and fibers that were not isolated for 1–5 days.
To further validate the ability of GhHVA22s to respond to salt stress, six GhHVA22s in the TM-1 database that responded to salt stress were randomly selected for qRT-PCR validation. All six genes were able to rapidly up-regulate their expression after salt stress (Figure 6). Among them, GhHVA22K1D (GH_D02G2206), GhHVA22I1D (GH_D05G1906), GhHVA22J1D (GH_D01G2283), and GhHVA22J1A (GH_A01G2201) had the highest expression at 1 h after salt treatment (250 mM NaCl). The expression level of GhHVA22K1D increased to the highest level at 1 h after salt stress and then gradually decreased until the expression level was similar to the control. The expression of GhHVA22J3D (GH_D09G1472) was gradually up-regulated after salt stress, and the expression level was the highest at 12h. GhHVA22I1D and GhHVA22J1D maintained high levels of expression within 24 h after salt stress. In contrast, GhHVA22J1A was up-regulated only at 1 h and 12 h and showed a down-regulated expression pattern at 6 h after salt treatment. GhHVA22E1A (GH_A07G0559) was up-regulated only at 3 h and 6 h after salt treatment. From these results, most of the genes in the HVA22 family were able to respond to salt stress through expression regulation.

Figure 6 Expression patterns of six GhHVA22 genes in response to salt treatment. Each sample was sampled at 0, 3, 6, 12 and 24h after treatment. The results are the mean of three replicates. Significant differences are indicated by * (P < 0.05) and ** (P < 0.01, Student T-test).
Expression pattern of GhHVA22E1D
Previously, whole-genome association and differential expression analysis of salt tolerance at germination stage in upland cotton were performed in our laboratory. The expression of the GhHVA22E1D (GH_D07G0564) gene, which is related to salt tolerance, was detected to be up-regulated under salt stress (Yuan et al., 2019). Therefore, we performed a more detailed expression pattern detection for GhHVA22E1D. GhHVA22E1D was up-regulated in leaves, stems, and roots under exogenous ABA, salt stress, and drought stress (Figure 7). GhHVA22E1D showed the highest sensitivity to exogenous ABA and salt stress in root tissue. The expression of GhHVA22E1D was up-regulated to 8.2-fold of the control at 24 h after ABA treatment, and the expression level of GhHVA22E1D was up-regulated to 18.6-fold in the roots after salt treatment for 12 h. GhHVA22E1D in leaf tissue and stem tissue was more sensitive to drought stress, reaching 33.7-fold and 19.9-fold that of the control at 12 h and 24 h after drought stress, respectively.
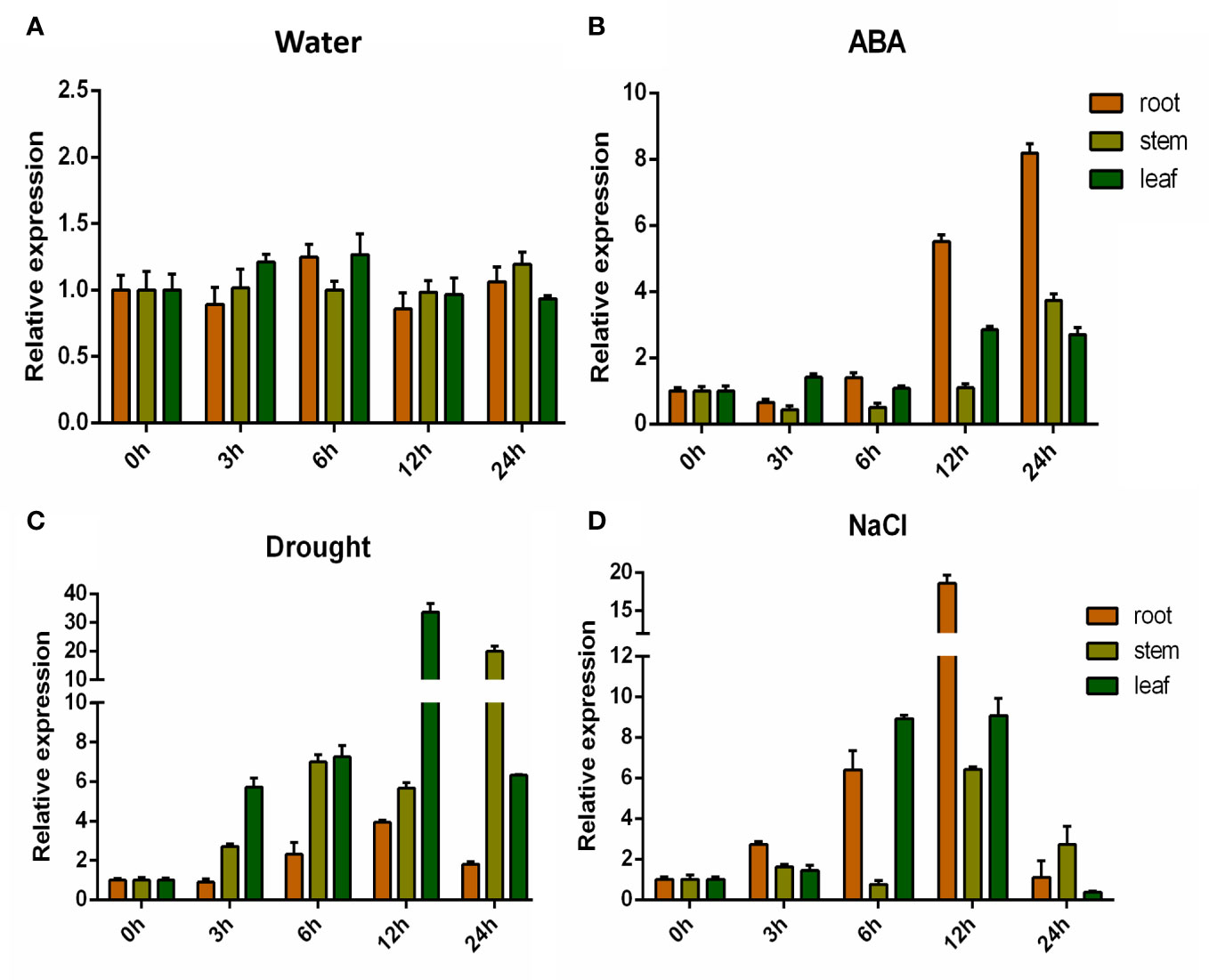
Figure 7 Expression pattern of GhHVA22E1D. Expression pattern of GhHVA22E1D in G hirsutum after 500 μM ABA (B), 10% PEG6000 (C) and 250 mM NaCl (D) stresses, and the water treatment group was the control (A). Each sample was sampled at 0, 3, 6, 12, and 24 h after treatment. The results are the mean of three replicates.
Ectopic overexpression of GhHVA22E1D enhances tolerance of Arabidopsis thaliana to abiotic stresses
GhHVA22E1D was overexpressed in A. thaliana and a homozygous T3 generation was obtained. The OE22 line with the highest gene expression was selected to identify the salt and drought tolerance of the plant (Figure S2). After 7 days of irrigation with 200 mM NaCl solution, the leaves of OE plants died less, while the rosette leaves of WT plants had yellowed and died in large areas (Figure 8A). Chlorophyll content in the leaves of OE plants was significantly higher than that of WT plants after salt stress treatment (Figure 8B), and conductivity was significantly lower than that of control plants (Figure 8C). After watering was stopped for 7 days, almost all the leaves of the WT plants were wilted, while OE plants still had some healthy leaves (Figure 8C). After re-watering, the survival rate of OE plants was 62.26%, which was significantly higher than that of wild-type plants (27.42%) (Figure 8D). These results indicated that the overexpression of GhHVA22E1D could significantly improve the salinity and drought resistance in A. thaliana plants.
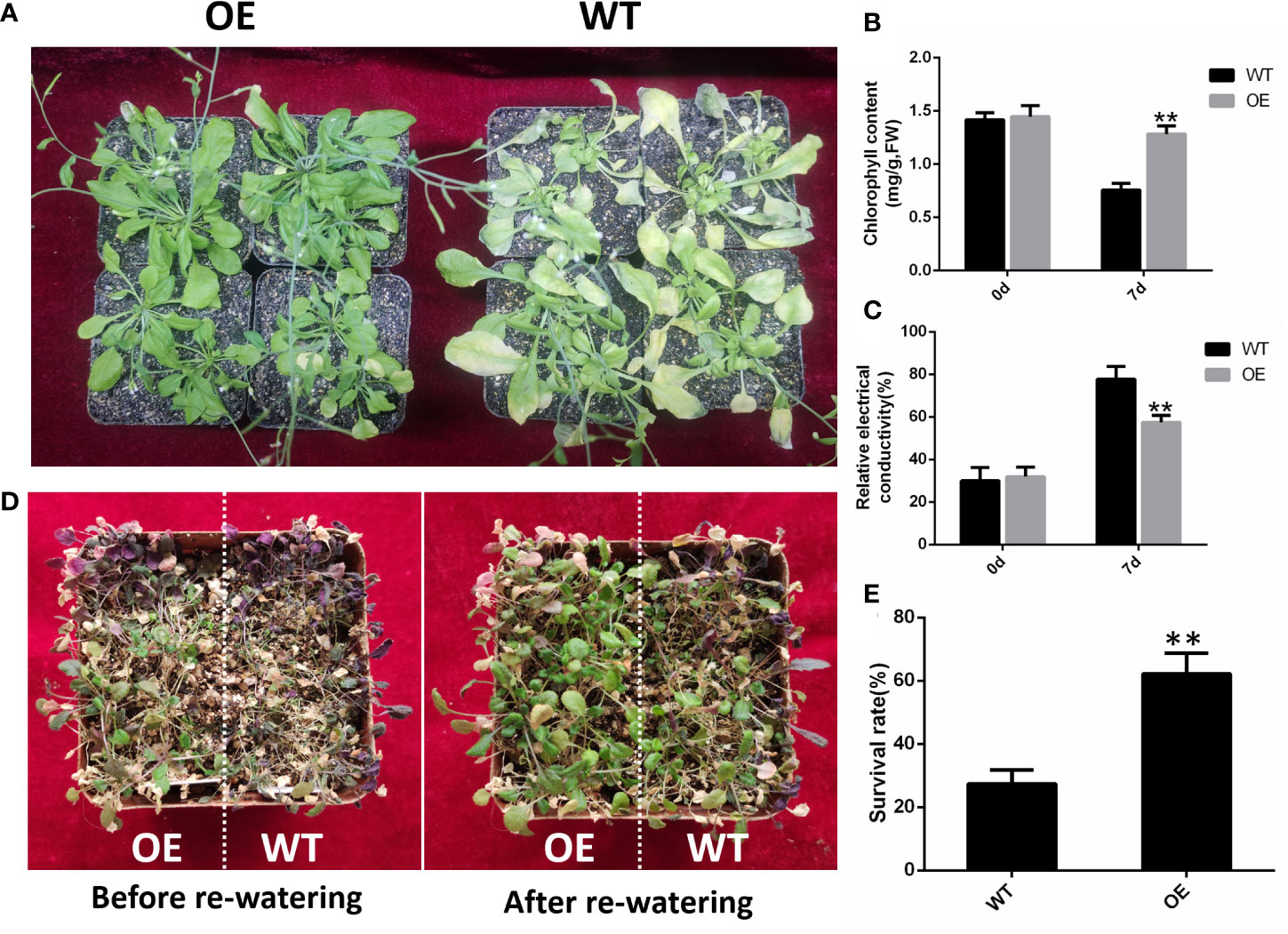
Figure 8 The overexpression of GhHVA22E1D increased salt tolerance and drought resistance of Arabidopsis thaliana. (A) Phenotype of Arabidopsis plants after 7 days of watering with 200 mM NaCl solution. (B) Total chlorophyll content in Arabidopsis leaves after 0 and 7 days of salt stress. (C) Relative electrical conductivity of Arabidopsis leaves after 0 and 7 days of salt stress. (D) Phenotype of Arabidopsis after 7 days of natural drought and 2 days of re-watering. (E) Survival rate of Arabidopsis after 2 days of re-watering. Results are from three replicate experiments. Significant differences are indicated by **(p < 0.01, Student t-test).
Silencing GhHVA22E1D by VIGS reduced salt and drought tolerance in G. hirsutum
CLCrV-based virus-induced gene silencing (VIGS) was used to silence the GhHVA22E1D gene in G. hirsutum. After 2 weeks of A. tumefaciens injection, the GhHVA22E1D transcript level of CLCrV-GhHVA22E1D plants decreased by 62.79%, indicating that this gene was effectively silenced (Figure 9E). CLCrV-GhHVA22E1D plants wilted most of the whole plant after 10 days of 400 mM NaCl stress, while only a few leaves of the control plants wilted (Figure 9A). The activities of POD and SOD in root tissues of CLCrV-GhHVA22E1D plants were significantly lower, and the MDA content was significantly higher than that of the control under salt stress (Figure 9C). In the leaves, the MDA content of the silenced plants was significantly higher than that of the control after salt treatment, while the activities of POD and SOD were similar to those of the control plants (Figure 9B). It suggests that the role of GhHVA22E1D in root and leaf tissues was not exactly the same when plants were subjected to salt stress. Both plants were severely wilted after 14 days of natural drought (Figure 9D). But after 2 days of re-watering, most of the empty vector control plants were able to recover, while most of the CLCrV-GhHVA22E1D plants wilted and died (Figure 9F). From the statistics of the survival rate, it could be seen that the survival rate of the silenced group was 7.31%, which was significantly lower than that of the control group, which was 67.97% (Figure 9G). The above results showed that the silencing of GhHVA22E1D significantly reduced the salt tolerance and drought resistance of cotton.
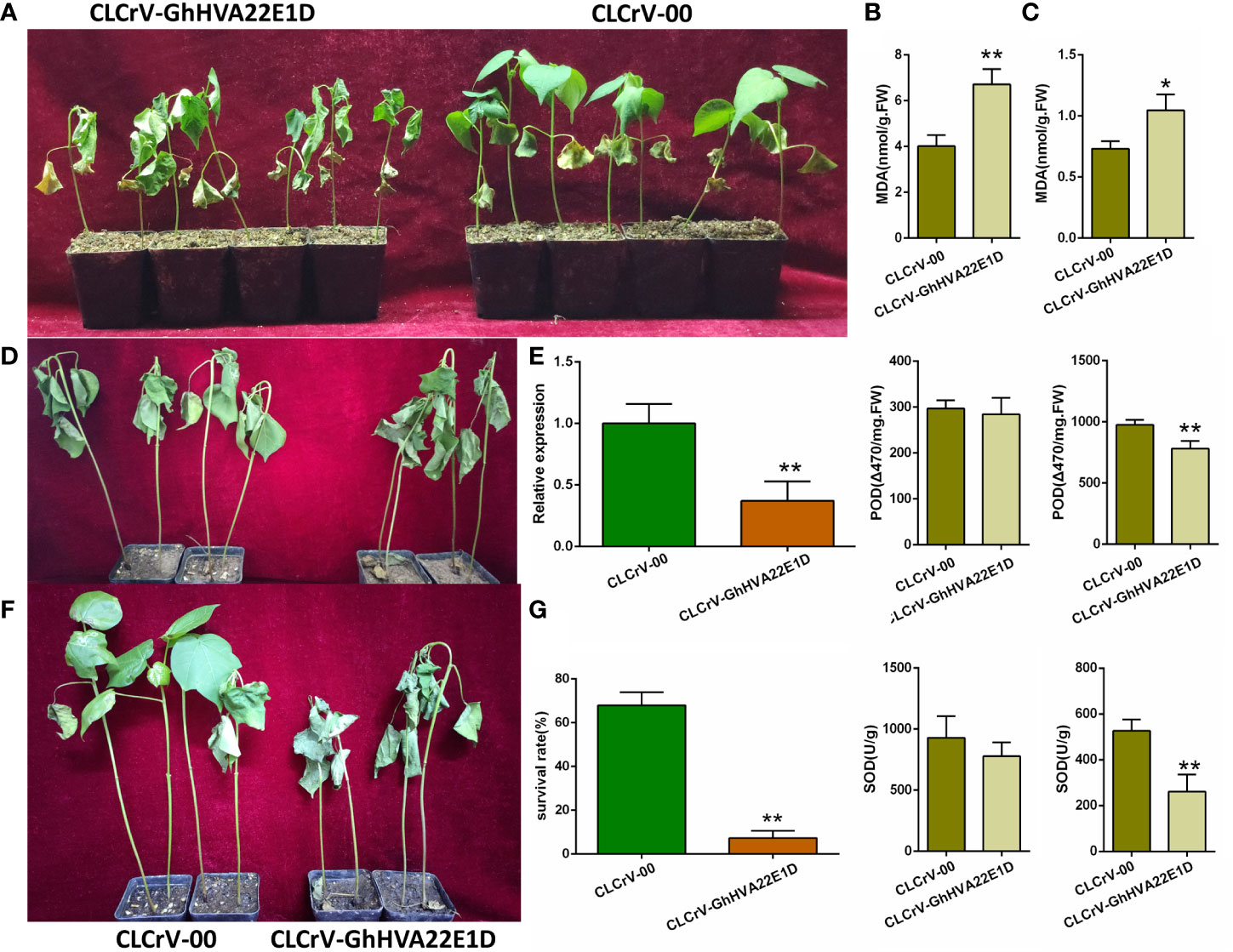
Figure 9 Silencing of GhHVA22E1D leads to reduced salt and drought tolerance in G hirsutum. (A) Phenotype of cotton after soaked in 400 mM NaCl solution for 10 days. Detection of POD and SOD activities and MDA content in cotton leaves (B) and roots (C) after salt stress. (D) The phenotype of cotton after 14 days of natural drought. (E) Silencing efficiency of CLCrV- GhHVA22E1D plants. Phenotype (F) and survival rate (G) of cotton plants after 2 days of re-watering. The results are the mean of three replicates. *p ≤ 0.05, **p ≤ 0.01 (Student t-test).
Discussion
Here, 16 GaHVA22s, 17 GrHVA22s, 32 GhHVA22s, and 34 GbHVA22s genes were identified. The HVA22 genes in cotton were clustered into nine lineages, which were different from five lineages in A. thaliana (Chen et al., 2002), 15 lineages in tomato (Wai et al., 2022) and six lineages in citrus (Gomes Ferreira et al., 2019), indicating that HVA22 classes were different among different species. In each lineage, the number of HVA22 genes in tetraploid cotton was almost twice that of diploid cotton. And in the same chromosome number, the tetraploid HVA22 and the diploid HVA22 clustered together. A0 and D5 genomes hybridized, doubled, and differentiated to form the AD1 and AD2 genomes of G. hirsutum and G. barbadense, and A0 differentiated to the A2 genome of G. arboreum (Huang et al., 2020). The genome of the four cotton species was traced back to the common A0 and D5 ancestor, and the distribution pattern of HVA22 family genes also confirmed this conclusion.
The similarity in gene structure and distribution of conserved motifs in the same subclade suggested that they had similar roles in plant growth (Zafar et al., 2022). The distribution of motifs varied among different subclades. Some motifs were even specific to a particular subclade; for example, motif 12 was specific to J lineage. This suggested that gene functions differed between subgroups (Sun et al., 2020). Cis-acting elements on gene promoters play an active role in the stress response of plants, and the type of cis-acting element determines the signals to which a gene can respond (Zhao et al., 2016). Multiple cis-acting elements that respond to environmental stress (ARE, STRE, MBS, LTR, and WRE3, etc.), phytohormones (ABRE, AAGAA-motif, TGACG-motif, CGTCA-motif, TCA-element and P-box, etc.), and those responsible for growth and development (RY-element, HD-Zip 1, CCGTCC-box and CAT-box, etc.), respectively, were found in the promoter sequences of the HVA22 family of cotton (Zhu et al., 2005; Faraji et al., 2018; Yang et al., 2019; Zhang et al., 2019). In addition, a large number of MYC and MYB transcription factor binding sites existed in most HVA22 gene promoters. There was evidence that MYC and MYB transcription factors play an important role in plant resistance to stress (Wang et al., 2021; Kim et al., 2022; Wang et al., 2022). Previous studies showed that the inducible HVA22 promoter could respond to salt, drought, low temperature, and abscisic acid induction, which supported the accuracy of our results (Chen et al., 2002; Xiao et al., 2009; Chen et al., 2014; Cheng et al., 2021; Lyu et al., 2021). The distribution of these cis-acting elements on the promoter indicates that HVA22s was likely to respond to adversity stress.
The study of gene expression patterns was usually used to predict gene function. The expression of HVA22 was different in different crops. For example, the HVA22 gene in maize was down-regulated after cold stress (Chen et al., 2014), while the CmHVA22 gene in pumpkin (Cucurbita moschata) was up-regulated after cold stress (Wang, 2021). The HVA22 gene response patterns of different subgroups in G. hirsutum differed in response to environmental stress. For example, the genes of the K lineage responded rapidly and showed a down-regulated expression pattern in response to cold stress, while the genes of I lineage were up-regulated under heat stress. GhHVA22Is tended to be expressed in ovules and fibers, which was consistent with the previous detection of differential expression of the HVA22 during seed development in Jatropha curcas (Maghuly et al., 2020). On the contrary, GhHVA22Es were more favored in vegetative tissues, consistent with the pattern previously found in citrus (Gomes Ferreira et al., 2019). Several duplication events of HVA22 family genes observed in the cotton genome, such as GhHVA22C1A and GhHVA22C1D, showed different expression patterns. It seems that during evolution, some intentional point mutations occurred in the coding sequence and promoter region of the gene, resulting in a change in the expression pattern of duplicated genes (Heidari et al., 2022; Yaghobi and Heidari, 2023). Further qRT-PCR analysis demonstrated that GhHVA22s could be rapidly upregulated in response to salt stress. In our qRT-PCR results, the level of up-regulated expression of GhHVA22s in response to salt stress appeared to be more significant than the differences in expression profiles. This may be because the data in the expression profile came from the whole cotton seedling after salt treatment, while the root tissue after salt stress was adopted in this experiment. The expression trend of GhHVA22s in root in response to salt stress may be slightly different from that in other tissues. This point was confirmed by qRT-PCR analysis of root, stem, and leaf tissues of GhHVA22E1D in response to salt stress.
The GhHVA22E1D of the E lineage was previously identified as a differentially expressed gene under salt stress in the association analysis of the salt-tolerant traits and salt-tolerance transcriptome in G. hirsutum seedling stage (Yuan et al., 2019). In this experiment, GhHVA22E1D was up-regulated under salt stress and drought stress, which verified the previous findings. In addition, GhHVA22E1D was also up-regulated in cotton under exogenous ABA stress, which confirmed that HVA22 gene was an ABA responsive gene (Chen et al., 2002). Arabidopsis overexpressed GhHVA22E1D exhibited significant salt tolerance and drought resistance, while cotton silenced GhHVA22E1D showed significantly reduced salt and drought tolerance. These results strongly support the conclusion that GhHVA22E1D is a salt tolerance responsive gene. In addition, the mechanism of GhHVA22E1D response to stress in different tissues of cotton seems to be different. For example, in roots, differential expression of GhHVA22E1D under salt stress regulated the activity of POP and SOD and affected the accumulation of MDA content, whereas in leaves, differential expression of GhHVA22E1D only seemed to cause differences in the accumulation of MDA content. From the perspective of gene expression pattern, GhHVA22E1D gene was most sensitive to salt stress in roots and drought stress in leaves. This might be related to the different functions of different organs in response to stress. Roots were first subjected to salt stress, and plants can resist salt stress by regulating the absorption of ions and water in root tissues (Hines, 2008; Rajabi et al., 2014; Deolu-Ajayi et al., 2019). When the plant was suffered from drought stress, they first reduce water loss through stomatal regulation (Tardieu et al., 2018; Visentin et al., 2020; Ghimire et al., 2021). Finally, whether GhHVA22E1D had a crucial role in these processes needs further verification.
Conclusion
In the present study, there were 16 GaHVA22s, 17 GrHVA22s, 32 GhHVA22s, and 34 GbHVA22s genes identified, and these HVA22s were divided into nine lineages. The gene structures and conserved motifs of these HVA22s had significant similarity in the same lineage, and thus genes of the same lineage had similar functions. Six GhHVA22s could be rapidly up-regulated in response to salt stress. Overexpression of GhHVA22E1D in A. thaliana improved salt tolerance and drought resistance of Arabidopsis. Conversely, silencing GhHVA22E1D in cotton decreased salt tolerance and drought resistance of cotton. These results provide a basis for further understanding the role of HVA22s in plant resistance to high salt and drought stress. These results would be helpful in the future for genomics-assisted breeding programs for salt and drought tolerance in cotton.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XLS and ZW: Designed experiments, revised manuscripts, and obtained funds. HZ: Completed gene function identification and manuscript writing. YY and QW: Performed the bioinformatics analysis. HX and MX: Data statistical analysis. MS: Corrected the grammar of the manuscript. TZ, XZ, LM, and XZS: Plant material management. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Modern Agro-industry Technology Research System of Shandong Province (SDAIT-03-02/03/05) and the Agricultural Seed Projects of Shandong Province (2020LZGC002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1139526/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37 (suppl_2), 202–208. doi: 10.1093/nar/gkp335
Benny, J., Pisciotta, A., Caruso, T., Martinelli, F. (2019). Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-seq data. BMC Plant Biol. 19 (1), 194. doi: 10.1186/s12870-019-1794-y
Campos-Rivero, G., Osorio-Montalvo, P., Sánchez-Borges, R., Us-Camas, R., Duarte-Aké, F., De-la-Peña, C. (2017). Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 214, 16–27. doi: 10.1016/j.jplph.2017.03.018
Chen, Q., Chen, Q. J., Sun, G. Q., Zheng, K., Yao, Z. P., Han, Y. H., et al. (2019). Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 20 (2), 349. doi: 10.3390/ijms20020349
Chen, C. N., Chen, H. R., Yeh, S. Y., Vittore, G., Ho, T. H. (2009). Autophagy is enhanced and floral development is impaired in AtHVA22d RNA interference arabidopsis. Plant Physiol. 149 (4), 1679–1689. doi: 10.1104/pp.108.131490
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C. N., Chu, C. C., Zentella, R., Pan, S. M., Ho, T. H. (2002). AtHVA22 gene family in arabidopsis: Phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol. Biol. 49 (6), 631–642. doi: 10.1023/a:1015593715144
Chen, Z. J., Sreedasyam, A., Ando, A., Song, Q., De Santiago, L. M., Hulse-Kemp, A. M., et al. (2020b). Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52 (5), 525–533. doi: 10.1038/s41588-020-0614-5
Chen, J. J., Wei, M. U., Zhou, S. F., Wan-Chen, L. I., Feng-Ling, F. U., University, S. A. (2014). Cloning and functional validation of HVA22 gene promoter in maize. J. Nucl. Agric. Sci. 28 (04), 560–568. doi: 10.11869/j.issn.100-8551.2014.04.0560
Cheng, H., Zhang, Y., Zhang, J., Gu, Z., Zeng, T., Zhang, J. (2021). ABA affects the coloration of ornamental begonia and functional verification of McHVA22. J. Beijing Agric. Univ. 36 (02), 58–62. doi: 10.13473/j.cnki.issn.1002-3186.2021.0212
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium -mediated transformation of arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collin, A., Daszkowska-Golec, A., Kurowska, M., Szarejko, I. (2020). Barley ABI5 (Abscisic acid INSENSITIVE 5) is involved in abscisic acid-dependent drought response. Front. Plant Sci. 11 1138. doi: 10.3389/fpls.2020.01138
Courtney, A. J., Xu, J., Xu, Y. (2016). Responses of growth, antioxidants and gene expression in smooth cordgrass (Spartina alterniflora) to various levels of salinity. Plant Physiol. Biochem. 99, 162–170. doi: 10.1016/j.plaphy.2015.12.016
Deolu-Ajayi, A. O., Meyer, A. J., Haring, M. A., Julkowska, M. M., Testerink, C. (2019). Genetic loci associated with early salt stress responses of roots. iScience 21, 458–473. doi: 10.1016/j.isci.2019.10.043
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50 (6), 796–802. doi: 10.1038/s41588-018-0116-x
Faraji, S., Rasouli, S. H., Kazemitabar, S. K. (2018). Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. durum): insights into the roles in biological processes especially stress response. Biometals 31 (6), 1019–1042. doi: 10.1007/s10534-018-0146-y
Ghimire, K., Gupta, S., Geng, S., Chen, S., Boe, A., Wu, Y. (2021). Identification of physiological and morphological traits governing high water use efficiency in alfalfa. J. Agron. Crop Sci. 207 (4), 644–653. doi: 10.1111/jac.12483
Gomes Ferreira, M. D., Araújo Castro, J., Santana Silva, R. J., Micheli, F. (2019). HVA22 from citrus: A small gene family whose some members are involved in plant response to abiotic stress. Plant Physiol. Biochem. 142, 395–404. doi: 10.1016/j.plaphy.2019.08.003
Grover, C. E., Gallagher, J. P., Jareczek, J. J., Page, J. T., Udall, J. A., Gore, M. A., et al. (2015). Re-evaluating the phylogeny of allopolyploid Gossypium l. Mol. Phylogenet. Evol. 92, 45–52. doi: 10.1016/j.ympev.2015.05.023
Gu, Z., Huang, C., Li, F., Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12 (5), 638–649. doi: 10.1111/pbi.12169
Guo, W.-J., David Ho, T.-H. (2008). An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol. 147 (4), 1710–1722. doi: 10.1104/pp.108.120238
Heidari, P., Puresmaeli, F., Mora-Poblete, F. (2022). Genome-wide identification and molecular evolution of the magnesium transporter (MGT) gene family in Citrullus lanatus and Cucumis sativus. Agronomy 12 (10) 2253. doi: 10.3390/agronomy12102253
Hines, P. J. (2008). Plant responses to salt stress. Sci. Signaling 1 (20), ec192–ec192. doi: 10.1126/stke.120ec192
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51 (4), 739–748. doi: 10.1038/s41588-019-0371-5
Huang, G., Wu, Z., Percy, R. G., Bai, M., Li, Y., Frelichowski, J. E., et al. (2020). Genome sequence of gossypium herbaceum and genome updates of gossypium arboreum and gossypium hirsutum provide insights into cotton a-genome evolution. Nat. Genet. 52 (5), 516–524. doi: 10.1038/s41588-020-0607-4
Kim, J., Kim, D. H., Lee, J. Y., Lim, S. H. (2022). The R3-type MYB transcription factor BrMYBL2.1 negatively regulates anthocyanin biosynthesis in Chinese cabbage (Brassica rapa l.) by repressing MYB-bHLH-WD40 complex activity. Int. J. Mol. Sci. 23 (6) 3382. doi: 10.3390/ijms23063382
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Liu, H., Ma, X., Liu, S., Du, B., Cheng, N., Wang, Y., et al. (2020). The nicotiana tabacum l. major latex protein-like protein 423 (NtMLP423) positively regulates drought tolerance by ABA-dependent pathway. BMC Plant Biol. 20 (1), 475. doi: 10.1186/s12870-020-02690-z
Liu, Y., Wang, L., Li, X., Luo, M. (2022). Detailed sphingolipid profile responded to salt stress in cotton root and the GhIPCS1 is involved in the regulation of plant salt tolerance. Plant Sci. 316, 111174. doi: 10.1016/j.plantsci.2021.111174
Lyu, J. I., Ramekar, R., Kim, J. M., Hung, N. N., Seo, J. S., Kim, J.-B., et al. (2021). Unraveling the complexity of faba bean (Vicia faba l.) transcriptome to reveal cold-stress-responsive genes using long-read isoform sequencing technology. Sci. Rep. 11 (1), 21094. doi: 10.1038/s41598-021-00506-0
Ma, Y., Cao, J., He, J., Chen, Q., Li, X., Yang, Y. (2018). Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 19 (11), 3643. doi: 10.3390/ijms19113643
Maghuly, F., Deák, T., Vierlinger, K., Pabinger, S., Tafer, H., Laimer, M. (2020). Gene expression profiling identifies pathways involved in seed maturation of Jatropha curcas. BMC Genomics 21 (1), 290. doi: 10.1186/s12864-020-6666-1
Magwanga, R. O., Kirungu, J. N., Lu, P., Cai, X., Xu, Y., Wang, X., et al. (2019a). Knockdown of ghAlba_4 and ghAlba_5 proteins in cotton inhibits root growth and increases sensitivity to drought and salt stresses. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01292
Magwanga, R. O., Kirungu, J. N., Lu, P., Yang, X., Dong, Q., Cai, X., et al. (2019b). Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiologia Plantarum 167 (3), 447–464. doi: 10.1111/ppl.12920
Meng, F., Zhao, Q., Zhao, X., Yang, C., Liu, R., Pang, J., et al. (2022). A rice protein modulates endoplasmic reticulum homeostasis and coordinates with a transcription factor to initiate blast disease resistance. Cell Rep. 39 (11), 110941. doi: 10.1016/j.celrep.2022.110941
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492 (7429), 423–427. doi: 10.1038/nature11798
Peng, R., Xu, Y., Tian, S., Unver, T., Liu, Z., Zhou, Z., et al. (2022). Evolutionary divergence of duplicated genomes in newly described allotetraploid cottons. Proc. Natl. Acad. Sci. 119 (39), e2208496119. doi: 10.1073/pnas.2208496119
Rajabi, A., Khayamim, S., Abbasi, Z., Ober, E. (2014). “Salt stress and sugar beet improvement: Challenges and opportunities,” in Improvement of crops in the era of climatic changes: Volume 2. Eds. Ahmad, P., Wani, M. R., Azooz, M. M., Phan Tran, L.-S. (New York, NY: Springer New York), 121–150.
Sharon, K. (2017). Cloning of HVA22 homolog from aloe vera and preliminary study of transgenic plant development. Int. J. Pure Appl. Bioscience 5 (6), 1113–1121. doi: 10.18782/2320-7051.6033
Shen, Q., Chen, C. N., Brands, A., Pan, S. M., Ho, T. H. (2001). The stress- and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 45 (3), 327–340. doi: 10.1023/a:1006460231978
Shen, Q., Uknes, S. J., Ho, T. H. (1993). Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J. Biol. Chem. 268 (31), 23652–23660. doi: 10.1016/S0021-9258(19)49512-4
Sun, J., Bie, X. M., Wang, N., Zhang, X. S., Gao, X.-Q. (2020). Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in common wheat. BMC Plant Biol. 20 (1), 351. doi: 10.1186/s12870-020-02505-1
Tardieu, F., Simonneau, T., Muller, B. (2018). The physiological basis of drought tolerance in crop plants: A scenario-dependent probabilistic approach. Annu. Rev. Plant Biol. 69 (1), 733–759. doi: 10.1146/annurev-arplant-042817-040218
Ullah, A., Sun, H., Hakim, Yang, X., Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant 162 (4), 439–454. doi: 10.1111/ppl.12651
Visentin, I., Pagliarani, C., Deva, E., Caracci, A., Turečková, V., Novák, O., et al. (2020). A novel strigolactone-miR156 module controls stomatal behaviour during drought recovery. Plant Cell Environ. 43 (7), 1613–1624. doi: 10.1111/pce.13758
Wai, A. H., Waseem, M., Cho, L. H., Kim, S. T., Lee, D. J., Kim, C. K., et al. (2022). Comprehensive genome-wide analysis and expression pattern profiling of the SlHVA22 gene family unravels their likely involvement in the abiotic stress adaptation of tomato. Int. J. Mol. Sci. 23 (20), 12222. doi: 10.3390/ijms232012222
Wang, M. (2021). Cold-resistant function and mechanism of pumpkin rootstock genes CmRCC1 and CmHVA22 (Master, Huazhong Agricultural University).
Wang, W., Cheng, Y., Chen, D., Liu, D., Hu, M., Dong, J., et al. (2019). The catalase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Cells 8 (2), 86. doi: 10.3390/cells8020086
Wang, L., Deng, R., Bai, Y., Wu, H., Li, C., Wu, Q., et al. (2022). Tartary buckwheat R2R3-MYB gene FtMYB3 negatively regulates anthocyanin and proanthocyanin biosynthesis. Int. J. Mol. Sci. 23 (5) 2775. doi: 10.3390/ijms23052775
Wang, X., Niu, Y., Zheng, Y. (2021). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22 (11) 6152. doi: 10.3390/ijms22116125
Wendel, J. F., Cronn, R. C. (2003). “Polyploidy and the evolutionary history of cotton,” in Advances in agronomy (Academic Press) 139–186. doi: 10.1016/S0065-2113(02)78004-8
Xiao, B.-Z., Chen, X., Xiang, C.-B., Tang, N., Zhang, Q.-F., Xiong, L.-Z. (2009). Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant 2 (1), 73–83. doi: 10.1093/mp/ssn068
Yaghobi, M., Heidari, P. (2023). Genome-wide analysis of aquaporin gene family in Triticum turgidum and its expression profile in response to salt stress. Genes 14 (1), 202. doi: 10.3390/genes14010202
Yang, R., Chen, M., Sun, J. C., Yu, Y., Min, D. H., Chen, J., et al. (2019). Genome-wide analysis of LIM family genes in foxtail millet (Setaria italica l.) and characterization of the role of SiWLIM2b in drought tolerance. Int. J. Mol. Sci. 20 (6) 1303. doi: 10.3390/ijms20061303
Yuan, Y., Xing, H., Zeng, W., Xu, J., Mao, L., Wang, L., et al. (2019). Genome-wide association and differential expression analysis of salt tolerance in Gossypium hirsutum l at the germination stage. BMC Plant Biol. 19 (1), 394. doi: 10.1186/s12870-019-1989-2
Zafar, M. M., Rehman, A., Razzaq, A., Parvaiz, A., Mustafa, G., Sharif, F., et al. (2022). Genome-wide characterization and expression analysis of erf gene family in cotton. BMC Plant Biol. 22 (1), 134. doi: 10.1186/s12870-022-03521-z
Zhang, H., Liu, D., Yang, B., Liu, W.-Z., Mu, B., Song, H., et al. (2019). Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 71 (1), 188–203. doi: 10.1093/jxb/erz432
Zhang, Q., Song, T., Guan, C., Gao, Y., Ma, J., Gu, X., et al. (2021). OsANN4 modulates ROS production and mediates Ca2+ influx in response to ABA. BMC Plant Biol. 21 (1), 474. doi: 10.1186/s12870-021-03248-3
Zhao, W., Liu, Y.-W., Zhou, J.-M., Zhao, S.-P., Zhang, X.-H., Min, D.-H. (2016). Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica l.). Plant Cell Tissue Organ Culture (PCTOC) 127 (2), 335–346. doi: 10.1007/s11240-016-1053-y
Zhu, T., Liang, C., Meng, Z., Sun, G., Meng, Z., Guo, S., et al. (2017). CottonFGD: an integrated functional genomics database for cotton. BMC Plant Biol. 17 (1), 101. doi: 10.1186/s12870-017-1039-x
Keywords: cotton, HVA22s, salt, overexpression, VIGS, drought, stress
Citation: Zhang H, Yuan Y, Xing H, Xin M, Saeed M, Wu Q, Wu J, Zhuang T, Zhang X, Mao L, Sun X, Song X and Wang Z (2023) Genome-wide identification and expression analysis of the HVA22 gene family in cotton and functional analysis of GhHVA22E1D in drought and salt tolerance. Front. Plant Sci. 14:1139526. doi: 10.3389/fpls.2023.1139526
Received: 07 January 2023; Accepted: 21 February 2023;
Published: 06 March 2023.
Edited by:
Linghe Zeng, Crop Genetics Research Unit (USDA), United StatesReviewed by:
Parviz Heidari, Shahrood University of Technology, IranXingxing Wang, State Key Laboratory of Cotton Biology (CAAS), China
Richard Odongo Magwanga, Institute of Cotton Research (CAAS), China
Jalaja Naravula, Vignan University, India
Copyright © 2023 Zhang, Yuan, Xing, Xin, Saeed, Wu, Wu, Zhuang, Zhang, Mao, Sun, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianliang Song, c29uZ3hsOTk5QDE2My5jb20=; Zongwen Wang, d2FuZ3p3NjZAMTYzLmNvbQ==
†These authors share first authorship
 Haijun Zhang
Haijun Zhang Yanchao Yuan
Yanchao Yuan Huixian Xing1,3†
Huixian Xing1,3† Ming Xin
Ming Xin Muhammad Saeed
Muhammad Saeed Jing Wu
Jing Wu Xuezhen Sun
Xuezhen Sun Xianliang Song
Xianliang Song