- 1School of BioSciences, University of Melbourne, Parkville, VIC, Australia
- 2School of Biosciences, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany
- 3Faculty of Science, University of Nottingham, Leicestershire, United Kingdom
- 4La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC, Australia
- 5Mechano(bio)chemistry Department, Max Planck Institute of Colloids and Interfaces, Potsdam, Germany
- 6Institute of Experimental Physics, Johannes Kepler University, Linz, Austria
The cell wall is one of the defining features of plants, controlling cell shape, regulating growth dynamics and hydraulic conductivity, as well as mediating plants interactions with both the external and internal environments. Here we report that a putative mechanosensitive Cys-protease DEFECTIVE KERNEL1 (DEK1) influences the mechanical properties of primary cell walls and regulation of cellulose synthesis. Our results indicate that DEK1 is an important regulator of cellulose synthesis in epidermal tissue of Arabidopsis thaliana cotyledons during early post-embryonic development. DEK1 is involved in regulation of cellulose synthase complexes (CSCs) by modifying their biosynthetic properties, possibly through interactions with various cellulose synthase regulatory proteins. Mechanical properties of the primary cell wall are altered in DEK1 modulated lines with DEK1 affecting both cell wall stiffness and the thickness of the cellulose microfibril bundles in epidermal cell walls of cotyledons.
Introduction
The polysaccharide rich extracellular matrix of plant cells has crucial roles in providing mechanical support, enabling water transport, cell-to-cell communication, and is a determining factor for turgor driven morphogenesis (Cosgrove, 2005). Cell walls vary in chemical composition and mechanical properties to support the different functional requirements of the cell. In primary walls of Arabidopsis the major components are cellulose, xyloglucan and pectins. Changes in cell wall composition and interaction between its component polymers greatly affect its mechanical properties and in turn impact growth and development of the plant (Zhang et al., 2019; Wang et al., 2020). Cellulose is a significant component of most types of cell walls, organized into cellulose microfibrils (CMF) that impart mechanical strength to the cell wall (Johnson et al., 2018; Zhang et al., 2019). Cellulose is synthesized by membrane bound CELLULOSE SYNTHASE (CESA) complexes (CSCs) organized in heterotrimers (Persson et al., 2007; Nixon et al., 2016; Polko and Kieber, 2019). Cellulose chains synthesized by CESAs form CMFs in the apoplast and become entangled in the cell wall matrix (Johnson et al., 2018). Mutants for either different CESA genes, such as prc1-1, or for regulatory proteins, such as csi1/pom2 and korrigan (kor), have decreased cellulose content and growth defects (Fagard et al., 2000; Lane et al., 2001; Bringmann et al., 2012; Vain et al., 2014). Mutants for pectin and hemicellulose synthesis , such as either quasimodo2 or xyloglucan xylosyltransferase 1, 2 (xxt1xxt2) have less stiff cell walls than wild type (WT) plants. Lack of either pectin or xyloglucans (XGs) in these mutants decreases bundling of CMFs, changing the mechanical properties of cell walls and initiating complex signaling cascades that result in developmental defects (Xiao and Anderson, 2016; Du et al., 2020). Decreased synthesis of pectins and XGs also results in slower CSCs and less cellulose being produced, demonstrating that all components of the cell wall contribute to maintenance of its biochemical composition and integrity, in a continuous feedback loop-like manner (Xiao and Anderson, 2016; Du et al., 2020).
Proteins capable of sensing changes in biomechanical properties/mechanical signals at the level of the apoplast (PM/cell wall) are important for maintenance of cell wall integrity (CWI) (Engelsdorf and Hamann, 2014; Vaahtera et al., 2019). Previous studies have identified several protein families involved in CWI sensing and monitoring the mechanical status of the cell wall. These proteins belong to a family of Catharanthus roseus receptor like kinases (CrRLK), wall associated kinases (WAKs) and mechanosensitive (MS) ion channels (Vaahtera et al., 2019). Some of these proteins (eg. WAK2 and FERONIA (FER)) interact directly with pectins via their extracellular domains, while others (eg. MS ion channels), sense mechanical changes in the PM caused by mechanical perturbations of the cell wall. Changes in cell wall mechanics caused by either different developmental processes or environmental stress factors are sensed by these CWI sensors, eliciting different intra- and extra-cellular responses (Kohorn, 2009; Feng et al., 2018; Gjetting et al., 2020). Rapid mechanically activated (RMA) calcium currents are known to be activated by mechanical signaling. A functioning DEFECTIVE KERNEL1 (DEK1) protein has been shown to be crucial for RMA activity in Arabidopsis (Tran et al., 2017). DEK1 has been identified as a regulator of cell wall biochemical composition and structure (Johnson et al., 2008; Demko et al., 2014; Galletti et al., 2015; Amanda et al., 2016; Amanda et al., 2017).
DEK1 is a major regulator of plant development and growth (Johnson et al., 2005; Johnson et al., 2008). Loss-of-function mutants of dek1 result in embryo lethal phenotypes in both Arabidopsis and maize (Becraft et al., 2002; Johnson et al., 2005). Plants with reduced levels of DEK1 such as dek1 RNA interference and artificial microRNA (amiRNA) lines show a range of severe phenotypes, such as an almost complete absence and misspecification of the epidermal layer (Johnson et al., 2005), an irregular epidermal layer made of large cells (Johnson et al., 2008) or a loss of cell-to-cell adhesion and occasional gaps in the epidermal layer (Galletti et al., 2015). In addition, DEK1 indirectly controls HD-ZIP IV transcription factors which are involved in the regulation of epidermal identity (Galletti et al., 2015). It was proposed that DEK1 could act as a mediator of cellular adhesion by receiving and integrating signals from the apoplast and facilitating cell-to-cell contact through maintenance of epidermal identity (Galletti et al., 2015).
DEK1 belongs to the calpain superfamily of regulatory proteases. Calpains function in a range of cellular signaling pathways by modulating their targets through proteolytic processing resulting in altered protein activity, localization, substrate specificity or stability (Ono and Sorimachi, 2012). DEK1 is the only identified member of the calpain family in land plants (Lid et al., 2002; Demko et al., 2014) and its overall structure is distinct from the mostly cytosolic animal calpains (Lid et al., 2002; Wang et al., 2003). DEK1 is an approx. 240 kDa transmembrane protein with a complex structure including 21-23 transmembrane domains (TM), a loop domain, a cytoplasmic regulatory juxtamembrane domain and a proteolytically active CALPAIN domain close to its C-terminus (Lid et al., 2002; Johnson et al., 2008). DEK1 has been shown to undergo autocatalytic cleavage to release the CALPAIN domain into the cytoplasm where it is proposed to proteolytically cleave target proteins to alter their activity (Johnson et al., 2008; Amanda et al., 2016). The catalytic activity of DEK1 CALPAIN domain is dependent on calcium (Wang et al., 2003), as shown for animal calpains (García Díaz et al., 2006).Complementing dek1 mutant lines of either Arabidopsis or Physcomitrella patens with only the CALPAIN domain is sufficient to rescue mutants, confirming the CALPAIN region as the catalytically active domain of the protein (Johnson et al., 2008; Demko et al., 2014). Over-expression of CALPAIN (OE CALPAIN) results in altered cell division and expansion, unlike over-expression of the full length DEK1, suggesting the TM and juxtamembrane regions of DEK1 act as regulatory domains (Johnson et al., 2008).
Studies performed on an over-expressor of CALPAIN (OE CALPAIN) and on amiRNA DEK1 lines have shown that DEK1 is a major regulator of cell wall composition and structure in the epidermis of Arabidopsis leaves (Amanda et al., 2016). Immunohistochemical studies on OE CALPAIN lines showed a significant increase in antibody density for both cellulose, high- and low-methyl esterified homogalacturonan (HG) and arabinan while the same cell wall components were less abundant in epidermal cell walls of amiRNA DEK1 compared with the WT (Amanda et al., 2016).
Increased labelling of epitopes for crystalline cellulose were specifically observed in epidermal cell walls of leaves and stems with no change in overall cellulose content in leaves of OE CALPAIN-GFP in a dek1-3 mutant background and an amiDEK1 line (Amanda et al., 2016). In this study, we investigated the role of DEK1 in the regulation of cellulose synthesis at a molecular level. An overexpressor of CALPAIN domain of DEK1 in WT background (pRPS5A:CALPAIN-6XHIS, OE CALPAIN, Galletti et al., 2015) and an EMS single point mutant, dek1-4, which has a C to T nucleotide substitution in the CALPAIN domain and has mild phenotypes (Roeder et al., 2012) were used in this study to enable more direct comparisons to WT plants. Results of this study demonstrate that DEK1 is involved in cellulose synthesis in Arabidopsis cotyledon pavement cells by regulating the dynamics of CSCs. Our data also indicate that DEK1 influences cell wall mechanics.
Results
DEK1 modulated lines show altered responses to isoxaben-induced cellulose perturbations
To investigate the possible involvement of DEK1 in regulation of cellulose biosynthesis, WT and DEK1 modulated lines were treated with isoxaben. Isoxaben is a plant-specific herbicide which inhibits the synthesis of cellulose through interacting with CESAs and causing them to be removed from the PM and internalized into the cell (Heim et al., 1990; Shim et al., 2018). Isoxaben-induced disruption of cellulose synthesis causes reduced root growth and cell swelling. Cellulose deficient mutants have been shown to display altered responses to isoxaben treatment (Engelsdorf and Hamann, 2014; Vaahtera et al., 2019). We measured root lengths of 6-day old WT and DEK1 modulated plants grown on media supplemented with either 2 nM isoxaben or DMSO as a control (Figure 1A). In response to isoxaben treatment WT plants showed a significant reduction of root length (Figure 1B). For DMSO control plants no differences in root lengths between WT and OE CALPAIN seedlings were observed. In contrast, we observed significant differences between the mean root length of WT and OE CALPAIN when grown on isoxaben, shown by the 95% CIs of the mean differences of these two groups not overlapping (Figure 1B). Estimation statistics analysis showed high mean differences in mean root length between OE CALPAIN plants grown on 2 nM isoxaben and the DMSO controls (Figure 1B). In dek1-4 mutants grown on DMSO media the mean difference in root length compared to WT indicated significantly shorter root growth (Figure 1A). In response to isoxaben-induced mechanical perturbation of the cell wall dek1-4 mutants resulted in no change in root growth compared to controls (Figures 1A, B). In addition, no significant differences in the mean difference were observed between dek1-4 and WT (Figure 1B, 95% CI-s are overlapping). Changes in cellulose levels are known to influence plant tolerance to abiotic stresses such as salt (Endler et al., 2015). The response of DEK1 modulated lines to salt treatment was investigated. WT showed significantly reduced root growth after transfer to salt treatment compared to plants transferred to control media (Supplementary Figure 1). The response of OE CALPAIN plants to salt was more moderate with a smaller reduction in root growth than WT (Supplementary Figure 1). A similar insensitive response to that of isoxaben treatment was observed for dek1-4, with a small mean difference between root length in control and salt-grown plants (Supplementary Figure 1).
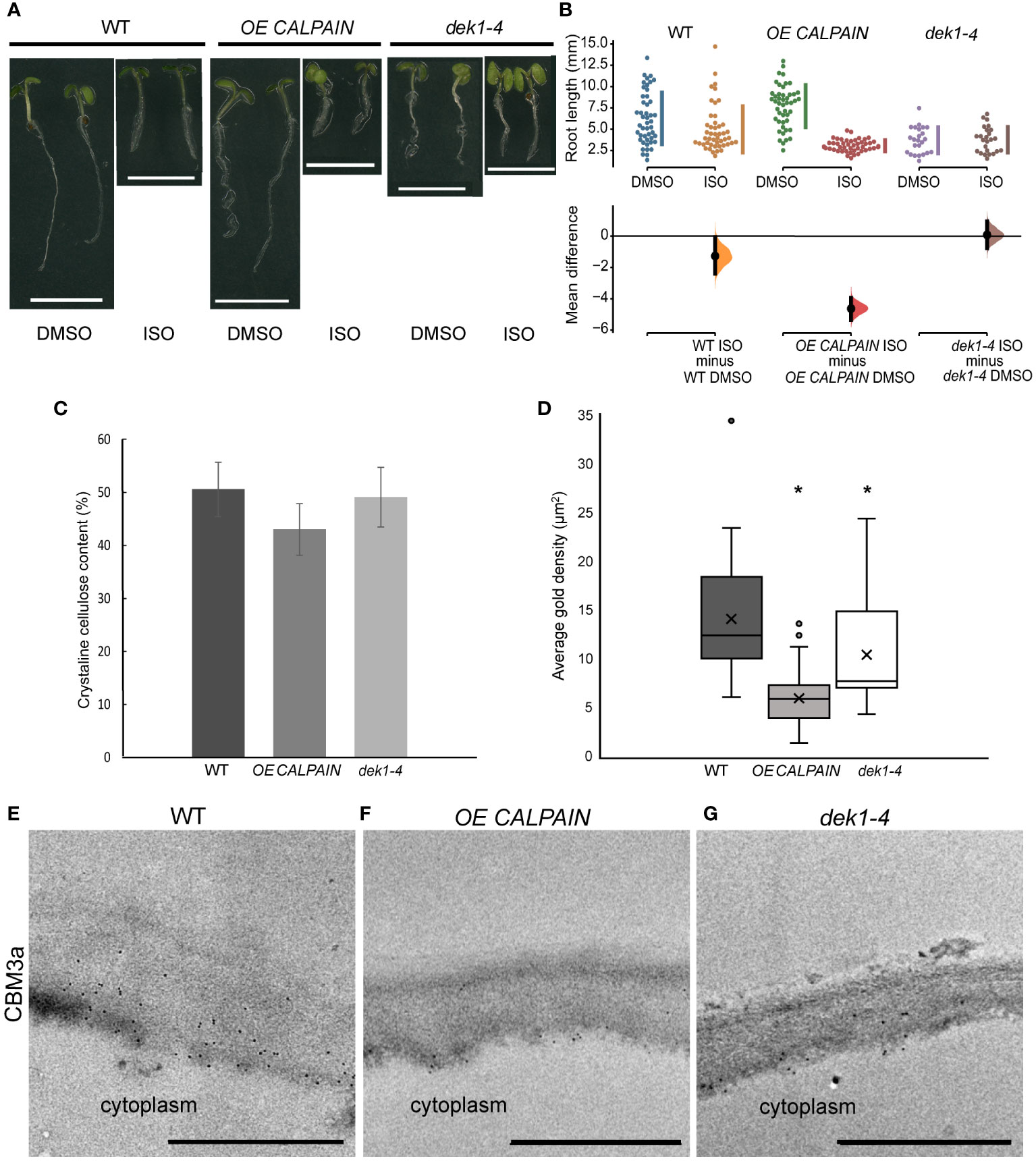
Figure 1 Isoxaben treatment and cell wall analysis of WT and DEK1 modulated Arabidopsis lines. (A) Representative images of 6-day old WT, OE CALPAIN and dek1-4 seedlings grown on media with either DMSO (control) or 2 nM isoxaben (ISO), scale bar = 5 mm. (B) Multiple two-group estimation analysis of root length for each genotype between plants grown on media with DMSO and isoxaben. The mean differences are shown in the Cumming estimation plot (lower panel). The raw data are plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The difference between ISO/DMSO plants was WT -2.7 mm (95% CI-2.39, -0.0585, p = 0.0332), OE CALPAIN -4.63 mm (95% CI -5.35, -3.95, p = <0.0001) and dek1-4 0.0824 mm (95% CI -0.762, 0.94, p = 0.848). N=47 WT DMSO, WT ISO, OE CALPAIN DMSO, 50 OE CALPAIN ISO, 24 dek1-4 DMSO, 25 dek1-4 ISO (C) Crystalline cellulose contents of AIR cell wall preparations of 10-day old seedlings. Analysis performed on two biological replicates and two technical replicates. Error bars are representing standard error. No statistical significance observed p>0.05, Students t-test. (D) Average gold particle density (indicated by 'x') for CBM3a labelled cellulose in outer periclinal epidermal walls was WT 14.09 ±1.01 µm-2, OE CALPAIN 5.93±1.01 µm-2 and dek1-4 10.41±1.44 µm-2. Asterisk represents statistical significance, p<0.0001 for OE CALPAIN, p<0.05 for dek1-4, unpaired Student t-test. n=36 WT, 25 OE CALPAIN and 16 dek1-4 epidermal cell walls. Analysis performed on biological duplicates. (E–G) Transmission electron micrographs of outer epidermal cell walls of 10-day old cotyledons showing immunogold labelled CBM3a cellulose binding. Scale bars = 500 nm.
DEK1 is involved in fine tuning of epidermal cell wall composition
Studies of DEK1 modulated lines grown on isoxaben and salt suggest cellulose content and/or assembly is altered. Cell wall linkage analysis was undertaken on alcohol insoluble residue (AIR) wall preparations to investigate potential wall compositional differences in 10-day old seedlings of DEK1 modulated lines compared to WT. No statistically significant differences were found (Supplementary Figure 2; Supplementary Table 1). A trend towards reduced levels of cellulose in OE CALPAIN lines was observed and this was investigated further using the Updegraff (1969) method which determines crystalline cellulose content. Of the AIR, crystalline cellulose constituted 51% (w/w) in WT, 42% in OE CALPAIN and 48% in dek1-4 (Figure 1C). Previous studies have shown DEK1-related changes in cellulose and pectin content predominantly occurs in the outer epidermal cell walls of both young and mature leaves and inflorescence stems (Amanda et al., 2016; Amanda et al., 2017).
Immunofluorescence and immuno-gold labelling of cell wall epitopes was used to determine if wall composition differs at the tissue/cellular scale that may be beyond the resolution of the whole seedling chemical analyses. CBM3a labelling of cellulose epitopes showed slightly lower fluorescence intensity in the outer periclinal epidermal cell wall of both DEK1 modulated lines compared to WT (Supplementary Figures 3A–C). TEM imaging of the cotyledon epidermal cell walls showed significantly decreased immuno-gold labelling of cellulose epitopes with CBM3a in both DEK1 modulated lines compared with WT (Figures 1D–G). Cotyledon sections labelled with Calcofluor white to detect cellulose, JIM5 and JIM7 to detect partially and highly methyl-esterified HG pectin, respectively, and LM15 to detect XG revealed no obvious differences in fluorescence signal between WT and DEK1 modulated lines (Supplementary Figure 4). We performed immuno-gold labelling with JIM5 and JIM7 antibodies that detect HG pectins in cotyledon epidermal cell walls (Supplementary Figure 5). Our results show that dek1-4 had significantly higher average density of JIM5 immuno-gold particles compared with WT, while OE CALPAIN has a significantly lower count of JIM5 immuno-gold particles (Supplementary Figures 5A–C, G). Levels of partially-methyl esterified HG detected by the JIM7 antibody were significantly higher in OE CALPAIN and significantly lower in dek1-4 compared to WT (Supplementary Figures 5D–F, H).
CESA transcript levels show no differences between WT and DEK1 modulated lines
Isoxaben growth assays, immuno-labelling and cell wall compositional analysis suggest DEK1 influences levels of cellulose, particularly in epidermal walls. To determine how changes in cellulose may have come about we investigated CSCs at both the transcriptional and post-transcriptional levels. Previous studies have shown that modulation of DEK1 affects expression of numerous cell wall-related genes, however CESA levels were unaffected (Johnson et al., 2008; Amanda et al., 2016). CESA transcript levels were investigated in 6-day old seedlings of WT, OE CALPAIN and dek1-4 lines using qRT-PCR. No significant differences in transcript levels of CESA1, CESA3 and CESA6, encoding primary wall CESAs, were observed between WT and DEK1 modulated lines (p>0.05) using fold change analysis (Supplementary Figure 6). These data suggest DEK1 influences cellulose biosynthesis at the post-transcriptional level.
DEK1 modulated lines influence mobility and not density of CSCs
Changes in cellulose content could be explained by differences in the number of CSCs at the PM. OE CALPAIN and dek1-4 lines were crossed with a pCESA3:GFP : CESA3 mCh : TUA5 (GFP : CESA3) reporter line and the number of GFP : CESA3 complexes on pavement cell PM patches quantified (Figure 2A). No statistically significant differences were detected in CSC particle density between WT and DEK1 modulated lines (Figure 2B). We then proceeded to investigate if DEK1 could affect the velocity of the CSC. Investigation of CSC velocity is commonly used as a proxy for determining cellulose synthesis rates at the cellular level. In brief, the more cellulose CESAs synthesize, the faster CSCs will move along within the plane of the PM (Paredez et al., 2006; Diotallevi and Mulder, 2007; Fujita et al., 2011). CSCs at the PM of the periclinal epidermal cells of 3-day old cotyledons were tracked during 10-min time lapses (Figure 2C) and kymographs of GFP-labelled CESA3 particles (Figure 2D) were used to obtain velocities of CSCs (Figure 2E). CSC velocities in both OE CALPAIN and dek1-4 mutant lines were significantly reduced compared to WT (Figure 2E). A reduction of CSC velocities in the periclinal cell walls of cotyledon pavement cells is consistent with the findings of immuno-histochemical assays that indicate significantly reduced cellulose levels in OE CALPAIN compared to WT (Figure 1D).
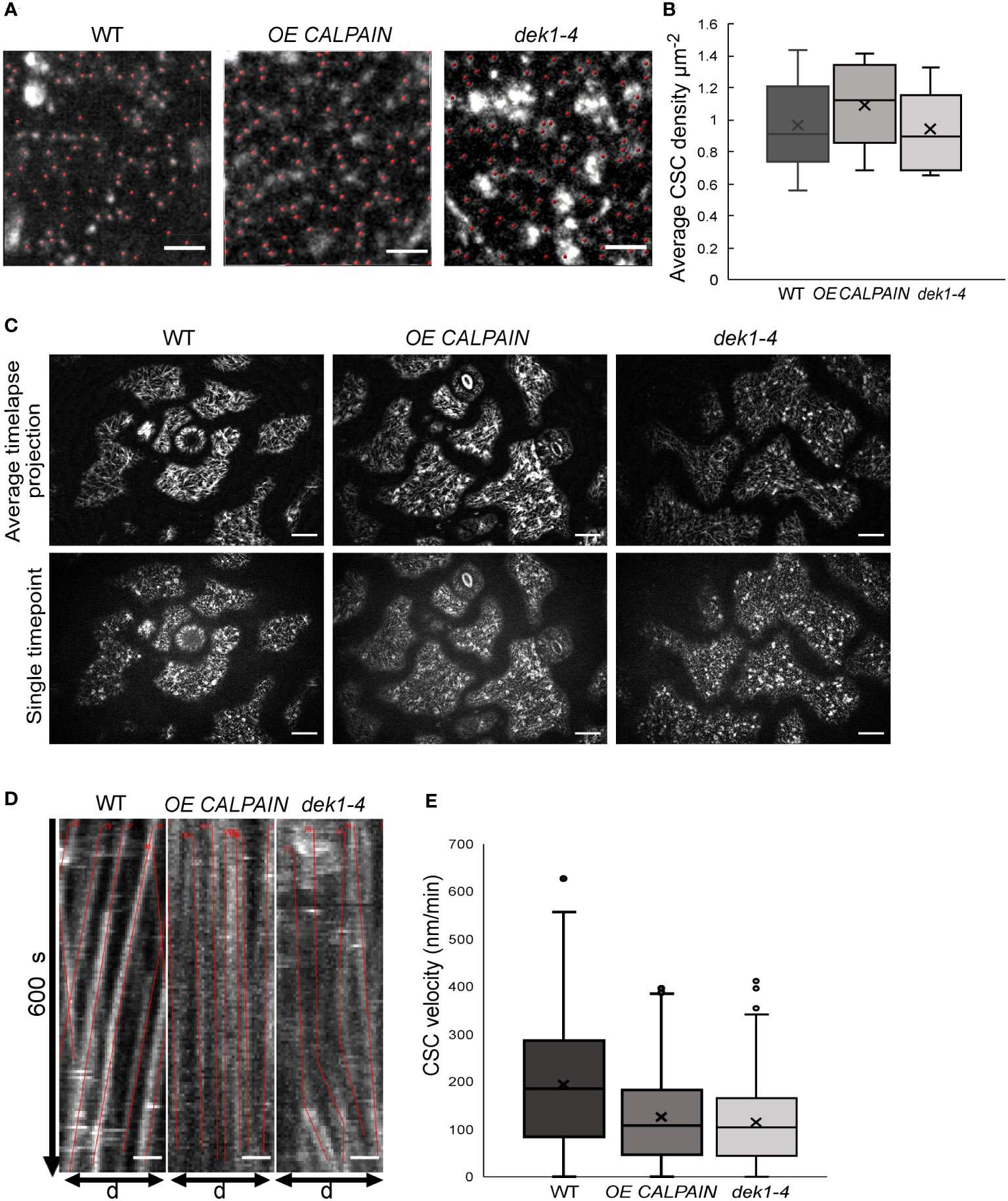
Figure 2 Analysis of cellulose synthase complex (CSC) migration at the plasma membrane in WT and DEK1 modulated Arabidopsis lines. (A) Representative micrographs of 3-day old Arabidopsis cotyledon epidermal pavement cells' membrane patches expressing GFP.CESA3 fluorescent marker used for CSC (red dots) density calculation in Col-0 and DEK1 modulated Iines, Scale bars = 2 µm, (B) Box plots showing average density of CESA particles in 3-days old cotyledon pavement cells. WT 0.89 ± 0.04 µm-2, OE CALPAIN 0.93 ± 0.04 µm-2 and dek1-4 0.82 ± 0.04 µm-2, N=6 plants for WT, 31 cells; 6 plants for OE CALPAIN, 33 cells; 6 plants for dek1-4, 27 cells. No statistical significance observed between WT and DEK1 modulated lines, p=0.5647 for OE CALPAIN, p=0.2705 for dek1-4. (C) Representative images of 3 day old cytoledon epidermal pavement cells used for generation of kymographs. Upper panel represents average projections of time lapses, bottom panel represents a single frame of a corresponding time lapse, N=6 plants per genotype. Scale bars=10 µm. (D) Representative kymographs showing migration of CESA complexes in 3-day old cotyledon pavement cells of Col-0 and DEK1 modulated lines. Red lines represent trajectories of individual CESA particles. WT, N=6 plants, 937 CSC particles tracked; OE CALPAIN, N=6 plants, 982 CSC particles tracked; dek1-4, N-6 plants, 692 CSC particles tracked. Analysis performed on biological duplicates. Scale bars=10 µm. (E) Box plots of CESA velocities obtained from kymographs WT 192.41 ± 4.16 nm min-1, OE CALPAIN 124.11 ± 3.01 nm min-1 and dek1-4 113.91 ± 3.09 nm min-1. Asterisk represent statistical significance Student (p<0.0001, Student t-test).
CSC membrane trafficking is significantly altered in DEK1 modulated lines
To investigate if DEK1 might be regulating other functional properties of CSC complexes, delivery rate of CSCs and their lifetime at the PM was examined. Fluorescence recovery after photobleaching (FRAP) experiments were performed on OE CALPAIN and dek1-4 plants crossed into a GFP : CESA3 reporter line to determine if the rate of exocytosis of CSCs into the PM was altered. Imaging of GFP : CESA3 particles during the 10 min following photobleaching (Figure 3A) showed CESAs in OE CALPAIN had significantly faster recovery after FRAP than WT and dek1-4 had significantly slower recovery (Figures 3B, C). Studies of the lifetime of CSC particles at the PM showed the opposite trends (Figure 3D). OE CALPAIN CSCs spent significantly shorter time in the PM than WT CSCs, while dek1-4 CSCs stayed significantly longer in the PM before undergoing endocytosis (Figure 3D).
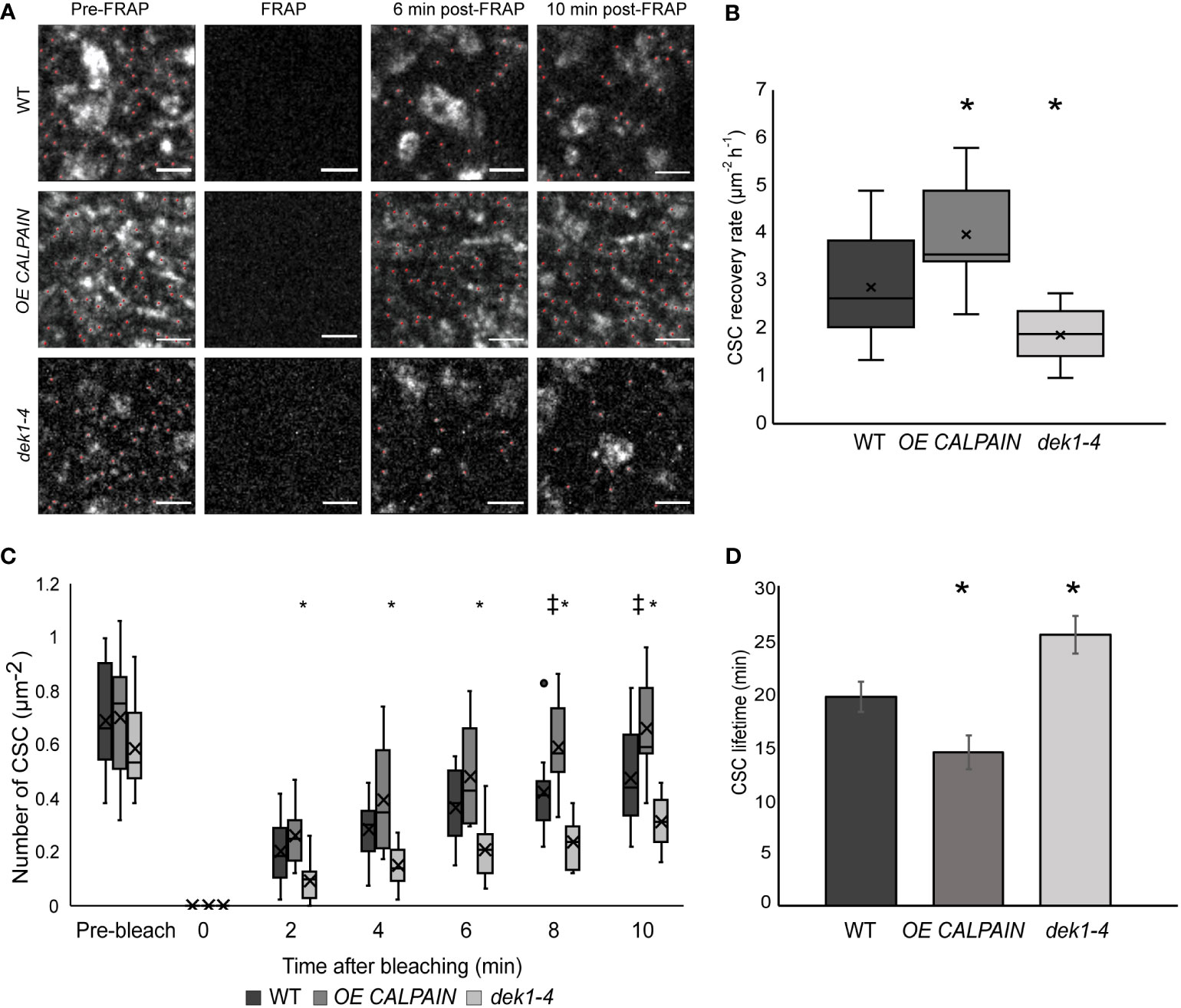
Figure 3 Delivery and resident time of CESAs at the plasma membrane of WT and DEK1 modulated lines in Arabidopsis using fluorescent-recovery after photo-bleaching (FRAP). (A) Representative images showing delivery of new CESA complexes (red dots) after photo-bleaching in 3-day old cotyledon pavement cells of WT, OE CALPAIN, dek1-4. Scale bars = 2 µm. (B) Box plots showing average recovery rate of CESA particles following FRAP in cotyledon pavement cells WT 2.86 ± 0.25 CSC µm-2h-1 , OE CALPAIN 3.96 ± 0.36. CSC µm-2h-1, dek1-4 1.87 ± 0.15 CSC µm-2h-1. Asterisk represent statistical significance (p<0.0188 for OE CALPAIN, p<0.0033 for dek1-4, Student's t-test ). (C) Box plots representing recovery of CESA particles after photo-bleaching at each time-point. Symbols above box plots represent statistical significance (p=0.0102 and 0.0188 for OE CALPAIN, p=0.0001 and -0.0055 for dek1-4, Student's t-test). (D) Histogram representing average lifetime of CESA complexes at the PM, WT 19.56 ± 1.42 min, OE CALPAIN 14.33 ± 4.69 min and dek1-4 25.36 ± 1.73 min. Asterisk above boxplot represents statistical significance (p<0.05, Student's t-test), error bars represent standard error. (B–D) Analysis was performed in two biological replicates for WT and dek1-4, and one replicate for OE CALPAIN. N = 7 cotyledons for WT, 3 cotyledons for OE CALPAIN, 6 cotyledons for dek1-4;N =16 cells for WT, 9 cells for OE CALPAIN, 14 cells for dek1-4.
DEK1 affects cell wall mechanics of cotyledon pavement cells
The defects in CESA dynamics observed in cotyledon pavement cells prompted us to examine the possible role of DEK1 in influencing structure and physical properties of the CMFs and consequently the mechanical properties of the cell wall. We used atomic force microscopy (AFM) to image and mechanically characterize native cell walls.
We performed AFM imaging on dry cell wall monolayers of isolated abaxial epidermal cell walls from cotyledons of 10-day old Arabidopsis seedlings (Figures 4A–C; Supplementary Figures 7, 8). Imaging was done on the inner surface of the abaxial epidermis outer periclinal walls, which is the newest deposited layer of the cell wall, abutting the PM (Supplementary Figure 7). Cell wall images of WT clearly show the CMF network (Figures 4A–C; Supplementary Figure 9), as reported for onion epidermal cell wall monolayers imaged by AFM (Kafle et al., 2014; Zhang et al., 2016). Differences in the thickness of CMFs were observed in OE CALPAIN and dek1-4 (Figures 4A–C) compared to WT. In OE CALPAIN, thicker bundles of CMFs were observed as well as amorphous deposits on the cell wall surface (Figure 4B). Diameters of the CMF bundles were quantified from height channel images (Supplementary Figure 9). Results showed that the CMF bundles of both OE CALPAIN and dek1-4 had significantly higher mean diameters than WT CMFs (Figure 4D). Analysis of the distribution of CMF dimeters shows that both DEK1 modulated lines have a higher frequency of bundled CMFs in the range of 15-40 nm whereas WT CMFs are mostly represented in the range of 1-15 nm, implying that WT has either an abundance of single CMFs (~3.5 nm is the diameter of a typical CMF (Zhang et al., 2016)) or smaller bundles of CMFs (Figure 4E).
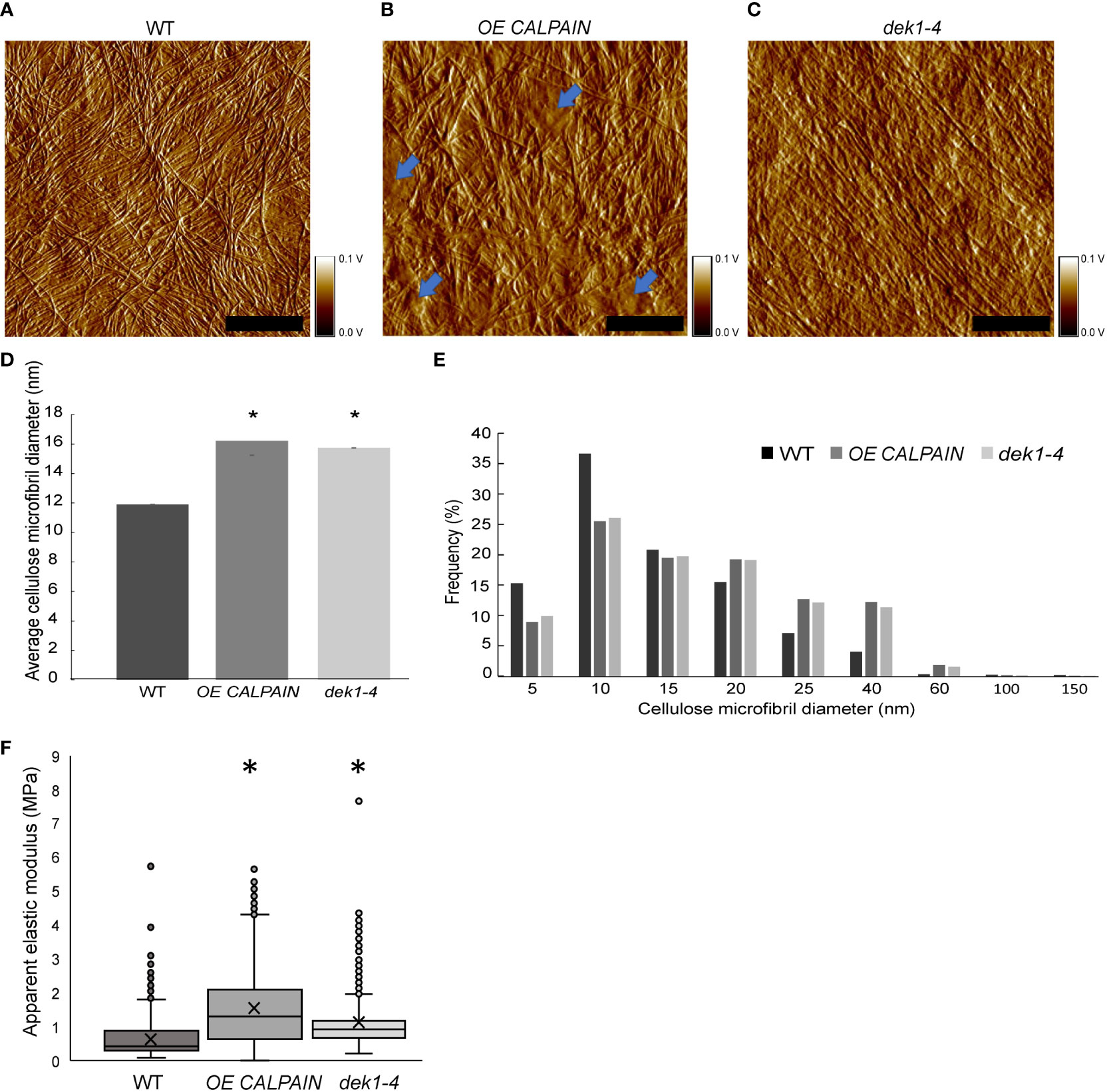
Figure 4 AFM imaging and and nanomechanical characterization of outer periclinal cell walls of Arabidopsis. (A–C) AFM amplitude channel images of the inner face (juxtaposed to the PM) of periclinal primary cell walls in abaxial cotyledon pavement cells of 10-days old seedlings of WT, OE CALPAIN and dek1-4. Samples were imaged using tapping mode in air. Arrows indicate amorphous cell wall regions. Scale bars=250 nm. Corresponding height channel AFM images are shown in Supplementary Figure 8. (D) Histograms representing average thickness of cellulose microfibrils in cotyledon epidermal cell walls, WT 11.09 ± 0.03 nm, OE CALPAIN 16.21 ± 0.03 nm and dek1-4 15.74 ± 0.03 nm. Asterisk represents statistical significance, p<0.0001, unpaired Student t-test. N=5 cotyledons, 10 cells, 1-2 regions per cell for WT; N=6 plants, 6 cells, 1-2 regions per cell for OE CALPAIN; N=4 cotyledons, 7 cells, 1-3 regions per cell for dek1-4. (E) Distribution of CMF thickness and frequency in WT, OE CALPAIN and dek1-4. (F) Box plots representing distribution of cell wall apparent elastic moduli generated after nanoindentation of extracted cotyledon epidermal cell walls in a direction perpendicular to CMFs of 10-day old cotyledons of WT 0.71 ± 0.01 MPa, OE CALPAIN 1.52 ± 0.03 MPa and dek1-4 1.11 ± 0.02 MPa. N=12 indented cotyledons from 12 different plants for WT (1427 indentation points), 9 cotyledons from 9 different plants for OE CALPAIN (1553 indentation points), and 12 cotyledons from 12 different plants for dek1-4 (1509 indentation points). One cell per one cotyledon was indented in all genotypes. Asterisk represents statistical significance, p<0.0001, Student t-test. (A–F) All experiments were performed in two biological replicates.
Previous studies have shown that changes of cell wall composition in cell wall mutants of Arabidopsis can affect mechanical behavior of the cell wall and CMF properties (Xiao and Anderson, 2016). To examine cell wall mechanical properties in DEK1 modulated lines, we performed AFM nanoindentation on the isolated abaxial epidermal cell walls of 10-day old Arabidopsis cotyledons. Nanoindentation generated force-displacement curves, were used to calculate cell wall elastic moduli (Supplementary Figures 10A, C, E). It is worth noting that within all analyzed genotypes, a large range of elastic moduli was observed (Figure 4F; Supplementary Figures 10B, D, F). In WT, values ranged from 0.1 Mpa to 5 Mpa, which is consistent with previous data showing highly heterogenous distribution of cell wall mechanical properties (Yakubov et al., 2016). Analysis of the apparent elastic modulus in OE CALPAIN and dek1-4 suggests that both lines had significantly stiffer cell walls compared to WT (Figure 4F). These results suggest DEK1 is involved in regulating and maintaining mechanical properties of the cell wall.
Discussion
In this study we show that DEK1 affects the synthesis of both cellulose and pectins in early developmental stages of Arabidopsis. Both DEK1 modulated lines show decreased crystalline cellulose in outer epidermal cell wall of cotyledons. Velocity of CSCs in the epidermal pavement cell PMs, which is used as a proxy for the biosynthetic activity, is significantly reduced in both DEK1 modulated lines, while the density of CSC complexes is similar to that of the WT. PM dynamics of CSCs is significantly altered in both lines, with OE CALPAIN CSCs having significantly slower CSC exocytosis and faster endocytosis, while these features of CSC dynamics were reversed in dek1-4 plants. Epidermal cell wall stiffness was significantly increased in both DEK1 modulated lines compared to WT.
DEK1 in cellulose assembly
DEK1 is proposed to act as a mechano-sensor protein at the PM (Amanda et al., 2016; Tran et al., 2017). Upon perception of mechanical stimuli, originating either from other growing cells in the tissue or from a cell’s own turgor pressure, or an external environmental signal, the CALPAIN domain of DEK1 is proposed to be auto-catalytically released into the cytoplasm (Amanda et al., 2016; Tran et al., 2017). DEK1 CALPAIN likely acts as a regulatory protease (Wang et al., 2003; Johnson et al., 2008) and downstream signaling initiates changes in cell wall biosynthesis/remodeling and epidermal specification (Demko et al., 2014; Galletti et al., 2015; Amanda et al., 2016; Amanda et al., 2017). In this study we show DEK1 influences CESA dynamics, cellulose content, pectin re-structuring and mechanical properties of epidermal walls.
It is currently understood that slower movement of CSCs across the PM, a proxy for a lower rate of cellulose biosynthesis, results in less cellulose synthesized (Diotallevi and Mulder, 2007). The significant reduction of CSC velocity in OE CALPAIN and dek1-4 (Figures 2D, E) support a role for DEK1 in regulation of CESA dynamics. Mutants for CESA6/prc1-1, and CESA1/any1, have significantly reduced CESA velocities in dark grown hypocotyls (Bischoff et al., 2011) that is proposed to be the major cause of a 30% reduction in growth (Fagard et al., 2000). There is no data in the literature that CESA proteins undergo proteolytic processing after insertion (by exocytosis) into the PM. RT-qPCR analysis of CESA1,3 and 6 expression levels, and measurement of CESA density at the cotyledon pavement epidermal cell PMs showed no difference between WT and DEK1 modulated lines. These data support DEK1 regulation of CSCs at the post-translational level. An interesting possibility is that DEK1 regulates CSC activity indirectly, potentially through interactions with either CESA regulatory proteins or the cytoskeleton components that guide CSCs. Candidates would include CSI1/POM2 that facilitate binding between CSCs and CMTs (Gu et al., 2010; Bringmann et al., 2012), the endo-1,4-beta-glucanase KORRIGAN (KOR) involved in CESA regulation (His et al., 2001; Vain et al., 2014) and PATROL1 (PTL1) that interacts with exocyst complex proteins and CS1/POM2 to deliver CSCs to the PM (Zhu et al., 2018). Mutants kor1-1 and kor1-3 display a similar reduction in CSC mobility to DEK1 modulated lines when compared to WT plants (Vain et al., 2014). In addition kor mutants have been shown to have decreased cellulose content, but also increased pectin abundance, which was interpreted as a compensatory mechanism and part of feedback loops connecting cellulose and pectin synthesis to changes in the mechanical properties of the cell wall (His et al., 2001).
It could be conceivable that DEK1 is involved in CSC guidance along CMTs, either by modifying the activity and properties of CSI1/POM2 or some other CESA-CMT interface proteins. Reduced CSCs velocities and cellulose content is observed in pom2-1 mutants compared to WT plants as well as spiral twisting of the entire rosette and leaves. Previous studies of lines with reduced levels/activity of DEK1, including dek1-4, showed epinastic curling of cotyledons and leaves (Roeder et al., 2012; Galletti et al., 2015). CMTs are responsive to mechanical stress and assumed to align in the direction of maximal stress (Hamant et al., 2008). During salt stress, CMTs are rapidly disassembled, and CSCs removed from the PM (Endler et al., 2015). COMPANION OF CELLULOSE SYNTHASE (CC) proteins bind to CMTs and promote salt tolerance by stabilizing CMTs and enabling cellulose synthesis. CMTs were shown to re-organize normally in dek1-4 mutants in response to ablation in cotyledons, however, were more highly orientated than in WT pavement cells due to changes in cell shape (Galletti et al., 2015). A study in animals has shown that a non-proteolytic variant of CALPAIN, CALPAIN6 (CAPN6), stabilizes microtubules by binding to them and preventing their de-polymerization (Tonami et al., 2011). Unlike CAPN6, DEK1 CALPAIN has proteolytic activity, and it would be interesting to investigate if DEK1 CALPAIN can interact with CMT and potentially disrupt binding of CESA complexes (Tonami et al., 2011).
Upregulation of the exocytotic module of the CSC trafficking machinery in OE CALPAIN (Figures 3B, C) could be one explanation for the faster exocytosis of the CESAs, as observed by faster post-FRAP recovery of CSC particles (Figures 3A, B). Exocytosis of CSCs is regulated by a large protein complex (Polko and Kieber, 2019), which interacts with CSI1 and PTL1 to deliver CSCs to the PM (Zhu et al., 2018). The frequency of CSC insertions and alignment with the CMT was highly decreased and the delivery rate of CSC severely diminished in csi1-3/ptl1-2 double mutants. In addition, ptl mutants also have lower CSC velocities, and both ptl1-2 and csi1-3/ptl1-2 lines have significant decreases in cellulose content (Zhu et al., 2018). The contrasting effect which OE CALPAIN and dek1-4 have on CSC PM trafficking could mean that DEK1 is interacting with several components involved in CSC trafficking to (and in) the PM, and that its role in signaling involves multiple target proteins.
DEK1 and cell wall mechanics
The epidermis is a key regulator of plant growth and this is influenced by the mechanical properties of the cell wall (Galletti et al., 2016; Moulia et al., 2021). In DEK1 modulated lines, pectins and cellulose are altered yet the dynamics of these changes are unclear and if direct or indirect. Previous studies (Amanda et al., 2016; Amanda et al., 2017), have shown that DEK1 affects cell wall composition almost exclusively on the level of leaf outer epidermal cell walls. In leaf 4 of 17 day old Arabidopsis plants, increased labelling of CBM3a recognizing cellulose and JIM5 detecting low-methyl esterified HG was found in dek1-3 OE CALPAIN GFP compared to WT and an amiRNA DEK1 line. In this study, decreased levels of CBM3a and JIM5 labelling was found in epidermal walls of cotyledons of OE CALPAIN. DEK1 potentially acts in different regulatory networks and/or on tissue specific targets during early and late developmental stages of Arabidopsis. Alternatively, changes in cell walls at later stages could be indirect and result from compensatory changes initiated by cell wall integrity pathways (Vaahtera et al., 2019). Further analysis is required to uncover the dynamics of DEK1-regulated wall changes in future.
Compensatory mechanisms to counteract changes in cell wall composition and biomechanics are known to occur, for example, pectins have been shown to increase in response to reduced levels in cellulose and lead to increased stiffness of the cell wall (His et al., 2001). DEK1 has previously been shown to regulate expression of pectin-related genes, including GAUTs and pectin methyltransferases as well as altered deposition of pectic polysaccharides in leaf epidermal cell walls of OE CALPAIN GFP (Amanda et al., 2016; Amanda et al., 2017). Mutants in pectin methyltransferase, quasimodo2 and tumorous shoot development2, have decreased levels of both HGs and cellulose, decreased CESA velocities, thinner and more fragmented CMFs (Mouille et al., 2007; Du et al., 2020), while mutants in putative galacturonic acid transferase, quasimodo1, experience severe defects in cell-to-cell adhesion due to low HG-content (Bouton et al., 2002; Verger et al., 2018). Increased partially methyl-esterified HG in dek1-4 is consistent with a role for pectin-related increased bundling of CMFs in this line. Stiffer cell walls in OE CALPAIN, with reduced levels of partially methyl-esterified HG and increased levels of highly methyl-esterified HG requires further investigation. The relationship between the degree of HG methyl-esterification and cell wall stiffness is unresolved. It could depend upon local concentrations of Ca2+ in the apoplast (Wang et al., 2020), orientation of the cell wall, such as anticlinal vs periclinal cell walls (Haas et al., 2020), activity of polygalacturonases (Yang et al., 2018) and many as yet unknown factors. Changes in interactions between pectins and CMF could also influence mechanical properties of the cell wall in DEK1 modulated lines (Figure 4E). NMR studies of ‘never dried’ onion epidermal cell walls have shown that HGs make the majority of contacts with CMFs, which could mean that they act as primary mechanical tethers (Wang et al., 2015). Pectins affect cell wall mechanical properties through poro-elastic effects, without impacting the structural properties of cellulose (Lopez-Sanchez et al., 2015).
In future it will be essential to identify the proteolytic targets of DEK1 CALPAIN and the dynamics of cell wall changes in response to altered DEK1 to determine direct vs indirect effects. In this study, pectin composition and root inhibition by isoxaben have opposite patterns in OE CALPAIN and dek1-4 lines, whereas cellulose composition and cell wall stiffness show similar trends. Further studies are required to determine if, for example, the CALPAIN domain is required for the former effects, whereas the latter effects are a response to the induced cell wall modifications. Different stress conditions are known to lead to similar responses of plants in terms of growth and morphogenesis (Tenhaken, 2014). However, the underlying molecular mechanism of these changes and their associated signaling cascades can differ greatly. CWI sensors of the CrRLK family, including FER and THE1 are involved in responses to stress, mediated by RALF peptide binding (Feng et al., 2018; Blackburn et al., 2020; Zhang et al., 2020). RALF peptides are known to be processed by substilin-like proteases (Stegmann et al., 2017), however, other proteases such as DEK1 may be involved in generating mature RALF ligands. DEK1 signaling cascades are still completely unknown and further work is required to determine if cross-talk between CrRLK signaling and DEK1 pathways occur. Identification of CALPAIN targets through the use of targeted proteomics approaches is essential for understanding the pathways through which DEK1 acts, and its precise role in plant growth and development.
Material and methods
Plant resources
Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used as wild type (WT) control. Dek1-4 is an EMS single point mutant (Roeder et al., 2012) backcrossed into Col-0 as described in (Galletti et al., 2015). The CALPAIN overexpression construct pRPS5A:CALPAIN:6X HISTIDINE (OE CALPAIN), with hygromycin resistance in Col-0 wild type background is described in Galletti et al., 2015. For CSC dynamics analysis we used OE CALPAIN and dek1-4 crossed into CSC reporter line pCESA3:GFP : CESA3 (Desprez et al., 2007).
Plants were grown vertically in petri dishes, on MS 1% (w/w) sucrose media, 0.8% agar, pH 5.8 (Murashige and Skoog, 1962), without vitamins. Plants were grown in Percival growth cabinets, model CU36L/LT (Percival Scientific, Perry, Iowa, USA), at a temperature of 21°C and light intensity of 95-111 mol m–2 s–1. Growth conditions used in different experiments are detailed in sections below.
F2 DEK1 modulated lines crossed with GFP : CESA3 were used for CESA dynamics experiments. Non-genotyped segregating F2 plants were grown on MS 1% sucrose media for 3 days, imaged and then genotyped for identification of DEK1 modulated lines containing pCESA3:GFP : CESA3 constructs. After confocal imaging, OE CALPAIN pCESA3:GFP : CESA3 F2 plants were transferred onto MS 1% sucrose media supplemented with 10 µg ml-1 hygromycin. Plants were grown horizontally for 10-14 days together with Col-0 and GFP CESA3 plants as positive controls. Only imaging data for OE CALPAIN pCESA3:GFP : CESA3 plants which survived the hygromycin selection were used for CESA dynamics analysis.
After CESA imaging, dek1-4 pCESA3:GFP : CESA3 plants were transferred to MS 1% sucrose media supplemented with Plant Preservative Mixture (Plant Cell Technology, Washington DC, USA) diluted in ratio 1:1000. Plants were grown until they developed the first true leaves (7-10 days post imaging). A leaf was used to isolate DNA using Edwards buffer method (Edwards et al., 1991) and the DNA analyzed by dCAPS (Neff et al., 1998) (Supplementary Table 2) or KASP genotyping (LGC Genomics, Berlin, Germany) to identify dek1-4 homozygous plants. Only imaging data for dek1-4 homozygous plants was used in further CESA dynamics analysis.
Analysis of CESA transcript levels
RNA was isolated from 6-day old Arabidopsis plants using the QIAGEN Rneasy Mini Kit (ID: 74104, QIAGEN, Hilden, Germany). RNA quality was determined using a Nanodrop 2000/2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For relative quantification (RT)-qPCR experiments, cDNA synthesis was performed using Maxima First Strand cDNA Synthesis Kit (K1641, Thermo Fisher Scientific, Waltham, MA, USA) using 1 μg of RNA. Power SYBR Green Mastermix (4367659, Thermo Fisher Scientific, Waltham, MA, USA) was used for RT-qPCR and runs performed in 384 well plates (AB3384, Thermo Fisher Scientific, Waltham, MA, USA) in 5 μl reactions. Primers used for RT-qPCR are outlined in Supplementary Table 2. Lightcycler ABI 7900 HT (Applied Biosystems, Foster City, California, United States) lightcycler was used for RT-qPCR using amplification, conditions: 50° C – 2 min; 95° C –10 min; 40 cycles of 95° C for 15 s, 60° C for 1 min; 95° C for 15 s, 60° C for 15 s, 95° for 15 s. qPCR data was analyzed using SDS v 2.4.1 analysis software (Thermo Fisher Scientific, Waltham, MA, USA). CESA expression levels were normalized against GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GADPH) which was used as a reference gene.
Relative gene expression values expressed as fold changes were calculated using the method of (Livak and Schmittgen, 2001). CESA expression levels were treated as 1 in Col-0 WT and values in DEK1 modulated lines were calculated compared to Col-0. Gene expression fold changes lower than 0.5 and higher than 2.5, compared to Col-0, were treated as significant.
Cell wall composition analysis
Cell wall composition analysis was performed on pooled 10-day old whole seedlings. Analysis of the cell wall polysaccharide composition of alcohol insoluble cell wall residues (AIR) from Col-0 and DEK1 modulated lines was performed according to (Pettolino et al., 2012). Analysis of the total crystalline cellulose in seedlings was performed using Updegraff assay (Updegraff, 1969) of AIR (10-50 mg).
Immunofluorescence labelling
Cotyledon tissues from 10 day old seedlings were fixed and embedded according to (Wilson and Bacic, 2012). A Leica Ultracut R microtome (Leica Microsystems, Germany) was used to obtain 250 nm thin sections and labelled with JIM5 [low methyl esterified HG; (Knox et al., 1990)], JIM7 [high methyl esterified HG; (Knox et al., 1990)], and LM15 [xyloglucan; (Marcus et al., 2008)] antibodies at 1:10 dilution, secondary antibody was Alexa Fluor 488 goat anti-rat IgG (H+L) (Life Technology; # A48262) with 1:100 dilutions. For crystalline cellulose labelling, CBM3a (Blake et al., 2006) was used at 1:50 dilution, the secondary antibody used was anti-6x-His tag monoclonal (Invitrogen, # MA1-21315) with 1:100 dilutions, the third antibody was Alexa Fluor 488 goat anti-mouse IgG (H+L) (Life Technology; # A11006) with 1:100 dilutions. Images were acquired with an Olympus BX53 microscope under GFP channel. Calcofluor white (0.02%) was used to stain cellulose and images were acquired under UV channel. Two biological replicates from two independent lines were performed.
Transmission-electron microscopy and immunolabelling
Thin sections (~80 nm) from cotyledon tissues were acquired as stated above. Antibody labelling and post-staining were performed according to (Wilson and Bacic, 2012). For pectin labelling, JIM5 and JIM7 antibodies were used at 1:15 dilution, secondary antibodies used was goat anti-rat 18 nm gold conjugated secondary antibody (Jackson Immuno Research #112-215-167) at 1:20 dilutions. For crystalline cellulose labelling, CBM3a at 1:50 dilution, secondary anti-6x-His tag monoclonal antibody (Invitrogen, # MA1-21315) at 1:175 dilutions, and the third antibody was goat anti-mouse 12 nm gold conjugated secondary antibody (Jackson Immuno Research #115-205-166) at 1:35 dilution. Grids were post-stained using 2% uranyl acetate for 10 min and Reynold’s lead citrate for 1 min. Grids were imaged using a Jeol 2100 EM equipped with a Gatan Orius SC 200 CCD camera. Two biological replicates from two independent lines were performed.
Spinning disc confocal microscopy of CSC
Three-day old seedlings were mounted on microscope object slides and imaged using instruments and settings as described in (Sampathkumar et al., 2013). Photobleaching used to access CSC trafficking dynamics was performed using a FRAP/PA system (Roper Scientific, Acton, MA, USA) integrated into the spinning disc confocal imaging system, as described in (Sampathkumar et al., 2013).
CSC timelapse processing
Image processing was performed using ImageJ software (Rasband, W., NIH, Bethesda, MD, USA). Brightness and contrast of the raw time-lapses were modified. Subtract background function (rolling ball size 30-40 pixels) and Walking average plugin (three frames averaged) were used to remove background noise and to obtain clearer images.
Density of CSC particles was determined from confocal time-lapse images using IMARIS 7.4 image analysis software (Bitplane, Oxford Instruments, Oxford, United Kingdom). Time-lapse image stacks were pre-processed in ImageJ as described above and imported into IMARIS. Spot detection and tracking function of the IMARIS was used to detect and label CSC particles. CSC particle numbers used to calculate mean CSC density were taken from three random timeframes from each analyzed pavement cell form every imaged cotyledon for each of the analyzed genotypes. Density was calculated as mean of CSC particles density per one μm2 for each cell, plant and genotype.
Velocity analysis of CESA particles imaged using confocal microscopy was performed using Fluorescent Image Evaluation Software for Tracking and Analysis (FIESTA) software (Ruhnow et al., 2011), which is available under open license from FIESTA, University of Dresden Fusion Forge Wiki webpage (https://fusionforge.zih.tu-dresden.de/plugins/mediawiki/wiki/fiesta/index.php/FIESTA). Raw confocal time-lapse images were imported into FIESTA. Maximum signal intensity projections were used to generate kymographs. Kymographs were generated from the CSC trajectories manually drawn using segmented line tool in FIESTA.
Analysis of the data obtained by FRAP was performed using a modified version of the CSC density measurement method described above. CSC particles detection and quantification was performed on 9 x 9 μm cell patches rather than 10 x 10 μm, which were the dimensions of bleached surface. After performing detection of fluorescently labelled CSC particles using IMARIS software, CSC post-FRAP density, recovery rates and average plasma membrane life-time of the complexes was calculated as described in (Sampathkumar et al., 2013).
Cell wall isolation for AFM topographical imaging and nanoindentation
Cotyledon pavement epidermal cell walls were isolated for analysis with the atomic force microscope (imaging and nanoindentation). Rectangular microscope coverslips (Roth, Karlsruhe, Germany) were covered with 200 μl of 0.01% poly-D-lysine-hydrobromide (PDL; P6407, Sigma Aldrich, St. Luis, MO, USA) in deionized water. Coverslips were left for 15-20 min to allow the PDL to polymerize, after which coverslips were washed thoroughly with deionized water and dried in a stream of nitrogen.
We used 10-day old seedlings of Arabidopsis to obtain cotyledons for cell wall isolation. The larger of the two cotyledons was cut off using surgical tweezers, placed on a PDL coated coverslip (with the abaxial side facing the coverslip) and the following steps were performed under a stereomicroscope (Zeiss Stemi 508, Zeiss, Jena, Germany). The cotyledon was pressed gently on one side using surgical tweezers and then a vertical cut was made using a micro-surgery scalpel (Fine Scientific Tools, Vancouver, Canada), across the middle of the cotyledon in a direction perpendicular to its longer axis. After the vertical cut, a second cut was performed in parallel to the longer cotyledon axis to remove all leaf tissues and expose the abaxial epidermis with patches of the cell wall monolayers on its edge (Supplementary Figure 6). Regions with cell wall monolayers were observable under 50x magnification as small opaque patches of material on the edge of the abaxial epidermis (Supplementary Figure 6).
The remaining parts of the dissected cotyledon were carefully attached to the surface with nitrocellulose based red stained adhesive (nail polish) to immobilize the sample during imaging and mechanical probing. Adhesive was applied with an injection syringe needle, carefully avoiding regions with cell wall monolayers and left to cure for 5-10 seconds. Samples were then incubated with 500 μl of 1% SDS for 20-30 s to remove organelles, proteins and other cellular components released during the cotyledon dissection. SDS was removed by washing with deionized water, ensuring that the dissected cotyledon does not curl up on the coverslip. For high-resolution AFM topological imaging the samples were left overnight to completely dry while, samples used for mechanical measurements were stored in water to prevent loss of native mechanical properties.
AFM topographical imaging
For measuring the CMF diameter, a Dimension 3100 AFM (Veeco, Plainview, NY, USA) was used. Imaging was performed in tapping mode in air, using silicon ARROW-NCR cantilevers (tip radius <10 nm; Nanoworld, Neuchâtel, Switzerland).The optical camera integrated into the AFM setup was used to identify regions of interest for imaging. Regions with a size of 2 x 2 μm2 were imaged with a resolution of 512 x 512 pixels. We recorded height channel images (Supplementary Figure 8) and amplitude channel images (Figures 4A–C). Imaging of all genotypes was performed in biological duplicate; that is, the same experiments were repeated twice with two sets of plants grown independently at different times.
Analysis of the cellulose microfibril images
High-resolution, high-quality cell wall monolayers imaged in dry conditions were analyzed using custom made MATLAB script to determine the diameter of the imaged CMFs. For the analysis of CMF diameter, we used the height channel AFM images (Supplementary Figure 8) as obtained from the AFM software. The extracted values of the CMF diameter were pooled for each genotype and CMF diameter values.
Mechanical characterization of isolated cell walls using atomic force microscopy
To determine the apparent elastic modulus of the isolated cell wall monolayers, we performed AFM-based nanoindentation (manual force mapping mode), applying a method previously described (Yakubov et al., 2016) and modified for our experimental conditions. Nanoindentation experiments were performed using a JPK Nanowizard 3 AFM (Bruker Nano GmbH, Berlin, Germany) mounted on an Olympus IX71 inverted phase-contrast microscope (Olympus Corporation, Shinjuku City, Tokyo, Japan). Coverslips with prepared samples were mounted onto the AFM sample holder as described in (Yakubov et al., 2016).
An optical CCD camera integrated into the AFM setup was used during the entire measurement; first to align the cantilever and then to perform constant checks of the sample and cantilever properties. We used DNP-10 cantilevers (cantilever A) with a nominal spring constant of 0.35 N m-1 (Bruker Corporation, Billerica, MA, USA). The optical sensitivity of the cantilevers was determined from contactless oscillations after which the thermal noise method was used to obtain the spring constant. The used cantilevers had spring constants ranging from 0.30-0.38 N m-1.
Before starting the mechanical characterization, topological imaging of the cell wall monolayer was performed to identify the thinnest regions of the sample. First, using intermittent contact mode, we imaged an area of 5 x 5 μm2 with a resolution of 512 x 512 pixels. Imaging of a 5 x 5 μm2 region was always performed near the edge of the dissected cotyledon, so that part of imaged area represented the bare glass slide to which the cotyledon was attached. The flat glass slide surface, which contained some tissue debris after dissection, was used as a reference (position 0 nm), to determine the thickness of the imaged sample (Supplementary Figure 7). If sample thickness was in the 0.5-2.0 μm range, we selected a smaller 1 x 1 μm2 region within the original 5 x 5 μm area and repeated the topography imaging to re-confirm cell wall thickness (Supplementary Figure 7). If the thickness of the topological features inside this region remained in the 0.5-2 μm range, we performed nanoindentation on that region. This range of sample thicknesses values was empirically chosen. During method development, we determined that regions thicker than 2.0 μm were either wrinkled cell wall patches or cell wall bilayers (i.e. from both adaxial and abaxial sides; thickness 2.5-4 μm). Samples thinner than 0.5 μm were not used to avoid non-specific surface interactions and to minimize the contribution of the underlying glass surface to the elastic modulus.
For nanoindentation, the 1 x 1 μm2 region of interest was further divided into a 16 x 16 array. Each position was indented once to obtain a force map consisting of 256 force-displacement curves. For each approach-retract cycle, the contact force was set to 2.5 nN and the z-piezo speed was 1 μm s-1. The average indentation depth was 180 nm for Col-0, 164 nm for OE CALPAIN and 147 nm for dek1-4, while indentation depth values where mostly in the range of 100-350 nm (Supplementary Figure 9G). While the indentation depths partially exceeded 10% of the sample thickness, the values lie in a similar range. Contributions from the underlying glass surface can thus be expected to be similar for all three samples.
One force map was obtained from each dissected cotyledon. Imaging of all genotypes was performed in biological duplicate; the same experiments were repeated two times, with two sets of plants grown independently at different times. From each replicate, cotyledons from 4-6 different plants were imaged, one cotyledon per plant. Force maps were analyzed using JPK SPM processing software, version 6.1.9 (Bruker Nano GmbH). As adhesion was observed in the retract segments of the force-displacement curves, the approach segments were fit with the Hertz model to obtain the apparent elastic modulus. The Poisson ratio was set to 0.5 and the baseline value was unpinned and set to 0. The extracted apparent elastic moduli from different cotyledons for the same genotype were pooled and dek1-4 and OE CALPAIN values were compared with Col-0.
Isoxaben and salt assays
Plants used for the isoxaben treatments were sterilized and grown for 6 days on MS media containing 1% sucrose, with the addition of 2 nM isoxaben (Sigma-Aldrich, St. Louis, MO, USA) dissolved in DMSO (Honeywell, Charlotte, NC, USA), and on MS 1% sucrose containing media supplemented with 2nM DMSO as a control. Experiments were performed in biological duplicates and technical triplicates. Salt stress experiments were performed as described in (Feng et al., 2018).
Plants were imaged using a Keyence VHX-6000 digital microscope (Keyence Corporation of America, Itasca, IL, USA). Whole plates were placed on a motorized microscope stage and series of images were acquired in sequence covering the entire surface of the plate, and then automatically stitched by the software to generate a single whole-plate image. We used objective ZS20, 20X magnification, on black field, with epi-illumination, for imaging 6-day isoxaben grown and salt-treated plants. Root lengths were quantified using Measure length function of ImageJ software (Rasband, W., NIH, Bethesda, MD, USA).
Statistical analysis
Unless stated otherwise, all statistical analysis in this study were performed using unpaired Student t-test, results were treated statistically significant if p<0.05. We used online Student t-test calculator at the webpage https://www.graphpad.com/quickcalcs/ttest1.cfm.
Analysis of data from isoxaben growth and salt assays was performed using estimation statistics. After obtaining root length data, statistical significance of the results was assessed using the online, open-access Estimation Stats software (https://www.estimationstats.com/#/) and data are plotted using Cumming estimation plot as described in (Ho et al., 2019).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
LN performed CSC imaging, AFM measurements and imaging and CWI sensing experiments, and analyzed all the data from these experiments. KJ, TB, and AS conceptualized and designed the research. LN, GY, KJ, and KB conceptualized, developed, and validated cell wall isolation method and AFM imaging and force spectroscopy protocols. KJ and AS made DEK1 crosses with CSC reporter line. YM performed immunofluorescences and immunohistochemical imaging experiments and analyzed the data. GY wrote the MATLAB script for analysis of AFM cellulose images. LN wrote the original draft of the article manuscript. KJ, TB, KB, and AS edited, curated and corrected the original draft of the manuscript. KB, GY, and YM gave valuable input and comments on the article manuscript.
Funding
LN was funded by University of Melbourne Research Scholarship, Max Planck Society, Norma Hilda Schuster neé Swift Scholarship and Albert Shimmins Postgraduate Writing-up Award. YM acknowledges the support of a University of Melbourne Research Scholarship. KJ and AB would like to acknowledge the support of funds from the ARC Centre of Excellence in Plant Cell Walls (CE1101007) and start-up funds from La Trobe University.
Acknowledgments
Authors would like to thank and acknowledge Mr. Pengfei (Alfie) Hao of La Trobe University, Australia, for performing cell wall composition and data analysis and the La Trobe BioImaging facility for assistance with electron microscopy. Authors would like to acknowledge A/Prof Monika S Doblin of La Trobe University, Australia and Dr. René Schneider of University of Potsdam, Germany for critical discussion and advice on method development and data interpretation. Finally, authors would like to acknowledge Ms. Reinhild Dünnebacke and Ms. Irina Berndt of the Max Planck Institute of Colloids and Interfaces, Germany, for their invaluable support and assistance during the AFM imaging and mechanical characterization of the isolated cell walls.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1150202/full#supplementary-material
References
Amanda, D., Doblin, M. S., Galletti, R., Bacic, A., Ingram, G. C., Johnson, K. L. (2016). DEFECTIVE KERNEL1 (DEK1) regulates cell walls in the leaf epidermis. Plant Physiol. 172, 2204–2218. doi: 10.1104/pp.16.01401
Amanda, D., Doblin, M. S., Macmillan, C. P., Galletti, R., Golz, J. F., Bacic, A., et al. (2017). Arabidopsis DEFECTIVE KERNEL1 regulates cell wall composition and axial growth in the inflorescence stem. Plant Direct 1, e00027. doi: 10.1002/pld3.27
Becraft, P. W., Li, K., Dey, N., Asuncion-Crabb, Y. (2002). The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129 (22), 5217–5225. doi: 10.1242/dev.129.22.5217
Bischoff, V., Desprez, T., Mouille, G., Vernhettes, S., Gonneau, M., Höfte, H. (2011). Phytochrome regulation of cellulose synthesis in arabidopsis. Curr. Biol. 21, 1822–1827. doi: 10.1016/j.cub.2011.09.026
Blackburn, M. R., Haruta, M., Moura, D. S. (2020). Twenty years of progress in physiological and biochemical investigation of RALF Peptides1. Plant Physiol. 182, 1657–1666. doi: 10.1104/pp.19.01310
Blake, A. W., Mccartney, L., Flint, J. E., Bolam, D. N., Boraston, A. B., Gilbert, H. J., et al. (2006). Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J. Biol. Chem. 281, 29321–29329. doi: 10.1074/jbc.M605903200
Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M. T., Talbotec, J., Granier, F., et al. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in arabidopsis. Plant Cell 14, 2577–2590. doi: 10.1105/tpc.004259
Bringmann, M., Li, E., Sampathkumar, A., Kocabek, T., Hauser, M.-T., Persson, S. (2012). POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in arabidopsis. Plant Cell 24 (1), 163–177. doi: 10.1105/tpc.111.093575
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 (11), 850–861. doi: 10.1038/nrm1746
Demko, V., Perroud, P.-F., Johansen, W., Delwiche, C. F., Cooper, E. D., Remme, P., et al. (2014). Genetic analysis of DEFECTIVE KERNEL1 loop function in three-dimensional body patterning in physcomitrella patens. Plant Physiol. 166, 903–919. doi: 10.1104/pp.114.243758
Desprez, T., Juraniec, M., Crowell, E. F., Jouy, H., Pochylova, Z., Parcy, F., et al. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in arabidopsis thaliana. Proc. Natl. Acad. Sci. United States America 104, 15572–15577. doi: 10.1073/pnas.0706569104
Diotallevi, F., Mulder, B. (2007). The cellulose synthase complex: a polymerization driven supramolecular motor. Biophys. J. 92, 2666–2673. doi: 10.1529/biophysj.106.099473
Du, J., Kirui, A., Huang, S., Wang, L., Barnes, W. J., Kiemle, S. N., et al. (2020). Mutations in the pectin methyltransferase QUASIMODO2 influence cellulose biosynthesis and wall integrity in arabidopsis. Plant Cell 32, 3576–3597. doi: 10.1105/tpc.20.00252
Edwards, K., Johnstone, C., Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349 doi: 10.1093/nar/19.6.1349
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
Engelsdorf, T., Hamann, T. (2014). An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann. Bot. 114, 1339–1347. doi: 10.1093/aob/mcu043
Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., et al. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of arabidopsis. Plant Cell 12, 2409–2424. doi: 10.1105/tpc.12.12.2409
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca signaling. Curr. Biol. 28, 1–10. doi: 10.1016/j.cub.2018.01.023
Fujita, M., Himmelspach, R., Hocart, C. H., Williamson, R. E., Mansfield, S. D., Wasteneys, G. O. (2011). Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in arabidopsis. Plant J. 66, 915–928. doi: 10.1111/j.1365-313X.2011.04552.x
Galletti, R., Johnson, K. L., Scofield, S., San-Bento, R., Watt, A. M., Murray, J., et al. (2015). DEFECTIVE KERNEL 1 promotes and maintains plant epidermal differentiation. Development 142, 1978–1983. doi: 10.1242/dev.122325
Galletti, R., Verger, S., Hamant, O., Ingram, G. C. (2016). Developing a 'thick skin': a paradoxical role for mechanical tension in maintaining epidermal integrity? Development 143, 3249–3258. doi: 10.1242/dev.132837
García Díaz, B. E., Gauthier, S., Davies, P. L. (2006). Ca2+ dependency of calpain 3 (p94) activation. Biochemistry 45 (11), 3714–3722. doi: 10.1021/bi051917j
Gjetting, S. K., Mahmood, K., Shabala, L., Kristensen, A., Shabala, S., Palmgren, M., et al. (2020). Evidence for multiple receptors mediating RALF-triggered Ca2+ signaling and proton pump inhibition. Plant J. 104 (2), 433–446. doi: 10.1111/tpj.14935
Haas, K. T., Wightman, R., Meyerowitz, E. M., Peaucelle, A. (2020). Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science 367, 1003–1007. doi: 10.1126/science.aaz5103
Hamant, O., Heisler, M. G., Jönsson, H., Krupinski, P., Uyttewaal, M., Bokov, P., et al. (2008). Developmental patterning by mechanical signals in arabidopsis. Science 322, 1650–1655. doi: 10.1126/science.1165594
Heim, D. R., Skomp, J. R., Tschabold, E. E., Larrinua, I. M. (1990). Isoxaben inhibits the synthesis of acid insoluble cell wall materials in arabidopsis thaliana. Plant Physiol. 93, 695–700. doi: 10.1104/pp.93.2.695
His, I., Driouich, A., Nicol, F., Jauneau, A., Höfte, H. (2001). Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212, 348–358. doi: 10.1007/s004250000437
Ho, J., Tumkaya, T., Aryal, S., Choi, H., Claridge-Chang, A. (2019). Moving beyond p values: data analysis with estimation graphics. Nat. Methods 16, 565–566. doi: 10.1038/s41592-019-0470-3
Johnson, K. L., Faulkner, C., Jeffree, C. E., Ingram, G. C. (2008). The phytocalpain defective kernel 1 is a novel arabidopsis growth regulator whose activity is regulated by proteolytic processing. Plant Cell 20, 2619–2630. doi: 10.1105/tpc.108.059964
Johnson, K. L., Degnan, K. A., Ross Walker, J., Ingram, G. C. (2005). AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44 (1), 114–127. doi: 10.1111/j.1365-313X.2005.02514.x
Johnson, K. L., Gidley, M. J., Bacic, A., Doblin, M. S. (2018). Cell wall biomechanics: a tractable challenge in manipulating plant cell walls 'fit for purpose'! Curr. Opin. Biotechnol. 49, 163–171. doi: 10.1016/j.copbio.2017.08.013
Kafle, K., Xi, X., Lee, C. M., Tittmann, B. R., Cosgrove, D. J., Park, Y. B., et al. (2014). Cellulose microfibril orientation in onion (Allium cepa l.) epidermis studied by atomic force microscopy (AFM) and vibrational sum frequency generation (SFG) spectroscopy. Cellulose 21, 1075–1086. doi: 10.1007/s10570-013-0121-2
Knox, J. P., Linstead, P. J., King, J., Cooper, C., Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181, 512–521. doi: 10.1007/BF00193004
Kohorn, B. D., Johansen, S., Shishido, A., Todorova, T., Martinez, R., Defeo, E., et al. (2009). Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60 (6), 974–982. doi: 10.1111/j.1365-313X.2009.04016.x
Lane, D. R., Wiedemeier, A., Peng, L., Höfte, H., Vernhettes, S., Desprez, T., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in arabidopsis. Plant Physiol. 126 (1), 278–288. doi: 10.1104/pp.126.1.278
Lid, S. E., Gruis, D., Jung, R., Lorentzen, J. A., Ananiev, E., Chamberlin, M., et al. (2002). The defective kernel1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U.S.A. 99 (8), 5460–5465. doi: 10.1073/pnas.042098799
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCt) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Sanchez, P., Cersosimo, J., Wang, D., Flanagan, B., Stokes, J. R., Gidley, M. J. (2015). Poroelastic mechanical effects of hemicelluloses on cellulosic hydrogels under compression. PloS One 10, e0122132. doi: 10.1371/journal.pone.0122132
Marcus, S. E., Verhertbruggen, Y., Herve, C., Ordaz-Ortiz, J. J., Farkas, V., Pedersen, H. L., et al. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60. doi: 10.1186/1471-2229-8-60
Mouille, G., Ralet, M. C., Cavelier, C., Eland, C., Effroy, D., Hématy, K., et al. (2007). Homogalacturonan synthesis in arabidopsis thaliana requires a golgi-localized protein with a putative methyltransferase domain. Plant J. 50, 605–614. doi: 10.1111/j.1365-313X.2007.03086.x
Moulia, B., Douady, S., Hamant, O. (2021). Fluctuations shape plants through proprioception. Science 372, eabc6868. doi: 10.1126/science.abc6868
Nixon, B. T., Mansouri, K., Singh, A., Du, J., Davis, J. K., Lee, J.-G., et al. (2016). Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci. Rep. 6, 28696. doi: 10.1038/srep28696
Neff, M. M., Neff, J. D., Chory, J., Pepper, A. E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in arabidopsis thaliana genetics. Plant J. 14, 387–392. doi: 10.1046/j.1365-313X.1998.00124.x
Ono, Y., Sorimachi, H. (20121824). Calpains — an elaborate proteolytic system. Biochim. Biophys. Acta 1), 224–236. doi: 10.1016/j.bbapap.2011.08.005
Paredez, A. R., Somerville, C. R., Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in arabidopsis. Proc. Natl. Acad. Sci. U S A. 104 (39), 15566–15571. doi: 10.1073/pnas.0706592104
Pettolino, F. A., Walsh, C., Fincher, G. B., Bacic, A. (2012). Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 7, 1590–1607. doi: 10.1038/nprot.2012.081
Polko, J. K., Kieber, J. J. (2019). The regulation of cellulose biosynthesis in plants. Plant Cell 31, 282–296. doi: 10.1105/tpc.18.00760
Roeder, A. H. K., Cunha, A., Ohno, C. K., Meyerowitz, E. M. (2012). Cell cycle regulates cell type in the arabidopsis sepal. Development 139, 4416–4427. doi: 10.1242/dev.082925
Ruhnow, F., Zwicker, D., Diez, S. (2011). Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100, 2820–2828. doi: 10.1016/j.bpj.2011.04.023
Sampathkumar, A., Gutierrez, R., Mcfarlane, H. E., Bringmann, M., Lindeboom, J., Emons, A.-M., et al. (2013). Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in arabidopsis interphase cells. Plant Physiol. 162, 675–688. doi: 10.1104/pp.113.215277
Shim, I., Law, R., Kileeg, Z., Stronghill, P., Northey, J. G. B., Strap, J. L., et al. (2018). Alleles causing resistance to isoxaben and flupoxam highlight the significance of transmembrane domains for CESA protein function. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01152
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. doi: 10.1126/science.aal2541
Tenhaken, R. (2014). Cell wall remodeling under abiotic stress. Front. Plant Sci. 5, 771. doi: 10.3389/fpls.2014.00771
Tonami, K., Kurihara, Y., Arima, S., Nishiyama, K., Uchijima, Y., Asano, T., et al. (2011). Calpain-6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J. Cell Sci. 124, 1214–1223. doi: 10.1242/jcs.072561
Tran, D., Galletti, R., Neumann, E. D., Dubois, A., Sharif-Naeini, R., Geitmann, A., et al. (2017). A mechanosensitive Ca2+ channel activity is dependent on the developmental regulator DEK1. Nat. Commun. 8, 1009. doi: 10.1038/s41467-017-00878-w
Updegraff, D. M. (1969). Semimicro determination of cellulose in biological materials. Analytical Biochem. 32, 420–424. doi: 10.1016/S0003-2697(69)80009-6
Vaahtera, L., Schulz, J., Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932. doi: 10.1038/s41477-019-0502-0
Vain, T., Crowell, E. F., Timpano, H., Biot, E., Desprez, T., Mansoori, N., et al. (2014). The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165, 1521–1532. doi: 10.1104/pp.114.241216
Verger, S., Long, Y., Boudaoud, A., Hamant, O. (2018). A tension-adhesion feedback loop in plant epidermis. eLife 7, e34460. doi: 10.7554/eLife.34460
Wang, C., Barry, J. K., Min, Z., Tordsen, G., Rao, A. G., Olsen, O. A. (2003). The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J. Biol. Chem. 278, 34467–34474. doi: 10.1074/jbc.M300745200
Wang, T., Park, Y. B., Cosgrove, D. J., Hong, M. (2015). Cellulose-pectin spatial contacts are inherent to never-dried arabidopsis primary cell walls: Evidence from solid-state nuclear magnetic resonance. Plant Physiol. 168, 871–884. doi: 10.1104/pp.15.00665
Wang, X., Wilson, L., Cosgrove, D. J. (2020). Pectin methylesterase selectively softens the onion epidermal wall yet reduces acid-induced creep. J. Exp. Bot. 71, 2629–2640. doi: 10.1093/jxb/eraa059
Wilson, S. M., Bacic, A. (2012). Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat. Protoc. 7, 1716–1727. doi: 10.1038/nprot.2012.096
Xiao, C., Anderson, C. T. (2016). Interconnections between cell wall polymers, wall mechanics, and cortical microtubules: Teasing out causes and consequences. Plant Signaling Behav. 11, e1215396–e1215396. doi: 10.1080/15592324.2016.1215396
Yakubov, G. E., Bonilla, M. R., Chen, H., Doblin, M. S., Bacic, A., Gidley, M. J., et al. (2016). Mapping nano-scale mechanical heterogeneity of primary plant cell walls. J. Exp. Bot. 67, 2799–2816. doi: 10.1093/jxb/erw117
Yang, Y., Yu, Y., Liang, Y., Anderson, C. T., Cao, J. (2018). A profusion of molecular scissors for pectins: Classification, expression, and functions of plant polygalacturonases. Front. Plant Sci. 9, 1208. doi: 10.3389/fpls.2018.01208
Zhang, T., Zheng, Y., Cosgrove, D. J. (2016). Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J. 85, 179–192. doi: 10.1111/tpj.13102
Zhang, X., Yang, Z., Wu, D., Yu, F. (2020). RALF–FERONIA signaling: Linking plant immune response with cell growth. Plant Commun. 1, 100084. doi: 10.1016/j.xplc.2020.100084
Zhang, T., Tang, H., Vavylonis, D., Cosgrove, D. J. (2019). Disentangling loosening from softening: insights into primary cell wall structure. Plant J. 100 (6), 1101–1117. doi: 10.1111/tpj.14519
Keywords: DEFECTIVE KERNEL1, cellulose, cell wall, mechanical properties, cellulose synthase complex
Citation: Novaković L, Yakubov GE, Ma Y, Bacic A, Blank KG, Sampathkumar A and Johnson KL (2023) DEFECTIVE KERNEL1 regulates cellulose synthesis and affects primary cell wall mechanics. Front. Plant Sci. 14:1150202. doi: 10.3389/fpls.2023.1150202
Received: 23 January 2023; Accepted: 28 February 2023;
Published: 14 March 2023.
Edited by:
Michael A. Held, Ohio University, United StatesReviewed by:
David Collings, The University of Newcastle, AustraliaTimo Engelsdorf, University of Marburg, Germany
Copyright © 2023 Novaković, Yakubov, Ma, Bacic, Blank, Sampathkumar and Johnson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kim L. Johnson, ay5qb2huc29uQGxhdHJvYmUuZWR1LmF1; Arun Sampathkumar, U2FtcGF0aGt1bWFyQG1waW1wLWdvbG0ubXBnLmRl
†Present addresses: Lazar Novaković, School of Biology, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom
Yingxuan Ma, Key Laboratory of Forest Genetics & Biotechnology of Ministry of Education, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, Jiangsu, China
 Lazar Novaković
Lazar Novaković Gleb E. Yakubov
Gleb E. Yakubov Yingxuan Ma
Yingxuan Ma Antony Bacic
Antony Bacic Kerstin G. Blank
Kerstin G. Blank Arun Sampathkumar
Arun Sampathkumar Kim L. Johnson
Kim L. Johnson