- 1Department of Biological Science and Institute of Women’s Health, Sookmyung Women’s University, Seoul, Republic of Korea
- 2Department of Systems Biology, Yonsei University, Seoul, Republic of Korea
The shade avoidance syndrome (SAS) is a collective adaptive response of plants under shade highlighted by characteristic phenotypes such as hypocotyl elongation, which is largely mediated by concerted actions of auxin and GA. We identified ATHB2, a homeodomain-leucine zipper (HD-Zip) domain transcription factor known to be rapidly induced under shade condition, as a positive regulator of GA biosynthesis necessary for the SAS by transactivating the expression of GA20ox2, a key gene in the GA biosynthesis pathway. Based on promoter deletion analysis, EMSA and ChIP assay, ATHB2 appears to regulate the GA20ox2 expression as a direct binding target. We also found that the GA20ox2 expression is under negative control by TCP13, the effect of which can be suppressed by presence of ATHB2. Considering a rapid induction kinetics of ATHB2, this relationship between ATHB2 and TCP13 may allow ATHB2 to play a shade-specific activator for GA20ox by derepressing a pre-existing activity of TCP13.
1 Introduction
Plants make use of light for sustaining their lives through photosynthetic processes and transduction of light signals. In crowded plant canopies, the lower layers of plants receive reduced amounts of photosynthetically active radiation (PAR, 400-700 nm), over a specific range of wavelengths of light in particular. The upper leaves preferentially absorb the red (R) and blue (B) wavelengths of sunlight and transmit and reflect the far-red (FR) wavelengths (Casal, 2013; Roig-Villanova and Martínez-García, 2016; Fernández-Milmanda and Ballaré, 2021; Pierik and Ballaré, 2021). This reduces the R:FR ratio of light in shaded areas, as a result, and induces a series of responses that enable the shaded plants to capture unfiltered light. Primarily, it promotes the elongation of petioles, internodes and hypocotyls and the hyponastic growth of leaves, while it inhibits the expansion of cotyledons and mature leaves. In addition, early flowering and reduced seed yield as well as reduced defense against plant pathogens are observed. These effects are collectively known as the shade avoidance syndrome (SAS). Although the SAS is a strategy for the shaded plants to adapt to natural environments, it is at the same time considered to be detrimental to crop productivity as it diverts resources for growth to support stem growth (Pierik and Testerink, 2014; Wang et al., 2020; Fernández-Milmanda and Ballaré, 2021).
The roles of plant hormones on the SAS have been extensively examined as they the key mediators for the most developmental processes of plants (Bou-Torrent et al., 2014). Auxin, a principal regulator of cell elongation well documented in the case of hypocotyl elongation through relevant transcription factors, has been identified as the major player modulating the hypocotyl elongation in response to shade as well (Tao et al., 2008; Oh et al., 2014; Chaiwanon et al., 2016; Procko et al., 2016; Yang and Li, 2017; Reed et al., 2018; Ma and Li, 2019). Tao et al. (2008) described TAA1, an enzyme which catalyzes the first step in auxin biosynthesis pathway, is rapidly deployed to synthesize auxin at the high levels to initiate the changes in plant associated with shade avoidance. Under the prolonged shade condition, auxin contents decrease while the auxin sensitivity increases, resulting in the enhanced auxin perception and signaling (Pucciariello et al., 2018).
A number of evidence indicate that gibberellins are also involved in the shade-induced changes in plants (Casal, 2013). FR-irradiated epicotyls exhibited higher amount of GA1 than R-irradiated ones (Martínez-García et al., 2000). Petiole elongation by a FR stimulus was reduced in AtGA20ox2-silenced plants (Hisamatsu et al., 2005), and petiole elongation rates coincided with RGA breakdown, showing the strong correlation of GA with shade responses (Djakovic-Petrovic et al., 2007). Shade treatment for 4 h resulted in increased levels of auxin but rather decreased levels of BR, and mildly increased levels of GA, while a prolonged shade for 24 h exhibited increased levels of GA but the same level of auxin and BR as non-shaded seedlings, showcasing a complex dynamics of hormone balance during the entire stage of shade response, which may be interpreted as that in the long-term shade treatment gibberellins might be the one contributing to the hypocotyl elongation rather than auxin (Bou-Torrent et al., 2014). GA has been known as an essential hormone for hypocotyl and stem elongation (Yamaguchi, 2008), with its major contribution recognized in shade responses as described above, but how shade increases active GA remains obscure.
Arabidopsis thaliana homeobox 2 (ATHB2), a member of the homeodomain- leucine zipper family II (HD-Zip II), is induced by a low R:FR (Carabelli et al., 1993; Carabelli et al., 2013). Its HD-Zip domain has been identified to bind to a 9-bp dyad symmetric consensus sequence (Sessa et al., 1993). ATHB2 was demonstrated to be a direct target of PIF4/PIF5, major transcriptional regulators of SAS (Kunihiro et al., 2011; Favero et al., 2020; Zhang et al., 2020). In addition, PIF7 was also reported to strongly bind to the upstream region of ATHB2 (Willige et al., 2021). Overexpression of ATHB2 resulted in longitudinal cell elongation in hypocotyls (Steindler et al., 1999), while mutant analyses of ATHB2 indicated that ATHB2 and its close paralogues (ATHB4 and HAT3) together control apical embryo (Turchi et al., 2013). In athb-2 mutant, YUCCAs, genes encoding a rate-limiting enzyme for auxin biosynthesis, were still found to be fully induced by low R:FR treatment, indicating that ATHB2 is not required for auxin biosynthesis during shade responses (Müller-Moulé et al., 2016). Rather, ATHB2 induction resulted in reduced expression of some auxin biosynthetic genes including YUCCAs (He et al., 2020). The three HD-Zip II protein genes, ATHB2, ATHB4 and HAT3, exhibit a common functional characteristic including being inducible by a low R:FR and conferring shade response-specific phenotypes in transgenic plants overexpressing them (Steindler et al., 1999; Sorin et al., 2009; Turchi et al., 2015). ATHB2, ATHB4, and HAT3 were also shown to interact with TCP13, TCP17 and TCP5, members of the CIN-like TCP family which are transcriptional regulators of plant development in a reciprocal manner (Martin-Trillo and Cubas, 2010; Hur et al., 2015; Lopez et al., 2015), implying their functional association as modular regulatory component in coordinating the shade avoidance response, since these TCPs have been also implicated in the process. However, how exactly the function of ATHB2 is played in shade avoidance, especially in regulating hypocotyl elongation in response to shade, remain unknown.
In the present study, we explored a possibility of ATHB2 as a regulator of GA biosynthesis for promoting hypocotyl elongation under shade condition. We found that rapid induction of GA20ox2 in response to shade could be attributed to ATHB2-mediated transcriptional activation as evidenced from GA20ox2 expression analyses made after protoplast transfection as well as in transgenic plants expressing ATHB2. The stimulatory function of ATHB2 on the expression of GA20ox2 was found to be tightly associated with a cis-regulatory sequence element present on the promoter of GA20ox2 and a direct binding of ATHB2 was also demonstrated by EMSA and ChIP analyses, suggesting that GA20ox2 is likely to be a direct target of the ATHB2-mediated transactivation. On the contrary, we found that TCP13 functioned as a negative regulator of GA20ox2 expression and the site of its action on the promoter is distantly located from that of the ATHB2, but its effect is still affected by the presence of ATHB2, suggesting ATHB2 and TCP13 are playing opposite roles in regulating GA20ox2 expression and subsequent GA synthesis under shade condition to provide a balance in coordinating the hypocotyl elongation response.
2 Materials and methods
2.1 Plant material and growth condition
Arabidopsis seeds (Col-0) were surface-sterilized in 70% ethanol and 0.01% Tween-20 for 15 min, rinsed twice in 95% ethanol, and once again in 100% ethanol. Arabidopsis plants were grown on half-strength Murashige and Skoog (MS) medium supplemented with 1% sucrose and 0.8% plant agar in a growth chamber at 22°C under long-day conditions (16-h light/8-h dark photoperiods) at 50 µmol m-2 s-1.
2.2 Plant treatments
Seven-day-old seedlings grown under continuous white light (W; 24 µmol m-2 s-1, R:FR ratio > 2.1) were treated with simulated shade (W+FR; R:FR ratio of 0.05, W enriched with supplementary FR provided by QB1310CS-670–735 LED hybrid lamps of Quantum Devices Inc.) in time course (0 h, 1 h, 2 h, 3 h, and 4 h), and then used for RNA extraction. Four-day-old seedlings grown under continuous white light (W) were treated with simulated shade (W+FR) for 3 days, or white light for 3 days as a control, and then used for the observation of shade avoidance phenotypes. For the dexamethasone (DEX) treatment, 7-day-old seedlings were transferred to the liquid MS medium containing 1 or 10 μM DEX, or 0.01% ethanol alone during the indicated times (Hur et al., 2015).
2.3 Vector construction and plant transformation
To generate transgenic plants containing GVG::ATHB2-GFP, ATHB2-GFP fusion fragment was amplified by PCR using a vector containing P35S::ATHB2-GFP (Hur et al., 2019) as template, and cloned into the pTA7002 vector (Hiratsu et al., 2003) using XhoI and SpeI. The resulting vector was introduced into Agrobacterium tumefaciens strain GV3101, and used for floral dip transformation of Arabidopsis (Col-0). The primers used for making constructs were shown in Supplementary Table S1.
2.4 Measurement of hypocotyl lengths
Five-day-old seedlings were exogenously treated with MOCK solution, 4 μmol 3- nitropropionic acid (NPA), and 1 μmol paclobutrazol (PAC) for 24 h before transferring to growth chambers with different light conditions for 5 days. Hypocotyl lengths were measured with Image J software (http://rsb.info.nih.gov/ij) using the pictures of seedlings (Ince and Galvão, 2021). All data were statistically evaluated using one-way ANOVA.
2.5 Transient gene expression assay
Protoplast isolation and transfection from Arabidopsis leaves were performed according to Yoo et al. (Yoo et al., 2007). Plasmid DNAs were extracted using cesium chloride (CsCl)/ethidium bromide (EB) centrifugation. Protoplasts (1 x 104) were transfected with a total of 10 μg of plasmid DNAs and incubated for 16 h in the dark. Protoplasts were harvested and subsequently used in real-time quantitative PCR or luciferase assay.
2.6 Real-time quantitative PCR
RNA was extracted from leaves or seedlings using RNeasy plant mini kit (Qiagen) and reverse transcription of RNA was performed using Moloney Murine Leukemia Virus (M- MLV) reverse transcriptase (Promega). Real-time quantitative PCR was performed with SYBR Green PCR Master Mix (Takara) using a LightCycler 96 (Roche, Mannheim, Germany). mRNA levels were normalized with EF1α and ACTIN2. The primers used for real-time quantitative PCR were shown in Supplementary Table S1.
2.7 Dual luciferase assay
The cauliflower mosaic virus 35S promoter (35S pro)::ATHB2-GFP or 35S pro::TCP13- GFP or 35S pro::TCP13-VP16 were used as effector plasmids. Different lengths of upstream region of GA20ox2 (1.5-kb, 1.0-kb, 0.5-kb, 0.2-kb, respectively) were fused to luciferase gene to make GA20ox2 pro::LUC as reporter genes, and Renilla luciferase (35S pro::RLUC) was used as internal control. Mutagenized versions of 1.5-kb upstream region of GA20ox2 (one at ABS1 and the other at ABS2) were fused to LUC; ABS1(m) and ABS2(m). Arabidopsis protoplasts were isolated and transfected by the polyethylene glycol (PEG)-method as previously described (Yoo et al., 2007). Luciferase activity was measured with a dual-luciferase reporter assay system according to the manufacturer’s instruction (Promega) in a SpectraMax i3X microplate reader (Molecular Devices). Firefly luciferase (LUC) activity was normalized with Renilla luciferase (RLUC) activity.
2.8 EMSA
Electrophoretic mobility shift assay (EMSA) was performed using a LightShift® Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction. Oligonucleotides (31-mer) flanking the ABS1 were labeled by 3’ end biotin, and unlabeled oligonucleotides were used as competitors as shown in Table S1. His- tagged ATHB2 protein was isolated from E. coli transformed with ATHB2-containing pET-28a using Ni-NTA agarose (Qiagen) and further purified using Amicon® Ultra-15 centrifugal filters (Millipore).
2.9 Yeast two-hybrid assay
Yeast strain and methods used in the yeast two-hybrid assay were essentially the same as in Son et al., 2016. TCP13 partial cDNAs were subcloned into pGBKT7 as baits, while GAF ORF cDNA was subcloned into pGADT7 as a prey, respectively (Clontech Inc.). Primers used were shown in Supplementary Table S1.
2.10 Chromatin immunoprecipitation (ChIP) assay
For quantitative chromatin immunoprecipitation (ChIP)-PCR assays, 10-day-old seedlings with either GVG::ATHB2-GFP or GVG::3HA-GFP were harvested after treating with10 μM DEX for 48 h. ChIP was performed as described by Gendrel et al. (2002). Briefly, leaves were cross-linked with 1% (v/v) formaldehyde solution by vacuum infiltration for 10 min. Chromatin complexes were sonicated with a Ultrasonic Processor (VC-505, SONICS) in a nuclei lysis buffer (50 mM Tris-HCl, 10 mM EDTA, 1% SDS, 1 mM PMSF, 1 X Protease inhibitor cocktail, pH 8.0), and the protein-DNA complexes were incubated with an anti-GFP antibody (ab290, Abcam) overnight at 4°C. Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) was added and incubated for 2 h. The fractions were washed and eluted with elution buffer (1% SDS and 0.1 M NaHCO3) at 65°C for 30 min. The fractions were reverse cross-linked with NaCl (final concentration 0.2 M) at 65°C overnight. The precipitated DNAs were purified using QIAquick PCR Purification Kits (Qiagen), and analyzed using the real-time quantitative PCR. The primers were designed to amplify fragments within GA20ox2 promoter (Supplementary Table S1).
3 Results
3.1 GA biosynthetic genes are rapidly induced upon shade treatment
To obtain an insight into a relative contribution of GA in mediating shade response, hypocotyl elongation of Arabidopsis seedlings grown under shade were monitored after treating with inhibitors of auxin and GA metabolism. Reflecting the well-established role of auxin during the onset and the early stage of the shade avoidance response (Casal, 2013; Wang et al., 2020), seedlings treated with an auxin transport inhibitor, naphthylphthalamic acid (NPA) (Abas et al., 2020), failed to exhibit the characteristic hypocotyl elongation under shade as expected. Similarly, the seedlings treated with an inhibitor of GA biosynthesis, paclobutrazol (PAC/PBZ) (Rademacher, 2000), also showed a noticeably reduced hypocotyl length under shade, indicating that GA also plays a role in mediating the shade response as much as auxin does (Figures 1A, B).
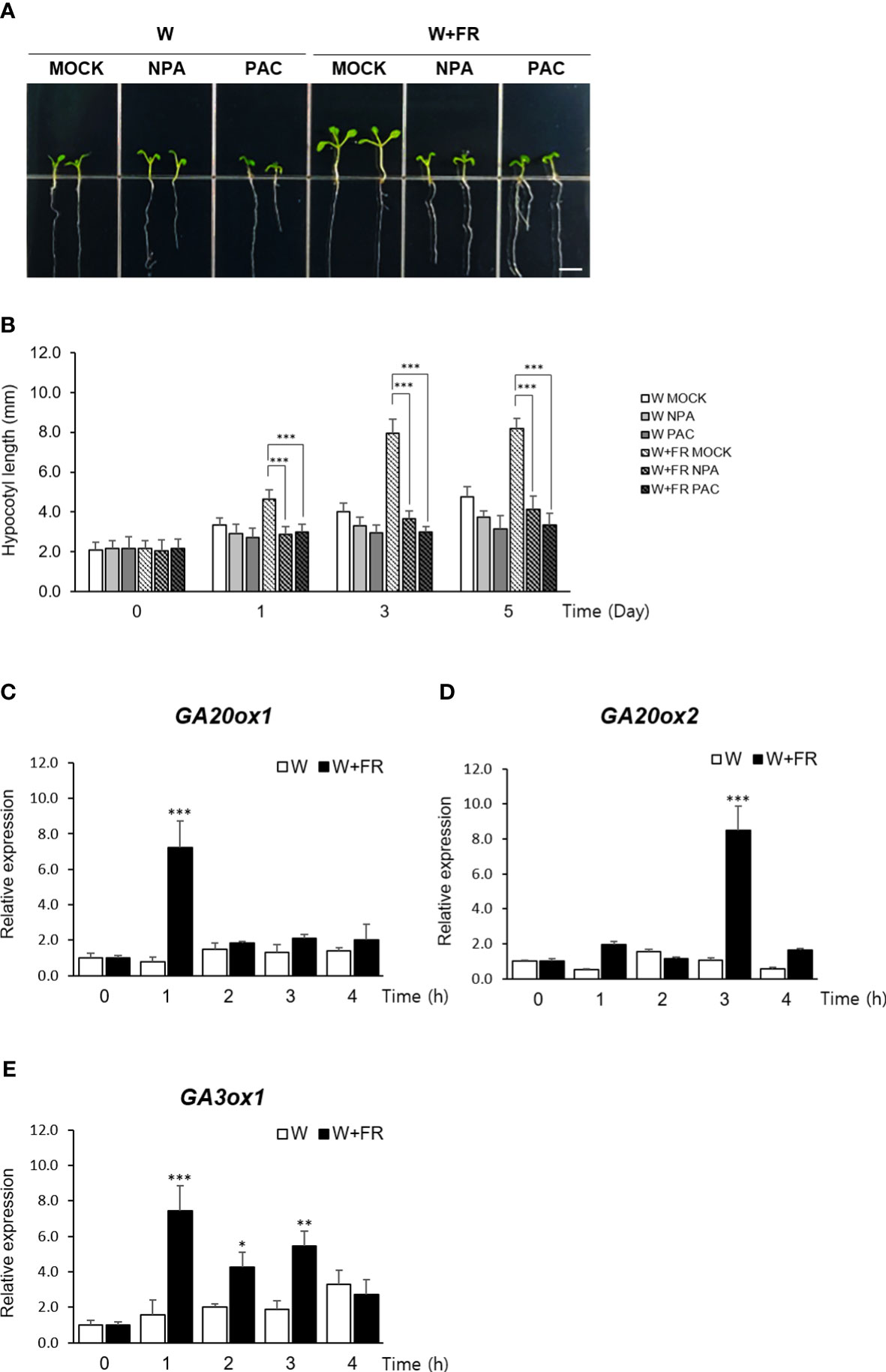
Figure 1 Expression of GA biosynthetic genes under the shade condition. (A) Effects of auxin transport inhibitor NPA or GA biosynthetic inhibitor PAC on hypocotyl lengths of Arabidopsis thaliana under shade condition was observed. Light condition was as follows: W, continuous white light; W+FR, continuous white light supplemented with far-red light. Five-day-old seedlings grown under either W or W+FR condition were treated with MOCK, NPA, or PAC for 5 days on 1/2 MS media. Scare bar, 5 mm. (B) Hypocotyl lengths were measured at the indicated times: 0 day, 1 day, 3 days, and 5 days after NPA or PAC treatment. Data shown are mean ± SD (n > 15). (C–E) Expression of GA20ox1 (C), GA20ox2 (D), and GA3ox1 (E) under W and W+FR in time course using 7-day-old seedlings were analyzed by real-time quantitative PCR. Results are representative of three independent experiments. Data shown are mean ± SD. All data were statistically evaluated using one-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.005.
Consistent with this observation, expressions of GA20ox1, GA20ox2, and GA3ox1, three key regulators of GA biosynthesis in Arabidopsis (Hedden et al., 2001; Rieu et al., 2008), were found to be induced over a 4-hr period of shade treatment (Figures 1C–E). GA20 oxidase (GA20ox) mediates several conversion steps in the late stage of GA biosynthetic pathway occurring in the cytoplasm and GA3 oxidase (GA3ox) catalyzes the final step of producing biologically active form of GA, GA1 and GA4 (Hedden, 2020). Of the two major isoforms of GA20 oxidase in Arabidopsis, our result revealed that the induction of GA20ox1 expression preceded that of GA20ox2, reaching at the peak within an hour of shade treatment, which was followed by the induction of GA20ox2 expression within next couple of hours.
3.2 ATHB2 acts as a positive regulator of GA20ox2 expression
The rapid transcriptional activation of the GA biosynthetic genes under shade condition observed in our result was reminiscent of the quick induction kinetics of ATHB2 during the onset of the shade avoidance response (Carabelli et al., 1996; Steindler et al., 1999; Carabelli et al., 2013). We therefore tested if any of these genes’ expression is regulated by ATHB2 as part of the shade avoidance response. Protoplasts derived from Arabidopsis leaf mesophyll cells were transfected with a plasmid construct expressing the ATHB2 and the transcript levels of GA20ox1, GA20ox2, and GA3ox1 were measured by real-time quantitative PCR analysis. To a little surprise, the ectopic expression of ATHB2 resulted in an increased expression of the endogenous GA20ox2 only, while those of GA20ox1 and GA3ox1 remain largely unchanged (Figure 2A), indicating that the regulatory effect of ATHB2 is limited to the control of GA20ox2 expression despite all these three genes were induced under shade condition.
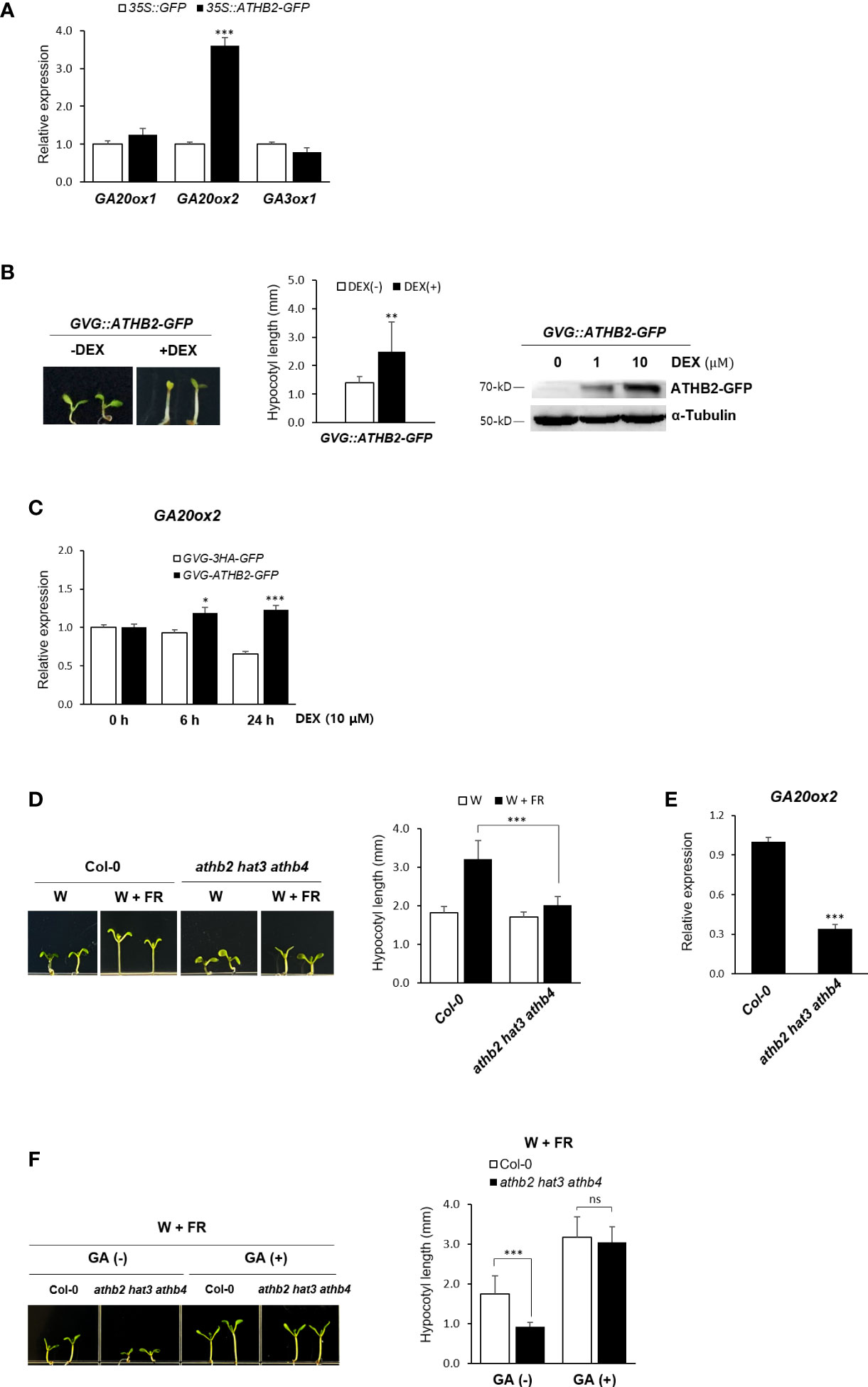
Figure 2 Regulation of GA20ox2 expression in ATHB2-expressing protoplasts and transgenic plants. (A) Expression of GA20ox1, GA20ox2, and GA2ox3 was analyzed in Arabidopsis protoplasts transfected with plasmids containing 35S::ATHB2-GFP or 35S::GFP by real-time quantitative PCR. (B) Expression of ATHB2 in transgenic plants with GVG::ATHB2-GFP was examined after treatment with dexamethasone (DEX). Seven-day-old seedlings were grown on 1/2 MS media treated with 0.01% ethanol (-DEX) or 10 μM DEX (+DEX) (left) and their hypocotyl lengths were measured (center). Data shown are mean ± SD (n > 15). Western blot analysis using 7-day-old plants treated with DEX was performed with antisera against GFP of α tubulin (right). (C) GA20ox2 expression was examined in transgenic plants with GVG::ATHB2-GFP treated with DEX for 0 h, 6 h, and 24 h by real-time quantitative PCR. Results are representative of three independent experiments. Data shown are mean ± SD (n > 3). (D) A triple-knockout mutant of ATHB2 paralogues (athb2 hat3 athb4) was grown under W or W+FR conditions and their hypocotyl lengths were measured. Data shown are mean ± SD (n > 20). (E) GA20ox2 expression was examined in 7-day-old seedlings of athb2 hat3 athb4. Results are representative of three independent experiments. (F) A triple-knockout mutant of ATHB2 paralogues (athb2 hat3 athb4) was grown under W+FR condition in the absence/presence of GA and their hypocotyl lengths were measured. Data shown are mean ± SD (n > 20). Data shown are mean ± SD (n > 3). All data were statistically evaluated using one-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.005. ns, no significant difference.
The observed transcriptional activation of GA20ox2 by ATHB2 was also confirmed in transgenic Arabidopsis plants expressing a dexamethasone (DEX)-inducible ATHB2-GFP fusion construct as well as in a triple-knockout mutant of ATHB2 paralogues (athb2 hat3 athb4). Upon DEX treatment, the transgenic plants displayed a characteristic hypocotyl elongation phenotype of the shade response even under light condition, demonstrating a central role of ATHB2 in maintaining the shade avoidance response (Figure 2B). Accordingly, expression of GA20ox2 was found to be increased along with the ATHB2 induction in response to the DEX treatment in the transgenic plants (Figures 2C, S1). In addition, the hypocotyl elongation phenotype in DEX-treated transgenic plants expressing ATHB2-GFP was not visible with PAC treatment (Supplementary Figure S2). On the other hand, noticeably reduced hypocotyl elongation under shade and the GA20ox2 expression was observed with the triple mutant of ATHB2 paralogues (athb2 hat3 athb4) (Figures 2D, E). However, the hypocotyl elongation of the triple mutant plants under shade condition was equivalent to that of the wild-type seedlings when they were provided with exogenous GA treatment (Figure 2F), revealing that the observed mutant phenotype was mainly due to a defect in GA biosynthesis. While there might be other unidentified mechanism by which GA biosynthesis is regulated and under the control of the ATHB2, at least our results may indicate that one of the roles of ATHB2 in coordinating the shade response is to stimulate GA biosynthesis through the transcriptional activation of GA20ox2, thereby contributing to the hypocotyl elongation.
3.3 ATHB2 transactivates GA20ox2 by directly binding to a unique cis element on the promoter
It has been reported that the transcription of GA20ox2 in Arabidopsis is mainly regulated through a GA-feedback mechanism mediated by DELLA-GAF1 complex, in which GAF1 recruits DELLA that functions as a GA concentration-dependent transcriptional co-activator in this occasion (Fukazawa et al., 2014; Fukazawa et al., 2017). As multiple GAF1-binding sites have been identified on the GA20ox2 promoter (Fukazawa et al., 2017), it is possible that the positive effect of ATHB2 on the GA20ox expression could be mediated as a component of the DELLA-GAF1 complex. Thus, we tried to locate approximate site of ATHB2 action on GA20ox2 promoter by comparing relative transcriptional activity of several deletion versions of the GA20ox2 promoter fragments driving the luciferase reporter construct after transient expression in Arabidopsis protoplasts. The result showed that a region of the GA20ox2 promoter occupying between -1,000 ~ -1,500 bp upstream was specifically responsible for the ATHB2-dependent transcriptional activation of GA20ox2 (Figures 3A, B). As this region is outside of the area where all the GAF1-binding elements (cisA – cisE) have been located (Fukazawa et al., 2017), the stimulatory effect of ATHB2 may operates independently of that of well-established DELLA-GAF1 transactivating complex. Thus, in order to determine if this region alone was sufficient for conferring the stimulatory effect of ATHB2 on the GA20ox2 promoter, this fragment was directly fused to a LUC reporter with the minimal promoter region of the GA20ox2 (-0.2 kb fragment) and tested for its transcriptional activity along with other partial fragments of GA20ox2 promoter-reporter constructs (Figure 3C). Based upon the result, it appears that although the region is necessary for ATHB2 to implement the stimulatory effect on the GA20ox2 expression, but not sufficient for exerting the effect alone, suggesting that it may require additional positive regulator(s) acting on other regions of the promoter for an optimal activity.
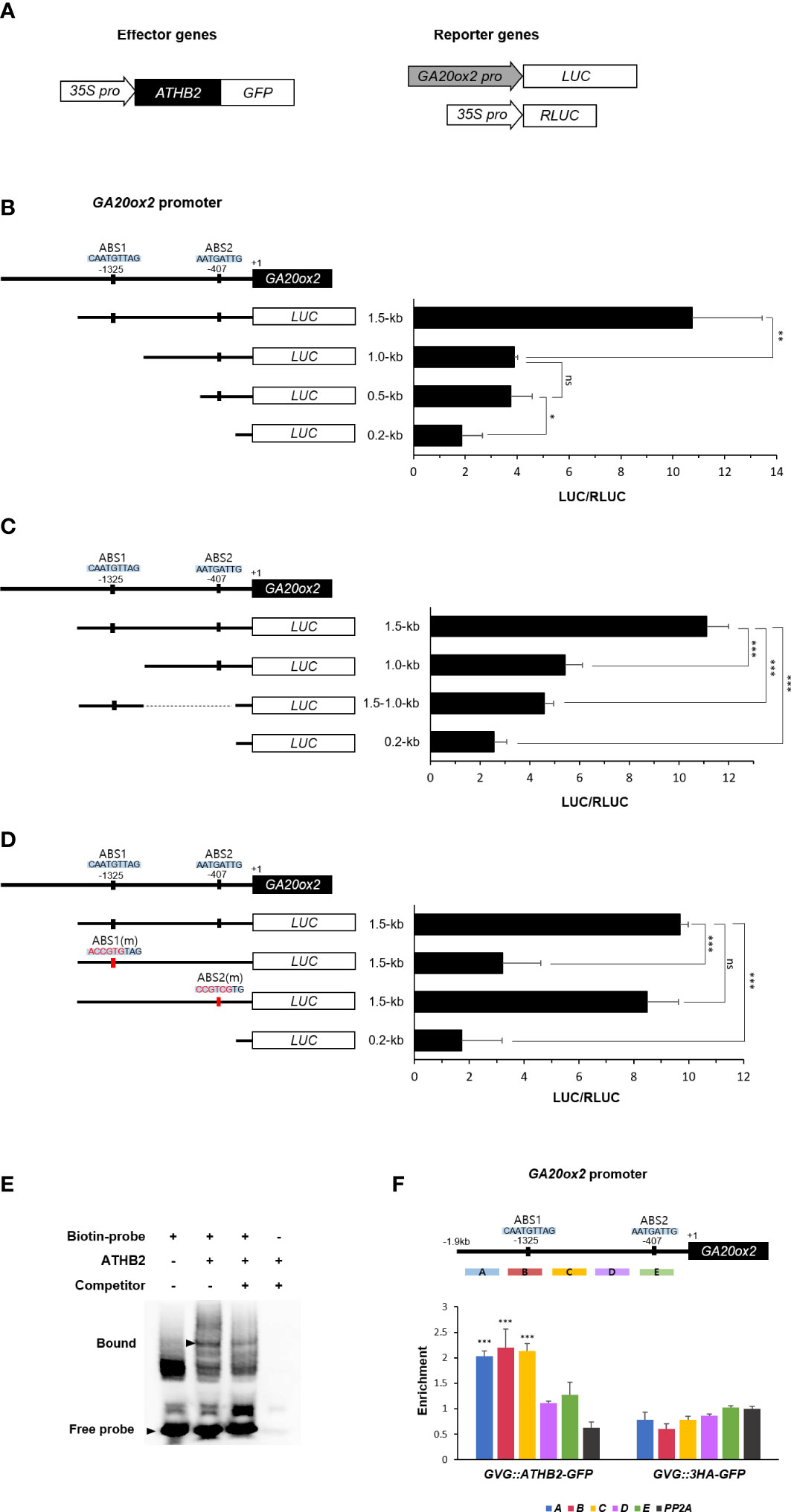
Figure 3 Activation of GA20ox2 by ATHB2 binding to an upstream region of GA20ox2. (A) Luciferase assays using different lengths of GA20ox2 upstream region fused with firefly luciferase (GA20ox2 pro::LUC) as reporters and 35S pro::ATHB2-GFP as an effector construct were performed. (B) Different lengths of GA20ox2 upstream region (1.5-kb, 1.0-kb, 0.5-kb, and 0.2-kb upstream regions) fused with firefly luciferase were used as reporter constructs, and 35S pro::RLUC was used as internal control. Firefly luciferase (LUC) activity was normalized with Renilla luciferase (RLUC) activity. The diagram shows two putative ATHB2 binding sites (ABS1 and ABS2) in GA20ox2 upstream region. (C) A region of the GA20ox2 promoter occupying between -1.0-kb ~ -1.5-kb upstream was fused to a LUC reporter with the minimal promoter region of GA20ox2 (-0.2 kb fragment) tested for the activation activity. (D) ABS1 and ABS2 on GA20ox2 promoter were mutated, respectively; ABS1(m) and ABS2(m). The resulting constructs were used for luciferase assay. (E) An electrophoretic mobility shift assay (EMSA) was performed using a 31-mer of biotin-labeled oligonucleotides flanking the ABS1 sequence. Purified 6X His-tagged ATHB2 protein was incubated with labelled oligonucleotides (second lane). The same EMSA experiment with 100X molar excess of unlabeled oligonucleotides as competitor was examined as well for indicating the specificity of ATHB2 binding to ABS1. Arrowheads indicate free and bound oligonucleotides separated in a native gel. (F) ChIP assay showing enrichments of GA20ox2 promoter fragments by ATHB2. Schematic diagram indicates ATHB binding sites (ABS1 and ABS2) and ChIP amplicons (A-E) in the upstream region of GA20ox2. The enrichment of each DNA fragment was normalized by the level of input DNA and the ChIP enrichment value of the control PP2A promoter. Asterisks indicate significant differences from the enrichment of PP2A genomic fragments Results are representative of three independent experiments. Data shown are mean ± SD (n=3). All data were statistically evaluated using one-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.005; ns, no significant difference.
A putative binding site for plant HD-zip domain transcription factors (Sessa et al., 1993) was identified within the region (CAATGTTAG), which we tentatively named ABS1, and altering its sequence into ACCGTGTAG abolished the ATHB2-dependent transcriptional activation of the reporter construct conferred by the fragment. On the other hand, a mutation of a similar sequence found at more proximal end of the promoter (-407 from the transcription start site), which also contains a conserved core sequence of the HD-zip domain binding motif (named ABS2), did not affect the reporter gene expression, suggesting that the observed effect is likely to be mediated through the ABS1 sequence element (Figure 3D).
To test if ATHB2 can directly bind to the ABS1 sequence motif, we carried out an electrophoretic mobility shift assay (EMSA) using biotin-labeled oligonucleotides of the ABS1 sequence and recombinant ATHB2 protein expressed in and purified from E. coli. As shown in Figure 3E, an ATHB2-specific electrophoretic mobility shift of ABS1-containing oligonucleotides was detected, demonstrating that ATHB2 can directly bind to the DNA having the ABS1 sequence motif (GAATGTTAG) at least in vitro. This was further verified in vivo by chromatin immunoprecipitation (ChIP) – PCR analysis with the DNA samples isolated from transgenic Arabidopsis expressing ATHB2-GFP, the result of which showed the ABS1-containing area of the GA20ox2 promoter as the most strongly amplified region (Figure 3F).
3.4 GA20ox2 expression is negatively regulated by TCP13
Another key regulator of shade-specific gene expression we have followed for years is TCP13, a member of plant-specific transcription factor with a unique helix-loop-helix motif (Aggarwal et al., 2010). TCP13 and its close paralogues of the CIN-like TCPs, TCP5 and TCP17, have been implicated in a number of developmental and physiological processes of plants including shade avoidance response, thermomorphogenesis, GA biosynthesis and cell expansion, many of which are accomplished through functional association with other regulators such as PIFs (Zhou et al., 2018; Han et al., 2019; Zhou et al., 2019; Rath et al., 2022). Therefore, we considered a possibility of ATHB2 regulating the GA20ox2 transcription as a functional complex with TCP13 and examined if there was any additive effect of TCP13 on the expression of GA20ox2 together with ATHB2. On the contrary to what we have anticipated, however, GA20ox2 transcript level was found to be significantly lower in our transgenic Arabidopsis constitutively expressing TCP13-GFP construct, which was also corroborated in triple mutant of tcp5 tcp13 tcp17 that showed significantly elevated level of the GA20ox2 expression (Figure 4A). Of particular interest regarding the relative roles of these TCP13 paralogues in the regulation of GA20ox2 expression uncovered from this result was that only TCP13, but not TCP5 or TCP17, appears to be acting as a repressor of GA20ox2 expression, as evidenced in the result of the transgenic plants expressing TCP5 (TCP5-myc) or TCP17 (TCP17-myc) as well as in the result of the TCP13 knockout mutant (tcp13) in comparison with the triple mutant (Figure 4A). We attempted to confirm this negative role of TCP13 in a transient assay of protoplast transfection. Although ectopic expressions of both TCP13 and ATHB2 were examined (Figure S3), they may not show their respective effects on GA20ox2 expression under the real physiological condition, considering their expression pattern. Thus, we tried to observe the effect of TCP13 on the GA20ox2 expression in an ATHB2-expressing background since the two proteins have shown a specific interaction in yeast two-hybrid analysis (Hur et al., 2019), thus it could potentially result in a confounding effect. When protoplasts made from the TCP13-GFP transgenic plants were transfected with a plasmid expressing ATHB2 and assayed for the expression of endogenous GA20ox2, derepression of GA20ox2 transcription by the ectopic expression of the ATHB2 gene was evident, indicating an antagonistic regulatory effect of the two on the regulation of GA20ox2 expression (Figure 4B).
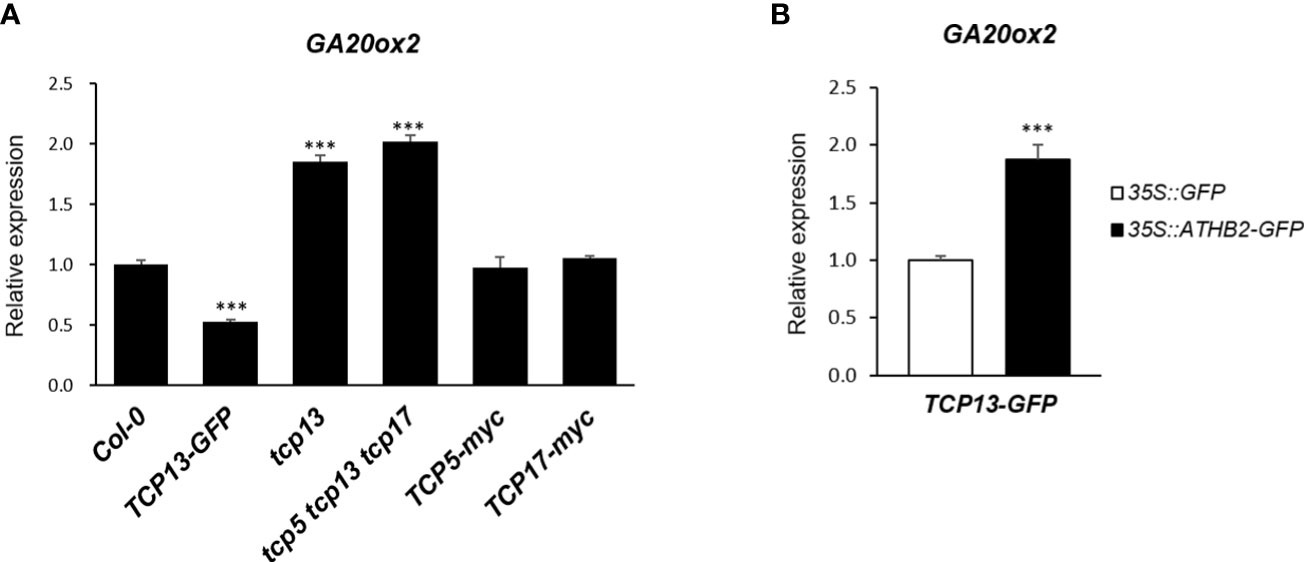
Figure 4 Regulation of GA20ox2 expression in TCP13-expressing transgenic plants, protoplasts, and mutants. (A) Expression of GA20ox2 in Arabidopsis seedlings with 35S promoter-driven TCP13-GFP fusion construct (TCP13-GFP), tcp13 mutant, triple KO mutant for TCP13 and its paralogs, TCP5 and TCP17 (tcp5 tcp13 tcp17), seedlings with 35S promoter-driven TCP5-myc fusion construct (TCP5-myc), and those with 35S promoter-driven TCP17-myc fusion construct (TCP17-myc) examined by real-time quantitative PCR. (B) Expression of GA20ox2 in Arabidopsis protoplasts of TCP13-GFP transgenic plants that were subsequently transfected with 35S promoter-driven ATHB2-GFP fusion construct (35S pro::ATHB2-GFP) examined by real-time quantitative PCR. More than 3 independent transgenic plants expressing TCP5, TCP13 or TCP17, respectively. Results are representative of more than three independent experiments. Data shown are mean ± SD (n > 3). All data were statistically evaluated using one-way ANOVA: ***P < 0.005.
3.5 The site of repression by TCP13 on GA20ox2 promoter does not overlap with the ATHB2-specific element
It was intriguing to contemplate the mechanism of transcriptional repression of GA20ox2 brought about by TCP13. For instance, if it requires formation of heterodimer with ATHB2 it is conceivable that the inhibitory effect of TCP13 is mainly achieved through titrating the ATHB2 out of the GA20ox2 promoter. Therefore, we attempted to determine whether TCP13 indeed binds to the promoter of GA20ox2, and if so, the binding region coincides with the area where the ABS1 sequence is present. A chimeric construct having the activation domain of VP16 (Herpes Simplex Virion Protein 16) fused to TCP13 ORF at its C-terminus was generated and Arabidopsis protoplasts were transfected with this construct together with the plasmid expressing the luciferase under GA20ox2 promoter (Figure 5A). Truncated versions of the GA20ox2 promoter fragments were used to examine the presence of any region that displays a TCP13-dependent (TCP13-VP16) transactivation of the reporter. The result revealed that a promoter fragment spanning to - 500 bp upstream of the transcription start site was the only one conferred a significant activation of the reporter gene expression (0.5-kb fragment in the Figure 5B). The originally observed repressive function of the TCP13 on the GA20ox2 expression was also evident in this assay, as shown with the TCP13 construct fused with GFP instead of the VP16 cotransfected with the reporter driven by the full-length 1.5-kb promoter (Figure 5B). With this result, therefore, it could be concluded that TCP13 acts as a repressor of GA20ox2 binding to a specific promoter region which does not overlap with the area containing the putative ATHB2 binding site, ABS1.
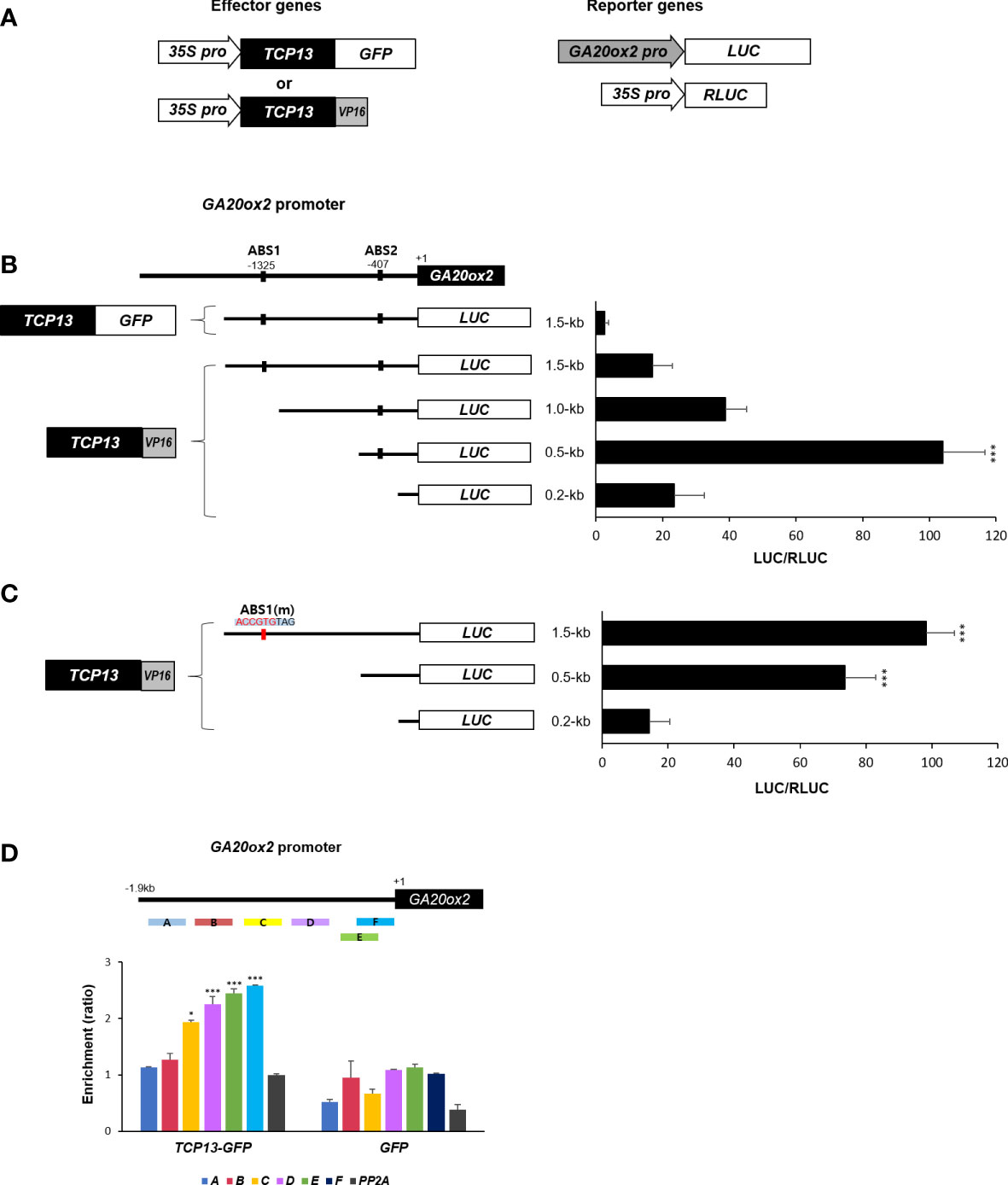
Figure 5 Regulation of GA20ox2 by TCP13. (A) Luciferase assays using different lengths of GA20ox2 upstream region fused with firefly luciferase (GA20ox2 pro::LUC) as reporters and 35S pro::TCP13-GFP or 35S pro::TCP13-VP16 as effector constructs were performed. (B) Different lengths of GA20ox2 upstream region (1.5-kb, 1.0-kb, 0.5-kb, and 0.2-kb upstream regions) fused with firefly luciferase were used as reporter constructs, and 35S pro::RLUC was used as internal control. Firefly luciferase (LUC) activity was normalized with Renilla luciferase (RLUC) activity. (C) ABS1 on GA20ox2 promoter was mutated; ABS1(m). The resulting construct was used for luciferase assay. (D) ChIP assay showing enrichments of GA20ox2 promoter fragments by TCP13. Schematic diagram indicates ChIP amplicons (A–F) in the upstream region of GA20ox2. The enrichment of each DNA fragment was normalized by the level of input DNA and the ChIP enrichment value of the control PP2A promoter. Asterisks indicate significant differences from the enrichment of PP2A genomic fragments. Results are representative of three independent experiments. Data shown are mean ± SD (n=3). All data were statistically evaluated using one-way ANOVA: *P < 0.05; ***P < 0.005.
One puzzling finding out of this experiment, however, was that the full-length promoter did not show as much transcriptional activation as with the 0.5-kb promoter fragment which was still included in the full-length version of the promoter. We speculated that there might be a sequence-specific protein factor interfering the operation of TCP13 elsewhere on the promoter region of the GA20ox2. A possible candidate for such interference could be the formation of heterodimer between the endogenous ATHB2 and the ectopically expressed TCP13-VP16 proteins being recruited onto the ABS1 sequence on the GA20ox2 promoter. To test this possibility, we repeated the same transfection assay with the GA20ox2 promoter having the ABS1 sequence (CAATGTTAG) changed into ABS1(m) (ACCGTGTAG), which abolished the stimulatory effect of ATHB2 on GA20ox2 expression in the result summarized in the Figure 3C. We thought that by abolishing possible influence of the endogenous ATHB2 by eliminating its binding site on the promoter of the reporter gene, the stimulatory effect of TCP13-VP16 on the full-length promoter would be restored. In support of our hypothesis, a dramatic increase in the activation of the reporter gene expression was observed with the mutant full-length promoter of GA20ox2, even higher than with that of the 0.5-kb fragment of the wild-type promoter (Figure 5C). To clearly test if TCP13 can directly bind to the 0.5-kb fragment of GA20ox2 promoter, we performed chromatin immunoprecipitation (ChIP) – PCR analysis with the DNA samples isolated from transgenic Arabidopsis expressing TCP13-GFP. Result showed possibly the TCP13 binding to the 0.5-kb fragment of GA20ox2 promoter (Figure 5D) although the binding region needs further specification. Therefore, it appears that ATHB2 and TCP13 are acting antagonistically in regulating the GA20ox2 expression, playing a stimulatory and a repressive role, respectively.
3.6 TCP13 can specifically interact with GAF1
Since the GAF1-DELLA trans-activator complex has been reported as the principal regulator for the expression of GA20ox2 in Arabidopsis (Fukazawa et al., 2017), the inhibitory role of TCP13 on GA20ox2 disclosed in our present study made us wonder if this activity might involve a physical interaction between TCP13 and GAF1. In line with the presence of the intrinsically disordered region (IDR) at the C-terminus (Dunker et al., 2001), a characteristic feature of all TCP proteins that is thought to be responsible for establishing interactions with multiple different proteins by providing flexible structural platform (Valsecchi et al., 2013), a number of proteins have been identified as capable of forming a specific interaction with TCP13 (http://interactome.dfci.harvard.edu, http://zzdlab.com/plappisite). As GAF1 is not included in the list of these interacting proteins, however, we inquired a possible interaction between GAF1 and TCP13 by conducting yeast two-hybrid analyses, in which several partial fragments as well as the full-length TCP13 protein were probed with the full-length GAF1 protein for the interaction. The result indicated that TCP13 can interact with GAF1 and the region between the amino acid residues 179 and 240, which includes the C-terminal portion of the TCP domain and a few more amino acids beyond, were found to be responsible for the interaction (Figure 6A). This interaction between TCP13 and GAF1 was also confirmed in planta by bimolecular fluorescence complementation (BiFC) analysis, in which full-length fragments of GAF1 and TCP13 were fused to N-terminal and C-terminal halves of YFP, respectively, and demonstrated their specific interaction (Figure 6B). Therefore, it seems probable that the repression of GA20ox2 by TCP13 may be achieved through inhibition of GAF1-dependent transactivation of the gene, by interfering the formation of an active DELLA-GAF1 complex. However, one caveat of this idea is that the site of TCP13 action on the GA20ox2 promoter, as we have determined by the results of Figure 5, does not overlap with any of the GAF1-binding sites identified on the GA20ox2 promoter, cis A through E (Fukazawa et al., 2017).
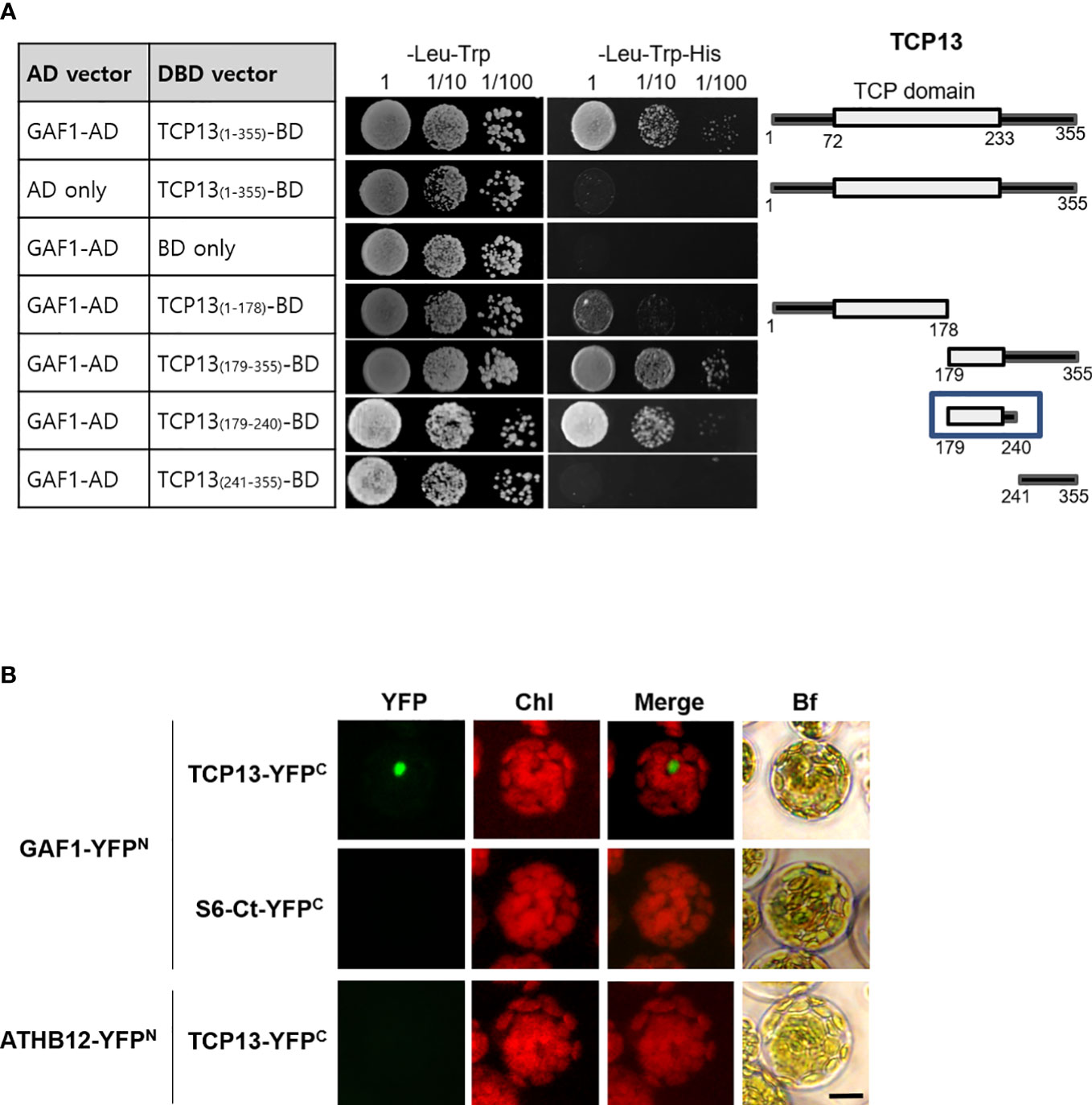
Figure 6 Interaction of TCP13 with GAF1. (A) Different regions of TCP13 were tested for the interaction with GAF1 in yeast two-hybrid assay. The DNA-binding domain (DBD) of GAL4 was fused to different regions of TCP13 while the activation domain (AD) of GAL4 was fused to GAF1. Co-transformed yeasts were selected on media lacking Leu and Trp (-Leu-Trp), and selected on media lacking Leu and Trp and His (-Leu-Trp-His) with or without 1 mM 3-amino-1,2,4 triazole (3-AT). The minimal region of TCP13 required for the interaction with GAF1 was indicated as a box. (B) Bimolecular fluorescence complementation (BiFC) analysis of interaction between TCP13 and GAF1. Arabidopsis protoplasts were co-transfected with several combinations of the constructs of P35S::GAF1-YFPN and P35S::TCP13-YFPC. S6-Ct-YFPC: truncated RPS6 fused to YFP. A transcription factor ATHB12 fused to the N-terminal fragment of YFP (ATHB12-YFPN) was used as negative control. Bf, bright field. BiFC experiments were replicated three times with similar results. Scale bar = 10 μm.
4 Discussion
Our present study uncovered the role of ATHB2 as a positive regulator of GA biosynthesis by transactivating the expression of GA20ox2. Thus, as a downstream target of the PIF4/PIF5 activity, ATHB2 appears to coordinate the synthesis of GA which is an essential component of the early-stage shade avoidance response. While it is not clear whether the induction of ATHB2 is a result of direct activation by PIF4/PIF5 during the procedure or the one requiring the auxin biosynthesis as a prerequisite as was recently proposed (Küpers et al., 2023), ATHB2 seems to play a key role in bridging the auxin and GA signaling together to facilitate the shade avoidance response.
4.1 ATHB2 activates GA20ox2 expression via a specific sequence element
Our result identified a cis-regulatory sequence element specifically associated with the stimulatory effect of ATHB2 on GA20ox2 promoter. The sequence motif, which we tentatively named ABS1, is highly homologous to the consensus sequence motif known for the plant HD- domain transcription factors (Sessa et al., 1993). Presence of the ABS1 appears to be essential for the ATHB2-dependent transactivation of the GA20ox2 gene, as revealed from our promoter deletion analysis together with the effect of mutating its sequence on the reporter gene expression. Moreover, direct binding of ATHB2 to ABS1 sequence was shown by EMSA and ChIP assay (Figures 3D, E), thus ATHB2 is likely to act directly on the GA20ox2 promoter through ABS1 sequence in regulating its expression in response to the onset of shade condition.
Even though both the GA20ox1 and GA20ox2 are induced under shade, our result showed that only the GA20ox2 expression was regulated by ATHB2, which is in consistence with that no ABS1 sequence motif is found on the promoter of GA20ox1. By contrast, both genes have been reported to be under the positive control by the DELLA-GAF1 complex, which incorporates the GA-feedback regulation of these genes (Fukazawa et al., 2017; Fukazawa et al., 2021). Therefore, while it has not been determined if the GAF1/DELLA regulatory machinery is also in operation under the shade condition, the shade-specific expression of GA20ox2 seems to have obtained a unique element of control from ATHB2 and thereby delineates from the control of GA20ox1 expression under the same condition.
4.2 TCP13 plays as a repressor of GA20ox2 and acts independently of ATHB2
As ATHB2 has been identified to be interacting with TCP13 (Hur et al., 2019), a member of a plant-specific transcription factor family having noncanonical bHLH domain (Martin-Trillo and Cubas, 2010), we contemplated possible formation of a functional heterodimer between ATHB2 and TCP13 in transactivating the GA20ox2 expression. However, our results showed that TCP13 functioned as a repressor of GA20ox2, casting a scrutiny regarding the biological relevance and possible mechanism of the observed repression brought up by TCP13. The GA20ox2-specific repressor activity of TCP13 was initially detected in the protoplast transfection experiment and was also confirmed in transgenic plants over-expressing TCP13-GFP construct as well as in the single (tcp13) and triple knock-out mutant (tcp5 tcp13 tcp17), validating its biological relevance.
The question remained as to whether the repressor function of TCP13 involved in the change in the ATHB2 activity, reducing its stimulatory effect by titrating it out of the GA20ox2 promoter, for example. In such a scenario, the site of TCP13 action on the promoter would be mapped to be overlapping with that of ATHB2. Thus, to map the site of TCP13 activity on the GA20ox2 promoter we designed a chimeric construct having the VP16 transcription activation domain fused to the C-terminus of TCP13 and used it as an effector in protoplast transfection assays with deletion fragments of GA20ox2 promoter driving the reporter luciferase expression (Figure 5A). The result indicated that TCP13 exercises its effect on a location of the GA20ox2 promoter different from the area where the ATHB2 plays its stimulatory action through its putative binding site, ABS1 (Figure 5B).
4.3 TCP13 appears to act on the promoter GA20ox2 through direct binding
An additional important understatement of this result is that TCP13 appears to bind directly to a promoter region of GA20ox2. In contrast to the original inhibitory activity of TCP13 on the GA20ox2 expression, our chimeric construct was designed to confer a stimulatory effect through the fused VP16 activation domain. If TCP13 did not directly bind to the promoter of GA20ox2, no positive effect of VP16 could have been possibly detected in our reporter assays in this scheme. A putative TCP13-specific element presumably serving as a direct binding site for the TCP13 may be present within the 0.5 kb-promoter fragment most proximal to the TCP13 ORF based on our result. The consensus sequence motif for binding of the TCP-family transcription factors has been known as GGNCCCAG for the class I TCPs and G(G/T)GGNCC(A/C) for the class II TCPs (Kosugi and Ohashi, 2002; Aggarwal et al., 2010), to which the TCP13 belongs. Although no sequences exactly matching to this consensus motif were found within the 0.5-kb promoter fragment, a sequence similar to the consensus, GTTGGCCA, was present within the fragment at about 400 bp upstream of a putative TATA element. Further detailed analyses on this promoter region, with respect to both the cis-elements and protein factors associated with them would be able to provide more comprehensive insight into the mechanism of the transcriptional repression of GA20ox2 mediated by TCP13.
One highly speculative but potentially interesting scenario is a possibility of TCP13 playing an inhibitory role against the DELLA-GAF1 transactivation complex. Although there is no concrete evidence for the DELLA-GAF1 in operation to control GA20ox gene expression under shade, the induction kinetics of GA20ox1 and GA20ox2 under shade condition appears to be reminiscent of the GA feedback inhibition. As the activity of DELLA-GAF1 complex is known to be repressed by a transcriptional corepressor TPR (TOPLESS-Related) (Fukazawa et al., 2014) and it has been also reported that TCP13 can specifically interact with TPR in an earlier study (Tao et al., 2013), it may be possible for TCP13 to participate in this regulatory complex as the one mediating the recruitment of the repressive factor of TPR to the promoter of GA20ox2. In support of this idea, we detected a specific interaction between TCP13 and GAF1 in our yeast two-hybrid and BiFC assays (Figure 6). However, at this point this idea of interdependency of ATHB2 and TCP13 to the GAF1 function has not been supported by any experimental evidence and can only be further tested and evaluated in later studies involving gaf1 mutant plants and necessary rigorous molecular analyses.
4.4 Possible role of ATHB2 in regulating GA20ox2 expression with respect to TCP13
One peculiar result obtained from the promoter deletion experiment with our TCP13- VP16 chimeric construct was that the luciferase activity obtained from the full-length promoter was significantly lower than that of the smaller, - 0.5-kb fragment which contained the putative TCP13 binding sequence. A straightforward account for this observation would be a presence of an inhibitory activity for the function of the TCP13 outside of the - 0.5-kb promoter region. One immediate suspect for this activity that came to our mind was ATHB2, as it is capable of having a specific interaction with TCP13. Having the transfection assay with the chimeric TCP13-VP16 construct with ABS1(m), the mutated ABS1 sequence, in the GA20ox2 promoter, we were able to demonstrate that ABS1 sequence was responsible for suppressing the activity of TCP13, suggesting that ATHB2, either directly or indirectly through other unknown protein factor(s), plays as a derepressor for the expression of GA20ox2 under shade response in Arabidopsis. TCP13 may bind to 0.5-kb region of the GA20ox2 promoter and then as ATHB2 is rapidly induced upon shade treatment, such functional relation between ATHB2 and TCP13 would allow the induction of GA20ox2 expression in response to the onset of the shade condition. Therefore, the role of ATHB2 in the regulation of GA20ox2 expression may be in a shade-specific lifting of the inhibitory effect of TCP13 to enable the stimulatory role of GAF1-DELLA complex to initiate the early-stage activation of GA20ox2, followed by GA-dependent feedback inhibition (Fukazawa et al., 2017). Further studies regarding molecular details of the interaction between TCP13 and GAF1 and investigation of its biological significance, including possible involvement of GAF1 in the shade avoidance response as well, should be able to provide much clearer understanding of complex regulatory networks coordinating the adaptive response of plant under shade condition.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
OS and CZ designed the experiments and made and analyzed transgenic plants; XY, LD, and Y-SH performed assays and yeast works; CZ, KN, and S-YC analyzed the transgenic plants and interpreted the data. C-IC and SK coordinated the study and finalized the article. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A1A01056744, 2020R1I1A1A01074533) and by Sookmyung Women’s University Research Cluster Program Grants (3-2203-0004).
Acknowledgments
We thank Dr. Eshed for his kind gift of the tcp5 tcp13 tcp17 triple mutant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1158288/full#supplementary-material
References
Abas, L., Kolb, M., Stadlmann, J., Janacek, D. P., Lukic, K., Schwechheimer, C., et al. (2020). Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc. Natl. Acad. Sci. USA 118, e2020857118. doi: 10.1073/pnas.2020857118
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Bou-Torrent, J., Galstyan, A., Gallemí, M., Cifuentes-Esquivel, N., Molina-Contreras, M. J., Salla-Martret, M., et al. (2014). Plant proximity perception dynamically modulates hormone levels and sensitivity in arabidopsis. J. Exp. Bot. 65, 2937–2947. doi: 10.1093/jxb/eru083
Carabelli, M., Morelli, G., Whitelam, G., Ruberti, I. (1996). Twilight-zone and canopy shade induction of the athb-2 homeobox gene in green plants. Proc. Natl. Acad. Sci. USA 93, 3530–3535. doi: 10.1073/pnas.93.8.3530
Carabelli, M., Sessa, G., Baima, S., Morelli, G., Ruberti, I. (1993). The arabidopsis athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J. 4, 469–479. doi: 10.1046/j.1365-313x.1993.04030469.x
Carabelli, M., Turchi, L., Ruzza, V., Morelli, G., Ruberti, I. (2013). Homeodomain-leucine zipper II family of transcription factors to the limelight: central regulators of plant development. Plant Signal Behav. 8, e25447. doi: 10.4161/psb.25447
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Chaiwanon, J., Wang, W., Zhu, J. Y., Oh, E., Wang, Z. Y. (2016). Information integration and communication in plant growth regulation. Cell 164, 1257–1268. doi: 10.1016/j.cell.2016.01.044
Djakovic-Petrovic, T., de Mieke, W., Voesenek, L. A., Pierik, R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51, 117–126. doi: 10.1111/j.1365-313X.2007.03122.x
Dunker, A. K., Lawson, J. D., Brown, C. J., Williams, R. M., Romero, P., Oh, J. S., et al. (2001). Intrinsically disordered protein. J. Mol. Graph Model. 19, 26–59. doi: 10.1016/s1093-3263(00)00138-8
Favero, D. S., Kawamura, A., Shibata, M., Takebayashi, A., Jung, J. H., Suzuki, T., et al. (2020). AT-hook transcription factors restrict petiole growth by antagonizing PIFs. Curr. Biol. 30, 1454–1466. doi: 10.1016/j.cub.2020.02.017
Fernández-Milmanda, G. L., Ballaré, C. L. (2021). Shade avoidance: expanding the color and hormone palette. Trends Plant Sci. 26, 509–523. doi: 10.1016/j.tplants.2020.12.006
Fukazawa, J., Miyamoto, C., Ando, H., Mori, K., Takahashi, Y. (2021). DELLA-GAF1 complex is involved in tissue-specific expression and gibberellin feedback regulation of GA20ox1 in arabidopsis. Plant Mol. Biol. 107, 147–158. doi: 10.1007/s11103-021-01195-z
Fukazawa, J., Mori, M., Watanabe, S., Miyamoto, C., Ito, T., Takahashi, Y. (2017). DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 175, 1395–1406. doi: 10.1104/pp.17.00282
Fukazawa, J., Teramura, H., Murakoshi, S., Nasuno, K., Nishida, N., Ito, T., et al. (2014). DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in arabidopsis. Plant Cell 26, 2920–2938. doi: 10.1105/tpc.114.125690
Gendrel, A. V., Lippman, Z., Yordan, C., Colot, V., Martienssen, R. A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the arabidopsis gene DDM1. Science 297, 1871–1873. doi: 10.1126/science.1074950
Han, X., Yu, H., Yuan, R., Yang, Y., An, F., Qin, G. (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15, 611–622. doi: 10.1016/j.isci.2019.04.005
He, G., Liu, P., Zhao, H., Sun, J. (2020). The HD-ZIP II transcription factors regulate plant architecture through the auxin pathway. Int. J. Mol. Sci. 21, 3250. doi: 10.3390/ijms21093250
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hedden, P., Phillips, A. L., Rojas, M. C., Carrera, E., Tudzynski, B. (2001). Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20, 319–331. doi: 10.1007/s003440010037
Hiratsu, K., Matsui, K., Koyama, T., Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313x.2003.01759.x
Hisamatsu, T., King, R. W., Helliwell, C. A., Koshioka, M. (2005). The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of arabidopsis. Plant Physiol. 138, 1106–1116. doi: 10.1104/pp.104.059055
Hur, Y. S., Kim, J., Kim, S., Son, O., Kim, W. Y., Kim, G. T., et al. (2019). Identification of TCP13 as an upstream regulator of ATHB12 during leaf development. Genes (Basel) 10, 644. doi: 10.3390/genes10090644
Hur, Y. S., Um, J. H., Kim, S., Kim, K., Park, H. J., Lim, J. S., et al. (2015). Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 205, 316–328. doi: 10.1111/nph.12998
Ince, Y. Ç., Galvão, V. C. (2021). Analysis of shade-induced hypocotyl elongation in arabidopsis. Methods Mol. Biol. 2297, 21–31. doi: 10.1007/978-1-0716-1370-2_3
Kosugi, S., Ohashi, Y. (2002). DNA Binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Kunihiro, A., Yamashino, T., Nakamichi, N., Niwa, Y., Nakanishi, H., Mizuno, T. (2011). Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 52, 1315–1329. doi: 10.1093/pcp/pcr076
Küpers, J. J., Snoek, B. L., Oskam, L., Pantazopoulou, C. K., Matton, S. E. A., Reinen, E., et al. (2023). Local light signaling at the leaf tip drives remote differential petiole growth through auxin-gibberellin dynamics. Curr. Biol. 33, 75–85. doi: 10.1016/j.cub.2022.11.045
Lopez, J. A., Sun, Y., Blair, P. B., Mukhtar, M. S. (2015). TCP Three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 20, 238–245. doi: 10.1016/j.tplants.2015.01.005
Ma, L., Li, G. (2019). Auxin-dependent cell elongation during the shade avoidance response. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00914
Martínez-García, J. F., Huq, E., Quail, P. H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. doi: 10.1126/science.288.5467.859
Martin-Trillo, M., Cubas, P. (2010). TCP Genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Müller-Moulé, P., Nozue, K., Pytlak, M. L., Palmer, C. M., Covington, M. F., Wallace, A. D., et al. (2016). YUCCA auxin biosynthetic genes are required for arabidopsis shade avoidance. PeerJ 4, e2574. doi: 10.7717/peerj.2574
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the arabidopsis hypocotyl. Elife 3, e03031. doi: 10.7554/eLife.03031
Pierik, R., Ballaré, C. L. (2021). Control of plant growth and defence by photoreceptors: from mechanisms to opportunities in agriculture. Mol. Plant 14, 61–76. doi: 10.1016/j.molp.2020.11.021
Pierik, R., Testerink, C. (2014). The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol. 166, 5–22. doi: 10.1104/pp.114.239160
Procko, C., Burko, Y., Jaillais, Y., Ljung, K., Long, J. A., Chory, J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30, 1529–1541. doi: 10.1101/gad.283234.116
Pucciariello, O., Legris, M., Costigliolo Rojas, C., Iglesias, M. J., Hernando, C. E., Dezar, C., et al. (2018). Rewiring of auxin signaling under persistent shade. Proc. Natl. Acad. Sci. USA 115, 5612–5617. doi: 10.1073/pnas.1721110115
Rademacher, W. (2000). Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 501–531. doi: 10.1146/annurev.arplant.51.1.501
Rath, M., Challa, K. R., Sarvepalli, K., Nath, U. (2022). CINCINNATA-like TCP transcription factors in cell growth - an expanding portfolio. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.825341
Reed, J. W., Wu, M. F., Reeves, P. H., Hodgens, C., Yadav, V., Hayes, S., et al. (2018). Three auxin response factors promote hypocotyl elongation. Plant Physiol. 178, 864–875. doi: 10.1104/pp.18.00718
Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S. J., Gong, F., et al. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the arabidopsis life cycle. Plant J. 53, 488–504. doi: 10.1111/j.1365-313X.2007.03356.x
Roig-Villanova, I., Martínez-García, J. F. (2016). Plant responses to vegetation proximity: a whole life avoiding shade. Font. Plant Sci. 7. doi: 10.3389/fpls.2016.00236
Sessa, G., Morelli, G., Ruberti, I. (1993). The athb-1 and -2 HD-zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12, 3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x
Son, O., Kim, S., Hur, Y. S., Cheon, C. I. (2016). Identification of the raptor-binding motif on arabidopsis S6 kinase and its use as a TOR signaling suppressor. Biochem. Biophys. Res. Commun. 472, 83–87. doi: 10.1016/j.bbrc.2016.02.068
Sorin, C., Salla-Martret, M., Bou-Torrent, J., Roig-Villanova, I., Martínez-García, J. F. (2009). ATHB4, a regulator of shade avoidance, modulates hormone response in arabidopsis seedlings. Plant J. 59, 266–277. doi: 10.1111/j.1365-313X.2009.03866.x
Steindler, C., Matteucci, A., Sessa, G., Weimar, T., Ohgishi, M., Aoyama, T., et al. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126, 4235–4245. doi: 10.1242/dev.126.19.4235
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tao, Q., Guo, D., Wei, B., Zhang, F., Pang, C., Jiang, H., et al. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in arabidopsis. Plant Cell 25, 421–437. doi: 10.1105/tpc.113.109223
Turchi, L., Baima, S., Morelli, G., Ruberti, I. (2015). Interplay of HD-zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 66, 5043–5053. doi: 10.1093/jxb/erv174
Turchi, L., Carabelli, M., Ruzza, V., Possenti, M., Sassi, M., Peñalosa, A., et al. (2013). Arabidopsis HD-zip II transcription factors control apical embryo development and meristem function. Development 140, 2118–2129. doi: 10.1242/dev.092833
Valsecchi, I., Guittard-Crilat, E., Maldiney, R., Habricot, Y., Lignon, S., Lebrun, R., et al. (2013). The intrinsically disordered c-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol. Biosyst. 9, 2282–2295. doi: 10.1039/c3mb70128j
Wang, X., Gao, X., Liu, Y., Fan, S., Ma, Q. (2020). Progress of research on the regulatory pathway of the plant shade-avoidance syndrome. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00439
Willige, B. C., Zander, M., Yoo, C. Y., Phan, A., Garza, R. M., Trigg, S. A., et al. (2021). PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 53, 955–961. doi: 10.1038/s41588-021-00882-3
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yang, C., Li, L. (2017). Hormonal regulation in shade avoidance. Front. Plant Sci. 8, 1527. doi: 10.3389/fpls.2017.01527
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, Y., Pfeiffer, A., Tepperman, J. M., Dalton-Roesler, J., Leivar, P., Gonzalez Grandio, E., et al. (2020). Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. USA 117, 3261–3269. doi: 10.1073/pnas.1918317117
Zhou, Y., Xun, Q., Zhang, D., Lv, M., Ou, Y., Li, J. (2019). TCP Transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15, 600–610. doi: 10.1016/j.isci.2019.04.002
Keywords: ATHB2, TCP13, shade avoidance syndrome, GA20ox2, GAF1
Citation: Son O, Zhang C, Yang X, Duc LT, Hur Y-S, Nam KH, Choi S-Y, Cheon C-I and Kim S (2023) Identification of GA20ox2 as a target of ATHB2 and TCP13 during shade response. Front. Plant Sci. 14:1158288. doi: 10.3389/fpls.2023.1158288
Received: 06 February 2023; Accepted: 03 April 2023;
Published: 21 April 2023.
Edited by:
Ping Wang, Iowa State University, United StatesReviewed by:
Trevor M. Nolan, Duke University, United StatesZhongpeng Li, Iowa State University, United States
Copyright © 2023 Son, Zhang, Yang, Duc, Hur, Nam, Choi, Cheon and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunghan Kim, a2ltc2hAc29va215dW5nLmFjLmty; Choong-Ill Cheon, Y2NoZW9uQHNvb2tteXVuZy5hYy5rcg==
†These authors share first authorship
 Ora Son1†
Ora Son1† Chaoyue Zhang
Chaoyue Zhang Xiaoyu Yang
Xiaoyu Yang Kyoung Hee Nam
Kyoung Hee Nam Choong-Ill Cheon
Choong-Ill Cheon