- 1Engineering Research Center of Agricultural Microbiology Technology, Ministry of Education & Heilongjiang Provincial Key Laboratory of Plant Genetic Engineering and Biological Fermentation Engineering for Cold Region & Key Laboratory of Molecular Biology, College of Heilongjiang Province & School of Life Sciences, Heilongjiang University, Harbin, China
- 2Department of Biology, University of Mississippi, Oxford, MS, United States
- 3Department of Natural Sciences, Novosibirsk State University, Novosibirsk, Russia
- 4Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia
Sugar beet is one of the most important sugar crops in the world. It contributes greatly to the global sugar production, but salt stress negatively affects the crop yield. WD40 proteins play important roles in plant growth and response to abiotic stresses through their involvement in a variety of biological processes, such as signal transduction, histone modification, ubiquitination, and RNA processing. The WD40 protein family has been well-studied in Arabidopsis thaliana, rice and other plants, but the systematic analysis of the sugar beet WD40 proteins has not been reported. In this study, a total of 177 BvWD40 proteins were identified from the sugar beet genome, and their evolutionary characteristics, protein structure, gene structure, protein interaction network and gene ontology were systematically analyzed to understand their evolution and function. Meanwhile, the expression patterns of BvWD40s under salt stress were characterized, and a BvWD40-82 gene was hypothesized as a salt-tolerant candidate gene. Its function was further characterized using molecular and genetic methods. The result showed that BvWD40-82 enhanced salt stress tolerance in transgenic Arabidopsis seedlings by increasing the contents of osmolytes and antioxidant enzyme activities, maintaining intracellular ion homeostasis and increasing the expression of genes related to SOS and ABA pathways. The result has laid a foundation for further mechanistic study of the BvWD40 genes in sugar beet tolerance to salt stress, and it may inform biotechnological applications in improving crop stress resilience.
Introduction
WD40 proteins are evolutionarily conserved and widely distributed in eukaryotic organisms. They tend to consist of 4 to 16 WD40 domains, which are also known as the WD40 repeats (WDR) (Smith et al., 1999). The WDR is generally composed of 40 to 60 amino acid residues, with a GH (glycine-histidine) dipeptide at the N-terminus and WD (tryptophan-aspartate) dipeptide at the C-terminus (Neer et al., 1994). The WDR typically folds into a highly stable seven-bladed β-propeller (Stirnimann et al., 2010), connected by an N-terminal amino acid residue in a closed loop, which determines specific protein functions (Mishra et al., 2012).
Members of the WD40 protein family have been identified in many plants. For example, the reference plant Arabidopsis has 230 WD40 proteins (Li et al., 2014). Among the cash crops reported, there were 225 WD40 proteins in red sage (Salvia miltiorrhiza) (Liu et al., 2020), 187 WD40 proteins in Rosaceae (Rosa chinensis ‘old blush’) (Sun et al., 2020), 42 WD40 proteins in walnut (Juglans regia), and 204 WD40s in fig (Ficus carica) (Chen et al., 2022a; Fan et al., 2022). Furthermore, among the food crops, wheat (Triticum aestivum) has 743 WD40 proteins (Hu et al., 2018), potato (Solanum tuberosum) has 168 WD40 proteins (Tao et al., 2019), and rice (Oryza sativa) has 200 WD40 proteins (Ouyang et al., 2012). However, the WD40 protein family in sugar beet (Beta vulgaris L) has not been reported. As one of the largest protein families, WD40 proteins were once regarded as scaffolds for recruiting other molecules to form functional complexes or participate in protein-protein interactions (Li and Roberts, 2001). In recent years, a large number of studies have shown that WD40 proteins have a variety of biological functions. In animals, they are involved in many biological processes, including signal transduction (Liang et al., 2022), histone modification (Lorton et al., 2020), DNA damage response (Choi et al., 2022), transcriptional regulation (Mo et al., 2023), ribosome biosynthesis (Barandun et al., 2018), protein degradation, and apoptosis (An et al., 2022; Cai et al., 2022). In plants, WD40 protein is generally considered to be an important regulator of several biological processes, e.g., anthocyanin biosynthesis (Ji et al., 2023), flowering meristem development (Park et al., 2019), gametogenesis (Shi et al., 2005), embryogenesis (Kim et al., 2021), and yield (Chen et al., 2022b). In addition, genes encoding WD40 proteins also play important roles in plant response to abiotic stresses. For instance, overexpression of a TaPUB1 gene in tobacco enhanced tobacco salt tolerance by reducing Na+ accumulation and reactive oxygen species (ROS) in the transgenic plants and increasing the expression of antioxidant-related genes (Zhang et al., 2017). In addition, an AtXIW1 gene in Arabidopsis plays a positive role in ABA response. Mutation of AtXIW1 inhibited the induction of ABA-responsive genes and the accumulation of ABI5, and led to rapid proteasome degradation of the ABI5 (Xu et al., 2019). Furthermore, inhibition of a rice OsRACK1A expression enhanced the rice salt tolerance through maintaining high K+/Na+ and reducing the accumulation of malondialdehyde (MDA). OsRACK1A was found to interact with many salt stress response proteins to decrease the salt tolerance of rice (Zhang et al., 2018). Recently, overexpression of another TaWD40-4B.1 gene increase the biomass of transgenic wheat under drought stress. The TaWD40-4B.1 protein interacts with TaCAT3 protein to promote their oligomerization and catalase activity under drought stress, leading to improved drought tolerance of the transgenic wheat (Tian et al., 2023).
Sugar beet is an Amaranthaceae biennial herbaceous plant, one of the world’s important sugar crops, accounting for 20%-25% of the world’s annual sugar production (Khan et al., 2019). Sugar beet is also an important cash crop in northeast China, and its root has high economic value (Thiruvengadam et al., 2022). Sugar beet is also a halophyte tolerant to salt and alkali stresses. With the increase of saline-alkali land in the world, crop production and food security have become a grand challenge (Kopecká et al., 2023). Systematic identification of plant salt tolerance genes toward improving crop stress resilience and increasing yield is urgently needed (Ma et al., 2017; Ji et al., 2019). Since the WD40 proteins are involved in many biological processes including plant growth and stress response, we hypothesize that some WD40 protein encoding genes in halophyte sugar beet play important roles in plant salt stress tolerance. In this study, we analyzed the sequences of the WD40 family proteins in the sugar beet genome. The expression profiles of the BvWD40 genes under salt stress were characterized. One of the genes, BvWD40-82, was found to confer plant salt stress tolerance. The result not only highlights the utility of gene functional analysis informed by genome-wide informatics, but also provides important resources for the community to further explore the roles of WD40 genes in crop stress resilience and yield.
Materials and methods
Identification of BvWD40 proteins in sugar beet
The seed file for the WD40 domain was downloaded from the InterPro database (www.ebi.ac.uk/interpro/entry/pfam/PF00400/). A Hidden Markov Model (HMM) of WD40 domain was constructed using the HMMER program (Mistry et al., 2013), and the NCBI protein database of sugar beet (www.ncbi.nlm.nih.gov/genome) was searched and compared. E-value was used to screen candidate proteins (E-Value<0.05) and SMART (http://smart.embl-heidelberg.de/) was used to verify and confirm that all the BvWD40 members contain the WD40 domain. The conserved motifs of BvWD40s were predicted by the MEME program (meme-suite.org/tools/meme). The domain and conserved motifs of BvWD40s were visualized by TBtools (Chen et al., 2020). The theoretical molecular weight and isoelectric point of each BvWD40 were analyzed by using the Expasy tool (web.expasy.org/compute_pi/).
Phylogenetic analysis
The sequences of 177 BvWD40 proteins were downloaded from the NCBI, and then multi-sequence alignment of the BvWD40s was performed by the Clustal W (Larkin et al., 2007). The trimAl was used to trim the sequences (Capella-Gutiérrez et al., 2009). The parameter was set as the fraction of sequences with allowed gaps of 0.8, the minimum average similarity of 0.001, and the minimum percentage of positions retained in the original route of 80 percent. The pruned files were imported into IQ-TREE 2 (Minh et al., 2020), and the phylogenetic tree was established by the maximum likelihood method with 1000 replicates of bootstrapping.
Gene structure, chromosomal location, and gene duplication analysis of the BvWD40 genes
The genome annotation file, coding sequence (CDS), and the sequences of BvWD40 genes were downloaded from NCBI. The gene structure pattern map of BvWD40s was obtained from the GSDS website (gsds.gao-lab.org/). We used TBtools to map BvWD40s chromosomal location information. Based on the previous data (Li et al., 2014), we supplemented the Arabidopsis WD40 genes with newly identified WD40 genes: AT1G05631.1, AT1G51690.1, AT1G655801.1, AT2G31830.1, AT2G439001, AT3G56990.1, and a total of 236 AtWD40s were obtained. The collinearity relationship between BvWD40s and AtWD40s was analyzed by MCScanX (Wang et al., 2012), and the collinearity map of sugar beet and Arabidopsis was generated by Circos (Krzywinski et al., 2009).
Interaction networking, expression profiling, and gene ontology analysis
The BvWD40 proteins interaction network was obtained from the String website (cn.string-db.org/) (Szklarczyk et al., 2015) and visualized using the Cytoscape software (Shannon et al., 2003). The RNA-seq data of B. vulgaris were downloaded from the NCBI SRA database (Accession: PRJNA666117). The transcriptome data of the BvM14 line under salt stress were generated and stored in the Li laboratory. The expression profile of BvWD40 genes was used by TBtools software. The basic data of gene ontology analysis were obtained from the GO network database (geneontology.org/), and GO annotation and enrichment of the BvWD40 genes were performed using TBtools and visualized by an online tool (www.bioinformatics.com.cn).
Plant materials and salt stress treatment
The BvM14 line is a monomeric additional line obtained by crossing and backcrossing between diploid cultivated sugar beet and tetraploid wild sugar beet, which was created and propagated by the Li lab (Beta vulgaris L., VV+1C, 2n=18 + 1) (Li et al., 2022b). The seeds were disinfected and cultured in a hydroponic system as previously reported (Ma et al., 2017; Ji et al., 2019). After growing to the third pair of fully expanded leaves, leaves and roots were sampled to extract total RNA using the TRIzol method (Meng and Feldman, 2010). Three biological replicates were conducted. Primer 3 plus (www.primer3plus.com) was used to design specific primers, and 18sRNA was used as the reference. qRT-PCR was performed using SYBR, and the relative gene expression was calculated using the 2-ΔΔCt method (Schmittgen and Livak, 2008).
A. thaliana Columbia ecotype (Col-0) seeds were obtained from the ABRC (abrc.org), and were germinated in 1/2 MS medium under 300 µmol/m2 s light intensity, 14 h light and 10 h darkness. After eight days, the seedlings were transferred to a new MS medium (with or without 150 mM NaCl) for 7 days to further screen transgenic plants, or observe their growth phenotypes under salt stress. After soil and vermiculite were mixed at 2:1, and sterilized at 180 °C in a dryer, the seeds were sowed in the mixed soil. After 21 days of culture (relative humidity 65%-75%, 22 °C, light 16 h/dark 8 h), the seedings were treated with 150 mM NaCl salt stress, and three plants with consistent growth were selected from each line for subsequent physiological/biochemical analyses and RNA extraction.
Subcellular localization of the BvWD40-82 and generation of transgenetic lines
The BvWD40-82 gene was constructed in the pCAMVBIA2300-35S-eYFP vector and its subcellular localization was observed by laser scanning confocal microscopy (FV1200, Olympus). The nucleotide sequence of AtUTP18, the homologous gene of the BvWD40-82 gene in Arabidopsis, was obtained by NCBI online BLASTN software. In order to construct utp18 mutation plants to characterize the function of this gene, the online tool CRISPR-P2.0 (cbi.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR) was used to select appropriate targets and construct CRISPR/Cas9 vectors (pNGG2F vector). After infecting the flowers of wild-type Arabidopsis, the utp18 homozygous mutant line was obtained by Hygromycin B (30 mg/L) screening and TA clone sequencing. The BvWD40-82 gene was constructed into the pCAMBIA1300-35S-3xFLAG vector, the wild-type Arabidopsis and utp18 mutant lines were transformed by floral infestation, and the seeds of each plant were collected and placed in MS medium which contains Hygromycin B (30 mg/L), and the seedlings with normal growth were placed in the soil for culture after about ten days. The DNA and RNA from the leaves of each transgenic plant were extracted after four weeks for DNA verification and RT-PCR. The T3 generation homozygous transgenic lines (Heterologous overexpression, OE#16, OE#17, OE#18; Heterologous complementation, CO#1, CO#2, CO#5) were used for further analysis.
Physiological and biochemical index determination
Root length of each transgenic seedling and wild-type seedling with or without 150 mM NaCl treatment in the MS medium was determined. The dry weight and fresh weight (with or without 150 mM NaCl in the soil) were also analyzed. The MDA content, SOD and POD enzyme activities were analyzed using previously published methods (Ma et al., 2017; Ji et al., 2019); Betaine content was determined at 525 nm (Swarna et al., 2013); Na+, K+, and Ca2+ contents were analyzed by a flame atomic absorption method, as previously described (Gao et al., 2016).
Results
Identification of WD40 proteins in sugar beet
A total of 177 BvWD40s were obtained by removing redundant proteins and repetitive sequences. Based on the location of the corresponding genes of these proteins on the nine chromosomes of the sugar beet, they were named BvWD40-1 to BvWD40-177. In silico analysis showed that the physicochemical properties and sequence composition of the BvWD40 proteins varied largely, e.g., the molecular weights (MW, 13.79-398.63 kDa) and isoelectric points (pI, 4.25-9.67) spanned a wide range. The sequence length of BvWD40s ranges from 126 to 3599 amino acids, with an average length of 627.7 amino acids (Supplementary Table 1). SMART indicated that the BvWD40s contained 1 to 13 WD repeats.
Conservative domain and motif analysis of BvWD40s
The results of protein structure analysis showed that all the BvWD40 proteins contained the WD40 domain, and more than 35 additional functional domains in total (Supplementary Table 2). Most (49) BvWD40s had six WD40 repeats, followed by seven WD40 repeats in 43 BvWD40s. Thirty-six BvWD40s had their WD40 domains at the C-terminus of their other domains. Except for the WD40 domain, the frequency of the LisH domain was the highest (12 BvWD40s). Studies have shown that the LisH domain affects rice growth and reproduction (Gao et al., 2012), suggesting that BvWD40 proteins with the LisH domain may also be involved in similar processes. Various other domains were present but infrequently, such as the ATG16 domain only in BvWD40-106 and the BCAS3 domain only in BvWD40-57 and BvWD40-64. These two domains are involved in cell autophagy of yeast (Xiong et al., 2018; Yamada and Schaap, 2021). The UTP15 domain (only in BvWD40-20), was a component of the UtpA complex involved in the assembly of small ribosomal subunits (Kornprobst et al., 2016). However, the Arabidopsis AtMSI4 gene encoding a WD40 protein contains six WD40 domains and a CAF1C domain, and may be involved in nucleosome assembly, but some studies have shown that AtMSI4 is involved in regulation of flowering in Arabidopsis (Pazhouhandeh et al., 2011). As another example, the RNAi-AtATG18a plants are more sensitive to salt and mannitol and defective in autophagosome formation in Arabidopsis, it indicated that AtATG18a may plays an important role in plant autophagy and abiotic stress. It should be noted that the AtATG18a contains only the WD40 domain (Liu et al., 2009).
We predicted 50 conserved motifs for 177 BvWD40s using the MEME tool (Supplementary Figure 1), among which the most highly conserved motifs were Motif 1 (173 times), Motif 2 (171 times) and Motif 4 (171 times). The frequencies of tryptophan (W) and aspartate (D) were the highest in Motif 1 and Motif 2, and the frequencies of glutamate (G) and histidine (H) were the highest in Motif 4, the results also showed that the amino acid composition of the WD40 domain was quite different. Interestingly, some motifs occur less frequently, such as Motif 6 (8 times) and Motif 18 (61 times), which have the WDxR motif (Supplementary Figure 2). The WDxR motifs are often found in DCAFs, which have been proposed as substrate recruiting proteins for the E3 ubiquitin ligase complex Cullin4-DDB1 (Mistry et al., 2020). In addition, previous studies have shown that the WDxR motif of the DCAF WD40 domain in plants is a critical motif for interacting with the DDB1 protein. It may also be involved in a variety of plant developmental pathways (Zhang et al., 2008).
Subfamily classification and phylogenetic analysis
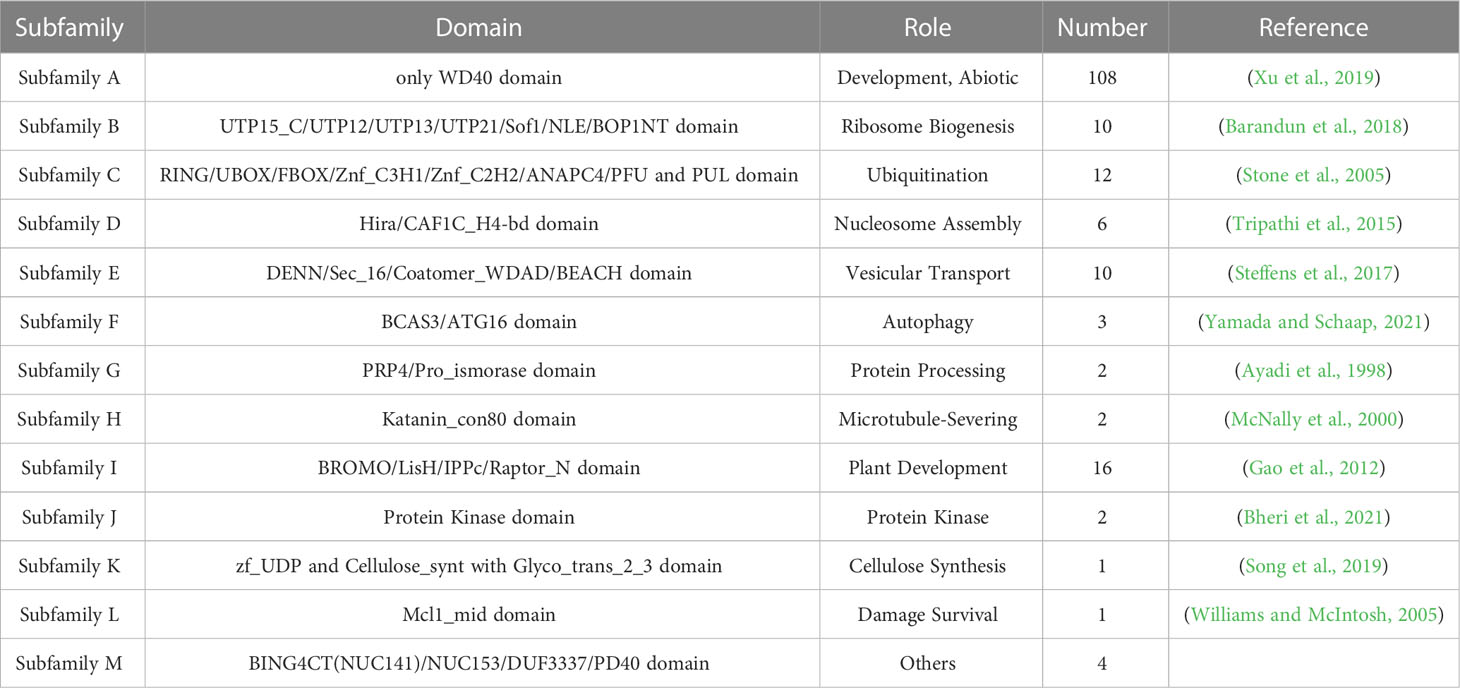
We divided the BvWD40 proteins into 13 subfamilies based on their domain composition and the roles of different domains in biological processes (Table 1). Subfamily A is the largest in sugar beet with 108 members. It contains only the WD40 domain. Subfamily I is the second largest family with 16 members that contain LisH and IPPc domains associated with plant growth and development. Other subfamilies also have functional domains that play different key roles, such as ribosome synthesis, ubiquitination, nucleosome assembly, and vesicle transport, suggesting that BvWD40s may be master regulators in various processes. The BvWD40 proteins were divided into 14 different clusters (G1-G14) based on their sequence homology, among which G1 has only 3 members and was the smallest cluster and G14 was the largest cluster with 21 members (Figure 1). The structure similarity of the members in each cluster is high, and the composition and order of the conserved domain are consistent (Supplementary Figure 1). It is worth noting that the clusters are not the same as subfamilies, possibly due to the large differences in amino acid composition of domains that maintain similar functions.
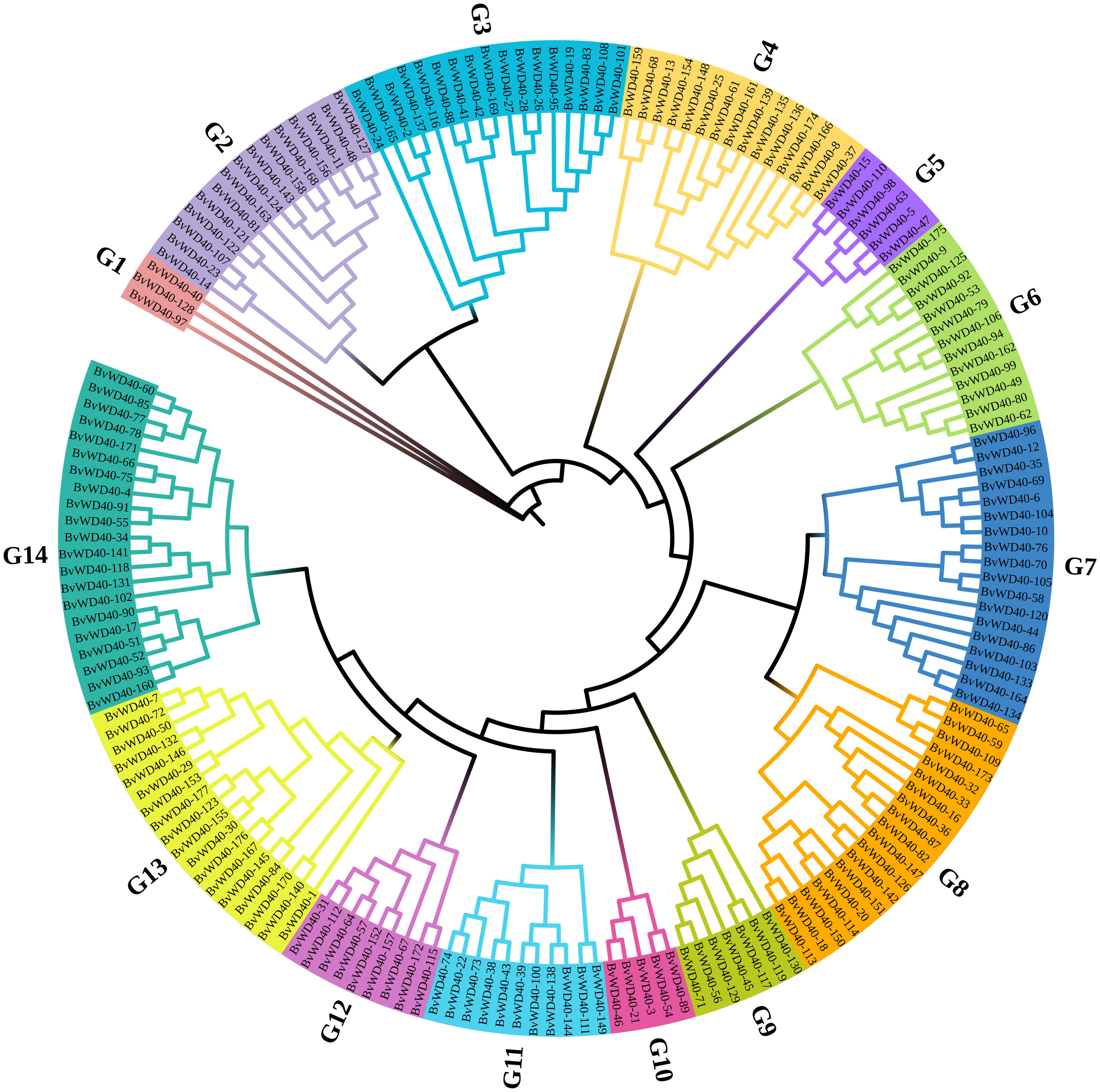
Figure 1 Phylogenetic tree of the WD40 proteins from sugar beet. The phylogenetic tree was constructed using ClustalW, trimAl, and IQ-Tree software with the maximum likelihood method and 1000 bootstrap replicates. Branch lines in different colors represented different groups (G1 to G14).
Gene structure, chromosomal location, and duplication of BvWD40 genes
The gene structure of the 177 BvWD40 genes was analyzed by the GSDS software (Supplementary Figure 3). The number of exons in the BvWD40s varied greatly. BvWD40-10 has 39 exons, while 10 BvWD40s have only one exon. On average, the BvWD40s have 10.8 exons and 9.8 introns.
Since 13 BvWD40s were not fully assembled, we mapped the remaining 164 BvWD40s to the sugar beet chromosomes using positional information. The BvWD40s were widely distributed on the nine chromosomes (Figure 2). Chromosome 9 has 22, the highest number of BvWD40s, while chromosome 3 and chromosome 6 each has 15, the lowest number.
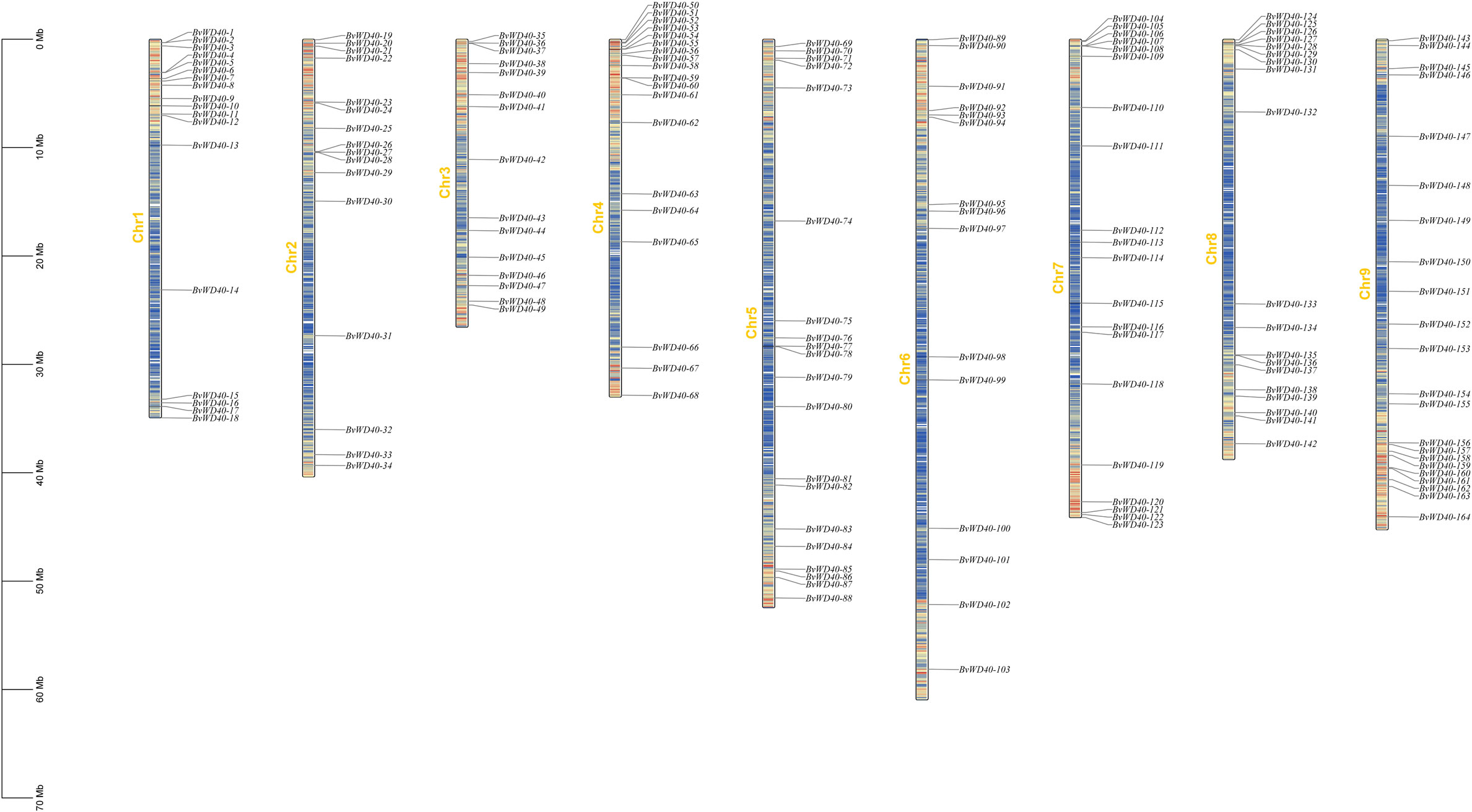
Figure 2 Locations of BvWD40 genes on the sugar beet chromosome. According to the location information of the BvWD040s, BvWD40s was marked on each chromosome of sugar beet. There was a number marker on the left side of each chromosome, the length was synchronized with the scale, and the gene density was distributed according to the color (low density: blue, high density: red).
To study the occurrence of the WD40 genes in sugar beet and its evolutionary relationship among different species, we plotted the collinearity map of the WD40 genes in sugar beet and Arabidopsis (Figure 3). The result showed that the BvWD40 genes had fewer duplication events, only 4 pairs (BvWD40-34/BvWD40-141, BvWD40-7/BvWD40-72, BvWD40-17/BvWD40-90, BvWD40-11/BvWD40-156) were found. In contrast, Arabidopsis had 28 duplication events. However, 58 pairs of duplication events existed between sugar beet and Arabidopsis, more than the two species had on their own.
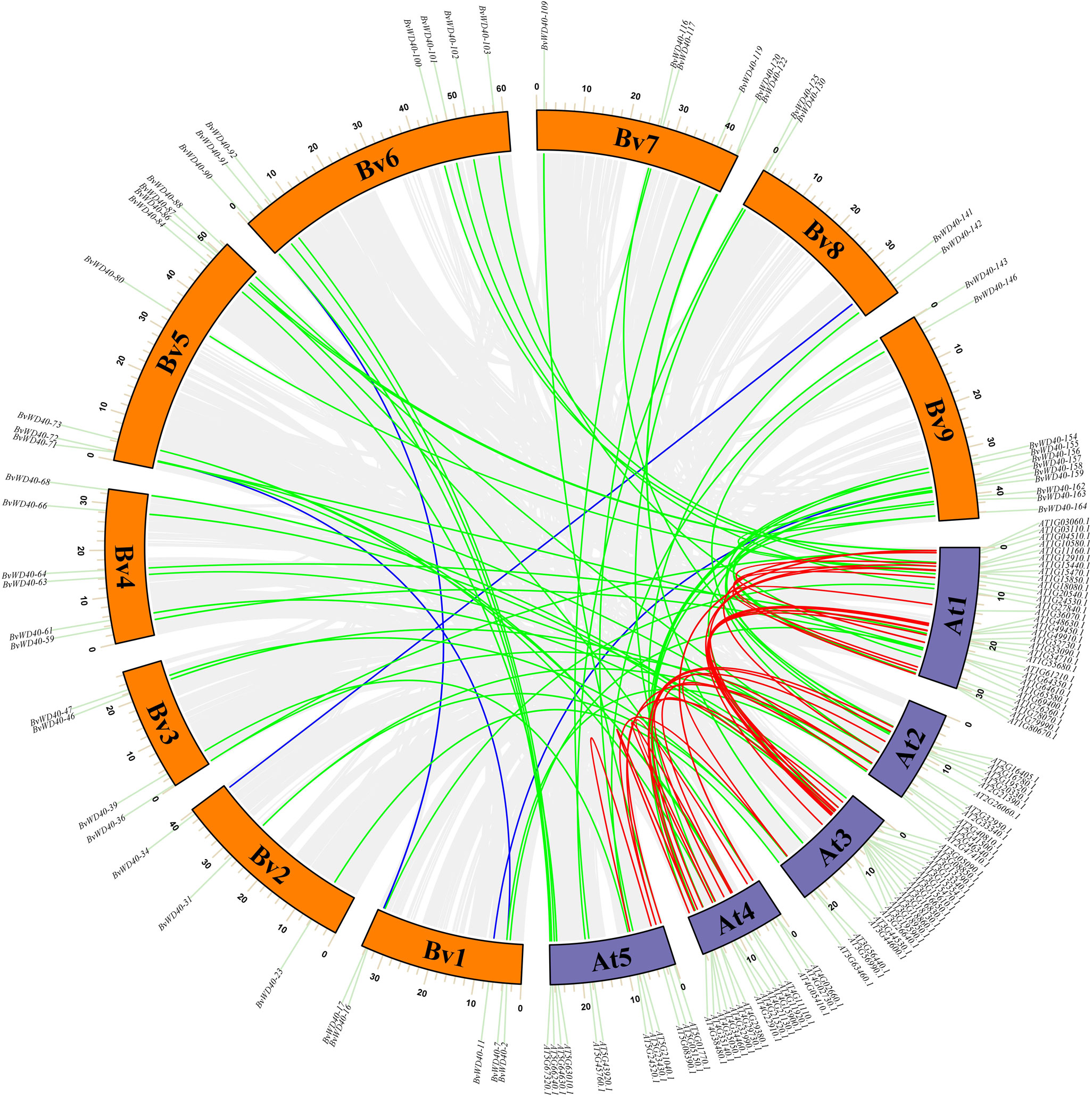
Figure 3 Collinearity analysis of the WD40 genes in sugar beet and Arabidopsis. The sugar beet and Arabidopsis chromosomes were indicated by the orange and purple block, respectively. Grey lines indicate all existing genes for a linear relationship, the blue line represents the sugar beet with collinearity of WD40 genes, the red line represents the Arabidopsis have collinearity of WD40 genes, the green line represents sugar beet and Arabidopsis have collinearity WD40 genes.
Gene ontology analysis of BvWD40 genes
GO functional enrichment results showed (Figure 4C) that a large number of BvWD40s were enriched in organ composition, plant development process, regulation of biological processes, catalytic activity, protein binding, signal transduction, nucleotide binding, and other basic biological functions. In addition, a large number of BvWD40s were enriched in response to stress such as stimulus-response, heat response, and salt response.
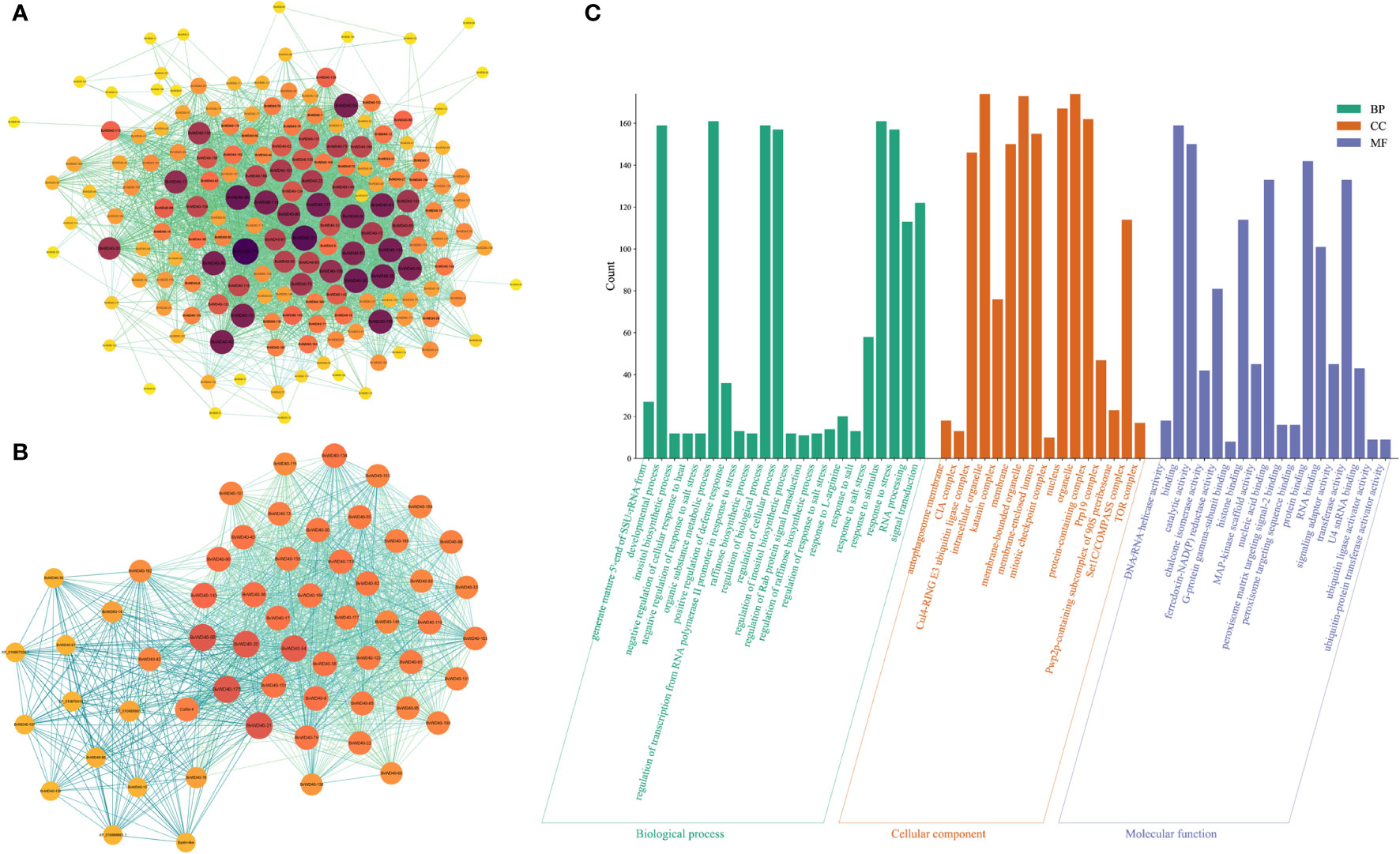
Figure 4 Interaction network of BvWD40 proteins and GO analysis of BvWD40 genes. (A) Interaction network among the BvWD40s. (B) Network of interactions between BvWD40s and other proteins. In both images, the sphere size and fill color represent the number of nodes (yellow – dark purple: 1-100), and the line color represents the Combined Score (light green - light blue: 0.4-1) calculated from the String tool. (C) BvWD40 genes ontology analysis, where the abscissa represents the GO item, the ordinate represents the number of genes enriched in GO, with BP, CC and MF representing the biological process, cell composition, and molecular function, respectively.
Analysis of BvWD40 protein interaction network
We obtained the interaction network relationship of the BvWD40 proteins using the String. There were abundant interactions among the BvWD40s (Figure 4A). A total of 167 proteins had 3337 interaction relationships, and 50 BvWD40s had more than 50 interaction relationships. BvWD40-175 may interact with 60% of other BvWD40s (100 BvWD40s). It is a homolog of root initiation defect protein 3 (RID3), which was involved in the apical meristem (SAM) regeneration as a negative regulator of the CUC-STM pathway (Tamaki et al., 2009). In order to obtain the interaction network between WD40s and other proteins, we expanded the String network node and found a total of 207 proteins with 4418 interactions. We conducted MCODE analysis on this network and found a key network, which was composed of 53 BvWD40 proteins and 6 non-BvWD40 proteins, with 1160 interactions (Figure 4B). In the network, BvWD40-175 and Cullin4 were the BvWD40 and non-WD40 protein with the most interactions (117 and 110 nodes, respectively). Cullin-4 is the scaffold subunit of E3 ligase, which binds to DDB1 and DCAF to play the role of E3 ligase. Some studies have shown that the Arabidopsis DCAF protein ABD1 negatively regulates abscisic acid signaling in Arabidopsis (Mistry et al., 2020). The tomato DCAF protein DDI1 acts as a substrate receptor for CUL4-DDB1 ubiquitin ligase and actively regulates abiotic stress tolerance in tomato (Miao et al., 2014). It is speculated that BvWD40s may interact with each other to participate in plant development and growth. Most of them may be the substrates of Cullin-4 and may be involved in abiotic stress responses.
Expression pattern of BvWD40 genes under salt stress
To further explore the response of BvWD40 genes under salt stress, we plotted the expression profile of all the BvWD40s in cultivated B. vulgaris under salt stress (Supplementary Figure 4). We studied the expression patterns of BvWD40s in two tissues at different times under 200 mM, 300 mM or 400 mM NaCl treatment. Between 12 h and 72 h under 300 mM NaCl treatment, there were a large number of BvWD40s changed expression in leaves and roots. The number of genes responding to salt stress in the roots reached the maximum at 72 h (35 genes), and those in the leaf reached the maximum at 24 h (39 genes). We found that six BvWD40s in the roots and five BvWD40s in the leaves consistently responded to salt stress for 12 to 72 hours. In the leaves of the BvM14 line, there were most 49 BvWD40s responded to 200 mM salt stress and 32 BvWD40s responded to both 200 mM and 400 mM salt stress. In the roots, there were most 92 BvWD40s responded to 200 mM salt stress and 58 BvWD40s changed in response to salt stress at different concentrations. In both tissues, 15 BvWD40s (BvWD40-6, 11, 38, 44, 62, 66, 72, 82, 83, 87, 109, 126, 141, 152, and 162) simultaneously responded to different concentrations of salt stress. These results indicate that many BvWD40s in different tissues are responsive to salt stress conditions. In different sugar beet lines, there are unique and shared salt-stress responses of the BvWD40s at the spatiotemporal level. Clearly, many BvWD40s may play important roles in sugar beet salt tolerance, and those unique to the BvM14 and changed in both leaves and roots may possess high value for biotechnological applications.
Cloning, tissue specific expression, and subcellular localization of BvWD40-82
According to the expression analysis of BvWD40 genes, we found that the BvWD40-82 gene was up-regulated in both the leaves and roots of the BvM14 line under the 200 mM and 400 mM NaCl. To the best of our knowledge, no previous studies on the salt tolerance function of the BvWD40-82 were reported. Here we cloned the BvWD40-82 gene from the BvM14 line (Supplementary Figure 5). The tissue-specific expression analysis showed that the expression level of the BvWD40-82 gene in leaves was 12.3-fold higher than in roots (Figure 5A). GFP imaging showed that BvWD40-82 protein was localized to the nucleus (Figure 5B).
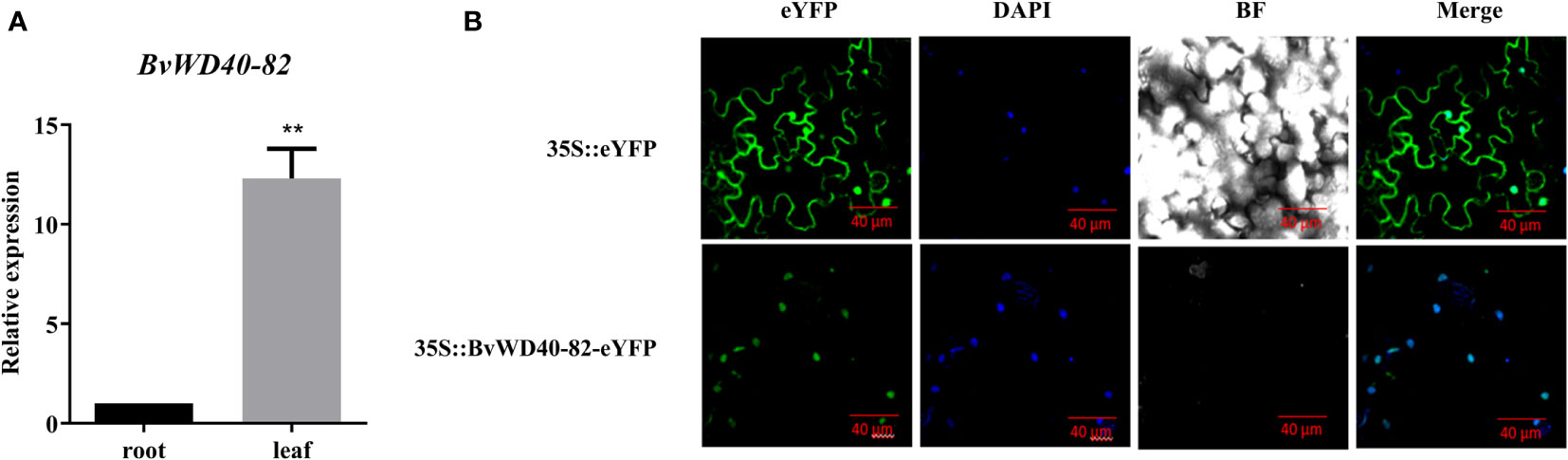
Figure 5 Tissue specificity analysis of BvWD40-82 gene and subcellular localization of BvWD40-82 protein. (A) qRT-PCR analysis of BvWD40-82 expression levels in different tissues, using 18sRNA gene as the reference gene, the data are the mean ± SD (n = 3), ** represents the chi-square test at p<0.01 significant difference. (B) Subcellular localization of BvWD40-82 protein in tobacco with 35S::eYFP as blank control.
Generation of BvWD40-82 CRISPR mutant and overexpression transgenic Arabidopsis
For functional characterization, we identified the most homologous gene of BvWD40-82 in A. thaliana, AtUTP18 (AT5G14050). Due to the lack of AtUTP18 mutants in public repositories, we generated a mutant of AtUTP18 using a CRISPR/Cas9 method (Pawluk et al., 2016). Sequencing results showed that the utp18 mutant had a single peak indicative of an inserted thymine at 872 bp of the CDS of the AtUTP18 gene (only one exon), causing a frameshift mutation and termination at 879 bp (knockout, KO) (Supplementary Figure 5). In addition, we have also created BvWD40-82 overexpression (OE) Arabidopsis and complementation line of the utp18 mutant (CO). Genotyping and RT-PCR results showed that the BvWD40-82 gene was detected at the DNA group level and RNA level in OE and CO, but not in WT and KO, and AtUTP18 was knocked out in KO (Supplementary Figure 5). The results proved that transgenic plants and the knockout mutant are reliable and can be used for subsequent functional analysis.
Phenotypic analysis of transgenic plants, WT and KO under salt stress
The roots of OE and CO lines were longer than those of WT and KO under both the control and salt treatment conditions. These results indicate that the BvWD40-82 gene could promote the root growth of plants at the seedling stage, and at the same time, increase the tolerance of plants to salt stress by promoting root growth (Figures 6A, B). Surprisingly, the KO and WT exhibited similar root growth under the control and salt stress conditions. This may be attributed to potentially redundant functions of AtUTP18 homologs in the Arabidopsis roots.
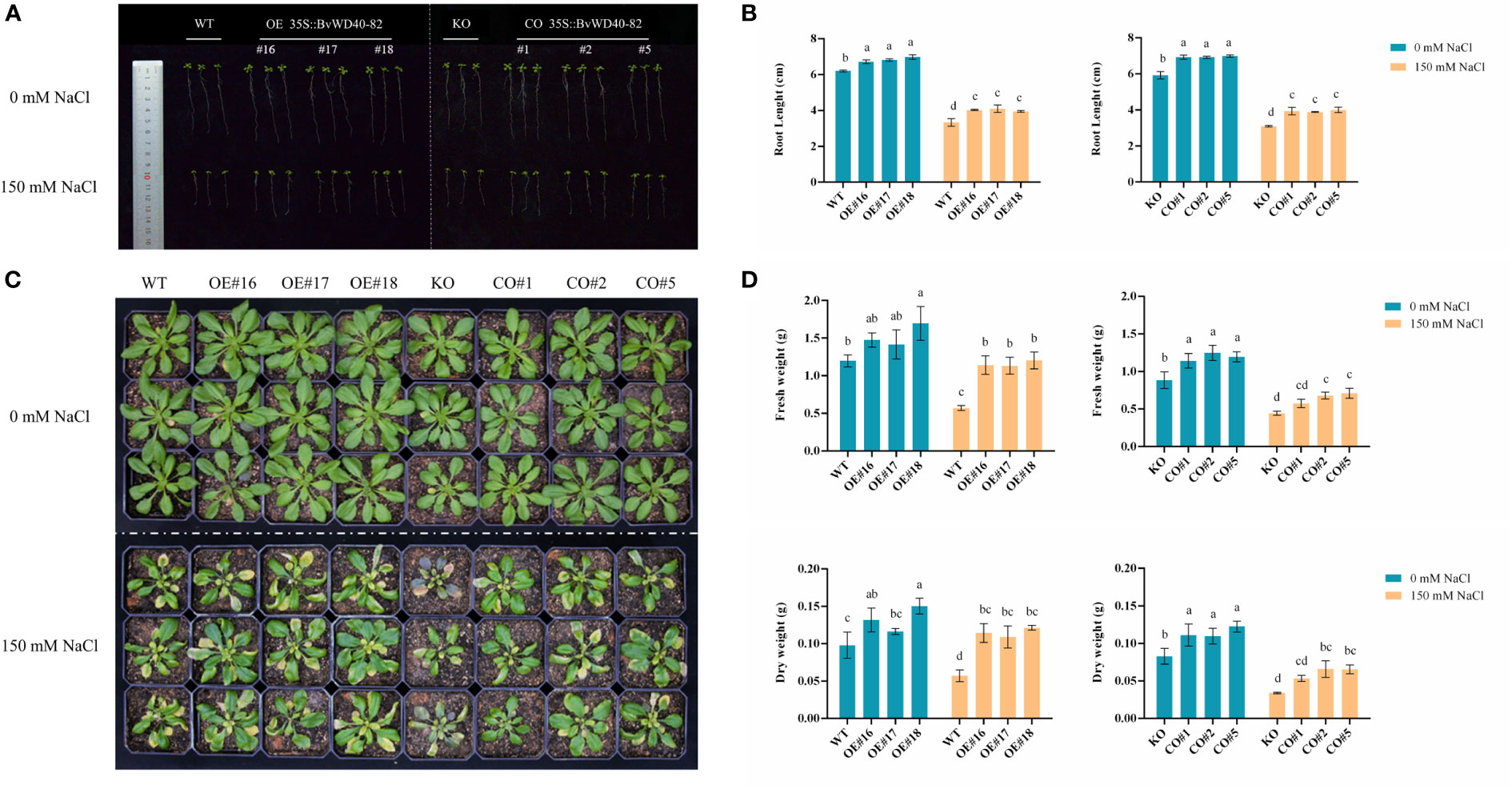
Figure 6 Phenotype analysis of BvWD40-82 gene in Arabidopsis salt tolerance. (A, B), Root length analysis of transgenic Arabidopsis seedlings before and after salt treatment. (C, D). Comparison of growth status, fresh weight, and dry weight of transgenic Arabidopsis before and after salt treatment. OE represents overexpression lines, CO represents complementation lines, KO represents knockout mutant, and WT represents wild-type plants.
In contrast to the early seedling stage of Arabidopsis, at the rosette stage the growth parameters (fresh weight (FW) and dry weight (DW)) of the mutant line showed significant differences from the WT and transgenic lines. The leaves of the KO line withered the most, and showed a stress-related black-purple color (Figure 6C), while the FW and DW of WT, OE and CO lines were higher than the mutant line under both the control and salt stress conditions (Figure 6D). Compared to WT plants, the FW and DW of OE and CO lines were significantly higher (Figure 6D). These results showed that BvWD40-82 may increase the biomass of transgenic lines under salt stress.
Physiological, biochemical and salt-stress gene expression analyses
In addition to growth phenotypes, here we profile physiological and biochemical changes, as well as salt-stress responsive pathways in the transgenic, WT and CO plants. There were no significant differences in Ca2+ contents and K+/Na+ content ratios between the transgenic lines (Figures 7A, B), WT, and KO lines under control. Under salt stress, the Ca2+ contents and K+/Na+ ratios of the OE lines and CO lines were significantly higher than those of WT and KO lines. Under control, the MDA and betaine contents in the WT and KO did not show significant differences (Figures 7C, D). Under salt stress, the MDA contents of the OE and CO lines were significantly lower than those of the WT and KO, and the differences of betaine contents were opposite to those of the MDA in the different plants. Under control, the SOD and POD activities of the transgenic lines, WT and KO lines were not significantly different. Under the salt stress, the SOD and POD activities of OE lines and CO lines were significantly higher than those of WT and KO plants (Figures 7E, F).
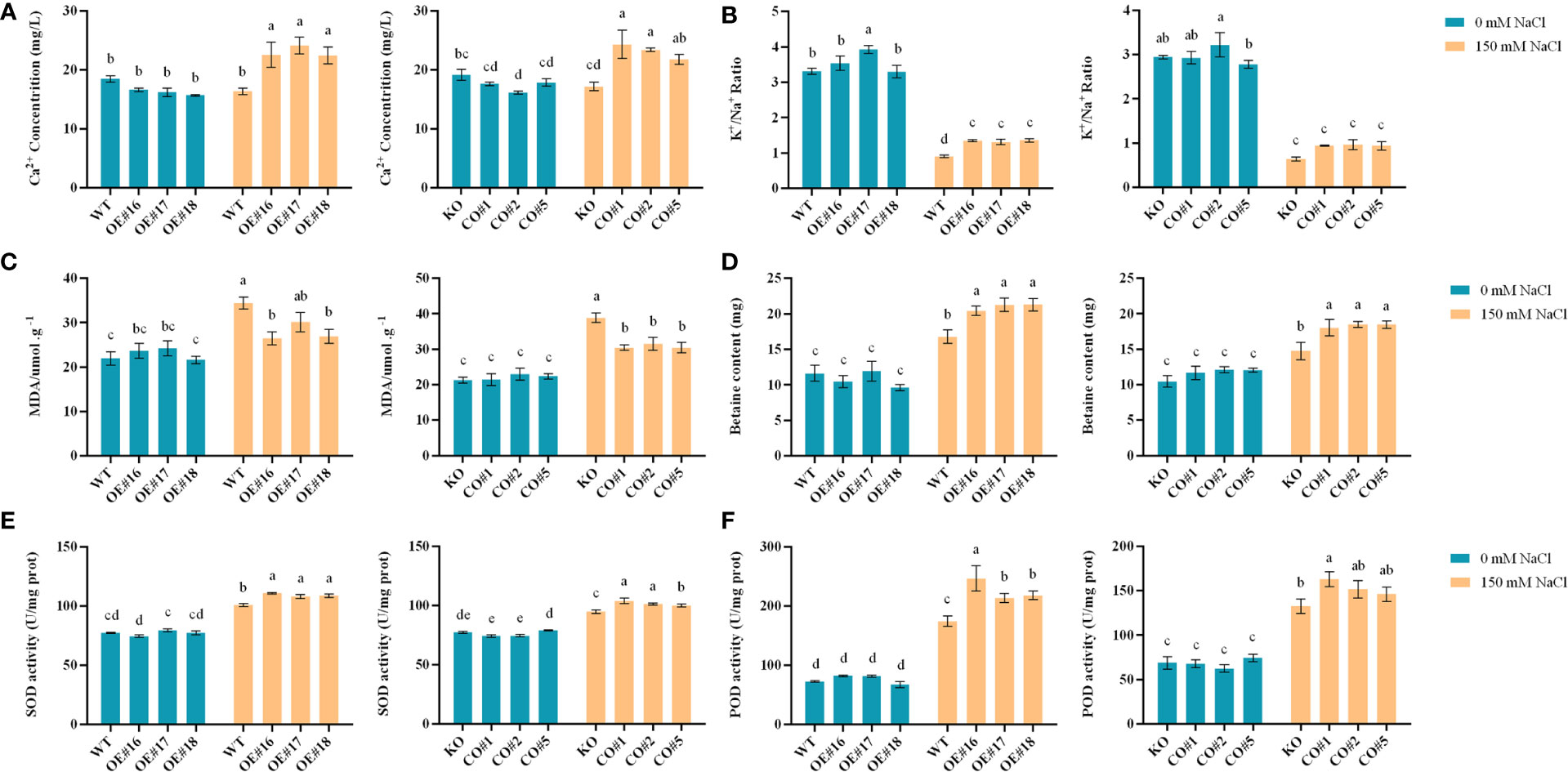
Figure 7 Physiological and biochemical analyses of different plants including OE, CO, KO and WT. (A-F). The Ca2+ content, K+/Na+ content ratio, MDA content, betaine, SOD, and POD enzyme activity of each plant. Lowercase letters indicate significant difference between different groups.
To test the potential salt-stress response pathways affected by the BvWD40-82 gene in plant response to salt stress, we selected the salt-overly-sensitive (SOS) pathway and ABA pathway and measured the expression levels of key genes in the pathways. Under control, the expression levels of relevant genes in each line were similar, while under salt stress the expression levels of SOS pathway-related genes SOS1, SOS2, and SOS3 (Figure 8A) in OE lines and CO lines were significantly higher than those of the WT and KO lines. With the exception of PYL6, the expression levels of PYL4 and PYL5 in the ABA pathway-related genes were induced by salt stress, and they were significantly higher in the OE and CO lines than those in the WT and KO (Figure 8B).
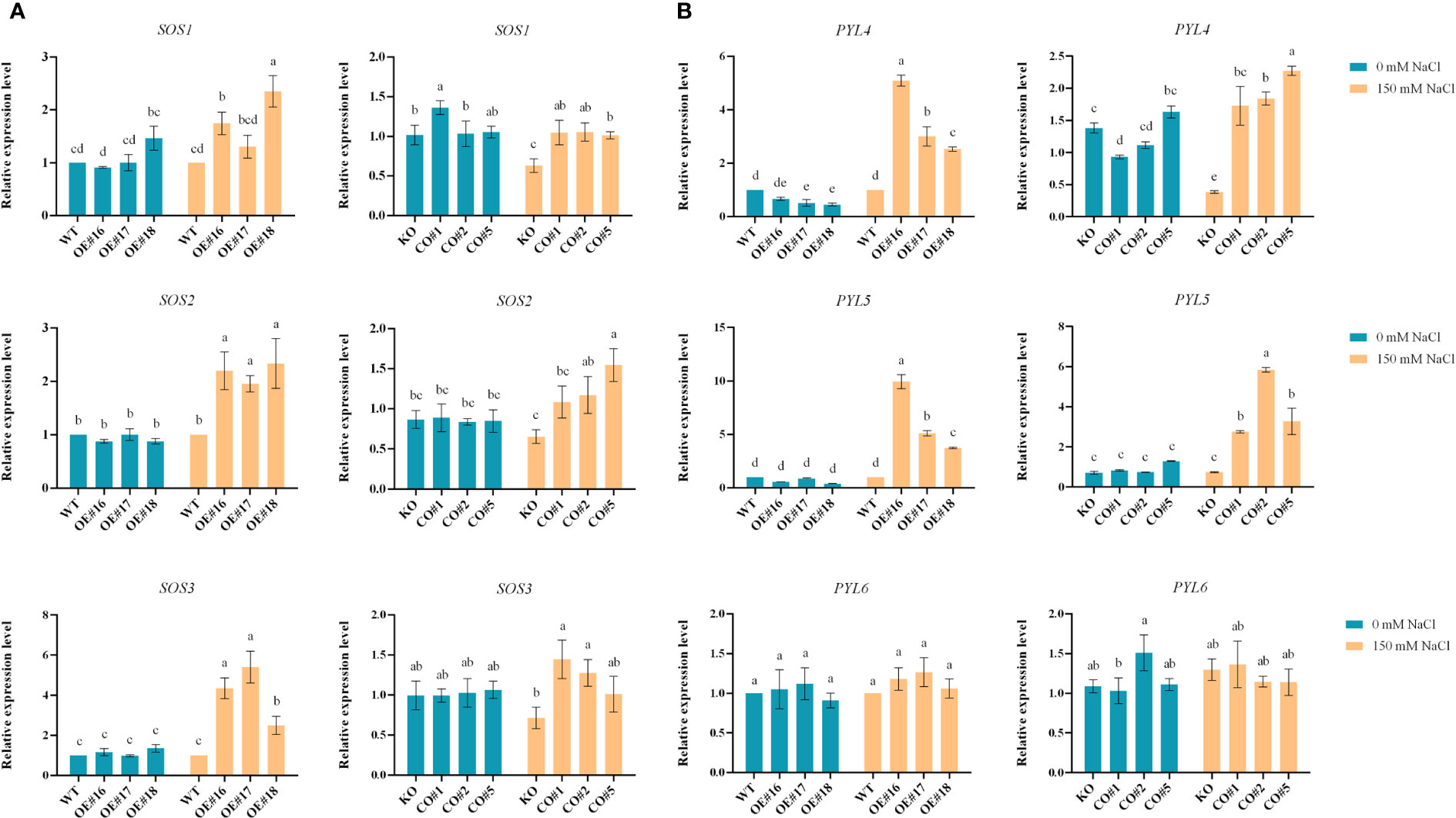
Figure 8 Gene expression analysis of potential BvWD40-82 mediated salt stress pathways. (A) SOS-related pathway expression analysis. (B) ABA-related pathway expression analysis. Lowercase letters indicate significant difference between different groups.
Discussion
Evolution of the WD40 protein family in sugar beet
Sugar beet is an important sugar crop in the world and salt stress can seriously compromise its yield. Therefore, the specific roles of WD40 protein in sugar beet and whether WD40 protein affects its salt tolerance and yield remain to be investigated. In this study, bioinformatics tools and public databases were used to conduct a whole genome analysis of the WD40 protein family in sugar beet. A total of 177 BvWD40s were obtained, and the number was smaller than most plant WD40 protein families, such as rice (Ouyang et al., 2012), cucumber, and Arabidopsis (Li et al., 2014). Although their genome sizes are smaller than sugar beet, they have more WD40 proteins. Previous studies have shown that duplication events are the main reason for the large family size (Cannon et al., 2004). Thus, it may be logical to deduce that the sugar beet genome had fewer duplication events than those three species (Supplementary Figure 6). The topological structure of the evolutionary tree divided the BvWD40 proteins into 14 groups. The composition of the domains in the same group was similar, indicating that the closely related BvWD40s may have redundant or cooperative functions (Feng et al., 2019). The phylogenetic tree also reflects the differences in physicochemical properties of BvWD40s, which are similar to WD40 proteins in other species (Sun et al., 2020; Chen et al., 2023).
To explore the evolutionary relationship between WD40 genes, Arabidopsis WD40 genes and sugar beet WD40 genes were used to construct a collinearity map. In both sugar beet and Arabidopsis, only a small amount of WD40 genes is produced by duplication events (4 pairs and 28 pairs, respectively), while most WD genes did not undergo gene duplication events, indicating that WD40 is a relatively old gene family. It also indicated that the WD40 genes were present before the differentiation between sugar beet and Arabidopsis, and has a certain diversity due to a series of duplication events after differentiation (Yang et al., 2020). This result is consistent with the results of other studies (Ouyang et al., 2012; Hu et al., 2018).
The BvWD40 genes are involved in many biological processes
To further investigate the possible function of BvWD40 proteins in sugar beet, its gene ontology (GO) was annotated. GO annotation and enrichment results indicate that the functions of BvWD40s were diverse, including response to salt stress. The enrichment results were similar to those of potato (Tao et al., 2019), barley (Chen et al., 2023), cotton (Salih et al., 2018), and rose (Sun et al., 2020), indicating species conservation. Functions in plant development, ubiquitination, organ composition, microtubule cleavage, signal transduction, protein binding, nucleotide binding, stimulation response, and other biological functions were enriched, indicating the functional diversity of the WD40 genes. Compared with WD40 genes of other species, a large number of BvWD40s were enriched in heat response, salt response, as well as in raffinose synthesis and inositol synthesis. Accumulation of raffinose was reported to contribute to the salt tolerance of sugar beet (Naguib et al., 2021), and Populus inositol transporter gene PtINT1b can enhance the salt tolerance of the transgenic plants (Zhang et al., 2023). Thus, BvWD40s may have a unique role in salt stress tolerance. Other enriched functions of BvWD40s also showed correlation with salt stress, such as peroxisome biogenesis, TOR signal transduction, MAPK complex, Katanin complex, RNA binding, autophagy, and ubiquitination. Previous studies have shown that OsPEX11, a peroxisome biogenesis factor in rice, contributes to salt stress tolerance in rice (Cui et al., 2016), TOR signaling is necessary during plant stress (Haq et al., 2022), and MAPK can mediate salt stress signal transduction in plants (Wu et al., 2023). AtKATANIN1, which encodes a microtubule cutting protein, regulates microtubule depolymerization in response to salt stress in Arabidopsis (Yang et al., 2019). OsRGG1, a gene coding for a γ subunit of G protein, promotes salt tolerance in rice by promoting ROS removal (Swain et al., 2017). An RNA binding protein MUG13.4 can interact with AtAGO2, and the MUG13.4-AtAGO2 complex plays an important role in the salt tolerance of Arabidopsis (Wang et al., 2019). Previous reports have also shown that autophagy plays an important role in plants coping with adversity, and plants adapt to environmental stresses by selective protein degradation through ubiquitination (Xu and Xue, 2019; Raffeiner et al., 2023). The GO results of BvWD40 proteins are exciting, and consistent with other studies (Sun et al., 2020; Chen et al., 2023). They not only show the potential functional diversity of the BvWD40 proteins, but also provide important insight into understanding the roles of the BvWD40 proteins in sugar beet salt tolerance.
Subfamily classification supporting the role of BvWD40s in salt stress response and tolerance
It has been widely recognized that the protein domains are highly correlative to their functions (Lees et al., 2016). Therefore, studying the domain compositions of the BvWD40 proteins has potential in predicting their functions and generating testable hypotheses. In this study, 177 BvWD40s were divided into 13 subfamilies according to the composition of their domains. It was noted that subfamily C possesses typical ubiquitination domains such as UBOX, FBOX and RING, indicating involvement in ubiquitination. Previous studies have shown that a soybean GmPUB21 containing UBOX domain negatively regulates drought and salt stress tolerance in soybean (Yang et al., 2022). The RING zinc finger protein with RING domain as an E3 ligase plays an important role in plant growth and abiotic stress tolerance (Han et al., 2022). In addition, AtSDR containing FBOX domain is involved in abiotic stress in Arabidopsis (Li et al., 2022a). Ten BvWD40s were found to contain ubiquitination domains in sugar beet. We hypothesized that these BvWD40s may play a role in abiotic stress tolerance through the activities of proteasomes. It was also noted that subfamily K has only one member, annotated as cellulose synthesis based on its domain function. Plants under salt stress may cope with the damage caused by salt stress by regulating the synthesis and deposition of cell wall (Dabravolski and Isayenkov, 2023). In addition, subfamily A has the largest number of BvWD40s, and previous reports have confirmed that the WD40 protein containing only the WD40 domain positively responds to salt stress in plants. For example, mango MiTTG1 coding WD40 protein plays an important role in promoting the development of root length and root hair, and the transgenic line has a stronger ability to adapt to salt and drought stresses (Tan et al., 2021). An LbTTG1 of L. bicolor can promote the growth of Arabidopsis trichome and actively exudate salt through salt glands to enhance the plant tolerance to salt stress (Yuan et al., 2019). Overexpression of TaWD40D could increase the expression of genes related to the SOS pathway in transgenic plants under salt stress, thus enhancing the tolerance of wheat to salt stress (Kong et al., 2015), and GbLWD1-like gene in Ginkgo biloba can improve the growth of transgenic poplar and increase the expression of salt-stress-related transcription factors, thus improving the salt tolerance of transgenic poplar (Xin et al., 2021). Overexpression of another OsABT gene improved rice salt stress tolerance through preventing excessive ROS accumulation, increasing intracellular K+/Na+, decreasing ABA synthesis, and activating ABA responsive gene expression and ABA signaling pathway (Wen et al., 2022).The WD40 protein REBC in quinoa is involved in the formation of epidermal bladder cells, and mutation of REBC led to salt stress sensitivity (Imamura et al., 2020). Different subfamily members with different domains may participate in multiple biological functions. Studies on some domains indicate that they may also be associated with salt stress, while studies on members of subfamily A indicate that this subfamily has a higher correlation with salt stress than other subfamilies. These results support that BvWD40 proteins may play a key role in plant salt stress tolerance.
BvWD40-82 enhanced salt tolerance in transgenic Arabidopsis
BvM14 line is an excellent germplasm resource independently created in Profession Li’s laboratory. It grows normally under high salt concentration, while the cultivated sugar beet cannot. At present, some excellent salt tolerance genes have been isolated from the BvM14 line, and they are considered to be important biomarkers for salt tolerance of sugar beet (Ma et al., 2017; Ji et al., 2019). However, no WD40-related genes have been characterized to be related to the salt tolerance of sugar beet. First, expression analysis of the BvWD40s under salt stress showed that a large number of BvWD40s responded to different concentrations of salt stress in different tissues of sugar beet, and BvWD40-82 had a significant response. Second, the BvWD40-82 contains only the WD40 domain, and belong to subfamily A, indicating that the BvWD40-82 may be involved in plant salt tolerance. Therefore, BvWD40-82 was cloned for functional studies using reverse genetics in Arabidopsis. Under salt stress, MDA can be increased significantly under salt stress, thus damaging the cell membranes (Liu et al., 2023a). The BvWD40-82 gene can promote root growth of the BvWD40-82 transgenic plants, reduce the accumulation of MDA under salt stress, thereby reducing the plant damage under salt stress, and improve the accumulation of betaine in transgenic plants to maintain the osmotic pressure of plant cells (Annunziata et al., 2019). Third, heterologous overexpression and complementation of the BvWD40-82 gene promoted Na+ efflux and inhibited K+ efflux, maintained a high K+/Na+ ratio, increase Ca2+ content, and improved plant salt tolerance (Zhao et al., 2021; Liu et al., 2023b). Fourth, BvWD40-82 positively regulated SOD and POD activities under salt stress, thus maintaining the ROS homeostasis in plants (Yang and Guo, 2018). Additionally, under salt stress, the BvWD40-82 enhanced salt tolerance through regulating the SOS signal pathways related-gene expression, activated the Na+/H+ transport channels and K+ intake (Xu et al., 2023), and increased the expression of ABA receptor genes (PYL4, PYL5) in the ABA signaling pathway (Wen et al., 2022). Functional characterization of BvWD40-82 clearly showed that the BvWD40-82 played an important role in plant response to salt stress. It can improve the salt tolerance of Arabidopsis. However, further investigation is needed to elucidate the molecular mechanisms underlying the BvWD40-82 function in plant salt tolerance.
Conclusion
This study identified 177 BvWD40 proteins from the sugar beet genome and described their gene and protein structures, chromosome distribution, and evolutionary characteristics. The response of BvWD40s to salt stress was characterized and a potential salt tolerance gene BvWD40-82 was isolated. The functions of the BvWD40-82 gene in salt tolerance were studied through a series of molecular, physiological and biochemical analyses. The BvWD40-82 gene can improve the salt tolerance of transgenic plants by increasing the contents of osmolytes and antioxidant enzyme activities, maintaining intracellular ion homeostasis and increasing the expression of genes related to SOS and ABA pathways. This study revealed the important role of BvWD40s in the response to salt stress in sugar beet and provided a theoretical basis for improving sugar beet tolerance to salt stress, and the BvWD40s may also serve as important resources for the genetic breeding of other crops.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Writing and editing: ZW and SC. Molecular experiment and assistance in editing: TZ and JL. Idea conception and supervision: BY and HL. Document check: IG and DZ. Conception and modification: SC and HL. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by International Cooperation and Exchange of the National Science Foundation of China Project (32261133530) and the National Science Foundation of China Project (32072122) and the Russian Science Foundation (23-44-00050). This work was supported by Heilongjiang Provincial Key Laboratory of Plant Genetic Engineering and Biological Fermentation Engineering for Cold Region.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1185440/full#supplementary-material
References
An, H.-J., Lee, C.-J., Lee, G.-E., Choi, Y., Jeung, D., Chen, W., et al. (2022). FBXW7-mediated ERK3 degradation regulates the proliferation of lung cancer cells. Exp. Mol. Med. 54, 35–46. doi: 10.1038/s12276-021-00721-9
Annunziata, M. G., Ciarmiello, L. F., Woodrow, P., Dell’Aversana, E., Carillo, P. (2019). Spatial and temporal profile of glycine betaine accumulation in plants under abiotic stresses. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00230
Ayadi, L., Callebaut, I., Saguez, C., Villa, T., Mornon, J.-P., Banroques, J. (1998). Functional and structural characterization of the Prp3 binding domain of the yeast Prp4 splicing factor. J. Mol. Biol. 284, 673–687. doi: 10.1006/jmbi.1998.2183
Barandun, J., Hunziker, M., Klinge, S. (2018). Assembly and structure of the SSU processome - a nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 49, 85–93. doi: 10.1016/j.sbi.2018.01.008
Bheri, M., Mahiwal, S., Sanyal, S. K., Pandey, G. K. (2021). Plant protein phosphatases: what do we know about their mechanism of action? FEBS J. 288, 756–785. doi: 10.1111/febs.15454
Cai, J., Su, L., Luo, W. (2022). WD repeat domain 62 (WDR62) promotes resistance of colorectal cancer to oxaliplatin through modulating mitogen-activated protein kinase (MAPK) signaling. Bioengineered 13, 14450–14459. doi: 10.1080/21655979.2022.2086381
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Chen, L., Zhang, X., Yang, N., Guo, J., Wang, M., et al. (2022b). Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375, eabg7985. doi: 10.1126/science.abg7985
Chen, L., Cui, Y., Yao, Y., An, L., Bai, Y., Li, X., et al. (2023). Genome-wide identification of WD40 transcription factors and their regulation of the MYB-bHLH-WD40 (MBW) complex related to anthocyanin synthesis in qingke (Hordeum vulgare l. var. nudum hook. f.). BMC Genomics 24, 166. doi: 10.1186/s12864-023-09240-5
Chen, S., Li, D., Chen, S., He, J., Wang, Z., Yang, G., et al. (2022a). Identifying and expression analysis of WD40 transcription factors in walnut. Plant Genome 15, e20229. doi: 10.1002/tpg2.20229
Choi, S. H., Cho, K., Kim, E. S., Yoo, H. Y. (2022). Proline-serine-threonine-repeat region of MDC1 mediates Chk1 phosphorylation and the DNA double-strand break repair. Int. J. Biochem. Cell Biol. 143, 106152. doi: 10.1016/j.biocel.2021.106152
Cui, P., Liu, H., Islam, F., Li, L., Farooq, M. A., Ruan, S., et al. (2016). OsPEX11, a peroxisomal biogenesis factor 11, contributes to salt stress tolerance in Oryza sativa. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01357
Dabravolski, S. A., Isayenkov, S. V. (2023). The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1118313
Fan, Z., Zhai, Y., Wang, Y., Zhang, L., Song, M., Flaishman, M. A., et al. (2022). Genome-wide analysis of anthocyanin biosynthesis regulatory WD40 gene FcTTG1 and related family in Ficus carica l. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.948084
Feng, R., Zhang, C., Ma, R., Cai, Z., Lin, Y., Yu, M. (2019). Identification and characterization of WD40 superfamily genes in peach. Gene 710, 291–306. doi: 10.1016/j.gene.2019.06.010
Gao, X., Chen, Z., Zhang, J., Li, X., Chen, G., Li, X., et al. (2012). OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta 235, 713–727. doi: 10.1007/s00425-011-1532-7
Gao, Y., Lu, Y., Wu, M., Liang, E., Li, Y., Zhang, D., et al. (2016). Ability to remove na+ and retain k+ correlates with salt tolerance in wwo maize inbred lines seedlings. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01716
Han, G., Qiao, Z., Li, Y., Yang, Z., Wang, C., Zhang, Y., et al. (2022). RING zinc finger proteins in plant abiotic stress tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.877011
Haq, S. I. U., Shang, J., Xie, H., Qiu, Q.-S. (2022). Roles of TOR signaling in nutrient deprivation and abiotic stress. J. Plant Physiol. 274, 153716. doi: 10.1016/j.jplph.2022.153716
Hu, R., Xiao, J., Gu, T., Yu, X., Zhang, Y., Chang, J., et al. (2018). Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum l.). BMC Genomics 19, 803. doi: 10.1186/s12864-018-5157-0
Imamura, T., Yasui, Y., Koga, H., Takagi, H., Abe, A., Nishizawa, K., et al. (2020). A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun. Biol. 3, 513. doi: 10.1038/s42003-020-01249-w
Ji, M., Wang, K., Wang, L., Chen, S., Li, H., Ma, C., et al. (2019). Overexpression of a s-adenosylmethionine decarboxylase from sugar beet M14 increased Arabidopsis salt tolerance. IJMS 20, 1990. doi: 10.3390/ijms20081990
Ji, X. L., Zhang, M., Wang, D., Li, Z., Lang, S., Song, X. S. (2023). Genome-wide identification of WD40 superfamily in Cerasus humilis and functional characteristics of ChTTG1. Int. J. Biol. Macromol. 225, 376–388. doi: 10.1016/j.ijbiomac.2022.11.074
Khan, I., Iqbal, M., Hashim, M. M. (2019). Physicochemical characteristics and yield of sugar beet (Beta vulgaris l.) cv. “California-kws” influenced with irrigation intervals. SJA 35, 57–69. doi: 10.17582/journal.sja/2019/35.1.57.69
Kim, B., Piao, R., Lee, G., Koh, E., Lee, Y., Woo, S., et al. (2021). OsCOP1 regulates embryo development and flavonoid biosynthesis in rice (Oryza sativa l.). Theor. Appl. Genet. 134, 2587–2601. doi: 10.1007/s00122-021-03844-9
Kong, D., Li, M., Dong, Z., Ji, H., Li, X. (2015). Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 34, 395–410. doi: 10.1007/s00299-014-1717-1
Kopecká, R., Kameniarová, M., Černý, M., Brzobohatý, B., Novák, J. (2023). Abiotic stress in crop production. Int. J. Mol. Sci. 24, 6603. doi: 10.3390/ijms24076603
Kornprobst, M., Turk, M., Kellner, N., Cheng, J., Flemming, D., Koš-Braun, I., et al. (2016). Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380–393. doi: 10.1016/j.cell.2016.06.014
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lees, J. G., Dawson, N. L., Sillitoe, I., Orengo, C. A. (2016). Functional innovation from changes in protein domains and their combinations. Curr. Opin. Struct. Biol. 38, 44–52. doi: 10.1016/j.sbi.2016.05.016
Li, B. W., Gao, S., Yang, Z. M., Song, J. B. (2022a). The f-box E3 ubiquitin ligase AtSDR is involved in salt and drought stress responses in Arabidopsis. Gene 809, 146011. doi: 10.1016/j.gene.2021.146011
Li, D., Roberts, R. (2001). WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 58, 2085–2097. doi: 10.1007/pl00000838
Li, J., Yu, B., Ma, C., Li, H., Jiang, D., Nan, J., et al. (2022b). Functional characterization of sugar beet M14 antioxidant enzymes in plant salt stress tolerance. Antioxid. (Basel) 12, 57. doi: 10.3390/antiox12010057
Li, Q., Zhao, P., Li, J., Zhang, C., Wang, L., Ren, Z. (2014). Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genomics 289, 103–124. doi: 10.1007/s00438-013-0789-x
Liang, R.-P., Zhang, X.-X., Zhao, J., Lu, Q.-W., Zhu, R.-T., Wang, W.-J., et al. (2022). RING finger and WD repeat domain 3 regulates proliferation and metastasis through the wnt/β-catenin signalling pathways in hepatocellular carcinoma. World J. Gastroenterol. 28, 3435–3454. doi: 10.3748/wjg.v28.i27.3435
Liu, X., Chen, S., Du, F., Sun, L., Huang, Q., Gao, X., et al. (2023b). Insights into adaptive regulation of the leaf-petiole system: strategies for survival of water lily plants under salt stress. Int. J. Mol. Sci. 24, 5605. doi: 10.3390/ijms24065605
Liu, M., Lv, Y., Cao, B., Chen, Z., Xu, K. (2023a). Physiological and molecular mechanism of ginger (Zingiber officinale Roscoe) seedling response to salt stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1073434
Liu, Y., Ma, W., Niu, J., Li, B., Zhou, W., Liu, S., et al. (2020). Systematic analysis of SmWD40s, and responding of SmWD40-170 to drought stress by regulation of ABA- and H2O2-induced stomal movement in Salvia miltiorrhiza bunge. Plant Physiol. Biochem. 153, 131–140. doi: 10.1016/j.plaphy.2020.05.017
Liu, Y., Xiong, Y., Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Lorton, B. M., Harijan, R. K., Burgos, E. S., Bonanno, J. B., Almo, S. C., Shechter, D. (2020). A binary arginine methylation switch on histone H3 arginine 2 regulates its interaction with WDR5. Biochemistry 59, 3696–3708. doi: 10.1021/acs.biochem.0c00035
Ma, C., Wang, Y., Gu, D., Nan, J., Chen, S., Li, H. (2017). Overexpression of s-Adenosyl-l-Methionine synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. IJMS 18, 847. doi: 10.3390/ijms18040847
McNally, K. P., Bazirgan, O. A., McNally, F. J. (2000). Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113 (Pt 9), 1623–1633. doi: 10.1242/jcs.113.9.1623
Meng, L., Feldman, L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from arabidopsis siliques and dry seeds. Biotechnol. J. 5, 183–186. doi: 10.1002/biot.200900211
Miao, M., Zhu, Y., Qiao, M., Tang, X., Zhao, W., Xiao, F., et al. (2014). The tomato DWD motif-containing protein DDI1 interacts with the CUL4–DDB1-based ubiquitin ligase and plays a pivotal role in abiotic stress responses. Biochem. Biophys. Res. Commun. 450, 1439–1445. doi: 10.1016/j.bbrc.2014.07.011
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mishra, A. K., Puranik, S., Prasad, M. (2012). Structure and regulatory networks of WD40 protein in plants. J. Plant Biochem. Biotechnol. 21, 32–39. doi: 10.1007/s13562-012-0134-1
Mistry, B. V., Alanazi, M., Fitwi, H., Al-Harazi, O., Rajab, M., Altorbag, A., et al. (2020). Expression profiling of WD40 family genes including DDB1- and CUL4- associated factor (DCAF) genes in mice and human suggests important regulatory roles in testicular development and spermatogenesis. BMC Genomics 21, 602. doi: 10.1186/s12864-020-07016-9
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121–e121. doi: 10.1093/nar/gkt263
Mo, D., Liu, C., Chen, Y., Cheng, X., Shen, J., Zhao, L., et al. (2023). The mitochondrial ribosomal protein mRpL4 regulates notch signaling. EMBO Rep., e55764. doi: 10.15252/embr.202255764
Naguib, W. B., Divte, P. R., Chandra, A., Sathee, L., Singh, B., Mandal, P. K., et al. (2021). Raffinose accumulation and preferential allocation of carbon (14 c) to developing leaves impart salinity tolerance in sugar beet. Physiol. Plantarum 173, 1421–1433. doi: 10.1111/ppl.13420
Neer, E. J., Schmidt, C. J., Nambudripad, R., Smith, T. F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. doi: 10.1038/371297a0
Ouyang, Y., Huang, X., Lu, Z., Yao, J. (2012). Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genomics 13, 100. doi: 10.1186/1471-2164-13-100
Park, H. J., Baek, D., Cha, J.-Y., Liao, X., Kang, S.-H., McClung, C. R., et al. (2019). HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31, 37–51. doi: 10.1105/tpc.18.00721
Pawluk, A., Amrani, N., Zhang, Y., Garcia, B., Hidalgo-Reyes, Y., Lee, J., et al. (2016). Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9. doi: 10.1016/j.cell.2016.11.017
Pazhouhandeh, M., Molinier, J., Berr, A., Genschik, P. (2011). MSI4/FVE interacts with CUL4–DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 3430–3435. doi: 10.1073/pnas.1018242108
Raffeiner, M., Zhu, S., González-Fuente, M., Üstün, S. (2023). Interplay between autophagy and proteasome during protein turnover. Trends Plant Sci. 28, 698–714. doi: 10.1016/j.tplants.2023.01.013
Salih, H., Gong, W., Mkulama, M., Du, X. (2018). Genome-wide characterization, identification, and expression analysis of the WD40 protein family in cotton. Genome 61, 539–547. doi: 10.1139/gen-2017-0237
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, D.-Q., Liu, J., Xiang, Y.-H., Ye, D., Sundaresan, V., Yang, W.-C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17, 2340–2354. doi: 10.1105/tpc.105.033563
Smith, T. F., Gaitatzes, C., Saxena, K., Neer, E. J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. doi: 10.1016/S0968-0004(99)01384-5
Song, X., Xu, L., Yu, J., Tian, P., Hu, X., Wang, Q., et al. (2019). Genome-wide characterization of the cellulose synthase gene superfamily in Solanum lycopersicum. Gene 688, 71–83. doi: 10.1016/j.gene.2018.11.039
Steffens, A., Jakoby, M., Hülskamp, M. (2017). Physical, functional and genetic interactions between the BEACH domain protein SPIRRIG and LIP5 and SKD1 and its role in endosomal trafficking to the vacuole in Arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01969
Stirnimann, C. U., Petsalaki, E., Russell, R. B., Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Stone, S. L., Hauksdóttir, H., Troy, A., Herschleb, J., Kraft, E., Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30. doi: 10.1104/pp.104.052423
Sun, Y.-B., Zhang, X.-J., Zhong, M.-C., Dong, X., Yu, D.-M., Jiang, X.-D., et al. (2020). Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in rosaceae. Plant Mol. Biol. 104, 81–95. doi: 10.1007/s11103-020-01026-7
Swain, D. M., Sahoo, R. K., Srivastava, V. K., Tripathy, B. C., Tuteja, R., Tuteja, N. (2017). Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 245, 367–383. doi: 10.1007/s00425-016-2614-3
Swarna, J., Lokeswari, T. S., Smita, M., Ravindhran, R. (2013). Characterisation and determination of in vitro antioxidant potential of betalains from Talinum triangulare (Jacq.) willd. Food Chem. 141, 4382–4390. doi: 10.1016/j.foodchem.2013.06.108
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tamaki, H., Konishi, M., Daimon, Y., Aida, M., Tasaka, M., Sugiyama, M. (2009). Identification of novel meristem factors involved in shoot regeneration through the analysis of temperature-sensitive mutants of arabidopsis. Plant J. 57, 1027–1039. doi: 10.1111/j.1365-313X.2008.03750.x
Tan, L., Salih, H., Htet, N. N. W., Azeem, F., Zhan, R. (2021). Genomic analysis of WD40 protein family in the mango reveals a TTG1 protein enhances root growth and abiotic tolerance in Arabidopsis. Sci. Rep. 11, 2266. doi: 10.1038/s41598-021-81969-z
Tao, N., Zhu, W., Gan, M., Chen, M., Li, T., Tendu, A., et al. (2019). Genome-wide identification and functional analysis of the WDR protein family in potato. Biotech. 9, 432. doi: 10.1007/s13205-019-1965-4
Thiruvengadam, M., Chung, I.-M., Samynathan, R., Chandar, S. R. H., Venkidasamy, B., Sarkar, T., et al. (2022). A comprehensive review of beetroot (Beta vulgaris l.) bioactive components in the food and pharmaceutical industries. Crit. Rev. Food Sci. Nutr., 1–33. doi: 10.1080/10408398.2022.2108367
Tian, G., Wang, S., Wu, J., Wang, Y., Wang, X., Liu, S., et al. (2023). Allelic variation of TaWD40-4B.1 contributes to drought tolerance by modulating catalase activity in wheat. Nat. Commun. 14, 1200. doi: 10.1038/s41467-023-36901-6
Tripathi, A. K., Singh, K., Pareek, A., Singla-Pareek, S. L. (2015). Histone chaperones in arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 15, 42. doi: 10.1186/s12870-015-0414-8
Wang, H., Liu, C., Ren, Y., Wu, M., Wu, Z., Chen, Y., et al. (2019). An RNA-binding protein MUG13.4 interacts with AtAGO2 to modulate salinity tolerance in Arabidopsis. Plant Sci. 288, 110218. doi: 10.1016/j.plantsci.2019.110218
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wen, D., Bao, L., Huang, X., Qian, X., Chen, E., Shen, B. (2022). OsABT is involved in abscisic acid signaling pathway and salt tolerance of roots at the rice seedling stage. IJMS 23, 10656. doi: 10.3390/ijms231810656
Williams, D. R., McIntosh, J. R. (2005). Mcl1p is a polymerase α replication accessory factor important for s-phase DNA damage survival. Eukaryot. Cell 4, 166–177. doi: 10.1128/EC.4.1.166-177.2005
Wu, J., Liang, X., Lin, M., Lan, Y., Xiang, Y., Yan, H. (2023). Comprehensive analysis of MAPK gene family in Populus trichocarpa and physiological characterization of PtMAPK3-1 in response to MeJA induction. Physiol. Plant 175, e13869. doi: 10.1111/ppl.13869
Xin, Y., Wu, Y., Han, X., Xu, L. (2021). Overexpression of the Ginkgo biloba WD40 gene GbLWD1-like improves salt tolerance in transgenic Populus. Plant Sci. 313, 111092. doi: 10.1016/j.plantsci.2021.111092
Xiong, Q., Li, W., Li, P., Yang, M., Wu, C., Eichinger, L. (2018). The role of ATG16 in autophagy and the ubiquitin proteasome system. Cells 8, 2. doi: 10.3390/cells8010002
Xu, X., Wan, W., Jiang, G., Xi, Y., Huang, H., Cai, J., et al. (2019). Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Mol. Plant 12, 1598–1611. doi: 10.1016/j.molp.2019.07.001
Xu, F.-C., Wang, M.-J., Guo, Y.-W., Song, J., Gao, W., Long, L. (2023). The Na+/H+ antiporter GbSOS1 interacts with SIP5 and regulates salt tolerance in Gossypium barbadense. Plant Sci. 330, 111658. doi: 10.1016/j.plantsci.2023.111658
Xu, F., Xue, H. (2019). The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 42, 2931–2944. doi: 10.1111/pce.13633
Yamada, Y., Schaap, P. (2021). The proppin Bcas3 and its interactor KinkyA localize to the early phagophore and regulate autophagy. Autophagy 17, 640–655. doi: 10.1080/15548627.2020.1725403
Yang, J., An, B., Luo, H., He, C., Wang, Q. (2019). AtKATANIN1 modulates microtubule depolymerization and reorganization in response to salt stress in Arabidopsis. IJMS 21, 138. doi: 10.3390/ijms21010138
Yang, Y., Guo, Y. (2018). Unraveling salt stress signaling in plants: salt stress signaling. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yang, Y., Karthikeyan, A., Yin, J., Jin, T., Ren, R., Fang, F., et al. (2022). The E3 ligase GmPUB21 negatively regulates drought and salinity stress rsponse in soybean. IJMS 23, 6893. doi: 10.3390/ijms23136893
Yang, L., Su, D., Chang, X., Foster, C. S. P., Sun, L., Huang, C.-H., et al. (2020). Phylogenomic insights into deep phylogeny of angiosperms based on broad nuclear gene sampling. Plant Commun. 1, 100027. doi: 10.1016/j.xplc.2020.100027
Yuan, F., Leng, B., Zhang, H., Wang, X., Han, G., Wang, B. (2019). A WD40-repeat protein from the recretohalophyte Limonium bicolor enhances trichome formation and salt tolerance in Arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01456
Zhang, Y., Feng, S., Chen, F., Chen, H., Wang, J., McCall, C., et al. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20, 1437–1455. doi: 10.1105/tpc.108.058891
Zhang, L., Guo, W., Lu, Y., Zhou, T., Wang, Y., Tang, X., et al. (2023). Genome-wide characterization of the inositol transporters gene family in Populus and functional characterization of PtINT1b in response to salt stress. Int. J. Biol. Macromol. 228, 197–206. doi: 10.1016/j.ijbiomac.2022.12.233
Zhang, D., Wang, Y., Shen, J., Yin, J., Li, D., Gao, Y., et al. (2018). OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 11, 45. doi: 10.1186/s12284-018-0232-3
Zhang, M., Zhang, G.-Q., Kang, H.-H., Zhou, S.-M., Wang, W. (2017). TaPUB1, a putative E3 ligase gene from wheat, enhances salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Physiol. 58, 1673–1688. doi: 10.1093/pcp/pcx101
Keywords: sugar beet (Beta vulgaris L.), WD40 proteins, expression profile, salt stress, BvWD40-82, function
Citation: Wu Z, Zhang T, Li J, Chen S, Grin IR, Zharkov DO, Yu B and Li H (2023) Genome-wide analysis of WD40 protein family and functional characterization of BvWD40-82 in sugar beet. Front. Plant Sci. 14:1185440. doi: 10.3389/fpls.2023.1185440
Received: 13 March 2023; Accepted: 10 May 2023;
Published: 02 June 2023.
Edited by:
Rupesh Tayade, Kyungpook National University, Republic of KoreaReviewed by:
Dinakaran Elango, Iowa State University, United StatesGhazala Nawaz, Kohat University of Science and Technology, Pakistan
Javaid Akhter Bhat, Nanjing Agricultural University, China
Copyright © 2023 Wu, Zhang, Li, Chen, Grin, Zharkov, Yu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Yu, eWJnaXJsMTIzNEBzaW5hLmNvbQ==; Haiying Li, bHZ6aDMwMDBAc2luYS5jb20=
 Zhirui Wu1
Zhirui Wu1 Inga R. Grin
Inga R. Grin Bing Yu
Bing Yu Haiying Li
Haiying Li