- 1ICAR-Indian Institute of Wheat and Barley Research, Regional Station, Shimla, Himachal Pradesh, India
- 2Division of Crop Improvement, ICAR-Directorate of Cashew Research, Puttur, Karnataka, India
- 3ICAR-Indian Agricultural Research Institute, Indore, Regional Station, Madhya Pradesh, India
- 4Genetics and Plant Breeding Group, Agharkar Research Institute, Pune, India
- 5All India Coordinated Research Project on Wheat & Barley, University of Agricultural Sciences, Dharwad, Karnataka, India
- 6Mahatma Phule Krishi Vidyapeeth, Rahuri, Agricultural Research Station, Niphad, Maharashtra, India
- 7JNKVV, Zonal Agricultural Research Station, Powarkheda, Narmadapuram, Madhya Pradesh, India
- 8Crop Improvement Division, ICAR-Indian Institute of Wheat and Barley Research, Karnal, Haryana, India
Wheat stem rust, caused by Puccinia graminis f. sp. tritici (Pgt), has re-emerged as one of the major concerns for global wheat production since the evolution of Ug99 and other virulent pathotypes of Pgt from East Africa, Europe, Central Asia, and other regions. Host resistance is the most effective, economic, and eco-friendly approach for managing stem rust. Understanding the virulence nature, genetic diversity, origin, distribution, and evolutionary pattern of Pgt pathotypes over time and space is a prerequisite for effectively managing newly emerging Pgt isolates through host resistance. In the present study, we monitored the occurrence of stem rust of wheat in India and neighboring countries from 2016 to 2022, collected 620 single-pustule isolates of Pgt from six states of India and Nepal, analyzed them on Indian stem rust differentials, and determined their virulence phenotypes and molecular genotypes. The Ug99 type of pathotypes did not occur in India. Pathotypes 11 and 40A were most predominant during these years. Virulence phenotyping of these isolates identified 14 Pgt pathotypes, which were genotyped using 37 Puccinia spp.-specific polymorphic microsatellites, followed by additional phylogenetic analyses using DARwin. These analyses identified three major molecular groups, demonstrating fewer lineages, clonality, and long-distance migration of Pgt isolates in India. Fourteen of the 40 recently released Indian wheat varieties exhibited complete resistance to all 23 Pgt pathotypes at the seedling stage. Twelve Sr genes were postulated in 39 varieties based on their seedling response to Pgt pathotypes. The values of slow rusting parameters i.e. coefficient of infection, area under disease progress curve, and infection rates, assessed at adult plant stage at five geographically different locations during two crop seasons, indicated the slow rusting behavior of several varieties. Six Sr genes (Sr2, Sr57, Sr58, Sr24, Sr31, and Sr38) were identified in 24 wheat varieties using molecular markers closely linked to these genes. These findings will guide future breeding programs toward more effective management of wheat stem rust.
Introduction
Wheat (Triticum spp.) with over 760 million tons of annual global production from a total area of more than 219 million hectares is one of the major food crops that provide the majority of calories worldwide (FAO, 2021). Several biotic and abiotic stresses, reducing cultivable fertile land and rapid climate change conditions, are the major limiting factors for global food production. Therefore, ensuring global food and nutrition security is frightening with a soaring world population, which is expected to reach 9 billion by 2050 (Rosegrant and Agcaoili, 2010). Among several constraints to wheat production, biotic stresses including fungal and insect damage could cause up to 21.5% yield reduction (Savary et al., 2019). Furthermore, climate change effects on temperature, relative humidity, rainfall, photoperiod, wind speed, and direction could potentially upset global wheat production directly or indirectly by maneuvering the growth, multiplication, pathogenesis, dissemination, and survival of plant pathogens (Prasad et al., 2021b).
Wheat rusts caused by Puccinia spp. are among the most threatening and economically important crop diseases. Stem or black rust caused by Puccinia graminis (Pers.) f. sp. tritici Eriks. & E. Henn. (Pgt) is one of the most detrimental diseases of wheat and has continued to threaten global wheat production since time immemorial. It can destroy all the crops if the infection happens in the early crop season and could cause 100% yield loss (Singh et al., 2011). Several stem rust epidemics were documented in the major wheat-growing regions of the world until the late 1950s (Saari and Prescott, 1985). After that era, this disease was efficiently managed through the extensive use of host resistance and eradication of alternate host, barberry (Berberis vulgaris L.), especially in Europe and the USA. One of the most important historical events during this period was the introduction of rye (Secale cereale L.) gene (1B/1R translocation), carrying Lr26/Sr31/Yr9/Pm8, into bread wheat (Mettin et al., 1973; Zeller, 1973). The 1B/1R translocation contributed 12%–20% yield jump and imparted resistance to major biotic and abiotic stresses (Cox et al., 1995). This introduction, popularly known as Sr31 alone or with other Sr genes, kept the stem rust fungus under control for several decades. However, such a contentment did not last for too long and an Sr31 virulent isolate, named TTKSK based on the North American nomenclature system and widely known as Ug99, was detected from Uganda during 1998 (Pretorius et al., 2000). The emergence of Ug99 race, virulent on at least 80% of world’s wheat varieties grown during that period, has augmented the risk of stem rust epidemics worldwide (Singh et al., 2011). The Ug99 and 15 known variants in its lineage have been reported from 14 countries in East Africa and the Middle East (https://rusttracker.cimmyt.org/?page_id=22; Singh et al., 2011; Patpour et al., 2016; Prasad et al., 2016; Nazari et al., 2021). The occurrence and movement of Ug99, its variants, and other new races caused severe localized stem rust outbreaks and epidemics in East Africa (Olivera et al., 2015), Europe (remained wheat stem rust-free since the mid- 20th century; Lewis et al., 2018), and Central Asia (Shamanin et al., 2020), indicating that stem rust is re-emerging as a challenging menace to wheat production worldwide.
Wheat stem rust is largely prevalent in nearly 7 million hectares of Central and Southern Indian states (Bhardwaj et al., 2019). India has witnessed several stem rust outbreaks in the past. Some notable epidemics were reported from Central India during 1786, 1805, 1827, 1828–1829, 1831–1832, 1879, 1894–1895, and 1907. Subsequently, a devastating stem rust outbreak was reported from the southern district of Bombay in 1946–1947 and again from Central India during 1948–1949 (Nagarajan and Joshi, 1975). Since then, stem rust was kept under control through the effective monitoring of pathogen variability and deployment of stem rust-resistant wheat cultivars and no epidemic was witnessed during last seven decades (Bhardwaj et al., 2019). Nevertheless, efforts must continue to combat possible future stem rust outbreaks due to continuous evolution in local Pgt races and/or exotic incursions.
Developing and deploying stem rust-resistant wheat cultivars complemented by understanding the pathogen variability and distribution is an effective, sustainable, and environmentally safe approach to manage this disease (Prasad et al., 2020a). Monitoring the pathogen variability and distribution through continuous surveillance is critical for breeding rust-resistant wheat. The Puccinia spp. causing wheat rusts are highly dynamic and continue to evolve, through mutation, somatic recombination, introduction, or selection, to produce new isolates periodically (Patpour et al., 2016). Such evolving nature of these pathogens renders resistant varieties susceptible in a short span. Therefore, continuous breeding for rust resistance including anticipatory breeding, search for new resistance sources, and monitoring of pathogen variability, structure, and distribution are prerequisite and continuous approaches for effective stem rust management (Abebe et al., 2012). The present study reports the following: (i) the distribution pattern of Pgt isolates in the Indian sub-continent from 2016 to 2022, (ii) virulence-based phenotypic and microsatellite marker-based genotypic diversity among these isolates, (iii) stem rust response of recently developed wheat varieties at seedling and adult plant stages at five different geographical locations, and (iv) the presence of Sr genes in these varieties postulated through gene matching technique and genetic analysis using closely linked molecular markers to these genes.
Materials and methods
Pathogenic and genetic diversity of Puccinia graminis tritici
Sampling and isolation of single-pustule fungal isolates
Wheat stem rust surveys were conducted from 2016 to 2022 in geographically different locations of Gujarat, Karnataka, Madhya Pradesh, Maharashtra, Tamil Nadu, and Uttarakhand states in India and Nepal. A total of 620 individual uredinial isolates of Pgt were collected from commercial wheat fields, trap plots, and experimental plots across these locations. All isolates were inoculated and multiplied on a stem rust-susceptible wheat cultivar, Agra Local, as described previously (Prasad et al., 2021a). To ensure the purity of individual isolates and to minimize the chances of heterogeneity within individual isolates, a well-demarcated pustule from inoculated Agra Local was excised aseptically and uredospores from it were used to inoculate new plants using a lancet needle. The inoculated plants, after 48 h of incubation in high-humidity chambers, were transferred to separate glass house chambers maintained at 25 ± 2°C. Uredospores from each isolate were collected 15 days post-inoculation (dpi) for further studies.
Identification of virulence phenotypes and pathotype designation
To determine the virulence phenotypes and pathotype designation of Pgt isolates, Indian stem rust differentials (Bahadur et al., 1985; Nayar et al., 1997; Prasad et al., 2018) were used. Indian stem rust differentials comprised 24 wheat genotypes including a universal susceptible, a line resistant to all pathotypes that serves as a control, currently cultivated wheat lines, individual Sr genes, and lines from the international stem rust differential set. Three to five seedlings of these differentials were raised to the single-leaf stage and spray inoculated with a uredospore suspension containing 5 mg uredospores/ml of non-phytotoxic isoparaffinic mineral oil Soltrol 170 (Chevron Phillips Chemical Company, USA). The inoculated seedlings were sprayed with a fine mist of water, and incubated in high-humidity chambers for 24 h. Subsequently, inoculated plants were transferred to the glasshouse chambers as described previously. The virulence phenotypes of each isolate on stem rust differentials were determined at 15 dpi using a 0–4 infection rating scale (Stakman et al., 1962) with some modifications (Nayar et al., 1997), where scores 0–2 and 3–4 were considered as avirulent and virulent reactions, respectively. Repeated virulence phenotyping of each Pgt isolate was done to confirm their identity. Pathotypes were designated according to their avirulence/virulence profiles on Indian stem rust differentials (Nagarajan et al., 1983). All 14 pathotypes identified in 620 Pgt isolates detected during the study were mass multiplied on Agra Local in separate chambers. The harvested uredospores were shade dried and stored at −20°C for genetic analysis.
SSR genotypes and phylogenetic analysis
Genetic analysis of 14 Pgt pathotypes (11, 11A, 15-1, 21, 21-1, 21A-2, 34-1, 40A, 40-2, 40-3, 42, 117, 117-6, and 122), identified during 2016–2022, was performed using Puccinia spp.-specific microsatellite/simple sequence repeat (SSR) markers. The genomic DNA was isolated from dried uredospores (~50 mg) of each pathotype using the cetyltrimethylammonium bromide (CTAB) method (Kiran et al., 2017). The quality and quantity of genomic DNA were determined using NanoDrop 2000® UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The quality of genomic DNA was also assessed through agarose gel electrophoresis (1% agarose). A set of 100 Puccinia spp.-specific SSR markers designed previously (Karaoglu et al., 2013; Prasad et al., 2017; Prasad et al., 2018; Savadi et al., 2020), synthesized in Agile Life Science Technologies (New Delhi, India), were used for genotyping 14 Pgt pathotypes. Thirty-seven markers (Supplementary Table 1), identified as polymorphic in our earlier study (Prasad et al., 2022), were selected to characterize Pgt pathotypes. Each 20 μl of the polymerase chain reaction (PCR) reaction mixture contained 25 ng of template DNA, 800 μM of dNTPs, 1X PCR buffer (10 mM Tris, pH 9.0, and 50 mM KCl), 1.5 mM MgCl2, 0.5 U Taq polymerase (HiMedia Laboratory Pvt. Ltd., Mumbai, Maharashtra, India), and 10 pmol of both forward and reverse primers. The reaction programs were set at 94°C for 2 min, followed by 35 cycles of 30 s at 94°C, 30 s at 48°C, and 1 min at 72°C, with a final extension at 72°C for 10 min in a thermal cycler (Applied Biosystems Veriti™, California, USA). After the completion of amplification, the amplified DNA was analyzed on 3% Super MT4 Agarose gel (Life Technologies India Pvt. Ltd., New Delhi, India) in 1X TBE buffer at 65–70 V for 2–3 h. DNA fragments were visualized under UV light and photographed using the gel documentation system (Bio-Rad Laboratories Inc., Hercules, California, USA).
Assessment of Indian wheat varieties for stem rust resistance
Plant and pathogen material
A set of 40 wheat varieties (Bread and Durum) recently released for commercial cultivation in India was evaluated. The central varietal release committee identified and notified these varieties based on several parameters, including yield advantage, disease resistance, and quality. The parentage, year of release, and characteristic features of these varieties are presented in Table 1. Twenty-three pathotypes (Supplementary Table 2) of Pgt with different avirulence/virulence formula were used to assess the seedling stage stem rust resistance and postulate stem rust resistance genes among selected 40 wheat varieties (Table 1). These pathotypes are being maintained in the national cereal rusts pathogen repository at the Indian Council of Agricultural Research–Indian Institute of Wheat and Barley Research (ICAR-IIWBR), Regional Station, Shimla, India.
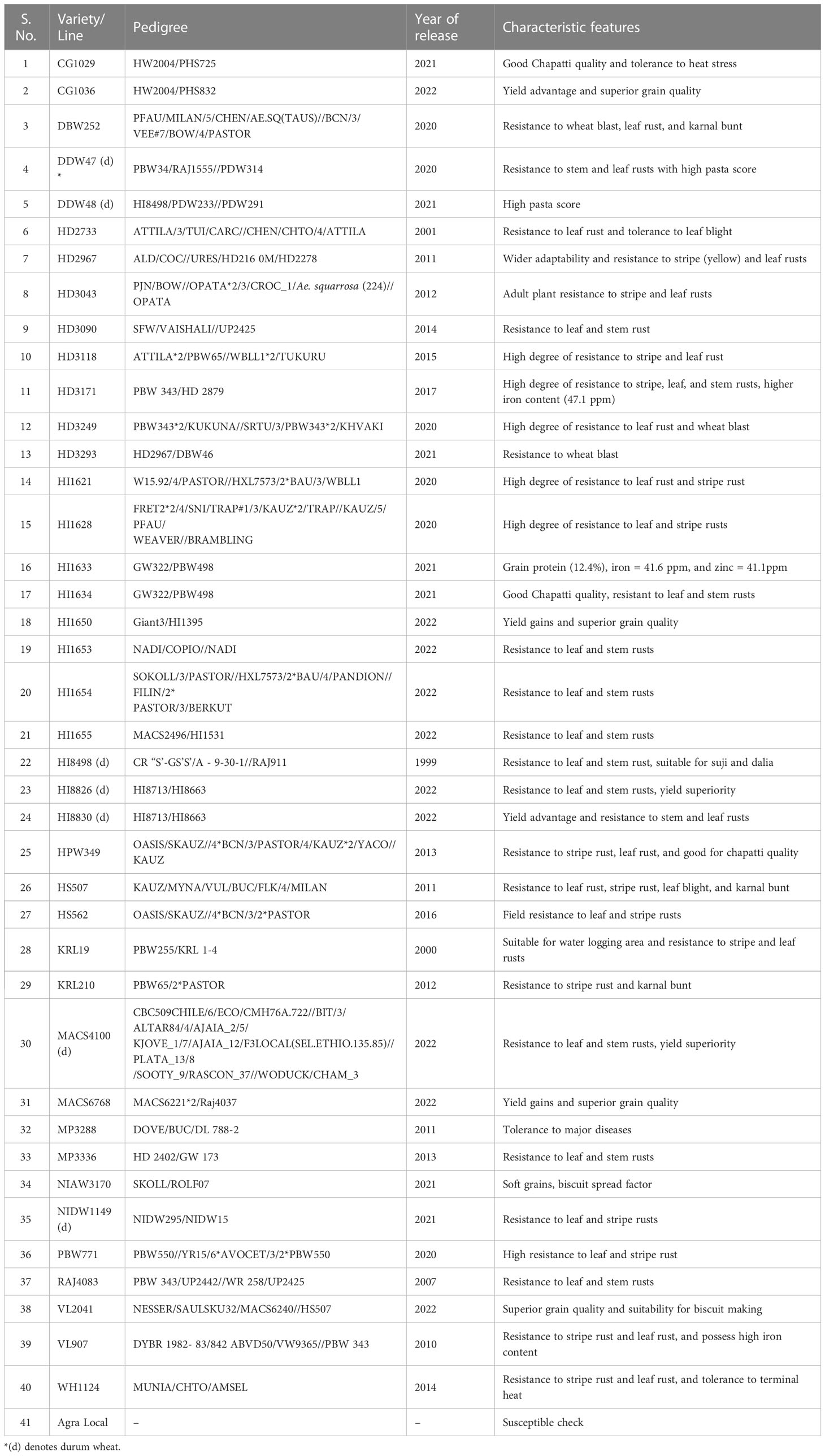
Table 1 List of the tested Indian wheat varieties and their pedigrees, year of release, and characteristic features.
Seedling response and postulation of stem rust resistance genes
Four to six seeds of each variety and a standard set of 24 stem rust differentials were sown in 10-cm pots filled with a mixture of fine loam and farmyard manure (3:1) in an environmentally controlled greenhouse at ICAR-IIWBR, Regional Station, Shimla. Stem rust differentials were included to determine the purity of all pathotypes. Each pot had four varieties in a clockwise circle. After sowing, pots were kept in isolated rust-free greenhouses at 20°C. All wheat varieties and 24 stem rust differentials were screened against 23 pathotypes of Pgt (Supplementary Table 2). The uredospore suspension (10 mg uredospores/5 ml of Soltrol) was used to inoculate 1-week-old seedlings using a glass atomizer. The inoculated seedlings were then transferred to dew chambers at 25 ± 2°C and 90%–100% relative humidity (RH) and incubated for 48 h. Subsequently, the inoculated plants were transferred to temperature and RH-controlled separate greenhouse benches maintained at 25 ± 2°C and 60%–70% RH. Seedling infection types (ITs) were recorded at 14 dpi using a 0–4 scale (Stakman et al., 1962) with slight modifications as proposed by Luig (1983). IT 0 (immune or fleck), 1 (small uredia with necrosis), and 2 (small to medium uredia with chlorosis or necrosis) indicate incompatibility, whereas IT 3 (medium-size uredia with/without chlorosis) and 4 (large uredia without chlorosis or necrosis) specify compatible interaction between a specific host and pathotype. This experiment was repeated to validate the response of these varieties against all Pgt pathotypes. The seedling response (ITs) of all 40 varieties to 23 pathotypes of Pgt was utilized to postulate Sr genes. The gene postulation was done using the gene matching technique by comparing IT arrays of all the pathotypes on test variety with those of controls with known resistance genes (Browder, 1973; Nayar et al., 1997). In case of Sr31, linkage with Lr26/Yr9 was used to infer resistance. Furthermore, micro flecking, the morphological marker linked to Sr2, was also utilized for postulating Sr2 gene (Khan et al., 2017).
Adult plant response
Adult plant stem rust response (APR) of 40 test wheat varieties and a susceptible check (Agra Local) was investigated during two crop seasons 2020–2021 and 2021–2022 at the following five geographically different stem rust hot spot locations (experimental farm): (i) ICAR-Indian Agriculture Research Institute, Regional Station, Indore, Madhya Pradesh (GPS coordinates: 22°42′31.7′′N, 75°53′29.0′′E); (ii) Jawaharlal Nehru Krishi Vishwavidyalaya, Zonal Agricultural Research Station, Powarkheda, Madhya Pradesh (GPS coordinates: 22°42′03.8′′N, 77°44′49.8′′E); (iii) Agharkar Research Institute, Hol farm, Baramati, Maharashtra (GPS coordinates: 18°07′07.8′′N, 74°18′26.8′′E); (iv) Mahatma Phule Krishi Vidyapeeth, Agricultural Research Station, Niphad, Maharashtra (GPS coordinates: 20°06′20.7′′N, 74°04′26.7′′E); and (v) University of Agricultural Sciences, Dharwad, Karnataka (GPS coordinates: 15°29′29.6′′, 74°58′34.0′′E). The well-prepared and drained fields were used and recommended doses of fertilizers and other agronomic practices for respective wheat production areas were followed. Approximately 30 g of seeds/row of each test entry was sown during the regular sowing period in three rows of 3 m with 20-cm row spacing. The experiments were laid out in randomized block design with three replicates during both the crop seasons at all five locations. A double row of a spreader mixture of two universal stem rust-susceptible wheat cultivars Agra Local and Lal Bahadur was planted perpendicular to all three replicate blocks and a single row after every five test entries for each block. The uredospores suspension of most virulent and predominant Pgt pathotypes 11, 21A-2, 40A, 117-6, and 122, collected from ICAR-IIWBR, Shimla, was prepared in distilled water mixed with 0.05% Tween 20 for inoculation. Artificial stem rust epiphytotic was initiated by spray inoculation of disease spreaders twice at three and four leaf stages, respectively, with freshly prepared uredospores suspension. A fine distilled water spray was frequently done on and around inoculated spreader plants until 48 hours to get desired RH for optimum spore germination and infection.
The adult plant stem rust response of all test entries was scored as infection type and disease severity when the spreaders had 50% disease severity. Disease response and severity were scored three times at weekly intervals. The response of the test entries was expressed in six infection types (Johnston and Browder, 1966) as follows: 0 (No infection, score 0.0), R (Resistant, necrotic areas with or without minute uredia, 0.2), MR (Moderately resistant, small uredia surrounded by necrotic areas, 0.4), X (Intermediate, variable size uredia, some with necrosis or chlorosis, 0.6), MS (Moderately susceptible, medium uredia with no chlorosis, 0.8), and S (Susceptible, large uredia, no necrosis or chlorosis, 1.0). A modified Cobb scale was used to determine the percentage coverage of stem/leaves with stem rust pustules (Peterson et al., 1948).
Identification of Sr genes using molecular markers
Genomic DNA was extracted from fresh leaves of 40 wheat varieties and positive and negative controls for all screened Sr genes using the CTAB method (Bansal et al., 2010). The quality and quantity of genomic DNA were determined using NanoDrop 2000® UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The presence of four slow rusting/adult plant response (APR) (Sr2/Yr30, Sr57/Lr34/Yr18, Sr58/Lr46/Yr29, and Sr55/Lr67/Yr46) and three all stage (ASR) Sr genes (Sr24/Lr24, Sr31/Lr26/Yr9, and Sr38/Lr37/Yr17) was determined in 40 wheat varieties using closely linked molecular markers to these genes (Table 2). The PCR was performed in 20 μl of reaction mixture containing 25 ng of template DNA, 200 μM of dNTPs, 1X PCR buffer (10 mM Tris, pH 9.0, and 50 mM KCl), 1.5 mM MgCl2, 2.5 U Taq polymerase (Hi Media Lab, Mumbai, India), and 10 pmol of both forward and reverse primers. The PCR programs were set at 94°C for 5 min (initial denaturation), followed by 35 cycles of 30 s at 94°C, 30 s at marker specific annealing temperature (Table 2), and 1 min at 72°C, with a final extension at 72°C for 7 min in a thermal cycler (Applied Biosystems Veriti™, California, USA). A touchdown PCR profile (annealing temperature reduced by 1°C/cycle for first 5 cycles) was used for Xgwm533 marker (Spielmeyer et al., 2003). The PCR products were resolved on 3% high-resolution Agarose (Hi Media Lab, Mumbai, India) gel using 1× TAE buffer and pre-stained with ethidium bromide. Gel imaging was done in gel documentation system (Bio-Rad Laboratories Inc., Hercules, California, USA). The allele scoring was performed using Gene Mapper v 4.0 software (Applied Biosystems, California, USA). Separate negative and positive controls were included for each marker analysis and observations were repeated to confirm the results.
Data analysis
Based on SSR allele sizes, a multilocus allelic data matrix for each pathotype–primer combination was generated as binary data for the alleles’ presence (1) or absence (0). Polymorphic information content (PIC) value, distinguishing two locus or loci, was assessed following Anderson et al. (1993):
where n and are the numbers of alleles and frequency of the jth allele for marker f, respectively.
Based on the SSR genotype data, the genetic relationship among the Pgt pathotypes was determined using an unrooted dendrogram generated by unweighted neighbor-joining (NJ) cluster analysis in DARwin 6.0.14 (Perrier and Jacquemoud-collet, 2006). A total of 3,000 bootstraps were used to analyze the data set in DARwin 6.0.14 with no missing data option.
Stem rust resistance behavior of all test entries was assessed through the average coefficient of infection (ACI), area under disease progress curve (AUDPC), and infection rate (r-value). The average coefficient of infection (ACI) was calculated following Saari and Wilcoxson (1974). AUDPC was calculated by using the formula given by Wilcoxson et al. (1975).
where Xi = the average coefficient of infection of the ith record, Xi+1 = the average coefficient of infection of the i+1th record, t i+1 − ti = number of days between the ith record and i+1th record, and n = number of observations. The ACI and AUDPC scores were used to perform the Wilcoxon signed-rank test to rank wheat varieties based on their APR to stem rust (Xia, 2020).
The infection rate was estimated using the following formula proposed by Van der Plank (1963).
where X1 = the proportion of the infected leaf area (disease severity) at date t1, X2 = the proportion of the infected leaf area (disease severity) at date t2, and t2 − t1 = the interval in days between these dates.
Results
Occurrence of wheat stem rust from 2016 to 2022
The systematic monitoring conducted during main and off-seasons in major stem rust-prone areas indicated that the distribution of wheat stem rust was not uniform. The natural infection of stem rust was observed only in six Indian states (Gujarat, Madhya Pradesh, Maharashtra, Karnataka, Tamil Nadu, and Uttarakhand) and Nepal (Figure 1). The occurrence of stem rust was usually observed towards the crop maturity, and therefore, no stem rust epidemic was reported during this period from India or neighboring countries. The stem rust severity was low (up to 30 MS to S) on advanced wheat varieties, whereas off-types, old wheat genotypes, and other susceptible varieties suffered up to 80 S severity. Six-hundred twenty samples of wheat stem rust were collected during this period and analyzed on Indian stem rust differentials. The incidence of stem rust, reported only from Gujarat, Madhya Pradesh, Maharashtra, and Tamil Nadu, was very low during 2021–2022. The overall prevalence of wheat stem rust was maximum in Madhya Pradesh, followed by Karnataka, Gujarat, and Tamil Nadu as 33.4%, 25.4%, 17.8%, and 12.1% stem rust-infected samples, respectively, were collected from these states (Figure 2). Ug99 types of pathotypes were not observed in any sample. The pathotype analysis resulted in the identification of 14 Pgt pathotypes (11, 11A, 15-1, 21, 21-1, 21A-2, 34-1, 40A, 40-2, 40-3, 42, 117, 117-6, and 122) in these samples. Pathotypes 11 and 40A were the most predominant and were reported in 60.8% and 18.5% of samples (Figure 3), respectively. These two pathotypes were observed during all six crop seasons. Other pathotypes were missing at least in one crop season. The frequency (%) of pathotype 11, ranging from 43.05 (2016–2017) to 88.18 (2019–2020), was maximum during all six crop seasons. It was observed in all the states where stem rust appeared, whereas pathotype 40A was present in Madhya Pradesh, Karnataka, and Tamil Nadu only. The frequency (%) of pathotype 40A ranged between 4.72 (2019–2020) and 47.22 (2016–2017). Pathotypes 34-1, 42, 117, and 117-6 were observed only in one sample during 2021–2022, 2019–2020, 2017–2018, and 2017–2018, respectively (Table 3). Pathotypes 15-1, 40-2, and 40-3 were identified in 6%, 4.7%, and 3.4% of samples, respectively.
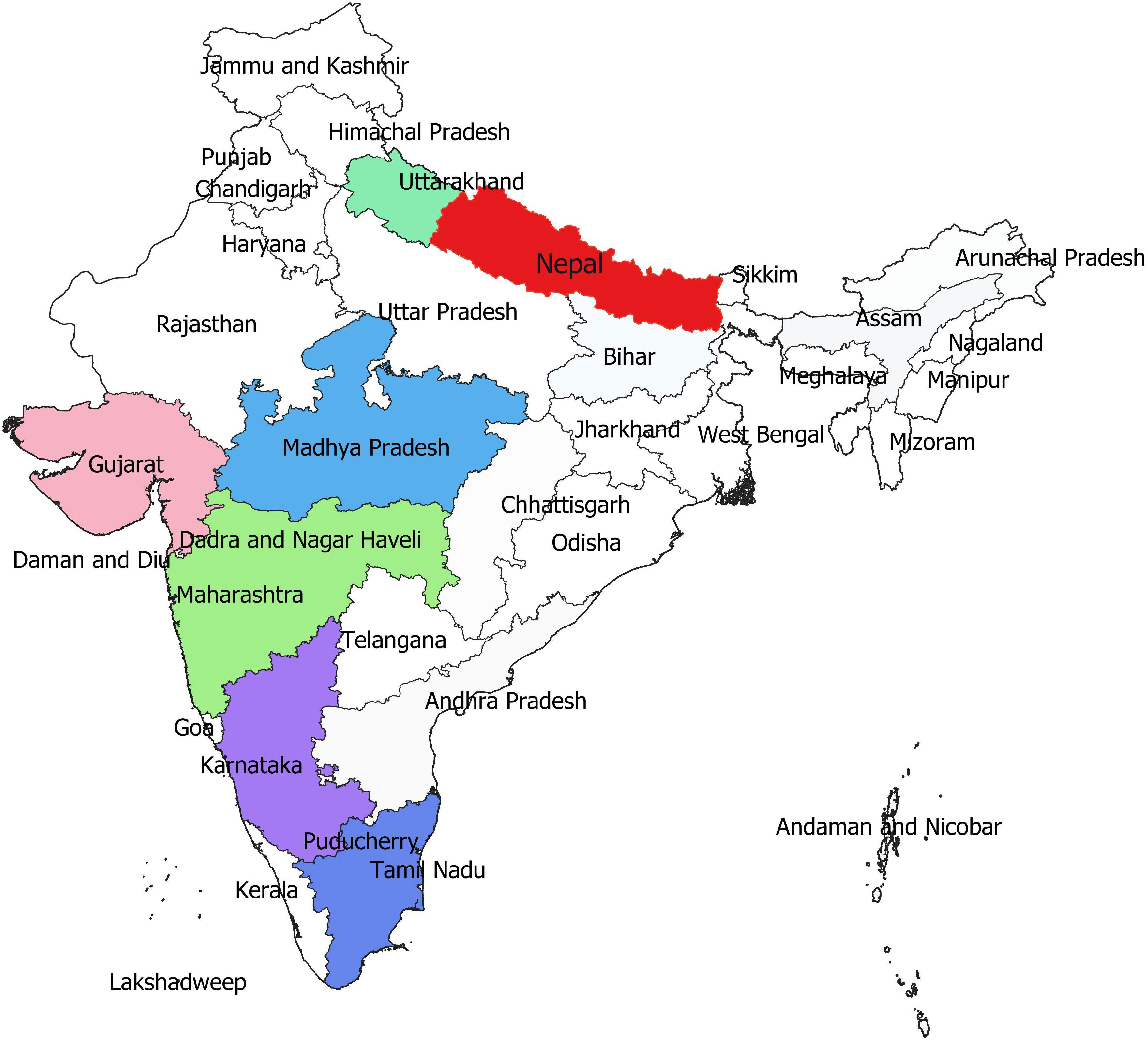
Figure 1 The distribution of wheat stem rust (highlighted in colors) in different wheat-growing areas of India and Nepal during 2016–2022.
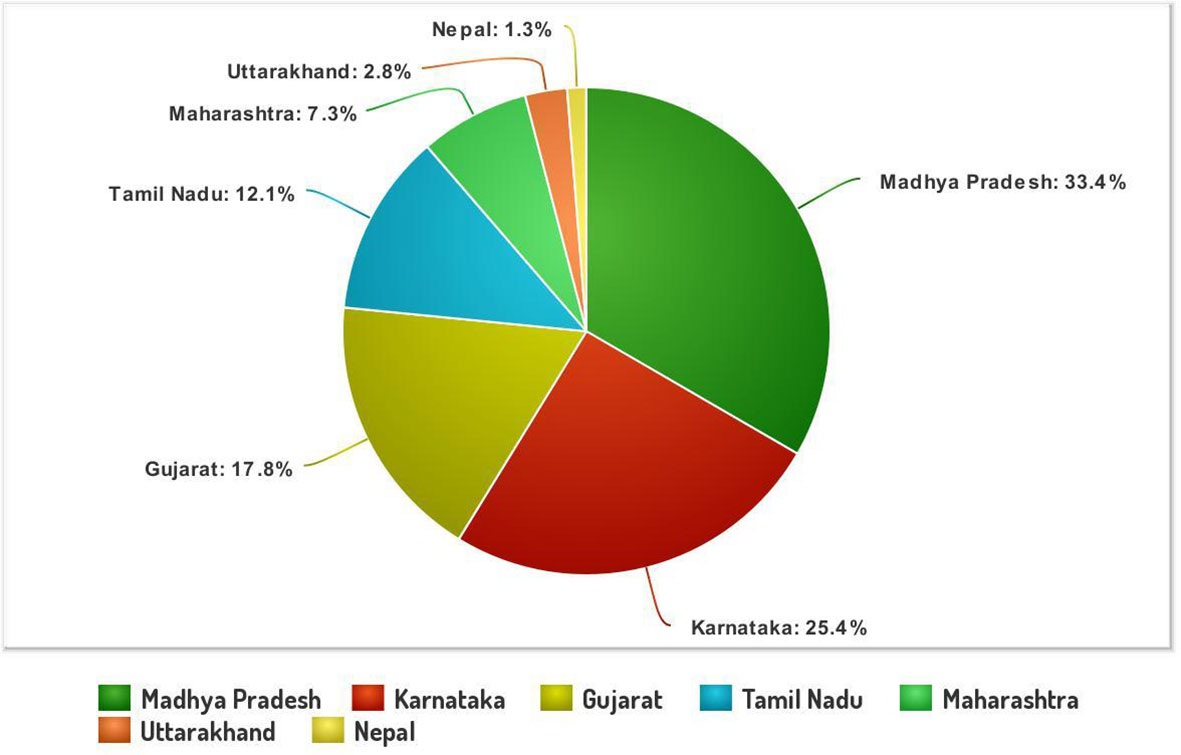
Figure 2 The stem rust samples (%) collected and analyzed from different Indian states and Nepal during 2016–2022.
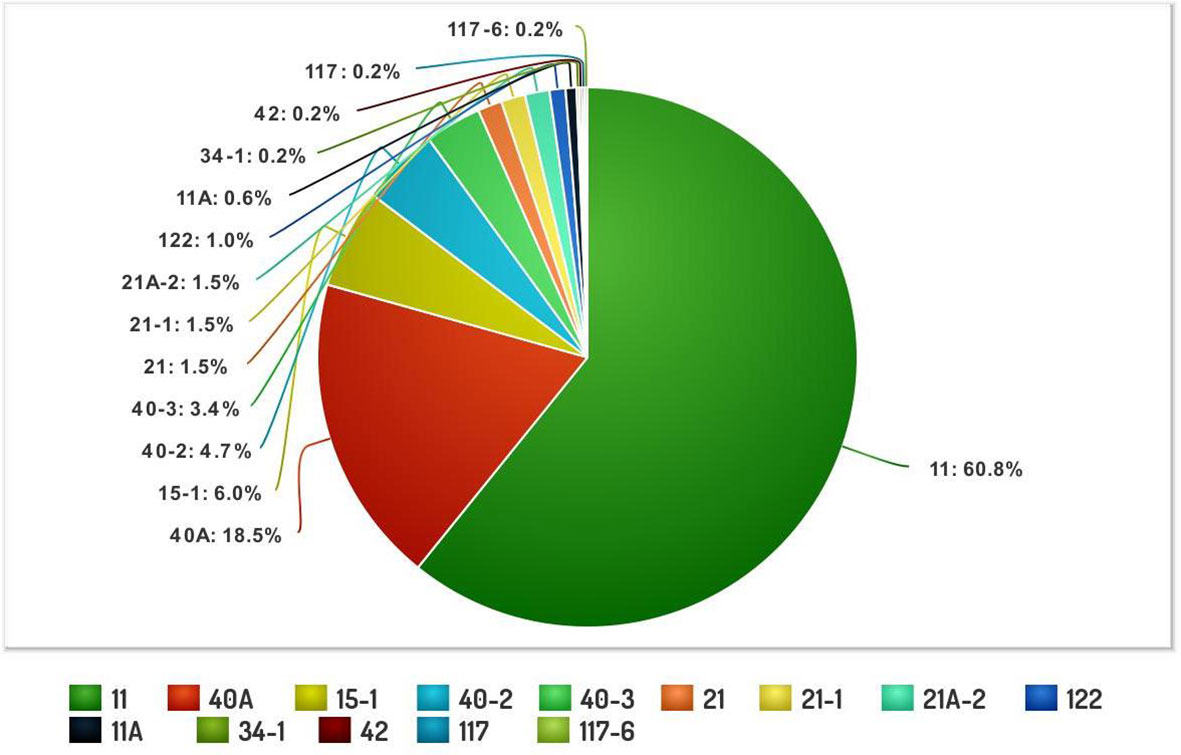
Figure 3 The composite frequency (%) of Puccinia graminis f. sp. tritici pathotypes identified from major wheat-growing regions of India and Nepal during 2016–2022.
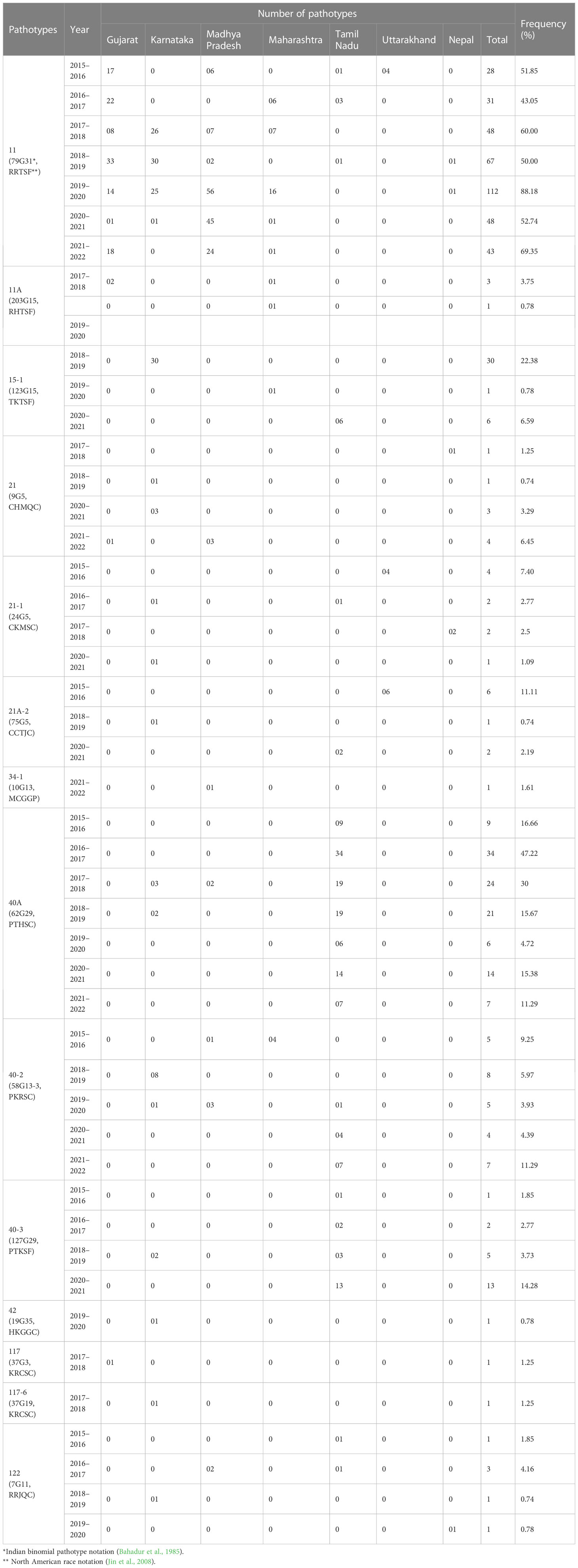
Table 3 Year-wise distribution and frequency of Puccinia graminis f. sp. tritici pathotypes identified from major wheat-growing regions of India and Nepal during 2016–2022.
SSR genotypes and phylogenetic analysis
One hundred seventy-five alleles were amplified by 37 Puccinia sp.-specific SSRs among 14 pathotypes. The number of alleles varied from two (PgSUN27, PGTG 00856, PGTG 07438, SSR-P AG-40, SSR-P CAAC- 44, and SSR-P TCCG-36) to eight (SSR-P CT-36, PtESSR22, PtESSR26, and PtESSR33) with an average of 4.8 alleles per locus. The heterozygosity of these SSRs ranged from 0.24 (SSR-P CAA-60) to 0.5 (PgSUN27 and PtESSR33). The heterozygosity of 21 SSRs was ≥48. The average polymorphic information index (PIC) value of these primers was 0.34 (Supplementary Table 1). An unweighted neighbor-joining (NJ) cluster analysis produced three distinct molecular groups (MG I to III). Seven and six pathotypes were grouped in MG I and II, respectively, while pathotype 42 with poor genetic similarity with the rest of the pathotypes formed a distinct MG III (Figure 4). The MG I and II were further divided into two subgroups (Ia, Ib; and IIa, IIb). Pathotypes 21, 21A-2, 21-1, and 40A were clustered in subgroup Ia, while pathotypes 40-2, 40-3, and 34-1 occupied subgroup Ib. Three pathotypes were clustered in subgroup IIa (117, 117-6, and 122) and subgroup IIb (11, 11A, and 15-1). Interestingly, pathotypes with nearly similar virulence patterns were grouped in one molecular group/subgroup. For example, pathotypes 21, 21A-2, and 21-1 in subgroup Ia, 11 and 11A in subgroup IIb, 117 and 117-6 in IIa, and 40A, 40-2, and 40-3 were grouped in MG I.
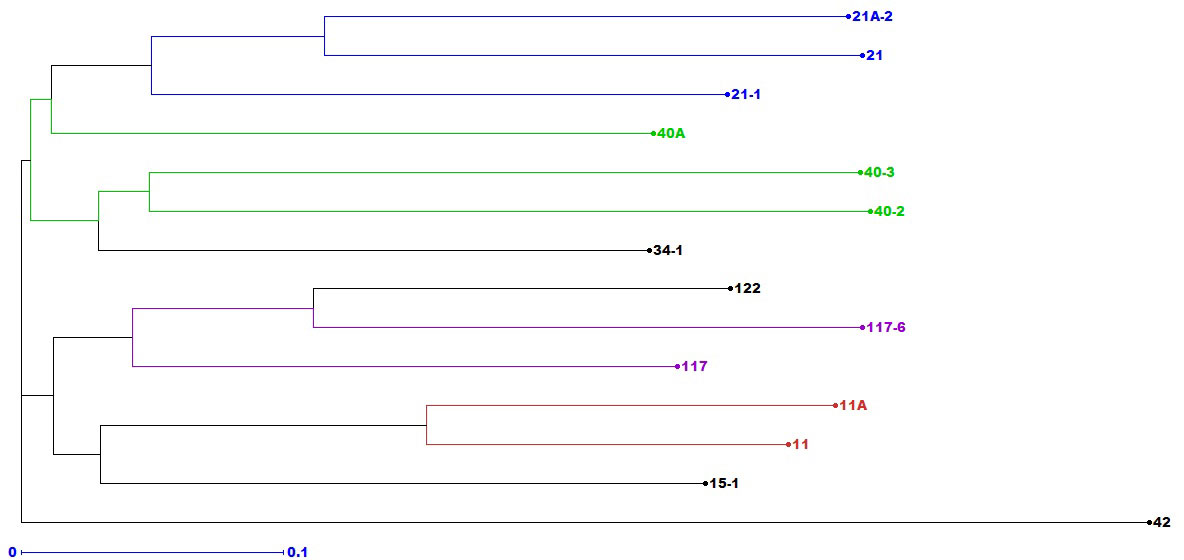
Figure 4 The simple sequence repeats-based unrooted neighbor-joining phylogenetic tree of 14 Puccinia graminis f. sp. tritici pathotypes detected during 2016–2022.
Seedling response and postulation of stem rust resistance genes
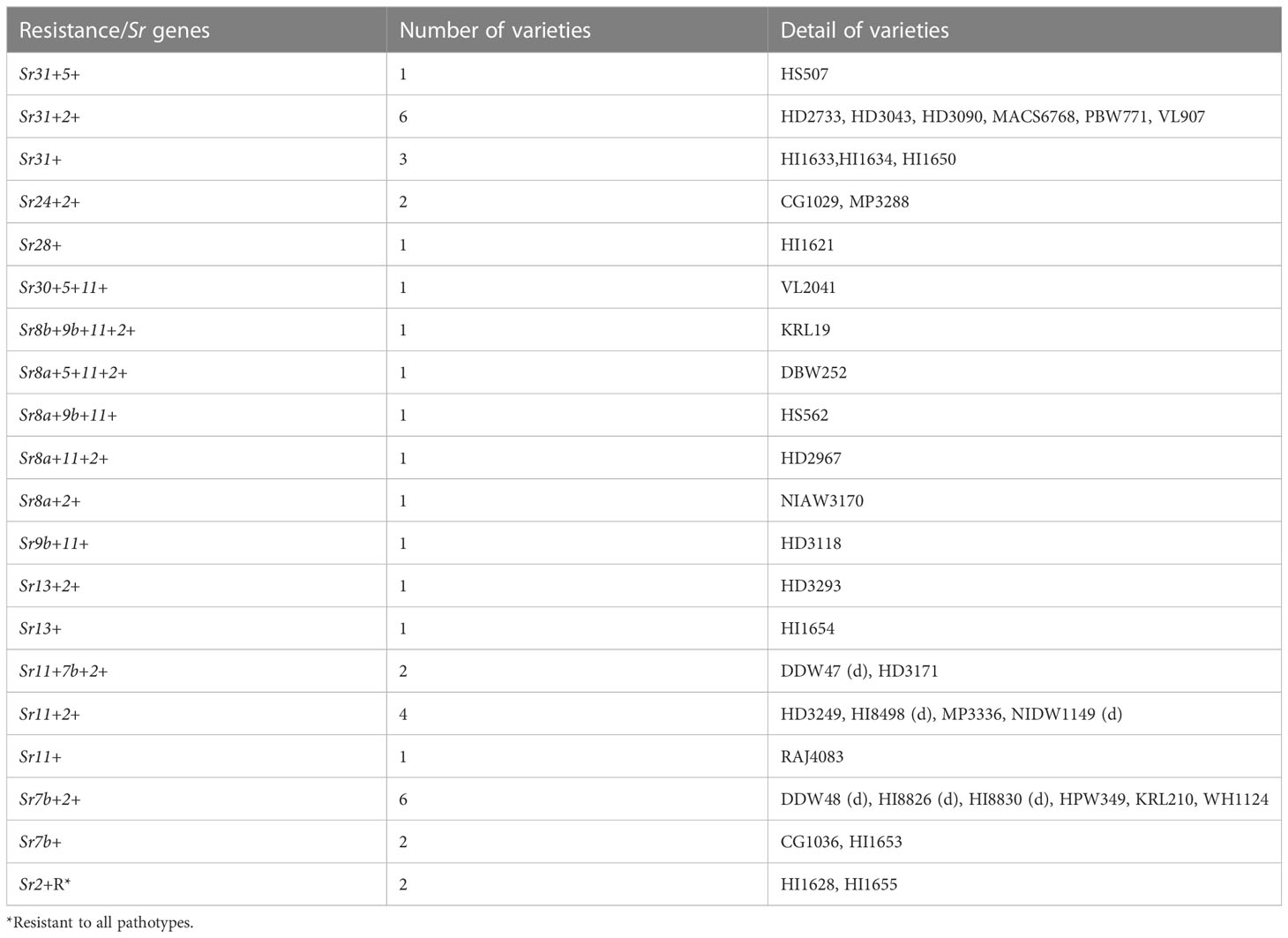
The seedling stem rust response of 40 wheat varieties against 23 Pgt pathotypes indicated substantial resistance in these varieties (Supplementary Table 3). Pathotype 40-3 with compatible reactions in 15 varieties was the most virulent. Likewise, pathotypes 11, 24A, 40-2, 117-1, 117-2, and 117-6 were virulent on six varieties, while 117A-1 was virulent on five varieties and 40-1, 184-1, and 295 on four varieties (Supplementary Table 3). Fourteen varieties exhibited resistant response to all 23 pathotypes, including the stem rust resistance conferred by Sr31 and Sr24 in 10 and 2 varieties, respectively (Table 4). Two varieties (HI1628 and HI1655), exhibiting resistance to all pathotypes, had Sr2, which was postulated based on micro flecking, an Sr2-linked morphological marker and identified through genetic analysis. We could postulate 12 Sr genes (Sr2, Sr11, Sr7b, Sr31, Sr8a, Sr5, Sr9b, Sr13, Sr24, Sr28, Sr30, and Sr8b), individually or in combination, in 39 wheat varieties. No Sr gene was postulated in MACS4100 as its resistance response to 23 pathotypes did not match with any known Sr genes. The frequency of Sr2 was highest and postulated in 67.5% of varieties. It was followed by Sr11, Sr7b, Sr31, and Sr8a, which were postulated in 32.5%, 30%, 25%, and 10% varieties, respectively. The Sr genes Sr28, Sr30, and Sr8b were postulated only in HI1621, VL2041, and KRL19 (Table 4), respectively.
Adult plant responses of wheat varieties against stem rust
The ACI, AUDPC, and apparent infection rate (r) of wheat varieties were calculated based on their screening at stem rust hot spot locations consecutively for 2 years (Table 5). There were substantial variations in stem rust severity among different varieties across the locations. According to the Wilcoxon signed-rank test, the top 10 best-performing wheat varieties had ≤3.72 ACI and ≤103.40 AUDPC scores. The ACI, AUDPC, and r-values of all wheat varieties from all five locations ranged from 1.12 (HI1655) to 38.28 (KRL210), from 33.60 (HI1655) to 1,203.40 (KRL210), and from 0.009 (HI1655 and HI8826) to 0.063 (KRL19), respectively. Based on ACI values, these wheat varieties were grouped into highly resistant (ACI ≤ 5), resistant (ACI >5 to 10), moderately resistant (ACI >10 to 15), and susceptible (ACI>15). Among 40 varieties, 18 were highly resistant, 12 were resistant, 5 were moderately resistant, and the remaining 5 were classified as susceptible to stem rust (Table 5). Contrary to test entries, the stem rust severity on Agra Local was very high (>40S) at all locations and crop seasons. It demonstrated 58.0 and 2,240.0 ACI and average AUDPC scores, respectively, which were significantly high from test varieties. Wheat variety HI1655, with the lowest AUDPC and “r” values was the most resistant among all varieties, while KRL210 was highly susceptible with high AUDPC and “r” values. HI1655 has shown immune response at multiple locations during both the years. Some durum cultivars, viz., HI8826 and HI8830, were also immune to stem rust at multiple locations. The cultivars with lower ACI values were also strong to moderate slow rusters as these had lower AUDPC values and infection rates. The KRL210 was the most vulnerable to stem rust with the highest ACI (38.28) and AUDPC (1203.4) values.
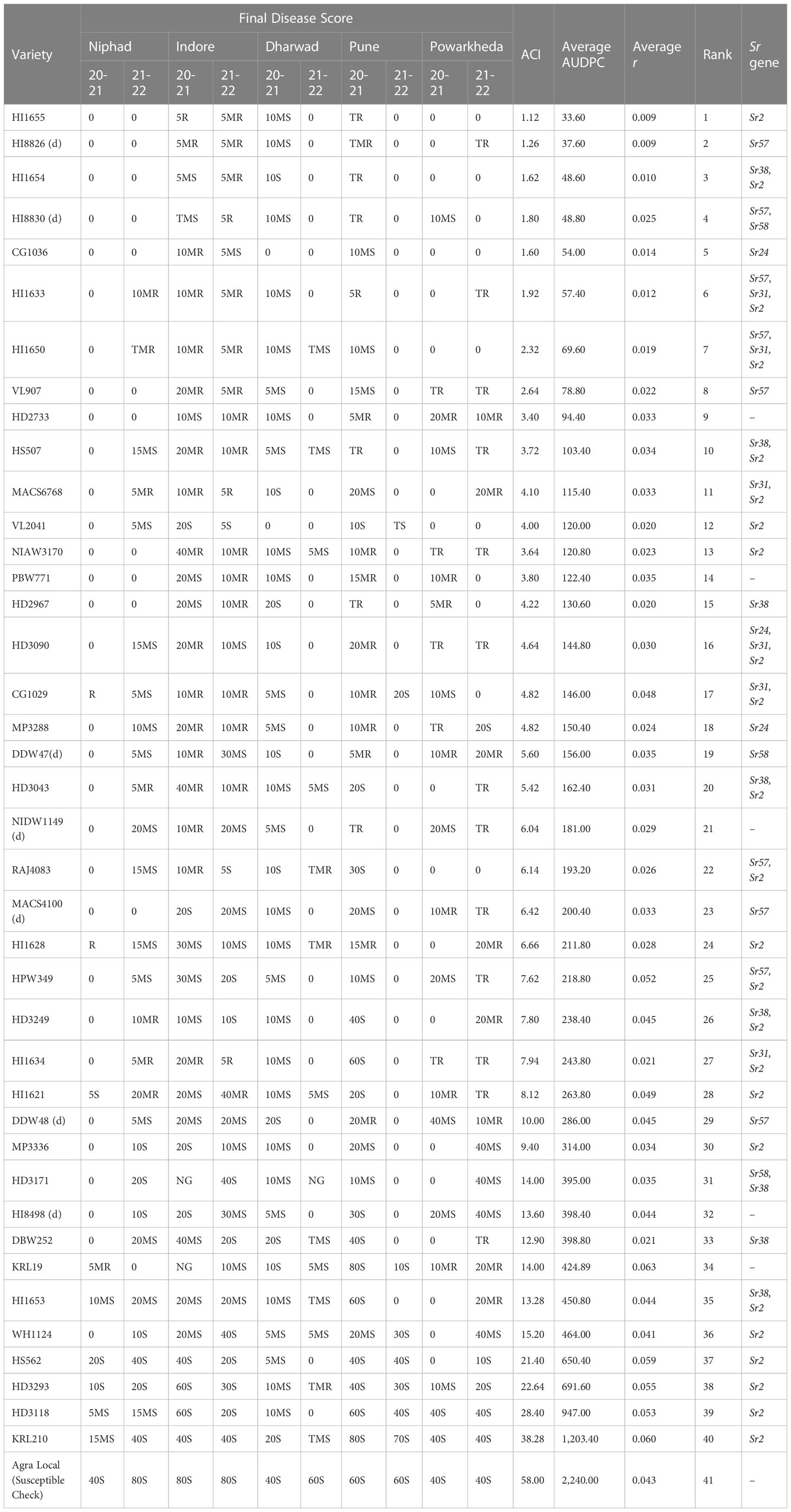
Table 5 Stem rust severity, coefficient of infection (ACI), area under disease progress curve (AUDPC), and apparent infection rate (r) of wheat varieties arranged in increasing Wilcoxon signed-rank; and detail of Sr genes identified in these varieties using molecular markers.
Identification of Sr genes using molecular markers
Genetic analysis of 40 wheat varieties using six molecular markers closely linked to selected Sr genes indicated the presence of six Sr genes (Sr2, Sr57, Sr58, Sr24, Sr31, and Sr38) in 35 varieties. The APR gene Sr55 was not identified in any of the varieties with the CFD71 marker. Sr2 gene linked marker (Xgwm533) indicated the presence of this gene in 24 varieties (Supplementary Figure 1). Other APR genes, i.e., Sr58 and Sr57, were confirmed in three and nine varieties, respectively (Table 5), while the ASR genes Sr24, Sr31, and Sr38 were identified in three, six, and eight varieties, respectively. A combination of two/three Sr genes was marked in CG1029 (Sr31 and Sr2), HD3043 (Sr38 and Sr2), HD3090 (Sr24, Sr31, and Sr2), HD3171 (Sr58 and Sr38), HD3249 (Sr38 and Sr2), HI1633 (Sr57, Sr31, and Sr2), HI1634 (Sr31 and Sr2), HI1650 (Sr57, Sr31, and Sr2), HI1653 (Sr38 and Sr2), HI1654 (Sr38 and Sr2), HI8830 (Sr57 and Sr58), HPW349 (Sr57 and Sr2), HS507 (Sr38 and Sr2), MACS6768 (Sr31 and Sr2), and RAJ4083 (Sr57 and Sr2).
Discussion
Wheat stem rust has re-emerged as a serious menace to wheat production globally since the evolution of Ug99 in Uganda. To date, about 15 variants in the Ug99 lineage have been identified within its lineage. Some of these variants have spread to several wheat-growing areas in different countries including Zimbabwe, South Africa, Sudan, Yemen, Egypt, and Iran. The Ug99 variants and other new races reported from East Africa (Digalu race; Olivera et al., 2015), Europe (Olivera Firpo et al., 2017; Lewis et al., 2018), and Central Asia (Shamanin et al., 2020) have caused severe localized stem rust outbreaks and endangered wheat productivity and food security (Lewis et al., 2018). The further spread of the races in the Ug99 lineage to major wheat-growing countries in Asia, including India where almost 7 million hectares area in Central and Southern India is highly prone to stem rust outbreaks, and beyond could devastate wheat production in the affected areas (Olivera et al., 2018). Therefore, survey for monitoring the evolution or introduction of Pgt virulences supported by virulence phenotypes and molecular genotypes is crucial and must be undertaken regularly. Moreover, replacing stem rust-susceptible wheat varieties with resistant ones is a systematic approach to minimize yield losses and thwart pathogen survival (Randhawa et al., 2018; Prasad et al., 2020b). The present study was designed to monitor the variability and distribution pattern of Pgt isolates in India and neighboring countries from 2016 to 2022 through exhaustive surveillance, virulence phenotyping, and SSR genotyping, evaluate recently released wheat varieties for stem rust response at seedling and adult plant stages during two crop seasons at five stem rust-prone geographically different areas, and to undertake genetic analysis of wheat varieties to identify Sr genes in them.
The occurrence of wheat stem rust was sporadic during the crop years 2016 to 2022. Stem rust incidence was extremely low during 2015–2016 and 2021–2022 as we could detect only a few isolates of pathotypes 11, 21-1, 21A-2, 40A, 40-2, 40-3, and 122 from 54 samples during 2015–2016, and 11, 21, 34-1, 40A, and 40-2 from 62 samples during 2021–2022 (Table 3). Most of the advanced wheat material was stem rust-free while some old genotypes and admixtures had up to 80S severity under natural conditions. The availability of Sr31 and other known and unknown Sr genes in recently developed wheat varieties, effective against all known pathotypes of stem rust in India and neighboring countries, accompanied by poor rainfall and a significant reduction in relative humidity over the years could have hampered the survival and infection rate of the natural population of Pgt and, therefore, poor stem rust incidence (Ayliffe et al., 2008; Prank et al., 2019). As revealed previously (Prasad et al., 2018; Prasad et al., 2022), the Pgt population reported from India and Nepal remained avirulent to Sr7a, Sr26, Sr27, Sr31, Sr32, Sr33, Sr39, Sr40, Sr43, SrTmp, and SrTt3 to date. Some of these genes, for instance, Sr26, have been found to be effective against the Pgt population including Ug99 globally. Hence, these genes can be targeted for prudent wheat breeding for stem rust resistance through their pyramiding with other effective major and minor genes (Singh et al., 2015). The Sr26 has been extensively used in the Australian wheat breeding program. Still, its use in India is seldom and no Indian wheat variety, including the tested varieties in this study, is reported to contain it.
A comparison of Pgt pathotypes identified in the present study revealed some dissimilarity with previous reports (Jain et al., 2013; Prasad et al., 2018). Pathotypes 42, 117, and 117-6, which were not detected during 2009–2015, reappeared during this period, whereas pathotype 40-1, virulent on Sr24, was present during 2009–2015 but did not appear afterward (Prasad et al., 2018). Pathotypes 42 (2019–2020, Karnataka), 117 (2017–2018, Gujarat), and 117-6 (2017–2018, Karnataka) made their reappearance after a long span of more than 20, 19, and 9 years, respectively (Bhardwaj et al., 2003; Bhardwaj et al., 2006; Jain et al., 2013). The Pgt pathotypes in virulence group 21, earlier used to dominate northern India and parts of Nepal, were also detected from Gujarat, Madhya Pradesh, Karnataka, and Tamil Nadu (Table 3). Likewise, a primitive Sr24 virulent pathotype 34-1, first identified from Madhya Pradesh in 1991 and prevalent on some local wheat landraces only in far north Leh Ladakh region afterwards, was identified again from Madhya Pradesh during 2021–2022. Such a shift in virulence pattern over time is not surprising, as the prevalence of some pathotypes in a particular crop season and their establishment in a new region depend on the diversity and resistance level of wheat varieties cultivated during that season (Singh, 1991), and to some extent on the prevailing environmental conditions such as temperature, rainfall, and relative humidity in particular (Roelfs et al., 1992). A similar shift in virulence pattern for stem rust of wheat has also been reported in many other countries including Canada, Mexico, the USA, Australia, and South Africa (Le Roux and Rijkenberg, 1987; Singh, 1991; Park and Wellings, 1992; Fetch, 2005; Jin, 2005; Terefe et al., 2016); for leaf (brown) rust from USA, Pakistan, India, and Egypt (Kolmer et al., 2017; Khadegah et al., 2019; Kolmer, 2019; Bhardwaj et al., 2021); and for stripe (yellow) rust from Canada, India, and other Asian countries (Hovmoller et al., 2011; Gangwar et al., 2022). There was a major shift in the frequency of predominant pathotypes of Pgt in India from 2016 to 2022. For instance, the frequency of pathotype 40A, which was the most predominant pathotype for almost three decades (Bhardwaj, 2012), reduced significantly after 2013–2014. Pathotype 11 remained most predominant since 2013–2014 throughout the current study period except during 2016–2017, when 40A surpassed pathotype 11 again (Figure 5). Although both pathotypes 40A and 11 are thought to have more virulence and better fitness potential over others, the modifications in the environmental conditions and altered varietal spectrum could have better-supported pathotype 11 over 40A during these years. Similar assumptions were made to justify the dominance of one pathotype over another in the case of sunflower rust caused by Puccinia helianthi Schw. (Prud'homme and Sackston, 1990), wheat leaf rust (Bhardwaj et al., 2014), and stripe rust (Hovmoller et al., 2011). The optimum temperature, sufficient moisture, and cultivation of diverse wheat genotypes supported maximum virulence diversity in Pgt populations during 2018–2019 and 2020–2021 (Prasad et al., 2019; Prasad et al., 2021c). Eight pathotypes with different virulence structures were identified during these two crop seasons in Gujarat, Himachal Pradesh, Karnataka, Madhya Pradesh, Maharashtra, Tamil Nadu, and Nepal.
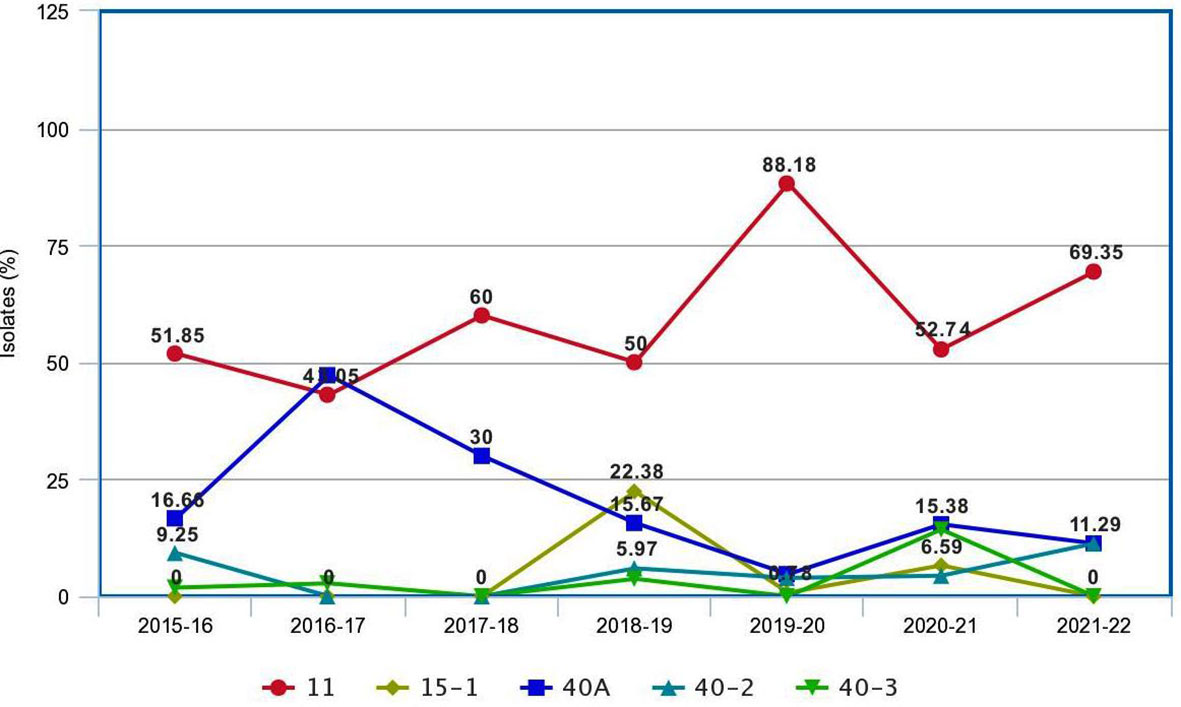
Figure 5 The pattern of occurrence of predominant pathotypes of Puccinia graminis f. sp. tritici on wheat in the Indian subcontinent during 2016–2020.
The phylogenetic analysis of 14 Pgt pathotypes, using 37 Puccinia sp.-specific polymorphic SSRs, clustered them in three molecular groups (Figure 4), indicating the existence of very few lineages in the whole Pgt population. The heterozygosity and PIC values of these SSRs suggested that these markers had sturdy differentiating power as suggested by Shete et al. (2000) and Botstein et al. (1980). The molecular grouping of Pgt pathotypes, generated through unweighted neighbor-joining (NJ) cluster analysis in DARwin 6.0.14, corroborated, up to some extent, virulence-based grouping of these pathotypes, which was in agreement with our previous findings (Prasad et al., 2022). Pathotypes 21, 21-1, and 21A-2; 11 and 11A; 117 and 117-6; and 40A, 40-2, and 40-3, sharing almost matching avirulence/virulence structure as reported previously (Prasad et al., 2018; Prasad et al., 2022), were clustered in one MG/subgroup (Figure 4). Such an association of virulence phenotypes and SSR genotypes among P. triticina (Wang et al., 2010) and P. striiformis tritici (Hovmoller et al., 2002) isolates is also reported, which is not uncommon if the genes responsible for virulence/pathogenicity are extensively distributed in pathogen genome (Ordonez and Kolmer, 2009). The comprehensive evolutionary pattern of all these Pgt pathotypes and others reported from the Indian sub-continent is already discussed previously (Prasad et al., 2022). Lower genetic diversity among Pgt pathotypes may be attributed to the clonal multiplication and long-distance migratory nature of the pathogen in the Indian sub-continent (Wingen et al., 2013).
Stem rust response of 40 wheat varieties revealed substantial resistance in these varieties against 23 pathotypes of Pgt (Supplementary Table 2) at seedling and against 11, 21A-2, 40A, 117-6, 122, and other naturally occurring isolates at adult plant stages from five geographically different locations (Supplementary Table 3; Table 5). At the seedling stage, 14 varieties exhibited resistant response against all 23 pathotypes, which included resistance conferred by Sr31 in 10 varieties and by Sr24 in 2 varieties. Despite the fact that 2 (34-1 and 40-1) of the 23 pathotypes are particularly virulent on Sr24 (Jain et al., 2013), these pathotypes were unable to establish a compatible relationship with two Sr24-containing wheat varieties (CG1029 and MP3288). Such interactions could result from the presence of some additional unknown effective Sr gene other than Sr24, which might impede pathotypes 34-1 and 40-1 from establishing compatible interactions with these varieties (Dakouri et al., 2013). The Indian population of Pgt possesses very low virulence frequencies on Sr31, Sr24, and Sr30 (Prasad et al., 2022), which are among 12 Sr genes postulated in 39 varieties (Table 4), and therefore, these varieties could be targeted for skillful gene deployment. Generally, Sr2, Sr5, Sr6, Sr7a, Sr7b, Sr8a, Sr8b, Sr9b, Sr9e, Sr11, Sr12, Sr13, Sr17, Sr21, Sr24, Sr25, Sr30, and Sr31 are frequently occurring Sr genes in Indian wheat varieties. Among these, Sr7b, Sr9e, Sr11, and Sr13 are more common in durum wheat, while Sr2, Sr11, and Sr31 are mostly postulated in bread wheat (Jain et al., 2013). The frequency of Sr2, postulated in 67.5% of varieties based on micro flecking: an Sr2-linked morphological marker (Khan et al., 2017), was the highest. The Sr2, along with other slow rusting Sr genes such as Sr55, Sr56, Sr57, and Sr58, could be a valuable resource to have and utilize in combination with other effective major genes, i.e., Sr26, for achieving durable stem rust resistance (Singh et al., 2015).
The slow rusting/APR/field response of 40 wheat varieties and a susceptible check (Agra Local) were assessed based on the valuation of the final disease score (FDS), ACI, AUDPC, and apparent infection rate. The findings showed that the tested wheat varieties had more available resistance than Agra Local, which was severely infected during both the crop years at all five locations. Generally, the slow rusting behavior in wheat has been estimated and reported through assessment of disease severity, AUDPC, CI values, and apparent infection rates (Broers et al., 1996; Pathan and Park, 2006). The FDS is also used as a parameter to assess the slow rusting character of wheat genotypes by many earlier researchers (Ali et al., 2009); however, ACI, AUDPC, and apparent infection rate are considered most appropriate and preferred. Among 40 test varieties, 30 had ACI values ≤ 17.24% and 23 had AUDPC values ≤ 8.94% of the check (Table 5), which specify high to moderate levels of slow rusting resistance character of these varieties (Ali et al., 2009). Furthermore, most of the varieties had MR/MS-type infection responses, which are the characteristics of the slow rusting genes, conferring partial field resistance (Parlevliet, 1988). The Wilcoxon signed-rank test categorized HI1655, HI8826, HI1654, HI8830, CG1036, HI1633, HI1650, VL907, HD2733, and HS507, with ≤3.72 ACI and ≤103.40 AUDPC scores, as the 10 best-performing varieties for stem rust APR. Among these, the Sr31 was postulated in HI1633, HI1650, VL907, HD2733, and HS507, based on their response to 23 Pgt pathotypes at the seedling stage and its linkage to Lr26/Yr9. Lower ACI and AUDPC values in these five varieties are implicit since Sr31 virulences are not reported from South Asian countries (Bhardwaj et al., 2019). Although Sr31 lost its effectiveness in East Africa and the Middle East due to Ug99 and other variants in its lineage, yet it is valued plus owing to its contribution to yield advantage and effectiveness to stem rust and other diseases in many countries including those in South Asia (Cox et al., 1995; Yu et al., 2014). Some weak stem rust resistance genes, i.e., Sr13+, Sr7b+2+, Sr7b+, and Sr2+, were postulated in the remaining (5) top 10 varieties HI1654, HI8826 (d) and HI8830 (d), CG1036, and HI1655, respectively. The genetic analysis of wheat germplasm using rust resistance gene linked molecular markers is another substitute to traditional gene postulation and could help in the swift and precise identification of resistance genes (Goutam et al., 2013). The molecular markers closely linked to rust resistance genes have frequently been identified and utilized in marker-assisted breeding for rust resistance (Mago et al., 2013). Besides being quick and unambiguous, this method could also circumvent the problems associated with traditional gene postulation such as plant stage-dependent gene expression, temperature, and other abiotic factor-dependent gene interactions. In the present study, genetic analysis of wheat varieties using molecular markers closely linked to Sr31 and Sr24 confirmed the gene postulation results in the majority of the cases except for Sr24 in the wheat variety CG1036 and Sr31 in CG1029. The gene postulation using the gene matching technique listed the presence of Sr7b and Sr24 in CG1036 and CG1029, respectively (Table 4). None of the six Sr genes could be identified in five varieties (HD2733, PBW771, NIDW1149, HI8498, and KRL19) through genetic analysis, indicating the presence of some other known or unknown ASR/APR genes. All these markers have been extensively used to identify Sr genes or Yr and Lr genes linked to Sr genes. For instance, marker iag95 was effectively used to identify Sr31 in some Chinese wheat cultivars (Xu et al., 2017; Xu et al., 2018). Similarly, Sr38 linked to Yr17 and Lr37 was reported from wheat cultivars in China and Nebraska using VENTRIUP-LN2, an Sr38/Yr17/Lr37 linked marker (Peng et al., 2013; Xue et al., 2014; Xu et al., 2017; Mourad et al., 2019). Yang et al. (2008) and Alma et al. (2012) identified the Sr57 gene in some Chinese and Central Asian wheat cultivars, respectively, using the csLV34 marker. The WMC44 marker was applied for the molecular analysis of slow rusting resistance gene Sr58/Lr46/Yr29 in triticale cultivars (Skowrońska et al., 2020). Herrera-Foessel et al. (2014) used CFD71 marker to detect Sr55 in wheat varieties that conferred APR to stem rust and powdery mildew in wheat. The marker Sr24#50 was used to evaluate and identify Sr24 in wheat lines from Gansu province in China (Xu et al., 2017). This marker indicated the presence of Sr24 in CG1036 contrary to gene postulation, which indicated the presence of a relatively weak Sr7b in the same variety (Tables 4, 5). The susceptibility of CG1036 to several Pgt pathotypes, other than Sr24 virulences (34-1 and 40-1, Supplementary Table 3), also ruled out the presence of Sr24 in it (Prasad et al., 2018). The SSR marker Xgwm533 has been extensively used for the identification of Sr2/Yr30 in wheat worldwide (Mago et al., 2011; Khan et al., 2017; Rahmatov et al., 2019). We could identify Sr2 in 24 wheat varieties using this marker. The Sr2 was confirmed in 13 varieties using both gene postulation and marker analysis approaches. However, when looking at specific methods individually, it was identified in 11 varieties through marker analysis and in 14 varieties through gene postulation (Tables 4, 5 and Supplementary Figure 1). Similar observations were reported previously (Khan et al., 2017). The failure of the Xgwm533 marker to validate the micro- flecking-based postulation of Sr2 in some varieties could be justified by the fact that many of these varieties might lack Sr2 and micro-flecking could have occurred due to external environmental factors influencing host–pathogen interactions (Mago et al., 2011). Another probability could be that the variety with micro-flecking might have separated from Sr2 or that the marker was not diagnostic (Rahmatov et al., 2019). Similar observations were reported for Lr34/Yr18/Pm38 linked to more accurate molecular markers, where markers reported the presence of these genes even in susceptible wheats (Lagudah et al., 2009). The collective gene postulation and marker analysis results indicated that wheat varieties HI1655 and HI1654 might harbor some effective known/unknown Sr genes other than Sr2 to confer adequate APR. Therefore, these varieties could be appraised for durable stem rust resistance breeding in wheat. More importantly, further genetic analysis of these varieties could reveal some interesting loci that confer resistance to all Indian Pgt pathotypes. The locus contributing to stem rust APR, which includes the Sr2 gene, has been found to be effective and recommended for use in breeding programs (Rutkoski et al., 2014; Singh et al., 2015). The deployment of Sr2 is largely advised in combination with other effective Sr genes as it (alone) could not offer adequate protection under epidemic situations (Haile and Roder, 2013).
In conclusion, 14 Pgt pathotypes were identified as a result of surveys on the prevalence of wheat stem rust from 2016 to 2022 and pathotype analysis of the samples gathered during the surveys. Pathotypes 11 and 40A remained the most predominant from the previous reports, while pathotypes 42, 117, and 117-6 made their reappearance after more than 9 years. Remarkable stem rust resistance, conferred by some effective known ASR (Sr24, Sr31 and Sr38) and APR (Sr2, Sr57, and Sr58) genes, as revealed by gene postulation and genetic analysis using closely linked markers to these genes, was observed among most wheat varieties. The values of adult plant slow rusting parameters in these varieties were highly encouraging and could prove useful for stem rust management through their deployment or as durable stem rust resistance sources for future breeding program. These findings would be useful in deploying appropriate resistant varieties and streamlining wheat breeding approaches to combat stem rust. These conclusions provide good opportunity to explore some novel resistances where already known slow resisting loci are not identified. In view of the dynamic nature, virulence diversity, and long-distance migration of Pgt isolates, their monitoring the shifting virulence pattern, searching for new sources of resistance, and their transfer into high-yielding cultivars must be sustained to mitigate the impact of stem rust menace on wheat.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
PP and SB conceived the experiment. PP, RT, SS, OG, CL, SA, SuK, SoK, and AM conducted the lab and greenhouse experiments. TP, SN, GH, BG, and KM conducted field experiments. PP, HK, VG, CM, and StK conducted statistical analysis. PP wrote the first draft of the article. SB, SudK, and GS edited and improved the article. All authors read the final version of the manuscript and approved it.
Acknowledgments
We acknowledge the generous funding provided by the Director, ICAR-Indian Institute of Wheat and Barley Research, Karnal, Haryana, for the rust monitoring work. We sincerely appreciate all of the cooperators’ assistance in monitoring and reporting on wheat rust.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1196808/full#supplementary-material
References
Abebe, T., Woldeab, G., Dawit, W. (2012). Distribution and physiologic races of wheat stem rust in tigray, Ethiopia. J. Plant Pathol. Microbiol. 3, 1–6. doi: 10.4172/2157-7471.1000142
Ali, S., Shah, S. J. A., Khalil, I. H., Raman, H., Maqbool, K., Ullah, W. (2009). Partial resistance to yellow rust in introduced winter wheat germplasm at the north of Pakistan. Aust. J. Crop Sci. 3, 37–43.
Alma, K., Gulzat, Y., Alex, M., Francis, O. (2012). The screening of wheat germplasm for resistance to stripe and leaf rust in Kazakhstan using molecular markers. J. Life Sci. 6, 353–362.
Anderson, J. A., Churchil, G. A., Autrique, J. E., Tanksley, S. O., Sorrels, M. E. (1993). Optimizing parent selection for genetic linkage maps. Genome 36, 181–186. doi: 10.1139/g93-024
Ayliffe, M., Singh, R., Lagudah, E. (2008). Durable resistance to wheat stem rust needed. Curr. Opin. Plant Biol. 11, 187–192. doi: 10.1016/j.pbi.2008.02.001
Bahadur, P., Nagarajan, S., Nayar, S. K. (1985). Identification of new virulence of Puccinia graminis f. sp. tritici in India during 1985. Cereal rusts Powdery Mildew Bull. 16 (2), 6–7.
Bansal, U. K., Hayden, M. J., Gill, M. B., Bariana, H. S. (2010). Molecular mapping of a seedling stripe rust resistance gene in wheat cultivar rubric. Euphytica 171, 121–127. doi: 10.1007/s10681-009-0007-4
Bhardwaj, S. C. (2012). “Wheat rust pathotypes in Indian subcontinent then and now,” in Wheat-productivity enhancement under changing climate. Eds. Singh, S. S., Hanchinal, R. R., Singh, G., Sharma, R. K., Saharan, M. S., Sharma, I. (New Delhi, India: Narosa Publishing House), 227–238.
Bhardwaj, S. C., Gupta, N., Sharma, T. R., Pal, D., Prasad, P. (2014). Competitive ability and fitness potential among the pathotypes of Puccinia triticina on wheat in India. Ind. Phytopath 67 (1), 33–37.
Bhardwaj, S. C., Kumar, S., Gangwar, O. P., Prasad, P., Kashyap, P. L., Khan, H., et al. (2021). Physiologic specialization and genetic differentiation of Puccinia triticina causing leaf rust of wheat in the Indian subcontinent during 2016-2019. Plant Dis. 105, 1992–2000. doi: 10.1094/PDIS-06-20-1382-RE
Bhardwaj, S. C., Nayar, S. K., Prashar, M., Jain, S. K., Singh, S. B. (2003). Physiologic specialization of Puccinia graminis tritici on wheat (Triticum aestivum) in India during 2002-2004. Ind. Phytopathol. 56 (1), 75–77.
Bhardwaj, S. C., Prashar, M., Singh, S. B. (2006). Physiologic specialization of Puccinia graminis tritici on wheat (Triticum aestivum) in India during 2002-2004. Indian J. Agr Sci. 76 (6), 386–388.
Bhardwaj, S. C., Singh, G. P., Gangwar, O. P., Prasad, P., Kumar, S. (2019). Status of wheat rust research and progress in rust management-Indian context. Agronomy 9 (12), :892. doi: 10.3390/agronomy9120892
Botstein, D., White, R. L., Skolnick, M., Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 (3), 314–331.
Broers, L. H. M., Cuesta-Subias, X., Lopez-Atilano, R. M. (1996). Field assessment of quantitative resistance to yellow rust in ten spring bread wheat cultivars. Euphytica 90, 9–16. doi: 10.1007/BF00025154
Browder, L. E. (1973). Probable genotype of some Triticum aestivum ‘Agent’ derivatives for reaction to Puccinia recondita f. sp. tritici. Crop Sci. 13, 203–206. doi: 10.2135/cropsci1973.0011183X001300020016x
Cox, T. S., Gill, B. S., Sears, R. G. (1995). Notice and release of KS94WGRC32 leaf rust resistant hard red winter wheat germplasm. Annu. Wheat Newslett. 41, 241.
Dakouri, A., McCallum, B. D., Radovanovic, N., Cloutier, S. (2013). Molecular and phenotypic characterization of seedling and adult plant leaf rust resistance in a world wheat collection. Mol. Breed 32, 663–677. doi: 10.1007/s11032-013-9899-8
FAO (2021) Crop prospects and food situation - quarterly global report no. 1 (Rome) (Accessed March 2021).
Fetch, T. G. (2005). Races of Puccinia graminis on wheat, barley, and oat in Canada, in 2002 and 2003. Can. J. Plant Pathol. 27, 572–580. doi: 10.1080/07060660509507258
Gangwar, O. P., Kumar, S., Bhardwaj, S. C., Prasad, P., Lata, C., Adhikari, S., et al. (2022). Elucidating the population structure and genetic diversity of Indian Puccinia striiformis f. sp. tritici pathotypes based on microsatellite markers. Phytopathol 112, 1444–1453. doi: 10.1094/PHYTO-10-21-0422-R
Goutam, U., Kukreja, S., Tiwari, R., Chaudhary, A., Gupta, R. K., Dholakia, B. B., et al. (2013). Biotechnological approaches for grain quality improvement in wheat: present status and future possibilities. Aust. J. Cereal Science. 7, 469–483.
Haile, J. K., Roder, M. (2013). Status of genetic research for resistance to Ug99 race of Puccinia graminis f. sp. tritici: a review of current research and implications. Afr J. Agric. Res. 8, 6670–6680. doi: 10.5897/AJAR2013.7257
Helguera, M., Khan, I. A., Kolmer, J., Lijavetzky, D., Zhong-qi, L., Dubcovsky, J. (2003). PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci. 43, 1839–1847. doi: 10.2135/cropsci2003.1839
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789. doi: 10.1007/s00122-013-2256-9
Hiebert, C. W., Thomas, J. B., McCallum, B. D., Humphreys, G. D., DePauw, R. M., Hayden, M. J., et al. (2010). An introgression on wheat chromosome 4DL in RL6077 (Thatcher*6/PI 250413) confers adult plant resistance to stripe rust and leaf rust (Lr67). Theor. Appl. Genet. 121, 1083–1091. doi: 10.1007/s00122-010-1373-y
Hovmoller, M. S., Justesen, A. F., Brown, J. K. M. (2002). Clonality and long-distance migration of Puccinia striiformis f sp. tritici in northwest Europe. Plant Pathol. 51, 24–32. doi: 10.1046/j.1365-3059.2002.00652.x
Hovmoller, M. S., Sorensen, C. K., Walter, S., Justesen, A. F. (2011). Diversity of Puccinia striiformis on cereals and grasses. Annu. Rev. Phytopathol. 49, 197–217. doi: 10.1146/annurev-phyto-072910-095230
Jain, S. K., Bhardwaj, S. C., Prashar, M., Singh, S. B. (2013). Physiologic specialization and new virulences of Puccinia graminis f. sp. tritici causing black rust of wheat (Triticum aestivum) in India during 2005-2009. Ind. J. Agric. Sci. 83 (10), 1058–1063.
Jin, Y. (2005). Races of Puccinia graminis identified in the united states in 2003. Plant Dis. 89, 1125–1127. doi: 10.1094/PD-89-1125
Jin, Y., Szabo, L. J., Pretorius, Z. A., Singh, R. P., Ward, R., TJr, F. (2008). Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 92, 923–926. doi: 10.1094/PDIS-92-6-0923
Johnston, C. O., Browder, L. E. (1966). Seventh revision of the international register of physiologic races of Puccinia recondita f. sp. tritici. Plant Dis. Rep. 50, 756–760.
Karaoglu, H., Lee, C. M. Y., Park, R. (2013). Simple sequence repeats in Puccinia graminis: abundance, cross-formae speciales and intra-species utility, and development of novel markers. Australas. Plant Pathol. 42, 271–281. doi: 10.1007/s13313-013-0199-x
Khadegah, N. M. A., Thabet, M., Negm, S. S., El-Deeb, S. H. (2019). Monitoring of Puccinia triticina eriks. physiologic races and effectiveness of lr-genes in Egyptian wheat during 2014-2016 growing seasons. Int. J. Agric. Technol. 15 (1), 35–54.
Khan, H., Bhardwaj, S. C., Gangwar, O. P., Prasad, P., Kashyap, P. L., Savadi, S. (2017). Identifying some additional rust resistance genes in Indian wheat varieties using robust markers. Cer Res. Comm. 45 (4), 633–646. doi: 10.1556/0806.45.2017.041
Kiran, K., Rawal, H. C., Dubey, H., Jaswal, R., Bhardwaj, S. C., Prasad, P., et al. (2017). Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 7, 42419. doi: 10.1038/srep42419
Kolmer, J. A. (2019). Virulence of Puccinia triticina, the wheat leaf rust fungus, in the united states in 2017. Plant Dis. 103, 2113–2120. doi: 10.1094/PDIS-09-18-1638-SR
Lagudah, E. S., Mcfadden, H., Singh, R. P., Heurta-Espino, J., Bariana, H. S., Spielmyer, W. (2006). Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor. Appl. Genet. 114, 21–30. doi: 10.1007/s00122-006-0406-z
Le Roux, J., Rijkenberg, F. H. J. (1987). Pathotypes of Puccinia graminis f. sp. tritici with increased virulence for Sr24. Plant Dis. 71, 1115–1119. doi: 10.1094/PD-71-1115
Lewis, C. M., Persoons, A., Bebber, D. P., Kigathi, R. N., Maintz, J., Kim Findlay, K., et al. (2018). Potential for re-emergence of wheat stem rust in the united kingdom. Comm Biol. 1, 13. doi: 10.1038/s42003-018-0013-y
Luig, N. H. (1983). A survey of virulence genes in wheat stem rust Puccinia graminis f. sp. tritici. advances in plant breeding (Suppl. 11 to plant breeding) (Berlin, Germany: Verlag Paul Parey).
Mago, R., Bariana, H. S., Dundas, L. S., Speilmeyer, W., Lawrence, G. J., Pryor, A. J., et al. (2005). Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504. doi: 10.1007/s00122-005-2039-z
Mago, R., Brown-Guedira, G., Dreisigacker, S., Breen, J., Jin, Y., Singh, R., et al. (2011). An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor. Appl. Genet. 122, 735–744. doi: 10.1007/s00122-010-1482-7
Mago, R., Verlin, D., Zhang, P., Bansal, U., Bariana, H., Jin, Y., et al. (2013). Development of wheat-aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2s1 chromosome. Theor. Appl. Genet. 126 (12), 2943–2955. doi: 10.1007/s00122-013-2184-8
Mettin, D., Bluthner, W. D., Schlegel, R. (1973). “Additional evidence on spontaneous 1B/1R, wheat rye substitutions and translocations,” in Proceedings of the 4th international wheat genetics symposium. Eds. Sears, E. R., Sears, L. M. S. (Columbia, MO: Agricultural Experimental Station, University of Missouri), pp 179–pp 184.
Mourad, A. M., Sallam, A., Belamkar, V., Wegulo, S., Bai, G., Mahdy, E., et al. (2019). Molecular marker dissection of stem rust resistance in Nebraska bread wheat germplasm. Sci. Rep. 9 (1), 1–0. doi: 10.1038/s41598-019-47986-9
Nagarajan, S., Joshi, L. M. (1975). Historic account of wheat rust epidemics in India and their significance. Cer Rusts Bull. 3, 25–33.
Nagarajan, S., Nayar, S. K., Bahadur, P. (1983). The proposed brown rust of wheat (Puccinia recondia f. sp. tritici) virulence monitoring system. Curr. Sci. 52 (9), 413–416.
Nayar, S. K., Prashar, M., Bhardwaj, S. C. (1997). Manual of current techniques in wheat rusts. research bulletin No.2 (Shimla, Himachal Pradesh: Regional Station, DWR, Flowerdale), p 32.
Nazari, K., Al-Maaroof Emad, M., Kurtulus, E., Kavaz, H., Hodson, D., Ozseven, I. (2021). First report of Ug99 race TTKTT of wheat stem rust (Puccinia graminis f sp. tritici) in Iraq. Plant Dis. 105 (9), 2719. doi: 10.1094/PDIS-02-21-0404-PDN
Olivera, P. D., Newcomb, M., Szabo, L. J., Rouse, M., Johnson, J., Gale, S., et al. (2015). Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013/14. Phytopathol 105, 917–928. doi: 10.1094/PHYTO-11-14-0302-FI
Olivera, P. D., Rouse, M. N., Jin, Y. (2018). Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01719
Olivera Firpo, P., Newcomb, M., Flath, K., Sommerfeldt-Impe, N., Szabo, L., Carter, M., et al. (2017). Characterization of Puccinia graminis f. sp. tritici isolates derived from an unusual wheat stem rust outbreak in Germany in 2013. Plant Pathol. 66, 1258–1266. doi: 10.1111/ppa.12674
Ordonez, M. E., Kolmer, J. A. (2009). Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in north America. Phytopathol 99, 750–758. doi: 10.1094/PHYTO-99-6-0750
Park, R. F., Wellings, C. R. (1992). Pathogenic specialization of wheat rusts in Australia and new Zealand in 1988 and 1989. Austral Plant Pathol. 21, 61–69. doi: 10.1071/APP9920061
Parlevliet, J. E. (1988). “Strategies for the utilization of partial resistance for the control of cereal rust0,” in Breeding strategies for resistance to the rusts of wheat. Eds. Rajaram, S., Simmonds, N. W. (Mexico, D.F.: CIMMYT), pp 48–pp 62.
Pathan, A. K., Park, R. F. (2006). Evaluation of seedling and adult plant resistance to leaf rust in European wheat cultivars. Euphytica 149, 327–342. doi: 10.1007/s10681-005-9081-4
Patpour, M., Hovmøller, M. S., Justesen, A. F., Newcomb, M., Olivera, P., Jin, Y., et al. (2016). Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 100, 522. doi: 10.1094/PDIS-06-15-0668-PDN
Peng, X., Ren, M. J., Zhang, S. S., Xu, R. H. (2013). Molecular detection of wheat leaf rust resistance gene Lr34 and Lr37. Guizhou Agric. Science. 41, 13–16.
Perrier, X., Jacquemoud-collet, J. P. (2006) DARwinsoftware. Available at: http://darwin.cirad.fr (Accessed 28.08.2021).
Peterson, R. F., Campbell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Prank, M., Kenaley, S. C., Bergstrom, G. C., Acevedo, M., Mahowald, N. M. (2019). Climate change impacts the spread potential of wheat stem rust, a significant crop disease. Environm Res. Lett. 14 (12), 124053. doi: 10.1088/1748-9326/ab57de
Prasad, P., Bhardwaj, S. C., Gangwar, O. P., Kumar, S. (2020a). “Wheat rust research: impact, thrusts, and roadmap to sustained wheat production,” in Improving cereal productivity through climate smart practices. Eds. Sindhu Sareen, S., Sharma, P., Singh, C., Jasrotia, P., Singh, G. P., Sarial, A. K. (UK: Elsevier), 177–203.
Prasad, P., Bhardwaj, S. C., Gangwar, O. P., Kumar, S., Khan, H., Kumar, S., et al. (2017). Population differentiation of wheat leaf rust fungus Puccinia triticina in south Asia. Curr. Sci. 112 (10), 2073–2084. doi: 10.18520/cs/v112/i10/2073-2084
Prasad, P., Bhardwaj, S. C., Khan, H., Gangwar, O. P., Kumar, S., Singh, S. B. (2016). Ug99: saga, reality and status. Curr. Sci. 110 (9), 1614–1616.
Prasad, P., Bhardwaj, S. C., Savadi, S., Kashyap, P. L., Gangwar, O. P., Khan, H., et al. (2018). Population distribution and differentiation of Puccinia graminis tritici detected in the Indian subcontinent during 2009-2015. Crop Prot. 108, 128–36. doi: 10.1016/j.cropro.2018.02.021
Prasad, P., Bhardwaj, S. C., Thakur, R. K., Adhikari, S., Gangwar, O. P., Lata, C., et al. (2021b). Prospects of climate change effects on crop diseases with particular reference to wheat. J. Cer Res. 13 (2), 117–134. doi: 10.25174/2582-2675/2021/112817
Prasad, P., Gangwar, O. P., Bhardwaj, S. C., Kumar, S. (2019). Mehtaensis: six-monthly newsletter. (Shimla, Himachal Pradesh, India: ICAR-Indian Institute of Wheat and Barley Research, Regional Station) 39 1–31.
Prasad, P., Gangwar, O. P., Kumar, S., Lata, C., Adhikari, S., Bhardwaj, S. C. (2021c). Mehtaensis: six-monthly newsletter. (Shimla, Himachal Pradesh, India: ICAR-Indian Institute of Wheat and Barley Research, Regional Station) 41 1–35.
Prasad, P., Khan, H., Bhardwaj, S. C., Savadi, S., Gangwar, O. P., Kumar, S.. (2021a). Practical manual on protocols and methodologies in wheat rusts research (New Delhi, India: Directorate of Knowledge Management in Agriculture, I.C.A.R.), 1–76.
Prasad, P., Savadi, S., Bhardwaj, S. C., Gupta, P. K. (2020b). The progress of leaf rust research in wheat. Fungal Biol. 124 (6), 537–550. doi: 10.1016/j.funbio.2020.02.013
Prasad, P., Thakur, R. K., Savadi, S., Bhardwaj, S. C., Gangwar, O. P., Lata, C., et al. (2022). Genetic diversity and population structure reveal cryptic genetic variation and long distance migration of Puccinia graminis f. sp. tritici. Indian subcontinent. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.842106
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84 (2), 203. doi: 10.1094/PDIS.2000.84.2.203B
Prud'homme, A., Sackston, W. E. (1990). Relative fitness of races 1 (0) and 3 (0, 1) of rust (Puccinia helianthi) in mixtures on susceptible sunflower (Helianthus annuus). Can. J. Bot. 68, 1602–1608. doi: 10.1139/b90-206
Rahmatov, M., Otambekova, M., Muminjanov, H., Rouse, M. N., Hovmøller, M. S., Nazari, K., et al. (2019). Characterization of stem, stripe and leaf rust resistance in tajik bread wheat accessions. Euphytica 215, 1–22. doi: 10.1007/s10681-019-2377-6
Randhawa, M. S., Singh, R. P., Dreisigacker, S., Bhavani, S., Huerta-Espino, J., Rouse, M. N., et al. (2018). Identification and validation of a common stem rust resistance locus in two bi-parental populations. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01788
Roelfs, A. P., Singh, R. P., Saari, E. E. (1992). Rust diseases of wheat: concepts and methods of disease management (Mexico City, Mexico: CIMMYT).
Rosegrant, M. W., Agcaoili, M. (2010). Global food demand, supply, and price prospects to 2010 (Washington, D.C. USA: International Food Policy Research Institute).
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., William, H. M., Bouchet, S., Cloutier, S., et al. (2006). Leaf tip necrosis, molecular markers and β1- proteasome subunits associated with the slow rusting resistance genes Lr46/Yr29. Theor. Appl. Genet. 112, 500–508. doi: 10.1007/s00122-005-0153-6
Rutkoski, J. E., Poland, J. A., Singh, R. P., Huerta-Espino, J., Bhavani, S., Barbier, H., et al. (2014). Genomic selection for quantitative adult plant stem rust resistance in wheat. Plant Genome 7, 3. doi: 10.3835/plantgenome2014.02.0006
Saari, E. E., Prescott, J. M. (1985). “World distribution in relation to economic losses. wheat and rye stem rust,” in The cereal rusts vol. II: diseases, distribution, epidemiology and control. Eds. Roelfs, A. P., Bushnell, W. R. (London, United Kingdom: Academic Press), (pp. 259–298).
Saari, E. E., Wilcoxson, R. D. (1974). Plant disease situation of high yielding durum wheat in Asia and Africa. Annu. Rev. Phytopathol. 2, 49–68. doi: 10.1146/annurev.py.12.090174.000405
Savadi, S., Prasad, P., Sharma, K., Rathore, R., Bhardwaj, S. C., Gangwar, O. P., et al. (2020). Development of novel transcriptome-based SSR markers in Puccinia triticina and their potential application in genetic diversity studies. Trop. Plant Pathol. 45, 499–510. doi: 10.1007/s40858-020-00347-8
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3 (3), 430–439. doi: 10.1038/s41559-018-0793-y
Shamanin, V. P., Pototskaya, I. V., Shepelev, S. S., Pozherukova, V. E., Salina, Е. А., Skolotneva, Е. S., et al. (2020). Stem rust in western Siberia – race composition and effective resistance genes. Vavilov J. Genet. Breed 24, 131–138. doi: 10.18699/VJ20.608
Shete, S., Tiwari, H., Elston, R. C. (2000). On estimating the heterozygosity and polymorphism information content value. Theor. Popul Biol. 57 (3), 265–271. doi: 10.1006/tpbi.2000.1452
Singh, R. P. (1991). Pathogenicity variation of Puccinia recondita f. sp. tritici and P. graminis f. sp. tritici in wheat-growing areas of Mexico during 1988 and 1989. Plant Dise 75, 790–794. doi: 10.1094/PD-75-0790
Singh, R. P., Hodson, D. P., Huerta-Espino, J., Jin, Y., Bhavani, S., Njau, P., et al. (2011). The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49, 465–481. doi: 10.1146/annurev-phyto-072910-095423
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathol 105, 872–874. doi: 10.1094/PHYTO-01-15-0030-FI
Skowrońska, R., Tomkowiak, A., Nawracała, J., Kwiatek, M. T. (2020). Molecular identification of slow rusting resistance Lr46/Yr29 gene locus in selected triticale (× triticosecale wittmack) cultivars. J. Appl. Genet. 61 (3), 359–366. doi: 10.1007/s13353-020-00562-8
Spielmeyer, W., Sharp, P. J., Lagudah, E. S. (2003). Identification and validation of markers linked to broad spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci. 43, 333–336. doi: 10.2135/cropsci2003.3330
Stakman, E. C., Stewart, D. M., Loegering, W. Q. (1962). “Identification of physiologic races of Puccinia graminis var. tritici,” (USDA, Washington, USA.: U.S, Agricultural Research Service, ARS E617), 1–53.
Terefe, T. G., Visser, B., Pretorius, Z. A. (2016). Variation in Puccinia graminis f. sp. tritici detected on wheat and triticale in south Africa from 2009 to 2013. Crop Prot 86, 9–16. doi: 10.1016/j.cropro.2016.04.006
Wang, X., Bakkeren, G., McCallum, B. (2010). Virulence and molecular polymorphisms of the wheat leaf rust fungus Puccinia triticina in Canada from 1997 to 2007. Amer J. Bot. 88, 575–589. doi: 10.1139/B10-034
Wilcoxson, R. D., Skovmand, B., Atif, A. H. (1975). Evaluation of wheat cultivars ability to retard development of stem rust. Annu. Appl. Biol. 80, 275–281. doi: 10.1111/j.1744-7348.1975.tb01633.x
Wingen, L. U., Shaw, M. W., Brown, J. K. (2013). Long-distance dispersal and its influence on adaptation to host resistance in a heterogeneous landscape. Plant Pathol. 62 (1), 9–20. doi: 10.1111/j.1365-3059.2012.02621.x
Xia, Y. (2020). Correlation and association analyses in microbiome study integrating multiomics in health and disease. Prog. Mol. Biol. Transl. Sci. 171, 309–491. doi: 10.1016/bs.pmbts.2020.04.003
Xu, X. F., Li, D. D., Liu, Y., Gao, Y., Wang, Z. Y., Ma, Y. C., et al. (2017). Evaluation and identification of stem rust resistance genes Sr2, Sr24, Sr25, Sr26, Sr31 and Sr38 in wheat lines from gansu province in China. PeerJ 5, e4146. doi: 10.7717/peerj.4146
Xu, X., Yuan, D., Li, D., Gao, Y., Wang, Z., Liu, Y., et al. (2018). Identification of stem rust resistance genes in wheat cultivars in China using molecular markers. PeerJ 6, e4882. doi: 10.7717/peerj.4882
Xue, W. B., Xu, X., Mu, J. M., Wang, Q. L., Wu, J. H., Huang, L. L., et al. (2014). Evaluation of stripe rust resistance and genes in Chinese elite wheat varieties. J. Triticeae Crops. 34, 1054–1060.
Yang, W., Yang, F., Liang, D., He, Z., Shang, X., Xia, X. (2008). Molecular characterization of slow-rusting genes Lr34/Yr18 in Chinese wheat cultivars. Acta Agron. Sin. 34, 1109–1113. doi: 10.3724/SP.J.1006.2008.01109
Yu, L., Barbier, H., Rouse, M., Singh, S., Singh, R., Bhavani, S., et al. (2014). A consensus map for Ug99 stem rust resistance loci in wheat. Theor. Appl. Genet. 127, 1561–1581. doi: 10.1007/s00122-014-2326-7
Keywords: wheat, stem rust, Puccinia graminis tritici, virulence diversity, genetic diversity, rust resistance
Citation: Prasad P, Thakur R, Bhardwaj SC, Savadi S, Gangwar OP, Lata C, Adhikari S, Kumar S, Kundu S, Manjul AS, Prakasha TL, Navathe S, Hegde GM, Game BC, Mishra KK, Khan H, Gupta V, Mishra CN, Kumar S, Kumar S and Singh G (2023) Virulence and genetic analysis of Puccinia graminis tritici in the Indian sub-continent from 2016 to 2022 and evaluation of wheat varieties for stem rust resistance. Front. Plant Sci. 14:1196808. doi: 10.3389/fpls.2023.1196808
Received: 30 March 2023; Accepted: 20 June 2023;
Published: 14 July 2023.
Edited by:
Peng Zhang, The University of Sydney, AustraliaReviewed by:
Naeela Qureshi, International Maize and Wheat Improvement Center, MexicoShyam Solanki, South Dakota State University, United States
Copyright © 2023 Prasad, Thakur, Bhardwaj, Savadi, Gangwar, Lata, Adhikari, Kumar, Kundu, Manjul, Prakasha, Navathe, Hegde, Game, Mishra, Khan, Gupta, Mishra, Kumar, Kumar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. C. Bhardwaj, c2NiZmRsQGhvdG1haWwuY29t
 Pramod Prasad
Pramod Prasad Rajnikant Thakur
Rajnikant Thakur S. C. Bhardwaj
S. C. Bhardwaj Siddanna Savadi
Siddanna Savadi O. P. Gangwar
O. P. Gangwar Charu Lata
Charu Lata Sneha Adhikari
Sneha Adhikari Subodh Kumar1
Subodh Kumar1 T. L. Prakasha
T. L. Prakasha Sudhir Navathe
Sudhir Navathe K. K. Mishra
K. K. Mishra Vikas Gupta
Vikas Gupta C. N. Mishra
C. N. Mishra Satish Kumar
Satish Kumar Sudheer Kumar
Sudheer Kumar Gyanendra Singh
Gyanendra Singh