- Vegetable Research Institute, Gansu Academy of Agricultural Sciences, Lanzhou, China
Citric acid is the most important organic acid in melon and has a great influence on fruit flavor quality. ATP-citrate (pro-S) lyase (ACL) is a key regulator in the acetyl-CoA pathway and plays an important role in citric acid metabolism. In this study we analyzed the structure and phylogenetics of CmACL genes and their functions in sugar and acid accumulation in melon. A total of four CmACL genes were identified in the melon genome, and phylogenetic analysis assigned these genes into the α subfamily (CmACLα1 and CmACLα2) and the β subfamily (CmACLβ1 and CmACLβ2). Conserved motif and gene structure analyses showed that members of the same subfamily shared identical conserved motifs and gene structures, and probably have similar biological functions. Analysis of cis-acting elements revealed that CmACL promoter sequences contained regulatory elements related to light, stress, phytohormones, and growth and development, indicating that CmACL genes may be involved in melon growth and stress responses. The prediction of protein interaction network showed that CmACL proteins were closely related to the proteins belonging to tricarboxylic acid cycle, glyoxylic acid cycle and glycolytic pathway, suggesting that CmACL proteins may play an important role in sugar and acid metabolism. The expression of CmACLβ1 was significantly and positively correlated with sucrose content, and CmACLβ2 expression was significantly positively correlated with citric acid content, suggesting that CmACLβ1 and CmACLβ2 have important roles in sugar and acid accumulation in melon. Our results offer novel insights and avenues for the regulation of sugar and acid levels in melon and provide a theoretical foundation for breeding high-quality melon cultivars.
1 Introduction
Melon (Cucumis melo L.) is an annual herbaceous trailing plant in the Cucurbitaceae family. Melon fruit is popular and has a fragrant, sweet, and juicy flesh. With the improvement of living standards, high-quality melon is favored by breeders and consumers, and flavor is the most important trait for cultivar improvement and cultivation. Organic acids are a crucial factor controlling the flavor of melon. Moderate levels of organic acids can enhance the flavor of melon, whereas excessive quantities of these acids can adversely affect fruit quality. However, the mechanisms governing organic acid metabolism in melon fruit have not been fully elucidated.
The organic acids in ripe fruits can be dominated by citric acid, malic acid, and tartaric acid (Zhou et al., 2015). The fruits of horticultural plants such as melon (Tang, 2010; Hu, 2018; Tang et al., 2019), citrus (Li et al., 2020a), pineapple (Lu et al., 2013), blueberry (Yu et al., 2016), passion fruit (Zhang et al., 2021), strawberry (Li et al., 2019), and tomato (Wang et al., 2017) mainly contain citric acid. Therefore, the study of citric acid metabolism holds great value for regulating the quality of melon and the fruits of horticultural plants with citric acid as the primary organic acid. Citric acid degradation can occur through three primary pathways: the γ-aminobutyric acid pathway, glutamine pathway, and acetyl-CoA pathway. The acetyl-CoA pathway has been extensively investigated in recent years. ATP-citrate (pro-S) lyase (ACL) is a key regulator in citric acid degradation through the acetyl-CoA pathway and converts citric acid into oxaloacetate and acetyl-CoA. The role of ACL in citric acid metabolism has only been documented in a small number of horticultural plants, such as citrus and blueberry (Hu, 2015; Guo, 2020; Li et al., 2020a; Li et al., 2020b). The majority of studies investigating the physiological functions and regulatory mechanisms of ACL have focused on heterologous expression and the model plant Citrus reticulata, with no reports on melon. Herein we used bioinformatics analyses to identify CmACL genes in melon. We analyzed their phylogenetic relationships, conserved motifs, gene structures, cis-acting elements, chromosomal locations, and subcellular locations. The content of soluble sugars and organic acids was measured and correlated to the relative expression of CmACL genes. Our results lay a foundation for further investigation into the roles of these genes in sugar and acid accumulation in melon.
2 Materials and methods
2.1 Data acquisition and identification of CmACL proteins
The AtACL protein sequences of Arabidopsis thaliana was retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) and the CitACL protein sequences of Citrus reticulata were from the Citrus Pan-genome to Breeding Database of Huazhong Agricultural University (citrus.hzau.edu.cn/orange/) (Hu, 2015; Hu et al., 2015). These sequences were aligned using Blast against the Melon Genomics Database (http://cucurbitgenomics.org/) to search for candidate melon proteins (CmACL proteins). The CmACL proteins were also identified using the conserved domains of ACL proteins. The proteins identified using these two methods were deduplicated and verified by NCBI CDD (http://www.ncbi.nlm.nih.gov/cdd) with an e-value of 10−4. The CmACL protein sequences were obtained.
2.2 Physicochemical properties of CmACL proteins
The physicochemical properties of the CmACL proteins were determined. Amino acid number, molecular weight, isoelectric point, and total average hydrophilicity were calculated using Expasy (https://www.expasy.org/). The subcellular localization of the CmACL proteins was predicted online using the Cell-PLoc 2.0 package (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
2.3 Phylogenetic analysis and chromosomal localization of CmACL genes
Phylogenetic analysis was performed using the identified CmACL protein sequences and the ACL protein sequences of A. thaliana and C. reticulata. Based on multiple sequence alignment, a phylogenetic tree was constructed using the Neighbor-Joining method in MEGA v11.0.13.
The data on the chromosomal locations of the CmACL genes were downloaded from the Melon Genomics Database and visualized using an online tool (http://mg2c.iask.in/mg2c_v2.1/).
2.4 Conserved motifs and gene structure of CmACL
The DNA and CDS sequences of the CmACL genes were downloaded from the Melon Genomics Database. The structure of the CmACL genes was analyzed using the online tool Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/). The conserved motifs of the CmACL proteins were predicted using MEME 5.5.1 (https://meme-suite.org/meme/tools/meme). The length of the conserved motifs was set to 6–50 amino acids and the number of conserved motifs was set to 20. The 2-kb sequences upstream of the four genes were used as the promoter sequences, and the cis-acting elements were identified using PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Conserved motifs and cis-acting elements were visualized with TBtools.
2.5 Construction of a CmACL protein interaction network
The interactions of the CmACL proteins were predicted online using STRING (https://cn.string-db.org). The target genes with the highest default Blast scores were selected to construct the protein interaction network. Optimising protein-protein interation networks with Cytoscape (Doncheva et al., 2019; Yu et al., 2022).
2.6 Plant materials and growth condition
The melon selfing line Bailangua P7 was provided by Vegetable Research Institute, Gansu Academy of Agricultural Sciences. The plant material were planted in the greenhouse with the day/night temperature of 23–28°C/18–22°C at the melon breeding base in Gaolan county (36°16′49″, 103°37′57″), China, on March 18, 2022. The plant were grown with a 50 cm distance between seedlings and a 60 cm distance between rows and were under the same local management practices (soil management, irrigation, fertilization, and disease control).
Fruit samples were collected at 10, 20, 30, 35, and 40 d after pollination. At each time point, three fruits were harvested from three different plants and used as a replicate, and three replicates were used for the following analyses. The flesh was obtained from the center-equatorial portion of each fruit after removing the pericarp. Then the samples were immediately transferred to liquid nitrogen and stored at –80°C.
2.7 Determination of soluble sugars and organic acids in melon
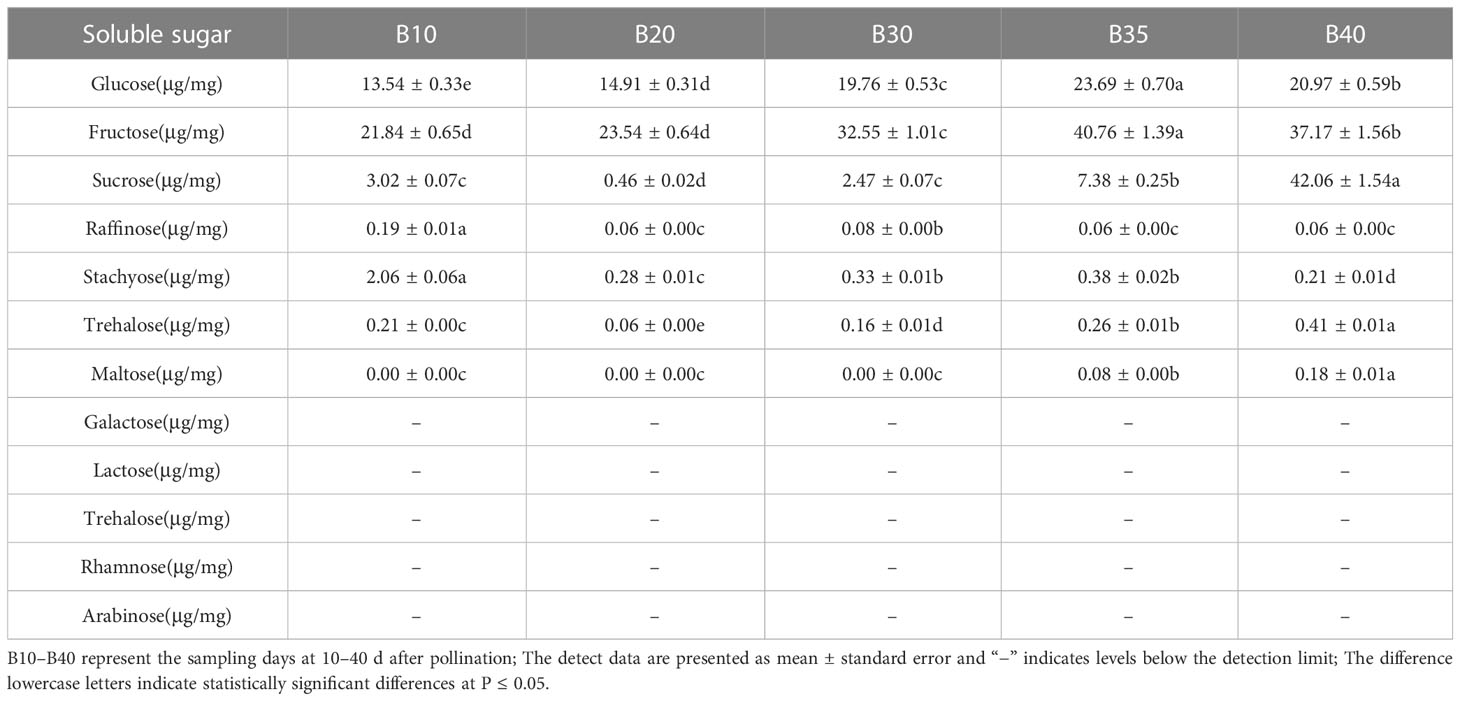
The content of soluble sugars (glucose, fructose, sucrose, raffinose, stachyose, trehalose, maltose, galactose, lactose, trehalose, rhamnose, and arabinose) at different fruit developmental stages was determined using ion chromatography (Thermo iCS5000 ion chromatography system and electrochemical detector).
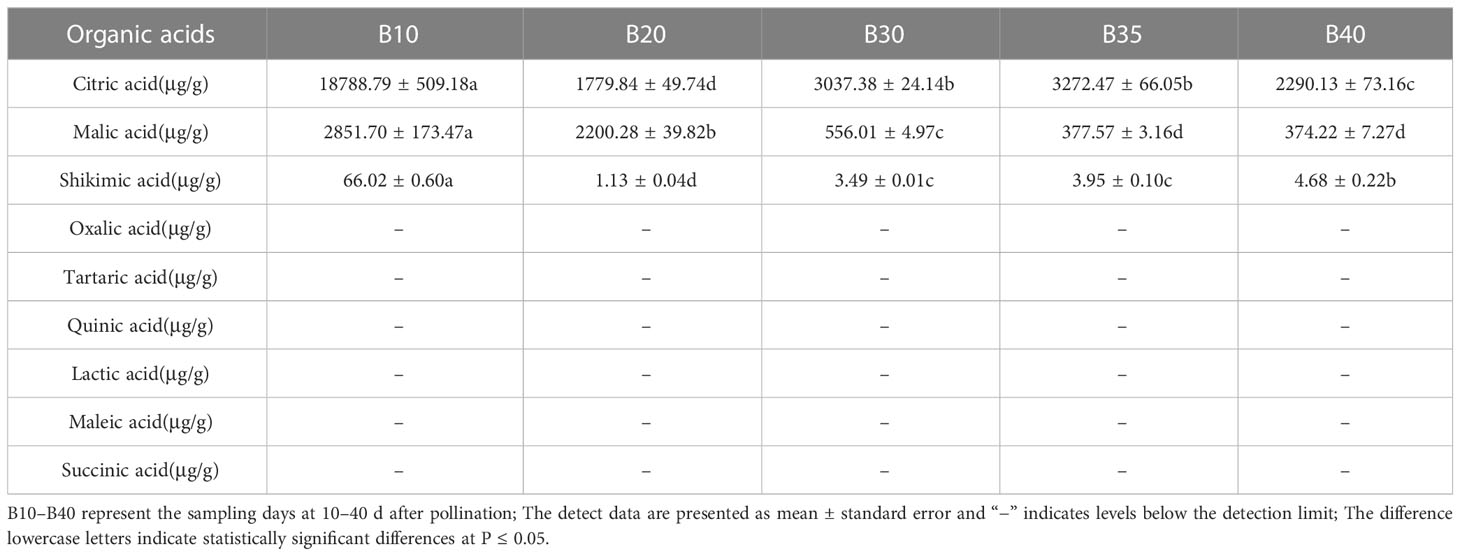
The content of organic acids (citric acid, malic acid, shikimic acid, oxalic acid, tartaric acid, quinic acid, lactic acid, maleic acid, and succinic acid) at different fruit developmental stages was determined using high-performance liquid chromatography (Waters e2695 separation module and Waters 2998 detector).
2.8 Determination of the relative expression of CmACL genes in melon
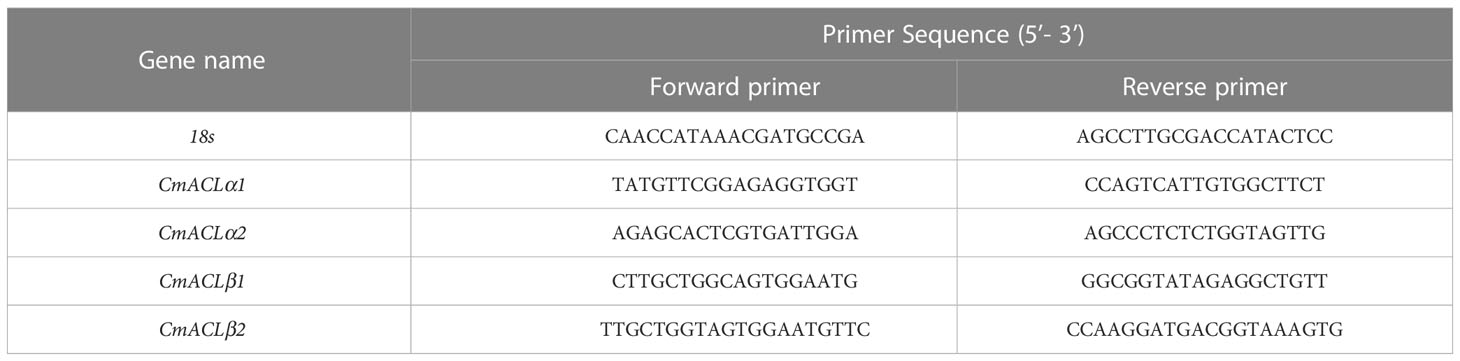
Quantitative real-time (qRT)-PCR was used to determine the relative expression of the CmACL genes at different developmental stages in the melon material. Primers were designed using the PrimerQuest Tool with the 18S gene as an internal reference. The primer sequences are shown in Table 1. The RNA reverse transcription was performed using the TUREscript 1st Stand cDNA Synthesis Kit (Aidlab), and fluorescence quantification was performed using the abm® EvaGreen qPCR Master Mix-ROX Kit. Each treatment had three technical replicates, and the 2−ΔΔCt method was used to calculate relative gene expression.
2.9 Data analysis
IBM SPSS Statistics 26 software was used for T-test significance analysis and Pearson correlation analysis. Heat map and column chart were generated using GraphPad Prism 9 software.
3 Results
3.1 Identification and phylogenetic analysis of CmACL proteins in melon
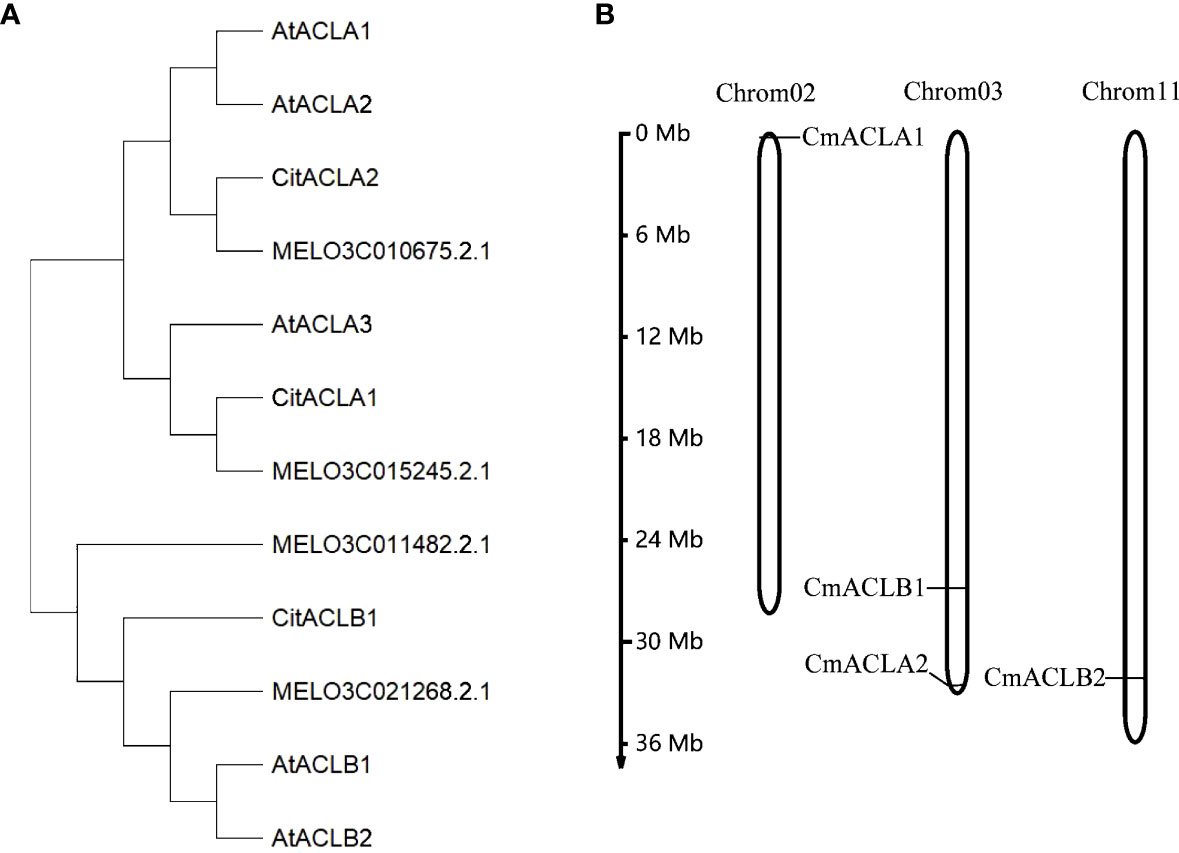
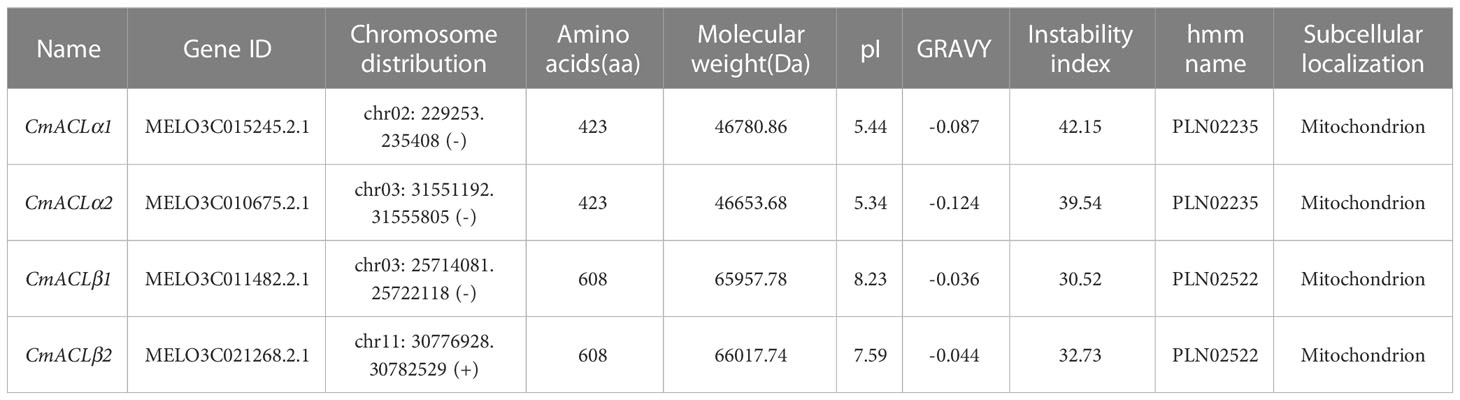
A total of four CmACL proteins were identified in the melon genome using Blast with the ACL protein sequences of A. thaliana and C. reticulata (Table 1). To elucidate the phylogenetic relationships of the ACL proteins, a phylogenetic tree was constructed using the Neighbor-Joining method for 12 ACL protein sequences from A. thaliana, C. reticulata, and C. melo (Figure 1A). The ACL proteins were divided into α and β subfamilies. MELO3C015245.2.1 clustered with CitACLα1, and MELO3C010675.2.1 clustered with CitACLα2. Therefore, MELO3C015245.2.1 and MELO3C010675.2.1 were renamed as CmACLα1 and CmACLα2, respectively. MELO3C011482.2.1 and MELO3C021268.2.1 clustered with the ACL proteins in the β subfamily and were renamed as CmACLβ1 and CmACLβ2.
Chromosomal localization (Figure 1B) showed one CmACL gene on Chr2, one on Chr11, and two on Chr3. The number of amino acids was 423 and 608 in the α and β subfamilies, respectively (Table 2). The relative molecular weight was 46653.68–66017.74 Da, and the isoelectric point was 5.34–8.23. The hydrophilicity ranged between −0.124 and −0.036, and the four CmACL proteins were hydrophilic. The instability index ranged between 30.52 and 42.15, indicating that the CmACL proteins were stable except for CmACLα1. The four CmACL proteins were localized in the nucleus.
3.2 Conserved motifs and gene structure of CmACL
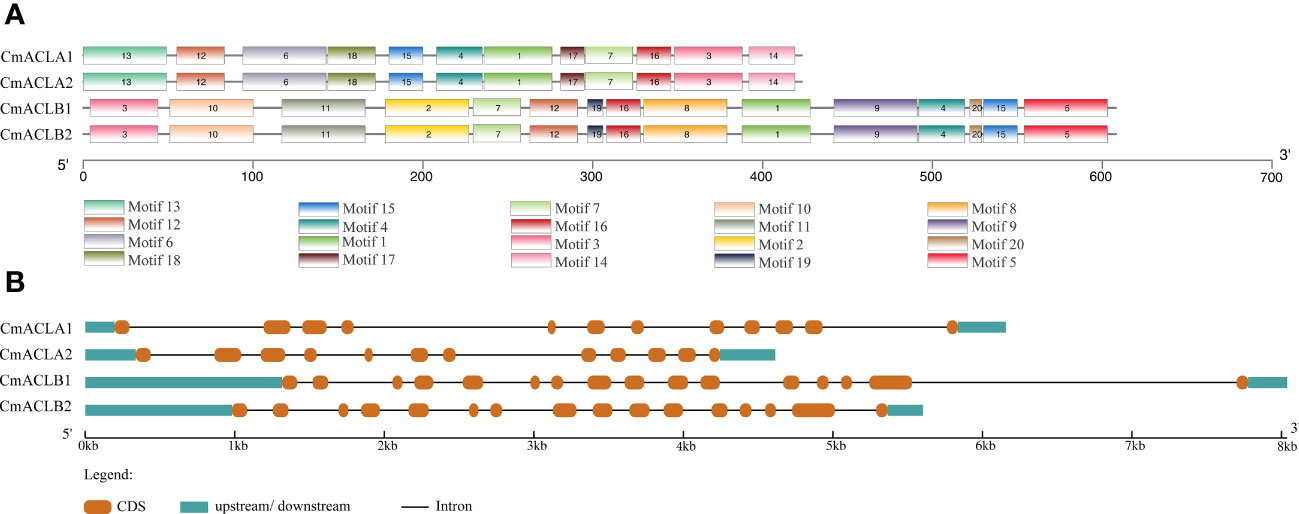
The function of a protein is determined by its number and type of conserved motifs. The conserved motifs of the CmACL proteins were predicted by MEME and visualized using TBtools. As shown in Figure 2A, both CmACLα1 and CmACLα2 of the α subfamily contained 12 conserved motifs with similar positions, including motif1, motif3, motif4, motif6, motif7, motif12, motif13, motif14, motif15, motif16, motif17, and motif18. Both CmACLβ1 and CmACLβ2 of the β subfamily contained 15 conserved motifs with similar positions, including motif1, motif2, motif3, motif4, motif5, motif7, motif8, motif9, motif10, motif11, motif12, motif15, motif16, motif19, and motif20. Analysis of the conserved motifs in the CmACL proteins showed that the members in the same subfamily shared identical conserved motifs indicating that the same subfamily might have similar biological functions.
The structures of four CmACL genes were analyzed to further reveal their phylogenetic diversity (Figure 2B). The results showed that the genes in the same subfamily shared similar structures. CmACLα1 and CmACLα2 contained 11 introns and 12 exons, while CmACLβ1 and CmACLβ2 contained 15 introns and 16 exons. The variances in intron and exon distribution may be attributed to evolution, as distinct gene structures have led to different gene functions.
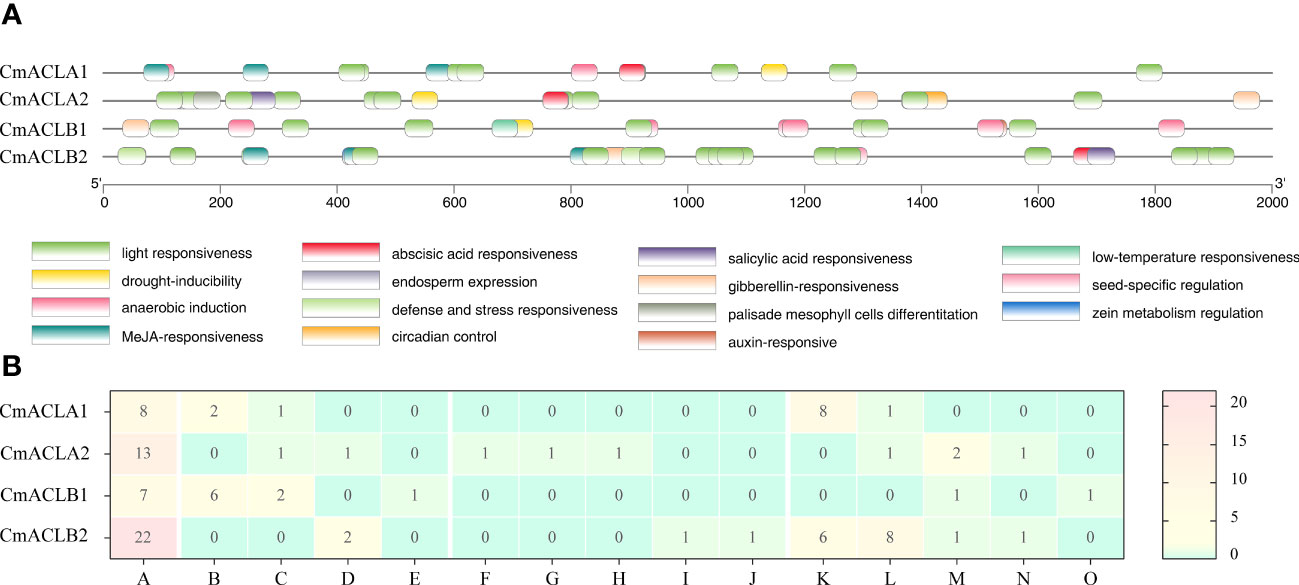
3.3 Analysis of CmACL cis-acting elements and protein interaction network
The cis-acting elements of the CmACL genes are shown in Figure 3. These elements were related to light, abiotic and biotic stress, plant growth and development, and phytohormone. Light responsiveness elements were most abundant and present in the four CmACL genes. Plant abiotic and biotic stress elements included anaerobic induction (CmACLα1 and CmACLβ1), drought-inducibility (CmACLα1/2 and CmACLβ1), defense and stress responsiveness (CmACLα2 and CmACLβ2), and low-temperature responsiveness (CmACLβ1). Plant growth and development elements included circadian control (CmACLα2), palisade mesophyll cells differentiation (CmACLα2), endosperm expression (CmACLα2), seed-specific regulation (CmACLβ2), and zein metabolism regulation (CmACLβ2). Phytohormone elements included MeJA-responsiveness (CmACLα1 and CmACLβ2), abscisic acid responsiveness (CmACLα1/2 and CmACLβ2), gibberellin-responsiveness (CmACLα2 and CmACLβ1/2), salicylic acid responsiveness (CmACLα2 and CmACLβ2), and auxin-responsive (CmACLβ1). These results suggest that CmACL genes may have important roles in plant growth, development, and the stress response.
The protein interaction network was constructed using the online tool STRING (Figure 4). Results showed that all four CmACL proteins interacted with one another. Notably, several proteins such as Aco, XP_008467310.1, XP_008455442.1, XP_008456386.1, XP_008461234.1, XP_008456567.1, XP_008440462.1, XP_008460481.1 and XP_008446580.1, belonging to tricarboxylic acid cycle, glyoxylate cycle, and glycolytic pathway, were showing tight relationships with CmACL, suggesting that CmACL may play an important role in sugar and acid metabolism.
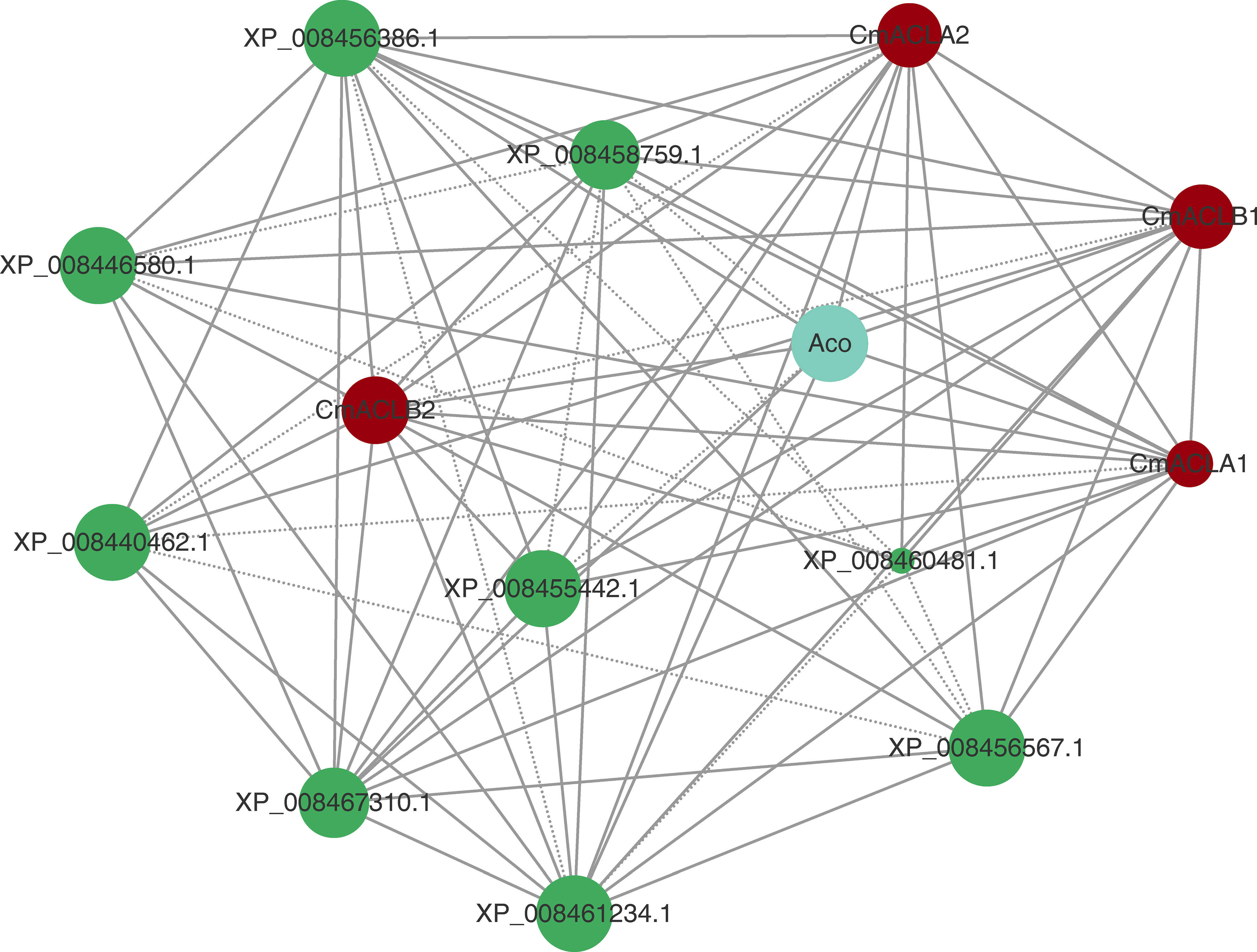
Figure 4 Protein-protein interaction network. The area of the circles represents the number of interacting proteins, the larger area of the circles indicate more interacting proteins. The dashed lines represent protein interaction score range from 0.7 to 0.9, and solid lines means protein interaction score>0.9.
3.4 Soluble sugar and organic acid content in melon fruit
The levels of soluble sugars and organic acids in Bailangua P7 were determined at 10, 20, 30, 35, and 40 d after pollination. Table 3 shows that the melon contained high levels of sucrose, fructose, and glucose; low levels of raffinose, stachyose, trehalose, and maltose; and no detectable levels of galactose, lactose, trehalose, rhamnose, and arabinose. Fructose and glucose increased as the fruit ripened, maximized at 35 d after pollination, and then decreased slightly at 40 d post-pollination. Sucrose was low from 10 to 30 d and accumulated rapidly from 35 to 40 d after pollination, peaking at 40 d post-pollination. Table 4 shows that the melon fruit contained high levels of citric acid and malic acid; low levels of shikimic acid; and no detectable levels of oxalic acid, tartaric acid, quinic acid, lactic acid, maleic acid, and succinic acid. Citric acid was the highest at 10 d after pollination, decreased rapidly from 10 to 20 d, and then increased and decreased from 20 to 40 d. Malic acid decreased with fruit ripening.
3.5 Relative expression of CmACL genes in melon fruit
The relative expression of CmACL genes was determined at different fruit developmental stages (Figure 5). The expression of four CmACL genes decreased and then increased with fruit ripening. The expression of CmACLα2 decreased from 10 to 20 d and then gradually increased from 20 to 40 d. The expression of CmACLα1, CmACLβ1, and CmACLβ2 decreased from 10 to 30 d and then increased from 30 to 40 d. The expression of CmACLα1 at 40 d after pollination was 2.36-, 2.65-, 2.95-, and 1.33-fold that at 10, 20, 30, and 35 d. The expression of CmACLβ1 at 40 d after pollination was 5.46-, 11.38-, 13.65-, and 4.40-fold that at 10, 20, 30, and 35 d.
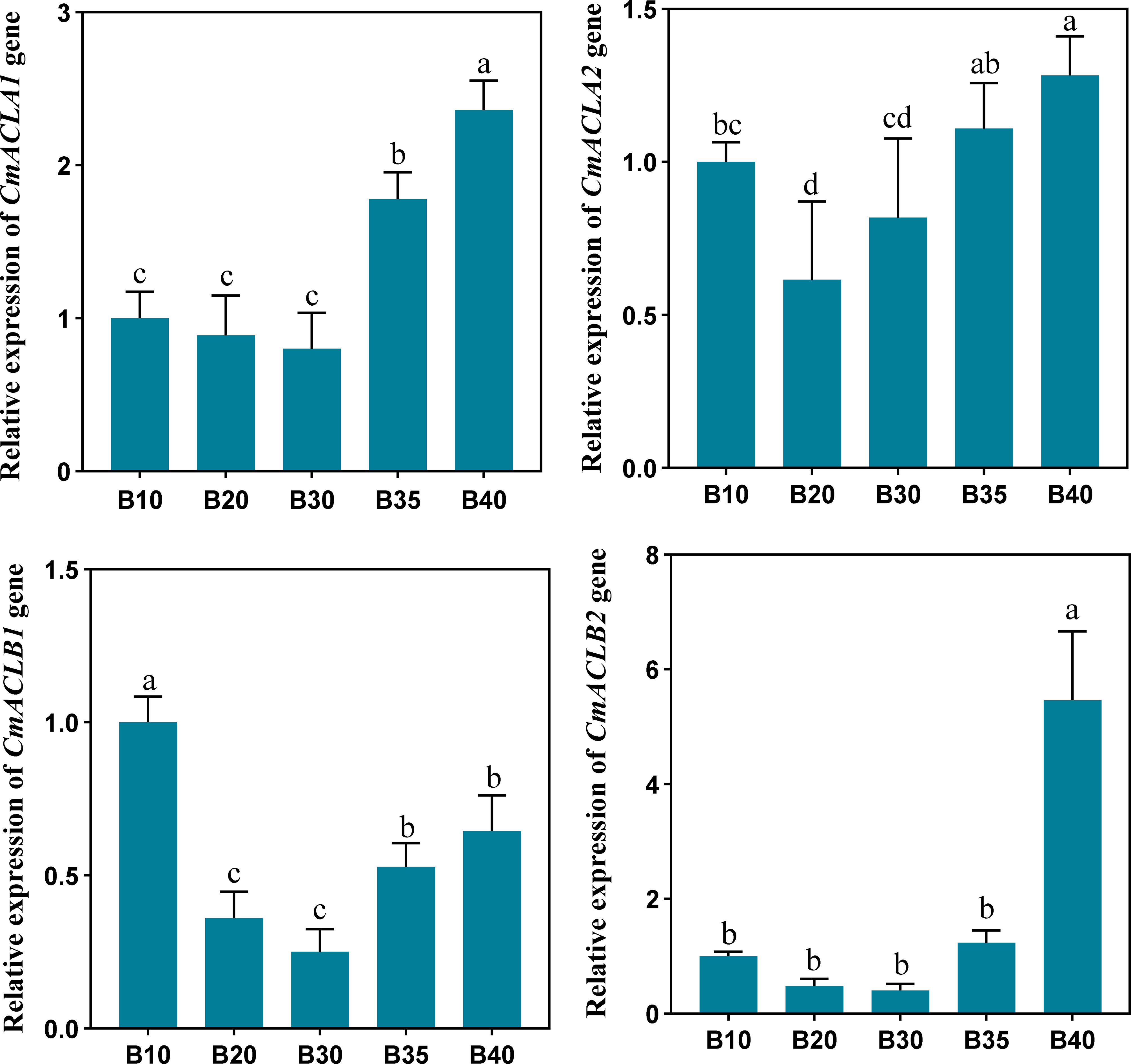
Figure 5 Relative expression of CmACL genes. The difference lowercase letters indicate statistically significant differences at P ≤ 0.05.
3.6 Correlation of CmACL gene expression with sugar and acid content
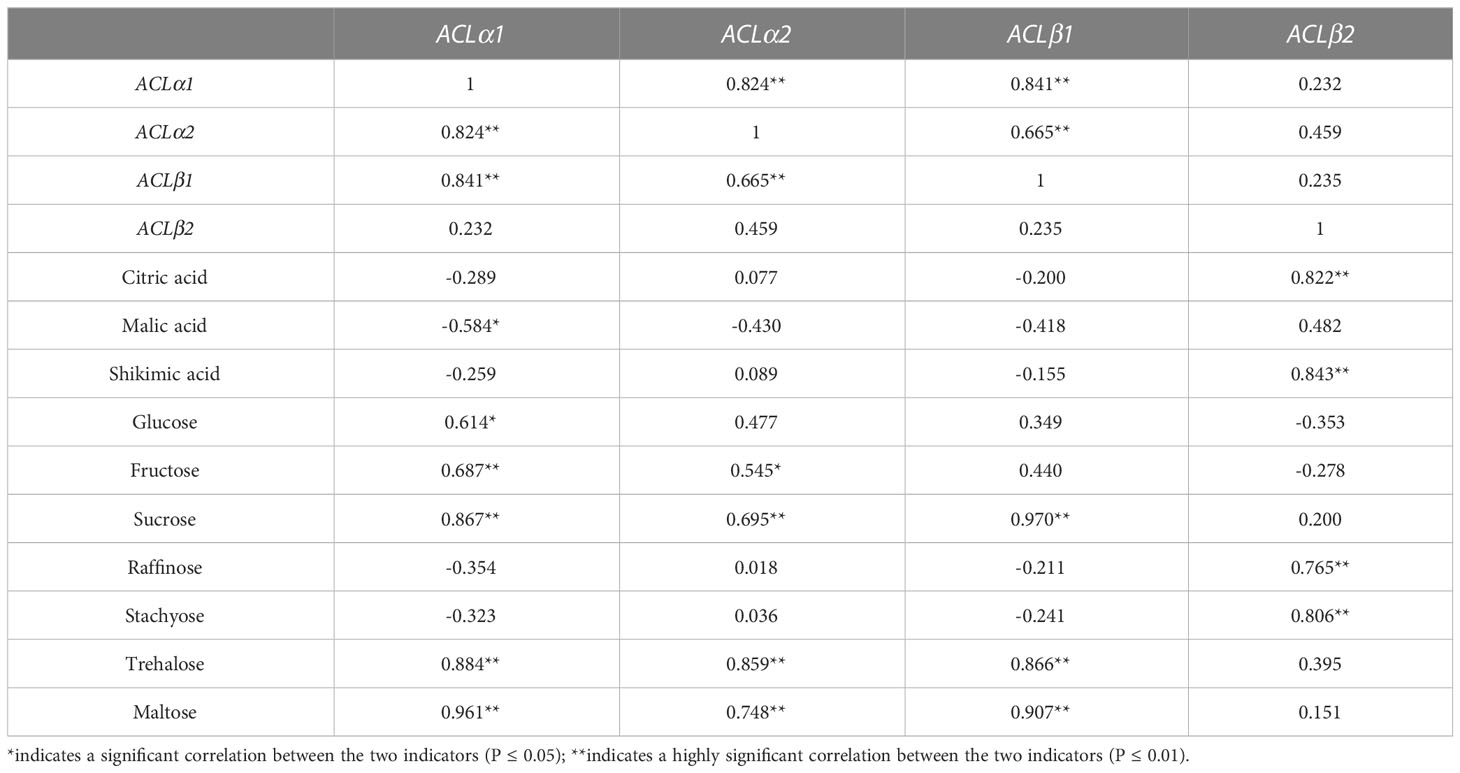
The expression of CmACL genes was correlated with the content of sugars and acids in melon (Table 5). The results showed significant correlations between the expression of four genes. For example, the expression of CmACLα1 was significantly and positively correlated with CmACLα2 and CmACLβ1 (correlation coefficients of 0.824 and 0.841, respectively), and the expression of CmACLα2 was significantly and positively correlated with CmACLβ1 (0.665).
There were significant correlations between the expression of CmACL genes and sugar and acid content. The expression of CmACLα1 was significantly negatively correlated with malic acid (−0.584) and significantly positively correlated with glucose, fructose, sucrose, trehalose, and maltose (0.614, 0.687, 0.867, 0.884, and 0.961). The expression of CmACLα2 was significantly positively correlated with fructose, sucrose, trehalose, and maltose (0.545, 0.695, 0.859, and 0.748). The expression of CmACLβ1 was significantly positively correlated with sucrose, trehalose, and maltose (0.970, 0.866, and 0.907). The expression of CmACLβ2 was significantly positively correlated with citric acid, shikimic acid, raffinose, and stachyose (0.822, 0.843, 0.765, and 0.806).
4 Discussion
The ACL proteins can be divided into α and β subfamilies based on conserved domains. In Lupinus albus and Saccharum officinarum, two genes were identified that encode proteins in the ACLα and ACLβ subfamilies, respectively (Langlade et al., 2002; Li et al., 2012). In A. thaliana, three genes encode ACLα proteins and two genes encode ACLβ proteins (Fatland et al., 2002). In C. reticulata, two genes encode ACLα proteins and one gene encodes an ACLβ protein (Wang, 2018). In the present study, four CmACL genes, including two α subfamily genes (CmACLα1 and CmACLα2) and two β subfamily genes (CmACLβ1 and CmACLβ2), were identified in melon by whole-genome analysis using the ACL protein sequences of A. thaliana and C. reticulata. Physicochemical characterization suggested that proteins in the same subfamily had identical numbers of amino acids (423 in the α subfamily and 608 in the β subfamily) and similar molecular weights (46653.68–46780.86 Da in the α subfamily and 65957.78–66017.74 Da in the β subfamily). These findings indicate different structures of proteins in different subfamilies. Subcellular localization showed that the four CmACL proteins were localized in the mitochondria, which differs from previous studies whereby ACL proteins were found to be localized in the plastid, cytoplasm, and nucleus (Rangasamy and Ratledge, 2000; Guo, 2020), which might be explained by the different species used in these studies. Analyses of gene structure and protein conserved motifs showed identical gene structures and conserved motifs in the same subfamily, indicating that they may have similar biological functions.
Plant ACL proteins are involved in many biological processes and have different functions in plants. These ACL proteins have important roles in plant growth and development. For instance, ACL overexpression promoted the accumulation of wax in A. thaliana stems, rubber and triterpenes in Taraxacum brevicorniculatum roots, and anthocyanins in Paeonia lactiflora petals, by increasing the abundance of their precursor acetyl-CoA (Go et al., 2014; Xing et al., 2014; Luan et al., 2022). The ACL proteins also play important regulatory roles in plant responses to stresses such as drought. The SoACL genes in S. officinarum and CitACL genes in C. reticulata were reported to respond to drought and abscisic acid (Crifò et al., 2012; Li et al., 2012; Lo Piero et al., 2014; Hu et al., 2015; Phan et al., 2017). Promoter sequences usually contain cis-acting elements with specific functions that reflect the potential functions of genes (Zhang et al., 2019). Analysis of the CmACL gene promoters identified elements related to light, stress response, plant growth and development, and phytohormone response. Moreover, the functions of ACL proteins in plant growth, development, and stress response have been widely documented in other plants. Therefore, we speculate that CmACL genes may have important roles in regulating melon growth, development, and the stress response. Furthermore, the prediction of protein interaction network showed that CmACL proteins were closely related to the proteins belonging to tricarboxylic acid cycle, glyoxylic acid cycle and glycolytic pathway, suggesting that CmACL protein may play an important role in sugar and acid metabolism.
The sugars and acids in melon determine fruit flavor quality. We measured the soluble sugar and organic acid contents in melon fruit. The results showed that the fruit mainly contained sucrose and citric acid, which is consistent with previous studies (Hu, 2018; Zhang et al., 2022). We also measured the relative expression of CmACL genes and found that their expression decreased and then increased with fruit ripening. The expression of CmACLα1, CmACLα2, and CmACLβ1 was significantly and positively correlated with sucrose and the highest correlation was CmACLβ1, with a correlation coefficient of 0.970. The expression of CmACLβ2 was significantly and positively correlated with citric acid, and its correlation coefficient was 0.843. Therefore, CmACLβ1 and CmACLβ2 may serve important roles in sugar and acid accumulation in melon fruit. CitACLα1 in C. reticulata (Li et al., 2020a) and VcACL in Vaccinium corymbosum (Li et al., 2020b) were reported to be involved in citric acid metabolism. The role of ACL in fruit sucrose regulation has not been reported. The present study provides a reference and new approach for further investigations of CmACL genes in sugar and acid metabolism in melon, providing a theoretical foundation for breeding high-quality melon cultivars.
5 Conclusion
In this study, four CmACL genes were identified in melon genome and divided into two subfamily according to phylogenetic relationships, and members of the same subfamily had similar gene structure and conserved domain. The promoter sequences of CmACL genes contained regulatory elements related to light, stress, phytohormones, and growth and development, and the protein interaction network prediction showed that CmACL proteins were closely related to the proteins belonging to tricarboxylic acid cycle, glyoxylic acid cycle and glycolytic pathway. Correlation analysis was conducted between CmACL genes expression and sugar/acid content. Taken together, the CmACL genes that potentially regulated sugar and acid metabolism were mined. The results expand the understanding of CmACL genes in melon and provide a reference for further investigation of their functions in sugar and acid accumulation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KR and HC designed the experiments. KR, WK and TT performed the experiments. KR analyzed the data, and wrote the manuscript. HC helped revise the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The paper is supported by Biological Breeding Project of Gansu Academy of Agricultural Sciences (Grant No. 2022GAAS05) and the National Natural Science Foundation of China (Grant No. 31960523).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1239482/full#supplementary-material
References
Crifò, T., Petrone, G., Lo Cicero, L., Lo Piero, A. R. (2012). Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges. J. Agric. Food Chem. 60 (1), 476–481. doi: 10.1021/jf203891e
Doncheva, N. T., Morris, J. H., Gorodkin, J., Jensen, L. J. (2019). Cytoscape stringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 18 (2), 623–632. doi: 10.1021/acs.jproteome.8b00702
Fatland, B. L., Ke, J., Anderson, M. D., Mentzen, W. I., Cui, L. W., Allred, C. C., et al. (2002). Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol. 130 (2), 740–756. doi: 10.1104/pp.008110
Go, Y. S., Kim, H., Kim, H. J., Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell. 26 (4), 1666–1680. doi: 10.1105/tpc.114.123307
Guo, L. X. (2020). Studying the role of citrus ATP-citrate lyase (ACL) in the accumulation of citrate and its relative metabolites, as well as ACL action mechanism and ACL-affecting factors (Wuhan: Huazhong Agricultural University). doi: 10.27158/d.cnki.ghznu.2020.000799
Hu, X. M. (2015). Genome-wide identification of ATP-citrate lyase genes in citrus, and its role in the metabolism of citric acid (Wuhan: Huazhong Agricultural University). doi: 10.7666/d.Y2803467
Hu, K. Y. (2018). The relationship between organic acid accumulation and related metabolic enzymes in melon fruit development (Zhengzhou: Henan Agricultural University). doi: 10.27117/d.cnki.ghenu.2018.000092
Hu, X. M., Shi, C. Y., Liu, X., Jin, L. F., Liu, Y. Z., Peng, S. A. (2015). Genome-wide identification of citrus ATP-citrate lyase genes and their transcript analysis in fruits reveals their possible role in citrate utilization. Mol. Genet. Genomics 290 (1), 29–38. doi: 10.1007/s00438-014-0897-2
Langlade, N. B., Messerli, G., Weisskopf, L., Plaza, S., Massonneau, A. (2002). ATP citrate lyase: Cloning, heterologous expression and possible implication in root organic acid metabolism and excretion. Plant Cell Environment. 25 (11), 1561–1569. doi: 10.1046/j.1365-3040.2002.00936.x
Li, X., Li, C., Sun, J., Jackson, A. (2020b). Dynamic changes of enzymes involved in sugar and organic acid level modification during blueberry fruit maturation. Food Chem. 309, 125617. doi: 10.1016/j.foodchem.2019.125617
Li, C. N., Nong, Q., Tan, Q. L., Manoj Kumar, S., Yang, L. T., Li, Y. R. (2012). Cloning and expression analysis of ATP-citrate lyase genes from sugarcane. Acta Agronomica Sinica. 38 (11), 2024–2033. doi: 10.3724/sp.j.1006.2012.02024
Li, S. J., Wang, W. L., Ma, Y. C., Liu, S. C., Grierson, D., Yin, X. R., et al. (2020a). Citrus citERF6 contributes to citric acid degradation via upregulation of citAclα1, encoding ATP-citrate lyase subunit α. J. Agric. Food Chem. 68 (37), 10081–10087. doi: 10.1021/acs.jafc.0c03669
Li, J. X., Zhang, C. L., Liu, H., Chen, D. L., Liu, C. J., Jiao, Z. G. (2019). Composition analysis of soluble sugars and organic acids in strawberry juice and its application for authentication. Sci. Technol. Food Industry. 40 (21), 268–273. doi: 10.13386/j.issn1002-0306.2019.21.044
Lo Piero, A. R., Lo Cicero, L., Puglisi, I. (2014). The metabolic fate of citric acid as affected by cold storage in blood oranges. J. Plant Biochem. Biotechnol. 23, 161–166. doi: 10.1007/s13562-013-0197-7
Lu, X. H., Sun, D. Q., Wu, Q. S., Liu, S. H., Chen, J., Sun, G. M. (2013). Components and contents of organic acids in pineapple germplasm. Chin. J. Trop. Crops. 34 (05), 915–920. doi: 10.3969/j.issn.1000-2561.2013.05.020
Luan, Y., Chen, Z., Wang, X., Zhang, H., Tao, J., Zhao, D. (2022). Herbaceous peony PlACLB2 positively regulates red petal formation by promoting anthocyanin accumulation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.992529
Phan, T. T., Li, J., Sun, B., Liu, J. Y., Zhao, W. H., Huang, C., et al. (2017). Atp-citrate lyase gene (soacla-1), a novel acla gene in sugarcane, and its overexpression enhance drought tolerance of transgenic tobacco. Sugar Tech. 19 (3), 258–269. doi: 10.1007/s12355-016-0464-8
Rangasamy, D., Ratledge, C. (2000). Compartmentation of ATP: citrate lyase in plants. Plant Physiol. 122 (4), 1225–1230. doi: 10.1104/pp.122.4.1225
Tang, M. (2010). Formation mechanism for the high sugar and high acid contents in the fruits of ‘flavor melon’ series (Wuhan: Huazhong Agricultural University). doi: 10.7666/d.Y1805333
Tang, M., Bie, Z. L., Wu, M. Z., Yin, H. P., Feng, J. X. (2019). Changes in organic acids and acid metabolism enzymes in melon fruit during development. China Cucurbits Vegetables 32 (08), 233–234. doi: 10.16861/j.cnki.zggc.2019.0210
Wang, W. L. (2018). Identification and transcriptional regulation of ACL genes in citrus furits (Hangzhou: Zhejiang University). doi: 10.27461/d.cnki.gzjdx.2018.000198
Wang, R., Tian, Y., Yang, L., Zhang, X. C. (2017). Analysis of organic acid components in tomato fruit during different development stages. China Vegetables 10, 58–62. doi: 10.19928/j.cnki.1000-6346.2017.10.010
Xing, S., van Deenen, N., Magliano, P., Frahm, L., Forestier, E., Nawrath, C., et al. (2014). ATP citrate lyase activity is post-translationally regulated by sink strength and impacts the wax, cutin and rubber biosynthetic pathways. Plant J. 79 (2), 270–284. doi: 10.1111/tpj.12559
Yu, T., Cen, Q., Kang, L., Mou, W., Zhang, X., Fang, Y., et al. (2022). Identification and expression pattern analysis of the OsSnRK2 gene family in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1088281
Yu, M., Ruan, C. J., Li, H., Zhang, S. F., Xu, H. H., Liu, X. Q. (2016). Components and contents of sugar and acid in 10 blueberry cultivars from the North American using HPLC. Sci. Technol. Food Industry. 37 (10), 77–81. doi: 10.13386/j.issn1002-0306.2016.10.006
Zhang, W., Abdelrahman, M., Jiu, S., Guan, L., Han, J., Zheng, T., et al. (2019). VvmiR160s/VvARFs interaction and their spatio-temporal expression/cleavage products during GA-induced grape parthenocarpy. BMC Plant Biol. 19 (1), 111. doi: 10.1186/s12870-019-1719-9
Zhang, X., Wei, X., Ali, M. M., Rizwan, H. M., Li, B., Li, H., et al. (2021). Changes in the content of organic acids and expression analysis of citric acid accumulation-related genes during fruit development of yellow (Passiflora edulis f. flavicarpa) and purple (Passiflora edulis f. edulis) passion fruits. Int. J. Mol. Sci. 22 (11), 5765. doi: 10.3390/ijms22115765
Zhang, K. L., Zhang, M. M., Kong, W. P., Tang, T. X., Ren, K. L., Cheng, H. (2022). Correlation analysis of sugar accumulation and physiological indexes in different developmental stages of muskmelon. Northern Horticulture. 14, 24–30. doi: 10.11937/bfyy.20220319
Keywords: melon, ACL gene family, ATP-citrate lyase, citric acid, fruit
Citation: Ren K, Kong W, Tang T and Cheng H (2023) Identification of CmACL genes in melon and analysis of their potential functions in fruit sugar and acid accumulation. Front. Plant Sci. 14:1239482. doi: 10.3389/fpls.2023.1239482
Received: 13 June 2023; Accepted: 01 August 2023;
Published: 15 August 2023.
Edited by:
Cristina Cruz, University of Lisbon, PortugalReviewed by:
Yuanyue Shen, Beijing University of Agriculture, ChinaSagar Datir, Naoroji Godrej Centre for Plant Research (NGCPR), India
Copyright © 2023 Ren, Kong, Tang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Cheng, Y2hlbmdqbkB5ZWFoLm5ldA==
 Kaili Ren
Kaili Ren Weiping Kong
Weiping Kong Hong Cheng
Hong Cheng