- 1ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2Bidhan Chandra Krishi Viswavidyalaya, Mohanpur, West Bengal, India
- 3Department of Crop and Soil Sciences, University of Georgia, Griffin, GA, United States
- 4School of Agriculture, GIET University, Gunupur, Rayagada, Odisha, India
- 5ICAR- National Institute of Biotic Stress Management, Raipur, India
- 6WBAS (Research), Government of West Bengal, Field Crop Research Station, Burdwan, India
- 7Centurion University of Technology and Management, Paralakhemundi, Odisha, India
- 8Indian Institute of Pulses Research, Kanpur, India
Improper use of water resources in irrigation that contain a significant amount of salts, faulty agronomic practices such as improper fertilization, climate change etc. are gradually increasing soil salinity of arable lands across the globe. It is one of the major abiotic factors that inhibits overall plant growth through ionic imbalance, osmotic stress, oxidative stress, and reduced nutrient uptake. Plants have evolved with several adaptation strategies at morphological and molecular levels to withstand salinity stress. Among various approaches, harnessing the crop genetic variability across different genepools and developing salinity tolerant crop plants offer the most sustainable way of salt stress mitigation. Some important major genetic determinants controlling salinity tolerance have been uncovered using classical genetic approaches. However, its complex inheritance pattern makes breeding for salinity tolerance challenging. Subsequently, advances in sequence based breeding approaches and functional genomics have greatly assisted in underpinning novel genetic variants controlling salinity tolerance in plants at the whole genome level. This current review aims to shed light on physiological, biochemical, and molecular responses under salt stress, defense mechanisms of plants, underlying genetics of salt tolerance through bi-parental QTL mapping and Genome Wide Association Studies, and implication of Genomic Selection to breed salt tolerant lines.
1 Introduction
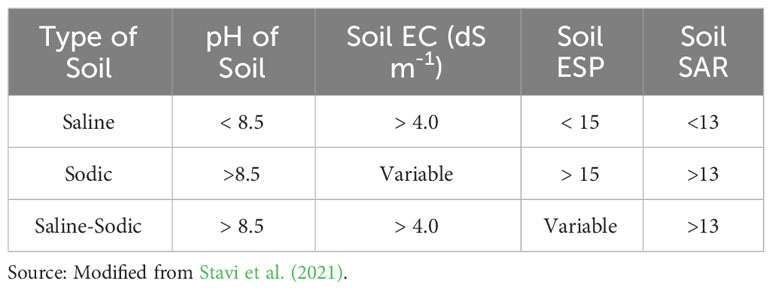
An abiotic or biotic constraint that reduces a plant’s ability to convert energy to biomass can be called plant stress (Grime, 1977). Reduction in crop yield due to various abiotic stresses such as excessive salt, drought, cold, and heat is a major challenge to meet rising food demand (Vorasoot et al., 2003; Kaur et al., 2008; Shanker and Venkateswarlu, 2011; Ahmad et al., 2012; Mantri et al., 2012; He et al., 2018). Crop cultivation is facing many challenges due to soil salinity which is not only limited to coastal areas, but also induced by other factors like faulty agronomic practices or the use of recycled water in irrigation that may contain large amounts of salts (Kumar and Sharma, 2020). Salinity stress and thereby the ensued damages to the plant may arise due to an excess accumulation of soluble ions (Na+, Ca+, K+, Mg2+) in the root zone (Tester and Davenport, 2003; Munns, 2005; Atta et al., 2022a; Atta et al., 2022b). Saline soils inevitably have salt concentrations high enough in their solutions to impair plant growth. Sulphates and chlorides of calcium, sodium, and magnesium are commonly associated with the development of soil salinity. However, the detrimental effects of Na salts on plants were reported to be more pronounced than that of calcium (Bryla et al., 2021; Wu et al., 2023). As the electrical conductivity (EC) of any system increases with the increasing abundance of neutral salt species, it is the most widely used determinant for assessing the degree of soil salinity. Sodic soils, on the other hand, are formed by the excessive accumulation of sodium ions in the soil exchange complex. These are often characterized by high pH and dispersibility (and consequent poor soil transmissibility). Carbonates and bicarbonates of sodium play major role in the development of soil sodicity. Exchangeable Sodium Percentage (ESP) and Sodium Adsorption Ratio (SAR) are the most useful parameters to assess the hazard of sodium in soil and soil solution, respectively. Classification of salt affected soils based on soil pH, Electrical Conductivity (EC), ESP, and SAR is given in Table 1.
There are two types of salinity based upon salt accumulation: dry land salinity and irrigation salinity (Chakraborty et al., 2013; McFarlane et al., 2016; Stavi et al., 2021). Dry land salinity refers to the accumulation of salts in the soil surface of non-irrigated lands. There are three general processes which are associated with dryland salinity: deep drainage, groundwater movement, and groundwater discharge. Dry land salinity may be classified into two categories: Primary, where salinity occurs naturally, and secondary, where salinity is caused by human activities, such as agriculture. Irrigation salinity occurs when rigorous irrigation with groundwater builds up salinity on the surface of soil through repeated salt accumulation, due to the leaching of irrigation water but salts (Chakraborty et al., 2013; McFarlane et al., 2016 and Zaman et al., 2018).
Plants are amenable to the detrimental effects of salinity throughout their life-cycle but are most vulnerable during the germination and seedling stage. The negative effects of salt on plant growth are related to a reduction in the osmotic potential of growing media, specific ion toxicity, and nutrient imbalance (Greenway and Munns, 1980; Askari-Khorasgani et al., 2021 and Lu et al., 2023). The level of salt tolerance in plants is determined by osmotic adjustment and ionic homeostasis (Acosta-Motos et al., 2017). Excess ions in the soil water lower the solute potential (ψs) and thereby the total water potential (ψw) of the soil. To maintain water uptake and turgor under such conditions, plants need to keep their internal water potential (ψw) below that of the soil (Taiz et al., 2015) Osmotic adjustment is mediated by the accumulation of osmolytes viz., organic acids, sugars, and amino acids in plant cells under salt stress. Increased accumulation of osmolytes helps plants lower their water potential to facilitate water uptake from saline soils (Acosta-Motos et al., 2017; Ma et al., 2020; Munns et al., 2020; Zhao et al., 2021). Plants need to maintain a balance between the accumulation of sodium (Na+) and the loss of potassium (K+) from the cell through ion homeostasis to ensure proper cellular functions. A high potassium-to-sodium (K+: Na+) ratio in tissues serves as an important indicator of higher salt tolerance in plants. Key strategies to maintain a higher K+:Na+ involve selective ion uptake and transport mechanisms that allow plants to either exclude or compartmentalize excess Na+, and retain K+ (Tester and Davenport, 2003; Hasanuzzaman et al., 2018; Ketehouli et al., 2019; Kumari et al., 2021). Therefore, a clear understanding of the mechanisms of salt tolerance at physiological, biochemical, and molecular levels, underlying genetics, and chromosomal regions associated with salt tolerance helps in the identification and further exploitation of tolerant genotypes. The impact of salinity on plants, their stress tolerance mechanisms, and the deployment of modern genomic and breeding approaches to understanding the genetics and mitigation of salt stress are explored in this review article.
2 Current global scenario of soil salinity
The increase in soil salinity poses a serious threat to agriculture production worldwide. Here, Figure 1A shows the gradual increase of total salt affected land area over thirty years. Globally, more than one billion hectares of land is affected in more than 100 countries and these numbers are constantly growing (Szabolcs, 1989; Squires and Glenn, 2004; Abbas et al., 2013; FAO and ITPS, 2015; Hossain, 2019; Ivushkin et al., 2019). According to Hossain (2019), about 1125 million hectares of land are affected by salinity at the present time, of which 76 million are affected by human-induced salinization and sodification, and 1.5 million hectares become unsuitable for agricultural production each year due to rising salinity levels. Recently published data by Food and Agricultural Organization (FAO) in 2021 showed global distribution of saline land area at topsoil (0-30 cm) and subsoil (30-100 cm) profile (Figure 1B). The salinization of the soil will result in the loss of 50% of cultivable lands by 2050 if it continues increasing at the present rate (Kumar and Sharma, 2020).
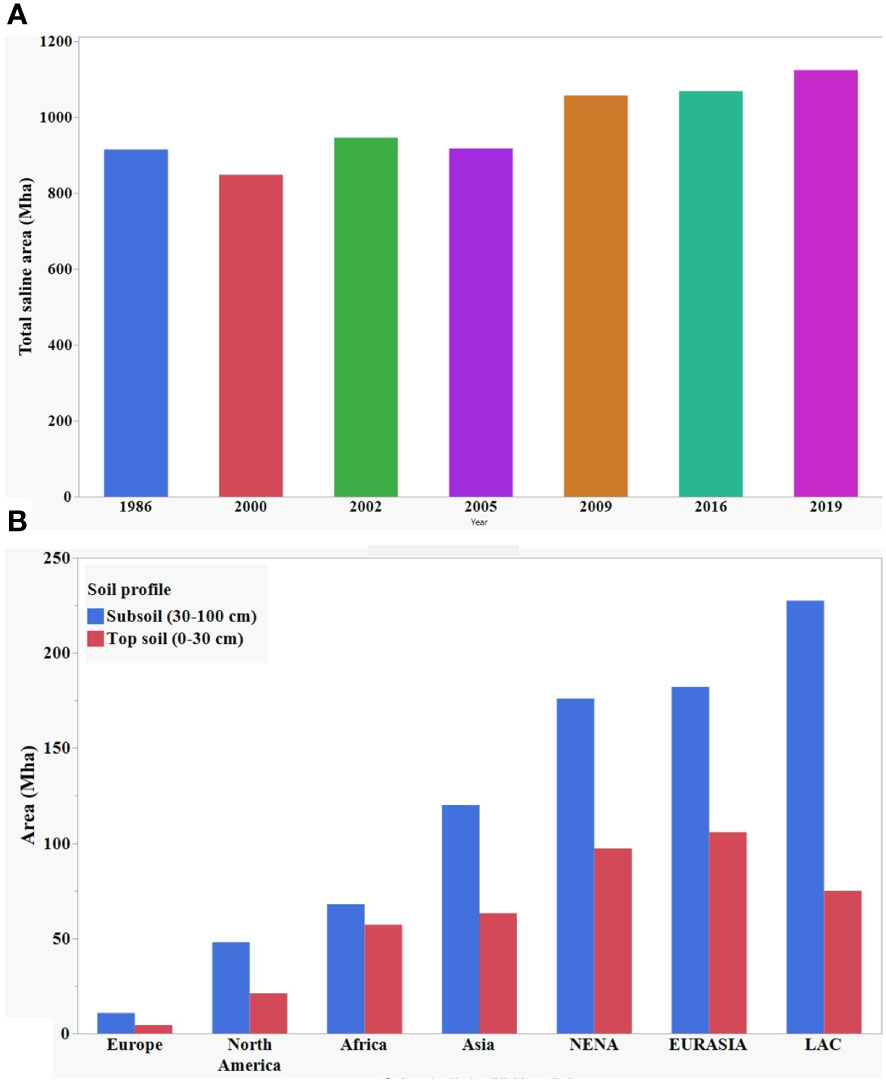
Figure 1 (A) Year wise distribution of global total saline area. (B) Global distribution of salt affected area at topsoil and subsoil. Data source: Ivushkin et al. (2019); Hossain (2019), and FAO (2021).
3 Brief account of physiological and biochemical alterations under salinity stress
3.1 Effect on germination and seedling stage
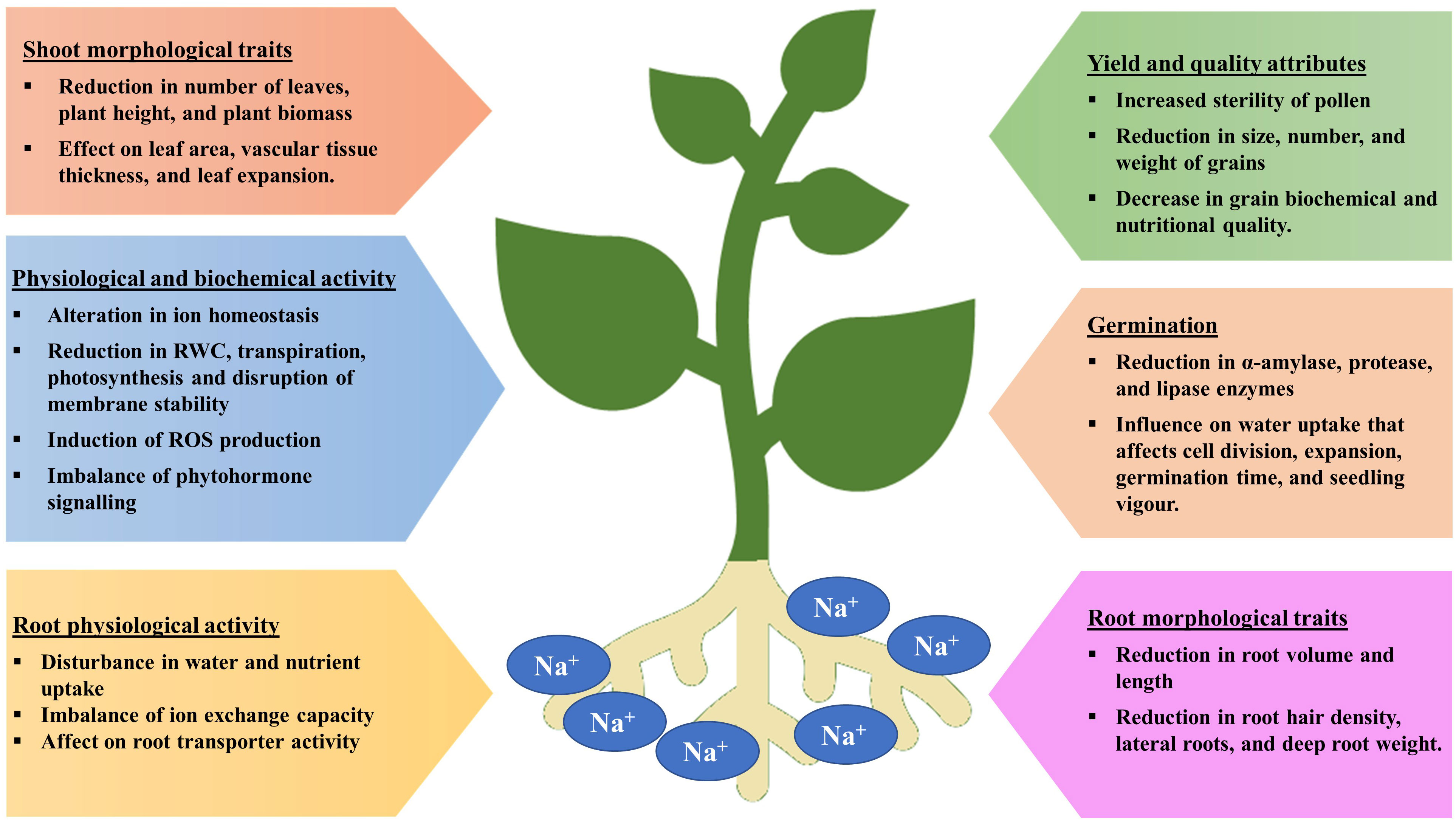
Seed germination is a multi-stage developmental process that is influenced by both internal and external factors. Impact on seed germination under salinity stress may be attributed to the delayed absorption of water and a decline in the activity of α-amylase, an enzyme involved in starch hydrolysis. Salinity lowers the soil osmotic potential relative to the internal osmotic potential of seed, which inhibits the absorption of water during seed imbibition (Figure 2) (Munns, 2002 and Munns et al., 2020). As a consequence, the seed germination rate is reduced and the seed germination period is delayed. Even after germination, salinity may also have detrimental effect on embryo viability due to the excess accumulation of Na+ and Cl- ions (Daszkowska-Golec, 2011; EL Sabagh et al., 2021). Furthermore, salt stress increases the generation of reactive oxygen species (ROS) and oxidative damages, which disrupt different macromolecules. Therefore, a decrease in α-amylase activity results in a significant reduction in the transfer of sugars, which restricts the embryo’s growth and development. (Hasanuzzaman et al., 2021).
Under normal conditions, seed germination occurs in three phases. Dry seeds absorb water rapidly (imbibition) during Phase I of germination. In phase II, metabolic processes are reactivated and water uptake is limited. Phase III is the post-germination phase, during which continuous water uptake occurs until full germination. Phase I osmotic stress and phase II ionic stress are generally attributed to inhibiting seed germination or delaying germination under salinity stress (Yadav et al., 2011). However, even after seed germination, the development of embryonic organs, seedling growth, and vigor are affected at phase III by both ionic and osmotic stress (Wahid et al., 2014; El-Hendawy et al., 2019).
Salinity impose deleterious effects on seed germination by decreasing gibberellic acid levels, increasing abscisic acid levels, altering membrane permeability, and reducing water absorption in seeds. (Lee and Luan, 2012). Germination of seeds occurs when it is catalyzed by hydrolytic enzymes such as α-amylase which subsequently breaks down the starch stored in the endosperm into metabolizable sugars that provide energy to the growing embryo and radicle. (Weiss and Ori, 2007). For most crops, although seed may be germinated under a certain limit of salinity, it significantly delays the seed germination. The major causes of a delay in germination time may include a delay in water intake and a decrease in α-amylase activity with an increase in NaCl content (Kaneko et al., 2002). It has been noted that the salt-sensitive genotypes exhibit a greater decline in α-amylase activity than the salt-tolerant genotypes. This decrease in α-amylase activity has a substantial impact on the translocation of sugars, which is crucial for the development of the embryo (Uçarlı, 2020).
3.2 Ionic imbalance and salinity mediated nutritional deficiencies
It is also important to note that some ions can also act as plant nutrients, such as K+ and SO42-, while Na+ is not considered to be an essential plant nutrient. Thus, soil salinity is often measured in terms of Na+ and Cl− (Stavi et al., 2021). Plants are affected by salinity in three ways. Because of its low osmotic potential, salt makes it difficult for plants to extract water from the soil, subjecting plants to osmotic stress, which limits growth and reduces yield. The Na+ and Cl- ions when absorbed and accumulated into the tissues by plants at excess concentrations from soil cause cytotoxicity which eventually result in leaf firing, reduced growth, and finally plant death. Moreover, high levels of Na+ decrease the availability of other ions such as K+, Ca2+, and Mg2+ due to the cation competition, which can lead to nutrient deficiencies (Munns and Tester, 2008; Atta et al., 2019; Yildiz et al., 2020; Atta et al., 2021). During salinity, the plant takes up more Na+ than K+ as the amount of Na+ in the growth medium increases, increasing K+ efflux from the cell and raising the Na/K ratio (Figure 3) (Parvin et al., 2016; Rahman et al., 2016). During salt stress, excessive Na+ influx encourages ion channel disruption, nutrient replacement and membrane depolarization, leading to abnormalities in nutrient uptake and assimilation (Shabala et al., 2007; Zhao et al., 2007; Nahar et al., 2016; Gaikwad et al., 2022). According to a study in rice by Farooq et al. (2022), the concentrations of all measured nutrients (Fe, K, Mn, Mg, P, and Zn) in the roots and shoots declined under salinity stress. Salinity stress significantly reduces the root surface area by lowering root hair density and root hair length which are directly proportional to nutrient uptake (Robin et al., 2016 and Arif et al., 2019). Essential elements including Ca2+, Mg2+, Fe2+, and Zn2+, which are impacted by salt stress, are required for normal root growth. Therefore, the decrease in root growth can further affect the intake of Ca2+, Mg2+, Fe2+, and Zn2+ (EL Sabagh et al., 2021; Robin et al., 2016). According to certain studies, corn shoots under salinity stress had lower Mn2+ levels (Rahman et al., 1993). Nahar et al. (2016) reported, salt stress decreased the amounts of Ca2+, Mg2+, and Zn2+ in the leaves of mung bean seedlings.
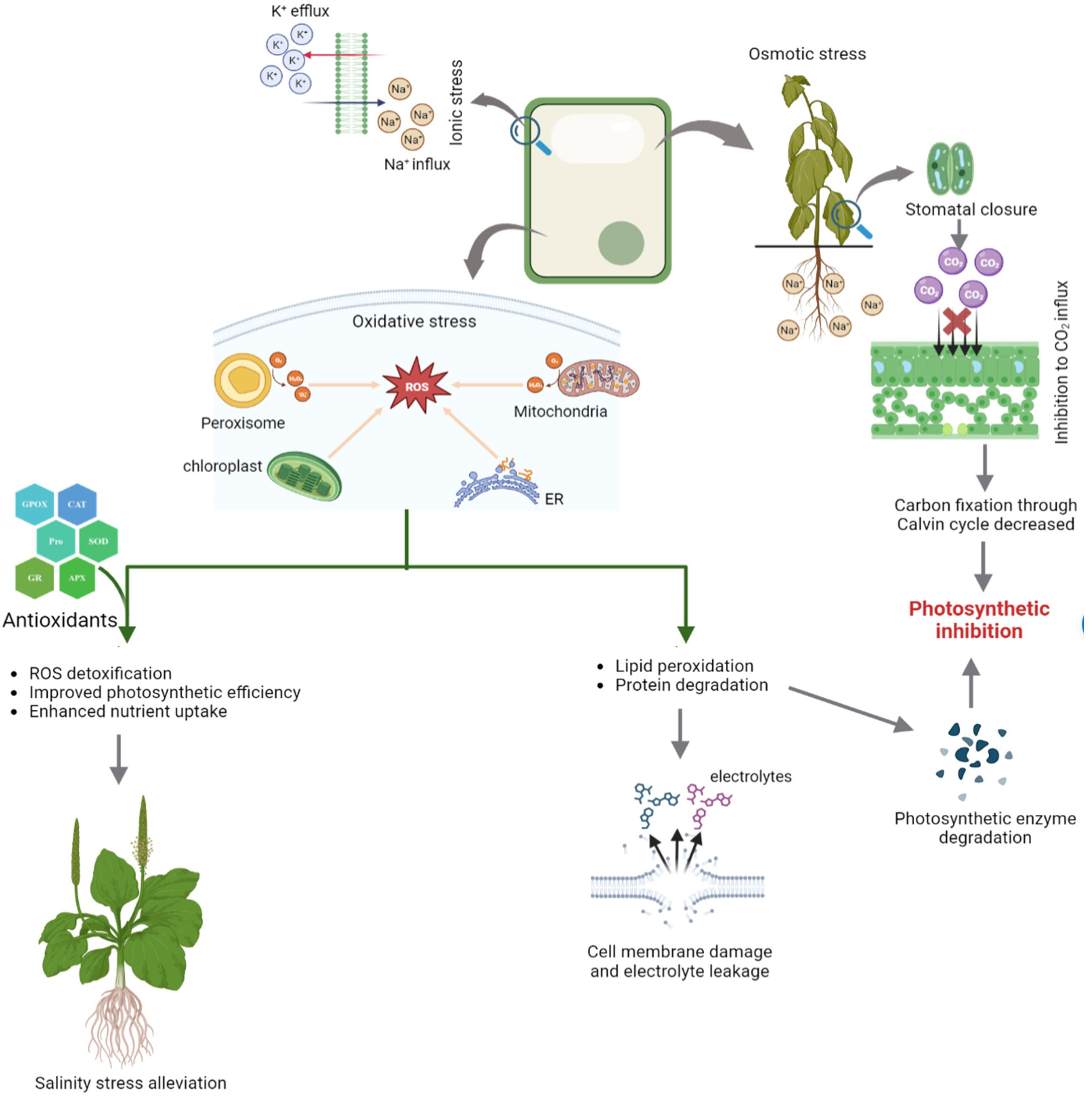
Figure 3 Different physiological alterations in plants under salinity and role of antioxidants in stress alleviation.
3.3 Oxidative stress caused by salinity
Along with its direct effects on plants, salinity frequently results in an excessive build-up of reactive oxygen species (ROS), which can interact with other essential components of plant cells and cause oxidative damage in plants, such as DNA damage, lipid peroxidation, enzyme inactivation, protein oxidation, hormone and nutritional imbalances (Hasanuzzaman et al., 2021). ROS are primarily produced in the chloroplasts, mitochondria, endoplasmic reticulum, cytosol, and peroxisome (Figure 3). Stomatal closure caused by salt stress can decrease the amount of available carbon dioxide in the leaves and thus, induces photosynthetic inhibition (Kamran et al., 2020). The light reactions in the chloroplast have a pivotal role in the production of a majority of ROS such as superoxide (O2 •-), hydrogen peroxide (H2O2), hydroxyl radical (OH•), and singlet oxygen (1O2) (Parida and Das, 2005; Ahmad and Sharma, 2008; Ahmad et al., 2010a; Khorobrykh et al., 2020).
The complex nature of salt stress coupled with water deficit have a variety of detrimental effects on plant metabolic process which in turn produce ROS that affect plant systems. Previous studies have shown oxidative damage caused by salt in many crops, viz. in Oryza sativa (Xu et al., 2011; Rahman et al., 2016), Zea mays (Khodarahmpour et al., 2012), Vigna radiata (Nahar et al., 2016) and Solanum lycopersicum (Kaveh et al., 2011). They also reported reduced ascorbate (AsA) to dehydroascorbate (DHA) ratio, superoxide dismutase (SOD) and catalase (CAT) activity, along with an increase of methylglyoxal (MG) content, which collectively contribute to oxidative damage. Greater levels of H2O2, as well as higher MDA and overproduced MG, were also reported in rice seedlings under salt stress, where Lipoxygenase (LOX) and SOD activity increased while CAT activity decreased (Rahman et al., 2016).
3.4 Effect on yield and yield components
There have been numerous studies demonstrating that abiotic stresses, like salinity, cause significant yield losses in major crops during the reproductive stage (Kalhoro et al., 2016; Ehtaiwesh and Rashed, 2020). During the reproductive stage, Na+ is excluded from leaf blades by the class I high-affinity K+ transporter (HKT) family, affecting sodium ion homeostasis under salinity stress (Suzuki et al., 2016). It was also found that grain dry matter and the K+/Na+ ratio was significantly correlated with grain filling rate and duration under salt stress (Poustini and Siosemardeh, 2004).
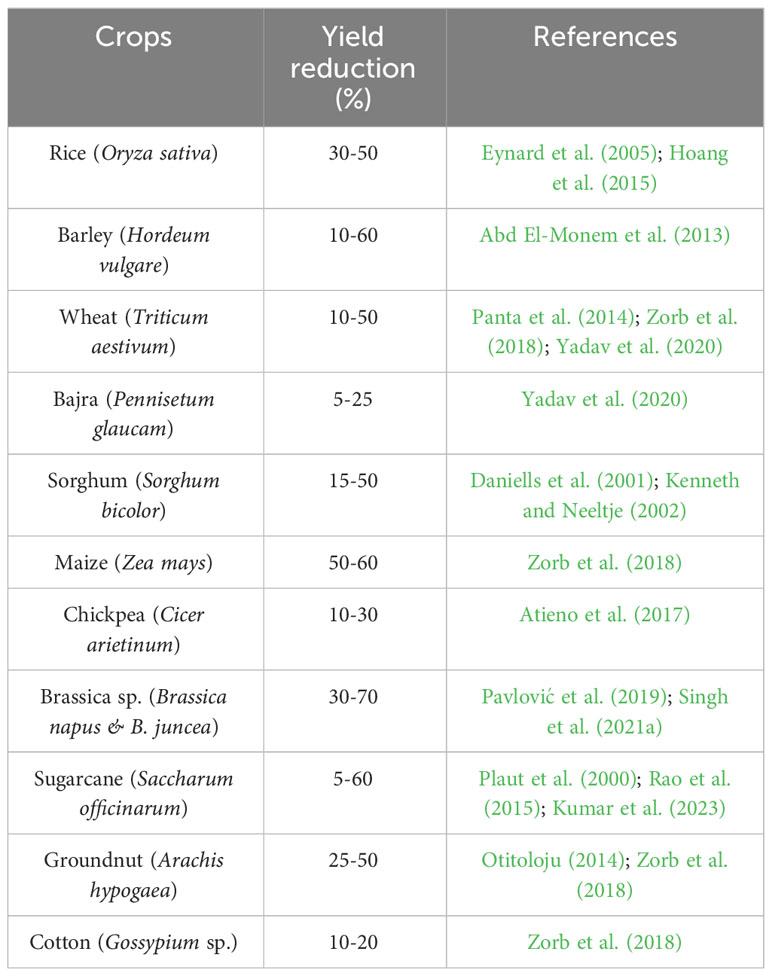
Changes in water relations, transpiration, nutritional imbalances, stomatal conductance, and oxidative damage due to salt stress all contribute to a drop in yield. By altering morpho-physiological and biochemical processes, salinity reduces agricultural yield and production (Kaveh et al., 2011). Additionally, it slows down photosynthetic activity, biomass accumulation, and source-sink activity, which has a negative impact on yield response variables and speeds up the senescence of reproductive organs (Khataar et al., 2018). Similar to this, throughout the reproductive phase, changes in water potential result in decreased flag leaf and vascular tissue thickness, mesophyll cell size, cell elongation, and epidermal cell size, which affect the leaf area, flag leaf turgidity, assimilate synthesis, and ultimately yield potential (Farouk, 2011). However, Ashraf and Ashraf (2016) hypothesized that changes in such biochemical and physiological characteristic are stage-specific and related to yield attributes. Salinity, for instance, reduces grain output by 39.1, 24.3, and 13.4%, respectively, at various stages of wheat like anthesis, early booting, and mid grain filling (Ashraf and Ashraf, 2016; Sabagh et al., 2021). Nadeem et al. (2020) found that salinity had a detrimental effect on wheat crop yield (yield, test weight and grain length), nutritional quality features (gluten content, fiber, fat, ash and moisture), and mineral element content (Mg, P, Ca, Zn, K and Fe). At 8 dSm−1, yield per plant in Brassica oleracea var. capitata can drop by up to 62% (Parvin et al., 2017). Salinity has a considerable impact on crop reproductive responses and yield contributing qualities, resulting in yield loss (Parvin et al., 2015a). Various crop species, including moong bean (Nahar and Hasanuzzaman, 2009), B. oleracea var. capitata (Parvin et al., 2017), tomato (Parvin et al., 2015a) and B. oleracea var. italica (Parvin and Haque, 2017), showed reduced yield component and yield under salt stress. Table 2 illustrates the extent to which salinity stress reduces crop yields.
4 Plant adaptations to salinity stress and mechanisms
In high salinity soils, plants develop a range of physiological and biochemical adaptations. In addition to ion homeostasis and compartmentalization, the principal responses include biosynthesis of compatible solutes, osmo-protectants, antioxidant compounds, polyamines, and nitrous oxide, as well as regulation of phyto-hormones. The following sections discuss recent advances in elucidating these mechanisms.
4.1 Ion uptake, transport, compartmentalization, and homeostasis
Plant organs are unable to function when their tissues or cells contain excessive Na+ or Cl– concentrations. Increased Na+ concentration usually results in a decrease in K+, and it may be critical for tolerance to maintain cytosolic K+ levels at an acceptable level or to maintain ‘homeostasis’ (Shabala and Pottosin, 2014). The toxic effect of Na+ may also be a result of its competition with K+ for enzymes that require K+, such that the ratio of cytoplasmic Na+ to K+ may be more important than the concentration of Na+ itself (Shabala and Cuin, 2008).
Neither glycophytes nor halophytes can tolerate high salt concentrations in their cytoplasm; however halophytes have developed mechanisms to sequester these ions. Na+ ion exclusion or compartmentalization’s are essential for normal plant growth under salinity stress (Serrano et al., 1999; Hasegawa, 2013). In saline soils, the most abundant salt form is NaCl, therefore most research has been on how Na+ ions are transported and compartmentalized in cells. During salinity stress, cell membranes and their associated components regulate ion uptake and transport within the cytosol (Sairam and Tyagi, 2004). This results in either excess salt being transported to the vacuole or sequestered in older tissues that eventually senesce, thereby protecting plants from salt stress (Reddy et al., 1992; Zhu, 2003). Carrier proteins, channel proteins, antiporters, and symporters all take part in ion transport. Some well characterized transporters include Na+ transporter AtHKT1:1 (Rus et al., 2004), Na+/H+ antiporter AtNHX1 (Apse et al., 1999), K+/Na+ symporter TaHKT1 (Schachtman and Schroeder, 1994) and Na+/H+ antiporter SOS1 (Zhu, 2003). These antiporters facilitate the compartmentalization of excess ions through the movement of Na+ ions into vacuoles from the cytoplasm (Gupta and Huang, 2014; Segami et al., 2018). After Na+ ions enter the cytoplasm in excess, antiporters transport them into vacuoles. Vacuolar membrane H+ pumps exist in two forms: the vacuolar type H+-ATPase (V-ATPase) and the vacuolar pyro-phosphatase (V-PPase); V-ATPase being the predominant form which generate motive forces across the vacuolar membrane. The ability of the plant to survive under high salinity depends to a great extent on its V-ATPase activity (Dietz et al., 2001).
4.2 Osmoprotection via compatible solutes
Compatible solutes, also called osmolytes are uncharged, polar, and soluble molecules which usually do not interfere with the cellular metabolism even at high concentrations. Organic osmolytes are synthesized and accumulated in varying amounts amongst different plant species to adjust osmotic potentials and protect cells. They are most commonly proline (Ahmad et al., 2010b; Hossain et al., 2011; Nounjan et al., 2012; Tahir et al., 2012), glycine betaine (Khan et al., 2000; Wang and Nii, 2000), sugars (Bohnert et al., 1995; Kerepesi and Galiba, 1995), and polyols (Ashraf and Foolad, 2007; Saxena et al., 2013). The amino acid proline has been found in diverse taxonomical groups of plants (Saxena et al., 2013), while accumulations of beta alanine betaine have been observed in plants of the Plumbaginaceae family which are glycophytes (Hanson et al., 1994 Matysik et al., 2002; Ben Ahmed et al., 2010). Under salinity stress, the concentrations of cysteine, arginine, and methionine, which represent about 55% of total free amino acids, decreased, whereas proline concentration increased (El-Shintinawy and El-Shourbagy, 2001). Additionally, proline accumulated in the intracellular space during salt stress also serves as an organic nitrogen reserve during stress recovery. In salt-stressed plants, sugars like glucose, fructose, fructans, and trehalose are also accumulated (Parida et al., 2004). These carbohydrates facilitate stress mitigation via, osmoprotection, and neutralization of reactive oxygen species. Many plant species have been reported to increase their levels of reducing and non-reducing sugars (sucrose and fructans) when they are under salinity stress (Kerepesi and Galiba, 1995). Trehalose accumulation is not only a carbohydrate reserve but also a protective mechanism against several stresses including salinity (Ahmad et al., 2013). A reduction in sucrose content was observed in tomato (Solanum lycopersicum) when exposed to salinity; due to an increase in saccharophosphate synthase activity (Gao et al., 1998). Different rice genotypes have been reported to both increase and decrease their sugar content under salinity stress (Alamgir and Yousuf Ali, 1999). Compatible solutes stabilize cellular structures and enzymes, act as metabolic signals, and scavenge ROS. Osmoprotection through compatible solutes is thus an important mechanism for plants to mitigate the negative effects of salinity stress.
4.3 Antioxidant regulation
Abiotic stresses including salinity result in electron overflow, deregulation, and even disruption of electron transport chains (ETCs) in chloroplasts and mitochondria. ROS produced under salinity stress are scavenged by enzymatic oxidants (SOD- Superoxide dismutase, APX- Ascorbate peroxidise, GPX- glutathione reductase, CAT- catalase, PPO- polyphenol oxidase, MDHA- monodehydroascorbate, MDHAR- monodehydroascorbate reductase etc.) as well as non-enzymatic antioxidants (reduced glutathione, flavanoids, phenolics, α-tocopherol, alkaloids etc.) which protect the plants from oxidative damage (Figure 3) (Hasanuzzaman et al., 2021). Among these, Ascorbate peroxidase (APX) and glutathione reductase (GR) are important antioxidant enzymes positively related to salt tolerance (Asada, 1999; Gupta et al., 2005). At lower concentration, ROS act as signaling molecules which initiates complex cascade of pathways and interactions (Golldack et al., 2014). Of these, MAPK (mitogen-activated protein kinase) and salt overly sensitive (SOS) signaling pathway cascades are important mediators of osmotic, ionic and ROS homoeostasis (Huang et al., 2019). ROS signals, through these pathways, trigger antioxidant defense mechanisms and scavenging of ROS (Kurusu et al., 2015).
4.4 Role of polyamines
Polyamines (PA) are cationic, aliphatic, and low molecular weight molecules which play a variety of roles in normal growth and development, including cell proliferation, morphogenesis, and growth of flowers and fruits. Plants show higher tolerance to stresses when polyamine levels rise (Yang et al., 2007; Groppa and Benavides, 2008; Kovács et al., 2010; Gupta et al., 2013a; Gupta et al., 2013b). Under salinity stress, polyamines contribute to cellular responses by modulating ROS homeostasis (Takahashi and Kakehi, 2010). Some research has suggested exogenous application of polyamines helps to alleviate salinity stress (Minocha et al., 2014; Rathinapriya et al., 2020) whereas it has also been suggested that catabolism products of polyamines (like H2O2) limit the ability of plants to tolerate stress (Saha et al., 2015). Further investigations on the role of polyamines are needed in this regard before conclusive remarks can be made.
4.5 Roles of nitric oxide
The molecule Nitric Oxide (NO) is a small volatile gas that plays an important role in many important plant processes, including regulating root growth, respiration, stomata closure, flowering, cell death, seed germination, and stress responses (Crawford, 2006; Besson-Bard et al., 2008; Zhao et al., 2009). Many redox-regulated genes are induced by NO directly or indirectly. By interacting with lipid radicals, NO prevents lipid oxidation, scavenging superoxide radicals and forming peroxynitrite that can be neutralized by other cellular processes. Additionally, it activates antioxidant enzymes (SOD, CAT, GPX, APX, and GR) (Bajgu, 2014). There is evidence that exogenous NO application can mitigate stress but the effects are dependent on NO concentration (Hossain et al., 2010; Sung and Hong, 2010; Xiong et al., 2010). NO has been demonstrated to mediate salt stress tolerance in plants (Wang et al., 2012) by counteracting germination inhibition (Zheng et al., 2009), negating inhibition of growth (Ageeva-Kieferle et al., 2019) and by its role in ion-homeostasis (Zhao et al., 2004).
4.6 Hormonal regulation of salinity tolerance
Among the well characterized plant hormones, abscisic acid (ABA), salicylic acid, jasmonic acid, and ethylene are considered as stress response hormones (Verma et al., 2016). Many studies have elucidated that these phytohormones have sophisticated roles in plant systems and their action is growth stage, tissue and environment specific (Ku et al., 2018). As a consequence of osmotic stress and water deficit, salt stress increases ABA production in vascular tissues and its distribution in roots and shoots (Popova et al., 1995; Jeschke et al., 1997; Cramer and Quarrie, 2002; Kang et al., 2005; Cabot et al., 2009). There is evidence that the positive association between ABA accumulation and salinity tolerance is at least partially due to the accumulation of potassium, calcium, and compatible solutes, such as proline and sugars, within the vacuoles of roots, which counteract Na+ and Cl- uptake (Golldack et al., 2014). In rice seedlings, the level of endogenous salicylic acid increased under salinity stress (Sawada et al., 2006). Furthermore, salinity can be mitigated by brassinosteroids (Krishna, 2003; Ashraf et al., 2010; El-Mashad and Mohamed, 2012). As a result of brassinosteroid application, the antioxidant enzymes SOD, POX, APX, and GPX were increased and non-enzymatic antioxidant compounds (tocopherol, ascorbate, and reduced glutathione) were accumulated (El-Mashad and Mohamed, 2012). Studies on Arabidopsis provide most of our information about hormone-mediated salt stress tolerance, but rice and maize might not conserve the same regulatory mechanisms (Yu et al., 2020). In order to guide their application and translation into agricultural production, further research on hormones in crop plants is required in order to elucidate their role in salt tolerance.
5 Phenomics
Irrespective of the breeding approach adopted, be it conventional or modern, precise phenotyping stands as the pivotal determinant of success in any breeding program. The expeditious advancement of crop breeding programs heavily rely on the adoption of sensor-based automated high throughput phenotyping (HTP). Phenomics aids in getting an extra genetic gain when it is used along with genomic studies by increasing selection intensity and accuracy (Lozada and Carter, 2020; Sandhu et al., 2021b). Recent advances in phenomics facilitate acquiring robust, non-invasive, high throughput phenotyping that quantify different morphological and physiological parameters of plants. In case of osmotic stress (due to salt stress), plants try to reduce transpiration loss by reducing stomatal conductance. Low transpiration leads to increase leaf surface temperature. Thermal Infra-Red (IR) imaging can sense the change in leaf surface temperature more precisely between stressed and non-stressed plants under increasing stress level. Sirault et al. (2009) optimized a high throughput screening protocol to quantify osmotic stress response in barley, grown in a range of salt concentrations, using IR thermography based on visualizing the leaf temperature differences due to the variation in stomatal conductance. In another study, plant temperature captured by thermal images showed negative correlation with stomatal conductance and relative water content and no relation with photosynthetic quantum yield in rice under salt stress environment Siddiqui et al., 2014).
Another tool used for extracting information on plant structural and physiological trait is optical imaging. Among these, Red-Green-Blue (RGB) imaging and hyper spectral imaging (HSI) are two most widely used and promising phenotyping approaches. A number of plant morphological changes like change in plant canopy area, compactness, leaf colour are associated with salt stress. Dissanayake et al. (2020) screened 276 accessions of lentil for salt tolerance using RGB imaging where the discrimination between tolerant and susceptible genotypes were done based on projected shoot area, leaf colour, height, convex hull area, and compactness. In a different study, genetic variation of 245 diverse chickpea accessions under elevated salt was assessed using image-based phenotyping (projected shoot area, senescence) and analytic measurements (leaf Na+ and K+ content, biomass, pod number, and 100-seed weight) where pod and seed numbers found to be the most important traits under consideration for breeding chickpea with improved salinity tolerance (Atieno et al., 2017).
Chlorophyll fluorescence (ChlF) is a potential indicator of stress induced changes in photosynthetic mechanisms (PSII activity, photochemical and non-photochemical quenching, electron transport rate) of plants. Tissue tolerance to Na+ ion toxicity in rice was determined by steady state ChlF imaging. The fluorescent images captured can measure chlorotic or necrotic area and thus can separate healthy plant parts from senescent parts (Hairmansis et al., 2014). Similar studies have been conducted in soybean (Kim et al., 2014), sunflower (Neto et al., 2011), acer (Percival et al., 2003), and grape (Dunlevy et al., 2022) for the development and optimization of salt tolerance screening protocol. However, steady state ChlF imaging cannot provide insight into the photosynthetic activity of plants, so kinetic ChlF has been introduced that can quantify the photosynthetic performance under stress condition. Awlia et al. (2016) studied rosette area, rosette colour and photosynthetic performance of Arabidopsis thaliana under salt stress using kinetic ChlF imaging in conjunction with RGB imaging. Fluorescence imaging revealed that non-photochemical quenching and quantum yield are two important traits related to salt stress that induced Arabidopsis plant performance for early and late phase salt stress, respectively. In another study Tsai et al. (2019) reported that in a salt stress sensitive rice variety, the maximum quantum efficiency of PSII was much lower than a tolerant one after NaCl treatment. Al-Tamimi et al. (2016) used RGB based phenotyping to conduct a genome wide association study (explained later in section 7) in rice and identified loci related to transpiration use efficiency. Similarly, in a separate study two novel candidate genes, BnCKX5 and BnERF3, linked to salt stress in Brassica napus were discovered by high-throughput phenotyping-based QTL mapping. (Zhang et al., 2022a). Consequently, to achieve a better grasp of the genetic mechanisms governing stress tolerance and to accelerate breeding endeavors targeting these traits, it is imperative to prioritize the incorporation of phenomic tools within breeding programs.
6 Mapping of salinity tolerance genes
Use of existing genetic variation within or among the species or the creation of variations through mutation or other genetic engineering approaches is a prerequisite to crop improvement. Efficient utilization of such tolerant genotypes as donors in crop improvement programs either through conventional breeding or genetic engineering necessitates unravelling the underlying genetics of salt tolerance. Incorporation of tolerance genes from tolerant lines to agronomically superior but susceptible lines through Marker Assisted Selection (MAS), is a widely followed crop improvement scheme. Mapping those genes is the elementary and crucial step for successful MAS. The genomic regions that contain genes which influence the expression of quantitative characters are referred to as Quantitative Trait Loci (QTL) (Tanksley et al., 1996; Collard et al., 2005). Bi-parental mapping of QTL has been reported to provide valuable information for further map-based cloning of salt tolerance genes and MAS in many economically important crops like rice, maize, pearl millet etc. (Sharma et al., 2011; Tiwari et al., 2016; Luo et al., 2017).
The major constraints in research progress for salinity tolerance are attributed to the polygenic inheritance pattern and significant influence by genotype and environment interactions (Zhu, 2000; Singh et al., 2007; Wang et al., 2014). The basic principle behind the QTL analysis aims to detect an association between phenotype and genotype within the population (Collard et al., 2005). However, mapping of salt tolerance is highly influenced by screening protocol and phenotyping accuracy of the traits, size of the mapping population, linkage between markers and QTL, and parental specificity. Phenotypic response of the plants under salinity stress depends on the age of the plant, duration and level of the salt treatment (Singh et al., 2007). Therefore, a reliable mapping of QTL necessitates accurate phenotyping as well as genotyping of the population.
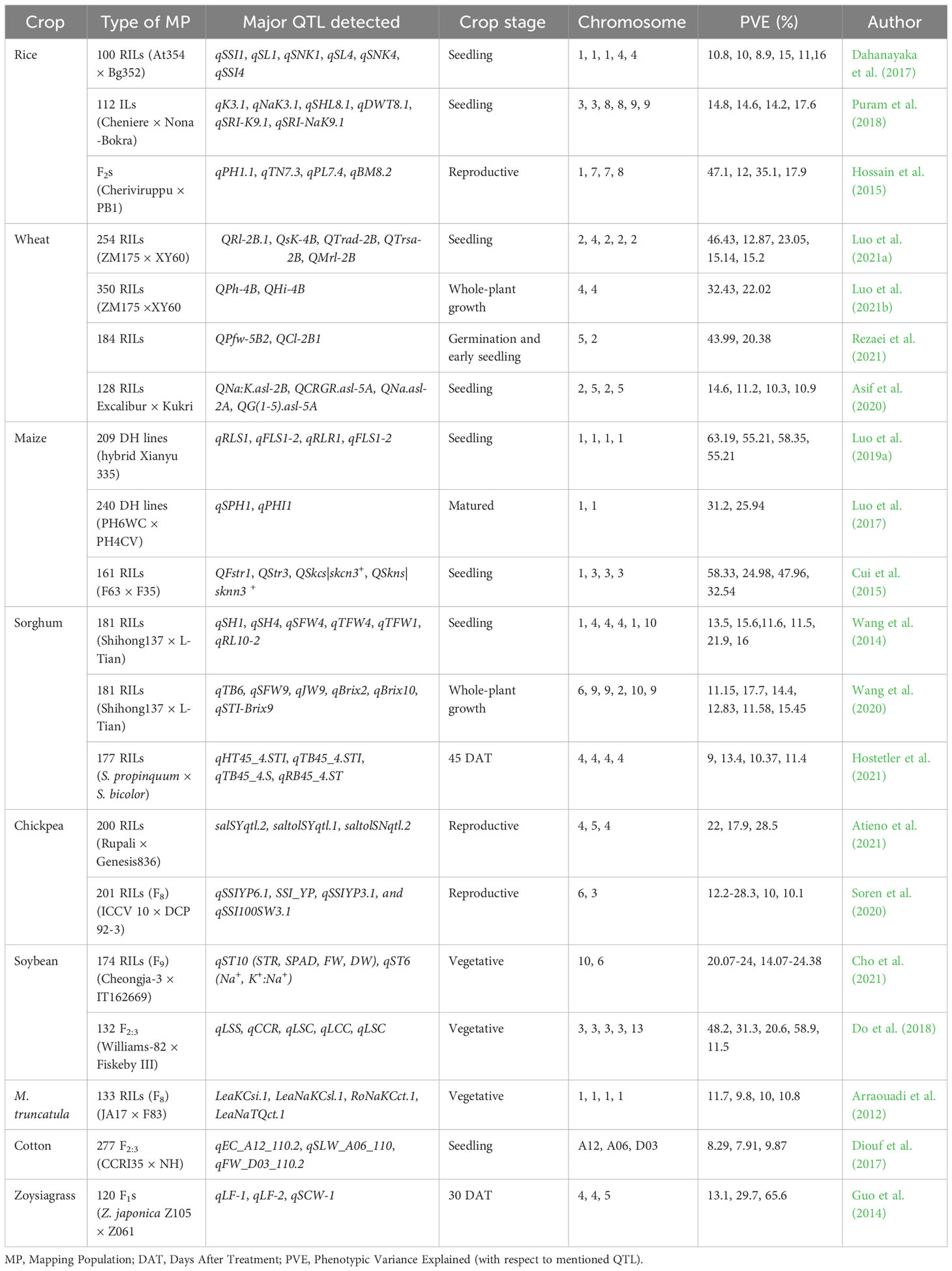
Major crop-specific achievements in QTL mapping for salinity tolerance through bi-parental populations in recent years, valuable stress indicative parameters, and the importance of crop growth stage while screening are discussed in the following section.
6.1 Rice
Several mapping studies in regard to salinity tolerance have been conducted on rice. Although the reproductive stage is considered more critical than the seedling stage as it directly affects grain yield, major reports on QTL are limited to the seedling stage to avoid cumbersome phenotyping efforts (Singh et al., 2021b). Saltol is a widely known QTL in rice at the seedling stage, which has been reported by several groups of researchers (Bonilla et al., 2002; Niones et al., 2006; Thomson et al., 2010). Mapping of an indica Recombinant Inbred Lines (RIL) population identified six QTL at the seedling stage distributed on chromosomes 1 and 4 (Dahanayaka et al., 2017). Puram et al. (2018) used a set of introgressed Lines (IL) from donor parent ‘Nona Bokra’ and identified 18 QTL for salt tolerance indices. This study suggested shoot Na+ exclusion, Na+: K+ homeostasis, and compartmentation of Na+ as probable salt tolerance mechanisms in ‘Nona Bokra’. Another QTL mapping study conducted on an F2 population at the reproductive stage, detected sixteen QTL related to salinity stress on four linkage groups (Hossain et al., 2015).
6.2 Wheat
QTL mapping on 350 RILs identified 90 stable QTL for 15 traits with a genome-wide distribution except for chromosomes 4D, 6B, and 7D. Out of four QTL clusters that were located on chromosomes 2D, 3D, 4B, and 6A, eight notable QTL were validated in a collected natural population. Among them, one QTL was found to be associated with the dwarfing gene Rht-B1 (Rht- Reduced height) which is responsible for reduced plant height and increased seed yield. Additionally, three Kompetitive Allele-Specific PCR (KASP) markers derived from SNPs were successfully designed for three QTL clusters (Luo et al., 2021b). Another study in a RIL population comprising of 254 individuals revealed a total of 158 stable additive QTL for 27 morpho-physiological traits distributed over all the wheat chromosomes except 3A and 4D. 78 out of the 158 QTL, were mapped in nine QTL clusters and seven QTL were validated in two unique populations to check the reliability and potential use in MAS, leading to the development of KASP markers closely linked to stable QTL (Luo et al., 2021a). Rezaei et al. (2021) detected a total of 61 main effect QTL distributed over 15 chromosomes from a study of 186 F10 RILs during the germination and early-seedling stages. Two major QTLs for primary-leaf fresh weight and coleoptile fresh weight were detected on chromosomes 5 and 2, respectively (Rezaei et al., 2021). Asif et al. (2020) identified six QTL for salt tolerance traits: sodium accumulation, chloride accumulation, K+-Na+ ratio, and maintenance of shoot growth under salinity in a RIL population of wheat developed from biparental mating of Excalibur × Kukri. GBS data of the mapping population was associated with both non-destructive high-throughput imaging data [projected shoot area (PSA), relative growth rate (calculated from PSA)], and destructive data such as Na+, K+, and Cl- ion content in the leaves.
6.3 Maize
In a salt tolerance study of 209 doubled haploid (DH) lines, 41 QTL out of a total of 61 were found to be associated with salt tolerance for biomass related traits. These salt tolerance-specific QTL clustered on chromosomes 1, 3, 7, and 9, among which 13 major effect QTL on chromosome 1 contributed the most to the phenotypic variance (Luo et al., 2019a). Another study by Luo et al. (2017) on a DH population of 240 individuals at maturity stage revealed a major QTL for plant height on Chromosome 1 under salinity. In addition, the major QTL influencing plant height-based salt tolerance index was also mapped at the same position on Chromosome 1, and two candidate genes related to ion homeostasis were identified within the confidence interval of this QTL. Using 161 F2:5 RILs, a field as well as a hydroponic experiment were conducted for QTL analysis (Cui et al., 2015). A total of 29 QTL, clustered on chromosomes 1, 3, and 5, were detected. Among those 14 showed significant QTL by treatment (Q × T) interaction effects.
6.4 Sorghum
In a biparental mapping population of 181 RILs, three traits at the germination stage and nine traits at the seedling stage were analyzed, where a total of 12 QTL [(PVE range of 5.4 to 6.0%) and 29 QTL (PVE range 5.3–21.9%) were identified, respectively. Six major QTL at the seedling stage were identified with the positive effects being majorly from the maternal parent. Further extension of this study at the whole plant growth stages detected a total of 53 QTL for six characters for both salt and control conditions. Out of which, six QTL were declared as major QTL (Wang et al., 2020). In a recent study, using a population of 177 F3:5 interspecific RILs, a high-density genetic map was generated covering the 10 Sorghum chromosomes with 1991 markers. The genetic map was used to identify 10 salt stress-specific QTL related to plant growth and overall plant health. Four of them that affected plant height, total biomass, and root biomass, were found to colocalize on chromosome 4. These salt-responsive QTL contained genes related to osmotic and ionic tolerance along with many aquaporins (Hostetler et al., 2021).
6.5 Chickpea
In a recent study in both glasshouse and field conditions, 200 RILs from a cross between two Cicer arietinum varieties Rupali (sensitive) and Genesis836 (tolerant), 42 QTL were detected as having effects on different growth parameters. Among them six major QTL on chromosomes four, five, and six were related to salt tolerance. However, in total, 21 QTL were mapped to two distinct regions on chromosome 4 (Atieno et al., 2021). Soren et al. (2020) screened a set of 201 RILs at the reproductive stage under field conditions developed by crossing two diverse parental lines ICCV 10 (salt-tolerant) and DCP 92-3 (salt-sensitive). An association between genotypic and phenotypic data identified 28 QTL in the population, among which one individual QTL on chromosome 6 related to yield contributed to the maximum phenotypic variance (28.4%). Major QTL associated with yield and yield-related components under salinity stress were found to be clustered on CaLG03 and CaLG06. Candidate genes related to salinity tolerance included histidine kinase, Ca-dependent protein kinases, antiporter genes, and transcription factors such as WRKY and MYB.
6.6 Soybean
An F9 generation RIL population (salt-sensitive cultivar, Cheongja 3 × salt-tolerant landrace, IT162669) consisting of 174 individuals, was screened for salt tolerance at the vegetative stage (Cho et al., 2021). Phenotypic data taken after two weeks of salt stress included major stress-indicative physiological traits such as vegetative damage and Na+-K+ ion contents. Two novel major QTL on chromosomes 6 and 10 were identified as related to ionic stress and other major physiological parameters, respectively under salinity. Analyses of differential gene expression patterns between parents and functional annotation revealed two potential candidate genes in qST6 and six in qST10, which included a phosphoenolpyruvate carboxylase and an ethylene response factor. Do et al. (2018) developed a population of 132 F2:3 (moderately sensitive cultivar Williams 82 × tolerant cultivar Fiskeby III (PI 438471)) and identified major chromosomal loci related to salt stress. Plants were phenotyped with vegetative parameters and ion contents after two weeks of salt stress. A major QTL derived from Fiskeby III located on Chr-03 was found to be significantly associated with leaf scorch, chlorophyll content ratio, and sodium and chloride ion contents. Additionally, another allele related to leaf sodium content was detected and mapped on Chr-13.
6.7 Medicago truncatula
In a study on the model legume crop Medicago truncatula, a set of 133 RILs (Jemalong A17 (JA17) × F83005.5 (F83)) were screened for salt tolerance (Arraouadi et al., 2012). Dry biomass and accumulation of Na+ and K+ ion in roots, stems, and leaves were considered for the phenotyping of the plants. Out of 13 QTL spanning eight linkage groups, six QTL in control, two in salt, and five for salt sensitivity index were mapped. Most of the QTL were found to be clustered on Chr-1, however, no QTL were detected on Chr-5 and 6. Identification of non-overlapping QTL for root and leaf traits suggested the involvement of different genes in ion transportation between roots and leaves.
6.8 Cotton
In an SNP-based QTL mapping study, a total of 66 QTL were detected from an F2-derived F3 population of tolerant cotton line CCRI35 and susceptible Nan Dan (NH) (Diouf et al., 2017). Plants were screened with morpho-physiological parameters at the seedling stage under three salt concentrations. Out of all detected QTL, only 14 (10 from the male parent and four from the female parent) for six traits showed consistency across three salt environments, which accounted for 2.72 to 9.87% of PVE. Five and nine QTL were found to be located in the At and Dt sub-genomes, respectively. Further analysis detected eight clusters that were associated with 12 putative key genes related to salinity.
6.9 Zoysiagrass
Guo et al. (2014) conducted a study in an intraspecific F1 mapping population comprising of 120 progeny derived from a cross between salt-tolerant accession Zoysia japonica Z105 and salt-sensitive accession Z061. Two QTL having a significant impact on leaf firing were detected on chromosome 4. Another major QTL for reduced shoot clipping dry weight was detected on chromosome 5.
Major findings of above-mentioned QTL studies are summarized in the Table 3.
7 Genome-wide association mapping approach for investigating chromosomal regions related to salinity tolerance
Despite higher efficiency and strong detection power to detect a major variant that influences phenotype, mapping of genes using a bi-parental mapping population faces several challenges, especially where cross incompatibility is a major barrier to developing a successful mapping population. With recent advancements in the Next Generation Sequencing (NGS) and HTP, alternate strategies for mapping genes and QTL have become possible. Genome Wide Association Study (GWAS) is a widely used approach to overcome the shortcomings of bi-parental mapping by utilizing the large genetic variation present in a diverse panel of plants (Tibbs Cortes et al., 2021). GWAS rely on Linkage Disequilibrium (LD), the correlation structure that exists among DNA variants in the candidate genome as a result of historical evolutionary forces, particularly finite population size, mutation, recombination rate, and natural selection (Visscher et al., 2017).
GWAS approaches have been implemented in different plant species to find out the underlying genetics of several abiotic stresses which include drought, salinity, temperature, and Boron toxicity tolerance (Challa and Neelapu, 2018). Major crop-specific achievements in identifying potential QTL related to salt tolerance, important stress indicative parameters, and crop growth stage while screening through GWAS, in recent years are discussed in the following section (Table 4).
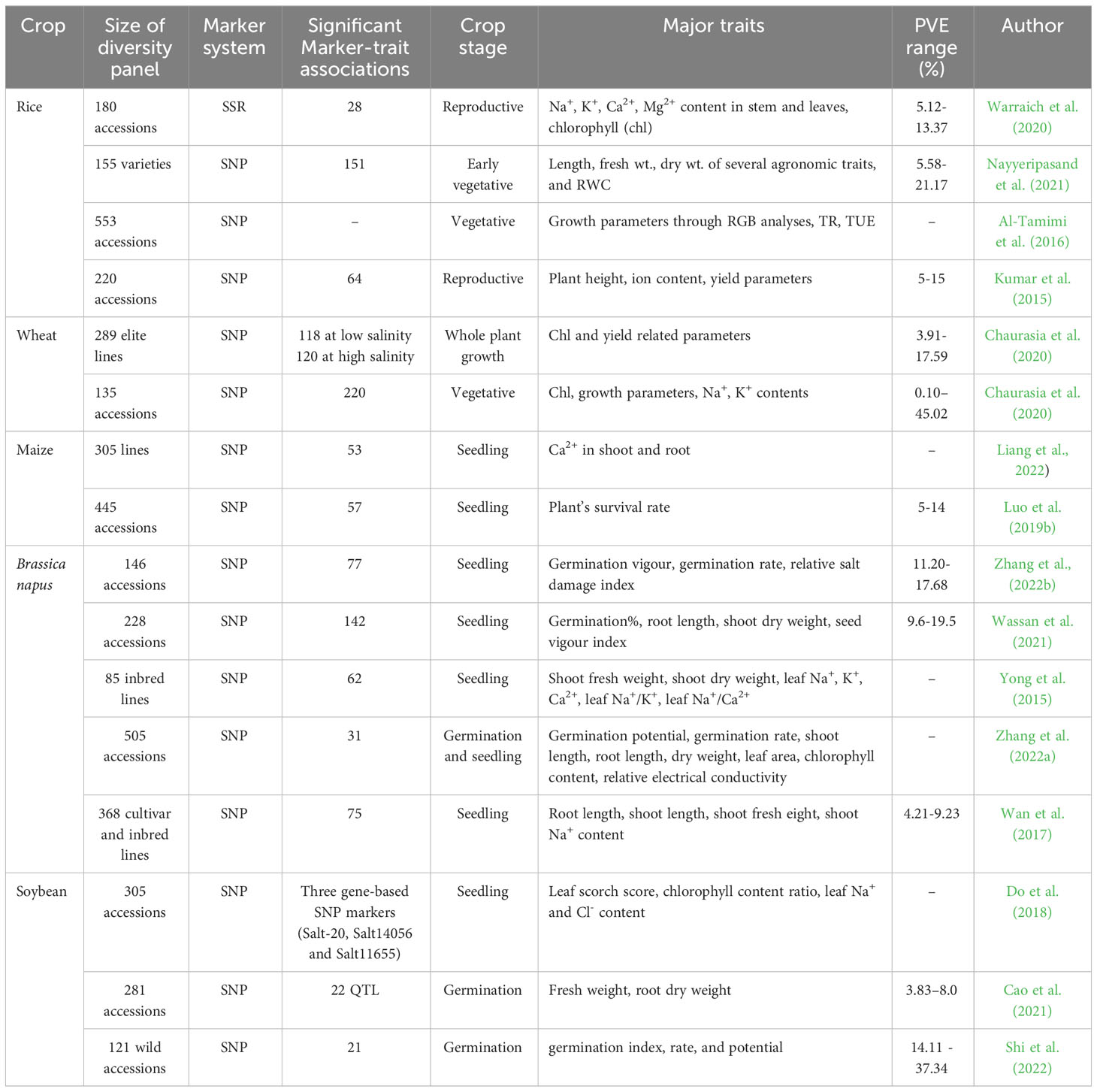
Table 4 Summary of major genome-wide association studies carried out in different crops in recent years.
7.1 Rice
Warraich et al. (2020) conducted GWAS in rice at the reproductive stage where 180 diverse accessions were evaluated for 13 morpho-physiological parameters (including ion contents in stem and leaves, grain yield, and salt injury score) and genotyped by genome-wide SSR markers. This study identified 28 significant marker-trait associations, out of which 19 associations were related to ion homeostasis in stems and leaves. GWAS with 155 rice varieties at the early vegetative stage identified 151 significant marker trait associations scattered on 10 chromosomes (Nayyeripasand et al., 2021). The diverse panel was phenotyped with several agronomic parameters such as shoot and root length, the biomass of above and below ground parts, and RWC. A genomic region of 11.26 Mbp on chromosome 1 was found to be colocalised with the QTL region SalTol1. A number of candidate genes involved in ion transportation and encoding transcription factors were identified on different chromosomes. Al-Tamimi et al. (2016) used 533 rice accessions (297 indica and 257 aus) to investigate early response of rice to soil salinity where they phenotyped the accessions for projected shoot area (PSA), absolute and relative growth rate (derived from PSA) using RGB imaging, transpiration rate (TR), and transpiration use efficiency (TUE). Through GWAS, combining these phenotyping data identified new loci affecting TUE on chromosome 11. Another GWAS was successfully implemented in rice to identify SNPs related to 12 different salt tolerance related traits during the reproductive stage under field grown conditions (Kumar et al., 2015). Twenty and 44 SNP loci were identified to be associated with Na+: K+ ratio and other yield parameters (Table 4) observed under salinity, respectively. Widely reported QTL at the seedling stage present on chromosome 1: Saltol, was identified to be associated with ion homeostasis at the reproductive stage. Other potential QTL were detected on chromosomes 4, 6, and 7 (Kumar et al., 2015).
7.2 Wheat
A recent genome wide association study by Alotaibi et al. (2022) in 289 elite lines of the Wheat Association Mapping Initiative (WAMI) population was conducted under low and high salinity conditions using 15,737 SNP markers at the whole plant growth stage. Seven yield related traits were evaluated and 118 significant marker trait associations at low salinity and 120 at high salinity were found. A multi-locus GWAS in 135 diverse lines at the vegetative stage of wheat revealed novel genomic regions for salinity tolerance (Chaurasia et al., 2020). Out of a total 220 Quantitative Trait Nucleotides (QTNs) identified for 12 salt tolerance related traits, 42 QTNs were found to have significant impacts on 10 salt tolerance traits. Further studies identified 58 candidate genes for the associated genomic regions.
7.3 Maize
An association panel comprising 305 maize inbred lines were phenotyped for above and below ground Ca2+ concentrations and transport coefficients, and genotyped by SNPs to perform GWAS. Fifty three significant SNPs along with 544 associated genes in the linkage disequilibrium regions were identified under salt treatment. Further expression and genetic variation effects by gene-based association analysis revealed a significant association of a pentatricopeptide repeat protein coding gene GRMZM2G123314 with Ca2+ transport (Liang et al., 2022). Another study by Luo et al. (2019b), investigated the survival rates of 445 maize accessions under salinity stress at the seedling stage. GWAS detected 57 loci significantly associated with salt tolerance, which contained 49 candidate genes.
7.4 Brassica
To know the genetic basis of salt tolerance in rapeseed a GWAS was conducted on a diverse panel of 146 accessions using 10,658 high quality SNP markers (Zhang et al., 2022b). In this study 77 SNPs were identified having significant associations with salt tolerance traits, out of which 36 SNPs were associated with three salt tolerance traits (germination vigor, germination rate, relative salt damage index) and gene ontology annotations revealed 19 candidate genes having putative roles in response to salt stress. Using 2,01,817 SNP markers in a GWAS, Wassan et al. (2021) identified 142 SNPs significantly associated with salt tolerance in a diverse panel of 228 lines of Brassica napus. They obtained 117 candidate genes associated with 40 SNPs mostly encoding transcription factors, DNA binding proteins, and aquaporins. Further differential expression between salt tolerant and susceptible lines validated ten candidate genes. Sequence analysis of putative candidate genes by Yong et al. (2015) revealed the loss-of-function in coding regions was due to frame shift mutations or formation of premature stop codons. This resulted in differential response in salt tolerant and sensitive lines under stressed condition. Similarly, QTL and candidate genes related to salt stress in both germination and seedling stage in rapeseed were identified by Zhang et al. (2022a). This study reported overexpression of two candidate genes BnCKX5 and BnERF3 were associated with increased sensitivity to salt stress. Another GWAS experiment identified 75 SNPs and 38 putative candidate genes associated with salt stress tolerance traits under multiple environments at the seedling stage of brassica (Wan et al., 2017).
7.5 Soybean
Do et al. (2018) conducted two GWAS on a diverse panel of soybean accessions to map and validate genomic regions for salt tolerance at the seedling stage. SNPs derived through SoySNP50K (Song et al., 2013) from 305 accessions and Whole Genome Resequencing (WGR) of a subset of 234 accessions confirmed a major locus for salt tolerance on chromosome 3. The highest association of three gene-based SNP markers of the known gene, Glyma03g32900, on Chr-3: Salt-20, Salt14056, and Salt11655 were found to be consistent in both studies (Do et al., 2018). Another GWAS on 281 diverse soybean accessions at the germination stage by Cao et al. (2021) identified a total of 22 QTL associated with salt tolerance. Four salt tolerance indices related to germination and biomass were used for phenotyping. Two major QTL were identified on chromosomes 5 and 16. Shi et al. (2022) evaluated 121 wild soybean accessions during seed germination under salt stress conditions. A total of 21 SNPs with significant associations with salt tolerance were found to be distributed in Chr- 2, 3, 10, 18, and 19, where Chr- 10 registered the highest number of associations.
8 Role of Whole Genome Resequencing for investigating structural variations related to salinity tolerance
Our understanding of the genetic mechanisms underlying salt stress has become even more effective with the recent developments in Whole Genome Resequencing (WGR) and pangenomics. The goal of a WGR experiment is often to identify the differences between a specific individual’s genome and the reference genome. By comparing the sequenced genomes to the reference, a list of mutations unique to each sequenced individual is acquired. These mutations are typically single nucleotide polymorphisms (SNPs) and insertions-deletions (InDels). Substantial rearrangements, including translocations, inversions, and large copy number changes can also be identified using WGR. Findings of noteworthy information related to salt tolerance through WGR studies in various crops are discussed in the following section.
8.1 Rice
Jain et al. (2014) performed WGR to analyze two rice cultivars with contrasting responses to salinity stress and identified 401,683 SNPs and 57,656 InDels. They found a total of 614 genes with a higher density of nonsynonymous SNPs and 576 large-effect SNPs in 1247 genes, which might have a role in the contrasting stress response of those rice cultivars. Furthermore, 266 potential genes were detected that could be validated and utilized for the improvement of salt tolerance. Another study with WGR in rice genotypes having differences in salt tolerance identified 2347 nonsynonymous SNPs and 51 frameshift mutations (Subudhi et al., 2020). Additionally, 396 differentially expressed genes with large-effect variants in the coding regions were identified that are associated with various salt tolerance mechanisms.
8.2 Chickpea
A WGR study of chickpea genotypes having contrasting tolerance to salinity was carried out by Rajkumar et al. (2021). In total, 920 InDels and 6173 SNPs were found that could distinguish the chickpea genotypes with differing salinity stress responses. Chromosome 4 and chromosome 1 were found to have the highest number and frequency of DNA polymorphisms. However, the least amount of DNA polymorphisms was found on chromosome 5. DNA polymorphisms were discovered in the cis-regulatory motifs of genes related to abiotic stress, which might affect the response to salinity stress by modifying the binding affinity of transcription factors.
8.3 Soybean
Further extension of the previously mentioned study by Do et al. (2019) using WGR with a subset of 234 accessions of soybean facilitated the detection of some additional regions on Chr- 1, 8, and 18 which are related to salt tolerance. The region identified on Chr- 8 was predicted as a new minor locus for salt tolerance in soybean. In another study, Patil et al. (2016) conducted a WGR experiment on 106 diverse soybean lines and identified three major structural variants (SV-1, SV-2, and SV-3) and allelic variation in the promoter and genic regions of the GmCHX1 ion transporter gene associated with salinity tolerance. The presence of Ty1/copia retrotransposon in the given locus was found in SV1, which was manifested by salt-sensitive genotypes as reported by Qi et al. (2014). In contrast, salt-tolerant SV-2 lacked this retrotransposon. Interestingly, the SV-3, which did not contain any retrotransposon, showed a salt-sensitive reaction.
8.4 Medicago
The study by Friesen et al. (2014) used WGR to investigate the genetic basis of adaptation under salinity stress in 39 wild accessions of Medicago truncatula. They identified candidate genes such as Medtr3g098090.1, which is orthologous to AtCIPK21 in Arabidopsis thaliana. AtCIPK21 is a calcium-dependent protein kinase that plays a role in abscisic acid and jasmonic acid signal transduction pathways. These pathways play critical roles in the regulation of plant responses to various abiotic stresses, including salinity stress. The researchers found that Medtr3g098090.1 showed differential expression in response to salt stress, with higher expression levels in salt-tolerant accessions. This gene could be a potential target for the genetic improvement of crop plants for increased salt tolerance.
8.5 Linseed
To understand the genetic basis of salt tolerance in linseed, WGR has been used to identify QTL for salt tolerance and candidate genes associated with salt tolerance. Zhao et al. (2022) performed WGR on salt-tolerant and salt-sensitive varieties of linseed and a total of 15 candidate genes related to salt tolerance were identified within a 2.597 Mb region on chromosome 1. The study identified two candidate genes for salt tolerance, Lus.o.m. scaffold91.141 and Lus.o.m. Scaffold1.14, which encode WD40 and cytochrome P450, respectively. Previous studies have demonstrated that overexpression of the Ginkgo biloba WD40 gene improved salt tolerance in poplar (Xin et al., 2021). In addition, Ahmad et al. (2019) found that the candidate gene Lus o.m. Scaffold1.14 in linseed encodes a cytochrome P50 protein which promotes flavonoid biosynthesis and enhances plant cell resistance by changing osmotic pressure. These findings suggest that these two candidate genes could potentially be targeted for improving salt tolerance in linseed.
9 Genomic selection for developing salinity tolerant crops
In nature, most of the quantitative traits of economic importance like- yield, biotic and abiotic stress tolerance are controlled by a large number of minor effect QTL and MAS fails to capture these small effect alleles (Xu and Crouch, 2008; Bernardo, 2016). To overcome this challenge, a prediction model-based marker strategy called genomic selection (GS) was introduced (Meuwissen et al., 2001). GS uses a training population to develop a prediction model based on its phenotypic and genotypic data and this model in turn used to obtain genomic estimated breeding value (GEBV) of all the individuals of a large breeding population using its genotypic information only (Poland et al., 2012). There are several methods to develop parametric and non-parametric prediction models like- best linear unbiased prediction (BLUP), Bayesian regression, ridge regression, kernel regression, machine learning methods (random forest, support vector machine) (de Los Campos et al., 2013) and their prediction accuracy depends on several factors like- size of the training population (Technow et al., 2013; Endelman et al., 2014), heritability of the trait (Isidro et al., 2015; Duangjit et al., 2016), and marker density for the genotyping population (Zhang et al., 2015). Medina et al. (2020) used eight different GS models to predict the performance of alfalfa under salt stress condition. Prediction accuracy and root mean square due to error (RMSE) values were the criteria to select the best-fitting model. Among rrBLUP, BayesA, BayesB, BayesC, Bayesian Ridge Regression (BRR), Bayesian LASSO (BL), Support Vector Machine (SVM), and Random Forest (RF), SVM outperformed the others with a prediction accuracy of 0.793 for yield. With a negative correlation value r2= -0.64 between accuracy and RMSE, SVM proved to be the best for predicting breeding values with high accuracy and low RMSE. Other studies also found machine learning methods to be better over others, as it identifies both additive and non-additive (dominant and epistatic) variance of the trait by capturing SNPs with major effects and complex SNP-SNP interactions (Li et al., 2018; Sandhu et al., 2021a; Enoma et al., 2022). Bartholomé et al. (2022) attempted genomic selection in rice for salt stress related morphological traits and ion mass fractions where they trained the GS model using SNP arrays in a training population of 241 japonica accessions. A high prediction accuracy of 0.25-0.64 for morphological traits and moderate to high accuracy (0.05-0.40) for stress indices were found when this model was used to predict a breeding population. Further cross validation resulted in a strong correlation (r2 = 0.69) in the predictive abilities of a subset of a breeding population with the reference panel under salinity stress conditions. All of these points to the possibility that breeding efforts for developing new lines for salinity-prone environments could become more effective by integrating genomic selection in breeding programs (Figure 4).
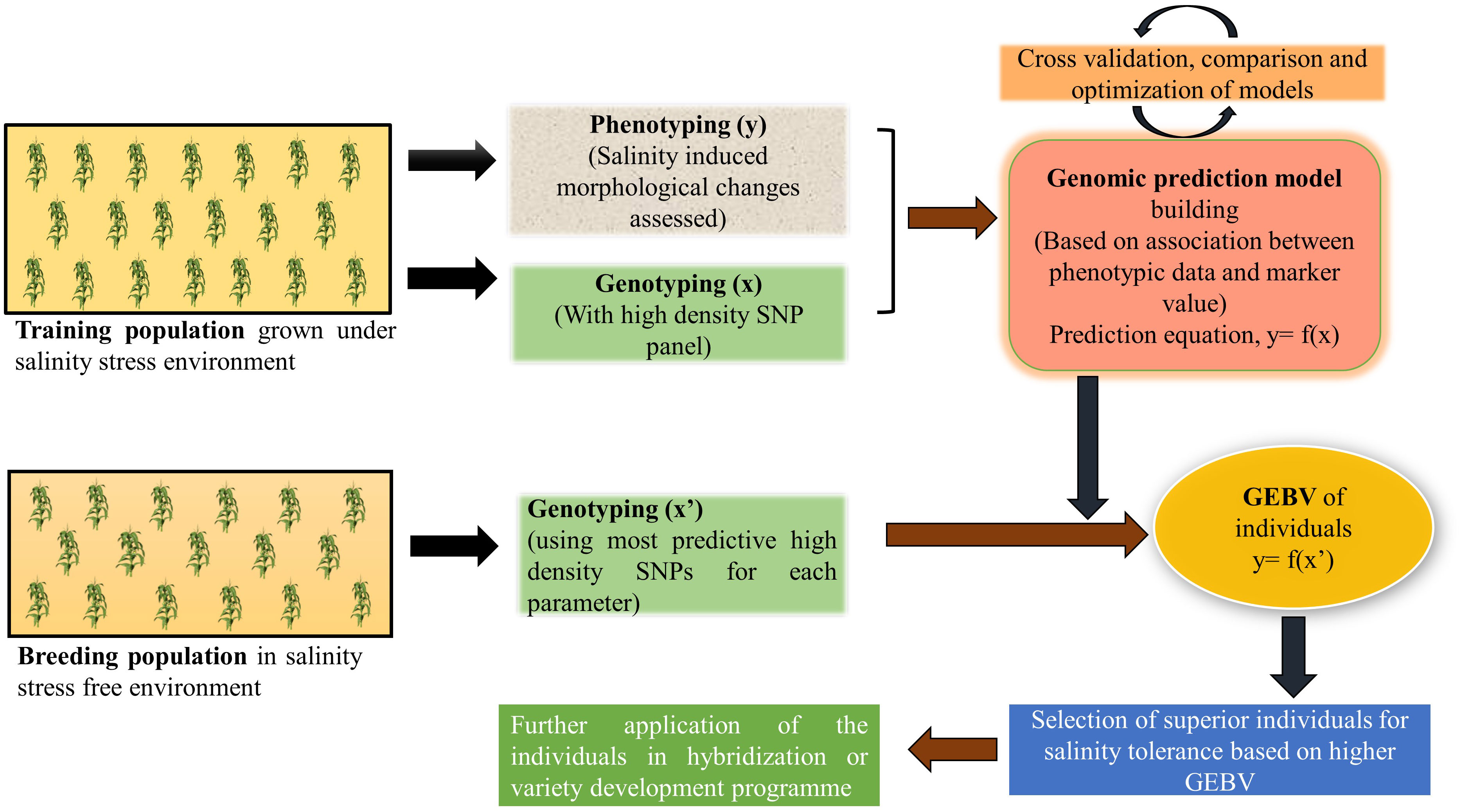
Figure 4 A schematic illustration of genomic selection to improve salinity stress tolerance in crop plants.
10 Scope of pangenomics
WGR studies based on a single reference genome may not be good enough in identifying Structural Variations (SVs) and do not provide sufficient details for extensive understanding of genetic variants due to the lack of diversity within a species (Bayer et al., 2020; Danilevicz et al., 2020). Considering these limitations, the concept of pangenomics was put forward by the researchers. Pangenomes not only represent more diversity of a species or identify core genes those are present in all individuals, but also identify distinct and variable genes that are missing in the reference genome and in some individuals, respectively (Zhao et al., 2018; Bayer et al., 2020). It also facilitates a multidimensional analysis of polymorphism and SVs within and across genomes to simultaneously investigate the genomic variations among individuals of a species or higher taxonomic groups.
Pangenomes have been compiled for many major crop species such as maize, rice, hexaploid wheat, soybean, and Brassica oleracea, but have yet to be fully explored in terms of connecting genetic and phenotypic variations, particularly about salt tolerance (Hirsch et al., 2014; Li et al., 2014; Golicz et al., 2016; Montenegro et al., 2017; Zhao et al., 2018). To combat harsh environmental conditions, researchers are still working to mine certain superior genes related to salinity tolerance from wild relatives through forward genetics or reverse genetics approaches. The identification and mapping of salinity-tolerant genes in important crop species have not yet been reported from a pangenome analysis (Ullah et al., 2022). This pangenome idea along with high-throughput third-generation sequencing offers a reliable platform to recapture deleted genes, identify new genes, and increase our understanding of the dynamics and architecture of the genome. Therefore, considering the power of pangenome analysis to address salinity stress, research efforts are required to mine novel genes in exotic germplasms.
11 Conclusions and future prospects
Irrigation with recycled water, improper fertilization, and deterioration of agricultural lands are gradually increasing the threat of soil salinization (Jha et al., 2019). Consequently, salinity being one of the major abiotic stresses, causes significant yield loss in agricultural crops worldwide. To mitigate crop yield loss caused by the rising challenge of salinity stress, various strategies have been embraced. Interdisciplinary research advances in various model plants as well as crop plants have improved our understanding of the complex molecular mechanisms and underlying genetics of salinity tolerance. Similarly, advances in genomic resources have allowed for the identification of salinity tolerance QTLs and genes through bi-parental and genome wide association mapping studies. Furthermore, availability of crop genome sequences, genome re-sequencing approaches, and pangenome assemblies can greatly facilitate in discovery of genomic regions, structural genomic variants or haplotypes contributing to salinity tolerance across the whole genome.
Increasing facilities capable of HTP has improved the screening of germplasms with enhanced phenotypic accuracy and efficiency which has been proven highly effective for selecting potential salinity tolerant crop plants (Al-Tamimi et al., 2016). Emerging genome editing techniques, such as CRISPR/Cas9, are powerful tools for the validation of candidate genes and knocking out negative alleles associated with salt susceptibility (Kumar and Sharma, 2020; Luo et al., 2021). Future research studies may explore special traits and structures such as salt glands, papillae etc. present across various plant gene pools and harness their potential towards improving salinity tolerance (Marcum et al., 1998; Yamamoto et al., 2016; Jha et al., 2019; Spiekerman and Devos, 2020; Mondal et al., 2023). Deciphering different pathways involved in salt tolerance, understanding genetics through molecular mapping, embracing conventional breeding approaches along with emerging breeding tools can help improve genetic gain under increasing salinity stress, leading to more resilient crops for the future.
Author contributions
All authors contributed through writing and editing the manuscript and they approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, A., Khan, S., Hussain, N., Hanjra, M. A., Akbar, S. (2013). Characterizing soil salinity in irrigated agriculture using a remote sensing approach. Phys. Chem. Earth Parts A/B/C. 55-57, 43–52. doi: 10.1016/j.pce.2010.12.004
Abd El-Monem, A. A., El-Habbasha, S. F., Hozayn, M. (2013). Mitigation salinity stress effects on barley (Hordeum vulgare L.) growth, yield and some physiological aspects by hemin. J. Appl. Sci. Res. 9 (3), 2411–2421.
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy. 7 (1), 18. doi: 10.3390/agronomy7010018
Ageeva-Kieferle, A., Rudolf, E. E., Lindermayr, C. (2019). Redox-dependent chromatin remodeling: a new function of nitric oxide as architect of chromatin structure in plants. Front. Plant Sci. 10, 625. doi: 10.3389/fpls.2019.00625
Ahmad, P., Hakeem, K. R., Kumar, A., Ashraf, M., Akram, N. A. (2012). Salt induced changes in photosynthetic activity and oxidative defence system of three cultivars of mustard (Brassica juncea L.). Afr. J. Biotechnol. 11, 2694–2703.
Ahmad, P., Jaleel, C. A., Salem, M. A., Nabi, G., Sharma, S (2010a). Roles of enzymatic and non-enzymatic antioxidants in plants during abiotic stress”. Crit. Rev. Biotechnol. 30 (3), 161–175. doi: 10.3109/07388550903524243
Ahmad, P., Jaleel, C. A., Sharma, S. (2010b). Antioxidative defence system, lipid peroxidation, proline metabolizing enzymes and biochemical activity in two genotypes of Morus alba L. subjected to NaCl stress. Russ. J. Plant Physiol. 57, 509–517. doi: 10.1134/S1021443710040084
Ahmad, R., Lim, C. J., Kwon, S. Y. (2013). Glycine betaine: a versatile compound with great potential for gene pyramiding to improve crop plant performance against environmental stresses. Plant Biotechnol. Rep. 7, 49–57. doi: 10.1007/s11816-012-0266-8
Ahmad, N., Liu, J. Y., Tian, X., Noman, M., Jameel, A., Yao, N., et al. (2019). Overexpression of a novel cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 10, 756. doi: 10.3390/genes10100756
Ahmad, P., Sharma, S. (2008). Salt stress and phyto-biochemical responses of plants. Plant Soil Environ. 54, 89–99. doi: 10.17221/2774-PSE
Alamgir, A. N. M., Yousuf Ali, M. (1999). Effect of salinity on leaf pigments, sugar and protein concentrations and chloroplast ATPase activity of rice (Oryza sativa L.). Bangladesh J. Botany. 28 (2), 145–149.
Alotaibi, F. S., Al-Qthanin, R. N., Aljabri, M., Shehzad, T., Albaqami, M., Abou-Elwafa, S. F. (2022). Identification of genomic regions associated with agronomical traits of bread wheat under two levels of salinity using GWAS. Plant Mol. Biol. Reporter 40 (3), 595–609. doi: 10.1007/s11105-022-01341-x
Al-Tamimi, N., Brien, C., Oakey, H., Berger, B., Saade, S., Ho, Y. S., et al. (2016). Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 7 (1), 1–11. doi: 10.1038/ncomms13342
Apse, M. P., Aharon, G. S., Snedden, W. A., Blumwald, E. (1999). Salt tolerance conferred by over-expression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 285 (5431), 1256–1258.
Arif, M. R., Islam, M. T., Robin, A. H. K. (2019). Salinity stress alters root morphology and root hair traits in Brassica napus. Plants. 8 (7), 192. doi: 10.3390/plants8070192
Arraouadi, S., Badri, M., Abdelly, C., Huguet, T., Aouani, M. E. (2012). QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula Recombinant Inbred Lines. Genomics. 99 (2), 118–125. doi: 10.1016/j.ygeno.2011.11.005
Asada, K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygen’s and dissipation of excess photons. Annu. Rev. Plant Biol. 50, 601–639. doi: 10.1146/annurev.arplant.50.1.601
Ashraf, M., Akram, N. A., Arteca, R. N., Foolad, M. R. (2010). The physiological, biochemical and molecular roles of brassinosteroids and salicylic acid in plant processes and salt tolerance. Crit. Rev. Plant Sci. 29 (3), 162–190. doi: 10.1080/07352689.2010.483580
Ashraf, M. A., Ashraf, M. (2016). Growth stage-based modulation in physiological and biochemical attributes of two genetically diverse wheats (Triticum aestivum L.) cultivars grown in salinized hydroponic culture. Environ. Sci. pollut. Res. 23, 6227–6243. doi: 10.1007/s11356-015-5840-5
Ashraf, M., Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Botany. 59 (2), 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Asif, M. A., Garcia, M., Tilbrook, J., Brien, C., Dowling, K., Berger, B., et al. (2020). Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping. Funct. Plant Biol. 48 (2), 131–140.
Askari-Khorasgani, O., Rehmani, M. I. A., Wani, S. H., Kumar, A. (2021). 23 osmotic stress. Handb. Plant Crop Physiol., 445–455. doi: 10.1201/9781003093640
Atieno, J., Colmer, T. D., Taylor, J., Li, Y., Quealy, J., Kotula, L., et al. (2021). Novel salinity tolerance loci in chickpea identified in glasshouse and field environments. Front. Plant Science. 12. doi: 10.3389/fpls.2021.667910
Atieno, J., Li, Y., Langridge, P., Dowling, K., Brien, C., Berger, B., et al. (2017). Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci. Rep. 7 (1), 1–11. doi: 10.1038/s41598-017-01211-7
Atta, K., Sen, J., Chettri, P., Pal, A. K. (2022a). Antioxidant responses of ricebean [Vigna umbellata (Thunb.) Ohwi and Ohashi] seedling under iso-osmotic potential of salinity and drought Stress. Legume Res. 45 (4), 429–434. doi: 10.18805/LR-4551
Atta, K., Karmakar, S., Dutta, D., Pal, A. K., Jana, K. (2019). Comparative physiology of salinity, drought and heavy metal stress during seed germination in ricebean [Vigna umbellata (Thunb.) Ohwi and Ohashi]. J. Crop Weed 15 (3), 145–149. doi: 10.22271/09746315.2019.v15.i3.1251
Atta, K., Pal, A. K., Jana, K. (2021). Effects of salinity, drought and heavy metal stress during seed germination stage in ricebean [Vigna umbellata (Thunb.) Ohwi and Ohashi]. Plant Physiol. Rep. 26, 109–115. doi: 10.1007/s40502-020-00542-4
Atta, K., Adhikary, S., Mondal, S., Mukherjee, S., Pal, A., Mondal, S., et al. (2022b). A review on stress physiology and breeding potential of an uderutilized, multipurpose legume: rice bean (Vigna umbellata). In: Jha, U. C., Nayyar, H., Agrawal, S. K., Siddique, K. H. M. (eds) Developing climate resilient grain and forage legumes. Springer, Singapore. doi: 10.1007/978-981-16-9848-4_11
Awlia, M., Nigro, A., Fajkus, J., Schmoeckel, S. M., Negrão, S., Santelia, D., et al. (2016). High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front. Plant Science. 7, 1414. doi: 10.3389/fpls.2016.01414
Bajgu, A. (2014). “Nitric oxide: role in plants under abiotic stress,” in Physiological mechanisms and adaptation strategies in plants under changing environment (Springer), 137–159.
Bartholomé, J., Frouin, J., Brottier, L., Tuong-Vi, C. A. O., Boisnard, A., Ahmadi, N., et al. (2022). Genomic selection for salinity tolerance in japonica rice. doi: 10.21203/rs.3.rs-2007613/v1
Bayer, P. E., Golicz, A. A., Scheben, A., Batley, J., Edwards, D. (2020). Plant pan-genomes are the new reference. Nat. Plants. 6, 914–920. doi: 10.1038/s41477-020-0733-0
Ben Ahmed, C., Ben Rouina, B., Sensoy, S., Boukhriss, M., Ben Abdullah, F. (2010). Exogenous proline effects on photosynthetic performance and antioxidant defense system of young olive tree. J. Agric. Food Chem. 58 (7), 4216–4222. doi: 10.1021/jf9041479
Bernardo, R. (2016). Bandwagons I, too, have known. Theor. Appl. Genet. 129 (12), 2323–2332. doi: 10.1007/s00122-016-2772-5
Besson-Bard, A., Pugin, A., Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
Bohnert, H. J., Nelson, D. E., Jensen, R. G. (1995). Adaptations to environmental stresses. Plant Cell. 7 (7), 1099–1111.
Bonilla, P., Mackell, D., Deal, K., Gregorio, G. (2002). RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philippine Agric. Scientist (Philippines).
Bryla, D. R., Scagel, C. F., Lukas, S. B., Sullivan, D. M. (2021). Ion-specific Limitations of Sodium Chloride and Calcium Chloride on Growth, Nutrient Uptake, and Mycorrhizal Colonization in Northern and Southern Highbush Blueberry. J. Am. Soc. Hortic. Sci. 146 (6), 399–410. doi: 10.21273/jashs05084-21
Cabot, C., Sibole, J. V., Barcel´o, J., Poschenrieder, C. (2009). Abscisic acid decreases leaf Na+ exclusion in salt treated Phaseolus vulgaris L. J. Plant Growth Regulation. 28 (2), 187–192. doi: 10.1007/s00344-009-9088-5
Cao, Y., Zhang, X., Jia, S., Karikari, B., Zhang, M., Xia, Z., et al. (2021). Genome-wide association among soybean accessions for the genetic basis of salinity-alkalinity tolerance during germination. Crop Pasture Science. 72, 255–267. doi: 10.1071/CP20459
Chakraborty, K., Singh, A. L., Bhaduri, D., Sairam, R. K. (2013). “Mechanism of salinity stress tolerance in crop plants and recent developments,” in Advances in plant physiology, vol. 14 . Ed. Hemantaranjan, A. (Scientific Publishers, Jodhpur), 466–496.
Challa, S., Neelapu, N. R. R. (2018). “Chapter –9 - genome-wide association studies (GWAS) for abiotic stress tolerance in plant”s,” in Biochemical, physiological and molecular avenues for combating abiotic stress tolerance in plants. Ed. Wani., S. H. (Academic Press), 135–150. doi: 10.1016/B978-0-12-813066-7.00009-7
Chaurasia, S., Singh, A. K., Songachan, L. S., Sharma, A. D., Bhardwaj, R., Singh, K. (2020). Multi-locus genome-wide association studies reveal novel genomic regions associated with vegetative stage salt tolerance in bread wheat (Triticum aestivum L.). Genomics. 112 (6), 4608–4621. doi: 10.1016/j.ygeno.2020.08.006
Cho, K.-H., Kim, M. Y., Kwon, H., Yang, X., Lee, S.-H. (2021). Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.). Plant Science. 313, 111085. doi: 10.1016/j.plantsci.2021.111085
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica. 142 (1), 169–196. doi: 10.1007/s10681-005-1681-5
Cramer, G. R., Quarrie, S. A. (2002). Abscisic acid is correlated with the leaf growth inhibition of four genotypes of maize differing in their response to salinity. Funct. Plant Biol. 29 (1), 111–115. doi: 10.1071/PP01131
Crawford, N. M. (2006). Mechanisms for nitric oxide synthesis in plants. J. Exp. Botany. 57 (3), 471–478. doi: 10.1093/jxb/erj050
Cui, D., Wu, D., Somarathna, Y., Xu, C., Li, S., Li, P., et al. (2015). QTL mapping for salt tolerance based on snp markers at the seedling stage in maize (Zea mays L.). Euphytica 203 (2), 273–283. doi: 10.1007/s10681-014-1250-x
Dahanayaka, B. A., Gimhani, D. R., Kottearachchi, N. S., Samarasighe, W. L. G. (2017). QTL mapping for salinity tolerance using an elite rice (oryza sativa) breeding population. SABRAO J. Breed. Genet. 49 (2), 123–134.
Daniells, I. G., Holland, J. F., Young, R. R., Alston, C. L., Bernardi, A. L. (2001). Relationship between yield of grain sorghum (Sorghum bicolor) and soil salinity under field conditions. Aust. J. Exp. Agriculture. 41, 211–217. doi: 10.1071/EA00084
Danilevicz, M. F., Fernandez, C. G. T., Marsh, J. I., Bayer, P. E., Edwards, D. (2020). Plant pangenomics: approaches, applications and advancements. Curr. Opin. Plant Biol. 54, 18–25. doi: 10.1016/j.pbi.2019.12.005
Daszkowska-Golec, A. (2011). Arabidopsis seed germination under abiotic stress as a concert of action of phytohormones. OMICS: J. Integr. Biol. 15 (11), 763–774. doi: 10.1089/omi.2011.0082
de Los Campos, G., Hickey, J. M., Pong-Wong, R., Daetwyler, H. D., Calus, M. P. (2013). Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics. 193 (2), 327–345. doi: 10.1534/genetics.112.143313
Dietz, K. J., Tavakoli, N., Kluge, C., Mimura, T., Sharma, S. S., Harris, G. C., et al. (2001). Significance of the V type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J. Exp. Botany. 52 (363), 1969–1980. doi: 10.1093/jexbot/52.363.1969
Diouf, L., Pan, Z., He, S.-P., Gong, W.-F., Jia, Y. H., Magwanga, R. O., et al. (2017). High-density linkage map construction and mapping of salt-tolerant qtl at seedling stage in upland cotton using genotyping by sequencing (GBS). Int. J. Mol. Sci. 18 (12), 2622. doi: 10.3390/ijms18122622
Dissanayake, R., Kahrood, H. V., Dimech, A. M., Noy, D. M., Rosewarne, G. M., Smith, K. F., et al. (2020). Development and application of image-based high-throughput phenotyping methodology for salt tolerance in lentils. Agronomy. 10 (12), 1992. doi: 10.3390/agronomy10121992
Do, T. D., Vuong, T. D., Dunn, D., Clubb, M., Valliyodan, B., Patil, G., et al. (2019). Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genomics 20 (1), 1–16. doi: 10.1186/s12864-019-5662-9
Do, T. D., Vuong, T. D., Dunn, D., Smothers, S., Patil, G., Yungbluth, D. C., et al. (2018). Mapping and confirmation of loci for salt tolerance in a novel soybean germplasm, Fiskeby III. Theor. Appl. Genet. 131 (3), 513–524. doi: 10.1007/s00122-017-3015-0
Duangjit, J., Causse, M., Sauvage, C. (2016). Efficiency of genomic selection for tomato fruit quality. Mol. Breeding. 36 (3), 1–16. doi: 10.1007/s11032-016-0453-3
Dunlevy, J. D., Blackmore, D. H., Betts, A., Jewell, N., Brien, C., Berger, B., et al. (2022). Investigating the effects of elevated temperature on salinity tolerance traits in grapevine rootstocks using high-throughput phenotyping. Aust. J. Grape Wine Res. 28 (2), 276–291. doi: 10.1111/ajgw.12549
Ehtaiwesh, F. A., Rashed, H. F. (2020). Growth and yield responses of libyan hard wheat (Triticum durum Desf) genotypes to salinity stress. Zawia Univ. Bull. 22, 33–58.
El-Hendawy, S., Elshafei, A., Al-Suhaibani, N., Alotabi, M., Hassan, W., Dewir, Y. H., et al. (2019). Assessment of the salt tolerance of wheat genotypes during the germination stage based on germination ability parameters and associated SSR markers. J. Plant Interactions. 14 (1), 151–163. doi: 10.1080/17429145.2019.1603406
El-Mashad, A. A. A., Mohamed, H. I. (2012). Brassinolide alleviates salt stress and increases antioxidant activity of cowpea plants (Vigna sinensis). Protoplasma. 249 (3), 625–635. doi: 10.1007/s00709-011-0300-7
El Sabagh, A., Islam, M. S., Skalicky, M., Ali Raza, M., Singh, K., Hossain, M., et al. (2021). Salinity stress in wheat (Triticum aestivum L.) in the changing climate: adaptation and management strategies. Front. Agron. 3. doi: 10.3389/fagro.2021.661932
El-Shintinawy, F., El-Shourbagy, M. N. (2001). Alleviation of changes in protein metabolism in NaCl-stressed wheat seedlings by thiamine. Biol. Plantarum. 44 (4), 541–545. doi: 10.1023/A:1013738603020
Endelman, J. B., Atlin, G. N., Beyene, Y., Semagn, K., Zhang, X., Sorrells, M. E., et al. (2014). Optimal design of preliminary yield trials with genome-wide markers. Crop Science. 54 (1), 48–59. doi: 10.2135/cropsci2013.03.0154
Enoma, D. O., Bishung, J., Abiodun, T., Ogunlana, O., Osamor, V. C. (2022). Machine learning approaches to genome-wide association studies. J. King Saud University-Science. 101847. doi: 10.1016/j.jksus.2022.101847
Eynard, A., Lal, R., Wiebe, K. (2005). Crop response in salt-affected soils. J. Sustain. Agric. 27, 5–50. doi: 10.1300/J064v27n01-03
FAO (2021). Global map of salt-affected soils. Available at: https://www.fao.org/3/cb7247en/cb7247en.pdf.
FAO, ITPS (2015). Status of the wor’ld's soil resources (SWSR) – main report (Rome, Italy: Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils).
Farooq, M., Asif, S., Jang, Y.-H., Park, J.-R., Zhao, D.-D., Kim, E.-G., et al. (2022). effect of different salts on nutrients uptake, gene expression, antioxidant, and growth pattern of selected rice genotypes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.895282
Farouk, S. (2011). Ascorbic acid and α-tocopherol minimize salt-induced wheat leaf senescence. J. Stress Physiol. Biochem. 7, 58–79.
Friesen, M. L., von Wettberg, E. J., Badri, M., Moriuchi, K. S., Barhoumi, F., Chang, P. L., et al. (2014). The ecological genomic basis of salinity adaptation in Tunisian Medicago truncatula. BMC Genomics 15 (1), pp.1–pp18. doi: 10.1186/1471-2164-15-1160
Gaikwad, D. J., Ubale, N. B., Pal, A., Singh, S., Ali, M. A., Maitra, S. (2022). Abiotic stresses impact on major cereals and adaptation options - A review. Res. Crop 23, 896–815. doi: 10.31830/2348-7542.2022.ROC-913
Gao, Z., Sagi, M., Lips, S. H. (1998). Carbohydrate metabolism in leaves and assimilate partitioning in fruits of tomato (Lycopersicum esculentum L.) as affected by salinity. Plant Science. 135 (2), 149–159.
Golicz, A. A., Bayer, P. E., Barker, G. C., Edger, P. P., Kim, H., Martinez, P. A., et al. (2016). The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7, 13390. doi: 10.1038/ncomms13390
Golldack, D., Li, C., Mohan, H., Probst, N. (2014). Tolerance to drought and salt stress in plants: unravelling the signaling networks. Front. Plant Sci. 5, 151. doi: 10.3389/fpls.2014.00151
Greenway, H., Munns, R. (1980). Mechanisms of salt tolerance in non-halophytes. Annu. Rev. Plant Phy. 31, 149–190. doi: 10.1146/annurev.pp.31.060180.001053
Grime, J. P. (1977). Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194. doi: 10.1086/283244
Groppa, M. D., Benavides, M. P. (2008). Polyamines and abiotic stress: recent advances. Amino Acids 34 (1), 35–45. doi: 10.1007/s00726-007-0501-8
Guo, H., Ding, W., Chen, J., Chen, X., Zheng, Y., Wang, Z., et al. (2014). Genetic linkage map construction and QTL mapping of salt tolerance traits in Zoysiagrass (Zoysia japonica). PloS One 9 (9), e107249. doi: 10.1371/journal.pone.0107249
Gupta, K., Dey, A., Gupta, B. (2013a). Plant polyamines in abiotic stress responses. Acta Physiologiae Plantarum. 35 (7), 2015–2036. doi: 10.1007/s11738-013-1239-4
Gupta, K., Dey, A., Gupta, B. (2013b). “Polyamines and their role in plant osmotic stress tolerance,” in Climate change and plant abiotic stress tolerance. Eds. Tuteja, N., Gill, S. S. (Weinheim, Germany: Wiley-VCH), 1053–1072.
Gupta, B., Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014.
Gupta, K. J., Stoimenova, M., Kaiser, W. M. (2005). In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Botany. 56 (420), 2601–2609. doi: 10.1093/jxb/eri252
Hairmansis, A., Berger, B., Tester, M., Roy, S. J. (2014). Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice. 7 (1), 1–10. doi: 10.1186/s12284-014-0016-3
Hanson, A. D., Rathinasabapathi, B., Rivoal, J., Burnet, M., Dillon, M. O., Gage, D. A. (1994). Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc. Natl. Acad. Sci. United States America. 91 (1), 306–310. doi: 10.1073/pnas.91.1.306
Hasanuzzaman, M., Bhuyan, M. B., Nahar, K., Hossain, M. S., Mahmud, J. A., Hossen, M. S., et al. (2018). Potassium: A vital regulator of plant responses and tolerance to abiotic stresses. Agronomy. 8 (3), 31.
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22 (17), 9326. doi: 10.3390/ijms22179326
Hasegawa, PM. (2013). Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Botany. 92, 19–31.
He, M., He, C. Q., Ding, N. Z. (2018). Abiotic stresses: general defenses of land plants and chances for engineering multi-stress tolerance. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01771
Hirsch, C. N., Foerster, J. M., Johnson, J. M., Sekhon, R. S., Muttoni, G., Vaillancourt, B., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell. 26 (1), 121–135. doi: 10.1105/tpc.113.119982
Hoang, T. M. L., Moghaddam, L., Williams, B., Khanna, H., Dale, J., Mundree, S. G. (2015). Development of salinity tolerance in rice by constitutive-overexpression of genes involved in the regulation of programmed cell death. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00175
Hossain, M. S. (2019). Present scenario of global salt affected soils, its management and importance of salinity research. Int. Res. J. Biol. Sci. 1 (1), 1–3.
Hossain, K. K., Itoh, R. D., Yoshimura, G., Tokuda, G., Oku, H., Cohen, M. F., et al. (2010). Effects of nitric-oxide scavengers on the rachiarmo-inhibition of seed germination in Arabidopsis thaliana. Russian J. Plant Physiol. 57 (2), 222–232.
Hossain, M. A., Munemasa, S., Uraji, M., Nakamura, Y., Mori, I. C., Murata, Y. (2011). Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol. 156 (1), 430–438. doi: 10.1104/pp.111.172254
Hossain, H., Rahman, M. A., Alam, M. S., Singh, R. K. (2015). Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Science. 201 (1), 17–31. doi: 10.1111/jac.12086
Hostetler, A. N., Govindarajulu, R., Hawkins, J. S. (2021). QTL mapping in an interspecific sorghum population uncovers candidate regulators of salinity tolerance. Plant Stress. 2, 100024. doi: 10.1016/j.stress.2021.100024
Huang, S., Waadt, R., Nuhkat, M., Kollist, H., Hedrich, R., roelfsema, M. R. G. (2019). Calcium signals in guard cells enhance the efficiency by which abscisic acid triggers stomatal closure. New Phytologist. 224 (1), 177–187. doi: 10.1111/nph.15985
Isidro, J., Jannink, J. L., Akdemir, D., Poland, J., Heslot, N., Sorrells, M. E. (2015). Training set optimization under population structure in genomic selection. Theor. Appl. Genet. 128 (1), 145–158. doi: 10.1007/s00122-014-2418-4
Ivushkin, K. B., Bregt, H., AK, P. A., Kempen, B., de Sousa, L. (2019). Global mapping of soil salinity change. Remote Sens. environment. 231, 111260. doi: 10.1016/j.rse.2019.111260
Jain, M., Moharana, K. C., Shankar, R., Kumari, R., Garg, R. (2014). Genome wide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol. J. 12, 253–264. doi: 10.1111/pbi.12133
Jeschke, W. D., Peuke, A. D., Pate, J. S., Hartung, W. (1997). Transport, synthesis and catabolism of abscisic acid (ABA) in intact plants of castor bean (Ricinus communis L.) under phosphate deficiency and moderate salinity. J. Exp. Botany. 48 (314), 1737–1747.
Jha, U. C., Bohra, A., Jha, R., Parida, S. K. (2019). Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 38, 255–277. doi: 10.1007/s00299-019-02374-5
Kalhoro, N. A., Rajpar, I., Kalhoro, S. A., Ali, A., Raza, S., Ahmed, M., et al. (2016). Effect of salts stress on the growth and yield of wheat (Triticum aestivum L.). Am. J. Plant Sci. 7, 2257. doi: 10.4236/ajps.2016.715199
Kamran, M., Parveen, A., Ahmar, S., Malik, Z., Hussain, S., Chattha, M. S., et al. (2020). An Overview of hazardous impacts of soil salinity in crops, tolerance mechanisms, and amelioration through selenium supplementation. Int. J. Mol. Sci. 21 (1), 148. doi: 10.3390/ijms21010148
Kaneko, M., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Matsuoka, M. (2002). The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 128 (4), 1264–1270. doi: 10.1104/pp.010785
Kang, D. J., Seo, Y. J., Lee, J. D., Ishii, R., Kim, K. U., Shin, D. H., et al. (2005). Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Science. 191 (4), 273–282. doi: 10.1111/j.1439-037X.2005.00153.x
Kaur, G., Kumar, S., Nayyar, H., Upadhyaya, H. D. (2008). Cold stress injury during the pod- fillingphase in chickpea (Cicer arietinum L.) effects on quantitative and qualitative components of seeds. J. Agron. Crop Sci. 194, 457–464.
Kaveh, H., Nemati, H., Farsi, M., Jartoodeh, S. V. (2011). How salinity affect germination and emergence of tomato lines. J. Biol. Environ. Sci. 5, 159–163.
Kenneth, K., Neeltje, C. (2002). Agricultural drainage water management in arid and semi-arid areas (Rome: FAO Irrigation and Drainage Paper), 61.
Kerepesi, I., Galiba, G. (1995). Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Science. 40 (2), 482–487.
Ketehouli, T., Idrice Carther, K. F., NOman, M., Wang, F. W., Li, X. W., Li, H. Y. (2019). Adaptation of plants to salt stress: characterization of Na+ and K+ transporters and role of CBL gene family in regulating salt stress response. Agronomy. 9 (11), 687. doi: 10.3390/agronomy9110687
Khan, M. A., Ungar, I. A., Showalter, A. M. (2000). Effects of sodium chloride treatments on growth and ion accumulation of the halophyte haloxylon recurvum. Commun. Soil Sci. Plant Analysis. 31 (17-18), 2763–2774. doi: 10.1080/00103620009370625
Khataar, M., Mohammadi, M. H., Shabani, F. (2018). Soil salinity and matric potential interaction on water use, water use efficiency and yield response factor of bean and wheat. Sci. Rep. 8, 2679. doi: 10.1038/s41598-018-20968-z
Khodarahmpour, Z., Ifar, M., Motamedi, M. (2012). Effects of NaCl salinity on maize (Zea mays L.) at germination and early seedling stage. Afr. J. Biotechnol. 11, 298–304.
Khorobrykh, S., Havurinne, V., Mattila, H., Tyystjärvi, E. (2020). Oxygen and ROS in photosynthesis. Plants. 9 (1), 91. doi: 10.3390/plants9010091
Kim, J. W., Lee, T. Y., Nah, G., Kim, D. S. (2014). Potential of thermal image analysis for screening salt stress-tolerant soybean (Glycine max). Plant Genet. Resources. 12 (S1), S134–S136. doi: 10.1017/S1479262114000422
Kovács, Z., Simon-Sarkadi, L., Szucs, A., Kocsy, G. (2010). Differential effects of cold, osmotic stress and abscisic acid on polyamine accumulation in wheat. Amino Acids 38 (2), 623–631. doi: 10.1007/s00726-009-0423-8
Krishna, P. (2003). Brassinosteroid-mediated stress responses. J. Plant Growth Regulation. 22 (4), 289–297. doi: 10.1007/s00344-003-0058-z
Ku, Y. S., Sintaha, M., Cheung, M. Y., Lam, H. M. (2018). Plant hormone signaling cross talks between biotic and abiotic stress responses. Int. J. Mol. Sci. 19, 3206. doi: 10.3390/ijms19103206
Kumar, R., Dhansu, P., Kulshreshtha, N., Meena, M. R., Kumaraswamy, M. H., Appunu, C., et al. (2023). Identification of salinity tolerant stable sugarcane cultivars using AMMI, GGE and some other stability parameters under multi environments of salinity stress. Sustainability 15, 1119. doi: 10.3390/su15021119
Kumar, P., Sharma, P. K. (2020). Soil salinity and food security in India. Front. Sustain. Food Systems. 4. doi: 10.3389/fsufs.2020.533781
Kumar, V., Singh, A., Mithra, S. V. A., Krishnamurthy, S. L., Parida, S. K., Jain, S., et al. (2015). Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 22 (2), 133–145. doi: 10.1093/dnares/dsu046
Kumari, S., Chhillar, H., Chopra, P., Khanna, R. R., Khan, M. I. R. (2021). Potassium: A track to develop salinity tolerant plants. Plant Physiol. Biochem. 167, 1011–1023. doi: 10.1016/j.plaphy.2021.09.031
Kurusu, T., Kuchitsu, K., Tada, Y. (2015). Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Science. 6. doi: 10.3389/fpls.2015.00427
Lee, S. C., Luan, S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell environment. 35 (1), 53–60. doi: 10.1111/j.1365-3040.2011.02426.x
Li, B., Zhang, N., Wang, Y. G., George, A. W., Reverter, A., Li, Y. (2018). Genomic prediction of breeding values using a subset of SNPs identified by three machine learning methods. Front. Genet. 9, 237. doi: 10.3389/fgene.2018.00237
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32 (10), 1045–1052. doi: 10.1038/nbt.2979
Liang, T., Qing, C., Liu, P., Zou, C., Yuan, G., Pan, G., et al. (2022). Joint GWAS and WGCNA uncover the genetic control of calcium accumulation under salt treatment in maize seedlings. Physiologia Plantarum. 174 (1), e13606. doi: 10.1111/ppl.13606
Lozada, D. N., Carter, A. H. (2020). Genomic selection in winter wheat breeding using a recommender approach. Genes 11 (7), 779–793. doi: 10.3390/genes11070779
Lu, K., Guo, Z., Di, S., Lu, Y., Muhammad, I. A. R., Rong, C., et al. (2023). OsMFT1 inhibits seed germination by modulating abscisic acid signaling and gibberellin biosynthesis under salt stress in rice. Plant Cell Physiol. 64 (6), 674–685. doi: 10.1093/pcp/pcad029
Luo, Q., Hu, P., Yang, G., Li, H., Liu, L., Wang, Z., et al. (2021a). Mapping QTL for seedling morphological and physiological traits under normal and salt treatments in a RIL wheat population. Theor. Appl. Genet. 134 (9), 2991–3011. doi: 10.1007/s00122-021-03872-5
Luo, X., Wang, B., Gao, S., Zhang, F., Terzaghi, W., Dai, M. (2019b). Genome-wide association study dissects the genetic bases of salt tolerance in maize seedlings. J. Integr. Plant Biol. 61 (6), 658–674. doi: 10.1111/jipb.12797
Luo, M., Zhang, Y., Chen, K., Kong, M., Song, W., Lu, B., et al. (2019a). Mapping of quantitative trait loci for seedling salt tolerance in maize. Mol. Breeding. 39 (5), 64. doi: 10.1007/s11032-019-0974-7
Luo, M., Zhang, Y., Li, J., Zhang, P., Chen, K., Song, W., et al. (2021c). Molecular dissection of maize seedling salt tolerance using a genome-wide association analysis method. Plant Biotechnol. J. doi: 10.1111/pbi.13607
Luo, M., Zhao, Y., Zhang, R., Xing, J., Duan, M., Li, J., et al. (2017). Mapping of a major QTL for salt tolerance of mature field-grown maize plants based on SNP markers. BMC Plant Biol. 17 (1), 140. doi: 10.1186/s12870-017-1090-7
Luo, Q., Zheng, Q., Hu, P., Liu, L., Yang, G., Li, H., et al. (2021b). Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor. Appl. Genet. 134 (1), 171–189. doi: 10.1007/s00122-020-03689-8
Ma, Y., Dias, M. C., Freitas, H. (2020). Drought and salinity stress responses and microbe-induced tolerance in plants. Front. Plant Science. 11, 591911. doi: 10.3389/fpls.2020.591911
Mantri, N., Patade, V., Penna, S., Ford, R., Pang, E. (2012). “Abiotic stress responses in plants: present and future,” in Abiotic stress responses in plants: metabolism, productivity and sustainability. Eds. Ahmad, P., Prasad, M. N. V. (New York: Springer), 1–19.
Marcum, K. B., Anderson, S. J., Engelke, M. C. (1998). Salt gland ion secretion: A salinity tolerance mechanism among five zoysiagrass species. Crop Sci. 38 (3), 806–810. doi: 10.2135/cropsci1998.0011183X003800030031x
Matysik, J., Alia, A., Bhalu, B., Mohanty, P. (2002). Molecular mechanisms of quenching of reactive oxygen species by proline under stress in plants. Curr. Science. 82 (5), 525–532.
McFarlane, D. J., George, R. J., Barrett-Lennard, E. G., Gilfedder, M. (2016). Salinity in dryland agricultural systems: challenges and opportunities. Innov. In Dryland Agriculture. 521-547. doi: 10.1007/978-3-319-47928-6_19
Medina, C. A., Hawkins, C., Liu, X. P., Peel, M., Yu, L. X. (2020). Genome-wide association and prediction of traits related to salt tolerance in autotetraploid alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 21 (9), 3361. doi: 10.3390/ijms21093361
Meuwissen, T. H., Hayes, B. J., Goddard, M. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics. 157 (4), 1819–1829. doi: 10.1093/genetics/157.4.1819
Minocha, R., Majumdar, R., Minocha, S. C. (2014). Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00175
Mondal, S., Atta, K., Mukherjee, S., Chowdhury, S. R., Pal, A., Maitra, S., et al. (2023). Advancement of transgenic wheat (Triticum aestivum L.) to survive against abiotic stresses in the era of the changing climate. In Khan, M. K., Pandey, A., Hamurcu, M., Gupta, O. P., Gezgin, S. (Eds) Abiotic stresses in wheat unfolding the challenges Academic Press, Elsevier Inc. 1, 150–159. doi: 10.1016/B978-0-323-95368-9.00021-7
Montenegro, J. D., Golicz, A. A., Bayer, P. E., Hurgobin, B., Lee, H., Chan, C. K. K., et al. (2017). The pangenome of hexaploid bread wheat. Plant J. 90, 1007–1013. doi: 10.1111/tpj.13515
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environment. 25 (2), 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Munns, R., Passioura, J. B., Colmer, T. D., Byrt, C. S. (2020). Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 225, 1091–1096. doi: 10.1111/nph.15862
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 (1), 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nadeem, M., Tariq, M. N., Amjad, M., Sajjad, M., Akram, M., Imran, M., et al. (2020). Salinity induced changes in the nutritional quality of bread wheat (Triticum aestivum L.) genotypes. Agrivita. 42, 1–12. doi: 10.17503/agrivita.v42i1.2273
Nahar, K., Hasanuzzaman, M. (2009). Germination, growth, nodulation and yield performance of three mungbean varieties under different levels of salinity stress. Green Farming. 2, 825–829.
Nahar, K., Hasanuzzaman, M., Rahman, A., Alam, M. M., Mahmud, J., Suzuki, T., et al. (2016). Polyamines confer salt tolerance in mung bean (Vigna radiata L.) by reducing sodium uptake, improving nutrient homeostasis, antioxidant defense, and methylglyoxal detoxification systems. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01104
Nayyeripasand, L., Garoosi, G. A., Ahmadikhah, A. (2021). Genome wide association study (GWAS) to identify salt-tolerance QTL carrying novel candidate genes in rice during early vegetative stage. Rice. 14 (1), 9. doi: 10.1186/s12284-020-00433-0
Neto, A. D. A., Pereira, P. P., Costa, D. P., dos Santos, A. C. (2011). Chlorophyll fluorescence as a possible tool for salt-stress tolerance screening in the sunflower. Rev. Ciec. Agronômica. 42 (4), 893.
Niones, J. M., Gregorio, G., Tumimbang, E., Adorada, D., Hernandez, J. (2006). Fine mapping of the salinity tolerance gene in chromosome 1 of rice (Oryza sativa L.) using near isogenic lines. In: Proceedings of the 36th CSSP Scientific Conference "Value Added Crop Science for Sustainable Agricultural Development", 8-12 May 2006 (Puerto Princesa City, Philippines: Crop Science Society of the Philippines) 31 (supplement no. 1), 46.
Nounjan, N., Nghia, P. T., Theerakulpisut, P. (2012). Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J. Plant Physiol. 169 (6), 596–604. doi: 10.1016/j.jplph.2012.01.004
Otitoloju, K. (2014). Growth, yield and seed nutritional composition of groundnut (Arachis hypogaea LINN) under elevated level of soil salinity. Mol. Soil Biol. 5 (5). doi: 10.5376/msb.2014.05.0005
Panta, S., Flowers, T., Lane, P. A., Doyle, R., Haros, G., Shabala, S. N. (2014). Halophyte agriculture: Success stories. Environ. Exp. Botany. 107, 71–83. doi: 10.1016/j.envexpbot.2014.05.006
Parida, A. K., Das, A. B. (2005). Salt tolerance and salinity effect on plants: a review. Ecotoxicology Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Parida, A. K., Das, A. B., Mohanty, P. (2004). Investigations on the antioxidative defense responses to NaCl stress in a mangrove, Bruguiera parviflora: differential regulations of isoforms of some antioxidative enzymes. Plant Growth Regulation. 42, 213–226. doi: 10.1023/B:GROW.0000026508.63288.39
Parvin, K., Ahmed, K. U., Islam, M. M., Haque, M. N. (2015a). Response of tomato plant under salt stress: role of exogenous calcium. J. Plant Sci. 10 (6), 222–233. doi: 10.3923/jps.2015.222.233
Parvin, K., Ahmed, K. U., Islam, M. M., Haque, M. N. (2016). Modulation of ion uptake in tomato (Lycopersicon esculentum L.) plants with exogenous application of calcium under salt stress. Poljoprivreda/Agriculture. 22 (2), 40–49.
Parvin, K., Haque, M. N. (2017). Protective role of salicylic acid on salt affected broccoli plant. J. Agric. Ecol. Res. 10 (2). doi: 10.9734/JAERI/2017/30142
Parvin, K., Hasanuzzaman, M., Fujita, M. (2017). Salicylic acid enhances growth and productivity in cabbage (Brassica oleracea var. Capitata L.) grown under saline condition. Focus Sci. 3 (1). doi: 10.21859/focsci-030162
Patil, G., Do, T., Vuong, T. D., Valliyodan, B., Lee, J. D., Chaudhary, J., et al. (2016). Genomic assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 6 (1), 1–13. doi: 10.1038/srep19199
Pavlović, I., Mlinarić, S., Tarkowská, D., Oklestkova, J., Novák, O., Lepeduš, H., et al. (2019). Early Brassica crops responses to salinity stress: a comparative analysis between chinese cabbage, white cabbage, and kale. Front. Plant Science. 10. doi: 10.3389/fpls.2019.00450
Percival, G. C., Fraser, G. A., Oxenham, G. (2003). Foliar salt tolerance of Acer genotypes using chlorophyll fluorescence. Arboriculture Urban Forestry. 29 (2), 61. doi: 10.48044/jauf.2003.008
Plaut, Z., Meinzer, F. C., Federman, E. (2000). Leaf development, transpiration and ion uptake and distribution in Sugarcane cultivars grown under salinity. Plant Soil. 218, 59–69. doi: 10.1023/A:1014996229436
Poland, J., Endelman, J., Dawson, J., Rutkoski, J., Wu, S., Manes, Y., et al. (2012). Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome. 5 (3). doi: 10.3835/plantgenome2012.06.0006
Popova, L. P., Stoinova, Z. G., Maslenkova, L. T. (1995). Involvement of abscisic acid in photosynthetic process in Hordeum vulgare L. during salinity stress. J. Plant Growth Regulation. 14 (4), 211–218. doi: 10.1007/BF00204914
Poustini, K., Siosemardeh, A. (2004). Ion distribution in wheat cultivars in response to salinity stress. Field Crops Res. 85, 125–133. doi: 10.1016/S0378-4290(03)00157-6
Puram, V. R. R., Ontoy, J., Subudhi, P. K. (2018). Identification of QTLsQTL for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol. Biol. Reporter. 36 (5), 695–709. doi: 10.1007/s11105-018-1110-2
Qi, X., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5, 4340. doi: 10.1038/ncomms5340
Rahman, A., Nahar, K., Hasanuzzaman, M., Fujita, M. (2016). Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00609
Rahman, S., Vance, G., Munn, L. (1993). Salinity induced effects on the nutrient status of soil, corn leaves and kernels. Commun. Soil Sci. Plant Anal. 24, 2251–2269. doi: 10.1080/00103629309368953
Rajkumar, M. S., Jain, M., Garg, R. (2021). Discovery of DNA polymorphisms via whole genome resequencing and their functional relevance in salinity stress response in chickpea. Physiologia Plantarum. 173 (4), 1573–1586. doi: 10.1111/ppl.13507
Rao, V. P., Sengar, R. S., Singh, S., Sharma, V. (2015). Molecular and metabolic perspectives of sugarcane under salinity stress pressure. Progress. Agric. 15, 77–84.
Rathinapriya, P., Pandian, S., Rakkammal, K., Balasangeetha, M., Alexpandi, R., Satish, L., et al. (2020). The protective effects of polyamines on salinity stress tolerance in foxtail millet (Setaria italica L.), an important C4 model crop. Physiol. Mol. Biol. Plants. 26, 1815–1829. doi: 10.1007/s12298-020-00869-0
Reddy, M. P., Sanish, S., Iyengar, E. R. R. (1992). Photosynthetic studies and compartmentation of ions in different tissues of Salicornia brachiata Roxb. under saline conditions. Photosynthetica 26, 173–179.
Rezaei, E., Hervan, E. M., Azadi, A., Etminan, A., Ramshini, H. (2021). Prioritisation of candidate genes in QTL regions for seed germination and early seedling growth in bread wheat (Triticum aestivum) under salt-stress conditions. Crop Pasture Science. 72 (1), 1–16. doi: 10.1071/CP20319
Robin, A. H. K., Matthew, C., Uddin, M. J., Bayazid, K. N. (2016). Salinity induced reduction in root surface area and changes in major root and shoot traits at the phytomer level in wheat. J. Exp. Bot. 67, 3719–3729. doi: 10.1093/jxb/erw064
Rus, A., Lee, B. H., Munoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J. K., et al. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136 (1), 2500–2511. doi: 10.1104/pp.104.042234
Saha, J., Brauer, E. K., Sengupta, A., Popescu, S. C., Gupta, K., Gupta, B. (2015). Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Science. 3. doi: 10.3389/fenvs.2015.00021
Sairam, R. K., Tyagi, A. (2004). Physiology and molecular biology of salinity stress tolerance in plants. Curr. Science vol. 86 (3), 407–421.
Sandhu, K. S., Lozada, D. N., Zhang, Z., Pumphrey, M. O., Carter, A. H. (2021a). Deep learning for predicting complex traits in spring wheat breeding program. Front. Plant Science. 11, 613325. doi: 10.3389/fpls.2020.613325
Sandhu, K. S., Mihalyov, P. D., Lewien, M. J., Pumphrey, M. O., Carter, A. H. (2021b). Combining genomic and phenomic information for predicting grain protein content and grain yield in spring wheat. Front. Plant Science. 170. doi: 10.3389/fpls.2021.613300
Sawada, H., Shim, I. S., Usui, K. (2006). Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis modulation by salt stress in rice seedlings. Plant Science. 171 (2), 263–270. doi: 10.1016/j.plantsci.2006.03.020
Saxena, S. C., Kaur, H., Verma, P., Petla, B. P., Andugula, V. R., Majee, M. (2013). “Osmo-protectants: potential for crop improvement under adverse conditions,” in Plant acclimation to environmental stress (New York, NY, USA: Springer), 197–232.
Schachtman, D. P., Schroeder, J. I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 370 (6491), 655–658.
Segami, S., Asaoka, M., Kinoshita, S., Fukuda, M., Nakanishi, Y., Maeshima, M. (2018). Biochemical, structural, and physiological characteristics of vacuolar H+- pyro phosphatase. Plant Cell Physiol. 59, 1300–1308.
Serrano, R., Mulet, J. M., Rios, G., Marquez, J. A., De Larrinoa, I. F., Leube, M. P., et al. (1999). A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Botany. 50, 1023–1036. doi: 10.1093/jxb/50.Special_Issue.1023
Shabala, S., Cuin, T. (2008). Potassium transport and plant salt tolerance. Physiologia Plantarum. 133, 651–669. doi: 10.1111/j.1399-3054.2007.01008
Shabala, S., Cuin, T. A., Pottosin, I. (2007). Polyamines prevent NaCl- induced K+ efflux from pea mesophyll by blocking non-selective cation channels. FEBS Lett. 581, 1993–1999. doi: 10.1016/j.febslet.2007.04.032
Shabala, S., Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiologia Plantarum. 151, 257–279. doi: 10.1111/ppl.12165
Shanker, A. K., Venkateswarlu, B. (2011). “Abiotic stress in plants mechanisms and adaptations,” in TechJaneza trdine. (51000 Rijeka, Croatia), 9.
Sharma, P. C., Sehgal, D., Singh, D., Singh, G., Yadav, R. S. (2011). A major terminal drought tolerance QTL of pearl millet is also associated with reduced salt uptake and enhanced growth under salt stress. Mol. Breeding. 27 (2), 207–222. doi: 10.1007/s11032-010-9423-3
Shi, M. Q., Xi-Liang, L., Qian, Y., Zhang, W., Ya-Kai, L., Bhat, A. J., et al. (2022). Linkage and association mapping of wild soybean (Glycine soja) seeds germinating under salt stress. J. Integr. Agriculture 21 (10), 2833–2847. doi: 10.1016/j.jia.2022.07.031
Siddiqui, Z. S., Cho, J. I., Park, S. H., Kwon, T. R., Ahn, B. O., Lee, G. S., Park, S. C. (2014). Phenotyping of rice in salt stress environment using high-throughput infrared imaging. Acta Botanica Croatica. 73 (1), 149–158.
Singh, J., Chander Sharma, P., Singh, V. (2021a). Breeding mustard (Brassica juncea) for salt tolerance: problems and prospects. Brassica Breed. Biotechnol 3, 69–85. doi: 10.5772/intechopen.94551
Singh, R., Gregorio, G., Jain, R. K. (2007). QTL mapping for salinity tolerance in rice. Physiol. Mol. Biol. Plants. 13, 87–99.
Singh, R. K., Kota, S., Flowers, T. J. (2021b). Salt tolerance in rice: seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 134 (11), 3495–3533. doi: 10.1007/s00122-021-03890-3
Sirault, X. R., James, R. A., Furbank, R. T. (2009). A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct. Plant Biol. 36 (11), 970–977. doi: 10.1071/FP09182
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2013). Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PloS One 8 (1), e54985. doi: 10.1371/journal.pone.0054985
Soren, K. R., Madugula, P., Kumar, N., Barmukh, R., Sengar, M. S., Bharadwaj, C., et al. (2020). Genetic dissection and identification of candidate genes for salinity tolerance using axiom®CicerSNP array in chickpea. Int. J. Mol. Sci. 21 (14), 5058. doi: 10.3390/ijms21145058
Spiekerman, J. J., Devos, K. M. (2020). The halophyte seashore paspalum uses adaxial leaf papillae for sodium sequestration. Plant Physiol. 184 (4), 2107–2119. doi: 10.1104/pp.20.00796
Squires, V. R., Glenn, E. P. (2004). “Salination, desertification, and soil erosion,” in The role of food, agriculture, forestry and fisheries in human nutrition. Ed. Squires, V. R. (Oxford, UK: UNESCO, EOLSS Publishers).
Stavi, I., Thevs, N., Priori, S. (2021). Soil salinity and sodicity in drylands: a review of causes, effects, monitoring, and restoration measures. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.712831
Subudhi, P. K., Shankar, R., Jain, M. (2020). Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 10 (1), 1–13. doi: 10.1038/s41598-020-78256-8
Sung, C. H., Hong, J. K. (2010). Sodium nitroprusside mediates seedling development and attenuation of oxidative stresses in Chinese cabbage. Plant Biotechnol. Rep. 4 (4), 243–251. doi: 10.1007/s11816-010-0138-z
Suzuki, K., Yamaji, N., Costa, A., Okuma, E., Kobayashi, N. I., Kashiwagi, T., et al. (2016). OsHKT1; 4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 16, 22. doi: 10.1186/s12870-016-0709-4
Tahir, M. A., Farooq, T., M. and Sarwar, G. (2012). Silicon induced changes in growth, ionic composition, water relations, chlorophyll contents and membrane permeability in two salt stressed wheat genotypes. Arch. Agron. Soil Science. 58 (3), 247–256. doi: 10.1080/03650340.2010.518959
Taiz, L., Zeiger, E., Møller, I. M., Murphy, A. (2015). Plant physiology and development (No. Ed. 6) (Sinauer Associates Incorporated).
Takahashi, T., Kakehi, J. (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 105 (1), 1–6. doi: 10.1093/aob/mcp259
Tanksley, S. D., Grandillo, S., Fulton, T. M., Zamir, D., Eshed, Y., Petiard, V., et al. (1996). Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92 (2), 213–224. doi: 10.1007/BF00223378
Technow, F., Bürger, A., Melchinger, A. E. (2013). Genomic prediction of northern corn leaf blight resistance in maize with combined or separated training sets for heterotic groups. G3: Genes| Genomes| Genet. 3 (2), 197–203. doi: 10.1534/g3.112.004630
Tester, M., Davenport, R. (2003). Na+ tolerant and Na+ transport in higher plants. Ann. Botany. 91, 503–527. doi: 10.1093/aob/mcg058
Thomson, M. J., de Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice. 3 (2), 148–160. doi: 10.1007/s12284-010-9053-8
Tibbs Cortes, L., Zhang, Z., Yu, J. (2021). Status and prospects of genome-wide association studies in plants. Plant Genome 14 (1), e20077. doi: 10.1002/tpg2.20077
Tiwari, S., Sl, K., Kumar, V., Singh, B., Rao, A. R., Mithra Sv, A., et al. (2016). Mapping QTLsQTL for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PloS One 11 (4), e0153610. doi: 10.1371/journal.pone.0153610
Tsai, Y. C., Chen, K. C., Cheng, T. S., Lee, C., Lin, S. H., Tung, C. W. (2019). Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 19 (1), 1–17. doi: 10.1186/s12870-019-1983-8
Uçarlı, C. (2020). “Effects of salinity on seed germination and early seedling stage,” in Abiotic stress in plants. Eds. Fahad, S., Saud, S., Chen, Y., Wu, C., Wang, D. (IntechOpen). doi: 10.5772/intechopen.93647
Ullah, M. A., Abdullah-Zawawi, M. R., Zainal-Abidin, R. A., SukIran, N. L., Uddin, M. I., Zainal, Z. (2022). A review of integrative omic approaches for understanding rice salt response mechanisms. Plants. 11 (11), 1430. doi: 10.3390/plants11111430
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Visscher, P. M., Wray, N. R., Zhang, Q., Sklar, P., McCarthy, M. I., Brown, M. A., et al. (2017). 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101 (1), 5–22. doi: 10.1016/j.ajhg.2017.06.005
Vorasoot, N., Songsri, P., Akkasaeng, C., Jogloy, S., Patanothai, A. (2003). Effect of water stress on yield and agronomic characters of peanut. Songklanakarin J. Sci. Technology. 25, 283–288.
Wahid, A., Ahmad, S. S., Ahmad, M. N., Khaliq, B., Nawaz, M., Shah, S. Q., et al. (2014). Assessing the effects of hydrogen fluoride on mango (Mangifera indica L.) in the vicinity of a brick kiln field in southern Pakistan. Fluoride. 47, 307–314.
Wan, H., Chen, L., Guo, J., Li, Q., Wen, J., Yi, B., et al. (2017). Genome-wide association study reveals the genetic architecture underlying salt tolerance-related traits in rapeseed (Brassica napus L.). Front. Plant Science. 8, 593. doi: 10.3389/fpls.2017.00593
Wang, H., Chen, G., Zhang, H., Liu, B., Yang, Y., Qin, L., et al. (2014). Identification of QTLsQTL for salt tolerance at germination and seedling stage of Sorghum bicolor L. Moench. Euphytica. 196 (1), 117–127. doi: 10.1007/s10681-013-1019-7
Wang, Y., Li, L., Cui, W., Xu, S., Shen, W., Wang, R. (2012). Hydrogen sulfide enhances alfalfa (Medicago sativa) tolerance against salinity during seed germination by nitric oxide pathway. Plant Soil. 351, 107–119. doi: 10.1007/s11104-011-0936-2
Wang, Y., Nii, N. (2000). Changes in chlorophyll, ribulose bisphosphate carboxylase-oxygenase, glycine betaine content, photosynthesis and transpiration in Amaranthus tricolor leaves during salt stress. J. Hortic. Sci. Biotechnol. 75 (6), 623–627. doi: 10.1080/14620316.2000.11511297
Wang, H., Wang, R., Liu, B., Yang, Y., Qin, L., Chen, E., et al. (2020). QTL analysis of salt tolerance in Sorghum bicolor during whole-plant growth stages. Plant Breed. .139 (3), 455–465. doi: 10.1111/pbr.12805
Warraich, A. S., Krishnamurthy, S. L., Sooch, B. S., Vinaykumar, N. M., Dushyanthkumar, B. M., Bose, J., et al. (2020). Rice GWAS reveals key genomic regions essential for salinity tolerance at reproductive stage. Acta Physiologiae Plantarum. 42 (8), 134. doi: 10.1007/s11738-020-03123-y
Wassan, G. M., Khanzada, H., Zhou, Q., Mason, A. S., Keerio, A. A., Khanzada, S., et al. (2021). Identification of genetic variation for salt tolerance in Brassica napus using genome-wide association mapping. Mol. Genet. Genomics 296, 391–408. doi: 10.1007/s00438-020-01749-8
Weiss, D., Ori, N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144 (3), 1240–1246. doi: 10.1104/pp.107.100370
Wu, D., Chen, C., Liu, Y., Yang, L., Yong, J. W. H. (2023). Iso-osmotic calcium nitrate and sodium chloride stresses have differential effects on growth and photosynthetic capacity in tomato. Scientia Hortic. 312, 111883. doi: 10.1016/j.scienta.2023.111883
Xin, Y., Wu, Y. Q., Han, X., Xu, L. (2021). Overexpression of the Ginkgo biloba WD40 gene GbLWD1-like improves salt tolerance in transgenic Populus. Plant Science. 313, 111092. doi: 10.1016/j.plantsci.2021.111092
Xiong, J., Fu, G., Tao, L., Zhu, C. (2010). Roles of nitric oxide in alleviating heavy metal toxicity in plants. Arch. Biochem. Biophysics. 497 (1-2), 13–20. doi: 10.1016/j.abb.2010.02.014
Xu, Y., Crouch, J. H. (2008). Marker-assisted selection in plant breeding: from publications to practice. Crop Sci. 48, 391–407. doi: 10.2135/cropsci2007.04.0191
Xu, S., Hu, B., He, Z., Ma, F., Feng, J., Shen, W., et al. (2011). Enhancement of salinity tolerance during rice seed germination by pre-soaking with haemoglobin. Int. J. Mol. Sci. 12, 2488–2501. doi: 10.3390/ijms12042488
Yadav, T., Kumar, A., Yadav, R. K., Yadav, G., Kumar, R., Kushwaha, M. (2020). Salicylic acid and thiourea mitigate the salinity and drought stress on physiological traits governing yield in pearl millet- wheat. Saudi J. Biol. Sci. 27 (8), 2010–2017. doi: 10.1016/j.sjbs.2020.06.030
Yadav, P. V., Kumari, M., Ahmed, Z. (2011). Seed priming mediated germination improvement and tolerance to subsequent exposure to cold and salt stress in Capsicum. Res. J. Seed Sci. 4, 125–136. doi: 10.3923/rjss.2011.125.136
Yamamoto, A., Hashiguchi, M., Akune, R., Masumoto, T., Muguerza, M., Saeki, Y., et al. (2016). The relationship between salt gland density and sodium accumulation/secretion in a wide selection from three Zoysia species. Aust. J. Botany. 64 (4), 277–284. doi: 10.1071/BT15261
Yang, J., Zhang, J., Liu, K., Wang, Z., Liu, L. (2007). Involvement of polyamines in the drought resistance of rice. J. Exp. Botany. 58 (6), 1545–1555. doi: 10.1093/jxb/erm032
Yildiz, M., Poyraz, İ., Çavdar, A., Özgen, Y., Beyaz, R. (2020). Plant responses to salt stress. Plant Breed. Curr. Future Views. doi: 10.5772/intechopen.93920
Yong, H. Y., Wang, C., Bancroft, I., Li, F., Wu, X., Kitashiba, H., et al. (2015). Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta. 242, 313–326. doi: 10.1007/s00425-015-2310-8
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant science. 25 (11), 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zaman, M., Shahid, S. A., Heng, L., Shahid, S. A., Zaman, M., Heng, L. (2018). Introduction to soil salinity, sodicity and diagnostics techniques. Guideline salinity assessment mitigation adaptation using Nucl. related techniques, 1–42. doi: 10.1007/978-3-319-96190-3_1
Zhang, Y., Li, P., Zhang, J., Li, Y., Xu, A., Huang, Z. (2022b). Genome wide association studies of salt tolerance at the seed germination stage and yield related traits in Brassica napus L. Int. J. Mol. Sci. 23 (24), 15892. doi: 10.3390/ijms232415892
Zhang, X., Pérez-Rodríguez, P., Semagn, K., Beyene, Y., Babu, R., López-Cruz, M. A., et al. (2015). Genomic prediction in biparental tropical maize populations in water-stressed and well-watered environments using low-density and GBS SNPs. Heredity. 114 (3), 291–299. doi: 10.1038/hdy.2014.99
Zhang, G., Zhou, J., Peng, Y., Tan, Z., Li, L., Yu, L., et al. (2022a). Genome-wide association studies of salt tolerance at seed germination and seedling stages in Brassica napus. Front. Plant Science. 12, 772708. doi: 10.3389/fpls.2021.772708
Zhao, M. G., Chen, L., Zhang, L. L., Zhang, W. H. (2009). Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol. 151 (2), 755–767. doi: 10.1104/pp.109.140996
Zhao, Q., Feng, Q., Lu, H., Li, Y., Wang, A., Tian, Q., et al. (2018). Pangenome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50 (2), 278–284. doi: 10.1038/s41588-018-0041-z
Zhao, F., Song, C. P., He, J., Zhu, H. (2007). Poly amines improve K+/Na+ homeostasis in barley seedlings by regulating root ion channel activities. Plant Physiol. 145, 1061–1072. doi: 10.1104/pp.107.105882
Zhao, L., Zhang, F., Guo, J., Yang, Y., Li, B., Zhang, L. (2004). Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 134, 849–857. doi: 10.1104/pp.103.030023
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, P. (2021). Regulation of plant responses to salt stress. Int. J. Of Mol. Sci. 22 (9), 4609. doi: 10.1016/j.envexpbot.2009.05.002
Zhao, W., Zhang, Y., Zhang, J., Qi, Y., Wang, L., Dang, Z., et al. (2022). QTL mapping by whole genome re-sequencing and analysis of candidate genes for salt tolerance in linseed (Linum usitatissmum L.). Oil Crop Science. 7 (2), 80–85. doi: 10.1016/j.ocsci.2022.05.004
Zheng, C., Jiang, D., Liu, F., Dai, T., Liu, W., Jing, Q., et al. (2009). Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environ. Exp. Bot. 67, 222227. doi: 10.1016/j.envexpbot.2009.05.002
Zhu, J. K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124 (3), 941–948. doi: 10.1104/pp.124.3.941
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 (5), 441–445. doi: 10.1016/S1369-5266(03)00085-2
Keywords: salinity stress, oxidative stress, antioxidants, phenomics, QTL mapping, GWAS, genomic selection
Citation: Atta K, Mondal S, Gorai S, Singh AP, Kumari A, Ghosh T, Roy A, Hembram S, Gaikwad DJ, Mondal S, Bhattacharya S, Jha UC and Jespersen D (2023) Impacts of salinity stress on crop plants: improving salt tolerance through genetic and molecular dissection. Front. Plant Sci. 14:1241736. doi: 10.3389/fpls.2023.1241736
Received: 17 June 2023; Accepted: 14 August 2023;
Published: 15 September 2023.
Edited by:
Lorenzo Barbanti, University of Bologna, ItalyReviewed by:
Muhammad Ishaq Asif Rehmani, Ghazi University, PakistanPooja Choudhary, Jaypee Institute of Information Technology, India
Copyright © 2023 Atta, Mondal, Gorai, Singh, Kumari, Ghosh, Roy, Hembram, Gaikwad, Mondal, Bhattacharya, Jha and Jespersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouvik Gorai, OS5zaG91dmlrLjlAZ21haWwuY29t; David Jespersen, ZGplc3BlckB1Z2EuZWR1
†These authors have contributed equally to this work and share first authorship
 Kousik Atta
Kousik Atta Saptarshi Mondal
Saptarshi Mondal Shouvik Gorai
Shouvik Gorai Aditya Pratap Singh
Aditya Pratap Singh Amrita Kumari2
Amrita Kumari2 Arkaprava Roy
Arkaprava Roy Dinkar Jagannath Gaikwad
Dinkar Jagannath Gaikwad Sudip Bhattacharya
Sudip Bhattacharya Uday Chand Jha
Uday Chand Jha David Jespersen
David Jespersen