- 1Shandong Academy of Agricultural Sciences, Jinan, China
- 2State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan, China
- 3Key Laboratory of Plant Genetics and Molecular Breeding, Zhoukou Normal University, Zhoukou, China
- 4State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, China
The Bph15 gene, known for its ability to confer resistance to the brown planthopper (BPH; Nilaparvata lugens Stål), has been extensively employed in rice breeding. However, the molecular mechanism by which Bph15 provides resistance against BPH in rice remains poorly understood. In this study, we reported that the transcription factor OsWRKY71 was highly responsive to BPH infestation and exhibited early-induced expression in Bph15-NIL (near-isogenic line) plants, and OsWRKY71 was localized in the nucleus of rice protoplasts. The knockout of OsWRKY71 in the Bph15-NIL background by CRISPR-Cas9 technology resulted in an impaired Bph15-mediated resistance against BPH. Transcriptome analysis revealed that the transcript profiles responsive to BPH differed between the wrky71 mutant and Bph15-NIL, and the knockout of OsWRKY71 altered the expression of defense genes. Subsequent quantitative RT-PCR analysis identified three genes, namely sesquiterpene synthase OsSTPS2, EXO70 family gene OsEXO70J1, and disease resistance gene RGA2, which might participate in BPH resistance conferred by OsWRKY71 in Bph15-NIL plants. Our investigation demonstrated the pivotal involvement of OsWRKY71 in Bph15-mediated resistance and provided new insights into the rice defense mechanisms against BPH.
Introduction
The brown planthopper (BPH; Nilaparvata lugens Stål) is a major rice (Oryza sativa) pest and a representative piercing-sucking insect. The most effective strategy for managing BPH is the cultivation of insect-resistant rice varieties (Zhang, 2007). Research has identified and characterized a total of 34 major BPH resistance genes, with fifteen of them having been isolated (Du et al., 2020; Chen et al., 2022). Among these genes, Bph14 encodes a nucleotide-binding and leucine-rich repeat (NLR) protein, which interacts with OsWRKY46 and OsWRKY72, ultimately leading to the activation of downstream defensive genes in rice (Du et al., 2009; Hu et al., 2017). A recent discovery has revealed that Bph14 can directly bind BPH effector BISP, leading to the activation of effector-triggered immunity (ETI) (Guo et al., 2023). Bph6 encodes a unique leucine-rich repeat (LRR) protein that is specifically localized within the exocyst complex and exhibits interactions with OsEXO70E1 and OsEXO70H3. This interaction promotes protein secretion and strengthens the plant cell wall (Guo et al., 2018; Wu et al., 2022). Bph30 encodes a novel protein comprising two leucine-rich domains, which is expressed in sclerenchyma cells and enhances the deposition of hemicellulose (Shi et al., 2021). Bph3, which was discovered in Rathu Heenati (RH), encodes three lectin receptor kinases (OsLecRK1-OsLecRK3) that are located on the plasma membrane and function as receptors in plant immunity (Liu et al., 2014). The Bph15 locus, derived from Oryza officinalis, is a compound locus consisting of two genes. One of these genes is identical to Bph3 (Cheng et al., 2013a; Xiao et al., 2016), while the other major genetic locus has been localized to the adjacent 580-kb recombination cold-spot region (Lv et al., 2014). The resistance to BPH is mediated through a molecular mechanism that shares similarities with defense mechanisms against pathogens (Cheng et al., 2013b; Du et al., 2020; Chen et al., 2022). Bph15 and Bph3 potentially function as receptors for recognizing herbivore-associated molecular patterns (HAMPs), thereby initiating pattern-triggered immunity (PTI). However, the precise molecular mechanisms underlying the functions of Bph15 and Bph3 remain poorly elucidated.
In the realm of plants, WRKY transcription factors (TFs) represent a substantial assemblage of TFs, characterized by a discernible affinity for the W-box sequence situated within the promoter regions of their respective target genes (Rushton et al., 2010). The influence of WRKY transcription factors on the immune responses of plants is both significant and extensive (Eulgem and Somssich, 2007). In rice, numerous WRKY TFs have been identified as positive regulators of plant pathogen resistance, exemplified by the enhanced resistance against diverse pathogens observed in OsWRKY13, OsWRKY30, OsWRKY45, and OsWRKY89 (Qiu et al., 2007; Shimono et al., 2007; Wang et al., 2007; Peng et al., 2012). However, the role of OsWRKY45 is of significant interest due to its documented importance in providing the rice R protein Pb1 against fungal pathogens, albeit with a negative impact on resistance against the BPH (Inoue et al., 2013; Huangfu et al., 2016). Additionally, another WRKY transcription factor, OsWRKY53, has been discovered to act as a positive regulator of plant pathogen and BPH resistance. Moreover, it has been identified as a negative feedback regulator of MPK3 in response to the herbivorous striped stem borer (SSB) (Chujo et al., 2007; Hu et al., 2015; Hu et al., 2016). The activation of OsWRKY70 in response to BPH attack has been demonstrated to result in a reduction in gibberellic acid (GA) production in rice, leading to an increased susceptibility to BPH (Li et al., 2015). In general, WRKY TFs play a significant role in regulating plant immune response and can have both positive and negative effects on defense against various pests and pathogens in rice.
Among the members of the OsWRKY IIa subfamily, OsWRKY28, OsWRKY62, OsWRKY71, and OsWRKY76 are encompassed (Peng et al., 2010). Among these genes, OsWRKY28 and OsWRKY76 function as negative regulators of basal defense responses against blast fungus (Chujo et al., 2013; Yokotani et al., 2013). OsWRKY62 functions as a suppressor of basal and Xa21-mediated defense against the bacterial pathogen (Peng et al., 2008). In the absence of Pi9, OsWRKY62 assumes a beneficial role in regulating blast resistance; however, in the presence of Pi9, it assumes an adverse role (Shi et al., 2023). The involvement of OsWRKY71, which is activated by biotic elicitors and pathogen invasion, plays a significant role in the defense response of rice. Overexpression of OsWRKY71 enhances rice’s resistance to Xoo13751, resulting in the constitutive expression of two marker genes, OsNPR1 and OsPR1b (Liu et al., 2007). Furthermore, OsWRKY71 can interact with the promoter region of the blast resistance gene GF14b, thereby influencing its expression (Liu et al., 2016). In addition, OsWRKY71 also functions as a transcriptional inhibitor (Chujo et al., 2008). Previous studies showed that the overexpression of OsWRKY71 leads to an increase in H2O2 accumulation, resulting in enhanced resistance against the small brown planthopper (Laodelphax striatellus, SBPH) in rice (Ji et al., 2021). However, its role in regulating resistance against the BPH remains unclear.
Our previous research indicated that the expression of OsWRKY71 is up-regulated in the Bph15 introgression line following BPH feeding (Lv et al., 2014). This study demonstrated that OsWRKY71 plays a critical role in the Bph15-mediated resistance against BPH. The knockout of OsWRKY71 resulted in an impaired BPH resistance in Bph15-NIL. Transcriptome sequencing analysis demonstrated distinct transcript profiles between the OsWRKY71 knockout plants and Bph15-NIL plants, suggesting that OsWRKY71 is involved in regulating the expression of plant defense genes. Further investigation using qRT-PCR identified three genes, namely sesquiterpene synthase OsSTPS2, EXO70 family gene OsEXO70J1, and disease resistance gene RGA2, which may participate in the BPH resistance conferred by OsWRKY71 in Bph15-NIL plants.
Materials and methods
Plant materials and insects
As previously stated by Lv et al. (2014), the near-isogenic line Bph15-NIL (XF07-151) was found to possess the resistance gene Bph15 in the Taichung Native 1 (TN1) background. This particular line, Bph15-NIL, was employed as a control in the study due to its demonstrated resistance. In contrast, the varieties TN1 and Nipponbare (NIP) were selected as susceptible rice controls. The BPH insects, which were reared on the TN1 variety, were utilized for plant feeding during the research conducted at Wuhan University in China.
Development of mutants and overexpression transgenic plants
The wrky71 mutants were generated using CRISPR-Cas9 technology (Ma et al., 2015). The target sequence of WRKY71 was designed and used to generate sgRNA expression cassettes driven by the OsU6a promoter. Using an Agrobacterium-mediated technique, the resulting structure was transformed into Bph15-NIL plants. The confirmation of T0 transgenic plants was achieved through genomic targeting by directly sequencing PCR products. The identification of homozygous mutant plants was conducted using PARMS (Penta-primer Amplification Refractory Mutation System) genotyping technology, and these plants were subsequently cultivated for bioassay purposes (Lv et al., 2019).
To construct plasmids for the overexpression of OsWRKY71, a 1047 bp coding sequence (CDS) fragment of OsWRKY71 (accession no. AY676927) was amplified from Bph15-NIL (Supplementary Table S1). The PCR product was subsequently introduced into the binary vector pCXUN (accession no. FJ905215). Using an Agrobacterium-mediated technique, the resulting structure was transformed into Nipponbare and Bph15-NIL plants. The T2 homozygous plants were grown for the following bioassay.
BPH resistance evaluation
A progeny test was conducted to obtain the rice resistance scores. The seedling stage of rice varieties was evaluated for resistance against BPH, with each cultivar or line being replicated a minimum of three times. Eighteen seeds were collected from individual plants and planted in plastic cups, or fifty seeds were randomly planted in plastic boxes. At the three-leaf stage, eight nymphs of BPH (second- or third-instar) were introduced to infest the seedlings. The plant condition was assessed and assigned a resistance score ranging from 0 to 9, with 9 indicating the highest susceptibility and 0 indicating the highest resistance level, as explained in previous research (Huang et al., 2001).
BPH weight gain assays
The assessment of BPH insect performance on rice encompassed the utilization of assays to quantify BPH weight gain and weight gain rate. BPH was introduced to seedlings at the four-leaf stage after the planting of each line’s seeds in a plastic cup with a diameter of 10 cm. Newly emerged female BPH insects were weighed and enclosed in parafilm sachets, which were then affixed to the leaf sheath of a seedling at the four-leaf stage. The BPH instars were allowed to feed on the rice crops for 48 hours. After this feeding period, each insect was removed from the sachet and reweighed. The BPH weight gain was determined by measuring the difference between the initial and final weights of the BPH insect. Furthermore, we computed the weight gain rate, which is determined by dividing the weight gained by the insect’s initial weight before feeding. For each cultivar or line, a minimum of three replicates, each containing 10 insects, were utilized.
RNA-seq and data analysis
Stems of three-leaf stage Bph15-NIL and wrky71 mutant wrky71-5-9 were harvested for 0 (uninfested control), 3, and 24 HAI (hours after BPH infestation). All treatments were terminated at the same time, as previously described (Lv et al., 2014). The Bph15-NIL samples were designated as R0, R3, and R24, while the wrky71-5-9 samples were labeled as S0, S3, and S24. The RNA-seq analysis was conducted using an Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd.).
Using the Illumina paired-end RNA-seq technique, transcriptome sequencing was performed, resulting in a total of 305 million paired-end reads with a length of 2×150 bp. Following the application of Cutadapt, the reads underwent filtration and purification, resulting in 42 Gbp of reads with high quality. Subsequently, the cleaned reads were aligned to the rice reference genome using HISAT2. The estimation of transcript expression levels was conducted using the StringTie and Ballgown software, and the expression abundance was determined by calculating the FPKM values. The DESeq2 software was employed for performing differential expression analysis between distinct groups. Genes displaying a p-value below 0.05 and an absolute fold change exceeding 2 were categorized as differentially expressed genes (DGEs). Furthermore, the DGEs underwent subsequent analysis to ascertain the enrichment of functions associated with Gene Ontology (GO) and KEGG pathways. The bioinformatics analysis was carried out utilizing the OmicStudio tools accessible at https://www.omicstudio.cn/tool.
Quantitative RT-PCR
The stems of three-leaf stage Bph15-NIL, wrky71-5-9, and TN1 were collected after BPH infestation for 0, 3, 6, 12, 24, and 48 HAI. All treatments were terminated simultaneously. The tissue expression profile samples included different rice tissues: stem and leaf of 10 days seedling; stem, sheath, and leaf at the heading stage; stem, sheath, and leaf at 7 days after pollination. The TRIzol reagent (Invitrogen) was employed for the extraction of total RNA. Subsequently, the PrimeScript RT Reagent Kit (Takara) was utilized to perform reverse transcription of 1 μg of total RNA from each sample into cDNA. The qRT-PCR analysis was performed using the QuantStudio™ 5 (Applied Biosystems) and gene-specific primers (Supplementary Table S1) in conjunction with the TB Green (TaKaRa) in the PCR system. The internal control TBP (LOC_Os03g45410) was utilized to standardize the results, and the gene expression levels were determined using the 2-ΔΔCt method. Three biological replicates were conducted for each gene in the qRT-PCR analysis.
Subcellular localization analysis
The coding sequences of OsWRKY71 were amplified through PCR utilizing the primers OsWRKY71-YFP (Supplementary Table S1). Subsequently, the sequences were cloned downstream of a ubiquitin promoter, in frame with YFP, within the vector pCAMBIA1300. The resulting construct was designated as OsWRKY71-YFP. To serve as a nucleus marker, a bZIP63-CFP expressing construct was developed and cloned into the pGWB17 vector. The expression constructs were simultaneously introduced into rice protoplasts. The fluorescence was detected using a confocal laser-scanning microscope (FV1000, Olympus).
Statistical analyses
The data were subjected to statistical analysis using a one-way ANOVA in either Microsoft Excel or SPSS software (version 20).
Results
OsWRKY71 is early induced by BPH in Bph15-NIL
In our previous study, it was observed that the expression of OsWRKY71 increased in response to BPH feeding in the Bph15 introgression line (Lv et al., 2014). To further investigate the role of OsWRKY71 in the response to BPH, we conducted an analysis of its expression in both the Bph15-NIL and TN1 rice plants at various time points following BPH infestation (0, 3, 6, 12, 24, and 48 HAI). The expression level of OsWRKY71 in the Bph15-NIL was significantly higher than in TN1 at 0, 3, and 6 HAI, but this expression pattern was reversed at 12 and 24 HAI (Figure 1A). This suggests that the expression profile of OsWRKY71 in Bph15-NIL differs from that of TN1. Specifically, OsWRKY71 is early induced by BPH in Bph15-NIL. Tissue expression analysis demonstrated that the expression level of OsWRKY71 is higher in the leaf and sheath tissues at the heading stage compared to other tissues (Figure 1B). Therefore, OsWRKY71 may play a role in Bph15-mediated BPH resistance.
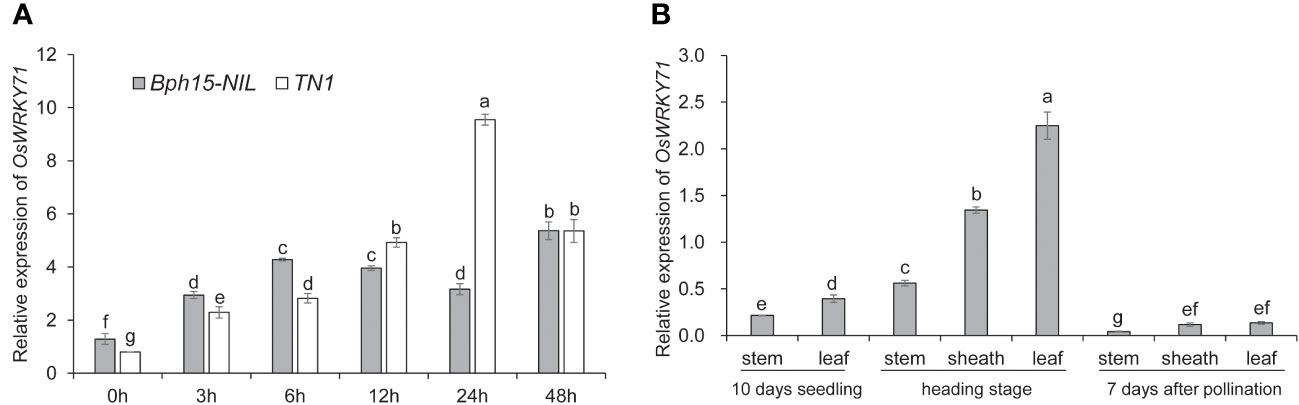
Figure 1 The expression profile of OsWRKY71. (A) Time-dependent expression of OsWRKY71 in the resistant plant Bph15-NIL and susceptible plant TN1 after BPH infestation. (B) Tissue expression profile of OsWRKY71 in various organs. Data represent the means ± SD. The average is determined by calculating the mean of three biological repeats. Significant differences (p < 0.05) are indicated by varying letters above the bars, as determined by a one-way ANOVA.
OsWRKY71 is localized in the nucleus
Inconsistent subcellular localization of OsWRKY71 has been reported in previous studies (Liu et al., 2007; Kim et al., 2016; Ji et al., 2021). The present study reveals a significant colocalization between OsWRKY71 and the nucleus marker bZIP63 in rice protoplasts, thereby providing substantial evidence supporting the nuclear localization of OsWRKY71 (Figure 2). This localization pattern is consistent with the well-established role of transcription factors, which primarily function within the nucleus to regulate gene expression.
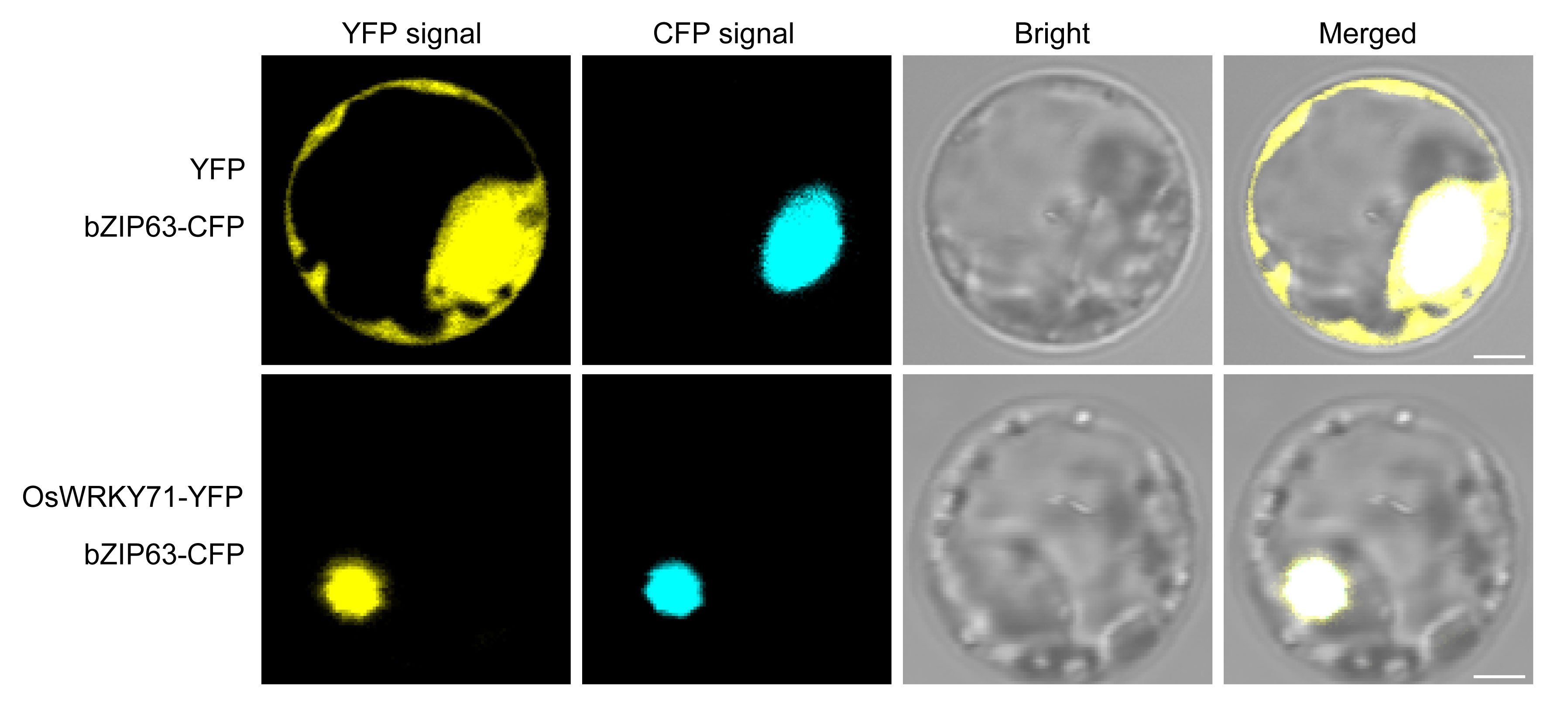
Figure 2 OsWRKY71 subcellular localization. The OsWRKY71-YFP and nucleus marker bZIP63-CFP were co-expressed in rice protoplasts. Scale bars, 2.5 μ m.
Knockout of OsWRKY71 impairs the Bph15-mediated resistance against BPH
To further understand the role of OsWRKY71 in Bph15-mediated resistance, we used CRISPR-Cas9 technology to knockout OsWRKY71 in the Bph15-NIL background. Consequently, we achieved the creation of four distinct homozygous mutant plants, denoted as wrky71-5-9, wrky71-2-1, wrky71-3-3, and wrky71-6-2, within the T2 generation. Subsequent DNA sequencing analysis unveiled that each mutant exhibited distinct genetic alterations within the targeted region of the OsWRKY71 gene. It was observed that wrky71-5-9 had an insertion of A, wrky71-2-1 had a deletion of 4 base pairs, wrky71-3-3 had a deletion of 5 base pairs, and wrky71-6-2 had a deletion of 13 base pairs (Figure 3A). The BPH resistance scores of the wrky71 mutants were evaluated, revealing that wrky71-5-9, wrky71-2-1, wrky71-3-3, and wrky71-6-2 plants were susceptible. Conversely, the Bph15-NIL plants, which possessed intact OsWRKY71, exhibited resistance and survived the BPH attack (Figure 3B). The mean resistance score for Bph15-NIL plants was determined to be 3.27, indicating their resistance to BPH. Conversely, the wrky71 mutants exhibited significantly higher resistance scores, ranging from 7.42 to 8.51, thereby suggesting their susceptibility to BPH (Figure 3C). Additionally, BPH insects that consumed the wrky71 mutants displayed a greater increase in weight compared to those that consumed Bph15-NIL plants (Figures 3D, E). These findings provide evidence that the knockout of OsWRKY71 impairs the resistance of Bph15-NIL plants to BPH, thereby emphasizing the crucial role of OsWRKY71 in Bph15-mediated resistance.
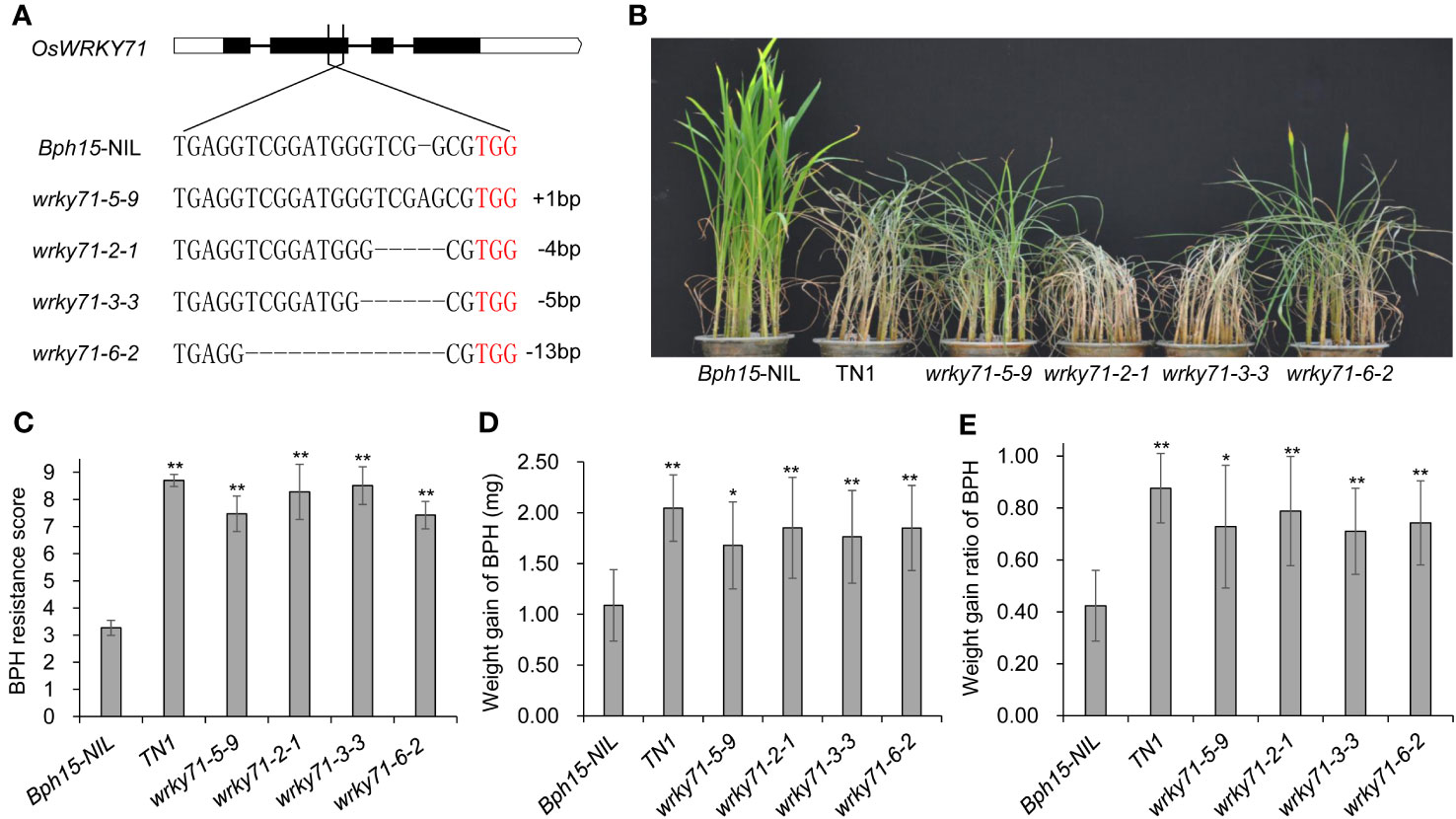
Figure 3 BPH resistance test of the OsWRKY71 knockout plants. (A) Four representative homozygous wrky71 mutants in the Bph15-NIL background. TGG (red) is the PAM sequence. (B) BPH resistance evaluation of wrky71 mutants. (C) BPH average resistance scores of wrky71 mutants. Data represent the means ± SD from three separate experiments, each consisting of 15 seedlings per rice line. Significant differences in comparison with Bph15-NIL are indicated by asterisks above the bars (**p < 0.01 by one-way ANOVA). (D, E) BPH weight gain and weight gain rate assays of wrky71 mutants. Data represent the means ± SD from three separate experiments, each consisting of 10 BPH insects per replicate. Significant differences in comparison with Bph15-NIL are indicated by asterisks above the bars (**p < 0.01, *p < 0.05 by one-way ANOVA).
We also generated overexpression lines of OsWRKY71 in both the Nipponbare and Bph15-NIL backgrounds. However, the overexpression of OsWRKY71 in both backgrounds did not result in significant differences in BPH resistance compared to the control plants. Specifically, the Bph15-NIL-OE plants, which overexpressed OsWRKY71 in the Bph15-NIL background, exhibited BPH resistance scores similar to those of the Bph15-NIL control plants. This suggests that the overexpression of OsWRKY71 did not enhance the resistance provided by Bph15 (Supplementary Figure S1A). In a similar vein, the NIP-OE plants, characterized by the overexpression of OsWRKY71 in the Nipponbare background, exhibited susceptibility to BPH and were killed by BPH at the same time as the susceptible Nipponbare plants (Supplementary Figure S1B). These observations imply that increasing the expression of the OsWRKY71 gene does not exert any discernible influence on BPH resistance, irrespective of the presence or absence of Bph15. Consequently, it can be deduced that OsWRKY71 does not function as a positive regulator of basal resistance in the absence of Bph15.
The transcript profiles that respond to BPH are different in the wrky71 mutant and Bph15-NIL
To investigate the potential mechanism underlying the impact of the wrky71 mutant on Bph15-mediated resistance to BPH, we conducted an analysis of the transcriptomes of wrky71-5-9 and Bph15-NIL using RNA-seq. The RNA-seq analysis identified a total of 1387 differentially expressed genes (DEGs) among seven comparisons: R0_S0, R3_S3, R24_S24, R0_R3, R0_R24, S0_S3, and S0_S24 (Supplementary Table S2). Significant disparities in gene expression were observed between the wrky71 mutant and Bph15-NIL. Specifically, in the comparisons of R0_S0, R3_S3, and R24_S24, the wrky71 mutant displayed significant alterations in 390, 208, and 153 genes, respectively, when compared to Bph15-NIL. Moreover, the wrky71 mutant displayed 248 and 339 DEGs in the S0_S3 and S0_S24 comparisons, respectively. Conversely, in Bph15-NIL, there were 607 and 529 DEGs in the R0_R3 and R0_R24 comparisons, respectively. Notably, both the R and S comparisons exhibited a higher abundance of up-regulated genes compared to down-regulated genes (Figure 4A). Subsequent analysis revealed significant disparities in the majority of the up-regulated and down-regulated genes between the wrky71 mutant and Bph15-NIL (Figure 4B). Additionally, hierarchical cluster analysis of 1387 DEGs further confirmed substantial distinctions between the R and S comparisons (Supplementary Figure S2).
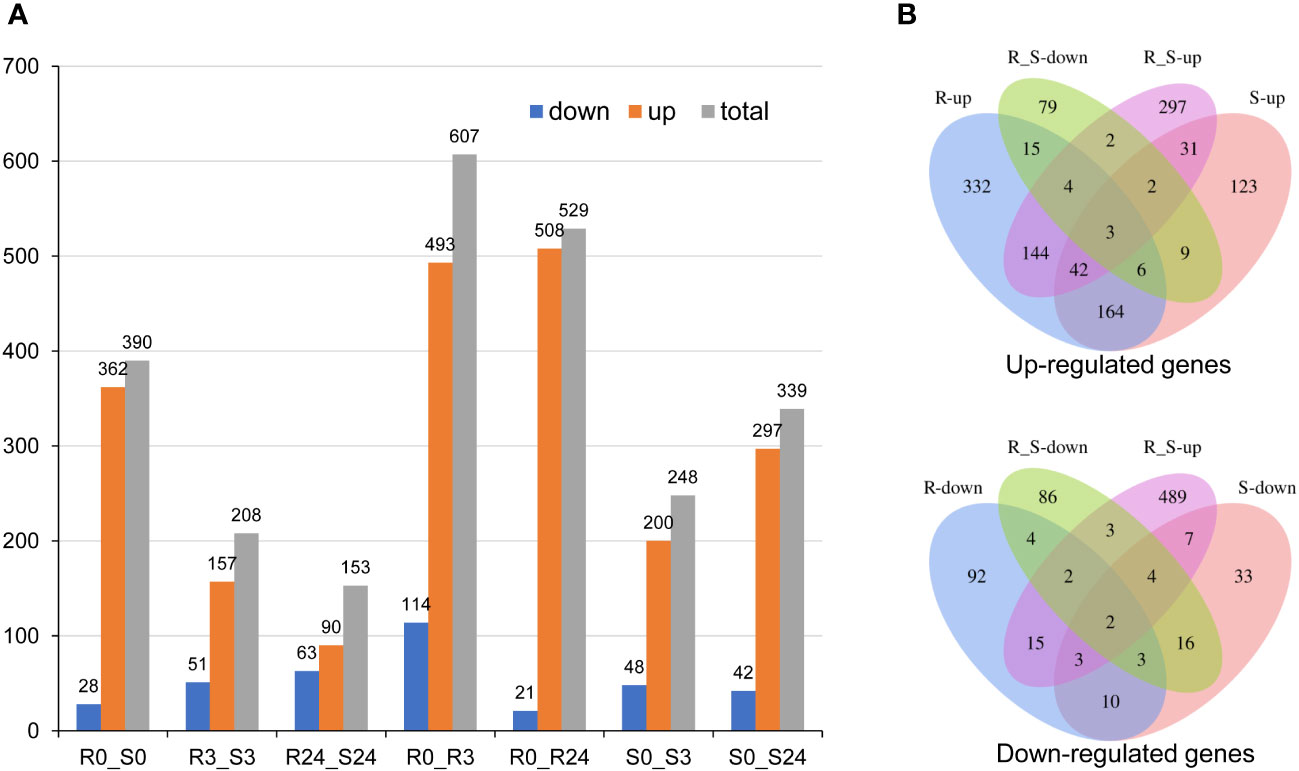
Figure 4 DEGs of two rice varieties (Bph15-NIL and wrky71-5-9). (A) The DEGs in all comparisons. (B) Venn diagrams of the up-regulated and down-regulated DEGs.
The enriched Gene Ontology (GO) terms identified in this study suggest that the response to BPH feeding in both the wrky71 mutant and Bph15-NIL rice involves multiple key biological processes. Specifically, the common GO terms observed in the up-regulated DEGs (Supplementary Figure S3; Table S3_1) in the S0_S3, S0_S24, R0_R3, and R0_R24 comparisons include wounding response, chitin response, fungus defense, and salicylic acid (SA) response. These shared GO terms indicate that both the wrky71 mutant and Bph15-NIL rice employ similar defense mechanisms in response to BPH feeding. Significant disparities in enriched GO terms were observed when comparing gene expression between the wrky71 mutant and Bph15-NIL rice. Specifically, the R0_R3 and R0_R24 comparisons revealed enriched GO terms related to water deprivation response, cell wall, autophagy, and trehalose phosphatase. Conversely, the S0_S3 and S0_S24 comparisons exhibited specifically enriched GO terms associated with ethylene (ET) response, jasmonic acid (JA) response, gibberellin metabolic, bacterium defense, virus defense, and herbivore response (Figure 5; Supplementary Tables S3_2-S3_5). The enriched GO term “sesquiterpene biosynthetic” in both the R0_R24 and S0_S3 comparisons suggests that this biological process may play a crucial role in conferring resistance to BPH in both the wrky71 mutant and Bph15-NIL rice (Figure 5). These results emphasize the potential contribution of diverse biological processes and signaling pathways in the resistance mechanisms of the wrky71 mutant and Bph15-NIL rice, providing valuable insights for future investigations on BPH resistance in rice.
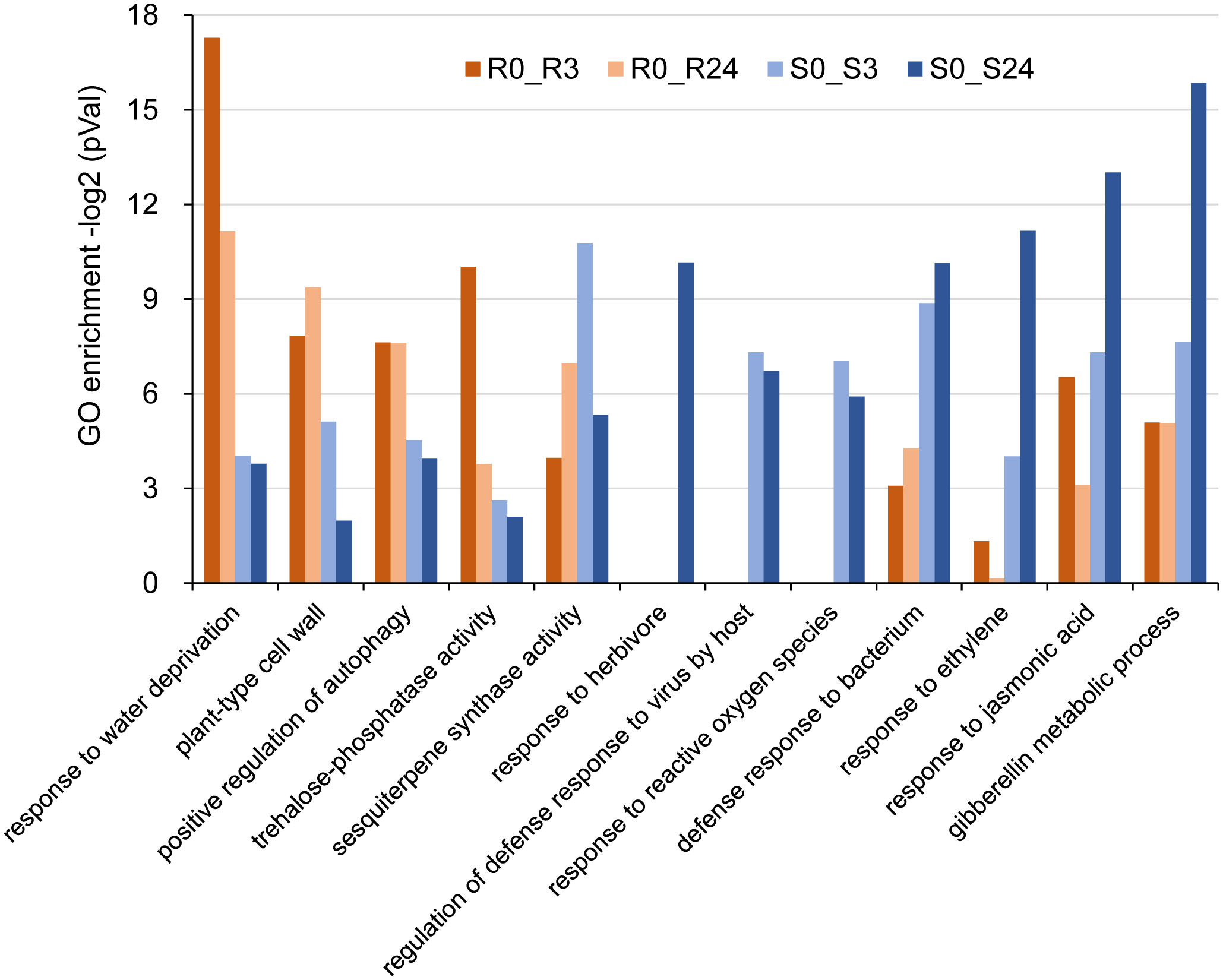
Figure 5 Representative GO terms of up-regulated DEGs in R0_R3, R0_R24, S0_S3, and S0_S24 comparisons. vertical coordinate: -log2(P-value). -log2(P-value) > 6.64 = p < 0.01, -log2(P-value) > 4.32 = p < 0.05.
The KEGG pathway enrichment analysis demonstrated significant enrichment in multiple pathways within the transcript profiles of both the wrky71 mutant and Bph15-NIL plants upon exposure to BPH infestation. Notably, these pathways encompassed plant-pathogen interaction, plant hormone signal, autophagy, et al. (Supplementary Figure S4; Table S4_1). Intriguingly, the R0_R3 and R0_R24 comparisons exhibited a higher number of genes involved in these pathways compared to the S0_S3 and S0_S24 comparisons (Supplementary Figure S5; Tables S4_2-S4_5). This observation implies the existence of discrete regulatory mechanisms functioning in the two genotypes in response to BPH infestation. Overall, these findings provide insights into the molecular mechanisms of BPH resistance in rice and underscore the importance of OsWRKY71 in governing gene expression during BPH infestation.
Knockout of OsWRKY71 alters the expression of plant defense genes
The study revealed a significant down-regulation of the sesquiterpene synthase gene OsSTPS2 in the R3_S3 and R24_S24 comparisons (Figure 6). Subsequent analysis using qRT-PCR demonstrated a notable decrease in expression levels of OsSTPS2 in both the wrky71 mutant and TN1 compared to Bph15-NIL at all five time points (Figure 7). Terpenes, a class of secondary metabolites, possess diverse functions, including defense against herbivores. Previous research has demonstrated the differential expression of OsSTPS2 in rice cultivars RH (carrying Bph3) and KD (sensitive to BPH), and its involvement in BPH antixenosis resistance (Kamolsukyunyong et al., 2013; Kamolsukyeunyong et al., 2021). This suggests that OsSTPS2 may participate in BPH resistance conferred by OsWRKY71 in Bph15-NIL plants.
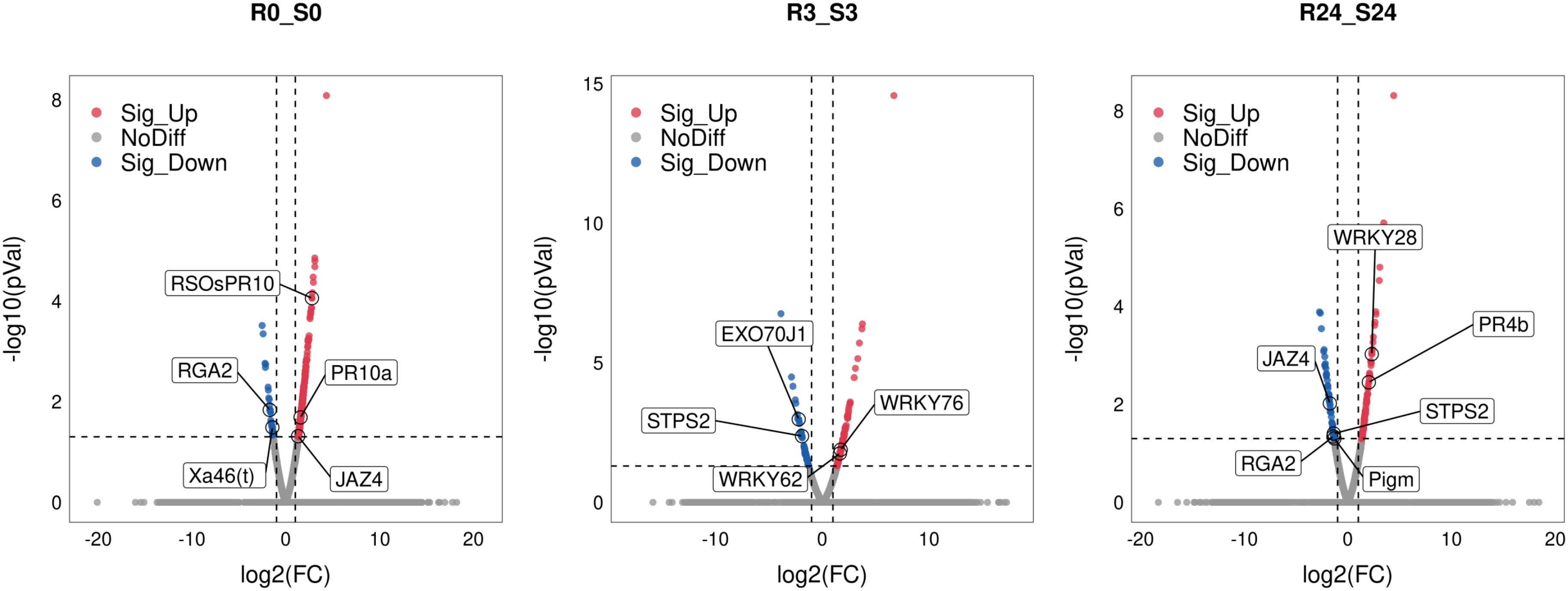
Figure 6 Volcano plots of R0_S0, R3_S3, and R24_S24 comparisons. The representative DEGs were marked as boxes.
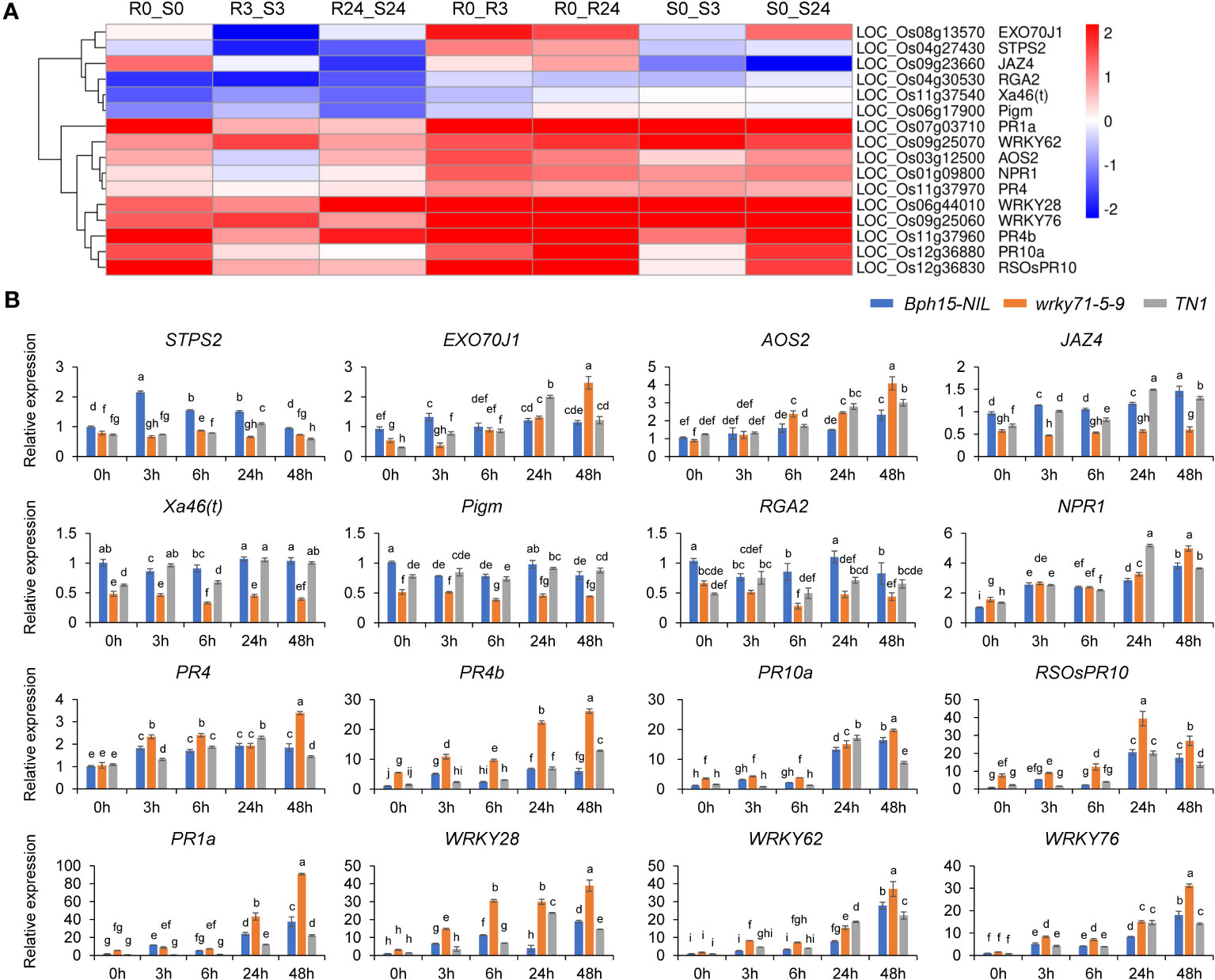
Figure 7 Heat map and qRT-PCR analysis of representative DGEs. (A) Heat map of representative DGEs studied in this work. (B) qRT-PCR analysis of representative DGEs. Data represent the means ± SD. Significant differences (p < 0.05) are indicated by varying letters above the bars, as determined by a one-way ANOVA.
The excretion of proteins associated with resistance and the plant’s resistance to BPH are influenced by the trafficking pathway that relies on the presence of EXO70. Specifically, the functions of OsEXO70E1 and OsEXO70H3 are involved in Bph6-mediated BPH resistance (Guo et al., 2018; Wu et al., 2022). Our investigation led to the discovery of the OsEXO70J1 gene (LOC_Os08g13570), a member of the EXO70 family, which exhibited significant up-regulation in the R0_R3 and R0_R24 comparisons, but down-regulation in the R3_S3 comparison (Figure 6; Supplementary Figure S6). Additionally, the results of qRT-PCR analysis showed that the expression levels of OsEXO70J1 were reduced in both the wrky71 mutant and TN1 cultivars compared to Bph15-NIL at the onset of BPH infestation (Figure 7). This suggests that OsEXO70J1 may participate in BPH resistance conferred by OsWRKY71 in Bph15-NIL plants.
Interestingly, the down-regulation of three R genes, namely Xa46(t) (LOC_Os11g37540), Pigm (LOC_Os06g17900), and RGA2 (LOC_Os04g30530), was observed in the R_S comparisons (Figure 6). It was found that BPH feeding did not induce the up-regulation of these genes in any of the rice lines. However, the expression levels of these genes were significantly reduced in the wrky71 mutant compared to Bph15-NIL. Additionally, the expression level of RGA2 was also decreased in TN1 when compared to Bph15-NIL plants (Figure 7). These findings suggest that OsWRKY71 may be involved in controlling the expression of these three R genes and contribute to disease resistance in rice. It is also suggested that the R gene RGA2 may have a dual function in both disease and BPH resistance.
The influence of the SA and JA pathways on plant responses to insects is of significant importance. However, our previous study suggested that the SA signaling pathway may not be triggered in Bph15-mediated resistance (Lv et al., 2014). In this current investigation, we focused on the JA pathway and found that the expression levels of the AOS2 gene, which is involved in JA synthesis, were increased in both the wrky71 mutant and TN1 compared to Bph15-NIL. Furthermore, the transcript levels of the JA repressor JAZ4 were lower in the wrky71 mutant compared to Bph15-NIL and TN1 (Figure 7). The results indicate that OsWRKY71 may inhibit the JA pathway in Bph15-NIL plants. Hence, the Bph15-mediated BPH resistance may not be associated with the SA and JA signaling pathways.
The present study observed an increase in the expression of NPR1 and five PR genes (PR1a, PR4, PR4b, PR10a, and RSOsPR10) in all three rice lines. However, the expression levels of these genes were higher in the wrky71 mutant compared to Bph15-NIL plants (Figure 7). This suggests that these genes may be involved in a common defense mechanism in both resistant and susceptible rice varieties. These results are consistent with our previous study (Lv et al., 2014). It was observed that the transcript levels of OsWRKY28, OsWRKY62, and OsWRKY76, members of the OsWRKY IIa subfamily, exhibited a significant increase in all three rice lines. However, in the wrky71 mutant, their expression levels were even higher compared to Bph15-NIL plants (Figure 7). This suggests that in the absence of Bph15, these genes may act as promoters of BPH resistance, but in the presence of Bph15, they could exert an inhibitory effect. These findings are consistent with the previous studies conducted by Chujo et al. (2013) and Shi et al. (2023).
Discussion
BPH resistance genes Bph15 and Bph3 may act as pattern recognition receptors (PRRs) in the activation of PTI for plant immunity (Du et al., 2020; Chen et al., 2022). The present study has revealed the significant contribution of OsWRKY71 in the Bph15-mediated resistance against BPH. The knockout of OsWRKY71 has been observed to impair the resistance to BPH in Bph15-NIL plants, as depicted in Figure 3. These findings underscore the role of OsWRKY71 in the underlying molecular mechanisms of Bph15-mediated resistance against BPH. In our previous studies, we successfully cloned the first BPH resistance gene Bph14, and observed the interaction between the BPH14 protein and OsWRKY46 and OsWRKY72 (Du et al., 2009; Hu et al., 2017). Further research about how OsWRKY71 is induced by BPH15 and BPH infestation will enhance our comprehension of the signaling pathways implicated in rice defense against BPH.
The study by Liu et al. (2007) and Ji et al. (2021) reported that OsWRKY71 enhances resistance to bacterial blight and SBPH in the Nipponbare variety of rice. However, our study found that overexpression of OsWRKY71 did not increase resistance to BPH in both the Nipponbare and Bph15-NIL backgrounds. This suggests that OsWRKY71 may have varying effects on resistance to BPH and other pathogens or piercing-sucking insects. Similar contrasting roles have been observed for another gene, OsWRKY45, in rice pathogens and BPH resistance (Huangfu et al., 2016).
Interestingly, the OsWRKY71 gene displayed an early induced expression pattern in Bph15-NIL, while its level of gene expression was comparatively lower than that of the TN1 at 12 and 24 HAI (Figure 1A). However, little is known about the underlying mechanism, and more research needs to be done. The induced expression pattern in TN1 suggests that OsWRKY71 may potentially contribute to a basal defense in susceptible rice varieties. However, it is important to note that the NIP-OE plants, which overexpress OsWRKY71, exhibited susceptibility to BPH infestation (Supplementary Figure S1B). This indicates that OsWRKY71 does not function as a promoter of basal resistance in the absence of Bph15. Previous studies have demonstrated that OsWRKY62 can play opposite roles depending on the existence or nonexistence of the blast-resistance gene Pi9 (Shi et al., 2023). Similarly, it is plausible that OsWRKY71, akin to OsWRKY62, possesses a dual capacity in governing the basal resistance against BPH in rice. Nevertheless, further research is required to ascertain whether OsWRKY71 acts as a negative modulator of BPH basal defense.
Previous research has provided evidence for the involvement of the plant sesquiterpene synthase OsSTPS2 in an antixenosis mechanism of BPH resistance mediated by Bph3 (RH variety) (Kamolsukyunyong et al., 2013; Kamolsukyeunyong et al., 2021). In this study, we observed that the expression of OsSTPS2 was suppressed in the wrky71 mutant within the Bph15-NIL genetic background (Figure 7). This finding suggests that OsSTPS2 may play a part in the antixenosis process of BPH resistance regulated by Bph15, and its regulation is mediated by the Bph15-OsWRKY71 pathway. Our future investigations aim to identify the specific plant volatile compounds released by the wrky71 mutant and Bph15-NIL following BPH infestation. Furthermore, our previous research has demonstrated the involvement of OsEXO70E1 and OsEXO70H3 in the Bph6-mediated resistance against planthoppers, as they facilitate exocytosis and enhance the strength of the cell wall (Guo et al., 2018; Wu et al., 2022). Notably, the analysis revealed a significant enrichment of the GO term “plant-type cell wall” in the R comparison (Figure 5). Moreover, the expression of OsEXO70J1 was found to be downregulated in both the wrky71 mutant and TN1 varieties, in contrast to Bph15-NIL, during the initial infestation of BPH (Figure 7). This finding suggests that OsEXO70J1 may contribute to the resistance against BPH conferred by OsWRKY71 in Bph15-NIL plants. Interestingly, the study found that the disease-resistance gene RGA2 may also participate in BPH resistance conferred by OsWRKY71 in Bph15-NIL plants. This implies that the plant immune system, traditionally recognized for its role in protecting against pathogens, may also be involved in plant-insect interactions. There may be shared downstream signaling mechanisms within the plant immune system that are relevant to these diverse types of interactions.
The SA and JA plant hormone signals are essential for defending rice against insects. However, the effectiveness of SA and JA in regulating rice defense against BPH is dependent on the specific genotype of the rice. The SA pathway is implicated in BPH resistance mediated by Bph6, Bph9, and Bph14 (Du et al., 2009; Zhao et al., 2016; Guo et al., 2018). Moreover, the synergistic impact of SA and JA has been observed to enhance BPH resistance in Bph6 plants, as reported by Guo et al. (2018). However, it has been discovered that Bph15-mediated BPH immunity is not influenced by the SA and JA pathways. This suggests the existence of additional mechanisms contributing to Bph15-mediated resistance. Additionally, NPR1, PR1a, PR4, PR4b, PR10a, and RSOsPR10 are involved in a shared fundamental protective mechanism in both susceptible and resistant varieties of rice, except for Bph15-mediated BPH resistance.
Based on the findings of the study, it was observed that BPH insects that fed on the wrky71 mutants exhibited a slight reduction in weight in comparison to those that fed on TN1 plants (Figures 3C–E). This implies that the wrky71 mutants possess little resistance against BPH. Thus, it can be inferred that there exist other pathways implicated in Bph15-mediated BPH resistance signaling, and OsWRKY71 does not exclusively regulate this resistance. The Bph15-NIL harbors two BPH resistance genes (Bph3 and Bph15) (Lv et al., 2014; Xiao et al., 2016), thereby indicating the presence of multiple mechanisms contributing to BPH resistance. Further research should be undertaken to clone and characterize the resistance gene located in the recombination cold-spot region (Lv et al., 2014) to gain a more comprehensive understanding of their molecular mechanism against BPH. One potential approach involves the reconstruction of the genetic population capable of producing crossover in the recombination cold-spot region. This necessitates the identification of susceptible parents exhibiting a greater degree of chromosomal homology within the specified interval. Additionally, the deficiency in the Bph15 physical map (Lv et al., 2014) could be addressed through the utilization of third-generation PacBio long-read sequencing to achieve completion.
In summary, the most significant finding to emerge from this study is that OsWRKY71 plays a critical role in the Bph15-mediated BPH resistance in rice. This investigation significantly enhances our comprehension of the molecular mechanisms underlying Bph15-mediated BPH resistance. Further investigations should be conducted to successfully clone and identify all resistance genes in the Bph15 locus.
Data availability statement
The raw sequence files were deposited in the GSA database of the National Genomics Data Center (https://ngdc.cncb.ac.cn) with the accession number CRA011634 (https://ngdc.cncb.ac.cn/gsa/browse/CRA011634).
Author contributions
XL: Investigation, Writing – original draft, Writing – review & editing. JZ: Investigation, Writing – original draft, Writing – review & editing. XS: Investigation, Writing – review & editing. JY: Investigation, Writing – review & editing. LZ: Investigation, Writing – review & editing. JH: Investigation, Writing – review & editing. BD: Investigation, Writing – review & editing. WL: Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 31601650 and 32001921), the Young Talents Training Program of Shandong Academy of Agricultural Sciences, the Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (Grant No. CXGC2018E16), the Open Research Fund of the State Key Laboratory of Hybrid Rice (Wuhan University) (Grant No. KF201902), the Agricultural Variety Improvement Project of Shandong Province (Grant No. 2022LZGC024), and the Key Research and Development Plan of Shandong Province (Grant No. 2022TZXD0040).
Acknowledgments
We thank Prof. Guangcun He (Wuhan University) for providing the Bph15-NIL. We also thank Prof. Xianzhi Xie (Shandong Academy of Agricultural Sciences) and Dr. Jianping Guo (Wuhan University) for providing helpful suggestions in this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1260526/full#supplementary-material
Supplementary Figure 1 | BPH-resistance scores of the OsWRKY71 overexpression lines in the Bph15-NIL and Nipponbare backgrounds. (A) BPH-resistance scores of the OsWRKY71 overexpression lines in the Bph15-NIL backgrounds. Bph15NIL-OE1, -OE2, and -OE3 are homozygous T2 progeny from independent transformants. (B) BPH-resistance scores of the OsWRKY71 overexpression lines in the Nipponbare backgrounds. NIP-OE1, -OE2, and -OE3 are homozygous T2 progeny from independent transformants. Data represent the means ± SD from three separate experiments, each consisting of 15 seedlings per rice line. Significant differences (p < 0.05) are indicated by varying letters above the bars, as determined by a one-way ANOVA.
Supplementary Figure 2 | Hierarchical cluster analysis of 1387 DEGs based on the log2(FC).
Supplementary Figure 3 | GO enrichment analysis of 1387 DEGs.
Supplementary Figure 4 | KEGG pathway enrichment analysis of 1387 DEGs.
Supplementary Figure 5 | Representative KEGG pathway numbers of R0_R3, R0_R24, S0_S3, and S0_S24 comparisons.
Supplementary Figure 6 | Volcano plots of R0_R3, R0_R24, S0_S3, and S0_S24 comparisons. Horizontal coordinate: log2(fold change), vertical coordinate: -log10(P-value). The representative DEGs were marked as boxes.
References
Chen, R., Deng, Y., Ding, Y., Guo, J., Qiu, J., Wang, B., et al. (2022). Rice functional genomics: decades’ efforts and roads ahead. Sci. China Life Sci. 65, 33–92. doi: 10.1007/s11427-021-2024-0
Cheng, X., Wu, Y., Guo, J., Du, B., Chen, R., Zhu, L., et al. (2013a). A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 76, 687–698. doi: 10.1111/tpj.12328
Cheng, X., Zhu, L., He, G. (2013b). Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant 6, 621–634. doi: 10.1093/mp/sst030
Chujo, T., Kato, T., Yamada, K., Takai, R., Akimoto-Tomiyama, C., Minami, E., et al. (2008). Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci. Biotechol. Biochem. 72, 240–245. doi: 10.1271/bbb.70553
Chujo, T., Miyamoto, K., Shimogawa, T., Shimizu, T., Otake, Y., Yokotani, N., et al. (2013). OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. doi: 10.1007/s11103-013-0032-5
Chujo, T., Takai, R., Akimoto-Tomiyama, C., Ando, S., Minami, E., Nagamura, Y., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta (BBA) - Gene Structure Expression 1769, 497–505. doi: 10.1016/j.bbaexp.2007.04.006
Du, B., Chen, R., Guo, J., He, G. (2020). Current understanding of the genomic, genetic, and molecular control of insect resistance in rice. Mol. Breed. 40, 24. doi: 10.1007/s11032-020-1103-3
Du, B., Zhang, W., Liu, B., Hu, J., Wei, Z., Shi, Z., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168. doi: 10.1073/pnas.0912139106
Eulgem, T., Somssich, I. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Guo, J., Wang, H., Guan, W., Guo, Q., Wang, J., Yang, J., et al. (2023). A tripartite rheostat controls self-regulated host plant resistance to insects. Nature 618, 799–807. doi: 10.1038/s41586-023-06197-z
Guo, J., Xu, C., Wu, D., Zhao, Y., Qiu, Y., Wang, X., et al. (2018). Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 50, 297–306. doi: 10.1038/s41588-018-0039-6
Hu, L., Wu, Y., Wu, D., Rao, W., Guo, J., Ma, Y., et al. (2017). The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell 29, 3157–3185. doi: 10.1105/tpc.17.00263
Hu, L., Ye, M., Li, R., Lou, Y. (2016). OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant Signal Behav. 11, e1169357. doi: 10.1080/15592324.2016.1169357
Hu, L., Ye, M., Li, R., Zhang, T., Zhou, G., Wang, Q., et al. (2015). The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol. 169, 2907–2921. doi: 10.1104/pp.15.01090
Huang, Z., He, G., Shu, L., Li, X., Zhang, Q. (2001). Identification and mapping of two brown planthopper resistance genes in rice. Theor. Appl. Genet. 102, 929–934. doi: 10.1007/s001220000455
Huangfu, J., Li, J., Li, R., Ye, M., Kuai, P., Zhang, T., et al. (2016). The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 17, 697. doi: 10.3390/ijms17060697
Inoue, H., Hayashi, N., Matsushita, A., Xinqiong, L., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 9577–9582. doi: 10.1073/pnas.1222155110
Ji, R., Fu, J., Shi, Y., Li, J., Jing, M., Wang, L., et al. (2021). Vitellogenin from planthopper oral secretion acts as a novel effector to impair plant defenses. New Phytol. 232, 802–817. doi: 10.1111/nph.17620
Kamolsukyeunyong, W., Sukhaket, W., Pitija, K., Thorngkham, P., Mahatheeranont, S., Toojinda, T., et al. (2021). Rice sesquiterpene plays important roles in antixenosis against brown planthopper in rice. Plants 10, 1049. doi: 10.3390/plants10061049
Kamolsukyunyong, W., Sukhaket, W., Ruanjaichon, V., Toojinda, T., Vanavichit, A. (2013). Single-feature polymorphism mapping of isogenic rice lines identifies the influence of terpene synthase on brown planthopper feeding preferences. Rice 6, 18. doi: 10.1186/1939-8433-6-18
Kim, C., Vo, K., Nguyen, C., Jeong, D., Lee, S., Kumar, M., et al. (2016). Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 10, 13–23. doi: 10.1007/s11816-015-0383-2
Li, R., Zhang, J., Li, J., Zhou, G., Wang, Q., Bian, W., et al. (2015). Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife 4, e04805. doi: 10.7554/eLife.04805
Liu, X., Bai, X., Wang, X., Chu, C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, Y., Wu, H., Chen, H., Liu, Y., He, J., Kang, H., et al. (2014). A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 33, 301. doi: 10.1038/nbt.3069
Liu, Q., Yang, J., Zhang, S., Zhao, J., Feng, A., Yang, T., et al. (2016). OsGF14b positively regulates panicle blast resistance but negatively regulates leaf blast resistance in rice. Mol. Plant Microbe Interact. 29, 46–56. doi: 10.1094/mpmi-03-15-0047-r
Lv, W., Du, B., Shangguan, X., Zhao, Y., Pan, Y., Zhu, L., et al. (2014). BAC and RNA sequencing reveal the brown planthopper resistance gene BPH15 in a recombination cold spot that mediates a unique defense mechanism. BMC Genomics 15, 674. doi: 10.1186/1471-2164-15-674
Lv, W., Yin, J., Yin, X., Xu, G., Wu, X., Li, X., et al. (2019). Application of PARMS technology in genotyping of rice gene edited progeny. Shandong Agric. Sci. 51, 8–13.
Ma, X., Zhang, Q., Zhu, Q., Liu., W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Peng, Y., Bartley, L., Canlas, P., Ronald, P. (2010). OsWRKY IIa transcription factors modulate rice innate immunity. Rice 3, 36–42. doi: 10.1007/s12284-010-9039-6
Peng, Y., Bartley, L., Chen, X., Dardick, C., Chern, M., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant Microbe Interact. 1, 446–458. doi: 10.1093/mp/ssn024
Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X., Wang, H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. doi: 10.1007/s00425-012-1698-7
Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20, 492–499. doi: 10.1094/mpmi-20-5-0492
Rushton, P., Somssich, I., Ringler, P., Shen, Q. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shi, S., Wang, H., Nie, L., Tan, D., Zhou, C., Zhang, Q., et al. (2021). Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol. Plant 14, 1714–1732. doi: 10.1016/j.molp.2021.07.004
Shi, X., Xiong, Y., Zhang, K., Zhang, Y., Zhang, J., Zhang, L., et al. (2023). The ANIP1-OsWRKY62 module regulates both basal defense and Pi9-mediated immunity against Magnaporthe oryzae in rice. Mol. Plant 16, 739–755. doi: 10.1016/j.molp.2023.03.001
Shimono, M., Sugano, S., Nakayama, A., Jiang, C., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Wang, H., Hao, J., Chen, X., Hao, Z., Wang, X., Lou, Y., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wu, D., Guo, J., Zhang, Q., Shi, S., Guan, W., Zhou, C., et al. (2022). Necessity of rice resistance to planthoppers for OsEXO70H3 regulating SAMSL excretion and lignin deposition in cell walls. New Phytol. 234, 1031–1046. doi: 10.1111/nph.18012
Xiao, C., Hu, J., Ao, Y., Cheng, M., Gao, G., Zhang, Q., et al. (2016). Development and evaluation of near-isogenic lines for brown planthopper resistance in rice cv. 9311. Sci. Rep. 6, 38159. doi: 10.1038/srep38159
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Zhang, Q. (2007). Strategies for developing green super rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Keywords: rice, brown planthopper, Bph15, OsWRKY71, defense mechanism, transcriptome
Citation: Li X, Zhang J, Shangguan X, Yin J, Zhu L, Hu J, Du B and Lv W (2023) Knockout of OsWRKY71 impairs Bph15-mediated resistance against brown planthopper in rice. Front. Plant Sci. 14:1260526. doi: 10.3389/fpls.2023.1260526
Received: 18 July 2023; Accepted: 09 October 2023;
Published: 02 November 2023.
Edited by:
Shengli Jing, Xinyang Normal University, ChinaReviewed by:
Ruifeng He, Agricultural Research Service (USDA), United StatesXianjin Qiu, Yangtze University, China
Hao Zhou, Sichuan Agricultural University, China
Copyright © 2023 Li, Zhang, Shangguan, Yin, Zhu, Hu, Du and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wentang Lv, bHZ3ZW50YW5nQHdodS5lZHUuY24=
†These authors have contributed equally to this work
 Xiaozun Li
Xiaozun Li Jian Zhang
Jian Zhang Xinxin Shangguan2,3
Xinxin Shangguan2,3 Bo Du
Bo Du Wentang Lv
Wentang Lv