Abstract
Introduction:
The TGA transcription factors, plays a crucial role in regulating gene expression. In cultivated peanut (Arachis hypogaea), which faces abiotic stress challenges, understanding the role of TGAs is important.
Methods:
In this study, we conducted a comprehensive in analysis of the TGA gene family in peanut to elucidate their regulatory mechanisms and expression patterns under abiotic stress and hormone treatments. Furthermore, functional studies on the representative AhTGA gene in peanut cultivars were conducted using transgenic Arabidopsis and soybean hair roots.
Results:
The genome-wide analysis revealed that a total of 20 AhTGA genes were identified and classified into five subfamilies. Collinearity analysis revealed that AhTGA genes lack tandem duplication, and their amplification in the cultivated peanut genome primarily relies on the whole-genome duplication of the diploid wild peanut to form tetraploid cultivated peanut, as well as segment duplication between the A and B subgenomes. Promoter and Protein-protein interaction analysis identified a wide range of cis-acting elements and potential interacting proteins associated with growth and development, hormones, and stress responses. Expression patterns of AhTGA genes in different tissues, under abiotic stress conditions for low temperature and drought, and in response to hormonal stimuli revealed that seven AhTGA genes from groups I (AhTGA04, AhTGA14 and AhTGA20) and II (AhTGA07, AhTGA11, AhTGA16 and AhTGA18) are involved in the response to abiotic stress and hormonal stimuli. The hormone treatment results indicate that these AhTGA genes primarily respond to the regulation of jasmonic acid and salicylic acid. Overexpressing AhTGA11 in Arabidopsis enhances resistance to cold and drought stress by increasing antioxidant activities and altering endogenous hormone levels, particularly ABA, SA and JA.
Discussion:
The AhTGA genes plays a crucial role in hormone regulation and stress response during peanut growth and development. The findings provide insights into peanut's abiotic stress tolerance mechanisms and pave the way for future functional studies.
Introduction
Transcription factors (TFs) play a critical role in mediating the response of plants to changes in their external environment (Javed et al., 2020; Hrmova and Hussain, 2021; Song et al., 2022). By binding to specific DNA sequences, transcription factors can activate or repress the expression of genes (Latchman, 2001). They can also interact with other signaling molecules and proteins to modulate the expression of target genes, enabling plants to quickly and effectively respond to environmental challenges such as drought, heat, cold, or pathogen attack (Fraire-Velázquez et al., 2011; Nakashima et al., 2014; Innes, 2018; Srivastava et al., 2023). The ability of transcription factors to integrate diverse signals and coordinate gene expression makes them essential components of the complex regulatory networks that control plant responses to environmental stress (Valliyodan and Nguyen, 2006; Borrill et al., 2019; Kidokoro et al., 2022). Understanding the mechanisms by which transcription factors regulate gene expression in response to environmental cues is crucial for developing strategies to enhance plant resilience and improve agricultural productivity.
The basic leucine zipper (bZIP) gene family, one of the largest transcription factor families in plants, is categorized into ten groups (A, B, C, D, E, F, G, H, I, and S), along with two additional groups, J and K. This classification is based on the similarity in the basic region and additional conserved motifs (Jakoby et al., 2002; Nijhawan et al., 2008; Wang et al., 2015). TGA (TGACG motif-binding factor) transcription factors are part of group D, which recognizes as-1-type cis-elements found in the promoter regions of target genes (Gatz, 2013; Liu et al., 2022b). The bZIP domain’s primary structure in TGA proteins is highly conserved and includes an invariant motif N-x7R/K-x9-L-x6-L-x6-L in the N-terminus. In the C-terminus, the motif Yx2RL[RQ]ALSS[LS]W represents the signature domain of group D (Tomaz et al., 2022). Since the first TGA gene TGA1a was identified in tobacco, this gene family has been isolated and identified in various species, including Arabidopsis, rice and soybean (Johnson et al., 2003; Fitzgerald et al., 2005; Li et al., 2022). In the Arabidopsis genome, there are a total of 10 TGA transcription factors that can be divided into five groups based on their sequence similarity (Gatz, 2013). Group I comprise TGA1 and TGA4, which are the most comparable to tobacco TGA1a (Budimir et al., 2021; Kim et al., 2022). Group II consists of TGA2, TGA5, and TGA6, which are closely related and have functional overlap with Group I (Zhang et al., 2003; Fonseca et al., 2022). Group III includes TGA3 and TGA7, Group IV includes TGA9 and TGA10, while Group V has only one member, PERIANTHIA (PAN) (Murmu et al., 2010; Noshi et al., 2016). Numerous studies have been conducted to understand the role of TGA genes, TGA1-TGA7 have been widely demonstrated to enhance plant resistance to biotic and abiotic stresses, while TGA9, TGA10 and PERIANTHIA (PAN) were proved to be involved in the development of plant floral organs (Murmu et al., 2010; Li et al., 2019b; Budimir et al., 2021; Liu et al., 2022a). The TGA gene participates in the regulation of multiple hormone signaling pathways, including salicylic acid, jasmonic acid, ethylene, and cytokinin, by interacting with key regulatory factors such as NPR1, GRX480, ERF72, SCL14 and BIN2 in plants (Johnson et al., 2003; Fode et al., 2008; Choi et al., 2010; Zander et al., 2010; Jin et al., 2018; Kim et al., 2022).
Cultivated peanuts (Arachis hypogaea), a vital cash crop cultivated in tropical and subtropical regions globally, serve as a primary source of both oil and protein on a global scale. Peanuts hold considerable economic and nutritional importance worldwide and play a crucial role in the agricultural and food industries of numerous countries (Syed et al., 2021). Nevertheless, a significant portion of the world’s peanut cultivation takes place on suboptimal soils with limited resources in many developing nations, leading to a substantial disparity between demand and supply (Zhang et al., 2023). In the face of climate change, various abiotic factors such as drought and temperature fluctuations impose constraints on both the quality and productivity of peanut crops (Dwivedi et al., 2013; Puppala et al., 2023). Hence, there is an urgent necessity to pinpoint pivotal genes capable of conferring tolerance to abiotic stress, which can then be harnessed in biotechnological initiatives aimed at developing enhanced peanut varieties (Baillo et al., 2019; Dormatey et al., 2020). Cultivated peanuts are natural allotetraploids originating from the hybridization of two diploid species, A. duranensis and A. ipaensis. The complete sequencing of the genomes of A. duranensis, A. ipaensis, and A. hypogaea has opened up new avenues for genomic research focusing on functional genes within the peanut (Bertioli et al., 2016; Bertioli et al., 2019). Presently, recent studies have shed light on the potential roles of TGA transcription factors in responding to abiotic stress. However, there have been no reports regarding their role in cultivated peanuts (Li et al., 2019a). In this study, we focused on TGA genes from A. hypogaea genome.
Our analysis included their phylogenetic relationships, conserved domains, gene structures, expansion patterns, cis-regulatory elements, protein-protein interactions, and expression profiles in various tissues and under different abiotic stresses. Furthermore, we conducted initial functional validation of key TGA genes in Arabidopsis and soybean. Our findings provide a comprehensive understanding of TGA genes in peanut, and offer a foundation for future functional studies to investigate their roles in regulating peanut’s tolerance to abiotic stress.
Materials and methods
Plant materials and treatment
The peanut varieties Nonghua5 (NH5, drought-tolerant and cold-tolerant genotype) and Fuhua18 (FH18, sensitive genotype) were selected as the plant material (Jiang et al., 2020; Zhang et al., 2020; Ren et al., 2022). The young peanut plant seedlings were grown in vermiculite with a light cycle of 16 hours of light (28°C) followed by 8 hours of dark (25°C). After 20 days from sowing, the seedlings were used to investigate their response to various hormones and abiotic stresses.
For cold stress treatment, the temperature in the climate chamber was reduced to 6°C while maintaining other growth conditions. For drought stress treatment, the seedlings were first allowed to recover in hydroponic cultures for 3 days before being subjected to the stress treatment. Subsequently, the seedlings were incubated in a 20% (w/v) solution of polyethylene glycol (PEG-6000). For various abiotic stress treatments, the second leaves were harvested at 0, 6, 12, 24, and 48 hours post-treatment, with three independent replicates. These collected leaves were rapidly frozen in liquid nitrogen and stored at -80°C. To investigate responses to different hormones, including methyl salicylate (MeSA) (0.1 mmol/L), methyl jasmonate (MeJA) (0.1 mmol/L), gibberellin (GA) (0.1 mmol/L), and abscisic acid (ABA) (0.1 mmol/L), they were applied as sprays onto the leaves of cultivated peanut seedlings, while sterile water was utilized as a control. Leaf samples were then collected at 0, 6, 12, 24, and 48 hours after treatment and stored at -80°C for RNA extraction. The phenotypic changes of the plants were observed on the fourth day after treatment.
Identification of TGA family members in cultivated and wild peanut
To identify the members of the TGA gene family in cultivated and wild peanut species, we obtained ten reported protein sequences of Arabidopsis TGA members from the Arabidopsis information resource TAIR (https://www.Arabidopsis.org/). These sequences served as queries to conduct searches for potential TGA genes within the genomes of cultivated peanut and two wild peanut species, utilizing the peanut genome database accessible at https://peanutbase.org/ using BLAST (E-value ≤ 10-5). The identification of TGA genes was carried out by predicting protein domains using the Pfam website (https://pfam.xfam.org/) and the SMART website (https://smart.embl.de/), followed by the removal of redundant sequences. Finally, the members of the TGA gene family were determined. To further analyze the characteristics of the peanut TGA gene family, we predicted their molecular weight, isoelectric point, and other physicochemical properties using the online software ExPASy (https://www.expasy.org/). Additionally, the subcellular localization of the peanut TGA gene family members was predicted using the online software WoLF PSORT (https://www.genscript.com/wolf-psort.html).
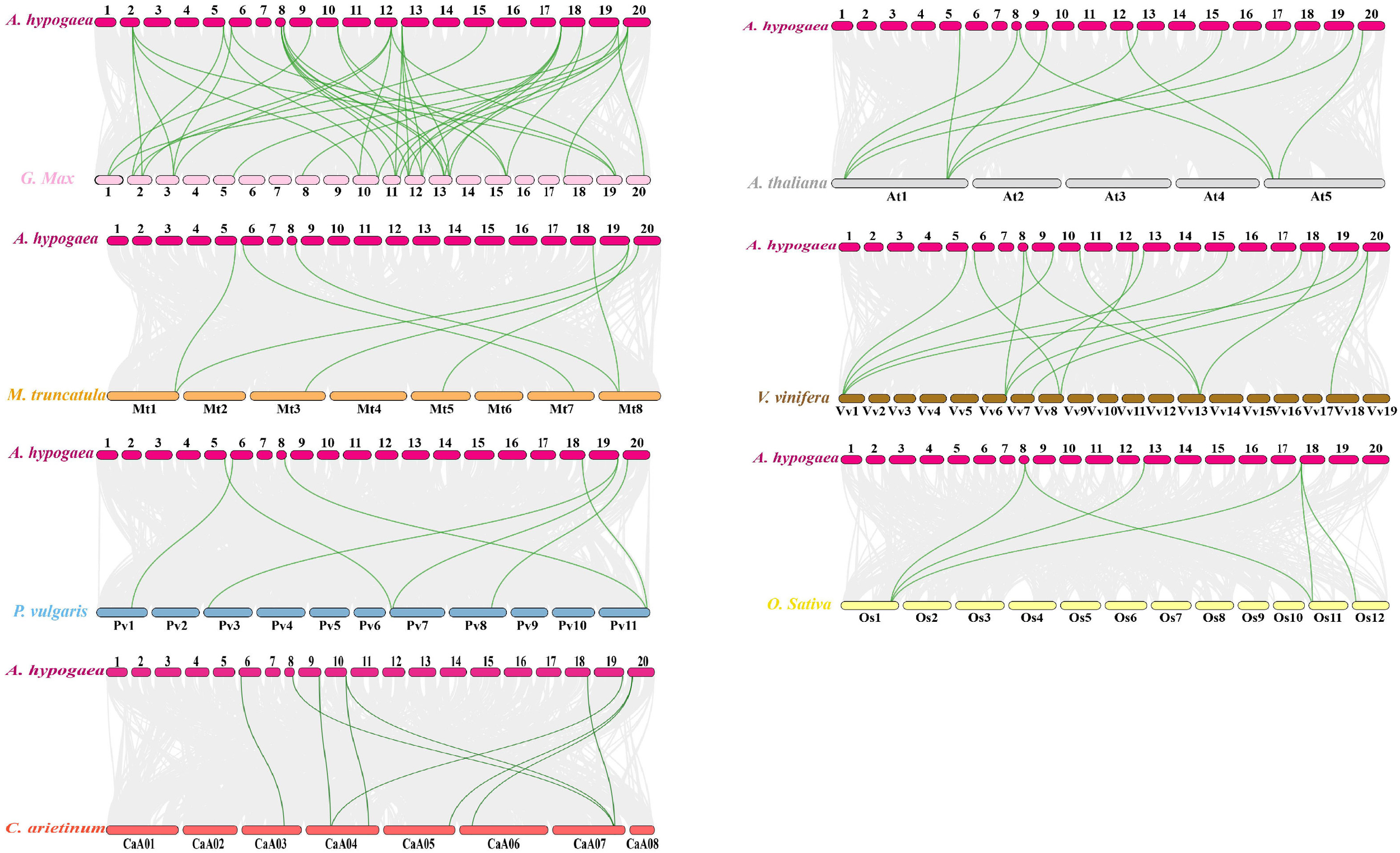
Phylogenetic, conserved motif and gene structure analysis of AhTGA genes
Sequence alignment was performed using ClustalW with default parameters (Larkin et al., 2007) to align all candidate AhTGA amino acid sequences with TGA family members from other species, including Arabidopsis thaliana, Phaseolus vulgaris, Glycine Max, Cicer arietinum, Oryza Sativa, Zea mays, Sorghum bicolor, Vitis vinifera, and Medicago truncatula. For phylogenetic analysis, MEGA 11.0 software was employed, applying the neighbor-joining (NJ) method with the Poisson model, pairwise deletion, and 1,000 bootstrap replications (Tamura et al., 2021). The resulting phylogenetic tree was visualized using the Evolview v2 webserver (He et al., 2016). To analyze the protein structure domains of TGA family proteins, we utilized the SMART online tool (https://smart.embl.de/), and a functional domain diagram was created using the Conserved Domain Database (CDD) from NCBI (https://www.ncbi.nlm.nih.gov/cdd/). The conserved amino acid motifs within the candidate AhTGA gene sequences were predicted using the MEME Suite 5.4.1 (https://meme-suite.org/meme/doc/meme.html) with default parameters. The resulting conserved motifs were visualized, and the gene structure diagrams were generated using the Gene Structure View program in TBtools (Chen et al., 2020).
Chromosomal locations, gene duplications, and synteny analysis
Chromosome length and position information of the 40 AhTGA members were extracted from the peanut genome and annotation files. Gene visualization was performed using the MG2C online software (http://mg2c.iask.in/mg2c_v2.1/), while gene duplication events were analyzed using MCScanX with default parameters. To demonstrate the collinearity of the TGA gene family, we employed the Advanced Circos function in TBtools. Additionally, the Multiple Synteny Plot was used to illustrate the synteny relationships between A. hypogaea and nine other species. The Ka (nonsynonymous substitution rate) and Ks (synonymous substitution rate) were examined using TBtools software, and the calculation of selection pressure was based on the Ka/Ks ratio, as detailed in the work by Wang et al. (2010).
Protein-protein and microRNAs-AhTGAs interaction network
The interaction relationships between AhTGAs and other proteins were analyzed using the STRING database (https://string-db.org), with a confidence score threshold set at > 0.4. Arabidopsis thaliana was used as the query organism. the visualization of the predicted protein-protein interaction (PPI) network was accomplished using Cytoscape 3.9.1 software (Shannon et al., 2003). To predict miRNA-target relationships for AhTGA, the psRNA Target Server (http://plantgrn.noble.org/psRNATarget/) was used, with an expected value set to the default value of 5, using the CDS sequence of AhTGAs as the candidate target. The predicted miRNA and their corresponding target genes were displayed using Cytoscape 3.9.1 software.
Conserved cis-element analysis in promoters
The promoter region was defined by extracting the upstream 2000 bp sequences of the TGA genes from the peanut genome database. To analyze and quantify the presence of cis-acting elements associated with growth, development, hormones, and stress response in this promoter region, we utilized the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Transcriptome-based expression pattern of AhTGAs
The orthologous genes for the 20 AhTGA genes were obtained through BLAST and the gene’s chromosomal locations using http://peanutgr.fafu.edu.cn/index.php (Zhuang et al., 2019). Furthermore, the FPKM values of these genes under hormone, low-temperature, and drought treatments were obtained. Transcriptome data, provided by Clevenger et al. (2016), of 22 different tissues in peanut were obtained from the Phytozome 13 database. The FPKM values of TGA genes in cultivated peanut tissues were converted to log2FPKM and standardized. Heatmap clustering was performed using the HeatMap function in TBtools software.
Quantitative RT−PCR validation
RNA extraction was performed on leaves and roots obtained using the Plant Total RNA Extraction Kit from Tiangen Biotech, Beijing, China. The resulting RNA was used to create cDNA using the PrimeScript™ RT Kit from TaKaRa, Japan, following the manufacturer’s instructions. Primers to identify AhTGA genes with differential expression were obtained from PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), and Actin11 was used as the internal control. The gene expression analysis was carried out using the SYBR Premix Ex TaqII kit (TliRNaseH Plus) from TaKaRa, Japan, and fluorescence quantitative reactions were detected using ABI7500 from Applied Biosystems, United States. The relative expression analysis was calculated using the 2−ΔΔCT approach (Livak and Schmittgen, 2001).
The AhTGA11 function analysis under chilling and drought stress conditions in Arabidopsis Plants
Gene-specific primers were designed using Primer Premier 6.0 to amplify the cDNA sequence of AhTGA11 in FH18. PCR amplification of AhTGA11’s coding sequence was conducted using the TransTaq DNA Polymerase High Fidelity Amplification Kit (Transgen Biotech). The resulting PCR products were visualized on a 1% agarose gel, purified using a DNA purification recovery kit, and then inserted into the pBWA (V) BS cloning vector driven by the CaMV35S promoter. The ligated DNA was introduced into Escherichia coli Top10 competent cells, and positive clones were selected and confirmed by sequencing. The recombinant plasmid pBWA (V) BS- AhTGA11 was subsequently transferred into Agrobacterium tumefaciens EHA105, which was used to transform wild-type (WT) Arabidopsis plants via the floral dip method. After screening with antibiotics and verifying the transgenic seedlings through PCR, homozygous transgenic lines were successfully obtained in the T2 generation. Subsequently, homozygous T3 progeny were examined and selected for further experimental procedures. The Col-0 seeds and empty half MS medium as control. 12 d old seedlings were transferred to new plates containing no or 8% (w/v) PEG for 5 days. The plant phenotypes were measured, and leaf and root tissues were collected for the measurement of relevant physiological and hormone indicators. For cold treatment, the 18 d old seedlings were treated at 4°C for 5 days, and the phenotypes, physiology, and hormones analyzed.
Generation of soybean hairy roots using Agrobacterium Rhizogenes transformation
The AhTGA11 gene was amplified and cloned into the pCAMBIA3301 vector under the control of the CaMV35S promoter to generate the pCAMBIA3301-AhTGA11 overexpression vector. Transgenic hairy roots were induced following the method described by Kereszt et al. (2007). The pCAMBIA3301-AhTGA11 vector was transformed into Agrobacterium strain K599, and soybean cultivar Williams seedlings at 7 days old were inoculated with the transformed K599 cells for treatment, while soybean plants induced with the K599 strain carrying pCAMBIA3301 were used as controls. The induction of hairy roots was carried out under 90% humidity conditions, and when the hairy roots reached a length of 3-4 cm, the plants were placed in a solution of 8% (w/v) PEG to promote their stable growth. After 4 days of treatment, phenotypic observations and physiological indicators in the roots and leaves were measured.
Physiological and hormone measurements
Physiological indicators and hormone levels were determined in both overexpressed Arabidopsis plants and soybean transgenic plants containing roots. The H2O2 content was measured following the method described by Sagisaka (1976), malondialdehyde (MDA) content and superoxide dismutase (SOD) activity were determined following the method of Zhang H. et al., 2019, and peroxidase (POD) activity was measured according to the method of Do et al. (2003). Quantitative analysis of hormones in plant samples was performed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) technology, following the procedures outlined in studies (Pan et al., 2010; Cai et al., 2014; Li et al., 2016).
Statistical analysis
The analyses of statistically significant data were processed by a one-way analysis of variance (ANOVA). In the case of multiple comparisons between different groups, Dunnett’s multiple comparison test or the Student’s t-test method was used. All data were analyzed using R language (version 4.3.0), and all statistical analyses in this study were conducted using the respective R packages. All the values were calculated as the means ± standard deviation (SD). Asterisks indicate significant differences. *P < 0.05. * *P < 0.01. Error bars represent the standard deviation from triplicate values.
Results
Genome-wide identification of TGA genes in cultivated and wild peanut
We conducted a comprehensive search in the peanut genome database using ten Arabidopsis TGA protein sequences as references, resulting in the identification of 40 candidate genes (Supplementary Table 1). Subsequently, structural domain analysis was performed on all sequences, revealing that the gene sequences of cultivated peanut and the two wild peanut species, A. duranensis and A. ipaensis, all contained intact bZIP and DOG1 domains (Supplementary Figure 1). Specifically, the cultivated peanut genome encompassed 20 AhTGA genes, designated as AhTGA01-AhTGA20 based on their respective chromosomal locations (Table 1). Similarly, the wild peanut genomes harbored 20 TGA genes, denoted as AdTGA01-AdTGA09 and AiTGA01-AiTGA11, distributed across eight chromosomes. We further analyzed the candidate TGA protein sequences (pI) (Table 1). The coding lengths of the 20 AhTGA protein sequences ranged from 331 to 531 amino acids. The predicted MW and pI of AhTGA proteins varied between 37.02 (AhTGA09 and AhTGA18) and 59.17 (AhTGA13) kDa, and 5.84 (AhTGA05) and 8.34 (AhTGA04 and AhTGA14), respectively. In the wild peanut species, the coding lengths of the 20 TGA protein sequences ranged from 331 to 538 amino acids in A. duranensis and from 331 to 550 amino acids in A. ipaensis. The expected MW of these TGA proteins ranged from 37.02 kDa (AdTGA07) to 59.63 kDa (AdTGA05) in A. duranensis, and from 37.02 kDa (AiTGA08) to 61.13 kDa (AiTGA06) in A. ipaensis. Moreover, the pI values ranged from 5.84 (AdTGA04) to 8.63 (AdTGA07) in A. duranensis, and from 5.97 (AiTGA04) to 8.63 (AiTGA08) in A. ipaensis. Subcellular localization predictions indicated that AhTGA, AdTGA, and AiTGA genes were primarily localized in the nucleus. Notably, the AhTGA genes in cultivated peanut and the wild diploid peanut species exhibited similar lengths and physical properties.
Table 1
| GENE NAME | Locus ID | Chromosomal Location | DNA(bp) | mRNA(bp) | Protein(aa) | MWb(kDa) | pI | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| AhTGA01 | ArahyR4ID1P | Arahy.02:74466805-74478269 | 11464 | 1892 | 503 | 56.63 | 6.69 | nucl: 13 |
| AhTGA02 | ArahyLTZY89 | Arahy.03:245406-250717 | 5311 | 2244 | 467 | 51.96 | 6.32 | nucl: 14 |
| AhTGA03 | ArahyS1DMKE | Arahy.03:279111-284423 | 5312 | 2244 | 467 | 51.96 | 6.32 | nucl: 14 |
| AhTGA04 | ArahyRIV6ZB | Arahy.03:123676732-123681028 | 4296 | 1944 | 368 | 41.63 | 8.34 | nucl: 11, chlo: 3 |
| AhTGA05 | Arahy0GP62G | Arahy.04:47164073-47171787 | 7714 | 1421 | 388 | 43.62 | 5.84 | nucl: 11, cyto: 1, pero: 1 |
| AhTGA06 | Arahy661MCQ | Arahy.05:109518217-109525080 | 6863 | 1584 | 459 | 51.24 | 6.21 | nucl: 10, chlo: 4 |
| AhTGA07 | ArahyKZ9VWQ | Arahy.06:10302739-10309738 | 6999 | 2211 | 450 | 49.51 | 7 | nucl: 14 |
| AhTGA08 | ArahyJ1CFJJ | Arahy.08:30570146-30577442 | 7296 | 1932 | 487 | 53.62 | 7.38 | nucl: 13 |
| AhTGA09 | ArahyRKP385 | Arahy.08:43602879-43611829 | 8950 | 1998 | 331 | 37.02 | 8.63 | nucl: 12, nucl_plas: 8 |
| AhTGA10 | ArahyW15JJQ | Arahy.09:113914413-113918157 | 3744 | 2553 | 456 | 50.92 | 6.31 | nucl: 11, chlo: 3 |
| AhTGA11 | ArahyP1JBJN | Arahy.10:115248749-115254745 | 5996 | 2141 | 463 | 51.15 | 6.53 | nucl: 14 |
| AhTGA12 | Arahy5F9GBV | Arahy.12:88825865-88838186 | 12321 | 1899 | 504 | 56.71 | 6.56 | nucl: 13 |
| AhTGA13 | ArahyEH3L7F | Arahy.13:2283773-2288910 | 5137 | 2213 | 531 | 59.17 | 6.49 | nucl: 14 |
| AhTGA14 | Arahy6K442L | Arahy.13:126982024-126986332 | 4308 | 1944 | 368 | 41.59 | 8.34 | nucl: 10, chlo: 4 |
| AhTGA15 | ArahyH6GQLR | Arahy.15:121748615-121755797 | 7182 | 1694 | 469 | 52.08 | 5.97 | nucl: 10, chlo: 4 |
| AhTGA16 | ArahyPHPT4F | Arahy.16:9377082-9384326 | 7244 | 3502 | 431 | 47.17 | 6.21 | nucl: 14 |
| AhTGA17 | Arahy980X4K | Arahy.18:6346988-6353680 | 6692 | 2011 | 492 | 54.38 | 8.27 | nucl: 12, cyto: 1 |
| AhTGA18 | ArahySERN02 | Arahy.18:122380117-122388818 | 8701 | 2013 | 331 | 37.02 | 8.63 | nucl: 12, nucl_plas: 8 |
| AhTGA19 | ArahyK04A4E | Arahy.19:154421962-154425784 | 3822 | 2494 | 456 | 50.97 | 6.34 | nucl: 10.5, cyto_nucl: 6, chlo: 3 |
| AhTGA20 | ArahyAAI19J | Arahy.20:23833950-23839123 | 5173 | 1848 | 362 | 41.03 | 6.63 | nucl: 6, chlo: 5, mito: 3 |
| AdTGA01 | AraduX4N5C | Aradu.A02:67538591-67544697 | 6106 | 6106 | 416 | 46.84 | 5.98 | nucl: 14 |
| AdTGA02 | AraduU5DG8 | Aradu.A03:114621519-114625756 | 4237 | 4237 | 368 | 41.63 | 8.34 | nucl: 11, chlo: 3 |
| AdTGA03 | AraduM9L15 | Aradu.A04:72882430-72890856 | 8426 | 8426 | 373 | 42 | 5.97 | nucl: 11, cyto: 2 |
| AdTGA04 | AraduC716W | Aradu.A05:103557314-103564234 | 6920 | 6920 | 476 | 53 | 5.81 | nucl: 14 |
| AdTGA05 | AraduBQ4IU | Aradu.A06:9768086-9774583 | 6497 | 6497 | 538 | 59.63 | 8.32 | chlo: 7, nucl: 6 |
| AdTGA06 | AraduI6GFF | Aradu.A08:28342298-28348900 | 6602 | 6602 | 492 | 54.28 | 7.1 | nucl: 14 |
| AdTGA07 | AraduGUH8G | Aradu.A08:41608257-41616928 | 8671 | 8671 | 331 | 37.02 | 8.63 | nucl: 12, nucl_plas: 8 |
| AdTGA08 | Aradu1JQ3A | Aradu.A09:114398072-114400850 | 2778 | 2778 | 485 | 54.25 | 6.37 | chlo: 6, nucl: 5, mito: 2 |
| AdTGA09 | AraduCM60U | Aradu.A10:15794805-15799916 | 5111 | 5111 | 414 | 47.24 | 6.53 | nucl: 11, chlo: 1, plas: 1 |
| AiTGA01 | AraipI0X2E | Araip.B02:78769485-78775072 | 5587 | 5587 | 397 | 44.45 | 6.17 | nucl: 13 |
| AiTGA02 | AraipHE4VX | Araip.B03:2236977-2240401 | 3424 | 3424 | 448 | 50.14 | 8.41 | nucl: 13 |
| AiTGA03 | AraipW8KP6 | Araip.B03:116847342-116851500 | 4158 | 4158 | 368 | 41.59 | 8.34 | nucl: 10, chlo: 4 |
| AiTGA04 | AraipNZ32C | Araip.B04:65308191-65317689 | 9498 | 9498 | 373 | 42.05 | 5.97 | nucl: 11, cyto: 2 |
| AiTGA05 | AraipX0N4G | Araip.B05:112439034-112446285 | 7251 | 7251 | 479 | 53.05 | 6.06 | nucl: 12, chlo: 2 |
| AiTGA06 | AraipW3SNY | Araip.B06:6055755-6061740 | 5985 | 5985 | 550 | 61.13 | 7.64 | chlo: 9, nucl: 4 |
| AiTGA07 | Araip1FG0Z | Araip.B08:5553954-5560404 | 6450 | 6450 | 467 | 51.27 | 6.92 | nucl: 14 |
| AiTGA08 | AraipD5QVM | Araip.B08:117483523-117491929 | 8406 | 8406 | 331 | 37.02 | 8.63 | nucl: 12, nucl_plas: 8 |
| AiTGA09 | AraipUIF5W | Araip.B09:143024347-143027150 | 2803 | 2803 | 456 | 50.97 | 6.34 | nucl: 10.5, cyto_nucl: 6, chlo: 3 |
| AiTGA10 | AraipS8WR7 | Araip.B10:22665260-22670590 | 5330 | 5330 | 356 | 40.35 | 6.63 | nucl: 6, chlo: 5, mito: 3 |
| AiTGA11 | Araip7QE9U | Araip.B10:134367929-134373817 | 5888 | 5888 | 433 | 48.1 | 6.04 | nucl: 13 |
The genomic and biochemical information for TGA genes identified in cultivated peanut and two wild.
Phylogenetic analysis of the TGA genes in cultivated peanut
The phylogenetic tree was constructed for peanut, Arabidopsis, common bean, soybean, grape, alfalfa, chickpea, rice, maize, and sorghum (Supplementary Table 1). Based on the classification of 133 TGA proteins with reference to Arabidopsis TGA proteins, the TGA proteins were divided into five groups (Figure 1), namely Group I-Group V, each of which contained 41, 25, 15, 18, and 31 members, respectively. AhTGA04, AhTGA14, and AhTGA20 belonged to branch I, AhTGA07, AhTGA09, AhTGA11, AhTGA16, and AhTGA18 belonged to branch II, AhTGA05 belonged to branch III, AhTGA01, AhTGA02, AhTGA03, AhTGA08, AhTGA12, AhTGA13, and AhTGA17 belonged to branch IV, and AhTGA06, AhTGA10, AhTGA15, and AhTGA19 belonged to branch V. Phylogenetic analysis showed that peanut TGA proteins had high similarity and genetic distance with the protein sequences of other leguminous plants, indicating that they may have similar functions.
Figure 1
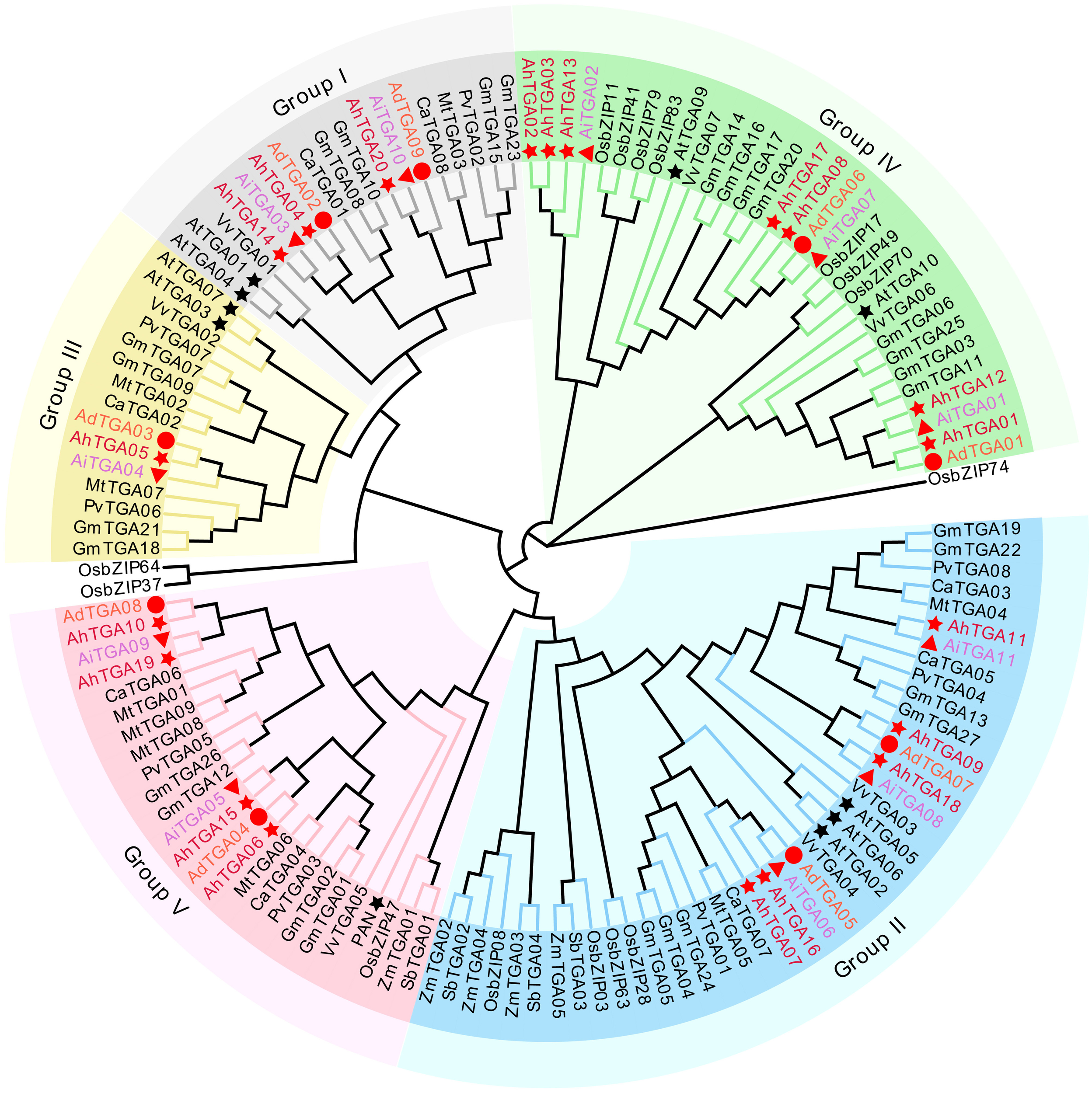
A neighbor-joining phylogenetic tree was created using 133 TGA proteins from various plant species, including Arabidopsis (At), chickpea (Ca), alfalfa (Mt), common beans (Pv), soybean (Gm), grapes (Vv), rice (Os), sorghum (Sb), maize (Zm), and three Arachis species. The TGA proteins were categorized into five clades, distinguished by distinct background colors. TGA proteins found in cultivated and wild peanuts are denoted by red stars, circles, and triangles, respectively.
Gene structure and conserved motifs analysis
In cultivated peanut proteins, up to 8 motifs were identified, with a total length of 21-50 amino acids (Figures 2A, B; Supplementary Figure 2; Supplementary Table 2). The number of motifs in TGA proteins ranged from 5 to 7, all of which contained the 5 conserved motifs, Motif 1 to Motif 5. There were differences in motif distribution among different branches, with Motif 6 found only in branches II, IV, and V; Motif 7 only in branches II, and III and V; and Motif 8 only in groups I and III. Differences in motif distribution may lead to changes in TGA gene structure and function (Figure 2B). To analyze the relationship between genome evolution and functional differentiation, the gene structure of AhTGAs was further analyzed. The number of exons in the AhTGA gene family ranged from 9 to 16. TGA genes with close evolutionary relationships not only had the same number of exons, but also had similar structures (Figure 2C). According to conserved domain analysis, both bZIP and DOG1 domains were found in each AhTGA protein, and these domains were located in similar positions in different sequences (Figure 2D). These results suggest that AhTGA genes have conserved structural domains in gene structure, but specific sequence structures exist in TGA genes in different clades.
Figure 2
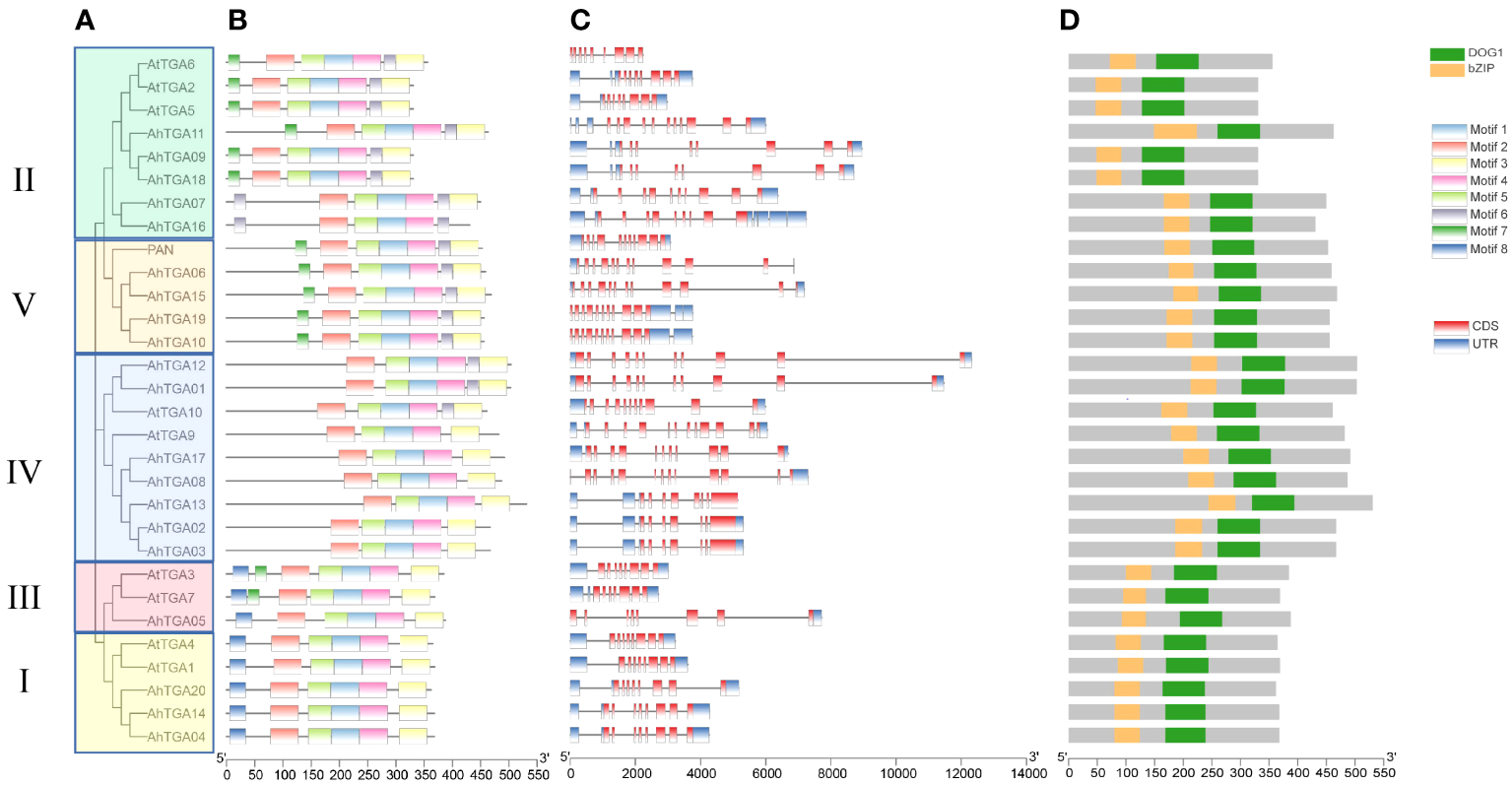
Phylogenetic analysis, conserved motifs, gene structures, and conserved domains of TGA genes in both cultivated peanut and Arabidopsis were examined. (A) A phylogenetic tree was generated using the Neighbor-Joining (NJ) method, illustrating the relationships among TGA protein sequences in cultivated peanut and Arabidopsis. (B) Conserved motifs within AhTGAs were identified using MEME, with different colors representing distinct motifs. (C) The structural characteristics of twenty AhTGA genes were analyzed. (D) A comparison of conserved domains between AtTGAs and AhTGAs was conducted, with the ruler at the bottom indicating sequence lengths.
Chromosomal locations, gene duplication and syntenic analysis of AhTGAs
The AhTGA genes, comprising a total of 20 genes, are distributed across 15 chromosomes in the cultivated peanut genome. Similarly, the wild peanut species A. duranensis and A. ipaensis possess 20 TGA genes, which are located on eight chromosomes, respectively (Figure 3). While the chromosomal locations of most TGAs in cultivated peanut remain consistent with those in wild peanut species, some TGA genes have undergone changes in their genomic positions, likely attributed to segmental duplication events within the cultivated peanut genome (Figure 4A). These genetic rearrangements contribute to the diversification of the TGA gene family in cultivated peanut. To gain insights into the evolutionary mechanisms governing the AhTGA gene family, gene duplication events within the Arachis species were analyzed using MCScanx, leading to the identification of several duplication events (Figure 4; Supplementary Table 3). Notably, cultivated peanut lacks tandem duplications among its TGA genes, but it does exhibit 14 gene pairs for segment duplication. In contrast, only one pair of segmental duplication was detected in the wild peanut species A. ipaensis, while A. duranensis did not display any duplication events (Figure 4A). Furthermore, collinearity analysis revealed that 23 gene pairs were shared between A. hypogaea and A. duranensis, while 28 gene pairs were common to A. hypogaea and A. ipaensis. Additionally, 12 collinear gene pairs were identified between A. duranensis and A. ipaensis (Figure 4B). These findings emphasize a robust collinearity relationship between wild and cultivated peanut species.
Figure 3
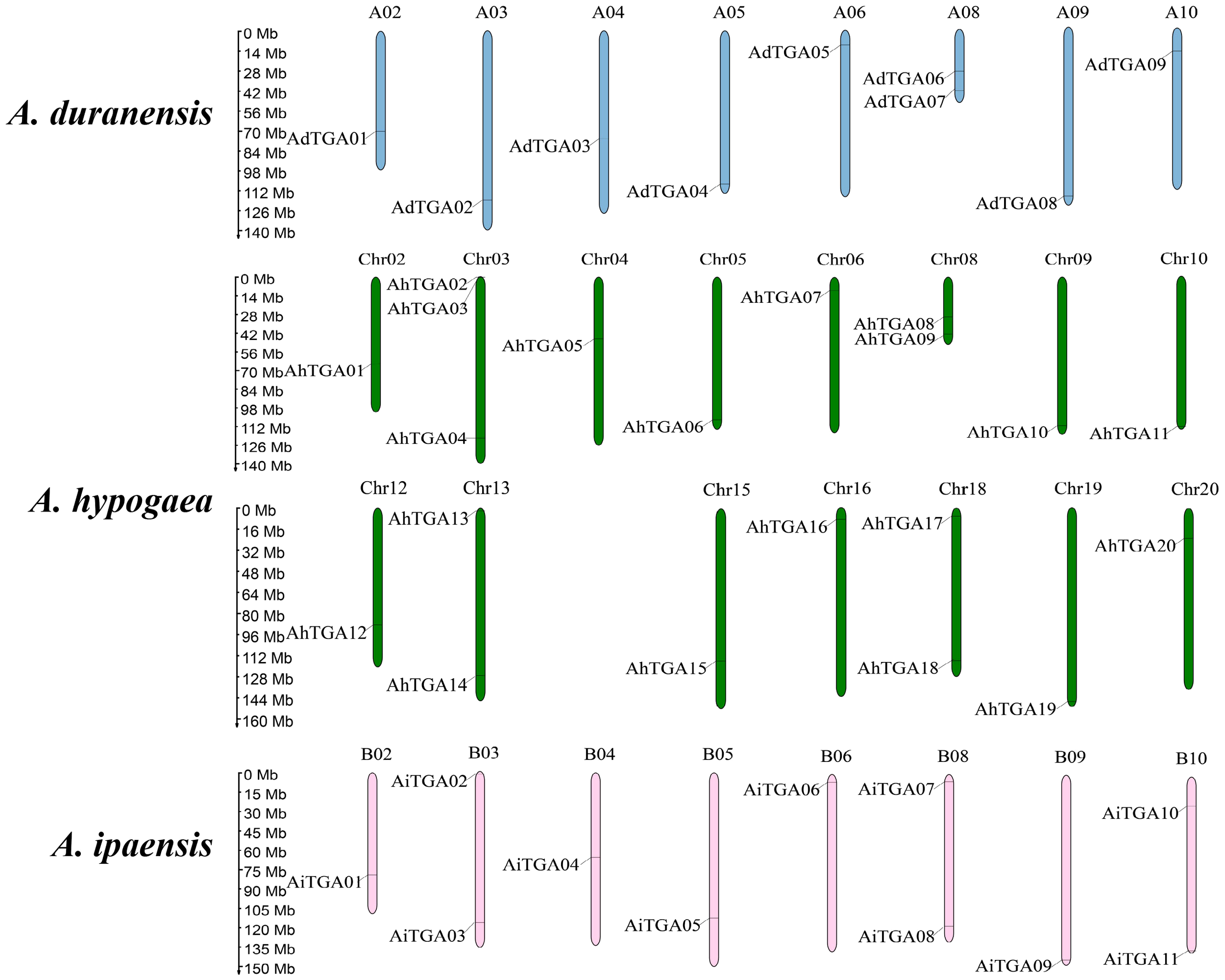
Distribution of TGA genes in three Arachis species genome. The chromosomes of A. duranensis, A. ipaensis and A. hypogaea were shown with pink, green and blue colors, respectively. Chromosome size is indicated by its relative length. The scale on the left is shown in megabases (Mb).
Figure 4
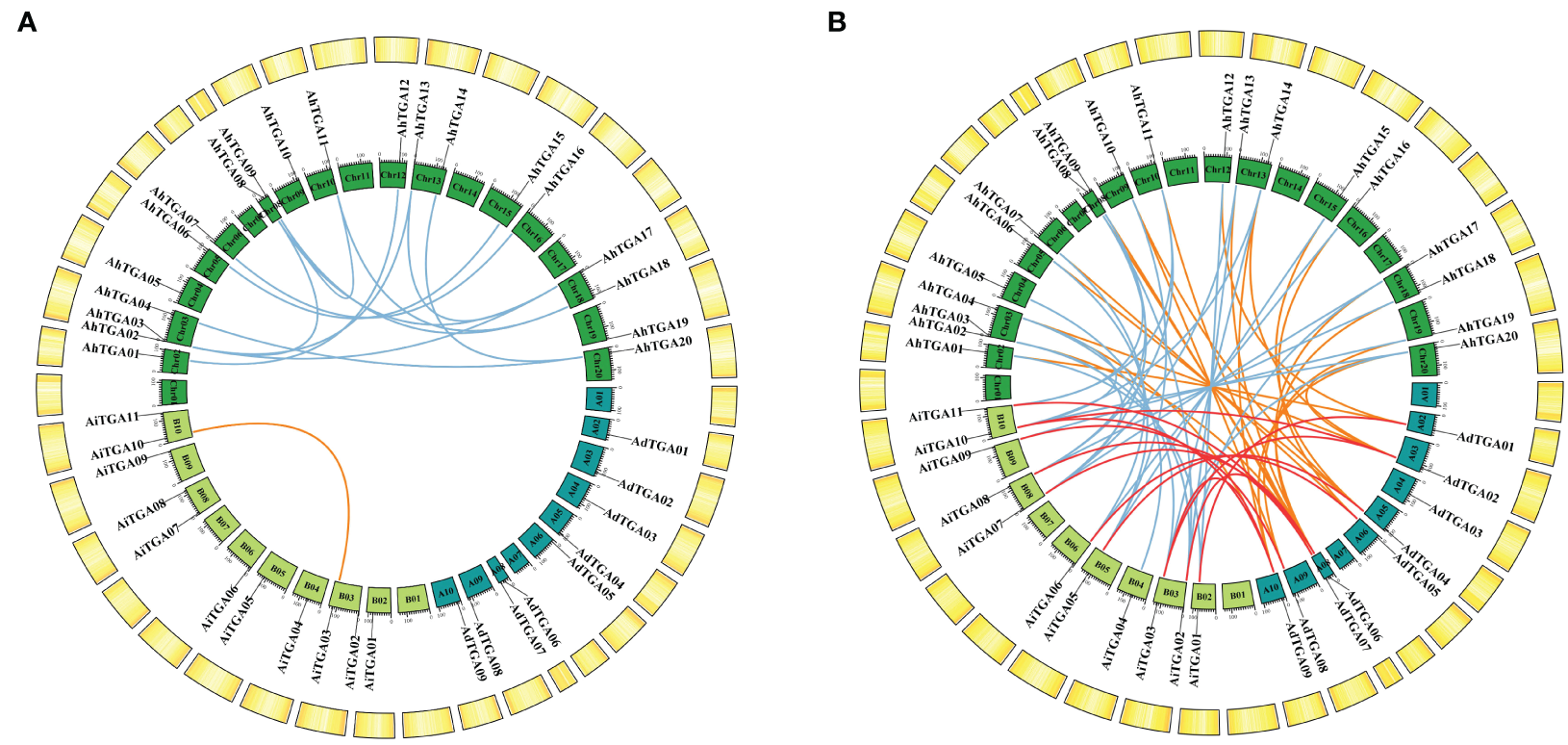
Syntenic analysis of TGA genes between cultivated peanuts and other plants. (A) Syntenic relationships of TGA genes with in cultivated peanut and two wild diploid peanuts, respectively. (B) Syntenic connections among TGA genes across A. duranensis, A. ipaensis, and A. hypogaea were examined. The chromosomes of A. duranensis, A. ipaensis, and A. hypogaea are depicted in dark green, light green, and green colors, respectively. Putative homologous TGA genes are indicated by lines of varying colors.
Synteny analysis of TGA genes among various species
Orthologous gene pairs were identified between TGA genes in cultivated peanut and those in other plants, including soybean, chickpea, alfalfa, common bean, Arabidopsis, grape, rice, sorghum, and maize (Figure 5; Supplementary Table 3). There were 39, 7, 9, and 7 gene pairs between cultivated peanut and other legumes, including soybean, alfalfa, chickpea and common bean, respectively; 14 and 10 gene pairs were found between cultivated peanut and the dicotyledonous plants grape and Arabidopsis, respectively; 6 gene pairs were identified between cultivated peanut and the monocotyledonous plant rice, while there were no orthologous gene pairs between cultivated peanut and the monocotyledonous plants sorghum and maize. Collinearity analysis revealed that the relationship of TGA genes between cultivated peanut and dicotyledonous plants was closer than that with monocotyledonous plants, with the closest relationship found with soybean.
Figure 5
Syntenic analysis of TGA genes between cultivated peanuts and other plants, including soybean, chickpea, alfalfa, common beans, Arabidopsis, grapes, rice, sorghum and maize. Gray lines in the background indicated the collinear blocks with in A. hypogaea and other plant genomes, while the syntenic TGA gene pairs are linked with green lines.
Comparison of orthologous gene pairs revealed differences in the synteny relationships of AhTGA genes with various species. For example, AhTGA20 in Group I showed the highest number of orthologous gene pairs (15 pairs) with other species, followed by AhTGA17 in Group IV and AhTGA19 in Group V, each having 12 pairs of orthologous gene pairs. AhTGA08, AhTGA09, and AhTGA13 in had 10-11 pairs of orthologous gene pairs with other species. These results indicate that the synteny modules containing these genes are highly conserved in different species genomes. There were differences in the synteny of AhTGAs, as some TGA genes’ synteny modules were only present within the three peanut genomes, such as AhTGA16, AhTGA05, AhTGA04, AhTGA14, and AhTGA02. Other AhTGAs showed synteny relationships with other legume crops, while cultivated peanuts and rice, a monocot, showed synteny only in Group V. These results suggest that different AhTGA genes gradually evolved with the replication of synteny modules during the evolutionary process.
To analyze the evolutionary selection pressure on AhTGA genes, we calculated the Ka/Ks ratio of TGA gene pairs. Except for 14 collinear gene pairs that could not be calculated, the Ka/Ks ratios of the remaining 131 collinear gene pairs between cultivated peanut and other species were all less than 1, indicating that the AhTGA genes were mainly subject to purifying selection during the evolutionary process (Supplementary Table 4).
Promoter analysis of AhTGAs
Promoter analysis plays a crucial role in unraveling the transcriptional regulation and potential functions of peanut TGA genes. To gain insights into these aspects, we conducted an investigation by submitting the 2000 bp regulatory region upstream of the ATG (promoter) to the PlantCARE database, which allowed us to detect cis-acting elements. Remarkably, a total of 860 cis-acting elements, encompassing 43 different types with potential functions, were successfully predicted (Figure 6A; Supplementary Table 5). Among the predicted cis-acting elements, 29 types were associated with growth and development, 9 were related to hormones, and 5 were associated with stress responses. Notably, a significant portion of the growth and development-related elements were light-responsive components, with a total count of 223. Hormone-responsive elements accounted for 102 instances, including 42 JA-responsive elements, 20 SA-responsive elements, 16 ABA-responsive elements, 10 GA-responsive elements, and 14 auxin-responsive elements. Furthermore, the promoter regions of AhTGA genes exhibited a clustering pattern into five distinct clades, which was consistent with the protein sequence analysis. Intriguingly, variations were observed in the composition of cis-acting elements present among the different clusters (Figure 6B). A thorough analysis of cis-acting elements in the AhTGA genes responding to hormones and abiotic stresses such as low temperature and drought has been conducted (Figure 6C). The hormone responses are primarily focused on MeJA, SA, ABA, GA, and auxins. Variations in the distribution of cis-acting elements in different genes have been observed; for instance, MeJA-related elements are found to be more abundant in AhTGA04, AhTGA11, and AhTGA14, while SA-responsive elements are more prevalent in AhTGA02, AhTGA03, and AhTGA13. Concerning stress responses, it has been noted that most AhTGA genes contain 1-3 cis-acting elements related to low temperature or drought, but some genes, such as AhTGA08, AhTGA17, and AhTGA20, are not associated with these elements related to low temperature and drought.This finding suggests that different types of TGA genes interact with specific transcription factors, enabling their participation in diverse regulatory pathways.
Figure 6
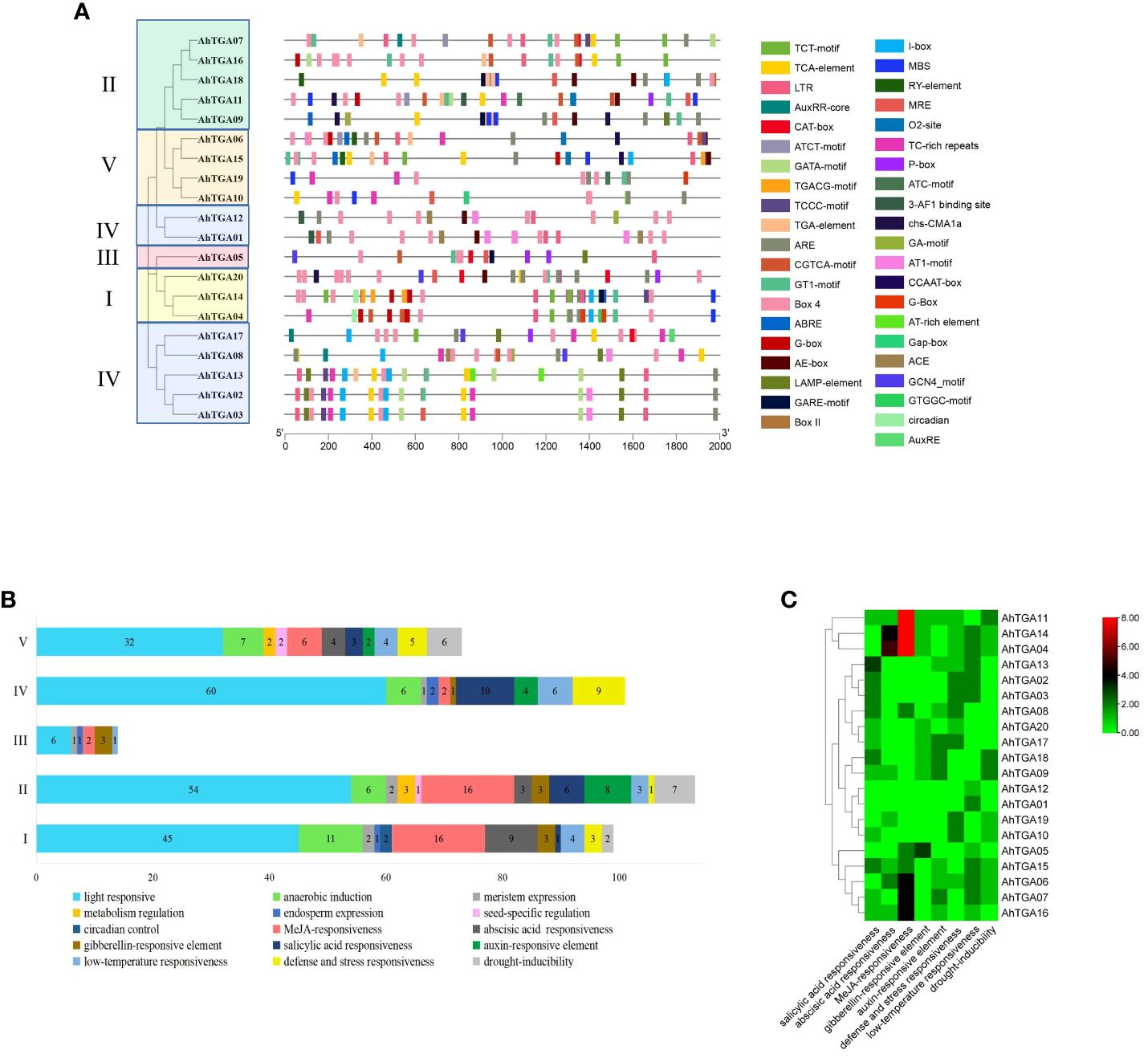
Analysis of the cis-acting elements in the promoter regions of 20 AhTGA genes. (A) Distribution of cis-elements in the promoters of AhTGAs. Gray lines indicate the promoters. Cis-elements differing in function are color-coded accordingly. (B) Functional statistics of cis-elements in the promoters within different groupings. (C)Cis-acting elements of AhTGA genes responded to cold, drought stress and hormone treatment.
Protein-protein and miRNA-genes regulatory networks prediction for AhTGAs
To gain further insights into the potential functions, signal transduction, and metabolic pathways of AhTGA members, we constructed a protein-protein interaction (PPI) network (Figure 7A). The analysis revealed that all 20 AhTGA genes exhibited orthologous relationships with 10 Arabidopsis TGAs and interacted with 40 functional proteins (Figure 7A; Supplementary Table 6). Notably, the proteins interacting with AhTGAs encompassed stress-responsive and pathogen defense-related proteins, such as NPR1, PR1, NIMIN1, and WRKY70. Additionally, several proteins involved in reproductive growth and flower organ development, including COI1, ROXY1, BOP1, and BOP2, were identified based on gene ontology (GO) information (Supplementary Figure 3).
Figure 7
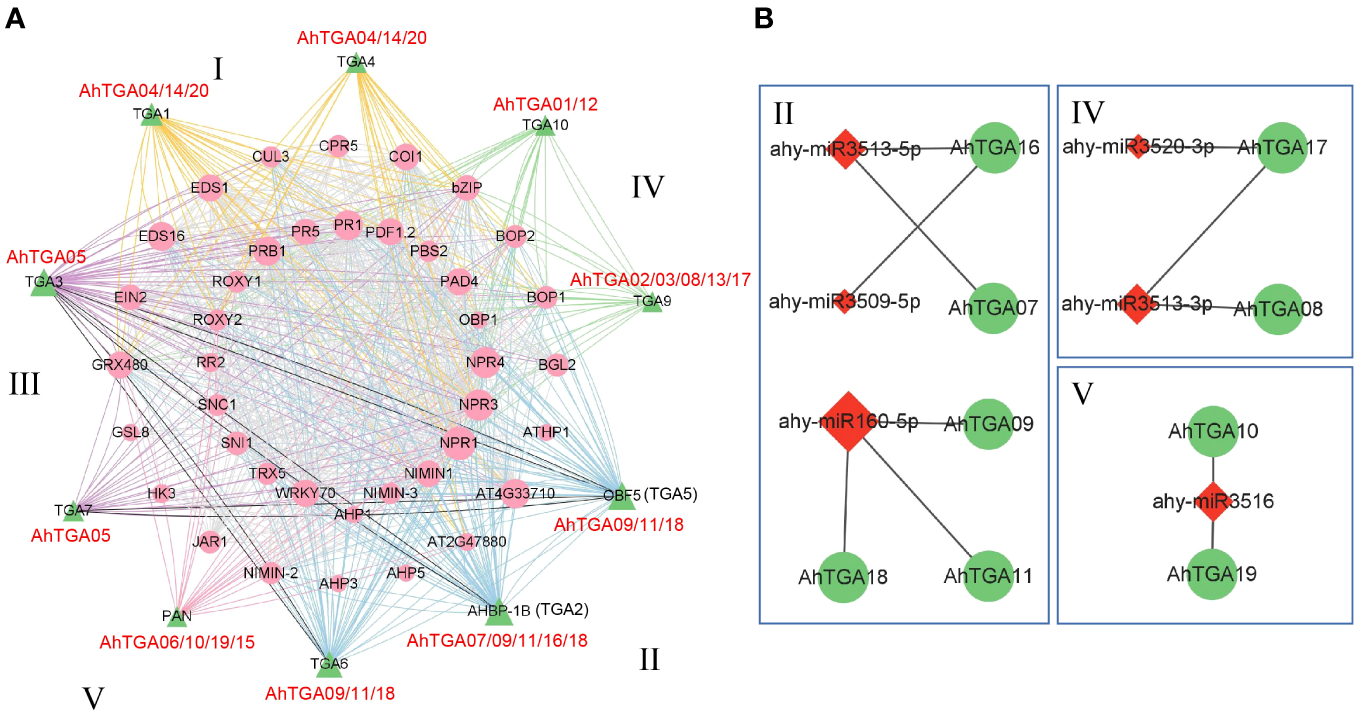
Interactions and regulatory associations involving peanut TGA genes with other proteins and miRNAs were explored. (A) A protein-protein interaction network involving peanut TGA proteins and other interacting proteins is presented. Peanut TGA proteins are represented by green triangles, while other proteins interacting with AhTGAs are depicted as pink circles. Interaction relationships are categorized into five groups based on evolutionary connections, denoted by different colored lines. (B) A regulatory network illustrates the potential miRNAs and their corresponding targeted AhTGAs. MiRNAs are represented by red lozenges, and the targeted AhTGAs are denoted by green circles. The presumed regulatory associations between miRNAs and their targeted AhTGAs are depicted as grey lines.
Based on the functional classification of genes, all 20 AhTGA members, across the five groups, were found to be involved in the interaction with stress-responsive proteins. Furthermore, 16 AhTGA members in the four groups, except Group V, interacted with proteins associated with plant pathogen defense. Moreover, 9 members from Group I, II, and Group III were implicated in responding to environmental stress. These findings collectively indicate that the majority of AhTGA genes play a crucial role in disease resistance and stress response.
In addition to protein-protein interactions, we performed miRNA target prediction for AhTGA genes. The analysis revealed that 9 AhTGA genes were targeted by 6 miRNAs belonging to 6 different families (Figure 7B; Supplementary Table 7). Most miRNAs were found to target only one or two AhTGA genes, except for ahy-miR160-5p, which targeted 3 genes (AhTGA11, AhTGA18, and AhTGA09). Interestingly, specific groups of AhTGA genes were regulated by distinct miRNAs. For instance, in Group II, 5 TGA genes were targeted by 3 miRNAs, namely ahy-miR3513-5p, ahy-miR3509-5p, and ahy-miR160-5p. Additionally, AhTGA17 and AhTGA08 in Group IV were regulated by ahy-miR3509-5p and ahy-miR3513-3p, respectively. AhTGA10 and AhTGA19 in Group V were targeted by ahy-miR3516. Notably, no miRNA regulation was observed in Group III and Group I. These results suggest that miRNAs may play a crucial role in the molecular regulation of AhTGA genes. Moreover, the regulation of AhTGA genes by different types of miRNAs in different groups may contribute to variations in their expression levels, thereby influencing their functions.
Expression patterns of AhTGA genes in various tissues and under different hormone and abiotic stress conditions
AhTGA gene expression patterns were investigated using RNA-Seq data, encompassing various developmental stages and tissues. Unique expression profiles for 20 AhTGA genes were revealed in the analysis of 22 tissues. Similar expression in orthologous pairs from A and B subgenomes was observed due to mRNA and promoter sequence similarity (Supplementary Figure 4). Expression was detected in all 22 tissues for AhTGA07, AhTGA16, AhTGA11, AhTGA09, AhTGA18, and AhTGA20, indicating their involvement throughout the peanut life cycle. Certain genes, such as AhTGA01, AhTGA06, AhTGA08, AhTGA12, AhTGA13, AhTGA15, and AhTGA17, showed higher expression in roots, nodules, and reproductive organs, suggesting roles in peanut reproductive development and underground growth. Similar expression levels among homologous genes indicated functional redundancy. Orthologous AhTGAs were identified in the reference genome Shitouqi (Supplementary Table 8) (Zhuang et al., 2019). The transcriptome-based expression patterns of orthologs under hormone, low-temperature, and drought treatments were determined. The results showed that, regardless of low-temperature or drought treatments, the expression levels of five TGA genes in subgroup II, AhTGA07, AhTGA09, AhTGA11, AhTGA16, and AhTGA18, significantly increased. In subgroup I and III, AhTGA04, AhTGA05, AhTGA14, and AhTGA20 also exhibited significant upregulation under stress conditions but at lower expression levels than genes in subgroup II (Figure 8). In contrast, under different hormone treatments, genes in subgroups I, II, and III showed significantly higher expression compared to others. Combining tissue-specific expression with stress and hormone responses, the five TGA genes in subgroup II, AhTGA07, AhTGA09, AhTGA11, AhTGA16, and AhTGA18, displayed higher expression levels throughout the entire developmental stages of peanut and significantly increased expression under stress and hormone treatments such as salicylic acid, indicating their crucial roles in peanut growth, development, and stress regulation.
Figure 8
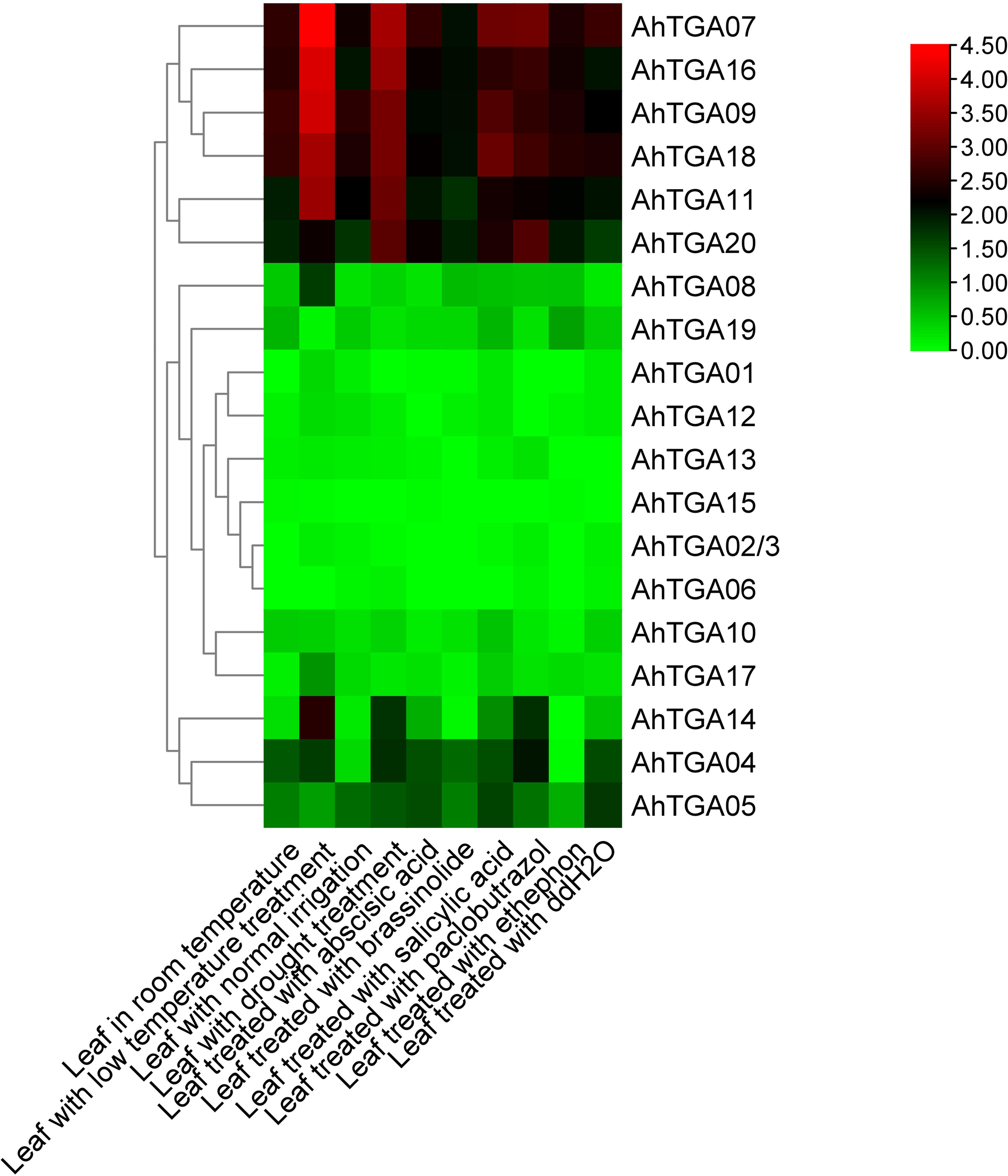
Expression profiles of 20 AhTGAs under different hormone and abiotic stress conditions. Heatmap clustering was based on the FPKM values of AhTGA genes following hormone and stress condition treatments. Log2-transformed values are used in a color-coded heatmap, with bars representing normalized FPKM (Log2) expression levels, and red for higher expression, green means low.
Expression patterns of AhTGAs under abiotic stresses and diverse hormone treatment
To investigate the response of AhTGA genes to abiotic stresses and hormone treatments, we performed qPCR analysis on two peanut varieties, NH5 (tolerant) and FH18 (sensitive), under low temperature and drought stress conditions (Figure 9). The expression levels of selected TGA genes were measured at five different time points. The primers used for qPCR are listed in Supplementary Table 9. Our results revealed that several genes, including AhTGA01, AhTGA02, AhTGA03, AhTGA06, AhTGA08, AhTGA09, AhTGA10, AhTGA12, AhTGA13, AhTGA15, AhTGA17, and AhTGA19, exhibited relatively low expression levels in both NH5 and FH18 under low temperature and drought stress treatments. Furthermore, no significant changes in expression levels were observed across the five time points, suggesting that these genes may not be involved in stress responses or may lack functional roles under these conditions.
Figure 9
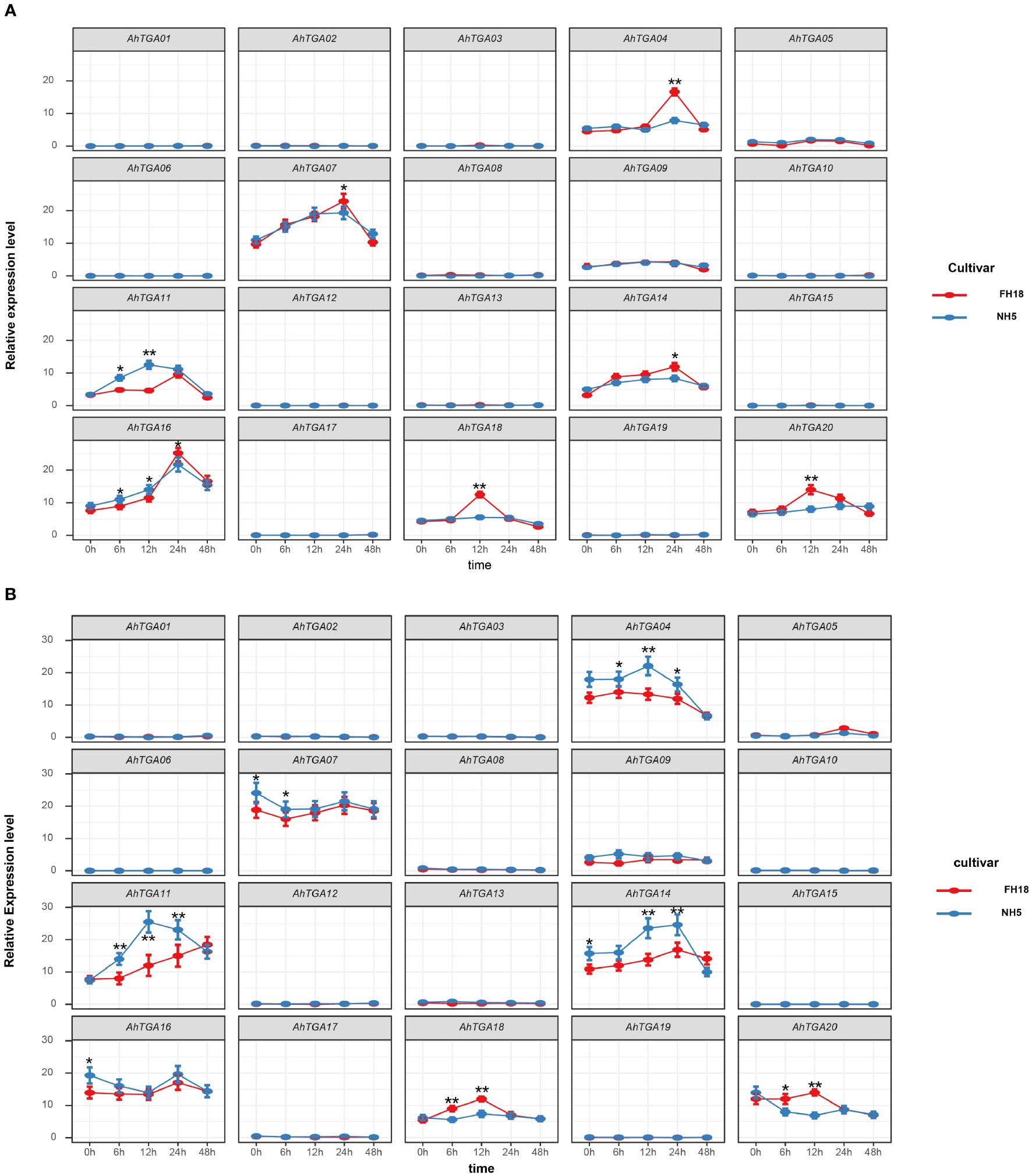
Expression patterns of the AhTGA genes in NH5 and FH18 under drought and low-temperature treatments. (A) Relative expression levels of the AhTGA gene in the cold-sensitive variety FH18 and the cold-tolerant variety NH5. (B) Relative expression levels of the AhTGA genes in the drought-sensitive variety FH18 and the drought-tolerant variety NH5. Student’s t-test was used to detect differences between the two varieties at the same time point (*, P < 0.05; **, P < 0.01).
Under low temperature conditions, the expression levels of AhTGA04, AhTGA07, AhTGA11, AhTGA14, AhTGA16, AhTGA18, and AhTGA20 exhibited significant changes over time. Some genes showed differential expression patterns between cold-tolerant and cold-sensitive varieties. For instance, AhTGA04 and AhTGA14 exhibited significantly higher expression levels in FH18 than in NH5 at 24 hours, while AhTGA18 and AhTGA20 displayed significantly higher expression levels in FH18 than in NH5 at 12 hours. These findings suggest that these genes have distinct expression patterns between cold-tolerant and cold-sensitive varieties and may play roles in the molecular regulation of peanut cold tolerance (Figure 9A). Similar expression patterns were observed under drought stress conditions. AhTGA04, AhTGA07, AhTGA11, AhTGA14, AhTGA16, AhTGA18, and AhTGA20 also displayed higher expression levels, but with differences in expression patterns between FH18 and NH5. For instance, AhTGA04 and AhTGA14 exhibited opposite expression patterns compared to those under low temperature conditions, showing significantly upregulated expression in FH18. On the other hand, AhTGA18 and AhTGA20 displayed similar expression patterns to those under low temperature conditions, with significantly higher expression levels in FH18 than in NH5 at 12 hours (Figure 9B).
To elucidate the involvement of different hormone regulatory mechanisms in AhTGA genes, we treated the low temperature and drought-sensitive variety FH18 with exogenous hormones and analyzed the expression characteristics of selected TGA genes (Figure 10). Seven Genes that displayed significant expression level differences under low temperature and drought conditions were selected for analysis under hormone treatments. Within 48 hours of distilled water spray treatment, the expression levels of seven genes did not show significant changes. However, after hormone treatments, the expression levels of AhTGA genes exhibited significant alterations, particularly in response to MeJA and MeSA treatments compared to ABA and GA treatments. With the exception of AhTGA, the expression levels of other AhTGA genes showed an increase followed by a decrease. Notably, the expression level of AhTGA treated with salicylic acid decreased significantly, whereas the expression levels of AhTGA genes treated with other hormones initially increased and then decreased. Following exogenous ABA treatment, the expression levels of AhTGA11 and AhTGA14 decreased, while exogenous GA treatment led to decreased expression levels of AhTGA genes, with AhTGA11 displaying an initial increase followed by a decrease. These findings suggest that AhTGA genes are involved in the regulation of different hormone pathways, with the salicylic acid and jasmonic acid pathways showing the most significant regulation (Figure 10).
Figure 10
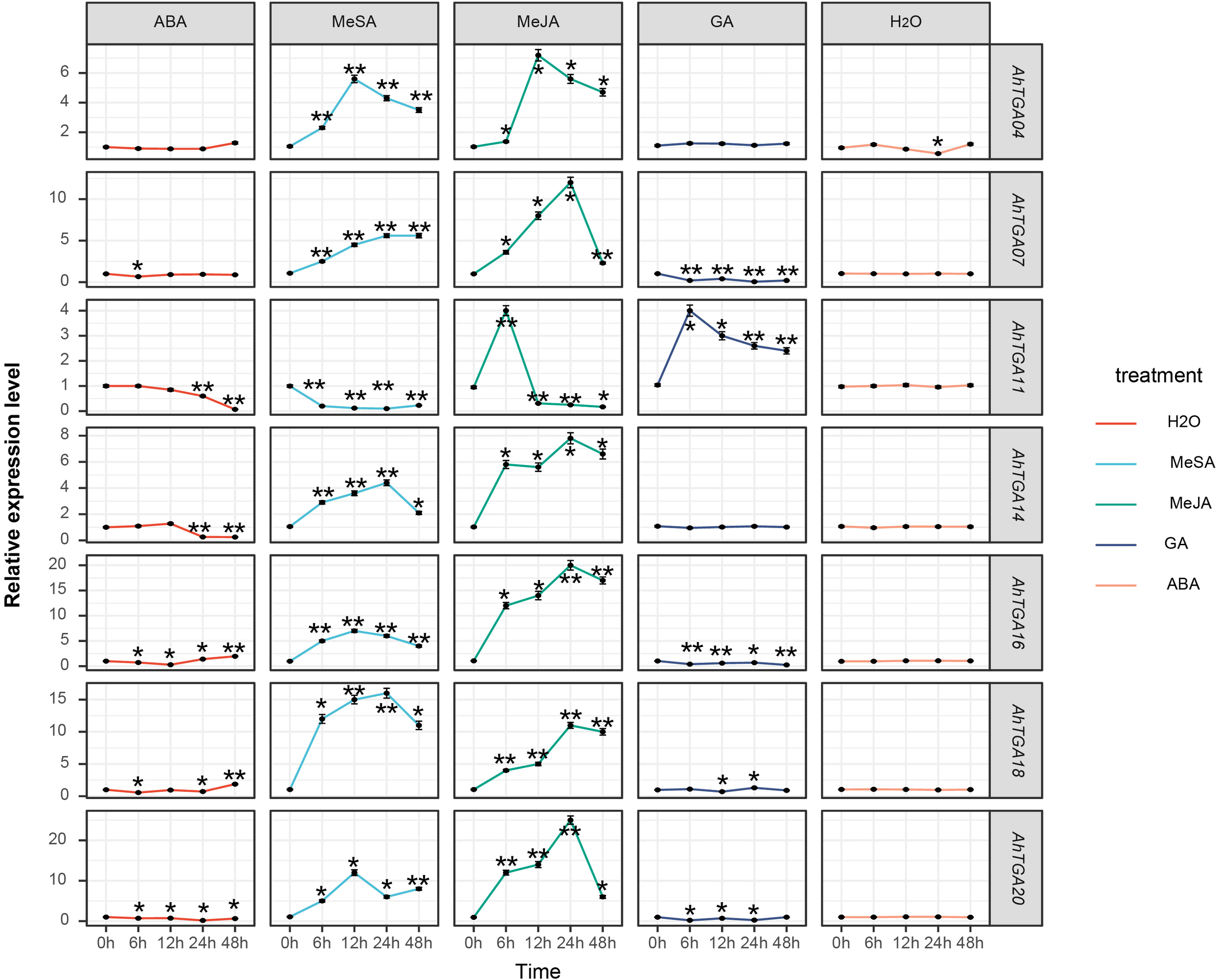
The expression levels of the AhTGA genes under different hormone treatments were examined in. The differential expression levels of AhTGA in FH18 after hormone treatment were analyzed using ANOVA and Dunnett’ s multiple comparison method, with the 0h time point used as the control group. Significant differences were indicated by ** (P < 0.01).
Overexpression of AhTGA11 confers cold and drought stress in transgenic Arabidopsis
Due to its significant responses under both abiotic stress and hormonal treatments, as well as differential expression patterns observed between cold and drought-tolerant genotypes versus sensitive genotypes, AhTGA11 is hypothesized to be a key factor in abiotic stress and hormone regulatory pathways. Therefore, AhTGA11 was transformed into Arabidopsis and its gene function was identified in transgenic plants of the T3 generation. After amplification with the primers AhTGA11-F/R, the CDS sequence of AhTGA11 was sequenced and found to be consistent with the reference genome sequence (Supplementary Table 10; Supplementary Figure 5). Following insertion into a vector under the control of the 35S promoter, Arabidopsis was transformed. Under non-PEG treatment conditions, the wild-type and AhTGA11-OE phenotypes were similar, whereas under 8% PEG treatment, the wild-type Arabidopsis plants exhibited wilting, with significantly reduced root length compared to the overexpressing AhTGA11-OE plants (Figure 11A). Under low-temperature conditions, the leaves of wild-type plants appeared water-soaked (Figure 11B). Physiological and endogenous hormone measurements of wild-type and AhTGA11-overexpressing plants under low-temperature and drought treatment conditions revealed that in overexpressing plants, root tissue H2O2 content, POD activity, and SOD activity were significantly higher than in wild-type plants, while MDA content was significantly lower than in wild-type (Figure 11C). Hormone measurements for four stress-related endogenous hormones showed that in overexpressing plants, JA content was significantly higher than in the wild-type, whereas ABA content was significantly lower than in the wild-type, with no significant differences in SA and GA. These results indicate that AhTGA11 likely plays a positive regulatory role in drought and low-temperature stress in plants and is involved in mediating ABA and JA-related pathways.
Figure 11
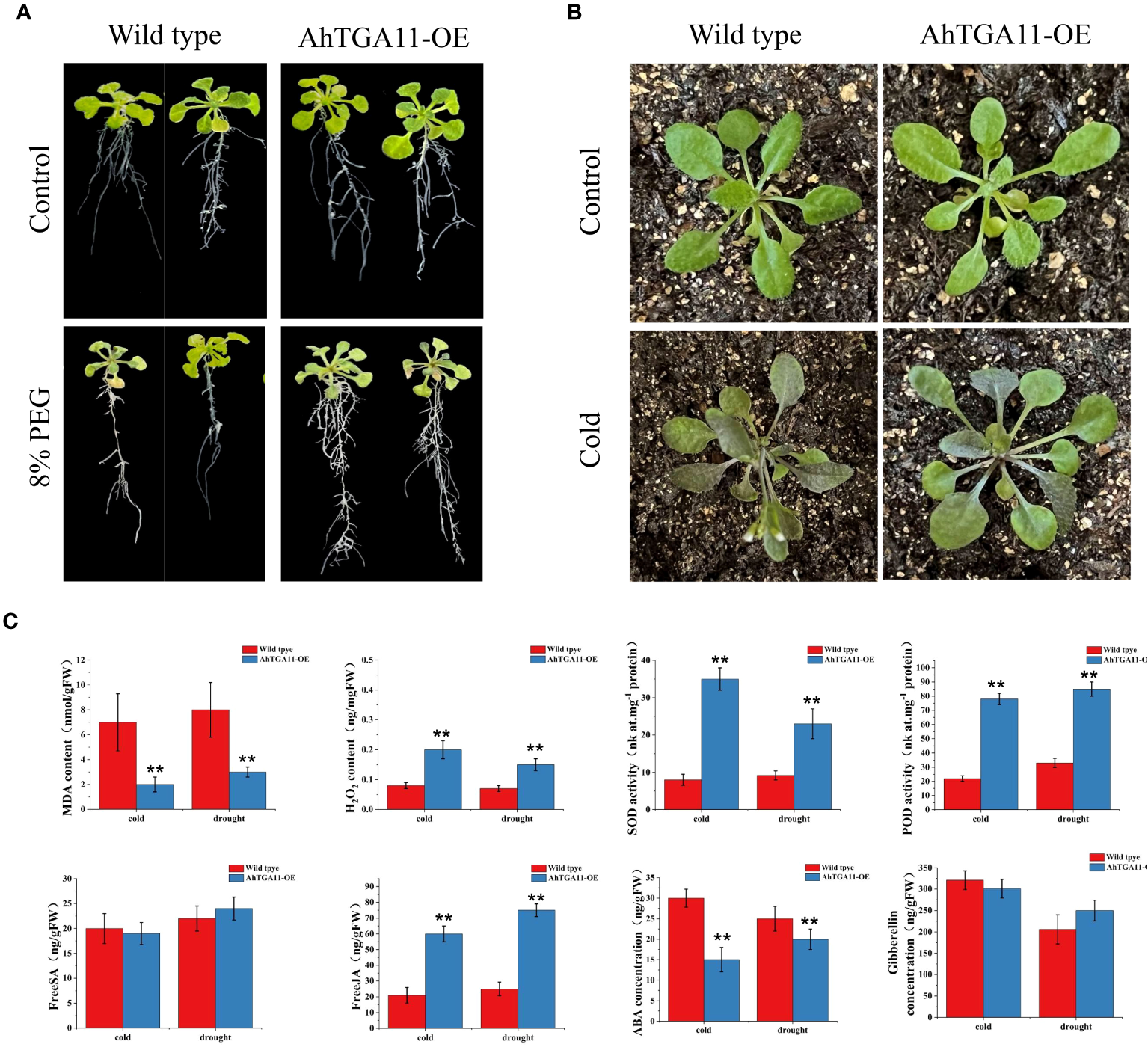
The phenotypic and physiological changes in AhTGA11-overexpressing Arabidopsis plants (AhTGA11-OE) and their wild-type under low temperature and drought stress conditions. (A) Phenotypic changes in AhTGA11-OE plants and their wild-type controls following 5 days of treatment with 8% PEG. (B) Eighteen-day-old seedlings of AhTGA11-OE plants and their wild types were subjected to 5 days of treatment at 4°C. (C) Physiological parameters and endogenous hormone alterations in AhTGA11-OE and wild-type plants under low-temperature and drought treatments. The data is expressed as the mean ± standard deviation (n = 3), and asterisks denote significant distinctions between transgenic plants and wild-type plants subjected to the same treatment, as determined by t-tests (** P < 0.01).
AhTGA11 enhances drought resistance in transgenic soybean hairy roots
AhTGA11’s sensitivity to drought stress was assessed with a focus on the root region, and this assessment was conducted through Agrobacterium rhizogenes-mediated transformation of soybean hairy roots. After 4 days of exposure to 8% PEG treatment, it was observed that overexpressing AhTGA11 did not exhibit significant changes when compared to transgenic hairy soybean roots containing an empty vector. In contrast, transgenic hairy roots containing an empty vector displayed symptoms of drought stress under PEG treatment (Figure 12A). Root tissue of overexpressing plants showed significantly higher levels of H2O2, POD activity, and SOD activity compared to control plants, while MDA content was significantly lower in overexpressing plants (Figure 12B). These trends were consistent between root tissue and non-transgenic leaf tissue. Furthermore, both root and leaf tissues of overexpressing plants exhibited significantly higher levels of endogenous SA and JA compared to control plants. However, there was no significant difference in GA content between overexpressing and wild-type plants in both root and leaf tissues.
Figure 12
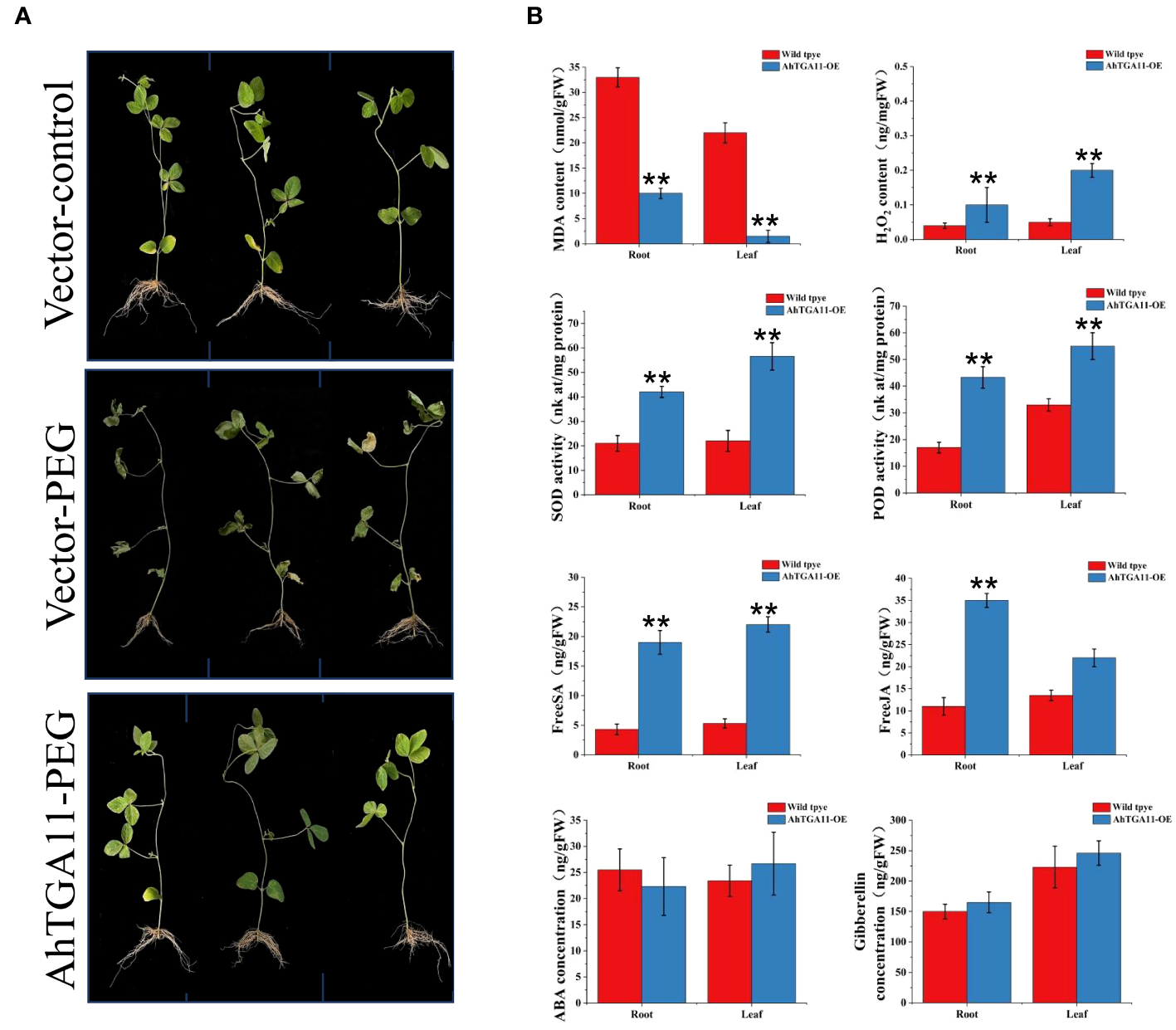
Phenotypes of AhTGA11 transgenic soybean hairy roots under drought stress. (A) Phenotypic differences between AhTGA11 transgenic soybean hair roots, empty vector transgenic soybean hair roots under PEG treatment, and their empty vector controls. (B) Physiological and endogenous hormone changes in AhTGA11 transgenic soybean hair roots and leaves without transgenic components under drought treatment. The data is expressed as the mean ± standard deviation (n = 3), and asterisks denote significant distinctions between transgenic plants and wild-type plants subjected to the same treatment, as determined by t-tests (* P < 0.05; ** P < 0.01).
Discussion
Investigating gene families of different transcription factors is of great significance for uncovering the functions of key plant genes and understanding the intricate regulatory networks governing plant processes (Feng et al., 2020; Dong et al., 2023). In recent years, extensive studies have focused on elucidating the functions of transcription factor families in peanut (Arachis hypogaea), such as the AP2/ERF, WRKY and NAC families (Song et al., 2016; Yuan et al., 2020; Cui et al., 2021). These studies have shed light on the roles of these transcription factors in peanut growth, development, and stress responses. However, despite the importance of the TGA transcription factor in other plant species, such as Arabidopsis (Kim et al., 2022), rice (Ueda et al., 2020) and soybean (Li et al., 2019a), little is known about the TGA transcription factors in peanut. To address this knowledge gap, the present study aimed to identify and characterize the TGA transcription factors in peanut and explore their potential roles in peanut growth, development, and stress responses.
A total of 20 TGA genes were identified in cultivated peanut, while 9 and 11 TGA genes were identified in wild diploid peanuts, respectively. The majority of wild peanuts exhibit a correspondence between the number of TGA genes, their chromosomal locations, and physicochemical properties with the TGA genes in the subgenomes A and B of cultivated peanut. This alignment significantly improves the accuracy of AhTGA gene identification. The identified AhTGA genes all contained representative domains, bZIP and DOG1 (Figure 2D). TGA genes belong to the D branch of the bZIP gene family, and the number of genes identified in the D branch of bZIP in wild diploid peanuts differed from the findings of this study (Wang et al., 2019c). In contrast, our study employed the latest version of the tetraploid cultivated peanut and diploid wild peanut genomes (Genome Assembly 2) data, allowing for a more efficient identification of peanut gene family members. This approach likely led to more comprehensive genome annotations (Bertioli et al., 2019) (https://www.peanutbase.org/download/). When comparing the identification results of the D group of bZIP genes in wild diploid peanuts from previous studies, we identified two additional AdTGA genes, AdTGA02 and AdTGA03, and four AiTGA genes, AiTGA03, AiTGA04, AiTGA08, and AiTGA09. These genes were also retained in the cultivated peanut genome as AhTGA04, AhTGA05, AhTGA14, AhTGA18, and AhTGA19. Notably, AhTGA04 and AhTGA18 exhibited changes in expression levels under stress and hormone treatments in this study (Figure 9), while other genes also showed expression level variations in different tissues (Supplementary Figure 4). This suggests that these genes may have corresponding functions in the growth, development, and stress response of cultivated peanuts. Thus, the utilization of the updated cultivated peanut genome data in this study facilitated the exploration of AhTGA genes. The targeted exploration of TGA genes in the bZIP gene family using newly released genome data has also been reported in other plants such as soybean and kiwifruit (Li et al., 2019a; Jin et al., 2021; Liu et al., 2022b; Yue et al., 2023).
Understanding the gene structure is of significant importance in elucidating gene function (Wang et al., 2019a). The structural features of a gene, such as the arrangement of exons and introns, promoter regions, and conserved domains, provide valuable insights into its regulatory mechanisms and functional roles (Weake and Workman, 2010). The non-coding regions of AhTGA genes exhibit significant variation, particularly in the UTR regions and introns, while the coding structure remains relatively conserved (Figure 2C). The five most conserved motifs, Motif 1-5, are present in every AhTGA gene, comprising the key structural domains of bZIP and DOG1 (Figures 2B, D). Specific motifs unique to different groups are mainly located at the N-terminus, indicating a higher diversity in the N-terminal region of TGA genes, while the C-terminal region is more conserved (Figure 2). bZIP proteins are characterized by their conserved bZIP domain, responsible for DNA binding and dimerization. The bZIP domain shows high sequence identity among Arabidopsis TGAs and is conserved across plant species (Jindrich and Degnan, 2016). The DOG1 domain, found in the C-terminus of TGAs, plays a role in seed dormancy control and potentially modulates TGA activity (Magnani et al., 2014). The presence of conserved residues implicated in Calmodulin (CaM) binding suggests a potential interaction between DOG1 and CaM, linking TGA transcriptional regulation to calcium signaling (Sall et al., 2019). The N-terminus of TGAs exhibits high variability in sequence and length, likely contributing to their functional specificity in transcriptional regulation (Katagiri et al., 1989; Tomaz et al., 2022). The TGA genes in peanut also exhibit these structural characteristics, indicating the conservation of TGA gene structure and function across different species.
Gene duplication is a key driver of gene family formation (Flagel and Wendel, 2009). No tandem duplications were found in both wild diploid and cultivated peanuts. In the genome of wild diploid peanuts, only one segment duplication was identified. However, within A. hypogaea genome, 14 pairs of segment duplications were detected. The occurrence of segment duplications is primarily attributed to the homologous genes derived from the A. duranensis and A. ipaensis, resulting in transpositions between the A and B subgenomes of cultivated peanut. Consequently, this leads to locational differences, gene duplications, or deletions among orthologous genes from A and B subgenomes within cultivated peanuts (Figure 3). Similar results have been documented in other polyploid plant species, including hexaploid wheat (Triticum aestivum) and cotton (Gossypium hirsutum) (Yousafzai et al., 2010; Paterson et al., 2012). Homologous exchange, also known as recombination, plays a vital role in creating genetic diversity and facilitating the evolution of species. Therefore, the origin of TGA genes in cultivated peanuts is primarily due to whole-genome duplication during the formation of tetraploid cultivated peanuts, as well as transposition events between the A and B subgenomes. TGA genes in peanuts lack tandem duplication and primarily originate from the retention of orthologous genes during the species evolution. The results of the synteny analysis between AhTGA genes and other species showed that AhTGA genes within the same group share the same collinearity modules (Figure 5; Supplementary Table 3). There are significant differences in collinearity of AhTGA genes across different species. For instance, AhTGA20 exhibits collinearity relationships in all dicotyledonous plants studied in this research, while AhTGA02, AhTGA04, AhTGA05, AhTGA14, and AhTGA16 show collinearity only in Arachis species. This suggests that different AhTGA genes have diverged in their formation time during evolution. In addition, all AhTGA genes have undergone purifying selection, as indicated by the Ka/Ks ratio calculation resulting in values less than 1. Combined phylogenetic analysis, gene structure, and collinearity analysis reveal that AhTGA genes have similar structures within the same clade, showing close phylogenetic and collinearity relationships with other species. Therefore, the sequence and structural analysis of AhTGA genes suggest that they may have similar functions within the same clade while exhibiting significant differences between clusters.
The expression patterns of the AhTGA genes under hormonal and abiotic stress in peanuts were studied using transcriptome data and qPCR. Significant differences in the expression of three AhTGA genes (AhTGA04, AhTGA14, and AhTGA20) in Group I and four AhTGA genes (AhTGA7, AhTGA11, AhTGA16, and AhTGA18) in Group II were observed across different peanut tissues, hormonal treatments, and stress responses (Figures 8–10). AhTGA04, AhTGA14, and AhTGA20, belonging to the same branch as the Arabidopsis TGA genes AtTGA01 and AtTGA04, are involved in SA signaling and contribute to plant resistance against pathogens through their interaction with NPR1 (Gatz, 2013; Shearer et al., 2021). In terms of abiotic stress, overexpression of AtTGA4 in Arabidopsis has been shown to enhance drought tolerance by improving nitrate transport and absorption (Zhong et al., 2015). Genetic transformation of soybean with the overexpressing group I GmTGA15 gene has been found to increase drought resistance (Chen et al., 2021). AhTGA04 and AhTGA14 are homologous genes from the A and B subgenomes, and under low-temperature conditions, the cold-tolerant genotype NH5 exhibits lower expression levels compared to the sensitive genotype FH18, whereas under drought conditions, the expression pattern is reversed (Figures 8–10). In Clade II, AhTGA07, AhTGA11, AhTGA16, and AhTGA18 exhibit higher expression levels than other AhTGA genes in peanut, and their expression patterns are altered under low temperature, drought, and hormone treatments. AtTGA2, AtTGA5, and AtTGA6, as Clade II genes, are essential for systemic acquired resistance in Arabidopsis, as they interact with NPR1 and regulate the expression of pathogenesis-related (PR) genes and pathogen resistance (Zhang et al., 2003). Low temperature and drought stress can disrupt membrane lipid metabolism (Gigon et al., 2004; Zhang X. et al., 2019). AtTGA2, AtTGA5, and AtTGA6 play critical roles in lipid stress responses (Mueller et al., 2008). These transcription factors are involved in both the SA-dependent SAR pathway against biotrophic pathogens and the JA-ethylene-dependent defense mechanism against necrotrophic pathogens (Zander et al., 2010; Zander et al., 2014). Interestingly, in this study, besides being regulated by JA and SA, we also found significant changes in the expression levels of TGA genes in response to exogenous ABA and GA treatments. While the ability of TGA genes to respond to ABA induction has been reported in soybean, the involvement of TGA genes in GA-related gene regulation is less explored (Li et al., 2019a). Therefore, further investigations are needed to elucidate the regulatory mechanisms of AhTGA11 in response to GA induction (Figure 10).
AhTGA11 was selected for functional validation in transgenic Arabidopsis and soybean hairy roots. The results demonstrated that AhTGA11 effectively alleviates plant antioxidant activity under low-temperature and drought conditions, thereby mitigating abiotic stress. However, in terms of hormone regulation, there were differences in the overexpression of AhTGA11 between transgenic Arabidopsis and soybean hairy roots. For instance, salicylic acid did not accumulate significantly in Arabidopsis, whereas ABA levels were significantly identified. Additionally, gibberellin levels did not accumulate in either transgenic soybean or Arabidopsis. This discrepancy may be attributed to the different genetic backgrounds of the two species, highlighting the complexity of TGA transcription factor regulation and the significant interspecies differences involved. The functional roles of TGAs from different clades have traditionally been associated with plant immunity for clades I, II, and III, while clades IV and V were initially implicated in developmental processes (Gatz, 2013). However, emerging evidence suggests that the functional division between clades is not as clear-cut, as studies have revealed their involvement in various biological processes. For instance, clade I TGAs have been shown to play a role in growth and development regulation (Li et al., 2019b; Wang et al., 2019b), and clade IV TGAs have been implicated in biotic stress responses (Venturuzzi et al., 2021). Functional analyses of TGAs across different clades have demonstrated their significance not only in biotic stress responses (Kesarwani et al., 2007; Sun et al., 2018) but also in the regulation of gene expression related to abiotic stress responses (Fang et al., 2017; Herrera-Vasquez et al., 2021), developmental processes (Maier et al., 2011), circadian rhythm (Zhou et al., 2015), detoxification (Fode et al., 2008; Mueller et al., 2008; Herrera-Vasquez et al., 2021), nitrate signaling (Alvarez et al., 2014; Canales et al., 2017), flowering (Thurow et al., 2005; Xu et al., 2021), and autophagy (Wang et al., 2020). Although we conducted in silico and expression patterns analysis of AhTGA genes, and functional identification of AhTGA11 in other plants species, it is important to note that TGA genes hold a pivotal position in various facets of plant regulatory pathways, encompassing growth, development, and stress responses. As peanuts are a tetraploid crop with a complex genetic backgroud, this complexity may result in increased functional redundancy and evolutionary adaptations. Consequently, additional research efforts are warranted to thoroughly characterize the functions of each AhTGA gene in cultivated peanut.
Conclusion
In our study, a comprehensive in silico analysis was conducted to investigate the AhTGA gene family in cultivated peanut, including evolutionary analysis, gene structure examination, identification of regulatory elements, prediction of protein-protein interactions, and identification of miRNA targets. A total of 20 AhTGA genes were identified and classified into five groups. Differential expression patterns of AhTGA genes in different tissues, under abiotic stress conditions such as low temperature and drought, and in response to hormonal stimuli were analyzed. AhTGA11 was chosen for functional validation in transgenic Arabidopsis plants and soybean transgenic hairy roots. It was found to have a positive role on both low-temperature and drought responses, involving regulation by SA, JA, and ABA, mitigating the oxidative stress generated in plants due to stress. These findings provide valuable insights into the functional characterization of AhTGA genes and can guide the breeding of novel abiotic-resistant peanut varieties.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
CZ: Conceptualization, Data curation, Funding acquisition, Software, Validation, Writing – original draft, Writing – review & editing. YL: Data curation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ZL: Data curation, Software, Visualization, Writing – review & editing. XW: Methodology, Supervision, Writing – review & editing. CJ: Investigation, Methodology, Supervision, Writing – review & editing. XZ: Data curation, Investigation, Supervision, Writing – review & editing. SK: Formal Analysis, Investigation, Project administration, Writing – review & editing. XL: Formal Analysis, Investigation, Project administration, Supervision, Writing – review & editing. SZ: Project administration, Supervision, Writing – review & editing. JW: Data curation, Formal Analysis, Methodology, Writing – review & editing. HZ: Data curation, Software, Writing – review & editing. YH: Formal Analysis, Investigation, Supervision, Writing – review & editing. HY: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. RX: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the earmarked fund for CARS-13, Liaoning Provincial Natural Science Foundation (2023-MS-214), Science and Technology Program of Shenyang (No.21-110-3-17) and Disciplinary construction project of Liaoning Academy of Agricultural Sciences (2022DD030905; 2022-HBZ-1010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1269200/full#supplementary-material
References
1
Alvarez J. M. Riveras E. Vidal E. A. Gras D. E. Contreras-Lopez O. Tamayo K. P. et al . (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J.80 (1), 1–13. doi: 10.1111/tpj.12618
2
Baillo E. H. Kimotho R. N. Zhang Z. Xu P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (Basel)10 (10), 771. doi: 10.3390/genes10100771
3
Bertioli D. J. Cannon S. B. Froenicke L. Huang G. Farmer A. D. Cannon E. K. et al . (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet.48 (4), 438–446. doi: 10.1038/ng.3517
4
Bertioli D. J. Jenkins J. Clevenger J. Dudchenko O. Gao D. Seijo G. et al . (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet.51 (5), 877–884. doi: 10.1038/s41588-019-0405-z
5
Borrill P. Harrington S. A. Simmonds J. Uauy C. (2019). Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. Plant Physiol.180 (3), 1740–1755. doi: 10.1104/pp.19.00380
6
Budimir J. Treffon K. Nair A. Thurow C. Gatz C. (2021). Redox-active cysteines in TGACG-BINDING FACTOR 1 (TGA1) do not play a role in salicylic acid or pathogen-induced expression of TGA1-regulated target genes in Arabidopsis thaliana. New Phytol.230 (6), 2420–2432. doi: 10.1111/nph.16614
7
Cai B. D. Zhu J. X. Gao Q. Luo D. Yuan B. F. Feng Y. Q. (2014). Rapid and high-throughput determination of endogenous cytokinins in Oryza sativa by bare Fe3O4 nanoparticles-based magnetic solid-phase extraction. J. Chromatogr. A1340, 146–150. doi: 10.1016/j.chroma.2014.03.030
8
Canales J. Contreras-Lopez O. Alvarez J. M. Gutierrez R. A. (2017). Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J.92 (2), 305–316. doi: 10.1111/tpj.13656
9
Chen C. Chen H. Zhang Y. Thomas H. R. Frank M. H. He Y. et al . (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
10
Chen Z. Fang X. Yuan X. Zhang Y. Li H. Zhou Y. et al . (2021). Overexpression of transcription factor gmTGA15 enhances drought tolerance in transgenic soybean hairy roots and arabidopsis plants. Agronomy11 (1), 170. doi: 10.3390/agronomy11010170
11
Choi J. Huh S. U. Kojima M. Sakakibara H. Paek K. H. Hwang I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell19 (2), 284–295. doi: 10.1016/j.devcel.2010.07.011
12
Clevenger J. Chu Y. Scheffler B. Ozias-Akins P. (2016). A developmental transcriptome map for allotetraploid arachis hypogaea. Front. Plant Sci.7. doi: 10.3389/fpls.2016.01446
13
Cui M. Haider M. S. Chai P. Guo J. Du P. Li H. et al . (2021). Genome-wide identification and expression analysis of AP2/ERF transcription factor related to drought stress in cultivated peanut (Arachis hypogaea L.). Front. Genet.12. doi: 10.3389/fgene.2021.750761
14
Do H. M. Hong J. K. Jung H. W. Kim S. H. Ham J. H. Hwang B. K. (2003). Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Mol. Plant-Microbe Interact.16 (3), 196–205. doi: 10.1094/MPMI.2003.16.3.196
15
Dong X. Han B. Yin X. Mao P. Luo D. Zhou Q. et al . (2023). Genome-wide identification of the GRAS transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under drought stress. Ind. Crops Products194, 116379. doi: 10.1016/j.indcrop.2023.116379
16
Dormatey R. Sun C. Ali K. Coulter J. A. Bi Z. Bai J. (2020). Gene pyramiding for sustainable crop improvement against biotic and abiotic stresses. Agronomy10 (9), 1255. doi: 10.3390/agronomy10091255
17
Dwivedi S. Sahrawat K. Upadhyaya H. Ortiz R. (2013). Food, nutrition and agrobiodiversity under global climate change. Adv. Agron.120, 1–128. doi: 10.1016/B978-0-12-407686-0.00001-4
18
Fang H. Liu Z. Long Y. Liang Y. Jin Z. Zhang L. et al . (2017). The Ca(2+) /calmodulin2-binding transcription factor TGA3 elevates LCD expression and H(2) S production to bolster Cr(6+) tolerance in Arabidopsis. Plant J.91 (6), 1038–1050. doi: 10.1111/tpj.13627
19
Feng K. Hou X. L. Xing G. M. Liu J. X. Duan A. Q. Xu Z. S. et al . (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol.40 (6), 750–776. doi: 10.1080/07388551.2020.1768509
20
Fitzgerald H. A. Canlas P. E. Chern M. S. Ronald P. C. (2005). Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J.43 (3), 335–347. doi: 10.1111/j.1365-313X.2005.02457.x
21
Flagel L. E. Wendel J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol.183 (3), 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
22
Fode B. Siemsen T. Thurow C. Weigel R. Gatz C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell20 (11), 3122–3135. doi: 10.1105/tpc.108.058974
23
Fonseca A. Urzua T. Jelenska J. Sbarbaro C. Seguel A. Duarte Y. et al . (2022). The TGA transcription factors from clade II negatively regulate the salicylic acid accumulation in arabidopsis. Int. J. Mol. Sci.23 (19), 11631. doi: 10.3390/ijms231911631
24
Fraire-Velázquez S. Rodríguez-Guerra R. Sánchez-Calderón L. (2011). “Abiotic and biotic stress response crosstalk in plants,” in Abiotic stress response in plants—physiological, biochemical and genetic perspectives. (Croatia: InTech), 3–26. doi: 10.5772/23217
25
Gatz C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant-Microbe Interact.26 (2), 151–159. doi: 10.1094/MPMI-04-12-0078-IA
26
Gigon A. Matos A. R. Laffray D. Zuily-Fodil Y. Pham-Thi A. T. (2004). Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot.94 (3), 345–351. doi: 10.1093/aob/mch150
27
He Z. Zhang H. Gao S. Lercher M. J. Chen W. H. Hu S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res.44 (W1), W236–W241. doi: 10.1093/nar/gkw370
28
Herrera-Vasquez A. Fonseca A. Ugalde J. M. Lamig L. Seguel A. Moyano T. C. et al . (2021). TGA class II transcription factors are essential to restrict oxidative stress in response to UV-B stress in Arabidopsis. J. Exp. Bot.72 (5), 1891–1905. doi: 10.1093/jxb/eraa534
29
Hrmova M. Hussain S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci.22 (11), 5662. doi: 10.3390/ijms22115662
30
Innes R. (2018). The positives and negatives of NPR: A unifying model for salicylic acid signaling in plants. Cell173 (6), 1314–1315. doi: 10.1016/j.cell.2018.05.034
31
Jakoby M. Weisshaar B. Dröge-Laser W. Vicente-Carbajosa J. Tiedemann J. Kroj T. et al . (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci.7 (3), 106–111. doi: 10.1016/S1360-1385(01)02223-3
32
Javed T. Shabbir R. Ali A. Afzal I. Zaheer U. Gao S. J. (2020). Transcription factors in plant stress responses: challenges and potential for sugarcane improvement. Plants (Basel)9 (4), 491. doi: 10.3390/plants9040491
33
Jiang C. Zhang H. Ren J. Dong J. Zhao X. Wang X. et al . (2020). Comparative transcriptome-based mining and expression profiling of transcription factors related to cold tolerance in peanut. Int. J. Mol. Sci.21 (6), 1921. doi: 10.3390/ijms21061921
34
Jin H. Choi S. M. Kang M. J. Yun S. H. Kwon D. J. Noh Y. S. et al . (2018). Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res.46 (22), 11712–11725. doi: 10.1093/nar/gky847
35
Jin M. Gan S. Jiao J. He Y. Liu H. Yin X. et al . (2021). Genome-wide analysis of the bZIP gene family and the role of AchnABF1 from postharvest kiwifruit (Actinidia chinensis cv. Hongyang) in osmotic and freezing stress adaptations. Plant Sci.308, 110927. doi: 10.1016/j.plantsci.2021.110927
36
Jindrich K. Degnan B. M. (2016). The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol.16, 28. doi: 10.1186/s12862-016-0598-z
37
Johnson C. Boden E. Arias J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell15 (8), 1846–1858. doi: 10.1105/tpc.012211
38
Katagiri F. Lam E. Chua N.-H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature340, 727–730. doi: 10.1038/340727a0
39
Kereszt A. Li D. Indrasumunar A. Nguyen C. D. Nontachaiyapoom S. Kinkema M. et al . (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc.2 (4), 948–952. doi: 10.1038/nprot.2007.141
40
Kesarwani M. Yoo J. Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol.144 (1), 336–346. doi: 10.1104/pp.106.095299
41
Kidokoro S. Shinozaki K. Yamaguchi-Shinozaki K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci.27 (9), 922–935. doi: 10.1016/j.tplants.2022.01.008
42
Kim Y. W. Youn J. H. Roh J. Kim J. M. Kim S. K. Kim T. W. (2022). Brassinosteroids enhance salicylic acid-mediated immune responses by inhibiting BIN2 phosphorylation of clade I TGA transcription factors in Arabidopsis. Mol. Plant15 (6), 991–1007. doi: 10.1016/j.molp.2022.05.002
43
Larkin M. A. Blackshields G. Brown N. P. Chenna R. McGettigan P. A. McWilliam H. et al . (2007). Clustal W and clustal X version 2.0. Bioinformatics23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
44
Latchman D. S. (2001). Transcription factors: bound to activate or repress. Trends Biochem. Sci.26 (4), 211–213. doi: 10.1016/S0968-0004(01)01812-6
45
Li B. Liu Y. Cui X. Y. Fu J. D. Zhou Y. B. Zheng W. J. et al . (2019a). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci.10. doi: 10.3389/fpls.2019.00549
46
Li N. Muthreich M. Huang L. J. Thurow C. Sun T. Zhang Y. et al . (2019b). TGACG-BINDING FACTORs (TGAs) and TGA-interacting CC-type glutaredoxins modulate hyponastic growth in Arabidopsis thaliana. New Phytol.221 (4), 1906–1918. doi: 10.1111/nph.15496
47
Li X. Wang Q. Guo C. Sun J. Li Z. Wang Y. et al . (2022). NtNAC053, A novel NAC transcription factor, confers drought and salt tolerances in tobacco. Front. Plant Sci.13. doi: 10.3389/fpls.2022.817106
48
Li Y. Zhou C. Yan X. Zhang J. Xu J. (2016). Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Separation Sci.39 (10), 1804–1813. doi: 10.1002/jssc.201501239
49
Liu C. Liu Q. Mou Z. (2022a). A direct link between BR and SA signaling: Negative regulation of TGA4 by BIN2. Mol. Plant15 (8), 1254–1256. doi: 10.1016/j.molp.2022.06.006
50
Liu W. Zhao C. Liu L. Huang D. Ma C. Li R. et al . (2022b). Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection. Int. J. Biol. Macromol.222, 101–113. doi: 10.1016/j.ijbiomac.2022.09.154
51
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25 (4), 402–408. doi: 10.1006/meth.2001.1262
52
Magnani E. de Klein N. Nam H. I. Kim J. G. Pham K. Fiume E. et al . (2014). A comprehensive analysis of microProteins reveals their potentially widespread mechanism of transcriptional regulation. Plant Physiol.165 (1), 149–159. doi: 10.1104/pp.114.235903
53
Maier A. T. Stehling-Sun S. Offenburger S. L. Lohmann J. U. (2011). The bZIP transcription factor PERIANTHIA: A multifunctional hub for meristem control. Front. Plant Sci.2. doi: 10.3389/fpls.2011.00079
54
Mueller S. Hilbert B. Dueckershoff K. Roitsch T. Krischke M. Mueller M. J. et al . (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell20 (3), 768–785. doi: 10.1105/tpc.107.054809
55
Murmu J. Bush M. J. DeLong C. Li S. Xu M. Khan M. et al . (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol.154 (3), 1492–1504. doi: 10.1104/pp.110.159111
56
Nakashima K. Yamaguchi-Shinozaki K. Shinozaki K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci.5. doi: 10.3389/fpls.2014.00170
57
Nijhawan A. Jain M. Tyagi A. K. Khurana J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol.146 (2), 333–350. doi: 10.1104/pp.107.112821
58
Noshi M. Mori D. Tanabe N. Maruta T. Shigeoka S. (2016). Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flg22. Plant Sci.252, 12–21. doi: 10.1016/j.plantsci.2016.06.019
59
Pan X. Welti R. Wang X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc.5 (6), 986–992. doi: 10.1038/nprot.2010.37
60
Paterson A. H. Wendel J. F. Gundlach H. Guo H. Jenkins J. Jin D. et al . (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature492 (7429), 423–427. doi: 10.1038/nature11798
61
Puppala N. Nayak S. N. Sanz-Saez A. Chen C. Devi M. J. Nivedita N. et al . (2023). Sustaining yield and nutritional quality of peanuts in harsh environments: Physiological and molecular basis of drought and heat stress tolerance. Front. Genet.14. doi: 10.3389/fgene.2023.1121462
62
Ren J. Guo P. Zhang H. Shi X. Ai X. Wang J. et al . (2022). Comparative physiological and coexpression network analyses reveal the potential drought tolerance mechanism of peanut. BMC Plant Biol.22 (1), 460. doi: 10.1186/s12870-022-03848-7
63
Sagisaka S. (1976). The occurrence of peroxide in a perennial plant, Populus gelrica. Plant Physiol.57 (2), 308–309. doi: 10.1104/pp.57.2.308
64
Sall K. Dekkers B. J. W. Nonogaki M. Katsuragawa Y. Koyari R. Hendrix D. et al . (2019). DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. Plant J.100 (1), 7–19. doi: 10.1111/tpj.14485
65
Shannon P. Markiel A. Ozier O. Baliga N. S. Wang J. T. Ramage D. et al . (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13 (11), 2498–2504. doi: 10.1101/gr.1239303
66
Shearer H. L. Yu T. C. Lipu W. Jinman L. Patrick B. Charles D. et al . (2021). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant-Microbe Interact.25 (11), 1459–1468. doi: 10.1094/MPMI-09-11-0256
67
Song H. Wang P. Lin J. Y. Zhao C. Bi Y. Wang X. (2016). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci.7. doi: 10.3389/fpls.2016.00534
68
Song L. Li W. Chen X. (2022). Transcription factor is not just a transcription factor. Trends Plant Sci.27 (11), 1087–1089. doi: 10.1016/j.tplants.2022.08.001
69
Srivastava R. Kobayashi Y. Koyama H. Sahoo L. (2023). Cowpea NAC1/NAC2 transcription factors improve growth and tolerance to drought and heat in transgenic cowpea through combined activation of photosynthetic and antioxidant mechanisms. J. Integr. Plant Biol.65 (1), 25–44. doi: 10.1111/jipb.13365
70
Sun T. Busta L. Zhang Q. Ding P. Jetter R. Zhang Y. (2018). TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol.217 (1), 344–354. doi: 10.1111/nph.14780
71
Syed F. Arif S. Ahmed I. Khalid N. (2021). “Groundnut (peanut) (Arachis hypogaea),” in Oilseeds: health attributes and food applications. (Singapore: Springer Nature Singapore Pte Ltd.), 93–122. doi: 10.1007/978-981-15-4194-0_4
72
Tamura K. Stecher G. Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38 (7), 3022–3027. doi: 10.1093/molbev/msab120
73
Thurow C. Schiermeyer A. Krawczyk S. Butterbrodt T. Nickolov K. Gatz C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J.44 (1), 100–113. doi: 10.1111/j.1365-313X.2005.02513.x
74
Tomaz S. Gruden K. Coll A. (2022). TGA transcription factors-Structural characteristics as basis for functional variability. Front. Plant Sci.13. doi: 10.3389/fpls.2022.935819
75
Ueda Y. Ohtsuki N. Kadota K. Tezuka A. Nagano A. J. Kadowaki T. et al . (2020). Gene regulatory network and its constituent transcription factors that control nitrogen-deficiency responses in rice. New Phytol.227 (5), 1434–1452. doi: 10.1111/nph.16627
76
Valliyodan B. Nguyen H. T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol.9 (2), 189–195. doi: 10.1016/j.pbi.2006.01.019
77
Venturuzzi A. L. Rodriguez M. C. Conti G. Leone M. Caro M. D. P. Montecchia J. F. et al . (2021). Negative modulation of SA signaling components by the capsid protein of tobacco mosaic virus is required for viral long-distance movement. Plant J.106 (4), 896–912. doi: 10.1111/tpj.15268
78
Wang S. Alseekh S. Fernie A. R. Luo J. (2019a). The structure and function of major plant metabolite modifications. Mol. Plant12 (7), 899–919. doi: 10.1016/j.molp.2019.06.001
79
Wang Z. Cheng K. Wan L. Yan L. Jiang H. Liu S. et al . (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics16, 1053. doi: 10.1186/s12864-015-2258-x
80
Wang P. Nolan T. M. Yin Y. Bassham D. C. (2020). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy16 (1), 123–139. doi: 10.1080/15548627.2019.1598753
81
Wang Y. Salasini B. C. Khan M. Devi B. Bush M. Subramaniam R. et al . (2019b). Clade I TGACG-motif binding basic leucine zipper transcription factors mediate BLADE-ON-PETIOLE-dependent regulation of development. Plant Physiol.180 (2), 937–951. doi: 10.1104/pp.18.00805
82
Wang Z. Yan L. Wan L. Huai D. Kang Y. Shi L. et al . (2019c). Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genomics20 (1), 1–14. doi: 10.1186/s12864-019-5434-6
83
Wang D. Zhang Y. Zhang Z. Zhu J. Yu J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf.8 (1), 77–80. doi: 10.1016/S1672-0229(10)60008-3
84
Weake V. M. Workman J. L. (2010). Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet.11 (6), 426–437. doi: 10.1038/nrg2781
85
Xu X. Xu J. Yuan C. Hu Y. Liu Q. Chen Q. et al . (2021). Characterization of genes associated with TGA7 during the floral transition. BMC Plant Biol.21 (1), 367. doi: 10.1186/s12870-021-03144-w
86
Yousafzai F. K. Al-Kaff N. Moore G. (2010). The molecular features of chromosome pairing at meiosis: the polyploid challenge using wheat as a reference. Funct. Integr. Genomics10 (2), 147–156. doi: 10.1007/s10142-010-0171-6
87
Yuan C. Li C. Lu X. Zhao X. Yan C. Wang J. et al . (2020). Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol.20 (1), 454. doi: 10.1186/s12870-020-02678-9
88
Yue L. Pei X. Kong F. Zhao L. Lin X. (2023). Divergence of functions and expression patterns of soybean bZIP transcription factors. Front. Plant Sci.14, 1150363. doi: 10.3389/fpls.2023.1150363
89
Zander M. La Camera S. Lamotte O. Metraux J. P. Gatz C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J.61 (2), 200–210. doi: 10.1111/j.1365-313X.2009.04044.x
90
Zander M. Thurow C. Gatz C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol.165 (4), 1671–1683. doi: 10.1104/pp.114.243360
91
Zhang H. Gao X. Zhi Y. Li X. Zhang Q. Niu J. (2019). A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol.223 (4), 1918–1936. doi: 10.1111/nph.15925
92
Zhang H. Jiang C. Ren J. Dong J. Shi X. Zhao X. et al . (2020). An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Front. Plant Sci.11. doi: 10.3389/fpls.2020.01110
93
Zhang Y. Tessaro M. J. Lassner M. Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell15 (11), 2647–2653. doi: 10.1105/tpc.014894
94
Zhang X. Xu Y. Huang B. . (2019). Lipidomic reprogramming associated with drought stress priming-enhanced heat tolerance in tall fescue (Festuca arundinacea). Plant Cell Environ.42 (3), 947–958. doi: 10.1111/pce.13405
95
Zhang H. Yu Y. Wang S. Yang J. Ai X. Zhang N. et al . (2023). Genome-wide characterization of phospholipase D family genes in allotetraploid peanut and its diploid progenitors revealed their crucial roles in growth and abiotic stress responses. Front. Plant Sci.14. doi: 10.3389/fpls.2023.1102200
96
Zhong L. Chen D. Min D. Li W. Xu Z. Zhou Y. et al . (2015). AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun.457 (3), 433–439. doi: 10.1016/j.bbrc.2015.01.009
97
Zhou M. Wang W. Karapetyan S. Mwimba M. Marques J. Buchler N. E. et al . (2015). Redox rhythm reinforces the circadian clock to gate immune response. Nature523 (7561), 472–476. doi: 10.1038/nature14449
98
Zhuang W. Chen H. Yang M. Wang J. Pandey M. K. Zhong C. et al . (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet.51 (5), 865–876. doi: 10.1038/s41588-019-0402-2
Summary
Keywords
TGA genes, peanut (Arachis hypogaea), abiotic stress, hormone signaling, transcription factors
Citation
Zhong C, Liu Y, Li Z, Wang X, Jiang C, Zhao X, Kang S, Liu X, Zhao S, Wang J, Zhang H, Huang Y, Yu H and Xue R (2023) Genome-wide analysis reveals regulatory mechanisms and expression patterns of TGA genes in peanut under abiotic stress and hormone treatments. Front. Plant Sci. 14:1269200. doi: 10.3389/fpls.2023.1269200
Received
29 July 2023
Accepted
06 November 2023
Published
21 November 2023
Volume
14 - 2023
Edited by
Sushil Satish Chhapekar, University of Missouri, United States
Reviewed by
Kusum Khatri, Ben-Gurion University of the Negev, Israel; Yan Liying, Chinese Academy of Agricultural Sciences, China
Updates
Copyright
© 2023 Zhong, Liu, Li, Wang, Jiang, Zhao, Kang, Liu, Zhao, Wang, Zhang, Huang, Yu and Xue.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqiu Yu, yuhaiqiu@syau.edu.cn; Renfeng Xue, xuerf82@163.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.