- 1Northeast Agricultural University, Agricultural College, Harbin, China
- 2Heilongjiang Academy of Green Food Science/National Soybean Engineering Technology Research Center, Harbin, China
Soybean is an important food and oil crop widely cultivated globally. However, water deficit can seriously affect the yield and quality of soybeans. In order to ensure the stability and increase of soybean yield and improve agricultural water use efficiency (WUE), research on improving drought tolerance and the efficiency of water utilization of soybeans under drought stress has become particularly important. This study utilized the drought-tolerant variety Heinong 44 (HN44) and the drought-sensitive variety Suinong 14 (SN14) to analyze physiological responses and transcriptome changes during the gradual water deficit at the early seed-filling stage. The results indicated that under drought conditions, HN44 had smaller stomata, higher stomatal density, and lower stomatal conductance (Gs) and transpiration rate as compared to SN14. Additionally, HN44 had a higher abscisic acid (ABA) content and faster changes in stomatal morphology and Gs to maintain a dynamic balance between net photosynthetic rate (Pn) and Gs. Additionally, drought-tolerant variety HN44 had high instantaneous WUE under water deficit. Further, HN44 retained a high level of superoxide dismutase (SOD) activity and proline content, mitigating malondialdehyde (MDA) accumulation and drought-induced damage. Comprehensive analysis of transcriptome data revealed that HN44 had fewer differentially expressed genes (DEGs) under light drought stress, reacting insensitivity to water deficit. At the initial stage of drought stress, both varieties had a large number of upregulated DEGs to cope with the drought stress. Under severe drought stress, HN44 had fewer downregulated genes enriched in the photosynthesis pathway than SN14, while it had more upregulated genes enriched in the ABA-mediated signaling and glutathione metabolism pathways than SN14. During gradual water deficit, HN44 demonstrated better drought-tolerant physiological characteristics and water use efficiency than SN14 through key DEGs such as GmbZIP4, LOC100810474, and LOC100819313 in the major pathways. Key transcription factors were screened and identified, providing further clarity on the molecular regulatory pathways responsible for the physiological differences in drought tolerance among these varieties. This study deepened the understanding of the drought resistance mechanisms in soybeans, providing valuable references for drought-resistant soybean breeding.
1 Introduction
Soybean (Glycine max L.) is a globally cultivated oilseed crop and serves as a significant source of protein and fat (Liu et al., 2020). As an important source of protein and edible oil, soybean consumption demand has skyrocketed, and the supply and demand gap is widening. Therefore, improving soybean yield and increasing benefits are still important directions for soybean research. However, due to global climate change and increasing water scarcity, drought has emerged as one of the primary abiotic stress factors, which inhibits the potential for a high yield of soybeans (Lian et al., 2021). Thus, it is urgent to study the stress tolerance and resistance of soybeans to enhance their ability to cope with drought.
Drought has a range of impacts on plants, encompassing alterations in the internal structure of leaves and photosynthesis, variation in osmotic regulation systems, disruption of hormone homeostasis, production of reactive oxygen species (ROS), and damage to cell membranes (Xu et al., 2010). Drought stress inhibits leaf photosynthesis by affecting stomatal movement, resulting in reduced photosynthetic rate, stomatal conductance, and intercellular CO2 concentration. Therefore, plants have evolved complex physiological and molecular mechanisms to adapt to drought stress (Chimungu et al., 2014). Phytohormones, particularly abscisic acid (ABA), play a pivotal role in defending against abiotic stresses (Vishwakarma et al., 2017). ABA levels significantly escalate under water-deficient conditions, and ABA receptors PYR1/PYL/RCAR bind to ABA and target protein phosphatase type 2C (PP2C). This release activates the kinase activity of Snf1-related kinase 2 (SnRK2), which then phosphorylates downstream transcription factors and transporters to activate ABA reactions and improve plant stress resistance (Ohashi et al., 2006; Wang et al., 2021). Plants integrate multiple environmental signals to optimize stomatal aperture and density in different environments to maintain high water use efficiency (WUE) (Bertolino et al., 2019). Research has demonstrated that targeted changes in stomatal development can improve the WUE and drought resistance of crop varieties (Yin et al., 2017). In response to drought stress, plants also activate antioxidant and osmoregulation systems, mitigating drought-induced damage by increasing antioxidant enzyme activity and osmoregulation protectants such as proline (Dien et al., 2019; Wang et al., 2022a). Additionally, the increase in superoxide dismutase (SOD) activity and peroxidase (POD) activity enhances the metabolism of glutathione, which can eliminate excessive ROS in the plant, inhibit the accumulation of malondialdehyde, and reduce the damage caused by drought (Zhang et al., 2020). These physiological changes are coordinated by drought-related functional and regulatory genes. Functional genes, such as those associated with photosynthesis and antioxidant enzymes, are direct response genes that respond to perceived micro-environmental changes under stress. Conversely, regulatory genes are indirect response genes that facilitate stress adaptation through signaling pathways. These genes include transcription factors and protein kinases (Shinozaki and Yamaguchi-Shinozaki, 2006). Some genes and metabolites have been demonstrated to play roles in drought regulation. For example, under drought stress, soybeans enhanced their prevention of reactive oxygen damage and tolerance to abiotic stress by upregulating or downregulating the expression of ascorbic acid and glutathione-related proteins at the protein level (Zhou et al., 2008). Additionally, some transcription factors have been confirmed to be the main players in water stress signals, including MYB, bHLH, AP2/EREBP, bZip, and NAC. These transcription factors respond to drought through various pathways, including signaling cell motility pathways and metabolic synthesis (Lata and Prasad, 2011; Baldoni et al., 2015; Joshi et al., 2016; Ariyarathne and Wone, 2022).
Various omics have shed light on the responses of plants to abiotic stresses (Basim et al., 2021). For instance, RNA sequencing (RNA-seq) is a high-throughput sequencing technology with high accuracy and low cost and has been widely used in gene expression analysis in plants for evaluating the interactions between plants and abiotic stresses (Zhang and Song, 2017). Yang et al (2023) conducted a transcriptome analysis of leaf tissues under drought stress in the drought resistance soybean SS-2 and the drought-sensitive soybean Taekwang, and they discovered a significant increase in the expression of phosphatidylinositol 4-phosphate 5-kinase (PIP5K), which is involved in lipid signaling pathways, in SS2-2 (Yang et al., 2023). Additionally, Wang et al (2022b) carried out transcriptome and metabolome analyses on the drought-resistance variety Heinong 44 (HN44) and the drought-sensitive variety Heinong 65 (HN65), and they discovered that in the soybean drought resistance pathway, the tricarboxylic acid (TCA) cycle was key in mediating processes such as hormone transport and amino acid synthesis (Wang et al., 2022b). However, the complex network regulation of soybeans requires further exploration to discover more genes for drought tolerance and water use efficiency.
Previously, much research has been conducted on the transcriptomics of soybeans under drought stress. However, most of these studies have been limited to individual varieties, with less emphasis on the differences in drought tolerance among different varieties. A study on HN44 and Suinong 14 (SN14) soybean varieties selected by Jiang et al. (2022) was conducted to understand their response mechanisms under gradual water deficit during the early seed-filling stage (Jiang et al., 2022). Thus, building on the aforementioned research background and research foundation of our research group, this study analyzed the physiology characteristics and transcriptome responses of different soybean varieties under water deficit conditions. Specifically, we aimed to identify key differentially expressed genes involved in regulatory pathways related to drought tolerance and explore potential transcription factors that could improve drought resistance and water use efficiency in soybeans. This study will improve our understanding of the physiological responses and molecular mechanisms of different soybean varieties under water deficit conditions.
2 Materials and methods
2.1 Plant materials and stress treatments
In this study, the drought-tolerant variety HN44 and the drought-sensitive SN14 were tested in 2021 at the glass canopy of Northeast Agricultural University. Soybean seeds with full grains and uniform size were selected for sowing in 1.5-gallon pots with a dimension of 17.5 cm top diameter × 14 cm bottom diameter × 16 cm height with three biological replicates for each treatment. Before sowing, the weight of each empty pot was recorded. Subsequently, the soil in each pot was saturated with deionized water and allowed to drain overnight. The weight of the pot soil water capacity (PSWC) was the total weight of the soil mixture and water and was recorded and considered as 100% of the PSWC (100%PSWC) at that time (Bramley et al., 2022). Two seeds were sown in each pot, and one seedling was retained in each pot after germination. The pots were bundled with black plastic bags to prevent surface moisture evaporation and loss. From sowing to the early seed-filling stage, the pots’ weight was measured daily and watered to 70%PSWC to maintain normal plant moisture (Jiang et al., 2020). At the early stage of seed-filling (R5), a completely randomized experimental design consisted of drought treatment for each variety, and the PSWC of each pot was monitored and adjusted daily by weighing. Drought treatments included normal water (NW; 70%PSWC), light drought (LD; 60%PSWC), moderate drought (MD; 55%PSWC), and severe drought (SD; 50%PSWC), forming a gradual water deficit. Each water deficit treatment lasted for 1 day (Figure 1).
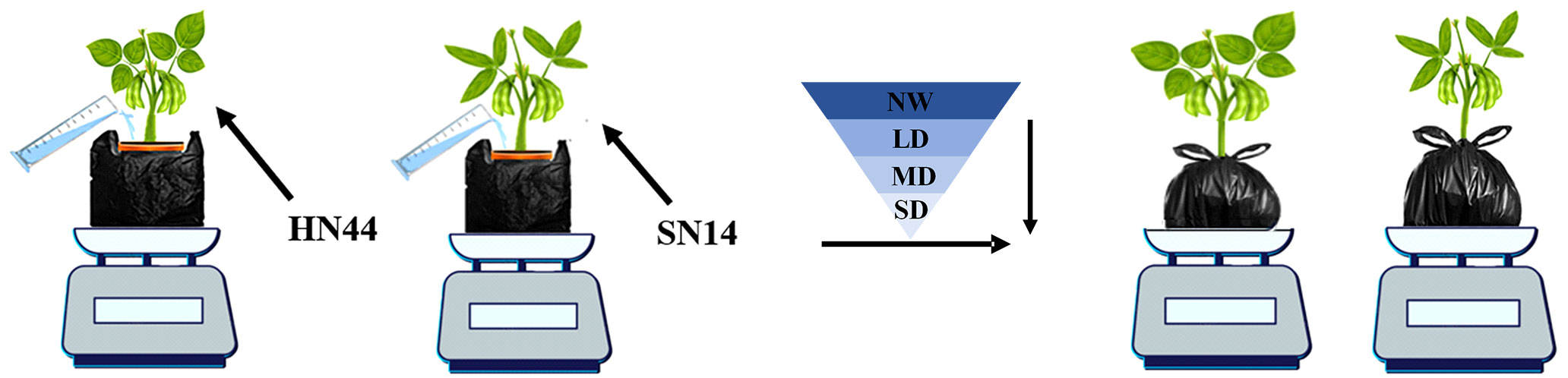
Figure 1 Gradual water deficit treatments. HN44, drought-tolerant variety Heinong 44; SN14, drought- sensitive Suinong 14; NW, normal water, 70%PSWC; LD, light drought, 60%PSWC; MD, moderate drought, 55%PSWC; SD, severe drought, 50%PSWC. The same as below.
2.2 Sampling and measurement
2.2.1 Photosynthetic characteristics
After treatment, the third fully expanded trifoliate leaves at the tip of the main stem were measured. These measurements were conducted between 8:00 AM and 10:00 AM on a sunny day using a Cl-340 photosynthesizer (CID Bio-Science, Inc., Camas, WA, USA). The photosynthetic characteristics assessed included net photosynthetic rate (Pn), stomatal conductance (Gs), transpiration rate (Tr), and intercellular carbon dioxide concentration (Ci), with five replicates for each treatment. The instantaneous water use efficiency (WUEi) was calculated as the ratio of Pn to Tr.
2.2.2 Stomatal parameters
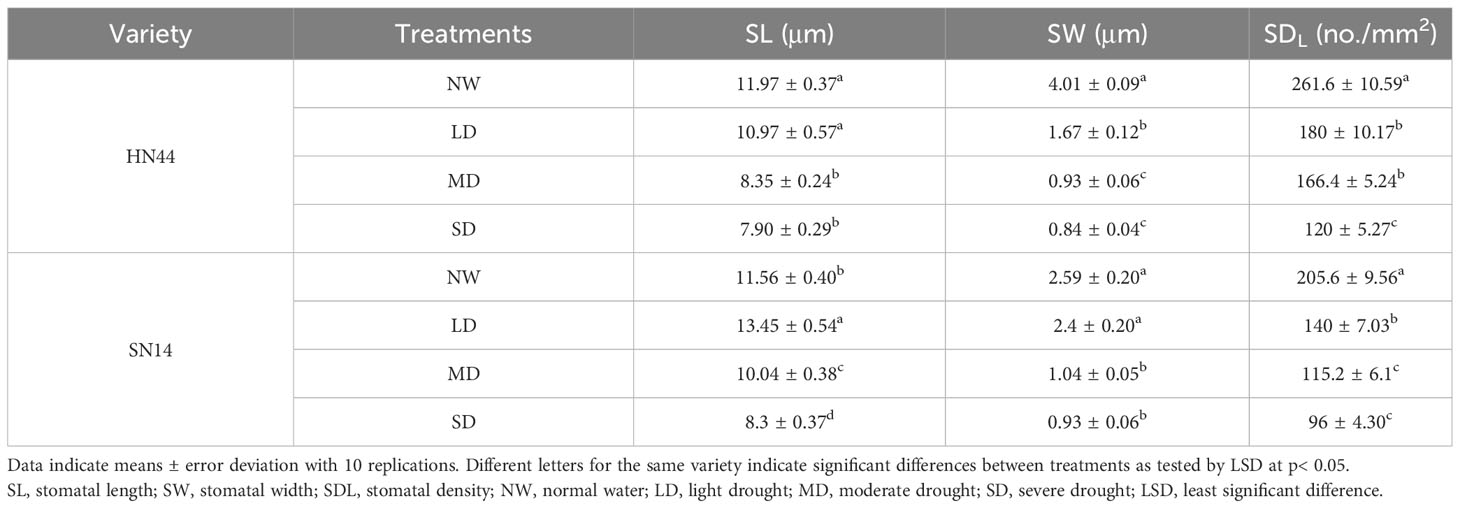
Blotting and microscopy techniques were employed to visualize stomatal characteristics. On the abaxial surface of the fourth fully expanded trifoliate leaves at the tip of the main stem, five 0.5-cm2 areas were coated with transparent nail polish. After the nail polish dried, the stomatal characteristics of the leaves in different treatments were observed with a fluorescence microscope (Olympus BX53, Tokyo, Japan); the stomatal length (SL), stomatal width (SW), area of the field of view (S), and number of stomata in each field of view (Q) (0.5 mm × 0.5 mm) were measured; stomatal density (SDL) was calculated based on the above indexes. Ten replicates were performed for each treatment..
2.2.3 Abscisic acid content
A frozen sample with a mass of 0.1 g (the second fully expanded trifoliate leaves at the tip of the main stem) was taken and added to 0.9 ml of phosphate-buffered saline (PBS) solution with a concentration of 0.01 mol/L for ice bath grinding. The supernatant was collected after centrifugation at 4,000 rpm for 15 min at 4°C. Then, using the Plant Hormone Abscisic Acid ELISA kit (Shanghai Enzyme Link Biotechnology, Shanghai, China), ABA concentration was measured.
2.2.4 Leaf relative water content
After treatment, the third fully expanded trifoliate leaves at the tip of the main stem were harvested, and their fresh weight was recorded. The leaves were immersed in water and placed in a dark room overnight. Afterward, the excess surface moisture on the leaves was removed using filter paper, the saturated weight was measured, and the leaves were then carefully placed in an 80°C oven and dried until a constant weight was achieved, at which point the dry weight was recorded.
2.2.5 Determination of superoxide dismutase activity, malondialdehyde content, and proline content
The SOD activity, malondialdehyde (MDA) content, and proline content were measured using the SOD Activity Assay Kit (SOD-1-W, Comin, Hangzhou, China), MDA Quantitative Assay Kit (MDA-1-Y, Comin), and Proline Quantitative Assay Kit (PRO-1-Y, Comin), respectively. All testing and analyses were conducted according to the manufacturer’s instructions.
2.2.6 RNA extraction, library construction, and sequencing
Based on the physiological analysis results under different treatments, transcriptome sequencing was performed on the leaves subjected to LD, SD, and NW conditions. After 1 day of stress treatment, the first fully expanded trifoliate leaves at the tip of the main stem were taken as samples for total plant RNA extraction. The plant total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the instructions provided by the manufacturer. RNA quality identification, library construction, and sequencing were performed by Frasergen (Wuhan, China).
2.2.7 RNA-seq analysis and gene expression analysis
Clean reads were compared to the Wm82.a2.v1 reference genome using Hisat2 (v2.2.1) software, and the number of reads compared to each gene was counted, using transcripts per thousand bases per million mapped reads transcripts (TPM) values per thousand bases to determine gene expression levels. The transcriptome raw sequencing data from this study were submitted to the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) database as individual BioProjects: PRJNA1011827. Differentially expressed genes (DEGs) were screened using DESeq2 software, and Padj< 0.01 and |log2FoldChange ≥ 2| were used as the differential gene screening conditions. Differentially communicated qualities were explained to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway datasets to acquire comments on the data.
2.2.8 Quantitative real-time PCR analysis
Total RNA extracted from the two leaves of soybean plants 1 day after the seed-filling period (R5) was isolated with total RNA using the total RNA rapid extraction kit (BioTeke Corporation, Beijing, China) and reverse transcribed using PrimeScript™ RT reagent Kit (TaKaRa, Mountain View, CA, USA). The qRT-PCR was performed on an AriaMx quantitative PCR instrument using SYBR Premix Ex Taq™ (TaKaRa), and each reaction was performed in triplicate. The actin gene Glyma.02G091900 was used as the internal reference (Liu et al., 2016). Gene relative expression levels were calculated according to the 2−ΔΔCT method. The primer sequences are shown in Supplementary Table 1.
2.3 Statistical analysis
All data were recorded in Excel 2016 (Microsoft Corp., Redmond, WA, USA). Analysis of variance (ANOVA) for relative leaf water content (RLWC), ABA content, photosynthetic parameters, and stomatal parameters was performed using SPSS 26.0 (IBM Corp., Armonk, NY, USA). Significance among gradual water deficit treatments was determined by the least significant difference (LSD) test, p< 0.05. GO enrichment analysis with KEGG enrichment analysis of transcriptome data was performed with R software (4.2.2). Figures were plotted using Origin 2021pro (OriginLab Co., Northampton, MA, USA).
3 Results
3.1 Effects of gradual water deficit on physiological traits
3.1.1 Effect of gradual water deficit on photosynthetic indexes
The Pn, Tr, Gs, and Ci of HN44 and SN14 all decreased with water stress increasing (Figure 2). Each treatment was significantly different from the control (p< 0.05). Compared to NW, Pn respectively decreased by 50.67%, 54.51%, and 56.92% for HN44 and by 41.50%, 53.47%, and 57.93% for SN14 under LD, MD, and SD conditions; Tr respectively decreased by 61.44%, 72.14%, and 73.63% for HN44 and by 50.8%, 53.48%, and 59.62% for SN14 under LD, MD, and SD conditions. The greater decrease in Tr for HN44 under gradual water stress reduced water loss and improved drought resistance. Similarly, Gs respectively decreased by 24.71%, 28.91%, and 46.74% for HN44 and by 15.52%, 21.51%, and 41.58% for SN14 under LD, MD, and SD conditions; Ci respectively decreased by 60.98%, 65.29%, and 66.24% for HN44 and by 55.91%, 65.23%, and 73.75% for SN14 under LD, MD, and SD conditions. The larger decrease in Gs for HN44 reduced water transpiration by rapidly reducing Gs. The reduction in Gs caused a decrease of Ci, which conversely inhibited the assimilation of CO2 and reduced its photosynthetic capacity. As the stress severity increased, the photosynthetic parameters of both varieties were suppressed to a certain extent, affecting the photosynthetic system of soybean plants. Pn, Gs, and Tr of HN44 decreased greater than those of SN14 under different water stress treatments; HN44 rapidly adjusted its photosynthetic gas channels, changed gas parameters, reduced water transpiration, and maintained its water status.
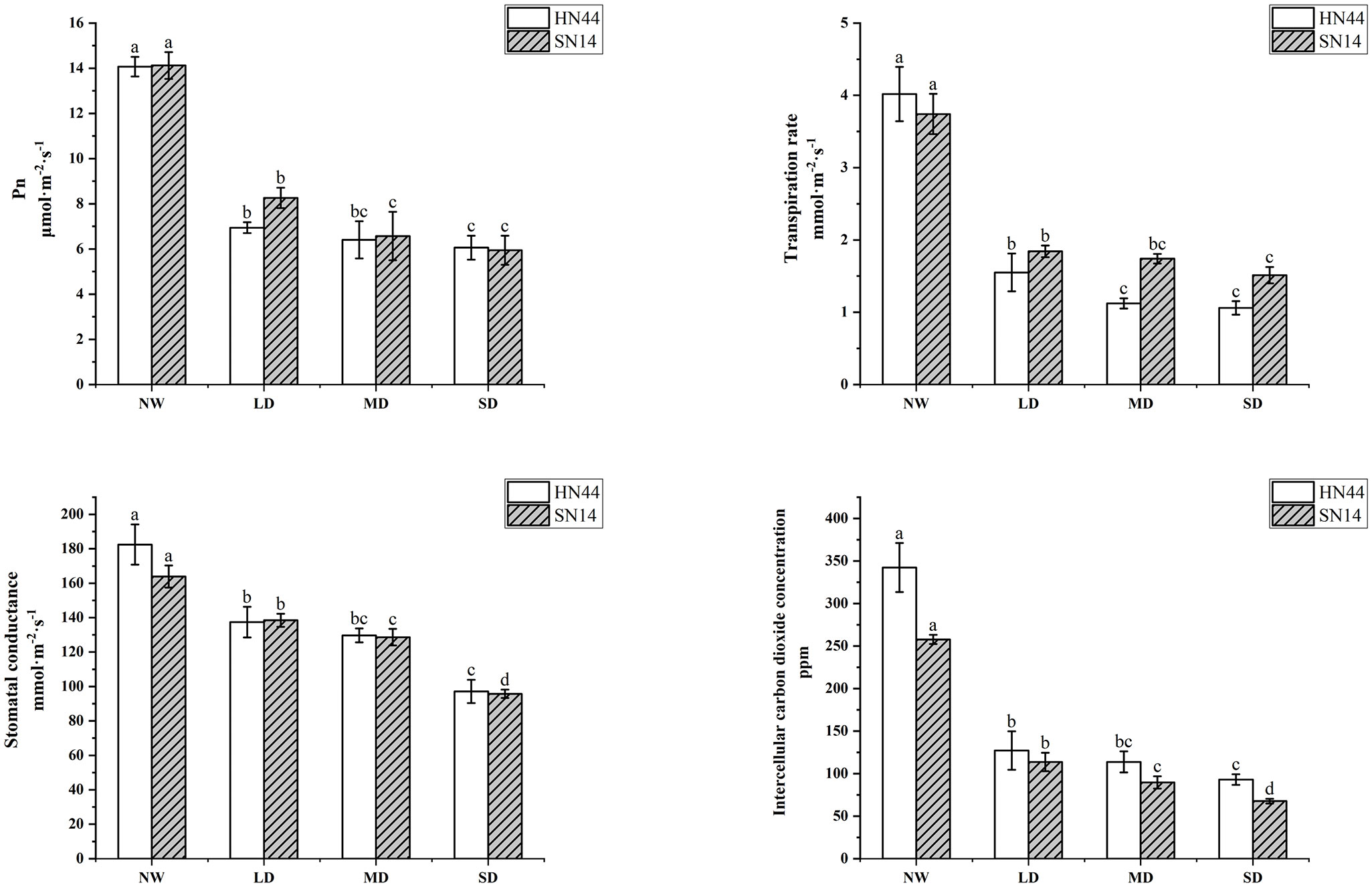
Figure 2 Effect of gradual water deficit on photosynthetic parameters. Data indicate means ± standard deviation with five replications. Different letters for the same variety indicate significant differences between treatments as tested by LSD at p< 0.05. LSD, least significant difference.
3.1.2 Effect of gradual water deficit on stomatal characteristics
In response to water scarcity, plants activate their drought response mechanisms, including the modification of stomatal morphology. As the degree of drought stress increased, notable changes were observed in the length, width, and density of the stomata (Table 1 and Figures 3A, B). Under LD, MD, and SD conditions, the stomatal length and width of HN44 decreased significantly compared to the control treatment, while only SW decreased significantly for SN14 under MD and SD conditions.
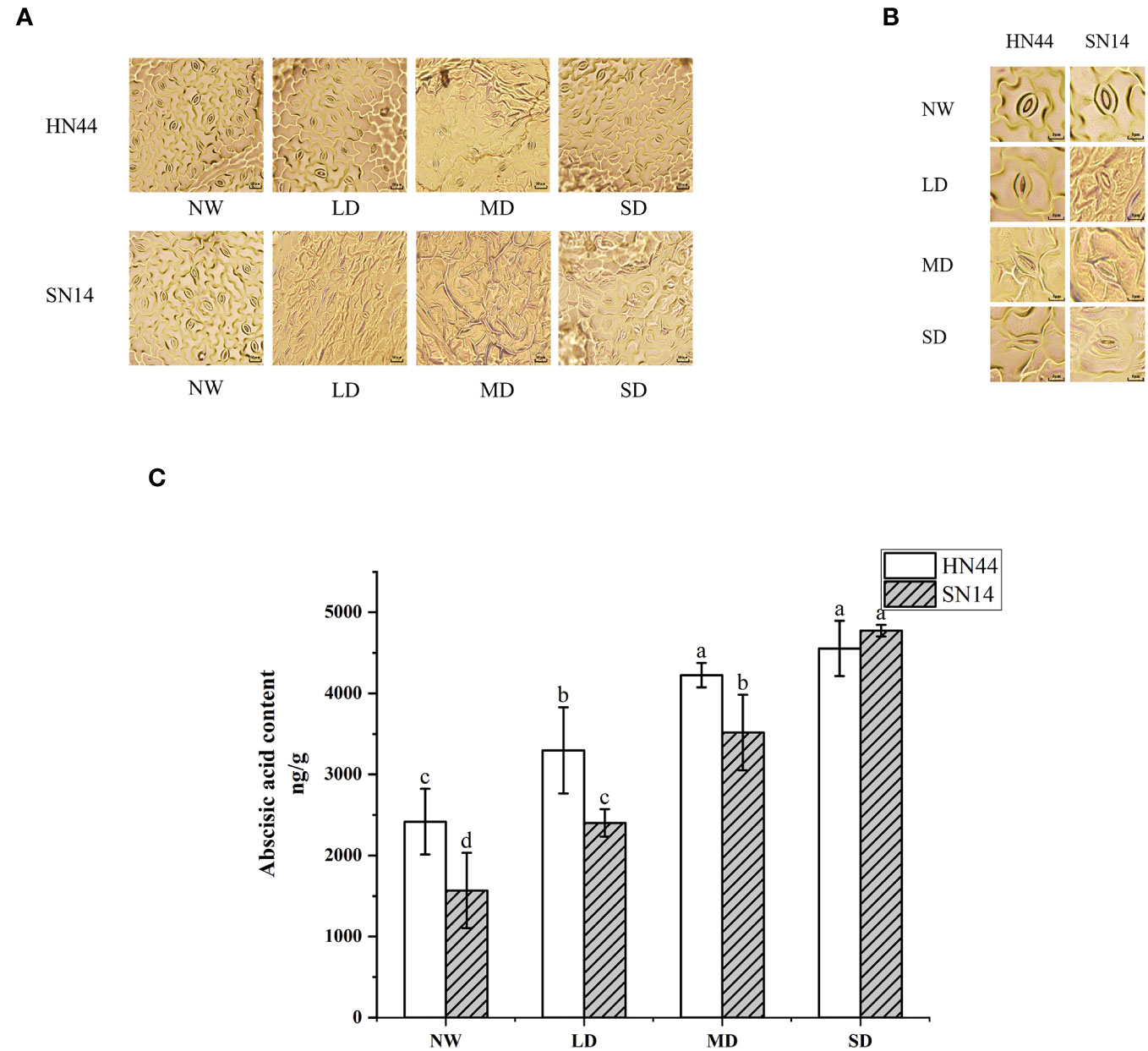
Figure 3 Effect of gradual water deficit on stomatal characteristics: (A) standard scale was 50 μm; (B) standard scale was 5 μm. (C) Abscisic acid content. Data indicate means ± standard deviation with three replications. Different letters for the same variety indicate significant differences between treatments as tested by LSD at p< 0.05. LSD, least significant difference.
ABA is closely associated with plant responses to environmental stress and stomatal movement. ABA content increased with increasing water stress for HN44 and SN14 (Figure 3C). According to the ABA content level under NW treatment, HN44 had characteristics of drought and stress resistance. The ABA contents of HN44 under LD and MD conditions were higher than those of SN14, which promoted stomatal closure in HN44 leaves; this rapidly regulated stomatal morphology in the initial stage of water shortage led to reduced Gs and transpiration and improved drought resistance and tolerance.
3.1.3 Effect of gradual water deficit on WUEi and RLWC
Under water deficit conditions, plants actively avoid drought stress by closing the stomata, reducing transpiration, and maintaining efficient water use. With the increase of water deficit in HN44, WUEi increased significantly (p< 0.05); under LD, MD, and SD, it increased by 28.81%, 61.30%, and 62.71% respectively, but WUEi of SN14 increased insignificantly (Figure 4B). During the water deficit, the drought-tolerant variety HN44 showed high WUEi.
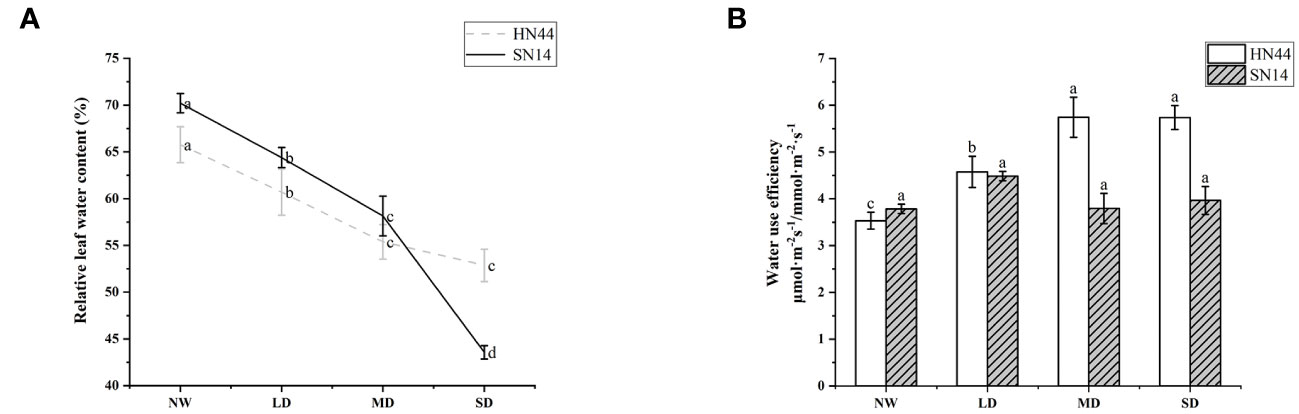
Figure 4 (A) Effects of gradual water deficit on the leaf relative water content. Data indicate means ± standard deviation with three replications. Different letters for the same variety indicate significant differences between treatments as tested by LSD at p< 0.05. (B) Effects of gradual water deficit on the instantaneous water use efficiency. Data indicate means ± standard error with five replications. Different letters for the same variety indicate significant differences between treatments as tested by LSD at p< 0.05. LSD, least significant difference.
RLWC can well reflect the water condition in plant cells. With increasing severity of water deficit, RLWC of both varieties was significantly reduced (p< 0.05). For HN44, RLWC decreased by 7.71%, 15.80%, and 19.61% under LD, MD, and SD conditions, respectively. In comparison, SN14 showed reductions of 8.28%, 17.17%, and 37.94% under LD, MD, and SD conditions, respectively, in RLWC (refer to Figure 4A). In the occurrence of drought stress, the elevated ABA content of HN44 regulated the stomatal movement and morphological structure, leading to timely and effective reductions in photosynthetic indexes such as Tr, and this maintained adequate water levels and normal physiological functions within the plant.
3.1.4 Effect of gradual water deficit on antioxidant enzyme activities, MDA content, and proline content
SOD plays a pivotal role in catalyzing the dismutation of superoxide anion radicals (O2−), eliminating O2−, maintaining the balance of reactive oxygen metabolism, and protecting membrane structures. Its activity level serves as an essential indicator of plant stress tolerance. As drought stress intensified, in comparison with NW conditions, the SOD activity of HN44 increased by 42.33%, 60.65%, and 68.07% in LD, MD, and SD treatments, respectively, reaching significant differences (p< 0.05); the content of SN14 increased by 9.8%, 24.56%, and 11.62% in LD, MD, and SD treatments, respectively (Figure 5A). With the increase in the water stress degree, the increase in SOD activity of HN44 in each treatment was greater than that of SN14. The SOD content of HN44 showed a gradual upward trend, and that of SN14 showed a trend of first increasing and then decreasing. It was indicated that HN44 can maintain its balance of active oxygen metabolism and reduce the damage of free radicals to itself during drought stress. These results suggest that HN44 can maintain its reactive oxygen metabolism balance well in the process of increasing drought stress, reducing the damage of free radicals to itself. However, the activity of SOD of SN14 was inhibited severely under SD, disrupting the balance of reactive oxygen metabolism, and its ability to resist oxidative stress was weaker than that of HN44.
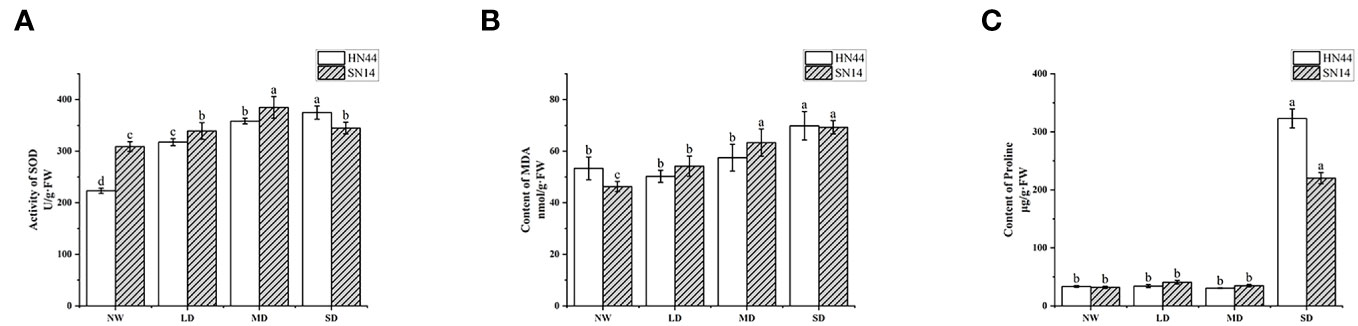
Figure 5 Effect of gradual water deficit on antioxidant enzyme activity and osmoregulatory substances. (A) The activity of SOD. (B) The content of MDA. (C) The content of proline. Data indicate means ± standard deviation with three replications. Different letters for the same variety indicate significant differences between treatments as tested by LSD at p< 0.05. SOD, superoxide dismutase; MDA, malondialdehyde; LSD, least significant difference.
MDA is a significant product of plant membrane lipid peroxidation and is often used as an indicator of the degree of cell membrane lipid peroxidation, cell damage, and the intensity of plant responses to adverse conditions. The MDA content of HN44 and SN14 significantly increased under SD treatment (p< 0.05). Compared with NW, the MDA content of HN44 decreased by 5.81% under LD and increased by 7.74% and 30.97% under MD and SD, respectively. The MDA content of SN14 increased by 17.12%, 36.81%, and 47.45% under LD, SD, and MD treatments, respectively, compared with NW (Figure 5B). The increase in MDA content in HN44 under LD, MD, and SD treatments was less than that in SN14. It suggested that with the increasing drought stress, the degree of membrane damage in SN14 was greater than that in HN44.
Plants often accumulate significant amounts of proline under various adversities. Beyond its role in osmotic regulation, proline plays a crucial role in preventing excessive dehydration and cell death. In the early stage of stress, the proline content did not significantly differ from the control. However, under SD, the proline content significantly increased (p< 0.05). Compared with the NW, the proline content of HN44 and SN14 under SD increased by 865.85% and 587.45%, respectively (Figure 5C). The increase in proline content in HN44 was much higher than that in SN14, and HN44 accumulated more proline to maintain its osmotic regulation balance and tissue water retention ability, thereby enhancing its adaptability to drought stress.
3.2 Transcriptome analysis
3.2.1 RNA-seq data revealed DEGs in soybeans under gradual water deficit conditions
A total of 99.2 GB of raw data was initially obtained, and following processing with fastp (v0.23.1), 86.9 GB of clean data was obtained. In the comparison between LD and NW, 479 and 3,317 DEGs were identified in HN44 and SN14, respectively. Among these, 377 and 1,999 genes were upregulated, and 102 and 1,318 genes were downregulated in HN44 and SN14, respectively. Notably, HN44 exhibited significantly fewer DEGs under LD as compared to SN14. In the comparison between SD and NW, 3,042 and 3,231 DEGs were identified in HN44 and SN14, respectively. Among these, 1,018 and 852 genes were upregulated, and 2,024 and 2,379 genes were downregulated in HN44 and SN14, respectively (Figure 6).
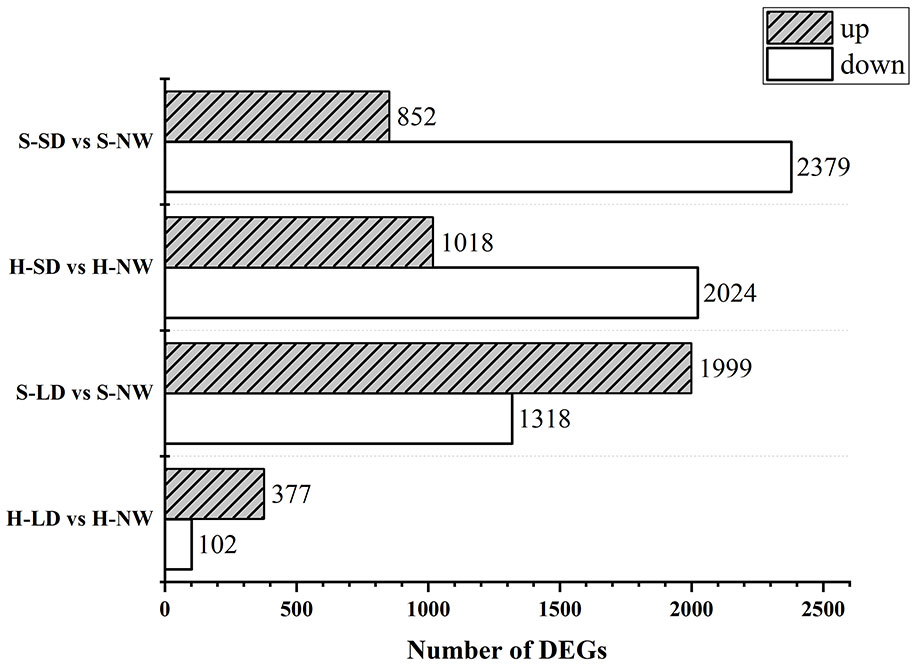
Figure 6 Number comparison of differentially expressed genes between both varieties under drought stress. H-NW, HN44 under normal Water; H-LD, HN44 under light drought; H-SD, HN44 under severe drought; S-NW, SN14 under normal water; S-LD, SN14 under light drought; S-SD, SN14 under severe drought. The same as below.
3.2.2 GO enrichment analysis of the DEGs
Under LD treatment, the DEGs of HN44 and SN14 were enriched in stress-related terms such as response to hydrogen peroxide (GO:0042542), reactive oxygen species (GO:0000302), light stimulus (GO:0009416), and signaling (GO:0007165), with most being upregulated (Figures 7A, B). It indicates that plants induced upregulation genes in these pathways to resist the effects of mild stress in the early stage of stress. SN14 was associated with more pathways and genes, requiring more energy consumption to resist water stress.
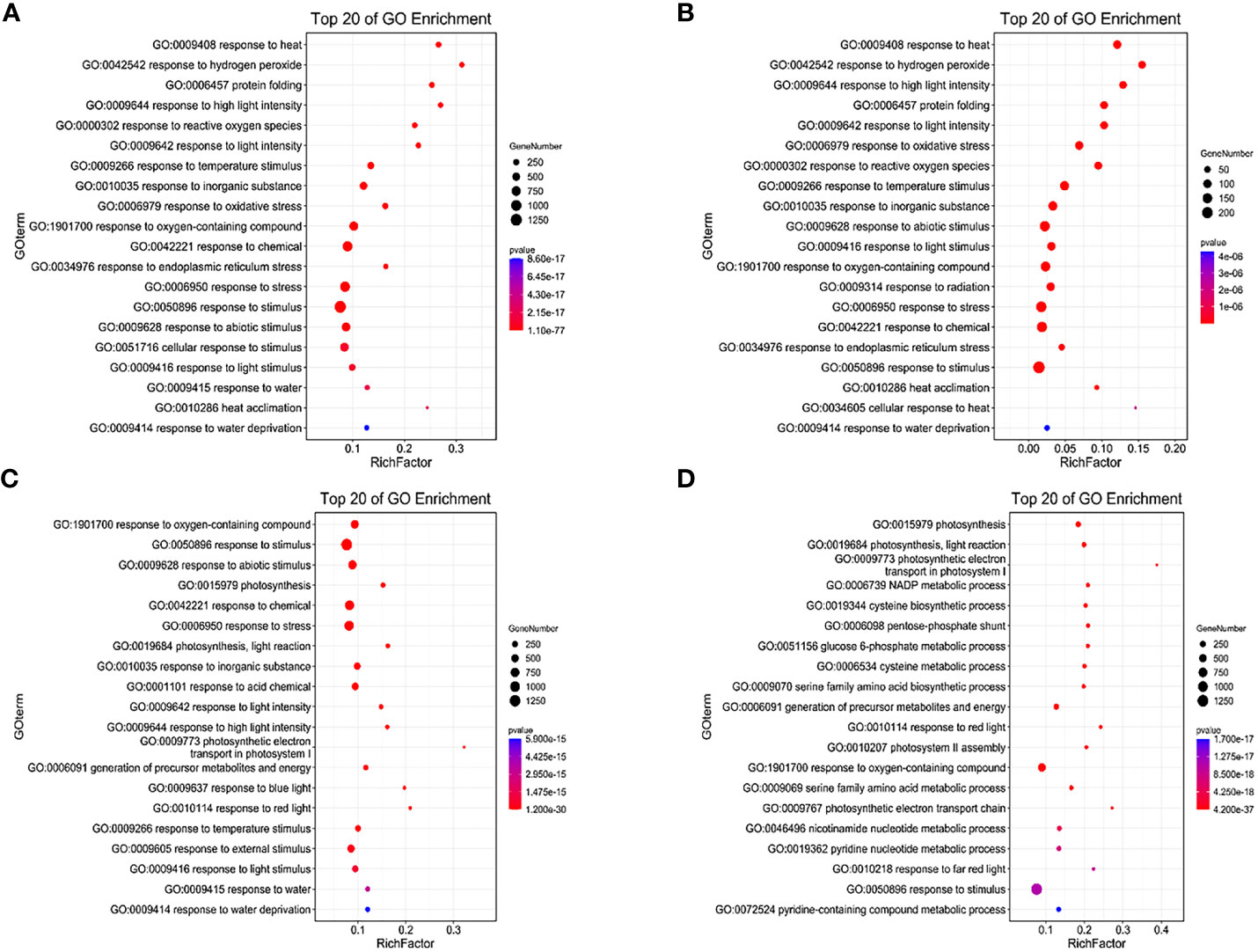
Figure 7 GO enrichment of differentially expressed genes between both varieties under water deficit. (A) H-LD vs. H-NW; (B) S-LD vs. S-NW; (C) H-SD vs. H-NW; (D) S-SD vs. S-NW. GO, Gene Ontology.
Under SD treatment, in addition to being enriched in the same terms as under LD, more DEGs were enriched in photosynthesis-related terms such as photosynthesis (GO:0015979), chloroplast thylakoid membrane (GO:0009535), and plastid thylakoid membrane (GO:0031976), with most being downregulated (Figures 7C, D). This suggests that cellular components, particularly the photosynthetic membrane system, were highly sensitive to stress conditions. As drought stress intensified, photosynthesis was inhibited, and the membrane system suffered severe damage.
3.2.3 KEGG enrichment analysis of the DEGs
For LD treatment, HN44 and SN14 had DEGs enriched in 56 and 115 KEGG pathways, respectively, with 2 and 11 pathways showing significant enrichment (p< 0.05). Among these pathways, two were co-enriched in both varieties, namely, “protein processing in the endoplasmic reticulum” (ko04141) and “galactose metabolism” (ko00052) (as illustrated in Figures 8A, B). Under LD conditions, most DEGs enriched in “protein processing in the endoplasmic reticulum” (ko04141) were upregulated, potentially playing a crucial role in ensuring proper protein folding or positively regulating drought tolerance. Under SD treatment, both varieties exhibited co-enriched in multiple pathways, including photosynthesis-antenna proteins (ko00196), photosynthesis (ko00195), glyoxylate and dicarboxylic acid metabolism (ko00630), carbon fixation in photosynthetic organisms (ko00710), galactose metabolism (ko00052), glycolysis/gluconeogenesis (ko00010), ABC transporter proteins (ko02010), carotenoid biosynthesis (ko00906), glycine, serine, and threonine metabolism (ko00260), and other pathways related to amino acid metabolism (Figures 8C, D), and most of the genes showed downregulated expression, indicating that both photosynthesis and energy metabolism were differently affected under SD.
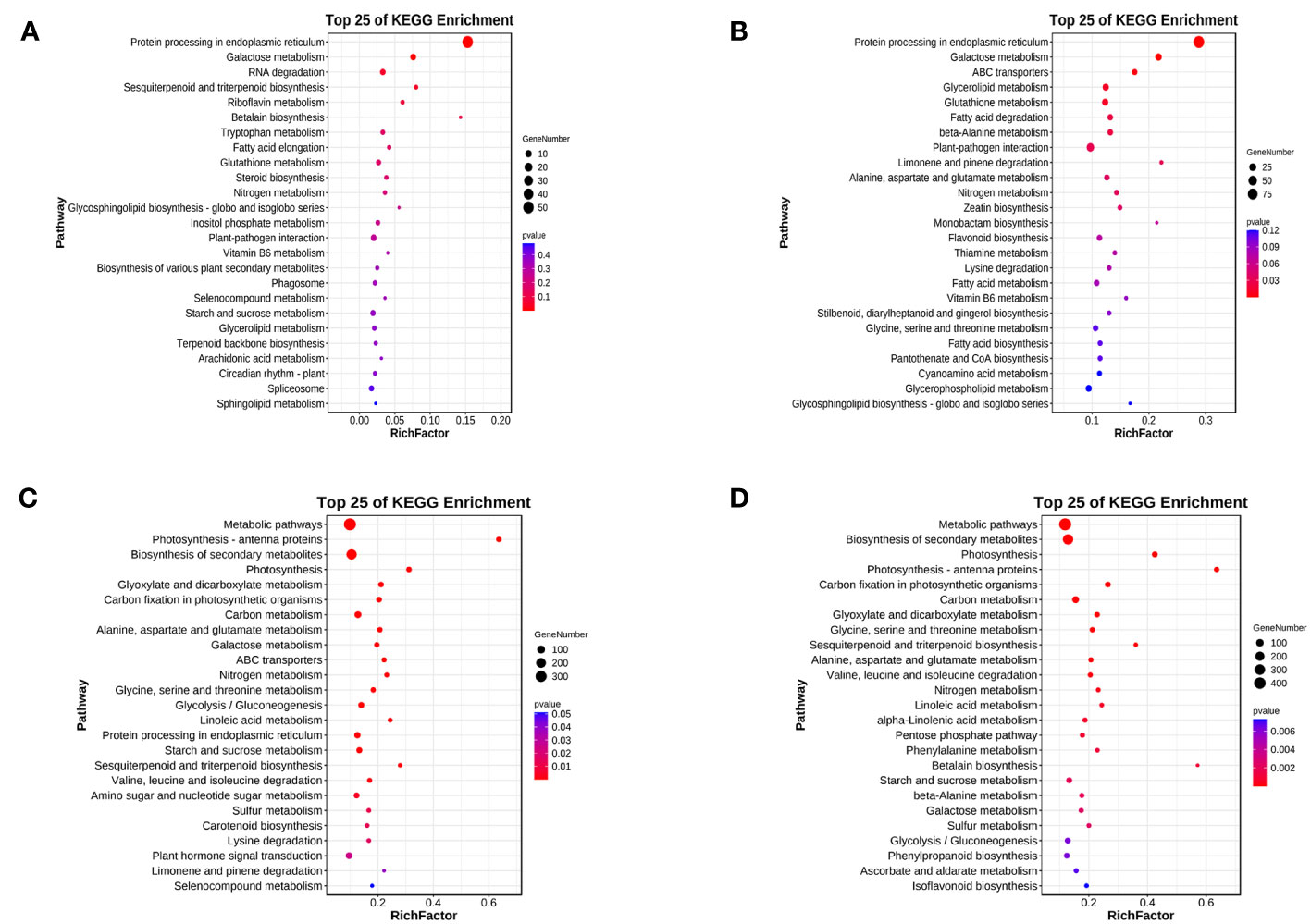
Figure 8 Top 25 KEGG pathway enrichment of differentially expressed genes between both varieties under water deficit. (A) H-LD vs. H-NW; (B) S-LD vs. S-NW; (C) H-SD vs. H-NW; (D) S-SD vs. S-NW. KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.2.3.1 Expression of differential photosynthetic pathways (ko00195)
Photosynthesis is a vital process responsible for the conversion of materials and energy, serving as the foundation for the survival and development of nearly all life forms on Earth. During periods of severe drought stress, both HN44 and SN14 exhibited downregulation of genes associated with the regulation of the photosynthetic pathway, as illustrated in Figure 9. Notably, HN44 had fewer DEGs enriched in photosystem I and photosystem II as compared to SN14, and the extent of downregulation in HN44 was also lower than that observed in SN14. These findings suggest that under drought stress, HN44 experienced less inhibition of photosynthesis in comparison to SN14, ultimately resulting in a stronger carbon assimilation capacity in HN44 when compared to SN14.
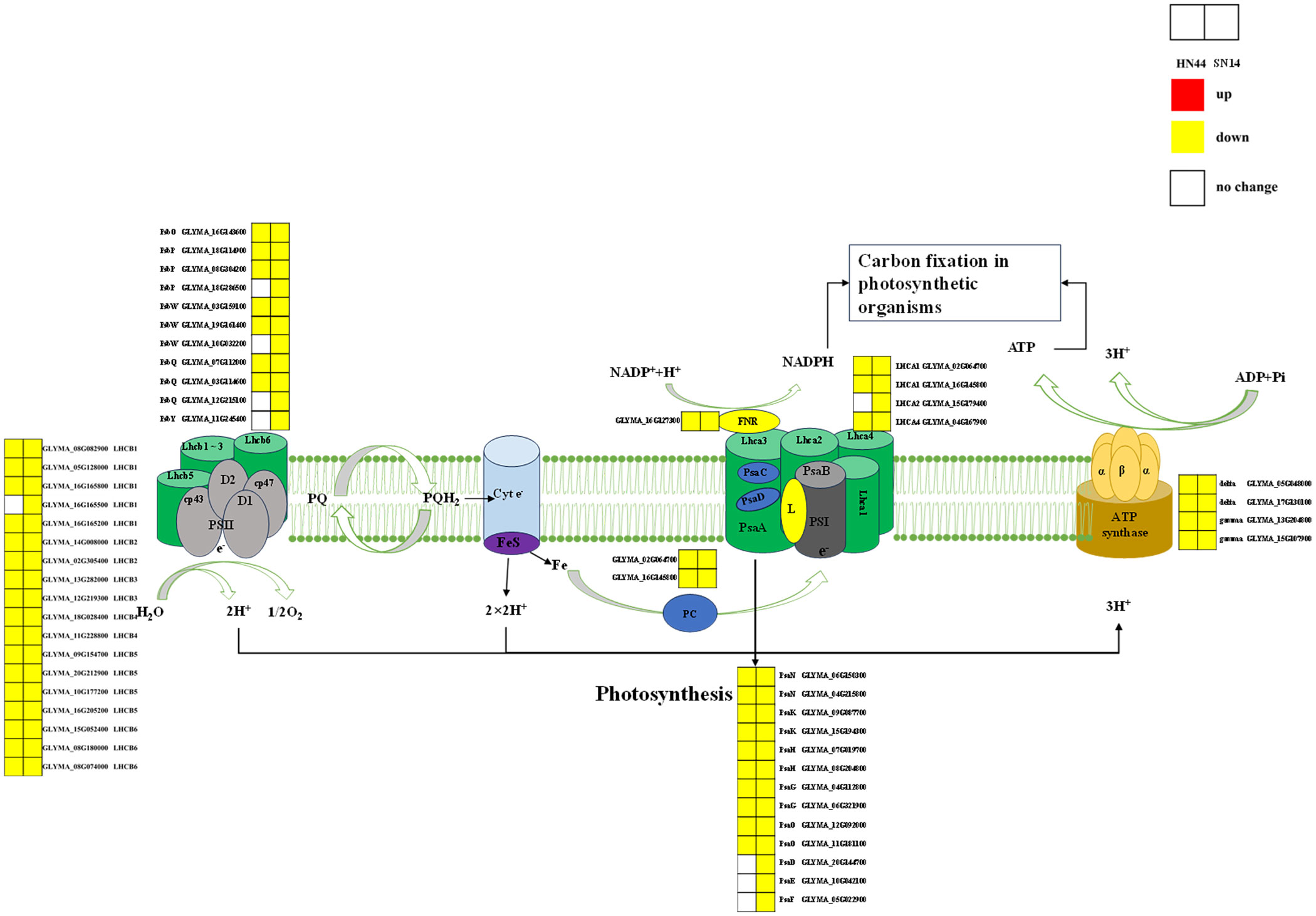
Figure 9 Photosynthetic pathway diagram. PSII, optical system II. PSII proteins included PsbO, PsbP, PsbQ, PsbY, and PsbW. PSI proteins included PsaD, PsaE, PsaF, PsaN, PsaK, PsaH, PsaG, and PsaO. Antenna proteins included Lhca1, Lhca2, Lhca4, Lhcb1, Lhcb2, Lhcb3, Lhcb4, Lhcb5, and Lhcb6. Type F atp enzyme protein included atp enzyme gamma, delta. Red represents the upregulated gene, yellow represents the downregulated genes, and white represents no significant change in the gene. The same as below.
3.2.3.2 Analysis of phytohormone signal transduction pathways (ko04075)
Phytohormones play a crucial role in growth, development, and adaptation to biotic and abiotic stresses. ABA plays an important role in phytohormone osmotic stress regulation, rapidly accumulating and regulating survival when plants are subjected to a variety of stresses as a stress hormone. Under severe drought stress, in both HN44 and SN14, genes regulating the ABA receptor proteins (PYR/PYL/RCAR) exhibited upregulated expression, while genes governing PP2C and SnRK2 showed downregulated expression (Figure 10), which allowed the regulation of the ABA response binding element ABF to promote stomatal closure, reduce water evaporation, and improve the stress tolerance of soybean.
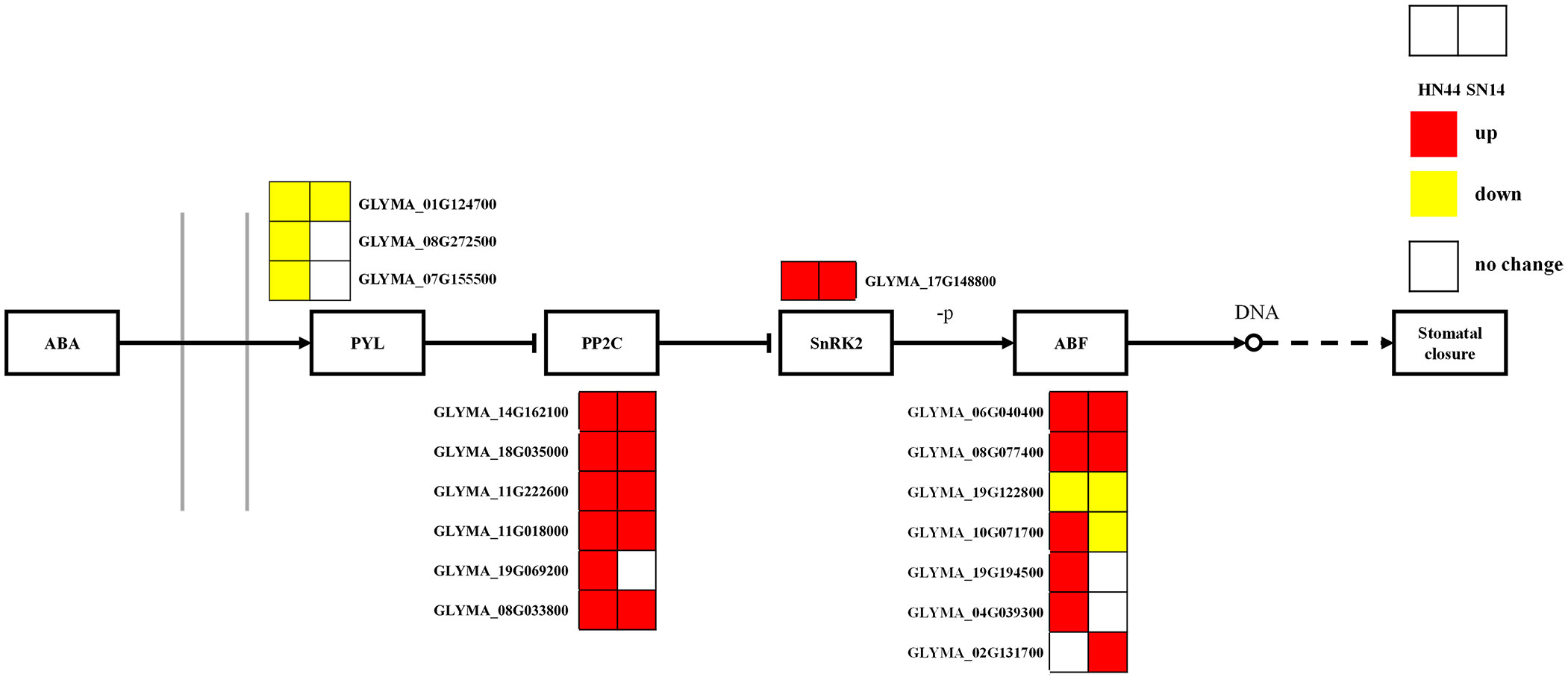
Figure 10 ABA-mediated signaling pathways. Red represents the upregulated gene, yellow represents the downregulated genes, and white represents no significant change in the gene. ABA, abscisic acid.
3.2.3.3 Analysis of glutathione metabolic pathways (ko00480)
Glutathione (GSH) is a pivotal antioxidant in plants and plays an important role in various abiotic stresses. Under SD, the genes regulating glutathione peroxidase (GPX) in HN44 and SN14 were upregulated, while the genes regulating glutathione S-transferase (GST) were both partially upregulated and partially downregulated. In HN44, genes regulating γ-glutamyl transpeptidase (GGT) were upregulated, and the number of upregulated genes regulating GST was greater than the number of downregulated genes. SN14 had more genes downregulating GST than upregulating GST (Figure 11). These findings indicate that genes related to glutathione metabolism were activated, and the glutathione metabolic pathway in HN44 exhibited superior performance compared to SN14, reflecting a heightened ability to effectively mitigate ROS.
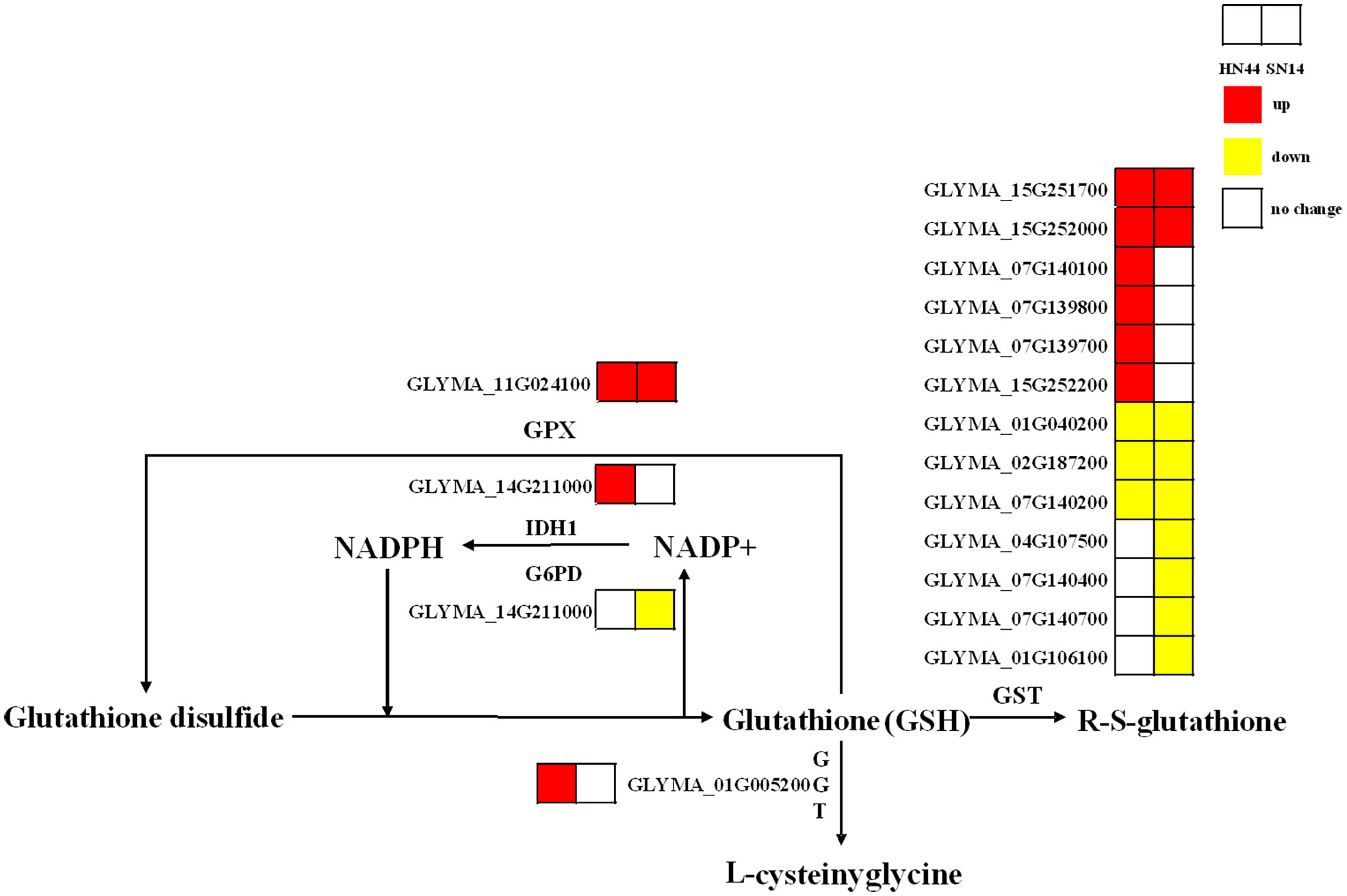
Figure 11 Glutathione metabolic pathway. Red represents the upregulated gene, yellow represents the downregulated genes, and white represents no significant change in the gene.
3.2.4 qRT-PCR
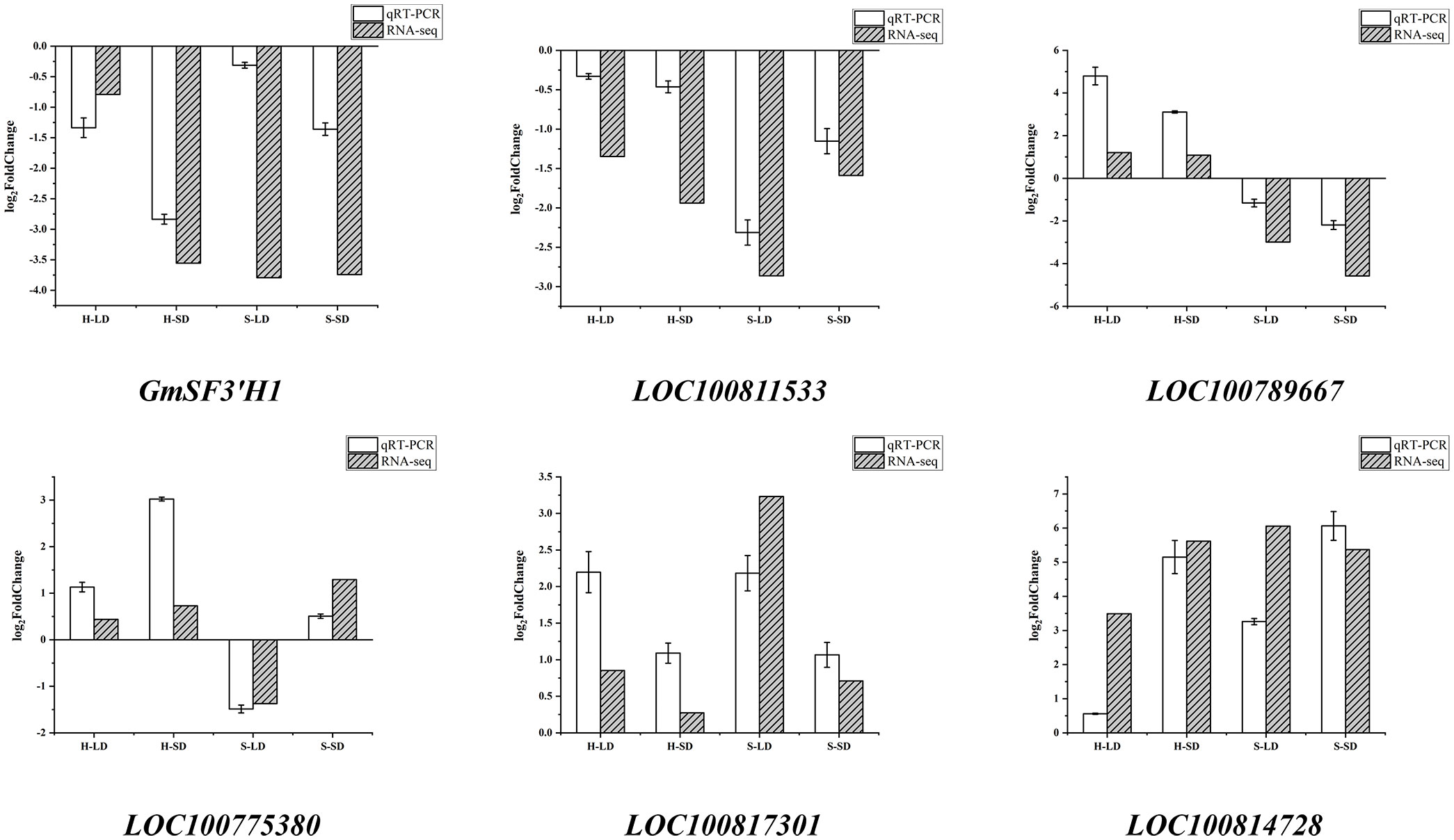
Based on expression levels and related pathways, six key genes with significant differences in major drought stress response pathways were selected: the flavonoid 3′-hydroxylase gene (GmSF3′H1), the transcription activation factor PTI5 (LOC100811533), a stomatal movement-related gene (LOC100789667), a gene encoding SCR protein (LOC100775380), the phosphatidylinositol 4-kinase gamma 4 gene (LOC100817301), and an abscisic acid-mediated signaling pathway-related gene (LOC100814728). These genes were validated using qRT-PCR. As shown in Figure 12, the trends in gene changes were consistent with the RNA-seq results, indicating the reliability of the RNA-seq data.
3.2.5 Screening of key transcription factors
Transcription factors (TFs) play important roles in regulating plant adaptation to abiotic stresses, including drought. Under LD treatment, 32 and 209 transcription factors were identified for HN44 and SN14, respectively, which indicated that SN14 activated more transcription factors to regulate pathways to resist drought stress damage. Under SD, 233 and 208 transcription factors were identified for HN44 and SN14, respectively (Supplementary Figure 1), with little difference in the number and type of transcription factors between the two varieties. The major transcription factor families were bHLH, ERF, WRKY, NAC, MYB, HD-ZIP, MYB_related, and bZIP, with both varieties significantly enriched in phytohormone signaling (ko04075), circadian rhythm (ko04712), and MAPK signaling pathway (ko04016).
While the number and type of transcription factors were similar between HN44 and SN14 under SD treatment, there were differences in their expression patterns. For example, the genes of the WRKY transcription factor family were partially upregulated and partially downregulated in HN44, whereas only downregulated expression was observed in SN14, and the genes of the ERF transcription factor family were expressed as mostly upregulated in HN44, and vice versa in SN14. A number of genes were screened based on the different expression patterns of the two transcription factors (Table 2). LOC100808190, GmWRKY49, LOC100798375, LOC100804384, LOC100817546, GmbZIP4, and LOC100819313 genes were upregulated for expression in HN44 with no significant changes in SN14, and GmbZIP1 was downregulated for expression in SN14 with no significant changes in HN44.
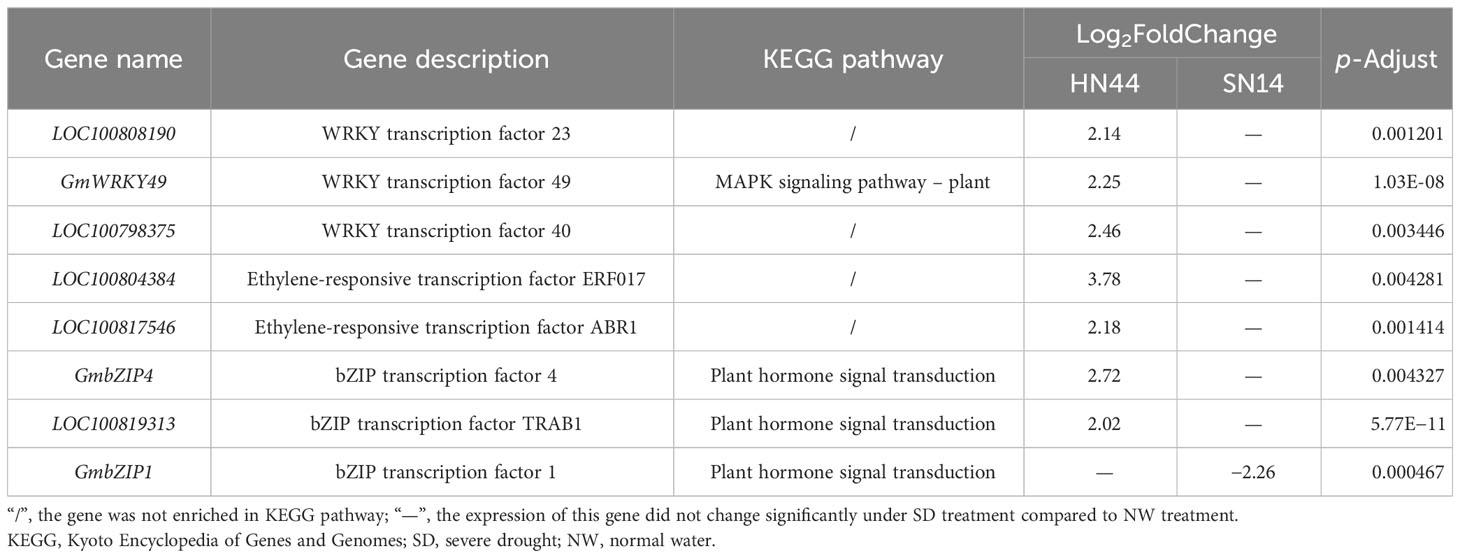
Table 2 Key candidate genes identified through differential expression analysis between the two soybean varieties.
4 Discussion
With global climate change and the increasing scarcity of water resources, drought has become one of the major abiotic stresses affecting soybean yield and quality (Chen et al., 2023). Therefore, studying the drought resistance of soybeans to improve their ability to adapt to drought is of great importance. RNA-seq is an indispensable tool for whole transcriptome differential gene expression analysis (Stark et al., 2019). This study compared the transcriptional responses of different soybean varieties under gradual water deficit conditions through RNA-seq transcriptome analysis to better understand the gene regulation patterns of soybeans under gradual water deficit conditions. Additionally, we analysed the physiological responses, in order to corroborate the factors causing differences in drought tolerance between the contrasting soybean varieties.
4.1 Change of physiology in soybean under gradual water deficit
As the degree of drought stress increased, both HN44 and SN14 experienced a reduction in their photosynthetic capacity, and all photosynthetic parameters were gradually reduced. However, HN44 had a greater reduction in Gs to promote stomatal closure and rapidly reduce transpiration, a greater reduction in Ci, and more CO2 participation in the Calvin cycle to increase HN44’s carbon assimilation to improve WUE compared with SN14. Research by McAusland et al. (2016) highlighted the importance of dynamic WUEi as well as the underlying Pn and Gs as light intensity changes. Gs responded more slowly than Pn to decreasing light intensity, leading to the desynchronization of a dynamic Pn and Gs, such as when switching from high to low light, resulting in excess transpiration and reduced WUEi. It indicated that WUEi could be improved by accelerating the rate of change in Gs (Way and Pearcy, 2012; McAusland et al., 2016). Leaves with more rapid changes in Gs often had many smaller stomata, likely due to larger surface-to-volume ratios, allowing faster solute transport to manipulate guard cells (Drake et al., 2013). With increasing stress, the stomatal length and width of HN44 were smaller than those of SN14, while the decrease in stomatal density was lower than that of SN14. HN44 rapidly changed in Gs to maintain a dynamic balance between Pn and Gs and improve instantaneous WUEi. As drought stress intensified, the WUEi of the drought-tolerant variety HN44 gradually increased, while that of SN14 showed a wavy trend with an increase under LD followed by a decrease under MD and SD. The drought-tolerant variety was able to maintain high WUEi despite being under SD, while the drought-sensitive variety showed an increase in WUEi under LD followed by a decreasing trend under SD. It was consistent with the results of a study by Fletcher et al. (Fletcher et al., 2007). Drought stress triggers the accumulation of excess ROS in plants, necessitating the activation of antioxidant systems for their removal. If these excess ROS are not promptly cleared, they can damage plant tissues and accelerate plant aging, a notable symptom of which is the accumulation of MDA (Baldoni et al., 2016). In addition to activating antioxidant systems, plants also employ osmoregulatory strategies to mitigate the damage caused by drought stress (Ashraf and Foolad, 2007). In this study, as the degree of stress increased, the activity of SOD and the content of MDA in HN44 gradually increased, while in SN14, the content of MDA gradually increased and SOD activity peaked under MD conditions before subsequently declining. The proline content in both did not change significantly under LD or MD conditions but increased dramatically under SD conditions. Compared to SN14, HN44 had a lower increase in MDA accumulation and a higher increase in SOD activity and proline content. It showed that under drought stress, HN44 had superior antioxidant systems and osmoregulatory strategies to reduce the damage caused by drought stress.
4.2 Photosynthetic pathways under gradual water deficit
As drought stress intensified, the expression of many genes encoding photosystem I (PsaN, PsaK, PsaH, PsaG, and PsaO) and photosystem II (PsbO, PsbP, PsbQ, and PsbW) reaction center subunit proteins were downregulated in HN44 and SN14. These specific proteins had multiple functions in photosynthesis, and their downregulation under drought stress inhibited photosynthesis and reduced carbon assimilation (Calderone et al., 2003; García-Cerdán et al., 2011; Wang et al., 2020). The downregulation of genes enriched in the Pet family with ATP synthase led to a decrease in electron transport rates in PSII and PSI and a decrease in ATP synthesis, thus inhibiting plant photosynthesis (Caruso et al., 2009; Awasthi et al., 2014). The experiment identified downregulated expression of genes regulating ATP synthase activity in soybeans under drought stress (Das et al., 2016). In addition to the above co-regulated genes, SN14 also had downregulated expression of genes encoding PsaD, PsaE, PsaF, and PsbY proteins. PsaD and PsaE were important components of the PSI reduction site involved in electron transfer and binding of ferredoxin and NADP+ oxidoreductase (Scheller et al., 2001). PsaF was a peripheral subunit of PSI involved in the assembly and interaction of light-harvesting pigment proteins (Jensen et al., 2000). PsbY was important for the redox characterization of Cytb559 and might interact directly or indirectly with its heme motif. found that electron transfer on the acceptor side involving QA and QB was significantly slowed in the absence of PsbY, making Arabidopsis more susceptible to photoinhibition von Sydow et al. (2016). Compared to those in HN44, electron transport and photosynthesis were more strongly inhibited in SN14, potentially causing a variety of differences in photosynthetic and WUEi.
4.3 ABA-mediated signaling pathways under gradual water deficit
The PYL-PP2C-SnRK2 family is a core component of ABA signaling, with PYLs as ABA receptors and PP2C and SnPKs as important negative and positive regulators of ABA signaling, respectively. As ABA receptors, PYR/PYL/RCAR proteins can hinder the activity of PP2C. In the presence of ABA, the negative regulation mediated by PP2C is released. Subclass III SnRK2 (SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1, and SRK2I/SnRK2.3) phosphorylates ABA response elements (ABRE binding protein/ABRE binding factors (AREB/ABFs)), thereby activating the expression of many ABA response genes in an ABRE-dependent manner (Ma et al., 2009; Park et al., 2009; Huang et al., 2019). The AREB/ABF-SnRK2 pathway, as a major positive regulator of ABA/stress signaling, can directly regulate the expression of ABRE-mediated genes to cope with osmotic stress. Subclass III SnRK2 is a protein kinase that acts as an upstream control factor for AREB/ABFs through phosphorylation activity and plays a global regulatory role in the ABA/stress signaling pathway (Geiger et al., 2010). In this study, the downregulation of genes regulating ABA receptor (PYR/PYL/RCAR) proteins and the upregulation of PP2C and SnRK2 genes in HN44 and SN14 are consistent with the above results. It promotes ABF regulation of soybean stomatal closure, reduces water evaporation, and enhances soybean resistance through the ABA response binding element. Overall, HN44 was superior to SN14 in the ABA-mediated signaling pathway, with more enriched genes regulating the ABA pathway and a greater degree of upregulation. It allowed HN44 to better reduce its water loss, improve water use efficiency, and enhance its adaptability to drought.
4.4 Glutathione metabolic pathways under gradual water deficit
GSH is a primary antioxidant that can bind with electrophilic compounds and promote the reduction of peroxides (Li et al., 2022). In this study, under SD, upregulated genes in HN44 and SN14 regulated GPX, but the genes regulating GST were partially upregulated and partially downregulated. These enzymes collectively participated in the function of plants to clear excessive ROS in the body (Anderson and Davis, 2004; Kumar et al., 2013). Additionally, upregulated genes in HN44 regulated GGT, which can decompose extracellular GSH and provide cysteine (rate-limiting substrate) for de novo synthesis of GSH in cells. It was a key enzyme in glutathione metabolism and played a key role in GSH homeostasis (Zhang et al., 2005). The number of upregulated genes regulating GGT in HN44 was more than the number of downregulated genes. The number of downregulated genes regulating GST in SN14 was more than the number of upregulated genes. This suggests that the antioxidant system in SN14 was compromised under severe stress, resulting in reduced resistance to damage caused by drought stress.
4.5 The selection and differential analysis of key transcription factors
During long-term selection and evolution, plants have gradually developed the ability to rapidly perceive and adapt to external stress stimuli, in which the transmission of multiple adversity signals in plants plays a very important role in the process of abiotic stress in plants. The mechanisms of plant response to adversity are very complex, and the effect of improving plant resistance through the expression of single functional genes is insignificant, while transcription factors are involved in many different processes in plants, including growth, development, and stress signaling. These transcription factors have the ability to synergistically regulate multiple adversity-responsive genes or networks and can establish a complex network to regulate their metabolic responses, which is conducive to the timely response of plants to adversity stress and enhance tolerance.
In this study, we identified several members of the WRKY, ERF, and bZIP transcription factor families. These transcription factors exhibited different expression patterns, and some were involved in the ABA-mediated signaling pathway, regulating the ABA-responsive binding element ABF. Among them, WRKY and ERF differential transcription factors played more important roles in stress tolerance. The AP2/ERF family of transcription factors had multiple modes of regulation in response to drought stress, with hormonal regulation being the main mode. ABR1 was a negative regulator of ABA-regulated gene expression, and ABR1 mutants showed high sensitivity to ABA, osmotic stress, and salt stress (Pandey et al., 2005). ERF17 was able to increase iron deficiency tolerance; played a key role in photosynthesis, chlorophyll (Chl) biosynthesis, and respiratory electron transport; and was able to attenuate the effects of stress on plant photosynthesis and reduce membrane damage (Cheng et al., 2020). SlWRKY23 transcription factor in tomato was able to increase tomato tolerance to osmotic stress and salt stress by inducing the interaction of growth hormone and ethylene pathway (Singh et al., 2023).GmWRKY49 transcription factor was strongly induced by ABA, and its expression was significantly increased when exogenous ABA treatment was applied, which may act on the plant stress tolerance pathway (Zhou et al., 2008). WRKY18 and WRKY60 were able to increase plant sensitivity to ABA, osmotic stress, and salt stress, while WRKY40, WRKY18, and WRKY60 acted antagonistically to each other, and plants overexpressing WRKY40 were able to reduce the plant sensitivity to ABA and enhance the tolerance to osmotic stress and salt stress (Chen et al., 2010). These transcription factors were significantly upregulated in HN44 and downregulated in SN14, indicating that HN44 was better adapted to drought stress than SN14. In conclusion, HN44 promoted stomatal closure and reduced Gs and transpiration by regulating the upregulated expression of more ABA-responsive binding protein transcription factors to achieve efficient utilization of limited water.
5 Conclusion
Through a comparative analysis of the physiological characteristics of two soybean varieties under gradually increasing water deficit conditions, we observed distinct traits in the drought-tolerant variety, HN44. Notably, HN44 exhibited smaller stomata, higher stomatal density, and lower stomatal conductance and transpiration rate, as well as elevated abscisic acid content. These characteristics allowed HN44 to rapidly adapt its stomatal morphology and stomatal conductance, maintaining a dynamic equilibrium between net photosynthesis and stomatal conductance. As a result, HN44 retained higher relative leaf water content, and intrinsic water use efficiency was enhanced. Furthermore, HN44 sustained higher SOD activity and proline, reducing the accumulation of MDA and the damage caused by drought stress. Through transcriptome analysis, HN44 had fewer DEGs under LD and insensitivity to water deficit as compared to SN14. Under SD, the expression of genes encoding proteins in the photosynthesis pathway (including subunit proteins in photosystems I and II, Pet family proteins, and ATP synthase) was inhibited, resulting in the reduction of photosynthetic capacity and carbon assimilation ability and affecting growth and development. In terms of glutathione metabolism, genes regulating GPX and GGT in glutathione metabolism were upregulated, and the genes of the glutathione metabolic pathway in HN44 were activated with a stronger ability to clear ROS. The expression of genes encoding PYR/PYL/RCAR proteins in the ABA regulatory signaling pathway was downregulated, while the expression of PP2C and SnRK2 genes was upregulated. These genes activated the regulation of downstream ABFs, thereby controlling stomatal closure, reducing water loss, improving WUE, and enhancing drought resistance. The drought-tolerant variety HN44 exhibited better drought adaptability and WUE through differential expression of key genes in these major pathways. Based on the predicted differences in transcription factor expression regulation patterns between the two varieties, eight candidate drought resistance transcription factors had been identified: LOC100808190, GmWRKY49, LOC100798375, LOC100804384, LOC100817546, GmbZIP4, LOC100819313, and GmbZIP1. These genes were related to the MAPK signaling pathway and plant hormone signal transduction. The DEGs and key protectants involved in regulatory pathways identified in this study could provide important insights into the complex drought resistance mechanisms in plants. They could provide a theoretical basis for molecular breeding for high drought resistance and high water use efficiency of soybeans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Bioproject accession number: PRJNA1011827.
Author contributions
YX: Writing – review & editing, Data curation, Formal Analysis, Investigation, Validation, Writing – original draft. DS: Validation, Writing – review & editing. XQ: Validation, Writing – review & editing. MA: Writing – review & editing. SuW: Software, Validation, Writing – review & editing. XT: Validation, Visualization, Writing – review & editing. YJ: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. ShW: Funding acquisition, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The experiment was supported by the Fourteenth Five-Year Key R&D Program of the Ministry of Science and Technology of China (2021YFD1201103-01-04) and the National Modern Soybean Industry Technology System Construction Project (CARS-04-PS06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1269884/full#supplementary-material
Supplementary Figure 1 | The number of transcription factors under water deficit.
References
Anderson, J. V., Davis, D. G. (2004). Abiotic stress alters transcript profiles and activity of glutathione S-transferase, glutathione peroxidase, and glutathione reductase in Euphorbia esula. Physiologia Plantarum 120, 421–433. doi: 10.1111/j.0031-9317.2004.00249.x
Ariyarathne, M. A., Wone, B. W. M. (2022). Overexpression of the Selaginella lepidophylla bHLH transcription factor enhances water-use efficiency, growth, and development in Arabidopsis. Plant Sci. 315, 111129. doi: 10.1016/j.plantsci.2021.111129
Ashraf, M., Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59, 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Awasthi, R., Kaushal, N., Vadez, V., Turner, N. C., Berger, J., Siddique, K. H. M., et al. (2014). Individual and combined effects of transient drought and heat stress on carbon assimilation and seed filling in chickpea. Funct. Plant Biol. 41, 1148. doi: 10.1071/FP13340
Baldoni, E., Bagnaresi, P., Locatelli, F., Mattana, M., Genga, A. (2016). Comparative leaf and root transcriptomic analysis of two rice japonica cultivars reveals major differences in the root early response to osmotic stress. Rice 9. doi: 10.1186/s12284-016-0098-1
Baldoni, E., Genga, A., Cominelli, E. (2015). Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 16, 15811–15851. doi: 10.3390/ijms160715811
Basim, H., Basim, E., Tombuloglu, H., Unver, T. (2021). Comparative transcriptome analysis of resistant and cultivated tomato lines in response to Clavibacter michiganensis subsp. Michiganensis. Genomics 113, 2455–2467. doi: 10.1016/j.ygeno.2021.05.033
Bertolino, L. T., Caine, R. S., Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00225
Bramley, H., Ranawana, S., Palta, J. A., Stefanova, K., Siddique, K. (2022). Transpirational leaf cooling effect did not contribute equally to biomass retention in wheat genotypes under high temperature. Plants (Basel) 11. doi: 10.3390/plants11162174
Calderone, V., Trabucco, M., Vujičić, A., Battistutta, R., Giacometti, G. M., Andreucc, F.. (2003). Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep. 4, 900–905. doi: 10.1038/sj.embor.embor923
Caruso, G., Cavaliere, C., Foglia, P., Gubbiotti, R., Samperi, R., Lagana, A. (2009). Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci. 177, 570–576. doi: 10.1016/j.plantsci.2009.08.007
Chen, X., Jiang, X., Niu, F., Sun, X., Hu, Z., Gao, F., et al. (2023). Overexpression of lncRNA77580 regulates drought and salinity stress responses in soybean. Plants 12, 181. doi: 10.3390/plants12010181
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X.. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Cheng, L., Zhao, T., Wu, Y., Wang, H., Zhang, Z., Zhang, D., et al. (2020). Identification of AP2/ERF genes in apple (Malus × domestica) and demonstration that MdERF017 enhances iron deficiency tolerance. Plant Cell Tissue Organ Culture (PCTOC) 143, 465–482. doi: 10.1007/s11240-020-01925-z
Chimungu, J. G., Brown, K. M., Lynch, J. P.. (2014). Reduced root cortical cell file number improves drought tolerance in maize. Plant Physiol. 166, 1943–1955. doi: 10.1104/pp.114.249037
Das, A., Eldakak, M., Paudel, B., Kim, D., Hemmati, H., Basu, C., et al. (2016). Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed. Res. Int. 2016, 1–23. doi: 10.1155/2016/6021047
Dien, D. C., Mochizuki, T., Yamakawa, T. (2019). Effect of various drought stresses and subsequent recovery on proline, total soluble sugar and starch metabolisms in Rice (Oryza sativa L.) varieties. Plant Production Sci. 22, 530–545. doi: 10.1080/1343943X.2019.1647787
Drake, P. L., Froend, R. H., Franks, P. J. (2013). Smaller, faster stomata: Scaling of stomatal size, rate of response, and stomatal conductance. J. Exp. Bot. 64, 495–505. doi: 10.1093/jxb/ers347
Fletcher, A. L., Sinclair, T. R., Allen, L. H. (2007). Transpiration responses to vapor pressure deficit in well watered ‘slow-wilting’ and commercial soybean. Environ. Exp. Bot. 61, 145–151. doi: 10.1016/j.envexpbot.2007.05.004
García-Cerdán, J. G., Kovács, L., Tóth, T., Kereïche, S., Aseeva, E., Boekema, J., et al. (2011). The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65, 368–381. doi: 10.1111/j.1365-313X.2010.04429.x
Geiger, D., Scherzer, S., Mumm, P., Marten, I., Ache, P., Matschi, S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. 107, 8023–8028. doi: 10.1073/pnas.0912030107
Huang, Y., Jiao, Y., Xie, N., Guo, Y., Zhang, F., Xiang, Z., et al. (2019). OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 287, 110188. doi: 10.1016/j.plantsci.2019.110188
Jensen, P. E., Gilpin, M., Knoetzel, J., Scheller, H. V. (2000). The PSI-K subunit of photosystem i is involved in the interaction between light-harvesting complex i and the photosystem i reaction center core. J. Biol. Chem. 275, 24701–24708. doi: 10.1074/jbc.M000550200
Jiang, Y., King, C. A., Purcell, L. C., Wang, S. (2020). Nitrogen fixation sensitivity related to water use efficiency at reproductive development in soybean. Plant Soil Environ. 66, 345–350. doi: 10.17221/271/2020-PSE
Jiang, Y., Wang, X. X., Meng, H., Xu, Y. W., Wang, S., Wang, S. D.. (2022). Photosynthetic physiology performance and expression of transcription factors in soybean of water use efficiency difference. Russian J. Plant Physiol. 69. doi: 10.1134/S102144372201006X
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A.A., et al. (2016). Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01029
Kumar, S., Asif, M. H., Chakrabarty, D., Tripathi, R. D., Dubey, R. S., Trivedi, P. K., et al. (2013). Differential expression of rice lambda class GST gene family members during plant growth, development, and in response to stress conditions. Plant Mol. Biol. Rep. 31, 569–580. doi: 10.1007/s11105-012-0524-5
Lata, C., Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Li, M., Li, H., Sun, A., Wang, L., Ren, C., Liu, J., et al. (2022). Transcriptome analysis reveals key drought-stress-responsive genes in soybean. Front. Genet. 13. doi: 10.3389/fgene.2022.1060529
Lian, X., Piao, S., Chen, A., Huntingford, C., Fu, B., Li, L. Z. X., et al. (2021). Multifaceted characteristics of dryland aridity changes in a warming world. Nat. Rev. Earth Environ. 2, 232–250. doi: 10.1038/s43017-021-00144-0
Liu, W., Deng, Y., Zhou, Y., Chen, H., Dong, Y., Wang, N., et al. (2016). Normalization for relative quantification of mRNA and microRNA in soybean exposed to various abiotic stresses. PloS One 11, e155606. doi: 10.1371/journal.pone.0155606
Liu, S., Zhang, M., Feng, F., Tian, Z. (2020). Toward a “green revolution” for soybean. Mol. Plant 13, 688–697. doi: 10.1016/j.molp.2020.03.002
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
McAusland, L., Vialet Chabrand, S., Davey, P., Baker, N. R., Brendel, O., Lawson, T., et al. (2016). Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol. 211, 1209–1220. doi: 10.1111/nph.14000
Ohashi, Y. H. U. J., Nakayama, N. H. U. J., Saneoka, H. H. U. J., Fujita, K. H. U. J. (2006). Effects of drought stress on photosynthetic gas exchange, chlorophyll fluorescence and stem diameter of soybean plants. Biol. Plantarum 50, 138–141. doi: 10.1007/s10535-005-0089-3
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., Luan, S. (2005). ABR1, an APETALA2-Domain transcription factor that functions as a repressor of ABA response in arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Scheller, H. V., Jensen, P. E., Haldrup, A., Lunde, C., Knoetzel, J. (2001). Role of subunits in eukaryotic Photosystem I. Biochim. Biophys. Acta 1507, 41–60. doi: 10.1016/s0005-2728(01)00196-7
Shinozaki, K., Yamaguchi-Shinozaki, K. (2006). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Singh, D., Debnath, P., Sane, A. P., Sane, V. A. (2023). Tomato (Solanum lycopersicum) WRKY23 enhances salt and osmotic stress tolerance by modulating the ethylene and auxin pathways in transgenic Arabidopsis. Plant Physiol. Biochem. 195, 330–340. doi: 10.1016/j.plaphy.2023.01.002
Stark, R., Grzelak, M., Hadfield, J. (2019). RNA sequencing: The teenage years. Nat. Rev. Genet. 20, 631–656. doi: 10.1038/s41576-019-0150-2
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R.K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 08. doi: 10.3389/fpls.2017.00161
von Sydow, L., Schwenkert, S., Meurer, J., Funk, C., Mamedov, F., Schröder, W. P.. (2016). The PsbY protein of Arabidopsis Photosystem II is important for the redox control of cytochrome b559. Biochim Biophys Acta 1857, 1524–1533. doi: 10.1016/j.bbabio.2016.05.004
Wang, K., Bu, T., Cheng, Q., Dong, L., Su, T., Chen, Z., et al. (2021). Two homologousLHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 229, 2660–2675. doi: 10.1111/nph.17019
Wang, X., Song, S., Wang, X., Liu, J., Dong, S. (2022b). Transcriptomic and metabolomic analysis of seedling-stage soybean responses to peg-simulated drought stress. Int. J. Mol. Sci. 23, 6869. doi: 10.3390/ijms23126869
Wang, X., Wu, Z., Zhou, Q., Wang, X., Song, S., Dong, S.. (2022a). Physiological response of soybean plants to water deficit. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.809692
Wang, H., Zhou, Q., Mao, P. (2020). Ultrastructural and photosynthetic responses of pod walls in alfalfa to drought stress. Int. J. Mol. Sci. 21, 4457. doi: 10.3390/ijms21124457
Way, D. A., Pearcy, R. W. (2012). Sunflecks in trees and forests: From photosynthetic physiology to global change biology. Tree Physiol. 32, 1066–1081. doi: 10.1093/treephys/tps064
Xu, Z., Zhou, G., Shimizu, H. (2010). Plant responses to drought and rewatering. Plant Signaling Behav. 5, 649–654. doi: 10.4161/psb.5.6.11398
Yang, X., Kwon, H., Kim, M. Y., Lee, S. (2023). Rna-seq profiling in leaf tissues of two soybean (glycine max [l.] Merr.) Cultivars that show contrasting responses to drought stress during early developmental stages. Mol. Breed. 43. doi: 10.1007/s11032-023-01385-1
Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757. doi: 10.1007/s00299-017-2118-z
Zhang, H., Jay Forman, H., Choi, J. (2005). “G-glutamyl transpeptidase in glutathione biosynthesis,” in Elsevier science & Technology(United States: Elsevier science & Technology), 468–483.
Zhang, H., Song, B. (2017). RNA-seq data comparisons of wild soybean genotypes in response to soybean cyst nematode ( Heterodera glycines ). Genomics Data 14, 36–39. doi: 10.1016/j.gdata.2017.08.001
Zhang, H., Xiang, Y., He, N., Liu, X., Liu, H., Fang, L., et al. (2020). ). Enhanced vitamin c production mediated by an aba-induced ptp-like nucleotidase improves plant drought tolerance in arabidopsis and maize. Mol. Plant 13, 760–776. doi: 10.1016/j.molp.2020.02.005
Zhou, Q., Tian, A., Zou, H., Xie, Z., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: drought stress, soybean, photosynthesis, stomata, water use efficiency, transcriptome
Citation: Xu Y, Song D, Qi X, Asad M, Wang S, Tong X, Jiang Y and Wang S (2023) Physiological responses and transcriptome analysis of soybean under gradual water deficit. Front. Plant Sci. 14:1269884. doi: 10.3389/fpls.2023.1269884
Received: 31 July 2023; Accepted: 09 October 2023;
Published: 26 October 2023.
Edited by:
Junliang Fan, Northwest A&F University, ChinaReviewed by:
Waqar Shafqat, Mississippi State University, United StatesTinashe Zenda, Hebei Agricultural University, China
Copyright © 2023 Xu, Song, Qi, Asad, Wang, Tong, Jiang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Jiang, WWppYW5nQG5lYXUuZWR1LmNu; Shaodong Wang, d3NkaGxqQG5lYXUuZWR1LmNu
 Yuwen Xu
Yuwen Xu Di Song1
Di Song1 Sui Wang
Sui Wang Yan Jiang
Yan Jiang