- 1College of Resources, Hunan Agricultural University, Changsha, China
- 2Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
Several mechanisms have been proposed to explain NH4+ toxicity. However, the core information about the biochemical regulation of plants in response to NH4+ toxicity is still lacking. In this study, the tissue NH4+ concentration is an important factor contributing to variations in plant growth even under nitrate nutrition and NH4+ tolerance under ammonium nutrition. Furthermore, NH4+ led to the reprogramming of the transcriptional profile, as genes related to trehalose-6-phosphate and zeatin biosynthesis were downregulated, whereas genes related to nitrogen metabolism, camalexin, stilbenoid and phenylpropanoid biosynthesis were upregulated. Further analysis revealed that a large number of genes, which enriched in phenylpropanoid and stilbenoid biosynthesis, were uniquely upregulated in the NH4+- tolerant ecotype Or-1. These results suggested that the NH4+-tolerant ecotype showed a more intense response to NH4+ by activating defense processes and pathways. Importantly, the tolerant ecotype had a higher 15NH4+ uptake and nitrogen utilization efficiency, but lower NH4+, indicating the tolerant ecotype maintained a low NH4+ level, mainly by promoting NH4+ assimilation rather than inhibiting NH4+ uptake. The carbon and nitrogen metabolism analysis revealed that the tolerant ecotype had a stronger carbon skeleton production capacity with higher levels of hexokinase, pyruvate kinase, and glutamate dehydrogenase activity to assimilate free NH4+, Taken together, the results revealed the core mechanisms utilized by plants in response to NH4+, which are consequently of ecological and agricultural importance.
Introduction
Nitrogen (N) is an essential plant macronutrient that is required for growth and is thus important in agricultural production. NH4+-based fertilizers contain commonly used N forms synthesized via the Haber–Bosch process (Halvorson et al., 2014). While NH4+ is the preferred form of N for plants at low concentrations, it is toxic to plants at high concentrations (von Wirén et al., 2000; Britto and Kronzucker, 2002). As NH4+ escapes little with water in soil and elevated CO2 reduces nitrate reductions in C3 species, a “more NH4+ solution” was considered to mitigate nitrogen pollution and improve crop yields (Rubio-Asensio and Bloom, 2016; Subbarao and Searchinger, 2021). However, excess NH4+/NH3 atmospheric depositions have attracted attention in recent decades as they caused environmental problems related to species richness and composition (Stevens et al., 2004; Clark and Tilman, 2008; Duprè et al., 2010). The mechanisms underlying NH4+ toxicity in plants are consequently of ecological and agricultural importance.
NH4+ toxicity causes a retardation in plant growth, shortened roots, reduced root gravitropism, and leaf chlorosis (Britto and Kronzucker, 2002; Li et al., 2014). Several mechanisms have been proposed to explain NH4+ toxicity, including the depletion of organic acids (Hachiya et al., 2010), deficiency of cations (Hoopen et al., 2010), futile transmembrane NH4+ cycling (Britto et al., 2001; Li et al., 2022), photodamage to photosystem II (Dai et al., 2014), reactive oxygen and nitrogen species (RONS) induced oxidative stress (Liu et al., 2022), and the disruption of hormonal homeostasis (Li et al., 2012). The reactions of NH4+ conjugation to glutamic acid and the synthesis of glutamic acid are critical for the detoxification of NH4+, and are catalyzed by glutamine synthase (GS), glutamate synthase (GOGAT), and glutamate dehydrogenase (GDH) (Masclaux-Daubresse et al., 2006). Studies have shown that GS activity is upregulated by high external NH4+ levels and that NH4+-tolerant plants have higher GS activity and lower levels of NH4+ accumulation in their tissues (Cruz et al., 2006; Guan et al., 2016). In addition, NH4+ uptake is tightly controlled through the NH4+-dependent inhibition of AMT1 by the phosphorylation of Thr-460 (Straub et al., 2017).
In recent years, numerous genetic loci controlling NH4+ toxicity have been identified by screening for mutants. The Arabidopsis mutant hsn1-1 and its alelic mutant vtc1 are hypersensitive to NH4+ because of point mutations in the gene encoding GDP-mannose pyrophosphorylase (GMPase). Defects in protein N-glycosylation of hsn1-1 have been linked to the root hypersensitivity phenotype (Qin et al., 2008). Based on leaf hypersensitivity to NH4+, ammonium overly sensitive 1 (amos1) and amos2 were identified. amos1 was found to be an allelic mutation of EGY1 encoding a plastid metalloprotease. AMOS1/EGY1 mediated plastid retrograde signaling regulates NH4+-responsive genes to maintain chloroplast functionality (Li et al., 2012). An ammonium tolerance mutant (amot1) was identified as allelic to EIN3, which positively regulates ROS production and induces oxidative stress under NH4+ stress (Li et al., 2019a). Moreover, other genetic loci that control root development and gravitropism have been identified in Arabidopsis, including auxin resistant 1 (aux1), tiny root hair 1 (trh1), dolichol phosphate mannose synthase1 (dpms1), and gravitropism sensitive to ammonium 1 (gsa1) (Li et al., 2014).
Although the numerous genetic loci were found to associate with the toxicity of NH4+ nutrition, the function studies of individual genes cannot fully reflect the natural responses of plants to NH4+ toxicity. The tolerance of plants to NH4+ depends on the species and variety. For example, some rice cultivars have adapted to NH4+, while Arabidopsis thaliana and the Brassicaceae family are sensitive to NH4+ (Esteban et al., 2016; Li et al., 2021). In this study, we explore the cellular threshold of NH4+ concentration by investigated the A. thaliana natural accessions. More importantly, the core information about the biochemical regulation of plants in response to NH4+ toxicity was identified by comparative assessments of the sensitive and tolerant species.
Materials and methods
Plant material and culture conditions
A natural accessions of Arabidopsis thaliana were used in this study (Table S1). Surface-sterilized seeds were sown onto nutritional soil in a greenhouse (300 μmol photons m-2 s-1, 16 h photoperiod, 22°C) for 7 d. Then, eight uniformly growing plants of each ecotype were grown hydroponically with 4 L of nutritional media per pot, containing 1 mM Ca(NO3)2 or 1 mM (NH4)2SO4. The other essential nutrients in the two different nitrogen nutrient solutions were the same as our previous study (Chen et al., 2021), specially 1.25 mM KCl, 0.625 mM KH2PO4, 0.5 mM MgSO4, 25 μM Fe-EDTA, 17.5 μM H3BO3, 3.5 μM MnCl2, 0.25 μM ZnSO4, 0.05 μM NaMoO4, and 0.125 μM CuSO4. It is of note that the concentration of Ca2+ in the solution was uniformly set to 1.5 mM, and The pH was buffered to 6.0 using 2.5 mM MES. The thaliana natural accessions was grown under the NH4+ and NO3-.
The seedlings were first grown in 1 mM Ca(NO3)2 nutrient solution for 5 d and then transferred to nitrogen-free nutrient solution for 3 d. Subsequently, the thaliana natural accessions were grown under the NH4+ and NO3- treatments for 8 days to investigate the NH4+ concentration and fresh weight. The culture solution was refreshed every 4 days.
Measurement of the NH4+ and total nitrogen concentrations
NH4+ was extracted with deionized water using fresh rosette leaves. The supernatant was used to determine the NH4+ concentration after the samples were centrifuged at 12,000 × g. Briefly, 20 μL supernatant was mixed with 0.5 mL phenol solution (10 g/L Phenol, 100 mg/L sodium nitrosoferricyanide) and 5 mL sodium hypochlorite alkaline solution (Sodium hydroxide 5 g, sodium hydrogen phosphate 3.53 g, sodium phosphate 15.9 g, sodium hypochlorite solution (ω= 5.25%) 5 mL, dissolved in 500 ml deionized water). The NH4+ concentrations were measured colorimetrically using phenol hypochlorite (Berthelot reaction) colorimetry at 630 nm, and (NH4)2SO4 was used as a standard.
For total nitrogen (TN) measurement, the plants were sampled and then dried at 105°C for half an hour and further dried at 65°C until a constant weight. The samples were weighed 0.100g using a thousand balance, packed in 150 mL narrow-mouth triangular flasks, boiled with H2SO4-H2O2 until transparent and clear. The solution was transferred to a 50 mL volumetric flask and made up to volume. The solution was then filtered and analyzed using a continuous-flow analyzer AA3 (Autoanalyzer 3; SEAL, Germany). NUtE (Nitrogen utilization efficiency) was calculated as dry shoot biomass/shoot N.
Enzyme activity
Fresh samples (100 mg) were homogenized in 3 mL of 50 mM Tris-HCl buffer (pH 8.0) containing 2 mM Mg2+, 2 mM DTT, and 0.4 M sucrose. After centrifuged at 10000 × g and 4°C for 10 min, the supernatant was then used to determine the activity of glutamine synthase. The activities of GS were measured according to the reference (Chen et al., 2018). Briefly, 0.5 mL supernatant was mixed with 1.6 mL reagent①, which is composed of 100 mM Tris-HCl (pH 7.4), 80 mM MgSO4, 20 mM glutamate, 20 mM cysteic acid, 2 mM EGTA and 80 mM hydroxyl-amine hydrochloride, and 0.7 mL reagent②, which is composed of 40 mM ATP. The 3 mL reaction mixture was incubated for 30 min at 37°C and was terminated by adding 1 mL FeCl3 reagent (88 mM FeCl3, 670 mM HCl and 200 mM TCA). After 10 min, the mixture was centrifuged at 4000 × g for 10 min, and the absorption value of supernatanta solution was determined at 540 nm wavelength.
NADH-dependent glutamate dehydrogenase was measured according to the reference (Groat and Vance, 1981). Briefly, fresh samples (100 mg) were homogenized in 3 mL of 50 mM Tris-HCl buffer (pH 8.0). 50 μL of the supernatant was mixed with the 950 μL reagent, which contained 100 μM NADH, 2.5 mM 2-oxoglutarate (2-OG), 200 mM NH4Cl. The enzyme activity was defined as the reduction of absorbance due to NADH at 340 nm. The assay of enzyme activity was performed according the instruction of commercial kit (NADH-GDH kit, BC1465, Solarbio).
Pyruvate (Pyr) concentrations were measured according to the reference with some modifications (Kachmar and Boyer, 1953). Pyr reacts with 2, 4-dinitrophenylhydrazine to form pyruvate-2, 4-dinitrophenylhydrazone, which could be determined by calorimetry. Briefly, 0.1 g Fresh samples (100 mg) were homogenized in 1 mL trichloroacetic acid (8%) and placed on ice for 3 min. After centrifuged at 8000 × g and 4°C for 10 min, 75 μL the supernatant was added to the enzyme label plate and added 25 μL 2, 4-dinitrophenylhydrazine (0.05%), to react 2 min. Finally, 125 μL 1.5 M NaOH was added, and the absorption value of the tube was determined at 520 nm wavelength. The assay was performed using the Micro Pyruvate Assay Kit (BC2205; Solarbio).
Pyruvate kinase (PK) activity was measured according to the reference (Lepper et al., 2010). The assay was performed according to the instructions of the Pyruvate Kinase Assay Kit (BC0540; Solarbio). Briefly, 0.1 g Fresh samples (100 mg) were homogenized in 3 mL extracting solution, which contained 100 mM Tris-HCl (pH 7.5), 10 mM β-mercaptoethanol, 12.5% (v/v) glycerine, l mM EDTA-Na2, 10 mM MgCl2, and 1% (m/v) PVP-40. After centrifuged at 8000 × g and 4°C for 10 min, 0.1 mL of the supernatant was mixed with 0.9 mL reagent, which contained 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.16 mM NADH, 75 mM KCl, 5.0 mM ADP, 7.0 units L-lactate dehydrogenase (LDH), and 1.0 mM phosphoenolpyruvate (PEP). The absorption values at 340 nm wavelength were immediately recorded at 20s and 140s.
Hexose kinase (HXK) activity was measured according to the reference (Pancera et al., 2006). The assay was performed according to the instructions of the Hexose Kinase Assay Kit (BC0740; Solarbio). Briefly, 0.1 g Fresh samples (100 mg) were homogenized in 1 mL extracting solution. After centrifuged at 8000 × g and 4°C for 10 min, 10 μL the supernatant was added to the enzyme label plate, followed by 10 μL G-6-PDH solution (0.12 g/L) and 180 μL reagent, which 5 μl of a 2.0 mM NADP+ solution, 15 μl of a 0.1 M ATP solution, 50 μl of a 1 M glucose solution, and 110 μL of a 100 mM Tris–HCl buffer (pH 7.5). The absorption values at 340 nm wavelength were immediately recorded at 20s and 320s.
15N-NH4+ isotope tracing
The objective of 15N-labeling experiment after 3d of N starvation is investigate the transport capacity of both genotypes. Seedlings were first grown in 1 mM Ca(NO3)2 nutrient solution for 5 d, and then transferred to a nitrogen-free nutrient solution for 3 d. Subsequently, the plants were transplanted into 1 mM (15NH4)2SO4 with a 15N abundance of 5%. Samples were taken at 3, 6, and 24 h to determine the 15N content.
Comparative transcriptome analysis
The seedlings were grown in a NO3- nutrient solution for 5 d, and then transferred to a nitrogen-free nutrient solution for 3 d to deplete the stored NO3-. Subsequently, the plants were transferred to 1 mM Ca(NO3)2 or 1 mM (NH4)2SO4 for 1 d. The total RNA was then extracted from the roots of the NH4+-tolerant and -sensitive ecotypes. Three biological replicates were used for each ecotype under each treatment. The sequencing library was generated using the NEBNext Ultra™ RNA Library Prep Kit from Illumina (New York, NEB, USA), followed by sequencing on an Illumina HiSeq 2500 platform (San Diego, CA, USA). Gene expression levels were calculated using the FPKM method (Fragments Per Kilobase per Million mapped reads. Differentially expressed genes (DEGs) were defined as genes with |log2 (fold change) | >1 and a false discovery rate (FDR) < 0.05. The comparative transcriptome between NH4+ and NO3- was identified through comparisons of the FPKM values for each gene between NH4+ and NO3-. The comparative transcriptome between the two genotypes was identified through comparisons of the FPKM values for each gene between Or-1 and the Rak-2. GO enrichment analysis was performed by the agriGo program (http://bioinfo.cau.edu.cn/agriGO/). The significantly enriched GO terms were defined with corrected P < 0.05.
Statistical analysis
Graphical values represent the mean ± SD. Statistical analyses were conducted using two-tailed Student’s t-tests or one-way analysis of variance (ANOVA). Least significant difference (LSD) tests for different treatments were used for multiple comparisons with P < 0.05. The significance level was set at P < 0.05 (*) and P < 0.01 (**).
Results
The tissue NH4+ concentration was negatively linked to Arabidopsis growth
A panel of A. thaliana natural accessions was grown under identical concentrations of NH4+ and NO3-. The growth of each ecotype was significantly inhibited by NH4+ (Figures 1A, B). Conversely, nitrogen concentration was significantly higher in plants grown under NH4+ than under NO3- conditions (Figure 1C). However, the free NH4+ concentration in the shoots was significantly higher than that under NO3-, and its concentration varied widely among ecotypes (Figure 1D). The two-segment linear model accurately simulated the relationship between fresh weight and tissue NH4+ concentration under different nitrogen sources (Figure 1E, Table S1). The fresh weight sharply decreased with the tissue NH4+ concentration (<50 μg/g) under NO3- conditions, and the pace of decline slowed when the NH4+ concentration was > 50 g/g under NH4+ conditions (Figure 1E). Our data revealed that the tissue NH4+ concentration was negatively linked to Arabidopsis growth, even under NO3- culture conditions.
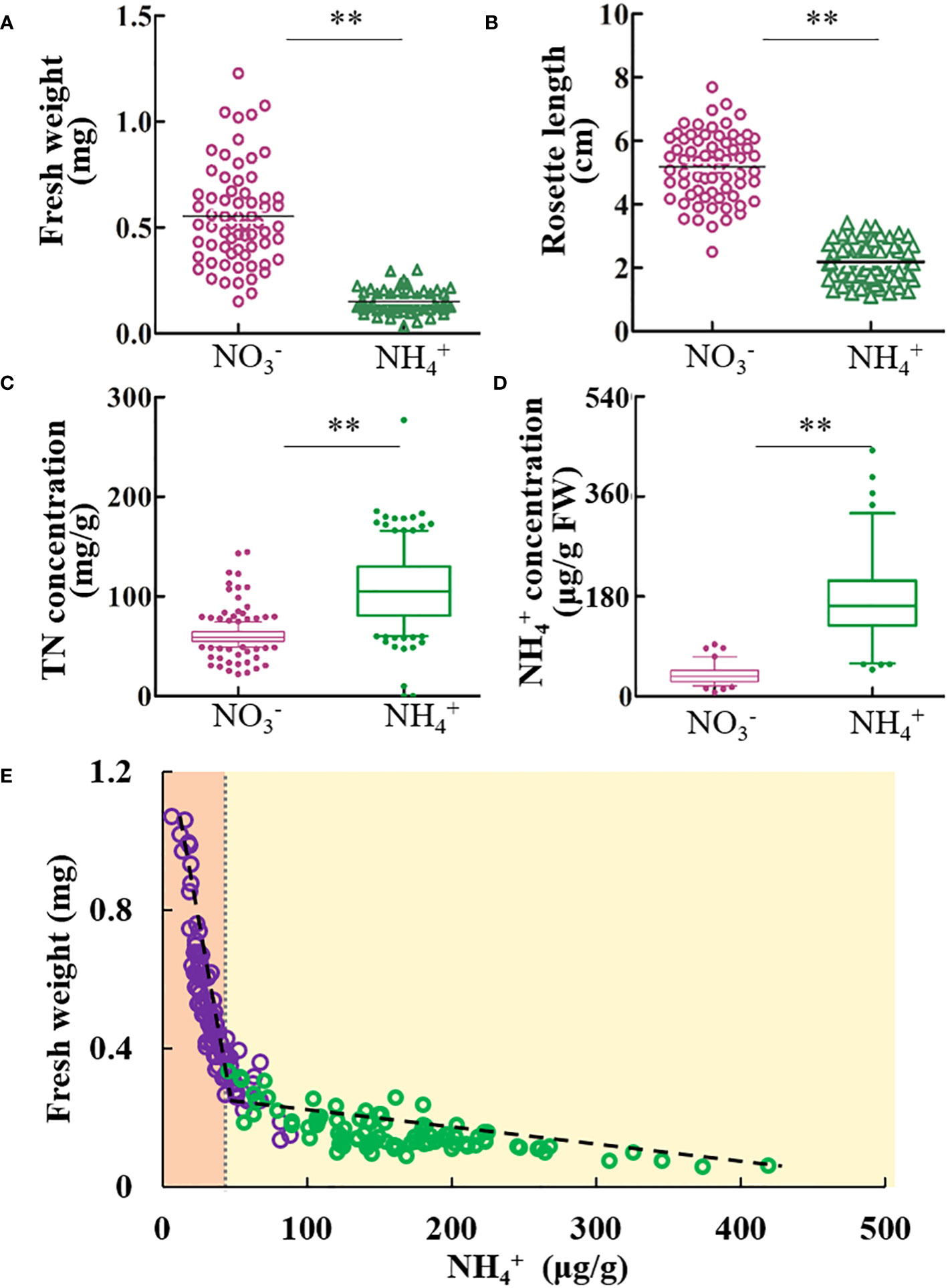
Figure 1 The tissue NH4+ concentration is negatively linked to the growth of Arabidopsis. After 8 d cultivation with NO3- or NH4+ nutrition, the physiological parameters including the fresh weight, rosette length, nitrogen concentration and free NH4+ were investigated. (A) The fresh weight, and (B) rosette length under nitrate or NH4+ nutrition. (C) Total nitrogen concentration, and (D) free NH4+ concentration in the plants. (E) The correlation analysis of free NH4+ with fresh weight. Dashed lines indicated two linear models. Pearson R2 values are given when p <.05. Student’s t test (**p <.01) was used to analyze statistical significance. For fresh weight and rosette length, results of each ecotype are means of eight biological replicates, and for TN and free NH4+ concentration, results of each ecotype are means of three biological replicates.
Sensitivity to NH4+ between Or-1 and the Rak-2
We comprehensively considered the biomass under ammonium nitrogen and the biomass under normal conditions, and excluded the ecotypes with poor growth under nitrate nitrogen. Additionally, the Or-1 and Rak-2 showed similar growth period. Thus, ecotypes Or-1 and Rak-2 were selected for further investigation. Or-1 grew better under both NO3- and NH4+ nutrition (Figure 2A), which was also supported by its biomass (Figure 2B). With NO3- nutrition, the biomass of Rak-2 was 18.4% smaller than that of Or-1. However, the difference in biomass between the two ecotypes was 53% under NH4+ nutrition (Figure 2B). The free NH4+ concentration in Rak-2 was significantly higher than that in Or-1 under both nitrate and NH4+ nutrition conditions (Figure 2C). Thus, Or-1 was characterized as an NH4+-tolerant ecotype with high biomass and low NH4+ concentration, while Rak-2 was characterized as an NH4+ -sensitive ecotype.
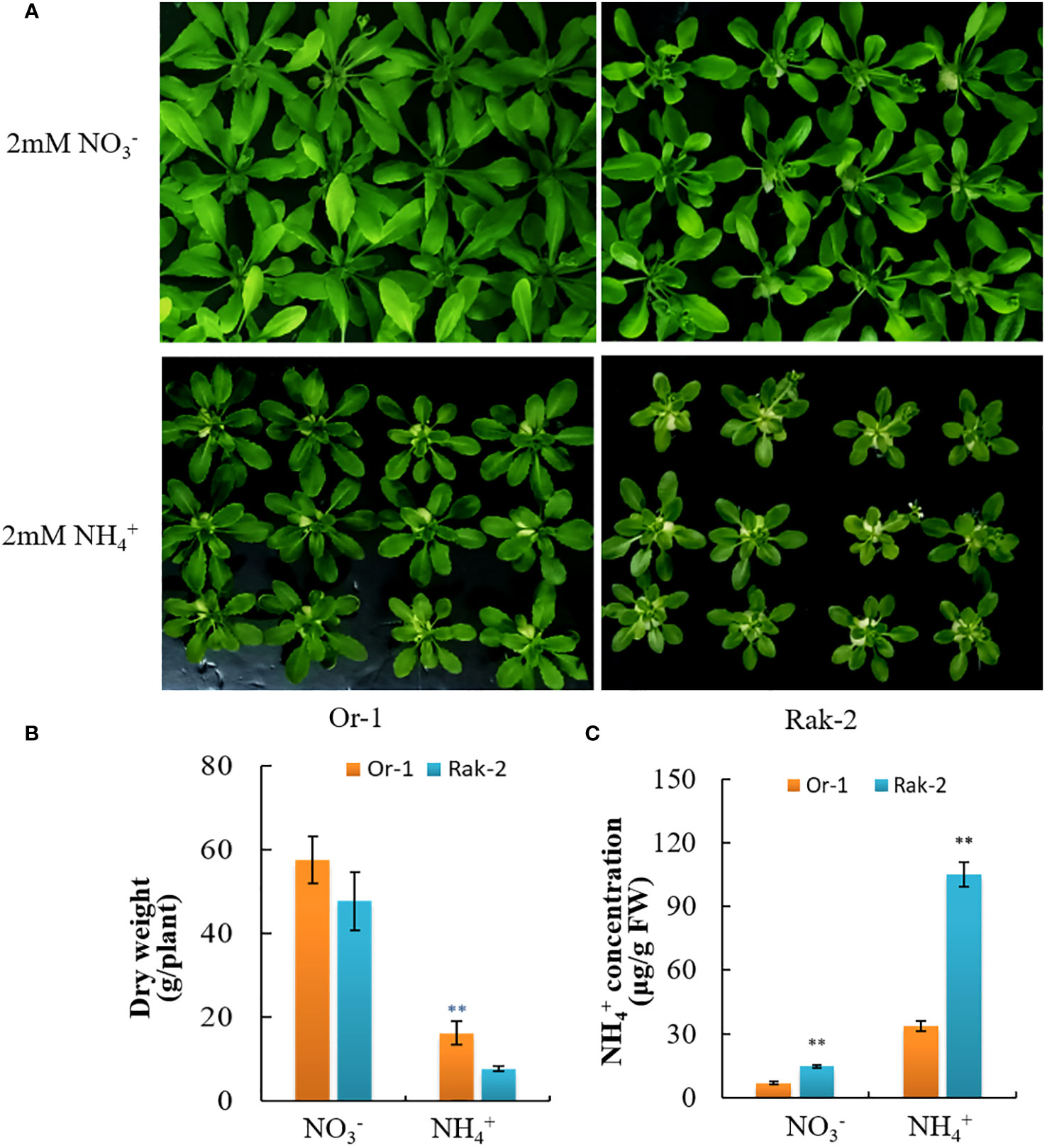
Figure 2 Different sensitivity to NH4+ nutrition between the Or-1 and the Rak-2. (A) Photos of the two ecotypes under NO3- or NH4+ nutrition. (B) The dry weight of whole plants, and (C) free NH4+ concentration of the two ecotypes under NO3- or NH4+ nutrition. For dry weight, results of each ecotype are means ± SD of eight biological replicates, and for free NH4+ concentration, results of each ecotype are means ± SD of three biological replicates. **p < .01.
The response of the transcriptome to NH4+ was analyzed to explore the underlying molecular mechanisms. In Or-1, 1016 genes were upregulated and 687 genes were downregulated by NH4+ (Figure S1A). In Rak-2 cells, 483 and 637 genes were upregulated and downregulated by NH4+, respectively (Figure S1B). NO3–responsive genes (e.g., NRT2.1, NIA1, G6PD3, and FNR2) were activated by NO3- in both ecotypes (Figure S1). Based on the fold change and FDR of the differentially expressed genes (DEGs), the genes (CLE4, PMEI4, FAR3, HHO1) were strongly activated by NO3-, and the genes (ALMT1, CCX1, ALD1, PBS3, DUR3) were strongly activated by NH4+ (Figure S1).
Core biological processes and pathways involved in the responses to NH4+
The DEGs in the two ecotypes were used to analyze the core biological processes and pathways in response to NH4+. The Gene Ontology (GO) enrichment analysis indicated that genes involved in hydrogen peroxide transport, camalexin biosynthesis, trehalose biosynthesis, and glutamate dehydrogenase activity were the most enriched terms regulated by NH4+ (Figure 3A). The genes (PIP2.1, PIP2.4, TIP1.1, TIP1.2), annotated to transport H2O and H2O2, respectively, were repressed by NH4+ (Figure 3B). However, the genes (NAC042, CYP71B15, AtCYP71B2, MPK9, WRKY33, PAD4) that participate in camalexin biosynthesis, were activated by NH4+ (Figure 3B). The trehalose-6-phosphate synthesis genes (TPS8, TPS 10) were repressed by NH4+, while the trehalose-6-phosphate phosphatase genes (TPPD, TPPG) were activated (Figure 3B). The results suggest that NH4+ represses the process of water transport and trehalose-6-phosphate synthesis but induces camalexin synthesis.
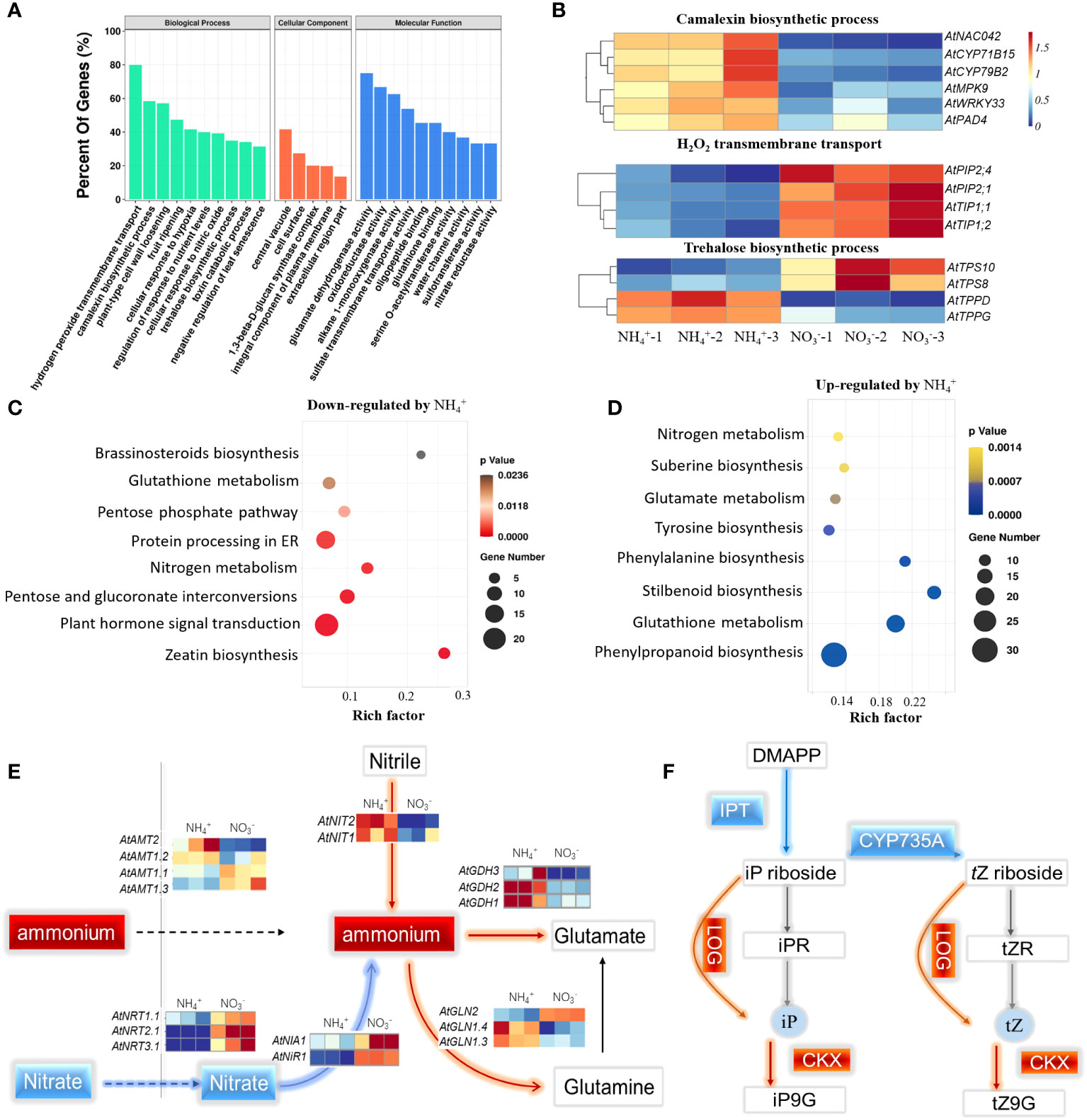
Figure 3 Overview of the transcriptome and the DEGs that were highly enriched in the core biological process and pathways in response to NH4+. (A) Gene ontology (GO) enrichment analysis of the DEGs under NH4+ stress. (B) The expression profiling of the genes involving in camalexin biosynthesis, H2O2 transmembrane transport and trehalose biosynthesis. (C) The enrichment pathways of the down-regulated genes by NH4+. (D) The enrichment pathways of the up-regulated genes by NH4+. (E) The effects of NH4+ nutrition on the pathway of nitrogen assimilation. (F) The effects of NH4+ nutrition on the pathway of zeatin biosynthesis. The red lines indicated the process was facilitated and the blue lines indicated the process was inhibited by NH4+.
By analyzing the up- or downregulated gene sets, the KEGG results showed that the most enriched pathways were activated or repressed by NH4+. The zeatin biosynthesis pathway was the most enriched process and was repressed by NH4+ (Figure 3C). The isopentenyl-transferase gene (IPT) and the cytochrome P450 monooxygenase CYP735A encodes the rate-limiting enzyme in CK synthesis. Surprisingly, the transcript levels of IPT3, 5, 7 with NH4+ nutrients were 6%, 39%, and 22%, respectively, of those under NO3- nutrients, and the transcript levels of CYP735A1, 2 under NH4+ nutrients were only 10% and 15%, respectively, of those under NO3- nutrient conditions (Figure 3E; Figure S2). However, the CK oxidase genes (CKX1, 5, 6) under NH4+ nutrient were 1.6 and 3.1 times higher than that under nitrate nutrient. These results support the conclusion that NH4+ represses CK synthesis but reduced the level of active CK.
The most enriched pathway activated by NH4+ was stilbenoid biosynthesis, followed by phenylalanine biosynthesis, glutathione metabolism, phenylpropanoid biosynthesis, and glutamate metabolism (Figure 3D). Nitrogen metabolism was simultaneously enriched in the upregulated and downregulated pathways, as the nitrate transporters (NRT1.1, NRT2.1, NRT3.1) and reduction genes (NIA1, NiR1) were strongly repressed by NH4+, and NH4+ assimilation genes (GLN1.4, GLN1.3, GDH3, GDH2, GDH1) were highly activated by NH4+ (Figure 3E).
Stilbenoid, phenylpropanoid, and sucrose metabolism enriched in the tolerant ecotype were up-regulated by NH4+
To explore the molecular mechanism of NH4+ sensitivity in Or-1 and Rak-2, we compared the transcriptomes of the NH4+-sensitive ecotype Rak-2 and the NH4+-tolerant ecotype Or-1, under NH4+ nutrient conditions. Overall, there were 2460 DEGs between the two ecotypes (Figure 4A; Figure S3). Among them, the expression of 1291 genes were higher in the NH4+-tolerant ecotype Or-1, which was more than the number of genes with higher expression in the NH4+-sensitive ecotype Rak-2 (Figure 4A). The functions of these DEGs were predicted using GO enrichment analysis. Some enriched GO terms between the two genotypes overlapped with those that responded to NH4+, such as camalexin biosynthesis, water channel activity, and nitrilase activity (Figure 4B; Figure 3A).
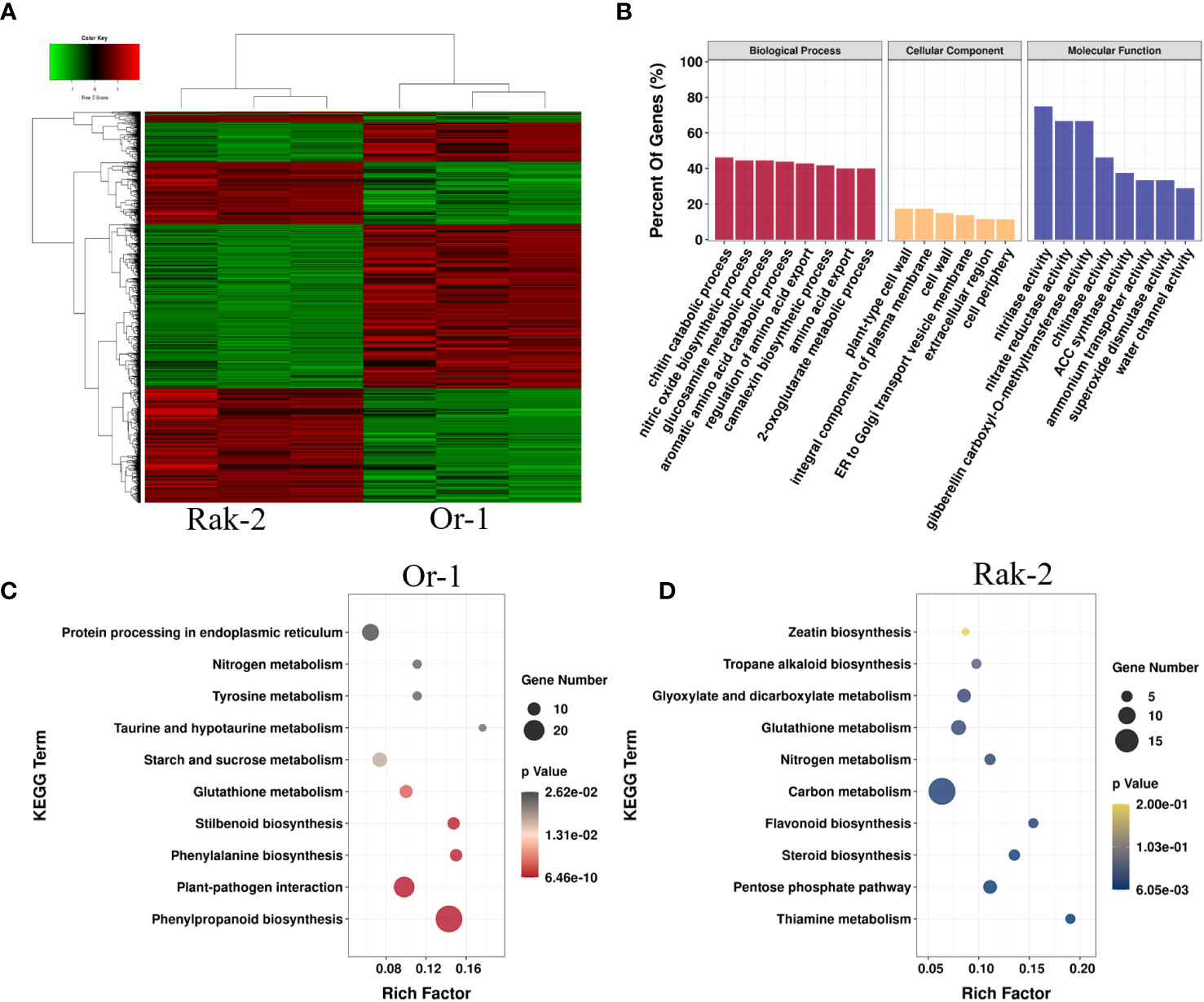
Figure 4 Transcriptional characterization of the DEGs between the NH4+-tolerant ecotype and the NH4+-sensitive ecotype. (A) The expression profiling of DGEs between the NH4+-tolerant ecotype and the NH4+-sensitive ecotype. (B) GO enrichment analysis of the DGEs. (C) The enrichment pathways of the DGEs with higher expression in the NH4+-tolerant ecotype Or-1. (D) The enrichment pathways of the DGEs with higher expression in the NH4+-sensitive ecotype Rak-2. The circle size indicates the number of DEGs, and the rich factor indicates the degree of enrichment of the KEGG pathways.
By analyzing the higher expression genes in the Rak-2 compared with the Or-1, The most enriched pathways were found to be thiamine metabolism, pentose phosphate pathway (Figure 4D). Analysis of genes with higher expression in Or-1 than in Rak-2 under NH4+-rich conditions. The most enriched pathways included stilbenoid, phenylpropanoid, glutathione, and sucrose metabolism (Figure 4C). Importantly, these pathways enriched in the Or-1 were up-regulated by NH4+ (Figure S4).
The NH4+-tolerant ecotype Or-1 exhibits a more intensive response to NH4+ by activating defense processes and pathways
As shown in the VENN diagram, 148 DEGs between the two ecotypes were simultaneously responsive to NH4+ (Figure 5A). In addition, 391 DEGs between the two ecotypes specifically responded to NH4+ in the tolerant ecotype Or-1 (Figure 5A). Similarly, 194 DEGs between the two ecotypes specifically responded to NH4+ in the sensitive ecotype Rak-2 (Figure 5A). The core 148 genes were classified into upregulated and downregulated genes following NH4+ treatment (Figures 5B, C; Table S2). Among the core 148 genes, a higher proportion of genes were upregulated by NH4+, especially Or-1 (Figure 5C). The cytochrome P450 genes CYP86A4, CYP71B22, CYP706A2, and CYP709B3 were strongly induced by NH4+ and showed higher Or-1 expression (Figure 5C). The transcription factors WRKY55, WRKY41, bZIP8, and ZAT8 were more strongly induced in the NH4+-tolerant ecotype Or-1 than that in the NH4+-sensitive ecotype Rak-2 (Figure 5C).
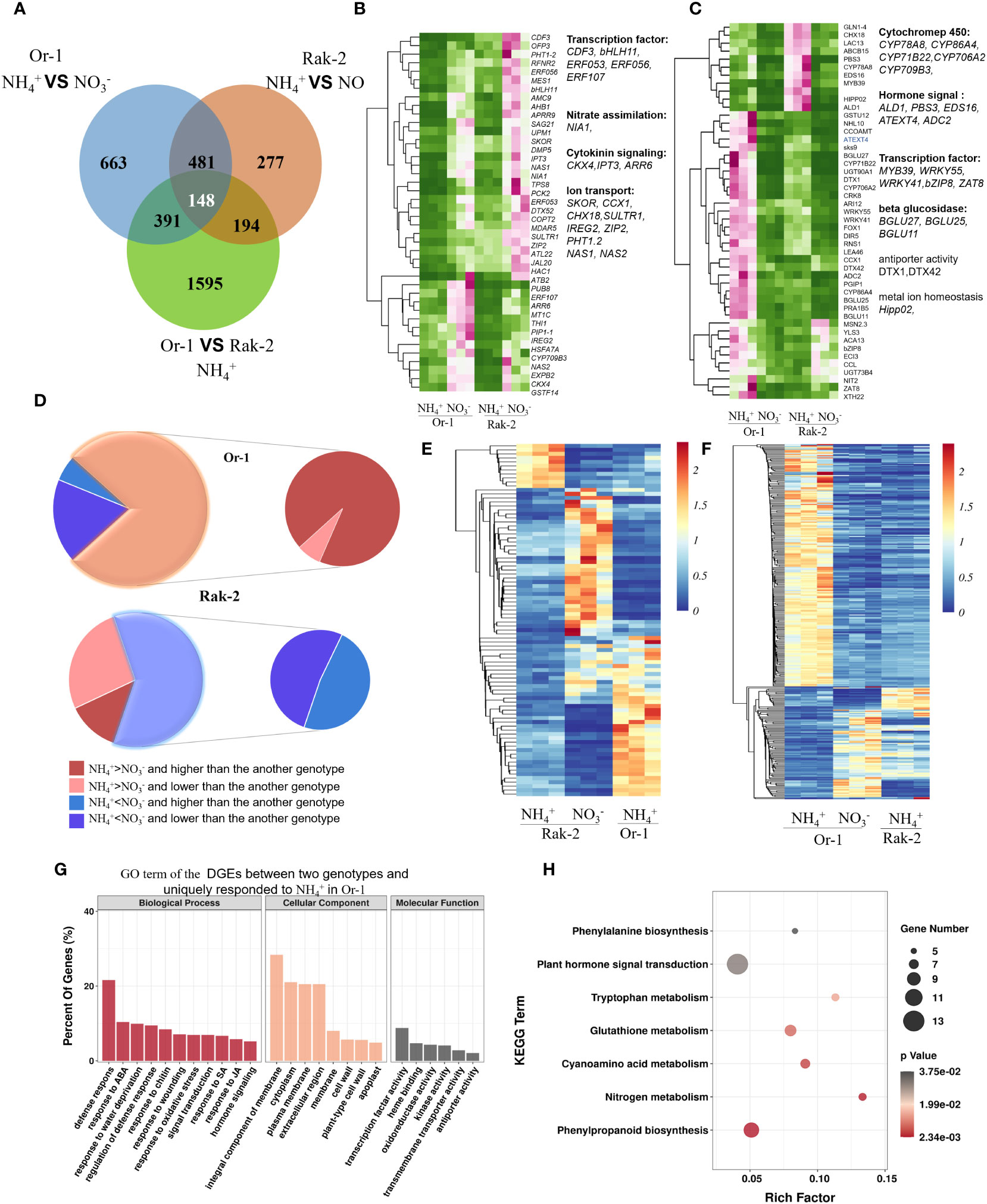
Figure 5 Analysis of the common and specific NH4+-responsive genes between the two ecotypes. (A) The VENN diagram of the DEGs. (B) The commonly down-regulated DEGs between the two ecotypes. (C) The commonly down-regulated DEGs between the two ecotypes. (D) The DEGs between the two ecotypes specifically responded to NH4+ only in one ecotype. (E) The expression profiling of the DEGs specifically responded to NH4+ in the sensitive ecotype Rak-2. (F) The expression profiling of the DEGs specifically responded to NH4+ in the tolerant ecotype Or-1. (G) GO enrichment analysis of the DGEs specifically responded to NH4+ in the tolerant ecotype Or-1. (H) The enrichment pathways of the DGEs specifically responded to NH4+ in the tolerant ecotype Or-1.
Surprisingly, 194 DEGs between the two ecotypes that specifically responded to NH4+ in Rak-2 were mostly downregulated by NH4+ (Figures 5D, E; Table S2). Conversely, 391 DEGs between the two ecotypes specifically responded to NH4+ in Or-1 plants (Figures 5D, F; Table S2). These genes were mostly upregulated by NH4+ and showed higher expression levels in Or-1 cells (Figures 5D, F). These results suggest that a large number of genes were uniquely upregulated in the NH4+- tolerant ecotype Or-1. The function of these unique NH4+-responsive genes in the Or-1 were predicted using GO enrichment analysis and KEGG pathways. The most highly enriched biological processes were defense responses, including responses to ABA, water deprivation, chitin, wounding, and oxidative stress (Figure 5G). Similarly, the KEGG pathway analysis indicated that nitrogen metabolism, phenylpropanoid biosynthesis, glutathione metabolism, and plant hormone signal transduction were the most highly enriched pathways (Figure 5H). Collectively, these results suggest that the NH4+-tolerant ecotype Or-1 exhibits a more intensive response to NH4+ by activating defense processes and pathways, including phenylpropanoid biosynthesis, nitrogen metabolism, and glutathione metabolism.
The tolerant ecotype maintained a low NH4+ level, mainly by promoting NH4+ assimilation rather than inhibiting NH4+ uptake
As the NH4+-sensitive ecotype, Rak-2 accumulated more NH4+ than the NH4+-tolerant ecotype Or-1 (Figure 2C), their uptake and assimilation of NH4+ were investigated. Both the concentration and accumulation of 15N were significantly higher in the NH4+-tolerant ecotype Or-1 (Figures 6A, B). The higher nitrogen utilization efficiency (NUtE) in the tolerant ecotype indicated the higher efficiency of the conversion of shoot N into shoot biomass (Figure 6C). The expression of AMT2 and AMT1.2 were also higher in the NH4+-tolerant ecotype (Figure 6D), which supports the results for 15NH4+ assimilation. Although the GS activity of both ecotypes increased with increasing NH4+ concentration, no difference was observed between the two (Figure 6E). The expression of Gln1s was significantly induced by NH4+, and only the expression of Gln1.4 slightly higher in the NH4+ -sensitive ecotype (Figure 6D). However, GDH activity was strongly induced by NH4+ nutrition and was significantly higher in the tolerant ecotype when compared with that in the sensitive ecotype (Figure 6F). Accordingly, the expression of GDH1 and GDH2 was more significantly induced in the roots of the tolerant ecotype than in those of the sensitive ecotype (Figure 6D).
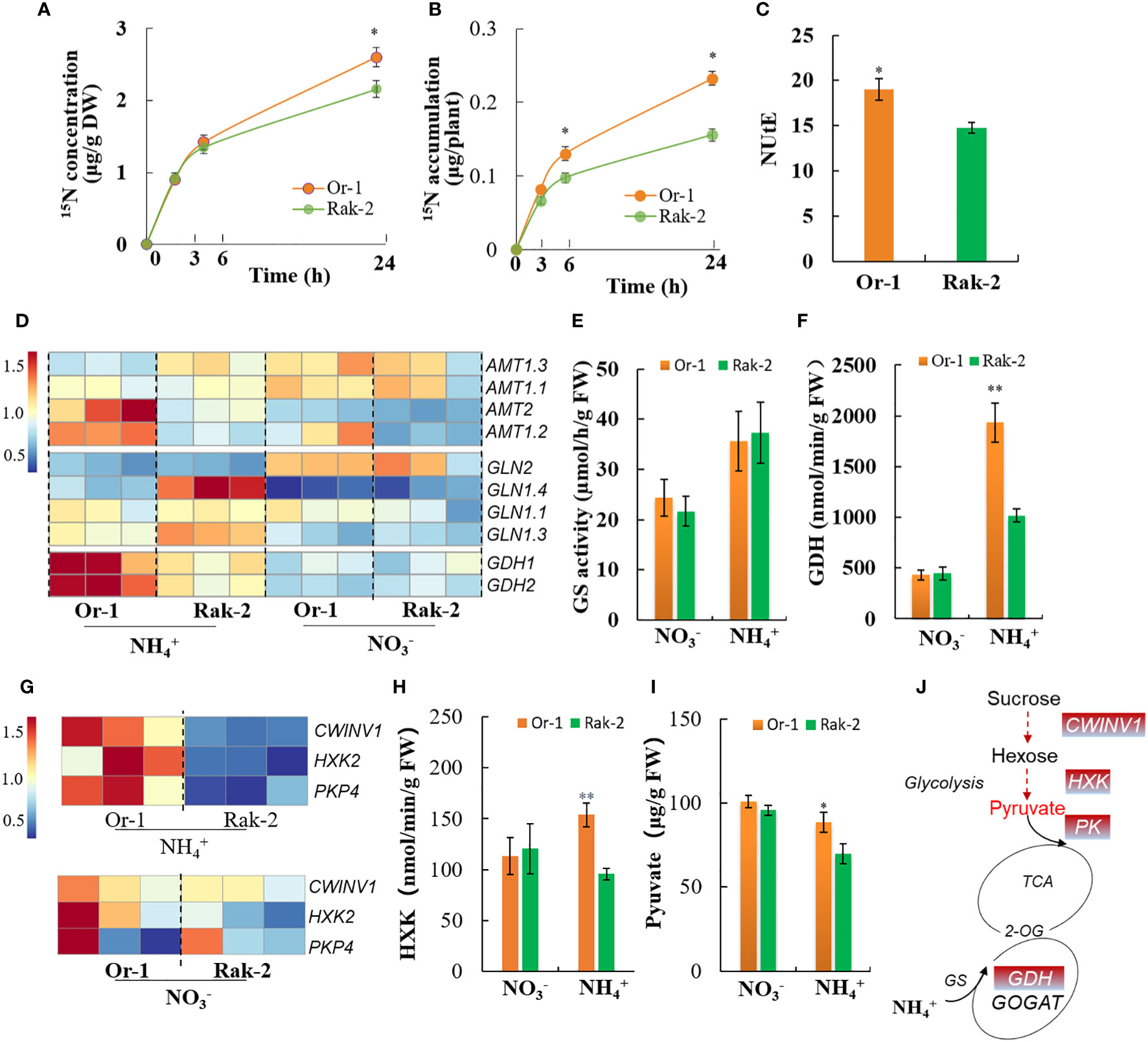
Figure 6 Analysis of NH4+ assimilation capacity between the two varieties. (A, B) The short-term of 15NH4+ uptake for 3 h, 6 h and 24 h. (C) The nitrogen utilization efficiency (NUtE) between the two ecotypes under NH4+ nutrition. NUtE is calculated by biomass/nitrogen. (D) Differential expression profiling of the NH4+ uptake and assimilation genes, including the NH4+ transporter genes AMTs, glutamine synthesis genes GLN and glutamate dehydrogenase genes GDH. (E) The activity of glutamine synthesis in the roots of two ecotypes. (F) The activity of GDH in the roots of two ecotypes. (G) Differential expression profiling of the carbohydrate metabolism genes, including the cell wall-bound invertase gene CWINV1, hexokinase gene HXK2 and pyruvate kinase gene PK4. (H) The activity of hexokinase in the roots of two ecotypes. (I) The content of pyruvate in the roots of two ecotypes. (J) The schematic diagram of carbon and nitrogen metabolism. The enzymes and metabolite highlighted in red font represent higher activity or content in the root of tolerant ecotype. Results are means ± SD of three biological replicates. *p < .05 and **p < .01.
The vital nodes of C and N metabolism were also investigated. The expression of hexokinase gene (HXK2) and pyruvate kinase gene (PK4) were much higher in the roots of the tolerant ecotype than the sensitive ecotype with NH4+ nutrition (Figure 6G). These results are consistent with the enzymatic activities of HXK and PK (Figure 6H; Figure S5). Importantly, pyruvate content, which is essential for N metabolism, was reduced by NH4+ nutrition, whereas its content was significantly higher in the roots of the tolerant ecotype than in the sensitive ecotype under NH4+ nutrition (Figure 6I). These results indicated that the tolerant ecotype maintained a low NH4+ level, mainly by promoting NH4+ assimilation rather than inhibiting NH4+ uptake.
Discussion
The tissue NH4+ concentration is an important factor contributing to variations in plant growth and NH4+ tolerance
NH4+ is toxic to plants, even at micromolar concentrations (von Wirén et al., 2000). Many hypotheses have been proposed to explain NH4+ toxicity, such as the depletion of organic acids, deficiency of cations, futile transmembrane NH4+ cycling and so on (Li et al., 2014). Previous studies have explored the medium NH4+ concentration toxic to different crops (Britto and Kronzucker, 2002). A previous study has expounded the critical role of NH4+ in Arabidopsis natural variability in NH4+ tolerance (Sarasketa et al., 2014). In this study, we found the two-segment linear model accurately simulated the relationship between fresh weight and tissue NH4+ concentration under different nitrogen sources (Figure 1E). When the tissue NH4+ concentration was below 50 μg/g under nitrate conditions, the fresh weight rapidly decreased with the tissue NH4+ concentration. Whereafter, the pace of declines slowed, when the tissue NH4+ concentration was over 50 μg/g under NH4+ conditions, (Figure 1E). This phenomenon possibly enlightened that low concentrations of NH4+ do not cause obvious symptoms of NH4+ toxicity, such as leaf chlorosis, but affect plant growth in an imperceptible way. However, the visible symptoms of NH4+ toxicity appeared over 50 μg/g under NH4+ conditions. A previous study revealed that the shoot K+ was positively correlated with the growth and NH4+ tolerance (Chen et al., 2021), and this correlation may be indirectly caused by the inhibition of K+ transport. Thus, the tissue NH4+ concentration is an important factor contributing to variations in plant growth and NH4+ tolerance.
Plants deal with NH4+ overload by using different strategies, including NH4+ transporters and NH4+ assimilation regulation. Inactivation of AMT1;1 and AMT1;2 by interaction with CIPK23 is important for NH4+ uptake regulation; Thus, the Loss of CIPK23 increases root NH4+ uptake and confers hypersensitivity to NH4+ (Straub et al., 2017). In the present study, the NH4+-tolerant ecotype Or-1 accumulated more 15N-labeled NH4+ (Figures 6A, B), thus the repression of AMT transporter cannot explain the tolerance mechanism of Or-1. Instead, the NH4+-tolerant ecotype displayed higher expression of AMT2 and AMT1.2 than the NH4+ -sensitive ecotype Rak-2, and its expression were induced by NH4+ (Figure 6D). Recently study found the expression of ZmAMT1s were induced by NH4+ nutrition and discovered glutamine rather than NH4+ regulated ZmAMT1s (Hui et al., 2022). The higher content of assimilated 15N-labeled H4+ and NUtE indicated that the tolerant ecotype Or-1 had a stronger NH4+ assimilation capacity, thus induced the expression of AMTs through a positive feedback by the NH4+ assimilate - glutamine.
The tolerant ecotype had a stronger NH4+ assimilation capacity to alleviating NH4+ toxicity
In plants, NH4+ assimilation generally occurs via the GS/GOGAT cycle. Cytosolic (GS1) and chloroplast (GS2) isoforms play opposing roles in NH4+ stress. Root Gln1.2 alleviates NH4+ toxicity, whereas shoot Gln2 aggravates NH4+ toxicity in a pH-dependent manner (Hachiya et al., 2021). Indeed, the expression of Gln1s was significantly induced by NH4+, whereas the expression of Gln2 was repressed (Figure 6D), suggesting an interesting regulation of GS to adapt NH4+ stress in plants. Although the GS activity of both ecotypes increased with increasing NH4+ concentration, no difference was observed between the two ecotypes (Figure 6E). Another study found no correlation between GS activity and shoot biomass in Arabidopsis under NH4+ nutrition (Sarasketa et al., 2014). Thus, variations in GS activity may not be crucial for NH4+ tolerance in different Arabidopsis ecotypes. GDH can incorporate NH4+ independently of the GS/GOGAT cycle and plays a critical role in the detoxification of NH4+ in stress conditions (Xian et al., 2020). Although the capacity of GDH to synthesize Glu in vivo has not been clearly demonstrated, heterologous expression of fungal GDH in plants could alleviates NH4+ toxicity and improve nitrogen assimilation (Tang et al., 2018; Yan et al., 2021). In this study, GDH activity and its encoding genes in both ecotypes were induced by NH4+ nutrition, and the activity was much higher in the tolerant ecotype than in the sensitive ecotype (Figure 6F). Moreover, GDH1 and GDH2 were more strongly induced in the roots of the tolerant ecotype than in those of the sensitive ecotype (Figure 6D). Overall, these results possibly indicate the critical role of GDH in Arabidopsis natural variation in NH4+ tolerance.
The sucrose metabolism pathway was enriched in the tolerant ecotype, indicating that this process was more active in the NH4+-tolerant ecotype (Figure 4C; Figure S4). Hexokinase (HXK) and Pyruvate kinase (PK) function crucial roles in Glycolysis and TCA cycle, providing 2-OG and energy for N metabolism (Stitt, 1999). In the present work, the expression of HXK2 and PK4 were much higher in the root of tolerant ecotype than the sensitive ecotype under NH4+ nutrition (Figure 6G). These results are consistent with the enzymatic activities of HXK and PK (Figure 6H; Figure S5). Importantly, the crucial carbon metabolite pyruvate was reduced by NH4+ nutrition, whereas its content was significantly higher in the roots of the tolerant ecotype than in the sensitive ecotype under NH4+ nutrition (Figure 6I). These results suggest that the tolerant ecotype had a stronger carbon skeleton (2-OG) production capacity for NH4+ assimilation (Figure 6J).
Coordination of carbon (C) and nitrogen (N) metabolism is essential for plant growth and stress tolerance (Reguera et al., 2013; Liang et al., 2020). The higher nitrogen utilization efficiency (NUtE) in the tolerant ecotype indicated the higher efficiency of the conversion of shoot N into shoot biomass (Figure 6C). The NUtE was similar among the accessions with different nitrogen use efficiency and uniformly decreased with high N supply, but the accessions differed in their NUtE under N restriction (Menz et al., 2018). The nitrogen was not limited and the concentration was even higher under NH4+ nutrition than that under nitrate nutrition. Thus, the higher NUtE in the tolerant ecotype may suggest better-coordinated C/N metabolism compared with the sensitive ecotype under NH4+ stress.
The tolerant ecotype displayed stronger defense responses by activating phenylpropanoids and the derived stilbenoids
NH4+ induces ROS formation, leading to oxidative damage in plants (Podgorska et al., 2013; Liu et al., 2022). Here, stilbenoid biosynthesis, phenylpropanoid biosynthesis, and glutathione metabolism pathways were commonly activated by NH4+ in all ecotypes (Figure 3D). Stilbenoids, which are hydroxylated derivatives of stilbene belonging to the phenylpropanoid family, have been shown to have a wide spectrum of biological functions, such as antioxidant and antimicrobial activities (Dong and Lin, 2021). Many other secondary metabolites, such as anthocyanins and flavonols, share a common origin in the phenylpropanoid biosynthetic pathway and functions as an ROS scavenger induced by abiotic stress (Dong and Lin, 2021). Treatment with flavonoids, such as naringenin, reduces oxidative damage under Cd stress in rice (Chen et al., 2022). Moreover, phenylpropanoid metabolism is associated with the reinforcement of cell walls under NH4+ nutrition, which was considered as an important NH4+ tolerance mechanism (Royo et al., 2019; Yang et al., 2022). Importantly, the NH4+-tolerant ecotype Or-1 showed a more intense response to NH4+ by activating pathways, including phenylpropanoid and stilbenoid biosynthesis (Figure 4C). Therefore, the biosynthesis of phenylpropanoids and stilbenoids derived from NH4+ may play a positive role in the defense response to NH4+ toxicity, and this thus warrants further investigation.
The tolerant ecotype was more responsive to NH4+ stress than the sensitive ecotype. These genes were highly enriched in defense responses, including their responses to ABA, water deprivation, chitin, wounding, and oxidative stress (Figure 5G). ABA signaling plays a key role in oxidative stress responses and is associated with the induction of antioxidant defense systems (Yang et al., 2015; Li et al., 2019b). Interestingly, the NH4+- tolerant ecotype has stronger ABA responses and phenylpropanoid metabolism for ROS scavenging (Figures 5G, H). Recently, a research investigated the genetic variation underlying differential ammonium and nitrate responses in Arabidopsis thaliana, and found the preferring ammonium or nitrate, appeared to be generated by combinations of loci rather than a few large-effect loci, which most are specific to a developmental or defense trait under specific nitrogen source (Katz et al., 2022). In our results, we also found only few genes were simultaneously responsive to NH4+ between the two genotypes (Figure 5A). This suggested that ammonium tolerance genotypes are a combination of multiple mini-effect genes.
Conclusion
In this study, the tissue content of NH4+ was found the main cause for NH4+ toxicity, and the two-segment linear model accurately simulated the relationship between fresh weight and tissue NH4+ concentration under different nitrogen sources. we revealed that the tolerant ecotype maintained a low NH4+ level, mainly by promoting NH4+ assimilation rather than inhibiting NH4+ uptake. The carbon and nitrogen metabolism analysis revealed that the tolerant ecotype had a stronger carbon skeleton (2-OG) production capacity with higher levels of hexokinase (HXK), pyruvate kinase (PK), and GDH activity to assimilate free NH4+. Furthermore, the core information about the biochemical regulation of plants in response to NH4+ toxicity was identified. The most enriched pathways included nitrogen metabolism, camalexin, stilbenoid and phenylpropanoid biosynthesis were upregulated by NH4+. Interestingly, a large number of genes, which enriched in phenylpropanoid and stilbenoid biosynthesis, were uniquely upregulated in the NH4+- tolerant ecotype. These results suggested that the NH4+-tolerant ecotype showed a more intense response to NH4+ by activating defense processes and pathways.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Gene Expression Omnibus, GSE243624.
Author contributions
HC: Conceptualization, Data curation, Formal analysis, Writing – original draft. WL: Data curation, Formal analysis, Methodology, Writing – original draft. WZ: Investigation, Software, Validation, Writing – review & editing. JZ: Data curation, Formal analysis, Project administration, Writing – review & editing. QZ: Writing – review & editing. ZZ: Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported in part by the National Natural Science Foundation of China (Grant No. 32172669; U21A20236); the Scientific Research Fund of Hunan Provincial Education Department (Grant No. 22A0157).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1286174/full#supplementary-material
References
Britto, D. T., Kronzucker, H. J. (2002). NH4+ toxicity in higher plants: a critical review. J. Plant Physiol. 159, 567–584. doi: 10.1078/0176-1617-0774
Britto, D. T., Siddiqi, M. Y., Glass, A. D., Kronzucker, H. J. (2001). Futile transmembrane NH4+ cycling: a cellular hypothesis to explain ammonium toxicity in plants. PNAS 98, 4255–4258. doi: 10.1073/pnas.061034698
Chen, H., Zhang, Q., Lu, Z., Xu, F. (2018). Accumulation of ammonium and reactive oxygen mediated drought-induced rice growth inhibition by disturbed nitrogen metabolism and photosynthesis. Plant Soil 431, 107–117. doi: 10.1007/s11104-018-3752-
Chen, H., Zhang, Q., Lv, W., Yu, X., Zhang, Z. (2022). Ethylene positively regulates Cd tolerance via reactive oxygen species scavenging and apoplastic transport barrier formation in rice. Environ. pollut. 302, 119063. doi: 10.1016/j.envpol.2022.119063
Chen, H., Zhang, Q., Wang, X., Zhang, J., Ismail, A. M., Zhang, Z. (2021). Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell Environ. 44, 3806–3818. doi: 10.1111/pce.14182
Chen, H, Zhang, Q., Lu, Z., Xu, F. (2018). Accumulation of ammonium andreactive oxygen mediated drought-induced rice growth inhibition by disturbednitrogen metabolism and photosynthesis. Plant and Soil 431, 107-117. doi: 10.1007/s11104-018-3752-0
Clark, C. M., Tilman, D. (2008). Loss of plant species after chronic low-level nitrogen deposition to prairie grasslands. Nature 451, 712–715. doi: 10.1038/nature06503
Cruz, C., Bio, A. F. M., Domínguezvaldivia, M. D., Apariciotejo, P. M., Lamsfus, C., Martinsloução, M. A. (2006). How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223, 1068–1080. doi: 10.1007/s00425-005-0155-2
Dai, G. Z., Qiu, B. S., Forchhammer, K. (2014). Ammonium tolerance in the cyanobacterium S ynechocystis sp. strain PCC 6803 and the role of the psbA multigene family. Plant Cell Environ. 37, 840–851. doi: 10.1111/pce.12202
Dong, N. Q., Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Duprè, C., Stevens, C. J., Ranke, T., Bleeker, A., Gowing, D. J., Diekmann, M. (2010). Changes in species richness and composition in European acidic grasslands over the past 70 years: the contribution of cumulative atmospheric nitrogen deposition. Global Change Biol. 16, 344–357. doi: 10.1111/j.1365-2486.2009.01982.x
Esteban, R., Ariz, I., Cruz, C., Moran, J. F. (2016). Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Groat, R. G., Vance, C. P. (1981). Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L.) developmental patterns and response to applied nitrogen. Plant Physiol. 67 (6), 1198–1203. doi: 10.1104/pp.67.6.1198
Guan, M., de Bang, T., Pedersen, C., Schjoerring, J. K. (2016). Cytosolic glutamine synthetase Gln1;2 is the main isozyme contributing to GS1 activity in Arabidopsis shoots and can be up-regulated to relieve ammonium toxicity. Plant Physiol. 171, 1921–1933. doi: 10.1104/pp.16.01195
Hachiya, T., Inaba, J., Wakazaki, M., Sato, M., Toyooka, K., Miyagi, A., et al. (2021). Excessive ammonium assimilation by plastidic glutamine synthetase causes ammoniumNH4+ toxicity in Arabidopsis thaliana. Nat. Commun. 12, 4944. doi: 10.1038/s41467-021-25238-7
Hachiya, T., Watanabe, C. K., Boom, C., Tholen, D., Takahara, K., Kawai-yamada, M. (2010). Ammonium-dependent respiratory increase is dependent on cytochrome pathway in Arabidopsis thaliana shoots. Plant Cell Environ. 33, 1888–1897. doi: 10.1111/j.1365-3040.2010.02189.x
Halvorson, A. D., Snyder, C. S., Blaylock, A. D., Del Grosso, S. J. (2014). Enhanced efficiency nitrogen fertilizers: potential role in nitrous oxide emission mitigation. Agron. J. 106, 715–722. doi: 10.2134/agronj2013.0081
Hoopen, F. T., Cuin, T. A., Pedas, P., Hegelund, J. N., Shabala, S., Schjoerring, J. K., et al. (2010). Competition between uptake of ammonium and potassium in barley and Arabidopsis roots: molecular mechanisms and physiological consequences. J. Exp. Bot. 61, 2303–2315. doi: 10.1093/jxb/erq057
Hui, J., Liu, Z., Duan, F. Y., Zhao, Y., Li, X. L., Xia, A. N., et al. (2022). Ammonium-dependent regulation of ammonium transporter ZmAMT1s expression conferred by glutamine levels in roots of maize. J. Integr. Agric. 21, 2413–2421. doi: 10.1016/S2095-3119(21)63753-X
Kachmar, J. F., Boyer, P. D. (1953). Kinetic analysis of enzyme reactions: II. The potassium activation and calcium inhibition of pyruvic phosphoferase. J. Biol. Chem. 200 (2), 669–682. doi: 10.1016/S0021-9258(18)71413-0
Katz, E., Knapp, A., Lensink, M., Keller, C. K., Stefani, J., Li, J. J., et al. (2022). Genetic variation underlying differential ammonium and nitrate responses in Arabidopsis thaliana. Plant Cell 34 (12), 4696–4713. doi: 10.1093/plcell/koac279
Lepper, T. W., Oliveira, E., Koch, G. D., Berlese, D. B., Feksa, L. R. (2010). Lead inhibits in vitro creatine kinase and pyruvate kinase activity in brain cortex of rats. Toxicol. In Vitro 24, 1045–1051. doi: 10.1016/j.tiv.2009.11.012
Li, B., Li, G., Kronzucker, H. J., Baluška, F., Shi, W. (2014). Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci. 19, 107–114. doi: 10.1016/j.tplants.2013.09.004
Li, B., Li, Q., Xiong, L., Kronzucker, H. J., Krämer, U., Shi, W. (2012). Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 160, 2040–2051. doi: 10.1104/pp.112.206508
Li, G., Zhang, L., Wang, M., Di, D., Kronzucker, H. J., Shi, W. (2019a). The Arabidopsis AMOT1/EIN3 gene plays an important role in the amelioration of ammonium toxicity. J. Exp. Bot. 70, 1375–1388. doi: 10.1093/jxb/ery457
Li, G., Zhang, L., Wu, J., Yue, X., Wang, M., Sun, L., et al. (2022). OsEIL1 protects rice growth under NH4+ nutrition by regulating OsVTC1-3-dependent N-glycosylation and root NH4+ efflux. Plant Cell Environ. 45, 1537–1553. doi: 10.1111/pce.14283
Li, S., Wang, N., Ji, D., Zhang, W., Wang, Y., Yu, Y., et al. (2019b). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31, 2107–2130. doi: 10.1105/tpc.18.00662
Li, S., Yan, L., Riaz, M., White, P. J., Yi, C., Wang, S., et al. (2021). Integrated transcriptome and metabolome analysis reveals the physiological and molecular responses of allotetraploid rapeseed to ammonium toxicity. Environ. Exp. Bot. 189, 104550. doi: 10.1016/j.envexpbot.2021.104550
Liang, Y., Wang, J., Zeng, F., Wang, Q., Zhu, L., Li, H., et al. (2020). Photorespiration regulates carbon–nitrogen metabolism by magnesium chelatase d subunit in rice. J. Agric. Food Chem. 69, 112–125. doi: 10.1021/acs.jafc.0c05809
Liu, Y., Maniero, R. A., Giehl, R. F., Melzer, M., Steensma, P., Krouk, G., et al. (2022). PDX1.1-dependent biosynthesis of vitamin B6 protects roots from ammonium-induced oxidative stress. Mol. Plant 15 (5), 820–839. doi: 10.1016/j.molp.2022.01.012
Masclaux-Daubresse, C., Reisdorf-Cren, M., MPageau, K., Lelandais, M., Grandjean, O., Kronenberger, J., et al. (2006). Glutamine synthetase/glutamate synthase pathway and glutamate dehydrogenase play distinct roles for sink/source nitrogen cycle in tobacco (Nicotiana tabacum L.). Plant Physiol. 140, 444–456. doi: 10.1104/pp.105.071910
Menz, J., Range, T., Trini, J., Ludewig, U., Neuhäuser, B. (2018). Molecular basis of differential nitrogen use efficiencies and nitrogen source preferences in contrasting Arabidopsis accessions. Sci. Rep. 8 (1), 3373. doi: 10.1038/s41598-018-21684-4
Pancera, S. M., Gliemann, H., Schimmel, T., Petri, D. F. S. (2006). Adsorption behavior and activity of hexokinase. J. colloid Interface Sci. 302 (2), 417–423. doi: 10.1016/j.jcis.2006.06.066
Podgorska, A., Gieczewska, K., Łukawska-Kuźma, K., Rasmusson, A. G., Gardeström, P., Szal, B. (2013). Long-term ammonium nutrition of Arabidopsis increases the extrachloroplastic NAD (P) H/NAD (P)+ ratio and mitochondrial reactive oxygen species level in leaves but does not impair photosynthetic capacity. Plant Cell Environ. 36, 2034–2045. doi: 10.1111/pce.12113
Qin, C., Qian, W., Wang, W., Wu, Y., Yu, C., Jiang, X., et al. (2008). GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. PNAS 105, 18308–18313. doi: 10.1073/pnas.0806168105
Reguera, M., Peleg, Z., Abdel-Tawab, Y. M., Tumimbang, E. B., Delatorre, C. A., Blumwald, E. (2013). Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 163, 1609–1622. doi: 10.1104/pp.113.227702
Royo, B., Esteban, R., Buezo, J., Santamaría, E., Fernández-Irigoyen, J., Becker, D., et al. (2019). The proteome of Medicago truncatula in response to ammonium and urea nutrition reveals the role of membrane proteins and enzymes of root lignification. Environ. Exp. Bot. 162, 168–180. doi: 10.1016/j.envexpbot.2019.02.010
Rubio-Asensio, J. S., Bloom, A. J. (2016). Inorganic nitrogen form: a major player in wheat and Arabidopsis responses to elevated CO2. J. Exp. Bot. 68, 2611–2625. doi: 10.1093/jxb/erw465
Sarasketa, A., González-Moro, M. B., González-Murua, C., Marino, D. (2014). Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J. Exp. Bot. 65, 6023–6033. doi: 10.1093/jxb/eru342
Stevens, C. J., Dise, N. B., Mountford, J. O., Gowing, D. J. (2004). Impact of nitrogen deposition on the species richness of grasslands. Science 303, 1876–1879. doi: 10.1126/science.1094678
Stitt, M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186. doi: 10.1016/S1369-5266(99)80033-8
Straub, T., Ludewig, U., Neuhäuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29, 409–422. doi: 10.1105/tpc.16.00806
Subbarao, G. V., Searchinger, T. D. (2021). A “more ammonium solution” to mitigate nitrogen pollution and boost crop yields. PNAS 118, e2107576118. doi: 10.1073/pnas.2107576118
Tang, D., Peng, Y., Lin, J., Du, C., Yang, Y., Wang, D., et al. (2018). Ectopic expression of fungal EcGDH improves nitrogen assimilation and grain yield in rice. J. Integr. Plant Biol. 60 (2), 85–88. doi: 10.1111/jipb.12519
von Wirén, N., Gazzarrini, S., Gojon, A., Frommer, W. B. (2000). The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 3, 254–261. doi: 10.1016/S1369-5266(00)00073-X
Xian, L., Zhang, Y., Cao, Y., Wan, T., Dai, C., Liu, F. (2020). Glutamate dehydrogenase plays an important role in ammonium detoxification by submerged macrophytes. Sci. Total Environ. 722, 137859. doi: 10.1016/j.scitotenv.2020.137859
Yan, L. U., Gong, Y., Luo, Q., Dai, G. X., Teng, Z., He, Y., et al. (2021). Heterologous expression of fungal AcGDH alleviates NH4+ toxicity and suppresses photorespiration, thereby improving drought tolerance in rice. Plant Sci. 305, 110769. doi: 10.1016/j.plantsci.2020.110769
Yang, T., Zhang, L., Hao, H., Zhang, P., Zhu, H., Cheng, W., et al. (2015). Nuclear-localized at HSPR links abscisic acid-dependent salt tolerance and antioxidant defense in Arabidopsis. Plant J. 84, 1274–1294. doi: 10.1111/tpj.13080
Keywords: nitrogen, NH4+ toxicity, NH4+ assimilation, phenylpropanoid biosynthesis, transcriptome analysis
Citation: Chen H, Lv W, Zhang W, Zhao J, Zhang Q and Zhang Z (2023) Integrated comparative transcriptome and physiological analysis reveals the metabolic responses underlying genotype variations in NH4+ tolerance. Front. Plant Sci. 14:1286174. doi: 10.3389/fpls.2023.1286174
Received: 05 September 2023; Accepted: 16 November 2023;
Published: 13 December 2023.
Edited by:
Marco Betti, Sevilla University, SpainReviewed by:
Idoia Ariz, Public University of the Navarre, SpainDaniel Marino, University of the Basque Country, Spain
Copyright © 2023 Chen, Lv, Zhang, Zhao, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhua Zhang, emh6aDE0NjhAMTYzLmNvbQ==
 Haifei Chen
Haifei Chen Wei Lv1
Wei Lv1 Quan Zhang
Quan Zhang Zhenhua Zhang
Zhenhua Zhang