- Genomics Division, Department of Agricultural Bio-Resources, National Institute of Agricultural Sciences, Jeonju, Republic of Korea
Salt stress is an ever-increasing stressor that affects both plants and humans. Therefore, developing strategies to limit the undesirable effects of salt stress is essential. Sodium ion exclusion is well known for its efficient salt-tolerance mechanism. The High-affinity K+ Transporter (HKT) excludes excess Na+ from the transpiration stream. This study identified and characterized the HKT protein family in Bienertia sinuspersici, a single-cell C4 plant. The HKT and Salt Overly Sensitive 1 (SOS1) expression levels were examined in B. sinuspersici and Arabidopsis thaliana leaves under four different salt stress conditions: 0, 100, 200, and 300 mM NaCl. Furthermore, BsHKT1;2 was cloned, thereby producing stable transgenic Brassica rapa. Our results showed that, compared to A. thaliana as a glycophyte, the HKT family is expanded in B. sinuspersici as a halophyte with three paralogs. The phylogenetic analysis revealed three paralogs belonging to the HKT subfamily I. Out of three copies, the expression of BsHKT1;2 was higher in Bienertia under control and salt stress conditions than in A. thaliana. Stable transgenic plants overexpressing 35S::BsHKT1;2 showed higher salt tolerance than non-transgenic plants. Higher biomass and longer roots were observed in the transgenic plants under salt stress than in non-transgenic plants. This study demonstrates the evolutionary and functional differences in HKT proteins between glycophytes and halophytes and associates the role of BsHKT1;2 in imparting salt tolerance and productivity.
1 Introduction
Salt stress, the second most important abiotic stress, is a global concern that challenges food security by affecting crop growth and yield factors (Rengasamy, 2006; Munns and Tester, 2008; Zhu, 2016). Salt stress affects more than 6% of the total land area worldwide and 20% of irrigated land, restraining global food production (Rengasamy, 2006; Munns and Tester, 2008). The Mediterranean, semi-arid, and arid areas have experienced severe salinity effects. In South Korea, reclaimed tidelands (RTLs) are used to cultivate crops such as rice. However, cultivating crops such as Brassica in the RTL is still impossible due to high salinity and poor soil physical properties (Hong et al., 2020; Lim et al., 2020). Soil containing 40 mM NaCl potentially affects crop yield by inducing osmotic and ionic stress (Flowers and Colmer, 2008; Huang et al., 2008; Munns and Tester, 2008). In plants, three major mechanisms have been demonstrated to cope with salinity stress: Na+ extrusion into the soil involving salt overly sensitive 1 (SOS1) (Shi et al., 2000; Shi et al., 2003; Zhu, 2016); Na+ ion exclusion from the xylem stream involving high-affinity K+ transporter 1 (HKT1) (Munns and Tester, 2008; Møller et al., 2009; Munns et al., 2012); and Na+ sequestration into vacuoles involving Na+/H+ exchangers 1-4 (NHX1-4) antiporters (Zhang et al., 2019; Van Zelm et al., 2020). In addition to HKT and NHX transporters, studies have demonstrated that the high-affinity K+ (HAK) transporter located in the plasma membrane of stele cells shows salt tolerance by selectively transporting Na+ from the xylem to the stele regions (Zhang et al., 2012; Chen et al., 2015; Zhang et al., 2019). Interestingly, halophytes living in high-salinity lands exclude sodium and chloride ions from their cells and endure up to 450 mM NaCl. In contrast, glycophytes cannot sustain more than 50 mM NaCl (Munns and Tester, 2008). Halophytes possess a salt-tolerant (exclusion) trait that exploits the HKT family of transporter proteins to prevent the delivery of Na+ to the plant shoots, whereas glycophytes do not possess such an efficient functional mechanism (Zhu, 2016; Van Zelm et al., 2020).
HKT belongs to the Trk/Ktr/HKT transporter family (Platten et al., 2006). The HKT transporter family proteins are located in the plasma membrane of xylem parenchyma cells (XPCs) and transport Na+ ions from the transpiration stream to the cells, where the ions are sequestered into vacuoles by NHX transporters located in the tonoplast membrane (Horie et al., 2009). Møller et al. (2009) showed that HKT alone reduced the transport of Na+ from the roots to shoots by up to 64%. In plants, the HKT protein family has been classified into two subfamilies based on their structure and ion preference (Platten et al., 2006; Horie et al., 2009). Subfamily I preferentially transports Na+ in one direction, that is, from the xylem stream to the XPCs. In contrast, subfamily II transports both K+ and Na+ in a unidirectional direction to the XPCs (Horie et al., 2009). The variation in amino acid residues in the pore loop (PA) determines the type, that is, serine in subfamily I and glycine in subfamily II (Horie et al., 2009). These transporters maintain Na+ homeostasis in the crop plants. TaHKT2;1 was first discovered as an HKT transporter in wheat in 1994 (Schachtman and Schroeder, 1994). Plants possess more than one HKT transporter gene in a single genome. However, Arabidopsis possesses only one AtHKT1;1 transporter gene (Horie et al., 2009). The overexpression of the HKT gene in plants improves salt tolerance compared to wild-type plants (Møller et al., 2009; Mian et al., 2011; Xu et al., 2018; Zhang et al., 2019). Natural variations in salinity tolerance have also been demonstrated in HKT transporters in plants. Zhang et al. (2019) explored the natural variation in the HKT1 (ZmHKT1) gene in Zea mays L. and its salinity tolerance. This study showed that a favorable HKT1 allele imparts greater salinity tolerance. In addition, the Na+ Content 3 (ZmNC3) gene exhibits natural allelic variation and function. Moreover, they identified two haplotypes, HapA with adenine- and HapC with cytosine-associated high and low salt tolerance, respectively. Furthermore, variations associated with gene function were demonstrated in transgenic plants carrying the HapA alleles (Zhang et al., 2019).
Brassica is one of the most economically important crops worldwide, contributing to approximately 12% of vegetable oil production globally (Mcalvay et al., 2021). B. rapa belongs to the A genome of the Brassica genus, and its diploid genome was first sequenced in 2011 (Wang et al., 2011). B. rapa is a natural glycophyte (Yepes et al., 2018). With increasing salinity stress worldwide owing to natural calamities and human intervention, developing salt-tolerant crops is important. B. rapa is salt-sensitive compared to B. oleracea, as B. rapa, has a higher Na+/K+ ratio in leaves than in roots (Pavlović et al., 2019; Hong et al., 2020). Similarly, B. rapa had a higher Na+/K+ ratio in the shoot than B. oleracea under salt stress (Pavlović et al., 2019). Overexpression of salt-responsive genes such as RING-H2 finger (BrATL30) and zinc-finger homeodomain protein 10 (BrZHD10) from B. rapa in B. juncea has exhibited better salt tolerance (Hong et al., 2020). Under salt stress, B. napus showed reduced shoot length and leaf area, which is attributed to salt sensitivity (Hussain Wani et al., 2013). Similarly, B. juncea exhibited decreased plant growth and photosynthetic parameters (Hayat et al., 2012).
B. sinuspersici is a single-cell C4 plant with non-Kranz photosynthetic anatomy (Akhani et al., 2005; Offermann et al., 2015; Koteyeva et al., 2016; Soundararajan et al., 2019). B. sinuspersici possesses dimorphic chloroplasts in a single cell, namely central and peripheral chloroplasts, unlike other C4 plants (Offermann et al., 2015; Koteyeva et al., 2016; Mai et al., 2019; Han et al., 2023; Won et al., 2023). B. sinuspersici is a salt-tolerant halophyte belonging to the family Amaranthaceae sensu lato (Kapralov et al., 2012; Han et al., 2023) and possesses salt glands in its leaves. It commonly grows in the saline areas surrounding the Persian Gulf (Akhani et al., 2005; Park et al., 2009; Uzilday et al., 2023). As halophytes complete their life cycle in saline environments, they could be a source of salt-tolerant genes for developing transgenic plants. Moreover, halophytes possess an efficient Na+ compartmentalization mechanism. A few studies have been conducted on the salt response of B. sinuspersici and have demonstrated salt tolerance (Park et al., 2009; Uzilday et al., 2023). However, no studies have investigated the role of HKT1 from B. sinuspersici in developing salt-tolerant crops.
This study utilized information from an ongoing B. sinuspersici genome annotation to identify and characterize HKT genes. We studied the HKTs of B. sinuspersici and their relationships with HKTs from other plant lineages. In addition, this study suggests the number of paralogs and their expression patterns in leaves. Then, overexpression of BsHKT1;2 in the B. rapa crop, a glycophyte, with the cauliflower mosaic virus 35S promoter and its characterization was performed. We found that transgenic plants expressing 35S::BsHKT1;2 showed significantly increased salt tolerance compared to non-transgenic plants. This study provides insights into the HKT gene from B. sinuspersici and its role in salt tolerance.
2 Materials and methods
2.1 Plant growth and conditions
Seeds of B. sinuspersici (BioProject Accession no. PRJNA273351) obtained from Prof. Sascha Offermann (Institute for Botany, Leibniz University Hannover, 136 Germany), mature B. rapa and A. thaliana (Col-0), were used in the experiments. All seeds were surface-sterilized using 2% (v/v) sodium hypochlorite for 15 min in a shaker, followed by 20 washes with sterile water. Seeds were sown and grown in Murashige and Skoog (MS) medium with 1% sucrose and 0.8% agar or in a soil mixture under a 16-hour/8-hour photoperiod at 25 °C and 70% humidity. A. thaliana seeds were germinated and grown in MS for up to two weeks, and then the plants were used for the salt experiments.
2.2 Identification of HKT1
To identify HKT gene families in the B. sinuspersici genome, the hidden Markov model (HMM) file of the cation transport protein TrkH (PF02386; https://www.ebi.ac.uk/interpro/entry/pfam/PF02386/) was employed and searched against whole-genome peptides and the transcriptome of B. sinuspersici (genome annotation is underway; BS790c1g1i2 in the transcriptome) (Han et al., 2023). Repeated and truncated proteins were removed. Furthermore, the results were validated by BLAST with well-characterized AtHKT1 (NP567354.1; AT4G10310) proteins from A. thaliana against B. sinuspersici (Supplementary Table 1).
2.3 Phylogenetic tree construction and subcellular localization
A phylogenetic tree was constructed for HKT proteins from various plants (Supplementary Table 2), A. thaliana, and B. sinuspersici, using MEGA 11 (Tamura et al., 2021) with ClustalW using the maximum likelihood statistics of 1,000 bootstrap replicates, partial deletion (95), and nearest-neighbor-interchange (NNI) settings. Published HKT proteins belonging to subfamilies 1 and 2 were obtained from the literature and used in the analysis. Furthermore, the PA, PB, PC, and PD domains were examined for subfamily-specific amino acid residues, such as serine (Ser) or glycine (Gly). The DeepLoc-2.0 online tool (https://services.healthtech.dtu.dk/services/DeepLoc-2.0/) was used to predict the subcellular localization of BsHKT proteins (Almagro Armenteros et al., 2017) (Supplementary Table 3). For the phylogenetic tree analysis gene, coding sequence, and amino acid sequences in FASTA format were used as inputs (Supplementary Files. 1-4).
2.4 Gene structural arrangement and motif discovery
To determine the gene structural organization, the genomic position of the HKT1 genes in B. sinuspersici was determined using tBLASTn. Contigs encoding HKT1 were retrieved using the SAM tools. The region encoding HKT1 was identified using the FGENESH+HMM profile and extracted using BEDtools. The intron-exon arrangement was mapped using the Gene Structure Display Server 2.0 (GSDS 2.0) online tool (http://gsds.gao-lab.org/index.php) (Hu et al., 2015). Gene and coding sequences (CDS) were used as input into the GSDS 2.0 (Supplementary Files. 1 and 2). Conserved HKT1 motifs were identified using the Multiple Em for Motif Elicitation (MEME) Suite web server (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2009) with the zero or one occurrence (ZOOPS) mode, 15 maximum motifs, and 6–200 motif width. Protein sequences were used as inputs (Supplementary File 3). Along with BsHKT1;1, BsHKT1;2, and BsHKT1;3, the HKT1 orthologs available in the literature were included in the analysis for comparison (Supplementary File 4).
2.5 Salt stress imposition
Half-MS medium with 1% sucrose, 0.8% agar, and different salt concentrations (0, 100, 200, and 300 mM NaCl) was prepared. B. sinuspersici plants were grown on half-MS media with different salt concentrations in a magenta box for three weeks. The plants were exposed to salt stress on the day of germination. Observations and leaf samples were collected two and three weeks after stress imposition (WAI). Two-week-old A. thaliana Col-0 seedlings were also exposed to salt stress. Then, leaf samples were collected one and two days after stress imposition (DAI), snap-frozen in liquid N2, and stored at –80 °C for RNA isolation. For transgenic plants, transgenic seeds were grown on MS media plates for one week. Then, plants were transferred to liquid MS media with different salt concentrations. Wild-type and transgenic plants were subjected to salt stress for seven days. The observations and sample collection were performed at 7 DAI.
2.6 Morpho-physiological parameters
Plants subjected to salt stress treatments were observed for variations in root length and foliar canopy growth. At 2 and 3 WAI, plant images were captured using a Canon EOS M50 Mark II (Canon Europe, Amstelveen, the Netherlands), and root length was measured for both treated and untreated plants. Canopy size was also measured. ImageJ software (https://imagej.nih.gov/ij/download/) was used to measure the canopy size.
2.7 Gene expression analysis
Total RNA was isolated using the cetyltrimethylammonium bromide (CTAB) method, precipitated in LiCl, and washed with ethanol (Soundararajan et al., 2019). Isolated RNA was dissolved in 30 µL of diethylpyrocarbonate (DEPC)-treated water and stored at –70°C for further analysis. 1 µg of RNA was used for cDNA synthesis (amfiRivert cDNA Synthesis Master Mix; GenDEOPT, USA). Five-fold diluted cDNA was used for real-time quantitative polymerase chain reaction (qPCR) analysis (iQTM SYBR Green® Supermix, Biorad, USA) using the primers mentioned (Supplementary Table 4). A two-step SYBR method with 58°C amplification was used with the following conditions in a thermocycler (CFX96 TouchTM Real-Time PCR Detection System; Bio-Rad, USA). Denaturation at 95°C for 3 min, 40 cycles at 95°C for 15 s, 58°C for 30 s, and a melting curve at 95°C for 10 s, 65°C for 5 s, and 60°C for 50 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for relative expression analysis. All samples were analyzed using three biological replicates. The relative expression was calculated using the 2-ΔΔCt method.
2.8 Construction of the overexpression vector, Brassica transformation, and development of stable lines
A full-length gene of BsHKT1;2 was amplified from the B. sinuspersici genome and cloned into the pCAMBIA1390 vector with a 35S promoter derived from the cauliflower mosaic virus (CaMV35S) using the conventional cloning method in a custom cloning service (http://www.bionicsro.co.kr/). pCAMBIA1390_35SP_BsHKT1;2 was transformed into Brassica using Agrobacterium tumefaciens GV3101. Positive transgenic plants were screened using hygromycin in MS media, and the PCR confirmed the presence of hygromycin phosphotransferase (hptII). T1-generation seeds were backcrossed to T3 generations to produce homozygous plants for BsHKT1;2.
2.9 Flanking DNA sequencing and analysis
Genomic DNA from transgenic and non-transgenic plants was isolated and digested with HaeIII. The digested products were ligated using an adaptor. PCR amplification was performed using primers specific to the T-DNA and adaptor sequences. PCR products were isolated and sequenced using T-DNA-specific primers. The analysis was performed using the flanking sequence tag validator online tool (http://bioinfo.mju.ac.kr/fstval/) (Kim et al., 2012).
2.10 Statistical analysis
Statistical analysis was performed using SPSS v16.0 software (https://www.ibm.com/spss) and GraphPad Prism (v5 and 9; https://www.graphpad.com/). One-way or two-way analysis of variance (ANOVA) was used to analyze the significance of differences between wild-type and transgenic lines and treatments. In the two-way ANOVA, a Bonferroni post-hoc test was performed to compare the replicate means of non-transgenic plants.
3 Results
3.1 BsHKTs belong to the HKT subfamily I
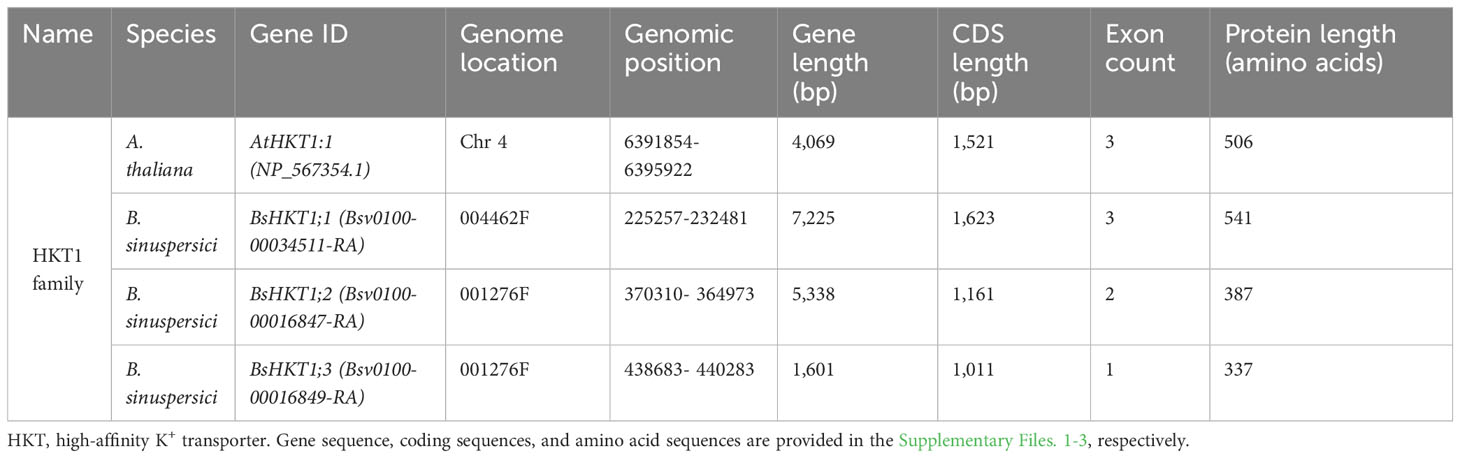
Genome-wide identification and a literature survey showed that A. thaliana has one copy of AtHKT1, which was later renamed AtHKT1;1 following the subfamily classification (Uozumi et al., 2000). In contrast, three copies of HKT were identified in the B. sinuspersici genome (Table 1). Basic information regarding the three copies of the gene is provided (Supplementary Table 1). For phylogenetic analysis, we used protein sequences designated as subfamily I (HKT1) and subfamily II (HKT2) from the literature (Supplementary Tables 1, 2). The analysis showed that all three HKT genes, BsHKT1a, BsHKT1b, and BsHKT1c, were classified under HKT subfamily I (Figure 1). Multiple sequence alignment (MSA) analysis showed that AtHKT1;1 possessed four predominant pore domains. Pore domain A contained a Ser residue at a conserved position. Further analysis at the amino acid sequence level showed that all three BsHKT proteins possessed Ser-Gly-Gly-Gly residues in their PA, PB, PC, and PD domains, respectively. The Ser residues in the PA domain further confirmed that BsHKT1a, BsHKT1b, and BsHKT1c belong to the HKT subfamily I (Supplementary Figure 1A). Genomic information on HKT1 genes is presented in Table 1. Based on homology and topology, we assigned the following names to the identified orthologs: BsHKT1;1, BsHKT1;2, and BsHKT1;3.
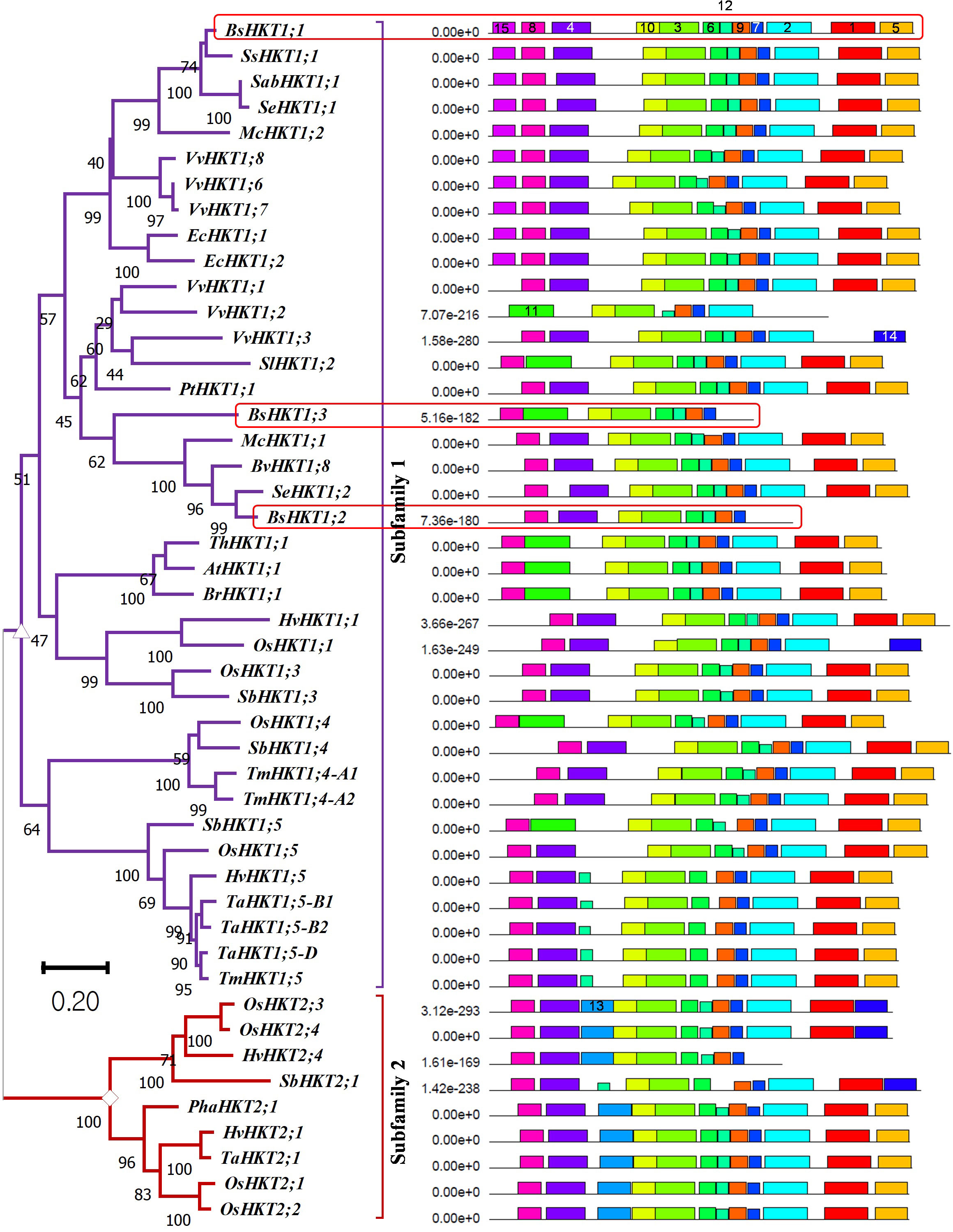
Figure 1 Evolutionary relationships, nomenclature, and motif locations in BsHKTs. The unrooted maximum likelihood tree shows an evolutionary relationship with HKTs from higher plants. The name was given based on their similarity to existing HKT proteins. The tree was developed from the data obtained from Platten et al. (2006) and the literature with the addition of HKT protein sequences from B. sinuspersici. Evolutionary analyses were conducted in MEGA11 with 1000 bootstrap replicates, and the percentage of replicate trees is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Light red square boxes indicate HKTs in B. sinuspersici. Scale bar = 0.2 substitutions per site. Species expansion, protein length, and ID details are provided in the Supplementary Tables 1, 2. Image (right side) shows the locations of conserved motifs (15) in AtHKT1;1, BsHKT1;1, BsHKT1;2, BsHKT1;3 and other proteins recognized by the MEME motif analysis. Different colors and lengths represent motifs and their lengths, respectively. Amino acid details of identified motifs labeled with numbers and represented by individual colors are provided in Supplementary Figure 1B.
3.2 BsHKTs possess domains similar to HKTs belonging to halophytes
MEME is a motif-based sequence analysis tool used to identify conserved motifs (Bailey et al., 2009). The MEME analysis revealed that all three orthologous copies of BsHKT1s from B. sinuspersici shared conserved motifs with AtHKT1;1 (Figure 1; Supplementary Figure 1B). The MEME analysis, with a total of 15 motifs used as input parameters, showed that BsHKT1;1 possessed 12 conserved motifs, except for motifs 11, 13, and 14. In contrast, AtHKT1;1 and BrHKT1;1 had 11 motifs that did not include the motifs 4 and 15, and BsHKT1;2 and BsHKT1;3 possessed eight motifs, lacking the motifs 1, 2, and 5, compared to BsHKT1;1. Moreover, MEME analysis showed the following amino acid sequence variations: HKT subfamily I possessed 14 motifs, except for motif 13, whereas HKT subfamily II contained 13 conserved motifs, except for motifs 11 and 15. BsHKT1;1 possessed several motifs similar to those of EcHKT1;1, EcHKT1;2, McHKT1;2, SabHKT1;1, SeHKT1;1, SsHKT1;1, VvHKT1;6, VvHKT1;7, and VvHKT1;8. In contrast, BsHKT1;2, BsHKT1;3, and VvHKT1;3 possessed fewer motifs. The least number of motifs were found in VvHKT1;2 (N=7) and HvHKT2;4 (N=9) in the HKT1 and HKT2 families, respectively. All HKTs contained motifs 3, 7, 9, and 10 (Figure 1; Supplementary Figure 1B). Subcellular localization prediction analysis showed that BsHKT1;1, BsHKT1;2, and BsHKT1;2 were localized in the plasma membrane, similar to AtHKT1;1 (Almagro Armenteros et al., 2017) (Supplementary Table 3).
3.3 BsHKTs contain variable numbers of introns and exons
The GSDS 2.0 online tool (Hu et al., 2015) was used to investigate the arrangement of introns and exons in the BsHKT genes. The analysis showed that BsHKT1;1 had three exons and two introns, similar to AtHKT1;1 structure (Supplementary Figure 1C). However, the length of BsHKT1;1 was almost double that of AtHKT1;1. Notably, the other two paralogs, BsHKT1;2 and BsHKT1;3, contained only two and one exon, respectively. Moreover, BsHKT1;2 has only one intron, whereas BsHKT1;3 has no introns. Furthermore, the length of BsHKT1;2 was similar to AtHKT1;1, whereas BsHKT1;3 was shorter than AtHKT1;1 (Supplementary Figure 1C).
3.4 Bienertia grows better in salt medium with higher BsHKT1;2 expression
To investigate the expression patterns of HKT genes under salt stress conditions, wild-type plants were treated with 0, 100, 200, and 300 mM NaCl. As the growth of A. thaliana was severely affected over two days under in vitro conditions, samples were collected one and two DAI. In A. thaliana, reductions in growth, leaf scorching symptoms, and root inhibition have been observed under salt stress. Overall, the salt treatment diminished the growth of A. thaliana (Figure 2A). For B. sinuspersici, salt treatment was conducted for two and three weeks.
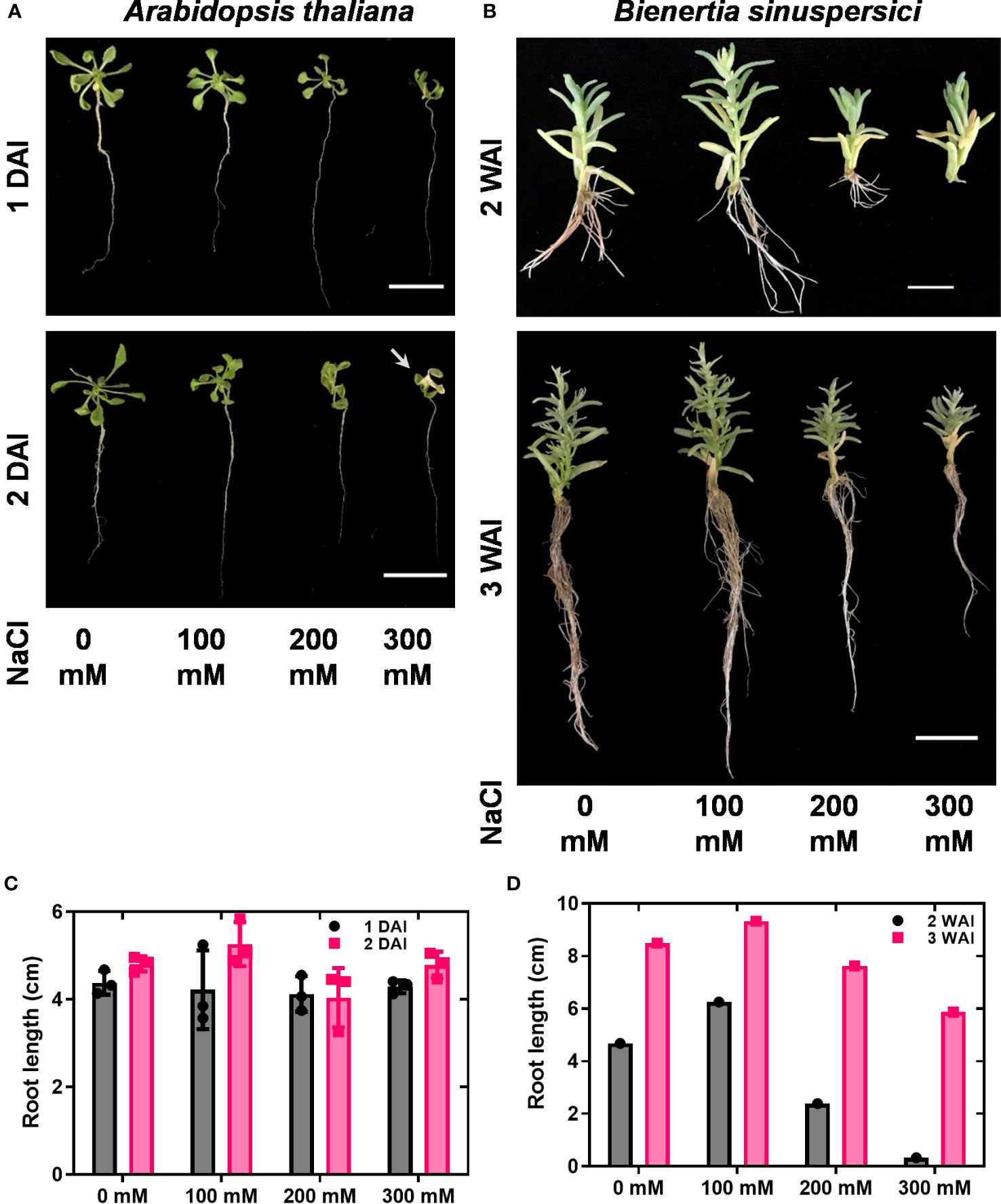
Figure 2 Shoot and root phenotypes under salt stress. A. thaliana and B. sinuspersici were subjected to salt stress (0, 100, 200, and 300 mM). NaCl was used as a salt component. A. thaliana and B. sinuspersici plants were transferred to ½ MS media containing different salt concentrations two weeks after sowing and subjected to salt stress. A. thaliana plants were observed for phenotype changes 1 and 2 DAI, whereas B. sinuspersici plants were observed for phenotype changes 2 and 3 WAI. (A) Images show the A. thaliana phenotype at 1 DAI and 2 DAI under different salt concentrations. (B) Images show the B. sinuspersici phenotype at 2 and 3 WAI. (C, D) graphs show the root lengths of A. thaliana and B. sinuspersici under different salt concentrations, respectively. Scale bar: 2 cm. Error bar= standard error of the mean. N=4 for A. thaliana and N=1 for B. sinuspersici. Note: the white arrow points out the foliar yellowing caused by salt stress. DAI, days after stress imposition; WAI, weeks after stress imposition.
Improved growth of B. sinuspersici was observed in the 100 mM NaCl treatment compared to the control. In contrast, 200 and 300 mM NaCl reduced growth and decreased root length in plants compared to the control (Figures 2B–D). In this study, the expression levels of three orthologous copies, BsHKT1;1, BsHKT1;2, and BsHKT1;3 from the HKT1 family and SOS1 from the NHX family, under salt stress conditions were determined. Decreased expression of AtHKT1;1 was observed at all salt concentrations. The BsHKT1;1, BsHKT1;2, and BsHKT1;3 expression levels were strongly increased in B. sinuspersici treated with 100 mM NaCl. Higher expression of BsHKT1;2 was observed at moderate and high NaCl concentrations (0, 100, and 200 mM) at 3 WAI than at 2 WAI, except at 300 mM NaCl. In contrast, a higher expression of BsHKT1;3 was observed at exceptionally high NaCl concentrations (200 and 300 mM) at 3 WAI than at 2 WAI. Importantly, while increased expression of BsSOS1 in B. sinuspersici was observed at 200 and 300 mM NaCl, AtSOS1 in A. thaliana did increase its expression level at 300 mM NaCl at 2 DAI. In addition, extremely high expression of AtSOS1 was detected in the presence of 200 mM NaCl at 2 DAI. Interestingly, BsHKT1;3 and BsSOS1 showed similar gene expression patterns under salt stress. The maximum level of BsSOS1 expression was observed at 100 mM NaCl (Figure 3).
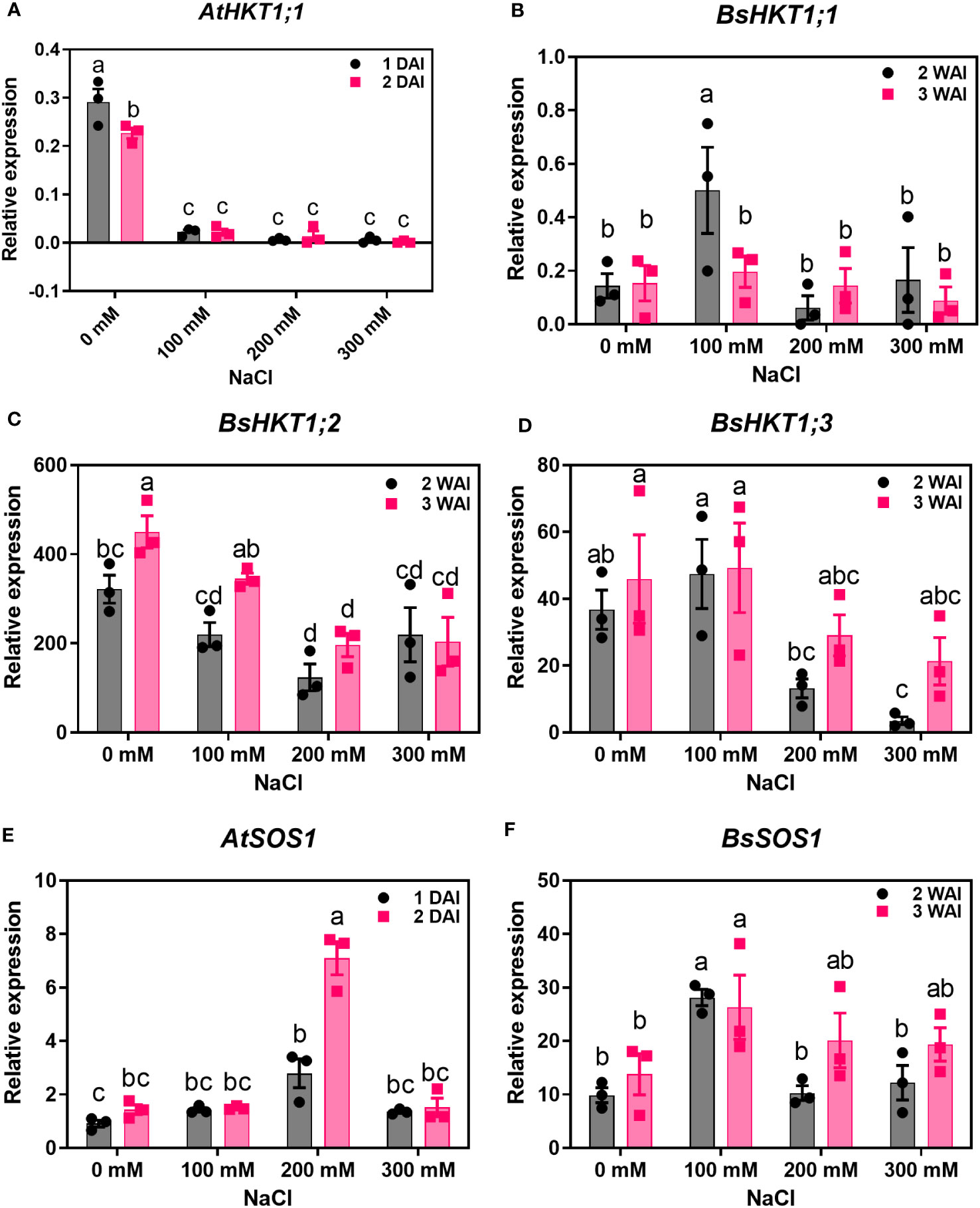
Figure 3 Quantitative real-time PCR results of HKTs and SOS1 genes under salt stress. A. thaliana and B. sinuspersici were subjected to salt stress (0, 100, 200, and 300 mM), and RNA was isolated from shoots 1 and 2 DAI for A. thaliana, whereas for B. sinuspersici plants, at 2 and 3 WAI. Graphs show the relative expression levels of (A), AtHKT1;1; (B), BsHKT1;1; (C), BsHKT1;2; (D), BsHKT1;3; (E), AtSOS1 and (F), BsSOS1 to the internal control BrGAPDH under 0 mM NaCl. Error bar= standard error of the mean. N=3. The letters above the column indicate the significant difference among the treatments. DAI, days after stress imposition; WAI, weeks after stress imposition.
3.5 Homozygous transgenic B. rapa plants overexpress BsHKT1;2 under the 35S promoter
Since BsHKT1;2 showed increased expression levels (Figure 3; Supplementary Figure 2A), to further investigate its efficiency in salt tolerance, the full-length gene was amplified from cDNA and cloned into pCAMBIA1390 using the cauliflower mosaic virus 35S promoter (CaMV35S) (Figures 4A, B). The primer details are shown in Supplementary Table 4. B. rapa was transformed with 35S::BsHKT1;2 using A. tumefaciens. Transformed plants were initially screened under hygromycin antibiotics in MS medium for four rounds of plant transfer, during which the chimeric plants were removed. PCR amplification and plant mortality in salt media confirmed that transgenic B. rapa contained the 35S::BsHKT1;2 segments (Figures 4B, D). Then, the expression levels were measured. Positive plants were confirmed by PCR, and T1-positive plants were backcrossed for up to two generations (Supplementary Figure 3). From the segregation analysis, stable lines (T2 generation) were selected and used in salt stress experiments.
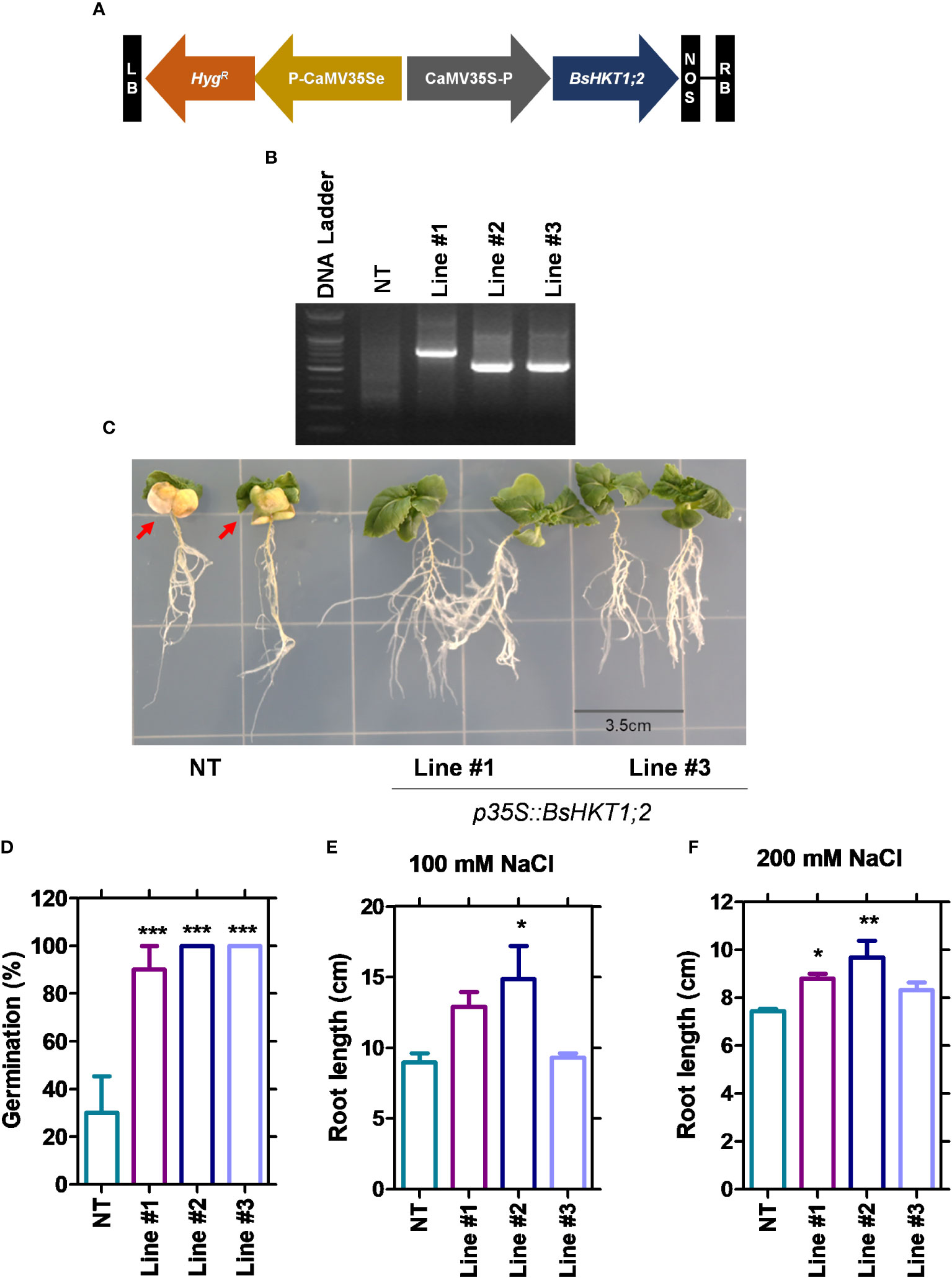
Figure 4 Morpho-physiological responses of transgenic plants under salt stress. NT and transgenic plants were subjected to salt stress (150 mM), and observation was carried out on 7 DAI. (A) The diagram shows the BsHKT1;2 gene construct in vector pCAMBIA1390. (B) The traditional cloning result is confirmed by restriction enzymes. (C) The image panel shows the foliar and root phenotypes of NT and transgenic plants subjected to 100 and 200 mM salt stress, respectively. (D) The graph shows the percentage of germination of the NT and transgenic plants. (E, F) The graphs show the root length of the NT and transgenic plants under 100 and 200 mM NaCl. The error bar indicates the standard error of the mean. A one-way ANOVA was performed, followed by Tukey’s multiple comparison test. Asterisks indicate the significance (* p < 0.05; **p < 0.01; ***p < 0.001). N=3. The experiment was at least repeated twice. Note: Line #1, 4-4; Line #2, 5-8; Line #3, 5-9; NT, non-transgenic plant. Red arrows indicate the yellowing foliar symptom in NT plants. DAI, days after stress imposition.
3.6 BsHKT1;2 is inserted into the 5’ upstream and intron regions
Flanking DNA sequencing analysis was performed (Supplementary Figure 4) and showed that BsHKT1;2 genes were inserted at the 1 kb 5’ upstream of the DnaJ domain in line #1 (4-4) and at the intron regions of a protein with unknown function in line #2 (5-8) and #3 (5-9) (Table 2). Irrespective of the insertion location, transgenic plants were healthy and did not show a slight change in plant morphology, similar to non-transgenic plants.
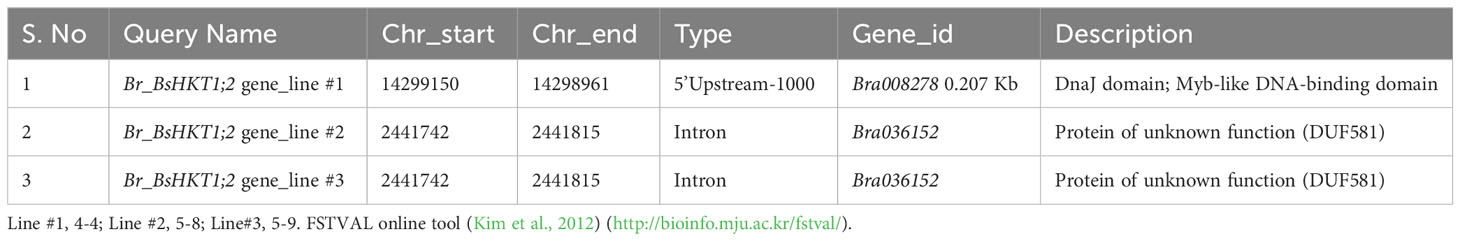
Table 2 Genomic details on BsHKT1;2 insertion locations in the B. rapa genome based on flanking sequence tag validator (FSTVAL) analysis.
3.7 BsHKT1;2 in B. rapa imparts salt tolerance under salt stress
Stable transgenic and non-transgenic lines were exposed to 0, 100, or 200 mM salt concentrations (NaCl) in the MS medium (Supplementary Figure 5). The growth of the transgenic plants was similar to that of non-transgenic plants under 0 mM salt (non-salt conditions). However, at 100 mM NaCl, the growth of the transgenic plants was greater than that of non-transgenic plants. Transgenic plants had green canopies, relatively larger leaves, longer roots, and grew faster than non-transgenic plants (Figure 4C). However, root length decreased in both the non-transgenic and transgenic lines with increasing salt concentrations (100 and 200 mM) (Figures 4E, F). The BsHKT1;2 expression levels in the transgenic plants were quantified, along with BrNHX1.1, BrNHX1.2, BrNHX1.3, BrNHX2, BrNHX3.2, and BrNHX7.
The results showed that the expression level of BsHKT1;2 was 2–4-fold higher at 100 mM salt stress and up to 5-fold higher under 200 mM salt stress in transgenic plants than in non-transgenic plants. Moreover, the BrNHX1.2, BrNHX1.3, BrNHX3.2, and BrNHX7 expression levels were higher under 100 mM NaCl, whereas their expression levels were not different at 200 mM NaCl compared to non-transgenic plants (Figures 5A, B).
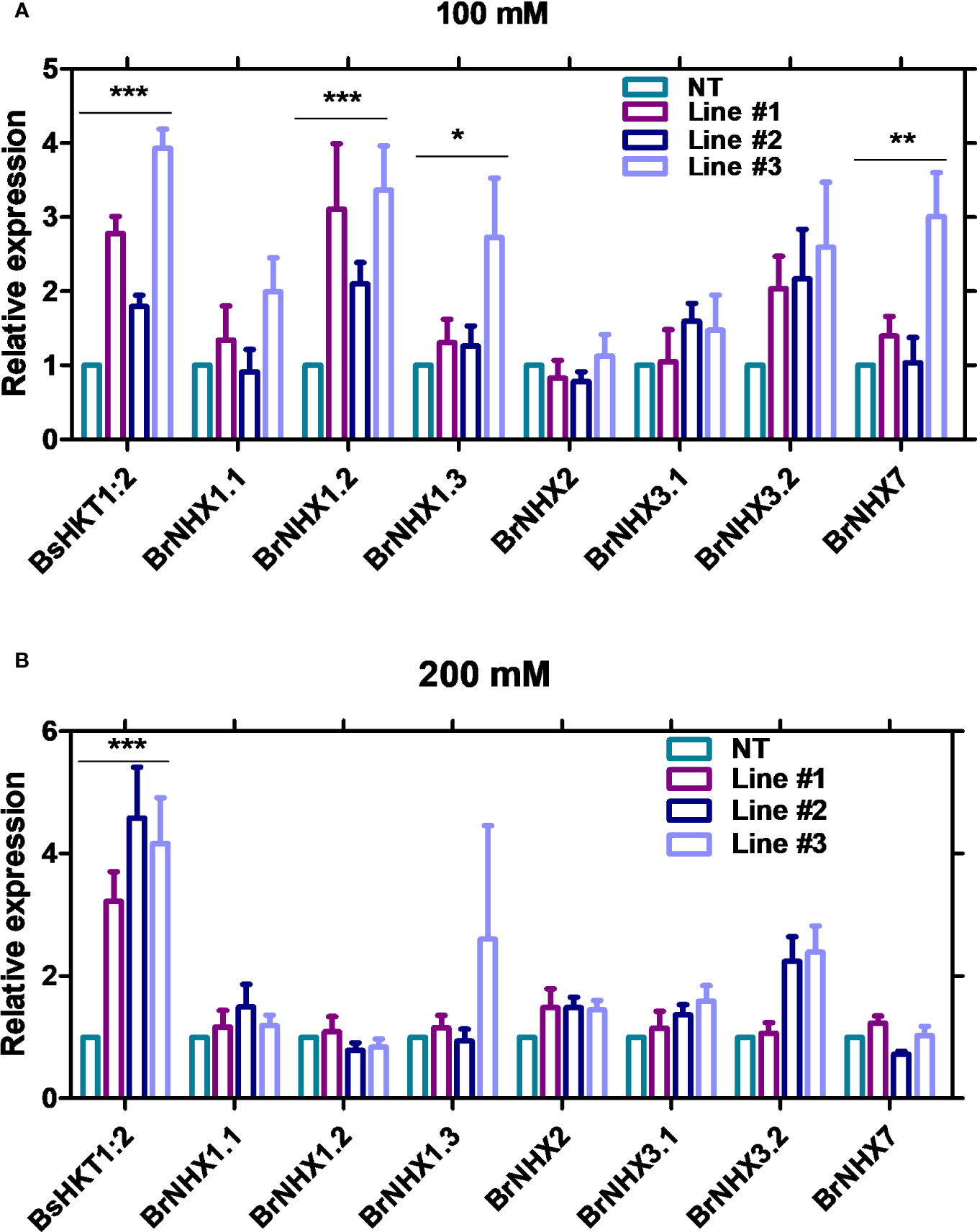
Figure 5 Quantitative real-time PCR results of salt stress-associated genes in transgenic B. rapa. Non-transgenic plants (NT) and 35S::BsHKT1;2 transgenic plants (lines#1-3) were subjected to salt stress (0, 100, and 200 mM NaCl). Plant samples were collected 7 DAI, and gene expression was quantified. Graphs show the relative gene expression level of BsHKT1;2 and salt stress-associated genes under 100 (A) and 200 mM salt stress (B). The error bar indicates the standard error of the mean. A two-way ANOVA was performed, followed by a Bonferroni post-test. Asterisks indicate the significance (* p < 0.05; **p < 0.01; ***p < 0.001). N=3. The experiment was at least repeated twice. Note: Line #1, 4-4; Line #2, 5-8; Line#3, 5-9; NT, non-transgenic plant expression levels. DAI, days after stress imposition.
4 Discussion
Plants experience salt stress in soils with electrical conductivity of saturated soil paste extract (ECe) of 4 deciSiemens per metre (dS/m). Crop plants, especially glycophytes, exhibit salt stress symptoms in soils with ECe <4 dS/m (Munns and Tester, 2008). Plants possess HKT transporters that decrease the transport of Na+ from the root to the shoot. The heterologous expression of AtHKT1;1 from Arabidopsis in oocytes has been shown to be a Na+-selective uniporter (Uozumi et al., 2000). The HKT transporter was explored in the B. rapa genome. BrHKT1;1 (XP_009134189.2) was predicted through automated computational analysis from a genomic sequence (NC_024797.2) (Zhang et al., 2018). However, no further analysis was available. This may be attributed to the glycophyte nature of B. rapa. Several studies have strengthened the potential of HKT transporters to decrease and increase Na+ content in shoots and roots, respectively (Uozumi et al., 2000; Xu et al., 2018; Wang et al., 2019). Interestingly, HKT expression was observed in both root and shoot tissues. However, their functions varied. In the root, it sequesters Na+ ions into XPCs, whereas in the shoot, HKT transports excess Na+ from the leaf parenchymal cells to the phloem stream (Mian et al., 2011), which is attributed to salt tolerance. In this study, we investigated the role of the salt-responsive BsHKT1:2 in a terrestrial halophyte plant B. sinuspersici. Particularly, we studied the structural classification and functional characteristics of the BsHKT transporters. B. sinuspersici is an SCC4 plant that thrives in salt-affected areas and is commonly found around the Persian Gulf and Oman Sea, in southwestern Pakistan, southern Iran, Kuwait, southern Iraq, Saudi Arabia, Qatar, and the UAE (Akhani et al., 2005; Akhani et al., 2012; Uzilday et al., 2023). As Bienertia is a halophyte (Park et al., 2009), we assumed that it has efficient mechanisms for coping with salt stress. Therefore, we performed genome annotation and identified the HKT and salt overly sensitive 1 (SOS1) genes in the Bienertia genome. Three paralogs of the HKT family were identified: BsHKT1;1, BsHKT1;2, and BsHKT1;3 (Figure 1, Table 1, and Supplementary Table 1). Studies have demonstrated that the rice genome contains nine paralogs of HKT genes (Garciadeblás et al., 2003; Jabnoune et al., 2009). Of these, five were classified as HKT subfamily II and four as HKT subfamily I (Garciadeblás et al., 2003). Similarly, barley possesses five paralogs of HKT (Hazzouri et al., 2018). In addition, variations in the number of paralogs were demonstrated between durum wheat and bread wheat genomes (Huang et al., 2008). Most monocot genomes contain HKT proteins belonging to subfamily II, whereas dicots contain HKT proteins belonging to subfamily I (Platten et al., 2006). Furthermore, monocot genomes contain more than one copy of the HKT protein (Garciadeblás et al., 2003; Platten et al., 2006; Jabnoune et al., 2009). Based on the phylogenetic clusters of Platten et al. (2006), our phylogenetic analysis, which included HKT proteins representing two subfamilies, showed that all three HKTs from B. sinuspersici were classified as HKT subfamily I (Figure 1; Supplementary Table 2). Most HKT from diploid plants contains the Ser residue at pore domain A, which transports Na+ from the transpiration stream to XPCs (Horie et al., 2009; Møller et al., 2009; Wang et al., 2020). Particularly, HKT proteins in the family Amaranthaceae sensu lato are classified into subfamily I because they have Ser-Gly-Gly-Gly residues in four major pore domains (Flowers and Colmer, 2008; Wang et al., 2020).
Moreover, motif analysis showed that BsHKT1;1 had motifs similar to those of AtHKT1;1 and BrHKT1;1; however, it had one motif replacement and one extra motif compared with AtHKT1;1 (Figure 1). However, fewer motifs were identified in BsHKT1;2 and BsHKT1;3. HKT proteins from halophytes possess complete motifs. However, the function of BsHKT1;1 may differ from those of BsHKT1;2 and BsHKT1;3. Interestingly, although most glycophytes contained a large number of motifs, a few HKTs contained a small number of motifs and were barely similar to Bienertia and grapevine. A study reported that Criolla grapevine cultivars exhibited greater salt resistance than other cultivars, and AtNHX1 transgenic grapevines showed enhanced salt tolerance (Venier et al., 2018). Therefore, we assumed that the co-regulation of HKT and NHX may be important for fine-tuning the mechanism of salt resistance. In addition, both OsHKT1;1 and OsHKT2;1 lacked the motif 15. The arrangement and reduction in the number of motifs were attributed to the evolution of the genes. BsHKT1;1 possesses three exons and two introns (Supplementary Figure 1C), similar to AtHKT1;1 (Platten et al., 2006). However, BsHKT1;2 and BsHKT1;3 contained two exons and one exon, respectively. Most HKT genes contain three exons (Uozumi et al., 2000; Platten et al., 2006; Jabnoune et al., 2009). However, HKT genes from Vitis vinifera (VvHKT3a2) possess four exons (Li et al., 2019). Similarly, CsHKT3b from Cucumis sativus contains 11 exons. Moreover, variation in the number of introns was observed. For example, GmHKT3c4 from Glycine max contains only one intron (Li et al., 2019). The reason behind the loss of exons and introns may be attributed to convergent evolution.
Salt tolerance is a complex process. Entering Na+ in plant root cells triggers an elevation in Ca2+ in the cytoplasm. In turn, SOS3 perceives the increased Ca2+ levels and activates SOS2, a protein kinase. Consequently, SOS2 translocates to the plasma membrane and phosphorylates SOS1, facilitating the extrusion of Na+ into the soil (Ali et al., 2023; Liu et al., 2023). Under salt stress conditions, the differential expression of BsHKT1;1, BsHKT1;2, BsHKT1;3, and BsSOS1 in B. sinuspersici under different concentrations of NaCl showed that the increased expression level of BsHKTs slightly improved plant growth at 100 and 200 mM NaCl, whereas decreased expression of BsHKTs at 300 mM NaCl arrested plant growth. Overall, increasing salt concentrations affected the HKT expression. However, BsHKT1;2 was highly expressed among the three BsHKTs. Probably, the extremely increased expression of BsHKT1;2 is due to BsHKT1;2 transports more Na+ into the xylem parenchyma. Moreover, increased BsHKT1;2 expressions (CT value=22.6 ± 0.2 under control conditions) indicates that it may participate in salt stress signaling. Further detailed research on BsHKT1;2 will support plant salt stress signaling.
Decreased expression of AtHKT1;1 and BrHKT1;1 was observed at all salt concentrations, which may be because both AtHKT1;1 and BrHKT1;1 is salt sensitive due to the A. thaliana and B. rapa being a glycophyte (Shi et al., 2003). HKT overexpression improves salinity tolerance in models and crop plants (Shi et al., 2003; Xu et al., 2018; Wang et al., 2019). When transgenic plants were exposed to salt stress, all plants showed higher expression levels than the control plants, indicating that BsHKT1;2 is functional in B. rapa (Figure 6). Morpho-physiological parameters, such as biomass, canopy size, and root length, showed that B. rapa with 35S::BsHKT1;2 was salt tolerant. In addition, BsHKT1;2 in the cell membrane strengthens the idea that the highly expressed BsHKT1;2 in transgenic plants under salt stress transports more Na+ into XPC. In addition, under 200 mM NaCl, the high expression of BsHKT1;2 and low expression of NHX transporters indicate that BsHKT1;2 may play a role in transporting Na+ into vascular organelles. However, studies have proven that HKT proteins are generally located in the XPC plasma membrane (Uozumi et al., 2000; Horie et al., 2009; Mian et al., 2011). Further studies need to be conducted to understand the interaction between SOS2 and BsHKT1;2 under salt stress. Hence, the results of this study imply that BsHKT1;2 is more efficient in transporting Na+ and imparts salt tolerance to B. sinuspersici. In addition, when BsHKT1;2 was overexpressed in B. rapa, salt tolerance was increased to maintain its productivity.
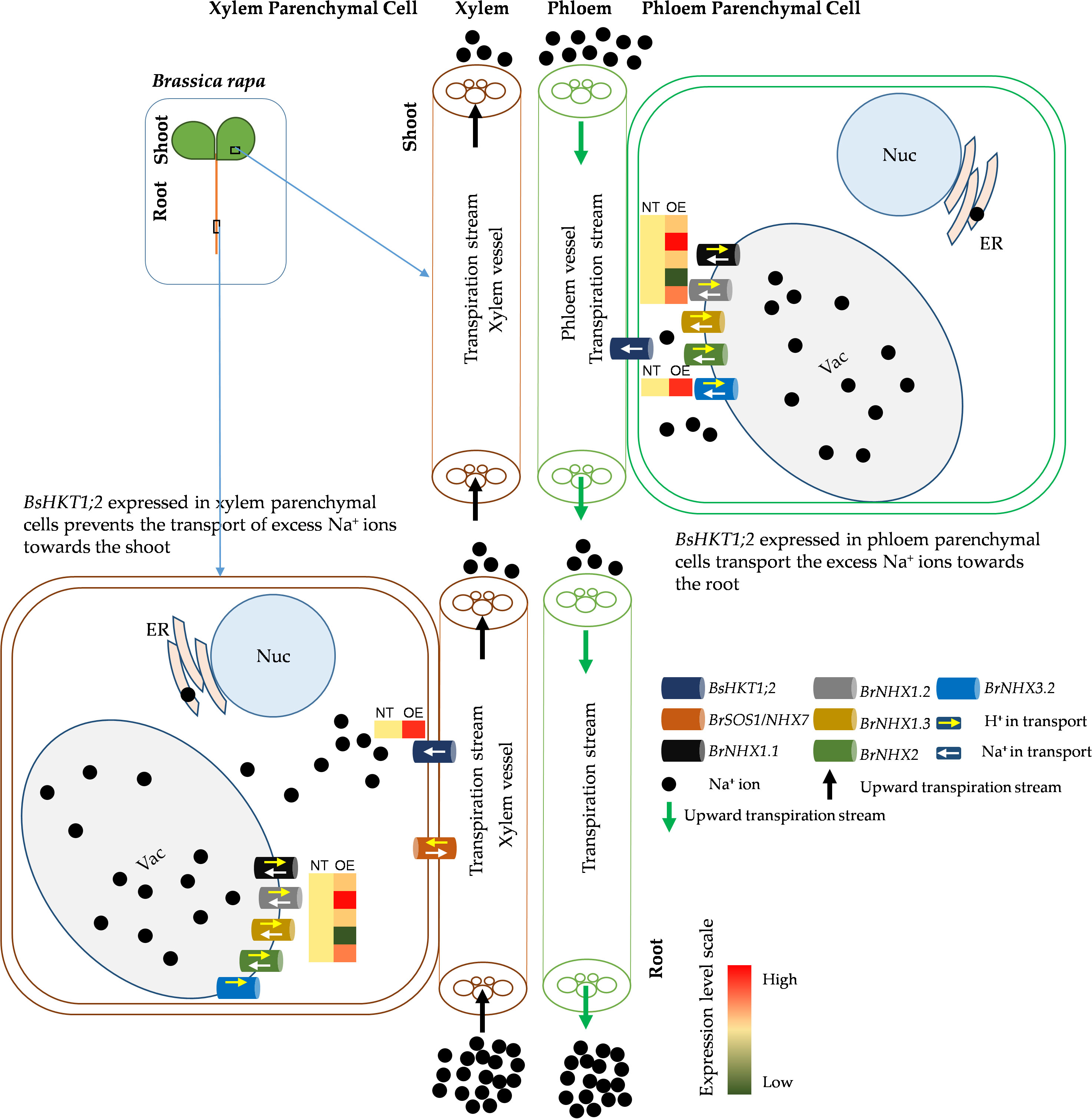
Figure 6 Schematic diagram of improved salt stress tolerance in transgenic BsHKT1;2 plant. Plants uptake Na+ ions through non-selective cation channel transporters located in the root epidermal cells. Then, Na+ ions travel toward the shoot in the xylem transpiration stream. HKT transporters present in the xylem parenchymal cell (XPC) membranes transport the Na+ ions into the XPC in a unidirectional way. Na+/H+ exchangers (NHXs) further transport the excess Na+ ions into vacuoles bidirectionally with the exchange of H+ ions. In this study, we found that increased expression of BsHKT1;2 in the transgenic B. rapa imparted salt tolerance by transporting more Na+ into XPC from the transpiration stream. Also, BsHKT1;2 located in phloem parenchymal cells in the foliar transport excess Na+ ions into the phloem vessel, which in turn are extruded out by the NHX7 transporter. Vac, vacuole; Nuc, nucleus; ER, endoplasmic reticulum; NT, non-transgenic; OE, overexpressing transgenic lines; Na+, sodium ions; H+, protons.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/protein/WOK82374.1, https://www.ncbi.nlm.nih.gov/protein/WOK82375.1, https://www.ncbi.nlm.nih.gov/protein/WOK82376.1.
Author contributions
VI: Data curation, Investigation, Validation, Visualization, Writing – original draft. HP: Investigation, Resources, Validation, Writing – review & editing. SH: Data curation, Investigation, Writing – review & editing. MK: Data curation, Validation, Writing – review & editing. JK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by “The Cooperative Research Program for Agriculture Science & Technology Development (PJ016737022023)” of the Rural Development Administration (RDA), Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1302315/full#supplementary-material
References
Akhani, H., Barroca, J., Koteeva, N., Voznesenskaya, E., Franceschi, V., Edwards, G., et al. (2005). Bienertia sinuspersici (Chenopodiaceae): a new species from Southwest Asia and discovery of a third terrestrial C4 plant without Kranz anatomy. System. Bot. 30, 290–301. doi: 10.1600/0363644054223684
Akhani, H., Chatrenoor, T., Dehghani, M., Khoshravesh, R., Mahdavi, P., Matinzadeh, Z. (2012). A new species of Bienertia (Chenopodiaceae) from Iranian salt deserts: a third species of the genus and discovery of a fourth terrestrial C4 plant without Kranz anatomy. Plant Biosystems An Int. J. Dealing all Aspects Plant Biol. 146, 550–559. doi: 10.1080/11263504.2012.662921
Ali, A., Petrov, V., Yun, D.-J., Gechev, T. (2023). Revisiting plant salt tolerance: novel components of the SOS pathway. Trends Plant Sci. 28 (9), 1060–1069. doi: 10.1016/j.tplants.2023.04.003
Almagro Armenteros, J. J., Sønderby, C. K., Sønderby, S. K., Nielsen, H., Winther, O. (2017). DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33, 3387–3395. doi: 10.1093/bioinformatics/btx431
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Chen, G., Hu, Q., Luo, L., Yang, T., Zhang, S., Hu, Y., et al. (2015). Rice potassium transporter OsHAK 1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. doi: 10.1111/pce.12585
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Garciadeblás, B., Senn, M. E., Bañuelos, M. A., Rodríguez-Navarro, A. (2003). Sodium transport and HKT transporters: the rice model. Plant J. 34, 788–801. doi: 10.1046/j.1365-313X.2003.01764.x
Han, S.-Y., Kim, W.-Y., Kim, J. S., Hwang, I. (2023). Comparative transcriptomics reveals the role of altered energy metabolism in the establishment of single-cell C4 photosynthesis in Bienertia sinuspersici. Front. Plant Sci. 14, 1202521. doi: 10.3389/fpls.2023.1202521
Hayat, S., Maheshwari, P., Wani, A. S., Irfan, M., AlYemeni, M. N., Ahmad, A. (2012). Comparative effect of 28 homobrassinolide and salicylic acid in the amelioration of NaCl stress in Brassica juncea L. Plant Physiol. Biochem. 53, 61–68. doi: 10.1016/j.plaphy.2012.01.011
Hazzouri, K. M., Khraiwesh, B., Amiri, K. M., Pauli, D., Blake, T., Shahid, M., et al. (2018). Mapping of HKT1; 5 gene in barley using GWAS approach and its implication in salt tolerance mechanism. Front. Plant Sci. 9, 156. doi: 10.3389/fpls.2018.00156
Hong, J. K., Lim, M.-H., Suh, E. J., Yoon, H.-J., Park, J., Lee, Y.-H. (2020). Functional Screening of Salt Stress Tolerance Genes Using Transgenic Arabidopsis thaliana Lines Overexpressing Brassica rapa Full-length Genes and Brassica napus Transformation. Korean Soc. Breed. Sci. 52, 297–309. doi: 10.9787/KJBS.2020.52.4.297
Horie, T., Hauser, F., Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668. doi: 10.1016/j.tplants.2009.08.009
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, S., Spielmeyer, W., Lagudah, E. S., Munns, R. (2008). Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot. 59, 927–937. doi: 10.1093/jxb/ern033
Hussain Wani, S., Brajendra Singh, N., Haribhushan, A., Iqbal Mir, J. (2013). Compatible solute engineering in plants for abiotic stress tolerance-role of glycine betaine. Curr. Genomics 14, 157–165. doi: 10.2174/1389202911314030001
Jabnoune, M., Espeout, S., Mieulet, D., Fizames, C., Verdeil, J.-L., Conéjéro, G., et al. (2009). Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 150, 1955–1971. doi: 10.1104/pp.109.138008
Kapralov, M. V., Smith, J., Filatov, D. A. (2012). Rubisco evolution in C4 eudicots: an analysis of Amaranthaceae sensu lato. PloS One 7, e52974. doi: 10.1371/journal.pone.0052974
Kim, J. S., Kim, J., Lee, T.-H., Jun, K. M., Kim, T. H., Kim, Y.-H., et al. (2012). FSTVAL: a new web tool to validate bulk flanking sequence tags. Plant Methods 8, 1–9. doi: 10.1186/1746-4811-8-19
Koteyeva, N. K., Voznesenskaya, E. V., Berry, J. O., Cousins, A. B., Edwards, G. E. (2016). The unique structural and biochemical development of single cell C4 photosynthesis along longitudinal leaf gradients in Bienertia sinuspersici and Suaeda aralocaspica (Chenopodiaceae). J. Exp. Bot. 67, 2587–2601. doi: 10.1093/jxb/erw082
Li, H., Xu, G., Yang, C., Yang, L., Liang, Z. (2019). Genome-wide identification and expression analysis of HKT transcription factor under salt stress in nine plant species. Ecotoxicol. Environ. Saf. 171, 435–442. doi: 10.1016/j.ecoenv.2019.01.008
Lim, S.-S., Yang, H. I., Park, H.-J., Park, S.-I., Seo, B.-S., Lee, K.-S., et al. (2020). Land-use management for sustainable rice production and carbon sequestration in reclaimed coastal tideland soils of South Korea: A review. Soil Sci. Plant Nutr. 66, 60–75. doi: 10.1080/00380768.2019.1674121
Liu, X., Yu, X., Shi, Y., Ma, L., Fu, Y., Guo, Y. (2023). Phosphorylation of RhoGDI1, a Rho GDP dissociation inhibitor, regulates root hair development in Arabidopsis under salt stress. Proc. Natl. Acad. Sci. 120, e2217957120. doi: 10.1073/pnas.2217957120
Mai, K. K. K., Yeung, W.-T., Han, S.-Y., Cai, X., Hwang, I., Kang, B.-H. (2019). Electron tomography analysis of thylakoid assembly and fission in chloroplasts of a single-cell C4 plant, Bienertia sinuspersici. Sci. Rep. 9, 19640. doi: 10.1038/s41598-019-56083-w
Mcalvay, A. C., Ragsdale, A. P., Mabry, M. E., Qi, X., Bird, K. A., Velasco, P., et al. (2021). Brassica rapa domestication: untangling wild and feral forms and convergence of crop morphotypes. Mol. Biol. 38, 3358–3372. doi: 10.1093/molbev/msab108
Mian, A., Oomen, R. J., Isayenkov, S., Sentenac, H., Maathuis, F. J., Véry, A. A. (2011). Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 68, 468–479. doi: 10.1111/j.1365-313X.2011.04701.x
Møller, I. S., Gilliham, M., Jha, D., Mayo, G. M., Roy, S. J., Coates, J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type–specific alteration of Na+ transport in Arabidopsis. Plant Cell 21, 2163–2178. doi: 10.1105/tpc.108.064568
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Offermann, S., Friso, G., Doroshenk, K. A., Sun, Q., Sharpe, R. M., Okita, T. W., et al. (2015). Developmental and subcellular organization of single-cell C4 photosynthesis in Bienertia sinuspersici determined by large-scale proteomics and cDNA assembly from 454 DNA sequencing. J. Proteome Res. 14, 2090–2108. doi: 10.1021/pr5011907
Park, J., Okita, T. W., Edwards, G. E. (2009). Salt tolerant mechanisms in single-cell C4 species Bienertia sinuspersici and Suaeda aralocaspica (Chenopodiaceae). Plant Sci. 176, 616–626. doi: 10.1016/j.plantsci.2009.01.014
Pavlović, I., Mlinarić, S., Tarkowská, D., Oklestkova, J., Novák, O., Lepeduš, H., et al. (2019). Early Brassica crops responses to salinity stress: a comparative analysis between Chinese cabbage, white cabbage, and kale. Front. Plant Sci. 10, 450. doi: 10.3389/fpls.2019.00450
Platten, J. D., Cotsaftis, O., Berthomieu, P., Bohnert, H., Davenport, R. J., Fairbairn, D. J., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11, 372–374. doi: 10.1016/j.tplants.2006.06.001
Rengasamy, P. (2006). World salinization with emphasis on Australia. J. Exp. Bot. 57, 1017–1023. doi: 10.1093/jxb/erj108
Schachtman, D. P., Schroeder, J. I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658. doi: 10.1038/370655a0
Shi, H., Ishitani, M., Kim, C., Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Lee, B.-H., Wu, S.-J., Zhu, J.-K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Soundararajan, P., Won, S. Y., Park, D. S., Lee, Y.-H., Kim, J. S. (2019). Comparative analysis of the YABBY gene family of Bienertia sinuspersici, a single-cell C4 plant. Plants 8, 536. doi: 10.3390/plants8120536
Tamura, K., Stecher, G., Kumar, S., Evolution (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Uozumi, N., Kim, E. J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., et al. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122, 1249–1260. doi: 10.1104/pp.122.4.1249
Uzilday, B., Ozgur, R., Yalcinkaya, T., Sonmez, M. C., Turkan, I. (2023). Differential regulation of reactive oxygen species in dimorphic chloroplasts of single cell C4 plant Bienertia sinuspersici during drought and salt stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1030413
Van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Venier, M., Agüero, C., Bermejillo, A., Filippini, M., Hanana, M., Walker, M. A., et al. (2018). Analysis of salinity tolerance of Vitis vinifera'Thompson Seedless' transformed with AtNHX1. Vitis 57 (4), 143-150. doi: 10.5073/vitis.2018.57.143-150
Wang, W.-Y., Liu, Y.-Q., Duan, H.-R., Yin, X.-X., Cui, Y.-N., Chai, W.-W., et al. (2020). SsHKT1; 1 is coordinated with SsSOS1 and SsNHX1 to regulate Na+ homeostasis in Suaeda salsa under saline conditions. Plant Soil 449, 117–131. doi: 10.1007/s11104-020-04463-x
Wang, L., Liu, Y., Li, D., Feng, S., Yang, J., Zhang, J., et al. (2019). Improving salt tolerance in potato through overexpression of AtHKT1 gene. BMC Plant Biol. 19, 1–15. doi: 10.1186/s12870-019-1963-z
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Won, S. Y., Soundararajan, P., Irulappan, V., Kim, J. S. (2023). In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici—a comparative study across C3, C4, CAM, and SCC4 model plants. PeerJ 11, e15696. doi: 10.7717/peerj.15696
Xu, M., Chen, C., Cai, H., Wu, L. (2018). Overexpression of PeHKT1; 1 improves salt tolerance in populus. Genes 9, 475. doi: 10.3390/genes9100475
Yepes, L., Chelbi, N., Vivo, J.-M., Franco, M., Agudelo, A., Carvajal, M., et al. (2018). Analysis of physiological traits in the response of Chenopodiaceae, Amaranthaceae, and Brassicaceae plants to salinity stress. Plant Physiol. 132, 145–155. doi: 10.1016/j.plaphy.2018.08.040
Zhang, L., Cai, X., Wu, J., Liu, M., Grob, S., Cheng, F., et al. (2018). Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5, 50. doi: 10.1038/s41438-018-0071-9
Zhang, M., Liang, X., Wang, L., Cao, Y., Song, W., Shi, J., et al. (2019). A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 5, 1297–1308. doi: 10.1038/s41477-019-0565-y
Zhang, Z., Zhang, J., Chen, Y., Li, R., Wang, H., Wei, J. (2012). Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 39, 8465–8473. doi: 10.1007/s11033-012-1700-2
Keywords: Bienertia sinuspersici, Brassica rapa, high-affinity K+ transporter (HKT), salt stress, abiotic stress, Na+/H+ exchangers (NHX), glycophyte, halophyte
Citation: Irulappan V, Park HW, Han S-Y, Kim M-H and Kim JS (2023) Genome-wide identification of a novel Na+ transporter from Bienertia sinuspersici and overexpression of BsHKT1;2 improved salt tolerance in Brassica rapa. Front. Plant Sci. 14:1302315. doi: 10.3389/fpls.2023.1302315
Received: 26 September 2023; Accepted: 24 November 2023;
Published: 12 December 2023.
Edited by:
Sajid Masood, Bahauddin Zakariya University, PakistanReviewed by:
Marzena Małgorzata Kurowska, University of Silesia in Katowice, PolandMohammed Ali Abd Elhammed Abd Allah, Desert Research Center, Egypt
Copyright © 2023 Irulappan, Park, Han, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung Sun Kim, anNua2ltQGtvcmVhLmty
 Vadivelmurugan Irulappan
Vadivelmurugan Irulappan Hyun Woo Park
Hyun Woo Park Sang-Yun Han
Sang-Yun Han Myung-Hee Kim
Myung-Hee Kim Jung Sun Kim
Jung Sun Kim