- 1College of Horticulture and Landscape Architecture, Yangzhou University, Yangzhou, China
- 2Jiangsu Academy of Forestry, Nanjing, China
- 3Jiangsu Yangzhou Urban Forest Ecosystem National Observation and Research Station, Yangzhou, China
Dendrobium is a perennial herb found in Asia that is known for its medicinal and ornamental properties. Studies have shown that the stem is the primary medicinal component of Dendrobium spp. To investigate the effect of the species and age of Dendrobium (in years) on the content of its medicinal components, we collected the stems of 1-to-4-year-old D. officinale, D. moniliforme, and D. huoshanense, sequenced the transcriptome, metabolome, and microbiome, and analyzed the data in a comprehensive multi-omics study. We identified 10,426 differentially expressed genes (DEGs) with 644 differentially accumulated metabolites (DAMs) from 12 comparative groups and mapped the flavonoid pathway based on DEGs and DAMs. Transcriptomic and metabolomic data indicated a general trend of the accumulation of flavonoids exhibiting pharmacological effects in the three Dendrobium species. In addition, joint metabolome and microbiome analyses showed that actinobacteria was closely associated with flavonoid synthesis with increasing age. Our findings provide novel insights into the interactions of flavonoids of Dendrobium with the transcriptome and microbiome.
1 Introduction
Dendrobium is a perennial herb valued as an herbal medicine. It was first identified by Olaf Swartz in 1799 (Puchooa, 2004). It belongs to the second-largest genus in the orchid family. There are several species of Dendrobium, and the Dendrobium genera discovered thus far include approximately 1100 species, among which 74 species are found in subtropical China, including D. huoshanense, D. nobile, D. fimhriatum, and D. officinale (Pharmacopoeia, 2010; Deng et al., 2018). In China, Dendrobium has a long history of medicinal use and has proven hypoglycemic, hypolipidemic, antitumor, pain-relieving, and antipyretic effects (Teoh, 2016). The chemical components of Dendrobium spp. are diverse, some of which include polysaccharides, alkaloids, bibenzoids, phenanthrenes, sesquiterpenes, and inulinones (Chen, 2013). The key medicinal components are polysaccharides, alkaloids, and flavonoids, which impart the unique pharmacological properties of Dendrobium (Wang et al., 2021).
With the development of high-throughput sequencing technologies, multi-omics studies have deepened our understanding of molecular mechanisms underlying the synthesis of the major components of medicinal plants. At present, multi-omics studies of Dendrobium primarily focus on the combined transcriptome and metabolome analysis. Combined transcriptomics and metabolomics analysis is an important tool for identifying variations in metabolites and gene regulation mechanisms in different tissues of different plants. The method is suitable for establishing synergistic networks for active ingredient biosynthesis and exploring key enzyme-synthesizing genes involved in metabolite synthesis (Trikka et al., 2015). Zhan et al. found that flavanone 3-hydroxylase (F3H) and leucoanthocyanidin dioxygenase (LDOX) were the key genes responsible for color changes in purple D. officinale (Zhan et al., 2020). Zhang et al. used multi-omics methods to determine the adaptation of D. officinale leaf tissue to salt stress response through the enhanced synthesis of secondary metabolites (Zhang et al., 2021b). Wu et al. used transcriptomic and metabolomic techniques based on physiological characterization to determine the response to low temperature domestication during the growth of D. officinale (Wu et al., 2016). Zhang et al. showed that purine and phenylpropanoid biosynthetic pathway may play an important role in the response to drought stress in D. sinense (Zhang et al., 2021a). Overall, multi-omics studies on Dendrobium spp. primarily focus on stress responses, growth and development, and secondary metabolism, with D. officinale being the most studied species.
Several factors, such as rainfall, temperature, light, humidity, and other abiotic factors, and biotic factors such as insects, herbivores, and microorganisms, affect the synthesis of plant secondary metabolites (Borges et al., 2017). Researchers have identified an inextricable link between the plant microbiome and secondary metabolism, especially the endophytic bacteria in plants (Etalo et al., 2018). In brief, plant metabolites can shape the microbiome of plants, which can influence the synthesis of related metabolites in plants or even directly synthesize metabolites in plants (Jacoby et al., 2021; Koprivova and Kopriva, 2022). For example, the endophytic fungus Taxomycesan dreanae in Taxus brevifolia can synthesize paclitaxel, a key anticancer substance in Taxus plants (Stierle et al., 1993). Many bacteria can act as inducers to enhance the biosynthesis of secondary metabolites, such as alkaloids, flavonoids, and terpenoids, in medicinal plants (Chamkhi et al., 2021). At present, studies on the Dendrobium microbiome primarily focus on the composition and diversity of microbial communities. Few studies have analyzed the metabolome and microbiome collectively (Chen et al., 2020; Zhao et al., 2023). Here, we aimed to use an integrated transcriptome-metabolome-microbiome analysis method to further elucidate the reciprocal regulatory relationship among host gene expression, microbial composition, and metabolites.
In this study, three annual, biennial, triennial, and quadrennial plants each from the species D. huoshanense, D. officinale, and D. moniliforme were collected. The metabolites of Dendrobium stems were identified by widely targeted metabolomics techniques. Transcripts from the stem samples were sequenced by RNA sequencing, and the endophytess of the stems were sequenced by amplicon 16S and internal transcribed spacer (ITS) rRNA. We established the metabolic profiles, transcripts, and microbial communities of the stems of the different Dendrobium species with different growth years, described the synthetic pathways of important secondary metabolites of Dendrobium, screened key genes in the synthetic pathways of important secondary metabolites of Dendrobium, and explored the microbial composition among endophytic bacteria in Dendrobium stems. This study focused on the following key factors: (1) differences in the composition of major secondary metabolites in different species of Dendrobium, and whether the accumulation patterns of these secondary metabolites are related to the expression of corresponding key genes; (2) the effects of age and species on the accumulation of major medicinal components of Dendrobium; (3) and the exploration of potential microbiota affecting the synthesis of major medicinal components of Dendrobium. Our findings are relevant for cultivation and research on Dendrobium.
2 Materials and methods
2.1 Plant material and sample collection
Three different Dendrobium species (D. huoshanense, D. officinale, and D. moniliforme) were cultivated artificially and housed in a greenhouse at Anhui Tongjisheng Biotechnology Co., Ltd. The cultivation conditions for the samples were similar to those used in our previous study (Yuan et al., 2019). Plant stems from one-year D. huoshanense (1Dh_1, 1Dh_2, 1Dh_3), D. officinale (1Do_1, 1Do_2, 1Do_3), and D. moniliforme (1Dm_1, 1Dm_2, 1Dm_3); two-year D. huoshanense (2Dh_1, 2Dh_2, 2Dh_3), D. officinale (2Do_1, 2Do_2, 2Do_3), and D. moniliforme (2Dm_1, 2Dm_2, 2Dm_3); three-year D. huoshanense (3Dh_1, 3Dh_2, 3Dh_3), D. officinale (3Do_1, 3Do_2, 3Do_3), and D. moniliforme (3Dm_1, 3Dm_2, 3Dm_3); and four-year D. huoshanense (4Dh_1, 4Dh_2, 4Dh_3), D. officinale (4Do_1, 4Do_2, 4Do_3), and D. moniliforme (4Dm_1, 4Dm_2, 4Dm_3) were used for the study. We selected the first and second sections of the uppermost main stems of Dendrobium at each instance (Figure 1), for specific picking locations. To extract RNA, DNA, and metabolites, a part of the material was frozen in liquid nitrogen at -80°C. Before the material was used for evaluating total alkaloids and flavonoids, it was washed and dried. In the current study, we included three biological replicates for all tests.
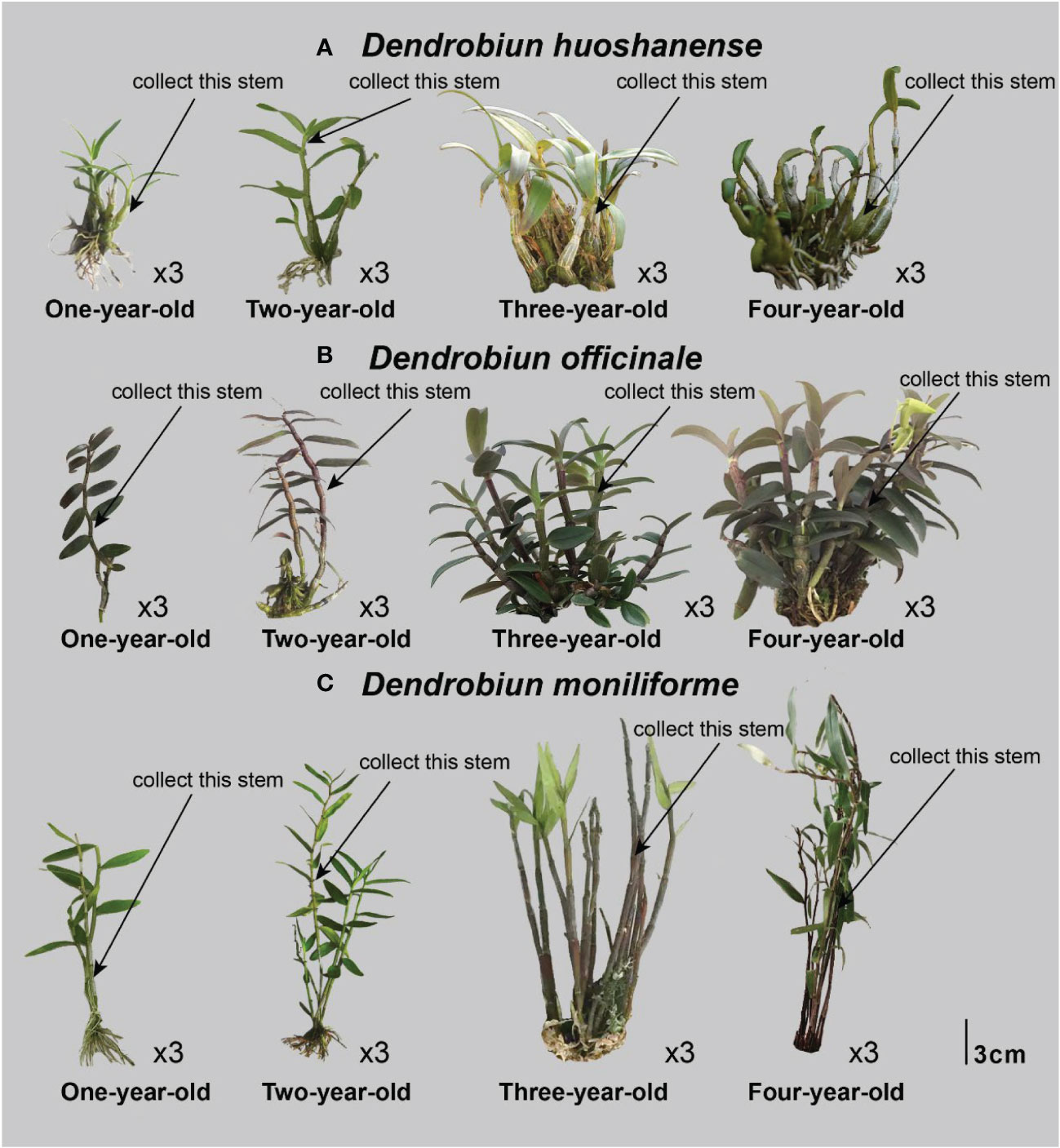
Figure 1 Stem samples collected from Dendrobium of different years. (A) Dendrobiun huoshanense; (B) Dendrobiun officinale; (C) Dendrobiun moniliforme.
2.2 Widely-targeted metabolomic profiling
To investigate metabolite variations in the different stem samples, we conducted widely targeted metabolomics experiments on samples with three biological duplicates for each growth year and species. Metware Biotechnology Ltd. conducted metabolite analysis of the stem samples (Wuhan, China). The Analyst 1.6.3 program was used to analyze metabolic data. The differences between the metabolites of the two samples were maximized using OPLS-DA to identify differential metabolites (orthogonal projections to latent structures-discriminant analysis). Based on the OPLS-DA results, the derived Variable Importance in Projection (VIP) of the OPLS-DA model for multivariate analysis was used to conduct a preliminary screening of differential metabolites. Foldchange ≥ 2, foldchange ≤ 0.5, and VIP ≥ 1 were the differential metabolites in our inquiry for the next step of the analysis. Using R and principal component analysis (PCA), we examined the accumulation of Dendrobium metabolites in different growth years. All samples were evaluated using a cluster heatmap developed once the data were standardized.
2.3 Total RNA extraction, transcriptome sequencing, and differential expression analysis
Total RNA was extracted from the stem tissue samples. The complete frozen stems were crushed in liquid nitrogen, and RNA was extracted using an Omni Plant RNA kit (CWBIO, China) and checked for purity. A TruSeq™ RNA sample preparation kit (Illumina, USA) was used to prepare the cDNA library for each sample. The cDNA libraries were sequenced at high throughput using the Illumina HiSeq 2500 platform. The raw data of all transcriptomes were stored in the NCBI database under the codes PRJCA007253, PRJNA776680, and PRJNA776418.
Cutadapt was used to filter the data and obtain clean reads (Martin, 2011). The filtered clean reads were mapped to the reference genome using HISAT2 (http://dx.doi.org/10.1038/srep19029) (Sirén et al., 2014; Zhang et al., 2016a). Gene expression levels were assessed using Fragments Per Kilobase Million (FPKM) values. Genes showing an absolute log2 fold change (FC) > 1 with a P-value < 0.05 were considered to be differentially expressed in D. huoshanense, D. officinale, and D. moniliforme. TopGo and clusterProfiler packages were used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the annotated differentially expressed genes (DEGs), respectively.
2.4 DNA extraction, high-throughput sequencing, and microbial data analysis
Microbial DNA from 72 samples (three species of 1-to-4-year-old Dendrobium, with triplicates of each species of Dendrobium with different growth years) was extracted using the CTAB/SDS method (Niemi et al., 2001). The DNA concentration and purity were tested on a 1% agarose gel. Depending on the concentration, DNA was diluted to 1 ng/μL with sterile water. Primer sets 515F (5′-GTGCCAGCMGCCGCGG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to generate bacterial libraries with a unique 6-nt barcode at 5’ of the forward primer to amplify the V4 region of the 16S rRNA gene for each sample. Similar to the process used for constructing the bacterial library, the ITS1 region of the fungus was amplified using ITS1-1F-F (CTTGGTCATTTAGGAAGTAA) and ITS1-1F-R (GCTGCGTTCTTCATCGATGC) (Caporaso et al., 2012). Phusion® High-Fidelity PCR Master Mix (New England Biolabs) was used for PCR. Next, the PCR products were mixed with an equal volume of 1× loading buffer (containing SYB green), and their purity was tested using 2% agarose gel electrophoresis. PCR products were mixed at equal density ratios. The mixed PCR products were purified using the GeneJETTM Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated and index codes were added using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina) according to the manufacturer’s recommendations. The library quality was evaluated on a Qubit@ 2.0 fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. After purification and quantification, the libraries were sequenced on an Illumina NovaSeq 6000 platform according to standard protocols.
Quality control processing of raw data from the 16S V4 bacterial and fungal ITS1 regions was performed using QIIME (V1.9.1, http://qiime.org/scripts/split libraries fastq.html) (Caporaso et al., 2010). Paired reads were processed using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) (Magoč and Salzberg, 2011). After quality control, bacteria and fungi were annotated by matching the Silva sequences with the UCHIME algorithm and the Unite database (ITS: http://unite.ut.ee/) (UCHIME, http://www.drive5.com/usearch/manual/uchime algo.html) (Quast et al., 2012; Kõljalg et al., 2013). The calculation was performed using Usearch and Vsearch to assign sequences to the same amplicon sequence variants (ASVs) (Edgar, 2010; Rognes et al., 2016).
2.5 Statistical analysis
Quantitative data were statistically analyzed using the SPSS software (version 25.0; SPSS Inc., Chicago. IL, USA) using one-way ANOVA to determine significant differences at a p-value of > 0.05. If significant differences were observed, Duncan’s post-hoc test was performed to determine the values that differed from all other values. An infrared spectrogram was drawn using Origin 2019b (OriginLab, Northampton. MA, USA). Other analyses were conducted and figures were constructed using R (version 3.6.1; https://www.r-project.org/). The “dplyr” software was used for data cleaning. “vegan,” “phyloseq,” and “ggplot2” packages were used to calculate alpha diversity indexes such as Shannon, Pielou, Chao1, and ACE (McMurdie and Holmes, 2013; Oksanen, 2013; Wickham, 2011). Beta diversity was calculated using the “phyloseq” 154 and visualized using “ggplot2.” Histograms of the top 10 relative abundances at the phylum and genus levels were generated using “ggplot2.” The “ggClusterNet” package was used to create correlation network diagrams of bacterial communities (https://github.com/taowenmicro/ggClusterNet).
3 Results
3.1 Comparative transcriptomics of three Dendrobium species with different growth years
We used transcriptome sequencing to analyze the differential expression of genes in the stems of three 1-to-4-year-old Dendrobium cultivars. The screening criteria for DEGs were crucial. We identified DEGs based on of |log2 (fold change)| > 1 and P value < 0.05. Owing to the large number of comparison groups among the 36 samples, we only focus on the DEGs of the stems with the same growth age from the three cultivars of Dendrobium. Twelve comparison groups were constructed in this study as follows: 1Dh vs. 1Dm, 1Dh vs. 1Do, 1Do vs. 1Dm, 2Dh vs. 2Dm, 2Dh vs. 2Do, 2Do vs. 2Dm, 3Dh vs. 3Dm, 3Dh vs. 3Do, 3Do vs. 3Dm, 4Dh vs. 4Dm, 4Dh vs. 4Do, and 4Do vs. 4Dm. A total of 10,426 DEGs were identified in 12 comparison groups. The 2Do vs. 2Dm comparison group had the highest number of DEGs, with 1665 upregulated and 2900 downregulated DEGs. In contrast, the 2Dh vs. 2Dm comparison group had the fewest DEGs, with 264 up-regulated and 345 down-regulated DEGs (Supplementary File Figure S1).
To further analyze the biological function of DEGs, we performed KEGG enrichment analysis. The biosynthesis of secondary metabolites is significantly enriched in all comparison groups, indicating that genes related to secondary metabolites play a key regulatory role in the stems of the three Dendrobium species (Figure 2). Since secondary metabolites were the active components of many medicinal plants, we would next focus on the synthetic pathways associated with secondary metabolites. Phenylpropanoid biosynthesis was significantly enriched in several comparison groups except for the four comparison groups: 2Do vs. 2Dm, 3Dh vs. 3Do, 3Do vs. 3Dm, 4Do vs. 4Dm (Figure 2). As one of the important secondary metabolic pathways of plants, phenylpropane metabolism can produce more than 8,000 metabolites such as lignin, flavonoids and phenolic acids. In addition, we found that flavonoid biosynthesis downstream of the phenylpropyl pathway was significantly enriched in the comparison groups of 1Dh vs. 1Dm, 1Dh vs. 1Do, 2Dh vs. 2Dm, 2Do vs. 3Dm, 3Dh vs. 3Dm, 3Dh vs. 3Do, 4Dh vs. 4Dm, 4Dh vs. 4Do and 4Do vs. 4Dm. Flavone and flavonol biosynthesis was significantly enriched in the comparison group of 1Dh vs. 1Do, 2Dh vs. 2Dm, 2Dh vs. 2Do, 3Dh vs. 3Dm, 3Do vs. 3Dm, 4Dh vs. 4Dm, 4Dh vs. 4Do. Meanwhile, tropane, piperidine and pyridine alkaloid biosynthesis in the comparison group 1Dh vs. 1Do, 2Dh vs. 2Dm, 2Dh vs. 2Do, 2Do vs. 2Dm, 3Dh vs. 3Do, 4Dh vs. 4Do, 4Do vs. 4Dm were significantly enriched. These results indicated that genes related to the synthesis of flavonoids and alkaloids played key regulatory roles in the stems of three Dendrobium species. Flavonoids and alkaloids are one of the main medicinal components of Dendrobium, therefore, we focus on these two metabolites in the metabolome analysis below.
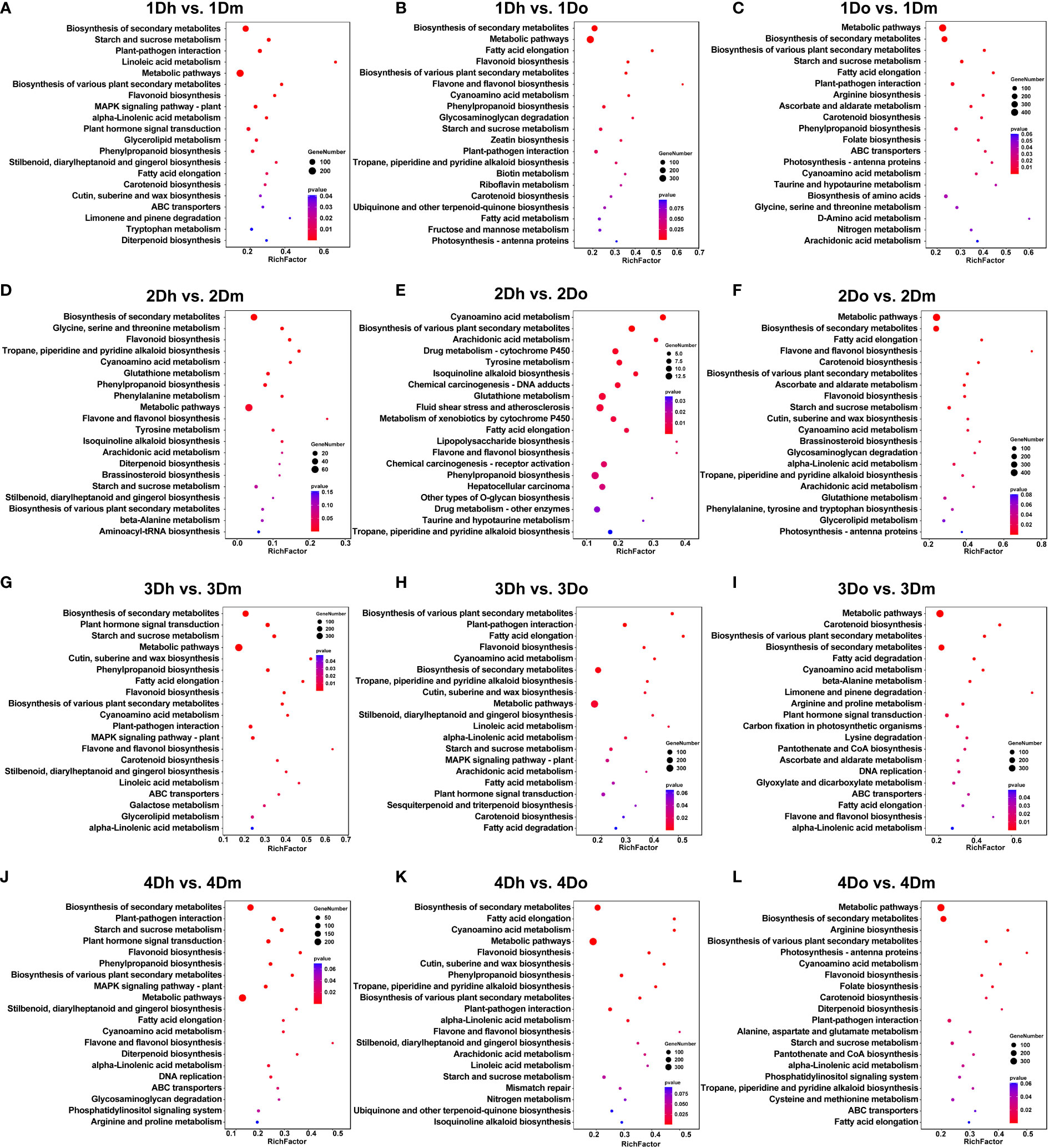
Figure 2 The KEGG pathway analysis of DEGs. (A) 1Dh vs 1Dm; (B) 1Dh vs 1Do; (C) 1Do vs 1Dm; (D) 2Dh vs 2Dm; (E) 2Dh vs 2Do; (F) 2Do vs 2Dm; (G) 3Dh vs 3Dm; (H) 3Dh vs 3Do; (I) 3Do vs 3Dm; (J) 4Dh vs 4Dm; (K) 4Dh vs 4Do; (L) 4Do vs 4Dm.
3.2 Comparative metabolomics of three Dendrobium species in different years
Based on Fold change ≥ 2, fold change ≤ 0.5, and VIP ≥ 1, we identified 644 DAMs in 12 comparison groups (Supplementary File Figure S2). The number of DAMs in the 2Do vs. 2Dm comparison group was the highest (310), whereas the number of DAMs in the 4Dh vs. 4Dm comparison group was the lowest (199). To further explore the function of these DAMs, KEGG enrichment analysis was conducted (Supplementary File Figure S3). Flavone and flavonol biosynthesis were significantly enriched in all comparison groups. Flavonoid biosynthesis was enriched in the comparison group of 1Dh vs. 1Dm, 3Dh vs. 3Do, 3Do vs. 3Dm, 4Dh vs. 4Dm. It indicated that the content of flavonoids especially flavones and flavonols in the three kinds of stems of Dendrobium may vary widely. Tropane, piperidine, and pyridine alkaloid biosynthesis were significantly enriched only in two comparison groups: 2Dh vs. 2Do and 2Do vs. 2Dm. Therefore, we focused on flavonoid biosynthesis in our subsequent combined transcriptome-metabolome analysis.
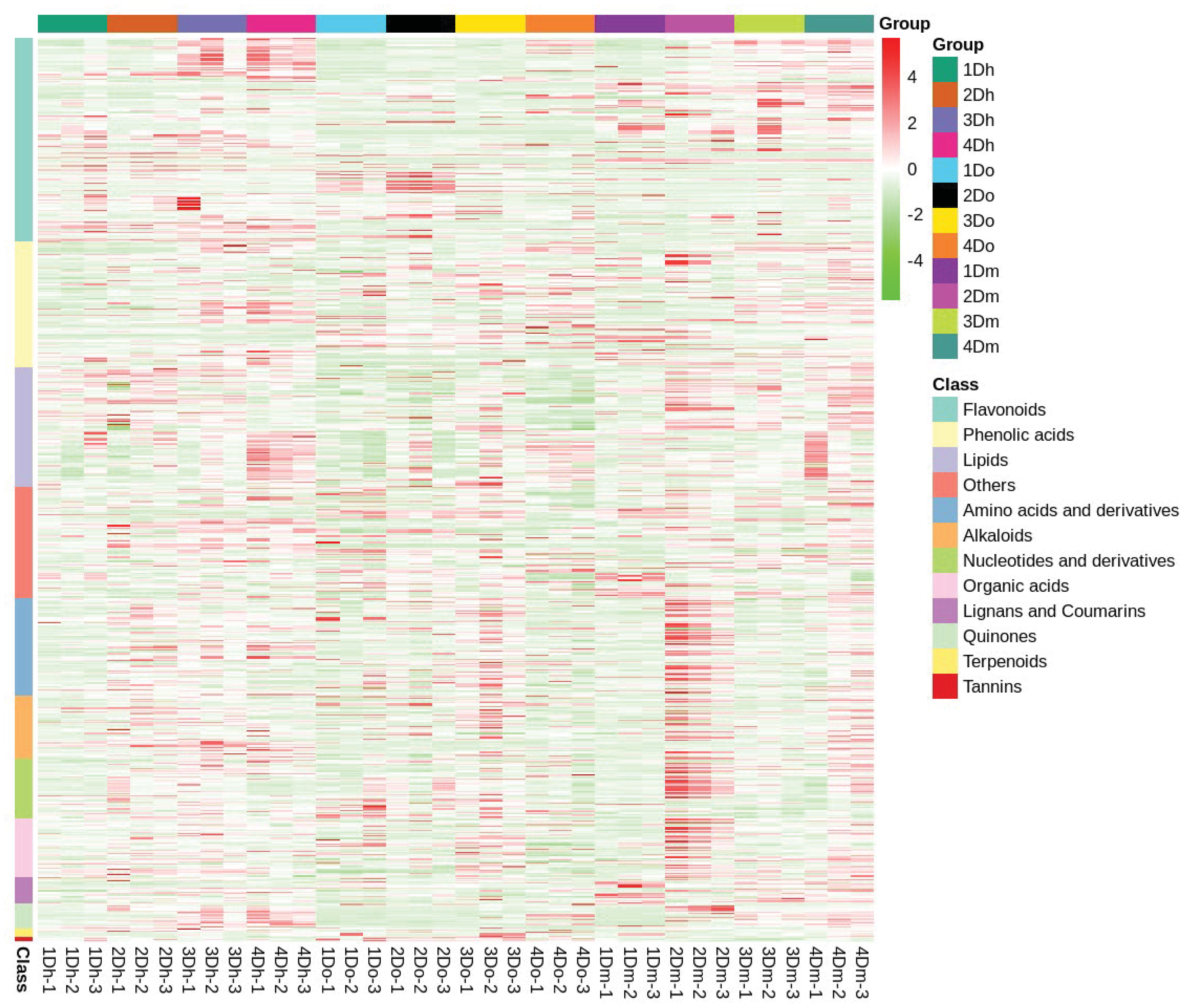
Using LC-MS analysis, we identified 767 compounds, including 173 flavonoids (65 flavonoids, 42 flavonols, 3 chalcones, 12 dihydroflavonoids, 7 dihydroflavonols, 4 isoflavones, 4 flavanols, and 36 flavonoid carboglycosides), systematically and comprehensively identified the composition of flavonoids in the stems of the three Dendrobium species, and confirmed that the stems of D. huoshanense, D. officinale, and D. moniliforme were rich in flavonoids. Further, based on hierarchical cluster analysis, a heatmap of metabolite abundance clustering was drawn. The heatmap revealed an upward trend in the relative expression of most flavonoids in D. huoshanense and D. moniliforme as the years progressed (Figure 3). In D. officinale, the relative expression of most flavonoids reached the highest in the second year. In D. huoshanense, the relative expressions of phenolic acids, lignans and coumarins and quinones increased with age. In D. officinale, except flavonoids and quinones, the relative expression of other metabolites reached the highest in the third year. In D. moniliforme, the relative expression levels of most metabolites, such as lipids, organic acids, quinones and terpenoids, reached their maximum in the second year and then gradually decreased. The findings suggest that the accumulation pattern of metabolites varies among different varieties of Dendrobium, potentially leading to a decline in the content of certain medicinal ingredients as the plant ages.
3.3 Changes in metabolites and genes related to flavonoid biosynthesis
KEGG enrichment analysis of the transcriptome and metabolome showed that the DEGs and DAMs of the three Dendrobium species with different growth years were associated with flavonoid biosynthesis. We further analyzed the expression of related metabolites and genes involved in flavonoid biosynthesis in the selected Dendrobium species. We identified 37 DEGs associated with flavonoid biosynthesis in the transcriptome data (Figure 4). In all, we identified phenylalanine, cinnamic acid, naringenin chalcone, naringenin, isoflavones, apigenin, dihydrokaempferol, kaempferol, dihydroquercetin, quercitrin, eriodictyol, luteolin, and tricetin as 13 DAMs related to the flavonoid biosynthetic pathway in the metabolomic data from the samples.
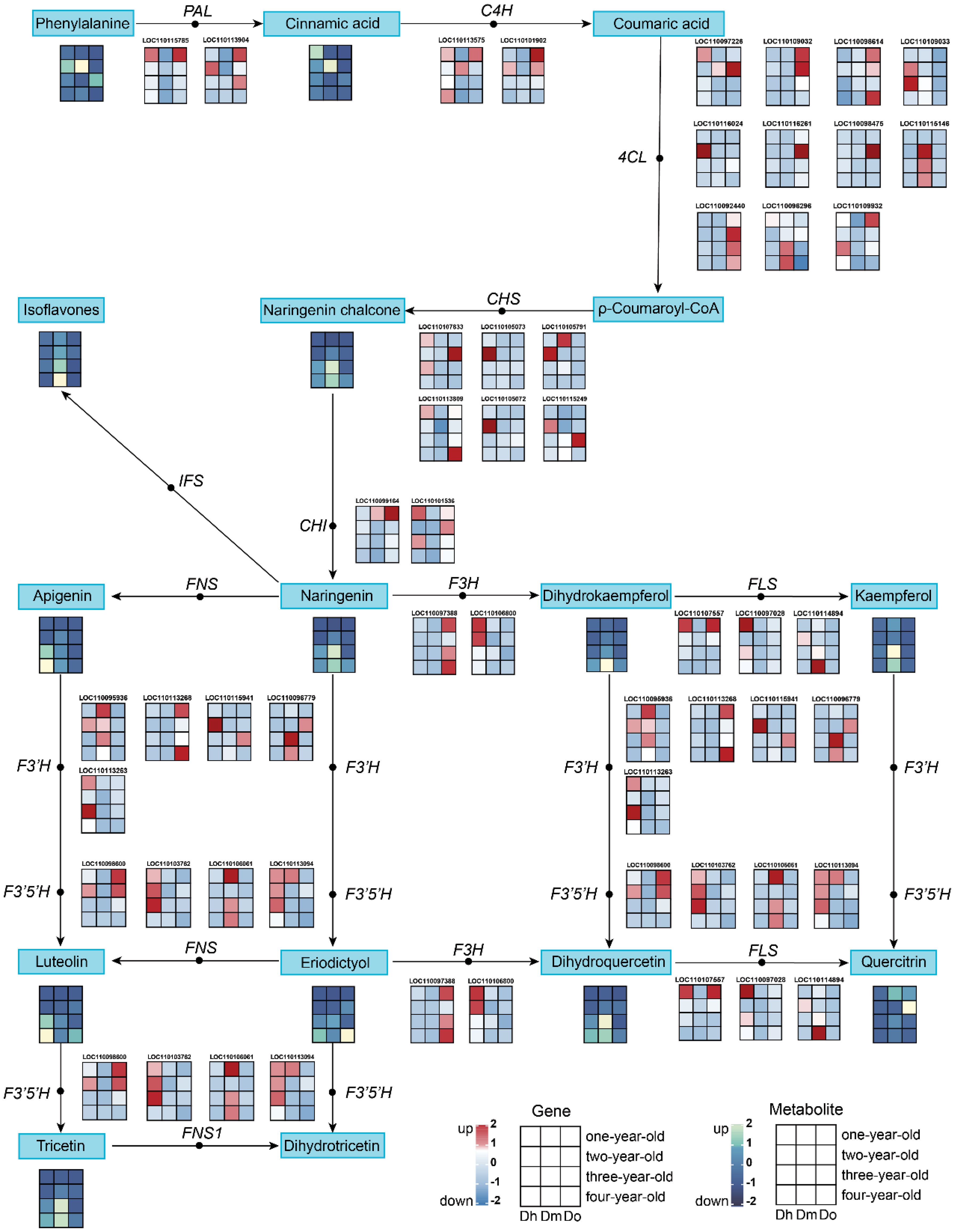
Figure 4 The pathway associated with flavonoid biosynthesis. The expression heatmap of the key metabolites and transcripts for flavonoid synthesis between four years of three Dendrobium species was shown by the colored cell on the bottom. Key enzyme gene abbreviation: PAL, phenylalanine ammonia lyase; 4CL, 4-coumarate: CoA ligase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; CHI, chalcone isomerase; IFS, isoflavone synthase; F3H, flavonoid 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; F3’5’H, flavonoid 3’,5’-hydroxylase; FNS, flavone synthase; FNS1, flavone synthase I; FLS, flavonol synthase.
The results showed that the relative expression levels of most flavonoids, such as naringenin, isoflavones, apigenin, luteolin, and tricetin, in D. huoshanense and D. moniliforme were higher in the third and fourth years. This is consistent with the trend observed in the relative expression of structural genes downstream of the flavonoid synthesis pathway. In D. officinale, the relative expression levels of many flavonoids (such as isoflavones, tricetin, quercitrin, and dihydroquercetin) peaked in the second year. This was consistent with the expression trend of structural genes upstream of the flavonoid biosynthesis pathway in D. officinale. The expression of PAL, C4H, 4CL, CHI, and other genes was higher in the first- and second-year stems. When comparing the relative expression of flavonoids in the stems of different kinds of Dendrobium of the same growing age, we found that compared with the other two kinds of Dendrobium, the relative contents of isoflavones, naringenin chalcone, naringenin, kaempferol, tricetin were the highest in D. moniliforme. This shows the potential of D. moniliforme as a medicinal plant in addition to an ornamental plant.
3.4 Taxonomic characteristics of endophytic microbes of the three Dendrobium species
Many studies have shown that the accumulation of secondary metabolites in medicinal plants is closely related to the metabolism of endophytes. To investigate the community composition of endophytes in the stems of three Dendrobium species aged 1-4 year, 16S rRNA and ITS sequencing analyses were used to analyze the stem samples. We used an Upset diagram to show the number of ASVs shared by endophytic bacteria in the stems of Dendrobium of the same variety and different years, where Figures 5A–C is bacteria and Figures 5D–F is fungi. The results showed that each Dendrobium had a large number of unique ASVs, indicating that the endophytes in the stems of the three Dendrobium species were very different.
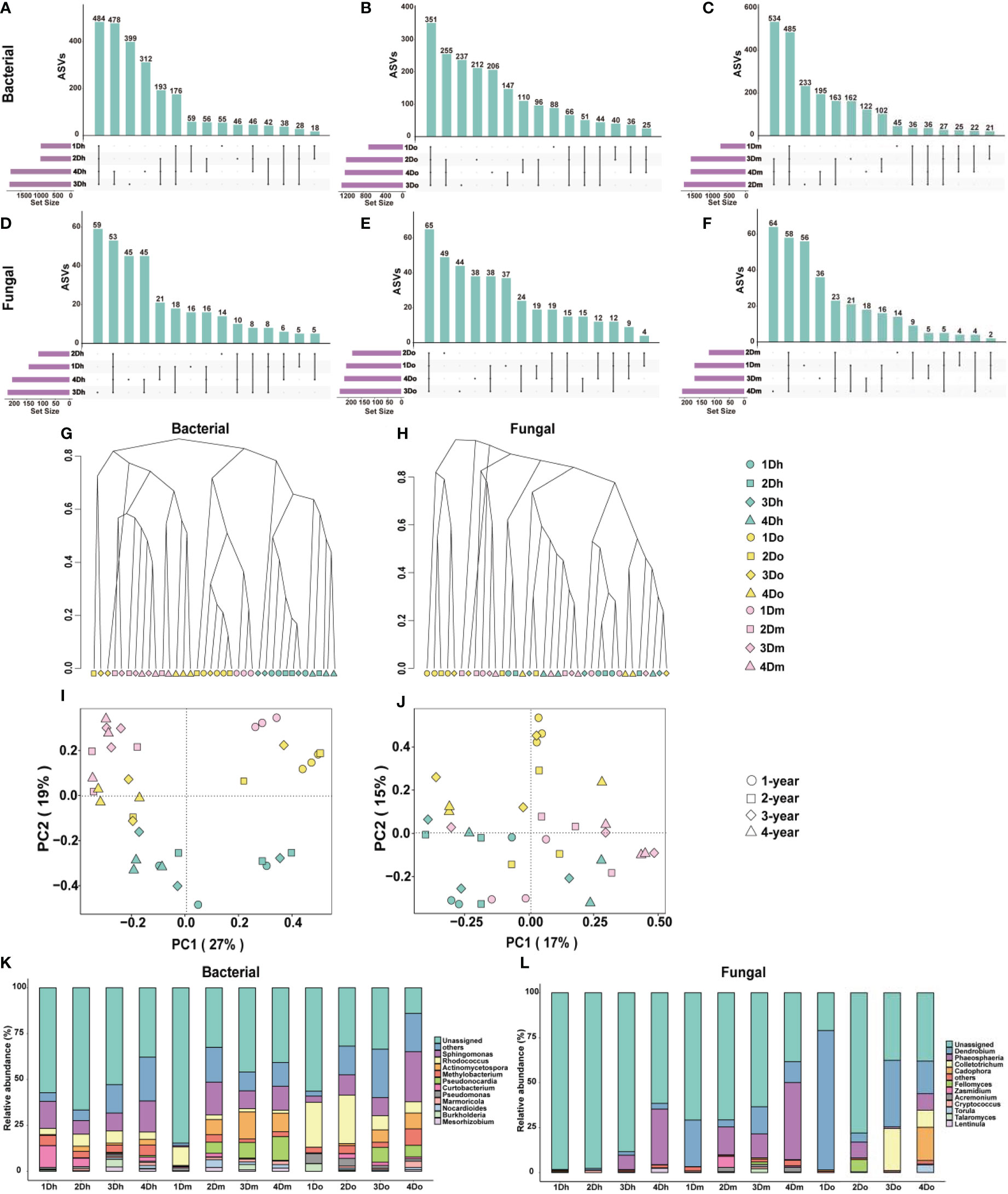
Figure 5 Microbial community differences in three kinds of Dendrobium of 1–4-year-old. (A–F) Upset diagram representing bacterial (A–C) and fungal (D–F) amplicon sequence variants (ASVs) associated with the endophytes from the stems of three kinds of Dendrobium of 1-4-year-old. (G, H) Hierarchical clustering of bacterial (G) and fungal (H) communities showing the Bray-Curtis dissimilarity of the whole ASV count data. (I, J) Principal component analysis (PCA) of bacterial (I) and fungal (J) communities. (K, L) Top 10 relative abundances of bacterial (K) and fungal (L) communities classified at genus.
To compare the diversity of the microbial communities of the endophytes of the stems, we performed the α-diversity analysis, and the results are shown in Supplementary File Tables S1, S2. The results showed that the differences in the diversity of the fungal communities in the samples were not significant, whereas the bacterial communities were more diverse. Shannon, Chao1, and ACE indices indicated that the alpha diversity of two-year-old D. moniliforme was significantly higher than that of one-year-old D. moniliforme and D. officinale, indicating that the diversity of the bacterial community in two-year-old D. moniliforme was significantly higher than that of one-year-old D. moniliforme and D. officinale. In addition, the alpha diversity of two-year-old D. moniliforme was the highest among all samples, indicating that two-year-old D. moniliforme had the highest bacterial community diversity. To compare bacterial and fungal community compositions between samples, we calculated the Bray-Curtis metric, which is related to beta diversity. Hierarchical clusters of bacteria showed that samples of the same species were mostly clustered together, whereas samples of different species and the same age were in close proximity (Figure 5G). Hierarchical clusters of the fungus showed the evolutionary distance of the phylogeny between one-year-old D. officinale and other samples (Figure 5H). In the PCoA figure for bacteria, PCo1 and PCo2 accounted for 27% and 19% of the total variance, respectively (Figure 5I). Meanwhile, one-year-old D. moniliforme and D. officinale were positioned farther away from the other samples, indicating that the endophytic communities in their stems were more different from each other. In the PCoA of fungi (Figure 5J), PCo1 and PCo2 accounted for 17% and 15% of the total variance, respectively. The differences between the endophytic bacterial communities in the stems of different Dendrobium species (aged from 1 to 4 years) were greater than the differences between the endophytic fungal communities.
Taxonomy at the genus level showed different patterns between bacterial and fungal communities in different samples, with different compositions of endophytic communities in stems of the same species but different growth years. As shown in Figure 5K, in terms of bacterial community composition, actinomycetospora accounted for a higher proportion of endophytic bacterial community in D. moniliforme compared to the other two Dendrobium species. The proportion of pseudonocardia in the endophytic bacterial community of D. moniliforme gradually increased with age. Among all the samples, curtobacterium occupies the largest proportion in 1Dh. The proportion of rhodococcus in the three species of Dendrobium showed a decreasing trend with increasing age. As shown in Figure 5L, colletotrichum accounted for the largest proportion in 3Do, while torula and cadophora accounted for the largest proportion in 4Do. The proportion of phaeosphaeria increased with age in both D. huoshanense and D. moniliforme. In D. officinale, the proportion of Phaeosphaeria reached the largest in the second year. Overall, the community composition of the endophytes of the stems of the three 1–4-year-old Dendrobium species differed, with their fungal community composition being less complex than their bacterial community composition. At the same time, the endophyte community composition of the three Dendrobium species became more complex with increasing age.
3.5 Joint analyses of metabolome and microbiome
To further understand the regulatory relationship between endophytess and secondary metabolites and their effects on metabolism in D. officinale, D. moniliforme, and D. huoshanense, we analyzed the correlation between the microbiome and metabolome using a multi-omics analysis. We performed correlation analyses based on differentially expressed metabolites, differential flora (genus level), and chord diagrams of Spearman correlations between microbes and metabolites. Correlation analysis revealed potential interactions between endophytess and secondary metabolites in Dendrobium stems from different years. Based on the analysis of the transcriptome and metabolome above, we will focus on the correlation between endophytes and flavonoids.
At the bacterial level, in 1Dh vs. 1Dm and 1Dh vs. 1Do, flavonoids (Genkwanin, Syringetin, Apigenin-8-C-Arabinoside, Tricin, Tricin-7-O-(2’’-Sinapoyl) glucoside, Apigenin-6,8-di-C-glucoside-4’-O-glucoside, Diosmetin-6-C-glucoside), flavonols (6-Hydroxykaempferol-7-O-glucoside) and flavonoid carbonoside (Apigenin-8-C-glucoside-7-O-(6’’-sinapoyl) glucoside, Isoorientin-7-O-glucoside, Hispidulin-8-C-glucoside) that showed significant positive correlation with williamsia. Zymomonas was significantly negatively correlated with several flavonoids, such as apigenin-7-O-glucoside (cosmosiin), kaempferol-3,7-di-O-glucoside, and luteolin-7-O-gentiobioside, in 1Dh vs. 1Dm. In 2Do vs. 2Dm, lapillicoccus was positively correlated with most flavonoids (rhamnocitrin, diosmetin, chrysoeriol, and tricin, among others). In 3Dh vs. 3Dm, actinomycetospora and actinomycetospora were positively associated with flavonols (Avicularin, Hyperin, Quercetin-7-O-(6’’-malonyl) glucosyl-5-O-glucoside). In 4Dh vs. 4Dm, propionibacterium was significantly negatively correlated with genkwanin, rhamnocitrin, diosmetin, chrysoeriol, hispidulin, and tamarixetin. Ninety percent of the bacteria significantly correlated with flavonoid levels were found to belong to the phyla actinobacteria and proteobacteria (Figure 6).
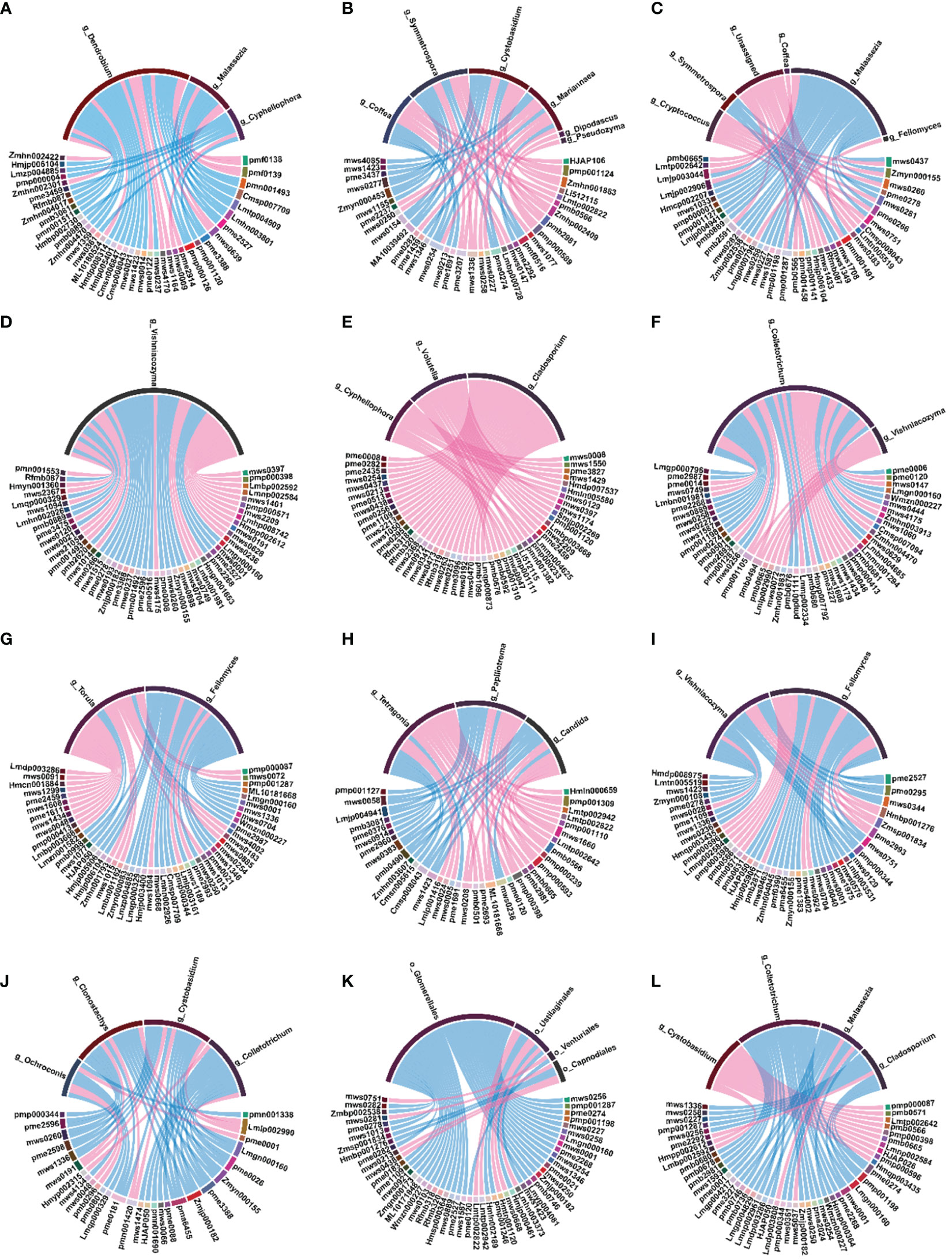
Figure 6 Spearman’s correlation chord diagram of differential microorganisms (bacteria) with DAMs at the genus level. (A) 1Dh vs 1Dm; (B) 1Dh vs 1Do; (C) 1Do vs 1Dm; (D) 2Dh vs 2Dm; (E) 2Dh vs 2Do; (F) 2Do vs 2Dm; (G) 3Dh vs 3Dm; (H) 3Dh vs 3Do; (I) 3Do vs 3Dm; (J) 4Dh vs 4Dm; (K) 4Dh vs 4Do; (L) 4Do vs 4Dm.
At the fungal level, cyphellophora was significantly negatively correlated with flavonoids in 1Dh vs. 1Dm. However, cyphellophora was significantly positively correlated with flavonoids in 2Do vs. 2Dm. Vishniacozyma was positively correlated with kaempferol-3,7-di-O-glucoside, luteolin-7-O-gentiobioside and apigenin in 2Dh vs. 2Dm. In 3Dh vs. 3Do, vishniacozyma was negatively correlated with 3’,4’,7-trihydroxyflavone, luteolin-3’-O-glucoside and genkwanin. Colletotrichum was negatively correlated with flavonoids in both 4Dh vs. 4Do and 4Do vs. 4Dm. Ninety-four percent of the fungi that were significantly associated with the levels of flavonoids belonged to ascomycota and basidiomycota (Supplementary File Figure S4).
4 Discussion
We detected differentially synthesized metabolites in the stems of D. huoshanense, D. officinale, and D. moniliforme from four different growth years. A total of 644 DAMs were identified, including 170 flavonoid metabolites, primarily flavonoids and flavonols. Notably, some important flavonoids, such as kaempferol, eriodictyol, luteolin, and tricetin, were synthesized at significantly higher levels in three- and four-year-old stems than in one- and two-year-old stems, regardless of the species; these have been reported to have high medicinal value. Kaempferol has been widely used in the treatment of multiple diseases because of its antioxidant and anti-inflammatory properties (Rajendran et al., 2014). Eriodictyol exhibits significant antioxidant (Rossato et al., 2011) and anti-inflammatory properties (Lee, 2011). Luteolin exhibits antioxidant, anti-inflammatory, anti-allergic, and anticancer properties (Seelinger et al., 2008; Imran et al., 2019; Gendrisch et al., 2021). Tricetin exhibits anti-inflammatory properties, in addition to inhibiting the proliferation of human breast cancer cells by blocking cell cycle progression and inducing apoptosis (Hsu et al., 2009; Sun et al., 2019). The accumulation of active ingredients is a potential factor affecting the complete utilization of medicinal plants, and it is of great significance to understand the synthetic mechanism. In summary, multiple flavonoids showed significant accumulation in the stems of D. huoshanense, D. officinale, and D. moniliforme, indicating that flavonoids play an important role in the growth of these plants. Therefore, the stems of D. huoshanense, D. officinale, and D. moniliforme have significant medicinal value.
Furthermore, we constructed a regulatory network for flavonoid biosynthesis in D. huoshanense, D. officinale, and D. moniliforme stems to concisely visualize the role of genes involved in flavonoid synthesis in this pathway. Our data showed 37 DEGs involved in flavonoid biosynthesis in six comparison groups, including PAL, C4H, 4CL, CHS, CHI, F3H, F3’H, F3’5’H and FLS genes. In addition, we identified key metabolites and genes by analyzing metabolite-gene interactions in the flavonoid biosynthetic pathway. CHS was reported to be a key enzyme in the flavonoid biosynthetic pathway, involved in the upstream step of the pathway (Winkel-Shirley, 2001), and is the first rate-limiting factor in the flavonoid biosynthetic pathway (Austin and Noel, 2003). CHS also drives the production of phytoalexins in plants in response to different forms of stress and is involved in a series of defense responses (Dao et al., 2011; Ye et al., 2017). Notably, in plants such as Citrus unshiu (Wang et al., 2010), Grewia asiatica (Wani et al., 2017), Syringa oblata (Wang et al., 2017), and Ginkgo biloba (Xu et al., 2007), CHS expression levels were reported to be highly correlated with the accumulation of flavonoids. Consistent with the results of previous studies, the pathway diagram clearly showed that the expression of CHS promoted the accumulation of flavonoids. The expression levels of two CHS genes (LOC110115249 and LOC119195073) were upregulated in the third and fourth years of D. moniliforme growth, and the corresponding flavonoid naringin chalcone also accumulated to a certain extent. Naringin chalcone has received widespread attention owing to its numerous pharmacological activities. Reportedly, it has antibacterial, anti-cancer, anti-tumor (Zhang et al., 2016b), anti-inflammatory (Ur Rashid et al., 2019), anxiolytic and anticonvulsant (Ferreira et al., 2021) properties, with high applicability and market value. Therefore, the accumulation of flavonoids in D. moniliforme may be closely related to the expression of CHS.
The literature has demonstrated that curtobacterium represents a group of Gram-positive phytopathogens, primarily associated with infections in legume and sugar beet (Chen et al., 2007; Osdaghi et al., 2015). The pathogenicity of curtobacterium in different host plants is different. Curtobacterium can cause bacterial wilt in legumes, but it has not been pathogenic to solanaceae (Osdaghi et al., 2018). Currently, there is no scientific evidence indicating the pathogenicity of curtobacterium towards Dendrobium. However, it is crucial to exercise caution and implement effective control measures in order to safeguard the quality of the harvest of Dendrobium. Through the previous analysis, we found that with the increase of growth years, the proportion of some pathogenic fungi gradually increased. Phaeosphaeria is a pathogen of leaf spot disease (Do Amaral et al., 2005). At present, leaf spot is mainly reported in D. officinale (Cao et al., 2022). It is noteworthy that phaeosphaeria accounted for a larger proportion in the D. moniliforme and D. huoshanense in this study. Therefore, cultivators of D. moniliforme and D. huoshanense need to pay extra attention to leaf spot, especially if they have been planted continuously for many years. Colletotrichum is the cause of anthrax, a common pathogen found in vegetables and fruits (Liu et al., 2016). Sarsaiya et al. isolated five endophytic bacteria from D. nobile and found that colletotrichum was highly pathogenic and even caused host death by studying the pathogenicity of these five bacteria (Sarsaiya et al., 2020). Our results show that colletotrichum accounts for a significant proportion of the composition of the fungal community of triennial D. officinale. Therefore, the growth period of D. officinale should not be too long, otherwise it will accumulate a large number of harmful endophytic bacteria, which will lead to its quality decline.
Lastly, based on the results of the joint metabolome and microbiome analysis, the highest correlation between microorganism abundance and flavonoid levels was observed in bacteria, specifically in actinobacteria and proteobacteria. The abundances of ascomycota and basidiomycota showed the highest correlation with flavonoid levels. Xie et al. investigated the metabolites, transcriptome, and microbiome of 1-35-year-old poplar roots and found that the abundance of actinobacteria was strongly correlated with flavonoid biosynthesis with the increase in tree age (Xie et al., 2023). The study conducted by Natsagdorj et al. utilizing HPLC analysis revealed that endophytic actinobacteria, which were isolated from desert plants, demonstrated the ability to synthesize flavonoids and phenolic compounds (Natsagdorj et al., 2021). The findings of these studies suggest a strong correlation between actinobacteria and the biosynthesis of flavonoids in the stems of Dendrobium. In our previous analysis, we observed a significant correlation between williamsia, lapillicoccus, actinomycetospora and actinomycetospora with flavonoid synthesis. However, there is currently no existing literature on the correlation between these bacteria and flavonoid biosynthesis. We hypothesize that these microorganisms have potential flavonoid production capabilities, but further experimental verification is needed.
5 Conclusion
We used multi-omics to analyze the different growth years of three types of medicinal dendrobium and found that the flavonoid content of Dendrobium huoshanensis and Dendrobium moniliforme increased with the increasing years. Furthermore, we analyzed the endophytes that affect medicinal ingredients and found that as the cultivation years increased, the pathogenic microbial in the stems of the three types of Dendrobium also gradually increased. Although the four-year-old Dendrobium also had higher flavonoids, the pathogenic microbial also increased. Therefore, after comprehensive consideration, it is recommended to harvest flavonoids in the third year. And based on the results of previous analysis, it was also found that there is a close connection between actinobacteria and the synthesis of flavonoid compounds in Dendrobium stems, which provides some valuable references for our subsequent research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YY: Conceptualization, Investigation, Writing – original draft. JZ: Software, Writing – original draft. XW: Data curation, Supervision, Validation, Conceptualization, Writing – original draft. RZ: Data curation, Methodology, Writing – original draft. WX: Supervision, Writing – review & editing. SL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project funded by the Forestry science and Technology Innovation and promotion Project of Jiangsu Province ‘Long-term Research Base of Forest and Wetland Positioning Monitoring in Jiangsu Province’ (Grant No. LYKJ[2020]21), Natural Science Foundation of Jiangsu Province, China (Grant No. BK20210800), the National Natural Science Foundation of China (Grant No. 32001341 and 32202523). Construction model of Efficient Farmland Protection Forest Network in Jiangsu Province (No. LYKJ[2021]38); and efficiency management technology of carbon sequestration forest in Jiangsu coast (No. LYKJ[2021]25).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1333989/full#supplementary-material
References
Austin, M. B., Noel, J. P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod Rep. 20, 79–110. doi: 10.1039/b100917f
Borges, C. V., Minatel, I. O., Gomez-Gomez, H. A., Lima, G. P. P. (2017). “Medicinal plants: Influence of environmental factors on the content of secondary metabolites,” in Medicinal plants and environmental challenges. (Springer, Cham), 259–277.
Cao, P., Fang, Y., Zheng, Z., Han, X., Zou, H., Yan, X. (2022). Occurrence of Neopestalotiopsis clavispora causing leaf spot on Dendrobium officinale in China. Plant Dis. 106, 1761. doi: 10.1094/PDIS-11-21-2432-PDN
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Knight, R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chamkhi, I., Benali, T., Aanniz, T., El Menyiy, N., Guaouguaou, F.-E., El Omari, N., et al. (2021). Plant-microbial interaction: The mechanism and the application of microbial elicitor induced secondary metabolites biosynthesis in medicinal plants. Plant Physiol. Biochem. 167, 269–295. doi: 10.1016/j.plaphy.2021.08.001
Chen, S., Dai, J., Song, X., Jiang, X., Zhao, Q., Sun, C., et al. (2020). Endophytic microbiota comparison of Dendrobium huoshanense root and stem in different growth years. Planta Med. 86, 967–975. doi: 10.1055/a-1046-1022
Chen, X.-M. (2013). Research progress on chemical composition and chemical analysis of Dendrobium officinale. Chin. Pharm. J., 1634–1640.
Chen, Y.-F., Yin, Y.-N., Zhang, X.-M., Guo, J.-H. (2007). Curtobacterium flaccumfaciens pv. beticola, a new pathovar of pathogens in sugar beet. Plant Dis. 91, 677–684. doi: 10.1094/PDIS-91-6-0677
Dao, T. T., Linthorst, H. J., Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi: 10.1007/s11101-011-9211-7
Deng, Y., Li, M., Chen, L.-X., Chen, X.-Q., Lu, J.-H., Zhao, J., et al. (2018). Chemical characterization and immunomodulatory activity of acetylated polysaccharides from Dendrobium devonianum. Carbohydr. Polymers 180, 238–245. doi: 10.1016/j.carbpol.2017.10.026
Do Amaral, A., Dal Soglio, F., De Carli, M., Neto, J. B. (2005). Pathogenic fungi causing symptoms similar to Phaeosphaeria leaf spot of maize in Brazil. Plant Dis. 89, 44–49. doi: 10.1094/PD-89-0044
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Etalo, D. W., Jeon, J.-S., Raaijmakers, J. M. (2018). Modulation of plant chemistry by beneficial root microbiota. Natural product Rep. 35, 398–409. doi: 10.1039/C7NP00057J
Ferreira, M. K. A., Da Silva, A. W., Dos Santos Moura, A. L., Sales, K. V. B., Marinho, E. M., Do Nascimento Martins Cardoso, J., et al. (2021). Chalcones reverse the anxiety and convulsive behavior of adult zebrafish. Epilepsy Behav. 117, 107881. doi: 10.1016/j.yebeh.2021.107881
Gendrisch, F., Esser, P. R., Schempp, C. M., Wölfle, U. (2021). Luteolin as a modulator of skin aging and inflammation. Biofactors 47, 170–180. doi: 10.1002/biof.1699
Hsu, Y.-L., Uen, Y.-H., Chen, Y., Liang, H.-L., Kuo, P.-L. (2009). Tricetin, a dietary flavonoid, inhibits proliferation of human breast adenocarcinoma mcf-7 cells by blocking cell cycle progression and inducing apoptosis. J. Agric. Food Chem. 57, 8688–8695. doi: 10.1021/jf901053x
Imran, M., Rauf, A., Abu-Izneid, T., Nadeem, M., Shariati, M. A., Khan, I. A., et al. (2019). Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharmacother. 112, 108612. doi: 10.1016/j.biopha.2019.108612
Jacoby, R. P., Koprivova, A., Kopriva, S. (2021). Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J. Exp. Bot. 72, 57–69. doi: 10.1093/jxb/eraa424
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi (Wiley Online Library, Hoboken).
Koprivova, A., Kopriva, S. (2022). Plant secondary metabolites altering root microbiome composition and function. Curr. Opin. Plant Biol. 67, 102227. doi: 10.1016/j.pbi.2022.102227
Lee, J. K. (2011). Anti-inflammatory effects of eriodictyol in lipopolysaccharide-stimulated raw 264.7 murine macrophages. Arch. Pharm. Res. 34, 671–679. doi: 10.1007/s12272-011-0418-3
Liu, F., Tang, G., Zheng, X., Li, Y., Sun, X., Qi, X., et al. (2016). Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Sci. Rep. 6, 32761. doi: 10.1038/srep32761
Magoč, T., Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
McMurdie, P. J., Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8, e61217. doi: 10.1371/journal.pone.0061217
Natsagdorj, O., Bekh-Ochir, D., Baljinova, T., Iizaka, Y., Fukumoto, A., Kato, F., et al. (2021). “Bioactive compounds and molecular diversity of endophytic actinobacteria isolated from desert plants,” in IOP Conference Series: Earth and Environmental Science (IOP Publishing, Bristol), 012008.
Niemi, R. M., Heiskanen, I., Wallenius, K., Lindström, K. (2001). Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J. microbiol. Methods 45, 155–165. doi: 10.1016/S0167-7012(01)00253-6
Osdaghi, E., Taghavi, S. M., Fazliarab, A., Elahifard, E., Lamichhane, J. R. (2015). Characterization, geographic distribution and host range of Curtobacterium flaccumfaciens: an emerging bacterial pathogen in Iran. Crop Prot. 78, 185–192. doi: 10.1016/j.cropro.2015.09.015
Osdaghi, E., Taghavi, S., Hamzehzarghani, H., Fazliarab, A., Harveson, R., Tegli, S., et al. (2018). Epiphytic Curtobacterium flaccumfaciens strains isolated from symptomless solanaceous vegetables are pathogenic on leguminous but not on solanaceous plants. Plant Pathol. 67, 388–398. doi: 10.1111/ppa.12730
Pharmacopoeia, C. (2010). Editorial Committee of Chinese Pharmacopoeia (China Medical Science and Technology Press Beijing, Beijing).
Puchooa, D. (2004). Comparison of different culture media for the in vitro culture of Dendrobium (Orchidaceae). Int. J. Agric. Biol. 6, 884–888.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rajendran, P., Rengarajan, T., Nandakumar, N., Palaniswami, R., Nishigaki, Y., Nishigaki, I. (2014). Kaempferol, a potential cytostatic and cure for inflammatory disorders. Eur. J. Med. Chem. 86, 103–112. doi: 10.1016/j.ejmech.2014.08.011
Rognes, T., Flouri, T., Nichols, B., Quince, C., Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Rossato, M. F., Trevisan, G., Walker, C. I., Klafke, J. Z., De Oliveira, A. P., Villarinho, J. G., et al. (2011). Eriodictyol: a flavonoid antagonist of the TRPV1 receptor with antioxidant activity. Biochem. Pharmacol. 81, 544–551. doi: 10.1016/j.bcp.2010.11.004
Sarsaiya, S., Jain, A., Jia, Q., Fan, X., Shu, F., Chen, Z., et al. (2020). Molecular identification of endophytic fungi and their pathogenicity evaluation against Dendrobium nobile and Dendrobium officinale. Int. J. Mol. Sci. 21, 316. doi: 10.3390/ijms21010316
Seelinger, G., Merfort, I., Schempp, C. M. (2008). Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 74, 1667–1677. doi: 10.1055/s-0028-1088314
Sirén, J., Välimäki, N., Mäkinen, V. (2014). Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinf. 11, 375–388. doi: 10.1109/TCBB.2013.2297101
Stierle, A., Strobel, G., Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260, 214–216. doi: 10.1126/science.8097061
Sun, F.-F., Hu, P.-F., Xiong, Y., Bao, J.-P., Qian, J., Wu, L.-D. (2019). Tricetin protects rat chondrocytes against IL-1β-induced inflammation and apoptosis. Oxid. Med. Cell. Longevity 2019, 4695381. doi: 10.1155/2019/4695381
Teoh, E. S. (2016). “Secondary metabolites of plants,” in Medicinal orchids of Asia (Springer, Cham), 59–73.
Trikka, F. A., Nikolaidis, A., Ignea, C., Tsaballa, A., Tziveleka, L. A., Ioannou, E., et al. (2015). Combined metabolome and transcriptome profiling provides new insights into diterpene biosynthesis in S. pomifera glandular trichomes. BMC Genomics 16, 935. doi: 10.1186/s12864-015-2147-3
Ur Rashid, H., Xu, Y., Ahmad, N., Muhammad, Y., Wang, L. (2019). Promising anti-inflammatory effects of chalcones via inhibition of cyclooxygenase, prostaglandin E2, inducible NO synthase and nuclear factor kappab activities. Bioorg. Chem. 87, 335–365. doi: 10.1016/j.bioorg.2019.03.033
Wang, Z., Jiang, W., Liu, Y., Meng, X., Su, X., Cao, M., et al. (2021). Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between Protocorm-like bodies and leaves. BMC Genomics 22, 1–17. doi: 10.1186/s12864-021-07887-6
Wang, Y., Li, J., Xia, R. (2010). Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow). Sci. Hortic. 125, 110–116. doi: 10.1016/j.scienta.2010.02.001
Wang, S., Wang, B., Hua, W., Niu, J., Dang, K., Qiang, Y., et al. (2017). De novo assembly and analysis of polygonatum sibiricum transcriptome and identification of genes involved in polysaccharide biosynthesis. Int. J. Mol. Sci. 18, 1950. doi: 10.3390/ijms18091950
Wani, T. A., Pandith, S. A., Gupta, A. P., Chandra, S., Sharma, N., Lattoo, S. K. (2017). Molecular and functional characterization of two isoforms of chalcone synthase and their expression analysis in relation to flavonoid constituents in Grewia asiatica L. PloS One 12, e0179155. doi: 10.1371/journal.pone.0179155
Wickham, H. (2011). ggplot2. Wiley interdisciplinary reviews: computational statistics. 3, 180–185. doi: 10.1002/wics.147
Winkel-Shirley, B. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127, 1399–1404. doi: 10.1104/pp.010675
Wu, Z.-G., Jiang, W., Chen, S.-L., Mantri, N., Tao, Z.-M., Jiang, C.-X. (2016). Insights from the cold transcriptome and metabolome of Dendrobium officinale: global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 7, 1653. doi: 10.3389/fpls.2016.01653
Xie, J., Ma, Y., Li, X., Wu, J., Martin, F., Zhang, D. (2023). Multifeature analysis of age-related microbiome structures reveals defense mechanisms of Populus tomentosa trees. New Phytol. 238, 1636–1650. doi: 10.1111/nph.18847
Xu, F., Cheng, S. Y., Cheng, S. H., Wang, Y., Du, H. W. (2007). Time course of expression of chalcone synthase gene in Ginkgo biloba. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33, 309–317.
Ye, Z., Dai, J.-R., Zhang, C.-G., Lu, Y., Wu, L.-L., Gong, A. G. W., et al. (2017). Chemical Differentiation of Dendrobium officinale and Dendrobium devonianum by Using HPLC Fingerprints, HPLC-ESI-MS, and HPTLC Analyses. Evidence-Based Complement. Altern. Med. 2017, 8647212. doi: 10.1155/2017/8647212
Yuan, Y., Zhang, J., Kallman, J., Liu, X., Meng, M., Lin, J. (2019). Polysaccharide biosynthetic pathway profiling and putative gene mining of Dendrobium moniliforme using RNA-Seq in different tissues. BMC Plant Biol. 19, 521. doi: 10.1186/s12870-019-2138-7
Zhan, X., Qi, J., Zhou, B., Mao, B. (2020). Metabolomic and transcriptomic analyses reveal the regulation of pigmentation in the purple variety of Dendrobium officinale. Sci. Rep. 10, 17700. doi: 10.1038/s41598-020-74789-0
Zhang, C., Chen, J., Huang, W., Song, X., Niu, J. (2021a). Transcriptomics and metabolomics reveal purine and phenylpropanoid metabolism response to drought stress in dendrobium sinense, an endemic orchid species in Hainan island. Front. Genet. 12, 692702. doi: 10.3389/fgene.2021.692702
Zhang, S., Jiang, Z.-F., Pan, Q., Song, C.-Y., Zhang, W.-H. (2016b). Anti-cancer effect of naringenin chalcone is mediated via the induction of autophagy, apoptosis and activation of PI3K/Akt signalling pathway. Bangladesh J. Pharmacol. 11, 684–690. doi: 10.3329/bjp.v11i3.27518
Zhang, G.-Q., Xu, Q., Bian, C., Tsai, W.-C., Yeh, C.-M., Liu, K.-W., et al. (2016a). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 1–10. doi: 10.1038/srep19029
Zhang, M., Yu, Z., Zeng, D., Si, C., Zhao, C., Wang, H., et al. (2021b). Transcriptome and metabolome reveal salt-stress responses of leaf tissues from dendrobium officinale. Biomolecules 11, 736. doi: 10.3390/biom11050736
Keywords: Dendrobium, transcriptome, metabolome, microbiome, flavonoid
Citation: Yuan Y, Zuo J, Wan X, Zhou R, Xing W and Liu S (2024) Multi-omics profiling reveal responses of three major Dendrobium species from different growth years to medicinal components. Front. Plant Sci. 15:1333989. doi: 10.3389/fpls.2024.1333989
Received: 08 November 2023; Accepted: 09 February 2024;
Published: 23 February 2024.
Edited by:
Sheng-Hong Li, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Chen Li, Hubei University of Medicine, ChinaJihua Wang, Guangdong Academy of Agricultural Sciences, China
Copyright © 2024 Yuan, Zuo, Wan, Zhou, Xing and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Xing, bGt5eGluZ3dlaUAxNjMuY29t; Sian Liu, c2lhbmxpdUB5enUuZWR1LmNu
†These authors have contributed equally to this work
 Yingdan Yuan
Yingdan Yuan Jiajia Zuo1†
Jiajia Zuo1† Sian Liu
Sian Liu