- 1College of Agriculture, Anhui Science and Technology University, Fengyang, Anhui, China
- 2International Joint Research Center of Forage Bio-Breeding in Anhui Province, Chuzhou, China
- 3Department of Biology, University of Louisiana at Lafayette, Lafayette, LA, United States
Introduction: A whole plant can be regenerated through tissue culture from an embryogenic callus in a process referred to as plant regeneration. Regeneration ability of embryogenic callus is a quantitative trait and the main limiting factor for genetic studies in sorghum.
Methods: We evaluated 236 sorghum mini core varieties for callus induction rate, embryogenic callus rate, callus browning rate and differentiation rate and performed a multi-locus genome-wide association study (GWAS) of the four traits with 6,094,317 SNPs.
Results: We found five mini core varieties most amenable to tissue culture manipulations: IS5667, IS24503, IS8348, IS4698, and IS5295.Furthermore, we mapped 34 quantitative trait loci (QTLs) to the four traits and identified 47 candidate genes. Previous studies provided evidence for the orthologs of 14 of these genes for their role in cellular function and embryogenesis and that the ortholog of WIND1 (WOUND INDUCED DEDIFFERENTIATION 1) identified in this study promotes callus formation and increases de novo shoot regeneration.
Conclusion: These candidate genes will help to further understand the genetic basis of plant embryonic callus regeneration.
1 Introduction
Sorghum [Sorghum bicolor (L.) Moench] is a highly valuable and adaptable crop, playing key roles in food, feed, biofuels, and even traditional alcoholic beverages like baijiu in China (Zheng et al., 2023). The challenges in sorghum’s tissue culture and genetic transformation, particularly the browning of calli, highlight the complexity of working with this plant for biotechnological advancements (Zheng et al., 2023; Silva et al., 2022). Factors like genotype-dependent phenolic production contribute to the difficulty, making it a recalcitrant species for genetic modification (Flinn et al., 2020). This presents an interesting area of study for overcoming tissue culture barriers to improve sorghum’s seed quality, which is crucial for its various uses, especially in baijiu production. Sorghum recalcitrance to tissue culture has been demonstrated by several studies. An evaluation of five sorghum varieties has showed a plantlet regeneration frequency of 0-2.8% from immature embryo (Ahmed et al., 2021). Another study evaluated 10 sorghum varieties and attained embryogenic callus rate of 13.4-47.5% across media also from immature embryo (Assem et al., 2014). Culturing immature embryos from Keller, Kansas, Atlas and Fortuna found callus induction rate of 1-84% in their best media (Espinoza-Sánchez et al., 2018). An embryogenic callus rate of 10-89% was reported for immature embryos from 32 sorghum varieties, depending on varieties and media (Raghuwanshi and Birch, 2010).
Although immature embryos were frequently used, their isolation is labor-intensive and transformation efficiency using immature embryos harvested in winter can be reduced by 1/3 (Do et al., 2016). In contrast, mature seeds represent an attractive alternative because they are independent of seasonal effects and convenient to access (Wang et al., 2021a). Using mature seeds from sorghum varieties GK Emese, GK Zsófia, and Róna 1, a plantlet regeneration rate of 6.7-24.4% was recorded (Chege et al., 2019). Although callus induction rate from mature seeds can be very high (10-87%), none of the calli regenerated (Murty et al., 1990). In a screening of 250 sorghum varieties, although mature seeds from 245 varieties produced calli, only calli from 47 varieties regenerated shoot and root (Li et al., 2017). We have previously cultured mature seeds from 120 mini core landrace varieties and found that 25 (21%) of the varieties could be induced to produce callus and 23 of the 25 showed low callus browning rate (Li et al., 2017).
Previous studies have mapped callus induction (Tuskan et al., 2018)and callus and shoot regeneration (Nagle et al., 2024) in poplar, callus and shoot regeneration in barley (Tyagi et al., 2010), callus formation in rice (Zhang et al., 2019), and callus induction rate and regeneration in maize (Ma et al., 2018) and identified possible genes underlying embryogenesis. These are important as they may facilitate breeding for genotypes more amenable to in vitro plant regeneration and genetic transformation, in addition to elucidating genetic control of induced embryogenesis. We evaluated 236 sorghum landrace varieties of the globally sampled mini core collection (Upadhyaya et al., 2009) for callus induction, callus browning, embryogenic callus induction and callus differentiation from mature seeds. The sorghum mini core collection has been previously used to clone a gene pleiotropic for sorghum biomass and sugar yield (Upadhyaya et al., 2022) and to map sorghum panicle architecture (Wang et al., 2021b) and sorghum plant color (Wang et al., 2024). To our knowledge, there have been no reports on mapping tissue-culture-based genetic transformation related traits in sorghum.
In this study, we have two objectives. First, we want to find highly competent sorghum varieties for genetic transformation from a global germplasm collection. Second, we want to understand the genetic mechanism behind embryogenesis even though the process can be affected by hormones. We performed a GWAS on the four traits with 6,094,317 SNP markers using the GWAS method as previously described (Li et al., 2018) and mapped eight callus induction loci, one locus each for callus browning and embryogenic callus induction, and 24 differentiation loci, identified 47 candidate genes from the loci and provided evidence that orthologs for 14 of the genes play roles in cellular function and embryogenesis. To our knowledge, this is the first such report on mapping tissue-culture-based genetic transformation related traits in sorghum.
2 Materials and methods
2.1 Plant material
A total of 236 sorghum mini core varieties (Supplementary Table S1) were evaluated for the traits described below Each variety was planted in a row with 10 plants in each row. The row spacing was 50 cm and the plant spacing was 20 cm. In order to avoid cross-pollination, all panicles were wrapped in paper bags before flowering. After seeds grew mature, they were harvested for tissue culture.
2.2 Tissue culture method
We selected 90 mature seeds from each of the 236 varieties for three replicates. The seeds were washed with 70% alcohol for 10 minutes, rinsed with sterile water once, then washed with 0.1% HgCl2 solution and 2-3 drops of Tween 20® for 15 minutes, rinsed with sterile water for 5-6 times in an ultra-clean bench until no foam was observed, and placed in a mature seed induction medium (Supplementary Table S2) for 10-14 days. The sample was incubated at 28°C with 70% humidity(PGX-250, Shanghai, China) without light, and then the varieties that could produce callus were selected. After subculture, callus was transferred to regeneration medium with 16h light and 8h dark at 28°C.The embryo surface of sorghum seeds was oriented downward, with 30 seeds placed per Petri dish. If callus tissue developed, the seed and bud parts were excised and transferred to a subculture medium. After 7-10 days, the medium was replaced with a differentiation medium (Supplementary Table S2). The seeds and embryonic callus were then cut and subcultured. Within seven days of subculture, the ability to generate embryonic tissue and the browning rate were recorded (Li et al., 2018).
2.3 Phenotypic measurement
We evaluated the callus induction rate of mature seeds from 236 sorghum varieties. Varieties that produced translucent callus were considered capable of generating inducible embryonic calli. During the tissue culture of sorghum mature seeds, we found that some varieties lacked callus induction ability, resulting in water-soaked callus that could not form embryonic callus. These varieties also exhibited low callus differentiation rates and browning. The callus most suitable for sorghum genetic transformation was a granular, compact, yellowish, embryogenic callus. Therefore, washed seeds were placed on the induction medium (Supplementary Table S2), and those that produced translucent callus were considered inducible material. After 7 days of subculture, a relatively hard, pale yellow, granular callus formed, which was identified as embryogenic callus. During this period, although embryogenic callus could grow, brown phenolic substances accumulated around the callus, leading to browning (Flinn et al., 2020). Callus that could differentiate on the differentiation medium (Supplementary Table S2) was considered differentiated materially. The induction rate was calculated as the number of induced calli divided by 90. The browning rate was the proportion of browning calli relative to the total number of induced calli. The embryogenic callus rate was the proportion of induced calli that developed into pale yellow granular callus, relative to the total number of calli. The differentiation rate was the proportion of regenerated plants relative to the total number of calli.
2.4 Data processing
The steps of DNA extraction are as follows: Firstly, the fresh leaves were grinded with liquid nitrogen, and the DNA lysis solution(Supplementary Table S3)was added. The cytoplasm was broken and dissolved in 65 °C water bath for 10 minutes, so that the DNA was released into the solution. Centrifugation at 12000 rpm for 4 minutes to absorb the supernatant, and then the supernatant was added to the magnetic beads, DNA binding solution(Supplementary Table S3) and isopropanol. After the oscillation was uniform, it was placed on the magnetic frame for 2 minutes to absorb the supernatant. By adding 70% alcohol, the DNA is separated and precipitated from the mixture, and washed twice. After drying for 10 minutes, double distilled water was added, fully vortexed, and bathed in water at 65 °C for 3 minutes. After standing on a magnetic frame for 2 minutes, the supernatant was rich in DNA.
DNA size of 350 bp were generated using the Covaris E210 Ultrasonicator (Woburn, MA). A sequencing library was constructed with DNBSEQ-T7RS. The library was sequenced with BGI’ PCR DNBSEQ™ (Shenzhen, China). Raw data was filtered by fastp (Chen et al., 2018). Clean data were mapped to the reference genome using BWA-MEM version 0.7.17 (Li, 2013). The BWA-MEM algorithm was robust to sequencing errors and was suitable for a wide range of sequence lengths from 70 bp to several megabases. Sort by SAMtools version 1.10 (Li et al., 2009). Using Picard version 2.0.1 (http://broadinstitute.github.io/picard/)), you removed duplicate reads. Sequence variation detection and SNP detection were performed using GATK 4.17 version functions HaplotypeCaller and SelectVariants (McKenna et al., 2010). SNP was invoked using parameters”QD < 2.0, MQ < 40.0, FS > 60, SOR > 3.0, MQRankSum < − 12.5, ReadPosRankSum < − 8.0.” VCFtools version 1.16 (Li, 2013) was used to filter SNPs using parameters”max-missing 0.1, maf 0.05, maxDP 50 and minDP 10.”The identification of SNPs was based on the criteria of secondary allele frequency ≥ 0.05 and missing data rate ≥ 10% in the population. A total of 6,094,317 SNPs were identified on chromosome 1-10. The genetic relationship matrix (K) was generated using EMMAX (Kang et al., 2010) and used for GWAS analysis of Q matrices calculated using STRUCTURE 2.3.4 (Pritchard et al., 2000) as covariates.
The induction rate, embryogenic callus rate, browning rate and differentiation rate of mature seeds from all the 236 sorghum varieties under the same culture conditions were calculated. The GWAS mixed linear model (MLM) was used with the population structure as covariate, and the genetic relationship matrix was added as a random effect (Upadhyaya et al, 2022). For MLM (Q + K), the Bonferroni threshold of the significantly associated marker is calculated as -log10 (p) >= 8.1 (Upadhyaya et al, 2022).Manhattan maps were drawn by R package CMplot (Yin et al, 2021). Genes closest to or containing the linked SNPs are considered candidate genes based on the reference genome Sorghum bicolor v3.1.1 curated in Phytozome 13 (https://phytozome-next.jgi.doe.gov/). When reported candidate genes, we used Phytozome 13 to identify candidate genes closest from linked SNP markers. This ranges from 0-75 kb.Orthologs were identified as the best match by protein sequence BLAST in Phytozome 13.The default parameter settings were used for CMplot and BLAST.
3 Results
3.1 Phenotypic analysis
In the process of tissue culture of sorghum mature seeds, we found that some varieties had no callus induction ability, and/or the generated callus appeared to be water-soaked and could not become embryogenic calli, and/or low callus differentiation rate and browned. Regeneration-competent calli were granular compact yellowish as previously described (Belide et al., 2017).
We evaluated the callus induction rate, embryonic callus rate, browning rate and differentiation rate of mature seeds from the 236 sorghum varieties. Among these, 116 varieties were able to grow calli with an induction rate ranging from 1.1% to 68.97%. Out of these 116 varieties, 74 exhibited browning, producing brown phenolic substances with a browning rate ranging from 9% to 100%. There were 51 varieties whose callus tissues were embryonic, with an embryonic callus rate ranging from 9% to 100% (Table 1).Through statistical analysis of phenotypic data, we found that the induction rate, embryonic callus rate and differentiation rate basically conform to the normal distribution, while the browning rate does not conform to the normal distribution (Supplementary Figure S1).
After evaluating these varieties and based on high induction and differentiation rate but low browning rate, we identified five varieties which were efficient in plantlet regeneration using tissue culture (Table 2). This regeneration efficiency was not related to either seed coat color, agronomic types (races), or country of origin (Table 2). This agrees with a previous study using eight sorghum varieties of various seed coat color, countries of origin and agronomic races (Flinn et al., 2020). The examples of high/low callus induction rate, embryonic callus rate, browning rate and differentiation rate are presented in Figure 1.
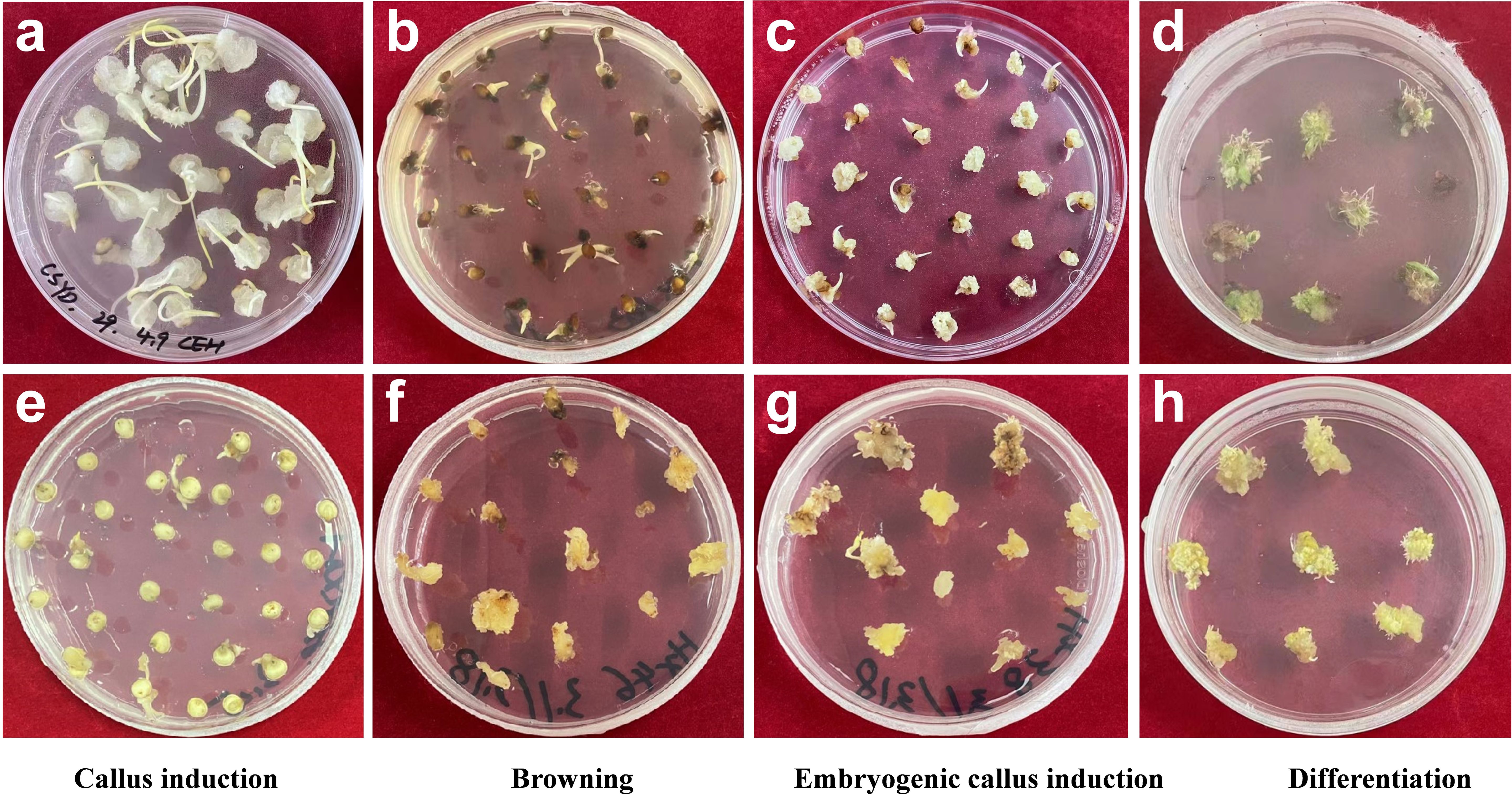
Figure 1. Examples of high/low callus induction rate (a, e), browning rate (b, f), embryonic callus rate (c, g), and differentiation rate (d, h).The corresponding varieties of a, b, c, d, e, f, g, h are IS4581, IS29233, IS7250, IS4698, IS608, IS7310, IS5667, IS2205, respectively.
3.2 Association analysis
Manhattan plots for callus induction rate, embryonic callus rate, browning rate and differentiation rate are presented in Figure 2. To identify quantitative trait loci (QTLs) underlying the traits, we sought for loci with -log(p) value above the Bonferroni but found no loci mapped in browning and embryonic callus rate. We decided to use a -log(p) value of 6.0 (Nguyen et al., 2020). With this value and that the linkage had to be with multiple markers as in previous studies (Upadhyaya et al., 2022; Wang et al., 2021b; Wang et al., 2024), we identified eight loci for callus induction rate, one locus each for browning and embryogenic callus rate, and 24 for differentiation rate (Figure 2). Detailed locus distribution and position on each chromosome are listed in Table 3.
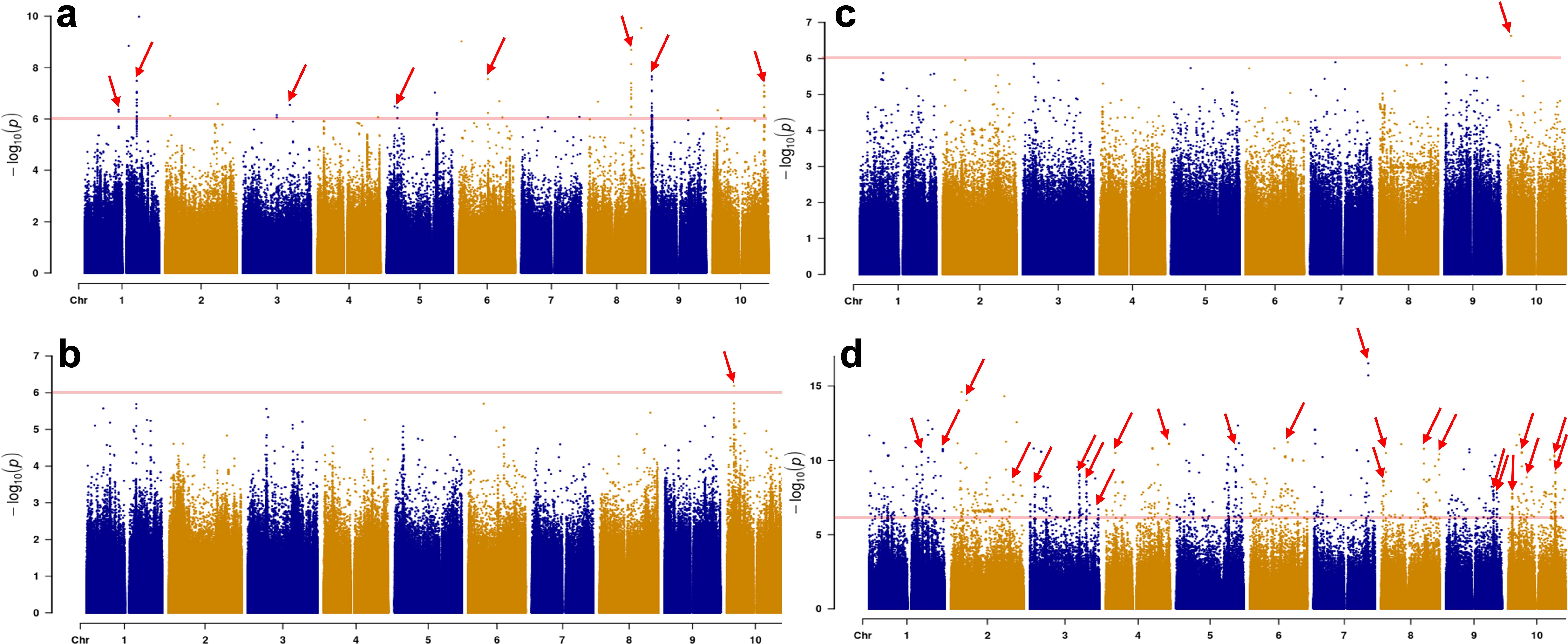
Figure 2. Manhattan plot for induction rate (a), browning rate (b), embryogenic callus rate (c) and differentiation rate (d). Arrows indicate loci mapped to the four traits on each sorghum chromosome. Horizontal lines indicate the threshold -log(p) value.
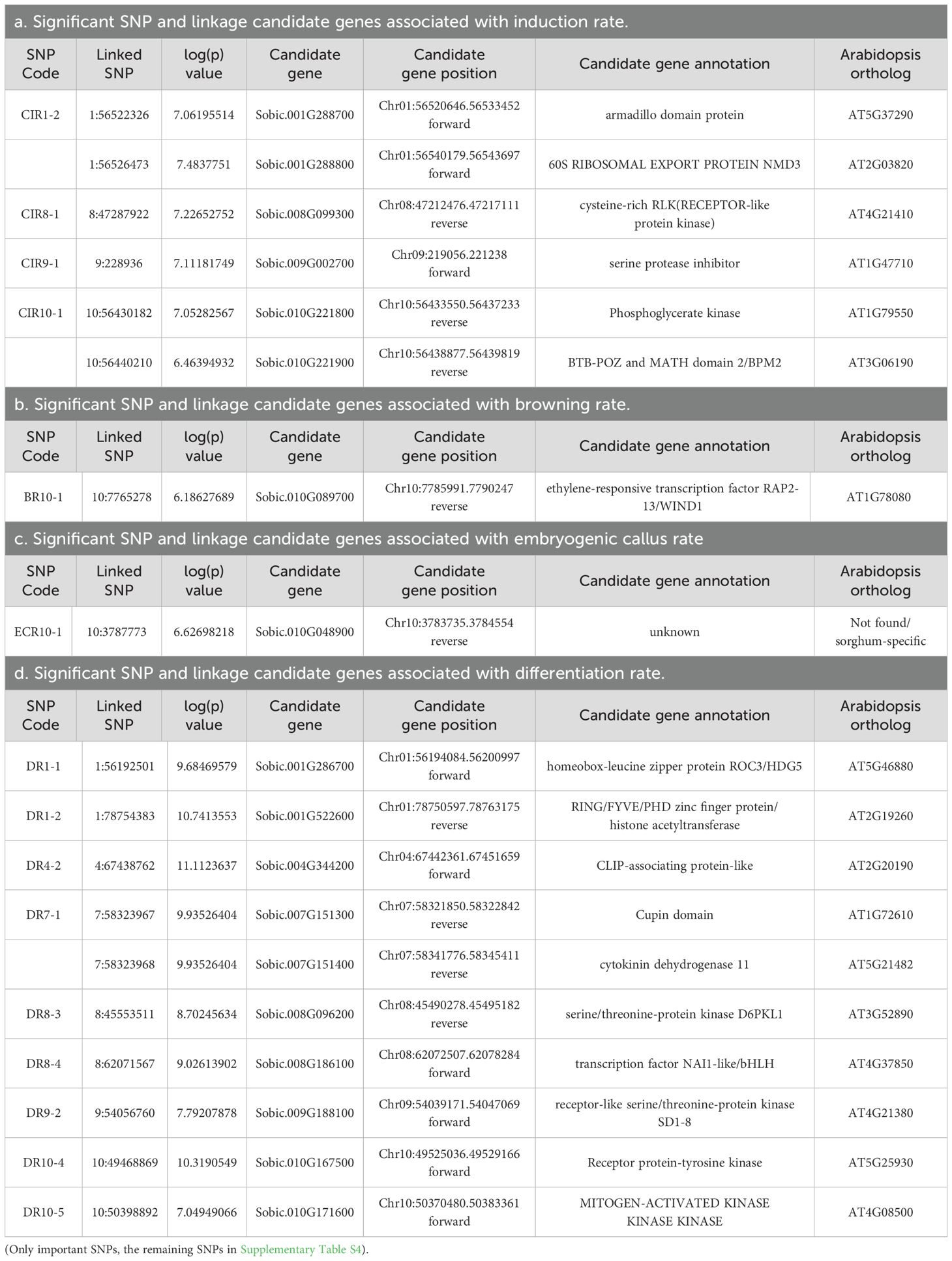
Table 3. Significant SNPs linked to induction rate (a), browning rate (b), embryogenic callus rate (c), and differentiation rate (d) and candidate genes.
In total, eight callus induction loci were mapped onto seven chromosomes with one each on chromosomes 3, 5, 6, 8, 9, 10 and two on chromosome 1. One locus was mapped each for callus browning and embryogenic callus induction on chromosome 10. Twenty-four differentiation loci were mapped for differentiation rate, with one each to chromosomes 5, 6, and 7, two each to chromosomes 1, 2, 4, and 9, four each to chromosomes 3 and 8, and five to chromosome 10 (Figure 2; Table 3). The DR1-2 locus contained the tightest linked markers with an average -log(p) of 10.69 from six markers (Table 3).
3.3 Candidate gene identification
To identify candidate gene for each locus, either gene(s) encompassing linked SNPs, closest or flanking genes were listed in Table 3. In total, 47 candidate genes were found from the 34 loci. To break it down, one candidate gene was found for 22 of the 34 mapped loci while two flanking genes were found for nine and 0, 3, and 4 genes were each found in one locus (Table 3). Six of the 22 loci (CIR6-1, ECR10-1, DR2-1, DR4-1, DR6-1, and DR8-2) contained a gene with unknown function and specific to sorghum - no Arabidopsis ortholog was found. CIR1-1 and CIR5-1 housed ZINC FINGER CCHC DOMAIN and AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, respectively, and neither ortholog were found in Arabidopsis (Table 3). Figure 3 shows the DR1-2 locus and its candidate gene, Sobic.001G522600 coding for RING/FYVE/PHD zinc finger protein/histone acetyltransferase. Among all seven significant SNPs six were located in the 6th intron and one in the last intron (the 8th) (Figure 3). Based on studies of their orthologs in Arabidopsis, quite a few our candidate genes play roles in embryogenesis which are further detailed in the Discussion.
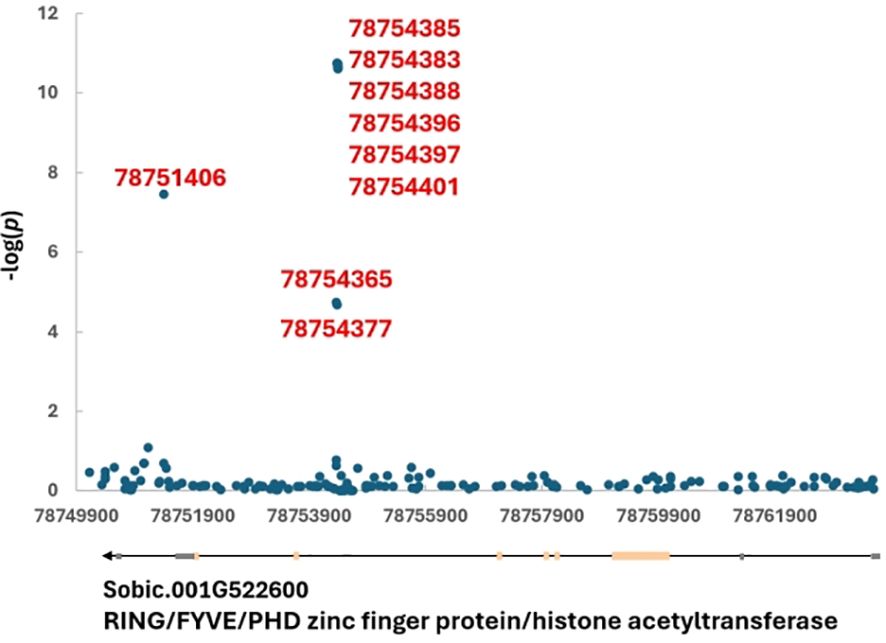
Figure 3. DR1-2 locus and its candidate gene, Sobic.001G522600 coding for RING/FYVE/PHD zinc finger protein/histone acetyltransferase on chromosome 1. The linked SNP positions are listed.
4 Discussion
In this study, we set out to find highly competent sorghum varieties for genetic transformation and to understand the genetic architecture of embryogenesis. We have found five highly competent sorghum varieties for genetic transformation. They are IS5667, IS24503, IS8348, IS4698, and IS5295 (Table 2). These varieties will be valuable to future sorghum transgenic research. We mapped 34 QTLs to the four traits and identified 47 candidate genes, suggesting a complex mechanism underlying embryogenesis. Fourteen of the candidate genes are known to play roles in tissue culture and plant embryonic regeneration in other species. These are discussed in the following sections and will aid our understanding of the genetic mechanism behind embryogenesis.
Previous studies research on callus regeneration ability has primarily focused on plants such as Arabidopsis, rice, wheat, and maize and have identified callus formation and embryogenesis related candidate genes (Tuskan et al., 2018; Nagle et al., 2024; Zhang et al., 2019; Ma et al., 2018) However, almost all candidate genes identified in each study are unique suggests that a large number of genes are involved in the processes. Since overexpressing WUSCHEL-related homeobox 2 (WOX2) transcription factor in maize increased transformation frequencies in previously nontransformable maize inbred lines (Lowe et al., 2016), we examined transcription factors reported by each study using Arabidopsis orthologs to compare across the studies. While AT1G03800 and AT3G14230 [both coding for ethylene response factor (ERF)/Apetala (AP) subfamily] are identified in rice (Zhang et al., 2019), in poplar (Nagle et al., 2024) they are AT3G12250 (TGA6), AT5G35550 (R2R3 MYB), At1g75250 (RADIALIS-LIKE SANT/MYB 3), AT4G16110 (a pollen-specific transcription factor), and AT4G27950 (another ERF/AP) and in maize (Ma et al., 2018) AT5G59340 (WOX2), AT1G62310 (JIMONJI 21), AT5G50670 (SBP 18), AT5G61620 (MYB 32), AT4G04890 (HDG2) and AT1G09530 (bHLH 43) are reported. These are 13 transcription factors reported in one eudicot and two monocots with two in rice, five in poplar and six in maize. Yet there is no transcription factor found in more than one study. In this study, we identified three transcription factors: AT1G78080 [WIND1 (ERF/AP), Sobic.010G089700], AT5G46880 (HDG5, Sobic.001G286700) and AT4G37850 (bHLH, Sobic.008G186100) (Table 3). Again, there is no overlap with genes discovered in poplar (Nagle et al., 2024), rice (Zhang et al., 2019) and maize (Ma et al., 2018). We downloaded the top 50 coexpressed genes of each sorghum transcription factor from Sorghum bicolor v3.1.1 at Phytozome 13 (McCormick et al., 2018) and found that while WIND1 (WOUND INDUCED DEDIFFERENTIATION 1) is coexpressed with seven other transcription factors (no homeobox genes) and two transporters (Supplementary Table S5), HDG5 is coexpressed with 14 other transcription factors (five of them homeobox genes) and also two transporters (Supplementary Table S6), but bHLH is coexpressed with five other transcription factors (no homeobox genes) and 11 transporters (Supplementary Table S7).This non-overlap in transcription factors during embryogenesis of different species could be due to two reasons. One is that the combination of species and tissue culture environments such as temperature, light, and medium composition could require different genes for embryogenesis. The other reason could be that embryogenesis starts a new plant and there are many transcription factors involved (Umehara et al., 2007; Yuan et al., 2024), as indicated by many coexpressed ones in our study, and only a few are mapped.
As in the case of overexpressing WOX2 increasing maize transformation frequencies (Lowe et al., 2016), transcription factors play an essential role in embryogenesis. Arabidopsis WIND1 (WOUND INDUCED DEDIFFERENTIATION 1) is an ERF/AP transcription factor and is rapidly induced at the wound site, promoting cell dedifferentiation and subsequent cell proliferation to form a mass of pluripotent cells termed callus (Iwase et al., 2011). Ectopic expression of WIND1 can bypass both wounding and auxin pre-treatment and increase <i>de novo</i> shoot regeneration from root explants cultured on shoot-regeneration promoting media (Iwase et al., 2015), suggesting that WIND1 can reprogram somatic cells to be pluripotent. WIND1 promotes callus formation and shoot regeneration by directly binding the promoter the ENHANCER OF SHOOT REGENERATION1 (ESR1-AT1G12980) gene, which encodes another AP2/ERF transcription factor in Arabidopsis thaliana (Iwase et al., 2017). Therefore, WIND1 from BR10-1 could promote regeneration and reduce callus browning rate in sorghum.
The four ERF/AP transcription factors (two from rice, WIND1 and ESR1) are potentially all involved in regeneration. It turns out that the BPM1 and BPM2 identified in CIR10-1 (AT5G19000 and AT3G06190; Table 3) assemble with CUL3-based E3 ligases using their BTB/POZ domains (Weber et al., 2005; Gingerich et al., 2005). BPM proteins can also assemble/interact with members of the ERF/AP transcription factor family (Weber and Hellmann, 2009) and the assembled CUL3-based E3 ligases interact with a broad range of ERF/AP2 transcription factors, mediated by MATH-BTB/POZ proteins (Chen et al., 2013). The assembly with an E3 ligase causes degradation of the ERF/AP2 transcription factors via the 26S proteasome and loss of MATH-BTB/POZ proteins significantly affects plant development (Chen et al., 2013). All these have consequences for embryogenesis.
Other candidate genes related to embryogenesis are also found. A locus with the most tightly linked markers landed in the introns of Sobic.001G522600 which encodes RING/FYVE/PHD zinc finger protein/histone acetyltransferase. Both Sobic.001G522600 and AT2G19260 (its Arabidopsis ortholog) proteins contain an ELM2 domain towards their C-terminal (Supplementary Figure S2). ELM2 domain has been shown to recruit histone deacetylase (HDAC) 1 in humans (Ding et al., 2003). Histone deacetylation can prevent the initiation of somatic embryogenesis. Inhibiting HDAC by using an HDAC inhibitor, trichostatin A (TSA), triples embryogenic callus size in sorghum although it does not change embryogenic callus rate (Wu et al., 2024). TSA treatment also has been shown to repress seedling growth but upregulate expression of embryo-specific transcription factors such as LEC1 (Tanaka et al., 2008) which initiates formation of embryo-like structures in vegetative cells in Arabidopsis (Lotan et al., 1998).
Some candidate genes may perform cellular maintenance related functions. Sobic.001G288800 in CIR1-2 encodes 60S RIBOSOMAL EXPORT PROTEIN NMD3 which in Arabidopsis participates in ribosome assembly and nuclear export of ribosomal subunits (Chen et al., 2012) and ribosome biogenesis is a fundamental process that provides cells with molecular factories for cellular protein production (Kressler et al., 2010). The ortholog of Sobic.009G002700 from CIR9-1, AT1G47710, encodes a serine protease inhibitor (AtSerpin1) which inhibits AtMC9 activity in vitro (Vercammen et al., 2006). AtMC9 can target and destroy multiple transcription and translation elongation factors, actins, ribosomal proteins (Tsiatsiani et al., 2013), all important players in basic cellular maintenance.
Some candidate genes are related to hormone-activated cell division. The ortholog of Sobic.004G344200 from DR4-2, AT2G20190, encodes a CLIP-associated protein (CLASP). In Arabidopsis CLASP promotes microtubule stability during mitosis, hence involved in cell division (Ambrose et al., 2007). Mutation in CLASP produces dwarf plants and significantly reduces cell number in cell division zone (Ambrose et al., 2007). The ortholog of Sobic.008G096200 from DR8-3 (Arabidopsis ortholog: AT3G52890) is paralogous to Sobic.006G067700 (Dw2/SbKIPK), belonging to the AGCVIII subgroup of the AGC protein kinase family (Hilley et al., 2017). Known members of the AGCVIII subgroup have been reported to regulate auxin efflux transporters (Barbosa and Schwechheimer, 2014). AT3G52890 protein KIPK (KCBP-interacting protein kinase) interacts with the myosin tail region of KCBP (kinesin-like calmodulin-binding protein) (Day et al., 2000) which is known to be involved in cell division through its interaction with microtubules (Narasimhulu et al., 1997). Another hormone related gene is Sobic.007G151400. The ortholog of Sobic.007G151400 from DR7-1, AT5G21482, encodes cytokinin oxidase (CKX 7) which is expressed in the mature embryo sac and catalyzes the degradation of cytokinins (Köllmer et al., 2014).It is not clear if Sobic.008G096200 and Sobic.007G151400 impact the balance of auxin and cytokinin whose ratios determine how regeneration occurs (Ikeuchi et al., 2016).
We found three receptor protein kinases, Sobic.010G167500, Sobic.008G099300, and Sobic.009G188100 and a mitogen-activated kinase kinase kinase in Sobic.010G171600 (MAP KKK) (Table 3). A receptor-like protein kinase was identified as a major quantitative trait locus (QTL) that affects shoot regeneration in Arabidopsis (Motte et al., 2014). A nonsense mutation (rpk1-5) in the gene produces a protein truncated after the first 1/3 of LRR and completely cuts off the last 2/3 of LRR, transmembrane and kinase domains (Nodine et al., 2007). The mutant is almost completely recalcitrant to shoot regeneration (Motte et al., 2014). A MAP KKK was previously identified as one of the four callus formation genes in poplar (Tuskan et al., 2018).
In conclusion, in this study we evaluated 236 landrace varieties from the sorghum mini core collection and identified the top five varieties most amenable for in vitro embryogenesis and consequently genetic transformation. Using the results for GWAS allows us to map eight callus induction loci, one each for callus browning and embryogenic callus induction, and 24 differentiation loci, identified 47 candidate genes from the loci and provided evidence from the literature that orthologs for 14 of the genes play roles in cellular function and embryogenesis. Future projects will focus on functional characterization of selected genes and utilization of the five sorghum varieties for genetic transformation studies with TSA.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JX: Writing – original draft, Writing – review & editing. LW: Methodology, Writing – review & editing. YL: Data curation, Project administration, Writing – review & editing. QS: Data curation, Software, Visualization, Writing – review & editing. WT: Data curation, Writing – review & editing. ZC: Data curation, Writing – review & editing. LH: Data curation, Project administration, Writing – review & editing. YW: Data curation, Software, Writing – review & editing. JL: Data curation, Funding acquisition, Project administration, Software, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China(32372134), Chuzhou "Star of Innovation and Entrepreneurship" Industrial Innovation Team, Postgraduate Innovation and Entrepreneurship Practice Project of Anhui Province (2023cxcysj179), Student Innovation and Entrepreneurship Project of Anhui Science and Technology University (S202310879188), and Key Discipline Construction Funds of Anhui Sciences and Technology University(No.XK-XJGF001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1430141/full#supplementary-material
References
Ahmed, I., Ur Rehman, S., Akhtar, L. H., Mahmood, K., Ahmad, R. T. (2021). Optimization of in vitro responses of various explants sources in sorghum (Sorghum bicolor). Asian J. Agric. Biol. 1–8. doi: 10.35495/ajab.2021.02.102
Ambrose, J. C., Shoji, T., Kotzer, A. M., Pighin, J. A., Wasteneys, G. O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19, 2763–2775. doi: 10.1105/tpc.107.053777
Assem, S., Zamzam, M., Hussein, B. A. (2014). Evaluation of somatic embryogenesis and plant regeneration in tissue culture of ten sorghum (Sorghum bicolor L.) genotypes. Afr. J. Biotechnol. 13, 3672–3681. doi: 10.5897/AJB2014.13924
Barbosa, I. C., Schwechheimer, C. (2014). Dynamic control of auxin transport-dependent growth by AGCVIII protein kinases. Curr. Opin. Plant Biol. 22, 108–115. doi: 10.1016/j.pbi.2014.09.010
Belide, S., Vanhercke, T., Petrie, J. R., Singh, S. P. (2017). Robust genetic transformation of sorghum (Sorghum bicolor L.) using differentiating embryogenic callus induced from immature embryos. Plant Methods 13, 109. doi: 10.1186/s13007-017-0260-9
Chege, P., Palágyi, A., Lantos, C. (2019). Improved culture media for embryogenic callus generation in sorghum [Sorghum bicolor (L.) Moench. Phyton Int. J. Exp. Botany. 89, 111–119. doi: 10.32604/phyton.2020.07554
Chen, L., Lee, J. H., Weber, H., Tohge, T., Witt, S., Roje, S., et al. (2013). Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25, 2253–2264. doi: 10.1105/tpc.112.107292
Chen, M. Q., Zhang, A. H., Zhang, Q., Zhang, B. C., Nan, J., Li, X., et al. (2012). Arabidopsis NMD3 is required for nuclear export of 60S ribosomal subunits and affects secondary cell wall thickening. PloS One 7, e35904. doi: 10.1371/journal.pone.0035904
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Day, I. S., Miller, C., Golovkin, M., Reddy, A. S. (2000). Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J. Biol. Chem. 275, 13737–13745. doi: 10.1074/jbc.275.18.13737
Ding, Z., Gillespie, L. L., Paterno, G. D. (2003). Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol. Cell Biol. 23, 250–258. doi: 10.1128/MCB.23.1.250-258.2003
Do, P. T., Lee, H., Mookkan, M., Folk, W. R., Zhang, Z. J. (2016). Rapid and efficient Agrobacterium-mediated transformation of sorghum (Sorghum bicolor) employing standard binary vectors and bar gene as a selectable marker. Plant Cell Rep. 35, 2065–2076. doi: 10.1007/s00299-016-2019-6
Espinoza-Sánchez, E. A., Sánchez-Peña, Y. A., Torres-Castillo, J. A., García-Zambrano, E. A., Treviño Ramírez, J. E., Zavala-Garc, F., et al. (2018). Somatic embryogenesis induction from immature embryos of sorghum bicolor L. (Moench). Phyton-International J. Exp. Bot. 87, 105–112. doi: 10.32604/phyton.2018.87.105
Flinn, B., Dale, S., Disharoon, A., Kresovich, S. (2020). Comparative analysis of in vitro responses and regeneration between diverse bioenergy sorghum genotypes. Plants 9, 248. doi: 10.3390/plants9020248
Gingerich, D. J., Gagne, J. M., Salter, D. W., Hellmann, H., Estelle, M., Ma, L., et al. (2005). Cullins 3a and 3b assemble with members of the broad complex ⁄tramtrack ⁄ bric-a-brac (BTB) protein family to form essential ubiquitin–protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280, 18810–18821. doi: 10.1074/jbc.M413247200
Hilley, J. L., Weers, B. D., Truong, S. K., McCormick, R. F., Mattison, A. J., McKinley, B. A., et al. (2017). Sorghum Dw2 encodes a protein kinase regulator of stem internode length. Sci. Rep. 7, 4616. doi: 10.1038/s41598-017-04609-5
Ikeuchi, M., Ogawa, Y., Iwase, A., Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M., Komaki, S., et al. (2017). WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in arabidopsis. Plant Cell. 29, 54–69. doi: 10.1105/tpc.16.00623
Iwase, A., Mita, K., Nonaka, S., Ikeuchi, M., Koizuka, C., Ohnuma, M., et al. (2015). WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 128, 389–397. doi: 10.1007/s10265-015-0714-y
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Kang, H. M., Sul, J. H., Service, S. K., Zaitlen, N. A., Kong, S. Y., Freimer, N. B., et al. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354. doi: 10.1038/ng.548
Köllmer, I., Novák, O., Strnad, M., Schmülling, T., Werner, T. (2014). Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX 7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 78, 359–371. doi: 10.1111/tpj.12477
Kressler, D., Hurt, E., Baβler, J. (2010). Driving ribosome assembly. Biochim. Biophys. Acta(BBA)-Molecular Cell Res. 1803, 673–683. doi: 10.1016/j.bbamcr.2009.10.009
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. 1–3. doi: 10.48550/arXiv.1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennel, T., Ruan, J., Homer, N., et al. (2009). 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, J., Tang, W., Zhang, Y. W., Chen, K. N., Wang, C., Liu, Y., et al. (2018). Genome-wide association studies for five forage quality-related traits in sorghum (Sorghum bicolor L.). Front. Plant Sci. 9, 1146. doi: 10.3389/fpls.2018.01146
Li, G., Wang, L., Liu, Y., Li, Y., Yang, X., Zhan, Q., et al. (2017). Construction of an efficient tissue culture system for sorghum using mature embryos. Pakistan J. Bot. 49, 995–1000.
Lotan, T., Ohto, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/s0092-8674(00)81463-4
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell. 28, 1998–2015. doi: 10.1105/tpc.16.00124
Ma, L., Liu, M., Yan, Y., Qing, C., Zhang, X., Zhang, Y., et al. (2018). Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00561
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi: 10.1111/tpj.2018.93.issue-2
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20 (9), 1297–303. doi: 10.1101/gr.107524.110
Motte, H., Vercauteren, A., Depuydt, S., Landschoot, S., Geelen, D., Werbrouck, S., et al. (2014). Combining linkage and association mapping identifies RECEPTOR-LIKE PROTEIN KINASE1 as an essential Arabidopsis shoot regeneration gene. Proc. Natl. Acad. Sci. U.S.A. 111, 8305–8310. doi: 10.1073/pnas.1404978111
Murty, U. R., Visarada, A., Annapurna, A., Bharathi, H. (1990). Developing tissue culture system for sorghum, Sorghum bicolor (L) Moench. I. Embryogenic callus induction from elite genotypes. Cereal Res. Commun. 18, 257–262.
Nagle, M. F., Yuan, J., Kaur, D., Ma, C., Peremyslova, E., Jiang, Y., et al. (2024). GWAS supported by computer vision identifies large numbers of candidate regulators of in planta regeneration in Populus trichocarpa. G3: Genes Genomes Genet. 14, jkae026. doi: 10.1093/g3journal/jkae026
Narasimhulu, S. B., Kao, Y. L., Reddy, A. S. (1997). Interaction of Arabidopsis kinesin-like calmodulin-binding protein with tubulin subunits: modulation by Ca(2+)-calmodulin. Plant J. 12, 1139–1149. doi: 10.1046/j.1365-313x.1997.12051139.x
Nguyen, D. Q., Van Eck, J., Eamens, A. L., Grof, C. P. L. (2020). Robust and reproducible agrobacterium-mediated transformation system of the C4 genetic model species Setaria viridis. Front. Plant Sci. 11, 281. doi: 10.3389/fpls.2020.00281
Nodine, M. D., Yadegari, R., Tax, F. E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev. Cell. 12, 943–956. doi: 10.1016/j.devcel.2007.04.003
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Raghuwanshi, A., Birch, R. G. (2010). Genetic transformation of sweet sorghum. Plant Cell Rep. 29, 997–1005. doi: 10.1007/s00299-010-0885-x
Silva, T. N., Thomas, J. B., Dahlberg, J., Rhee, S. Y., Mortimer, J. C. (2022). Progress and challenges in sorghum biotechnology, a multipurpose feedstock for the bioeconomy. J. Exp. Bot. 73, 646–664. doi: 10.1093/jxb/erab450
Tanaka, M., Kikuchi, A., Kamada, H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. doi: 10.1104/pp.107.111674
Tsiatsiani, L., Timmerman, E., De Bock, P. J., Vercammen, D., Stael, S., van de Cotte, B., et al. (2013). The Arabidopsis metacaspase9 degradome. Plant Cell. 25, 2831–2847. doi: 10.1105/tpc.113.115287
Tuskan, G. A., Mewalal, R., Gunter, L. E., Palla, K. J., Carter, K., Jacobson, D. A., et al. (2018). Defining the genetic components of callus formation: A GWAS approach. PLoS One 13, e0202519. doi: 10.1371/journal.pone.0202519
Tyagi, N., Dahleen, L. S., Bregitzer, P. (2010). Candidate genes within tissue culture regeneration QTL revisited with a linkage map based on transcript-derived markers. Crop science. 50, 1697–1707. doi: 10.2135/cropsci2009.10.0624
Umehara, M., Ikeda, M., Kamada, H. (2007). Endogenous factors that regulate plant embryogenesis: recent advances. Japanese J. Plant Sci. 1, 1–6.
Upadhyaya, H. D., Pundir, R. P., Dwivedi, S. L., Gowda, C. L., Reddy, V. G., Singh, S. (2009). Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 49, 1769–1780. doi: 10.2135/cropsci2009.01.0014
Upadhyaya, H. D., Wang, L., Prakash, C. S., Liu, Y., Gao, L., Meng, R., et al. (2022). Genome-wide association mapping identifies an SNF4 ortholog that impacts biomass and sugar yield in sorghum and sugarcane. J. Exp. Bot. 73, 3584–3596. doi: 10.1093/jxb/erac110
Vercammen, D., Belenghi, B., Cotte van de, B., Beunens, T., Gavigan, J. A., De Rycke, R., et al. (2006). Serpin1 of Arabidopsis thaliana is a suicide inhibitor for Metacaspase 9. J. Mol. Biol. 364, 625–636. doi: 10.1016/j.jmb.2006.09.010
Wang, L., Gao, L., Liu, G., Meng, R., Liu, Y., Li, J. (2021a). An efficient sorghum transformation system using embryogenic calli derived from mature seeds. PeerJ 9, e11849. doi: 10.7717/peerj.11849
Wang, L., Tu, W., Jin, P., Liu, Y., Du, J., Zheng, J., et al. (2024). Genome wide association study of plant color in Sorghum bicolor. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1320844
Wang, L., Upadhyaya, H. D., Zheng, J., Liu, Y., Singh, S. K., Gowda, C. L., et al. (2021b). Genome-wide association mapping identifies novel panicle morphology loci and candidate genes in sorghum. Front. Plant Sci. 12, 743838. doi: 10.3389/fpls.2021.743838
Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., et al. (2005). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137, 83–93. doi: 10.1104/pp.104.052654
Weber, H., Hellmann, H. (2009). Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276, 6624–6635. doi: 10.1111/j.1742-4658.2009.07373.x
Wu, H., Zhang, K., Li, J., Wang, J., Wang, Y., Yu, J., et al. (2024). Somatic embryogenesis from mature sorghum seeds: An underutilized genome editing recipient system. Heliyon 10, e23638. doi: 10.1016/j.heliyon.2023.e23638
Yin, L., Zhang, H., Tang, Z., Xu, J., Yin, D., Zhang, Z., et al. (2021). rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association Study. Genom. Proteom. Bioinform. 19 (4), 619–628. doi: 10.1016/j.gpb.2020.10.007
Yuan, H. Y., Kagale, S., Ferrie, A. M. R. (2024). Multifaceted roles of transcription factors during plant embryogenesis. Front. Plant Sci. 14, 1322728. doi: 10.3389/fpls.2023.1322728
Zhang, Z., Zhao, H., Li, W., Wu, J., Zhou, Z., Zhou, F., et al. (2019). Genome-wide association study of callus induction variation to explore the callus formation mechanism of rice. J. Integr. Plant Biol. 61, 1134–1150. doi: 10.1111/jipb.12759
Keywords: tissue culture, regeneration, GWAS, HDG5, WIND1
Citation: Xu J, Wang L, Liang Y, Shen Q, Tu W, Cheng Z, Hu L, Wang Y-H and Li J (2025) Association mapping and identification of candidate genes for callus induction and regeneration using sorghum mature seeds. Front. Plant Sci. 16:1430141. doi: 10.3389/fpls.2025.1430141
Received: 09 May 2024; Accepted: 31 March 2025;
Published: 24 April 2025.
Edited by:
Andrés J. Cortés, Colombian Corporation for Agricultural Research (AGROSAVIA), ColombiaReviewed by:
Felipe López-Hernández, Colombian Agricultural Research Corporation (CORPOICA), ColombiaSebastián Arenas, National Autonomous University of Mexico, Mexico
Copyright © 2025 Xu, Wang, Liang, Shen, Tu, Cheng, Hu, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jieqin Li, d2xobGpxQDE2My5jb20=; Yi-Hong Wang, eWlob25nLndhbmdAbG91aXNpYW5hLmVkdQ==
 Jingyi Xu
Jingyi Xu Lihua Wang
Lihua Wang Yuan Liang1,2
Yuan Liang1,2 Yi-Hong Wang
Yi-Hong Wang Jieqin Li
Jieqin Li