- College of Horticulture and Landscape Architecture, Henan Institute of Science and Technology, Xinxiang, Henan, China
The SBP-box gene family, an exclusively plant transcription factor, is critical for plant growth, development, and adaptive responses to both biotic and abiotic stresses. However, its role under non-biological stresses, specifically drought, remains overlooked in pepper plants. In our previous work, we isolated an SBP-box gene, CaSBP11, from the pepper genomic database. Subsequently, we employed virus-induced gene silencing, overexpression, and protein interaction experiments to investigate the function of CaSBP11 under drought stress. Our results revealed that drought conditions significantly upregulated CaSBP11 expression, whereas ABA treatment suppressed it. Silencing CaSBP11 enhanced drought resistance in pepper, with increased stomatal aperture and ABA levels, and reduced stomatal density, water loss rates, and reactive oxygen species (ROS) accumulation compared to control plants. Conversely, overexpression of CaSBP11 in Nicotiana benthamiana decreased drought tolerance, with CaSBP11-overexpressing plants showing reduced ABA sensitivity, lower stomatal aperture and ABA levels, and increased stomatal density and ROS production compared to wild-type plants. Interestingly, under non-stress conditions, core ABA signaling genes (CaPP2C, CaPYL9, CaSNRK2.4, CaAREB) exhibited lower expression in CaSBP11-silenced plants compared to controls, whereas this trend was reversed in CaSBP11-overexpressing lines (NbPP2C, NbAREB, NbSNRK2.4, NbSRK2E). Additionally, CaSBP11 was found to interact with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB in nucleus. These data suggest that CaSBP11 negatively regulates plant responses to drought stress and may interact with these key genes in the ABA signaling pathway to mediate this response.
Introduction
Pepper, an important solanaceous vegetable, is highly nutritious and financially rewarding. However, it is susceptible to variousbiological and environmental stresses, including Phytophthora capsici infection, salinity, cold, and drought, all of which adversely affect crop yield (Feng et al., 2019). Drought, in particular, significantly impacts both the production and quality of pepper (Mahmood et al., 2021). To combat these biotic and abiotic stresses, plants activate intricate physiological and biochemical responses for adaptation (Hura et al., 2022; Zhang et al., 2023). Notably, stress responsive genes are triggered under most of such defense mechanisms and most importantly, regulated by transcription factors (Hirayama and Shinozaki, 2010). Numerous transcription factors in pepper have been identified as playing roles in the plant’s response to drought stress. For instance, the MYB transcription factor CaDIM1 positively regulates the response to drought stress by modulating ABA-mediated gene expression (Lim et al., 2022). Additionally, the bZIP transcription factor CaATBZ1, a target protein of CaASRF1 whose stability is regulated by CaASRF1, negatively regulates the ABA signaling pathway and the pepper’s response to drought stress (Joo et al., 2019). Furthermore, CaATBZ1 can be ubiquitinated by CaATIR1 and interacts with it. Silencing of CaATBZ1 can alleviate the sensitivity of CaATIR1-silenced plants to ABA and drought stress (Joo et al., 2020). Moreover, CaDILZ1, another member of the bZIP transcription factor family, shows increased expression levels following ABA treatment and interacts with CaDSR1 to positively regulate the pepper’s response to drought stress (Lim et al., 2018a). The pepper dehydration-responsive homeobox domain transcription factor CaDRHB1 interacts with CaDSIZ1 and positively regulates plant responses to drought stress by modulating ABA-mediated stomatal closure (Lim et al., 2018b; Joo et al., 2022). The squamosa promoter Binding protein (SBP-box), a unique transcription factor in plants first discovered in Antirrhinum majus, containsa conserved SBP domain composed ofapproximately 76 amino acid residues, incorporating two canonical zinc finger proteins (Klein et al., 1996). Within the SBP-box gene family, some SBP genes harbor highly conserved miR156 recognition sites (Li et al., 2020). The SBP-box gene family is crucial for plant growth, morphogenesis, metabolic regulation, and stress adaptation, including hormone response, shoot branching, flower and fruit development, as well as biotic stress. For instance, during photoperiod-induced growth cessation in Populus, the photoperiodic perception mediated by the FyB-PIF8 module correlates with the miR156-SPL16/23-FT2/BRC1 regulatory cascade (Wei et al., 2024). FHY3 and FAR1 integrate photonic signals into the miR156-SPL module-modulated aging pathway, impacting Arabidopsis floral transition (Xie et al., 2020). In addition, Arabidopsis thaliana AtSPL14 exhibits sensitivity to fumonisin B1 (Stone et al., 2005). Furthermore, VpSBP5 is likely to be involved in regulating resistance to Erwinia necator in grape by inducing salicylic acid (SA) and methyl jasmonate (MeJA) molecular signals. The level of disease resistance in grapevine genotypes may correlate with the timing of the peak appearance of these signals (Hou et al., 2013). Additionally, the phosphorylated IPA1/OsSPL14 triggers the expression of WRKY45, which in turn enhances resistance to rice blast (Wang et al., 2018). Besides, the SBP-box gene is also pivotal in abiotic stress responses. For instance, overexpression of SPL6/7/9 genes in Sugarbeet confers enhanced drought resistance in transgenic plants (Wang et al., 2024). Overexpresion of MiSPL3a/b in Arabidopsis enhanced drought and ABA tolerance, yet Pro-Ca responsiveness remained high (Zhu et al., 2024). Silencing the MsSPL9 gene in alfalfa enhances drought resistance by potentially regulating anthocyanin biosynthesis (Hanly et al., 2020). Besides, moderate miR156 expression attenuates SPL13 gene expression, increasing WD40–1 expression for drought resistance, while elevated miR156 expression impairs alfalfa’s drought resistance (Arshad et al., 2017; Feyissa et al., 2019). Moreover, in rice, knocking out the OsSPL10 gene enhances the plant’s tolerance to salt stress (Lan et al., 2019). Overexpression of VpSBP16 in Arabidopsis enhances the plant’s tolerance to both salt and drought stress (Hou et al., 2018). Furthermore, in Arabidopsis, SPL9 regulates the plant’s response to freezing stress by modulating the expression of CBF2 (Zhao et al., 2022). In wheat, silencing the TaSPL6 gene enhancesdrought stress tolerance, while overexpressing TaSPL6 reduces drought stress tolerance (Zhao et al., 2024). Additionally, the overexpression of AhSPL5 in transgenic Arabidopsis can enhance salt tolerance by boosting its ROS-scavenging capability and positively regulating the expression of stress-responsive genes (Sun et al., 2024).There are 15 SBP-box genes in pepper, among which CaSBP08, CaSBP11, and CaSBP12 negatively regulate the defense response of pepper to Phytophthora capsici infection (Zhang et al., 2016, 2018, 2020a, 2020b). Additionally, CaSBP12 negatively regulates the defense response of pepper to salt stress (Zhang et al., 2020c). Furthermore, CaSBP13 negatively regulates the defense response of pepper to drought stress (Zhang et al., 2024).
However, it appears that, beyond our research, the role of pepper SBP-box genes in drought stress is unreported (Zhang et al., 2024). Therefore, we explored the functionality of CaSBP11 (Accession No. Capana10g000709) in pepper’s drought tolerance, building on our previous research (Zhang et al., 2016). The findings indicate that CaSBP11 negatively regulates pepper’s response to drought stress.
Materials and methods
Plant materials and growing environments
The seeds for pepper cultivar AA3 and Nicotiana benthamiana(N. benthamiana)were obtained from the School of Horticulture Landscape Architecture at Henan Institute of Science and Technology, Xinxiang 453003, China. The pepper plants were grown in a controlled environment with a photoperiod of 16 hours of light and 8 hours of darkness, at temperatures of 22°C during dayand 18°C at night, and maintained at an optimal humidity level of 80%. Similarly, N. benthamiana was maintained at 25°C during the day and 18°C at night, with a preferred relative humidity of 60%.
Silencing of CaSBP11 in pepper
In compliance with the tobacco rattle virus-induced gene silence (VIGS) technology elucidated by Wang in 2013, the CaSBP11 gene of pepper was silenced (Wang, 2013). The development of a virus-induced gene silencing test vector was based on the methodology by Zhang et al. (2020b). Specifically, a 224bp-specific fragment from the CaSBP11 CDS region was amplified by specific primers (Supplementary Table 1), followed by cloning into TRV2 vector via a double enzyme digestion technique. The recombinant vector was sequenced by Shanghai Sangon Biotechnology Co., Ltd. and transferred to Agrobacterium strain GV3101 for storage.
During the formation of two true leaves in pepper seedlings, the silencing of the CaSBP11 gene was performed using Zhang’s methodology with a prepared bacterial suspension containing the CaSBP11 gene, and cultivated at 28°C until 0D600 = 0.8 (Zhang et al., 2013). It was mixed with an equal amount of TRV1 and inserted into the pepper cotyledon using a 1ml syringe without needles. After injection, the plants were maintained at 28°Cin darkness for 2 days.They were then cultured with a photoperiod of 16/8 hours, at temperatures of 22°C during the day and 18°C at night. Upon observing photo-bleaching in positive plants (TRV2:CaPDS), five random leaves from CaSBP11-silenced plants and control plants (TRV2:00) were chosen to assess the efficiency of silencing.
Overexpression of CaSBP11 in N. benthamiana
The vector used for CaSBP11 N. benthamiana overexpression was constructed based on the protocol outlined by Zhang et al. (2020b). Then, the re-engineered vector (pVBG2307:CaSBP11:GFP) was then employed for CaSBP11 overexpression in N. benthamiana. CaSBP11 overexpressing transgenic N. benthamiana were created through Agrobacterium tumefaciens-assisted leaf disc transformation (Oh et al., 2005). RNA analysis confirmed two kanamycin-resistant CaSBP11 transformants. The T1 progeny originated from T0 plant regeneration, with T2 progeny deriving from T1 plants. T3 progeny were chosen for subsequent research.
Stress treatments and samples collection
To evaluate the expression of the CaSBP11 gene in peppers under drought stress, 6–8 true leaf seedlings were obtained from substrate (constructed with a 3:1:1 blend of matrix, perlite, and vermiculite).The seedlings were then cultivated in 1/2 Hoagland’s solution, with 20% Polyethylene glycol (PEG6000) applied for three days. Control cultures remained solely in 1/2 Hoagland’s solution. Leaves were collected at 0 h, 3 h, 6 h, 12 h, and 24 h, and stored at −80°C.
For drought treatments of CaSBP11-silenced plants and CaSBP11 overexpression plants, procedures outlined by Zhang et al. (2024) are applied. CaSBP11-silenced and control plants, as well as CaSBP11 overexpression and wild-type plants, were cultivated under identical controlled conditions prior to and during drought treatment. The conditions for CaSBP11-silenced and control plants were 16 hours of light, 8 hours of darkness, 22°C during the day, and 18°C at night, with a humidity of 80%. For CaSBP11 overexpression and wild-type plants, the conditions were 16 hours of light, 8 hours of darkness, 25°C during the day, and 18°C at night, with a humidity of 60%. Prior to drought treatment of CaSBP11-silenced and control plants, sufficient irrigation is implemented one month ahead with watering repeated every third day until the final watering, marking the onset of drought stress (Day 0) three days past the treatment commencement. Sampling is performed on days 0, 1, 2, 3, and 4, with samples are stored at −80°C for future use. For drought treatment in CaSBP11 overexpressed and wild-type plants, the identical procedures involve pre-treating with sufficient irrigation, followed by watering every four days until the final watering, marking the onset of drought stress (day 0). Sampling is conducted on days 0, 2, and 4, with samples being stored at −80°C for future use.
For ABA treatment, seedlings were exposed to 20μM ABA in accordance with Yin et al. (2014). Controls consisted of a 0.5% tween and 0.1% ethanol solution. Leaves were harvested at 0h, 3h, 6h, 12h, 24h, and 48h, and stored at −80°C for future use. The statistical data for germination and root length of CaSBP11 overexpressed plants are performed according to the procedure delineated by Ma et al. (2011). Four ABA concentration gradients (0g/L ABA, 0.1g/L ABA, 0.5g/L ABA, and 1.0g/L ABA) were used for these experiments. The seeds of CaSBP11-overexpression plants were soaked in these ABA solutions for 24 hours, then placed on a culture dish with two layers of moistened filter paper under 25°C, 16 hours light, and 8 hours dark conditions. Ventilation was performed daily for 10 minutes, and water was added to maintain moisture. During the third, fifth, and tenth days, germination data were documented. For the root length statistics of CaSBP11-overexpression plants, the same method was employed, with the root length being counted on the tenth day.
RNA extraction and quantitative real-time PCR
Total RNA was extracted via the RNAprep Pure Micro Kit (Tiangen Beijing, China), in accordance with manufacturer’s guidelines. Reverse transcription was accomplished with the transcriptor First Strand cDNA Synthesis Kit (Tiangen Beijing, China). Diluted cDNA was used for quantitative real-time (qPCR) analysis at a concentration of 50 ng/L. Utilizing the iQ5.0 Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA, USA), we implemented real-time Qpcr according to the methodology outlined by Zhang et al. (2020a). This involved a pre-denaturation phase at 95 °C for 1 min, followed by 40 cycles of denaturation (95 °C, 10 s), annealing (56 °C, 30 s), and extension (72°C, 30s). End-of-cycle fluorescence detection ensured PCR primer accuracy through post-PCR profile analysis ranging from 56 to 95 °C. The specificity of all primers was rigorously validated using NCBI Primer BLAST (Supplementary Table 1). Gene expression was assessed and standardized against CaUBI3 (GenBank: AY486137.1), and Nbactin-97 (GenBank: XM_019369243.1) (Schmittgen and Livak, 2008; Du et al., 2015; Zhang et al., 2016).
Drought index percentage analysis
Percentage drought index analysis employed Zhang’s methodology (Zhang et al., 2020b). Upon 5 days of drought stress, the drought phenotypes of CaSBP11-silenced and control plants were classified, and the percentage drought indices of both groups were calculated. The classification criteria were as follows: level 0, no symptoms; level 1, lower leaves wither; level 2, all leaves except the growing point wither; level 3, entire plant withers. For drought stress of 13 days, the drought phenotypes of CaSBP11 overexpression and wild-type plants were classified, and the percentage drought indices of both groups were calculated. The classification criteria were as follows: level 0, no symptoms; level 1, lower leaves wither or yellow; level 2, lower leaf death; level 3, entire plant death except the growing point.
Determination of physiological indicators
Malondialdehyde (MDA), relative water content, and relative electrical conductivity evaluations followed Zhang et al. (2018) and Pan et al. (2012) protocols. Water loss rate was calculated based on Ma et al. (2021) method. Total chlorophyll content was determined via Arkus et al. (2005) methodology. Assays for peroxidase (POD), catalase (CAT) and superoxide dismutase (SOD) activities were conducted following Zhang et al. (2018); Stewart and Bewley (1980) methodology.
For hydrogen peroxide (H2O2) and oxygen (O2-) radical analysis, diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT) stains were utilized as per Choi and Hwang (2012) and Kim et al. (2012) protocol, respectively. Quantification of these stained regions was executed based on Sekulska-nalewajko et al. (2016),while H2O2 content determination utilized Liu et al. (2010). Superoxide anion (O2-) detection is accomplished in accordance with the Solarbio Superoxide Anion kit guidelines (Solarbio, Beijing, China).
The analysis of ABA content utilizes the Plant Hormone Abscisic Acid (ABA) Enzyme Immuno Fluorometric Assay Kit (KeLu, Wuhan, China). The procedure is carried out as per the enclosed instructions.
The stomatal morphologies were observed at high magnification using a scanning electron microscope (FEI Quanta 200, USA). The stomatal density was accurately measured via the NIH (National Institutes of Health)-endorsed Image J software Stomatal apertures were determined via SEM, with the aperture dimensions encompassing both pore length (dumbbell-shaped aperturae) and width (maximum perpendicular to the dumbbell-shaped aperturae). Besides, SEM examination unveiled the dimensions of the stomatal apertures. The stomatal aperture encompasses both the pore length and width of the pore. The dumbbell-shaped aperturaeis identified as the latter, while its maximal perpendicular value represents the former.
Bimolecular fluorescence complementation and co-immunoprecipitation experiments
The vectors of pSPYCE-35S and pSPYNE-35S were used for these experiments. Besides, the target gene was ligated into the vector using the homologous recombination method, and the primers for vector construction are listed in Supplementary Table 1. The constructed recombinant vectors were transferred into Agrobacterium (GV3101) for the BiFC and Co-IP experiments. For the BiFC experiment, the above transformed Agrobacterium bacterial solution was cultured overnight at 28°C and 200 rpm until the optical density (OD) reached 0.5. The culture was then centrifuged, and the supernatant was discarded. The pellet was re-suspended in an equal volume of suspension buffer. After resuspension, the bacterial solution was left to stand at room temperature for 3 hours. The bacterial suspension was then injected into Nicotiana Benthamiana leaves using a 1 ml syringe without a needle. After injection, the plants were maintained in darkness at 28°C for one day, followed by cultivation under a photoperiod of 16/8 hours, with temperatures of 22°C during the day and 18°C at night. Fluorescence was observed using a confocal laser scanning microscope (FV10-ASW, Olympus Corporation, Japan) three days after infection. The excitation wavelengths used were 515 nm for the yellow fluorescence field, 488 nm for the chloroplast auto fluorescence field, and 358 nm for the DAPI field (nuclear staining). The Co-IP experiments were conducted according to the method described by Song et al. (2023).
Statistical analysis
The SPSS 22.0 statistical software was utilized to evaluate the effects of different treatment alterations using a one-way ANOVA test. Post hoc Tukey analysis revealed potential significant differences at P ≤ 0.05 and P ≤ 0.01. Data is presented as mean ± standard deviation (SD). Experiments necessitate at least three biological replicas in rigorously designed protocols.
Results
Expression of the CaSBP11 gene during drought and ABA stress in pepper
To investigate the role of CaSBP11 in resilience to drought and ABA stress its expression pattern in pepper was analyzed. As shown in Supplementary Figure 1, CaSBP11 transcripts initially decreased at 3h post drought stress, subsequently significantly increasing at 12h. Besides, the expression of CaSBP11 was suppressed during ABA treatment (Supplementary Figure 1). These findings indicating that CaSBP11 responds to both drought and ABA stress.
CaSBP11 gene silencing enhances pepper plant drought resilience
Virus-induced gene silencing was applied for elucidation of CaSBP11’s function in pepper’s drought stress response (Wang, 2013).The established positive control,TRV2:CaPDS, silenced the CaPDS gene, resulting in photo-bleached leaf symptoms, while TRV2:00 served as the negative control. Upon observing photo-bleaching in positive control plants, the silencing efficiency of TRV2:CaSBP11 was evaluated (Supplementary Figure 2). The results in Supplementary Figure 2 show that CaSBP11-silenced (TRV2:CaSBP11) and control (TRV2:00) plants displayed no remarkable phenotypic variations under regular conditions. Additionally, the silencing efficiency of the CaSBP11 gene exceeds 84%. Therefore, both CaSBP11-silenced and control plants were used for subsequent analyses.
After enduring three drought days, CaSBP11-silenced plants demonstrated negligible phenotypic modifications while controls exhibited leaf wilting symptoms, including chlorosis in lower foliage (Figure 1A). Comparatively, there were no discernible differences in the phenotype of non-stressed CaSBP11-silenced and control plants (Figure 1A). Upon drought exposure, chlorophyll content decreased in both CaSBP11-silenced and control plants, yet the former retained significantly more (Figure 1B).Additionally, the rate of water loss in CaSBP11-silenced plants showed a decreasing trend after drought, notably outperforming controls at two and three days (Figure 1C). The electrical conductivity of CaSBP11-silenced plants varied consistently below the control plants, particularly on Days 1 and 3 (Figure 1C). Both plant types exhibited an overall increase in MDA content, with control plants showing higher values on Days 1 and 2 (Figure 1C). Besides, CaSBP11-silenced plant samples exhibited superabundant CAT and POD activities on Day 1, achieving statistical significance (Figure 1C). SOD activity exhibits a trend of initial increase and subsequent decline, while the peak time point in control plants precedes that of CaSBP11-silenced plants (Figure 1C). Upon drought stress for 5 days, we classified the drought phenotypes of CaSBP11-silenced and control plants to calculate their percent drought indices. The classifying criteria were: level 0, no symptoms; level 1, lower leaves of plant wilted; level 2, all plant leaves except the growing point wilted; level 3, entire plant wilted (Supplementary Figure 3A). Subsequently, we statistically analyzed the percent drought indices of CaSBP11-silenced and control plants. As demonstrated in Supplementary Figure 3B, at 5 days of drought stress, the drought index of CaSBP11-silenced plants was notably lower than that of control plants. These results indicated improved drought endurance in the CaSBP11-silenced plants. Subsequently, to assess reactive oxygen species (ROS) accumulation in drought-stressed plants with CaSBP11-silenced and control groups, H2O2 and O2- were detected via DAB and NBT staining (Figures 2A, D). After four days of drought exposure, significant DAB and NBT staining area was observed in control leaves compared to CaSBP11-silenced leaves (Figures 2B, E), with elevated H2O2 and O2-content (Figures 2C, F). This evidence suggests reduced ROS accumulation in CaSBP11-silenced foliage relative to control leaves. Furthermore, gene expression levels pertinent to ROS removal (CaAPX1, CaPOD, CaSOD, and CaCAT2) were evaluated (Figure 2G). On the fourth day of drought stress, except for elevated CaPOD expression, gene expression in CaSBP11-silenced and control plants decreased (Figure 2F). Nonetheless, these genes displayed significantly higher expression in CaSBP11-silenced plants compared to controls (Figure 2G).
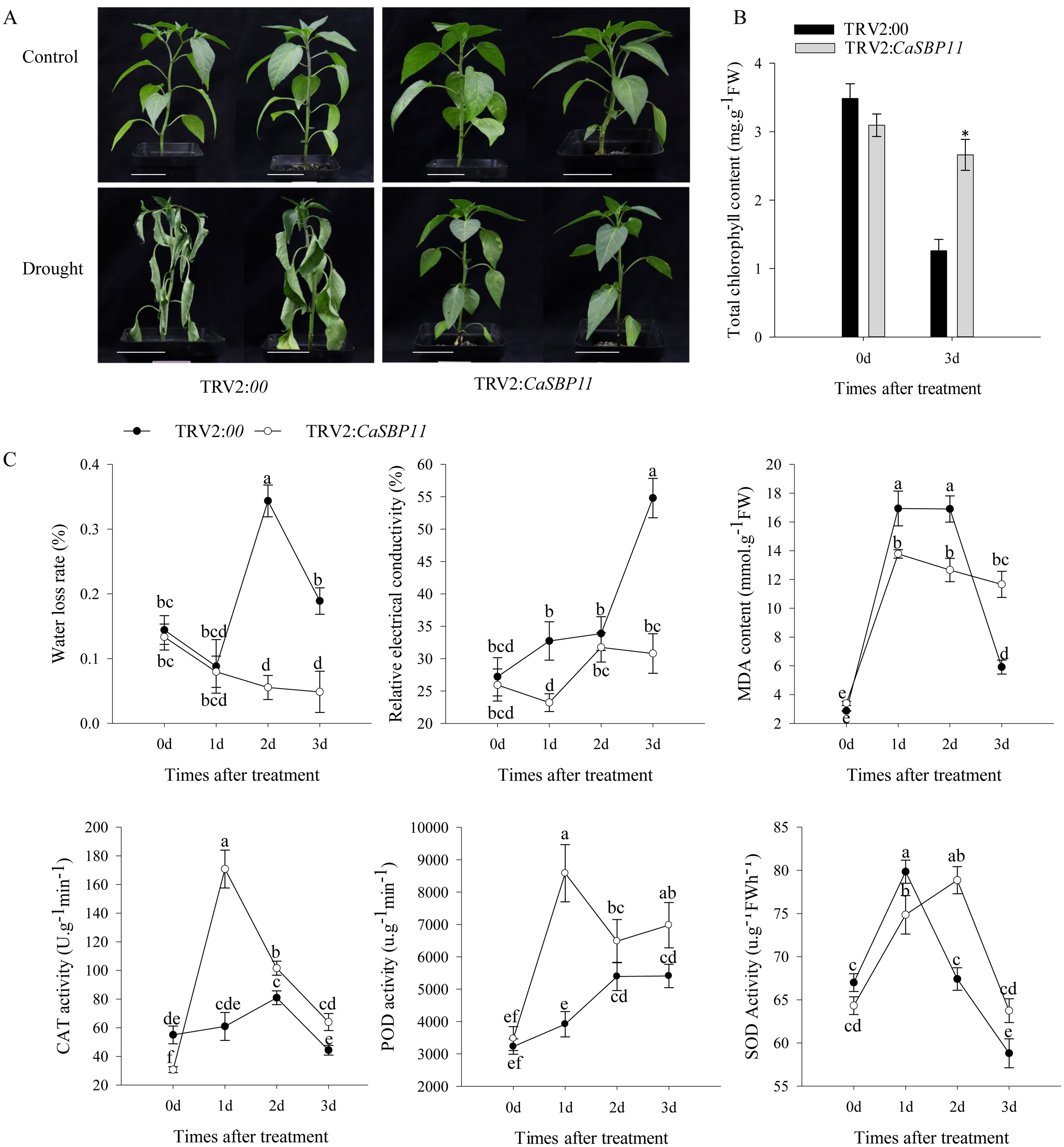
Figure 1. Investigating drought resilience in CaSBP11-silenced plants. (A) Drought-induced morphological changes of CaSBP11-silenced and control plants at three days post-drought stress, alongside their respective non-stressed conditions. Scale bar, 3.5 cm. (B) Chlorophyll content of CaSBP11-silenced versus control plants under drought stress. (C) Post-drought stress, the water loss rate, relative electrical conductivity, MDA concentration, SOD, CAT, and POD activities of CaSBP11-silenced and control plants d: day. * Denotes significance at P ≤ 0.05. Letters denote significant differences at P ≤ 0.05. Mean values and SDs for three replicates are displayed.
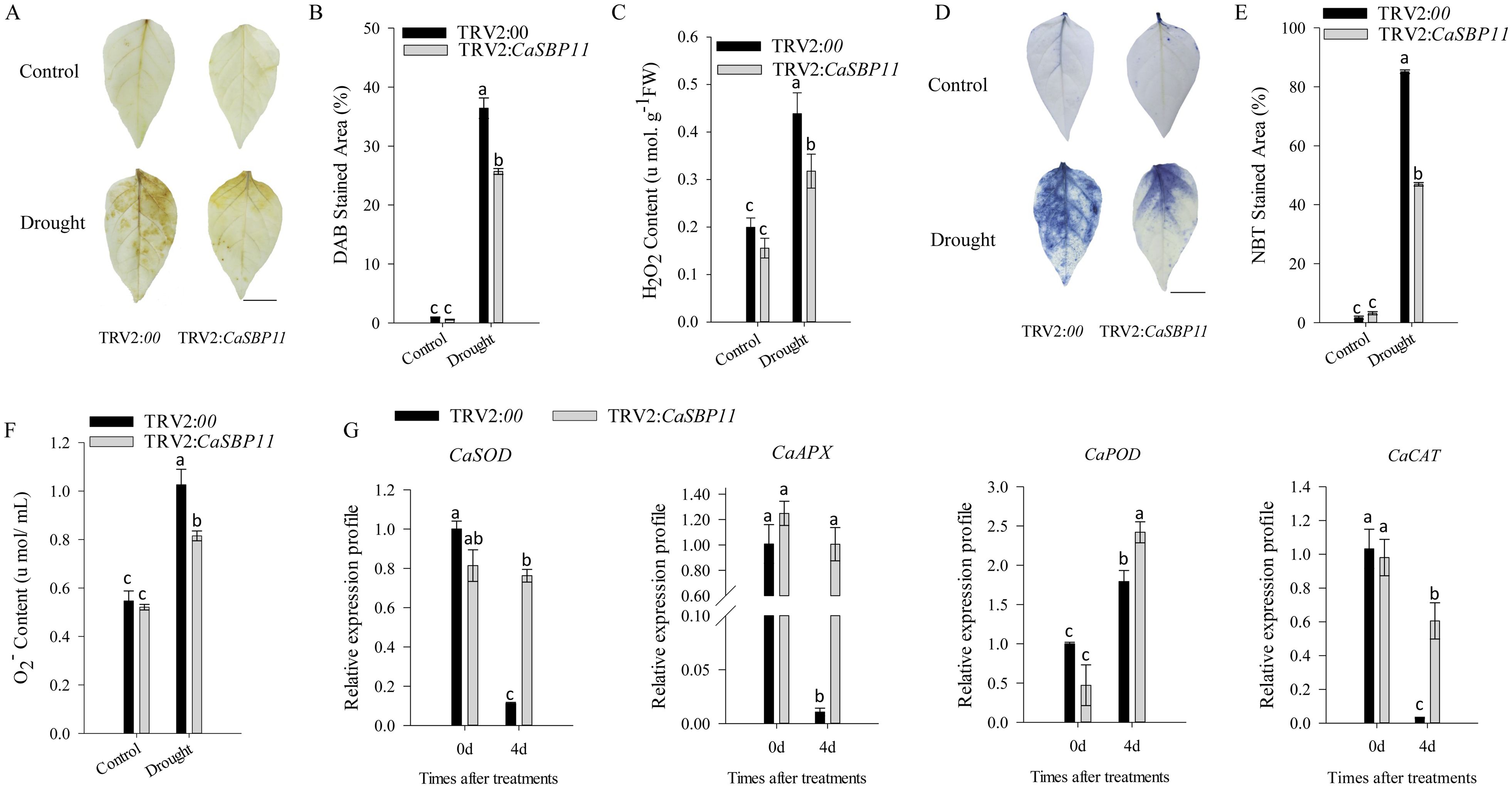
Figure 2. Characterization of CaSBP11-silenced versus control plants through DAB and NBT staining, evaluation of hydrogen peroxide and superoxide anion levels, and monitoring of ROS-scavenging enzyme gene expression. (A) DAB staining in drought-stressed leaf tissue after four days. Scale bar, 0.6 cm. (B) DAB-stained region in leaves following drought stress. (C) Post-drought stress hydrogen peroxide content in CaSBP11-silenced versus control plants.(D) NBT staining in drought-stressed leaf tissue after four days. Scale bar, 0.6 cm. (E) NBT-stained region in leaves post-drought stress. (F) Post-drought stress superoxide anion content in CaSBP11-silenced versus control plants. (G) Expression of ROS-scavenging enzyme genes post-drought stress in CaSBP11-silenced and control plants. Different letters denote significant differences at P ≤ 0.05. Mean values and SDs for three replicates are displayed.
The stomatal density of control plants was significantly higher than that of CaSBP11-silenced plants (Figures 3A, B). During four-day drought stress, ABA content increased in both CaSBP11-silenced and control plants; however, the increase was significantly greater in the former (Figure 3C). Additionally, post-drought stress, stomatal length and width decreased, but CaSBP11-silenced plants still exhibited noticeably larger dimensions compared to the controls (Figures 3D, E). Furthermore, key genes in the ABA signaling pathway (CaSNRK2.4, CaPYL9, CaAREB, and CaPP2C) were analyzed as shown in Figure 3F. These genes displayed elevated expression levels in CaSBP11-silenced plants under drought stress after four days (Figure 3F). Notably, even when untreated, their expression levels were lower in the CaSBP11-silenced plants than in the controls (Figure 3F).
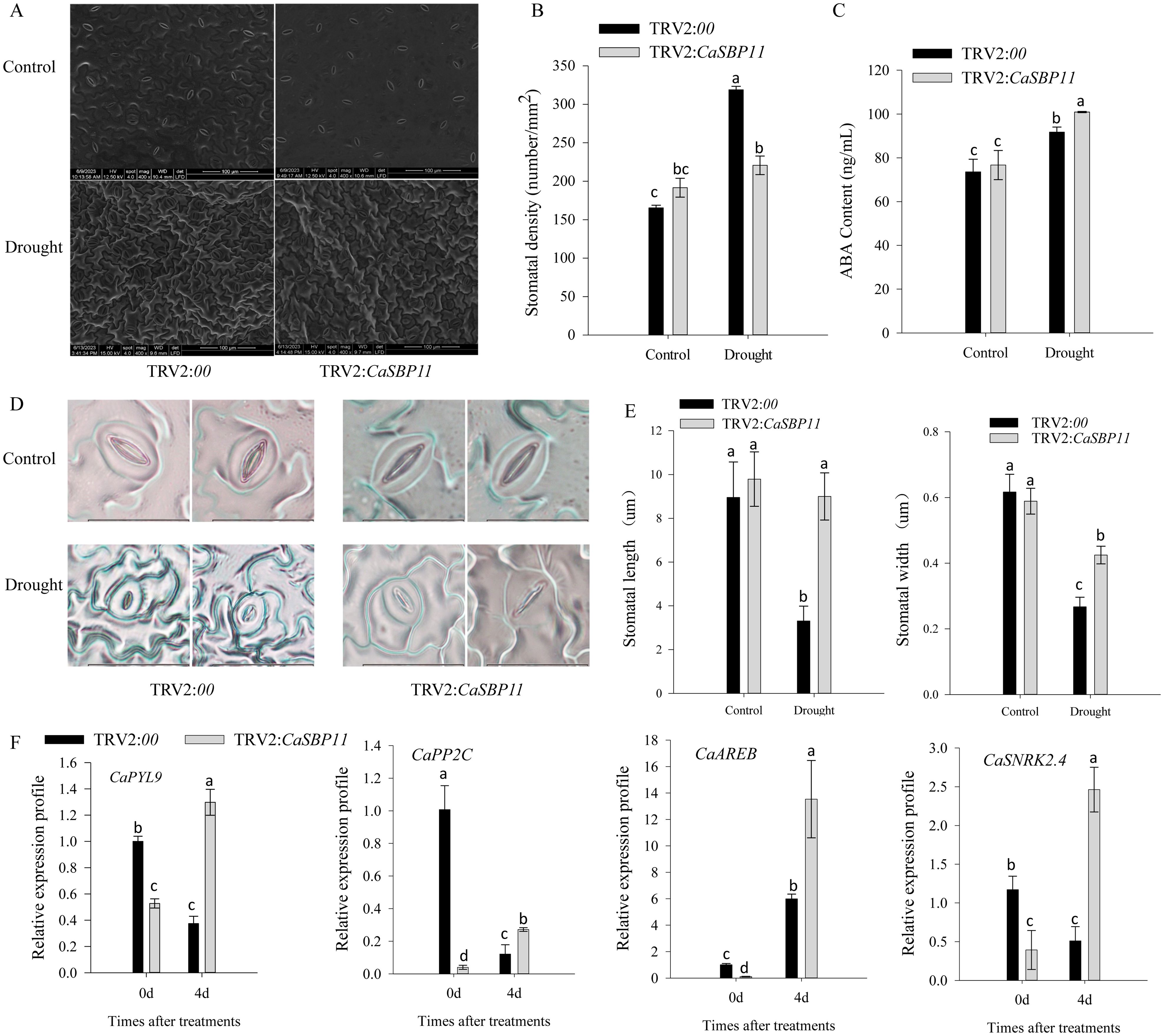
Figure 3. The stomatal state, ABA concentration, and key gene expression of the ABA signaling pathway in drought-stressed CaSBP11-silenced and control plants. (A) Stomatal morphometric data for both groups. (B) Estimation of stomatal density. Scale bar, 100 µm. (C) ABA content of both groups. (D) Stomatal morphologic traits. Scale bar, 20 µm. (E) Stomatal length/width. (F) Key gene expression of ABA signaling pathway post drought in both groups. Different letters denote significant differences at P ≤ 0.05. Mean values and SDs for three replicates are displayed.
CaSBP11 overexpression notably diminishes drought resistance in N. benthamiana
To better understand CaSBP11’s role in plant drought resilience, we generated two transgenic lines (line 9 and line10) overexpressing this gene in N. benthamiana. The gene expression level in these lines is illustrated in Supplementary Figure 4. In the absence of ABA, there was no discernible difference in germination rates between wild-type and CaSBP11 overexpression seeds (Supplementary Table 2). However, under ABA treatment, the germination rates of CaSBP11 overexpression seeds exceeded those of wild-type seeds (Supplementary Table 2). Specifically, at 0.1g/L ABA, CaSBP11 overexpression seeds demonstrated a distinctly increased germination rate at day 3 when compared to wild-types. Similarly, at 0.5g/L ABA, CaSBP11 overexpression seeds displayed a substantially superior germination rate when compared to wild-types by the third day. Lastly, under 1g/L ABA, CaSBP11 overexpression seeds demonstrated markedly enhanced germination rates compared to wild-types by day 5 (Supplementary Table 2). Moreover, root length of CaSBP11 overexpression plants and wild-types diminished as the treatment intensity elevated across different concentration gradients. However, exposure to 0.1g/L ABA, 0.5g/L ABA, and 1g/L ABA significantly enhanced the root length of CaSBP11 overexpression plants compared to wild-types (Supplementary Figures 5A, B). These data show overexpression of CaSBP11 in N.benthamiana diminishing plant sensitivity to ABA.
Additionally, post-seven-day drought exposure, both CaSBP11 overexpression and wild-type plants displayed wilt (Figure 4A). However, the transgenic plants exhibited pervasive wilt and severe yellowing of lower leaves, while the wild-type ones only experienced wilt of lower leaves (Figure 4A). After 13 days of drought, when rewatering occurred, nearly all CaSBP11 overexpression plants, apart from the growing point, had died. In contrast wild-type plants displayed no growth anomalies except for leaf yellowing (Figure 4A). Besides, we categorized the drought phenotypes of CaSBP11 overexpression plants and wild-type plants after 13 days of drought stress. The classification criteria were as follows: level 0, no symptoms; level 1, wilting or yellowing of lower leaves; level 2, sublethal leaf death; level 3, plant death excluding the growing point (Supplementary Figure 6A). Subsequently, we calculated the percentage of drought index of CaSBP11 overexpression plants and wild-type plants. As shown in Supplementary Figure 6B, the percentage of drought index of CaSBP11 overexpression plants was notably elevated compared to wild-types. Additionally, at day 4 of drought, CaSBP11 overexpression plants displayed significantly reduced chlorophyll content compared to wild-types (Figure 4B). Similarly, their MDA content and relative electrical conductivity showed a significant increase (Figure 4C). Additionally, plants overexpressing CaSBP11 displayed a noticeable decrease in relative water content on day 9 of drought stress exposure, as depicted in Figure 4C. These data underscore improved drought stress sensitivity in CaSBP11 overexpressing plants.
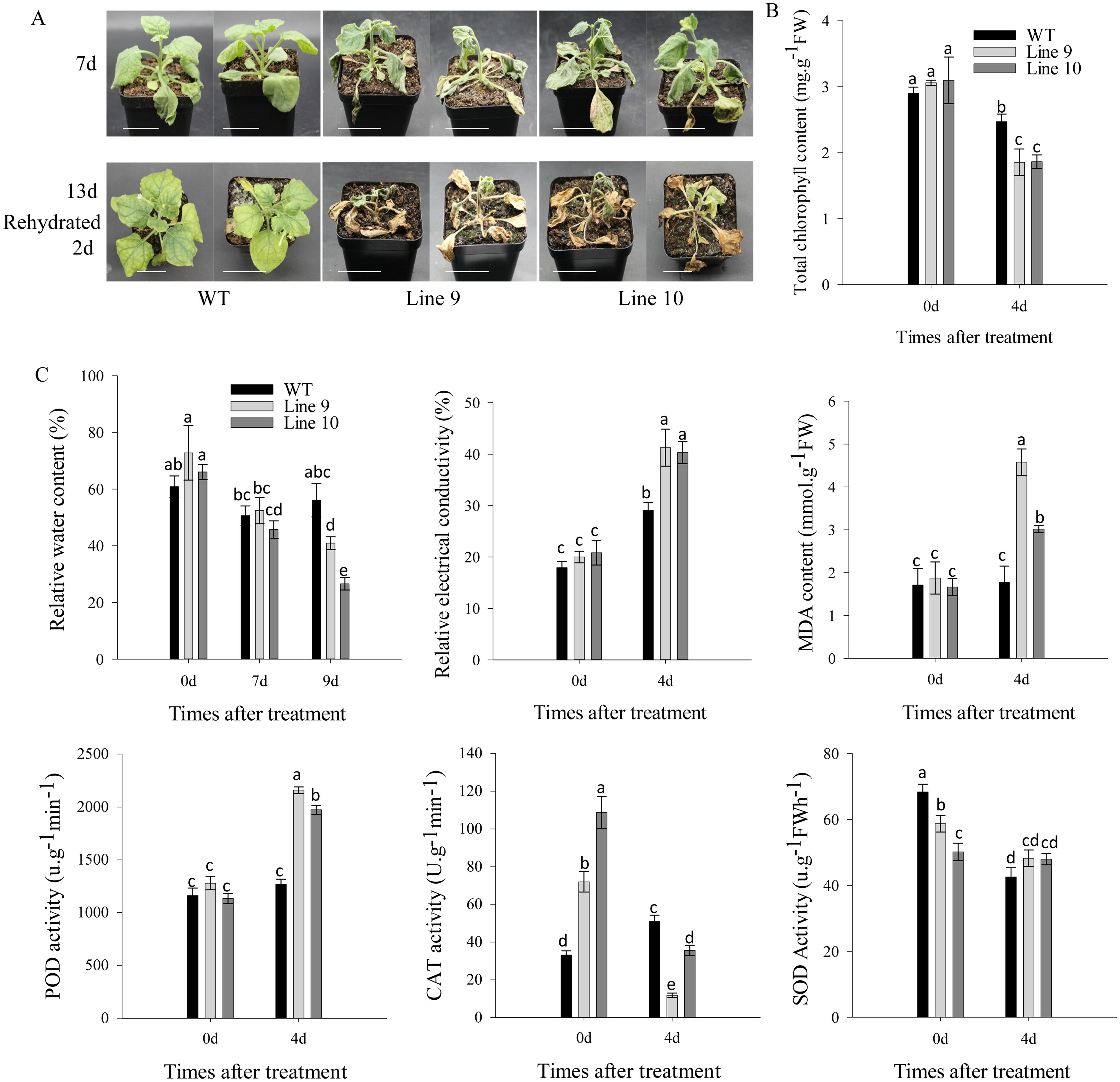
Figure 4. CaSBP11 overexpression in Nicotiana Benthamiana heightens drought susceptibility. (A) Upper layer illustrates the phenotype of CaSBP11 overexpressed and wild-type (WT) lines during seven days of drought, lower layer depicts their status after two days post rehydration following a thirteen days drought period. Scale bar, 3.5 cm. (B) Four days post-drought stress, CaSBP11 overexpression and wild-type total chlorophyll content. (C) Four days post -drought stress, CaSBP11 overexpression versus wild type relative water content, relative electrical conductivity, and malondialdehyde levels. Different letters denote significant differences at P ≤ 0.05. Mean values and SDs for three replicates are displayed.
In addition, on day 4 of drought stress, the CaSBP11 overexpression plants exhibited larger DAB and NBT staining regions than wild-types (Figures 5A, B, D, E). Their H2O2 and O2-content was appreciably higher than wild-type plants (Figures 5C, F). These data suggest a potential role for CaSBP11 in ROS signaling pathways during drought stress. Therefore, gene expressions associated with the ROS pathway (NbPOD, NbCAT, NbAPX, NbSOD) were examined. During drought, NbPOD expression was induced and notably elevated compared to wild-type (Figure 5G). Conversely, NbCAT expression in the CaSBP11 overexpression plants was notably reduced (Figure 5G). Moreover, during the fourth day of drought stress, NbAPX expression in CaSBP11 overexpressing plants displayed notably higher levels compared to wild-types (Figure 5G). However, NbSOD expression in these overexpressing plants is notably reduced (Figure 5G)
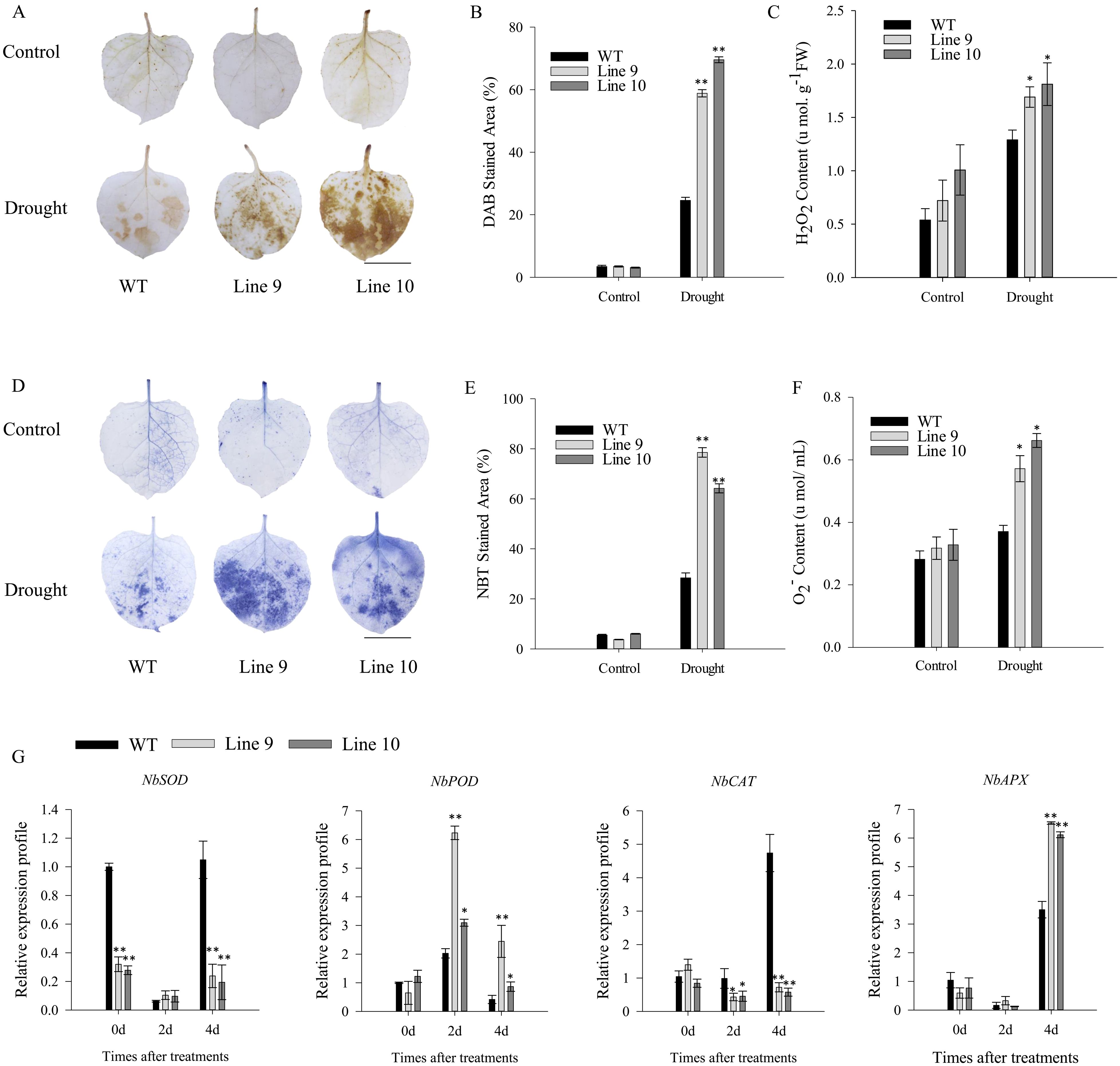
Figure 5. Assessing CaSBP11 overexpressed and wild-type lines via DAB and NBT staining, coupled with analysis of hydrogen peroxide and superoxide anion contents, and expression of ROS-scavenging enzyme genes following drought stress. (A) DAB staining in leaf tissues exposed to four-day drought. Scale bar, 1.4 cm. (B) DAB stain area for these lines after drought. (C) H2O2 content for both lines post-drought stress. (D) NBT staining in leaf tissues post-drought stress. Scale bar, 1.4 cm. (E) NBT stain area for both lines post drought-stress. (F) O2- content for both lines post-drought stress. (G) Expression of ROS-scavenging enzyme genes post-drought stress. * and ** denote significant differences at P ≤ 0.05 and P ≤0.01 respectively. Mean values and SDs for three replicates are displayed.
Besides, the stomatal density of CaSBP11 overexpression plants significantly exceeded that of their wild-type counterparts (Figures 6A, B). Similarly, the ABA concentration in CaSBP11 overexpression plants significantly declined compared to wild-types (Figure 6C). Notably, drought stress resulted in diminished pore size in both CaSBP11 overexpression and wild-type plants after four days, with a remarkable disparity in pore length and width between these strains (Figures 6D–F). During this time, significant reductions in the transcript levels of key ABA signaling pathway genes like NbPYL9, NbAREB, NbPP2C, and NbSNRK2.4 were observed in the CaSBP11 overexpression plants compared to wild-types (Figure 6G). Notably, without treatment, the CaSBP11 overexpression plants exhibited higher expressions of NbPP2C, NbSNRK2.4, and NbSRK2E versus wild-types (Figure 6G). These results indicate that overexpression of CaSBP11 in N.benthamiana accentuates plant drought stress susceptibility, potentially linked to ROS and ABA signaling pathways.
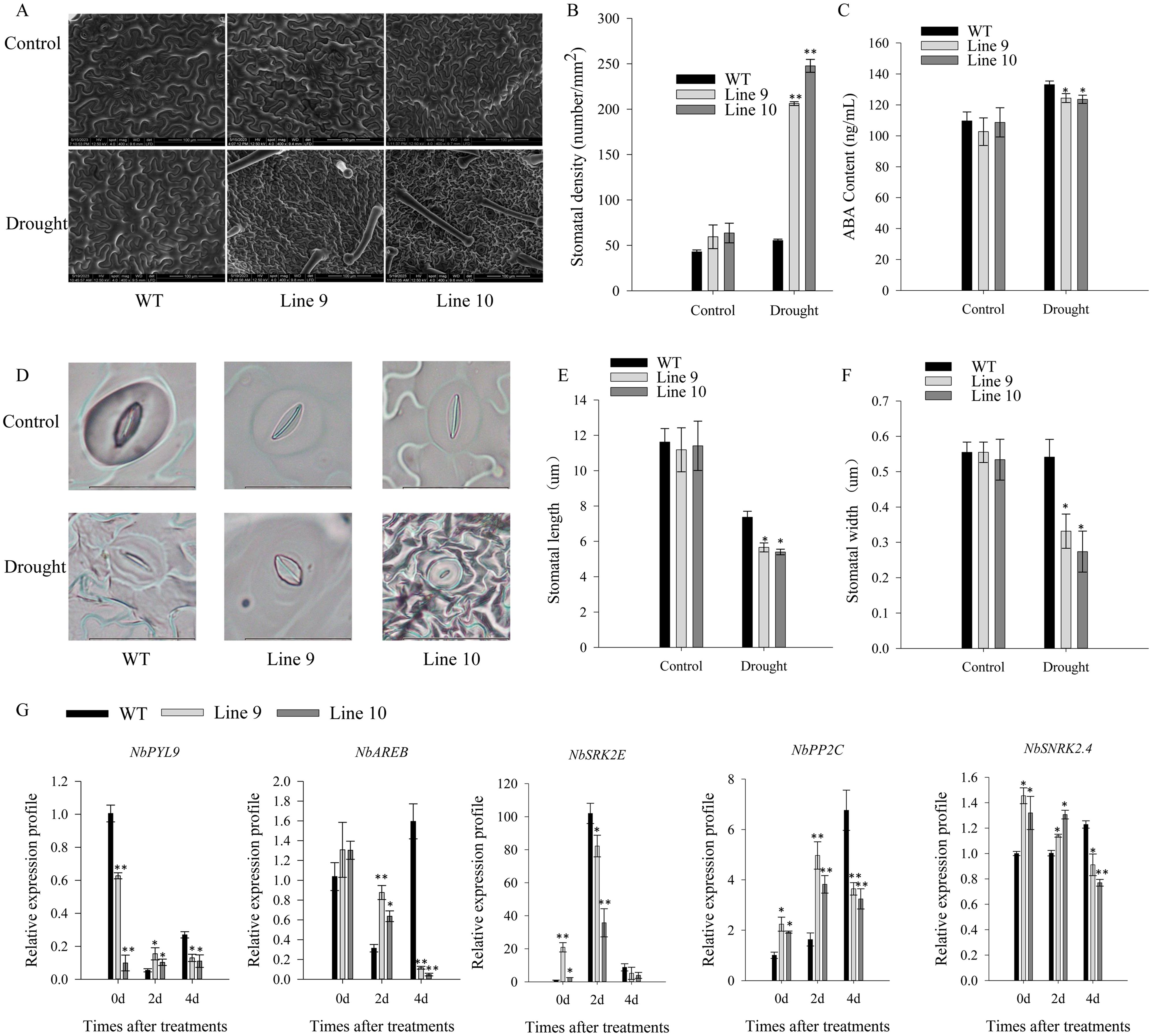
Figure 6. Stomatal state, ABA concentration, and key gene expression of the ABA signaling pathway in drought-stressed CaSBP11 overexpressed and wild-type plants. (A) Stomatal morphologies of CaSBP11 overexpressed plants and wild types. (B) Stomatal density evaluation. Scale bar, 100 µm. (C) ABA levels in CaSBP11 overexpressed plants and wild types. (D) Stoma morphologies. Scale bar, 20 µm. (E), (F) Stoma length/width. (G) ABA signaling pathway key genes’ expression of CaSBP11 overexpressed plants and wild-types post-drought stress. * and ** denote significance at P ≤ 0.05 and P ≤ 0.01 respectively. Mean values and SDs for three replicates are displayed.
CaSBP11 interacts with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB
In the previous study, it was found that under non-stress conditions, core ABA signaling cascade genes (CaPP2C, CaPYL9, CaSNRK2.4, CaAREB) exhibited lower expression levels in CaSBP11-silenced plants when compared to controls. Conversely, this trend was reversed for CaSBP11 overexpressed lines (NbPP2C, NbAREB, NbSNRK2.4, NbSRK2E). Therefore, we speculate that CaSBP11 may regulate the plant’s response to drought stress through interactions with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB. Further research was conducted on this hypothesis. The BiFC experiments demonstrated that CaSBP11 interacts with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB in the nucleus. This was evidenced by a significant YFP signal in the nucleus, while no YFP signal was observed in the control samples (Figure 7, Supplementary Figures 7-9). We also performed Co-IP assays by co-expressing CaPYL9-MYC and CaSBP11-CE, CaAREB-MYC and CaSBP11-CE, CaSnRK2.4-MYC and CaSBP11-CE, CaPP2C-MYC and CaSBP11-CE in N. benthamiana leaves respectively (Figure 8). These results indicated that CaSBP11 interacts with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB. Additionally, CaSBP11 may participate in the response of pepper to drought stress by interacting with these genes.
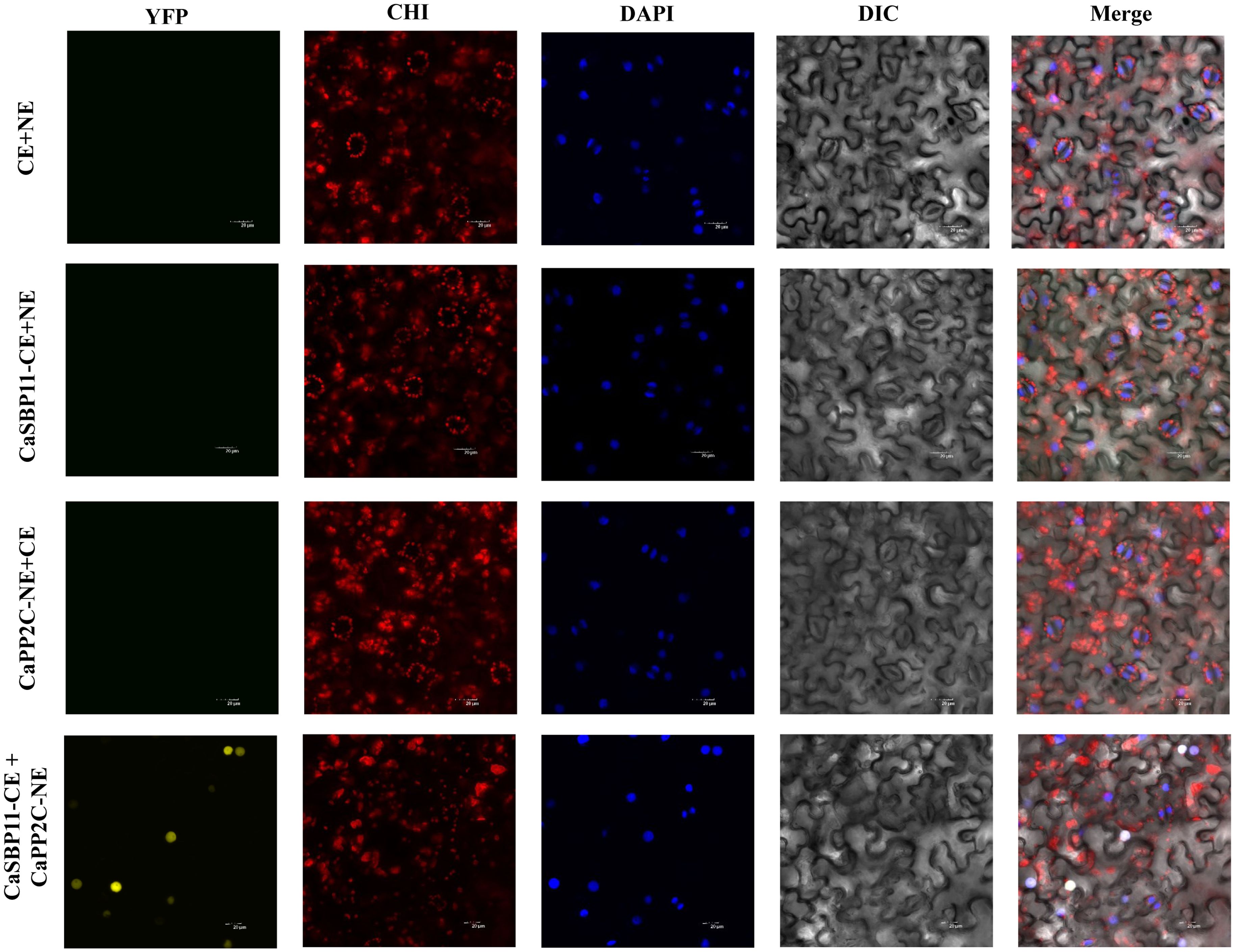
Figure 7. BiFC assay of CaSBP11 and CaPP2C. YFP represents the yellow fluorescent field, CHI represents the chloroplast autofluorescence field, DAPI represents the DAPI field (nuclear staining), DIC represents the bright field, and Merge represents the overlay field. Excitation wavelengths: YFP field (515 nm), CHI field (488 nm), DAPI field (358 nm). Bar = 20 µM.
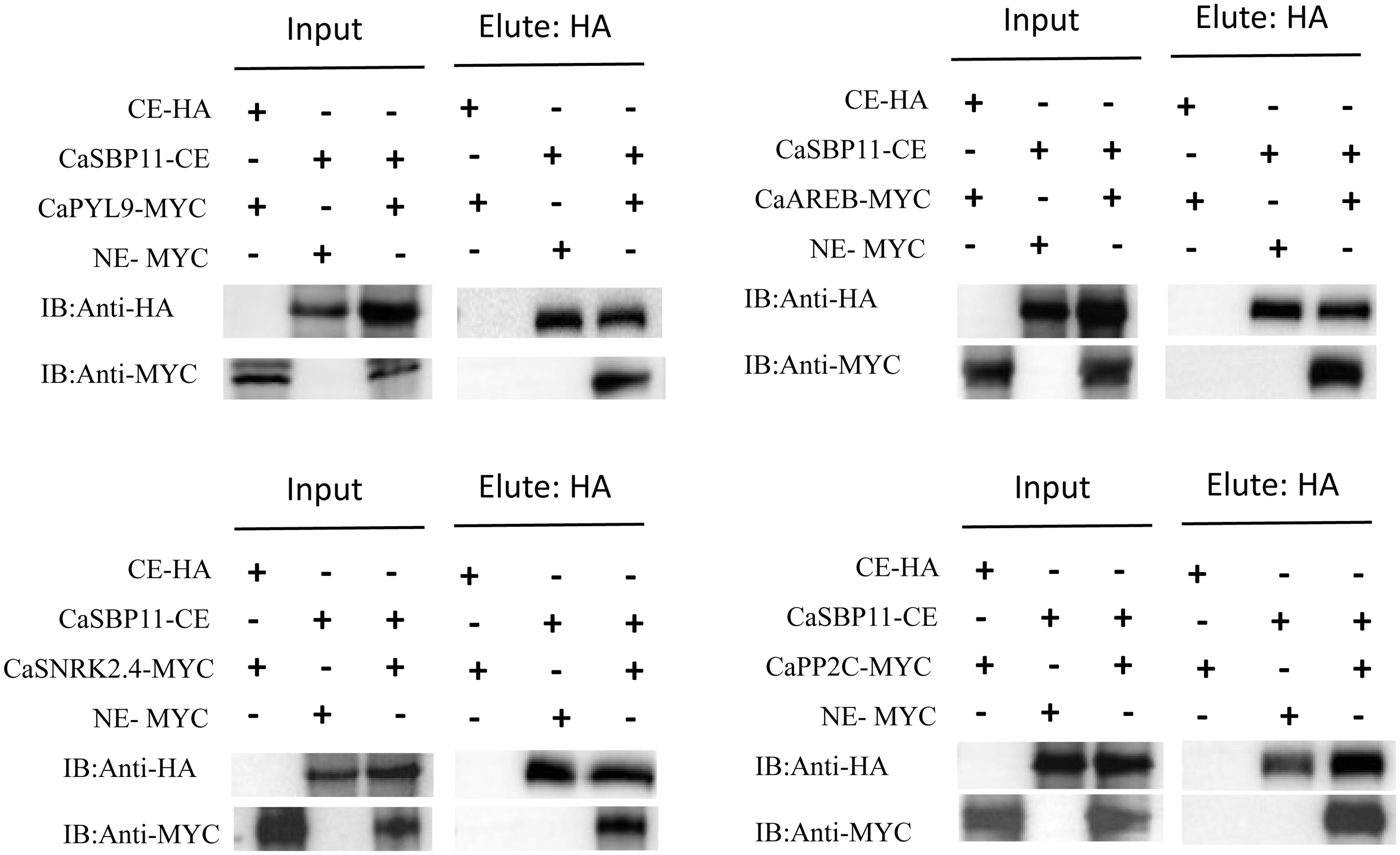
Figure 8. Co-immunoprecipitation (Co-IP) assays of CaSBP11 and CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB. N.benthamiana leaves co-expressing CaPYL9-MYC and CaSBP11-CE, CaAREB-MYC and CaSBP11-CE, CaSnRK2.4-MYC and CaSBP11-CE, CaPP2C-MYC and CaSBP11-CE respectively were subjected to protein immunoprecipitation using the anti-MYC antibody.
Discussion
The plant-specific SBP-box gene family comprises 15 members in pepper, including CaSBP11. Our previous research indicated that CaSBP11 is located in the nucleus and contributes to the pepper’s defense against Phytophthora capsici infection via the salicylic acid (SA) signaling pathway (Zhang et al., 2020b). Nevertheless, its role in drought stress in peppers remains unclear.
Our previous study discovered that the transgenic N. benthamiana line overexpressing CaSBP11 was sensitive to drought stress, prompting us to investigate CaSBP11’s function under drought conditions. Here, we found that the expression level of CaSBP11 gene increases at 12 hours under drought stress (Supplementary Figure 1). To explore whether CaSBP11 influences drought stress responses, we conducted gene silencing experiments. Reportedly, most SBP-Box genes are associated with plant morphogenesis and development. For example, the knockout of SPL13 in Solanum lycopersicum increases lateral bud growth. Additionally, SPL13 directly represses the transcription of IPT1, inhibiting bud growth in spl13 mutants (Chen et al., 2023). In maize, knockout of Zmspl13 and Zmspl29 delays the vegetative phase change and flowering time, while overexpression of ZmSPL29 accelerates these processes, leading to early flowering (Yang et al., 2023a). ARF3 binds to elements P1 and P2 of the SPL promoter, efficiently blocking AG-induced SPL activation and causing abnormal phenotypes in the mutants (Yang et al., 2023b). However, CaSBP11-silenced plants and control plants exhibited no discernible phenotype (Supplementary Figure 2). This lack of phenotype may be due to silencing not completely abolishing the gene’s function as a mutation would, or to CaSBP11 not affecting pepper growth and development. Nonetheless, this necessitates further empirical validation. Silencing CaSBP11resulted in increased drought tolerance in plants (Figure 1A). Previous research has shown that silencing CaSBP13 also enhances drought tolerance in pepper plants (Zhang et al., 2024). Both CaSBP11 and CaSBP13 belong to the SBP-box gene family in pepper (Zhang et al., 2016). Additionally, CaSBP11 and CaSBP13 negatively regulate pepper’s defense response to Phytophthora capsici infection (Zhang, 2020). Evolutionary analysis of SBP-box family genes in Arabidopsis, rice, and tomato revealed that CaSBP11 and CaSBP13 belong to different groups and are evolutionarily distant. Furthermore, an analysis of tandem duplications within the pepper SBP-box gene family showed that there are no interchromosomal segmental duplications between CaSBP11 and CaSBP13 (Zhang et al., 2016). Silencing either CaSBP11 or CaSBP13 genes improved the plant’s response to drought stress (Zhang et al., 2024). Hence, it is speculated that there is no functional redundancy between CaSBP11 and CaSBP13. It has been demonstrated that drought induces excessive ROS production, leading to structural damage (Sewelam et al., 2016). ROS scavengers such as SOD, POD, and CAT efficiently convert surplus and detrimental ROS to harmless water under stress conditions (Noctor and Foyer, 1998; Xu et al., 2016).
In this investigation, the activity of CAT and POD was found to be significantly higherin CaSBP11-silenced plants compared to control plants. Conversely, the peak time for SOD activity was observed to be earlier in control plants than in CaSBP11-silenced plants (Figure 1C). Concurrently, the accumulation of H2O2 and O2- in CaSBP11-silenced plants significantly declined compared to controls (Figures 2A–F). Additionally, the expression level of CaAPX1, CaPOD, CaCAT2, and CaSOD, which are linked to ROS-scavenging enzymes was detected post-drought stress after 4 days. Specifically, these genes were more highly expressed in CaSBP11-silenced plants compared to controls (Figure 2G). It has been reported that exposing soybeans to varying degrees of drought stress during the early flowering phase leads to an enhancement in H2O2 levels alongside a fluctuating pattern for SOD, POD, and CAT levels (Song et al., 2022). Similarly, inducing overexpression of TaFDL2-1A in wheat resulted in elevated SOD and GPX activity after drought stress, indicating a superior ability to counteract ROS in wheat plants (Wang et al., 2022). Furthermore, studies suggest that emphasized expression of BpSPL9 can reinforce ROS scavenging by stimulating POD and SOD enzymes, thereby enhancing plant resistance to drought stress (Ning et al., 2017). OsSPL10 overexpression is crucial for drought tolerance by managing ROS generation in rice (Li et al., 2023). Conversely, TaSPL6-A over-expression in wheat compromised drought resilience, presenting a significant surge in ROS (Zhao et al., 2024).
Furthermore, this research reveals that the number of stomata increases in both CaSBP11-silenced plants and controls, with notably higher numbers in the controls (Figures 3A, B). Additionally, the aperture of the stomata becomes smaller, but it is significantly higher in CaSBP11-silenced plants compared to the control plants (Figures 3D, E). Moreover, the ABA levels augmented in both CaSBP11-silenced and control plants, with significantly greater levels evident in CaSBP11-silenced plants (Figure 3C). Besides, it has been reported that ABA participates in plant responses to drought stress by inducing stomatal closure to minimize transpirational water loss and activating drought responsive genes, ultimately enhancing drought tolerance (Koh et al., 2023). During drought stress, ABA levels rise, causing a decrease in stomatal aperture and an increase in stomatal density (Jalakas et al., 2018; Geng et al., 2023; Hasanuzzaman et al., 2023; Lim et al., 2024). For instance, in CaJAZ1–03 gene-silenced plants, ABA can induce stomatal closure, resulting in a decrease in stomatal aperture. Moreover, CaJAZ1–03 dampens abscisic acid (ABA) signaling and drought stress responses (Koh et al., 2023). CaDeSI2 reduces the stability of the PP2C protein CaAITP1, a core component of ABA signaling, through deSUMOylation, thereby positively regulating drought stress tolerance and ABA-induced stomatal closure (Joo et al., 2024). Under drought stress, both the RcNAC091-silenced plants and the control plants exhibited an increase in stomatal density, and RcNAC091 improves drought tolerance in an ABA-dependent manner. Furthermore, RcNAC091 can bind to the promoter of RcWRKY71, regulating its function during drought stress. Similarly, both the RcWRKY71-silenced plants and the control plants showed an increase in stomatal density. RcWRKY71 positively regulates the plant’s response to drought stress by modulating genes related to the ABA signaling pathway. Additionally, RcWRKY71 might facilitate the ABA-dependent pathway during drought stress (Geng et al., 2023). Besides, CaMEKK17 can interact with PP2C, a core component of the ABA signaling pathway, and positively regulate plant tolerance to drought stress by impairing ABA-mediated stomatal closure (Lim et al., 2024). Furthermore, OsSPL10 in rice negatively regulates drought stress responses by controlling stomatal movements (Li et al., 2023). Based on these results, we hypothesized that CaSBP11’s role in drought tolerance may be linked to the ABA signaling pathway. We subsequently examined these pivotal gene expressions, and discovered elevated expression of CaPYL9, CaAREB, CaPP2C and CaSNRK2.4 in drought-stressed CaSBP11-silenced plants (Figure 3F). Of note was the significant reduction of these gene expressions in CaSBP11-silenced plants, which was uninfluenced by other factors, suggesting that the function of CaSBP11 under drought stress may be related to the ABA signaling pathway (Figure 3F). Reportedly, when plants are challenged by adversity, ABA levels within plants rise. This ABA binds to its receptor PYL, initiating a reaction with PP2C that dephosphorylates SnRK2, thus resetting the inhibitory influence of PP2C on SnRK2. Phosphorylated SnRK2 in turn modulates downstream transcription factors like AREB, triggering ABA signal transduction (Kim et al., 2022).
Besides, CIPK1 interacts with and phosphorylates most ABA receptors at the evolutionary conserved site equivalent to PYL4 Ser129, thereby attenuating their activation and enhancing PP2C activity under normal conditions. During drought stress, ABA inhibits CIPK1-induced phosphorylation of PYLs, preventing them from fully responding to ABA signaling and enabling survival within challenging environmental conditions (You et al., 2023). Moreover, CaDeSI2 interacts with CaAITP1, a member of Group A PP2C proteins, enhancing drought resilience in pepper (Joo et al., 2024). The CaSnRK2.4 protein binds and phosphorylates the CaNAC035 protein, which enhancing cold tolerance via CaSnRK2.4 dependence in pepper (Zhang et al., 2023). Additionally, MicroRNA miR156 is known to regulate SBP family genes in plants. Using the psRNATarget website (http://plantgrn.noble.org/psRNATarget/), we predicted that the CaSBP11 gene contains miR156 target sequences. It has been reported that the interaction between miRNAs and transcription factors coordinates various signaling pathways in plants, including those mediated by ABA and non-ABA (Singroha et al., 2021). For instance, the potential involvement of miRNAs in ABA-dependent drought responses was evident when it was observed that the hyl1 mutant showed hypersensitivity to both ABA and drought (Lu and Fedoroff, 2000). The miRNA156 inhibits the transcription of miRNA172b through SPL9 and, redundantly, through SPL10. Moreover, miRNA172 can reduce plants sensitivity to drought stress (Wu et al., 2009; Han et al., 2013; Singroha et al., 2021). Additionally, in alfalfa, SPL3 is regulated by miR156. Moderate levels of miR156 transcripts are sufficient to enhance drought tolerance in alfalfa by silencing SPL13 and increasing WD40–1 expression. However, excessive overexpression of miR156 results in drought susceptibility. Furthermore, SPL13 acts as a direct regulator of DFR, which is itself regulated by WD40-1 (Arshad et al., 2017; Feyissa et al., 2019). Transgenic plants show enhanced tolerance to drought and salt stress through the expression of the miR156-SPLs-DFR network (Cui et al., 2014; Jeyakumar et al., 2020).
In order to validate CaSBP11’s involvement in plant drought stress response, we engineered N. benthamiana overexpressing CaSBP11. These CaSBP11 overexpression lines exhibited increased drought sensitivity compared to their wild-type counterparts, with elevated ROS accumulation, MDA content, and relative electrical conductivity (Figures 4, 5). Similarly, the expression levels of NbSOD and NbCAT in the CaSBP11 overexpressors were significantly reduced after drought stress, corroborating findings from previous studies on pepper (Figures 2G, 5G). In contrast, the expression of NbPOD and NbAPX increased; with the CaSBP11 overexpression lines showing higher levels than wild-type plants (Figure 5G). This expression pattern is inconsistent with the results observed in pepper ((Figures 2G, 5G)). It has been reported that under severe salt stress, the APX in the roots of Leymus chinensis increases significantly, while POD activity in the leaves increases, jointly eliminating ROS (Li et al., 2017). The expression of ROS-scavenging related genes is closely associated with the activity of ROS-scavenging enzymes. Thus, the contrasting expression patterns of NbPOD and NbAPX compared to those in pepper may be attributed to differences in crop species and the varying degrees of drought stress experienced by the plants. Moreover, under drought stress, both CaSBP11 overexpression and wild-type plants showed an increase in stomatal density in their leaves compared to the control. However, the increase in stomatal density in wild-type plants was not significant (Figure 6B). In contrast, in pepper plants, the stomatal density of TRV2 plants under drought stress was significantly higher than that of the control TRV2 plants (Figure 3B). This difference may be attributed to the varying drought tolerance among different crop species. Furthermore, studies by Wang et al. (2010) have demonstrated that drought stress results in an increase in stomatal density and a decrease in stomatal aperture in the leaves of most plants, while stomatal distribution varies according to the species and the severity of the stress. Additionally, on day 4 of drought stress, the transcript levels of NbPYL9, NbPP2C, NbAREB and NbSNRK2.4 in the CaSBP11 overexpressing plants markedly declined compared to wild-type, corroborating pepper research predictions (Figures 3F, 6G). Beyond this, under non-treatment conditions, the expressions of NbAREB, NbPP2C, NbSNRK2.4, and NbSRK2E in the CaSBP11 overexpressors significantly exceeded those in the wild-type plants, echoing prior studies on pepper (Figures 6G, 3F). Furthermore, ABA levels were elevated in both CaSBP11 overexpressing and wild-type plants; however, the ABA content in the CaSBP11 overexpressors was significantly lower than that in wild-type plants (Figure 6C). Notably, seed germination percentage and root length both decreased under fluctuating ABA conditions. Nevertheless, the wild-type exhibited a stronger ABA response than the plants overexpressing CaSBP11 (Supplementary Table 2, Supplementary Figure 5). Moreover, we found that CaSBP11 interacts with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB in the nucleus (Figures 7, 8, Supplementary Figures 7-9). Therefore, according to Fidler et al.’s findings regarding the ABA signaling pathway’s core components (PYL, PP2C, SnRK2, and AREB) and their roles therein (Fidler et al., 2022), combined with our research data, we speculated that under drought stress, the ABA content in the plant increases. Besides, ABA can inhibit the expression of the CaSBP11 gene, thereby may reducing the interaction between CaSBP11 and CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB. This inhibition promotes the expression of the CaPYL9, CaSNRK2.4, and CaAREB genes, initiating the ABA signaling pathway response and enhancing plant drought tolerance. However, further experimental verification is required.
Conclusions
In summary, the expression level of CaSBP11 gene is upregulated by drought stress at 12 hours post-treatment in pepper. Silencing CaSBP11 enhances drought tolerance, correlating with reduced ROS content compared to control plants. Conversely, overexpressing CaSBP11 in N. benthamiana increases the plant’s susceptibility to drought stress and the ROS accumulation compared to wild-types. Remarkably, drought-induced upregulations of CaAPX1, CaCAT2, CaSOD, and CaPOD transcripts in CaSBP11-silenced plants surpasses control levels. Conversely, post-stress, expression levels of NbCAT1and NbSOD are significantly reduced in CaSBP11 overexpressors. Notably, under non-stress conditions, core ABA signaling genes (CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB) exhibited lower expression in CaSBP11-silenced plants compared to the controls. Conversely, this trend was reversed in CaSBP11-overexpressing lines (NbPP2C, NbAREB, NbSNRK2.4, and NbSRK2E). Besides, CaSBP11 interacts with CaPP2C, CaPYL9, CaSNRK2.4, and CaAREB in the nucleus. These results suggest thatCaSBP11 plays a negatively role in plant drought tolerance, likely tied to ABA and ROS signaling. Nonetheless, additional research is imperative to elucidate these mechanisms.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
H-XZ: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. YZ: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Writing – review & editing. B-WZ: Data curation, Investigation, Methodology, Project administration, Writing – review & editing. F-FP: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported through funding from the National Natural Science Foundation of China (No.32202492) and Henan Province Science and Technology Research Projects (No.242102111159).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1497425/full#supplementary-material
References
Arkus, K. A. J., Cahoon, E. B., Jez, J. M. (2005). Mechanistic analysis of wheat chlorophyllase. Arch. Biochem. Biophys. 438, 146–155. doi: 10.1016/j.abb.2005.04.019
Arshad, M., Feyissa, B. A., Amyot, L., Aung, B., Hannoufa, A. (2017). MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 258, 122–136. doi: 10.1016/j.plantsci.2017.01.018
Chen, S., Song, X., Zheng, Q., Liu, Y., Yu, J., Zhou, Y., et al. (2023). The transcription factor SPL13 mediates strigolactone suppression of shoot branching by inhibiting cytokinin synthesis in Solanum lycopersicum. J. Exp. Bot. 74, 5722–5735. doi: 10.1093/jxb/erad303
Choi, H. W., Hwang, B. K. (2012). The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta. 235, 1369–1382. doi: 10.1007/s00425-011-1580-z
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Du, Y., Berg, J., Govers, F., Bouwmeester, K. (2015). Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 207, 735–747. doi: 10.1111/nph.13355
Feng, X. H., Zhang, H. X., Ali, M., Gai, W. X., Cheng, G. X., Yu, Q. H., et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 142, 151–162. doi: 10.1016/j.plaphy.2019.07.001
Feyissa, B. A., Arshad, M., Gruber, M. Y., Kohalmi, S. E., Hannoufa, A. (2019). The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa. BMC Plant Biol. 19, 1–19. doi: 10.1186/s12870-019-2059-5
Fidler, J., Graska, J., Gietler, M., Nykiel, M., Prabucka, B., Rybarczyk-Płońska, A., et al. (2022). PYR/PYL/RCAR receptors play a vital role in the abscisic-acid-dependent responses of plants to external or internal stimuli. Cells 11, 1–21. doi: 10.3390/cells11081352
Geng, L., Yu, S., Zhang, Y., Su, L., Lu, W., Zhu, H., et al. (2023). Transcription factor RcNAC091 enhances rose drought tolerance through the abscisic acid-dependent pathway. Plant Physiol. 193, 1695–1712. doi: 10.1093/plphys/kiad366
Han, Y., Zhang, X., Wang, Y., Ming, F. (2013). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8, 1–16. doi: 10.1371/journal.pone.0073541
Hanly, A., Karagiannis, J., Lu, Q. S. M., Tian, L., Hannoufa, A. (2020). Characterization of the role of SPL9 in drought stress tolerance in medicago sativa. Int. J. Mol. Sci. 21, 1–14. doi: 10.3390/ijms21176003
Hasanuzzaman, M., Zhou, M., Shabala, S. (2023). How does stomatal density and residual transpiration contribute to osmotic stress tolerance? Plants 12, 1–19. doi: 10.3390/plants12030494
Hirayama, T., Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Hou, H., Jia, H., Yan, Q., Wang, X. (2018). Overexpression of a SBP-box gene (VpSBP16) from chinese wild vitis species in arabidopsis improves salinity and drought stress tolerance. Int. J. Mol. Sci. 19, 1–17. doi: 10.3390/ijms19040940
Hou, H., Yan, Q., Wang, X., Xu, H. (2013). A SBP-box gene vpSBP5 from chinese wild vitis species responds to erysiphe necator and defense signaling molecules. Plant Mol. Biol. Rep. 31, 1261–1270. doi: 10.1007/s11105-013-0591-2
Hura, T., Hura, K., Ostrowska, A. (2022). Drought-stress induced physiological and molecular changes in plants. Int. J. Mol. Sci. 23, 4698. doi: 10.3390/ijms23094698
Jalakas, P., Merilo, E., Kollist, H., Brosché, M. (2018). ABA-mediated regulation of stomatal density is OST1-independent. Plant Direct 2, 1–7. doi: 10.1002/pld3.82
Jeyakumar, J. M. J., Ali, A., Wang, W. M., Thiruvengadam, M. (2020). Characterizing the role of the miR156-SPL network in plant development and stress response. Plants 9, 1–15. doi: 10.3390/plants9091206
Joo, H., Baek, W., Lim, C. W., Lee, S. C. (2024). Pepper SUMO protease CaDeSI2 positively modulates the drought responses via deSUMOylation of clade A PP2C CaAITP1. New Phytol. 243, 1361–1373. doi: 10.1111/nph.19920
Joo, H., Lim, C. W., Lee, S. C. (2019). Roles of pepper bZIP transcription factor CaATBZ1 and its interacting partner RING-type E3 ligase CaASRF1 in modulation of ABA signalling and drought tolerance. Plant J. 100, 399–410. doi: 10.1111/tpj.14451
Joo, H., Lim, C. W., Lee, S. C. (2020). The pepper RING-type E3 ligase, CaATIR1, positively regulates abscisic acid signalling and drought response by modulating the stability of CaATBZ1. Plant Cell Environ. 43, 1911–1924. doi: 10.1111/pce.13789
Joo, H., Lim, C. W., Lee, S. C. (2022). Pepper SUMO E3 ligase CaDSIZ1 enhances drought tolerance by stabilizing the transcription factor CaDRHB1. New Phytol. 235, 2313–2330. doi: 10.1111/nph.18300
Kim, G., Ryu, H., Sung, J. (2022). Hormonal crosstalk and root suberization for drought stress tolerance in plants. Biomolecules 12, 1–16. doi: 10.3390/biom12060811
Kim, J. M., Woo, D. H., Kim, S. H., Lee, S. Y., Park, H. Y., Seok, H. Y., et al. (2012). Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 31, 217–224. doi: 10.1007/s00299-011-1157-0
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity geneSQUAMOSA. Mol. Gen. Genet. MGG 250, 7–16. doi: 10.1007/BF02191820
Koh, H., Joo, H., Lim, C. W., Lee, S. C. (2023). Roles of the pepper JAZ protein CaJAZ1–03 and its interacting partner RING-type E3 ligase CaASRF1 in regulating ABA signaling and drought responses. Plant Cell Environ. 46, 3242–3257. doi: 10.1111/pce.14692
Lan, T., Zheng, Y., Su, Z., Yu, S., Song, H., Zheng, X., et al. (2019). OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). Genes|Genomes|Genetics 9, 4107–4114. doi: 10.1534/g3.119.400700
Li, J., Gao, X., Zhang, X., Liu, C. (2020). Dynamic expansion and functional evolutionary profiles of plant conservative gene family SBP-box in twenty two flowering plants and the origin of miR156. Biomolecules 10, 757. doi: 10.3390/biom10050757
Li, Y., Han, S., Sun, X., Khan, N. U., Zhong, Q., Zhang, Z., et al. (2023). Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice. J. Integr. Plant Biol. 65, 918–933. doi: 10.1111/jipb.13414
Li, J. D., Jin, H., Piao, S. L., Zou, J. X., Guo, P., Li, L. L., et al. (2017). Physiological response of leaves and roots of Leymus chinensis under drought and salt stress. Pratacultural Sci. 34, 1705–1710. doi: 10.11829/j.issn.1001G0629.2016G0528
Lim, C. W., Baek, W., Lee, S. C. (2018a). Roles of pepper bZIP protein CaDILZ1 and its interacting partner RING-type E3 ligase CaDSR1 in modulation of drought tolerance. Plant J. 96, 452–467. doi: 10.1111/tpj.14046
Lim, C. W., Hong, E., Bae, Y., Lee, S. C. (2018b). The pepper dehydration-responsive homeobox 1, CaDRHB1, plays a positive role in the dehydration response. Environ. Exp. Bot. 147, 104–115. doi: 10.1016/j.envexpbot.2017.11.015
Lim, C. W., Jeong, S., Baek, W., Choi, H., Lee, S. C. (2024). A positive role for caMEKK17 in response to drought stress, modulated by clade A PP2Cs. Plant Cell Environ. 1–14. doi: 10.1111/pce.15223
Lim, J., Lim, C. W., Lee, S. C. (2022). Role of pepper MYB transcription factor CaDIM1 in regulation of the drought response. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1028392
Liu, X. W., Chen, Z. L., Shen, J. M., Ye, M. M., Chen, W. H. (2010). Spectrophotometric determination of low concentration of hydrogen peroxide in O3/H2O2 system using titanium sulfate. China Water Wastewater 26, 126–129. doi: 10.19853/j.zgjsps.1000-4602.2010.16.035
Lu, C., Fedoroff, N. (2000). A mutation in the arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351. doi: 10.2307/3871234
Ma, W., Cui, H., Li, Y., Zheng, Y., Hu, J. (2011). Effects of Seed Soaking with Different Agents on Seed Germination and Seedling Growth in Tobacco ( Nicotiana tabacum L. ) under Low Temperature Stress. Bull. Sci. andTechnology 27, 873–880. doi: 10.13774/j.cnki.kjtb.2011.06.008
Ma, X., Li, Y., Gai, W. X., Li, C., Gong, Z. H. (2021). The CaCIPK3 gene positively regulates drought tolerance in pepper. Hortic. Res. 8, 1–14. doi: 10.1038/s41438-021-00651-7
Mahmood, T., Rana, R. M., Ahmar, S., Saeed, S., Gulzar, A., Khan, M. A., et al. (2021). Effect of drought stress on capsaicin and antioxidant contents in pepper genotypes at reproductive stage. Plants 10, 1286. doi: 10.3390/plants10071286
Ning, K., Chen, S., Huang, H., Jiang, J., Yuan, H., Li, H. (2017). Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell. Tissue Organ Cult. 130, 469–481. doi: 10.1007/s11240-017-1226-3
Noctor, G., Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Oh, S. K., Jeong, M. P., Young, H. J., Lee, S., Chung, E., Kim, S. Y., et al. (2005). A plant EPF-type zinc-finger protein, CaPIF1, involved in defence against pathogens. Mol. Plant Pathol. 6, 269–285. doi: 10.1111/j.1364-3703.2005.00284.x
Pan, Y., Seymour, G. B., Lu, C., Hu, Z., Chen, X., Chen, G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Sekulska-nalewajko, J., Gocławski, J., Chojak-koz, J. (2016). Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 109, 114–122. doi: 10.1016/j.ymeth.2016.05.018
Sewelam, N., Kazan, K., SchenK, P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00187
Singroha, G., Sharma, P., Sunkur, R. (2021). Current status of microRNA-mediated regulation of drought stress responses in cereals. Physiol. Plant 172, 1808–1821. doi: 10.1111/ppl.13451
Song, S., Qu, Z., Zhou, X., Wang, X., Dong, S. (2022). Effects of weak and strong drought conditions on physiological stability of flowering soybean. Plants 11, 2708. doi: 10.3390/plants11202708
Song, J., Sun, P., Kong, W., Xie, Z., Li, C., Liu, J. H. (2023). SnRK2.4-mediated phosphorylation of ABF2 regulates ARGININE DECARBOXYLASE expression and putrescine accumulation under drought stress. New Phytol. 238, 216–236. doi: 10.1111/nph.18526
Stewart, R. R. C., Bewley, J. D. (1980). Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 65, 245–248. doi: 10.1104/pp.65.2.245
Stone, J. M., Liang, X., Nekl, E. R., Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 1, 744–754. doi: 10.1111/j.1365-313X.2005.02334.x
Sun, X., Zhang, L., Xu, W., Zheng, J., Yan, M., Zhao, M., et al. (2024). A comprehensive analysis of the peanut SQUAMOSA promoter binding protein-like gene family and how ahSPL5 enhances salt tolerance in transgenic arabidopsis. Plants 13, 1057. doi: 10.3390/plants13081057
Wang, J. E. (2013). Expression analysis and functional identification of CaRGA1 and CaPOD genes induced by phytophthora capsici in pepper (Yangling, China: Institute of Horticulture, Northwest A&F University).
Wang, B., Li, L., Liu, M., Peng, D., Wei, A., Hou, B., et al. (2022). TaFDL2-1A confers drought stress tolerance by promoting ABA biosynthesis, ABA responses, and ROS scavenging in transgenic wheat. Plant J. 112, 722–737. doi: 10.1111/tpj.15975
Wang, B. X., Zeng, Y. H., Yong, W., Zhao, Yong, Xu, X. (2010). Responses of leaf stomata to environmental stresses in distribution and physiological characteristics. Agric. Res. Arid Areas 28, 122–126. doi: 10.3724/SP.J.1143.2010.00439
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Sci. (80-.). 361, 1026–1028. doi: 10.1126/science.aat7675
Wang, H., Zhu, S., Yang, C., Zeng, D., Luo, C., Dai, C., et al. (2024). Expression and functional identification of SPL6/7/9 genes under drought stress in sugarbeet seedlings. Int. J. Mol. Sci. 25, 1–18. doi: 10.3390/ijms25168989
Wei, H., Luo, M., Deng, J., Xiao, Y., Yan, H., Liu, H., et al. (2024). SPL16 and SPL23 mediate photoperiodic control of seasonal growth in Populus trees. New Phytol. 241, 1646–1661. doi: 10.1111/nph.19485
Wu, G., Park, M. Y., Conway, S. R., Wang, J.-W., Weige, D., Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.molp.2020.01.013
Xie, Y., Zhou, Q., Zhao, Y., Li, Q., Liu, Y., Ma, M., et al. (2020). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering. Mol. Plant. 13, 483–498. doi: 10.1016/j.molp.2020.01.013
Xu, Y., Burgess, P., Zhang, X., Huang, B. (2016). Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 67, 1979–1992. doi: 10.1093/jxb/erw019
Yang, Q., Wang, J., Zhang, S., Zhan, Y., Shen, J., Chang, F. (2023b). ARF3-mediated regulation of SPL in early anther morphogenesis: maintaining precise spatial distribution and expression level. Int. J. Mol. Sci. 24, 11740. doi: 10.3390/ijms241411740
Yang, J., Wei, H., Hou, M., Chen, L., Zou, T., Ding, H., et al. (2023a). ZmSPL13 and ZmSPL29 act together to promote vegetative and reproductive transition in maize. New Phytol. 239, 1505–1520. doi: 10.1111/nph.19005
Yin, Y. X., Guo, W. L., Zhang, Y. L., Ji, J. J., Xiao, H. J., Yan, F., et al. (2014). Cloning and characterisation of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell. Tissue Organ Cult. 118, 431–444. doi: 10.1007/s11240-014-0495-3
You, Z., Guo, S., Li, Q., Fang, Y., Huang, P., Ju, C., et al. (2023). The CBL1/9-CIPK1 calcium sensor negatively regulates drought stress by phosphorylating the PYLs ABA receptor. Nat. Commun. 14, 1–14. doi: 10.1038/s41467-023-41657-0
Zhang, H.-X. (2020). Functional Identification and Regulation Mechanism of Pepper SBP-box Gene in Response to Phytophthora Capsici Infection. (Yangling, China: Institute of Horticulture, Northwest A&F University).
Zhang, H., Ali, M., Feng, X., Jin, J., Huang, L., Khan, A., et al. (2018). A Novel Transcription Factor CaSBP12 Gene Negatively Regulates the Defense Response against Phytophthora capsici in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 20, 1–20. doi: 10.3390/ijms20010048
Zhang, H. X., Feng, X. H., Ali, M., Jin, J. H., Wei, A. M., Khattak, A. M., et al. (2020a). Identification of pepper caSBP08 gene in defense response against phytophthora capsici infection. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00183
Zhang, H. X., Feng, X. H., Jin, J. H., Khan, A., Guo, W. L., Du, X. H., et al. (2020b). CaSBP11 participates in the defense response of pepper to phytophthora capsici through regulating the expression of defense-related genes. Int. J. Mol. Sci. 21, 1–22. doi: 10.3390/ijms21239065
Zhang, Y. L., Jia, Q. L., Li, D. W., Wang, J. E., Yin, Y. X., Gong, Z. H. (2013). Characteristic of the pepper CaRGA2 gene indefense responses against phytophthora capsici leonian. Int. J. Mol. Sci. 14, 8985–9004. doi: 10.3390/ijms14058985
Zhang, H.-X., Jin, J.-H., He, Y.-M., Lu, B.-Y., Li, D.-W., Chai, W.-G., et al. (2016). Genome-Wide Identification and Analysis of the SBP-Box Family Genes under Phytophthora capsici Stress in Pepper (Capsicum annuum L.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00504
Zhang, H., Pei, Y., Zhu, F., He, Q., Zhou, Y., Ma, B., et al. (2023). CaSnRK2.4-mediated phosphorylation of CaNAC035 regulates abscisic acid synthesis in pepper (Capsicum annuum L.) responding to cold stress. Plant J. 117, 1–15. doi: 10.1111/tpj.16568
Zhang, H., Zhang, Y., Zhang, B. (2024). Pepper SBP-box transcription factor, CaSBP13, plays a negatively role in drought response. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1412685
Zhang, H. X., Zhu, W. C., Feng, X. H., Jin, J. H., Wei, A. M., Gong, Z. H. (2020c). Transcription factor CaSBP12 negatively regulates salt stress tolerance in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 21, 1-16. doi: 10.3390/ijms21020444
Zhao, Y., He, J., Liu, M., Miao, J., Ma, C., Feng, Y., et al. (2024). The SPL transcription factor TaSPL6 negatively regulates drought stress response in wheat. Plant Physiol. Biochem. 206, 108264. doi: 10.1016/j.plaphy.2023.108264
Zhao, J., Shi, M., Yu, J., Guo, C. (2022). SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol. 22, 1–13. doi: 10.1186/s12870-022-03445-8
Keywords: pepper, CaSBP11, drought stress, stomatal, ABA signaling pathway
Citation: Zhang H-X, Zhang Y, Zhang B-W and Pan F-F (2025) The CaSBP11 gene functions as a negative regulator in pepper drought stress. Front. Plant Sci. 16:1497425. doi: 10.3389/fpls.2025.1497425
Received: 17 September 2024; Accepted: 10 April 2025;
Published: 28 April 2025.
Edited by:
Surekha Katiyar-Agarwal, University of Delhi, IndiaReviewed by:
Paola Punzo, National Research Council (CNR), ItalyZhonglin Shang, Hebei Normal University, China
Copyright © 2025 Zhang, Zhang, Zhang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huai-Xia Zhang, aHVhaXhpYUBud2FmdS5lZHUuY24=
 Huai-Xia Zhang
Huai-Xia Zhang Yuan Zhang
Yuan Zhang Bo-Wen Zhang
Bo-Wen Zhang Fei-Fei Pan
Fei-Fei Pan