- 1School of Horticulture and Landscape Architecture, Henan Institute of Science and Technology, Xinxiang, China
- 2Henan Province Engineering Research Center of Horticultural Plant Resource Utilization and Germplasm Enhancement, Henan Institute of Science and Technology, Xinxiang, China
- 3Horticulture Research and Extension, The Postharvest Education Foundation, La Pine, OR, United States
The miR159 gene family plays an essential role in plant growth and development, and stress response. Nevertheless, there are no reports defining its specific function in cucumber fruit expansion and response to abiotic stresses. In this study, we retrieved six Csa-miR159 sequences from the EnsemblPlants database, which were located on chromosome 1, chromosome 3, and chromosome 5 of cucumber, respectively. Phylogenetic analysis showed that Csa-miR159c/d/e/f belonged to one branch and Csa-miR159a/b to another. Cis-acting regulatory elements (CREs) including light response elements, phytohormone response elements, stress response elements, regulatory elements associated with plant growth and development were distributed unevenly in the promoter regions of Csa-miR159s, which indicated that Csa-miR159s might mediate the stress response, and growth and development. Moreover, it was determined that CsMYBs were the target genes of Csa-miR159s through psRNA-Target prediction and qRT-PCR analysis. Further findings suggested that Csa-miR159b might negatively regulate cucumber fruit expansion by targeting Cs1RMYB9, Cs1RMYB31, Cs2RMYB37 and Cs2RMYB64. Similarly, Csa-miR159d might negatively regulate cucumber fruit expansion by targeting Cs2RMYB27 and Cs2RMYB32. In addition, the differential expression of Csa-miR159s suggested their potential response to abiotic stresses and plant phytohormones. This study would provide valuable information on the molecular characterization of Csa-miR159s and establish a foundation for further research on the mechanisms of Csa-miR159s in regulating fruit expansion and stress response.
1 Introduction
Cucumber (Cucumis sativus L.) is a widely cultivated vegetable worldwide, with China being the largest producer and consumer. In 2023, China cultivated approximately 1.37 million hectares of cucumber, with a total yield of 80.21 million tons. This accounted for 60.40% of the global cucumber cultivation area and 82.01% of the total production (Food and Agriculture Organization of the United Nations, 2023). However, unfavorable environmental conditions such as inappropriate temperature, weak light, drought and salt stresses, have a significant impact on cucumber fruit expansion, leading to the decreased yield and quality. Therefore, it is important to understand the molecular mechanism of cucumber fruit expansion in order to improve yield and stress tolerance in cucumber.
MicroRNA (miRNA) is a class of highly conserved endogenous non-coding small RNA, regulating the expression of its target genes at both the levels of transcription and post-transcription by directly cleaving or inhibiting the translation of target mRNA (Jones-Rhoades et al., 2006). It plays crucial roles in various physiological and metabolic processes, such as plant growth and development, and stress response (Jeong and Green, 2013; Liu et al., 2007; Zhang et al., 2019). miR159 has been extensively studied in plants (Montes et al., 2014) and the studies reveal how it influences plant growth and development by targeting MYB family genes (Dubos et al., 2010). For instance, miR159-GAMYB pathway has been widely implicated in plant growth, stress response, and phytohormone signaling in various species such as Arabidopsis (Allen et al., 2010, 2007; Alonso-Peral et al., 2010), tomato (Zhang et al., 2020) and rice (Zhao et al., 2017). In Gloxinia (Sinningia speciosa), expression patterns of miR159 and GAMYB were negatively correlated during flower development (Li et al., 2013). In addition, some studies have demonstrated the important role of the miR159-GAMYB in fruit development. For instance, in tomato, overexpression of Sl-MIR159 led to the down-regulation of SlGAMYB, thereby inducing parthenocarpy and early fruit ripening (da Silva et al., 2017). Similarly, Sly-miR159-SlGAMYB2 was also found to control fruit growth, as the inhibition of Sly-miR159 and overexpression of SlGAMYB2 resulted in the larger fruit, while the loss of function of SlGAMYB2 led to the smaller fruit (Zhao et al., 2022). In the case of grape, exogenous application of gibberellin (GA) promoted parthenocarpy, accompanied by the up-regulation of Vvi-miR159c and the down-regulation of VvGAMYB (Wang et al., 2018).
The miR159-GAMYB pathway is known to play a crucial role in the response to drought and salt stresses. Studies have shown that miR159 was induced by drought stress in plants such as Arabidopsis (Reyes and Chua, 2007), maize (Wei et al., 2009), wheat (Akdogan et al., 2016), barley (Hackenberg et al., 2015) and poplar (Fu et al., 2023). However, in potato, the expression level of miR159 decreased under drought treatment, while the expression level of GAMYB-like homologues increased (Pieczynski et al., 2013). SlMYB33, the target gene of Sly-miR159, was associated with the accumulation of proline and putrescine, which enhanced plant tolerance to drought stress (López-Galiano et al., 2019). Furthermore, it has been reported that miR159 can be induced by salt stress in Arabidopsis (Liu et al., 2008) and soybean (Li et al., 2023). Additionally, miR159-GAMYB plays a crucial role in some plant phytohormone signaling pathways, such as abscisic acid (ABA) (Reyes and Chua, 2007) and GA (Wang et al., 2017). For instance, in ‘Zuijinxiang’ grape, the expression level of VvimiR159 increased after GA treatment, while the expression level of VvGAMYB significantly decreased (Wang et al., 2018). In ‘Rosario Bianco’ grape, the expression of miR159 was up-regulated in the pulp after GA treatment, whereas the expression of miR159a/c was down-regulated in the pulp and pericarp (Han et al., 2014). Overall, the miR159-GAMYB pathway plays a role in response to abiotic stresses and plant phytohormones. In our previous study, differential expression of Csa-miR159b was observed between the ovary and expanded fruit using small RNA sequencing, which suggested that Csa-miR159b was involved in cucumber fruit expansion (Sun et al., 2019). However, there was a scarcity of studies on the functions of Csa-miR159s in relation to cucumber fruit expansion and stress response.
This study aims to characterize Csa-miR159s in cucumber and to investigate their roles in fruit expansion and stress response. In the present study, multiple sequence alignment, chromosomal location, secondary structure, phylogenetic relationship, cis-regulatory elements (CREs), and the target genes of Csa-miR159s were studied in detail. Additionally, expression profiles of Csa-miR159s were analyzed in the ovary and expanded fruit, and in response to different stresses and plant phytohormones. Our findings will provide valuable information for further functional analysis of Csa-miR159s in cucumber, and also provide references for improving cucumber yield and resilience.
2 Materials and methods
2.1 Identification of Csa-miR159s
A search for miR159 family members in cucumber was conducted using EnsemblPlants database (http://plants.ensembl.org/). The mature sequences of miR159s from various crop species (zucchini, watermelon, pumpkin, cucumber, melon, tomato, rice and Arabidopsis) were obtained from the PmiREN database (Guo et al., 2020). Multiple sequence alignments of Csa-miR159s were performed using ClustalW software (Thompson et al., 1994), and were used to generate a sequence logo diagram through the online website (https://weblogo.berkeley.edu/). TBtools software (Chen et al., 2023) was employed to visualize the distribution of Csa-miR159s on cucumber chromosomes. The RNA secondary structure of pre-MIR159s was predicted using the RNAstructure web server (http://rna.urmc.rochester.edu/RNAstructureWebServers/Predictl/Predictl.html). Mature sequences of miR159s were submitted to MEGA v5.1 software (Kumar et al., 2018) to construct phylogenetic relationships using the neighbor-joining (NJ) method with 1000 bootstrap replicates to assess branch confidence. The 2000 bp promoter sequences upstream from the initiation codon of Csa-MIR159s were extracted from EnsemblPlants database. The putative cis-regulatory elements (CREs) were identified and analyzed using the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
2.2 Prediction of target genes
psRNA-Target uses sequence complementarity and energy-based scoring to predict miRNA-target interactions. A score threshold of ≤5.0 was chosen based on established standards for psRNA-Target to ensure high confidence in predicted interactions. To predict the potential target relationships of Csa-miR159s and CsMYBs, their gene sequences were submitted to the psRNA-Target online website (https://www.zhaolab.org/psRNATarget/) (Liu et al., 2015), and target genes with a score ≤5.0 was selected and submitted to Cucurbit Genomics Database (CuGenDB) for further analysis.
2.3 Plant growth conditions and stress treatments
Seeds of cucumber (cv. Jinyou No. 1) were soaked in water at a temperature of 55°C for 15 min and then incubated at 28°C for 2 days to germinate. The germinated seeds were cultivated in a pot filled with a medium consisting of peat soil, perlite, and vermiculite in a 2:1:1 ratio, and placed in a climate-controlled chamber at a temperature of 28°C with a light period of 16 h and a dark period of 8 h. Cucumber seedlings at the three-leaf stage were transferred to the plastic greenhouse for continuous growth. Samples from ovary (on the day of anthesis), and expanded fruit (5 days after anthesis) were collected for gene expression analysis. For drought and NaCl stresses, cucumber seedlings at the three-leaf stage with a similar size and height were cultured into 40 L (113 cm × 73 cm × 5 cm) hydroponic pots. The control group was cultured in Hoagland nutrient solution. Drought stress was induced using a Hoagland nutrient solution containing 10% PEG-6000, while NaCl stress was induced using a Hoagland nutrient solution containing 150 mmol/L NaCl (Lu et al., 2022). Leaves were collected at 0, 3, 6, 12, and 24 h after treatment for gene expression analysis. For plant phytohormone treatments, the cucumber seedlings at the three-leaf stage were sprayed with 100 µmol L-1 ABA, 100 µmol L-1 salicylic acid (SA), 100 µmol L-1 jasmonic acid (JA), 50 µmol L-1 ethephon (ETH), 50 µmol L-1 2,4-dichlorophenoxyacetic acid (2,4-D), and 50 µmol L-1 GA, respectively, while the control condition was sprayed with double distilled water (Li et al., 2019). Plant phytohormone treatments were conducted once a day. After three consecutive days of treatment, cucumber leaves were collected for gene expression analysis. All treatments were performed with three biological replicates.
2.4 qRT-PCR analysis
RNA was isolated from various cucumber tissues using the TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). Isolated RNA was stored at -80°C until further use to prevent degradation. The Mir-X miRNA First-Strand Synthesis Kit (TaKaRa, Dalian, China) was then utilized for first-strand complementary DNA (cDNA) synthesis. qRT-PCR was conducted using the TB Green® Premix Ex TaqTM II (Tli RnaseH Plus) (TaKaRa, Dalian, China), using U6 snRNA as the endogenous control for Csa-miR159s, and 18S as the endogenous control for CsMYBs. Stem-loop of mature Csa-miR159s was used for qRT-PCR. The specific primer sequences utilized in this study were provided in detail in Supplementary Table S1. Gene expression levels were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001), each expression level was evaluated using three biological replicates.
3 Results
3.1 Identification of Csa-miR159s
Six Csa-miR159 sequences were identified from the EnsemblPlants database. The mature sequences of Csa-miR159s were 21-22 nt in length and highly conserved (Figure 1A). These sequences were mapped to chromosome 1 (Csa-miR159a), chromosome 3 (Csa-miR159b), and chromosome 5 (Csa-miR159c/d/e/f), respectively, based on their physical positions (Figure 1B). Although Csa-miR159c/d/e/f were located on the same chromosome, their mature sequences showed lower similarity. In contrast, Csa-miR159a and Csa-miR159b, which were located on the different chromosomes, shared higher similarity.
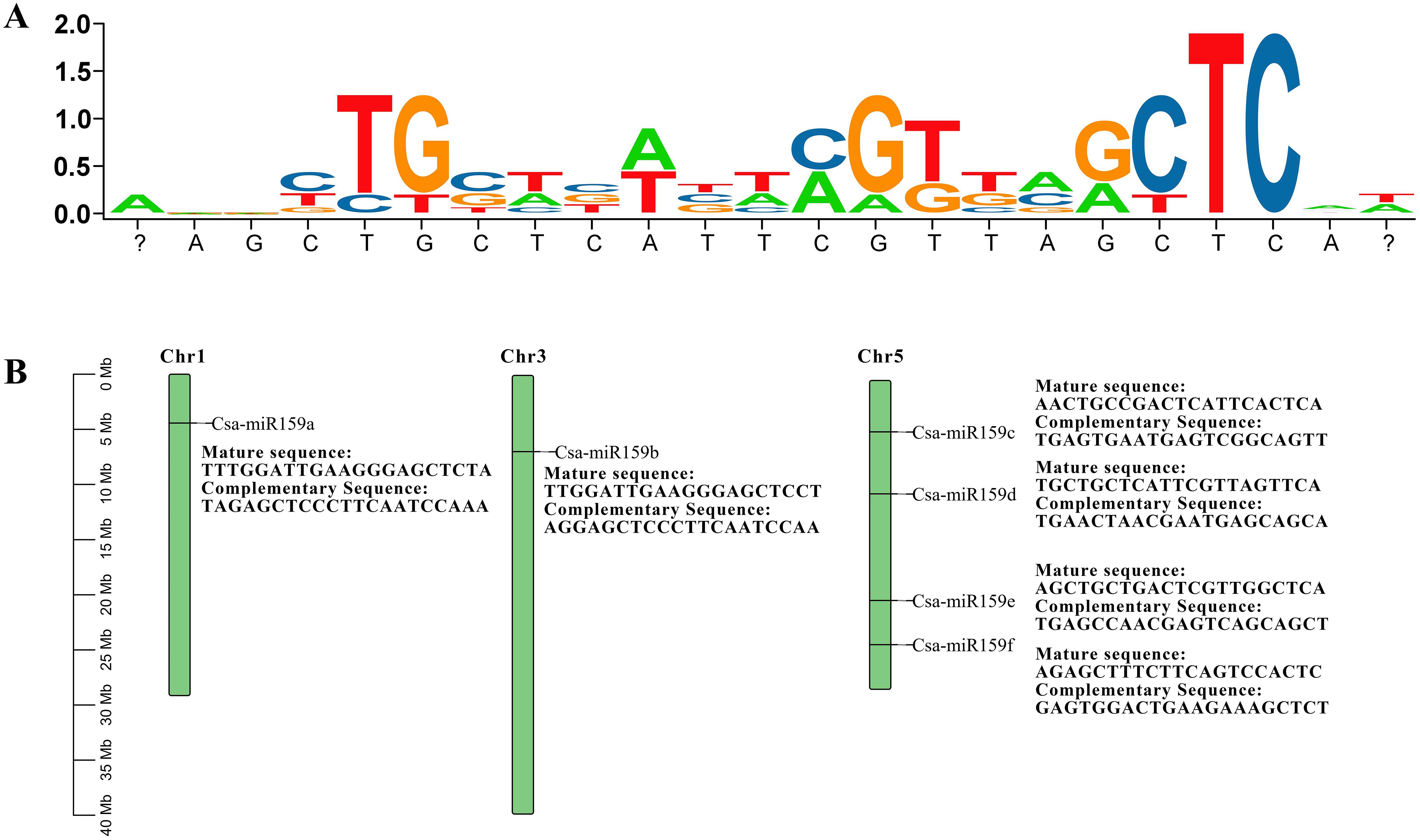
Figure 1. Mature sequences and chromosomal positions of Csa-miR159s. The ratio is measured in megabases (Mb). (A) Mature sequences; (B) Chromosomal positions.
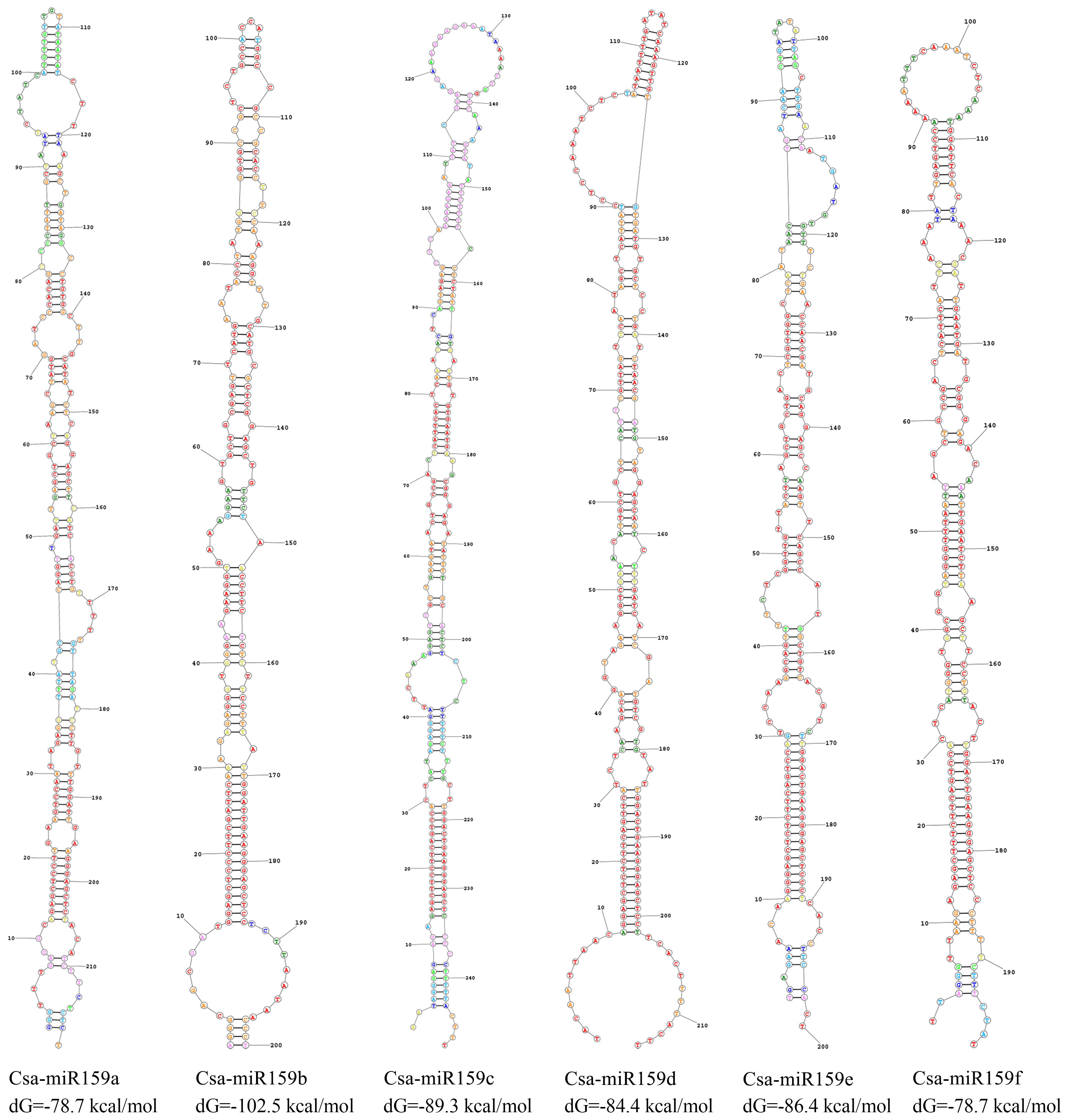
The prediction result of secondary structure showed that all pre-miR159s demonstrated a typical stem-loop structures (Figure 2). The number of sub-loops varied from 12 (Csa-miR159f) to 18 (Csa-miR159a), and the stem-loop folding free energy ranged from -102.5 kcal/mol (pre-miR159b) to -78.7 kcal/mol (Csa-miR159a/f) (Figure 2).
3.2 Phylogenetic relationship of miR159s
To better understand the evolutionary relationships among miR159s, we further analyzed the mature sequences from cucumber (Csa-miR159a/b/c/d/e/f), melon (Cme-miR159a/b), zucchini (Cma-miR159a/b/c/d), pumpkin (Cmo-miR159a/b), watermelon (Cla-miR159a/b), tomato (Sly-miR159a/b), rice (Osa-miR159a/b/c/d/e/f), and Arabidopsis (Ath-miR159a/b/c) (Figure 3). Twenty-seven miR159s were classified into two branches based on the evolutionary divergence. Csa-miR159c/d/e/f and Cma-miR159a/b belonged to one branch. Csa-miR159a/b were classified into another branch with remaining members. The phylogenetic tree revealed that cucumber miR159s share closer evolutionary relationships with those of zucchini.
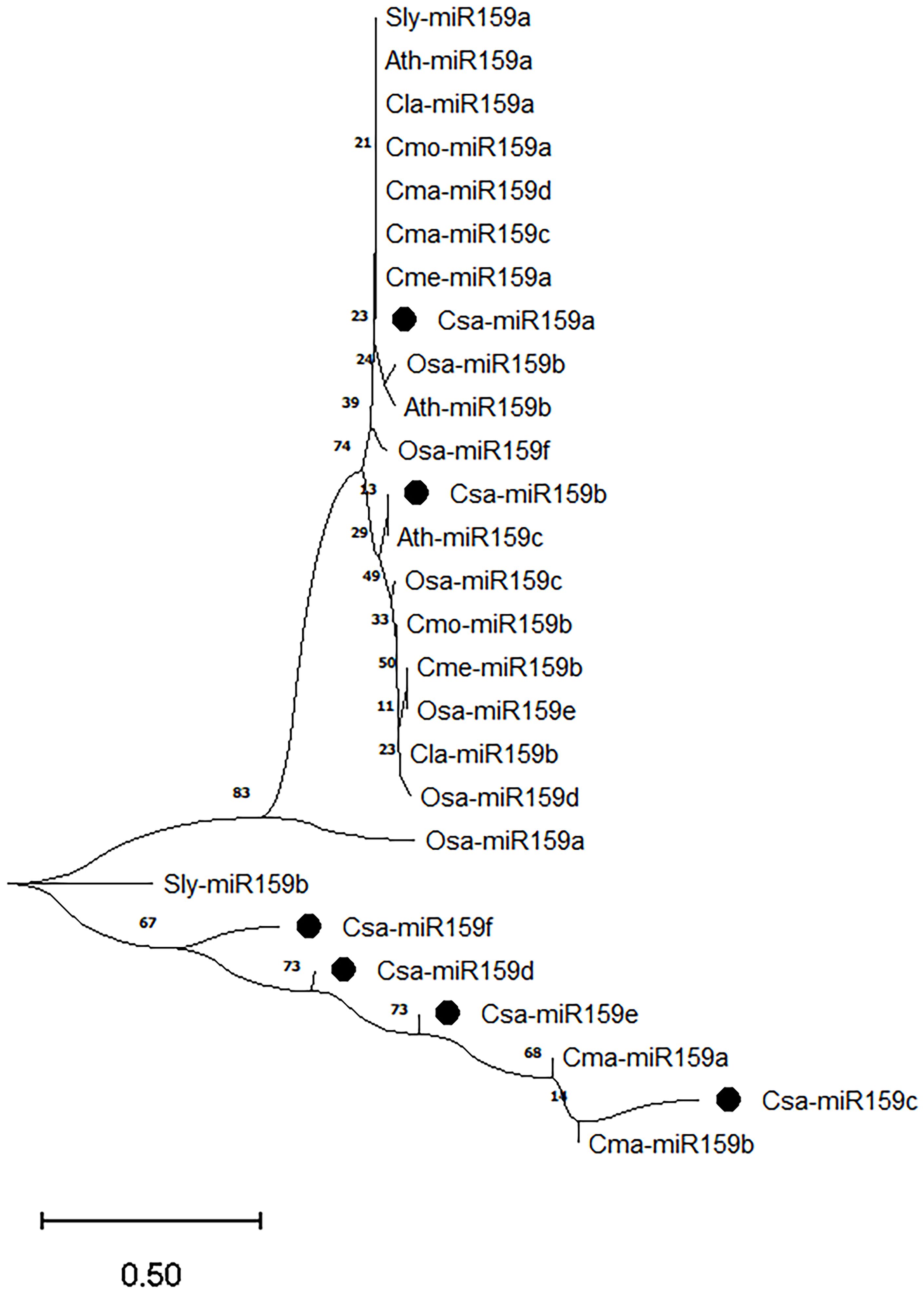
Figure 3. Phylogenetic analysis of miR159s from cucumber, melon, zucchini, pumpkin, watermelon, tomato, rice and Arabidopsis. Csa, cucumber; Cme, melon; Cma, zucchini; Cmo, pumpkin; Cla, watermelon; Sly, tomato; Osa, rice; Ath, Arabidopsis.
3.3 Cis-regulatory elements analysis of Csa-miR159s
To investigate the potential functions of Csa-miR159s, we analyzed the CREs in the promoter regions. As shown in Figure 4, these CREs were grouped into four functional categories. The most abundant category was light response elements, which included Box 4, AAAC-motif, G-Box, TCT-motif, AE-box, GATA-motif, GT1-motif, I-box, GA-motif, TCCC-motif, ATCT-motif, chs-CMA2a, ATC-motif, and MRE. We also detected various phytohormone response elements, such as abscisic acid responsiveness (ABRE), gibberellin responsiveness (TATC-box and P-box), ethylene responsiveness (ERE), MeJA responsiveness (CGTCA-motif, TGACG-motif) and salicylic acid responsiveness (TCA-element, SARE). Furthermore, stress response elements were identified, including anaerobic induction (ARE), drought inducibility (MBS), heat induction (STRE), low temperature responsiveness (LTR), wound responsiveness (WUN-motif), and defense and stress responsiveness (TC-rich). Additionally, regulatory elements related to plant growth and development were also identified, such as zein metabolism regulation (O2-site), meristem expression (CAT-box), endosperm expression (GCN4-motif) and circadian control (circadian).
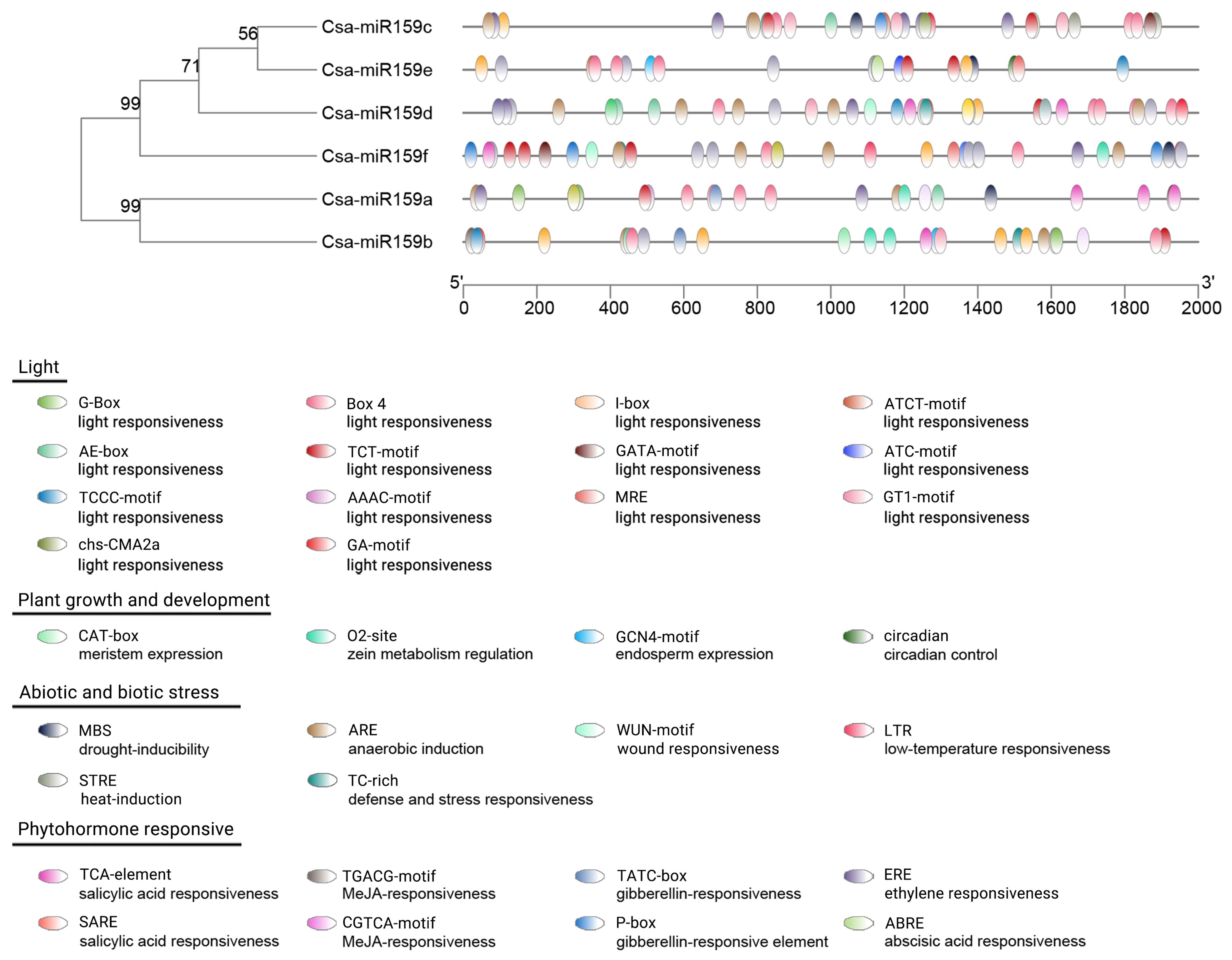
Figure 4. CREs in the promoter regions of Csa-miR159s. Different CRE was presented in the different color shape.
3.4 Prediction of target genes of Csa-miR159s
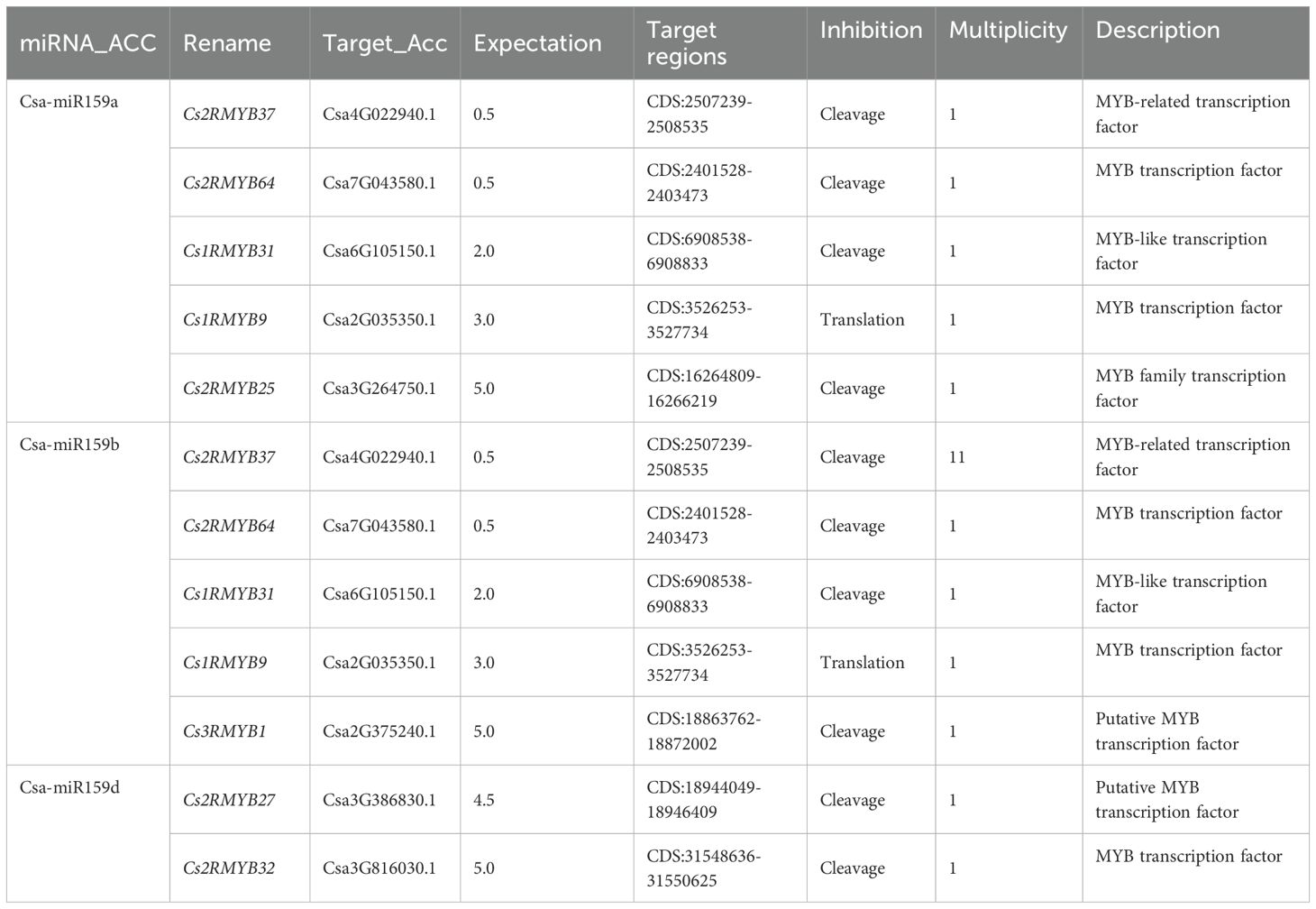
To investigate the regulatory mechanisms of Csa-miR159s, potential target genes were predicted using the psRNA-Target tool (Table 1). All target genes were named according to their subgroups and chromosomal positions from top to bottom (from Cs1RMYB to Cs4RMYB). The results revealed that Cs2RMYB37, Cs2RMYB64, Cs1RMYB31, Cs1RMYB9 and Cs2RMYB25 were recognized as the target genes of Csa-miR159a. Similarly, Cs2RMYB37, Cs2RMYB64, Cs1RMYB31, Cs1RMYB9 and Cs3RMYB1 were predicted as the target genes of Csa-miR159b. Additionally, Cs2RMYB27 and Cs2RMYB32 were found to be the target genes of Csa-miR159d. However, no target genes were detected for Csa-miR159c, Csa-miR159e and Csa-miR159f. Notably, all predicted target genes were classified as MYB or MYB-like transcription factors.
3.5 Expression profiles of Csa-miR159s and their target genes
In this study, qRT-PCR was used to confirm the expression profiles of Csa-miR159s and their target genes in the ovary and expanded fruit (Figure 5). The expression of Csa-miR159b in the ovary was 2.44-fold higher than that in the expanded fruit, while the expression levels of Csa-miR159a/c/e/f in the ovary were lower than those in the expanded fruit. Notably, Csa-miR159d was only expressed in the ovary and was not detected in the expanded fruit. In terms of the target genes, Cs1RMYB9, Cs1RMYB31, Cs2RMYB37, Cs2RMYB64, Cs2RMYB27 and Cs2RMYB32 showed lower expression in the ovary compared to the expanded fruit, except for Cs3RMYB1 which exhibited the opposite trend. These findings suggested that Csa-miR159b might negatively regulate cucumber fruit expansion by targeting Cs1RMYB9, Cs1RMYB31, Cs2RMYB37 and Cs2RMYB64. Additionally, Csa-miR159d might negatively regulate cucumber fruit expansion by targeting Cs2RMYB27 and Cs2RMYB32. Taken together, these results highlighted the tissue specificity and functional diversity of Csa-miR159s.
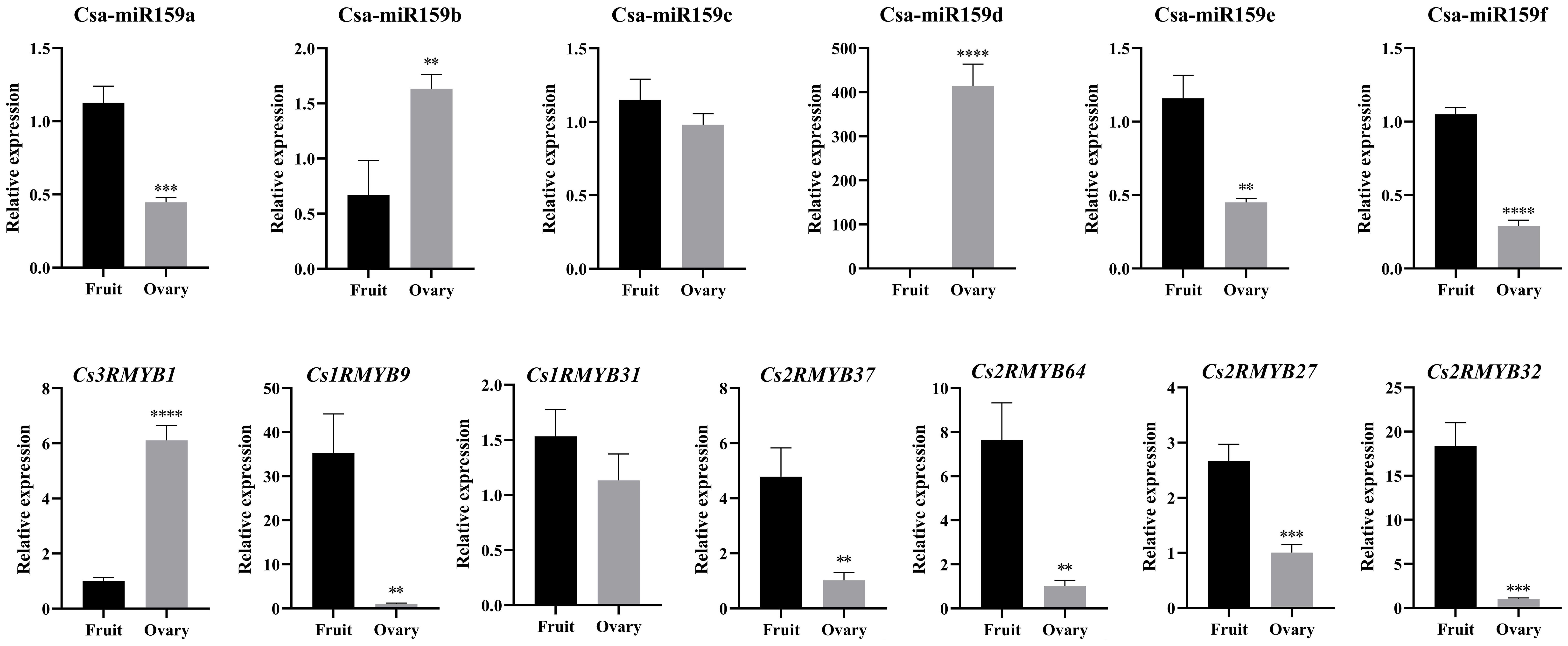
Figure 5. Relative expression of Csa-miR159s and CsMYBs in the ovary and the expanded fruit of cucumber by qRT-PCR. The X-axis indicated the tested tissue samples. Error bars represented ± standard deviation (SD) with three biological replicates. Different asterisks above the bars indicated significant differences. (** p < 0.01, *** p < 0.001, **** p < 0.0001).
3.6 Csa-miR159s response to abiotic stresses and plant phytohormones
The study investigated the expression patterns of Csa-miR159s under different treatments including PEG, NaCl and plant phytohormones. Under PEG stress, the expression levels of Csa-miR159a/f increased gradually, reaching the top at 24 h after treatment. Specifically, Csa-miR159a was 24.4-fold higher than the control, and Csa-miR159f was 174.1-fold higher. Conversely, Csa-miR159c exhibited the increased expression, peaking at 12 h, and then decreasing to the lowest level at 24 h after treatment, 1.3-fold lower than the control. However, Csa-miR159b/d/e were significantly down-regulated under PEG stress. Compared to the control, their expression levels decreased by 76.9, 2.4 and 4.3-fold, respectively (Figure 6). Under NaCl stress, Csa-miR159a/d/e were all significantly up-regulated at 6 h, with increases of 88.5, 33.2 and 3.9-fold compared to the control. While Csa-miR159b was remarkably down-regulated from 3 h to 24 h. In contrast, the expression levels of Csa-miR159c initially decreased at 3 h, then increased at 6 h, and peaked at 24 h. Csa-miR159f showed the increased expression, peaking at 12 h, and then decreasing at 24 h (Figure 7). In relation to plant phytohormones, it was observed that Csa-miR159a/b/c were significantly induced by SA, ETH and 2,4-D. On the other hand, Csa-miR159d showed significant up-regulation in response to ABA, GA, SA, ETH, and 2,4-D. Furthermore, Csa-miR159e exhibited a remarkable down-regulation when exposed to ABA, GA, JA, ETH, and 2,4-D. Additionally, Csa-miR159f displayed significant up-regulation specifically in response to SA, ETH, and 2,4-D (Figure 8). These findings indicated that Csa-miR159s might be involved in plant stress response and phytohormone regulation.
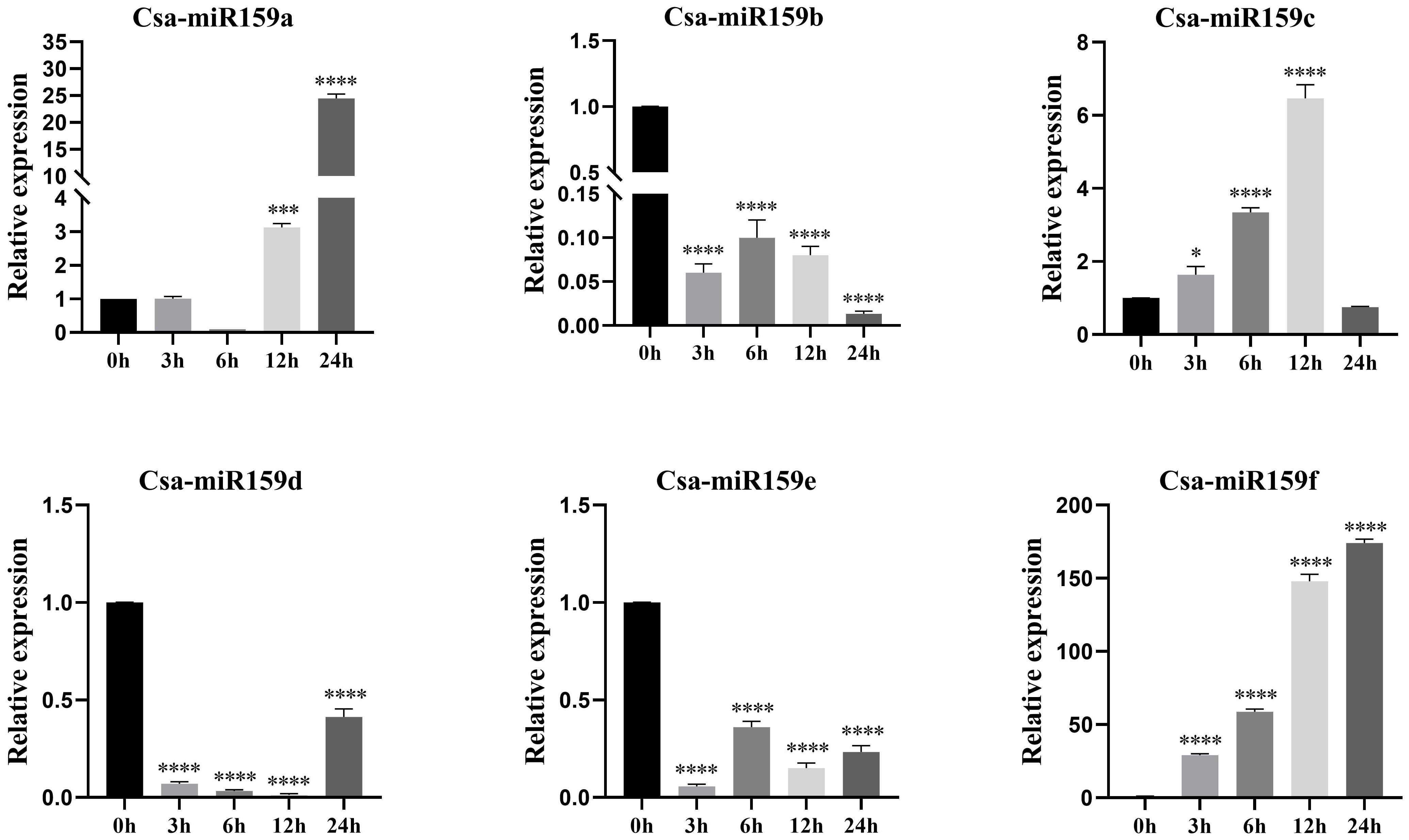
Figure 6. Relative expression of Csa-miR159s in response to 10% PEG-6000 treatment (0, 3, 6, 12 and 24 h). Error bars represented ± standard deviation (SD) with three biological replicates. Different asterisks above the bars indicated significant differences. (* p < 0.05, *** p < 0.001, **** p < 0.0001).
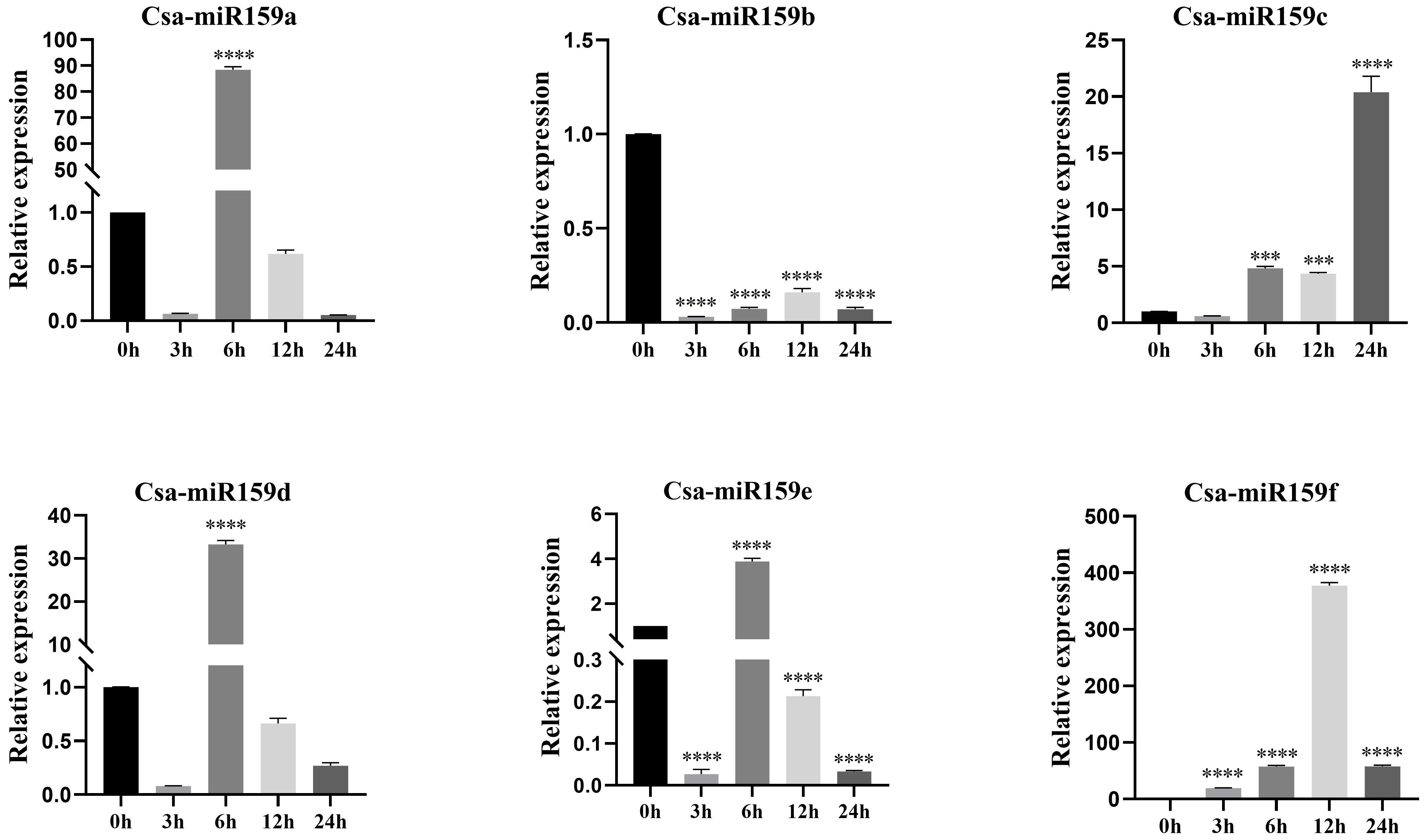
Figure 7. Relative expression of Csa-miR159s in response to 150 mmol/L NaCl treatment (0, 3, 6, 12 and 24 h). Error bars represented ± standard deviation (SD) with three biological replicates. Different asterisks above the bars indicated significant differences. (*** p < 0.001, **** p < 0.0001).
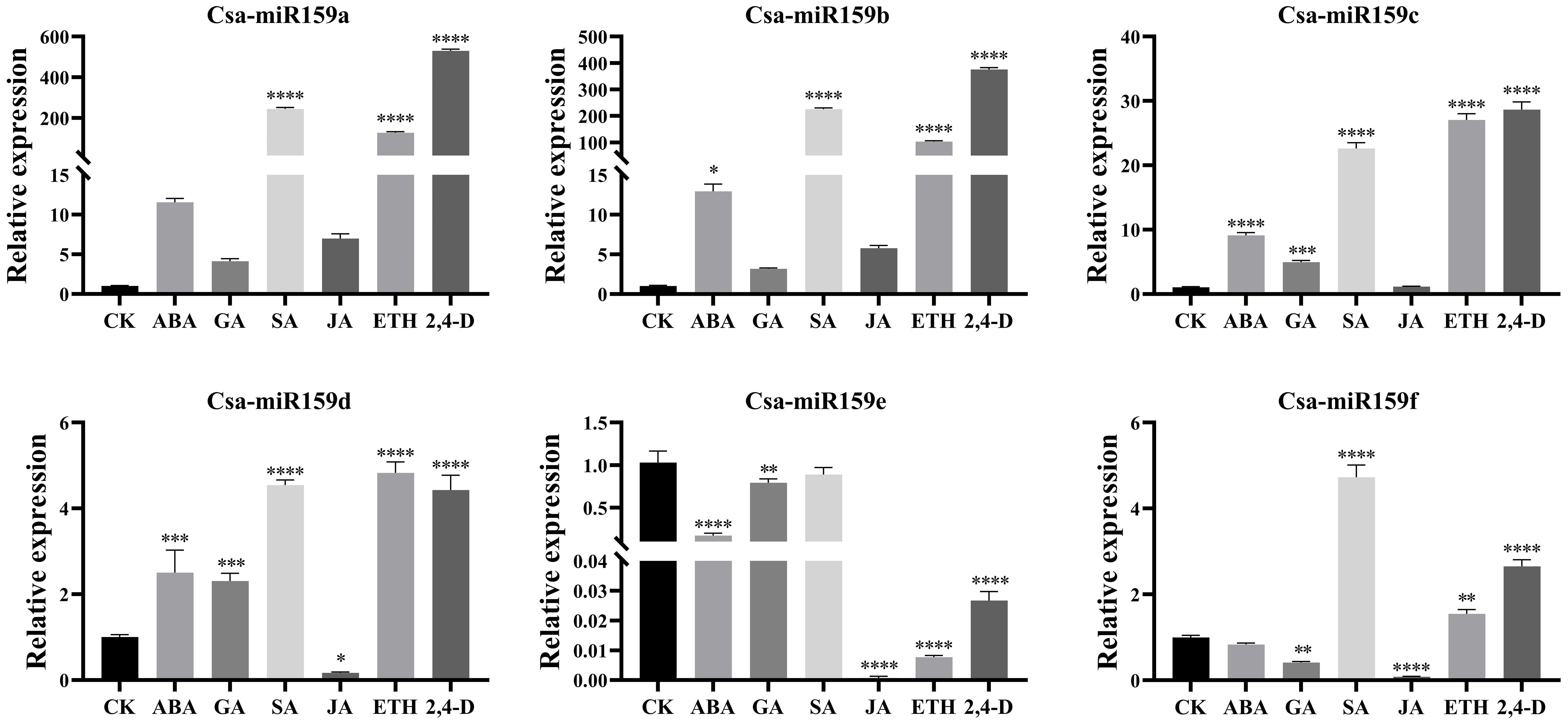
Figure 8. Relative expression of Csa-miR159s in response to different plant phytohormones, such as, ABA, GA, SA, JA, ETH and 2,4-D. Error bars represented ± standard deviation (SD) with three biological replicates. Different asterisks above the bars indicated significant differences. (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
4 Discussion
miRNA plays a crucial role in plant growth and development, and stress response by regulating the expression of their target genes. Among these miRNAs, miR159 has been extensively identified and characterized in numerous plant species, such as Arabidopsis (Palatnik et al., 2007), grape (Zhang et al., 2019), soybean (Li et al., 2023) and Dendrobium officinale (Hao and Zhang, 2022). However, there is limited research on the functions of Csa-miR159s regarding fruit expansion and abiotic stress response in cucumber. In this study, we identified six Csa-miR159s and their target genes. Csa-miR159s were unevenly distributed on chromosome 1, chromosome 3, and chromosome 5. Interestingly, Csa-miR159c/d/e/f, which had different mature sequences, were located on the same chromosome. Conversely, Csa-miR159a/b, which had higher sequence homology, were distributed on the different chromosomes. This suggested that the divergent chromosomal localization of Csa-miR159s could be a result of gene duplication events or evolutionary pressures that had caused their dispersion across different chromosomes, and this dispersion might have promoted the diversified functions of Csa-miR159s. Phylogenetic analysis revealed a close relationship between miR159s in cucumber and those in zucchini, possibly due to common evolutionary processes as the members of the Cucurbitaceae family.
The promoter region typically contains specific CREs with distinct functions. Analysis of CREs can offer insights into the potential functions of genes in the growth and development, and stress response. In our study, we identified stress-related elements in Csa-miR159s, including MBS, ARE, LTR, STRE and TC-rich, which suggested that Csa-miR159s might be associated with stress response in cucumber. Previous studies have demonstrated that up-regulated expression of miR159 enhanced stress tolerance in Arabidopsis (Reyes and Chua, 2007; Liu et al., 2008) and sweet potato (Yang et al., 2020). Conversely, some reports indicated that miR159 was down-regulated under salt stress and drought stress (Yang et al., 2020), and overexpression of miR159 increased stress sensitivity in rice (Wang et al., 2012) and potato (Pieczynski et al., 2013). In addition, Peng et al. (2018) observed that the expression of miR159 in rice was down-regulated after 3 h of salt stress treatment, followed by up-regulation. Our results confirmed that Csa-miR159s could respond to PEG and NaCl stresses by qRT-PCR, and significant differences were observed in their expression patterns. Additional experimental validation is necessary to elucidate the transcriptional regulation mechanisms of Csa-miR159s under PEG and NaCl stresses.
Several studies have indicated that miR159 can respond to some plant phytohormones, including ABA (Reyes and Chua, 2007) and GA (Wang et al., 2017). For instance, the application of exogenous GA led to a significant decrease in the expression level of Fa-miR159a, while the expression level of Fa-miR159b remained unchanged in strawberry (Csukasi et al., 2012). Similarly, exogenous GA treatment resulted in the up-regulated expression of VvmiR159c during flowering, whereas VvmiR159a/b showed no significant changes in grape (Wang et al., 2018). In this study, CREs of six Csa-miR159s included various plant phytohormone response elements such as ABA, GA, SARE, ABRE, MeJA and ETH. Csa-miR159s exhibited distinct expression patterns under ABA, GA, SA, JA, ETH and 2,4-D treatments. This suggested that Csa-miR159s might play a significant role in plant phytohormone signaling pathways with varying response mechanisms.
Fruit development is a crucial stage in the life cycle of plants, encompassing a variety of intricate physiological and molecular processes. miR159 has been found to be crucial for ovule development and fruit set in tomato. Overexpression of Sly-miR159 caused abnormal ovule development, premature maturation, and seedless fruit in tomato (da Silva et al., 2017; Deng, 2020). Furthermore, Sly-miR159-SlGAMYB2 pathway has been identified to regulate fruit morphology, whereby inhibition of Sly-miR159 led to larger fruit and a reduced length/width ratio (Zhao et al., 2022). Here, we discovered that the expression of Csa-miR159b/d was significantly higher in the ovary than that in the expanded fruit. Conversely, the expression levels of Csa-miR159a/c/e/f in the ovary were lower than those in the expanded fruit. These findings suggested that Csa-miR159s could be involved in fruit expansion in cucumber. miRNA regulates the growth and development in plants by inhibiting the expression of its target genes. In this study, Cs2RMYB37, Cs2RMYB64, Cs1RMYB31, Cs1RMYB9 and Cs3RMYB1 were predicted as the target genes of Csa-miR159b based on the PsRNA-Target results. While, Cs2RMYB27 and Cs2RMYB32 were found to be the target genes of Csa-miR159d. Furthermore, we found that Cs1RMYB9, Cs1RMYB31, Cs2RMYB37, Cs2RMYB64, Cs2RMYB27 and Cs2RMYB32 showed lower expression in the ovary compared to the expanded fruit by qRT-PCR. These findings suggested that Csa-miR159b might negatively regulate cucumber fruit expansion by targeting Cs1RMYB9, Cs1RMYB31, Cs2RMYB37 and Cs2RMYB64. Similarly, Csa-miR159d might negatively regulate cucumber fruit expansion by targeting Cs2RMYB27 and Cs2RMYB32. Taken together, our results suggested a vital role of Csa-miR159s in fruit expansion and stress response in cucumber. Further research is required to comprehend the functions of Csa-miR159s by performing a gain of function or loss of function assay.
5 Conclusions
In this study, six miR159 family members were identified in cucumber. Bioinformatics and expression profiles of Csa-miR159s were performed to discover their potential functions. The results showed that Csa-miR159s played a crucial role in the response to PEG, NaCl and plant phytohormones. Additionally, it was found that Csa-miR159b/d might inhibit the cucumber fruit expansion by targeting their target genes. Our study provided a theoretical foundation for further investigation into the roles of Csa-miR159s under fruit expansion and abiotic stresses in cucumber.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZZ: Formal Analysis, Investigation, Methodology, Writing – original draft. WA: Formal Analysis, Investigation, Methodology, Writing – original draft. WL: Data curation, Formal Analysis, Software, Writing – review & editing. YAS: Data curation, Formal Analysis, Software, Writing – review & editing. VT: Data curation, Formal Analysis, Software, Writing – review & editing. JL: Data curation, Formal Analysis, Software, Writing – review & editing. SZ: Data curation, Formal Analysis, Software, Writing – review & editing. YOS: Conceptualization, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Key Science and Technology Project of Henan Province (No. 242102111125; No. 232102110177), Year 2024 Research Funding Program Based on Merit for Overseas Persons in Henan Province (No. 202404).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1518406/full#supplementary-material
References
Akdogan, G., Tufekci, E. D., Uranbey, S., Unver, T. (2016). miRNA-based drought regulation in wheat. Funct. Integr. Genomics 16, 221–233. doi: 10.1007/s10142-015-0452-1
Allen, R. S., Li, J., Alonso-Peral, M. M., White, R. G., Gubler, F., Millar, A. A. (2010). MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence. 1, 18. doi: 10.1186/1758-907x-1-18
Allen, R. S., Li, J., Stahle, M. I., Dubroue, A., Gubler, F., Millar, A. A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. U.S.A. 104, 16371–16376. doi: 10.1073/pnas.0707653104
Alonso-Peral, M. M., Li, J., Li, Y., Allen, R. S., Schnippenkoetter, W., Ohms, S., et al. (2010). The MicroRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 154, 757–771. doi: 10.1104/pp.110.160630
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Csukasi, F., Donaire, L., Casanal, A., Martinez-Priego, L., Botella, M. A., Medina-Escobar, N., et al. (2012). Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 195, 47–57. doi: 10.1111/j.1469-8137.2012.04134.x
da Silva, E. M., Silva, G., Bidoia, D. B., Azevedo, M. D., de Jesus, F. A., Pino, L. E., et al. (2017). microRNA159-targeted SIGAMYB transcription factors are required for fruit set in tomato. Plant J. 92, 95–109. doi: 10.1111/tpj.13637
Deng, Y. (2020). Mechanism of Sly-miR159a regulating tomato fruit shape (Shenzhen Univeristy: Shenzhen).
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Food and Agriculture Organization of the United Nations (2023). Available online at: http://www.faoorg/faostat/ (Accessed 25 August 2023).
Fu, T., Wang, C., Yang, Y., Yang, X., Wang, J., Zhang, L., et al. (2023). Function identification of miR159a, a positive regulator during poplar resistance to drought stress. Hortic. Res. 10, 17. doi: 10.1093/hr/uhad221
Guo, Z., Kuang, Z., Wang, Y., Zhao, Y., Tao, Y., Cheng, C., et al. (2020). PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 48, D1114–D1121. doi: 10.1093/nar/gkz894
Hackenberg, M., Gustafson, P., Langridge, P., Shi, B. (2015). Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol. J. 13, 2–13. doi: 10.1111/pbi.12220
Han, J., Fang, J., Wang, C., Yin, Y., Sun, X., Leng, X., et al. (2014). Grapevine microRNAs responsive to exogenous gibberellin. BMC Genomics 15, 17. doi: 10.1186/1471-2164-15-111
Hao, L., Zhang, Y. (2022). Genome-wide analysis of miR159 gene family and predicted target genes associated with environmental stress in dendrobium officinale: A bioinformatics study. Genes. 13, 13. doi: 10.3390/genes13071221
Jeong, D. H., Green, P. J. (2013). The role of rice microRNAs in abiotic stress responses. J. Plant Biol. 56, 187–197. doi: 10.1007/s12374-013-0213-4
Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, X., Bian, H., Song, D., Ma, S., Han, N., Wang, J., et al. (2013). Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann. Bot. 111, 791–799. doi: 10.1093/aob/mct034
Li, S., Miao, L., Huang, B., Gao, L., He, C., Yan, Y., et al. (2019). Genome-wide identification and characterization of cucumber BPC transcription factors and their responses to abiotic stresses and exogenous phytohormones. Int. J. Mol. Sci. 20, 5048. doi: 10.3390/ijms20205048
Li, B., Tao, P., Xu, F., He, P., Wang, J. (2023). Function of Soybean miR159 family members in plant responses to low phosphorus, high salinity, and abscisic acid treatment. Agron. (Basel) 13, 16. doi: 10.3390/agronomy13071798
Liu, S., Li, J., Wu, J., Zhou, K., Zhou, H., Yang, J., et al. (2015). StarScan: a web server for scanning small RNA targets from degradome sequencing data. Nucleic Acids Res. 43, W480–W486. doi: 10.1093/nar/gkv524
Liu, P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H., Carrington, J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146. doi: 10.1111/j.1365-313X.2007.03218.x
Liu, H., Tian, X., Li, Y., Wu, C., Zheng, C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 14, 836–843. doi: 10.1261/rna.895308
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
López-Galiano, M. J., García-Robles, I., Gonzalez-Hernandez, A. I., Camañes, G., Vicedo, B., Real, M. D., et al. (2019). Expression of miR159 is altered in tomato plants undergoing drought stress. Plants-Basel. 8, 11. doi: 10.3390/plants8070201
Lu, L., Luo, W., Yu, W., Zhou, J., Wang, X., Sun, Y. (2022). Identification and characterization of Csa-miR395s reveal their involvements in fruit expansion and abiotic stresses in cucumber. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.907364
Montes, R. A. C., Rosas-Cardenas, F. D., De Paoli, E., Accerbi, M., Rymarquis, L. A., Mahalingam, G., et al. (2014). Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 5, 15. doi: 10.1038/ncomms4722
Palatnik, J. F., Wollmann, H., Schommer, C., Schwab, R., Boisbouvier, J., Rodriguez, R., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis MicroRNAs miR159 and miR319. Dev. Cell. 13, 115–125. doi: 10.1016/j.devcel.2007.04.012
Peng, T., Wen, H., Zhao, Y., Wang, B., Jin, Y., Sun, H., et al. (2018). Identification and expression analysis of rice miRNA related to salt and drought stresses. Acta Agric. Boreali-Sin. 33, 20–27.
Pieczynski, M., Marczewski, W., Hennig, J., Dolata, J., Bielewicz, D., Piontek, P., et al. (2013). Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 11, 459–469. doi: 10.1111/pbi.12032
Reyes, J. L., Chua, N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Sun, Y., Luo, W., Chang, H., Li, Z., Zhou, J., Li, X., et al. (2019). Identification of miRNAs and their target genes involved in cucumber fruit expansion using small RNA and degradome sequencing. Biomolecules. 9, 15. doi: 10.3390/biom9090483
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment throug sequence weighting, position-specific gap penalties and weight marix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, C., Jogaiah, S., Zhang, W., Abdelrahman, M., Fang, J. (2018). Spatio-temporal expression of miRNA159 family members and their GAMYB target gene during the modulation of gibberellin-induced grapevine parthenocarpy. J. Exp. Bot. 69, 3639–3650. doi: 10.1093/jxb/ery172
Wang, Y., Sun, F., Cao, H., Peng, H., Ni, Z., Sun, Q., et al. (2012). TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PloS One 7, 10. doi: 10.1371/journal.pone.0048445
Wang, M., Xie, Z., Sun, X., Li, X., Zhu, X., Wang, C., et al. (2017). Function analysis of miR159 and its target gene VvGAMYB in grape flower development. Acta Hortic. Sin. 44, 1061–1072. doi: 10.16420/j.issn.0513-353x.2016-0816
Wei, L., Zhang, D., Xiang, F., Zhang, Z. (2009). Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int. J. Plant Sci. 170, 979–989. doi: 10.1086/605122
Yang, Z., Zhu, P., Kang, H., Liu, L., Cao, Q., Sun, J., et al. (2020). High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genomics 21, 16. doi: 10.1186/s12864-020-6567-3
Zhang, W., Han, X., Zhu, X., Xie, Z., Jiu, S., Huang, Y., et al. (2019). Identification of the target genes of VvmiR159s and their regulation in response to GA in different tissues of grape berry. Sci. Agric. Sin. 52, 2858–2870.
Zhang, Y., Zhang, B., Yang, T., Zhang, J., Liu, B., Zhan, X., et al. (2020). The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato. Hortic. Res. 7, 133. doi: 10.1038/s41438-020-00366-1
Zhao, P., Wang, F., Deng, Y., Zhong, F., Tian, P., Lin, D., et al. (2022). Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 20, 833–845. doi: 10.1111/pbi.13762
Keywords: cucumber (Cucumis sativus. L), Csa-miR159s, fruit expansion, abiotic stress, phytohormones
Citation: Zhao Z, Ao W, Luo W, Sun Y, Tokala VY, Liu J, Zhi S and Sun Y (2025) Identification and characterization of Csa-miR159s and their expression patterns under different abiotic stresses in cucumber (Cucumis sativus L.). Front. Plant Sci. 16:1518406. doi: 10.3389/fpls.2025.1518406
Received: 28 October 2024; Accepted: 01 April 2025;
Published: 24 April 2025.
Edited by:
Xuming Li, Hugo Biotechnologies Co., Ltd., ChinaReviewed by:
Gograj Singh Singh Jat, Division of Vegetable Science - IARI, IndiaMamy Jayne Nelly Rajaofera, Hainan Medical University, China
Copyright © 2025 Zhao, Ao, Luo, Sun, Tokala, Liu, Zhi and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongdong Sun, c3VueWRAaGlzdC5lZHUuY24=
†These authors have contributed equally to this work
 Zhenxiang Zhao
Zhenxiang Zhao Wenhong Ao1,2†
Wenhong Ao1,2† Yongdong Sun
Yongdong Sun