Abstract
Mesocotyl length is a key trait affecting seedling emergence and establishment in dry direct-seeded rice, with longer mesocotyls promoting rapid and uniform emergence, thereby forming larger effective populations. Therefore, mining genes associated with mesocotyl length will facilitate the development of rice varieties suitable for dry direct seeding. In this study, 300 rice germplasm resources with a wide range of sources were selected as experimental materials. Phenotypic traits such as mesocotyl length and seedling emergence rate were systematically determined in each variety by setting different mulch depth treatments. Genome-wide association analysis (GWAS) was used to locate QTL controlling mesocotyl length and predict candidate genes. The results showed that mesocotyl length increased significantly with greater soil cover depth, while excessively deep sowing treatments inhibited seedling emergence. The GWAS analysis identified four QTLs associated with mesocotyl length and two QTLs associated with seedling emergence, with phenotypic contributions of 6.96-8.48%. Among them, the mesocotyl length-related QTL qML3 located at 28.03-28.43 Mb on chromosome 3 was detected at both sowing depths. Gene annotation analysis identified nine candidate genes related to plant hormones and transcription factors for qML3. Further investigation revealed three genes (LOC_Os03g49250, LOC_Os03g49400, and LOC_Os03g49510) exhibiting distinct haplotypes with significant differences in mesocotyl length, suggesting they may be causal genes for qML3. The results provide new clues to elucidate the molecular mechanism of rice mesocotyl development and lay an important foundation for subsequent gene function verification and molecular breeding. In the future, the functions of these candidate genes will be verified by transgenic and other methods, and molecular markers will be developed for genetic improvement of drought-tolerant rice varieties.
1 Introduction
Rice (Oryza sativa L.) is a major global food crop that is traditionally grown by transplanting seedlings into paddy fields (Zhao et al., 2018). However, conventional transplanted rice production systems face significant challenges because of climate change, urbanization, water scarcity, and labor shortages in many regions (Sagare et al., 2020; Zhang et al., 2023). Dry direct seeding, as an alternative to transplanting, saves time and labor while reducing water and energy consumption (Liu et al., 2019; Ohno et al., 2018; Zhan et al., 2019). Additionally, direct dry seeding eliminates the transplanting step, not only accelerating seedling cultivation but also increasing yield (Ishfaq et al., 2018; Muhammad et al., 2020). Although direct seeding of rice has multiple advantages over traditional planting methods, its popularization still faces several challenges, such as a low seedling emergence rate (ER), poor seedling establishment, weed infestation, and susceptibility to lodging (Mahender et al., 2015; Lee et al., 2017). Therefore, a key challenge at this stage is to identify effective methods to promote rapid and high-quality germination and growth of direct-seeded rice.
The mesocotyl, which forms when rice seeds germinate in the dark, is the organ that connects the germinal sheath to the seed root (Schillinger et al., 1998; Zhang et al., 2012). Light is a key factor influencing mesocotyl length (ML). Typically, light inhibits ML, whereas darkness significantly promotes its elongation. The study revealed that under light-free conditions, the cells of the mesocotyl not only actively divided but also exhibited particularly pronounced elongation and growth. This process is accompanied by increased levels of phytohormones such as gibberellins (GA), auxins (IAA), and cytokinins (CK), which act synergistically to strongly promote mesocotyl elongation (Li et al., 2012, 2013). In addition, temperature also affects the elongation of the ML. The optimal temperature range is 25-30°C, within which the ML increases with the rise in temperature. Beyond the suitable temperature range, as the temperature continues to rise, the growth rate of the rice mesocotyl slows down (Cao et al., 2002). Temperature is another factor that regulates the elongation of the mesodermal axis. The optimal temperature promotes growth, while both high and low temperatures are detrimental to the elongation of the mesodermal axis. Thus, the mesocotyl is not only a central feature of seed germination adapted to dark conditions but also a crucial factor in regulating the initial morphogenesis and growth strategy of rice seedlings. Rice seeds with longer mesocotyls demonstrate superior seedling emergence and uniformity (Turner et al., 1982; Mgonja et al., 1993; Chung, 2010). ML is recognized as a key trait for breeding direct-seeded rice varieties (Zhan et al., 2019). Therefore, an economical and effective method to promote the adoption of direct seeding technology is to investigate the locus ML, analyze the underlying genetic mechanisms, and screen and innovate germplasm with long mesocotyls.
ML can be influenced by external environmental factors but is also determined by intrinsic genetic mechanisms (Gray et al., 1998; Kato and Katsura, 2014; Liu et al., 2023). ML is typically a quantitative inherited trait controlled by multiple genes (Wu et al., 2005; Wang et al., 2021; Zhang et al., 2023). Therefore, mining of ML-related genes and molecular breeding could facilitate the development of direct-seeded rice varieties. With the rapid development of molecular marker technology and genome sequencing technology, many ML-related QTLs have been identified in various natural populations through biparental QTL analysis and genome-wide association studies (Lee et al., 2017; Liu et al., 2019; Zhan et al., 2019; Liu et al., 2023; Wang et al., 2023);. Eleven QTLs were identified using a population of recombinant inbred lines (RILs) from an interspecific cross between O. sativa and O. rufipogon (Cai and Morishima, 2002). In a population of backcrossed inbred lines (BILs) from a cross between Kasalath and Nipponbare, five QTLs for ML were detected (Lee et al., 2011). Three major QTLs, designated qMel-1, qMel-3, and qMel-6, located on chromosomes 1, 3, and 6, respectively, were identified through the analysis of 98 self-crossed lines from the Kasalath and Nipponbare crosses (Lee et al., 2017). In addition, 9, 17, 16, and 11 QTLs for ML were identified using GWAS (Wu et al., 2015; Lu et al., 2016; Liu et al., 2019; Kwon, 2021). To date, several ML-related genes have been cloned. Xiong et al. (2017) reported that ethylene (ETH) regulates ML by repressing GY1 gene expression and controlling the biosynthesis of jasmonic acid (JA). Sun et al. (2018) reported that the OsGSK2 gene regulates ML by coordinating the signaling of strigolactone (SL) and brassinolide (BR). Zheng et al. (2020) reported that the karrikin signaling pathway and the BR signaling pathway may jointly regulate mesoembryo axis elongation.
ML as a crucial indicator of rice growth and development, demonstrates significant pleiotropic effects through its regulatory genes in crop breeding. Choi (2003) reported that OsEXP4 overexpression produced a biphasic phenotype with 12% of plants showing increased height and 88% displaying reduced stature compared to controls; the overexpressing lines exhibited 31% and 97% increases in coleoptile and ML, respectively, while antisense plants showed 28% and 43% reductions. Ma et al. (2013) found that ethylene treatment significantly promoted coleoptile and mesocotyl elongation in etiolated MHZ7 overexpressing plants while increasing grain length in transgenic plants, whereas mhz7 mutants exhibited reduced thousand-grain weight. Lv et al. (2021) demonstrated that OsPAO5 knockout across different genetic backgrounds significantly enhanced mesocotyl elongation, improved emergence rate and speed under deep-sowing conditions, and concurrently increased grain length, grain weight, and grains per panicle, ultimately boosting single-plant yield. Through analysis of 165 rice accessions, Meng et al. (2023) further identified the superior gene OsML1, which not only markedly improved emergence rate in deep sowing but also conferred comprehensive enhancements in agronomic traits including plant height, panicle length, and grain width in overexpression lines.
Discovering rice resources with long mesocotyls, identifying and cloning the genes that control ML, and analyzing their molecular regulatory mechanisms are key steps in breeding rice varieties with long mesocotyls. In this study, a GWAS was conducted on a rice population consisting of 300 genotypes to assess mesocotyl phenotypes. Four new QTLs were ultimately identified, and three new candidate genes for the ML were initially predicted. This study lays the foundation for the cloning of new ML genes and analyzing the genetic mechanisms of rice ML.
2 Materials and methods
2.1 Plant materials
In this study, we selected 300 rice varieties, all of which were derived from the National Gene Bank of China. Based on previous research results, we selected samples including local varieties, superior varieties, foundation varieties and their derivatives to ensure a wide distribution and high genetic diversity. (Supplementary Table S1) (Cui et al., 2022). These accessions were grown in Hainan Province, China, in 2022. The seeds were harvested in April 2023, air-dried, and stored in the laboratory of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China.
2.2 Mesocotyl length measurement and emergence rate evaluation
Seeds were placed in an oven at 45°C for one week to break dormancy. For each treatment, 50 full and uniform seeds were independently selected and sterilized with 1.4% sodium hypochlorite solution and then placed in an oven at 35°C for 2 days to promote germination. To assess the effect of different sowing depths on mesocotyl length (ME), three mulching depth treatments of 2, 5 and 7 cm were set up in 84-well plastic frames, with three biological replicates at each depth, and 20 high-quality germinated seeds were sown in each replicate, and then incubated for 7 days in the dark at 28°C. The mesocotyl length of each seedling was measured using a ruler (distance from root base to the node of the embryonic sheath) (Huang et al., 2022). The plates were incubated in a 28°C dark growth chamber for 7 days, after which the mesocotyl length (ML) was measured. For the emergence rate (ER), germinated seeds were sown at depths of 5 cm and 7 cm respectively, and cultivated under conditions of 30°C and 75% relative humidity with 20,000 lux white fluorescent lighting (14 h light/10 h dark photoperiod) for 7 days. All experiments included three biological replicates, with at least 10 seedlings measured per replicate. Phenotypic variation analysis, correlation analysis, and Student’s t-tests were performed using IBM SPSS v26.
2.3 Genome-wide association study and candidate genes identification
SNPs with a minor allele frequency (MAF) greater than 5% and deletion under 40% were selected for association analysis, and 711,268 variants were used for GWAS. The Yeo–Johnson transformation was applied to normalize the data for GWAS analysis (Yeo and Johnson, 2000). The GWAS was performed using a mixed linear model (MLM) and the script run_pipeline.pl in the TASSEL software (Bradbury et al., 2007). The significance threshold was set at 1.0 × 10-7. Principal component analysis (PCA) was used to characterize and differentiate the samples using Plink software (Purcell et al., 2007). SNP density heatmaps and Manhattan and Q–Q plots were generated using the CMplot package in R software (Yin et al., 2021).
QTLs were screened based on the results of the Manhattan plot, where column peaks exceeding the threshold line were considered as candidate QTLs. According to existing studies, genome-wide linkage disequilibrium (LD) attenuation in cultivated rice usually occurs in the range of 100–200 kb. Therefore, we designated the 200 kb region upstream and downstream of the QTL peak SNP locus as the QTL region. We extracted the genes encoding the target intervals from the Rice Expression Database (http://expression.ic4r.org/) and screened them using gene function annotation information from the China Rice data Center (https://ngdc.cncb.ac.cn/red/index). First, transposon- and reverse transcript-transposon-related genes were excluded, and then candidate genes related to the target QTL were preliminarily screened based on the gene functional annotations in combination with existing research reports on rice mesocotyl and internode elongation genes (Kawahara et al., 2013).
2.4 Haplotype analysis
Haplotype analyses were performed on the QTL candidate genes that were repeatedly detected under two different environmental treatments. High-confidence SNPs in the coding regions of the candidate genes within the QTL regions were extracted from the Rice Variation Atlas v2.0 database (http://ricevarmap.ncpgr.cn/) (Zhao et al., 2021), and the haplotypes of the candidate genes within the QTL regions were analyzed. The significance of the differences between the phenotypic values and haplotypes was determined by independent samples t-test (P<0.05), and genes showing significant differences in phenotypic value between the different haplotypes were screened by comparing the phenotypic values of the different haplotypes (≥10 materials).
3 Results
3.1 Phenotypic variation
Firstly, the ML of 300 rice materials was evaluated under different overburden conditions (2, 5, 7cm). The results showed that the ML exhibited a continuous distribution with considerable genetic variation, indicating that this diverse population is an ideal material for GWAS studies (Figure 1). At a seeding depth of 2 cm, the ML ranged from 0 to 16.21 mm, with an average of 4.77 mm. At a seeding depth of 5 cm, the ML ranged from 0 to 33.90 mm, with an average of 5.61 mm. At a seeding depth of 7 cm, the ML ranged from 0 to 46.00 mm, with an average of 8.36 mm. In addition, we observed the ER. The results showed that under at 5 cm and 7 cm soil coverings, there were differences in the ERs among the different varieties. At a seeding depth of 5 cm, the ER ranged from 20% to 100%, with an average of 82.36%. At a seeding depth of 7 cm, the ER ranged from 5% to 100%, with an average of 68.88%.
Figure 1

Correlation and differential analysis of mesocotyl length and emergence rate under three deep sowing depths. (A) Phenotypic distribution and correlation analysis of mesocotyl length and emergence rate under 2cm, 5 cm and 7 cm deep sowing conditions. Numbers in upper right corner are correlation coefficients (r), * indicates the significance at p ≤ 0.05. (B) Phenotypic variations of phenotypic traits: mesocotyl length and emergence rate under 5 cm and 7 cm deep sowing conditions. Significance levels are indicated by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed t-test). ML_2, ML_5 and ML_7: mesocotyl length under deep sowing depths of 2cm, 5 cm and 7 cm; ER_5 and ER_7: emergence rate under deep sowing depths of 5 cm and 7 cm.
Correlation analyses showed that ML exhibited a significant positive correlation among the three sowing depths: the correlation coefficients were 0.60 between 2 cm and 5 cm soil depth, 0.67 between 5 cm and 7 cm soil depth, and 0.71 between 2 cm and 7 cm soil depth. Additionally, the ERs at 5 cm and 7 cm sowing depths also exhibited a positive correlation, with a correlation coefficient of 0.46 (Figure 1A). In addition, independent samples t-test results indicated that there were significant differences in ML and ER under different sowing depths. A comparison of ML and ER under different sowing depths revealed that the ML followed the order of 7 cm > 5 cm > 2 cm, while the ER was higher at 5 cm than at 7 cm (P < 0.05) (Figure 1B). This indicates that as the sowing depth increases, the ML also increases, but the seedling ER decreases.
3.2 Genome-wide association analysis and candidate gene identification
The sequencing data of the materials were input and filtered, yielding 711,268 high-quality SNP markers distributed across 12 chromosomes (Supplementary Figure S1). The PCA results indicated that the first 20 principal components accounted for a substantial proportion of the genetic variance, exhibiting a decreasing trend, whereas the 10th and subsequent components explained less than 2% of the genetic variance (Supplementary Figure S2). Therefore, in this study, the group structure was corrected by principal component analysis, using the first 10 principal components as covariates. Calculations were performed using the MLM.
Genome-wide linkage disequilibrium (LD) attenuation in cultivated rice has been reported to occur within a range of 100–200 kb (Huang et al., 2010). We defined the region 200 kb upstream and downstream of the significant SNP as the QTL region associated. A total of six unique loci were detected, which explained the phenotypic variations ranged from 6.96% to 8.48% (Table 1). For ML, two significant loci were detected on chromosomes 3 and 10 at a sowing depth of 2 cm, one significant locus was detected on chromosome 3 at a sowing depth of 5 cm, and 1 significant locus were detected on chromosome 6 at a sowing depth of 7 cm, collectively explaining 6.96% to 7.54% of the phenotypic variation (Figure 2, Table 1). For ER, one significant locus was detected on chromosomes 5 at a sowing depth of 5 cm, one significant locus was detected on chromosome 4 at a sowing depth of 7 cm, with explanation rates of 8.4% and 8.48%, respectively (Supplementary Figure S3; Table 1). Among them, qML3 detected under the 2 cm sowing depth and qML3 detected under the 5 cm sowing depth are located at the same position in the genome, representing co-localized loci. For ease of description, we will refer to them collectively as qML3 in the following text.
Table 1
| Treatment | QTL | Chr | Position (Mb) | P value | R² (%) | Co-location QTL | Reference |
|---|---|---|---|---|---|---|---|
| Stress (2cm) | qML3 | 3 | 28.03-28.43 | 1.5059e-10 | 7.54 | qml3, qML3, qML3 | Cao et al., 2002; Niu et al., 2019; Huang et al., 2022 |
| Stress (2cm) | qML10 | 10 | 3.29-3.69 | 9.9445e-10 | 6.96 | ||
| Stress (5cm) | qML3 | 3 | 28.03-28.43 | 5.6478e-08 | 7.19 | qml3, qML3, qML3 | Cao et al., 2002; Niu et al., 2019; Huang et al., 2022 |
| Stress (7cm) | qML6 | 6 | 3.14-3.54 | 7.9207e-09 | 7.09 | ||
| Stress (5cm) | qER5 | 5 | 2.61-3.01 | 3.3156e-08 | 8.4 | ||
| Stress (7cm) | qER4 | 4 | 0-0.21 | 1.5602e-08 | 8.48 |
Candidate QTLs identified from genome-wide association analysis.
ML, Mesocotyl length; ER, Emergence Rate; QTLs, quantitative trait loci; Chr, chromosome; R2, phenotypic variation rate; Position, 200 kb upstream and downstream of the significant SNP locus used to define the QTL interval; Co-location QTLs, QTLs for shoot length identified by the previous studies.
Figure 2
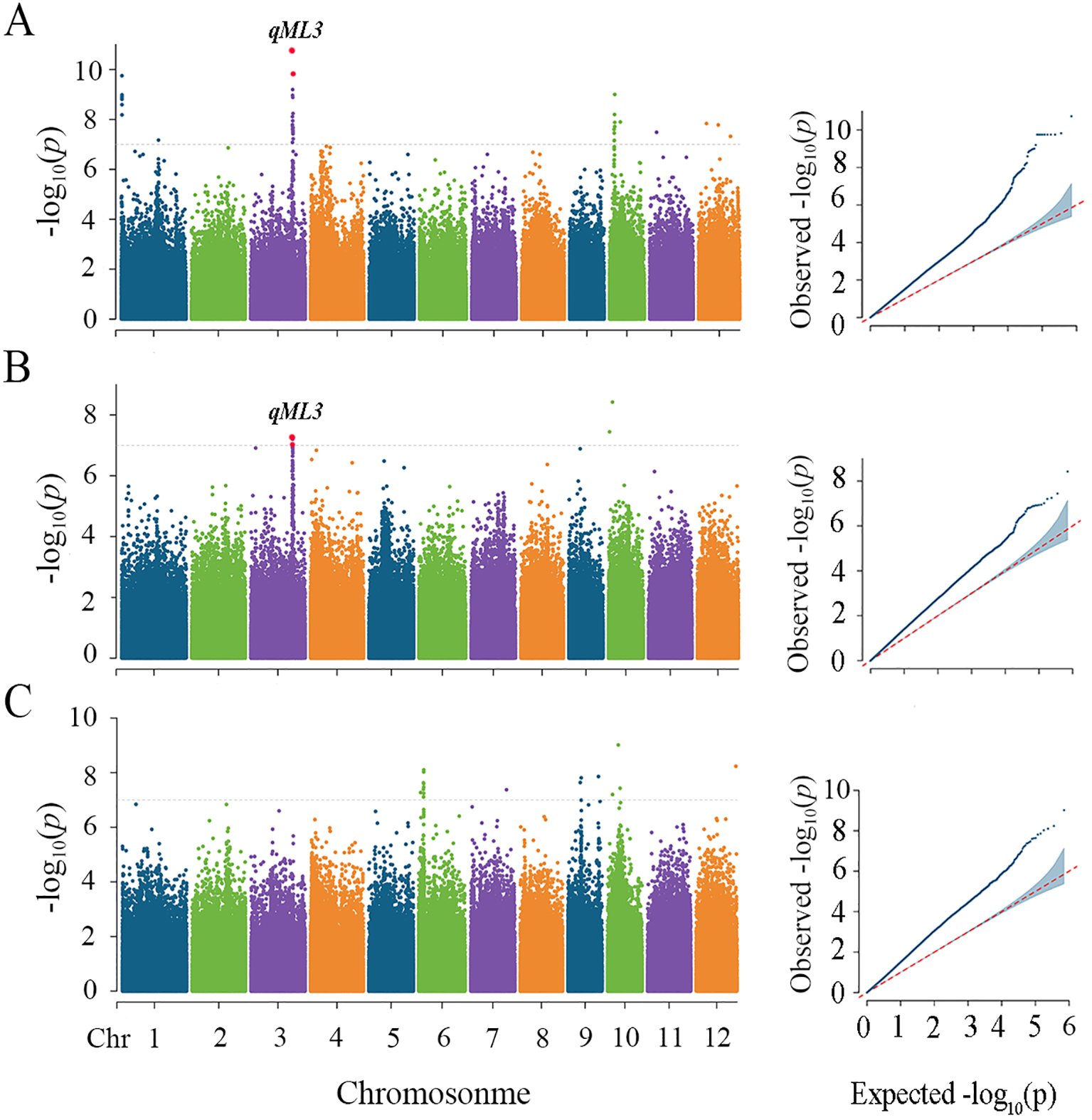
Genome-wide association studies for mesocotyl length among three deep sowing depths. Manhattan plots and Quantile-quantile plots of mesocotyl lengths in soil culture with a sowing depth of 2 cm (A), 5cm (B) and 7cm (C). Negative log10-transformed p values from a genome-wide scan are plotted against position on each of 12 chromosomes. The gray horizontal dashed lines indicate the genome-wide significance threshold. The red dots represent SNPs that were detected at both depths.
3.3 Haplotype analysis of candidate genes
By analysing the functional variants within 200 kb upstream and downstream of the peak SNPs and their LD block characteristics (Figure 3A), we performed a preliminary screening of candidate genes for the qML3 locus. The genetic information of 300 rice varieties has been annotated with reference to the 7th version of the MSU Rice Genome Annotation Project (Rice 7) (http://rice.plantbiology.msu.edu/) (Kawahara et al., 2013) on the rice IRGSP-1.0 genome. There were 68 genes in the qML3 candidate QTL interval, with 44 genes remaining after the removal of putative proteins, reduced transposons, and transposon-encoding genes (Supplementary Table S3). Many studies have reported the association between the activities of phytohormones such as abscisic acid (ABA), BR, SL, CK, ETH, JA, GA and IAA with ML. Therefore, we focused on genes encoding phytohormone-related genes within the specified intervals, resulting in the identification of 9 candidate genes (Table 2).
Table 2
| Gene | Functional annotation |
|---|---|
| LOC_Os03g49050 | possible lysine decarboxylase domain containing protein |
| LOC_Os03g49132 | ZOS3-16 - C2H2 zinc finger protein |
| LOC_Os03g49170 | zinc finger family protein |
| LOC_Os03g49250 | OsFBO16 - F-box and other domain containing protein |
| LOC_Os03g49400 | ethylene-insensitive protein |
| LOC_Os03g49480 | elongation of fatty acids protein 2 |
| LOC_Os03g49500 | ethylene receptor |
| LOC_Os03g49510 | phosphatidylinositol-4-phosphate 5-kinase |
| LOC_Os03g49620 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 |
Candidate genes for gene annotation.
High-confidence candidate genes for mesocotyl lengths in qML3.
To identify potential candidate genes, we performed haplotype analysis of nine genes based on non-synonymous mutations in the coding region of each gene. Among these genes, 2 genes showed no significant phenotypic differences between their different haplotypes (Supplementary Figure S4), and 4 genes had only one haplotype. Of particular interest, 3 genes (LOC_Os03g49250, LOC_Os03g49400 and LOC_Os03g49510) showed significant phenotypic differences (P < 0.05) between their different haplotypes (Figure 3). For LOC_Os03g49250, one major single-base variations in the CDS region that cause amino acid substitution. The lines carrying Hap_2LOC_Os03g49250 exhibited significantly longer ML than those carrying Hap_1LOC_Os03g49250 under the two sowing depth conditions (P < 0.05) (Figures 3B, E). For LOC_Os03g49400, there were eight major single-base variations in the CDS region that cause amino acid substitution. The lines carrying Hap_3LOC_Os03g49400 exhibited significantly longer ML than those carrying Hap_1LOC_Os03g49400 and Hap_2 LOC_Os03g49400 under the two sowing depth conditions (P < 0.05) (Figures 3C, E). For LOC_Os03g49510, there were three major single-base variation in its CDS region that caused amino acid substitution, and ML exhibited significantly different between the two haplotypes identified based on this variation. Under the two sowing depth conditions, the lines carrying Hap_2 LOC_Os03g49510 exhibited significantly longer ML than those carrying Hap_1 LOC_Os03g49510 (P < 0.05) (Figures 3D, E).
Figure 3
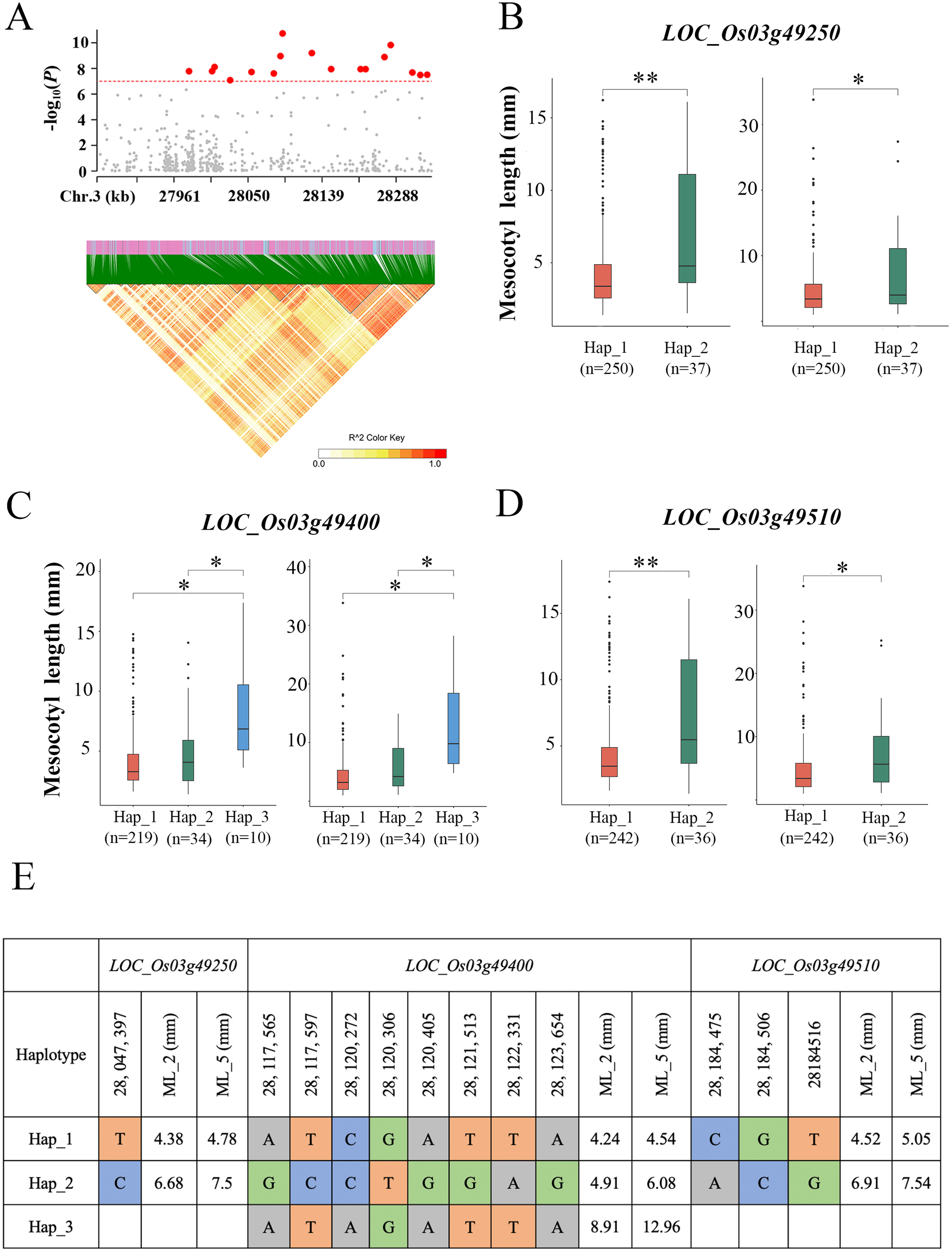
Gene structure and haplotype analysis of the candidate genes underlying qML3. (A) Local Manhattan plot (top) and linkage disequilibrium heatmap (bottom). (B–D) The boxplot illustrates the distribution of mesocotyl length across haplotype groups of LOC_Os03g49250, LOC_Os03g49400 and LOC_Os03g49510 under different sowing depths (2 cm, right; 5 cm, left). The middle line in each boxplot represents the median. P < 0.05. Significance levels are indicated by asterisks: *P < 0.05, **P < 0.01 (two-tailed t-test). (E) Haplotype analysis of the three genes in the region. ML_2 and ML_5: mesocotyl length under deep sowing depths of 2cm and 5cm.
4 Discussion
4.1 Sowing depth affects mesocotyl length and seedling emergence rate
Soil covering depth significantly influences the elongation of rice mesocotyl and its ER. Studies have shown that as sowing depth increases, ML exhibits a significant growth trend. This adaptive elongation mechanism contributes to improving rice ERs (Zhao et al., 2018; Lu et al., 2016). Research by Lee et al. (1999) demonstrated that at sowing depths of 5, 7, and 10 cm, ML significantly increased with depth, a finding consistent with the trend observed in our experiment at sowing depths of 2, 5, and 7 cm. In addition, this pattern was also confirmed by Liu et al. (2019) who observed a significant increase in ML with increasing sowing depth at three mulch depths of 2 cm, 4 cm and 6 cm. In direct-seeded rice, deeper soil covering promotes mesocotyl elongation to facilitate seedling emergence. However, significant differences in mesocotyl elongation ability exist among different varieties: varieties with strong elongation ability can maintain a high ER under deep soil covering, while those with weak elongation ability may struggle to emerge, ultimately affecting yield. In this study, statistical analysis of seedling ERs under different sowing depths revealed that the ER at a 7 cm sowing depth was significantly lower than that at a 5 cm depth. These results indicate that sowing depth is a critical factor influencing ML and ER. Therefore, selecting an appropriate sowing depth is crucial for the practical production of direct-seeded rice.
4.2 qML3 exhibits significant potential value in rice direct seeding breeding
Previous studies have identified QTLs and candidate genes associated with mesocotyl elongation in rice using linkage mapping or GWAS techniques. In this study, through GWAS analysis, we identified four QTLs related to ML and two QTLs associated with ER. Among these, a significant co-localized locus for ML, named qML3, was detected on chromosome 3 under both 2 cm and 5 cm sowing depths. This locus overlaps with previously identified ML QTLs (Cao et al., 2002; Niu et al., 2019; Huang et al., 2022), underscoring the authenticity and reliability of qML3. Furthermore, qML3 may be closely related to the adaptability of mesocotyl elongation in shallow soil conditions, providing important insights into the genetic regulatory mechanisms of ML in such environments. Regarding ER, we did not detect any significant loci co-localized with ML, suggesting that the genetic regulation of ER may be more complex or significantly influenced by other environmental factors.
4.3 Candidate mesocotyl length genes
Using GWAS, we initially identified 68 candidate genes within the qML3 interval that might be related to mesocotyl elongation. From these, nine candidate genes with homology to known functional genes were screened by functional annotation analysis and combined with previous studies. Further haplotype analysis showed that the haplotype variants of three genes, LOC_Os03g49250, LOC_Os03g49400 and LOC_Os03g49510, were significantly associated with mesocotyl elongation, suggesting that they may be potential candidate genes for qML3.
All three candidate genes screened in this study regulate mesocotyl and internode elongation through the phytohormone signalling pathway. LOC_Os03g49250 encodes an F-box protein, a family of proteins that play important roles in plant signalling (Yan et al., 2017). Its homologous gene, D3, was shown to specifically recognise the OsGSK2-phosphorylated CYCLIN U2 protein and promote its degradation through the ubiquitin-proteasome pathway, thereby negatively regulating rice mesocotyl elongation (Sun et al., 2018). The ethylene-insensitive protein (EIN) encoded by LOC_Os03g49400 is a core component of the ethylene signalling pathway, and it has been shown that ethylene affects mesocotyl development by regulating the expression of JA and BR related genes via EIN2/EIL2 (Xiong et al., 2017; Jun et al., 2004). LOC_Os03g49510 encodes phosphatidylinositol 4-phosphate 5-kinase, a family of enzymes involved in growth hormone signalling and plant developmental processes through the regulation of phospholipid metabolism (Liang et al., 2018). It was found that OsPIP5K1 and DWT1/DWL2 synergistically regulate rice growth and development through phosphatidylinositol signalling, and its functional deficiency leads to disturbances in phospholipid metabolism, triggering typical phenotypes such as plant dwarfing and internode shortening (Fang et al., 2020).
During plant growth, the gradual increase in mesocotyl length is typically regulated by phytohormones through complex signaling pathways. Generally, phytohormones control mesocotyl elongation by modulating either cell division or cell expansion (Zhan et al., 2019). In this study, the three identified candidate genes were found to be associated with the biosynthesis and signalling pathways of plant hormones, suggesting their potential involvement in mesocotyl elongation by regulating the expression of hormone-related proteins. The molecular pathways by which the proteins encoded by these genes regulate mesocotyl elongation growth and the correlation (positive or negative) between changes in their expression levels (up- or down-regulation) and the rate of mesocotyl elongation need to be further investigated. In the following work, we will use molecular biology techniques such as gene editing, genetic transformation and DNA insertion to systematically verify the biological functions of these candidate genes in mesocotyl elongation.
5 Conclusions
This study utilized GWAS to analyze the ML and seedling ER of 300 rice germplasm resources. By analyzing two traits, ML and ER, a total of six QTLs were identified. Among them, the ML-related locus qML3 was detected under two different sowing depths and contributed the most to phenotypic variation, indicating its stable genetic effects across multiple environmental conditions. Further prediction of candidate genes for qML3 identified three potential candidate genes associated with ML: LOC_Os03g49250, LOC_Os03g49400, and LOC_Os03g49510. The functions of these genes require further validation, but their potential mechanisms provide new research directions for the regulation of mesocotyl elongation. In the future, these genes are expected to be applied in molecular breeding for ideal rice plant types, offering theoretical foundations and genetic resources for the improvement of direct-seeded rice.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LX: Investigation, Software, Writing – original draft. SW: Writing – original draft. QZ: Writing – review & editing. BH: Writing – review & editing. DC: Writing – review & editing. LH: Writing – review & editing. JD: Writing – review & editing. XM: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Biological Breeding-National Science and Technology Major Project (2022ZD04017), the National Crop Germplasm Resources Center (NCGRC-2023-02) and the Agricultural Science and Technology Innovation Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1546580/full#supplementary-material
References
1
BradburyP. J.ZhangZ.KroonD. E.CasstevensT. M.RamdossY.BucklerE. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics23, 2633–2635. doi: 10.1093/bioinformatics/btm308
2
CaiH. W.MorishimaH. (2002). QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet.104, 1217–1228. doi: 10.1007/s00122-001-0819-7
3
CaoL. Y.ZhuJ.YanQ. C.HeL. B.WeiX. H.ChengS. H. (2002). Mapping QTLs with epistasis for mesocotyl length in a DH population from indica-japonica cross of rice (Oryza sativa). Chin. J. Rice Sci.03), 24–27. doi: 10.16819/j.1001-7216.2002.03.005
4
ChoiD. (2003). Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell15, 1386–1398. doi: 10.1105/tpc.011965
5
ChungN. J. (2010). Elongation habit of mesocotyls and coleoptiles in weedy rice with high emergence ability in direct-seeding on dry paddy fields. Crop Pasture Sci.61, 911–917. doi: 10.1071/CP10099
6
CuiD.ZhouH.MaX.LinZ.SunL.HanB.et al. (2022). Genomic insights on the contribution of introgressions from Xian/Indica to the genetic improvement of Geng/Japonica rice cultivars. Plant communications3, 100325. doi: 10.1016/j.xplc.2022.100325
7
FangF.YeS.TangJ.BennettM. J.LiangW. (2020). DWT1/DWL2 act together with OsPIP5K1 to regulate plant uniform growth in rice. New Phytol.225, 1234–1246. doi: 10.1111/nph.16216
8
GrayW. M.OstinA.SandbergG.RomanoC. P.EstelleM. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. United States America95, 7197–7202. doi: 10.1073/pnas.95.12.7197
9
HuangQ.JuC. Y.ChengY. B.CuiD.HanB.ZhaoZ. W.et al. (2022). QTL mapping of mesocotyl elongation and confirmation of a QTL in dongxiang common wild rice in China. Agronomy12, 1800. doi: 10.3390/agronomy12081800
10
HuangX.WeiX.SangT.ZhaoQ.FengQ.ZhaoY.et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet.42, 961–967. doi: 10.1038/ng.695
11
IshfaqM.ZulfiqarU.AnjumS. A.AhmadM. (2018). Optimizing row spacing for direct seeded aerobic rice under dry and moist fields. Pakistan J. Agric. Res.31, 291–299. doi: 10.17582/journal.pjar/2018/31.4.291.299
12
JunS. H.HanM. J.LeeS. (2004). OsEIN2 is a positive component in ethylene signaling in rice. Plant Cell Physiol.45, 281–289. doi: 10.1093/pcp/pch033
13
KatoY.KatsuraK. (2014). Rice adaptation to aerobic soils: Physiological considerations and implications for agronomy. Plant Production Sci.17, 1–12. doi: 10.1626/pps.17.1
14
KawaharaY.BastideM.HamiltonJ. P.KanamoriH.McCombieW. R.OuyangS.et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (New York NY)6, 4. doi: 10.1186/1939-8433-6-4
15
KwonS. W. (2021). Genome-wide association study (GWAS) of mesocotyl length for direct seeding in Rice. Agronomy11, 1–17. doi: 10.3390/agronomy11122527
16
LeeS. S.KimJ. H.HongS. B. (1999). Effects of priming and growth regulator treatment of seed on emergence and seedling growth of rice. Korean J. Crop Sci.44, 134–137. Available at: https://koreascience.kr/article/JAKO199911922913759.page (Accessed May 3, 2025)
17
LeeH. S.SasakiK.HigashitaniA.AhnS. N.SatoT. (2011). Mapping and characterization of quantitative trait loci for mesocotyl elongation in rice (Oryza sativa L.). Rice5, 13. doi: 10.1186/1939-8433-5-13
18
LeeH. S.SasakiK.KangJ. W.SatoT.SongW. Y.AhnS. N. (2017). Mesocotyl elongation is essential for seedling emergence under deep-seeding condition in rice. Rice10, 32. doi: 10.1186/s12284-017-0173-2
19
LiL.LiangQ.WangC.ChengW. F. (2013). Effect of auxin on weedy rice mesocotyl cell wall oxidase. Appl. Mechanics Materials448-453, 69–73. doi: 10.4028/www.scientific.net/AMM.448-453
20
LiL.MaD. R.SunJ.LiangQ.ChengW. F. (2012). Observation of mesocotyl cell morphology of weed rice. J. Shenyang Agric. Univ.43, 749–753. Available at: http://en.cnki.com.cn/Article_en/CJFDTOTAL-SYNY201206020.htm (Accessed May 3, 2025).
21
LiangT. N.ZhaoY.HeZ. B.CaoQ. G.ZhangY. J.ZhangY. X.et al. (2018). Bioinformatics analysis and fluorescence quantitative PCR results of Ricinus communis PIP5Ks. Chin. Traditional Herbal Drugs49, 5892–5900. Available at: http://en.cnki.com.cn/Article_en/CJFDTotal-ZCYO201824025.htm (Accessed May 3, 2025).
22
LiuJ. D.WangY. M.LiuH. Y. (2023). Genome wide association study of candidate genes underlying rice mesocotyl elongation. J. Plant Genet. Resour.24, 1702–1735. doi: 10.13430/j.cnki.jpgr.20230326001
23
LiuH.ZhanJ.LiJ.LuX.LiuJ.WangY.et al. (2019). Genome-wide Association Study (GWAS) for Mesocotyl Elongation in Rice (Oryza sativa L.) under Multiple Culture Conditions. Genes11, 49. doi: 10.3390/genes11010049
24
LuQ.ZhangM.NiuX.WangC.XuQ.FengY.et al. (2016). Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping. Planta243, 645–657. doi: 10.1007/s00425-015-2434-x
25
LvY. S.ShaoG. N.JiaoG. A.ShengZ. H.XieL. H.HuS. K.et al. (2021). Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol. Plant14, 344–351. doi: 10.1016/j.molp.2020.11.007
26
MaB.HeS. J.DuanK. X.YinC. C.ChenH.YangC.et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant6, 1830–1848. doi: 10.1093/mp/sst087
27
MahenderA.AnandanA.PradhanS. K. (2015). Early seedling vigour, an imperative trait for direct-seeded rice: an overview on physio-morphological parameters and molecular markers. Planta241, 1027–1050. doi: 10.1007/s00425-015-2273-9
28
MengY.ZhanJ. H.LiuH. Y.LiuJ. D.WangY. M.GuoZ.et al. (2023). Natural variation of OsML1, a mitochondrial transcription termination factor, contributes to mesocotyl length variation in rice. Plant Journal: Cell Mol. Biol.115, 910–925. doi: 10.1111/tpj.16267
29
MgonjaM. A.LadeindeT. A. O.Aken’OvaM. E. (1993). Genetic analysis of mesocotyl length and its relationship with other agronomic characters in rice (Oryza sativa L.). Euphytica72, 189–195. doi: 10.1007/BF00034157
30
MuhammadI.NadeemA.ShakeelA. A.MuhammadA. I. H. (2020). Growth, yield and water productivity of dry direct seeded rice and transplanted aromatic rice under different irrigation management regimes. J. Integr. Agric.19 (11), 2656–2673. doi: 10.1016/S2095-3119(19)62876-5
31
NiuS. P.LvY. S.WuY. W.WeiX. J.ShengZ. H.JiangG. A.et al. (2019). QTLs mapping for mesocotyl length in rice. China Rice25, 55–59. Available at: https://link.cnki.net/urlid/33.1201.s.20191112.0913.022 (Accessed May 3, 2025).
32
OhnoH.Banayo NinoP. M. C.Bueno CrisantaS.KashiwagiJ.-i.NakashimaT.Corales AuroraM.et al. (2018). Longer mesocotyl contributes to quick seedling establishment, improved root anchorage, and early vigor of deep-sown rice. Field Crops Res.228, 84–92. doi: 10.1016/j.fcr.2018.08.015
33
PurcellS.NealeB.Todd-BrownK.ThomasL.FerreiraM. A.BenderD.et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575. doi: 10.1086/519795
34
SagareD. B.AbbaiR.JainA.JayadevappaP. K.DixitS.SinghA. K.et al. (2020). More and more of less and less: Is genomics-based breeding of dry direct-seeded rice (DDSR) varieties the need of hour? Plant Biotechnol. J.18, 2173–2186. doi: 10.1111/pbi.13454
35
SchillingerW. F.DonaldsonE.AllanR. E.JonesS. S. (1998). Winter wheat seedling emergence from deep sowing depths. Agron. J.90, 582–586. doi: 10.2134/agronj1998.00021962009000050002x
36
SunS.WangT.WangL.LiX.JiaY.LiuC.et al. (2018). Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat. Communication9, 2523. doi: 10.1038/s41467-018-04952-9
37
TurnerF. T.ChenC. C.BollichC. N. (1982). Coleoptile and mesocotyl lengths in semi-dwarf rice seedlings. Crop Sci.22, 43–46. doi: 10.2135/cropsci1982.0011183X002200010010x
38
WangY.LiuJ.MengY.LiuH.LiuC.YeG. (2021). Rapid identification of QTL for mesocotyl length in rice through combining QTL-seq and genome-wide association analysis. Front. Genet.12. doi: 10.3389/fgene.2021.713446
39
WangY.LiuH.MengY.LiuJ.YeG. (2023). Validation of genes affecting rice mesocotyl length through candidate association analysis and identification of the superior haplotypes. Front. Plant sci.14. doi: 10.3389/fpls.2023.1194119
40
WuJ.FengF.LianX.TengX.WeiH.YuH.et al. (2015). Genome-wide Association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol.15, 1–10. doi: 10.1186/s12870-015-0608-0
41
WuM. G.ZhangG. H.LinJ. R.ChengS. H. (2005). Screening for rice germplasms with specially elongated mesocotyl. Rice Sci.12, 226–228. Available at: http://www.ricesci.org/EN/Y2005/V12/I3/226 (Accessed May 3, 2025)
42
XiongQ.MaB.LuX.HuangY. H.HeS. J.YangC.et al. (2017). Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell.29, 1053–1072. doi: 10.1105/tpc.16.00981
43
YanJ.LiaoX.HeR.ZhongM.FengP.LiX.et al. (2017). Ectopic expression of GA 2-oxidase 6 from rapeseed (Brassica napus L.) causes dwarfism, late flowering and enhanced chlorophyll accumulation in Arabidopsis thaliana. Plant Physiol. Biochem.111, 10–19. doi: 10.1016/j.plaphy.2016.11.008
44
YeoI. K.JohnsonR. A. (2000). A new family of power transformations to improve normality or symmetry. Biometrika87, 954–959. doi: 10.1093/biomet/87.4.954
45
YinL.ZhangH.TangZ.XuJ.YinD.ZhangZ.et al. (2021). rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom Proteom Bioinform19, 10. doi: 10.1016/j.gpb.2020.10.007
46
ZhanJ.LuX.LiuH.ZhaoQ.YeG. (2019). Mesocotyl elongation, an essential trait for dry-seeded rice (Oryza sativa l.): a review of physiological and genetic basis. Planta251, 27. doi: 10.1007/s00425-019-03322-z
47
ZhangX. J.LaiY. C.MengY.TangA.DongW. J.LiuY. H.et al. (2023). Analyses and identifications of quantitative trait loci and candidate genes controlling mesocotyl elongation in rice. J. Integr. Agric.22, 325–340. doi: 10.1016/j.jia.2022.08.080
48
ZhangH.MaP.ZhaoZ.ZhaoG.TianB.WangJ.et al. (2012). Mapping QTL controlling maize deep-seeding tolerance-related traits and confirmation of a major QTL for mesocotyl length. Theor. Appl. Genet.124, 223–232. doi: 10.1007/s00122-011-1700-y
49
ZhaoH.LiJ. C.YangL.QinG.XiaC. J.XuX. B.et al. (2021). An inferred functional impact map of genetic variants in rice. Mol. Plant14, 1584–1599. doi: 10.1016/j.molp.2021.06.025
50
ZhaoY.ZhaoW. P.JiangC. H.WangX. N.XiongH. Y.TodorovskaE. G.et al. (2018). Genetic architecture and candidate genes for deep-sowing tolerance in rice revealed by non-syn GWAS. Front. Plant Sci.9. doi: 10.3389/fpls.2018.00332
51
ZhengJ.HongK.ZengL.WangL.KangS.QuM.et al. (2020). Karrikin signaling acts parallel to and additively with Strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell32, 2780–2805. doi: 10.1105/tpc.20.00123
Summary
Keywords
rice, mesocotyl length, genome-wide association analysis, haplotype, candidate gene
Citation
Xue L, Wang S, Zhang Q, Han B, Cui D, Han L, Deng J and Ma X (2025) Identification of key genes associated with mesocotyl length through a genome-wide association study in rice. Front. Plant Sci. 16:1546580. doi: 10.3389/fpls.2025.1546580
Received
17 December 2024
Accepted
21 April 2025
Published
13 May 2025
Volume
16 - 2025
Edited by
Yuqing He, Huazhong Agricultural University, Wuhan, China
Reviewed by
Gurjeet Singh, Texas A and M University, United States
Ujjawal Kumar Singh Kushwaha, Nepal Agricultural Research Council, Nepal
Qiaojun Lou, Shanghai Agrobiological Gene Center, China
Updates
Copyright
© 2025 Xue, Wang, Zhang, Han, Cui, Han, Deng and Ma.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longzhi Han, hanlongzhi@caas.cn; Jianxin Deng, djxin555@hotmail.com; Xiaoding Ma, maxiaoding@caas.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.