- 1Department of Plant Pathology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 2Senior Research Coordinator, IFFCO- Nanoventions Private Limited, Coimbatore, Tamil Nadu, India
- 3Directorate of Research, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 4Department of Rice, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 5Institute of Agriculture, Tamil Nadu Agricultural University, Kumulur, Trichy, Tamil Nadu, India
Sheath blight disease is accountable for substantial loss in rice production worldwide. Endophytic bacteria are exploited as biocontrol agents due to their effectiveness in antagonizing a wide range of phytopathogens through a multifaceted approach. In the present study, the potentiality of deploying endophytic bacteria for the sustainable management of rice sheath blight was investigated. Over 40 bacterial endophytes were obtained and screened for their antagonistic activity against Rhizoctonia solani by a dual-culture assay. Among them, B. velezensis B13 exhibited higher mycelial inhibition (77.33%) against R. solani. A scanning electron microscopic study of the interaction of R. solani with B13 revealed distorted and deformed mycelia of R. solani. An analysis of secondary metabolites produced by B. velezensis B13 at their zone of interaction with R. solani confirmed the presence of various bioactive compounds of an antifungal and antimicrobial nature. A molecular docking study revealed that the compound 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone exhibited the highest binding affinity for Actin like protein (−7.6 kcal/mol), β-1,3 glucan synthase (−7.7 kcal/mol), Pectinesterase (−4.2 kcal/mol) and Polygalacturonase (−6.5 kcal/mol) protein targets of R. solani compared to the commercial fungicide carbendazim. In vivo experiments also proved the efficacy of B. velezensis B13 in suppressing rice sheath blight disease reduction upto 16.8± 0.2 besides enhancing the growth of the plant. Furthermore, B. velezensis B13 upregulated the expression of rice transcription factors and defense genes, viz., WRKY, PR1, PAL, LOX, FLS2 and CERK1, by several folds related to the inoculated and healthy control, leading to the suppression of R. solani. Our results suggest that B. velezensis (B13) could be a potential candidate for developing a bioconsortia for the sustainable management of rice sheath blight.
Introduction
Sheath blight of rice, caused by Rhizoctonia solani, is a destructive disease hindering rice production. Anastomosis group AG1-IA of R. solani is the dominant group infecting rice and is considered the primary causal agent of sheath blight, a serious disease impacting rice yield globally (Chen et al., 2023). AG1-IA exhibits high strain variation, including differences in virulence, genetic diversity and host adaptation. Multiple pathotypes and haplotypes have been identified within AG1-IA populations, reflecting its substantial genetic variability and adaptability, which pose challenge for breeding resistant rice cultivars (Zheng et al., 2013). The disease causes significant annual yield loss, ranging from 10% to 30%, and it is estimated to increase up to 50% in the coming years (Jamali et al., 2020). To date, only a few rice varieties resistant to sheath blight have been identified. Although several cultivars have exhibited varying levels of resistance, no variety has displayed complete immunity to the disease. The screening of resistant varieties against rice sheath blight is still in progress (Abbas et al., 2023). Commercial fungicides have been exploited for controlling the disease, but they are expensive and leave chemical residues in food materials, leading to health and environmental problems. An eco-friendly method for controlling the plant disease is the use of biocontrol agents, which is a sustainable agricultural practice (Kumar et al., 2020). Endophytic microorganisms are now widely used as biocontrol agents in agriculture due to their innate ability to colonize and reside within plant tissues, boosting their physiological and developmental processes. They also help overcome biotic and abiotic stresses encountered by the host plant (Wani et al., 2015).
Identifying potential endophytes for promoting plant growth and controlling diseases requires exploration of a range of bacterial endophytes found in landraces and rice varieties. Several research works have documented the isolation of bacterial endophytes of rice from traditional varieties of rice and wild accessions (Elbeltagy et al., 2000). Various approaches, including culture-based and culture-independent methods, have been employed to isolate endophytes from plant tissues, ovule and seed endosphere to determine their plant growth-promoting activities (Hardoim et al., 2012; Walitang et al., 2017).
Several endophytic bacterial genera, including Bacillus, Strenotrophomonas, Pseudomonas, Burkholderia, Enterobacter, Micrococcus and Serratia, are well known for producing lytic enzymes, such as chitinase, protease and β-glucanase, which are accountable for the degradation of the fungal cell wall during antagonistic interactions (Khare et al., 2018). Endophytic bacteria play a crucial role in promoting plant growth and minimizing pathogen infection through direct and indirect mechanisms. Their direct impact is primarily due to their antagonistic behavior against the pathogen by competing with and parasitizing them, exhibiting antibiosis and producing extracellular digestive enzymes. In contrast, the indirect impact is through the activation of plant defense mechanisms in response to various diseases (Soliman et al., 2022).
Endophytic Bacillus species have been investigated to combat sheath blight disease as they possess beneficial properties, such as stimulating plant growth and eliciting an immune response (Nagendran et al., 2014; Islam et al., 2020; Zheng et al., 2021). These bacteria can produce various antimicrobial compounds, viz., siderophores (bacillibactin), polyketides (bacillaene) and lipopeptides (fengycin, surfactin, bacillomycin D and iturin) (Nicholson, 2002). It is imperative to identify these secondary metabolites and their related antimicrobial nature to ascertain the biological control activity of Bacillus species. Bacillus spp. acts as plant safeguards against phytopathogens through the induction of systemic resistance and subsequent upregulation of the plant defense-linked genes (Seo et al., 2012; Rais et al., 2017). In the current investigation, the endophytic bacteria B. velezensis B13 was evaluated for its antagonistic potential, secondary metabolite production, determining the potential of the metabolites used in silico docking studies, and its efficacy for inducing the defense system against rice sheath blight disease.
Materials and methods
Collection and isolation of Rhizocontia solani
The plants showing typical sheath blight symptoms were collected during 2021–2022 from various rice growing regions of Tamil Nadu. The samples were brought to the laboratory and washed under running water to eliminate dirt particles and blot dried. A lesion of 2–3 mm portion of the advancing region was carefully excised; the surface was sterilized with 1% sodium hypochlorite (NaOCl) for 1 min and then rinsed thrice in sterile distilled water. The sterilized leaf tissues were dried with sterile filter paper and then transferred to a Petri plate containing potato dextrose agar (PDA) supplemented with streptomycin sulphate (PCT1120 Himedia, Mumbai, Maharashtra, India), followed by incubation at 28 ± 2°C for 5–7 days. The pure culture of isolates was transferred to the potato dextrose agar (PDA) slants by the single hyphal tip method and maintained at 25 ± 2°C for further studies. A glycerol stock of all the isolates was also preserved at −80°C for long-term storage (Sandoval and Cumagun, 2019).
Isolation of endophytic bacteria
For endophytic bacteria isolation, healthy rice plants and wild rice species grown in Paddy Breeding Station, Tamil Nadu Agricultural University, Coimbatore, were used (Table 1). Plant tissues, such as leaves, stems and roots, were collected in a polythene bag and were brought to the laboratory. One gram of tissue samples was surface sterilized using a 0.5% sodium hypochlorite solution. The samples were then washed three times with sterile distilled water and dried on sterile filter paper to ensure sterility. The samples were homogenized using a pestle and mortar with 1 mL of 0.9% peptone saline buffer. A total of 1 ml of tissue suspension was subjected to serial dilution, and 10−4–10−5 dilutions were inoculated on Luria Bertani agar (M1151, HIMEDIA) plates and incubated at 28 ± 2°C for bacterial growth. The morphologically different bacterial colonies were re-streaked on LB agar plates. The purified bacteria were maintained in an LB broth of 20% sterile glycerol and preserved at −80 °C for further studies (Kumar et al., 2020).
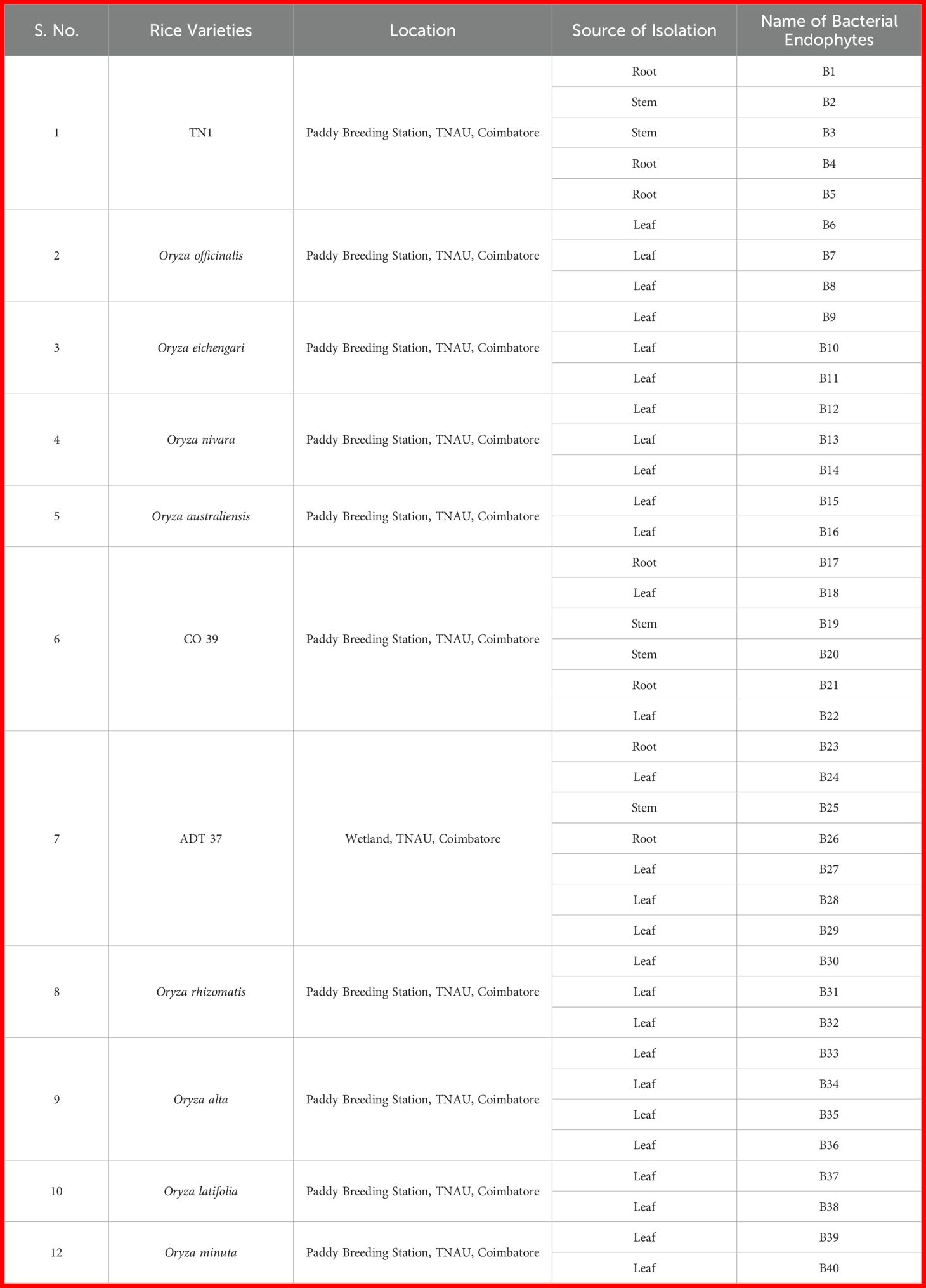
Table 1. List of endophytic bacterial isolates obtained from commercial rice varieties and wild rice species.
In vitro screening of endophytic bacteria against R. solani
The antagonistic activity of forty endophytic bacterial isolates was tested in vitro against a virulent isolate of R. solani AG1-1A which was isolated from var. ADT 43 (NCBI accession No. OQ940459.1). A freshly grown mycelial disc (9 mm) of a five-day old culture of R. solani was placed on one side, 1 cm away from the edge of the sterilized Petri plate. The bacterial endophytes were streaked perpendicular to the mycelial disc on the other side of the Petri plate (Jamali et al., 2020). The experiment was performed with three replications for each bacterial isolate, and a control plate was maintained by inoculating the pathogen alone at the end of the Petri dish containing the PDA medium. The plates were then incubated for seven days at room temperature (28 ± 2°C). The efficacy of the endophytic bacteria against the pathogen was determined based on the size of their inhibition zone. The radial growth of the pathogen and percent inhibition relative to the control were calculated using the following formula (Shakeel et al., 2015):
where C is the mycelial growth of the pathogen in the control and T is the mycelial growth of the pathogen in the dual-culture treatment. On the basis of the higher percentage inhibition of R. solani, Bacillus strain B13 was selected for further studies.
Molecular characterization of effective endophytic bacteria
For the molecular identification of strain B13, the 1.5 kb full-length 16S rRNA gene was amplified by polymerase chain reaction (PCR) with a universal forward and reverse primer: 27F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGACTT-3′). Polymerase chain reaction (PCR) was performed in a 25 µL reaction mixture that included 10 µL of master mix (RR310 EmeraldAmp), 1 µL of bacterial genomic DNA at a concentration of 20 ng, 1 µL of each primer at a concentration of 10 µM and 12 µL of sterilized deionized water. The PCR amplification conditions of the thermocycler (Nexus Gradient, Eppendorf, Hamburg, Germany) were as follows: an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min; primer annealing at 55°C for 1 min; extension at 72°C for 40 s and final extension at 72°C for 10 min. The amplified 16S rRNA gene product was visualized on 1% agarose gel with a UV transilluminator, photographed using the gel documentation system and sequenced at Biokart India Pvt. Ltd., Bangalore, India. The sequence similarities were determined using Basic Local Alignment Search Tool (BLAST) analysis (https://www.ncbi.nlm.nih.gov). The sequences with the greatest homology and highest similarity were obtained from the NCBI GenBank database, and multiple sequence alignment was conducted using the ClustalW algorithm. A neighbor joining (NJ) phylogenetic tree was constructed with bootstrap test (1000 replicates) using MEGA 11 software (Kumar et al., 2018).
Scanning electron microscopy
SEM analysis was performed to further validate the effect of the extracellular secondary metabolites of Bacillus strain B13 on R. solani (Zhu et al., 2023). A small piece of freshly grown mycelia of R. solani regarded as the control and the mycelia and interaction region with B13 in the dual-culture plate were cut using a sterile scalpel and placed in perforated capsules. The samples were then fixed in 1.5% glutaraldehyde in phosphate buffer for 4 h. After fixation, the mycelia were rinsed with 0.2 M sodium cacodylate buffer (pH 6.2) and dehydrated in a series of ethanol concentrations (30%, 50%, 70%, 80%, 90% and 100%) for 15 min each. The dehydrated mycelia were then mounted onto aluminum stubs using conductive double-sided carbon tape, followed by subsequent sputter coating with gold in a rotary vacuum pump for 40 s for complete and uniform coating over the sample surface. Finally, the morphological changes in the mycelium of the pathogen were observed under a scanning electron microscope.
GC/MS analysis of secondary metabolites extracted from the zone of inhibition of R. solani and bacterial strain B13
The secondary metabolites produced by the highly efficient bacterial endophyte (B13) and their di-trophic interaction with R. solani were characterized through gas chromatography–mass spectrophometry (GC–MS). The bioactive compounds produced by strain B13 during their di-trophic interaction with R. solani in PDA from the zone of inhibition were extracted by excising the agar using a sterile scalpel. The excised agar was blended with HPLC-grade acetonitrile in a 1:4 ratio (5 g agar in 20 mL of HPLC grade acetonitrile). For homogenization, the mixture was sonicated twice for 30 s at 30% of the power of the sonicator. After homogenization, the samples were centrifuged and filtered to eliminate the solid particles. A vacuum flash evaporator (Roteva Equitron Make, Mumbai, India) was used for drying the samples. The final product was dissolved in 1 mL of HPLC-grade methanol following the removal of the eluent (Kumar et al., 2018). The variation in the secondary metabolite profile generated during the interaction of B13 with R. solani was analyzed using uninoculated control, pathogen-inoculated control and bacterial antagonist-inoculated control via GC/MS (GC Clarus 500 Perkin Elmer, USA) with reference to the NIST 2005 MS data library.
In silico molecular docking study of R. solani
Protein targets of R. solani utilized for molecular modeling
A molecular docking study was performed to predict the potential biomolecules that bind with the protein targets of R. solani. Four potential protein targets of R. solani, namely Polygalacturonase (UniProt ID:L8X539) (Chen et al., 2017), β1,3 glucan synthase (UniProt ID: L8WXK6) (Bhaskar Rao et al., 2020), Pectinesterase (UniProt ID:L8X224) (Prabhukarthikeyan et al., 2022) and Actin like protein ARP6 (UniProt ID: A0A8H3CCY1) (Dallakyan and Olson, 2015), that may have a pathogenesis role were chosen, and the protein sequences were retrieved from the Uniprot database (https://www.uniprot.org/, 2024-05). The sequences were compared with other organisms using the Basic Local Alignment Search Tool (BLAST) in the NCBI database. Based on the similarity of the sequences from the BLAST analysis, templates for homology modeling were selected. Homology modelling was employed for the hypothetical protein structures from the SWISS-MODEL server (https://swissmodel.expassy.org/). The protein targets were validated utilizing the Ramachandra plot of the PROCHECK tool from the Structural Analysis and Verification Server (SAVES v6.1, Meta server) (http://saves.mbi.ucla.edu/) in order to ensure the accuracy of the protein model.
Identification of biomolecule ligands for molecular docking
The bioactive compounds produced by the strain B13 alone and its di-trophic interaction with R. solani showing high peak areas were specifically chosen as the ligands for docking. The three-dimensional (3D) structures of the ligands were retrieved in SDF format from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The commercial fungicide carbendazim was used as a respective reference ligand molecule.
Molecular docking and virtual screening
Molecular docking was carried out employing the PyRx 0.8 AutoDock vina module (Singh et al., 2002). Using PyRx software 0.8 version “make macromolecule option”, protein preparation was performed. All ligand structures underwent minimization using a conjugate gradient, involving 200 steps of the first-order derivative optimization process and commercial molecular mechanics parameters Unified Force Field (UFF). The binding site pockets for the targets were identified using the Computed Atlas Topography of Proteins CASTp 3.0 server. During the docking protocol execution, the ligands were permitted to create flexible conformations and orientations with an exhaustiveness value of 8. The interactions of the docked conformations of the protein–ligand complexes were visualized using BIOVIA Discovery studio client 2021 (https://www.3ds.com/products-services/biovia/) for visualization. To distinguish between the receptor, ligand and interacting atoms, different colors were assigned to each of them.
In vivo challenge experiment
An in vivo challenge experiment was conducted to test the efficacy of the isolated bacterial strain B13 under glass-house conditions (temperature range of 25–30°C and a relative humidity of 60–90%). The experiment was performed in a completely randomized block (CRD) design, with the three treatments replicated thrice with three plants per replication. The treatments included (1) pathogen inoculation alone (R. solani), (2) seed treatment and Foliar spraying with B13 along with a challenge inoculation of R. solani (8 × 108 cfu/mL) and (3) healthy control.
Seeds of the rice cultivar Co43 were surface sterilized and raised in an earthen pot. When the plants were 50 days old, a mycelial disc of R. solani (diameter 2 mm) was inoculated beneath the rice leaf sheath covered with absorbent cotton (IRRI, 2002). Strain B13 was utilized by spraying after ten days of pathogen inoculation on the rice plants. The disease severity was assessed on the 10th day post-treatment using the 0–9 standard (Yang et al., 2017), and the disease index and control efficacy were calculated with the following formula (Azizi et al., 2017):
Study of defense gene expression in rice plant through RT-qPCR
To understand the induction of the resistance response against R. solani infection, differential expression of some antifungal defensive genes, namely WRKY 45 (transcription factor), WAK 85 (wall associated kinases), CERK 1 (chitin elicitor receptor kinase 1), LOX (lipoxygenases), PR1 (pathogenesis related protein), JAZ (Jasmonate zim domain protein), FLS 2 (receptor like kinase for flagellin) and PAL (phenylalanine ammonia lyase), of rice susceptible cv. CO43 by the endophyte B13 in the presence of R. solani, a glass-house experiment was conducted with four treatments: (1) interaction (spraying of rice plants with B13 and challenge inoculated with R. solani), (2) biocontrol (spraying of rice plants with B13 alone), (3) inoculated control (R. solani alone) and (4) healthy control (Mock). One hundred milligrams of rice plant tissue were collected separately from all the sets of the in vivo experiment and carefully brought to the laboratory for RNA extraction. Total RNA was extracted from the rice plants treated with B. velezensis challenged with R. solani using Trizol (Sigma Aldrich, St. Louis, MO, USA) at 0 h, 24 h, 48 h, 72 h and 96 h post-inoculation with R. solani. Likewise, RNA was extracted from untreated healthy control, plants treated with B. velezensis B13 alone and R. solani-inoculated control (Livak and Schmittgen, 2001). The concentration and purity of total RNA were quantified using a NanoDrop ND1000 spectrophotometer.
RNA was converted to cDNA utilizing the Thermo Fischer Scientific-Revert Aid First Strand cDNA Synthesis Kit (cat. #RR820A). An optimal nucleic acid quality was indicated by a ratio of 1.8 + 0.2. Subsequently, the cDNA was diluted 10-fold and utilized for qRT-PCR analysis, which was carried out in a BIO RAD CFX manager system. For qRT-PCR, the reaction mixture included 1.4 µL of cDNA template, 5 µL of SYBR Green master mix (KAPA SYBR@FAST for Light Cycler 480), 0.8 µL of 10 µM forward primer and 0.8 µL of 10 µM reverse primer. Using nuclease-free water, the final volume was adjusted to 10 µL. The PCR program included denaturation at 95°C for 1 min, followed by 50 cycles of amplification at 95°C for 10 s, 60°C for 30 s and 72°C for 30 s. Subsequently, a standard melting temperature analysis was performed. Actin was used as a housekeeping gene to normalize the gene expression. For each defense gene expression analysis, three biological replicates and two technical replicates were consistently maintained.
Statistical analysis
Fold changes in gene expression were determined utilizing the formula ΔΔCt = ΔCt sample–ΔCt reference. The relative fold changes in transcript levels were graphically represented by converting the ΔΔCt value to 2-ΔΔCt (Khaskheli et al., 2020).
Results
Isolation and screening of endophytic bacteria
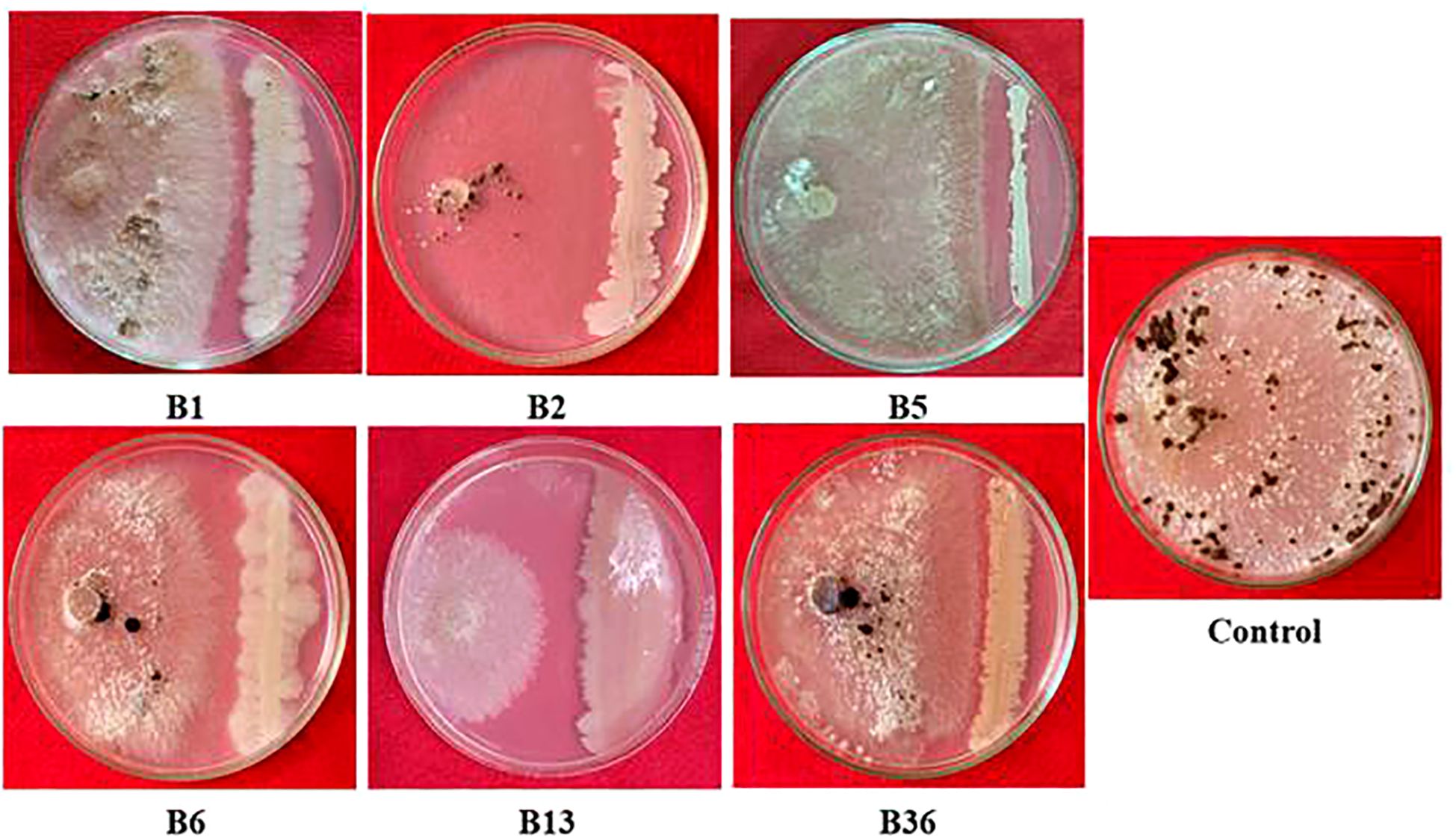
A total of forty endophytic bacteria were isolated and evaluated for their antagonistic activity against R. solani in a dual-culture assay (Supplementary Figure S1). Among them, six endophytic bacterial isolates showed antagonism against R. solani. The results showed that strain B13 displayed the highest inhibitory and antagonistic effects against the pathogen with the highest percentage of mycelia inhibition (77.33 (± 1.27) %) (Table 2). Therefore, based on the results obtained in the screening, isolate B13 was utilized for further study (Figure 1).
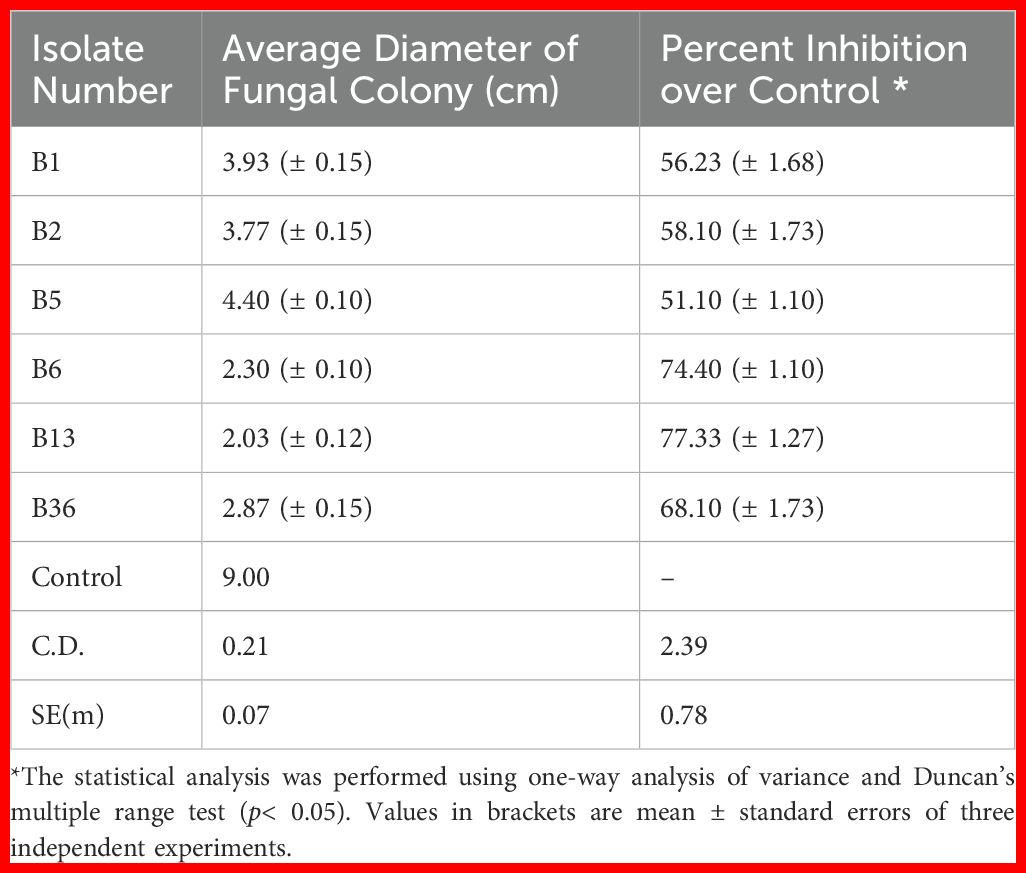
Table 2. Antagonistic activity of effective endophytic bacteria against R. solani by dual-culture method.
Molecular identification
Upon 16S rRNA gene sequencing, strain B13 was identified as B. velezensis. The 1500 bp sequence of the strain B13 was submitted to the NCBI database and provided with the accession number OQ941779.1. It showed 99.63% homology from the NCBI database with B. velezensis strain R-71003 (ON358418.1). A neighbor joining phylogenetic tree was constructed based on the 16S rRNA, which revealed that the isolate B13 formed a cluster with the B. velezensis group (Supplementary Figure S2).
Scanning electron microscopy
The morphological alterations in the mycelia of R. solani caused by volatile compounds from B. velezensis B13 were analyzed using scanning electron microscopy. The untreated mycelia were compared with the mycelia inoculated with B13. The results showed that the untreated mycelium exhibited a smooth and structurally intact appearance. Conversely, the mycelium of R. solani treated with B13 displayed a distorted and deformed morphology with an uneven thickness (Figure 2). Based on the results of the observed hyphal malformation under SEM, strain B13 had a significant impact on the mycelium of R. solani.
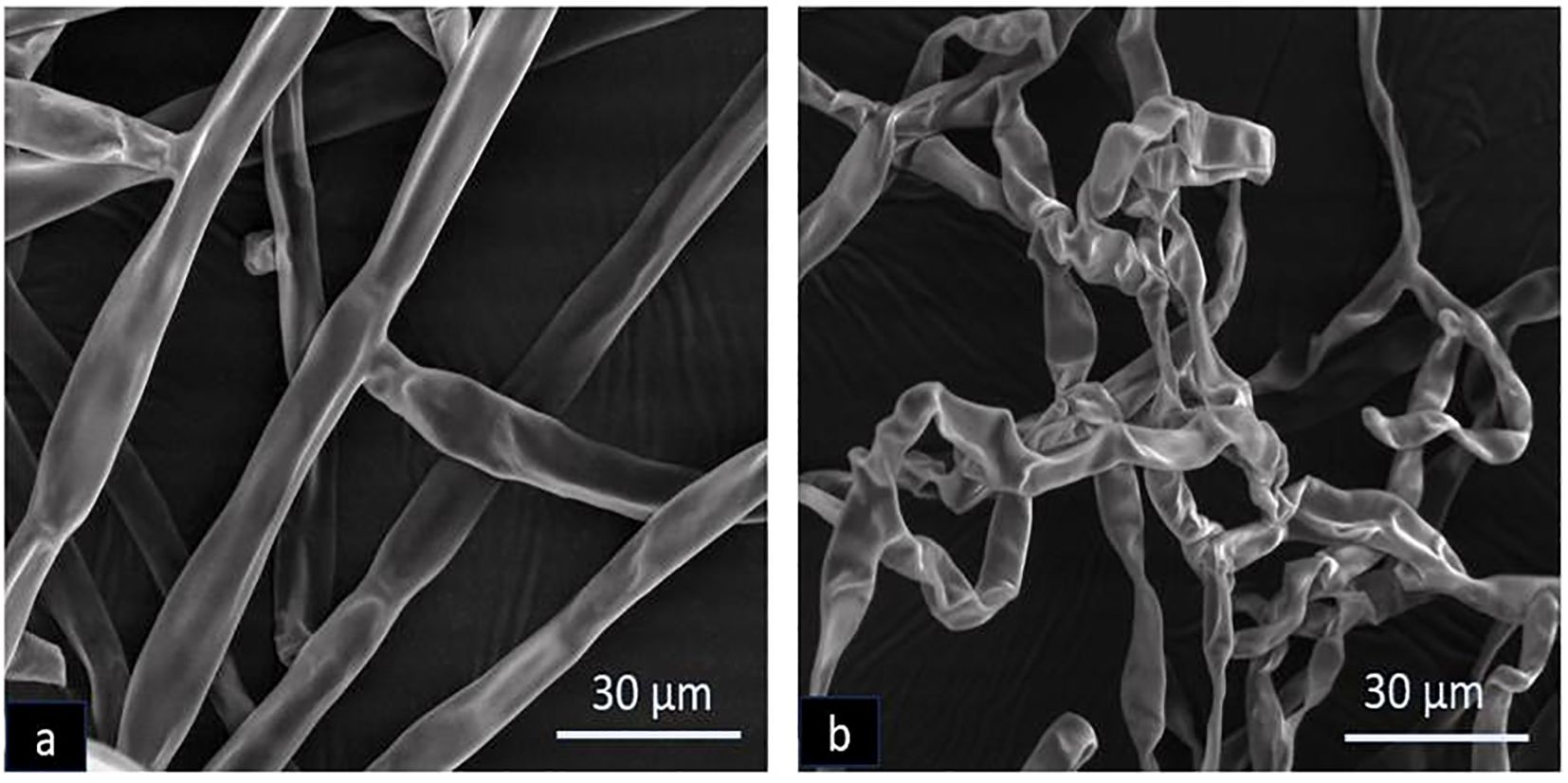
Figure 2. Scanning electron microscopy of morphological deformation of R. solani caused by B. velezensis B13: (a) untreated control and (b) treated with B13.
GC/MS analysis of secondary metabolites produced by B. velezensis B13, R. solani and their di-trophic interaction
B. velezensis B13, R. solani and their di-trophic interaction were profiled for a total of 39 biomolecules upon elimination of compounds in the PDA medium (control). A total of twelve bioactive metabolites produced by R. solani in PDA medium were identified: Naphthalene, squalene, Trans-geranylgeranio, Oleic acid, Hexadecanoic acid 1 4- methyl ester, 1-Hexadecanol 2-methyl, Phenol, Tetradecanoic acid, 1 2 -methyl methyl ester, Dichloroacetic acid, Trichloroacetic acid tridecyl ester and 1-Hexadecane (Supplementary Table S1; Figure 3a).
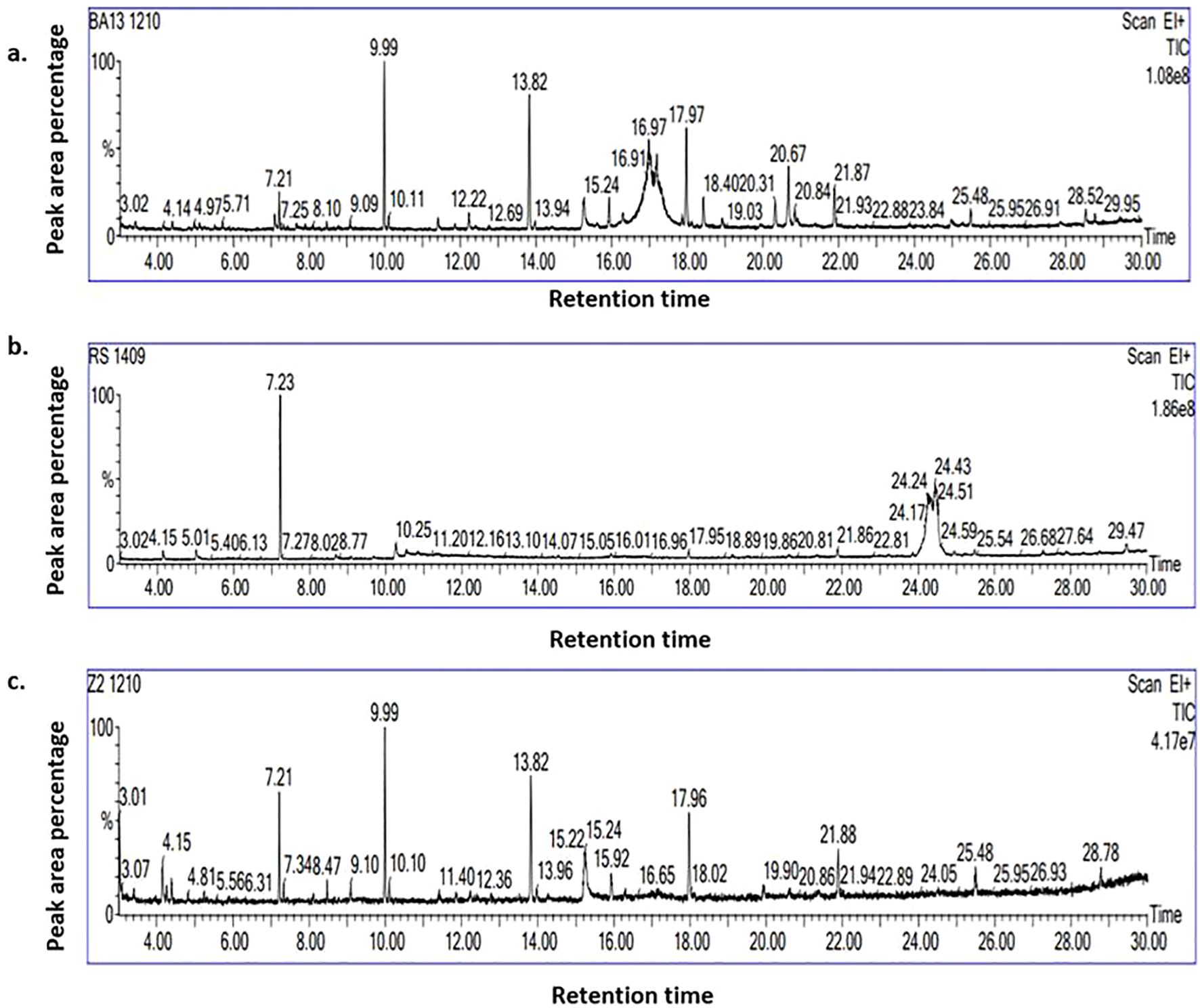
Figure 3. GC–MS chromatogram of biomolecules produced by (a) B. velezensis B13; (b) R. solani and (c) during the interaction of B. velezensis B13 and R. solani.
In the absence of R. solani, B. velezensis B13 generated a total of 17 compounds, 14 of which were unique to B. velezensis B13, which included Bis(2-ethylhexyl) phthalate, Octahydro-2H-pyrido(1,2-a) pyrimidin-2-one, 1-Undecanol, 4a(2H)-Naphthalenol octahydro-trans, Hexadecen-1-ol trans-9, Furan 2-ethenyl, Phthalic acid di (6-methylhept-2-yl) ester, Octadecanoic acid 2-propenyl ester, Dodecyl acrylate, Formamide N-(4,6-diamino-5-pyrimidinyl), Ergotamine, 9-Eicosene (E), Phenol 3,5-bis(1,1-dimethylethyl) and Di-Aspartic acid (Supplementary Table S2; Figure 3b).
During the interaction of B. velezensis B13 with R. solani, 14 biomolecules were identified, among which 12 molecules were profiled during interaction: Benzoic acid 3-amino-6-(1-pyrrolidinyl), Methyl 5,9-heptadecadienoate, 1-Octene 3,7-dimethyl, 5,8-Dimethoxyquinoxaline, Cyclotetradecane, Formic acid phenyl ester, 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone,4-(2,4,4-Trimethyl-cyclohexa-1,5-dienyl)-but-3-en-2-one, 2-Propenoic acid, butyl ester, 1-Docosene, Octadecane 1-chloro- and 1-Dodecane (Supplementary Table S3; Figure 3c). None of the biomolecules were produced in common by R. solani and its interaction with B. velezensis B13. The Venn diagram of the differentially expressed bioactive metabolites revealed phenol as a common compound during the interaction of the endophytic bacteria B. velezensis B13 with R. solani, B. velezensis B13 alone and R. solani alone (Supplementary Figure S3).
Molecular modeling and validation of protein targets
The protein targets, like Actin like protein ARP6, β1,3 glucan synthase, Pectinesterase and Polygalacturonase, were chosen as receptors due to their significant physiological functions (Supplementary Table S4). The three-dimensional structures of the proteins were predicted using the SWISS-MODEL for molecular docking studies. The sequence similarity between the template and 3D-modelled structure of the proteins was validated through the Ramachandran plot (Supplementary Table S5). The Ramachandran plot analysis of the 3D structure of modeled Actin like protein ARP6, β-1,3 glucan synthase, Pectinesterase and Polygalacturonase revealed 90.5%, 92.3%, 91% and 88.5% of amino acid residues in the most favored region, respectively (Supplementary Figure S4).
Molecular docking and virtual screening
Four protein targets of R. solani were docked with 12 compounds and exhibited different binding affinity towards the 12 compounds. The results showed that, out of 12 compounds, 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone interacted well with all four protein targets and displayed a higher binding affinity than the reference ligand carbendazim. The compound had a binding affinity value of −7.6 kcal/mol with the target Actin like protein ARP6 (H bonds: ASN 379), −7.7 kcal/mol with the target β-1,3 glucan synthase (H bonds: LYS 735), −7 kcal/mol with the target Pectinesterase (H bonds: GLN 76, ARG 152) and −6.5 kcal/mol with the target Polygalacturonase (H bonds: LYS 242) (Figure 4a). Similarly, compound 4-(2,4,4-Trimethyl-cyclohexa-1,5-dienyl)-but-3-en-2-one showed a good interaction of −6.9 kcal/mol for β-1,3 glucan synthase (H bonds: ASN 1096), −6.1 kcal/mol for Actin like protein ARP6 (H bonds: LYS 174, THR 188) and −5.7 kcal/mol for Pectinesterase (H bonds: ILE 219) (Figure 4b). Compound Hexadecen-1-ol, trans-9 showed a good interaction of −5.2 kcal/mol with Actin like protein ARP6 (H bonds: SER 202). The compound Phthalic acid, di (6-methylhept-2-yl) ester showed a good binding energy of −6.2 kcal/mol for Actin like protein ARP6 (H bonds: ASN 379) and −5.4 kcal/mol for pectinerase (H bonds: GLN 54, GLN 76). The binding energy for the control carbendazim was −5.0 kcal/mol for Actin like protein ARP6 (H bonds: ARG 374), −6.8 kcal/mol for β-1,3 glucan synthase (H bonds: GLU 1049), −5.5 kcal/mol for polygalacturonase (H bonds: TYR 251) and −5.3 kcal/mol for pectinesterase (H bonds: GLY 111, THR 141, SER 112, THR 113) (Table 3; Figures 4c, d).
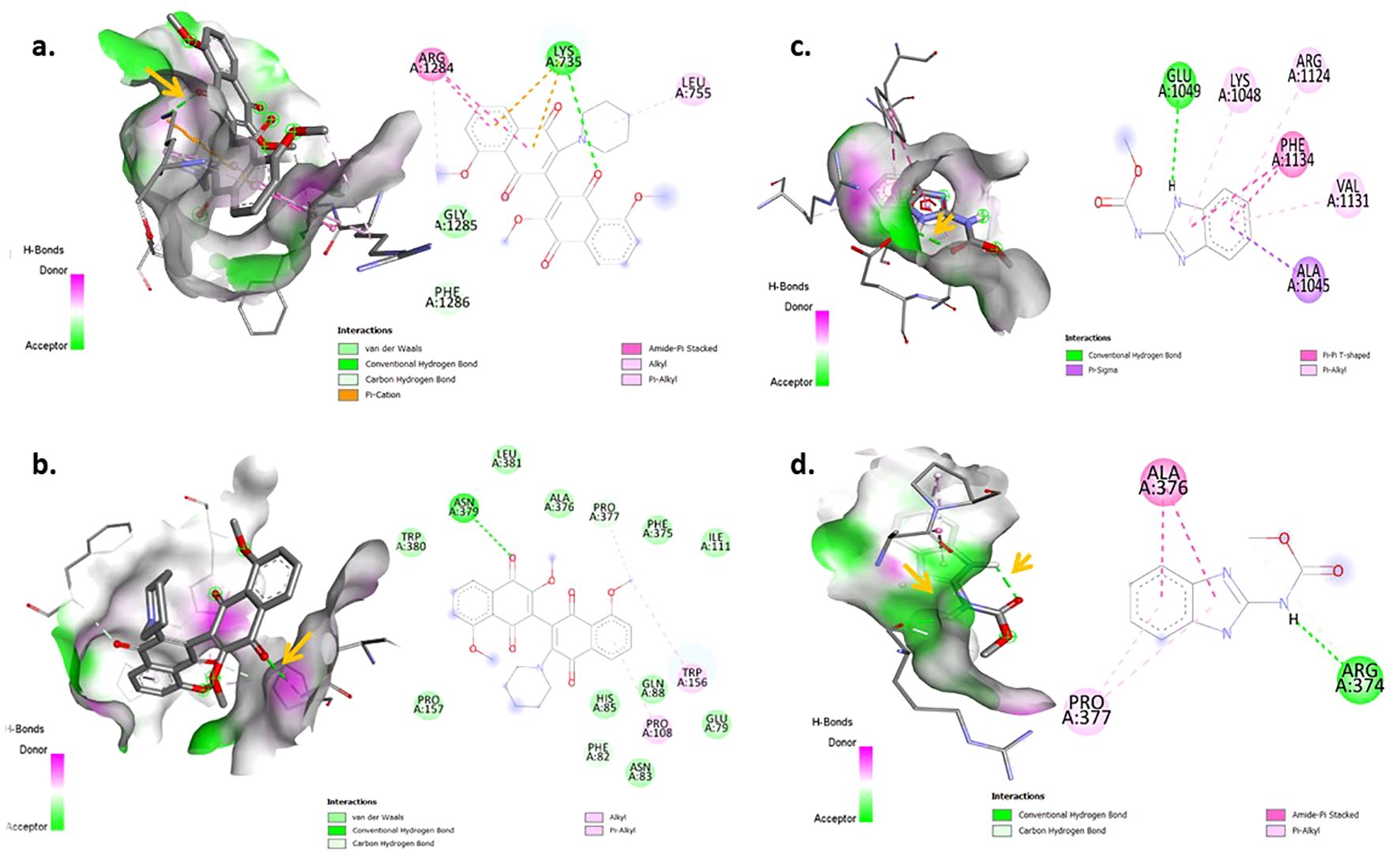
Figure 4. Visualization of Molecular docking interaction in 3D (left) and 2D (right): (a, b) Docked complex of 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene- 1,1′,4,4′-tetron with active site residues of β-1,3 glucan synthase and Actin like protein ARP 6 and x (c, d) Docked complex of carbendazim with active site residues of β-1,3 glucan synthase and Actin like protein ARP6. Arrow represents the conventional hydrogen bond.
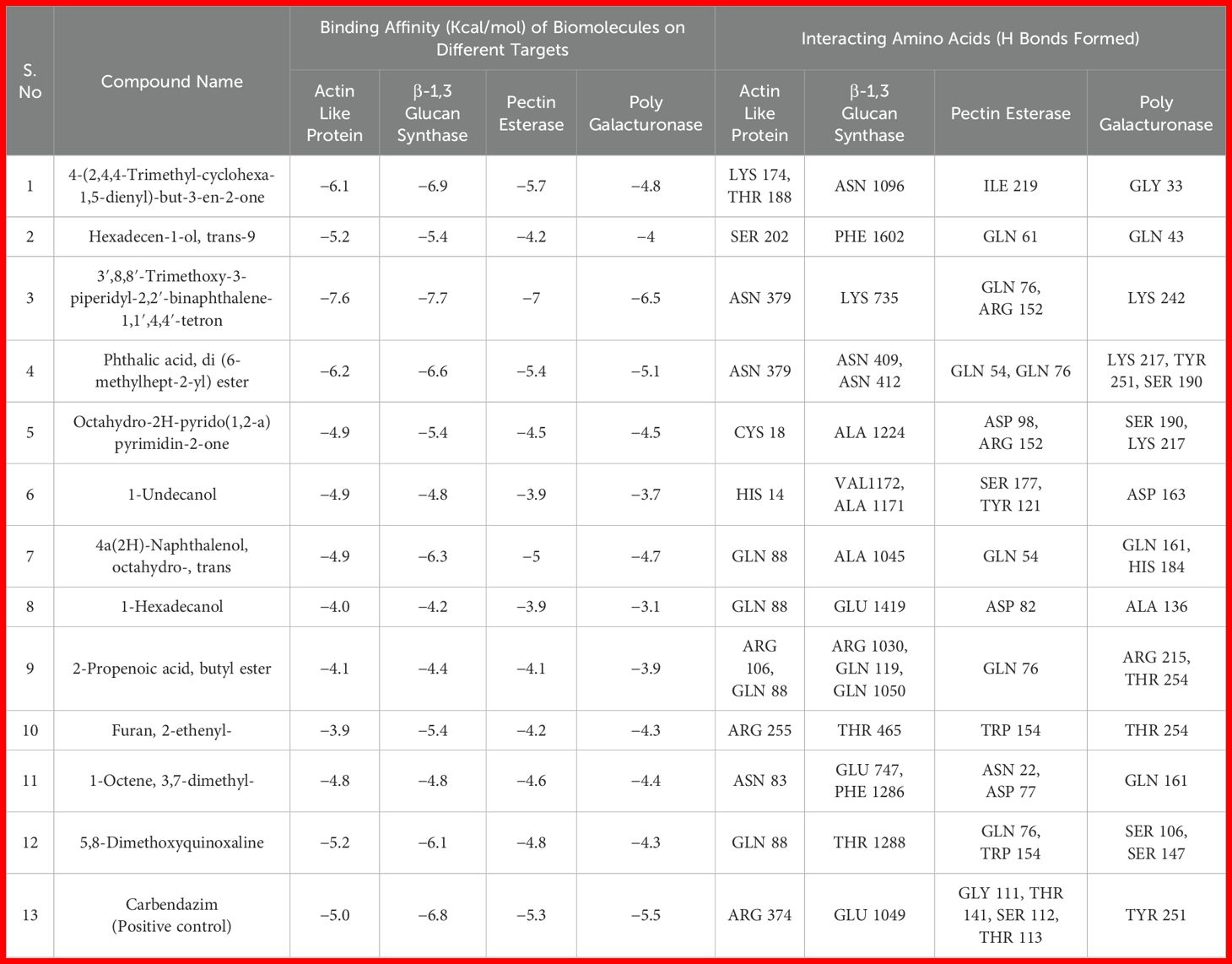
Table 3. Molecular docking for the interaction of metabolites of B. velezensis B13 with the protein targets of R. solani.
In vivo evaluation of B. velezensis B13 against R. solani infected rice plants
The antagonistic nature of B. velezensis B13 was studied under glass-house conditions for determining its effect in suppressing sheath blight disease caused by R. solani. The rice plants inoculated with R. solani showed a disease index of 65.46 ± 1.12, while application of B. velezensis B13 resulted in a reduced disease index of 54.43 ± 0.03 (Table 4). Both seed treatment and foliar spraying of B. velezensis B13 resulted in a disease reduction of 16.8 ± 0.2 compared to the control (Figure 5).
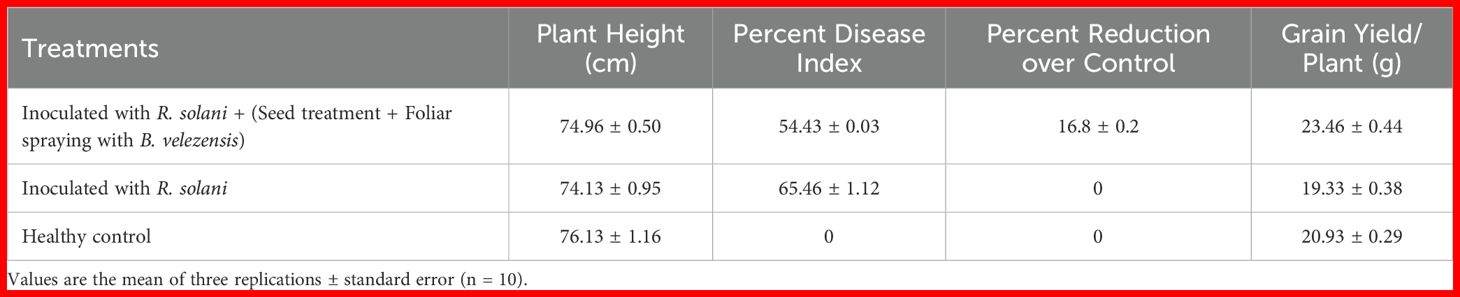
Table 4. In Vivo antagonistic effect of B. velezensis B13 against rice sheath blight under glass-house conditions.

Figure 5. In vivo antagonistic activity of B. velezensis B13 against R. solani infection of rice. (Arrows indicate the development of lesions upon inoculation of R. solani).
Induction of plant defense gene transcripts by endophytic bacterium
The rice plants treated with B. velezensis B13 challenged with or without R. solani altered the expression of transcription factor (WRKY45), wall associated kinases 85 (WAK 85), chitin elicitor receptor kinase 1 (CERK1), lipoxygenases (LOX), jazmonate zim domain protein (JAZ), receptor like kinase for flagellin (FLS2), phenylalanine ammonia lyase (PAL) and pathogenesis related protein 1 (PR1) genes responsible for plant defense.
The expression of WRKY transcript was upregulated in all the treatments from 0 to 24 h. However, upregulation was more significant in the plants treated with B. velezensis B13 challenged with R. solani in all the intervals, and a 0.8-fold increase in WRKY 45 transcript was observed at 96 h. In addition, the plants treated with B. velezensis showed an increase in expression of WRKY transcript up to 0.7-fold after 72 h, which was reduced to 0.1-fold after 72 h. The transcript level in the R. solani inoculated control declined after 48 h. Furthermore, the expression level of WRKY transcript in the untreated healthy control increased up to 0.3-fold and declined after 24 h (Figure 6a).
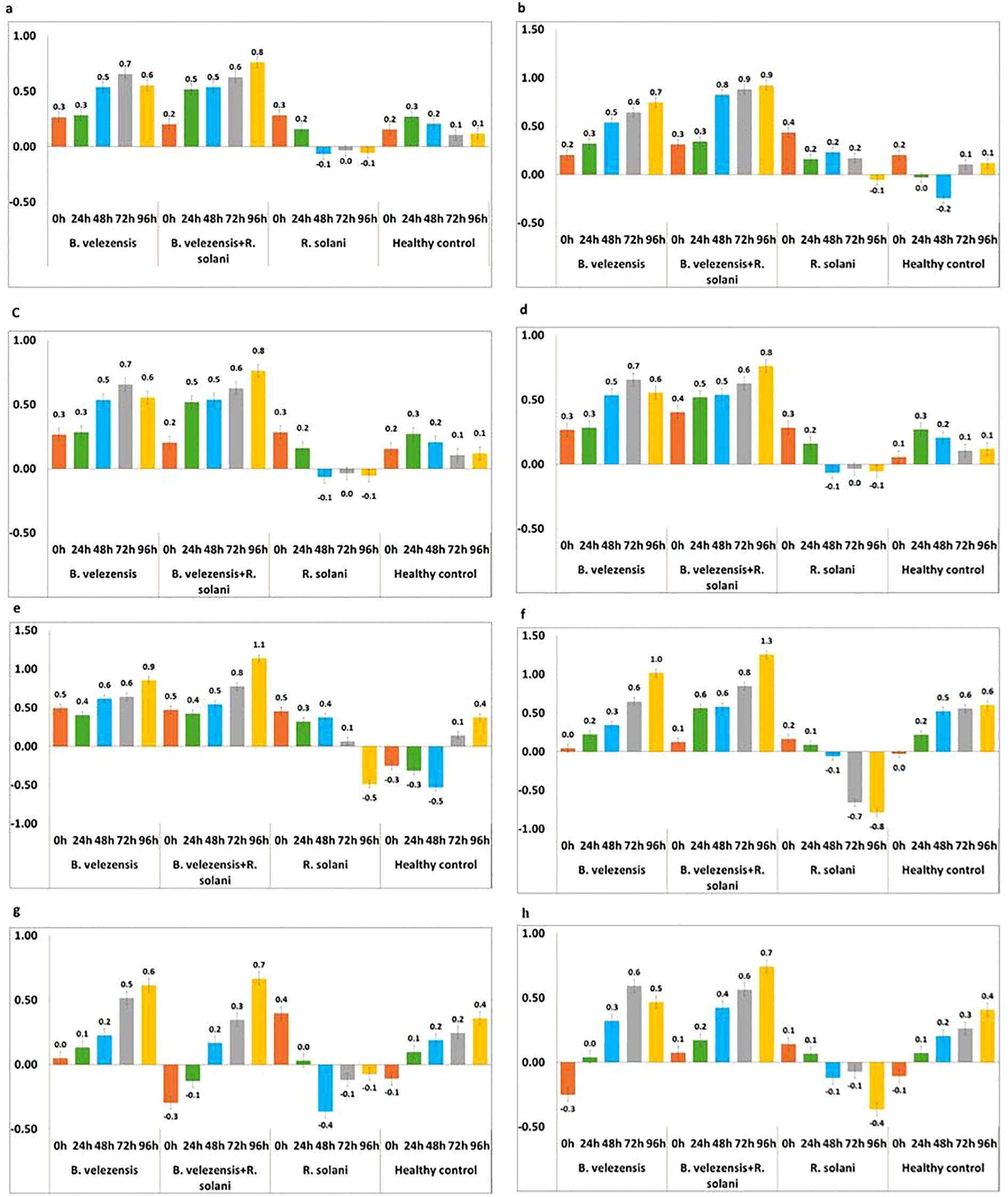
Figure 6. Expression pattern of defense genes in rice plants treated with B. velezensis B13 under mono-, di- and tri-trophic interactions at different time intervals. (a) Expression pattern of WRKY; (b) Expression pattern of JAZ; (c) Expression pattern of WAK; (d) Expression pattern of PR1; (e) Expression pattern of PAL; (f) Expression pattern of LOX; (g) Expression pattern of CERK1; (h) Expression pattern of FLS2.
JAZ gene was a key regulator in the plant defense response through the JA signaling cascade. Upregulation of JAZ was noticed at 0 h in all the treated and untreated plants. The level of JAZ was more pronounced in the plants treated with B. velezensis B13 challenged with R. solani after 24 h, which showed a 0.9-fold increase after 72 to 96 h. The expression level of JAZ was upregulated in the B. velezensis-treated plants, which showed a 0.7-fold increase after 72 h. The plants inoculated with R. solani showed an upregulation from 0 to 72 h and displayed downregulation after 72 h (Figure 6b).
Assessing the expression of WAK transcript revealed a significant increase in the plants treated with B. velezensis challenged with R. solani, which showed up to a two-fold increase after 72 h. Similarly, the plants treated with B. velezensis also showed an increase in WAK transcript from 0 h to 96 h, which showed up to a 1.9-fold increase after 72 h. The expression of WAK transcript was downregulated after 24 h in the plants inoculated with R. solani, while in the healthy control, there was an upregulation from 0 h to 96 h (Figure 6c).
PR1 gene is regarded as a key regulator gene for systemic acquired resistance. The expression of PR1 was upregulated from 0 to 24 h in all the treatments, and a characteristic downregulation of the PR1 transcript was noticed in the R. solani-inoculated plants after a 24 h interval. The significant upregulation of PR1 was observed in the plants treated with B. velezensis challenged with R. solani, which increased the transcript level to 0.8-fold after 72 h. The plants treated with B. velezensis showed an increase in the expression level from 0.3 to 0.7-fold in 0 to 72 h and showed a decline to 0.1-fold after 72 h. The expression of PR1 was upregulated from 0 h to 96 h in the untreated control plants (Figure 6d).
The expression level of PAL was upregulated in both the plants treated with B. velezensis alone and plants treated with B. velezensis challenged with R. solani. The level of induction of PAL was increased to 1.1-fold after 72 h in the plants treated with B. velezensis challenged with R. solani. A comparison of the expression of PAL in the B. velezensis-treated plants reflected a 0.9-fold increase after 72 h, while the pathogen-inoculated control showed an upregulation of PAL from 0 h to 72 h and a downregulation after 72 h. The PAL expression level in the untreated healthy control showed a downregulation from 0 h to 48 h and an upregulation after 48 h (Figure 6e).
The upregulation of LOX was noticed in the plants treated with B. velezensis and also in the plants treated with B. velezensis co-challenged with R. solani. The level of induction of LOX was increased to 1.3-fold after 72 h in the plants treated with B. velezensis challenged with R. solani. A comparison of the expression of LOX in the B. velezensis-treated plants reflected a one-fold increase after 72 h, while in the pathogen-inoculated control, the expression of LOX was downregulated after 24 h. The untreated plants showed an upregulation after 24 h, and a 0.6-fold increase was noticed from 72 h to 96 h (Figure 6f).
The expression level of CERK transcript varied between the different treatments. The expression of the CERK gene was upregulated in the B. velezensis-treated plants, and an increase of 0.6-fold was observed after 72 h. The rice plants treated with B. velezensis B13 challenged with R. solani showed upregulation after 24 h, and an increase of up to 0.7-fold was noticed after 72 h, which declined to 0.3-fold after 72 h. The activity of CERK in the R. solani-inoculated control was significantly downregulated after 24 h to 96 h. The untreated control plants showed an upregulation of the CERK gene after 24 h and an increase to 0.4-fold after 72 h (Figure 6g).
The transcription rate of the FLS2 gene was upregulated in the B. velezensis-treated plants after 24 h and showed a 0.6-fold increase after 48 h and a decrease to 0.1-fold after 72 h. The plants treated with B. velezensis challenge inoculated with R. solani showed an increase in FLS2 from 0.1-fold to 0.7-fold in 0 h to 96 h. The plants inoculated with R. solani showed downregulation of FLS after 24 h intervals. The untreated healthy control showed an upregulation of the FLS2 gene after 24 h and an increase to 0.4-fold after 72 h (Figure 6h).
Discussion
Biological control is considered a sustainable strategy for the management of plant diseases by reducing the need for harmful pesticides and, thereby, promoting a healthier ecosystem. Recently, the endophytic bacteria suppression of plant diseases has received much attention (Kumar et al., 2020). The diversity of the different endophytic bacteria from the rice plant that are antagonistic to R. solani, which causes sheath blight in rice, is well established (Jeong et al., 2017; Khaskheli et al., 2020). Among the different endophytic bacteria, Bacillus species quench the plant pathogens by producing antifungal biomolecules and antimicrobial peptides and by inducing an immune response (Aloo et al., 2019; Nakkeeran et al., 2019). Because of their wide range of bioactive substances, the Bacillus species were considered a more effective and eco-friendlier supplement when it comes to suppressing soil-borne diseases (Sun et al., 2017). They are also a source of secondary metabolites of biotechnological interest with pharmaceutical applications (Balderas-Ruíz et al., 2020).
In our current investigation, approximately forty endophytic bacteria were isolated from the rice plants. Among them, six isolates showed significant mycelial growth suppression against R. solani. The strain B13 showed the highest inhibition activity both under in vitro and in vivo conditions. The strain B13 was identified as Bacillus velezensis based on 16S rRNA gene sequencing. The potentiality of B. velezensis has been extensively studied as a biological control agent for controlling numerous fungal plant pathogens (Saravanan et al., 2022; Soliman et al., 2022). B. velezensis NKG-2 was able to produce hydrolytic enzymes associated with the breakdown of the fungal cell wall and was effective in suppressing phytopathogens (Myo et al., 2019). The interaction of antifungal biomolecules produced during the di-trophic interaction with R. solani through scanning electron microscopy revealed distorted and deformed mycelium with an uneven thickness. Similar ultrastructural changes in R. solani were observed during its di-trophic interaction with B. safensis Y246 (Zhu et al., 2023).
Bacillus velezensis harbors numerous biosynthetic gene clusters with the potential to produce a wide variety of metabolites (Su et al., 2024). The secondary metabolites produced by B. velezensis and their di-trophic interaction with R. solani were analyzed using GC–MS chromatography. Our study identified a diverse range of bioactive secondary metabolites of B. velezensis B13, which exhibit strong antifungal activity, effectively inhibiting the mycelial growth of R. solani. The antifungal property of these bacteria may be related to several chemical classes, including esters, alcohols, fatty acids, aldehydes, alkaloids, tertiary amines and ketones. Similarly, co-culturing B. velezensis B13 with R. solani stimulated the production of secondary metabolites, viz., Benzoic acid 3-amino-6-(1-pyrrolidinyl), Methyl 5,9-heptadecadienoate, 1-Octene 3,7-dimethyl, 5,8-Dimethoxyquinoxaline, Cyclotetradecane, Formic acid phenyl ester, 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone,4-(2,4,4-Trimethyl-cyclohexa-1,5-dienyl)-but-3-en-2-one, 2-Propenoic acid, Cetyl alcohol, Isobutyl acrylate and 1-Dodecane. The biosynthetic pathway of these compounds involves non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS), enabling the production of antimicrobial lipopeptides (e.g., surfactin, fengycin, bacillomycin D and bacillibactin) and polyketides (e.g., macrolactin, difficidin, and bacillaene) (Cawoy et al., 2015; Rabbee et al., 2019). Various secondary metabolites produced by the EA fraction of B. velezensis Lle-9, viz., cyclopeptides, linear peptides and some antibiotics were reported which possessed antifungal properties (Khan et al., 2020).
In this study, molecular docking was performed to identify the potential of 12 secondary metabolites produced by B. velezensis against the four targeted proteins (Actin like protein ARP6, Polygalacturonase, β1,3 glucan synthase and Pectinesterase) of R. solani. All the metabolites were found to interact with the target proteins. Among those, 3′,8,8′-Trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone exhibited the highest binding energy with all the protein targets. Several studies carried out on the interaction between protein and ligand provided valuable insights into the mechanisms by which these compounds combat pathogens (Malik et al., 2019; Gorai et al., 2023; Yasmin et al., 2023). The compound 3′,8,8′-trimethoxy-3-piperidyl-2,2′-binaphthalene-1,1′,4,4′-tetrone derived from the B. cereus strain KSAS17 exhibited antifungal properties against phytopathogenic fungus, Sclerotium bataticola (Al-Askar et al., 2024). It also shows broad-spectrum potential in antimicrobial, immunomodulatory and anti-inflammatory activities (Amer et al., 2019). Phthalic acid, a dicarboxylic compound derived from benzoic acid, is recognized for its antifungal properties. Its ester derivatives demonstrate insecticidal, antibacterial, and allelopathic effects, indicating their potential to enhance the resilience of plants, algae, and microorganisms against both biotic and abiotic stresses, thereby supporting their competitive survival (Awan et al., 2023). The antifungal activity of the Phthalic acid, di (6-methylhept-2-yl) ester produced by B. amyloliquefaciens against B. cinerea was reported by (Nakkeeran et al., 2020). Compound 4-(2,4,4-Trimethyl-cyclohexa-1,5-dienyl)-but-3-en-2-one is a naturally occurring terpenoid ketone which exhibits antimicrobial activity, potentially by disrupting bacterial cell membranes, leading to increased permeability and cell lysis (Idan et al., 2015). Volatile compound hexadecen-1-ol, trans-9 produced by Serratia plymuthica affected the mycelial growth of the soil-borne fungus R. solani (Kai et al., 2007).
Bacillus spp. protects the plants against phytopathogens by triggering systemic resistance, which subsequently results in the upregulation of defense-linked genes (Sahu et al., 2019). Further, bacterial endophytes stimulate the immune system of plants by releasing macromolecules and MAMP molecules in response to host signals (Wei et al., 1991). Our study demonstrated that the rice plants treated with bacterial endophyte B. velezensis B13 showed a significant upregulation of defense genes WRKY45 transcription factor, WAK 85, CERK1, LOX, JAZ, FLS 2, PAL and PR1 than in the inoculated control and healthy control plants. Transcription factors play a vital role in triggering plant defense mechanisms by controlling a wide array of signal transduction pathways responsible for carrying out diverse functions. A group of transcription regulators called WRKY can attach to the box in specific promoter regions in target genes and regulate transcription (Eulgem et al., 2000). Proteins of the WRKY family are important regulators of pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (Zhang and Wang, 2005; Chen et al., 2019). WRKY 45 transcription factor in the rice plants treated with B. velezensis B13 increased by multiple folds, which might have triggered the constitutive defense response against R. solani. Plant cell wall-associated kinases (WAKs) are receptor-like kinases present in multiple plant species possessing an extracellular domain and an intercellular domain that span the plasma membrane (Yang et al., 2019). They play a role in monitoring cell wall integrity and are implicated in pathogen responses (Rui and Dinneny, 2020). Several studies have shown that increased PAL activity, which is a crucial enzyme in the phenylpropanoid pathway of higher plants, is primarily associated with the ability to resist pathogens (Tonnessen et al., 2015). PR1 proteins were the first pathogenesis-related proteins identified in the PR family, associated with plant resistance to pathogens (Showmy and Yusuf, 2020). Tomato plants treated with B. subtilis CBR05 systemically induced the enzyme activities of PAL and β-1,3-glucanase, and significant upregulation was observed in PAL transcript of 0.49-fold expression at 72 h post-inoculation (hpi) as compared to its expression at 12 h post-inoculation (hpi) (Chandrasekaran and Chun, 2016). As a cell surface receptor, CERK 1 is essential for triggering innate immunity in response to biotic and abiotic stresses (Shinya et al., 2014). In rice, Chitin Elicitor Receptor Kinase 1 (CERK1) recognizes several elicitor compounds that have similar elicitor motifs, such as chitin as well as peptidoglycan and its derivatives. CERK1 is essential for chitin-mediated signaling and plays a crucial role in fungal resistance. Notably, pathogen effector proteins may interfere with CERK1 function, thereby evading CERK1-mediated recognition. Plants lacking functional CERK1 are not only unresponsive to chitin treatment but also exhibit increased susceptibility to fungal pathogens (Zhang and Zhou, 2010). These multifunctional receptors are essential for plant defense against pathogen invasion (Liu et al., 2012). LOX mediates jasmonic acid (JA) biosynthesis, which is essential for plant development and resists biotic and abiotic stresses (Singh et al., 2022). The rice ortholog receptor-like kinase FLAGELLIN-SENSITIVE 2 (FLS2) can identify the flagellar peptide flg22 and activate a plant immune response (Li et al., 2014). Ultimately, the present investigation highlighted the potential of endophytic B. velezensis in preventing pathogenic infections by triggering the WRKY 45 transcription factor and activating defense genes, such as WAK, PAL, LOX, PR1, JAZ, FLS2 and CERK1.
Conclusion
The present study highlights the potential of Bacillus velezensis B13 as an effective biocontrol agent against Rhizoctonia solani, the pathogen responsible for rice sheath blight. Strain B13 exhibited strong antagonistic activity through the production of novel secondary metabolites, mycoparasitic interactions, and induction of plant defense mechanisms. Molecular docking studies further supported the antifungal potential of metabolites with high binding affinity to key fungal pathogenicity proteins. Therefore, B. velezensis B13, after successful formulation and field trials across different rice ecosystems, can be used as a potential bio-control agent for the sustainable management of rice sheath blight.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Author contributions
SN: Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft. CG: Conceptualization, Project administration, Supervision, Visualization, Writing – review & editing. RL: Conceptualization, Formal Analysis, Investigation, Methodology, Supervision, Writing – review & editing. MR: Writing – review & editing. RP: Writing – review & editing. PL: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We gratefully acknowledge the DST-FIST Lab, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1554867/full#supplementary-material
References
Abbas, A., Mubeen, M., Iftikhar, Y., Shakeel, Q., Imran Arshad, H. M., Carmen Zuñiga Romano, M. D., et al. (2023). Rice sheath blight: A comprehensive review on the disease and recent management strategies. Sarhad J. Agric. 39, 111–125. doi: 10.17582/journal.sja/2023/39.1.111.125
Al-Askar, A., Al-Otibi, F. O., Abo-Zaid, G. A., and Abdelkhalek, A. (2024). Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-3-(2-methylpropyl), as the primary secondary metabolite of Bacillus spp., could be an effective antifungal agent against the soil-borne fungus, Sclerotium bataticola. Egypt. J. Chem. 67, 1009–1022. doi: 10.21608/ejchem.2024.325664.10571
Aloo, B. N., Makumba, B. A., and Mbega, E. R. (2019). The potential of Bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiol. Res. 219, 26–39. doi: 10.1016/j.micres.2018.10.011
Amer, M. S., Abd Ellatif, H. H., Hassan, S. W. M., Aboelela, G. M., and Gad, A. M. (2019). Characterization of some fungal strains isolated from the Eastern coast of Alexandria, Egypt, and some applications of Penicillium crustosum. Egypt J. Aquat. Res. 45, 211–217. doi: 10.1016/j.ejar.2019.06.006
Awan, Z. A., Shoaib, A., Schenk, P. M., Ahmad, A., Alansi, S., and Paray, B. A. (2023). Antifungal potential of volatiles produced by Bacillus subtilis BS-01 against Alternaria solani in Solanum lycopersicum. Front. Plant Sci. 13, 1089562. doi: 10.3389/fpls.2022.1089562
Azizi, P., Rafii, M. Y., Mahmood, M., Abdullah, S. N. A., Hanafi, M. M., Latif, M. A., et al. (2017). Evaluation of RNA extraction methods in rice and their application in expression analysis of resistance genes against Magnaporthe oryzae. Biotechnol. Biotechnol. Equip. 31, 75–84. doi: 10.1080/13102818.2016.1259015
Balderas-Ruíz, K. A., Bustos, P., Santamaria, R. I., González, V., Cristiano-Fajardo, S. A., Barrera-Ortíz, S., et al. (2020). Bacillus velezensis 83 a bacterial strain from mango phyllosphere, useful for biological control and plant growth promotion. AMB Express 10, 163. doi: 10.1186/s13568-020-01101-8
Bhaskar Rao, T., Chopperla, R., Prathi, N. B., Balakrishnan, M., Prakasam, V., Laha, G. S., et al. (2020). A comprehensive gene expression profile of pectin degradation enzymes reveals the molecular events during cell wall degradation and pathogenesis of rice sheath blight pathogen Rhizoctonia solani AG1-IA. J. Fungi 6, 71.
Cawoy, H., Debois, D., Franzil, L., De Pauw, E., Thonart, P., and Ongena, M. (2015). Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 8, 281–295. doi: 10.1111/1751-7915.12238
Chandrasekaran, M. and Chun, S. C. (2016). Expression of PR-protein genes and induction of defense-related enzymes by Bacillus subtilis CBR05 in tomato (Solanum lycopersicum) plants challenged with Erwinia carotovora sub sp. carotovora. Biosci. Biotechnol. Biochem. 80, 2277–2283. doi: 10.1080/09168451.2016.1206811
Chen, J., Xuan, Y., Yi, J., Xiao, G., Yuan, D. P., and Li, D. (2023). Progress in rice sheath blight resistance research. Front. Plant Sci. . 14, 1141697. doi: 10.3389/fpls.2023.1141697
Chen, X., Li, C., Wang, H., and Guo, Z. (2019). WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Chen, X., Lili, L., Zhang, Y., Zhang, J., Ouyang, S., and Zhang, Q. (2017). Functional analysis of polygalacturonase gene RsPG2 from Rhizoctonia solani, the pathogen of rice sheath blight. Eur. J. Plant Pathol. 149, 491–502. doi: 10.1007/s10658-017-1198-5
Dallakyan, S. and Olson, A. J. (2015). Small-molecule library screening by docking with PyRx. Chem. Biol. Methods Protoc. 1263, 243–250.
Elbeltagy, A., Nishioka, K., Suzuki, H., Sato, T., Sato, Y. I., Morisaki, H., et al. (2000). Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. J. Soil Sci. Plant Nutr. 46, 617–629. doi: 10.1080/00380768.2000.10409127
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Gorai, P. S., Ghosh, R., Ghosh, S., Samanta, S., Sen, A., Panja, S., et al. (2023). Management of Black Root Disease-Causing Fungus Fusarium solani CRP1 by Endophytic Bacillus siamensis CNE6 through Its Metabolites and Activation of Plant Defense Genes. Microbiol. Spectr. 11, e0308222. doi: 10.1128/spectrum.03082-22
Hardoim, P. R., Hardoim, C. C., van Overbeek, L. S., and van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PloS One 7, 30438. doi: 10.1371/journal.pone.0030438
Idan, S. A., Al-Marzoqi, A. H., and Hameed, I. H. (2015). Spectral analysis and anti-bacterial activity of methanolic fruit extract of Citrullus colocynthis using gas chromatography-mass spectrometry. Afr. J. Biotechnol. 14, 3131–3158. doi: 10.5897/AJB2015.14957
IRRI (2002). Standard Evaluation System for Rice (Manila, Philippines: Philippines International Rice Research Institute).
Islam, M. S., Mahmud, S., Sultana, R., and Dong, W. (2020). Identification and in silico molecular modelling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani. Arab J. Chem. 13, 8600–8612. doi: 10.1016/j.arabjc.2020.09.044
Jamali, H., Sharma, A., Roohi, N., and Srivastava, A. K. (2020). Biocontrol potential of Bacillus subtilis RH5 against sheath blight of rice caused by Rhizoctonia solani. J. Basic Microbiol. 60, 268–280. doi: 10.1002/jobm.201900347
Jeong, M. H., Lee, Y. S., Cho, J. Y., Ahn, Y. S., Moon, J. H., Hyun, H. N., et al. (2017). Isolation and characterization of metabolites from Bacillus licheniformis MH48 with antifungal activity against plant pathogens. Microb. Pathog. 110, 645–653. doi: 10.1016/j.micpath.2017.07.027
Kai, M., Effmert, U., Berg, G., and Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Khan, M. S., Gao, J., Chen, X., Zhang, M., Yang, F., Du, Y., et al. (2020). The endophytic bacteria Bacillus velezensis Lle-9, isolated from Lilium leucanthum, harbors antifungal activity and plant growth-promoting effects. J. Microbiol. Biotechnol. 30, 668. doi: 10.4014/jmb.1910.10021
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 9, 2732. doi: 10.3389/fmicb.2018.02732
Khaskheli, M. A., Wu, L., Chen, G., Chen, L., Hussain, S., Song, D., et al. (2020). Isolation and characterization of root-associated bacterial endophytes and their biocontrol potential against major fungal phytopathogens of rice (Oryza sativa L.). Pathogens 9, 172. doi: 10.3390/pathogens9030172
Kumar, V., Jain, L., Jain, S. K., Chaturvedi, S., and Kaushal, P. (2020). Bacterial endophytes of rice (Oryza sativa L.) and their potential for plant growth promotion and antagonistic activities. S Afr. J. Bot. 134, 50–63. doi: 10.1016/j.sajb.2020.02.017
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Malik, A., Afaq, S., El-Gamal, B., Abd Ellatif, M., Hassan, W. N., and Dera, A. (2019). Molecular docking and pharmacokinetic evaluation of natural compounds as targeted inhibitors against Crz1 protein in Rhizoctonia solani. Bioinformation 15, 277–286. doi: 10.6026/97320630015277
Myo, E. M., Liu, B., Ma, J., Shi, L., Jiang, M., Zhang, K., et al. (2019). Evaluation of Bacillus velezensis NKG-2 for bio-control activities against fungal diseases and potential plant growth promotion. Biol. Control 134, 23–31. doi: 10.1016/j.biocontrol.2019.03.017
Nagendran, K., Karthikeyan, G., Mohammed Faisal, P., Kalaiselvi, P., Raveendran, M., Prabakar, K., et al. (2014). Exploiting endophytic bacteria for the management of sheath blight disease in rice. Biol. Agric. Hortic. 30, 8–23. doi: 10.1080/01448765.2013.841099
Nakkeeran, S., Priyanka, R., Rajamanickam, S., and Sivakumar, U. (2020). Bacillus amyloliquefaciens alters the diversity of volatile and non-volatile metabolites and induces the expression of defence genes for the management of Botrytis leaf blight of Lilium under protected conditions. J. Plant Pathol. 102, 1179–1189. doi: 10.1007/s42161-020-00602-6
Nakkeeran, S., Vinodkumar, S., Renukadevi, P., Rajamanickam, S., and Jogaiah, S. (2019). “Bioactive molecules from Bacillus spp.: An effective tool for plant stress management,” in Bioactive Molecules in Plant Defense: Signaling in Growth and Stress (Springer, Cham, Switzerland), 1–23.
Nicholson, W. L. (2002). Roles of Bacillus endospores in the environment. Cell Mol. Life Sci. 59, 410–416. doi: 10.1007/s00018-002-8433-7
Prabhukarthikeyan, S. R., Parameswaran, C., Sawant, S. B., Naveenkumar, R., Mahanty, A., Keerthana, U., et al. (2022). Comparative proteomic analysis of Rhizoctonia solani isolates identifies the differentially expressed proteins with roles in virulence. J. Fungi 8, 370. doi: 10.3390/jof8040370
Rabbee, M. F., Ali, M. S., Choi, J., Hwang, B. S., Jeong, S. C., and Baek, K. H. (2019). Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24, 1046. doi: 10.3390/molecules24061046
Rais, A., Jabeen, Z., Shair, F., Hafeez, F. Y., and Hassan, M. N. (2017). Bacillus spp., a bio-control agent enhances the activity of antioxidant defense enzymes in rice against Pyricularia oryzae. PloS One 12, 0187412.
Rui, Y. and Dinneny, J. R. (2020). A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.16166
Sahu, P. K., Singh, S., Gupta, A., Singh, U. B., Brahmaprakash, G. P., and Saxena, A. K. (2019). Antagonistic potential of bacterial endophytes and induction of systemic resistance against collar rot pathogen Sclerotium rolfsii in tomato. Biol. Control 137, 104014. doi: 10.1016/j.biocontrol.2019.104014
Sandoval, R. F. C. and Cumagun, C. J. R. (2019). Phenotypic and molecular analyses of Rhizoctonia spp. associated with rice and other hosts. Microorganisms 7, 88.
Saravanan, R., Nakkeeran, S., Saranya, N., Kavino, M., Ragapriya, V., Varanavasiappan, S., et al. (2022). Biohardening of Banana cv. Karpooravalli (ABB; Pisang Awak) With Bacillus velezensis YEBBR6 Promotes Plant Growth and Reprograms the Innate Immune Response Against Fusarium oxysporum f. sp. cubense. Front. Sustain Food Syst. 6, 845512.
Seo, D. J., Nguyen, D. M. C., Song, Y. S., and Jung, W. J. (2012). Induction of defense response against Rhizoctonia solani in cucumber plants by endophytic bacterium Bacillus thuringiensis GS1. J. Microbiol. Biotechnol. 22, 407–415. doi: 10.4014/jmb.1107.07027
Shakeel, M., Rais, A., Hassan, M. N., and Hafeez, F. Y. (2015). Root associated Bacillus sp. improves growth, yield and zinc translocation for basmati rice (Oryza sativa) varieties. Front. Microbiol. 6, 1286.
Shinya, T., Yamaguchi, K., Desaki, Y., Yamada, K., Narisawa, T., Kobayashi, Y., et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL 27. Plant J. 79, 56–66. doi: 10.1111/tpj.12535
Showmy, K. S. and Yusuf, A. (2020). Characterization of disease resistance in nine traditional rice (Oryza sativa L.) cultivars and expression of chennellu PR1 gene in response to Xanthomonas oryzae pv. oryzae. Indian Phytopathol. 73, 281–291. doi: 10.1007/s42360-020-00220-3
Singh, P., Arif, Y., Miszczuk, E., Bajguz, A., and Hayat, S. (2022). Specific roles of lipoxygenases in development and responses to stress in plants. Plants 11, 979. doi: 10.3390/plants11070979
Singh, A., Rohilla, R., Singh, U. S., Savary, S., Willocquet, L., and Duveiller, E. (2002). An improved inoculation technique for sheath blight of rice caused by Rhizoctonia Solani. Can. J. Plant Pathol. 24, 65–68. doi: 10.1080/07060660109506973
Soliman, S. A., Khaleil, M. M., and Metwally, R. A. (2022). Evaluation of the antifungal activity of Bacillus amyloliquefaciens and B. velezensis and characterization of the bioactive secondary metabolites produced against plant pathogenic fungi. Biology 11, 1390.
Su, T., Shen, B., Hu, X., Teng, Y., Weng, P., Wu, Z., et al. (2024). Research advance of Bacillus velezensis: Bioinformatics, characteristics, and applications. Food Sci. Hum. Wellness 13, 1756–1766. doi: 10.26599/FSHW.2022.9250148
Sun, G., Yao, T., Feng, C., Chen, L., Li, J., and Wang, L. (2017). Identification and biocontrol potential of antagonistic bacteria strains against Sclerotinia sclerotiorum and their growth-promoting effects on Brassica napus. Biol. Control 104, 35–43. doi: 10.1016/j.biocontrol.2016.10.008
Tonnessen, B. W., Manosalva, P., and Lang, J. M. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Walitang, D. I., Kim, K., Madhaiyan, M., Kee Kim, Y., Kong, Y., and Sa, T. (2017). Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of rice. BMC Microbiol. 17, 209. doi: 10.1186/s12866-017-1117-0
Wani, Z. A., Ashraf, N., Mohiuddin, T., and Riyaz-Ul-Hassan, S. (2015). Plant-endophyte symbiosis, an ecological perspective. Appl. Microbiol. Biotechnol. 99, 2955–2965. doi: 10.1007/s00253-015-6487-3
Wei, G., Kloepper, J. W., and Tuzun, S. (1991). Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 81, 1508–1512. doi: 10.1094/Phyto-81-1508
Yang, P., Praz, C., Li, B., Singla, J., Robert, C. A., Kessel, B., et al. (2019). Fungal resistance mediated by maize wall-associated kinase Zm WAK-RLK 1 correlates with reduced benzoxazinoid content. New Phytol. 221, 976–987. doi: 10.1111/nph.15419
Yang, J. H., Zhang, W. W., Zhuang, Y. Q., and Xiao, T. (2017). Biocontrol activities of bacteria from cowdung against the rice sheath blight pathogen. J. Plant Dis. Prot 124, 131–141. doi: 10.1007/s41348-017-0080-1
Yasmin, H., Shah, Z. A., Mumtaz, S., Ilyas, N., Rashid, U., Alsahli, A. A., et al. (2023). Alleviation of banded leaf and sheath blight disease incidence in maize by bacterial volatile organic compounds and molecular docking of targeted inhibitors in Rhizoctonia solani. Front. Plant Sci. 14, 1218615. doi: 10.3389/fpls.2023.1218615
Zhang, Y. and Wang, L. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhang, J. and Zhou, J. M. (2010). Plant immunity triggered by microbial molecular signatures. Mol. Plant 3, 783–793. doi: 10.1093/mp/ssq035
Zheng, A., Lin, R., Zhang, D., Qin, P., Xu, L., and Ai, P. (2013). The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 4, 1424. doi: 10.1038/ncomms2427
Zheng, T. W., Liu, L., Nie, Q. W., Hsiang, T., Sun, Z. X., and Zhou, Y. (2021). Isolation, identification and biocontrol mechanisms of endophytic bacterium D61-A from Fraxinus hupehensis against Rhizoctonia solani. Biol. Control 158, 104621. doi: 10.1016/j.biocontrol.2021.104621
Keywords: Bacillus velezensis, Rhizoctonia solani, secondary metabolites, molecular docking, defense genes expression
Citation: Naveena S, Gopalakrishnan C, Logeshwari R, Raveendran M, Pushpam R and Lakshmidevi P (2025) Metabolomic profiling of Bacillus velezensis B13 and unveiling its antagonistic potential for the sustainable management of rice sheath blight. Front. Plant Sci. 16:1554867. doi: 10.3389/fpls.2025.1554867
Received: 03 January 2025; Accepted: 13 June 2025;
Published: 25 July 2025.
Edited by:
Amitava Rakshit, Banaras Hindu University, IndiaReviewed by:
Waheed Akram, University of the Punjab, PakistanSrayan Ghosh, Durham University, United Kingdom
Tasvina R. Borah, The ICAR Research Complex for North Eastern Hill Region (ICAR RC NEH), India
Runmao Lin, Hainan University, China
Copyright © 2025 Naveena, Gopalakrishnan, Logeshwari, Raveendran, Pushpam and Lakshmidevi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chellappan Gopalakrishnan, cGNnb3BhbGFncmlAZ21haWwuY29t
 Sirivella Naveena1
Sirivella Naveena1 Chellappan Gopalakrishnan
Chellappan Gopalakrishnan Rajendran Logeshwari
Rajendran Logeshwari Muthurajan Raveendran
Muthurajan Raveendran Ramamoorthy Pushpam
Ramamoorthy Pushpam Paranthaman Lakshmidevi
Paranthaman Lakshmidevi