- College of Life Sciences, Shanghai Normal University, Shanghai, China
The integration of novel components into functional multi-subunit protein complexes is a key evolutionary strategy for enhancing stability, activity, and adaptation to oxidative stress. This is exemplified by the evolution of the conserved photosynthetic NDH-1 (cpNDH-1) complex, though its precise evolutionary history remains unresolved. In this study, we constructed a time-calibrated phylogenetic tree of cyanobacteria to trace the evolutionary trajectory of cpNDH-1. By mapping the orthologous of oxygenic photosynthesis-specific (OPS) subunits onto this tree, we found that the cpNDH-1 complex progressively acquired OPS subunits. Specifically, during the transition from non-photosynthetic to thylakoid-less photosynthetic cyanobacteria, cpNDH-1 incorporated OPS subunits NdhM, NdhN, NdhO, NdhP, and NdhS. Subsequently, NdhL, NdhQ, and NdhV were added as thylakoid-bearing photosynthetic cyanobacteria evolved. Our analysis reveals that the emergence of oxygenic photosynthesis was closely linked with the progressive incorporation of OPS subunits into cpNDH-1. We propose a two-step model for the evolution of these subunits, identifying potential driving factors behind this process. Genome-wide sequence analysis and structural predications further suggest that the OPS cpNDH-1 genes either evolved de novo or arose from modifications of existing genes. Collectively, these findings provide a robust framework for understanding the evolutionary emergence of OPS subunits in cyanobacterial cpNDH-1, underscoring the acquisition of new subunits as a critical adaptation to oxidative environments during the evolution of oxygenic photosynthesis.
Introduction
Approximately 2.4 billion years ago, cyanobacteria revolutionized Earth’s atmosphere by initiating oxygenic photosynthesis, which led to the Great Oxidation Event (GOE) and significantly altered the planet’s environment (Bekker et al., 2004). The rise of atmospheric O2 exposed cyanobacteria to unprecedented oxidative stress, driving evolutionary adaptations that were essential for their survival and the continued evolution of oxygenic photosynthesis (Fischer et al., 2016; Khademian and Imlay, 2021). In this challenging environment, maintaining the stability and functionality of multi-subunit protein complexes became crucial. The addition of subunits is a well-documented mechanism by which such complexes adapt to oxidative stress (Marsh and Teichmann, 2015; Urano et al., 2016). For example, prior to the rise of oxygen, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acquired an accessory subunit to enhance its specificity for CO2 over O2 (Schulz et al., 2022). Similarly, superoxide dismutase diversified into four distinct forms, each with different catalytic metals, to better manage the oxidative challenges posed by elevated O2 levels (Boden et al., 2021). Thus, the gain of subunits represents a fundamental evolutionary strategy that enhanced the functionality of multi-subunit protein complexes, enabling cyanobacteria to adapt to oxidative stress.
In cyanobacteria, the photosynthetic NDH-1 (pNDH-1) complex is a critical multi-subunit enzyme involved in cyclic electron transport around photosystem I (PSI CET), a process that reduces reactive oxygen species (ROS) production by providing additional ATP for photosynthetic carbon assimilation. This activity is essential for efficient photosynthesis and serves as a key antioxidant mechanism (Zhang et al., 2020; Miller et al., 2021). Notably, pNDH-1 has undergone evolutionary diversification, with four distinct complex types—pNDH-1L, pNDH-1L’, pNDH-1MS, and pNDH-1MS’ (Battchikova et al., 2011a; Ma and Ogawa, 2015; Laughlin et al., 2020)—each fulfilling a common role in PSI CET (Bernát et al., 2011). The conserved pNDH-1L form (hereafter referred to as cpNDH-1) is considered the ancestral form and the precursor to the other three types (Peltier et al., 2016).
The structure of cyanobacterial cpNDH-1 is L-shaped (Zhang et al., 2020), a feature shared with its archaeal group 4 membrane-bound [NiFe] hydrogenase (Yu et al., 2018) and its bacterial homolog, respiratory complex I (Baradaran et al., 2013). This architecture enables the efficient coupling of electron transfer, mediated by a hydrophilic arm extending into the cytoplasm, with proton translocation carried out by the membrane-embedded arm. The structural stability of cpNDH-1 is crucial for this coupling and, consequently, for PSI CET activity. Unlike its ancestral and homologous counterparts, cyanobacterial cpNDH-1 incorporates eight additional subunits, termed oxygenic photosynthesis-specific (OPS) subunits (NdhL to NdhQ, NdhS, and NdhV), which are critical for the structural integrity and function of the complex (Ma and Ogawa, 2015; Zhang et al., 2020). These OPS subunits are involved in various processes, such as electron donor binding (NdhO, NdhS and NdhV), assembly of the hydrophilic arm (NdhM and NdhN), stabilization of the membrane arm (NdhP and NdhQ), and formation of the plastoquinone (PQ) channel (NdhL). The absence of these OPS subunits destabilizes cpNDH-1 (Battchikova et al., 2005, 2011b; Zhang et al., 2014; Zhao et al., 2014, 2015; Gao et al., 2016; He et al., 2016; He and Mi, 2016), resulting in reduced PSI CET activity, compromised ATP production, and a disrupted ATP/NADPH ratio, which in turn impairs carbon fixation (Zhang et al., 2020; Zheng et al., 2025). Under aerobic stress conditions, this disruption leads to increased ROS production.
In 2007, a study of 17 cyanobacterial strains focused on the evolutionary history of OPS subunits NdhL, NdhM, NdhN, and NdhO (Ogawa and Mi, 2007). However, the incomplete identification of other OPS subunits and the lack of comprehensive evolutionary data at that time limited our understanding of their evolution. These OPS subunits are generally small and structurally simple (Supplementary Figure S1). Moreover, with the exception of NdhV, they lack unique domains, with NdhV belonging to the DUF2996 domain-containing protein group. Despite significant progress in understanding cpNDH-1 evolution (Peltier et al., 2016; Kato et al., 2021; Yu et al., 2021, 2022), the precise evolutionary trajectory of these OPS subunits remains unresolved.
Recent discoveries of sister groups to cyanobacteria, including the non-photosynthetic species like Candidatus Melainabacteria and Candidatus Sericytochromatia (Di Rienzi et al., 2013; Soo et al., 2017), as well as new thylakoid-less species such as Candidatus Aurora vandensis and Anthocerotibacter panamensis (Grettenberger et al., 2020; Rahmatpour et al., 2021), have provided critical insights into the early evolution of cyanobacteria. These findings, along with expanding genomic data, offer an unparalleled opportunity to further elucidate the evolutionary history of OPS subunits. In this study, we constructed a time-calibrated phylogenetic tree of cyanobacteria to trace the evolution of cpNDH-1. Our analysis reveals that cpNDH-1 progressively acquired OPS subunits throughout cyanobacterial evolution, coinciding with the emergence and diversification of oxygenic photosynthesis. Based on these findings, we propose a two-step model for the acquisition of OPS subunits, suggesting that their incorporation was crucial for the adaptation of cpNDH-1 to oxygenic photosynthesis. Furthermore, we investigate potential OPS homologs in both non-photosynthetic and thylakoid-less photosynthetic cyanobacteria through protein sequence and structural analyses, providing new insights into the origins of OPS genes. Together, our findings illuminate the timing and mechanisms underlying the evolution of OPS subunits in cpNDH-1.
Materials and methods
Time-calibrated phylogenetic analysis
A time-calibrated phylogenetic tree for the phylum Cyanobacteria was constructed using BEAST v1.10.4 (Drummond and Rambaut, 2007) based on 16S rDNA sequences. These sequences were retrieved from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) and Cydrasil 3 (https://www.cydrasil.org) (Roush et al., 2021). The species names and their corresponding 16S rDNA sequences used in this study are provided in Supplementary Data 1. Sequence alignment and gap removal were performed using MUSCLE (Edgar, 2004). The original phylogenetic tree employed for time calibration is presented in Supplementary Figure S2.
Three calibration points, previously established in the literature, were applied (Blank and Sánchez-Baracaldo, 2010; Sánchez-Baracaldo et al., 2014). The prior distribution for Nostocales was modeled as a normal distribution with a mean of 2,250 million years ago (Mya) and a standard deviation of 100, truncated between 2,100 and 2,450 Mya (Tomitani et al., 2006; Sánchez-Baracaldo et al., 2014). For Pleurocapsales, the prior distribution was set as a normal distribution with a mean of 2,050 Mya and a standard deviation 200, truncated between 1,700 and 2,450 Mya (Sánchez-Baracaldo et al., 2014). The cyanobacterial root age was constrained to a uniform distribution between 2,320 and 2,700 Mya (Bekker et al., 2004).
The analysis employed a Yule process speciation model with an uncorrelated log-normal relaxed clock model. A total of 50 million iterations were run, with sampling every 5,000 iterations. Convergence and posterior distributions were assessed using Tracer v1.6, and the maximum clade credibility tree was generated with TreeAnnotator v1.10.4 from the BEAST package. The resulting phylogenetic tree was visualized and edited using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree).
Conserved ndh genes analysis
The analysis of conserved ndh gene was conducted across a range of cyanobacterial species, including non-photosynthetic cyanobacteria Candidatus Melainabacteria (Melaina) and Candidatus Sericytochromatia (Seri); thylakoid-less cyanobacteria Anthocerotibacter panamensis (A. pana) and Gloeobacter violaceus PCC7421 (Gloeo7421); and thylakoid-bearing cyanobacteria Synechococcus sp. PCC7336 (SynPCC7336), Synechococcus sp. JA-3-3Ab (SynJA33Ab), Synechococcus sp. PCC7502 (SynPCC7502), Pseudanabaena sp. PCC7367 (Pseud7367), Synechocystis sp. PCC6803 (SynPCC6803), Thermosynechococcus elongatus BP-1 (T. elongatus), Synechococcus sp. PCC7942 (SynPCC7942), Synechococcus sp. WH5701 (SynWH5701), Synechococcus sp. WH8102 (SynWH8102), and Prochlorococcus marinus CCMP1375 (ProCCMP1375). These species were selected based on the availability of complete or nearly complete genome sequences. The GenBank accession numbers for all species are provided in Supplementary Data 2.
Orthologous protein sequences corresponding to conserved subunits of the cpNDH-1 complex from Synechocystis sp. PCC6803 were identified in the aforementioned species using BLASTP searches on NCBI. The top hits for each sequence were verified through reciprocal BLASTP against Synechocystis sp. PCC6803. The NCBI reference sequence accession numbers for these proteins are listed in Supplementary Data 2.
The genomic positions of the ndh genes for each species were retrieved to construct the ndh gene cluster architecture. Comparative structural analyses of these ndh gene clusters were visualized by ChiPlot (https://www.chiplot.online).
Sequence analysis of OPS cpNDH-1 subunits
To investigate the evolutionary history of OPS cpNDH-1 subunits, we used the protein sequence of OPS cpNDH-1 subunits from Synechocystis sp. PCC6803 to identify homologous sequences in the aforementioned cyanobacterial species via BLASTP searches, applying a stringent E-value threshold of 1.0 × 10-6. The sequence data and their similarities are provided in Supplementary Data 3. To confirm the evolutionary history, phylogenetic trees of each OPS cpNDH-1 subunit were constructed. Detailed sequences information is available in Supplementary Data 4.
To trace the origins of OPS cpNDH-1 subunits, we employed two approaches: one based on sequence information and the other on structural data. In the sequence-based approach, we identified potential homologous fragments by comparing the amino acid sequences of OPS cpNDH-1 subunits to sequences from non-photosynthetic and thylakoid-less cyanobacteria in local nucleotide databases using TBLASTN. Protein sequences of OPS subunits from cyanobacterial cpNDH-1 served as templates. To enable a broader search, the TBLASTN expectation value threshold was set to 10. The raw alignment data are available in Supplementary Data 5. In the structure-based approach, the AlphaFold server (Abramson et al., 2024) was used to predict the structures of the earliest OPS subunits. These predicted structures were then employed as templates to identify distant orthologs using Foldseek (van Kempen et al., 2024).
Data availability
All data supporting the findings of this study are included in the main text and Supplementary Information files.
Results and discussion
Time-calibrated phylogenetic tree reveals cyanobacterial evolution
Recent advancements in genome and metagenome sequencing have significantly expanded our understanding of cyanobacterial evolution and the diversification of their multi-subunit protein complexes (Di Rienzi et al., 2013; Soo et al., 2017; Grettenberger et al., 2020; Rahmatpour et al., 2021). To explore the evolutionary origins of cpNDH-1 within the phylum Cyanobacteria, we reconstructed a time-calibrated phylogenetic tree (Figure 1). This analysis underscores key evolutionary transitions within cyanobacteria.
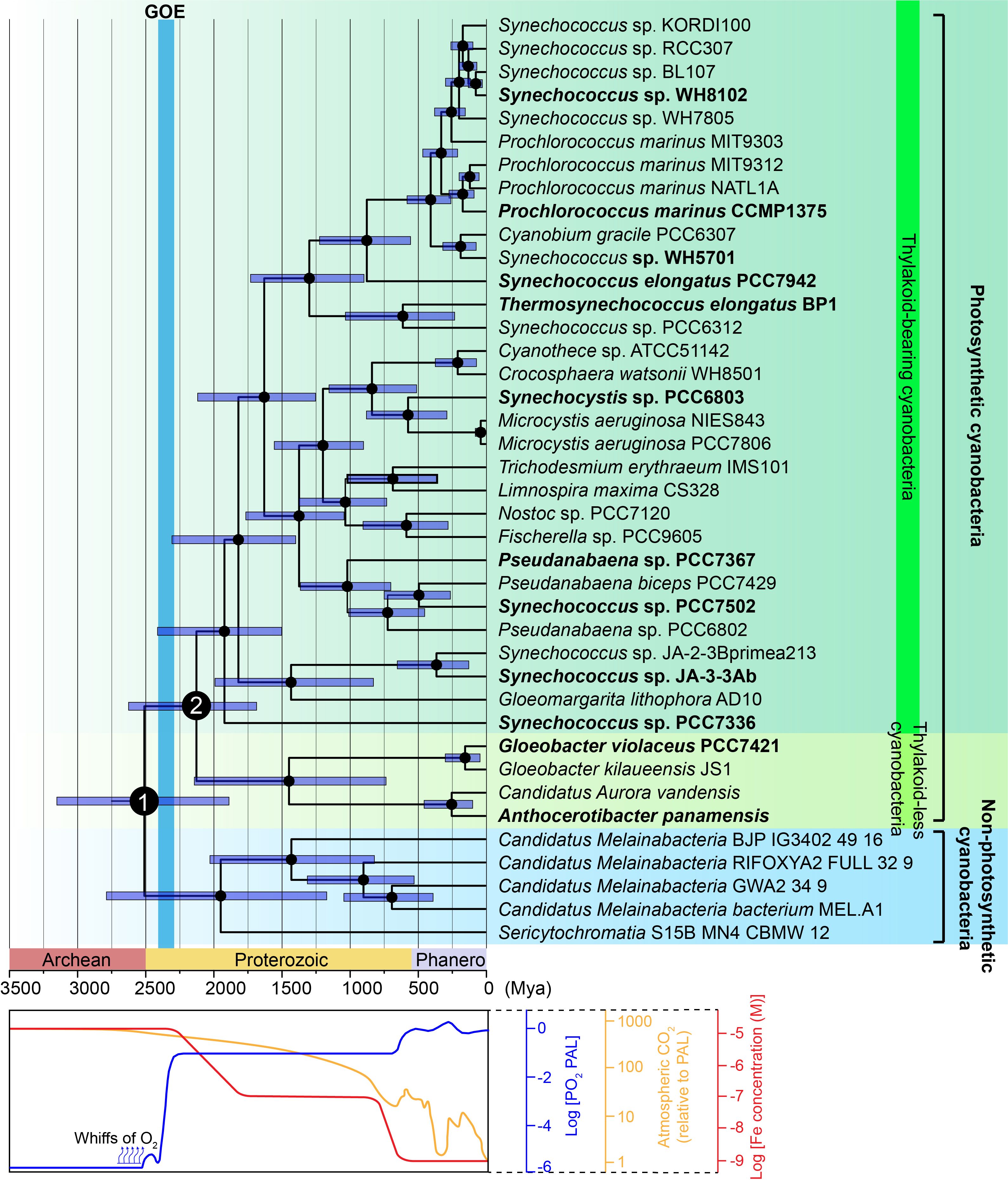
Figure 1. Time-calibrated phylogenic tree of the phylum Cyanobacteria. The phylogenetic tree was generated using Bayesian analysis of aligned 16S rDNA sequences. (Upper panel) Bayesian relaxed molecular clock analyses, implemented in BEAST, were used to estimate divergence times. Three calibrations points were applied (see Materials and Methods). Dark blue bars represent the posterior 95% confidence intervals for node ages. Non-photosynthetic cyanobacteria (blue background) form the sister group to photosynthetic cyanobacteria. Photosynthetic cyanobacteria are subdivided into the class Gloeobacteria (thylakoid-less, chartreuse background) and crown cyanobacteria (thylakoid-bearing, green background) based on the presence or absence of thylakoid membrane. Node 1 marks the divergence between non-photosynthetic and photosynthetic cyanobacteria, while Node 2 represents the divergence between thylakoid-less Gloeobacteria and thylakoid-bearing cyanobacteria. The Great Oxidation Event (light blue bar) occurred approximately 2,400 Mya. Photosynthetic cyanobacteria species selected for detailed analysis are highlighted in bold. The original phylogenetic tree used for time calibration is presented in Supplementary Figure S2. (Lower panel) Timeline of major evolutionary and geochemical transitions. Changes in atmospheric O2 (blue line), CO2 (yellow line), and Fe (red line) concentrations were adapted from He et al. (2021); Flamholz and Shih (2020), and Anbar (2008), respectively.
The base of the tree is occupied by non-photosynthetic cyanobacteria (indicated by the blue background in Figure 1), including Candidatus Sericytochromatia and Candidatus Melainabacteria, which are regarded as the closest living relatives of photosynthetic cyanobacteria (Blankenship, 2017). The divergence between non-photosynthetic and photosynthetic cyanobacteria is estimated to have occurred around 2,500 million years ago (Mya) (Node 1, Figure 1).
Within photosynthetic cyanobacteria, the class Gloeobacteria, comprising Candidatus Aurora vandensis, Anthocerotibacter panamensis, and the genus Gloeobacter, is characterized by the absence of thylakoid membranes, positioning it as a critical lineage for understanding the early stages of cyanobacterial evolution (Rahmatpour et al., 2021) (indicated by the chartreuse background in Figure 1). Thylakoid-bearing cyanobacteria, in which thylakoid membranes evolved, form a distinct, more derived group (indicated by the green background in Figure 1). The divergence between thylakoid-less and thylakoid-bearing cyanobacteria is estimated to have occurred between 2,500 and 2,000 Mya (Node 2, Figure 1). This trajectory—spanning from non-photosynthetic to photosynthetic thylakoid-less cyanobacteria, and ultimately to photosynthetic thylakoid-bearing cyanobacteria—aligns with prevailing models of cyanobacterial evolution (Garcia-Pichel et al., 2020; Sánchez-Baracaldo and Cardona, 2020; Sánchez-Baracaldo et al., 2022). However, discrepancies persist regarding the placement of Gloeomargarita lithophora AD10 and Pseudanabaena (Fournier et al., 2021; Strunecký et al., 2023).
In non-photosynthetic cyanobacteria, the genes for the 11 conserved subunits of cpNDH-1 (NdhA-NdhK) are already present (Supplementary Figure S3), suggesting that the core components of cpNDH-1 evolved in the last common ancestor of photosynthetic cyanobacteria. However, genes encoding the OPS subunits are absent in these non-photosynthetic cyanobacteria (Figure 2), supporting the hypothesis that the acquisition of these subunits coincided with the transition to photosynthetic functions such as PSI CET in cyanobacteria. Future investigations will focus on pinpointing the timing and mechanisms by which these OPS subunits were integrated into cpNDH-1.
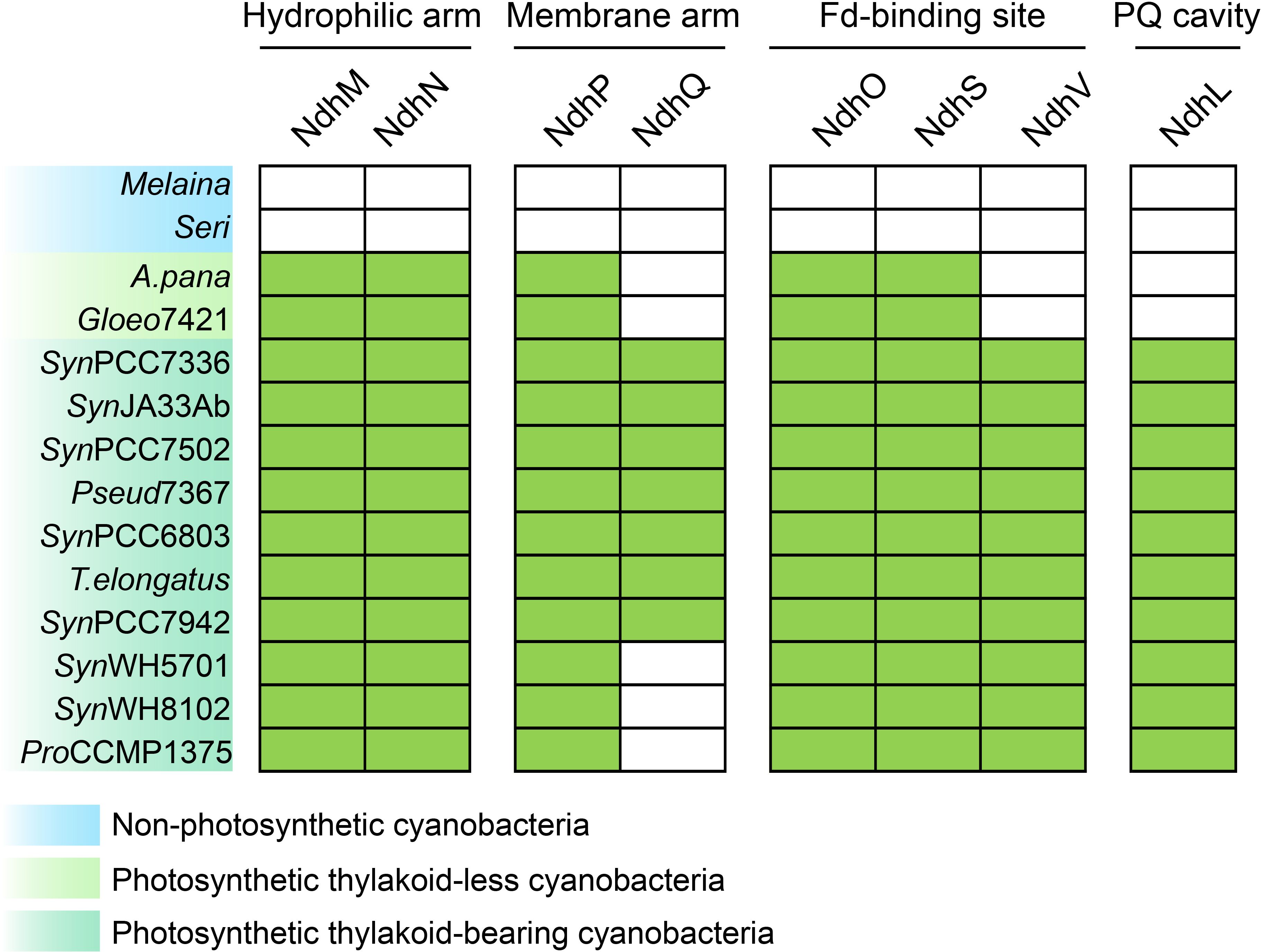
Figure 2. Distribution of OPS cpNDH-1 subunits in non-photosynthetic and photosynthetic cyanobacteria. The presence and absence of homologs of OPS cpNDH-1 subunits, identified through BLASTP searches, are mapped onto the evolutionary tree of selected cyanobacteria shown in Figure 1. Filled boxes denote the presence of genes, while empty boxes indicate their absence. The sequence information and their similarity are provided in Supplementary Data 3. Strain abbreviations are provided in the Materials and Methods section.
The emergence of OPS cpNDH-1 subunits
To explore the evolutionary integration of OPS subunits into cpNDH-1, we conducted BLASTP searches to identify orthologous genes, followed by mapping these data onto the reconstructed cyanobacterial phylogeny. Phylogenetic trees of these OPS subunits were also constructed (Supplementary Figure S4), providing insights into the progressive acquisition of these subunits throughout cyanobacterial evolution (Figure 2).
Following the emergence of photosynthetic thylakoid-less cyanobacteria (Node 1, Figure 1), five OPS subunits were integrated to cpNDH-1 (Figure 2; Supplementary Figure S5). These include NdhM and NdhN in the hydrophilic arm, NdhP in the membrane arm, and NdhO and NdhS near the ferredoxin (Fd)-binding site (Laughlin et al., 2019; Schuller et al., 2019; Pan et al., 2020; Zhang et al., 2020) (Supplementary Figure S1). These subunits are essential for stabilizing cpNDH-1, as demonstrated in Synechocystis sp. PCC6803, where the absence of NdhM, NdhN, or NdhP destabilizes cpNDH-1, while NdhO and NdhS are critical for Fd binding (Battchikova et al., 2011b; Zhang et al., 2014; Zhao et al., 2014; He and Mi, 2016; He et al., 2016; He and Mi, 2016).
As thylakoid-bearing cyanobacteria, such as Synechococcus sp. PCC7336, emerged (Node 2, Figure 1), additional OPS subunits were integrated into cpNDH-1 (Laughlin et al., 2019; Schuller et al., 2019; Pan et al., 2020; Zhang et al., 2020). These included NdhV at the Fd-binding site, enhancing Fd-binding efficiency (Gao et al., 2016), and NdhQ in the membrane arm (Figure 2; Supplementary Figures S1, S5), which works synergistically with NdhP to stabilize the cpNDH-1 structure (Zhao et al., 2015). The incorporation of NdhL further reinforced the PQ cavity, ensuring efficient PSI CET (Mi et al., 1992; Zhang et al., 2020).
The acquisition of OPS subunits aligns with two key transitions in cyanobacterial photosynthesis, providing a potential evolutionary scenario for these subunits (Figure 3). During the shift from non-photosynthetic to thylakoid-less cyanobacteria (Step I, Figure 3), early oxygenic photosynthesis occurred a geochemically constrained environment, resulting in slow oxygen evolution and intermittent oxygen generation (Rahmatpour et al., 2021), setting the stage prior to the GOE (Figure 1). Rising oxygen levels and ROS likely triggered oxidative stress, disrupting the stabilization of cpNDH-1. The recruitment of NdhM, NdhN, NdhP, and NdhS would have been critical for this adaptation, potentially ensuring the stabilization and functionality of cpNDH-1 in an increasingly oxidative environment.
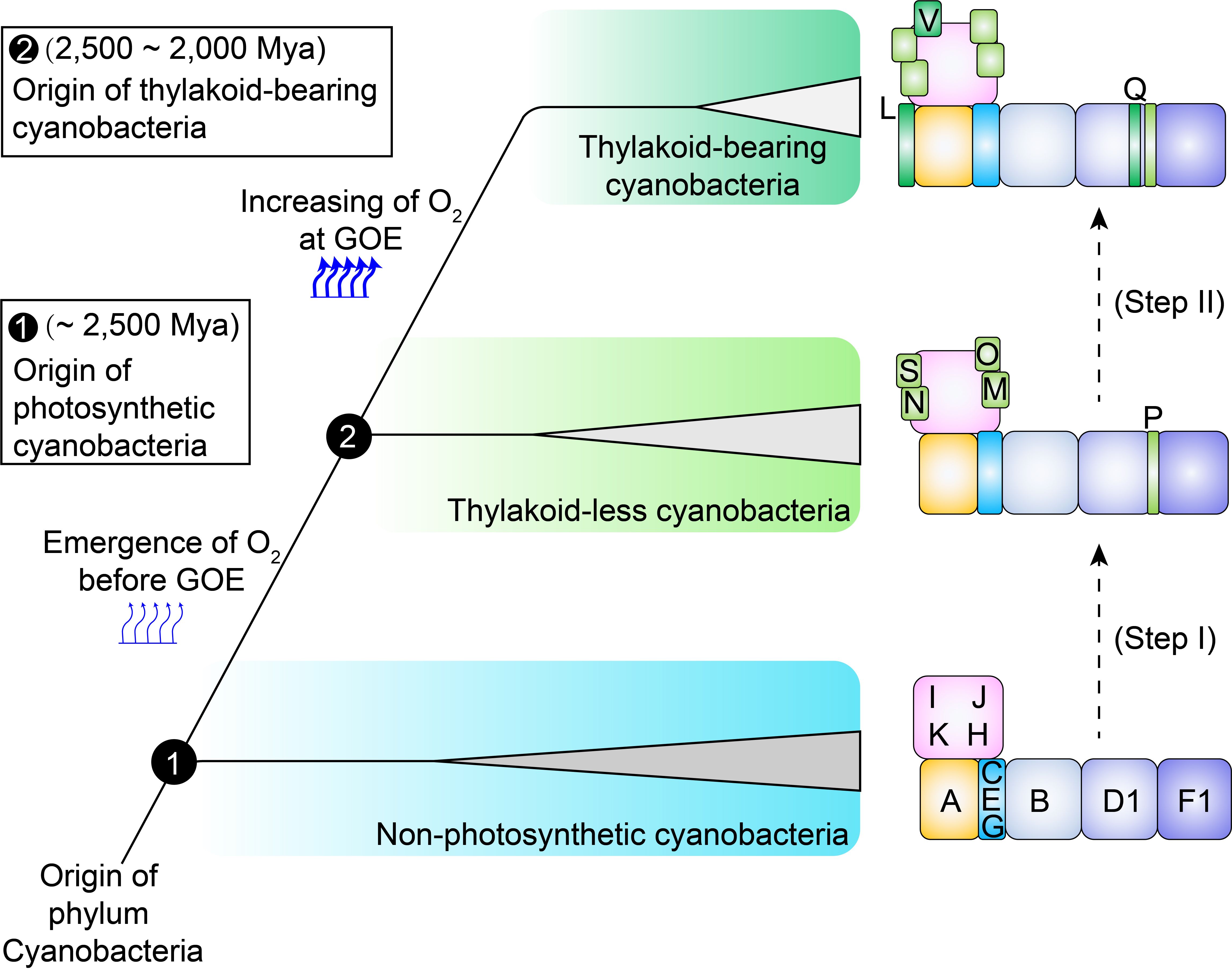
Figure 3. A two-step schematic model illustrating the evolutionary history of OPS subunits in cyanobacterial cpNDH-1. In non-photosynthetic cyanobacteria, 11 conserved ndh genes are present. The acquisition of OPS cpNDH-1 genes occurred in two distinct evolutionary phases during the transition to photosynthetic cyanobacteria. Step I: During the evolution from non-photosynthetic to photosynthetic cyanobacteria, several OPS subunits (NdhM to NdhP, NdhS) were incorporated, enhancing the stability and activity of cpNDH-1. Step II: As thylakoid-less Gloeobacteria evolved into crown cyanobacteria, additional OPS subunits (NdhL, NdhQ and NdhV) were acquired, further stabilizing and optimizing cpNDH-1 activity. Subunits are represented by black text, labeled according to their designations in cpNDH-1. The emergence and rise of oxygen levels are depicted with thin and thick arrows, respectively.
As cyanobacteria transitioned to thylakoid-bearing species (Step II, Figure 3), the evolution of thylakoid membranes accelerated oxygenic photosynthesis, producing a large amount of oxygen (Rast et al., 2015), driving the GOE (Guéguen and Maréchal, 2022) (Figure 1). Thylakoid-less cyanobacterial species, such as Anthocerotibacter panamensis and Gloeobacter violaceus PCC7421, exhibited limited photosynthetic capacity (Rahmatpour et al., 2021; Raven and Sánchez-Baracaldo, 2021), with a cell doubling time of 3-8 days (Rippka et al., 1974; Rahmatpour et al., 2021). In contrast, the thylakoid-bearing cyanobacterium Synechococcus sp. PCC7336, the closest relative of thylakoid-less cyanobacteria (Tan et al., 2024), has a cell doubling time of approximately 1 day (Herrmann et al., 2021), while model species, such as Synechocystis sp. PCC6803, can divide within hours (Rahmatpour et al., 2021). This increased photosynthetic efficiency led to heightened ATP demand, further driving the need for cpNDH-1 stabilization.
To meet these demands, cpNDH-1 acquired NdhL, NdhQ, and NdhV, which are critical for maintaining high-efficiency PSI CET. The deletion of ndhL severely impairs PSI CET activity (Mi et al., 1992; Zhang et al., 2020), while the loss of ndhV disrupts Fd interaction (Gao et al., 2016; Zhang et al., 2020). These adaptations enabled cpNDH-1 to support its high levels of stabilization, potentially driving the evolution of robust oxygenic photosynthesis and carbon assimilation in thylakoid-bearing cyanobacteria. This analysis provides a hypothesis for the driving factors behind the evolution of OPS subunits, which will require further experimental validation.
The evolutionary origin of OPS cpNDH-1 subunits
The evolutionary origins of OPS cpNDH-1 subunits provide valuable insights into their emergence and functional development. New genes can arise through various mechanisms, including de novo origination of genetic sequences or the modification of existing genes (Long et al., 2013).
To investigate the origins of OPS cpNDH-1 genes, we performed TBLASTN searches to identify homologous gene fragments corresponding to specific OPS cpNDH-1 subunits in early-branching cyanobacteria. Our analysis revealed that NdhM, NdhN, NdhO, NdhP, and NdhS—subunits that first emerged in thylakoid-less cyanobacteria—are represented by only partial homologous fragments, rather than complete homologous sequences, in non-photosynthetic cyanobacteria of the class Sericytochromatia (Figure 4A; Supplementary Data 5 and 6). These results suggest that the genetic sequences giving rise to these OPS subunits were likely absent in non-photosynthetic cyanobacteria, implying that these subunits evolved through de novo origination.
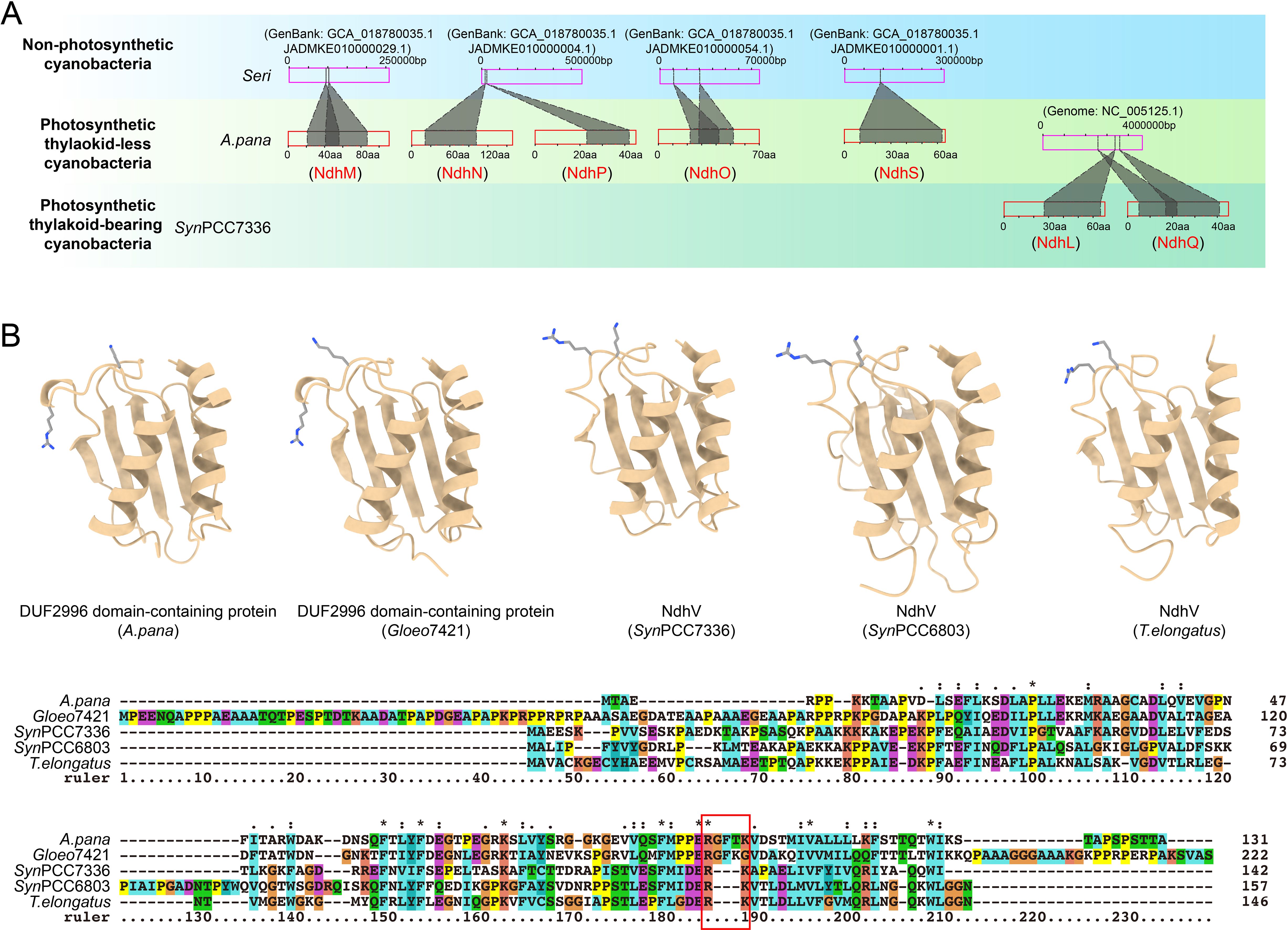
Figure 4. Gene origins of OPS subunits in cyanobacterial cpNDH-1. (A) TBLASTN and BLASTP analyses were performed to identify potential gene origins of OPS cpNDH-1 subunits. For each OPS cpNDH-1 subunit, the upper pink rectangle represents genomes or metagenome-assembled genomes, and the lower red rectangle represents the protein sequence of the OPS subunits as it first emerged in cyanobacteria. Shaded areas indicate regions of potential homology. The sequence data used for plotting are provided in Supplementary Data 6. Possible homologous fragments, rather than complete homologous sequences, identified in cyanobacteria with closely related evolutionary backgrounds suggest gene origination through de novo evolution. (B) Structure prediction (Upper panel) and sequence alignment (Lower panel) between DUF2996 domain-containing proteins from thylakoid-less cyanobacteria and NdhV from thylakoid-bearing cyanobacteria. Asterisks denote identical amino acids, while colons and periods indicate conserved and semi-conserved substitutions, respectively. The key residues involved in binding to the electron donor Fd in NdhV are highlighted by red box. The similar structures and changes in important amino acid sites suggest a hypothesis that NdhV may be generated by modifying existing gene of DUF2996 domain-containing proteins.
Similarly, NdhL and NdhQ, which arose in thylakoid-bearing cyanobacteria, are also represented by only partial homologous fragments in thylakoid-less cyanobacteria (Figure 4A; Supplementary Data 5 and 6). This further supports the hypothesis that these subunits, too, may have evolved via de novo origination. Moreover, structure-based Foldseek analysis failed to identify any structurally similar homologs in cyanobacteria lacking OPS subunits, reinforcing the idea that these OPS subunits likely did not have ancestral counterparts in early cyanobacteria, further supporting their de novo evolution. However, the possibility remains that ancestral sequences of these OPS subunits existed, through verifying this would require more extensive genetic data from early-branching cyanobacteria lineages.
Interestingly, NdhV appears to have a distinct evolutionary origin compared to the other OPS subunits. NdhV contains a DUF2996 domain (Gao et al., 2016), and BLASTP and TBALSTN searches revealed no homologous fragments in thylakoid-less cyanobacteria (Figure 2; Supplementary Data 5). While DUF2996 domain-containing proteins are present in these cyanobacteria, their sequences show minimal similarity to NdhV from thylakoid-bearing cyanobacteria (Supplementary Data 3). Nevertheless, Foldseek analysis revealed significant structural similarity between the DUF2996 domain-containing proteins and NdhV (Sll0272) from Synechocystis sp. PCC 6803 (Supplementary Figure S6), which was further confirmed through structural alignments (Figure 4B). Sequence alignment of these structural homologs revealed that the two consecutive basic amino acids, critical for binding the electron donor Fd in NdhV (Zhang et al., 2020), are separated in the DUF2996 domain-containing proteins from thylakoid-less cyanobacteria (Figure 4B). Notably, this distance appears to shorten in the evolutionary trajectory of thylakoid-free cyanobacteria (indicated by the red box in Figure 4B). Collectively, these analyses suggest that NdhV may have evolved through the modification of an existing DUF2996 domain-containing proteins.
Conclusions
In this study, we utilized extensive genomic data from cyanobacteria to investigate, for the first time, the evolutionary trajectory of OPS subunits in the cyanobacterial cpNDH-1 complex and proposed a possible evolutionary scenario for its development. Our findings suggest that, during the transition from non-photosynthetic to thylakoid-less photosynthetic cyanobacteria, the cpNDH-1 complex incorporated the OPS subunits NdhM, NdhN, NdhO, NdhP, and NdhS. Subsequently, during the shift from thylakoid-less to thylakoid-bearing cyanobacteria, additional OPS subunits—NdhL, NdhQ, and NdhV—were acquired. This proposed evolutionary trajectory not only reflects the adaptation of cyanobacteria to changing environmental stresses, but also offers a potential hypothesis for the driving forces behind the evolution of these subunits. Specifically, the rise of oxygen and the resulting oxidative stress, combined with the increased efficient oxygenic photosynthesis, likely intensified the demand for antioxidant defenses and PSI CET activity. Such evolutionary pressures may have driven the acquisition of OPS subunits to enhance the stability of cpNDH-1, a critical complex involved in these processes. Furthermore, protein sequence and structural analyses provide insights into the possible evolutionary origins of these OPS subunits. In conclusion, our findings highlight the potential role of oxygenic photosynthesis in shaping the evolution of cpNDH-1 in cyanobacteria and lay a groundwork for future studies exploring the functional and evolutionary significance of OPS subunits.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
XP: Formal Analysis, Writing – review & editing, Investigation. YJ: Formal Analysis, Writing – review & editing, Investigation. JY: Formal Analysis, Investigation, Writing – original draft. ZR: Conceptualization, Formal Analysis, Writing – original draft, Investigation. WM: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the National Key Research and Development Program of China (2023YFA0914600), National Natural Science Foundation of China (32170257), Shanghai Science and Technology Committee (22010503500).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1561629/full#supplementary-material
Abbreviations
cpNDH-1, conserved photosynthetic NDH-1; GOE, Great Oxidation Event; Mya, million years ago; OPS, oxygenic photosynthesis-specific; PSI, photosystem I; PSI CET, cyclic electron transport around PSI; pNDH-1, photosynthetic NDH-1; ROS, reactive oxygen species; Rubisco, Ribulose-1,5-bisphosphate carboxylase/oxygenase.
References
Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., et al. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500. doi: 10.1038/s41586-024-07487-w
Baradaran, R., Berrisford, J. M., Minhas, G. S., Sazanov, L. A. (2013). Crystal structure of the entire respiratory complex I. Nature 494, 443–448. doi: 10.1038/nature11871
Battchikova, N., Eisenhut, M., Aro, E. M. (2011a). Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta Bioenerg. 1807, 935–944. doi: 10.1016/j.bbabio.2010.10.017
Battchikova, N., Wei, L., Du, L., Bersanini, L., Aro, E. M., Ma, W. (2011b). Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH: plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 286, 36992–37001. doi: 10.1074/jbc.M111.263780
Battchikova, N., Zhang, P., Rudd, S., Ogawa, T., Aro, E. M. (2005). Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 280, 2587–2595. doi: 10.1074/jbc.M410914200
Bekker, A., Holland, H. D., Wang, P.-L., Rumble, D., III, Stein, H. J., Hannah, J. L., et al. (2004). Dating the rise of atmospheric oxygen. Nature 427, 117–120. doi: 10.1038/nature02260
Bernát, G., Appel, J., Ogawa, T., Rögner, M. (2011). Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various ndh-deficient mutants of Synechocystis sp. strain PCC6803. J. Bacteriol. 193, 292–295. doi: 10.1128/jb.00984-10
Blank, C. E., Sánchez-Baracaldo, P. (2010). Timing of morphological and ecological innovations in the cyanobacteria–a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23. doi: 10.1111/j.1472-4669.2009.00220.x
Blankenship, R. E. (2017). How Cyanobacteria went green. Science 355, 1372–1373. doi: 10.1126/science.aam9365
Boden, J. S., Konhauser, K. O., Robbins, L. J., Sánchez-Baracaldo, P. (2021). Timing the evolution of antioxidant enzymes in cyanobacteria. Nat. Commun. 12, 4742. doi: 10.1038/s41467-021-24396-y
Di Rienzi, S. C., Sharon, I., Wrighton, K. C., Koren, O., Hug, L. A., Thomas, B. C., et al. (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2, e01102. doi: 10.7554/eLife.01102
Drummond, A. J., Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fischer, W. W., Hemp, J., Valentine, J. S. (2016). How did life survive Earth’s great oxygenation? Curr. Opin. Chem. Biol. 31, 166–178. doi: 10.1016/j.cbpa.2016.03.013
Flamholz, A., Shih, P. M. (2020). Cell biology of photosynthesis over geologic time. Curr. Biol. 30, R490–R494. doi: 10.1016/j.cub.2020.01.076
Fournier, G. P., Moore, K. R., Rangel, L. T., Payette, J. G., Momper, L., Bosak, T. (2021). The Archean origin of oxygenic photosynthesis and extant cyanobacterial lineages. Proc. Biol. Sci. 288, 20210675. doi: 10.1098/rspb.2021.0675
Gao, F., Zhao, J., Wang, X., Qin, S., Wei, L., Ma, W. (2016). NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. strain PCC 6803. Plant Physiol. 170, 752–760. doi: 10.1104/pp.15.01430
Garcia-Pichel, F., Zehr, J. P., Bhattacharya, D., Pakrasi, H. B. (2020). What’s in a name? The case of cyanobacteria. J. Phycol. 56, 1–5. doi: 10.1111/jpy.12934
Grettenberger, C. L., Sumner, D. Y., Wall, K., Brown, C. T., Eisen, J. A., Mackey, T. J., et al. (2020). A phylogenetically novel cyanobacterium most closely related to Gloeobacter. ISME J. 14, 2142–2152. doi: 10.1038/s41396-020-0668-5
Guéguen, N., Maréchal, E. (2022). Origin of cyanobacterial thylakoids via a non-vesicular glycolipid phase transition and their impact on the Great Oxygenation Event. J. Exp. Bot. 73, 2721–2734. doi: 10.1093/jxb/erab429
He, Z., Mi, H. (2016). Functional characterization of the subunits N, H, J, and O of the NAD(P)H dehydrogenase complexes in Synechocystis sp. strain PCC 6803. Plant Physiol. 171, 1320–1332. doi: 10.1104/pp.16.00458
He, H., Wu, X., Xian, H., Zhu, J., Yang, Y., Lv, Y., et al. (2021). An abiotic source of Archean hydrogen peroxide and oxygen that pre-dates oxygenic photosynthesis. Nat. Commun. 12, 6611. doi: 10.1038/s41467-021-26916-2
He, Z., Xu, M., Wu, Y., Lv, J., Fu, P., Mi, H. (2016). NdhM subunit is required for the stability and the function of NAD(P)H dehydrogenase complexes involved in CO2 uptake in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 291, 5902–5912. doi: 10.1074/jbc.M115.698084
Herrmann, A. J., Sorwat, J., Byrne, J. M., Frankenberg-Dinkel, N., Gehringer, M. M. (2021). Diurnal Fe(II)/Fe(III) cycling and enhanced O2 production in a simulated Archean marine oxygen oasis. Nat. Commun. 12, 2069. doi: 10.1038/s41467-021-22258-1
Kato, Y., Odahara, M., Shikanai, T. (2021). Evolution of an assembly factor-based subunit contributed to a novel NDH-PSI supercomplex formation in chloroplasts. Nat. Commun. 12, 3685. doi: 10.1038/s41467-021-24065-0
Khademian, M., Imlay, J. A. (2021). How microbes evolved to tolerate oxygen. Trends Microbiol. 29, 428–440. doi: 10.1016/j.tim.2020.10.001
Laughlin, T. G., Bayne, A. N., Trempe, J. F., Savage, D. F., Davies, K. M. (2019). Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–414. doi: 10.1038/s41586-019-0921-0
Laughlin, T. G., Savage, D. F., Davies, K. M. (2020). Recent advances on the structure and function of NDH-1: The complex I of oxygenic photosynthesis. Biochim. Biophys. Acta Bioenerg. 1861, 148254. doi: 10.1016/j.bbabio.2020.148254
Long, M., VanKuren, N. W., Chen, S., Vibranovski, M. D. (2013). New gene evolution: Little did we know. Annu. Rev. Genet. 47, 307–333. doi: 10.1146/annurev-genet-111212-133301
Ma, W., Ogawa, T. (2015). Oxygenic photosynthesis–specific subunits of cyanobacterial NADPH dehydrogenases. IUBMB Life 67, 3–8. doi: 10.1002/iub.13411
Marsh, J. A., Teichmann, S. A. (2015). Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 84, 551–575. doi: 10.1146/annurev-biochem-060614-034142
Mi, H., Endo, T., Schreiber, U., Ogawa, T., Asada, K. (1992). Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 33, 1233–1237. doi: 10.1093/oxfordjournals.pcp.a078378
Miller, N. T., Vaughn, M. D., Burnap, R. L. (2021). Electron flow through NDH-1 complexes is the major driver of cyclic electron flow-dependent proton pumping in cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1862, 148354. doi: 10.1016/j.bbabio.2020.148354
Ogawa, T., Mi, H. (2007). Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93, 69–77. doi: 10.1007/s11120-006-9128-y
Pan, X., Cao, D., Xie, F., Xu, F., Su, X., M, H., et al. (2020). Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase. Nat. Commun. 11, 610. doi: 10.1038/s41467-020-14456-0
Peltier, G., Aro, E. M., Shikanai, T. (2016). NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu. Rev. Plant Biol. 67, 55–80. doi: 10.1146/annurev-arplant-043014-114752
Rahmatpour, N., Hauser, D. A., Nelson, J. M., Chen, P. Y., Ho, M. Y., Li, F. W. (2021). A novel thylakoid-less isolate fills a billion-year gap in the evolution of Cyanobacteria. Curr. Biol. 31, 2857–2867. doi: 10.1016/j.cub.2021.04.042
Rast, A., Heinz, S., Nickelsen, J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophys. Acta Bioenerg. 1847, 821–830. doi: 10.1016/j.bbabio.2015.01.007
Raven, J. A., Sánchez-Baracaldo, P. (2021). Gloeobacter and the implications of a freshwater origin of cyanobacteria. Phycologia 60, 402–418. doi: 10.1080/00318884.2021.1881729
Rippka, R., Waterbury, J., Cohen-Bazire, G. (1974). A cyanobacterium which lacks thylakoids. Arch. Microbiol. 100, 419–436. doi: 10.1007/BF00446333
Roush, D., Giraldo-Silva, A., Garcia-Pichel, F. (2021). Cydrasil 3, a curated 16S rRNA gene reference package and web app for cyanobacterial phylogenetic placement. Sci. Data. 8, 230. doi: 10.1038/s41597-021-01015-5
Sánchez-Baracaldo, P., Bianchini, G., Wilson, J. D., Knoll, A. H. (2022). Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 30, 143–157. doi: 10.1016/j.tim.2021.05.008
Sánchez-Baracaldo, P., Cardona, T. (2020). On the origin of oxygenic photosynthesis and Cyanobacteria. New Phytol. 225, 1440–1446. doi: 10.1111/nph.16249
Sánchez-Baracaldo, P., Ridgwell, A., Raven, J. A. (2014). A neoproterozoic transition in the marine nitrogen cycle. Curr. Biol. 24, 652–657. doi: 10.1016/j.cub.2014.01.041
Schuller, J. M., Birrell, J. A., Tanaka, H., Konuma, T., Wulfhorst, H., Cox, N., et al. (2019). Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363, 257–260. doi: 10.1126/science.aau3613
Schulz, L., Guo, Z., Zarzycki, J., Steinchen, W., Schuller, J. M., Heimerl, T., et al. (2022). Evolution of increased complexity and specificity at the dawn of form I Rubiscos. Science 378, 155–160. doi: 10.1126/science.abq1416
Soo, R. M., Hemp, J., Parks, D. H., Fischer, W. W., Hugenholtz, P. (2017). On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 355, 1436–1440. doi: 10.1126/science.aal3794
Strunecký, O., Ivanova, A. P., Mareš, J. (2023). An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 59, 12–51. doi: 10.1111/jpy.13304
Tan, S., Liu, L., Jiao, J. Y., Li, M. M., Hu, C. J., Lv, A. P., et al. (2024). Exploring the origins and evolution of oxygenic and anoxygenic photosynthesis in deeply branched cyanobacteriota. Mol. Biol. Evol. 41, msae151. doi: 10.1093/molbev/msae151
Tomitani, A., Knoll, A. H., Cavanaugh, C. M., Ohno, T. (2006). The evolutionary diversification of cyanobacteria: molecular–phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. U.S.A. 103, 5442–5447. doi: 10.1073/pnas.0600999103
Urano, D., Maruta, N., Trusov, Y., Stoian, R., Wu, Q., Liang, Y., et al. (2016). Saltational evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Sci. Signal. 9, ra93. doi: 10.1126/scisignal.aaf9558
van Kempen, M., Kim, S., Tumescheit, C., Mirdita, M., Lee, J., Gilchrist, C. L. M., et al. (2024). Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246. doi: 10.1038/s41587-023-01773-0
Yu, J., Ran, Z., Zhang, J., Wei, L., Ma, W. (2022). Genome-wide insights into the organelle translocation of photosynthetic NDH-1 genes during evolution. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.956578
Yu, H., Schut, G. J., Haja, D. K., Adams, M. W. W., Li, H. (2021). Evolution of complex I–like respiratory complexes. J. Biol. Chem. 296, 100740. doi: 10.1016/j.jbc.2021.100740
Yu, H., Wu, C. H., Schut, G. J., Haja, D. K., Zhao, G., Peters, J. W., et al. (2018). Structure of an ancient respiratory system. Cell 173, 1636–1649. doi: 10.1016/j.cell.2018.03.071
Zhang, J., Gao, F., Zhao, J., Ogawa, T., Wang, Q., Ma, W. (2014). NdhP is an exclusive subunit of large complex of NADPH dehydrogenase essential to stabilize the complex in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 289, 18770–18781. doi: 10.1074/jbc.M114.553404
Zhang, C., Shuai, J., Ran, Z., Zhao, J., Wu, Z., Liao, R., et al. (2020). Structural insights into NDH-1 mediated cyclic electron transfer. Nat. Commun. 11, 888. doi: 10.1038/s41467-020-14732-z
Zhao, J., Gao, F., Zhang, J., Ogawa, T., Ma, W. (2014). NdhO, a subunit of NADPH dehydrogenase, destabilizes medium size complex of the enzyme in Synechocystis sp. PCC 6803. J. Biol. Chem. 289, 26669–26676. doi: 10.1074/jbc.M114.553925
Zhao, J., Rong, W., Gao, F., Ogawa, T., Ma, W. (2015). Subunit Q is required to stabilize the large complex of NADPH dehydrogenase in Synechocystis sp. strain PCC 6803. Plant Physiol. 168, 443–451. doi: 10.1104/pp.15.00503
Keywords: cyanobacteria, conserved photosynthetic NDH-1, OPS subunits, evolutionary history, cyclic electron transfer
Citation: Pang X, Jiang Y, Yu J, Ran Z and Ma W (2025) Genome-wide insights into the evolutionary history of conserved photosynthetic NDH-1 in cyanobacteria. Front. Plant Sci. 16:1561629. doi: 10.3389/fpls.2025.1561629
Received: 16 January 2025; Accepted: 28 March 2025;
Published: 15 April 2025.
Edited by:
Yan Lu, Western Michigan University, United StatesReviewed by:
Minting Liang, Chinese Academy of Sciences (CAS), ChinaGeorg Hochbergm, University of Marburg, Germany
Debanjan Mitra, University of Burdwan, India
Copyright © 2025 Pang, Jiang, Yu, Ran and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoxing Ran, cmFuemhhb3hpbmdAMTI2LmNvbQ==; Weimin Ma, d21hQHNobnUuZWR1LmNu
†These authors have contributed equally to this work
 Xiaoqin Pang
Xiaoqin Pang Yuanyuan Jiang
Yuanyuan Jiang Jie Yu
Jie Yu Zhaoxing Ran
Zhaoxing Ran Weimin Ma
Weimin Ma