- 1Maize Research Institute, Shandong Academy of Agricultural Sciences, Jinan, China
- 2Plant Protection Research Institute, Rizhao Academy of Agricultural Sciences, Rizhao, China
Improving nitrogen use efficiency (NUE) is one of the major objectives for crop breeding. As nitrate signaling plays pivotal roles in nitrogen use of plants, factors in this pathway might be valuable for improving the NUE of maize. In this research, we performed Gene Set Enrichment Analysis (GSEA) of maize transcriptomes in response to nitrate and found that the ethylene action pathway might participate in nitrate signaling. Through a modified reciprocal best hit approach, we obtained 16 maize aminocyclopropane-1-carboxylic acid (ACC) oxidase (ACO) and four ACC synthase (ACS) homologs in maize genome. In silico analyses and the reverse transcription quantitative PCR assays demonstrated that ZmACCO7, ZmACCO5, ZmACCO15, ZmACCO35, and ZmACCO31 are the top five highly expressed ACO genes, and ZmACS1 is the most highly expressed ACS gene in the primary and seminal roots of maize. We discovered that ACO and ACS genes have different regulatory modes in response to nitrate provision. Some ACO genes, which are mainly expressed in root regions far from the root tip like ZmACCO7, are repressed by nitrate, while the others, which are mainly expressed in root regions near the root tip like ZmACCO5, are induced by nitrate. ZmACS1, which has more uniform expression across maize roots, is induced in root regions near the root tip and repressed in regions far from the root tip. A phenotypic analysis indicated that upregulation of ACO and ACS genes by nitrate is linked to repression of axial root elongation by nitrate while the downregulation of these genes is associated with the promotion of growth of lateral roots of the axial roots. In addition, differences in regulation of ACO and ACS genes by nitrate were observed between genotypes, which is related to the differences in the responses of their primary root growth to nitrate. These results suggested that the ethylene synthesis pathway is involved in the responses of maize roots to nitrate, which is associated with the remodeling of maize root architecture by nitrate.
1 Introduction
Nitrogen fertilizer is one of the major investments for crop production due to the high amount of requirement of nitrogen of plants and limited nitrogen available in the soil. Moreover, application of fertilizer leads to the eutrophication of aquatic ecosystems (Raun and Johnson, 1999), and the production of fertilizer also causes air pollution. Therefore, improving nitrogen use efficiency (NUE) becomes one of the major objectives of crop breeding programs. However, as a complex trait, it is a challenge at present. Since nitrate is the predominant nitrogen form available to plants in aerated soil (Dechorgnat et al., 2011), nitrate uptake, transport, metabolism, and their regulation are the key processes to enhance the NUE of arable crops like maize.
The root architecture and the activity of nitrate transporters of root cells play critical roles in the NUE of crops as roots are the main organs for plants to absorb nitrate from soils (Aluko et al., 2023; Hawkesford and Griffiths, 2019). A comparison between hybrids from different eras showed that the remodeled root architectures in the breeding processes enhanced the NUE of maize (Li et al., 2024). Genes in the pathway of auxin signaling were found to be key elements in regulating maize root development (Hochholdinger et al., 2018). A total of 81 high-priority candidate genes associated with root angle were identified by integrating genome-wide association analysis with co-expression network analysis (Ren et al., 2022). Research in rice demonstrated that NRT1.1B-indica allele improved the grain yield and NUE when introduced into varieties without this allele (Hu et al., 2015). In maize, 78 Nitrate Transporter1/Peptide Transporter Family (NPF), seven Nitrate Transporters2 (NRT2), and two NRT3 genes were reported (Jia et al., 2023). Transcriptional factors involved in the nitrate signaling were also valuable in improving the NUE of crops. For example, OsTCP19 is associated with a high tillering response to nitrogen (Liu et al., 2021).
Ethylene is a plant hormone regulating numerous physiological and morphological responses of plants by interacting with other signaling molecules (Khan et al., 2015). Generally, ethylene is regarded as a stress hormone (Wang et al., 2002). Suboptimal N status, either deficiency or excess, acts as a stress to stimulate ethylene biosynthesis and signaling to remodel root architectures, nitrogen uptake, and translocation (Ma et al., 2023). High nitrate concentration (10 mmol/L) induces ethylene synthesis in the root of Arabidopsis (Tian et al., 2009). Low nitrate treatment also induces a rapid burst of ethylene production (Zheng et al., 2013). Research in Arabidopsis showed that ethylene interacts with cytokinin through ethylene receptor ETR1 to control primary root growth (Zdarska et al., 2019) and modulates the transcription of auxin carrier genes to inhibit lateral root formation and elongation (Lewis et al., 2011). Ethylene also regulates the growth of root hairs through transcriptional factor ethylene insensitive3 (EIN3) (Xiao et al., 2021). In addition, the production of ethylene represses the expression of the low-affinity nitrate transporter coding gene NRT2.1 but upregulates NRT1.1 expression in Arabidopsis (Tian et al., 2009).
The synthesis of ethylene from its general precursor S-adenosyl-l-methionine (SAM) in plants mainly involves two processes. The first step is the conversion of SAM to 1-aminocyclopropane-1-carboxylic acid (ACC), which is catalyzed by ACC synthase (ACS). The second step is the production of ethylene from ACC, which is catalyzed by ACC oxidase (ACO) (Wang et al., 2002). The pathway of ethylene production is precisely tuned at both transcriptional and post-translational levels (Houben and Van de Poel, 2019; Pattyn et al., 2021). The conversion of SAM to ACC was considered as the rate-limiting step in ethylene biosynthesis (Tsuchisaka et al., 2009). Evidence also showed that the activities of ACOs are the rate-limiting factors during some processes (Houben and Van de Poel, 2019). Maize ZmACS7 over-expressors displayed accelerated leaf senescence in response to N deficiency and improved NUE (Xing et al., 2024). However, how ethylene synthesis and signaling pathways are involved in the nitrate-regulated plant growth in maize remains poorly understood.
Here we reported that public data mining indicated that the ethylene action pathway is disturbed by nitrate in maize roots, and experiments showed that transcriptions of ACO and ACS genes are regulated by nitrate with different modes in maize seedling roots, which is related to nitrate-modulated root growth.
2 Materials and methods
2.1 Plant materials and growth condition
The inbred lines B73 and Z58 were used in this study. Z58 (Zheng58) is a parent of Zhengdan958, a widely planted hybrid in North China. Plant seeds were sterilized using 1:20 dilution of sodium hypochlorite for 15 min, washed six times with deionized water, and germinated on a wet towel for 2 days at 25°C. The seedlings were grown in paper rolls containing 0.1 mmol/L CaCl2 for 3 days at 25°C and then fixed to boxes full of 0.1 mmol/L CaCl2 solution to grow in a chamber with controlled climate conditions (25°C) for 1 day. Subsequently, the seedlings were supplied with nutrient solution containing different concentrations of nitrogen to grow after their endosperms were removed.
The base nutrient solution contained 0.75 mmol/L K2SO4, 0.1 mmol/L KCl, 0.25 mmol/L KH2PO4, 0.65 mmol/L MgSO4, 0.2 mmol/L EDTA-Fe, 1.0 μmol/L H3BO3, 1.0 μmol/L MnSO4, 1.0 μmol/L ZnSO4, 0.1 μmol/L CuSO4, and 0.005 μmol/L (NH4)6Mo7O24. For high nitrogen solution (HN), Ca(NO3)2 was added into the base nutrient solution at a concentration of 2.0 mmol/L, and for low nitrogen solution (LN), CaCl2 was added at a concentration of 2.0 mmol/L. The pH of the growth solutions was adjusted to 6.0 with NaOH. For cobalt treatments, nutrient solutions containing 3.0 μmol/L CoCl2 were used. For hydrophonic culture, the solutions were ventilated for 20 min each 2 h by electric pumps.
For the analysis of gene expression in response to nitrate, seedlings were grown in LN for 1 day, and then the solutions were replaced with fresh LN (as the control) or HN solutions. To examine nitrate’s effects on the phenotype of plants, seedlings were grown in LN or HN for 9 days, and the solutions were renewed every 2 days.
2.2 RNA extraction, reverse transcription, and quantitative PCR
Seedling roots were harvested for RNA extraction. The total RNA was prepared using Aidlab’s plant RNA extraction kit RN09 (Aidlab, Beijing, China) and reverse transcribed to cDNA using Accurate Biology’s RT Kit (Accurate Biology, Changsha, China). Quantitative PCRs (qPCR) were performed with SYBR Green Real-Time PCR Master Mix (Accurate Biology, Changsha, China) using Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) following the comparative CT experiment protocol of the manufacturer. The number of biological repeats were described in figures in detail. For the analysis of root-region-specific mRNA expression of genes in response to nitrate provision, 14 plant samples were bulked for RNA extraction unless otherwise stated in the text. For the mRNA expression analysis in other cases, six to seven biological replicates, each of which contained one plant sample, were performed. Two to three technical replicates were done and folypolyglutamate synthase (FPGS) was used as the reference gene (Manoli et al., 2012) in qPCR assays. Relative gene expression was calculated as described by Livak and Schmittgen (2001). The primers used in the research are listed in Supplementary Table S1.
2.3 Measurement of root length, number of lateral roots and plant height, and statistical analysis
Plant height, the length from the node where the coleoptile grows to the tip of the longest leaf of a seedling, was measured with a ruler. Each primary and seminal root was measured separately. After being detached from the plant at the base, an axial root with branched lateral roots was floated in a transparent plastic tray (20 cm × 15 cm) full of water and next scanned with Epson Perfection V700 Photo (EPSON, Beijing, China), followed by measuring the axial root length with a ruler. The resulting images were analyzed using the software WinRHIZO Pro 5.0 (Quebec City, Canada). The number of root tips obtained with WinRHIZO was considered as the number of lateral roots. The total lateral root length was calculated by subtracting the axial root length from the total root length derived from WinRHIZO. Each group of treatments was conducted in an independent experiment and was analyzed separately with two-tailed t-test in the phenotype evaluation. For seminal roots, the mean of values of a trait per plant was used for further analysis.
2.4 Analysis of RNA−Seq data
Transcriptomic data from PRJNA283053 and PRJNA304223 were used for Gene Set Enrichment Analysis (GSEA). In silico organ- or tissue-specific expression patterns of ACS and ACO genes were estimated with transcriptomic data from PRJNA217053 and PRJNA171684. These data were downloaded from https://www.ebi.ac.uk/ (accessed on January 20, 2020). Reads with poor quality were filtered with Trimmomatic V0.39 (Bolger et al., 2014). The clean data passing the quality control were evaluated with FastQC V 0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and used to quantify transcripts with Salmon V1.0.0 (Patro et al., 2017) based on the gene models of Zm-B73-REFERENCE-GRAMENE-4.0 (https://www.maizegdb.org/, accessed on December 1, 2019). Gene expression changes were determined using the R package DESeq2 V1.42.1 (Love et al., 2014). GSEA (Subramanian et al., 2005) was performed by using the software GSEA V4.3.3 with Signal2Noise being selected as metric for ranking genes and gene sets being used for permutation. Gene counts calculated with transcript counts derived from Salmon analysis and then normalized with EdgeR package V3.20 (Robinson et al., 2010) were used for further GSEA. Gene sets were constructed according to the maize gene pathway annotation of MapMan (Thimm et al., 2004) based on gene models of Zm-B73-REFERENCE-GRAMENE-4.0.
2.5 Maize ACS and ACO homolog searching and phylogenetic analysis
A modified reciprocal best hit (RBH) (Bork et al., 1998; Tatusov et al., 1997) method was used to obtain maize ACS and ACO homologs. In brief, Arabidopsis ACO (Houben and Van de Poel, 2019) and ACS proteins (Yamagami et al., 2003) were used to blast against predicted maize proteins from the gene models of Zm-B73-REFERENCE-GRAMENE-4.0. Top hits, with e-value being less than 1 × 10−20 as well as both query and subject coverage being more than 50%, were kept for reverse blast against the Arabidopsis protein database (TAIR10). A maize homolog was considered as a true homolog when a positive reciprocal blast search was obtained. The canonical transcript for a maize gene and the representative transcript for an Arabidopsis gene were considered for further analyses where there were several transcripts for a gene. Sequences were aligned by ClustalW, and phylogenetic trees were constructed with the maximum likelihood method implemented in MEGAX V10.0.5 (Kumar et al., 2018). A total of 1,000 replicates of bootstraps were performed in the phylogenetic analyses.
3 Results
3.1 The ethylene action pathway was enriched in the root transcriptomic response to nitrate in maize
As nitrate signaling plays pivotal roles in the use of nitrogen in plants, factors in this process might be valuable to improve the NUE of maize. Thus, we conducted the public transcriptomic data mining in order to identify key genes in nitrate signaling as candidates to enhance the NUE of maize. PRJNA283053 (Yu et al., 2015) was a project of transcriptomic assays of stele tissues extracted from the regions between 5 and 25 mm of shoot-borne roots of B73 in response to 24-h local high-nitrate stimulation. The project of PRJNA304223 included comparative transcriptome data of roots of two genotypes, the rtcs (rootless concerning crown and seminal roots) mutant and the wild type B73, in response to different concentrations of nitrate (He et al., 2016). The B73 transcriptomes in these two data sets were downloaded for analyses. GSEA is considered as a robust method to identify a priori defined gene set differentially affected between experiment groups (Subramanian et al., 2005). Thus, we performed GSEA of the expression data with MapMan pathway (Thimm et al., 2004) used as gene sets. In the analysis of data from PRJNA283053, we found that ethylene action (11.5 in MapMan) was the third significantly enriched pathway (Figure 1; Supplementary Table S2). A detailed examination showed that 10 were annotated as ACO or ACS genes, and five of them were significantly upregulated by high nitrate treatment among 20 genes in the core enrichment (Table 1). However, this result was not confirmed in the analysis of B73 transcriptomic data from PRJNA304223. It was reported that ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana (Tian et al., 2009). These results indicated that it is possible that the ethylene synthesis pathway contributes to nitrate signaling in maize.
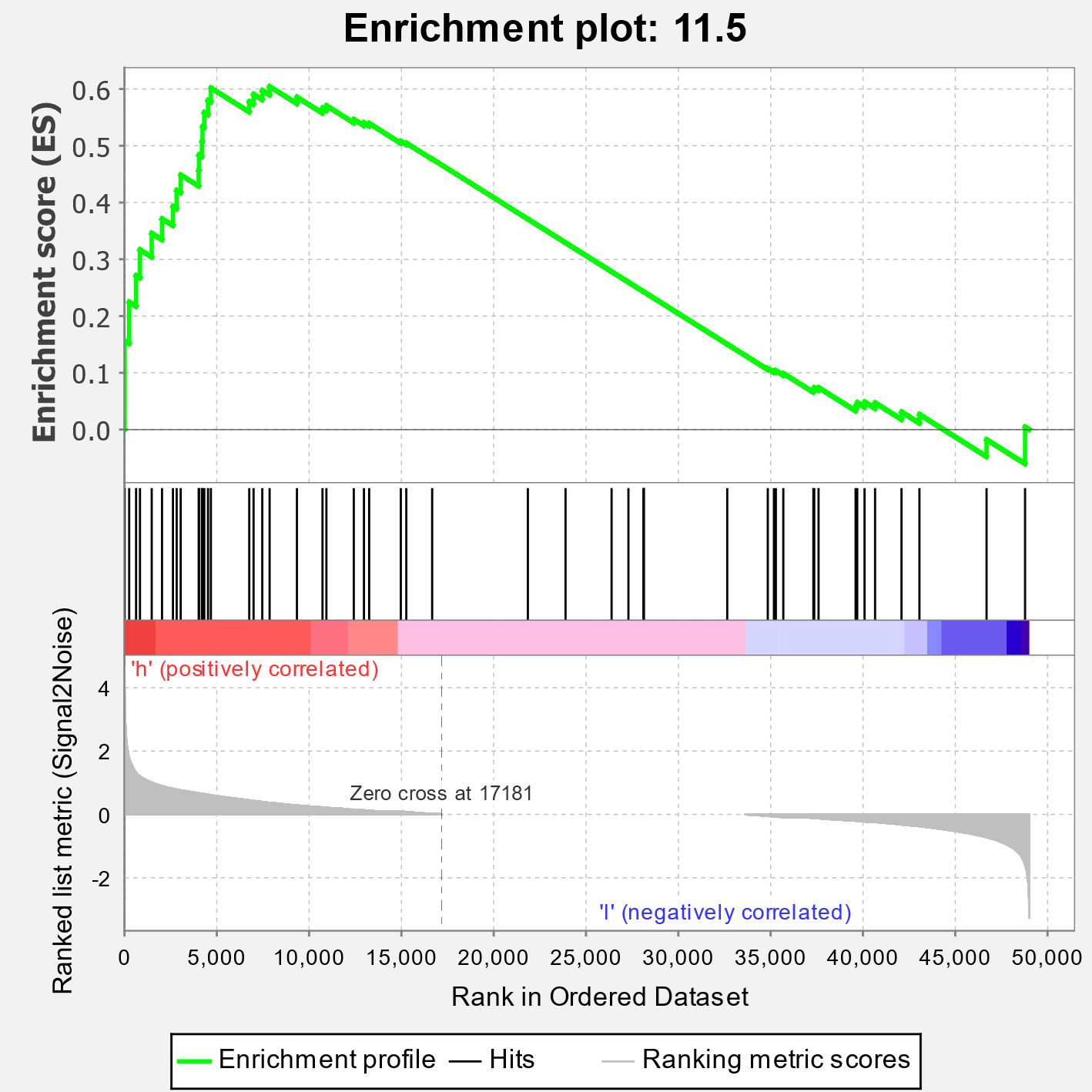
Figure 1. Enrichment of the ethylene action pathway (pathway 11.5 in MapMan) in maize transcriptomic responses to nitrate treatment. Clean data derived from transcriptomic data of PRJNA283053 were subjected to quantify transcripts with Salmon-SMEM to get gene counts. Enrichment scores were calculated using the software GSEA-MSigDB with edgeR-normalized gene counts used as expression data and MapMan-derived gene pathways used as gene sets.
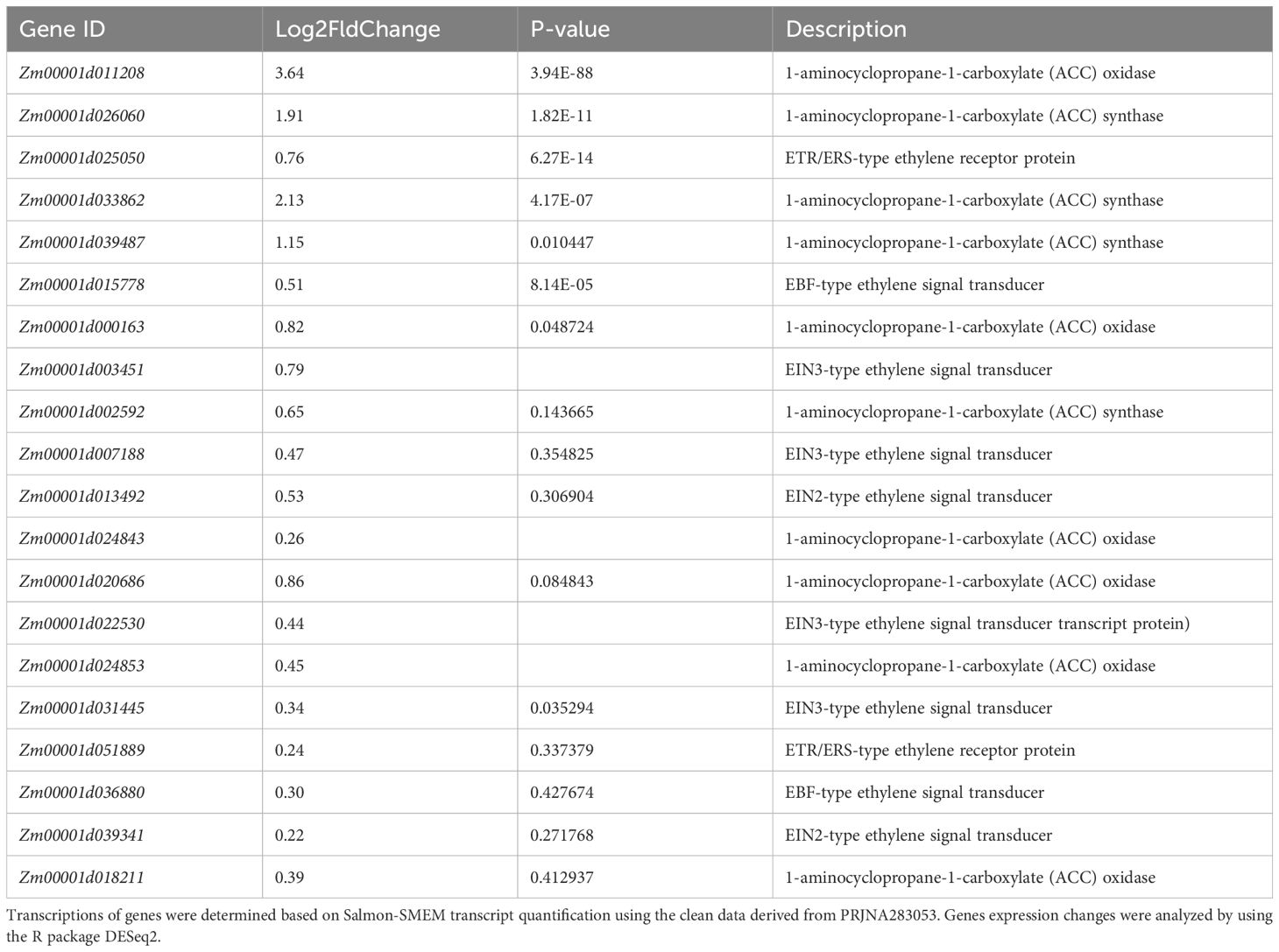
Table 1. Transcriptional responses to nitrate of genes in the leading-edge subset of the ethylene action pathway in GSEA.
3.2 ACO and ACS gene families in maize
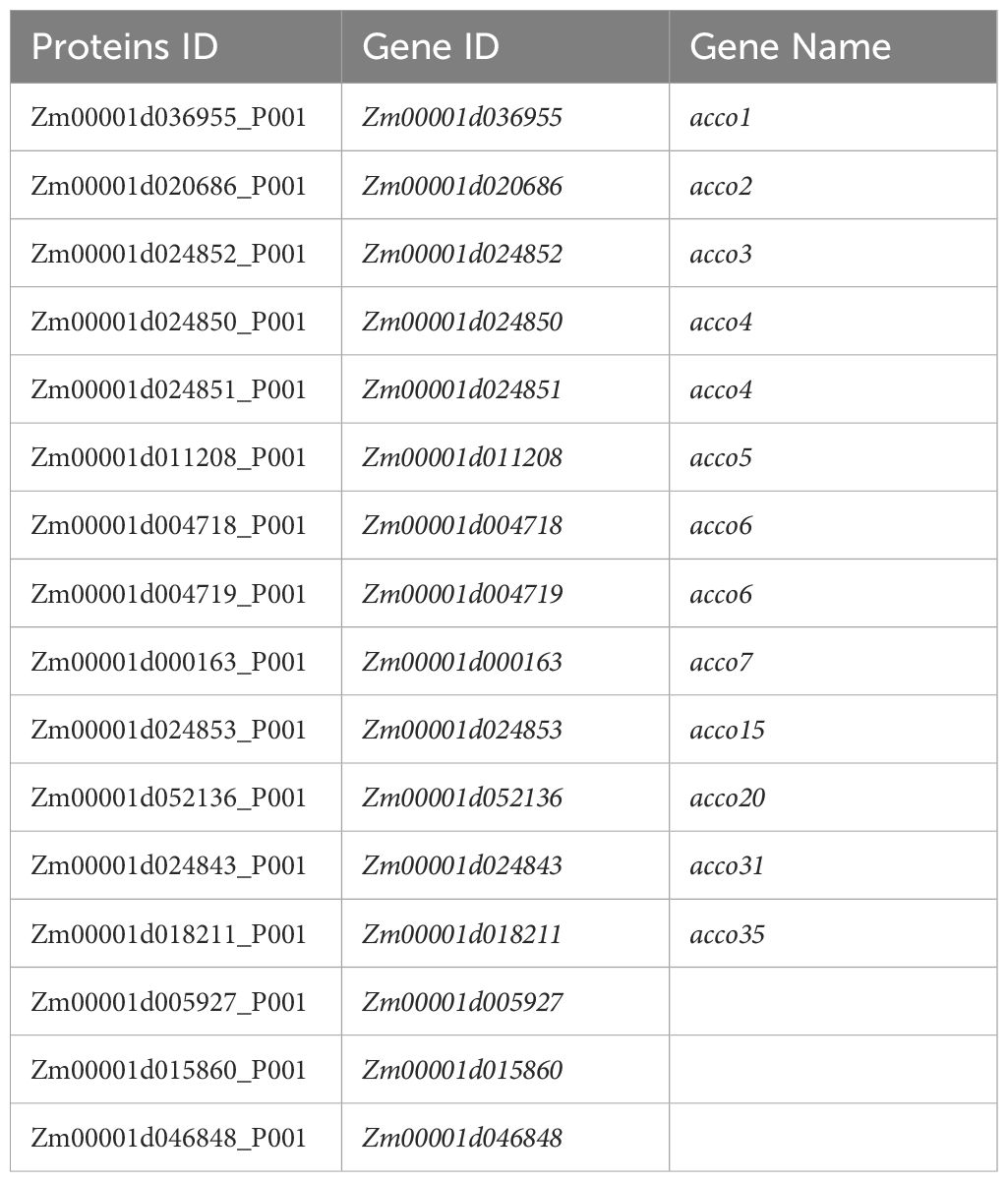
Given that ACO and ACS proteins were the only two enzymes responsible for ethylene biosynthesis, we aimed to identify the master ACO and ACS genes involved in root response to nitrate. First, we retrieved all ACO and ACS candidate genes in maize B73 genome through RBH search using ACO and ACS protein sequences of Arabidopsis as queries. ACOs were encoded by five genes in Arabidopsis genome, including AtACO1 (AT2G19590.1), AtACO2 (AT1G62380.1), AtACO3 (AT1G12010.1), AtACO4 (AT1G05010.1), and AtACO5 (AT1G77330.1) (Houben and Van de Poel, 2019), while 16 homologs were found in maize genome (Table 2), of which Zm00001d004718 and Zm00001d004719 have the same sequences and were named as the same gene ACCO6 in the database, and Zm00001d024850 and Zm00001d024851 were named as the same gene ACCO4 (https://maizegdb.org/, accessed on March 11, 2025).
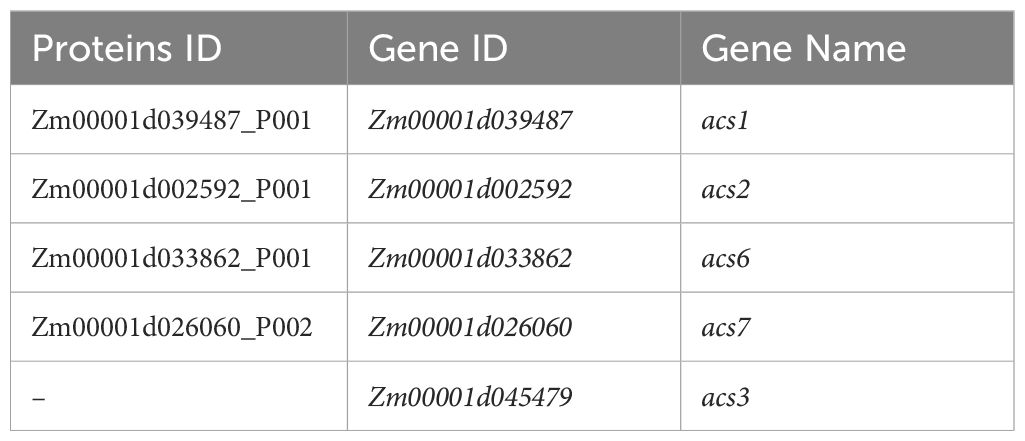
Eight authentic ACS genes including AtACS2 (AT1G01480), AtACS4 (AT2G22810), AtACS5 (AT5G65800), AtACS6 (AT4G11280), AtACS7 (AT4G26200), AtACS8 (AT4G37770), AtACS9 (AT3G49700), and AtACS11 (AT4G08040) were reported in Arabidopsis (Yamagami et al., 2003). It was interesting that only four ACS homologs were identified in the B73 genome, which included ZmACS1, ZmACS2, ZmACS6, and ZmACS7. ZmACS3 was not identified in this research, but it was also annotated as ACS in the database (https://maizegdb.org/, accessed on March 11, 2025) (Table 3). These homologs were subjected to further analyses except that ZmACS3 was not included in the phylogenetic tree analyses.
We next inspected the evolution relationships and expression correlations between these ACOs and ACSs from maize. The phylogenetic tree showed that most of these genes might result from duplicate events after the divergence between the ancestors of maize and Arabidopsis. In these processes, more ACO genes were kept in the maize genome, while more ACS genes were kept in the Arabidopsis genome (Figures 2A,B). As expected, there were no evolution distance between ZmACCO4 (Zm00001d024850) and ZmACCO4 (Zm00001d024851) and between ZmACCO6 (Zm00001d004718) and ZmACCO6 (Zm00001d004719). It was observed that the relationships between ZmACCO3, ZmACCO4, ZmACCO15, and ZmACCO31 and between ZmACCO1 and Zm000015860 were close in the evolution. The ACS genes were shown to be more divergent compared with ACO genes in maize, and ZmACS2 and ZmACS7 are closer to each other in the evolution among all of the ACS genes.
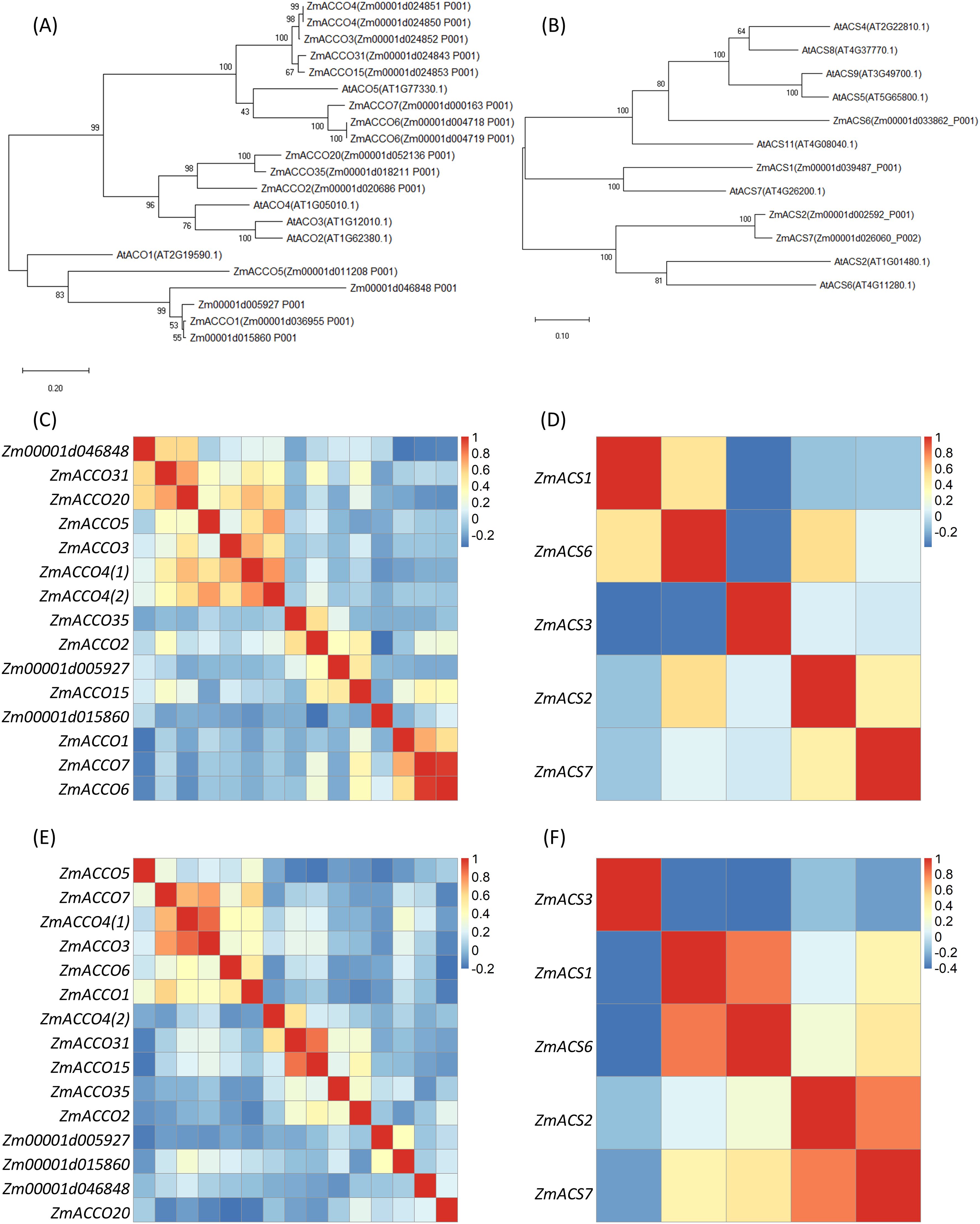
Figure 2. Relationships between Maize ACOs and between ACSs. (A) A phylogenetic tree of ACO proteins of maize and Arabidopsis. (B) A phylogenetic tree of ACS proteins of maize and Arabidopsis. (C, D) Expression correlations between ACO (C) and ACS (D) genes calculated with data from PRJNA2170531. (E, F) Expression correlations between ACO (E) and ACS (F) genes calculated with data from PRJNA171684. Maize ACO and ACS homologs were obtained by search maize database with known Arabidopsis ACO and ACS proteins using RBH method. Protein sequences were aligned by ClustalW and phylogenetic trees were constructed with maximum likelihood method. Transcriptions of genes were determined based on Salmon-SMEM transcript quantification using the clean data. The colors in (C-F) indicates the values of correlation coefficients. ZmACCO4(1) and ZmACCO4(2) are corresponding to Zm00001d024850 and Zm00001d024851, respectively.
The expression correlations between ACO genes and that between ACS genes were investigated with RNA-Seq data from PRJNA217503 (Walley et al., 2016) and PRJNA171684 (Stelpflug et al., 2016), respectively. These two projects included profiling data of gene expression in multiple tissues of B73. ZmACCO6 (Zm00001d004719) was not included in the analysis as it has the same sequence as ZmACCO6 (Zm00001d004718). Data from PRJNA217503 demonstrated that similar expression patterns existed between ZmACCO6 (Zm00001d004718), ZmACCO7, and ZmACCO1, between ZmACCO20, ZmACCO31, and Zm00001d046848, between ZmACCO4(1) (Zm00001d024850), ZmACCO4(2) (Zm00001d0024851), ZmACCO3, and ZmACCO5 in ACO genes (Figure 2C; Supplementary Figure S1A). A high expression correlation between ZmACCO3 and ZmACCO4(1) (Zm00001d024850) was also observed in the data from PRJNA171684 (Figure 2E; Supplementary Figure S1C). The high expression correlations between ZmACCO6 (Zm00001d004718) and ZmACCO7 and between ZmACCO4(1) (Zm00001d0024850), ZmACCO4(2) (Zm00001d0024851) and ZmACCO3 were consistent with their evolutional relationships observed in the phylogenetic tree (Figure 2A). The high expression correlation between ZmACCO15 and ZmACCO31 only observed in the data from PRJNA171684 (Figure 2E; Supplementary Figure S1C) was also consistent with their evolutional relationship (Figure 2A). On the whole, no clear links between the expression correlations and the evolutionary distances between these genes were observed.
For ACS genes, both data sets supported the distinctive expression pattern of ZmACS3 compared with the other four genes (Figures 2D, F; Supplementary Figures S1B,D), which was in line with the result that ZmACS3 was not identified in our homolog search. Relatively high expression correlations between ZmACS1 and ZmACS6 and between ZmACS2 and ZmACS7 were observed in both data sets (Figures 2D, F; Supplementary Figures S1B, D). The latter was consistent with their evolutionary relationship (Figure 2B).
We further examined the mRNA levels of these ACO and ACS genes in maize roots in detail. Data from PRJNA171684 demonstrated that the transcription profiles of ACO genes in different types of roots were similar (Figures 3A–D). The expression profiles of ACO genes in brace roots showed a higher similarity to that of crown roots (Figure 3A, B), while that in primary roots exhibited a higher similarity to that in seminal roots (Figure 3C, D). Similar profiles of ACO genes were also observed in the data from PRJNA217503 (Figures 3E, F). A high relative expression of ZmACCO2, ZmACCO5, ZmACCO7, ZmACCO15, ZmACCO31, and ZmACCO35 was observed in both primary roots (Figure 3C) and seminal roots (Figure 3D) data from PRJNA171684 which were also perceived in the seminal roots data (Figure 3F) from PRJNA217503. Each of the maize ACO gene clusters in the phylogenetic tree has at least one member being relatively highly expressed in roots except for the two clusters to which ZmACCO1 and Zm00001d046848 belong, respectively (Figure 2A). The much higher expression of ZmACCO7 and ZmACCO35 observed in the primary root data from PRJNA171684 was not well confirmed by the data from PRJNA217503 (Figures 3C, E). No expression of Zm00001d005927 was detected in all types of root data from both projects.
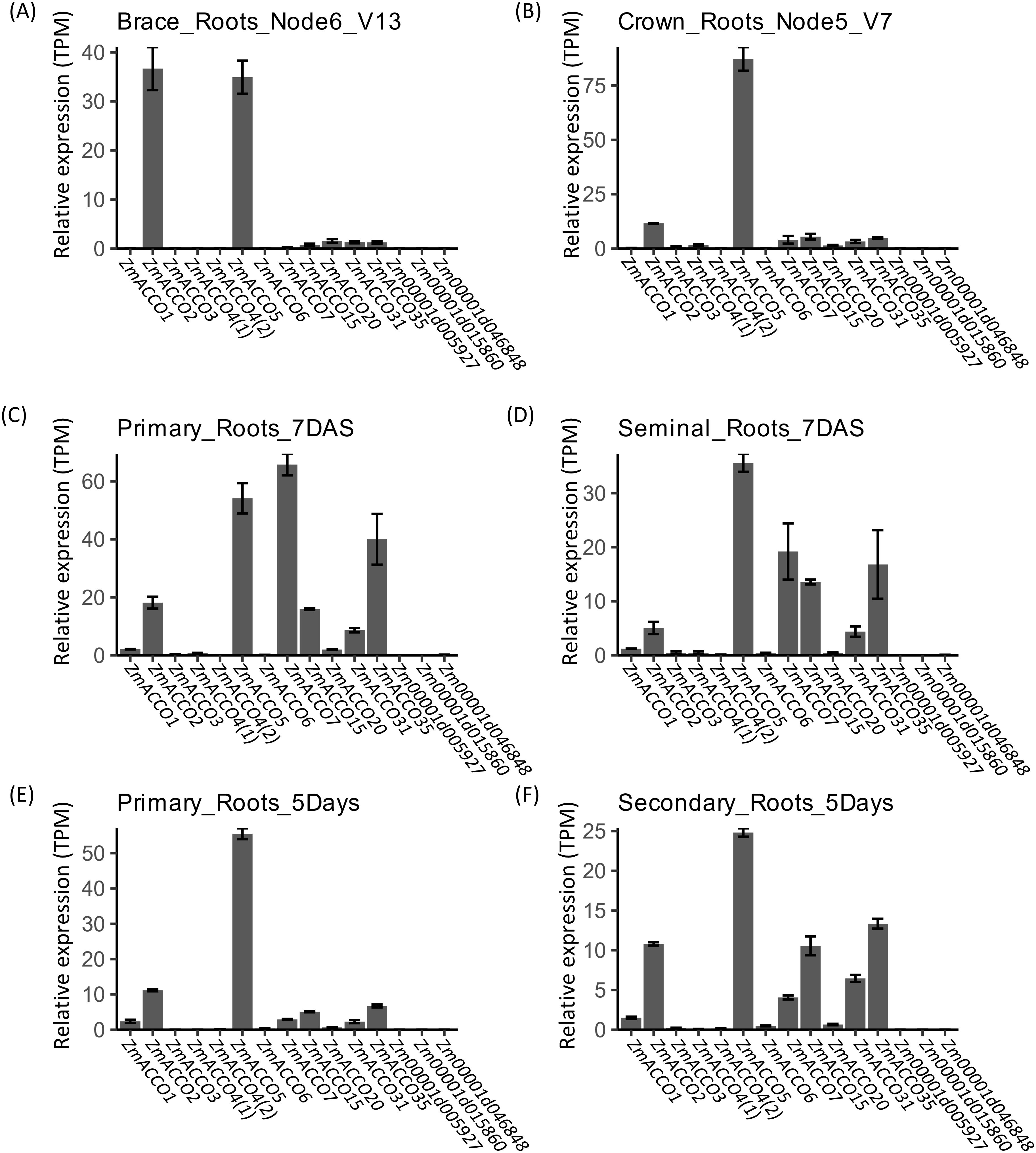
Figure 3. In silico assay of transcription of ACO genes in different type of roots of B73. (A–D) Expression of ACO genes estimated with the data from PRJNA171684. (E, F) Expression of ACO genes estimated with the data from PRJNA217053. Relative gene expression was calculated based on results from Salmon-SMEM quantification of transcripts and indicated with TPM (Transcripts Per Kilobase Million). Data were means ± SE (n = 3). ZmACCO4(1) and ZmACCO4(2) are corresponding to Zm00001d024850 and Zm00001d024851, respectively.
As observed for ACO genes, the expression profile of ACS genes in brace roots exhibited a higher similarity to that in crown roots (Figures 4A, B), while the expression profile of ACS genes in the primary roots showed a higher similarity to that in seminal roots (Figures 4C–F). However, differences were obvious between the expression profiles of ACS genes in primary roots or seminal roots from these two projects (Figures 4C–F). ZmACS1, ZmACS3, and ZmACS7 were the top three highly expressed genes in both primary and seminal roots according to the data from PRJNA171684 (Figures 4C, D), each of which belongs to one of the three clusters of expression mentioned above (Figures 2D, F), while ZmACS1 was the main expressed ACS gene based on the data from PRJNA217503 (Figures 4E, F).
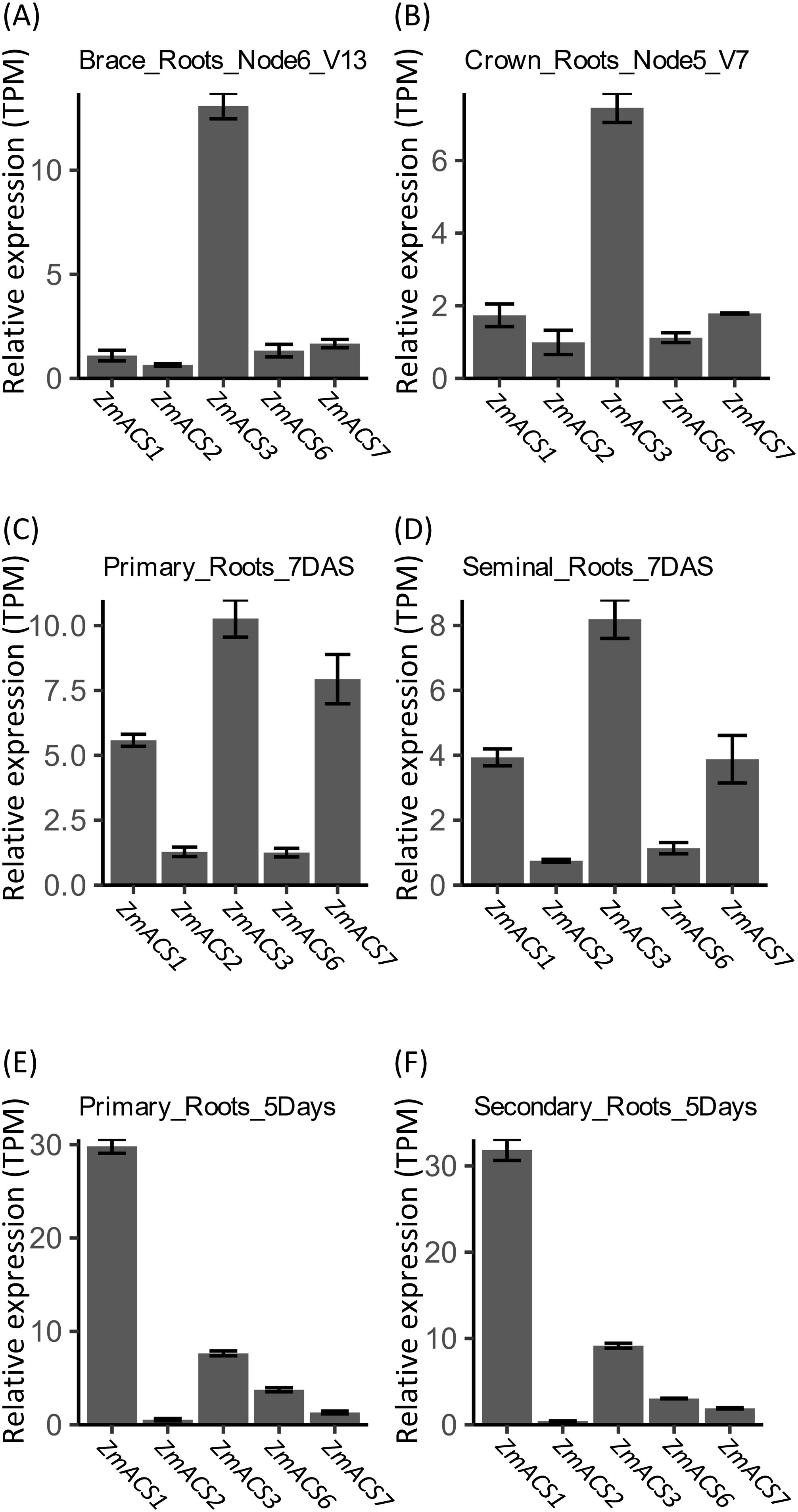
Figure 4. In silico assay of expression of ACS genes in different type of roots of B73. (A–D) Expression of ACS genes estimated with the data from PRJNA171684; (E, F) Expressions of ACS genes estimated with the data from PRJNA217053. Relative gene expression was calculated based on results from Salmon-SMEM quantification of transcripts and indicated with TPM (Transcripts Per Kilobase Million). Data were means ± SE (n = 3).
Taken together, ZmACCO2, ZmACCO5, ZmACCO7, ZmACCO15, ZmACCO31, and ZmACCO35 among ACO genes and ZmACS1, ZmACS3, and ZmACS7 among the ACS genes in the ethylene synthesis pathway might be the master ACO or ACS genes function in maize roots.
3.3 Expression changes of ACO and ACS genes in response to nitrate
We examined the transcription responses of all of these ACO and ACS genes in the roots of B73 seedlings to nitrate provision with qPCR. Since no specific primers for ZmACCO4(2) (Zm00001d024851) were obtained, the total expression of ZmACCO4(1) (Zm00001d024850) and ZmACCO4(2) (Zm00001d024851) was investigated with a common primer pair. Amplification of the ACO homolog Zm00001d005927 is not observed, and ZmACCO5, ZmACCO15, ZmACCO35, ZmACCO31, and ZmACCO7 are highly expressed in the roots of B73 seedlings, which is consistent with the results of in silico analysis (Figures 3, 5A, B). However, our data did not reveal a high expression of ZmACCO2 relative to other ACO genes as in silico analysis demonstrated (Figures 3C–F, 5B). The much high expression of ZmACCO7 relative to other ACO genes observed in the data from PRJNA171684 is in line with our results (Figures 3C, D, 5B). Some different from the results of the GSEA (Table 1), these ACO genes can be categorized into two groups according to their transcriptional responses to nitrate provision, with expression of genes in the first group being up-regulated by nitrate (Figure 5A) and that in the other group being down-regulated (Figure 5B). Expression of five genes was significantly up-regulated (Figure 5A) and expression of two was significantly down-regulated by nitrate provision (Figure 5B). Expression of all 7 the top high expressed ACO genes except ZmACCO31, which have relative expressions > 0.01 before treatment, was significantly regulated by nitrate provision (Figures 5A, B).
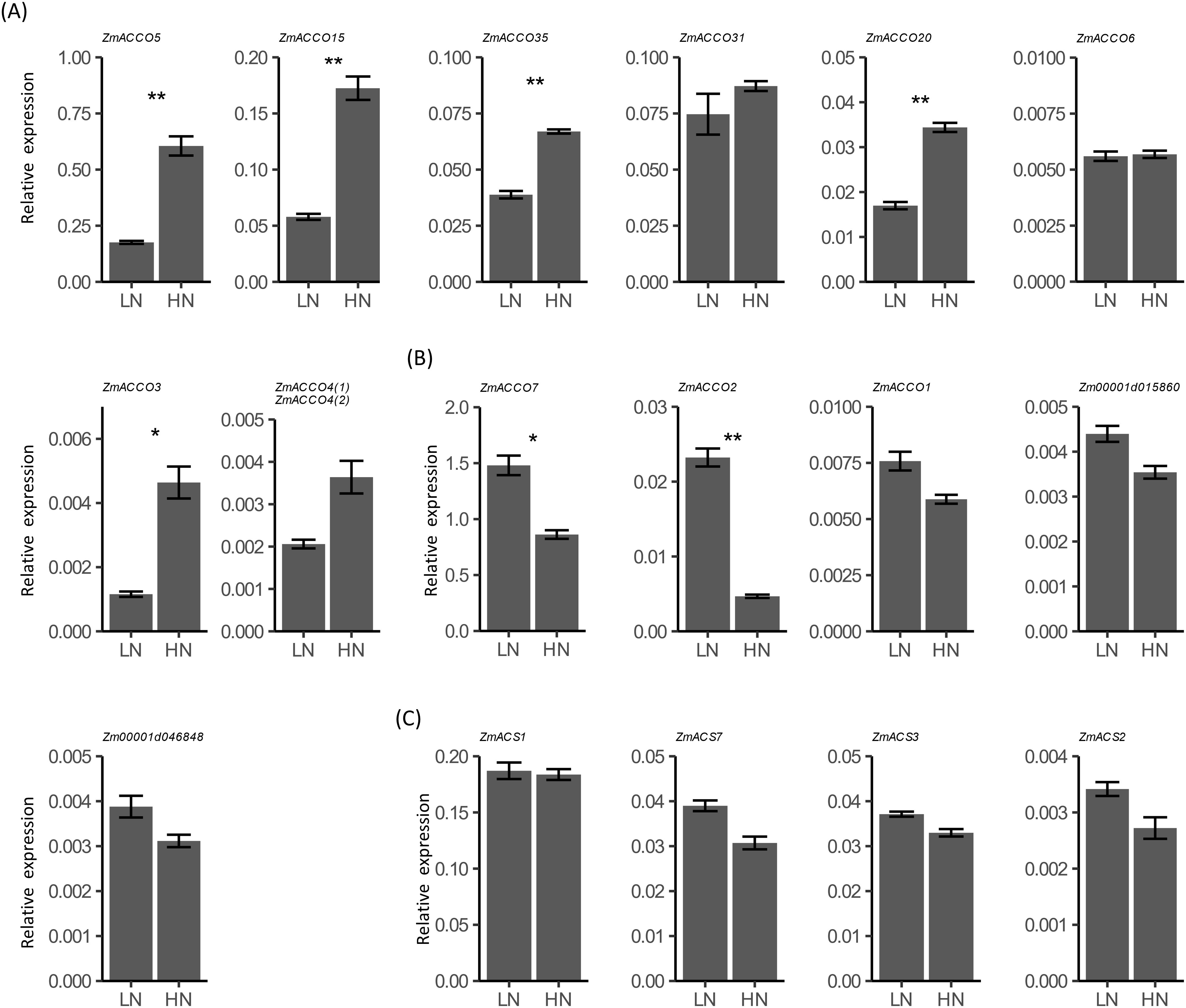
Figure 5. Relative mRNA expression of maize ACO and ACS homologs in response to nitrate provision. (A) Relative expression of ACO homologs of which expression is induced by nitrate in the roots of 6-day-old seedlings of B73 deficient in nitrate treated with 2.0 mmol/L Ca(NO3)2 (HN) or 2.0 mmol/L CaCl2 (as the mock treatment, LN) for 24 hours. (B) Relative e expression of ACO homologs of which expression is repressed by nitrate. (C) Relative expression of ACS homologs. Values are means ± SE (n = 7, 2 technical replicates were performed). ** and * indicate P < 0.01, P < 0.05 with Student’s t-test, respectively. FPGS was used as the reference gene. Expression of ZmACCO4(1) (Zm00001d024850) and ZmACCO4(2) (Zm00001d024851) was investigated with a common primer pair.
For ACS homologs, we failed to detect expression of ZmACS6. Results of qPCR showed that ZmACS1 has much higher transcription relative to other ACS genes in roots of maize seedlings, which is in agreement with the result from the data of PRJNA217053 (Figures 4E, F, 5C). Unlike ACO homologs, expression of all of the four ACS genes was slightly down-regulated by nitrate provision, and no significant regulations of these genes by nitrate were observed, which were also distinct from the results achieved from GSEA (Figure 5C; Table 1).
To determine whether the time of treatments affected the detected regulatory modes of ACO and ACS genes in response to nitrate, we further examined the dynamic expression of the top five highly expressed ACO genes including ZmACCO7, ZmACCO5, ZmACCO15, ZmACCO31, and ZmACCO35 as well as the most highly expressed ACS genes ZmACS1 in response to the nitrate provision. The data of dynamic expression confirmed that four of the five ACO genes are induced by nitrate provision and that ZmACCO7 is repressed (Figure 6). It was observed that regulations of these five genes by nitrate last longer than 2 days. No high extent of expression changes of ZmACS1 in response to the nitrate treatment was observed at most time points (Figure 6), which was also in line with the results mentioned before.
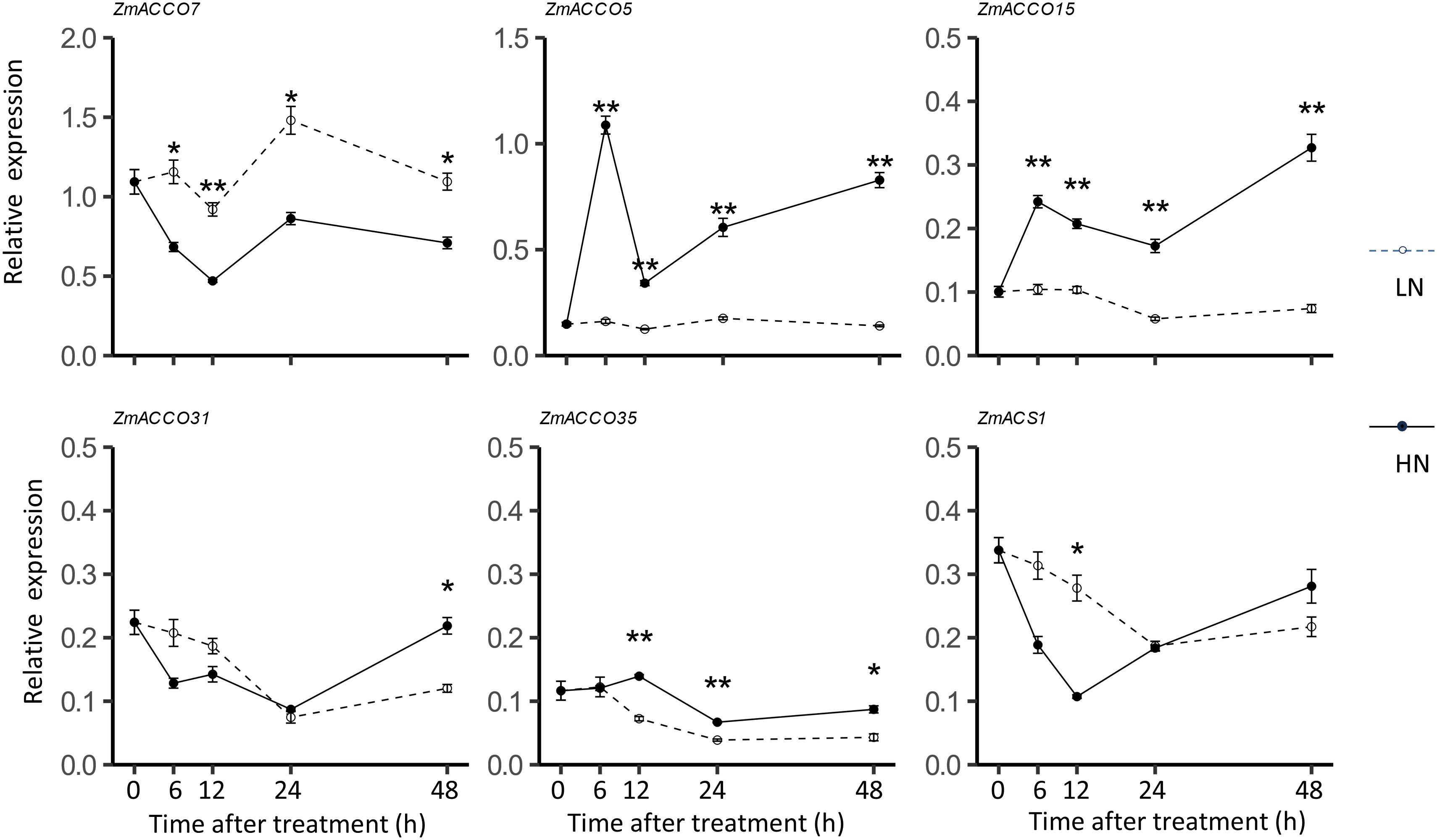
Figure 6. Dynamic mRNA expression of maize ACO and ACS homologs in response to nitrate provision. Relative mRNA levels of genes were detected in roots of 6-day-old B73 seedlings deficient in nitrate treated with 2.0 mmol/L Ca(NO3)2 (HN) or 2.0 mmol/L CaCl2 (as the mock treatment, LN). Values are means ± SE (n =6 or 7, 2 technical replicates were performed). ** and * indicate P < 0.01, P < 0.05 with Student’s t-test, respectively. FPGS was used as the reference gene.
Together, our experiment results and the in silico analyses suggested that ZmACCO7, ZmACCO5, ZmACCO15, ZmACCO31, and ZmACCO35 are the top five highly expressed ACO genes in the roots of maize seedlings, and ZmACS1 is the most highly expressed ACS gene. The expression of ACO and ACS genes is affected by nitrate provision in seedling roots of inbred line B73, but different regulatory modes exist for ACO genes and the expression response of ACS genes to nitrate provision is not obvious, which was not completely consistent with the results from GSEA.
3.4 Tissue-specific expression of ACO and ACS genes determines their particular regulatory modes in response to nitrate
We speculated that the differences between regulatory modes of ACO and ACS genes in response to nitrate might be related to their tissue-specific expression. To test the hypothesis, we inspected the expression of three representative genes, ZmACCO7, ZmACCO5, and ZmACS1, in different segments of both primary roots and seminal roots of B73 in response to nitrate, including the root segments of 0–1 cm (Zone1), 1–2 cm (Zone2), 2–3 cm (Zone3), and >3 cm (Zone4) from root tips (Figures 7A, B). It was intriguing that ZmACCO7 has a low expression in the root tips of primary roots, and its expression increases with distances from the root tip in primary roots while the expression pattern of ZmACCO5 is opposite to that of ZmACCO7 under both nitrogen conditions (Figure 7A). Similar results were obtained in seminal roots, except that the highest expression of ZmACCO5 was detected in Zone2 root segments under high nitrogen condition (Figure 7B). ZmACS1 has more uniform expression across root segments compared with the two ACO genes under both conditions (Figures 7A, B).
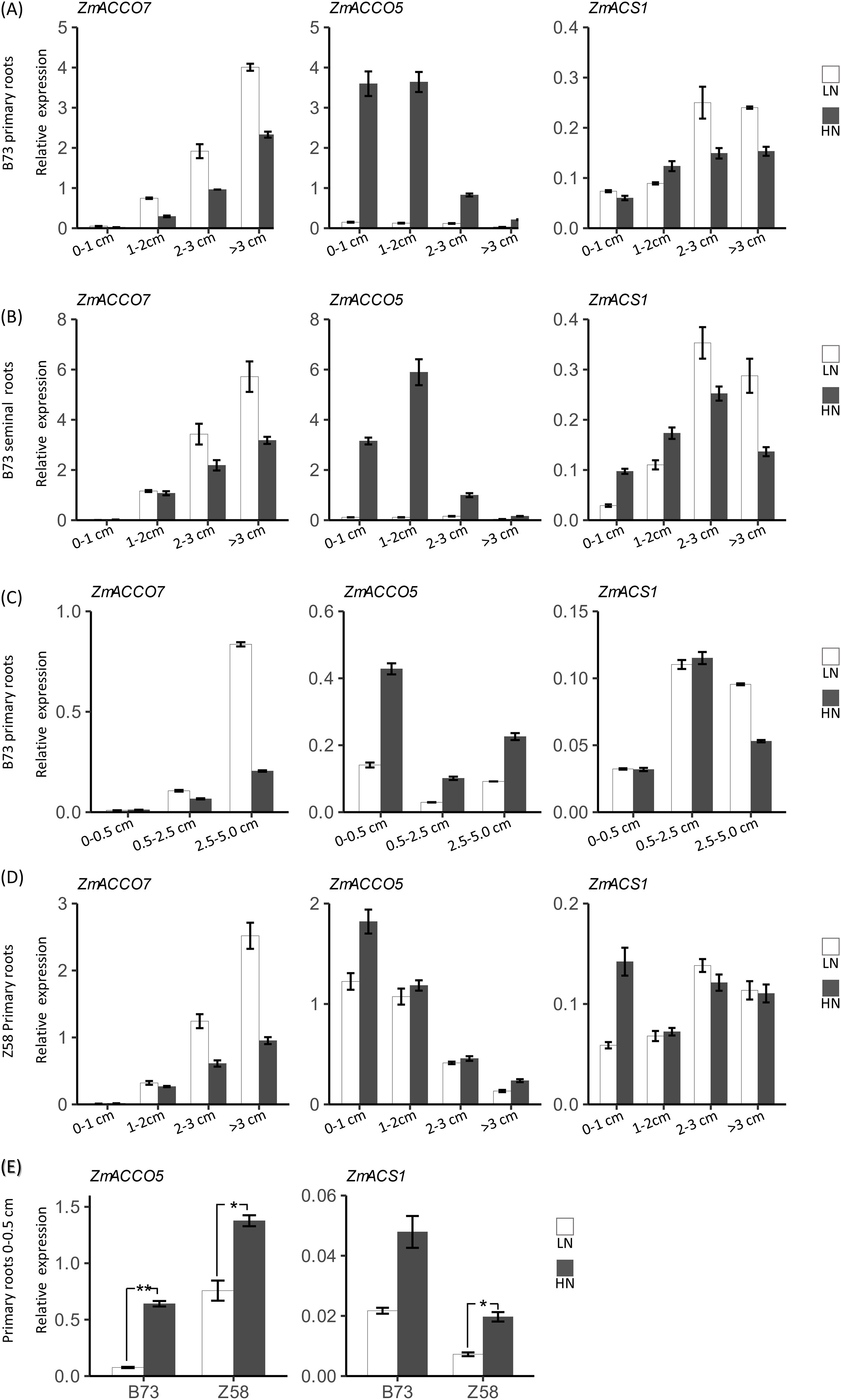
Figure 7. Root region-specific mRNA expression of maize ACO and ACS genes in response to nitrate provision. (A) Relative expression of ZmACCO7, ZmACCO5 and ZmACS1 in primary root segments of B73. (B) Relative expression of ZmACCO7, ZmACCO5 and ZmACS1 in seminal root segments of B73. (C) Relative expression of ZmACCO7, ZmACCO5 and ZmACS1 in primary root segments of B73. (D) Relative expression of ZmACCO7, ZmACCO5 and ZmACS1 in primary root segments of Z58. (E) Relative expression of ZmACCO5 and ZmACS1 in tips of primary roots of B73 and Z58. mRNA levels of genes were detected in roots of 6-day-old seedlings deficient in nitrate treated with 2.0 mmol/L Ca(NO3)2 (HN) or 2.0 mmol/L CaCl2 (as the mock treatment, LN) for 24 hours. For (A–D), values are means ± SE, and 14 plants samples were bulked for RNA extraction and qPCR analysis. 3 technical replicates were performed. For (E), values are means ± SE (n=6, 2 technical replicates were performed), and ** and * indicate P < 0.01, P < 0.05 with Student’s t-test, respectively. FPGS was used as the reference gene. The length in horizontal axes indicated start or end positions of sampled root segments relative to the root tip.
In agreement with the results mentioned before, the expression of ZmACCO7 is repressed by nitrate provision, while the expression of ZmACCO5 is induced (Figures 7A, B). These results indicated that the tissue-specific expression indeed determines their regulatory modes in response to nitrate. It was interesting that the expression of ZmACS1 is induced by nitrate in root segments near the root tip and repressed in root segments far from the root tip, although its expression changes in response to nitrate provision are obviously lower than that of the two ACO genes (Figures 7A, B), which interpreted the result that no obvious expression response to nitrate provision was detected for ZmACS1 in the samples of whole roots. The more uniform expression of ZmACS1 across root segments might determine that its expression is upregulated in root regions near the root tip and downregulated in root regions far from the root tip by nitrate provision. In addition, it was also observed that the expression of ZmACCO5 is extremely highly regulated by nitrate provision compared with ZmACCO7.
To confirm the results and inspect the effects of root regions on the regulations of ACO and ACS genes by nitrate in more detail, we examined the expression of these three genes in primary root segments sampled at positions with some differences from that we used above, i.e., 0–0.5 cm (Zone1), 0.5–2.5 cm (Zone2), and 2.5–5 cm (Zone3) (Figure 7C). As expected, the regulatory modes of ZmACCO7 and ZmACCO5 in response to nitrate in this experiment are similar to the results mentioned above (Figure 7C). ZmACS1 still exhibits a more uniform expression across root segments and lower expression changes in response to nitrate provision in Zone1 and Zone2 segments (Figure 7C).
Taken together, ACO and ACS genes have different tissue-specific expression patterns across maize root regions which determine their particular regulatory modes in response to nitrate. ACO genes like ZmACCO7 which are mainly expressed in root regions far from the root tip and are repressed by nitrate, while those like ZmACCO5 which are mainly expressed in root regions close to the root tip are induced. ACS genes have relatively uniform expression across root regions and are induced in root regions close to the root tip but repressed by nitrate in root regions far from the root tip. As a result, nitrate provision might promote the production of ethylene in regions near the root tip but repress the production of ethylene in regions far from the root tip through the regulation of ACO and ACS genes at mRNA levels.
3.5 Differences in the expression responses of ACO and ACS genes to nitrate exist between genotypes
To know whether different genotypes share similar regulatory modes of expression of ACO and ACS genes in response to nitrate in roots, we next explored the mRNA expression of the three representative genes in response to nitrate provision in the primary roots of another maize inbred line Z58. The relative expression levels of the three genes in Z58 were comparable to that in B73, and their expression patterns across the primary root segments in Z58 were also similar to that in B73 (Figures 7A, D). The regulatory modes of these three genes in response to nitrate in Z58 were likewise similar to that in B73. However, obvious differences in expression change extents for both ZmACCO5 and ZmACS1 existed between the two genotypes (Figures 7A, D). The repression extent of ZmACCO7 by nitrate in B73 was comparable to that in Z58, but the extreme induction of ZmACCO5 by nitrate in B73 was not observed in Z58. The expression of ZmACCO5 under the conditions of nitrate provision was 24.0, 29.0, 7.1, and 7.0 folds of that under the mock treatment in the zone1 to zone4 of B73 seedling primary roots, respectively (Figure 7A), while its expression under the conditions of nitrate provision was 1.5, 1.1, 1.1, and 1.8 folds of that under the mock treatment in the four corresponding root segments of Z58 plants, respectively (Figure 7D). Interestingly, induction of the ZmACS1 by nitrate in Zone1 of Z58 roots (with a HN/LN expression ratio of 2.4) was stronger than that observed in B73 roots (Figures 7A, D). To confirm the differences in regulations of ZmACCO5 and ZmACS1 by nitrate between the two genotypes, we reexamined their expression responses to nitrate provision in the root tips of these two lines with an independent experiment. It was demonstrated that the expression of ZmACCO5 under the nitrate treatment was 8.5 and 1.8 folds of that under the mock treatment in B73 and Z58, respectively (Figure 7E). Although the induction of ZmACS1 by nitrate was more obvious in B73 compared with that observed before, it was still lower than that detected in Z58 (Figure 7E).
These data suggested that differences exist between genotypes in the regulation of ACO and ACS genes by nitrate. We suspect that it might be the defect in the transcriptional regulation of ACS gene(s) by nitrate in the regions near the root tip in B73 that results in the extreme induction of ACO gene(s) by nitrate due to a feedback effect as ACSs function upstream of ACOs.
3.6 Relationship between ethylene synthesis and nitrate-dependent root growth
To reveal the role of ethylene synthesis in nitrogen-regulated plant growth in maize, we investigated B73 seedling growth under hydroponic conditions containing different concentrations of nitrate. As expected, plants with supply of 4 mmol/L nitrate were stronger and had more abundant biomass than that without nitrate supply after 9 days of treatments (Figures 8A, B). The total number of lateral roots and the total length of lateral roots per root of plants with nitrate supply were also significantly higher than that of plants without nitrate supply for both primary and seminal roots (Figure 8B). The observed nitrate-promoted lateral root growth is in line with other reports in maize (Gao et al., 2015) but different from the results from Arabidopsis where a high concentration of nitrate inhibits lateral root growth (Tian et al., 2009). However, the length of primary roots and seminal roots of B73 plants with nitrate supply was shorter than that of plants without nitrate supply (Figures 8A, B), which is in agreement with both the results from maize (Chun et al., 2005; Tian et al., 2008) and Arabidopsis (Linkohr et al., 2002; Ma et al., 2014) although the difference was not significant. These data indicated that maize has a specific regulatory mode of root growth in response to nitrate that high nitrate represses the elongation of primary and seminal roots but stimulates the growth of lateral roots on axial roots. High ethylene generally represses root growth (Khan et al., 2015). In Arabidopsis, high nitrate promotes ethylene synthesis to repress lateral root growth by upregulating the expression of ACO and ACS genes (Tian et al., 2009). The regulatory mode of root growth in response to nitrate in maize is also consistent with the regulations of ACO and ACS genes by nitrate that high nitrate upregulates the mRNA levels of ACO and ACS genes on the whole in root regions near the root tip of axial roots but downregulates their mRNA levels in the root regions far from the root tip.
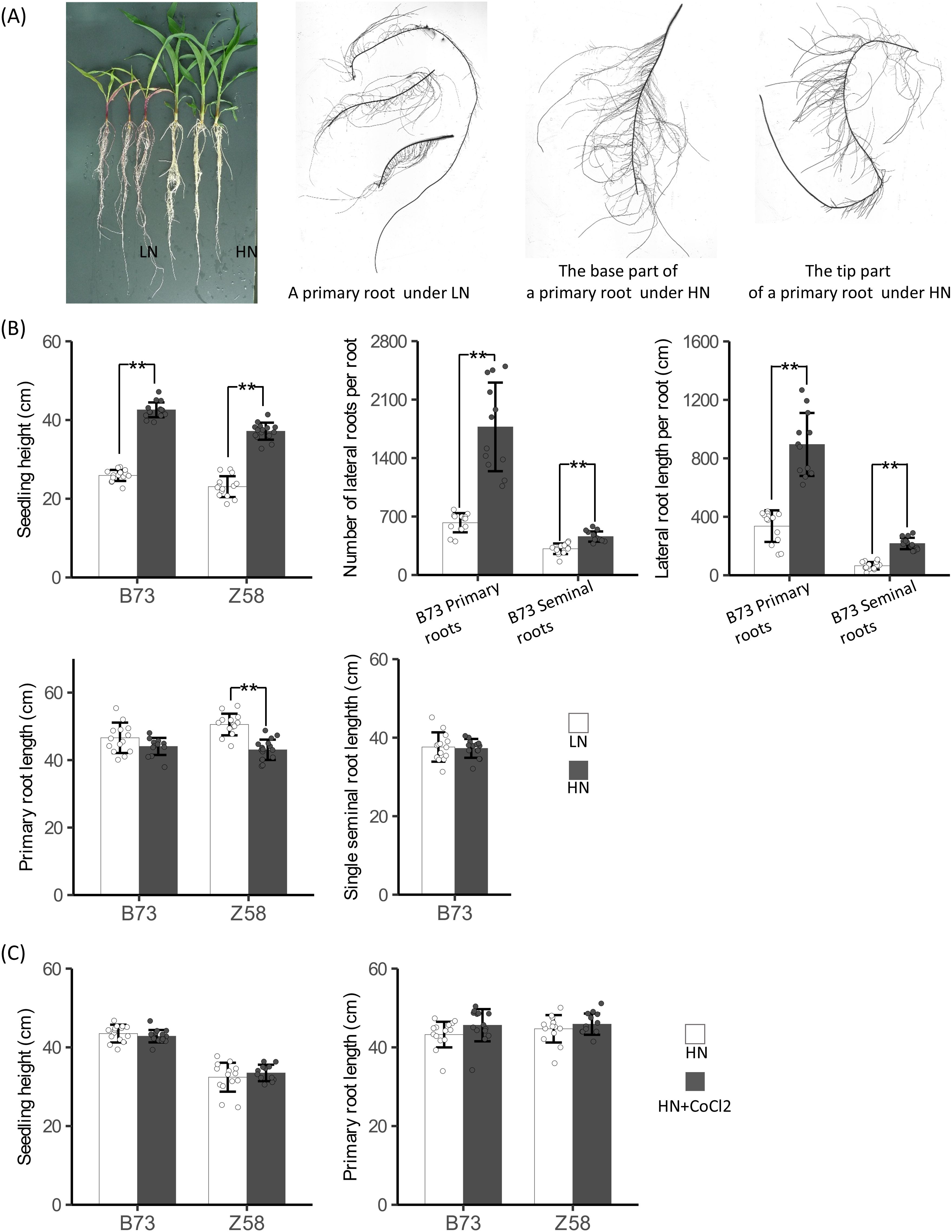
Figure 8. Relationships between ethylene synthesis and root growth. (A) Seedling growth of B73 under low nitrogen condition (LN) and high nitrogen condition (HN). (B) Quantitative analysis of seedling height and root growth of B73 and Z58 plants under LN and HN. (C) Quantitative analysis of seedling height and primary root length of B73 and Z58 plants grown under HN and HN + 3 µmol/L CoCl2. Five-day-old seedlings were subjected to hydroponic culture with 2.0 mmol/L Ca(NO3)2 (HN) or 2.0 mmol/L CaCl2 (LN) or 2.0 mmol/L Ca(NO3)2 plus 3 µmol/L CoCl2 (HN + CoCl2) for 9 days. For (B) and (C), values are means ± SD (n ≥ 14 for seedling height; n ≥ 12 for primary related traits; n ≥ 14 for seminal roots related traits), and ** indicates P < 0.01 with Student’s t-test, respectively.
To know whether the differences in regulations of ACO and ACS genes by nitrate between genotypes are associated with their root growth responses to nitrate provision, we next investigated the growth responses of Z58 seedlings to nitrate under hydroponic conditions. High nitrate stimulated shoot growth and repressed the primary root growth in Z58 as observed in B73 (Figure 8B). However, the primary roots of Z58 exhibited a higher extent of response to nitrate provision compared with that of B73 (Figure 8B). Z58 plants with nitrate supply demonstrated 8.5% reduction of primary root length compared to the control, while B73 plants with nitrate supply exhibited 5.5% reduction relative to the control (Figure 8B). These results might indicate that the defect in regulation of ZmACS1 by nitrate in B73 is linked to its weaker primary root growth responses to nitrate where less effective regulation of ACS genes by nitrate might result in less ethylene production, although ACO genes are extremely induced by nitrate as ACSs act upstream ACOs in the ethylene synthesis pathway. To test the hypothesis that high nitrate inhibits elongation of primary and seminal roots in maize through upregulating ACO and ACS genes to elevate the production of ethylene, we examined root growth under a condition containing both the ethylene synthesis repressor CoCl2, an antagonist for ACO (Lau and Yang, 1976), and nitrate. As expected, 3 µmol/L CoCl2 relieved the inhibition of primary root growth of both B73 and Z58 by nitrate, although the effects were not significant (Figure 8C).
4 Discussion
In this research, we found that the ethylene synthesis pathway is evolved in nitrate signaling in maize seedling roots and participates in nitrate-dependent root growth. Moreover, we observed that there exist differences in the transcriptional regulations of ACO and ACS genes by nitrate between genotypes.
A wealth of information is contained in the size-increasing public next-generation sequencing (NGS) database. GSEA is a robust approach to identify valuable clues linked to a defined gene set in these data (Subramanian et al., 2005). However, in this study, we found enrichment of the ethylene action pathway in only one of the two explored transcriptomic data. Our further experiments showed that members of the same gene family have different regulatory modes in different tissues. These results indicate that it is more possible to identify interesting results using data of a particular type of tissues or cells in NGS data mining with methods like GSEA.
The expression of genes in the ethylene synthesis pathway was upregulated by nitrate (10 mmol/L NO3-) in Arabidopsis (Tian et al., 2009). We found that, in maize seedlings, the expression of ACO and ACS genes on the whole is stimulated in root regions close to the root tip but repressed in root regions far from the root tip by nitrate provision (4 mmol/L NO3-). These results indicated that maize has a nitrate regulatory mode for the ethylene synthesis pathway different from that of Arabidopsis. However, the relationships of ethylene synthesis pathway with root growth might be similar between species, i.e., upregulation of the ethylene synthesis pathway by nitrate is accompanied by inhibited root growth, while its downregulation promotes root growth, which agrees with the general view that ethylene is a stress hormone (Le et al., 2001; Swarup et al., 2007; Wang et al., 2002). Thus, it is possible to remodel the regulatory modes of particular members of gene families in the ethylene synthesis pathway in response to nitrate to obtain an ideotype of root architecture, such as to relieve the induction of ZmACCO5 by nitrate in regions near the root tip to promote axial root growth in order to gain a deep rooting genotype for enhancing NUE. In this research, we only investigated relationships between root growth and transcription regulations of ACO and ACS genes by nitrate. Further works are required to unravel the underlying mechanisms. Additionally, how ethylene affects nitrate transport and metabolism in maize roots remains to be investigated.
We found that ZmACCO7, ZmACCO5, ZmACCO15, ZmACCO31, and ZmACCO35 are the top five highly expressed ACO genes, and ZmACS1 is the most highly expressed ACS gene in the roots of maize seedlings, which is consistent with some results of in silico analysis (Figures 3-5). However, an in silico expression analysis demonstrated that the master ACO and ACS genes in brace roots and crown roots might not be master genes in primary and seminal roots. Our analysis also exhibited tissue-specific expression of ACO and ACS genes in same roots. Crown roots and brace roots are most important root systems for maize during vegetative growth and reproductive development (Lynch, 2013). Therefore, it is necessary to confirm which members of this gene family are master ACO and ACS genes and investigate their regulatory modes in response to nitrate in crown roots and brace roots. In fact, data from the project of PRJNA283053 showed that ZmACCO7 was upregulated by nitrate in brace roots, which is different from the result observed in this research.
Our analyses showed differences in the regulations of ACO and ACS genes existing between B73 and Z58 which might be associated with the differences in remodeling of root architectures by the nitrate between the two genotypes, i.e., weaker induction of ZmACS1 in root regions near the root tips by nitrate might result in less reduction of primary root elongation by nitrate in B73 compared with Z58. Further experiments are needed to find the factors determining the regulatory differences in response to nitrate between genotypes and, moreover, to identify more diversity of regulation of ACO and ACS genes by nitrate in maize germplasm and to explore their application in improving NUE in maize.
In conclusion, we found that the ethylene synthesis pathway is involved in the responses to nitrate of maize seedling roots in a more complex mode relative to that in Arabidopsis, which is associated with remodeling of the root architecture by nitrate, and there exist differences in regulations of ACO and ACS genes between genotypes, which might be valuable in improving the NUE of maize varieties.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
GY: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. ZY: Methodology, Writing – original draft, Writing – review & editing. SM: Visualization, Writing – original draft, Writing – review & editing. XL: Funding acquisition, Resources, Writing – original draft, Writing – review & editing. JS: Project administration, Writing – original draft, Writing – review & editing. BC: Funding acquisition, Writing – original draft, Writing – review & editing. XC: Investigation, Writing – original draft, Writing – review & editing. BL: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. CM: Conceptualization, Funding acquisition, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Salt-alkali Tolerant Crop Cultivation Positions of Shandong Province Agricultural Industry Technology System for Saline-Alkali Land (grant number SDAIT-29-04) and the Agricultural Science and Technology Innovation Projects of Shandong Academy of Agricultural Sciences, China (grant number CXGC2025C02 and CXGC2025F02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1566213/full#supplementary-material
Abbreviations
ACC, aminocyclopropane-1-carboxylic acid; ACS, ACC synthase; ACO, ACC oxidase; ETR1, ethylene receptor1; EIN3, ethylene insensitive3; FPGS, folypolyglutamate synthase; GSEA, Gene Set Enrichment Analysis; NGS, next-generation sequencing; NPF, nitrate transporter1/peptide transporter family; NRT, nitrate transporters; NUE, nitrogen use efficiency; RBH, reciprocal best hit; qPCR, quantitative PCR; SAM, S-adenosyl-l-methionine; TPM, transcripts per kilobase million.
References
Aluko, O. O., Kant, S., Adedire, O. M., Li, C., Yuan, G., Liu, H., et al. (2023). Unlocking the potentials of nitrate transporters at improving plant nitrogen use efficiency. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1074839
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bork, P., Dandekar, T., Diaz-Lazcoz, Y., Eisenhaber, F., Huynen, M., Yuan, Y. (1998). Predicting function: from genes to genomes and back. J. Mol. Biol. 283, 707–725. doi: 10.1006/jmbi.1998.2144
Chun, L., Mi, G., Li, J., Chen, F., Zhang, F. (2005). Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant Soil 276, 369–382. doi: 10.1007/s11104-005-5876-2
Dechorgnat, J., Nguyen, C. T., Armengaud, P., Jossier, M., Diatloff, E., Filleur, S., et al. (2011). From the soil to the seeds: the long journey of nitrate in plants. J. Exp. Bot. 62, 1349–1359. doi: 10.1093/jxb/erq409
Gao, K., Chen, F., Yuan, L., Zhang, F., Mi, G. (2015). A comprehensive analysis of root morphological changes and nitrogen allocation in maize in response to low nitrogen stress. Plant Cell Environ. 38, 740–750. doi: 10.1111/pce.12439
Hawkesford, M. J., Griffiths, S. (2019). Exploiting genetic variation in nitrogen use efficiency for cereal crop improvement. Curr. Opin. Plant Biol. 49, 35–42. doi: 10.1016/j.pbi.2019.05.003
He, X., Ma, H., Zhao, X., Nie, S., Li, Y., Zhang, Z., et al. (2016). Comparative RNA-seq analysis reveals that regulatory network of maize root development controls the expression of genes in response to N stress. PloS One 11, e0151697. doi: 10.1371/journal.pone.0151697
Hochholdinger, F., Yu, P., Marcon, C. (2018). Genetic control of root system development in maize. Trends Plant Sci. 23, 79–88. doi: 10.1016/j.tplants.2017.10.004
Houben, M., Van de Poel, B. (2019). 1-aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00695
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Jia, L., Hu, D., Wang, J., Liang, Y., Li, F., Wang, Y., et al. (2023). Genome-wide identification and functional analysis of nitrate transporter genes (NPF, NRT2 and NRT3) in maize. Int. J. Mol. Sci. 24. doi: 10.3390/ijms241612941
Khan, M. I. R., Trivellini, A., Fatma, M., Masood, A., Francini, A., Iqbal, N., et al. (2015). Role of ethylene in responses of plants to nitrogen availability. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00927
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lau, O. L., Yang, S. F. (1976). Inhibition of Ethylene Production by Cobaltous Ion. Plant Physiol. 58, 114–117. doi: 10.1104/pp.58.1.114
Le, J., Vandenbussche, F., van der Straeten, D., Verbelen, J. P. (2001). In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiol. 125, 519–522. doi: 10.1104/pp.125.2.519
Lewis, D. R., Negi, S., Sukumar, P., Muday, G. K. (2011). Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Dev. Camb. Engl. 138, 3485–3495. doi: 10.1242/dev.065102
Li, Y., Bai, L., Wei, S., Wu, H., Li, R., Wang, Y., et al. (2024). Integrating heterosis for root architecture and nitrogen use efficiency of maize: A comparison between hybrids from different decades. Agronomy 14. doi: 10.3390/agronomy14092018
Linkohr, B. I., Williamson, L. C., Fitter, A. H., Leyser, H. M. O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. Cell Mol. Biol. 29, 751–760. doi: 10.1046/j.1365-313x.2002.01251.x
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lynch, J. P. (2013). Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112, 347–357. doi: 10.1093/aob/mcs293
Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. Cell Mol. Biol. 78, 70–79. doi: 10.1111/tpj.12448
Ma, B., Ma, T., Xian, W., Hu, B., Chu, C. (2023). Interplay between ethylene and nitrogen nutrition: How ethylene orchestrates nitrogen responses in plants. J. Integr. Plant Biol. 65, 399–407. doi: 10.1111/jipb.13355
Manoli, A., Sturaro, A., Trevisan, S., Quaggiotti, S., Nonis, A. (2012). Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 169, 807–815. doi: 10.1016/j.jplph.2012.01.019
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pattyn, J., Vaughan-Hirsch, J., Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229, 770–782. doi: 10.1111/nph.16873
Raun, W. R., Johnson, G. V. (1999). Improving nitrogen use efficiency for cereal production. Agron. J. 91, 357–363. doi: 10.2134/agronj1999.00021962009100030001x
Ren, W., Zhao, L., Liang, J., Wang, L., Chen, L., Li, P., et al. (2022). Genome-wide dissection of changes in maize root system architecture during modern breeding. Nat. Plants 8, 1408–1422. doi: 10.1038/s41477-022-01274-z
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 26, 139–140. doi: 10.1093/bioinformatics/btp616
Stelpflug, S. C., Sekhon, R. S., Vaillancourt, B., Hirsch, C. N., Buell, C. R., de Leon, N., et al. (2016). An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome. doi: 10.3835/plantgenome2015.04.0025
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Swarup, R., Perry, P., Hagenbeek, D., van der Straeten, D., Beemster, G. T. S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Tatusov, R. L., Koonin, E. V., Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. Cell Mol. Biol. 37, 914–939. doi: 10.1111/j.1365-313x.2004.02016.x
Tian, Q., Chen, F., Liu, J., Zhang, F., Mi, G. (2008). Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 165, 942–951. doi: 10.1016/j.jplph.2007.02.011
Tian, Q.-Y., Sun, P., Zhang, W.-H. (2009). Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol. 184, 918–931. doi: 10.1111/j.1469-8137.2009.03004.x
Tsuchisaka, A., Yu, G., Jin, H., Alonso, J. M., Ecker, J. R., Zhang, X., et al. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003. doi: 10.1534/genetics.109.107102
Walley, J. W., Sartor, R. C., Shen, Z., Schmitz, R. J., Wu, K. J., Urich, M. A., et al. (2016). Integration of omic networks in a developmental atlas of maize. Science 353, 814–818. doi: 10.1126/science.aag1125
Wang, K. L.-C., Li, H., Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl, S131–S151. doi: 10.1105/tpc.001768
Xiao, F., Gong, Q., Zhao, S., Lin, H., Zhou, H. (2021). MYB30 and ETHYLENE INSENSITIVE3 antagonistically modulate root hair growth in Arabidopsis. Plant J. Cell Mol. Biol. 106, 480–492. doi: 10.1111/tpj.15180
Xing, J., Feng, Y., Zhang, Y., Wang, Y., Li, Z., Zhang, M. (2024). Ethylene accelerates maize leaf senescence in response to nitrogen deficiency by regulating chlorophyll metabolism and autophagy. Crop J. 12, 1391–1403. doi: 10.1016/j.cj.2024.05.006
Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W. F., Harden, L. A., Theologis, A. (2003). Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. doi: 10.1074/jbc.M308297200
Yu, P., Eggert, K., von Wirén, N., Li, C., Hochholdinger, F. (2015). Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol. 169, 690–704. doi: 10.1104/pp.15.00888
Zdarska, M., Cuyacot, A. R., Tarr, P. T., Yamoune, A., Szmitkowska, A., Hrdinová, V., et al. (2019). ETR1 integrates response to ethylene and cytokinins into a single multistep phosphorelay pathway to control root gxrowth. Mol. Plant 12, 1338–1352. doi: 10.1016/j.molp.2019.05.012
Keywords: maize, nitrate, ethylene synthesis pathway, ACO, ACS, root growth
Citation: Yao G, Yan Z, Ma S, Liu X, Shan J, Cao B, Chen X, Leng B and Mu C (2025) Transcriptions of ACO and ACS genes are involved in nitrate-dependent root growth of maize seedlings. Front. Plant Sci. 16:1566213. doi: 10.3389/fpls.2025.1566213
Received: 31 January 2025; Accepted: 07 April 2025;
Published: 02 May 2025.
Edited by:
Gwendolyn Kristin Kirschner, The James Hutton Institute, United KingdomReviewed by:
Kangyu Wang, Jilin Agriculture University, ChinaBilal Beig, National University of Sciences and Technology (NUST), Pakistan
Copyright © 2025 Yao, Yan, Ma, Liu, Shan, Cao, Chen, Leng and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingying Leng, bGVuZ2Jpbmd5aW5nQHNhYXMuYWMuY24=; Chunhua Mu, bWFpemVzZEAxNjMuY29t
 Guoqi Yao
Guoqi Yao Zhenwei Yan1
Zhenwei Yan1