Abstract
Introduction:
Finger millet (Eleusine coracana) is gaining increasing recognition as a functional food and a promising source of nutraceuticals for mitigating metabolic disorders, owing to its abundance of bioactive compounds. Despite its nutritional and therapeutic potential, comprehensive metabolomic profiling of its primary and secondary metabolites remains limited. This study aimed to perform an in-depth metabolomic analysis of finger millet landraces cultivated in the temperate region of Padder Valley, District Kishtwar, Union Territory of Jammu and Kashmir, and to assess the therapeutic relevance of these metabolites in preventing metabolic diseases.
Methods:
Comprehensive phytochemical profiling was conducted using liquid chromatography coupled with high-resolution tandem mass spectrometry (LC-HRMS/MS) to identify and characterize primary and secondary metabolites in finger millet grains. Inductively coupled plasma mass spectrometry (ICP-MS) was employed to quantify macro- and microelemental contents.
Results:
Metabolomic analysis identified a total of 50 primary metabolites, including derivatives of amino acids, fatty acids, and carbohydrates such as dehydroascorbic acid, niacin, xanthine, orotic acid, nicotinuric acid, gluconic acid, propionic acid, decanoic acid, and palmitic acid. Additionally, 135 secondary metabolites were characterized, encompassing heterocyclic compounds, phenolics, flavonoids, alkaloids, and terpenes such as 4-hydroxycyclohexylcarboxylic acid, 2-furoic acid, methyl cinnamate, mesitol, 4-hydroxybenzoic acid, heptalactone, viburtinal, and geranic acid. Elemental analysis revealed the presence of 10 macro- and microelements, with magnesium (Mg), calcium (Ca), potassium (K), and phosphorus (P) being the most abundant.
Discussion:
The comprehensive metabolite profiling demonstrates that finger millet is a rich source of bioactive primary and secondary compounds with potential therapeutic benefits. The diversity of metabolites and essential minerals highlights its value as a functional food ingredient for the prevention and management of metabolic disorders. These findings provide a biochemical basis for the development of value-added nutraceutical products derived from finger millet landraces.
Highlights
-
53 phenolic compounds have been identified for the first time in finger millet in Jammu and Kashmir.
-
Coumarin, and eugenol is the major phenolic compound in finger millet.
-
Phenolic compounds significantly contribute to the antioxidant capacity.
Introduction
Food insecurity and malnutrition are serious health problems in developing countries especially India and underdeveloped countries that impact millions of people. Currently, over 1.4 billion people are living in the country which has been increasing progressively, it is critical to address these issues to ensure nutritional equity and self-sufficiency (Baduni et al., 2024). Being the world’s primary producer of millet, India has a lot of potential to combat malnutrition and food insecurity. Millets, belong to the Poaceae family of small-seeded edible grasses and are found in arid and marginal areas of both tropical and temperate climates (Yadav et al., 2024). India accounts for 41% of the world’s production of millet, according to global data. (Vishal et al., 2024). Millets significantly improve nutritional and food security and increase genetic diversity of the global food basket (Mallick et al., 2024). Millets such as pearl millet (Pennisetum glaucum), foxtail millet (Setaria italica), porso millet (Panicum miliaceum), finger millet (Eleusine coracana), barnyard millet (Echinochlo aesculenta), kodo millet (Paspalum scrobiculatum), sorghum (Sorghum bicolor), and little millet (Panicum sumatrense) are rich in vital nutrients, gluten-free and low glycaemic index, to make them ideal for people with diabetes, degenerative, and coeliac diseases (Singh et al., 2024). As staple grains for centuries, millets played a significant role in Indian diets, especially in hilly and rural areas due to their low water requirement, drought tolerance, resistance to harsh weather conditions, climate resilience and also reduced input requirement (Vikash et al., 2024). However, with the advent of modern agricultural practices and the widespread cultivation of rice and wheat millet usage has declined sharply, contributing less than 10 per cent to the human diet (Ghosh et al., 2024).
Eleusine coracana, sometimes referred to as ragi, nachni, kodra or finger millet, is widely grown in China, India, and other parts of Africa. The highlands of Eastern Africa, especially Ethiopia and Uganda, are believed to be its birthplace (Tamilselvan et al., 2023). About 5,000 years ago, E. coracana was domesticated and then spread to Asia and other parts of Africa. It has the efficacy for addressing food security and malnutrition because of its high content of nutrition. Due to significant amounts of minerals, protein, carbohydrates, and dietary fiber, finger millet has a well-established nutritional value that draws a lot of interest in the present era (Deme et al., 2019). Additionally, micro and macronutrients, is a virtuous source of phytoconstituents, especially phenolic compounds, which help in lowering chronic diseases including cardiovascular diseases, diabetes, and cancer. Proanthocyanidins, hydroxybenzoic (p-hydroxybenzoic and, protocatechuic) acids, flavonoids (apigenin, quercetin, epicatechin, and catechin), and hydroxycinnamic (p-coumaric acid, ferulic acid) are predominant polyphenols present in finger millet (Kalsi et al., 2023). Finger millet has been identified as a prominent source of minerals and protein, although little is known about its phytochemical makeup. Understanding finger millet’s phytochemicals and nutritional value may enhance its application as a source of functional food material and help comprehend the associated health benefits (Abioye et al., 2022). In humans, severe chronic diseases include heart disease, cancer, diabetes, and cognitive dysfunction have been linked to the oxidation of cellular molecules by reactive species (Sharma et al., 2023). Dietary antioxidants play a crucial role in protection against oxidative damage and maintaining a healthy metabolic balance. Recent research has increasingly concentrated on plant bioactive compounds due to their several health benefits (Pandey et al., 2023).
Metabolomics as a relatively new field, is capable of analyzing every low molecular weight molecule present in particular organism or tissue. Liquid Chromatography Mass Spectrometer (LC-MS) technology has been employed in several research to detect and assess expression levels of various metabolites. These studies span multiple dimensions, including plant development (Theodoridis et al., 2023), plant-microbe interactions (Spina et al., 2021), human diseases (Zhou and Zhong, 2022), and plant nutrition (Zhong et al., 2022).
The phytochemical composition of Eleusine coracana remains inadequately explored in existing literature, highlighting the need for a comprehensive analysis of its beneficial compounds. In the current investigation, phytochemical analysis was conducting utilizing LC-MS/MS, while elemental analysis was performed through ICP-MS. The goal of this study was to examine the profiling of phytochemical compounds present in the local landraces of finger millet cultivated in the temperate region of District Kishtwar (Padder Valley) within the Union Territory of Jammu and Kashmir. The development of innovative food products and nutraceutical applications could benefit from an enhanced comprehensive of phytochemical components. This strategy may also contribute to the sustainable advancement of the Indian biome by harnessing the untapped potential of this underutilized crop and promoting biodiversity conservation. These findings of the present study offer a thorough and understandable explanation of the phytochemical and nutritional components of finger millet as well as the metabolic pathways connected to the metabolic compounds that were identified through differential analysis.
Materials and methods
Plant material
Seeds of finger millet were collected from local habitat found in Chasoti village (33°34′ N latitude, 75°53′58″ E longitude, at an altitude of 1638 m), of Padder Valley (District Kishtwar), Jammu and Kashmir, India (Figure 1). The collected specimens were transported to Division of Plant Pathology, Faculty of Agriculture, SKUAST Jammu, within 48 hours of collection. Further, the samples were thoroughly rinsed with distilled water to eliminate soil particles and extraneous materials. The seeds were subsequently separated manually for further processing and analysis.
Figure 1

(A) Global distribution of finger millet (B) Local habitat for collection of finger millet in the UT of Jammu and Kashmir used in current study, (C) samples of finger millet seeds from Chasoti village of Padder Valley (District Kishtwar), Jammu and Kashmir, India.
Characterization of finger millet
Molecular characterization of the collected finger millet samples was performed to ensure precise validation. The seeds were germinated under controlled conditions in plant growth chamber on 30°C with 55% humidity and 12 h light, and the resulting seedling tissues were utilized for genomic DNA extraction to facilitate further molecular analyses (Ibrahim et al., 2023). Purified DNA is used as the template for PCR to amplify segments of matK and rbcL gene sequences. The primers matK-F (5′-CTTTGCATTTATTACGGCTC-3′), matK-R (5′-GATTGGTTACGGGAGAAAAAG-3′) and rbcL-F (5′-GAAGTAGTAGGATTGATTCT-3′), rbcL-R (5′-CATCATTATTGTATACTCTTTC-3′) were used to amplification (Molla et al., 2023). The PCR mix (20 µl), which included 10 µl of master mixture, 1 µl of 10 nm each primer, 1 µl of DNA, and 7 µl of milli Q, was supplemented with total DNA (50–500 ng). The following circumstances was performed when the reaction was carried out: 1 min of 94 °C, followed by 32 cycles for 10 sec at 98°C, 56°C and 55°Cfor 15 sec, 68°C for 30 sec, and final elongation of 3 min at 68°C. 1.5% agarose gel electrophoresis was used to isolate and visualise the amplified DNA products under UV light (Supplementary Figure S1). Biologia Research India Pvt Ltd., Karnal, Haryana sequenced the purified partial amplicons. Before being compared to those in the GenBank database using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi), the sequences were constructed, altered, and aligned in MEGA11 to determine the sequence homology with closely related taxa. In this research, the plant species exhibiting the highest sequence identity (100%) were selected as closest taxonomic match for molecular identification (Almutairi, 2021).
Preparation of extract
Finger millet dried seeds were coarsely grounded for extraction. 50 g of the powered seed sample from three independent seed batches (n = 3) was placed in a cellulose thimble and extracted using 400 mL of 95% methanol from Merck (Darmstadt, Germany) (65°C) in a Soxhlet apparatus. Exhaustive extraction was applied with solvent for 10–12 hours to ensure a complete extraction procedure (Figure 2). The extract was concentrated at 40°C under reduced pressure by using a rotary evaporator and was stored at 4°C until further use (Malathi et al., 2024).
Figure 2
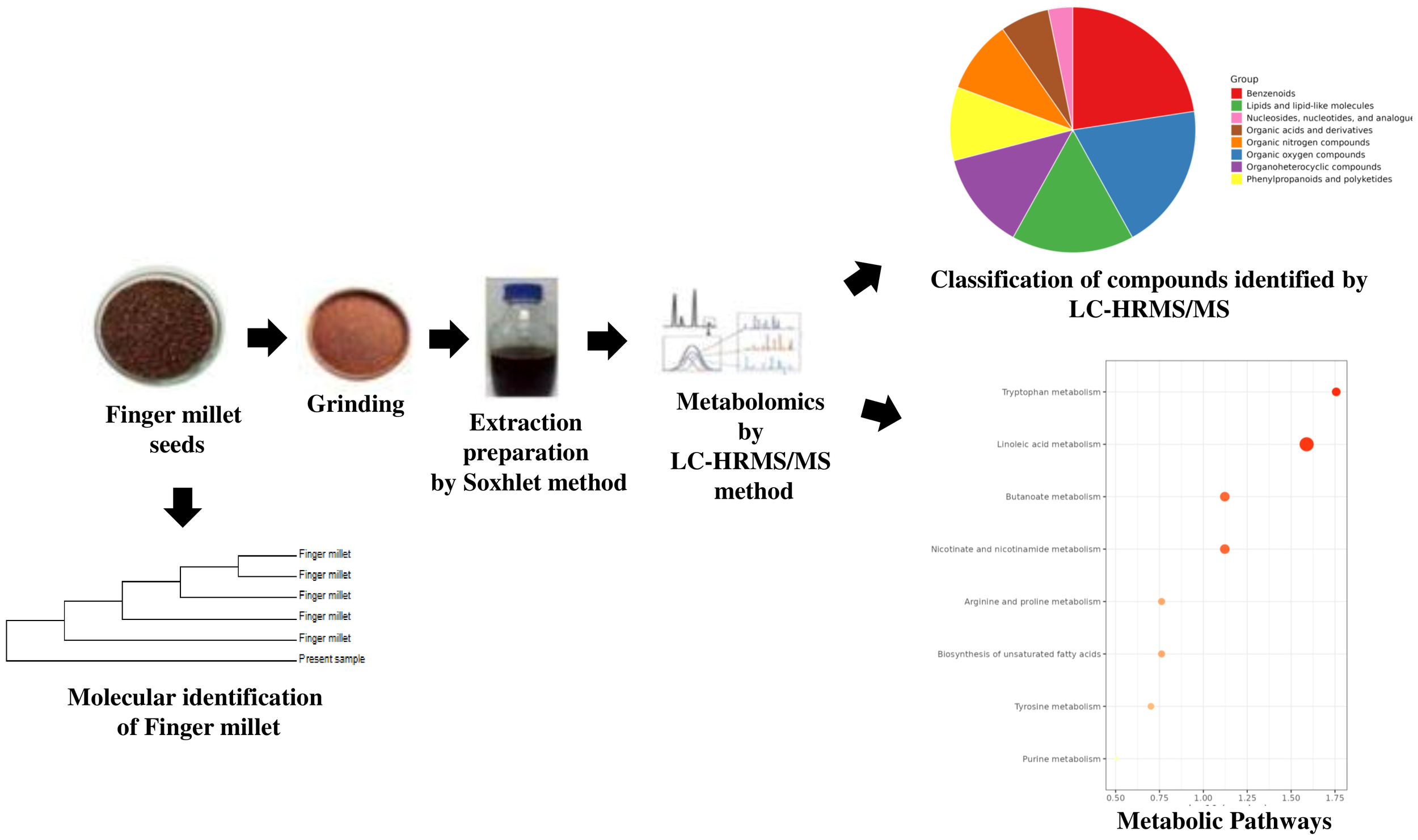
Workflow of LC–HRMS based untargeted metabolomic analysis of finger millet.
Elemental composition by ICP-MS
0.5-1.0 gm of homogenized powder of sample was weighed using weighing paper and transferred into polytetrafluoroethylene-Teflon (PTFE-TFM) vessels, performed in triplicate. Add 15 mL dilute nitric acid 70% (v/v), was purchased from Merck (Darmstadt, Germany) into the same vessel. The pre-digestion reactions were allowed to proceed for 20 mins. Then, the vessels were tightly closed and inserted into the 20SVT50 rotor. The rotor was subjected to Multiwave 5000 radiation system at 140°C for 30 mins. After completion of the digestion, the vessels were taken out of the microwave and were allowed to cool down to room temperature (25 °C) for 20 mins. The digests produced were quantitatively transferred into 25 mL volumetric flasks and the flasks were then filled up to mark with ultrapure water. The aqueous solutions were then passed through polyvinylidene difluoride (PVDF) microfilters, prior to metal analysis by ICP-OES. Each sample was digested in triplicates with a blank as the fourth sample (Mafukata et al., 2024).
Liquid chromatography-high resolution mass spectrometry identification
LC-HRMS was used to identify secondary metabolites in the methanol extract of finger millet, using a previously described method with some modifications (Anggraeni et al., 2025). This study represents a preliminary profiling of finger millet. The analysis was performed in three independent replicates to ensure consistency. Biologia Research India Pvt Ltd., Karnal, Haryana performed the LC-HRMS/MS analysis. Extracted metabolite samples were analyzed for identifying and relatively quantifying using LC-HESI-MS/MS method on Vanquish Flex UHPLC coupled with Orbitrap Fusion™ Lumos™ Tribrid™ MS from Thermo Scientific. The polar metabolites were eluted by running the samples on Hypersil GOLD VANQUISH column (150mm x 2.1mm; 1.9 µ) column at column temperature of 40 °C. For UHPLC run, 0.1% formic acid in water and 0.1% formic acid in acetonitrile were used as mobile phase A and mobile phase B, respectively. The gradient run includes %B from 1 to 25 in 3.5 minutes, 25 to 35 in 4 minutes, 35 to 95 in 2.8 minute, a stable 95% for 3.7 minutes and re-equilibration at 1% for 6 minutes. The H-ESI was used for ionization at static spray voltage of 3400 (v) for positive ions. The full scans and MS2 scans were acquired at 120000 at scan range 50–1500 m/z. The ddMS2 spectra were acquired using Orbitrap detector at 60000 resolutions with HCD fragmentation using stepped collision energy at 20, 40, 75%. The raw spectral data were processed using Compound discoverer 3.3.2.3.1 Software and mzCloud server and ChemSpider database search.
Kyoto encyclopedia of genes and genomes annotation and metabolic pathway analyses of differential identified compounds
Identified metabolites were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) compound database (http://www.kegg.jp/kegg/compound/) and annotated metabolites were mapped to the KEGG Pathway database (http://www.kegg.jp/kegg/pathway.html) using software MetaboAnalyst. Pathways with significantly regulated metabolites were then fed into metabolite set enrichment analysis, and their significance was determined by hypergeometric test p-values (Supplementary Tables S1, S2). The KEGG pathways with corrected p-values of less than 0.05 were considered significantly enriched by differentially expressed genes (Cao et al., 2022; Han et al., 2024).
Statistical analysis
Every experiment was conducted three times, and the means of the results are provided. To compute the mean and standard deviation, SPSS-22 statistical software (SPSS, Inc., Chicago, IL, USA) was used.
Results
Molecular identification
The matK and rbcL gene sequencing was utilized to identify finger millet sample, and a phylogenetic tree was made using MEGA 11 software. The Tamura 3-parameter model with lowest BIC and highest AIC values served as the basis for the maximum likelihood tree of the present millet sample, which was built using MEGA 11 and based on the study of matK and rbcL gene sequences. Every area with lacking information and gaps was eliminated. With gaps filled by pairwise deletion, the estimated transition/transversion bias (R) was 2.2. The bootstrap consensus tree was inferred from 1,000 to 3,000 iterations, and the maximum likelihood approach was used to reconstruct the evolutionary history. Eleusine coracana, the current millet sample, is well depicted in the dendrogram (Figure 3) that illustrates the evolutionary relationship. The sequences utilized in this investigation have been added to GenBank with accession numbers PQ753526 (rbcL) and PQ728252 (matK).
Figure 3
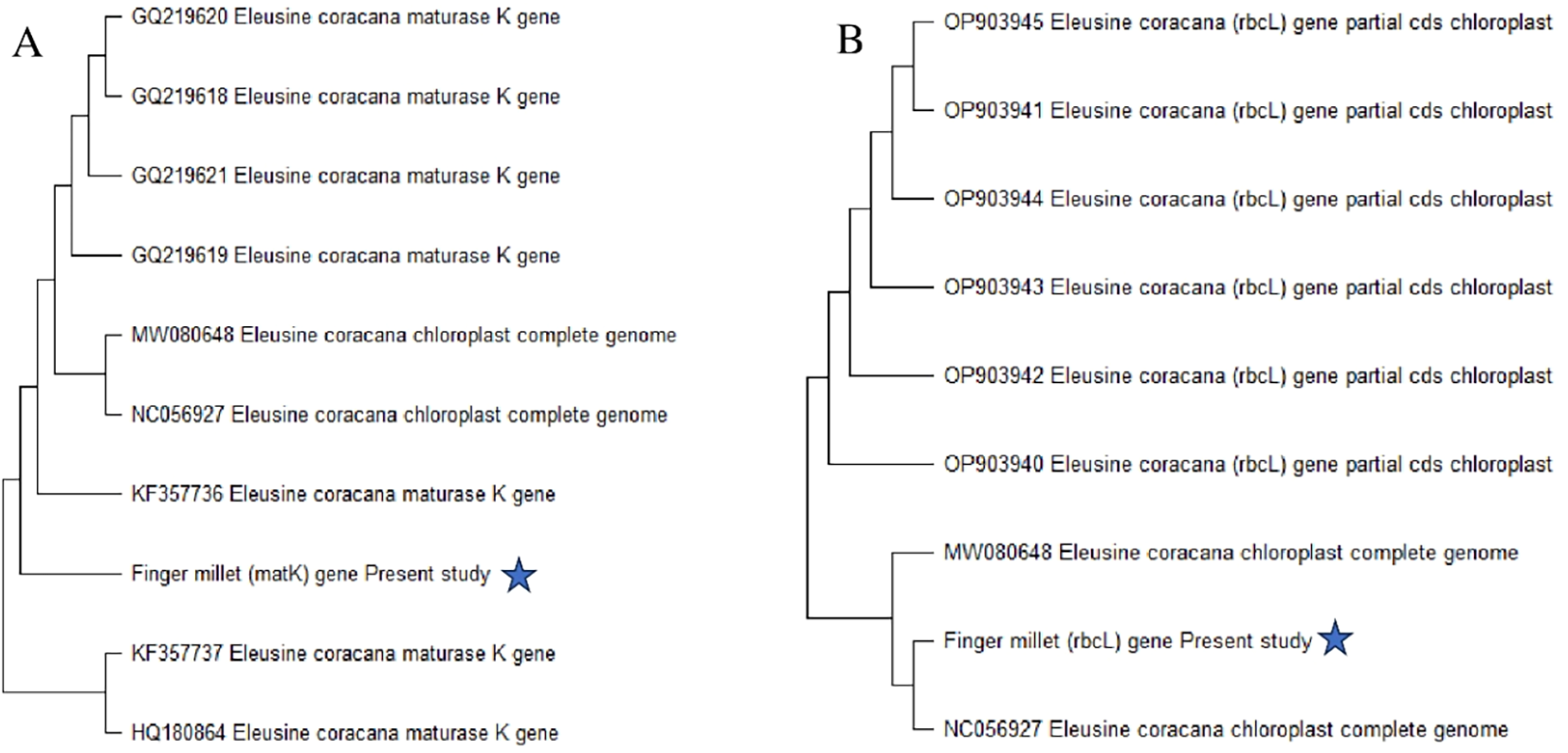
Maximum likelihood phylogenetic tree on basis of matK and rbcL sequence of Eleusine coracana:(A) depicts phylogenetic tree on basis of matK and (B) depicts according to rbcL sequence of Eleusine coracana.
ICP-MS analysis
Elemental analysis was carried out for the finger millet. A total of 10 macroelements and microelements were present at different concentrations (Table 1). The major elements found were calcium (Ca), potassium (K), phosphorus(P), and magnesium (Mg). Among all the analyzed elements, potassium was found to have the highest concentration. It is an essential mineral for the efficient functioning of tissues, the body’s cells, and organs. Increased potassium intake in diets can help maintain healthy blood pressure levels. The elements that are present in low concentrations are manganese (Mn), copper (Cu), iron (Fe), zinc (Zn), sodium (Na), and selenium (Se).
Table 1
| S. No. | Minerals | Sample concentration (mg/100gram) |
|---|---|---|
| 1 | Ca | 326.17 ± 0.8 |
| 2 | Mg | 138.24 ± 0.2 |
| 3 | Zn | 2.62 ± 1.4 |
| 4 | Mn | 2.13 ± 0.4 |
| 5 | Fe | 3.73 ± 0.4 |
| 6 | Cu | 0.67 ± 0.8 |
| 7 | Se | 0.04 ± 0.6 |
| 8 | P | 249.47 ± 0.8 |
| 9 | K | 416.93 ± 0.2 |
| 10 | Na | 1.27 ± 0.2 |
ICP-MS analysis of finger millet.
Phytochemical profile of Eleusine coracana by LC-HRMS/MS
A developed and broad untargeted metabolomics method was conducted to the comprehensive phytochemical profile investigation of Eleusine coracana seed metabolites utilizing LC-HRMS/MS. Peaks were examined manually for signal/noise (s/n) > 10 in resulting data matrix, which included around 4000 signals with an MS2 spectrum overall. Duplicate signals were then eliminated as previously mentioned. Ultimately, the product ion spectra (MS2) yielded 521 very reproducible metabolite signatures. 185 were allegedly detected with the commercial standards based on fragmentation pattern, retention time (RT), and mass-to-charge-ratio (m/z) values of each metabolite. Aside from a few primary metabolites, the results included 13 fatty acid derivatives, 19 amino acids and their derivatives, 6 nucleotide derivatives, 7 carbohydrate derivatives, and 5 vitamin-related substances. An overview of all annotated compounds is given in Tables 2, 3. Our study also putatively identified several secondary metabolites, including 53 phenolic compounds, 5 flavonoids, 13 terpenoids, 10 alkaloids, 13 heterocyclic compounds, 24 organic compounds, 6 ester compounds, 5 sugar alcohols, 4 quinoline compounds, and 2 acid derivatives. Significantly, in case of primary metabolites, four vitamin biosynthesis-related metabolites, including dehydroascorbic acid, niacin, vitamin C, D-pantothenic acid, 5′-O-β-D-glucosyl pyridoxine; various nucleotide derivatives including xanthine, orotic acid, nicotinuricacid, epiguanine, 7-carboxy-7-deazaguanine, allantoin; carbohydrate derivatives such as gluconic acid δ-lactone, methyl α-D-mannoside, xylitol, 2-deoxyhexopyranose; various fatty acid derivatives such as propionic acid, decanoic acid, palmitic acid, 6-oxohexanoic acid, triacetic acid, 6-oxohexanoic acid were detected in the seeds of Eleusine coracana.
Table 2
| Name | RT [min] | m/z | Molecular weight | Molecular formula | Delta mass [PPM] | Activity |
|---|---|---|---|---|---|---|
| Sugar alcohols | ||||||
| Volemitol | 5.18 | 211.082 | 212.089 | C7 H16 O7 | 1.5 | Antioxidant |
| D-(-)-Mannitol | 5.07 | 181.072 | 182.079 | C6 H14 O6 | 1.88 | Antioxidant |
| D-(+)-Arabitol | 4.77 | 151.061 | 152.068 | C5 H12 O5 | 2.43 | Antibiotic |
| 1-Deoxy-L-mannitol | 3.30 | 165.077 | 166.084 | C6 H14 O5 | 2.64 | Antioxidant |
| 6-Methyltetrahydro-2H-pyran-2,5-diol | 8.41 | 287.148 | 132.079 | C6 H12 O3 | 4.33 | – |
| Quinoline | ||||||
| Xanthurenic acid | 1.28 | 206.045 | 205.038 | C10 H7 N O4 | 2.77 | Neuroactive |
| Hydroxy-1,4-benzoquinone | 1.80 | 123.009 | 124.016 | C6 H4 O3 | 4.58 | Cytotoxic |
| Hydroxynaphthoic acid | 7.74 | 377.104 | 188.048 | C11 H8 O3 | 3.84 | Allelopathic |
| Benzoquinone | 0.68 | 167.035 | 108.021 | C6 H4 O2 | 3.83 | Antimicrobial |
| Phenolic aldehyde | ||||||
| β-Resorcylaldehyde | 0.54 | 137.024 | 138.032 | C7 H6 O3 | 2.98 | Anti-inflammatory |
| Benzaldehyde | 2.28 | 276.100 | 106.042 | C7 H6 O | 4.38 | Antimicrobial |
| 3-(4-Hydroxyphenyl)-2-oxiranecarbaldehyde | 0.54 | 163.040 | 164.047 | C9 H8 O3 | 1 | – |
| Phenolic compounds | ||||||
| Vanilpyruvic acid | 3.50 | 209.045 | 210.053 | C10 H10 O5 | 1.86 | – |
| Vanillin | 0.53 | 151.040 | 152.047 | C8 H8 O3 | 2.62 | Antioxidant |
| Urolithin B | 8.91 | 447.085 | 212.047 | C13 H8 O3 | 1.95 | Anti-inflammatory |
| Umbelliferone | 0.74 | 161.024 | 162.032 | C9 H6 O3 | 2.33 | Antioxidant |
| Scopoletin | 0.78 | 191.035 | 192.042 | C10 H8 O4 | 2.18 | Antioxidant |
| Schisandrin C | 12.17 | 385.166 | 384.158 | C22 H24 O6 | 4.03 | Antioxidant |
| Salicylic acid | 0.74 | 137.024 | 138.032 | C7 H6 O3 | 2.99 | Antimicrobial |
| Protocatechuic acid | 0.72 | 153.019 | 154.027 | C7 H6 O4 | 2.93 | Antioxidant |
| Phloroglucinol | 4.08 | 125.024 | 126.032 | C6 H6 O3 | 4.42 | Phloroglucinol |
| Phenylpropiolic acid | 1.80 | 145.030 | 146.037 | C9 H6 O2 | 4.08 | Anticancer |
| Phenylglyoxylic acid | 0.49 | 149.024 | 150.032 | C8 H6 O3 | 2.93 | – |
| Phenylethyl alcohol | 0.66 | 181.087 | 122.073 | C8 H10 O | 3.65 | Antimicrobial |
| Phenol | 0.73 | 93.034 | 94.042 | C6 H6 O | 3.32 | Antioxidant |
| p-Cresol | 0.38 | 107.050 | 108.057 | C7 H8 O | 3.2 | Antimicrobial |
| Naphthalene-2,3-diol | 0.94 | 159.045 | 160.05 | C10 H8 O2 | 2.49 | – |
| N-[2-Hydroxy-4(sulfooxy)phenyl]acetamide | 6.15 | 145.028 | 247.015 | C8 H9 N O6 S | 0.85 | – |
| Methyl cinnamate | 1.79 | 161.061 | 162.068 | C10 H10 O2 | 4.15 | Antimicrobial |
| Mesitol | 0.66 | 135.081 | 136.089 | C9 H12 O | 3.17 | Antioxidant |
| Meconic acid | 0.35 | 198.988 | 199.996 | C7 H4 O7 | 2.22 | – |
| Lithospermoside | 6.99 | 330.119 | 329.111 | C14 H19 N O8 | 1.57 | Antioxidant |
| Isovanillic acid | 0.41 | 167.035 | 168.042 | C8 H8 O4 | 3.18 | – |
| Hydroquinone | 0.73 | 109.029 | 110.037 | C6 H6 O2 | 3.86 | – |
| Homovanillic acid | 0.81 | 181.051 | 182.058 | C9 H10 O4 | 2.24 | – |
| Guaietolin | 0.65 | 211.098 | 212.105 | C11 H16 O4 | 2.79 | Antimicrobial |
| Guaiacol | 0.94 | 123.045 | 124.052 | C7 H8 O2 | 3.55 | Antimicrobial |
| Eugenol | 1.75 | 163.077 | 164.084 | C10 H12 O2 | 3.82 | Antimicrobial |
| Dihydrophloroglucinol | 0.92 | 187.061 | 128.047 | C6 H8 O3 | 3.56 | – |
| Coumarin | 0.71 | 145.030 | 146.037 | C9 H6 O2 | 3.65 | Antimicrobial |
| Benzoic acid | 10.11 | 121.029 | 122.037 | C7 H6 O2 | 1.8 | Antimicrobial |
| Benzeneacetamide-4-O-sulphate | 7.65 | 273.054 | 231.020 | C8 H9 N O5 S | 1.46 | – |
| Aceturic acid | 1.18 | 98.025 | 117.043 | C4 H7 N O3 | 3.45 | – |
| 5-carboxyvanillic acid | 0.35 | 211.025 | 212.032 | C9 H8 O6 | 2.22 | Antioxidant |
| 4-Hydroxyphthalic acid | 0.41 | 181.014 | 182.022 | C8 H6 O5 | 3.38 | – |
| 4-Hydroxycyclohexylcarboxylic acid | 1.12 | 189.077 | 144.079 | C7 H12 O3 | 4.39 | – |
| 4-Hydroxybenzoic acid | 1.12 | 137.024 | 138.032 | C7 H6 O3 | 4.13 | Antimicrobial |
| 4-Hydroxy-3-nitrophenylacetic acid | 0.54 | 196.025 | 197.032 | C8 H7 N O5 | 2.64 | – |
| 4-Ethylguaiacol | 0.49 | 151.076 | 152.084 | C9 H12 O2 | 2.93 | Antimicrobial |
| 4,5-Dihydroxyphthalic acid | 0.36 | 197.009 | 198.016 | C8 H6 O6 | 2.18 | – |
| 3-Hydroxybenzoic acid | 3.37 | 137.024 | 138.031 | C7 H6 O3 | 1.24 | Antimicrobial |
| 3-(4-Hydroxy-3-methoxyphenyl)-2-methylpropanoic acid | 0.48 | 209.082 | 210.089 | C11 H14O4 | 2.72 | – |
| 3,4-Dihydroxyphthalic acid | 0.43 | 197.009 | 198.016 | C8 H6O6 | 2.44 | – |
| 3,4-Dihydroxyphenylglycol | 7.21 | 171.064 | 170.057 | C8 H10O4 | 0.44 | Antioxidant |
| 2-Furoic acid | 0.98 | 111.009 | 112.016 | C5 H4 O3 | 3.6 | Antimicrobial |
| 2,4-Dihydroxybenzoic acid | 0.40 | 153.019 | 154.027 | C7 H6 O4 | 3.23 | Antimicrobial |
| 1,2-Benzoquinone | 0.53 | 153.019 | 108.021 | C6 H4 O2 | 3.57 | Cytotoxic |
| (E)-p-coumaric acid | 2.63 | 163.040 | 164.047 | C9 H8 O3 | 2.25 | Antioxidant |
| (E)-Isoferulic acid | 3.46 | 193.051 | 194.058 | C10 H10 O4 | 2.26 | Anti-inflammatory |
| (E)-Ferulic acid | 0.70 | 193.051 | 194.058 | C10 H10 O4 | 2.34 | Antimicrobial |
| (2Z)-3-(3-Hydroxyphenyl)-2-methylacrylaldehyde | 1.12 | 161.061 | 162.068 | C10 H10 O2 | 3.89 | – |
| (2E)-3-(3,4-Dimethoxyphenyl)acrylic acid | 0.77 | 207.066 | 208.074 | C11 H12 O4 | 2.38 | – |
| Alkaloids | ||||||
| Roquefortine L | 12.01 | 202.590 | 403.166 | C22 H21 N5 O3 | 4.98 | Antimicrobial |
| Putaminoxin | 0.59 | 211.134 | 212.141 | C12 H20 O3 | 2.75 | Antimicrobial |
| Naphthalen-2-amine | 4.81 | 142.066 | 143.073 | C10 H9 N | 2.22 | – |
| D-(-)-Quinic acid | 5.60 | 191.056 | 192.068 | C7 H12 O6 | 2.08 | Antioxidant |
| Chaksine | 13.61 | 473.286 | 450.297 | C22 H38 N6 O4 | 3.92 | – |
| 6-hydroxypseudooxynicotine | 8.87 | 452.227 | 194.105 | C10 H14 N2 O2 | 0.47 | – |
| 4-O-α-D-Glucopyranosylmoranoline | 6.45 | 326.144 | 325.137 | C12 H23 N O9 | 1.14 | – |
| 3-Methyloxindole | 0.67 | 146.061 | 147.068 | C9 H9 N O | 2.8 | – |
| 3-hydroxy-3-methyloxindole | 0.96 | 162.056 | 163.063 | C9 H9 N O2 | 2.57 | – |
| 2-pyridone | 0.72 | 140.035 | 95.037 | C5 H5 N O | 4.45 | – |
| Terpenoids | ||||||
| γ-Heptalactone | 1.14 | 127.076 | 128.084 | C7 H12 O2 | 3.98 | – |
| Viburtinal | 0.71 | 159.045 | 160.052 | C10 H8 O2 | 3.26 | Antimicrobial |
| Tulipalin A | 0.53 | 195.066 | 98.037 | C5 H6 O2 | 2.96 | Cytotoxic |
| Trans-geranic acid | 0.47 | 167.108 | 168.115 | C10 H16 O2 | 3.02 | Antimicrobial |
| Maraniol | 0.95 | 203.071 | 204.079 | C12 H12 O3 | 2.67 | – |
| Glycyrin | 12.00 | 383.149 | 382.142 | C22 H22 O6 | 2.37 | Anti-inflammatory |
| Geranyl formate | 12.81 | 183.137 | 182.130 | C11 H18 O2 | 0.23 | – |
| Geranyl acetate | 0.47 | 195.139 | 196.146 | C12 H20 O2 | 2.1 | Antimicrobial |
| Gallic acid | 0.53 | 169.014 | 170.021 | C7 H6 O5 | 2.32 | Antioxidant |
| DL-Mevalonic acid | 0.43 | 129.056 | 148.073 | C6 H12 O4 | 2.93 | – |
| Abietatriene | 13.80 | 271.241 | 270.234 | C20 H30 | 0.3 | – |
| 6-Methylhept-5-en-2-one | 0.48 | 185.118 | 126.104 | C8 H14 O | 3.93 | – |
| 4-Methyl-2-propyltetrahydro-2H-pyran-4-yl acetate | 0.50 | 199.134 | 200.141 | C11 H20 O3 | 2.52 | – |
| Heterocyclic compounds | ||||||
| Thiophene | 0.46 | 82.996 | 84.003 | C4 H4 S | 3.04 | Anti-inflammatory |
| Thiazole | 0.40 | 83.991 | 84.998 | C3 H3 N S | 1.95 | – |
| Quinolone | 4.17 | 144.046 | 145.053 | C9 H7 N O | 3.87 | Antibacterial |
| Pyrrole | 2.97 | 66.035 | 67.042 | C4 H5 N | 2.09 | – |
| Lumazine | 1.12 | 163.026 | 164.034 | C6 H4 N4 O2 | 3.94 | Antimicrobial |
| Isoxazolin-5-one | 10.10 | 84.009 | 85.016 | C3 H3 N O2 | 1.44 | Antimicrobial |
| Isoplumbagin | 0.71 | 187.040 | 188.047 | C11 H8 O3 | 3.3 | Anticancer |
| Indole | 1.83 | 116.051 | 117.058 | C8 H7 N | 4.55 | – |
| Furoic acid | 1.85 | 202.038 | 169.004 | C3 H7 N O5 S | 1.89 | Antimicrobial |
| Furan | 1.12 | 135.045 | 68.026 | C4 H4 O | 3.73 | Anticancer |
| 4,5-Dihydroxy-3-oxo-1-cyclohexene-1-carboxylic acid | 0.66 | 171.030 | 172.037 | C7 H8 O5 | 2.63 | – |
| 2-(Methylthio)benzothiazole | 2.58 | 404.039 | 181.002 | C8 H7 N S2 | 2.53 | – |
| 2-(3-Hydroxy-3,4,5,6-tetrahydro-1H-cyclopenta-furan-4-yl)-3-methoxy-3-oxopropanoic acid | 9.65 | 243.087 | 242.079 | C11 H14 O6 | 3.62 | – |
| Organic compounds | ||||||
| 1-Acetylcyclohexyl acetate | 10.18 | 183.102 | 184.110 | C10 H16 O3 | 1.37 | – |
| 2,5-Dimethyl-4-ethoxy-3(2H)-furanone | 1.15 | 215.093 | 156.079 | C8 H12 O3 | 4.05 | – |
| Propenal | 0.84 | 55.019 | 56.026 | C3 H4 O | 2.88 | – |
| Phenylacetaldehyde | 1.66 | 119.050 | 120.057 | C8 H8 O | 3.32 | – |
| Methyl Phenyl Disulfide | 1.06 | 157.014 | 156.007 | C7 H8 S2 | 3.84 | – |
| Isatin | 10.22 | 146.061 | 147.032 | C8 H5 N O2 | 1.34 | Antibacterial |
| Indole-3-carbidol | 2.96 | 146.025 | 147.068 | C9 H9 N O | 2.63 | – |
| Heptenal | 0.59 | 171.103 | 112.089 | C7 H12 O | 3.63 | Antimicrobial |
| Glyoxylic acid | 5.08 | 133.014 | 74.0 | C2 H2 O3 | 4.17 | – |
| Furfuryl acetone | 3.51 | 137.061 | 138.068 | C8 H10 O2 | 2.06 | Antimicrobial |
| Furfuranol | 2.10 | 97.029 | 98.037 | C5 H6 O2 | 3.94 | Antimicrobial |
| Formylpyruvate | 5.50 | 115.004 | 116.011 | C4 H4 O4 | 2.67 | – |
| Ethylpropyldisulfide | 1.63 | 137.045 | 136.038 | C5 H12 S2 | 1.8 | Antifungal |
| Cumene | 0.66 | 119.087 | 120.094 | C9 H12 | 3.37 | – |
| Butyronitrile | 1.83 | 70.065 | 69.057 | C4 H7 N | 0.2 | – |
| Acetophenone | 0.46 | 119.050 | 120.057 | C8 H8 O | 3.06 | – |
| 4-Oxo-2-propylpentanoic acid | 0.52 | 157.087 | 158.094 | C8 H14 O3 | 2.16 | – |
| 4-Methylthio-4-methyl-2-pentanone | 1.41 | 147.084 | 146.077 | C7 H14 O S | 3.16 | – |
| 4-Hydroxy-5-methyl-3-furanone | 4.86 | 113.024 | 114.032 | C5 H6 O3 | 3.14 | – |
| 4,7-Dihydroxycoumarin | 0.50 | 179.033 | 178.026 | C9 H6 O4 | 1.81 | Anti-inflammatory |
| 4,5,7-Trihydroxycoumarin | 2.97 | 193.014 | 194.021 | C9 H6 O5 | 2.39 | Antimicrobial |
| 3,4-Dimethylthiophene | 2.04 | 113.042 | 112.035 | C6 H8 S | 4.88 | – |
| 2-methylcitric acid | 4.96 | 205.035 | 206.043 | C7 H10 O7 | 2.27 | – |
| Dhurrin | 7.01 | 334.09 | 311.100 | C14 H17 N O7 | 0.31 | – |
| Flavonoids | ||||||
| Catechol | 0.41 | 109.043 | 110.058 | C6 H6 O2 | 4.47 | Antioxidant |
| Flaviolin | 0.46 | 205.012 | 206.083 | C10 H6 O5 | 2.17 | Antioxidant |
| Pyrogallol | 1.18 | 125.058 | 126.063 | C6 H6 O3 | 3.61 | Antioxidant |
| Kumarone | 0.76 | 161.073 | 162.023 | C11 H14 O | 2.13 | Anti-inflammatory |
| Purpurogallin | 0.47 | 219.048 | 220.093 | C11 H8 O5 | 2.89 | Antioxidant |
| Esters | ||||||
| Tetrahydrofurfuryl acetate | 0.67 | 143.071 | 144.079 | C7 H12 O3 | 2.72 | – |
| Methyl sorbate | 1.14 | 125.061 | 126.068 | C7 H10 O2 | 3.51 | Antimicrobial |
| Ethyl levulinate | 3.69 | 189.077 | 144.079 | C7 H12 O3 | 3.51 | Antioxidant |
| Ethyl benzoylacetate | 0.60 | 191.071 | 192.079 | C11 H12 O3 | 2.77 | – |
| Benzyl formate | 3.99 | 135.045 | 136.053 | C8 H8 O2 | 4.2 | – |
| Acevaltrate | 11.25 | 241.107 | 480.200 | C24 H32 O10 | 1.42 | – |
| Acid derivatives | ||||||
| Suberic acid | 1.00 | 174.089 | 173.082 | C8 H14 O4 | 2.93 | – |
| Crotonic acid | 0.50 | 86.037 | 84.024 | C4 H6 O2 | 2.62 | – |
| Hormones | ||||||
| Indoleacetylaspartate | 6.467 | 290.057 | 288.036 | C14 H14 N2 O5 | 1.59 | – |
Secondary metabolite phytochemical profiling of the Eleusine coracana seed extract analyzed by LC-HRMS/MS.
PPM is a widely accepted unit used in high-resolution mass spectrometry to express the mass deviation. This value reflects the instrumental mass accuracy, which is critical for high-confidence metabolite identification. A low Delta Mass (typically within ±5 PPM) is considered indicative of a high-confidence match.
Table 3
| Name | RT [min] | m/z | Molecular weight | Molecular formula | Delta mass [PPM] |
|---|---|---|---|---|---|
| Carbohydrate derivatives | |||||
| δ-Gluconic acid δ-lactone | 1.892 | 177.041 | 178.048 | C6 H10 O6 | 3.46 |
| Xylitol | 4.297 | 151.061 | 152.069 | C5 H12 O5 | 3.87 |
| Methyl α-D-mannoside | 3.563 | 193.072 | 194.079 | C7 H14 O6 | 2.85 |
| L-(-)-Malic acid | 4.212 | 133.014 | 134.022 | C4 H6 O5 | 4.02 |
| Hydroxyacetone phosphate | 5.239 | 152.996 | 154.003 | C3 H7 O5 P | 2.13 |
| 6-Deoxy-3-O-methyl-β-D-galactopyranose | 4.163 | 177.077 | 178.084 | C7 H14 O5 | 3.66 |
| 2-Deoxyhexopyranose | 4.993 | 163.061 | 164.068 | C6 H12 O5 | 2.45 |
| Nucleotide derivatives | |||||
| Xanthine | 3.782 | 151.026 | 152.033 | C5 H4 N4 O2 | 2.79 |
| Orotic acid | 0.49 | 155.010 | 156.017 | C5 H4 N2 O4 | 2.23 |
| Nicotinuric acid | 1.133 | 179.046 | 180.054 | C8 H8 N2 O3 | 3.12 |
| Epiguanine | 4.002 | 164.058 | 165.065 | C6 H7 N5 O | 3.4 |
| 7-Carboxy-7-deazaguanine | 2.081 | 193.037 | 194.044 | C7 H6 N4 O3 | 3.23 |
| (S)-(+)-allantoin | 3.831 | 157.037 | 158.044 | C4 H6 N4 O3 | 2.15 |
| Vitamins | |||||
| Vitamin C | 5.13 | 175.025 | 176.032 | C6 H8 O6 | 2.42 |
| Niacin | 2.903 | 122.058 | 123.024 | C6 H5 N O2 | 3.08 |
| Dehydroascorbic acid | 1.762 | 173.067 | 174.017 | C6 H6 O6 | 3.6 |
| D-pantothenic acid | 12.238 | 439.230 | 219.111 | C9 H17 N O5 | 2.22 |
| 5′-O-β-D-Glucosylpyridoxine | 6.998 | 332.134 | 331.127 | C14 H21 N O8 | 2.22 |
| Amino acid derivatives | |||||
| N-Acetyl-L-phenylalanine | 0.713 | 206.082 | 207.090 | C11 H13 N O3 | 3.28 |
| N-Acetyl-L-leucine | 1.351 | 172.098 | 173.105 | C8 H15 N O3 | 2.55 |
| N-Acetyl-L-glutamic acid | 7.051 | 379.135 | 189.064 | C7 H11 N O5 | 4.63 |
| N-Acetyl-D-quinovosamine | 4.919 | 204.088 | 205.095 | C8 H15 N O5 | 1.33 |
| N-(3-Carboxypropanoyl)-5-hydroxynorvaline | 7.223 | 467.188 | 233.090 | C9 H15 N O6 | 1.19 |
| Maleamic acid | 0.509 | 114.020 | 115.063 | C4 H5 N O3 | 3.14 |
| L-Pyroglutamic acid | 3.458 | 188.056 | 129.042 | C5 H7 N O3 | 2.91 |
| L-Proline | 1.17 | 114.056 | 115.063 | C5 H9 N O2 | 4.26 |
| L-Histidinol phosphate | 8.502 | 222.064 | 221.057 | C6 H12 N3 O4 P | 2.74 |
| L-(−)-threo-3-Hydroxyaspartic acid | 1.981 | 150.039 | 149.032 | C4 H7 N O5 | 0.64 |
| L-(+)-Aspartic acid | 1.134 | 132.030 | 133.038 | C4 H7 N O4 | 3.67 |
| Isovalerylglycine | 1.783 | 218.104 | 159.090 | C7 H13 N O3 | 4.95 |
| DL-Glutamine | 4.999 | 145.062 | 146.032 | C5 H10 N2 O3 | 2.87 |
| D-(+)-Tryptophan | 4.818 | 203.082 | 204.090 | C11 H12 N2 O2 | 1.83 |
| Aminolevulinic acid | 1.125 | 130.051 | 131.058 | C5 H9 N O3 | 4.04 |
| γ-Thiomethyl glutamate | 3 | 194.248 | 193.163 | C6 H11 N O4 S | 3.71 |
| 6-Acetamido-2-oxohexanoic acid | 4.035 | 168.324 | 187.012 | C8 H13 N O4 | 3.33 |
| 4-Methyleneglutamic acid | 4.972 | 158.027 | 159.086 | C6 H9 N O4 | 2.12 |
| 3-Sulfamoylalanine | 8.028 | 186.034 | 168.063 | C3 H8 N2 O4 S | 2.37 |
| Fatty acid derivatives | |||||
| Triacetic acid | 4.947 | 143.035 | 144.042 | C6 H8 O4 | 2.49 |
| Sorbic acid | 3.913 | 93.034 | 112.052 | C6 H8 O2 | 3.39 |
| Propionic acid | 0.792 | 73.029 | 74.036 | C3 H6 O2 | 2.41 |
| Palmitic Acid | 15.237 | 535.471 | 256.240 | C16 H32 O2 | 0.11 |
| Decanoic acid | 10.209 | 171.139 | 172.146 | C10 H20 O2 | 0.89 |
| 8-Hydroxy-5,6-octadienoic acid | 0.624 | 215.093 | 156.079 | C8 H12 O3 | 3.59 |
| 6-Oxohexanoic acid | 0.883 | 189.077 | 130.063 | C6 H10 O3 | 3.07 |
| 3E-octenoic acid | 0.46 | 141.092 | 142.099 | C8 H14 O2 | 2.46 |
| 3-Hydroxysebacic acid | 0.811 | 217.108 | 218.115 | C10 H18 O5 | 2.27 |
| 2-Hydroxyundecanoate | 13.933 | 425.288 | 201.149 | C11 H21 O3 | 3.5 |
| 1-Nonanoic acid | 0.662 | 157.123 | 158.131 | C9 H18 O2 | 2.35 |
| 1-Hexanal | 0.531 | 99.081 | 100.089 | C6 H12 O | 3.5 |
| 9-Oxononanoic acid | 0.719 | 171.103 | 172.110 | C9 H16 O3 | 3.18 |
Primary metabolite phytochemical profiling of the Eleusine coracana seed extract analyzed by LC-HRMS/MS.
PPM is a widely accepted unit used in high-resolution mass spectrometry to express the mass deviation. This value reflects the instrumental mass accuracy, which is critical for high-confidence metabolite identification. A low Delta Mass (typically within ±5 PPM) is considered indicative of a high-confidence match.
The phenolic compounds in the seed were coumarin, guaiacol, eugenol, phloroglucinol, 4-hydroxycyclohexylcarboxylic acid, 2-furoic acid, 1,2-benzoquinone, 4-hydroxybenzoic acid, phenylpropiolic acid, methyl cinnamate, schisandrin C, mesitol, dihydrophloroglucinol. Similarly, terpenoids compounds detected in seeds of Eleusine coracana were γ-heptalactone, viburtinal, geranic acid, 6-methylhept-5-en-2-one, maraniol, gallic acid, glycyrinetc; alkaloids compounds including roquefortine L, 2-pyridone, chaksine, putaminoxin, 3-methyloxindole; phytohormone include indoleacetylaspartate; acid derivatives such as suberic acid and crotonic acid.
Classification of metabolites detected using LC-MS/MS
Untargeted metabolomics of Eleusine coracana collected from the temperate region of District Kishtwar (Padder Valley), revealed putative compounds belonging to 9 different superclasses of compounds such as hydrocarbon derivatives, nitrogen, lipid, and oxygen compounds, heterocyclic compounds, phenylpropanoids, benzenoids, organosulfur compounds, organic acids and its derivatives (Figure 4). Further, detected metabolites were classified into 25 different main classes such as coumarin, cinnamic acid, indole and keto acid derivatives, fatty acyls, pyridine derivatives, furans, lactones, heteroaromatic compounds, carboxylic acid derivatives among others.
Figure 4
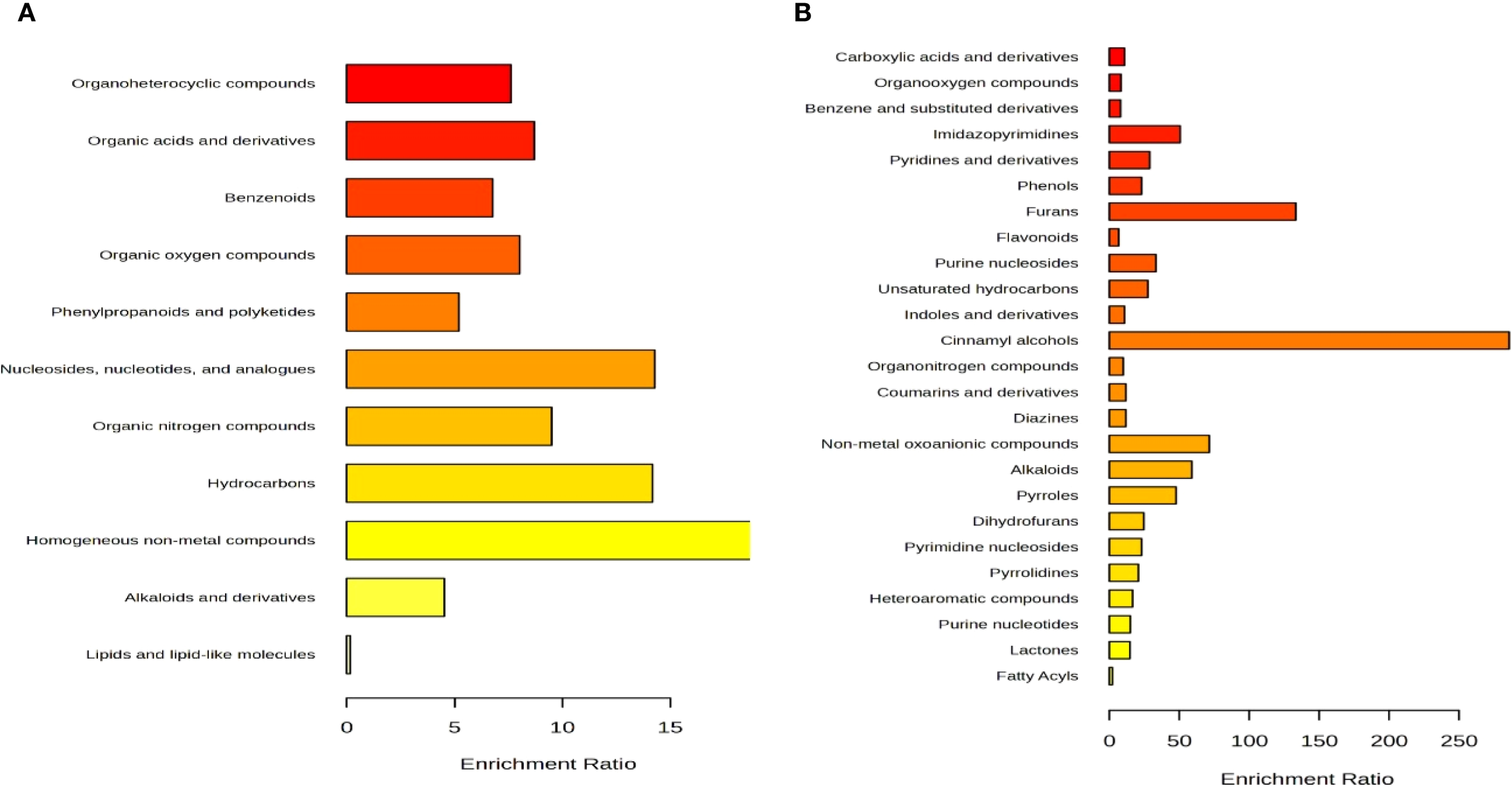
Detection and analysis of compounds according to LC–HRMS/MS. (A) indicate bar graph of the superclass of 185 differential identified metabolites; (B) indicate bar graph of the main class of 185 differential identified metabolites detected in the Eleusine coracana seed extract analyzed by LC-HRMS/MS.
KEGG annotation analysis of identified metabolites
KEGG analyses were carried out to classify the functions of differentially identified metabolites and clarify biological pathways and functions in finger millet seeds. KEGG analysis results demonstrated that the metabolic pathways were mapped to identified metabolites in finger millet seed. Based on the findings presented in Figure 5, 185 differential metabolites were identified as participating in the biosynthesis of amino acids, carbohydrate metabolism, citric acid pathway, metabolism of nitrogen, glutathione and porphyrin, biosynthesis of fatty acid, terpenoid biosynthesis, metabolism of propanoate. Notably, pathways involved in carbohydrate metabolism highlighting the grain’s role as a rich source of complex carbohydrates. Additionally, enrichment in amino acid biosynthesis pathways underscores its protein content, while phenylpropanoid biosynthesis points to the presence of antioxidant compounds contributing to finger millet’s health-promoting properties. Phenylpropanoids help plants withstand pests and diseases, and research on Arabidopsis shows that the phenylpropanoid metabolic pathway is activated by salt and drought stress. Amino acid production is essential to an organism’s growth and development. Plants under LT stress typically had higher levels of free amino acids than control plants, suggesting that amino acid production responds favorably to stress (Pandohee et al., 2023). Research has demonstrated that in transgenic alfalfa lines containing myoinositol phosphate synthase (MIPS), abiotic stressors including cold, drought, and salt stressors cause IP1 to be generated, while H2O2 and NO stressors cause high expression of MIPS. In plant tissues, glutathione metabolism is essential for preserving antioxidant qualities and controlling redox-sensitive signal transduction (Yang et al., 2024). Plant tolerance to range of biotic and abiotic environmental stressors is directly correlated with glutathione levels (Ye et al., 2024).
Figure 5
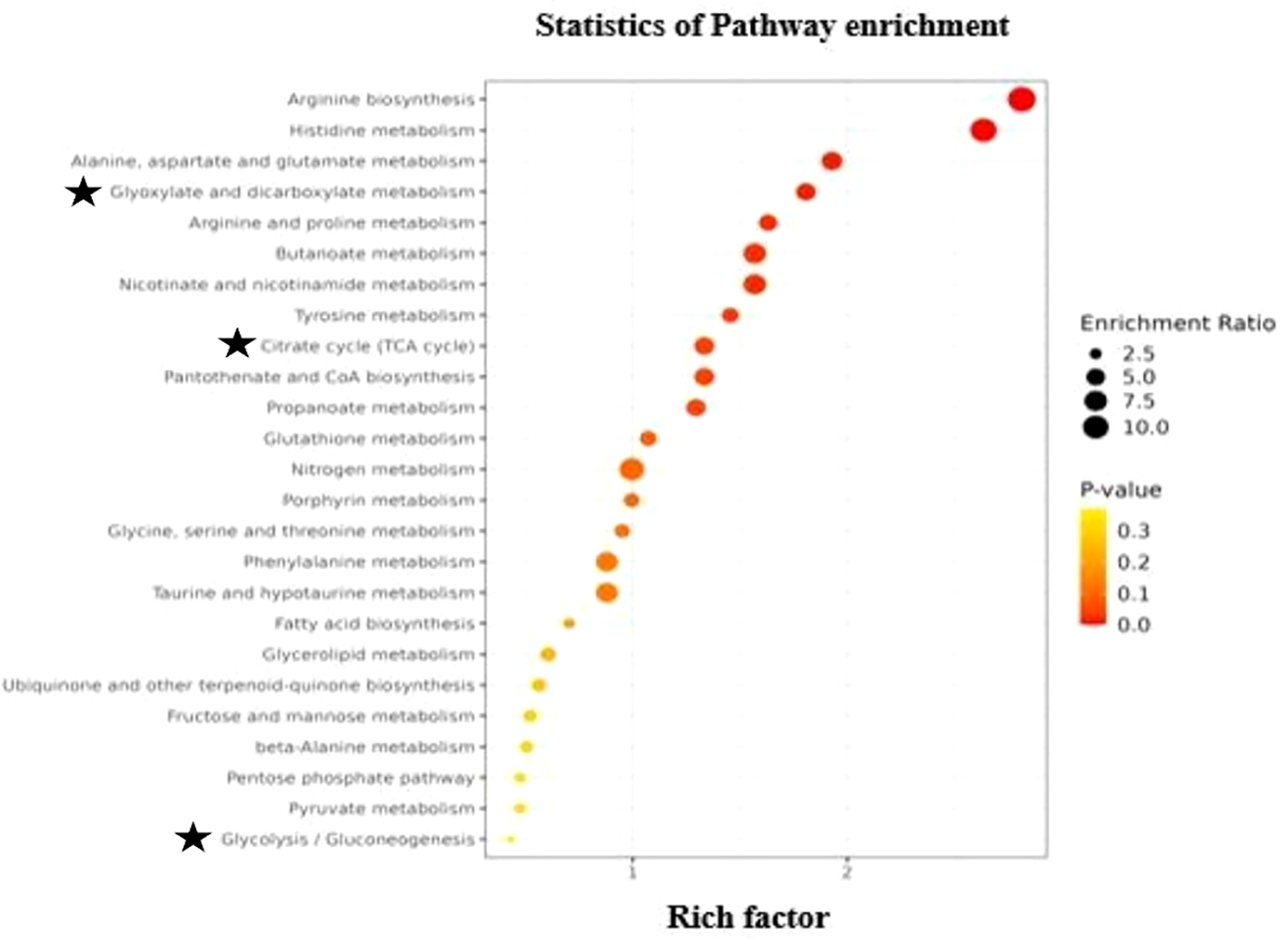
Scatter plot of top 25 KEGG pathways for differential identified metabolites in the seeds of Eleusine coracana. Abscissa, or rich factor, indicates the percentage of metabolites in a certain route that have been differentially identified. Circle color denoted ranges of corrected P value, while circle area showed number of metabolites that were differentially identified. The ratio of the number of metabolites in the associated pathway that were differentially identified to total number of metabolites that the pathway detected and annotated is known as the rich factor. The degree of enrichment increases with the value.
Discussion
The assessment of food and crop products as well as functional genomic research have made extensive use of metabolite profiling and chemometrics, which further guides breeding methods for enhancing and optimizing balance of food components (Li et al., 2021). Several metabolites can be identified and quantified in a single extract by applying the broad target metabolomics technique with LC-HRMS/MS (Dan et al., 2021). Metabolomics has been widely used to examine various compounds found in food products. Numerous significant metabolites with a range of physiological and bioactive roles have been identified by metabolomics investigation of several cereal crops, including millets (Wu et al., 2022). During the barnyard millet’s developmental stages, a total of 35 metabolites were found (Meniya et al., 2023). Sugar and sugar alcohols were most abundant of these compounds, followed by amino acids, organic acids, and sterols, in that order. Putative metabolites from 15 different groups of chemicals, including flavonoids, amino acids, organic acid and its derivatives, were identified by untargeted metabolomics of two distinct foxnut kinds that were gathered from various geographical locations (Bao et al., 2022). Numerous physiological effects on the human body, including antioxidative, antimicrobial, neuro-protective, anti-diabetic, antipyretic, anti-inflammatory, anti-depressive, anti-septic, antibiotic, anti-angiogenic, immunomodulatory activities, and anti-carcinogenic, have been demonstrated by number of these compounds that were identified in the current study (Saxena et al., 2024). Plant products include coumarin, which has been shown to have anti-inflammatory, antibacterial, antioxidant, and neuroprotective properties (Vishal et al., 2024). Pervious study observed that catechin and epicatechin were found to be the main polyphenols in all the finger millet samples (Xiang et al., 2019). Quinic acid is an alkaloid molecule with anti-inflammatory, antibacterial, and antioxidant properties that is found in plant metabolites (Deme et al., 2019). The current study also identified abietatriene, a diterpene with range of biological characteristics, such as anti-inflammatory, antibacterial, antiparasitic, antifungal, antioxidant, and antiviral actions (Kuźma and Gomulski, 2022). Results from several studies using animal models and cell cultures indicate that abietatriene has significant advantages by halting cellular DNA damage over time (Sena et al., 2024). Because of its flavour, glycyrin, also referred to as sugar alcohol, is frequently employed as a sweetener in the food sector. Furthermore, studies have shown that glycerin is useful as a skin protectant and to treat glaucoma (Pan et al., 2024).The antioxidant and anti-inflammatory potential of several phenolic compounds found in nutritional bar’s metabolome, including ferulic acid, guaiacol, eugenol, and guaietolin, have been thoroughly investigated (Pan et al., 2020).
Phenolic chemicals can withstand gastrointestinal digestion and stay intact as they pass through the colon because they are found in bound form in cereals, such as millet. These substances may have protective effects on the colon after being released by microbial fermentation (Sun et al., 2022). This phenomenon may help explain how whole grains can prevent colon cancer. Ferulic acid was found to significantly decrease production and expression of inflammatory markers include tumour necrosis factor-α (TNF-α), nitric oxide (NO), and interleukin-1β (IL-1β) while increasing levels of anti-inflammatory factor β-endorphin (β-EP) in a human HMC-3 microglial cell model (Vishal et al., 2024). Previous study showed that phenolic compounds such as syringic acid, hydroxyl caffeic acid, caffeic acid, coumaric acid ethyl ester, ellagic acid glucoside, 3-hydroxycoumarin, and feruloyl tartaric acid were identified in seed coat of finger millet (Soujanya et al., 2024). Antibacterial, anti-inflammatory, and antioxidant capabilities are mainly possessed by the hydroxybenzoic acid. Most fruits have a low hydroxybenzoic acid concentration, however, berries including blackberries, blackcurrants, strawberries, raspberries, and pomegranates can have very high levels of this compound (Carrillo et al., 2023; Kumar et al., 2024a). Coumarin, a naturally occurring phenolic compound identified in finger millet, exhibits multiple therapeutic activities. It acts as an anticoagulant by inhibiting vitamin K epoxide reductase, thereby reducing blood clot formation. Additionally, coumarin and its derivatives have been shown to possess anti-inflammatory and antioxidant properties by modulating inflammatory cytokines and scavenging reactive oxygen species (ROS), which are critical in the prevention of chronic diseases such as cardiovascular disorders and cancer (Sharma and Mallubhotla, 2022; Hussain et al., 2019; Kumar et al., 2024b). Eugenol, another key metabolite, is well known for its potent antimicrobial, analgesic, and anti-inflammatory effects. It exerts these effects primarily through the inhibition of COX-2 and NF-κB signaling pathways, which are involved in inflammation and pain signaling. Eugenol also disrupts microbial membranes, making it effective against a broad range of pathogens. Its antioxidant capacity further contributes to its role in protecting cells from oxidative damage, supporting its application in managing oxidative stress-related conditions (Ulanowska et al., 2021; Sharma et al., 2024).
Furthermore, flavonoids were identified as the primary phenolic compound class present in Eleusine coracana seeds by the LC-MS approach for phenolic compound quantification (Buelvas et al., 2021), the most abundant phenolic components in the crude extract of Eugenia pollicina leaves were flavonoids. Seeds of Eugenia uniflora primarily contain phenolics from the flavonoid class, including quercitrin and kaempferol pentoside (Fidelis et al., 2022). Both internal and external factors, including physical, chemical, and biological ones, affect the quantity of phenolic compounds. Moreover, the amount of total phenolic chemicals in plant tissues is similarly influenced by photosynthesis and carbon production (Araujo et al., 2021).
By analyzing the millet’s primary and secondary metabolites, we have been able to examine their detailed profiles and try to better understand their roles (Girardelo et al., 2020). The current study employed UHPLC-ESI-QTOF-MS, a potent, dependable, and combinative analytical approach, to quickly characterize a number of phytochemicals in extracts from two Lamiaceae plants (Cocan et al., 2018). It is evident from comparing our results with those of relevant literature that the phenolic compounds (gallic acid, ferulic acid) of porso and foxtail millets was comparable to that of the study suggested by (Pujari and Hoskeri, 2022). Thus, Eleusine coracana proved to be a prominent source of phenolic, flavonoid, and terpene chemicals, and its exploitation can find intriguing uses in pharmaceutical, cosmetic, functional food, food supplement, and novel product creation (Dey et al., 2022).
Conclusion
This study provides a comprehensive nutritional and bioactive profiling of underutilized finger millet (Eleusine coracana) landraces from Padder Valley, District Kishtwar, Jammu and Kashmir, India. LC-HRMS/MS analysis identified key metabolites, including catechol, flaviolin, pyrogallol, ferulic acid, protocatechuic acid, coumarin, eugenol, cumene, and guaietolin, alongside significant fatty acids, alkaloids, terpenes, and hydrocarbons. These compounds exhibit diverse bioactivities, such as antioxidant, anti-diabetic, antibacterial, and anti-obesity effects, underscoring the potential of finger millet as a functional food ingredient and source of nutraceuticals. The findings highlight the crop’s capacity to contribute to dietary diversification, address chronic illnesses, and combat malnutrition. While the results demonstrate substantial therapeutic potential, they are based on metabolomic and in silico analyses. Therefore, future studies should focus on in vivo validation, mechanistic investigations, and targeted quantification to confirm the physiological relevance of these bioactive compounds and explore their applications in nutrition, health, and functional food development.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
VG: Conceptualization, Data curation, Formal Analysis, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing. MS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. SK: Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing. SJ: Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. ZA: Investigation, Visualization, Writing – original draft, Writing – review & editing. SA: Validation, Visualization, Writing – original draft, Writing – review & editing. JK: Data curation, Funding acquisition, Visualization, Writing – review & editing. RU: Formal analysis, Funding acquisition, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors are grateful to the Ongoing Research Funding Program, (ORF-2025-360), King Saud University, Riyadh, Saudi Arabia.
Acknowledgments
The authors express our appreciation to the HADP Project No. 8, titled “Promotion of Millets and Nutricereals in the Union Territory of Jammu & Kashmir,” for their logistical support. Special thanks to Dr. Sanjay Guleria, Professor and Head, Division of Biochemistry, SKUAST-Jammu, for his critical review of the manuscript and constructive feedback, which greatly enhanced the quality of this work. The authors are grateful to the Ongoing Research Funding Program, (ORF-2025-360), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1570787/full#supplementary-material
Abbreviations
ACN, acetonitrile; FA, formic acid; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC-HRMS, Liquid Chromatography-High Resolution Mass Spectrometry; m/z, mass-to-charge-ratio; MEGA, Molecular Evolutionary Genetics Analysis; mL, Milliliter; RT, retention time.
References
1
Abioye V. Babarinde Ogunlakin G. Adejuyitan G. Olatunde J. Abioye S. (2022). Varietal and processing influence on nutritional and phytochemical properties of finger millet: A review. Heliyon8, 1–15. doi: 10.1016/s2405.03598.8
2
Almutairi (2021). Molecular characterization and expression analysis of ribosomal L18/L5e gene in Pennisetum glaucum (L.) R. Br. Saudi J. Biol. Sci.28, 3585–3593. doi: 10.1016/j.sjbs.2021.03.035
3
Anggraeni A. Alam L. Utami I. Khasanah Y. Nurfahmi L. Noviana I. et al . (2025). Untargeted metabolomic LC-HRMS combined with chemometric reveal metabolites change from sorghum flakes affected by food processing. Case Stud. Chem. Environ. Eng.11, 101034. doi: 10.1016/j.cscee.2024.101034
4
Araujo N. Arruda H. Paulo Farias D. Molina G. Pereira G. Pastore G. (2021). Plants from the genus Eugenia as promising therapeutic agents for the management of diabetes mellitus: A review. Food Res. Int.142, 110182. doi: 10.1016/j.foodres.2021.110182
5
Baduni P. Maikhuri R. Bhatt G. C. Rawat H. Singh R. Semwal C. et al . (2024). Contribution of Millets in food and nutritional security to human being: Current status and future perspectives. Natural Resour. Conserv. Res.7, 1–21. doi: 10.24294/nrcr.v7i1.5479
6
Bao K. Jing Z. Wang Q. Huang Z. Han D. Dai et al . (2022). Quality analysis of Euryales Semen from different origins and varieties based on untargeted metabolomics. J. Chromatogr. B1191, 123114. doi: 10.1016/j.jchromb.2022.123114
7
Buelvas L. Franco G. Martínez H. Ballesteros D. Sánchez A. Miranda D. et al . (2021). Supercritical fluid extraction of phenolic compounds from mango (Mangifera indica L.) seed kernels and their application as an antioxidant in an edible oil. Molecules26, 7516. doi: 10.3390/molecule26247516
8
Cao X. Hu Y. Song J. Feng H. Wang J. Chen L. et al . (2022). Transcriptome sequencing and metabolome analysis reveals the molecular mechanism of drought stress in millet. Int. J. Mol. Sci.23, 10792. doi: 10.3390/ijms231810792
9
Carrillo C. Tomasevic I. Barba F. Kamiloglu S. (2023). Modern analytical techniques for berry authentication. Chemosensors11, 500. doi: 10.3390/chemosensors11090500
10
Cocan I. Alexa E. Danciu C. Radulov I. Galuscan A. Obistioiu D. (2018). Phytochemical screening and biological activity of Lamiaceae family plant extracts. Exp. Ther. Med.15, 1863–1870. doi: 10.3892/etm.2017.5640
11
Dan Z. Chen Y. Li H. Zeng Y. Xu W. Zhao W. et al . (2021). The metabolomic landscape of rice heterosis highlights pathway biomarkers for predicting complex phenotypes. Plant Physiol.187, 1011–1025. doi: 10.3892/etm.2017.5640
12
Deme P. Aluganti C. Parthasarathy S. (2019). Evaluation of anti-inflammatory properties of herbal aqueous extracts and their chemical characterization. J. medicinal Food22, 861–873. doi: 10.1089/jmf.2019.0009
13
Dey S. Saxena A. Kumar Y. Maity T. Tarafdar A. (2022). Understanding the antinutritional factors and bioactive compounds of kodo millet (Paspalum scrobiculatum) and little millet (Panicum sumatrense). J. Food Qual.2022, 1578448. doi: 10.1155/2022/1578448
14
Fidelis E. Savall A. Oliveira Pereira F. Quines C. Ávila D. Pinton S. (2022). Pitanga (Eugenia uniflora L.) as a source of bioactive compounds for health benefits: A review. Arabian J. Chem.15, 103691. doi: 10.1016/j,arabjc.2022.103691
15
Ghosh K. Vinodkumar T. Singh R. Singh A. Ilavarasan R. (2024). Promising potential of millets as the staple of our plate: An Indian perspective. Discover Food4, 103. doi: 10.1007/s44187-024-00176-7
16
Girardelo J. Munari E. Dallorsoleta J. Cechinel G. Goetten A. Sales L. et al . (2020). Bioactive compounds, antioxidant capacity and antitumoral activity of ethanolic extracts from fruits and seeds of Eugenia involucrata DC. Food Res. Int.137, 109615. doi: 10.1016/j.foodres.2020.109615
17
Han Y. Zhao P. Zhao Y. Liu M. Guo E. Wang G. et al . (2024). Transcriptome sequencing and metabolome analysis reveals the regulatory and molecular mechanisms of the grain filling rate in foxtail millet (Setaria italica L.). Agronomy14, 1114. doi: 10.3390/agronomy14061114
18
Hussain M. Syed Q. Khattak M. Hafez B. Reigosa M. Keblawy A. (2019). Natural product coumarins: biological and pharmacological perspectives. Biologia74, 863–888. doi: 10.2478/s11756-019-00242-x
19
Ibrahim M. Detroja A. Bishoyi A. (2023). A rapid, efficient and cost-effective DNA isolation protocol for various herbal products which is appropriate for different downstream genomics applications. Braz. J. Bot.46, 873–880. doi: 10.1007/s40415-023-00923-7
20
Kalsi R. Bhasin J. Goksen G. Kashyap P. (2023). Exploration of nutritional, pharmacological, and the processing trends for valorization of finger millet (Eleusine coracana): A review. Food Sci Nutr.11, 6802–6819. doi: 10.1002/fsn3.3659
21
Kumar V. Kumar A. Singh M. Dhyani P. Mishra H. Rai D. (2024a). Bioactive metabolites identification of the foxnut and broken millet-based nutritional bar using HR-MS. Food Chemistry: Mol. Sci.9, 100214. doi: 10.1016/j.fochms.2024.100214
22
Kumar V. Yadav M. Awala S. Valombola J. Saxena M. Ahmad F. et al . (2024b). Millets: A nutritional powerhouse for ensuring food security. Planta260(4), 101. doi: 10.1007/s00425-024-04533-9
23
Kuźma Ł. Gomulski J. (2022). Biologically active diterpenoids in the Clerodendrum genus—a review. Int. J. Mol. Sci.23, 11001. doi: 10.3390/ijms231911001
24
Li S. Tian Y. Jiang P. Lin Y. Liu X. Yang H. (2021). Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci Nutr.61, 1448–1469. doi: 10.1080/10408398.2020.1761287
25
Mafukata Z. Bamidele O. Ramashia S. Mashau M. (2024). Nutritional composition, protein digestibility and consumer acceptability of sorghum-based mahewu enriched with Moringa oleifera leaf powder. Int. J. Food Sci Technol.59, 6150–6162. doi: 10.1111/ijfs.17349
26
Malathi V. Jacob J. Venkateswarlu R. Kannababu N. Ratnavathi C. (2024). Nutritional aspects, phytochemical composition and potential health benefits of small millets. Genet. improvement Small Millets4, 129–152. doi: 10.1007/978-981-99-7232-6_7
27
Mallick A. Rajnish K. Mondal K. Hazra R. Samal A. (2024). Millet-based food adoption for environmental sustainability and nutritional security. Ecosystem Management: Climate Change Sustainability1, 629–658. doi: 10.1002/9781394231249.ch20
28
Meniya V. Padhiyar S. Desai H. Kheni J. Tomar R. (2023). Devel-opment and validation of microsatellite markers for Barnyard Millet obtained by partial genome assembly. Ann. Arid Zone62, 83–89. doi: 10.59512/aaz.2023.62.1.9
29
Molla M. Saha M. Serba D. Talukdar S. Salam M. Ahmed M. et al . (2023). Phylogenetic Relationship among Grasses, Wheat and Pearl Millet using matK and rbcR Genes. Plant Tissue Culture Biotechnol.33, 57–70. doi: 10.3329/ptcb.v33i1.66904
30
Pan J. Li Z. Dai S. Ding H. Wang Q. Li X. et al . (2020). Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep.10, 13660. doi: 10.1038/s41598-020-70520-1
31
Pan L. Zhou T. Chen C. Xu H. Wang W. (2024). Phytochemistry, pharmacology, and traditional medicine applications of juniperus sabina L.: A comprehensive overview. Molecules29, 5876. doi: 10.3390/molecules29245876
32
Pandey S. Mittal P. Singh G. (2023). Phyto-pharmacological importance with nutritional potential of eleusine coracana linn.: A review. Int. J. Pharm. Invest.13, 1–10. doi: 10.5530/ijpi.13.4.086
33
Pandohee J. Kyereh E. Kulshrestha S. Xu B. Mahomoodally M. (2023). Review of the recent developments in metabolomics-based phytochemical research. Crit. Rev. Food Sci Nutr.63, 3734–3749. doi: 10.1080/10408398.2021.1993127
34
Pujari N. Hoskeri J. (2022). Minor millet phytochemicals and their pharmacological potentials. Pharmacognosy Rev.16, 101. doi: 10.5530/phrev.2022.16.15
35
Saxena A. Srivastava D. Srivastava A. Garg S. (2024). Suitability of underutilized aquatic cash crop Euryale ferox Salisb. for low-land agriculture and enhancing farmer’s income. Vegetos1, 1–8. doi: 10.1007/s42535-024-01030-y
36
Sena J. Rodrigues S. Sakumoto K. Inumaro R. González P. Mendez E. et al . (2024). Antioxidant activity, antiproliferative activity, antiviral activity, NO production inhibition, and chemical composition of essential oils and crude extracts of leaves, flower buds, and stems of tetradenia riparia. Pharmaceuticals17, 888. doi: 10.3390/ph17070888
37
Sharma V. Tomar M. S. Sahoo S. Pradhan R. C. (2024). Development of barnyard millet flour rich in bioactive compounds with enhanced functional properties by application of multipin atmospheric cold plasma. J. Food Process Eng.47 (6), e14667. doi: 10.1111/jfpe.14667
38
Sharma S. Bharti A. Goswami S. Mallubhotla (2023). Diversity, antimicrobial, antioxidant, and anticancer activity of culturable fungal endophyte communities in cordia dichotoma. Molecules28, 6926. doi: 10.3390/molecules28196926
39
Sharma M. Mallubhotla S. (2022). Diversity, antimicrobial activity, and antibiotic susceptibility pattern of endophytic bacteria sourced from Cordia dichotoma L. Front. micro13. doi: 10.3389/fmicb.2022.879386
40
Singh P. Singh R. Bhadauria V. Singh H. (2024). The functionality and extraction of protein from sorghum, finger millet, and Kodo millet: a review. Int. J. Food Sci Technol.59, 512–521. doi: 10.1111/ijfs.16747
41
Soujanya K. Gurusiddaiah S. Jayadeep A. (2024). UPLC-ESI-HRMS/MS phenolic characterization, antioxidant activity, and proximate composition of drum-dried finger millet (Eleusine coracana) seed coat-and endosperm-rich fractions. ACS Food Sci Technol.5, 85–94. doi: 10.1021/acsfoodscitech.4c00581
42
Spina R. Saliba S. Dupire F. Ptak A. Hehn A. Piutti S. Laurain-Mattar D. (2021). Molecular identification of endophytic bacteria in Leucojum aestivum in vitro culture, NMR-based metabolomics study and LC-MS analysis leading to potential Amaryllidaceae alkaloid production. Int. J. Mol. Sci.22 (4), 1773. doi: 10.3390/ijms22041773
43
Sun L. Liu L. Wang Y. Feng Y. Yang W. Wang D. et al . (2022). Integration of metabolomics and transcriptomics for investigating the tolerance of foxtail millet (Setaria italica) to atrazine stress. Front. Plant Sci13. doi: 10.3389/fpls.2022.890550
44
Tamilselvan T. Sharma S. Prabhasankar P. (2023). Finger Millet (Eleusine coracana): Phytochemical Profile, Potential Health Benefits, and Techno-Functional Properties. In: Nutri-Cereals (119–139). ( CRC Press).
45
Theodoridis G. Gika H. Raftery D. Goodacre R. Plumb R. S. Wilson I. D. (2023). Ensuring fact-based metabolite identification in liquid chromatography–mass spectrometry-based metabolomics. Anal. Chem.95 (8), 3909–3916. doi: 10.1021/acs.analchem.2c05192
46
Ulanowska M. Olas B. (2021). Biological properties and prospects for the application of eugenol—a review. Int. J. Mol. Sci.22 (7), 3671. doi: 10.3390/ijms22073671
47
Vikash K. Yadav M. Awala S. K. Valombola J. S. Saxena M. S. Ahmad F. et al . (2024). Millets: A nutritional powerhouse for ensuring food security. Planta260(4), 101.
48
Vishal S. Tomar M. S. Sahoo S. Pradhan R. C. . (2024). Development of barnyard millet flour rich in bioactive compounds with enhanced functional properties by application of multipin atmospheric cold plasma. J. Food Process Engineer. 46(6), e14667. doi: 10.1111/jfpe.14667
49
Wu W. Zhang L. Zheng X. Huang Q. Farag M. Zhu R. et al . (2022). Emerging applications of metabolomics in food science and future trends. Food Chemistry: X16, 100500. doi: 10.1016/j.fochx.2022.100500
50
Xiang J. Apea F. Ndolo V. Katundu M. Beta T. (2019). Profile of phenolic compounds and antioxidant activity of finger millet varieties. Food Chem.275, 361–368. doi: 10.1016/j.foodchem.2018.09.120
51
Yadav O. P. Singh D. V. Kumari V. Prasad M. Seni S. Singh R. K. et al . (2024). Production and cultivation dynamics of millets in India. Crop Sci.64(5), 2459–2484. doi: 10.1002/csc2.21207
52
Yang X. Zhang W. Lan Y. Zhang J. Zheng W. Wu J. et al . (2024). An investigation into the effects of various processing methods on the characteristic compounds of highland barley using a widely targeted metabolomics approach. Food Res. Int.180, 114061. doi: 10.1016/j.foodres.2024.114061
53
Ye L. Zhang B. Zhou J. Yang X. Zhang X. Tan W. et al . (2024). LC-MS/MS-based targeted amino acid metabolic profile of Auricularia cornea grown on pinecone substrate. Food Chem.432, 137247. doi: 10.1016/j.foodchem.2023.137247
54
Zhou J. Zhong L. (2022). Applications of liquid chromatography-mass spectrometry based metabolomics in predictive and personalized medicine. Front. Mol. Biosci.9, 1049016. doi: 10.3389/fmolb.2022.1049016
55
Zhong P. Wei X. Li X. Wei X. Wu S. Huang W. et al . (2022). Untargeted metabolomics by liquid chromatography‐mass spectrometry for food authentication: A review. Compre. Rev. Food Sci. Food Safety21(3), 2455–2488. doi: 10.1111/1541-4337.12938
Summary
Keywords
metabolomic profiling, bioactive compounds, ragi, nutraceutical, phenolics
Citation
Gupta V, Sharma M, Gupta SK, Javeria S, Amin Z, Ashraf S, Khan JM and Udavant RN (2025) Metabolomic profiling of finger millet: unlocking the secrets of a nutritious staple food. Front. Plant Sci. 16:1570787. doi: 10.3389/fpls.2025.1570787
Received
05 February 2025
Accepted
28 September 2025
Published
06 November 2025
Volume
16 - 2025
Edited by
Wricha Tyagi, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Reviewed by
Wei Cai, Hunan University of Medicine, China
Rasmieh Hamid, Education and Extension Organization (AREEO), Iran
Updates
Copyright
© 2025 Gupta, Sharma, Gupta, Javeria, Amin, Ashraf, Khan and Udavant.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahima Sharma, mahisharma2403@gmail.com
†These authors have contributed equally to this work
ORCID: Vishal Gupta, orcid.org/0000-0002-6353-0792; Mahima Sharma, orcid.org/0009-0000-5220-1961; Sushil Kumar Gupta, orcid.org/0009-0009-9745-9523; Shaily Javeria, orcid.org/0000-0003-3815-9896; Zakir Amin, orcid.org/0000-0003-2702-6043; Suhail Ashraf, orcid.org/0000-0002-5851-3832
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.