- 1State Key Laboratory of Crop Stress Resistance and High-Efficiency Production, College of Agronomy, Northwest A&F University, Yangling, China
- 2Hainan Research Institute of Northwest A&F University, Sanya, China
- 3State Key Laboratory of Crop Stress Adaptation and Improvement, Henan Joint International Laboratory for Crop Multi-Omics Research, School of Life Sciences, Henan University, Kaifeng, China
Gray leaf spot (GLS), caused by Cercospora zeae-maydis and Cercospora zeina, is a devastating foliar disease affecting maize production worldwide. However, the defense mechanisms underlying GLS resistance are poorly understood. A major quantitative trait locus (QTL), Qgls8, associated with GLS resistance, was previously identified on maize chromosome 8 bin 8.06. Here we conducted transcriptome analyses on leaves from a pair of B73-teosinte near-isogenic lines (NILs) with contrasting Qgls8 alleles infected with C. zeina at 0, 4, 8, and 12 hours post inoculation (hpi). A total of 1225 up-regulated genes (URGs) were identified in the resistant line Qgls8-R compared with the susceptible line Qgls8-S across the four time points. By contrast, 908 URGs were identified in Qgls8-S. The URGs in Qgls8-R were significantly enriched in metabolic processes, phytohormone signaling, and response to biotic stress, while the URGs in Qgls8-S were mostly involved in plant growth and developmental processes. Additionally, C. zeina-induced URGs were consistently enriched in terpenoid metabolism and jasmonic acid (JA) signaling. Terpene- and JA-related genes showed increased expression at least at one time point after C. zeina infection which were confirmed by RT-qPCR. Furthermore, metabolite quantification indicated higher levels of JA and its isoleucine conjugate (JA-Ile) in Qgls8-R compared to Qgls8-S. Weighted gene co-expression network analysis (WGCNA) identified the module “turquoise”, which exhibited the highest positive correlation with Qgls8-R and was related to JA signaling. These findings suggest that the defense response mediated by terpenoid metabolism and the JA signaling pathway plays crucial roles in enhancing GLS resistance following C. zeina infection.
1 Introduction
Plants often initiate their innate immune systems to respond to pathogen attacks, including pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006; Couto and Zipfel, 2016; Yuan et al., 2021; Ngou et al., 2022a, 2022). PTI is triggered by the perception of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) via pattern recognition receptors (PRRs) on the surface of plant cells. ETI is triggered by the recognition of effectors via intracellular nucleotide-binding domain, leucine-rich-repeat containing receptors (NLRs), thereby leading to hypersensitive response (HR) (Tsuda and Katagiri, 2010; Cui et al., 2015; Couto and Zipfel, 2016; Peng et al., 2018). Plant responses to pathogens involve the production of phytohormones and phytoalexins (Tsuda and Katagiri, 2010; Yuan et al., 2021; Zhang and Dong, 2022). Upon the perception of pathogen infection, the phytohormone jasmonic acid (JA) conjugates with isoleucine to activate jasmonoyl-isoleucine (JA-Ile)-triggered signal transduction (Chini et al., 2007; Thines et al., 2007; Fonseca et al., 2009; Sheard et al., 2010; Zhang et al., 2015). In maize, the major phytoalexins belong to terpenoids, including sesquiterpenes such as zealexins and α/β-costic acids, as well as diterpenes such as kauralexins and dolabralexins, which usually restrict the growth of pathogens (Huffaker et al., 2011; Ding et al., 2017; Mafu et al., 2018; Ding et al., 2019).
Maize (Zea mays L.) is one of the key staple crops for feeding the growing global population. Gray leaf spot (GLS), one of the most devastating foliar diseases in maize worldwide, is caused by the necrotrophic fungi Cercospora zeae-maydis and Cercospora zeina. Since its first discovery in Alexander County, Illinois, USA, GLS has been prevalent in the United States, South Africa, Brazil and China due to the expansion of the causal fungal pathogens, resulting in a yield loss of up to 60% (Ward et al., 1997, 1999; Dunkle and Levy, 2000; Liu and Xu, 2013; Nsibo et al., 2019). GLS resistance is predominantly inherited as a quantitative trait, and a number of quantitative trait loci (QTLs) have been identified on all the 10 maize chromosomes (Zhang et al., 2012; Berger et al., 2014; Xu et al., 2014; Benson et al., 2015; Zhang et al., 2017; Du et al., 2020; Lv et al., 2020). Among them, only five genes have been cloned, including ZmWAKL (Cell-wall-associated receptor kinase-like protein gene) (Zhong et al., 2024) and ZmWAK02 (RD wall-associated kinase gene) (Dai et al., 2024), as well as three multiple disease resistance genes, ZmCCoAOMT2 (Caffeoyl-CoA O-methyltransferase gene (Yang et al., 2017), ZmMM1 (Mexicana lesion mimic 1) (Wang et al., 2021), and ZmCPK39 (Calcium-dependent protein kinase gene) (Zhu et al., 2024).
Teosinte, as the ancestor of modern maize, possesses potential rare novel alleles for maize improvement (Piperno et al., 2009; Kistler et al., 2018; Yang N. et al., 2023), such as UPA2 (Upright Plant Architecture2) for leaf angle (Tian et al., 2019), and THP9 (TEOSINTE HIGH PROTEIN 9) for seed protein content (Huang et al., 2022). In maize disease resistance, the teosinte-derived ZmMM1 allele confers enhanced resistance to northern leaf blight, GLS, and southern corn rust (Wang et al., 2021). Six QTLs for GLS resistance have been identified using a population of 693 teosinte introgressed near-isogenic lines (NILs) in maize inbred line B73 background, which are located in bins 2.04, 3.06, 4.07, 5.03, 8.06, and 9.03 (Lennon et al., 2016; Liu et al., 2016). The QTL in bin 8.06 has been fine-mapped to an approximately 130-kb interval, which were designated as Qgls8 (Zhang et al., 2017). A pair of B73-teosinte NILs, hereafter named as Qgls8-R/Qgls8-S, carrying different teosinte accession introgressions in the Qgls8 region, exhibited a significant difference for GLS resistance (Lennon et al., 2016; Zhang et al., 2017).
This study aimed to elucidate the molecular mechanisms underlying GLS resistance in maize. We conducted a comprehensive transcriptome analysis using the two NILs Qgls8-R and Qgls8-S, which were inoculated with the fungal pathogen C. zeina. The comparative transcriptome analysis was performed at four time points after C. zeina infection to identify up-regulated genes (URGs) specific to each line. Functional analysis of these URGs revealed several critical pathways involved in defense signaling against C. zeina. The activation of these signaling pathways was confirmed by quantitative analyses of representative genes expression and metabolites accumulation levels during pathogen infection. Furthermore, weighted gene co-expression network analysis (WGCNA) was performed across 24 samples. These findings contribute to a deeper understanding of the molecular mechanisms underlying GLS resistance.
2 Materials and methods
2.1 Plant materials
Qgls8, a QTL for GLS resistance located in maize bin 8.06, has been previously reported (Zhang et al., 2017). A pair of NILs was developed by introgressing teosinte accessions into the B73 background, which carry different teosinte alleles at Qgls8 region, termed as Qgls8-R and Qgls8-S (Lennon et al., 2016; Liu et al., 2016). Qgls8-R and Qgls8-S were provided by Dr. Peter Balint-Kurti at the Department of Entomology and Plant Pathology of North Carolina State University. Qgls8-R exhibited increased GLS resistance, while Qgls8-S exhibited decreased GLS resistance. Transcriptome sequencing was carried out on both Qgls8-R and Qgls8-S. The genomic DNA was extracted from both Qgls8-R and Qgls8-S. The Maize6H-60K SNP array was used to detect the genetic background of the two NILs.
2.2 GLS resistance evaluation in field trials
In the summers of 2023 and 2024, Qgls8-R and Qgls8-S were planted in Tongchuan, Shaanxi Province of China, where GLS occurs naturally each year. The field experiment was set up with two replicates of each line in both years. In all trials, each replicate was made up of three rows. Fifteen seeds were sown in a single row which was 3 m in length and spaced 0.6 m apart. Disease scoring was performed three times after anthesis. The interval between each disease scoring was approximately 10 days. The GLS symptoms were scored on a scale of 1 to 9, where “1” for the most susceptible score indicated that everything was completely dead; 2 indicated that almost all of the plant tissues had withered; 3 indicated that the leaf above the ear was withered; 4 indicated that spots of the leaf above the ear had merged together; 5 indicated that spots on the ear leaf had merged together; 6 indicated that many spots appeared on the ear leaf and was separate from each other; 7 indicated that a few spots appeared on the ear leaf; 8 indicated that a few spots appeared on the leaf below the ear; and “9” for the most resistant score indicated that plants showed no disease symptom (Lennon et al., 2016; Sermons and Balint-Kurti, 2018).
2.3 Artificial inoculation and sample collection
The two NILs, Qgls8-R and Qgls8-S, were grown in greenhouse at 25 °C under 14 h light/10 h dark conditions. C. zeina was isolated from maize leaves with GLS symptoms in the fields of Baoji, Shaanxi Province, China. The pathogenic fungi were cultured for ten days on MLPCA medium (20 g/L maize leaf powder + 2 g/L CaCO3 + 15 g/L agar) in the dark at 25 °C. The spores of C. zeina were collected using the elution solution (0.5 g/L agar + 0.5 mL/L Tween 20; autoclaved at 121°C). The spore suspension was diluted to 5×104 conidia/mL and sprayed on both sides of the fourth leaf of inoculated plants at the four-leaf stage. Subsequently, the inoculated plants were humidified for 24 hours using humidifier. C. zeina-inoculated leaves were sampled at 0, 4, 8, and 12 hours post inoculation (hpi) for transcriptome sequencing.
2.4 Transcriptome sequencing and analysis
The fourth leaves of maize plants inoculated with C. zeina were sampled at 0, 4, 8, and 12 hpi. Three biological replicates were set for each time point, and each biological replicate was derived from three independent plants. All samples were sequenced based on Illumina X plus platform using a 150 bp pair-end sequencing strategy in Biomarker Technologies. Clean reads were mapped to B73_v4 reference genome (Zm-B73-reference-4.0 genome. fasta) using STAR 2.7.10a. The number of counts per gene was obtained using StringTie. Gene expression was normalized to transcripts per million (TPM). DEseq2 was used to screen URGs with a significance threshold of Fold Change > 1.5 and padj ≤ 0.05 (Lecourieux et al., 2020). URGs were identified in Qgls8-R compared with Qgls8-S or in Qgls8-S compared with Qgls8-R. URGs responding to C. zeina infection at 4, 8, and 12 hpi were selected by comparing the expression levels at three time points with those 0 hpi for either Qgls8-R or Qgls8-S (4 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 8 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 12 hpi vs 0 hpi in Qgls8-R or Qgls8-S).
Weighted gene co-expression network analysis (WGCNA) was performed on all expressed genes across 24 samples using the R package. The analyzed genes met the following criteria: an average TPM value exceeding 1 and a ranking within the top 20,000 genes in descending order according to Mean Absolute Deviation (MAD) across 24 samples. A soft thresholding power of 22 was used to calculate the adjacency matrix and topological overlap matrix (TOM) using Pearson’s correlation. Gene modules with at least 30 genes were detected using the dynamic cutting algorithm and merged with a default cutoff value (0.25).
2.5 Quantitative analysis of jasmonic acid, salicylic acid and their derivatives
The C. zeina-inoculated fourth leaves of Qgls8-R and Qgls8-S were collected at 0 hpi and 24 hpi. For each sample, three biological replicates were set up, and each biological replicate consisted of five plants. Salicylic acid (SA), salicylic acid 2-O-β-glucoside (SAG), jasmonic acid (JA), jasmonoyl-L-isoleucine (JA-Ile), cis(+)-12-oxophytodienoic acid (OPDA), 2, 3-dihydroxybenzoic acid (2,3-DHBA), 2, 5-dihydroxybenzoic acid (2,5-DHBA), and L-phenylalanine (Phe) were extracted and analyzed in Henan University. They were quantified by ultra-high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Samples were analyzed using a Xevo TQ-XS system (Waters, USA) equipped with an ESI ion source. Data analysis was performed using the spectrometer software (Masslynx v.4.2).
2.6 Gene expression analysis by RT‐qPCR
Total RNAs of maize leaves were extracted using E.Z.N.A.™ Plant RNAKit (OMEGA) and converted into cDNA using FastKing RT SuperMix (Tiangen) following the manufacturer’s instructions. The cDNA of all samples was diluted six times as a template. Gene-specific primers were designed using Primer 5.0 (Supplementary Table S14). RT‐qPCR was performed using RealUniversal Color PreMix (Tiangen) and run on QuantStudio™ 7 Flex system. The maize ZmEF1α was used as internal control.
3 Results
3.1 Phenotypic evaluation of Qgls8-R and Qgls8-S for GLS resistance
Using NIL populations in which segments of the teosinte genome had been introgressed into the maize B73 background, the QTL Qgls8 for GLS resistance was previously mapped into a ~130-kb region in bin 8.06 (Lennon et al., 2016; Zhang et al., 2017). A pair of NILs Qgls8-R and Qgls8-S was developed from segregating populations derived from the NIL parents Z032E0081 and Z033E0056 with contrasting teosinte alleles at Qgls8. The allele from Z032E0081 confers increased GLS resistance, while allele from Z033E0056 confers decreased GLS resistance compared with B73 (Zhang et al., 2017). Assessment of the genetic backgrounds using the Maize6H-60K array revealed that the similarity between Qgls8-R and Qgls8-S was about 95.75% (Supplementary Table S1). To evaluate the GLS resistance, Qgls8-R and Qgls8-S were infected by fungal pathogens of GLS naturally in the field trials in Tongchuan, Shaanxi Province in 2023 and 2024. Based on the 1–9 scale assessment, the GLS score of Qgls8-R was significantly higher than that of Qgls8-S (Figures 1A, B). These results indicate that the allele from Qgls8-R confers enhanced resistance to GLS in the field.
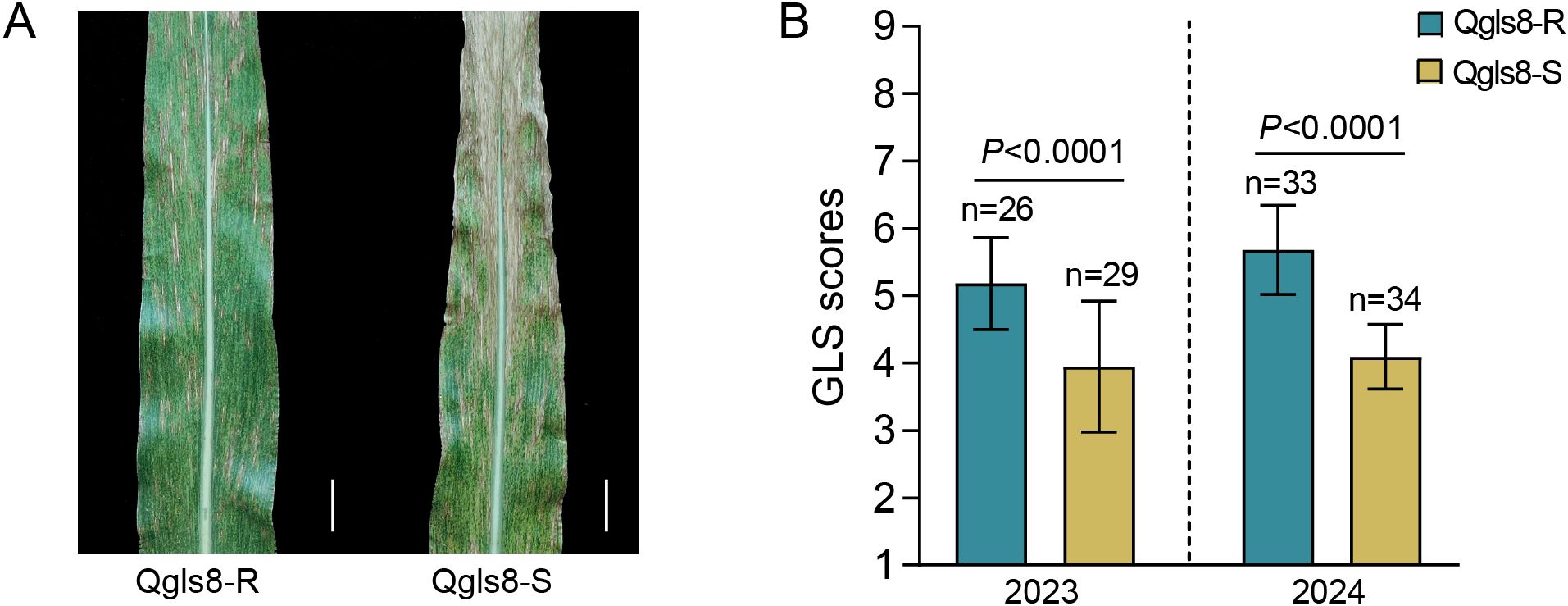
Figure 1. Phenotype evaluation of Qgls8-R and Qgls8-S for GLS resistance. (A) GLS symptom of Qgls8-R and Qgls8-S in field trials. Scale bars corresponds to 2.5 cm. (B) GLS scores of Qgls8-R and Qgls8-S. Disease scoring, 1–9 scales; 1 indicates the most susceptible score and 9 indicates the most resistant score. Data are presented as all the mean values ± SD. Significant differences were determined by Student’s t-test. The “n” indicates the number of plants for GLS assessment.
3.2 Transcriptome profiling of the NILs pair in response to C. zeina infection
A total of 24 samples were collected from Qgls8-R and Qgls8-S at 0, 4, 8, and 12 hpi for transcriptome profiling. The mapping rates of the 24 transcriptome datasets of both Qgls8-R and Qgls8-S ranged from 93.94% to 96.61% (Supplementary Table S2). Principal Component Analysis (PCA) revealed that the biological replicates of each sample grouped together (Supplementary Figure S1). Differential expression was conducted to explore the transcriptome differences between Qgls8-R and Qgls8-S. A total of 1,225 URGs were identified in Qgls8-R compared with Qgls8-S, which included 350, 639, 535, and 339 URGs at 0 hpi, 4 hpi, 8 hpi, 12 hpi, respectively. While there were 908 URGs in Qgls8-S compared with Qgls8-R, including 205, 561, 229, and 279 URGs at 0 hpi, 4 hpi, 8 hpi, and 12 hpi, respectively (Figures 2A, B). The highest number of URGs was found at 4 hpi in both Qgls8-R and Qgls8-S, indicating that 4 hpi might be a key time point for Qgls8-R and Qgls8-S to defend against C. zeina attack.
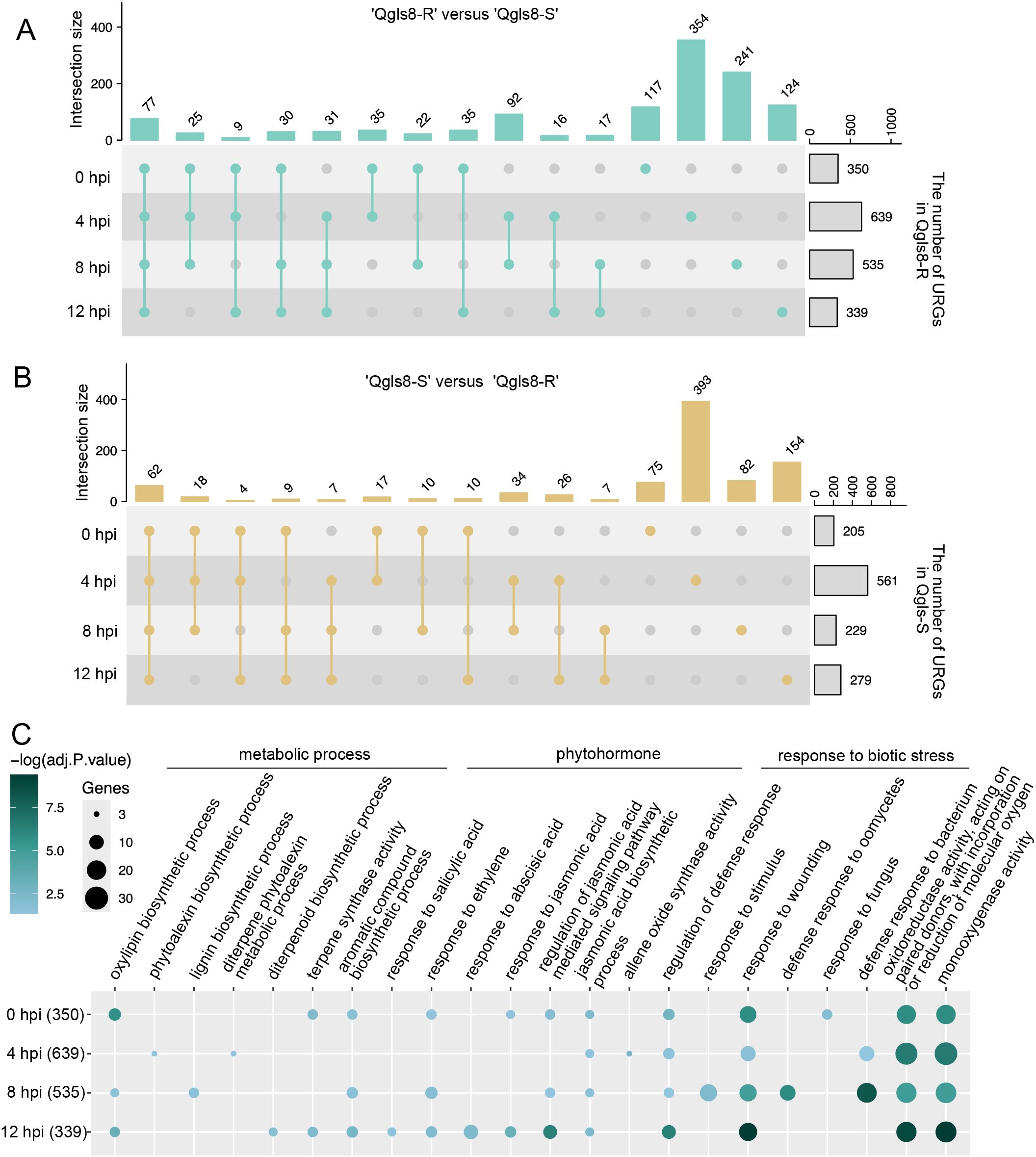
Figure 2. Comparison analysis of gene expression at the four time points after inoculation with C. zeina between Qgls8-R and Qgls8-S. (A) The number of up-regulated genes (URGs, |Log2FoldChange| > 0.58, padj ≤ 0.05) from ‘Qgls8-R versus Qgls8-S’ among the four comparison groups (Qgls8-R vs Qgls8-S at 0 hpi, 4 hpi, 8 hpi and 12 hpi). (B) The number of URGs from ‘Qgls8-S versus Qgls8-R’ among the four comparison groups (Qgls8-S vs Qgls8-R at 0 hpi, 4hpi, 8 hpi and 12 hpi). (C) GO analysis of the URGs from ‘Qgls8-R versus Qgls8-S’ at four time points. All the GO terms were shown with a P-value less than 0.05. URGs were annotated into three categories: biological processes, cellular components, and molecular functions.
Gene Ontology (GO) analysis was performed to identify enriched GO terms for URGs in Qgls8-R and Qgls8-S, respectively. The 350 URGs at 0 hpi in Qgls8-R were enriched for defense response associated processes including response to fungus, JA biosynthesis process, and response to wounding (Figure 2C; Supplementary Table S3). Similarly, the URGs at 4, 8, and 12 hpi in Qgls8-R were related to the JA biosynthesis process and response to wounding as well (Figure 2C, Supplementary Table S3). In contrast, the 205 URGs at 0 hpi in Qgls8-S did not show significant enrichment in any GO terms. Notably, the URGs at 4, 8, and 12 hpi in Qgls8-S were mostly involved in plant growth-associated processes including photosynthesis, chloroplasts, and DNA replication (Supplementary Table S3). The GO enrichment analysis revealed that the URGs in Qgls8-R were mainly involved in defense response, which may contribute to its GLS resistance.
3.3 C. zeina-induced URGs displayed significantly higher expression in Qgls8-R than in Qgls8-S and were associated with defense signaling pathways
A total of 350 URGs were identified in Qgls8-R compared with Qgls8-S at 0 hpi (Figure 2A). To further identify C. zeina-induced URGs, we compared the expression changes of both Qgls8-R and Qgls8-S at the three time points (4 hpi, 8 hpi, and 12 hpi) to their respective 0 hpi (4 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 8 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 12 hpi vs 0 hpi in Qgls8-R or Qgls8-S) (Figure 3A). We found 19 out of the 350 URGs were up-regulated by C. zeina infection exclusively in Qgls8-R and 137 URGs were induced in both Qgls8-R and Qgls8-S (Figure 3B; Supplementary Table S4). In total, 156 URGs which displayed higher expression levels in Qgls8-R and were induced by C. zeina were performed for GO enrichment analysis. These URGs were mostly related to monooxygenase activity, response to wounding, terpene synthase activity, lignin biosynthetic process, and oxylipin biosynthetic process (Figure 3C; Supplementary Table S5).
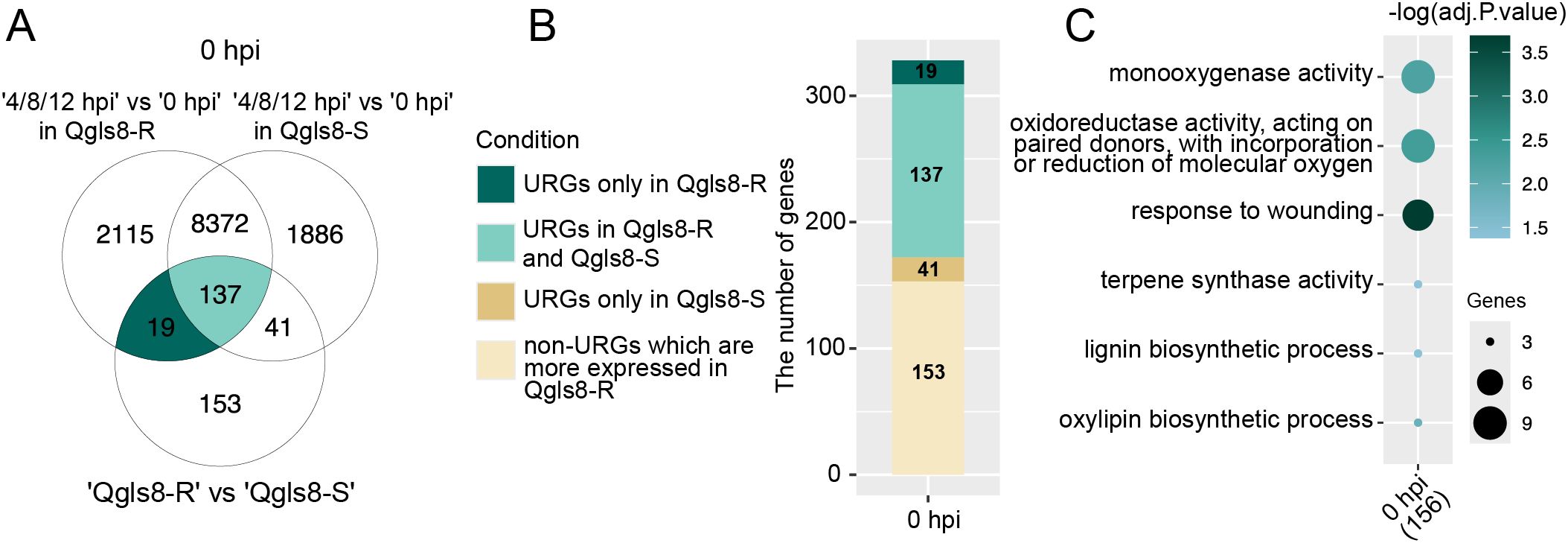
Figure 3. Identification of C. zeina-induced URGs that were more highly expressed in Qgls8-R at 0 h after C. zeina infection. (A) Upset plots displaying the number of overlapping URGs in seven comparison groups. (B) The number of URGs and non-URGs induced by C. zeina among 350 URGs identified in Qgls8-R compared with Qgls8-S at 0 hpi. (C) GO enrichment analysis of 156 genes that were not only up-regulated induced by C. zeina but also expressed at a higher level in Qgls8-R compared with Qgls8-S at 0 hpi. All the GO terms were shown with a P-value less than 0.05. URGs were annotated into three categories: biological processes, cellular components, and molecular functions.
To further analyze C. zeina-induced URGs, we identified genes exhibiting up-regulated response to the pathogen and higher expression after C. zeina inoculation in Qgls8-R compared with Qgls8-S. C. zeina-induced URGs at 4, 8, and 12 hpi for either Qgls8-R or Qgls8-S were selected by comparing the expression levels at three time points to 0 hpi (4 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 8 hpi vs 0 hpi in Qgls8-R or Qgls8-S, 12 hpi vs 0 hpi in Qgls8-R or Qgls8-S). Furthermore, by comparing expression at the three time points between Qgls8-R and Qgls8-S (Qgls8-S 4 hpi vs Qgls8-R 4 hpi, Qgls8-S 8 hpi vs Qgls8-R 8 hpi, Qgls8-S 12 hpi vs Qgls8-R 12 hpi), the URGs displaying higher expression in Qgls8-R at 4 hpi, 8 hpi, and 12 hpi were identified. There were 135, 129, and 44 URGs exclusive to Qgls8-R at 4 hpi, 8 hpi, and 12 hpi, respectively (Figures 4A, B; Supplementary Tables S6-S8). A total of 175, 137, and 93 URGs were identified in Qgls8-R and Qgls8-S at 4 hpi, 8 hpi, and 12 hpi, respectively (Figure 4A, B; Supplementary Tables S6-S8). The expression profiling of these genes revealed that all URGs had the lowest expression at 0 hpi of Qgls8-R and Qgls8-S (Figure 4C; Supplementary Tables S6-S8). URGs exclusive to Qgls8-R were up-regulated at 4 hpi, 8 hpi, and 12 hpi, reaching higher expression levels in Qgls8-R after C. zeina infection (Figure 4C, Supplementary Tables S6-S8), whereas the expression of those genes remained unchanged at 4, 8, and 12 hpi in Qgls8-S after C. zeina infection (Figure 4C, Supplementary Tables S6-S8). The URGs in both Qgls8-R and Qgls8-S were up-regulated to significantly higher expression levels at 4, 8, and 12 hpi in Qgls8-R. In contrast, those genes showed slight up-regulation exclusive to Qgls8-S after C. zeina infection (Figure 4C; Supplementary Tables S6-S8). All URGs were functionally classified using GO analysis. The URGs of Qgls8-R were enriched in metabolic processes such as monooxygenase activity, oxidoreductase activity, terpene synthase activity, diterpenoid biosynthetic process, sterol metabolic process, and defense responses including against bacterium, fungus, oomycetes, wounding, regulation of defense response, JA biosynthetic process, regulation of JA mediated signaling pathway, etc. (Figure 5; Supplementary Table S9). Furthermore, we identified genes exhibiting specifically down-regulated in Qgls8-R response to C. zeina inoculation and higher expression in Qgls8-R compared with Qgls8-S at four time points. A few genes were found (Supplementary Figure S2). Given the limited number of genes in the subset, GO enrichment analysis was not carried out. These results indicate that Qgls8-R might enhance GLS resistance by upregulating genes related to terpenoid metabolism, the JA signaling pathway, and defense responses against C. zeina infection.
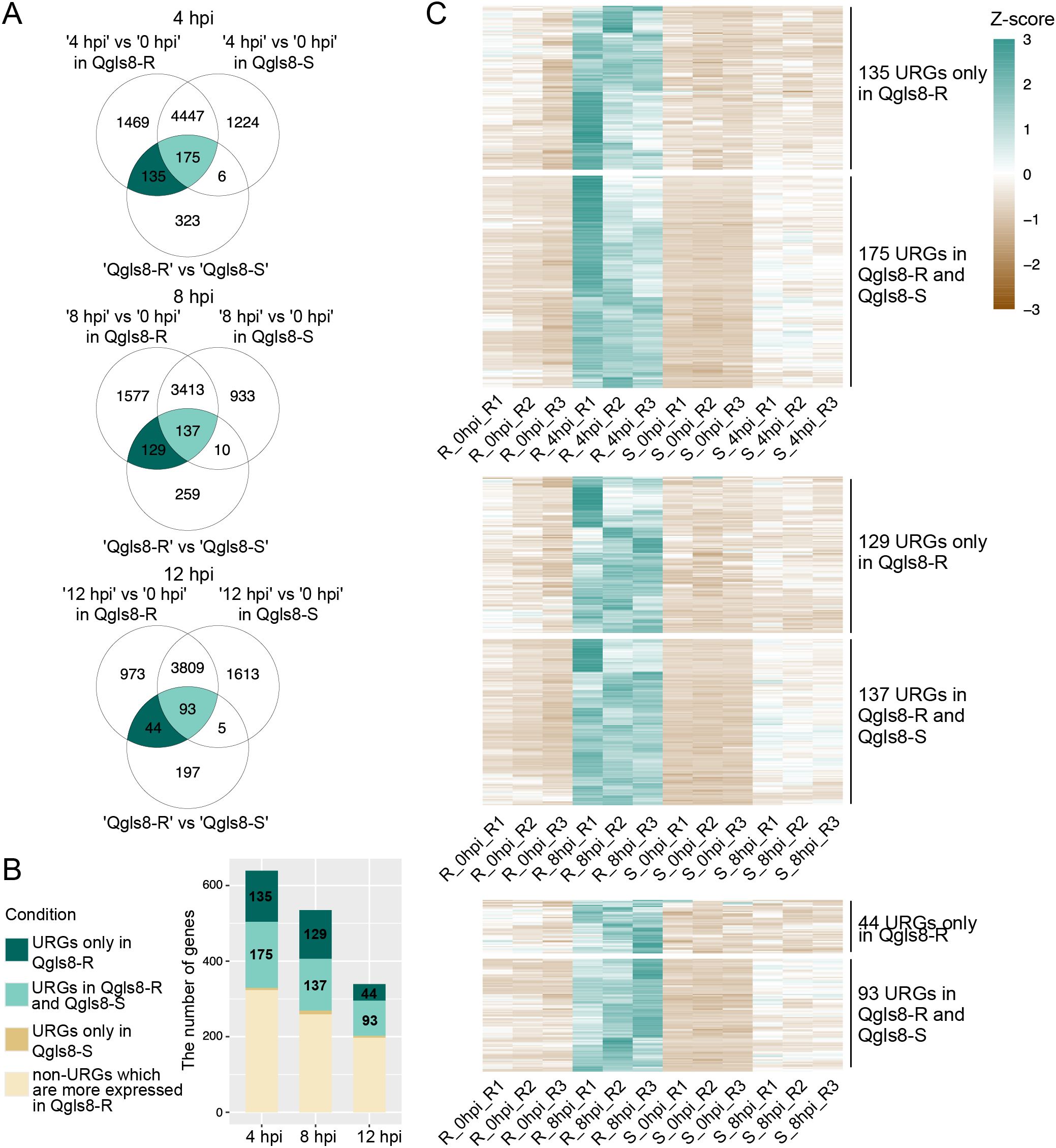
Figure 4. Identification of C. zeina-induced URGs that were expressed at a higher level in Qgls8-R compared with Qgls8-S at 4 h, 8 h, and 12 h post inoculation (hpi) with C. zeina. (A) Vene diagram showing the number of overlapping URGs in three comparison groups at 4 hpi, 8 hpi, and 12 hpi, respectively. (B) The number of URGs and non-URGs induced by C. zeina among 639, 535, and 339 URGs at 4 hpi, 8 hpi, and 12 hpi, respectively. (C) Expression profiles of URGs after C. zeina infection using transcripts per million (TPM). R, Qgls8-R; S, Qgls8-S.
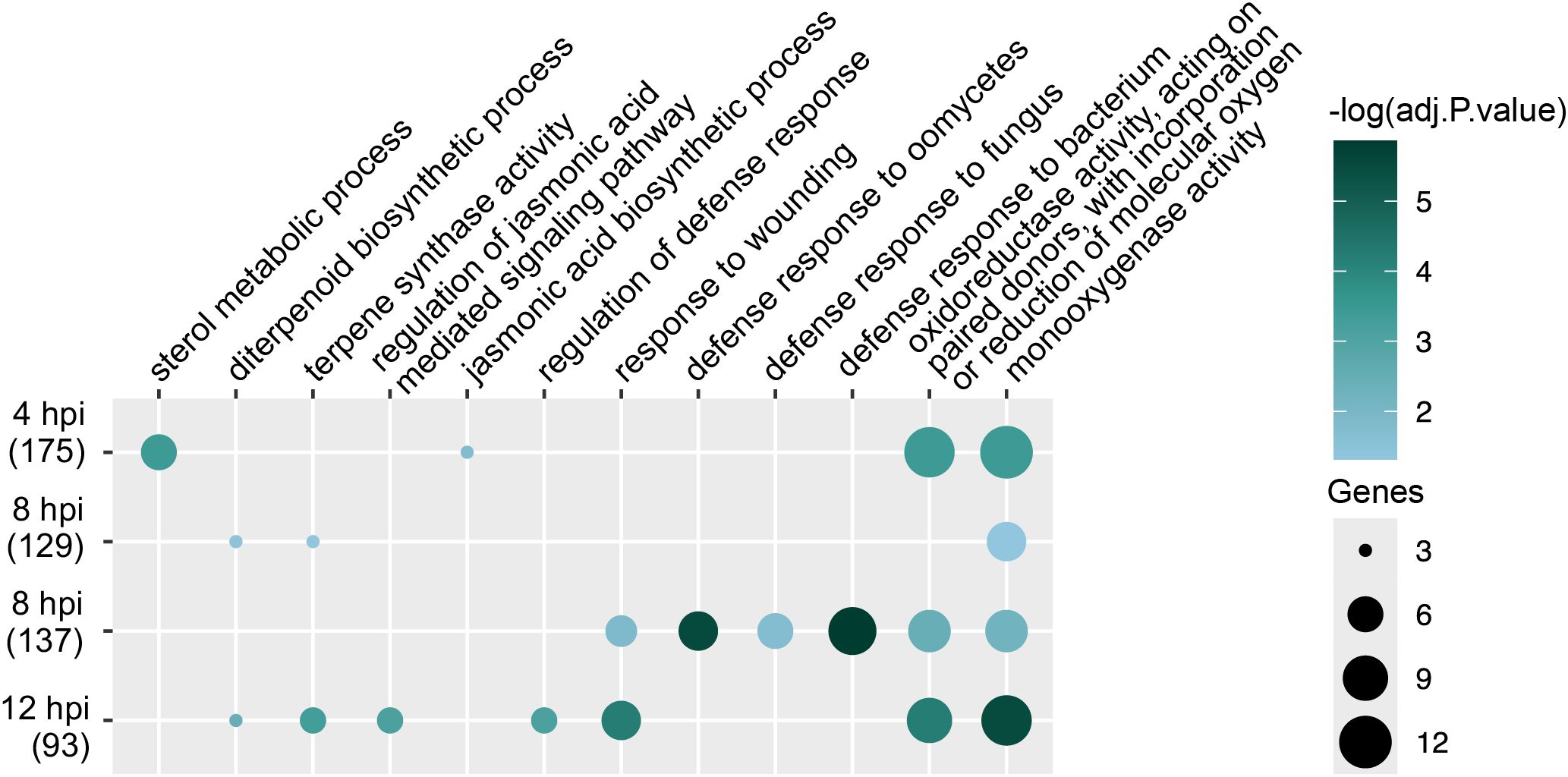
Figure 5. GO enrichment analysis of C. zeina-induced URGs at 4 hpi, 8 hpi, and 12 hpi, respectively. These genes were induced by C. zeina and expressed more highly in Qgls8-R compared with Qgls8-S at 4 hpi, 8 hpi, and 12 hpi, respectively. All the GO terms were shown with a P-value less than 0.05. URGs were annotated into three categories: biological processes, cellular components, and molecular functions.
3.4 Terpene-related genes were activated in response to C. zeina
GO enrichment analysis revealed that eight URGs were involved in terpenoid metabolic pathway, including ZmTPS1, ZmTPS7, ZmKSL2, ZmDLS, ZmCYP71Z18, ZmCYP92C5, Zm00001d034516, and Zm00001d043165. At the four time points in the NILs, ZmKSL2 and ZmCYP71Z18 showed higher expression at 4 hpi in Qgls8-R. The expression of genes including ZmTPS1, ZmTPS7, ZmDLS, ZmCYP92C5, Zm00001d034516, and Zm00001d043165 peaked at 12 hpi in Qgls8-R with ZmTPS1, ZmTPS7, Zm00001d034516, and Zm00001d043165 displaying a gradual transcriptional increase after C. zeina infection. In the susceptible line Qgls8-S, these genes did not exhibit specific response patterns (Figure 6A). All eight terpene-related genes were significantly induced at least at one infection time point in Qgls8-R and Qgls8-S with a significant increase at 12 hpi in both lines (Figure 6B). Furthermore, five genes involved in diterpenoid (kauralexins) pathways were induced by C. zeina (Figure 6C). The expression of ZmKSL2, ZmTPS7, and ZmCYP92C5 was also verified by RT‐qPCR (Figure 6D). These results suggest that Qgls8-R effectively responds to C. zeina infection through terpenoid metabolism.
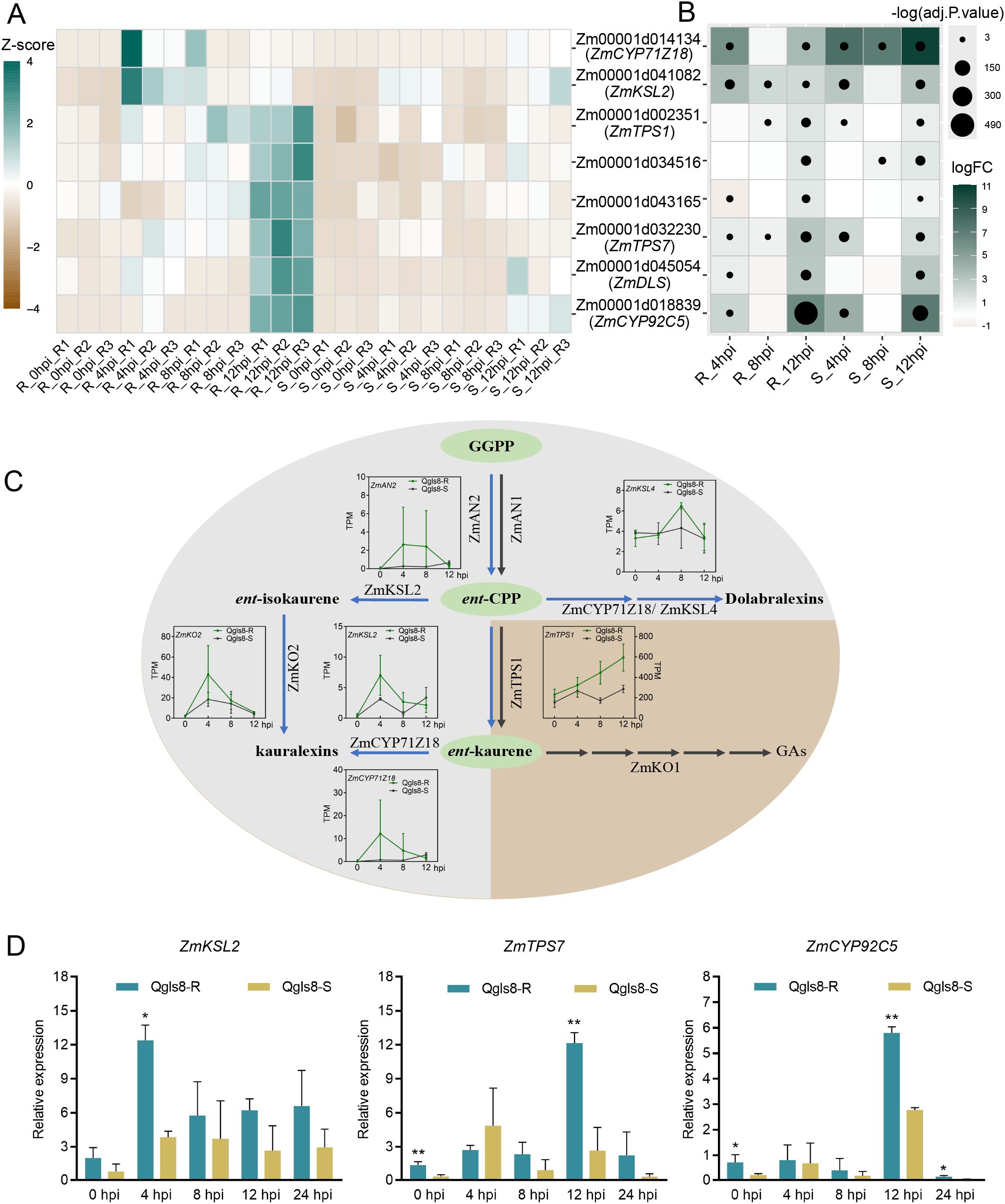
Figure 6. The expression of genes involved in terpenoid metabolism after C. zeina infection. (A) Expression of terpene-related genes at four time points using transcripts per million (TPM). (B) The expression analysis of terpene-related genes induced by C. zeina. Log2 fold-change of transcripts per million (TPM) at 4 hpi, 8 hpi, and 12 hpi relative to 0 hpi in Qgls8-R or Qgls8-S. The upregulated expression of genes is labeled with blue, the downregulated expression with yellow, and the unchanged expression with white, respectively. (C) Expression pattern analysis of genes involved in the biosynthesis of kauralexin and gibberellic acid (GA) in maize. Blue arrows, kauralexins pathways; gray arrows, GA pathways; TPM, transcripts per million. (D) The expression pattern analysis of ZmKSL2, ZmTPS7, and ZmCYP92C5 after C. zeina inoculation by RT-qPCR. Data are presented as the mean values ± SD from three repeats. Asterisks represent significant differences by Student’s t-test (*0.01 < P < 0.05, **P < 0.01).
3.5 Jasmonic acid-related genes were involved in resistance against C. zeina
GO analysis identified seven URGs associated with the JA signaling pathway, including ZmAOC1, ZmAOS2b, ZmAOS1c, ZmTIFY15, ZmJAZ3, ZmJAZ2, and ZmJAZ21. Among those, ZmAOC1, ZmAOS2b and ZmAOS1c displayed the highest expression levels at 4 hpi in both lines, with significantly higher levels in Qgls8-R (Figure 7A). Four genes showed a gradual increase in Qgls8-R, with ZmTIFY15 and ZmJAZ3 up-regulated from 4 hpi to 12 hpi, and ZmJAZ2 and ZmJAZ21 up-regulated from 8 hpi to 12 hpi (Figure 7A). In contrast, these four JA-related genes did not display specific expression patterns in the susceptible Qgls8-S (Figure 7A). Compared with 0 hpi, all seven genes were significantly up-regulated at least at one time point in both lines (Figure 7B). ZmAOC1, ZmAOS2b, and ZmAOS1c were significantly up-regulated at 4 hpi in both Qgls8-R and Qgls8-S, while ZmTIFY15, ZmJAZ3, ZmJAZ2, ZmJAZ21 were most significantly induced by C. zeina at 12 hpi with greater up-regulation in Qgls8-R (Figure 7B). Five of these genes including ZmAOS2b, ZmAOS1c, ZmAOC1, ZmJAZ3, and ZmTIFY15, were involved in three steps of the JA signaling pathway (Figure 7C). The expression of ZmAOC1, ZmTIFY15, and ZmJAZ3 was verified at 0 h, 4 h, 8 h, 12 h, and 24 h after C. zeina inoculation by RT-qPCR (Figure 7D).
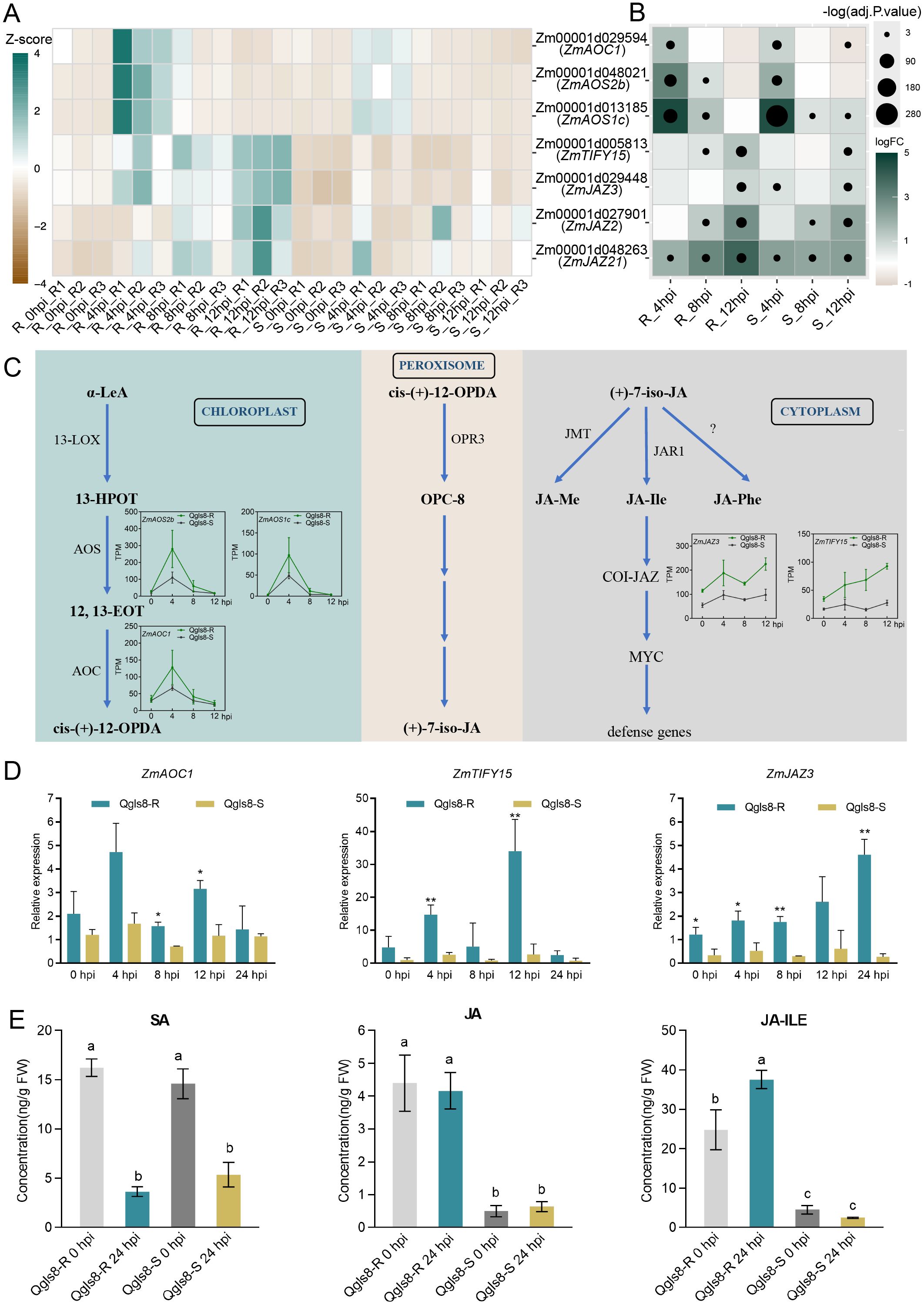
Figure 7. The expression of genes related to Jasmonic acid pathway after C. zeina infection. (A) Expression of Jasmonic acid-related genes at different infection time points using transcripts per million (TPM). (B) The expression analysis of Jasmonic acid-related genes induced by C. zeina. Log2 fold-change of TPM at 4 hpi, 8 hpi, and 12 hpi relative to 0 hpi in Qgls8-R or Qgls8-S. The upregulated expression of genes is labeled with blue, the downregulated expression with yellow, and the unchanged expression with white, respectively. (C) Expression patterns of up-regulated genes in jasmonic acid pathways. TPM, transcripts per million. (D) The expression pattern analysis of ZmAOC1, ZmTIFY15, and ZmJAZ3 after C. zeina inoculation by RT-qPCR. Data are presented as the mean values ± SD from three repeats. Asterisks represent significant differences by Student’s t-test (*0.01 < P < 0.05, **P < 0.01). Significant differences were determined by Student’s t-test. (E) The levels of SA, JA, and JA-Ile of both Qgls8-R and Qgls8-S at 0 hpi and 24 hpi. Data are presented as all the mean values ± SD. The letters indicate a significant difference (P< 0.05) by one-way ANOVA with Tukey’s test.
To further investigate the role of the JA pathway during C. zeina infection, we quantified JA, SA and their derivatives at 0 h and 24 h after C. zeina inoculation (Figure 7E, Supplementary Figure S3). SA showed no significant difference between Qgls8-R and Qgls8-S at both time points, but decreased after C. zeina infection (Figure 7E). Notably, JA and JA-Ile in Qgls8-R were significantly higher than in Qgls8-S at 0 hpi and 24 hpi with JA-Ile showing a significant increase at 24 hpi in Qgls8-R (Figure 7E). These results indicate that Qgls8-R positively responds to C. zeina through the JA signaling pathway.
3.6 Analysis of a gene co-expression network in response to C. zeina infection
Furthermore, gene co-expression network analysis was conducted to identify genes responsive to C. zeina infection using weighted gene co-expression network analysis (WGCNA). Twenty distinct modules were identified, with the largest containing 5,296 genes in the module “turquoise” and the fewest 47 genes in the module “royalblue” (Figures 8A, B, Supplementary Table S10). Among these, two modules (“midnightblue” with 208 genes and “salmon” with 273 genes) displayed opposite correlations with Qgls8-R and Qgls8-S at all four time points. The module “salmon” exhibited a positive correlation with Qgls8-R and a negative correlation with Qgls8-S (Figure 8B), associated with diverse enzyme activities, including monooxygenase activity, oxidoreductase activity, and negative regulation of endopeptidase activity (Supplementary Table S11). In contrast, the module “midnightblue” was not enriched in these terms, and showed a negative correlation with Qgls8-R and a positive correlation with Qgls8-S (Figure 8B, Supplementary Table S12). Notably, the module “turquoise” had the highest positive correlation with Qgls8-R at 4 hpi (Figure 8B), which was related to cell surface receptor signaling pathway, protein kinase activity and phosphorylation, as well as defense response related processes, including immune response, response to fungus, and JA biosynthetic process (Figure 8C, Supplementary Table S13). The genes in the module “turquoise” were regulated at 4 hpi in Qgls8-R and Qgls8-S (Supplementary Figure S4). Twelve genes involved in the JA signaling pathway were identified in the module “turquoise”, including three key enzyme-coding genes ZmAOS1c, ZmAOS2b, and ZmAOC1 (Supplementary Table S13). These results indicate that genes in the module “turquoise” might be an important component for the resistance of Qgls8-R against C. zeina.
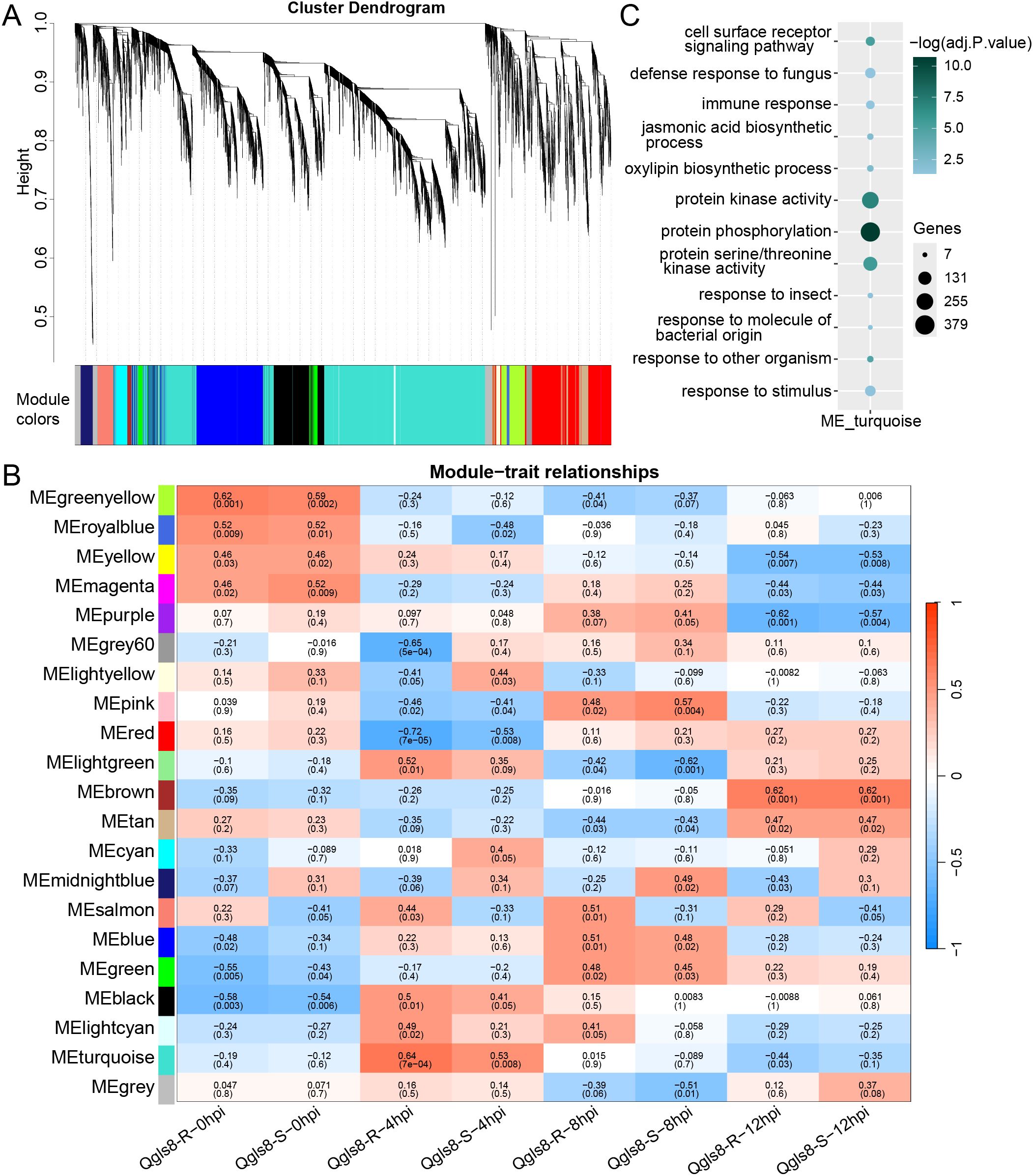
Figure 8. WGCNA of genes identified in both Qgls8-R and Qgls8-S at 0, 4, 8, and 12 hpi. (A) Hierarchical clustering tree of twenty distinct modules. Different colors are used to indicate each module. The genes that are not assigned to any module are indicated by the module “grey”. (B) Module-trait relationships. The module-trait correlation, ranging from -1 (blue) to 1 (red), is represented by the color scale. The corresponding correlation value is indicated on the top and the P-value is indicated on the bottom in each box. (C) GO enrichment analysis of genes identified in the module “turquoise”. All the GO terms were shown with a P-value less than 0.05. URGs were annotated into three categories: biological processes, cellular components, and molecular functions.
4 Discussion
GLS significantly threatens maize production, which is caused by C. zeae-maydis and C. zeina with necrotrophic lifestyles. Plant resistance to those necrotrophic pathogens relies on defense related secondary metabolites including camalexin, glucosinolates, kauralexins and zealexins (Kliebenstein et al., 2005; Ahuja et al., 2012; Burow and Halkier, 2017), as well as phytohormones, such as ethylene, JA, abscisic acid, and auxins (Berrocal-Lobo et al., 2002; Adie et al., 2007; Llorente et al., 2008; Chen et al., 2021). This study analyzed the transcriptional differences between Qgls8-R and Qgls8-S at 0, 4, 8, and 12 hpi in response to C. zeina. URGs of the resistant line Qgls8-R were related to metabolic processes, phytohormones, and response to biotic stresses, while those in the susceptible Qgls8-S were associated with plant growth and development (Figure 2). Furthermore, the C. zeina-induced URGs in Qgls8-R were involved in defense signaling pathways and secondary metabolism, such as terpenoid biosynthetic process, defense response, and regulation of JA (Figures 3-5). The URGs in Qgls8-R involved in terpenoid metabolism and JA signaling may play crucial roles in GLS defense response.
Terpenoid production is catalyzed by terpene synthases (TPSs) and cytochrome P450s (Banerjee and Hamberger, 2018; Karunanithi and Zerbe, 2019), which regulate the biosynthesis of terpenoids and influence plant disease resistance, including ZmCYP71Z16, ZmCYP71Z18, ZmTPS6, and ZmTPS11 (Huffaker et al., 2011; van der Linde et al., 2011; Ding et al., 2019). ZmTPS1, ZmTPS7, ZmDLS, and ZmKSL2 catalyze sesquiterpenes, monoterpenes, and diterpenes, while ZmCYP71Z18 converts ent-CPP into dolabradiene and downstream dolabralexins (Mafu et al., 2018). ZmCYP92C5, a dimethylnonatriene/trimethyltetradecatetraene synthase, functions in the rate-limiting terpene production (Richter et al., 2016; Block et al., 2019). The expression of these genes was significantly induced by C. zeina with some showing continuously increased expression in Qgls8-R, suggesting a strong defense response activated through terpenoid metabolism upon C. zeina infection (Figure 6).
JA-dependent defenses play an important role in response to necrotrophic pathogens (Glazebrook, 2005). Key enzymes in JA biosynthesis include allene oxide synthase (AOS) and allene oxide cyclase (AOC) (Stenzel et al., 2012; Yang L. et al., 2023). JAZ proteins negatively regulate transcription factors in JA signaling (Vanholme et al., 2007; Chini et al., 2016). The degradation of JAZ repressor releases MYC transcription factors, which in turn induces the expression of JA-responsive genes (Chini et al., 2016). The expression of JA-related genes was significantly upregulated and the accumulation levels of JA and JA-Ile were higher upon C. zeina infection in Qgls8-R compared with Qgls8-S (Figure 7). This indicates that the JA signaling pathway is activated in Qgls8-R upon C. zeina infection, which may contribute to enhanced GLS resistance.
Qgls8 has been fine-mapped to 130 kb, containing four predicted candidate genes based on B73_v4 reference genome, Zea mays subsp. mexicana genome (Zx-TIL18-REFERENCE-PanAnd-1.0; Zx-TIL25-REFERENCE-PanAnd-1.0), and Zea mays subsp. parviglumis genome (Zv-TIL01-REFERENCE-PanAnd-1.0; Zv-TIL11-REFERENCE-PanAnd-1.0) (Zhang et al., 2017; Stitzer et al., 2025). Among them, two genes are annotated as leucine-rich repeat receptor-like kinases (LRR-RLK), one as phosphoethanolamine N-methyltransferase, and one as ABC transporter. LRR-RLKs often act as PRRs to recognize pathogens in PTI, such as FLS2 (Gomez-Gomez and Boller, 2000), EFR (Zipfel et al., 2006), and Xa21 (Song et al., 1995; Ngou et al., 2022a). The perception of pathogen activates JA signal transduction (Chini et al., 2007; Thines et al., 2007; Fonseca et al., 2009; Sheard et al., 2010; Zhang et al., 2015). In addition, plants LRR-RLKs defend against pathogens or herbivores by regulating JA and terpenoids biosynthesis. Rice OsLRR-RLK1 functions upstream of the JA signaling pathway and regulates the biosynthesis of JA induced by chewing herbivores to initiate defense responses against herbivores (Hu et al., 2018). Maize FI-RLPK inhibits the growth of fungal pathogen C. heterostrophus by accumulating JA, kauralexins and zealexins in maize (Block et al., 2021). The phosphoethanolamine N-methyltransferase has not been reported in the JA signaling and terpenoid biosynthesis pathway. ABC transporters are shown to be involved in the transport of monoterpenoids, sesquiterpenes and diterpenes, such as VmABCG1 (Demessie et al., 2017), AaPDR3 (Fu et al., 2017), NbABCG1 and NbABCG2 (Shibata et al., 2016). The determination of the exact functional gene underling Qgls8 will be interesting for future work to uncover the comprehensive network relating to Qgls8-mediated GLS resistance.
5 Conclusion
GLS is a devastating foliar disease of maize. This study used a pair of NILs Qgls8-R and Qgls8-S for transcriptome profiling to explore the molecular mechanisms underlying GLS resistance. We found that URGs in the resistant line Qgls8-R were key to immune signaling, which may contribute to increasing resistance against C. zeina. C. zeina infection significantly induced terpene- and JA-related genes with the highest expression in Qgls8-R. JA and JA-Ile accumulation levels were also significantly higher in Qgls8-R than in Qgls8-S. Genes related to JA biosynthetic process were also identified by WGCNA analysis. These results indicate that terpenoid metabolism and JA signaling pathway may be activated in response to C. zeina, enhancing GLS resistance.
Data availability statement
All raw RNA-seq data generated in this study have been deposited into SRA (sequence read archive) of NCBI, with the accession number PRJNA1222306. The data supporting the conclusions of this article are in the Supplementary Files.
Author contributions
XS: Writing – original draft, Writing – review & editing. RZ: Writing – review & editing. ZoW: Writing – review & editing. HW: Writing – review & editing. JZ: Writing – review & editing. NY: Writing – review & editing. ZhW: Writing – review & editing. HT: Writing – original draft, Writing – review & editing. QY: Funding acquisition, Resources, Writing – original draft, Writing – review & editing. HZ: Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Key Research and Development Program of Shaanxi province (2021ZDLNY01-06 and 2024NC2-GJHX-33) and Chinese Universities Scientific Fund (1090222003).
Acknowledgments
We are thankful to Dr. Peter Balint-Kurti at the Department of Entomology and Plant Pathology of North Carolina State University for providing B73-teosinte near-isogenic lines, Qgls8-R and Qgls8-S. We also thank Dr. Chen Zhang at Institute for Integrative Systems Biology of Universitat de València-CSIC for her assistance in the RNA-Seq data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor CL declared a past co-authorship with the author QY.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1580016/full#supplementary-material
References
Adie, B. A. T., Perez-Perez, J., Perez-Perez, M. M., Godoy, M., Sanchez-Serrano, J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. doi: 10.1105/tpc.106.048041
Ahuja, I., Kissen, R., and Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. doi: 10.1016/j.tplants.2011.11.002
Banerjee, A. and Hamberger, B. (2018). P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 17, 81–111. doi: 10.1007/s11101-017-9530-4
Benson, J. M., Poland, J. A., Benson, B. M., Stromberg, E. L., and Nelson, R. J. (2015). Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PloS Genet. 11, e1005045. doi: 10.1371/journal.pgen.1005045
Berger, D. K., Carstens, M., Korsman, J. N., Middleton, F., Kloppers, F. J., Tongoona, P., et al. (2014). Mapping QTL conferring resistance in maize to gray leaf spot disease caused by Cercospora zeina. BMC Genet. 15, 60. doi: 10.1186/1471-2156-15-60
Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Block, A. K., Tang, H. V., Hopkins, D., Mendoza, J., Solemslie, R. K., du Toit, L. J., et al. (2021). A maize leucine-rich repeat receptor-like protein kinase mediates responses to fungal attack. Planta 254, 73. doi: 10.1007/s00425-021-03730-0
Block, A. K., Vaughan, M. M., Schmelz, E. A., and Christensen, S. A. (2019). Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta 249, 21–30. doi: 10.1007/s00425-018-2999-2
Burow, M. and Halkier, B. A. (2017). How does a plant orchestrate defense in time and space? Using glucosinolates in Arabidopsis as case study. Curr. Opin. Plant Biol. 38, 142–147. doi: 10.1016/j.pbi.2017.04.009
Chen, L., Zhang, L., Xiang, S., Chen, Y., Zhang, H., and Yu, D. (2021). The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489. doi: 10.1093/jxb/eraa529
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Couto, D. and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dai, Z., Pi, Q., Liu, Y., Hu, L., Li, B., Zhang, B., et al. (2024). ZmWAK02 encoding an RD-WAK protein confers maize resistance against gray leaf spot. New Phytol. 241, 1780–1793. doi: 10.1111/nph.19465
Demessie, Z., Woolfson, K. N., Yu, F., Qu, Y., and De Luca, V. (2017). The ATP binding cassette transporter, VmTPT2/VmABCG1, is involved in export of the monoterpenoid indole alkaloid, vincamine in Vinca minor leaves. Phytochemistry 140, 118–124. doi: 10.1016/j.phytochem.2017.04.019
Ding, Y., Huffaker, A., Kollner, T. G., Weckwerth, P., Robert, C., Spencer, J. L., et al. (2017). Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol. 175, 1455–1468. doi: 10.1104/pp.17.00879
Ding, Y., Murphy, K. M., Poretsky, E., Mafu, S., Yang, B., Char, S. N., et al. (2019). Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants 5, 1043–1056. doi: 10.1038/s41477-019-0509-6
Du, L., Yu, F., Zhang, H., Wang, B., Ma, K., Yu, C., et al. (2020). Genetic mapping of quantitative trait loci and a major locus for resistance to grey leaf spot in maize. Theor. Appl. Genet. 133, 2521–2533. doi: 10.1007/s00122-020-03614-z
Dunkle, L. D. and Levy, M. (2000). Genetic relatedness of African and United States populations of Cercospora zeae-maydis. Phytopathology 90, 486–490. doi: 10.1094/PHYTO.2000.90.5.486
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Fu, X., Shi, P., He, Q., Shen, Q., Tang, Y., Pan, Q., et al. (2017). AaPDR3, a PDR transporter 3, is involved in sesquiterpene beta-Caryophyllene transport in Artemisia annua. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00723
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gomez-Gomez, L. and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Hu, L., Ye, M., Kuai, P., Ye, M., Erb, M., and Lou, Y. (2018). OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 219, 1097–1111. doi: 10.1111/nph.15247
Huang, Y., Wang, H., Zhu, Y., Huang, X., Li, S., Wu, X., et al. (2022). THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 612, 292–300. doi: 10.1038/s41586-022-05441-2
Huffaker, A., Kaplan, F., Vaughan, M. M., Dafoe, N. J., Ni, X., Rocca, J. R., et al. (2011). Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol. 156, 2082–2097. doi: 10.1104/pp.111.179457
Jones, J. D. and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karunanithi, P. S. and Zerbe, P. (2019). Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01166
Kistler, L., Maezumi, S. Y., Gregorio, D. S. J., Przelomska, N., Malaquias, C. F., Smith, O., et al. (2018). Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 362, 1309–1313. doi: 10.1126/science.aav0207
Kliebenstein, D. J., Rowe, H. C., and Denby, K. J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44, 25–36. doi: 10.1111/j.1365-313X.2005.02508.x
Lecourieux, D., Kappel, C., Claverol, S., Pieri, P., Feil, R., Lunn, J. E., et al. (2020). Proteomic and metabolomic profiling underlines the stage- and time-dependent effects of high temperature on grape berry metabolism. J. Integr. Plant Biol. 62, 1132–1158. doi: 10.1111/jipb.12894
Lennon, J. R., Krakowsky, M., Goodman, M., Flint-Garcia, S., and Balint-Kurti, P. J. (2016). Identification of alleles conferring resistance to gray leaf spot in maize derived from its wild progenitor species teosinte. Crop Sci. 56, 209–218. doi: 10.2135/cropsci2014.07.0468
Liu, Z., Cook, J., Melia-Hancock, S., Guill, K., Bottoms, C., Garcia, A., et al. (2016). Expanding maize genetic resources with predomestication alleles: Maize-teosinte introgression populations. Plant Genome 9. doi: 10.3835/plantgenome2015.07.0053
Liu, K. J. and Xu, X. D. (2013). First report of gray leaf spot of maize caused by Cercospora zeina in China. Plant Dis. 97, 1656. doi: 10.1094/PDIS-03-13-0280-PDN
Llorente, F., Muskett, P., Sanchez-Vallet, A., Lopez, G., Ramos, B., Sanchez-Rodriguez, C., et al. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1, 496–509. doi: 10.1093/mp/ssn025
Lv, X., Song, M., Cheng, Z., Yang, X., Zhang, X., Zhou, Z., et al. (2020). qGLS1.02, a novel major locus for resistance to gray leaf spot in maize. Mol. Breed. 40, 59. doi: 10.1007/s11032-020-01134-8
Mafu, S., Ding, Y., Murphy, K. M., Yaacoobi, O., Addison, J. B., Wang, Q., et al. (2018). Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in maize. Plant Physiol. 176, 2677–2690. doi: 10.1104/pp.17.01351
Ngou, B., Ding, P., and Jones, J. (2022a). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Ngou, B., Jones, J., and Ding, P. (2022b). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Nsibo, D. L., Barnes, I., Kunene, N. T., and Berger, D. K. (2019). Influence of farming practices on the population genetics of the maize pathogen Cercospora zeina in South Africa. Fungal Genet. Biol. 125, 36–44. doi: 10.1016/j.fgb.2019.01.005
Peng, Y., van Wersch, R., and Zhang, Y. (2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 31, 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Piperno, D. R., Ranere, A. J., Holst, I., Iriarte, J., and Dickau, R. (2009). Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. U. S. A. 106, 5019–5024. doi: 10.1073/pnas.0812525106
Richter, A., Schaff, C., Zhang, Z., Lipka, A. E., Tian, F., Kollner, T. G., et al. (2016). Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 28, 2651–2665. doi: 10.1105/tpc.15.00919
Sermons, S. M. and Balint-Kurti, P. J. (2018). Large scale field inoculation and scoring of maize southern leaf blight and other maize foliar fungal diseases. Bio-Protocol 8, e2745. doi: 10.21769/BioProtoc.2745
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Shibata, Y., Ojika, M., Sugiyama, A., Yazaki, K., Jones, D. A., Kawakita, K., et al. (2016). The full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and postinvasion defense against Phytophthora infestans in Nicotiana benthamiana. Plant Cell 28, 1163–1181. doi: 10.1105/tpc.15.00721
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Stenzel, I., Otto, M., Delker, C., Kirmse, N., Schmidt, D., Miersch, O., et al. (2012). ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: tissue- and organ-specific promoter activities and in vivo heteromerization. J. Exp. Bot. 63, 6125–6138. doi: 10.1093/jxb/ers261
Stitzer, M. C., Seetharam, A. S., Scheben, A., Hsu, S. K., Schulz, A. J., AuBuchon-Elder, T. M., et al. (2025). Extensive genome evolution distinguishes maize within a stable tribe of grasses. Biorxiv. doi: 10.1101/2025.01.22.633974
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Tsuda, K. and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
van der Linde, K., Kastner, C., Kumlehn, J., Kahmann, R., and Doehlemann, G. (2011). Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol. 189, 471–483. doi: 10.1111/j.1469-8137.2010.03474.x
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Wang, H., Hou, J., Ye, P., Hu, L., Huang, J., Dai, Z., et al. (2021). A teosinte-derived allele of a MYB transcription repressor confers multiple disease resistance in maize. Mol. Plant 14, 1846–1863. doi: 10.1016/j.molp.2021.07.008
Ward, J. M. J., Laing, M. D., and Nowell, D. C. (1997). Chemical control of maize grey leaf spot. Crop Prot. 16, 265–271. doi: 10.1016/S0261-2194(96)00097-X
Ward, J. M. J., Stromberg, E. L., Nowell, D. C., and Nutter, F. W. (1999). Gray leaf spot: a disease of global importance in maize production. Plant Dis. 83, 884–895. doi: 10.1094/PDIS.1999.83.10.884
Xu, L., Zhang, Y., Shao, S., Chen, W., Tan, J., Zhu, M., et al. (2014). High-resolution mapping and characterization of qRgls2, a major quantitative trait locus involved in maize resistance to gray leaf spot. BMC Plant Biol. 14, 230. doi: 10.1186/s12870-014-0230-6
Yang, Q., He, Y., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., et al. (2017). A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–1372. doi: 10.1038/ng.3919
Yang, L., Sun, Q., Geng, B., Shi, J., Zhu, H., Sun, Y., et al. (2023). Jasmonate biosynthesis enzyme allene oxide cyclase 2 mediates cold tolerance and pathogen resistance. Plant Physiol. 193, 1621–1634. doi: 10.1093/plphys/kiad362
Yang, N., Wang, Y., Liu, X., Jin, M., Vallebueno-Estrada, M., Calfee, E., et al. (2023). Two teosintes made modern maize. Science 382, eadg8940. doi: 10.1126/science.adg8940
Yuan, M., Ngou, B., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030. doi: 10.1016/j.pbi.2021.102030
Zhang, X. and Dong, X. (2022). Life-or-death decisions in plant immunity. Curr. Opin. Immunol. 75, 102169. doi: 10.1016/j.coi.2022.102169
Zhang, Y., Xu, L., Fan, X., Tan, J., Chen, W., and Xu, M. (2012). QTL mapping of resistance to gray leaf spot in maize. Theor. Appl. Genet. 125, 1797–1808. doi: 10.1007/s00122-012-1954-z
Zhang, X., Yang, Q., Rucker, E., Thomason, W., and Balint-Kurti, P. (2017). Fine mapping of a quantitative resistance gene for gray leaf spot of maize (Zea mays L.) derived from teosinte (Z. mays ssp. parviglumis). Theor. Appl. Genet. 130, 1285–1295. doi: 10.1007/s00122-017-2888-2
Zhang, F., Yao, J., Ke, J., Zhang, L., Lam, V. Q., Xin, X. F., et al. (2015). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. doi: 10.1038/nature14661
Zhong, T., Zhu, M., Zhang, Q., Zhang, Y., Deng, S., Guo, C., et al. (2024). The ZmWAKL-ZmWIK-ZmBLK1-ZmRBOH4 module provides quantitative resistance to gray leaf spot in maize. Nat. Genet. 56, 315–326. doi: 10.1038/s41588-023-01644-z
Zhu, M., Zhong, T., Xu, L., Guo, C., Zhang, X., Liu, Y., et al. (2024). The ZmCPK39-ZmDi19-ZmPR10 immune module regulates quantitative resistance to multiple foliar diseases in maize. Nat. Genet. 56, 2815–2826. doi: 10.1038/s41588-024-01968-4
Keywords: maize, gray leaf spot, transcriptome, terpenoid metabolism, jasmonic acid
Citation: Sun X, Zhang R, Wang Z, Zhang H, Wen H, Zhang J, Yu N, Wang Z, Tai H and Yang Q (2025) Comparative transcriptomics in B73-teosinte near-isogenic maize lines reveals key defense signaling and phytoalexins in response to Cercospora zeina infection. Front. Plant Sci. 16:1580016. doi: 10.3389/fpls.2025.1580016
Received: 20 February 2025; Accepted: 19 May 2025;
Published: 05 June 2025.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Jianyu Wu, Henan Agricultural University, ChinaLanglang Ma, Maize Research Institute of Sichuan Agricultural University, China
Copyright © 2025 Sun, Zhang, Wang, Zhang, Wen, Zhang, Yu, Wang, Tai and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanhuan Tai, aGh0YWlAbndhZnUuZWR1LmNu; Qin Yang, cXlhbmdAbndhZnUuZWR1LmNu
 Xiaomeng Sun1,2
Xiaomeng Sun1,2 Junli Zhang
Junli Zhang Zhe Wang
Zhe Wang Qin Yang
Qin Yang