- 1Department of Biomedical Sciences, College of Health Sciences, QU Health, Qatar University, Doha, Qatar
- 2Department of Biological and Environmental Sciences, College of Arts and Sciences, Qatar University, Doha, Qatar
- 3Ministry of Environment and Climate Change, Doha, Qatar
- 4Agrico for Agricultural Development Company, Doha, Qatar
- 5Department of Nutrition Sciences, College of Health Sciences, QU Health, Qatar University, Doha, Qatar
- 6Biomedical Research Center, QU Health, Qatar University, Doha, Qatar
- 7Department of Basic Medical Sciences, College of Medicine, QU Health, Qatar University, Doha, Qatar
- 8The KINDI Center for Computing Research, College of Engineering, Qatar University, Doha, Qatar
Introduction: With the predicted 9-10 billion world population increase by 2050 and its accompanying need for sustainable food production, and with the harsh climate conditions challenging agriculture and food security in many countries world-wide, employing “horticultural protected cultivation practices” in farming for seasonal and off-seasonal crop production is on the rise, among which is the use of agricultural greenhouses. The importance of greenhouse farming has been, indeed, evident by the perceived increase in year-round crops production, curtail in production risks, upsurge in agricultural profits, outreaching food stability and security in many countries globally. Yet, and despite this acknowledged success of employing greenhouses in farming, many constraints, including the presence of insect pests, still chaperoned this practice over the years, significantly impacting crop quality and production.
Methods: As such, we assessed in this study the status of “insect pests” in the greenhouse model by collecting insects from different greenhouse sectors grown with tomatoes and cucumbers and identified the collected insects using relevant identification keys. To further explore the pest paradigm in greenhouses, we then focused on particularly studying Trialeurodes vaporariorum (TRIAVA), a key insect species among the collected and identified insects in the studied greenhouse model and a significant pest with an impactful effect on many crops worldwide. To do so, we traced the abundance of TRIAVA in the tomato and cucumber grown greenhouse sectors over the period of the study, analyzed its metagenome and associated its abundance with crop yield.
Results and discussion: Our findings revealed TRIAVA hosted microbes with aptitudes to either serve as symbiotic microorganisms and protect TRIAVA against pathogens or to potentially cause damage to crops. This work provides additional insight into the insect pests paradigm in greenhouses, an upshot that could serve integrated insect pest management strategies in greenhouses for optimal agricultural practices.
Highlights
● Landscape of “insect pests” in agricultural greenhouse systems.
● Trialeurodes vaporariorum abundance in agricultural greenhouses.
● Trialeurodes vaporariorum metagenomic analysis.
● Trialeurodes vaporariorum impact on crop yield.
Introduction
For the past decades, and despite the substantial progress made in combating global hunger, malnutrition and food insecurity persisted as far-reaching problems in many countries worldwide, mainly in Asia and Africa (Foley et al., 2011; Global Panel on Agriculture and Food Systems for Nutrition, 2016; International Food Policy Research Institute (IFPRI), 2016; FAO, I et al., 2017). While poverty classifies as a major motive, the ground for food insecurity outstretches far beyond poverty to encompass other additional motives including the rapidly growing human population worldwide, climate change, and weather conditions. These motives also exhibit serious threats to farming practices and agricultural sustainability (Farooq et al., 2019). Being a major food security pillar, the necessity to develop multidimensional farming approaches for agricultural sustainability without impacting economy, society, and environmental integrity becomes therefore imperative. Among various technological innovations, the use of “controlled-environment-agricultural-production-technologies” such as greenhouse farming has been promising in overcoming production related obstacles, increasing productivity rates and securing agricultural sustainability (Benke and Tomkins, 2017; Forkuor et al., 2022). Compared to open field farming practices, greenhouse cultivation has been shown to upsurge crop yields, decrease the release and accumulation of greenhouse gases, promote an efficient use of agricultural resources such as water, nutrients and energy, and curtail the stressing effect of some abiotic and biotic factors (Kanwar, 2011). Despite these advantages; however, several challenges still acquainted the use of this innovative technology. Among those challenges, pest manifestation has been considered a paramount factor in causing drastic losses in crop yield, if left uncontrolled. Agricultural pests span a broad-spectrum including insects/insect vectors, weeds, rodents, nematodes, mites, microorganism and viruses (Nriagu, 2011; Heck, 2018; Walker and Frederick, 2019; Vermelho et al., 2024). Indeed, insect vectors have been reported to cause one of the greatest threats not only to crops but also to animals and humans on a global scale (Kamareddine, 2012; Heck, 2018). Most insect vectors carrying plant pathogens classify as hemipteran insects, examples of which include psyllids, aphids and whiteflies (Heck, 2018). Interestingly, many of these insect vectors have been identified in agricultural greenhouses. The Trialeurodes vaporariorum (TRIAVA) white fly pest, for example, has been reported to pose serious threats to roses cultivated in greenhouses (Roditakis, 1990; Voigt et al., 2019) and to transmit pathogens like the Tomato torrado virus (Verbeek et al., 2014) and the Pepino mosaic virus (Noël et al., 2014) to tomato crops causing severe necrosis and crop loss (van der Vlugt et al., 2000; Wieczorek et al., 2020). Likewise, aphids have been shown to significantly impact different crops including cucumber, eggplant, tomato and potato grown in greenhouses (Eastop and Balckmann, 2007; Capinera, 2008; Tarek, 2013; Mohammed et al., 2018; Mohammed et al., 2022). Aphids have been, indeed, elegantly outlined for their roles as vector. Among various aphid-transmitted plant virus families, the family Potyviridae, genus Potyvirus, is probably considered the most important based on the exceptionally large number of virus species that it includes (Qin et al., 2020). Furthermore, the Bactericera cockerelli psyllid, known to transmit Candidatus Liberibacter solanacearum, has been also reported as solanaceous crops (tomato and potato) insect pest (Rosson et al., 2006; Yang and Liu, 2009; Munyaneza, 2012; Munyaneza, 2013; Castillo Carrillo, 2019; Tang et al., 2020; Sarkar et al., 2023). It is noteworthy here that despite classifying as vectors, many of those insect pests do not necessarily exclusively cause crop damage by means of the pathogen they carry, but could rather exhibit pathogenicity via other mechanisms or by the combination of multiple-damaging strategies. The impact of whiteflies, for instance, on crop damage or yield loss is not only attributed to the pathogens carried by the insect, but also to the insect’s direct feeding effect on the plant, along with the potentiality of this feeding behavior to attract other pathogens (Navas-Castillo et al., 2011; Saurabh et al., 2021).
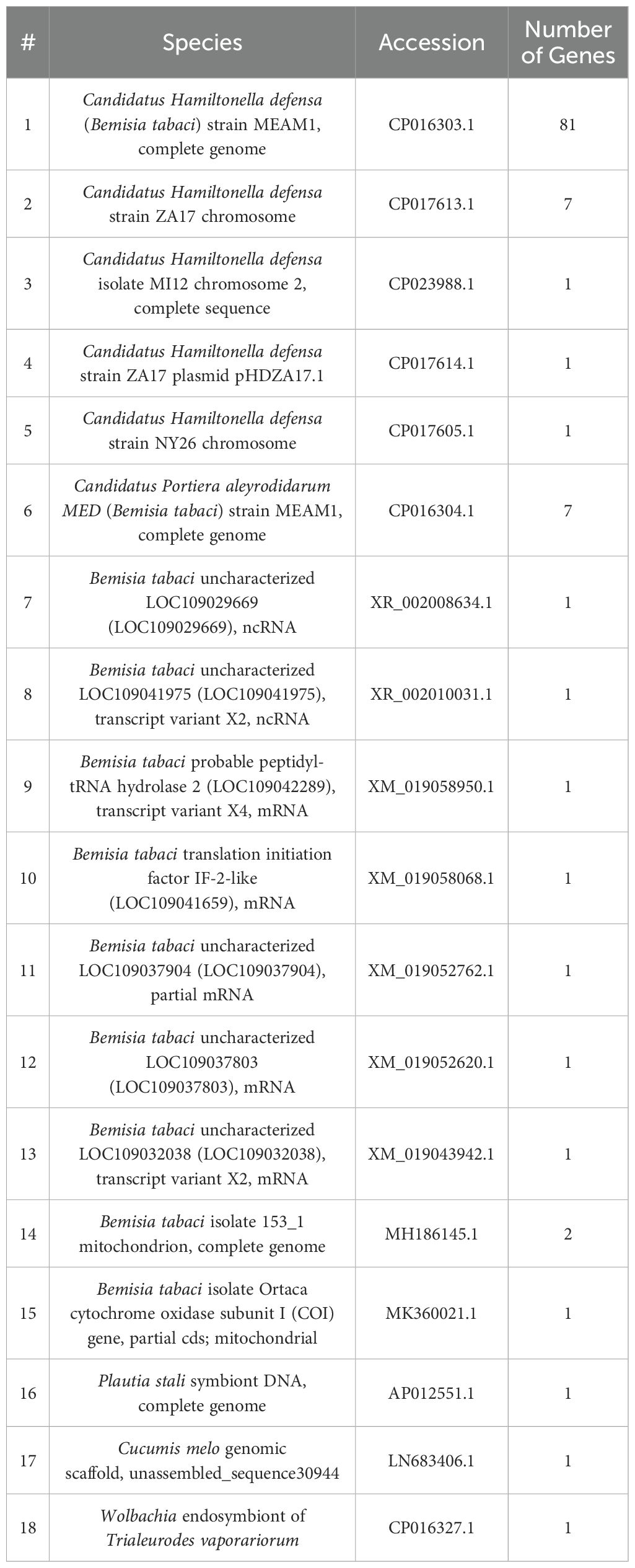
Owing to the effect of insect pests on greenhouse farming and to the existence of such multiple-plant/crop damaging mechanisms, a proper and comprehensive understanding of the landscape of insect pests in agricultural greenhouses and their virulence mechanisms is crucial for the development of either in isolation or in combination strategies for insect pests control. As such, we conducted in this study an “insects survey” to detect the present and most abundant insect pests in selected agricultural greenhouse sectors grown with tomato and cucumber. Being among the mostly detected key insect species in our studied greenhouse model and a major global pest, we further scored for the abundance of TRIAVA, in particular, and evaluated the association of its abundance with the greenhouse environmental conditions including temperature and humidity. We also conducted a metagenomics analysis to detect the richness and diversity of the microbial communities residing in TRIAVA and to assess the potential contribution of those microbes to vector protection and crop yield loss using computational predictions (Figure 1). Taken together, our findings provide novel insights into the insect pests paradigm in agricultural greenhouses, and its associated biotic and abiotic framework. By focusing on such paradigms, the agricultural community can work towards a more efficient, sustainable, and cost-effective future, securing a steady supply of crops while safeguarding the ecosystem.
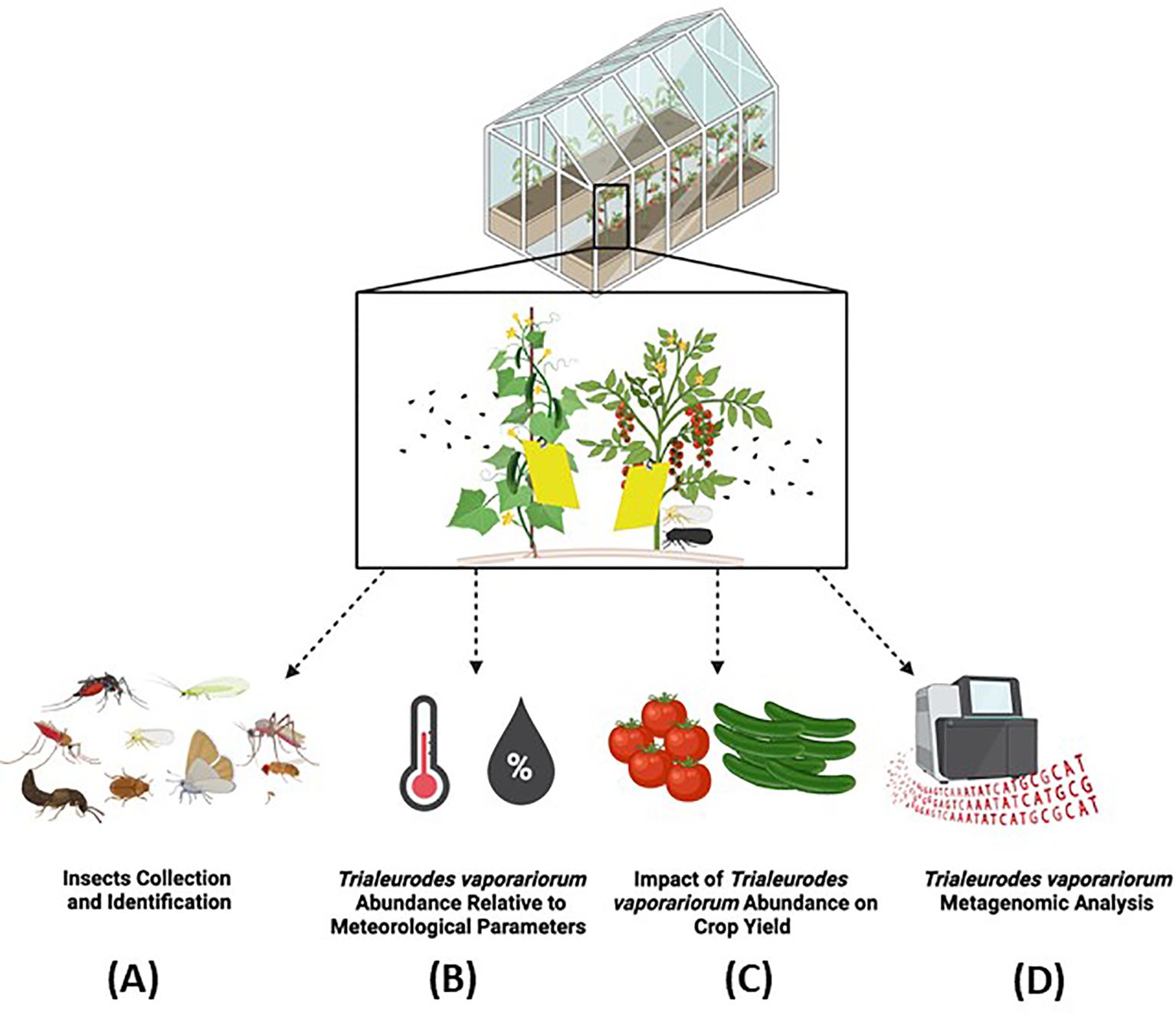
Figure 1. Study overview. In this study (A) agricultural greenhouse insects were collected and identified, Trialeurodes vaporariorum abundance was scored for and associated with (B) meteorological parameters and (C) cucumber and tomato crop yield, and (D) Trialeurodes vaporariorum metagenome was analyzed. Biorender was used to generate the study overview diagram.
Materials and methods
Agricultural greenhouses
The insects survey was conducted in three greenhouses located in the AGRICO Agricultural Development W.L.L. farm that is situated in Al-Khor city, 58 km away from Doha the capital of Qatar, and that covers an estimated area of 24 hectares (https://agrico.qa/). Cucumbers were grown in an arch-shaped, one side, UV coated greenhouse made of an 8mm clear polycarbonate hollow sheet. Cherry and Beef tomatoes were grown in arch-shaped greenhouses covered with 200 microns-white plastic sheets, with a 7% UV light penetration ability. For close monitoring of the environmental conditions inside the agricultural greenhouses, meteorological data including temperature (°C), relative humidity (%), vapor pressure deficit (kPa), evapotranspiration (mm d-1), solar radiation/photosynthetically active radiation (Wm-2), and wind velocity (km/h) were recorded at fixed intervals using the Crop Estimation through Resource and Environment Synthesis monitoring system (CERES, USA).
Insects collection and identification
The insects survey was conducted during the planting season, particularly during the period extending from the 19th of February to the 11th of June 2022. Over the indicated period, insects were collected on weekly basis from selected 3000-4000m2 agricultural greenhouse sectors grown with tomato or cucumber. The collection methods included UV light traps (BIONIVA, Malaysia), mechanical pooters/aspirators (Labitems, India), yellow sticky traps, and handpicking. Collected insects were placed in Eppendorf or Falcon conical tubes, transported on ice from the farm to the laboratory, and stored at -80°C for identification. Using available literature and taxonomic keys (Diarra et al; Kelton, 1975; Rattanarithikul, 1982; Abdu and Shaumar, 1985; Harbach, 1985; Aldryhim, 1996; Martin et al., 2000; Beadle and Leckie, 2012; Gibb and Oseto, 2019; Garba et al., 2020; Dioli et al., 2021; Lonsdale, 2021), and under a stereo microscope (Leica M125), insects were identified up to the species level whenever possible and representative images were taken.
Trialeurodes vaporariorum abundance monitoring
To monitor TRIAVA density throughout the planting season in the studied sectors, 15cm x 20cm (width x length) yellow sticky traps were distributed into five selected spots in each tomato and cucumber grown sector. To obtain unbiased mean density and ensure that sticky traps are spatially independent, traps were placed at 18m apart in 3000m2 sector and at 22.5m apart in 4000m2 sector, as recommended from previous studies (Kim et al., 2001). On weekly basis, old traps were replaced by new ones and TRIAVA counts were recorded.
Data analysis
Descriptive statistics were used to present the meteorological data, particularly temperature (°C) and relative humidity (%), recorded in the tomato and cucumber grown greenhouse sectors across the period of the study. Pearson’s correlation was used to associate TRIAVA abundance with meteorological data and crop yield. The IBM SPSS Statistics software V22 was used to conduct data analysis between compared groups and statistical significance was concluded for p-value ≤ 0.05.
Metagenomic analysis
The metagenome analysis workflow involved multiple steps including sample preparation, quality control and functional annotation, the details of each step are summarized as follows:
Sample preparation and DNA library construction and sequencing
During the study period, extending from the 19th of February to the 11th of June 2022, 3-5 TRIAVA insects per sample were collected at different times from different spots of the studied sectors using a mechanical aspirator. Samples were then pooled to make up a total of 16 insects per pooled sample and surface sterilized before DNA extraction. TRIAVA genomic DNA was then extracted using phenol-chloroform at Beijing Genomics Institute (BGI) (BGI Genomics, 2021). In brief, tissues samples were grinded with liquid nitrogen, suspended in 1 mL of lysis buffer, incubated at 56°C for 30-180 minutes with continuous mixing by inversion every 5-10 minutes, and centrifuged at 16700×g for 10 minutes. The supernatant was then transferred to a new tube and mixed with an equal volume of chloroform/isoamyl alcohol (24:1). The aqueous phase was then collected and transferred into a new tube, mixed with an equal volume of isopropyl alcohol, and centrifuged at 16700×g for 10 minutes. The aqueous phase was further collected and transferred into a new tube, mixed with 2/3 volume of isopropyl alcohol, incubated at -20°C for 2 hours for precipitation, and centrifuged at 16700×g for 10 minutes. The DNA pellet was then washed twice with 75% ethanol, centrifuged, air-dried, and resuspended in 25-100 µL of TE buffer. The quality and quantity of the extracted samples was then assessed using a Qubit fluorometer and agarose gel electrophoresis. To prepare the DNA libraries, 1μg of genomic DNA was randomly fragmented, and the fragmented DNA was size-selected using the Agencourt AMPure XP kit to an average of 200–400 bp. Fragments were end repaired and 3’ adenylated. Adaptors were further ligated to the DNA fragments then amplified. The PCR products were purified using the Agencourt AMPure XP kit. The double-stranded PCR products were heat-denatured and circularized using a splint oligonucleotide. The single-stranded circle DNA (ssCir DNA) was formatted as the final library. The Library was validated through quality control. The qualified libraries were then sequenced by BGISEQ-500_ DNBSEQ™ and through rolling-cycle replication, ssCir DNA molecule formed a DNA nanoball library (DNB). DNBs were next loaded onto the patterned nanoarray by using high-density DNA nanochip technology and pair-end 100 bp reads were finally obtained through combinatorial Probe-Anchor Synthesis (cPAS).
Quality control and preprocessing
To evaluate the overall quality, including base quality scores, sequence length distribution, and potential contamination, quality control of raw sequencing reads was performed using FastQC (Andrews et al., 2010). To expedite the process, the analysis was conducted using 30 threads, and the results were saved in the output directory. Following this, Fastp (Chen, 2023) was used to trim low-quality bases, remove adapter sequences, and improve read quality. The paired-end input files were processed with 30 threads, and the cleaned reads were output to the output directory for downstream analysis.
Removal of human reads
To ensure that only non-human reads were retained, the trimmed reads were mapped to the human reference genome (GRCh38) using Bowtie2 (Langmead and Salzberg, 2012). To maximize read alignment precision, mapping was performed with a very-sensitive alignment setting. Reads that aligned to the human genome were filtered out, while non-human reads were extracted and saved for subsequent taxonomic classification. The unmapped reads were considered for further downstream analysis.
Taxonomic classification
To determine the taxonomic composition of the metagenome, Kraken2 (Lu et al., 2022), a database-driven classifier that assigns taxonomic labels based on exact k-mer matches, was used. The analysis was performed with a minimum base quality threshold of 30 and a requirement for at least two hit groups to reduce false-positive classifications. The classified and unclassified reads were saved in respective files for later inspection. A detailed report (trv.k2report) was also generated to provide an overview of the classified taxa.
Abundance estimation and filtering
To estimate taxonomic abundances from Kraken2 output at various levels including kingdom (K), domain (D), phylum (P), class (C), order (O), family (F), genus (G), and species (S), we used Bracken (Lu and Salzberg, 2019). Using a custom Python script, filtering to remove specific taxa that were considered contaminants or non-target organisms was performed. The filtered output files were saved with relevant taxonomic breakdowns, ensuring that only taxa of interest were retained for analysis.
Taxonomic visualization
To visualize the taxonomic distribution of the metagenome, Krona (Ondov et al., 2011) plots were generated. The kreport2krona.py script converted the Bracken report into a Krona-compatible format, and the ktImportText tool was used to generate an interactive HTML visualization, providing an intuitive representation of the relative abundance of each taxonomic group.
Gene prediction
To conduct gene prediction, MetaGeneMark (Gemayel et al., 2022), a robust tool for identifying coding sequences (CDS) in metagenomic assemblies, was used. The analysis utilized the MetaGeneMark v1 model, and both nucleotide and protein-coding sequences were extracted. The predicted proteins and nucleotide sequences were output for subsequent functional annotation and downstream comparative analysis.
Functional annotation
The predicted nucleotide sequences were annotated using BLASTn (Geneious) by querying the NCBI nucleotide database. The search was performed with an e-value cutoff of 1×10−51 times to ensure significant alignments, and the output was formatted in tabular format (format 6) to facilitate interpretation. Only the top match for each query sequence was retained to avoid redundancy, and the analysis was parallelized using 30 threads to improve computational efficiency.
Gene Ontology mapping
For functional characterization, Gene Ontology (GO) terms associated with the annotated genes were retrieved. A custom Python script was employed to map GO terms by querying the QuickGO API (European Bioinformatics Institute) and fetching descriptions for each GO term. The input file containing GO terms was processed, and the output was saved as a tab-separated values (TSV) file. A brief delay was included between API requests to avoid exceeding the rate limit.
Results
Studied greenhouse sectors and recorded meteorological datasets
Although ambient conditions are relatively maintained in agricultural greenhouses as compared to open fields, variations in meteorological conditions, particularly in temperature and relative humidity, could still occur due to natural and static driven factors (van Tuijl et al., 2008; Balendonck et al., 2010). In this study, we chose to particularly focus on greenhouse sectors grown with cucumber and tomato, including Cucumber Mini Munch (Figure 2A), Cherry Tomato (Figure 2C), and Beef Tomato (Figure 2E), being essential horticultural crops grown in greenhouses globally (Xue et al., 2023). We also recorded the meteorological datasets, specifically temperature (°C) and relative humidity (%), in those sectors throughout the period of the study. In concordance with the reported from previous findings, slight variations in temperature and relative humidity readouts throughout the period of the study have been, indeed, detected in these monitored greenhouse sectors (Figures 2B, D, F). Along this, slight variations in the temperature and relative humidity readouts between the three studied sectors were also recorded. For example, in the Cucumber Mini Munch sector, the average temperature levels fluctuated from a minimum of 20.4°C to a maximum of 29.2°C, and the average relative humidity levels from a minimum of 67.9% to a maximum of 82.5% (Figure 2B), reflecting a slightly higher minimum average temperature and a slightly lower minimum average humidity levels compared to those recorded in the two other sectors. In the Cherry Tomato sector, on the other hand, average temperature ranged from a minimum reading of 17.6°C to a maximum reading of 28.3°C, while average relative humidity varied from a minimum of 69.3% to a maximum 85% (Figure 2D). Similarly, the recorded average temperature values in the Beef Tomato sector ranged from a minimum reading of 17.6°C to a maximum reading of 29.2°C, and the average relative humidity levels fluctuated from a minimum of 70% to a maximum of 88.1% (Figure 2F).
Figure 2. Greenhouse crops and meteorological conditions. Representative images of the studied agricultural greenhouse sectors grown with (A) Cucumber Mini Munch (C) Cherry Tomato and (E) Beef Tomato. Bar graphs representing the weekly average temperature and line charts demonstrating the weekly average relative humidity in (B) Cucumber Mini Munch (D) Cherry Tomato and (F) Beef Tomatc grown sectors recorded during the period of the study.
Collected and identified insects
Monitoring and controlling the dynamics of pest population in greenhouses is paramount in efficiently managing greenhouse farming. Herein, we performed greenhouse insects (beneficial or pests) investigation by collecting insects residing in the studied cucumber and tomato grown greenhouse sectors using readily available tools (Figure 3). Collected insects were further identified up to the species level expect for a few cases where identification was only achieved up to the genus level (Table 1; Figure 4). Failure to complete species-level classification was either due to the lack of local and regional taxonomic keys or to damages that have occurred to some samples during the collection, transferring and/or characterization processes. Our findings revealed a total of 42 insects belonging to different orders, including Diptera, Coleoptera, Hemiptera, Lepidoptera, and Neuroptera (Table 1; Figure 4). Among those insects, Liriomyza trifolii, Trialeurodes vaporariorum, Dicranomyia modesta, Nomophila noctuella, Spodoptera exigua, and Ceraeochrysa were the most prevalent during the study period. Interestingly, a remarkable increase in the insects’ overall abundance in general was also detected from May to June.

Figure 3. Insects collection tools. Images of the tools including (A) UV light trap, (B) mechanical pooter (aspirator), (C) yellow sticky trap, and (D) handpicking used for insects collection from Cucumber Mini Munch, Cherry Tomato, and Beef Tomato grown greenhouse sectors.
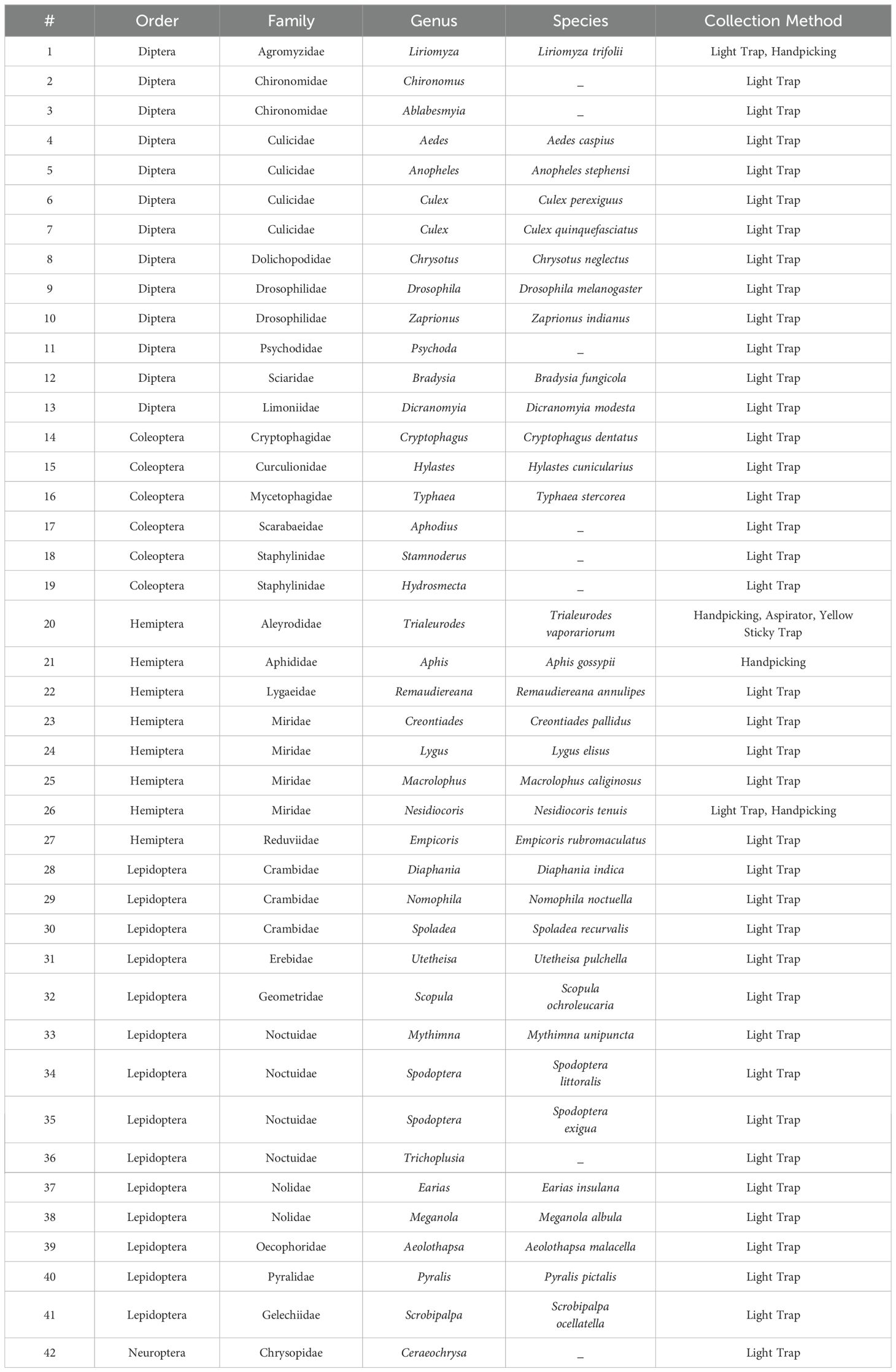
Table 1. List of collected insects from Cucumber Mini Munch, Cherry Tomato, and Beef Tomato grown greenhouse sectors during the period of the study.

Figure 4. Collected insects. Representative images of (1) Liriomyza trifolii, (2) Chironomus, (3) Ablabesmyia, (4) Aedes caspius, (5) Anopheles stephensi, (6) Culex perexiguus, (7) Culex quinquefasciatus, (8) Chrysotus neglectus, (9) Drosophile melanogaster, (10) Zaprionus indianus, (11) Psychoda, (12) Bradysia fungicola, (13) Dicranomyia modesta, (14) Cryptophagus dentatus, (15) Hylastes cunicularius, (16) Typhaea stercorea, (17) Aphodius, (18) Stamnoderus, (19) Hydrosmecta, (20) Trialeurodes vaporariorum, (21) Aphis gossypii, (22) Remaudiereana annulipes, (23) Creontiades pallidus, (24) Lygus elisus, (25) Macrolophus caliginosus, (26) Nesidiocoris tenuis, (27) Empicoris rubromaculatus, (28) Diaphania indica, (29) Nomophila noctuella. (30) Spoladea recurvalis, (31) Utetheisa pulchella, (32) Scopula ochroleucaria, (33) Mythimna unipuncta, (34) Spodoptera littoralis, (35) Spodoptera exigua, (36) Trichoplusia, (37) Earias insulana, (38) Meganola albula, (39) Aeolothapsa malacella, (40) Pyralis Pyralis pictalis, (41) Scrobipalpa ocellatella, and (42) Ceraeochrysa collected from Cucumber Mini Munch, Cherry Tomato, and Beef Tomato grown greenhouse sectors during the period of the study.
Trialeurodes vaporariorum abundance and impact on crop yield
Among the identified insects in the studied greenhouse sectors (Table 1, Figure 4), TRIAVA was one of the most abundant pest species known to affect vegetables and ornamental greenhouse crops and cause economical losses (Wraight et al., 2017). As such, we next evaluated the abundance of the TRIAVA whitefly in these sectors throughout the period of the study and correlated its abundance with meteorological parameters, particularly with temperature and relative humidity, being pivotal factors in dictating pest plentitude and distribution (Kamareddine, 2019; Khan, 2019). Subsequently, we further correlated TRIAVA abundance with cucumber and tomato crop yield. Our findings revealed an overall positive correlation between TRIAVA abundance and temperature (Figures 5A–C) and negative correlation between TRIAVA abundance and relative humidity in the three studied sectors (Figures 5D–F). Moreover, our findings also revealed that the total weekly crop yield fluctuates in an inversely proportional manner with TRIAVA abundance, as evidenced by the negative correlation between TRIAVA abundance and crop yield in the three studied sectors (Figures 5G–I). It is worth noting here that despite the clear correlation trends seen between TRIAVA abundance and the studied meteorological conditions and crop yield, boarded line or ≥ 0.05 p-values have been concluded in some occurrences. Failure to achieve statistical significance in these occurrences does not negate the existing trend and could rather relate to many reasons, one of which might be the low whitefly counts sometimes recorded in these studied sectors.
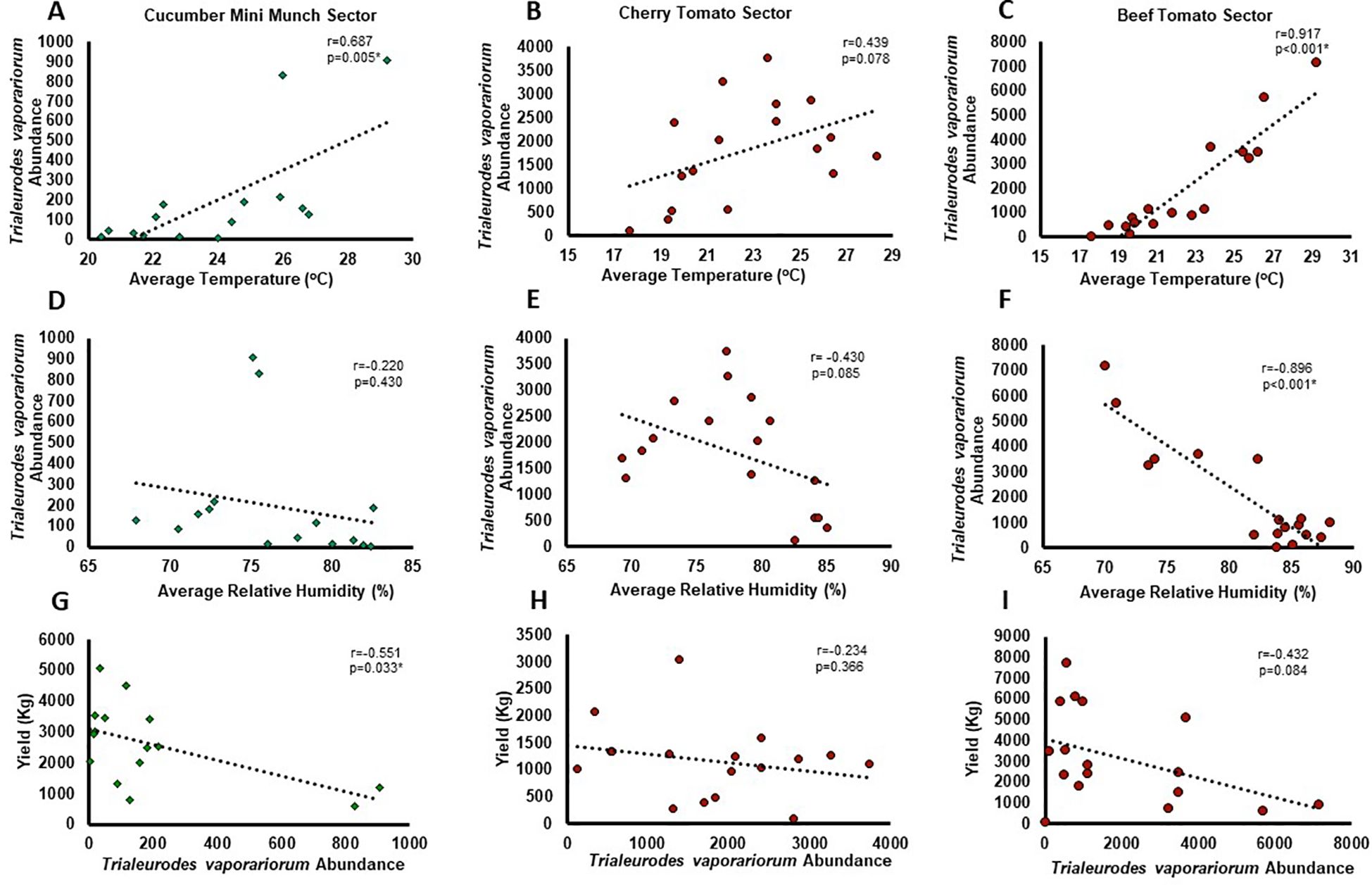
Figure 5. Correlation between Trialeurodes vaporariorum abundance and greenhouse meteorological conditions and crop yield. Scatter diagram and best-fit linear model representing the correlation between average Trialeurodes vaporariorum weekly abundance and (A-C) average weekly temperature, (D-F) average weekly relative humidity, and (G-I) average weekly crop yield recorded throughout the period of the study in (A, D, G) Cucumber Mini Munch, (B, E, H) Cherry Tomato, and (C, F, I) Beef Tomato grown sectors. Pearson correlation coefficient (r) and p-value are shown on each graph, with positive r-value signifying positive correlation, negative r-value signifying negative comelation, and p-value ≤ 0.05 indicating statistical significance.
Trialeurodes vaporariorum metagenomic analysis
Insects, and like other living beings, generally harbor diverse groups of microbes including bacteria, viruses, fungi, archaea, and protozoa (Engel and Moran, 2013). These microbes could associate with the insect host either permanently or transiently and could be beneficial or harmful, therefore dictating the insect’s overall adaptability, survival aptitude and vector competence in various ecological niches (Chellappan and Ranjith, 2021). Along this notion, and to identify potential symbiotic or pathogenic microorganisms harbored in/carried by TRIAVA for self-protection against pathogens or for insect pathogenicity, we conducted a metagenomic analysis of TRIAVA and particularly scored for its Archaeal (Figure 6, Supplementary Table 1), viral (Figure 7, Supplementary Table 2), and bacterial (Figure 8, Supplementary Table 3) harbored communities. The calculated percentage of the abundance of archaeal, viral, and bacterial groups was determined by mapping the reads of each class to the total number of reads within each respective domain. For the archaeal community, the composition in TRIAVA was mainly dominated by members of the Euryarchaeota phylum, accounting for 3.57k reads (68.3%). The most abundant genera were Thermococcus (456 reads, 8.7%) and Methanosarcina (391 reads, 7.5%). Among these, the predominant species were Thermococcus cleftensis (133 reads), followed by Thermococcus sp. (76 reads), and Thermococcus barophilus (38 reads). Similarly, within Methanosarcina, the predominant species included Methanosarcina bakeri (91 reads), Methanosarcina mazei (82 reads), and Methanosarcina sp. Kolksee (62 reads). Likewise, TRIAVA’s viral community was also diverse, and mainly dominated by large dsDNA viruses, with Nucleocytoviricota (826 reads, 42.8%) emerging as the most abundant phylum. Within the Mimiviridae family, the genus Mimivirus (316 reads, 16.4%) was the most dominant. At the species level, Mimivirus bradforsmassiliense was the most prevalent (316 reads). Additionally, Urovicota (499 reads, 26%) was the second most abundant phylum. Within Caudoviricetes, the genus Hiyaavirus and the species Hiyaavirus hiyaa (70 reads, 3.6%) were the most abundant. Additionally, the bacterial community in TRIAVA was primarily composed of members of the Pseudomonadota phylum, which accounted for 579K reads (80.9%). The most dominant genera and species were Candidatus Portiera aleyrodidarum (306k reads, 42.7%), Candidatus Hamiltonella defensa (171k reads, 23.8%), followed by Wolbachia endosymbionts of Bemisia tabaci (46.3k reads, 6.3%). Furthermore, and to predict genes/proteins from the classified metagenome reads and interpret these gene sets, we also performed functional annotation and GO enrichment analysis (Tables 2, 3; Supplementary Table 4; Figure 9). Our blastN findings revealed that the most prevalent species identified was Candidatus Hamiltonella defensa, with 81 genes aligned to the accession CP016303.1, 7 genes aligned to CP017613.1, 1 gene each for CP023988.1, CP017614.1, and CP017605.1. Additionally, Candidatus Portiera aleyrodidarum was also detected, CP016304.1 (7 genes). Various sequences matching Bemisia tabaci (another whitefly host) were also identified, including different RNA types: two non-coding RNAs (XR_002008634.1 and XR_002010031.1) and several mRNAs (XM_019058950.1, XM_019058068.1, XM_019052762.1, XM_019052620.1 and XM_019043942.1), each corresponding to one gene. Furthermore, we also identified complete and partial mitochondrial genome of Bemisia tabaci such as MH186145.1 (2 genes), XM_017766450.1 (1 gene), and MK360021.1 (1 gene). It is worth noting here that both TRIAVA and Bemisia tabaci are whiteflies and belong to the same family (Aleyrodidae), which means their genomes share significant sequence similarities. Since the genome of Bemisia tabaci is more extensively studied and well-annotated compared to TRIAVA’s, many of the sequences from TRIAVA could align to Bemisia tabaci simply due to evolutionary conservation rather than contamination. Additional sequences of interest included a match to Plautia stali symbiont (AP012551.1) with one gene. Plautia stali symbionts are typically associated with shield bugs, suggesting potential cross-contamination or shared microbial sequences. A sequence from Cucumis melo (LN683406.1) with one gene was also detected and this may indicate the presence of plant-originated sequences, potentially from environmental contamination. Lastly, Wolbachia endosymbiont of TRIAVA (CP016327.1) was also detected with one gene (Table 2). The percentage identity of the hits from the output for blastN (format 6) is provided in Supplementary Table 5.
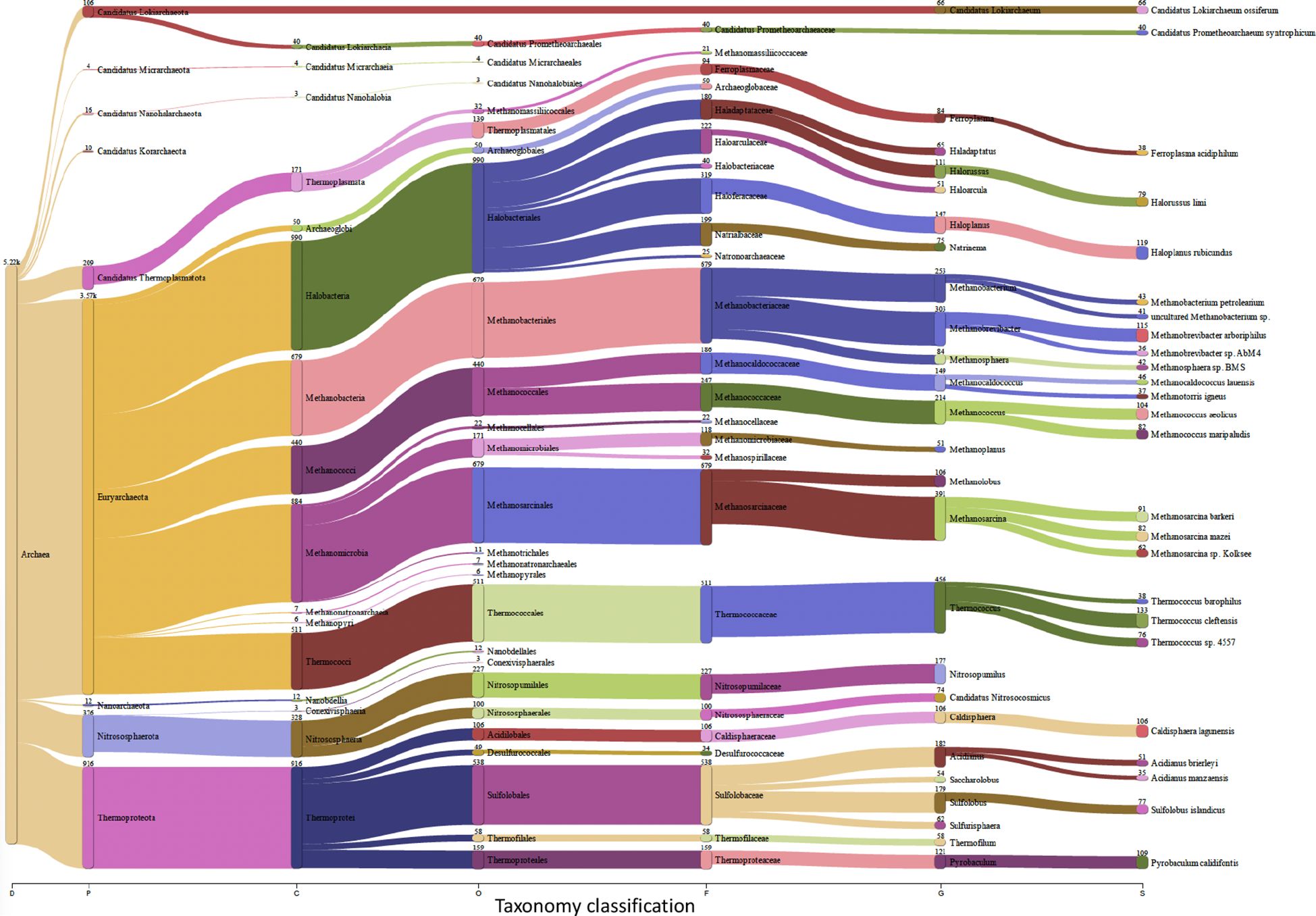
Figure 6. Archeal Communities in Trialeurodes vaporariorum. Sankey plot comprehensively illustrating, at various hierarchical taxonomic levels and after fitration at default threshold, the abundance and diversity of archeal communities harbored In Trialeurodes vaporariorum. D, Domain; P, Phylum; C, Class; O, Order; F, Family; G, Genus; S, Species.
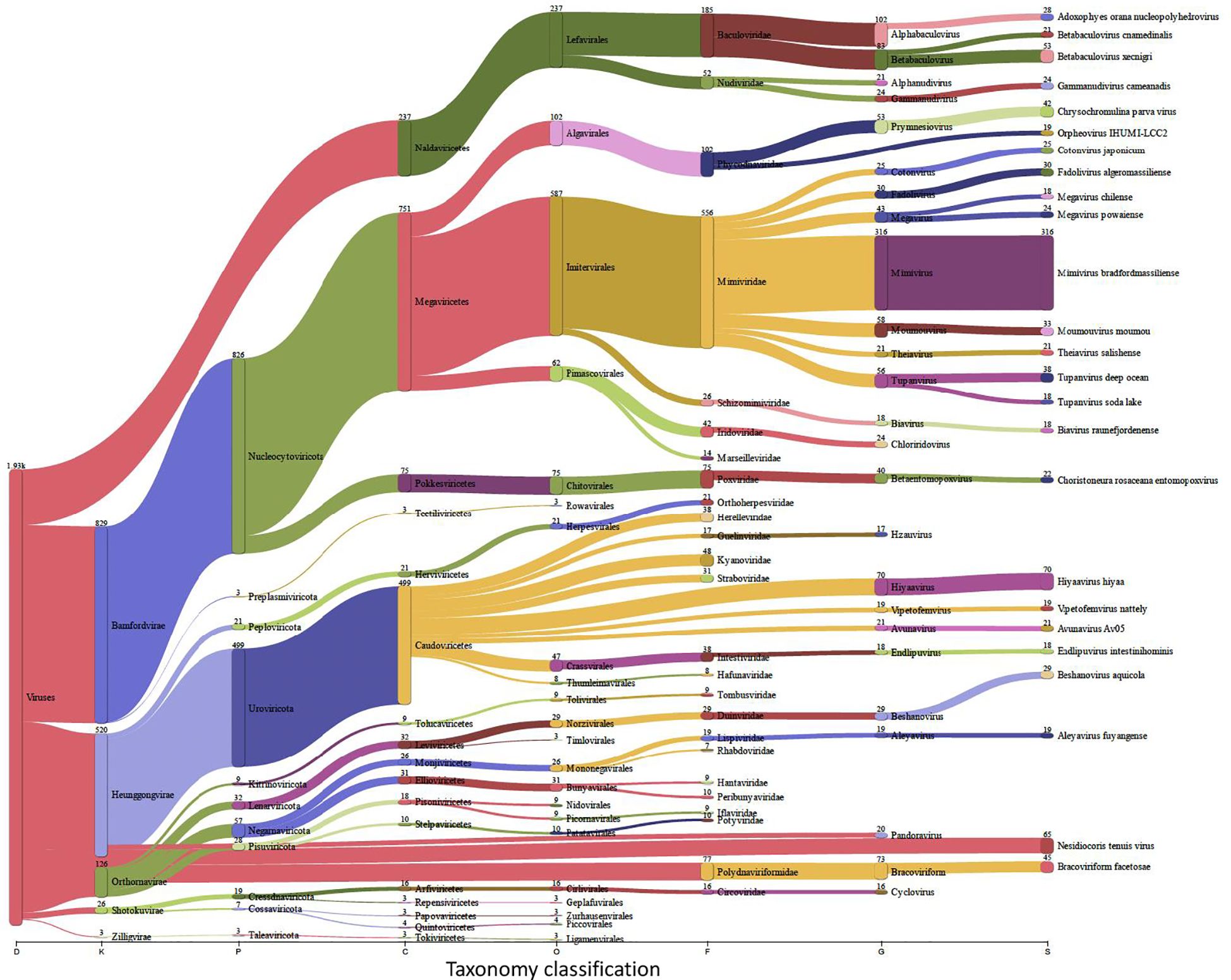
Figure 7. Viral Communities in Traleurodes vaporariorum. Sankey plot comprehensively illustrating, at various hierarchical taxonomic levels and after fitration at default threshold, the abundance and diversity of viral communities harbored in Trialeurodes vaporariorum. D, Domain; K, Kingdom; P, Phylum; C, Class; S, Species; O, Order; F, Family; G, Genus; S, Species.
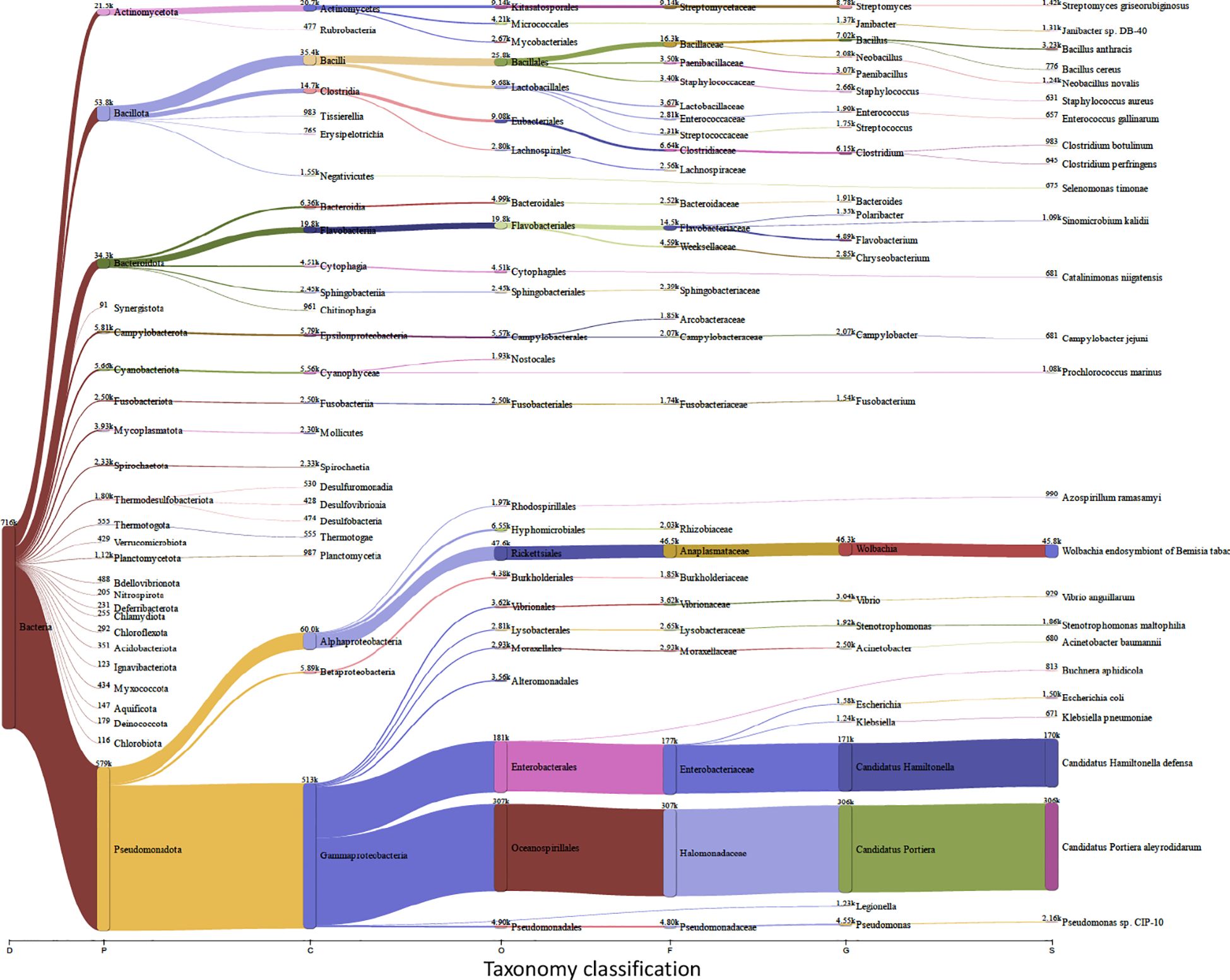
Figure 8. Bacterial Communities in Trialeurodes vaporariorum. Sankey plot comprehensively illustrating, at various hierarchical taxonomic levels and after fitration at default threshold, the abundance and diversity of harbored bacterial communities in Trialeurodes vaporaniorum. D, Domain; P, Phylum; C, Class; O, Order; F, Family; G, Genus; S, Species.
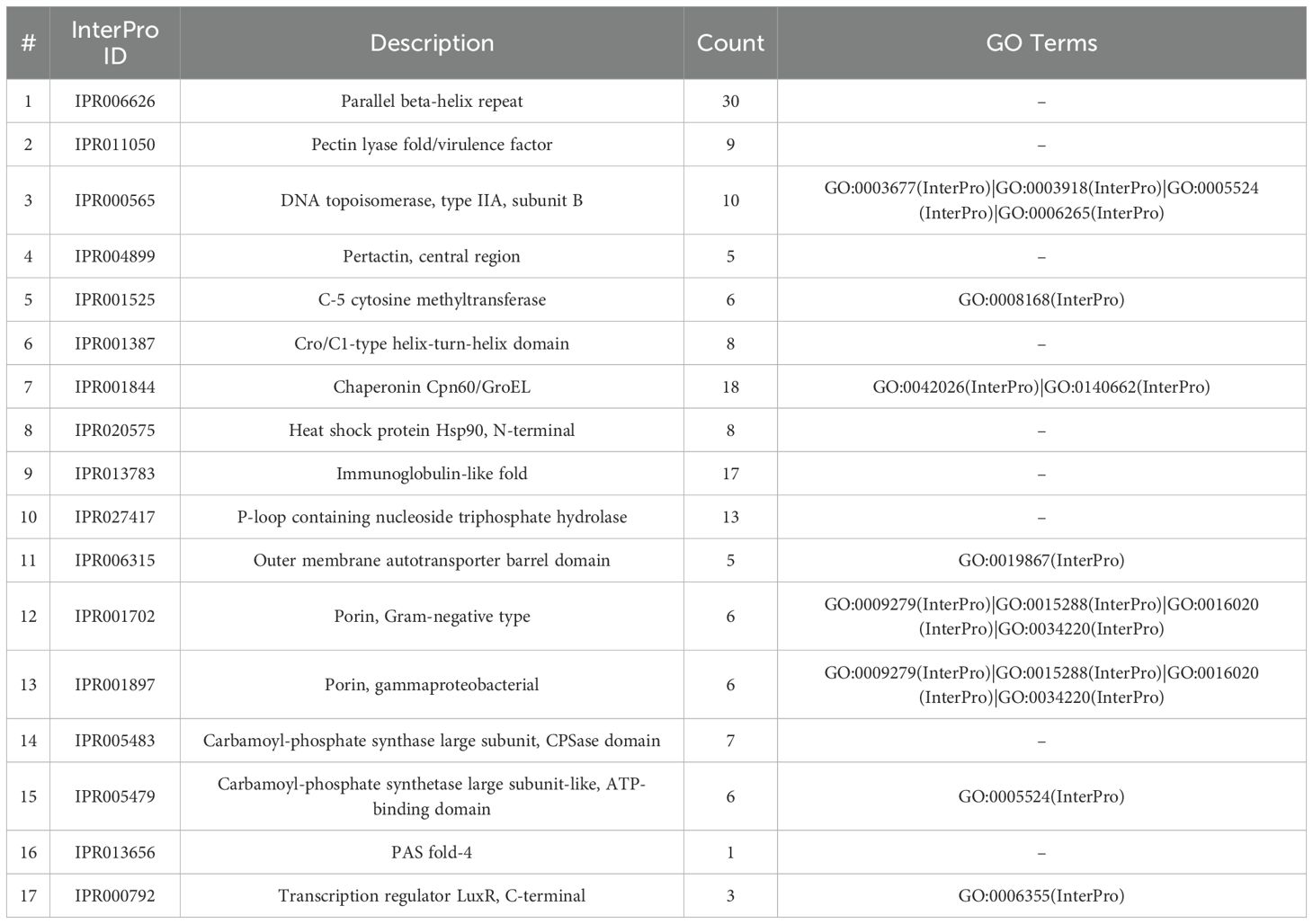
Table 3. Summarized list of identified protein domains in Trialeurodes vaporariorum microbiome linked to pathogenicity and symbiotic protection.
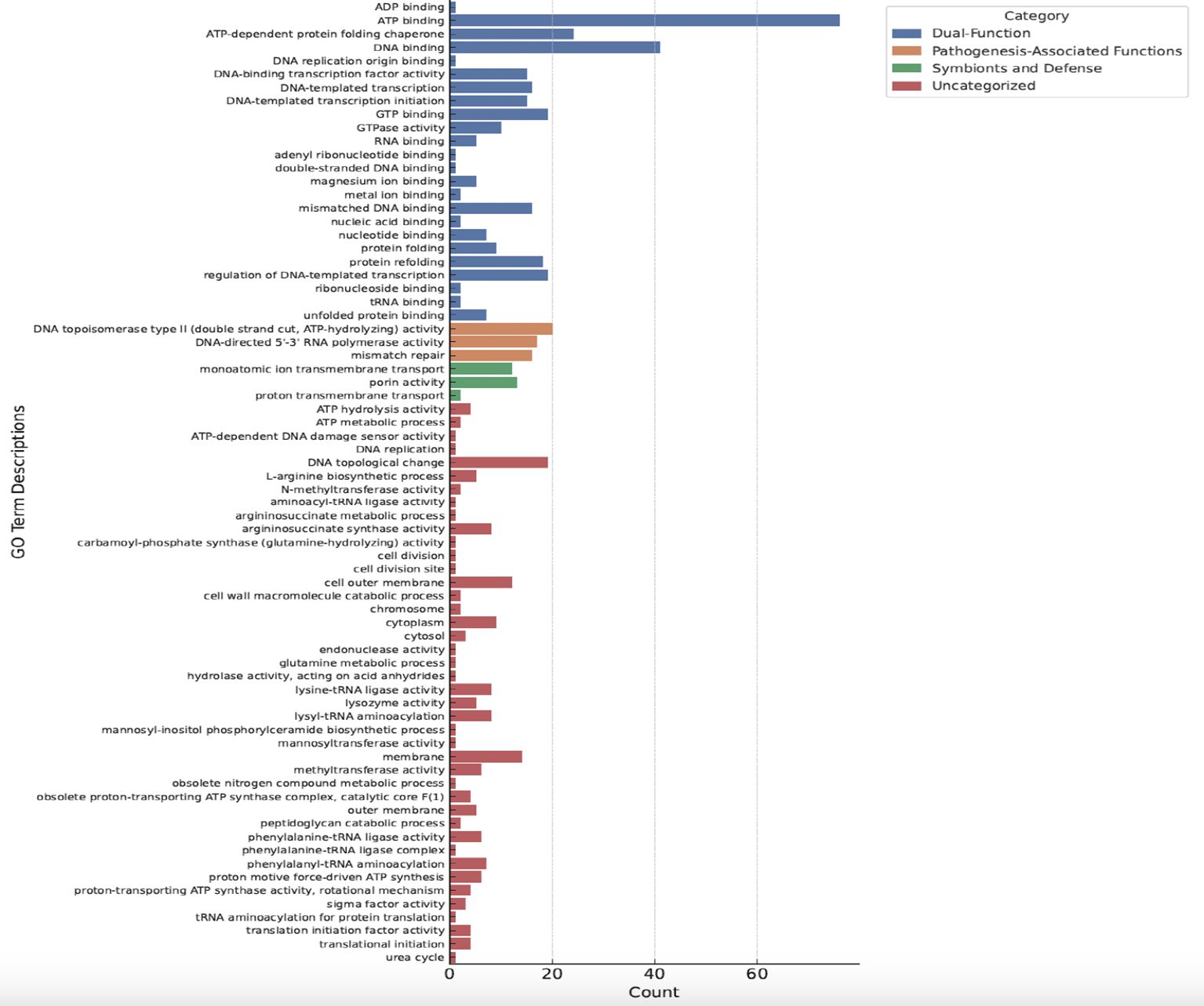
Figure 9. Gene ontology enrichment analysis of Trialeurodes vaporariorum metagenome. Histogram illustrates GO terms relevant to the genes in Trialeurodes vaporariorum metagenome.The y-axis represents the GO terms divided into four categories: blue (dual function),orange (pathogenesis-associated functions), green (symbionts and defense), and red (uncategorized). The x-axis represents the number of genes within each category. GO terms were considered enriched with a p-value cut-off of <0.05.
Along this, we identified a list of TRIAVA microbiome predicted protein domains encompassing a wide range of biological roles, including enzymes involved in various metabolic pathways, transporters facilitating the movement of molecules across cell membranes, DNA-binding domains participating in gene regulation, and chaperones assisting in protein folding and stability (Supplementary Table 4). Interestingly, several of those identified protein domains could be linked to pathogenicity and symbiotic protection (Table 3). The “Parallel beta-helix repeat” (IPR006626), a structural motif commonly found in enzymes such as pectate lyases that play a role in plant cell wall degradation, was among the most abundant domains detected in the TRIAVA microbiome. Furthermore, the “Chaperonin Cpn60/GroEL” (IPR001844), a molecular chaperone involved in protein folding and stress response, was also observed to be in high abundance. The “P-loop containing nucleoside triphosphate hydrolase” (IPR027417), an ATP-binding domain present in numerous enzymes, was also among the high abundant domains identified (Table 3). Along this, one of the most striking observations in the GO terms results (Figure 9) was the abundance of terms related to various DNA-binding and transcription-related processes. For instance, “DNA-binding transcription factor activity”, “regulation of DNA-templated transcription,” and “DNA-templated transcription initiation” were among the most highly represented terms. Another notable finding was also the presence of several GO terms associated with symbiosis and pathogenesis, such as “protein folding”, “ATP binding”, and “ADP binding”, highlighting further the intricate balance between beneficial and detrimental interactions within the TRIAVA microbiome. Remarkably as well, a significant number of GO terms that uniquely associate with symbiosis and defense mechanisms was detected. These include “lysozyme activity”, “lysyl-tRNA aminoacylation”, and “L-arginine biosynthetic process”. Also, several GO terms that could be linked to pathogenesis-associated functions exclusively like “phenylalanine-tRNA ligase activity,” “phenylalanine-tRNA ligase complex,” and “outer membrane” were also detected.
Discussion
Greenhouse farming has been recently presented as the trail towards “precision agriculture” (Karanisa et al., 2022). With this bright prospect; however, and despite the positive impact of greenhouse farming on enhancing production and quality as compared to the traditional practice in open field systems, agronomists are still facing significant challenges (Maraveas et al., 2023; Farvardin et al., 2024; Yan et al., 2024). These challenges could be either due to natural driven factors like climate setpoint, wind, solar radiation, or static driven ones like defects in ventilation, screen, greenhouse homogenous structure (van Tuijl et al., 2008; Balendonck et al., 2010), and natural enemies including insect pests, plant pathogens, and weed (Oerke, 2006). Although environmental parameters are relatively maintained in greenhouse systems, fluctuations could still occur. In the cucumber and tomato grown sectors that we have studied, the recorded weekly average temperature and relative humidity readouts did fluctuate; yet, within standard ranges generally adopted for ideal production outcomes, with only a few, if any, readouts marginally either below or above these standard values (Peet et al., 1997; Miao, 2000; Kittas et al., 2005; Cherie, 2010; Boote et al., 2012; Hochmuth and Hochmuth, 2012; Jones, 2013; Shamshiri et al., 2018; Nikolaou et al., 2019; Amin et al., 2021). This; therefore, ruled out any potential negative impact of greenhouse environmental conditions on crop quality and yield in our studied sectors. Though ideal, establishing a “model climate” in greenhouses for production efficiency could; on the other hand, potentially impact the growth and abundance of insects in greenhouses, among which are pests that could cause crop damage and disease. In support of this outlook, the “insects survey” that we have conducted in the tomato and cucumber grown greenhouse sectors did indeed confirm the presence of various insect pests, including insect vectors, previously reported to cause crop diseases. One example of those pests is the whitefly, TRIAVA, known to transmit viruses like the Tomato torrado virus (Verbeek et al., 2014) and the Pepino mosaic virus (Noël et al., 2014) to tomato crops leading to severe necrosis and yield loss (van der Vlugt et al., 2000; Wieczorek et al., 2020). Another example is the Aphis gossypii aphid, a vector for more than fifty plant viruses including the cucumber mosaic virus, the potato virus, the citrus tristeza virus, and the turnip virus (Kennedy et al., 1962; Shim et al., 1979; Eastop VF, 2000), and a broadly dispersed pest for agricultural crops (Ebert and Cartwright, 1997; Im et al., 2022). Other insect vectors including Creontiades pallidus (Soyer, 1942; Wheeler, 2001; Zamharir and Shameli, 2023) and Earias insulana (Nasr and Azab, 1969; Borker et al., 1980) that transmit phytopathogenic pathogens to plants causing disease were also detected in our studied greenhouse sectors. Furthermore, insect pests reported to infest horticultural crops in open field and/or greenhouses systems in different parts of the world including Liriomyza trifolii (Spencer, 1973; Kang, 1996; Gao et al., 2017), Hylastes cunicularius (Zhou et al., 2004), Bradysia species (Hamlen and Mead, 1979; Jagdale et al., 2004; Vaughan et al., 2011), Spoladea recurvalis (Batra and Bhattacherjee, 1960; Aderolu, 2013; Mureithi et al., 2017; Othim et al., 2017; Othim et al., 2018), Trichoplusia species (Franklin et al., 2010; Cervantes et al., 2011), and Diaphania indica (Hutson, 1931; Soares, 1945; May, 1947) were also present in these studied cucumber and tomato grown sectors. It is worth noting here that insect collection in this study was carried out using only four tools, opening up for the potentiality of the presence of additional insects in these sectors that could have been probably missed.
To establish figurative alliance between insect presence and greenhouse environmental conditions, we correlated the abundance of TRIAVA, one of the identified insect pests/vectors in the studied greenhouse sectors, with greenhouse temperature and relative humidity conditions. Our findings revealed an overall positive correlation between TRIAVA abundance and temperature and a negative correlation between TRIAVA abundance and relative humidity. Comparable with our findings, previous studies conducted on whiteflies also delineated an inverse correlation between whiteflies population and precipitation and a direct correlation between whiteflies population and high temperature, with a fluctuation in the correlation trend between whiteflies population and humidity over several months of the year (Saghafipour et al., 2020). Other similar studies revealed no significant correlation between temperature and TRIAVA population and a negative correlation between rainfall rates and TRIAVA population (Nasruddin et al., 2021). In Gamarra et al’s adjusted temperature-dependent phenology predicted model, a temperature ranging from 11.5°C to 35.5°C was considered the typical range that allows TRIAVA development, with ~ 24°C being the ideal value within the range for a growth peak (Gamarra et al., 2020). It is important to highlight in this context that the impact severity of climate pressure on the insect is dependable on the adaptability and tolerance ability of the insect itself as well as on the amount of environmental stress exhibited. In some occurrences, climate conditions could also impact the microbial community residing within the insect, including the pathogens carried by the insect vectors, affecting the overall epidemic of a plant disease in an agricultural setting (Aregbesola et al., 2019). Here, we show that TRIAVA abundance, which adjacently concords with temperature and inversely correlates with relative humidity, negatively associates with crop yield, a finding that aligns with that of previous studies (Gamarra et al., 2016; Park et al., 1998; Bi et al., 2002; Moreau and Isman, 2012; Prijovic et al., 2013; Nasruddin and Mound, 2016; Kaya Apak et al., 2017).
Today, metagenomics has made significant advancements in agricultural practices. With the vast amount of generated data from metagenomics, the use of computational models based on environmental facets and changes in the microbial communities, has been allowing for more proactive pest control approaches. As such, and to better understand TRIAVA’s microbial community in such a scenery, conclude causal relationships between microbial presence and insect fitness, and particularly unravel whether the insect’s residing microbes could be potentially involved in TRIAVA’s self-protection mechanisms or in its pathogenic effect on crops, we comprehensively analyzed the metagenome of TRIAVA insects collected from the studied cucumber and tomato grown greenhouse sectors. Interestingly, the bacterial taxa detected in the TRIAVA metagenome highlighted the dominance of symbionts such as Candidatus Portiera aleyrodidarum, which is essential for nutrient provision to the host vector (Jiang et al., 2013), and other secondary symbionts such as Candidatus Hamiltonella defensa and Wolbachia. Hamiltonella defensa is known to harbor bacteriophages that encode toxins, which inhibit the larvae of parasitoid wasps (Degnan et al., 2009; Oliver et al., 2009). This symbiosis provides TRIAVA with enhanced survival capabilities in environments with high parasitoid pressure. Wolbachia modulates immune responses and may interfere with viral replication, this could; therefore, provide an indirect protection too (Hoffmann et al., 2011; Joubert et al., 2016). Previous studies have also shown that Wolbachia can reduce the ability of insect hosts to transmit certain viruses, which may be relevant to limiting the spread of begomoviruses (Kamtchum-Tatuene et al., 2017). Taken together, these symbionts detected in the TRIAVA microbiome support our postulate that the presence of symbiotic bacteria provides protective effects to the insect host.
Pathogenic bacterial species were also detected in TRIAVA’s studied microbiome. Examples of which include Pseudomonas spp., which are commonly associated with vascular wilt, blight and root diseases (AUSVEG; Durrands and Cooper, 1988; Hayward, 1991), Stenotrophomonas spp., known to be opportunistic pathogens (Gherardi et al., 2015; Lira et al., 2017), and Flavobacterium spp. which are often associated with tissue degradation in fish and could potentially cause rot (Rochat et al., 2019). Streptomyces spp., that could produce antibiotics (Ohnishi et al., 2008; Patzer and Braun, 2010) and cause scab-like symptoms through certain strains (Ismail et al., 2020; Ünlü and Elçi, 2022), were also detected in TRIAVA’s bacterial microbiome community. The presence of antibiotic-producing genera, such as Streptomyces, indicates the potentiality of microbial interference with external pathogenic bacteria. It is worth highlighting here that while Streptomyces spp. are more commonly associated with soil microbiomes (Gopalakrishnan et al., 2020), their low abundance in TRIAVA may still contribute to localized pathogen suppression. These symbionts likely form part of a broader defense mechanism that balances pathogen suppression and nutrient acquisition.
Relevant to TRIAVA’s identified viral community, the presence of the Geminiviridae family, albeit at low abundance, supports previous reports of TRIAVA’s aptitude to potentially serve as a vector for viral pathogens that cause significant economic losses. This family, for example, includes begomoviruses, such as Tomato yellow leaf curl virus (TYLCV), which is a known pathogen for tomatoes (Wu et al., 2006; Stansly and Naranjo, 2010; Navas-Castillo et al., 2011; Yan et al., 2021). It is important to point out here that a key limitation in our metagenomic study is its exclusiveness in analyzing genomic DNA and not RNA samples which might have resulted in precluding the detection of RNA viruses present in TRIAVA’s viral community. Moreover, TRIAVA’s archaeal community was dominated by the phylum Euryarchaeota, which includes methanogenic and halophilic archaea. These archaea are typically found in saline and extreme environments (DasSarma et al., 2009). This is particularly relevant in Qatar, where the Persian Gulf has one of the highest salinity levels globally, reaching up to 50 PSU (practical salinity units) (Yao and Johns, 2010; Mohan et al., 2024). The detected halophilic archaea may originate from saline irrigation water used in greenhouses or from soil microbial communities affected by the environmental salinity. While archaea usually do not directly cause plant diseases (Cavicchioli et al., 2003), they may play a role in modulating the microbiome, affecting nutrient cycling and microbial interactions in the greenhouse ecosystem.
Along this, several of the identified protein domains linked to plant pathogenicity, insect virulence and invasion mechanisms, further support TRIAVA’s classification as a pest and a vector that can adversely affect plant health and crop yield. Among the identified domains, for example, the parallel beta-helix repeat (IPR006626) is commonly found in proteins such as pectin lyases known to degrade plant cell walls (Jenkins et al., 1998; Marín-Rodríguez et al., 2002) and is also reported to associate with bacteria that cause tissue degradation, such as Pseudomonas and Erwinia species (Weigele et al., 2003; Yamasaki et al., 2004; Abbott and Boraston Alisdair, 2008). Similarly, the pectin lyase fold/virulence factor (IPR011050) domain which indicates enzymatic activity to break down pectin, a major component of plant cell walls, potentially contributing to soft rot and vascular wilt diseases (Durrands and Cooper, 1988; Yoder et al., 1993; Mayans et al., 1997; de Sain and Rep, 2015), was also identified in our data set. The DNA topoisomerase, type IIA, subunit B (IPR000565) domain, which is essential for DNA replication and repair (Roca, 1995; Champoux, 2001; Wang, 2002; Corbett et al., 2005), was also identified. Its presence could potentially indicate bacterial replication mechanisms or prophage integration events, both of which can enhance virulence. Another significant domain is pertactin (IPR004899), which is commonly associated with bacterial adhesion and virulence (Charles et al., 1989; Li et al., 1992; Emsley et al., 1996). Pertactin-containing proteins help bacteria attach to host surfaces, facilitating colonization and infection (Locht, 1999). Additionally, the detection of the C-5 cytosine methyltransferase (IPR001525) domain suggests that certain bacteria in the metagenome may use DNA methylation to regulate gene expression or evade plant immune responses (Pósfai et al., 1988; Cheng et al., 1993; Goll et al., 2006; Ryazanova et al., 2012). Furthermore, the identification of the Cro/C1-type helix-turn-helix domain (IPR001387), typically found in regulatory proteins that control viral lytic and lysogenic cycles (Aggarwal et al., 1988; Steinmetzer et al., 2002), suggests the presence of prophages or bacteriophages that may contribute to horizontal gene transfer and enhance bacterial adaptability. Collectively, these findings highlight the potential presence of pathogenic bacteria and prophage elements that can contribute to plant damage. Our results also identified several protein domains that support the protective role of symbiotic bacteria in the whitefly microbiome. The chaperonin Cpn60/GroEL (IPR001844) domain was among the most common. Chaperonins are molecular chaperones that assist in protein folding and are crucial for maintaining protein stability under stressful conditions (Hemmingsen et al., 1988; Hill et al., 2004; Chaurasia and Apte, 2009). Symbionts like Hamiltonella defensa and Wolbachia may use these chaperonins to enhance their resilience and support the whitefly host during environmental stress. Another identified key protein domain is the heat shock protein Hsp90, N-terminal (IPR020575) domain, which plays a role in stabilizing regulatory proteins during stress responses (Zuehlke and Johnson, 2010; Johnson, 2012; Lavery et al., 2014). Its identification could suggest that symbionts or TRIAVA itself may rely on heat shock proteins to survive in certain environmental conditions, such as the greenhouse temperature conditions. The immunoglobulin-like fold (IPR013783), structurally similar to immune-related proteins (Bork et al., 1994; Halaby et al., 1999; Potapov et al., 2004), was also detected, indicating the presence of proteins that may participate in defense-like responses, potentially modulating interactions with pathogens. The P-loop containing nucleoside triphosphate hydrolase (IPR027417) domain, commonly associated with ATPases, was also pronounced in our results. ATPases play crucial roles in energy-dependent processes such as signal transduction and defense mechanisms (Walker et al., 1982; Leipe et al., 2002; Leipe et al., 2004). Symbionts may; therefore, use these ATP-dependent proteins to maintain their protective functions. Additionally, the presence of the outer membrane autotransporter barrel domain (IPR006315) suggests that symbionts may secrete effector proteins to modulate the host’s immune response or inhibit pathogenic bacteria (Henderson et al., 1998; Lee and Schneewind, 2001; Meuskens et al., 2019). Two porin-related domains, Gram-negative type porins (IPR001702) and gammaproteobacterial porins (IPR001897), were also detected in our results. Porins mediate the transport of molecules across bacterial membranes, which can regulate nutrient uptake and the release of molecules involved in microbial competition (Jap and Walian, 1990; Pauptit et al., 1991; Kreusch and Schulz, 1994; Galdiero et al., 2012). These porins may thus enable symbionts to control their internal environment or secrete factors that suppress pathogens. We also identified the carbamoyl-phosphate synthase/synthetase domains (IPR005483, IPR005479), involved in nitrogen metabolism (Stapleton et al., 1996; Raushel et al., 1999; Kim et al., 2002; Saha et al., 2007; Schoneveld et al., 2007), suggesting that symbionts may contribute to nitrogen cycling, potentially enhancing whitefly survival by supplementing essential nutrients. Another interesting protein domain detected in our findings is the Signal Transduction and Environmental Sensing, PAS Fold-4 (IPR013656). PAS domains are known for their roles in sensing environmental stimuli such as oxygen, redox potential, and light (Ponting and Aravind, 1997; Zhulin et al., 1997; Hefti et al., 2004). Therefore, the presence of these domains suggests that symbionts (like Hamiltonella and Portiera) may help the vector detect environmental stressors, such as reactive oxygen species (ROS), known to be part of a plant’s immune defense mechanisms. Proteins with PAS domains could regulate such stress responses, helping the symbionts and the whitefly vector adapt to harsh environments that could occur in greenhouses. Additionally, protein domains relevant to the Transcription regulator LuxR, C-terminal (IPR000792), involved in quorum sensing, were also detected (Ducros et al., 2001; Maris et al., 2002). Quorum sensing may allow symbionts like Hamiltonella to coordinate toxin production when parasitoids or other threats are present. LuxR proteins could also regulate genes involved in nutrient exchange between the symbionts and the whitefly. Taken together, the presence of transcriptional regulators (like LuxR family) and environmental sensors (like PAS domains) indicates that symbionts may modulate the host’s immune system, reducing susceptibility to pathogens. This could also explain how symbionts like Wolbachia interfere with viral replication (Chrostek et al., 2013; Rainey et al., 2016). The detection of PAS and LuxR domains could also support the postulate that symbionts regulate their defense mechanisms and potentially enhance TRIAVA’s resilience by sensing environmental stressors, coordinating protective responses through quorum sensing and modulating stress and immune responses to reduce ROS-related damage. Collectively, these computational predictions set the stage for further experimental validations to elucidate insects-microbial interactions in future studies.
Conclusion
In agro-ecological systems, the interaction between climate conditions, multi-species and the grown crops is complex and multifaceted, complicating in many occurrences the development of robust, smart, and controlled agricultural production strategies. In this study, we provide further insight into the rapports between meteorological circumstances, insects, and crop yield in greenhouses, the findings of which will serve ongoing efforts to sail towards an upsurge in agricultural production and surpass food insecurity.
Data availability statement
The metagenome assembly data for this study have been deposited in NCBI BioProject under accession number PRJNA1216413.
Ethics statement
All protocols followed in this study were approved by Qatar University-Institutional Biosafety Committee (Approval#QU-IBC-2022/006). The study adhered to national and international ethical standards and research guidelines and was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AQ: Methodology, Validation, Investigation, Formal analysis, Writing – review & editing. NA: Methodology, Validation, Investigation, Formal analysis, Writing – review & editing. FAA: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Writing – review & editing. MSA: Conceptualization, Resources, Methodology, Writing – review & editing. NA: Conceptualization, Resources, Methodology, Writing – review & editing. MB: Methodology, Software, Validation, Formal Analysis, Writing – review & editing. HMY: Conceptualization, Supervision, Writing – review & editing. AZ: Conceptualization, Resources, Methodology, Writing – review & editing. RR: Conceptualization, Visualization, Software, Methodology, Validation, Investigation, Formal analysis, Writing – review & editing. LK: Conceptualization, Visualization, Funding acquisition, Resources, Methodology, Investigation, Validation, Formal analysis, Data curation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Ministry of Environment and Climate Change and the Ministry of Municipality/Doha Municipality (Insects and Rodents Control Section) in the State of Qatar, by Agrico Agricultural Development W.L.L., and by Qatar University Research Fund (QUST-1-CHS-2022_323) awarded to Layla Kamareddine. The authors also thank the Health Sector at Qatar University for funding the article processing charges and BGI for providing the sequencing platform and technical support. The graphical abstract of the study overview was created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1581707/full#supplementary-material
References
Abbott, D. W. and Boraston Alisdair, B. (2008). Structural biology of pectin degradation by enterobacteriaceae. Microbiol. Mol. Biol. Rev. 72, 301–316. doi: 10.1128/MMBR.00038-07
Abdu, R. M. and Shaumar, N. F. (1985). A preliminary list of the insect fauna of Qatar. Qatar University - Science Bulletin, vol. 5, pp. 215–232.
Aderolu, I. (2013). Occurrence, abundance and control of the major insect pests associated with amaranths in ibadan, Nigeria. Entomology Ornithology Herpetology: Curr. Res., 02. doi: 10.4172/2161-0983.1000112
Aggarwal, A. K., Rodgers, D. W., Drottar, M., Ptashne, M., and Harrison, S. C. (1988). Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Sci. (New York N.Y.) 242, 899–907. doi: 10.1126/science.3187531
Amin, B., Atif, M. J., Wang, X., Meng, H., Ghani, M. I., Ali, M., et al. (2021). Effect of low temperature and high humidity stress on physiology of cucumber at different leaf stages. Plant Biol. (Stuttg) 23, 785–796. doi: 10.1111/plb.13276
Andrews, S., Krueger, F., Segonds-Pichon, A., Biggins, L., Krueger, C., and Wingett, S. (2010). FastQC. A quality control tool for high throughput sequence data. Babraham Bioinformatics. 370.
Aregbesola, O. Z., Legg, J. P., Sigsgaard, L., Lund, O. S., and Rapisarda, C. (2019). Potential impact of climate change on whiteflies and implications for the spread of vectored viruses. J. Pest Sci. 92, 381–392. doi: 10.1007/s10340-018-1059-9
AUSVEG Bacterial diseases – an overview. Available online at: https://ausveg.com.au/knowledge-hub/crop-protection/bacterial-diseases-in-vegetable-crops/ (Accessed January 23, 2025).
Balendonck, J., Van Os, E., van der Schoor, R., Van Tuijl, B., and Keizer, L. (2010). Monitoring spatial and temporal distribution of temperature and relative humidity in greenhouses based on wireless sensor technology. Int. Conf. Agric. Engineering-AgEng.
Batra, H. and Bhattacherjee, N. (1960). Occurrence of Hymenia recurvalis (Fabricius)(Lepidoptera: Pyalidae) as a bad pest of some leaf vegetables. Indian J. Entomol 22, 128–130.
Beadle, D. and Leckie, S. (2012). “Peterson field guide to moths of northeastern North America,” in Peterson field guide series, 1st ed (Houghton Mifflin Harcourt Pub. Company Boston, Boston).
Benke, K. and Tomkins, B. (2017). Future food-production systems: vertical farming and controlled-environment agriculture. Sustainability: Science Pract. Policy 13, 13–26. doi: 10.1080/15487733.2017.1394054
BGI Genomics (2021). DNA & RNA sample extraction. Available online at: https://services.bgi.com/dna-rna-sample-extraction-promotion1 (Accessed March 24, 2025).
Bi, J., Toscano, N., and Ballmer, G. (2002). Greenhouse and field evaluation of six novel insecticides against the greenhouse whitefly Trialeurodes vaporariorum on strawberries. Crop Prot. 21, 49–55. doi: 10.1016/s0261-2194(01)00063-1
Boote, K., Rybak, M., Scholberg, J., and Jones, J. (2012). Improving the CROPGRO-tomato model for predicting growth and yield response to temperature. HortScience 47, 1038–1049. doi: 10.21273/hortsci.47.8.1038
Bork, P., Holm, L., and Sander, C. (1994). The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242, 309–320. doi: 10.1016/s0022-2836(84)71582-8
Borker, S. G., Verma, J. P., and Singh, R. P. (1980). Transmission of Xanthomonas malvacearum (Smith) Dowson the incitant of bacterial blight of cotton through spotted bollworms. Indian J. Entomology 42, 390–397.
Capinera, J. L. (2008). Potato Aphid, Macrosiphum euphorbiae (Thomas) (Hemiptera: Aphididae), in Encyclopedia of Entomology. Ed. Capinera, J. L. (Dordrecht: Springer Netherlands), 3008–3011.
Castillo Carrillo, C. (2019). First record of the tomato potato psyllid Bactericera cockerelli from South America. Bull. Insectology 72, 85–91.
Cavicchioli, R., Curmi, P. M., Saunders, N., and Thomas, T. (2003). Pathogenic archaea: do they exist? Bioessays 25, 1119–1128. doi: 10.1002/bies.10354
Cervantes, V. M., Sarfraz, R. M., and Myers, J. H. (2011). Survival of cabbage looper, Trichoplusia ni (Lepidoptera: Noctuidae), through winter cleanups of commercial vegetable greenhouses: Implications for insecticide resistance management. Crop Prot. 30, 1091–1096. doi: 10.1016/j.cropro.2011.03.021
Champoux, J. J. (2001). DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413. doi: 10.1146/annurev.biochem.70.1.369
Charles, I. G., Dougan, G., Pickard, D., Chatfield, S., Smith, M., Novotny, P., et al. (1989). Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc. Natl. Acad. Sci. U. S. A/ 86, 3554–3558. doi: 10.1073/pnas.86.10.3554
Chaurasia, A. K. and Apte, S. K. (2009). Overexpression of the groESL Operon Enhances the Heat and Salinity Stress Tolerance of the Nitrogen-Fixing Cyanobacterium Anabaena sp. Strain PCC7120. Appl. Environ. Microbiol. 75, 6008–6012. doi: 10.1128/aem.00838-09
Chellappan, M. and Ranjith, M. T. (2021). “Metagenomic approaches for insect symbionts,” in Microbial Approaches for Insect Pest Management. Ed. Omkar (Springer Singapore, Singapore), 271–313.
Chen, S. (2023). Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2, e107. doi: 10.1002/imt2.107
Cheng, X., Kumar, S., Posfai, J., Pflugrath, J. W., and Roberts, R. J. (1993). Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell 74, 299–307. doi: 10.1016/0092-8674(93)90421-l
Cherie, E. (2010). The Complete Guide to Growing Tomatoes: A Complete Step-by-Step Guide Including Heirloom Tomatoes (Back-To-Basics Gardening) (Ocala, Florida, USA: Atlantic Publishing Group Inc).
Chrostek, E., Marialva, M. S., Esteves, S. S., Weinert, L. A., Martinez, J., Jiggins, F. M., et al. (2013). Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9, e1003896. doi: 10.1371/journal.pgen.1003896
Corbett, K. D., Schoeffler, A. J., Thomsen, N. D., and Berger, J. M. (2005). The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561. doi: 10.1016/j.jmb.2005.06.029
DasSarma, S., Coker, J. A., and DasSarma, P. (2009). “Archaea (overview),” in Encyclopedia of Microbiology (Third Edition). Ed. Schaechter, M. (Academic Press, Oxford), 1–23.
Degnan, P. H., Yu, Y., Sisneros, N., Wing, R. A., and Moran, N. A. (2009). Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. U. S. A. 106, 9063–9068. doi: 10.1073/pnas.0900194106
de Sain, M. and Rep, M. (2015). The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 16, 23970–23993. doi: 10.3390/ijms161023970
Diarra, K., Labou, B., Diatte, M., Sylla, S., Tendeng, E., Diamé, L., et al. Biological control in horticultural agro-ecosystems in Senegal. Acta Hortic. (1348), 129–134. doi: 10.17660/ActaHortic.2022.1348.18
Dioli, P., Hobson, E., and Salvetti, M. (2021). Creontiades pallidus (Rambur) (Hemiptera: Miridae) new true bug intercepted in England. Br. J. Entomol. Nat. Hist. 34, 266–267.
Ducros, V. M., Lewis, R. J., Verma, C. S., Dodson, E. J., Leonard, G., Turkenburg, J. P., et al. (2001). Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis. J. Mol. Biol. 306, 759–771. doi: 10.1006/jmbi.2001.4443
Durrands, P. K. and Cooper, R. M. (1988). The role of pectinases in vascular wilt disease as determined by defined mutants of Verticillium albo-atrum. Physiol. Mol. Plant Pathol. 32, 363–371. doi: 10.1016/s0885-5765(88)80030-4
Eastop, V. and Balckmann, R. (2007). Taxonomic issues. Aphids as crop pests. Eds. Van Emden, H. F. and Harrington, R. (UK: CABI publishing).
Eastop VF, B. R. (2000). Aphids on the World’s Crops: An Identification and Information Guide (Chichester, England: John Wiley).
Ebert, T. A. and Cartwright, B. O. (1997). Biology and ecology of Aphis gossypii glover (Homoptera: Aphididae). Southwestern Entomologist 22, 116–153.
Emsley, P., Charles, I. G., Fairweather, N. F., and Isaacs, N. W. (1996). Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381, 90–92. doi: 10.1038/381090a0
Engel, P. and Moran, N. A. (2013). The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
European Bioinformatics Institute QuickGO REST API: Example usage. Available online at: https://www.ebi.ac.uk/QuickGO/api/example.html (Accessed January 24, 2025).
FAO, I, UNICEF, WFP, and WHO (2017). The State of Food Security and Nutrition in the World 2017. Building Resilience for Peace and Food Security (Rome, Italy: The State of Food Security and Nutrition in the World (SOFI).
Farooq, M., Rehman, A., and Pisante, M. (2019). Sustainable Agriculture and Food Security. In Innovations in Sustainable Agriculture (Cham: Springer) pp. 3–24. doi: 10.1007/978-3-030-23169-9_1
Farvardin, M., Taki, M., Gorjian, S., Shabani, E., and Sosa-Savedra, J. C. (2024). Assessing the physical and environmental aspects of greenhouse cultivation: A comprehensive review of conventional and hydroponic methods. Sustainability 16, 1273. doi: 10.3390/su16031273
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Forkuor, G., Amponsah, W., Oteng-Darko, P., and Osei, G. (2022). Safeguarding food security through large-scale adoption of agricultural production technologies: The case of greenhouse farming in Ghana. Cleaner Eng. Technol. 6, 100384. doi: 10.1016/j.clet.2021.100384
Franklin, M. T., Ritland, C. E., and Myers, J. H. (2010). Spatial and temporal changes in genetic structure of greenhouse and field populations of cabbage looper, Trichoplusia ni. Mol. Ecol. 19, 1122–1133. doi: 10.1111/j.1365-294x.2010.04548.x
Galdiero, S., Falanga, A., Cantisani, M., Tarallo, R., Della Pepa, M. E., D’Oriano, V., et al. (2012). Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr. Protein Pept. Sci. 13, 843–854. doi: 10.2174/138920312804871120
Gamarra, H., Carhuapoma, P., Mujica, N., Kreuze, J., and Kroschel, J. (2016). 4.3. 4 Greenhouse whitefly, Trialeurodes vaporariorum (Westwood 1956). The International Potato Center (CIP). doi: 10.4160/9789290604761-12
Gamarra, H., Sporleder, M., Carhuapoma, P., Kroschel, J., and Kreuze, J. (2020). A temperature-dependent phenology model for the greenhouse whitefly Trialeurodes vaporariorum (Hemiptera: Aleyrodidae). Virus Res. 289, 198107. doi: 10.1016/j.virusres.2020.198107
Gao, Y., Reitz, S., Xing, Z., Ferguson, S., and Lei, Z. (2017). A decade of leafminer invasion in China: lessons learned. Pest Manag Sci. 73, 1775–1779. doi: 10.1002/ps.4591
Garba, M., Streito, J. C., and Gauthier, N. (2020). First report of three predatory bugs (Heteroptera: Miridae) in tomato fields infested by the invasive South American tomato pinworm, Tuta absoluta in Niger: an opportunity for biological control? Phytoparasitica 48, 215–229. doi: 10.1007/s12600-020-00788-6
Gemayel, K., Lomsadze, A., and Borodovsky, M. (2022). MetaGeneMark-2: improved gene prediction in metagenomes. BioRxiv, 2022.07. 25.500264. doi: 10.1101/2022.07.25.500264
Geneious Annotate by BLAST. Geneious manual. Available online at: https://assets.geneious.com/manual/2022.1/static/GeneiousManualsu83.htm.
Gherardi, G., Creti, R., Pompilio, A., and Di Bonaventura, G. (2015). An overview of various typing methods for clinical epidemiology of the emerging pathogen Stenotrophomonas maltophilia. Diagn. Microbiol. Infect. Dis. 81, 219–226. doi: 10.1016/j.diagmicrobio.2014.11.005
Gibb, T. J. and Oseto, C. (2019). Insect collection and identification: Techniques for the field and laboratory. Cambridge, Massachusetts, USA: Academic Press, 1–354.
Global Panel on Agriculture and Food Systems for Nutrition. (2016). Food systems and diets: Facing the challenges of the 21st century (London, UK: Global Panel on Agriculture and Food Systems for Nutrition).
Goll, M. G., Kirpekar, F., Maggert, K. A., Yoder, J. A., Hsieh, C.-L., Zhang, X., et al. (2006). Methylation of tRNAAsp by the DNA methyltransferase homolog dnmt2. Science 311, 395–398. doi: 10.1126/science.1120976
Gopalakrishnan, S., Srinivas, V., and Prasanna, S. L. (2020). “Chapter 5 - streptomyces,” in Beneficial Microbes in Agro-Ecology. Ed. Amaresan, N., Kumar Senthil, M., Annapurna, K., Kumar, K., and Sankaranarayanan, A. (London, United Kingdom: Academic Press), 55–71.
Halaby, D. M., Poupon, A., and Mornon, J. (1999). The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 12, 563–571. doi: 10.1093/protein/12.7.563
Hamlen, R. A. and Mead, F. W. (1979). Fungus gnat larval control in greenhouse plant production12. J. Economic Entomology 72, 269–271. doi: 10.1093/jee/72.2.269
Harbach, R. (1985). Pictorial keys to the genera of mosquitoes, subgenera of culex and the species of culex (Culex) occurring in southwestern asia and Egypt, with a note on the subgeneric placement of culex deserticola (Diptera: culicidae). Mosq. Syst. 17, 25.
Hayward, A. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
Heck, M. (2018). Insect transmission of plant pathogens: a systems biology perspective. mSystems 3 (2), e00168–00117. doi: 10.1128/msystems.00168-17
Hefti, M. H., Françoijs, K.-J., de Vries, S. C., Dixon, R., and Vervoort, J. (2004). The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 271, 1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x
Hemmingsen, S. M., Woolford, C., van der Vies, S. M., Tilly, K., Dennis, D. T., Georgopoulos, C. P., et al. (1988). Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333, 330–334. doi: 10.1038/333330a0
Henderson, I. R., Navarro-Garcia, F., and Nataro, J. P. (1998). The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6, 370–378. doi: 10.1016/s0966-842x(98)01318-3
Hill, J. E., Penny, S. L., Crowell, K. G., Goh, S. H., and Hemmingsen, S. M. (2004). cpnDB: a chaperonin sequence database. Genome Res. 14, 1669–1675. doi: 10.1101/gr.2649204
Hochmuth, G. J. and Hochmuth, R. C. (2012). Production of greenhouse tomatoes - Florida greenhouse vegetable production handbook. Selection Cultivars. Production 3, 1–18. doi: 10.32473/edis-cv266-1990
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457. doi: 10.1038/nature10356
Im, Y., Park, S. E., Lee, S. Y., Kim, J. C., and Kim, J. S. (2022). Early-Stage Defense Mechanism of the Cotton Aphid Aphis gossypii Against Infection With the Insect-Killing Fungus Beauveria bassiana JEF-544. Front. Immunol. 13, 907088. doi: 10.3389/fimmu.2022.907088
International Food Policy Research Institute (IFPRI) (2016). Global Nutrition Report: From Promise to Impact Ending Malnutrition by 2030. Washington, D.C.: International Food Policy Research Institute. https://globalnutritionreport.org/reports/2016-global-nutrition-report/ (Accessed January 24, 2025).
Ismail, S., Jiang, B., Nasimi, Z., Inam-Ul-Haq, M., Yamamoto, N., Danso Ofori, A., et al. (2020). Investigation of streptomyces scabies causing potato scab by various detection techniques, its pathogenicity and determination of host-disease resistance in potato germplasm. Pathogens 9. doi: 10.3390/pathogens9090760
Jagdale, G., Casey, M., Grewal, P., and Lindquist, R. (2004). Application rate and timing, potting medium, and host plant effects on the efficacy of Steinernema feltiae against the fungus gnat, Bradysia coprophila, in floriculture. Biol. Control - Biol. Control 29, 296–305. doi: 10.1016/s1049-9644(03)00164-6
Jap, B. K. and Walian, P. J. (1990). Biophysics of the structure and function of porins. Q. Rev. biophysics 23, 367–403. doi: 10.1017/s003358350000559x
Jenkins, J., Mayans, O., and Pickersgill, R. (1998). Structure and evolution of parallel beta-helix proteins. J. Struct. Biol. 122, 236–246. doi: 10.1006/jsbi.1998.3985
Jiang, Z. F., Xia, F., Johnson, K. W., Brown, C. D., Bartom, E., Tuteja, J. H., et al. (2013). Comparison of the genome sequences of “Candidatus Portiera aleyrodidarum” primary endosymbionts of the whitefly Bemisia tabaci B and Q biotypes. Appl. Environ. Microbiol. 79, 1757–1759. doi: 10.1128/aem.02976-12
Johnson, J. L. (2012). Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta 1823, 607–613. doi: 10.1016/j.bbamcr.2011.09.020
Jones, J. B. (2013). Instructions for Growing Tomatoes: in the garden and greenhouse Kindle Edition. (Scotts Valley, California, USA: CreateSpace Independent Publishing Platform).
Joubert, D. A., Walker, T., Carrington, L. B., De Bruyne, J. T., Kien, D. H., Hoang Nle, T., et al. (2016). Establishment of a wolbachia superinfection in aedes aEgypti mosquitoes as a potential approach for future resistance management. PLoS Pathog 12, e1005434. doi: 10.1371/journal.ppat.1005434
Kamareddine, L. (2012). The biological control of the malaria vector. Toxins (Basel) 4, 748–767. doi: 10.3390/toxins4090748
Kamareddine, L. (2019). “The impact of environmental and anthropogenic factors on the transmission dynamics of vector borne diseases,” in Encyclopedia of Environmental Health (Second Edition). Ed. Nriagu, J. (Elsevier, Oxford), 609–613.
Kamtchum-Tatuene, J., Makepeace, B. L., Benjamin, L., Baylis, M., and Solomon, T. (2017). The potential role of Wolbachia in controlling the transmission of emerging human arboviral infections. Curr. Opin. Infect. Dis. 30, 108–116. doi: 10.1097/qco.0000000000000342
Kang, L. (1996). Ecology and sustainable control of serpentine leafminers (Beijing, China: Science Press).
Kanwar, M. S. (2011). Performance of tomato under greenhouse and open field conditions in the trans-Himalayan region of India. Adv. Hortic. Sci. 25(1), 65–68. doi: 10.13128/ahs-12786
Karanisa, T., Achour, Y., Ouammi, A., and Sayadi, S. (2022). Smart greenhouses as the path towards precision agriculture in the food-energy and water nexus: case study of Qatar. Environ. Syst. Decisions 42, 521–546. doi: 10.1007/s10669-022-09862-2
Kaya Apak, F., Altuntas, O., and Poyraz, P. (2017). Population dynamics of the trialeurodes vaporariorum (Westw.) (Aleyrodidae, hemiptera) on broccoli under greenhouse condition, in Malatya, Turkey. Adnan Menderes Üniversitesi Ziraat Fakültesi Dergisi 20, 25–31. doi: 10.25308/aduziraat.1157134
Kelton, L. A. (1975). The lygus bugs (Genus lygus hahn) of North America (Heteroptera: miridae). Memoirs Entomol. Soc. Canada 107, 5–101.
Kennedy, J. S., Day, M. F., Eastop, V. F., and E. Commonwealth Institute of and C. Commonwealth Agricultural Bureaux Executive (1962). A conspectus of aphids as vectors of plant viruses. (London: Commonwealth Institute of Entomology London).
Khan, M. M. H. (2019). Effect of temperature and relative humidity on the population dynamics of brinjal and tomato infesting whitefly, Bemisia tabaci. Jahangirnagar Univ. J. Biol. Sci. 8, 83–86. doi: 10.3329/jujbs.v8i1.42471
Kim, J., Howell, S., Huang, X., and Raushel, F. M. (2002). Structural defects within the carbamate tunnel of carbamoyl phosphate synthetase. Biochemistry 41, 12575–12581. doi: 10.1021/bi020421o
Kim, J.-K., Park, J.-J., Park, H., and Cho, K. (2001). Unbiased estimation of greenhouse whitefly, Trialeurodes vaporariorum, mean density using yellow sticky trap in cherry tomato greenhouses. Entomologia Experimentalis Applicata 100, 235–243. doi: 10.1046/j.1570-7458.2001.00868.x
Kittas, C., Karamanis, M., and Katsoulas, N. (2005). Air temperature regime in a forced ventilated greenhouse with rose crop. Energy Buildings 37, 807–812. doi: 10.1016/j.enbuild.2004.10.009
Kreusch, A. and Schulz, G. E. (1994). Refined structure of the porin from Rhodopseudomonas blastica. Comparison with the porin from Rhodobacter capsulatus. J. Mol. Biol. 243, 891–905. doi: 10.1006/jmbi.1994.1690
Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lavery, L. A., Lavery, L. A., Partridge, J. R., Ramelot, T. A., Elnatan, D., Kennedy, M. A., et al. (2014). Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol. Cell 53, 330–343. doi: 10.1016/j.molcel.2013.12.023
Lee, V. T. and Schneewind, O. (2001). Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15, 1725–1752. doi: 10.1101/gad.896801
Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28. doi: 10.1016/j.jmb.2004.08.023
Leipe, D. D., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72. doi: 10.1006/jmbi.2001.5378
Li, J., Fairweather, N. F., Novotny, P., Dougan, G., and Charles, I. G. (1992). Cloning, nucleotide sequence and heterologous expression of the protective outer-membrane protein P.68 pertactin from Bordetella bronchiseptica. J. Gen. Microbiol. 138 Pt 8, 1697–1705. doi: 10.1099/00221287-138-8-1697
Lira, F., Berg, G., and Martínez, J. L. (2017). Double-face meets the bacterial world: the opportunistic pathogen stenotrophomonas maltophilia. Front. Microbiol 8, 2190. doi: 10.3389/fmicb.2017.02190
Locht, C. (1999). Molecular aspects of Bordetella pertussis pathogenesis. Int. Microbiol. 2, 137–144.
Lonsdale, O. (2021). Manual of North American Agromyzidae (Diptera, Schizophora), with revision of the fauna of the “Delmarva” states. ZooKeys 1051, 1–481. doi: 10.3897/zookeys.1051.64603
Lu, J., Rincon, N., Wood, D. E., Breitwieser, F. P., Pockrandt, C., Langmead, B., et al. (2022). Metagenome analysis using the Kraken software suite. Nat. Protoc. 17, 2815–2839. doi: 10.1038/s41596-022-00738-y
Lu, J. and Salzberg, S. L. (2019). Bracken: Bayesian re-estimation of abundance with Kraken. Available online at: https://ccb.jhu.edu/software/bracken (Accessed January 23, 2025).
Maraveas, C., Karavas, C.-S., Loukatos, D., Bartzanas, T., Arvanitis, K. G., and Symeonaki, E. (2023). Agricultural greenhouses: resource management technologies and perspectives for zero greenhouse gas emissions. Agriculture 13, 1464. doi: 10.3390/agriculture13071464
Marín-Rodríguez, M. C., Orchard, J., and Seymour, G. B. (2002). Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53, 2115–2119. doi: 10.1093/jxb/erf089
Maris, A. E., Sawaya, M. R., Kaczor-Grzeskowiak, M., Jarvis, M. R., Bearson, S. M. D., Kopka, M. L., et al. (2002). Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9, 771–778. doi: 10.1038/nsb845
Martin, J. H., Mifsud, D., and Rapisarda, C. (2000). The whiteflies (Hemiptera: aleyrodidae) of Europe and the mediterranean basin. Bull. Entomol Res. 90, 407–448. doi: 10.1017/s0007485300000547
Mayans, O., Scott, M., Connerton, I., Gravesen, T., Benen, J., Visser, J., et al. (1997). Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure 5, 677–689. doi: 10.1016/s0969-2126(97)00222-0
Meuskens, I., Saragliadis, A., Leo, J. C., and Linke, D. (2019). Type V secretion systems: an overview of passenger domain functions. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01163
Miao, M. (2000). Studies on the mechanism of heat injury and heat adaptation and summer cultivation techniques of cucumber (Cucumis sativus L.). Nanjing Agricultural University, Nanjing.
Mohammed, A. A., Ahmed, F. A., Younus, A. S., Kareem, A. A., and Salman, A. M. (2022). Molecular identification of two entomopathogenic fungus Clonostachys rosea strains and their efficacy against two aphid species in Iraq. J. Genet. Eng Biotechnol. 20, 67. doi: 10.1186/s43141-022-00347-y
Mohammed, A. A., Kadhim, J. H., and Kamaluddin, Z. N. A. (2018). Selection of highly virulent entomopathogenic fungal isolates to control the greenhouse aphid species in Iraq. Egyptian J. Biol. Pest Control 28, 71. doi: 10.1186/s41938-018-0079-3
Mohan, M., Dutta Roy, A., Montenegro, J. F., Watt, M. S., Burt, J. A., Shapiro, A., et al. (2024). Mangrove forest regeneration age map and drivers of restoration success in Gulf Cooperation Council countries from satellite imagery. Remote Sens. Applications: Soc. Environ. 36, 101345. doi: 10.1016/j.rsase.2024.101345
Moreau, T. L. and Isman, M. B. (2012). Combining reduced-risk products, trap crops and yellow sticky traps for greenhouse whitefly (Trialeurodes vaporariorum) management on sweet peppers (Capsicum annum). Crop Prot. 34, 42–46. doi: 10.1016/j.cropro.2011.11.011
Munyaneza, J. E. (2012). Zebra chip disease of potato: biology, epidemiology, and management. Am. J. potato Res. 89, 329–350. doi: 10.1007/s12230-012-9262-3
Munyaneza, J., Sengoda, V. G., Aguilar, E., Bextine, B., and McCue, K. F. (2013). First report of 'Candidatus Liberibacter solanacearum' associated with psyllid-infested tobacco in Nicaragua. Plant Dis. 9, 1244–1245. doi: 10.1094/PDIS-03-13-0247-PDN
Mureithi, D., Fiaboe, K. K. M., Ekesi, S., and Meyhöfer, R. (2017). Important arthropod pests on leafy amaranth (Amaranthus viridis, A. tricolor and A. blitum) and broad-leafed African nightshade (Solanum scabrum) with a special focus on host. Afr. J. Hortic. 11, 1–17.
Nasr, E. S. A. and Azab, A. K. (1969). Rhizopus nigricans Ehr. infection in cotton bolls in relation to bollworm infestation, age of bolls and the insecticides used on cotton plants (Lepidoptera). Bull. Entomological Soc. Egypt Economic Ser. 3, 97–102.
Nasruddin, A., Jumardi, J., and Melina, M. (2021). Population dynamics of Trialeurodes vaporariorum (Westwood) (Hemiptera: Aleyrodidae) and its populations on different planting dates and host plant species. Ann. Agric. Sci. 66, 109–114. doi: 10.1016/j.aoas.2021.08.001
Nasruddin, A. and Mound, L. (2016). First record of Trialeurodes vaporariorum Westwood (Hemiptera: Aleyrodidae) severely damaging field grown potato crops in South Sulawesi, Indonesia. J. Plant Prot. Res. 56, 199–202. doi: 10.1515/jppr-2016-0023
Navas-Castillo, J., Fiallo-Olivé, E., and Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nikolaou, G., Neocleous, D., Katsoulas, N., and Kittas, C. (2019). Effects of cooling systems on greenhouse microclimate and cucumber growth under Mediterranean climatic conditions. Agronomy 9, 300. doi: 10.3390/agronomy9060300
Noël, P., Hance, T., and Bragard, C. (2014). Transmission of the Pepino mosaic virus by whitefly. Eur. J. Plant Pathol. 138, 23–27. doi: 10.1007/s10658-013-0313-5
Oerke, E. C. (2006). Crop losses to pests. J. Agric. Sci. 144, 31–43. doi: 10.1017/s0021859605005708
Ohnishi, Y., Ishikawa, J., Hara, H., Suzuki, H., Ikenoya, M., Ikeda, H., et al. (2008). Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060. doi: 10.1128/JB.00204-08
Oliver, K. M., Degnan, P. H., Hunter, M. S., and Moran, N. A. (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. doi: 10.1126/science.1174463
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinf. 12, 385. doi: 10.1186/1471-2105-12-385
Othim, S., Agbodzavu, K., Kahuthia-Gathu, R., Akutse, K., Muchemi, S., Ekesi, S., et al. (2017). Performance of Apanteles hemara (Hymenoptera: Braconidae) on two Amaranth leaf-webbers: Spoladea recurvalis and Udea ferrugalis (Lepidoptera: Crambidae). Environ. Entomol. 46, 1284–1291. doi: 10.1093/ee/nvx156
Othim, S., Kahuthia-Gathu, R., Akutse, K., Caroline, F., and Fiaboe, K. (2018). Seasonal occurrence of amaranth Lepidopteran defoliators and effect of attractants and amaranth lines in their management. J. Appl. Entomol. 142, 637–645. doi: 10.1111/jen.12513
Park, J.-D., Kim, D.-I., and Park, U. (1998). Occurrence and within-plant distribution of Trialeurodes vaporariorum (Westwood) and Encarsia formosa (Gahan) in greenhouse. Korean J. Appl. Entomol. 37, 117–121.
Patzer, S. I. and Braun, V. (2010). Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 192(2), 426–435. doi: 10.1128/jb.01250-09
Pauptit, R. A., Schirmer, T., Jansonius, J. N., Rosenbusch, J. P., Parker, M. W., Tucker, A. D., et al. (1991). A common channel-forming motif in evolutionarily distant porins. J. Struct. Biol. 107, 136–145. doi: 10.1016/1047-8477(91)90017-q
Peet, M. M., Willits, D. H., and Gardner, R. (1997). Response of ovule development and post-pollen production processes in male-sterile tomatoes to chronic, sub-acute hight temperature stress. J. Exp. Bot. 48, 101–111. doi: 10.1093/jxb/48.1.101
Ponting, C. P. and Aravind, L. (1997). PAS: a multifunctional domain family comes to light. Curr. Biol. 7, R674–R677. doi: 10.1016/s0960-9822(06)00352-6
Pósfai, J., Bhagwat, A. S., and Roberts, R. J. (1988). Sequence motifs specific for cytosine methyltransferases. Gene 74, 261–265.
Potapov, V., Sobolev, V., Edelman, M., Kister, A., and Gelfand, I. (2004). Protein–protein recognition: juxtaposition of domain and interface cores in immunoglobulins and other sandwich-like proteins. J. Mol. Biol. 342, 665–679. doi: 10.1016/j.jmb.2004.06.072
Prijovic, M., Marcic, D., Drobnjakovic, T., Medjo, I., and Peric, P. (2013). Life history traits and population growth of greenhouse whitefly (Trialeurodes vaporariorum westwood) on different tomato genotypes. Pesticidi i fitomedicina 28, 239–245. doi: 10.2298/pif1304239p
Qin, L., Shen, W., Tang, Z., Hu, W., Shangguan, L., Wang, Y., et al. (2020). A newly identified virus in the family potyviridae encodes two leader cysteine proteases in tandem that evolved contrasting RNA silencing suppression functions. J. Virol 95 (1), e01414–01420. doi: 10.1128/jvi.01414-20
Rainey, S. M., Martinez, J., McFarlane, M., Juneja, P., Sarkies, P., Lulla, A., et al. (2016). Wolbachia blocks viral genome replication early in infection without a transcriptional response by the endosymbiont or host small RNA pathways. PLoS Pathog 12, e1005536. doi: 10.1371/journal.ppat.1005536
Rattanarithikul, R. (1982). “A guide to the genera of mosquitoes (diptera: culicidae) of Thailand with illustrated keys,” in Biological notes and preservation and mounting techniques. Am Mosq Control Assoc 14 (3), 139–208.
Raushel, F. M., Thoden, J. B., and Holden, H. M. (1999). The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry 38, 7891–7899. doi: 10.1021/bi990871p
Roca, J. (1995). The mechanisms of DNA topoisomerases. Trends Biochem. Sci. 20, 156–160. doi: 10.1016/s0968-0004(00)88993-8
Rochat, T., Pérez-Pascual, D., Nilsen, H., Carpentier, M., Bridel, S., Bernardet, J. F., et al. (2019). Identification of a novel elastin-degrading enzyme from the fish pathogen flavobacterium psychrophilum. Appl. Environ. Microbiol. 85 (6), e02535–02518. doi: 10.1128/aem.02535-18
Roditakis, N. E. (1990). Host plants of greenhouse whitefly Trialeurodes vaporariorum westwood (Homoptera: Aleyrodidae) in Crete. Attractiveness and impact on whitefly life stages. Agriculture ecosystems Environ. 31, 217–224. doi: 10.1016/0167-8809(90)90221-x
Rosson, P., Niemeyer, M., Palma, M., and Ribera, L. (2006). Economic impacts of zebra chips on the Texas potato industry (College Station, TX: Center for North American studies, department of agricultural economics, Texas A&M university).
Ryazanova, A. Y., Abrosimova, L. A., Oretskaya, T. S., and Kubareva, E. A. (2012). “Diverse domains of (Cytosine-5)-DNA methyltransferases: structural and functional characterization,” in Methylation. Ed. Anica, D. (IntechOpen, Rijeka), Ch. 2.
Saghafipour, A., Zahraei-Ramazani, A., Vatandoost, H., Asadollahi, A., Fouladi-Fard, R., Hamta, A., et al. (2020). Relationship between some environmental and climatic factors on outbreak of whiteflies, the human annoying insects. J. Arthropod Borne Dis. 14(1), 78–87. doi: 10.18502/jad.v14i1.2714
Saha, N., Datta, S., Kharbuli, Z. Y., Biswas, K., and Bhattacharjee, A. (2007). Air-breathing catfish, Clarias batrachus upregulates glutamine synthetase and carbamyl phosphate synthetase III during exposure to high external ammonia. Comparative biochemistry and physiology. Part B. Biochem. Mol. Biol. 147, 520–530. doi: 10.1016/j.cbpb.2007.03.007
Sarkar, S. C., Hatt, S., Philips, A., Akter, M., Milroy, S. P., and Xu, W. (2023). Tomato potato psyllid bactericera cockerelli (Hemiptera: triozidae) in Australia: incursion, potential impact and opportunities for biological control. Insects 14, 263. doi: 10.3390/insects14030263
Saurabh, S., Mishra, M., Rai, P., Pandey, R., Singh, J., Khare, A., et al. (2021). Tiny flies: A mighty pest that threatens agricultural productivity—A case for next-generation control strategies of whiteflies. Insects 12, 585. doi: 10.3390/insects12070585
Schoneveld, O. J. L. M., Hoogenkamp, M., Stallen, J. M., Gaemers, I. C., and Lamers, W. H. (2007). cyclicAMP and glucocorticoid responsiveness of the rat carbamoylphosphate synthetase gene requires the interplay of upstream regulatory units. Biochimie 89, 574–580. doi: 10.1016/j.biochi.2006.12.013
Shamshiri, R. R., Jones, J. W., Thorp, K. R., Ahmad, D., Man, H. C., and Taheri, S. (2018). Review of optimum temperature, humidity, and vapour pressure deficit for microclimate evaluation and control in greenhouse cultivation of tomato: a review. Int. Agrophysics 32, 287–302. doi: 10.1515/intag-2017-0005
Shim, J., Park, J., and Paik, W. (1979). Studies on the life history of cotton aphid, Aphis gossypii Glover (Homoptera). Korean J. Appl. Entomol. 18, 85–88.
Soyer, D. (1942). Miride du cotonnier Creontiades pallidus Ramb. Capsidae (Miridae). (Publications de l'Institut National pour l'Etude Agronomique du Congo Beige (INEAC)), 29, 1–14.
Spencer, K. A. (1973). Agromyzidae (Diptera) of economic importance Vol. 9 (Hague, Netherlands: Springer Science & Business Media).
Stansly, P. A. and Naranjo, S. E. (2010). Bemisia: bionomics and management of a global pest (Dordrecht, Netherlands: Springer).
Stapleton, M. A., Javid-Majd, F., Harmon, M. F., Hanks, B. A., Grahmann, J. L., Mullins, L. S., et al. (1996). Role of conserved residues within the carboxy phosphate domain of carbamoyl phosphate synthetase. Biochemistry 35, 14352–14361. doi: 10.1021/bi961183y
Steinmetzer, K., Behlke, J., Brantl, S., and Lorenz, M. (2002). CopR binds and bends its target DNA: a footprinting and fluorescence resonance energy transfer study. Nucleic Acids Res. 30, 2052–2060. doi: 10.1093/nar/30.9.2052
Tang, X.-T., Longnecker, M., and Tamborindeguy, C. (2020). Acquisition and transmission of two ‘Candidatus Liberibacter solanacearum’ haplotypes by the tomato psyllid Bactericera cockerelli. Sci. Rep. 10, 14000. doi: 10.1038/s41598-020-70795-4
Tarek, A. (2013). Compared the efficiency of botanical extract oxamatrin and bio-insecticicd abamectin to controlling myzus persicae aphid insect on hybrid eggplant solanum melongena in greenhouses. Anbar J Agric Sci. 11 (2), 359–369.
Ünlü, N. and Elçi, E. (2022). Determination of Streptomyces species causing common scab disease on potato in Niğde province. Turk J Agric Forestry 27 (3), 402–412. doi: 10.37908/mkutbd.1092635
van Tuijl, B., van Os, E., and Van Henten, E. J. (2008). Wireless sensor networks: State of the art and future perspective. Acta Hortic. 801, 547–554. doi: 10.17660/ActaHortic.2008.801.61
van der Vlugt, R. A. A., Stijger, C., Verhoeven, J. T. J., and Lesemann, D. E. (2000). First report of pepino mosaic virus on tomato. Plant Dis. 84, 103. doi: 10.1094/pdis.2000.84.1.103c
Vaughan, M. M., Tholl, D., and Tokuhisa, J. G. (2011). An aeroponic culture system for the study of root herbivory on Arabidopsis thaliana. Plant Methods 7, 5. doi: 10.1186/1746-4811-7-5
Verbeek, M., van Bekkum, P. J., Dullemans, A. M., and van der Vlugt, R. A. A. (2014). Torradoviruses are transmitted in a semi-persistent and stylet-borne manner by three whitefly vectors. Virus Res. 186, 55–60. doi: 10.1016/j.virusres.2013.12.003
Vermelho, A. B., Moreira, J. V., Akamine, I. T., Cardoso, V. S., and Mansoldo, F. R. P. (2024). Agricultural pest management: the role of microorganisms in biopesticides and soil bioremediation. Plants 13, 2762. doi: 10.3390/plants13192762
Voigt, D., Schrameyer, K., Kiefer, J., Zebitz, C. P. W., and Gorb, S. (2019). Anchoring of greenhouse whitefly eggs on different rose cultivars. Arthropod-Plant Interact. 13, 335–348. doi: 10.1007/s11829-019-09680-5
Walker, K. and Frederick, R. (2019). “Entomological risks of genetically engineered crops☆,” in Encyclopedia of Environmental Health (Second Edition). Ed. Nriagu, J. (Elsevier, Oxford), 339–346.
Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x
Wang, J. C. (2002). Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews. Mol. Cell Biol. 3, 430–440. doi: 10.1038/nrm831
Weigele, P. R., Scanlon, E., and King, J. (2003). Homotrimeric, beta-stranded viral adhesins and tail proteins. J. Bacteriol. 185(4), 4022–4030. doi: 10.1128/jb.185.14.4022-4030.2003
Wheeler, A. G. (2001). Biology of the Plant Bugs (Hemiptera: Miridae): Pests, Predators, Opportunists (New York, USA: Cornell University Press).
Wieczorek, P., Budziszewska, M., Frąckowiak, P., and Obrępalska-Stęplowska, A. (2020). Development of a new tomato torrado virus-based vector tagged with GFP for monitoring virus movement in plants. Viruses 12. doi: 10.3390/v12101195
Wraight, S. P., Lopes, R., and Faria, M. (2017). Microbial control of mite and insect pests of greenhouse crops. In Microbial Control of Insect and Mite Pests. Academic Press. pp. 237–252. doi: 10.1016/B978-0-12-803527-6.00016-0
Wu, J. B., Dai, F. M., and Zhou, X. P. (2006). First Report of Tomato yellow leaf curl virus in China. Plant Dis. 90, 1359. doi: 10.1094/pd-90-1359c
Xue, R., Zhang, C., Yan, H., Li, J., Ren, J., Akhlaq, M., et al. (2023). Physiological response of tomato and cucumber plants to micro-spray in high-temperature environment: A scientific and effective means of alleviating crop heat stress. Agronomy 13, 2798. doi: 10.3390/agronomy13112798
Yamasaki, M., Moriwaki, S., Miyake, O., Hashimoto, W., Murata, K., and Mikami, B. (2004). Structure and Function of a Hypothetical Pseudomonas aeruginosa Protein PA1167 Classified into Family PL-7: A NOVEL ALGINATE LYASE WITH A β-SANDWICH FOLD*. J. Biol. Chem. 279(30), 31863–31872. doi: 10.1074/jbc.M402466200
Yan, B., Deng, T., and Shi, L. (2024). Towards sustainable productivity of greenhouse vegetable soils: limiting factors and mitigation strategies. Plants 13, 2885. doi: 10.3390/plants13202885
Yan, Z., Wolters, A.-M. A., Navas-Castillo, J., and Bai, Y. (2021). The global dimension of tomato yellow leaf curl disease: Current status and breeding perspectives. Microorganisms 9, 740. doi: 10.3390/microorganisms9040740
Yang, X. B. and Liu, T. X. (2009). Life history and life tables of Bactericera cockerelli (Homoptera: Psyllidae) on eggplant and bell pepper. Environ. Entomol. 38(6), 1661–1667. doi: 10.1603/022.038.0619
Yao, F. and Johns, W. E. (2010). A HYCOM modeling study of the Persian Gulf: 1. Model configurations and surface circulation. J. Geophysical Research: Oceans 115. doi: 10.1029/2009jc005781
Yoder, M. D., Keen, N. T., and Jurnak, F. (1993). New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260, 1503–1507. doi: 10.1126/science.8502994
Zamharir, M. G. and Shameli, S. (2023). Study on soybean bud proliferation phytoplasma insect vector. Phytopathogenic Mollicutes 13, 37–38. doi: 10.5958/2249-4677.2023.00019.1
Zhou, X., De Beer, Z. W., Ahumada, R., Wingfield, B., and Wingfield, M. (2004). Ophiostoma and Ceratocystiopsis spp. associated with two pine-infesting bark beetles in Chile. Fungal Diversity 15, 261–274.
Zhulin, I. B., Taylor, B. L., and Dixon, R. (1997). PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22, 331–333. doi: 10.1016/s0968-0004(97)01110-9
Keywords: greenhouse systems, insects, Trialeurodes vaporariorum, meteorological conditions, metagenomics, agricultural yield
Citation: Qush A, Assaad N, Alkhayat FA, Al-Kuwari MS, Al-Khalaf N, Bassil M, Yassine HM, Zeidan A, Razali R and Kamareddine L (2025) Insects in agricultural greenhouses: a metagenomic analysis of microbes in Trialeurodes vaporariorum infesting tomato and cucumber crops. Front. Plant Sci. 16:1581707. doi: 10.3389/fpls.2025.1581707
Received: 22 February 2025; Accepted: 31 March 2025;
Published: 19 May 2025.
Edited by:
Mohamed Mannaa, Pusan National University, Republic of KoreaReviewed by:
Elena Gonella, University of Turin, ItalyMeena Pandey, University of California, Davis, United States
Copyright © 2025 Qush, Assaad, Alkhayat, Al-Kuwari, Al-Khalaf, Bassil, Yassine, Zeidan, Razali and Kamareddine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Layla Kamareddine, bGthbWFyZWRkaW5lQHF1LmVkdS5xYQ==
 Abeer Qush
Abeer Qush Nada Assaad2
Nada Assaad2 Maya Bassil
Maya Bassil Hadi M. Yassine
Hadi M. Yassine Asad Zeidan
Asad Zeidan Rozaimi Razali
Rozaimi Razali Layla Kamareddine
Layla Kamareddine