- 1College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2Technology Center, China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 3College of Agronomy, Hunan Agricultural University, Changsha, China
The ubiquitination pathway is extensively involved in the regulation of plant biological processes, such as growth, development, and disease resistance, among others. Our previous study showed that the Arabidopsis U-box protein PUB13 regulates plant cell death, immunity, and development. Here, we report that the E3 ubiquitin ligase activity of PUB13 is required for its regulation of the plant size, flowering time, and immunity based on the analysis of the biological processes on the PUB13 enzyme activity loss mutant. Furthermore, we identified the copine protein BON1 interacting with PUB13, which was ubiquitinated by PUB13. Interestingly, the knockout of BON1 in the pub13 mutant further strengthens its phenotypes of retarded growth and early flowering. In addition, the knockout of BON1 further enhanced the resistance of pub13 to the biotrophic pathogen. In contrast, the pub13bon1 double mutant was more susceptible to the necrotrophic pathogen compared with the pub13 and bon1 single mutants. The synergistic effect between PUB13 and BON1 was also observed in the regulation of pathogen-associated molecular pattern-triggered immunity (PTI). These results indicate that the E3 ubiquitin ligase activity is required for PUB13 regulating biological functions and that BON1 synergistically interacts with PUB13 to regulate plant growth, flowering, and immunity in Arabidopsis.
1 Introduction
Protein is constantly produced and degraded during the whole life cycle of an organism. One of the most important protein degradation strategies in eukaryotic cells is the ubiquitin–26S proteasome system (UPS). However, not all proteins are degraded by the UPS after ubiquitination: except for poly-ubiquitination, mono-ubiquitinated and multi-ubiquitinated proteins are not degraded by the UPS, but can alter localization, affect the protein–protein interaction, or regulate the protein activity (Mukhopadhyay and Riezman, 2007; Ye and Rape, 2009). The UPS involves the sequential action of three enzymes, namely, E1 (the ubiquitin-activating enzyme), E2 (the ubiquitin-conjugating enzyme), and E3 (the ubiquitin ligase enzyme), to ultimately ligate one or more ubiquitin molecules to specific target proteins (Sun et al., 2019; Xu and Xue, 2019). Plant U-box proteins (PUBs) are the earliest discovered E3 ubiquitin ligases, and their U-box domain is structurally conserved (Koegl et al., 1999; Azevedo et al., 2001). In Arabidopsis, the largest class of PUBs includes the Armadillo (ARM) repeat domain behind the U-box domain (Mudgil et al., 2004). The ARM domain generally exists in eukaryotes with three to eight repeats and plays a role mainly in protein–protein interaction (Peifer et al., 1994; Samuel et al., 2006).
E3 ubiquitin ligases play an important role in plant growth and development. For example, the rice U-box/ARM protein OsPUB15 regulates rice growth during the seedling stage [10]. With the knockout of OsPUB15, the plant primary roots were inhibited during germination, and the shoot development of the pub15 mutant was also significantly slower than that of the wild type (Park et al., 2011). In addition, the rice PUB protein Spotted Leaf 11 (SPL11) with ubiquitin ligase E3 activity mono-ubiquitinated and repressed SPL11-interacting Protein 1 (SPIN1) to positively regulate the flowering time (Vega-Sánchez et al., 2008). In contrast, Arabidopsis PUB13, an ortholog of SPL11, negatively regulated the flowering time (Li et al., 2012a). Under long-day conditions, the pub13 mutant exhibited a significant early flowering phenotype, with a decrease in the transcription level of the negative flowering regulator FLOWERING LOCUS C (FLC), while the transcription levels of the positive regulators FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) were increased (Lu et al., 2011; Li et al., 2012a; Antignani et al., 2015).
E3 ubiquitin ligases are also involved in plant disease resistance. For example, SPL11 negatively regulates the plant defense against the rice blast pathogen (Magnaporthe grisea) and the bacterial blight pathogen (Xanthomonas oryzae pv. oryzae). Similarly, the SPL11 orthologs PUB12 and PUB13 in Arabidopsis play an important role in the regulation of plant pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) (Lu et al., 2011; Li et al., 2012a; Antignani et al., 2015). Upon stimulation with flagellin, the receptor kinase BAK1 phosphorylated PUB12 and PUB13, thereby activating the ubiquitination and UPS degradation of the flagellin receptor FLS2 by PUB12 and PUB13. This signal transduction ultimately led to the negative regulation of FLS2 by PUB12 and PUB13 of the plant PTI (Lu et al., 2011; O’Neill and , 2011; Zhou et al., 2015). Similarly, the expression of the Arabidopsis E3 ubiquitin ligases PUB22/PUB23/PUB24 can be induced by flagellin, and the mutants pub22, pub23, and pub24 significantly improve plant resistance to pathogens (Trujillo et al., 2008; Stegmann et al., 2012; Chen et al., 2014; Wang et al., 2020). Furthermore, both Arabidopsis PUB20 and PUB21 negatively regulate plant innate immunity, negatively regulating plant resistance to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (Heise et al., 2002; Yee and Goring, 2009; Yi et al., 2024). In addition, PUB25 and PUB26 negatively regulate plant immunity by specifically targeting the non-activated BIK1 for 26S-UPS degradation in Arabidopsis (Wang et al., 2018). Interestingly, the E3 ubiquitin ligase PUB CMPG1 is required for cell death activation in Nicotiana benthamiana through Cf-9/Avr9, Cf-4/Avr4, Pto/AvrPto, or a cellulose-binding elicitor lectin (CBEL) (Gilroy et al., 2011). In addition, the orthologous tobacco ACRE276, the tomato ACRE276, and the Arabidopsis PUB17 are PUBs, and all of them regulate the plant effector-triggered immunity (ETI) (Yang et al., 2006; He et al., 2015). The transiently expressed Arabidopsis PUB17 can complement the silencing of ACRE276 in tobacco, restoring the Cf-9/Avr9- and Cf-4/Avr4-mediated hypersensitive response (HR); however, PUB17 lacking E3 ubiquitin ligase activity is not able to complement the silenced ACRE276, indicating that E3 ubiquitin ligase activity plays a key role in ETI signaling (Yang et al., 2006).
Copines are a group of Ca2+-dependent phospholipid-binding proteins that are widely distributed in eukaryotes (Warner et al., 2013; Tang et al., 2021). There are two calcium-dependent phospholipid-binding C2 domains at the amino (N)-terminus of copine proteins and a putative protein–protein interaction vWA (von Willebrand A) domain at the carboxyl (C)-terminus (Rizo and Südhof, 1998; Whittaker and Hynes, 2002). The ubiquitous conservation of copines among eukaryotes suggests that they play important roles in common biological pathways, which is supported by emerging studies that have demonstrated their roles in plant development, defense, and stress responses (Yang and Hua, 2004; Yang et al., 2006; Gou et al., 2015; Yang et al., 2017; Yin et al., 2018; Chen et al., 2020).
Arabidopsis BON1 is a copine protein, and the bon1 mutant has shown inhibited growth at low temperature (22°C), while the phenotype could be recovered at 28°C (Hua et al., 2001). In addition, BON1 Associated Protein 1 (BAP1) has similar functions in growth regulation. Under 22°C growth conditions, the overexpression of BAP1 can restore the bon1 mutant to the wild-type phenotype (Hua et al., 2001; Li et al., 2010). This suggests that BON1 is necessary for plants to maintain normal growth at low temperatures. Moreover, the Arabidopsis BON1, BON2, and BON3 exhibit a functional overlap, and they are necessary for the normal growth of Arabidopsis (Yang et al., 2006).
On the other hand, BON1, BON2, and BON3 have been reported to negatively regulate plant disease resistance (Yang and Hua, 2004; Yang et al., 2006). The bon1 mutant shows enhanced disease resistance to the bacterial pathogen Pst DC3000 and the oomycete pathogen Hyaloperonospora parasitica, largely resulting from the upregulation of the plant immune receptor NLR (NOD-like receptor) gene SNC1 (Yang and Hua, 2004). Furthermore, at least four R-like genes are induced in bon1 and bon3, indicating that BON1 and BON3 negatively regulate disease resistance mediated by R-like genes (Li et al., 2009). Similarly, both OsBON1 and OsBON3 are negative regulators of disease resistance in rice (Yin et al., 2018).
In this study, we show that the E3 ubiquitin ligase activity of PUB13 is required for the regulation of plant growth, development, and disease resistance. In addition, a PUB13 interacting protein, BON1, was screened using yeast two-hybrid assay, which was found to be ubiquitinated by PUB13. Moreover, our data indicate that BON1 enhances the function of PUB13 on the growth, flowering, and disease resistance.
2 Materials and methods
2.1 Plants and growth conditions
The Arabidopsis (Arabidopsis thaliana) wild type (Col-0), pub13 (Li et al., 2012a), PUB13V273R/pub13, PUB13/pub13, bon1 (Hua et al., 2001), and pub13(−/−)bon1(+/−) were used in this study. The pub13(−/−)bon1(+/−) mutant was constructed by genetic crosses. PUB13 and PUB13V273R, driven by a native promoter 2 kb upstream of the PUB13 start codon, were constructed into the binary vector pCAMMBIA1300. Subsequently, the Agrobacterium tumefaciens-mediated flower-dipping method of transformation was performed according to a standard protocol to obtain PUB13V273R/pub13 and PUB13/pub13 genetic plants. Arabidopsis plants were grown in nutrient soil at 22°C, 75% relative humidity (RH), and 8-h light/16-h dark as short day (regular condition) or 16-h light/8-h dark as long day in growth chambers.
2.2 Pathogen inoculation
Plants (4 weeks old) were inoculated with different pathogens at long-day conditions. Pseudomonas syringae pv. maculicola ES4326 and P. syringae pv. tomato DC3000 were sprayed on the plants at 1 × 108 CFU ml−1 for disease symptoms or injected at 5 × 105 CFU ml−1 for the bacterial growth assay according to (Li et al., 2012a). For Botrytis cinerea inoculation, the fungus was cultured on potato dextrose agar (PDA) media for 2 weeks at 24°C with a 12-h photoperiod. The fungal culture was washed with distilled water and filtered using sterile gauze. Subsequently, the concentration was adjusted to 3 × 104 conidia ml−1. The detached rosette leaves were placed in Petri dishes containing 0.8% agar, 5 μl of the conidia suspension was dropped onto the leaf surface, and the Petri dishes were sealed. The inoculated leaves were incubated at 22°C with a 12-h photoperiod. The diameter of the lesion was measured at 48 h post-inoculation.
2.3 Yeast two-hybrid assay
The full-length coding sequence (CDS) of the PUB13V273R and BON1/prey library was individually inserted into the BD vector (pDBLeu) and the AD vector (pPC86), respectively. The pDBLeu and pPC86 empty vectors were used as negative controls for the screening and confirmation in this assay. The constructs or the corresponding empty vector (control) was transformed into the MATα yeast strain MaV203. The transformed yeast cells were plated on SD/-Leu/-Trp and SD/-Leu/-Trp/-His dropout media and cultured at 30°C for 2–5 days. Positive yeast colonies with successful transformation were selected for the X-gal assay on YPDA medium (ProQuest™ Two-Hybrid System with Gateway™ Technology, Invitrogen, Carlsbad, CA, USA) (Li et al., 2012b).
2.4 GST pull-down
The full-length CDS of PUB13 and BON1 was cloned into the pGEX-6p-1 and pMAL-c4x vectors, respectively. The purified PUB13:MBP and BON1:GST fusion proteins were mixed, and 60 µl of pre-rinsed glutathione Sepharose beads (Promega, Madison, WI, USA) was co-incubated with the protein mixture for 4 h at 4°C. The beads were then washed five times with 1× TBST buffer. Finally, the protein was eluted from the beads and was used for the Western blot assay. The BON1 and PUB13 proteins were detected with anti-MBP and anti-GST antibodies, respectively, in the Western blot assay, with the antibodies diluted to 1/1,500.
2.5 E3 ubiquitination assay
The full-length CDS of PUB13 was cloned into the pGEX-6P-1::GST vector, while BON1 was cloned into the pET-28a::myc vector. The fusion protein was expressed in Escherichia coli BL21, with 1.0 μg of PUB13/V273R-GST and BON1-myc used for each reaction. The wheat (Triticum aestivum) E1 (GI:136632; approximately 40.0 ng) and the human E2 (UBCH5b; approximately 40.0 ng) were used in the in vitro E3 ligase activity assays as described (Xie et al., 2002). The reaction samples were separated by 10% SDS-PAGE and detected by Western blot using an anti-myc antibody.
2.6 Luciferase complementation imaging
The full-length CDS of PUB13 was cloned into the pCAMBIA-NLuc vector, while the full-length CDS of BON1 was cloned into the pCAMBIA-CLuc vector. Subsequently, they were transferred into the Agrobacterium strain GV3101 and the cells cultured at 28°C until ∽2.0 optical density (OD). The cells were harvested, resuspended, and then incubated with 10 mM MES solution containing 0.2 mM acetosyringone for 3 h at 28°C. The cells were harvested and resuspended in MES solution to yield 1.0 OD suspension. The NLuc and CLuc vectors were co-infiltrated into N. benthamiana leaves, and the RNAi suppressor P19 vector with 0.5 OD was also infiltrated. After storage at 26°C (dark) for 48 h, the leaves were sprayed with fluorescein potassium (CellGro, Manassas, VA, USA) and photographed using a chemiluminescence imaging system (Bio-Rad, Hercules, CA, USA).
2.7 Measurement of ROS accumulation
A luminol-based assay was employed to monitor the production of reactive oxygen species (ROS) (Li et al., 2021). Leaf discs were punched from the adult plant before flowering and soaked in sterilized distilled water in the dark overnight. Three leaf discs were placed in a 1.5-ml tube containing 1 μl of 100 nM flg22 or sterilized distilled water as a control, 1 μl of 10 μg/ml horseradish peroxidase (HRP), and 100 μl of 0.2 mM chemiluminescent probe L-012. Luminescence was detected immediately at 1-min intervals for 20 min with the Glomax 20/20 Luminometer. Each sample and treatment condition was repeated three times.
2.8 RNA extraction and RT-qPCR analysis
Total RNA was extracted using the TRIzol reagent (TransGen, Beijing, China) and was treated with a gDNA remover. Thereafter, the RNA was reverse-transcribed to cDNA using a cDNA synthesis kit (TransGen, Beijing, China). The cDNA was then diluted to 20 ng/μl before use as a template for real-time quantitative PCR (RT-qPCR). Three replicates were carried out per sample. Subsequently, RT-qPCR was conducted with the Bio-Rad CFX96 PCR amplifier. Three independent repeats were performed.
3 Results
3.1 The E3 ubiquitin ligase activity of PUB13 is required for regulating plant growth
PUB13 functions as a regulator of flowering, cell death, and immunity in Arabidopsis [12,14], and it is an active E3 ubiquitin ligase. The 273rd amino acid valine (V273) in the U-box domain is highly conserved and critical for PUBs. In this research, to determine the biological functions of the E3 ubiquitin ligase activity of PUB13, we created the PUB13V273R construct by substituting V273 to arginine, the mutation that suppresses the E3 ubiquitin ligase activity of PUB13 (Li et al., 2012a). The PUB13V273R construct was transformed into pub13 plant (PUB13V273R/pub13), with the wild-type PUB13 gene also transformed into pub13 (PUB13/pub13). When the plants were 45 days old, the plant size of the pub13 mutant was observed to be noticeably smaller than the wild-type Col-0 under short-day conditions (Figure 1). However, the complemented plant PUB13/pub13 recovered to the wild-type level. Interestingly, the plant size of PUB13V273R/pub13 was still smaller than the wild type (Figure 1). Subsequently, the rosette diameter, the leaf petiole length, and the leaf lamina length of Col-0, pub13, PUB13V273R/pub13, and PUB13/pub13 were compared. The numerical results of these parameters are consistent with the phenotypic observations, which showed no differences in Col-0 and PUB13/pub13, while both pub13 and PUB13V273R/pub13 exhibited relatively low values (Supplementary Figure S1). These results suggest that the E3 ubiquitin ligase activity of PUB13 is required for plant growth.

Figure 1. The E3 Ubiquitin Ligase Activity of PUB13 Is Required for Regulating Plant Growth. The indicated plants were 45 days old and grown under short day conditions.
3.2 PUB13 negatively regulates flowering time depending on the E3 ubiquitin ligase activity
Our previous results showed that PUB13 negatively regulates flowering time in Arabidopsis (Li et al., 2012a). We also examined whether the E3 ubiquitin ligase activity of PUB13 is required for the regulation of Arabidopsis flowering time. The pub13 mutant showed an early flowering phenotype under long-day conditions, as reported, while the complementary plants PUB13/pub13 did not show early flowering (Figure 2A). However, the PUB13V273R/pub13 plants also showed a noticeable early flowering phenotype, similar to pub13 plants (Figure 2A). Correspondingly, the number of rosette leaves before flowering of the pub13 and PUB13V273R/pub13 plants was significantly lower than that of Col-0, while the number of rosette leaves of PUB13/pub13 did not show significant differences compared with Col-0 (Figure 2B). To confirm the function of PUB13 enzyme activity on flowering, the negative flowering regulator FLC and the positive regulators FT and SOC1 in the above plants were detected using RT-qPCR. The results showed that the expression of FLC was suppressed in pub13 and PUB13V273R/pub13 compared with that in Col-0 (Figure 2C). In contrast, the expression of FT and SOC1 in pub13 and PUB13V273R/pub13 was enhanced compared with that in Col-0, and the expression of all three genes did not show significant differences between Col-0 and PUB13/pub13 (Figure 2C). Therefore, the results of the expression of the flowering regulator genes in the above plants are consistent with the flowering phenotypes in Figure 2A. Taken together, PUB13 regulates flowering time depending on its E3 ubiquitin ligase activity.
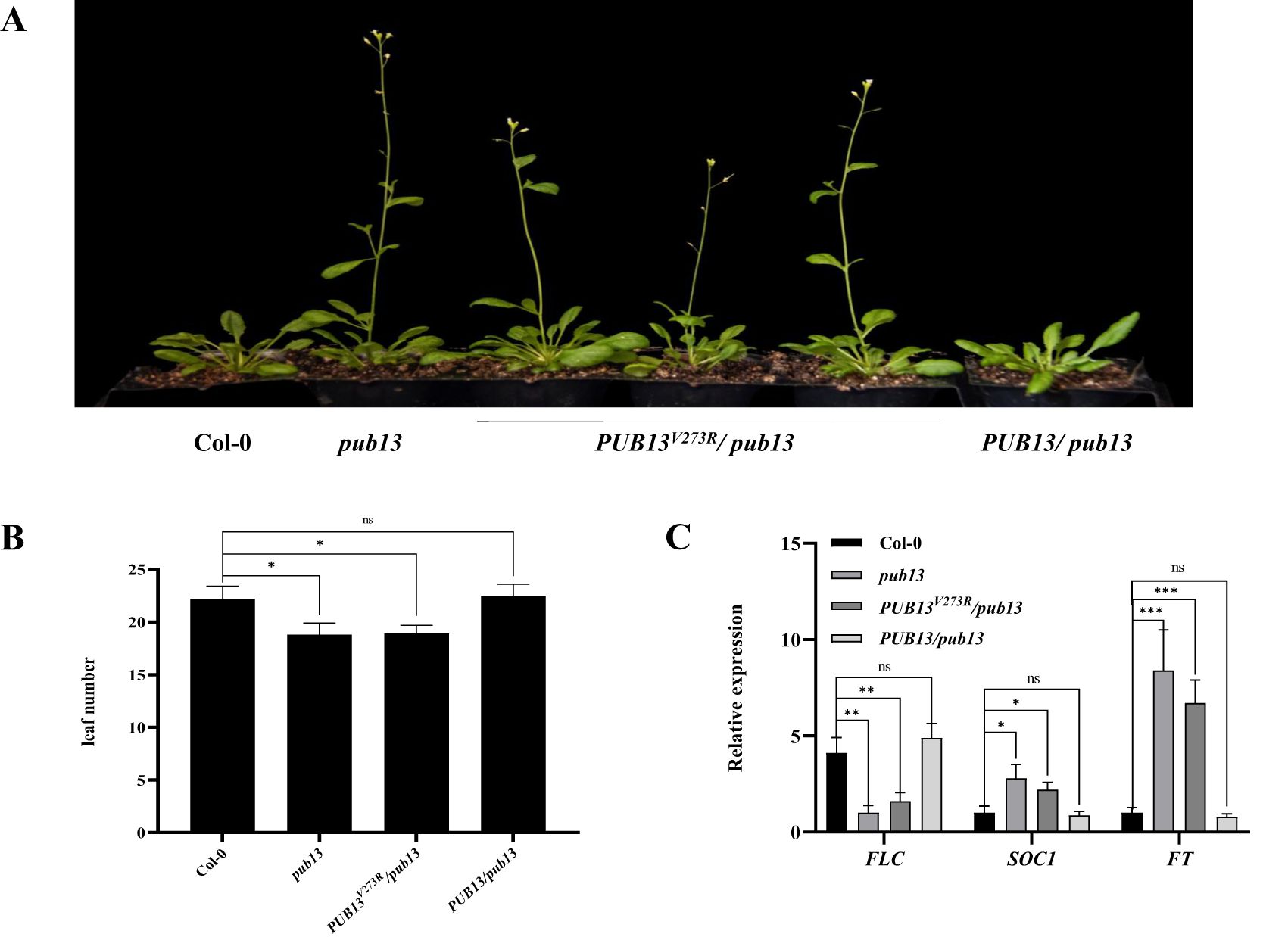
Figure 2. The E3 Ubiquitin Ligase Activity of PUB13 Is Required for Regulating Flowering Time. (A) Flowering phenotypes of Col-0, pub13, PUB13V273R/ pub13, and PUB13/ pub13 grown under long day conditions. (B) The rosette leaves number of indicated plants. The leaves number was counted when the first flower bud appeared. (C), Expression of flowering marker genes FLC, SOC1, and FT in indicated plants. Actin was used internal reference. Asterisks denote significant difference based on nested ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001). These experiments were repeated three times with similar results.
3.3 E3 ubiquitin ligase activity is required for PUB13 regulating disease resistance
As PUB13 regulates plant disease resistance based on our previous research (Li et al., 2012a), we determined whether E3 ubiquitin ligase activity is required for PUB13-regulated resistance. The Col-0, PUB13/pub13, PUB13V273R/pub13, and pub13 plants were inoculated with the bacterial pathogen P. syringae pv. maculicola ES4326. After 3 days post-inoculation (dpi), the leaves of both Col-0 and PUB13/pub13 showed leaf chlorosis, while the leaves of pub13 appeared healthy. PUB13V273R/pub13 was as resistant as pub13 (Figure 3A), which indicates that the E3 ubiquitin ligase activity-suppressed PUB13V273R cannot complement pub13 to the wild type. Furthermore, the bacterial growth in the above plants was assessed after infiltration with Psm ES4326 at a lower concentration. The results showed that the susceptibility of PUB13/pub13 recovered to that of the wild-type Col-0, while the resistance of PUB13/pub13 recovered to the wild-type level (Figure 3B). Taken together, the pathogen inoculation results demonstrated that E3 ubiquitin ligase activity is required for PUB13 negatively regulating plant resistance against bacterial pathogens.

Figure 3. E3 Ubiquitin Ligase Activity Is Required for PUB13 Regulated Disease Resistance. (A) Disease symptoms of Col-0, PUB13/pub13, PUB13V273R/pub13 and pub13 inoculated with Psm ES4326. Four weeks old plants growing under long day conditions were sprayed with 1×108 CFU mL-1 of Psm ES4326, and observed the symptoms at 3 dpi. (B) Bacterial growth in Col-0, PUB13/pub13, PUB13V273R/pub13 and pub13 inoculated with Psm ES4326. Four weeks old plants growing under long day conditions were injected with 5×105 CFU mL-1 Psm ES4326. The bacteria numbers in injected leaves were counted at 0 dpi and 3 dpi, respectively. Asterisks denote significant difference based on nested ANOVA (*P < 0.05, **P < 0.01). These experiments were repeated at least three times with similar results.
3.4 PUB13 interacts with and ubiquitinates BON1
It has been reported that PUB13 interacts with BAK1, HFR1, and ABI1 to regulate plant immunity (Li et al., 2009; Lu et al., 2011; Li et al., 2021). To further illustrate the molecular mechanism of PUB13 regulating growth, development, and immunity, PUB13V273R, which cannot degrade target proteins, was used as bait for the yeast two-hybrid assay to screen its interacting proteins (Supplementary Table S1). Among the screened interactors, the calcium-dependent phospholipid-binding protein BON1 was studied as it is involved in plant growth homeostasis and disease resistance. Firstly, we confirmed the interaction between BON1 and PUB13V273R in the yeast system (Figure 4A). Secondly, the GST pull-down experiment was conducted, which showed that PUB13 interacted with BON1 in vitro (Figure 4B). Lastly, the interaction between PUB13 and BON1 was proven through luciferase complementation imaging (LCI) experiment on N. benthamiana (Figure 4C). The results of these assays indicate that PUB13 physically interacts with BON1.
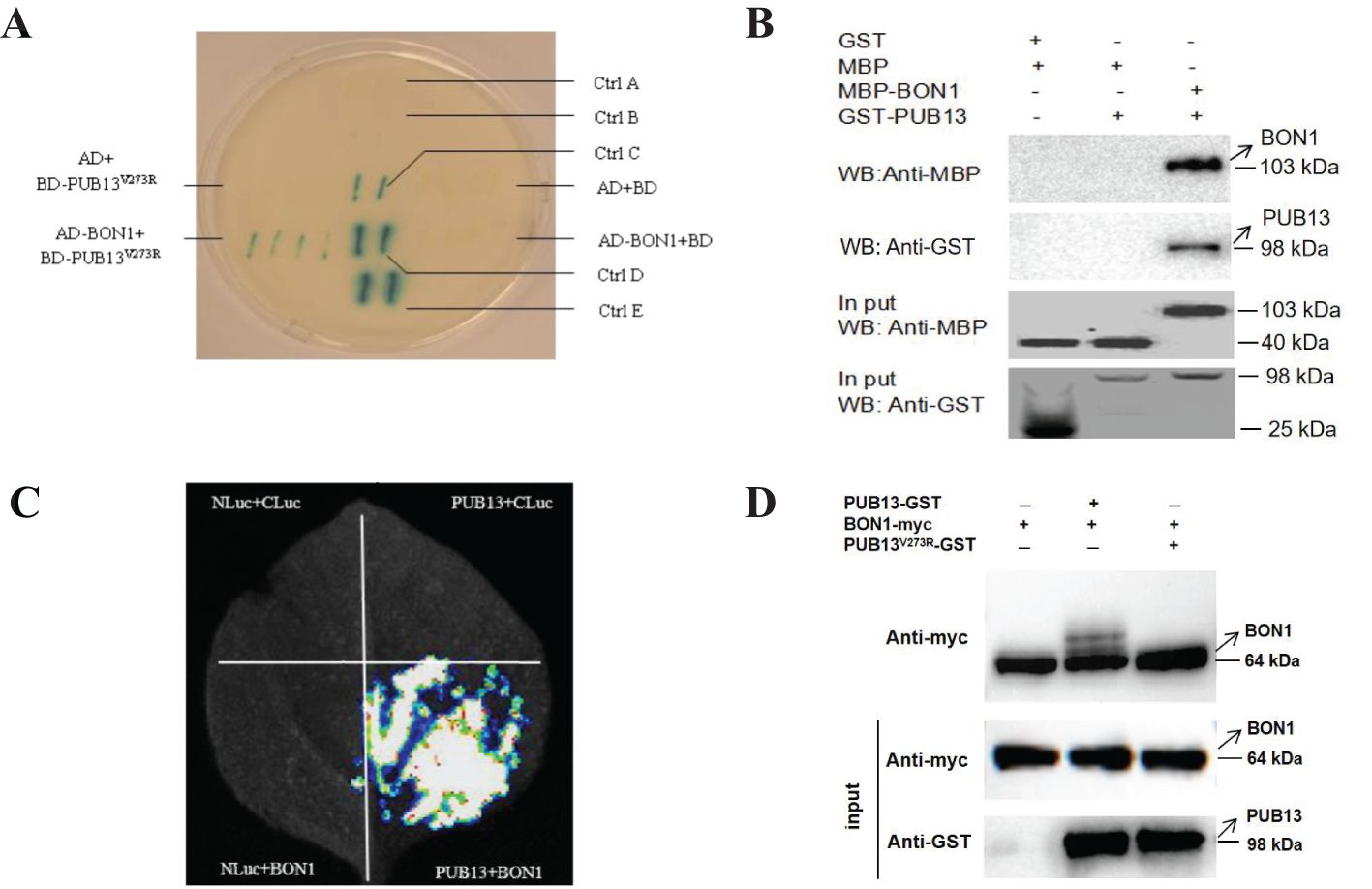
Figure 4. PUB13 Interacts with and Ubiquitinates BON1. (A) PUB13V273R interacted with BON1 in yeast. The full-length CDS of PUB13V273R and BON1 were cloned into pDBleu (BD) and pPC86 (AD), respectively, then transformed them into yeast strain MaV203. The yeast transformants were plated on YPDA medium for X-gal assay. Ctrl (A-E) represent A: pDEST™32; B: pDEST™22; C: pEXP™32/Krev1+pEXP™22/RalGDS-m1; D: Bait plasmid+Known prey; E: pEXP™32/Krev1+pEXP™22/RalGDS-wt. These five controls indicate the spectrum of interaction strengths. (B), The GST pull-down assay confirmed the interaction between BON1 and PUB13. MBP-BON1 and GST-PUB13 proteins were mixed and conducted GST pull down, then detected the eluted protein by Western blotting with the anti-MBP antibody; (C), LCI assay in N. benthamiana leaves proved the interaction between PUB13 and BON1. Each of the quadrants was infiltrated with PUB13-Nluc or BON1-Cluc and the indicated construct. (D), PUB13 ubiquitinates BON1. The upper panel is anti-myc WB to detecte BON1 treated with E1, E2, and indicated proteins, the middle panel is anti-myc WB to detect BON1 before treating with E1 and E2 as loading control, and the bottom panel is anti-GST WB to detect PUB13 and PUB13V273R. The ubiquitination of BON1-myc by PUB13-GST and PUB13V273R-GST in the presence of E1, E2. Ubiquitination was detected with anti-myc Western-blot.
In consideration of PUB13 functioning as an E3 ubiquitin ligase, we determined whether PUB13 and the enzyme inactive mutant PUB13V273R ubiquitinate the interactor BON1. According to the ubiquitination assay, in vitro, PUB13 ubiquitinated BON1 without obvious degradation in the presence of the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and ubiquitin, while PUB13V273R did not ubiquitinate BON1 in the same reaction conditions (Figure 4D).
3.5 BON1 synergizes with PUB13 to regulate plant growth
As previously reported, PUB13 regulates plant growth depending on its E3 ubiquitin ligase activity (12). Similarly, the bon1 mutant also showed a growth defect phenotype, such as leaf shrinkage and plant dwarfing (Figure 1). To analyze the interaction between BON1 and PUB13 on the regulation of physiological functions, we constructed bon1 and heterozygous pub13(−/−)bon1(+/−) mutants, with the homozygous pub13(−/−)bon1(−/−) mutant being lethal. As expected, both the bon1 and pub13 plants, including their leaves, were smaller than the Col-0 plants (Figure 5A). Furthermore, the pub13(−/−)bon1(+/−) plants, as well as their leaves, were even much smaller than pub13 and bon1 (Figure 5A). The statistics for the leaf size and rosette diameters of the above plants were consistent with the above plant growth phenotypes (Figures 5B, C), indicating that BON1 synergized with PUB13 to regulate plant growth.
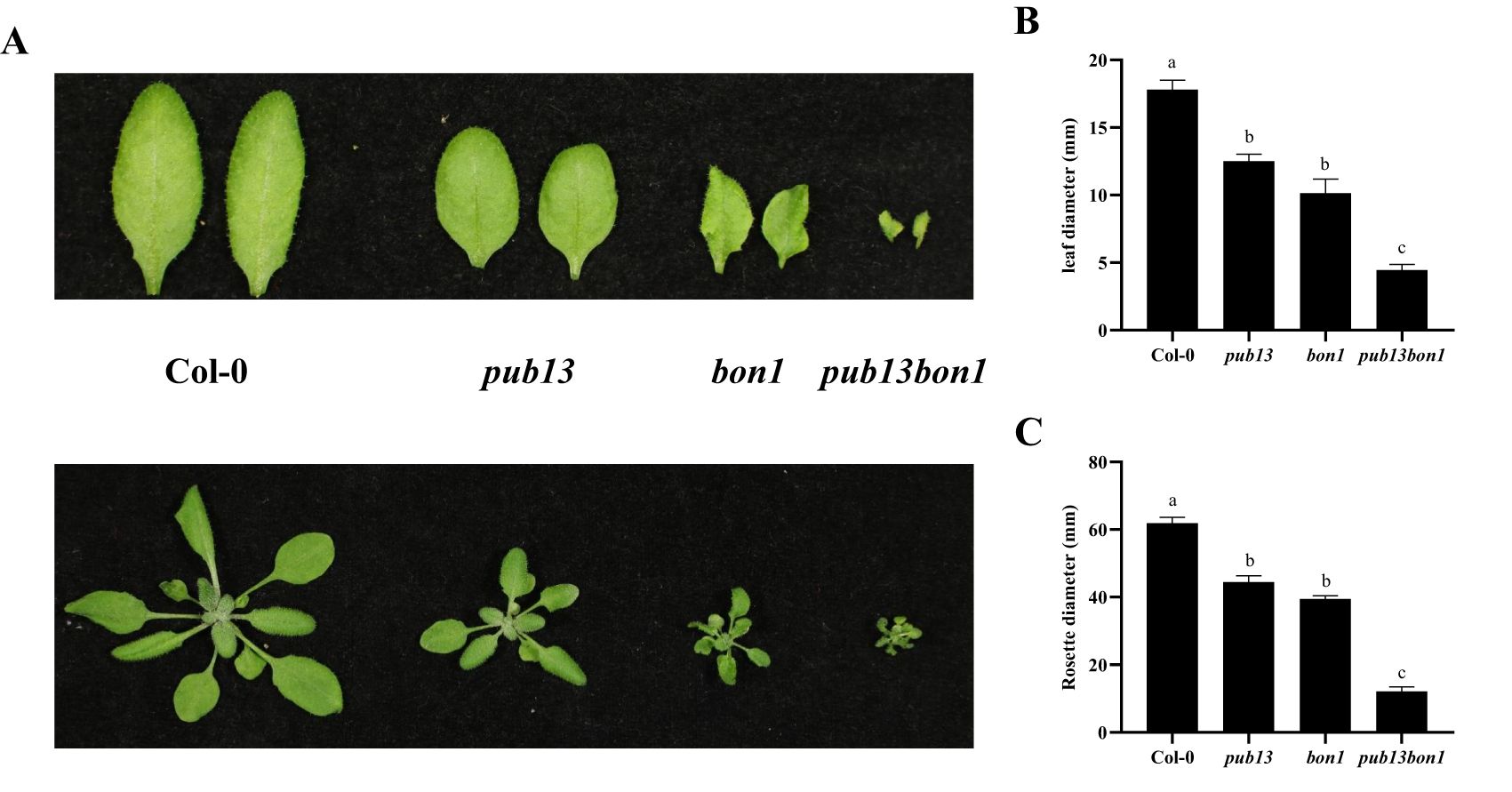
Figure 5. BON1 Synergizes with PUB13 to Regulate Plant Growth. (A), The leaves and plants of Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) under long day conditions. (B), The statistics of leaf width diameters of Col-0, pub13, bon1, and pub13(-/-)bon1(+/-). (C), The rosette diameters of Col-0, pub13, bon1, and pub13(-/-)bon1(+/-). Lowercase letters indicate significant difference at P<0.01. All experiments were repeated at least three times with similar results.
3.6 BON1 negatively regulates flowering time
PUB13 negatively regulates flowering time, particularly under long-day conditions (Li et al., 2012a). Therefore, the function of BON1 on flowering time was analyzed. The heterozygous pub13(−/−)bon1(+/−), as well as Col-0, pub13, and bon1, was planted under long-day conditions. As expected, the pub13 mutant showed an early flowering phenotype compared with Col-0 (Figure 6A). Interestingly, both the bon1 and pub13(−/−)bon1(+/−) mutants showed early flowering, while pub13(−/−)bon1(+/−) did not show a noticeably earlier flowering compared with the bon1 and pub13 single mutants (Figure 6A). To confirm the flowering phenotypes, the leaf number of the above plants was counted before flowering. The results showed that the rosette leaf numbers of pub13 and bon1 were significantly fewer than that of Col-0, while that of pub13(−/−)bon1(+/−) was extremely significantly fewer than that of Col-0 (Figure 6B). Moreover, the transcription levels of the flowering marker genes FT and FLC in these plants were examined with RT-qPCR. As shown in Figure 6C, the transcription levels of the negative flowering regulator FLC in pub13, bon1, and pub13(−/−)bon1(+/−) were noticeably lower than that in Col-0. In contrast, the expression of the positive flowering regulator FT was significantly higher in pub13, bon1, and pub13(−/−)bon1(+/−) compared with Col-0 (Figure 6C). These data indicate that BON1 negatively regulates flowering time, probably through a PUB13 independent pathway.
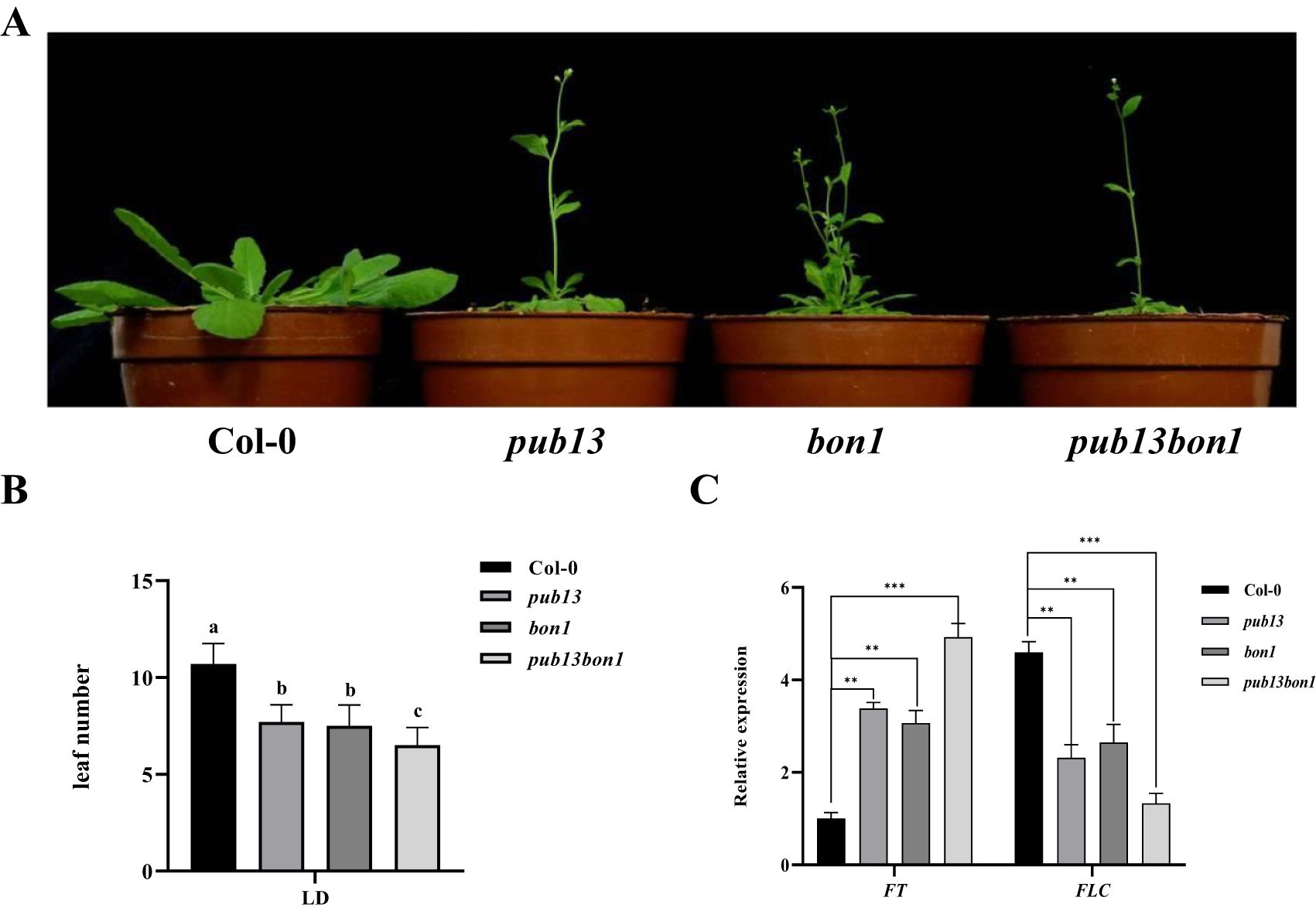
Figure 6. BON1 Negatively Regulates Flowering Time. (A), Flowering phenotypes of pub13, bon1, and pub13(-/-)bon1(+/-). Plants were grown under long day conditions. (B), The leaf numbers of Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) plants before flowering under long day conditions. Lowercase letters indicate significant difference at P<0.01. (C), Expression of flowering marker genes in Col-0、pub13、bon1 and pub13(-/-)bon1(+/-) under long day conditions. Gene expression of FLC and FT in indicated plants was determined by qRT-PCR. Actin was used as internal reference. Asterisks denote significant difference based on nested ANOVA (**P < 0.01, ***P < 0.001). All experiments were repeated at least three times with similar results.
3.7 BON1 synergizes with PUB13 to negatively regulate resistance to biotrophic pathogens
Considering PUB13 negatively regulating plant resistance to biotrophic pathogens, the role of BON1 in PUB13-regulated disease resistance was analyzed. Col-0, pub13, bon1, and pub13(−/−)bon1(+/−) plants were inoculated with P. syringae pv. tomato DC3000, which is a biotrophic pathogen at the early stage of infection and mainly defended by the salicylic acid (SA) resistance pathway. The bacterial growth results showed that both the pub13 and bon1 mutants were more resistant than Col-0. Furthermore, pub13(−/−)bon1(+/−) was found to be even more resistant than pub13 and bon1 (Figure 7A). We further analyzed the expression of the defense-related genes PR1 and PDF1.2 in the above plants before and after pathogen inoculation. At 0 and 48 h post-infection (hpi), the expression of the SA defense pathway gene PR1 in the pub13 and bon1 mutants was increased, and PR1 in pub13bon1 was higher than that in pub13 and bon1 (Figure 7B). In contrast, the expression of the PDF1.2 gene in the jasmonic acid (JA)/ethylene (ET) defense pathway, which is antagonistic to the SA defense pathway, was inhibited in pub13 and bon1 and was lowest in pub13(−/−)bon1(+/−) (Figure 7C). These results indicate that BON1 synergizes with PUB13 to negatively regulate resistance to Pst DC3000.
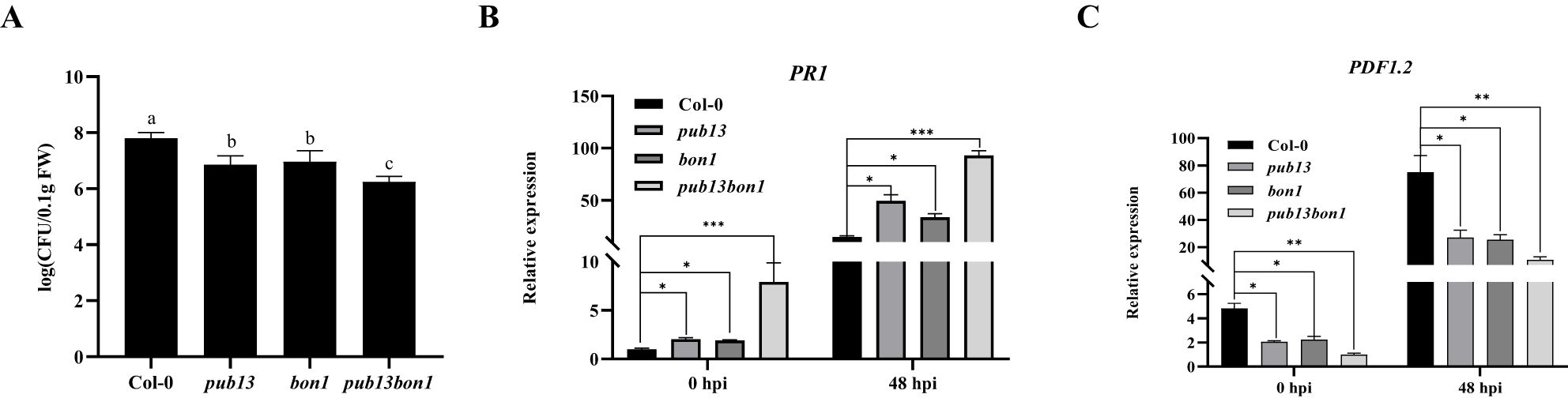
Figure 7. BON1 Synergizes with PUB13 to Negatively Regulate Resistance to Pst DC3000. (A), Bacterial growth of Pst DC3000 in Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) at 48 hpi. The leaves were injected with 5×105 CFU mL-1 Pst DC3000.Lowercase letters indicate significant difference at P<0.01. (B, C), The transcription level of PR1 and PDF1.2 in plants inoculated with DC3000. After injected the indicated plants with Pst DC3000, the RNA was extracted at 0 hpi and 48 hpi for qRT-PCR. Actin was used as internal reference. Asterisks denote significant difference based on nested ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001). These experiments were repeated three times with similar results.
3.8 Knockout of BON1 increases PUB13 susceptibility to necrotrophic pathogens
To determine the function of BON1 in the PUB13-regulated defense to necrotrophic pathogens, Col-0, pub13, bon1, and pub13(−/−)bon1(+/−) plants were inoculated with the necrotrophic pathogen B. cinerea BO5-10, which is mainly defended by the JA/ET resistance pathway. According to the observed symptoms, both pub13 and bon1 were more susceptible than Col-0, and pub13(−/−)bon1(+/−) was even more susceptible than pub13 and bon1 (Figure 8A). Correspondingly, the lesion diameters of pub13 and bon1 were significantly bigger than that of Col-0, with that of pub13(−/−)bon1(+/−) being the biggest (Figure 8B). The expression of B. cinerea Actin, which represents fungal biomass, in pub13 and bon1 was higher than that in Col-0 at 48 hpi and was highest in pub13(−/−)bon1(+/−) (Figure 8C). In addition, the transcription level of PR1 in these four plants at 48 hpi showed the same trend as that of B. cinerea Actin (Figure 8D). However, the expression of PDF1.2 was opposite to that of PR1, where PDF1.2 was inhibited in pub13 and bon1 at 48 hpi and was even lower in pub13(−/−)bon1(+/−) than in pub13 and bon1 (Figure 8E). These results further indicate that BON1 synergizes with PUB13 to positively regulate plant resistance to necrotrophic pathogens.
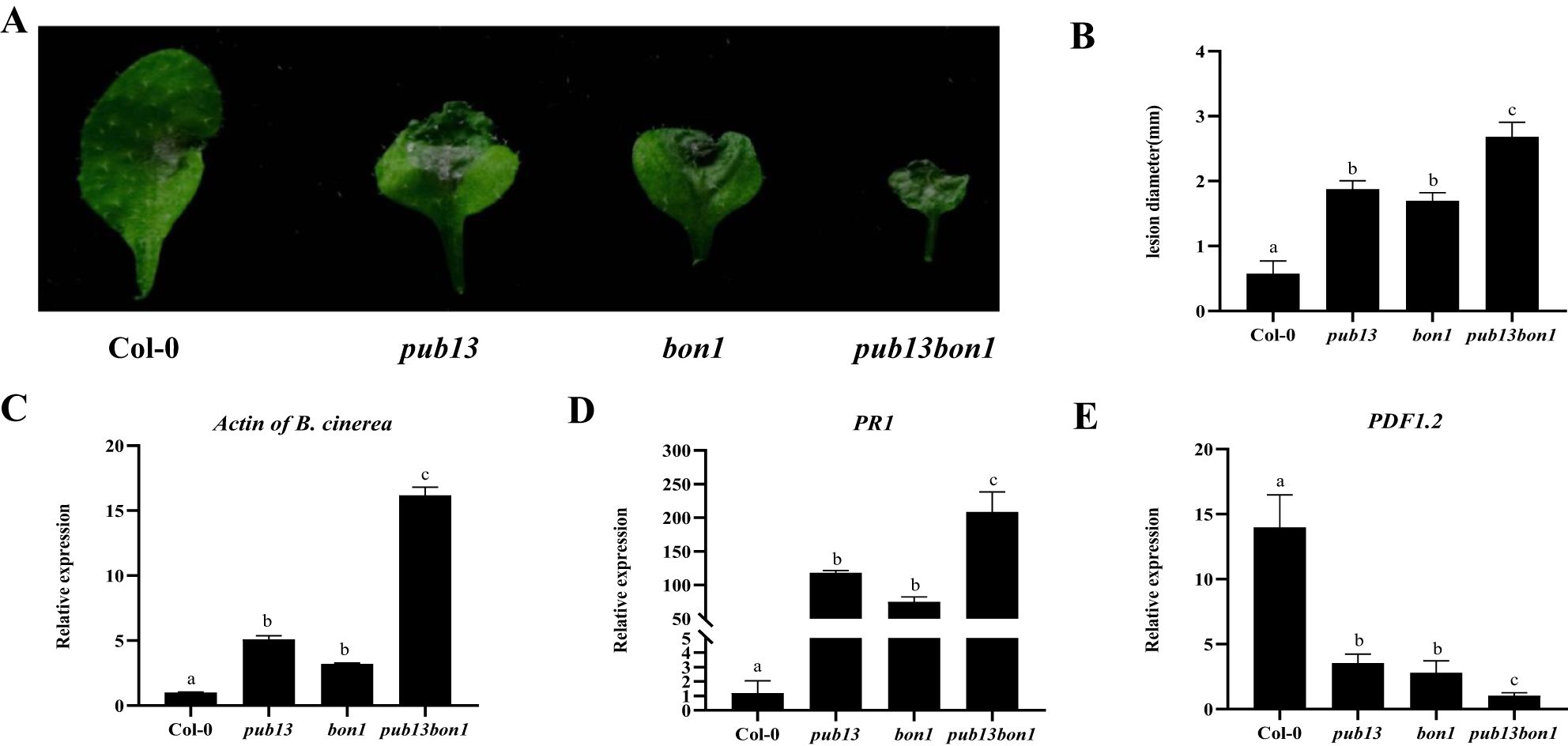
Figure 8. BON1 Synergizes with PUB13 to Positively Regulate Resistance to to Botrytis cinerea. (A), Disease symptoms of Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) infected with B. cinerea BO5-10. Detached leaves were inoculated with 5 µL of conidia suspension (3×104 conidia mL-1) of BO5-10, then kept the samples under high humidity conditions, and observe the symptoms at 48 hpi. B, Lesion size of plants inoculated with (B) cinerea BO5-10 at 48 hpi. (C-E), Transcription level of BO5-10 Actin, PR1 and PDF1.2 in indicated plants after inoculation with Botrytis cinerea BO5-10 at 48 hpi. Arabidopsis Actin was used as internal control. Lowercase letters indicate significant difference at P<0.01. All experiments were repeated at least three times with similar results.
3.9 BON1 is involved in the PTI response regulated by PUB13
To determine whether BON1 is involved in PUB13-regulated PTI signaling, the PAMP flg22-induced PTI responses were investigated in Col-0, pub13, bon1, and pub13(−/−)bon1(+/−) plants. Upon stimulation with flg22, the accumulation of ROS was enhanced in these plants (Figure 9A). Notably, pub13 and bon1 showed stronger induction of flg22-triggered ROS compared with Col-0, and the pub13(−/−)bon1(+/−) double mutant showed more ROS accumulation than the other three plants, indicating additive effects between PUB13 and BON1 on PTI response (Figure 9A). Furthermore, the expression of the marker gene FRK1 in the PTI signaling pathway was examined by RT-qPCR. The results showed that before (0 hpi) or after (5 hpi) flg22 treatment, the FRK1 expression in pub13, bon1, and pub13(−/−)bon1(+/−) was significantly upregulated compared with Col-0 and was highest in pub13(−/−)bon1(+/−) (Figure 9B). These data suggest that both PUB13 and BON1 function as negative regulators of PTI and that BON1 is involved in the PTI response regulated by PUB13.
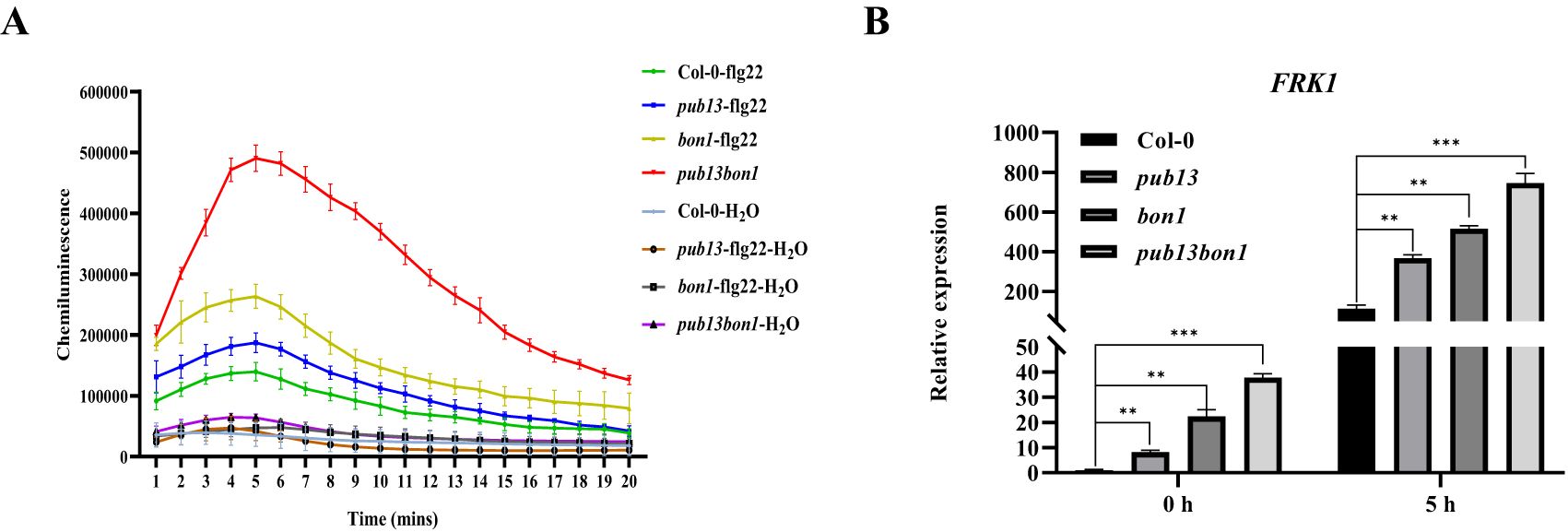
Figure 9. PUB13 Synergizes with BON1 to regulate PTI. (A), flg22 induced ROS accumulation in Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) leaves. ROS was determined using the luminol-based chemiluminescence. Three leaf discs for each sample were treated with 100 nM flg22 or sterilized distilled water as control. (B), FRK1 expression in Col-0, pub13, bon1, and pub13(-/-)bon1(+/-) upon flg22 induction. After injecting 100 nM flg22, RNA was extracted at 0 h and 5h for RT-qPCR. Actin was used as internal reference. Asterisks denote significant difference based on nested ANOVA (**P < 0.01, ***P < 0.001). These experiments were repeated three times with similar results.
4 Discussion
The Arabidopsis E3 ubiquitin ligase PUB13, as an important hub of several signals, regulates multiple plant biological processes, including growth, flowering, plant immunity, and cell death (Lu et al., 2011; Li et al., 2012a; Kong et al., 2015). In this study, it was found that PUB13 regulated plant growth, flowering time, and disease resistance. On the other hand, the copine protein BON1 was identified as an interactor and a ubiquitination substrate of PUB13, which enhanced its biological functions on growth, flowering, and immunity. Moreover, BON1 can be ubiquitinated, but not degraded, by PUB13. Here, we hypothesize that PUB13 activates BON1 through ubiquitination, enabling their cooperative regulation of plant stress responses through synergistic signaling.
On the other hand, PUB13 negatively regulated the plant flowering time and disease resistance depending on the SA and JA/ET signaling pathways (Li et al., 2012a). Based on the enhanced PR1 and the inhibited PDF1.2 expression in bon1 and pub13(−/−)bon1(+/−), conceivably, BON1 synergizes with PUB13 to negatively regulate disease resistance probably via the SA and JA/ET signaling pathways. SA and JA/ET are major phytohormones that mediate plant disease resistance against biotrophic and necrotrophic pathogens, respectively (Shigenaga et al., 2017; Costarelli et al., 2020). The increase in the transcription levels of PR1 indicates that the SA signaling pathway was activated in bon1 and pub13(−/−)bon1(+/−) mutants, as well as in pub13, leading to the enhanced resistance to biotrophic pathogens regulated by the SA signaling pathway. In contrast, the decrease in the transcription levels of PDF1.2 suggests that the JA/ET signaling pathway was suppressed in the pub13, bon1, and pub13(−/−)bon1(+/−) mutants, resulting in the increased susceptibility to necrotrophic pathogens regulated by the JA/ET signaling pathway.
Both PUB13 and BON1 simultaneously regulate plant disease resistance and growth, indicating the correlation between these two biological processes. According to numerous studies, the enhancement of disease resistance usually leads to the inhibition of plant growth by the resistance substances in the plant; hence, there is a certain antagonistic relationship between these two biological processes, which is a challenge in the field of disease resistance breeding. For example, OsBON1 or OsBON3, the orthologs of Arabidopsis BON1, suppresses disease resistance, but promotes plant growth (Yin et al., 2018). Moreover, a pair of homologous E3 ubiquitin ligases (APIP6 and IPI1) in rice enhances the resistance to rice blast by facilitating the degradation of the circadian clock-related protein OsELF3 (Xu et al., 2024). In addition, the OsELF3 protein is associated with the regulation of flowering time in rice; therefore, its degradation results in a delay in the flowering period of rice (Xu et al., 2024). On the other hand, the pub13 and bon1 mutants displayed inhibited growth, but they were early flowering, implying the phenomenon that plants will start their development stage in advance once their growth is inhibited, which is a self-preservation mechanism in plants.
In this study, it was found that the ubiquitination system protein PUB13 interacts with the Ca2+-dependent phospholipid-binding protein BON1 and that they can synergistically regulate the growth, development, and disease resistance in Arabidopsis. The interaction between calcium signaling and the ubiquitination pathway has been reported (Mukherjee et al., 2017). The ubiquitination pathway can regulate the stability and activity of proteins involved in calcium signaling transduction, thus influencing the transmission and response of calcium signals (Meng et al., 1999; Wang et al., 2010). For example, the calmodulin-regulated transcription factor AtSR1/CAMTA3 in Arabidopsis negatively regulates the SA-regulated plant disease resistance according to the ubiquitination system (Zhang et al., 2014). When the plant is invaded by pathogens, the ubiquitination and the degradation of AtSR1 are activated, leading to the suppression of the AtSR1-regulated calcium/calmodulin signaling, which results in the SA-mediated plant defense being enhanced (Zhang et al., 2014). Nevertheless, in-depth research is needed on the crosstalk between plant disease resistance and growth/development, as well as on the interaction mechanism between ubiquitination and calcium signaling in plant immune responses, which will provide a better understanding of the signal transduction network of plant disease resistance.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This article does not contain any studies with human participants or animals performed by any authors. All plant procedures were approved by the Tobacco Research Institute, Chinese Academy of Agricultural Sciences.
Author contributions
JY: Writing – original draft, Writing – review & editing. XZ: Writing – original draft, Writing – review & editing. YP: Resources, Writing – review & editing. SP: Data curation, Formal Analysis, Writing – review & editing. CH: Data curation, Methodology, Writing – review & editing. BW: Methodology, Validation, Writing – review & editing. LD: Conceptualization, Writing – review & editing. WL: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Changde Tobacco Company in Hunan (CD2024KJ04) and the National Natural Sciences Foundation of China (31300250, 32272559).
Conflict of interest
Author YP was employed by the company China Tobacco Hunan Industrial Co, LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1585221/full#supplementary-material
Supplementary Table 1 | Part of library screening results by yeast two-hybrid.
References
Antignani, V., Klocko, A. L., Bak, G., Chandrasekaran, S. D., Dunivin, T., and Nielsen, E. (2015). Recruitment. of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell. 27, 243–261. doi: 10.1105/tpc.114.134262
Azevedo, C., Santos-Rosa, M. J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant science. 6, 354–358. doi: 10.1016/s1360-1385(01)01960-4
Chen, K., Gao, J., Sun, S., Zhang, Z., Yu, B., Li, J., et al. (2020). BONZAI proteins control global osmotic. Stress responses in plants. Curr. biology: CB. 30, 4815–4825.e4. doi: 10.1016/j.cub.2020.09.016
Chen, Y. C., Wong, C. L., Muzzi, F., Vlaardingerbroek, I., Kidd, B. N., and Schenk, P. M. (2014). Root defense. analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci. Rep. 4, 5584. doi: 10.1038/srep05584
Costarelli, A., Bianchet, C., Ederli, L., Salerno, G., Piersanti, S., Rebora, M., et al. (2020). Salicylic acid. induced by herbivore feeding antagonizes jasmonic acid mediated plant defenses against insect attack. Plant Signaling behavior. 15, 1704517. doi: 10.1080/15592324.2019.1704517
Gilroy, E. M., Taylor, R. M., Hein, I., Boevink, P., Sadanandom, A., and Birch, P. R. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New phytologist. 190, 653–666. doi: 10.1111/j.1469-8137.2011.03643.x
Gou, M., Zhang, Z., Zhang, N., Huang, Q., Monaghan, J., Yang, H., et al. (2015). Opposing effects on two. Phases of defense responses from concerted actions of HEAT SHOCK COGNATE70 and BONZAI1 in arabidopsis. Plant Physiol. 169, 2304–2323. doi: 10.1104/pp.15.00970
He, Q., McLellan, H., Boevink, P. C., Sadanandom, A., Xie, C., Birch, P. R., et al. (2015). U-box E3. ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J. Exp. botany. 66, 3189–3199. doi: 10.1093/jxb/erv128
Heise, A., Lippok, B., Kirsch, C., and Hahlbrock, K. (2002). Two immediate-early pathogen-responsive. members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proc. Natl. Acad. Sci. United States America. 99, 9049–9054. doi: 10.1073/pnas.132277699
Hua, J., Grisafi, P., Cheng, S. H., and Fink, G. R. (2001). Plant growth homeostasis is controlled by the. Arabidopsis BON1 and BAP1 genes. Genes Dev. 15, 2263–2272. doi: 10.1101/gad.918101
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel. ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 96, 635–644. doi: 10.1016/s0092-8674(00)80574-7
Kong, L., Cheng, J., Zhu, Y., Ding, Y., Meng, J., Chen, Z., et al. (2015). Degradation of the ABA co-receptor. ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6, 8630. doi: 10.1038/ncomms9630
Li, W., Ahn, I. P., Ning, Y., Park, C. H., Zeng, L., Whitehill, J. G., et al. (2012a). The U-Box/ARM E3 ligase. PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159, 239–250. doi: 10.1104/pp.111.192617
Li, W., Dai, L., and Wang, G. L. (2012b). PUB13, a U-box/ARM E3 ligase, regulates plant defense, cell death, and flowering time. Plant Signaling behavior. 7, 898–900. doi: 10.4161/psb.20703
Li, Y., Gou, M., Sun, Q., and Hua, J. (2010). Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J. Biol. Chem. 285, 29884–29891. doi: 10.1074/jbc.M109.066100
Li, Y., Pennington, B. O., and Hua, J. (2009). Multiple R-like genes are negatively regulated by BON1 and BON3 in arabidopsis. Mol. Plant-Microbe interactions: MPMI. 22, 840–848. doi: 10.1094/MPMI-22-7-0840
Li, W., Xiong, Y., Lai, L. B., Zhang, K., Li, Z., Kang, H., et al. (2021). The rice RNase P protein. subunit Rpp30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol. J. 19, 1988–1999. doi: 10.1111/pbi.13612
Lu, D., Lin, W., Gao, X., Wu, S., Cheng, C., Avila, J., et al. (2011). Direct ubiquitination of pattern recognition. receptor FLS2 attenuates plant innate immunity. Sci. (New York N.Y.). 332, 1439–1442. doi: 10.1126/science.1204903
Meng, W., Sawasdikosol, S., Burakoff, S. J., and Eck, M. J. (1999). Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 398, 84–90. doi: 10.1038/18050
Mudgil, Y., Shiu, S. H., Stone, S. L., Salt, J. N., and Goring, D. R. (2004). A large complement of the predicted. Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134, 59–66. doi: 10.1104/pp.103.029553
Mukherjee, R., Das, A., Chakrabarti, S., and Chakrabarti, O. (2017). Calcium dependent regulation of protein. ubiquitination - Interplay between E3 ligases and calcium binding proteins. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1227–1235. doi: 10.1016/j.bbamcr.2017.03.001
Mukhopadhyay, D. and Riezman, H. (2007). Proteasome-independent functions of ubiquitin in endocytosis. and signaling. Sci. (New York N.Y.). 315, 201–205. doi: 10.1126/science.1127085
O’Neill, L. A. (2011). Plant science. Innate immunity in plants goes to the. Sci. (New York N.Y.). 332, 1386–1387. doi: 10.1126/science.1208448
Park, J. J., Yi, J., Yoon, J., Cho, L. H., Ping, J., Jeong, H. J., et al. (2011). OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant journal: Cell Mol. Biol. 65, 194–205. doi: 10.1111/j.1365-313X.2010.04416.x
Peifer, M., Berg, S., and Reynolds, A. B. (1994). A repeating amino acid motif shared by proteins with diverse. cellular roles. Cell. 76, 789–791. doi: 10.1016/0092-8674(94)90353-0
Rizo, J. and Südhof, T. C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882. doi: 10.1074/jbc.273.26.15879
Samuel, M. A., Salt, J. N., Shiu, S. H., and Goring, D. R. (2006). Multifunctional arm repeat domains in. plants. Int. Rev. cytology. 253, 1–26. doi: 10.1016/S0074-7696(06)53001-3
Shigenaga, A. M., Berens, M. L., Tsuda, K., and Argueso, C. T. (2017). Towards engineering of hormonal. crosstalk in plant immunity. Curr. Opin. Plant Biol. 38, 164–172. doi: 10.1016/j.pbi.2017.04.021
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Žársky, V, et al. (2012). The ubiquitin. ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell. 24, 4703–4716. doi: 10.1105/tpc.112.104463
Sun, J., Sun, Y., Ahmed, R. I., Ren, A., and Xie, A. M. (2019). Research progress on plant RING-finger. Proteins. Genes. 10, 973. doi: 10.3390/genes10120973
Tang, H., Pang, P., Qin, Z., Zhao, Z., Wu, Q., Song, S., et al. (2021). The CPNE family and their role in cancers. Front. Genet. 12. doi: 10.3389/fgene.2021.689097
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered. immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. biology: CB. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Vega-Sánchez, M. E., Zeng, L., Chen, S., Leung, H., and Wang, G. L. (2008). SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell. 20, 1456–1469. doi: 10.1105/tpc.108.058610
Wang, J., Grubb, L. E., Wang, J., Liang, X., Li, L., Gao, C., et al. (2018). A regulatory module controlling. Homeostasis of a plant immune kinase. Mol. Cell. 69, 493–504.e6. doi: 10.1016/j.molcel.2017.12.026
Wang, W., Liu, N., Gao, C., Cai, H., Romeis, T., and Tang, D. (2020). The Arabidopsis exocyst subunits. EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New phytologist. 227, 529–544. doi: 10.1111/nph.16515
Wang, J., Peng, Q., Lin, Q., Childress, C., Carey, D., and Yang, W. (2010). Calcium activates Nedd4 E3. ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J. Biol. Chem. 285, 12279–12288. doi: 10.1074/jbc.M109.086405
Warner, A., Xiong, G., Qadota, H., Rogalski, T., Vogl, A. W., Moerman, D. G., et al. (2013). CPNA-1, a.copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans. Mol. Biol. Cell. 24, 601–616. doi: 10.1091/mbc.E12-06-0478
Whittaker, C. A. and Hynes, R. O. (2002). Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell. 13, 3369–3387. doi: 10.1091/mbc.e02-05-0259
Xie, Q., Guo, H. S., Dallman, G., Fang, S., Weissman, A. M., and Chua, N. H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 419, 167–170. doi: 10.1038/nature00998
Xu, X., Shi, X., You, X., Hao, Z., Wang, R., Wang, M., et al. (2024). A pair of E3 ubiquitin ligases control. immunity and flowering by targeting different ELF3 proteins in rice. Dev. Cell. 59, 2731–2744.e4. doi: 10.1016/j.devcel.2024.06.013
Xu, F. Q. and Xue, H. W. (2019). The ubiquitin-proteasome system in plant responses to environments. Plant Cell environment. 42, 2931–2944. doi: 10.1111/pce.13633
Yang, C. W., González-Lamothe, R., Ewan, R. A., Rowland, O., Yoshioka, H., Shenton, M., et al. (2006). The. E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell. 18, 1084–1098. doi: 10.1105/tpc.105.039198
Yang, S. and Hua, J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates. temperature-dependent growth control in Arabidopsis. Plant Cell. 16, 1060–1071. doi: 10.1105/tpc.020479
Yang, D. L., Shi, Z., Bao, Y., Yan, J., Yang, Z., Yu, H., et al. (2017). Calcium pumps and interacting BON1. Protein modulate calcium signature, stomatal closure, and plant immunity. Plant Physiol. 175, 424–437. doi: 10.1104/pp.17.00495
Yang, S., Yang, H., Grisafi, P., Sanchatjate, S., Fink, G. R., Sun, Q., et al. (2006). The BON/CPN gene family. represses cell death and promotes cell growth in Arabidopsis. Plant journal: Cell Mol. Biol. 45, 166–179. doi: 10.1111/j.1365-313X.2005.02585.x
Ye, Y. and Rape, M. (2009). Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764. doi: 10.1038/nrm2780
Yee, D. and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators. to downstream target substrates. J. Exp. botany. 60, 1109–1121. doi: 10.1093/jxb/ern369
Yi, S. Y., Nekrasov, V., Ichimura, K., Kang, S. Y., and Shirasu, K. (2024). Plant U-box E3 ligases PUB20 and PUB21 negatively regulate pattern-triggered immunity in Arabidopsis. Plant Mol. Biol. 114, 7. doi: 10.1007/s11103-023-01409-6
Yin, X., Zou, B., Hong, X., Gao, M., Yang, W., Zhong, X., et al. (2018). Rice copine genes OsBON1 and. OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol. J. 16, 1476–1487. doi: 10.1111/pbi.12890
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through. ubiquitin-mediated modulation of Ca(2+) -calmodulin-AtSR1/CAMTA3 signaling. Plant journal: Cell Mol. Biol. 78, 269–281. doi: 10.1111/tpj.12473
Keywords: PUB13, E3 ubiquitin ligase, BON1, flowering, resistance, immunity
Citation: Yue J, Zou X, Peng Y, Pan S, Hu C, Wang B, Dai L and Li W (2025) The Arabidopsis E3 ubiquitin ligase PUB13 synergistically interacts with BON1 to regulate plant flowering and immunity. Front. Plant Sci. 16:1585221. doi: 10.3389/fpls.2025.1585221
Received: 28 February 2025; Accepted: 12 May 2025;
Published: 02 June 2025.
Edited by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Srayan Ghosh, Durham University, United KingdomDinesh Kumar Yadav, Allahabad University, India
So Young Yi, Kongju National University, Republic of Korea
Copyright © 2025 Yue, Zou, Peng, Pan, Hu, Wang, Dai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangying Dai, ZGFpbHlAaHVuYXUubmV0; Wei Li, bGl3ZWkzNTA1NTFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Jiani Yue
Jiani Yue Xiuwei Zou1†
Xiuwei Zou1† Bing Wang
Bing Wang Wei Li
Wei Li