- 1Hainan Institute, Zhejiang University, Sanya, China
- 2Institute of Crop Science, Zhejiang Key Laboratory of Crop Germplasm Innovation and Utilization, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 3Department of Biology, College of Science and Technology, Wenzhou-Kean University, Wenzhou, China
Heading date, a critical agronomic trait determining rice regional adaptation and yield potential, is regulated by complex genetic networks. Although stress-associated proteins (SAPs) are well-documented mediators of abiotic stress responses, their roles in reproductive development remain poorly characterized. Here we demonstrate that stress-associated protein 5 (OsSAP5) functions as a positive regulator of heading date in rice. Loss-of-function ossap5 mutants exhibited significant heading delay under natural short day condition, accompanied by marked downregulation of florigen genes Hd3a and RFT1 and subsequent suppression of MADS-box genes in the shoot apical meristem (SAM). A similar decrease in expression of florigen genes was observed in OsGF14c overexpression lines under short day condition. The molecular and biochemical assays confirmed that OsSAP5 interacts with OsGF14c. Notably, OsSAP5 had E3 ubiquitin ligase activity and might promote the ubiquitination of OsGF14c. Therefore, OsSAP5 regulates heading date by interacting with OsGF14c under short day condition.
Introduction
Rice (Oryza sativa) is one of the most important crops in the world, and heading date is one of the main factors affecting the yield (Cai et al., 2021). Rice is a typical short day (SD) plant, the heading date of which is promoted under SD condition and delayed under long day (LD) condition. Hd3a and RFT1, FT-like genes, as the florigen play critical roles in regulating heading date in rice (Komiya et al., 2008; Kojima et al., 2002). Hd3a acts as mobile florigens moving from leaves to the shoot apical meristem (SAM), where it forms the florigen activation complex (FAC) with 14-3–3 and OsFD1 under SD condition. RFT1, as the same florigen in LD condition, performs a similar function (Tamaki et al., 2007; Komiya et al., 2009; Taoka et al., 2011; Peng et al., 2021). FTL12, a member of FT-like family, interacts with OsGF14b and OsFD1 to form the florigen repression complex (FRC) by competing with Hd3a for binding OsGF14b (Zheng et al., 2023). The RICE CENTRORADIALIS (RCN) is a rice TFL 1-like protein that also competitively binds to 14-3–3 with Hd3a and form a FRC (Wang et al., 2020). OsbZIP42/HBF1 and OsbZIP9/HBF2, two paralogs of OsFD1, form a complex with Hd3a in leaf cells, which represses Hd3a expression under SD and LD conditions. HBFs could also interact with RFT1 via OsGF14c as the bridge to form a complex in leaf cells, which also represses RFT1 expression under SD and LD conditions (Brambilla et al., 2017).
Under SD condition, the pathway of OsGI-Hd1-Hd3a promotes rice heading, which is conserved with the GI-CO-FT pathway in Arabidopsis (Zhu et al., 2023). Hd1 promotes heading date by inducing directly Hd3a expression (Kojima et al., 2002). OsGI induces Hd1 expression under SD and LD conditions (Hayama et al., 2003). Under LD condition, the pathway Ghd7-Ehd1-Hd3a/RFT1 is the main pathway to promote rice heading (Zhu et al., 2023). Ehd1 is a special gene in rice and induces Hd3a and RFT1 directly in a special rice variety that lacks the Hd1 function under SD condition (Doi et al., 2004). The expression of Ghd7 increases as the day progresses, and Ghd7 represses the expression of Ehd1 (Xue et al., 2008; Itoh et al., 2010). Hd1 also suppresses Hd3a expression under LD condition (Hayama et al., 2003).
The A20/AN1-type zinc finger protein, also called stress-associated protein (SAP), is well known to play significant roles in stress response and plant development. In mammals, the A20 family proteins regulate NF-kB, a tumor necrosis factor, with activation through the ubiquitin–proteasome system (Heyninck et al., 1999). In plants, there are 18 SAP protein family members in rice and 14 members in Arabidopsis, namely, OsSAP1–18 and AtSAP1-14 (Vij and Tyagi, 2006). OsSAP1 is first reported and overexpression lines in tobacco conferred tolerance to cold, dehydration, and salt stress at seed germination/seedling stage (Mukhopadhyay et al., 2004). AtSAP5 shows a strong homology to OsSAP1 and has E3 ubiquitin ligase activity. Plant that overexpress AtSAP5 has increased tolerance to salt stress, osmotic stress, and water deficit (Kang et al., 2011). AtSAP5 promotes AtMBP-1, a c-myc binding protein, degradation by ubiquitin-dependent proteasome pathway (Choi et al., 2012; Feo et al., 2000; Kang et al., 2011). TaSAP5 overexpression in Arabidopsis and wheat seedlings increased their drought tolerance (Zhang et al., 2017). DREB2A INTERACTING PROTEIN1 (DRIP1) and DRIP2, as E3 ubiquitin ligase, interact with drought response transcription factor DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) (Qin et al., 2008). TaSAP5 interacts with TaDRIPs, leading to their subsequent degradation through the 26S proteasome pathway and then increasing the level of DREB2A protein and its downstream genes (Zhang et al., 2017). Plants that overexpress AtSAP9 delayed the flowering time by about 4 days under LD condition, and the expression analysis showed that the expression of CO, SOC, and FT significantly decreases (Kang et al., 2017). The effector of AtSAP8-overexpressing in tobacco showed early flowering time by about 9 days (Lin et al., 2023).
Ubiquitination is a posttranslational modification process. Ubiquitin/26S proteasome system (UPS) plays important roles in plants to cope with environmental changes or in plant development (Linden and Callis, 2020). This system needs ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). Ubiquitin binds to E1, and E1 transfers ubiquitin to E2, which binds with E3. Then, E3 facilitates the transfer of ubiquitin from E2 to the substrate. The consequences of polyubiquitinated substrate become a target for the 26S proteasome for subsequent degradation (Al-Saharin et al., 2022). The abundant E3 ubiquitin ligase determines the plant development and response to environment changes, as E3 can bind to the substrate directly, and different substrates have different functions in the plant (Mazzucotelli et al., 2006).
In our laboratory, the previous study proved that OsSAP5-overexpressed lines in Arabidopsis increase heat tolerance during seed germination (Chen et al., 2021a). In this study, we provided evidence that OsSAP5 is an E3 ubiquitin ligase and is involved in positively regulating heading date under natural short day condition. OsSAP5 might also promote OsGF14c degradation by UPS. Consistent with this, the florigen Hd3a and RFT1 expression was lower in OsSAP5 mutants and in the OsGF14c overexpression lines compared to NIP.
Materials and methods
Plant materials and growth conditions
NIP (Oryza sativa L. Japonica cv. Nipponbare) was used to generate transgenic lines. NIP, ossap5 mutants, and OsSAP5 overexpression lines were grown at the Sanya transgenic experimental base of Zhejiang University, China. The natural long day (NLD) was in Changxing, Zhejiang, China (119.91° E, 31.02° N). The natural short day (NSD) was in the Sanya, Hainan, China (109.5° E, 18.5° N). The germinated seed was grown for 15 days under control short day (CSD) (10-h light and 14-h dark at 28 °C) and control long day (CLD) (14-h light and 10-h dark at 28 °C), respectively.
Vector construction and rice transformation
To construct the CRISPR/Cas9 vector for OsSAP5 or OsGF14c, the predictions made by CRISPR-GE (http://cbi.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR) were used to select two guide sequences with a low off-target rate (Xie et al., 2017). The gRNA framework with the guide sequence was inserted into the pYLCRISPR/Cas9Pubi-H empty vector to produce the CRISPR/Cas9 recombinant vector. OsSAP5 or OsGF14c CDS sequences were amplified by PCR using cDNA as the template and then inserted into the pRHVcMYC for overexpression vector construction. The 2.0-kb OsSAP5 promoter sequences (i.e., sequences upstream of the ATG start codon) were amplified by PCR using genomic DNA as template and then inserted into pDX2181 vector to control the expression of the GUS-encoding gene. The promoter sequences replaced the 35s promoter sequences of pC1300-35s-EGFP vector by using restriction enzymes NcoI and SacI, and then the CDS sequences were inserted into the pC1300-proOsSAP5-EGFP vector. All primers used to construct the recombinant vectors are listed in Supplementary Table S3.
Histochemical analysis of GUS activity
For the GUS staining analysis, leaf blades, leaf sheath, SAM, stems, and young panicles were collected from transgenic plants of pOsSAP5::GUS at 30-day-old seedling and the heading stage. The GUS activity was detected using the GUSblue kit (Huayueyang, Beijing, China) according to the manufacturer’s manual. The samples were destained with pure ethanol and then examined using a dissecting microscope (Leica PLANPOFOV3.6). All primers used to construct recombinant vectors are listed in Supplementary Table S3.
Fluorescence microscope observation of the transgenic OsSAP5-EGFP
The leaf sheath of 15-day-old seedlings was cut 2 to 3 cm by using a scalpel, thus obtaining interesting tissue (the seedling leaf sheath of 2~3 cm). The tissue was put into the embedding cassette with the fixative (Tissue-Tek O. C. T. Compound) for frozen 30 min at -20°C, making sure that the tissue was fully surrounded by the fixative. Then, a machine (CryoStar NX50 OPD) was used to cut the section with 50-μm thickness. Finally, the section of the interact tissue was observed using a fluorescence microscope (LEICA DFC7000 T). All primers used to construct the recombinant vectors are listed in Supplementary Table S3.
Subcellular localization
OsSAP5 or OsGF14c coding sequence was inserted into pC1300-35s-EGFP for subcellular observation. AtPIPT2A-mCherry was used as cell membrane marker (Nelson et al., 2007). The nucleus marker was histone H2B-mCherry. The OsGF14c sequence was replaced with the H2B sequence of the H2B-mCherry nuclear marker vector by using restriction enzymes KpnI and BamHI. Agrobacterium tumefaciens GV3101 cells were transformed with the pC1300-35s-EGFP empty vector or the pC1300-35s-OsSAP5-EGFP recombinant vector for the subsequent infiltration of healthy N. benthamiana leaves. Fluorescent signals were detected using the Leica/STELLARIS 5 laser scanning confocal microscope. For green fluorescence observation, the excitation wavelength was 488 nm and the emission wavelengths were 520 to 540 nm; for mCherry fluorescence observation, the excitation wavelength was 561 nm and the emission wavelengths were 610 to 630 nm. The primers used for the subcellular vectors are listed in Supplementary Table S3.
Rhythmic expression analysis
The germinated seed was transferred to the soil matrix for growing 15 days in a growth chamber with SD condition (10 h light and 14 h dark at 28°C) or with LD condition (14 h light and 10 h dark at 28°C). The leaves of each line were harvested every 4 h for 20 h. For each time point, the leaves from three different individuals were collected as biological replicates.
RNA extraction and quantitative real-time PCR
Total RNA was isolated using Trizol solution (Vazyme, Najing, China). cDNA was synthesized from 1 μg of total RNA using the cDNA synthesis kit with a gDNA wiper (Vazyme, Najing, China). One microliter of cDNA was used for real-time-PCR analysis with gene-specific primers using SYBR Green PCR master mix in a LightCycler480 system (Roche). The 2−ΔΔCt method described was used for the analysis of relative gene expression (Livak and Schmittgen, 2001). Three biology replicates were performed. All of the qRT-PCR primers are listed in Supplementary Table S3.
Bimolecular fluorescence complementation assay
The OsSAP5 coding sequence was PCR-amplified and inserted into pSPYCE-35s vector, and the OsGF14c coding sequence was PCR-amplified and inserted into pSPYNE-35s vector (Wang et al., 2023). Agrobacterium tumefaciens GV3101 cells were transformed with the recombinant vector, which were combined (1:1, v/v) for the subsequent infiltration of healthy N. benthamiana leaves. Fluorescent signals were detected using Leica/STELLARIS 5 laser scanning confocal microscope. The laser used was similar to the methods of subcellular localization. The primers used for bimolecular fluorescence complementation (BiFC) vectors are listed in Supplementary Table S3.
Luciferase complementation imaging assay
The plasmids of pCAMBIA1300-cLUC and pCAMBIA1300-nLUC were described in the paper (Chen et al., 2008). The OsSAP5 coding sequence was PCR-amplified and inserted into pCAMBIA1300-cLUC vector, and the OsGF14c coding sequence was PCR-amplified and inserted into pCAMBIA1300-nLUC. Agrobacterium tumefaciens GV3101 cells were transformed with the recombinant vector, which were combined (1:1, v/v) for the subsequent infiltration of healthy N. benthamiana leaves, and incubated in the growth room (25°C light/23°C dark) for 48 h before the LUC activity measurement. For the CCD imaging and LUC activity measurement, 1 mM luciferin was sprayed onto the leaves. The cooled CCD imaging apparatus was used to capture the LUC image. Each data point contains at least four replicates, and three independent experiments were carried out. The primers used for LCI vectors are listed in Supplementary Table S3.
Pull-down assay
OsSAP5 and OsGF14c coding sequences were inserted into pET32a and pGEX-4T-1 vectors, respectively. Escherichia coli BL21 (DE3) cells were transformed with the recombinant vectors for the expression of GST-OsGF14c or His-OsSAP5 proteins. A pull-down assay was performed for 2 h at 4°C using GSTSep Glutathione Agarose Resin (GST) particles (YEASEN, Shanghai, China) in binding buffer (25 mM Tris-HCl [pH = 8.0]; 150 mM NaCl; 1/2,000 (v/v) NP-40; 1/20 (v/v) glycerinum). Finally, the samples were boiling at 100°C for 10 min and immunoblotted using anti-GST (Abmart, 1:1,000) and anti-His (Abmart, 1:1,000) mouse monoclonal antibody. The primers used for the pull-down vectors are listed in Supplementary Table S3.
Co-immunoprecipitation assay
OsSAP5 and OsGF14c coding sequences were inserted into pRTVcHA and pRTVcGFP vectors to generate p35s::OsSAP5-HA and p35s::OsGF14c-GFP, respectively. To transiently express these recombinant vectors, the vector combinations were co-transformed into 100 mL of rice protoplasts using a PEG-mediated transfection method as described previously (Zhang et al., 2011). After incubation at 25°C for 14 h, the transfected protoplasts were pelleted at 200 g for 3 min. The pellets were resuspended in 500vmL protein extraction buffer (50 mM Tris–HCl [pH 8], 500 mM sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 1× PMSF, and 1× protease inhibitor cocktail) and then incubated at 4°C for 1 h with vortexing. After centrifugation at 13,000 g for 10 min at 4°C, 20 μL anti-GFP affinity beads 4FF (Smart-Lifesciences, Changzhou, China) was added to the supernatant with rotation at 4°C for 2 h. The beads were then washed with protein extraction buffer three times and boiled with 100 μL 1× SDS-PAGE loading buffer (Beyotime, Shanghai, China) for 10 min at 100°C to elute bound proteins from the beads. The proteins were then separated in 10% SDS-PAGE gels and detected by western blot analysis using anti-GFP antibodies (Abmart 1:1,000) and anti-HA antibodies (Abmart, 1:1,000). The primers used for the Co-IP vectors are listed in Supplementary Table S3.
In vitro ubiquitination assay
Ubiquitination assay was performed as previously described with some modifications (Wang et al., 2017). Briefly, 1 μg His-OsSAP5 was incubated in a 20 μL reaction mixture containing 1 M Tris-HCl (pH 7.5), 200 mM MgCl2, 40 mM DTT, 100 mM ATP, 50 ng E1 (R&D systems, E-305-025), 150 ng E2 (Sigma-Aldrich, UbcH5b), and 1 μg ubiquitin (R&D systems, HA-Ub) and incubated at 30 °C for 1.5 h. The reaction was stopped by adding 1× SDS-PAGE loading buffer (Beyotime, Shanghai, China) and boiling at 100°C for 10 min. The proteins were then separated in 10% SDS–PAGE gels and detected by western blot analysis using anti-HA antibodies (Abmart, 1:1,000).
In vitro degradation assay
A cell-free protein degradation assay was performed as previously described (Spoel et al., 2009). The total protein of NIP 30-day-old seedling was extracted from cultured cells with proteolysis buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 10 mM ATP, and 5 mM DTT). Then, total protein and His-OsGF14c protein were incubated with the inhibitors PMSF (4 μM), MG132 (150 μM, Yuanye Biotechnology Co. Ltd., Shanghai, China), or organic solvent DMSO at 25°C, with the sample taken every 30 min for 2 h. Finally, the samples were boiling at 100 °C for 10 min after adding 1× SDS loading buffer; then, the proteins were separated in 10% SDS–PAGE gels and detected by western blot analysis using anti-His antibodies (Abmart, 1:1,000).
IP-MS analysis
The seedlings of NIP and OsSAP5-MYC were collected and ground into fine powder in liquid nitrogen. The total protein was isolated with Co-IP buffer, and then the homogenate was centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was collected and incubated with Anti-c-Myc Affinity Beads (Smart-Lifesciences, Changzhou, China) for 4 h at 4°C. After washing, the immunoprecipitated protein mixture was eluted and analyzed by LC-MS/MS using a Q Exactive HF-X System (Thermo Fisher). The spectrum data were searched against the RGAP database using Protein Prospector (Chen et al., 2021b).
Dual LUC reporter assays
The OsSAP5 coding sequence was inserted into pRT-BD-GAL4 plasmid. The empty BD-GAL4 served as negative control, and BD-GAL4-VP16 was the positive control. The reporter plasmid was the pGreenII-0800-5×UAS_35S_LUC vector. The effector and reporter plasmids were co-transformed into rice protoplasts. After 12 h of incubation in the dark, the protoplasts were collected and immediately utilized for LUC assays. Untreated protoplast populations were used as controls. The relative LUC activity was quantified using the Dual Luciferase Assay Kit (Promega) and then calculated as the ratio of fLUC to rLUC (fLUC/rLUC). Renilla LUC was used as an internal control. Three independent transformations were performed for each sample. The primers used for subcellular vectors are listed in Supplementary Table S3.
Expression of OsMADS genes in rice protoplasts
Promoter sequences of about 2,264 bp of OsGF14c replaced the Ubiq promoter sequences in pRTVcGFP vector by using restriction enzymes Pstl and EcoRV. Then, the OsFD1 and OsGF14c coding sequences were inserted into the vector with pOsGF14c sequences to generate pOsGF14c::OsFD1 and pOsGF14c::OsGF14c. The promoter sequences, about 2,060 bp of Hd3a, replaced the Ubiq promoter sequences in pRTVcGFP vector by using restriction enzymes Pstl and EcoRV. Then, the OsGF14c coding sequence was inserted into the vector with pHd3a sequences to generate pHd3a::OsGF14c. The Hd3a coding sequence was inserted into pRTVcMyc to generate p35s::Hd3a. The different combination of effector was transformed into rice protoplasts by the methods of CO-IP. After 60 h of incubation, the checking of OsMADS expression was similar to the methods in “RNA extraction and quantitative real-time PCR”. The primers used for subcellular vectors are listed in Supplementary Table S3.
Results
OsSAP5 positively regulates heading date in rice
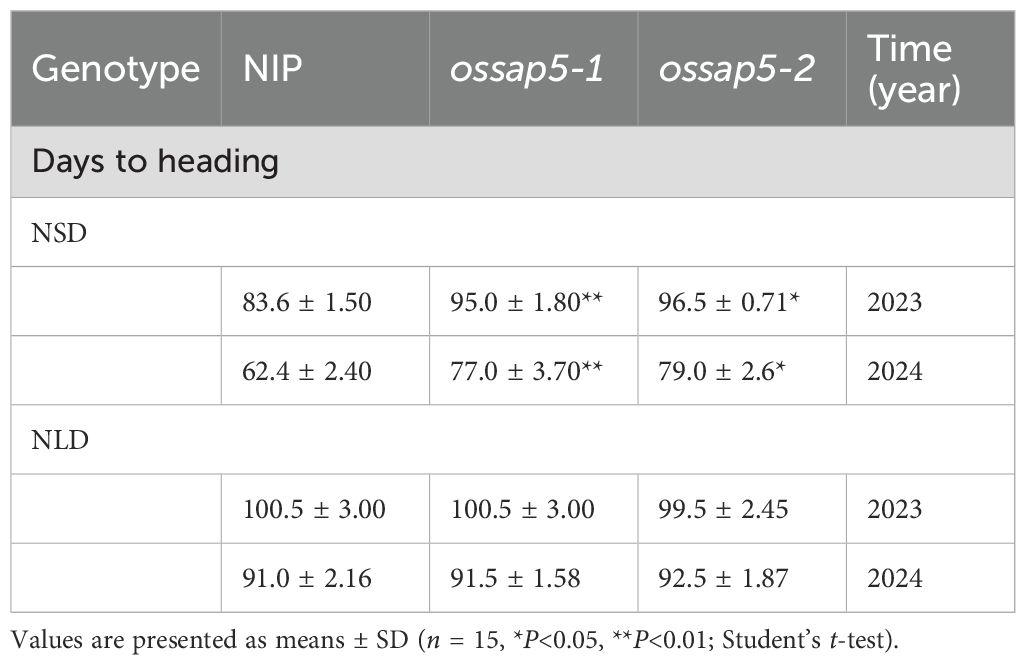
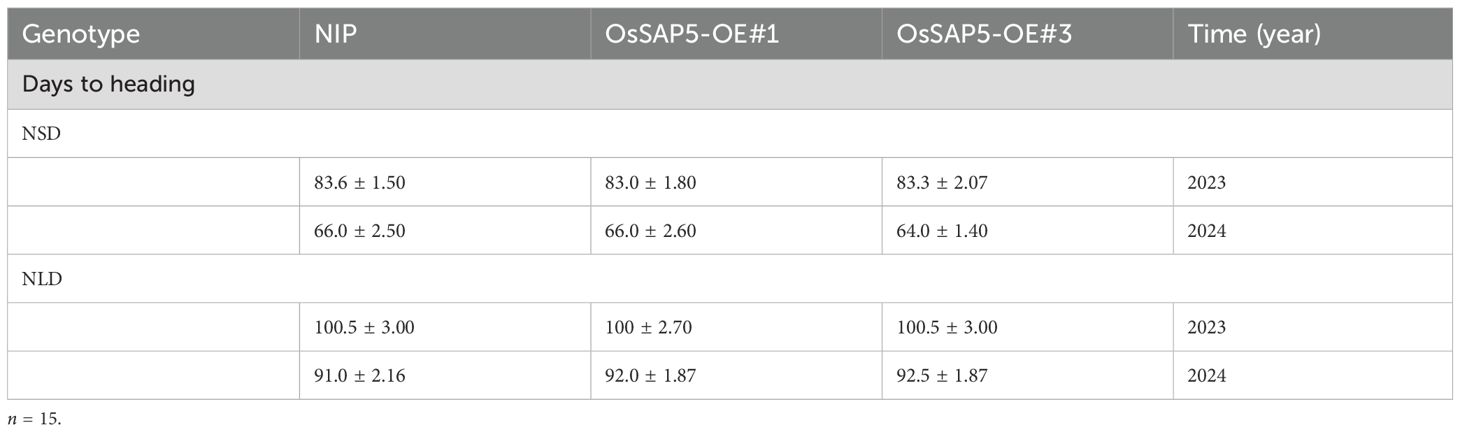
In our previous study, OsSAP5-overexpressed lines in Arabidopsis enhanced the tolerance to drought during seed germination (Chen et al., 2021a), but the function of OsSAP5 remains unknown in rice. The ossap5-knockout mutants were generated in the japonica rice cultivar Nipponbare background using the CRISPR/Cas9 genome editing approach. Two independent homozygous lines (ossap5-1 and ossap5-2) were selected for further study. Both mutant lines showed the deletion of long fragment bases in OsSAP5 coding sequence (Figure 1C). Therefore, OsSAP5 protein function was disrupted. The seedlings of two mutant lines showed lower tolerance to heat stress and drought compared to NIP (data not shown). In addition, in the natural short day (NSD) field, ossap5-1 and ossap5-2 delayed about 15 days in heading date compared to NIP, but no significant differences were observed under natural long day (NLD) condition (Figures 1A, B; Table 1). The overexpression construct (pUbi::OsSAP5-MYC) was transformed into Nipponbare, and two homozygous lines (OsSAP5-OE#1 and OsSAP5-OE#3) with MYC tag, which had increased OsSAP5 expression, were selected for further study (Figures 2A, B; Supplementary Figure S1). Both overexpression lines showed the same heading date compared to NIP under both NSD and NLD conditions (Figures 2C, D; Table 2).
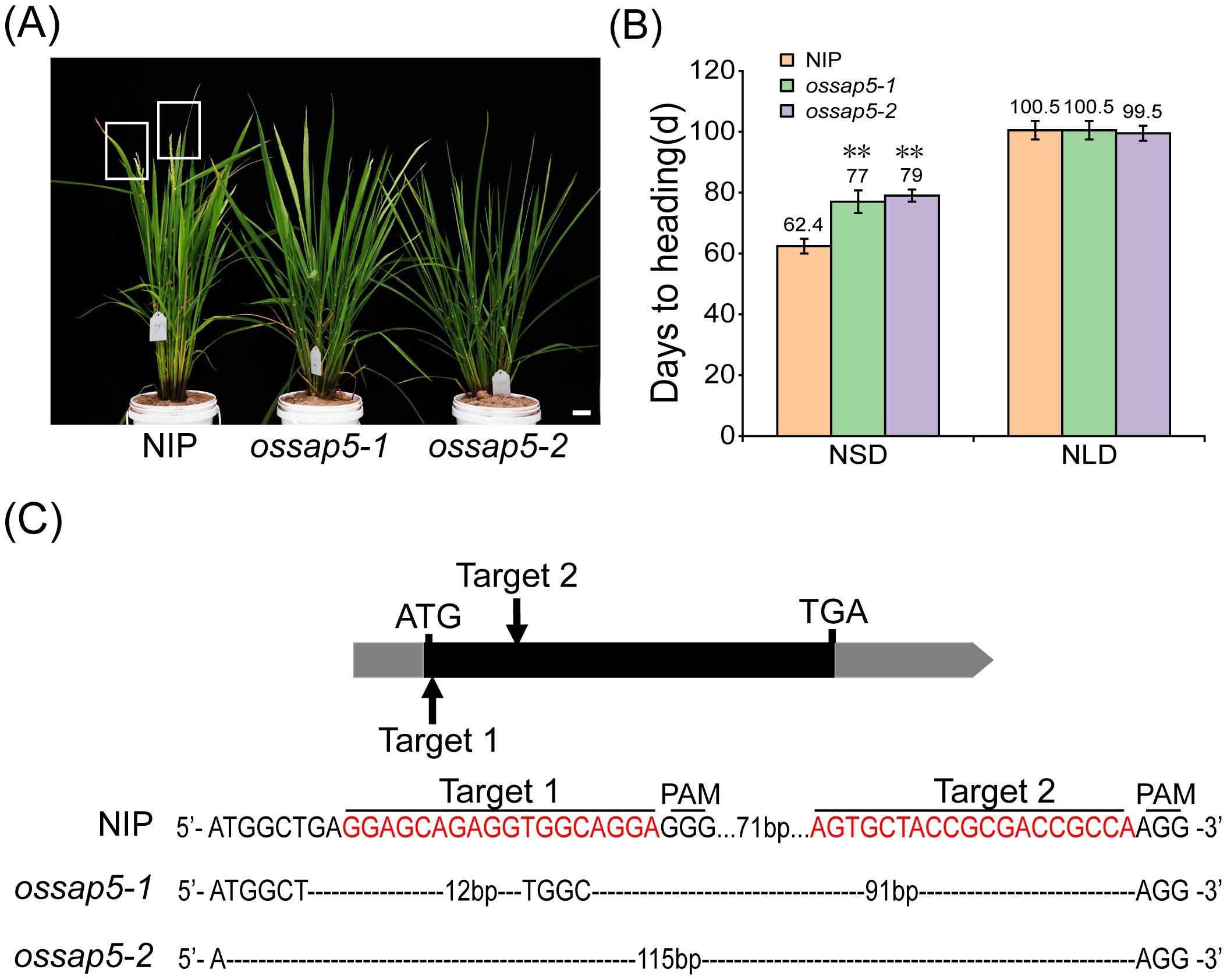
Figure 1. Heading date of OsSAP5 mutants. (A) Heading phenotype of NIP (Oryza sativa L.) and OsSAP5 mutants under NSD condition. Bar = 10 cm; ossap5–1 and ossap5–2 represent two OsSAP5 mutants. The rectangular box indicates rice heading. (B) Days to heading of NIP, OsSAP5 mutants under NLD and NSD conditions. Values are presented as means ± SD (n = 15, **P < 0.01; Student’s t-test). NSD means natural short day. NLD means natural long day. (C) Generation and identification of two OsSAP5 mutants generated by CRISPR/Cas9 genome-editing approach. The target 1 and target 2 sites and the protospacer adjacent motif (PAM) are indicated. The black dash indicates the deletion of a base pair.
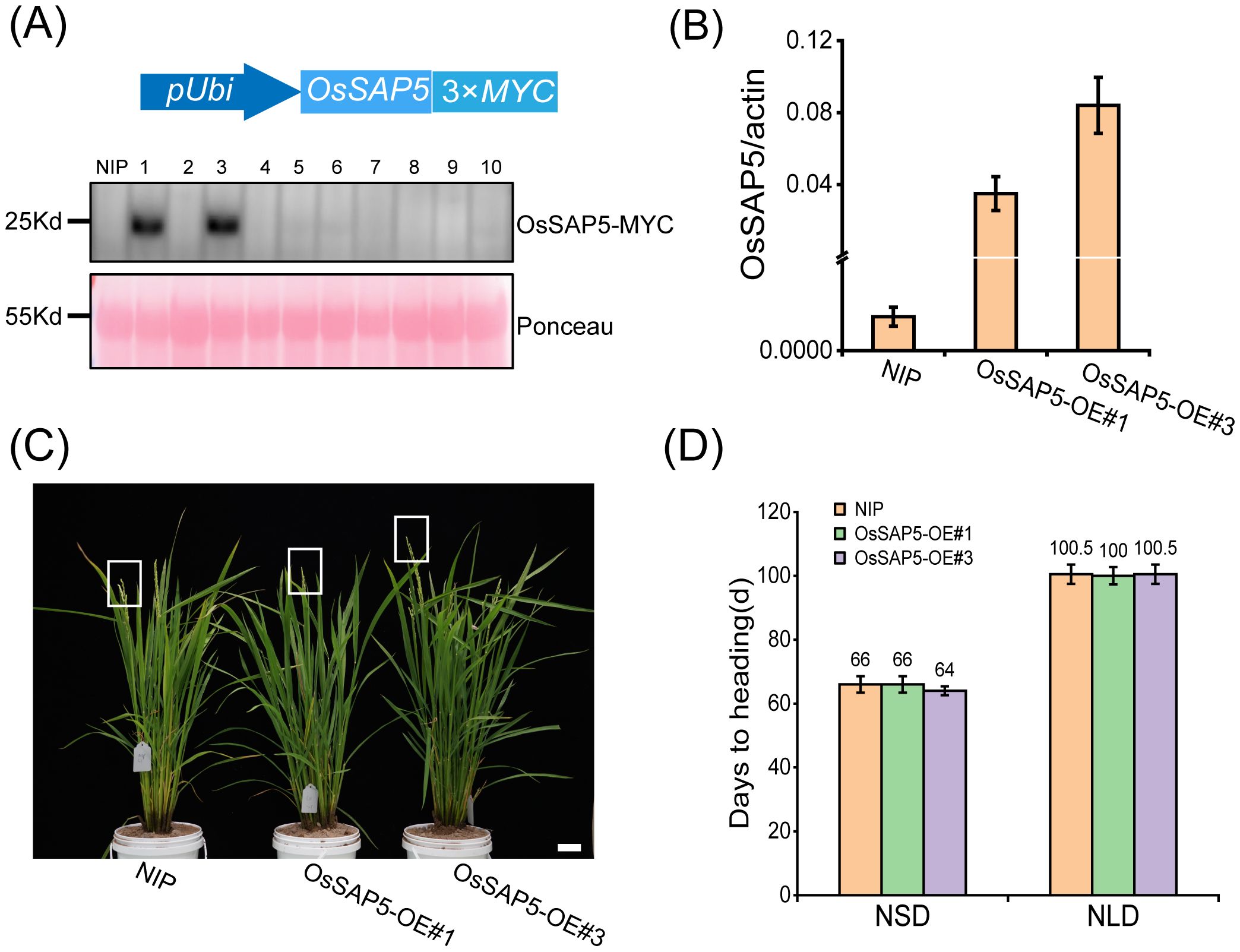
Figure 2. Heading date of OsSAP5 overexpression lines. (A) MYC-tag immunoprecipitate of OsSAP5 overexpression plants. A total of 10 protein samples are detected with anti-MYC antibodies with loading as shown by Ponceau S staining for RUBISCO. (B) qRT-PCR analysis of the expression levels of OsSAP5 in NIP and OsSAP5 overexpression lines. Values are presented as means ± SD (n = 3). OsSAP5-OE#1 and OsSAP5-OE#3 indicate two OsSAP5 overexpression lines, respectively. (C) Heading phenotype of NIP and OsSAP5 overexpression lines under NSD condition. Bar = 10 cm. The rectangular box indicates rice heading. (D) Days to heading of NIP, OsSAP5 overexpression lines under NLD and NSD conditions. Values are presented as means ± SD (n = 15).
ossap5-CRISPR exhibited a higher seed setting compared to NIP, while maintaining comparable plant height, tiller number, and yield per plant under NSD condition (Supplementary Table S1). However, OsSAP5-overexpressing lines showed significantly reduced seed setting and decreased yield per plant relative to NIP (Supplementary Table S1). These findings demonstrate that OsSAP5 positively regulates heading date and negatively regulates seed setting under NSD condition.
OsSAP5 influences florigen Hd3a and RFT1 expression
Under the chamber conditions, the ossap5-CRISPR delayed heading date about 7 days and 9 days compared to NIP under control SD (CSD) and control LD (CLD) conditions, respectively (Figures 3A, B). However, the OsSAP5-overexpression lines showed no significant difference in heading date (Figures 3A, B). To investigate the effect of OsSAP5 on the flowering-related genes, the expression of two key pathway genes (GI, Ghd7, Ehd1, Hd1, Hd3a, RFT1) related to heading date and FAC member genes (OsGF14b, OsGF14c, OsFD1) was examined in NIP, ossap5–2 and OsSAP5-OE#1. The expression of these genes did not change significantly, but florigen genes Hd3a and RFT1 were obviously decreased in ossap5–2 under CSD condition, with no difference in OsSAP5-OE#1 (Figures 3C, E; Supplementary Figure S2). The expression of Hd3a and RFT1 was too low to be detected at each ZT (Zeitgeber time) points under CLD condition, so we examined changes in the expression of Hd3a and RFT1 at two points (bright and dark). The results showed the expression of Hd3a and RFT1 in ossap5 was significantly lower than that in NIP at bright and dark points, excepting the RFT1 expression had no remarkable changes at the dark point (Figures 3D, F).
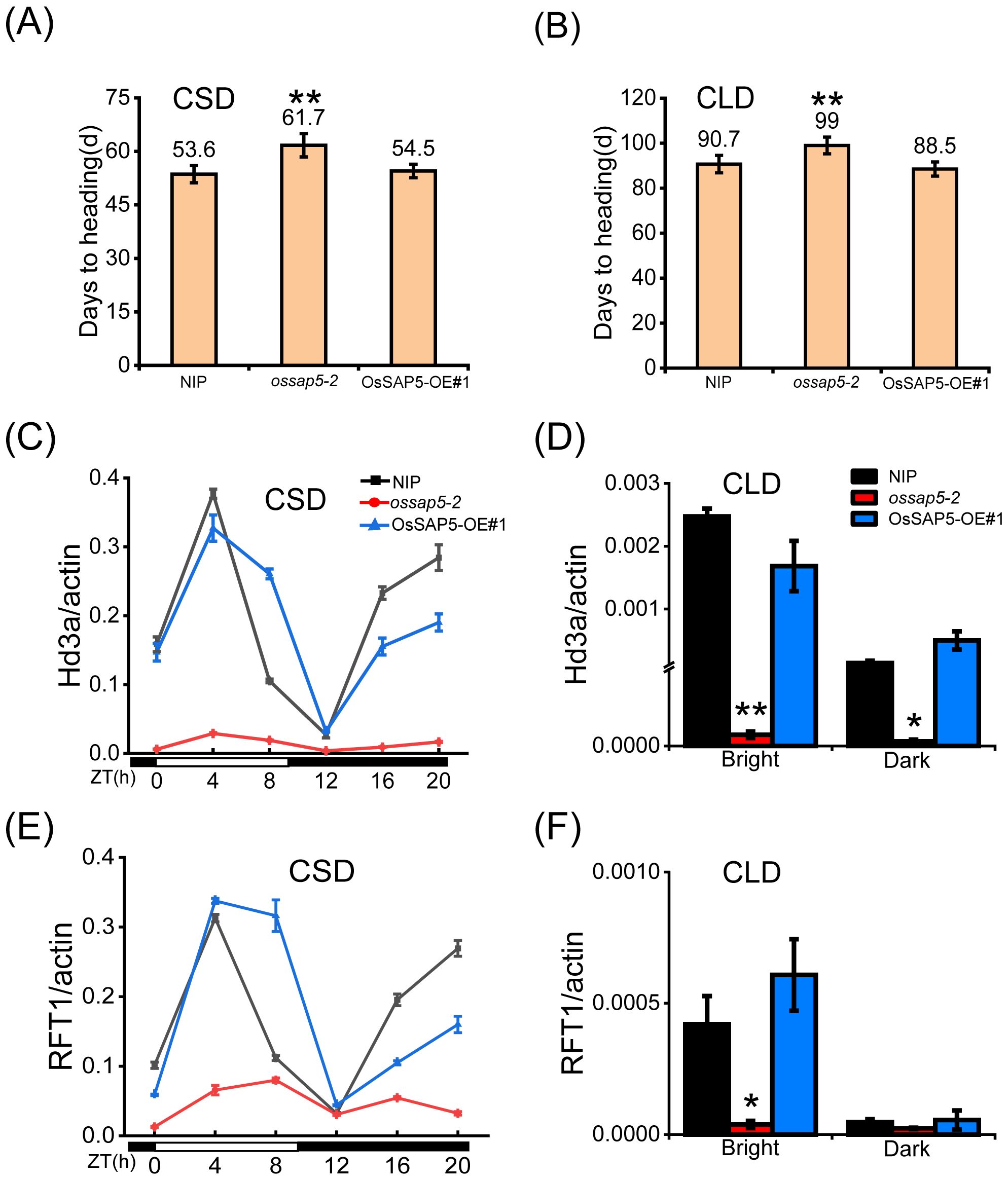
Figure 3. Heading date of OsSAP5 transgenic material under CSD and CLD conditions. (A, B) Days to heading of NIP, ossap5–2 and OsSAP5-OE#1 under CSD and CLD conditions. Values are presented as means ± SD ((n = 15, **P < 0.01; Student’s t-test). CSD means control short day. CLD means control long day. (C, E) Rhythmic expression of Hd3a (C) and RFT1 (E) in NIP, ossap5-1, and OsSAP5-OE#1 under CSD conditions. ZT is the means of Zeitgeber time. (D, F) Expression of Hd3a (D) and RFT1 (F) in NIP, ossap5-1, and OsSAP5-OE#1 under CLD conditions. Values are presented as means ± SD ((n = 3, *P < 0.05; **P < 0.01; Student’s t-test).
The Hd3a and RFT1 are crucial factors for OsFD1 inducing MADS-box (OsMADS14, OsMADS15, OsMADS18 and OsMADS34) genes expression, which regulate the rice heading date (Kobayashi et al., 2012). Therefore, the expressions of MADS genes were checked in the SAM of NIP and OsSAP5 transgenic materials. The results showed that four MADS genes were reduced obviously in ossap5-2, but not significantly changed in OsSAP5-OE#1 under CSD condition (Figures 4A–D). Under CLD condition, the OsMADS14 and OsMADS15 expression did not differ markedly compared with NIP (Figures 4E, F). However, the OsMADS18 and OsMADS34 expression declined significantly in ossap5-2 (Figures 4G, H). The expression change of these genes related to heading was consistent with the late-heading phenotype of ossap5-CRISPR mutants.
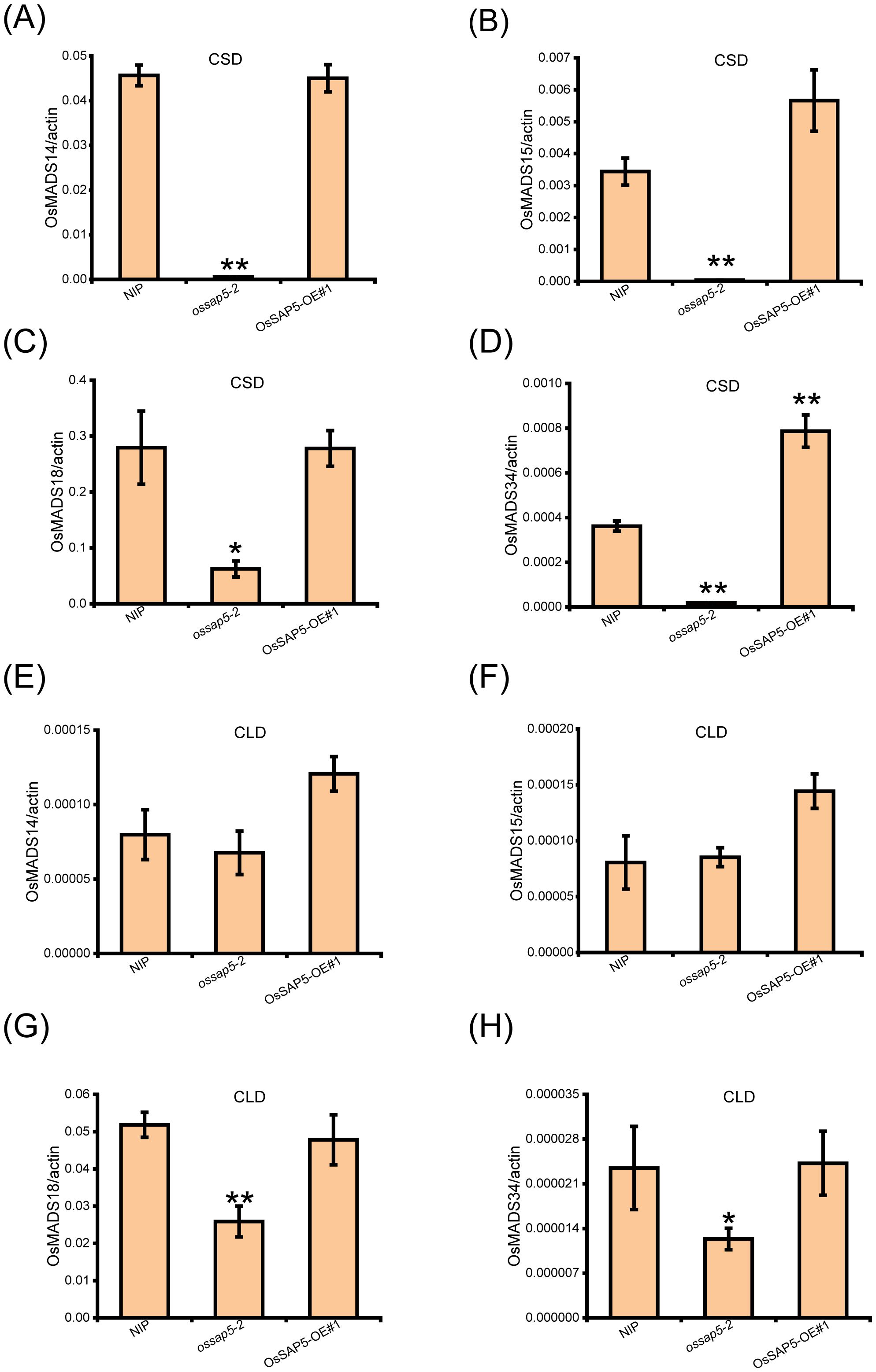
Figure 4. Expression analysis of OsMADS genes in the SAM of transgenic plants of OsSAP5. (A–D) Expression levels of OsMADS14 (A), OsMADS15 (B), OsMADS18 (C), and OsMADS34 (D) in NIP, ossap5-2, OsSAP5-OE#1 under CSD condition. Values are presented as means ± SD (n = 3, *P < 0.05; **P < 0.01; Student’s t-test). (E–H). Expression levels of OsMADS14 (E), OsMADS15 (F), OsMADS18 (G), and OsMADS34 (H) in NIP, ossap5-2, OsSAP5-OE#1 under CLD condition. Values are presented as means ± SD (n = 3, *P < 0.05; **P < 0.01; Student’s t-test). Samples are prepared from SAM of 15 different plants of every transgenic line.
OsSAP5 expression pattern and subcellular localization
To further understand the biological function of OsSAP5, we detected the spatiotemporal expression pattern of OsSAP5 in different tissues. Expression analysis showed that OsSAP5 exhibited rhythmic expression patterns under both CSD and CLD conditions (Figure 5A). OsSAP5 was expressed in the leaf blade, leaf sheath and SAM of 30-day seedling (Figure 5C). Histochemical assays showed that the GUS activity was detected in the same tissue with qRT-PCR (Figure 5D). GUS staining assay also showed that OsSAP5 was expressed in the spikelet with different sizes (Figure 5B). A transgenic material containing pOsSAP5::OsSAP5-EGFP was constructed for checking the expression pattern of OsSAP5 as well. Fluorescence microscope observation assays showed that OsSAP5 expressed in the phloem and xylem cells of vessel of the leaf sheath (Supplementary Figure S3). We co-expressed OsSAP5-EGFP with membrane-labeled AtPIPT2A-mCherry and nucleus-labeled H2B-mCherry in tobacco leaves. The results showed that the green overlapped with the red fluorescence in nucleus of tobacco cells, and not in the plasma membrane of tobacco cells (Figure 5E). It was speculated that OsSAP5 was localized in the cytoplasm and nucleus.
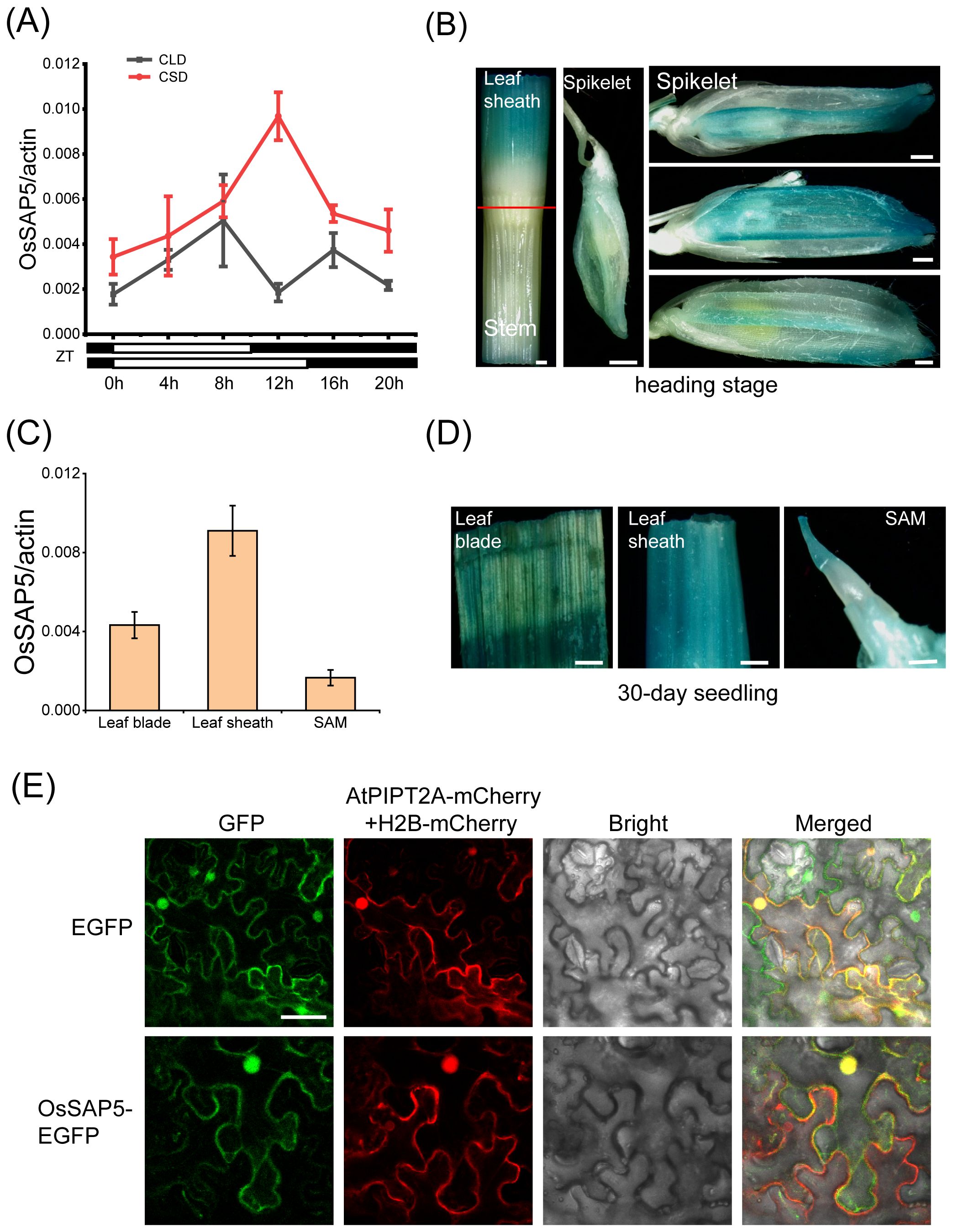
Figure 5. Expression pattern analysis of OsSAP5. (A) qRT-PCR analysis of the rhythmic expression of OsSAP5. Values are presented as means ± SD (n = 3). (B) Gus histochemical staining of pOsSAP5::gus transgenic lines in heading stage. Bar = 500 μm. (C) Expression pattern analysis of OsSAP5 in different tissues of NIP. Values are presented as means ± SD (n = 3). (D) Gus histochemical staining of 30-day-old pOsSAP5::gus transgenic lines in leaf blade, leaf sheath, and shoot apical meristem (SAM). Bar = 300 μm. (E) Subcellular localization of the OsSAP5-EGFP fusion protein in tobacco leaf. AtPIPT2A-mCherry is used as a cytomembrane marker. H2B-mCherry is used as a nuclear marker. Free EGFP is used as the control. Bar = 50 μm.
OsSAP5 interacts with OsGF14c
To study the molecular mechanism of OsSAP5-mediated heading date, we performed a coimmunoprecipitation assay followed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). The candidate protein, OsGF14c (LOC_Os08g33370), showed late-heading phenotype in its overexpression lines under SD condition (Purwestri et al., 2009; Supplementary Table S2). The pull-down assay showed that GST-OsGF14c was bound to His-OsSAP5, while no binding was detected between GST alone and His-OsSAP5 (Figure 6A). In addition, Co-IP assay indicated that OsGF14c interacted with OsSAP5 in the rice protoplasts (Figure 6B). The LCI assay also showed that co-transformation with OsGF14c-nLuc and cLuc-OsSAP5 displayed a considerable interaction signal in tobacco cells, but the controls did not (Figure 6C). The BiFC assay confirmed again that only the co-expression of OsGF14c and OsSAP5 could detect the YFP fluorescence signal in tobacco cells (Figure 6D). OsGF14c was localized in the cytoplasm (Supplementary Figure S4). The co-expression of OsGF14c-mCherry and OsSAP5-EGFP assay showed that the interaction of OsGF14c and OsSAP5 was only in the cytoplasm (Figure 6E), and the results were consistent with the localization of OsGF14c.
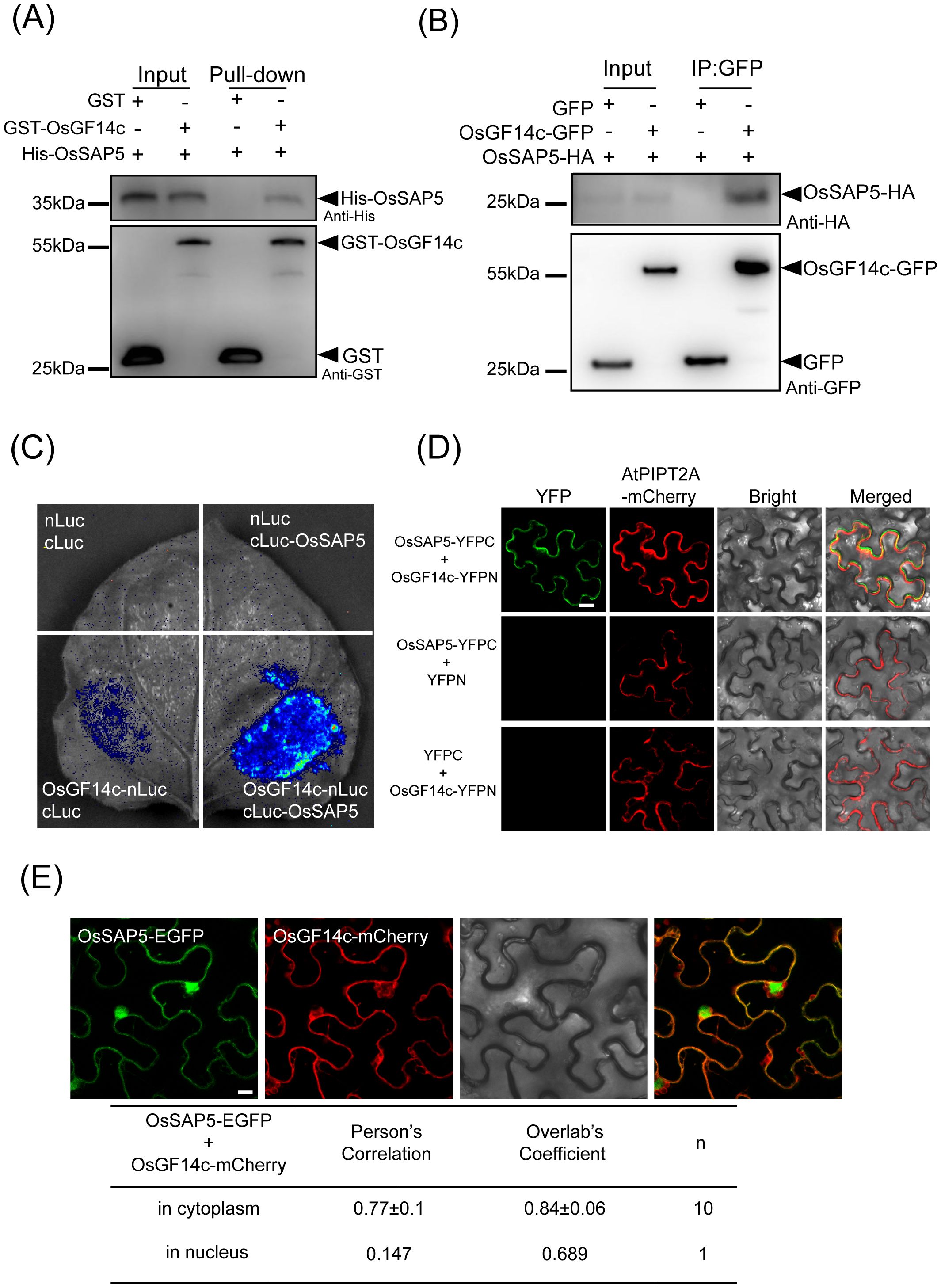
Figure 6. OsSAP5 interacts with OsGF14c. (A) An in vitro pull-down assay validates the interaction between OsSAP5 and OsGF14c. Induced His-OsSAP5 is incubated with the GST-OsGF14c or GST protein. Protein samples are immunoprecipitated with anti-GST beads and immunoblotted with anti-His and anti-GST antibodies. The symbols “-” and “+” represent the absence and presence of the corresponding proteins. (B) An in vivo Co-IP assay validates the interaction between OsSAP5 and OsGF14c in rice protoplasts. Proteins are isolated from protoplasts co-transfecting p35s::OsGF14c-GFP or p35s::GFP with p35s::OsSAP5-HA. Co-immunoprecipitation is performed using anti-GFP beads and immunoblotted with anti-HA and anti-GFP antibodies. (C) LCI assay verifies that OsSAP5 interacts with OsGF14c in the leaf epidermal cells of N. benthamiana. (D) BiFC assay shows that OsSAP5 interacts with OsGF14c in the leaf epidermal cells of N. benthamiana (n = 4 plants). Bar = 20 μm. (E) OsSAP5-GFP and OsGF14c-mCherry are co-localized in the leaf epidermal cells of N. benthamiana. Taking a statistic of fluorescence of GFP and mCherry in the cytoplasm (n = 10) and nucleus (n = 1) by Leica confocal microscopy. Bar = 10 μm.
The 14-3–3 family proteins OsGF14b, OsGF14d, and OsGF14e were the homologous proteins of OsGF14c that regulated the heading date of rice (Taoka et al., 2011). The pull-down assay confirmed that OsSAP5 also interacts with OsGF14b, OsGF14d, and OsGF14e (Supplementary Figure S5). Taken together, OsSAP5 interacts with OsGF14c in vivo and in vitro.
OsSAP5 has E3 ubiquitin ligase activity and may promote OsGF14c degradation
OsSAP5 contains two zinc-finger domains, A20 and AN1. A20 and AN1 were reported to have the E3 ubiquitin ligase activity (Heyninck et al., 1999). Therefore, the purified His-OsSAP5 protein was incubated with E1, E2, and ubiquitin-HA (Ub-HA) proteins in an in vitro ubiquitination assay, and the results were analyzed by immunoblotting using anti-HA antibodies. A series of molecular masses was detected only when all reaction components were added, whereas lacking any one of the components E1, E2 Ub-HA, or His-OsSAP5 failed to produce a positive result (Figure 7A), which indicated that OsSAP5 was an E3 ubiquitin ligase. The co-transformation of OsSAP5-HA, OsGF14c-GFP, and GFP-HA or OsGF14c-GFP and GFP-HA into rice protoplast was carried out, and the results showed that OsGF14c-GFP was more degraded and ubiquitinated when OsSAP5-HA was expressed together (Figure 7B). Substrate ubiquitination modification had different polyubiquitin chains, and lysine-48 (K48)-linked chains were the signal for 26S proteasomes (Al-Saharin et al., 2022). Thus, the OsGF14c level in the presence or absence of MG132, a chemical inhibitor of the 26S proteasome, was examined. Immunoblot analysis showed that OsGF14c abundance decreased slowly in the presence of MG132 but decreased quickly in the absence of MG132 (Figure 7C). Collectively, we confirmed that OsSAP5 is an E3 ubiquitin ligase and may promote the ubiquitination of OsGF14c.
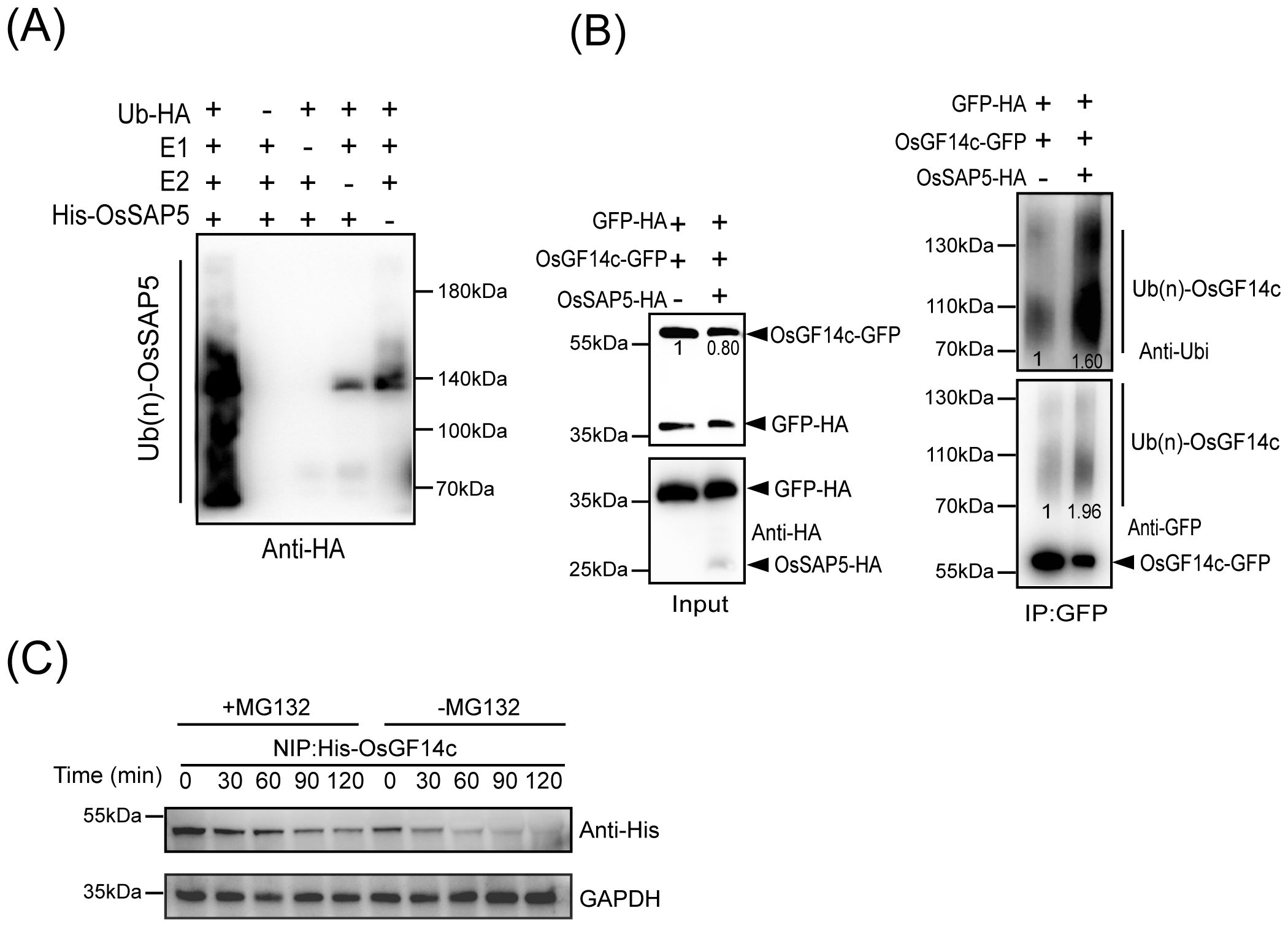
Figure 7. OsSAP5 ubiquitinates OsGF14c. (A) OsSAP5 is an E3 ubiquitin ligase. E1 (Ub-activating enzyme1, human), E2 (UbcH5b), HA-tagged Ub protein (Ub-HA), and His-OsSAP5 are added to reactions in the presence of ATP. Ubiquitin-bound protein of OsSAP5 is detected by immunoblotting with anti-Ub antibodies. His-OsSAP5 is 35.83 kDa. (B) OsSAP5 promotes the ubiquitination of OsGF14c in vivo. Proteins are isolated from protoplasts co-transfected with p35s::OsGF14c-GFP or p35s::GFP-HA with p35s::OsSAP5-HA. Co-immunoprecipitation is performed using anti-GFP beads and immunoblotted with anti-HA, anti-ubi, and anti-GFP antibodies. In the input results, the ratio of immunoblot values of OsGF14c-GFP and GFP-HA was set as 1. In co-immunoprecipitation results, the band of left was set as 1. (C) OsGF14c is degraded through the 26S proteasome pathway. Purified His-OsGF14c is incubated with equal amounts of total proteins extracted from NIP plant leaf at the indicated times at 25°C. +MG132 means 300 μm MG132 is added to NIP samples to examine protein degradation through the 26S proteasome pathway. –MG132 means DMSO is added to NIP samples. His-OsGF14c is immunoblotted with anti-His antibodies. Loading of protein is shown by the GAPDH antibodies.
OsGF14c negatively regulates heading date by reducing the Hd3a and RFT1 expression
OsGF14c-overexpression lines were later than NIP in heading date (Purwestri et al., 2009). However, the detailed reason is not understood. The two CRISPR mutants, osgf14c-1 and osgf14c-2, were constructed by the CRISPR/Cas9 genome editing approach. The osgf14c-1 showed the deletion of 55 bases in OsGF14c coding sequence, and the osgf14c-2 lost one base and led to a premature stop codon (Supplementary Figure S6D). Two CRISPR-mutants did not have extremely different changes in heading date compared to NIP (Supplementary Figures S6A–C). The qRT-PCR and immunoblotting with anti-MYC assays showed that the two overexpressing lines of OsGF14c (OsGF14c-OE#2 and OsGF14c-OE#4) were constructed successfully (Supplementary Figures S7A, B). OsGF14c-OE#2 and OsGF14c-OE#4 showed the delayed heading date for about 11 days and 6 days compared to NIP under CSD condition (Figure 8A). The expression of Hd3a and RFT1 declined extremely in the OsGF14c-OE#2 than in NIP under bright and dark conditions (Figures 8B, C). OsGF14c-OE#4 also showed a significant decrease of Hd3a and RFT1 expression compared to NIP, except the Hd3a expression under dark condition (Figures 8B, C). These results were consistent with the results of ossap5-CRISPR mutants influencing the heading date by reducing Hd3a and RFT1 expression.
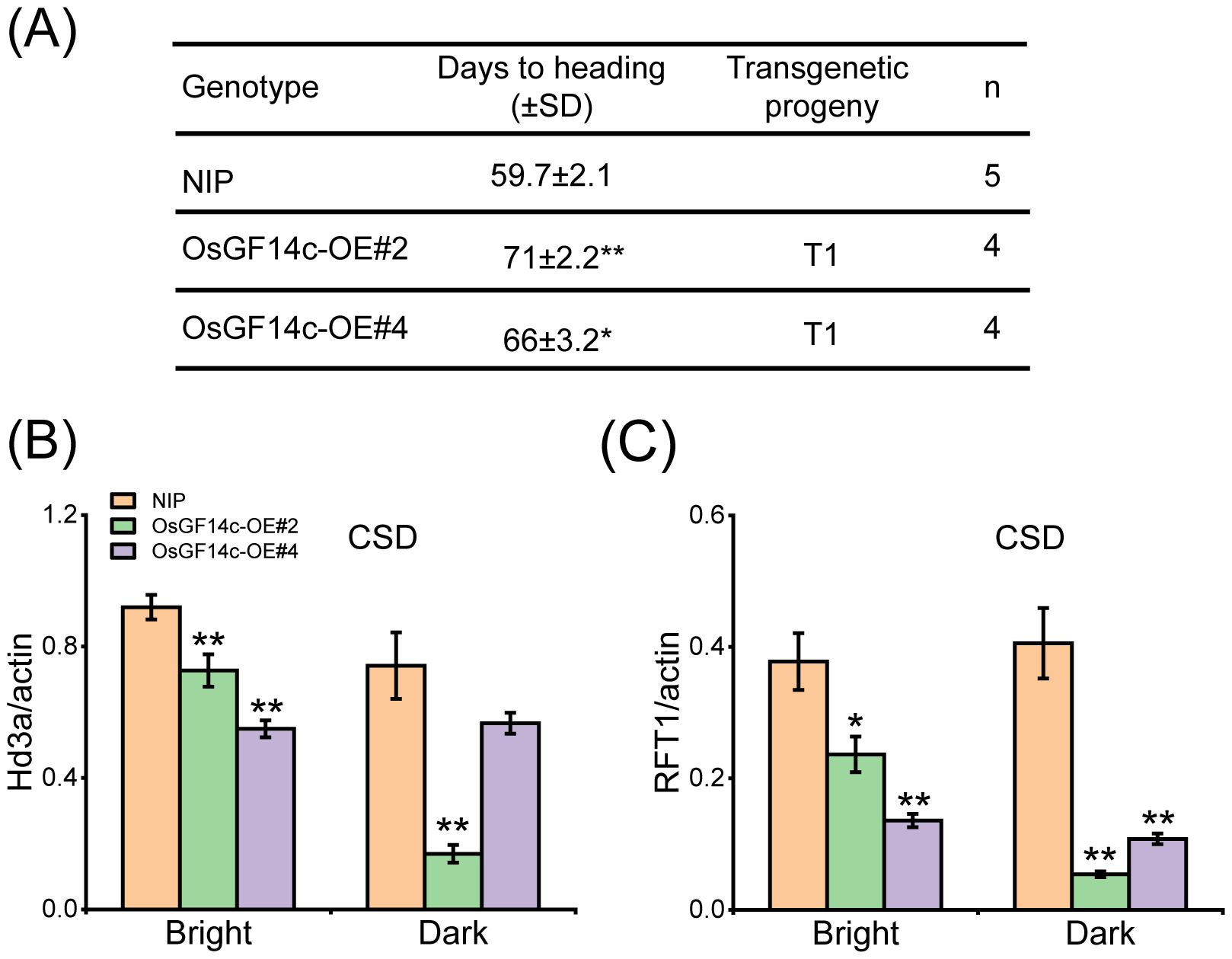
Figure 8. Heading date of OsGF14c overexpression transgenic material under CSD condition. (A) Days to heading of NIP, OsGF14c-OE#2, and OsGF14c-OE#4 under CSD condition. Values are presented as means ± SD (*P < 0.05; **P < 0.01; Student’s t-test). (B, C) The expression of Hd3a (B) and RFT1 (C) in NIP, OsGF14c-OE#2, and OsGF14c-OE#4 under CSD condition. Values are presented as means ± SD (n = 3, *P < 0.05; **P < 0.01; Student’s t-test).
Discussion
OsSAP5 positively regulates heading date in rice
This study firstly reported that SAP family protein was involved in the reproductive growth of rice. In Arabidopsis, only AtSAP9 was reported to have the function in flowering time (Kang et al., 2017). In this study, the ossap5-CRISPR mutants delayed the heading date under NSD, CSD, and CLD conditions (Figures 1A, B, 3A, B). However, the mutants had no changes in heading date under NLD, the reason of that may be that OsSAP5 was associated with temperature in the wild field. SAP family proteins were also involved in biotic and abiotic stresses (Mukhopadhyay et al., 2004; Kang et al., 2011, 2017). The ossap5-CRISPR mutants showed weak tolerance to heat stress in the seedling, but the molecular mechanism needed to be further explored.
A cotton protein, GaZnF, contains an A20 domain and had been reported to bind to a MYB-box element present in the GUSP1 (cotton stress responsive gene) promoter, suggesting a possible transcription regulator activity of SAP protein (Zahur et al., 2012). Thus, OsSAP5 transcriptional activity in rice protoplasts was performed by dual luciferase reporter (DLR) system, and VP16 was used as the transcriptional activator (Supplementary Figure S8). OsSAP5 expression resulted in a similar firefly luciferase/Renilla luciferase (LUC/REN) activity as did the empty GAL4BA vector (Supplementary Figure S8). The results indicated that OsSAP5 did not have transcriptional activity. OsSAP5 could not regulate the florigen Hd3a and RFT1 expression directly either, although Hd3a and RFT1 showed a remarkable decline in ossap5-CRISPR mutants (Figures 3C–F).
OsGF14c overexpression has a delay in heading date
Overexpression of Hd3a or OsFD1 promoted heading date in rice (Kojima et al., 2002; Peng et al., 2021); however, OsGF14c-overexpressed plants delayed heading date by approximately 15 days under SD condition (Purwestri et al., 2009). This phenotype of genetic material shows that FAC components may be balanced for FAC activity. The expressions of OsMADS genes could be induced by effector Hd3a and FD1 together in the rice protoplast system (Cai et al., 2021). OsGF14c had a higher transcription level than Hd3a in rice tissue (Supplementary Figure S9A). Therefore, the effector of p35s::Hd3a, pGF14c::OsFD1, and pHd3a::OsGF14c was compared with those of p35s::Hd3a, pGF14c::OsFD1, and pGF14c::OsGF14c in rice protoplasts. The results revealed no significant changes in OsMADS expression in response to altered OsGF14c protein levels (Supplementary Figure S9B). It may be identified that FAC components do not have a balance for each other.
FTL12 interacted with OsGF14b and OsFD1 to form the florigen repression complex (FRC) by competing with Hd3a for binding OsGF14b (Zheng et al., 2023), but Hd3a could not compete with FTL12 for binding OsGF14b (Zheng et al., 2023). Thus, OsGF14c was more easily interacted with FTL12 to form more FRC than FAC. This may be another possible reason for the OsGF14c-overexpressed lines prohibiting heading date. OsFTL4 was also reported to compete with Hd3a interacting with 14-3–3 and negatively regulates the heading date (Gu et al., 2022). HBF1/HBF2, two paralogs with OsFD1, are mainly expressed in rice leaves (Brambilla et al., 2017). HBFs influenced the Hd3a/RFT1 expression by regulating Ehd1 directly and also interacted with Hd3a directly, with RFT1 by OsGF14c as the bridge (Brambilla et al., 2017). In our study, Hd3a and RFT1 expression was lower in OsGF14c-overexpression than NIP under CSD condition (Figures 8B, C). Thus, HBFs may be another reason to explain why OsGF14c overexpression has a late-heading phenotype. How the OsGF14c-overexpression lines play in heading under long day conditions needs further study.
OsSAP5 has E3 ubiquitin activity and interacts with OsGF14c
OsSAP5 had E3 ubiquitin ligase activity in our study (Figure 7A). A lot of SAP proteins have been reported to have E3 ubiquitin ligase activity in wheat and Arabidopsis, such as TaSAP5, AtSAP5, AtSAP9, and so on (Choi et al., 2012; Kang et al., 2017; Zhang et al., 2017).
We got the OsGF14c protein by IP-MS and confirmed that OsSAP5 interacts with OsGF14c (Figure 6). OsGF14c-overexpression plants and ossap5-CRISPR mutants showed a similar heading phenotype in rice under short day condition (Figures 1B, 8A). OsGF14c was degraded by UPS system (Figure 7C). OsSAP5 may promote the ubiquitination of OsGF14c (Figure 7B). Thus, OsSAP5 may positively regulate heading date by promoting the degradation of OsGF14c ubiquitination in rice. The OsGF14c-CRISPR mutants showed a comparable phenotype in heading date compared to NIP. The reason may be that OsGF14b, OsGF14d, and OsGF14e have functional redundancy in regulating the heading date. That might also be the reason why OsSAP5-overexpression lines showed the similar heading date with NIP, as 14-3–3 protein could not be degraded by OsSAP5 completely.
Conclusion
The study first demonstrates that ossap5-CRISPR mutants delay the heading date by reducing the Hd3a and RFT1 expression. Similarly, the OsGF14c-overexpression displayed comparable phenotypic characteristics with delayed heading and downregulation of Hd3a/RFT1 expression. Further molecular characterization revealed that OsSAP5 functions as an E3 ubiquitin ligase that may facilitate the ubiquitination of OsGF14c. Thus, OsSAP5 may mediate the proteasomal degradation of OsGF14c through the 26S proteasome pathway. A schematic model summarizing the regulatory relationship between OsSAP5 and OsGF14c in controlling rice heading date is presented in Figure 9.
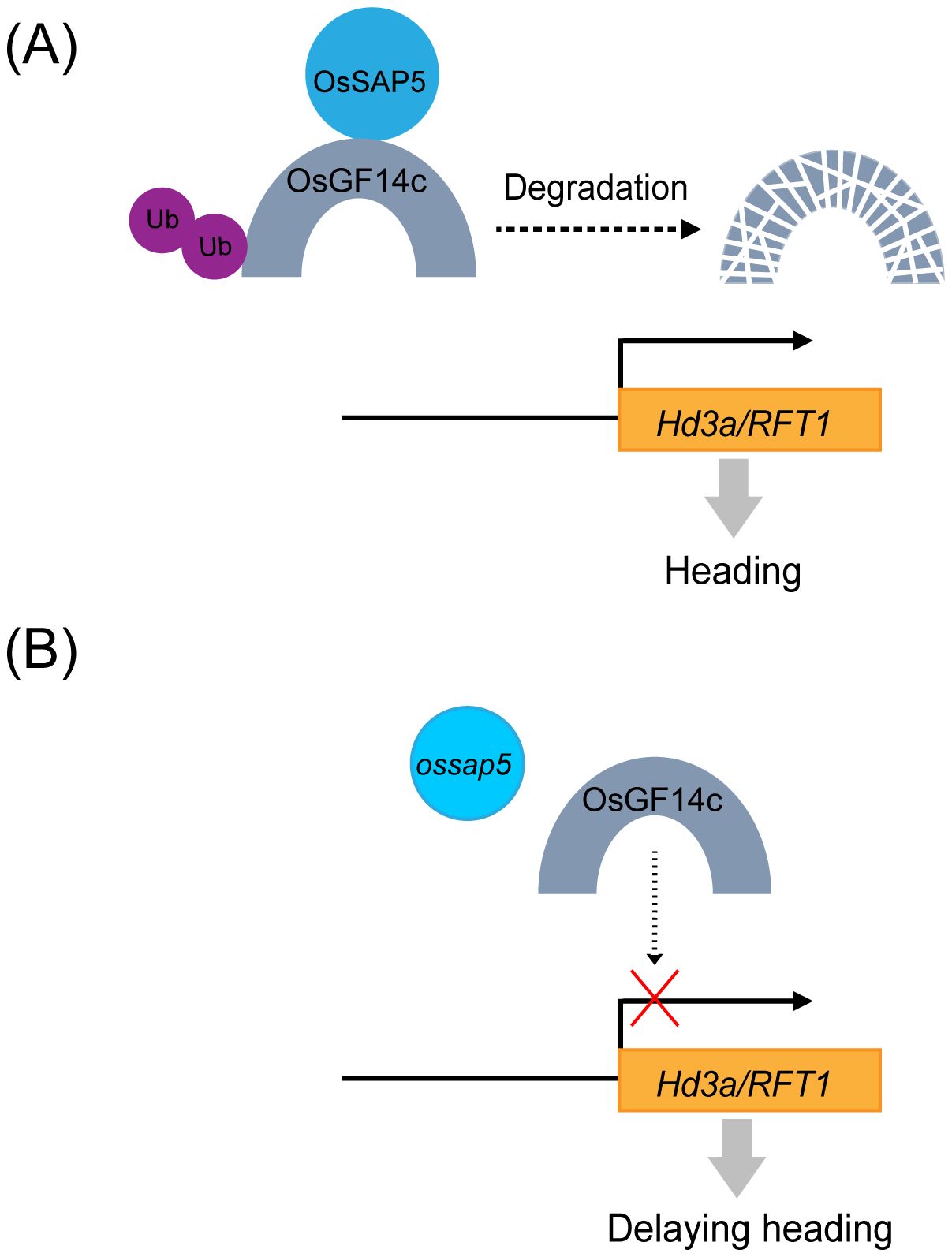
Figure 9. A proposed working model for the regulation of rice heading by OsSAP5. (A) In wild type, OsSAP5 may promote OsGF14c ubiquitination and OsGF14c may be degraded by 26S proteasome, and Hd3a/RFT1 is normally expressed. (B) In the ossap5 mutant, more OsGF14c decrease the expression of Hd3a/RFT1, which then delays heading date. UPS, ubiquitin/26S proteasome system.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
XZ: Data curation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. MC: Resources, Writing – review & editing. ZD: Data curation, Visualization, Writing – review & editing. YL: Investigation, Writing – review & editing. QM: Investigation, Writing – review & editing. FB: Writing – review & editing. JL: Writing – review & editing. JH: Writing – review & editing. YG: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by the Hainan Provincial Natural Science Foundation of China (322CXTD522), the Natural Science Foundation of Zhejiang Province (LQ24C130003), the National Natural Science Foundation of China (32072127), the PhD Scientific Research and Innovation Foundation of Sanya Yazhou Bay Science and Technology City (HSPHDSRF-2023-04-017) and Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP).
Acknowledgments
Thanks to Professor Yuanwen Teng and Jie Dong from Zhejiang University for providing the BiFC and LCI assay vector. Thanks to Professor Xuewei Chen from Sichuan Agricultural University for providing the Co-IP assay vector. Thanks to Professor Jiangfeng Huang from Guangxi University for providing the marker vector. Thanks to Yu Fang, Jun Xiong, and Hui Shi from Sichuan Agricultural University and to Qi Li from Zhejiang University for their assistance in the molecular assays.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fpls.2025.1712287.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1589989/full#supplementary-material
Supplementary Figure 1 | Electrophoresis diagram of HYG in OsSAP5 overexpression lines. The positive band is 481 bp; the primer is HYG-F/HYG-R.
Supplementary Figure 2 | Rhythmic expression of heading-related genes in NIP, ossap5-2, and OsSAP5-OE#1 under CSD condition. RNA is extracted from 15-day seedling. ZT is the means of Zeitgeber time.
Supplementary Figure 3 | Microscope observation of pOsSAP5::OsSAP5-EGFP. Microscope images of pOsSAP5::OsSAP5-EGFP transgenic plants. Bar = 50 μm. GFP fluorescence observed in the cell of the phloem (Phl) and xylem (Xy).
Supplementary Figure 4 | Subcellular localization of the OsGF14c protein. Subcellular localization of the OsGF14c-EGFP in tobacco leaf. AtPIPT2A-mCherry is used as a plasma membrane marker. H2B-mCherry is used as a nuclear marker. The free EGFP is used as the control. Bar = 50 μm.
Supplementary Figure 5 | OsSAP5 interacts with OsGF14b, OsGF14d, and OsGF14e. An in vitro pull-down assay validates the interaction between OsSAP5 and OsGF14b, d, e. The induced His-OsSAP5 is incubated with the GST-OsGF14b or GST-OsGF14d or GST-OsGF14e or GST protein. Protein samples are immunoprecipitated with anti-GST beads and immunoblotted with anti-His and anti-GST antibodies. The symbols “-” and “+” represent the absence and presence of the corresponding proteins.
Supplementary Figure 6 | Heading date of OsGF14c mutants. (A) Heading phenotype of NIP and OsGF14c mutants under NSD condition. Bar = 10 cm; osgf14c-1 and osgf14c-2 represent two OsGF14c mutants. The rectangular box indicates rice heading. (B) Days to heading of NIP, OsGF14c mutants under NSD and NLD conditions. Values are presented as means ± SD (n = 15). (C) Days to heading of NIP, OsGF14c mutants under CSD and CLD conditions. Values are presented as means ± SD (n = 13). (D) Generation and identification of two OsGF14c mutants generated by CRISPR/Cas9 genome-editing approach. The target 1 site and protospacer adjacent motif (PAM) are indicated. The black dash indicates the deletion of a base pair.
Supplementary Figure 7 | Construction of OsGF14c overexpression lines. (A) MYC-tag immunoprecipitate of OsGF14c overexpression plants. A total of 10 protein samples are detected with anti-MYC antibodies with loading shown by Ponceau S staining for RUBISCO. (B) The qRT-PCR analysis of the expression levels of OsGF14c in NIP and OsGF14c overexpression lines. Values are presented as means ± SD (n = 3, *P < 0.05; **P < 0.01; Student’s t-test). OsGF14c-OE#2 and OsGF14c-OE#4 indicate two OsGF14c overexpression lines.
Supplementary Figure 8 | Transcriptional activity analysis of OsSAP5 using the DLR (dual-luciferase reporter) assay system. (A) Schematic representation of reporter and effectors. (B) Measurement of relative luciferase activity (LUC/REN). Values are presented as means ± SD (n = 3).
Supplementary Figure 9 | The protein level of OsGF14c does not influence the function of FAC. (A) Expression pattern of Hd3a and OsGF14c in leaf blade, leaf sheath, and SAM. (B) The levels of OsMADS14, OsMADS15, and OsMADS18 are detected by qRT-PCR. Values are presented as means ± SD (n = 3). The different protein levels of OsGF14c are supported by pOsGF14c::OsGF14c and pHd3a::OsGF14c in rice protoplasts.
Supplementary Table 1 | Agronomic traits of NIP, OsSAP5 mutants, and overexpression lines under NSD condition.
References
Al-Saharin, R., Hellmann, H., and Mooney, S. (2022). Plant E3 ligases and their role in abiotic stress response. Cells 11, 890. doi: 10.3390/cells11050890
Brambilla, V., Martignago, D., Goretti, D., Cerise, M., Somssich, M., de Rosa, M., et al. (2017). Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell. 29, 2801–2816. doi: 10.1105/tpc.17.00645
Cai, M., Zhu, S., Wu, M., Zheng, X., Wang, J., Zhou, L., et al. (2021). DHD4, a CONSTANS-like family transcription factor, delays heading date by affecting the formation of the FAC complex in rice. Mol. Plant 14, 330–343. doi: 10.1016/j.molp.2020.11.013
Chen, M., Fu, Y., Mou, Q., An, J., Zhu, X., Ahmed, T., et al. (2021a). Spermidine induces expression of stress associated proteins (SAPs) genes and protects rice seed from heat stress-induced damage during grain-filling. Antioxid (Basel) 10, 1544. doi: 10.3390/antiox10101544
Chen, Y., Shen, J., Zhang, L., Qi, H., Yang, L., Wang, H., et al. (2021b). Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol. Plant 14, 1297–1311. doi: 10.1016/j.molp.2021.05.001
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Choi, H., Han, S., Shin, D., and Lee, S. (2012). Polyubiquitin recognition by AtSAP5, an A20-type zinc finger containing protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 419, 436–440. doi: 10.1016/j.bbrc.2012.02.044
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Feo, S., Arcuri, D., Piddini, E., Passantino, R., and Giallongo, A. (2000). ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473, 47–52. doi: 10.1016/s0014-5793(00)01494-0
Gu, H., Zhang, K., Chen, J., Gull, S., Chen, C., Hou, Y., et al. (2022). OsFTL4, an FT-like gene, regulates flowering time and drought tolerance in rice (Oryza sativa L.). Rice 15, 47. doi: 10.1186/s12284-022-00593-1
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Heyninck, K., De Valck, D., Vanden Berghe, W., Van Criekinge, W., Contreras, R., Fiers, W., et al. (1999). The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J. Cell Biol. 145, 1471–1482. doi: 10.1083/jcb.145.7.1471
Itoh, H., Nonoue, Y., Yano, M., and Izawa, T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42, 635–638. doi: 10.1038/ng.606
Kang, M., Fokar, M., Abdelmageed, H., and Allen, R. D. (2011). Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 75, 451–466. doi: 10.1007/s11103-011-9748-2
Kang, M., Lee, S., Abdelmageed, H., Reichert, A., Lee, H. K., Fokar, M., et al. (2017). Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 40, 702–716. doi: 10.1111/pce.12892
Kobayashi, K., Yasuno, N., Sato, Y., Yoda, M., Yamazaki, R., Kimizu, M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 24, 1848–1859. doi: 10.1105/tpc.112.097105
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., and Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–5340. doi: 10.1242/dev.040170
Lin, C., Huang, Q., Liu, Z., Brown, S. E., Chen, Q., Li, Y., et al. (2023). AoSAP8-P encoding A20 and/or AN1 type zinc finger protein in asparagus officinalis L. Improving stress tolerance in transgenic Nicotiana sylvestris. Gene 862, 147284. doi: 10.1016/j.gene.2023.147284
Linden, K. J. and Callis, J. (2020). The ubiquitin system affects agronomic plant traits. J. Biol. Chem. 295, 13940–13955. doi: 10.1074/jbc.REV120.011303
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001
Mazzucotelli, E., Belloni, S., Marone, D., De Leonardis, A., Guerra, D., Di Fonzo, N., et al. (2006). The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genomics 7, 509–522. doi: 10.2174/138920206779315728
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U. S. A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Peng, Q., Zhu, C., Liu, T., Zhang, S., Feng, S., and Wu, C. (2021). Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice. Mol. Plant 14, 1135–1148. doi: 10.1016/j.molp.2021.04.003
Purwestri, Y. A., Ogaki, Y., Tamaki, S., Tsuji, H., and Shimamoto, K. (2009). The 14-3–3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50, 429–438. doi: 10.1093/pcp/pcp012
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 20, 1693–1707. doi: 10.1105/tpc.107.057380
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872. doi: 10.1016/j.cell.2009.03.038
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3–3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Vij, S. and Tyagi, A. K. (2006). Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genomics 276, 565–575. doi: 10.1007/s00438-006-0165-1
Wang, Y., Lu, Y., Guo, Z., Ding, Y., and Ding, C. (2020). RICE CENTRORADIALIS 1, a TFL1-like gene, responses to drought stress and regulates rice flowering transition. Rice 13, 70. doi: 10.1186/s12284-020-00430-3
Wang, X., Wei, J., Wu, J., Shi, B., Wang, P., Alabd, A., et al. (2023). Transcription factors BZR2/MYC2 modulate brassinosteroid and jasmonic acid crosstalk during pear dormancy. Plant Physiol. 194, 1794–1814. doi: 10.1093/plphys/kiad633
Wang, J., Yu, H., Xiong, G., Lu, Z., Jiao, Y., Meng, X., et al. (2017). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell. 29, 697–707. doi: 10.1105/tpc.16.00879
Xie, X., Ma, X., Zhu, Q., Zeng, D., Li, G., and Liu, Y. G. (2017). CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10, 1246–1249. doi: 10.1016/j.molp.2017.06.004
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Zahur, M., Maqbool, A., Irfan, M., Jamal, A., Shahid, N., Aftab, B., et al. (2012). Identification and characterization of a novel gene encoding myb-box binding zinc finger protein in Gossypium arboreum. Biol. Plant 56, 641–647. doi: 10.1007/s10535-012-0255-3
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30. doi: 10.1186/1746-4811-7-30
Zhang, N., Yin, Y., Liu, X., Tong, S., Xing, J., Zhang, Y., et al. (2017). The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol. 175, 1878–1892. doi: 10.1104/pp.17.01319
Zheng, R., Meng, X., Hu, Q., Yang, B., Cui, G., Li, Y., et al. (2023). OsFTL12, a member of FT-like family, modulates the heading date and plant architecture by florigen repression complex in rice. Plant Biotechnol. J. 21, 1343–1360. doi: 10.1111/pbi.14020
Keywords: rice (Oryza sativa), OsSAP5, heading date, OsGF14C, E3 ubiquitination ligase
Citation: Zhu X, Chen M, Dong Z, Liu Y, Mu Q, Basit F, Liu J, Hu J and Guan Y (2025) Stress-associated protein OsSAP5 regulates rice heading date through interacting with OsGF14c in rice. Front. Plant Sci. 16:1589989. doi: 10.3389/fpls.2025.1589989
Received: 08 March 2025; Accepted: 31 July 2025;
Published: 09 September 2025; Corrected: 15 October 2025.
Edited by:
Aliki Kapazoglou, Hellenic Agricultural Organization -DEMETER (ELGO-DIMITRA), GreeceReviewed by:
Jianping Liu, Fujian Agriculture and Forestry University, ChinaChanghui Sun, Sichuan Agricultural University, China
Copyright © 2025 Zhu, Chen, Dong, Liu, Mu, Basit, Liu, Hu and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yajing Guan, dmNndWFuQHpqdS5lZHUuY24=
 Xiaobo Zhu1,2
Xiaobo Zhu1,2 Farwa Basit
Farwa Basit Yajing Guan
Yajing Guan