Abstract
Introduction:
Land cultivation is the cornerstone of national food security. However, with the development of non-grain production on cultivated land, China has to use less cultivated land to feed a larger population of the world. To effectively resolve issues caused by non-grain production on cultivated land, Zhejiang Province has initiated efforts to restore non-grain-converted land back to grain cultivation. Whereas, the discovery and application of plant growth-promoting fungi (PGPF) can offer promising solutions to these challenges.
Methods:
PGPF was isolated and identified from soil converted from non-grain lands based on bioassays for plant growth promoting traits, and then their impacts on soil properties and microbial community structure were also investigated.
Results:
In this study, 15 fungal isolates from 108 soil samples were considered as potential PGPF due to their ability to solubilize phosphate (11.91 to 31.65 mm), produce both siderophores (17.09 to 24.66 mm) and indole-3-acetic acid (8.79 to 50.23 μg/mL or 36.72 to 96.50 μg/mL). Results of in vivo assays showed that isolates TL-B31f and FY-R41f could cause a great increase in plant height (15.30% and 13.84%), root length (33.62% and 43.31%), seedling fresh weight (78.58% and 89.77%) and dry weight (9.31% and 28.12%) of rice compared to the control. Based on morphological and molecular analyses, isolates TL-B31f and FY-R41f were identified as Aspergillus tubingensis and Talaromyces veerkampii, respectively. Furthermore, after 55 days of inoculation with the two isolates, the soil content of available phosphate was significantly increased by 42.52% and 48.51%, respectively, compared to the control. In addition, high-throughput sequencing analysis showed that compared with the control, the microbial community composition of the two isolates treatments was reconstructed by increasing or decreasing some specific microbes, while soil properties, such as pH, soil organic matter (SOM), total phosphorus (TP), and available phosphate (AP) might play important roles in modulating rice growth by influencing the composition of microbial communities.
Conclusions:
Overall, our findings highlight the potential of these isolates to be developed into novel biofertilizers for crop growth in non-grain lands.
1 Introduction
Rice (Oryza sativa L.) is the critical staple food for nearly half the world’s population, and global rice consumption is projected to reach about 550 million tons by 2030 (Yuan et al., 2021). Maintaining a steady increase in rice production is vital to ensure social stability and sustainability (Hashim et al., 2024). However, with rapid socio-economic development, dietary patterns in China have partially transformed from rice toward diversified diets with more energy and macronutrients (Yuan et al., 2019). To accommodate this change and obtain more financial benefits, lots of cultivated lands have been used for non-grain production, such as planting fruits, vegetables, bamboo, etc., in recent years (Zhang et al., 2024). Nevertheless, excessive non-grain production on cultivated land not only seriously threatens national grain security, but also triggers a series of problems, such as soil quality and ecosystem degradation due to heavy chemical inputs (Yang and Zhang, 2021; Zhu et al., 2022b; He et al., 2024). To solve these problems, Zhejiang Province has initiated efforts to restore non-grain converted land back to grain cultivation. During this conversion process, soil health is critical in determining agricultural productivity, ecosystem health, and sustainable development.
Among many factors contributing to healthy soil, microbes stand out as an important part of the soil ecosystem by improving soil structure and fertility, driving nutrient cycling, suppressing soil-borne disease, and supporting plant growth (Wang et al., 2023; Chen et al., 2024; Romão et al., 2025). Among the diverse microbes, PGPF have attracted more attention in recent years. Varieties of PGPF belonging to genera Aspergillus, Arbuscular, Fusarium, Penicillium, Phoma, Podospora, and Trichoderma have been widely studied (Hossain and Sultana, 2020; Adedayo and Babalola, 2023). PGPF have been found to be able to promote crop growth and improve soil conditions through mechanisms such as phosphate solubilization (Chuang et al., 2007; Radhakrishnan et al., 2015; Li et al., 2016), organic matter mineralization (Yadav et al., 2009), production of plant growth-promoting hormones and enzymes (Abou Alhamed and Shebany, 2012; Sofo et al., 2012), generation of volatile organic compounds (Naznin et al., 2013; Bitas et al., 2015), and induction of disease resistance (Murali et al., 2013; Murali and Amruthesh, 2015; Naziya et al., 2019). Plant growth promotion processes by PGPF are often complex, and arrays of interconnected mechanisms are involved in these processes, which help PGPF maintain rhizosphere competence and stability in performance (Hossain and Sultana, 2020). Thus, it is important to find novel PGPF that can be used as bioinoculants with multiple traits for improving plant growth and soil fertility in non-grain converted lands.
Besides the direct effects on plant growth, PGPF can also change the physiological features of soil bacteria (Akinola and Babalola, 2020). Bacteria and fungi in soil generally account for >90% of total soil microbes, and soil microbial communities play several important roles in preserving ecosystem function and soil health (Banerjee and van der Heijden, 2023). Previous research has shown that Aspergillus brunneoviolaceus HZ23 can significantly increase the richness of bacteria in the rhizosphere soil of pakchoi in newly reclaimed land (Li et al., 2023). Soil microbial diversity plays an important role in resisting and restoring degraded ecosystems (Pedrinho et al., 2024). Diversity and asynchrony in soil microbial communities stabilize ecosystem functioning (Wagg et al., 2021). However, the response patterns of bacterial and fungal communities to agricultural practices and environmental factors may differ, thus leading to variations in their ecological functions (Zhou et al., 2025). For example, bacteria not only exhibited greater sensitivity to fire disturbance than fungi, but also recovered faster than fungi following one month of burning (Arunrat et al., 2024). Soil bacteria are also more sensitive than fungi to the fertilization practices, while fungi are more active in response to crop conversion from wheat-maize to wheat-soybean rotation (Ai et al., 2018). Converting upland with maize to paddy with rice fields alters soil nitrogen (N) microbial functions at different depths in the black soil region (Li et al., 2024). Yet, soil ecosystem multifunctionality is strongly linked with crop yield and environmental sustainability (Deng et al., 2024). Therefore, it is essential to investigate further how different PGPF affect bacterial and fungal communities of rice in non-grain converted land after inoculation, as well as their respective ecological roles in soil health. However, at present, a limited number of researches are available on PGPF improving the soil conditions of non-grain converted lands and enhancing their transformation back to grain production.
In order to enhance soil fertility of typical non-grain converted lands, this study was carried out to isolate fungal isolates from non-grain converted lands, screen novel PGPF by evaluating their plant growth promoting traits, and identify them based on combined analysis of morphological and molecular data. Furthermore, the potential of the obtained PGPF to be developed into novel microbial control products was determined by investigating their effect on soil properties and microbial community structure of rice in non-grain converted land and clarifying their respective ecological roles in soil health.
2 Methods
2.1 Fungal isolation
One hundred and eight soil samples were collected with a hand auger from a 5–20 cm soil layer around the crown of plants in Jiande (loquat-rice), Chun’an (mulberry-rice), Tonglu (blueberry-rice), Fuyang (grape-rice), Lin’an (bamboo-rice), and Yuhang (seedling-rice), Zhejiang province, China. For each conversed mode, six samples were taken from each field (such as loquat-rice converted mode in Jiande, every six samples were collected from loquat garden, paddy field converted from loquat garden, and perennial paddy field, respectively) using the five-point sampling method (Panagos et al., 2014), and a total of eighteen samples were collected. Then, each sample was packed individually on-site into a sterilized and sealed polythene bag, and transported to the laboratory using a portable cooler for further analysis. Indeed, after dissolving 10 g fresh soil in 90 mL sterile water, mixing well by vortex, and serially diluting with 9 mL of sterile water, 100 μL of 10–3 dilution was inoculated on potato dextrose agar (PDA, potato extract 200 g, dextrose 20 g, agar 20 g, ddH2O 1000 mL) medium and then incubated at 25°C for 3–5 days. After three times of repetitive purification on PDA medium, the colonies with different morphological characteristics and higher growth rate were selected to further purify via single-spore isolation, and then stored in 20% glycerol at -70°C.
2.2 Identification of fungal isolates
The fungal isolates were identified as described before (Samson et al., 2014; Tian et al., 2021), which was carried out by incubating them on PDA, Czapek yeast autolysate agar (CYA, czapek concentrate 10 mL, sucrose 30 g, yeast extract 5 g, K2HPO41 g, CuSO4·5H2O 0.005 g, ZnSO4·7H2O 0.01 g, agar 20 g, ddH2O 1000 mL), and male extract autolysate (MEA, malt extract 50 g, CuSO4·5H2O 0.005 g, ZnSO4·7H2O 0.01 g, agar 20 g, ddH2O 1000 mL) medium for 7 days at 25°C, and observing colony morphology of each isolate. Molecular identification of the fungal isolates was carried out using the ITS, TUB, CaM, and RPB2 combined dataset with the following steps: mycelia of each test isolate were harvested from the surface of PDA, frozen in liquid nitrogen, and extracted for genomic DNA using the Rapid Fungi Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, China).
Primer sets ITS5/ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3’ and 5’-GGA AGT AAA AGT CGT AAC AAG G-3’), Bt2a/Bt2b (5’-GGT AAC CAA ATC GGT GCT GCT TTC-3’ and 5’-ACC CTC AGT GTA GTG ACC CTT GGC-3’), CMD5/CMD6 (5’-CCG AGT ACA AGG ARG CCT TC-3’ and 5’-CCG ATR GAG GTC ATR ACG TGG-3’), and 5F/7CR (5’-GAY GAY MGW GAT CAY TTY GG-3’ and 5’-CCC ATR GCT TGY TTR CCC AT-3’) were used for amplification of rDNA ITS, TUB, CaM, and RPB2 genes, respectively (Hong et al., 2005; Nasri et al., 2015; Tomaha et al., 2020; Haituk et al., 2021). PCR amplification reaction mixture (50 μL) included ddH2O (18 μL), 2 × Hieff® PCR Master Mix (25 μL), 10 μM each primer (2 μL), and DNA (3 μL). Then, PCR was carried out as follow: for ITS gene, 94°C 5 minutes; 94°C 45 s, 52°C 45 s, 72°C 60 s, 35 cycles; 72°C 10 minutes; for TUB and CaM genes, 94°C 5 minutes; 94°C 60 s, 55°C 60 s, 72°C 90 s, 35 cycles; 72°C 10 minutes; for RPB2 gene, 94°C 5 minutes; 94°C 30 s, 51°C 30 s, 72°C 60 s, 5 cycles; 94°C 30 s, 49°C 30 s, 72°C 60 s, 5 cycles; 94°C 30 s, 47°C 30 s, 72°C 60 s, 30 cycles; 72°C 10 minutes.
PCR products were visualized on 1.0% agarose gels, then purified and submitted to Tsingke Biotechnology Co., Ltd. (Hangzhou, China) for sequencing in both directions. Sequences for each region were assembled and edited using DNASTAR Lasergene (v7.0.1) (DNAStar Inc., Madison, USA) and BioEdit (v7.0.9) (North Carolina St. University, Raleigh, USA) (Tippmann, 2004), and then analyzed by BLAST search in the GenBank database. After the reference sequences of closely related species for test strains were downloaded from the GenBank database, maximum likelihood (ML) phylogenetic trees were constructed using Mega 7.0 (Kumar et al., 2016). Bootstrap replicates were performed 1000 times, and bootstrap values above 50% were indicated on the cladogram to indicate the significance of the separation.
2.3 Bioassays for plant growth promoting traits
The ability of the selected isolates to solubilize phosphate was evaluated in National Botanical Research Institute’s phosphate (NBRIP, glucose 10 g, Ca3(PO4)25 g, MgCl25 g, KCl 0.2 g, MgSO4·7H2O 0.25 g, (NH4)2·SO4 0.1 g, agar 25 g, ddH2O 1000 mL, pH 7.0) medium (Abdallah et al., 2019). In brief, a 6-mm agar disc of test fungus was placed on NBRIP medium at 25°C for 3 days, the phosphate solubility was assessed by observing the formation of a clear halo zone surrounding the colony, and measuring the size of the phosphate-solubilizing zone. Each fungal isolate was performed in triplicate.
Fungal isolates with high phosphate solubility (producing a clear halo zone surrounding the colony) were further used to evaluate their siderophore-producing ability, which was carried out using the chrome azurol S (CAS) assay as previously described (Murugappan et al., 2011; Li et al., 2021). In brief, a 6-mm agar disc of the test fungus was placed on CAS-agar medium and incubated at 25°C for 3 days. Colonies of siderophore-producing fungus were surrounded by an orange halo in the CAS plate. Each fungal isolate was performed in triplicate.
Fungal isolates with high phosphate solubility (producing a clear halo zone surrounding the colony) and siderophore-producing (producing an orange halo surrounding the colony) ability were further analyzed for indole acetic acid (IAA) production, which was performed as described before (Li et al., 2021). In brief, a fungal piece (6 mm in diameter) was separately added to a test tube containing 10 mL PDB (PDA without agar) containing 0.1% or 1.0% tryptophan, respectively, while PDB without tryptophan was used as a control. After shaking at 25°C, 150 rpm for 7 days, approximately 2 mL of culture solution was collected and centrifuged at 4°C, 8000 rpm for 5 minutes, respectively. Finally, 1 mL of supernatant for each isolate was taken and separately mixed with 4 mL Salkowski’s reagent (0.5 M FeCl3, 15 mL, H2SO4, 300 mL, ddH2O, 500 mL). After the mixtures were incubated at room temperature for 20 minutes in the dark, the observation of pink–red color changes indicated IAA production. Absorbance was read at 530 nm using a spectrophotometer (Perkin Elmer Lambda35, Waltham, MA, USA), and IAA concentrations were calculated on the basis of the standard curve prepared by standard IAA solutions (0, 10, 20, 30, 40, 50, 100 μg/mL). Each experiment was performed in triplicate.
Plant growth promotion (PGP) activity of the selected fungal isolates was determined by evaluating their effect on growth of rice cv. Xiushui131 (purchased from Jiaxing Academy of Agricultural Sciences, China). In details, after sterilization with 75% alcohol for 1 minutes, rinsing three times with sterile water, and pre-germination on wet sterile filter papers at 25°C for two days, rice seeds were sowed into planting pots (8 cm × 8 cm × 12.5 cm) containing soil previously planted with vegetables, and then put in a greenhouse with a relative humidity of 70% and temperature of 25°C. Two days after sowing, 10 mL conidial suspension of each tested isolate (prepared by scraping the conidial masses on a 7-day-old culture grown on PDA at 25°C to sterile distilled water, and diluting to 106 spores/mL) was poured into the soil surrounding the plants. Plants irrigated with 10 mL sterile water were used as controls. After 55 days of inoculation, the PGP ability was determined by measuring the seedlings height, root length, fresh and dry weight (dried in an oven at 65°C for two days), and the growth promotion efficacy (GPE) was calculated using the formula: GPE% = (treatment – control)/control × 100%. Each treatment consisted of three replicates with four pots (six plants per pot) per replicate (total 72 plants per treatment).
2.4 Impacts of PGPF on soil properties and microbial community structure
At harvest of rice, about 1.0 kg of soil from each treatment was used to detect soil pH, SOM, TP, and AP as previously described. Indeed, after drying at room temperature and passing through a 0.45-mm sieve, soil pH was measured using a pH meter (FE28, Mettle-Toledo, Zurich, Switzerland); SOM content was measured by the potassium dichromate volumetric method; TP was determined by automated colorimetric analysis after persulfate digestion; AP was measured by molybdenum-antimony anti-spectrophotometry following extraction with hydrochloric acid ammonium fluoride (Jackson, 1973; Brookes et al., 1985; Vance et al., 1987; Baran et al., 2019).
Simultaneously, 10 g of root-zone soil around rice plants was sampled from each treatment and stored it at -80°C for further genome sequencing. In detail, genomic DNA was extracted from 1 g of soil samples using the E.Z.N.ATM Mag–Bind Soil DNA Kit (Omega, USA) following the manufacturer’s instructions, and then the DNA quality was assessed using a Qubit 4.0 (Thermo, USA). Here, the V3-V4 region of bacterial 16S rRNA genes and the ITS1 region of fungal ITS genes were amplified using the universal primers 341F (5′−CCT ACG GGN GGC WGC AG−3′) and 806R (5′−GGA CTA CHV GGG TWT CTA AT−3′) (Wu et al., 2015), and ITS1F (5′−CTT GGT CAT TTA GGA AGT AA−3′) and ITS2 (5′−GCT GCG TTC TTC ATC GAT GC−3′) (Adams et al., 2013), respectively. PCR mixture consisted of 2×Hieff® Robust PCR Master Mix (15 μL), 10 μM universal primer (1 μL of each primer), DNA template (1 μL), and ddH2O (12 μL). PCR thermal protocol consisted of an initial 3 minutes denaturation step at 94°C, 25 amplification cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final extension step of 5 minutes at 72°C. PCR products were purified using Hieff NGS™ DNA selection beads (Yeasen, China), and then pooled in equimolar concentrations and sequenced using the 2 × 250 bp pair-end sequencing protocol on an Illumina MiSeq system (Sangon Biotechnology Co., Ltd., Shanghai, China).
After sequencing, raw data were assembled using PEAR (v0.9.8) and preprocessed using PRINSEQ to remove low-quality reads (average quality score < 20) to ensure high data quality (Schmieder and Edwards, 2011; Zhang et al., 2014). After primers were trimmed with Cutadapt (v1.18), clean reads were clustered into operational taxonomic units (OTUs) of ≥97% similarity using Usearch (v11.0.667) (Martin, 2011; Edgar, 2013, 2016). After selection of the representative read of each OTU using the QIIME package (v2020.06), all bacterial and fungal OTU representative sequences were classified taxonomically by blasting against the RDP database and UNITE database, respectively (Altschul et al., 1997; Wang et al., 2007; Caporaso et al., 2010; Quast et al., 2013; Urmas et al., 2013). Among them, relative abundances (RAs) of Aspergillus and Talaromyces species, and their relationships with environmental factors were especially focused using Origin software (v2023, Hampton, USA) and redundancy discriminant analysis (RDA), as well as correlation network.
2.5 Statistical analysis
One-way variance analysis (ANOVA) was performed using SPSS (v16, SPSS, USA). OTUs and alpha diversity indices (Chao1 and Shannon index) were analyzed by Origin software. The difference of microbial communities among different samples was measured by principal component analysis (PCA) based on beta diversity metrics from the Bray-Curtis metrics (Ramette, 2007), and the significant difference was tested using permutational multivariate ANOVA (PERMANOVA) with 999 permutations used to calculate p-values (Dixon, 2003). RAs (at the phylum and genus level, respectively) and heat map (at the family level) of the dominant microbes were calculated using Origin software. Differential biomarkers between groups were discovered by linear discriminant analysis effect size (LEfSe) (Segata et al., 2011). Different environmental factors on microbial community structure were calculated using RDA. Diagrams of the correlation network between soil properties and microbial taxa were performed using R software (v4.1.3).
3 Results
3.1 Assessment of plant growth promoting traits
The isolated fungi were screened to determine their plant growth-promoting traits. Results showed that 15 isolates displayed the ability of phosphate solubilization, siderophore production, and IAA production (Figures 1, 2). Indeed, as shown in Table 1, the diameter of the phosphate-solubilizing halo ranged from 11.91 to 31.65 mm, among which, isolates JD-L31f (31.65 mm), TL-B21f (28.96 mm), and FY-R71f (28.43 mm) exhibited the highest phosphate solubilization ability (Figure 1 upper). Moreover, the siderophore production ability was tested by growth of fungal isolates on CAS media. Results showed that all 15 isolates could secrete iron carriers. Among them, isolates YH-R21f, TL-B31f, LA-R21f, and YH-R31f displayed the siderophore-containing area of 24.66, 24.41, 24.16, and 23.66 mm, respectively, which were significantly (p < 0.05) higher than those of the other isolates (Figure 1 lower). Finally, all isolates had the ability to produce IAA in PDB medium amended with 0.1% or 1.0% tryptophan. At 0.1% tryptophan, the highest IAA production was obtained in isolates TL-B21f (50.23 μg/mL) and YH-R21f (31.18 μg/mL), while at 1.0% tryptophan, the highest IAA production was achieved in isolates FY-G-R31f (96.50 μg/mL) and TL-B31f (95.29 μg/mL) (Figure 2).
Figure 1
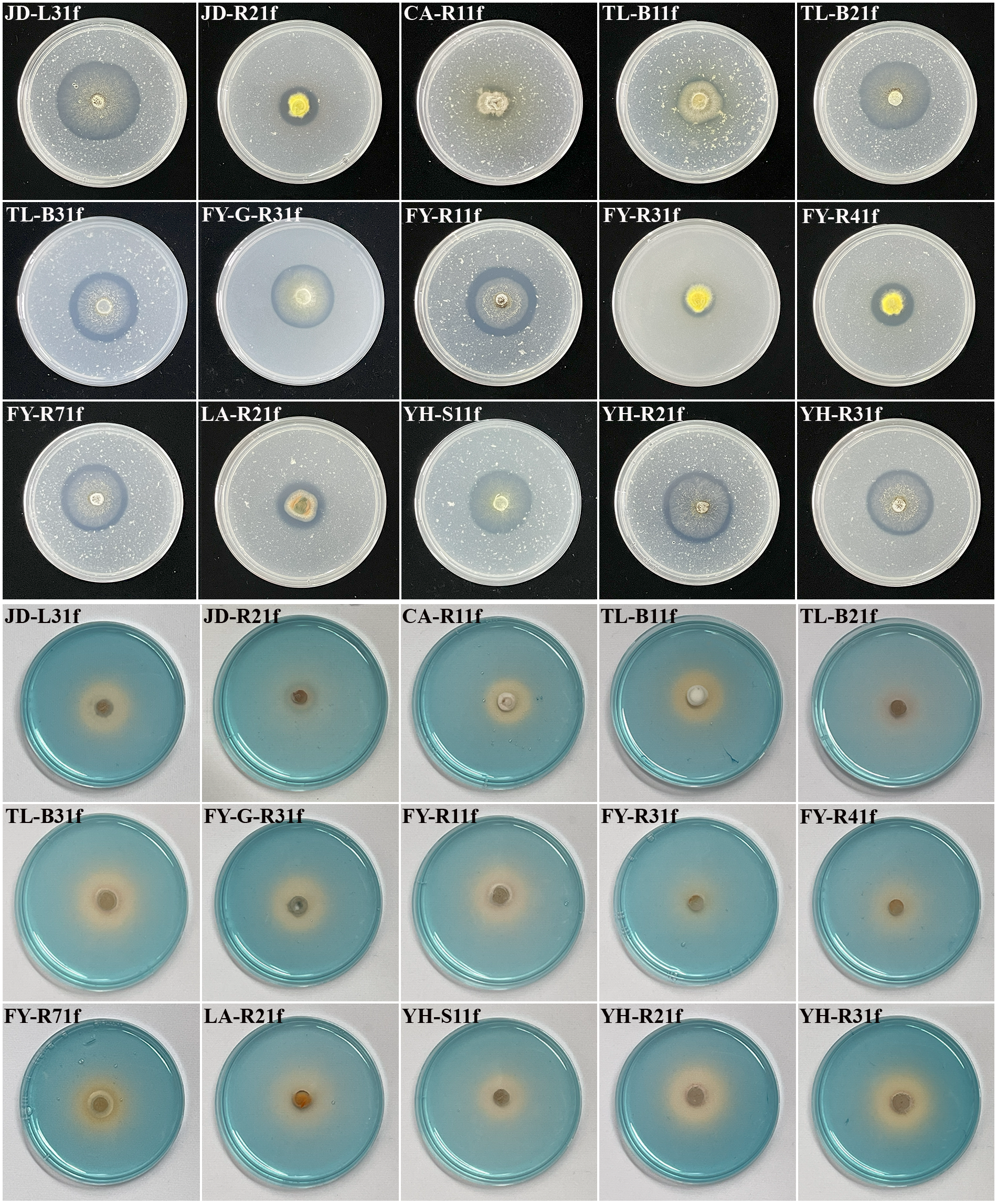
Assessment of phosphate solubilization and siderophore production ability of 15 fungal isolates on NBRIP medium (upper), and on CAS medium (lower), respectively.
Figure 2
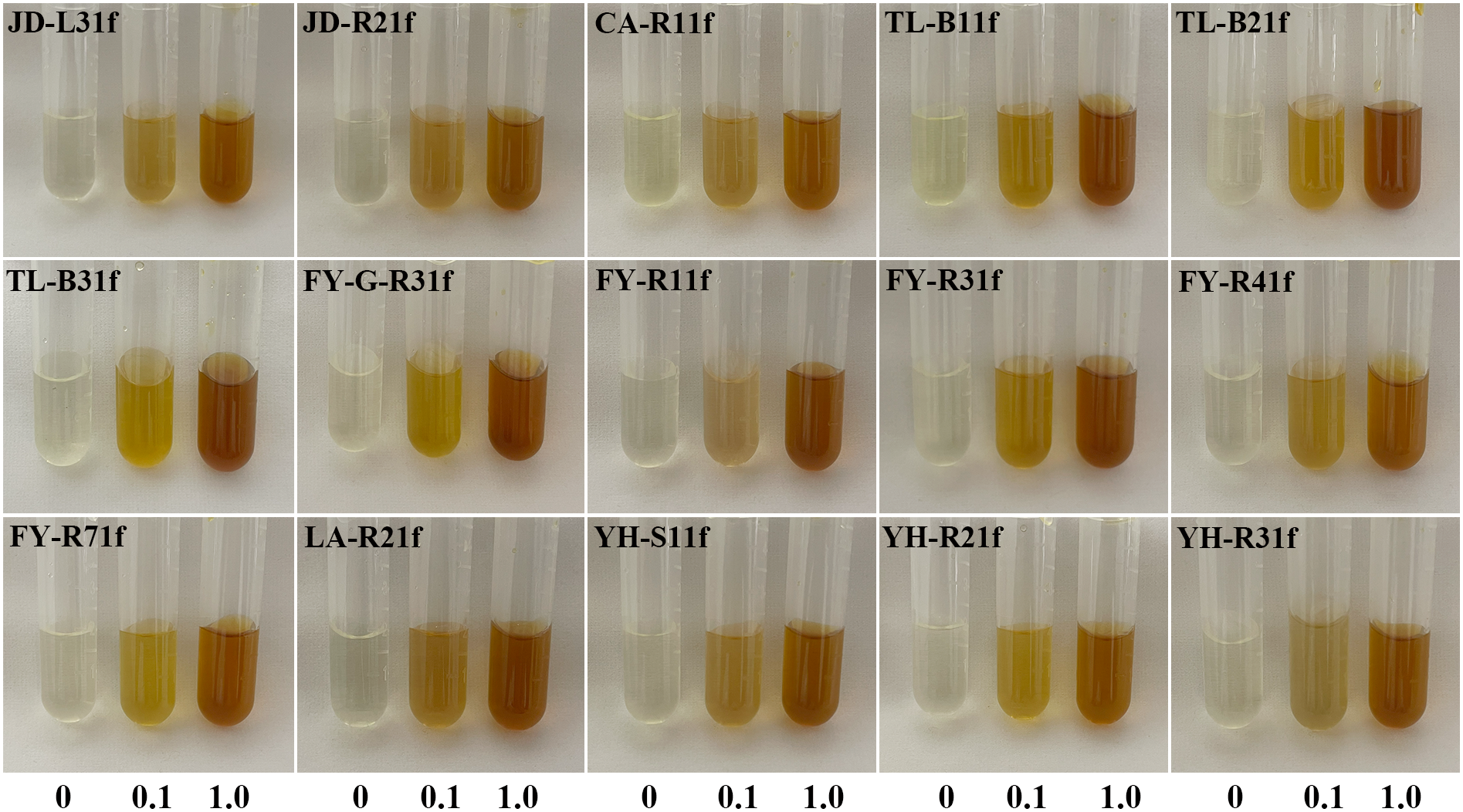
Ability of 15 fungal isolates to produce IAA in PDB medium amended with 0.1% or 1.0% tryptophan.
Table 1
| Strains | Phosphate solubilization (mm) | Siderophore production (mm) | IAA (μg/mL) | |
|---|---|---|---|---|
| 0.1% | 1.0% | |||
| JD-L31f | 31.65 ± 1.90 a | 17.69 ± 1.77 e | 10.14 ± 0.95 e | 36.72 ± 2.51 h |
| JD-R21f | 15.89 ± 1.23 g | 19.72 ± 2.40 cd | 15.43 ± 1.47 d | 40.91 ± 4.77 gh |
| CA-R11f | 15.44 ± 1.29 g | 17.57 ± 0.74 e | 11.89 ± 1.05 e | 39.84 ± 0.70 gh |
| TL-B11f | 20.05 ± 0.32 e | 21.74 ± 1.72 b | 18.23 ± 3.30 d | 77.42 ± 1.63 b |
| TL-B21f | 28.96 ± 1.57 b | 17.09 ± 1.52 e | 50.23 ± 3.96 a | 70.45 ± 2.09 c |
| TL-B31f | 27.63 ± 1.10 bcd | 24.41 ± 0.77 a | 25.36 ± 1.91 c | 95.29 ± 4.44 a |
| FY-G-R31f | 26.86 ± 0.67 cd | 21.87 ± 1.29 b | 17.04 ± 1.77 d | 96.50 ± 2.72 a |
| FY-R11f | 26.19 ± 2.05 d | 21.23 ± 1.34 bc | 11.04 ± 2.14 e | 59.57 ± 1.44 e |
| FY-R31f | 11.91 ± 1.46 h | 18.29 ± 0.62 de | 17.32 ± 0.87 d | 58.63 ± 0.83 e |
| FY-R41f | 17.53 ± 1.38 f | 17.37 ± 2.42 e | 25.12 ± 1.83 c | 64.95 ± 2.98 d |
| FY-R71f | 28.43 ± 1.32 bc | 21.06 ± 1.52 bc | 9.82 ± 0.55 e | 53.44 ± 1.12 f |
| LA-R21f | 18.81 ± 0.80 ef | 24.16 ± 0.62 a | 16.27 ± 0.74 d | 52.03 ± 1.08 f |
| YH-S11f | 27.12 ± 1.09 cd | 17.78 ± 1.19 e | 15.76 ± 1.58 d | 43.41 ± 2.76 g |
| YH-R21f | 27.00 ± 1.69 cd | 24.66 ± 1.39 a | 31.18 ± 0.30 b | 65.19 ± 5.07 d |
| YH-R31f | 26.51 ± 1.05 d | 23.66 ± 1.00 a | 8.79 ± 1.45 e | 38.51 ± 0.80 gh |
Assessment of the growth promotion ability of 15 selected fungal isolates.
Values that are separated by distinct lowercase letters within the same column indicate a significant difference at p < 0.05.
3.2 Effect of PGPF on rice growth
All 15 fungal isolates were further examined for their growth-promoting activity in rice seedlings. After 55 days of inoculation, all 15 fungal isolates could cause a noticeable increase in rice growth compared to the control based on the phenotypic observation (Figure 3). The measured data further demonstrated that the fungal 15 isolates significantly (p < 0.05) affected the growth and biomass accumulation of rice seedlings compared to the control (Table 2). Indeed, among all treatments, rice seedlings inoculated with various fungal isolates exhibited different increases in plant height. Isolate TL-B31f exhibited the highest increase in plant height (359.59 mm), which was about 1.15 times that of the control. Isolates FY-R11f and FY-R31f followed, with 14.58% and 13.90% increases in plant height, respectively, as compared to that of control seedlings. Moreover, the fungal strains exhibited significantly (p < 0.05) different growth-promoting effects on root length of seedlings. Isolate FY-R41f showed the highest increase in root length (205.47 mm), with an increase of 43.31% compared to that of the control, followed by isolates FY-G-R31f and TL-B11f with an increase of 37.41% and 33.98% compared to that of the control, respectively. Furthermore, compared to the control, the fresh weights of seedlings were significantly (p < 0.05) increased by isolates CA-R11f, FY-R41f, and TL-B31f. In particular, the highest fresh weight of seedlings was achieved by isolate CA-R11f, with an increase of 95.09% with respect to that of the control. In addition, the dry weight of seedlings treated by isolates FY-R41f, FY-R31f, and FY-R11f was significantly (p < 0.05) higher than that of the control, with increases of 28.12%, 19.58%, and 16.39%, respectively. In general, the quality of rice seedlings treated with different fungal isolates was higher than that of the control.
Figure 3

Effect of 15 fungal isolates on rice growth in a greenhouse, 55 days after inoculation.
Table 2
| Treatments | PH (mm) | GPE% | RL (mm) | GPE% |
|---|---|---|---|---|
| JD-L31f | 316.48 ± 8.08 | 1.48 g | 158.83 ± 12.92 | 10.78 g |
| JD-R21f | 344.11 ± 11.65 | 10.34 de | 166.45 ± 12.63 | 16.09 f |
| CA-R11f | 348.21 ± 12.37 | 11.65 cd | 189.19 ± 9.71 | 31.95 c |
| TL-B11f | 349.32 ± 13.88 | 12.01 cd | 192.10 ± 8.58 | 33.98 bc |
| TL-B21f | 346.09 ± 8.79 | 10.98 d | 161.45 ± 12.55 | 12.60 fg |
| TL-B31f | 359.59 ± 12.49 | 15.30 a | 191.57 ± 14.02 | 33.62 bc |
| FY-G-R31f | 346.04 ± 10.39 | 10.96 d | 197.01 ± 10.68 | 37.41 b |
| FY-R11f | 357.35 ± 11.68 | 14.58 ab | 185.04 ± 10.35 | 29.06 cd |
| FY-R31f | 355.21 ± 10.04 | 13.90 abc | 185.44 ± 9.88 | 29.33 cd |
| FY-R41f | 355.02 ± 12.15 | 13.84 abc | 205.47 ± 6.92 | 43.31 a |
| FY-R71f | 337.51 ± 13.36 | 8.22 e | 159.38 ± 13.31 | 11.16 fg |
| LA-R21f | 350.17 ± 6.05 | 12.28 bcd | 184.60 ± 10.93 | 28.75 cd |
| YH-S11f | 326.45 ± 9.88 | 4.68 f |
|
8.63 g |
| YH-R21f | 324.82 ± 8.35 | 4.15 f | 176.35 ± 10.60 | 23.00 e |
| YH-R31f | 338.43 ± 12.78 | 8.52 e | 179.71 ± 10.50 | 25.34 de |
| Control | 311.86 ± 10.24 | – | 143.38 ± 8.52 | – |
| Treatments | SFW (g) | GPE% | SDW (g) | GPE% |
| JD-L31f | 1.14 ± 0.09 | 59.39 de | 0.32 ± 0.04 | -1.04 def |
| JD-R21f | 1.04 ± 0.12 | 45.03 f | 0.29 ± 0.04 | -8.21 f |
| CA-R11f | 1.40 ± 0.08 | 95.09 a | 0.37 ± 0.06 | 15.22 abc |
| TL-B11f | 1.24 ± 0.09 | 72.63 bc | 0.34 ± 0.08 | 7.11 bcde |
| TL-B21f | 1.25 ± 0.10 | 74.42 bc | 0.35 ± 0.07 | 10.26 bcd |
| TL-B31f | 1.28 ± 0.15 | 78.58 b | 0.35 ± 0.06 | 9.31 bcd |
| FY-G-R31f | 1.24 ± 0.18 | 72.09 bc | 0.35 ± 0.08 | 8.66 bcde |
| FY-R11f | 1.18 ± 0.17 | 63.93 cde | 0.37 ± 0.07 | 16.39 abc |
| FY-R31f | 1.26 ± 0.12 | 75.63 bc | 0.38 ± 0.06 | 19.58 ab |
| FY-R41f | 1.36 ± 0.16 | 89.77 a | 0.41 ± 0.09 | 28.12 a |
| FY-R71f | 1.09 ± 0.15 | 52.24 ef | 0.33 ± 0.08 | 2.25 cdef |
| LA-R21f | 1.22 ± 0.14 | 69.65 bcd | 0.34 ± 0.05 | 5.92 bcde |
| YH-S11f | 1.09 ± 0.08 | 51.97 ef | 0.29 ± 0.03 | -8.11 f |
| YH-R21f | 0.87 ± 0.10 | 21.41 g | 0.34 ± 0.04 | 5.38 bcdef |
| YH-R31f | 1.11 ± 0.11 | 54.66 ef | 0.30 ± 0.05 | -5.12 ef |
| Control | 0.72 ± 0.09 | – | 0.32 ± 0.05 | – |
Effects of 15 fungal isolates on rice growth.
PH, plant height; RL, root length; SFW, seedling fresh weight; SDW, seedling dry weight; GPE, growth promotion efficacy. Means are averages ± SD. Different lowercase letters within the same columns reveal the significance among different treatments (p < 0.05). The two treatments with the best growth-promoting properties are shown in bold.
3.3 Fungal identification
After being cultured on PDA, CYA, and MEA medium at 25°C for 7 days, it was revealed that 15 fungal isolates belong to Aspergillus sp., Penicillium sp., and Talaromyces sp. based on the obvious distinctions of colony morphology (Figure 4). Indeed, eight isolates including JD-L31f, YH-S11f, TL-B21f, YH-R21f, YH-R31f, TL-B31f, FY-R71f, and FY-R11f were attributed to the genus Aspergillus due to the coffee brown to black colonies. Three isolates including FY-G-R31f, TL-B11f, and CA-R11f were attributed to the genus Penicillium in view of the irregular, green, velvety texture of colonies. Four isolates including LA-R21f, FY-R41f, FY-R31f, and JD-R21f were attributed to the genus Talaromyces due to the yellow-green colonies and a characteristic red pigment (Tsang et al., 2018; Houbraken et al., 2020). To further identify the species of all isolates, phylogenetic analysis was conducted by using the maximum likelihood method based on sequences of several conserved genes including ITS, TUB, CaM, and RPB2 of the tested isolates and closely related species (Table 3). The phylogenetic trees with the highest log likelihood were drawn, with branch lengths reflecting evolutionary distance and bootstrap proportions beside the branches indicating credibility.
Figure 4
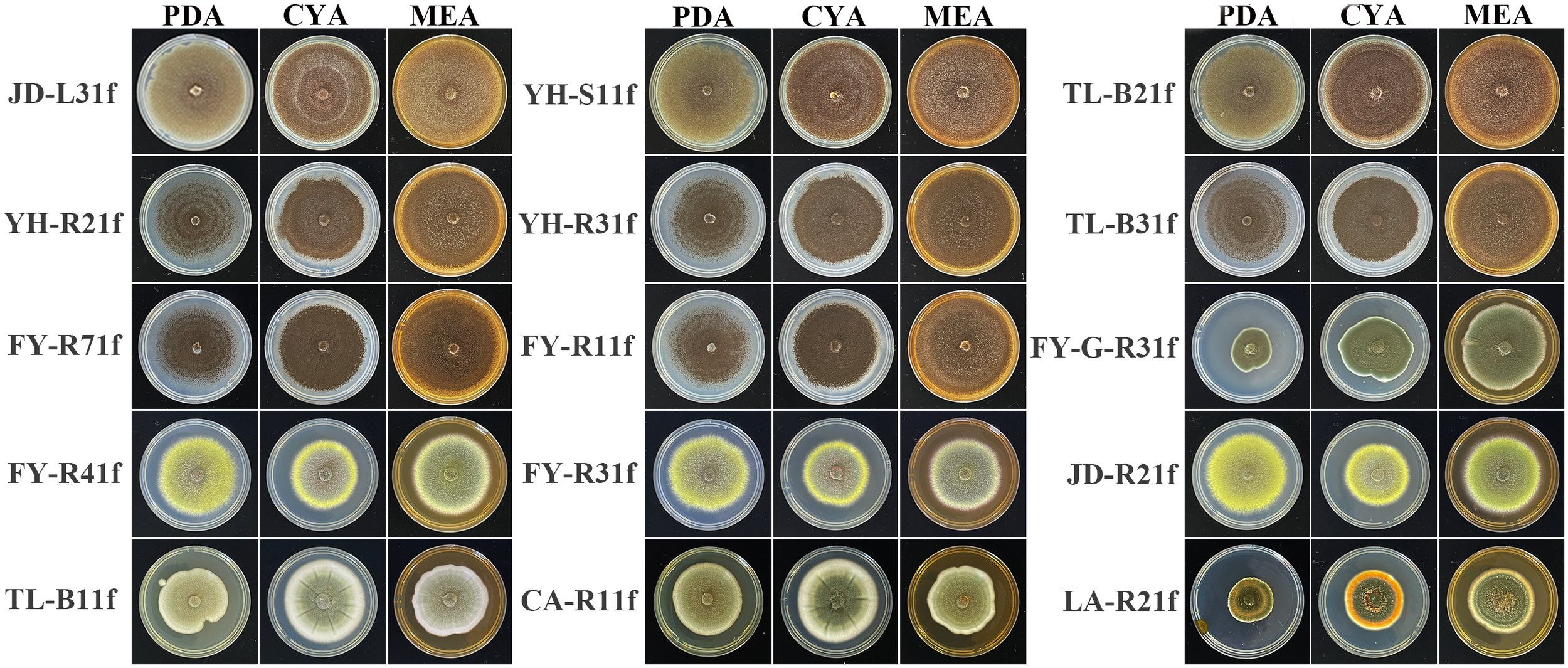
Colony features on PDA (potato dextrose agar), CYA (czapek yeast autolysate agar), and MEA (malt extract agar) after 7 days at 25°C.
Table 3
| Species | Isolates | GenBank accession number | |||
|---|---|---|---|---|---|
| ITS | TUB | CaM | RPB2 | ||
| Talaromyces aculeatus | NRRL2129 | KF741995 | KF741929 | KF741975 | MH793099 |
| T. apiculatus | CBS312.59 | JN899375 | JX091378 | KF741950 | KM023287 |
| T. aurantiacus | CBS314.59 | JN899380 | KF741917 | KF741951 | KX961285 |
| T. derxii | CBS412.89 | JN899327 | JX494306 | KF741959 | KM023282 |
| T. flavovirens | CBS102801 | JN899392 | JX091376 | KF741933 | KX961283 |
| T. flavus | CBS310.38 | MH867464 | JX494302 | KF741949 | JF417426 |
| T. helicus | CBS335.48 | MH856373 | KJ865725 | KJ885289 | KM023273 |
| T. muroii | CBS756.96 | MK450747 | KJ865727 | KJ885274 | KX961276 |
| T. pinophilu | NRRL62183 | MH909497 | MH909388 | MH909444 | MH909550 |
| T. purpureogenus | B195_0419 | OR192900 | OR233627 | OR327660 | OR211408 |
| LA-R21f | PQ269783 | PQ516235 | PQ516220 | PQ516268 | |
| T. ruber | CBS195.88 | JX965240 | JX965350 | JX965204 | JX965310 |
| T. rubicundus | CBS342.59 | JN899384 | JX494309 | KF741956 | KM023296 |
| T. sayulitensis | NRRL62272 | MH793083 | MH792956 | MH793020 | MH793147 |
| T. stollii | CBS624.93 | JX315676 | JX315636 | JX315644 | JX965315 |
| T. veerkampii | NRRL 6095 | MH793040 | MH792912 | MH792976 | MH793103 |
| FY-R31f | PQ269785 | PQ516237 | PQ516222 | PQ516270 | |
| FY-R41f | PQ269784 | PQ516236 | PQ516221 | PQ516269 | |
| JD-R21f | PQ269786 | PQ516238 | PQ516223 | PQ516271 | |
| T. viridulus | CBS 252.87 | JN899314 | JX091385 | KF741943 | JF417422 |
| Aspergillus aculeatus | CBS172.66 | FJ629320 | HE577806 | FN594542 | JN121448 |
| A. aculeatinus | CBS121060 | MH863102 | EU159220 | EU159241 | HF559233 |
| A. amoenus | NRRL4838 | EF652480 | JN853946 | JN854035 | JN853824 |
| A. austroafricanus | NRRL233 | JQ301891 | JN853963 | JN854025 | JN853814 |
| A. baeticus | CCF4226 | HE615086 | HE615092 | HE615117 | HE615124 |
| A. brevijanus | NRRL1935 | EF669582 | EU014078 | EF669540 | EF669624 |
| A. brunneoviolaceus | NRRL4912 | EF661220 | EF661105 | EF661147 | EF661045 |
| JD-L31f | PQ269772 | PQ516224 | PQ516209 | PQ516257 | |
| YH-S11f | PQ269773 | PQ516225 | PQ516210 | PQ516258 | |
| TL-B21f | PQ269774 | PQ516226 | PQ516211 | PQ516259 | |
| A. campestris | NRRL13001 | EF669577 | EU014091 | EF669535 | EF669619 |
| A. candidus | NRRL303 | EF669592 | EU014089 | EF669550 | EF669634 |
| A. capensis | DTO179E6 | KJ775550 | KJ775072 | KJ775279 | KP987020 |
| A. creber | NRRL58592 | JQ301889 | JN853980 | JN854043 | JN853832 |
| A. cvjetkovicii | NRRL227 | EF652440 | EF652264 | EF652352 | EF652176 |
| A. flavipes | NRRL302 | EF669591 | EU014085 | EF669549 | EF669633 |
| A. fructus | NRRL239 | EF652449 | EF652273 | EF652361 | EF652185 |
| A.iizukaelo | NRRL3750 | EF669597 | EU014086 | EF669555 | EF669639 |
| A. janus | NRRL1787 | EF669578 | EU014076 | EF669536 | EF669620 |
| A. jensenii | NRRL58600 | JQ301892 | JN854007 | JN854046 | JN853835. |
| A. protuberus | NRRL3505 | EF652460 | EF652284 | EF652372 | EF652196 |
| A. puniceus | NRRL5077 | EF652498 | EF652322 | EF652410 | EF652234 |
| A. puulaauensis | NRRL35641 | JQ301893 | JN853979 | JN854034 | JN853823 |
| A. saccharolyticus | CBS127449 | HM853552 | HM853553 | HM853554 | HF559235 |
| A. subalbidus | NRRL5214 | LT908114 | LT908034 | LT908035 | LT908036 |
| A. subversicolor | NRRL58999 | JQ301894 | JN853970 | JN854010 | JN853799 |
| A. sydowii | NRRL250 | EF652450 | EF652274 | EF652362 | EF652186 |
| A. tabacinus | NRRL4791 | EF652478 | EF652302 | EF652390 | EF652214 |
| A. tennesseensis | NRRL13150 | JQ301895 | JN853976 | JN854017 | JN853806 |
| A. tritici | CBS266.81 | MN431381 | MN969368 | MN969233 | MN969098 |
| A. tubingensis | NRRL62644 | KC796398 | KC796370 | KC796386 | KC796437 |
| YH-R21f | PQ269775 | PQ516227 | PQ516212 | PQ516260 | |
| YH-R31f | PQ269776 | PQ516228 | PQ516213 | PQ516261 | |
| TL-B31f | PQ269777 | PQ516229 | PQ516214 | PQ516262 | |
| FY-R71f | PQ269778 | PQ516230 | PQ516215 | PQ516263 | |
| FY-R11f | PQ269779 | PQ516231 | PQ516216 | PQ516264 | |
| A. ustus | NRRL4991 | EF652492 | EF652316 | EF652404 | EF652228 |
| A. venenatus | NRRL13147 | JQ301896 | JN854003 | JN854014 | JN853803 |
| A. versicolor | NRRL238 | EF652442 | EF652266 | EF652354 | EF652178 |
| P. alfredii | DTO269A4 | KJ775684 | KJ775177 | KJ775411 | KJ834520 |
| P. brefeldianum | NRRL710 | AF033435 | EU021669 | EU021683 | EU021658 |
| P. incoloratum | CBS101753 | KJ834508 | KJ834457 | KJ866984 | JN406651 |
| P. jamesonlandense | IBT21985 | KY989164 | KY989039 | KY989101 | KY989214 |
| P. janthinellum | CBS340.48 | GU981585 | GU981625 | MN969268 | GU981625 |
| P. javanicum | CBS341.48 | GU981613 | GU981657 | MN969269 | JN121498 |
| P. limosum | CBS339.97 | NR_111496 | GU981621 | MN969271 | KF296433 |
| TL-B11f | PQ269780 | PQ516232 | PQ516217 | PQ516265 | |
| CA-R11f | PQ269781 | PQ516233 | PQ516218 | PQ516266 | |
| P. malacaense | NRRL35754 | EU427300 | EU427268 | KF932944 | EU427261 |
| P. nodulum | CBS227.89 | KC411703 | KJ834475 | KJ867003 | JN406603 |
| P. oxalicum | CBS219.30 | MH855125 | KF296462 | MN969283 | JN121456 |
| FY-G-R31f | PQ269782 | PQ516234 | PQ516219 | PQ516267 | |
| P. penarojense | CBS113178 | GU981570 | GU981646 | MN969287 | KF296450 |
| P. piscarium | CBS362.48 | GU981600 | GU981668 | MN969288 | KF296451 |
| P. raistrickii | CBS261.33 | JN617697 | KJ834485 | KJ867006 | JN606589 |
| P. ribeum | CBS127809 | MH864716 | MN969395 | KJ866995 | JN406631 |
| P. sajarovii | CBS277.83 | KC411724 | MN969397 | KJ867007 | JN406588 |
| P. soppii | CBS226.28 | MH854995 | MN969399 | KJ867002 | JN406606 |
| P. vanderhammenii | CBS126216 | MH863982 | GU981647 | MN969308 | KF296458 |
| P. virgatum | CBS114838 | AJ748692 | KJ834500 | KJ866992 | JN406641 |
| P. wotroi | CBS118171 | GU981591 | GU981637 | MN969313 | KF296460 |
| P. zonatum | CBS992.72 | GU981581 | GU981651 | MN969315 | KF296461 |
Fungal isolates used in this study.
Species and sequences obtained from this study are shown in bold.
The phylogenetic trees showed that all 15 isolates were separated into six distant clades (Figure 5). Indeed, isolates JD-L31f, YH-S11f, and TL-B21f were clustered with A. brunneoviolaceus isolate NRRL4912 as a distinct clade with a bootstrap of 96%, suggesting the three isolates should be considered as A. brunneoviolaceus. Isolates YH-R21f, YH-R31f, TL-B31f, FY-R71f, and FY-R11f were clustered with A. tubingensis isolate NRRL62644 as a distinct clade with a bootstrap of 100%, suggesting the five isolates should be considered as A. tubingensis (Figure 5a). Isolate FY-G-R31f was clustered with Penicillium oxalicum isolate CBS219.30 as a distinct clade with a bootstrap of 100%, suggesting this isolate should be considered as P. oxalicum. Isolates TL-B11f and CA-R11f were clustered with Penicillium limosum isolate CBS339.97 as a distinct clade with a bootstrap of 100%, suggesting the two isolates should be considered as P. limosum (Figure 5b). Isolate LA-R21f clustered with Talaromyces purpureogenus isolate B195_0419 as a distinct clade with a bootstrap of 100%, suggesting this isolate should be considered as T. purpureogenus. Isolates FY-R41f, FY-R31f, and JD-R21f were clustered with T. veerkampii isolate NRRL6095 as a distinct clade with a bootstrap of 100%, suggesting the three isolates should be considered as T. veerkampii (Figure 5c).
Figure 5
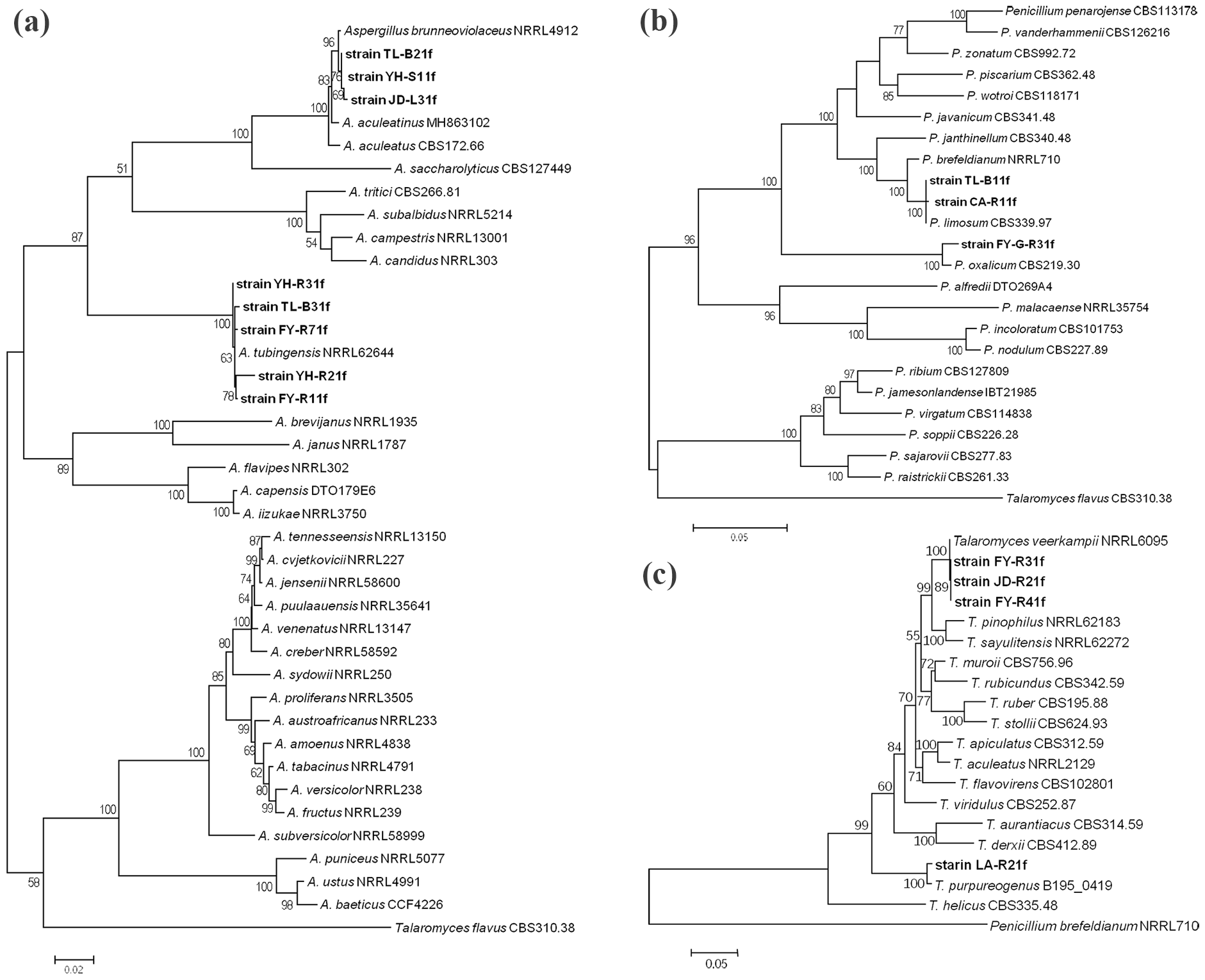
Maximum likelihood (ML) tree generated with MEGA 7.0 from the combined ITS, TUB, CaM, and RPB2 gene sequences of Aspergillus(a), Penicillium(b), and Talaromyces(c), respectively. The reference sequences of closely related species were gotten from NCBI, and the isolates obtained from soil in this study were shown in bold letters. Bootstrap values after 1000 replicates were expressed as percentages.
3.4 Improvement of soil quality, microbial community structure, and function by PGPF
Two PGPF isolates TL-B31f and FY-R41f, with the best growth-promoting properties, were selected for analyzing their effects on soil quality, microbial community composition, and function after 55 days of inoculation. As shown in Table 4, compared with the control, the soil pH and TP were generally unaffected by At-TLB31f (inoculated with isolate TL-B31f) and Tv-FYR41f (inoculated with isolate FY-R41f) treatments. Meanwhile, At-TLB31f treatment caused a 4.38% decrease (p < 0.05) in SOM compared to the control, while there was no significant difference between Tv-FYR41f treatment and the control. Furthermore, AP was significantly (p < 0.05) increased by At-TLB31f (42.52%) and Tv-FYR41f (48.51%) treatments compared to the control.
Table 4
| Treatments | pH | SOM (g/kg) | TP (g/kg) | AP (mg/kg) |
|---|---|---|---|---|
| At-TLB31f | 8.00 ± 0.05 ab | 8.55 ± 0.15 b | 0.88 ± 0.00 a | 27.33 ± 0.71 b |
| Tv-FYR41f | 8.06 ± 0.03 a | 9.01 ± 0.12 a | 0.88 ± 0.01 a | 28.47 ± 0.32 a |
| Control | 7.97 ± 0.05 b | 8.94 ± 0.29 a | 0.87 ± 0.01 a | 19.17 ± 0.63 c |
Impacts of two different PGPF isolates on soil quality properties.
Values that are separated by distinct lowercase letters within the same column indicate a significant difference at p < 0.05. SOM is soil organic matter; TP is total phosphorus; AP is available phosphorus. At-TLB31f, treatment inoculated with treatment inoculated with isolate TL-B31f; Tv-FYR41f, treatment inoculated with treatment inoculated with isolate FY-R41f.
Based on high-throughput amplicon sequencing of DNA extracted from soil samples, bacterial and fungal taxonomic diversity (α-diversity) and composition (β-diversity) were compared between treatments inoculated by isolate TL-B31f or FY-R41f and the control. Indeed, the average bacterial Chao1 index was 2,087 (1,977–2,169), 2,123 (2,000–2,262), 2,133 (2,043–2,228), and the average bacterial Shannon index was 7.07 (7.00–7.16), 7.14 (7.02–7.26), and 7.07 (6.97–7.13) in At-TLB31f, Tv-FYR41f treatments, and the control, respectively (Figures 6A1, A2). In general, the bacterial Chao1 index was lower (2.17% and 0.48%) in At-TLB31f and Tv-FYR41f treatments, along with a higher Shannon index (0.13% and 1.00%) than that in the control. Meanwhile, the average fungal Chao1 index was 223 (199–243), 195 (187–202), 191 (175–207), and the average fungal Shannon index was 3.89 (3.71–4.03), 3.70 (3.53–3.83), and 3.85 (3.82–3.88) in At-TLB31f, Tv-FYR41f treatments, and the control, respectively (Figures 6B1, B2). In other words, isolate TL-B31f inoculation caused a 16.49% (p < 0.05) and 1.00% increase in the fungal Chao1 and Shannon index of soil, while isolate FY-R41f inoculation caused a 2.21% increase and 4.00% reduction, respectively, compared to the control. Overall, there was no significant (p < 0.05) difference in the α-diversity of soil bacteria between two PGPF isolates treatments and the control, but the α-diversity of soil fungi was significantly (p < 0.05) changed by inoculation of isolates TL-B31f and FY-R41f.
Figure 6
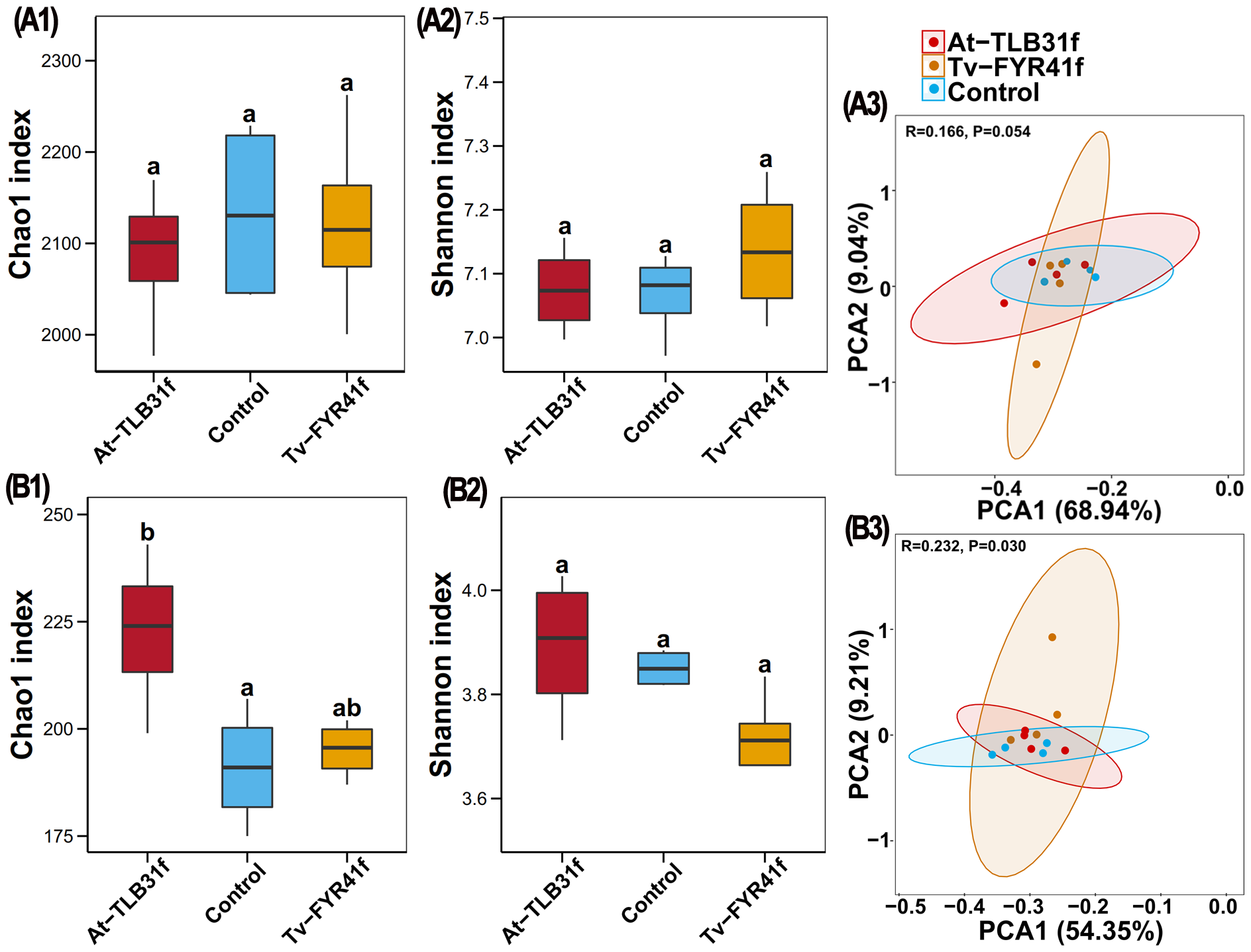
The effect of two different PGPF isolates on the Chao1 and Shannon index of bacteria (A1, A2) and fungi (B1, B2). Different lowercase letters reveal the significance among different treatments (p < 0.05). Principal component analysis (PCA) results of soil bacterial (A3) and fungal (B3) communities based on OTUs abundance. Ellipses have been drawn for each treatment with a confidence limit of 0.95.
Results of PCA revealed that the OTU abundance from 12 soil samples of At-TLB31f, Tv-FYR41f treatments, and the control comprised three different groups, but there was noticeable overlap among all three different treatments, regardless of bacterial or fungal data (Figures 6A3, B3). Indeed, the two principal components accounted for 77.98% (PC1 68.94%, PC2 9.04%, respectively) and 63.56% (PC1 54.35%, PC2 9.21%, respectively) of the total variance in the bacterial and fungal communities, respectively. PERMANOVA performed on all samples also indicated that different PGPF explained 16.6% (p = 0.054) and 23.2% (p = 0.030) of the variation, respectively. These findings showed that the bacterial and fungal communities in soil inoculated by isolate TL-B31f or FY-R41f were generally similar to the control, albeit with some differences.
The compositional differences of soil bacterial and fungal communities among the three different treatments were compared at the phylum and genus levels. Indeed, in the bacterial community, Pseudomonadota, Chloroflexota, Acidobacteriota, Bacteroidota, Bacillota, Actinomycetota, Cyanobacteriota, Gemmatimonadota, Myxococcota, and Thermodesulfobact were the top 10 dominant phyla (Figure 7A1), and OLB13, Luteitalea, Aggregatilinea, Subgroup 10, Vibrionimonas, MND1, Gemmatimonas, Sphingomonas, Vicinamibacter, and Lysobacter were the top 10 dominant genera (Figure 7A2). Compared with the control, the RAs of Vibrionimonas (22.12%) and Vicinamibacter (10.13%) were increased, while Lysobacter (62.56%), Gemmatimonas (11.76%), Sphingomonas (8.17%, p < 0.05), and MND1 (7.64%) were decreased in At-TLB31f treatment; the RAs of Vibrionimonas (30.15%), OLB13 (17.32%), Aggregatilinea (11.83%), Lysobacter (10.29%), and MND1 (6.79%) were decreased, while Sphingomonas (19.68%, p < 0.05), Vicinamibacter (7.92%), and Luteitalea (5.73%) were increased in Tv-FYR41f treatment (Figure 7A2).
Figure 7
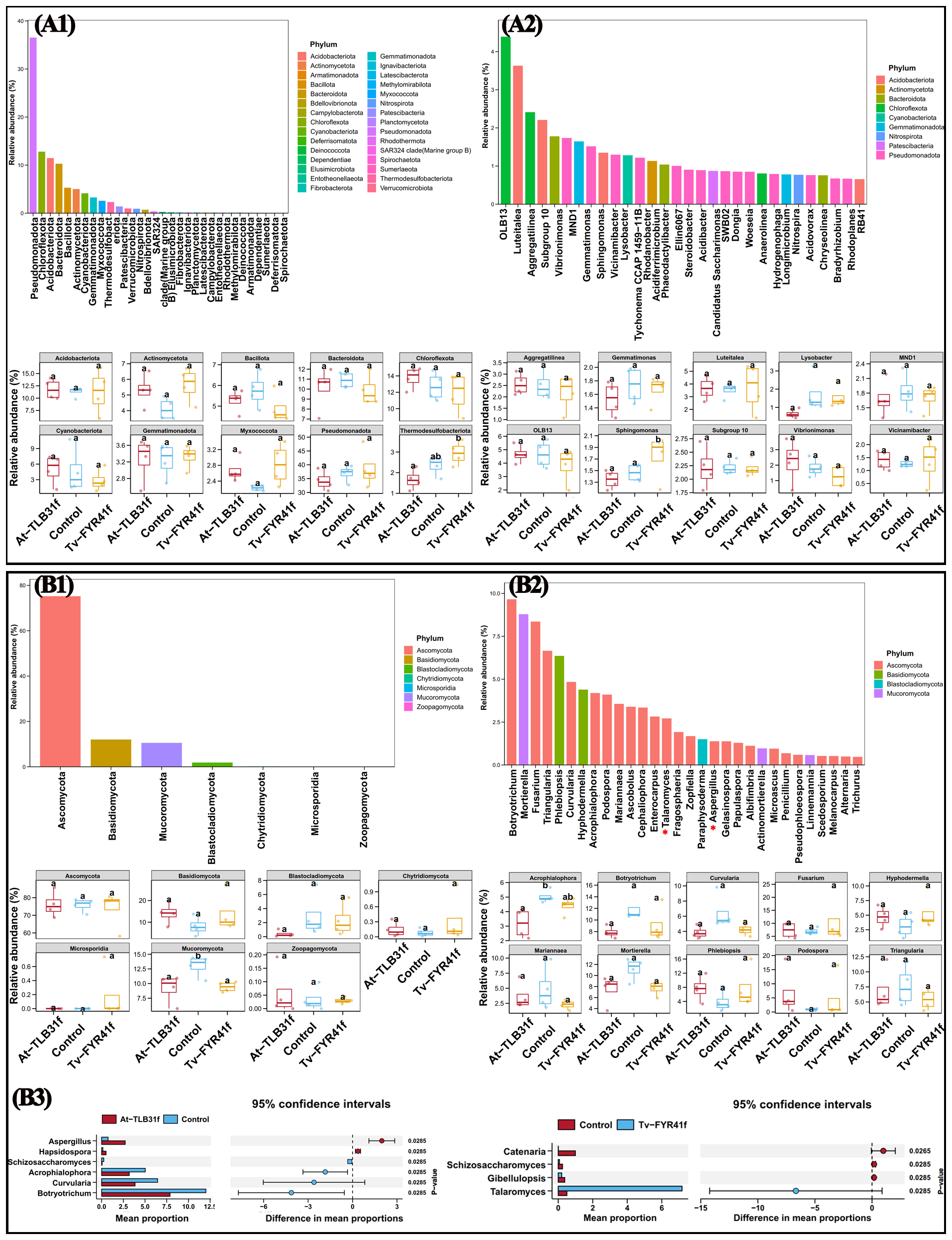
Relative abundance of dominant bacteria (A) and fungi (B) at the phylum (A1, B1) and genus (A2, B2) level. The abundance of fungal biomarkers at the genus level between At-TLB31f (or Tv-FYR41f) treatments and the control (B3).
Similarity, according to the distribution and RAs of fungi among all three different treatments at the phylum level, Ascomycota, Basidiomycota, Mucoromycota, and Blastocladiomycota were the main fungal phyla, with abundance of 73.80–76.11%, 8.12–14.09%, 9.20–13.03%, and 0.40–2.93%, respectively (Figure 7B1). Furthermore, at the genus level, Botryotrichum, Mortierella, Fusarium, Triangularia, Phlebiopsis, Curvularia, Hyphodermella, Acrophialophora, Podospora, and Mariannaea were the top 10 dominant genera (Figure 7B2). Compared with the control, the At-TLB31f treatment caused a 40.29%, 36.68%, 34.39%, 33.16%, 24.37%, and 9.45% reduction in Curvularia, Acrophialophora (p < 0.05), Botryotrichum, Mortierella, Mariannaea, and Triangularia, respectively, while a 641.74%, 98.87%, 49.74%, and 6.07% increase in Podospora, Phlebiopsis, Hyphodermella, and Fusarium, respectively (Figure 7B2). In addition, the RAs of Aspergillus were significantly increased by 264.68% in At-TLB31f treatment, while Talaromyces were significantly increased by 1328.35% in Tv-FYR41f treatment (Figure 7B3). In other words, compared to the control, the microbial community composition of the two PGPF isolates treatments was reconstructed with an increase or decrease of some specific microbes.
Furthermore, LEfSe was carried out to identify specific bacterial and fungal biomarkers distinguishing soil microbial communities among At-TLB31f, Tv-FYR41f treatments, and the control (Figure 8A). Results showed that 14 bacterial biomarkers (LDA > 3.0) and 18 fungal biomarkers (LDA > 3.5) were present in all treatments. In fact, communities of At-TLB31f treatment were enriched with Agaricales, Aspergillus, Aspergillaceae, Fragosphaeria, Pezizales, Pezizomycetes, Ophiostomataceae, and Ophiostomatales. Communities of Tv-FYR41f treatment were enriched with Caldilineaceae, Caldilineales, Desulfobulbia, Desulfuromonadia, Ellin6067, Geobacteraceae, Geobacterales, Geomonas, Lysobacteraceae, Pseudolabrys, Ramlibacter, Thermodesulfobacteriota, and Cystobasidiomycetes, Eurotiales, Eurotiomycetes, Trichocomaceae, Talaromyces. The controls were enriched with Hydrogenophaga, Lysobacter, and Blastocladiomycetes, Blastocladiales, Boudiera, Catenariaceae, and Catenaria. Overall, these microbial taxa might play important roles in modulating rice growth.
Figure 8
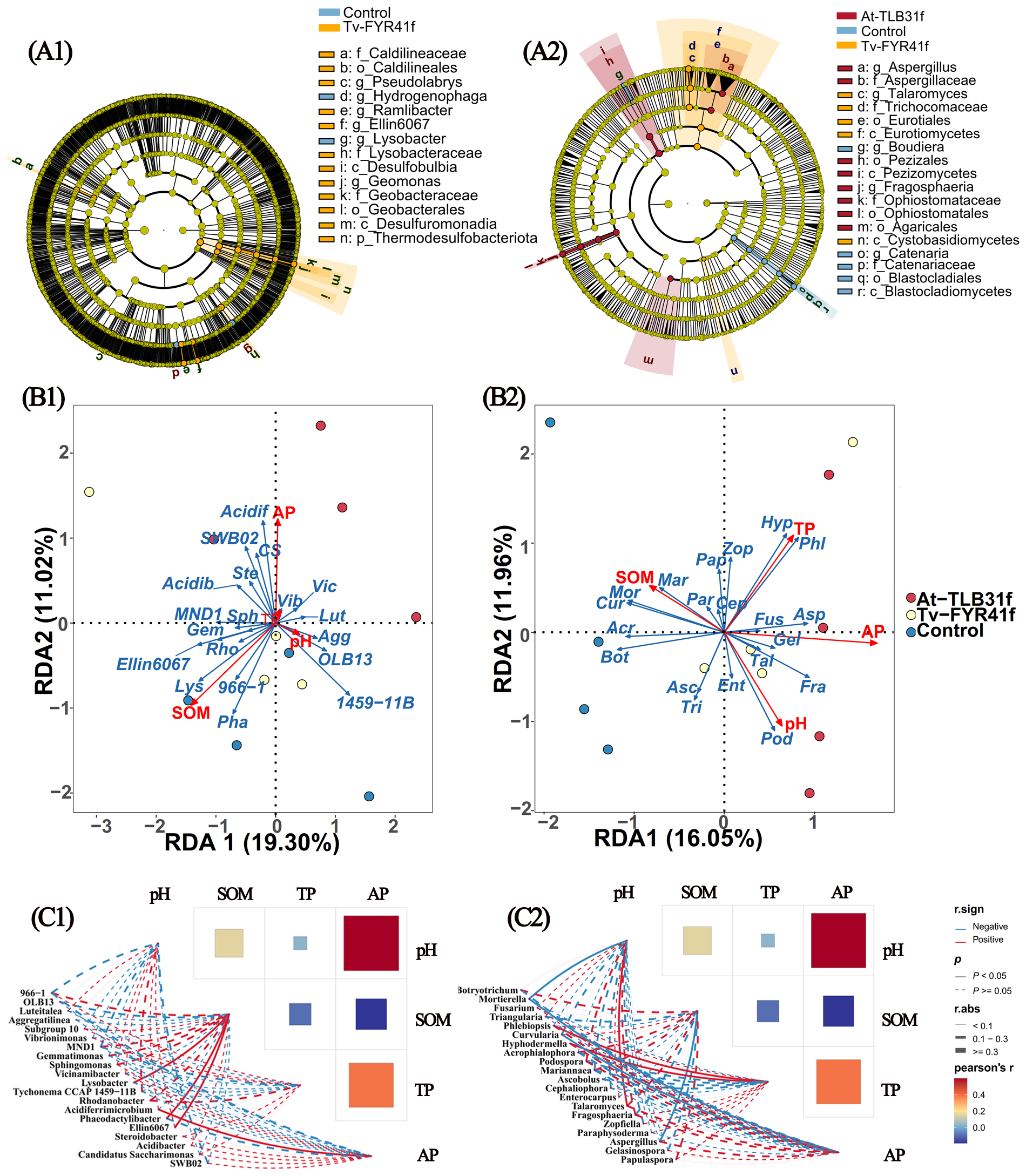
Liner discriminant analysis (LDA) effect size evaluation of bacterial (A1) or fungal (A2) taxa revealed the most differentially abundant taxa among At-TLB31f, Tv-FYR41f treatments, and the control soil communities. Only bacterial taxa with LDA > 3 (p < 0.05) and fungal taxa with LDA > 3.5 (p < 0.05) were shown. RDA (redundancy discriminant analysis) comparing soil characteristics to bacterial (B1) or fungal (B2) populations at the genus level. Diagrams of correlation network between soil properties and bacterial taxa (C1) or fungal taxa (C2) among three different treatments. Lut, Luteitalea, Agg, Aggregatilinea, Vib, Vibrionimonas, Gem, Gemmatimonas, Sph, Sphingomonas, Vic, Vicinamibacter, Lys, Lysobacter, 1459-11B, Tychonema CCAP 1459-11B, Rho, Rhodanobacter, Acidif, Acidiferrimicrobium, Pha, Phaeodactylibacter, Ste, Steroidobacter, Acidib, Acidibacter, CS, Candidatus Saccharimonas, Bot, Botryotrichum, Mor, Mortierella, Fus, Fusarium, Tri, Triangularia, Phl, Phlebiopsis, Cur, Curvularia, Hyp, Hyphodermella, Acr, Acrophialophora, Pod, Podospora, Mar, Mariannaea, Asc, Ascobolus, Cep, Cephaliophora, Ent, Enterocarpus, Tal, Talaromyces, Fra, Fragosphaeria, Zop, Zopfiella, Par, Paraphysoderma, Asp, Aspergillus, Gel, Gelasinospora, Pap, Papulaspora. SOM, soil organic matter; TP, total phosphorus; AP, available phosphorus.
RDA was performed to analyze how soil microbial community composition correlated with soil environmental factors. Indeed, SOM (r2 = 0.74, p = 0.004) and AP (r2 = 0.38, p = 0.117) were the most important factors in explaining the variation in the composition of the bacterial community, with axes 1 (19.30%) and axes 2 (11.02%) accounting for 30.32% of total variations (Figure 8B1). Meanwhile, AP (r2 = 0.80, p = 0.004), TP (r2 = 0.41, p = 0.080), pH (r2 = 0.37, p = 0.135), and SOM (r2 = 0.26, p = 0.281) were vital factors in explaining the variation in the composition of the fungal community, with axes 1 (16.05%) and axes 2 (11.96%) accounting for 28.01% of total variations (Figure 8B2). Simultaneously, correlation network analysis also indicated that soil properties obviously influence the composition of microbial communities at the genus level (Figure 8C). Results showed that Ellin6067, Lysobacter, and Phaeodactylibacter were significantly (p < 0.05) positively correlated with SOM. Acidiferrimicrobium was significantly (p < 0.05) positively correlated with AP, while Curvularia was significantly (p < 0.05) negatively correlated with AP (Figure 8C1). Mortierella was significantly (p < 0.05) negatively correlated with pH and AP. Phlebiopsis and Hyphodermella were significantly (p < 0.05) positively correlated with TP. Talaromyces was significantly (p < 0.05) positively correlated with pH, while Aspergillus was significantly (p < 0.05) negatively correlated with pH (Figure 8C2).
4 Discussion
Food security is a critical global issue that affects both the economy and livelihood, constituting a fundamental pillar of national food security (Liu et al., 2021). The foundation of food production greatly depends on the availability of quality and quantity of arable land. Unregulated expansion of non-grain crops not only jeopardizes food security by leading to shortages and imbalances in food supply but also results in the degradation of soil quality and environmental issues due to changes in land use practices (Cheng et al., 2022; Jiang et al., 2024). The artificial introduction of PGPF into soils represents a cost-effective and eco-friendly strategy for improving soil nutrient conditions and promoting crop growth (Hassan, 2017; Murali et al., 2021).
4.1 Isolation, identification and characterization of PGPF
In order to further enhance the productivity of land converted from non-grain to grain cultivation and facilitate the growth of food crops on such soils, we conducted an isolation of soil fungi by collecting soil samples from six sites where land use had been converted from non-grain to grain cultivation, yielding a total of 215 fungal isolates from 108 soil samples. These obtained fungal isolates were further screened and characterized based on their ability to solubilize phosphate, to produce siderophores, and to synthesize IAA, which has been widely reported to play an important role in growth promotion of plants (Figure 9).
Figure 9
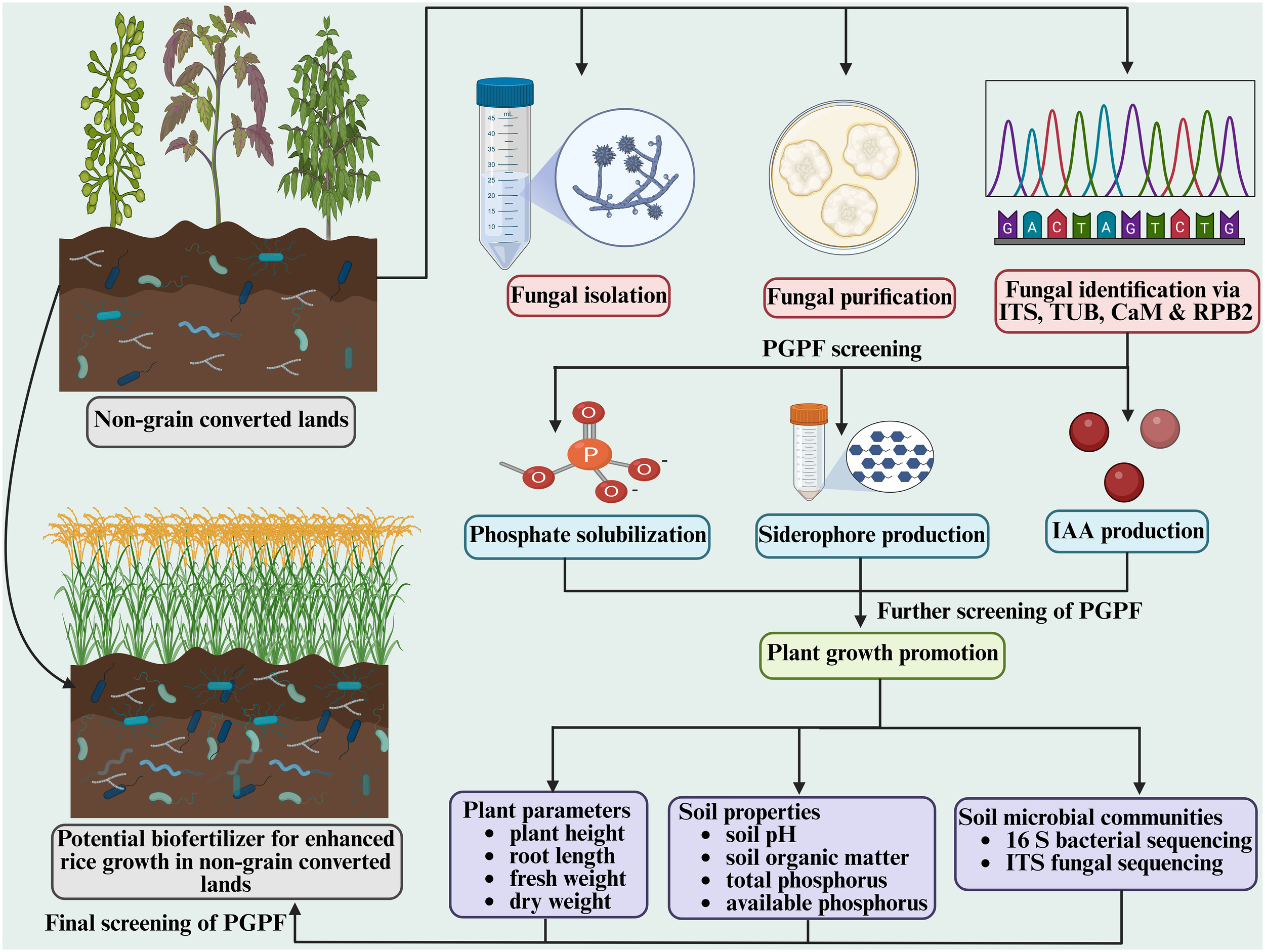
Schematic illustration of isolation, identification and characterization of multiple plant growth-promoting fungi (PGPF) and their effect on rice growth improvement on non-grain converted land.
Phosphorus is a key component of many vital substances and structures within plant cells, such as nucleoproteins, phospholipids, and nucleic acids. It plays a critical role in the physiological and biochemical processes throughout the plant’s life cycle and is indispensable for plant growth and development (Han et al., 2022). However, most phosphorus in the soil exists in insoluble inorganic or organic forms, making it difficult for plants to absorb and utilize (Wang et al., 2021). In this study, initial screening showed that 15 fungal isolates were capable of producing clear phosphate solubilization zones on NBRIP medium, demonstrating a broad ability to solubilize phosphate. Phosphate-solubilizing PGPF can dissolve insoluble forms of phosphorus or mineralize organic phosphorus through mechanisms such as the secretion of organic acids, excretion of protons, or enzymatic production, thereby making phosphorus available for plant uptake (Chen et al., 2019). In agreement with the result of this study, some fungal species including A. brunneoviolaceus, Aspergillus niger, Nigrospora sphaerica, Penicillium corylophilum, and Penicillium chrysogenum have been reported to enhance plant growth by participating in phosphate solubilization (Hassan, 2017; Li et al., 2023).
Additionally, previous studies showed that PGPF that produce siderophores also play a significant role in plant growth and development by improving plant resilience to stress and iron limitation (Sharma et al., 2024). Similarly, our study confirmed that all 15 fungal isolates possess the ability to produce siderophores. PGPF not only promotes crop growth by producing siderophores but also enhances the iron content of food crops, hence addressing nutritional deficiencies in human diets (Kaur et al., 2020). In addition, various phytohormones (particularly IAA) could be produced from PGPF, which could either directly enhance plant growth through regulating root growth, or indirectly induce tolerance to stress situation by modifying growth inhibitors (Ansabayeva et al., 2025). For instance, fungi-produced IAA has been reported to positively influence the root development of eggplant plants (Li et al., 2021). In agreement with previous results, our study indicated that IAA can be produced by the selected PGP fungal isolates in the presence of 0.1% and 1.0% tryptophan, indicating that the fungi isolated from the converted farmland have the potential to stimulate root growth and development. However, the IAA production of these fungal isolates can be affected by the concentration of tryptophan.
Interestingly, the bioassays indicated that all 15 fungal isolates screened in this study differed in their performance in promoting rice growth. For instance, isolates FY-R41f and FY-R31f did not display particularly strong capabilities in phosphate solubilization, siderophore production, or IAA synthesis during in vitro assays, yet they significantly enhanced rice seedling growth in bioassays, substantially increasing seedling height, root length, fresh and dry weight. The discrepancies between in vitro and in vivo results may be mainly due to differences in fungal metabolites or colonization abilities of fungal species in soil. Additionally, several plant growth-promoting isolates, including TL-B31f, FY-R11f, TL-B11f, and CA-R11f were non-pathogenic to the host rice, but significantly increased rice seedling height, root length, fresh weight, and dry weight. The PGP effect on rice seedlings depends on fungal isolates.
Based on a combination of morphological characteristics and phylogenetic analysis of multiple fungal gene sequences, all 15 fungal isolates were classified into six species: A. brunneoviolaceus, A. tubingensis, P. oxalicum, P. limosum, T. veerkampii, and T. purpureogenus. Previous studies have highlighted the potential applications of several of these isolates in agriculture through various mechanisms (Table 5). For example, P. oxalicum isolate UOM PGPF 16 had been used as PGPF to enhance plant growth and induce resistance in pearl millet against downy mildew disease (Murali and Amruthesh, 2015). P. limosum AK-7, isolated from soil, has demonstrated significant antifungal activity, making it a promising resource for developing biological pesticides (Basavarajappa et al., 2023). T. purpureogenus isolate M13026-2, isolated from mushroom substrate, could colonize in the rhizosphere soil stably, and significantly improve the growth of cucumber seedlings (Zhao et al., 2021). A. tubingensis demonstrated the capacity of increasing the bioavailability of phosphorus and potassium in the rhizosphere, and the ability of controlling Fusarium spp (Zapata et al., 2024). Similarly, in our earlier research, we isolated A. tubingensis HZ123, which showed potential as a PGPF by improving eggplant growth and soil fertility (Li et al., 2021).
Table 5
| PGPF | Plants | Mechanisms | References |
|---|---|---|---|
| Aspergillus | |||
| A. brunneoviolaceus (HZ23) | Pakchoi | Improved soil properties (especially phosphorus), rhizosphere bacterial community structure, and metabolites. | Li et al., 2023 |
| A. caespitosus (DS-3) | Fenugreek | Produced IAA. Induced desirable physiological properties (with higher protein content, carbohydrate content, total phenolic content and antioxidant activity than control). | Thakor et al., 2023 |
| A. elegans | Cucumber | Had a zinc solubilizing capacity. | Sidhoum et al., 2024 |
| A. falvus | Wheat | Induced plant resistance against wilt disease. | El-Maraghy et al., 2020 |
| A. niger (9-P) | Common bean | Produced IAA, siderophores, and hogh phosphorus solubilizing activity. Synthesized ACC deminase. | Galeano et al., 2021 |
| A. tubingensis | Common bean | Increased the bioavailability of phosphorus and potassium in the rhizosphere. Promoted nutrition, chlorophyll content, and biometric parameters of plant. Controlled Fusarium spp. | Zapata et al., 2024 |
| Penicillium | |||
| P. buchwaldii | Tomato | Enhanced tomato immunity against root-knot nematodes. | Attia et al., 2025 |
| P. chrysogenum (PenC-JSB9) | Pearl millet | Enhanced seed germination, root length, and shoot length. Induced resistance to downy mildew. | Murali et al., 2013 |
| P. citrinum | Wheat | Induced plant resistance against a pathogen. | El-Maraghy et al., 2020 |
| P. janthinellim | Melon | Elicited IAA. Induced resistance against stem rot caused R. solani. | Murali et al., 2021 |
| P. limosum (AK-7) | / | Exhibited antifungal activities. | Basavarajappa et al., 2023 |
| P. olsonii (A3) | Tobacco | Stimulated tobacco plant growth, enhanced its salt tolerance, and reduced by half the required chemical fertilizer inputs in a hydroponic farming system. | Tarroum et al., 2022 |
| P. oxalicum (UOMPGPF16) | Pearl millet | Enhanced seed germination and seedling vigor. Enhanced NPK uptake. Induced resistance against downy mildew disease. | Murali and Amruthesh, 2015 |
| P. simplicissimum (GP17-2) | Cucumber | Involved multiple defense mechanisms. | Hossain et al., 2007 |
| Talaromyces | |||
| T. purpureogenus (M13026-2) | Cucumber | Colonized in the rhizosphere soil stably. Phosphorus dissolving and siderophores production. | Zhao et al., 2021 |
| T. islandicus | Common bean | Increased the bioavailability of phosphorus and potassium in the rhizosphere. Promoted nutrition, chlorophyll content, and biometric parameters of common bean. Controlled Fusarium spp. | Zapata et al., 2024 |
| T. veerkampii | / | / | / |
| T. wortmannii (FS2) | Cabbage | Promoted growth and induced resistance. | Yamagiwa et al., 2011 |
Mechanism of plant growth promotion by PGPF on different plants.
4.2 Effect of PGPF on rice growth
The novelty in this study is the significant enhancement of rice growth and yield by T. veerkampii. Notably, there is limited literature on the agricultural application of T. veerkampii. In other words, our research further confirms the plant growth-promoting capabilities of A. tubingensis and P. limosum, isolated from non-grain soils, and highlights the potential of T. veerkampii to improve non-grain soils and promote crop growth. These findings provide valuable fungal isolates for the development of novel fungi-based products, such as bio-organic fertilizers and bio-amendments, grounded in PGPF. Furthermore, this study lays a theoretical foundation for improving non-grain soil conditions and enhancing crop growth. Further research should be conducted to elucidate the growth-promoting mechanisms of these fungal isolates.
4.3 Change of soil properties contributes to rice growth by PGPF
Among all 15 PGPF isolates, isolates TL-B31f (identified as A. tubingensis) and FY-R41f (identified as T. veerkampii) with the best growth-promoting properties were selected for further study. In fact, results showed that the soil nutrient elements could be significantly improved by inoculation of isolates TL-B31f and FY-R41f, in particular, the AP of 27.33 and 28.47 mg/kg, respectively, were greater than that of the control (19.17 mg/kg). Aligning with this result, our previous studies indicated that the application of A. brunneoviolaceus isolate HZ23 caused a significant increase in AP compared to the control (854.09 mg/kg vs 38.82 mg/kg) (Li et al., 2023). Phosphate application in farmland could enhance crop yield, while to increase phosphate use efficiency, and improve the fertility of low phosphate soils, which makes it a promising approach for sustainable agriculture (Li et al., 2015; Wang et al., 2024). Therefore, it can be inferred that isolates TL-B31f and FY-R41f may have a great effect on the improvement of the soil quality of non-grain cultivated land.
4.4 Change of microbial communities contributes to rice growth by PGPF
According to previous research, the effect of PGPF on various aspects of plant growth was variable, depending mostly on their successful survival, colonization, growth, and efficient adaptation to environmental conditions. Root colonization was an important strategy of PGPF for plant growth promotion. Upon colonization of the host plant, PGPF can change the microbial community features in the soil, thereby improving the plants’ well-being and productivity through a beneficial association with plants (Adedayo and Babalola, 2023). After 55 days of inoculation, isolates TL-B31f and FY-R41f had an obvious effect in increasing the richness of fungal communities, while the bacterial communities were not significantly changed. In all treatments, Pseudomonadota, Chloroflexota, Acidobacteriota, Bacteroidota, OLB13, Luteitalea, Aggregatilinea, Subgroup 10, Vibrionimonas, MND1, Gemmatimonas, and Sphingomonas were the main bacteria, while Ascomycota, Basidiomycota, Mucoromycota, Botryotrichum, Mortierella, Fusarium, Triangularia, and Phlebiopsis were the main fungi. In fact, more attention should be paid to Pseudomonadota, Chloroflexota, Acidobacteriota, Sphingomonas, Ascomycota, Basidiomycota, Botryotrichum, Mortierella, Fusarium, Triangularia, and Phlebiopsis. Pseudomonadota can mediate nutritional and growth-promotional activities for sustainable food security (Sah et al., 2021). Chloroflexota members may play a role in the nitrogen cycle, thereby improving the nitrogen removal performance in anammox bioreactors and activated sludge systems (Bovio-Winkler et al., 2024). Acidobacteriota is often an important contributor to the nutrient cycling system in soil microhabitats (Zhu et al., 2022a). Sphingomonas possess multifaceted functions ranging from the remediation of environmental contaminants to the production of highly beneficial phytohormones (Asaf et al., 2020). Ascomycota and Basidiomycota have the ability to degrade soil organic matter by producing cellulolytic enzymes (Manici et al., 2024). Mucoromycota can produce lipids, ethanol, organic acids, pigments, and enzymes. Thus, it has been considered a powerful cell factory for modern biorefinery (Dzurendova et al., 2022). Botryotrichum can produce some secondary metabolites that are unfavorable for plant growth (Elkhateeb and Daba, 2022). Mortierella has the ability to increase nutrient uptake efficiency, which has a positive effect on crop protection against adverse conditions, and reduces the use of chemical fertilizers and pesticides (Ozimek and Hanaka, 2020). Fusarium can cause severe economic damage in different agricultural productions such as rice, wheat, potato, etc (Ekwomadu and Mwanza, 2023). Triangularia often grows as saprophytes in the ground among leaf litter or in association with plant roots (Huang et al., 2021). Phlebiopsis has been used as a biocontrol agent against Heterobasidion annosum (Terhonen et al., 2013).
Compared with the control, the RAs of all these microbes were changed by the application of isolates TL-B31f and FY-R41f to potentially increase soil health and decrease rice morbidity. It was particularly worth mentioning that the RAs of Aspergillus were significantly increased by 264.68% in At-TLB31f treatments, while Talaromyces were significantly increased by 1328.35% in Tv-FYR41f treatments. In addition, RDA and correlation network analysis of microbes with soil properties indicated that SOM was significantly positively correlated with Ellin6067, Lysobacter, and Phaeodactylibacter; AP was significantly positively correlated with Acidiferrimicrobium and Mortierella, but was significantly negatively correlated with Curvularia; TP was significantly positively correlated with Phlebiopsis and Hyphodermella; pH was significantly negatively correlated with Mortierella and Aspergillus, but was significantly positively correlated with Talaromyces. In agreement with the results of this study, Guo et al. (2021) showed that SOC, TN, and TP were the most important factors explaining variations in the bacterial community structure. Li et al. (2023) reported that the composition of bacterial communities in packchoi rhizosphere soil was affected significantly by AP, pH, OMC, and TN. Arunrat et al. (2023) found that soil bacterial community was strongly influenced by organic matter and organic carbon in Maize field. The growth of microbes in bayberry rhizosphere soil was affected by many environmental factors, including pH, OMC, AP, and exchangeable magnesium (Ren et al., 2021). Taken overall, compared to the control, the microbial community composition of the two PGPF isolates treatments was reconstructed with an increase or decrease of some specific microbes, and soil properties might play important roles in influencing the growth of microbial communities to modulate rice growth.
5 Conclusions
Overall, 15 PGPF isolated from soils of different non-grain converted land exhibited great ability to solubilize phosphate, to secrete siderophore and to produce IAA, while the effect varied with different fungal strains. Furthermore, all 15 fungal isolates were separated into six distant clades including A. brunneoviolaceus, A. tubingensis, P. oxalicum, P. limosum, T. purpureogenus and T. veerkampii through morphological identification and phylogenetic analysis. Notably, our results indicated that both A. tubingensis and T. veerkampii had significant growth-promoting effects on rice. Based on microbial communities’ analysis of rice root-zone soil after 55 days of inoculation, our findings further highlight their potential to be developed into novel microbial formulations, such as biofertilizers, aimed at improving soil conditions and enhancing crop growth in non-grain production lands.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
XL: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. XR: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. HC: Methodology, Writing – review & editing. YX: Investigation, Writing – review & editing. TZ: Methodology, Writing – review & editing. JY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Validation, Writing – original draft, Writing – review & editing. JX: Data curation, Funding acquisition, Methodology, Project administration, Writing – review & editing. MI: Investigation, Writing – review & editing. TA: Supervision, Visualization, Writing – review & editing. BL: Conceptualization, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. QA: Conceptualization, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Hangzhou Science and Technology Development Plan Project (20231203A05), Science and Technology Innovation and Promotion Demonstration Project of Hangzhou Academy of Agricultural Sciences (2025HNCT-09), Hangzhou City Agricultural Science and Technology Collaboration and Innovation Project (202409SX16), Zhejiang Province Key Research and Development Program of China (2019C02035).
Acknowledgments
We thank United Arab Emirates University for providing a postdoctoral grant on climate action to Qurban Ali (#12S140).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. During the preparation of this work the author(s) used ChatGpt tool to improve language and readability. After using this tool, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abdallah Y. Yang M. Zhang M. Masum M. M. Ogunyemi S. O. Hossain A. et al . (2019). Plant growth promotion and suppression of bacterial leaf blight in rice by Paenibacillus polymyxa Sx3. Lett. Appl. Microbiol.68, 423–429. doi: 10.1111/lam.13117
2
Abou Alhamed M. F. Shebany Y. M. (2012). Endophytic Chaetomium globosum enhances maize seedling copper stress tolerance. Plant Biol.14, 859–863. doi: 10.1111/j.1438-8677.2012.00608.x
3
Adams R. I. Miletto M. Taylor J. W. Bruns T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J.7, 1262–1273. doi: 10.1038/ismej.2013.28
4
Adedayo A. A. Babalola O. O. (2023). Fungi that promote plant growth in the rhizosphere boost crop growth. J. Fungi9, 239. doi: 10.3390/jof9020239
5
Ai C. Zhang S. Zhang X. Guo D. Zhou W. Huang S. (2018). Distinct response of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Genderma319, 156–166. doi: 10.1016/j.geoderma.2018.01.010
6
Akinola S. A. Babalola O. O. (2020). The fungal and archaeal community within plant rhizosphere: A review on their contribution to crop safety. J. Plant Nutr.44, 600–618. doi: 10.1080/01904167.2020.1845376
7
Altschul S. F. Madden T. L. Schäffer A. A. Zhang J. Zhang Z. Miller W. et al . (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.25, 3389–3402. doi: 10.1093/nar/25.17.3389
8
Ansabayeva S. Makhambetov M. Rebouh N. Y. Abdelkader M. Saudy H. S. Hassan K. M. et al . (2025). Plant growth-promoting microbes for resilient farming systems: mitigating environmental stressors and boosting crops productivity—a review. Horticulturae11, 260. doi: 10.3390/horticulturae11030260
9
Arunrat N. Sansupa C. Sereenonchai S. Hatano R. (2023). Stability of soil bacteria in undisturbed soil and continuous maize cultivation in Northern Thailand. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1285445
10
Arunrat N. Sansupa C. Sereenonchai S. Hatano R. (2024). Short-term response of soil bacterial and fungal communities to fire in rotational shifting cultivation, northern Thailand. Appl. Soil Ecol.196, 105303. doi: 10.1016/j.apsoil.2024.105303
11
Asaf S. Numan M. Khan A. L. Al-Harrasi A. (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol.40, 138–152. doi: 10.1080/07388551.2019.1709793
12
Attia M. S. Abdelaziz A. M. Abdelglil M. I. Kandil E. K. Ergun M. Elsayed S. M. et al . (2025). Enhancing tomato immunity against root-knot nematodes using PGPF and Melithorin® through biochemical and molecular approaches. Physiol. Mol. Plant P.139, 102791. doi: 10.1016/j.pmpp.2025.102791
13
Banerjee S. van der Heijden M. G. A. (2023). Soil microbiomes and one health. Nat. Rev. Microbiol.21, 6–20. doi: 10.1038/s41579-022-00779-w
14
Baran A. Mierzwa-Hersztek M. Gondek K. Tarnawski M. Szara M. Gorczyca O. et al . (2019). The influence of the quantity and quality of sediment organic matter on the potential mobility and toxicity of trace elements in bottom sediment. Environ. Geochem. Health41, 2893–2910. doi: 10.1007/s10653-019-00359-7
15
Basavarajappa D. S. Niazi S. K. Bepari A. Assiri R. A. Hussain S. A. Muzaheed et al . (2023). Efficacy of Penicillium limosum strain AK-7 derived bioactive metabolites on antimicrobial, antioxidant, and anticancer activity against human ovarian teratocarcinoma (PA-1) cell line. Microorganisms11, 2480. doi: 10.3390/microorganisms11102480
16
Bitas V. McCartney N. Li N. Demers J. Kim J. E. Kim H. S. et al . (2015). Fusarium oxysporum volatiles enhance plant growth via affecting auxin transport and signaling. Front. Microbiol.6. doi: 10.3389/fmicb.2015.01248
17
Bovio-Winkler P. Cbezas A. Etchebehere C. (2024). Unveiling the hidden diversity and functional role of Chloroflexota in full-scale wastewater treatment plants through genome-centric analyses. ISME Commun.4, ycae050. doi: 10.1093/ismeco/ycae050
18
Brookes P. C. Landman A. Pruden G. Jenkinson D. S. (1985). Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil. Boil. Biochem.17, 837–842. doi: 10.1016/0038-0717(85)90144-0
19
Caporaso J. G. Kuczynski J. Stombaugh J. Bittinger K. Bushman F. D. Costello E. K. et al . (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods7, 335–336. doi: 10.1038/nmeth.f.303
20
Chen H. Zhang J. Tang L. Su M. Tian D. Zhang L. et al . (2019). Enhanced Pb immobilization via the combination of biochar and phosphate solubilizing bacteria. Environ. Int.127, 395–401. doi: 10.1016/j.envint.2019.03.068
21
Chen Q. Song Y. An Y. Lu Y. Zhong G. (2024). Soil microorganisms: their role in enhancing crop nutrition and health. Diversity16, 734. doi: 10.3390/d16120734
22
Cheng X. Tao Y. Huang C. Yi J. Yi D. Wang F. et al . (2022). Unraveling the causal mechanisms for non-grain production of cultivated land: An analysis framework applied in Liyang, China. Land11, 1888. doi: 10.3390/land11111888
23
Chuang C. C. Kuo Y. L. Chao C. C. Chao W. L. (2007). Solubilization of inorganic phosphates and plant growth promotion by Aspergillus Niger. Biol. Fertil. Soils44, 415–416. doi: 10.1007/s00374-007-0236-4
24
Deng H. Ma X. Liu Z. Hu H. Di H. Liu Y. et al . (2024). Soil ecosystem multifunctionality is strongly linked with crop yield after four decades chemical fertilization in black soil. Agr. Ecosys. Environ.368, 109007. doi: 10.1016/j.agee.2024.109007
25
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci.14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
26
Dzurendova S. Losada C. B. Dupuy-Galet B. X. Fjær K. Shapaval V. (2022). Mucoromytota fungi as powetful cell factories for modern biorefinery. Appl. Microbiol. Biotechnol.106, 101–115. doi: 10.1007/s00253-021-11720-1
27
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods10, 996–998. doi: 10.1038/nmeth.2604
28
Edgar R. C. (2016). SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv.2016, 74161. doi: 10.1101/074161
29
Ekwomadu T. I. Mwanza M. (2023). Fusarium fungi pathogens, identification, adverse effects, disease management, and global food security: a review of the latest research. Agriculture13, 1810. doi: 10.3390/agriculture13091810
30
Elkhateeb W. A. Daba G. M. (2022). Botryotrichum and scopulariopsis secondary metabolites and biological activities. Biotechnol. Bioproces.3, 67. doi: 10.31579/2766-2314/067
31
El-Maraghy S. S. Tohamy T. A. Hussein K. A. (2020). Role of plant-growth promoting fungi (PGPF) in defensive genes expression of Triticum aestivum against wilt disease. Rhizosphere15, 100223. doi: 10.1016/j.rhisph.2020.100223
32
Galeano R. M. S. Franco D. G. Chaves P. O. Giannesi G. C. Masui D. C. Ruller R. et al . (2021). Plant growth promoting potential of endophytic Aspergillus Niger 9-p isolated from native forage grass in Pantanal of Nhecolândia region, Brazil. Rhizosphere18, 100332. doi: 10.1016/j.rhisph.2021.100332
33
Guo J. Wu Y. Wu X. Ren Z. Wang G. (2021). Soil bacterial community composition and diversity response to land conversion is depth-dependent. Glob. Ecol. Conerv.32, e01923. doi: 10.1016/j.gecco.2021.e01923
34
Haituk S. Suwannarach N. Hongsanan S. Senwanna C. Cheewangkoon R. (2021). New genus of epiphytic sooty mold: Alloscorias syngonii (Readerielliopsidaceae) from Thailand. Phytotaxa507, 271–282. doi: 10.11646/phytotaxa.507.4.1
35
Han Y. White P. J. Cheng L. (2022). Mechanisms for improving phosphorus utilization efficiency in plants. Ann. Bot.129, 247–258. doi: 10.1093/aob/mcab145
36
Hashim N. Ali M. M. Mahadi M. R. Abdullah A. F. Wayayok A. Kassim M. S. M. et al . (2024). Smart farming for sustainable rice production: an insight into application, challenge, and future prospect. Rice Sci.31, 47–61. doi: 10.1016/j.rsci.2023.08.004
37
Hassan S. E. D. (2017). Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res.8, 687–695. doi: 10.1016/j.jare.2017.09.001
38
He H. Peng M. Hou Z. Li J. (2024). Organic amendment substitution improves the sustainability of wheat fields changed from cotton and vegetable fields. Agr. Ecosys. Environ.359, 108769. doi: 10.1016/j.agee.2023.108769
39
Hong S. B. Go S. J. Shin H. D. Frisvad J. C. Samson R. A. (2005). Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia97, 1316–1329. doi: 10.3852/mycologia.97.6.1316
40
Houbraken J. Kocsubé S. Visagie C. M. Yilmaz N. Wang X-C. Meijer M. et al (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud. Mycol.95, 5–169. doi: 10.1016/j.simyco.2020.05.002
41
Hossain M. Sultana F. (2020). “Application and machanisms of plant growth promoting fungi (PGPF) for phytostimulation,” in Organic Agriculture (Rijeka, Croatia: IntechOpen), 1–31. doi: 10.5772/intechopen,92338
42
Hossain M. Sultana F. Kubota M. Koyama H. Hyakumachi M. (2007). The plant growth-promoting fungus Penicillium simplicissimum GP17–2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol.48, 1724–1736. doi: 10.1093/pcp/pcm144
43
Huang S. K. Hyde K. D. Mapook A. Maharachchikumbura S. S. N. Bhat J. D. McKenzie E. H. C. et al . (2021). Taxonomic studies of some often over-looked Diaporthomycetidae and Sordariomycetidae. Fungal Divers.111, 443–572. doi: 10.1007/s13225-021-00488-4
44
Jackson M. L. (1973). Soil Chemical Analysis (New Delhi: Prentice Hall of India Pvt. Ltd.), 498.
45
Jiang C. Wang L. Guo W. Chen H. Liang A. Sun M. et al . (2024). Spatio-temporal evolution and multi-scenario simulation of non-grain production on cultivated land in Jiangsu Province, China. Land13, 670. doi: 10.3390/land13050670
46
Kaur T. Rana K. L. Kour D. Sheikh I. Yadav N. Kumar V. et al . (2020). “Chapter 1 - Microbe-mediated biofortification for micronutrients: Present status and future challenges,” in New and Future Developments in Microbial Biotechnology and Bioengineering. Eds. Ali Asghar RastegariA. A.YadavA. N.YadavN. (Leiden, Netherlands: Elsevier), 1–17. doi: 10.1016/B978-0-12-820528-0.00002-8
47
Kumar S. Stecher G. Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol.33, 1870–1874. doi: 10.1093/molbev/msw054
48
Li Z. Bai T. Dai L. Wang F. Tao J. Meng S. et al . (2016). A study of organic acid production in contrasts between two phosphate solubilizing fungi: Penicillium oxalicum and Aspergillus Niger. Sci. Rep.6, 25313. doi: 10.1038/srep25313
49
Li X. Li D. Jiang Y. Xu J. Ren X. Zhang Y. et al . (2023). The effects of microbial fertilizer based Aspergillus brunneoviolaceus HZ23 on pakchoi growth, soil properties, rhizosphere bacterial community structure, and metabolites in newly reclaimed land. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1091380
50
Li X. Li D. Yan J. Zhang Y. Wang H. Zhang J. et al . (2021). Effect of plant-growth-promoting fungi on eggplant (Solanum melongena L.) in new reclamation land. Agriculture11, 1036. doi: 10.3390/agriculture11111036
51
Li H. G. Liu J. Li G. H. Shen J. B. Zhang F. S. (2015). Past, present, and future use of phosphorus in Chinese agriculture and its influence on phosphorus losses. Ambio44, 274–285. doi: 10.1007/s13280-015-0633-0
52
Li B. Zhu D. Li J. Liu X. Yan B. Mao L. et al . (2024). Converting upland to paddy fields alters soil nitrogen microbial functions at different depths in black soil region. Agr. Ecosys. Environ.372, 109089. doi: 10.1016/j.aee.2024.109089
53
Liu Z. Zhong H. Li Y. Wen Q. Liu X. Jian Y. (2021). Change in grain production in China and its impacts on spatial supply and demand distributions in recent two decades. J. Nat. Resour.36, 1413. doi: 10.31497/zrzyxb.20210605
54
Manici L. M. Caputo F. De Sabata D. Formasier F. (2024). The enzyme patterns of Ascomycota and Basidiomycota fungi reveal their different functions in soil. Appl. Soil Ecol.196, 105323. doi: 10.1016/j.apsoil.2024.105323
55
Martin M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12. doi: 10.14806/ej.17.1.200
56
Murali M. Amruthesh K. N. (2015). Plant growth-promoting fungus Penicillium oxalicum enhances plant growth and induces resistance in pearl millet against downy mildew disease. J. Phytopathol.163, 743–754. doi: 10.1111/jph.12371
57
Murali M. Naziya B. Ansari M. A. Alomary M. N. AlYahya S. Almatroudi A. et al . (2021). Bioprospecting of rhizosphere-resident fungi: their role and importance in sustainable agriculture. J. Fungi7, 314. doi: 10.3390/jof7040314
58
Murali M. Sudisha J. Amruthesh K. N. Ito S. I. Shetty H. S. (2013). Rhizosphere fungus Penicillium chrysogenum promotes growth and induces defence-related genes and downy mildew disease resistance in pearl millet. Plant Biol.15, 111–118. doi: 10.1111/j.1438-8677.2012.00617.x
59
Murugappan R. Aravinth A. Karthikeyan M. (2011). Chemical and structural characterization of hydroxamate siderophore produced by marine Vibrio harveyi. J. Ind. Microbiol. Biotechnol.38, 265–273. doi: 10.1007/s10295-010-0769-7
60
Nasri T. Hedayati M. T. Abastabar M. Pasqualotto A. C. Armaki M. T. Hoseinnejad A. et al . (2015). PCR-RFLP on beta-tubulin gene for rapid identification of the most clinically important species of Aspergillus. J. Microbiol. Methods117, 144–147. doi: 10.1016/j.mimet.2015.08.007
61
Naziya B. Murali M. Amruthesh K. N. (2019). Plant growth-promoting fungi (PGPF) instigate plant growth and induce disease resistance in Capsicum annuum L. upon infection with Colletotrichum capsici (Syd.) Butler & Bisby. Biomolecules10, 41. doi: 10.3390/biom10010041
62
Naznin H. A. Kimura M. Miyazawa M. Hyakumachi M. (2013). Analysis of volatile organic compounds emitted by plant growth-promoting fungus Phoma sp. GS8–3 for growth promotion effects on tobacco. Microbes Environ.28, 42–49. doi: 10.1264/jsme2.ME12085
63
Ozimek E. Hanaka A. (2020). Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture11, 7. doi: 10.3390/agriculture1101007
64
Panagos P. Meusburger K. Ballabio C. Borrelli P. Alewell C. (2014). Soil erodibility in Europe: a high-resolution dataset based on LUCAS. Sci. Total Environ.479, 189–200. doi: 10.1016/j.scitotenv.2014.02.010
65
Pedrinho A. Mendes L. W. de Araujo Pereira A. P. Araujo A. S. F. Vaishnav A. Karpouzas D. G. et al . (2024). Soil microbial diversity plays an important role in resisting and restoring degraded ecosystems. Plant Soil500, 325–349. doi: 10.1007/s11104-024-06489-x
66
Quast C. Pruesse E. Yilmaz P. Gerken J. Schweer T. Yarza P. et al . (2013). The SILVA ribosomal RNA gene data base project: improved data processing and web-based tools. Nucleic Acids Res.41, 590–596. doi: 10.1093/nar/gks1219
67
Radhakrishnan R. Khan A. L. Kang S. M. Lee I. J. (2015). A comparative study of phosphate solubilization and the host plant growth promotion ability of Fusarium verticillioides RK01 and Humicola sp. KNU01 under salt stress. Ann. Microbiol.65, 585–593. doi: 10.1007/s13213-014-0894-z
68
Ramette A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol.62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
69
Ren H. Wang H. Yu Z. Zhang S. Qi X. Sun L. et al . (2021). Effect of two kinds of fertilizers on growth and rhizosphere soil properties of bayberry with decline disease. Plants10, 2386. doi: 10.3390/plants10112386
70
Romão I. R. do Carmo Gomes J. Silva D. Vilchez J. I. (2025). The seed microbiota from an application perspective: an underexplored frontier in plant–microbe interactions. Crop Health3, 12. doi: 10.1007/s44297-025-00051-6
71
Sah S. Krishnani S. Singh R. (2021). Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr. Res. Microb. Sci.2, 100084. doi: 10.1016/j.crmiccr.2021.100084
72
Samson R. A. Visagie C. M. Houbraken J. Hong S. B. Hubka V. Klaassen C. H. W. et al . (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol.78, 141–173. doi: 10.1016/j.simyco.2014.07.004
73
Schmieder R. Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics27, 863–864. doi: 10.1093/bioinformatics/btr026
74
Segata N. Izard J. Waldron L. Gevers D. Miropolsky L. Garrett W. S. et al . (2011). Metagenomic biomarker discovery and explanation. Genome Biol.12, R60. doi: 10.1186/gb-2011-12-6-r60
75
Sharma M. Devi S. Manorma K. Kesta K. Chand S. Sharma R. et al . (2024). “Chapter 18 - Plant growth-promoting fungi: a tool for agriculturally important industrial production,” in Developments in Applied Microbiology and Biotechnology, Microbial Essentialism. Eds. SinghR. P.ManchandaG.SarsanS.KumarA.PanosyanH. (New York, USA: Academic Press), 93–418. doi: 10.1016/B978-0-443-13932-1.00016-7
76
Sidhoum W. Dib S. Alim Y. Anseur S. Benlatreche S. Belaidouni Z. M. et al . (2024). Gowwth-promoting effects of Aspergillus elegans and the dark septate endophyte (DSE) periconia macrospinosa on cucumber. Arch. Microbiol.206, 226. doi: 10.1007/s00203-024-03958-w
77
Sofo A. Tataranni G. Xiloyannis C. Dichio B. Scopa A. (2012). Direct effects of Trichoderma harzianum strain T-22 on micropropagated shoots of GiSeLa6® (Prunus cerasus × Prunus canescens) rootstock. Environ. Exp. Bot.76, 33–38. doi: 10.1016/j.envexpbot.2011.10.006
78
Tarroum M. Romdhane W. B. Al-Qurainy F. Abdelrahim A. Ali M. Al-Doss A. et al . (2022). A novel PGPF Penicillium olsonii isolated from the rhizosphere of Aeluropus littoralis promotes plant growth, enhances salt stress tolerance, and reduces chemical fertilizers inputs in hydroponic system. Front. Microbiol.13. doi: 10.3389/fmicb.2022.996054
79
Terhonen E. Sun H. Buée M. Kasanen R. Paulin L. Asiegbu F. O. (2013). Effects of the use of biocontrol agent (Phlebiopsis gigantean) on fungal communities on the surface of Picea abies stumps. Forest. Ecol. Manage.310, 428–433. doi: 10.1016/j.foreco.2013.8.044
80
Thakor R. Mistry H. Bariya H. (2023). Efficacy of indole-3-acetic acid-producing PGPFs and their consortium on physiological and biochemical parameters of Trigonella foenum-graecum L. Hortic. Environ. Biotechnol.64, 533–546. doi: 10.1007/s13580-023-00512-3
81
Tian Y. Zhao Y. Fu X. Yu C. Gao K. Liu H. (2021). Isolation and identification of Talaromyces sp. strain Q2 and its biocontrol mechanisms involved in the control of Fusarium Wilt. Front. Microbiol.12. doi: 10.3389/fmicb.2021.724842
82
Tippmann H. F. (2004). Analysis for free: Comparing programs for sequence analysis. Brief. Bioinform.5, 82–87. doi: 10.1093/bib/5.1.82
83
Tomaha A. A. Abd Alamera I. S. Li B. Zhang J. (2020). A new species of Trichoderma and gliotoxin role: A new observation in enhanc ing biocontrol potential of T. virens against Phytophthora capsici on chili pepper. Biol. Control.145, 104261. doi: 10.1016/j.biocontrol.2020.104261
84
Tsang C. C. Tang J. Y. M. Lau S. K. P. Woo P. C. Y. (2018). Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era– Past, present and future. Comput. Struct. Biotec. J.16, 197–210. doi: 10.1016/j.csbj.2018.05.003
85
Urmas K. Nilsson R. H. Abarenkov K. Tedersoo L. Tayloy A. F. S. Bahram M. et al . (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol.22, 5271–5277. doi: 10.1111/mec.12481
86
Vance E. D. Brookes P. C. Jenkinson D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil. Boil. Biochem.19, 703–707. doi: 10.1016/0038-0717(87)90052-6
87
Wagg C. Hautier Y. Pellkofer S. Banerjee S. Schmid B. van der Heijden M. G. A. (2021). Diversity and asynchrony in soil microbial communities stabilizes ecosystem functioning. eLife10, e62813. doi: 10.7554/eLife.62813
88
Wang L. Fang H. Xue Z. De J. Guo X. F. (2023). Agrochemical exposure-induced seed microbiome response in barley. Crop Health1, 16. doi: 10.1007/s44297-023-00013-w
89
Wang Q. Garrity G. M. Tiedje J. M. Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol.73, 5261–5267. doi: 10.1128/AEM.00062-07
90
Wang L. Rengel Z. Cheng L. Shen J. (2024). Coupling phosphate type and placement promotes maize growth and phosphorus uptake by altering root properties and rhizosphere processes. Field Crop Res.306, 109225. doi: 10.1016/j.fcr.2023.109225
91
Wang L. Sheng M. Li S. Wu J. (2021). Patterns and dynamics of plant diversity and soil physical-chemical properties of the karst rocky desertification ecosystem, SW China. Pol. J. Environ. Stud.30, 1393–1408. doi: 10.15244/pjoes/124225
92
Wu L. Y. Wen C. Q. Qin Y. J. Yin H. Q. Tu Q. C. Nostrand J. D. V. et al . (2015). Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol.15, 125. doi: 10.1186/s12866-015-0450-4
93
Yadav R. L. Shukla S. K. Suman A. Singh P. N. (2009). Trichoderma inoculation and trash management effects on soil microbial biomass, soil respiration, nutrient uptake and yield of ratoon sugarcane under subtropical conditions. Biol. Fertil. Soils45, 461–468. doi: 10.1007/s00374-009-0352-4
94
Yamagiwa Y. Inagaki Y. Ichinose Y. Toyoda K. Hyakumachi M. Shiraishi T. (2011). Talaromyces wortmannii FS2 emits β -caryphyllene, which promotes plant growth and induces resistance. J. Gen. Plant Pathol.77, 336–341. doi: 10.1007/s10327-011-0340-z
95
Yang Q. Zhang D. (2021). The influence of agricultural industrial policy on non-grain production of cultivated land: a case study of the “one village, one product” strategy implemented in Guanzhong Plain of China. Land Use Pol.108, 105579. doi: 10.1016/j.landusepol.2021.105579
96
Yuan S. Linquist B. A. Wilson L. T. Cassman K. G. Stuart A. M. Pede V. et al . (2021). Sustainable intensification for a larger global rice bowl. Nat. Commun.12, 7163. doi: 10.1038/s41467-021-27424-z
97
Yuan M. Seale J. L. Jr. Wahl T. Bai J. (2019). The changing dietary patterns and health issues in China. China Agric. Econ. Rev.11, 143–159. doi: 10.1108/CAER-12-2017-0254
98
Zapata D. López J. E. Saldarriaga J. F. (2024). Plant growth-promoting and biocontrol potential of Aspergillus tubingensis and Talaromyces islandiccus. J. Soil Sci. Plant Nutr.24, 2354–2370. doi: 10.1007/s42729-024-01633-z
99
Zhang J. Kobert K. Flouri T. Stamatakis A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics30, 614–620. doi: 10.1093/bioinformatics/btt593
100
Zhang W. Ma L. Wang X. Chang X. Zhu Z. (2024). The impact of non-rain conversion of cultivated land on the relationship between agricultural carbon supply and demand. Appl. Geogr.162, 103166. doi: 10.1016/j.apgeog.2023.103166
101
Zhao L. Zhao W. Deng H. (2021). Effects of Talaromyces purpureogenus on cucumber growth promotion and it mechanism. J. Bacteriol Mycol.8, 1173. doi: 10.26420/jbacteriolmycol.2021.1173
102
Zhou X. Zhaang Q. Yan Y. Qu J. Zhou J. Zhao J. et al . (2025). Effects of soil management strategies based on different principles on soil microbial communities and the outcomes for plant health. Biol. Control201, 105708. doi: 10.1016/j.biocontrol.2025.105708
103
Zhu Z. Duan J. Li R. Feng Y. (2022b). Spatial evolution, driving mechanism, and patch prediction of grain-producing cultivated land in China. Agriculture12, 860. doi: 10.3390/agriculture12060860
104
Zhu Y. Wang L. You Y. Cheng Y. Ma J. Chen F. (2022a). Enhancing network complexity and function of soil bacteria by thiourea-modified biochar under cadmium stress in post-mining area. Chemosphere302, 134811. doi: 10.1016/j.chemosphere.2022.13481
Summary
Keywords
non-grain converted lands, plant growth promoting fungi, identification, soil properties, microbial community
Citation
Li X, Ren X, Chen H, Xin Y, Zhou T, Yan J, Xu J, Ijaz M, Ahmed T, Li B and Ali Q (2025) Isolation of multiple plant growth-promoting fungi and their effect on rice growth improvement on non-grain converted land. Front. Plant Sci. 16:1618073. doi: 10.3389/fpls.2025.1618073
Received
25 April 2025
Accepted
14 July 2025
Published
13 August 2025
Volume
16 - 2025
Edited by
Muhammad Qadir, Hunan University, China
Reviewed by
Murali M., University of Mysore, India
Miral Javed, University of Guelph, Canada
Updates
Copyright
© 2025 Li, Ren, Chen, Xin, Zhou, Yan, Xu, Ijaz, Ahmed, Li and Ali.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianli Yan, yanjianli00@gmail.com; Jun Xu, 15292165@qq.com; Qurban Ali, rattarqurban@uaeu.ac.ae
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.