- 1College of Life Sciences, College of Tea Sciences, The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou University, Guiyang, China
- 2Plant Conservation and Breeding Technology Center, Institute of Crop Germplasm Resources, Guizhou Academy of Agricultural Sciences, Guiyang, China
Eucommia ulmoides Oliv., a Tertiary period relict tree species endemic to China, is a rubber-producing plant valued for both medicinal and edible applications. E.ulmoides rubber is a high-quality natural rubber prized for its excellent elasticity, abrasion resistance, and insulation properties, leading to broad industrial applications. Previous research identified the EuUSP16 gene, encoding a protein containing E.ulmoides rubber particle protein peptides. While overexpression of EuUSP16 in tobacco enhanced drought tolerance, its role in E.ulmoides rubber biosynthesis remained undefined. In this study, we identified 29 EuUSP genes at the whole-genome level in E.ulmoides. Following low-temperature and drought treatments, the expression level of the EuUSP16 gene was found to be positively correlated with changes in rubber content (p<0.05), suggesting its potential regulatory role in rubber synthesis. In E.ulmoides subjected to Agrobacterium-mediated EuUSP16 gene overexpression or silencing, the expression levels of key E.ulmoides rubber biosynthesis enzyme genes, such as EuFPS1, exhibited corresponding increases and decreases, respectively. Furthermore, rubber content in EuUSP16-overexpressing callus increased by 254.51% compared to wild-type callus. These findings indicate that EuUSP16 regulates E.ulmoides rubber biosynthesis by modulating the expression of these genes. The 1,967 bp promoter region upstream of the EuUSP16 ATG start codon contains several responsive elements, including MBS (MYB-binding site; CAACTG), LTR (low-temperature responsive element; CCGAAA), ABRE (ABA-responsive element; ACGTG), and a Dof transcription factor binding motif (AAAG). Promoter activity assays showed that EuUSP16 promoter activity was induced by low temperature and drought but repressed by abscisic acid (ABA) treatment. Furthermore, using yeast one-hybrid screening, we identified a Cys2-Cys2 zinc finger domain-containing transcription factor, designated EuDof. Interaction analysis revealed that the EuDof transcription factor enhances the activity of the EuUSP16 promoter. The binding of EuDof to the EuUSP16 promoter was enhanced under low temperature and drought stress but inhibited by ABA. Collectively, this study provides crucial insights into the regulatory mechanism of EuUSP16 in E.ulmoides rubber biosynthesis.
1 Introduction
Eucommia ulmoides Oliv. is a unique Tertiary relict tree species endemic to China, valued for over a millennium for its medicinal properties and, critically, as the source of E.ulmoides rubber (Wei et al., 2021; Zhu and Sun, 2018). This natural rubber, chemically defined as trans-1,4-polyisoprene (TPI), serves as the structural isomer of the cis-1,4-polyisoprene (CPI) from Hevea brasiliensis (Wei et al., 2021). Derived from secondary metabolism, E.ulmoides rubber is a vital natural polymer material with significant applications in aerospace, national defense, healthcare, and other advanced industries (Du et al., 2011). Its biosynthesis originates from isopentenyl pyrophosphate (IPP), produced via the mevalonic acid (MVA) or methylerythritol phosphate (MEP) pathways, central to isoprenoid metabolism which also generates terpenoids and other crucial bioactives (Miziorko, 2011; Men et al., 2019; Rohmer, 1999).
Universal Stress Proteins (USPs) represent a class of highly conserved, stress-induced proteins ubiquitous across diverse organisms (Ren et al., 2022). They play significant roles in plant responses to abiotic stresses. Characterized by a conserved C-terminal UspA domain (140–160 amino acids), USP proteins interact with various functional motifs, enabling diverse functions including substance transport, signal transduction, cell defense, and antioxidant responses (Aravind et al., 2002; Gustavsson et al., 2002). Numerous studies demonstrate their importance in stress tolerance, for instance, specific USP genes modulate salt tolerance in Arabidopsis thaliana, enhance growth and ion homeostasis under stress in transgenic tobacco, and overexpression of the VyUSPA3 gene can improve drought tolerance in Chinese wild grapevine (Vitis yeshanensis), often interacting with hormone signaling (e.g., ethylene, abscisic acid), reactive oxygen species (ROS) scavenging, and ubiquitination pathways (Bhuria et al., 2022; Cui et al., 2022; Udawat et al., 2016).
While the isoprenoid metabolic pathway underpinning E.ulmoides rubber synthesis is known to be modulated by environmental stresses, the specific molecular regulators linking stress perception to enhanced rubber accumulation remain poorly understood. Intriguingly, our laboratory previously identified that the amino acid sequence of EuUSP16 protein contains a conserved rubber particle protein motif (-WALDNLADKGDTLYVLHLK-) (Hu and Zhao, 2024). This suggests a potential, yet unexplored, role for specific USPs, particularly those containing such motifs, in regulating E.ulmoides rubber biosynthesis in response to environmental cues. We hypothesize that EuUSPs gene containing this rubber particle motif, such as EuUSP16, regulate rubber synthesis under abiotic stress conditions.
Building upon the prior cloning of EuUSP16 and the discovery of its rubber particle motif, this study aims to: systematically identify and characterize the EuUSPs gene family within E.ulmoides; analyze changes in E.ulmoides rubber content under adverse environmental conditions; and investigate the specific role of EuUSP16 in E.ulmoides rubber synthesis and elucidate its underlying regulatory mechanisms. This work provides a crucial foundation for understanding the functional role of USPs in stress-induced rubber biosynthesis in E.ulmoides.
2 Materials and methods
2.1 Plant materials, growing conditions and treatment methods
E.ulmoides seedlings were cultivated in the following conditions: 16h light/8h dark photoperiod at 25°C and 70% relative humidity. Leaves, roots, and stem segments were collected from three-month-old E.ulmoides seedlings. Additionally, 15-month-old E.ulmoides seedlings subjected to natural drought or low-temperature stress for 10 days were sampled, with untreated 15-month-old seedlings serving as controls. All the samples were frozen in liquid nitrogen after collection and stored at −80°C until RNA isolation. Total RNA was extracted using Plant RNA Extraction Kit. All samples were processed in three biological replicates.
2.2 Analysis of the EuUSPs gene family
The genomic annotation data of E.ulmoides (Accession Number: CNA0148166), previously assembled by our research group from Guizhou Province, China, was downloaded from the CNGBdb database (https://db.cngb.org/). Concurrently, 42 A.thaliana USP gene and protein sequences were retrieved from the Arabidopsis Information Resource (TAIR) and UniProt databases. Putative E.ulmoides USP sequences were identified through local BLAST searches against the E.ulmoides genome using the A.thaliana sequences as queries. Additionally, protein sequences containing the USP (PF00582) domain were searched using HMMER 3.0 based on the InterPro-derived Hidden Markov Model (HMM) profile. Candidate E.ulmoides USP proteins obtained from these methods were combined into a non-redundant set. The conserved domains of these candidates were further validated using the NCBI Conserved Domain Database (CDD) and SMART. This comprehensive analysis ultimately identified the members of the E.ulmoides USP (EuUSPs) gene family. Genome-wide USP family sequences for A.thaliana, Oryza sativa (rice), H.brasiliensis (rubber tree), and Taraxacum kok-saghyz (rubber dandelion) were acquired from the Ensembl Plants database (http://plants.ensembl.org/index.html). A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method in MEGA 12.0 software, based on the alignment of corresponding amino acid sequences performed with ClustalW (1000 bootstrap replicates). The resulting phylogenetic tree was visualized and refined using the iTOL web platform (https://itol.embl.de/). Subcellular localization of the EuUSPs proteins was predicted using WoLF PSORT, CELLO, and Plant-mPLoc. Conserved motifs were identified using the online MEME suite. Genomic BLAST alignments between E.ulmoides, A.thaliana, and H.brasiliensis were performed using TBtools to generate synteny and tandem duplication files, which were used to plot syntenic gene relationships. Gene Ontology (GO) analysis for the EuUSPs genes was conducted using eggNOG-mapper.
2.3 The influence of environmental treatments on the expression levels of EuUSPs genes
qRT-PCR primers for 11 genes in the EuUSPs gene family were designed using Premier 5.0 and Primer-BLAST (Supplementary Table 1) and synthesized by Sangon Biotech. qRT-PCR was performed using SYBR GreenI dye-based qPCR premix. The reaction mixture (10 μL) consisted of: 5 μL of 2× Universal Blue SYBR Green qPCR Master Mix, 1 μL of cDNA, 0.2 μL each of forward and reverse primers (10 μmol·L-¹), and 3.6 μL of ddH2O. The reaction program was as follows: initial denaturation at 95°C for 30 s; followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), with the EuActin gene as the internal reference. At least three independent biological replicates and three technical replicates were performed for each sample.
2.4 Functional analysis of EuUSP16 gene in E.ulmoides rubber synthesis
The previously constructed pSH737-EuUSP16 overexpression vector and Virus-Induced Gene Silencing (VIGS) vector (pTRV2-EuUSP16) were genetically transformed into E.ulmoides via Agrobacterium-mediated transformation. The expression levels of E.ulmoides rubber biosynthesis-related genes and the rubber content were then analyzed. The primer sequences for the E.ulmoides rubber biosynthesis-related genes are listed in Supplementary Table 1, and the reaction system was the same as described in section 2.3.
2.5 E.ulmoides rubber content determination
Extraction of E.ulmoides rubber was performed with modifications based on the methods described by Lu et al. (2004) and Xie et al. (2013): Leaves and stem segments (approximately 10 g) of E.ulmoides after 10 days of drought and low-temperature treatments respectively were placed in an oven at 120°C for enzyme deactivation (15 min). After drying, the sample mass was recorded. The samples were ground into powder, mixed with 10% NaOH solution (200 mL), and subjected to a 3-hour water bath at 100°C, followed by 20 min of high-temperature/high-pressure treatment at 120°C. Residual NaOH solution was removed by rinsing under running water. The samples were then transferred to bottles containing 200 mL of purified water and incubated in a 60°C water bath for 3 h, followed by drying in an oven at 65°C. Petroleum ether (boiling range: 60-90°C) was added to a Soxhlet extractor at a solid-to-liquid ratio of 1:15 for 20 h of extraction. The petroleum ether solution was decanted into pre-weighed 50 mL centrifuge tubes and evaporated in a 37°C oven. Anhydrous ethanol was added to wash the precipitate repeatedly until white refined rubber was obtained. After oven-drying, the mass of the refined rubber was recorded, and the rubber content was calculated using the formula: Rubber content (%) = (Mass of refined rubber/Sample mass)×100%. Three biological replicates for all experiments.
2.6 Cloning and sequence analysis of EuUSP16 promoter
Specific primers were designed to amplify EuUSP16 promoter sequences of different lengths, and the cloned promoter fragments were constructed with the pCAMBIA1381Z vector. The constructed vectors were subsequently sent to BGI for sequencing. The primers used are shown in Supplementary Table 1.
2.7 Construction of the plant transformation vector and genetic transformation of tobacco
EuUSP16 promoter DNA fragments of different lengths(1967 bp/1286 bp/1193 bp/273 bp) were connected to the EcoRI and BamHI enzyme restriction sites of the pCAMBIA1381Z vector to obtain the following four vectors: PEuUSP16-1,PEuUSP16-2, PEuUSP16-3,PEuUSP16-4. These vectors were used to transform Agrobacterium GV3101, which was subsequently genetically transformed into tobacco. PCR was used to identify the transgenic tobacco plants for further study. The primers used are shown in Supplementary Table 1.
2.8 Yeast one-hybrid pairwise validation
The prey vector pGADT7 was double-digested with restriction enzymes BamHI and EcoRI. The coding sequence (CDS) of EuDof was then cloned into the linearized pGADT7 vector using homologous recombination. This generated the pGADT7-EuDof prey construct for subsequent pairwise yeast one-hybrid validation of the interaction between pGADT7-EuDof and pHIS2-PEuUSP16.
2.9 Dual-luciferase reporter assay
The reporter vector pGreenII 62-SK and the effector vector pGreenII 0800-LUC were double-digested with BamHI and KpnI, respectively. The CDS of EuDOf was cloned into the linearized pGreenII 62-SK vector, while the PEuUSP16–1 promoter fragment was cloned into the linearized pGreenII 0800-LUC vector, using homologous recombination. Plasmid DNA from sequence-verified clones was extracted and transformed into Agrobacterium tumefaciens GV3101 (pSoup) competent cells. The bacterial suspensions were infiltrated into young, fully expanded leaves of 3–4-week-old tobacco plants using a needleless syringe. Infiltrated plants were maintained under standard growth conditions for 48 h. Luminescence was then visualized and quantified using a plant in vivo imaging system.
2.10 Construction of the pSH737-EuDof overexpression vector and genetic transformation of PEuUSP16–1 transgenic tobacco
The pSH737 overexpression vector plasmid was double-digested with XbaI and EcoRI. The CDS of the EuDof gene was cloned into the plant overexpression vector pSH737 using homologous recombination. Transgenic tobacco line PEuUSP16–1 was used as the starting material. After genetic transformation of tobacco, the expression level of the β-glucuronidase (GUS) gene was assessed.
2.11 Statistical analysis
Statistical analysis and graphing were performed using Excel 2010 and GraphPad Prism 9.5 software. Data differences were assessed by one-way ANOVA, with statistical significance denoted as follows: different lowercase letters indicate significant differences (P<0.05), while asterisks represent *P<0.05, **P<0.01, ***P<0.001, and**P<0.0001.
3 Results
3.1 Identification of the EuUSPs gene family and screening of genes associated with E.ulmoides rubber biosynthesis
A total of 29 EuUSPs genes were identified and named EuUSP1 to EuUSP29 according to their chromosomal positions (Figure 1A). The physicochemical properties of the encoded proteins are shown in Supplementary Table 2. The 29 EuUSPs proteins contain 99 to 892 amino acid residues with relative molecular masses ranging from 11.28 to 98.86 kDa and theoretical isoelectric points (pI) between 4.81 and 10.37. These results indicate substantial variation in protein size and molecular weight within the EuUSPs family, suggesting diverse functional domains and structural features. The instability index ranged from 24.1 to 68.26, while the aliphatic index varied between 77.01 and 104.55 (23 proteins>80). This suggests moderate to high thermostability in most EuUSPs proteins. Twenty- five EuUSPs proteins exhibited negative grand average of hydropathicity values, indicating predominantly hydrophilic characteristics. Subcellular localization predictions revealed primary localization in the cytoplasm, chloroplasts, and mitochondria, with secondary localization in the nucleus, golgi apparatus, and endoplasmic reticulum.
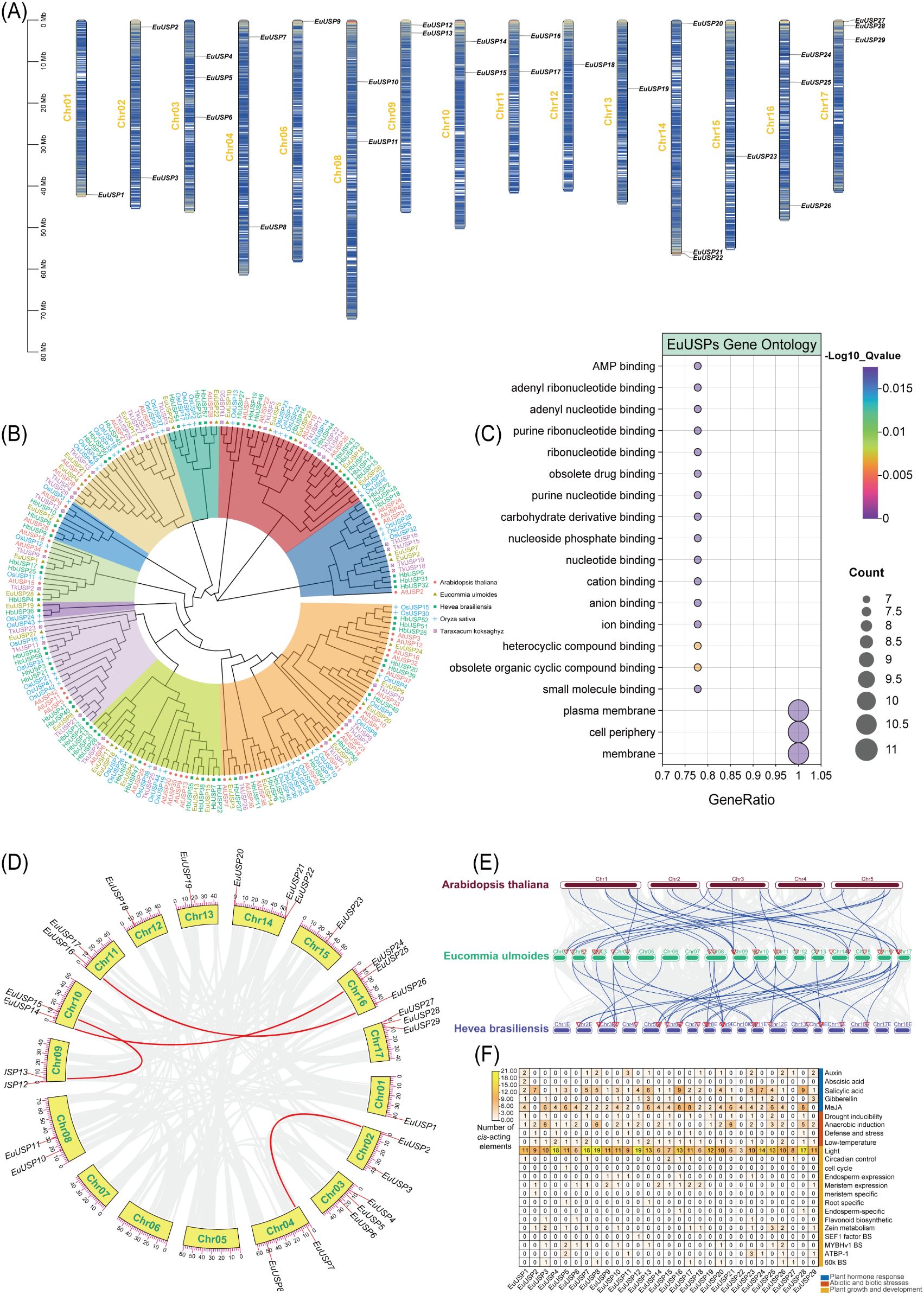
Figure 1. Identification of EuUSPs genes in E.ulmoides. (A) Chromosomal mapping. (B) Phylogenetic relationship among the EuUSPs proteins of the 5 plant species. (C) Gene ontology analysis. (D) Collinearity analysis of EuUSPs gene family in E. ulmoides. (E) Collinearity relation of USP genes in the genomes of A.thaliana and H.brasiliensis. (F) Analysis on cis-acting elements.
Phylogenetic analysis was conducted between 29 EuUSPs gene family members from E.ulmoides and USP genes from four representative species: A.thaliana (42 genes), H.brasiliensis (58 genes), Oryza sativa (46 genes), and T.kok-saghyz (29 genes). The resulting phylogenetic tree is shown in Figure 1B. The 204 USP protein sequences clustered into nine subgroups (I-IX), containing 50, 49, 3, 12, 8, 32, 5, 26, and 19 members respectively. Phylogenetic tree analysis revealed that EuUSP5, EuUSP16, EuUSP23, and EuUSP26 (all containing rubber particle protein peptides; unpublished data) consistently clustered with USPs from latex-producing plants (either H.brasiliensis or T.kok-saghyz). This suggests that EuUSPs harboring rubber particle peptides share high homology with those from laticiferous plants (H.brasiliensis and T.kok-saghyz), potentially indicating functional conservation in E.ulmoides rubber biosynthesis. Analysis of conserved motifs in EuUSPs proteins revealed that 27 members (all except EuUSP20 and EuUSP29) contain Motif2 and Motif8. Notably, Motif9 was exclusively present in EuUSP5,16,23, and 26. These distinctive motif architectures provide the theoretical foundation for gene classification and functional prediction. All 29 EuUSPs proteins contain the USP domain. Gene structure analysis demonstrated: the 29 EuUSPs genes contained 0–10 introns, with only EuUSP7 being intron-free (Supplementary Figure 1). To investigate the collinear relationships between EuUSPs and USP genes from other species, this study analyzed syntenic gene pairs among E.ulmoides, A.thaliana, and H.brasiliensis. As shown in Figure 1E, 17 collinear gene pairs were identified between EuUSPs and AtUSPs, while 21 collinear pairs existed between EuUSPs and HbUSPs. The higher frequency of syntenic events with the rubber-producing plant compared to the non-laticiferous species suggests evolutionary conservation of these USP genes within the EuUSPs family. This collinearity advantage reflects both the phylogenetic proximity between E. ulmoides and rubber tree, and underscores the potential functional importance of these genes in rubber biosynthesis. Analysis of duplication events in EuUSPs genes revealed that collinearity primarily occurs on chromosomes 2, 4, 9, 10, 11, and 16 (Figure 1D). Four segmental duplication events were identified within the EuUSPs family, involving 8 genes forming 4 duplicated pairs: EuUSP2-EuUSP7, EuUSP12-EuUSP15, EuUSP14-EuUSP25, and EuUSP16-EuUSP26. These gene pairs represent collinear genes within syntenic blocks. While EuUSP14 and EuUSP15 exhibit adjacent duplication in close chromosomal proximity, no evidence of tandem duplication events was detected among EuUSPs genes. Analysis of cis-regulatory elements in EuUSPs gene family promoters is presented in Figure 1F. Promoter regions of E.ulmoides EuUSPs are enriched with diverse regulatory elements. Major cis-elements include phytohormone-responsive motifs: abscisic acid (ABA), salicylic acid (SA), auxin (IAA), methyl jasmonate(MeJA), and gibberellin response elements. Four types of stress-related cis-elements were identified: anaerobic induction, low-temperature response, defense/stress response, and drought-inducible elements. Additionally, promoters contain elements associated with cell cycle regulation, circadian rhythm, meristem expression, and flavonoid biosynthesis. GO analysis was performed on EuUSPs genes, with the results shown in Figure 1C. Molecular Function: Eleven EuUSPs genes (EuUSP1, EuUSP2, EuUSP5, EuUSP7, EuUSP8, EuUSP10, EuUSP12, EuUSP19, EuUSP22, EuUSP27, and EuUSP29) showed significant enrichment in molecular functions. These include:AMP binding (GO:0016208), Adenyl ribonucleotide binding (GO:0032559), Adenyl nucleotide binding (GO:0030554) and other terms. Four genes (EuUSP2, EuUSP7, EuUSP8, and EuUSP27) exhibited significant enrichment in cellular components including:Plasma membrane (GO:0005886), cell periphery (GO:0071944) and membrane (GO:0016020). These findings indicate that EuUSPs genes participate in energy metabolism, signal transduction, and molecular recognition processes. No significant enrichment was observed for any Biological Process terms.
3.2 Tissue-specific expression analysis of EuUSPs
To validate the expression patterns of EuUSPs genes in the roots, stems, and leaves of E.ulmoides seedlings, we analyzed their expression in three-month-old seedlings. The results are presented in Figure 2. EuUSP1, EuUSP16, EuUSP20, EuUSP26, and EuUSP28 are highly expressed primarily in stems with high rubber content.
Figure 2. Tissue-specific expression pattern of the EuUSPs gene in E.ulmoides. Different lowercase letters indicate significant differences (P<0.05).
3.3 Effects of drought and low-temperature treatments on EuUSPs gene expression and rubber content in E.ulmoides seedlings
After 10 days of drought treatment in 15-month-old E.ulmoides seedlings, phenotypic changes are shown in Figure 3A. The absolute water content was 84.592% in the control group versus 73.191% in the drought-treated group, showing a significant decrease compared to the control (Figure 3B). Expression levels of EuUSPs in roots, stems, and leaves and rubber content were analyzed. EuUSP1, EuUSP6, EuUSP7, EuUSP16, EuUSP18, EuUSP20, EuUSP22, EuUSP23, EuUSP26, and EuUSP28 exhibited significant upregulation (2.28-to 108.19-fold increase) in stems compared to the control. EuUSP1, EuUSP16, EuUSP22, and EuUSP23 showed significant upregulation (1.38- to 1.80-fold increase) in leaves (Figure 4). Measurement of rubber content in E.ulmoides stems and leaves revealed the following: In control samples, stem and leaf rubber contents were 7.605% and 4.326%, respectively. In the experimental group, stem and leaf rubber contents were 8.989% and 5.426%, respectively. Rubber content in both stems and leaves was significantly higher than in the control group (Figure 3C). EuUSP1, EuUSP16, EuUSP22, and EuUSP23 upregulation in stems/leaves positively correlated with increased laticifer content (P<0.05) (Figures 3D, E). These results indicate that expression of EuUSP1, EuUSP16, EuUSP22, and EuUSP23 influences rubber biosynthesis in stems and leaves.
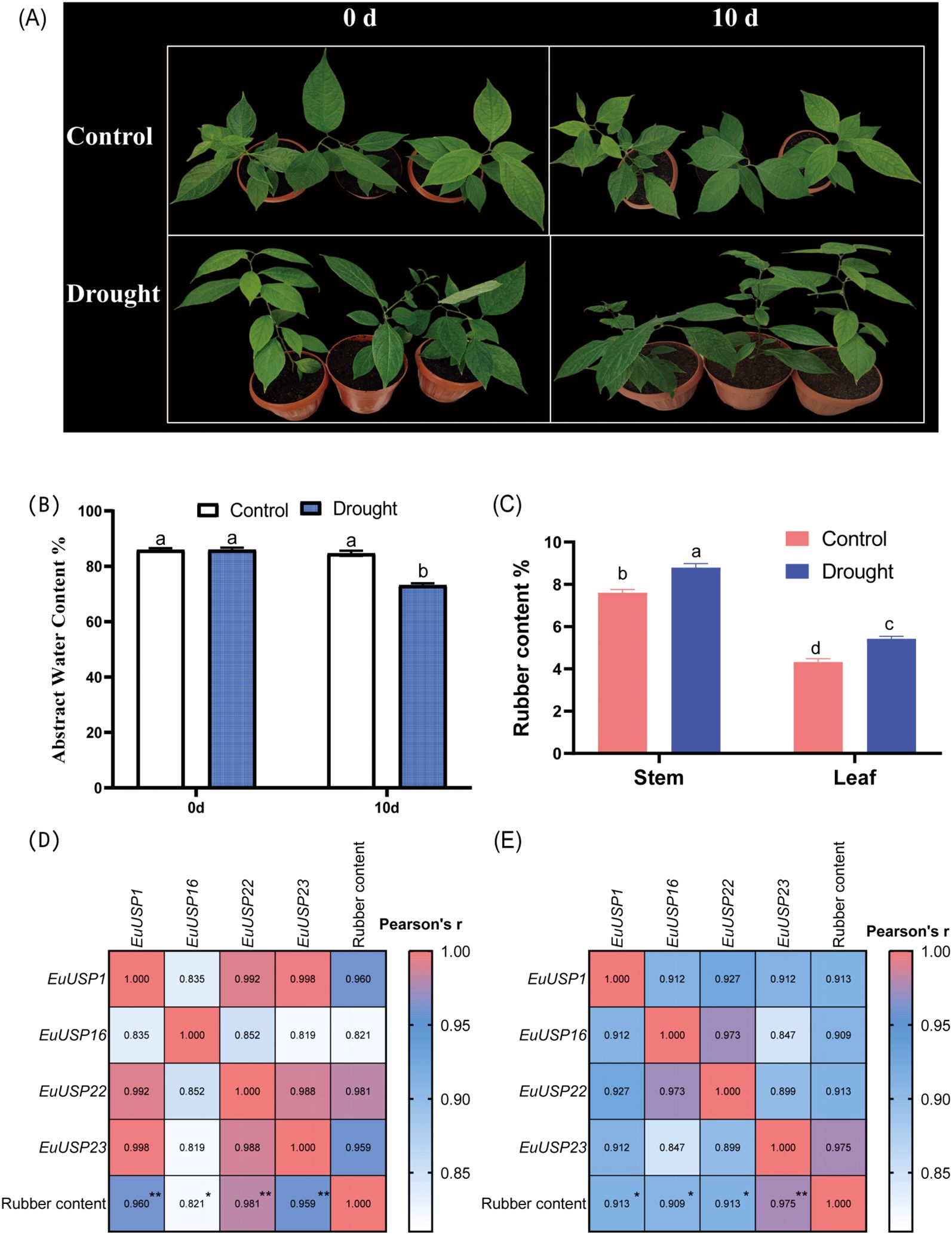
Figure 3. Pearson correlation analysis between EuUSPs gene expression and rubber content under 10-day drought stress. (A) Drought-treated E.ulmoides seedlings. (B) The absolute water content in E. ulmoides leaves before and after drought treatment. (C) Rubber content. (D) Correlation between EuUSPs expression in stems and rubber content. (E) Correlation between EuUSPs expression in leaves and rubber content. Control: negative control. Different lowercase letters indicate significant differences (P<0.05). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
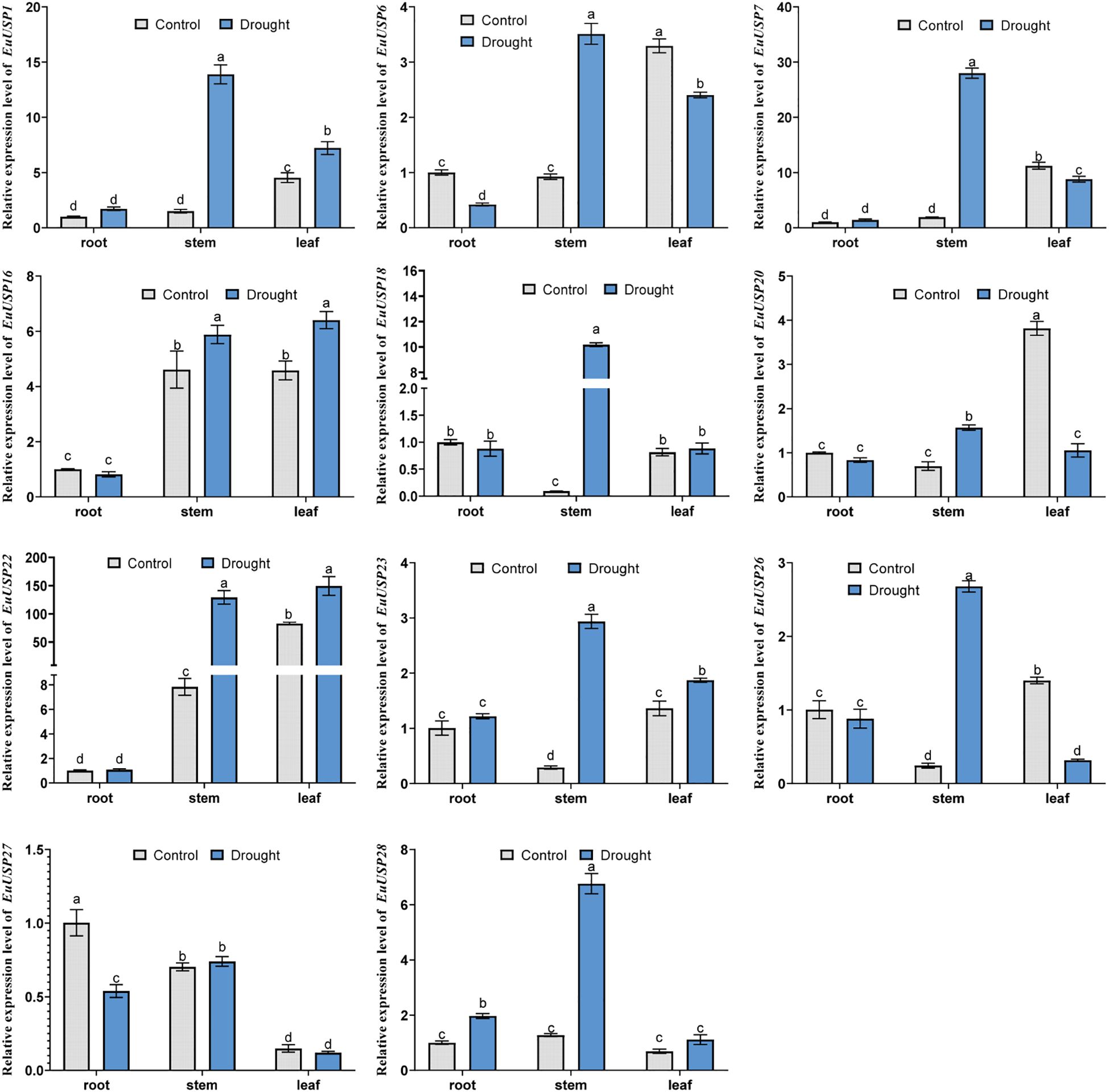
Figure 4. The expression analysis of EuUSPs gene in E.ulmoides after drought treatment. Control: negative control. different lowercase letters indicate significant differences (P<0.05).
Analysis of EuUSPs gene expression and rubber content was conducted after 10 days of low-temperature treatment (4°C) in 15-month-old seedlings (Figure 5A). The results showed that in stems, the relative expression levels of EuUSP1, EuUSP6, EuUSP7, EuUSP16, EuUSP18, EuUSP20, EuUSP26, and EuUSP28 increased by 1.46- to 13.29-fold compared to the control group. In leaves, the expression levels of EuUSP1, EuUSP6, EuUSP7, EuUSP16, EuUSP18, EuUSP20, EuUSP22, and EuUSP23 significantly decreased to 0.089-0.79 times that of the control group (Figure 6). Measurement of rubber content in E.ulmoides stems and leaves revealed: Control group: Stem content 7.605%, leaf content 4.326%. Low-temperature treatment group: Stem content 9.764%, leaf content 3.349%. After 10 days of 4°C treatment, stem rubber content significantly increased while leaf content significantly decreased compared to controls (Figure 5B). Additionally, expression levels of six genes (EuUSP1, EuUSP6, EuUSP7, EuUSP16, EuUSP18, and EuUSP20) in stems and leaves positively correlated with changes in rubber content (P<0.05) (Figures 5C, D). This indicates that expression of these six EuUSPs genes influences E.ulmoides rubber biosynthesis.
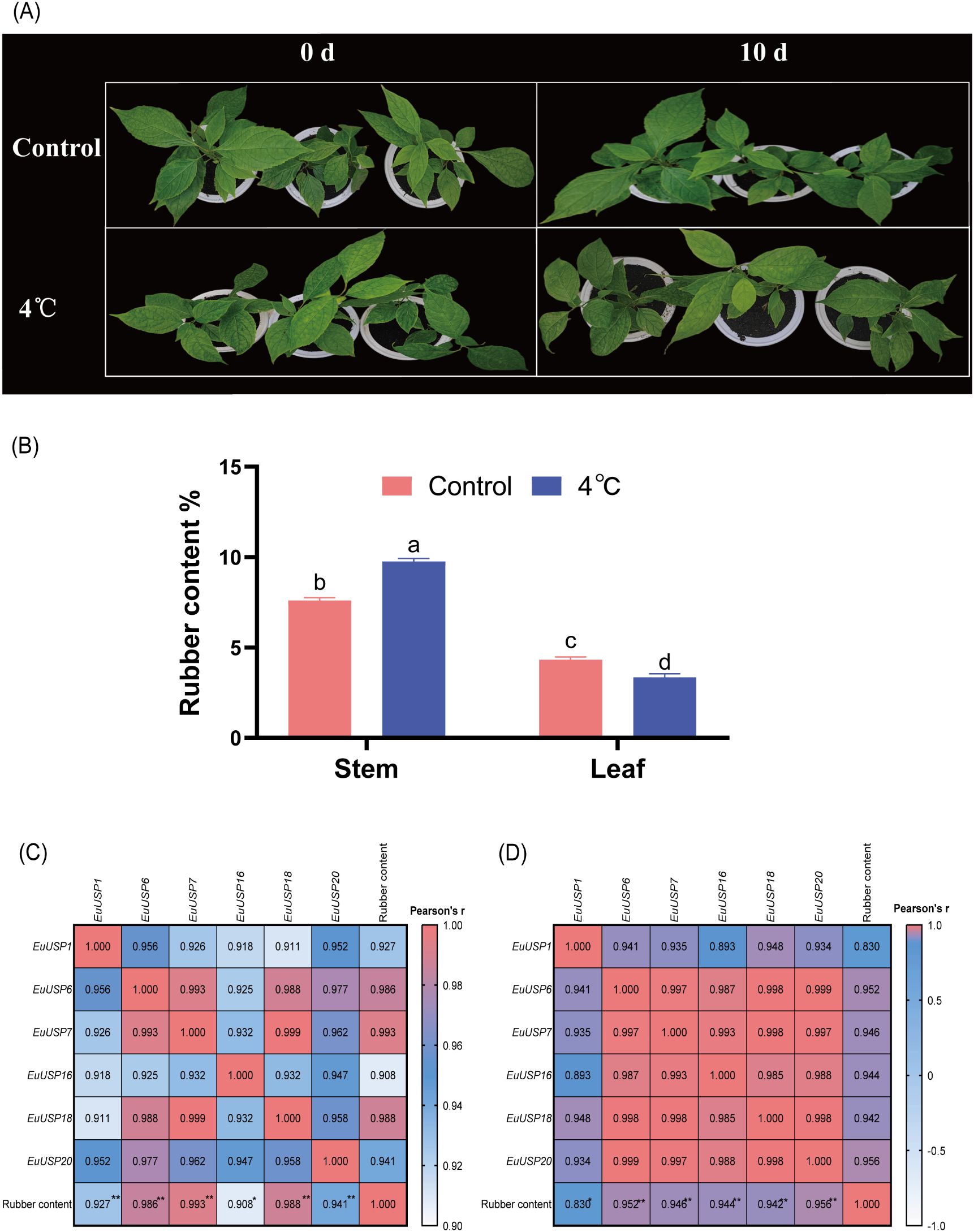
Figure 5. Pearson correlation between EuUSPs gene expression and rubber content under 4°C treatment. (A) 4°C-treated E.ulmoides seedlings (B) Rubber content. (C) Correlation between EuUSPs expression in stems and rubber content. (D) Correlation between EuUSPs expression in leaves and rubber content. Note: Control, negative control. Different lowercase letters indicate significant differences (P<0.05). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001..
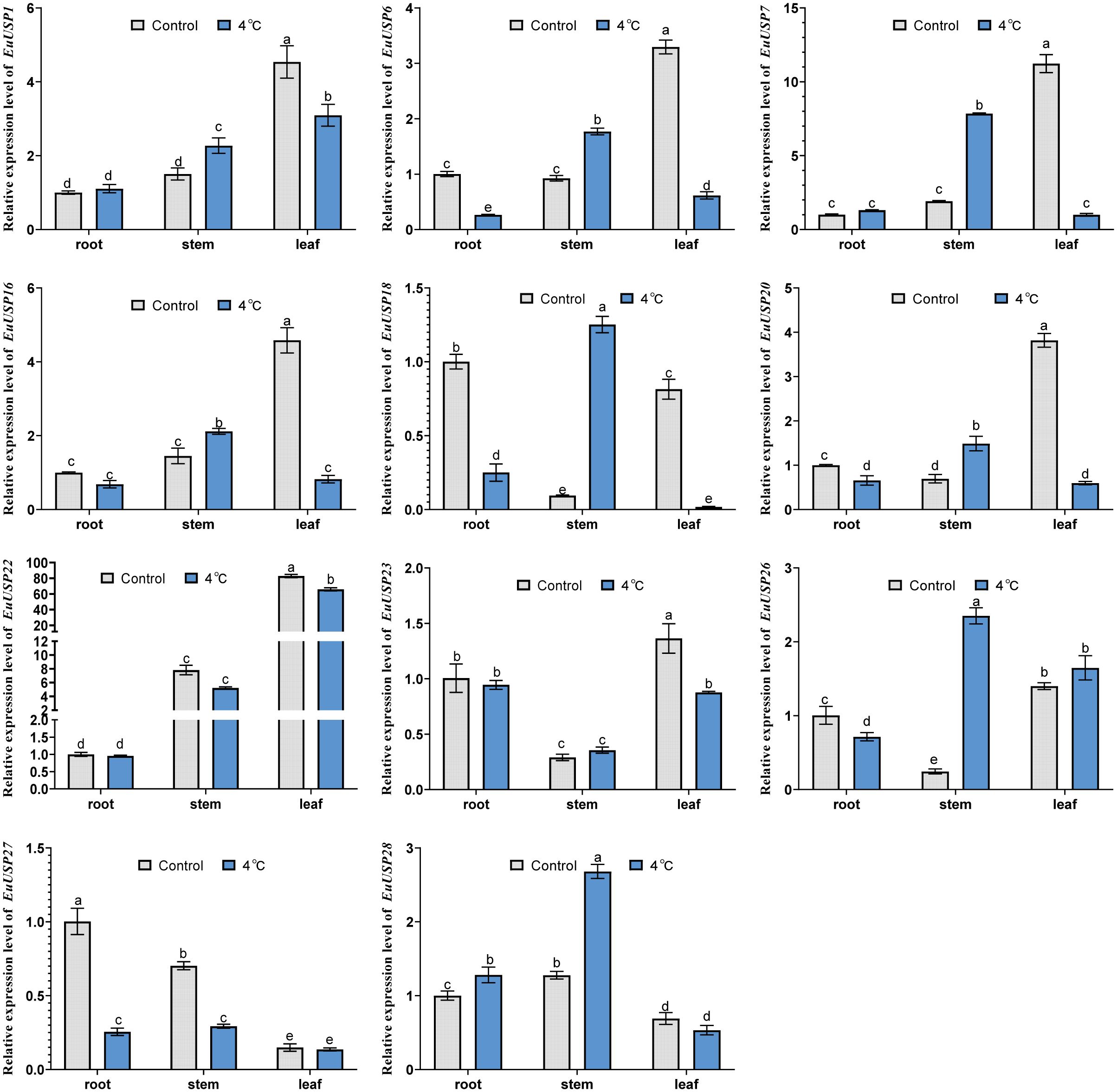
Figure 6. The expression analysis of EuUSPs gene in after 4°C treatment. Control: 25°C treatment. Different lowercase letters indicate significant differences (P<0.05).
Under low-temperature and drought stress treatments, the expression levels of multiple EuUSPs genes were altered. Notably, EuUSP16 expression consistently exhibited a positive correlation with rubber content. Furthermore, this gene encodes a protein containing a rubber particle protein motif. We therefore propose that EuUSP16 plays a significant role in E.ulmoides rubber biosynthesis under stress conditions. Consequently, EuUSP16 was selected for in-depth functional characterization.
3.4 The role of EuUSP16 in the E.ulmoides rubber synthesis
The pSH737-35S-EuUSP16 was genetically transformed into E.ulmoides hypocotyls (Figures 7 A–G). Six transgenic EuUSP16 adventitious buds of E.ulmoides were obtained following GUS staining and PCR verification, as shown in Figures 7G, H. In the transgenic EuUSP16 buds, the relative expression levels of EuGGPPS, EuFPS1, EuREF, and EuSRPP1 genes were significantly higher than those in wild type (WT) and transgenic empty vector(EV) adventitious buds, measuring 2.60-, 2.12-, 16.71-, and 4.51-fold of WT, respectively, while the relative expression of EuIPI was significantly reduced to 0.43-fold of WT (Figures 7J, K). E.ulmoides rubber was extracted from (WT) and Over-Expression (OE) callus, with results presented in Figure 7M. The rubber content in WT callus was 0.288%, whereas that in EuUSP16-overexpressing callus was 0.733%, indicating a significant increase. These findings demonstrate that EuUSP16 overexpression enhances the expression of key E.ulmoides rubber synthesis genes such as EuFPS1, further promoting E.ulmoides rubber synthesis.
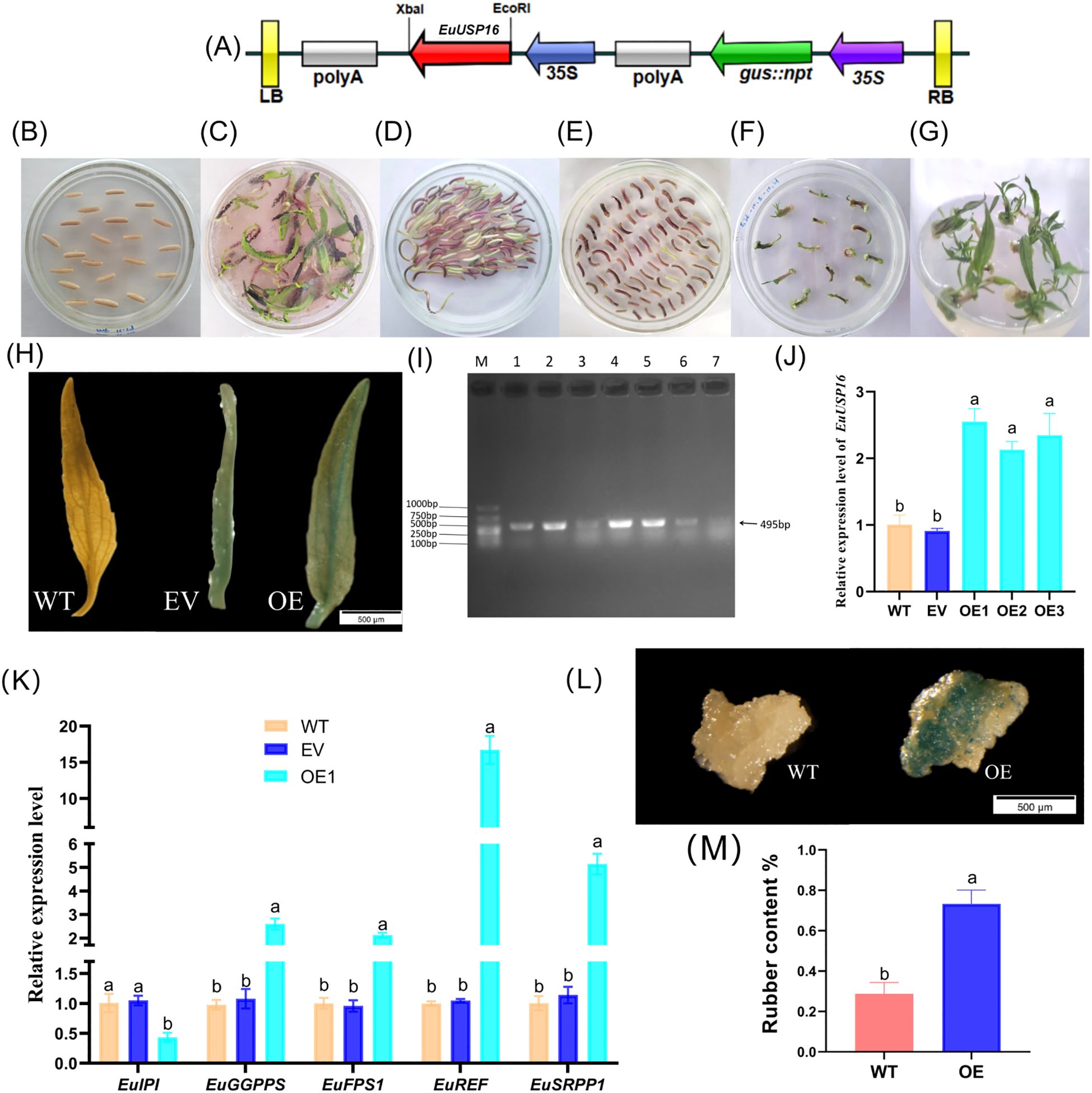
Figure 7. Functional analysis of the EuUSP16 gene. (A) T-DNA structure of the pSH737-35S-EuUSP16 vector. (B-C) Germination process of E.ulmoides seeds. (D) Co-cultivation. (E) Screening of resistant tissues. (F-G) Induction of resistant buds (H) GUS staining identification. (I) PCR verification of transgenic EuUSP16 plants. (J) Gene expression levels in transgenic EuUSP16 buds. (K) Expression of rubber synthesis-related genes in EuUSP16-overexpressing buds. (L) GUS staining of callus. (M) E.ulmoides rubber content in callus. Note: M, DL2000 marker; Different lowercase letters indicate significant differences at P<0.05.
The construction process of the EuUSP16 gene silencing vector is shown in Figures 8A–C. As shown in Figures 8D-F, following qRT-PCR validation, six E.ulmoides seedlings with reduced relative expression levels of the EuUSP16 gene were obtained. The line with the lowest expression, namely pTRV2-EuUSP16-1, was selected to analyze the relative expression levels of genes related to E.ulmoides rubber synthesis: EuGGPPS, EuFPS1, EuREF, and EuSRPP1. The results showed that in the pTRV2-EuUSP16–1 plants, the relative expression levels of the EuFPS1, EuREF, and EuSRPP1 genes were significantly reduced compared with WT and TRV2 plants, reaching 0.12-fold, 0.13-fold, and 0.59-fold that of WT, respectively. In contrast, the relative expression levels of the EuGGPPS and EuIPI genes were significantly increased, reaching 26.50-fold and 2.21-fold that of WT, respectively. This indicates that EuUSP16 silencing affects E.ulmoides rubber synthesis by reducing the expression of key enzyme genes such as EuFPS1.
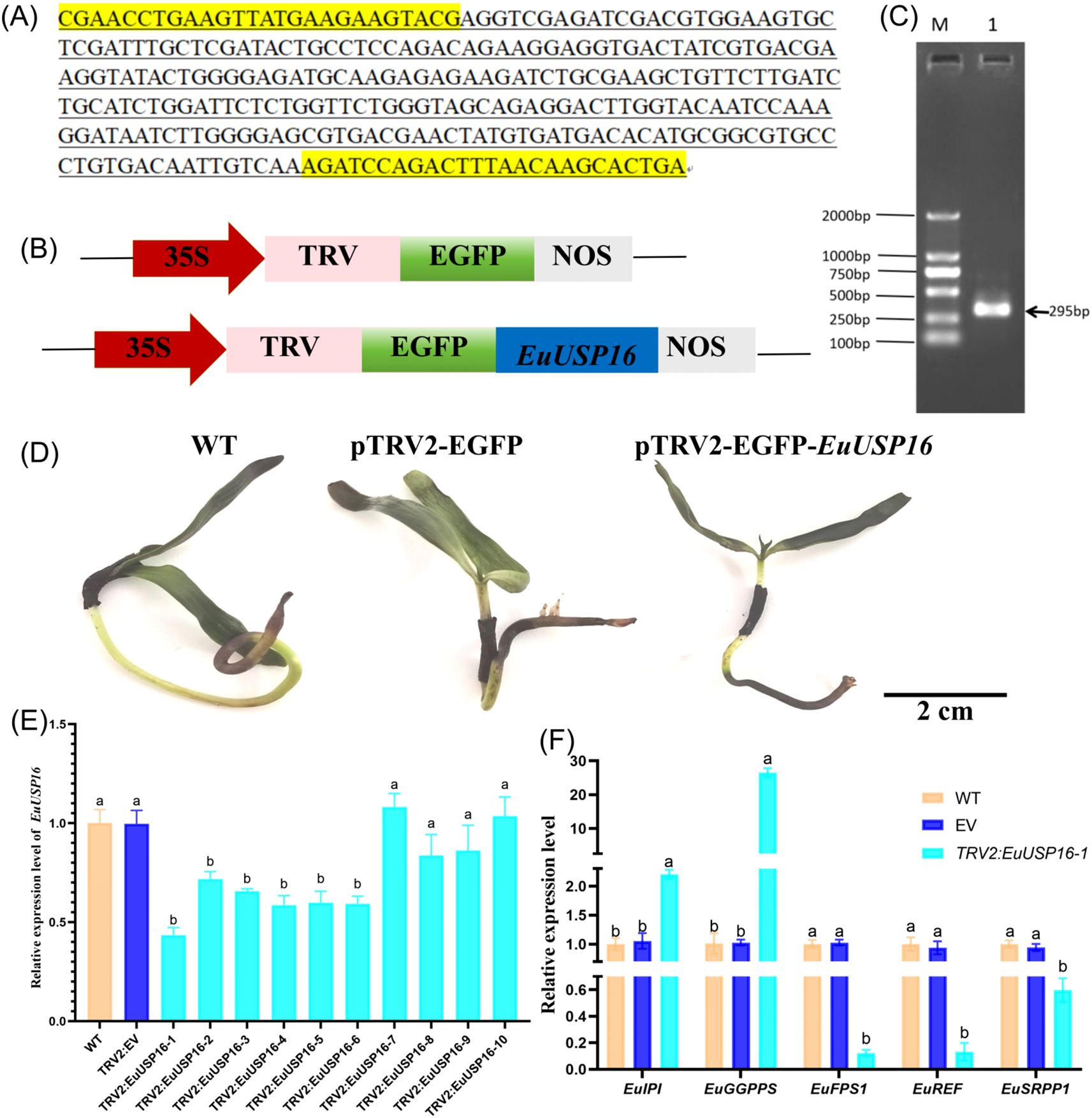
Figure 8. Effects of EuUSP16 gene silencing on the expression of rubber biosynthesis-related genes in E.ulmoides. (A) Virus-Induced Gene Silencing (VIGS) fragment. (B) Schematic diagram of pTRV2-GFP vector. (C) PCR detection results. M: DL 2000 DNA Marker Lane 1:pTRV2-GFP-EuUSP16 plasmid. (D) Phenotype of EuUSP16-silenced E.ulmoides seedlings. (E) Gene expression levels in EuUSP16-silenced E.ulmoides. (F) Expression levels of rubber biosynthesis-related genes in EuUSP16-silenced E.ulmoides. Different lowercase letters indicate significant differences (P<0.05).
3.5 Effects of hormone and environmental treatments on the activity of the EuUSP16 promoter
This study cloned a 1967 bp promoter sequence upstream of the ATG start codon of the EuUSP16 gene. The positions of response elements were analyzed, and four plant expression vectors driving GUS gene expression were constructed: the full-length promoter PEuUSP16-1 (1967 bp), and three deletion constructs, PEuUSP16-2 (1286 bp) lacking the drought response element, PEuUSP16-3 (1193 bp) lacking the low-temperature response element, and PEuUSP16-4 (273 bp) lacking the ABA response element. Genetic transformation was performed in tobacco. GUS gene expression analysis revealed that the EuUSP16 promoter functions as a constitutive promoter (Figures 9A, B). Transgenic tobacco harboring PEuUSP16–1 exhibited the highest GUS activity, while those with PEuUSP16–4 showed the lowest. No significant difference in GUS expression was observed between transgenic lines carrying PEuUSP16–2 and PEuUSP16-3, although both differed significantly from lines harboring either PEuUSP16–1 or PEuUSP16-4. These results indicate that the core promoter region of EuUSP16 resides within the -273 bp to -1 bp fragment.
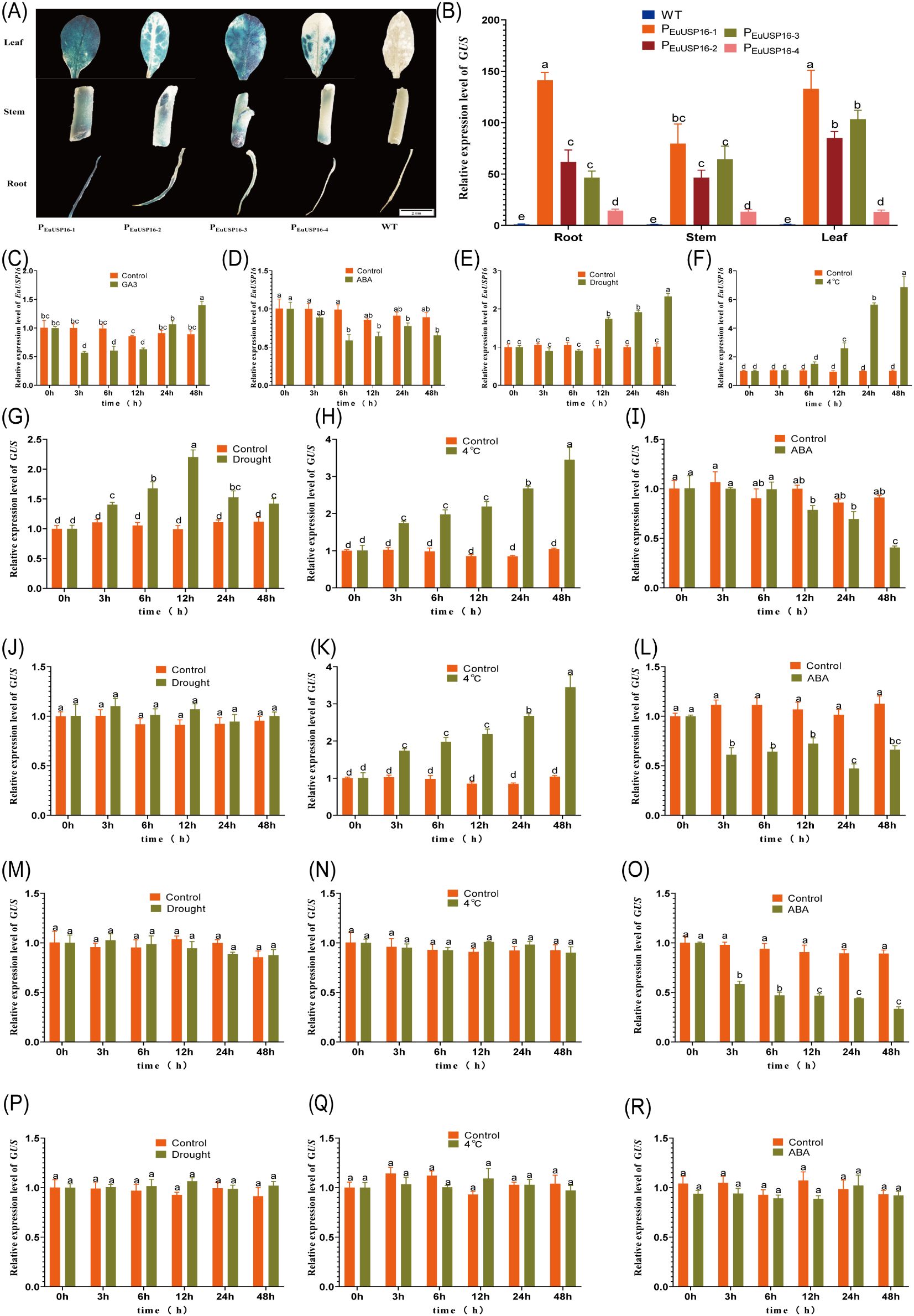
Figure 9. Effects of hormone and environmental treatments on the activity of the EuUSP16 promoter. (A) GUS histochemical staining in roots, stems, and leaves of transgenic tobacco. (B) Relative expression levels of the GUS gene in transgenic tobacco harboring four EuUSP16 promoter fragments. (C-F) Relative expression levels of EuUSP16 in E.ulmoides seedlings treated with 100 μmol·L-¹GA3, 100 μmol·L-¹ ABA, drought, and low temperature, respectively. (G-R) Relative GUS expression in transgenic tobacco carrying four EuUSP16 promoter fragments under treatments:(G-I), PEuUSP16–1 transgenic tobacco treated with 300 mmol·L-¹ mannitol (simulated drought), 4°C, or 100 μmol·L-¹ ABA spray, (J-L) PEuUSP16–2 transgenic tobacco treated identically, (M-O) PEuUSP16–3 transgenic tobacco treated identically, (P-R) PEuUSP16–4 transgenic tobacco treated identically. Control, Untreated group; 0 h: Pre-treatment baseline; Different lowercase letters indicate significant differences (P < 0.05).
In PEuUSP16–1 transgenic tobacco plants subjected to drought stress, the relative GUS expression level progressively increased from 3 h to 12 h of treatment, reaching a 2.2-fold induction at 12 h (Figure 9G). Following 4°C treatment, the relative GUS expression in leaves of PEuUSP16–1 transgenic plants increased over time, peaking at 48 h with a 3.4-fold increase compared to the pre-treatment level (Figure 9H). Conversely, treatment with ABA solution significantly reduced GUS expression in PEuUSP16–1 transgenic leaves between 12 h and 48 h, reaching levels 0.4- to 0.78-fold of the control, indicating that ABA significantly suppresses GUS expression (Figure 9I). These findings demonstrate the presence of response elements associated with MBS, LTR, and ABRE within the PEuUSP16–1 promoter fragment. Drought treatment of PEuUSP16–2 transgenic plants resulted in no significant change in relative GUS expression (Figure 9J). However, 4°C treatment induced a significant increase in GUS expression, reaching 5.8-fold of the control at 48 h (Figure 9K). Spray treatment with ABA solution caused a significant decrease in GUS expression, with levels dropping to 0.42-fold of the control at 24 h (Figure 9L). These results indicate that the PEuUSP16–2 promoter fragment contains functional LTR and ABRE response elements. No significant alterations in relative GUS expression were observed in leaves of PEuUSP16–3 transgenic plants following either drought or 4°C treatment (Figures 9M, N). ABA treatment, however, led to a gradual decline in GUS expression in PEuUSP16–3 transgenic leaves, reaching a minimum of 0.34-fold of the control at 48 h (Figures 9O). This suggests the presence of an ABRE-related response element within PEuUSP16-3. Treatments with drought, 4°C, or ABA solution elicited no significant differences in relative GUS expression in leaves of PEuUSP16–4 transgenic plants (Figures 9P-R). This indicates the absence of functional MBS, LTR, and ABRE response elements within the PEuUSP16–4 promoter fragment.
In summary, the EuUSP16 gene promoter harbors MBS (CAACTG), LTR (CCGAAA), and ABRE (ACGTG) response elements. GUS gene expression driven by this promoter can be induced by low temperature and drought stress, but is repressed by ABA treatment. This expression pattern is largely consistent with that of EuUSP16 observed in E.ulmoides seedlings (Figures 9C-F).
3.6 The EuDof transcription factor controls EuUSP16 gene expression through binding to its promoter
To identify upstream regulators of the EuUSP16 gene, a yeast one-hybrid (Y1H) screening assay was performed. A total of 36 initial positive clones were screened (Supplementary Table 3). BLAST analysis revealed one clone encoding a transcription factor, specifically a DNA-binding with one finger (Dof) protein, which is a plant-specific transcription factor. The CDS of the EuDof gene was amplified by PCR, yielding a 1272 bp fragment encoding 423 amino acids. The encoded protein belongs to the zf-Dof superfamily; thus, the gene was designated EuDof.
To determine whether the EuUSP16 promoter interacts with EuDof, yeast one-on-one assays and dual-luciferase reporter assays confirmed that the EuDof transcription factor directly binds to the EuUSP16 promoter and activates EuUSP16 expression (Figures 10A, B). Concurrently, the EuDof gene was cloned into the pSH737 plant overexpression vector. Agrobacterium-mediated genetic transformation was used to introduce EuDof into transgenic tobacco harboring the PEuUSP16–1 promoter construct (transgenic lines designated PD) (Figures 10C-I). Analysis of relative GUS gene expression revealed a significant increase in GUS levels in PEuUSP16–1 transgenic tobacco overexpressing EuDof (Figures 10J), further supporting the interaction between the EuUSP16 promoter and the EuDof transcription factor.
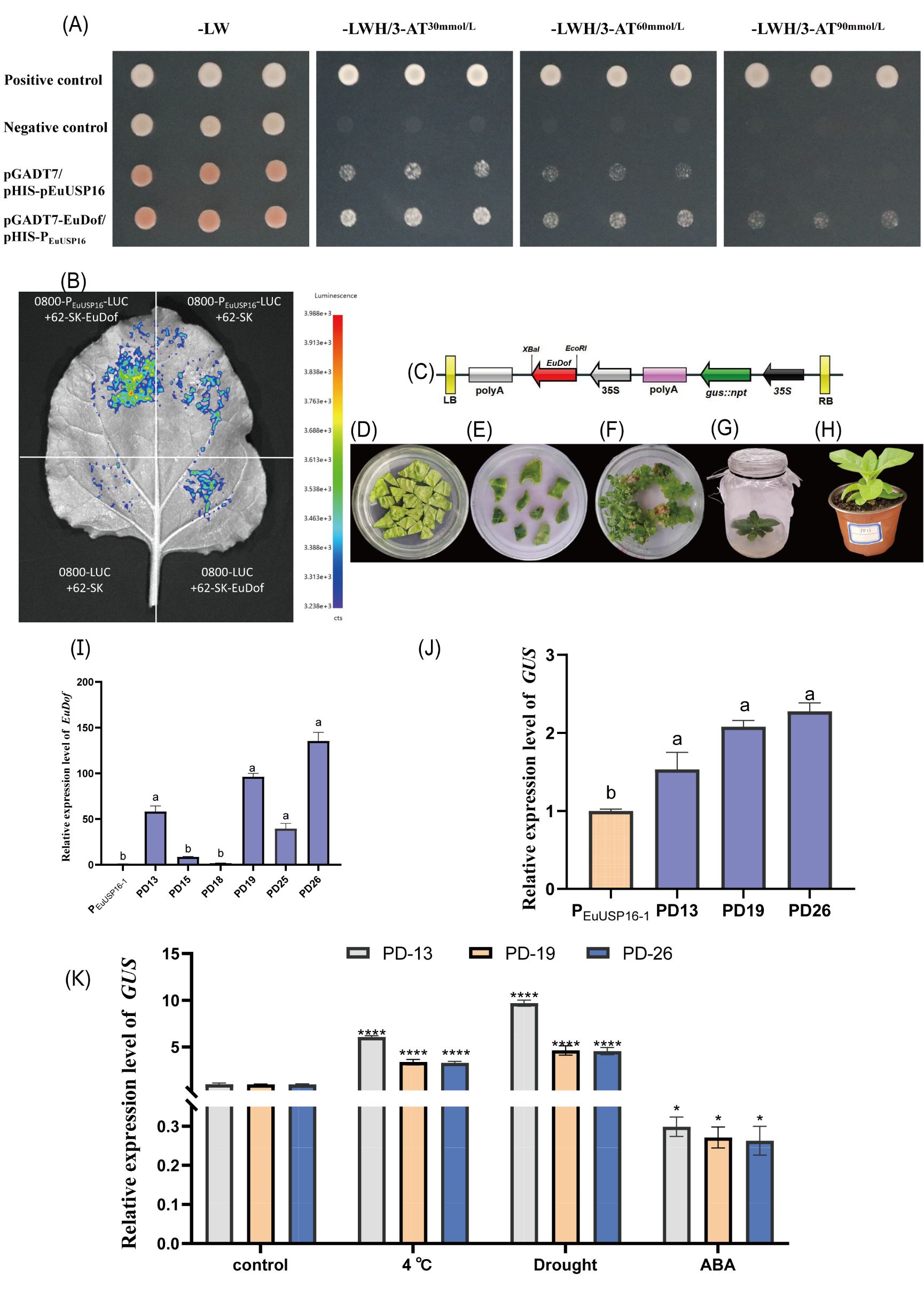
Figure 10. Interaction analysis between the EuUSP16 promoter and EuDof transcription factor. (A) Y1H one-on-one validation of PEuUSP16–1 and EuDof interaction. (B) Dual-luciferase (LUC) reporter assay. Note for b: Negative controls: 0800-PEuUSP16-LUC + 62-SK empty vector, 0800-LUC empty vector + 62-SK empty vector, 0800-LUC empty vector + 62-SK-EuDof; Experimental group: 0800-PEuUSP16-LUC + 62-SK-EuDof (C) T-DNA structure of the pSH737-35S-EuDof overexpression vector. (D) Co-cultivation of Agrobacterium with tobacco leaf discs (E). Selection of antibiotic-resistant callus. (F) Shoot induction from transgenic callus. (G) PCR identification of positive transgenic plants. (H) Potted transgenic seedlings. (I) Expression analysis of the EuDof transgene in PD transgenic tobacco lines. (J) GUS gene expression analysis in PD transgenic tobacco lines. (K) GUS expression in PD transgenic tobacco lines following 4°C, drought, or 100μmol·L-¹ABA treatments. Note: Different lowercase letters indicate significant differences (P<0.05). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001..
Additionally, transgenic lines PD13, PD19, and PD26 were subjected to low temperature (4°C), drought (simulated drought), or 100 µmol/L ABA treatment. Samples were collected at 48 h, and GUS expression was quantified (Figure 10K). Following low-temperature and drought treatments, GUS expression in all three lines was significantly higher than in untreated controls, reaching levels 3.30- to 6.06-fold and 4.57- to 9.69-fold of the control, respectively. Conversely, ABA spray treatment significantly reduced relative GUS expression to 0.26- to 0.29-fold of the control. These results indicate that low-temperature and drought stress promote the binding of the EuDof transcription factor to the EuUSP16 promoter, while ABA treatment has the opposite effect.
4 Discussion
4.1 Characterization of the EuUSPs gene family
Plant USPs play important roles in responding to abiotic and biotic stresses (Nabi et al., 2024; Tkaczuk et al., 2013). This study identified 29 EuUSPs gene family members from the E.ulmoides genome. The EuUSPs genes are unevenly distributed across the 15 chromosomes of E. ulmoides, and their distribution shows no clustering phenomenon, similar to the chromosomal distribution of the USP gene family in potato (Qi et al., 2024; Wang et al., 2024). Among the EuUSPs family members, 25 are hydrophilic proteins, and their subcellular localization is primarily in the cytoplasm, chloroplasts, mitochondria, and nucleus, consistent with the localization reported for USP gene family members in other plants (Arabia et al., 2021).
A phylogenetic tree revealed that EuUSP5, EuUSP16, EuUSP23, and EuUSP26 from E.ulmoides show close phylogenetic relationships with USPs from H.brasiliensis and T.kok-saghyz, and have been detected in rubber particles (unpublished data). This suggests that these EuUSPs members might share similar evolutionary trajectories with other rubber-producing plants like H.brasiliensis and T.kok-saghyz, and could potentially participate in the regulation of rubber biosynthesis. The number and composition of motifs vary among the EuUSPs family members. Except for EuUSP20 and EuUSP29, the other 27 EuUSPs all contain Motif 2 and Motif 8, indicating high conservation within the EuUSPs gene family during evolution. Among the EuUSPs family members, EuUSP7 lacks introns, while the other 27 EuUSPs possess 1 to 10 introns. Over half of the members contain either 2 introns or 3 introns. Gilbert found that introns in ancient gene structures correlate with gene evolution (Gilbert and Glynias, 1993). In Gossypium hirsutum, the U and F subfamilies of the GST family appeared first, followed by the evolution of other subfamilies (Xu et al., 2017). In this study, EuUSP7 (with no introns) and EuUSP2, EuUSP6, EuUSP19, EuUSP26 (each with only one intron) may represent the earliest evolved genes. The other members, with varying numbers of introns, likely appeared later during gene evolution. Intron-derived motifs can enhance mRNA accumulation and post-transcriptional gene regulation through both splicing-dependent and splicing-independent mechanisms (Gallegos and Rose, 2019). Therefore, it is hypothesized that the other 28 intron-containing EuUSPs evolved different numbers of introns to regulate gene expression. However, the possibility that the intronless EuUSP7 resulted from intron loss due to retrotransposition cannot be excluded and requires further analysis and validation.
Synteny can be used to predict homologous gene sequences during evolution. Synteny analysis at the whole-genome level revealed that all 21 EuUSPs genes analyzed show syntenic homology with HbUSPs genes from H.brasiliensis, indicating conservation of EuUSPs genes during the domestication of E.ulmoides. Within the EuUSPs family, four segmental duplication events involving eight EuUSPs genes were identified, but no tandem duplication events were found. Gene duplication, as a major driver of plant genome evolution, provides the material basis for gene family expansion through the generation of genetic redundancy (Lawton-Rauh, 2003). The newly duplicated genes in E.ulmoides, under natural selection, can acquire biological functions divergent from the original genes through functional mutations (Conant and Wolfe, 2008). Notably, the functional diversity arising from such duplication events profoundly influences the dynamics of gene expression regulatory networks. This is manifested as spatio-temporal adjustments in mRNA expression levels and adaptive changes in the physicochemical properties of proteins (Zhu et al., 2022). Such multi-layered molecular innovation mechanisms ultimately shape the evolutionary plasticity of plants in responding to environmental changes.
4.2 Expression characteristics and functional role of EuUSPs genes in E.ulmoides rubber biosynthesis
We found that the promoter regions of the EuUSPs gene family in E.ulmoides are rich in regulatory elements, primarily including plant hormone response elements. Additionally, four types of cis-acting elements related to abiotic and biotic stresses were identified, involving plant responses to abiotic stresses such as anaerobic conditions, low temperature, defense stress, and drought. Studies have shown that the environment can affect the content of plant secondary metabolites (Copolovici et al., 2012; Omokhafe and Emuedo, 2006). When E.ulmoides seedlings were subjected to drought treatment for 10 days, the expression levels of EuUSP1, EuUSP16, EuUSP22, and EuUSP23 were significantly upregulated. This change coincided with the alteration in E.ulmoides rubber content observed after 10 days of drought treatment. Furthermore, research indicates that temperature can influence E.ulmoides rubber synthesis (Yao and Zhao, 2023). In this study, during low-temperature treatment for 10 days, the expression levels of EuUSP1, EuUSP6, EuUSP7, EuUSP16, EuUSP18, and EuUSP20 were upregulated in the stems of E.ulmoides seedlings but downregulated in the leaves. This expression pattern aligns with the observed increase in rubber content in the stems and the decrease in rubber content in the leaves following treatment. Collectively, these findings suggest that EuUSP16 may be involved in E.ulmoides rubber synthesis under stress conditions.
4.3 Relationship between EuUSP16 gene expression and E.ulmoides rubber biosynthesis
E.ulmoides rubber biosynthesis primarily occurs via two metabolic pathways: the MVA pathway and the MEP pathway. Numerous key enzymes function within these pathways. Geranylgeranyl diphosphate synthase (GGPPS) and farnesyl diphosphate synthase (FPS) synthesize GGPP and FPP, respectively, providing the isoprenoid precursor isopentenyl diphosphate (IPP) for rubber chain elongation (Miziorko, 2011). Rubber elongation factor (REF) binds to the rubber synthase complex and directly participates in isoprenoid chain elongation, while small rubber particle protein (SRPP) stabilizes rubber particle structure, preventing polymer degradation. In E.ulmoides adventitious buds overexpressing EuUSP16, the relative expression levels of EuGGPPS, EuFPS1, EuREF, and EuSRPP1 genes were significantly increased. Conversely, in EuUSP16-silenced E.ulmoides plants, the relative expression levels of EuFPS1, EuREF, and EuSRPP1 genes were significantly decreased. Furthermore, rubber content was significantly elevated in EuUSP16-overexpressing callus tissues. These findings strongly indicate that EuUSP16 positively regulates the expression of core rubber biosynthesis genes (EuFPS1, EuREF, EuSRPP1). Whether EuUSP16 promotes precursor accumulation needs to be verified by detecting IPP levels in EuUSP16-transgenic overexpression plants. We also observed that Isopentenyl diphosphate isomerase(IPI)gene EuIPI expression was significantly downregulated in EuUSP16-overexpressing adventitious buds, while its expression was significantly upregulated in EuUSP16-silenced plants. IPI catalyzes the interconversion of IPP and dimethylallyl diphosphate (DMAPP) (Liao et al., 2016). The downregulation of the EuIPI gene may reduce the conversion of IPP to DMAPP, thereby diverting more IPP towards rubber synthesis rather than the synthesis of other metabolic products. Whether EuUSP16 promotes precursor accumulation needs to be verified by detecting IPP levels in EuUSP16-transgenic overexpression plants. Simultaneously, the protein-protein interaction between EuUSP16 and rubber synthase requires verification via yeast two-hybrid (Y2H) assay.
4.4 The EuDof transcription factor regulates the expression of the EuUSP16 gene
Studies have shown that the presence of specific cis-acting elements within promoter regions can regulate tissue-specific gene expression (Chen et al., 2014, 2015; Rusconi et al., 2013). With the exception of PEuUSP1-4, GUS expression levels were highest in leaves. For PEuUSP16-1, GUS expression showed no significant difference between roots and leaves. This may be associated with the presence of drought-inducible response elements in PEuUSP16-1, as drought primarily results from soil water deficit. Plant roots, being the primary organs for water and nutrient uptake from soil, play a crucial role under drought conditions (Kim et al., 2020). The GUS activity driven by PEuUSP16–1 was significantly higher than that by PEuUSP16-2, indicating that the region from -1967 bp to -1286 bp positively regulates GUS expression. Alterations in cis-elements within promoter regions can modulate the activity of downstream gene expression. The distance of cis-acting elements from the transcription start site (TSS) can influence promoter activity; cis-elements located further upstream of the TSS may exhibit reduced activity due to steric hindrance limiting transcription factor binding (Imoto et al., 2008; Kiesenhofer et al., 2018; Rushton, 2016). Following low-temperature treatment of transgenic tobacco plants harboring PEuUSP16–1 or PEuUSP16-2, the relative GUS expression increased in both, but the increase was more pronounced in PEuUSP16-2. Conversely, ABA treatment of transgenic tobacco plants carrying PEuUSP16-1, PEuUSP16-2, or PEuUSP1–3 resulted in decreased relative GUS expression, with a more significant reduction observed in PEuUSP16–3 compared to PEuUSP16–1 and PEuUSP16-2. These differential responses may be related to the positions of low-temperature and ABA-responsive elements within their respective promoter regions.
Research indicates that Dof transcription factors participate in abiotic stress responses in numerous plant species (Waschburger et al., 2023; Yanagisawa, 2004; Zou and Sun, 2023). ZmDof22 enhances drought tolerance in maize by positively regulating genes involved in stomatal closure to reduce water loss and activating antioxidant enzymes such as SOD and POD (Cao et al., 2024). In cotton, overexpression of GhDof1 improves salt and cold tolerance (Su et al., 2017). VaDof17d enhances cold tolerance in grapevine (Wang et al., 2021). Overexpression of tomato SlCDF1 and SlCDF3 genes in Arabidopsis confers enhanced drought and salt tolerance (Corrales et al., 2014). Furthermore, Dof transcription factors play significant regulatory roles in plant secondary metabolism (Guo et al., 2019). The Cys2/Cys2 zinc finger domain is a common DNA-binding domain widely involved in transcriptional regulation. Under drought/cold stress, EuDof directly binds the EuUSP16 promoter to activate expression, whereas ABA suppresses this activation. This EuDof-EuUSP16 regulatory pathway contrasts with established TF networks, such as the ethylene-induced HbERF1-mediated upregulation of HbREF/HbSRPP in H.brasiliensis, as it operates independently of Jasmonic Acid (JA)/ethylene signaling (Shi et al., 2016; Wu et al., 2010). This identification of a EuDof-EuUSP16 regulatory axis represents a novel mechanism within plant secondary metabolism. However, the identification of the EuDof transcription factor and its interaction with the EuUSP16 promoter suggest the potential existence of a complex transcriptional regulatory network. Future studies should further explore interactions between EuDof and other transcription factors, as well as its specific functional role in E.ulmoides rubber biosynthesis.
5 Conclusion
This study identified 29 EuUSPs genes at the genomic level. Among them, EuUSP16 plays a critical role in E.ulmoides rubber biosynthesis. The EuDof transcription factor enhanced the promoter activity of EuUSP16. Furthermore, binding of EuDof to the EuUSP16 promoter was enhanced under low temperature and drought conditions, but inhibited by ABA treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SL: Writing – original draft, Writing – review & editing. JY: Writing – review & editing, Methodology. HW: Writing – review & editing. XC: Writing – review & editing. D-GZ: Writing – original draft, Resources, Writing – review & editing, Funding acquisition. YZ: Writing – review & editing, Data curation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant number 31870285), Guizhou Academy of Agricultural Sciences Talent Special Project, grant number ((2023)02, (2024)02 and (2025)02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fpls.2025.1689576.
Generative AI statement
The author(s) declare no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1655155/full#supplementary-material
References
Arabia, S., Sami, A. A., Akhter, S., Sarker, R. H., and Islam, T. (2021). Comprehensive in silico characterization of Universal Stress Proteins in Rice (Oryza sativa L.) with insight into their stress-specific transcriptional modulation. Frontier Plant Sci. 12. doi: 10.3389/fpls.2021.712607
Aravind, L., Anantharaman, V., and Koonin, E. V. (2002). Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA world. Proteins: Structure Function Bioinf. 48, 1–14. doi: 10.1002/prot.10064
Bhuria, M., Goel, P., Kumar, S., and Singh, A. K. (2022). AtUSP17 negatively regulates salt stress tolerance through modulation of multiple signaling pathways in Arabidopsis. Physiologia Plantarum 174, e13635. doi: 10.1111/ppl.13635
Cao, L. R., Ye, F. Y., Fahim, A. M., Ma, C. C., Pang, Y. Y., and Zhang, X. (2024). Transcription factor ZmDof22 enhances drought tolerance by regulating stomatal movement and antioxidant enzymes activities in maize (Zea mays L.). Theor. Appl. Genet. 137, 132. doi: 10.1007/s00122-024-04625-w
Chen, L., Jiang, B. J., Wu, C. X., Sun, S., Hou, W. S., and Han, T. (2014). GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant Biol. 14, 245. doi: 10.1186/s12870-014-0245-z
Chen, L., Jiang, B. J., Wu, C. X., Sun, S., Hou, W. S., and Han, T. (2015). The characterization of GmTIP, a root-specific gene from soybean, and the expression analysis of its promoter. Plant Cell Tissue Organ Culture (PCTOC) 121, 259–274. doi: 10.1007/s11240-014-0682-2
Conant, G. and Wolfe, K. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Copolovici, L., Kännaste, A., Pazouki, L., and Niinemets, Ü. (2012). Emissions of green leaf volatiles and terpenoids from Solanum lycopersicum are quantitatively related to the severity of cold and heat shock treatments. J. Plant Physiol. 169, 664–672. doi: 10.1016/j.jplph.2011.12.019
Corrales, A. R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Cui, X. Y., Zhang, P. Y., Chen, C. C., and Zhang, J. X. (2022). VyUSPA3, a universal stress protein from the Chinese wild grape Vitis yeshanensis, confers drought tolerance to transgenic V.vinifera. Plant Cell Rep. 42, 181–196. doi: 10.1007/s00299-022-02943-1
Du, H. Y., Liu, C. Y., Li, Q., and Liu, P. F. (2011). Seasonal variation of the content of three main active ingredients in Du Zhong leaves. J. Cent. South Univ. Forestry Technol. 31, 6–9. doi: 10.14067/j.cnki.1673-923x.2011.08.024
Gallegos, J. E. and Rose, A. B. (2019). An intron-derived motif strongly increases gene expression from transcribed sequences through a splicing independent mechanism in Arabidopsis thaliana. Sci. Rep. 9, 13777. doi: 10.1038/s41598-019-50389-5
Gilbert, W. and Glynias, M. (1993). On the ancient nature of introns. Gene 135, 137–144. doi: 10.1016/0378-1119(93)90058-b
Guo, Y. X., Chen, J., Wang, Y. F., Sun, C. Y., Wang, Y., and Zhang, M. P. (2019). The regulatory role of Dof transcription factor in plants. Biotechnol. Bull. 35, 146–156. doi: 10.13560/j.cnki.biotech.bull.1985.2018-1062
Gustavsson, N., Diez, A., and Nyström, T. (2002). The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43, 107–117. doi: 10.1046/j.1365-2958.2002.02720.x
Hu, P. and Zhao, D. G. (2024). Cloning and functional analysis of universal stress protein gene EuUSP1 in Eucommia ulmoides. Plant Physiol. J. 60, 130–140. doi: 10.13592/j.cnki.ppj.100741
Imoto, Y., Yamada, T., Kitamura, K., and Kanazawa, A. (2008). Spatial and temporal control of transcription of the soybean beta-conglycinin alpha subunit gene is conferred by its proximal promoter region and accounts for the unequal distribution of the protein during embryogenesis. Genes Genet. Syst. 83, 469–476. doi: 10.1266/ggs.83.469
Kiesenhofer, D. P., Mach, R. L., and Mach-Aigner, A. R. (2018). Influence of cis-element arrangement on promoter strength in Trichoderma reesei. Appl. Environ. Microbiol. 84, e01742–e01717. doi: 10.1128/AEM.01742-17
Kim, Y., Chung, Y. S., Lee, E., Tripathi, P., Heo, S., and Kim, K. H. (2020). Root response to drought stress in Rice (Oryza sativa L.). Int. J. Mol. Sci. 21, 1513. doi: 10.3390/ijms21041513
Lawton-Rauh, R. A. (2003). Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 29, 396–409. doi: 10.1016/j.ympev.2003.07.004
Liao, P., Hemmerlin, A., Bach, T. J., and Chye, M. L. (2016). The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 34, 697–713. doi: 10.1016/j.bioteChadv.2016.03.005
Livak, k. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Z. K., Xie, B. X., and Du, H. Y (2004). Study on the extraction method of Eucommia Rubber. J. Fujian Univ. Forestry 04, 353–356. doi: 1001-389X(2004)04-353-04
Men, X., Wang, F., Chen, G. Q., Zhang, H. B., and Xian, M. (2019). Biosynthesis of natural rubber: current state and perspectives. Int. J. Mol. Sci. 20, 50. doi: 10.3390/ijms20010050
Miziorko, H. M. (2011). Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophysics 505, 131–143. doi: 10.1016/j.abb.2010.09.028
Nabi, B., Kumawat, M., Ahlawat, N., and Ahlawat, S. (2024). Molecular, structural, and functional diversity of universal stress proteins (USPs) in bacteria, plants, and their biotechnological applications. Protein J. 43, 437–446. doi: 10.1007/s10930-024-10192-2
Omokhafe, O. K. and Emuedo, A. O. (2006). Evaluation of influence of five weather characters on latex yield in Hevea brasiliensis. Int. J. Agric. Res. 5, 619–624. doi: 10.3923/ijar.2006.234.239
Qi, T. S., He, F. M., Zhang, X. Q., Wang, J., Zhang, Z. L., Jiang, H. R., et al. (2024). Genome-wide identification and expression profiling of potato (Solanum tuberosum L.) universal stress proteins reveal essential roles in mechanical damage and deoxynivalenol stress. Int. J. Mol. Sci. 25, 1341. doi: 10.3390/ijms25021341
Ren, Z. Q., Bi, X. Q., Zhang, L. W., Chen, Y., and Luo, Q. X. (2022). A preliminary study on expression characteristics and functions of recombinant amUspA protein from amygdalusmira. J. Shenyang Agric. Univ. 53, 24–33. doi: 10.3969/j.issn.1000-1700.2022.01.004
Rohmer, M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Natural Product Rep. 16, 565–574. doi: 10.1039/a709175c
Rusconi, F., Simeoni, F., Francia, P., Cominelli, E., Conti, L., Riboni, M., et al. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64, 3361–3371. doi: 10.1093/jxb/ert180
Rushton, P. J. (2016). What have we learned about synthetic promoter construction? Methods Mol. Biol. 1482, 1–13. doi: 10.1007/978-1-4939-6396-6_1
Shi, M. J., Cai, F. G., and Tian, W. M. (2016). Ethrel-stimulated prolongation of latex flow in the rubber tree (Hevea brasiliensis Muell. Arg.): an Hev b 7-like protein acts as a universal antagonist of rubber particle aggregating factors from lutoids and C-serum. J. Biochem. 159, 209–216. doi: 10.1093/jb/mvv095
Su, Y., Liang, W., Liu, Z. J., Wang, Y. M., Zhao, Y. P., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Tkaczuk, K. L., Shumilin., I., Chruszcz, M., Evdokimova, E., Savchenko, A., and Minor, W. (2013). Structural and functional insight into the universal stress protein family. Evolutionary Appl. 6, 434–449. doi: 10.1111/eva.12057
Udawat, P., Jha, R. K., Sinha, D., Mishra, A., and Jha, B. (2016). Overexpression of a cytosolic abiotic stress responsive Universal Stress Protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00518
Wang, Z. M., Wang, Y., Tong, Q., Xu, G., Xu, M., Li, H., et al. (2021). Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 253, 5. doi: 10.1007/s00425-021-03574-8
Wang, Z. M., Wang, Y., Tong, Q., Xu, G. Z., Xu, M. L., Li, H. Y., et al. (2024). Identification of the StUSP gene family of potato universal stress proteins and analysis of stress expression patterns. Agric. Res. arid areas 42, 32–42. doi: 10.3390/ijms25021341
Waschburger, E. L., Filgueiras, J. P. C., and Turchetto-Zolet, A. C. (2023). DOF gene family expansion and diversification. Genet. Mol. Biol. 46, e20230109. doi: 10.1590/1678-4685-GMB-2023-0109
Wei, X. N., Peng, P., Peng, F., and Dong, J. (2021). Natural polymer Eucommia Ulmoides rubber: A Novel Material. J. Agric. Food Chem. 69, 3797–3821. doi: 10.1021/acs.jafc.0c07560
Wu, H. L., Yu, B., Cheng, Q. Q., Zeng, R. Z., Duan, C. F., Nie, Z. Y., et al. (2010). Cloning and expression analysis of an AP2/EREBP gene, a jasmonic acid-inducible related factor in laticifer cells of rubber tree (Hevea brasiliensis). Chin. Agric. Sci. Bull. 26, 287–293.
Xie, J., Li, W., Chen, H., Dang, P. F., Liu, S. S., and Hu, T. F. (2013). Extraction and purification of Eucommia ulmoides gum in Eucommia ulmoides leaves. J. Ankang Coll. 25, 44–46. doi: 10.16858/j.issn.1674-0092.2013.06.014
Xu, L., Chen, W., Si, G. Y., Huang, Y. Y., Lin, Y., Cai, Y. P., et al. (2017). Genome-wide analysis of the GST gene families in upland cotton. Genetic 39, 737–752. doi: 10.16288/j.yczz.16-435
Yanagisawa, S. (2004). Dof Domain Proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Yao, W. Q. and Zhao, D. G. (2023). Effects of temperature on growth and isoprene metabolism pathway in eucommia ulmoides oliv. Horticulturae 9, 1298. doi: 10.3390/horticulturae9121298
Zhu, Q. B., Gan, Z. C., Li, X. C., Zhang, Y. J., Zhao, H. M., and Huang, X. Z. (2022). Genome-wide identification and evolution and expression of the MAPKKK gene family in mouse ear mustard. Genetic 44, 1044–1055. doi: 10.16288/j.yczz.22-261
Zhu, M. Q. and Sun, R. C. (2018). Eucommia ulmoides oliver: A potential feedstock for bioactive products. J. Agric. Food Chem. 66, 5433–5438. doi: 10.1021/acs.jafc.8b01312
Keywords: Eucommia ulmoides Oliv., EuUSPs gene family, environmental stress, rubber biosynthesis, EuDof
Citation: Long S, Yang J, Wang H, Chen X, Zhao D-g and Zhao Y (2025) Genome-wide identification of EuUSPs in Eucommia ulmoides and the role of EuUSP16 in rubber biosynthesis. Front. Plant Sci. 16:1655155. doi: 10.3389/fpls.2025.1655155
Received: 27 June 2025; Accepted: 30 July 2025;
Published: 20 August 2025; Corrected: 03 September 2025.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Dejun Li, Chinese Academy of Tropical Agricultural Sciences, ChinaWei Yao, Guangxi University, China
Copyright © 2025 Long, Yang, Wang, Chen, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yichen Zhao, eWN6aGFvQGd6dS5lZHUuY24=; De-gang Zhao, ZGd6aGFvQGd6dS5lZHUuY24=
 Shangmei Long
Shangmei Long Jiang Yang1
Jiang Yang1 De-gang Zhao
De-gang Zhao Yichen Zhao
Yichen Zhao