- 1Plant Molecular Biology Lab (DBT-BioCARe), Department of Biotechnology & Crop Improvement, College of Horticulture, Bagalkot, University of Horticultural Sciences, Bagalkot, Karnataka, India
- 2Kittur Rani Channamma College of Horticulture, Arabhavi, University of Horticultural Sciences, Bagalkot, Karnataka, India
- 3Indian Council of Forestry Research and Education (ICFRE)-Institute of Forest Biodiversity, Hyderabad, India
- 4ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
- 5Department of Plant & Agroecosystem Sciences, University of Wisconsin, Madison, WI, United States
- 6USDA-ARS Vegetable Crops Research Service, Madison, WI, United States
Carrot (Daucus carota L.) is a globally cultivated root vegetable with significant genetic diversity. This first study generated and validated carrot InDels to unravel the genetic divergence between Eastern and Western gene pools, integrating agro-morphometric traits with functional InDel markers. Eastern accessions exhibited larger plants, bigger roots with diverse colors, while Western accessions were more uniform orange color and compact in architecture. From RNA-seq data, 271 agarose-resolvable functional InDels (>15bp length difference) were identified, of which 48 validated markers showed high polymorphism (84.21%) across two gene pools supporting secondary domestication changes. Located in coding and UTR regions, these InDels likely regulate gene expression and may have contributed to significant genetic modifications among carrot gene pools. Genetic diversity in the Western gene pool indicated more intense selection and domestication. Population structure and phylogenetic analysis revealed clear gene pool differentiation (Fst = 0.181) with potential gene flow (Nm = 1.716). Functional annotation of linked InDels to key biological processes, highlighted their role in domestication. Key InDels (DcFInDel32, DcFInDel28, and DcFInDel55) were associated with multiple traits, underscoring their utility in marker-assisted selection (MAS). These findings provide insights for developing improved carrot cultivars with high yield and quality adapted to diverse climates.
Introduction
Carrot (Daucus carota subsp. sativus L.) is an ancient, cool-season root vegetable of the Apiaceae family with a diploid genome of 473 Mb and 2n = 2x = 18 (Iorizzo et al., 2016). Native to Afghanistan region, carrots are now cultivated globally for their rich nutritional profile, which includes high levels of carotenoids, antioxidants, vitamins, and minerals, contributing to their well-known immune-boosting, anti-inflammatory, and antioxidant properties (Kulkarni et al., 2023). Originally, carrots were valued more for their flavorful leaves and seeds than their roots. Over time, however, selective breeding has led to the domestication of the root, producing varieties with orange, purple, white, yellow, and red, with the orange type being commercially dominant (Bradeen and Simon, 2007).
Carrot germplasm exhibits considerable phenotypic diversity, with cultivated varieties broadly classified into Eastern and Western types based on morphological and evolutionary characteristics (Coe et al., 2023; Cholin et al., 2024). The Eastern varieties, often referred to as Asiatic carrots (Daucus carota ssp. sativus var. atrorubens Alef.), were domesticated over 1,100 years ago in Central and Eastern Asia from wild carrot progenitors such as Queen Anne’s Lace. These carrots are typically annual, red colored, and characterized by a juicier, coarser texture with larger core sizes. They also require little to no vernalization for flowering (Ellison, 2019; Kulkarni et al., 2023). In contrast, Western carrots (Daucus carota ssp. sativus var. sativus), originated in Northern Europe, are typically biennial, exhibit a deep orange color, and require vernalization for flowering. These carrots have been bred for higher carotene content and possess a more consistent phloem pattern (Ellison, 2019; Kulkarni et al., 2023). Domestication led to significant changes in root morphology, physiology, and biochemical composition, including improved carotenoid content, anthocyanin accumulation, and sugar levels, root size, and reducing lateral root branching. Molecular studies suggest Western varieties likely evolved from Eastern types, highlighting the evolutionary trajectory of carrot domestication (Iorizzo et al., 2013; Baranski et al., 2012; Coe et al., 2023).
In modern plant breeding, molecular markers are crucial in tracking genetic diversity and identifying genes linked to important agronomic traits (Anderson and Lubberstedt, 2003). While random DNA markers, such as AFLPs, detect polymorphisms, recombination events over generations limit their utility. Functional markers (FMs), derived from characterized sequence motifs, offer greater precision in identifying allelic diversity and linking genetic variation to phenotypic expression. Recombination does not affect these markers, making them more reliable for long-term genetic studies (Salgotra and Stewart, 2020). Among these Expressed Sequence Tag-Simple Sequence Repeat (EST-SSR) and Insertion-Deletion (InDel), are PCR-based and co-dominant. InDels, which arise from nucleotide insertions or deletions, play a major role in molecular and chromosomal evolution; second in frequency only after SNPs (Andersson and Purugganan, 2022; Redelings et al., 2024; Savino et al, 2022), providing higher polymorphism than SSRs, enabling detection of subtle genetic variation (Pan et al., 2021). Functionally characterized InDel markers, particularly in or near key genes provide information on key traits such as root size, flowering time, disease resistance, and stress tolerance, aiding marker-assisted selection (Thakur and Randhawa, 2018; Zhao et al., 2018; Shi et al., 2023; Sahu et al., 2017; Cui et al., 2021).
In carrots, InDel-mediated mutations played significant role in domestication and nutritional traits (Rolling et al., 2022). For example, ‘Y’ gene affecting carotenoid accumulation involve 216bp, 2bp, and 60bp in various alleles of the DCAR_032551 gene (Iorizzo et al., 2016), carotene hydroxylase is conditioned with both high total carotenoid content and high alpha-carotene content of carrot roots by an 8bp InDel (Arango et al., 2014; Coe et al., 2021). Sugar composition in edible root is also influenced by a 2.5kb InDel, in the invertase isozyme II Rs gene (Yau and Simon, 2003) and numerous InDels from transposable elements have been associated with intron-length polymorphism in carrots (Grzebelus et al., 2006; Stelmach et al, 2017).
Understanding the genetic diversity within and between cultivated carrot populations is essential for breeding to improve agronomic traits. Population structure analysis, which investigates genetic composition and gene flow, is a powerful tool for understanding the evolutionary forces shaping genetic diversity within a species. Previous studies using SSR markers have revealed significant genetic diversity within cultivated carrot germplasm, with moderate differentiation between Eastern and Western carrot gene pools (Clotault et al., 2010; Baranski et al., 2012; Iorizzo et al., 2013; Chaitra et al., 2020). In this study, we analyzed morphological and molecular diversity between the Eastern and Western carrot gene pools using transcriptome-derived, functionally characterized InDel markers from root and floral tissues (Cholin et al., 2024). This represents the first global report on transcriptome-based InDel marker development and validation in carrots, providing insights into genetic differences among the gene pools and enhancing the use of functional markers in breeding programs.
Materials and methods
Plant material
The study included 98 accessions, with 47 accessions representing the Western gene pool from the United States Department of Agriculture (USDA), USA, and 46 representing the Eastern gene pool of Indian origin. Additionally, five check varieties viz., three Eastern types Pusa Rudhira (C1), Pusa Asita (C2), Krishna Prabha Vriddhi (C3), and two Western carrot types Super Kuroda Improved (C4), and Kuroda (C5) were included for agro-morphometric analysis (Supplementary Table S1).
Location of the experiment
Agro-morphometric traits were assessed at two experimental stations in Karnataka, India: (a) the College of Horticulture, Bagalkot (CoH Bagalkot) and (b) the Kittur Rani Channamma College of Horticulture, Arabhavi (KRCCH, Arabhavi). CoH Bagalkot is situated in the northern dry zone at coordinates 16°12′N, 75°45′E, with an average elevation of approximately 610 m and annual rainfall of 552 mm. KRCCH Arabhavi, located in the northern transition zone, is positioned at 16°15′N, 75°45′E, with an elevation of 612.05 m and experiences an average annual precipitation of 554 mm. Both locations have black soil with a loamy texture, ideal for crop growth. The experiment was conducted during the Rabi season (winter cropping season- September to December 2023) using an augmented block design to evaluate 98 accessions, including five check varieties, distributed across 12 blocks.
Agro-morphometric trait evaluation
Phenotypic observations were recorded on 23 traits, including 10 qualitative (IPGRI,1998 descriptor) and 13 quantitative (Table 1), from five randomly selected plants. Biometrical analysis for quantitative traits was performed with the mean data of five plants in each accession.
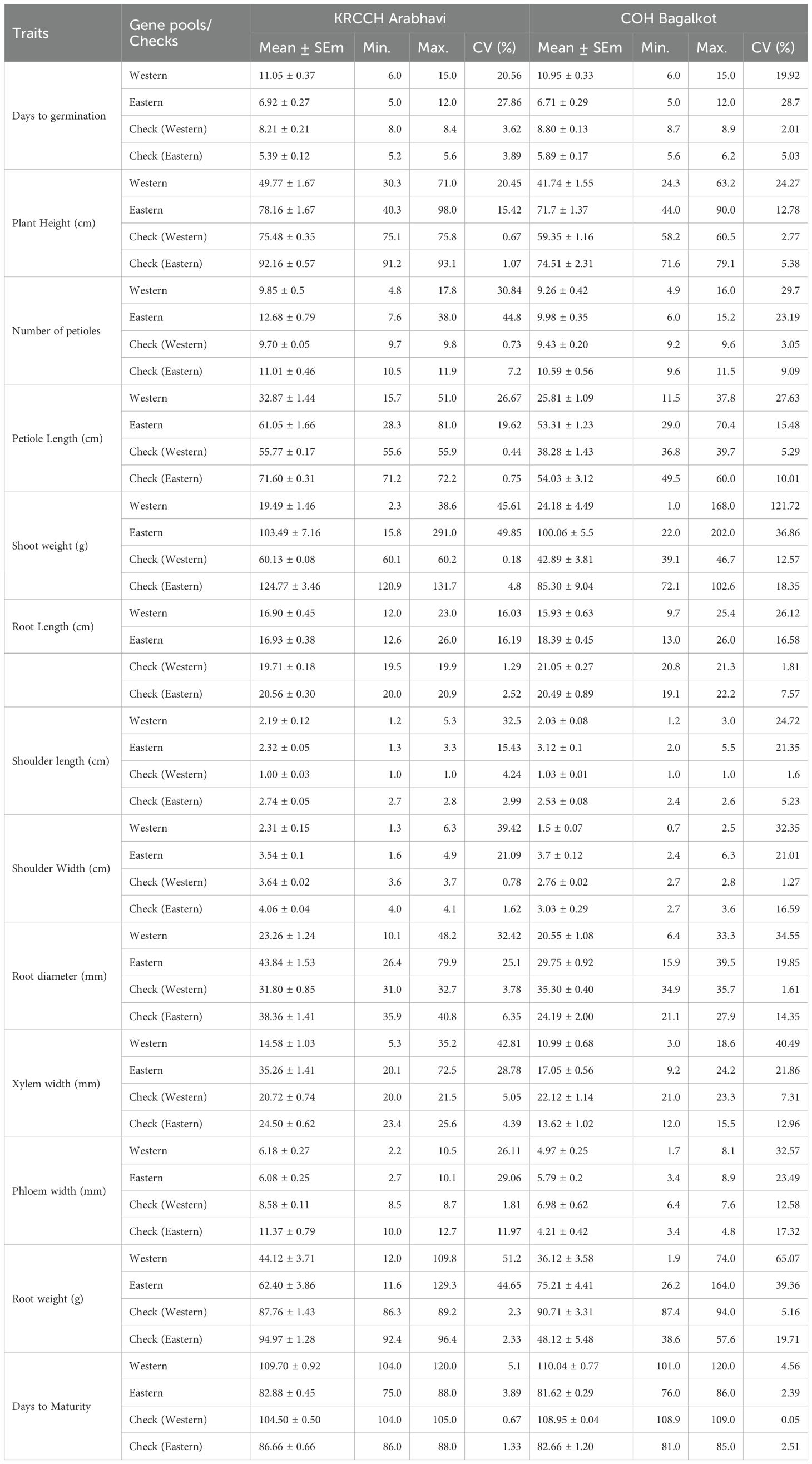
Table 1. Comparison of descriptive statistics across Western and Eastern accession and corresponding check varieties across locations.
Development of InDel markers
For the identification of InDels, twelve RNAseq libraries generated in a previous study (Cholin et al., 2024) for Eastern and Western representative accessions were utilized. Clean reads were obtained by trimming low-quality reads from the raw paired-end FASTQ files, and the reads were then aligned to the DH1 v3.0 carrot reference RNA assembly (Coe et al., 2023; NCBI RefSeq assembly GCF_001625215.2_rna) using the BWA-MEM tool (Li, 2013). BAM files from all 12 libraries were used to generate a BCF file using bcftools mpileup (Li et al., 2009). SNP calling was performed using bcftools call (Li et al., 2009), which generated SNP and InDel information for all 12 samples at each position of the reference RNA. The resulting VCF file, containing InDel information, was used for further analysis. Initially based on the length difference between reference and alternate alleles, 19,817 InDels having >6bp were extracted and functionally annotated (Supplementary Table S2). Further, InDel size variations less than 15 bp and heterozygotes were excluded. InDel lengths of 15 bp or longer were considered potential candidates and selected for further evaluation via agarose gel electrophoresis (Supplementary Table S3). Flanking sequences for each InDel (0–200 bp upstream and downstream) were extracted from the DH1 v3.0 reference RNA, depending on the position of the InDel. PCR primers were designed using the NCBI Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the Primer3Plus web tool (https://www.primer3plus.com/). The following criteria viz., primer length of 18–25 bp, GC content of 40-60%, melting temperature difference between forward and reverse primers of not more than 3°C, and an expected PCR product size of 100–500 bp (Shivaprasad et al., 2024) was considered for designing primer pairs. InDels across 57 important genes of carrots spread across 9 chromosomes were selected for designing primers discussed in the results section (Supplementary Table S4). The detailed functions of 57 genes across different organisms along with the references are presented in Supplementary Table S4 (Aber et al., 2019; Astegno et al., 2017; Baurle et al., 2007; Casson et al., 2009; Chatukuta et al., 2018; Cheng et al., 2022; Dieterle et al., 2005; Eda et al., 2016; Farrow and Facchini, 2014; Ferreyra et al., 2010; Goralogia et al., 2017; Grant et al., 2000; Gruber et al., 2021; Guo et al., 2021; Han et al., 2021, Han et al., 2022; Harshavardhan et al., 2014; Hayama et al., 2019; Inoue et al., 2022; Israel et al., 2022; Janssens and Goris, 2001; Jun et al., 2015; Kotera et al., 2023; Kumar Meena et al., 2019; Li et al., 2021; Liu et al., 2023; Lopez-Ortiz et al., 2020; Marchetti et al., 2020; Masri and Kiss, 2023; Megha et al., 2022; Mishra et al., 2017; Mitsuda et al., 2005; Mousavi et al., 2021; Murray et al., 2006; Nedelyaeva et al., 2020; Nongpiur et al., 2012; North et al., 2007; Oelmuller et al., 1996; Paniagua et al., 2017; Plant et al., 2021; Rempola et al., 2006; Saiga et al., 2008; Saini and Nandi, 2022; Schulz et al., 2013; Seemann et al., 1990; Song et al., 2005; Thanh Ha Thi Do et al., 2018; Thomas et al., 2009; Tzafrir et al., 2002; Valmonte et al., 2014; Waite and Dardick, 2018; Wang et al., 2024; Wang et al., 2005; Wei et al., 2021; Wudick et al., 2018; Yip Delormel and Boudsocq, 2019; Yusa et al., 2019; Zhang et al., 2019; Zhao M. et al., 2022; Zhao Z, 2022).
Validation of InDel markers
DNA was extracted from the young leaves of a single plant from each of 98 carrot accessions using a modified CTAB extraction protocol (Doyle, 1991). Polymerase chain reaction (PCR) was performed in a reaction mixture containing 50 ng DNA, 2x PCR master mix (AMPLIQON), 0.25 µl each of forward and reverse primer (10 pM), and nuclease-free water. PCR was performed in a thermocycler (Eppendorf India Pvt. Ltd.) with the following conditions: an initial denaturation at 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 57–61°C (based on primer GC content) for 45 seconds, and 72°C for 45 seconds, with a final extension at 72°C for 5 minutes. A total of 57 InDel markers designed in the present study were used for genotyping 98 accessions. The amplified fragments were subjected to electrophoresis fractionation in 3.5% agarose gel and visualized by ethidium bromide (EtBr) staining. Clear bands within the expected size range were used for genotypic data generation, with allele sizes (in base pairs) determined using a reference ladder (GeneDirex® 100 bp DNA Ladder RTU).
Statistical analysis
The mean data of five plants from each accession were used to record agro-morphometric observations at two locations (CoH Bagalkot and KRCCH, Arabhavi) in Karnataka, India. Biometrical analysis was performed for 13 quantitative traits, and the trend was compared across Eastern and Western gene pools. The descriptive statistics analysis and heritability were estimated in the R tool package augmented RCBD (v. 4.4.1). Pearson’s correlation analysis, principal component analysis (PCA), and Euclidean genetic distance-based phylogenetic analysis were performed using PAST 4.03 (Hammer et al., 2001). Twelve qualitative traits were subjected to frequency distribution across Western and Eastern accessions.
The allele scores of polymorphic InDel markers across 98 accessions were subjected to various analyses. Marker diversity parameters and allelic frequencies (Na, Ne, Ho, He, I, F, and PIC), analysis of molecular variance (AMOVA), and principal coordinate analysis (PCoA) were computed using GenAlEx 6.51b2 (Peakall and Smouse, 2012). Each gene pool was treated as a separate population, population-wise marker diversity parameters were compared, and unique or private alleles were extracted from each gene pool. Phylogenetic relationships among the Western and Eastern carrot gene pools were assessed by constructing a neighbor-joining tree using the DARwin 6.0.021 software package (Perrier and Jacquemoud-Collet, 2006) based on Dice’s dissimilarity coefficient. Population structure and gene flow among accessions were examined using a model-based Bayesian clustering approach implemented in STRUCTURE v.2.3.4 (Pritchard et al., 2000). The analysis involved a burn-in period of 10,000 iterations, followed by 10,000 Markov Chain Monte Carlo (MCMC) repetitions. The optimal number of populations (K) was determined by testing K values ranging from 1 to 10, with five independent runs for each K. The true number of populations was identified using the Δ K method (Evanno et al., 2005) by submitting the structure results files to the STRUCTURE SELECTOR website (Earl, 2012), which returned a Delta K value of two. The optimum K value of two was used to assign accessions to populations based on Q values.
Marker-trait association analysis was conducted using phenotypic data from diverse accessions, polymorphic InDel markers and population structure data (Q matrix). The analysis was performed in TASSEL 3.0 (Bradbury et al., 2007) using a general linear model (GLM), with Q as a covariate to account for population structure. Associations were considered highly significant at a threshold of P < 0.001.
Functional characterization of polymorphic InDels
The Gene Ontology (GO) classification analysis was performed in BLAST2GO v6.03 tools using 48 polymorphic markers. The position of InDels was searched for individual genes in NCBI (https://www.ncbi.nlm.nih.gov accessed on 18th November 2024). Further, InDels in the coding region were compared for their amino acid changes in reference and alternate alleles by pairwise alignment in T-COFFEE sequence alignment server (https://tcoffee.crg.eu/apps/tcoffee/do:regular accessed on 29.11.2024).
Results
Agro-morphometric traits comparison of Eastern and Western gene pools
Qualitative traits such as color, shape, and texture are important parameters when selecting superior accessions for consumer acceptance and breeding programs. Among the 10 qualitative parameters, the expression was consistent across both locations (Figures 1A, B). The frequency of medium root position was highest in Western types, while deep root position was most common in Eastern types (Figure 1A). For root shape, the majority of the accessions exhibited tapering root shapes (Figure 1A). For shoulder shape, Western accessions displayed a predominance of flat to rounded shapes, whereas, Eastern accessions mostly had flat shapes (Figure 1A). Western accessions had fern-type leaves, while Eastern types exhibited a normal leaf type (Figure 1A). Root texture, hairiness, and cracking are important parameters in carrots influenced by soil conditions and genetic architecture and decide the quality of carrots for a higher price in the market. Among Western accessions, dimpled root texture was predominant, followed by coarse root texture, while in Eastern types, coarse-textured roots were the most common (Figure 1A). Cracking was absent in Western accessions, and a few Eastern accessions showed the presence of cracking (Figure 1A). Low to medium hairiness was observed in Eastern accessions while Western accessions had low hairiness (Figure 1B). Attractive root color is an important parameter in determining consumer acceptance in the market, and carrot carotenoids contribute to consumer health. It is an important parameter that has undergone many genetic changes during primary and secondary domestication events. Western accessions represent the outcome of crop improvement following the domestication event. Consequently, uniformity and consistent expression of both internal and external root colors were key characteristics of Western accessions (Figure 1B). The majority of the Western accessions were of orange color with uniform internal and external color, followed by white, yellow, red, and purple hues. In contrast, light orange was the predominant external root color in Eastern accessions, with yellow being the most common internal xylem and phloem color. Few white, purple, and red roots were also found in Eastern accessions, with the lighter intensity of the respective colors in the xylem and phloem showing their distinct vascular tissue pattern (Figure 1B).
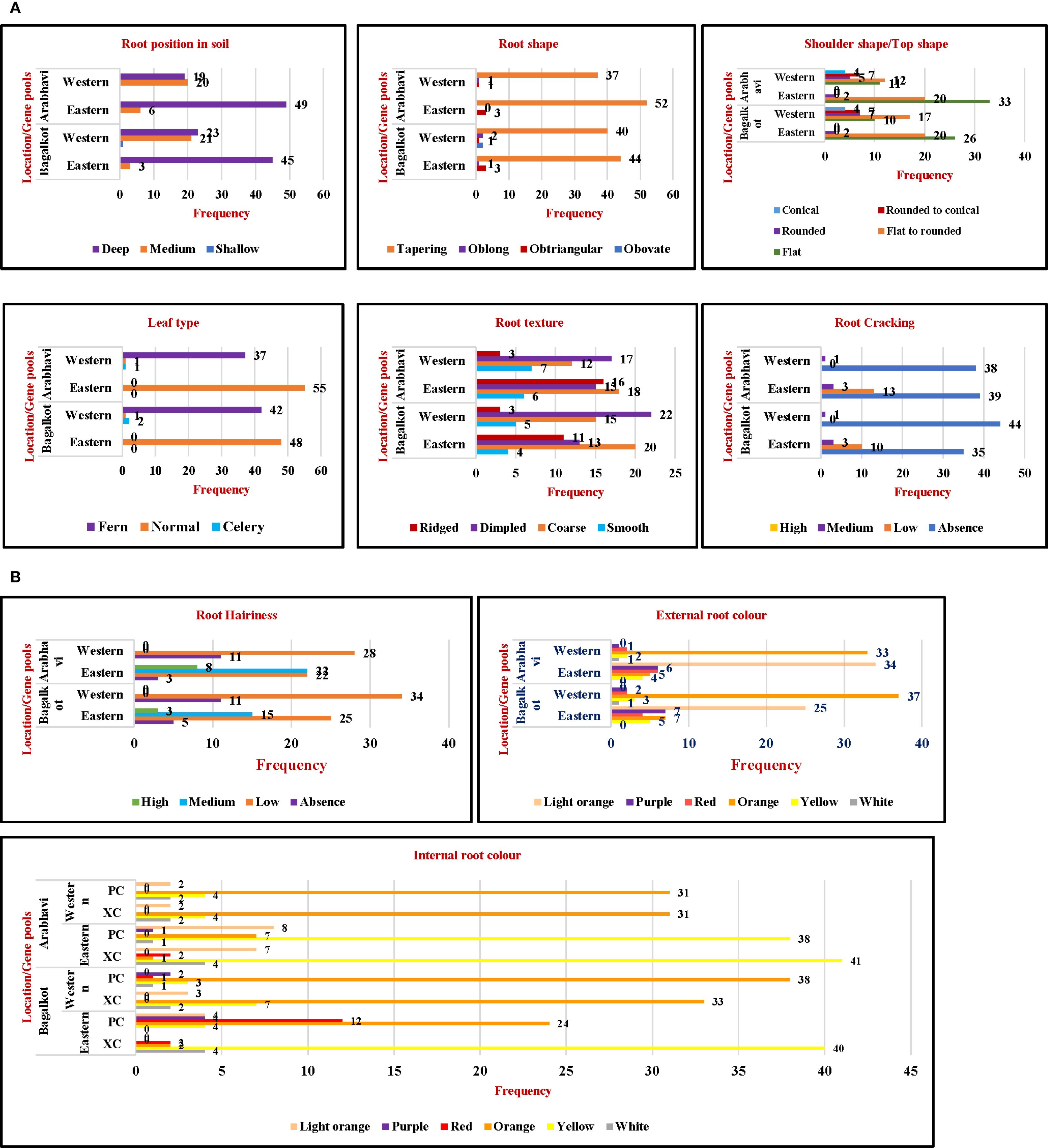
Figure 1. (A) Comparative frequency distribution of qualitative traits for two gene pools across locations. (B) Comparative frequency distribution of qualitative traits for two gene pools across locations. PC, Phloem color; XC, xylem color.
Descriptive statistics for quantitative traits revealed considerable variation among the 13 quantitative traits across the gene pools, with checks exhibiting less variability (Table 1). Among the above-ground traits, the Eastern gene pool demonstrated significantly higher values for plant height (PH), NP, TL, and SW at both locations. Root traits, including root length (RL), top length (TL), TW, RD, XW, and root weight (RW), were also significantly higher in the Eastern gene pool (Table 1). Notably, PW, a desirable trait, was higher in Western types at the Arabhavi location. Western carrot types require more time for both days to germination (DG) and days to maturation (DM). Heritability (h²) estimates were higher for most traits, except for root width, root length, phloem width, and root weight in Arabhavi, where these traits exhibited moderate heritability. While in Bagalkot location, higher heritability was observed for the majority of the traits, except for phloem width, number of petioles, and root length, which showed moderate heritability (Supplementary Table S5).
Pearson’s correlation analysis was performed to examine the relationships between quantitative traits across the Western and Eastern gene pools. The results revealed distinct association patterns between traits in the two gene pools (Figures 2A, B). In the Western gene pool, plant height (PH) exhibited a strong positive correlation with petiole length (PL), followed by root length (RL), and a moderate positive correlation with root weight (RW) and days to maturity (DM) (Figure 2A). In contrast, in the Eastern gene pool, PH showed a strong positive correlation with PL and was significantly positively correlated with most traits, except for the NP and TL, and DM (Figure 2B). SW was positively correlated with RD and XW in the Western gene pool, while in the Eastern gene pool, SW was positively correlated with RW. Additionally, a strong positive correlation was observed between TW, XW, and RD in the Western gene pool. In contrast, TW was positively correlated with RW in the Eastern gene pool. Notably, in the Western gene pool, TW was significantly positively correlated with RD, XW, PW, and RW, whereas in the Eastern gene pool, TW was positively associated with RW and RD. In the Eastern gene pool, RD showed a stronger positive correlation with XW, followed by PW and RW, while in the Western gene pool, XW and PW were similarly positively correlated with RD and RW. For both gene pools, XW showed significant positive correlations with PW and RW, and PW exhibited a positive correlation with RW. These correlation patterns highlight both the shared and distinct relationships between traits across the two gene pools.
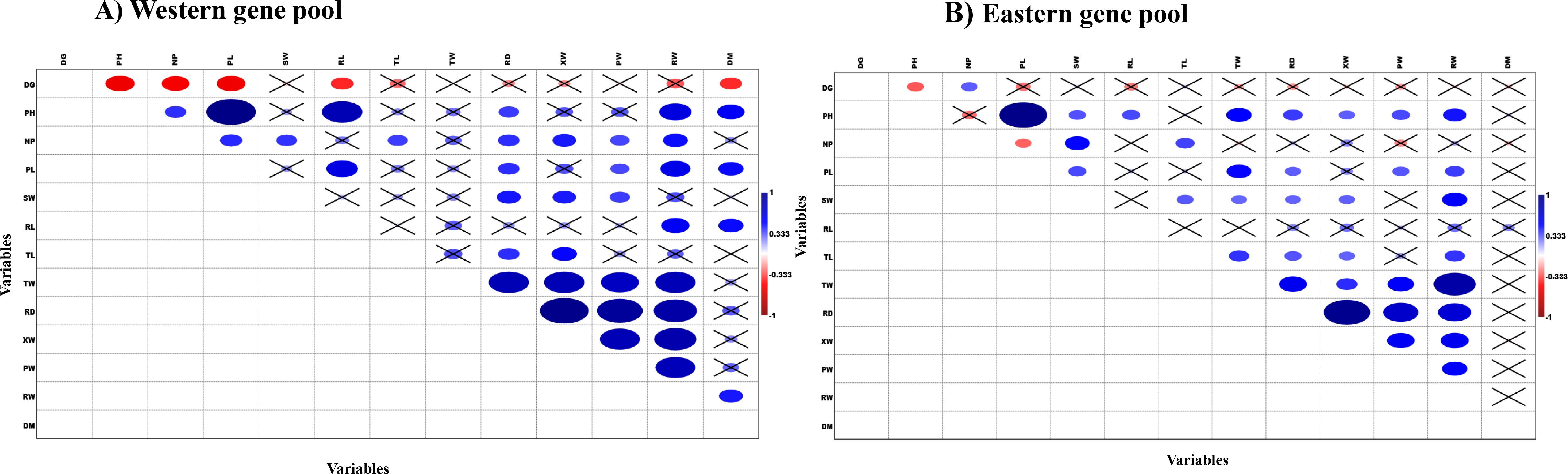
Figure 2. Comparison of correlation pattern among the phenotypic traits for Western and Eastern carrot gene pools. (A) Western gene pool. (B) Eastern gene pool (DG, Days to germination; PH, Plant height; NP, Number of petioles; PL, Petiole length; SW, Shoot weight; RL, Root length; TL, Top length; TW, Top width; RD, Root diameter; XW, Xylem width; PW, Phloem width; RW, Root weight; DM, Days to maturity).
Principal component analysis (PCA) of the 13 quantitative traits yielded 13 principal components (Supplementary Table S6). The first two components explained more than 80% of the total variance, as shown in the scree plot (Supplementary Table S6 & Supplementary Figure S1). Specifically, the first two components accounted for 74.16% and 9.22% of the variance in Arabhavi, and 81.12% and 7.74% of the variance in Bagalkot, respectively. The first principal component (PC1) explained with majority of variation as observed by the loading scores of 0.986 for shoot weight, 0.825 for plant height, 0.807 for petiole length, 0.787 for xylem width, 0.782 for root diameter, and 0.745 for top length at the Arabhavi location. While, in Bagalkot, PC1 accounted for the majority of variation by shoot weight (0.968), petiole length (0.882), top width (0.881), plant height (0.866), root weight (0.817), and top length (0.803) with their respective loading scores (Supplementary Tables S6B, C). The genotype-by-trait (GT) biplot, based on the first two principal components, effectively separated the Eastern and Western accessions at both locations (Figures 3A, B). The traits were distributed on the biplot according to their loadings and correlations. In Arabhavi, traits such as PW, RW, RL, TW, RD, DG, and DM formed small vectors with acute angles, indicating strong positive correlations among these traits (Figure 3A). In Bagalkot, XW showed positive correlations with these traits, and other vectors with acute angles also reflected positive correlations (Figure 3B). Vectors forming angles greater than 90° (e.g., PW and XW, TW and TW in Arabhavi, and SW and PW in Bagalkot) indicated negative correlations. The clear separation of Eastern and Western accessions in distinct quadrants of the biplot highlights their genetic differentiation, with the Western gene pool exhibiting greater genetic diversity than the Eastern gene pool.
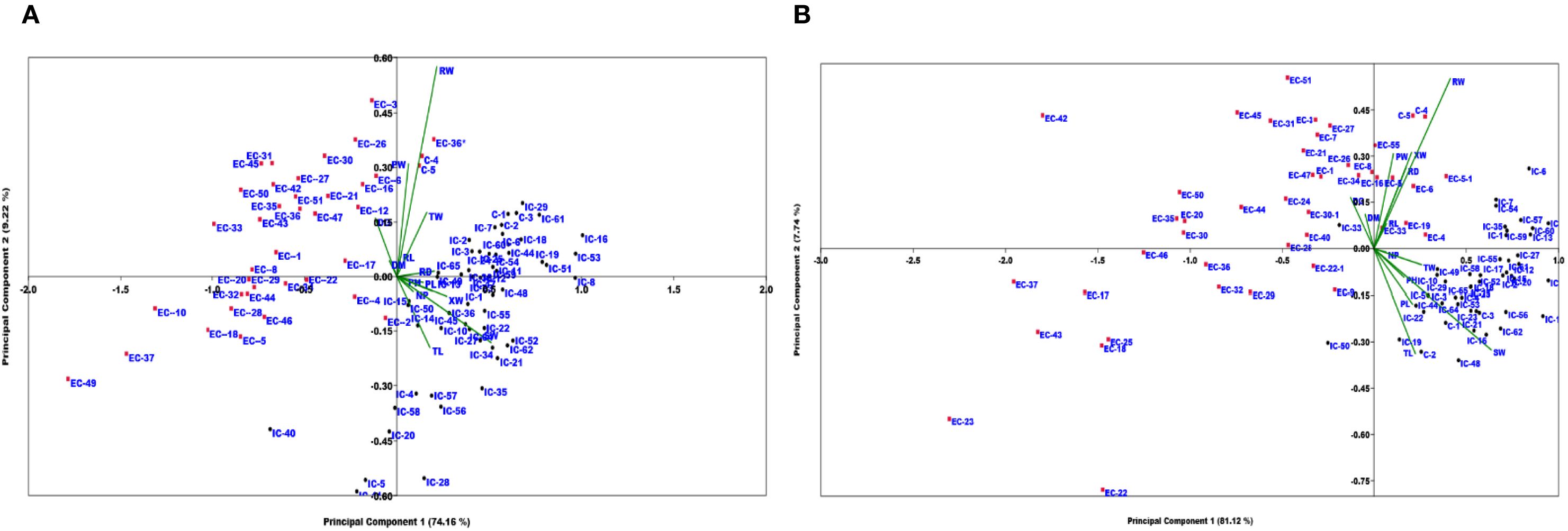
Figure 3. Genotype by trait biplot analysis based on the first two principal components across (A) KRCCH Arabhavi location and (B) COH Bagalkot location.
Cluster analysis based on Euclidean distance revealed a clear separation between the Western and Eastern gene pools at both locations (Figures 4A, B). At both sites, the phylogenetic tree grouped the accessions into two primary populations, Eastern (cluster1) and Western (cluster2), indicating that substantial variation exists for the two gene pools before and after secondary domestication for these quantitative traits. However, few Eastern accessions (IC-56, IC-57, IC-58, IC-59, IC-60) were observed to be admixtures in the Western group at Arabhavi. This indicates potential gene flow or overlapping traits between the gene pools at this location. The subgroups within the Eastern and Western clusters are due to the genetic variation for these plant growth and root traits among the accessions of each population. This pattern suggests substantial diversity within the individual gene pool.
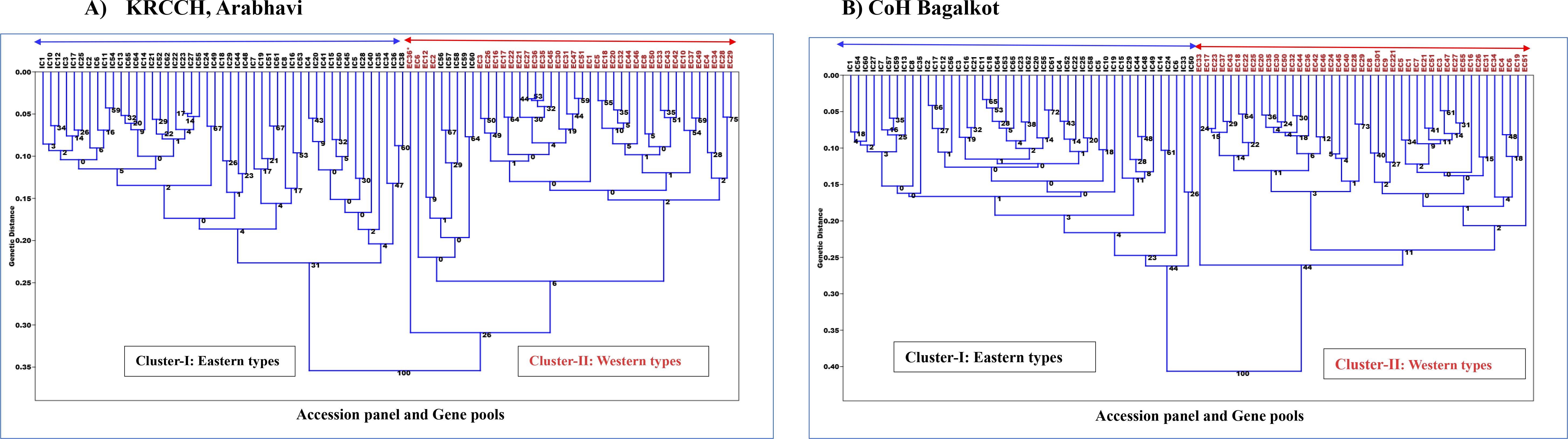
Figure 4. Phylogenetic relationship among Western and Eastern accessions based on quantitative traits in (A) KRCCH Arabhavi location and (B) COH Bagalkot location (Method: Neighbor-joining tree).
Development of InDel markers, primer design, and validation
The RNAseq data were analyzed using reference-based alignment to identify genetic variants, for insertions and deletions (InDels). From 12 paired-end (PE) RNA seq libraries obtained from Eastern and Western representative accessions, a total of 79,468 InDels were identified (data not shown) and initial filtering with the InDel length of >6bp yielded 19,817 InDels and they were functionally annotated with DH1V.3 reference annotation (Supplementary Table S2). Further, InDels were filtered to remove all heterozygous variants based on heterozygosity. We then applied a minimum length filter of 15 base pairs (bp), resulting in a refined dataset of 271 homozygous InDels, each 15 bp or longer. Supplementary Table S3 presents these 271 InDels along with their functional annotations. From these, 57 InDels were chosen for primer design and validation based on their chromosome distribution and association. The developed InDel markers and their corresponding chromosome location, primer sequences, and melting temperature (Tm) are detailed in Supplementary Table S3. Detailed information on the position of designed InDels and their function are depicted in Table 2.
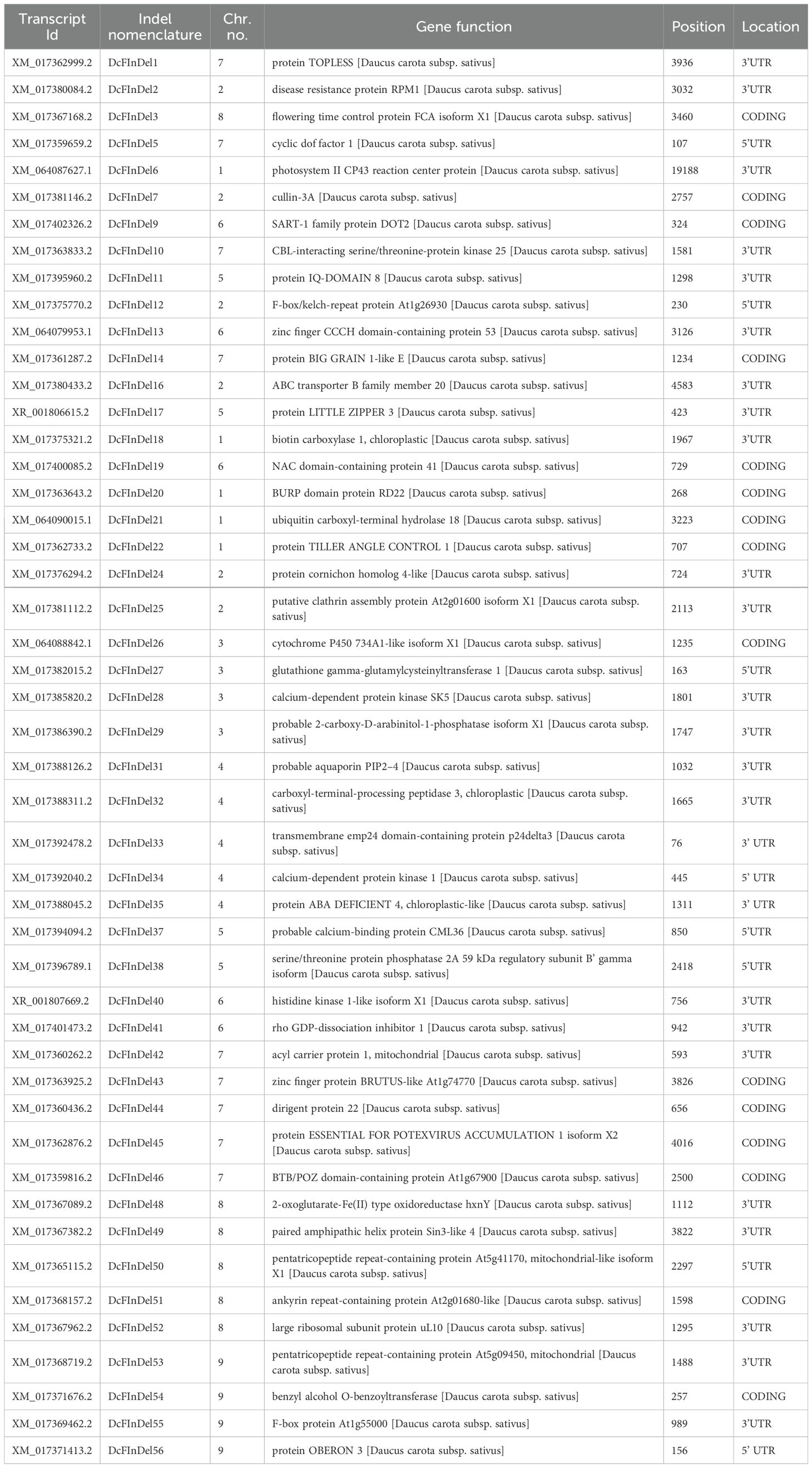
Table 2. Details of list of functionally characterized InDels used for genotyping the diverse set of gene pools with annotated functions, positions and locations in a gene.
These markers were further validated and utilized for diversity analysis across 98 accessions, representing both the Eastern and Western gene pools. Among these, 48 InDels were found to be polymorphic (84.21%), and they were used for assessing diversity and population structure across the gene pools. The physical locations of the 271 functional InDels identified in this study are depicted along with the mapped genes, QTLs associated with domestication traits, and selective sweeps across nine haploid chromosomes of carrot (Supplementary Figure S1). These QTLs helped to select the genes associated with the domestication traits and further aid in de novo domestication studies.
Gene ontology of polymorphic InDels
GO classification of the 48 polymorphic InDels provided insights into their potential biological functions (Supplementary Table S6). A total of 72 biological processes (BP), 25 cellular components (CC), and 40 molecular function (MF) terms were assigned to 26, 24, and 24 genes, respectively. In BP terms, ‘Cellular’ and ‘metabolic’ processes were most common. While in CC ‘cellular anatomical structure’ and ‘intercellular anatomical structure’ were predominant and in MF terms ‘binding’ and ‘activity’ terms were most common. This functional annotation suggests that these InDels may be involved in a diverse range of biological processes, including metabolic processes, cellular component organization, and catalytic activity. These findings highlight the potential of these InDels as valuable markers for improving carrot breeding programs. For instance, InDels in Y locus genes regulating carotenoid biosynthesis alter enzyme expression and metabolite accumulation, affecting root color, antioxidant capacity, and nutrition (Iorizzo et al., 2016). Indels in cellulose or pectin biosynthetic genes modify cell wall properties, impacting cell expansion and root morphology. Changes in enzyme or motif-binding sites due to InDels disrupt carotenoid accumulation and further influence root structure (Liu et al., 2019).
Molecular diversity parameters
Out of 57 validated InDels, 48 markers demonstrated polymorphism across 98 accessions, yielding a polymorphic percentage of 84.21%. This indicates that the in-house transcript data utilized to develop these InDels is highly effective for investigating the Eastern and Western gene pools. The remaining 9 InDels were found to be monomorphic across the panel but different from the DH reference genome. The representative polymorphic InDel variations across gene pools are presented in Figure 5.
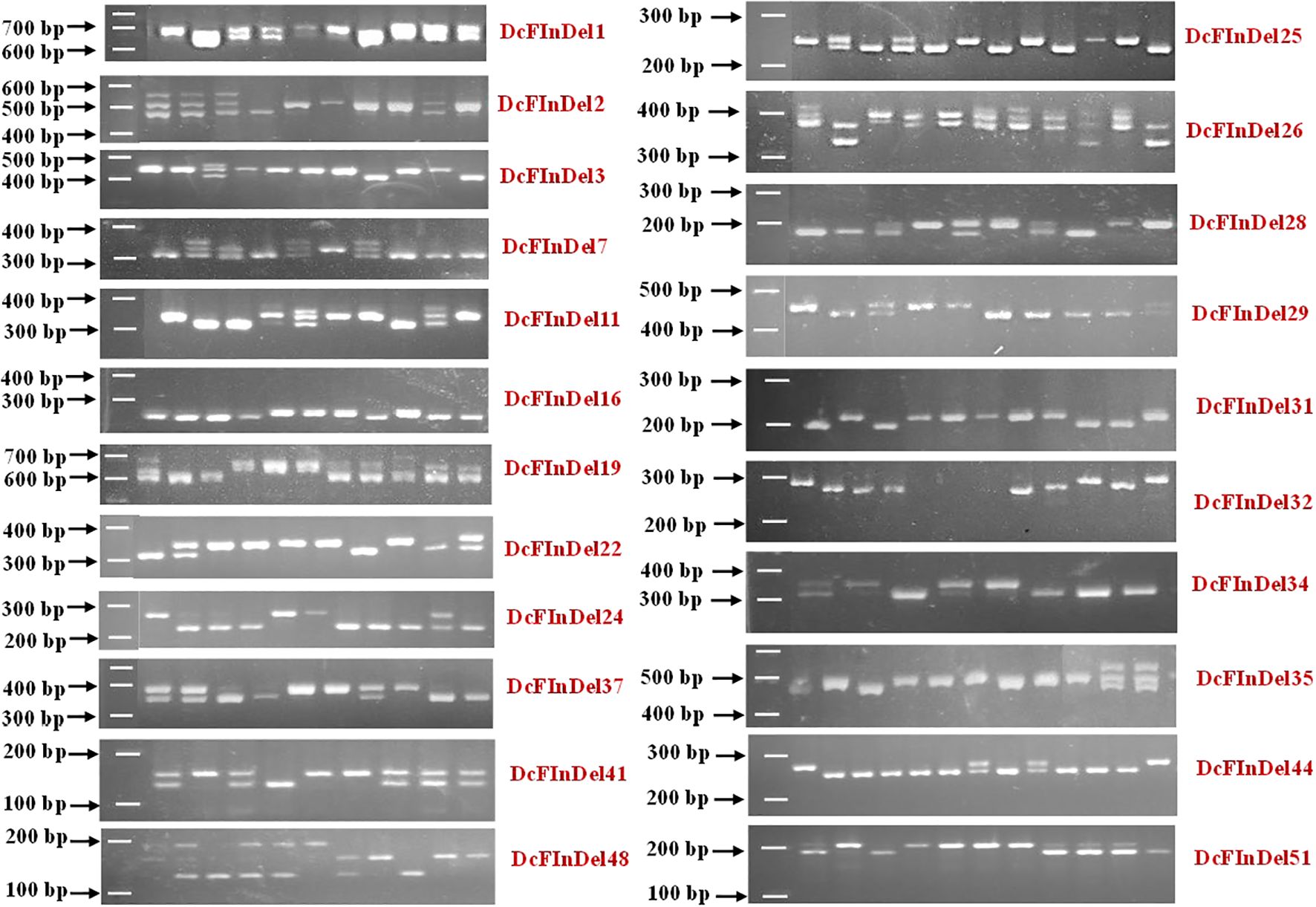
Figure 5. Representative profiles of InDel markers developed and validated across gene pools. Control images are shown here for illustrative purposes with original gels shown in Figure 3A.
A total of 224 alleles were identified from 98 accessions across the 48 markers, with the Western gene pool contributing a higher number of alleles (113) in comparison to the Eastern population (111). This difference highlights changes that may have occurred after domestication, likely resulting in improved traits within the Western population (Table 3). The Western population harbored a higher number of private alleles (13 alleles) compared to the Eastern population (11 alleles) (Table 4). These unique alleles reflect genetic modifications that may have influenced the function of corresponding genes both before and after domestication within these ancestrally related gene pools.

Table 3. Comparison of allelic diversity of 48 polymorphic InDels across Western and Eastern gene pools.
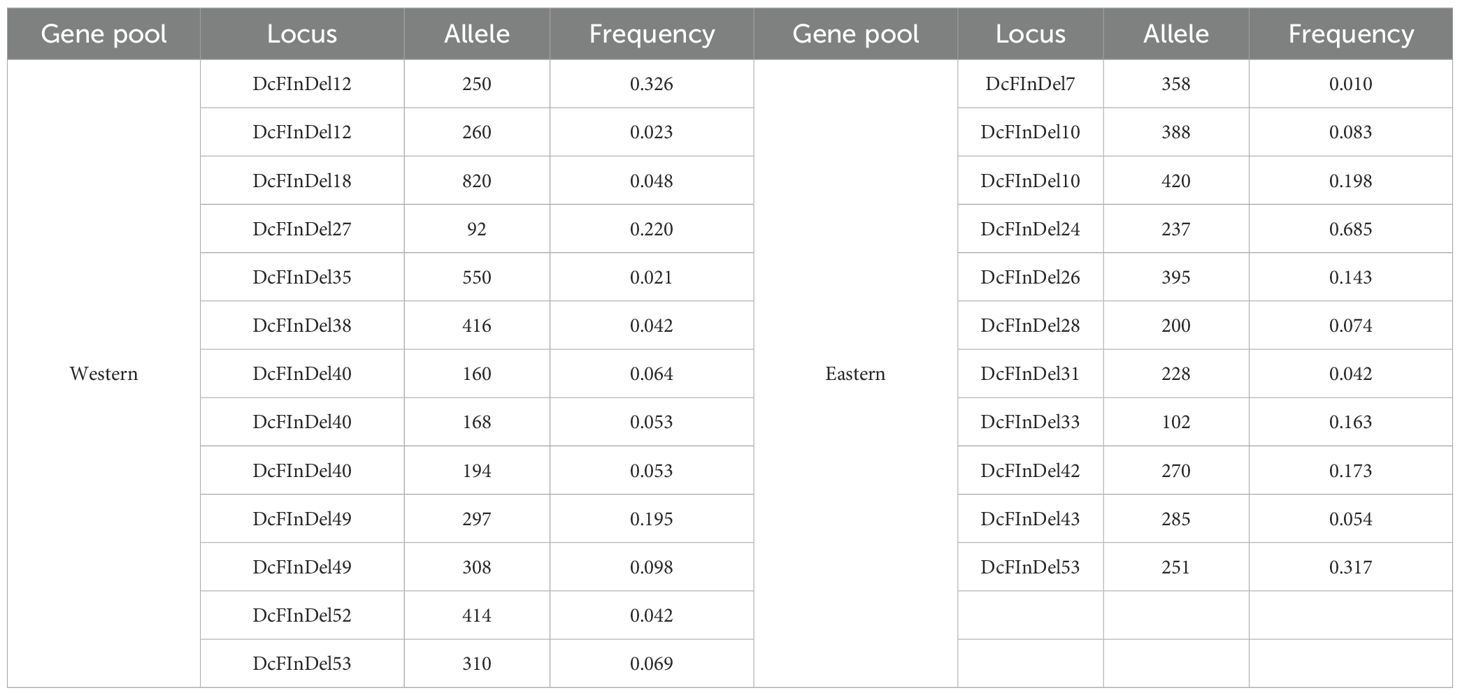
Table 4. Private/unique alleles and their frequency across Western and Eastern genepools from 48 functional InDels.
The Western population also exhibited a higher average number of effective alleles (1.76 ± 0.09) compared to the Eastern population (1.59 ± 0.06). Similarly, the Western population showed a higher average Shannon-Weaver index (‘I’) of 0.59 ± 0.05, compared to 0.52 ± 0.04 in the Eastern population, suggesting greater overall genetic diversity. Observed heterozygosity (Ho) was also higher in the Western gene pool (0.22 ± 0.03) than in the Eastern gene pool (0.21 ± 0.03), further indicating greater genetic variation within the Western population. Expected heterozygosity (He) was higher in the Western population (0.37 ± 0.03) compared to the Eastern population (0.33 ± 0.03), reinforcing the trend of greater diversity in the Western gene pool. The fixation index (F) ranged from -0.53 (DcFInDel24), indicating an excess of heterozygotes, to 1.00 (DcFInDel33), revealing complete fixation. In contrast, loci DcFInDel12 and DcFInDel27 showed no variation in the Eastern population, suggesting domestication-driven diversification in the Western types.
The polymorphic information content (PIC) values for the polymorphic markers ranged from 0.043 to 0.698, with an average PIC of 0.335. The highest PIC value was observed in the marker DcFInDel6, indicating its higher informativeness. Markers with a PIC greater than 0.50, such as DcFInDel6 and DcFInDel28, were highly informative and polymorphic, making them particularly effective for distinguishing individual accessions. The distinct allelic diversity was evident in both the gene pools for 16 InDel markers (DCFInDel7, 10, 12, 18, 20, 26, 27, 28, 33, 35, 38, 40, 42, 43, 49, and 52), suggesting their utility in the study of domestication. These results highlight the complex genetic landscape of carrots and provide valuable insights into the genetic differentiation and domestication of the species.
Population structure, AMOVA and molecular phylogenetic analysis
Population structure analysis was performed on the 98 accessions using the STRUCTURE 2.3.3 tool to gain insights into the genetic relationships among gene pools. The optimal K value of two, based on ΔK, indicated a clear division of accessions into two primary populations (Supplementary Figure S3). Cluster-1 represented the Western gene pool, while cluster-2 represented the Eastern gene pool (Figure 6). Within the Western population, few accessions (EC-5, EC-5-1, EC-9, EC-10, EC-17, EC-19, EC-24, and EC-26) were found as admixtures with Eastern types, suggesting that these accessions may have the potential to adapt to tropical conditions and exhibit minimal genetic dissimilarity to the Eastern gene pool. Conversely, within the Eastern population, accessions IC-33, IC-34, IC-38, and IC-40 were major admixtures with the Western types, indicating that these accessions likely originated as selections from the Western pool as confirmed by their phenotypic observations similar to Western cultivars.
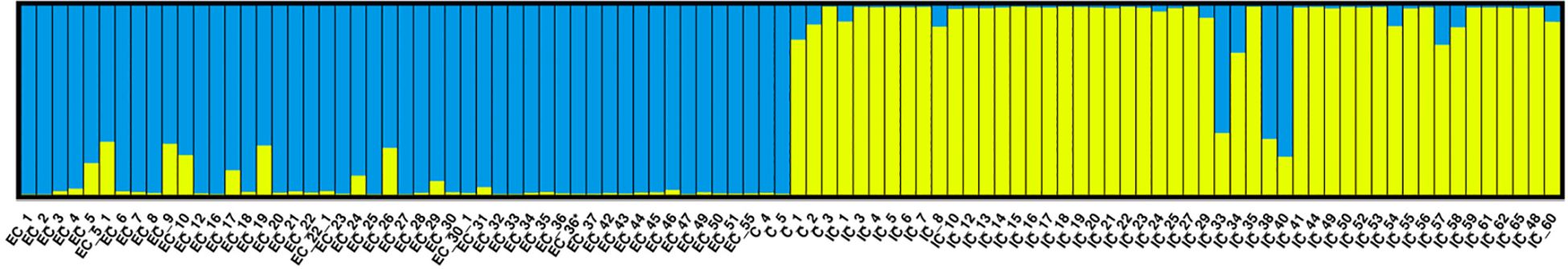
Figure 6. Population structure analysis showing the classification of 98 accessions of Western and Eastern gene pools and checks considering K = 2 allowing admixture.
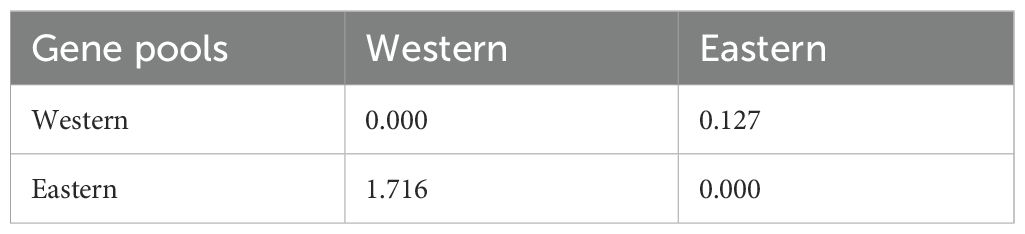
AMOVA revealed significant genetic differentiation among the accessions, with 18% of the total variance attributed to differences between populations. The majority of the variance (44%) was observed among individuals within the two populations, while 38% of the variance occurred within individuals across the entire population (Figure 7). F-statistics further indicated substantial genetic diversity within the population, with a fixation index relative to the total population (Fit) of 0.621. The fixation indices for population differentiation (Fst) and within-population genetic structure (Fis) were 0.181 and 0.537, respectively. The pairwise Fst value of 0.127 and pairwise Nm value of 1.716 respectively suggest a moderate genetic differentiation and higher gene flow between the populations as expected for the highly outcrossing nature of carrots (Table 5). Principal coordinate analysis (PCoA) effectively separated the accessions into distinct Western and Eastern groups, with the first two principal coordinates explaining 15.36% and 5.15% of the variance, respectively (Figure 8). Eastern accessions IC-33, IC-38, and IC-40 were identified as admixtures within the Western population, suggesting that these accessions likely originated from selections within the Western gene pool.
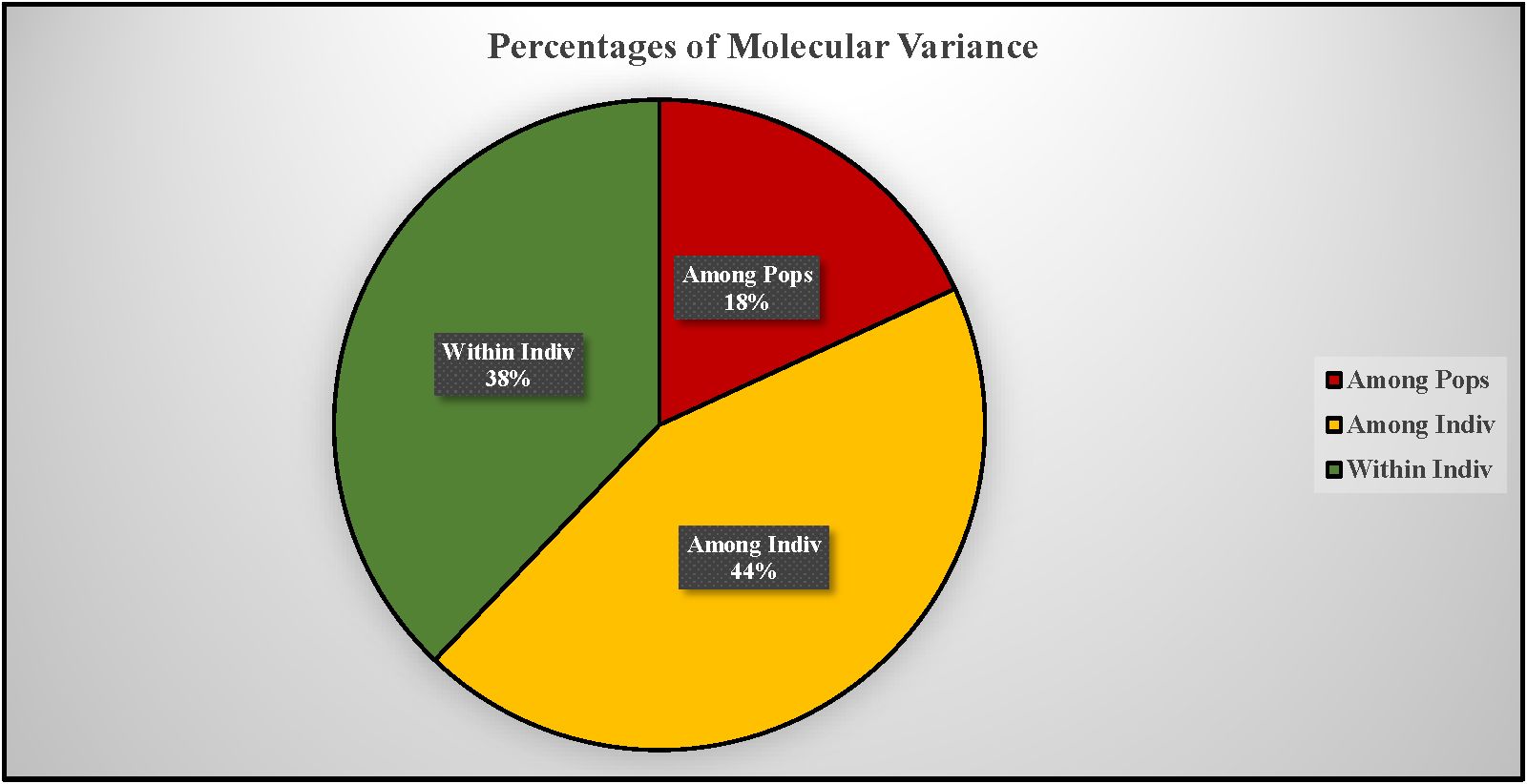
Figure 7. Analysis of molecular variance (AMOVA) showing the contribution of various components to total molecular variance from two populations/gene pools.
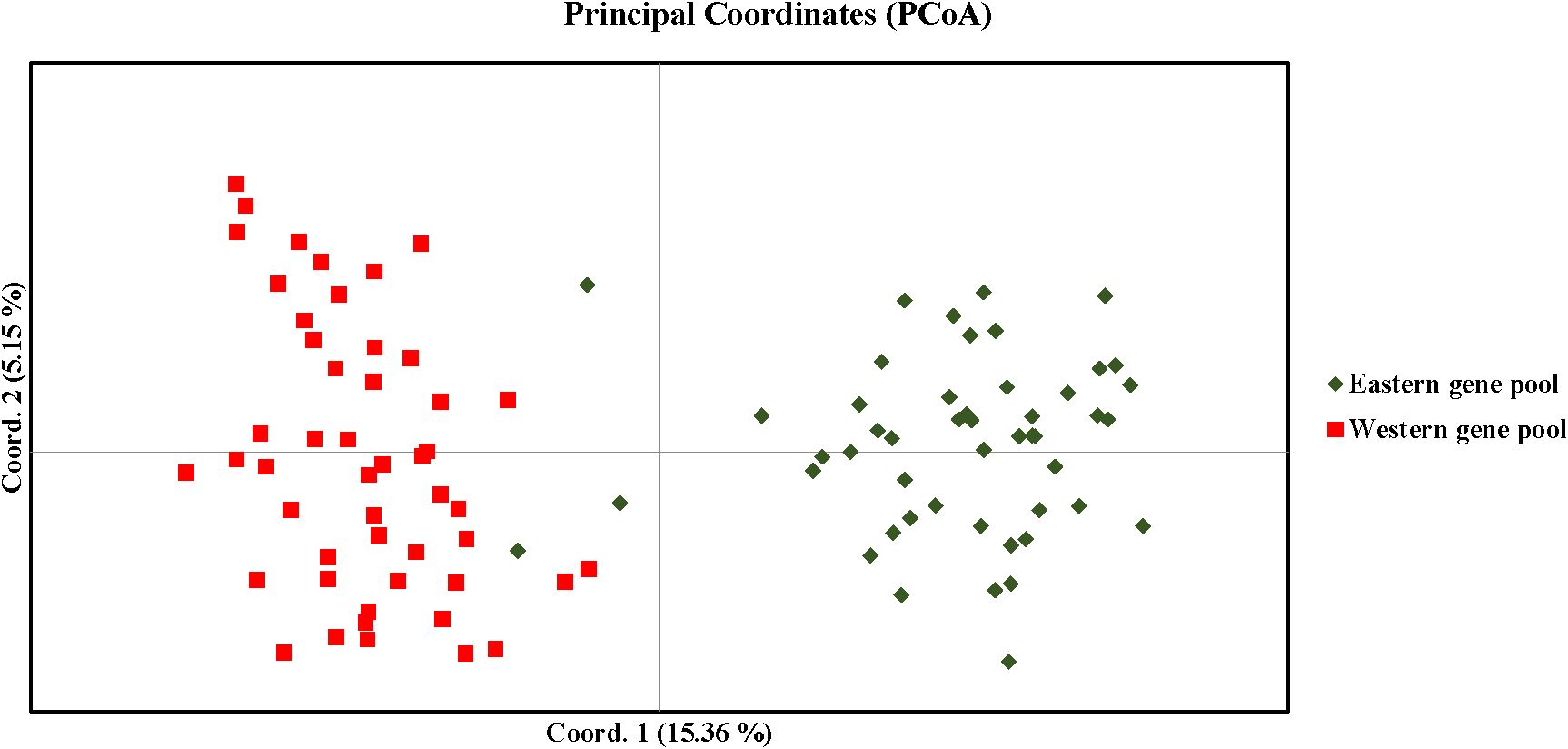
Figure 8. Distribution of accessions in different co-ordinates of PCoA (Principal co-ordinate analysis).
Phylogenetic analysis, based on Dice’s genetic dissimilarity matrix, revealed that the 98 accessions were grouped into three distinct clusters (Figure 9). The first cluster consisted of Eastern accessions and Eastern checks, while the second cluster included Western accessions and Western checks. Notably, the Eastern accession IC-40 was classified as an admixture within the Western population, suggesting that it shares phenotypic similarities with Western types and may have originated from selections within the Western gene pool. The third cluster comprised the Eastern genotype IC-38, which formed a solitary group, indicating that it is genetically distinct from both the Eastern and Western populations. This analysis highlights the complex genetic relationships and potential gene flow between the two gene pools, as well as the presence of genetically unique accessions.
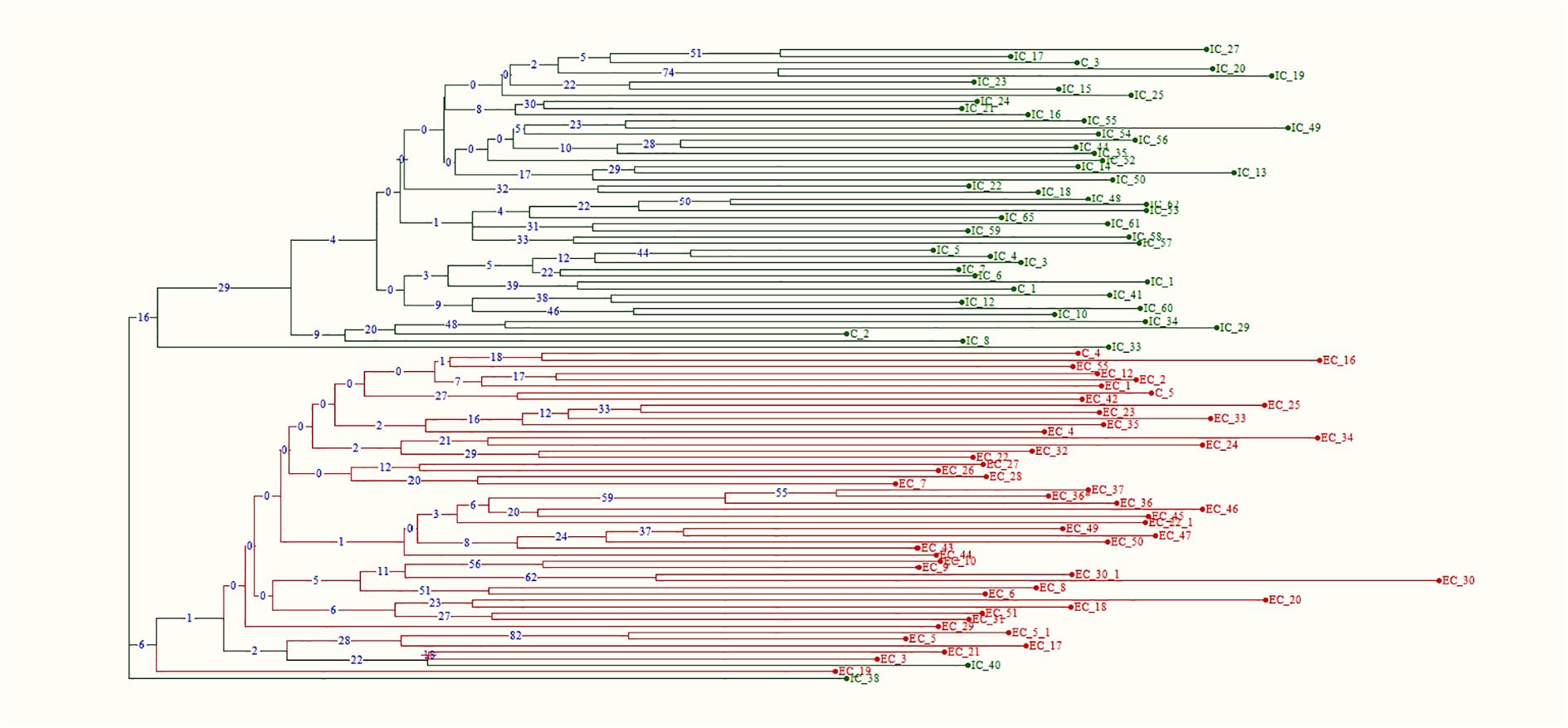
Figure 9. Neighbor-joining tree depicting phylogenetic relationship among Western and Eastern gene pool using 48 polymorphic functional markers (boot strap-1000).
Marker-trait association
At the Arabhavi location, significant marker-trait associations were identified for five quantitative and five qualitative parameters. Markers DcFInDel55, DcFInDel28, and DcFInDel38 exhibited substantial contributions to RW (R² = 0.21), SW (R² = 0.18), and SL (R² = 0.10), respectively, with high phenotypic variance explained (PVE). For qualitative traits, marker DcFInDel32 showed strong associations with RP (R² = 0.21) and RS (R² = 0.30), while DcFInDel28 also significantly influenced RS with a higher R² of 0.32. In addition, markers DcFInDel10, DcFInDel55, and DcFInDel38 contributed to RC (R² = 0.26), LT (R² = 0.13), and SS (R² = 0.21), respectively, highlighting their role in these traits.
At CoH Bagalkot, a significant association was found for three quantitative and five qualitative traits. DcFInDel7 and DcFInDel55 demonstrated notable contributions to PH (R² = 0.05) and XW (R² = 0.13), respectively. Moreover, markers DcFInDel26 and DcFInDel28 together accounted for 0.18 PVE in TW. Among qualitative traits, DcFInDel32 was associated with both RP (R² = 0.34) and RS (R² = 0.35) with high PVE, while DcFInDel10, DcFInDel28, and DcFInDel49 were significantly associated with RC, RS, and SS, with R² values of 0.24, 0.29, and 0.17, respectively (Table 6). Several markers had consistent associations across both locations for the same traits. Notably, DcFInDel10 was associated with RC, DcFInDel32 with both RP and RS, and DcFInDel28 with RS. These markers, particularly DcFInDel32, which was associated with both RP and RS at both locations, are promising candidates for further validation and application in marker-assisted selection (MAS) to improve carrot breeding programs.
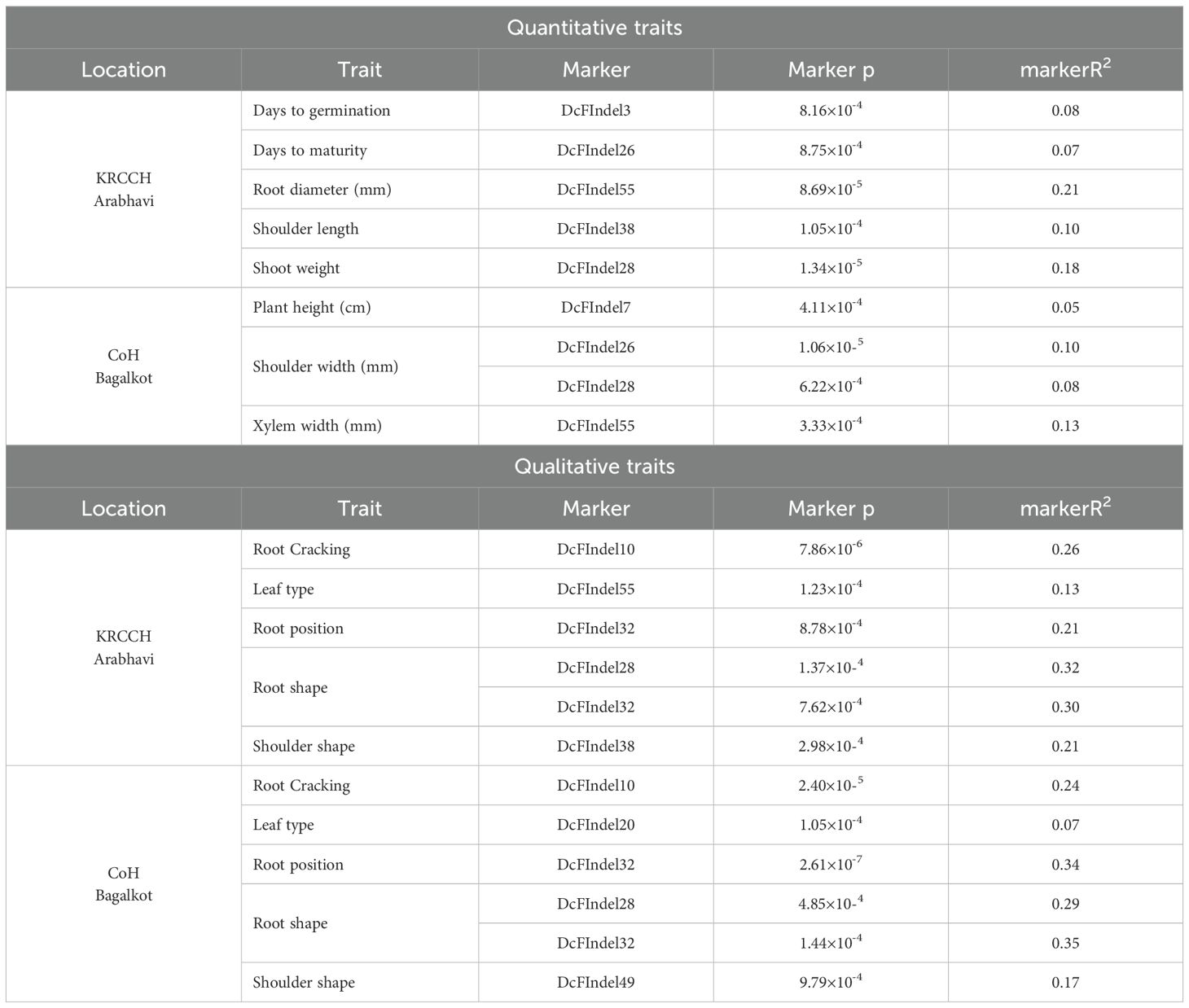
Table 6. Significant marker-trait associations for root traits (both quantitative and qualitative traits) across both the locations using general linear model.
Functional characterization of validated InDels
Among 48 polymorphic InDels assessed for their position in the gene, 15 InDels were found in the coding region, eight were in 5’UTR, and 25 were in 3’UTR (Table 2). The amino acid changes due to insertions or deletions in the coding sequences of 15 genes were assessed (Supplementary Figure S4). The changes in the nucleotide sequences at InDel regions of coding sequences significantly changed the amino acid composition, predicting the change in gene function and/or expression level across the gene pools. Further validation through gene expression analysis confirms their functionality and role in evolutionary changes during secondary domestication.
Discussion
Eastern carrots originated in Central Asia, while Western carrots were introduced to Europe and underwent further improvements during the post-domestication era (Ellison, 2019). During domestication, specific alleles were differentially selected in Eastern and Western carrots. Hence, Western and Eastern accessions vary in many domesticated traits, including plant growth habits, root development characteristics, flowering behavior, carotenoid accumulation, vernalization requirement, root quality (Ellison, 2019). Previous studies elucidated domestication changes and allelic diversity among diverse accessions (Iorizzo et al., 2013; Rong et al., 2014; Coe et al., 2023). However, the specific genetic mechanisms driving domestication changes and the genotypic variability causing phenotypic changes are still unclear. In the present investigation, morphological and molecular variation was evaluated and characterized using agro-morphometric and functional InDel markers to understand the genetic variations of important phenotypic and molecular changes across evolutionarily related Eastern and Western gene pools of two diverse geographic regions of the world.
Diversity in agro-morphometric patterns
Agro-morphological traits exhibited distinct variations between Western and Eastern carrot gene pools. Western accessions predominantly displayed tapering roots with dimpled textures, while Eastern types exhibited coarser textures and varied root positioning. Leaf type also differed, with Western accessions showcasing a fern leaf type and Eastern accessions exhibiting broad, typical carrot leaves. These phenotypic divergences are likely adaptations to specific regional growing conditions. Temperate climates often have colder winters and shorter growing seasons. We speculate that fern-type leaves may have evolved as a way to reduce the plant’s surface area, more efficiently capture light, minimize heat and enable water loss during colder periods. Root color, a crucial trait influencing market value and consumer preference, varied significantly between the two pools. Western accessions exhibited a wide range of colors, including orange, white, yellow, red, and purple, while Eastern accessions predominantly displayed light orange roots. These color variations are associated with genetic differences in carotenoid and anthocyanin biosynthesis and accumulation pathways.
Vegetative parameters also differed, with Eastern types exhibiting higher plant height, number of petioles, and petiole length, indicative of a stronger source-sink relationship. However, this increased vegetative growth was linked to a larger top length and width, a less desirable trait. In contrast, Western types, more sensitive to day length, showed delayed germination, and growth however, a more compact plant architecture with limited shoulder development. Root characters revealed further distinctions. Eastern types displayed a larger shoulder (top or crown), contributing significantly to root weight. Additionally, this was accompanied by a higher xylem width, negatively impacting consumer preference. Western types, on the other hand, had a negligible shoulder and a higher proportion of phloem tissue, resulting in a more desirable root shape and smooth texture (Luby et al., 2016). Genetic factors influencing vascular tissue development, such as MYB15, WRKY46, and AP2/ERF TF, may be crucial targets for improving root quality in Eastern carrot varieties (Kulkarni et al., 2023). Heritability estimates for most traits were notably high (> 60%) as depicted in Supplementary Table S5, indicating a strong genetic influence on their expression suggesting their high amenability to genetic improvement through selective breeding (Luby et al., 2016).
Pearson’s correlation analysis revealed distinct trait associations within the Western and Eastern gene pools. While both pools shared some common relationships like positive correlation between plant height and petiole length, they also exhibited unique patterns. For instance, Western accessions had a stronger link between shoot weight and root diameter, while Eastern accessions exhibited a stronger association between shoot weight and root weight and distinct xylem and phloem contribution towards root weight. Higher top width, length, plant height, and petiole length in Eastern accessions may have contributed to increased xylem width, facilitating a more efficient reproductive switch. In contrast, Western accessions, with their higher photosynthetic efficiency and lower xylem content, exhibited superior root quality and enhanced beta-carotene accumulation (Cholin et al., 2024).
PCA and cluster analysis further underscored the genetic divergence between the two gene pools. The GT-biplot separated Eastern and Western accessions, highlighting their distinct genetic makeup. The cluster analysis corroborated this finding, grouping accessions into distinct clusters based on genetic similarity (Singh et al., 2021). These findings suggest that the Western and Eastern gene pools have undergone distinct evolutionary trajectories, leading to the development of unique trait combinations (Stelmach et al., 2021; Hadagali et al., 2025). The presence of greater phenotypic variation among the Western gene pool makes it amenable to crop improvement (Luby et al., 2016). The identification of these genetic differences among the phenotypic traits provides valuable insights into the genetic architecture of carrots and has significant implications for future breeding programs. By understanding the genetic basis of these traits, breeders can develop cultivars with improved yield, quality, and stress tolerance. For instance, targeting the genetic loci associated with root length and phloem width, root texture, early germination, and maturity could lead to the development of high-yielding, superior-quality carrot cultivars.
Functional characterization of InDels and their diversity across gene pools
Exploration of the transcriptome data from Eastern and Western carrot accessions has led to the identification of a significant number of InDel variants. The distribution of these InDels across different chromosomes suggests a complex genetic architecture underlying carrot domestication. These InDels, particularly those associated with key genes related to domestication, offer valuable insights into the genetic diversity and evolutionary history of carrots. Important carrot genes, such as root development, flowering, physiological processes, biotic stress, and abiotic stress tolerance genes that played a critical role in trait modification during domestication, were selected. Eastern and western carrot accessions can be easily distinguished by a few important domestication traits/markers.
Using 57 InDel markers, molecular analysis revealed 48 polymorphic markers, with a high percentage of polymorphism (84.21%), suggesting that these markers are highly informative and suitable for assessing genetic diversity across carrot accessions. The validation of a set of 48 polymorphic InDel markers provides powerful and efficient tools for genetic diversity and population structure studies. The Western gene pool exhibited higher genetic diversity, as evidenced by the greater number of alleles, unique alleles, and higher values of genetic diversity indices. Higher allele count in the Western gene pool may be due to the introduction of new alleles after the domestication event, owing to evolutionary forces. This suggests that the Western gene pool has undergone a more intense selection and genetic diversification during the domestication, and crop improvement period (Ellison, 2019; Luby et al., 2016; Coe et al., 2023; Kulkarni et al., 2023). The results of the present study contrast with earlier reports by Iorizzo et al. (2013); Coe et al. (2023) and Chaitra et al. (2020), which found higher genetic divergence in Eastern than in the Western gene pool, highlighting the domestication bottleneck. The difference may be attributed to marker type, as SNPs primarily detect single-nucleotide substitutions and SSRs target tandem repeats, in contrast, the functional InDels employed here are more effective in capturing domestication-associated structural changes. The present study underscores the functional significance of genetic variations, specifically InDels, across ancestrally related Eastern and Western carrot gene pools. By analyzing these variations, we hypothesize that the Western gene pool, having undergone more intensive selection and improvement programs, exhibits greater genetic diversity and phenotypic variation. Even though the accessions of the two diverse gene pools were collected from geographically distinct locations (India and the USA), the moderate Fst and high Nm values suggest a moderate level of genetic differentiation and significant gene flow between the populations. This indicates a shared evolutionary history and potential for genetic exchange between the two gene pools.
Molecular phylogeny, population structure analysis, and PCoA further clarified the genetic relationships between the accessions. The STRUCTURE analysis, revealing two distinct populations corresponding to the Western and Eastern gene pools, aligns with phenotypic observations. This underscores the importance of integrating both morphological and molecular data in assessing genetic diversity. The observed admixture in certain accessions, such as IC-40 and EC-5, suggests potential gene flow between the two gene pools, likely due to historical crossbreeding or the movement of accessions across regions. Previous studies have utilized InDel markers to genetically group accessions in other crops like Cannabis (Pan et al., 2021), and radish (Wang et al., 2015; Li et al., 2023).
The identification of significant marker-trait associations provides valuable insights into the genetic architecture of key carrot traits. Markers such as DcFInDel10, DcFInDel28, DcFInDel32, DcFInDel38, and DcFInDel55 were consistently associated with multiple traits across different environments with significant phenotypic variance, highlighting their potential as robust markers for marker-assisted selection (MAS). For instance, DcFInDel28, linked to root shape and shoot weight, is annotated as calcium-dependent protein kinase SK5 (CDPKSK5), which regulates root growth, development, and stress response. DcFInDel32, associated with carboxyl-terminal-processing peptidase 3 (ctp3), plays a vital role in chloroplast development, photosynthesis, and overall plant development. Together, these markers explain over 50% of the variance (R²) in root shape, indicating their potential for carrot improvement. DcFInDel10, a marker for root cracking, is annotated as CBL-interacting serine/threonine-protein kinase 25 (CIPK25), involved in regulating abiotic stress responses. DcFInDel55, associated with root width and leaf type, is annotated as F-box protein At1g55000, which regulates cell cycle, development, flowering, stress responses, and hormonal signaling. These markers offer valuable tools for improving root shape, reducing cracking, and enhancing yield in carrot breeding.
Identifying InDels within coding and regulatory regions highlights their potential impact on gene expression and protein function in carrots. We further hypothesize that these genetic variations have played a crucial role in the diversification of carrot cultivars. GO classification also revealed their association with key biological functions, including cellular and metabolic processes, growth, development, and responses to stresses. Understanding the functional implications of these genetic variants provides opportunities for breeders to exploit them as functional markers in targeted breeding aimed at enhancing carrot yield, quality, and stress tolerance.
Conclusion
This is the first study to generate and validate carrot InDels, and apply them to dissect agro-morphometric and molecular diversity between Eastern and Western gene pools. Using transcriptome-derived InDels, we effectively demonstrated significant genotypic differences across two distinct gene pools. A clear genotypic difference was observed, with Eastern varieties exhibiting larger plant size and root dimensions, while Western varieties possessed deeper root position and uniform deep orange color. The InDel markers displayed high polymorphism, revealing strong genetic differentiation between the two gene pools, which underscores their potential for utilizing both gene pools in breeding programs. These functional InDels represent valuable molecular resources for understanding the evolutionary divergence and accelerating breeding through marker-assisted selection, enabling the development of high-yielding, climate-resilient cultivars with superior quality.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
TM: Data curation, Investigation, Formal analysis, Validation, Writing – review & editing, Writing – original draft. SC: Conceptualization, Supervision, Resources, Methodology, Formal analysis, Writing – review & editing, Writing – original draft, Project administration, Funding acquisition. CK: Investigation, Data curation, Writing – review & editing, Writing – original draft. KS: Formal analysis, Software, Writing – original draft, Writing – review & editing. PK: Writing – review & editing, Writing – original draft. WR: Formal analysis, Software, Writing – review & editing. PS: Supervision, Project administration, Funding acquisition, Formal analysis, Resources, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Department of Biotechnology (DBT), Govt of India (DBT-BIOCARe- 102/IFD/SAN/3308/2014-15), Vision Group of Science and Technology, Govt of Karnataka (KSTePS/VGST-RGS-F/2018-19/GRD No.840/315), UHS Bagalkot staff research funds, and USDA-ARS. Funders contributed to salaries, materials, supplies, facility use and administrative fees.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1658653/full#supplementary-material
References
Aber, R., Chan, W., Mugisha, S., and Jerome-Majewska, L. A. (2019). Transmembrane emp24 domain proteins in development and disease. Genet. Res. 10, e14. doi: 10.1017/S0016672319000090
Anderson, J. R. and Lubberstedt, T. (2003). Functional markers in plants. Trends. Plant Sci. 8, 554–560. doi: 10.1016/j.tplants.2003.09.010
Andersson, L. and Purugganan, L. (2022). Molecular genetic variation of animals and plants under domestication. Proc. Natl. Acad. Sci. 119, e2122150119. doi: 10.1073/pnas.2122150119
Arango, J., Jourdan, M., Geoffriau, E., Beyer, P., and Welsch, R. (2014). Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell 26, 2223–2233. doi: 10.1105/tpc.113.122127
Astegno, A., Bonza, M. C., Vallone, R., La Verde, V., D'Onofrio, M., Luoni, L., et al. (2017). Arabidopsis calmodulin-like protein CML36 is a calcium (Ca2+) sensor that interacts with the plasma membrane Ca2+-ATPase isoform ACA8 and stimulates its activity. J. Biol. Chem. 292, 15049–15061. doi: 10.1074/jbc.M117.787796
Baranski, R., Maksylewicz-Kaul, A., Nothnagel, T., Cavagnaro, P. F., Simon, P. W., and Grzebelus, D. (2012). Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genet.Resour. Crop Ev. 59, 163–170. doi: 10.1007/s10722-011-9777-3
Baurle, I., Smith, L., Baulcombe, D. C., and Dean, C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318, 109–112. doi: 10.1126/science.1146565
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Bradeen, J. M. and Simon, P. W. (2007). “Carrot,” in Genome mapping & Molecular Breeding, vol. 5 . Ed. Kole, C. (Springer, Berlin), 161–184.
Casson, S. A., Topping, J. F., and Lindsey, K. (2009). MERISTEM-DEFECTIVE, an RS domain protein, is required for the correct meristem patterning and function in Arabidopsis. Plant J. 57, 857–869. doi: 10.1111/j.1365-313X.2008.03738.x
Chaitra, K. C., Sarvamangala, C., Manikanta, D. S., Chaitra, P. A., and Fakrudin, B. (2020). Insights into genetic diversity and population structure of Indian carrot (Daucus carota L.) accessions. J. Appl. Genet. 61, 303–312. doi: 10.1007/s13353-020-00556-6
Chatukuta, P., Dikobe, T. B., Kawadza, D. T., Sehlabane, K. S., Takundwa, M. M., Wong, A., et al. (2018). An Arabidopsis clathrin assembly protein with a predicted role in plant defense can function as an adenylate cyclase. Biomolecules 8, 15. doi: 10.3390/biom8020015
Cheng, X., Liu, X., He, J., Tang, M., Li, H., and Li, M. (2022). The genome wide analysis of Tryptophan Aminotransferase Related gene family, and their relationship with related agronomic traits in Brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1098820
Cholin, S. S., Kulkarni, C. C., Grzebelus, D., Jakaraddi, R., Hundekar, A., Chandan, B. M., et al. (2024). Deciphering carotenoid and flowering pathway gene variations in Eastern and Western carrots (Daucus carota L.). Genes 15, 1462. doi: 10.3390/genes15111462
Clotault, J., Geoffriau, E., Lionneton, E., Briard, M., and Peltier, D. (2010). Carotenoid biosynthesis genes provide evidence of geographical subdivision and extensive linkage disequilibrium in the carrot. Theor. Appl. Genet. 121, 659–672. doi: 10.1007/s00122-010-1338-1
Coe, K., Bostan, H., Rolling, W., Turner-Hissong, S., Macko-Podgorni, A., Senalik, D., et al. (2023). Population genomics identifies genetic signatures of carrot domestication and improvement and uncovers the origin of high-carotenoid orange carrots. Nat. Genet. 9, 1643–1658. doi: 10.1038/s41477-023-01526-6
Coe, K., Ellison, S., Senalik, D., Dawson, J., and Simon, P. W. (2021). The influence of the Or and Carotene Hydroxylase genes on carotenoid accumulation in orange carrots [Daucus carota (L.). Theor. Appl. Genet. 134, 3351–3362. doi: 10.1007/s00122-021-03901-3
Cui, J., Peng, J., Cheng, J., and Hu, K. (2021). Development and validation of genome-wide InDel markers with high levels of polymorphism in bitter gourd (Momordica charantia). BMC Genom. 22, 1–9. doi: 10.1186/s12864-021-07499-1
Dieterle, M., Thomann, A., Renou, J. P., Parmentier, Y., Cognat, V., Lemonnier, G., et al. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41, 386–399. doi: 10.1111/j.1365-313X.2004.02302.x
Doyle, J. (1991). “DNA protocols for plants,” in Molecular techniques in taxonomy. Eds. Hewitt, G. M., Johnston, A., and Young, J. P. (Springer, Berlin), 283–293. doi: 10.1007/978-3-642-83962-7_18
Earl, D. A. (2012). Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Eda, M., Matsumoto, T., Ishimaru, M., and Tada, T. (2016). Structural and functional analysis of tomato β-galactosidase 4: insight into the substrate specificity of the fruit softening-related enzyme. Plant J. 86, 300–307. doi: 10.1111/tpj.13160
Ellison, S. (2019). “Carrot domestication,” in The Carrot Genome. Eds. Simon, P. W., Iorizzo, M., Grzebelus, D., and Baranski, R. (Springer, Berlin), 77–91. doi: 10.1007/978-3-030-03389-7_5
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Farrow, S. C. and Facchini, P. J. (2014). Functional diversity of 2-oxoglutarate/Fe (II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00524
Ferreyra, M. L. F., Pezza, A., Biarc, J., Burlingame, A. L., and Casati, P. (2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153, 1878–1894. doi: 10.1104/pp.110.157057
Goralogia, G. S., Liu, T. K., Zhao, L., Panipinto, P. M., Groover, E. D., Bains, Y. S., et al. (2017). CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 92, 244–262. doi: 10.1111/tpj.13649
Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. doi: 10.1046/j.1365-313x.2000.00804
Gruber, A. V., Kosty, M., Jami-Alahmadi, Y., Wohlschlegel, J. A., and Long, J. A. (2021). The dynamics of HD-ZIP III-ZPR protein interactions play essential roles in embryogenesis, meristem function and organ development. bioRxiv, 2021–2011. doi: 10.1101/2021.11.24.469949
Grzebelus, D., Yau, Y.-Y., and Simon, P. W. (2006). Master – a novel family of PIF/Harbinger-like transposable elements identified in carrot (Daucus carota L.). Molec. Genet. Genomics 275, 450–459. doi: 10.1007/s00438-006-0102-3
Guo, C., Zhou, J., and Li, D. (2021). New insights into functions of IQ67-domain proteins. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.614851
Hadagali, S., Stelmach-Wityk, K., Macko-Podgórni, A., Cholin, S., and Grzebelus, D. (2025). Polymorphic insertions of DcSto miniature inverted-repeat transposable elements reveal genetic diversity structure within the cultivated carrot. J. Appl. Genet. 66 (2), 293–303. doi: 10.1007/s13353-024-00916-6
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeont. Electron. 4, 9. Available online at: http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
Han, G., Qiao, Z., Li, Y., Wang, C., and Wang, B. (2021). The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. Int. J. Mol. Sci. 22, 8327. doi: 10.3390/ijms221583272
Han, G., Qiao, Z., Li, Y., Yang, Z., Wang, C., Zhang, Y., et al. (2022). RING zinc finger proteins in plant abiotic stress tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.877011
Harshavardhan, V. T., Van Son, L. E., Seiler, C., Junker, A., Weigelt-Fischer, K., Klukas, C., et al. (2014). AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS One 9, e110065. doi: 10.1371/journal.pone.0110065
Hayama, R., Yang, P., Valverde, F., Mizoguchi, T., Furutani-Hayama, I., Vierstra, R. D., et al. (2019). Ubiquitin carboxyl-terminal hydrolases are required for period maintenance of the circadian clock at high temperature in Arabidopsis. Sci. Rep. 9, 17030. doi: 10.1038/s41598-019-53229-8
Inoue, R., Nakamura, N., Matsumoto, C., Takase, H., Sekiya, J., and Prieto, R. (2022). Characterization of γ-glutamyltransferase-and phytochelatin synthase-mediated catabolism of glutathione and glutathione S-conjugates in Arabidopsis thaliana. Plant Biotechnol. 39, 381–389. doi: 10.5511/plantbiotechnology.22.1003a
Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., et al. (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asteroid genome evolution. Nat. Genet. 48, 657–666. doi: 10.1038/ng.3565
Iorizzo, M., Senalik, D. A., Ellison, S. L., Grzebelus, D., Cavagnaro, P. F., Allender, C., et al. (2013). Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae). American J. Bot. 100, 930–938. doi: 10.3732/ajb.1300055
Israel, D., Lee, S. H., Robson, T. M., and Zwiazek, J. J. (2022). Plasma membrane aquaporins of the PIP1 and PIP2 subfamilies facilitate hydrogen peroxide diffusion into plant roots. BMC Plant Biol. 22, 566. doi: 10.1186/s12870-022-03962-6
Janssens, V. and Goris, J. (2001). Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439. doi: 10.1042/0264-6021:3530417
Jun, X. U., Wang, X. Y., and Guo, W. Z. (2015). The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 14, 1673–1686. doi: 10.1016/S2095-3119(14)60980-1
Kotera, Y., Komori, H., Tasaki, K., Takagi, K., Imano, S., and Katou, S. (2023). The peroxisomal β-oxidative pathway and benzyl alcohol O-benzoyltransferase HSR201 cooperatively contribute to the biosynthesis of salicylic acid. Plant Cell Physiol. 64, 758–770. doi: 10.1093/pcp/pcad034
Kulkarni, C. C., Cholin, S. S., Bajpai, A. K., Ondrasek, G., Mesta, R. K., Rathod, S., et al. (2023). Comparative root transcriptome profiling and gene regulatory network analysis between Eastern and Western carrot (Daucus carota L.) cultivars reveals candidate genes for vascular tissue patterning. Plants. 12, 3449. doi: 10.3390/plants12193449
Kumar Meena, M., Kumar Vishwakarma, N., Tripathi, V., and Chattopadhyay, D. (2019). CBL-interacting protein kinase 25 contributes to root meristem development. J. Exp. Bot. 70, 133–147. doi: 10.1093/jxb/ery334
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303, 3997. doi: 10.48550/arXiv.1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, X., Sun, M., Liu, S., Teng, Q., Li, S., and Jiang, Y. (2021). Functions of PPR proteins in plant growth and development. Int. J. Mol. Sci. 22, 11274. doi: 10.3390/ijms222011274
Li, Y., Luo, X., Peng, X., Jin, Y., Tan, H., Wu, L., et al. (2023). Development of SNP and InDel markers by genome resequencing and transcriptome sequencing in radish (Raphanus sativus L.). BMC Genomics 24, 445. doi: 10.1186/s12864-023-09528-6
Liu, S., An, Y., Tong, W., Qin, X., Samarina, L., Guo, R., et al. (2019). Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis) and development of InDel markers for genetic research. BMC Genomics 20, 1–16. doi: 10.1186/s12864-019
Liu, X., Yu, X., Shi, Y., Ma, L., Fu, Y., and Guo, Y. (2023). Phosphorylation of RhoGDI1, a Rho GDP dissociation inhibitor, regulates root hair development in Arabidopsis under salt stress. Proc. Natl. Acad. Sci. 120, e2217957120. doi: 10.1073/pnas.2217957120
Lopez-Ortiz, C., Peña-Garcia, Y., Natarajan, P., Bhandari, M., Abburi, V., Dutta, S. K., et al. (2020). The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci. Rep. 10, 4044. doi: 10.1038/s41598-020-61057-4
Luby, C. H., Dawson, J. C., and Goldman, I. L. (2016). Assessment and accessibility of phenotypic and genotypic diversity of carrot (Daucus carota L. var. sativus) cultivars commercially available in the United States. PLoS One 11, e0167865. doi: 10.1371/journal.pone.0167865
Marchetti, F., Cainzos, M., Shevtsov, S., Córdoba, J. P., Sultan, L. D., Brennicke, A., et al. (2020). Mitochondrial pentatricopeptide repeat protein, EMB2794, plays a pivotal role in NADH dehydrogenase subunit nad2 mRNA maturation in Arabidopsis thaliana. Plant Cell Physiol. 61, 1080–1094. doi: 10.1093/pcp/pcaa028
Masri, R. and Kiss, E. (2023). The role of NAC genes in response to biotic stresses in plants. Physiol. Mol. Plant Pathol. 126, 102034. doi: 10.1016/j.pmpp.2023.102034
Megha, S., Wang, Z., Kav, N. N., and Rahman, H. (2022). Genome-wide identification of biotin carboxyl carrier subunits of acetyl-CoA carboxylase in Brassica and their role in stress tolerance in oilseed Brassica napus. BMC Genom. 23, 707. doi: 10.1186/s12864-022-08920-y
Mishra, B. S., Jamsheer K, M., Singh, D., Sharma, M., and Laxmi, A. (2017). Genome-wide identification and expression, protein–protein interaction and evolutionary analysis of the seed plant-specific BIG GRAIN and BIG GRAIN LIKE gene family. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01812
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mousavi, S. A., Dubin, A. E., Zeng, W. Z., Coombs, A. M., Do, K., Ghadiri, D. A., et al. (2021). PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 118, e2102188118. doi: 10.1101/2020.08.27.270355
Murray, J. W., Duncan, J., and Barber, J. (2006). CP43-like chlorophyll binding proteins: structural and evolutionary implications. Trends Plant Sci. 11, 152–158. doi: 10.1016/j.tplants.2006.01.007
Nedelyaeva, O. I., Shuvalov, A. V., and Balnokin, Y. V. (2020). Chloride channels and transporters of the CLC family in plants. Russ. J. Plant Physiol. 67, 767–784. doi: 10.1134/S1021443720050106
Nongpiur, R., Soni, P., Karan, R., Singla-Pareek, S. L., and Pareek, A. (2012). Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal. Behav. 7, 1230–1237. doi: 10.4161/psb.21516
North, H. M., Almeida, A. D., Boutin, J. P., Frey, A., To, A., Botran, L., et al. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50, 810–824. doi: 10.1111/j.1365-313X.2007.03094
Oelmuller, R., Herrmann, R. G., and Pakrasi, H. B. (1996). Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 271, 21848–21852. doi: 10.1074/jbc.271.36.21848
Pan, G., Li, Z., Huang, S., Tao, J., Shi, Y., Chen, A., et al. (2021). Genome-wide development of insertion-deletion (InDel) markers for Cannabis and its uses in genetic structure analysis of Chinese germplasm and sex-linked marker identification. BMC Genomics 22, 1–12. doi: 10.1186/s12864-021-07883-w
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301. doi: 10.1093/jxb/erx141
Peakall, R. and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Perrier, X. and Jacquemoud-Collet, J. P. (2006). DARwin software. Available online at: http://darwin.cirad.fr/darwin (Accessed November 30, 2024).
Plant, A. R., Larrieu, A., and Causier, B. (2021). Repressor for hire! The vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol. 231, 963–973. doi: 10.1111/nph.17428
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics. 155, 945–959. doi: 10.1093/genetics/155.2.945
Redelings, B. D., Holmes, I., Lunter, G., Pupko, T., and Anisimova, M. (2024). Insertions and deletions: computational methods, evolutionary dynamics, and biological applications. Molec. Biol. Evol. 41, msae177. doi: 10.1093/molbev/msae177
Rempola, B., Karkusiewicz, I., Piekarska, I., and Rytka, J. (2006). Fcf1p and Fcf2p are novel nucleolar Saccharomyces cerevisiae proteins involved in pre-rRNA processing. Biochem. Biophys. Res. Commun. 346, 546–554. doi: 10.1016/j.bbrc.2006.05.140
Rolling, W. R., Senalik, D., Iorizzo, M., Ellison, S., Van Deynze, A., and Simon, P. W. (2022). CarrotOmics: a genetics and comparative genomics database for carrot (Daucus carota). Database 2022, baac079. doi: 10.1093/database/baac079
Rong, J., Lammers, Y., Strasburg, J. L., Schidlo, N. S., Ariyurek, Y., De Jong, T. J., et al. (2014). New insights into domestication of carrot from root transcriptome analyses. BMC Genomics 15, 1–15. doi: 10.1186/1471-2164-15-895
Sahu, P. K., Mondal, S., Sharma, D., Vishwakarma, G., Kumar, V., and Das, B. K. (2017). InDel marker based genetic differentiation and genetic diversity in traditional rice (Oryza sativa L.) landraces of Chhattisgarh, India. PLoS One 12, e0188864. doi: 10.1371/journal.pone.0188864
Saiga, S., Furumizu, C., Yokoyama, R., Kurata, T., Sato, S., Kato, T., et al. (2008). The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development. 135, 1751–1759. doi: 10.1242/dev.014993
Saini, R. and Nandi, A. K. (2022). TOPLESS in the regulation of plant immunity. Plant Mol. Biol. 109, 1–12. doi: 10.1007/s11103-022-01258-9
Salgotra, R. K. and Stewart, C. N. (2020). Functional markers for precision plant breeding. Int. J. Mol. Sci. 21, 4792. doi: 10.3390/ijms21134792
Savino, S., Desmet, T., and Franceus, J. (2022). Insertions and deletions in protein evolution and engineering. Biotech. Adv. 60, 108010. doi: 10.1016/j.bioteChadv.2022.108010
Schulz, P., Herde, M., and Romeis, T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163, 523–530. doi: 10.1104/pp.113.222539
Seemann, J. R., Kobza, J., and Moore, B. D. (1990). Metabolism of 2-carboxyarabinitol 1-phosphate and regulation of ribulose-1, 5-bisphosphate carboxylase activity. Photosyn. Res. 23, 119–130. doi: 10.1007/BF00035005
Shi, Y., Zhu, H., Zhang, J., Bao, M., and Zhang, J. (2023). Development and validation of molecular markers for double flower of. Prunus mume. Sci. Hortic. 310, 111761. doi: 10.1016/j.scienta.2022.111761
Shivaprasad, K., Aski, M., Mishra, G. P., Sinha, S. K., Gupta, S., Mishra, D. C., et al. (2024). Genome-wide discovery of InDels and validation of PCR-Based InDel markers for earliness in a RIL population and accessions of lentil (Lens culinaris Medik.). PLoS One 19, 302870. doi: 10.1371/journal.pone.0302870
Singh, S., Kalia, P., Singh, S. K., Mishra, S., and Pandey, C. D. (2021). Agro-morphometric diversity analysis in carrot germplasm from Indian genebank. Indian J. Plant Genet. Resour. 34, 243–250. doi: 10.5958/0976-1926.2021.00023.1
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Stelmach, K., Macko-Podgórni, A., Allender, C., and Grzebelus, D. (2021). Genetic diversity structure of Western-type carrots. BMC Plant Biol. 21, 200. doi: 10.1186/s12870-021-02980-0
Stelmach, K., Macko-Podgórni, A., Machaj, G., and Grzebelus, D. (2017). Miniature inverted repeat transposable element insertions provide a source of intron length polymorphism markers in the carrot (Daucus carota L.). Front. Plant Sci. 8, 725. doi: 10.3389/fpls.2017.00725
Thakur, O. and Randhawa, G. S. (2018). Identification and characterization of SSR, SNP and InDel molecular markers from RNA-Seq data of guar (Cyamopsis tetragonoloba, L. Taub.) roots. BMC Genomics 19, 1–14. doi: 10.1186/s12864-018-5205-9
Thanh Ha Thi Do, T. H. T. D., Martinoia, E., and Lee YoungSook, L. Y. (2018). Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 41, 32–38. doi: 10.1016/j.pbi.2017.08.003
Thomas, C. L., Schmidt, D., Bayer, E. M., Dreos, R., and Maule, A. J. (2009). Arabidopsis plant homeodomain finger proteins operate downstream of auxin accumulation in specifying the vasculature and primary root meristem. Plant J. 59, 426–436. doi: 10.1111/j.1365-313X.2009.03874
Tzafrir, I., McElver, J. A., Liu, C. M., Yang, L. J., Wu, J. Q., Martinez, A., et al. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128, 38–51. doi: 10.1104/pp.010911
Valmonte, G. R., Arthur, K., Higgins, C. M., and MacDiarmid, R. M. (2014). Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 55, 551–569. doi: 10.1093/pcp/pct200
Waite, J. M. and Dardick, C. (2018). TILLER ANGLE CONTROL 1 modulates plant architecture in response to photosynthetic signals. J. Exp. Bot. 69, 4935–4944. doi: 10.1093/jxb/ery253
Wang, K., Guo, H., and Yin, Y. (2024). AP2/ERF transcription factors and their functions in Arabidopsis responses to abiotic stresses. Environ. Exp. Bot. 222, 105763. doi: 10.1016/j.envexpbot.2024.105763
Wang, Q., Zhang, L., and Zheng, P. (2015). Genetic diversity and evolutionary relationship analyses within and among Raphanus species using EST-SSR markers. Mol. Breed. 35, 1–12. doi: 10.1007/s11032-015-0261-1
Wang, Y. J., Yu, J. N., Chen, T., Zhang, Z. G., Hao, Y. J., Zhang, J. S., et al. (2005). Functional analysis of a putative Ca2+ channel gene TaTPC1 from wheat. J. Exp. Bot. 56, 3051–3060. doi: 10.1093/jxb/eri302
Wei, C., Zhao, W., Fan, R., Meng, Y., Yang, Y., Wang, X., et al. (2021). Genome-wide survey of the F-box/Kelch (FBK) members and molecular identification of a novel FBK gene TaAFR in wheat. PLoS One 16, e0250479. doi: 10.1371/journal.pone.0250479
Wudick, M. M., Portes, M. T., Michard, E., Rosas-Santiago, P., Lizzio, M. A., Nunes, C. O., et al. (2018). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 360, 533–536. doi: 10.1126/science.aar6464
Yau, Y. Y. and Simon, P. W. (2003). A 2.5-kb insert eliminates acid soluble invertase isozyme II transcript in carrot (Daucus carota L.) roots, causing high sucrose accumulation. Plant Molec. Biol. 53, 151–162. doi: 10.1023/B:PLAN.0000009272.44958.13
Yip Delormel, T. and Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604. doi: 10.1111/nph.16088
Yusa, A., Neriya, Y., Hashimoto, M., Yoshida, T., Fujimoto, Y., Hosoe, N., et al. (2019). Functional conservation of EXA1 among diverse plant species for the infection by a family of plant viruses. Sci. Rep. 9, 5958. doi: 10.1038/s41598-019-42400-w
Zhang, X., Gonzalez-Carranza, Z. H., Zhang, S., Miao, Y., Liu, C. J., and Roberts, J. A. (2019). F-Box proteins in plants. Annu. Plant Rev. 2, 307–328. doi: 10.1002/9781119312994.apr070
Zhao, Z., Fan, J., Yang, P., Wang, Z., Opiyo, S. O., Mackey, D., et al. (2022). Involvement of Arabidopsis acyl carrier protein 1 in PAMP-triggered immunity. Mol. Plant Microbe Interact. 35, 681–693. doi: 10.1094/MPMI-02-22-0049-R
Zhao, M., Ge, Y., Xu, Z., Ouyang, X., Jia, Y., Liu, J., et al. (2022). A BTB/POZ domain-containing protein negatively regulates plant immunity in Nicotiana benthamiana. Biochem. Biophys. Res. Commun. 600, 54–59. doi: 10.1016/j.bbrc.2022.02.050
Zhao, M., Yang, J. X., Mao, T. Y., Zhu, H. H., Xiang, L., Zhang, J., et al. (2018). Detection of highly differentiated genomic regions between lotus (Nelumbo nucifera Gaertn.) with contrasting plant architecture and their functional relevance to plant architecture. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01219
Keywords: carrot, gene pool, InDels, agro-morphometric, genetic diversity, coding region, population structure
Citation: Mudihal T, Cholin S, Kulkarni CC, Shivaprasad KM, Kumar P, Rolling W and Simon P (2025) Exploring functional InDels and genetic diversity: agro-morphometric and molecular insights into the Western and Eastern gene pools of carrot (Daucus carota L.). Front. Plant Sci. 16:1658653. doi: 10.3389/fpls.2025.1658653
Received: 02 July 2025; Accepted: 22 September 2025;
Published: 22 October 2025.
Edited by:
Miloslava Fojtova, Masaryk University, CzechiaReviewed by:
Shakeel Ahmad Jatoi, Pakistan Agricultural Research Council, PakistanMarcos Vinicius Bohrer Monteiro Siqueira, Universidade do Estado de Minas Gerais, Brazil
Copyright © 2025 Mudihal, Cholin, Kulkarni, Shivaprasad, Kumar, Rolling and Simon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarvamangala Cholin, c2FydWNob2xpbkBnbWFpbC5jb20=; Philipp Simon, cGhpbGlwcC5zaW1vbkB1c2RhLmdvdg==
 Trupthi Mudihal1
Trupthi Mudihal1 Sarvamangala Cholin
Sarvamangala Cholin K. M. Shivaprasad
K. M. Shivaprasad Philipp Simon
Philipp Simon