- 1Xi’an Botanical Garden of Shaanxi Province, Institute of Botany of Shaanxi Province, Xi’an, China
- 2College of Life Sciences, Engineering Research Center for High-Valued Utilization of Fruit Resources in Western China of Ministry of Education, Shaanxi Normal University, Xi’an, China
- 3College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, Zhejiang, China
- 4School of Horticulture, Ludong University, Yantai, China
Plant seeds have evolved diverse dormancy types and regulatory mechanisms to adapt to environmental conditions and seasonal changes. As a commonly used rootstock for cultivated pears, Pyrus betulaefolia faces challenges in seedling production and large-scale cultivation due to limited understanding of seed dormancy mechanisms. In this study, we report that Pyrus betulaefolia seeds exhibit non-deep physiological dormancy, with seed coats playing a pivotal regulatory role. Exogenous abscisic acid (ABA) treatment, fluridone application and seed coat bedding assay demonstrated that dormant seed coats actively synthesized ABA to inhibit embryo germination during imbibition. ABA in imbibed dormant seed coats stimulated ABA biosynthesis in embryos, leading to increased expression of genes involved in ABA biosynthesis (PbeNCED-3) and ABA-responsive (PbeABI3-1, PbeABI5-1, and PbeABI5-5). Importantly, PbeABI5-5 directly binds to the promoters of GIBBERELLIN 2-OXIDASE 3/4 (PbeGA2ox-3/4) to activate their transcription. We establish that in dormant Pyrus betulaefolia seeds, the seed coat controls embryo dormancy release through coordinated regulation of PbeABI5-GA2ox module, thereby maintaining the critical balance between ABA and GA.
1 Introduction
Pyrus betulaefolia is a forest tree species that belongs to Pyrus (Rosaceae). It exhibits remarkable environmental adaptability and thrives under various abiotic stresses, including drought, freezing temperatures, and waterlogged conditions. In horticultural cultivation, it is primarily used as a rootstock for pear trees and serves as a crucial parental material in breeding dwarfing rootstocks and disease-resistant pear varieties (Li et al., 2017; Thakur et al., 2024). It is also an exceptionally valuable germplasm resource for fruit trees (Li et al., 2015). Pyrus betulaefolia is seed-propagated but exhibits physiological dormancy. Seed dormancy represents a crucial adaptive strategy in plants, ensuring viable seeds remain temporarily incapable of germination period even when exposed to favorable environmental conditions such as optimal temperature, moisture, and light (Kim et al., 2024; Baskin and Baskin, 2007). This intrinsic biological mechanism, distinct from quiescence where seeds merely await suitable external conditions (Shu et al., 2016a), plays a fundamental role in plant life history strategies and ecological success. Cold stratification, the conventional dormancy-breaking technique, is constrained by mycotic susceptibility and viability decline during suboptimal treatment. Elucidating the mechanisms underlying seed dormancy—including its physiological basis and molecular regulation is critical for advancing propagation techniques. Such research will not only enhance seedling production efficiency but also support breeding initiatives aimed at improving pear rootstocks and cultivars (Bao and Zhang, 2010). Therefore, this study seeks to investigate the dormancy characteristics of Pyrus betulaefolia seeds, with the ultimate goal of developing optimized germination protocols for commercial and conservation applications.
Abscisic acid (ABA) serves as the primary germination inhibitor in seeds (Barrero et al., 2010). ABA is actively synthesized in seed coats and subsequently transported to embryos, where it effectively suppresses germination and maintains dormancy (Lee et al., 2010). Therefore, while the testa (outer seed coat) primarily imposes physical constraints, the endosperm (inner nutritive tissue) functions as a dynamic biochemical barrier that synthesizes and releases ABA to suppress germination. In Arabidopsis thaliana, seed coat removal triggers the plant embryo growth and greening (Debeaujon et al., 2000). ABA can inhibit the pre-harvest sprouting phenotype of seeds, and ABA content increases with the onset of dormancy (Aloryi et al., 2025). In the ABA signaling pathway, 9-cis-epoxycarotenoid dioxygenases (NCED) serve as the rate-limiting enzyme for ABA biosynthesis, which regulates ABA levels by catalyzing carotenoid cleavage. In Arabidopsis, triple mutant nced5/nced6/nced9 greatly reduced seed dormancy compared with aba2-2 (Frey et al., 2012). ABA-insensitive (ABI) transcription factors constitute core components of ABA signal transduction. Many allelic mutations of abi3 have been found in Arabidopsis thaliana, with the strongest phenotype being abi3-5, and the mutant seeds remain green and are not sensitive to ABA during germination (Ooms et al., 1993). ABI5, an ABA-responsive element-binding factor (AREB/ABF) that binds to ABA response elements (ABREs) in the promoters of ABA-regulated genes, negatively regulates seed germination (Shu et al., 2016b).
Gibberellins (GAs) represent another crucial class of phytohormones that play pivotal roles in breaking seed dormancy and promoting germination, primarily through their antagonistic interaction with ABA (Shu et al., 2018). During seed maturation, GA biosynthesis is actively suppressed, resulting in remarkably low GA levels in mature quiescent seeds (Jacobsen et al., 2002). However, upon imbibition, the scutellum epithelial cells of the embryo rapidly activate GA biosynthesis, leading to a significant accumulation of bioactive GAs in both the embryo and endosperm (Henderson et al., 2004). Gibberellin biosynthesis during seed germination is primarily regulated by GA20ox and GA3ox, which catalyze the production of bioactive GAs, while GA2ox plays a key role in GA deactivation to maintain dormancy (Hedden, 2020). GA burst triggers the production of various hydrolases, including α-amylase, which mobilize stored reserves in the endosperm by converting them into carbohydrates and other nutrients essential for germination (Hedden, 2025). It is the dynamic balance between ABA and GA, rather than their absolute concentrations, that serves as the critical determinant for seed dormancy and germination (Graeber et al., 2012). This balance is maintained through complex reciprocal regulation of hormone biosynthesis and catabolism. ABI4 can directly target the promoters of ABA metabolic genes CYP707A1 and CYP707A2 to promote ABA accumulation and impair GA accumulation during seed dormancy (Shu et al., 2013). Similarly, CHO1 (CHOTTO1), also regulates seed dormancy, but it plays a regulatory role in upstream of ABI4 (Yamagishi et al., 2009; Yano et al., 2009). Cantoro et al. found that SbABI4 and SbABI5 can directly bind to the promoter of SbGA2ox3 in vitro, which may activate GA degradation, thus preventing germination of dormant grains (Cantoro et al., 2013). However, further evidence is needed to confirm the occurrence of this interaction in vivo. Additionally, the GA signaling pathway is initiated when gibberellin binds to its receptor, GIBBERELLIN INSENSITIVE DWARF 1 (GID1), forming the GA-GID1-DELLA complex (Sun, 2011). This complex recruits the SCFSLY1/GID2 E3 ubiquitin ligase, leading to the degradation of DELLA proteins - a critical step in activating downstream GA-responsive genes (Wang and Deng, 2014). In Arabidopsis, five DELLA proteins have been identified: GA-INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), and three RGA-LIKE proteins (RGL1, RGL2, RGL3) (Bao et al., 2020). Among these, RGL2 plays a specialized role by forming a transcriptional complex with ABI4, which mediates the antagonistic interaction between ABA and GA, precisely controlling the shift from dormancy to germination (Liu et al., 2016). Under low GA conditions, DELLA proteins remain stable, exerting a dual regulatory function: they not only suppress GA signaling by modulating downstream targets but also inhibit the expression of key GA biosynthesis (GA20ox2, GA3ox1) and signaling (GID1) genes, thereby maintaining seed dormancy (Kim et al., 2023).
Despite evidence implicating seed coats in Pyrus betulaefolia dormancy, precise classification of its dormancy type and underlying regulatory mechanisms remains unresolved. In this study, we systematically investigated the role of seed coats in maintaining embryo dormancy in Pyrus betulaefolia. Our findings demonstrated that the seeds exhibit non-deep physiological dormancy. Through seed coat bedding assays, we identified that the seed coats of dormant Pyrus betulaefolia significantly suppress embryo germination while simultaneously upregulating key dormancy-related genes, including PbeNCED-3, PbeABI3-1, PbeABI5-1, PbeABI5-5, PbeGA2ox-3, and PbeGA2ox-4 in the embryo. Molecular and biochemical analyses further revealed that transcription factor PbeABI5-5 plays a pivotal role in dormancy maintenance by directly binding to the promoters of PbeGA2ox-3 and PbeGA2ox-4 to activate their transcription. These results provide compelling evidence that PbeABI5-GA2ox module serves as a central regulator of ABA/GA balance in Pyrus betulaefolia, offering new insights into the molecular basis of seed dormancy in this economically important species.
2 Results
2.1 Seed dormancy in Pyrus betulaefolia is imposed by seed coat
Cold stratification was used to examine the dormancy-breaking capacity of freshly harvested Pyrus betulaefolia seeds. Non-stratified seeds demonstrated strong dormancy (hereinafter referred to as “dormant seeds”) (Figure 1A). In contrast, seeds subjected to 2-week cold stratification showed significantly enhanced germination potential, reaching 93.33% after 5 days of incubation at 21°C. Maximal dormancy release was achieved following 5 weeks of cold stratification, resulting in 100% germination (hereinafter referred to as “nondormant seeds”), suggesting the experimental seeds possessed dormancy while maintaining high viability (Figure 1A). To elucidate the mechanisms underlying seed dormancy in Pyrus betulaefolia, germination assays were conducted using isolated embryos dissected from dormant seeds (hereinafter referred to as “coatless embryos”). Comparative analysis revealed that while excised embryos from dormant seeds exhibited delayed germination compared to non-dormant seeds, they ultimately achieved 100% germination within 6 days of incubation (Figures 1B, C). In stark contrast, intact dormant seeds showed a germination rate of 8.70%. These findings provided compelling evidence that the seed coat served as the primary constraint inhibiting germination in Pyrus betulaefolia seeds.
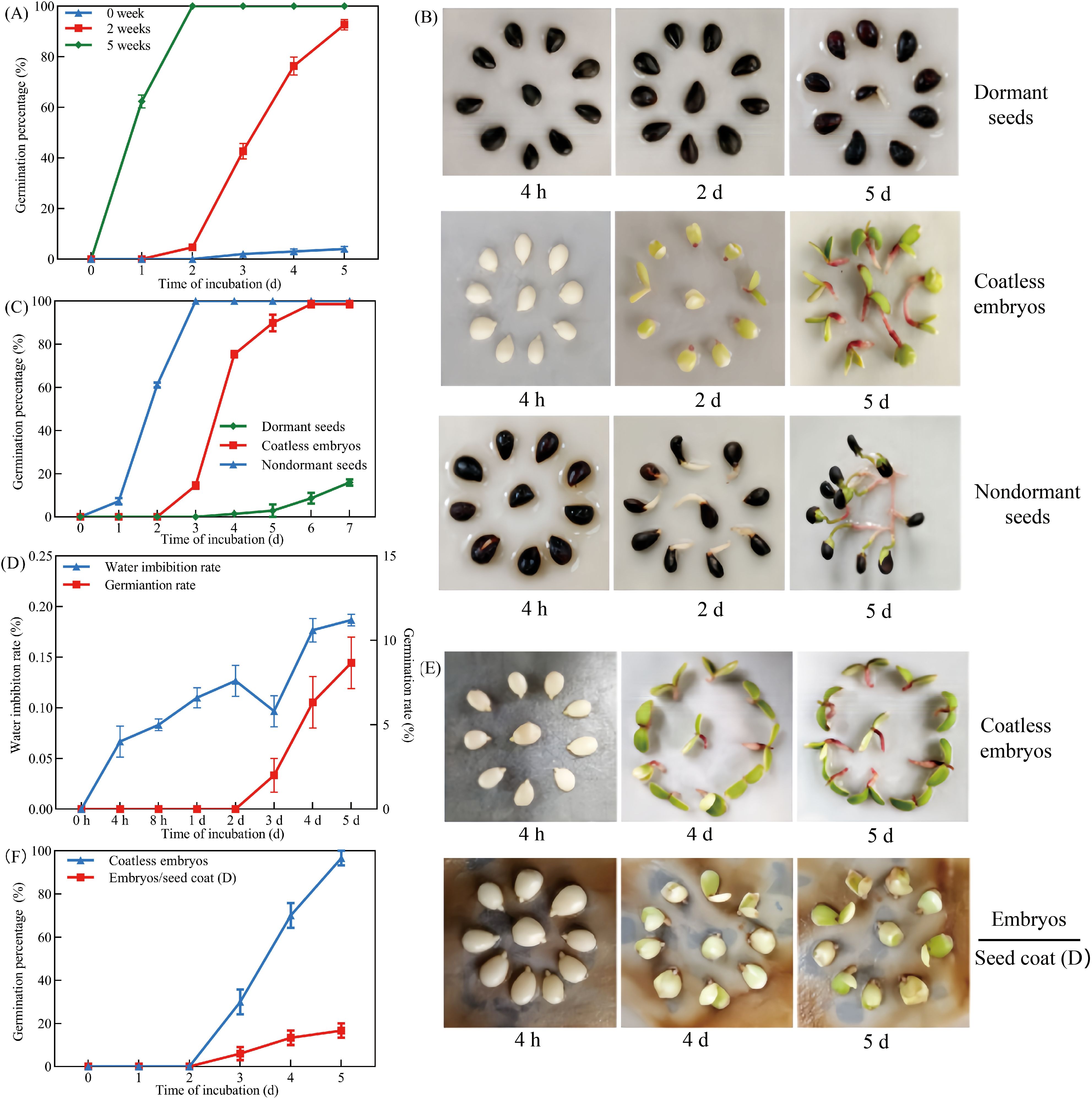
Figure 1. The dormant seed coats repressed germination of Pyrus betulaefolia embryos. (A) Percentage of seed germination after 0, 2 and 5 weeks of cold stratification. (B) Representative images of seed germination assays of untreated (Dormant seeds), coatless embryos dissected from dormant seeds (Coatless embryos), after 5 weeks of cold stratification (Nondormant seeds). (C) Quantification of the germination frequencies of seeds as shown in (B, D) The water imbibition rate and germination rate of dormant seeds. (E) Seed coats bedding assay using embryos dissected from dormant seeds and seed coats dissected from dormant seeds. Seed coats and coatless embryos were physically separated by filter paper. Pictures were taken 4 h, 4 and 5 d after imbibition. (F) Quantification of the germination frequencies of seeds as shown in (E). All data represent the mean ± SE of three biological replicates (> 30 seeds per replicate). Similar results were obtained from three different seed lots.
Seed coat-imposed dormancy in seeds involves three potential mechanisms, including physical impermeability to water, mechanical constraint, and endogenous inhibitors. To investigate the role of water impermeability, we measured water uptake kinetics in dormant seeds. As defined by Baskin and Baskin (2007), the seeds we used exhibited triphasic water uptake kinetics (Phase I: rapid 0–8 h absorption; Phase II: 8 h-3 d plateau; Phase III: post-3 d surge coupled with germination), confirming unimpeded water penetration through the seed coat (Figure 1D). The synchronized onset of Phase III and radicle emergence further verified that dormancy arrest occurs post-hydration, implicating physiological constraints. Using a seed coat bedding assay to physically separate seed coats from embryos (Figures 1E, F), germination rate of isolated embryos was 100% efficiency, while those cultured with dormant seed coats showed significantly suppressed germination rate (13.33% after 5 days of incubation). Together, these results demonstrated that dormancy of Pyrus betulaefolia seed was primarily mediated by inhibitors within the seed coats, which are gradually eliminated or degraded during cold stratification.
2.2 Endogenous ABA in dormant seed coats suppresses embryo germination in Pyrus betulaefolia
Given the well-established role of ABA in promoting seed dormancy and suppressing germination, the effects of fluridone, an ABA biosynthesis inhibitor, on dormant seeds in Pyrus betulaefolia were investigated. Treatment with 10 μM fluridone significantly promoted germination, achieving an 82.22% germination rate after 13 days under 16h/8h (light/dark) photoperiod at 22 ± 2°C (Figures 2A, B). To further examine the role of ABA in seed coat-mediated dormancy,seed coat bedding assays were conducted with or without 100 μM ABA supplementation (Figure 2C). Isolated embryos exhibited 100% germination, and non-dormant seed coats had no inhibitory effect. However, ABA completely suppressed germination in both isolated embryos and embryos co-cultured with non-dormant seed coats (Figure 2C), suggesting that dormant seed coats function similarly to exogenous ABA in preventing embryo germination. Besides, application of 10 μM fluridone in the seed coat bedding assay significantly attenuated the inhibitory effect of dormant seed coats, restoring embryo germination capacity (Figure 2C). These findings provide compelling evidence that dormant seed coats inhibit germination through endogenous ABA accumulation, and that this inhibition can be reversed by blocking ABA biosynthesis.
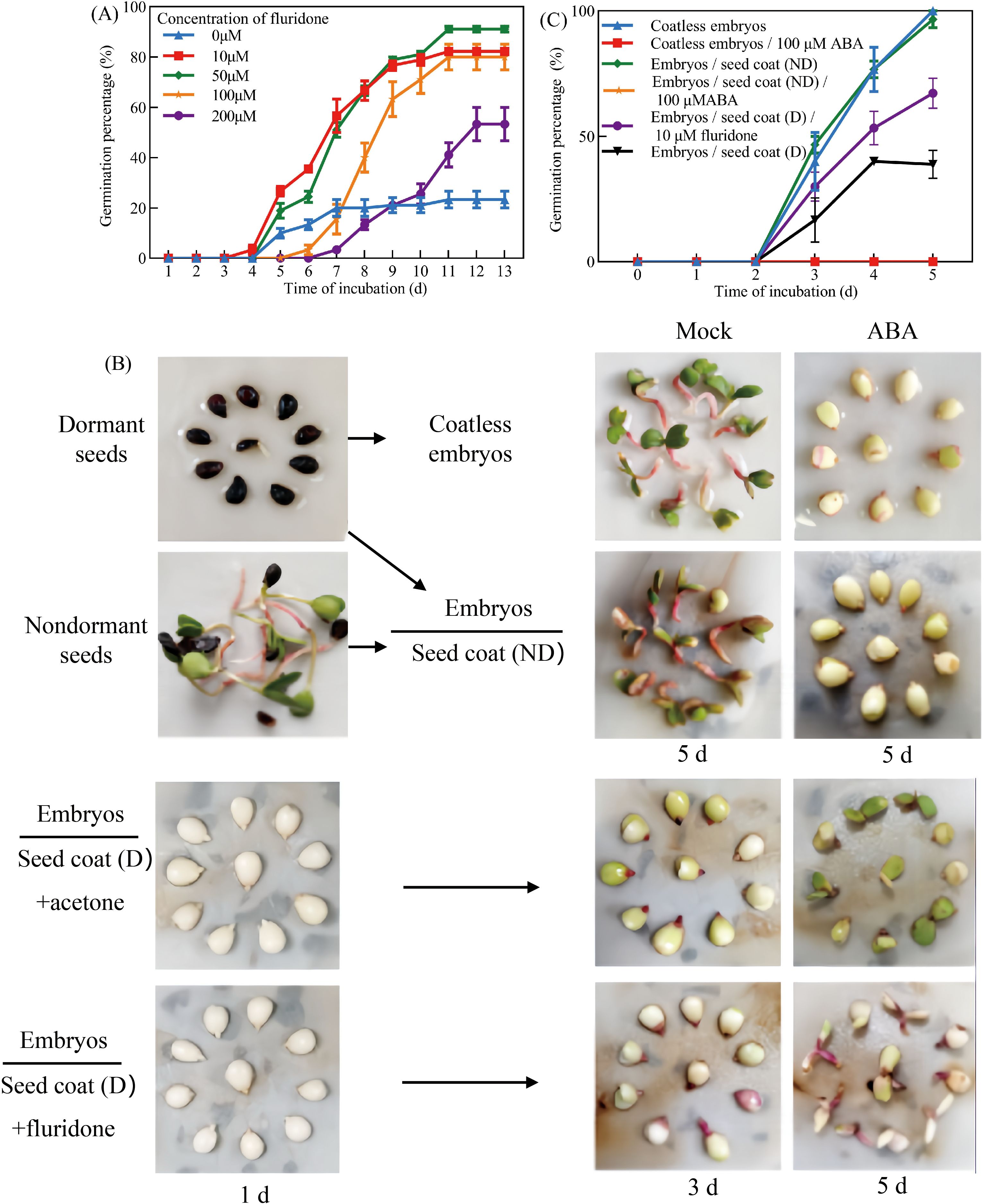
Figure 2. ABA repressed the germination of Pyrus betulaefolia embryos. (A) Germination percentage of dormant seed in the presence of various concentrations of exogenous fluridone. (B) Seed coats bedding assay in the absence or presence of 100 μM ABA using embryos dissected from dormant seeds and seed coats dissected from non-dormant seeds, and seed coats bedding assay in the absence or presence of 10 μM fluridone using embryos dissected from dormant seeds and seed coats dissected from dormant seeds, seed coats and coatless embryos were physically separated by filter paper. (C) Quantification of the germination frequencies of embryos as is shown in (B). All data represent the mean ± SE of three biological replicates (> 30 seeds per replicate). Similar results were obtained from three different seed lots.
2.3 Dormant seed coats regulate ABA biosynthesis and signaling in Pyrus betulaefolia embryos
Expression patterns of key ABA metabolic and signaling genes in coatless embryos treated with either exogenous ABA or dormant seed coats were analyzed (Figure 3; Supplementary Figure 1). Exogenous ABA treatment significantly increased transcript levels of the ABA biosynthesis gene PbeNCED-3 in both isolated embryos and embryos co-cultured with nondormant seed coats (Figure 3A). Similarly, dormant seed coats markedly enhanced the expression of ABA biosynthesis genes PbeNCED-1 and PbeNCED-3 (Figure 3B). Furthermore, we observed that exogenous ABA upregulated ABA signaling transcription factors PbeABI3-1, PbeABI4-1, PbeABI5-1, PbeABI5-2, and PbeABI5-5 (Figure 3A), while dormant seed coats specifically induced PbeABI3-1, PbeABI5-1 and PbeABI5-5 expression (Figure 3B). These transcriptional profiles implied that dormant seed coats modulated ABA homeostasis through coordinated regulation of NCED genes and selective activation of ABI3/ABI5-family signaling components, thereby maintaining seed dormancy.
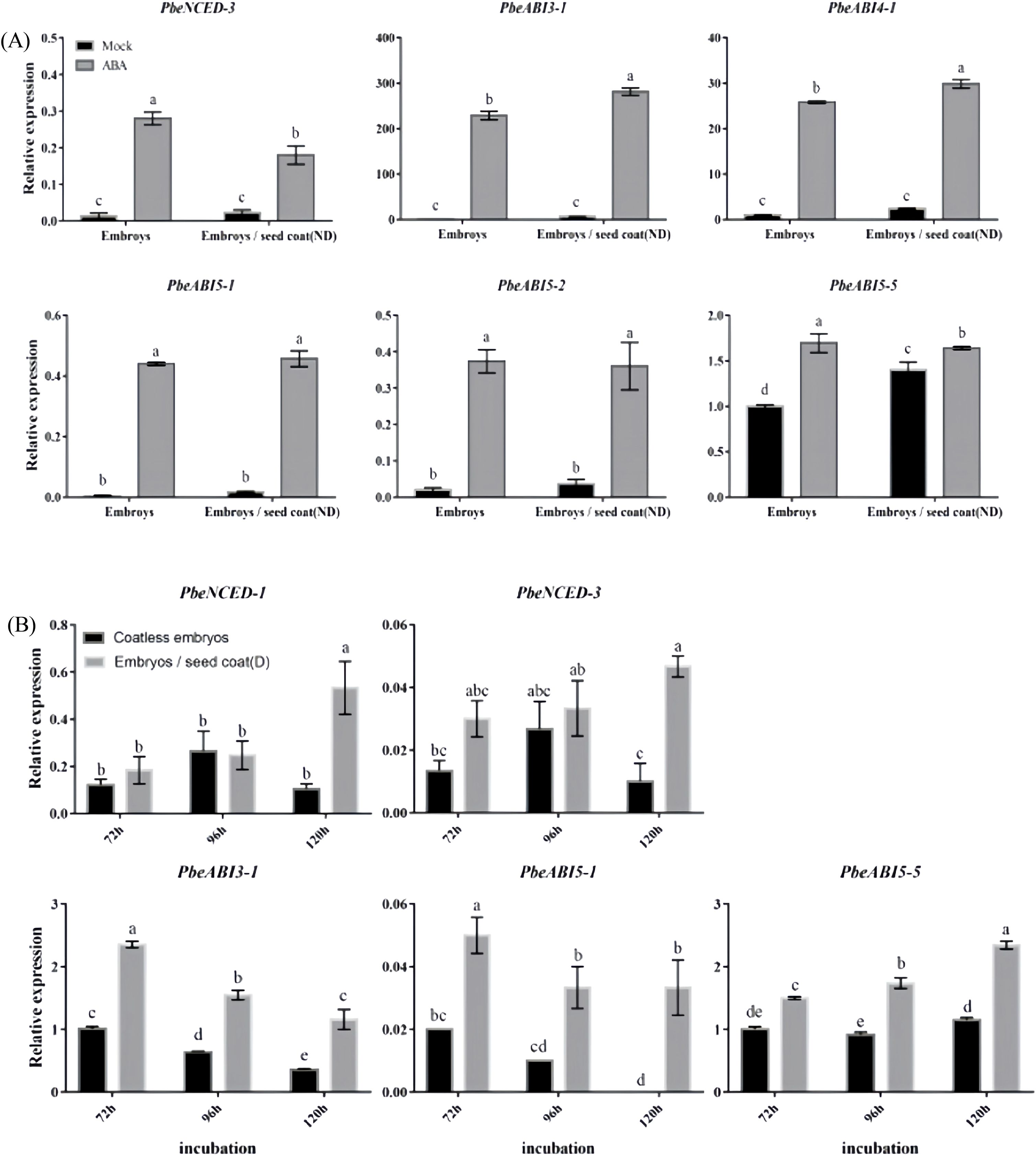
Figure 3. Expression profiles of ABA related genes at different conditions. (A) Total RNA was extracted from the coatless embryos and the embryos co-incubated with nondormant seed coats sampled after 120 h of incubation in the absence or presence of 100 μM ABA. (B) Total RNA was extracted from the coatless embryos and the embryos co-incubated with dormant seed coats sampled at 72, 96, and 120 h after beginning of incubation. Relative expression levels of ABA-related genes were determined by qRT-PCR as described in the Materials and Methods. All data represent the mean ± SE of three biological replicates (> 30 seeds per replicate). Similar results were obtained from three different seed lots. For all panels, different letters indicate significant differences between the data (p < 0.05) using one-way ANOVA with Dunnett’s test.
2.4 Seed coat-enforced dormancy involves ABA-mediated GA metabolism
Investigation of GA metabolic gene expression revealed that both exogenous ABA treatment and dormant seed coats significantly downregulated GA biosynthetic genes (PbeGA20ox-1, PbeGA20ox-2, and PbeGA3ox-1) while concurrently upregulating GA catabolic genes (PbeGA2ox-2 to PbeGA2ox-4, PbeGA2ox-6, and PbeGA2ox-9 to PbeGA3ox-12) in coatless embryos, with PbeGA2ox-3 and PbeGA2ox-4 showing strong induction (4-18-fold and 2-6-fold increases, respectively) under both treatments (Figure 4). Notably, qRT-PCR analysis demonstrated stable expression of DELLA protein family genes (PbeGAI-Like-1 to PbeGAI-Like-3 and PbeRGL1-Like-1 to PbeRGL1-Like-3) regardless of treatment (Supplementary Figure 2). Additionally, coatless embryos were highly sensitive to the GA biosynthesis inhibitor PAC, while exogenous GA4 + 7 could partially overcome dormancy, though both treatments exhibited similar early germination patterns that exceeded controls (Figure 5). These findings demonstrated that seed coat-imposed dormancy involved ABA-mediated suppression of GA biosynthesis coupled with enhanced GA catabolism.
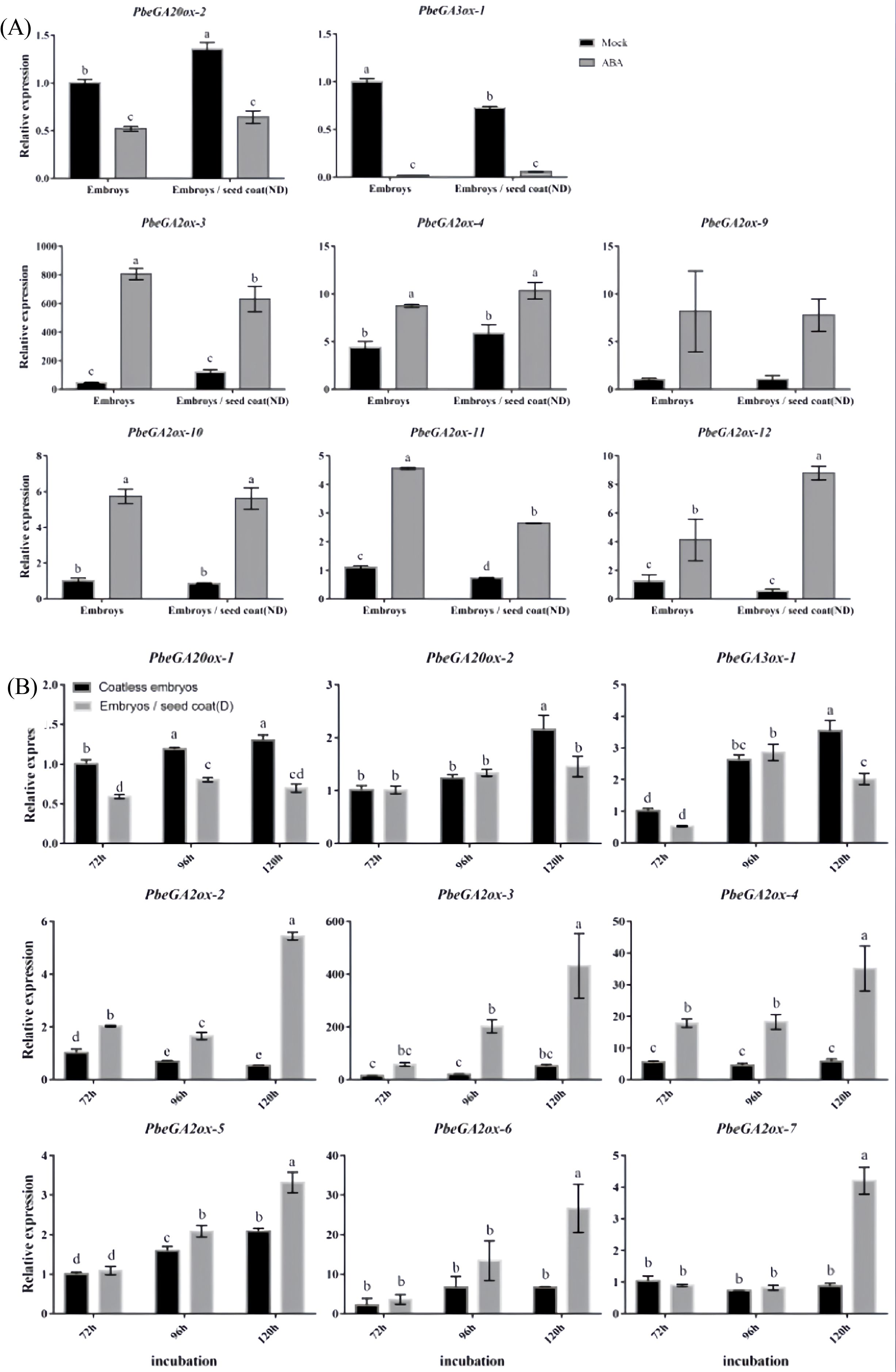
Figure 4. Expression profiles of GA related genes at different conditions. (A) Total RNA was extracted from the coatless embryos and the embryos co-incubated with nondormant seed coats sampled after 120 h of incubation in the absence or presence of 100 μM ABA. (B) Total RNA was extracted from the coatless embryos and the embryos co-incubated with dormant seed coats sampled at 72, 96, and 120 h after beginning of incubation. Relative expression levels of GA-related genes were determined by qRT-PCR as described in the Materials and Methods. All data represent the mean ± SE of three biological replicates (> 30 seeds per replicate). Similar results were obtained from three different seed lots. For all panels, different letters indicate significant differences between the data (p < 0.05) using one-way ANOVA with Dunnett’s test.
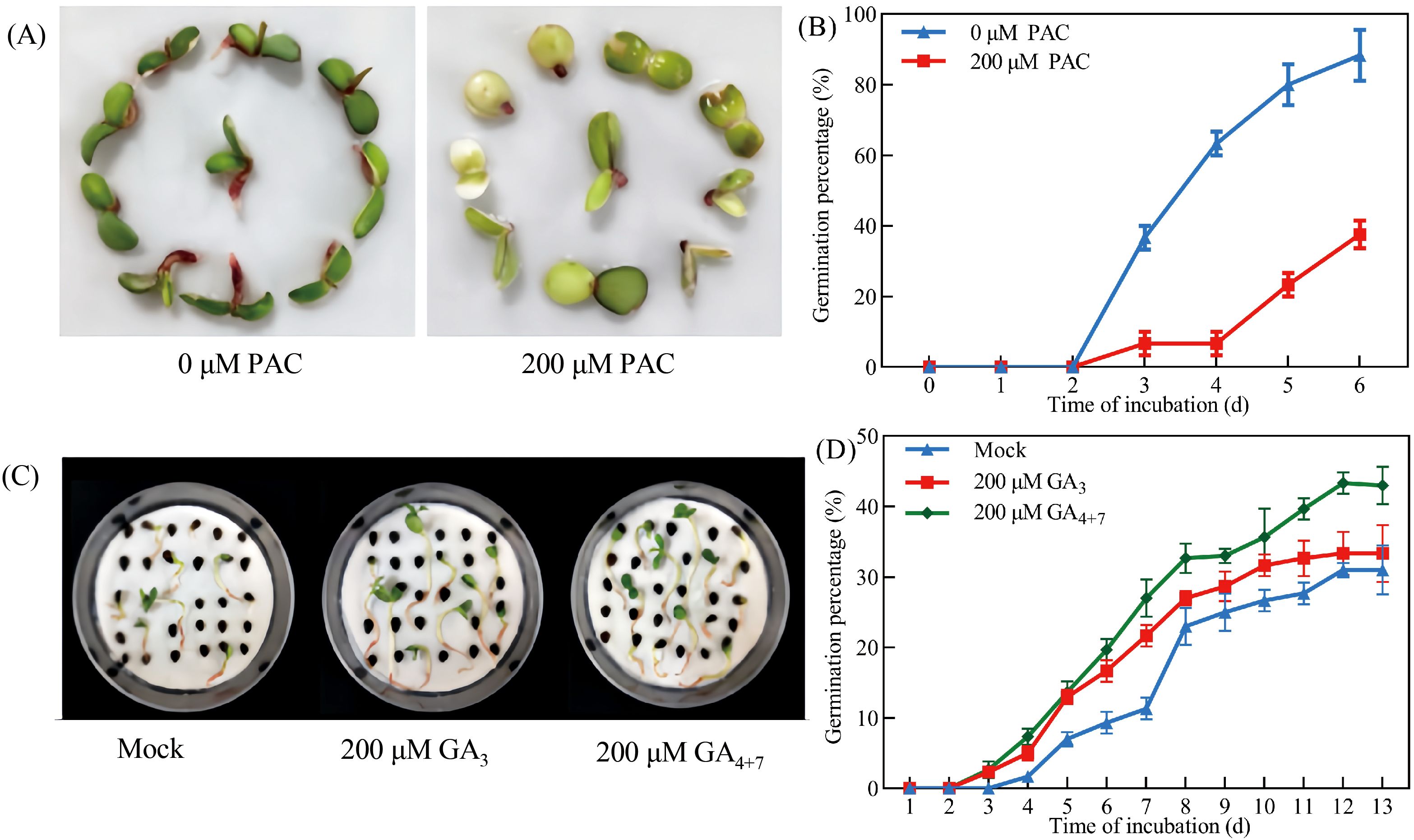
Figure 5. GA increased the germination of Pyrus betulaefolia seeds. (A) Effect of PAC on the germination of dormant seeds. Pictures were taken 6 d after imbibition. (B) Germination percentage of dormant seed in the absence or presence of 200 μM PAC. (C) Effect of GA on the germination of dormant seeds. Pictures were taken 13 days after imbibition. (D) Germination percentage of dormant seed in the absence or presence of 100 μM GA3 or GA4 + 7. The bars represent the mean ± SEM of three biological replicates. All data represent the mean ± SE of three biological replicates (> 30 seeds per replicate). Similar results were obtained from three different seed lots.
2.5 PbeABI5 directly binds to the PbeGA2ox promoters in vivo
Based on the observed co-expression patterns of PbeABI5-1, PbeABI5-5, PbeGA2ox-3 and PbeGA2ox-4 during seed germination, the potential transcriptional regulation of GA2ox genes by ABI5 transcription factors was investigated. Bioinformatics analysis identified three ABRE motifs in the 2-kb promoter regions of both PbeGA2ox-3 and PbeGA2ox-4 (Figure 6A). Yeast one-hybrid assays revealed that PbeABI5-1 and PbeABI5-5 bound to multiple regions of the PbeGA2ox-3 promoter, while only PbeABI5-5 interacted with the PbeGA2ox-4 promoter (Figures 6B, D). Transient expression assays in Nicotiana benthamiana leaves demonstrated that PbeABI5-5 significantly activated transcription from both the PbeGA2ox-3 and PbeGA2ox-4 promoters, as shown by both qualitative and quantitative LUC reporter assays (Figures 6E, F). These findings establish that PbeABI5-5 directly binds to and activates transcription of GA catabolic genes, providing a molecular mechanism for the coordinated regulation of ABA signaling and GA metabolism during seed germination.
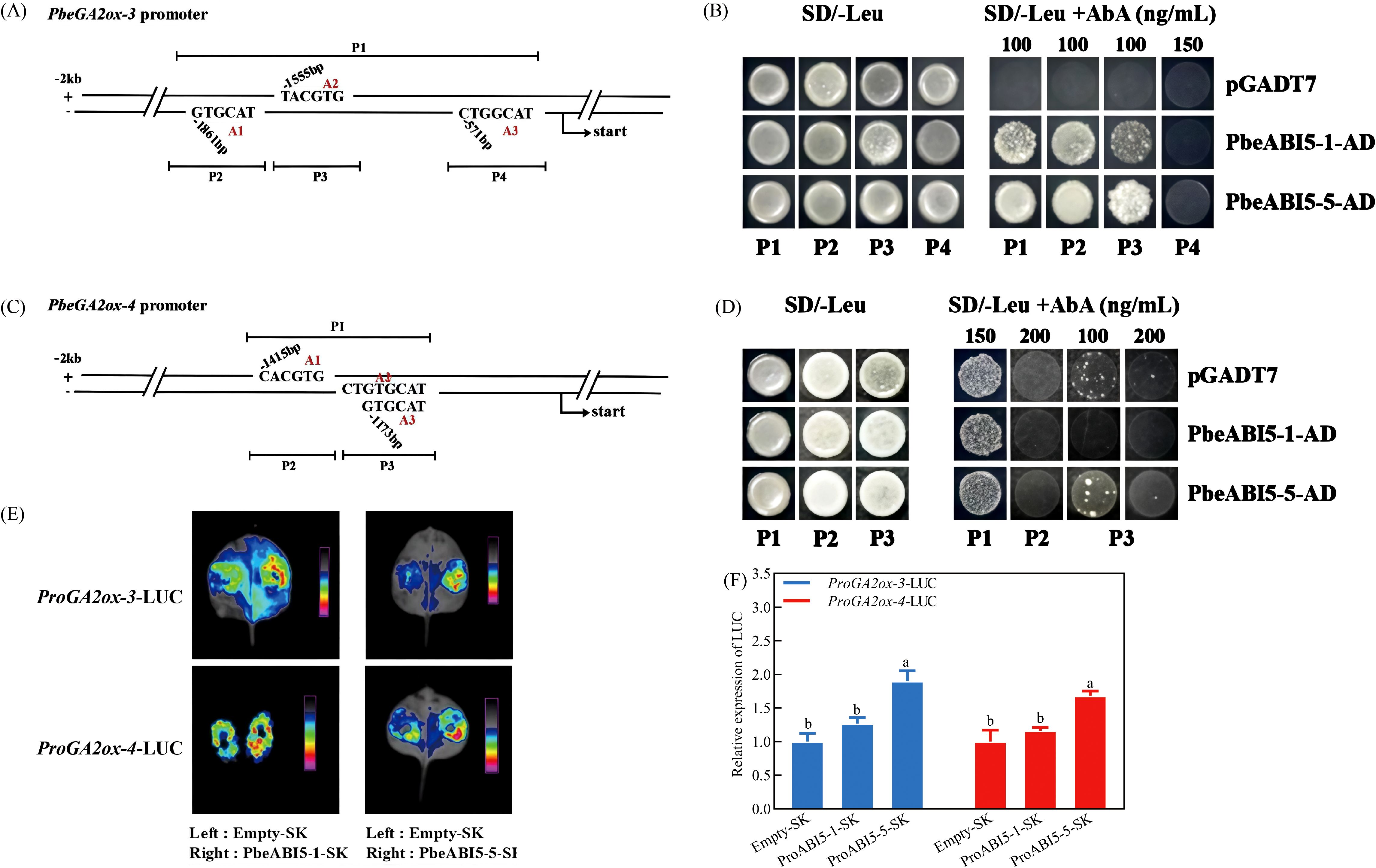
Figure 6. PbeABI5-5 directly bound to the promoter regions of the PbeGA2ox-3 and PbeGA2ox-4 to activate their expression. (A-D) The PbeGA2ox-3 gene promoter was divided into P1 fragment (containing A1, A2, and A3 sites, 1368 bp), and P2 fragment (containing A1 site, 202 bp), P3 fragment (containing A2 site, 252 bp), and P4 fragment (containing A3 site, 309 bp); and the promoter region of PbeGA2ox-4 was divided into P1 segment (containing A1, A2, and A3 sites, 341 bp), and P2 fragment (containing A1 site, 116 bp), P3 fragment (containing A2 and A3 site, 64 bp). The linearized constructs containing different promoter fragments (P1, P2, P3 or P4) in pAbAi were integrated into the genome of Y1HGold yeast strain and either PbeABI5-1-AD, PbeABI5-5-AD or empty AD vector were introduced into each. Aureobasidin A (AbA), an inhibitor of yeast cell growth, was used as a screening marker. Yeast growth on the SD/-Leu+AbA medium indicated binding. (E, F) The effects of PbeABI5-1/5 on the promoter activity of PbeGA2ox-3 and PbeGA2ox-4 in the luciferase reporter assay. These results are obtained through the NightSHADE LB 985 imaging system and Dual-Luciferase® Reporter Assay System. The experiment was performed independently six times with six leaves infiltrated with each plasmid. The bars represent the mean ± SEM of six biological replicates. Different letters indicate a significant difference among different groups (p < 0.05) using one-way ANOVA with Dunnett’s test.
3 Discussion
3.1 Pyrus betulaefolia seeds exhibit non-deep physiological dormancy with seed coat as ABA source
Seeds of Pyrus betulaefolia have a dormant stage, in which embryos with morphological dormancy are not yet fully developed, and they usually germinate after being stored for a period of time at room temperature (Zhang et al., 2022). Our study shows that the germination rate of Pyrus betulaefolia dormant seeds is low, and even if stored at room temperature for a period of time, the germination rate did not increase (Figure 1). Considering that the germination rate of coatless embryos reached 100% after 6 days of incubation, together with rapid germination of coatless embryos, fully developed embryos and permeable seed coats, we confirmed that Pyrus betulaefolia seeds exhibit physiological dormancy. Following Baskin’s classification (Baskin and Baskin, 2007), we identified non-deep physiological dormancy based on three key traits: (1) coatless embryos germinated like non-dormant seeds (Figure 1), (2) cold stratification (5 weeks) fully released dormancy (Figure 1), and (3) GA4 + 7 treatment significantly enhanced germination (Figure 5). Subsequent investigations could focus on performing detailed comparative analyses of dormancy depth throughout the Rosaceae family, with particular emphasis on identifying phylogenetic patterns and ecological correlates that may explain the evolutionary development and maintenance of these dormancy strategies.
In the mature seed of most angiosperms, the embryo is encased by two covering layers (coats): the living endosperm tissue and the outer layer of dead tissue (Topham et al., 2017). Experiments involving the removal of external testa layer while leaving the endosperm layer have shown that the endosperm provides most if not all of the germination repressive activity (Bethke et al., 2007). Previous reports have shown that seed coats surrounding the embryo contain compounds possessing germination-inhibitory activities, including ABA (Shimizu et al., 2022). Indeed, gene expression studies in isolated Arabidopsis seed coat tissue have shown that the endosperm expresses genes involved in ABA metabolism, and it has been demonstrated that the seed coat contains ABA (Barrero et al., 2010). While our data confirmed physiological dormancy in Pyrus betulaefolia (Figure 1 and Figure 2), the role of seed coat extends beyond a passive barrier. Crucially, dormant seed coats actively synthesize ABA to maintain dormancy, as demonstrated in Arabidopsis by Lee et al. (2010), seed coat bedding assay reveals that imbibed dormant coats release ABA to suppress even non-dormant embryos. This mechanism seems conserved across species - in capeweed (Arctotheca calendula), seed coat-derived ABA imposes shallow dormancy (Chapman, 2000), while in recalcitrant Avicennia marina seeds, the pericarp (homologous to seed coats) retains high ABA levels that constrain germination post-dehydration (Farrant et al., 1993).
The germination rate was significantly higher than that of the control group when the seeds and embryos were treated with fluridone (Figure 2). Consistently, in Arabidopsis, treating imbibed dormant wild-type seeds with fluridone will trigger their germination (Debeaujon and Koornneef, 2020; Millar et al., 2006), suggesting that repression of germination is an active process requiring de novo and constitutive production of the phytohormone ABA upon seed imbibition. Further supported by the observation that dormant and nondormant dry seeds contain similarly high amounts of ABA, which played an essential and well established developmental role during seed maturation, and upon seed imbibition, the high ABA levels present in both dormant and nondormant seeds drop drastically by about 10-fold within the first 12 h, and what indeed distinguishes dormant seeds from nondormant seeds is the capacity of dormant seeds to synthesize ABA de novo and constitutively once the initial drop of ABA levels has occurred upon seed imbibition (Ali-Rachedi et al., 2004; Piskurewicz et al., 2008), indicating that de novo ABA produced in the Pyrus betulaefolia seed coats and released toward the embryo to repress germination. Furthermore, the reported observations should not be meant to imply that the ABA derived from the seed coats is the only cause of dormancy. The strength of dormancy in a given seed, i.e., its capacity to restrain from germinating upon imbibition, is most likely related to the seed’s capacity to synthesize ABA as a whole: in both the endosperm and the embryo. Rather, we wish to propose that the endosperm is an essential contributor to sustain dormancy, i.e., to prevent germination over time upon imbibition.
3.2 ABA biosynthesis and catabolic feedback shape dormancy maintenance
That ABA synthesized in embryos also contributes to overall seed dormancy is most likely reflected by the observation that dormant embryos green and germinate slower than nondormant embryos upon dissection, and coatless embryos dissected from isolated dormant seeds 24 h after imbibition contained 2.5-fold more ABA than coatless embryos isolated from nondormant seeds (Lee et al., 2010). In agreement with this view, we found that the dormant seed coats promoted the expression of the ABA biosynthesis genes PbeNCED-1 and PbeNCED-3 in embryos (Figure 3). It should be pointed out that dormancy-promoting function of the seed coat is not strictly restricted to the endosperm. However, PbeABI3-1 was significantly up-regulated under exogenous ABA or dormant seed coat treatment, which was similar to the expression patterns of PbeABI5-1 or PbeABI5-5 (Figure 3, 4). Studies have also shown that ABI3 has a domain interacting with ABI5, and ABI3 can also positively regulate the expression of ABI5 (Lopez-Molina et al., 2001; Nakamura et al., 2001; Lopez-Molina et al., 2002), indicating the relationship between PbeABI3 and PbeABI5 need to be further studied. Even with a larger relative increase, the absolute amount of PbeABI5-1 transcript induced by exogenous ABA or dormant seed coat treatment might still be considerably less than the basal and induced levels of PbeABI5-5 (Figure 3, 4). Crucially, our functional assays clearly demonstrated that only the PbeABI5-5 protein possesses the specific ability to bind the identified cis-element in the PbeGA2ox promoter and activate its expression (Figure 6). While both are ABA-responsive bZIP transcription factors, they appear to have evolved distinct target gene specificities. In contrast, the expression of the ABA catabolic genes PbeCYP707A-1 to PbeCYP707A-5 were also significantly upregulated in the coatless embryos both treated with exogenous ABA and dormant seed coats (Figure 3). Considering the similar feedback regulation in Prunus persica and Arabidopsis (Wang and Deng, 2014; Shu et al., 2016a), we hypothesized that the upregulation could be due to feedback regulation by the higher ABA content in these embryos. Notably, although gene expression analysis revealed potential modulation of ABA/GA homeostasis during dormancy release, we emphasize that the absence of ABA and GA quantification constitutes a limitation. While established models indicate NCED upregulation typically elevates ABA levels and GA2ox induction promotes GA catabolism (Hedden, 2020; Aloryi et al., 2025), future direct hormone measurements will allow us to rule out potential decoupling between gene expression patterns and actual hormone concentrations caused by post-transcriptional regulation or compensatory metabolic pathways. The relative contributions of embryonic versus seed coat-derived ABA also deserves careful consideration. While qRT-PCR data showed rapid upregulation of de novo ABA biosynthesis genes in isolated embryos (Figure 3), potential priming effects from coat-translocated ABA prior to tissue isolation cannot be excluded. Future research will prioritize direct quantification of ABA levels to definitively resolve the contributions of seed coat-derived versus embryo-synthesized ABA in this system.
3.3 Molecular crosstalk between ABA and GA in regulating Pyrus betulaefolia seed dormancy
While studies have established the antagonistic relationship between abscisic acid and gibberellins in seed dormancy and germination (Lee et al., 2015; Hoang et al., 2014), the precise molecular mechanisms underlying their interaction remain incompletely understood. Crucially, seed coat-specific adaptations diverge across species: in Arabidopsis, endosperm-derived ABA transporters regulate embryonic ABI5 activation (Shimizu et al., 2022); in legumes, ABI5 directly promotes seed coat sclerification by repressing GA catabolism genes (Zinsmeister et al., 2016). AtABI4 has been identified as a crucial transcription factor in post-germination stages, which transcription and protein stability were enhanced by ABA, subsequently upregulating AtGA2ox7 expression to reduce active GA content; Conversely, GA signaling suppresses AtABI4 expression and accelerates its protein degradation, thereby inhibiting AtNCED6 transcription and ABA biosynthesis (Shu et al., 2016b). Cantoro et al. identified direct binding of SbABI4 and SbABI5 to the SbGA2ox3 promoter in vitro using electrophoretic mobility shift assay, suggesting these transcription factors may trigger GA catabolism to suppress germination in dormant grains (Cantoro et al., 2013).
Our research reveals another key player in this hormonal crosstalk - PbeABI5-5. Based on the induction of PbeGA2ox-3 and PbeGA2ox-4 genes expression by ABA (Figure 4), we demonstrated that PbeABI5-5 directly bound to and activates the promoters of the two genes (Figure 5). Through yeast one-hybrid and tobacco transient expression assays, we demonstrate that PbeABI5-5 directly binds to and activates the promoters of PbeGA2ox-3 and PbeGA2ox-4 genes (Figure 6). These findings support our proposed model (Figure 7), where in Pyrus betulaefolia seed coats enhance embryonic ABA biosynthesis. Elevated ABA levels upregulate PbeABI5-5 expression, which in turn activates PbeGA2ox genes through direct promoter binding. The resulting increase in GA catabolism reduces bioactive GA levels, maintaining embryonic dormancy by inhibiting germination. This regulatory cascade represents a novel mechanism through which ABA-dominated seed coat signaling influences embryonic GA metabolism to sustain dormancy.In conclusion, our study establishes that seed coat-derived ABA sustains dormancy in Pyrus betulaefolia by activating PbeABI5-5, which transcriptionally upregulates PbeGA2ox genes to enhance GA catabolism and suppress embryonic germination. This ABA-GA crosstalk mechanism, mediated through direct PbeABI5-GA2ox promoter binding, reveals a novel regulatory layer wherein seed coat signaling controls embryonic GA metabolism to maintain physiological dormancy.
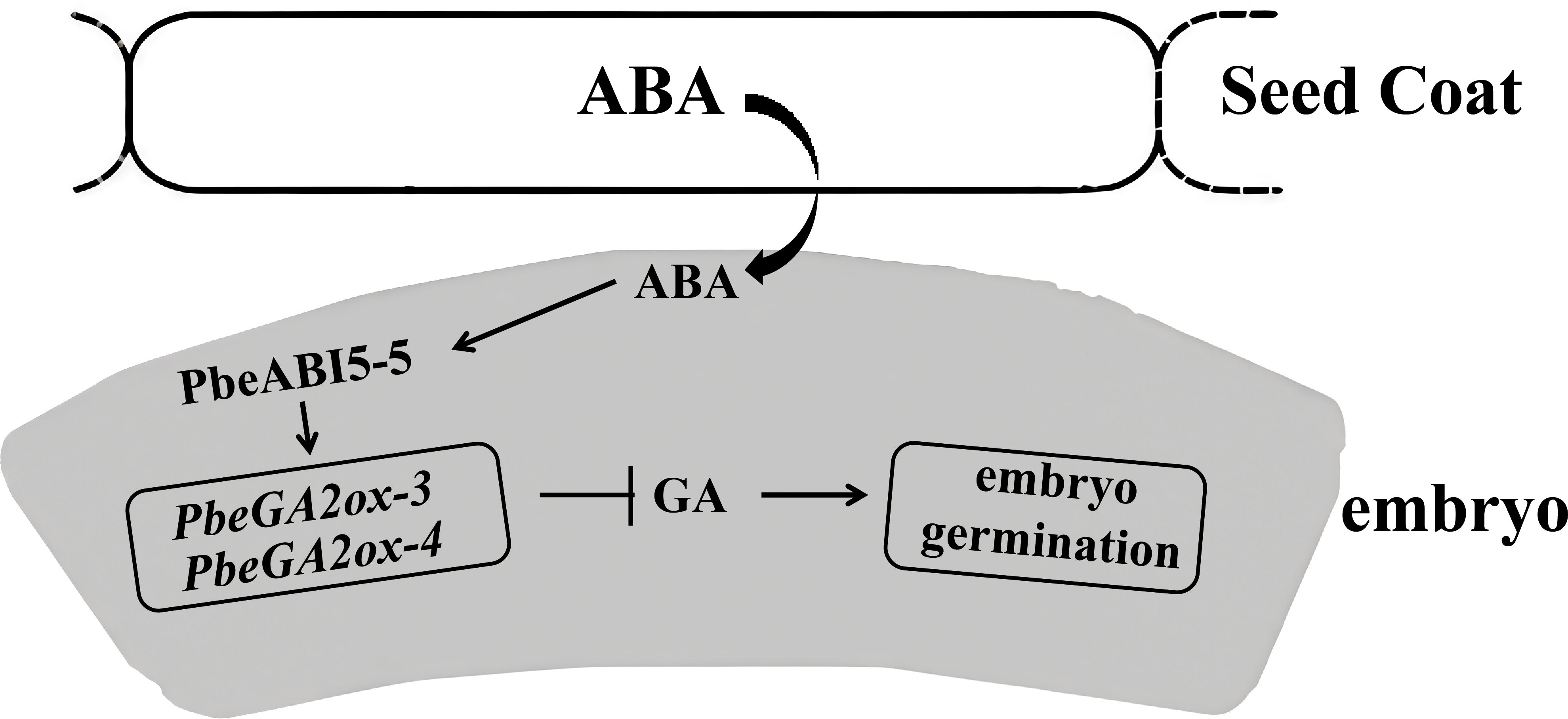
Figure 7. Proposed working model of seed coat- and ABA-dependent repression of dormant seed germination. Dormancy is a state where constitutive production of ABA in the coats maintains a proper level of PbeABI5 and repress GA biogenesis by the direct activation of PbeGA2ox-3/4, and therefore negatively regulates embryo germination, maintains dormancy. The lines ending with arrows denote positive regulation, while the lines ending with bars denote negative regulation. Note: GA promotes germination when present.
4 Materials and methods
4.1 Plant materials and growth conditions
Freshly harvested Pyrus betulaefolia seeds were collected from Xinjiang Uygur Autonomous Region, China. The seeds were rinsed with sterile water and air-dried in a shaded environment before storage in self-sealing bags at room temperature for subsequent experiments.
For germination assay, seeds were surface-sterilized with 5% NaClO for 5 min, followed by five washes with sterile water. After a 4-h imbibition in sterile water, seeds were placed on two layers of moist filter paper in Petri dishes. Each dish was treated with 2 mL of sterile water (control), ABA, fluridone, GA, or PAC, depending on the experimental conditions. The plates were incubated in a growth chamber (22 ± 2°C, 60–70% relative humidity, 16-h light/8-h dark cycle). Germination was monitored twice daily, and sterile water or treatment solutions were replenished as needed to maintain moisture. Seeds with a 2-mm radicle protrusion were scored as germinated. The germination rate was calculated as: Germination rate (%) = (Number of germinated seeds/Total seeds) × 100.
For cold stratification, sterilized seeds were mixed with moist sand and stored at 4°C. After stratification, seeds were transferred to germination conditions as described above.
The seed coat bedding assay was adapted from Lee et al (Lee et al., 2010). Briefly, imbibed seeds were dissected into embryos and seed coats using fine forceps. Ten coatless embryos were placed on filter paper overlying 30 seed coats, supplemented with 2 mL of sterile water, ABA, or fluridone, and incubated under standard conditions (Supplementary Figure 3).
Water imbibition rates were determined by weighing seeds before and after incubation. Surface moisture was removed prior to initial weighing. The imbibition rate was calculated as: Water imbibition rate (%) = [(Post-incubation weight − Initial weight)/Initial weight] × 100.
4.2 RNA extraction and qRT-PCR analysis
The reference genome used was Pyrus x bretschneideri genome sequence. Total RNA was extracted using the HiPure Plant RNA Mini Kit (Magen, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 µg of total RNA using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Japan) following the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Premix Ex Taq™ (Takara, Japan), on a CFX96 Real-Time PCR System (Bio-Rad, USA). Gene-specific primers (designed with Primer3, http://bioinfo.ut.ee/primer3-0.4.0/) are listed in Supplementary Table S1. Prior to analysis, primer efficiency was validated using a standard curve (80–120% efficiency required). Each reaction was run in three technical replicates, and gene expression levels were calculated using the 2-ΔΔCt method, with PpActin (JN684184) as the internal reference (Liu et al., 2012).
4.3 ABRE motifs analysis of PbeGA2ox promoters
The 2000bp region upstream from the start codon of PbeGA2ox genes were extracted from the Pyrus betulaefolia genome as a promoter. Then, the sequences of promoters were uploaded to the Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict ABRE motifs.
4.4 Yeast one-hybrid assay
Yeast one-hybrid (Y1H) assays were performed using the Matchmaker™ Gold Yeast One-Hybrid Library Screening System (Clontech, USA) following the manufacturer’s protocol. The primers used for constructing bait and prey are listed in Supplementary Table S2. Different promoter fragments of PbeGA2ox containing the ABRE sequence were cloned into the pAbAi vector. The recombinant plasmid was linearized and integrated into the genome of the Y1H Gold yeast strain. Subsequently, yeast cells were co-transformed with either PbeABI5-AD or the empty AD vector. Protein-DNA interactions were assessed based on yeast growth on SD/-Leu medium supplemented with 100–200 ng/mL aureobasidin A.
4.5 Dual-luciferase reporter assay
The dual-luciferase assay was performed in Nicotiana benthamiana leaves following the method of Niu et al (Niu et al., 2016). The full-length coding sequences of PbeABI5-1 and PbeABI5-5 were cloned into the pGreenII 0029 62-SK vector, while the promoter regions of PbeGA2ox-3 and PbeGA2ox-4 were inserted into pGreenII 0800-LUC. All constructs were transformed into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method. For infiltration, Agrobacterium cultures (OD600 = 1.0) carrying the effector and reporter constructs were mixed at a 10:1 ratio and incubated at room temperature for 2 h before infiltration into leaves. After 48-72 h post-infiltration, leaf samples were collected for observation. For qualitative detection, luciferase activity was visualized using the NightSHADE LB 985 imaging system (Berthold Technologies) after infiltration of 0.2 mM luciferin (Promega) 30 min prior to imaging. For quantitative analysis, firefly and Renilla luciferase activities were measured using the Dual-Luciferase® Reporter Assay System (Promega) on a Modulus™ Luminometer (Promega). Reporter gene expression was calculated as the ratio of LUCfirefly: RENRenilla. Each experiment included three independent biological replicates, with six technical replicates per biological replicate.
4.6 Statistical analysis
All experiments were arranged in a completely randomized design. Data were analyzed with SPSS Statistics v.18.0 (IBM, USA). Statistical significance was determined at p < 0.05 using appropriate tests. Data visualization was performed using GraphPad Prism 7.00. Values are presented as mean ± standard error (SE) from at least three independent biological replicates.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
XW: Formal Analysis, Funding acquisition, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. LH: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. JL: Formal Analysis, Investigation, Methodology, Software, Visualization, Writing – review & editing. SB: Project administration, Writing – review & editing. YT: Funding acquisition, Resources, Writing – review & editing. WH: Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32272679) to YT; Natural Science Research Plan of Shaanxi Province (No. 2025JC-YBMS-239), Shaanxi Academy of Science Research Funding Project (No.2024k-19) and Xi’an Innovation Capacity Foundation Strengthening Plan-Agricultural Technology Research and Development Project (No. 21NYYF0045) to XW.
Acknowledgments
Authors are grateful to the teachers and students who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1667946/full#supplementary-material
References
Ali-Rachedi, S., Bouinot, D., Wagner, M. H., Bonnet, M., Sotta, B., Grappin, P., et al. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta. 219, 479–488. doi: 10.1007/s00425-004-1251-4
Aloryi, K. D., Okpala, N. E., Amenyogbe, M. K., Bimpong, D., Karikari, B., Guo, H., et al. (2025). Whole-genome meta-analysis coupled with haplotype analysis reveal new genes and functional haplotypes conferring pre-harvest sprouting in rice. BMC Plant Biol. 25, 527. doi: 10.1186/s12870-025-06551-5
Bao, J. P. and Zhang, S. L. (2010). Effects of seed coat, chemicals and hormones on breaking dormancy in pear rootstock seeds (Pyrus betulaefolia Bge. and Pyrus calleryana Dcne.). Seed Sci. Technol. 38, 348–357. doi: 10.15258/sst.2010.38.2.08
Bao, S., Hua, C., Shen, L., and Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. doi: 10.1111/jipb.12892
Barrero, J. M., Millar, A. A., Griffiths, J., Czechowski, T., Scheible, W. R., Udvardi, M., et al. (2010). Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 61, 611–622. doi: 10.1111/j.1365-313X.2009.04088.x
Baskin, J. M. and Baskin, C. C. (2007). A classification system for seed dormancy. Seed Sci. Res. 14, 1–16. doi: 10.1111/j.1469-8137.2006.01787.x
Bethke, P. C., Libourel, I. G., Aoyama, N., Chung, Y. Y., Still, D. W., and Jones, R. L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143, 1173–1188. doi: 10.1104/pp.106.093435
Cantoro, R., Crocco, C. D., Benech-Arnold, R. L., and Rodríguez, M. V. (2013). In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 64, 5721–5735. doi: 10.1093/jxb/ert347
Chapman, A. J. E. (2000). Embryo and seed coat factors produce seed dormancy in capeweed (Arctotheca calendula). Crop Pasture Sci. 51, 849–854. doi: 10.1071/AR00050
Debeaujon, I. and Koornneef, M. (2020). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. doi: 10.1104/pp.122.2.415
Debeaujon, I., Léon-Kloosterziel, K. M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122, 403–414. doi: 10.1104/pp.122.2.403
Farrant, J. M., Pammenter, N. W., and Berjak, P. (1993). Seed development in relation to desiccation tolerance: A comparison between desiccation-sensitive (recalcitrant) seeds of Avicennia marina and desiccation-tolerant types. Seed Sci. Res. 3, 1–13. doi: 10.1017/S0960258500001513
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Graeber, K., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., and Soppe, W. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hedden, P. (2025). Induction of α-amylase and endosperm-imposed seed dormancy: two pioneering papers in gibberellin research. Planta. 261, 118. doi: 10.1007/s00425-025-04699-w
Henderson, J. T., Li, H. C., Rider, S. D., Mordhorst, A. P., Romero-Severson, J., Cheng, J. C., et al. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Hoang, H. H., Sechet, J., Bailly, C., Leymarie, J., and Corbineau, F. (2014). Inhibition of germination of dormant barley (Hordeum vulgare L.) grains by blue light as related to oxygen and hormonal regulation. Plant Cell Environ. 37, 1393–1403. doi: 10.1111/pce.12239
Jacobsen, J. V., Pearce, D. W., Poole, A. T., Pharis, R. P., and Mander, L. N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant 115, 428–441. doi: 10.1034/j.1399-3054.2002.1150313.x
Kim, S., Huh, S. M., Han, H. J., Lee, G. S., Hwang, Y. S., Cho, M. H., et al. (2023). A rice seed-specific glycine-rich protein OsDOR1 interacts with GID1 to repress GA signaling and regulates seed dormancy. Plant Mol. Biol. 111, 523–539. doi: 10.1007/s11103-023-01343-7
Kim, Y. J., Park, K., Jang, B. K., Kwon., S. P., and Cho, J. S. (2024). Classification of dormancy types and breakout conditions in Reynoutria sachalinensis exhibiting seed dormancy polymorphism. Hortic. Environ. Biotechnol. 65, 997–1007. doi: 10.1007/s13580-024-00626-2
Lee, H. G., Lee, K., and Seo, P. J. (2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 87, 371–381. doi: 10.1007/s11103-015-0283-4
Lee, K. P., Piskurewicz, U., Turecková, V., Strnad, M., and Lopez-Molina, L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U S A. 107, 19108–19113. doi: 10.1073/pnas.1012896107
Li, H., Cong, Y., Chang, Y. H., and Lin, J. (2015). Two AMT2-Type Ammonium transporters from Pyrus betulaefolia demonstrate distinct expression characteristics. Plant Mol. Biol. Rep. 34, 1–13. doi: 10.1007/s11105-015-0957-8
Li, K., Xing, C., Yao, Z., and Huang, X. (2017). PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 15, 1186–1203. doi: 10.1111/pbi.12708
Liu, G., Li, W., Zheng, P., Xu, T., Chen, L., Liu, D., et al. (2012). Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics 13, 700. doi: 10.1186/1471-2164-13-700
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun. 27, 12768. doi: 10.1038/ncomms12768
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313x.2002.01430.x
Millar, A. A., Jacobsen, J. V., Ross, J. J., Helliwell, C. A., Poole, A. T., Scofield, G., et al. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8’-hydroxylase. Plant J. 45, 942–954. doi: 10.1111/j.1365-313X.2006.02659.x
Nakamura, S., Lynch, T. J., and Finkelstein, R. R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. doi: 10.1046/j.1365-313x.2001.01069.x
Niu, Q., Li, J., Cai, D., Qian, M., Jia, H., Bai, S., et al. (2016). Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 67, 239–257. doi: 10.1093/jxb/erv454
Ooms, J., Leon-Kloosterziel, K. M., Bartels, D., Koornneef, M., and Karssen, C. M. (1993). Acquisition of Desiccation Tolerance and Longevity in Seeds of Arabidopsis thaliana (A Comparative Study Using Abscisic Acid-Insensitive abi3 Mutants). Plant Physiol. 102, 1185–1191. doi: 10.1104/pp.102.4.1185
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 20, 2729–2745. doi: 10.1105/tpc.108.061515
Shimizu, T., Kanno, Y., Watanabe, S., and Seo, M. (2022). Arabidopsis NPF5.1 regulates ABA homeostasis and seed germination by mediating ABA uptake into the seed coat. Plant Signal Behav. 17, 2095488. doi: 10.1080/15592324.2022.2095488
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016b). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016a). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PloS Genet. 9, e1003577. doi: 10.1371/journal.pgen.1003577
Shu, K., Zhou, W., Chen, F., Luo, X., and Yang, W. (2018). Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00416
Sun, T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, 338–345. doi: 10.1016/j.cub.2011.02.036
Thakur, M., Ajender, T. D., Thakur, K., and Rimpika, T. K. (2024). “Fruit and nut crops: pear genetic resources and utilization,” in Handbooks of crop diversity: conservation and use of plant genetic resources. Eds. Rajasekharan, P. E. and Rao, V. R. (Springer Press, Singapore), 119–140.
Topham, A. T., Taylor, R. E., Yan, D., Nambara, E., Johnston, I. G., and Bassel, G. W. (2017). Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc. Natl. Acad. Sci. U S A. 114, 6629–6634. doi: 10.1073/pnas.1704745114
Wang, Y. and Deng, D. (2014). Molecular basis and evolutionary pattern of GA-GID1-DELLA regulatory module. Mol. Genet. Genomics 289, 1–9. doi: 10.1007/s00438-013-0797-x
Yamagishi, K., Tatematsu, K., Yano, R., Preston, J., Kitamura, S., Takahashi, H., et al. (2009). CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol. 50, 330–340. doi: 10.1093/pcp/pcn201
Yano, R., Kanno, Y., Jikumaru, Y., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2009). CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 151, 641–654. doi: 10.1104/pp.109.142018
Zhang, Y., Liu, Y., Sun, L., Baskin, C. C., Baskin, J. M., Cao, M., et al. (2022). Seed dormancy in space and time: global distribution, paleoclimatic and present climatic drivers, and evolutionary adaptations. New Phytol. 234, 1770–1781. doi: 10.1111/nph.18099
Keywords: Pyrus betulaefolia, seed dormancy, seed coat, ABA, GA, ABI5, GA2ox
Citation: Wang X, He L, Li J, Bai S, Teng Y and Hui W (2025) Seed coat-derived ABA regulates seed dormancy of Pyrus betulaefolia by modulating ABA and GA balance. Front. Plant Sci. 16:1667946. doi: 10.3389/fpls.2025.1667946
Received: 17 July 2025; Accepted: 12 August 2025;
Published: 01 September 2025.
Edited by:
Fan Xu, Southwest University, ChinaReviewed by:
Alma Armenta-Medina, National Institute of Forestry and Agricultural Research (INIFAP), MexicoJuan Wang, Yangzhou University, China
Hue Xue, Beijing Forestry University, China
Wenguan Zhou, Northwestern Polytechnical University, China
Copyright © 2025 Wang, He, Li, Bai, Teng and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hui, aHVpaHVpQHNubnUuZWR1LmNu; Yuanwen Teng, eXd0ZW5nQHpqdS5lZHUuY24=
 Xiaoting Wang
Xiaoting Wang Lufang He2
Lufang He2 Jianzhao Li
Jianzhao Li Songling Bai
Songling Bai Yuanwen Teng
Yuanwen Teng Wei Hui
Wei Hui