- 1College of Plant Protection, Hebei Agricultural University, Technological Innovation Center for Biological Control of Crop Diseases and Insect Pests of Hebei Province, Baoding, Hebei, China
- 2Wheat Research Institute, Tangshan Academy of Agricultural Sciences, Tangshan, Hebei, China
- 3National Engineering Research Center for Agriculture in Northern Mountainous Areas, Hebei Agricultural University, Baoding, China
Puccinia triticina (Pt) races vary frequently, and new virulence races continue to emerge, leading to a lack of durable resistance in wheat cultivars. Therefore, monitoring avirulence gene composition and variation in Pt is crucial. In our previous study, we identified an avirulence protein AvrLr15 from Pt that induced Lr15-dependent immune responses and found that evasion of Lr15-mediated resistance in wheat was associated with a deletion and point mutations of amino acids in AvrLr15. In this study, TcLr15–Thatcher hybrid materials and Lr15 mutant materials were developed, which further confirmed the reliability of AvrLr15. A duplex PCR assay using two primer pairs (M1-F/M1-R and M2-F/M2-R) designed based on the differences between AvrLr15 and virulence gene avrLr15 successfully differentiated avirulent and virulent races, which can be used as the AvrLr15 marker. A total of 21 races with different virulence to Lr15 were tested to confirm the reliability of the AvrLr15 molecular marker. A total of 168 Pt isolates were collected from 15 provinces in 2024, and the AvrLr15 molecular marker can monitor the distribution of the V15 (virulent to the Lr15 gene) race and assess the durability of Lr15-mediated resistance effectively. Our findings developed the first Pt avirulence gene molecular marker to monitor natural Pt populations and guide the deployment of Lr15-resistant wheat cultivars in the field.
1 Introduction
Wheat leaf rust, caused by Puccinia triticina (Pt), is one of the most economically significant fungal diseases worldwide, leading to substantial yield losses in major wheat-producing regions (Eversmeyer and Kramer, 2000). The widespread cultivation of single resistant varieties exerts selective pressure on Pt populations, resulting in the emergence of new virulent races and shifts in dominant pathotypes. This dynamic leads to the breakdown of host resistance and subsequent disease epidemics, causing severe production losses (Aime et al., 2018; Ellis et al., 2014).
To mitigate the rapid erosion of resistance in Pt-resistant cultivars, continuous monitoring of Pt virulence in field populations is essential. To date, over 100 leaf rust resistance (Lr) genes have been identified in wheat and its wild relatives, with 85 formally designated (Sharma et al., 2024). The Lr15 gene, derived from common wheat (Triticum aestivum) cultivar Kenya and located on chromosome 2D, confers resistance to Pt races in Canada, Europe, and Central Asia. However, its effectiveness has declined in Germany, India, and South Africa (Huerta-Espino et al., 2011). Statistical analysis of the Pt physiological races in China’s major wheat-producing regions from 2007 to 2021 revealed that virulence frequency to Lr15 presented a general trend of fluctuation and decline (Li et al., 2024; Zhang et al., 2020). In addition, Lr15 is present in only a few commercial cultivars, indicating its limited deployment in field production. This low utilization indicates that Lr15 has application potential (Gao et al., 2019) in the future. Thus, developing effective management strategies to preserve Lr15 effectiveness and prevent large-scale epidemics is critical.
Gene-for-gene theory posits that effective resistance occurs only when a plant carries a functional R gene and the corresponding pathogen carries an Avr gene. In contrast, loss-of-function variations in Avr genes (deletions, point mutations) or Avr gene absence disrupts this recognition, allowing the pathogen to evade R-mediated resistance (Flor, 1971; Ellis et al., 2014). Molecular markers of the Avr gene are essential tools for detecting polymorphisms in Avr genes during pathogen surveillance (Panthee, 2023), as they enable rapid detection of pathogen avirulent/virulent races and support dynamic monitoring of field virulence. Single-nucleotide polymorphism (SNP) markers offer particular advantages due to their high stability, rapid detection, and suitability for high-throughput analysis (Terracciano et al., 2013; Al-Samarai and Al-Kazaz, 2015).
In our previous study, we identified the Pt effector Pt_19 as the avirulence gene AvrLr15 and characterized four nucleotide mutations in its coding sequence that differentiate the avirulent (AvrLr15) and virulent (avrLr15) alleles (Cui et al., 2024). Here, we further validated the function of AvrLr15 by generating TcLr15–Thatcher hybrids and EMS-induced TcLr15 mutant lines and developed an AvrLr15-specific molecular marker and applied it to systematically monitor the distribution of the V15 (virulent to the Lr15 gene) races across China’s major wheat-growing regions. Our findings support strategic deployment of Lr15-based wheat cultivars to improve durable resistance against Pt epidemics.
2 Materials and methods
2.1 Plant materials, Pt isolates, and sample collection
Wheat near-isogenic line TcLr15 (carrying the Lr15 gene in the Thatcher background) and its susceptible parent Thatcher, along with 21 Pt isolates with different virulence to TcLr15 from different years, were maintained at the Wheat Leaf Rust Laboratory of Hebei Agricultural University (Supplementary Table S1). A total of 168 Pt single-pustule isolates were generated in the greenhouse from field-collected leaves in 2024 (Supplementary Table S2). Each isolate was systematically numbered according to its geographic origin.
2.2 Generation and characterization of TcLr15–Thatcher hybrid materials
TcLr15 and Thatcher were hybridized to obtain the F1 generation (Li et al., 1992), which was then selfed to obtain the F2 generation of TcLr15–Thatcher hybrid material. The Lr15 molecular marker was used to detect Lr15 in TcLr15–Thatcher hybrid materials (F2 generation). TcLr15, Thatcher, and the F2 generation of TcLr15–Thatcher hybrid material were infiltrated with pure protein of AvrLr15, respectively, and the programmed cell death (PCD) on the infiltrated region was observed at 24 h. The test was repeated three to five times.
2.3 EMS mutagenesis and mutant screening
To obtain wheat materials with Lr15 gene mutation, mutagenesis was performed as previously described (Wang et al., 2023). In brief, 200 TcLr15 seeds were soaked in 100 mL of 0.4% EMS, mixed by 125 r min−1 shaking at 25 °C for 10 h, and washed with running water at 26°C for 4 h. Treated seeds were planted in a greenhouse maintained at 25 °C for 15 days to observe seedling growth and development. Putative mutants were screened using Lr15-specific molecular markers and avirulent Pt race PHNT inoculation.
2.4 Design and validation of the avirulence gene AvrLr15 molecular marker
Based on the four nucleotide variation sites previously identified between the avrLr15 and AvrLr15 sequences, a total of 30 primer pairs were designed targeting the CDS region to serve as molecular markers for AvrLr15 (Supplementary Table S3). The primers were designed using Primer 5.0 and subsequently synthesized by Sangon Biotech (Shanghai) Co., Ltd. Genomic DNA was extracted from urediniospores of 21 Pt races that exhibited different infection types on TcLr15, using a modified CTAB method based on the protocol by Doyle (Doyle and Doyle, 1987). PCR amplification was performed using specific primers in a total reaction volume of 25 μL, containing 12.5 μL of 1× Taq Master Mix, 1 μL each of 5 mmol/L forward and reverse primers, and 1 μL of 30-ng DNA template, with the remaining volume supplemented with ddH2O. The cycling conditions are provided in Supplementary Table S4 (the reaction system and program for the β-Actin internal reference were identical). The presence of an amplification product of the expected size was indicative of an avirulent isolate, whereas its absence indicated a virulent isolate.
2.5 Application of the avirulence gene AvrLr15 molecular marker
In 2024, a total of 168 Pt isolates were collected from 15 provinces across China, with Hebei and Henan provinces exhibiting relatively higher sampling densities compared with other regions. Samples were collected from three sites per province where natural infections had occurred and no fungicides had been applied. At each site, three biological replications were conducted within 10 m × 10 m experimental plots using the five-point sampling method for isolate collection (Saghai-Maroof et al., 1984). Following collection, Pt isolates were inoculated onto TcLr15 and Thatcher wheat cultivars, with three biological replicates per isolate, for virulence phenotyping under controlled environmental conditions (22 ± 2 °C, 70% relative humidity, 150–250 μmol·m−2·s−1 light intensity, and 12 h light/12 h dark). Genomic DNA was extracted from each isolate, and virulence was confirmed through PCR amplification using specific molecular markers (M1 and M2) (Supplementary Table S3). Statistical analysis was performed on the spatial distribution of V15 virulence frequency across different geographical regions. Chi-square tests were applied to the virulence frequency results, wherein the likelihood ratio method was adopted when the sample size (N) exceeded 40 and the minimum expected count fell between 1 and 5.
3 Results
3.1 The specific recognition between AvrLr15 and Lr15 was confirmed in hybrid plants
To provide the genetic proofs for the specific interaction between the avirulence gene AvrLr15 and the resistance gene Lr15, F2 progeny derived from the TcLr15–Thatcher hybrid were genotyped using an Lr15-specific molecular marker. Among 23 F2 hybrid plants, a fragment of 1,000-bp length was detected in 14 plants, which was identical to the TcLr15 as positive control, whereas nine plants exhibited a 500-bp fragment, matching Thatcher as negative control (Supplementary Figure S1), confirming the presence of Lr15 in 14 F2 hybrid plants. Chi-square test confirmed that the 14:9 segregation ratio matches the expected 3:1 mendelian ratio (χ2 = 2.43, df=1, P>0.05), supporting Lr15 as a single dominant gene.
To verify whether AvrLr15 could trigger an immune response in the TcLr15–Thatcher hybrid wheat, we infiltrated purified AvrLr15 protein into the F2 hybrid materials. Nine Lr15-negative hybrid plants did not display PCD response, consistent with the negative control (Thatcher infiltrated with AvrLr15). In contrast, 14 Lr15-positive hybrid plants developed PCD response upon AvrLr15 infiltration, identical to the positive control (TcLr15 infiltrated with AvrLr15) (Figure 1A). These results demonstrate that AvrLr15 can specifically induce an immune response in Lr15-containing TcLr15–Thatcher hybrid materials, providing genetic evidence for the specific recognition between AvrLr15 and Lr15.
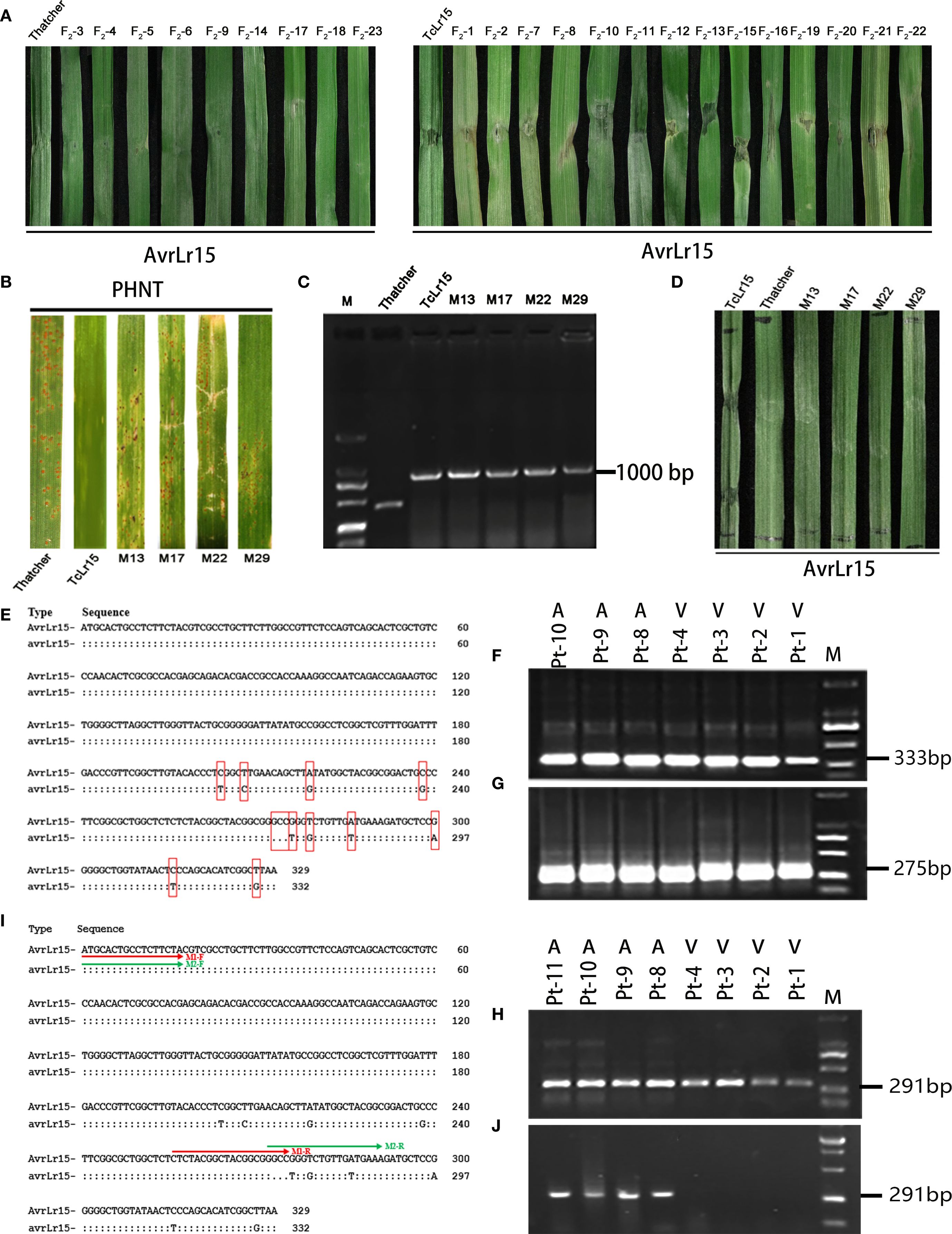
Figure 1. Validation of the avirulence gene AvrLr15 in Pt and molecular marker development. (A) Transient expression analysis of AvrLr15 in the hybrid wheat of TcLr15 and Thatcher, F2-1-F2-23: second hybrid generation of TcLr15–Thatcher. (B) Inoculation of TcLr15 mutants with the avirulent strain PHNT (against Lr15). (C) Detection of TcLr15 mutants using molecular markers for the Lr15 gene. (D) Transient expression analysis of AvrLr15 in TcLr15 mutants. (E) Schematic diagram of nucleotide sequence polymorphisms between the virulence (avrLr15) and avirulence (AvrLr15) alleles. (F) PCR amplification using primer M17 (333 bp) on four virulent and three avirulent races. (G) PCR amplification using primer M1 (275 bp) on four virulent and three avirulent races. (H) PCR amplification using primer M2 (291 bp) on four virulent and four avirulent races. (I) Primer positions are indicated by colored lines: M1 (red) and M2 (green). (J) Dual PCR amplification using primers M1 and M2, followed by electrophoresis, on four virulent and four avirulent races.
3.2 AvrLr15 could not trigger the immune response in the Lr15 mutants
Based on statistical analysis of germination rate and malformation frequency in TcLr15 wheat seeds treated with varying EMS concentrations (Supplementary Table S5), 0.4% EMS was identified as the optimal concentration for subsequent mutagenesis treatments. Phenotype assessments showed that four M0 plants (M13, M17, M22, and M29) displayed an intermediate susceptible phenotype (“3” infection type), whereas TcLr15 exhibited an immune response (“0” infection type), and Thatcher was highly susceptible (“4” infection type) (Figure 1B). Further PCR amplification using an Lr15-specific molecular marker confirmed that four M0 mutants retained the Lr15 gene, as evidenced by a 1,000-bp amplification product (Figure 1C). These results indicate that the Lr15 gene harbored mutations in the four mutant lines, M13, M17, M22, and M29.
To determine whether mutations in Lr15 affect its recognition with AvrLr15, purified AvrLr15 protein was infiltrated into the TcLr15 mutants. As expected, TcLr15 plants as positive control developed a PCD response, whereas four mutants (M13, M17, M22, and M29) did not, similar to the susceptible Thatcher as negative control (Figure 1D). These results demonstrate that mutations in Lr15 disrupt its recognition with AvrLr15.
3.3 Development of a molecular marker for avirulence gene AvrLr15
To develop molecular markers for AvrLr15, allele-specific primers were designed based on the 11 nucleotide variations identified between the AvrLr15 and avrLr15 alleles, including 10 single-nucleotide polymorphisms (SNPs: 204C→T, 208T→C, 219A→G, 238C→G, 276G→T, 278T→G, 286A→T, 300G→A, 316C→T, and 326T→G) and one trinucleotide deletion (273GCC→-) (Figure 1E). A total of 30 primer pairs were synthesized and initially screened using genomic DNA from seven to eight Pt races with defined virulence profiles. However, similar amplification products were observed between avirulence and virulence races to Lr15 (Figures 1F-H), suggesting that single SNP primer pairs could not distinguish the polymorphism between avirulence and virulence races to Lr15.
Subsequently, duplex PCR assays were performed using upstream primers (M1-F and M2-F) designed from the first 21 nucleotides of AvrLr15/avrLr15, coupled with deletion-flanking reverse primers: M1-R (3'-terminus anchored to the GCC deletion site) and M2-R (5'-terminus initiated at the deletion site) (Figure 1I). A 291-bp fragment exclusively was obtained in avirulent races to Lr15, whereas no amplification product appeared in the virulent races to Lr15 (Figure 1J). These results demonstrated the potential of M1-F/M1-R and M2-F/M2-R primer pairs for discriminating avirulent and virulent races to Lr15.
3.4 Validation of the AvrLr15-specific molecular marker
To elucidate sequence variations in AvrLr15 among different virulent races of Pt, we systematically assessed virulence profiles against Lr15. Physiological races Pt-1, Pt-2, Pt-3, Pt-4, Pt-5, Pt-6, and Pt-7 exhibited virulence toward TcLr15, developing abundant urediniospores with limited necrotic lesions—resembling disease susceptibility in Thatcher as controls. Conversely, races Pt-8, Pt-9, Pt-10, Pt-11, Pt-12, Pt-13, Pt-14, Pt-15, Pt-16, Pt-17, Pt-18, Pt-19, Pt-20, and Pt-21 showed avirulence to Lr15, inducing extensive necrotic responses without urediniospore formation on TcLr15 (Figures 2A, B; Supplementary Table S1). Collectively, 7 virulent races (Pt-1, Pt-2, Pt-3, Pt-4, Pt-5, Pt-6, Pt-7) and 14 avirulent races (Pt-8, Pt-9, Pt-10, Pt-11, Pt-12, Pt-13, Pt-14, Pt-15, Pt-16, Pt-17, Pt-18, Pt-19, Pt-20, Pt-21) were identified.
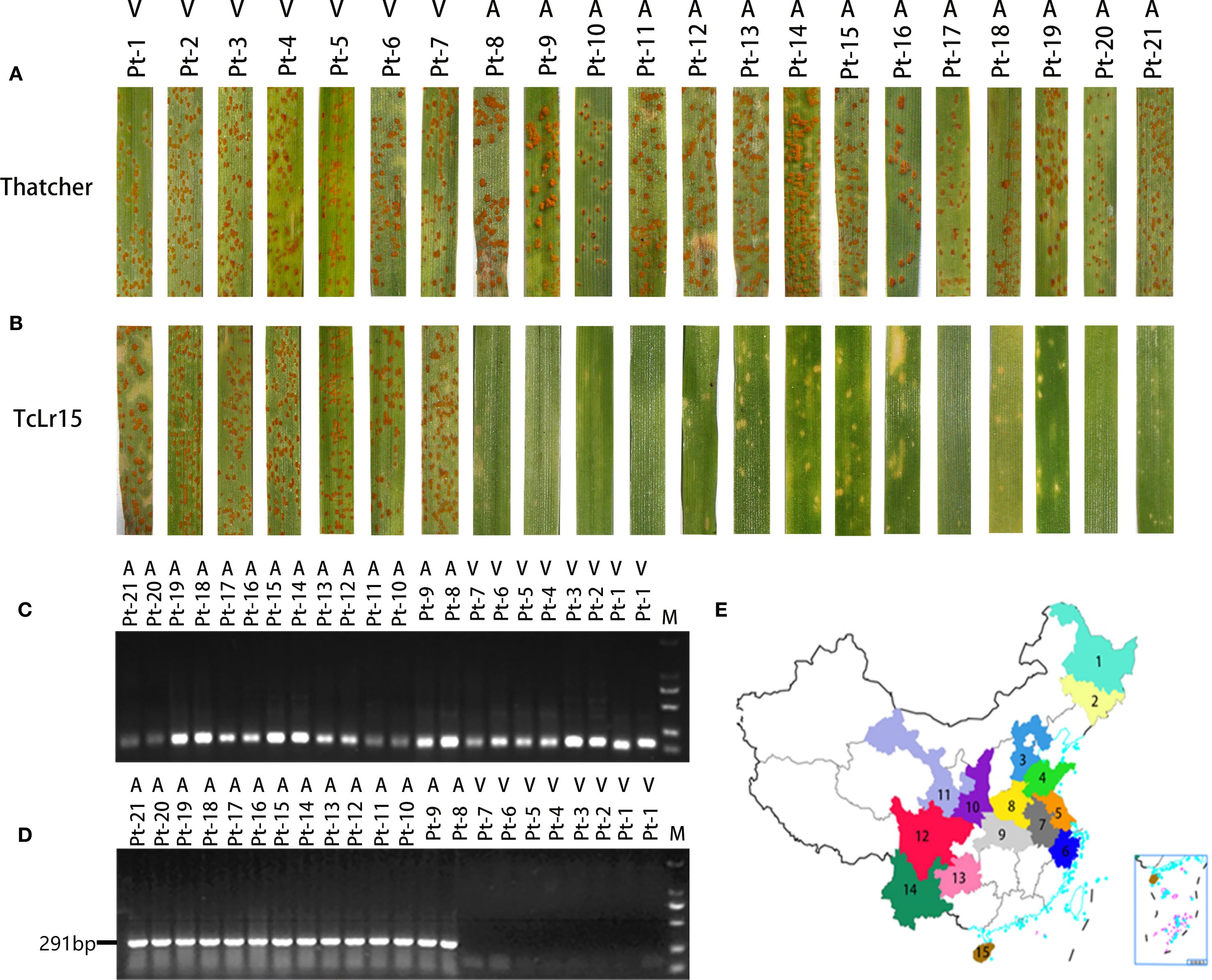
Figure 2. Validation and application of molecular markers for the wheat avirulence gene AvrLr15. (A) Phenotypic identification of 21 Pt physiological races inoculated on the susceptible material Thatcher. (B) Phenotypic identification of 21 Pt physiological races inoculated on TcLr15 wheat. (C) PCR detection of the β-actin reference gene in 21 Pt physiological races. (D) Dual PCR amplification using primers M1 and M2, followed by electrophoresis, on DNA from 21 Pt physiological races. (E) Sampling regions for 2024 Pt isolates. Colored areas indicate collection sites: 1: HLJ (Heilongjiang); 2: JL (Jilin); 3: HEB (Hebei); 4: SD (Shandong); 5: JS (Jiangsu); 6: ZJ (Zhejiang); 7: AH (Anhui); 8: HEN (Henan); 9: HUB (Hubei); 10: SX (Shanxi); 11: GS (Gansu); 12: SC (Sichuan); 13: GZ (Guizhou); 14: YN (Yunnan); 15: HAN (Hainan).
To further validate the reliability of the AvrLr15 molecular marker in distinguishing virulent and avirulent races, PCR amplification was conducted using genomic DNA from 21 characterized Pt races. Initial quality control was performed through β-actin gene amplification, confirming the integrity of all DNA templates (Figure 2C). Subsequent electrophoresis analysis revealed distinct patterns, a 291-bp amplification band appeared in 14 avirulent races, whereas 7 virulent races exhibited no amplification (Figure 2D). These results demonstrate that the AvrLr15 molecular marker can accurately differentiate Pt races with different virulence to Lr15.
3.5 Application of the AvrLr15 molecular marker
To monitor the distribution of the V15 race and assess the durability of Lr15-mediated resistance, 168 Pt isolates were collected from 15 provinces in 2024, predominantly in the Huang-Huai-Hai wheat region (Figure 2E; Supplementary Table S2). Phenotypic assessment after inoculation on TcLr15 classified 120 isolates as avirulent (immune response) and 48 isolates as virulent (highly susceptible). DNA quality verification followed by AvrLr15-specific marker testing confirmed that the 291-bp band was tested in 120 avirulent isolates, whereas the 291-bp band was absent in 48 virulent isolates (Supplementary Figure S2A, B). Lr15 virulent isolates were distributed as follows: Hebei (14), Henan (9), Shandong (6), Hubei (5), Zhejiang (5), Sichuan (4), Gansu (3), and Shanxi (2).
Statistical analysis of virulence frequency revealed that the V15 virulent strain was distributed at relatively high frequencies in Hubei (45.45%), Hebei (43.75%), Henan (39.13%), and Zhejiang (38.46%), whereas lower frequencies were observed in Gansu (37.50%) Shandong (35.29%), Shanxi (28.57%), and Sichuan (26.67%). The V15 isolates were not detected in any sampled isolates from Yunan, Jiangsu, Anhui, Heilongjiang, Hainan, Liaoning, and Guizhou. The chi-square test revealed significant differences in the V15 virulence frequency among regions (χ2 = 19.234, P = 0.007<0.05). Specifically, the Huang-Huai area exhibits significant differences in the V15 virulence frequency compared with the southwestern/northeastern provinces (Supplementary Table S6), supporting that V15 virulent strains show significant geographical different in China’s wheat regions.
4 Discussion
Plant resistance is generally conferred by immune receptors of the nucleotide-binding leucine-rich repeat (NLR) class, which recognize pathogen effector proteins delivered into the host cell during infection, often known as avirulence proteins. Phytophthora infestans avirulence genes Avramr1 was identified and verified by transiently co-expressed with potato late blight resistance gene Rpi-amr1 in Nicotiana benthamiana (Lin et al., 2020). Lin et al. (2022) identified P. infestans avirulence gene Avramr3 that is recognized by potato late blight Rpi-amr3. Wheat powdery mildew resistance gene Pm1a as an immune receptor gene specifically recognizes Blumeria graminis avirulence gene AvrPm1a (Hewitt et al., 2021). Müller et al. (2022) identified Blumeria graminis avirulence gene AvrPm17 by transient co-expression in Nicotiana benthamiana. However, the genome of Pt is relatively complex and dikaryotic, which has resulted in significantly lagging research on its avirulence genes in this field. Our previous study identified Pt avirulence genes AvrLr21 and AvrLr15 (Shen et al., 2024; Cui et al., 2024), finding that Lr21-breaking Pt isolates can suppress Lr21-mediated immunity. To provide strong genetic evidence for the specific recognition between AvrLr15 and Lr15, the hybrid plants of Thatcher and TcLr15 and Lr15 EMS mutants were constructed in this study, and the function of AvrLr15 as an avirulence gene was further verified.
The development of molecular markers for avirulence genes enables precise detection of gene variations, offering molecular evidence for understanding how pathogens evade host resistance recognition through genetic mutations (Dong et al., 2009). Tong et al. (2023) developed Sw-5b-specific markers to distinguish between Sw-5bR and Sw-5bS alleles, which enable precise selection and accelerating tomato spotted wilt virus (TSWV)-resistant tomato breeding to mitigate yield losses. Phytophthora sojae avirulence gene Avr3a exhibits two variation patterns, namely, coding sequence mutations and promoter-mediated transcriptional loss, facilitating the creation of two SNP markers and one PCR marker for virulence identification (Hu et al., 2021). Dussault-Benoit et al. (2020) developed a molecular assay method to define the pathotypes of Phytophthora sojae, based on seven Avr genes (Avr1a, Avr1b, Avr1c, Avr1d, Avr1k, Avr3a, and Avr6). The matching rate between the molecular assay method and the phenotyping assay was as high as 97%. Our previous study demonstrated that Lr15-breaking Pt isolates contained nucleotide deletions that led to one amino acid deletion located at amino acid residue P92 and multiple nucleotide changes that resulted in three amino acid substitutions at residues P80, M96, and P106, which provides a basis for the development of the AvrLr15 molecular marker. We tried to distinguish the avirulence and virulence races to Lr15 by 30 SNP-based primers, but no polymorphism was detected between the avirulence and virulence races to Lr15 (Figures 1F, G). SNP-based primer designs rely on single-base variation. However, such variation can be easily masked by non-specific primer binding during PCR (Zhang et al., 2023). This limitation was obvious in our study: All 30 SNP primers failed to differentiate AvrLr15 and avrLr15 alleles, resulting in cross-amplification in both virulent and avirulent Pt races. A duplex PCR assay enables the effective differentiate of the alleles in a single PCR reaction (Ma et al., 2003). In this study, the molecular marker for AvrLr15 was developed by duplex PCR assays and used to monitor the distribution of the V15 race. The virulence frequency of V15 was revealed in 168 Pt isolates collected from 15 provinces in China in 2024. The matching rate between the molecular assay and the phenotyping assay was as high as 100%. All these findings benefit the assess of the durability of Lr15-mediated resistance.
The AvrLr15 molecular marker was used to monitor the distribution of the V15 race collected from 15 provinces in 2024; ongoing monitoring is needed to track temporal shifts. All in all, the molecular marker of AvrLr15 as the first molecular marker for an avirulence gene of Pt was developed successfully, and it can rapidly detect Pt isolates with different virulence to Lr15. Our findings will provide a molecular tool for monitoring virulence in natural Pt populations and guiding the field deployment of Lr15-resistant wheat cultivars and contribute to maintaining the durable resistance endowed by wheat leaf rust resistance gene Lr15.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YJ: Validation, Methodology, Data curation, Investigation, Writing – review & editing, Conceptualization, Writing – original draft. ZC: Methodology, Investigation, Validation, Writing – original draft, Writing – review & editing, Data curation. MS: Validation, Methodology, Investigation, Data curation, Writing – review & editing. SHY: Methodology, Writing – review & editing, Investigation. DD: Writing – review & editing, Methodology, Investigation, Data curation. YL: Investigation, Methodology, Writing – review & editing. SY: Methodology, Writing – review & editing, Investigation. PX: Investigation, Writing – review & editing. CY: Investigation, Writing – review & editing. DL: Writing – review & editing, Supervision. ZW: Investigation, Writing – review & editing, Methodology. HW: Resources, Writing – review & editing, Project administration, Supervision, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by Hebei Province Postgraduate Innovation Ability Training Funding Project (CXZZBS2024077), the National Natural Science Foundation of China (32172384 and 31501623) and Local Science, the Technology Development Fund Projects of Hebei Province Guided by the Central Government (246Z6504G), and the Hebei Province Dry Alkali Wheat Industry Technology System (HBCT2024030206).
Acknowledgments
We thank the Wheat Leaf Rust Laboratory of Hebei Agricultural University. We also thank the Wheat Research Institute of Tangshan Academy of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1668725/full#supplementary-material
References
Aime, M. C., Bell, C. D., and Wilson, A. W. (2018). Deconstructing the evolutionary complexity between rust fungi (Pucciniales) and their plant hosts. Stud. Mycol. 89, 143–152. doi: 10.1016/j.simyco.2018.02.002
Al-Samarai, F. R. and Al-Kazaz, A. A. (2015). Molecular markers: an introduction and applications. Eur. J. Mol. Biotechnol. 19, 118–130. doi: 10.13187/ejmb.2015.9.118CrossRef
Cui, Z., Shen, S., Meng, L., Sun, X., Jin, Y., Liu, Y., et al. (2024). Evasion of wheat resistance gene Lr15 recognition by the leaf rust fungus is attributed to the coincidence of natural mutations and deletion in AvrLr15 gene. Mol. Plant Pathol. 25, e13490. doi: 10.1111/mpp.13490
Dong, S., Qutob, D., Tedman-Jones, J., Kuflu, K., Wang, Y., Tyler, B. M., et al. (2009). The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. Public Library Sci. One 4, e5556. doi: 10.1371/journal.pone.0005556
Doyle, J. J. and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Dussault-Benoit, C., Arsenault-Labrecque, G., Sonah, H., Belzile, F., and Bélanger, R. R. (2020). Discriminant haplotypes of avirulence genes of Phytophthora sojae lead to a molecular assay to predict phenotypes. Mol. Plant Pathol. 21, 318–329. doi: 10.1111/mpp.12898
Ellis, J. G., Lagudah, E. S., Spielmeyer, W., and Dodds, P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00641
Eversmeyer, M. G. and Kramer, C. L. (2000). Epidemiology of wheat leaf and stem rust in the central great plains of the USA. Annu. Rev. Phytopathol. 38, 491–513. doi: 10.1146/annurev.phyto.38.1.491
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Gao, P., Zhou, Y., Gebrewahid, T., Zhang, P., Yan, X., Li, X., et al. (2019). Identification of known leaf rust resistance genes in common wheat cultivars from Sichuan province in China. Crop Protect. 115, 122–129. doi: 10.1016/j.cropro.2018.09.012
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
Hu, Y., He, Z., Kang, Y., and Cui, L. (2021). Mutations in the promoter and coding regions of Avr3a cause gain of virulence of Phytophthora sojae to Rps3a in soybean. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.759196
Huerta-Espino, J., Singh, R. P., Germán, S., Mccallum, B. D., Park, R. F., Chen, W., et al. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica: Int. J. Plant Breeding 179, 143–160. doi: 10.1007/s10681-011-0361-x
Li, Y., Ding, S., Jia, J., and Zhang, H. (1992). A new technique of making crosses by clipping anther and bagging in wheat. Acta Agronom. Sinica 5, 387–390.
Li, H., Huang, L., Zhang, H., Liu, B., Gao, L., Chen, W., et al. (2024). Race and virulence dynamics of Puccinia triticina in China during 2007 to 2021. Plant Dis. 108, 256–263. doi: 10.1094/PDIS-04-23-0727-SR
Lin, X., Olave-Achury, A., Heal, R., Pais, M., Witek, K., Ahn, H. K., et al. (2022). A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol. Plant 15, 1457–1469. doi: 10.1016/j.molp.2022.07.012
Lin, X., Song, T., Fairhead, S., Witek, K., Jouet, A., Jupe, F., et al. (2020). Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen-enrichment sequencing (PenSeq). Mol. Plant Pathol. 11, 1502–1512. doi: 10.1111/mpp.12987
Ma, W., Zhang, W., and Gale, K. R. (2003). Multiplex-PCR typing of high molecular weight glutenin alleles in wheat. Euphytica 134, 51–60. doi: 10.1023/A:1026191918704
Müller, M. C., Kunz, L., Schudel, S., Lawson, A. W., Kammerecker, S., Isaksson, J., et al. (2022). Ancient variation of the AvrPm17 gene in powdery mildew limits the effectiveness of the introgressed rye Pm17 resistance gene in wheat. Proc. Natl. Acad. Sci. United States America 119, e2108808119. doi: 10.1073/pnas.2108808119
Panthee, D. R. (2023). Application of molecular markers in crop improvement and beyond. Agronomy 13, 2041. doi: 10.3390/agronomy13082041
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. United States America 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Sharma, D., Avni, R., Gutierrez-Gonzalez, J., Kumar, R., Sela, H., Prusty, M. R., et al. (2024). A single NLR gene confers resistance to leaf and stripe rust in wheat. Nat. Commun. 15, 9925. doi: 10.1038/s41467-024-54068-6
Shen, S., Wang, F., Cui, Z., Yuan, S., Meng, L., Liu, D., et al. (2024). Puccinia triticina avirulence protein AvrLr21 directly interacts with wheat resistance protein Lr21 to activate wheat immune response. Commun. Biol. 7, 1170. doi: 10.1038/s42003-024-06881-4
Terracciano, I., Maccaferri, M., Bassi, F., Mantovani, P., Sanguineti, M. C., Salvi, S., et al. (2013). Development of COS-SNP and HRM markers for high-throughput and reliable haplotype- based detection of Lr14a in durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 126, 1077–1101. doi: 10.1007/s00122-012-2038-9
Tong, C., Huang, S., Shi, Y., Wu, Q., Shangguan, L., Yu, H., et al. (2023). Development of novel specific molecular markers for the Sw-5b gene to assist with tomato spotted wilt virus-resistant tomato breeding. Phytopathol. Res. 5, 59. doi: 10.1186/s42483-023-00214-9
Wang, D., Li, Y., Wang, H., Xu, Y., Yang, Y., Zhou, Y., et al. (2023). Boosting wheat functional genomics via an indexed EMS mutant library of KN9204. Plant Commun. 4, 100593. doi: 10.1016/j.xplc.2023.100593
Zhang, L., Xiao, Y., Gao, Y., Zhao, N., An, Y., Yang, W., et al. (2020). Race and virulence analysis of Puccinia triticina in China during 2011 to 2013. Plant Dis. 104, 2095–2101. doi: 10.1094/PDIS-01-20-0047-RE
Zhang, J., Zhu, Y., Wheeler, T., and Dever, J. K. (2023). Development and validation of allele-specific PCR-based SNP typing in a gene on chromosome D03 conferring resistance to Fusarium wilt race 4 in upland cotton (Gossypium hirsutum). Mol. Genet. Genomics 298, 1579–1589. doi: 10.1007/s00438-023-02079-1
Keywords: Puccinia triticina, avirulence gene, molecular marker, virulence monitoring, AvrLr15
Citation: Jin Y, Cui Z, Song M, Yuan S, Ding D, Liu Y, Yin S, Xu P, Yan C, Liu D, Wu Z and Wang H (2025) Development and application of a Puccinia triticina avirulence gene AvrLr15-specific molecular marker. Front. Plant Sci. 16:1668725. doi: 10.3389/fpls.2025.1668725
Received: 22 July 2025; Accepted: 26 September 2025;
Published: 08 October 2025.
Edited by:
Bo Yang, Nanjing Agricultural University, ChinaReviewed by:
Anupam Singh, Shree Guru Gobind Singh Tricentenary University, IndiaVenancio Riella, Universidad de la República (UdelaR), Uruguay
Copyright © 2025 Jin, Cui, Song, Yuan, Ding, Liu, Yin, Xu, Yan, Liu, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Wu, d3poNDA2QHNpbmEuY24=; Haiyan Wang, bmR3YW5naGFpeWFuQDE2My5jb20=
†These authors have contributed equally to this work
 Yuqing Jin
Yuqing Jin Zhongchi Cui
Zhongchi Cui Maoxing Song2
Maoxing Song2 Haiyan Wang
Haiyan Wang