- National Engineering Research Center for Sugarcane, Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs, College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Introduction: Sugarcane mosaic virus (SCMV, Potyvirus) causes mosaic diseases and seriously threatens sugarcane production. Potyviral 6K2 protein plays a key role in viral infections. We previously screened a tetraspanin (TET)-like protein that interacts with SCMV-6K2 from a sugarcane cDNA yeast library. Although TETs have been extensively studied in response to viral infections in animals, the TET gene family in sugarcane and its role in SCMV infections remain largely unknown. This study aimed to identify the TET genes in sugarcane and determine their response to SCMV infection.
Methods: We employed genome-wide identification, phylogenetic analysis, real-time quantitative PCR (RT-qPCR), subcellular localization, and multiple protein–protein interaction assays to characterize TETs and their interactions with viral 6K2 proteins.
Results: We identified 35, 113, 73, and 17 TETs in the genomes of Saccharum spontaneum, sugarcane cultivar R570, sugarcane cultivar Xintaitang 22 (XTT22), and Nicotiana benthamiana, respectively. Phylogenetic tree analysis classified the TETs into nine distinct groups. Nine TET genes were cloned from XTT22 and designated ScTET2, ScTET8, ScTET13, ScTET23, ScTET34, ScTET55, ScTET67, ScTET78, and ScTET96. RT-qPCR demonstrated the differential expression of these genes following SCMV infection. Furthermore, subcellular localization assays revealed that they were mainly localized to the plasma membrane (PM), except for ScTET2 and ScTET8, which were localized in the cytoplasm and formed irregular spherical structures of different sizes. Yeast two-hybrid (Y2H), bimolecular fluorescent complementation, and luciferase complementation assays revealed extensive interactions between the ScTETs and SCMV-6K2, primarily in the PM. Y2H assays also showed that TETs of Arabidopsis and N. benthamiana extensively interacted with the 6K2 protein of turnip mosaic virus.
Discussion: This study reveals a potential mechanism by which potyviruses employ 6K2 to interact with TETs to establish infection in host plants, thus highlighting potential molecular targets for engineering sugarcane resistance against SCMV.
1 Introduction
Potyviruses represent the largest group of plant viruses and cause heavy yield losses in many crops worldwide (Urcuqui-Inchima et al., 2001; Revers and García, 2015; Yang et al., 2024). Potyviruses are flexuous rod-shaped particles at 680–900 nm long and 11–20 nm wide, and they contain approximately 10 kb of single-stranded positive-sense RNA that encodes two polyproteins (Urcuqui-Inchima et al., 2001; Yang et al., 2021; Pollari et al., 2024). These two polyproteins hydrolyze into 11 mature proteins: P1, HC-Pro, P3, P3N-PIPO, 6K1, CI, 6K2, Vpg, NIa-Pro, NIb, and CP (Valli et al., 2007; Chung et al., 2008; Cheng et al., 2017, 2020; Xiao et al., 2022; Hýsková et al., 2024). Among these, the 6K2 single-transmembrane protein localized in the endoplasmic reticulum (ER) participates in multiple biological processes during potyvirus infection (Xue et al., 2023; Zhang et al., 2024a; 2024). The 6K2 protein can induce the rearrangement of ER at ER exit sites (ERESs) to form virus replication complexes (Grangeon et al., 2013; Jiang et al., 2015; Cabanillas et al., 2018; Zhang et al., 2019; Xie et al., 2021; Solovyev et al., 2022) and fuse with other endomembrane systems, such as the outer membrane of chloroplasts, to facilitate efficient replication. Notably, it plays a role in the intra- or intercellular movement as well as long-distance transport of the virus (Wan et al., 2015; Movahed et al., 2019; Chai et al., 2020; He et al., 2023b) and immune responses, including reactive oxygen species (ROS) burst and autophagy (Wang, 2015; Hafrén et al., 2018; Li et al., 2020; Lin et al., 2024; Zhang et al., 2024; He et al., 2025; Rui et al., 2025). Interestingly, overexpressing of turnip mosaic virus (TuMV) 6K2 in Arabidopsis and Nicotiana benthamiana promotes salicylic acid accumulation and resistance to drought stress (Prakash et al., 2023). Given the significant role of 6K2 in potyvirus infection, we previously screened a sugarcane cDNA yeast library using the 6K2 protein of SCMV as bait and identified the tetraspanin (TET)-like protein ScTSPAN18 (Zhang H. et al., 2019).
TETs are evolutionarily conserved integral membrane proteins in cellular organisms (Hemler, 2003; Wang et al., 2012a; Green et al., 2019; Zhang et al., 2025a). TETs are composed of four transmembrane domains, one small extracellular loop, and one highly variable large extracellular loop (LEL) (Boucheix and Rubinstein, 2001; Hemler, 2003; Kovalenko et al., 2005; Reimann et al., 2017). In animals, LEL contains a conserved CCG motif, whereas in plants, it contains a GCCK/RP motif (Seigneuret et al., 2001; Huang et al., 2005; Wang et al., 2012a; Boavida et al., 2013). TETs can interact with themselves, other TETs, or other ligand proteins on the plasma membrane (PM) to form TET-enriched microdomains (TEMs), which are involved in signaling, cell adhesion, migration, proliferation, differentiation, fundamental immune response, and PM repair (Le Naour et al., 2000; Miyado et al., 2000; Boucheix and Rubinstein, 2001; Hemler, 2001; Charrin et al., 2009; Umeda et al., 2020; Huang et al., 2022; Zhang et al., 2025b). In mammals, TETs such as CD9, CD81, CD63, CD82, and CD151 are extensively involved in viral infections (Fast et al., 2017; Florin and Lang, 2018; Zhang et al., 2025b). CD9, CD63, and CD81 are components of extracellular vesicles (EVs) and respond to infection with the human immunodeficiency virus, Lujo virus, hepatitis B virus, or herpes simplex virus 1 (Florin and Lang, 2018; Ghossoub et al., 2020; Mathieu et al., 2021; Ninomiya et al., 2021; Poveda et al., 2022; Zhang et al., 2025b).
Plant TETs are involved in plant development and growth, including cell fate determination, hormonal regulation, plasmodesmata gating, and signaling (Cnops et al., 2006; Wang et al., 2015; Reimann et al., 2017). For instance, TET mutants have abnormal leaves or roots (Cnops et al., 2006; Qin et al., 2024; Zimmerman et al., 2024). Plant TETs are also involved in response to biotic and abiotic stresses and mutualistic interactions (Parra-Aguilar et al., 2023; Chen et al., 2025). In rice (Oryza sativa), OsTET5 regulates drought resistance by controlling ROS burst and ionic homeostasis (Mani et al., 2025), whereas in potato (Solanum tuberosum), StTET8 act as a positive immune regulator that inhibits Phytophthora infestans infection (Guo et al., 2022). Interestingly, PsTET1 and PsTET3 of soybean (Glycine max) root rot pathogen Phytophthora sojae are recognized by N. benthamiana, where they elicit immune responses (Zhu et al., 2023). In Capsicum, expression of the TET8-like gene is strongly correlated with the accumulation of capsicum chlorosis virus (CaCV) (Cnops et al., 2006; Wang et al., 2015; Reimann et al., 2017). In Arabidopsis, AtTET3 plays a key role in the cell-to-cell movement of cucumber mosaic virus (CMV) (Zhu et al., 2022). Plant TETs are also involved in the formation and signaling of EVs (Cui et al., 2020; He et al., 2021; Chen et al., 2022; Ruf et al., 2022; He et al., 2023a; Gao et al., 2024; Chen et al., 2025). AtTET8 and AtTET9 in Arabidopsis mediate the transport of EVs carrying RNA, including host-derived small RNAs (sRNAs), to fungal cells, thereby reducing fungal infection (Cai et al., 2018). Conversely, pathogenic fungi can also use EVs to deliver sRNAs into plant cells (He et al., 2023a). Moreover, TuMV-induced EVs are enriched in AtTET3, suggesting that TETs are involved in potyvirus infections (Movahed et al., 2019).
To date, the TET gene family has only been reported in Arabidopsis and rice (Wang et al., 2012a; Mani et al., 2015). Therefore, this study aimed to identify the TET gene family in sugarcane and determine its response to SCMV infection. This study provides valuable insights for the further exploration of TET functions and highlights the role of these proteins in the response to potyvirus infections.
2 Materials and methods
2.1 Plant materials and treatments
Tissue-cultured Xintaitang 22 (XTT22) plantlets were grown under a 14 h light/10 h dark cycle until reaching 15–25 cm in height with 4–5 fully expanded leaves and were individually inoculated with SCMV as previously described (Zhang H. et al., 2019; Yang et al., 2021). The plants were then inoculated with SCMV. XTT22 plantlets mock-inoculated with 0.01 M phosphate buffer (pH 7.0) served as negative controls. Sampling was conducted at 0 h, 12 h, 1 d, 3 d, 7 d, and 14 d, with three plantlets sampled at each time point. Roots, leaf rolls, +1 leaves, +7 leaves, +3 internodes, and +8 internodes were sampled from nine healthy 10-month-old XTT22 plants, which were divided into three groups of three plants each. N. benthamiana plants were cultured under a 16 h light/8 h dark cycle at 22°C and 60% humidity. All sampled plant materials were immediately frozen in liquid nitrogen and stored at −80°C.
2.2 Identification of putative sugarcane TETs
Genomic data of sugarcane cultivars XTT22 and R570 and Saccharum spontaneum AP85–441 were obtained from the Sugarcane Genome Database (https://sugarcane.gxu.edu.cn/scdb/download) (Zhang et al., 2018; Healey et al., 2024; Zhang et al., 2025c). Genomic data of N. benthamiana were obtained from an online website (http://lifenglab.hzau.edu.cn/Nicomics/Download/index.php) (Wang et al., 2024). Genomic data were obtained from an online website (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012). The Hidden Markov Model of TET (PF00335) was downloaded from an online database (http://pfam.xfam.org/) (Krogh et al., 2001) and used to query the genomes of S. spontaneum AP85-441, sugarcane cultivars XTT22 and R570, and N. benthamiana. The transmembrane domains of these TETs were analyzed on an online website (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). The identified TETs were then verified using the CDD tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
2.3 Phylogenetic tree, physicochemical properties, and subcellular localization of sugarcane TETs
The BLASTp tool was used to screen the Phytozome v13 database (https://phytozome-next.jgi.doe.gov/) for TET proteins of maize (Z. mays), sorghum (Sorghum bicolor), millet (Setaria italica), wheat (Triticum aestivum), soybean (G. max), and potato (S. tuberosum), as well as the identified TETs in rice (Mani et al., 2015) or Arabidopsis (Wang et al., 2012a). The above TET sequences and identified TET sequences in N. benthamiana (NbTETs) or XTT22 (XTT22TETs) were subjected to multiple sequence alignment using MUSCLE v3.7, and a phylogenetic tree was constructed using the maximum likelihood method (bootstrap = 1,000) of MEGA X (Kumar et al., 2018). Then, the online software EvolView (https://evolgenius.info//evolview-v2) (https://evolgenius.info//evolview-v2) (He et al., 2016) was employed to refine the phylogenetic tree. The online website ExPASy (https://web.expasy.org/compute_pi/) was used to predict the physicochemical properties of sugarcane TET proteins. The subcellular localization of the identified TETs was predicted using the online website WoLF PSORT (https://wolfpsort.hgc.jp/) (Horton et al., 2007). Additionally, AlphaFold3 software (https://alphafoldserver.com/) was used to simulate the protein structure.
2.4 Conserved motifs and gene structure analysis
The conserved TET motifs in S. spontaneum AP85-441 (SsTET) and sugarcane cultivars R570 (R570TET) and XTT22 were obtained using MEME (https://web.mit.edu/meme/current/share/doc/overview.html) (Bailey et al., 2009). The parameters were set to search for 10 conserved motifs, with the remaining parameters set to default values. The gene-finding format 3 (gff3) files of the above species were downloaded from the Sugarcane Genome Database (https://sugarcane.gxu.edu.cn/scdb/download) (Zhang et al., 2018; Healey et al., 2024; Zhang et al., 2025c). The conserved motifs and gene structures of SsTETs, R570TETs, and XTT22TETs were visualized using TBtools 2.0.
2.5 Cis-acting elements analysis
The 2,000 bp sequences upstream of the coding sequence (CDS) region of SsTETs, R570TETs, and XTT22TETs were acquired from their corresponding genomic data. The cis-acting elements were predicted using PlantCARE software (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and visualized using TBtools 2.0 (Chen et al., 2023).
2.6 Collinearity analysis
The collinearity and replication patterns of TETs were analyzed using MCScanX software (Wang et al., 2012b; Chen et al., 2023). Collinearity analysis of the genomes of sugarcane cultivars R570 and XTT22, sorghum (S. bicolor), maize (Z. mays), Arabidopsis, potato (S. tuberosum), millet (S. italica), and rice (O. sativa) was performed using TBtools 2.0 software. Genomic information of sorghum, maize, wheat, rice, Arabidopsis, and potato was downloaded from an online website (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012). In addition, the (Ka)/(Ks) value between homologous gene pairs was calculated based on the correlation of homology using TBtools 2.0.
2.7 Transcriptomic data analysis
The transcriptome data of AP85–441 and XTT22 are available in an online repository (https://sugarcane.gxu.edu.cn/scdb/download) (Hu et al., 2018; Zhang et al., 2025c). These data were collected at the seedling stage (35 d), early maturity stage (270 d), and mature stage (360 d). RNA-seq data from the leaves at four different developmental stages were collected to investigate the expression profiles of the TET family. The transcription fragments per million bases (FPKMs) of SsTETs or XTT22TET were used to generate heat maps and conduct cluster analysis using TBtools 2.0 (Chen et al., 2023).
2.8 RNA isolation, cDNA synthesis, and RT-qPCR
Total RNA was extracted from SCMV-infected sugarcane plants and healthy sugarcane plants using the TRIzol method. The PrimeScript® RT-PCR kit (TaKaRa Biotechnology Co., Ltd., Dalian, China) was used to synthesize the first-strand cDNA. Special primers (Supplementary Table S1) were designed to quantify the TET genes by RT-qPCR with eEF-1a and Actin used as internal references (Iskandar et al., 2004; Ling et al., 2014; Xue et al., 2014). The relative expression levels of TET genes were analyzed using the 2−ΔΔCt method. All primers used for RT-qPCR are listed in Supplementary Table S1.
2.9 Plasmid construction
Special primers (Supplementary Table S1) were designed to construct the plasmids. For the yeast two-hybrid (Y2H) experiments, DNA fragments and Y2H vectors were ligated individually at the Ecor I and Sam I sites. The bait vectors of TuMV-6K2 and SCMV-6K2 are from our previous work (Zhang H. et al., 2019; Zhang et al., 2024), and the target genes were cloned into the prey vector pPR3-N. In addition, three TET genes cloned from XTT22 (ScTETs) were inserted into the pBT-STE vector to investigate the interactions among ScTETs. Gateway technology was employed to construct the plasmids for the bimolecular fluorescence complementation (BiFC) assays. The 6K2-YN vector was generated in a previous research study (Zhang et al., 2024). For the subcellular localization experiments, all DNA fragments were inserted into the vectors via the Kpn I and Sal I sites. For the luciferase complementation assays (LCAs), DNA fragments and LCA vectors were ligated at the Kpn I and Sal I sites. All plasmids constructed in this study were verified through sequencing.
2.10 Y2H, BiFC, LCA, and subcellular localization assays
For the Y2H assays, paired prey and bait vectors were co-transformed into the yeast strain NMY51. Then the transformed yeast cells were spread onto the double dropout medium (DDO) SD/-Trp/-Leu solid medium and cultured at 30°C for 48–72 h. Yeast single colony grown on DDO solid medium were suspended in DDO liquid medium to OD600 = 0.6. Ten-fold serial dilutions of yeast were spotted onto DDO or quadruple dropout medium (QDO) SD/-Trp/-Leu/-His/-Ade solid medium and cultured at 30°C for 48–72 h. The yeast cells co-transformed with pNubG-Fe65 and pTSU2-APP served as positive controls, while those co-transformed with pNubG-Fe65 and pPR3-N served as negative controls, as previous report (Zhang H. et al., 2019).
For the BiFC experiments, complementary vectors containing the target genes for the identification of interactions were co-transformed into Agrobacterium tumefaciens GV3101 and cultured to an OD600 of 0.2. Equal volumes of each culture were mixed and infiltrated into N. benthamiana leaves using a needleless syringe. Agrobacterium-infiltrated plants were grown under normal conditions for 48–72 h (Yang et al., 2021).
For the LCA assays, the target genes were cloned into the pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors. Subsequently, these recombinant plasmids were introduced into different regions of the same N. benthamiana leaf via A. tumefaciens infiltration, with the final concentration of A. tumefaciens set at an OD600 of 0.4. Thereafter, a 0.2 mM luciferase substrate was infiltrated into the same regions, and imaging was performed 2 d post-infiltration (dpi) using a low-light-cooled CCD imaging system (Amersham Imager 680, GE, USA).
For the subcellular localization experiments, complementary vectors containing the target genes for the identification of interactions were transformed into A. tumefaciens GV3101 and cultured to an OD600 of 0.2. Equal volumes of each culture were mixed and agroinfiltrated into N. benthamiana leaves using a needleless syringe. AtCDPK9-mCherry was used as a PM marker (Cheng et al., 2017). Agrobacterium-infiltrated plants were grown under normal conditions for 48–72 h.
2.11 Confocal microscopy
Images were digitally acquired using a Leica SP8X confocal microscope (Leica, Wetzlar, Germany). Yellow fluorescent protein (YFP) was excited at 514 nm, and the emitted light was captured at 530–590 nm. The excitation wavelength of mCherry was 587 nm, and the collection wavelength was 610 nm. The excitation and emission wavelengths of green fluorescent protein (GFP) were 514 and 530–590 nm, respectively. Images were analyzed using Leica Microsystems.
3 Results
3.1 Identification and phylogenetic analysis of the TET gene family
We identified 35, 73, 113, and 17 members of the TET gene family in S. spontaneum AP85-441, sugarcane cultivar R570, sugarcane cultivar XTT22, and N. benthamiana, respectively (Supplementary Table S2). Phylogenetic analysis indicated that the TETs from sugarcane cultivars XTT22 and R570 were clustered into nine evolutionary groups, whereas none of SsTETs from S. spontaneum were distributed in Groups 5 and 9. Groups 7 and 8 contained only the TETs from monocotyledonous plants (Figure 1). The TETs of monocotyledonous and dicotyledonous plants were further clustered into different subgroups within the same group. AtTETs were clustered into seven groups, that is, Groups 1, 2, 3, 4, 5, 6, and 9, which is consistent with a previous report (Wang et al., 2012a). Surprisingly, ScTSPAN18, previously identified as a protein interacting with SCMV-6K2, was not included in the TET gene family (Zhang H. et al., 2019). To investigate the differences between ScTSPAN18 and the typical TETs AtTET1 and OsTET1, AlphaFold3 was used to simulate the protein structures. Notably, the LEL of ScTSPAN18 was small, and the conserved motifs were absent compared with typical TETs (Supplementary Figure S1).
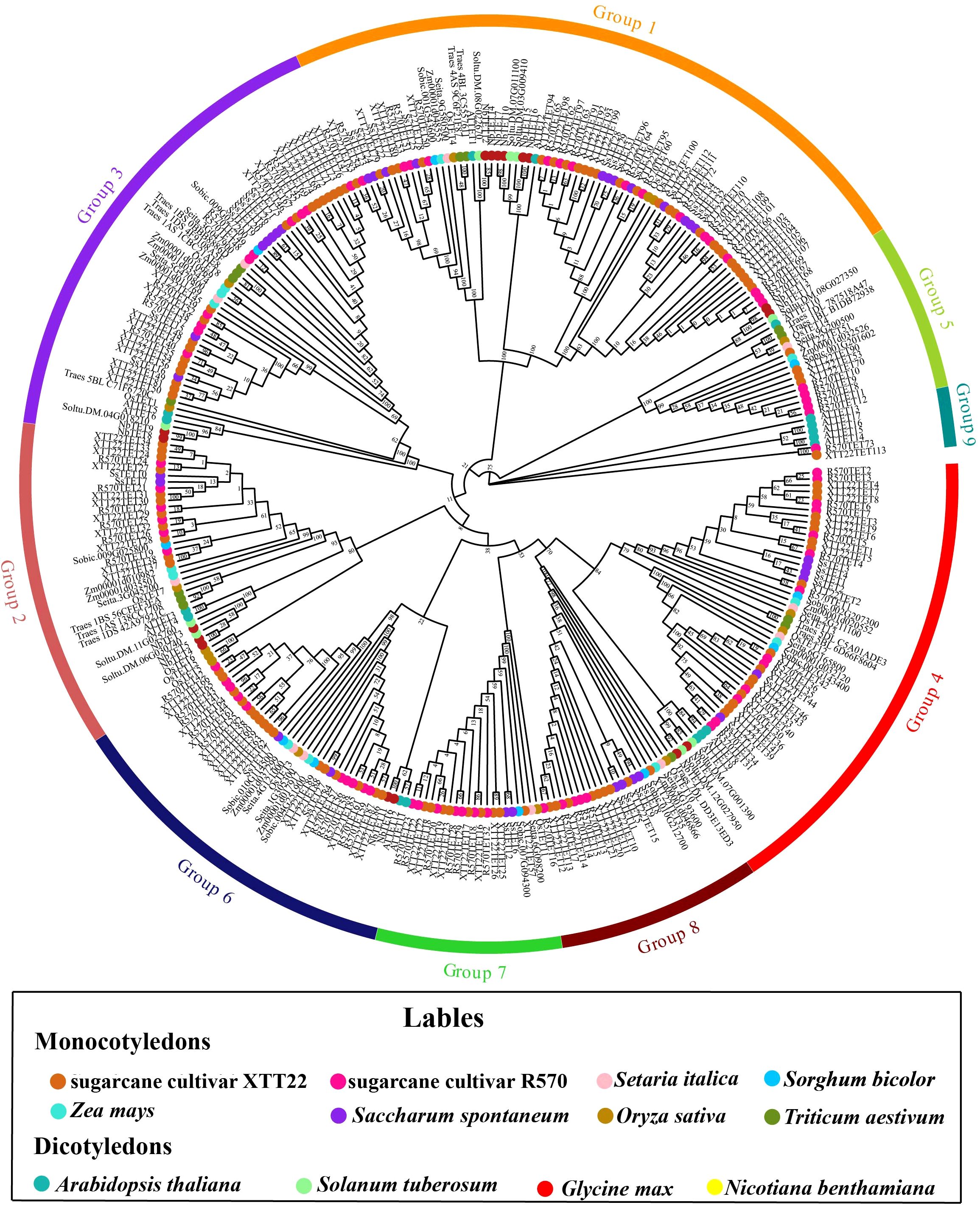
Figure 1. Phylogenetic tree analysis of Tetraspanins (TETs) of Saccharum species and other plant species. The phylogenetic tree was constructed using the maximum likelihood method with 1,000 bootstrap replicates. The TETs sequences are from the databases of 8 monocotyledons (sugarcane cultivar R570, sugarcane cultivar XTT22, Oryza sativa, Zea mays, Setaria italic, S. spontaneum, Triticum aestivum and Sorghum bicolor) and 4 dicotyledons (Arabidopsis thaliana. Nicotiana benthamiana, Glycine max, Solanum tuberosum). These TETs were grouped into 9 distinct groups and annotated with different colors.
3.2 Conserved motifs, gene structure, and physicochemical properties of TETs
Ten conserved motifs, distributed among Groups 1–8 in the same distribution order, were identified in the SsTETs, R570TETs, and XTT22TETs. For the TETs in Group 9, only motifs 1, 2, 5, 7, and 8 were detected (Supplementary Figure S2A). Gene structure analysis revealed that the ETs in Groups 1–4 and 6–9 contained 2–4 exons (Supplementary Figure S2B), whereas those in Group 5 contained more than 10 exons (Supplementary Figure S2B).
Physicochemical analysis of the sugarcane TET proteins revealed amino acid lengths ranging from 205 to 853, theoretical isoelectric points between 7.75 and 9.91, and instability coefficients between 29.53 and 58.03. Subcellular localization of the above proteins was predicted using a protein subcellular localization website (Supplementary Table S3). Most of the TETs in sugarcane were localized in the PM, although some were localized in the Golgi apparatus and ER (Supplementary Table S3).
3.3 Collinearity and chromosomal localization of the TET gene
To understand the evolution of the TET gene family, the intra- and inter-species collinearity of TET genes in XTT22, R570, and S. spontanerum AP85–441 was investigated. Intraspecies collinearity analysis revealed 266, 20, and 260 pairs of collinear TETs in XTT22 (Figure 2A), S. spontanerum AP85-441 (Figure 2B), and R570 (Supplementary Figure S3), respectively. Interspecies collinearity analysis revealed 96 and 86 pairs of collinear TETs between AP85–441 and R570 and XTT22, respectively (Figure 2C). To gain more evolutionary information of the TET gene family, we analyzed the synteny of TET genes from S. spontaneum with those from Setaria italica, sorghum, maize, rice, Arabidopsis, and Solanum lycopersicumt. The results showed that there are 19 pairs between Setaria italica and S. spontaneum, 19 pairs of homologous genes between sorghum and S. spontaneum, 15 pairs between maize and S. spontaneum, 18 pairs between rice and S. spontaneum, 3 pairs between Solanum lycopersicumt and S. spontaneum and no pairs between Arabidopsis and S. spontaneum (Figure 3). MCScanX analysis showed that whole-genome or segmental duplication was the primary origin of TETs in AP85-441 (45.7%), XTT22 (61.0%), and R570 (84.9%) (Supplementary Table S4). The Ka/Ks ratios of all the TET gene pairs were <1 (Supplementary Table S5), suggesting that homologous genes among rice, sorghum, S. italica, maize, and S. spontaneum, have undergone strong purifying selection.
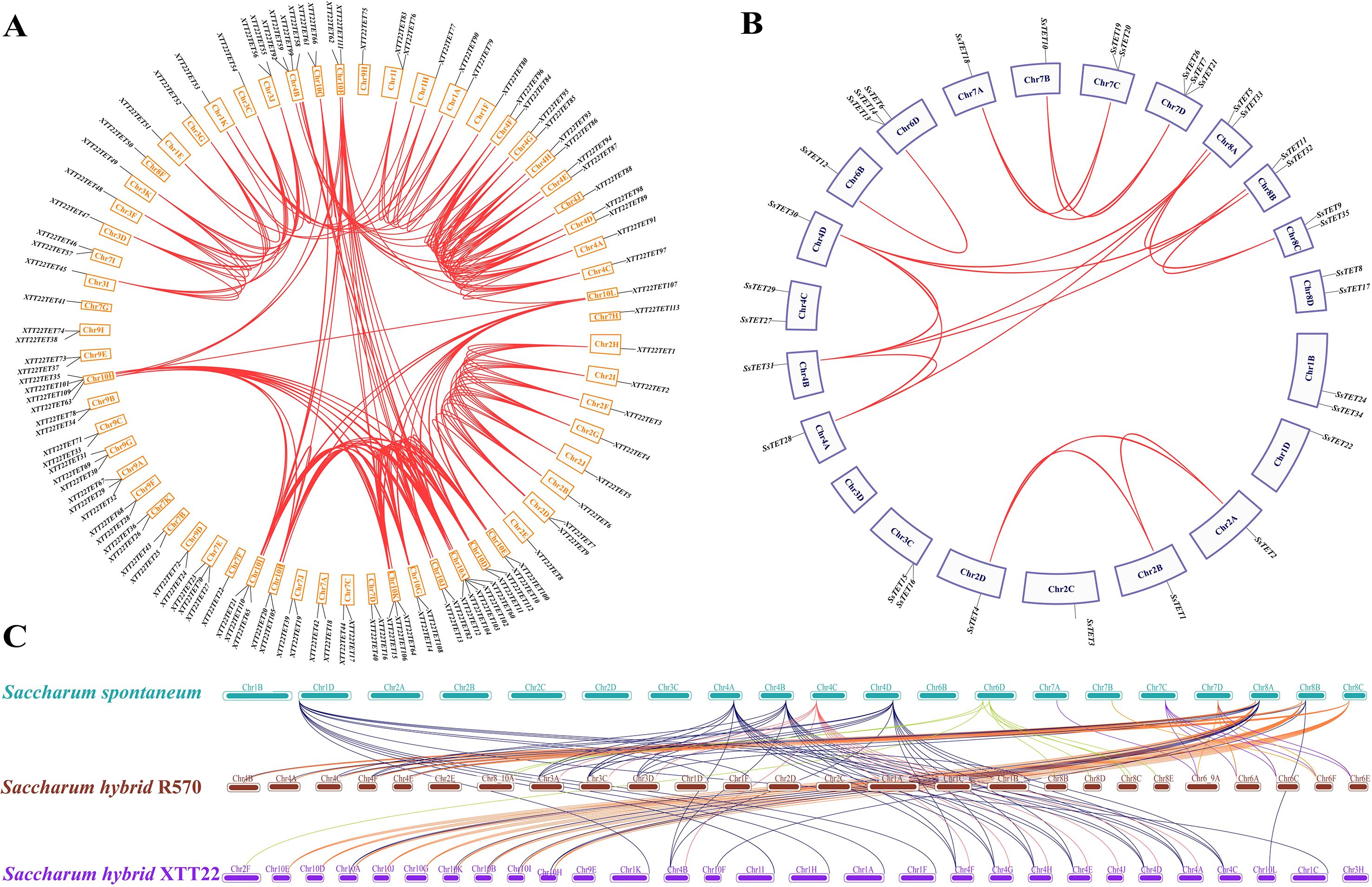
Figure 2. Interchromosomal and intrachromosomal collinearity relationship analysis of the TET gene families in Saccharum species. (A) Intrachromosomal collinearity analysis of the TET gene family in sugarcane cultivar XTT22. Red lines indicate duplicated TET gene pairs. (B) Intrachromosomal collinearity analysis of the TET gene family in S. spontaneum. Red lines indicate duplicated TET gene pairs. (C) Interchromosomal collinearity analysis of the TET gene families among in S. spontaneum, sugarcane cultivar R570, XTT22. The lines represent the TET homologous gene pairs.
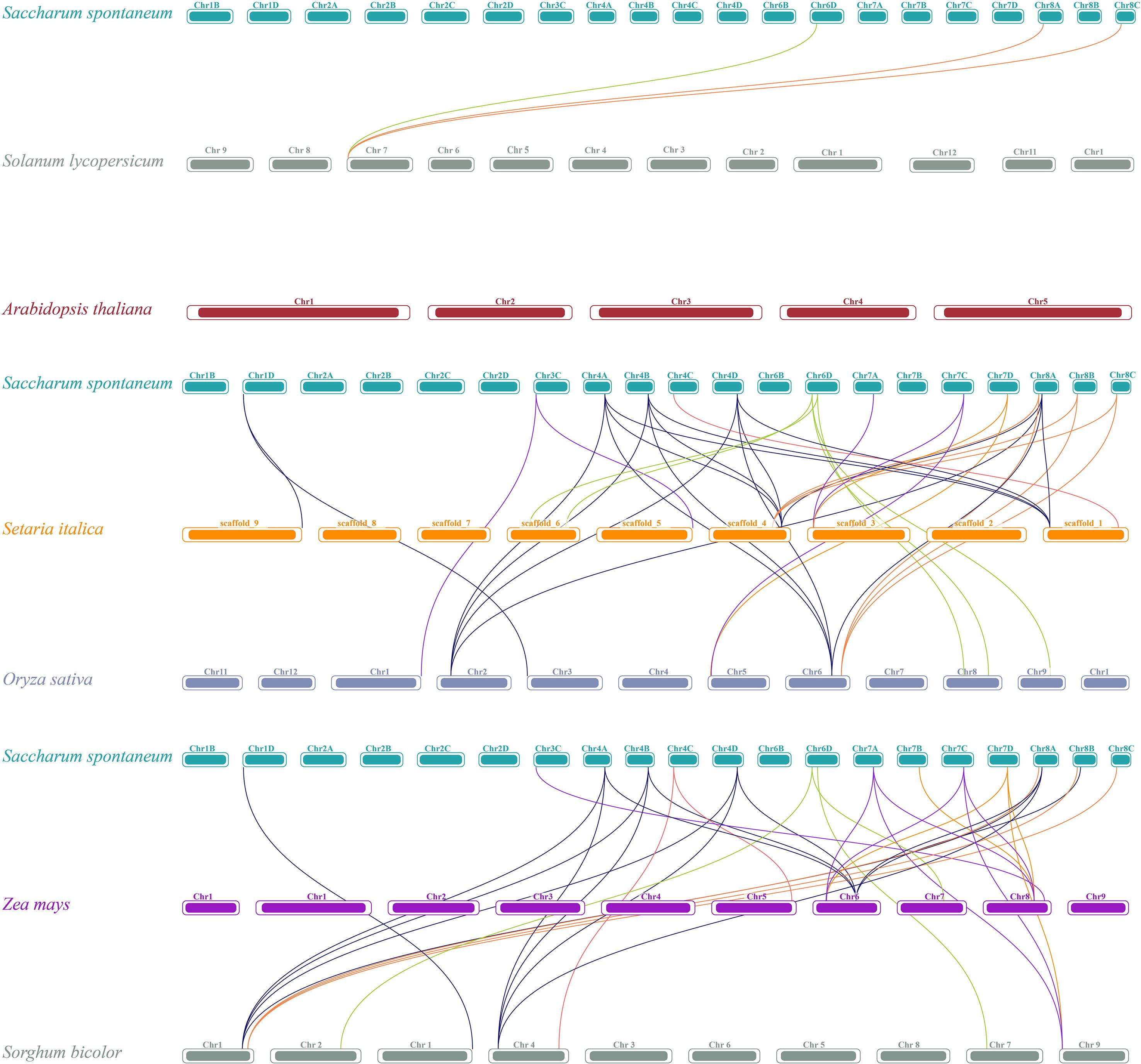
Figure 3. Synteny analysis of TET genes between S. spontaneum and other plant species (O. sativa, Z. may, S. italic, S. bicolor, A. thaliana, and S. Lycopersicum). The lines represent the TET homologous gene pairs.
Chromosomal mapping demonstrated uneven distribution of the TET genes in XTT22, with most chromosomes containing 1–2 and a few containing 3–5 genes (Supplementary Figure S4; Supplementary Table S6). The chromosomal distribution of TETs in AP85-441 (Supplementary Figure S5) and R570 (Supplementary Figure S6) was similar to that in XTT22 (Supplementary Table S6).
3.4 Cis-acting elements in TET genes
In total, 20 cis-acting elements were predicted in the promoter regions of the TETs (Supplementary Figure S7; Supplementary Table S7). ARE (94.6%), G-box (93.7%), ABRE (91.4%), TGACG-motif (90.0%), and CGTCA-motif (89.4%) were distributed in the promoter regions of more than 80% of the TETs, indicating their extensive involvement in responses to stress, light, and hormones. GATA box was only predicted in Group 3. In Group 6, the upstream region of the TET gene did not contain a CCAAT box, GC motif, or GA motif. In Group 8, the upstream region of the TET gene did not contain the GCN4 motif or GC motif (Supplementary Figure S7).
3.5 Expression patterns of TETs based on the transcriptomic data
Transcriptomic analysis of the continuous developmental gradient of leaves of XTT22 and S. spontaneum AP85–441 was performed to investigate the potential functions of TET genes in photosynthesis. Group 4 XTT22TETs were highly expressed in the mature zone, whereas Group 1 XTT22TETs were highly expressed in the transitional zone, with other XTT22TET genes exhibiting low or no expression (Figure 4A; Supplementary Table S8). For the SsTETs in the leaves of S. spontaneum AP85-441, Group 4 SsTETs were highly expressed in the transition and maturation zones. Groups 1, 2, and 8 SsTETs were expressed in the basal zone, whereas Group 3 SsTETs exhibited low expression in the leaves (Supplementary Figure S8A; Supplementary Table S8).
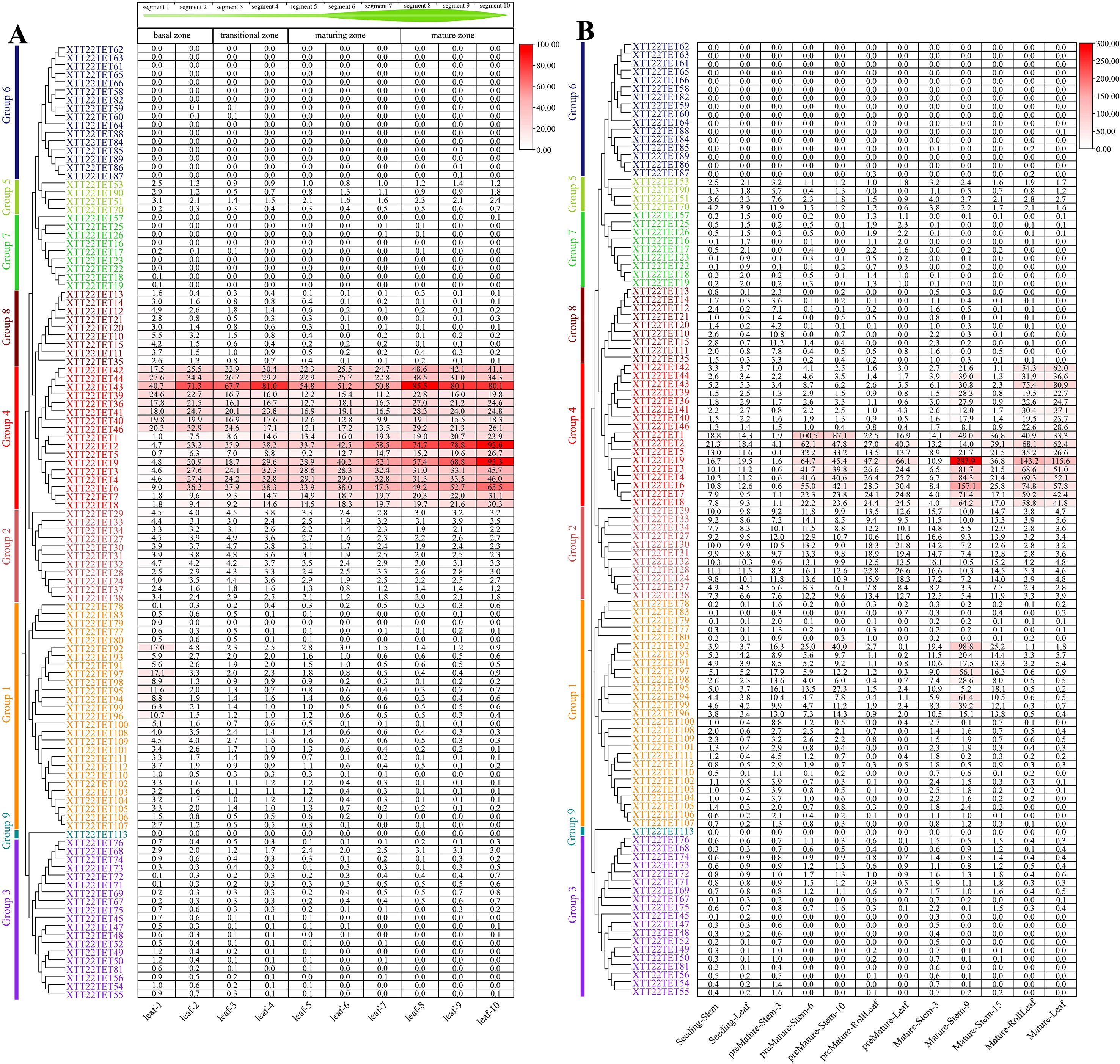
Figure 4. The expression patterns of TET genes in different tissues and across leaf gradients in sugarcane cultivar XTT22. (A) The expression patterns of XTT22TET genes based on FPKM across leaf gradients in sugarcane cultivar XTT22. (B) The expression patterns of XTT22TET gene family based on FPKM in different tissues of different stages in sugarcane cultivar XTT22.
The transcriptomic analysis of different tissues across different developmental stages of XTT22 revealed that Group 4 TETs were highly expressed in mature leaves and internodes (Figure 4B). Members of Groups 2 and 8 were expressed in different tissues in the seedling and premature stages, whereas members of Groups 3, 5, and 7 exhibited consistently low expression across all developmental stages, and members of Groups 6 and 9 were not expressed (Figure 4B; Supplementary Table S9). For S. spontaneum AP85-441, Group 4 SsTETs were highly expressed in the stems and leaves in the premature and mature stages, and four SsTETs in Group 1 showed relatively high expression in premature and mature stems (Supplementary Figure S8B; Supplementary Table S9).
3.6 Cloning and characterization of ScTETs
The results of bioinformatics and transcriptome data analyses were used to clone the following nine genes from Groups 1, 2, 3, 4, 7, and 8 of XTT22: XTT22TET2, XTT22TET8, XTT22TET13, XTT22TET23, XTT22TET34, XTT22TET55, XTT22TET67, XTT22TET78, and XTT22TET96, which were named ScTET2, ScTET8, ScTET13, ScTET23, ScTET34, ScTET55, ScTET67, ScTET78, and ScTET96, respectively (Supplementary Table S2). Sequence alignment and phylogenetic tree analysis revealed 98.53% similarity between ScTET8 and XTT22TET8 and 98.94% similarity between ScTET55 and XTT22TET55. The similarity between the other seven ScTETs was identical to the corresponding identified XTT22TETs. Subcellular localization assays showed that the fluorescence signals of ScTET13-GFP, ScTET23-GFP, ScTET34-GFP, ScTET55-GFP, ScTET67-GFP, ScTET78-GFP, and ScTET96-GFP overlapped with the fluorescence signal of AtCDPK9-mCherry (Figure 5), indicating that they were mainly located on the PM, aligning with the subcellular localization prediction results (Supplementary Table S3). The ScTET78-GFP and ScTET96-GFP exhibited punctate structures on the PM (Figure 5), whereas the ScTET2-GFP and ScTET8-GFP formed spherical structures of varying sizes ranging from 0.5 to 20 μm in diameter within the cells (Figure 5; Supplementary Figure S9).
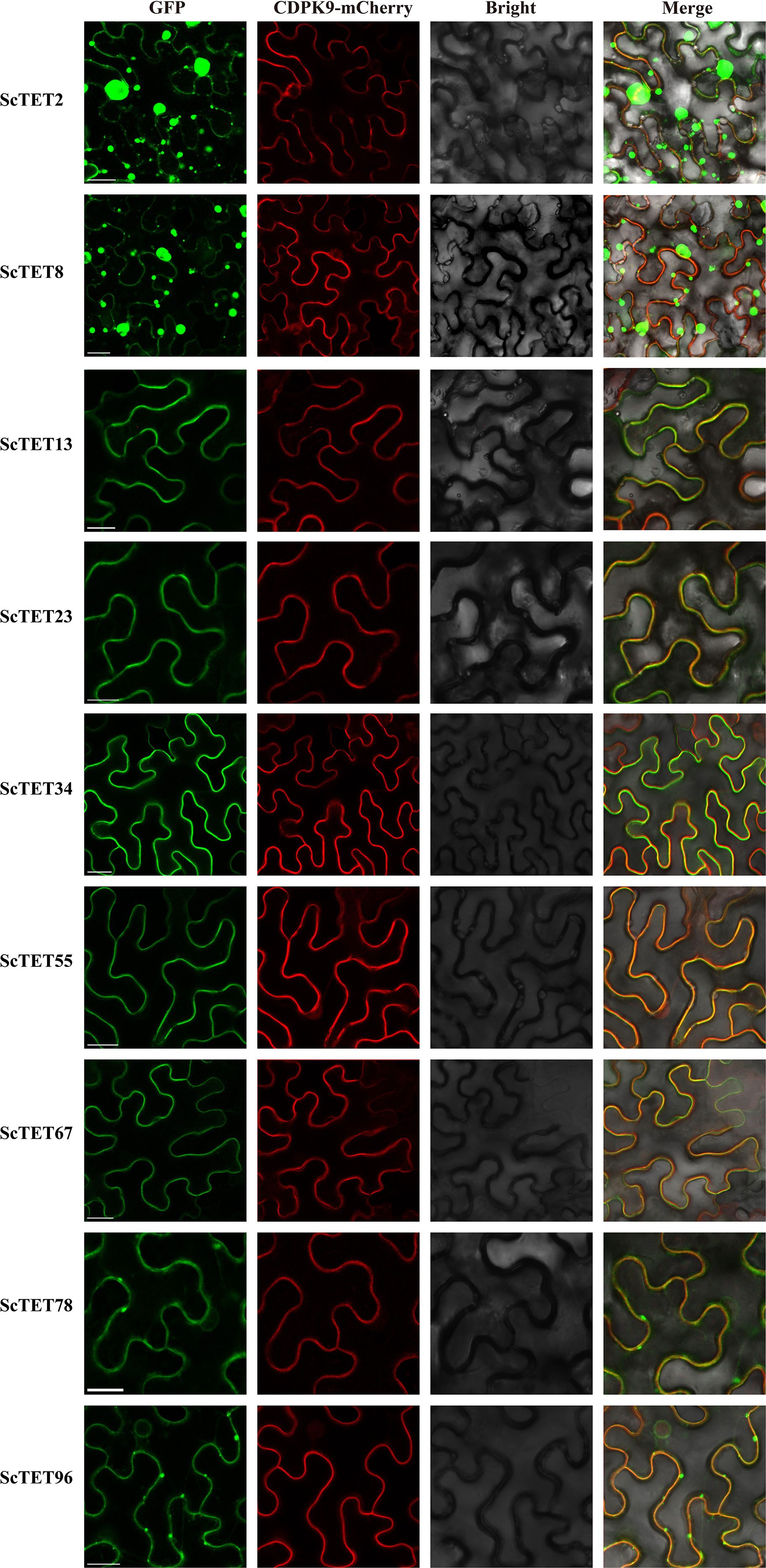
Figure 5. Subcellular localization of ScTETs. Agrobacteria harboring GFP fusion proteins were individually agroinfiltrated into N. benthamiana leaves. The images were captured at 48 h post infiltration. The plasma membrane was indicated using AtCDPK9-mCherry, Bar=20 μm.
The expression patterns of ScTETs in different tissues of the sugarcane cultivar XTT22 were analyzed via RT-qPCR. ScTET2, ScTET8, ScTET34, ScTET78, and ScTET96 exhibited significantly higher expression than the other genes (Figure 6A). ScTET2 and ScTET8 were highly expressed in the +7 leaf and +8 internode, ScTET34 was highly expressed in the +1 leaf and +3 internode, and ScTET78 and ScTET96 were highly expressed in the +8 internode (Figure 6A).
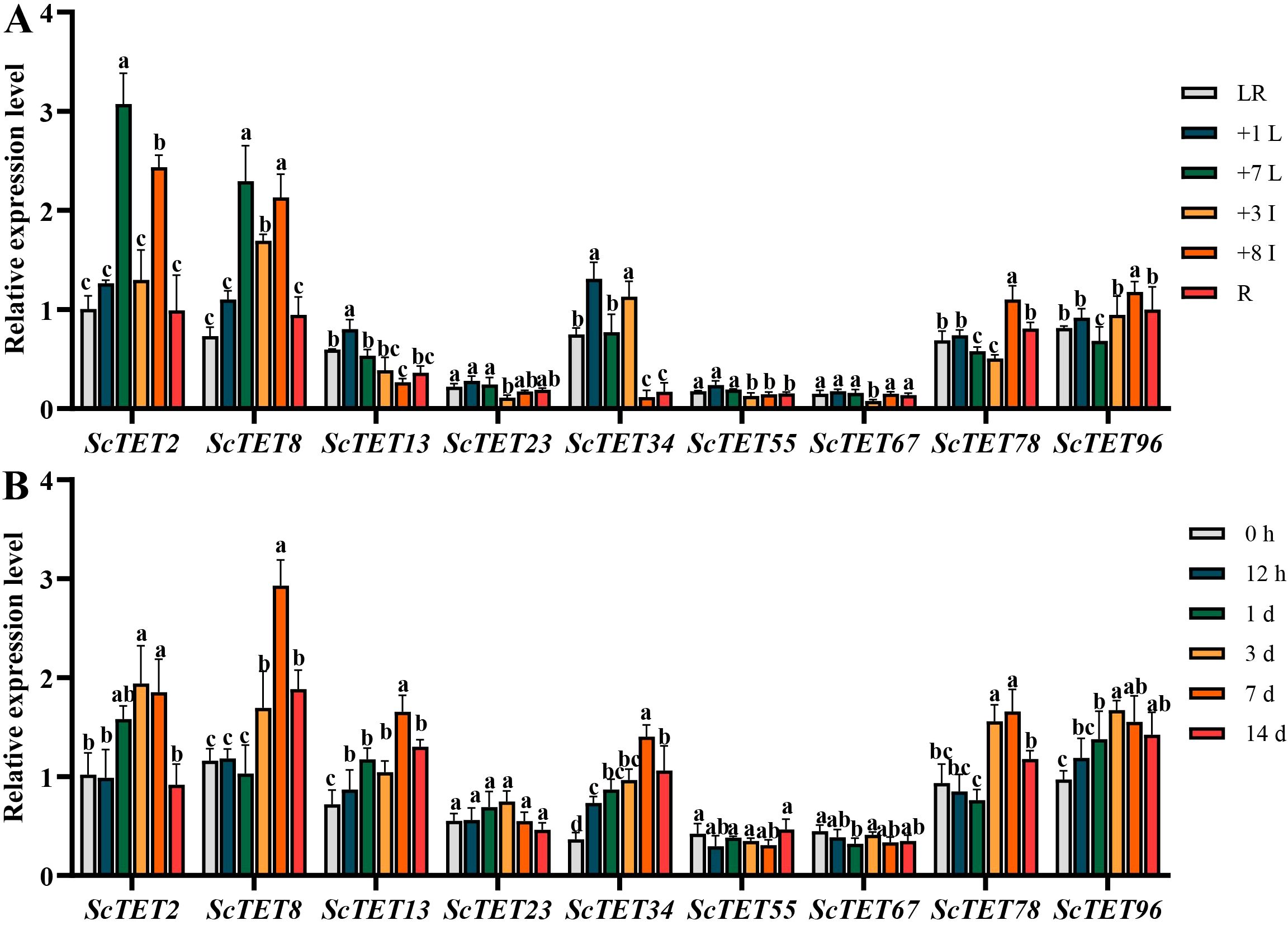
Figure 6. The expression profiles of nine ScTETs in different tissues of sugarcane cultivar XTT22, or under the challenge of sugarcane mosaic virus (SCMV). (A) The expression profiles of nine ScTETs in different tissues of sugarcane cultivar XTT22. LR: leaf roll; +1 L: the 1st leaf; +7 L: the 7st leaf; +3 I: the 3rd internode; +8 I: the 8th internode and R: root. (B) The expression profiles of nine ScTETs in the leaves of sugarcane cultivar XTT22 under the challenge of SCMV. Plants mock inoculated with 0.01 mM phosphate buffer (pH 7.0) were used as the negative controls. The Y axes indicates the relative expression levels of ScTETs. The X axes indicates the time point of materials collection. Error bars indicate SD (n = 3), a, b, c and e indicate significance at the corresponding time points, Student’s t-test, P < 0.05. Results were representative of three independent experiments.
All ScTETs except ScTET23 were differentially expressed upon SCMV infection (Figure 6B). Expressions of ScTET2, ScTET8, ScTET13, ScTET34, ScTET78, and ScTET96 increased significantly at 3 dpi and peaked 7 dpi (Figure 6B). All genes except ScTET96 were downregulated at day 14 (Figure 6B).
3.7 Interaction between ScTETs and SCMV-6K2
The interactions of the nine ScTETs with SCMV-6K2 were analyzed using Y2H, BiFC, and LCA assays. For the Y2H assays, pTUS2-APP- and pNUbG-Fe65-co-transformed NYM51 served as the positive controls, and pTUS2-APP- and pPR3-N-co-transformed NYM51 served as the negative controls. pPPR3-ScTET2, pPPR3-ScTET8, pPPR3-ScTET13, pPPR3-ScTET23, pPPR3-ScTET34, pPPR3-ScTET55, pPPR3-ScTET67, pPPR3-ScTET78, and pPPR3-ScTET96 were co-transferred with pBT-STE-SCMV-6K2 into the yeast NYM51. In DDO and QDO media containing X-gal, the yeast cells harboring pPPR3-ScTET2, pPPR3-ScTET8, pPPR3-ScTET13, pPPR3-ScTET23, pPPR3-ScTET34, pPPR3-ScTET78, or pPPR3-ScTET96 with pBT-STE-SCMV-6K2 grew normally as the positive controls (Figure 7A). In contrast, the yeast cells harboring pPPR3-ScTET55 or pPPR3-ScTET67 with pBT-STE-SCMV-6K2 grew only on DDO medium but not QDO (Figure 7A). These results demonstrated that all nine ScTETs, except ScTET55 and ScTET67, interacted with SCMV-6K2.
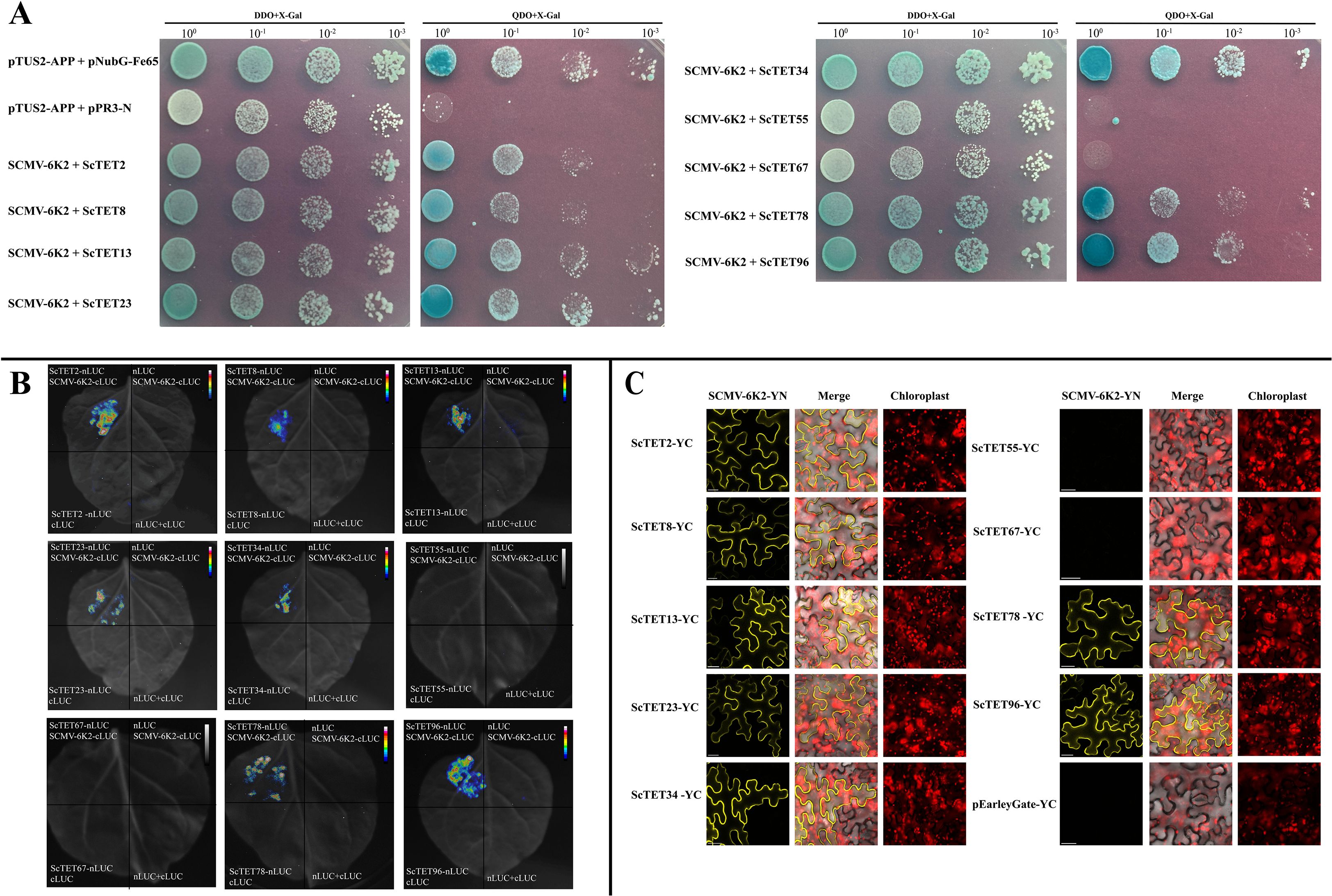
Figure 7. Interaction of 9 ScTETs with SCMV-6K2 by BiFC, Y2H and Luc assays. (A) Y2H assays. pPR3-ScTETs were individually pairwise co-transformed with the vector pBT-STE-SCMV-6K2 into the yeast NYM51 cells in a 10×dilution series of 10-μL aliquots which were then plated on a non-selective medium, SD/-Leu/-Trp or quadruple dropout medium, SD/-Leu/-Trp/-His/-Ade supplemented with X-Gal. Yeast cells co-transformed with pTUS2-APP and pNubG-Fe65 were used as a positive control. pTUS2-APP and pPR3-N were used as negative controls. (B) Luc assays. Agrobacteria harboring nLuc/cLuc fusion proteins were individually pairwise co-infiltrated into N. benthamiana leaves. The leaf epidermal cells pairwise co-transformed with ScTETs-nLUC and cLUC, cLuc-6K2 and nLuc, cLuc and nLuc were used as negative controls. (C) BiFC assays. Agrobacteria harboring YC/YN fusion proteins were individually pairwise co-infiltrated into N. benthamiana leaves. The images were captured at 48 h post infiltration. YC and SCMV-6K2-YN were used as negative controls. Bar=20 μm.
LCA (Figure 7B) and BiFC assays (Figure 7C) yielded results similar to those of the Y2H assays. Interestingly, the BiFC assays showed that all interactions occurred at the PM. These interactions resulted in the loss of intracellular localization and spherical structures of ScTET2 and ScTET8, as well as the disappearance of the punctate PM-associated structures formed by ScTET78 and ScTET96 (Figure 7C).
3.8 Interactions and self-interactions among ScTETs
Interactions and self-interactions among the TET members contribute to TEM formation (Charrin et al., 2002; Jimenez-Jimenez et al., 2019; Huang et al., 2022). We conducted Y2H assays to investigate the possible interactions and self-interactions among the nine ScTETs and observed extensive interactions among the ScTETs (Figure 8). ScTET2, ScTET13, ScTET23 and ScTET34 interacted with other ScTETs and could also self-interact. ScTET96 also interacted with other ScTETs but could not self-interact (Figure 8).

Figure 8. Self-interaction of ScTETs by Y2H assays. Combinations of pBT-STE-ScTET2 plus pPR3-ScTET2, pBT-STE-ScTET13 plus pPR3-ScTET13 and pBT-STE-ScTET96 plus pPR3-ScTET96 were individually transformed into the yeast NYM51. Yeast cells co-transformed with pTUS2-APP and pNubG-Fe65 were used as a positive control, while yeast cells co-transformed with pTUS2-APP and pPR3-N were used as negative controls.
3.9 Interaction of AtTETs or NbTETs with TuMV-6K2 assessed using Y2H
The intercellular movement of viruses is a key step in establishing systemic infection. AtTET3 is localized in the plasmodesmata and required for CMV to establish systemic infection (Fernandez-Calvino et al., 2011; Zhu et al., 2022). To investigate whether TET is widely involved in the interaction with potyvirus-6K2, we cloned four AtTET genes, AtTET1, AtTET3, AtTET7, and AtTET8, which are localized in the plasmodesmata of Arabidopsis (Fernandez-Calvino et al., 2011; Boavida et al., 2013). In addition, we cloned the 17 NbTET genes identified in this study (Supplementary Table S2). These genes were individually inserted into the pPR3-N vector and then individually co-transformed with pBT-STE-TuMV-6K2 into yeast NMY51. Notably, all these TETs interacted with TuMV-6K2 (Supplementary Figures S10, S11).
4 Discussion
Genome-wide identification of the TET gene family has only been previously performed in Arabidopsis and rice, which identified 17 and 15 TETs, respectively (Wang et al., 2012a; Mani et al., 2015). In the present study, we identified 35, 113, 73, and 17 TETs in S. spontaneum AP85-441, sugarcane cultivars XTT22 and R570, and N. benthamiana, respectively. We achieved the Phytozome database and screened 11, 10, 12, 14, 9, and 20 TETs for millet, sorghum, maize, wheat, potato, and soybean, respectively. The number of TETs in Saccharum species was much higher than that in other plant species, likely because of the highly polyploid characteristics of sugarcane (Zhang et al., 2022; Healey et al., 2024; Zhang et al., 2025c). XTT22TET and R570TET were clustered in nine phylogenetic groups, whereas SsTETs were absent in Groups 5 and 9 (Figure 1). Additionally, the TET genes in Group 5 contained a significantly higher number of exons at up to 10 or more compared with the other eight groups (Figure 1B). This feature is considered characteristic of the original TET genes (Garcia-España et al., 2008), as demonstrated by AtTET10 and OsTET14 (Mani et al., 2015). S. spontanerum and S. officinarum are recognized as the main progenitors of modern sugarcane cultivars, and S. officinarum is derived from S. robustum (Zhang J. et al., 2019; Zhang et al., 2022). We speculated that sugarcane TET genes in Groups 5 and 9 may have originated from S. robustum. Additionally, as only monocotyledonous plant TETs were present in Groups 7 and 8, we speculated that these groups are unique to monocotyledons (Figure 1), which is consistent with a previous report (Wang et al., 2012a). Interestingly, GATA-box is only present in the promoter region of Group 3 TETs (Supplementary Figure S7). As GATA-box usually responds to light signaling, we speculate that Group 3 TETs are involved in photosynthesis (Gangappa and Chattopadhyay, 2013). Expansion of the TET gene family in Saccharum species appears to have primarily occurred through WGD/fragmentation and dispersed replication (Supplementary Table S4), which is in line with previous reports (Zhang et al., 2018; Li et al., 2021; Wang et al., 2023). ScTSPAN18, which was identified in our previous study (Zhang H. et al., 2019), did not belong to the TET family (Supplementary Figure S1). In early studies on animal TETs, the L6D protein was mistakenly classified into the TET family (Wright et al., 2000; Boucheix and Rubinstein, 2001). Structural simulation using AlphaFold3 showed that the LEL of L6D lacked conserved motifs (Supplementary Figure S1). Homologous proteins with similar structural features were identified in Arabidopsis (four), rice (one), sorghum (two), and maize (three) using the Phytozome database. However, the biological functions of these proteins require further investigation.
Transcriptomic data analysis revealed that the 119 XTT22TET genes identified in this study were differentially expressed across various tissues and developmental stages (Figure 4; Supplementary Figure S8), indicating that they are extensively involved in sugarcane growth and development, which is consistent with the findings in Arabidopsis (Boavida et al., 2013). Plant viruses have undergone long-term co-evolution with their hosts and cannot establish systemic infections without interacting with host factors (Benitez-Alfonso et al., 2010; Niehl and Heinlein, 2011). Nine ScTET genes were cloned from the sugarcane cultivar XTT22, and RT-qPCR revealed their differential expression following SCMV infection (Figure 6), indicating that they all responded to the SCMV infection. Protein–protein interaction assays revealed interactions between all ScTETs and SCMV-6K2, except for ScTET55 and ScTET67 in Group 3 (Figure 7). Plasmodesmata are important channels for the intercellular movement of plant viruses (Valli et al., 2007; Chung et al., 2008; Wei et al., 2010; Cheng et al., 2017; Chai et al., 2020; Cheng et al., 2020; Xiao et al., 2022; Hýsková et al., 2024). Four plasmodesmata-localized AtTETs—AtTET1 (Group 1), AtTET3 (Group 2), AtTET7, and AtTET8 (Group 4) (Fernandez-Calvino et al., 2011; Boavida et al., 2013; Johnston et al., 2023)—interacted with TuMV-6K2 (Supplementary Figure S10). AtTET3 is essential for the intercellular movement of the CMV (Zhu et al., 2022). Based on this, we hypothesized that TETs may also play a role in the intercellular movement of potyviruses. Specifically, ScTET78 and ScTET96 (Group 1), ScTET34 (Group 2), and ScTET2 and ScTET8 (Group 4), which belong to the same phylogenetic groups as the AtTETs, may be involved in the intercellular movement of SCMV. Notably, ScTET78 and ScTET96 were localized in the PM and showed a punctate structure (Figure 5), resembling plasmodesmata. However, upon interaction with SCMV-6K2, the punctate structure disappeared (Figure 7C). Hence, further verification using plasmodesmata markers, such as aniline blue, is warranted. In addition, except for ScTET96, the other eight TETs exhibited both self-interaction and interaction with one another (Figure 8), indicating that they can form TEMs and perform various biological functions (Charrin et al., 2002; Boavida et al., 2013; Huang et al., 2022). ScTET13, ScTET23, ScTET34, ScTET55, and ScTET67 were localized in the PM, similar to some TETs in Arabidopsis and rice (Zhu et al., 2022). Interestingly, ScTET2 and ScTET8 form vesicular structures with diameters ranging from 0.5 to 20 μm in the cytoplasm, and both are associated with the PM (Figure 5). Under stressful conditions, plants produce stress granules (SGs) and P-bodies (Kearly et al., 2024). SGs are biphasic assemblies consisting of dense cores (∼0.2 μm in diameter) embedded within a less concentrated dynamic shell of variable size (Youn et al., 2019). P-bodies range from 0.8 to 1.0 μm and are located proximal to the PM (Xu and Chua, 2009), whereas MVBs range from 0.4 to 0.5 μm (Movahed et al., 2019). TET is involved in the formation of secretory vesicles and MVBs, which are employed by TuMV to move to neighboring cells (Movahed et al., 2019; Ghossoub et al., 2020; Jankovičová et al., 2020). Based on the vesicle size, we speculated that ScTET2 and ScTET8 are involved in the formation of secretory vesicles and MVBs and subsequently in cell-to-cell movement. As SG and P-bodies are membrane-less organelles, ScTET2 and ScTET8 may not be involved with them; however, further experiments are needed to verify this. AtTET8 in Arabidopsis serves as a marker for EVs (Zhang et al., 2020; He et al., 2021). Although ScTET2 and ScTET8 belong to Group 4 along with AtTET8, EVs range from 0.05 to 0.15 μm in diameter (Nolte-’t Hoen et al., 2016; Jeppesen et al., 2019), making them too small to be accurately distinguished by confocal microscopy due to the low resolution. Therefore, further studies using transmission electron microscopy or other techniques are necessary. ScTET2 and ScTET8 are highly expressed in sugarcane and show differential expression patterns upon SCMV infection. However, upon interaction with SCMV-6K2 on the PM, the vesicles induced by ScTET2 or ScTET8 disappeared. Plants can transport sRNAs through their vesicles (Li et al., 2024; Zhao et al., 2024). Therefore, we speculated that viral infection interferes with the secretory system, thereby influencing the growth and development of sugarcane plants.
Additionally, the Y2H assays showed that 17 NbTETs and 4 AtTETs interacted with TuMV-6K2 (Supplementary Figures S10, S11), indicating that the interaction of TET with 6K2 may represent a general mechanism employed by potyviruses to establish infection. Notably, ScTET55 and ScTET67 from sugarcane Group 3 did not interact with SCMV-6K2, indicating that TETs may be selectively utilized by different viruses.
5 Conclusion
In this study, 35, 113, 73, and 17 TETs were identified in S. spontaneum AP85-441, sugarcane cultivars XTT22 and R570, and N. benthamiana, respectively. These TETs clustered into nine phylogenetic groups, with Groups 7 and 8 being specific to monocotyledonous plants. The TET structure in Group 5 was significantly different from that of the other groups. Transcriptomic analysis revealed that Group 4 TETs were highly expressed in XTT22 and S. spontanerum. The nine cloned ScTETs from XTT22 showed different expression patterns upon SCMV infection. Subcellular localization analysis indicated that seven ScTETs were localized to the PM, with ScTET78 and ScTET96 forming punctate structures and ScTET2 and ScTET8 forming spherical structures of varying sizes. Interactions and self-interactions occurred extensively among the nine ScTETs. Seven of the nine ScTETs interacted with SCMV-6K2, and 17 N. benthamiana NbTETs and 4 Arabidopsis AtTETs interacted with the 6K2 protein of TuMV.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZC: Formal analysis, Investigation, Validation, Visualization, Writing – original draft. YL: Formal analysis, Investigation, Resources, Validation, Visualization, Writing – original draft. KZ: Formal Analysis, Investigation, Visualization, Writing – original draft. ZY: Formal Analysis, Investigation, Methodology, Resources, Writing – original draft. QY: Formal Analysis, Investigation, Validation, Writing – original draft. HL: Formal Analysis, Investigation, Writing – original draft. ZZ: Formal Analysis, Investigation, Writing – original draft. HZ: Formal Analysis, Investigation, Writing – original draft. GH: Resources, Validation, Writing – original draft. JX: Conceptualization, Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFD1200700, 2023YFD1200703), the National Natural Science Foundation of China (32472065), and the Science and Technology Innovation Project of Fujian Agriculture and Forestry University (KFB24033).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1684431/full#supplementary-material
Supplementary Figure 1 | Simulation of the structures of TETs from different species. AtTET1 (Arabidopsis thaliana, NP_199482.1), OsTET1 (Oryza sativa, LOC_Os01g74570), CD63 (Homo sapiens, AHI51903.1), as well as L6D (Homo sapiens, NP_004608.1) and ScTSPAN18 (Saccharum spp. Hybrid, QHD26891).
Supplementary Figure 2 | Gene structure and conserved motif analysis of TETs genes in Saccharum species. (A) Conserved motifs of TET genes (labeled 1–10) are indicated by differently colored boxes. (B) Structure analysis of TET genes. Green boxes represent CDS, yellow boxes represent UTR, and black lines represent intron.
Supplementary Figure 3 | Interchromosomal collinearity relationship analysis of TETs in sugarcane cultivar R570. Red lines represent the TET homologous gene pairs.
Supplementary Figure 4 | Chromosomal mapping analysis of TETs in sugarcane cultivar XTT22. Blue represents chromosomes and red represents the localization of TET genes on chromosomes of XTT22.
Supplementary Figure 5 | Chromosomal mapping analysis of TETs in S. spontaneum. Blue color represents chromosomes and red color represents the location of SsTET genes on chromosomes.
Supplementary Figure 6 | Chromosomal mapping analysis of TETs in sugarcane cultivar R570. Blue color represents chromosomes and red represents the localization of TET genes on chromosomes of R570.
Supplementary Figure 7 | Cis-acting analysis of the TETs in sugarcane cultivar XTT22, R570, and S. spontanerum. (A) Distribution of different cis-acting. (B) Statistic analysis of different cis-acting. Differently colored boxes represent different cis-acting.
Supplementary Figure 8 | The expression patterns of SsTETs in different tissues or across leaf gradients of S. spontanerum. (A) The expression patterns of SsTET genes based on FPKM across leaf gradients in S. spontaneum. (B) The expression patterns of SsTET gene family based on FPKM in different tissues of different developmental stages in S. spontaneum.
Supplementary Figure 9 | Size of vesicle like structures of localized by ScTET2-GFP in Figure 5. Image J was used to measure the diameter of the vesicle like structures as indicated by white arrows. Bar=20 μm.
Supplementary Figure 10 | Interaction of Arabidopsis AtTETs with the 6K2 protein of turnip mosaic virus (TuMV) by Y2H assays. pPR3-AtTETs were individually pairwise co-transformed with pBT-STE-TuMV-6K2 into the yeast NMY51 cells in a 10×dilution series of 10-μL aliquots which were then plated on a non-selective medium, SD/-Leu/-Trp or quadruple dropout medium, SD/-Leu/-Trp/-His/-Ade. Yeast cells co-transformed with pTUS2-APP and pNubG-Fe65 were used as a positive control, while yeast cells co-transformed with pTUS2-APP and pPR3-N were used as negative controls.
Supplementary Figure 11 | Interaction of N. benthamiana NbTETs with the 6K2 protein of TuMV by Y2H assays. pPR3-NbTETs were individually pairwise co-transformed with pBT-STE-TuMV-6K2 into the yeast NMY51 cells in a 10×dilution series of 10 μL aliquots which were then plated on a non-selective medium, SD/-Leu/-Trp or quadruple dropout medium, SD/-Leu/-Trp/-His/-Ade. Yeast cells co-transformed with pTUS2-APP and pNubG-Fe65 were used as a positive control, while yeast cells co-transformed with pTUS2-APP and pPR3-N were used as negative controls.
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Benitez-Alfonso, Y., Faulkner, C., Ritzenthaler, C., and Maule, A. J. (2010). Plasmodesmata: Gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23, 1403–1412. doi: 10.1094/MPMI-05-10-0116
Boavida, L. C., Qin, P., Broz, M., Becker, J. D., and McCormick, S. (2013). Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 163, 696–712. doi: 10.1104/pp.113.216598
Boucheix, C. and Rubinstein, E. (2001). Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205. doi: 10.1007/PL00000933
Cabanillas, D. G., Jiang, J., Movahed, N., Germain, H., Yamaji, Y., Zheng, H., et al. (2018). Turnip mosaic virus uses the SNARE protein VTI11 in an unconventional route for replication vesicle trafficking. Plant Cell. 30, 2594–2615. doi: 10.1105/tpc.18.00281
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 360, 1126–1129. doi: 10.1126/science.aar4142
Chai, M., Wu, X., Liu, J., Fang, Y., Luan, Y., Cui, X., et al. (2020). P3N-PIPO interacts with P3 via the shared N-terminal domain to recruit viral replication vesicles for cell-to-cell movement. J. Virol. 94, e01898–e01819. doi: 10.1128/JVI.01898-19
Charrin, S., le Naour, F., Silvie, O., Milhiet, P. E., Boucheix, C., and Rubinstein, E. (2009). Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154. doi: 10.1042/BJ20082422
Charrin, S., Manié, S., Oualid, M., Billard, M., Boucheix, C., and Rubinstein, E. (2002). Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516, 139–144. doi: 10.1016/s0014-5793(02)02522-x
Chen, A., He, B., and Jin, H. (2022). Isolation of extracellular vesicles from Arabidopsis. Curr. Protoc. 2, e352. doi: 10.1002/cpz1.352
Chen, H. C., Newton, C. J., Diaz, G., Zheng, Y., Kong, F., Yao, Y., et al. (2025). Proteomic landscape of pattern triggered immunity in the Arabidopsis leaf apoplast. bioRxiv. 123 (6), e70498. doi: 10.1101/2025.02.06.636724
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one“ bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cheng, G., Dong, M., Xu, Q., Peng, L., Yang, Z., Wei, T., et al. (2017). Dissecting the Molecular Mechanism of the Subcellular Localization and Cell-to-cell Movement of the Sugarcane mosaic virus P3N-PIPO. Sci. Rep. 7, 9868. doi: 10.1038/s41598-017-10497-6
Cheng, G., Yang, Z., Zhang, H., Zhang, J., and Xu, J. (2020). Remorin interacting with PCaP1 impairs Turnip mosaic virus intercellular movement but is antagonised by VPg. New Phytol. 225, 2122–2139. doi: 10.1111/nph.16285
Chung, B. Y. W., Miller, W. A., Atkins, J. F., and Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105, 5897–5902. doi: 10.1073/pnas.0800468105
Cnops, G., Neyt, P., Raes, J., Petrarulo, M., Nelissen, H., Malenica, N., et al. (2006). The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell. 18, 852–866. doi: 10.1105/tpc.105.040568
Cui, Y., Gao, J., He, Y., and Jiang, L. (2020). Plant extracellular vesicles. Protoplasma. 257, 3–12. doi: 10.1007/s00709-019-01435-6
Fast, L. A., Lieber, D., Lang, T., and Florin, L. (2017). Tetraspanins in infections by human cytomegalo- and papillomaviruses. Biochem. Soc Trans. 45, 489–497. doi: 10.1042/BST20160295
Fernandez-Calvino, L., Faulkner, C., Walshaw, J., Saalbach, G., Bayer, E., Benitez-Alfonso, Y., et al. (2011). Arabidopsis plasmodesmal proteome. PloS One 6, e18880. doi: 10.1371/journal.pone.0018880
Florin, L. and Lang, T. (2018). Tetraspanin assemblies in virus infection. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01140
Gangappa, S. N. and Chattopadhyay, S. (2013). MYC2 differentially regulates GATA-box containing promoters during seedling development in Arabidopsis. Plant Signal Behav. 8 (10), e25679. doi: 10.4161/psb.25679
Gao, J., Chen, J., Zhang, H., Jiang, L., and Cui, Y. (2024). Extracellular vesicle isolation and mass spectrometry-based proteomic analysis in Arabidopsis thaliana. Methods Mol. Biol. 2841, 75–83. doi: 10.1007/978-1-0716-4059-3_6
Garcia-España, A., Chung, P. J., Sarkar, I. N., Stiner, E., Sun, T. T., and Desalle, R. (2008). Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 91, 326–334. doi: 10.1016/j.ygeno.2007.12.005
Ghossoub, R., Chéry, M., Audebert, S., Leblanc, R., Egea-Jimenez, A. L., Lembo, F., et al. (2020). Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. U. S. A. 117, 5913–5922. doi: 10.1073/pnas.1922447117
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grangeon, R., Jiang, J., Wan, J., Agbeci, M., Zheng, H., and Laliberté, J. F. (2013). 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00351
Green, K. A., Eaton, C. J., Savoian, M. S., and Scott, B. (2019). A homologue of the fungal tetraspanin Pls1 is required for Epichloë festucae expressorium formation and establishment of a mutualistic interaction with Lolium perenne. Mol. Plant Pathol. 20, 961–975. doi: 10.1111/mpp.12805
Guo, L., Qi, Y., Mu, Y., Zhou, J., Lu, W., and Tian, Z. (2022). Potato StLecRK-IV.1 negatively regulates late blight resistance by affecting the stability of a positive regulator StTET8. Hortic. Res. 9, uhac010. doi: 10.1093/hr/uhac010
Hafrén, A., Üstün, S., Hochmuth, A., Svenning, S., Johansen, T., and Hofius, D. (2018). Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176, 649–662. doi: 10.1104/pp.17.01198
He, B., Cai, Q., Qiao, L., Huang, C. Y., Wang, S., Miao, W., et al. (2021). RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants. 7, 342–352. doi: 10.1038/s41477-021-00863-8
He, R., Li, Y., Bernards, M. A., and Wang, A. (2023b). Manipulation of the cellular membrane-cytoskeleton network for RNA virus replication and movement in plants. Viruses. 15, 744. doi: 10.3390/v15030744
He, R., Li, Y., Bernards, M. A., and Wang, A. (2025). Turnip mosaic virus selectively subverts a PR-5 thaumatin-like, plasmodesmal protein to promote viral infection. New Phytol. 245, 299–317. doi: 10.1111/nph.20233
He, B., Wang, H., Liu, G., Chen, A., Calvo, A., Cai, Q., et al. (2023a). Fungal small RNAs ride in extracellular vesicles to enter plant cells through clathrin-mediated endocytosis. Nat. Commun. 14, 4383. doi: 10.1038/s41467-023-40093-4
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Healey, A. L., Garsmeur, O., Lovell, J. T., Shengquiang, S., Sreedasyam, A., Jenkins, J., et al. (2024). The complex polyploid genome architecture of sugarcane. Nature. 628, 804–810. doi: 10.1038/s41586-024-07231-4
Hemler, M. E. (2001). Specific tetraspanin functions. J. Cell Biol. 155, 1103–1107. doi: 10.1083/jcb.200108061
Hemler, M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. doi: 10.1146/annurev.cellbio.19.111301.153609
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hu, W., Hua, X., Zhang, Q., Wang, J., Shen, Q., Zhang, X., et al. (2018). New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics. BMC Plant Biol. 18, 270. doi: 10.1186/s12870-018-1495-y
Huang, S., Yuan, S., Dong, M., Su, J., Yu, C., Shen, Y., et al. (2005). The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics. 86, 674–684. doi: 10.1016/j.ygeno.2005.08.004
Huang, Y., Zhang, X., Wang, H. W., and Yu, L. (2022). Assembly of Tetraspanin-enriched macrodomains contains membrane damage to facilitate repair. Nat. Cell Biol. 24, 825–832. doi: 10.1038/s41556-022-00920-0
Hýsková, V., Bělonožníková, K., Chmelík, J., Hoffmeisterová, H., Čeřovská, N., Moravec, T., et al. (2024). Potyviral helper-component protease: multifaced functions and interactions with host proteins. Plants (Basel). 13, 1236. doi: 10.3390/plants13091236
Iskandar, H. M., Simpson, R. S., Casu, R. E., Bonnett, G. D., Maclean, D. J., and Manners, J. M. (2004). Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol. Biol. Rep. 22, 325–337. doi: 10.1007/BF02772676
Jankovičová, J., Sečová, P., Michalková, K., and Antalíková, J. (2020). Tetraspanins, more than markers of extracellular vesicles in reproduction. Int. J. Mol. Sci. 21, 7568. doi: 10.3390/ijms21207568
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of exosome composition. Cell. 177, 428–445.e18. doi: 10.1016/j.cell.2019.02.029
Jiang, J., Patarroyo, C., Garcia Cabanillas, D., Zheng, H., and Laliberté, J. F. (2015). The vesicle-forming 6K2 protein of turnip mosaic virus interacts with the COPII coatomer Sec24a for viral systemic infection. J. Virol. 89, 6695–6710. doi: 10.1128/JVI.00503-15
Jimenez-Jimenez, S., Hashimoto, K., Santana, O., Aguirre, J., Kuchitsu, K., and Cárdenas, L. (2019). Emerging roles of tetraspanins in plant inter-cellular and inter-kingdom communication. Plant Signal. Behav. 14, e1581559. doi: 10.1080/15592324.2019.1581559
Johnston, M. G., Breakspear, A., Samwald, S., Zhang, D., Papp, D., Faulkner, C., et al. (2023). Comparative phyloproteomics identifies conserved plasmodesmal proteins. J. Exp. Bot. 74, 1821–1835. doi: 10.1093/jxb/erad022
Kearly, A., Nelson, A. D. L., Skirycz, A., and Chodasiewicz, M. (2024). Composition and function of stress granules and P-bodies in plants. Semin. Cell Dev. Biol. 156, 167–175. doi: 10.1016/j.semcdb.2022.11.008
Kovalenko, O. V., Metcalf, D. G., DeGrado, W. F., and Hemler, M. E. (2005). Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct. Biol. 5, 11. doi: 10.1186/1472-6807-5-11
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Le Naour, F., Rubinstein, E., Jasmin, C., Prenant, M., and Boucheix, C. (2000). Severely reduced female fertility in CD9-deficient mice. Science. 287, 319–321. doi: 10.1126/science.287.5451.319
Li, F., Lu, Y., Xi, K., Li, Y., Chen, X., Wang, P., et al. (2024). Interkingdom communication via extracellular vesicles: unraveling plant and pathogen interactions and its potential for next-generation crop protection. Microorganisms. 12, 2392. doi: 10.3390/microorganisms12122392
Li, Z., Wang, G., Liu, X., Wang, Z., Zhang, M., and Zhang, J. (2021). Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genomics 22, 456. doi: 10.1186/s12864-021-07799-5
Li, F., Zhang, C., Tang, Z., Zhang, L., Dai, Z., Lyu, S., et al. (2020). A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol. 228, 622–639. doi: 10.1111/nph.16716
Lin, J., Zhao, J., Du, L., Wang, P., Sun, B., Zhang, C., et al. (2024). Activation of MAPK-mediated immunity by phosphatidic acid in response to positive-strand RNA viruses. Plant Commun. 5, 100659. doi: 10.1016/j.xplc.2023.100659
Ling, H., Wu, Q., Guo, J., Xu, L., and Que, Y. (2014). Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PloS One 9, e97469. doi: 10.1371/journal.pone.0097469
Mani, B., Agarwal, M., and Katiyar-Agarwal, S. (2015). Comprehensive expression profiling of rice tetraspanin genes reveals diverse roles during development and abiotic stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01088
Mani, B., Kaur, I., Dhingra, Y., Saxena, V., Krishna, G. K., Kumar, R., et al. (2025). Tetraspanin 5 orchestrates resilience to salt stress through the regulation of ion and reactive oxygen species homeostasis in rice. Plant Biotechnol. J. 23, 51–71. doi: 10.1111/pbi.14476
Mathieu, M., Névo, N., Jouve, M., Valenzuela, J. I., Maurin, M., Verweij, F. J., et al. (2021). Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 12, 4389. doi: 10.1038/s41467-021-24384-2
Miyado, K., Yamada, G., Yamada, S., Hasuwa, H., Nakamura, Y., Ryu, F., et al. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science. 287, 321–324. doi: 10.1126/science.287.5451.321
Movahed, N., Cabanillas, D. G., Wan, J., Vali, H., Laliberté, J. F., and Zheng, H. (2019). Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 180, 1375–1388. doi: 10.1104/pp.19.00381
Niehl, A. and Heinlein, M. (2011). Cellular pathways for viral transport through plasmodesmata. Protoplasma. 248, 75–99. doi: 10.1007/s00709-010-0246-1
Ninomiya, M., Inoue, J., Krueger, E. W., Chen, J., Cao, H., Masamune, A., et al. (2021). The exosome-associated tetraspanin CD63 contributes to the efficient assembly and infectivity of the hepatitis B virus. Hepatol. Commun. 5, 1238–1251. doi: 10.1002/hep4.1709
Nolte-’t Hoen, E., Cremer, T., Gallo, R. C., and Margolis, L. B. (2016). Extracellular vesicles and viruses: are they close relatives? Proc. Natl. Acad. Sci. U. S. A. 113, 9155–9161. doi: 10.1073/pnas.1605146113
Parra-Aguilar, T. J., Sarmiento-López, L. G., Santana, O., Olivares, J. E., Pascual-Morales, E., Jiménez-Jiménez, S., et al. (2023). TETRASPANIN 8–1 from Phaseolus vulgaris plays a key role during mutualistic interactions. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1152493
Pollari, M. E., Aspelin, W. W. E., Wang, L., and Mäkinen, K. M. (2024). The molecular maze of potyviral and host protein interactions. Annu. Rev. Virol. 11, 147–170. doi: 10.1146/annurev-virology-100422-034124
Poveda, E., Tabernilla, A., Fitzgerald, W., Salgado-Barreira, Á., Grandal, M., Pérez, A., et al. (2022). Massive release of CD9+ microvesicles in human immunodeficiency virus infection, regardless of virologic control. J. Infect. Dis. 225, 1040–1049. doi: 10.1093/infdis/jiaa375
Prakash, V., Nihranz, C. T., and Casteel, C. L. (2023). The potyviral protein 6K2 from turnip mosaic virus increases plant resilience to drought. Mol. Plant Microbe Interact. 36, 189–197. doi: 10.1094/mpmi-09-22-0183-r
Qin, S., Li, W., Zeng, J., Huang, Y., and Cai, Q. (2024). Rice tetraspanins express in specific domains of diverse tissues and regulate plant architecture and root growth. Plant J. 117, 892–908. doi: 10.1111/tpj.16536
Reimann, R., Kost, B., and Dettmer, J. (2017). Tetraspanins in plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00545
Revers, F. and García, J. A. (2015). Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. doi: 10.1016/bs.aivir.2014.11.006
Ruf, A., Oberkofler, L., Robatzek, S., and Weiberg, A. (2022). Spotlight on plant RNA-containing extracellular vesicles. Curr. Opin. Plant Biol. 69, 102272. doi: 10.1016/j.pbi.2022.102272
Rui, P., Jia, Z., Fang, X., Yu, T., Mao, W., Lin, J., et al. (2025). A plant viral effector subverts FER-RALF1 module-mediated intracellular immunity. Plant Biotechnol. J. 23, 2734–2751. doi: 10.1111/pbi.70099
Seigneuret, M., Delaguillaumie, A., Lagaudrière-Gesbert, C., and Conjeaud, H. (2001). Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276, 40055–40064. doi: 10.1074/jbc.M105557200
Solovyev, A. G., Atabekova, A. K., Lezzhov, A. A., Solovieva, A. D., Chergintsev, D. A., and Morozov, S. Y. (2022). Distinct mechanisms of endomembrane reorganization determine dissimilar transport pathways in plant RNA viruses. Plants (Basel). 11, 2403. doi: 10.3390/plants11182403
Umeda, R., Satouh, Y., Takemoto, M., Nakada-Nakura, Y., Liu, K., Yokoyama, T., et al. (2020). Structural insights into tetraspanin CD9 function. Nat. Commun. 11, 1606. doi: 10.1038/s41467-020-15459-7
Urcuqui-Inchima, S., Haenni, A. L., and Bernardi, F. (2001). Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. doi: 10.1016/s0168-1702(01)00220-9
Valli, A., López-Moya, J. J., and García, J. A. (2007). Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88, 1016–1028. doi: 10.1099/vir.0.82402-0
Wan, J., Cabanillas, D. G., Zheng, H., and Laliberté, J. F. (2015). Turnip mosaic virus moves systemically through both phloem and xylem as membrane-associated complexes. Plant Physiol. 167, 1374–1388. doi: 10.1104/pp.15.00097
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Wang, F., Muto, A., Van de Velde, J., Neyt, P., Himanen, K., Vandepoele, K., et al. (2015). Functional analysis of the Arabidopsis TETRASPANIN gene family in plant growth and development. Plant Physiol. 169, 2200–2214. doi: 10.1104/pp.15.01310
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012b). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, F., Vandepoele, K., and Van Lijsebettens, M. (2012a). Tetraspanin genes in plants. Plant Sci. 190, 9–15. doi: 10.1016/j.plantsci.2012.03.005
Wang, T., Wang, B., Hua, X., Tang, H., Zhang, Z., Gao, R., et al. (2023). A complete gap-free diploid genome in Saccharum complex and the genomic footprints of evolution in the highly polyploid Saccharum genus. Nat. Plants. 9, 554–571. doi: 10.1038/s41477-023-01378-0
Wang, J., Zhang, Q., Tung, J., Zhang, X., Liu, D., Deng, Y., et al. (2024). High-quality assembled and annotated genomes of Nicotiana tabacum and Nicotiana benthamiana reveal chromosome evolution and changes in defense arsenals. Mol. Plant 17, 423–437. doi: 10.1016/j.molp.2024.01.008
Wei, T., Zhang, C., Hong, J., Xiong, R., Kasschau, K. D., Zhou, X., et al. (2010). Formation of complexes at plasmodesmata for Potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PloS Pathog. 6, e1000962. doi: 10.1371/journal.ppat.1000962
Wright, M. D., Ni, J., and Rudy, G. B. (2000). The L6 membrane proteins—a new four-transmembrane superfamily. Protein Sci. 9, 1594–1600. doi: 10.1110/ps.9.8.1594
Xiao, H., Lord, E., and Sanfaçon, H. (2022). Proteolytic processing of plant proteins by Potyvirus NIa proteases. J. Virol. 96, e0144421. doi: 10.1128/JVI.01444-21
Xie, J., Jiang, T., Li, Z., Li, X., Fan, Z., and Zhou, T. (2021). Sugarcane mosaic virus remodels multiple intracellular organelles to form genomic RNA replication sites. Arch. Virol. 166, 1921–1930. doi: 10.1007/s00705-021-05077-z
Xu, J. and Chua, N. H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 21, 3270–3279. doi: 10.1105/tpc.109.070078
Xue, M., Arvy, N., and German-Retana, S. (2023). The mystery remains: how do potyviruses move within and between cells? Mol. Plant Pathol. 24, 1560–1574. doi: 10.1111/mpp.13383
Xue, B., Guo, J., Que, Y., Fu, Z., Wu, L., and Xu, L. (2014). Selection of suitable endogenous reference genes for relative copy number detection in sugarcane. Int. J. Mol. Sci. 15, 8846–8862. doi: 10.3390/ijms15058846
Yang, Z., Ceng, G., Yu, Q., Jiao, W., Zeng, K., Luo, T., et al. (2024). Identification and characterization of the Remorin gene family in Saccharum and the involvement of ScREM1.5e-1/-2 in SCMV infection on sugarcane. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1365995
Yang, Z., Dong, M., Cheng, G., Liu, S., Zhang, H., Shang, H., et al. (2021). Selective interaction of sugarcane eIF4E with VPgs from sugarcane mosaic pathogens. Viruses. 13, 518. doi: 10.3390/v13030518
Youn, J. Y., Dyakov, B. J. A., Zhang, J., Knight, J. D. R., Vernon, R. M., Forman-Kay, J. D., et al. (2019). Properties of stress granule and P-body proteomes. Mol. Cell. 76, 286–294. doi: 10.1016/j.molcel.2019.09.014
Zhang, H., Cheng, G., Yang, Z., Wang, T., and Xu, J. (2019). Identification of sugarcane host factors interacting with the 6K2 protein of the sugarcane mosaic virus. Int. J. Mol. Sci. 20, 3867. doi: 10.3390/ijms20163867
Zhang, Y., Pan, C., Wang, S., Zhou, Y., Chen, J., Yu, X., et al. (2025b). Distinctive function of tetraspanins: implication in viral infections. Virulence. 16, 2474188. doi: 10.1080/21505594.2025.2474188
Zhang, J., Qi, Y., Hua, X., Wang, Y., Wang, B., Qi, Y., et al. (2025c). The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum. Nat. Genet. 57, 242–253. doi: 10.1038/s41588-024-02033-w
Zhang, Q., Qi, Y., Pan, H., Tang, H., Wang, G., Hua, X., et al. (2022). Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 54, 885–896. doi: 10.1038/s41588-022-01084-1
Zhang, J., Qiu, Y., and Xu, K. (2020). Characterization of GFP-AtPEN1 as a marker protein for extracellular vesicles isolated from Nicotiana benthamiana leaves. Plant Signal. Behav. 15, 1791519. doi: 10.1080/15592324.2020.1791519
Zhang, Z., Todeschini, T. C., Wu, Y., Kogay, R., Naji, A., Rodriguez., C., et al. (2025a). Kiwa is a membrane-embedded defense supercomplex activated at phage attachment sites. Cell. 8674 (25), 00791–3. doi: 10.1016/j.cell.2025.07.002
Zhang, H., Yang, Z., Cheng, G., Luo, T., Zeng, K., Jiao, W., et al. (2024). Sugarcane mosaic virus employs 6K2 protein to impair ScPIP2;4 transport of H2O2 to facilitate virus infection. Plant Physiol. 194, 715–731. doi: 10.1093/plphys/kiad567
Zhang, J., Zhang, Q., Li, L., Tang, H., Zhang, Q., Chen, Y., et al. (2019). Recent polyploidization events in three Saccharum founding species. Plant Biotechnol. J. 17, 264–274. doi: 10.1111/pbi.12962
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., Ma, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Zhao, Y., Zhou, Y., Xu, J., Fan, S., Zhu, N., Meng, Q., et al. (2024). Cross-kingdom RNA transport based on extracellular vesicles provides innovative tools for plant protection. Plants (Basel). 13, 2712. doi: 10.3390/plants13192712
Zhu, J., Qiao, Q., Sun, Y., Xu, Y., Shu, H., Zhang, Z., et al. (2023). Divergent sequences of tetraspanins enable plants to specifically recognize microbe-derived extracellular vesicles. Nat. Commun. 14, 4877. doi: 10.1038/s41467-023-40623-0
Zhu, T., Sun, Y., and Chen, X. (2022). Arabidopsis tetraspanins facilitate virus infection via membrane-recognition GCCK/RP motif and cysteine residues. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.805633
Keywords: sugarcane, sugarcane mosaic virus, tetraspanin, potyvirus, 6K2 protein
Citation: Cui Z, Li Y, Zeng K, Yang Z, Yu Q, Liu H, Zhan Z, Zhang H, Huang G and Xu J (2025) Identification of the tetraspanin gene family in sugarcane and its response to sugarcane mosaic virus infection. Front. Plant Sci. 16:1684431. doi: 10.3389/fpls.2025.1684431
Received: 12 August 2025; Accepted: 30 September 2025;
Published: 27 October 2025.
Edited by:
Ahmed Abdelkhalek, The National Institute of Horticultural Research, PolandReviewed by:
Hui Ling, Yulin Normal University, ChinaVed Prakash, The Ohio State University, United States
Wanwan Liang, Chinese Academy of Agricultural Sciences, China
Copyright © 2025 Cui, Li, Zeng, Yang, Yu, Liu, Zhan, Zhang, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingsheng Xu, eHVqaW5nc2hlbmdAMTI2LmNvbQ==
 Zhiyuan Cui
Zhiyuan Cui Yifei Li
Yifei Li Kang Zeng
Kang Zeng Zongtao Yang
Zongtao Yang Quanxin Yu
Quanxin Yu Haoming Liu
Haoming Liu Zhenhe Zhan
Zhenhe Zhan Hai Zhang
Hai Zhang Guoqiang Huang
Guoqiang Huang Jingsheng Xu
Jingsheng Xu