- 1Key Laboratory of Medicinal Plant Biology of Yunnan Province, National & Local Joint Engineering Research Center on Germplasms Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, China
- 2College of Agronomy & Biotechnology, Yunnan Agricultural University, Kunming, China
- 3Yunnan Characteristic Plant Extraction Laboratory, Kunming, Yunnan, China
This study identifies 148 bHLH transcription factors in Angelica sinensis and reveals four putative candidates associated with ferulic acid biosynthesis, providing a genetic foundation for metabolic engineering to enhance the plant’s medicinal value. The basic helix-loop-helix (bHLH) proteins regulate plant growth, development, stress responses, and secondary metabolites. While well-characterized in woody plants, they remain unexplored in Angelica sinensis (A. sinensis), a medicinal plant renowned for bioactive compounds including ferulic acid. Therefore, we systematically identified and characterized bHLH transcription factors in A. sinensis through whole-genome analysis and transcriptome profiling, identifying putative genes potentially regulating ferulic acid biosynthesis. Bioinformatic analyses were employed to characterize the physicochemical properties, gene structures, conserved motifs/domains, phylogenetic relationships, chromosome localization, collinearity, cis-acting elements, and transcriptome expression patterns of AsbHLHs. A total of 148 AsbHLH genes were annotated from the genomic database of A. sinensis, classified into 16 subfamilies based on the phylogenetic analysis. Results revealed that these transcription factors encode hydrophilic proteins (83–741 aa; 9.6–80.8 kDa), with nearly all localized to the nucleus. Gene structure analysis showed exon numbers ranging from 1 to 18, while MEME motif analysis identified five conserved motifs (1–5) shared across most AsbHLH proteins. Promoter analysis uncovered abundant cis-elements associated with growth, secondary metabolism (including ferulic acid biosynthesis), and stress responses. WGCNA revealed turquoise module contained 40 bHLH and five phenylpropanoid pathway-specific genes, from which PPI and phylogenetic analyses pinpointed four putative genes potentially associated with ferulic acid production. Quantitative RT-PCR validated these candidates, showing expression patterns consistent with transcriptome data. This study provides the first comprehensive genomic/transcriptomic resource for AsbHLH genes in A. sinensis, highlighting their secondary metabolic roles. Identified candidates enable genetic engineering strategies to boost ferulic acid production, enhancing A. sinensis’ medicinal value.
Introduction
A perennial herbaceous plant, Angelica sinensis (Oliv.) Diels belongs to the Apiaceae family, has been broadly used in traditional medicines due to the therapeutic properties of its dried root, which include blood nourishment, triggering blood circulation, regulating menstruation, relieving pain, and endorsing intestinal motility (Zhang et al., 2012; Shedoeva et al., 2019; Yuan et al., 2019; Guo et al., 2020; Chen et al., 2021; Han et al., 2023; Tian et al., 2024). These pharmacological properties are mainly attributed to bioactive compounds such as phenolic acids, flavonoids, coumarins, polysaccharides, and phthalides (Sarker and Nahar, 2004; Batiha et al., 2022; Sabeel et al., 2023; Wang et al., 2023; Mishra and Paliwal, 2025; Ren et al., 2025; Zhao et al., 2025). Recent studies have primarily focused on the chemical composition (Chen et al., 2022; Sun et al., 2024), pharmacological effects (Kim et al., 2023; Hu et al., 2024), and biological activities (Lee et al., 2023; Wang et al., 2023) of A. sinensis, while the systematic research on germplasm quality improvement remains limited. Therefore, there is an urgent need to advance breeding programs for quality improvement and to explore the putative regulatory genes associated with the biosynthesis of its secondary metabolites.
The genome-wide study is foundational for explicating gene families and their regulatory networks, and the reliability of such analysis depends upon the quality and wholeness of the reference genomic resource used. For Angelica sinensis, the genomic database has been pointedly improved in recent years. Subsequently, the initial draft genomes and several high-quality, chromosome-level assemblies have now been published (Han et al., 2022; Li et al., 2023; Dang et al., 2024; Wu et al., 2024). These databases provide complementary tools for the research community. It is extensively documented that exploiting the most contiguous and highly annotated assembly is the finest practice for exploiting the extensiveness of gene family identification (Dang et al., 2024; Wu et al., 2024). The existence of multiple genomes also suggests a unique chance for cross-validation of findings, confirming strength and correctness.
Ferulic acid is a major phenolic compound plentiful in Chinese herbal medicines, plays a key structural role in plant cell walls by esterifying with polysaccharides and facilitating cross-linking between lignin and hemicellulose (Mnich et al., 2020; Robert et al., 2022). In A. sinensis, ferulic acid is a principal bioactive compound (Giacomelli et al., 2017; Dong et al., 2022), contributing significantly to its traditional practices, particularly in treating gynecological disorders (Jiao et al., 2022; Pei et al., 2024). Its therapeutic benefits, especially improved blood circulation, menstrual regulation, and pain relief, have been largely attributed to its presence in the plant (Cheng et al., 2012; Li et al., 2020; Batiha et al., 2022; Yang X. et al., 2023; Shen et al., 2025). Current research has further tinted ferulic acid’s role in oxidative stress defense and its potential in averting chronic diseases such as cardiovascular disorders, diabetes, and cancer, making it valuable for clinical applications, including anti-inflammatory, anti-carcinogenic, and cardioprotective effects (Kiokias et al., 2020; Li et al., 2021; Afnan et al., 2022; Ma et al., 2022; Pandi et al., 2022; Bao et al., 2023).
At the transcriptional level, several compounds regarding the phenylpropanoid pathway, including ferulic acid biosynthesis, are primarily regulated by “bHLH (basic helix-loop-helix)” transcription factor (TF) (Zhao et al., 2023; Gao et al., 2024; Chen et al., 2024). The bHLH gene family signifies one of the largest TF families in plants, tangled in diverse biological processes including growth, development, and stress responses (Ke et al., 2020; Hao et al., 2021; Thoben and Pucker, 2023; Zuo et al., 2023; Zhang et al., 2025). These proteins contain a conserved bHLH domain of around 60 amino acids that simplifies DNA binding, classically identifying E-box motifs (CANNTG) in promoter regions (Yesudhas et al., 2017; de Martin et al., 2021; Guo et al., 2021; Zhang et al., 2025). Functionally, bHLH TFs regulate several physiological processes through homo- or heterodimer establishment, including hormone signaling, stress adaptation, and secondary metabolism (Feller et al., 2011; Goossens et al., 2016; Meraj et al., 2020; Jan et al., 2021; Khan et al., 2025). Numerous studies have predominantly focused on their role in secondary metabolism, particularly in the biosynthesis of phenylpropanoids such as flavonoids, ferulic acid, and lignins, which are necessary for plant defense mechanisms (Wang et al., 2018; Sharma et al., 2019; Guo et al., 2021; Hussain and Reigosa, 2021; Jan et al., 2021; Rabeh et al., 2025). For instance, MYC, a well-characterized bHLH TF, regulates the jasmonate signaling pathway essential for plant defense against herbivores and pathogens (Niu et al., 2011; Nakata et al., 2013; Du et al., 2018; Wei et al., 2022; Jia et al., 2025; Kim et al., 2025; Liu et al., 2025), while other bHLH TFs are involved in anthocyanin biosynthesis, iron homeostasis, and light-mediated growth (Li et al., 2020; Buti et al., 2020; Jing and Lin, 2020; Qian et al., 2021; Hao et al., 2021; Liu et al., 2021; LaFountain and Yuan, 2021).
Despite the long history of medicinal use of A. sinensis, the regulatory mechanisms underlying the biosynthesis of key compounds like ferulic acid remain poorly understood. Ferulic acid is synthesized via the phenylpropanoid pathway (Dong et al., 2022; Guo et al., 2023), yet the specific roles of bHLH TFs in modulating this pathway are unclear. Therefore, elucidating these potential regulatory mechanisms is crucial for enhancing the therapeutic potential of A. sinensis. Given the importance of ferulic acid and the central role of bHLH TFs in secondary metabolism, this study aims to conduct a genome-wide identification of bHLH transcription factors in A. sinensis to identify putative regulators associated with ferulic acid biosynthesis. To attain this, we used the chromosome-level genome assembly of Angelica sinensis (female ginseng) from Han et al., 2022; a resource selected for its inclusive annotation and established value in prior gene family studies. Based on this reference genome, we screened and identified bHLH transcription factors, followed by comprehensive bioinformatics analyses including protein physicochemical property assessment, phylogenetic and evolutionary analysis, gene structure, domains, and conserved motif identification, chromosomal localization, synteny analysis, and cis-acting element prediction. By using transcriptome data, gene expression patterns were further investigated, and qRT-PCR was performed to screen out bHLH TFs potentially associated with ferulic acid biosynthesis in A. sinensis. These efforts will provide a theoretical foundation for future studies on the molecular mechanisms of putative bHLH-regulated ferulic acid biosynthesis and support quality breeding programs for A. sinensis.
Materials and methods
Plant materials
The genomic sequence and GFF annotation file of A. sinensis were derived from the PlantGIR Genomic Database (http://plantgir.cn/Download) (Han et al., 2022). Genome-derived protein sequences were generated with GFFread (v0.12.7), followed by isolation of the longest isoform per gene via a Perl-based filtering script. The reference genome and gene family categorization data for A. thaliana were acquired from the TAIR public repository (https://www.arabidopsis.org/) (Swarbreck et al., 2007). The transcriptome data of A. sinensis from different tissues (root, leaf and stem) were sourced from our previous study (GSA: CRA016571), publicly accessible in the Genome Sequence Archive (GSA) at the CNCB-NGDC (https://ngdc.cncb.ac.cn/gsa) (Arshad et al., 2024).
Identification of bHLH family genes in A. sinensis
The bHLH transcription factor family genes in A. sinensis were identified through a comprehensive bioinformatics pipeline using the Pfam database (http://pfam.xfam.org/search) for domain prediction (PF00010) and TBtools (v2.096) for initial screening. We obtained reference sequences (225 A. thaliana bHLH proteins) from PlantTFDB (https://planttfdb.gao-lab.org/) and performed HMMER (v3.3.1) searches against the A. sinensis genome with sequential E-value thresholds (1×10-5 followed by 0.001), including domain alignment with ClustalW (v2.1) and HMM profile refinement (Finn et al., 2011). Putative genes were validated through BLAST comparison with A. thaliana sequences, SMART database screening for SANT domains, and NCBI-CDD verification (E-value ≤ 0.01), with final identification based on the intersection of HMMER and BLAST results to ensure both domain presence and evolutionary conservation.
Prediction of physicochemical properties, secondary structure, and subcellular localization of Angelica sinensis bHLH proteins
The physicochemical properties of identified bHLH proteins, including relative molecular weight, isoelectric point (pI), and grand average of hydropathicity (GRAVY), were predicted using ExPASy ProtParam (https://web.expasy.org/protparam/) (Dong et al., 2024). Subcellular localization was inferred with Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/), and secondary structure composition (α-helices, β-sheets, coils, etc.) was analyzed via SOPMA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). Furthermore, the amino acid configuration of the bHLH protein was examined using DNAMAN.
Phylogenetic and evolutionary analysis
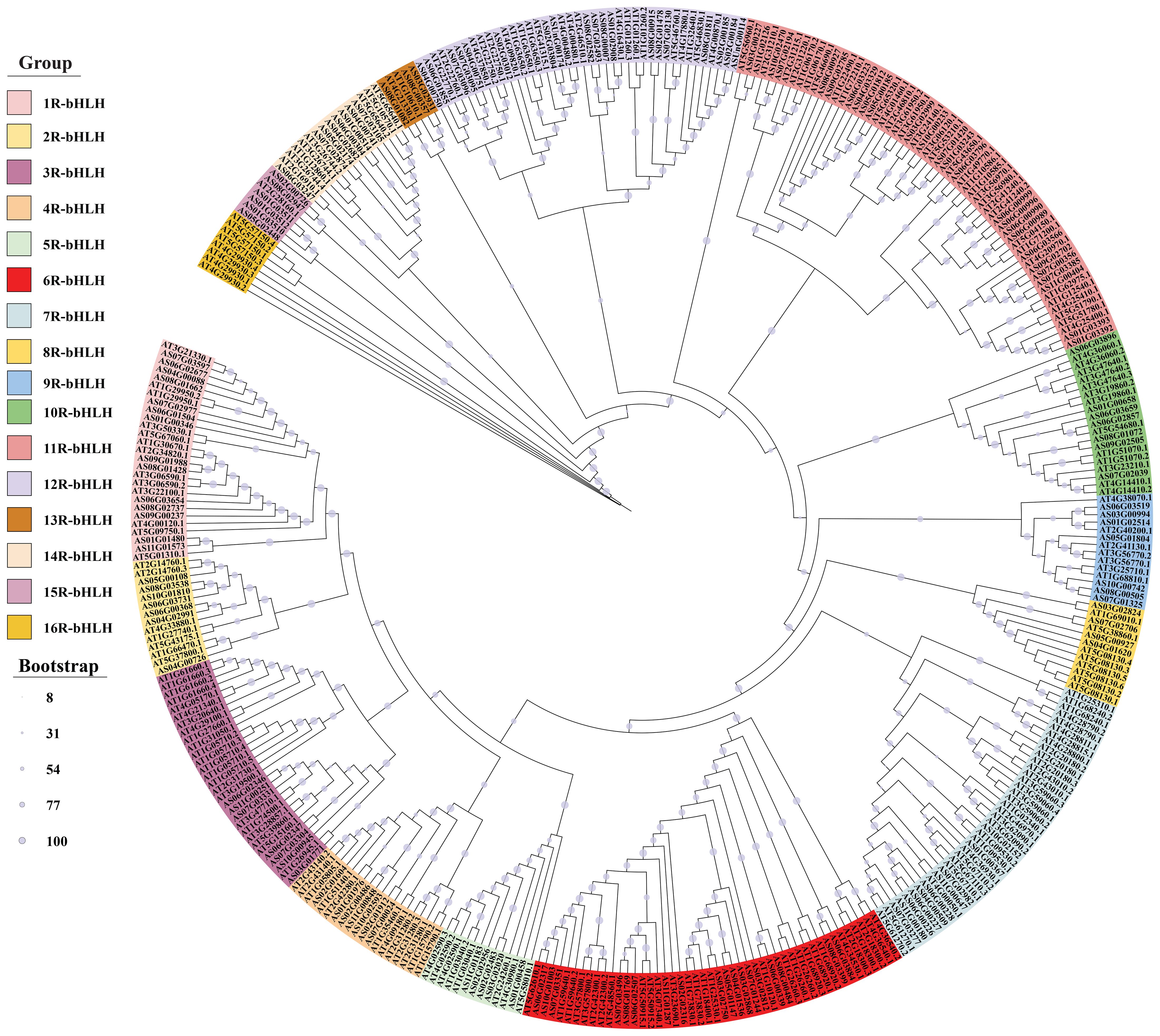
Phylogenetic analysis of AsbHLHs was performed using MEGA 11.0 (Tamura et al., 2021). The maximum likelihood (ML) tree was constructed with 225 A. thaliana bHLH sequences (from PlantTFDB) as reference, employing the VT+R6 model (automatically selected as optimal) and 1000 bootstrap replicates. Branch support values (%) are indicated by circles (proportional to bootstrap values) (Price et al., 2010; Horiike, 2016). The Newick-formatted tree (*.NWK) was visualized and annotated using the iTOL web tool (https://itol.embl.de/). Subfamily classification of A. sinensis bHLH TFs was inferred based on their clustering with Arabidopsis orthologs.
Structural analysis of AsbHLHs
The genomic coordinates and exon-intron structures were extracted from the genome’s GFF3 annotation file. TBtools was then used to generate visual representations of gene architectures, with coding sequences (CDS), untranslated regions (UTRs), and introns annotated (Chen and Xia, 2022; Wei et al., 2023; Rizwan et al., 2024).
Identification and visualization of conserved motifs
Conserved motifs within the A. sinensis bHLH protein sequences were identified using MEME Suite (v5.5.2; http://meme-suite.org) with the following parameters: maximum motif count = 10, E-value threshold < 0.01, and default settings for width and repetitions (Bailey et al., 2006). The motif architectures were graphically mapped to gene structures using TBtools (v2.096) to analyze positional conservation.
Conserved domains analysis
The conserved domains were identified using NCBI’s CD-search tool (Conserved Domain Database; https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), with default parameters for domain detection (Zhou et al., 2020). Domain architectures were visualized using TBtools to illustrate structural features such as the bHLH DNA-binding and dimerization regions.
Cis-acting element analysis in AsbHLH promoters
The 2,000 bp promoter regions upstream of transcription start sites (TSS) of A. sinensis bHLH genes were extracted using TBtools. Cis-acting regulatory elements were identified through the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot, 2002), and their results were statistically analyzed and visualized via ggplot2 (v3.5.1) in R to highlight element distribution and functional categories (Wickham, 2011; He et al., 2021; Li et al., 2024; Tobin et al., 2025).
Chromosomal localization and synteny analysis
Chromosomal positions were mapped using genome annotation data visualized with TBtools. Intraspecies synteny analysis was performed by self-aligning A. sinensis bHLH protein sequences using the “One Step MCScanX-SuperFast” tool in TBtools, with results displayed through “Advanced Circos” visualization. For interspecies analysis, the A. thaliana genome annotation (NCBI) was aligned with A. sinensis bHLH genes using the same MCScanX pipeline, with syntenic relationships illustrated via Circos plots (Zhang et al., 2025).
Expression pattern analysis and qRT-PCR
Gene expression analysis of A. sinensis bHLH transcription factors was performed using transcriptome data from our previous study and publicly available raw sequences deposited in the Genome Sequence Archive (GSA: CRA016571) at the National Genomic Data Center (https://ngdc.cncb.ac.cn/gsa). Tissue-specific expression patterns of bHLH genes, as well as the phenylpropanoid pathway-specific genes responsible for ferulic acid regulation in A. sinensis, were quantified using FPKM values in roots, leaves, and stems, and visualized as clustered heatmaps via TBtools, with color gradients representing expression intensity. Weighted gene co-expression network analysis (WGCNA) was performed to construct gene co-expression networks. The protein interaction analysis of differentially expressed genes is based on the STRING database of known and predicted protein–protein interactions. WGCNA was performed as described previously (Langfelder and Horvath, 2008). Protein-protein interaction (PPI) network was constructed using Cytoscape to analyze the interaction among bHLH and phenylpropanoid pathway-specific genes identified in the precise module to classify putative genes regarding ferulic acid biosynthesis. Further, we performed phylogenetic analysis to screen identified putative bHLH proteins potentially associated with ferulic acid biosynthesis. qRT-PCR validation was performed using primers designed with Primer3 (https://primer3.ut.ee/; Supplementary Table 1) in 20 μL reactions containing SYBR qPCR SuperMix Plus (10 μL), primers (0.4 μL each), cDNA (1 μL), and ddH2O (8.2 μL), with thermal cycling conditions of 95 °C for 30 s; 40 cycles of 95 °C for 10 s and 60 °C for 20 s; followed by melt curve analysis. For the calculation of specific gene expression differences, the previously reported ACTIN gene was used as an internal reference gene (Arshad et al., 2024). Relative expression (2-ΔΔCt method) was calculated and visualized using GraphPad Prism 9.5 ( (Livak and Schmittgen, 2001; Wu et al., 2025).
Results
Identification, physicochemical properties, and secondary structure prediction of AsbHLHs
A combined study of genome-wide and full-length transcriptome-wide was carried out to screen and identify bHLH genes in A. sinensis via publicly available genomic sequences and our previously published full-length transcriptome data (GSA: CRA016571; https://ngdc.cncb.ac.cn/gsa). We identified a total of 148 AsbHLH family members based on the A. sinensis genome (Supplementary Table 2). To further characterize these identified AsbHLHs, we analyzed the physicochemical properties of the putative proteins. These 148 AsbHLH proteins displayed a diversity in length, molecular weight, and theoretical isoelectric points (PIs). Results showed that the 148 AsbHLH proteins’ length ranged from 83 to 743 amino acid residues, with a relative molecular weight and PIs fluctuating from 9.6 kDa to 80.8 kDa, and 4.55 to 9.81, respectively (Supplementary Table 3). Notably, the gene AS10G02152 encoded the largest AsbHLH protein (743 aa, 80.8 kDa), while the AS06G02346 encoded the smallest (83 aa, 9.6 kDa). Furthermore, the fat coefficient of these proteins ranged from 53.21 to 107.95, signifying diverse lipid interaction potentials (Supplementary Table 3). The average PI of 148 AsbHLH proteins is about 6.7, ranging from a minimum PI of 4.5 (AS09G01988) to a maximum of 9.81 (AS11G00848). Among these, 47 bHLH proteins were basic (pI > 7.0), while the remaining were neutral or acidic. Additional analysis revealed that the average hydrophobicity index of all bHLH proteins is -0.56, indicating that the family inclusively has significant hydrophilic characteristics, with no hydrophobic protein members detected. The only AsbHLH proteins include AS03G00994, AS06G02317, AS02G00486, AS08G00915, AS06G03884, and AS06G03247 are stably present, while the other family members have an instability index of more than 40. The subcellular localization prediction of the 148 AsbHLHs revealed that 147 AsbHLH proteins are localized in the nucleus, while only one protein, AS07G00356, is located in the cytoplasm, indicating that these proteins have significantly nuclear localization characteristics. Moreover, an extrapolative analysis of the structural configuration of the Angelica bHLH family members was conducted. The results specified that the compositional ratio of the secondary structure of each Angelica bHLH protein differs dynamically, with the proportion of β-turns and extended chains being lower than that of random coils and α-helices (Supplementary Table 3).
Phylogenetic relationship between bHLH proteins of A. sinensis and Arabidopsis thaliana
To classify the A. sinensis bHLH protein subfamilies and recognize the evolutionary relationships among the bHLH proteins from A. sinensis and A. thaliana, a phylogenetic tree was built using the sequences of the 148 AsbHLH proteins and 225 A. thaliana bHLH proteins. Results indicated that the 148 AsbHLH proteins were clustered into 16 subfamilies (1R- to 16R-bHLH) according to the tree topology and classification of the bHLH superfamily in A. thaliana (Figure 1). The subfamilies showed the following gene distribution: 1R-bHLH (n=14), 2R-bHLH to 4R-bHLH, 9R-bHLH and 10R-bHLH (7 for each), 5R-bHLH (5), 6R-bHLH (18), 7R-bHLH (10), 8R-bHLH (4), 11R-bHLH (26), 12R-bHLH (19), 13R-bHLH (3), 14R-bHLH (8) and 15R-bHLH (6). There is no 16R-bHLH subfamily, suggesting that Angelica bHLHs closely related to Arabidopsis bHLH proteins may share similar functions. Among these groups, the 11R-bHLH subfamily was the most abundant with 26 gene members (39% of total bHLHs), followed by 12R-bHLH containing 19 proteins (28%), while 13R-bHLH contains only 3 bHLH proteins (4.4%). The small subfamilies (e.g., 13R-bHLH with 3 members and 5R-bHLH with 5 members) propose potential evolutionary specialization or conserved functions between A. sinensis and A. thaliana. This phylogenetic classification permits functional prediction of AsbHLHs based on characterized AtbHLH proteins within the same subgroups, mainly for processes like secondary metabolite biosynthesis and stress responses, though experimental validation remains necessary.
Gene structure and conserved motif analysis
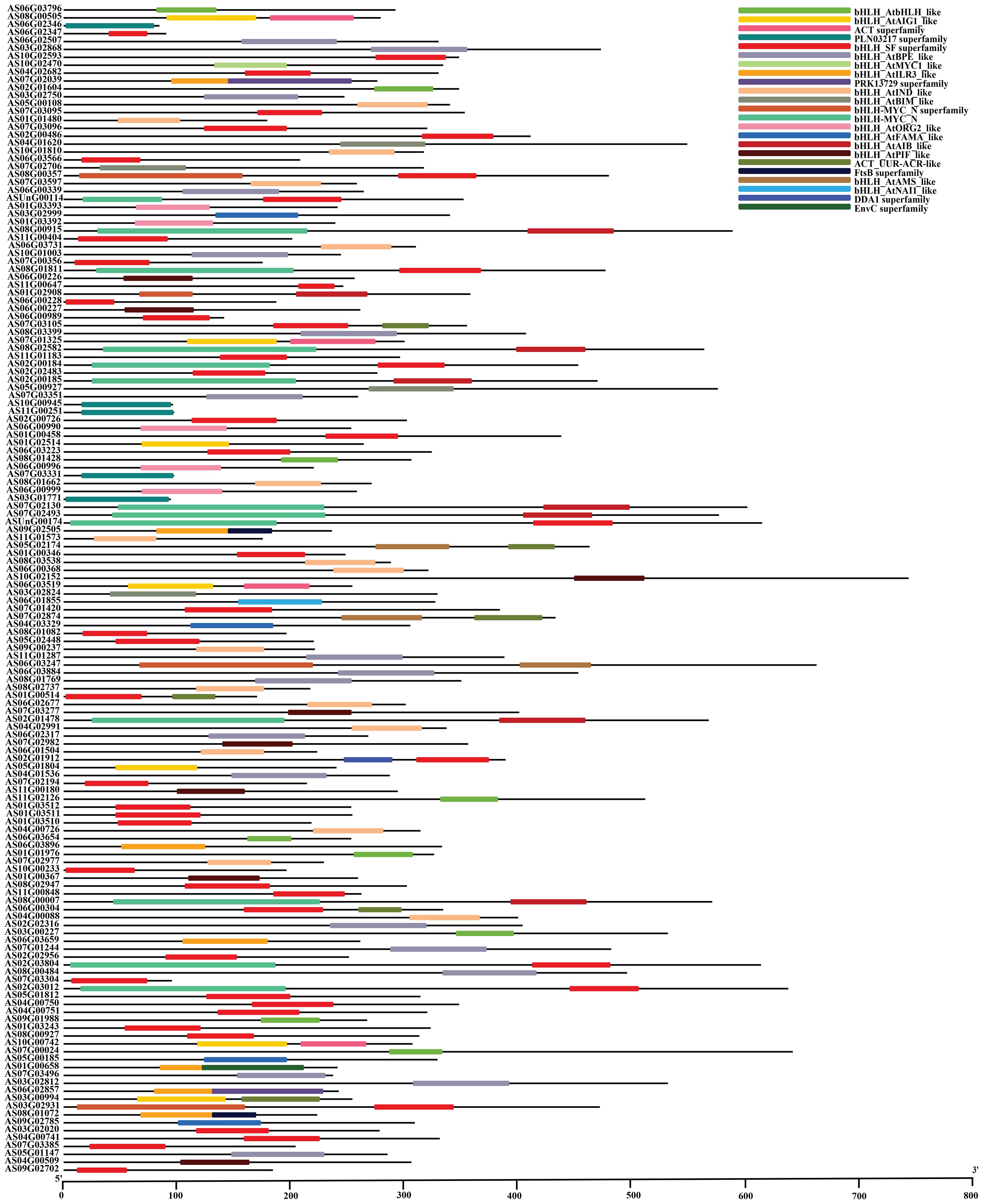
Phylogenetic analysis revealed that closely related bHLH members share similar motif compositions, demonstrating structural conservation (Figure 2A). MEME analysis identified 10 conserved motifs for bHLH proteins; motifs 1–5 were most prevalent, while motif 5 localized to the N-terminus and motifs 6/10 to the C-terminus (Supplementary Figure 1; Figure 2B). Gene structure analysis showed exon numbers ranging from 1 to 18 among the 148 bHLH genes, with 3–9 exons being most common; notably, AS02G01604 and AS02G03012 contained 18 exons and AS07G02706 contained 16 exons, while two genes, AS11G01573 and AS01G01480, possessed only one exon, suggesting potential functional diversification correlated with structural variation (Figure 2C).
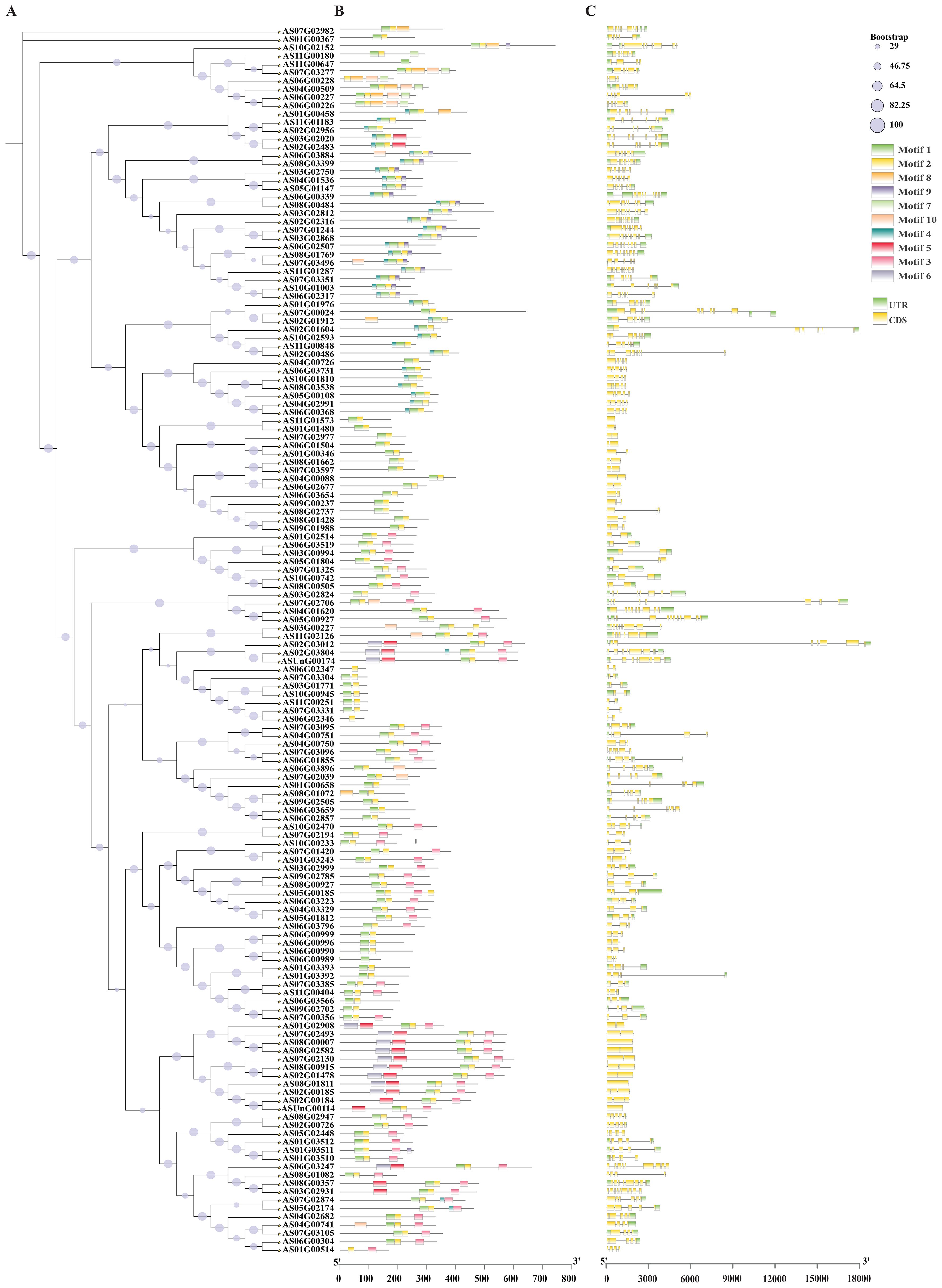
Figure 2. Evolutionary relationship, conserved motifs and genetic structures of bHLH genes in A. sinensis. (A) bHLH protein’s evolutionary relationship in A. sinensis; (B) Conserved motifs of bHLH genes in A. sinensis; (C) bHLH genes structure in A. sinensis.
Conserved domain analysis
The angelica bHLH proteins encompass a total of 23 distinct domain types (Figure 3). These 148 bHLH amino acid sequences contain 7 superfamilies, including PLN03217 superfamily, bHLH_SF superfamily, PRK13729 superfamily, bHLH_MYC_N superfamily, FtsB superfamily, DDA1 superfamily and EnvC superfamily. The amino acid sequences of 5 genes named AS06G02346, AS10G00945, AS11G00251, AS07G03331 and AS03G01771 belong to PLN03217 superfamily while only 4 genes AS08G00357, AS01G02908, AS06G03247 and AS03G02931, belong to bHLH_MYC_N superfamily. Other than superfamilies, the domain bHLH_MYC_N contains 13 gene sequences including ASUnG00114, AS08G00915, AS08G01811, AS08G02582, AS02G00184, AS02G00185, AS07G02130, AS07G02493, ASUnG00174, AS02G01478, AS08G00007, AS02G03804, and AS02G03012. Consequently, the multiple sequence alignment of the bHLH domain sequence of 148 AsbHLHs revealed that the basic region and two helixes were highly conserved in most of AsbHLH proteins, except that the basic region was absent in AS01G00154, AS06G00228, and AS06G02346 (Figure 4).
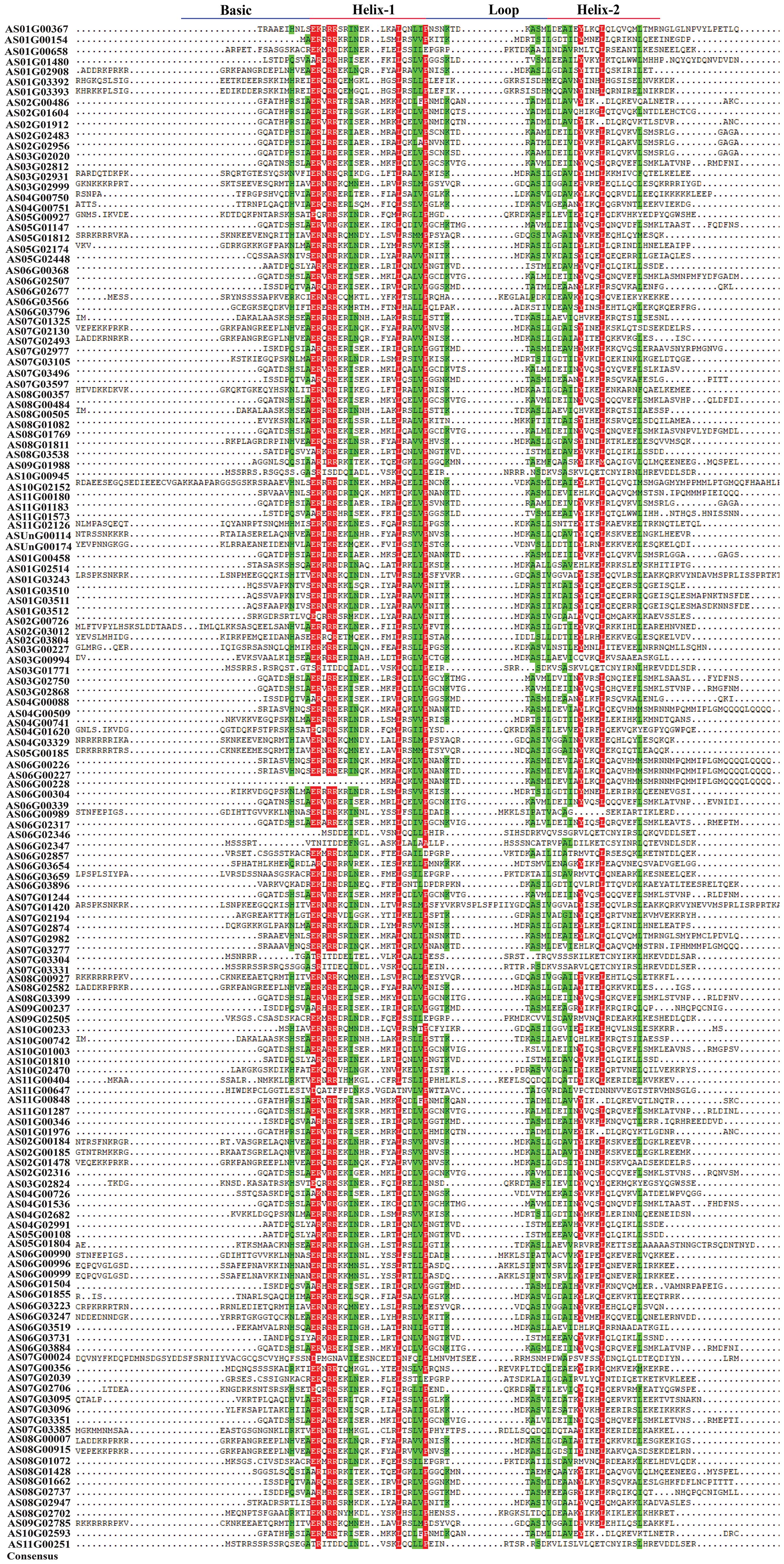
Figure 4. Multiple alignment of conserved domain amino acid sequences of multiple bHLH proteins from A. sinensis.
Chromosomal localization and synteny analysis
The results of the chromosomal localization of Angelica bHLH genes indicate that 148 Angelica bHLH genes are distributed on 11 chromosomes, with the highest number of bHLH genes (28) located on chr06 and the lowest (5) on chr09 (Figure 5A). Within the Angelica bHLH transcription factor family, there are 64 pairs of genes that have syntenic relationships (Figure 5B). Chromosomes chr01 to chr08 exhibit synteny relationships with bHLH genes located on chr02 to chr11, with chr02, chr03, and chr04 showing more distinct syntenic relationships with other chromosomes. Consequently, the synteny analysis of the AsbHLH and AtbHLH genomes reveals that there is a significant synteny relationship between the bHLH family members in A. sinensis and those in A. thaliana, with most of the genes on chromosomes 7, 8, and 11 of A. sinensis corresponding to genes on chromosomes 1 and 5 of A. thaliana (Figure 5C).
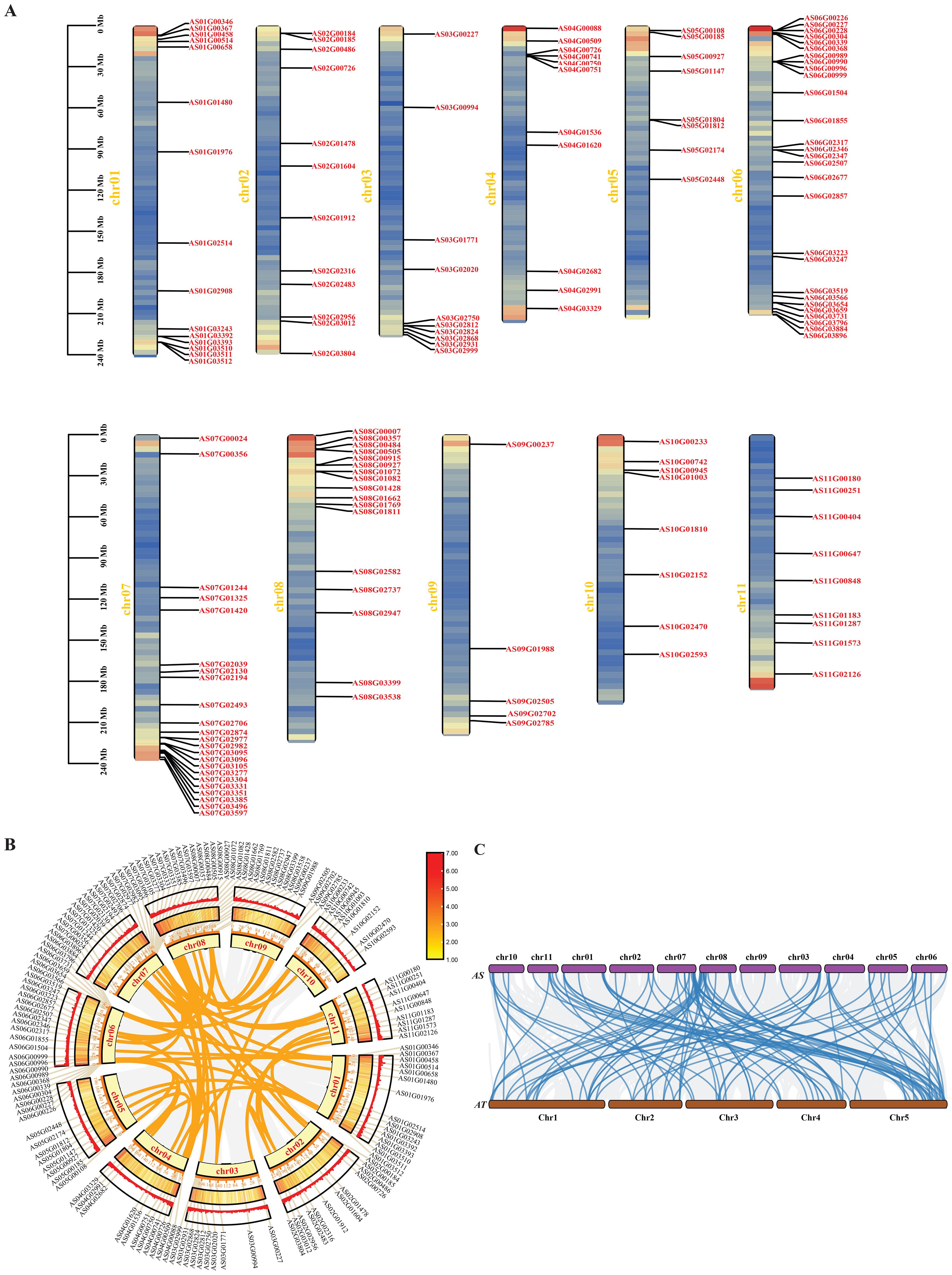
Figure 5. Chromosomal localization and synteny analysis. (A) Chromosomal distribution and regional duplication of 148 bHLH genes of A. sinensis. The scale bar on the left indicates the length (Mb) of chromosomes; (B) The collinearity circle diagram of the A. sinensis genome; (C) Collinearity analysis among the genomes of A. sinensis and A. thaliana; the blue line represents the bHLH gene pairs..
Cis-acting elements in the promoter region of the AsbHLHs
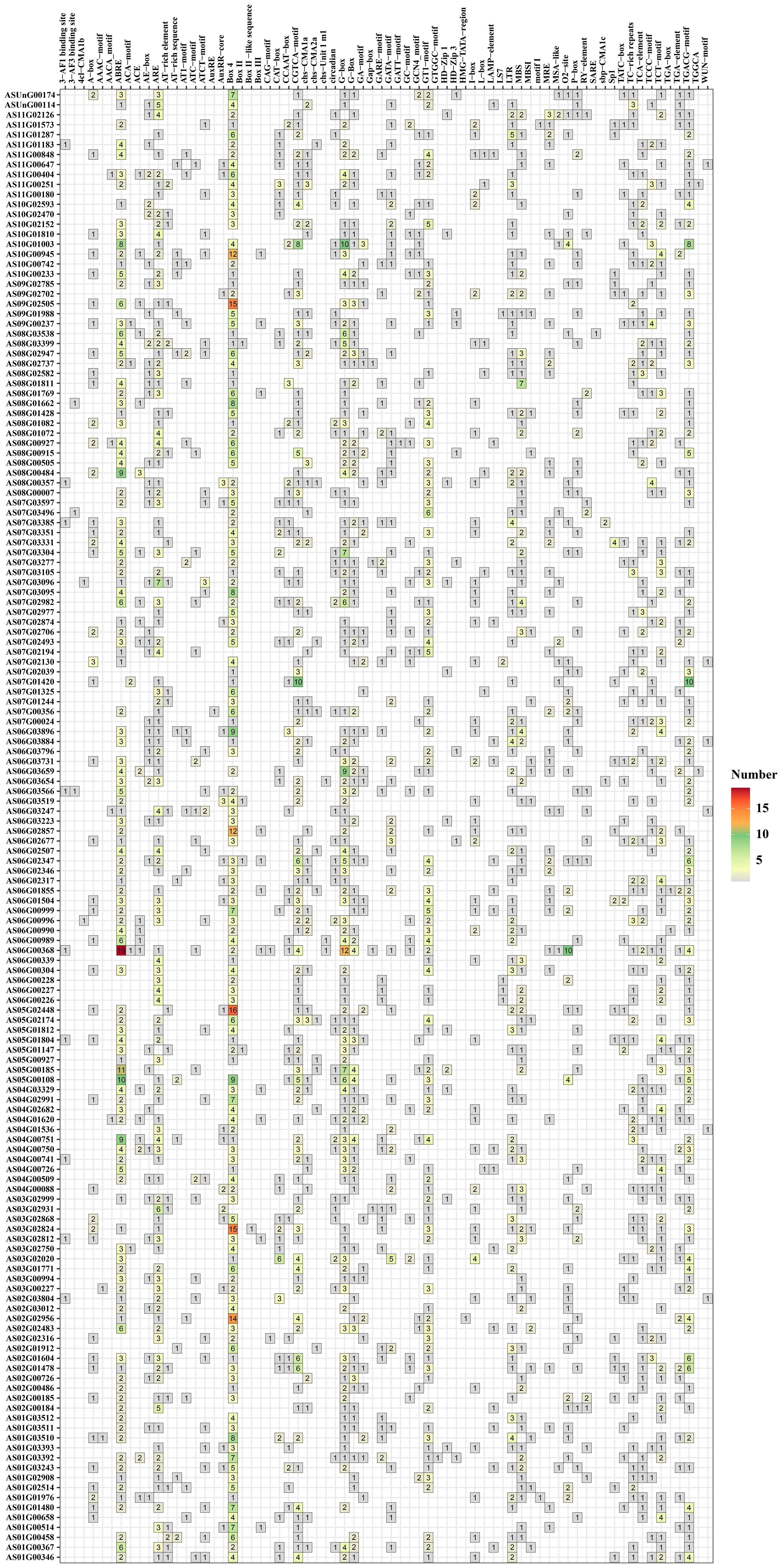
The upstream 2000 bp promoter region of the angelica bHLH gene sequence predicted 57 types of cis-acting elements and it was found that most of the angelica bHLH gene promoter regions contain hormone response elements, including abscisic acid, methyl jasmonate (MeJA), jasmonic acid (JA), auxin (AU), salicylic acid (SA), and gibberellin (GA) (Figure 6). Additionally, the majority of the angelica bHLH promoter regions also have light-responsive elements (Box 4, G-Box, GT1 motif, ATCT motif, AAAC motif, etc.), defense and stress response elements (LTR and TC-rich repeats), drought-induced bHLH binding sites (MBS), and cis-regulatory elements necessary for anaerobic induction (ARE), among other cis-acting elements. A small number contain bHLH binding sites (AF) involved in the regulation of flavonoid biosynthesis and wound response elements (WUN motif). It was found that although the angelica bHLH genes within the same group have close relationships, there are significant differences in the types and quantities of hormone response elements in their promoter regions, indicating that the expression regulatory mechanisms of the angelica bHLH family members exhibit specificity and complexity.
Identification of co-expression modules by WGCNA
To attain an inclusive understanding of genes expressed in the different tissues of A. sinensis and to identify the specific genes that are highly associated with ferulic acid biosynthesis in different plant tissue samples, WGCNA was performed (Langfelder and Horvath, 2008). To increase the accuracy of WGCNA, we used our previously published expression data of 18 samples (Supplementary Table 4) (Arshad et al., 2024). After filtering out the genes with a small coefficient of variation, 26,137 genes were selected for the WGCNA analysis, resulted in highly correlated gene clusters known as modules. Specifically, we identified 12 distinct modules of highly interconnected genes, shown in the dendrogram (Figure 7A), in which the major tree branches define the modules, based on soft threshold and mean connectivity (Figure 7B). Each module correlated with distinct samples due to sample-specific expression profiles (Supplementary Table 5). Gene co-expression modules showed distinct tissue specificity, with the turquoise module (10,008 genes) correlated with stems and the blue module (5,070 genes) with leaves (Figure 7C). The turquoise module and blue module were notably enriched for 32 and 8 bHLH transcription factors, and for three and one phenylpropanoid biosynthesis pathway-specific genes, respectively, associated with ferulic acid biosynthesis.
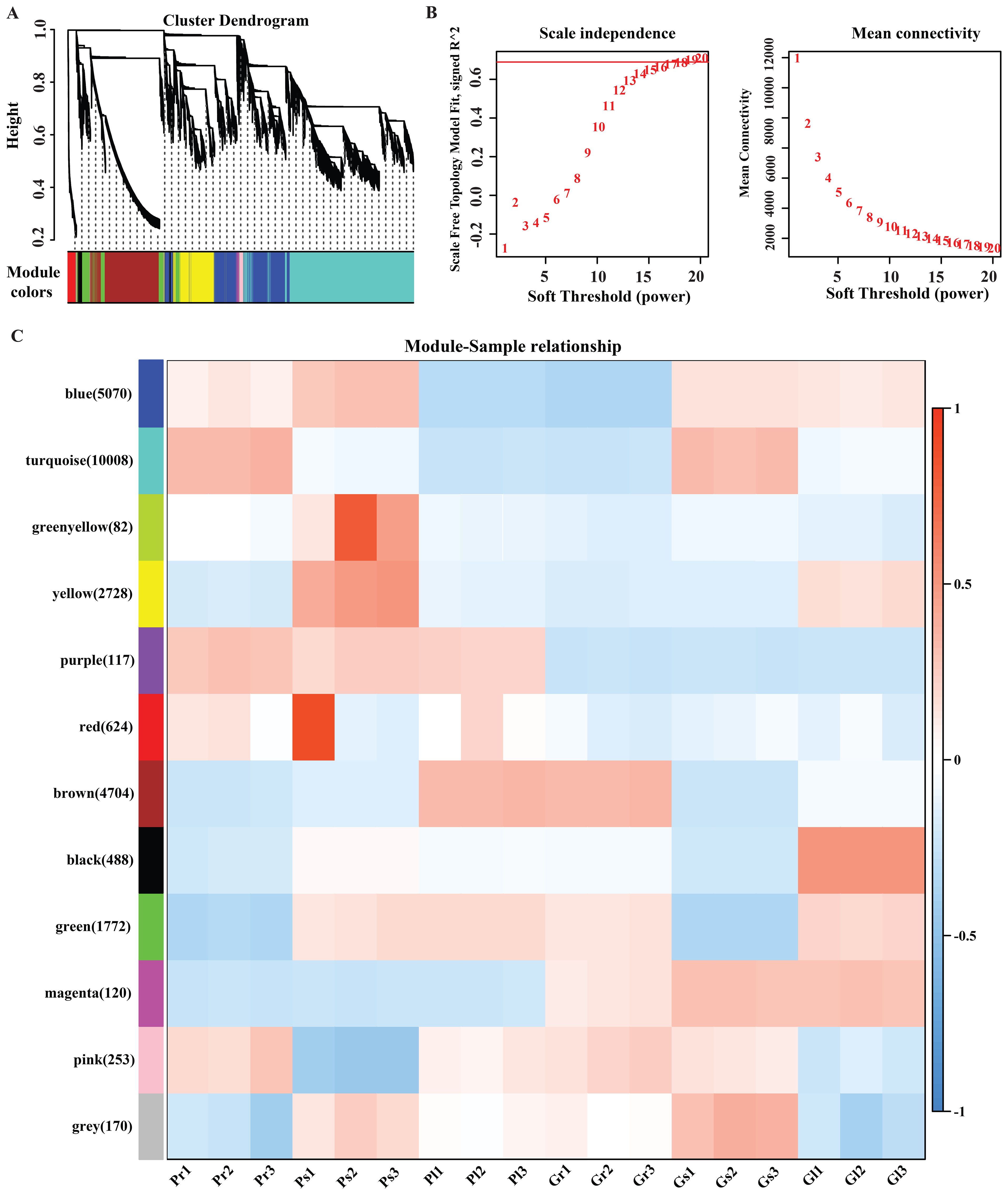
Figure 7. (A) Dendrogram of all expressed modules (each color represents a different module); (B) Soft threshold and mean connectivity; (C) Module trait correlation. The color key, ranging from blue to red, represents R2 values from -1 to 1.
Expression analysis of the AsbHLH genes and protein-protein interaction
The identified AsbHLH genes from turquoise and blue modules had differential expression levels, as determined by pairwise comparative analysis of each plant tissue sample (Supplementary Table 6). A heatmap of these identified AsbHLH genes from two modules was created by using their expression levels from three different tissues (leaf, stem and root). The results indicated that these AsbHLH genes have approximately the same expression levels in leaf and root samples as compared to the stem (Figure 8A). Therefore, using all 32 and 8 genes, we constructed a protein-protein interaction (PPI) network incorporating phenylpropanoid biosynthesis pathway-specific genes (responsible for ferulic acid biosynthesis) based on Spearman correlation coefficient (R > 0.8) and p-value < 0.05 (Supplementary Figure 2; Figures 8B, C). Results showed that the 8 AsbHLH genes in turquoise module, including, AS02G03804 (MYC6), AS06G01855 (bHLH-25), AS02G01478 (MYC2), AS08G01811 (MYC3), AS06G03566 (bHLH-162), AS01G03510 (bHLH-35), AS07G02982 (bHLH-24), and AS02G02316 (bHLH-62), were highly correlated to the pathway-specific enzymes AsPAL1, As4CL2 and AsCOMT. Similarly, in the blue module, 3 genes, including ASUnG00174 (MYC6-like), AS11G01183 (bHLH-59) and AS11G01287 (bHLH-62) were highly correlated with the core enzymes AsPAL3 and As4CL1 of the phenylpropanoid biosynthesis pathway, which are associated with the ferulic acid biosynthesis.
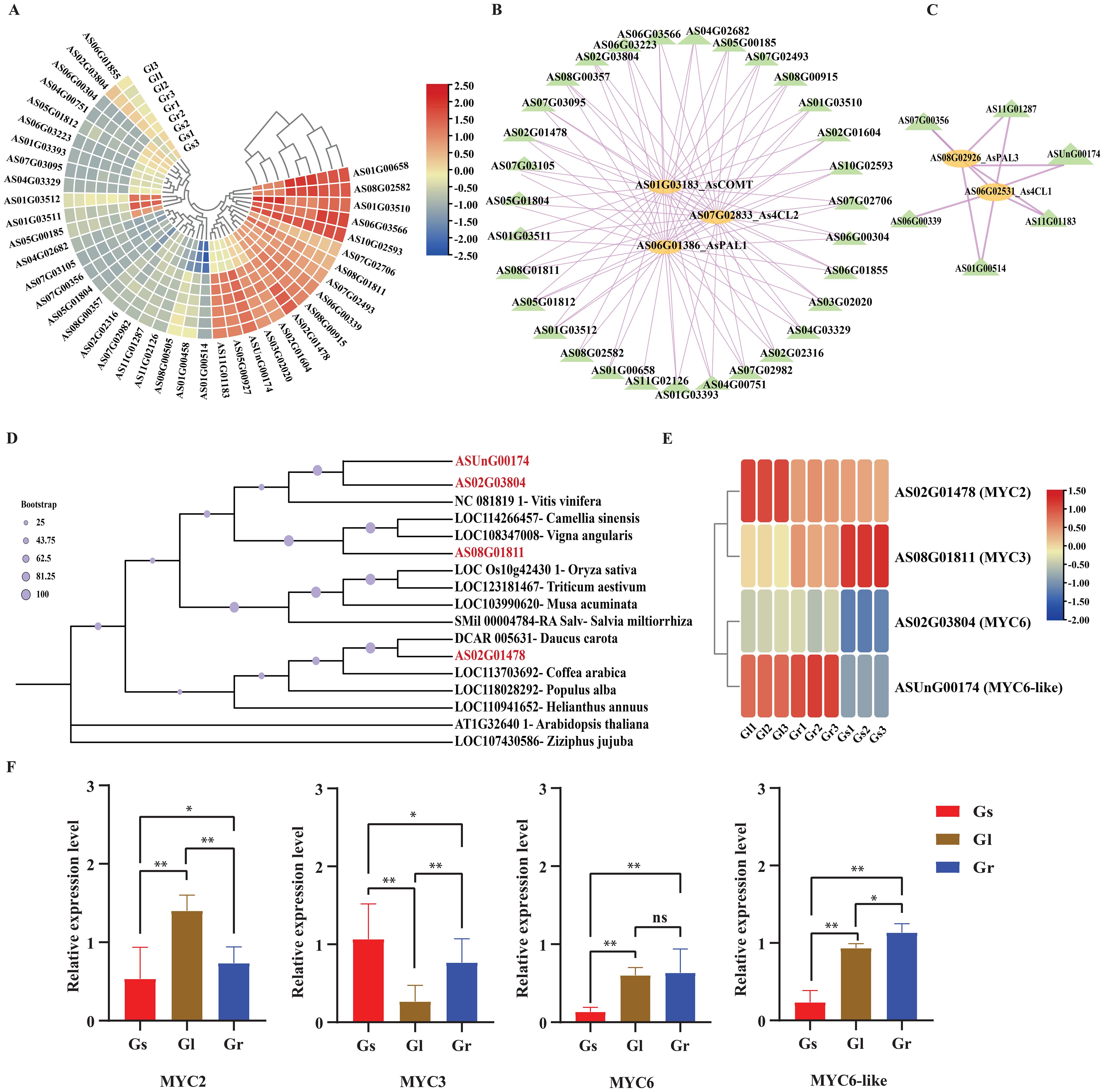
Figure 8. Identification of candidate bHLH genes regarding ferulic acid regulation. (A) Expression profiles analysis of bHLH genes in A. sinensis; Gr: Green root, Gl: Green leaf, and Gs: Green stem. Different colors in the figure represented different log2 (FPKM); (B) Protein-protein interaction among bHLH genes and phenylpropanoid pathway-specific genes of turquoise module; (C) Protein-protein interaction among bHLH genes and phenylpropanoid pathway-specific genes of blue module; (D) phylogenetic tree of AsbHLH in A. sinensis and bHLHs in other species; (E) heatmap presenting expression levels of candidate genes in three tissues; (F) qRT-PCR results of four candidate genes in three tissues.
Screening of putative AsbHLH genes regulating ferulic acid biosynthesis, and qRT-PCR verification
By selecting bHLH proteins known to be associated with regulating ferulic acid biosynthesis and performing homologous comparison and systematic analysis with bHLH proteins in Angelica, a total of 4 homologous AsbHLH proteins were screened from the genomic data of Angelica, which are potential homologs to known functional proteins (Vitis vinifera, Arabidopsis thaliana, Camellia sinensis, Oryza sativa, Triticum aestivum, Vigna angularis, Musa acuminata, Salvia miltiorrhiza, Daucus carota, Coffea arabica, Populus alba, Helianthus annuus, and Ziziphus jujuba) (Figure 8D). These 4 Angelica bHLH genes may be the potential genes for future research on ferulic acid biosynthesis. An expression pattern analysis was conducted on 4 putative genes with good expression reproducibility, revealing that AsMYC2 and AsMYC3 have stable expression levels in all three tissues as compared to AsMYC6 and AsMYC6-like (Figure 8E). Based on homologous comparison and systematic analysis, these 4 AsbHLH genes, homologous to those known to be associated with ferulic acid biosynthesis, were selected for quantitative fluorescent determination. Primers were designed using Primer3 (Supplementary Table 1). The results of these putative genes indicated that AsMYC2 presented a relatively higher expression level in leaves than in roots. At the same time, AsMYC3 and AsMYC6 had relatively lower expression levels in leaves compared to roots. Consequently, the AsMYC6-like represented a higher expression level as compared to all others and AsMYC6 did not show significant differences in expression levels between leaves and roots, which is consistent with transcriptome data (Figure 8F).
Discussion
Understanding the molecular mechanism that controls ferulic acid biosynthesis in A. sinensis is crucial for enhancing its therapeutic value. Ferulic acid biosynthesis is tightly regulated and highly coordinated by the phenylpropanoid pathway (Gao et al., 2023). Several studies reported that bHLH genes are important regulators of secondary metabolite production in plants (Zhou and Memelink, 2016; Wang et al., 2018, 2025; Guo et al., 2021). Therefore, in this study, we performed a genome-wide association study of the bHLH gene family in A. sinensis to find the putative genes potentially regulating ferulic acid production. The results provide comprehensive information on gene correlations involved in the regulation of ferulic acid biosynthesis. This study also shows that PPI is quite helpful in measuring the correlation between genes and transcription factors.
Genome-wide identification and evolutionary analysis of bHLH transcription factors in A. sinensis
bHLH is one of the biggest transcription factor families in plants (Sun et al., 2018; Wang et al., 2018; Guo et al., 2021; Hao et al., 2021). Based on the genomic data of Angelica sinensis, this study annotated 148 bHLH transcription factors, which is similar to the distribution of bHLH genes in Salvia miltiorrhiza (151 genes) (Zhang et al., 2015), Daucus carota (146 genes) (Chen et al., 2015), Oryza sativa (167 genes) (Li et al., 2006) and tomato (152 genes) (Wang et al., 2015). It is much lower than the Triticum aestivum (571 bHLH genes) (Wei and Chen, 2018), higher than the Cannabis sativa (99 bHLH genes) (Bassolino et al., 2020), and much higher than the Cyanidioschyzon merolae (1 bHLH genes) (Pires and Dolan, 2010). This may be related to plant evolution and development. The bHLH family genes were divided into 21 subfamilies in A. thaliana (Toledo-Ortiz et al., 2003), 23 in M. domestica (Yang et al., 2017), 17 in Ginkgo biloba (Zhou et al., 2020) and 19 in peach (Zhang et al., 2018). In this study, the evolutionary analysis identified 148 bHLH genes in A. sinensis, which were divided into 16 subfamilies (Supplementary Table 2; Figure 1). Our results on AsbHLHs showed similarities as well as differences compared to the classifications of A. thaliana. In general, the structures and functions of AsbHLH matched those of A. thaliana. These results indicate that certain bHLHs play strongly conserved roles in A. sinensis and A. thaliana, whereas other bHLHs may have specific functions. Consequently, the conserved motifs are crucial for proteins to perform biological functions. The results of conserved motif analysis of A. sinensis bHLHs showed that motifs 1–5 appeared frequently (Figure 2). bHLH members of the same subfamily share similar motif compositions. However, a few bHLH proteins clustered within a single subfamily exhibited distinct motif compositions, suggesting that this may be related to the functional differentiation of A. sinensis bHLH family members (Figure 2). Different subfamily members also contain distinct or specific motifs. These results suggest that the N-terminus of bHLH transcription factors is not completely conserved among different subfamily members, implying that these motifs may have different biological functions in A. sinensis. Furthermore, these AsbHLHs possess a highly conserved domain HLH at the C-terminus, which binds to the G-box cis-element in the MEJA and JA-responsive promoters (Figures 3, 4) (Fernández-Calvo et al., 2011; Figuora 2012). Further research into the functions and regulatory mechanisms of different motifs will provide a more comprehensive understanding of the functions and regulatory networks of bHLH transcription factors.
Identification and regulatory role of AsbHLH genes in ferulic acid biosynthesis
Taken together, the evolutionary analysis results and bHLH genes with known functions can be combined to predict the AsbHLH genes related to growth and development, secondary metabolism, and environmental responses in A. sinensis. The regulation of growth and development, stress resistance, and signal transduction by bHLH transcription factors has been reported in several plants (Zhou and Memelink, 2016; Sun et al., 2018; Wang et al., 2018, 2025; Mathesius, 2020; Guo et al., 2021). To date, at least 43 bHLH transcription factors have been identified as regulators of secondary metabolism in at least 21 distinct plants, and these active components include flavonoids, anthocyanins, alkaloids, tanshinone, and terpenoids (Zhang et al., 2015). bHLH transcription factor genes are expressed in stimulus-responsive, constitutive, or organ-specific manners. In addition, available functional information from Zea mays (Zhang et al., 2018), A. thaliana (Zhao et al., 2019), and Matthiola incana (Nuraini et al., 2020) suggests that bHLH transcription factors can regulate the phenylpropanoid biosynthesis pathway in both a positive and negative fashion.
The phenylpropanoid pathway is crucial for producing lignin (structural support), flavonoids (UV protection, pigments), ferulic acid and defense compounds (phytoalexins, antioxidants) (Dixon et al., 2002; Sharma et al., 2019; Yadav et al., 2020). Underlying this pathway, the Caffeic acid O-methyltransferase (COMT) is a key enzyme responsible for ferulic acid production (Eckardt, 2002; Narnoliya et al., 2021; Liang et al., 2022; Yang Y. et al., 2023). Previous studies have demonstrated that COMT catalyzes the methylation of caffeic acid to ferulic acid, a rate-limiting step in the pathway (Narnoliya et al., 2021; Shang et al., 2025). Our findings align with these reports, as we observed that COMT expression strongly correlates with ferulic acid accumulation in A. sinensis. Furthermore, phylogenetic, promoter analysis and protein-protein interactions of the identified genes in the turquoise module suggested that AsMYC2, AsMYC3, AsMYC6, and AsMYC6-like genes are highly correlated and potentially directly regulate COMT, implying a mechanistic link between these transcription factors and ferulic acid biosynthesis (Figure 8). This potential regulatory relationship is consistent with studies in other plants, where bHLH transcription factors have been shown to bind to the promoters of phenylpropanoid biosynthetic genes (Zhou and Memelink, 2016; Narnoliya et al., 2021; Shang et al., 2025).
Among the bHLH members, MYC genes have been extensively studied for their regulatory role in secondary metabolite biosynthesis. For instance, in A. thaliana, MYC2 is a well-characterized transcription factor that modulates jasmonate signaling and influences phenylpropanoid metabolism (Dombrecht et al., 2007; Kazan and Manners, 2013). In another study in A. thaliana reported the role of MYC3, along with MYC2 and MYC4, was reported in regulating JA-mediated defense responses (Dombrecht et al., 2007; Cheng et al., 2011; Fernández-Calvo et al., 2011; Wang et al., 2017). Similarly, in Medicago truncatula, MYC2 has been shown to regulate flavonoid biosynthesis by binding to the promoters of key biosynthetic genes (Zhou and Memelink, 2016; Mathesius, 2020; Naik et al., 2021). In Solanum lycopersicum, SIMYC3 regulates jasmonic acid biosynthesis (Wang et al., 2023). Consequently, in Catharanthus roseus (L.) G. Don, the bHLH transcription factor CrMYC2 regulates ORCA gene expression, which in turn regulates alkaloid biosynthesis genes (Colinas et al., 2021). Furthermore, in Salvia miltiorrhiza, SmMYC2 regulates tanshinone biosynthesis (Shi et al., 2020). Meanwhile, in rice and tomato, MYC1 and MYC6 (orthologs of AtMYC3/4) are involved in drought and JA signaling (Fu et al., 2017; Feng et al., 2023). Overall, in A. thaliana and other plants, MYC6 is less studied compared to MYC2/MYC3 but is implicated in JA-mediated stress responses and may have redundant or complementary roles with other MYC TFs. These findings highlight the conserved role of MYC genes in phenylpropanoid regulation across different plant species. In our study, we identified AsMYC2 and AsMYC3 as homologs of AtMYC3, suggesting their potential involvement in the phenylpropanoid pathway, particularly in ferulic acid biosynthesis. Further, the fluorescence quantitative results of the four selected AsbHLHs in this study were consistent with the transcriptome data (Figure 8D). Through comparative studies of MYC homologs across multiple plant species, our study suggests that AsMYC2, AsMYC3, AsMYC6, and AsMYC6-like may regulate ferulic acid accumulation in roots, leaves, and stems of A. sinensis. However, further functional characterization is required to confirm their direct binding to the COMT promoter, as well as it is vital to clearly state that the functional roles projected in this study for the identified AsbHLH genes, chiefly their regulatory influence on COMT and ferulic acid biosynthesis, are currently hypothetical and based on in silico analyses and correlation data. Conclusively, this study establishes a critical theoretical framework for understanding the regulatory network of bHLH transcription factors in ferulic acid biosynthesis in A. sinensis, providing a strong foundation for future gene functional studies and medicinal quality research.
Conclusion
This study systematically identified 148 bHLH transcription factors using whole-genome data from Angelica sinensis. Our findings demonstrate that AsMYC2, AsMYC3, AsMYC6, and AsMYC6-like genes are potential regulators of ferulic acid biosynthesis in A. sinensis, likely through their interaction with the COMT gene. This hypothesis is further supported by qRT-PCR analysis. These results strengthen the evidence that bHLH transcription factors play a central role in ferulic acid production in this medicinal plant. This study advances our understanding of the regulatory mechanisms governing ferulic acid biosynthesis. The identified genes and their potential COMT regulatory axis provide valuable tools for future research, enabling both functional validation and exploration of their applications for enhanced ferulic acid production in A. sinensis through genetic engineering or breeding approaches.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
KA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft. CL: Data curation, Formal analysis, Methodology, Software, Writing – original draft. LL: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. JW: Formal analysis, Investigation, Methodology, Writing – original draft. JC: Supervision, Writing – original draft, Writing – review & editing. YZ: Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82460743), the Major Special Science and Technology Project of Yunnan Province (202502AS100012), Yunnan Characteristic Plant Extraction Laboratory (2022YKZY001), Yunnan Province Youth Talent Support Program (XDYC-QNRC-2022-0219).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1718585/full#supplementary-material.
Supplementary Figure 1 | Meme predicted 10 motifs for the AsbHLH transcription factors.
Supplementary Figure 2 | Heatmap visualization and phenylpropanoid biosynthesis pathway. (A) Expression profile analysis of Phenylpropanoid biosynthesis pathway genes in A. sinensis; Gr: Green root, Gl: Green leaf, and Gs: Green stem. Different colors in the figure represented different log2 (FPKM); (B) Phenylpropanoid biosynthesis pathway.
References
Afnan, Saleem, A., Akhtar, M. F., Sharif, A., Akhtar, B., Siddique, R., et al. (2022). Anticancer, cardio-protective and anti-inflammatory potential of natural-sources-derived phenolic acids. Molecules 27, 7286. doi: 10.3390/molecules27217286
Arshad, K. T., Xiang, C., Yuan, C., Li, L., Wang, J., Zhou, P., et al. (2024). Elucidation of AsANS controlling pigment biosynthesis in Angelica sinensis through hormonal and transcriptomic analysis. Physiol. Plant 176, e14500. doi: 10.1111/ppl.14500
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bao, X., Li, W., Jia, R., Meng, D., Zhang, H., and Xia, L. (2023). Molecular mechanism of ferulic acid and its derivatives in tumor progression. Pharmacol. Rep. 75, 891–906. doi: 10.1007/s43440-023-00494-0
Bassolino, L., Buti, M., Fulvio, F., Pennesi, A., Mandolino, G., Milc, J., et al. (2020). In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L. Plants 9, 1540. doi: 10.3390/plants9111540
Batiha, G. E.-S., Shaheen, H. M., Elhawary, E. A., Mostafa, N. M., Eldahshan, O. A., and Sabatier, J.-M. (2022). Phytochemical constituents, folk medicinal uses, and biological activities of genus angelica: A review. Molecules 28, 267. doi: 10.3390/molecules28010267
Buti, S., Hayes, S., and Pierik, R. (2020). The bHLH network underlying plant shade-avoidance. Physiol. Plant 169, 312–324. doi: 10.1111/ppl.13074
Chen, Y., Li, Q., and Qiu, D. (2022). The dynamic accumulation rules of chemical components in different medicinal parts of angelica sinensis by GC-MS. Molecules 27, 4617. doi: 10.3390/molecules27144617
Chen, Y.-Y., Li, M.-Y., Wu, X.-J., Huang, Y., Ma, J., and Xiong, A.-S. (2015). Genome-wide analysis of basic helix–loop–helix family transcription factors and their role in responses to abiotic stress in carrot. Mol. Breed. 35, 125. doi: 10.1007/s11032-015-0319-0
Chen, C. and Xia, R. (2022). “Interactive data analyses using TBtools,” in Integrative bioinformatics (Springer Singapore, Singapore), 343–375. doi: 10.1007/978-981-16-6795-4_17
Chen, J., Xu, J., Wang, P., Wang, Y., Wang, Y., Lian, J., et al. (2024). Genome-Wide Characterization and Analysis of the bHLH Gene Family in Perilla frutescens. Int. J. Mol. Sci. 25, 13717. doi: 10.3390/ijms252413717
Chen, T.-T., Zou, L., Wang, D., Li, W., Yang, Y., Liu, X.-M., et al. (2021). Metabolomics study of Angelica sinensis (Oliv.) Diels on the abnormal uterine bleeding rats by ultra-performance liquid chromatography-quadrupole-time-of-flight mass spectrometry analysis. Food Sci. Nutr. 9, 6596–6609. doi: 10.1002/fsn3.2605
Cheng, C.-W., Chang, W.-L., Chang, L.-C., Wu, C.-C., Lin, Y.-F., and Chen, J.-S. (2012). Ferulic acid, an angelica sinensis -derived polyphenol, slows the progression of membranous nephropathy in a mouse model. Evidence-Based Complementary Altern. Med. 2012, 1–12. doi: 10.1155/2012/161235
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., et al. (2011). The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in arabidopsis. Mol. Plant 4, 279–288. doi: 10.1093/mp/ssq073
Colinas, M., Pollier, J., Vaneechoutte, D., Malat, D. G., Schweizer, F., De Milde, L., et al. (2021). Subfunctionalization of paralog transcription factors contributes to regulation of alkaloid pathway branch choice in catharanthus roseus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.687406
Dang, Z., Xu, Y., Zhang, X., Mi, W., Chi, Y., Tian, Y., et al. (2024). Chromosome-level genome assembly provides insights into the genome evolution and functional importance of the phenylpropanoid–flavonoid pathway in Thymus mongolicus. BMC Genomics 25, 291. doi: 10.1186/s12864-024-10202-8
de Martin, X., Sodaei, R., and Santpere, G. (2021). Mechanisms of Binding Specificity among bHLH Transcription Factors. Int. J. Mol. Sci. 22, 9150. doi: 10.3390/ijms22179150
Dixon, R. A., Achnine, L., Kota, P., Liu, C., Reddy, M. S. S., and Wang, L. (2002). The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Dong, H., Li, M., Jin, L., Xie, X., Li, M., and Wei, J. (2022). Cool temperature enhances growth, ferulic acid and flavonoid biosynthesis while inhibiting polysaccharide biosynthesis in angelica sinensis. Molecules 27, 320. doi: 10.3390/molecules27010320
Dong, J., Wu, Y.-W., Dong, Y., Pu, R., Li, X.-J., Lyu, Y.-M., et al. (2024). Genome-Wide Identification of the bHLH Gene Family in Rhododendron delavayi and Its Expression Analysis in Different Floral Tissues. Genes (Basel) 15, 1256. doi: 10.3390/genes15101256
Du, T., Niu, J., Su, J., Li, S., Guo, X., Li, L., et al. (2018). SmbHLH37 functions antagonistically with smMYC2 in regulating jasmonate-mediated biosynthesis of phenolic acids in salvia miltiorrhiza. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01720
Eckardt, N. A. (2002). Probing the mysteries of lignin biosynthesis. Plant Cell 14, 1185–1189. doi: 10.1105/tpc.140610
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, Y., Zeng, S., Yan, J., Li, K., and Xu, H. (2023). Genome-wide analysis and expression of MYC family genes in tomato and the functional identification of slmyc1 in response to salt and drought stress. Agronomy 13, 757. doi: 10.3390/agronomy13030757
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fu, J., Wu, H., Ma, S., Xiang, D., Liu, R., and Xiong, L. (2017). OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02108
Gao, P., Qi, Y., Li, L., Yang, S., Liu, J., Wei, H., et al. (2023). Amorphophallus muelleri activates ferulic acid and phenylpropane biosynthesis pathways to defend against Fusarium solani infection. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1207970
Gao, Q., Zhang, J., Cao, J., Xiang, C., Yuan, C., Li, X., et al. (2024). MetaDb: a database for metabolites and their regulation in plants with an emphasis on medicinal plants. Mol. Horticulture 4, 17. doi: 10.1186/s43897-024-00095-2
Giacomelli, N., Yongping, Y., Huber, F., Ankli, A., and Weckerle, C. (2017). Angelica sinensis (Oliv.) diels: influence of value chain on quality criteria and marker compounds ferulic acid and Z-ligustilide. Medicines 4, 14. doi: 10.3390/medicines4010014
Goossens, J., Mertens, J., and Goossens, A. (2016). Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 68 (6), 1333–1347. doi: 10.1093/jxb/erw440
Guo, M., Li, C., Huang, R., Qu, L., Liu, J., Zhang, C., et al. (2023). Ferulic acid enhanced resistance against blue mold of Malus domestica by regulating reactive oxygen species and phenylpropanoid metabolism. Postharvest Biol. Technol. 202, 112378. doi: 10.1016/j.postharvbio.2023.112378
Guo, J., Sun, B., He, H., Zhang, Y., Tian, H., and Wang, B. (2021). Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 22, 4921. doi: 10.3390/ijms22094921
Guo, H., Zheng, G., Chen, J., and Gao, Y. (2020). “Menstrual regulation delivered by angelicae sinensis radix,” in 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 1510–1515 (Piscataway, New Jersey, USA: IEEE). doi: 10.1109/BIBM49941.2020.9313147
Han, X., Li, C., Sun, S., Ji, J., Nie, B., Maker, G., et al. (2022). The chromosome-level genome of female ginseng (Angelica sinensis) provides insights into molecular mechanisms and evolution of coumarin biosynthesis. Plant J. 112, 1224–1237. doi: 10.1111/tpj.16007
Han, X., Li, M., Yuan, Q., Lee, S., Li, C., Ren, Y., et al. (2023). Advances in molecular biological research of Angelica sinensis. Medicinal Plant Biol. 2, 0–0. doi: 10.48130/MPB-2023-0016
Hao, Y., Zong, X., Ren, P., Qian, Y., and Fu, A. (2021). Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in arabidopsis. Int. J. Mol. Sci. 22, 7152. doi: 10.3390/ijms22137152
He, B., Zhang, C., Zhang, X., Fan, Y., Zeng, H., Liu, J., et al. (2021). Tissue-specific 5-hydroxymethylcytosine landscape of the human genome. Nat. Commun. 12, 4249. doi: 10.1038/s41467-021-24425-w
Horiike, T. (2016). AN INTRODUCTION TO MOLECULAR PHYLOGENETIC ANALYSIS. Rev. Agric. Sci. 4, 36–45. doi: 10.7831/ras.4.0_36
Hu, X., Liu, F., Yang, H., Qi, M., Ren, Y., Zhu, W., et al. (2024). Protective effect and related mechanism of modified danggui buxue decoction on retinal oxidative damage in mice based on network pharmacology. Curr. Pharm. Des. 30, 1912–1926. doi: 10.2174/0113816128293824240517113238
Hussain, M. I. and Reigosa, M. J. (2021). Secondary Metabolites, Ferulic Acid and p-Hydroxybenzoic Acid Induced Toxic Effects on Photosynthetic Process in Rumex acetosa L. Biomolecules 11, 233. doi: 10.3390/biom11020233
Jan, R., Asaf, S., Numan, M., Lubna, and Kim, K.-M. (2021). Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 11, 968. doi: 10.3390/agronomy11050968
Jia, Y., Meng, W., Chen, G., Fan, X., Zhang, Y., Ding, A., et al. (2025). The regulation mechanism of MYC on meJA-induced flavonoids synthesis in dendrobium officinale. J. Plant Growth Regul. 44, 217–232. doi: 10.1007/s00344-024-11388-7
Jiao, M., Liu, X., Ren, Y., Wang, Y., Cheng, L., Liang, Y., et al. (2022). Comparison of herbal medicines used for women’s menstruation diseases in different areas of the world. Front. Pharmacol. 12. doi: 10.3389/fphar.2021.751207
Jing, Y. and Lin, R. (2020). Transcriptional regulatory network of the light signaling pathways. New Phytol. 227, 683–697. doi: 10.1111/nph.16602
Kazan, K. and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Ke, Y.-Z., Wu, Y.-W., Zhou, H.-J., Chen, P., Wang, M.-M., Liu, M.-M., et al. (2020). Genome-wide survey of the bHLH super gene family in Brassica napus. BMC Plant Biol. 20, 115. doi: 10.1186/s12870-020-2315-8
Khan, A., Kanwal, F., Ullah, S., Fahad, M., Tariq, L., Altaf, M. T., et al. (2025). Plant secondary metabolites—Central regulators against abiotic and biotic stresses. Metabolites 15, 276. doi: 10.3390/metabo15040276
Kim, H.-S., Lee, D., Seo, Y.-H., Ryu, S.-M., Lee, A.-Y., Moon, B.-C., et al. (2023). Chemical constituents from the roots of angelica reflexa that improve glucose-stimulated insulin secretion by regulating pancreatic β-cell metabolism. Pharmaceutics 15, 1239. doi: 10.3390/pharmaceutics15041239
Kim, Y.-H., Park, C.-W., Lee, K. M., Hong, C.-O., Son, H.-J., Kim, K. K., et al. (2025). The roles of MYC2 transcription factor in JA-signaling pathway in plants. J. Plant Biol. 68, 113–131. doi: 10.1007/s12374-025-09462-y
Kiokias, S., Proestos, C., and Oreopoulou, V. (2020). Phenolic acids of plant origin—A review on their antioxidant activity in vitro (O/W emulsion systems) along with their in vivo health biochemical properties. Foods 9, 534. doi: 10.3390/foods9040534
LaFountain, A. M. and Yuan, Y. (2021). Repressors of anthocyanin biosynthesis. New Phytol. 231, 933–949. doi: 10.1111/nph.17397
Langfelder, P. and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Lee, D.-H., Kwak, H. J., Shin, Y., Kim, S. J., Lee, G. H., Park, I.-H., et al. (2023). Elucidation of phytochemicals affecting platelet responsiveness in dangguisu-san: active ingredient prediction and experimental research using network pharmacology. Plants 12, 1120. doi: 10.3390/plants12051120
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, S., Chiu, T.-Y., Jin, X., Cao, D., Xu, M., Zhu, M., et al. (2023). Integrating genomic and multiomic data for Angelica sinensis provides insights into the evolution and biosynthesis of pharmaceutically bioactive compounds. Commun. Biol. 6, 1198. doi: 10.1038/s42003-023-05569-5
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, D., Rui, Y., Guo, S., Luan, F., Liu, R., and Zeng, N. (2021). Ferulic acid: A review of its pharmacology, pharmacokinetics and derivatives. Life Sci. 284, 119921. doi: 10.1016/j.lfs.2021.119921
Li, Y.-Y., Sui, X.-Y., Yang, J.-S., Xiang, X.-H., Li, Z.-Q., Wang, Y.-Y., et al. (2020). A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco (Nicotiana tabacum L.). Biochem. Biophys. Res. Commun. 522, 233–239. doi: 10.1016/j.bbrc.2019.11.063
Li, R., Yao, J., Cai, S., Fu, Y., Lai, C., Zhu, X., et al. (2024). Genome-wide characterization and evolution analysis of miniature inverted-repeat transposable elements in Barley (Hordeum vulgare). Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1474846
Liang, S., Xu, S., Qu, D., Yang, L., Wang, J., Liu, H., et al. (2022). Identification and functional analysis of the caffeic acid O-methyltransferase (COMT) gene family in rice (Oryza sativa L.). Int. J. Mol. Sci. 23, 8491. doi: 10.3390/ijms23158491
Liu, M., Li, H., Chen, Y., Wu, Z., Wu, S., Zhang, J., et al. (2025). The MYC2 - JAMYB transcriptional cascade regulates rice resistance to brown planthoppers. New Phytol. 246, 1834–1847. doi: 10.1111/nph.70059
Liu, H., Liu, Z., Wu, Y., Zheng, L., and Zhang, G. (2021). Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. Int. J. Mol. Sci. 22, 8441. doi: 10.3390/ijms22168441
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, R., He, Y., Fang, Q., Xie, G., and Qi, M. (2022). Ferulic acid ameliorates renal injury via improving autophagy to inhibit inflammation in diabetic nephropathy mice. Biomedicine Pharmacotherapy 153, 113424. doi: 10.1016/j.biopha.2022.113424
Mathesius, U. (2020). “The role of the flavonoid pathway in Medicago truncatula in root nodule formation. A review,” in The model legume medicago truncatula (Taylor and Francis), 434–438. doi: 10.1002/9781119409144.ch54
Meraj, T. A., Fu, J., Raza, M. A., Zhu, C., Shen, Q., Xu, D., et al. (2020). Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes (Basel) 11, 346. doi: 10.3390/genes11040346
Mishra, A. K. and Paliwal, S. K. (2025). A deep insight into chemistry and pharmacology of genus angelica.: an up-to-date systematic review. Nat. Prod J. 15, 332–340. doi: 10.2174/0122103155287588240314065437
Mnich, E., Bjarnholt, N., Eudes, A., Harholt, J., Holland, C., Jørgensen, B., et al. (2020). Phenolic cross-links: building and de-constructing the plant cell wall. Nat. Prod Rep. 37, 919–961. doi: 10.1039/C9NP00028C
Naik, J., Rajput, R., Pucker, B., Stracke, R., and Pandey, A. (2021). The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol. Biol. 106, 157–172. doi: 10.1007/s11103-021-01135-x
Nakata, M., Mitsuda, N., Herde, M., Koo, A. J. K., Moreno, J. E., Suzuki, K., et al. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell 25, 1641–1656. doi: 10.1105/tpc.113.111112
Narnoliya, L. K., Sangwan, N., Jadaun, J. S., Bansal, S., and Sangwan, R. S. (2021). Defining the role of a caffeic acid 3-O-methyltransferase from Azadirachta indica fruits in the biosynthesis of ferulic acid through heterologous over-expression in Ocimum species and Withania somnifera. Planta 253, 20. doi: 10.1007/s00425-020-03514-y
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Nuraini, L., Ando, Y., Kawai, K., Tatsuzawa, F., Tanaka, K., Ochiai, M., et al. (2020). Anthocyanin regulatory and structural genes associated with violet flower color of Matthiola incana. Planta 251, 61. doi: 10.1007/s00425-020-03351-z
Pandi, A., Raghu, M. H., Chandrashekar, N., and Kalappan, V. M. (2022). Cardioprotective effects of Ferulic acid against various drugs and toxic agents. Beni Suef Univ J. Basic Appl. Sci. 11, 92. doi: 10.1186/s43088-022-00273-5
Pei, F., Zang, Y., Bhattacharya, P., and Liu, H. (2024). Application of angelica sinensis in gynecological diseases IGI Global. 155–170. doi: 10.4018/979-8-3693-2703-6.ch008
Pires, N. and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PloS One 5, e9490. doi: 10.1371/journal.pone.0009490
Qian, Y., Zhang, T., Yu, Y., Gou, L., Yang, J., Xu, J., et al. (2021). Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.677611
Rabeh, K., Hnini, M., and Oubohssaine, M. (2025). A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 5, 15. doi: 10.1007/s44154-024-00201-w
Ren, C., Luo, Y., Li, X., Ma, L., Wang, C., Zhi, X., et al. (2025). Pharmacological action of Angelica sinensis polysaccharides: a review. Front. Pharmacol. 15. doi: 10.3389/fphar.2024.1510976
Rizwan, H. M., He, J., Nawaz, M., Cheng, K.-W., and Wang, M. (2024). Comprehensive in silico characterization of soybean (Glycine max L.) isoflavone reductase genes and their expressions in response to spermidine and ultrasonication. Plant Stress 11, 100392. doi: 10.1016/j.stress.2024.100392
Robert, B., Chenthamara, D., and Subramaniam, S. (2022). Fabrication and biomedical applications of Arabinoxylan, Pectin, Chitosan, soy protein, and silk fibroin hydrogels via laccase - Ferulic acid redox chemistry. Int. J. Biol. Macromol 201, 539–556. doi: 10.1016/j.ijbiomac.2021.12.103
Sabeel, Z., Liang, Y., Hao, M., Ying, L., Guo, R., Chen, R., et al. (2023). A comprehensive review of antitumor properties of Angelica species and their antitumor-responsible constituents and the underlying molecular mechanisms involved in tumor inhibition. Phytotherapy Res. 37, 2187–2211. doi: 10.1002/ptr.7841
Sarker, S. and Nahar, L. (2004). Natural medicine: the genus angelica. Curr. Med. Chem. 11, 1479–1500. doi: 10.2174/0929867043365189
Shang, H. Q., Bo Yang, Q., Qiang, S., Zheng, R., Zhang, C. Q., Hu, C. Y., et al. (2025). Engineering caffeic acid O-methyltransferase for efficient de novo ferulic acid synthesis. Eng. Life Sci. 25, e70018. doi: 10.1002/elsc.70018
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., and Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24, 2452. doi: 10.3390/molecules24132452
Shedoeva, A., Leavesley, D., Upton, Z., and Fan, C. (2019). Wound healing and the use of medicinal plants. Evid Based Complement Alternat Med. 2019, 2684108. doi: 10.1155/2019/2684108
Shen, R., Zhao, W., Wang, Y., Sukamto, J., Li, G., Yang, X., et al. (2025). Multi-omics analysis reveals Angelica sinensis-Carthamus tinctorius herb pair ameliorates diabetic retinopathy comorbid with depressive symptoms via the gut-eye-brain axis. Phytomedicine 143, 156874. doi: 10.1016/j.phymed.2025.156874
Shi, M., Wang, Y., Wang, X., Deng, C., Cao, W., Hua, Q., et al. (2020). Simultaneous promotion of tanshinone and phenolic acid biosynthesis in Salvia miltiorrhiza hairy roots by overexpressing Arabidopsis MYC2. Ind. Crops Prod 155, 112826. doi: 10.1016/j.indcrop.2020.112826
Sun, X., Wang, Y., and Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Sun, Y., Zhou, J., Hu, Y., Chen, S., Ni, H., Feng, R., et al. (2024). Comprehensive analysis of phenylpropanoids and butylidenephthalides in Angelica sinensis (Oliv.) Diels from different regions and processing methods by high-performance liquid chromatography combined with chemometrics. Eur. Food Res. Technol. 250, 983–998. doi: 10.1007/s00217-023-04422-7
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T. Z., Garcia-Hernandez, M., Foerster, H., et al. (2007). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014. doi: 10.1093/nar/gkm965
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thoben, C. and Pucker, B. (2023). Automatic annotation of the bHLH gene family in plants. BMC Genomics 24, 780. doi: 10.1186/s12864-023-09877-2
Tian, Y., Shen, X., Hu, T., Liang, Z., Ding, Y., Dai, H., et al. (2024). Structural analysis and blood-enriching effects comparison based on biological potency of Angelica sinensis polysaccharides. Front. Pharmacol. 15. doi: 10.3389/fphar.2024.1405342
Tobin, L. A., Lam, M. M. C., and Hamidian, M. (2025). Pan-genus analysis and typing of antimicrobial resistance plasmids in Acinetobacter. NPJ Antimicrobials Resistance 3, 65. doi: 10.1038/s44259-025-00133-z
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The arabidopsis basic/helix-loop-helix transcription factor family[W. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Wang, Y., Gong, Y., Xiao, Y., Jiang, Y., Chen, J., Zhao, H., et al. (2023). Study on the dynamic metabolic characteristic of main active ingredients in Danggui Buxue Decoction by liquid chromatography-tandem mass spectrometry based on in situ sequential metabolism strategy. J. Sep Sci. 46, e2200941. doi: 10.1002/jssc.202200941
Wang, J., Hu, Z., Zhao, T., Yang, Y., Chen, T., Yang, M., et al. (2015). Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16, 39. doi: 10.1186/s12864-015-1249-2
Wang, H., Li, Y., Pan, J., Lou, D., Hu, Y., and Yu, D. (2017). The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in arabidopsis. Mol. Plant 10, 1461–1464. doi: 10.1016/j.molp.2017.08.007
Wang, T., Li, M., Tang, X., He, Y., Fang, Q., Fan, T., et al. (2025). Deciphering the role of a novel basic helix-loop-helix (bHLH) transcription factor: SmbHLH125 in Salvia miltiorrhiza secondary metabolism through transcriptomic and metabolomic insights. Int. J. Biol. Macromol 318, 145067. doi: 10.1016/j.ijbiomac.2025.145067
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00064
Wang, S., Wang, Y., Yang, R., Cai, W., Liu, Y., Zhou, D., et al. (2023). Genome-wide identification and analysis uncovers the potential role of JAZ and MYC families in potato under abiotic stress. Int. J. Mol. Sci. 24, 6706. doi: 10.3390/ijms24076706
Wei, K. and Chen, H. (2018). Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 18, 309. doi: 10.1186/s12870-018-1529-5
Wei, H., Liu, G., Qin, J., Zhang, Y., Chen, J., Zhang, X., et al. (2023). Genome-wide characterization, chromosome localization, and expression profile analysis of poplar non-specific lipid transfer proteins. Int. J. Biol. Macromol 231, 123226. doi: 10.1016/j.ijbiomac.2023.123226
Wei, X., Mao, L., Wei, X., Guan, W., Chen, R., and Luo, Z. (2022). AchMYC2 promotes JA-mediated suberin polyphenolic accumulation via the activation of phenylpropanoid metabolism-related genes in the wound healing of kiwifruit (Actinidia chinensis). Postharvest Biol. Technol. 188, 111896. doi: 10.1016/j.postharvbio.2022.111896
Wu, S., Da, L., Xiao, Q., Pan, Q., Zhang, J., and Yang, J. (2024). ASAP: a platform for gene functional analysis in Angelica sinensis. BMC Genomics 25, 96. doi: 10.1186/s12864-024-09971-z
Wu, L., Hu, Z., Lv, Y., Ge, C., Luo, X., Zhan, S., et al. (2025). Hericium erinaceus polysaccharides improve eggshell quality by improving uterine health and modulating uterine microbiota in aged hens. Anim. Nutr. 22, 72–87. doi: 10.1016/j.aninu.2025.03.008
Yadav, V., Wang, Z., Wei, C., Amo, A., Ahmed, B., Yang, X., et al. (2020). Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens 9, 312. doi: 10.3390/pathogens9040312
Yang, J., Gao, M., Huang, L., Wang, Y., van Nocker, S., Wan, R., et al. (2017). Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 7, 28. doi: 10.1038/s41598-017-00040-y
Yang, X., Hai, Y., Yang, Z., Wang, P., Li, J., Hao, D., et al. (2023). Pharmacodynamics, metabolomics and pathological studies on mechanisms of traditional benefits of Angelica sinensis in blood circulation. Indian J. Traditional Knowledge 22, 1510976. doi: 10.56042/ijtk.v22i1.53106
Yang, Y. H., Song, H. W., Lai, J. Y., Li, R. F., Wang, Z. C., Jia, H. C., et al. (2023). A Rehmannia glutinosa caffeic acid O -methyltransferase functional identification: Reconstitution of the ferulic acid biosynthetic pathway in Saccharomyces cerevisiae using Rehmannia glutinosa enzymes. Biotechnol. J. 18, 2300064. doi: 10.1002/biot.202300064
Yesudhas, D., Batool, M., Anwar, M. A., Panneerselvam, S., and Choi, S. (2017). Proteins recognizing DNA: structural uniqueness and versatility of DNA-binding domains in stem cell transcription factors. Genes (Basel) 8, 192. doi: 10.3390/genes8080192
Yuan, Z., Zhong, L., Hua, Y., Ji, P., Yao, W., Ma, Q., et al. (2019). Metabolomics study on promoting blood circulation and ameliorating blood stasis: Investigating the mechanism of Angelica sinensis and its processed products. Biomed. Chromatogr. 33, e4457. doi: 10.1002/bmc.4457
Zhang, H. Y., Bi, W. G., Yu, Y., and Liao, W. B. (2012). Angelica sinensis (Oliv.) Diels in China: distribution, cultivation, utilization and variation. Genet. Resour Crop Evol. 59, 607–613. doi: 10.1007/s10722-012-9795-9
Zhang, J., Liu, X., Yin, Z., Zhao, T., Du, D., Li, J., et al. (2025). Genome- and transcriptome-wide characterization and expression analyses of bHLH transcription factor family reveal their relevance to salt stress response in tomato. Plants (Basel) 14, 200. doi: 10.3390/plants14020200
Zhang, X., Luo, H., Xu, Z., Zhu, Y., Ji, A., Song, J., et al. (2015). Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 5, 11244. doi: 10.1038/srep11244
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18, 235. doi: 10.1186/s12870-018-1441-z
Zhao, Y., Liu, G., Yang, F., Liang, Y., Gao, Q., Xiang, C., et al. (2023). Multilayered regulation of secondary metabolism in medicinal plants. Mol. Horticulture 3, 11. doi: 10.1186/s43897-023-00059-y
Zhao, Y., Yang, S.-C., Arshad, K. T., Wang, J., Zhou, P.-H., and Li, C.-H. (2025). “Yunnan angelica sinensis (Oliv.) diels,” in A companion volume to medicinal plants and mushrooms of yunnan province of China (CRC Press, Boca Raton), 29–44. doi: 10.1201/9781003488552-3
Zhao, Y., Zhang, Y.-Y., Liu, H., Zhang, X.-S., Ni, R., Wang, P.-Y., et al. (2019). Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol. 19, 497. doi: 10.1186/s12870-019-2109-z
Zhou, X., Liao, Y., Kim, S.-U., Chen, Z., Nie, G., Cheng, S., et al. (2020). Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 10, 13723. doi: 10.1038/s41598-020-69305-3
Zhou, M. and Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.bioteChadv.2016.02.004
Keywords: Angelica sinensis, bHLH transcriptionfactors, whole-genome identification, bioinformatics analysis, gene expression analysis
Citation: Arshad KT, Li C, Li L, Wang J, Chen J and Zhao Y (2025) Genome-wide identification and expression profiling of bHLH transcription factors associated with ferulic acid biosynthesis in Angelica sinensis. Front. Plant Sci. 16:1718585. doi: 10.3389/fpls.2025.1718585
Received: 04 October 2025; Accepted: 30 October 2025;
Published: 26 November 2025.
Edited by:
Xin Fang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Xiaodong Zhang, Xuchang University, ChinaShiming Li, Hunan University of Medicine, China
Copyright © 2025 Arshad, Li, Li, Wang, Chen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junwen Chen, Y2p3MzE0MTJAMTYzLmNvbQ==; Yan Zhao, emhhb3lhbmttQDEyNi5jb20=
 Khadija Tehseen Arshad
Khadija Tehseen Arshad Chaohui Li1,2
Chaohui Li1,2 Junwen Chen
Junwen Chen Yan Zhao
Yan Zhao