- 1Department of Anesthesiology, Wake Forest University School of Medicine, Winston-Salem, NC, United States
- 2Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC, United States
- 3Department of Translational Neuroscience, Wake Forest University School of Medicine, Winston-Salem, NC, United States
- 4Krumware LLC, Columbia, SC, United States
- 5Department of Cancer Biology, Wake Forest University School of Medicine, Winston-Salem, NC, United States
- 6Clinical Translational Research Informatics Branch, National Cancer Institute, National Institutes of Health, Rockville, MD, United States
This work presents a framework for enhancing Gen3, an open-source data commons platform, with temporal visualization capabilities for clinical trial research. We describe the technical implementation of cloud-native architecture and integrated visualization tools that enable standardized analytics for longitudinal clinical trial data while adhering to FAIR principles. The enhancement includes Kubernetes-based container orchestration, Kibana-based temporal analytics, and automated ETL pipelines for data harmonization. Technical validation demonstrates reliable handling of varied time-based data structures, while maintaining temporal precision and measurement context. The framework's implementation in NIH HEAL Initiative networks studying chronic pain and substance use disorders showcases its utility for real-time monitoring of longitudinal outcomes across multiple trials. This adaptation provides a model for research networks seeking to enhance their data commons capabilities while ensuring findable, accessible, interoperable, and reusable clinical trial data.
Introduction
Clinical trial networks require sophisticated data commons platforms that support longitudinal analytics while following FAIR (Findable, Accessible, Interoperable, and Reusable) data principles (1). While existing data commons solutions excel at managing static datasets, they often lack native capabilities for tracking and visualizing the progression of clinical outcomes over time (2, 3). This gap presents a significant challenge for understanding treatment effectiveness in interventional studies.
Current data commons platforms face several technical limitations when applied to longitudinal clinical research. These specific technical requirements include: the need for temporal data modeling that preserves measurement timing and sequence, standardized harmonization of patient-reported outcome measures across multiple trials, real-time monitoring capabilities for trial management, and integration of diverse data types with varying collection frequencies from daily patient reports to monthly clinical assessments. Existing platforms typically require specialized informatics support and custom development for temporal analyses, limiting accessibility for clinical researchers.
Gen3, an open-source platform widely adopted across NIH-funded research networks, provides robust capabilities for data storage, access control, and basic querying (4). However, its implementation for clinical trial visualization has revealed important capability gaps, particularly in temporal analysis and standardized outcome measure harmonization (5). The complexity increases with requirements for integrating diverse data types from multiple concurrent trials, implementing granular access controls for multi-site studies, and supporting real-time monitoring of recruitment and outcomes (6, 7).
This paper addresses the following research questions: How can Gen3's architecture be enhanced to support temporal visualization of clinical trial data while maintaining FAIR principles? What technical implementations are required to enable real-time monitoring of longitudinal outcomes across multiple trials? How can standardized temporal analytics be achieved without compromising data security and access controls?
Our implementation maintains FAIR data principles while addressing the specific technical requirements of longitudinal clinical research (8). We demonstrate this framework's effectiveness through its deployment in NIH HEAL Initiative clinical trial networks studying chronic pain and substance use disorders (3). though the approach is broadly applicable across clinical domains (4, 9–11).
Methods
System architecture overview
The adaptation of Gen3 for clinical trial applications required significant architectural enhancements to support temporal data visualization and analysis. The core enhancement involved developing a cloud-agnostic implementation that freed the platform from vendor-specific constraints (5). This platform independence proved crucial for research networks that operate across multiple institutions with varying infrastructure requirements. The successful transition between cloud providers demonstrated the feasibility of cross-cloud deployment while maintaining full functionality.
Cloud-native infrastructure implementation
The implementation leverages comprehensive container orchestration through Kubernetes, with robust node pool configurations enabling automated scaling based on resource utilization metrics. The containerized environment is secured through NeuVector's zero-trust security model (12), providing Layer 7 firewall capabilities and continuous vulnerability scanning (13). This security framework integrates seamlessly with OAuth2 authentication patterns (14), ensuring consistent access controls across all microservices.
Temporal visualization framework
The integration of temporal visualization capabilities marked another crucial advancement. By incorporating Kibana-based analytics, the platform gained the ability to track longitudinal outcomes effectively. The visualization framework supports interactive filtering and temporal comparisons, enabling researchers to examine treatment effects across multiple timepoints and subgroups (15).
Our implementation integrates Kibana with ElasticSearch indices (16), enabling researchers to examine longitudinal patterns through both predefined and custom dashboard configurations. This implementation includes automated ETL pipelines that maintain data synchronization between PostgreSQL databases (17) and ElasticSearch indices, ensuring near real-time availability of temporal analytics (16, 18, 19).
Security architecture
Security enhancements formed a critical component of the adaptation (5). The implementation of NeuVector provided zero-trust container security, crucial for protecting sensitive clinical trial data (20). The security architecture implements comprehensive authentication and user management, specifically OAuth2 for external service authentication and includes comprehensive backup strategies across services, as illustrated in Figure 1 (14). The security model maintains compliance with regulatory requirements while enabling appropriate data sharing and collaboration across research sites.
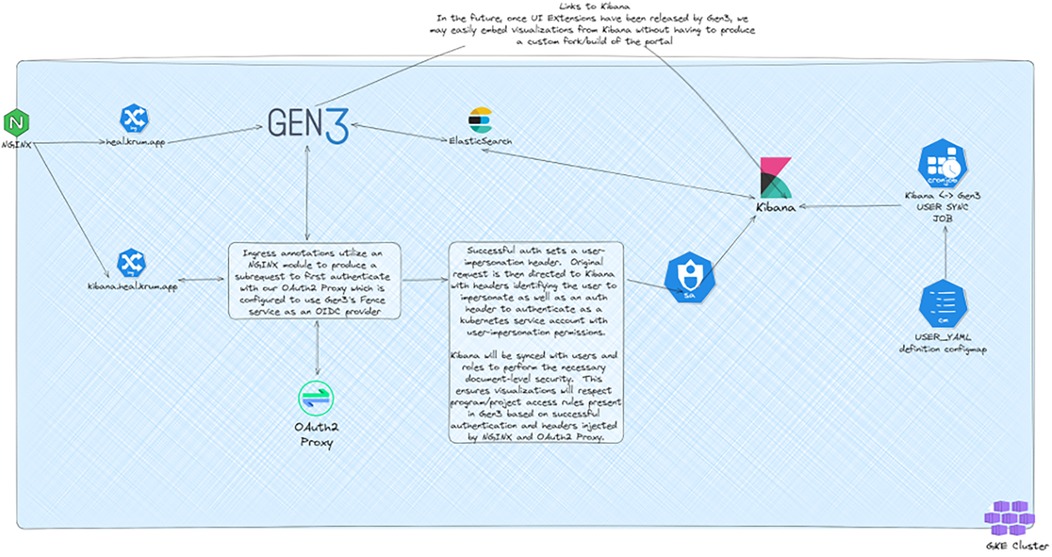
Figure 1. Gen3 authentication and user synchronization architecture. Architectural diagram showing the integration between Gen3, Kibana, and authentication services. The system utilizes NGINX modules for OAuth2 authentication and maintains user synchronization between Gen3 and Kibana to ensure consistent access controls across services. Created using excalidraw, https://excalidraw.com/.
ETL pipeline development
Data harmonization capabilities represent another significant enhancement. The platform now includes automated ETL pipelines for standardizing data across different trials and sites. To ensure consistent data processing and standardization, we developed an automated ETL pipeline architecture with specific adaptations for temporal clinical trial data (21). Our implementation, shown in Figure 2, extends their approach by incorporating clinical trial-specific data validation and temporal relationship preservation. The pipeline includes automated ETL processes that maintain data synchronization between PostgreSQL databases (17) and ElasticSearch indices, ensuring near real-time availability of temporal analytics (16, 18, 19). The enhanced data dictionary management system enables flexible adaptation to different clinical trial protocols while maintaining standardized data structures.
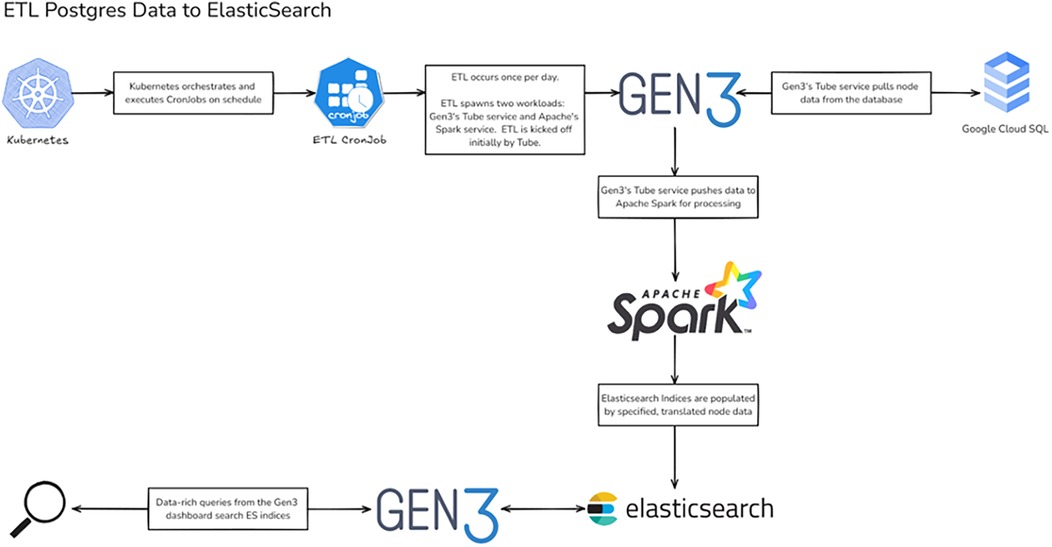
Figure 2. ETL pipeline architecture for Gen3 clinical trial data. Workflow diagram illustrating the ETL process from PostgreSQL to Elasticsearch. The pipeline leverages Kubernetes for orchestration, with Gen3's Tube service coordinating data extraction and Apache Spark handling transformation before loading into Elasticsearch indices. Created using excalidraw, https://excalidraw.com/.
The enhanced data dictionary management system enables flexible adaptation to different clinical trial protocols while maintaining standardized data structures. Validation focused on ensuring accurate representation of clinical trial trajectories and proper handling of temporal data harmonization across diverse measurement types and collection schedules.
Results
Temporal analytics validation
The enhanced Gen3 platform's temporal analytics capabilities required specific validation approaches to ensure reliability for clinical trial time series analysis. The integration of Kibana-based visualizations introduced new requirements for validating both data accuracy and analytical functionality across longitudinal datasets (16).
Validation of the temporal visualization framework demonstrated precise handling of varied time-based data structures, from regular visit schedules to irregular event-based capturing. Testing encompassed multiple temporal granularities, from daily patient-reported outcomes to monthly clinical assessments, while maintaining proper temporal relationships between different measurement types. The system successfully handled common clinical trial challenges including missing timepoints, out-of-window measurements, and protocol deviations.
ETL pipeline performance
The Elasticsearch ETL pipeline validation confirmed accurate transformation of diverse time-based measurements into standardized formats while preserving temporal precision and measurement context. Testing verified proper data synchronization between PostgreSQL databases and ElasticSearch indices, with successful handling of the data volume and complexity typical of multi-site clinical trials.
System performance and usability
Performance validation addressed the demands of temporal queries and real-time filtering. The platform maintained responsive performance when generating time-based visualizations across large datasets, with successful testing of concurrent users performing temporal analyses. Dashboard refresh rates remained within acceptable limits even when applying complex temporal filters and aggregations, ensuring practical utility for trial monitoring and interim analyses.
The platform's temporal visualization capabilities are demonstrated through a patient-reported outcomes (PROMIS) dashboard (Figure 3), which enables tracking of multiple outcome measures over time. Real-time trial monitoring capabilities are exemplified in the chronic pain clinical trial dashboard (Figure 4), which presents both temporal trends and distribution patterns of key outcome measures.
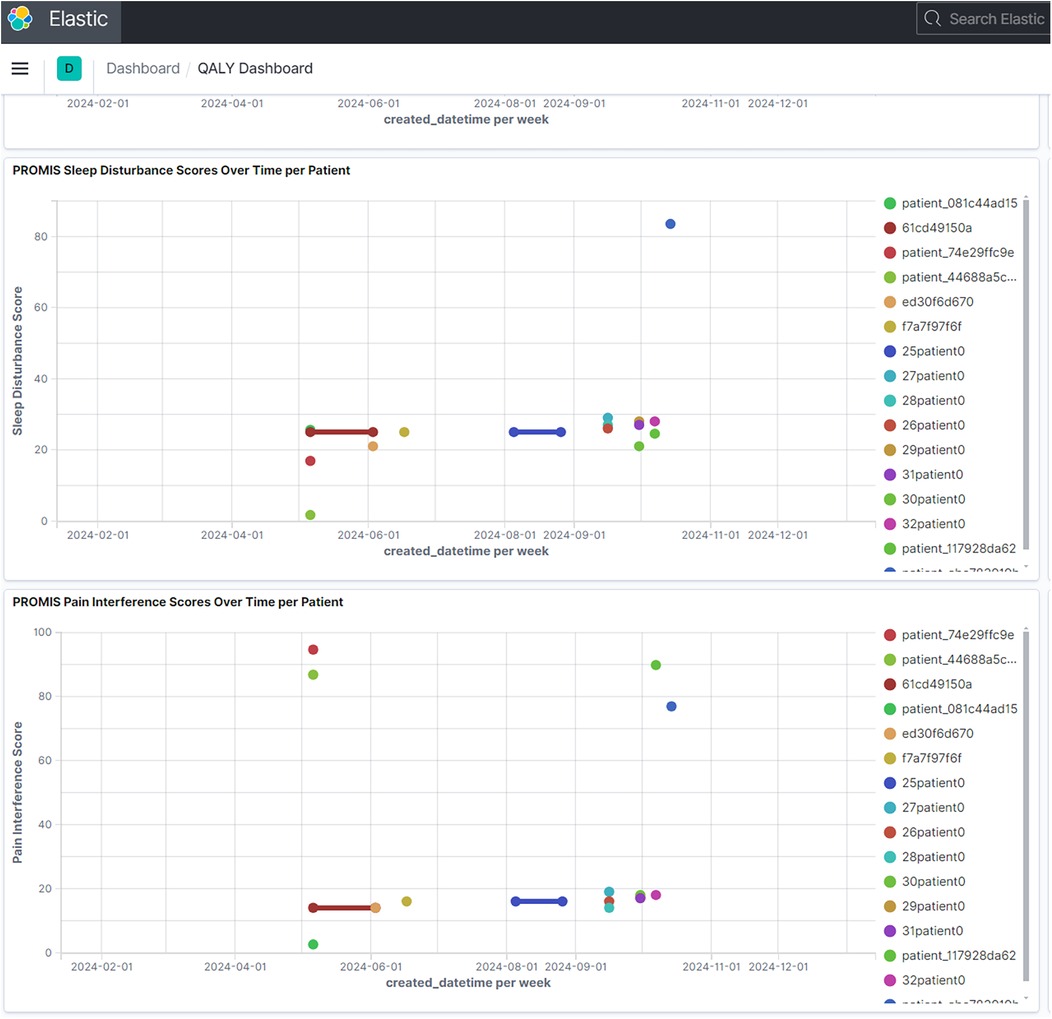
Figure 3. PROMIS patient-reported outcomes dashboard. Example of temporal visualization capabilities showing longitudinal patient-reported outcomes. Multiple measures including sleep disturbance, social activity, pain interference, and pain intensity are tracked over time, demonstrating the platform's ability to handle diverse temporal data streams. (Reproduced with permission from Wake Forest University, https://impowrgen3.wakehealth.edu/login).
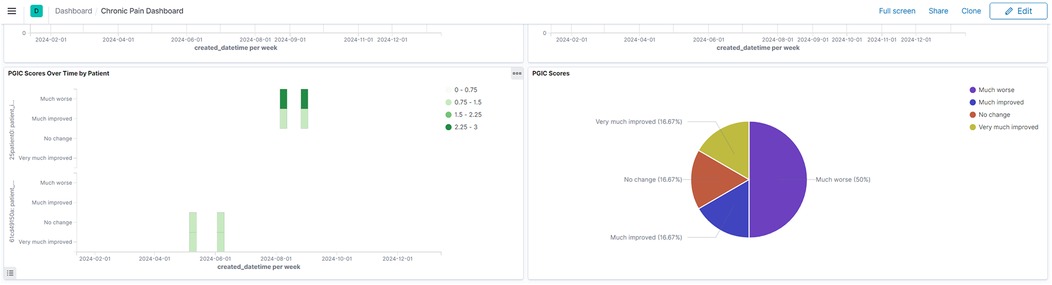
Figure 4. Chronic pain clinical trial monitoring dashboard. Interactive dashboard showing aggregated trial outcomes including PGIC scores and GAD-2 anxiety measures. The visualization demonstrates both temporal trends and distribution patterns, enabling real-time monitoring of trial progress and patient outcomes. (Reproduced with permission from Wake Forest University, https://impowrgen3.wakehealth.edu/login).
The platform's automated testing framework incorporates containerized test suites that validate both data integrity and temporal visualization accuracy. Performance validation demonstrated sustained responsiveness under concurrent user loads, with dashboard refresh rates remaining under acceptable thresholds even when applying complex temporal filters across large datasets (22).
Discussion
The integration of temporal analytics capabilities into the Gen3 data commons platform addresses a fundamental need in clinical research: the ability to understand and analyze treatment effects over time while maintaining the security and standardization benefits of a data commons architecture. Research teams can now track critical outcome measures across multiple timepoints without requiring specialized informatics support.
Technical contributions and enhancements
The integration of Kibana into the Gen3 stack significantly enhanced visualization capabilities beyond native architecture limitations. Our specific modifications to existing frameworks include: adaptation of ElasticSearch indexing for clinical trial temporal data structures, development of OAuth2 integration patterns for multi-service authentication, and implementation of automated ETL processes specifically designed for patient-reported outcome measures.
Broader research implications
The standardized approach to temporal visualization enables cross-trial comparisons and meta-analyses, fostering deeper understanding of treatment trajectories across different patient populations and interventions. In the NIH HEAL Initiative networks, researchers can now visualize patterns of patient-reported outcomes across multiple trials (Figures 3,4), leading to insights about treatment effectiveness that were previously difficult to obtain.
These enhancements align with evolving requirements for clinical trial transparency while maintaining appropriate privacy protections. The platform eliminates the need for specialized informatics support through Jupyter notebooks (9), democratizing data access and facilitating independent exploration of temporal patterns, evaluation of potential secondary analyses, and development of new research hypotheses (23).
The enhanced platform demonstrates particular utility in multi-site clinical trials, where real-time monitoring of longitudinal outcomes is essential for trial management. Research coordinators can track enrollment progress and outcome measure completion across sites, while investigators can examine temporal trends in key endpoints as they emerge.
Future directions
Integration with emerging clinical trial standards and data models presents a key opportunity for expanding interoperability. Development of more sophisticated statistical analysis tools integrated with temporal visualizations represents another promising direction, while maintaining the platform's emphasis on user-friendly interfaces.
The current implementation's cloud-agnostic architecture positions it well to incorporate emerging technologies for improved performance and scalability. Expansion of temporal visualization capabilities to support adaptive trial designs and integration with real-time data streams from wearable devices could extend the platform's utility for modern clinical trial designs.
The growing emphasis on patient-centered research suggests a need for developing interfaces that can effectively communicate temporal patterns to trial participants while maintaining appropriate data protections and scientific rigor.
Limitations
Several limitations should be acknowledged in this work. The technical implementation requires specific infrastructure and expertise that may not be readily available at all research institutions. The platform's dependency on the Kubernetes, Elasticsearch, and Kibana technology stack creates potential vendor lock-in despite efforts to maintain cloud-agnostic deployment.
From a clinical implementation perspective, the system requires training for clinical staff and may introduce workflow changes that could temporarily disrupt established data collection processes. The data migration process from existing clinical trial management systems presents potential challenges for ongoing studies.
The generalizability of this implementation may be limited by its specific design for NIH HEAL Initiative requirements. While the approach is broadly applicable, adaptation to other clinical trial contexts may require significant customization. The resource requirements for full implementation, including technical expertise and infrastructure costs, may limit adoption in resource-constrained research environments.
Finally, while the platform demonstrates improved temporal visualization capabilities, long-term usability studies and comprehensive user satisfaction assessments were not conducted as part of this implementation project.
Conclusion
The enhancement of the Gen3 data commons platform to support temporal analytics and dynamic data visualizations represents a crucial advancement for clinical trial infrastructure. Through the implementation of cloud-native architecture and integrated visualization capabilities, Gen3 now provides a framework that addresses fundamental needs in clinical trial data sharing and analysis while adhering to FAIR principles.
The specific technical contributions include: successful integration of temporal visualization tools with existing data commons architecture, development of automated ETL pipelines for clinical trial data harmonization, implementation of security frameworks suitable for multi-site clinical research, and creation of user-friendly interfaces that eliminate the dependency on specialized informatics support. These enhancements demonstrate measurable improvements in data accessibility and analytical capabilities for clinical trial networks.
The success of this implementation provides important lessons for the broader research community. It demonstrates that established data commons platforms can be effectively adapted for specialized research domains without compromising core data sharing capabilities. The cloud-agnostic approach ensures platform sustainability and broad applicability, while thoughtful integration of visualization tools enhances the utility of shared research data.
As clinical trials become increasingly complex and data-intensive, sophisticated yet accessible data commons platforms will continue to be essential. The enhancements described here provide a foundation for future developments in clinical trial data sharing infrastructure. By enabling robust temporal analytics while maintaining security and standardization, this approach advances the goal of making clinical trial data more findable, accessible, interoperable, and reusable for the broader research community.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
MA: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. CG: Conceptualization, Investigation, Methodology, Project administration, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. HA: Conceptualization, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SB: Conceptualization, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. RH: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. UT: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Research reported in this publication was supported by NIH HEAL Initiative under National Institute on Drug Abuse under grant numbers R24DA055306, U24DA057612, R25DA061740 and R24DA058606. This report does not represent the official view of the National Cancer Institute (NCI), the National Institutes of Health (NIH), or any part of the US Federal Government. No official support or endorsement of this article by the NCI or NIH is intended or should be inferred.
Conflict of interest
CG is the founder/owner of Krumware LLC. HA and SB are employees of Krumware LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Boeckhout M, Zielhuis GA, Bredenoord AL. The fair guiding principles for data stewardship: fair enough? Eur J Hum Genet. (2018) 26(7):931–6. doi: 10.1038/s41431-018-0160-0
2. Shitrit G, Tractinsky N, Moskovitch R. Visualization of frequent temporal patterns in single or two populations. J Biomed Inform. (2022) 134:104169. doi: 10.1016/j.jbi.2022.104169
3. Adams MCB, Hurley RW, Siddons A, Topaloglu U, Wandner LD. NIH heal clinical data elements (CDE) implementation: NIH heal initiative IMPOWR network idea-cc. Pain Med. (2023) 24(7):743–9. doi: 10.1093/pm/pnad018
4. Wyatt KD, Graglia L, Furner B, Kang B, Fitzsimons M, Grossman RL, et al. An open-source platform for pediatric cancer data exploration: a report from data for the common good. JAMIA Open. (2024) 7(1):ooae004. doi: 10.1093/jamiaopen/ooae004
5. Adams MCB, Griffin C, Adams H, Bryant S, Hurley RW, Topaloglu U. Adapting the open-source Gen3 platform and kubernetes for the NIH heal IMPOWR and MIRHIQL clinical trial data commons: customization, cloud transition, and optimization. J Biomed Inform. (2024) 159:104749. doi: 10.1016/j.jbi.2024.104749
6. Adams MCB, Hurley RW, Topaloglu U. Connecting chronic pain and opioid use disorder clinical trials through data harmonization: wake forest IMPOWR dissemination, education, and coordination center (IDEA-CC). Subst Use Addctn J. (2025) 46(1):141-145. doi: 10.1177/29767342241236287
7. Balise RR, Hu MC, Calderon AR, Odom GJ, Brandt L, Luo SX, et al. Data cleaning and harmonization of clinical trial data: medication-assisted treatment for opioid use disorder. PLoS One. (2024) 19(11):e0312695. doi: 10.1371/journal.pone.0312695
8. Lacey JV Jr., Chung NT, Hughes P, Benbow JL, Duffy C, Savage KE, et al. Insights from adopting a data commons approach for large-scale observational cohort studies: the California teachers study. Cancer Epidemiol Biomarkers Prev. (2020) 29(4):777–86. doi: 10.1158/1055-9965.EPI-19-0842
9. Trunnell M, Frankenberger C, Hota B, Hughes T, Martinov P, Ravichandran U, et al. The pandemic response commons. JAMIA Open. (2024) 7(2):ooae025. doi: 10.1093/jamiaopen/ooae025
10. Grossman RL, Dry JR, Hanlon SE, Johann DJ, Kolatkar A, Lee JSH, et al. Bloodpac data commons for liquid biopsy data. JCO Clin Cancer Inform. (2021) 5:479–86. doi: 10.1200/CCI.20.00179
11. Jin N, Li Z, Kettler C, Yang B, Tu W, Su J. Ardac common data model facilitates data dissemination and enables data commons for modern clinical studies. Stud Health Technol Inform. (2024) 310:3–7. doi: 10.3233/SHTI230916
12. Kang H, Liu G, Wang Q, Meng L, Liu J. Theory and application of zero trust security: a brief survey. Entropy. (2023) 25(12):1595. doi: 10.3390/e25121595
13. Xuan S, Yang W, Dong H, Zhang J. Performance evaluation model for application layer firewalls. PLoS One. (2016) 11(11):e0167280. doi: 10.1371/journal.pone.0167280
14. Choi J, Kim J, Lee DK, Jang KS, Kim DJ, Choi IY. The OAuth 2.0 web authorization protocol for the internet addiction bioinformatics (IABIo) database. Genomics Inform. (2016) 14(1):20–8. doi: 10.5808/GI.2016.14.1.20
15. Scheer J, Volkert A, Brich N, Weinert L, Santhanam N, Krone M, et al. Visualization techniques of time-oriented data for the comparison of single patients with multiple patients or cohorts: scoping review. J Med Internet Res. (2022) 24(10):e38041. doi: 10.2196/38041
16. Scott-Boyer MP, Dufour P, Belleau F, Ongaro-Carcy R, Plessis C, Perin O, et al. Use of elasticsearch-based business intelligence tools for integration and visualization of biological data. Brief Bioinform. (2023) 24(6). doi: 10.1093/bib/bbad348
17. Hackl WO, Neururer SB, Schweitzer M, Pfeifer B. Making a virtue of necessity - a highly structured clinical data warehouse as the source of assured truth in a hospital. Stud Health Technol Inform. (2023) 301:180–5. doi: 10.3233/SHTI230036
18. Banda JM, Halpern Y, Sontag D, Shah NH. Electronic phenotyping with aphrodite and the observational health sciences and informatics (ohdsi) data network. AMIA Jt Summits Transl Sci Proc. (2017) 2017:48–57.28815104
19. Bischof AY, Kuklinski D, Salvi I, Walker C, Vogel J, Geissler A. A collection of components to design clinical dashboards incorporating patient-reported outcome measures: qualitative study. J Med Internet Res. (2024) 26:e55267. doi: 10.2196/55267
20. Wang Z, Yu X, Xue P, Qu Y, Ju L. Research on medical security system based on zero trust. Sensors (Basel). (2023) 23(7):3774. doi: 10.3390/s23073774
21. Ong TC, Kahn MG, Kwan BM, Yamashita T, Brandt E, Hosokawa P, et al. Dynamic-Etl: a hybrid approach for health data extraction, transformation and loading. BMC Med Inform Decis Mak. (2017) 17(1):134. doi: 10.1186/s12911-017-0532-3
22. Gulden C, Macho P, Reinecke I, Strantz C, Prokosch HU, Blasini R. Recruit: a cloud-native clinical trial recruitment support system based on health level 7 fast healthcare interoperability resources (HL7 FHIR) and the observational medical outcomes partnership common data model (OMOP CDM). Comput Biol Med. (2024) 174:108411. doi: 10.1016/j.compbiomed.2024.108411
Keywords: data commons, cloud computing, opioid, chronic pain, Kubernetes, Gen3, timeseries, patient reported outcomes
Citation: Adams MCB, Griffin C, Adams H, Bryant S, Hurley RW and Topaloglu U (2025) Enhancing Gen3 for clinical trial time series analytics and data discovery: a data commons framework for NIH clinical trials. Front. Digit. Health 7:1570009. doi: 10.3389/fdgth.2025.1570009
Received: 2 February 2025; Accepted: 23 June 2025;
Published: 23 July 2025.
Edited by:
Eugenia Rinaldi, Charité Medical University of Berlin, GermanyReviewed by:
David Phillip Nickerson, University of Auckland, New ZealandParinaz Tabari, University of Salerno, Italy
Copyright: © 2025 Adams, Griffin, Adams, Bryant, Hurley and Topaloglu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meredith C. B. Adams, bWVyYWRhbXNAd2FrZWhlYWx0aC5lZHU=
†ORCID:
Meredith C. B. Adams
orcid.org/0000-0002-3969-4279
Robert W. Hurley
orcid.org/0000-0001-6591-9390
 Meredith C. B. Adams
Meredith C. B. Adams Colin Griffin
Colin Griffin Hunter Adams4
Hunter Adams4 Umit Topaloglu
Umit Topaloglu