- 1Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, China
- 2Department of Endocrinology and Metabolism, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 3Department of Gastroenterology and Haematology, Anning First People’s Hospital Affiliated to Kunming University of Science and Technology, Kunming, Yunnan, China
Introduction: Traditional fermented foods have recently been recognized for their potential benefits in managing hyperuricemia (HUA) and gout.
Methods: This study evaluated the purine degradation ability of seventy-eight lactic acid bacteria (LAB) isolated from traditional fermented foods in Yunnan Province, China, by HPLC. The possible mechanisms of in vitro purine degradation were explored through whole-genome sequencing, comparative genomics, and qRT-PCR, and the effect of the LAB on HUA in SD rats was verified.
Results and Discussion: In vitro results demonstrated that Limosilactobacillus fermentum MX-7 and GL-1-3L exhibited high degradation ratios for guanine, while Pediococcus acidilactici GJ09-3-7L showed good potential in degrading xanthine. All three strains were also effective in degrading inosine and guanosine. And the genomes of all three strains contained a high number of enzymes related to purine metabolism, transporter and permease. In vivo results suggested that the MX-7 strain not only lowering serum uric acid (UA) and urea nitrogen levels in HUA SD rats but also providing a protective effect on renal function. These findings indicate that the MX-7 strain could serve as a promising adjunctive therapy for treating HUA.
1 Introduction
Purine is a vital compound in the human body, serving as a key component of nucleic acids, coenzymes, signaling molecules, and energy transfer molecules (Nassogne et al., 2024). It is gradually broken down into uric acid (UA) through the action of xanthine oxidase (Bortolotti et al., 2021; Yang et al., 2022). Hyperuricemia (HUA), the second most common metabolic disorder after diabetes, is closely associated with conditions like gout, chronic kidney disease, and diabetes, posing a significant threat to human health (Lin et al., 2022; Yuan et al., 2022; Zheng et al., 2023). The global incidence of HUA has increased by approximately 20% (Chen-Xu et al., 2019; Cao et al., 2022). According to statistics, the prevalence of HUA among American adults was about 21.4, with 21.2 of men and 21.6% of women affected from 2007–2008 (Zhu et al., 2011). In East China, by 2019, the prevalence ratio for people aged 20–79 was approximately 18.7, with 24.8% of men and 5.6% of women affected, with young men (aged 20–29) emerging as the primary group affected by HUA (She et al., 2022). In the human body, UA is derived from both endogenous and exogenous sources (Li et al., 2018; Liu et al., 2018; Büsing et al., 2019). Endogenous UA, which accounts for 80–90% of the total UA in the body, results from the metabolism of nucleic acids (Li et al., 2018; Liu et al., 2018; Büsing et al., 2019). Exogenous UA, making up 10–20% of the body’s total UA, comes from dietary purine-rich foods, such as organ meats, seafood, meat broths, and sugary beverages (Li et al., 2018; Liu et al., 2018; Büsing et al., 2019). Therefore, modifying dietary habits to reduce the intake of purine-rich foods can help lower UA production, contributing to the prevention and management of HUA (Zhang et al., 2022).
Drug therapy remains the primary approach for lowering serum UA levels. Xanthine oxidase inhibitors, such as allopurinol and febuxostat, work by blocking the conversion of xanthine and hypoxanthine to UA, a process catalyzed by xanthine oxidase (Becker et al., 2005; Schumacher et al., 2008; Bardin and Richette, 2020; Liu G. et al., 2020; Mackenzie et al., 2020; Sekine et al., 2023). However, these drugs may be associated with adverse effects, including skin reactions, diarrhea, nausea, headaches, liver and kidney damage, and eosinophilia (Chen et al., 2019; Dong et al., 2023; Hali et al., 2023; Mohammad et al., 2023; Mao et al., 2025). Other drugs, such as benzbromarone (Benn et al., 2018; Strilchuk et al., 2019) and probenecid (Martínez-Reyes et al., 2020), help promote the excretion of UA and reduce its reabsorption. Nevertheless, benzbromarone has been linked to potential liver toxicity (Kaufmann et al., 2005), while probenecid may cause side effects like skin reactions, headaches, nausea, vomiting, loss of appetite, and gastrointestinal discomfort (Boger and Strickland, 1955; Colín-González Laura and Santamaría, 2013; Garcia-Rodriguez et al., 2023; Mazza et al., 2024). These challenges highlight the urgent need for new therapeutic approaches or alternative drugs (Zhang M. et al., 2023; Mao et al., 2025).
Lactic acid bacteria (LAB) are commonly found in naturally fermented foods (Soemarie et al., 2021). LAB has many benefits, for example, preventing food spoilage (Shehata et al., 2019; Riolo et al., 2024), removing mycotoxins from beverages (Nahle et al., 2022), and enhancing intestinal function and boosting immunity (Soemarie et al., 2021). Recent studies have shown that LAB can participate in the metabolism and degradation of purines, suggesting their potential for developing low-purine foods. For example, Li et al. (2014) demonstrated that Lactobacillus brevis DM9218-A can lower serum UA levels in male HUA Wistar rats (30 days old), indicating its potential to prevent and aid in the treatment of HUA. Similarly, Yamada et al. (2017) found that Lactobacillus gasseri PA-3 can bind and absorb inosine and related compounds in vitro, and in vivo studies showed that this strain reduces intestinal absorption of inosine 5’-monophosphate, inosine, and hypoxanthine in 8 weeks old Wistar rats. In another study, Lee et al. (2022) isolated and screened Lacticaseibacillus paracasei MJM60396, a strain capable of absorbing 100% of inosine and guanosine from fermented foods. After administering this strain to male HUA C57BL/6 mice (7 weeks old) for 3 weeks, their serum UA levels returned to normal. Further experiments revealed that L. paracasei MJM60396 prevents HUA by absorbing purines, reducing UA synthesis, and increasing UA excretion (Lee et al., 2022). Lu et al. (2022) isolated Limosilactobacillus fermentum 9–4 from Chinese fermented rice-flour noodles and demonstrated its efficient degradation of inosine and guanosine, suggesting its potential for low-purine food development. Wu et al. (2021) isolated Limosilactobacillus fermentum JL-3 from fermented food and showed its potential as an adjunctive treatment for HUA. However, the mechanisms by which LAB degrade purines remain unclear, and further research is needed, including whole-genome sequencing and studies on the expression of relevant enzymes. Yunnan Province is renowned for its diverse and vibrant traditional fermented foods (Liu et al., 2015, 2019; Liu X. F. et al., 2020; Zhang S. et al., 2023; Du et al., 2024), which offer a rich source of functional lactic acid bacteria. However, there has been limited research on the purines-degrading capabilities of lactic acid bacteria isolated from these foods.
The aim of this study was to isolate LAB capable of degrading purines from traditional fermented foods in Yunnan Province, and to explore the potential mechanisms of purines degradation in vitro through whole-genome sequencing and qRT-PCR. Additionally, the study sought to evaluate the effects of these LAB strains on HUA in SD rats. This research lays the foundation for further exploration of the microbial resources in Yunnan Province, offers potential candidate strains for developing low-purine foods, and presents a novel approach for the adjunctive treatment of HUA.
2 Materials and methods
2.1 Preparation of lactic acid bacteria
All LAB strains used in this experiment were from the microbial resource development and application research group, Kunming University of Science and Technology (Kunming, Yunnan, China). They were isolated from traditional fermented foods in Yunnan Province. 78 strains were randomly selected for the determination of purines degradation ability. 16S rRNA gene amplicon sequencing to identify bacteria were performed as described by Loong et al. (2016) (Supplementary Table 1), and primers 27F and 1492R were used for PCR amplification (Eden et al., 1991).
2.2 Determination of purines degradation ability of strains in vitro
In vitro assays, degradation ratios, quantitative standard curves and HPLC conditions were performed as described by Wu et al. (2021). In short, firstly, the washed LAB were incubated with 1 mL guanine, hypoxanthine, xanthine, inosine and guanosine (Yuanye, China) solution (20 μg/mL) respectively, at a concentration of 1 × 109CFU/mL, and incubated at 37°C and 180 rpm for 24 h (inosine and guanosine for 12 h). Secondly, purines were sampled at 0, 6, 12, 18 and 24 h, and nucleosides at 0 min, 20 min, 40 min and 60 min. Thirdly, High temperature and pressure (90°C, 10 min) to obtain the non-viable strains. Finally, sonication to obtain the cell-free extracts of strains (Li et al., 2014), and sonication as described by Feliu et al. (1998). The purines remaining contents were calculated by HPLC (Agilent LC1260, United States).
The conditions for HPLC were as follows: Agilent TC-C18(2) (4.6 mm × 250 mm × 5 μm; United States), the mobile phase for purines were ammonium acetate (LC-MS grade; Thermo Fisher, United States)/methanol (LC-MS grade; Thermo Fisher, United States), 90:10, v/v (nucleosides were 30:10, v/v), while column temperature was 30°C, with wavelength of 254 nm, injection volume of 20 μL and the retention time of 15 min. Three biological replicates were performed for all experiments.
2.3 Whole-genome sequencing, assembly, annotation and comparative genomics analysis
Limosilactobacillus fermentum MX-7 (NODE: OEZ00020971), GL-1-3L (NODE: OEZ00020969) and Pediococcus acidilactici GJ09-3-7L (NODE: OEZ00020970) were sent to Shanghai Yuanxu Biotechnology Co., Ltd. (Shanghai, China), and the second and third generations of whole-genome sequencing were performed using the Illumina NovaSeq PE150 and Oxford Nanopore ONT sequencing platforms, respectively (Chen et al., 2023). Assembly and annotation were performed as described by Chen et al. (2023), Niu et al. (2024), and Liu et al. (2024). Comparative genomic analysis of genes encoding nucleoside permease (Yamauchi et al., 2020), xanthine/uracil permeases, cytosine/uracil/thiamine/allantoin permeases, cytosine deaminase and related metal-dependent hydrolases, xanthine/uracil permeases, purine-nucleoside phosphorylase and guanine deaminase (GDA).
2.4 Validation by quantitive reverse transcription-PCR
The L. fermentum MX-7, GL-1-3L and P. acidilactici GJ09-3-7L were incubated into 50 mL MRS liquid medium at a concentration of 1 × 109CFU/mL, containing 40 μg/mL guanine. After incubated at 37°C and 180 rpm for 24 h, total RNA was extracted using EASYspin Plus Bacterial RNA Extraction Kit (Aidlab, China), and qRT-PCR experiments were performed using HiScript® II One Step qRT-PCR SYBR Green Kit (Vazyme, China). Primer sequences were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China). GDA gene (F: CACACCCTCTTAGCCAACGGA, R: GATTATCCATCGCCACACGC). With the 16S rRNA gene as the internal reference (Li et al., 2020), strains cultured without purines were used as the control group, and the 2–ΔΔCt method was utilized to normalize the expression results (Cao et al., 2022). Three biological replicates were performed for all experiments.
2.5 Potential probiotic characterization in vitro
Acid and bile tolerance, antimicrobial activity and the sensibility to the antibiotics of Limosilactobacillus fermentum MX-7 were performed as described by Li et al. (2014). In short, inoculating MX-7 strain in MRS liquid medium (HuanKai Microbial, China) with pH = 2.0 and 3.0. The bile salt (Yuanye, China) solutions at final concentrations of 0.3%. The pathogenic bacteria were Staphylococcus aureus, Escherichia coli, Salmonella enterica, Salmonella pullorum and Listeria monocytogenes. The contents of antibiotic discs (Binhe, China) were shown in Table 4. Three biological replicates were performed for all experiments.
2.6 Animals and experimental design
Thirty specific pathogen-free (SPF) 8–10 weeks old male SD rats were purchased from the Department of Experimental Animal Science, Kunming Medical University (Kunming, Yunnan, China), with one rat per cage. The rats were maintained at 23°C ± 2°C and 50% ± 10% relative humidity, with a 12 h/12 h light/dark cycle. All rats were fed a cobalt-60 irradiated basic feed purchased from Suzhou Shuangshi Experimental Animal Stall Food Technology Co., Ltd. (Suzhou, Jiangsu, China). The experiment was conducted in accordance with the animal management regulations of the Ministry of Science and Technology of China and approved by the Experimental Animal Ethics Committee of the Kunming University of Science and Technology (Approval No. PZWH -KUST-202501040001-1).
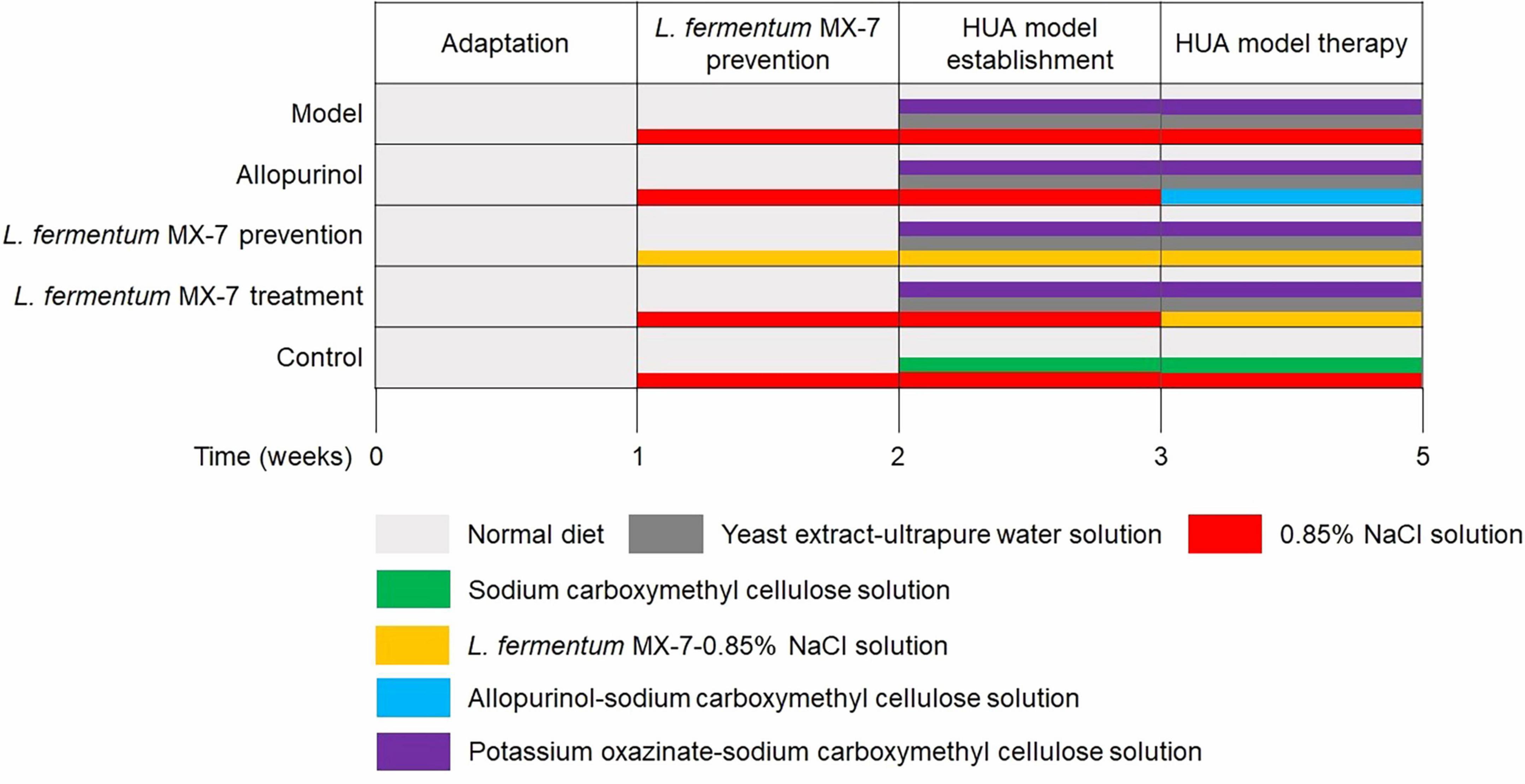
Establishment of HUA SD rats model as described by Cao et al. (2022) and Ni et al. (2021). In short, after a 1-week adaptation period, the rats were weighed and divided randomly into five groups (n = 6 per group). Figure 1 presents the details of the treatment protocol. In the second week, L. fermentum MX-7 prevention group was gavaged with 1 × 1010 CFU MX-7 strain-0.85% NaCl solution daily, and the other groups were gavaged with 0.85% NaCl solution. In the third week, the control group was added with sodium carboxymethyl cellulose solution (5 g/L; Yuanye, China), and the other groups were added with yeast extract (15 g/kg; Sangon, China)-ultrapure water solution and potassium oxazinate (250 mg/100 g; Yuanye, China)-sodium carboxymethyl cellulose solution. After successful modeling, allopurinol and MX-7 strain treatment groups replaced 0.85% NaCl solution with allopurinol (4.2 mg/100 g; Yuanye, China)-sodium carboxymethyl cellulose solution and 1 × 1010 CFU MX-7 strain-0.85% NaCl solution, respectively. On the 21st, 28th, and 35th days, 1 mL of tail vein blood was collected 60 min after gavage for serum isolation (Ni et al., 2021). After obtaining serum on the 35th day, two rats in each group were randomly selected to be anesthetized with 3% isoflurane and euthanized for cervical dislocation. One kidney per rats was fixed by 4% paraformaldehyde (Servicebio, China) for 24 h (Wu et al., 2021; Lee et al., 2022).
2.7 Serum biochemical analysis and histopathological assessment
The collected serum was measured for serum UA, serum urea nitrogen and serum creatinine by BECKMAN COULTER DxC 700 AU (BECKMAN COULTER, United States) automatic biochemical detector (Lee et al., 2022). The renal tissues were embedded in paraffin, sectioned and stained with Hematoxylin and Eosin (H&E), and observed under OLYMPUS BX51 microscope (OLYMPUS, Japan) (Wu et al., 2021; Lee et al., 2022). Six biological replicates were performed for serum biochemical analysis, and two for histopathological assessment.
2.8 Statistical analysis
The procedure of the animal experiment and representative arrangement of genomic regions from multiple organisms’ charts were generated using Microsoft PowerPoint 2016. Line and bar charts were generated using GraphPad Prism (v8). The KEGG purine metabolism maps were generated using the KEGG mapper visualization tool. Statistical significance was performed using one-way ANOVA with Tukey’s multiple comparison test with IBM SPSS Statistics (v27). p < 0.05 was considered statistically significant. The data was expressed as the mean ± SD.
3 Results
3.1 Isolation of purines degrading strains
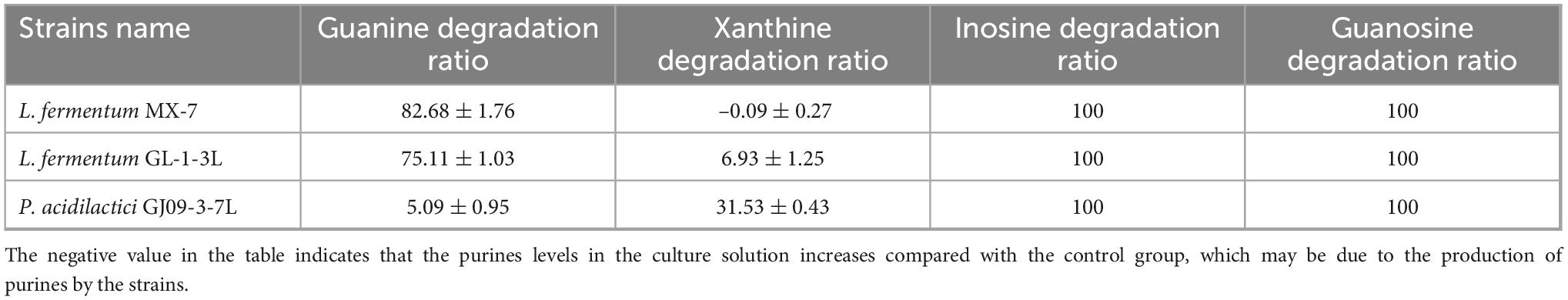
The quantitative standard curves and equations were shown in Supplementary Figure 1 and Supplementary Table 2. We measured the in vitro degradation ability of guanine, hypoxanthine and xanthine of seventy-eight strains of LAB (Supplementary Table 3). The best strains for guanine degradation were L. fermentum MX-7 and GL-1-3L, with degradation ratios of (82.68 ± 1.76)% and (75.11 ± 1.03)% respectively (Table 1). Limosilactobacillus fermentum MX-15 was the best strain to degrade hypoxanthine, but the degradation ratio was only (11.00 ± 0.28)%, almost no degradation ability. P. acidilactici GJ09-3-7L had the best degradation ability of xanthine, and the degradation ratio was (31.53 ± 0.43)% (Table 1). We selected three strains (MX-7, GL-1-3L and GJ09-3-7L) with the best degradation effect of guanine and xanthine to degrade inosine and guanosine. The three strains can degrade inosine and guanosine 100% (Table 1).
3.2 Purines degrading abilities undergo changes in vitro
3.2.1 Purines degradation abilities at different time points
The ability of L. fermentum MX-7 to degrade guanine was greater than that of L. fermentum GL-1-3L. The degradation ratios of MX-7 and GL-1-3L strains were (84.57 ± 0.01)% and (64.74 ± 0.01)%, respectively, at 24 h (Figure 2A). P. acidilactici GJ09-3-7L had no obvious degradation effect on xanthine within 0–12 h, and began to degrade xanthine after 12 h, but the degradation was slow, and the degradation ratio was only (41.59 ± 0.01)% at 24 h (Figure 2B). In addition, the three strains rapidly degraded inosine and guanosine from 0 to 40 min, and tended to be stable from 40 to 60 min. At 60 min, the degradation ratios of inosine and guanosine by the three strains were close to 100% (Figures 2C,D).
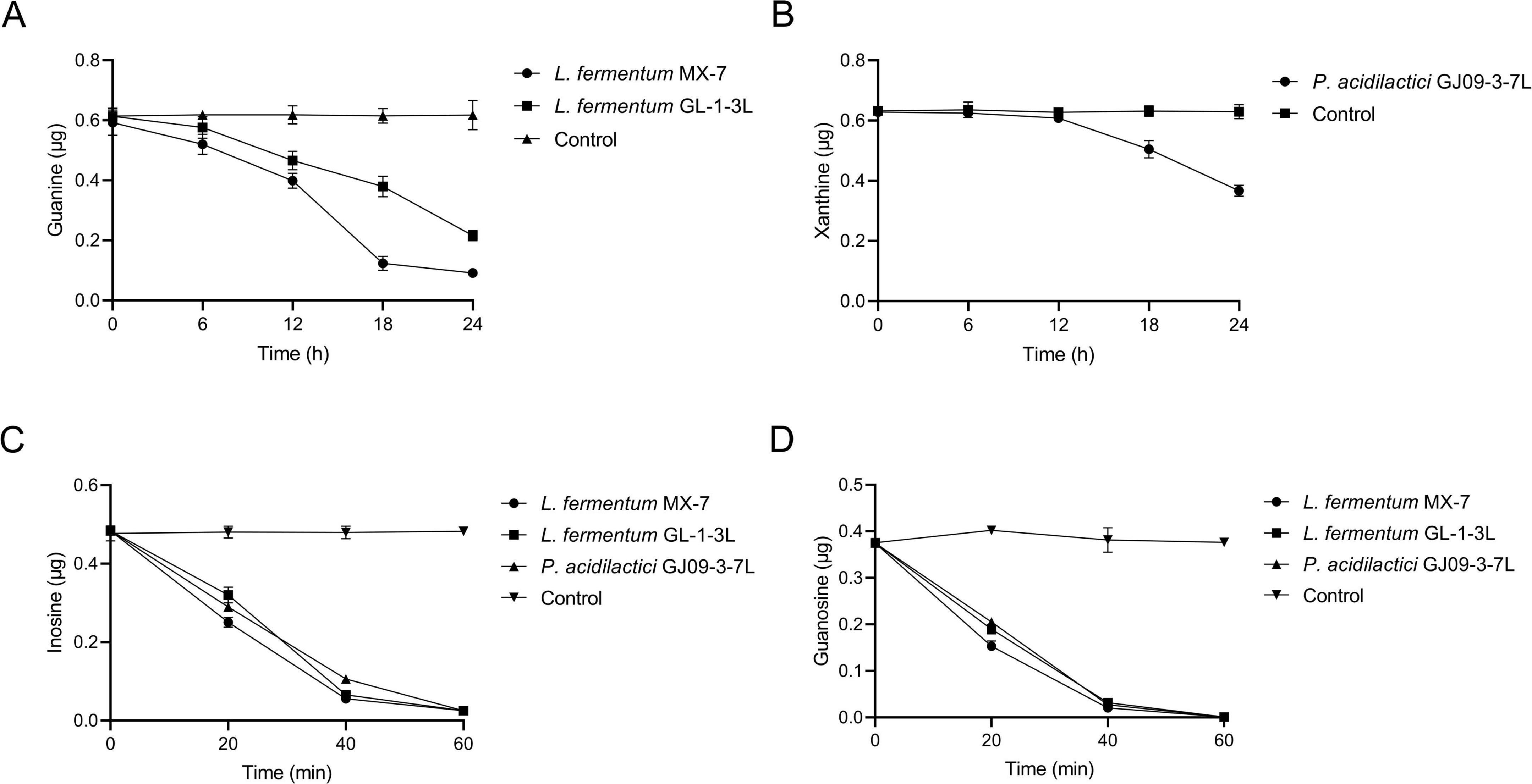
Figure 2. Purines degradation abilities of strains at different time points. (A) Guanine, (B) Xanthine, (C) Inosine, (D) Guanosine. Control: Control without strains.
3.2.2 Purines degradation abilities between non-viable and viable strains
The guanine degradation ability of non-viable L. fermentum MX-7 and GL-1-3L was significantly lower than that of viable cells (p < 0.001) (Figure 3A). The xanthine degradation ability of non-viable P. acidilactici GJ09-3-7L was significantly lower than that of viable cells (p < 0.001) (Figure 3B). In addition, compared with the viable strains, the degradation ability of the three non-viable strains to inosine (p < 0.001) and guanosine (p < 0.001, p < 0.01) was significantly reduced (Figures 3C,D).
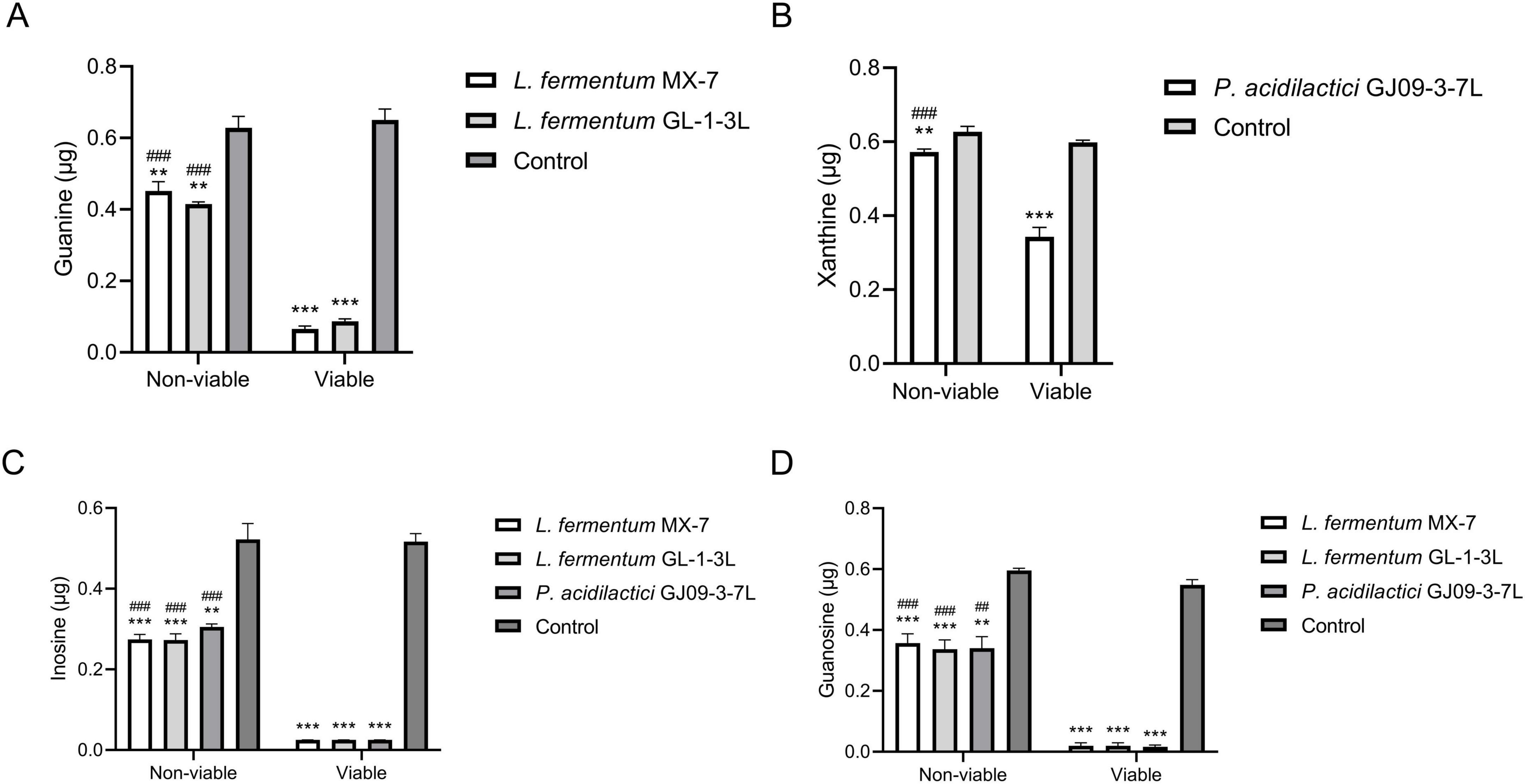
Figure 3. Purines degradation abilities between non-viable and viable strains. Non-viable and viable strains were incubated with guanine and xanthine for 24 h, inosine and guanosine for 60 min, respectively, and the remaining contents were calculated by HPLC. (A) Guanine, (B) Xanthine, (C) Inosine, (D) Guanosine. Control: Control without strains. ** p < 0.01, *** p < 0.001 compared with their respective control group. ## p < 0.01, ### p < 0.001 compared with their respective viable strains.
3.2.3 Purines degradation abilities of cell-free extracts
The precipitation of L. fermentum MX-7 and GL-1-3L had significantly higher degradation ability of guanine than their supernatant (p < 0.001) (Figure 4A). The precipitation of P. acidilactici GJ09-3-7L had significantly higher degradation ability of xanthine than its supernatant (p < 0.05) (Figure 4B). The degradation of inosine by the precipitation of MX-7 (p < 0.05) and GJ09-3-7L (p < 0.01) strains were significantly higher than that of their supernatant (Figures 4C,D).
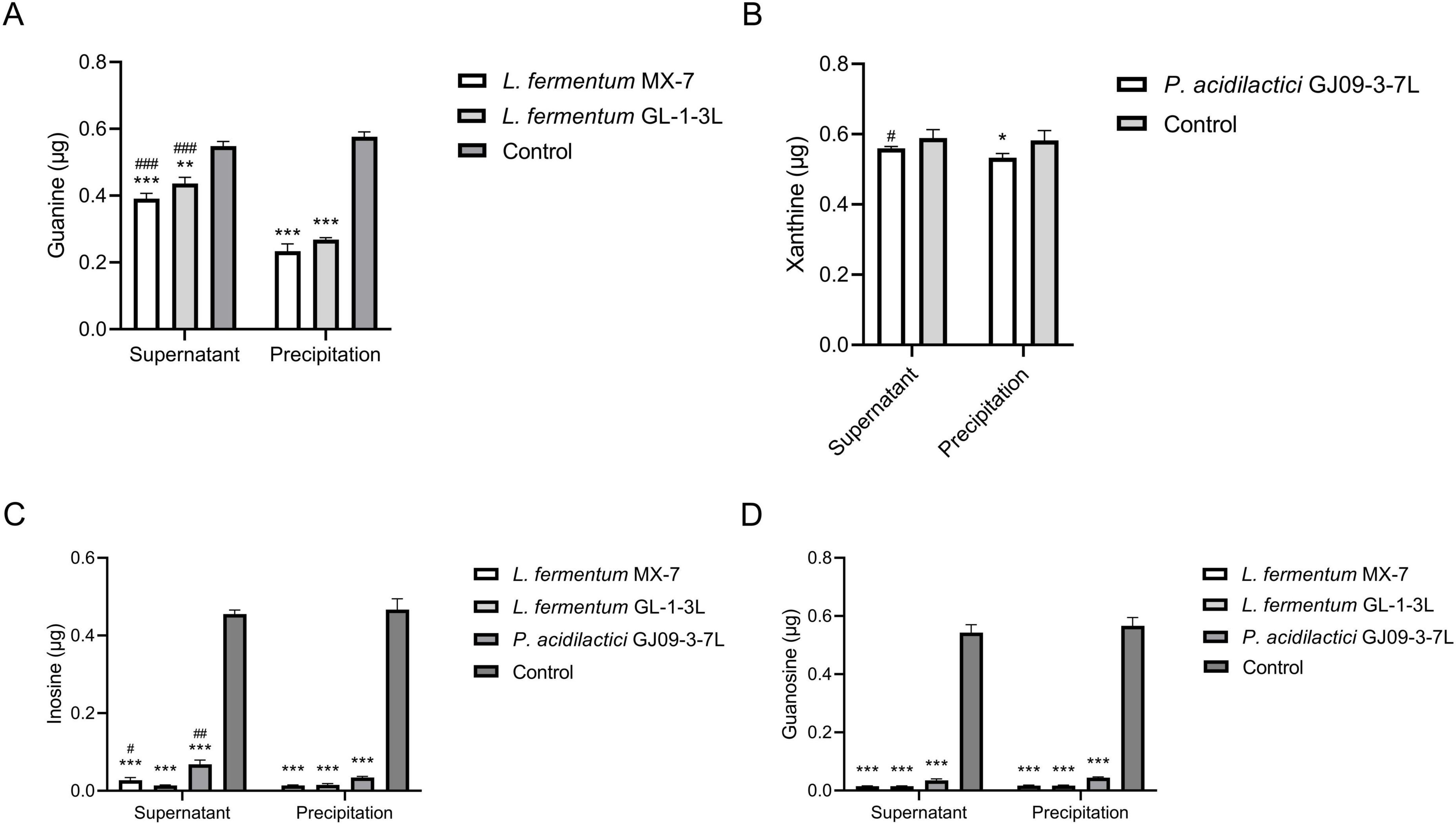
Figure 4. Purines degradation abilities of cell-free extracts. The washed 1 × 109 CFU strains were sonicated and centrifuged to obtain the supernatant and precipitation, respectively. They were incubated with guanine and xanthine for 24 h, inosine and guanosine for 60 min, respectively, and then and the remaining contents were calculated by HPLC. (A) Guanine, (B) Xanthine, (C) Inosine, (D) Guanosine. Control: Control without strains. *p < 0.05, **p < 0.01, ***p < 0.001 compared with their respective control group. #p < 0.05, ##p < 0.01, ###p < 0.001 compared with their respective precipitation.
3.3 Whole-genome and comparative genomics analysis
In order to further explore the molecular mechanism of purines degradation by LAB in vitro, we sequenced the whole-genome of L. fermentum MX-7, GL-1-3L and P. acidilactici GJ09-3-7L, which had good degradation effect on purines, in order to further clarify the related enzyme system of purines degradation. The genomic characteristics of the three strains was listed in Supplementary Table 4. As shown in the chromosome circle maps (Supplementary Figure 2), MX-7 strain contained 1 chromosome, GL-1-3L strain contained 1 chromosome and 2 plasmids and GJ09-3-7L strain contained 1 chromosome and 4 plasmids. Supplementary Figure 3 illustrated that, except for unknown functions, most of the COG functional categories genes were enriched in DNA replication, recombination, and repair, amino acid transport and metabolism, translation, ribosomal structure and biogenesis and carbohydrate transport and metabolism. According to the annotation result of KEGG (Supplementary Figure 4), genes responsible for amino acid metabolism or carbohydrate metabolism for the highest proportion. In addition, the KEGG purine metabolism maps showed that the GDA was present in all three strains and the purine-nucleoside phosphorylase was present only in GJ09-3-7L strain (Figure 5). There were five virulence genes in each of the genomes of the three strains (Supplementary Table 5), and there was also a resistance gene in MX-7 and GL-1-3L strains (Supplementary Table 6). Based on the current research results, the three strains could metabolize different types of carbon sources and had good propagation ability, and there were no obvious potential safety problems in the future application of the three strains.
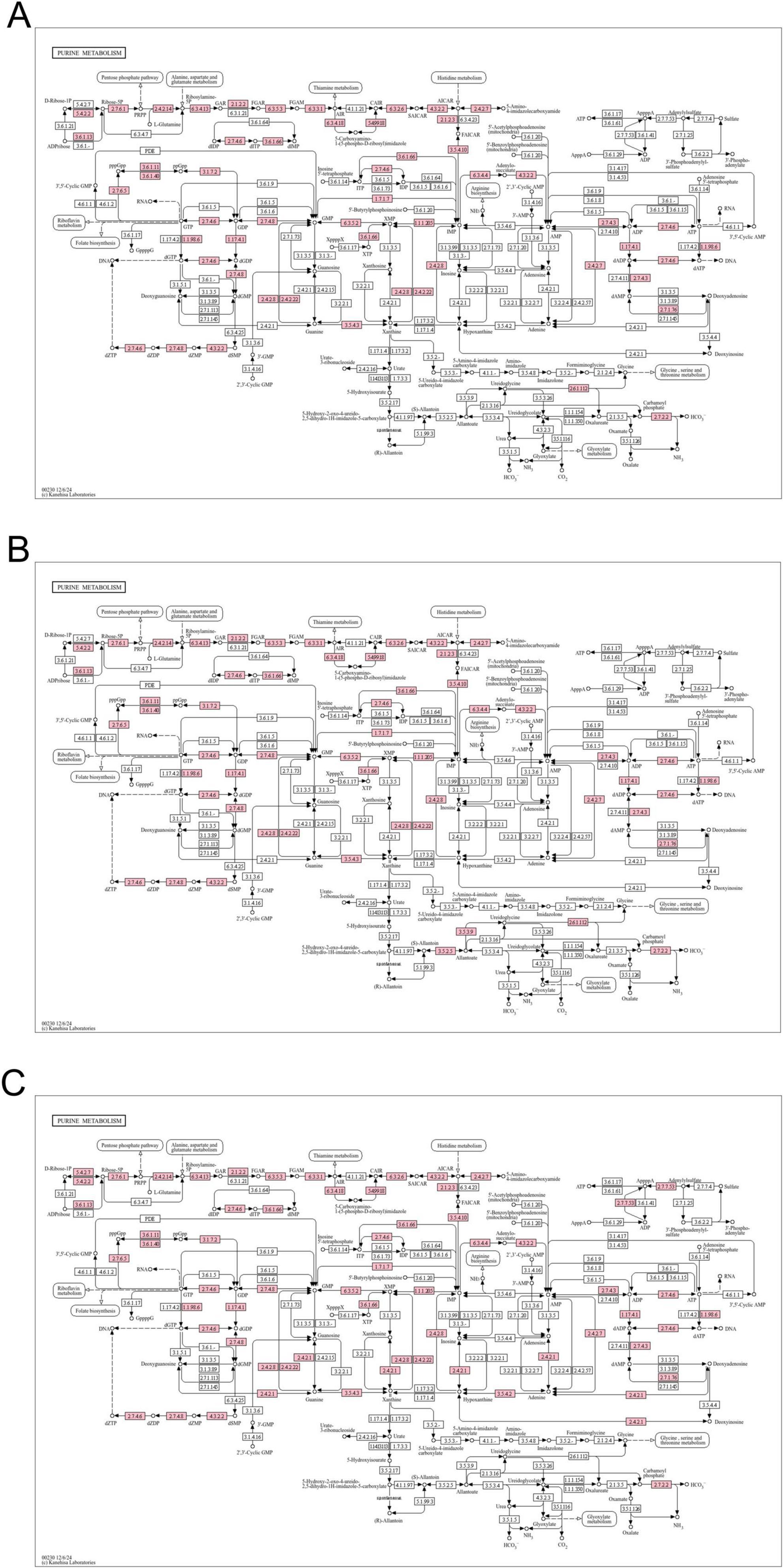
Figure 5. Purine metabolism maps. (A) L. fermentum MX-7, (B) L. fermentum GL-1-3L, (C) P. acidilactici GJ09-3-7L. Pink indicates the presence of the enzyme.
Comparative genomics revealed that MX-7, GL-1-3L, and GJ09-3-7L strains have a high number of enzymes related to purine transport and metabolism (Figure 6). The three strains all have cytosine deaminase, GDA and xanthine/uracil permease, which were largely absent in other strains. In addition, the presence of hydroxymethylpyrimidine transporter in the GL-1-3L strain and Limosilactobacillus fermentum 9-4, and the presence of allantoin permease only in the GL-1-3L strain demonstrate the unique ability of the GL -1-3L strain.
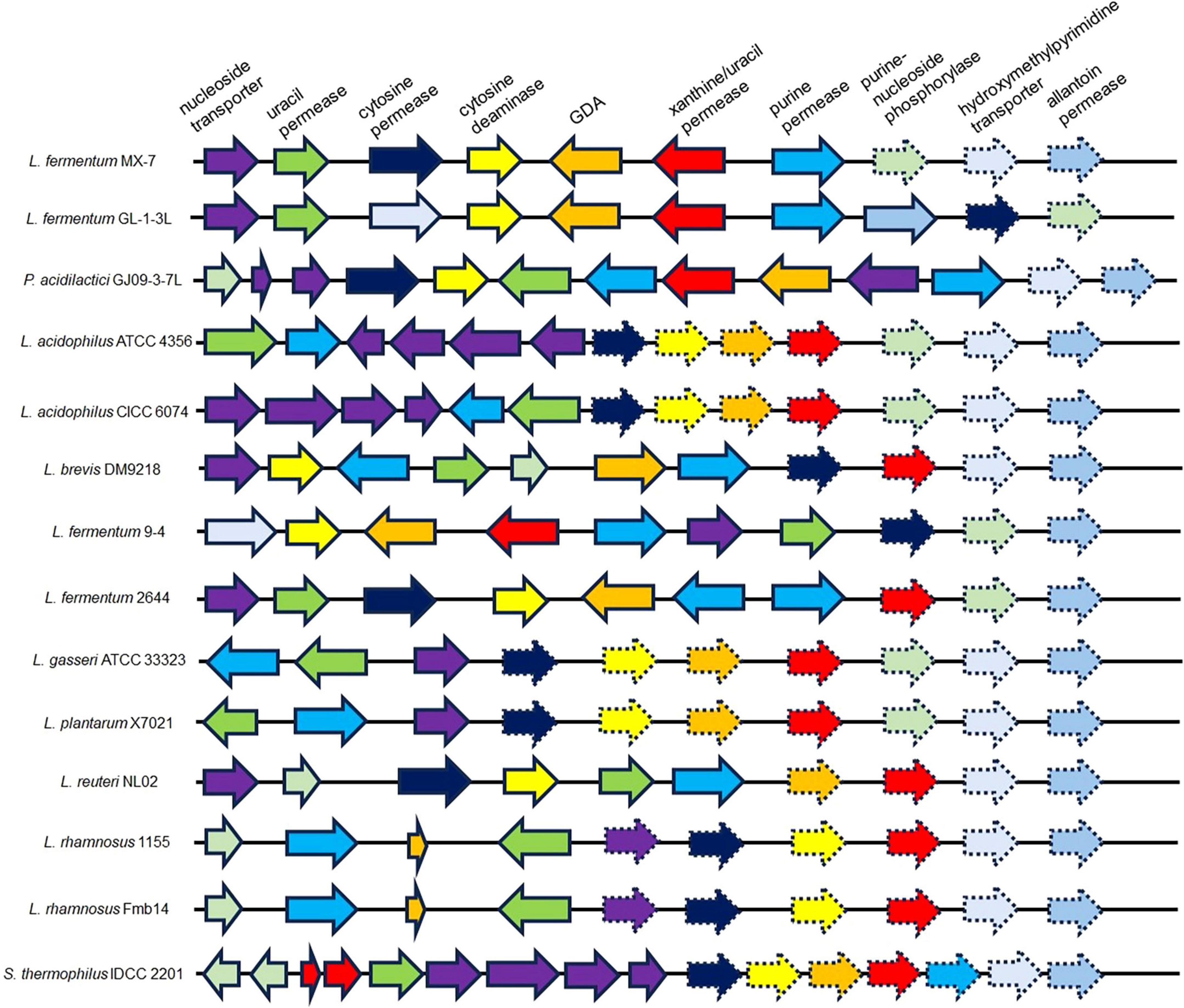
Figure 6. Representative arrangement of genomic regions from multiple organisms that catabolize purines and nucleosides. Solid arrows indicate presence; dashed arrows indicate absence. GDA: guanine deaminase. Lactobacillus acidophilus ATCC 4356 (Ma et al., 2023) (CP139575.1, NCBI), Lactobacillus acidophilus CICC 6074 (Li et al., 2024) (CP135998.1, NCBI), Levilactobacillus brevis DM9218 (Wang H. et al., 2019) (NZ_KZ793387.1, NZ_KZ793388.1, NZ_KZ793389.1, NZ_KZ793390.1, NZ_KZ793391.1, NZ_KZ793392.1, NZ_KZ793393.1, NZ_KZ793394.1, NZ_KZ793395.1 and NZ_KZ793396.1, NCBI), Limosilactobacillus fermentum 9-4 (Lu et al., 2022) (NZ_CP076082.1, NCBI), Limosilactobacillus fermentum 2644 (Li et al., 2022) (NZ_CP053312.1 and NZ_CP053313.1, NCBI), Lactobacillus gasseri ATCC 33323 (Yamada et al., 2017) (NC_008530.1, NCBI), Lactiplantibacillus plantarum X7021 (Du et al., 2022; Liu et al., 2022) (NZ_CP025412.1, NZ_CP025413.1, NZ_CP025414.1, NZ_CP025415.1, NZ_CP025416.1, NZ_CP025417.1 and NZ_CP025418.1, NCBI), Limosilactobacillus reuteri NL02 (Hu, 2022) (NZ_CP089962.1, NZ_CP089963.1 and NZ_CP089964.1, NCBI), Lacticaseibacillus rhamnosus 1155 (Li et al., 2022) (CP048623.1, CP048624.1 and CP048625.1, NCBI), Lacticaseibacillus rhamnosus Fmb14 (Zhao et al., 2023) (NZ_CP101845.1 and NZ_CP101846.1, NCBI) and Streptococcus thermophilus IDCC 2201 (Al Kassaa et al., 2024) (CP035306.1, NCBI).
3.4 Quantitive reverse transcription-PCR analysis of guanine deaminase
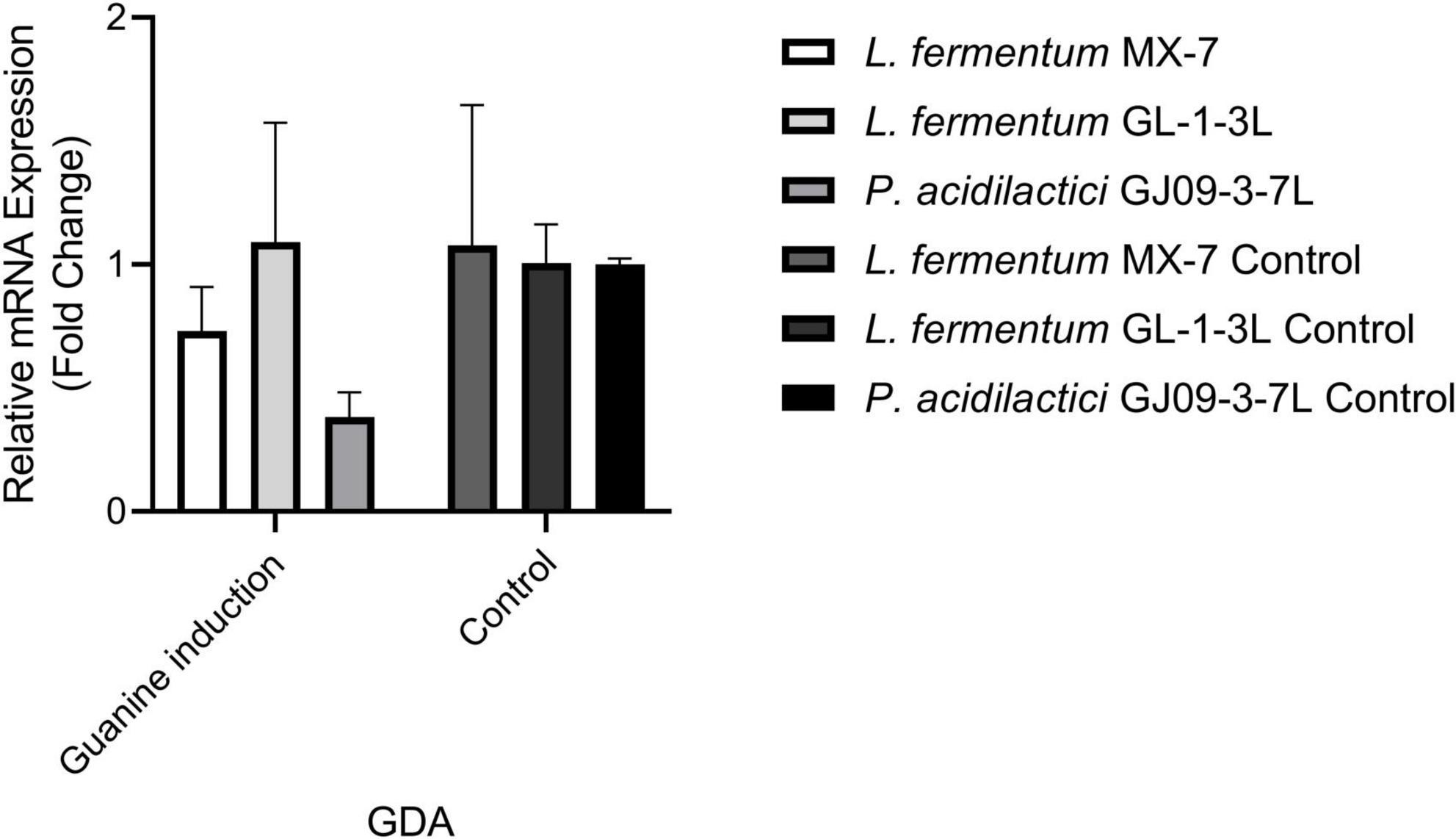
In order to further explore the effect of guanine deaminase on the degradation of guanine, we conducted qRT-RCR analysis. Under the induction of guanine, changes in GDA expression level were observed in L. fermentum MX-7, GL-1-3L, and P. acidilactici GJ09-3-7L. The GDA levels decreased in MX-7 and GJ09-3-7L strains (Figure 7).
3.5 Probiotic properties of L. fermentum MX-7
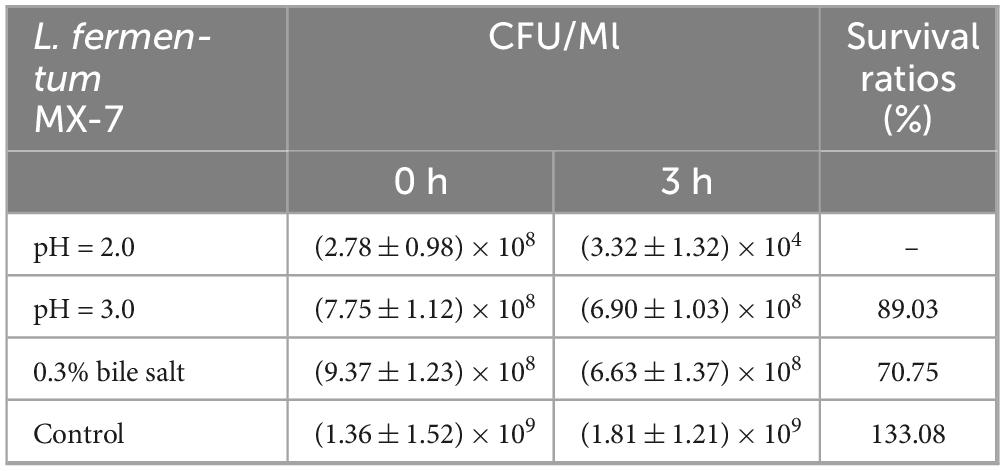
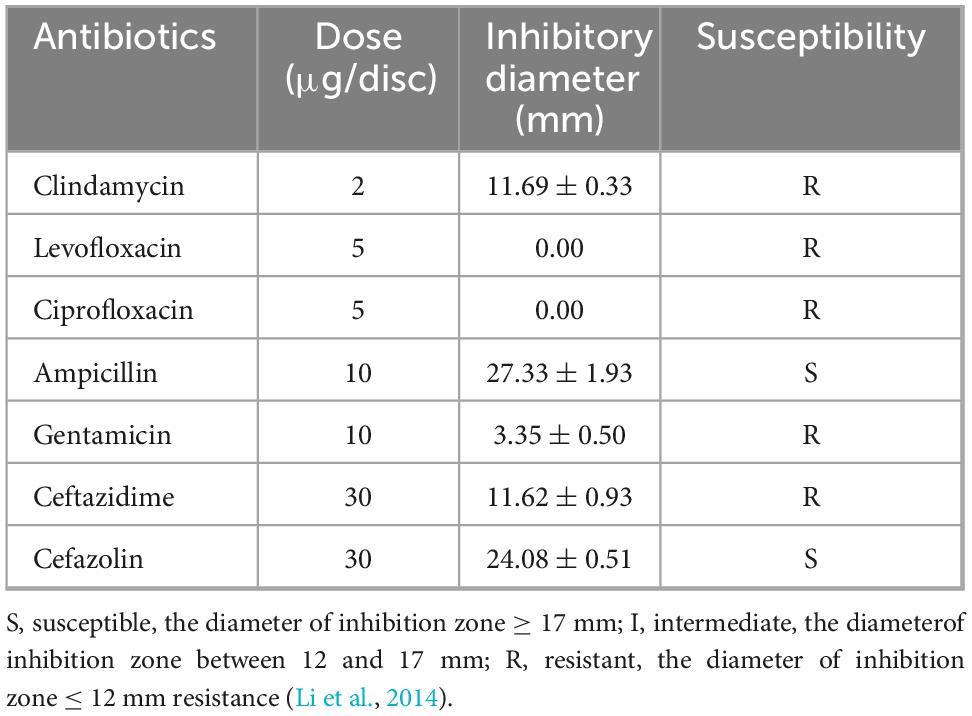
Because L. fermentum MX-7 had the highest degradation rate of guanine, in order to evaluate the possibility of MX-7 strain in fermented food, we studied its probiotic properties. The survival ratio of L. fermentum MX-7 in MRS liquid medium with pH = 3.0 for 3 h was 89.03%, and the survival ratio in MRS liquid medium with 0.3% bile salt concentration for 3 h was 70.75%, indicating that MX-7 strain has a certain ability of acid and bile salt tolerance (Table 2). Meanwhile, MX-7 strain had different degrees of inhibition on the five pathogenic bacteria, and had strong inhibition on S. aureus, E. coli, S. enterica and S. pullorum, but weak inhibition on L. monocytogenes (Table 3). In addition, MX-7 strain was resistant to clindamycin, levofloxacin, ciprofloxacin, gentamicin and ceftazidime (Table 4).
3.6 L. fermentum MX-7 restored impaired renal function
In order to verify the effect of L. fermentum MX-7 on HUA SD rats, we performed in vivo experiments. Figure 8 showed the changes of serum UA, serum urea nitrogen and serum creatinine in rats. For the serum UA of rats, the serum UA levels of the model (p < 0.01), allopurinol (p < 0.01), MX-7 strain prevention (p < 0.01) and treatment groups (p < 0.001) on the 21st day were significantly higher than those of the control group, indicating that the rats HUA model were successfully constructed, and the preventive effect of MX-7 strain on HUA in rats was not obvious. On the 28th and 35th days, we found that the serum UA levels of allopurinol (p < 0.001), MX-7 strain prevention (p < 0.01) and treatment groups (p < 0.01) were significantly lower than that of model group, and the serum UA level of allopurinol group was significantly lower than that of MX-7 strain prevention (p < 0.01) and treatment groups (p < 0.001, p < 0.01), indicating that MX-7 strain can reduce the serum UA level of HUA rats, but it was not as obvious as allopurinol. On the 28th day we also found that elevated serum UA levels in all groups, which could be due to feed and environmental effects. For the serum urea nitrogen of rats, the serum urea nitrogen levels of the model and MX-7 strain treatment groups on the 21st day were significantly higher than those of the control group (p < 0.05), indicating the damage effect of HUA on the kidney of rats, and the use of MX-7 strain prevention may reduce the kidney damage. On the 28th day, the level of serum urea nitrogen in the model group was significantly higher than that in the control group (p < 0.01). On the 35th day, the serum urea nitrogen levels in the model and allopurinol groups were significantly higher than those in the MX-7 strain prevention (p < 0.01, p < 0.05), treatment (p < 0.01, p < 0.05) and control groups (p < 0.001), indicating that MX-7 strain has a certain protective effect on the kidney of rats. For the serum creatinine of rats, the serum creatinine level of the model group was significantly higher than that of the control group on the 21st day (p < 0.05). On the 35th day, the serum creatinine levels in the model, allopurinol, MX-7 strain prevention and treatment groups were significantly higher than those in the control group (p < 0.01).
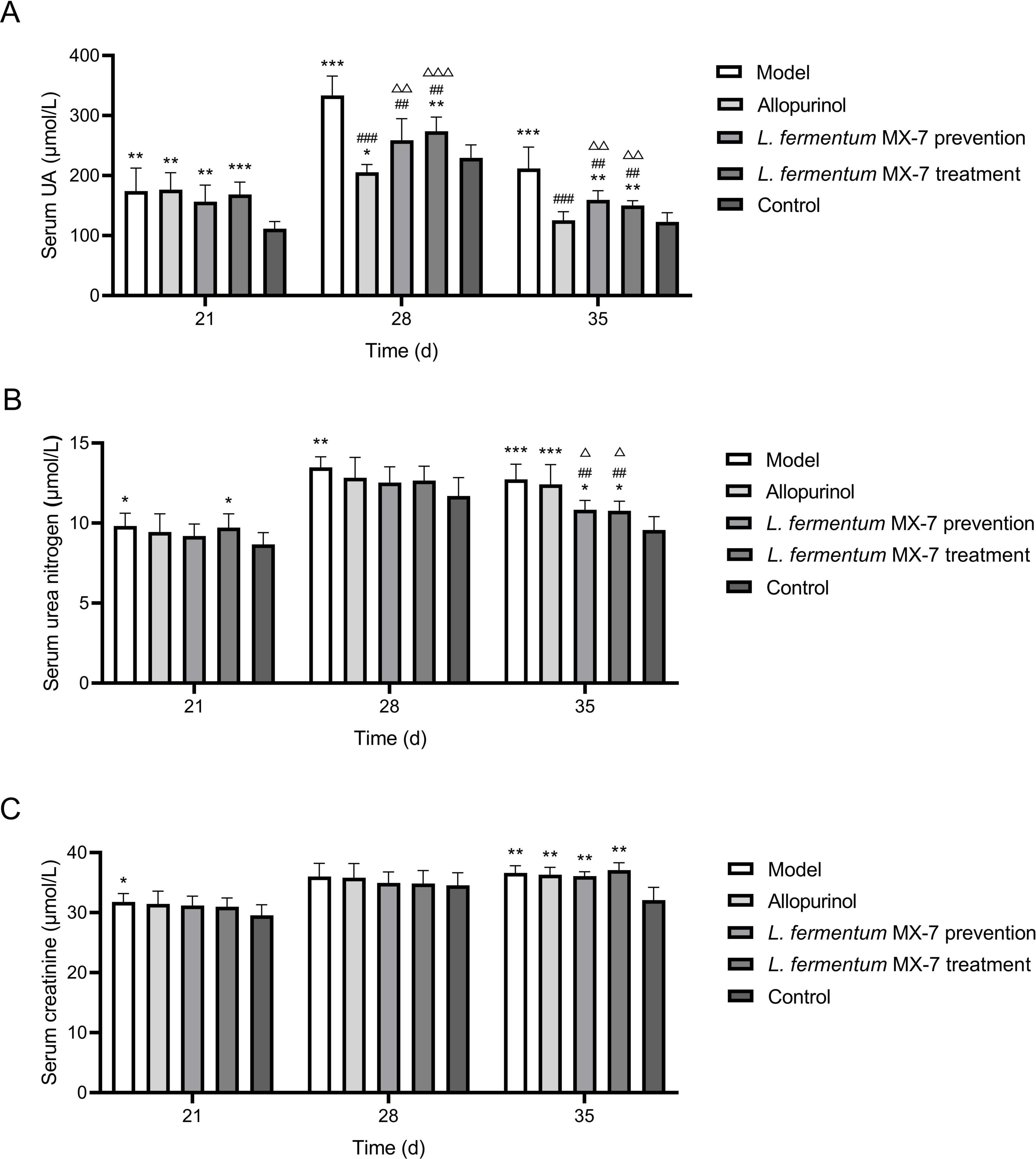
Figure 8. Serum biochemical indexes of rats. (A) Serum UA level, (B) serum urea nitrogenlevel, (C) serum creatinine level. UA, Uric acid. *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control group. ##p < 0.01, ###p < 0.001 compared with the model group. Δp < 0.05, ΔΔp < 0.01, ΔΔΔp < 0.001 compared with the allopurinol group.
Compared with the control group, the renal vascular wall in the model and allopurinol groups were significantly thickened, and there was inflammatory cell infiltration in the renal interstitium. The boundary of glomerulus was unclear, and the capillary endothelial cells swelled. The endothelial cells of renal tubules fell off. The MX-7 strain prevention and treatment groups can significantly improve these symptoms, but also observed a small amount of inflammatory cell infiltration in the renal interstitium, swelling of glomerular capillary endothelial cells, and a small amount of renal tubular endothelial cells falling off. Although the reduction of serum UA in the MX-7 strain prevention and the treatment groups were not as obvious as that in the allopurinol group, it can protect the kidney to a certain extent (Figure 9).
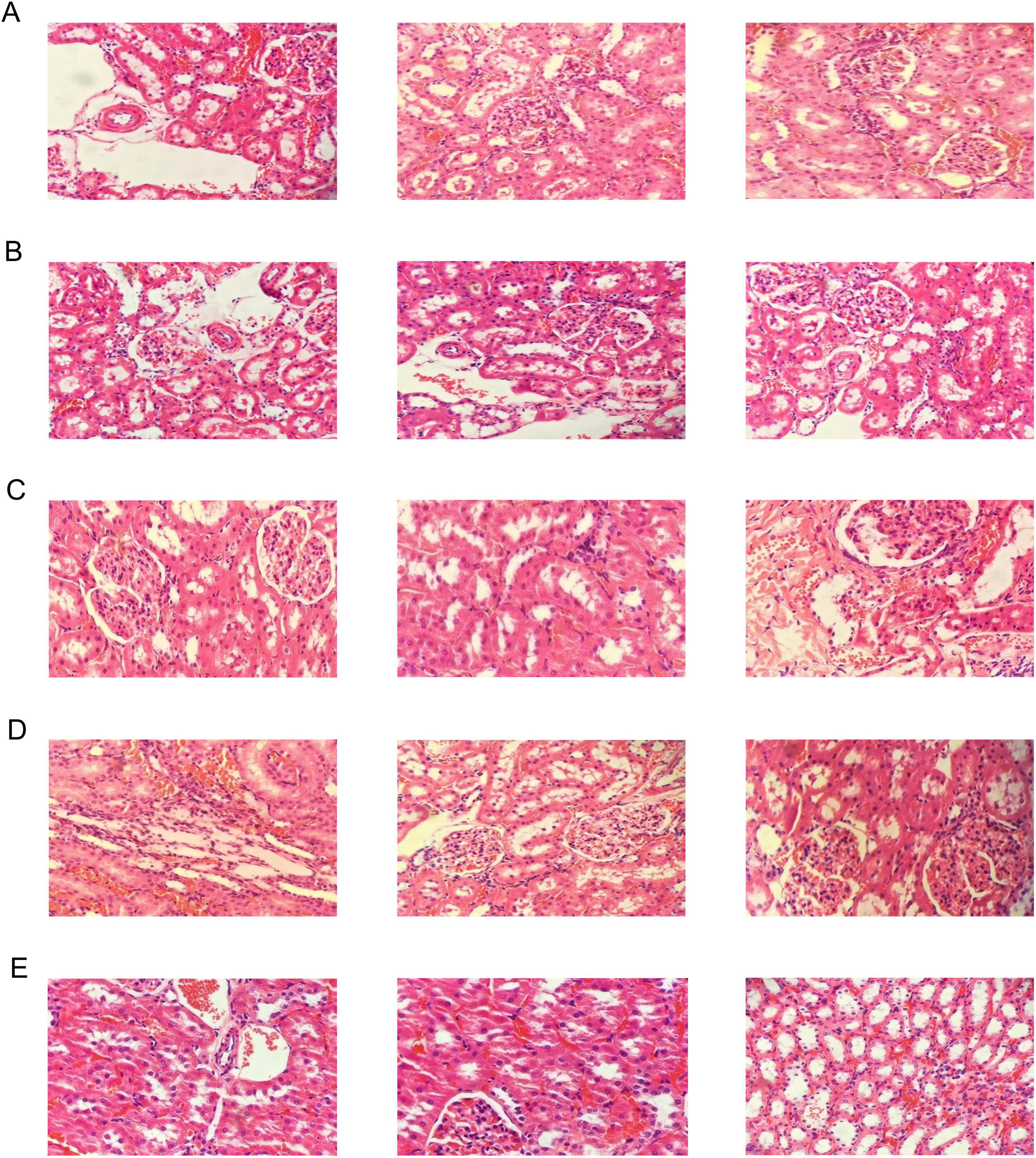
Figure 9. Histopathological analyses of H&E-stained kidney sections [the final magnification was 200X (only for the rightmost image in Figure 9E) or 400X (for the others)]. (A) Model group, (B) Allopurinol group, (C) L. fermentum MX-7 prevention group, (D) L. fermentum MX-7 treatment group, (E) Control group.
4 Discussion
In this study, the LAB isolated from traditional fermented foods in Yunnan Province were able to degrade purines in vitro (Table 1), and the three strains of LAB screened from them have the potential to develop low purine foods. Among them, L. fermentum MX-7 can not only reduce the serum UA and urea nitrogen levels of HUA rats (Figures 8A,B), but also reduce the damage of HUA to the kidney (Figure 9), indicating that MX-7 strain has the potential of adjuvant treatment of HUA. This study filled the gap of the study on the degradation of purines by LAB isolated from traditional fermented foods in Yunnan Province.
We found that the best strains for guanine degradation were L. fermentum MX-7 and GL-1-3L, and P. acidilactici GJ09-3-7L had the best degradation ability to xanthine (Supplementary Table 1). Our research results were similar to those of Wang et al. (2023), they screened Limosilactobacillus fermentum LF-1 from Huangjiu fermentation broth, and the degradation ratios of guanine, hypoxanthine, xanthine, adenine, inosine and guanosine were 95.0, 94.9, 65.9, 95.1, 97.4, and 83.4%, respectively. However, we have not screened LAB with good degradation effect of hypoxanthine, and the highest degradation ratio of xanthine was only (31.53 ± 0.43)% (Supplementary Table 1), which was similar to the research results of Ni et al. (2021). In addition, these three strains can degrade inosine and guanosine 100% (Table 1), which were higher than those of Lu et al. (2022). To further investigate the degradation mechanism, we performed genomic analysis and qRT-PCR. We found that GDA plays a huge role in degrading guanine by comparative genomics analysis and we found GDA in MX-7, GL-1-3L, and GJ09-3-7L strains (Figure 6). L. acidophilus CICC 6074 (Li et al., 2024) and L. plantarum X7021 (Du et al., 2022; Liu et al., 2022) were both deficient in GDA (Figure 6), and their guanine degradation ratios were less than 40%. In addition, transporter and permease may also play a huge role in purine metabolism in LAB (Martinussen et al., 2010; Liu et al., 2022), whereas many of them were found in MX-7, GL-1-3L, and GJ09-3-7L strains as compared to other (Figure 6). Nucleoside transporter plays a great role in the degradation of nucleosides (Martinussen et al., 2010), and we found it in MX-7, GL-1-3L, and GJ09-3-7L strains (Figure 6). L. rhamnosus Fmb14 (Zhao et al., 2023) deficient in nucleoside transporter (Figure 6), and it degrades nucleosides by less than 37% and at a slow ratio. In contrast, strains containing it (Figure 6) (such as L. acidophilus CICC 6074 (Li et al., 2024) and S. thermophilus IDCC 2201 (Al Kassaa et al., 2024) degraded nucleosides degradation ratios were greater than 50%; MX-7, GL-1-3L, GJ09-3-7L (Table 1), L. brevis DM9218 (Wang H. et al., 2019), L. fermentum 9-4 (Lu et al., 2022), and L. reuteri NL02 (Hu, 2022) greater than 90%) had a higher degradation ratio of nucleosides. Xanthine is a precursor substance for the synthesis of UA. We also found by KEGG purine metabolism map (data not shown) that although none of the 14 strains in the comparative genome (Figure 6) could continue to degrade xanthine, while MX-7 (Figure 8A), L. acidophilus ATCC 4356 (Ma et al., 2023), L. acidophilus CICC 6074 (Li et al., 2024), L. brevis DM9218 (Wang H. et al., 2019), L fermentum 2644 (Li et al., 2022), L. rhamnosus 1155 (Li et al., 2022), L. rhamnosus Fmb14 (Zhao et al., 2023), and S. thermophilus IDCC 2201 (Al Kassaa et al., 2024) all reduced serum UA levels in experimental animals, and we believed that the cause may be attributed to two points. The first point was that the strain competes with the intestinal epithelium for uptake of purines via transporter and permease, leading to a reduction in their uptake of purines, and thus a reduction in host UA production (Li et al., 2014; Hsu et al., 2019; Wang H. et al., 2019). This suggested an important role for transporter and permease. The second point is that these strains increased the level of UA in the feces, up-regulated the gene expression of UA transporter, ATP-binding cassette subfamily G-2 in colon and jejunum tissues, accelerated the excretion of UA from the intestines, inhibited the activity of xanthine oxidase in liver and serum (Li et al., 2022), restored intestinal flora and increased intestinal short-chain fatty acid content (Al Kassaa et al., 2024). It is the combination of these two reasons that leads to a decrease in serum UA levels. Besides, we also noted that the GDA expression level of MX-7 and GJ09-3-7L strains were decreased under guanine induction, and GL-1-3L strain expression level was similar to the control group (Figure 7), despite they all had the high degradation ratio of guanine except GJ09-3-7L strain (Supplementary Table 1). Although the GJ09-3-7L strain contained GDA (Figure 6), it did not degrade it at a high ratio (Supplementary Table 1), and one of the reasons may be that its GDA expression was inhibited. After the transfer of guanine to MX-7 and GL-1-3L strains via transporter and permease, there may be isoenzymes or other degradation pathways or utilization of guanine as an energy source in addition to the action of GDA, but this still needs to be verified by LC-MS. For example, high concentrations of inosine could activate compensatory metabolic pathways of L. rhamnosus Fmb14 to catalyze inosine as an energy source and produce intracellular folic acid and riboflavin (Zhao et al., 2023).
LAB can degrade and assimilate purines (Xiao et al., 2020; Kuo et al., 2021; Lu et al., 2022). For degradation, LAB degrade inosine to hypoxanthine and guanosine to guanine (Guimaraes et al., 2011; Kuo et al., 2021; Ni et al., 2021; Lu et al., 2022; Liao et al., 2024). Purines are not as easily absorbed by the intestine as nucleosides (Salati et al., 1984; Stow and Bronk, 1993; Yamada et al., 2016; Kuo et al., 2021), which may increase the excretion of purines in the intestine. LAB can further degrade guanine and hypoxanthine to xanthine (Wang H. et al., 2019; Kuo et al., 2021; Lu et al., 2022; James et al., 2023). For assimilation, LAB absorb purines in the environment and use them for their own growth without producing corresponding degradation products (Xiao et al., 2020; Kuo et al., 2021; Lu et al., 2022). In order to further determine whether purines were degraded or assimilated by the LAB in this study (Li et al., 2014), we analyzed the degradation ability of the cell-free extracts (supernatant and precipitation) of MX-7, GL-1-3L, and GJ09-3-7L strains to purines. The results showed that the cell-free extracts of MX-7 (p < 0.001) and GL-1-3L (p < 0.01, p < 0.001) strains were still able to significantly degrade guanine (Figure 4A), but the degradation ratio was lower than that of viable strains, suggesting that they had both degradation and assimilation effects on guanine. However, only the precipitation of GJ09-3-7L strain was able to significantly degrade xanthine (p < 0.05) (Figure 4B), but the degradation ratio was still lower than that of viable strain, indicating that xanthine mainly entered into the intracellular metabolism rather than being directly degraded by enzymes, as evidenced by the fact that the GJ09-3-7L strain degraded xanthine hardly at all from 0 to 12 h, and then degraded it slowly (Figure 2B). We observed that when MX-7, GL-1-3L and GJ09-3-7L strains were thermally inactivated, they could still significantly degrade purines (p < 0.01) and nucleosides (p < 0.001, p < 0.01), but significantly less than the viable strains (p < 0.001, p < 0.01) (Figure 3). This may indicate that there were still some viable strains under this thermally treatment (90°C, 10 min), which further emphasized the importance of active enzymes and cellular metabolism for purine degradation. At the same time, the cell-free extracts of these three strains were still able to significantly degrade inosine and guanosine (p < 0.001) (Figures 4C,D), suggesting that they mainly play a degrading role, i.e., the enzymes in the supernatants and precipitations contacted with inosine and guanosine to directly produce the corresponding degradation products, which is consistent with Li et al. (2014) using L. brevis DM9218. However, purine-nucleoside phosphorylase, which is capable of degrading inosine and guanosine, was present in the genome of DM9218 strain (Li et al., 2014; Figure 6), but the enzymes for degrading inosine and guanosine were deficiency in both MX-7 and GL-1-3L strains (Figure 5). From the KEGG purine metabolism map (data not shown), L. acidophilus CICC 6074 (Li et al., 2024) and L. fermentum 9-4 (Lu et al., 2022) were also both deficient in enzymes for degrading inosine and guanosine, but in vitro experiments showed that both strains were able to degrade them, which seems to suggest the presence of isozymes or other pathways that play a role in the degradation, but this needs further confirmation.
When selecting potential probiotic strains, it is important to detect their virulence genes and resistance genes (Papadimitriou et al., 2015; Dlamini et al., 2019). We found virulence genes in MX-7, GL-1-3L, and GJ09-3-7L strains (Supplementary Table 5), which was consistent with the existing studies on LAB (Togay et al., 2010; Dlamini et al., 2019). However, Bennedsen et al. (2011) did not find virulence genes in all LAB they studied. The existence of virulence genes does not guarantee the expression of genes, and there is a great possibility that these genes are not expressed or only weakly expressed (Dlamini et al., 2019). We also found resistance genes in MX-7 and GL-1-3L strains (Supplementary Table 6). Dlamini et al. (2019) found resistance genes in Limosilactobacillus reuteri VB4 and Streptococcus salivarius NBRC13956. But in many cases, resistance genes are not transferable (Dlamini et al., 2019). Therefore, we believe that MX-7, GL-1-3L and GJ09-3-7L strains have the potential to become probiotics.
Since MX-7 strain has the highest degradation ratio of guanine (Supplementary Table 1), its probiotic properties were evaluated to determine whether it can survive in the gastrointestinal tract and colonize successfully, laying the foundation for the development of low purine foods in the future. The gastrointestinal tract is rich in digestive enzymes and has strong acidity (Li et al., 2014). Bile is produced by the liver and contributes to the digestion of lipids in the small intestine (Li et al., 2014). Bile salts are present in bile (Li et al., 2014). Acid environment and bile salts can inhibit bacterial growth (Li et al., 2014). We found that MX-7 strain was better than most existing studies in acid resistance, but has poor bile salt resistance (Table 2; Li et al., 2014; Lu et al., 2022; Hussain et al., 2024; Liao et al., 2024). This indicates that MX-7 strain can survive in the intestine. In addition, the antibacterial ability of LAB mainly comes from two aspects. The first is to produce lactic acid and other acidic substances to reduce the environmental pH, making it unsuitable for the growth of other bacteria (Li et al., 2014). The second is the production of bacteriocin and other antibacterial substances (Li et al., 2014). Compared with existing studies (Li et al., 2014), we found that MX-7 strain has stronger ability to inhibit S. enterica and S. pullorum (Table 3). We also found that MX-7 strain was resistant to clindamycin, levofloxacin, ciprofloxacin, gentamicin and ceftazidime (Table 4), which was similar to the results of Li et al. (2014).
Since experimental animals (such as rats) can convert UA into allantoin for excretion, it is difficult to establish animal HUA model (Sánchez-Lozada et al., 2007; Sanchez-Lozada et al., 2008; Xiao et al., 2020). In this study, we established HUA rats model using yeast extract and potassium oxazinate (Figure 1; Yamada et al., 2017). Compared with the model group, the use of MX-7 strain for prevention and treatment can significantly reduce the serum UA level of HUA rats (p < 0.01), but it was still higher than that of allopurinol group (Figure 8A), which was consistent with the results of Li et al. (2014). Allopurinol can directly inhibit xanthine oxidase, thereby directly preventing the production of UA (Li et al., 2014). However, MX-7 strain may reduce UA production by competing with the intestinal epithelium for purine uptake and probiotic effects (Li et al., 2014; Hsu et al., 2019; Wang H. et al., 2019). Therefore, some purines can still be absorbed by the intestinal epithelium and eventually metabolized into UA (Li et al., 2014; Hsu et al., 2019; Wang H. et al., 2019). HUA can lead to the destruction of renal structure and function, and increase the levels of blood urea nitrogen and creatinine, which are indicators of renal toxicity (Lee et al., 2022). On the 35th day, we found that the serum urea nitrogen (p < 0.05) and serum creatinine (p < 0.01) levels in the MX-7 strain prevention and treatment groups were still significantly higher than those in the control group (Figures 8B,C), which is consistent with the research results of Lee et al. (2022). This may indicate that HUA will cause some damage to the kidney of rats in MX-7 strain prevention and treatment groups. Nevertheless, we still found that the prevention and treatment of MX-7 strain can reduce kidney damage to a certain extent through H&E staining of rats kidney (Figure 9), which was consistent with the existing results (Ni et al., 2021; Wu et al., 2021; Cao et al., 2022; Lee et al., 2022). Whether prolonging the intervention time of MX-7 strain can reverse the renal damage caused by HUA is also worth discussing. LAB have a protective effect on renal injury caused by acute HUA (Minelli et al., 2004; Xiao et al., 2020), so we speculate that MX-7 strain may repair the damaged kidney by reducing the production of serum UA and/or urea nitrogen (Ni et al., 2021), restoring intestinal flora and increasing intestinal short-chain fatty acid content (Al Kassaa et al., 2024).
5 Conclusion
Here, we screened L. fermentum MX-7 and GL-1-3L with high guanine efficiency and P. acidilactici GJ09-3-7L with high xanthine degradation efficiency from traditional fermented foods in Yunnan Province by HPLC. And these three strains can efficiently degrade inosine and guanosine. The presence of many purine metabolism, transporter and permease genes revealed the potential of these three strains to degrade purines. The use of MX-7 strain can successfully reduce the serum UA and urea nitrogen levels of HUA SD rats, and can reduce the damage of HUA to the kidney of rats. However, we did not study the extent of assimilation and degradation products of LAB to purines in vitro, nor did we set up a control group of non-degrading LAB in vivo. Our research provided three candidate strains for the development of low purine foods, and MX-7 strain had the potential of adjuvant therapy for HUA.
Data availability statement
The data that support the findings of this study are available in NODE at https://www.biosino.org/node, accession number OEZ00020969, OEZ00020970, and OEZ00020971.
Ethics statement
The animal study was approved by the Experimental Animal Ethics Committee of the Kunming University of Science and Technology (approval number: PZWH-KUST-202501040001-1). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZL: Conceptualization, Validation, Visualization, Writing – original draft. X-YZ: Investigation, Methodology, Validation, Writing – original draft. JY: Funding acquisition, Visualization, Writing – review & editing. SL: Writing – review and editing, Data curation, Formal analysis. XO: Investigation, Methodology, Writing – original draft. CH: Formal Analysis, Software, Writing – original draft. C-JL: Methodology, Resources, Writing – review and editing. X-RL: Conceptualization, Formal Analysis, Funding acquisition, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Kunming University of Science and Technology-Anning First People’s Hospital Medical Joint Special Project (KUST-AN2023001Z), Natural Science Foundation of Yunnan Province (202501AT070320), the Youth Promotion Program of the Peak Climbing Plan for Discipline Construction of Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine – “Challenge and Acceptance” Series Projects (RJTJ25-MS-060); and Yunnan (Kunming) Zhang Wenhong expert workstation (YSZJGZZ-2020051).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1627956/full#supplementary-material
References
Al Kassaa, I., Kim, D., Moon, J. S., Kim, J. E., Jang, Y.-J., Choi, H. S., et al. (2024). Evaluation of purine-nucleoside degrading ability and in vivo uric acid lowering of Streptococcus thermophilus IDCC 2201, a novel antiuricemia strain. PLoS One 19:e0293378. doi: 10.1371/journal.pone.0293378
Bardin, T., and Richette, P. (2020). FAST: New look at the febuxostat safety profile. Lancet 396, 1704–1705. doi: 10.1016/S0140-6736(20)32343-6
Becker, M. A., Schumacher, H. R., Wortmann, R. L., MacDonald, P. A., Eustace, D., Palo, W. A., et al. (2005). Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N. Engl. J. Med. 353, 2450–2461. doi: 10.1056/NEJMoa050373
Benn, C. L., Dua, P., Gurrell, R., Loudon, P., Pike, A., Storer, R. I., et al. (2018). Physiology of hyperuricemia and urate-lowering treatments. Front. Med. 5:160. doi: 10.3389/fmed.2018.00160
Bennedsen, M., Stuer-Lauridsen, B., Danielsen, M., and Johansen, E. (2011). Screening for antimicrobial resistance genes and virulence factors via genome sequencing. Appl. Environ. Microbiol. 77, 2785–2787. doi: 10.1128/AEM.02493-10
Boger, W. P., and Strickland, S. C. (1955). Probenecid (BENEMID): Its uses and side-effects in 2,502 patients. A.M.A. Arch. Internal Med. 95, 83–92. doi: 10.1001/archinte.1955.00250070099012
Bortolotti, M., Polito, L., Battelli, M. G., and Bolognesi, A. (2021). Xanthine oxidoreductase: One enzyme for multiple physiological tasks. Redox Biol. 41:101882. doi: 10.1016/j.redox.2021.101882
Büsing, F., Hägele, F. A., Nas, A., Döbert, L.-V., Fricker, A., Dörner, E., et al. (2019). High intake of orange juice and cola differently affects metabolic risk in healthy subjects. Clin. Nutr. 38, 812–819. doi: 10.1016/j.clnu.2018.02.028
Cao, J., Liu, Q., Hao, H., Bu, Y., Tian, X., Wang, T., et al. (2022). Lactobacillus paracasei X11 ameliorates hyperuricemia and modulates gut microbiota in mice. Front. Immunol. 13:940228. doi: 10.3389/fimmu.2022.940228
Chen, B., Zhou, Y., Duan, L., Gong, X., Liu, X., Pan, K., et al. (2023). Complete genome analysis of Bacillus velezensis TS5 and its potential as a probiotic strain in mice. Front. Microbiol. 14:1322910. doi: 10.3389/fmicb.2023.1322910
Chen, C. H., Chen, C. B., Chang, C. J., Lin, Y. Jr., Wang, C. W., Chi, C. C., et al. (2019). Hypersensitivity and Cardiovascular risks related to allopurinol and febuxostat therapy in Asians: A population-based cohort study and meta-analysis. Clin. Pharmacol. Therap. 106, 391–401. doi: 10.1002/cpt.1377
Chen-Xu, M., Yokose, C., Rai, S. K., Pillinger, M. H., and Choi, H. K. (2019). Contemporary prevalence of gout and hyperuricemia in the united states and decadal trends: The National Health and Nutrition Examination Survey, 2007–2016. Arthritis Rheumatol. 71, 991–999. doi: 10.1002/art.40807
Colín-González Laura, A., and Santamaría, A. (2013). Probenecid: An emerging tool for neuroprotection. CNS Neurol. Disord. Drug Targets 12, 1050–1065. doi: 10.2174/18715273113129990090
Dlamini, Z. C., Langa, R. L. S., Aiyegoro, O. A., and Okoh, A. I. (2019). Safety evaluation and colonisation abilities of four lactic acid bacteria as future probiotics. Probiot. Antimicrob. Proteins 11, 397–402. doi: 10.1007/s12602-018-9430-y
Dong, Y., Sun, N., Ge, Q., Lv, R., and Lin, S. (2023). Antioxidant soy peptide can inhibit xanthine oxidase activity and improve LO2 cell damage. Food Biosci. 52:102455. doi: 10.1016/j.fbio.2023.102455
Du, L., Ro, K.-S., Zhang, Y., Tang, Y.-J., Li, W., Xie, J., et al. (2022). Effects of Lactiplantibacillus plantarum X7021 on physicochemical properties, purines, isoflavones and volatile compounds of fermented soymilk. Process Biochem. 113, 150–157. doi: 10.1016/j.procbio.2021.12.028
Du, X., Yin, S., Wang, T., Chu, C., Devahastin, S., Yi, J., et al. (2024). Identification of proteolytic bacteria from Yunnan fermented foods and their use to reduce the allergenicity of β-lactoglobulin. J. Dairy Sci. 107, 8990–9004. doi: 10.3168/jds.2024-25055
Eden, P. A., Schmidt, T. M., Blakemore, R. P., and Pace, N. R. (1991). Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J. Syst. Bacteriol. 41, 324–325. doi: 10.1099/00207713-41-2-324
Feliu, J. X., Cubarsi, R., and Villaverde, A. (1998). Optimized release of recombinant proteins by ultrasonication of E-coli cells. Biotechnol. Bioeng. 58, 536–540. doi: 10.1002/(SICI)1097-0290(19980605)58:5<536::AID-BIT10>3.0.CO;2-9
Garcia-Rodriguez, C., Mujica, P., Illanes-Gonzalez, J., Lopez, A., Vargas, C., Saez, J. C., et al. (2023). Probenecid, an old drug with potential new uses for central nervous system disorders and neuroinflammation. Biomedicines 11:1516. doi: 10.3390/biomedicines11061516
Guimaraes, A. P., Oliveira, A. A., da Cunha, E. F. F., Ramalho, T. C., and Franco, T. C. C. (2011). Analysis of Bacillus anthracis nucleoside hydrolase via in silico docking with inhibitors and molecular dynamics simulation. J. Mol. Model. 17, 2939–2951. doi: 10.1007/s00894-011-0968-9
Hali, F., Chagraoui, H., Meftah, A., Marnissi, F., Chaabi, S., Chiheb, S., et al. (2023). Allopurinol induced severe cutaneous adverse reactions: Vigilance of prescription (31 cases). Revue Française d’Allergologie 63:103275. doi: 10.1016/j.reval.2022.103275
Hsu, C. L., Hou, Y. H., Wang, C. S., Lin, S. W., Jhou, B. Y., Chen, C. C., et al. (2019). Antiobesity and uric acid-lowering effect of Lactobacillus plantarum GKM3 in high-fat-diet-induced obese rats. J. Am. Coll. Nutr. 38, 623–632. doi: 10.1080/07315724.2019.1571454
Hu, J. (2022). Screening of Uric Acid-Lowering Lactic Acid Bacteria and Analysis of Its Whole Genome Sequence. Hohhot: Inner Mongolia Agricultural University.
Hussain, A., Rui, B., Ullah, H., Dai, P., Ahmad, K., Yuan, J., et al. (2024). Limosilactobacillus reuteri HCS02-001 attenuates hyperuricemia through gut microbiota-dependent regulation of uric acid biosynthesis and excretion. Microorganisms 12:637. doi: 10.3390/microorganisms12040637
James, A., Ke, H., Yao, T., and Wang, Y. (2023). The role of probiotics in purine metabolism, hyperuricemia and gout: Mechanisms and interventions. Food Rev. Int. 39, 261–277. doi: 10.1080/87559129.2021.1904412
Kaufmann, P., Török, M., Hänni, A., Roberts, P., Gasser, R., and Krähenbühl, S. (2005). Mechanisms of benzarone and benzbromarone-induced hepatic toxicity. Hepatology 41, 925–935. doi: 10.1002/hep.20634
Kuo, Y. W., Hsieh, S. H., Chen, J. F., Liu, C. R., Chen, C. W., Huang, Y. F., et al. (2021). Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ 9:e11209. doi: 10.7717/peerj.11209
Lee, Y., Werlinger, P., Suh, J.-W., and Cheng, J. (2022). Potential probiotic Lacticaseibacillus paracasei MJM60396 prevents hyperuricemia in a multiple way by absorbing purine, suppressing xanthine oxidase and regulating urate excretion in mice. Microorganisms 10:851. doi: 10.3390/microorganisms10050851
Li, H., Shen, N., Ren, J., Yang, S., Chen, Y., and Gao, Z. (2024). Biotransformation characteristics of urate-lowering probiotic fermented apple juice and potential regulatory mechanisms for ameliorating hyperuricemia via mediating gut microbiota and metabolic pathways. Food Chem. 460:140462. doi: 10.1016/j.foodchem.2024.140462
Li, M., Yang, D., Mei, L., Yuan, L., Xie, A., and Yuan, J. (2014). Screening and characterization of purine nucleoside degrading lactic acid bacteria isolated from chinese sauerkraut and evaluation of the serum uric acid lowering effect in hyperuricemic rats. PLoS One 9:e105577. doi: 10.1371/journal.pone.0105577
Li, Q., Liu, C., Luo, Y., Yang, E., and Li, X. (2020). Effects of different fermentation conditions on folate synthesis of Lactobacillus plantarum. Food Sci. 41, 114–118. doi: 10.7506/spkx1002-6630-20181229-356
Li, R., Yu, K., and Li, C. (2018). Dietary factors and risk of gout and hyperuricemia: A meta-analysis and systematic review. Asia Pac. J. Clin. Nutr. 27, 1344–1356. doi: 10.6133/apjcn.201811_27(6).0022
Li, Y., Zhu, J., Lin, G., Gao, K., Yu, Y., Chen, S., et al. (2022). Probiotic effects of Lacticaseibacillus rhamnosus 1155 and Limosilactobacillus fermentum 2644 on hyperuricemic rats. Front. Nutr. 9:993951. doi: 10.3389/fnut.2022.993951
Liao, J. J., Tan, C. Y., Liang, L., Luo, Y. Z., Li, H. L., Chen, Y. Y., et al. (2024). Screening and identification of selenium-enriched purine-lowering Lactobacillus plantarum and Lactococcus lactis with high stress tolerance and antioxidant capacity. Food Biosci. 60:104439. doi: 10.1016/j.fbio.2024.104439
Lin, J. X., Xiong, T., Peng, Z., Xie, M., and Peng, F. (2022). Novel lactic acid bacteria with anti-hyperuricemia ability: Screening and in vitro probiotic characteristics. Food Biosci. 49:101840. doi: 10.1016/j.fbio.2022.101840
Liu, C. J., Wu, B., Zhang, S. Y., Li, Q. K., Zeng, X. Q., Yang, E., et al. (2019). Transcriptomic analysis of de novo folate biosynthetic genes in Lactobacillus plantarum strain 4_3 in fermented soybean. Food Funct. 10, 2426–2438. doi: 10.1039/c8fo01952e
Liu, G., Chen, C., Jiang, Z., Liu, Y., Wang, X., Qiao, L., et al. (2024). Characterization and the first complete genome sequence of a novel strain of Bergeyella porcorum isolated from pigs in China. BMC Microbiol. 24:214. doi: 10.1186/s12866-024-03366-6
Liu, G., Chen, X., Lu, X., Zhao, J., and Li, X. (2020). Sunflower head enzymatic hydrolysate relives hyperuricemia by inhibiting crucial proteins (xanthine oxidase, adenosine deaminase, uric acid transporter1) and restoring gut microbiota in mice. J. Funct. Foods 72:104055. doi: 10.1016/j.jff.2020.104055
Liu, X. F., Liu, C. J., Zeng, X. Q., Zhang, H. Y., Luo, Y. Y., and Li, X. R. (2020). Metagenomic and metatranscriptomic analysis of the microbial community structure and metabolic potential of fermented soybean in Yunnan Province. Food Sci. Technol. 40, 18–25. doi: 10.1590/fst.01718
Liu, G., Liu, Y., Ro, K.-S., Du, L., Tang, Y.-J., Zhao, L., et al. (2022). Genomic characteristics of a novel strain Lactiplantibacillus plantarum X7021 isolated from the brine of stinky tofu for the application in food fermentation. Lwt 156:113054. doi: 10.1016/j.lwt.2021.113054
Liu, X., Huang, S., Xu, W., Zhou, A., Li, H., Zhang, R., et al. (2018). Association of dietary patterns and hyperuricemia: A cross-sectional study of the Yi ethnic group in China. Food Nutr. Res. 25:62. doi: 10.29219/fnr.v62.1380
Liu, X. F., Liu, C. J., Zhang, H. Y., Gong, F. M., Luo, Y. Y., and Li, X. R. (2015). The bacterial community structure of yond bap, a traditional fermented goat milk product, from distinct Chinese regions. Dairy Sci. Technol. 95, 369–380. doi: 10.1007/s13594-015-0216-7
Loong, S. K., Khor, C. S., Jafar, F. L., and AbuBakar, S. (2016). Utility of 16S rDNA sequencing for identification of rare pathogenic bacteria. J. Clin. Lab. Anal. 30, 1056–1060. doi: 10.1002/jcla.21980
Lu, L., Liu, T., Liu, X., and Wang, C. (2022). Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice-flour noodles. Food Sci. Hum. Wellness 11, 1402–1408. doi: 10.1016/j.fshw.2022.04.030
Ma, Q., Chen, M., Liu, Y., Tong, Y., Liu, T., Wu, L., et al. (2023). Lactobacillus acidophilus fermented dandelion improves hyperuricemia and regulates gut microbiota. Fermentation 9:352. doi: 10.3390/fermentation9040352
Mackenzie, I. S., Ford, I., Nuki, G., Hallas, J., Hawkey, C. J., Webster, J., et al. (2020). Long-term cardiovascular safety of febuxostat compared with allopurinol in patients with gout (FAST): A multicentre, prospective, randomised, open-label, non-inferiority trial. Lancet 396, 1745–1757. doi: 10.1016/s0140-6736(20)32234-0
Mao, Y., Xu, H., and Xia, P. (2025). Ilex cornuta leaves extracts ameliorate hyperuricemia by modulating uric acid transporters. J. Ethnopharmacol. 336:118618. doi: 10.1016/j.jep.2024.118618
Martínez-Reyes, C. P., Manjarrez-Reyna, A. N., Méndez-García, L. A., Aguayo-Guerrero, J. A., Aguirre-Sierra, B., Villalobos-Molina, R., et al. (2020). Uric acid has direct proinflammatory effects on human macrophages by increasing proinflammatory mediators and bacterial phagocytosis probably via URAT1. Biomolecules 10:576. doi: 10.3390/biom10040576
Martinussen, J., Sørensen, C., Jendresen, C. B., and Kilstrup, M. (2010). Two nucleoside transporters in Lactococcus lactis with different substrate specificities. Microbiology 156, 3148–3157. doi: 10.1099/mic.0.039818-0
Mazza, T., Roumeliotis, T. I., Garitta, E., Drew, D., Rashid, S. T., Indiveri, C., et al. (2024). Structural basis for the modulation of MRP2 activity by phosphorylation and drugs. Nat. Commun. 15:1983. doi: 10.1038/s41467-024-46392-8
Minelli, E. B., Benini, A., Marzotto, M., Sbarbati, A., Ruzzenente, O., Ferrario, R., et al. (2004). Assessment of novel problotic Lactobacillus casei strains for the production of functional dairy foods. Int. Dairy J. 14, 723–736. doi: 10.1016/j.idairyj.2004.01.007
Mohammad, C. M., Che Alhadi, S., Wan Fatimah, S. W. M., Salman, A., Mohd Zhafri, M. R., and Rasimah, I. (2023). Delayed hypersensitivity reaction to allopurinol: A case report. Malays. Family Phys. 18:11. doi: 10.51866/cr.65
Nahle, S., El Khoury, A., Assaf, J. C., Louka, N., Chokr, A., and Atoui, A. (2022). A promising innovative technique for mycotoxin detoxification from beverages using biofilms of lactic acid bacteria. Innovat. Food Sci. Emerg. Technol. 82:103165. doi: 10.1016/j.ifset.2022.103165
Nassogne, M. C., Marie, S., and Dewulf, J. P. (2024). Neurological presentations of inborn errors of purine and pyrimidine metabolism. Eur. J. Paediatr. Neurol. 48, 69–77. doi: 10.1016/j.ejpn.2023.11.013
Ni, C., Li, X., Wang, L., Li, X., Zhao, J., Zhang, H., et al. (2021). Lactic acid bacteria strains relieve hyperuricaemia by suppressing xanthine oxidase activityviaa short-chain fatty acid-dependent mechanism. Food Funct. 12, 7054–7067. doi: 10.1039/d1fo00198a
Niu, D., Feng, N., Xi, S., Xu, J., and Su, Y. (2024). Genomics-based analysis of four porcine-derived lactic acid bacteria strains and their evaluation as potential probiotics. Mol. Genet. Genomics 299:24. doi: 10.1007/s00438-024-02101-0
Papadimitriou, K., Zoumpopoulou, G., Foligne, B., Alexandraki, V., Kazou, M., Pot, B., et al. (2015). Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 6:58. doi: 10.3389/fmicb.2015.00058
Riolo, M., Villena, A. M., Calpe, J., Luz, C., Meca, G., Tuccitto, N., et al. (2024). A circular economy approach: A new formulation based on a lemon peel medium activated with lactobacilli for sustainable control of post-harvest fungal rots in fresh citrus fruit. Biol. Control 189:105443. doi: 10.1016/j.biocontrol.2024.105443
Salati, L. M., Gross, C. J., Henderson, L. M., and Savaiano, D. A. (1984). Absorption and metabolism of adenine, Adenosine-5’-Monophosphate, adenosine and hypoxanthine by the isolated vascularly perfused rat small intestine. J. Nutr. 114, 753–760. doi: 10.1093/jn/114.4.753
Sanchez-Lozada, L. G., Soto, V., Tapia, E., Avila-Casado, C., Sautin, Y. Y., Nakagawa, T., et al. (2008). Role of oxidative stress in the renal abnormalities induced by experimental hyperuricemia. Am. J. Physiol. Renal Physiol. 295, F1134–F1141. doi: 10.1152/ajprenal.00104.2008
Sánchez-Lozada, L. G., Tapia, E., Soto, V., Ávila-Casado, C., Franco, M., Zhao, L., et al. (2007). Treatment with the xanthine oxidase inhibitor febuxostat lowers uric acid and alleviates systemic and glomerular hypertension in experimental hyperuricaemia. Nephrol. Dial. Transplant. 23, 1179–1185. doi: 10.1093/ndt/gfm783
Schumacher, H. R. Jr., Becker, M. A., Wortmann, R. L., MacDonald, P. A., Hunt, B., Streit, J., et al. (2008). Effects of febuxostat versus allopurinol and placebo in reducing serum urate in subjects with hyperuricemia and gout: A 28-Week, Phase III, randomized, double-blind, parallel-group trial. Arth. Rheum. Arthritis Care Res. 59, 1540–1548. doi: 10.1002/art.24209
Sekine, M., Okamoto, K., Pai, E. F., Nagata, K., Ichida, K., Hille, R., et al. (2023). Allopurinol and oxypurinol differ in their strength and mechanisms of inhibition of xanthine oxidoreductase. J. Biol. Chem. 299:105189. doi: 10.1016/j.jbc.2023.105189
She, D., Wang, Y., Liu, J., Luo, N., Feng, S., Li, Y., et al. (2022). Changes in the prevalence of hyperuricemia in clients of health examination in Eastern China, 2009 to 2019. BMC Endocrine Disord. 22:202. doi: 10.1186/s12902-022-01118-z
Shehata, M. G., Badr, A. N., El Sohaimy, S. A., Asker, D., and Awad, T. S. (2019). Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Ann. Agricult. Sci. 64, 71–78. doi: 10.1016/j.aoas.2019.05.002
Soemarie, Y. B., Milanda, T., and Barliana, M. I. (2021). Fermented foods as probiotics: A review. J. Adv. Pharmaceut. Technol. Res. 12, 335–339. doi: 10.4103/japtr.japtr_116_21
Stow, R. A., and Bronk, J. R. (1993). Purine nucleoside transport and metabolism in isolated rat jejunum. J. Physiol. 468, 311–324. doi: 10.1113/jphysiol.1993.sp019773
Strilchuk, L., Fogacci, F., and Cicero, A. F. (2019). Safety and tolerability of available urate-lowering drugs: A critical review. Expert Opin. Drug Saf. 18, 261–271. doi: 10.1080/14740338.2019.1594771
Togay, S. O., Keskin, A. C., Acik, L., and Temiz, A. (2010). Virulence genes, antibiotic resistance and plasmid profiles of Enterococcus faecalis and Enterococcus faecium from naturally fermented Turkish foods. J. Appl. Microbiol. 109, 1084–1092. doi: 10.1111/j.1365-2672.2010.04763.x
Wang, H., Mei, L., Deng, Y., Liu, Y., Wei, X., Liu, M., et al. (2019). Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition 62, 63–73. doi: 10.1016/j.nut.2018.11.018
Wang, Y., Deng, M., Deng, B., Ye, L., Fei, X., and Huang, Z. (2019). Study on the diagnosis of gout with xanthine and hypoxanthine. J. Clin. Lab. Anal. 33:e22868. doi: 10.1002/jcla.22868
Wang, X., Yang, S., Lu, J., Xie, G., and Wu, D. (2023). Screening and application of purine degrading Limosilactobacillus fermentum LF-1 from Huangjiu fermentation broth. J. Sci. Food Agric. 103, 7921–7931. doi: 10.1002/jsfa.12881
Wu, Y., Ye, Z., Feng, P., Li, R., Chen, X., Tian, X., et al. (2021). Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes 13, 1–18. doi: 10.1080/19490976.2021.1897211
Xiao, Y., Zhang, C., Zeng, X., and Yuan, Z. (2020). Microecological treatment of hyperuricemia using Lactobacillus from pickles. BMC Microbiol. 20:195. doi: 10.1186/s12866-020-01874-9
Yamada, N., Iwamoto, C., Kano, H., Yamaoka, N., Fukuuchi, T., Kaneko, K., et al. (2016). Evaluation of purine utilization by Lactobacillus gasseri strains with potential to decrease the absorption of food-derived purines in the human intestine. Nucleosides Nucleotides Nucleic Acids 35, 670–676. doi: 10.1080/15257770.2015.1125000
Yamada, N., Saito-Iwamoto, C., Nakamura, M., Soeda, M., Chiba, Y., Kano, H., et al. (2017). Lactobacillus gasseri PA-3 uses the purines IMP, inosine and hypoxanthine and reduces their absorption in rats. Microorganisms 5:10. doi: 10.3390/microorganisms5010010
Yamauchi, T., Oi, A., Kosakamoto, H., Akuzawa-Tokita, Y., Murakami, T., Mori, H., et al. (2020). Gut bacterial species distinctively impact host purine metabolites during aging in drosophila. iScience 23:101477. doi: 10.1016/j.isci.2020.101477
Yang, K., Li, J., and Tao, L. (2022). Purine metabolism in the development of osteoporosis. Biomed. Pharmacother. 155:113784. doi: 10.1016/j.biopha.2022.113784
Yuan, Q., Cheng, Y., Sheng, R., Yuan, Y., and Hu, M. (2022). A brief review of natural products with urate transporter 1 inhibition for the treatment of hyperuricemia. Evid. Based Complement. Alternat. Med. 2022:5419890. doi: 10.1155/2022/5419890
Zhang, M., Cui, R., Zhou, Y., Ma, Y., Jin, Y., Gou, X., et al. (2023). Uric acid accumulation in the kidney triggers mast cell degranulation and aggravates renal oxidative stress. Toxicology 483:153387. doi: 10.1016/j.tox.2022.153387
Zhang, S., Shi, C., Liu, C., Zou, X., Gong, F., and Li, X. (2023). Dynamic analysis of the bacterial community and determination of antioxidant capacity during the fermentation of sour tea. Fermentation 9:161. doi: 10.3390/fermentation9020161
Zhang, X., Cui, J., Hou, J., and Wang, W. (2022). Research progress of natural active substances with uric-acid-reducing activity. J. Agric. Food Chem. 70, 15647–15664. doi: 10.1021/acs.jafc.2c06554
Zhao, H., Chen, X., Meng, F., Zhou, L., Pang, X., Lu, Z., et al. (2023). Ameliorative effect of Lacticaseibacillus rhamnosus Fmb14 from Chinese yogurt on hyperuricemia. Food Sci. Hum. Wellness 12, 1379–1390. doi: 10.1016/j.fshw.2022.10.031
Zheng, Z., Si, Z., Wang, X., Meng, R., Wang, H., Zhao, Z., et al. (2023). Risk prediction for the development of hyperuricemia: Model development using an occupational health examination dataset. Int. J. Environ. Res. Public Health 20:3411. doi: 10.3390/ijerph20043411
Keywords: purine, degrading, lactic acid bacteria, uric acid, hyperuricemia
Citation: Liu Z, Zou X-Y, Yue J, Li S, Ou X, Huang C, Liu C-J and Li X-R (2025) Screening and evaluation of purines-degrading lactic acid bacteria isolated from traditional fermented foods in Yunnan Province and their uric acid-lowering effects in vivo. Front. Microbiol. 16:1627956. doi: 10.3389/fmicb.2025.1627956
Received: 14 May 2025; Accepted: 23 June 2025;
Published: 18 July 2025.
Edited by:
Weiwei Dong, Hubei Normal University, ChinaCopyright © 2025 Liu, Zou, Yue, Li, Ou, Huang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen-Jian Liu, MjAwNjAxMjlAa3VzdC5lZHUuY24=; Xiao-Ran Li, c3RhcmtleXJhbkAxNjMuY29t
†These authors have contributed equally to this work
‡ORCID: Zhen Liu, orcid.org/0009-0002-1553-8228
 Zhen Liu
Zhen Liu Xing-Yuan Zou1†
Xing-Yuan Zou1† Xiao-Ran Li
Xiao-Ran Li