- 1 Department of Plant and Environmental Sciences, University of Gothenburg, Gothenburg, Sweden
- 2 Mer, Molécules, Santé, Faculté des Sciences et Techniques, Université du Maine à Le Mans, Le Mans, France
ATP is the common energy currency of cellular metabolism in all living organisms. Most of them synthesize ATP in the cytosol or on the mitochondrial inner membrane, whereas land plants, algae, and cyanobacteria also produce it on the thylakoid membrane during the light-dependent reactions of photosynthesis. From the site of synthesis, ATP is transported to the site of utilization via intracellular membrane transporters. One major type of ATP transporters is represented by the mitochondrial ADP/ATP carrier family. Here we review a recently characterized member, namely the thylakoid ATP/ADP carrier from Arabidopsis thaliana (AtTAAC). Thus far, no orthologs of this carrier have been characterized in other organisms, although similar sequences can be recognized in many sequenced genomes. Protein Sequence database searches and phylogenetic analyses indicate the absence of TAAC in cyanobacteria and its appearance early in the evolution of photosynthetic eukaryotes. The TAAC clade is composed of carriers found in land plants and some green algae, but no proteins from other photosynthetic taxa, such as red algae, brown algae, and diatoms. This implies that TAAC-like sequences arose only once before the divergence of green algae and land plants. Based on these findings, it is proposed that TAAC may have evolved in response to the need of a new activity in higher photosynthetic eukaryotes. This activity may provide the energy to drive reactions during biogenesis and turnover of photosynthetic complexes, which are heterogeneously distributed in a thylakoid membrane system composed of appressed and non-appressed regions.
Introduction
Oxygenic photosynthesis is a biophysicochemical process that converts carbon dioxide into organic compounds using sunlight as a source of energy. It occurs in the chloroplasts of land plants and algae, and also in the cytoplasm of cyanobacteria, and uses water as a source of electrons, releasing oxygen as a waste product (for a recent review, see Hohmann-Marriott and Blankenship, 2011). The chloroplast in algae and plants has evolved from a cyanobacterial ancestor via endosymbiosis with a primitive eukaryotic host. It is a highly compartmentalized organelle, with three membrane systems (outer envelope, inner envelope, and thylakoid) and three soluble spaces (intermembrane space, stroma, and thylakoid lumen). A wide variety of solute and metabolite transporters reside within the different types of chloroplast membranes and mediate communication between the cytosol, stroma, and lumen. Several excellent reviews on the identification and functional characterization of these transporters have recently become available (Spetea and Schoefs, 2010; Breuers et al., 2011; Flügge et al., 2011; Weber and Linka, 2011).
Regarding their evolution, the majority of the inner envelope metabolite transporters have been shown to have a host origin, driven by the requirement to establish communication between the host cytosol and the cyanobiont (Facchinelli and Weber, 2011). There are so far no evolutionary studies dedicated to solute transporters from the outer envelope or thylakoid membranes, but it is believed that they originate from proteins in the ancestral cyanobacterial outer and thylakoid membranes, respectively.
Mitochondria and chloroplasts are the two organelles able to synthesize ATP, which is the universal energy currency of cellular metabolism in all living organisms. The difference between these organelles in this respect is that in all eukaryotes mitochondria produce ATP via oxidative phosphorylation on the inner membrane, to be used during cell metabolism. Chloroplasts and also cyanobacteria use sunlight as a source of energy to produce ATP (photophosphorylation) on the thylakoid membrane, which is consumed during CO2 fixation in the stroma. In addition, ATP is used for energy-dependent reactions on the envelope and thylakoid membrane or inside the thylakoid lumen (Spetea and Thuswaldner, 2008).
ATP is the largest and most highly charged solute transported across organellar membranes. Two structurally and phylogenetically different types of ATP transporters are represented in chloroplasts, namely ATP/ADP antiporters (AAA, TC #2.1.12, according to Saier et al., 2009) and mitochondrial ADP/ATP carriers (AAC, TC #2.A.29.1.1). There are two ATP/ADP antiporters (AATPs) in the inner envelope membrane of photosynthetic and heterotrophic plastids, supplying cytosolic ATP to the stroma (for recent reviews, see Haferkamp et al., 2011; Traba et al., 2011). The transport is electroneutral since the counter-ions for ATP4− are ADP3− together with H2PO4− (Trentmann et al., 2008). AATPs possess 12 putative transmembrane helices, and share a common origin with the ATP/ADP antiporters found in the parasite bacteria Rickettsia prowazekii and Chlamydia psittaci (Haferkamp et al., 2011).
The mitochondrial ADP/ATP carriers are the first and most studied members of the mitochondrial carrier (MC) family and are present only in eukaryotic cells (Haferkamp et al., 2011; Traba et al., 2011). Like the members of the AAA family, AACs are exchangers, but the transport is electrogenic (ATP4−/ADP3−) and proceeds in the opposite direction, since they transport matrix ATP through the intermembrane space out into the cytosol. Yet another functional difference from AAA is that AACs are sensitive to specific inhibitors, such as bongkrekic acid and carboxyatractyloside (Klingenberg, 2008). The 3D structure of the bovine AAC has been resolved at 2.2 Å resolution in a conformation stabilized with carboxyatractyloside (Pebay-Peyroula et al., 2003). The structure revealed six transmembrane helices and a selectivity filter for adenine nucleotides, whose sequence could be used to predict other AACs and even other MCs (Nury et al., 2010). From a total of 58 MC in Arabidopsis thaliana, three classical mitochondrial AACs have been characterized, with at least four more paralogous sequences awaiting validation (Palmieri et al., 2011). In addition to the mitochondrion, AAC members have also been found in peroxisomes, endoplasmic reticulum, amyloplasts, and chloroplasts (Haferkamp et al., 2011; Traba et al., 2011).
Using western blotting and activity inhibition with an antibody against the bovine AAC, the activity of an AAC was reported in the spinach thylakoid membrane (Spetea et al., 2004). BLAST searches with the bovine AAC against the Arabidopsis protein database combined with prediction of chloroplast transit peptides revealed one putative chloroplast AAC, encoded by the At5g1500 gene. The corresponding protein was localized to the thylakoid membrane of Arabidopsis (Thuswaldner et al., 2007; Zybailov et al., 2008), and was annotated as the thylakoid ATP/ADP carrier (AtTAAC). Subfractionation as well as immunocytochemical experiments indicated the non-appressed regions as the precise location of TAAC within the thylakoids (Thuswaldner et al., 2007). Another chloroplast AAC is encoded by the At3g51870 gene and was initially localized to the envelope using western blotting (Spetea, C., unpublished data) and mass-spectrometry based proteomics (Ferro et al., 2010).
The AtTAAC sequence contains 415 amino acids that include a predicted transit peptide of 60 amino acids. The sequence of the processed form is 80 residues longer than that of bovine AAC, explaining the observed difference between the two proteins in the reported size in SDS gels (Spetea et al., 2004). The extra 80 residues are distributed as 50 in the N-terminus and 30 in the C-terminus – regions containing many charged residues and a five-glycine repeat that could play a role in the regulation of TAAC activity. TAAC shares about 30% identity with bovine AAC, which is concentrated in the six putative transmembrane helices and to a lesser degree in the connecting loops (Thuswaldner et al., 2007). The selectivity filter for adenine nucleotides, represented by residues K-130, R-186, Y-282, and K-369, is fully conserved, indicating adenine nucleotides as the most likely substrates for the transport activity of TAAC.
Arabidopsis TAAC was characterized in E. coli as an ATP importer in exchange for cytosolic ADP (Thuswaldner et al., 2007), and its activity was found sensitive to bongkrekic acid (Thuswaldner, S., and Spetea, C., unpublished data). Pi is not a substrate for transport by TAAC (Thuswaldner et al., 2007), implying the requirement for a separate thylakoid Pi transporter. Indeed, such a protein has been identified in Arabidopsis, and functionally characterized in yeast and E. coli (Guo et al., 2008; Pavón et al., 2008). When assessed in thylakoid membranes, TAAC transports stromal ATP into the thylakoid lumen in exchange for ADP. The direction of TAAC-mediated transport determined in both E. coli and thylakoids is opposite to the direction of transport by mitochondrial AACs (Thuswaldner et al., 2007). Therefore, to distinguish it from the mitochondrial ADP/ATP carrier, the thylakoid protein has been named ATP/ADP carrier. Through adenine nucleotide exchange, TAAC was proposed to supply ATP for nucleotide-dependent reactions in the thylakoid lumen (Spetea et al., 2004). An extensive review on the structure, function, and evolution of the MC family has become recently available and provides insights into their roles in plants (Palmieri et al., 2011). This review focuses on the evolutionary origin of the TAAC subfamily of the mitochondrial AACs, which aids in elucidating its function in the thylakoid membrane.
When and Where in the Tree of Life DID TAAC Originate?
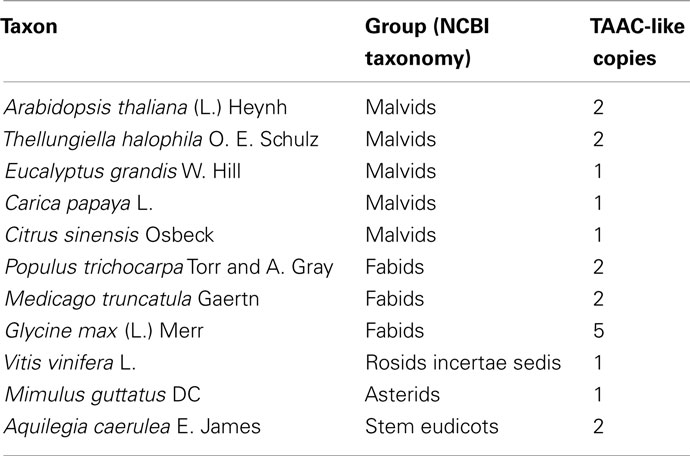
We assembled a set of protein sequences with which to place TAAC in an evolutionary context. We extracted protein sequences from the curated gene families at ARAMEMNON (Schwacke et al., 2003)1 and from search results to specific clades and genomes at NCBI2 and PHYTOZOME (Goodstein et al., 2011)3, respectively. We added the best BlastP matches to AtTAAC from each major eukaryotic division of life with an E-value <10−55. We also added all 17 members of the Arabidopsis MC family (including AtTAAC) that were listed as protein sequences related to TAAC at ARAMEMNON. We also added the best BlastP hits to proteins from several green plant clades, including Bryophyta, Chlorophyta, Lycopodiophyta, and Pinophyta. An alignment of these sequences was made using MUSCLE (Edgar, 2004, implemented at http://www.ebi.ac.uk). From the alignment (not shown) we could see that only two copies in Arabidopsis (encoded by the At5g01500 and At3g51870 genes) and all other non-Arabidopsis land plant proteins in this sample possessed a partly conserved N-terminal motif, consisting of 19 amino acid residues directly upstream from the first transmembrane helix delimited by Thuswaldner et al. (2007).
For the first phylogenetic analysis we included amino acid positions 121–369 in TAAC (spanning the six transmembrane domains), which resulted in 340 aligned positions. The analysis was carried out using a Bayesian approach. We used four parallel chains of a reverse model jump protein Bayesian analysis in MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001). Each chain was run for two million generations and sampled every 1,000 generations. The tree in Figure 1 shows a well-supported [posterior probability (PP) = 1.0] N-terminal containing clade (the “TAAC clade”) that includes land plants and some copies from Chlorophyta, but no proteins from other taxa. Sister to this clade with strong support (PP = 1.0) is a clade (also PP = 1.0) containing proteins from Chlorophyta that lack the N-terminal motif, including proteins from Chlamydomonas, Chlorella, and Volvox.
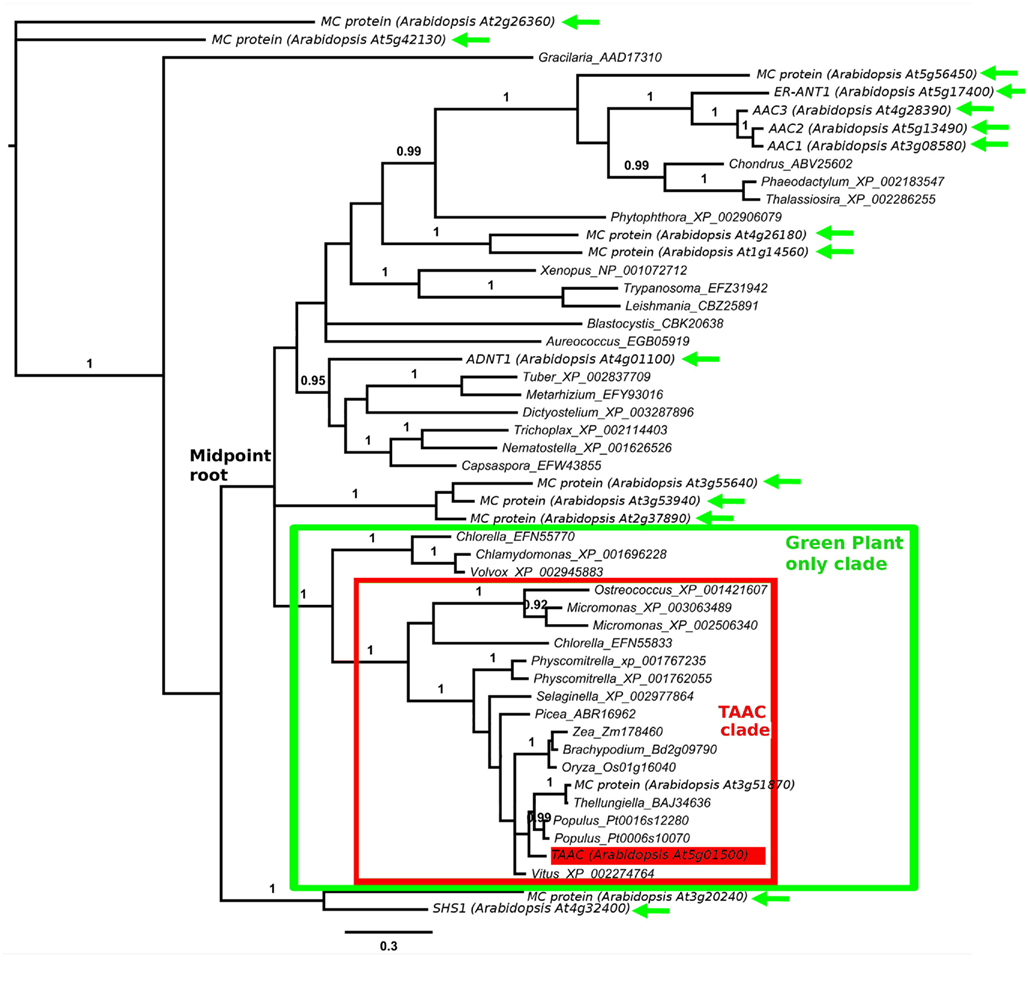
Figure 1. Protein phylogeny of 340 Muscle aligned positions of selected members of the MC family. The analysis involved 51 protein sequences, including 17 from Arabidopsis. The root position shown is arbitrary. The mid-point root position is marked, as is the clade containing all TAAC-like sequences (boxed in red). The clade of Green Plants is marked in green. All plant sequences outside this clade are indicated by green arrows. Accession numbers follow the gene annotation (where available) and names of genera within which the proteins are found. The position of Arabidopsis TAAC is highlighted in red. The scale bar indicates substitutions per site. Posterior probabilities ≥0.90 of clades summarizing two million Markov chain Monte Carlo generations (of a reversible model jump protein Bayesian analysis in MrBayes 3.1.2) are shown above branches. The results discussed are robust to an alternative alignment (MAFFT, not shown).
The best BlastP matches to proteins from the following taxa did not contain the N-terminal motif, although they each possessed the conserved six transmembrane domain structure: Amoebozoa, Animalia, Euglenozoa, Fungi, Opisthokonta, Rhodophyta, and Stramenopiles – the latter includes brown algae and diatoms (nomenclature based on NCBI’s taxonomy). All other eukaryote taxa at one hierarchical level below Eukaryota in NCBI’s taxonomy were searched, but did not contain BlastP matches to AtTAAC with E-values <10−55. Apart from TAAC, only one other protein from Arabidopsis is also a member of the TAAC clade and contains this motif, namely the one encoded by the At3g51870 gene. The amino acid sequences of these two proteins are 67% identical.
The TAAC clade contains relatively short branches in relation to the remainder of the phylogeny, making it unlikely that the root resides within the clade. This is also consistent with mid-point rooting (marked in Figure 1) as well as the phylogenetic pattern expected among the plants that carry these proteins. Together, this indicates that the TAAC clade contains several proteins orthologous to AtTAAC. This clade also contains duplicated copies (paralogs) from several plant taxa (more are seen in our expanded sample in Figure 2, details below), but these are inferred to have arisen only after green plants diverged from other eukaryotes and are expected due to the action of recent gene and/or genome duplication (e.g., in Arabidopsis, Bowers et al., 2003; legumes, Pfeil et al., 2005; Physcomitrella, Rensing et al., 2007).
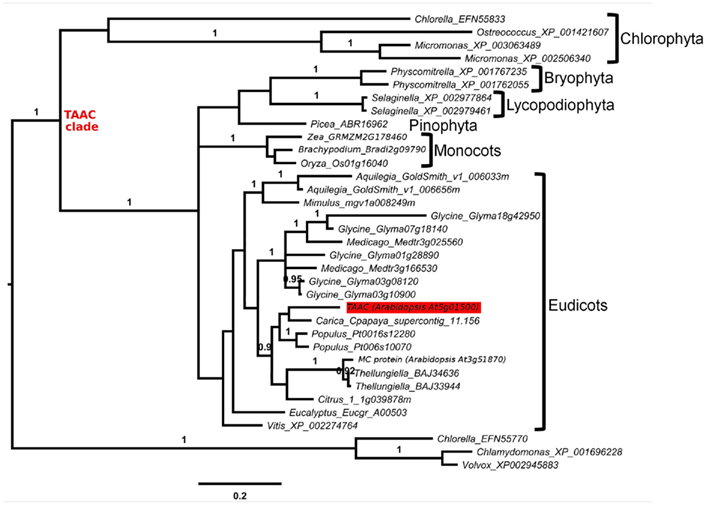
Figure 2. Protein phylogeny of 379 Muscle aligned positions of selected members of the MC family, focusing on the TAAC clade (marked). The analysis involved 35 protein sequences, including two from Arabidopsis. The root position is based on Figure 1. Accession numbers follow the names of genera within which the proteins are found. The taxon to which each subclade belongs is indicated. The position of Arabidopsis TAAC is highlighted in red. The scale bar indicates substitutions per site. Posterior probabilities ≥0.90 of clades, analyzed as in Figure 1, are shown above branches. The results discussed are robust to an alternative alignment (MAFFT, not shown).
The position of the remaining 15 Arabidopsis MC proteins (Figure 1) indicates that the MC protein family was present very early in eukaryote evolution. Scattered across the tree are, for example, homologs from Metazoa (Nematostella), Fungi (Tuber), Amoebozoa (Dictyostelium), Stramenopiles (Aureococcus), Euglenozoa (Leishmania), and Rhodophyta (Chondrus). However, none of these homologs are part of the TAAC clade, which contains only proteins from Chlorophyta and Streptophyta. No matches within the BlastP cut-off used were found to other photosynthetic organisms, such as Bacillariophyta (diatoms), Cyanophyta (cyanobacteria), Glaucocystophyceae, Phaeophyceae (brown algae), or Rhodophyta (red algae).
Therefore, we conclude that TAAC-like sequences arose only a single time, after the clade including both Chlorophyta and Streptophyta (the latter including land plants) diverged from the other taxa in our sample, but earlier than the divergence of Chlorophyta and Streptophyta. The phylogenetic results are robust to an alternative alignment based on MAFFT (Katoh et al., 2002), although this is not shown here.
How Did the TAAC Copies in Arabidopsis and other Eudicots Evolve?
We increased the sample of TAAC-like proteins in a more focused phylogenetic inference in order to place the two Arabidopsis members of the TAAC subfamily into context. For this purpose, we sampled all copies from the species listed in Table 1 that contain the N- and C-terminal motifs similar to TAAC and that were also part of the TAAC clade. We focused on eudicots, because preliminary analyses indicated that the two Arabidopsis TAAC subfamily members were placed in a clade containing proteins from eudicot species only.
We expanded our search for land plant sequences containing the N-terminal motif by using BlastP with AtTAAC as the query to specific genomes in Phytozome, as well as to Thellungiella (Brassicaceae), because in earlier BlastP searches a Thellungiella sequence closely related to TAAC was detected. We excluded possible allelic variants that differed by only a single amino acid from our enumeration. The alignment of this set of sequences was performed as before. From this alignment (Figure S1 in Supplementary Material), we used amino acids 107–482 in TAAC (corresponding to 375 aligned positions and including the N-terminal and C-terminal motifs) in our second phylogenetic analysis and also added the sister group of the TAAC clade (from Figure 1) as an outgroup. The phylogenetic analysis was performed as described for the data shown in Figure 1, with sample information summarized in Table 2 and the results presented in Figure 2.
Table 2. The presence (green) or absence (red) of TAAC-like proteins within the surveyed genomes is shown.
The copy number of TAAC-like proteins in the eudicots examined here appears to be rather conservative, ranging from one to five copies (Table 1). Arabidopsis, Medicago, and Populus each have only two copies, but these are perhaps the best-known eudicot genomes. Therefore, the copy number for the other species may be underestimated. The lack of support for some key nodes in the phylogeny (Figure 2) makes it difficult to pinpoint the timing of divergence of the two copies found in Arabidopsis. They almost certainly diverged sometime before this genus diverged from Thellungiella (perhaps 40–50 million years ago: Amtmann, 2009; Beilstein et al., 2010), but it is difficult to say how much earlier this divergence might have been.
Unlike the Arabidopsis copies, one of the copies found in Aquilegia differs from its closest relative by a long branch (Figure 2). In contrast, the rather recently diverged (in comparison to Arabidopsis) multiple copies found in Selaginella and Physcomitrella do not show this pattern. The long branch observed in Aquilegia may indicate functional shifts and positive selection may be involved, as has been shown in, e.g., monkey pancreatic ribonucleases (Zhang et al., 2002). These possibilities could be examined further and may lead to novel research questions. However, in the case of one Glycine sequence, poor assembly is a more likely explanation for the long branch observed in that taxon (NB: alignment positions 308–389 in Figure S1 in Supplementary Material), so caution needs to be taken in specific cases.
How Does the N-Terminal Motif Vary within the TAAC Subfamily?
The N-terminal motif may be helpful to identify potential members of the TAAC subfamily. Furthermore, this clade shows complete conservation of the ring of four positively charged residues (Figure S1 in Supplementary Material) proposed to act as a selectivity filter for adenine nucleotides (Thuswaldner et al., 2007), confirming that these aligned proteins are all AACs.
Potentially important is that significant variation in the N-terminal sequences (beyond just the 19 residue motif described earlier) occurs among the proteins sampled here. For example, several proteins contain many charged residues, including paired charged ones, and a five-glycine repeat (Figure S1 in Supplementary Material). These may be implicated in ligand (e.g., calcium) binding and large conformational changes that could regulate their activity, as in the case of Ca2+-dependent members of the MC family (Weber et al., 1997). However, some of these proteins do not contain either of these features. For example, the Selaginella sequences have only 13 residues upstream of the more conserved N-terminal motif, few charged residues, and lack the five-glycine repeat. One Micromonas protein appears to lack the N-terminal motif altogether (Figure S1 in Supplementary Material). Variation in the N-terminal motif among the land plant TAAC-like proteins is smaller (Table 2), but so is the phylogenetic distance. It is tempting to speculate as to what the precise function of the N-terminal residues might be, moreover, interesting research questions can be formulated around these sequences. However, the sequences (especially those apparently lacking the N-terminal motif) should of course be verified before pursuing these lines of investigation, as technical problems (e.g., with the assembly), deposition of incomplete sequences, etc., need to be ruled out first.
What are the Possible Functions of TAAC in Land Plants and Green Algae?
The phylogenetic analyses performed in this work indicate that TAAC is absent in cyanobacteria and that the most recent common ancestor of green plants (Streptophyta + Chlorophyta) was the earliest photosynthetic organism that we can identify to have carried a TAAC-like protein. Although TAAC is a thylakoid protein and the thylakoid membrane originated with the thylakoid membrane of an ancestral cyanobacterium, TAAC appears to be eukaryote-specific, as are all mitochondrial AACs (Palmieri et al., 2011). Nevertheless, in contrast to mitochondrial AACs, which are found in both photosynthetic and heterotrophic eukaryotes (Palmieri et al., 2011), only some photosynthetic eukaryotes, more specifically, Chlorophyta and Streptophyta, carry TAAC-like copies. Other photosynthetic eukaryotes (red algae, brown algae, and diatoms) and, interestingly, even some Chlorophyta (Chlamydomonas and Volvox) do not appear to have such a protein, i.e., not one with a similar N-terminus nor placed in the TAAC clade, whereas other Chlorophyta do (e.g., Chlorella; Table 2).
A potential answer to the question “why do not all photosynthetic organisms carry TAAC-like proteins?” could be that TAAC may fulfill specialized functions in the thylakoid membrane of land plants and most green algae. These membranes are organized in highly stacked (appressed) regions interconnected by stroma-exposed (non-appressed) regions (Figure 3). Other photosynthetic organisms (cyanobacteria, red algae, brown algae, and diatoms) display unstacked or weakly stacked thylakoids (for details and references, see the legend to Figure 3). To explain the apparent difference within green algae, it is relevant to consider that Chlamydomonas has appressed thylakoids, but they do not form regular grana stacks (de Vitry and Vallon, 1999). To our knowledge, the thylakoid structure has not, thus far, been studied in green algae other than Chlamydomonas, but they are expected to have a similar organization to the land plant thylakoids. The thylakoid organization depends on the type and arrangement of light-harvesting antennae and on the distribution of photosynthetic complexes (Figure 3). It was held for a long time that the different macrocomplexes comprising the photosynthetic apparatus were organized linearly along thylakoid membranes. This view is no longer valid since it has been established that these complexes are laterally distributed, i.e., localized exclusively in the appressed membranes (photosystem II), exclusively in the stroma-exposed thylakoids (photosystem I, ATP synthase) or in both types of membranes (cytochrome b6/f complex; Anderson, 2002). This heterogenous composition of thylakoids is restricted to the green algae and land plants (Figure 3).
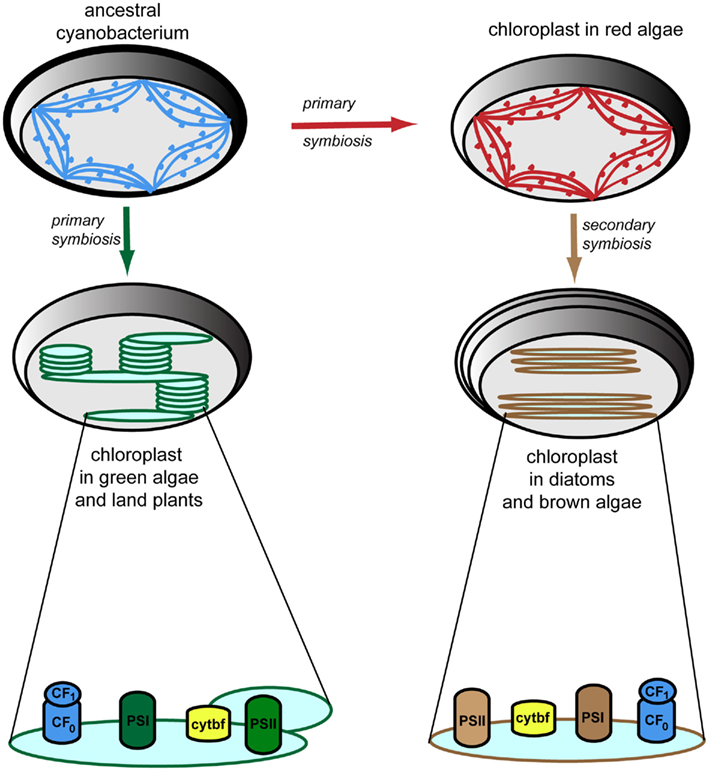
Figure 3. Evolution of thylakoid membrane organization and composition. Cyanobacterial thylakoid membranes (shown in blue) are located directly in the cytoplasm, are arranged in multiple (three to eight) layers, and make contacts with the plasma membrane. They have attached phycobilisomes for harvesting light, which protrude toward the cytoplasm, thus preventing membrane stacking (Liberton and Pakrasi, 2008). Chloroplasts have evolved via primary symbiosis from a cyanobacterial relative. Thus, similar arrangement of the thylakoid membranes is observed in red algae (shown in red), which also have attached phycobilisomes (Tsekos et al., 1996). Thylakoid membranes in diatoms and brown algae (shown in brown) are arranged in groups of three loosely appresed vesicles and contain fucoxanthin–chlorophyll-protein complexes for harvesting light (Bertrand, 2010). Note the four-layer envelopes surrounding the chloroplast as compared to the types found in other algae and plants, as a result of secondary symbiosis. Thylakoid membranes in green algae and higher plants (shown in green) are located inside the chloroplast, are organized in grana stacks (5–20 vesicles) interconnected by stroma-exposed lamellae, and contain chlorophyll–protein complexes for harvesting light (Mustardy and Garab, 2003). Various 3D models of the organization of these thylakoid membranes have been proposed and debated (Daum and Kühlbrandt, 2011). According to the most accepted model, based on electron tomography (Daum et al., 2010; Austin and Staehelin, 2011), grana should be seen as stacks of straight, exactly parallel pairs of thylakoid membranes, each pairs of grana disks merging with stroma lamellae by staggered membranes protrusions. Importantly, but not shown in this figure, the plane of the stromal thylakoids is tilted by 10–25° relatively to the plane of the grana stack so that the stromal thylakoids display a helix around the granum (Mustardy et al., 2008). The photosynthetic apparatus is composed of four multisubunit complexes, namely the water-oxidizing photosystem II (PSII), cytochrome b6/f (cytbf), photosystem I (PSI), and the H+-translocating ATP synthase (CF0F1; Nelson and Ben-Shem, 2004). These complexes are laterally distributed in green algae and plants, whereas in other algae and diatoms, they display a more uniform distribution.
The differences in thylakoid membrane organization among cyanobacteria, green algae, and land plants may have implications for biogenesis and turnover of photosynthetic complexes, such as photosystem II (PSII). The biosynthesis of complexes in cyanobacteria occurs at contact sites with the plasma membrane, whereas PSII repair takes place within thylakoid membranes (Zak et al., 2001). In green algae, the site of PSII assembly during de novo D1 synthesis is around a so-called pyrenoid, a specialized sub-compartment within the chloroplast for CO2 fixation that is different from the site of PSII repair – the stroma-regions of the thylakoid membrane (Uniacke and Zerges, 2007). In land plants, the location of thylakoid membrane complex biosynthesis is uncertain. A widely accepted model is that PSII subunits assemble in the non-appressed region of the thylakoid membrane (Baena-González and Aro, 2002). The mechanism of PSII repair in plants has been studied in detail and shown to have important differences from that in cyanobacteria, such as the shuttling of PSII subcomplexes between the grana and stroma membranes, which is accompanied by changes in the oligomeric structure of the complex (Mulo et al., 2011).
The question arising is whether TAAC plays a role in the biogenesis and turnover of plant photosynthetic complexes. Its tissue expression pattern has been studied in detail, and supports this possibility, as described below. AtTAAC was found highly expressed in young photosynthetic organs, such as developing leaves, flower buds, and green siliques. Furthermore, it was found expressed in etiolated seedlings, similar to the thylakoid HCF136 protein, which was proven to be required for biogenesis of thylakoids and PSII assembly (Meurer et al., 1998; Plücken et al., 2002). TAAC expression is strongly upregulated in leaves undergoing senescence or exposed to wounding, light stress, oxidative stress, salt stress, and desiccation, pointing to an additional role in supplying ATP for energy-dependent processes (e.g., proteolysis, folding) during turnover of photosynthetic complexes.
Thus far, no dedicated studies have been undertaken on the role of TAAC activity during thylakoid biogenesis. Nevertheless, its role during PSII repair has been studied in more detail by taking advantage of taac mutants. Remarkably, the mutants grew slower and were more sensitive to high light stress due to an inability to degrade the reaction center D1 protein (Yin et al., 2010). A model for its function during PSII repair cycle in plants under high light stress is presented in Figure 4. Briefly, ATP translocated by TAAC into the lumen is converted to GTP, which is then bound to the PsbO lumenal extrinsic subunit of the PSII dimeric complex (Spetea et al., 2004; Lundin et al., 2007a). The GTPase activity of this protein regulates the monomerization and partial disassembly of PSII, pre-requisite steps for the proteolytic degradation of the D1 protein (Lundin et al., 2007b). Replacement of the damaged D1 with a new copy requires coordination between its degradation, synthesis, insertion, and assembly of the new copy in the PSII complex. We propose that the novel mechanism of ATP transport and GTP signaling across thylakoid membranes, discussed above, may be a plant-specific feature for the following reasons: the D1 degradation is GTP dependent in plants but not in cyanobacterial thylakoids (Spetea et al., 1999); GTP binding to PsbO may be a plant-specific feature, since putative GTP binding sequence domains are not found conserved in cyanobacteria or the green alga Chlamydomonas PsbOs (Lundin et al., 2007a); TAAC belongs to the MC family, which is eukaryotic specific (Palmieri et al., 2011). Thus, we speculate that TAAC is associated with the need for highly controlled regulation of PSII repair in a highly stacked thylakoid membrane system requiring shuttling of complexes between the appressed and non-appressed regions.
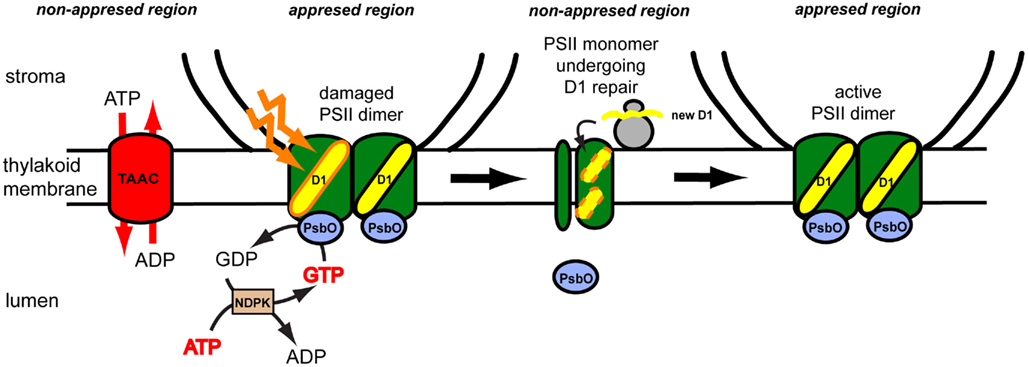
Figure 4. Proposed model for role of the thylakoid ATP/ADP carrier (TAAC) during photosystem II (PSII) repair cycle in plants. The plant thylakoid membrane is organized in grana stacks (appressed region) and stroma-exposed (non-appressed region) lamellae. The composition of the thylakoid membrane is heterogenous, although its lumenal space is continuous. TAAC is located in the stroma lamellae and exchanges stromal ATP for lumenal ADP. ATP is inter-converted to GTP by the lumenal nucleoside diphosphate kinase (NDPK). The active PSII dimer is located in the grana regions and contains lumenal extrinsic PsbO proteins. During illumination with excess light, the reaction center D1 protein may be oxidatively damaged and needs to be replaced. The PsbO subunit of the damaged monomer binds and hydrolyzes GTP, leading to its dissociation and partial disassembly of the monomeric complex on the way to the stroma-exposed regions. Here the D1 protein is degraded and replaced with a new copy synthesized by the chloroplast ribosomes and co-translationally inserted in the membrane. Monomers assemble into the dimers and migrate to the grana regions.
Conclusion
Study of the adenine nucleotide carrier TAAC in chloroplast thylakoids is interesting because it has revealed similarities but also differences from classical mitochondrial AACs. Most MC (including AACs) are present in both phototrophic and heterotrophic eukaryotes, suggesting functions required for basic eukaryotic processes that take place inside the mitochondria and other organelles. On the other hand, TAAC-like proteins have been found only in green plants, indicating at the earliest an appearance after the endosymbiosis of a phototrophic prokaryote with the most recent common ancestor of land plants and green algae. Essentially, TAAC is an AAC with additional N (and C) motifs, apart from the chloroplast targeting sequence. The pathway by which TAAC arrived in the chloroplast and the precise function of the N-motif in TAAC activity remain to be investigated. Nevertheless, based on the phylogenetic study presented here, we propose that TAAC may fulfill functions that may be required in the context of the more complex organization of thylakoid membranes in green algae and land plants, such as biogenesis and repair of photosynthetic complexes. Some of the lumenal enzymes participating in the biogenesis/turnover processes may require ATP or other nucleotides resulting from inter-conversion, and therefore ATP must be translocated. This would not be unexpected, taking into consideration the increasing evidence for a complex role of the thylakoid lumen in photosynthetic regulation and plant cell signaling, expanding its function beyond an energetic perspective (for a recent review, see Spetea, 2011). Thus, learning about TAAC evolution can be very important to understand its functional importance in green plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in the C.S. laboratory has been supported by grants from the Swedish Research Council and the Swedish Research Council for Environment, Agriculture and Space Planning (Formas). Bernard E. Pfeil acknowledge funding from: Swedish Research Council, Royal Swedish Academy of Sciences, Royal Physiographic Society in Lund, Lars Hiertas Minne fund, Helge Ax:son Johnsons Foundation, and Lundgrenska fund. Research in the Benoît Schoefs laboratory has been supported by grants from Université du Maine à Le Mans.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2011.00110/abstract
Figure S1| MUSCLE alignment of TAAC subfamily of full-length amino acid sequences.
Footnotes
References
Amtmann, A. (2009). Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2, 3–12.
Anderson, J. M. (2002). Changing concepts about the distribution of photosystems I and II between grana-appressed and stroma-exposed thylakoid membranes. Photosynth. Res. 73, 157–164.
Austin, J. R., and Staehelin, L. A. (2011). Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol. 155, 1601–1611.
Baena-González, E., and Aro, E. M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1451–1459.
Beilstein, M. A., Nagalingum, N. S., Clements, M. D., Manchester, S. R., and Mathews, S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 18724–18728.
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438.
Breuers, F., Braeutigam, A., and Weber, A. P. M. (2011). The plastid outer envelope – a highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2:97. doi:10.3389/fpls.2011.00097
Daum, B., and Kühlbrandt, W. (2011). Electron tomography of plant thylakoid membranes. J. Exp. Bot. 62, 2393–2402.
Daum, B., Nicastro, D., Austin, J. R., McIntosh, J. R., and Kühlbrandt, W. (2010). Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22, 1299–1312.
de Vitry, C., and Vallon, O. (1999). Mutants of Chlamydomonas: tools to study thylakoid membrane structure, function and biogenesis. Biochimie 81, 631–643.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Facchinelli, F., and Weber, A. P. M. (2011). The metabolite transporters of the plastid envelope. An update. Front. Plant Sci. 2:50. doi:10.3389/fpls.2011.00050
Ferro, M., Brugière, S., Salvi, D., Seigneurin-Berny, D., Court, M., Moyet, L., Ramus, C., Miras, S., Mellal, M., Le Gall, S., Kieffer-Jaquinod, S., Bruley, C., Garin, J., Joyard, J., Masselon, C., and Rolland, N. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell Proteomics 9, 1063–1084.
Flügge, U. I., Häusler, R. E., Ludewig, F., and Gierth, M. (2011). The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 62, 2381–2392.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., Mitros, T., Dirks, W., Hellsten, U., Putnam, N., and Rokhsar, D. S. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., and Versaw, W. K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898.
Haferkamp, I., Fernie, A. R., and Neuhaus, H. E. (2011). Adenine nucleotide transport in plants: much more than a mitochondrial issue. Trends Plant Sci. 16, 507–515.
Hohmann-Marriott, M. F., and Blankenship, R. E. (2011). Evolution of photosynthesis. Annu. Rev. Plant Biol. 62, 515–548.
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755.
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066.
Klingenberg, M. (2008). The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021.
Liberton, M., and Pakrasi, H. (2008). “Membrane systems in cyanobacteria,” in The Cyanobacteria: Molecular Biology, Genomics and Evolution, eds A. Herrero, and E. Flores (Norwich: Caister Academic Press), 271–289.
Lundin, B., Thuswaldner, S., Shutova, T., Eshaghi, S., Samuelsson, G., Barber, J., Andersson, B., and Spetea, C.. (2007a). Subsequent events to GTP binding by the plant PsbO protein: structural changes, GTP hydrolysis and dissociation from the photosystem II complex. Biochim. Biophys. Acta 1767, 500–508.
Lundin, B., Hansson, M., Schoefs, B., Vener, A. V., and Spetea, C. (2007b). The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 49, 528–539.
Meurer, J., Plücken, H., Kowallik, K. V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 7, 5286–5297.
Mulo, P., Sakurai, I., and Aro, E. M. (2011). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim. Biophys. Acta 1817, 247–257.
Mustardy, L., Buttle, K., Steinbach, G., and Garab, G. (2008). The three dimensional network of the thylakoid membranes in plants: quasihelical model of the granum–stroma assembly. Plant Cell 20, 2552–2557.
Mustardy, L., and Garab, G. (2003). Granum revisited. A three dimensional model–where things fall into place. Trends Plant Sci. 8, 117–122.
Nelson, N., and Ben-Shem, A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982.
Nury, H., Blesneac, I., Ravaud, S., and Pebay-Peyroula, E. (2010). Structural approaches of the mitochondrial carrier family. Methods Mol. Biol. 654, 105–117.
Palmieri, F., Pierri, C. L., De Grassi, A., Nunes-Nesi, A., and Fernie, A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181.
Pavón, R. L., Lundh, F., Lundin, B., Mishra, A., Persson, B. L., and Spetea, C. (2008). Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J. Biol. Chem. 283, 13520–13527.
Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trézéguet, V., Lauquin, G. J., and Brandolin, G. (2003). Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44.
Pfeil, B. E., Schlueter, J. A., Shoemaker, R. C., and Doyle, J. J. (2005). Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst. Biol. 54, 441–454.
Plücken, H., Müller, B., Grohmann, D., Westhoff, P., and Eichacker, L. A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532, 85–90.
Rensing, S. A., Ick, J., Fawcett, J. A., Lang, D., Zimmer, A., Van de Peer, Y., and Reski, R. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7, 130. doi:10.1186/1471-2148-7-130
Saier, M. H. Jr., Yen, M. R., Noto, K., Tamang, D. G., and Elkan, C. (2009). The transporter classification database: recent advances. Nucleic Acids Res. 37, D274–D278.
Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W. B., Flügge, U. I., and Kunze, R. (2003). Aramemnon, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26.
Spetea, C. (2011). “Role of chloroplast thylakoid lumen in photosynthetic regulation and plant cell signaling,” in Progress in Botany, Vol. 73, eds U. Lüttge, W. Beyschlag, B. Büdel, D. Francis (Heidelberg: Springer-Verlag), 207–230.
Spetea, C., Hundal, T., Lohmann, F., and Andersson, B. (1999). GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc. Natl. Acad. Sci. U.S.A. 96, 6547–6552.
Spetea, C., Hundal, T., Lundin, B., Heddad, M., Adamska, I., and Andersson, B. (2004). Multiple evidence for nucleotide metabolism in the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. U.S.A. 101, 1409–1414.
Spetea, C., and Schoefs, B. (2010). Solute transporters in plant thylakoid membranes: key players during photosynthesis and light stress. Commun. Integr. Biol. 3, 122–129.
Spetea, C., and Thuswaldner, T. (2008). “Update in nucleotide-dependent processes in plant chloroplasts,” in Plant Cell Compartments – Selected Topics, ed. B. Schoefs (Trivandrum: Research Signpost), 105–149.
Thuswaldner, S., Lagerstedt, J. O., Rojas-Stütz, M., Bouhidel, K., Der, C., Leborgne-Castel, N., Mishra, A., Marty, F., Schoefs, B., Adamska, I., Persson, B. L., and Spetea, C. (2007). Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J. Biol. Chem. 282, 8848–8859.
Traba, J., Satrústegui, J., and del Arco, A. (2011). Adenine nucleotide transporters in organelles: novel genes and functions. Cell. Mol. Life Sci. 68, 1183–1206.
Trentmann, O., Jung, B., Neuhaus, H. E., and Haferkamp, I. (2008). Nonmitochondrial ATP/ADP transporters accept phosphate as third substrate. J. Biol. Chem. 283, 36486–36493.
Tsekos, I., Reiss, H. D., Orfanidisi, S., and Orologas, N. (1996). Ultrastructure and supramolecular organization of photosynthetic membranes of some marine red algae. New Phytol. 133, 543–551.
Uniacke, J., and Zerges, W. (2007). Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19, 3640–3654.
Weber, A. P., and Linka, N. (2011). Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77.
Weber, F. E., Minestrini, G., Dyer, J. H., Werder, M., Boffelli, D., Compassi, S., Wehrli, E., Thomas, R. M., Schulthess, G., and Hauser, H. (1997). Molecular cloning of a peroxisomal Ca2+-dependent member of the mitochondrial carrier superfamily. Proc. Natl. Acad. Sci. U.S.A. 94, 8509–8514.
Yin, L., Lundin, B., Bertrand, M., Nurmi, M., Solymosi, K., Kangasjärvi, S., Aro, E. M., Schoefs, B., and Spetea, C. (2010). Role of thylakoid ATP/ADP carrier in photoinhibition and photoprotection of photosystem II in Arabidopsis. Plant Physiol. 153, 666–677.
Zak, E., Norling, B., Maitra, R., Huang, F., Andersson, B., and Pakrasi, H. B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 98, 13443–13448.
Zhang, J., Zhang, Y., and Rosenberg, H. F. (2002). Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415.
Keywords: green alga, chloroplast, plant, photosynthesis, ADP/ATP carrier, thylakoid, TAAC phylogeny
Citation: Spetea C, Pfeil BE and Schoefs B (2012) Phylogenetic analysis of the thylakoid ATP/ADP carrier reveals new insights into its function restricted to green plants. Front. Plant Sci. 2:110. doi: 10.3389/fpls.2011.00110
Received: 30 September 2011; Accepted: 17 December 2011;
Published online: 09 January 2012.
Edited by:
Heven Sze, University of Maryland, USAReviewed by:
Daniel Hofius, University of Copenhagen, DenmarkUener Kolukisaoglu, University of Tuebingen, Germany
Copyright: © 2012 Spetea, Pfeil and Schoefs. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Cornelia Spetea, Department of Plant and Environmental Sciences, University of Gothenburg, 40530 Gothenburg, Sweden. e-mail:Y29ybmVsaWEuc3BldGVhLndpa2x1bmRAZHBlcy5ndS5zZQ==