- Department of Biological Sciences, Pusan National University, Busan, South Korea
Peroxisomes play a critical role in many metabolic pathways during the plant life cycle. It has been proposed that the transition between different types of peroxisomes involves the degradation of obsolete peroxisomal enzymes via proteolytic activities in the peroxisome matrix, the cytosol, or the vacuole. Forward and reverse genetic studies recently provided evidence for autophagic degradation of peroxisomes in the vacuole of Arabidopsis seedlings. Here, we briefly review a model of pexophagy, or selective autophagy of peroxisomes, in plant cells.
Plant Peroxisomes: Types, Transition, and Protein Degradation
Plant peroxisomes are versatile organelles that participate in many metabolic pathways such as fatty acid β-oxidation and photorespiration (reviewed by Hu et al., 2012). In addition to the enzymes needed for these pathways, peroxisomes contain antioxidant enzymes, for example, catalase, to protect plants from oxidative damage, since hydrogen peroxide is generated from fatty acid β-oxidation and photorespiration and other oxidation reactions in the peroxisome.
Peroxisomes are dynamic organelles with the capacity to change their appearance, their association with other organelles, and their enzyme composition. These changes depend on the developmental program and metabolic needs of the cell. For example, when oil-storing seeds such as cucumber (Cucumis sativus) and Arabidopsis (Arabidopsis thaliana) germinate, peroxisomes contain the glyoxylate cycle enzymes. The enzymes are needed for the consumption of acetyl-CoA (the product of fatty acid β-oxidation) for synthesis of organic acids that can be used to generate sugars by gluconeogenesis (reviewed by Pracharoenwattana and Smith, 2008). These seedling peroxisomes, formerly called glyoxysomes, are closely associated with lipid bodies supplying fatty acids (Trelease et al., 1971). When the seedlings are exposed to light, peroxisomal glyoxylate cycle enzymes, such as isocitrate lyase (ICL) and malate synthase (MLS), are rapidly degraded and enzymes involved in photorespiration accumulate. These peroxisomes are referred to as leaf peroxisomes. The change in peroxisomal enzyme composition may result from the transition of seedling peroxisomes to leaf peroxisomes (the “one-population model”), rather than from the degradation of seedling peroxisomes and the formation of new leaf peroxisomes (the “two-population model”; Beevers, 1979; Nishimura et al., 1996). Light also triggers changes in the position of peroxisomes. Seedling peroxisomes are associated with lipid bodies, while leaf peroxisomes are positioned near chloroplasts (Trelease et al., 1971; Gruber et al., 1973) from which glycolate, a photorespiration intermediate, enters the peroxisome for oxidation. Interestingly, a reverse transition from leaf peroxisomes to peroxisomes containing ICL may occur during starvation and organ senescence (reviewed by Nishimura et al., 1996; Pracharoenwattana and Smith, 2008).
When seedling peroxisomes are transformed to leaf peroxisomes, obsolete ICL and MLS must be degraded. In recent studies, three mechanisms have been proposed for the degradation of these proteins during post-germinative growth of Arabidopsis seedlings (Figure 1). In one mechanism [herein designated intraperoxisomal degradation (IPD)], it is proposed that peroxisomal proteins are degraded by resident proteases. However, known peroxisomal proteases, which include Lon-related protease 2 (LON2), have not been implicated in full degradation of peroxisomal matrix proteins (Lingard and Bartel, 2009). This argument was mainly based on the observation that ICL and MLS levels in lon2 mutant were not higher than those in wild-type (Lingard and Bartel, 2009; Burkhart et al., 2013). According to a second mechanism, obsolete proteins are retranslocated from peroxisomes and degraded in the cytosol by the 26S proteasome. During this process, called peroxisome-associated protein degradation (PexAD), the proteins are polyubiquitylated before they are recognized by the proteasome, analogous to ER-associated protein degradation (ERAD; reviewed by Smith et al., 2012). The possibility of polyubiquitylation is supported by a survey of Arabidopsis ubiquitylome, in which ICL was identified as a ubiquitylated protein (Kim et al., 2013a). Furthermore, the PEROXIN4 (PEX4) gene, which may be involved in ubiquitylation, is necessary for the degradation of ICL and MLS (Zolman et al., 2005; Lingard et al., 2009). A third mechanism for peroxisomal degradation is pexophagy, a selective type of autophagy in which peroxisomes are targeted to the vacuole.
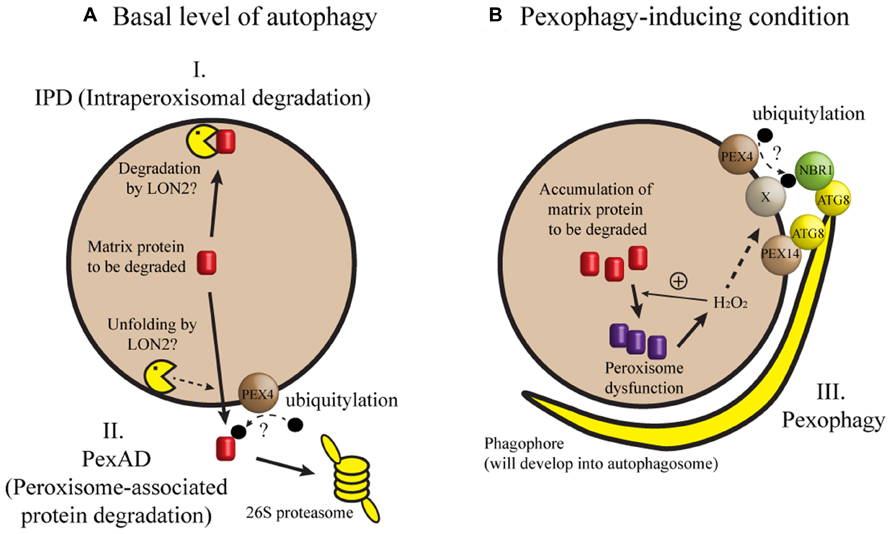
FIGURE 1. A model of peroxisomal protein degradation in plant cells. (A) When autophagy occurs at a basal level or when a core ATG gene is missing, degradation of matrix proteins like ICL and MLS may depend on intraperoxisomal degradation (IPD) or peroxisome-associated protein degradation (PexAD). (B) Pexophagy may be induced by loss of LON2, inactivation of IPD and PexAD, or a developmental process in which peroxisomes are past a critical level of oxidative stress (for example, intensive fatty acid β-oxidation during early seedling growth). Under pexophagy-inducing conditions, phagophore initiates, expands, and forms the autophagosome to sequester and target the peroxisome to the vacuole for degradation. In this model, hydrogen peroxide (H2O2) is shown as a sole matrix-derived induction signal for pexophagy, although we cannot exclude the possibility of additional induction signals. Refer to the main text for possible contributions of PEX4, PEX14, NBR1, and ATG8 proteins to target recognition. Broken arrows indicate highly speculative steps.
Pexophagy and its mechanism are well described in methylotrophic yeast and to a lesser extent in mammalian cells (reviewed by Till et al., 2012). Pexophagy typically removes obsolete or damaged peroxisomes. For example, peroxisomes are proliferated when methylotrophic yeast is grown in methanol, and excess peroxisomes are eliminated by pexophagy when methanol is replaced by other carbon source. Two types of pexophagy are known in yeast: in macropexophagy, peroxisomes are sequestered by the phagophore (Figure 1) and subsequently targeted to the vacuole, and micropexophagy occurs when peroxisomes are directly engulfed by the vacuolar membrane (Till et al., 2012). It was found that many core Autophagy-related (Atg) genes are required for pexophagy in yeast and mouse. Despite significant progress in our understanding of plant autophagy (reviewed by Floyd et al., 2012; Li and Vierstra, 2012), direct evidence for pexophagy in plant cells has not been available until recently and will be discussed herein.
Degradation of Peroxisomes by Autophagy in Arabidopsis
Early electron microscopy studies rarely include snapshots of autophagic degradation of peroxisomes in plant cells. However, there is a published example of autophagic vacuoles located near peroxisomes in castor bean endosperm, taken approximately 6 days after germination (Vigil, 1970). These snapshots alone were not sufficient for definitive evidence of plant pexophagy and required confirmation by immunoelectron microscopy and three-dimensional electron tomography.
A recent study employing a genetic suppressor screen provides evidence for pexophagy in plants. This was done by Dr. Bonnie Bartel’s group at Rice University in an attempt to identify the molecular function and targets of the LON2 protease (Farmer et al., 2013). The investigators screened for mutations suppressing lon2 phenotypes of defective β-oxidation activity and incomplete processing of peroxisome targeting signal (PTS) 2. In addition to the phenotypes, lon2 mutant cells had abnormally large spherical structures labeled by green fluorescent protein tagged with PTS (GFP-PTS), a widely used peroxisomal matrix marker. In contrast, the wild-type cells had small GFP-PTS puncta. Cloning of mutant genes that code for the lon2 suppressors resulted in the identification of several alleles of atg genes, specifically atg2, atg3, and atg7, two of which had been previously shown to cause defective autophagy (Doelling et al., 2002; Inoue et al., 2006). Double mutants of lon2 atg2, lon2 atg3, and lon2 atg7 all had normal β-oxidation and PTS2 processing, and had small GFP–PTS puncta. Moreover, endogenous ICL and MLS were stabilized in the double mutants, but not significantly in atg single-mutant seedlings. Farmer et al. (2013) presented a model in which autophagy removes a fraction of peroxisomes in wild-type Arabidopsis seedlings, while peroxisomal defects in lon2 mutation induce pexophagy (Figure 1B). Similar results were obtained by Dr. Mikio Nishimura’s group at National Institute for Basic Biology, Japan (Goto-Yamada et al., 2014). Lack of autophagy in the lon2 background appears to prevent the double-mutant seedlings from losing small peroxisomes, leading to the suppression of lon2 phenotypes (Bartel et al., 2014; Goto-Yamada et al., 2014). Although the precise function of LON2 in ICL and MLS degradation has yet to be defined, data described in Farmer et al. (2013), Goto-Yamada et al. (2014) indicate that LON2 protease plays a pivotal role in the IPD, unfolding, and/or translocation of misfolded peroxisomal proteins (Figure 1).
A role for pexophagy in ICL and MLS degradation was also demonstrated in a reverse genetics study performed in our laboratory (Kim et al., 2013b). 5-day-old wild-type hypocotyls had approximately 50% fewer peroxisomes than 3-day-old hypocotyls, while the reduction was about 20% in atg7 hypocotyls. Degradation of ICL and MLS was delayed in atg7 hypocotyls, but this stabilization effect was not obvious at the whole-seedling scale. Consistent with the observation that phenotypes were more obvious in hypocotyls than in the whole seedling, ATG7 transcription appeared to be induced preferentially in hypocotyls (Kim et al., 2013b). Thus, autophagy during seedling growth may be spatiotemporally controlled to promote degradation of peroxisomes. A relatively low level of autophagic activity in cotyledons and roots would explain why Farmer et al. (2013), Goto-Yamada et al. (2014) failed to detect stabilization of ICL and MLS in atg7 and atg2 single mutant seedlings. Finally, Kim et al. (2013b) reported ATG7- dependent degradation of peroxisomes in the central vacuole and observed autophagic puncta overlapping with peroxisomal markers.
As seedlings mature, pexophagy may have a role in peroxisomal quality control. This suggestion is supported by the results from another forward genetic screen performed by Dr. Mikio Nishimura’s group (Shibata et al., 2013). These authors identified mutants with aggregated peroxisomes and showed that the atg2, atg7, and atg18a mutations were responsible for aggregation. In line with the findings of Farmer et al. (2013), the atg2 mutation did not affect peroxisome function. However, leaves from 3-week-old atg2 mutant plants accumulated more peroxisomal proteins than the wild-type control, but had the same amount of mitochondrial and chloroplast proteins as leaves from wild-type plants, suggesting selective degradation of peroxisomal proteins by autophagy. Shibata et al. (2013) found that exogenously supplied hydrogen peroxide induced peroxisome aggregation. The aggregated peroxisomes in atg2 mutants were highly oxidized and contained a high level of inactive catalase. More recently, Yoshimoto et al. (2014) observed a phagophore-like structure that formed near the aggregated peroxisomes in atg2 leaves. These observations suggest that hydrogen peroxide is an induction signal for pexophagy, a process that aids in the disposal of damaged peroxisomes in the cell (Figure 1B).
Future Research Perspectives
We can identify several questions concerning pexophagy in plant cells. First, why have there been few ultrastructure images suggestive of pexophagy in plant cells? The scarcity may be due to rapid targeting of autophagic vesicles to the vacuole (Zhuang et al., 2013). This possibility is supported by the observation of the phagophore-like structures in atg2, where autophagosome formation may not be completed (Yoshimoto et al., 2014). In addition, the scarcity may result from a small developmental window in which seedling peroxisomes are rapidly transformed to leaf peroxisomes, as our study suggested (Kim et al., 2013b).
Another important question concerns the selectivity of autophagy. While autophagy in mature leaves may be selective for peroxisomes over mitochondria or plastids (Shibata et al., 2013), such selectivity was not clearly demonstrated in hypocotyls. A more quantitative tool to assess selectivity of autophagy in plant cells will be useful, and would apply to other types of selective autophagy, too.
Three of the studies mentioned here (Farmer et al., 2013; Kim et al., 2013b; Goto-Yamada et al., 2014) focused on peroxisome transition in young seedlings, while mature plants were used for the analysis of leaf peroxisomes in two other papers (Shibata et al., 2013; Yoshimoto et al., 2014). Nevertheless, these studies all underscore a role for autophagy in homeostasis of peroxisome number. Is there any unifying concept from the studies? It seems that the transition of seedling peroxisomes to leaf peroxisomes involves aggregation of small, highly oxidized peroxisomes that contain damaged or misfolded enzymes. In fact, aggregated peroxisomes were accumulated in the mesophyll cells of young atg2 seedlings (Goto-Yamada et al., 2014) and possibly in atg7 hypocotyl cells (Kim et al., 2013b). Hydrogen peroxide has been proposed as a signal for autophagy in plants (reviewed by Pérez-Pérez et al., 2012; Hackenberg et al., 2013). In support of this proposal, cat2 seedlings lacking a detectable level of catalase showed accelerated degradation of ICL and MLS (Lingard et al., 2009; see Figure 1B) compared to wild-type seedlings. Future work should clarify whether hydrogen peroxide acts as an upstream signal for both general and selective autophagy.
What proteins are necessary for recognizing peroxisomes targeted for autophagy? In methylotrophic yeast, a pexophagy receptor Atg30 bridges the molecular interaction between an autophagic complex and the peroxisomal proteins Pex3 and Pex14 (Farré et al., 2008; Zutphen et al., 2008). In mammalian cells, Pex14 interacts with LC3, an Atg8 homolog (Hara-Kuge and Fujiki, 2008). In addition, p62 and Neighbor of BRCA1 gene 1 (NBR1) may form a bridge between an ubiquitylated peroxisomal protein and LC3 (Kim et al., 2008; Deosaran et al., 2013). Intriguingly, an Arabidopsis ortholog of yeast Pex14 was identified from a genetic screen for mutants that showed stabilization of peroxisomal markers (Burkhart et al., 2013). It remains to be seen whether molecular interaction leading to pexophagy is conserved among distant eukaryotes (Figure 1).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work has not been mentioned because of space limitations. This work is supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (2011-0010683) and by a grant from the Next-Generation BioGreen 21 Program (No. PJ009004), Rural Development Administration, South Korea.
References
Bartel, B., Farmer, L. M., Rinaldi, M. A., Young, P. G., Danan, C. H., and Burkhart, S. E. (2014). Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy 10, 518–519. doi: 10.4161/auto.27565
Beevers, H. (1979). Microbodies in higher plants. Annu. Rev. Plant Physiol. 30, 159–193. doi: 10.1146/annurev.pp.30.060179.001111
Burkhart, S. E., Lingard, M. J., and Bartel, B. (2013). Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193, 125–141. doi: 10.1534/genetics.112.146100
Deosaran, E., Larsen, K. B., Hua, R., Sargent, G., Wang, Y., Kim, S., et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952. doi: 10.1242/jcs.114819
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Farmer, L. M., Rinaldi, M. A., Young, P. G., Danan, C. H., Burkhart, S. E., and Bartel, B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25, 4085–4100. doi: 10.1105/tpc.113.113407
Farré, J. C., Manjithaya, R., Mathewson, R. D., and Subramani, S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376. doi: 10.1016/j.devcel.2007.12.011
Floyd, B. E., Morriss, S. C., MacIntosh, G. C., and Bassham, D. C. (2012). What to eat: evidence for selective autophagy in plants. J. Integr. Plant Biol. 54, 907–920. doi: 10.1093/pcp/pcu017
Goto-Yamada, S., Mano, S., Nakamori, C., Kondo, M., Yamawaki, R., Kato, A., et al. (2014). Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 55, 482–496. doi: 10.1093/pcp/pcu017
Gruber, P. J., Becker, W. M., and Newcomb, E. H. (1973). The development of microbodies and peroxisomal enzymes in greening bean leaves. J. Cell Biol. 56, 500–518. doi: 10.1083/jcb.56.2.500
Hackenberg, T., Juul, T., Auzina, A., Gwizdz, S., Malolepszy, A., van der Kelen, K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25, 4616–4626. doi: 10.1105/tpc.113.117192
Hara-Kuge, S., and Fujiki, Y. (2008). The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp. Cell Res. 314, 3531–3541. doi: 10.1016/j.yexcr.2008.09.015
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303. doi: 10.1105/tpc.112.096586
Inoue, Y., Suzuki, T., Hattori, M., Yoshimoto, K., Ohsumi, Y., and Moriyasu, Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47, 1641–1652. doi: 10.1093/pcp/pcl031
Kim, D. Y., Scalf, M., Smith, L. M., and Vierstra, R. D. (2013a). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25, 1523–1540. doi: 10.1105/tpc.112.108613
Kim, J., Lee, H., Lee, H. N., Kim, S., Shin, K., and Chung, T. (2013b). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25, 4956–4966. doi: 10.1105/tpc.113.117960
Kim, P. K., Hailey, D. W., Mullen, R. T., and Lippincott-Schwartz, J. (2008). Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574. doi: 10.1073/pnas.0810611105
Li, F., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Lingard, M. J., and Bartel, B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151, 1354–1365. doi: 10.1104/pp.109.142505
Lingard, M. J., Monroe-Augustus, M., and Bartel, B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4561–4566. doi: 10.1073/pnas.0811329106
Nishimura, M., Hayashi, M., Kato, A., Yamaguchi, K., and Mano, S. (1996). Functional transformation of microbodies in higher plant cells. Cell Struct. Funct. 21, 387–393. doi: 10.1247/csf.21.387
Pérez-Pérez, M. E., Lemaire, S. D., and Crespo, J. L. (2012). Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 160, 156–164. doi: 10.1104/pp.112.199992
Pracharoenwattana, I., and Smith, S. M. (2008). When is a peroxisome not a peroxisome? Trends Plant Sci. 13, 522–525. doi: 10.1016/j.tplants.2008.07.003
Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. doi: 10.1105/tpc.113.116947
Smith, M. H., Ploegh, H. L., and Weissman, J. S. (2012). Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090. doi: 10.1126/science.1209235
Till, A., Lakhani, R., Burnett, S. F., and Subramani, S. (2012). Pexophagy: the selective degradation of peroxisomes. Int. J. Cell Biol. 2012, 18. doi: 10.1155/2012/512721
Trelease, R. N., Becker, W. M., Gruber, P. J., and Newcomb, E. H. (1971). Microbodies (glyoxysomes and peroxisomes) in cucunber cotyledons. Plant Physiol. 48, 461–475. doi: 10.1104/pp.48.4.461
Vigil, E. L. (1970). Cytochemical and developmental changes in microbodies (glyoxysomes) and related organelles of castor bean endosperm. J. Cell Biol. 46, 435–454. doi: 10.1083/jcb.46.3.435
Yoshimoto, K., Shibata, M., Kondo, M., Oikawa, K., Sato, M., Toyooka, K., et al. (2014). Quality control of plant peroxisomes in organ specific manner via autophagy. J. Cell Sci. 127, 1161–1168. doi: 10.1242/jcs.139709
Zhuang, X., Wang, H., Lam, S. K., Gao, C., Wang, X., Cai, Y., et al. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25, 4596–4615. doi: 10.1105/tpc.113.118307
Zolman, B. K., Monroe-Augustus, M., Silva, I. D., and Bartel, B. (2005). Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17, 3422–3435. doi: 10.1105/tpc.105.035691
Keywords: autophagy-related, Atg, peroxisome-associated protein degradation, hydrogen peroxide, glyoxylate cycle enzymes
Citation: Lee HN, Kim J and Chung T (2014) Degradation of plant peroxisomes by autophagy. Front. Plant Sci. 5:139. doi: 10.3389/fpls.2014.00139
Received: 21 January 2014; Paper pending published: 19 February 2014;
Accepted: 24 March 2014; Published online: 08 April 2014.
Edited by:
Diane C. Bassham, Iowa State University, USAReviewed by:
Jianping Hu, Michigan State University, USASigrun Reumann, University of Stavanger, Norway
Copyright © 2014 Lee, Kim and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taijoon Chung, Department of Biological Sciences, Pusan National University, 63 Beon-gil2, Busandaehag-ro, Geumjeong-gu, Busan 609-735, South Korea e-mail:dGFpam9vbkBwdXNhbi5hYy5rcg==
 Han Nim Lee
Han Nim Lee Jimi Kim
Jimi Kim Taijoon Chung
Taijoon Chung