- Department of Genetics, University of Georgia, Athens, GA, USA
The α/β-hydrolases KAI2 and D14 are paralogous receptors for karrikins and strigolactones, two classes of plant growth regulators with butenolide moieties. KAI2 and D14 act in parallel signaling pathways that share a requirement for the F-box protein MAX2, but produce distinct growth responses by regulating different members of the SMAX1-LIKE/D53 family. kai2 and max2 mutants share seed germination, seedling growth, leaf shape, and petiole orientation phenotypes that are not found in d14 or SL-deficient mutants. This implies that KAI2 recognizes an unknown, endogenous signal, herein termed KAI2 ligand (KL). Recent studies of ligand-specificity among KAI2 paralogs in basal land plants and root parasitic plants suggest that karrikin and strigolactone perception may be evolutionary adaptations of KL receptors. Here we demonstrate that evolutionarily conserved KAI2c genes from two parasite species rescue multiple phenotypes of the Arabidopsis kai2 mutant, unlike karrikin-, and strigolactone-specific KAI2 paralogs. We hypothesize that KAI2c proteins recognize KL, which could be an undiscovered hormone.
Karrikins (KARs) and strigolactones (SLs) are two classes of butenolide molecules that regulate diverse aspects of plant growth. KARs are bioactive components of smoke that stimulate germination of many species after fire, when competition for resources is reduced (Flematti et al., 2004, 2009; van Staden et al., 2004). However, responses to KARs or smoke are not limited to species from fire-prone environments, and are widely distributed among the angiosperms (Nelson et al., 2012). For example, KARs promote germination of dormant Arabidopsis thaliana seed (Nelson et al., 2009). KARs can also enhance the photomorphogenic growth of Arabidopsis seedlings, causing reduced hypocotyl elongation and enhanced cotyledon growth (Nelson et al., 2010). It is unclear why a KAR response mechanism is found in plants from environments with low fire frequencies, but one possibility is that plants produce KARs or KAR-like molecules. If KARs are similar to an endogenous plant compound, it might be relatively straightforward for high sensitivity to KARs to evolve independently in fire-adapted species.
Strigolactones (SLs) are a family of plant hormones that control shoot branching, root architecture, secondary growth, and senescence (Gomez-Roldan et al., 2008; Umehara et al., 2008; Agusti et al., 2011; Kapulnik et al., 2011; Rasmussen et al., 2012; Yamada et al., 2014; Ueda and Kusaba, 2015). SLs are also exuded from plant roots into the rhizosphere, particularly under low nutrient conditions, where they promote symbiotic associations with arbuscular mycorrhizal fungi (Akiyama et al., 2005). This fungal recruitment mechanism is exploited by root parasitic plants in the Orobanchaceae family, which use SLs as a way to detect a nearby host and germinate (Yoneyama et al., 2010). SLs are produced from carotenoids via a carlactone intermediate (Alder et al., 2012; Al-Babili and Bouwmeester, 2015). The basic structure of SLs consists of a tricyclic lactone (ABC ring) connected by an enol-ether bond to a butenolide moiety (D ring). The D ring is oriented in the 2′R configuration in all strigolactones that have been identified from plants (Xie et al., 2013; Zwanenburg and Pospisil, 2013).
KARs and SLs are not interchangeable signals. KARs do not rescue the excess branching phenotype of SL-deficient mutants (Nelson et al., 2011), and SLs and carlactone do not promote germination of Arabidopsis seed (Scaffidi et al., 2013, 2014). Some confusion about SL roles has arisen due to the widespread use of a synthetic strigolactone racemic mixture, rac-GR24. It has only recently been recognized that the two enantiomers that comprise rac-GR24 have different effects due to opposite orientations of the D ring: GR245DS mimics the configuration and activity of the natural strigolactone 5-deoxystrigol (5DS), but GR24ent-5DS has an unnatural 2′S configuration and primarily acts through the KAR signaling pathway (Scaffidi et al., 2014).
Mechanisms of KARRIKIN and STRIGOLACTONE Perception
Although KAR and SL have unique sources and effects on plant development, genetic studies have shown that both signals act through similar mechanisms. Growth responses to both KAR and SL require the F-box protein MAX2 (Nelson et al., 2011). Specific responses to karrikins and strigolactones respectively require the α/β-hydrolases KARRIKIN INSENSITIVE2 (KAI2) and DWARF14 (D14), which are ancient paralogs. D14 is only present in vascular plants, where it likely arose through duplication of KAI2 (Delaux et al., 2012; Waters et al., 2012). In Arabidopsis, kai2/htl mutants have increased seed dormancy, elongated hypocotyls, smaller cotyledons, hyponastic petioles, and altered leaf morphology, but have normal shoot branching (Sun and Ni, 2011; Waters et al., 2012). d14 mutants in rice, petunia, and Arabidopsis have increased axillary branching (or tillering), reduced stature, and delayed senescence; these phenotypes are shared with SL-deficient mutants, but are not rescued by SL application (Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012; Yamada et al., 2014; Ueda and Kusaba, 2015). Unlike kai2, however, d14 and SL-deficient mutants in Arabidopsis have wildtype germination and seedling growth (Nelson et al., 2011; Shen et al., 2012; Waters et al., 2012). max2 phenotypes are a complex union of kai2 and d14 phenotypes (Waters et al., 2012; Soundappan et al., 2015).
Substantial evidence supports the hypothesis that KAI2 and D14 are receptors for KAR and SL, respectively. Several types of in vitro assays have shown that KAI2 binds KAR1, and that D14 binds and hydrolyzes GR24 (Hamiaux et al., 2012; Guo et al., 2013; Kagiyama et al., 2013; Nakamura et al., 2013; Zhao et al., 2013, 2015). Protein-ligand complexes of KAI2-KAR1 and D14 with GR24 and its hydrolytic products have been resolved by X-ray crystallography (Guo et al., 2013; Nakamura et al., 2013; Zhao et al., 2013, 2015). D14 function requires an active catalytic site, but the byproducts of GR24 hydrolysis do not rescue the d14/dad2 mutant in petunia (Hamiaux et al., 2012). This indicates that signal transduction is achieved by the act of ligand binding or hydrolysis. Indeed, SL promotes the association of D14 with MAX2/D3 and DWARF53 (D53) in rice, which causes polyubiquitination and proteolysis of D53 (Jiang et al., 2013; Zhou et al., 2013, Zhao et al., 2014). Degradation of D53 is necessary for SL signal transduction (Jiang et al., 2013; Zhou et al., 2013). D14 does not show obvious conformational changes during SL binding or hydrolysis, but in the presence of GR24 it undergoes thermal destabilization that is enhanced by MAX2/D3 interactions (Hamiaux et al., 2012; Zhao et al., 2015). D14 interactions with MAX2/D3 and D53 are selectively activated by 2′R SL stereoisomers (Umehara et al., 2015; Zhao et al., 2015). Arabidopsis D14 destabilizes in the presence of either GR245DS or GR24ent-5DS, but it is primarily responsive to GR245DS in vivo (Scaffidi et al., 2014; Waters et al., 2015b). In Arabidopsis, the D53 orthologs SMXL6, SMXL7, and SMXL8 mediate growth responses to SL downstream of MAX2, interact with D14, and are also targeted for degradation in a D14-dependent manner following GR24 treatment (Soundappan et al., 2015; Umehara et al., 2015; Wang et al., 2015). KAR perception by KAI2 is hypothesized to trigger similar degradation of SMAX1, a D53/SMXL6/7/8 paralog in Arabidopsis that controls seed germination, seedling growth, and leaf development (Stanga et al., 2013; Bennett and Leyser, 2014; Soundappan et al., 2015).
kai2 Phenotypes Suggest Insensitivity to an Endogenous, Non-Strigolactone Ligand
In addition to KAR-insensitivity, kai2 and max2 mutants share phenotypes of increased seed dormancy and reduced photomorphogenic growth in seedlings. These phenotypes are opposite to the effects of KAR treatment, and are not observed in d14 or SL-deficient mutants (Nelson et al., 2011; Shen et al., 2012). This observation provided the first evidence that KAI2 and MAX2 are required for responses to an endogenous, non-SL signal (Flematti et al., 2013; Waters et al., 2014). In addition, Arabidopsis KAI2 hydrolyzes GR24ent-5DS and undergoes thermal destabilization in its presence, but KAI2 is not active toward GR245DS (Waters et al., 2015b). These data support observations that GR24ent-5DS, but not GR245DS or carlactone, enhances Arabidopsis germination, which is controlled by KAI2 but not D14 (Scaffidi et al., 2013, 2014). Thus Arabidopsis KAI2 has the capacity to respond to more than one type of non-SL butenolide compound, but is not involved in SL signaling. We propose that KARs and GR24ent-5DS can fortuitously substitute for its endogenous signal(s), which we herein refer to as KAI2 ligand (KL). Like strigolactones, KL is not necessarily a single compound and may be a class of similar molecules.
An alternative explanation for kai2 mutant phenotypes is that KAI2 is always active, but KAR enhances its activity. For example, KAI2 could have a weak physical interaction with a signaling partner that is strengthened by KAR. In this case kai2 phenotypes could result from a loss of constitutive, ligand-independent KAI2 activity rather than a lack of KL perception. Two observations counter this idea. First, a Ser-His-Asp catalytic triad is conserved throughout KAI2 proteins found in land plants, indicating selective pressure to retain catalytic activity. Indeed the catalytic Ser is required for KAI2 hydrolytic activity and rescue of kai2 mutants (Waters et al., 2014, 2015a,b). Thus hydrolysis-independent activity is unlikely to have functional significance. Second, overexpression of KAI2 does not induce constitutive KAR response phenotypes in seedlings, as would be expected if basal activity were increased (Waters and Smith, 2013). However, it does cause hypersensitivity to KAR treatment, which is consistent with an increased capacity for ligand-activated signaling. It must be noted that KAI2 overexpression does reduce seed dormancy (Waters and Smith, 2013); we propose that this observation reflects that KL is a limiting factor for KAI2 activity in seedlings but not in seed.
A Lycophyte Kai2 may be Kl-Specific
Further support for the existence of KL comes from the evolutionary history of KAR and SL signaling. KAI2 homologs are found in charophyte green algae and the basal land plants Physcomitrella patens and Marchantia polymorpha (Delaux et al., 2012; Waters et al., 2015b). D14 orthologs are found only in vascular plants, and most likely arose from a duplication of KAI2 (Delaux et al., 2012; Waters et al., 2012). This indicates that KAI2 signaling preceded strigolactone perception by D14, but does not clarify the ligand-specificities of basal KAI2. Indeed strigolactones have been identified in basal land plants, and rac-GR24 can induce growth responses in bryophytes and Charales (Proust et al., 2011; Delaux et al., 2012; Hoffmann et al., 2014). Waters et al. (2015b) recently investigated the biochemical activities and functions of KAI2 homologs in S. moellendorfii and M. polymorpha. SmKAI2b, one of two KAI2 paralogs in S. moellendorfii, has hydrolytic activity against GR245DS that suggests it might be a SL receptor, but it does not rescue Arabidopsis kai2 or d14 mutants. In contrast, SmKAI2a partially rescues kai2 seedling growth and fully rescues kai2 leaf morphology. The ability of SmKAI2a to rescue Arabidopsis kai2 is dependent upon an active catalytic site. Interestingly, SmKAI2a does not confer responses to KARs, GR24, or carlactone in transgenic seedling growth assays. Although SmKAI2a has in vitro hydrolase activity against a generic substrate, it does not hydrolyze either 2′R or 2′S GR24 stereoisomers. And unlike AtKAI2 and AtD14, it does not undergo thermal stability shifts in the presence of either GR24 stereoisomer (Waters et al., 2015b). As SmKAI2a partially rescues kai2 – despite having no apparent KAR or SL receptor activity – it likely recognizes endogenous substrates that are similar or the same in Selaginella and Arabidopsis, which we propose to be KL.
Kai2 Paralogs in Parasitic Plants have Diversified Ligand-Specificities
We also discovered evidence for KL-specificity among evolutionarily conserved KAI2 paralogs in root parasitic plants (Conn et al., 2015). A broad range of parasitism is found within the Orobanchaceae family, from a single non-parasitic genus, to facultative parasites that do not require a host, to obligate parasites that are dependent upon a host. The obligate parasite taxa include several highly destructive agricultural weeds, such as witchweed (Striga spp.) and broomrapes (Orobanche, Phelipanche spp.), which germinate in response to SLs but not KARs. As SLs indicate the presence of a host, and obligate parasites only survive if they attach to a host root within days of germination, this response is a critical adaptation. We found that KAI2 has undergone extensive gene duplication and diversification of ligand specificity in parasite genomes. KAI2 genes in the Lamiales order are grouped into three clades: conserved (KAI2c), intermediate (KAI2i), and divergent (KAI2d). KAI2c paralogs are the most broadly distributed in Lamiales species. The KAI2c clade is under the strongest purifying selection and has the highest similarity to KAI2 genes in non-parasitic angiosperms. KAI2i paralogs are under intermediate purifying selection, and are found in most Lamiales genomes. KAI2d paralogs comprise a parasite-specific clade that has undergone significant expansion and has evolved the most quickly. Homology modeling predicts a high degree of similarity between the ligand-binding pockets of KAI2c paralogs and AtKAI2, whereas KAI2d pockets are significantly enlarged and have diverse shapes. Our current evolutionary model is that KAI2c and KAI2i arose through KAI2 gene duplication in the Lamiids, and KAI2d arose through further duplication and neofunctionalization in the parasitic Orobanchaceae lineage (Conn et al., 2015) (Figure 1).
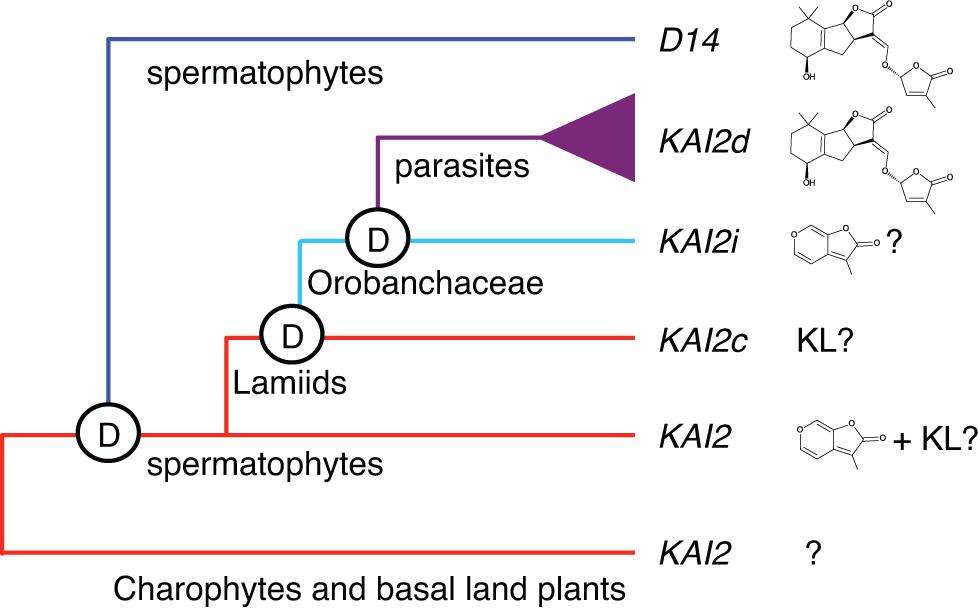
FIGURE 1. Model of KAI2 and D14 evolution. KAI2 orthologs are present in charophyte algae and basal land plants. D14 likely arose by duplication of KAI2 prior to the evolution of spermatophytes. Although the timing of this duplication is uncertain, a potential D14 ortholog is present in Selaginella moellendorfii. We hypothesize that most spermatophyte KAI2 are KL and/or KAR receptors. In parasitic plants, KAI2 duplication and evolution have produced three clades. KAI2c is hypothesized to be KL-specific, KAI2i may be KAR-specific, and KAI2d are SL-responsive. Reprinted from Conn et al. (2015) with permission from AAAS.
We investigated the ligand-specificity of KAI2 genes from Striga hermonthica and Phelipanche aegyptiaca by cross-species complementation of Arabidopsis kai2. KAI2 transgenes from the three clades confer different germination phenotypes: (1) KAI2c from P. aegyptiaca (PaKAI2c) rescues the Arabidopsis kai2 seed dormancy phenotype, but does not confer responses to KARs or rac-GR24; (2) KAI2i from S. hermonthica (ShKAI2i) does not rescue kai2 dormancy, but confers germination responses to KARs; and (3) three KAI2d paralogs do not rescue kai2 dormancy, but confer germination responses to rac-GR24. These KAI2d confer stronger germination responses to GR24 stereoisomers with SL-like 2′R configurations than those with unnatural 2′S configurations. Thus, KAI2d in parasitic weeds are likely to be SL receptors that function in host-responsive seed germination (Conn et al., 2015). An independent cross-species complementation analysis of KAI2/HTL paralogs from Striga hermonthica is consistent with our findings. The KAI2i class paralogs ShHTL2 and ShHTL3 confer KAR-specific germination to Arabidopsis kai2, and six KAI2d class paralogs (ShHTL4-ShHTL9) have SL-specific functions (Toh et al., 2015).
The conclusions from these functional approaches are supported by in vitro studies of ligand selectivity among KAI2/HTL paralogs in S. hermonthica (Tsuchiya et al., 2015). The KAI2c paralog (ShHTL1) does not hydrolyze rac-GR24, but eight KAI2d paralogs (ShHTL4 to ShHTL11) hydrolyze rac-GR24 and show a range of affinities for different strigolactones. KAI2i paralogs (ShHTL2 and ShHTL3) have weaker hydrolytic activities on rac-GR24 and lower in vitro affinities for strigolactones than any of the KAI2d paralogs (Tsuchiya et al., 2015).
PaKAI2c rescues Arabidopsis kai2 seed dormancy but does not confer KAR or GR24 responses, which suggests that this protein is specific for KL. ShKAI2c does not appear to be active in Arabidopsis seed; like SmKAI2a, however, it could encode a KL receptor that has reduced function in Arabidopsis compared to its native context. Reduced function could be due to inefficient interactions with Arabidopsis signaling partners, which is an inherent limitation of the cross-species complementation approach. Functional rescue at later stages of growth, as seen for SmKAI2a, could result from a lower signaling threshold, or changes in KAI2 signaling partner availability. Therefore we investigated whether PaKAI2c and ShKAI2c can rescue Arabidopsis kai2 phenotypes after germination.
New Evidence that Parasite Kai2c may be Kl-Specific Receptors
PaKAI2c completely restored, and ShKAI2c mostly restored, kai2 hypocotyl length and cotyledon size to wildtype (Figures 2A,B). ShKAI2i, which confers KAR-specific germination responses, partially rescued kai2 hypocotyl length but had little effect on cotyledon size. In comparison, the strigolactone-responsive genes PaKAI2d1, PaKAI2d3, and ShKAI2d1 had little or no effect on either aspect of kai2 seedling growth (Figures 2A,B). We tested the hypocotyl elongation responses of these transgenic lines to KAR1 and two GR24 stereoisomers (Figure 2C). All transgenic lines were expected to be GR24 responsive due to D14 activity (Scaffidi et al., 2014). PaKAI2c and ShKAI2c did not confer responses to KAR1 and were not more sensitive to GR245DS than kai2. ShKAI2i seedlings had a strong response to KAR1, consistent with this transgene’s KAR-specific germination responses (Conn et al., 2015). PaKAI2d1, PaKAI2d3, and ShKAI2d1 seedlings were insensitive to KAR1 and more responsive to GR24ent-5DS and GR245DS than kai2. Only ShKAI2d1 confers germination responses to GR24ent-5DS, but all three KAI2d lines responded to it at the seedling stage. Finally, we examined rosette phenotypes (Figure 2D). PaKAI2c and to a lesser extent ShKAI2c rescued kai2 leaf morphology, but KAI2d transgenes did not. As in seedlings, ShKAI2i partially rescued the kai2 leaf phenotype.
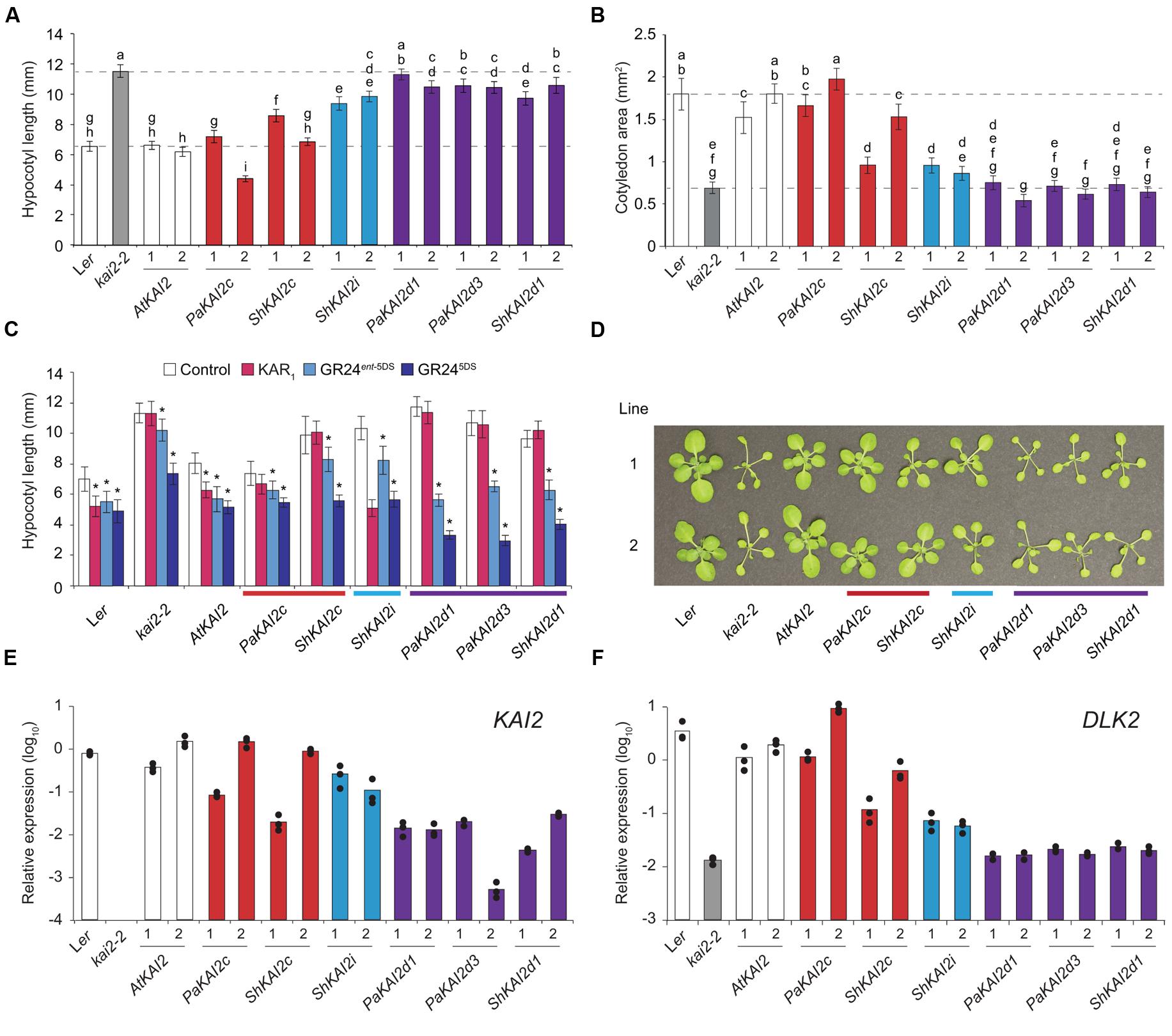
FIGURE 2. KAI2 from parasites confer different growth responses to Arabidopsis kai2. (A) Hypocotyl length and (B) cotyledon area of 10 day-old seedlings grown under short-day conditions. KAI2 transgenes from Arabidopsis thaliana (At), Striga hermonthica (Sh), and Phelipanche aegyptiaca (Pa) under control of an AtKAI2 promoter are in the Arabidopsis kai2-2 mutant background. Data are shown for two homozygous, independent transgenic lines per transgene. Mean ±99% CI, n = 45. Tukey–Kramer HSD test, p < 0.01. (C) Hypocotyl length of 10 day-old seedlings grown on 1 μM KAR1, GR24ent-5DS, and GR245DS as described above. Transgenic line 1 was tested. Mean ±99% CI, n = 15. Dunnett’s test comparing treatments to control, ∗p < 0.01. (D) 31 day-old rosettes grown under short-day conditions (white light, 65–136 μE). A representative plant from each of two independent transgenic lines is shown per transgene. (E) Expression of KAI2 and (F) DLK2 in 10 day-old seedlings, relative to the CACS reference gene. In (E), KAI2 expression is of wildtype KAI2 (Ler) or the respective parasite KAI2 transgene (all other lines). Columns represent the mean expression of three biological replicates (at least two technical replicates each), each of which is indicated by a black dot.
Thus PaKAI2c rescues all visible kai2 mutant phenotypes, and ShKAI2c – like SmKAI2a – can partially rescue kai2 at certain stages of development. These results suggest that receptors in the KAI2c clade in parasites recognize KL. To a lesser extent ShKAI2i, which has a stronger response to KAR1 than any other treatments, was able to partially rescue kai2. This may reflect a residual affinity for KL in these KAR-responsive receptors. However, the SL-specific KAI2d had little effect on kai2 phenotypes, even though they can function in an Arabidopsis context and confer sensitive seed and seedling responses to GR24. Toh et al. (2015) also found that strigolactone-specific KAI2d transgenes from Striga hermonthica do not rescue Arabidopsis kai2 seed dormancy.
The PaKAI2c and ShKAI2c transgenic lines rescued kai2 mutant phenotypes to different extents (Figures 2A,B). We investigated whether transgene expression level contributed to these differences. We found a correlation between transgene expression, rescue, and expression of the KAR/SL reporter gene DLK2 (Figure 2). Interestingly, KAI2d transgenes were expressed at a much lower level than KAI2c and KAI2i (Figure 2E). Thus, we cannot exclude the possibility that KAI2d do not rescue kai2 due to low transgene expression. However, even the lowest-expressing KAI2d line, PaKAI2d3 line 2, has a strong germination response to rac-GR24 (95% on 1 μM rac-GR24 vs. 12.5% on control media), indicating that its transgene expression levels are sufficient for function. Taken together with the biochemical studies of AtKAI2 described above, the lack of seed and seedling phenotypes in SL-deficient mutants (Nelson et al., 2011; Shen et al., 2012), and the inability of carlactone to stimulate Arabidopsis germination (Scaffidi et al., 2013), this supports that endogenous strigolactones do not contribute significantly to Arabidopsis seed germination or seedling growth.
Hypotheses and Future Directions
We hypothesize that the original function of KAI2 in land plants was KL perception, and that karrikin and strigolactone perception were derived through later adaptations of these receptors. SL-specific KAI2 paralogs likely evolved following KAI2 duplication in the vascular plant lineage, producing D14, and again within the Orobanchaceae family, producing KAI2d genes. Among basal vascular plants, KAI2b from S. moellendorfii hydrolyzes GR245DS in vitro. Cross-species complementation assays with SmKAI2b in Arabidopsis were not successful, so conclusions about its in vivo function will await reverse genetic analysis. The moss Physcomitrella patens has at least 10 KAI2 paralogs; perhaps some of these genes mediate SL responses. We propose that KAI2c is highly conserved in parasites, even though KAI2d enable host-responsive germination, because KL perception serves fundamental roles in plant growth. It remains to be determined if KAI2 in non-parasitic angiosperms like Arabidopsis gained or improved karrikin sensitivity in addition to KL perception, or if KAI2c in parasites subfunctionalized to exclude non-KL ligands.
A key question now facing the field is, what is KL? At this point we can only predict that KL has a butenolide moiety, a feature shared by both classes of compounds that signal via MAX2. The identification of KL would be a significant discovery for plant biology and agriculture alike, as this may represent a new plant hormone. Of practical significance to agriculture, the kai2 mutant demonstrates that KL promotes seed germination (or reduces seed dormancy) and seedling growth responses to light. Knockdown of KAI2/D14L in rice suggests a role for KL in reducing mesocotyl elongation in the dark (Kameoka and Kyozuka, 2015). In addition, KAR1 enhances the seedling vigor of some crops under stressful growth conditions (Jain et al., 2006; Kulkarni et al., 2006; van Staden et al., 2006), and KL may do the same. In the face of growing challenges to agriculture, KL may find uses as an agrichemical that enables more uniform germination and establishment of crops, or depletes the weed load in field seed banks. The discovery of likely receptors for KL is an important first step toward its identification. In the meantime, KARs will continue to serve as useful analogs to investigate KL biosynthesis and perception.
Methods
Transgenic lines were described in Conn et al. (2015). Seedling assays were performed on 10 day-old seedlings grown in 8 h white light (∼36 μE):16 h dark at 21°C on 0.5× Murashige and Skoog media supplemented with 1 μM treatments or an acetone control. Hypocotyls and cotyledons were measured with ImageJ (http://rsb.info.nih.gov/ij/). qRT-PCR was performed on 10 day-old seedlings according to Stanga et al. (2013). qPCR primers span introns and are listed in Supplementary Table S1. Statistical analyses were done in JMP (SAS Institute).
Author Contributions
CC performed the experiments, and collected and analyzed the data. CC and DN designed the research and wrote the manuscript.
Funding
Support for this work was provided by NSF grant IOS-1350561 to DN and an NSF Graduate Research Fellowship to CC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01219
References
Agusti, J., Herold, S., Schwarz, M., Sanchez, P., Ljung, K., Dun, E. A., et al. (2011). Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. U.S.A. 108, 20242–20247. doi: 10.1073/pnas.1111902108
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Al-Babili, S., and Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. doi: 10.1146/annurev-arplant-043014-114759
Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M., Bigler, P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. doi: 10.1126/science.1218094
Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., et al. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. doi: 10.1093/pcp/pcp091
Bennett, T., and Leyser, O. (2014). Strigolactone signalling: standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22C, 7–13. doi: 10.1016/j.pbi.2014.08.001
Conn, C. E., Bythell-Douglas, R., Neumann, D., Yoshida, S., Whittington, B., Westwood, J. H., et al. (2015). PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543. doi: 10.1126/science.aab1140
Delaux, P. M., Xie, X., Timme, R. E., Puech-Pages, V., Dunand, C., Lecompte, E., et al. (2012). Origin of strigolactones in the green lineage. New Phytol. 195, 857–871. doi: 10.1111/j.1469-8137.2012.04209.x
Flematti, G. R., Ghisalberti, E. L., Dixon, K. W., and Trengove, R. D. (2004). A compound from smoke that promotes seed germination. Science 305:977. doi: 10.1126/science.1099944
Flematti, G. R., Ghisalberti, E. L., Dixon, K. W., and Trengove, R. D. (2009). Identification of alkyl substituted 2H-Furo[2,3-c]pyran-2-ones as germination stimulants present in smoke. J. Agric. Food Chem. 57, 9475–9480. doi: 10.1021/jf9028128
Flematti, G. R., Waters, M. T., Scaffidi, A., Merritt, D. J., Ghisalberti, E. L., Dixon, K. W., et al. (2013). Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 6, 29–37. doi: 10.1093/mp/sss132
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pages, V., Dun, E. A., Pillot, J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi: 10.1038/nature07271
Guo, Y., Zheng, Z., La Clair, J. J., Chory, J., and Noel, J. P. (2013). Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 8284–8289. doi: 10.1073/pnas.1306265110
Hamiaux, C., Drummond, R. S., Janssen, B. J., Ledger, S. E., Cooney, J. M., Newcomb, R. D., et al. (2012). DAD2 Is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, Strigolactone. Curr. Biol. 22, 2032–2036. doi: 10.1016/j.cub.2012.08.007
Hoffmann, B., Proust, H., Belcram, K., Labrune, C., Boyer, F. D., Rameau, C., et al. (2014). Strigolactones inhibit caulonema elongation and cell division in the moss Physcomitrella patens. PLoS ONE 9:e99206. doi: 10.1371/journal.pone.0099206
Jain, N., Kulkarni, M. G., and van Staden, J. (2006). A butenolide, isolated from smoke, can overcome the detrimental effects of extreme temperatures during tomato seed germination. Plant Growth Regul. 49, 263–267. doi: 10.1007/s10725-006-9136-0
Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F., Wang, L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. doi: 10.1038/nature12870
Kagiyama, M., Hirano, Y., Mori, T., Kim, S. Y., Kyozuka, J., Seto, Y., et al. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160. doi: 10.1111/gtc.12025
Kameoka, H., and Kyozuka, J. (2015). Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. J. Genet. Genomics 42, 119–124. doi: 10.1016/j.jgg.2014.12.003
Kapulnik, Y., Delaux, P. M., Resnick, N., Mayzlish-Gati, E., Wininger, S., Bhattacharya, C., et al. (2011). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216. doi: 10.1007/s00425-010-1310-y
Kulkarni, M. G., Sparg, S. G., Light, M. E., and van Staden, J. (2006). Stimulation of rice (Oryza sativa L.) seedling vigour by smoke-water and butenolide. J. Agron. Crop Sci. 192, 395–398. doi: 10.1111/j.1439-037X.2006.00213.x
Nakamura, H., Xue, Y. L., Miyakawa, T., Hou, F., Qin, H. M., Fukui, K., et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4:2613. doi: 10.1038/ncomms3613
Nelson, D. C., Flematti, G. R., Ghisalberti, E. L., Dixon, K. W., and Smith, S. M. (2012). Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 63, 107–130. doi: 10.1146/annurev-arplant-042811-105545
Nelson, D. C., Flematti, G. R., Riseborough, J. A., Ghisalberti, E. L., Dixon, K. W., and Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 7095–7100. doi: 10.1073/pnas.0911635107
Nelson, D. C., Riseborough, J. A., Flematti, G. R., Stevens, J., Ghisalberti, E. L., Dixon, K. W., et al. (2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. doi: 10.1104/pp.108.131516
Nelson, D. C., Scaffidi, A., Dun, E. A., Waters, M. T., Flematti, G. R., Dixon, K. W., et al. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902. doi: 10.1073/pnas.1100987108
Proust, H., Hoffmann, B., Xie, X., Yoneyama, K., Schaefer, D. G., Nogue, F., et al. (2011). Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539. doi: 10.1242/dev.058495
Rasmussen, A., Mason, M., De Cuyper, C., Brewer, P. B., Herold, S., Agusti, J., et al. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987. doi: 10.1104/pp.111.187104
Scaffidi, A., Waters, M. T., Ghisalberti, E. L., Dixon, K. W., Flematti, G. R., and Smith, S. M. (2013). Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 76, 1–9. doi: 10.1111/tpj.12265
Scaffidi, A., Waters, M. T., Sun, Y. K., Skelton, B. W., Dixon, K. W., Ghisalberti, E. L., et al. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232. doi: 10.1104/pp.114.240036
Shen, H., Zhu, L., Bu, Q. Y., and Huq, E. (2012). MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 5, 750–762. doi: 10.1093/mp/sss029
Soundappan, I., Bennett, T., Morffy, N., Liang, Y., Stanga, J. P., Abbas, A., et al. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159. doi: 10.1105/tpc.15.00562
Stanga, J. P., Smith, S. M., Briggs, W. R., and Nelson, D. C. (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330. doi: 10.1104/pp.113.221259
Sun, X. D., and Ni, M. (2011). HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol. Plant 4, 116–126. doi: 10.1093/mp/ssq055
Toh, S., Holbrook-Smith, D., Stogios, P. J., Onopriyenko, O., Lumba, S., Tsuchiya, Y., et al. (2015). Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207. doi: 10.1126/science.aac9476
Tsuchiya, Y., Yoshimura, M., Sato, Y., Kuwata, K., Toh, S., Holbrook-Smith, D., et al. (2015). PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868. doi: 10.1126/science.aab3831
Ueda, H., and Kusaba, M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169, 138–147. doi: 10.1104/pp.15.00325
Umehara, M., Cao, M., Akiyama, K., Akatsu, T., Seto, Y., Hanada, A., et al. (2015). Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56, 1059–1072. doi: 10.1093/pcp/pcv028
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
van Staden, J., Jager, A. K., Light, M. E., and Burger, B. V. (2004). Isolation of the major germination cue from plant-derived smoke. S. Afr. J. Bot. 70, 654–659. doi: 10.1016/S0254-6299(15)30206-4
van Staden, J., Sparg, S. G., Kulkarni, M. G., and Light, M. E. (2006). Post-germination effects of the smoke-derived compound 3-methyl-2H-furo[2,3-c]pyran-2-one, and its potential as a preconditioning agent. Field Crops Res. 98, 98–105. doi: 10.1016/j.fcr.2005.12.007
Wang, L., Wang, B., Jiang, L., Liu, X., Li, X., Lu, Z., et al. (2015). Strigolactone Signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142. doi: 10.1105/tpc.15.00605
Waters, M. T., Nelson, D. C., Scaffidi, A., Flematti, G. R., Sun, Y. K., Dixon, K. W., et al. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. doi: 10.1242/dev.074567
Waters, M. T., Scaffidi, A., Flematti, G., and Smith, S. M. (2015a). Substrate-induced degradation of the alpha/beta-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8, 814–817. doi: 10.1016/j.molp.2014.12.020
Waters, M. T., Scaffidi, A., Moulin, S. L., Sun, Y. K., Flematti, G. R., and Smith, S. M. (2015b). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944. doi: 10.1105/tpc.15.00146
Waters, M. T., Scaffidi, A., Sun, Y. K., Flematti, G. R., and Smith, S. M. (2014). The karrikin response system of Arabidopsis. Plant J. 79, 623–631. doi: 10.1111/tpj.12430
Waters, M. T., and Smith, S. M. (2013). KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6, 63–75. doi: 10.1093/mp/sss127
Xie, X., Yoneyama, K., Kisugi, T., Uchida, K., Ito, S., Akiyama, K., et al. (2013). Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 6, 153–163. doi: 10.1093/mp/sss139
Yamada, Y., Furusawa, S., Nagasaka, S., Shimomura, K., Yamaguchi, S., and Umehara, M. (2014). Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408. doi: 10.1007/s00425-014-2096-0
Yoneyama, K., Awad, A. A., Xie, X., and Takeuchi, Y. (2010). Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol. 51, 1095–1103. doi: 10.1093/pcp/pcq055
Zhao, J., Wang, T., Wang, M., Liu, Y., Yuan, S., Gao, Y., et al. (2014). DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 55, 1096–1109. doi: 10.1093/pcp/pcu045
Zhao, L., Zhou, X., Wu, Z., Yi, W., Xu, Y., Li, S., et al. (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439. doi: 10.1038/cr.2013
Zhao, L. H., Zhou, X. E., Yi, W., Wu, Z., Liu, Y., Kang, Y., et al. (2015). Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236. doi: 10.1038/cr.2015.122
Zhou, F., Lin, Q., Zhu, L., Ren, Y., Zhou, K., Shabek, N., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. doi: 10.1038/nature12878
Keywords: karrikins, strigolactones, signaling, evolution, hormones
Citation: Conn CE and Nelson DC (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may Perceive an Unknown Signal that is not Karrikin or Strigolactone. Front. Plant Sci. 6:1219. doi: 10.3389/fpls.2015.01219
Received: 12 December 2015; Accepted: 17 December 2015;
Published: 08 January 2016.
Edited by:
Stefan A. Rensing, University of Marburg, GermanyReviewed by:
Caroline Gutjahr, Ludwig Maximilian University of Munich, GermanyPierre-Marc Delaux, John Innes Centre, France
Copyright © 2016 Conn and Nelson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David C. Nelson, ZGNuZWxzb25AdWdhLmVkdQ==
 Caitlin E. Conn
Caitlin E. Conn David C. Nelson
David C. Nelson